Abstract
Identification of potential therapeutic targets and biomarkers indicative of burden of early atherosclerosis that occur prior to advancement to life-threatening unstable plaques is the key to eradication of CAD prevalence and incidences. We challenged 16 baboons with a high cholesterol, high fat diet for 2 years and evaluated early-stage atherosclerotic lesions (fatty streaks, FS, and fibrous plaques, FP) in formalin-fixed common iliac arteries (CIA). We used small RNA sequencing to identify expressed miRNAs in CIA and in baseline blood samples of the same animals. We found 412 expressed miRNAs in CIA and 356 in blood samples. Eight miRNAs (miR-7975, -486-5p, -451a, -191-5p, -148a-3p, -17-5p, -378c, and -144-3p) were differentially expressed between paired fatty streak lesion and no-lesion sites of the tissue, and 27 miRNAs (e.g., miR-92a-3p, -5001, -342-3p, miR-28-3p, -21-5p, -221-3p, 146a-5p, and -16-5p) in fibrous plaques. The expression of 14 blood miRNAs significantly correlated with extent of lesions and the number of plaques. We identified coordinately regulated miRNA-gene networks in which miR-17-5p and miR-146a-5p are central hubs and miR-5001 and miR-7975 are potentially novel miRNAs associated with early atherosclerosis. In summary, we have identified miRNAs expressed in lesions and in blood that correlate with lesion burden and are potential therapeutic targets and biomarkers. These findings are a first step in elucidating miRNA regulated molecular mechanisms that underlie early atherosclerosis in a baboon model, enabling translation of our findings to humans.
Subject terms: Genetics, Non-coding RNAs, miRNAs
Introduction
Atherosclerotic cardiovascular disease (CVD) is the leading cause of morbidity and mortality globally, especially in developed countries1. Atherogenesis can be defined as a result of gene and protein expression changes leading to altered cellular processes and remodeling of local blood vessels. Atherogenesis is a complex progressive process involving many cellular processes in vascular cells, including endothelial cells, smooth muscle cells, and platelets, and in macrophages and innate immune cells2–5. Many central mechanisms are associated with atherogenesis, but the most compelling, include the modification of LDL via oxidation, desialylation of lipoproteins; and particle size alteration; activation and injury of vascular endothelial cells and infiltration of modified LDL into the intimal layer; formation of foam cells; production of cytokines and inflammation cascade; mitochondria dysfunction; dedifferentiation of smooth muscle cells; and autophagy impairment leading to accumulation of cell debris and plaque formation5–9.
Many human studies are focused on late-stage anatomical changes that are evident long after these initial molecular changes partially because of availability of arterial tissues after surgery10,11. The key to reducing CVD mortality and morbidity is early detection and treatment of atherosclerosis prior to irreversible tissue changes with advancement to life-threatening late-stage plaques12–14. To achieve this goal, there is a need to better understand the molecular mechanisms underlying the changes that lead to initiation and progression of atherosclerosis for prevention and treatment.
We posit that microRNAs (miRNAs) play a regulatory role in gene and protein expression changes observed during atherogenesis, and that miRNA related molecular mechanisms underlie initiation and progression of atherosclerosis. miRNAs are small non-coding RNAs that regulate gene expression post-transcriptionally by binding mRNA untranslated regions, resulting in degradation of the mRNA or inhibition of translation15. The first step in deciphering these miRNA related mechanisms linked to CVD is to compare miRNA expression in early atherosclerotic lesions with healthy sites of arteries and identify gene targets of these miRNAs as well as miRNA-gene networks involved.
The baboon model of atherosclerosis presents opportunities to investigate early molecular changes leading to development of atherosclerosis. It is not tenable to obtain arterial tissues from apparently healthy humans to decipher initial molecular changes accompanying early atherosclerosis. Baboons develop atherosclerosis naturally like humans8; unlike murine models, baboons do not require genetic manipulation to render them susceptible to atherosclerosis16. Non-human primates share similar genetic and physiological characteristics, including those involved in lipid metabolism and immunity with humans17. Therefore, understanding miRNA molecular mechanisms underlying atherosclerosis initiation in non-human primates is translatable to humans. The goal of our study is to provide novel insights to miRNA-related mechanisms in early atherosclerotic lesions in baboons. To our knowledge this is the first study to do so in non-human primates, and it provides a foundation to understanding atherosclerosis initiation as a basis for early detection and intervention in humans.
We previously challenged baboons with a high cholesterol, high fat (HCHF) diet for 2 years to stimulate early atherosclerosis (n = 112). We observed that HCHF diet stimulated increase of atherogenic markers and development of atherosclerotic lesions in common iliac arteries (CIA), a surrogate of systemic atherosclerosis18,19, with extensive variation in lesions among study animals19. Further, we evaluated the characteristics of these lesions, corresponding to fatty streaks and fibrous plaques20. In the current study, we quantified miRNAs expressed in the lesions and no-lesion sites of the CIA, and in blood samples. Comparing lesion and no-lesion sites of the same tissue of individual animals enables complete control of genetic and environmental factors that might influence miRNA expression. Using Ingenuity Pathway Analysis (IPA), we identified predicted miRNA targets and miRNA-gene networks that are central to deciphering molecular mechanisms underlying initiation and progression of atherosclerosis in primates. Finally, we discuss miRNAs expressed in atherosclerotic vasculature in humans and in baboons.
Materials and methods
Humane care guidelines
All research procedures involving animals for this study were conducted in facilities certified by the Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC) at the Southwest National Primate Research Center (SNPRC) in San Antonio, Texas by board certified veterinarians and veterinary staff. SNPRC veterinarian euthanized animals using ketamine and pentobarbital followed by exsanguination in accordance with American Veterinary Medical Association (AVMA) and Animal Research: Reporting of In Vivo Experiments (ARRIVE) guidelines and were approved by the Institutional Animal Care and Use Committee (IACUC) at the Texas Biomedical Research Institute, the host institution for the SNPRC.
Diet challenge and tissue collection
In previous work, 112 adult baboons (47 females and 65 males) were challenged with HCHF diet for 2 years and tissues were collected at the end of the study. Blood samples were collected at the beginning (baseline) and end of the study. Mean age of the females was 12.6 years (range = 8.7–17.0 years) and that of the males was 11.4 years (range = 8.1–14.1 years). Study details and diet composition have been described19.
CIA were collected and preserved in 10% formalin. The CIA were defatted and dissected longitudinally and stained as described19,21. The lesion type and the extent (proportion of tissue surface area covered by lesions) of atherogenic lesions was evaluated as described19. The lesion types were validated by immunohistochemistry20.
For the current study, a subset of CIA was selected from the 112 samples that represented the two categories of lesion type: fatty streaks (FS) and fibrous plaques (FP). The characteristics of the samples included 3 males and 5 females for FS, and 4 males and 4 females for FP. Mean age of the females was 12.0 years (range = 11.1–12.5 years) and that of the males was 11.2 years (range = 11.6–10.9 years).
Total RNA isolation
For the same animals, we isolated total RNA from baseline blood samples as described22 and from CIA using RecoverAll kit (Ambion). Briefly, we excised paired lesion and no-lesion sites of CIA from each tissue sample. We homogenized the tissues in lysis buffer using a Mini-Beadbeater-96 (Biospecproducts) for 5 min. The lysates were processed using the kit according to manufacturer’s protocol. RNA was quantified using Qubit Fluorimeter (DeNovix).
miRNA sequencing and sequence analysis
These steps were performed as described previously22. Briefly, for each sample, we used 500 ng of total RNA to generate cDNA libraries using NextFlex Small RNA-Seq Kit v3 (PerkinElmer). After cDNA quality check, two sets of 24 samples each were pooled and quantified. We multiplexed cDNA into two pools, each containing 24 samples, and hybridized the pool to an individual lane of the flow cell. We performed small RNA sequencing using Illumina HiSeq 2500 instrument and reagents, including HiSeq Rapid Cluster v2 and HiSeq Rapid SBS Kit v2 (50 cycles) for cluster generation. We used mirDeep2 pipeline to analyze fastq-formatted sequence reads with a Phred quality score of 30 or greater and an inferred base call accuracy of 99.9% to identify miRNA sequences and assess expression levels (read counts). miRNA read counts were normalized holistically using reads per million mapped to miRNA.
miRNA target identification, miRNA-gene networks, and pathway analysis
These steps were performed as described previously22. Briefly, differentially expressed miRNAs were imported to IPA and Core analysis performed to identify miRNA target genes related to cardiovascular system signaling and to generate networks. Putative miRNA target genes were identified using TargetScan embedded in IPA. Subsequently, we used miRNA targets predicted with high confidence and experimentally validated targets, to identify miRNA-gene networks.
Statistical analysis
We used the One-Way analysis of variance tool embedded in Partek Genomics Suite (Partek Inc.) to compare differences in variable distribution between lesion and no-lesion regions of CIA per lesion type and between lesion type. We used Prism 8 (GraphPad) to perform Pearson’s correlation analysis and identify circulating miRNAs expressed at baseline that associated with the extent of atherosclerosis measured after 2-year diet challenge. Statistical significance was tested with p < 0.05, and all tests were two-tailed. Multiple testing correction was performed using FDR q-value < 0.05.
Results
Number of expressed miRNAs
We analyzed miRNAs expressed in 8 pairs (lesion and no-lesion) of CIA with FS and 8 pairs with FP, collected after 2 years of diet challenge. In addition, we analyzed miRNAs expressed in blood samples from the same animals, collected at baseline prior to the 2 years diet challenge. We observed that 412 miRNAs were expressed in CIA (passing quality filters). Three hundred fifty-six miRNAs were expressed in blood samples (passing quality filters). Two hundred ninety-two miRNAs were expressed in both CIA and blood samples.
Differentially expressed miRNAs
We identified miRNAs with differing expression between lesion and non-lesion regions of CIA. In FS samples, 9 miRNAs were significantly up-regulated compared to the no-lesion site of the tissue (Table 1). For FP samples, 27 miRNAs (17 up-regulated and 10 down-regulated) were differentially expressed compared to no-lesion site of the tissue (Table 2).
Table 1.
Differentially expressed miRNAs between FS and no lesion regions in baboon CIA after 2-year diet challenge.
| ID | p-value | q-value | Fold-Change |
|---|---|---|---|
| pha-miR-7975 | 1.99E−10 | 7.26E−08 | 8.6 |
| pha-miR-486-5p | 8.27E−06 | 1.51E−03 | 4.6 |
| pha-miR-144-3p | 8.68E−04 | 4.52E−02 | 3.3 |
| pha-miR-191-5p | 3.28E−05 | 3.98E−03 | 3.2 |
| pha-miR-451a | 8.65E−04 | 4.52E−02 | 3.1 |
| pha-miR-106b-5p | 6.41E−04 | 4.52E−02 | 3.0 |
| pha-miR-148a-3p | 8.64E−04 | 4.52E−02 | 2.9 |
| pha-miR-93-5p | 1.18E−03 | 4.79E−02 | 2.5 |
| pha-miR-378c | 1.08E−03 | 4.79E−02 | 2.1 |
Fold-change is defined as the ratio of the average expression level of a miRNA in FP lesion and in no-lesion control and log2 transformed. Negative fold-change values were calculated as –(1/Fold-change). p < 0.05, q < 0.05.
Table 2.
Differentially expressed miRNAs between FP and no lesion regions in baboon CIA after 2-year diet challenge.
| ID | p-value | q-value | Fold-Change |
|---|---|---|---|
| pha-miR-451a | 2.12E−05 | 1.18E−03 | 7.8 |
| pha-miR-146a-5p | 5.55E−05 | 1.92E−03 | 6.8 |
| pha-miR-7975 | 1.15E−09 | 3.83E−07 | 6.7 |
| pha-miR-191-5p | 8.86E−07 | 1.46E−04 | 6.1 |
| pha-miR-486-5p | 1.32E−06 | 1.46E−04 | 5.9 |
| pha-miR-144-3p | 1.11E−05 | 7.42E−04 | 5.8 |
| pha-miR-106b-5p | 1.54E−04 | 4.32E−03 | 4.5 |
| pha-miR-17-5p | 5.74E−05 | 1.92E−03 | 4.2 |
| pha-miR-93-5p | 1.55E−04 | 4.32E−03 | 4.2 |
| pha-miR-342-3p | 1.47E−03 | 2.26E−02 | 3.9 |
| pha-miR-25-3p | 1.17E−03 | 2.05E−02 | 3.7 |
| pha-miR-222-3p | 4.19E−05 | 1.75E−03 | 2.8 |
| pha-miR-221-3p | 2.25E−03 | 2.78E−02 | 2.6 |
| pha-miR-21-5p | 8.19E−04 | 1.61E−02 | 2.4 |
| pha-miR-92a-3p | 1.66E−03 | 2.31E−02 | 2.4 |
| pha-let-7g-5p | 2.18E−03 | 2.78E−02 | 1.5 |
| pha-let-7f-5p | 9.71E−04 | 1.80E−02 | 1.3 |
| pha-miR-24-3p | 3.15E−04 | 7.01E−03 | − 1.4 |
| pha-miR-145-5p | 1.75E−03 | 2.33E−02 | − 1.5 |
| pha-miR-143-3p | 2.59E−04 | 6.16E−03 | − 1.5 |
| pha-miR-23b-3p | 4.02E−04 | 8.38E−03 | − 1.6 |
| pha-miR-28-3p | 1.49E−03 | 2.26E−02 | − 2.0 |
| pha-miR-143-3p | 1.04E−05 | 7.42E−04 | − 2.0 |
| pha-miR-5100 | 3.20E−05 | 1.52E−03 | − 2.1 |
| pha-let-7e-5p | 3.10E−03 | 3.69E−02 | − 2.1 |
| pha-miR-195-5p | 1.64E−03 | 2.31E−02 | − 2.3 |
| pha-miR-23c | 1.49E−03 | 2.26E−02 | − 4.1 |
Fold-change is defined as the ratio of the average expression level of a miRNA in FP lesion and in no-lesion control and log2 transformed. Negative fold-change values were calculated as –(1/Fold-change). p < 0.05, q < 0.05.
Lesion-specific expressed miRNAs
Two miRNAs (miR-148a-3p and miR-378c) up-regulated were unique to FS lesions. Twenty miRNAs (10 up-regulated; miR-222-3p, miR-221-3p, miR-17-5p, miR-21-5p, miR-7f-5p, miR-25-3p, miR-342-3p, miR-92a-3p, miR-7e-5p, miR-a-5p and 10 down-regulated; miR-143-5p, miR-5100, miR-1461-5p, miR-143-3p, miR-24-3p, miR-23b-3p, miR-23c, miR-28-3p, miR-195-5p, miR-145-5p) were uniquely expressed in FP lesions. Seven miRNAs (miR-7975, miR-486-5p, miR-144-3p, miR-191-5p, miR-106b-5p, miR-93-5p, miR-451a) were upregulated in both FS and FP when compared to the no-lesion site of the tissue (Fig. 1). miRNAs expressed in FS or in FP did not show significant sex differences after adjusting for multiple testing, q < 0.05.
Figure 1.
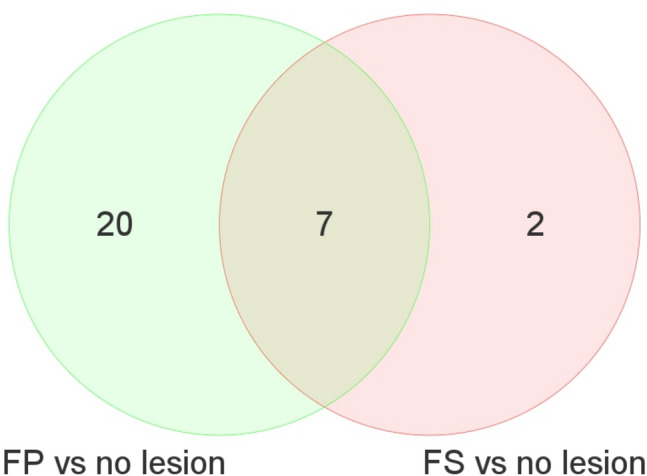
Number of miRNAs differentially expressed between lesion and no lesion sites of baboon CIA.
Circulating miRNAs that correlate with extent of lesions
We performed correlation analysis to identify circulating miRNAs in blood samples collected at baseline that correlated with extent of atherosclerosis. For the study cohort, the average extent of early atherosclerosis was 14% and ranged from 6 to 77%. The average number of plaques was 1, ranging from 1 to 7. We identified 14 miRNAs that significantly correlated with extent of CIA atherosclerotic lesions and the number of plaques (Table 3). Four miRNAs (miR-30d-5p, miR-21-5p, miR-339-5p, and miR-1301-3p) uniquely predicted extent of FS lesions, three miRNAs (miR-30a-5p, miR-30c-5p, and 5699-5p) uniquely predicted extent of FP, and miR-3200-3p predicted number of plaques. Two miRNAs (miR-30a-3p and miR-10527-5p) predicted both the extent and number of FP lesions.
Table 3.
Baseline expressed circulating miRNAs that significantly correlate with extent of fatty streaks, fibrous plaques, and the number of plaques in baboon CIA after 2-year diet challenge.
| Baseline blood miRNAs | Extent of FS | Extent of FP | Number of plaques |
|---|---|---|---|
| pha-miR-30a-5p | 0.06 (− 0.36) | 0.03 (− 0.40)* | 0.06 (− 0.36) |
| pha-miR-30c-5p | 0.19 (− 0.25) | 0.04 (− 0.39)* | 0.06 (− 0.35) |
| pha-miR-486-5p | 0.03 (0.41)* | 0.11 (0.31) | 0.03 (0.41)* |
| pha-miR-5699-5p | 0.29 (− 0.21) | 0.04 (− 0.38)* | 0.09 (− 0.32) |
| pha-miR-30a-3p | 0.14 (− 0.29) | 0.01 (− 0.46)* | 0.03 (− 0.41)* |
| pha-miR-3200-3p | 0.05 (− 0.29) | 0.1 (− 0.46) | 0.04 (− 0.41)* |
| pha-miR-10527-57 | 0.29 (− 0.21) | 0.01 (− 0.48)* | 0.02 (− 0.42)* |
| pha-miR-873-3p | 0.02 (0.43)* | 0.20 (− 0.48) | 0.04 (− 0.42)* |
| pha-miR-3155a | 0.02 (0.42)* | 0.14 (0.28) | 0.02 (− 0.42)* |
| pha-miR-3155b | 0.02 (0.42)* | 0.13 (0.29) | 0.02 (0.42)* |
| pha-miR-30d-5p | 0.02 (− 0.43)* | 0.42 (− 0.15) | 0.38 (− 0.17) |
| pha-miR-21-5p | 0.04 (− 0.26)* | 0.28 (− 0.21) | 0.33 (− 0.19) |
| pha-miR-339-5p | 0.02 (− 0.42)* | 0.09 (− 0.32) | 0.14 (− 0.28) |
| pha-miR-1301-3p | 0.04 (0.38)* | 0.05 (0.37) | 0.16 (0.27) |
The numerals in the second to fourth columns depict p values and in parathesis are Pearson’s correlation coefficient (r) values. The asterisk indicates a correlation that reached a significant level, p < 0.05.
miRNA targets and miRNA-gene networks
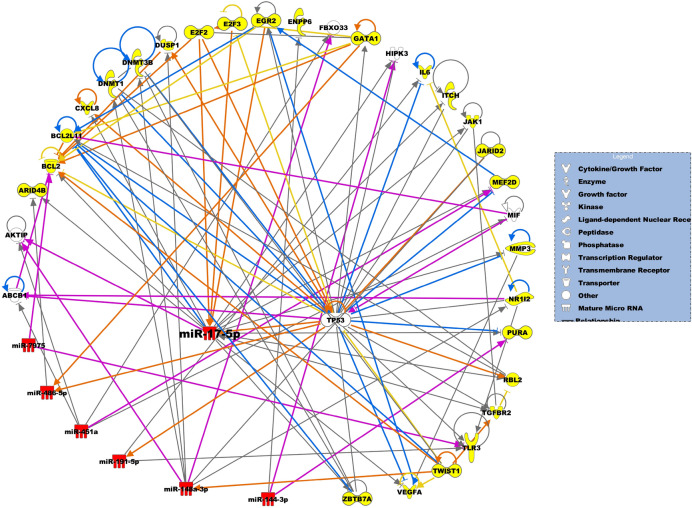
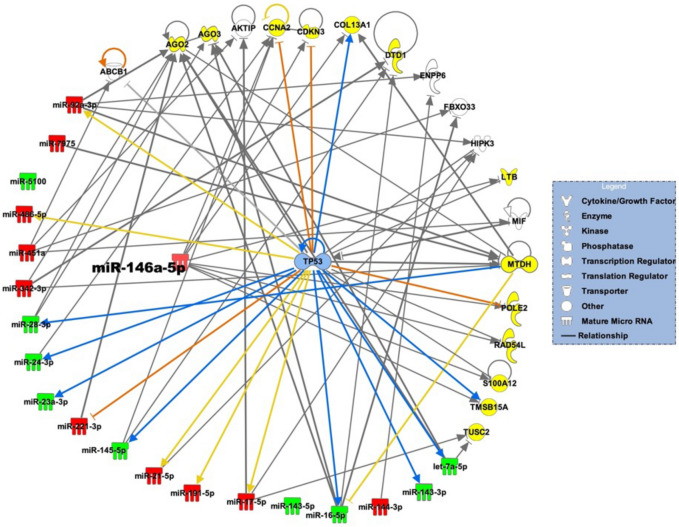
We identified genes predicted to be targeted by the differentially expressed miRNAs. Supplemental Table 1 shows the miRNA targets. Figures 2 and 3 show the miRNA-gene networks, respectively, related to FS and FP lesions. For the FS network, 25 putative genes are targeted by differentially expressed miRNAs, including IL6, MMP3, TGFBR2, TLR3, BCL2, CXCL8 and VGFA, and miR-17-5p is the central regulatory molecule. For the FP network, differentially expressed miRNAs target 13 genes, including CCNA2, CDKN3, COL13A1, DTD1, POLE2, and TUSC2, and miR-146a-5p is the central regulatory molecule. Seven genes are targeted by differentially expressed miRNAs in both FS and FP lesions, including TP53, ABCB1, FBXO33, AKTIP, ENPP6, HIPK3, and MIF.
Figure 2.
miRNA-gene network that underlie development of FS lesions. miRNAs and predicted miRNA target genes are represented as nodes. Red nodes indicate upregulated miRNAs. Yellow nodes depict miRNA target genes unique to FS lesions. The molecular relationships between nodes is presented as a line (edge); arrows indicate the direction of direct interaction.
Figure 3.
miRNA-gene network that underlie development of FP lesions. miRNAs and predicted miRNA target genes are represented as nodes. Red and green nodes indicate, respectively, upregulated and downregulated miRNAs. Yellow nodes depict miRNA target genes unique to FP lesions. The molecular relationships between nodes is presented as a line (edge); arrows indicate the direction of direct interaction.
Discussion
Identification of miRNAs expressed in relevant tissues and gene targets is essential for providing insights into molecular mechanisms underlying initiation and progression of atherosclerosis. In this study we analyzed baboon CIA samples with early-stage atherosclerotic lesions, including FS, which are initial lesions, and FP, which are relatively more advanced than FS20. We identified miRNAs differentially expressed between paired lesion and no-lesion sites in the vascular tissue, putative genes targeted by the differentially expressed miRNAs, and miRNA-gene networks specific to the lesion types. In addition, we identified potential circulating miRNA biomarkers predicting burden of atherosclerosis. To our knowledge, this is the first study to identify miRNAs expressed in vascular tissues with early atherosclerotic lesions and their putative targets in a non-human primate model of atherosclerosis. Our findings provide critical insights to identifying potential miRNA related molecular mechanisms that underlie early atherosclerosis in primates.
In our study we identified miRNAs that are differentially expressed between lesion and no-lesion sites of CIA and that which are lesion specific. Differentially expressed miRNAs include 8 miRNAs (miR-7975, miR-486-5p, miR-451a, miR-191-5p, miR-148a-3p, miR-17-5p, miR-378c, and miR-144-3p) in FS lesions, and 27 miRNAs (e.g., miR-92a-3p, -5001, -342-3p, miR-28-3p, -21-5p, -221-3p, and -16-5p) in FP lesions. Two miRNAs (miR-148a-3p and miR-378c) were uniquely expressed in FS lesions and 20 miRNAs (miR-143-5p, miR-5100, miR-222-3p, miR-1461-5p, miR-17-5 pm, miR-106a-5p, miR-143-3p, miR-24-3p, miR-23b-3p, miR-21-5p, miR-7f-5p, miR-25-3p, miR-342-3p, miR-23c, miR-28-3p, miR-195-5p, miR-92a-3p, miR-145-5p, miR-221-3p, miR-7e-5p) were uniquely expressed in FP lesions. Our findings show that some miRNAs are not only lesion-type specific but exhibit differential expression in different stages of the disease indicating relevance in disease initiation and progression.
Previous in vitro studies have showed that some of the differentially expressed, lesion-specific miRNAs identified in this study are involved in atherogenesis. For example, miR-143, miR-221 and miR-146a promote proliferation of vascular smooth cells23–25, a hallmark of initiation of atherogenesis. In contrast, miR-221 promotes apoptosis in endothelial cells by targeting PGC-1alpha resulting in mitochondria dysfunction and accumulation of RO species26. miR-92a is upregulated by shear stress and expressed in regions that are athero-susceptible27. In macrophages, miR-106, miR-378, miR-148a inhibit cholesterol efflux by cooperatively targeting ABCA/G128–30.
We have identified vascular miRNAs expressed in baboons and in humans and potentially novel miRNAs associated with atherosclerosis. For example, we observed that miR-144-3p, miR-146a-5p, miR-21, miR-221/222-3p are up-regulated in FP lesions and miR-195-5p was downregulated. In human studies, miR-21 and miR-146a-5p were up-regulated in carotid and aortic lesions11 while miR-144-5p and miR-221-3p were up-regulated and miR-195-5p down-regulated in tissues with aortic aneurysm compared to control31. Currently miR-195 is in preclinical trial for treatment of post-myocardial infarction32. These human studies collaborate our baboon findings. Moreover, we identified potentially novel miRNAs expressed in CIA early atherosclerotic lesions, including miR-7975and miR-5001. In our study miR-7975 exhibited high fold changes, respectively, 8.6 and 6.7 in FS and in FP lesions. Previous studies have shown that miR-7975 is involved in regulation of lung inflammation and in lung cancer33,34. In addition, a previous study indicated that miR-5001 expression was low in colorectal tumors and downregulation of miR-5001 that targets HES6 gene promoted cell proliferation35. Recent studies suggest miR-5001 is a potential biomarker for prostate and ovarian cancer36,37. Appreciating that the expression of miRNAs is tissue and/or cell specific, the role of miR-7975 and miR-5001 in development of atherosclerosis is not known and should be investigated in future studies.
miRNAs are potential biomarkers indicative of disease and health status and responsiveness to therapeutic interventions38–43. Findings from our study indicate that 14 circulating miRNAs are potential biomarkers of disease progression and lesion number, predicting the burden of CIA atherosclerosis. Of the 14 miRNAs, four (miR-21-5p, miR-339-5p, miR-30d-5p, and miR-1301-3p) correlated specifically with extent of FS lesions. Two miRNAs (miR-30a-3p and miR-10527-5p) correlated specifically with both the extent and number of fibrous plaques in CIA. Previous human studies have shown that miR-21-5p is overexpressed in serum of patients with coronary artery disease44–46. miR-339-5p and miR-1301-3p regulates smooth cell proliferation47,48 but to the best of our knowledge their role as biomarkers of human atherosclerosis is not known. Similarly, the role of miR-30d-5p, miR-30a-3p, and miR-10527-5p as biomarkers of human atherosclerosis is not known. Thus, our study presents potential novel biomarkers of early atherosclerosis.
We identified coordinately regulated miRNA-gene networks associated with early atherosclerotic lesions. The miRNA-gene networks play a critical role in putting large datasets into biological context for dissemination of potential hypothesis and molecular mechanisms related to disease status. For the FS network, among the seven miRNAs that target 25 genes, miR-17-5p is predicted to regulate most of the genes (13), including BCL2, VEGFA, CXCL8, IL6, MMPs, JAK1, ITCH, TLR3, and TGFBR2, which were previously demonstrated to be associated with atherosclerosis. For example, BCL2, encodes an outer mitochondrial membrane protein that is anti-apoptotic. The molecule is downregulated in aortic atherosclerotic lesions in rats49. VEGFA is a growth factor that induces proliferation and migration of vascular endothelial cells, and is essential for both physiological and pathological angiogenesis, resulting in plaque neovascularization, increased microvessel permeability, and intraplaque hemorrhage50,51. This process is linked to plaque progression through the accumulation of erythrocyte membranes, which are rich in free cholesterol and promote necrotic core enlargement52. CXCL8/IL-8 is a member of the CXC chemokine family and is a major mediator of the inflammatory response, a potent angiogenic factor, associated with development of atherosclerosis53. Although these genes are associated with atherogenesis, the mechanistic role of miR-17-5p on expression of these genes and involvement in the disease development is not known.
In addition, genes predicted to be targeted by the miR-17-5p, not previously associated with atherosclerosis, include ARID4B, E2F, GATA1, MEF2D, NR1I2, PURA, and RBL2. For example, ARID4B, encodes a protein with sequence similarity to retinoblastoma-binding protein-1, which functions in diverse cellular processes including proliferation, differentiation, apoptosis, oncogenesis, and cell fate in cancer cells. The expression of this gene was associated with air-pollution induced methylation in Multi-Ethic Study of Atherosclerosis (MESA) participants54. E2F, is a member of the E2F family of transcription factors. The E2F family plays a crucial role in the control of cell cycle and action of tumor suppressor proteins and is also a target of the transforming proteins of small DNA tumor viruses. This protein is involved in regulation of smooth muscle cell cycle55. We observe that these are potentially novel genes associated with development of atherosclerosis.
For the FP network, among the 19 miRNAs that target 13 genes, miR-146a-5p is the central hub predicted to regulate majority of the genes (8), including CCNA2, CDKN3, COL13A1, LTB, POLE2, RAD54L, S100A12, TUSC2, and TMSB15A. These genes have not previously been associated with development of atherosclerosis. The interactions of these genes with miR-146a-5p in relation to atherogenesis are not known. For example, CCNA2 protein encoded by this gene belongs to the highly conserved cyclin family, whose members function as regulators of the cell cycle. This protein binds and activates cyclin-dependent kinase 2 and thus promotes transition through G1/S and G2/M. CDKN3, is a cyclin-dependent kinase inhibitor, and has been shown to interact with, and dephosphorylate CDK2 kinase to regulate cell cycle. The gene was downregulated after exposure to cigarette smoke condensate56. Previous work demonstrated that miR-146a-5p interacts with CDKN3 in clear cell renal cell carcinoma57. COL13A1, is a collagen protein expressed in plasma membrane of connecting tissue-producing cells. Its function is not known. POLE2 is a DNA polymerase epsilon, which is involved in DNA repair and replication, is composed of a large catalytic subunit and a small accessory subunit. Defects in this gene have been linked to colorectal cancer and to combined immunodeficiency58. TUSC2 encodes a tumor suppressor 2 protein that is deleted in some carcinomas, including lung cancer. TUSC2 elicits its anti-tumor effects through regulating G1 cell cycle progression, apoptosis, calcium homeostasis, gene expression, and the activity of various protein tyrosine kinases and Ser/Thr kinases59.
Our findings suggest that miR-17-5p and miR-146a-5p, which are up regulated in FS and FP lesions, respectively, may be key molecules involved in molecular mechanisms that underlie initiation and progression of early atherosclerosis. Previous studies have demonstrated that miR-17-5p and miR-146a-5p are associated with atherosclerosis44,60–62. The miR-17-5p is activated in macrophages and promotes development of lesions. Silencing of this miRNA reduces lipid accumulation and production of inflammatory cytokines in vitro and in vivo to ameliorate atherosclerosis62. In addition, miR-17-5p is a circulatory biomarker of coronary atherosclerosis60. In contrast, miR-146a-5p has been shown to alleviate inflammation in endothelial cells and to reduce atherosclerosis in mice61. Our finding that miR-146a-5p is upregulated in lesions contradicts results from previous cellular and mouse studies but corroborate results of a study indicating that plasma miR-146a-5p is upregulated in human patients with myocardial infarction compared to control44. Our future studies will focus on the validation of the interactions between the miRNAs and miRNA targets identified in this study, and the identification of the molecular mechanisms by which miRNAs regulate the pathway leading to the development of atherosclerosis. These findings suggest new insights to potential roles of miRNAs in initiation and progression of atherosclerosis, and potential targets for early interventions.
Conclusion
For the first time, we have identified miRNAs that correlate with the types and extent of early atherosclerotic lesions in baboons and coordinated regulated miRNA-gene networks associated with early atherosclerosis. We identified 8 miRNAs that are differentially expressed in FS and 27 in FP lesions and novel miRNA targets associated with early atherosclerosis. We revealed that miR-17-5p and miR-146a-5p are potential gene expression regulatory hubs associated, respectively, with initiation and progression of early atherosclerotic lesions in a baboon model of atherosclerosis. In addition, we identified potentially novel therapeutic targets and biomarkers of early atherosclerosis. The findings provide insights to deepen the understanding of molecular mechanisms that underlie early atherosclerosis in baboons, potentiating translation to humans.
Limitations and future studies
Although our findings provide the first insights regarding miRNA and predicted miRNA targets associated with early atherosclerotic lesions, we reveal limitations that may hinder generalization. Because it was not feasible to perform RNA Seq on the FPEE samples to generate gene expression data (due to mRNA degradation), our study relied on computational prediction of miRNA targets with highly predicted confidence or experimental validation using public databases. Although we analyzed samples that are representative of each lesion type exhibited in the baboon cohort, future studies are needed to validate these findings in a larger cohort.
Our future studies will assess proteins from the CIA samples using mass spectrometry to identify expressed miRNA targets and perform molecular mechanistic studies to deepen our understanding of miRNA mechanistic roles in primate early atherosclerotic lesions.
Supplementary Information
Acknowledgements
We acknowledge the support of Samuel Galindo for staining the arteries. This work was supported by the American Heart Association (Grant Number: 4POST18150019) and the National Institutes of Health (Grant Number: K01HL130697 received by GMK and P01HL028972 received by JLV; NIH Office of the Director (Grant Number: P51 OD011133) received by JLV; NIH Office of the Director (Grant Number: P51 RR013986) received by JLV; (Grant Number: C06 RR14578; C06 RR15456; C06 RR013556; C06 RR017515). The funder supported the infrastructure that was used during the investigation of the work reported in the manuscript.
Author contributions
Conceptualization or design of the work, G.M.K., J.L.V., L.A.C. Acquisition and analysis of data, G.M.K., J.P.G., G.L., A.K.; Interpretation of data, G.M.K., L.A.C.; Drafting of the manuscript or substantial revision, G.M.K., J.L.V., L.A.C., J.P.G., G.L., A.K.
Data availability
For the current study, datasets generated during and/or analyzed are available in Sequence Read Achieve (SRA) repository (Accession Number PRJNA883251).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-023-29074-1.
References
- 1.Virani SS, Smith SC, Jr, Stone NJ, Grundy SM. Secondary prevention for atherosclerotic cardiovascular disease: Comparing recent US and European guidelines on dyslipidemia. Circulation. 2020;141(14):1121–1123. doi: 10.1161/CIRCULATIONAHA.119.044282. [DOI] [PubMed] [Google Scholar]
- 2.Bouhlel MA, Derudas B, Rigamonti E, Dievart R, Brozek J, Haulon S, Zawadzki C, Jude B, Torpier G, Marx N, et al. PPARgamma activation primes human monocytes into alternative M2 macrophages with anti-inflammatory properties. Cell Metab. 2007;6(2):137–143. doi: 10.1016/j.cmet.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 3.Hermansson A, Johansson DK, Ketelhuth DF, Andersson J, Zhou X, Hansson GK. Immunotherapy with tolerogenic apolipoprotein B-100-loaded dendritic cells attenuates atherosclerosis in hypercholesterolemic mice. Circulation. 2011;123(10):1083–1091. doi: 10.1161/CIRCULATIONAHA.110.973222. [DOI] [PubMed] [Google Scholar]
- 4.Khallou-Laschet J, Varthaman A, Fornasa G, Compain C, Gaston AT, Clement M, Dussiot M, Levillain O, Graff-Dubois S, Nicoletti A, et al. Macrophage plasticity in experimental atherosclerosis. PLoS ONE. 2010;5(1):e8852. doi: 10.1371/journal.pone.0008852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Libby P. Inflammation in atherosclerosis. Nature. 2002;420(6917):868–874. doi: 10.1038/nature01323. [DOI] [PubMed] [Google Scholar]
- 6.Ding Z, Wang X, Liu S, Shahanawaz J, Theus S, Fan Y, Deng X, Zhou S, Mehta JL. Corrigendum to: PCSK9 regulates expression of scavenger receptors and ox-LDL uptake in macrophages. Cardiovasc. Res. 2022;118(7):1849. doi: 10.1093/cvr/cvab354. [DOI] [PubMed] [Google Scholar]
- 7.Glanz V, Bezsonov EE, Soldatov V, Orekhov AN. Thirty-five-year history of desialylated lipoproteins discovered by Vladimir Tertov. Biomedicines. 2022;10(5):1174. doi: 10.3390/biomedicines10051174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kushwaha RS, McGill HC. Diet, plasma lipoproteins and experimental atherosclerosis in baboons (Papio sp.) Hum. Reprod. Update. 1998;4(4):420–429. doi: 10.1093/humupd/4.4.420. [DOI] [PubMed] [Google Scholar]
- 9.Mcgill HC, Mcmahan CA, Kruski AW, Mott GE. Relationship of lipoprotein cholesterol concentrations to experimental atherosclerosis in baboons. Arteriosclerosis. 1981;1(1):3–12. doi: 10.1161/01.ATV.1.1.3. [DOI] [PubMed] [Google Scholar]
- 10.He W, Zhu L, Huang Y, Zhang Y, Shen W, Fang L, Li J, Wang Z, Xie Q. The relationship of MicroRNA-21 and plaque stability in acute coronary syndrome. Medicine (Baltimore) 2019;98(47):e18049. doi: 10.1097/MD.0000000000018049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raitoharju E, Lyytikainen LP, Levula M, Oksala N, Mennander A, Tarkka M, Klopp N, Illig T, Kahonen M, Karhunen PJ, et al. miR-21, miR-210, miR-34a, and miR-146a/b are up-regulated in human atherosclerotic plaques in the Tampere Vascular Study. Atherosclerosis. 2011;219(1):211–217. doi: 10.1016/j.atherosclerosis.2011.07.020. [DOI] [PubMed] [Google Scholar]
- 12.Herrington DM, Mao C, Parker SJ, Fu Z, Yu G, Chen L, Venkatraman V, Fu Y, Wang Y, Howard TD, et al. Proteomic architecture of human coronary and aortic atherosclerosis. Circulation. 2018;137(25):2741–2756. doi: 10.1161/CIRCULATIONAHA.118.034365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ibanez B, Fernandez-Ortiz A, Fernandez-Friera L, Garcia-Lunar I, Andres V, Fuster V. Progression of early subclinical atherosclerosis (PESA) study: JACC focus seminar 7/8. J. Am. Coll. Cardiol. 2021;78(2):156–179. doi: 10.1016/j.jacc.2021.05.011. [DOI] [PubMed] [Google Scholar]
- 14.McGill HC, McMahan CA, Zieske AW, Sloop GD, Walcott JV, Troxclair DA, Malcom GT, Tracy RE, Oalmann MC, Strong JP, et al. Associations of coronary heart disease risk factors with the intermediate lesion of atherosclerosis in youth. Arterioscl. Throm. Vas. 2000;20(8):1998–2004. doi: 10.1161/01.ATV.20.8.1998. [DOI] [PubMed] [Google Scholar]
- 15.Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 16.Getz GS, Reardon CA. Animal models of atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 2012;32(5):1104–1115. doi: 10.1161/ATVBAHA.111.237693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cox LA, Olivier M, Spradling-Reeves K, Karere GM, Comuzzie AG, VandeBerg JL. Nonhuman primates and translational research-cardiovascular disease. ILAR J. 2017;58(2):235–250. doi: 10.1093/ilar/ilx025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clarkson TB, Anthony MS, Morgan TM. Inhibition of postmenopausal atherosclerosis progression: A comparison of the effects of conjugated equine estrogens and soy phytoestrogens. J. Clin. Endocrinol. Metab. 2001;86(1):41–47. doi: 10.1210/jcem.86.1.7151. [DOI] [PubMed] [Google Scholar]
- 19.Mahaney MC, Karere GM, Rainwater DL, Voruganti VS, Dick EJ, Jr, Owston MA, Rice KS, Cox LA, Comuzzie AG, VandeBerg JL. Diet-induced early-stage atherosclerosis in baboons: Lipoproteins, atherogenesis, and arterial compliance. J. Med. Primatol. 2018;47(1):3–17. doi: 10.1111/jmp.12283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karere GM, Dick EJ, Galindo S, Martinez JC, Martinez JE, Owston M, VandeBerg JL, Cox LA. Histological variation of early stage atherosclerotic lesions in baboons after prolonged challenge with high-cholesterol, high-fat diet. J. Med. Primatol. 2019;49:3–9. doi: 10.1111/jmp.12449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guzman MA, Mcmahan CA, Mcgill HC, Strong JP, Tejada C, Restrepo C, Teggen DA, Robertson WB, Solberg LA. Selected methodologic aspects of international atherosclerosis project. Lab. Investig. 1968;18(5):479. [PubMed] [Google Scholar]
- 22.Karere GM, Cox LA, Bishop AC, South AM, Shaltout HA, Mercado-Deane MG, Cuda S. Sex differences in MicroRNA expression and cardiometabolic risk factors in hispanic adolescents with obesity. J. Pediatr. 2021;235(138–143):e135. doi: 10.1016/j.jpeds.2021.03.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cordes KR, Sheehy NT, White MP, Berry EC, Morton SU, Muth AN, Lee TH, Miano JM, Ivey KN, Srivastava D. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature. 2009;460(7256):705–710. doi: 10.1038/nature08195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cordes KR, Srivastava D. MicroRNA regulation of cardiovascular development. Circ. Res. 2009;104(6):724–732. doi: 10.1161/CIRCRESAHA.108.192872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin Y, Liu X, Cheng Y, Yang J, Huo Y, Zhang C. Involvement of MicroRNAs in hydrogen peroxide-mediated gene regulation and cellular injury response in vascular smooth muscle cells. J. Biol. Chem. 2009;284(12):7903–7913. doi: 10.1074/jbc.M806920200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xue Y, Wei Z, Ding H, Wang Q, Zhou Z, Zheng S, Zhang Y, Hou D, Liu Y, Zen K, et al. MicroRNA-19b/221/222 induces endothelial cell dysfunction via suppression of PGC-1alpha in the progression of atherosclerosis. Atherosclerosis. 2015;241(2):671–681. doi: 10.1016/j.atherosclerosis.2015.06.031. [DOI] [PubMed] [Google Scholar]
- 27.Fang Y, Xiang J, Chen Z, Gu X, Li Z, Tang F, Zhou Z. miRNA expression profile of colon cancer stem cells compared to non-stem cells using the SW1116 cell line. Oncol. Rep. 2012;28(6):2115–2124. doi: 10.3892/or.2012.2054. [DOI] [PubMed] [Google Scholar]
- 28.Allen RM, Vickers KC. Coenzyme Q10 increases cholesterol efflux and inhibits atherosclerosis through microRNAs. Arterioscler. Thromb. Vasc. Biol. 2014;34(9):1795–1797. doi: 10.1161/ATVBAHA.114.303741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goedeke L, Rotllan N, Canfran-Duque A, Aranda JF, Ramirez CM, Araldi E, Lin CS, Anderson NN, Wagschal A, de Cabo R, et al. MicroRNA-148a regulates LDL receptor and ABCA1 expression to control circulating lipoprotein levels. Nat. Med. 2015;21(11):1280–1289. doi: 10.1038/nm.3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim J, Yoon H, Ramirez CM, Lee SM, Hoe HS, Fernandez-Hernando C, Kim J. MiR-106b impairs cholesterol efflux and increases Abeta levels by repressing ABCA1 expression. Exp. Neurol. 2012;235(2):476–483. doi: 10.1016/j.expneurol.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Plana E, Galvez L, Medina P, Navarro S, Fornes-Ferrer V, Panadero J, Miralles M. Identification of novel microRNA profiles dysregulated in plasma and tissue of abdominal aortic aneurysm patients. Int. J. Mol. Sci. 2020;21(13):4600. doi: 10.3390/ijms21134600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chakraborty C, Sharma AR, Sharma G, Lee SS. Therapeutic advances of miRNAs: A preclinical and clinical update. J. Adv. Res. 2021;28:127–138. doi: 10.1016/j.jare.2020.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pan Z, Yang G, He H, Gao P, Jiang T, Chen Y, Zhao G. Identification of cerebrospinal fluid MicroRNAs associated with leptomeningeal metastasis from lung adenocarcinoma. Front Oncol. 2020;10:387. doi: 10.3389/fonc.2020.00387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gohir W, Klement W, Singer LG, Palmer SM, Mazzulli T, Keshavjee S, Husain S. Identifying host microRNAs in bronchoalveolar lavage samples from lung transplant recipients infected with Aspergillus. J. Heart Lung Transpl. 2020;39(11):1228–1237. doi: 10.1016/j.healun.2020.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yan Y, Xuan B, Gao Z, Shen C, Cao Y, Hong J, Chen H, Cui Z, Ye G, Fang JY, et al. CCMAlnc promotes the malignance of colorectal cancer by modulating the interaction between miR-5001-5p and its target mRNA. Front Cell Dev. Biol. 2020;8:566932. doi: 10.3389/fcell.2020.566932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hamidi F, Gilani N, Belaghi RA, Sarbakhsh P, Edgunlu T, Santaguida P. Exploration of potential miRNA biomarkers and prediction for ovarian cancer using artificial intelligence. Front. Genet. 2021;12:724785. doi: 10.3389/fgene.2021.724785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mello-Grand M, Bruno A, Sacchetto L, Cristoni S, Gregnanin I, Dematteis A, Zitella A, Gontero P, Peraldo-Neia C, Ricotta R, et al. Two novel ceramide-like molecules and miR-5100 levels as biomarkers improve prediction of prostate cancer in gray-zone PSA. Front Oncol. 2021;11:769158. doi: 10.3389/fonc.2021.769158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Du S, Ling H, Guo Z, Cao Q, Song C. Roles of exosomal miRNA in vascular aging. Pharmacol. Res. 2021;165:105278. doi: 10.1016/j.phrs.2020.105278. [DOI] [PubMed] [Google Scholar]
- 39.Karere G, Bishop A, Cox L, Mercado-Deane MG, Cuda S. A potential MicroRNA signature of early atherosclerosis in obese adolescents. Arterioscl. Throm. Vas. 2019;39:A299. [Google Scholar]
- 40.Lu Y, Thavarajah T, Gu W, Cai J, Xu Q. Impact of miRNA in atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 2018;38(9):e159–e170. doi: 10.1161/ATVBAHA.118.310227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poller W, Dimmeler S, Heymans S, Zeller T, Haas J, Karakas M, Leistner DM, Jakob P, Nakagawa S, Blankenberg S, et al. Non-coding RNAs in cardiovascular diseases: Diagnostic and therapeutic perspectives. Eur. Heart J. 2018;39(29):2704–2716. doi: 10.1093/eurheartj/ehx165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang C, Wang J, Cui W, Liu Y, Zhou H, Wang Y, Chen X, Chen X, Wang Z. Serum Exosomal miRNA-1226 as potential biomarker of pancreatic ductal adenocarcinoma. Oncol. Targets Ther. 2021;14:1441–1451. doi: 10.2147/OTT.S296816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wei, T., Folkersen, L., Ehrenborg, E. & Gabrielsen, A. MicroRNA 486–3P as a stability marker in acute coronary syndrome. Biosci. Rep. 36(3) (2016). [DOI] [PMC free article] [PubMed]
- 44.Zhelankin AV, Stonogina DA, Vasiliev SV, Babalyan KA, Sharova EI, Doludin YV, Shchekochikhin DY, Generozov EV, Akselrod AS. Circulating extracellular miRNA analysis in patients with stable CAD and acute coronary syndromes. Biomolecules. 2021;11(7):962. doi: 10.3390/biom11070962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang F, Long G, Zhao C, Li H, Chaugai S, Wang Y, Chen C, Wang DW. Atherosclerosis-related circulating miRNAs as novel and sensitive predictors for acute myocardial infarction. PLoS ONE. 2014;9(9):e105734. doi: 10.1371/journal.pone.0105734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Velle-Forbord T, Eidlaug M, Debik J, Saether JC, Follestad T, Nauman J, Gigante B, Rosjo H, Omland T, Langaas M, et al. Circulating microRNAs as predictive biomarkers of myocardial infarction: Evidence from the HUNT study. Atherosclerosis. 2019;289:1–7. doi: 10.1016/j.atherosclerosis.2019.07.024. [DOI] [PubMed] [Google Scholar]
- 47.Jiang W, Zhao W, Ye F, Huang S, Wu Y, Chen H, Zhou R, Fu G. SNHG12 regulates biological behaviors of ox-LDL-induced HA-VSMCs through upregulation of SPRY2 and NUB1. Atherosclerosis. 2022;340:1–11. doi: 10.1016/j.atherosclerosis.2021.11.006. [DOI] [PubMed] [Google Scholar]
- 48.Huang T, Zhao HY, Zhang XB, Gao XL, Peng WP, Zhou Y, Zhao WH, Yang HF. LncRNA ANRIL regulates cell proliferation and migration via sponging miR-339-5p and regulating FRS2 expression in atherosclerosis. Eur. Rev. Med. Pharmacol. Sci. 2020;24(4):1956–1969. doi: 10.26355/eurrev_202002_20373. [DOI] [PubMed] [Google Scholar]
- 49.Long X, You G, Wu Q, Zhou Y, Xiao Y, Yu F, Deng S, Mo R, Song F, Huang J, et al. HomeoboxC6 affects the apoptosis of human vascular endothelial cells and is involved in atherosclerosis. J. Cell Physiol. 2021;236(3):1913–1925. doi: 10.1002/jcp.29974. [DOI] [PubMed] [Google Scholar]
- 50.Finn AV, Narula J. Intraplaque hemorrhage: most dangerous is the wound that bleedeth inwardly. JACC Cardiovasc. Imaging. 2012;5(8):856–858. doi: 10.1016/j.jcmg.2012.07.003. [DOI] [PubMed] [Google Scholar]
- 51.Jain RK, di Tomaso E, Duda DG, Loeffler JS, Sorensen AG, Batchelor TT. Angiogenesis in brain tumours. Nat. Rev. Neurosci. 2007;8(8):610–622. doi: 10.1038/nrn2175. [DOI] [PubMed] [Google Scholar]
- 52.Kolodgie FD, Gold HK, Burke AP, Fowler DR, Kruth HS, Weber DK, Farb A, Guerrero LJ, Hayase M, Kutys R, et al. Intraplaque hemorrhage and progression of coronary atheroma. N. Engl. J. Med. 2003;349(24):2316–2325. doi: 10.1056/NEJMoa035655. [DOI] [PubMed] [Google Scholar]
- 53.Meng Y, Zhang C, Liang L, Wei L, Wang H, Zhou F, Li R, Zou D, Huang X, Liu J. Identification of potential key genes involved in the carotid atherosclerosis. Clin. Interv. Aging. 2021;16:1071–1084. doi: 10.2147/CIA.S312941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chi GC, Liu Y, MacDonald JW, Barr RG, Donohue KM, Hensley MD, Hou L, McCall CE, Reynolds LM, Siscovick DS, et al. Long-term outdoor air pollution and DNA methylation in circulating monocytes: Results from the Multi-Ethnic Study of Atherosclerosis (MESA) Environ. Health. 2016;15(1):119. doi: 10.1186/s12940-016-0202-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Janjanam J, Zhang B, Mani AM, Singh NK, Traylor JG, Jr, Orr AW, Rao GN. LIM and cysteine-rich domains 1 is required for thrombin-induced smooth muscle cell proliferation and promotes atherogenesis. J. Biol. Chem. 2018;293(9):3088–3103. doi: 10.1074/jbc.RA117.000866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nordskog BK, Blixt AD, Morgan WT, Fields WR, Hellmann GM. Matrix-degrading and pro-inflammatory changes in human vascular endothelial cells exposed to cigarette smoke condensate. Cardiovasc. Toxicol. 2003;3(2):101–117. doi: 10.1385/CT:3:2:101. [DOI] [PubMed] [Google Scholar]
- 57.Wotschofsky Z, Gummlich L, Liep J, Stephan C, Kilic E, Jung K, Billaud JN, Meyer HA. Integrated microRNA and mRNA signature associated with the transition from the locally confined to the metastasized clear cell renal cell carcinoma exemplified by miR-146-5p. PLoS ONE. 2016;11(2):e0148746. doi: 10.1371/journal.pone.0148746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Belhadj S, Terradas M, Munoz-Torres PM, Aiza G, Navarro M, Capella G, Valle L. Candidate genes for hereditary colorectal cancer: Mutational screening and systematic review. Hum. Mutat. 2020;41(9):1563–1576. doi: 10.1002/humu.24057. [DOI] [PubMed] [Google Scholar]
- 59.Rimkus T, Sirkisoon S, Harrison A, Lo HW. Tumor suppressor candidate 2 (TUSC2, FUS-1) and human cancers. Discov. Med. 2017;23(128):325–330. [PMC free article] [PubMed] [Google Scholar]
- 60.Chen J, Xu L, Hu Q, Yang S, Zhang B, Jiang H. MiR-17-5p as circulating biomarkers for the severity of coronary atherosclerosis in coronary artery disease. Int. J. Cardiol. 2015;197:123–124. doi: 10.1016/j.ijcard.2015.06.037. [DOI] [PubMed] [Google Scholar]
- 61.Huang SF, Zhao G, Peng XF, Ye WC. The pathogenic role of long non-coding RNA H19 in atherosclerosis via the miR-146a-5p/ANGPTL4 pathway. Front Cardiovasc. Med. 2021;8:770163. doi: 10.3389/fcvm.2021.770163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tan L, Liu L, Jiang Z, Hao X. Inhibition of microRNA-17-5p reduces the inflammation and lipid accumulation, and up-regulates ATP-binding cassette transporterA1 in atherosclerosis. J. Pharmacol. Sci. 2019;139(4):280–288. doi: 10.1016/j.jphs.2018.11.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
For the current study, datasets generated during and/or analyzed are available in Sequence Read Achieve (SRA) repository (Accession Number PRJNA883251).