Abstract
Loss of the Y chromosome (LoY) is frequently observed in somatic cells of elderly men. However, LoY is highly increased in tumor tissue and correlates with an overall worse prognosis. The underlying causes and downstream effects of LoY are widely unknown. Therefore, we analyzed genomic and transcriptomic data of 13 cancer types (2375 patients) and classified tumors of male patients according to loss or retain of the Y chromosome (LoY or RoY, average LoY fraction: 0.46). The frequencies of LoY ranged from almost absence (glioblastoma, glioma, thyroid carcinoma) to 77% (kidney renal papillary cell carcinoma). Genomic instability, aneuploidy, and mutation burden were enriched in LoY tumors. In addition, we found more frequently in LoY tumors the gate keeping tumor suppressor gene TP53 mutated in three cancer types (colon adenocarcinoma, head and neck squamous carcinoma, lung adenocarcinoma) and oncogenes MET, CDK6, KRAS, and EGFR amplified in multiple cancer types. On the transcriptomic level, we observed MMP13, known to be involved in invasion, to be up-regulated in LoY of three adenocarcinomas and down-regulation of the tumor suppressor gene GPC5 in LoY of three cancer types. Furthermore, we found enrichment of a smoking-related mutation signature in LoY tumors of head and neck and lung cancer. Strikingly, we observed a correlation between cancer type-specific sex bias in incidence rates and frequencies of LoY, in line with the hypothesis that LoY increases cancer risk in males. Overall, LoY is a frequent phenomenon in cancer that is enriched in genomically unstable tumors. It correlates with genomic features beyond the Y chromosome and might contribute to higher incidence rates in males.
Keywords: Loss of Y, Y chromosome, Aneuploidy, TCGA, Pan-Cancer
Graphical Abstract
1. Introduction
With around 60 million base pairs, the Y chromosome is the third shortest human chromosome. While for most of the time its role was believed to be limited to sex determination and spermatogenesis there is now collected proof that its function is associated with male viability and differences between sexes in health and disease progression [7]. The Y chromosome has often been referred to as a genetic wasteland heading on its way to distinction from the human genome [34]. However, this perception has been challenged in recent years by identifying several ubiquitously expressed Y-linked genes as well as linkage to processes of the immune system and complex polygenic traits.
Twelve genes (KDM5D, CYorf15A, DDX3Y, EIF1AY, NLGN4Y, PRKY, RPS4Y1, TBL1Y, TMSB4Y, USP9Y, UTY, and ZFY) were previously described as essential for male viability [7]. All of those have a homolog on the X chromosome, suggesting that they are dosage-sensitive. Some of which already have been associated with susceptibility to complex diseases [34]. Genes of the Y chromosome are involved in coronary artery diseases [12], autoimmune diseases [11] and infectious diseases [11]. Furthermore, deficiency of KDM5D is involved in clear cell renal cell carcinoma [4]). Copy number variations in UTY have been associated with an increased risk for cardiovascular disease [41]. UTY and ZFY have been implied to be tumor suppressor genes [16].
Aging-related mosaic LoY has been known for half a century, but for the most part, it was considered a neutral karyotype related to normal aging [21]. LoY is the most commonly observed mosaic chromosomal alteration in healthy men and its occurrence strongly increases with age as it was detected in around 2.5% of men younger than 45 and around 40% of men at 70 years of age [47], [6]. There is no commonly accepted standard on how to detect and quantify LoY [23], [39], rendering it difficult to compare LoY frequencies across studies where methods with different sensitivities have been used. LoY might be seen as a quantitative trait starting at the level of an individual cell in a man, spanning over just detectable levels in bulk tissue analyses up to a level where the majority of cells within a tissue is affected. LoY has been detected in microglia on a single cell level with associated expression alterations of autosomal genes suggesting the possibility as a contributing factor in the pathogenesis of neurodegenerative disorders [48]. Multiple epidemiological studies identified LoY in blood cells as a significant risk factor for shortened lifespan and various diseases in males [22], including increased heart failure mortality and a mouse model for hematopoietic LoY showed macrophage and TGFβ1 mediated cardiac fibrosis [43].
In the context of cancer, LoY is increasingly observed in tumor tissue. However, the Y chromosome is usually overlooked in cancer genomics due to additional steps needed for copy number analysis. One of the major consequences associated with LoY is the correlation with reduced survival [19], [20], [31], [39]. An average decrease of 5.5 years was reported in patients with LoY in the blood [20], making it a better cancer predictor than age [5]. LoY is also associated with an overall higher susceptibility to cancer in men [19] and sex imbalance is characteristic for many cancer types, not adequately explained by differences in exposure to risk factors [15]. This raises the question whether differences on the genomic level, i.e., the sex-specific phenomenon of LoY, contribute to the disparity.
It has previously been described that LoY correlates with general genome-wide instability, a process that drives cancer evolution [32]. It remains unclear whether LoY causatively induces genomic instability or rather occurs as a "passenger". However, our study on 400 esophageal adenocarcinomas of male patients indicated that LoY is a prognostic marker for short overall survival, independent of TP53 mutation status, a strong indicator for genomic instability [30]. Overall, there is increasing evidence that LoY is a relevant factor in cancer biology but its molecular consequences are largely unknown.
In the present study, we investigated the genomic and transcriptomic architecture of tumors with LoY compared to RoY across various cancer types. To do so, we classified cancers based on single nucleotide polymorphism (SNP) array-derived copy number information into LoY and RoY, defined the frequency of LoY across cancer types, and used whole-exome and RNA sequencing data to test for differences between the two groups. We observed significant differences in frequencies of mutations, copy number alterations, mutation signatures, and expression levels, most of them cancer type-specific, and found a trend towards more frequent LoY in cancer types with higher incidence rates in males.
2. Material & methods
The results published here are, in whole, based on data generated by The Cancer Genome Atlas (TCGA) Research Network: https://www.cancer.gov/tcga. We retrieved whole-exome sequencing, RNA-seq, SNP-array copy number, and clinical data from the GDC portal using the TCGAbiolinks package (v2.24 [14]).
2.1. Investigated cancer types
For 21 of the 33 cancer types, tumor purity estimates were available [3], relevant for the copy number index calculation described below. From these projects, 13 cancer types presented a bimodal distribution of the Y chromosome copy number index (YCNI), indicating the presence of a sufficient fraction of LoY tumors, making them suitable for the following analyses. Multiple cancer types are subdivided into subtypes. For this reason, subtypes were either excluded or combined, as described in the tissue column of Table 1, but histologic distinctions between adenocarcinoma and squamous cell carcinoma were always maintained. Normal tissue was available for the majority of analyzed patients which was used as a reference for the YCNI threshold selection. All analyses were performed separately for the cancer types, and the results were compared.
Table 1.
Frequencies of LoY in TCGA cancer data sets. Selected thresholds for LoY classification, the absolute number of patients being classified as one of the four subgroups (LoY, RoY, outlier, microsatellite instability (MSI)), the two selected thresholds for low and high TAAI, and the absolute distributions of patients being classified TAAI subsets low, medium, high and LoY among TAAI low and high tumors (last two columns), respectively.
| TAAI threshold | TAAI subsets | LoY subsets | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Type | LoY threshold | LoY | RoY | Outlier | MSI | LoY frequency | low | high | low | mid | high | low | high |
| ACC | 0.6 | 12 | 15 | 0 | 28 | 0.22 | 9.44 | 11.8 | 9 | 11 | 10 | 4 | 4 |
| BLCA | 0.6 | 68 | 224 | 7 | 2 | 0.23 | 5.17 | 7.69 | 98 | 98 | 99 | 19 | 20 |
| COAD | 0.65 | 58 | 159 | 4 | 2 | 0.26 | 4.00 | 6.31 | 73 | 73 | 74 | 5 | 26 |
| EAC | 0.75 | 45 | 16 | 3 | 4 | 0.66 | 7.55 | 11.9 | 20 | 22 | 21 | 16 | 14 |
| ESC | 0.65 | 32 | 28 | 0 | 2 | 0.52 | 7.88 | 11.6 | 20 | 21 | 21 | 11 | 12 |
| HNSC | 0.75 | 139 | 231 | 0 | 0 | 0.38 | 2.63 | 3.86 | 123 | 125 | 124 | 16 | 67 |
| KICH | 0.7 | 22 | 17 | 4 | 0 | 0.51 | 10.58 | 12.48 | 13 | 14 | 14 | 5 | 10 |
| KIRC | 0.7 | 155 | 218 | 5 | 0 | 0.41 | 11.97 | 16.61 | 124 | 126 | 125 | 63 | 45 |
| KIRP | 0.7 | 173 | 46 | 8 | 2 | 0.76 | 4.71 | 7.09 | 73 | 74 | 74 | 58 | 52 |
| LUAD | 0.7 | 80 | 163 | 1 | 6 | 0.32 | 7.66 | 11.57 | 81 | 82 | 82 | 20 | 34 |
| LUSC | 0.75 | 190 | 184 | 5 | 1 | 0.50 | 7.12 | 9.57 | 125 | 125 | 126 | 59 | 60 |
| READ | 0.7 | 16 | 27 | 1 | 6 | 0.32 | 6.14 | 8.11 | 14 | 16 | 15 | 3 | 8 |
| SKCM | 0.75 | 20 | 37 | 8 | 2 | 0.30 | 3.68 | 5.7 | 19 | 20 | 20 | 5 | 9 |
2.2. Sample curation
Patients were excluded from the analyses if they either showed a copy number index for the Y chromosome higher than two or a very high mutational burden (upper boundary by taking two standard deviations from the mean) because these samples were expected to distort the results of the mutation analyses.
2.3. Copy number index of the Y chromosome
Copy number indices for the Y chromosome were calculated based on single nucleotide polymorphism (SNP) array data as previously described by Hollows and colleagues [23], with the exception of using only one separator at the lowest point between the two YCNI peaks to separate LoY from RoY instead of selecting extreme groups, due to the small sample size of a few cancer types. Adjustments for purity estimates were derived from Aran, Sirota, and Butte [3] and TCGA network (2017) for esophageal cancers. For most cancer types, the density curve of this calculation resulted in a bimodal distribution, representing ratios of LoY and RoY cells of a heterogeneous tumor, in concordance with previous findings for head and neck squamous carcinoma [23]. To increase clarity in the bimodal distribution of the YCNI density curve of a cancer type, we included normal tissue samples in the analysis to define the cutoff between LoY and RoY (Fig. 1 A).
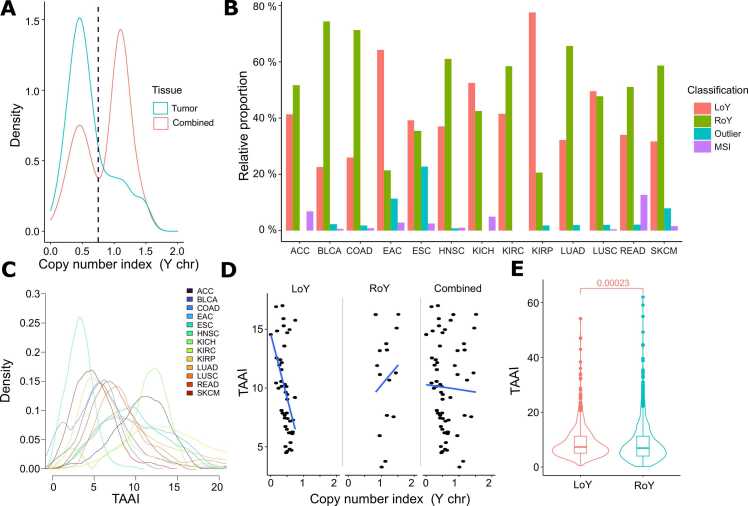
Fig. 1.
LoY Classification and distribution. (A) Threshold selection for esophageal adenocarcinoma (EAC) indicated as a dashed vertical line. Density is plotted for the Y chromosome copy number index (x-axis). (B) Cancer classification shown as distributions relative to the total number of each cancer type. (C) Density curves of the TAAI shown for the 13 cancer types. Each curve is colored as indicated in the legend. (D) Comparison of the copy number index for the Y chromosome and the respective patient’s TAAI shown for esophageal adenocarcinoma. (E) Combined TAAI of all 13 cancer entities. LoY (red) and RoY (turquoise). The abbreviated cancer types are: adrenocortical carcinoma (ACC), bladder urothelial carcinoma (BLCA), colon adenocarcinoma (COAD), esophageal adenocarcinoma (EAC), esophageal squamous cell carcinoma (ESC), head and neck squamous cell carcinoma (HNSC), kidney chromophobe (KICH), kidney renal clear cell carcinoma (KIRC), kidney renal papillary cell carcinoma (KIRP), lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), rectum adenocarcinoma (READ), and skin cutaneous carcinoma (SKCM).
2.4. Total autosomal aneuploidy index
Similar to the calculation for a single chromosome, the total autosomal aneuploidy index (TAAI) summarizes copy number indices of all autosomes. For this, the absolute difference between two and the copy number index for all autosomes was calculated, as previously described by Hollows and colleagues [23]. We tested for correlation between YCNI and the TAAI and noticed that both extremes of the YCNI values were associated with high TAAI. We therefore separated in a second step low and high YCNI (Fig. 1D).
2.5. Survival analyses
Patients were stratified into the two LoY groups, as well as the additional parameter of high and low TAAI to further visualize possible correlations between TAAI and LoY with regards to overall survival. For this, samples were subdivided into three sections of equal size, based on their TAAI. Samples with a TAAI in the middle section were then excluded. The analyses were implemented with the TCGAbiolinks package (v2.24 [14]) with default settings.
2.6. Mutation analyses
Each patient's tumor mutational burden (TMB, all variants, SNV, and indels) was derived from TCGAs mutation annotation format (MAF) files for each cancer type which, for all TCGA samples of the study, were derived from whole exome sequencing. The results were visualized with the maftools package (v2.12 [35]). Furthermore, the relative differences of the mean between LoY and RoY were highlighted. For TMB and gene-based analyses, only non-silent mutations were used.
2.7. Mutation signatures
For analyzing the single base substitutions (SBS), the mutSignatures package (v2.1.1 [18]) was used on the exome sequencing-derived variants with default settings. The first step consisted of comparing all patient's mutations from one cancer type against all 78 SBS COSMIC signatures. Signatures that contributed less than 5% to the overall burden were excluded. The analysis was then run again, but separately for LoY and RoY patients and only with signatures accounting for at least 5% of mutations in the respective cancer type. The distribution of the signatures between the two groups, LoY and RoY, was then tested with a paired t-test. The results were summarized in an upset plot using the ComplexUpset package (v1.3.3 [28]).
2.8. Differentially mutated genes
The number of non-silent mutations derived from the exome sequencing within both subgroups of LoY and RoY were compared for every mutated gene in each cancer type individually. A contingency table was created for each gene to compare the number of patients with at least one mutation in the respective gene versus the number of patients without a mutation for both, LoY and RoY. Results were analyzed with the fisher’s exact test to test for significant differences between LoY and RoY. Results were corrected for multiple testing [8].
2.9. Differential expression analyses
After classification into LoY and RoY, the patients’ gene expression derived from TCGAs RNA-sequencing data was compared in a differential expression analysis implemented with the R package DESeq2 (v3.15 [33]). Lists of significant up- or downregulated genes were derived and then compared across cancer types. Genes were declared as significantly up- or downregulated with a p-value below 0.05 and a log2 fold change above an absolute value of 1.
In addition to the transcriptome-wide expression analysis, an expression analysis exclusively for the Y chromosome was implemented. Here, we used the raw FPKM values of each gene along the Y chromosome to search for specific genomic regions that showed an enrichment or depletion for differentially expressed genes. Genes with an overall low expression (summed FPKM value below 1) were excluded. The results were then summarized in a series of boxplots for each gene according to the location on the Y chromosome and group (LoY and RoY), respectively. The expressions were then compared between LoY and RoY for each gene with a paired t-test and corrected for multiple testing [8].
2.10. Identification of frequent copy number alterations
GISTIC2.0 (v2.0.22 [36]) was used to analyze the SNP array-based copy number data via the Genepattern Cloud to identify genomic regions with copy number alterations of significant frequencies. Masked copy number data was used to exclude germline copy number variants, resulting in the exclusion of the sex chromosomes. From the copy number segments of each patient within a cancer type and Y chromosome-status subgroup, GISTIC2.0 identified genes most likely to be affected by amplification or deletion events. GISTIC2.0 was run at default settings with the most recent reference gene file GRCh38 (GISTIC default reference genome: Human_Hg38. UCSC.add_miR.160920). The confidence level that is used to calculate the region containing the driver was increased to 0.99, as suggested by the TCGA GISTIC pipeline, and the arm peel method was activated to better separate peaks. To further denoise the results, the analysis was done a second time for the normal tissue, and genes occurring in both sets were excluded. For each cancer type, four lists of genes were generated with either significantly amplified or deleted genes in the respective subgroup of LoY and RoY.
3. Results
3.1. Loss of Y classification
To reliably separate tumors into LoY and RoY, a copy number index for the Y chromosome (YCNI) was calculated (Methods). After retrieving the YCNIs for each cancer type separately, the results were visualized in density plots for both the tumor tissue and combined with normal tissue (Fig. 1A and Supp. Figs. 1–2). As expected, we observed two peaks, with the lower peak and indices below representing tumors with LoY and the higher peak and values above representing tumors with RoY. The thresholds between LoY and RoY ranged between 0.6 and 0.75 across the different cancer types (Table 1).
Despite the low variation of the LoY thresholds, the relative distributions of patients among the different cancer types being classified as LoY and RoY differed substantially (Fig. 1B). For example; approximately 25% of bladder urothelial carcinomas, colon-, and lung adenocarcinomas were LoY tumors, while up to 75% of tumors of other types, such as esophageal adenocarcinoma and kidney renal papillary cell carcinoma showed LoY. The ratios retrieved for esophageal adenocarcinoma and head and neck squamous cell carcinoma are in concordance with the literature [23], [30]. However, recent findings suggest a slightly different order with a higher LoY proportion for head and neck squamous cell carcinoma and lung adenocarcinoma [39].
In conclusion, the classification previously used exclusively on head and neck squamous cell carcinoma [23] can be transferred to the majority of TCGA datasets leading to differentiation into LoY and RoY tumors.
3.2. Loss of Y correlates with genome-wide instability
To further investigate the possibility that LoY reflects general genome instability, a total autosomal aneuploidy index (TAAI) for each sample was calculated. Unlike the bimodal distribution of the copy number index for the Y chromosome, the TAAI shows a Gaussian distribution (Fig. 1C). The level of TAAI reflecting general genomic instability varied greatly across cancer types, with head and neck squamous cell carcinoma showing low TAAI. In contrast, adrenocortical carcinoma, kidney chromophobe, and renal papillary cell carcinoma showed the highest levels.
After stratifying samples into RoY and LoY, we compared the YCNI with the TAAI (Fig. 1D and Supp. Fig. 3–6). Samples with YCNI< 0.75, corresponding to LoY (Table 1), showed an inverse correlation between LoY and TAAI, while samples with YNCI values equal to and greater than 0.75, reflecting the normal state in males and samples with gains of the Y chromosome, showed a positive correlation. This trend was observed for most cancer types, indicating that both non-normal copy states, loss and gain of the Y chromosome, are associated with a high level of genomic instability (Supp. Fig. 3–6). The combined analysis across all cancer types showed that LoY is associated with higher overall TAAI (Fig. 1E).
In summary, the TAAI varies greatly across the different cancer types. TAAI is higher in LoY compared to RoY tumors but also in tumors that gained an additional Y chromosome, in agreement with the hypothesis that an unstable genome is more likely to lose or gain a Y chromosome. However, it cannot be distinguished whether LoY is the consequence of genomic instability or vice versa genome instability is a consequence of LoY.
3.3. Differences in mutation profiles between LoY and RoY
Next, we tested for differences at the level of somatic single nucleotide variants (SNVs) between LoY and RoY, starting with each sample’s tumor mutational burden (TMB) defined by the number of non-silent somatic SNVs and INDELs/megabase (Mb) (Fig. 2 A).
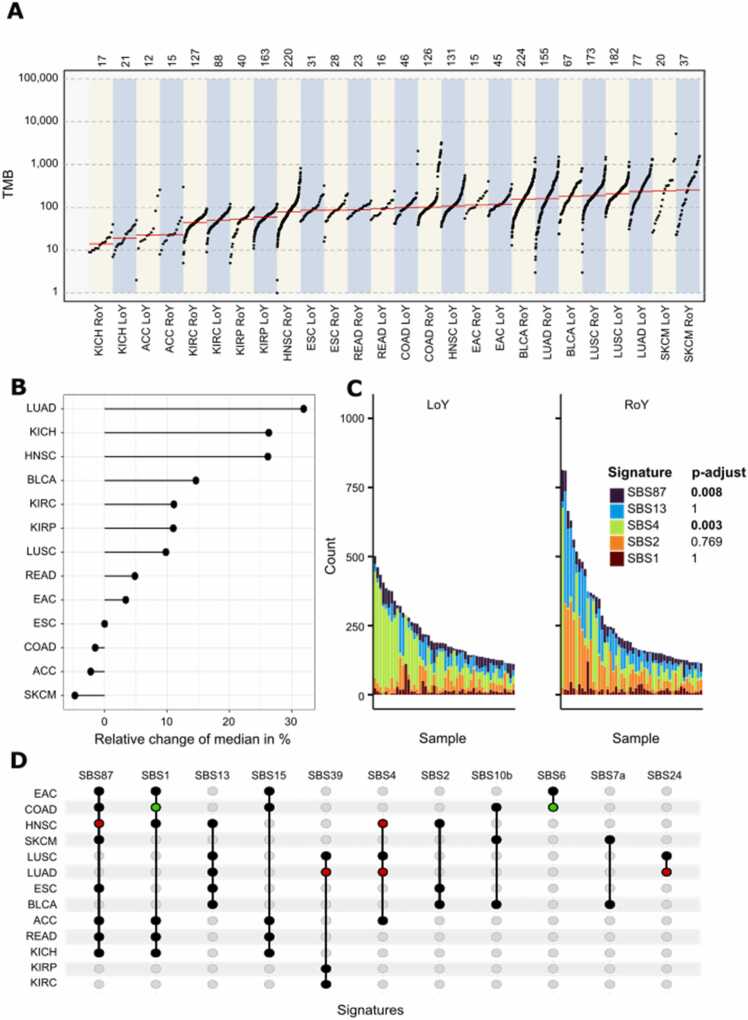
Fig. 2.
Comparison of somatic SNV features between LoY and RoY. (A) Cancer types separated in LoY and RoY tumors (x-axis) shown in an ascending order based on their median TMB (y-axis) marked in red. (B) Comparison of the relative differences between LoY and RoY with regards to the median TMB. The full names of the cancer types can be found in Fig. 1. (C) Mutation signature analysis for head and neck squamous cell carcinoma separated by Y chromosome status is shown with LoY on the left and RoY on the right. Patients are sorted by their absolute number of single base substitutions. The colors reflect COSMIC mutation signatures according to the panel on the right. Adjusted p-values of the t-test comparing the frequency of mutations assigned to a given signature between LoY and RoY are shown on the right. (D) Upset plot indicates the presence of a signature within a cancer type as a black dot. Significant enrichment in the subsets is highlighted in red for LoY and in green for RoY.
In general, LoY tumors tended to have higher TMB compared to RoY tumors (p = 0.01, t-test, Fig. 2B). Cancer types with relatively large differences were head and neck squamous cell carcinoma, kidney chromophobe, and lung adenocarcinoma. Exceptions from this trend were adrenocortical carcinoma, esophageal squamous cell carcinoma, colon adenocarcinoma, and skin cutaneous melanoma. This cancer-specific distribution is in concordance with previous findings [53]. The correlation of LoY with high TMB in the majority of cancers documents an association of LoY with mutation processes, DNA repair or general genome complexity, where the cause remains unclear.
To further analyze the different types of mutations, we assigned the respective SNVs (single base substitution or SBS) to the best fitting COSMIC mutation signature for each cancer type and Y chromosome group (Fig. 2C and Supp. Fig. 7–9). Out of 78 described signatures in COSMIC, the mutations were assigned to eleven signatures across all investigated cancer types (Fig. 2D). Interestingly, we observed significant depletion for signature 1 (SBS1, p-value = 0.012) and SBS6 (p-value = 0.001) in LoY tumors of colon adenocarcinoma. SBS1 is a clock-like signature as the number of mutations of this signature correlates with the patient's age [38]. Its lower level might suggest that LoY tumors develop earlier in the colon than RoY tumors. In LoY head and neck squamous cell carcinomas, SBS4 (p-value = 0.001) and SBS87 (p-value = 0.008) were significantly enriched. SBS87 is associated with thiopurine chemotherapy treatment [29] and was found in most cancer types (Fig. 2D). SBS4 is associated with tobacco smoking, a major risk factor for head and neck squamous cell carcinoma. The enrichment of SBS4 in LoY supports the reported enrichment of LoY in smokers. LoY tumors of kidney renal papillary cell carcinoma showed significant enrichment for SBS54 (p-value = 0.0002) and LoY lung adenocarcinomas for SBS24 (p-value = 0.016) and SBS39 (p-value = 0.017). SBS24 is associated with aflatoxin exposure [2] again connecting LoY to a mutagen. In a mutagenic-driven TMB high scenario, many premalignant cells accumulate weak driver gene mutations [37] and cells with LoY might bear the additional weak driver that could push a cell beyond the threshold of transformation.
Overall, LoY tumors showed a trend towards higher TMB resulting from higher genomic instability and a higher level of somatic SNVs. Interestingly, seven mutation signatures in four cancer types show significant differences between LoY and RoY, potentially reflecting differences in etiology.
3.4. TP53 is more frequently mutated in LoY tumors
After comparing the mutations at the signature level, we were interested in potential differences between LoY and RoY on the gene level. The number of genes that displayed significant differences in mutation frequencies in LoY tumors compared to RoY tumors ranged from 610 in lung adenocarcinoma to zero in adrenocortical carcinoma and kidney chromophobe (Supp. Table 1).
Classical oncogenes were among the differentially mutated genes (e.g., KRAS more frequent in RoY in colon adenocarcinoma) and tumor suppressor genes (e.g., BRCA1 more frequent in LoY in bladder cancer). Differentially mutated genes usually showed higher mutation counts for LoY tumors (2266 genes higher in LoY compared to 561 genes higher in RoY; Supp. Table 1), in agreement with an overall higher TMB in LoY tumors. However, the distribution of genes was heterogeneous across cancers, and we searched for genes with mutation frequency differences in more than one cancer type. Several genes showed differences in two cancer types and four genes in three cancer types (Supp. Table 2–3).
TP53 was more frequently mutated in LoY of three cancer types (colon adenocarcinoma, head and neck squamous carcinoma, lung adenocarcinoma) but less frequent in LoY of kidney renal papillary cell carcinoma. Loss of TP53 function correlates with enrichment of somatic genome copy number alterations and poor prognosis in several cancer types [53], again indicating an association of LoY with features of genomic instability. The other three genes more frequently mutated in LoY were PKHD1L1, AKAP9, and WDFY3. Interestingly, we found MMP13 (matrix metallopeptidase 13) and MUC16 (mucin 16) significantly more often mutated in LoY of two cancer types (lung adenocarcinoma/skin cutaneous carcinoma and head and neck squamous cell carcinoma/kidney renal papillary cell carcinoma, respectively). MMP13 has been implicated in invasion, metastasis, and angiogenesis [24], [27]. MUC16 is classified as an oncogene mediating proliferation and migration [13], [44]. Both enrichments in LoY tumors suggest more aggressive biology.
Overall, there are significant differences in gene-based mutation frequencies between LoY and RoY of heterogeneous gene sets across cancer types. For the majority of genes, the mutation frequencies are higher in LoY and TP53 is the most commonly enriched gene for mutations in LoY tumors, which is in agreement with recent findings [39].
3.5. Recurrent copy number alterations containing cancer genes are associated with LoY
Since LoY was associated with TAAI, we wondered whether more specific local copy number gains or losses were associated with LoY. We used GISTIC separately for LoY and RoY tumors of each cancer type and found largely overlapping copy number profiles of LoY and RoY (Supp. Fig. 10). Next, we searched for recurrent copy number alterations specific to LoY by subtracting regions also affected in RoY and searched for commonly affected genes across cancer types (Supp. Table 4–5). We found four classical oncogenes frequently affected by copy gains in LoY but not in RoY in different cancer types: MET in kidney renal papillary cell carcinoma and lung adenocarcinoma, CDK6 and KRAS in esophageal adeno- and squamous cell carcinoma, and EGFR in bladder urothelial carcinoma, lung adenocarcinoma, and esophageal squamous cell carcinoma. As recurrently deleted in LoY tumors of more than one cancer type, we observed the tumor suppressor genes FAT1 in colon adenocarcinoma, kidney renal papillary cell carcinoma, and esophageal adenocarcinoma and CASP3 in colon adenocarcinoma, kidney renal papillary cell carcinoma, lung squamous cell carcinoma, and esophageal adenocarcinoma and the oncogene NRAS in the adenocarcinoma of colon, lung, and rectum. While the enrichment of these events might reflect the association with genomically unstable tumors in general, it shows that LoY tumors can be enriched for the presence of clinically relevant targets by favoring specific copy number alterations.
3.6. Differential expression between LoY and RoY tumors
Following the hypothesis that LoY can impact the expression of autosomal genes through trans or epigenetic effects, we performed differential gene expression analyses (Fig. 3, Supp. Fig. 11–14, Supp. Table 6–7). The number of differentially expressed genes varied across cancer types. It ranged from 35 in esophageal adenocarcinoma to 726 in kidney renal papillary cell carcinoma for genes down-regulated in LoY tumors and from 8 in colon adenocarcinoma, kidney renal clear cell carcinoma, and lung squamous cell carcinoma to 173 in esophageal adenocarcinoma for up-regulated genes in LoY tumors (Supp. Table 1). Affected genes were heterogeneous across cancer types and ranged across a broad range of functional groups. We searched for genes with recurrent LoY-specific expression changes across several cancer types and found TMSB4Y downregulated in all cancer types and 33 other Y-linked genes, downregulated in at least five cancer types, further validating the LoY classification method (Supp. Fig. 11–14).
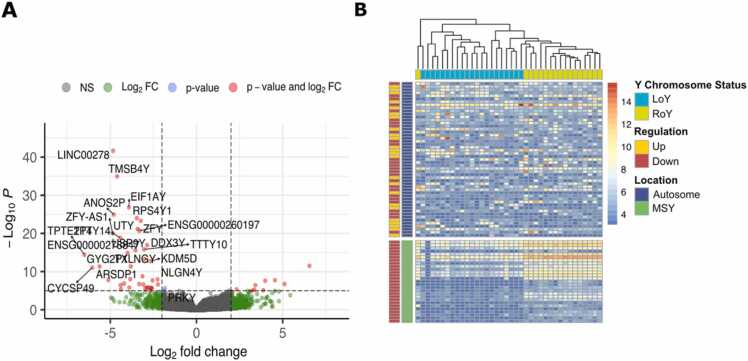
Fig. 3.
Differential gene expression analysis between LoY and RoY. (A) Volcano plot for kidney chromophobe with Y-linked genes highlighted. Genes that show a p-value smaller than 0.05 and an absolute log2 fold change> 2 are colored in red. (B) Heatmap shows genes that are either significantly up- or downregulated in LoY of kidney chromophobe compared to RoY (p-value smaller than 0.05 and log2FC above or below 2). Samples are arranged in columns, genes in rows. Top indicates Y chromosome status of each tumor, left indicates Y chromosomal or autosomal location of a gene and up or down regulation in LoY.
To explore how homogeneous the up- or down-regulation of differentially expressed genes was distributed in LoY and RoY of a given cancer type, we performed unsupervised clustering of the top differentially expressed genes for each cancer type (kidney chromophobe is shown as an example in Fig. 3B, other cancer types are shown in Supp. Fig. 15–18). A relatively clear distinction due to consistent LoY-correlated expression changes could be observed in adrenocortical carcinoma, kidney chromophobe, and rectum adenocarcinoma but less clear in other cancer types. The division was always more striking for the downregulated genes, dominated by the genes located on the Y chromosome. The data illustrates a high level of heterogeneity even within cancer types, suggesting that the potential epigenetic/trans effects of LoY are either modulated by many other factors or that the effect is moderate and/or the stroma dilutes the signal.
To gain confidence in LoY-associated autosomal gene expression changes, we searched for significant LoY/RoY expression differences of autosomal genes recurrently occurring in several cancer types. Interestingly, MMP13 was up-regulated in three LoY adenocarcinomas (colon, esophageal, and lung) while down-regulated in LoY kidney renal papillary cell carcinoma. In contrast, GPC5 (Glypican-5) was significantly down-regulated in LoY tumors of three cancer types (kidney renal papillary cell carcinoma, lung adenocarcinoma, and skin cutaneous melanoma). GPC5 down-regulation is associated with poor outcomes in lung adenocarcinoma patients [52]. Its down-regulation is based on promoter methylation, and overexpression of GPC5 inhibits proliferation, invasion, and tumor growth by suppressing the Wnt/beta-catenin axis [52]. Again, MMP13 up-regulation and GPC5 down-regulation in LoY tumors might indicate more aggressive tumor types.
In conclusion, the differences in gene expression between LoY- and RoY tumors further confirmed the valid classification based on differences in Y-linked genes. LoY-associated expression differences primarily affect different gene sets across cancers. However, recurrently affected genes are not restricted to the Y chromosome, potentially reflecting a functional connection to LoY.
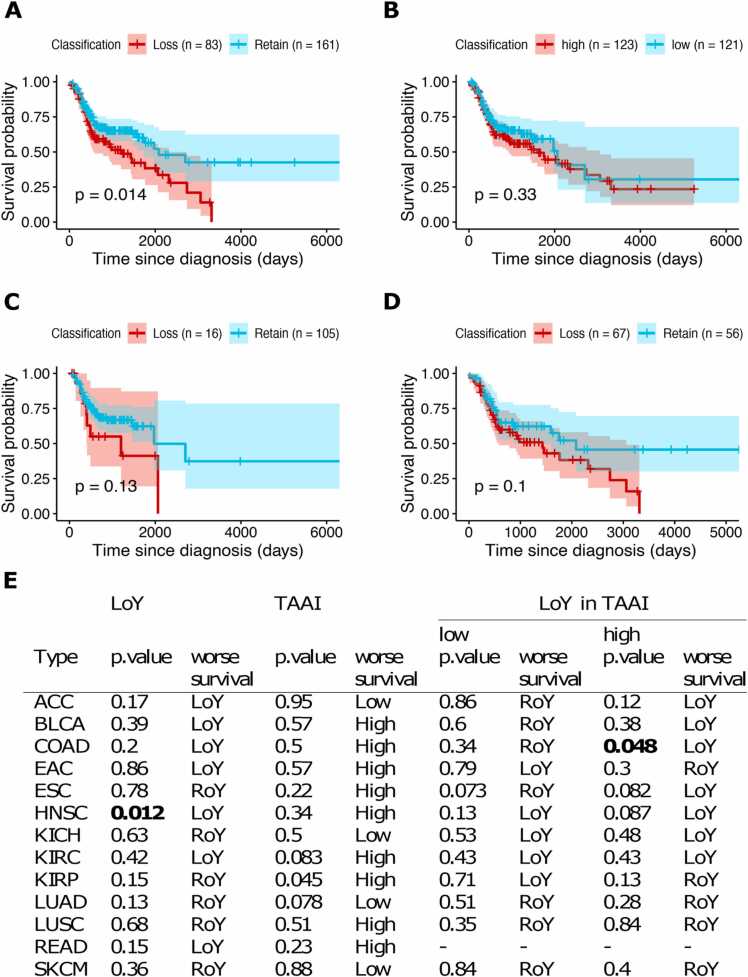
3.7. LoY is associated with shorter overall survival in head and neck squamous cell carcinoma
Further, we tested for association between LoY and overall survival. Since genome instability is correlated with LoY, we also tested for association between TAAI and overall survival to identify potential differences between LoY and TAAI. Only for head and neck squamous cell carcinoma, LoY was associated with shorter overall survival (Fig. 4 and Supp. Fig. 19–22). High TAAI, on the other hand, was associated with shorter overall survival in kidney renal papillary cell carcinoma. Next, we separated the effects of TAAI and LoY by subdividing the patient groups into TAAI high and TAAI low and tested within these subgroups for survival differences between patients with LoY or RoY tumors. LoY showed a borderline significance for association with shorter overall survival among the TAAI high colorectal adenocarcinoma (Fig. 4E). With these sample sizes, we could not identify strong effects of LoY with regards to survival even without correction for multiple testing. Therefore, the association between LoY and poorer outcome in tumors with LoY [39] could not be confirmed.
Fig. 4.
Survival analysis of the TCGA cohort stratified by LoY and TAAI. (A-D) Kaplan-Meier survival curves for HNSC patients. (A) Comparison between LoY (red) and RoY (turquoise). (B) Comparison between a high TAAI (red) and low TAAI (turquoise). (C) Comparison between LoY (red) and RoY (turquoise) for patients that show a low TAAI. (D) Comparison between LoY (red) and RoY (turquoise) for patients that show a high TAAI. (E) Overview of overall survival analyses for the 13 cancer cohorts showing p-values of Kaplan-Meier analyses for stratification based on LoY/RoY and low/high TAAI (left). Subanalyses were performed for patient groups with low and high TAAI, respectively, to test for an effect of LoY within these subgroups (right).
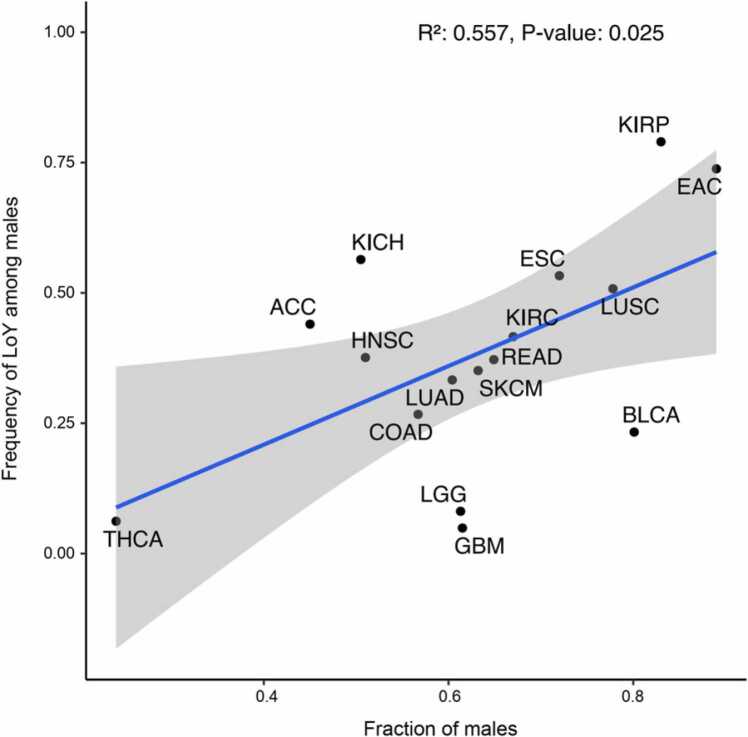
3.8. Male dominated cancer types show a trend for enrichment of LoY
Noting that LoY is associated with a number of features associated with aggressive tumor biology, we hypothesized that LoY contributes to tumor development. If this is true, we expect a higher incidence in males for cancer types with high rates of LoY. To investigate this, we used previously described sex bias in various cancers and tested for a possible correlation between the disparities in incidence rates between men and women and the proportions of LoY in the male patients’ tumors of the respective cancer types. For this analysis, we included cancer types that had meager proportions of LoY samples, which was the reason why they were excluded from previously described analyses (glioblastoma multiforme, brain lower grade glioma, and thyroid carcinoma). We did indeed observe a correlation between the rate of LoY and sex bias (R2 = 0.557, p-value = 0.025, Fig. 5) suggesting that LoY might contribute to cancer development and thereby influence the incidence rate. However, we observed similar results when comparing the incidence rates with the TAAIs, rendering it challenging to assign this effect to LoY.
Fig. 5.
Relation between the frequency of LoY and incidence rates of males. Comparison of the frequency of LoY among males against the incidence rate of males (fraction of males) relative to all patients of a cancer type is shown. Sex-specific incidence rates were derived from recent literature ([25], [1], [46], [40]). The full descriptions of the cancer types can be found in Fig. 1.
4. Discussion
Over the last years, our awareness of sex differences in cancer biology has increased. Besides sex hormone levels, genetic and epigenetic differences between males and females influence the clinical course [42]. While there are several examples of X chromosome-specific effects in females that can contribute to the development of cancer [45] or protect against it [26], the Y chromosome received less attention. This is in part based on the low gene content on the Y chromosome, the fact that males lose the Y chromosome in a fraction of cells during aging, and females do not have a Y chromosome at all. However, extreme down-regulation of Y chromosome gene expression (EDY) is associated with increased cancer risk in men and a driver model with EDY being the functional consequence of LoY has been proposed [10].
Further, we have shown that LoY is an independent prognostic marker for overall survival in esophageal cancer [30]. Additional evidence for a malignant effect of LoY comes from experiments where the introduction of the Y chromosome in a LoY prostate cancer cell line with LoY resulted in reduced tumor formation in a xenograft model [49]. To better understand the potential molecular consequences of LoY, we systematically characterized the genomic and transcriptomic landscape of cancers with LoY.
Based on genomic copy number information, we classified tumors into LoY and RoY in a pan-cancer data set. This approach can be used for the reanalysis of existing genome-wide data. The classification allowed us to compare LoY frequencies across cancer types, an advantage over the comparison of frequencies derived from individual cancer reports. LoY is a frequent genomic event. More than 20% of male patients’ derived tumors were affected in the 13 analyzed cancer types, with exceptionally high frequencies in kidney renal papillary cell carcinoma (76%) and esophageal adenocarcinoma (66%). These high frequencies agree with other reports [30], [39], [9]. We investigated the genomic and transcriptomic features that characterize LoY tumors to gain insight into the molecular context and potential value as a biomarker.
On the genomic level, we observed a correlation between altered Y chromosome copy number and the degree of aneuploidy (TAAI). It remains challenging to distinguish between LoY being the cause for or effect of high TAAI. However, despite the strong association with genome-wide instability, LoY might have unique effects specific to this event. Large autosomal aneuploidy events occur more frequently in men with LoY in somatic non-cancer cells, which implies that LoY precedes total autosomal aneuploidy [21]. This indicates that LoY could be considered a pre-disease state and could function as a biomarker for predicting genome-wide instability [6].
The somatic copy number alteration profiles between LoY and RoY overlap largely. However, we identified regions with enrichment for copy gains in LoY cancers, including amplification of EGFR and MET in lung adenocarcinoma, two driver genes that are targeted by tyrosine kinase inhibitors [50], [51]. We found LoY tumors to have more point mutations (higher TMB) than RoY. Therefore, the genomic profiles of copy number and single nucleotide alterations show a high level of genomic alterations in LoY tumors.
The distribution of somatic SNVs as mutation signatures can provide insight into the mutation mechanisms and etiology. Our analysis did not reveal a general association between LoY and a specific mutation signature which was not expected since the composition of mutation profiles across cancers differs. The smoking signature SBS4 is enriched in head and neck squamous cell carcinoma and lung adenocarcinoma with LoY suggesting that smoking and related mutation processes favor LoY or may more likely result in tumorgenesis from cells with LoY. On the gene level, TP53 was significantly more often mutated in LoY tumors. This is in agreement with the understanding that mutations in TP53 lead to genome instability [17]. Overall, LoY tumors have genomes with more alterations than RoY tumors, suggesting that the former might be more challenging to treat.
The transcriptomic analysis of autosomal genes displayed a high level of diversity with a lack of evidence for a strong trans-effect, e.g. through a Y-linked transcription factor. The LoY associated up-regulation of MMP13 and the downregulation of GPC5 in three cancer types are interesting and potentially cancer-relevant observations.
Earlier studies reported shorter overall survival for patients with LoY tumors [19], [20], [30]. We found significantly shorter overall survival for LoY compared to RoY head and neck squamous cell carcinoma patients. The absence of LoY/overall survival associations for other cancer types is possibly due to small sample sizes. Interestingly, we observed a correlation between the ratio of male to female incidence rates and the fraction of LoY tumors within a cancer type. This suggests that LoY increases the chance for cancer to develop in males and contributes to cancer development. It is in agreement with LoY being associated with poor prognosis and suggests that LoY is an early event in tumor evolution. Since it is known that an increasing number of normal somatic cells lose their Y chromosome during aging, it seems plausible that these cells are more likely than their RoY counterparts to develop cancer.
5. Conclusions
In this study, we conducted a systematic pan-cancer analysis to classify tumors of male patients into tumors that lost or retained their Y chromosome. LoY tumors show more genomic alterations than RoY tumors. A diversity of cancer-relevant molecular features differs between LoY- and RoY tumors and most of these characteristics are specific to individual cancer types. We found an association suggesting that LoY may contribute to the development of cancer and the higher incidence rates in males. Our study provides a resource and could aid future studies to investigate this new field of association of LoY with the disease in more depth.
CRediT authorship contribution statement
A.M.H. designed the study, O.V.C., A.Q., and A.M.H. supervised the study, P.M. performed analyses with help of O.V.C. and A.M.Y., data was interpreted by P.M., O.V.C., C.W., J.S., D.H., R.B., A.Q., and A.M.H., figures were prepared by P.M., manuscript was written by P.M. and A.M.H. with contributions from C.W. and D.H. All authors read and approved the manuscript.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
We thank Robert Hollows for help on YCNI calculation. The study was supported by DFG grant 418074181 and CRC1310 subproject “Predicting molecular mechanisms of adaptation to chemoradiation therapy in cancer” and BMBF grant 031L0267B “Deep Insight”. Graphical abstract was created with bioRender.
Supplement
Comparison of Y-linked RNA expression levels
Since the Y chromosome is not always completely lost, we searched for regions within the chromosome that are more (or less) frequently deleted. For this, a series of boxplots of the respective gene expression values was created for each cancer type. Each gene was then tested for significant differences between LoY and RoY with a paired t-test.
The locations of genes that showed significant differential expression between LoY and RoY were scattered across the Y chromosome with no obvious clustering. However, for some of the cancer types with a large number of samples, there was a recurring window of neighboring genes that were not significantly different between LoY and RoY that in turn were surrounded by significant ones. This window is located on q11.223 and includes the genes: TTTY10, EIF1AY, RPS4Y2, and PRORY (Supp. Fig. 23–26). The findings do not support the hypothesis that essential (tumor suppressing) genes are aggregated in a specific region on chromosome Y which is, therefore, less likely to be deleted.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.csbj.2023.02.024.
Appendix A. Supplementary material
Supplementary material Additional figures visualizing each cancer types results from the different analyses.
.
Supplementary material Additional tables comprising significant genes from the different analyses.
.
References
- 1.Alesina P.F., Walz M.K. Adrenal tumors: are gender aspects relevant? Visc Med. 2020;36(1):15–19. doi: 10.1159/000505788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexandrov L.B., Jones P.H., Wedge D.C., Sale J.E., Campbell P.J., Nik-Zainal S., Stratton M.R. Clock-like mutational processes in human somatic cells. Nat Genet. 2015;47(12):1402–1407. doi: 10.1038/ng.3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aran D., Sirota M., Butte A.J. Systematic pan-cancer analysis of tumour purity. Nat Commun. 2015;6:8971. doi: 10.1038/ncomms9971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arseneault M., Monlong J., Vasudev N.S., Laskar R.S., Safisamghabadi M., Harnden P., Egevad L., Nourbehesht N., Panichnantakul P., Holcatova I., Brisuda A., Janout V., Kollarova H., Foretova L., Navratilova M., Mates D., Jinga V., Zaridze D., Mukeria A., Jandaghi P., Brennan P., Brazma A., Tost J., Scelo G., Banks R.E., Lathrop M., Bourque G., Riazalhosseini Y. Loss of chromosome Y leads to down regulation of KDM5D and KDM6C epigenetic modifiers in clear cell renal cell carcinoma. Sci Rep. 2017;7:44876. doi: 10.1038/srep44876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Asim A., Agarwal S., Avasthi K.K., Sureka S., Rastogi N., Dean D.D., Mohindra S. Investigation of LOY in prostate, pancreatic, and colorectal cancers in males: a case-control study. Expert Rev Mol Diagn. 2020;20(12):1259–1263. doi: 10.1080/14737159.2020.1853528. [DOI] [PubMed] [Google Scholar]
- 6.Barros B., Morais M., Teixeira A.L., Medeiros R. Loss of chromosome Y and its potential applications as biomarker in health and forensic sciences. Cytogenet Genome Res. 2020;160(5):225–237. doi: 10.1159/000508564. [DOI] [PubMed] [Google Scholar]
- 7.Bellott D.W., Hughes J.F., Skaletsky H., Brown L.G., Pyntikova T., Cho T.J., Koutseva N., Zaghlul S., Graves T., Rock S., Kremitzki C., Fulton R.S., Dugan S., Ding Y., Morton D., Khan Z., Lewis L., Buhay C., Wang Q., Watt J., Holder M., Lee S., Nazareth L., Alföldi J., Rozen S., Muzny D.M., Warren W.C., Gibbs R.A., Wilson R.K., Page D.C. Mammalian Y chromosomes retain widely expressed dosage-sensitive regulators. Nature. 2014;508(7497):494–499. doi: 10.1038/nature13206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc: Ser B (Methodol) 1995;57(1):289–300. [Google Scholar]
- 9.Buscheck F., Fraune C., Garmestani S., Simon R., Kluth M., Hube-Magg C., Ketterer K., Eichelberg C., Hoflmayer D., Jacobsen F., Wittmer C., Wilczak W., Sauter G., Fisch M., Eichenauer T., Rink M. Y-chromosome loss is frequent in male renal tumors. Ann Transl Med. 2021;9(3):209. doi: 10.21037/atm-20-3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caceres A., Jene A., Esko T., Perez-Jurado L.A., Gonzalez J.R. Extreme downregulation of chromosome Y and cancer risk in men. J Natl Cancer Inst. 2020;112(9):913–920. doi: 10.1093/jnci/djz232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Case L.K., Wall E.H., Dragon J.A., Saligrama N., Krementsov D.N., Moussawi M., Zachary J.F., Huber S.A., Blankenhorn E.P., Teuscher C. The Y chromosome as a regulatory element shaping immune cell transcriptomes and susceptibility to autoimmune disease. Genome Res. 2013;23(9):1474–1485. doi: 10.1101/gr.156703.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Charchar F.J., Bloomer L.D., Barnes T.A., Cowley M.J., Nelson C.P., Wang Y., Denniff M., Debiec R., Christofidou P., Nankervis S., Dominiczak A.F., Bani-Mustafa A., Balmforth A.J., Hall A.S., Erdmann J., Cambien F., Deloukas P., Hengstenberg C., Packard C., Schunkert H., Ouwehand W.H., Ford I., Goodall A.H., Jobling M.A., Samani N.J., Tomaszewski M. Inheritance of coronary artery disease in men: an analysis of the role of the Y chromosome. Lancet. 2012;379(9819):915–922. doi: 10.1016/S0140-6736(11)61453-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen X., Li X., Wang X., Zhu Q., Wu X., Wang X. MUC16 impacts tumor proliferation and migration through cytoplasmic translocation of P120-catenin in epithelial ovarian cancer cells: an original research. BMC Cancer. 2019;19(1):171. doi: 10.1186/s12885-019-5371-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Colaprico A., Silva T.C., Olsen C., Garofano L., Cava C., Garolini D., Sabedot T.S., Malta T.M., Pagnotta S.M., Castiglioni I., Ceccarelli M., Bontempi G., Noushmehr H. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016;44(8) doi: 10.1093/nar/gkv1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cook M.B., Dawsey S.M., Freedman N.D., Inskip P.D., Wichner S.M., Quraishi S.M., Devesa S.S., McGlynn K.A. Sex disparities in cancer incidence by period and age. Cancer Epidemiol Biomark Prev. 2009;18(4):1174–1182. doi: 10.1158/1055-9965.EPI-08-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davoli T., Xu A.W., Mengwasser K.E., Sack L.M., Yoon J.C., Park P.J., Elledge S.J. Cumulative haploinsufficiency and triplosensitivity drive aneuploidy patterns and shape the cancer genome. Cell. 2013;155(4):948–962. doi: 10.1016/j.cell.2013.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eischen C.M. Genome stability requires p53. Cold Spring Harb Perspect Med. 2016;6(6) doi: 10.1101/cshperspect.a026096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fantini D., Vidimar V., Yu Y., Condello S., Meeks J.J. MutSignatures: an R package for extraction and analysis of cancer mutational signatures. Sci Rep. 2020;10(1):18217. doi: 10.1038/s41598-020-75062-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Forsberg L.A. Loss of chromosome Y (LOY) in blood cells is associated with increased risk for disease and mortality in aging men. Hum Genet. 2017;136(5):657–663. doi: 10.1007/s00439-017-1799-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Forsberg L.A., Rasi C., Malmqvist N., Davies H., Pasupulati S., Pakalapati G., Sandgren J., Diaz de Ståhl T., Zaghlool A., Giedraitis V., Lannfelt L., Score J., Cross N.C., Absher D., Janson E.T., Lindgren C.M., Morris A.P., Ingelsson E., Lind L., Dumanski J.P. Mosaic loss of chromosome Y in peripheral blood is associated with shorter survival and higher risk of cancer. Nat Genet. 2014;46(6):624–628. doi: 10.1038/ng.2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo X., Dai X., Zhou T., Wang H., Ni J., Xue J., Wang X. Mosaic loss of human Y chromosome: what, how and why. Hum Genet. 2020;139(4):421–446. doi: 10.1007/s00439-020-02114-w. [DOI] [PubMed] [Google Scholar]
- 22.Guo X., Li J., Xue J., Fenech M., Wang X. Loss of Y chromosome: an emerging next-generation biomarker for disease prediction and early detection? Mutat Res Rev Mutat Res. 2021;788 doi: 10.1016/j.mrrev.2021.108389. [DOI] [PubMed] [Google Scholar]
- 23.Hollows R., Wei W., Cazier J.B., Mehanna H., Parry G., Halford G., Murray P. Association between loss of Y chromosome and poor prognosis in male head and neck squamous cell carcinoma. Head Neck. 2019;41(4):993–1006. doi: 10.1002/hed.25537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang S.H., Law C.H., Kuo P.H., Hu R.Y., Yang C.C., Chung T.W., Li J.M., Lin L.H., Liu Y.C., Liao E.C., Tsai Y.T., Wei Y.S., Lin C.C., Chang C.W., Chou H.C., Wang W.C., Chang M.D., Wang L.H., Kung H.J., Chan H.L., Lyu P.C. MMP-13 is involved in oral cancer cell metastasis. Oncotarget. 2016;7(13):17144–17161. doi: 10.18632/oncotarget.7942. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25.Huret J.L., Ahmad M., Arsaban M., Bernheim A., Cigna J., Desangles F., Guignard J.C., Jacquemot-Perbal M.C., Labarussias M., Leberre V., Malo A., Morel-Pair C., Mossafa H., Potier J.C., Texier G., Viguie F., Yau Chun Wan-Senon S., Zasadzinski A., Dessen P. Atlas of genetics and cytogenetics in oncology and haematology in 2013. Nucleic Acids Res. 2013;41(Database issue):D920–D924. doi: 10.1093/nar/gks1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaneko S., Li X. X chromosome protects against bladder cancer in females via a KDM6A-dependent epigenetic mechanism. Sci Adv. 2018;4(6):eaar5598. doi: 10.1126/sciadv.aar5598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kudo Y., Iizuka S., Yoshida M., Tsunematsu T., Kondo T., Subarnbhesaj A., Deraz E.M., Siriwardena S.B., Tahara H., Ishimaru N., Ogawa I., Takata T. Matrix metalloproteinase-13 (MMP-13) directly and indirectly promotes tumor angiogenesis. J Biol Chem. 2012;287(46):38716–38728. doi: 10.1074/jbc.M112.373159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lex A., Gehlenborg N., Strobelt H., Vuillemot R., Pfister H. UpSet: visualization of intersecting sets. IEEE Trans Vis Comput Graph. 2014;20(12):1983–1992. doi: 10.1109/TVCG.2014.2346248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li B., Brady S.W., Ma X., Shen S., Zhang Y., Li Y., Szlachta K., Dong L., Liu Y., Yang F., Wang N., Flasch D.A., Myers M.A., Mulder H.L., Ding L., Liu Y., Tian L., Hagiwara K., Xu K., Zhou X., Sioson E., Wang T., Yang L., Zhao J., Zhang H., Shao Y., Sun H., Sun L., Cai J., Sun H.Y., Lin T.N., Du L., Li H., Rusch M., Edmonson M.N., Easton J., Zhu X., Zhang J., Cheng C., Raphael B.J., Tang J., Downing J.R., Alexandrov L.B., Zhou B.S., Pui C.H., Yang J.J., Zhang J. Therapy-induced mutations drive the genomic landscape of relapsed acute lymphoblastic leukemia. Blood. 2020;135(1):41–55. doi: 10.1182/blood.2019002220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loeser H., Wölwer C.B., Alakus H., Chon S.H., Zander T., Buettner R., Hillmer A.M., Bruns C.J., Schroeder W., Gebauer F., Quaas A. Y chromosome loss is a frequent event in Barrett's adenocarcinoma and associated with poor outcome. Cancers (Basel) 2020;12(7) doi: 10.3390/cancers12071743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loftfield E., Zhou W., Graubard B.I., Yeager M., Chanock S.J., Freedman N.D., Machiela M.J. Predictors of mosaic chromosome Y loss and associations with mortality in the UK Biobank. Sci Rep. 2018;8(1):12316. doi: 10.1038/s41598-018-30759-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Loftfield E., Zhou W., Yeager M., Chanock S.J., Freedman N.D., Machiela M.J. Mosaic Y loss is moderately associated with solid tumor risk. Cancer Res. 2019;79(3):461–466. doi: 10.1158/0008-5472.CAN-18-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maan A.A., Eales J., Akbarov A., Rowland J., Xu X., Jobling M.A., Charchar F.J., Tomaszewski M. The Y chromosome: a blueprint for men's health? Eur J Hum Genet. 2017;25(11):1181–1188. doi: 10.1038/ejhg.2017.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mayakonda A., Lin D.C., Assenov Y., Plass C., Koeffler H.P. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018;28(11):1747–1756. doi: 10.1101/gr.239244.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mermel C.H., Schumacher S.E., Hill B., Meyerson M.L., Beroukhim R., Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12(4):R41. doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nahar R., Zhai W., Zhang T., Takano A., Khng A.J., Lee Y.Y., Liu X., Lim C.H., Koh T.P.T., Aung Z.W., Lim T.K.H., Veeravalli L., Yuan J., Teo A.S.M., Chan C.X., Poh H.M., Chua I.M.L., Liew A.A., Lau D.P.X., Kwang X.L., Toh C.K., Lim W.T., Lim B., Tam W.L., Tan E.H., Hillmer A.M., Tan D.S.W. Elucidating the genomic architecture of Asian EGFR-mutant lung adenocarcinoma through multi-region exome sequencing. Nat Commun. 2018;9(1):216. doi: 10.1038/s41467-017-02584-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nik-Zainal S., Alexandrov L.B., Wedge D.C., Van Loo P., Greenman C.D., Raine K., Jones D., Hinton J., Marshall J., Stebbings L.A., Menzies A., Martin S., Leung K., Chen L., Leroy C., Ramakrishna M., Rance R., Lau K.W., Mudie L.J., Varela I., McBride D.J., Bignell G.R., Cooke S.L., Shlien A., Gamble J., Whitmore I., Maddison M., Tarpey P.S., Davies H.R., Papaemmanuil E., Stephens P.J., McLaren S., Butler A.P., Teague J.W., Jonsson G., Garber J.E., Silver D., Miron P., Fatima A., Boyault S., Langerod A., Tutt A., Martens J.W., Aparicio S.A., Borg A., Salomon A.V., Thomas G., Borresen-Dale A.L., Richardson A.L., Neuberger M.S., Futreal P.A., Campbell P.J., Stratton M.R., C. Breast Cancer Working Group of the International Cancer Genome Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149(5):979–993. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qi M., Pang J., Mitsiades I., Lane A., Rheinbay E. Loss of chromosome Y in primary tumors. bioRxiv. 2022 doi: 10.1016/j.cell.2023.06.006. 2022.08.22.504831. [DOI] [PubMed] [Google Scholar]
- 40.Rahbari R., Zhang L., Kebebew E. Thyroid cancer gender disparity. Future Oncol. 2010;6(11):1771–1779. doi: 10.2217/fon.10.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rogers M.J. Y chromosome copy number variation and its effects on fertility and other health factors: a review. Transl Androl Urol. 2021;10(3):1373–1382. doi: 10.21037/tau.2020.04.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rubin J.B., Lagas J.S., Broestl L., Sponagel J., Rockwell N., Rhee G., Rosen S.F., Chen S., Klein R.S., Imoukhuede P., Luo J. Sex differences in cancer mechanisms. Biol Sex Differ. 2020;11(1):17. doi: 10.1186/s13293-020-00291-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sano S., Horitani K., Ogawa H., Halvardson J., Chavkin N.W., Wang Y., Sano M., Mattisson J., Hata A., Danielsson M., Miura-Yura E., Zaghlool A., Evans M.A., Fall T., De Hoyos H.N., Sundstrom J., Yura Y., Kour A., Arai Y., Thel M.C., Arai Y., Mychaleckyj J.C., Hirschi K.K., Forsberg L.A., Walsh K. Hematopoietic loss of Y chromosome leads to cardiac fibrosis and heart failure mortality. Science. 2022;377(6603):292–297. doi: 10.1126/science.abn3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sondka Z., Bamford S., Cole C.G., Ward S.A., Dunham I., Forbes S.A. The COSMIC Cancer Gene Census: describing genetic dysfunction across all human cancers. Nat Rev Cancer. 2018;18(11):696–705. doi: 10.1038/s41568-018-0060-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spatz A., Borg C., Feunteun J. X-chromosome genetics and human cancer. Nat Rev Cancer. 2004;4(8):617–629. doi: 10.1038/nrc1413. [DOI] [PubMed] [Google Scholar]
- 46.Stoyanov G.S., Kitanova M., Dzhenkov D.L., Ghenev P., Sapundzhiev N. Demographics of head and neck cancer patients: a single institution experience. Cureus. 2017;9(7) doi: 10.7759/cureus.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thompson D.J., Genovese G., Halvardson J., Ulirsch J.C., Wright D.J., Terao C., Davidsson O.B., Day F.R., Sulem P., Jiang Y., Danielsson M., Davies H., Dennis J., Dunlop M.G., Easton D.F., Fisher V.A., Zink F., Houlston R.S., Ingelsson M., Kar S., Kerrison N.D., Kinnersley B., Kristjansson R.P., Law P.J., Li R., Loveday C., Mattisson J., McCarroll S.A., Murakami Y., Murray A., Olszewski P., Rychlicka-Buniowska E., Scott R.A., Thorsteinsdottir U., Tomlinson I., Moghadam B.T., Turnbull C., Wareham N.J., Gudbjartsson D.F., Kamatani Y., Hoffmann E.R., Jackson S.P., Stefansson K., Auton A., Ong K.K., Machiela M.J., Loh P.R., Dumanski J.P., Chanock S.J., Forsberg L.A., Perry J.R.B., (INTEGRAL-ILCCO) I.L.C.C., Consortium B.C.A., BRCA1/2 C. o I. o M. o, Consortium E.C.A., Consortium O.C.A., P. C. A. G. t. I. C. A. A. i. t. G. P. Consortium, K. C. G. M.-A. Project, e. Consortium, B.-b. I. O. S. B. Consortium and a. R. Team Genetic predisposition to mosaic Y chromosome loss in blood. Nature. 2019;575(7784):652–657. doi: 10.1038/s41586-019-1765-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vermeulen M.C., Pearse R., Young-Pearse T., Mostafavi S. Mosaic loss of Chromosome Y in aged human microglia. Genome Res. 2022;32(10):1795–1807. doi: 10.1101/gr.276409.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vijayakumar S., Garcia D., Hensel C.H., Banerjee M., Bracht T., Xiang R., Kagan J., Naylor S.L. The human Y chromosome suppresses the tumorigenicity of PC-3, a human prostate cancer cell line, in athymic nude mice. Genes Chromosomes Cancer. 2005;44(4):365–372. doi: 10.1002/gcc.20250. [DOI] [PubMed] [Google Scholar]
- 50.Wolf J., Seto T., Han J.Y., Reguart N., Garon E.B., Groen H.J.M., Tan D.S.W., Hida T., de Jonge M., Orlov S.V., Smit E.F., Souquet P.J., Vansteenkiste J., Hochmair M., Felip E., Nishio M., Thomas M., Ohashi K., Toyozawa R., Overbeck T.R., de Marinis F., Kim T.M., Laack E., Robeva A., Le Mouhaer S., Waldron-Lynch M., Sankaran B., Balbin O.A., Cui X., Giovannini M., Akimov M., Heist R.S., Investigators G. m- Capmatinib in MET Exon 14-Mutated or MET-amplified non-small-cell lung cancer. N Engl J Med. 2020;383(10):944–957. doi: 10.1056/NEJMoa2002787. [DOI] [PubMed] [Google Scholar]
- 51.Wu Y.L., Tsuboi M., He J., John T., Grohe C., Majem M., Goldman J.W., Laktionov K., Kim S.W., Kato T., Vu H.V., Lu S., Lee K.Y., Akewanlop C., Yu C.J., de Marinis F., Bonanno L., Domine M., Shepherd F.A., Zeng L., Hodge R., Atasoy A., Rukazenkov Y., Herbst R.S., Investigators A. Osimertinib in resected EGFR-mutated non-small-cell lung cancer. N Engl J Med. 2020;383(18):1711–1723. doi: 10.1056/NEJMoa2027071. [DOI] [PubMed] [Google Scholar]
- 52.Yuan S., Yu Z., Liu Q., Zhang M., Xiang Y., Wu N., Wu L., Hu Z., Xu B., Cai T., Ma X., Zhang Y., Liao C., Wang L., Yang P., Bai L., Li Y. GPC5, a novel epigenetically silenced tumor suppressor, inhibits tumor growth by suppressing Wnt/beta-catenin signaling in lung adenocarcinoma. Oncogene. 2016;35(47):6120–6131. doi: 10.1038/onc.2016.149. [DOI] [PubMed] [Google Scholar]
- 53.Zack T.I., Schumacher S.E., Carter S.L., Cherniack A.D., Saksena G., Tabak B., Lawrence M.S., Zhsng C.Z., Wala J., Mermel C.H., Sougnez C., Gabriel S.B., Hernandez B., Shen H., Laird P.W., Getz G., Meyerson M., Beroukhim R. Pan-cancer patterns of somatic copy number alteration. Nat Genet. 2013;45(10):1134–1140. doi: 10.1038/ng.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material Additional figures visualizing each cancer types results from the different analyses.
Supplementary material Additional tables comprising significant genes from the different analyses.