Abstract
Festuca rubra subsp. pruinosa is a perennial grass growing in sea cliffs where plants are highly exposed to salinity and marine winds, and often grow in rock fissures where soil is absent. Diaporthe species are one of the most abundant components of the root microbiome of this grass and several Diaporthe isolates have been found to produce beneficial effects in their host and other plant species of agronomic importance. In this study, 22 strains of Diaporthe isolated as endophytes from roots of Festuca rubra subsp. pruinosa were characterized by molecular, morphological, and biochemical analyses. Sequences of the nuclear ribosomal internal transcribed spacers (ITS), translation elongation factor 1-α (TEF1), beta-tubulin (TUB), histone-3 (HIS), and calmodulin (CAL) genes were analyzed to identify the isolates. A multi-locus phylogenetic analysis of the combined five gene regions led to the identification of two new species named Diaporthe atlantica and Diaporthe iberica. Diaporthe atlantica is the most abundant Diaporthe species in its host plant, and Diaporthe iberica was also isolated from Celtica gigantea, another grass species growing in semiarid inland habitats. An in vitro biochemical characterization showed that all cultures of D. atlantica produced indole-3-acetic acid and ammonium, and the strains of D. iberica produced indole 3-acetic acid, ammonium, siderophores, and cellulase. Diaporthe atlantica is closely related to D. sclerotioides, a pathogen of cucurbits, and caused a growth reduction when inoculated in cucumber, melon, and watermelon.
Keywords: grasses, Festuca rubra subsp. pruinosa, endophyte, multi-locus phylogeny, Diaporthe atlantica, Diaporthe iberica
1. Introduction
The genus Diaporthe is highly complex, according to Index Fungorum (2022) comprises nearly 1,000 fungal names without taking into account the 984 names attributed to its asexual state Phomopsis. Diaporthe is worldwide-distributed and associated with a broad range of host plants. Some members of this genus are important pathogens, responsible for several economically significant plant diseases including stem, root and fruit rots, gummosis, cankers, leaf spots, blights, diebacks, decay, and wilts on hosts such as citrus, grapevines, soybean, peach, or sunflower, to name just a few (Udayanga et al., 2011; Marin-Felix et al., 2019). They can also colonize decaying plant tissues as saprophytes (Thongkantha et al., 2008), or live endophytically inside healthy plant tissues (Gomes et al., 2013).
As endophytes, Diaporthe species have been isolated from agricultural, medicinal and ornamental plants growing in a wide variety of locations and habitats (Huang et al., 2015; Dos Santos et al., 2016, 2021; Yang et al., 2018). They have also been reported as dominant taxa in fungal microbiomes of plant species like Tectona grandis (Murali et al., 2006), the Brazilian medicinal plants Vochysia divergens and Stryphnodendron adstringens (Noriler et al., 2018), olive trees (Martins et al., 2016), Fagus crenata (Tateno et al., 2015) and the grass Festuca rubra subsp. pruinosa (Pereira et al., 2019). Non-pathogenic Diaporthe species have been recognized as plant growth promoters (Vázquez de Aldana et al., 2021; da Silva Santos et al., 2022; Toghueo et al., 2022), and biocontrol agents (Marak et al., 2002; Dos Santos et al., 2016; Dini-Andreote, 2020; Abramczyk et al., 2022). From an industrial and pharmaceutical point of view, they are well-known producers of enzymes (Lange et al., 2021; Baluyot et al., 2022) and secondary metabolites exhibiting a wide range of activities including antimicrobial, antiviral, antioxidant, anti-inflammatory and anticancer (Chepkirui and Stadler, 2017; Xu et al., 2021). Therefore, studies aiming to analyze and better characterize Diaporthe species are of interest, particularly to fully comprehend not only their role as ubiquitous species in nature, but also their potential as metabolite producers, plant growth promoters, biocontrol agents, or for control measures in case of potential pathogenicity.
Previously, Diaporthe species were identified on the basis of morphological characteristics (e.g., colony phenotype, size, shape, and type of spores) and host specificity (Rehner and Uecker, 1994; Udayanga et al., 2011). This methodology of Diaporthe identification resulted in a proliferation of names in the literature. However, several studies demonstrated that morphological characters are not sufficient for species level diagnoses, and that many species of Diaporthe have multihost capability (Rehner and Uecker, 1994; Thompson et al., 2011, 2015; Udayanga et al., 2012). Nowadays, Diaporthe species are being redefined based on a polyphasic approach, employing a combination of information including morphological characteristics and multi-locus sequence data (Udayanga et al., 2012, 2014a,b; Gomes et al., 2013; Norphanphoun et al., 2022). Currently, the taxonomy of Diaporthe species is being resolved by means of multigene phylogenetic analyses based on sequences of the 5.8S rDNA and internal transcribed spacers (ITS1-5.8S-ITS2), translation elongation factor 1-alpha (TEF1), beta-tubulin (TUB), histone H3 (HIS), and calmodulin (CAL) genes (Udayanga et al., 2012; Gomes et al., 2013; Marin-Felix et al., 2019; Norphanphoun et al., 2022). Using this methodological approach, the delimitation of species in the genus has improved. Lately, Diaporthe species occurring in several host plant families have been well-characterized and sometimes reclassified, resulting in an actively evolving taxonomy with numerous novel species being described each year (Gomes et al., 2013; Dissanayake, 2017; Guarnaccia et al., 2018; Yang et al., 2018; Guo et al., 2020; Wang et al., 2021; Norphanphoun et al., 2022). Despite the attention given to pathogenic Diaporthe species occurring in economically important crops, more effort is needed to understand the ecology and distribution of non-pathogenic species occurring in numerous plant species.
Festuca rubra, commonly known as red fescue, is a perennial grass distributed across a very diverse range of habitats in the Northern Hemisphere. Red fescues are cultivated and used as turfgrasses in ornamental and sports lawns, and some cultivars have been used for phytoremediation and rehabilitation of damaged soils (Braun et al., 2020). Among its several subspecies, Festuca rubra subsp. pruinosa (Festuca pruinosa thereafter) is native to the Atlantic coasts of Europe and North America, where often grows in sea cliffs as a chasmophyte in rock fissures where soil is absent (Castroviejo, 2020). In this highly unhospitable habitat, plants grow with a low nutrient availability and nearly continuous exposure to salinity and desiccating winds. A previous study revealed that the roots of Festuca pruinosa plants have a complex endophytic fungal microbiome, and Diaporthe is one of its most abundant components, occurring in 54% of the plants, and at all locations analyzed (Pereira et al., 2019). The main goal of this study was to identify the endophytic Diaporthe species associated to roots of Festuca pruinosa by means of genotypic, morphological, and biochemical analyses.
2. Materials and methods
2.1. Fungal isolates
Twenty-two Diaporthe isolates (Table 1) obtained from surface-disinfected roots of healthy Festuca pruinosa plants were analyzed (Pereira et al., 2019). Plants were collected at four locations in sea cliffs in the North Atlantic coast of Spain: Torre de Hércules (43°23’09” N 8°24’23”W), Cedeira (43°40’46”N 8°01’15”W), Estaca de Bares (43°47’25”N 7°41’16”W), and San Pedro de la Rivera (43°34’43”N 6°13’17”W). In addition, we included a Diaporthe isolate (T6) obtained from roots of Celtica gigantea (= Stipa gigantea), a tall grass growing in dry sandy soils collected in Cuatro Calzadas, Salamanca (40°49’03”N, 5°36’47”W) (Vázquez and Devesa, 1996; Vázquez de Aldana et al., 2021).
TABLE 1.
Diaporthe species, host plant, geographic origin, and GenBank accession numbers of strains used in the study. (Festuca rubra subsp. pruinosa = Festuca pruinosa).
| Species | Strain | Isolation source | Origin | GenBank accession number | ||||
| ITS | CAL | HIS | TEF1 | TUB | ||||
| Diaporthe ambigua | CBS 114015 (epitype) | Pyrus communis | South Africa | KC343010 | KC343252 | KC343494 | KC343736 | KC343978 |
| CBS 117167 | Aspalathus linearis | South Africa | KC343011 | KC343253 | KC343495 | KC343737 | KC343979 | |
| Diaporthe arecae | CBS 161.64 (ex-isotype) | Areca catechu | India | KC343032 | KC343274 | KC343516 | KC343758 | KC344000 |
| CBS 535.75 | Citrus sp. | Suriname | KC343033 | KC343275 | KC343517 | KC343759 | KC344001 | |
| Diaporthe foeniculacea | CBS 123208 (holotype of D. neotheicola) | Foeniculum vulgare | Portugal | KC343104 | KC343346 | KC343588 | KC343830 | KC344072 |
| CBS 123209; Di-C004/4 (ex-type of D. neotheicola) | Foeniculum vulgare | Portugal | KC343105 | KC343347 | KC343589 | KC343831 | KC344073 | |
| Diaporthe ganjae | CBS 180.91 = ILLS 43621 (ex-type) | Cannabis sativa | USA | KC343112 | KC343354 | KC343596 | KC343838 | KC344080 |
| Diaporthe inconspicua | CBS 133813 (type) | Maytenus ilicifolia | Brazil | KC343123 | KC343365 | KC343607 | KC343849 | KC344091 |
| LGMF922 = CPC 20298 | Spondias mombin | Brazil | KC343124 | KC343366 | KC343608 | KC343850 | KC344092 | |
| Diaporthe infecunda | CBS 133812 (type) | Schinus terebinthifolius | Brazil | KC343126 | KC343368 | KC343610 | KC343852 | KC344094 |
| LGMF918 | Schinus terebinthifolius | Brazil | KC343132 | KC343374 | KC343616 | KC343858 | KC344100 | |
| Diaporthe longispora | CBS 194.36 (type of D. strumella var. longispora | Ribes sp. | Canada | KC343135 | KC343377 | KC343619 | KC343861 | KC344103 |
| Diaporthe mayteni | CBS 133185 = CPC 20314 (ex-type) | Maytenus ilicifolia | Brazil | KC343139 | KC343381 | KC343623 | KC343865 | KC344107 |
| Diaporthe musigena | CBS 129519 (holotype) | Musa sp. | Australia | KC343143 | KC343385 | KC343627 | KC343869 | KC344111 |
| Diaporthe pseudophoenicicola | CBS 462.69 (ex-type) | Phoenix dactylifera | Spain | KC343184 | KC343426 | KC343668 | KC343910 | KC344152 |
| CBS 176.77 | Mangifera indica | Iraq | KC343183 | KC343425 | KC343667 | KC343909 | KC344151 | |
| Diaporthe sclerotioides | CBS 296.67 (type of P. sclerotioides) | Cucumis sativus | Netherlands | KC343193 | KC343435 | KC343677 | KC343919 | KC344161 |
| CBS 710.76 | Cucumis sativus | Netherlands | KC343194 | KC343436 | KC343678 | KC343920 | KC344162 | |
| Diaporthe atlantica sp. nov | TH2 | Festuca pruinosa | Spain | ON159897 | ON364024 | ON398814 | ON398836 | ON364045 |
| TH10 | Festuca pruinosa | Spain | ON159898 | ON364025 | ON398815 | ON398837 | ON364046 | |
| TH21 | Festuca pruinosa | Spain | ON159899 | ON364026 | ON398816 | ON398838 | ON364047 | |
| TH56 | Festuca pruinosa | Spain | ON159900 | ON364027 | ON398817 | ON398839 | ON364048 | |
| TH71 | Festuca pruinosa | Spain | ON159901 | ON364023 | ON398818 | ON398840 | ON364044 | |
| TH86 | Festuca pruinosa | Spain | ON159903 | ON364029 | ON398820 | ON398842 | ON364050 | |
| TH91 | Festuca pruinosa | Spain | ON159904 | ON364030 | ON398821 | ON398843 | ON364051 | |
| TH151 | Festuca pruinosa | Spain | ON159905 | ON364031 | ON398822 | ON398844 | ON364052 | |
| TH158 | Festuca pruinosa | Spain | ON159906 | ON364032 | ON398823 | ON398845 | ON364053 | |
| EB4 | Festuca pruinosa | Spain | OM944045 | ON018830 | ON018832 | ON018831 | ON018829 | |
| EB11 | Festuca pruinosa | Spain | ON159889 | ON364015 | ON398806 | ON398827 | ON364036 | |
| EB12 | Festuca pruinosa | Spain | ON159890 | ON364016 | ON398807 | ON398828 | ON364037 | |
| CD39 | Festuca pruinosa | Spain | ON159887 | ON364013 | ON398804 | ON398825 | ON364034 | |
| CD87 | Festuca pruinosa | Spain | ON159888 | ON364014 | ON398805 | ON398826 | ON364035 | |
| SP11 = CECT 21217 (holotype) | Festuca pruinosa | Spain | ON159893 | ON364019 | ON398810 | ON398831 | ON364040 | |
| SP45 | Festuca pruinosa | Spain | ON159894 | ON364020 | ON398811 | ON398832 | ON364041 | |
| SP130 | Festuca pruinosa | Spain | ON159895 | ON364021 | ON398812 | ON398833 | ON364042 | |
| SP131 | Festuca pruinosa | Spain | ON159896 | ON364022 | ON398813 | ON398834 | ON364043 | |
| Diaporthe iberica sp. nov | S32 | Festuca pruinosa | Spain | ON159892 | ON364018 | ON398809 | ON398830 | ON364039 |
| T6 | Celtica gigantea | Spain | MT645115 | ON364033 | ON398824 | ON398835 | ON364054 | |
| TH77 = CECT 21218 (holotype) | Festuca pruinosa | Spain | ON159902 | ON364028 | ON398819 | ON398841 | ON364049 | |
| Diaporthe sp.1 | EB73 = CECT 21219 | Festuca pruinosa | Spain | ON159891 | ON364017 | ON398808 | ON398829 | ON364038 |
| Diaporthella corylina | CBS 121124 = AR 4131 | Corylus sp. | China | KC343004 | KC343246 | KC343488 | KC343730 | KC343972 |
Type cultures are indicated in bold. CBS, CBS FUNGAL Biodiversity Centre, Utrecht, The Netherlands; CECT, Spanish type culture collection; CPC, Collection Pedro Crous, housed at CBS; ILLS, Illinois Natural History Survey Fungarium; LGMF, Culture collection of Laboratory of Genetics of Microorganisms, Federal University of Parana, Curitiba, Brazil; TUB, partial beta-tubulin gene; CAL, partial calmodulin gene; HIS, partial histone H3 gene; ITS, ITS1-5.8S rDNA-ITS2 internal transcribed spacer region; TEF1, partial translation elongation factor 1-alpha gene. Isolates marked with bold are ex-type or ex-epitype strains.
2.2. DNA isolation, PCR amplification, and sequencing
Fungal DNA was extracted and amplified from a small amount of mycelium scraped from seven-day-old potato dextrose agar (PDA) cultures using Extract-N-Amp Plant Tissue PCR kits (Merck). The oligonucleotide primers and PCR protocols used for the amplification of the different fungal genes are listed in Supplementary Table 1. After amplification, PCR amplicons were sequenced in both directions at the DNA sequencing service of the University of Salamanca (Spain). New sequences generated in this study were deposited at the GenBank nucleotide database (Table 1).
2.3. Phylogenetic analysis
To establish the identity of fungal isolates at species level, phylogenetic analyses based on sequences of five genes (ITS, TUB, CAL, TEF1, and HIS) were conducted. To determine which Diaporthe taxa were closest to our fungal isolates, a concatenated sequence of the five loci was aligned with a similar set of 243 sequences belonging to 95 Diaporthe species (TreeBASE, study S13943; Gomes et al., 2013). The alignment was performed using MAFFT (Katoh and Standley, 2013) with default settings, and manually adjusted with MEGA v. 7 (Kumar et al., 2016). Then, a Maximum Likelihood (ML) phylogenetic tree was made using MEGA v.7 to identify the clades to which our isolates belonged. Afterwards, more restricted phylogenetic trees were made using the sequences of the species included in these major clades. These phylogenetic analyses were based on ML for all the individual loci, and on both ML and Bayesian Inference (BI) for the concatenated sequence of the five loci. The best-fit models for each gene and the concatenated set were determined using MEGA v. 7 and incorporated into the analyses. For the BI, MrBayes v. 3.2.7 (Ronquist et al., 2012) was used to generate the phylogenetic trees under optimal criteria per data partition. The Markov Chain Monte Carlo analysis of four chains ran for 20 000 000 generations and started in parallel from a random tree topology and lasted until the average standard deviation of split frequencies was below 0.01. Trees were saved each 10 000 generations. The first 25% of saved trees were discarded as the burn-in phase and the posterior probabilities were determined from the remaining trees. The resulting phylogenetic tree was printed with Geneious v. 5.5.4 (Drummond et al., 2011). A ML tree for the concatenated sequence of the five loci was based on the Hasegawa-Kishino-Yano model with gamma correction. For other parameters, default settings were used. ML analyses were performed in MEGA v.7 with the tree bisection and reconnection algorithm, where gaps were treated as missing data. The robustness of the topology was evaluated by 1,000 bootstrap replications. Diaporthella corylina (CBS 121124) was used as an outgroup. Holotypes were preserved as metabolically inactive cultures at the Spanish Type Culture Collection (CECT). Nomenclatural novelties and descriptions were deposited at the CECT and MycoBank (MB).
2.4. Genetic diversity analysis
The 18 cultures of D. atlantica available were used for an analysis of its genetic diversity. For the samples investigated, diversity indices were calculated for each gene and the combined sequence dataset. Parameters such as the Tajima’s D (Tajima, 1989), the minimum numbers of recombination events (Rm) (Hudson and Kaplan, 1985), the total number of haplotypes (H), haplotype diversity (Hd), Watterson’s θ (θw), the number of segregating sites (S), and the average nucleotide diversity (pi) were calculated using the DnaSP v. 6.12 software (Librado and Rozas, 2009). To overcome the population size effects, Hd, θw, and pi were calculated after 1,000 repetitions, and the median estimate was recorded for each parameter.
2.5. Morphological characters
Agar plugs (6 mm diameter) from the edge of actively growing cultures on PDA were transferred to 9 cm diameter Petri dishes containing one of the following culture media: malt extract agar (MEA), PDA, water agar supplemented with sterile pine needles (PNA) (Smith et al., 1996) or with sterile pieces of Festuca pruinosa leaves (FLA). Plates were incubated at 21–22°C under a 12 h/12 h near-ultraviolet light/darkness cycle to induce sporulation as described by Gomes et al. (2013). Cultures were examined periodically for the development of ascomata or conidiomata. Colony diameters were determined on PDA cultures grown at 22–25°C in darkness after 3 days, while colony colors were described after 14 days using the charts of Rayner (1970). For microscopy, fungal structures were mounted in distilled water, and measurements determined for 30 conidia and other structures.
2.6. Biochemical properties of Diaporthe strains
The 22 Diaporthe strains were investigated for their metabolic activity in vitro as follows. Ammonium production was determined both qualitatively and quantitatively, as described by Cappuccino and Sherman (1998). Siderophore activity was determined using the chrome azurol S agar plate assay as described by Schwyn and Neilands (1987). The production of indole-3-acetic acid (IAA) was determined quantitatively using both spectrophotometric (Gordon and Weber, 1951) and chromatographic (Patel et al., 2018) assays. The ability of Diaporthe strains to produce amylase, cellulase and protease was analyzed in vitro in the supplemented media described by Hankin and Anagnostakis (1975). The phosphate solubilizing ability was determined using Pikovskaya’s agar medium, as described by Katznelson and Bose (1959).
2.7. Pathogenicity assays
Because of the close relatedness of the new Diaporthe species to D. sclerotioides, a well-known pathogen of cucurbits (Shishido et al., 2006, 2014), the pathogenicity of Diaporthe strains EB4 (D. atlantica) and S32 (D. iberica) was tested on cucumber (Cucumis sativus) cv. Ashley, melon (Cucumis melo) cv. Piñonet, and watermelon (Citrullus lanatus) cv. Crimson Sweet.
Mycelial inoculum of both Diaporthe strains was produced in 3-week old sugar beet pulp cultures (Vázquez de Aldana et al., 2020), and seedlings of the three cucurbits were obtained after seed germination in sterile vermiculite. Fourteen day old seedlings were transplanted to 200 mL pots containing a substrate consisting of seven volumes of a mixture of peat and perlite (1:1; v:v) previously treated at 80°C for 12 h in a forced air oven, and one volume of Diaporthe inoculum. Control seedlings were transplanted to substrate containing only the peat and perlite mixture. One seedling was transplanted to each pot and each treatment was replicated in six pots. The plants were maintained in a greenhouse, being tap watered on demand for a period of 40 days. After this time, the plants were harvested, the roots were cleaned with tap water and inspected for disease symptoms such as dark lesions. Dry weights of roots and aboveground tissues were measured, and differences among treatments were tested by means of an analysis of variance (ANOVA). Differences among means were tested with the Holm-Sidak method. Statistical calculations were made with SigmaPlot software v. 14.5.
3. Results
3.1. Phylogenetic analyses
A preliminary analysis of the concatenated sequences of the 22 Diaporthe strains from Festuca pruinosa aligned with 243 sequences belonging to 95 Diaporthe species (Gomes et al., 2013) allowed to identify the clades to which the Festuca isolates belonged (Supplementary Figure 1). Thereafter, we reviewed other multilocus taxonomic studies (Dissanayake et al., 2017; Gao et al., 2017; Guarnaccia et al., 2018; Guo et al., 2020; Norphanphoun et al., 2022), to search for other Diaporthe species closely related to the identified clades that were not included in Gomes et al. (2013). As a result, Diaporthe columnaris and D. cyatheae (Norphanphoun et al., 2022), were found to belong to the same clade as our endophytic isolates. Due to the lack of all five loci for these two species, they were not included in the multilocus tree. However, available loci were included in the single locus phylogeny to support the classification of our fungal isolates (Supplementary Figures 2–5).
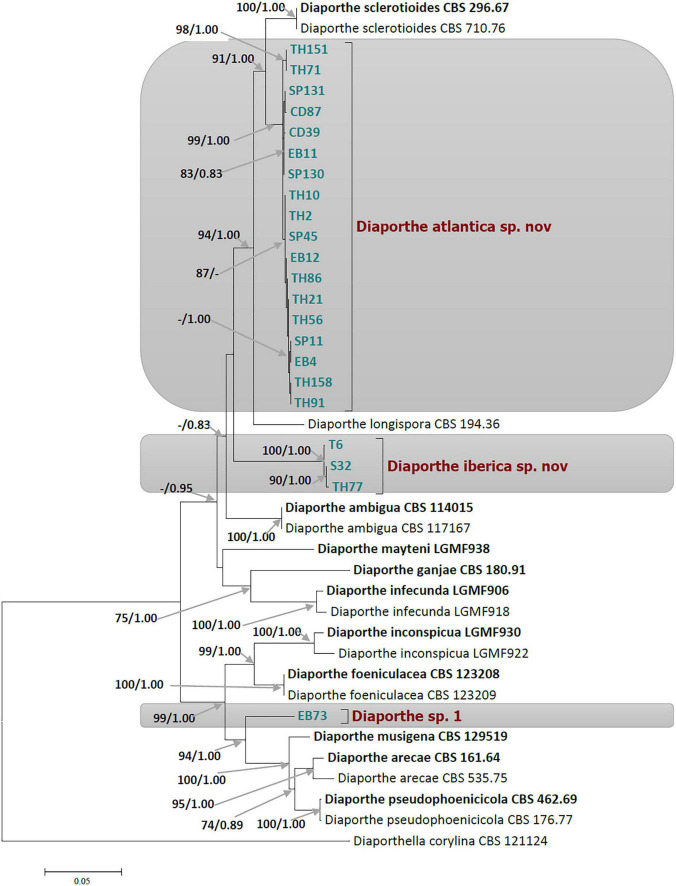
Ultimately, a set of 11 Diaporthe species closely related to the endophytic Festuca pruinosa isolates plus Diaporthella corylina (outgroup) were selected for a more restricted multilocus phylogenetic analysis (Table 1). A total of 2,786 characters including gaps were included in this phylogenetic analysis, 1,368 of these were conserved and 981 were variable, 649 of which were parsimony informative. The Bayesian analysis of the combined sequences of the five loci was based on the Hasegawa-Kishino-Yano substitution model with gamma distributed rate variation among sites (Table 2) and all partitions had Dirichlet base frequencies. The Bayesian analysis lasted 20 000 000 generations resulting in an average standard deviation of split frequencies of 0.001683. The consensus tree was generated and the posterior probabilities were calculated from the 27 443 trees left after discarding the first 25% for burn-in. The topologies resulting from ML and BI analyses of the concatenated dataset were congruent. Bayesian posterior probability (PP ≥ 0.8) and Maximum likelihood bootstrap values (ML ≥ 70) were obtained for the dendrogram nodes (Figure 1).
TABLE 2.
Nucleotide substitution models used in the phylogenetic analyses.
| Loci/Genes | Analysis | Best-fit model |
| ITS | ML | Kimura 2-parameter |
| TUB | ML | Hasegawa-Kishino-Yano |
| TEF1 | ML | Kimura 2-parameter |
| HIS | ML | Tamura-Nei |
| CAL | ML | Tamura 3-parameter |
| Concatenated loci | ML/BI | Hasegawa-Kishino-Yano |
BI, Bayesian inference; ML, maximum likelihood.
FIGURE 1.
Phylogram of Diaporthe resulting from a maximum likelihood analysis based on a combined matrix of ITS, TUB, CAL, TEF1, and HIS3. Numbers above the branches indicate ML bootstraps (left, ML BS ≥ 70%) and Bayesian Posterior Probabilities (right, PP ≥ 0.8). The tree is rooted with Diaporthella corylina. Isolates from present study are marked in colors. The scale bar represents the expected changes per site. The ex-type strains are Bold and strains from this study in green.
The 22 Diaporthe isolates from Festuca pruinosa were set apart from known species, and clustered together to create two novel clades statistically well-supported by ML and PP values. In the multilocus tree, 18 Festuca pruinosa isolates clustered together within a well-supported clade (ML/PP = 99/1.00) significantly distinct from D. sclerotioides, its closest known species (Figure 1). This new species was named Diaporthe atlantica sp. nov. Moreover, the single locus phylogeny of the five genes shows that D. atlantica was closer but separated from D. sclerotioides (Supplementary Figures 2–7) and also differed from D. columnaris in the ITS and TEF1 trees (Supplementary Figures 2, 4).
Three isolates (S32, TH77, and T6) clustered together to form a new clade (ML/PP = 100/1.00) unrelated to other known Diaporthe species, and was named Diaporthe iberica sp. nov. Two of these strains were isolated from maritime populations of Festuca pruinosa, but strain T6 was isolated several hundred km inland from roots of Celtica gigantea. Interestingly, Festuca pruinosa strains S32 and TH77 were genotypically closer than strain T6. In addition, the single locus analysis of each of the five genes consistently differentiated the clade formed by D. iberica strains (S32, TH77, and T6) from all known species included in the analysis (Supplementary Figures 2–6).
The single and multilocus phylogenetic analyses also showed that the strain EB73 was well-separated from closely related species (D. musigena, D. arecae, and D. foeniculacae), and might as well belong to a yet undescribed Diaporthe species. As we currently have only a single strain of this taxon, further investigations will have to await further collections.
3.2. Taxonomy
Based on the multi-locus phylogeny and their morphology, the 22 strains from Festuca pruinosa were assigned to two newly described taxa, plus a yet undescribed species. All species studied in culture are characterized below.
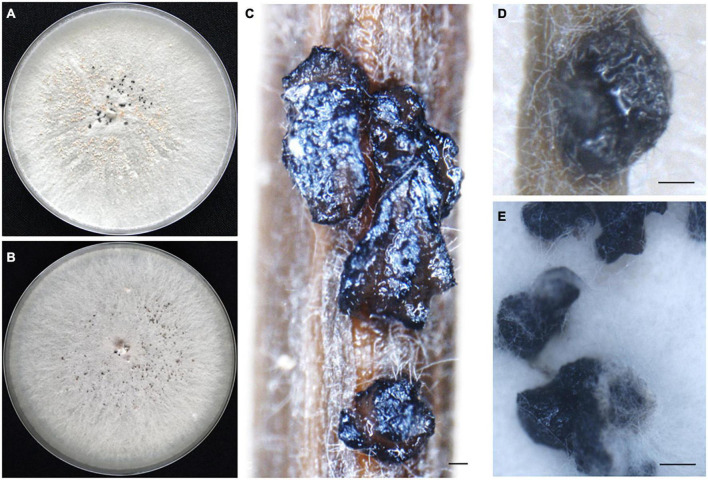
Diaporthe atlantica Toghueo, Vazq-Alda and Zabalgo, sp. nov.; Figure 2 and Supplementary Figure 8.
FIGURE 2.
Morphological characteristics of Diaporthe atlantica. Colony appearance on potato dextrose agar (PDA) (a) and malt extract agar (MEA) (b); conidiomata with conidial droplets on PDA (c,d); conidiomata on pine needle (e) and Festuca pruinosa leaves (f); conidiogenous cells (g); α-conidia (h,i). Scale bar: panels (d–f) = 100 μm; panels (c,g–i) = 10 μm.
MycoBank number: MB845433.
Etymology: Named after the Atlantic ocean, in whose coastal cliffs is found Festuca rubra subsp. pruinosa, the plant host of this species.
Description: Pycnidial conidiomata globose or irregular, black scattered or aggregated, and irregularly distributed over agar surfaces, with white or cream conidial droplets (50–160 μm) exuding from the ostioles. Conidiophores hyaline, terminal and lateral, cylindrical, 1-3-septate, densely aggregated, 14.3–30.9 × 2.5–4.1 μm, (mean ± SD = 21.84 ± 6.07×3.11 ± 0.61 μm). Conidiogenous cells [4.8–15.1 × 1.9–6.6 μm, (mean ± SD = 9.36 ± 3.84 × 4.44 ± 1.9μm)] hyaline, subcylindrical and filiform, straight to curved, terminal and lateral, with slight taper toward the apex. Alpha conidia were fusiform, hyaline, ellipsoidal, aseptate, rounded at each end, 8.7–12.3 × 3.3–5 μm, (mean ± SD = 10.29 ± 0.9 × 4.3 ± 0.42 μm). Beta and gamma conidia were not observed.
Culture characteristics: Colonies on PDA were white at first, with flattened mycelium and becoming dirty white and umber over time with dark black pycnidia bearing cream conidial droplets distributed over the agar surface. The colony margins had petaloidlike shapes. The reverse was graphite gray with black spots uniformly distributed. On MEA, colonies were white at first with flattened mycelium and becoming brownish over time. Dark black and sterile stromata abundantly distributed over the mycelium surface were observed. The colony diameter was 24–30 mm after 3 days at 25°C.
Specimens examined: SPAIN, Asturias, San Pedro de la Rivera (43°34’43”N, 6°13’17”W), from roots of Festuca rubra subsp. pruinosa, Mar. 2016, E. Pereira, holotype SP11 (CECT 21217); A Coruña, Cedeira, from roots of Festuca pruinosa, Mar. 2016, E. Pereira, strains CD39, CD87; A Coruña, Estaca de Bares, from roots of Festuca pruinosa, Mar. 2016, E. Pereira strains EB4, EB11, and EB12; Asturias, San Pedro de la Rivera, from roots of Festuca pruinosa, Mar. 2016, E. Pereira, strains SP45, SP130, and SP131; A Coruña, Torre de Hercules, from roots of Festuca pruinosa, Mar. 2016, E. Pereira, strains TH2, TH10, TH21, TH56, TH71, TH86, TH91, TH151, and TH158.
Host/Habitat: Festuca rubra subsp. pruinosa grows on rocky sea cliffs in the Atlantic coasts of Europe. In this habitat, soil and nutrients are very limited, and exposure to salinity is continuous.
Notes: Diaporthe atlantica was isolated as an endophyte from surface-disinfected roots of Festuca pruinosa. This species was described from a set of 18 strains among which only four produced spores, and this occurred on PNA medium. This species was found in plants from all the four locations investigated. In vitro, all 18 strains investigated produced indole-3-acetic acid (IAA), and ammonium, while 16 produced siderophores and cellulase, 13 solubilized phosphate and 11 produced amylase. Diaporthe atlantica is phylogenetically close but clearly differentiated from D. sclerotioides and D. columnaris two species known as pathogens of cucurbits (Shishido et al., 2014) and lingonberry (Farr et al., 2002), respectively. Morphologically D. atlantica cannot be reliably separated from D. sclerotioides due to overlaps in characteristics such as the culture phenotype or spore shape and size. However, D. atlantica is morphologically distant from D. columnaris by the size of alpha conidia, and the shape and size of conidiogenous cells (Farr et al., 2002). On the other hand, D. atlantica differs from D. sclerotioides by 110 nucleotides in the concatenated sequence alignment, 45 of which were distinct in the TUB region, 44 in the TEF1 region, 10 in the ITS region, 9 in the HIS region, and 2 in the CAL region and differs from D. columnaris by 2 and 13 nucleotides in the ITS and TEF1 regions respectively. Furthermore, a phylogenetic analysis based on the ITS sequences of all D. atlantica strains and those of thirteen isolates of D. sclerotioides retrieved from GenBank revealed a clear separation between both species. In the ML tree obtained, all D. atlantica strains clustered together in a well-supported clade (ML = 70; Supplementary Figure 7) clearly separated from the D. sclerotioides isolates (ML = 99; Supplementary Figure 7). Inoculation of D. atlantica strain EB4 in cucumber, melon, and watermelon plants did not cause root necrosis or seedling wilt, as D. sclerotioides does (Shishido et al., 2014), but shoot and root growth was delayed in the three plant species.
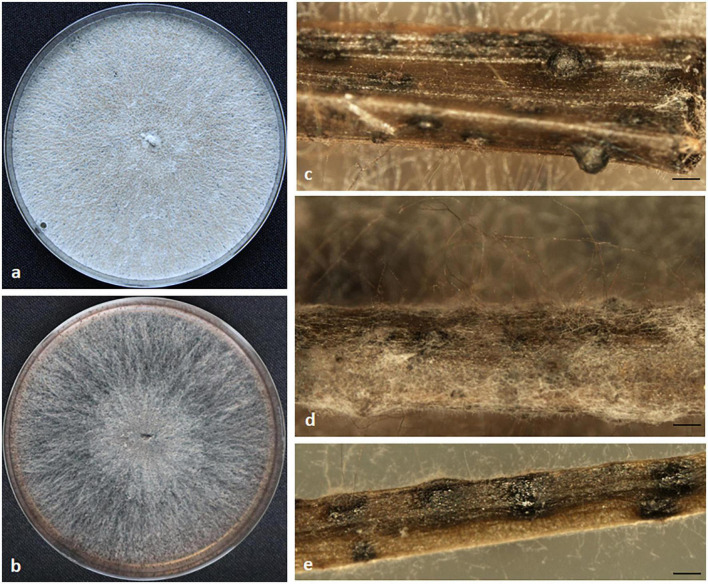
Diaporthe iberica Toghueo, Vazq-Alda and Zabalgo, sp. nov.; Figure 3 and Supplementary Figure 8.
FIGURE 3.
Morphological characteristics of Diaporthe iberica. Front view of colony appearance on potato dextrose agar (PDA) (A) and malt extract agar (MEA) (B); sterile stromata on pine needle (C) and F. pruinosa leaf (D) and on the surface of PDA (E); Scale bar: panels (C–E) = 100 μm.
MycoBank number: MB845435.
Etymology: Named after the Iberian Peninsula, where isolates were obtained in different habitats and host plant species.
Description: Sterile stromata were globose or irregular, dark black, irregularly distributed over the agar surface, scattered or aggregated and exposed on the surface of Festuca rubra leaves and pine needles. Alpha, beta and gamma conidia were not observed.
Culture characteristics: Colonies on PDA form flattened and white mycelium. Dark sterile stromata uniformly distributed over the agar plate emerged after 2 weeks at 25°C. On MEA, colonies with flattened and white gray mycelium with dark stromata were uniformly distributed over the agar plate. The colony diameter was 22–31 mm after 3 days at 25°C. The dark stromata remained sterile on PDA, MEA PNA, and FLA after 4 months in culture at 25°C, and after 2 weeks of continuous exposure to UV light.
Specimens examined: SPAIN, A Coruña, Torre de Hercules (43°23’09”N, 8°24’23”W), from roots of Festuca rubra subsp. pruinosa, Mar. 2016, E. Pereira, holotype TH77 (CECT 21218); Asturias, San Pedro de la Rivera, from roots of Festuca pruinosa, Mar. 2016, E. Pereira, strain S32; Salamanca, Cuatro Calzadas, from roots of Celtica gigantea, Mar. 2015, I. Zabalgogeazcoa, strain T6.
Host/Habitat: Festuca pruinosa grows on rocky sea cliffs, with poor nutrient availability, and continuous exposure to salinity, while Celtica gigantea grows in nutrient-poor sandy soils in semiarid habitats in the southwestern Iberian peninsula (Vázquez and Devesa, 1996).
Notes: The three isolates of Diaporthe iberica studied form a clade with high support (ML/PP = 100/1.00), distinct from other known Diaporthe species. This new taxon can colonize at least two distinct host grasses, Festuca pruinosa and Celtica gigantea, growing in very different habitats. In vitro, all three strains produced IAA, ammonium, siderophores and cellulase.
Diaporthe sp. 1
Specimen examined: SPAIN, A Coruña, Estaca de Bares (43°47’25”N, 7°41’16”W), from roots of Festuca pruinosa, Mar. 2016, E. Pereira, strain EB73 (CECT 21219).
Culture characteristics: Colonies on PDA form white gray mycelium at first and become dark over time. On MEA, light brown aerial mycelium, cottony and less abundant. The colony diameter was 26 mm after 3 day at 25°C (Figure 4).
FIGURE 4.
Morphological characteristics of Diaporthe sp. 1. Colony appearance on potato dextrose agar (PDA) (a) and malt extract agar (MEA) (b); sterile conidiomata on pine needle (c,d) and Festuca pruinosa leaves (e). Scale bar: panels (c–e) = 100 μm.
Host/Habitat: Festuca rubra subsp. pruinosa grows on rocky sea cliffs in the Atlantic coasts of Europe. Soil and nutrients are very limited, and exposure to salinity is continuous in this habitat.
Notes: Based on phylogenetic data, this endophytic isolate might belong to an undescribed species. Cultures were sterile on various media including PDA, MEA, PNA, and FLA at varied conditions. In vitro, strain EB73 produced IAA, ammonium, siderophores, cellulase and solubilized phosphate.
3.3. Genetic diversity of Diaporthe atlantica
The genetic diversity of Diaporthe atlantica was estimated on the basis of its 18 individuals (Table 3). The calculated haplotype diversity of D. atlantica was higher than 0.5 for TEF1, CAL, HIS, TUB and the combined data set, reflecting a high genetic diversity of the population analyzed (Nei and Tajima, 1981; Stumpf, 2004).
TABLE 3.
Polymorphism and genetic diversity parameters of Diaporthe atlantica strains associated with Festuca rubra subsp. pruinosa.
| Gene | n | bp | θ w | S | H | Hd | pi | k | TD | Rm |
| ITS | 18 | 423 | 0.581 | 2 | 2 | 0.209 | 0.00099 | 0.418 | −0.68482 | 0.0 |
| TUB | 18 | 709 | 2.035 | 7 | 7 | 0.869 | 0.00256 | 1.817 | −0.36375 | 0.0 |
| HIS | 18 | 386 | 2.907 | 10 | 9 | 0.895 | 0.00598 | 2.307 | −0.74337 | 0.0 |
| TEF1 | 18 | 230 | 4.070 | 14 | 11 | 0.922 | 0.01998 | 4.595 | 0.20324 | 2.0 |
| CAL | 18 | 312 | 0.581 | 2 | 3 | 0.569 | 0.00297 | 0.928 | 1.45481 | 0.0 |
| Combined | 18 | 2061 | 8.722 | 30 | 17 | 0.987 | 0.00451 | 9.294 | 0.12562 | 4.0 |
n, sample size; bp, total number of sites; S, number of segregating/polymorphic sites (parsimony informative sites); H, number of alleles; Hd, haplotype (allelic) diversity; pi, average nucleotide diversity; k, average number of nucleotide differences; TD, Tajima’s D (TD); θw, Watterson’s theta; Rm, minimum number of recombination events. TD was not significant (P > 0.10) for all the genes analyzed.
Overall, 17 haplotypes were detected in the concatenated gene sequence, while eleven, nine, seven, three, and two haplotypes were detected in TEF1, HIS, TUB, CAL, and ITS sequences respectively (Table 4). The only common haplotype, H-9, was shared by two strains obtained from different localities, Estaca de Bares (A Coruña) and San Pedro de la Rivera (Asturias). The haplotypes ITS-H1 (n = 16), CAL-H2 (n = 11), HIS-H6 (n = 5), TEF1-H4 (n = 4) and TUB-H5 (n = 5) were the most frequent for each gene. The highest degree of sequence polymorphism (pi = 0.01998) was observed for TEF1, with 11 haplotypes. Details of polymorphism at each locus and unique sequences types for each strain are available in Supplementary Table 2. In general, this analysis suggests that the populations of Diaporthe atlantica are genotypically diverse, but clonality exists in them.
TABLE 4.
Haplotypes of Diaporthe atlantica strains isolated from roots of Festuca rubra subsp. pruinosa.
| Strain | Location | Single locus haplotype | Five-locus haplotype | ||||
| ITS | TUB | HIS | TEF1-α | CAL | |||
| CD39 | CED | ITS-H1 | tub-H1 | his-H7 | tef1-H1 | Cal-H1 | Haplotype 1 |
| TH2 | TDH | ITS-H1 | tub-H5 | his-H9 | tef1-H7 | Cal-H2 | Haplotype 2 |
| TH10 | TDH | ITS-H1 | tub-H5 | his-H8 | tef1-H8 | Cal-H2 | Haplotype 3 |
| TH21 | TDH | ITS-H1 | tub-H5 | his-H6 | tef1-H8 | Cal-H2 | Haplotype 4 |
| TH86 | TDH | ITS-H1 | tub-H3 | his-H6 | tef1-H8 | Cal-H2 | Haplotype 5 |
| TH91 | TDH | ITS-H1 | tub-H7 | his-H4 | tef1-H11 | Cal-H2 | Haplotype 6 |
| SP45 | SPR | ITS-H1 | tub-H5 | his-H3 | tef1-H6 | Cal-H2 | Haplotype 7 |
| CD87 | CED | ITS-H1 | tub-H2 | his-H5 | tef1-H2 | Cal-H1 | Haplotype 8 |
| EB4 | EDB | ITS-H1 | tub-H4 | his-H6 | tef1-H3 | Cal-H2 | Haplotype 9 |
| SP11 | SPR | ITS-H1 | tub-H4 | his-H6 | tef1-H3 | Cal-H2 | Haplotype 9 |
| EB11 | EDB | ITS-H1 | tub-H1 | his-H1 | tef1-H4 | Cal-H1 | Haplotype 10 |
| SP130 | SPR | ITS-H1 | tub-H1 | his-H7 | tef1-H4 | Cal-H1 | Haplotype 11 |
| SP131 | SPR | ITS-H1 | tub-H1 | his-H5 | tef1-H3 | Cal-H1 | Haplotype 12 |
| TH71 | TDH | ITS-H2 | tub-H6 | his-H1 | tef1-H10 | Cal-H3 | Haplotype 13 |
| TH151 | TDH | ITS-H2 | tub-H6 | his-H1 | tef1-H4 | Cal-H3 | Haplotype 14 |
| TH56 | TDH | ITS-H1 | tub-H5 | his-H6 | tef1-H9 | Cal-H2 | Haplotype 15 |
| TH158 | TDH | ITS-H1 | tub-H7 | his-H4 | tef1-H4 | Cal-H2 | Haplotype 16 |
| EB12 | EDB | ITS-H1 | tub-H3 | his-H2 | tef1-H5 | Cal-H2 | Haplotype 17 |
The haplotypes observed at each sequenced locus for each strain analyzed (n = 18), and the resulting multilocus haplotype are shown. CED, Cedeira; EDB, Estaca de Bares; SPR, San Pedro de la Rivera; TDH, Torre de Hércules; CAL, partial sequence of the gene coding for calmodulin; HIS, Histone H3; ITS, ITS1-5.8S rDNA-ITS2 internal transcribed spacer region; TEF1, partial sequence of the gene coding for translation elongation factor 1-α; TUB, partial sequence of the gene coding for β-tubulin.
3.4. Biochemical characteristics of endophytic Diaporthe species
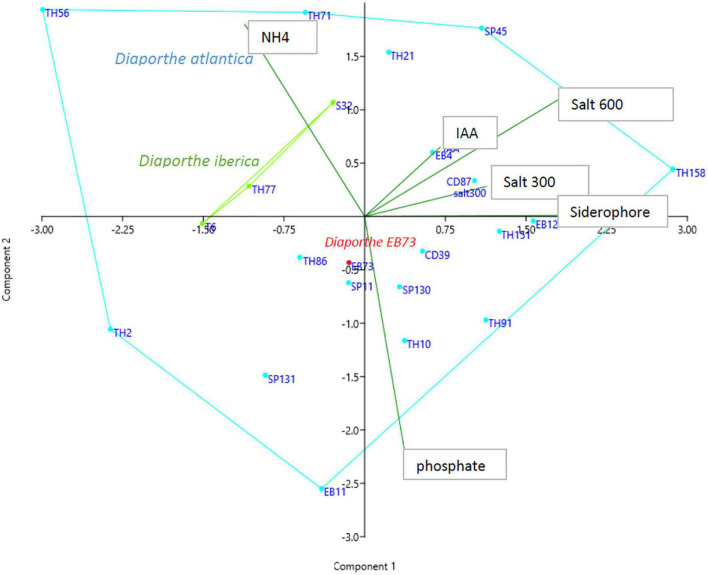
All 22 Diaporthe strains produced ammonium in peptone water and IAA in potato dextrose broth. On agar-supplemented media, 20 strains were able to produce siderophores and cellulase, 15 solubilized phosphate, 13 amylase, and 5 protease (Supplementary Table 3). A principal component analysis (PCA) was performed on the whole dataset to investigate if there is a relationship between Diaporthe species and some biochemical characteristics. The PCA showed that components I and II accounted for 62% of the total variance. The three D. iberica strains (T6, TH77, and S32) clustered in close proximity, within the sparse distribution of D. atlantica strains (Figure 5).
FIGURE 5.
Principal component analysis of in vitro characteristic of Diaporthe strains isolated from Festuca pruinosa.
3.5. Pathogenicity of Diaporthe
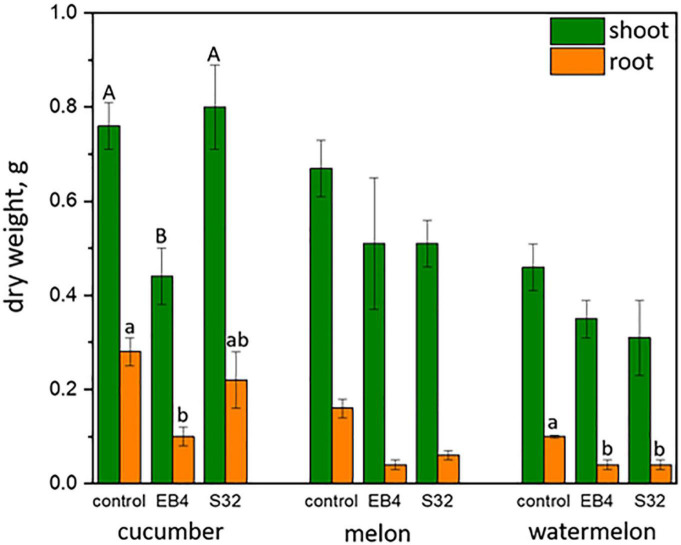
Neither dark lesions in roots or seedling wilt, symptoms reported for cucurbit root rot disease caused by D. sclerotioides (Shishido et al., 2006, 2014) were observed in plants inoculated with D. atlantica strain EB4 or D. iberica strain S32. However, a significant (P < 0.05) inhibition of shoot and root growth occurred in cucumber inoculated with strain EB4, and in roots of melon and watermelon inoculated with strains EB4 and S32 (Figure 6).
FIGURE 6.
Shoot and root dry weight (mean ± standard error) of cucurbitaceous plants inoculated with Diaporthe atlantica strain EB4 and Diaporthe iberica strain S32. For each plant species and organ, letters on columns indicate significantly different means.
4. Discussion
The association of Diaporthe species with many important plant diseases has stimulated a considerable interest in this genus. On the other hand, Diaporthe species are ubiquitous in nature as endophytes, and have been reported as dominant components of the microbiome of various plant species. Therefore, more efforts are still required to increase our understanding of their ecology and biology. In this study, a multi-locus phylogenetic analysis of endophytic Diaporthe strains associated with roots of Festuca pruinosa, a grass that lives in a very unhospitable habitat, revealed two new species, Diaporthe atlantica and Diaporthe iberica, and possibly a third undescribed species represented by Diaporthe sp. 1. Both species described in this study belong to the Diaporthe sojae species complex. This complex is composed of endophytes, saprophytes and pathogens, and is the largest of the thirteen species complexes of Diaporthe recently described by Norphanphoun et al. (2022).
Over the past decade, DNA sequence data have become paramount for the accurate definition of species boundaries, and the resolution of important taxonomic questions. In the genus Diaporthe, multi-locus phylogenies based on the combined analysis of five genes were found to be better suited for an accurate species delimitation than those based on four or less loci (Santos et al., 2017). Therefore, the five most commonly used loci (ITS, TUB, TEF1, CAL, and HIS) for the separation of species in this genus were used in the present study to resolve the taxonomy of endophytic Diaporthe strains from Festuca pruinosa (Gomes et al., 2013; Gao et al., 2017; Guarnaccia et al., 2018; Guo et al., 2020; Norphanphoun et al., 2022). Among the Diaporthe species identified, Diaporthe atlantica was the most abundant, occurred in roots of plants from all four locations examined, and its populations were genotypically diverse. Phylogenetically, D. atlantica was close to, but clearly differentiated from D. sclerotioides. However, the comparison of morphological characters such as the culture appearance, or the shape and size of alpha conidia make impossible to separate both species (Shishido et al., 2006). Indeed, the difficulties in morphological comparisons have historically made the taxonomy of Diaporthe very challenging (Gomes et al., 2013). Diaporthe species often have similar or overlapping morphological characteristics (Lawrence et al., 2015), and due to their plasticity and inability to properly reflect the evolutionary history of species, these cultural characteristics have been deemed unfit for species delimitation (Rehner and Uecker, 1994; Gao et al., 2015). Therefore, the five gene multi-locus phylogenetic analysis was the key approach used in the present study to classify accurately Diaporthe atlantica as new species.
The plants of Festuca pruinosa often grow in soilless fissures in the rock, where a compact mass of roots is associated to a complex microbiome (Pereira et al., 2019). In this habitat poor in nutrients, D. atlantica could be helping its host to survive by playing a role in the recycling of nutrients from dead Festuca roots and other organic debris. In connection with this, D. atlantica cultures had extracellular enzymatic activities capable of degrading protein to ammonium, cellulases which could reduce cell wall material to simpler carbohydrates, and siderophores which help to trap iron. Plant roots can readily absorb ammonium and some simple carbohydrates, thus, the acceleration of organic matter decay by this fungus could increase the nutrient availability in a nutrient poor environment. Therefore, in its host grass D. atlantica could be a latent saprophyte, an endophyte that becomes a saprophyte when host roots senesce or die. In addition, Diaporthe cultures produced IAA, which can stimulate root growth. This fungus is known to behave as a mutualist in other grasses than its original host, promoting leaf and root growth in tritordeum and perennial ryegrass (Lolium perenne) even under salinity conditions (Toghueo et al., 2022).
The other novel species identified in this study, Diaporthe iberica, was associated to two plant species inhabiting very different habitats. The strain T6 was isolated from roots of Celtica gigantea, a perennial grass that grows in nutrient-poor sandy soils in semiarid zones (Vázquez and Devesa, 1996; Vázquez de Aldana et al., 2021), while strains S32 and TH77 occurred in roots of the perennial grass Festuca pruinosa from sea cliffs. Like D. iberica, many Diaporthe species are known to be multi-host. For instance, Diaporthe eres, the type species of the genus has been reported as a pathogen and an endophyte in plant species belonging to several families (Udayanga et al., 2014a). Diaporthe iberica did not produce spores on any of the different agar media we tested. Diaporthe species like D. endophytica (Gomes et al., 2013), D. infertilis (Guarnaccia and Crous, 2017), and D. parvae (Guo et al., 2020) are also known to be sterile, at least under experimental conditions. Like Diaporthe atlantica, D. iberica could have a beneficial association with its original host, improving adaptability to nutrient poor environments by contributing to organic matter recycling. Amylase and cellulase activity, ammonium production from protein substrate, and siderophores were observed in its cultures. In addition, Diaporthe iberica strain T6 was previously found to exhibit the greatest plant growth promotion capability among 66 fungal endophytes from Celtica gigantea, increasing both leaf and root biomass, and the leaf content of several mineral elements in tritordeum (Vázquez de Aldana et al., 2021). Besides, we found that D. iberica strain S32 could significantly improve the growth of Festuca pruinosa clonal line CD8 in greenhouse conditions (data not shown).
In addition to D. atlantica and D. iberica, Festuca pruinosa hosts another undescribed species of Diaporthe, Diaporthe sp. 1. The single strain representing this taxon also showed in vitro characteristics consistent with potential plant growth promotion and nutrient recycling functions, like production of IAA, ammonium, siderophores, cellulase and phosphate solubilization. However, following the recommendations of the International Commission on the Taxonomy of Fungi on the description of new fungal species, the taxonomic status of this strain will have to await further collection, since only one member of this putative species is known so far (Aime et al., 2021).
As above mentioned, virulent Diaporthe pathogens can also occur as endophytes in alternative host plant species (Crous and Groenewald, 2005; Gomes et al., 2013). Because of the close relatedness of Diaporthe atlantica to D. sclerotioides, a well-known pathogen of cucurbits (Shishido et al., 2006), the potential pathogenicity of D. atlantica strain EB4 and D. iberica strain S32 were investigated on cucumber, melon and watermelon. None of these caused root necrosis or wilt symptoms on cucurbits, as reported for black root rot disease caused by D. sclerotioides on these species (Shishido et al., 2006, 2014). However, strains EB4 and S32 caused a decrease in the shoot and root biomass of the three cucurbits. This potentially harmful effect observed in cucurbits contrasts with the beneficial effects observed in plants of tritordeum, perennial ryegrass and tomato inoculated with the same D. atlantica strain (Pereira, 2021; Toghueo et al., 2022). A Diaporthe iberica strain also showed a beneficial response on tritordeum (Vázquez de Aldana et al., 2021). Therefore, it seems that the benefits imparted by D. atlantica and D. iberica may be host-dependent, with only particular plant–fungus interactions resulting in positive plant responses (Brader et al., 2017). The host plant species and genotype has been reported to have a significant influence on the symbiotic lifestyles of some Colletotrichum species, favoring in some cases a mutualistic, commensal, or pathogenic lifestyle (Redman et al., 2001). Further investigations in this direction are required to understand the bases of the change in lifestyle by these Diaporthe species.
5. Conclusion
This study revealed that at least three Diaporthe species are associated to roots of Festuca pruinosa as endophytes. We identified two new Diaporthe species, Diaporthe atlantica, Diaporthe iberica and a yet undescribed taxon Diaporthe sp.1. Diaporthe atlantica seems to be the most abundant taxon in Festuca pruinosa, while D. iberica is a multihost endophyte naturally associated to roots of at least two different species of grasses (Festuca pruinosa and Celtica gigantea) growing in very different habitats, marine sea cliffs and sandy soils in semiarid inland zones. We propose that these symbiotic fungi might improve nutrient availability for their hosts, living in a habitat where nutrient availability is very limited. Since some Diaporthe species are multihost, other plants sympatric with Festuca pruinosa in sea cliff habitats, like Armeria maritima or Crithmum maritimum (Lopez-Bedoya and Pérez-Alberti, 2009), might also be hosts of these Diaporthe species. However, the ecological role of the newly described species as well as their interaction with host plants still needs to be better understood. Descriptions, molecular data, in vitro activities, and host interactions of the Diaporthe species described in the present study could represent an important resource for agricultural scientists, plant pathologists and taxonomists.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
RT performed experiments, analyzed the data, and wrote the manuscript. IZ and BV supervised the research and wrote the manuscript. All authors designed the experiments, analyzed the data, and read and approved the final manuscript.
Acknowledgments
The authors would also like to thank Rebeca Alonso for technical help.
Funding Statement
This research was supported by grant PID2019-109133RB-I00 funded by MCIN/AEI/10.13039/501100011033 and by project “CLU2019-05–IRNASA/CSIC Unit of Excellence” funded by the Junta de Castilla y León and co-financed by EU (ERDF “Europe drives our growth”). RT was supported by grant FJC2018-03857-I funded by MCIN/AEI/10.13039/501100011033 and by “ESF Investing in your Future.”
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1105299/full#supplementary-material
References
- Abramczyk B., Marzec-Grządziel A., Grządziel J., Król E., Gałązka A., Oleszek W. (2022). Biocontrol potential and catabolic profile of endophytic Diaporthe eres strain 1420S from Prunus domestica L. in Poland—a preliminary study. Agronomy 12:165. 10.3390/agronomy12010165 [DOI] [Google Scholar]
- Aime M. C., Miller A. N., Aoki T., Bensch K., Cai L., Crous P. W., et al. (2021). How to publish a new fungal species, or name, version 3.0. IMA Fungus 12:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baluyot J. C., Santos H. K., Batoctoy D. C. R., Torreno V. P. M., Ghimire L. B., Joson S. E. A., et al. (2022). Diaporthe/Phomopsis longicolla degrades an array of bisphenol analogues with secreted laccase. Microbiol. Res. 257:126973. 10.1016/j.micres.2022.126973 [DOI] [PubMed] [Google Scholar]
- Brader G., Compant S., Vescio K., Mitter B., Trognitz F., Ma L.-J., et al. (2017). Ecology and genomic insights into plant-pathogenic and plant-nonpathogenic endophytes. Annu. Rev. Phytopathol. 55 61–83. 10.1146/annurev-phyto-080516-035641 [DOI] [PubMed] [Google Scholar]
- Braun R. C., Patton A. J., Watkins E., Koch P. L., Anderson N. P., Bonos S. A., et al. (2020). Fine fescues: A review of the species, their improvement, production, establishment, and management. Crop Sci. 60 1142–1187. 10.1002/csc2.20122 [DOI] [Google Scholar]
- Cappuccino J. C., Sherman N. (1998). Microbiology: A laboratory manual. California: Benjamin/Cummings Science. [Google Scholar]
- Castroviejo S. (2020). *Flora iberica. Plantas vasculares de la Peninsula Iberica e Islas Baleares Vol. XIX. Part I. Madrid: Real Jardín Botánico, Consejo Superior de Investigaciones Científicas (CSIC). [Google Scholar]
- Chepkirui C., Stadler M. (2017). The genus Diaporthe: A rich source of diverse and bioactive metabolites. Mycol. Prog. 16 477–494. 10.1007/s11557-017-1288-y [DOI] [Google Scholar]
- Crous P. W., Groenewald J. Z. (2005). Hosts, species and genotypes: Opinions versus data. Australas. Plant Pathol. 34:463. 10.1071/AP05082 [DOI] [Google Scholar]
- da Silva Santos S., da da Silva A. A., da, Polonio J. C., Polli A. D., Orlandelli R. C., Dos Santos Oliveira J. A. D. S., et al. (2022). Influence of plant growth-promoting endophytes Colletotrichum siamense and Diaporthe masirevici on tomato plants (Lycopersicon esculentum Mill.). Mycology 13 257–270. 10.1080/21501203.2022.2050825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dini-Andreote F. (2020). Endophytes: The second layer of plant defense. Trends Plant Sci. 25 319–322. 10.1016/j.tplants.2020.01.007 [DOI] [PubMed] [Google Scholar]
- Dissanayake A. (2017). Molecular phylogenetic analysis reveals seven new Diaporthe species from Italy. Mycosphere 8 853–877. 10.5943/mycosphere/8/5/4 [DOI] [Google Scholar]
- Dissanayake A., Phillips A. J. L., Hyde K. D., Yan J. Y., Li X. H. (2017). The current status of species in Diaporthe. Mycosphere 8 1106–1156. 10.5943/mycosphere/8/5/5 [DOI] [Google Scholar]
- dos Santos G. D., Gomes R. R., Gonçalves R., Fornari G., Maia B. H. L. N. S., Schmidt-Dannert C., et al. (2021). Molecular identification and antimicrobial activity of foliar endophytic fungi on the Brazilian pepper tree (Schinus terebinthifolius) reveal new species of Diaporthe. Curr. Microbiol. 78 3218–3229. 10.1007/s00284-021-02582-x [DOI] [PubMed] [Google Scholar]
- dos Santos T. T., de Souza Leite T., de Queiroz C. B., de Araújo E. F., Pereira O. L., de Queiroz M. V. (2016). High genetic variability in endophytic fungi from the genus Diaporthe isolated from common bean (Phaseolus vulgaris L.) in Brazil. J. Appl. Microbiol. 120 388–401. 10.1111/jam.12985 [DOI] [PubMed] [Google Scholar]
- Drummond A., Ashton B., Buxton S., Cheung M., Cooper A., Duran C., et al. (2011). Geneious v5.4. Available online at: http://www.geneious.com/ (accessed October 12, 2015). [Google Scholar]
- Farr D. F., Castlebury L. A., Rossman A. Y., Putnam M. L. (2002). A new species of Phomopsis causing twig dieback of Vaccinium vitis-idaea (lingonberry). Mycol. Res. 106 745–752. 10.1017/S095375620200583X [DOI] [Google Scholar]
- Gao Y., Liu F., Duan W., Crous P. W., Cai L., Htxdoo K. D. F., et al. (2017). Diaporthe is paraphyletic. IMA Fungus 8:35. 10.5598/imafungus.2017.08.01.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y., Su Y., Sun W., Cai L. (2015). Diaporthe species occurring on Lithocarpus glabra in China, with descriptions of five new species. Fungal Biol. 119 295–309. 10.1016/j.funbio.2014.06.006 [DOI] [PubMed] [Google Scholar]
- Gomes R. R., Glienke C., Videira S. I. R., Lombard L., Groenewald J. Z., Crous P. W. (2013). Diaporthe: A genus of endophytic, saprobic and plant pathogenic fungi. Persoonia 31 1–41. 10.3767/003158513X666844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon S. A., Weber R. P. (1951). Colorimetric estimation of indoleacetic acid. Plant Physiol. 26 192–195. 10.1104/pp.26.1.192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarnaccia V., Crous P. W. (2017). Emerging citrus diseases in Europe caused by species of Diaporthe. IMA Fungus 8 317–334. 10.5598/imafungus.2017.08.02.07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarnaccia V., Groenewald J. Z., Woodhall J., Armengol J., Cinelli T., Eichmeier A., et al. (2018). Diaporthe diversity and pathogenicity revealed from a broad survey of grapevine diseases in Europe. Persoonia 40 135–153. 10.3767/persoonia.2018.40.06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y. S., Crous P. W., Bai Q., Fu M., Yang M. M., Wang X. H., et al. (2020). High diversity of Diaporthe species associated with pear shoot canker in China. Persoonia 45 132–162. 10.3767/persoonia.2020.45.05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hankin L., Anagnostakis S. L. (1975). The use of solid media for detection of enzyme production by fungi. Mycologia 67:597. 10.2307/3758395 [DOI] [Google Scholar]
- Huang F., Udayanga D., Wang X., Hou X., Mei X., Fu Y., et al. (2015). Endophytic Diaporthe associated with citrus: A phylogenetic reassessment with seven new species from China. Fungal Biol. 119 331–347. 10.1016/j.funbio.2015.02.006 [DOI] [PubMed] [Google Scholar]
- Hudson R. R., Kaplan N. L. (1985). Statistical properties of the number of recombination events in the history of a sample of DNA sequences. Genetics 111 147–164. 10.1093/genetics/111.1.147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Index Fungorum (2022). Index fungorum. Index fungorum. Available online at: http://www.indexfungorum.org/names/Names.asp (accessed September, 2022) [Google Scholar]
- Katoh K., Standley D. M. (2013). MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katznelson H., Bose B. (1959). Metabolic activity and phosphate-dissolving capability of bacterial isolates from wheat roots, rhizosphere, and non-rhizosphere soil. Can. J. Microbiol. 5 79–85. 10.1139/m59-010 [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33 1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange L., Barrett K., Meyer A. S. (2021). New method for identifying fungal kingdom enzyme hotspots from genome sequences. J. Fungi 7:207. 10.3390/jof7030207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence D. P., Travadon R., Baumgartner K. (2015). Diversity of Diaporthe species associated with wood cankers of fruit and nut crops in northern California. Mycologia 107 926–940. 10.3852/14-353 [DOI] [PubMed] [Google Scholar]
- Librado P., Rozas J. (2009). DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25 1451–1452. 10.1093/bioinformatics/btp187 [DOI] [PubMed] [Google Scholar]
- Lopez-Bedoya J. L., Pérez-Alberti A. (2009). Bases ecologicas preliminares para la conservacion de los tipos de habitat de interes comunitario en españa. Madrid: Ministerio de Medio Ambiente. [Google Scholar]
- Marak H. B., Biere A., Van Damme J. M. M. (2002). Systemic, genotype-specific induction of two herbivore-deterrent iridoid glycosides in Plantago lanceolata L. in response to fungal infection by Diaporthe adunca (Rob.) Niessel. J. Chem. Ecol. 28 2429–2448. 10.1023/A:1021475800765 [DOI] [PubMed] [Google Scholar]
- Marin-Felix Y., Hernández-Restrepo M., Wingfield M. J., Akulov A., Carnegie A. J., Cheewangkoon R., et al. (2019). Genera of phytopathogenic fungi: GOPHY 2. Stud. Mycol. 92 47–133. 10.1016/j.simyco.2018.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins F., Pereira J. A., Bota P., Bento A., Baptista P. (2016). Fungal endophyte communities in above- and belowground olive tree organs and the effect of season and geographic location on their structures. Fungal Ecol. 20 193–201. 10.1016/j.funeco.2016.01.005 [DOI] [Google Scholar]
- Murali T. S., Suryanarayanan T. S., Geeta R. (2006). Endophytic Phomopsis species: Host range and implications for diversity estimates. Can. J. Microbiol. 52 673–680. 10.1139/w06-020 [DOI] [PubMed] [Google Scholar]
- Nei M., Tajima F. (1981). Genetic drift and estimation of effective population size. Genetics 98 625–640. 10.1093/genetics/98.3.625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noriler S. A., Savi D. C., Aluizio R., Palácio-Cortes A. M., Possiede Y. M., Glienke C. (2018). Bioprospecting and structure of fungal endophyte communities found in the Brazilian biomes, Pantanal, and Cerrado. Front. Microbiol. 9:1526. 10.3389/fmicb.2018.01526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norphanphoun C., Gentekaki E., Hongsanan S., Jayawardena R., Senanayake I. C., Manawasinghe I. S., et al. (2022). Diaporthe: Formalizing the species-group concept. Mycosphere 13 752–819. 10.5943/mycosphere/13/1/9 [DOI] [Google Scholar]
- Patel D., Patel A., Vora D., Menon S., Vadakan S., Acharya D., et al. (2018). A resourceful methodology to profile indolic auxins produced by rhizo-fungi using spectrophotometry and HPTLC. 3 Biotech 8:413. 10.1007/s13205-018-1428-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira E., Vázquez de Aldana B. R., San Emeterio L., Zabalgogeazcoa I. (2019). A survey of culturable fungal endophytes from Festuca rubra subsp. pruinosa, a grass from marine cliffs, reveals a core microbiome. Front. Microbiol. 9:3321. 10.3389/fmicb.2018.03321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira E. C. (2021). *Endophytic mycobiota of Festuca rubra subsp. pruinosa and its functionality. Dissertation Thesis. University of Salamanca, Salamanca, Spain. 10.14201/gredos.148559 [DOI] [Google Scholar]
- Rayner R. W. (1970). A mycological colour chart. Wallingford: Commonwealth Mycological Institute. [Google Scholar]
- Redman R. S., Dunigan D. D., Rodriguez R. J. (2001). Fungal symbiosis from mutualism to parasitism: Who controls the outcome, host or invader? New Phytol. 151 705–716. 10.1046/j.0028-646x.2001.00210.x [DOI] [PubMed] [Google Scholar]
- Rehner S. A., Uecker F. A. (1994). Nuclear ribosomal internal transcribed spacer phylogeny and host diversity in the coelomycete Phomopsis. Can. J. Bot. 72 1666–1674. 10.1139/b94-204 [DOI] [Google Scholar]
- Ronquist F., Teslenko M., van der Mark P., Ayres D. L., Darling A., Höhna S., et al. (2012). MrBayes 3.2: Efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61 539–542. 10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos L., Alves A., Alves R. (2017). Evaluating multi-locus phylogenies for species boundaries determination in the genus Diaporthe. PeerJ 5:e3120. 10.7717/peerj.3120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos P. J. C. D., Savi D. C., Gomes R. R., Goulin E. H., Da Costa Senkiv C., Tanaka F. A. O., et al. (2016). Diaporthe endophytica and D. terebinthifolii from medicinal plants for biological control of Phyllosticta citricarpa. Microbiol. Res. 186–187 153–160. 10.1016/j.micres.2016.04.002 [DOI] [PubMed] [Google Scholar]
- Schwyn B., Neilands J. B. (1987). Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 160 47–56. 10.1016/0003-2697(87)90612-9 [DOI] [PubMed] [Google Scholar]
- Shishido M., Ohashi T., Momma N. (2014). Diaporthe sclerotioides exhibits no host specificity among cucurbit species. Plant Pathol. 63 1357–1364. 10.1111/ppa.12201 [DOI] [Google Scholar]
- Shishido M., Yoshida N., Usami T., Shinozaki T., Kobayashi M., Takeuchi T. (2006). Black root rot of cucurbits caused by Phomopsis sclerotioides in Japan and phylogenetic grouping of the pathogen. J. Gen. Plant Pathol. 72 220–227. 10.1007/s10327-006-0273-0 [DOI] [Google Scholar]
- Smith H., Wingfield M. J., Coutinho T. A., Crous P. W. (1996). Sphaeropsis sapinea and Botryosphaeria dothidea endophytic in Pinus spp. and Eucalyptus spp. in South Africa. S. Afr. J. Bot. 62 86–88. 10.1016/S0254-6299(15)30596-2 [DOI] [Google Scholar]
- Stumpf M. P. (2004). Haplotype diversity and SNP frequency dependence in the description of genetic variation. Eur. J. Hum. Genet. 12 469–477. 10.1038/sj.ejhg.5201179 [DOI] [PubMed] [Google Scholar]
- Tajima F. (1989). Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123 585–595. 10.1093/genetics/123.3.585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tateno O., Hirose D., Osono T., Takeda H. (2015). Beech cupules share endophytic fungi with leaves and twigs. Mycoscience 56 252–256. 10.1016/j.myc.2014.07.005 [DOI] [Google Scholar]
- Thompson S. M., Tan Y. P., Shivas R. G., Neate S. M., Morin L., Bissett A., et al. (2015). Green and brown bridges between weeds and crops reveal novel Diaporthe species in Australia. Persoonia 35 39–49. 10.3767/003158515X687506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson S. M., Tan Y. P., Young A. J., Neate S. M., Aitken E. A. B., Shivas R. G. (2011). Stem cankers on sunflower (Helianthus annuus) in Australia reveal a complex of pathogenic Diaporthe (Phomopsis) species. Persoonia 27 80–89. 10.3767/003158511X617110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thongkantha S., Lumyong S., McKenzie E. H. C., Hyde K. D. (2008). Fungal saprobes and pathogens occurring on tissues of Dracaena lourieri and Pandanus spp. in Thailand. Fungal Divers. 30 149–169. [Google Scholar]
- Toghueo R. M. K., Zabalgogeazcoa I., Pereira E. C., Vázquez de Aldana B. R. (2022). A Diaporthe fungal endophyte from a wild grass improves growth and salinity tolerance of Tritordeum and Perennial Ryegrass. Front. Plant Sci. 13:896755. 10.3389/fpls.2022.896755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udayanga D., Castlebury L. A., Rossman A. Y., Chukeatirote E., Hyde K. D. (2014a). Insights into the genus Diaporthe: Phylogenetic species delimitation in the D. eres species complex. Fungal Divers. 67 203–229. 10.1007/s13225-014-0297-2 [DOI] [Google Scholar]
- Udayanga D., Castlebury L. A., Rossman A. Y., Hyde K. D. (2014b). Species limits in Diaporthe: Molecular re-assessment of D. citri, D. cytosporella, D. foeniculina and D. rudis. Persoonia 32 83–101. 10.3767/003158514X679984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udayanga D., Liu X., Crous P. W., McKenzie E. H. C., Chukeatirote E., Hyde K. D. (2012). A multi-locus phylogenetic evaluation of Diaporthe (Phomopsis). Fungal Divers. 56 157–171. 10.1007/s13225-012-0190-9 [DOI] [Google Scholar]
- Udayanga D., Liu X., McKenzie E. H. C., Chukeatirote E., Bahkali A. H. A., Hyde K. D. (2011). The genus Phomopsis: Biology, applications, species concepts and names of common phytopathogens. Fungal Divers. 50 189–225. 10.1007/s13225-011-0126-9 [DOI] [Google Scholar]
- Vázquez F. M., Devesa J. A. (1996). Revisión del género Stipa L. y nassella desv. (Poaceae) en la Península Ibérica e Islas Baleares. Acta Bot. Malacit. 21 125–189. 10.24310/abm.v21i0.8674 [DOI] [Google Scholar]
- Vázquez de Aldana B. R., Arellano J. B., Cuesta M. J., Mellado-Ortega E., González V., Zabalgogeazcoa I. (2021). Screening fungal endophytes from a wild grass for growth promotion in Tritordeum, an agricultural cereal. Plant Sci. 303:110762. 10.1016/j.plantsci.2020.110762 [DOI] [PubMed] [Google Scholar]
- Vázquez de Aldana B. R., Cuesta M. J., Zabalgogeazcoa I. (2020). Cultivation and growth dynamics of endophytic fungi in a solid culture medium based on sugar beet pulp. J. Sci. Food Agric. 100 441–446. 10.1002/jsfa.10030 [DOI] [PubMed] [Google Scholar]
- Wang X., Guo Y., Du Y., Yang Z., Huang X., Hong N., et al. (2021). Characterization of Diaporthe species associated with peach constriction canker, with two novel species from China. Mycokeys 80 77–90. 10.3897/mycokeys.80.63816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T.-C., Lu Y.-H., Wang J.-F., Song Z.-Q., Hou Y.-G., Liu S.-S., et al. (2021). Bioactive secondary metabolites of the genus Diaporthe and anamorph Phomopsis from terrestrial and marine habitats and endophytes: 2010–2019. Microorganisms 9:217. 10.3390/microorganisms9020217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q., Du Z., Tian C.-M. (2018). Phylogeny and morphology reveal two new species of Diaporthe from traditional Chinese medicine in Northeast China. Phytotaxa 336:159. 10.11646/phytotaxa.336.2.3 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.