Abstract
Objective
To investigate the effects of metabolic traits, lifestyle factors, and drug interventions on liver fat using the mendelian randomisation paradigm.
Design
Mendelian randomisation study.
Setting
Publicly available summary level data from genome-wide association studies.
Participants
Genome-wide association studies of 32 974 to 1 407 282 individuals who were predominantly of European descent.
Exposures
Genetic variants predicting nine metabolic traits, six lifestyle factors, four lipid lowering drug targets, three antihypertensive drug targets, and genetic association estimates formagnetic resonance imaging measured liver fat.
Main outcome measures
Mendelian randomisation analysis was used to investigate the effects of these exposures on liver fat, incorporating sensitivity analyses that relaxed the requisite modelling assumptions.
Results
Genetically predicted liability to obesity, type 2 diabetes, elevated blood pressure, elevated triglyceride levels, cigarette smoking, and sedentary time watching television were associated with higher levels of liver fat. Genetically predicted lipid lowering drug effects were not associated with liver fat; however, β blocker and calcium channel blocker antihypertensive drug effects were associated with lower levels of liver fat.
Conclusion
These analyses provide evidence of a causal effect of various metabolic traits, lifestyle factors, and drug targets on liver fat. The findings complement existing epidemiological associations, further provide mechanistic insight, and potentially supports a role for drug interventions in reducing the burden of hepatic steatosis and related disease. Further clinical study is now warranted to investigate the relevance of these genetic analyses for patient care.
WHAT IS ALREADY KNOWN ON THIS TOPIC
Fat accumulation in the liver contributes to the development of chronic liver disease and is implicated in adverse cardiometabolic outcomes
Several metabolic, lifestyle, and pharmacological factors have been implicated in the accumulation of liver fat, however, evidence for their causal effects is limited
WHAT THIS STUDY ADDS
Genetic evidence supports causal effects of increased adiposity, type 2 diabetes (including raised fasting insulin levels), systolic blood pressure, smoking, alcohol consumption, and sedentary time watching television on increasing liver fat
Genetic evidence supports protective effects on liver fat of higher low density lipoprotein cholesterol and high density lipoprotein cholesterol levels, but detrimental effects of higher triglyceride levels
No strong genetic evidence to support effects of lipid lowering drug targets on liver fat, but some evidence supports blood pressure lowering through β blocker and calcium channel blocker antihypertensive drugs might reduce liver fat
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE, OR POLICY
The findings complement existing epidemiological associations, further provide mechanistic insight, and support a potential role for drug interventions in reducing the burden of hepatic steatosis and related disease
Further clinical study is now warranted to investigate the relevance of these genetic analyses for patient care
Introduction
The accumulation of fat in the liver contributes to the development of chronic liver disease and is implicated in various adverse cardiometabolic outcomes.1 2 Hepatic steatosis, which is defined by a liver fat content of more than 5.5%, has an estimated prevalence of 25% globally, with rates rapidly increasing as the burden of diabetes and obesity also rises.3 Several metabolic, lifestyle, and pharmacological factors have been associated in the accumulation of liver fat. The most commonly encountered comorbidities are obesity, type 2 diabetes mellitus, hyperlipidaemia, and hypertension.4 In terms of lifestyle factors, alcohol consumption, cigarette smoking, physical inactivity, and caffeine consumption have most closely been linked with development of liver fat.5 Consensus guidance advocates alcohol cessation, weight loss, and specific drug treatments for reducing liver fat and the associated risk of hepatic steatosis.6 However, broader evidence of the causal effects of metabolic traits, lifestyle factors, and drug treatments on liver fat is limited. Furthermore, conventional epidemiological studies investigating these areas are limited in their ability to draw causal inferences because of the potential for spurious associations arising due to environmental confounding or reverse causation.
Use of mendelian randomisation leverages randomly allocated genetic variants as instrumental variables for investigating the effect of modifying an exposure on the risk of an outcome.7 The random distribution of genetic variants means that their associations with disease risk are not subject to confounding from environmental factors. Furthermore, the allocation of genetic variants at conception means that their associations with disease outcomes are unlikely to be attributable to reverse causation. In this study, we use two sample summary data mendelian randomisation to investigate the effects of metabolic traits, lifestyle factors, and lipid lowering and antihypertensive drug interventions on liver fat. The independent roles were explored by use of multivariable mendelian randomisation analysis for traits with genetic correlations. By identifying modifiable causal risk factors for hepatic steatosis, we offer insight into potential therapeutic strategies for lowering the burden of liver disease and related adverse cardiometabolic outcomes.
Methods
Study design
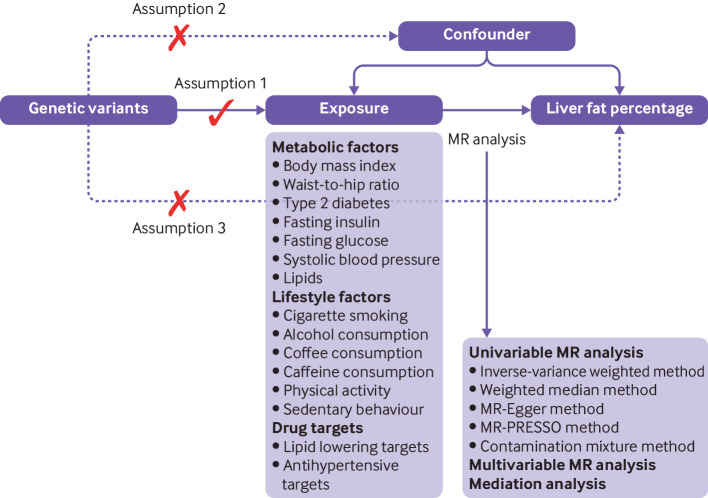
This study is a two sample, mendelian randomisation study based on publicly available summary level data for nine metabolic traits, six lifestyle factors, four lipid lowering drug targets, three antihypertensive drug targets, and liver fat measured by magnetic resonance imaging. Figure 1 shows the study design and three key assumptions of mendelian randomisation analysis: the genetic variants used as instrumental variables should be strongly associated with the exposure; the genetic variants for the exposure should not be associated with any confounders in the association between the exposure and outcome (independence assumption); and the genetic variants affect the outcome merely through their effects on the exposure, but not via other alternative pathways (exclusion restriction assumption). All studies that we used had been approved by corresponding ethical review committees.
Figure 1.
Directed acyclic graph and key assumptions of mendelian randomisation (MR) design. For details of assumptions, please refer to the study design part in the manuscript. MR-PRESSO=Mendelian Randomization Pleiotropy Residual Sum and Outlier
Genetic instrument selection
We selected single nucleotide polymorphisms associated at genome-wide significance (P<5×10–8) with nine metabolic traits (body mass index, waist-to-hip ratio, type 2 diabetes, fasting insulin, fasting glucose, systolic blood pressure, high density cholesterol (HDLC) and low density lipoprotein cholesterol (LDLC), and triglycerides) and six lifestyle factors (smoking initiation, alcohol, coffee and caffeine consumption, strenuous sports, and television watching) from corresponding genome-wide association studies (table 1). Linkage disequilibrium among these single nucleotide polymorphisms was estimated based on the 1000 Genomes European reference panel.8 Single nucleotide polymorphisms in linkage disequilibrium (r2 >0.01) were removed and the single nucleotide polymorphisms with the smallest P value for the genetic association were retained. Three additional smoking related traits (age of smoking initiation, cigarettes per day, and lifetime smoking index9) with selected single nucleotide polymorphisms based on the same approach as mentioned previously were included to further explore the smoking association. These smoking related traits were considered as supplementary exposures because of sample overlap in the population used to measure them with the liver fat outcome, and their smaller explained phenotypical variance as compared with the analysis consider smoking initiation, which could potentially introduce bias or reduce statistical power. We followed the same criteria for instrumental variable selection in multivariable mendelian randomisation analysis, first combining single nucleotide polymorphisms associated with either exposure at P<5×10–8 and then removing single nucleotide polymorphisms in high linkage disequilibrium.
Table 1.
Data sources on metabolic and lifestyle factors and drug targets
| Exposure | Instrumental variables | Unit | Participants included in analysis | Sample overlap (%) | Adjustments |
| Metabolic factor | |||||
| Body mass index33 | 312 | SD | 806 834 individuals of European descent | 4.1 | Age, sex, and genetic 1-5 principal components |
| Waist-to-hip ratio33 | 581 | SD | 697 734 individuals of European descent | 4.7 | Age, sex, and genetic 1-5 principal components |
| Type 2 diabetes34 | 497 | One unit in log-transformed odds | 228 499 type 2 diabetes cases and 1 178 783 non-cases of multi-ancestries | 0.0 | Age, sex, and the first 10 genetic principal components |
| Fasting insulin35 | 38 | log pmol/L | Up to 196 991 individuals of European descent | 0.0 | BMI, study specific covariates, and principal components |
| Fasting glucose35 | 71 | mmol/L | Up to 196 991 individuals of European descent | 0.0 | BMI index, study specific covariates, and principal components |
| Systolic blood pressure36 | 228 | 10 mm Hg | Up to 1 006 863 individuals of European descent | 3.3 | Age, sex, BMI, genotyping chips |
| HDL cholesterol37 | 473 | SD | 403 943 individuals of European descent | 8.2 | Age, sex, and genotyping chips |
| LDL cholesterol37 | 199 | SD | 440 546 individuals of European descent | 7.5 | Age, sex, and genotyping chips |
| Triglycerides37 | 392 | SD | 441 016 individuals of European descent | 7.5 | Age, sex, and genotyping chips |
| Lifestyle factor | |||||
| Smoking initiation38 | 314 | SD in log-transformer odds | 1 232 091 individuals of European descent | 2.7 | Age, sex, and the first 10 genetic principal components |
| Age of smoking initiation38 | 7 | SD | Up to 262 990 individuals of European descent | 12.5 | Age, sex, and the first 10 genetic principal components |
| Cigarettes per day38 | 19 | SD | Up to 263 954 individuals of European descent | 12.5 | Age, sex, and the first 10 genetic principal components |
| Lifetime smoking index9 | 126 | SD change of lifetime smoking index | 462 690 individuals of European descent | 7.1 | Genotyping chip and sex |
| Alcohol drinking38 | 84 | SD increase of log-transformed alcoholic drinks/week | 941 280 individuals of European descent | 3.5 | Age, sex, and the first 10 genetic principal components |
| Coffee consumption39 | 12 | 50% change | 375 833 individuals of European descent | 8.8 | Age, sex, BMI, total energy, proportion of typical food intake, and 20 genetic principal components |
| Caffeine consumption40 | 24 | SD | 362 316 individuals of European descent | 9.1 | Age, sex, genotyping array, and the first 30 genetic principal components |
| Strenuous sports41 | 6 | ≥ 2–3 v 0 days/week | 350 492 individuals of European descent | 9.4 | Age, sex, genotyping chip, first 10 genomic principal components, centre, and season |
| Television watching42 | 112 | Hours/day | 422 218 individuals of European descent | 7.8 | Age squared, age, sex, age-sex interaction, the first 30 principal components |
| Drug target | |||||
| Lipid lowering target10 | One for HMGCR, two for LDLR, one for NPC1L1, two for PCSK9 |
10 mg/dL LDL cholesterol | 80 959 to 295 826 individuals of European descent | 0 | Age, sex, study sample, and five principal components |
| Antihypertensive target11 | One for ACEi, six for β blockers, 23 for CCBs |
10 mm Hg systolic blood pressure | 757 601 individuals of European descent | 4.4 | Age squared, age, sex, BMI, genotyping chips |
Sample overlap was calculated by the sample size of the outcome (n=32 974) divided by the sample size of the exposure whose data sources included UK Biobank. ACEi=angiotensin converting enzyme inhibitor; BMI=body mass index; CCBs=calcium channel blockers; HDL=high density lipoprotein; LDL=low density lipoprotein; SD=standard deviation.
Genetic variants proxying the effects of lipid lowering and antihypertensive drug treatments were obtained by use of approaches similar to previous mendelian randomisation studies.10 11 For lipid lowering drugs, single nucleotide polymorphisms associated with LDLC concentrations at the genome-wide significance level (P<5×10–8) and located in gene regions corresponding to four drug targets (HMGCR (3-hydroxy-3-methyl-glutaryl-coenzyme A reductase), LDLR (low density lipoprotein receptor), NPC1L1 (Niemann-Pick C1-like 1), and PCSK9 (proprotein convertase subtilisin/kexin type 9)) were obtained from the Global Lipids Genetics Consortium.10 For antihypertensive drugs, genes encoding drug targets for ACEi (angiotensin converting enzyme inhibitor), β blockers, and calcium channel blockers were identified in DrugBank.11 Single nucleotide polymorphisms in identified gene regions associated with systolic blood pressure at the genome-wide significance level (P<5×10–8) were obtained from the International Consortium of Blood Pressure.11 Single nucleotide polymorphisms with r2 <0.01 were selected as instrumental variables.11 Details for data sources are displayed in table 1 and information about genetic instruments online supplemental table 1.
bmjmed-2022-000277supp001.pdf (526.3KB, pdf)
Outcome data source
Summary level data for hepatic fat measured by abdominal magnetic resonance imaging were obtained from a genome-wide association analysis of 32 974 generally healthy adults of European descent in the UK Biobank study.12 The UK Biobank study is an ongoing cohort study collecting phenotypic and genetic data from more than 500 000 individuals since its initiation in 2006-10.13 The genetic associations with liver fat were scaled to one standard deviation (SD) of liver fat percentage and one SD increase equals about 4.25 unit increase of absolute liver fat percentage points.12 The characteristics of the population are described in online supplemental table 2. Liver fat was quantified by a machine learning algorithm trained on a small subset (n=4511) with previously quantified liver fat values.12 Single nucleotide polymorphismsassociated with liver fat were adjusted for sex, year of birth, age at time of MRI, age at time of MRI squared, genotyping array, MRI device serial number, and the first 10 principal components of genetic variation. These adjustments were likely made in the original study to account for potential population stratification and confounding effects.12
Statistical analysis
We harmonised all variants serving as instrumental variables between the exposure and outcome data by effect allele. Given that a few single nucleotide polymorphisms were unavailable in the outcome data, we did not find proxies to replace missing single nucleotide polymorphisms. We calculate F statistic for metabolic and lifestyle factors in univariable mendelian randomisation analysis.14 Similarly, conditional F statistic was estimated to inform the instrument strength in multivariable mendelian randomisation analysis.
The inverse variance weighted mendelian randomisation method was used as the main analysis. The model under the multiplicative random effects was used for the traits with at least three single nucleotide polymorphisms and that model under the fixed effects was used for the traits with one or two single nucleotide polymorphisms. The weighted median,15 mendelian randomisation-Egger,16 mendelian randomisation pleiotropy residual sum and outlier (MR-PRESSO),17 and contamination mixture,18 are mendelian randomisation sensitivity analysis methods that were used to examine the robustness of the mendelian randomisation associations to pleiotropic variants that might be biasing the underlying assumptions of mendelian randomisation. The weighted median method can provide a robust mendelian randomisation estimate if 50% of the genetic instruments are valid.19 Mendelian randomisation-Egger regression can detect horizontal pleiotropy (violation of the exclusion restriction assumption that single nucleotide polymorphisms affect fat liver not merely through the exposure) by its intercept test and generate corrected mendelian randomisation estimates after adjusting for pleiotropic effects, although with a relatively low level of precision.16 MR-PRESSO can identify variants outlying in their mendelian randomisation estimates and provide a corrected estimate after removal of such outlier variants.17 The contamination mixture method can generate accurate mendelian randomisation estimates when a large number of variants are available, even if a proportion are invalid.18 To additionally minimise the direct influence of variants selected as instruments on the outcome, we performed a sensitivity analysis where we excluded single nucleotide polymorphisms that were associated with liver fat at the loci-wide significance level (P<1×10–5). We also conducted a Steiger directionality test to examine the possible reverse causality.19 Cochran’s Q value was used to assess heterogeneity among single nucleotide polymorphism estimates as an indicator of their pleiotropic effects.20
We performed multivariable mendelian randomisation analysis for the metabolic traits, with adjustment for genetically predicted body mass index. This analysis had two aims: to minimise potential pleiotropic effects from body mass index on the associations between metabolic factors and liver fat, and to examine for potential mediating roles of metabolic factors in the association between body mass index and liver fat. Network mendelian randomisation was used to estimate the proportion of the total effect of body mass index on liver fat that is mediated through other metabolic factors.21 Given correlations across three lipid biomarkers and correlation between smoking and alcohol consumption, we also conducted multivariable mendelian randomisation analyses with mutual adjustments for these sets of traits. The Benjamini-Hochberg false discovery rate correction was used to account for multiple testing.22 All tests were two sided and done using the TwoSampleMR,23 MR-PRESSO,17 and MendelianRandomization,19 packages in the R software (version 4.0.2).
Patients and public involvement
No patients or members of the public were involved in the design or reporting of this study. On publication, the study findings will be disseminated to the public through the authors’ institutional research media offices.
Results
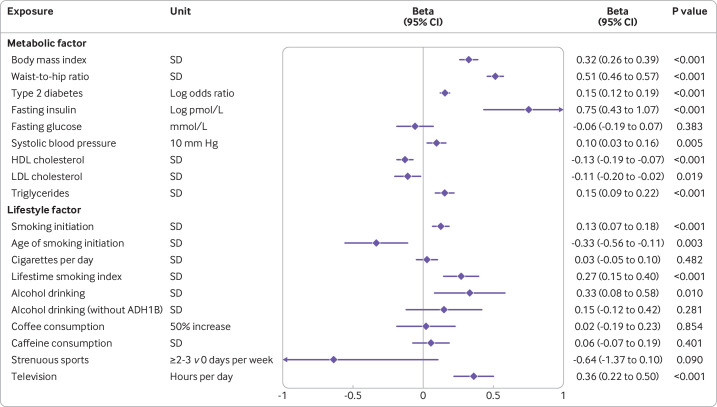
Sample overlap was 0-12.5% between the exposures and the outcome data sources (table 1). All estimated F statistics were more than 10, which indicated limited bias caused by sample overlap (online supplemental table 3). For eight of the nine metabolic factors and three of the six lifestyle factors considered, significant associations with hepatic fat were noted (figure 2). All associations persisted after the false discovery rate correction for multiple testing (online supplemental table 4). The change of liver fat was 0.32 SD units (95% confidence interval 0.26 to 0.39) per one SD increase in genetically predicted body mass index, 0.51 (0.45 to 0.57) per one SD increase in waist-to-hip ratio; 0.15 (0.12 to 0.19) per one unit increase in log-transformed odds ratio of type 2 diabetes; 0.75 (0.43 to 1.07) per one log-transformed pmol/L increase in fasting insulin levels; 0.10 (0.03 to 0.16) per 10 mm Hg increase in systolic blood pressure; −0.13 (−0.19 to –0.07), −0.11 (−0.20 to –0.02), and 0.15 (0.09 to 0.22) per one SD increase in levels of HDLC, LDLC, and triglycerides, respectively; 0.13 (0.07 to 0.18) per one SD increase in log-transformed odds ratio of smoking initiation; 0.33 (0.08 to 0.58) per one SD increase in log-transformed alcoholic drinks per week; and 0.36 (0.22 to 0.50) per one hour increase in television watching time. No strong associations with fat liver were recorded with genetically predicted fasting glucose concentrations, coffee consumption, caffeine (either from coffee or tea) consumption, or strenuous sports (figure 2). We observed an inverse association of genetically predicted age of smoking initiation and a positive association of genetically predicted lifetime smoking index with liver fat (figure 1). However, genetically predicted cigarettes smoked per day showed no strong association with liver fat (figure 1). The association for genetically predicted alcohol consumption did not remain significant after excluding single nucleotide polymorphisms in the ADH1B gene region (figure 2).
Figure 2.
Associations of genetically proxied metabolic and lifestyle factors with hepatic fat. The x axis unit is standard deviation (SD) change in liver fat percentage. One SD increase is approximately a 4.25 unit increase of absolute liver fat percentage points. CI=confidence interval; HDL=high density lipoprotein; LDL=low density lipoprotein
The associations for genetically predicted metabolic and lifestyle factors were overall consistent in sensitivity analyses, although with wider 95% confidence interval with the weighted median and mendelian randomisation-Egger regression analyses (online supplemental table 5). Moderate to high heterogeneity was observed in the analyses of waist-to-hip ratio, type 2 diabetes, the three lipid biomarkers, alcohol and coffee consumption, strenuous sports, and television watching (online supplemental table 5). We observed evidence of bias in the mendelian randomisation-Egger intercept test for type 2 diabetes, lipids, and alcohol consumption (P for intercept test<0.05). One to 15 outliers were identified in MR-PRESSO analyses; however, all observed associations remained consistent after removal of outliers (online supplemental table 5). The associations were also consistent in the sensitivity analysis after removing single nucleotide polymorphisms that were strongly associated with liver fat (online supplemental table 6). However, Steiger directionality tests indicated possible reverse causality for the associations of alcohol consumption and strenuous sports, respectively, with liver fat (online supplemental table 3).
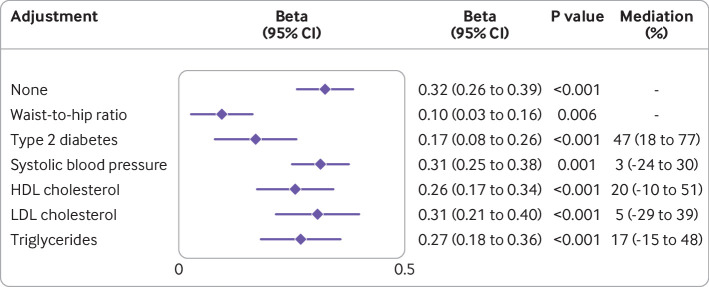
Conditional F statistics for traits included in multivariable mendelian randomisation analysis were more than 10 except for smoking initiation in the multivariable mendelian randomisation analysis with mutual adjustment for smoking initiation and alcohol consumption. The association attenuated for genetically predicted body mass index in a multivariable analysis that adjusted for genetically predicted levels of other metabolic factors (figure 3). Among five possible mediators, liability to type 2 diabetes appeared to mediate almost half (47% (95% confidence interval 18% to 77%)) of the effect of body mass index on liver fat. Adjustment for genetically predicted levels of other metabolic factors resulted in relatively little attenuation of the association for genetically predicted body mass index (table 2). Similarly, the pattern of the mendelian randomisation associations for lipid biomarkers only changed slightly in the multivariable model with mutual adjustment (table 2). The associations for genetically predicted smoking initiation and alcohol consumption also attenuated only slightly after mutual adjustment (table 2).
Figure 3.
Associations of genetically predicted body mass index with hepatic fat, with and without adjustment for other metabolic factors. CI=confidence interval; HDL=high-density lipoprotein; LDL=low-density lipoprotein
Table 2.
Multivariable associations of genetically proxied metabolic and lifestyle factors with hepatic fat
| Exposure | Adjustment | Without adjustment | With adjustment | ||
| Beta (95% CI) | P value | Beta (95% CI) | P value | ||
| Waist-to-hip ratio | Body mass index | 0.51 (0.46 to 0.57) | <0.001 | 0.49 (0.42 to 0.55) | <0.001 |
| Type 2 diabetes | Body mass index | 0.15 (0.12 to 0.19) | <0.001 | 0.14 (0.11 to 0.17) | <0.001 |
| Systolic blood pressure | Body mass index | 0.10 (0.03 to 0.16) | 0.005 | 0.10 (0.04 to 0.15) | 0.001 |
| HDL cholesterol | Body mass index | −0.13 (−0.19 to −0.07) | <0.001 | −0.11 (−0.16 to −0.06) | <0.001 |
| LDL cholesterol | Body mass index | −0.11 (−0.2 to −0.02) | 0.019 | −0.11 (−0.18 to −0.04) | 0.002 |
| Triglycerides | Body mass index | 0.15 (0.09 to 0.22) | <0.001 | 0.14 (0.08 to 0.21) | <0.001 |
| HDL cholesterol | LDL cholesterol and triglycerides | −0.13 (−0.19 to −0.07) | <0.001 | −0.11 (−0.18 to −0.04) | 0.004 |
| LDL cholesterol | HDL cholesterol and triglycerides | −0.11 (−0.20 to −0.02) | 0.019 | −0.13 (−0.20 to −0.06) | 0.001 |
| Triglycerides | LDL and HDL cholesterol | 0.15 (0.09 to 0.22) | <0.001 | 0.16 (0.07 to 0.24) | 0.001 |
| Smoking initiation | Alcohol drinking | 0.13 (0.07 to 0.18) | <0.001 | 0.09 (0.02 to 0.15) | 0.009 |
| Alcohol drinking | Smoking initiation | 0.33 (0.08 to 0.58) | 0.010 | 0.18 (0.01 to 0.35) | 0.036 |
| Alcohol drinking (without ADH1B) | Smoking initiation | 0.15 (−0.12 to 0.42) | 0.281 | −0.04 (−0.24 to 0.15) | 0.659 |
The unit for liver fat was standard deviation of liver fat percentage. HDL=high density lipoprotein; LDL=low density lipoprotein.
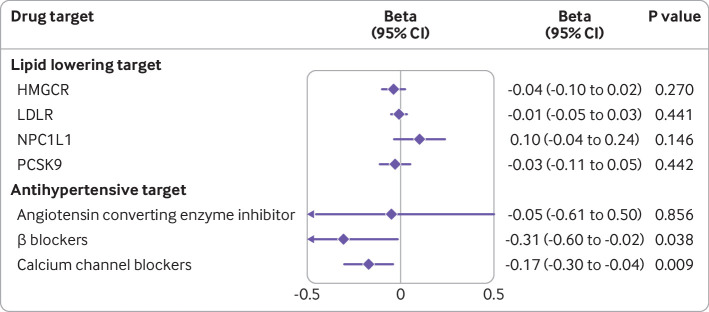
Genetically proxied β blocker and calcium channel blocker effects, but not lipid lowering or ACEi antihypertensive drug effects, were associated with lower levels of liver fat (figure 4). In genetically predicted systolic blood pressure via β blocker, a change in liver fat of −0.31 (95% confidence interval −0.60 to –0.02) per 10 mm Hg decrease was observed and via calcium channel blocker effect, −0.17 (−0.30 to –0.04) was observed. The associations were consistent in sensitivity analyses and unlikely to be biased by horizontal pleiotropy (online supplemental table 5). However, the association for genetically proxied β blockers did not pass the false discovery rate correction (online supplemental table 4).
Figure 4.
Associations of genetically proxied lipid lowering and antihypertensive therapies with hepatic fat. The associations for genetically proxied lipid lowering target was scaled to a decrease of 10 mg/dL low density lipoprotein cholesterol concentrations. The associations for genetically proxied antihypertensive target was scaled to a decrease of 10 mm Hg systolic blood pressure. The x axis unit is standard deviation (SD) change in liver fat percentage. One SD increase is approximately a 4.25 unit increase of absolute liver fat percentage points. CI=confidence interval; HMGCR=3-hydroxy-3-methyl-glutaryl-coenzyme A reductase; LDLR=low density lipoprotein receptor; NPC1L1=Niemann-Pick C1-like 1; PCSK9=proprotein convertase subtilisin/kexin type 9
Discussion
Principal findings
This mendelian randomisation analysis identified evidence supporting causal effects on increasing liver fat of increased adiposity, type 2 diabetes (including raised fasting insulin levels), systolic blood pressure, smoking, alcohol consumption, and sedentary time watching television. Additionally, genetic evidence supports the protective effects on liver fat of higher LDLC and HDLC concentrations, but detrimental effects of higher triglyceride levels. No strong evidence supported the effects of lipid lowering drug targets on liver fat but some evidence showed that blood pressure lowering through β blocker and calcium channel blocker antihypertensive drugs might reduce liver fat. Our findings are consistent with existing understanding on the determinants of fat accumulation in the liver; namely, increased uptake of free fatty acids into the liver, impaired metabolism within the liver, and increased de novo lipogenesis.24 In this way, the findings that genetically predicted higher LDLC and HDLC concentrations are associated with lower liver fat levels might be explained by their role in lipid cycling. Regarding our novel genetic evidence for a potential protective effect of β blocker and calcium channel blocker antihypertensive drugs on liver fat, the point estimates are consistent with the mechanism being through blood pressure reduction. Although the relation between blood pressure and liver fat is complicated and can be mediated through insulin resistance,25 the mendelian randomisation approach leveraged here supports a causal effect, which warrants further evaluation in clinical studies.
Strengths
These findings make important advances in our understanding of hepatic steatosis and related diseases.6 Firstly, the mendelian randomisation paradigm strengthens the evidence for causal effects of these risk factors, rather than only an association, which could also be attributable to environmental confounding factors and reverse causation. This factor is particularly important for identifying therapeutic targets that reduce disease risk and burden. Secondly, our findings offer novel mechanistic insight. For example, the evidence for a causal effect of insulin resistance on liver fat supports that this mediating mechanism is likely underlying the effect of type 2 diabetes. Previous work has used mendelian randomisation to find evidence supporting an effect of higher liver fat on increasing type 2 diabetes mellitus risk, supporting potential bi-directional effects in this relation.26 Similarly, the incorporation of multivariable mendelian randomisation provides evidence that effects of smoking on liver fat occur independently of alcohol consumption, and further evidence that body mass index is having direct effects even after accounting for its effects on other cardiometabolic risk factors, such as diabetes and blood pressure. The mendelian randomisation mediation analysis supported that approximately half of the effect of body mass index on liver fat was occurring through increased liability to type 2 diabetes mellitus, while around a fifth of the effect of body mass index can be mediated through dyslipidaemia. Thirdly, this work is consistent with safety of lipid lowering drugs, given that we did not identify any strong evidence for an effect on increasing liver fat. For the β blocker and calcium channel blocker antihypertensive drugs, evidence suggested that these agents might potentially be reducing liver fat, likely through their effects on lowering blood pressure.
Comparison with other studies
Our current work is largely consistent with previous epidemiological and genetic analyses. Existing epidemiological investigation has identified strong associations of fatty liver disease with obesity, type 2 diabetes, hyperlipidemia, hypertension, and metabolic syndrome.27 Considering lifestyle factors, observational associations have also previously been established for alcohol consumption, smoking, and physical activity, consistent with the pattern of our current findings. 5 Although evidence for a role of lipid lowering drugs in non-alcoholic fatty liver disease has been mixed, we did not identify any strong support in clinical studies,28 which is consistent with the findings of our genetic analyses. This finding might be explained by these drugs primarily targeting homeostasis of lipid levels in the blood and periphery, rather than liver fat. A previous mendelian randomisation analysis considering the outcome of non-alcoholic fatty liver disease similarly found associations for obesity traits, type 2 diabetes liability, blood pressure, and smoking similar to those associations observed in our current work considering liver fat as the outcome,29 thus supporting its conclusions. Our estimates for type 2 diabetes mediating approximately half of the effect of body mass index on liver fat are also similar to those obtained when using the mendelian randomisation paradigm to explore the mediating effects of type 2 diabetes for body mass index on cardiovascular disease outcomes.30 Our current mendelian randomisation investigation into the effects of drugs is novel and supports further investigation into the notion of β blocker and calcium channel blocker antihypertensive drug treatment for reducing liver fat, thus highlighting the possibility of interventions beyond lifestyle modification, which might also be difficult to implement and maintain in practice.31
Limitations
The main advantage of the mendelian randomisation approach taken in this work is that the analyses is time and cost efficient by use of pre-existing, large scale genetic association data. The random allocation of genetic variants at conception means that the approach is less susceptible to the bias from environmental confounding and reverse causation that can hinder causal inference in a traditional epidemiological study design. However, this approach also has weaknesses. Firstly, the analyses were largely restricted to individuals of European genetic ancestry, thus limiting generalisability to other ancestry groups. Secondly, the mendelian randomisation paradigm explores the effects of small, lifelong changes in the genetically predicted levels of a risk factor on an outcome, in this case liver fat. This differs from estimating the effect of a clinical intervention, which might have a larger magnitude but over a shorter period of life. Similarly, our approach could not inform on the dose-response association between these risk factors and levels of liver fat. Thirdly, the mendelian randomisation approach is susceptible to bias from variants that maybe having pleiotropic effects on the outcome through pathways unrelated to the exposure being considered. Although we incorporated a range of sensitivity analyses to prevent confounding affecting our conclusions, this possibility cannot be entirely excluded. Fourthly, we considered the outcome of liver fat in this work, which in itself, might not directly cause disease. Additionally, we quantified liver fat was by use of a machine learning algorithm,12 and the potential for misspecification with this approach should be acknowledged. Further work is required to investigate the relation between high levels of liver fat and consequent liver and cardiometabolic disease risk. Fifthly, even though around 10% of sample overlapped for certain exposures with the outcome, which might inflate type one error rate, F statistics for these associations were more than 10, which indicated that the bias caused by this mild sample overlap should have been limited. However, conditional F statistic for smoking initiation was less than 10, which might introduce weak instrument bias in multivariable analysis of smoking initiation and alcohol consumption in relation to liver fat. Sixthly, use of genetic instruments from the genome-wide association studies with additional adjustment except for age, sex, and population structure factors (eg, body mass index) might introduce collider bias in mendelian randomisation analysis.32 Thus, the associations for fasting insulin and glucose, coffee consumption, strenuous sports, and antihypertensive drugs need to be verified. Seventhly, one in four individuals were defined as excessive alcohol consumption according to the UK criteria in the outcome study,12 which might drive the observed associations specific to alcohol-related liver fat accumulation. However, the genetic associations for liver fat appeared to be largely unrelated of alcohol consumption with consistent findings in two supplementary liver fat genome-wide association analyses where individuals who reported having stopped drinking alcohol or who reported excessive alcohol intake were removed and where the self-reported number of alcoholic drinks consumed per week were adjusted for.12 Finally, a priori sample size calculations were not performed in this study. Instead, the statistical power of the various analyses can be interpreted through the confidence intervals of the point estimates.
Conclusion
In conclusion, we provided a wide angled investigation into the effects of metabolic traits, lifestyle factors, and pharmacological interventions on liver fat. The findings largely support the existing evidence on the role of cardiometabolic traits on hepatic steatosis, and further identify potential mediating mechanisms and pharmacological strategies for reducing the burden of related disease. Further work is now warranted to explore these findings in a clinical setting.
Acknowledgments
Genetic association estimates for liver fat were obtained from a genome-wide association analysis.12 Authors thank all researchers for sharing data.
Footnotes
Twitter: @Yuan_AS, @vujkovicm, @LarssonSC, @dpsg108
Contributors: DG and SY designed the study. SY performed statistical analysis. SY and DG drafted the manuscript. All authors edited the manuscript for intellectual content and approved the final version. DG is guarantor for this work. The corresponding author attests that all listed authors meet authorship criteria and that no others meeting the criteria have been omitted. Transparency: DG affirms that the manuscript is an honest, accurate, and transparent account of the study being reported; that no important aspects of the study have been omitted; and that any discrepancies from the study as planned (and, if relevant, registered) have been explained.
Funding: SCL is funded by the Swedish Heart-Lung Foundation (Hjärt-Lungfonden, 20210351), the Swedish Research Council for Health, Working Life and Welfare (Forte; grant No 2018‐00123), the Swedish Research Council (Vetenskapsrådet; grant no. 2019-00977), and the Swedish Cancer Society (Cancerfonden, funding number not applicable). DG is funded by the British Heart Foundation Centre of Research Excellence (RE/18/4/34215) at Imperial College London. This research was funded by United Kingdom Research and Innovation Medical Research Council (MC_UU_00002/7). The funders had no role in considering the study design or in the collection, analysis, interpretation of data, writing of the report, or decision to submit the article for publication.
Competing interests: All authors have completed the ICMJE uniform disclosure form at www.icmje.org/disclosure-of-interest/ and declare: support from the Swedish Heart-Lung Foundation, the Swedish Research Council for Health, Working Life and Welfare, the Swedish Research Council, the Swedish Cancer Society, the British Heart Foundation Centre of Research Excellence at Imperial College London, and the United Kingdom Research and Innovation Medical Research Council. DG is employed part-time by Novo Nordisk and has received consultancy fees from Policy Wisdom. The remaining authors report no support from any organisation for the submitted work; no financial relationships with any organisations that might have an interest in the submitted work in the previous three years; no other relationships or activities that could appear to have influenced the submitted work.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available in a public, open access repository. Data used are publicly available and the original sources are detailed in Table 1.
Ethics approval
Not applicable.
References
- 1. Loomba R, Friedman SL, Shulman GI. Mechanisms and disease consequences of nonalcoholic fatty liver disease. Cell 2021;184:2537–64. 10.1016/j.cell.2021.04.015 [DOI] [PubMed] [Google Scholar]
- 2. Speliotes EK, Massaro JM, Hoffmann U, et al. Fatty liver is associated with dyslipidemia and dysglycemia independent of visceral fat: the Framingham heart study. Hepatology 2010;51:1979–87. 10.1002/hep.23593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Loomba R, Sanyal AJ. The global NAFLD epidemic. Nat Rev Gastroenterol Hepatol 2013;10:686–90. 10.1038/nrgastro.2013.171 [DOI] [PubMed] [Google Scholar]
- 4. Younossi Z, Anstee QM, Marietti M, et al. Global burden of NAFLD and NASH: trends, predictions, risk factors and prevention. Nat Rev Gastroenterol Hepatol 2018;15:11–20. 10.1038/nrgastro.2017.109 [DOI] [PubMed] [Google Scholar]
- 5. Nivukoski U, Niemelä M, Bloigu A, et al. Combined effects of lifestyle risk factors on fatty liver index. BMC Gastroenterol 2020;20:109. 10.1186/s12876-020-01270-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chalasani N, Younossi Z, Lavine JE, et al. The diagnosis and management of nonalcoholic fatty liver disease: practice guidance from the American association for the study of liver diseases. Hepatology 2018;67:328–57. 10.1002/hep.29367 [DOI] [PubMed] [Google Scholar]
- 7. Smith GD, Ebrahim S. 'Mendelian randomization': can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol 2003;32:1–22. 10.1093/ije/dyg070 [DOI] [PubMed] [Google Scholar]
- 8. Clarke L, Zheng-Bradley X, Smith R, et al. The 1000 genomes project: data management and community access. Nat Methods 2012;9:459–62. 10.1038/nmeth.1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wootton RE, Richmond RC, Stuijfzand BG, et al. Evidence for causal effects of lifetime smoking on risk for depression and schizophrenia: a Mendelian randomisation study. Psychol Med 2020;50:1–9. 10.1017/S0033291719002678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ference BA, Ray KK, Catapano AL, et al. Mendelian Randomization Study of ACLY and Cardiovascular Disease. N Engl J Med 2019;380:1033–42. 10.1056/NEJMoa1806747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gill D, Georgakis MK, Koskeridis F, et al. Use of genetic variants related to antihypertensive drugs to inform on efficacy and side effects. Circulation 2019;140:270–9. 10.1161/CIRCULATIONAHA.118.038814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Haas ME, Pirruccello JP, Friedman SN, et al. Machine learning enables new insights into genetic contributions to liver fat accumulation. Cell Genom 2021;1:100066. 10.1016/j.xgen.2021.100066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sudlow C, Gallacher J, Allen N, et al. UK Biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med 2015;12:e1001779. 10.1371/journal.pmed.1001779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Burgess S, Davies NM, Thompson SG. Bias due to participant overlap in two-sample Mendelian randomization. Genet Epidemiol 2016;40:597–608. 10.1002/gepi.21998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bowden J, Davey Smith G, Haycock PC, et al. Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol 2016;40:304–14. 10.1002/gepi.21965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Burgess S, Thompson SG. Interpreting findings from Mendelian randomization using the MR-Egger method. Eur J Epidemiol 2017;32:377–89. 10.1007/s10654-017-0255-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Verbanck M, Chen C-Y, Neale B, et al. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet 2018;50:693–8. 10.1038/s41588-018-0099-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Burgess S, Foley CN, Allara E, et al. A robust and efficient method for Mendelian randomization with hundreds of genetic variants. Nat Commun 2020;11:376. 10.1038/s41467-019-14156-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yavorska OO, Burgess S. MendelianRandomization: an R package for performing Mendelian randomization analyses using summarized data. Int J Epidemiol 2017;46:1734–9. 10.1093/ije/dyx034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gill D. Heterogeneity between genetic variants as a proxy for pleiotropy in Mendelian randomization. JAMA Cardiol 2020;5:107–8. 10.1001/jamacardio.2019.4281 [DOI] [PubMed] [Google Scholar]
- 21. Gill D, Cameron AC, Burgess S, et al. Urate, blood pressure, and cardiovascular disease: evidence from Mendelian randomization and meta-analysis of clinical trials. Hypertension 2021;77:383–92. 10.1161/HYPERTENSIONAHA.120.16547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society: Series B 1995;57:289–300. 10.1111/j.2517-6161.1995.tb02031.x [DOI] [Google Scholar]
- 23. Hemani G, Zheng J, Elsworth B, et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife 2018;7:e34408. 10.7554/eLife.34408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Koo S-H. Nonalcoholic fatty liver disease: molecular mechanisms for the hepatic steatosis. Clin Mol Hepatol 2013;19:210–5. 10.3350/cmh.2013.19.3.210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Brookes MJ, Cooper BT. Hypertension and fatty liver: guilty by association? J Hum Hypertens 2007;21:264–70. 10.1038/sj.jhh.1002148 [DOI] [PubMed] [Google Scholar]
- 26. Martin S, Sorokin EP, Thomas EL, et al. Estimating the effect of liver and pancreas volume and fat content on risk of diabetes: a Mendelian randomization study. Diabetes Care 2022;45:460–8. 10.2337/dc21-1262 [DOI] [PubMed] [Google Scholar]
- 27. Younossi ZM, Koenig AB, Abdelatif D, et al. Global epidemiology of nonalcoholic fatty liver disease-Meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology 2016;64:73–84. 10.1002/hep.28431 [DOI] [PubMed] [Google Scholar]
- 28. Tzanaki I, Agouridis AP, Kostapanos MS. Is there a role of lipid-lowering therapies in the management of fatty liver disease? World J Hepatol 2022;14:119–39. 10.4254/wjh.v14.i1.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yuan S, Chen J, Li X, et al. Lifestyle and metabolic factors for nonalcoholic fatty liver disease: Mendelian randomization study. Eur J Epidemiol 2022;37:723–33. 10.1007/s10654-022-00868-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gill D, Zuber V, Dawson J, et al. Risk factors mediating the effect of body mass index and waist-to-hip ratio on cardiovascular outcomes: Mendelian randomization analysis. Int J Obes 2021;45:1428–38. 10.1038/s41366-021-00807-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Douketis JD, Macie C, Thabane L, et al. Systematic review of long-term weight loss studies in obese adults: clinical significance and applicability to clinical practice. Int J Obes 2005;29:1153–67. 10.1038/sj.ijo.0802982 [DOI] [PubMed] [Google Scholar]
- 32. Coscia C, Gill D, Benítez R, et al. Avoiding collider bias in Mendelian randomization when performing stratified analyses. Eur J Epidemiol 2022;37:671–82. 10.1007/s10654-022-00879-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pulit SL, Stoneman C, Morris AP, et al. Meta-analysis of genome-wide association studies for body fat distribution in 694 649 individuals of European ancestry. Hum Mol Genet 2019;28:166–74. 10.1093/hmg/ddy327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Vujkovic M, Keaton JM, Lynch JA, et al. Discovery of 318 new risk loci for type 2 diabetes and related vascular outcomes among 1.4 million participants in a multi-ancestry meta-analysis. Nat Genet 2020;52:680–91. 10.1038/s41588-020-0637-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chen J, Spracklen CN, Marenne G, et al. The trans-ancestral genomic architecture of glycemic traits. Nat Genet 2021;53:840–60. 10.1038/s41588-021-00852-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Evangelou E, Warren HR, Mosen-Ansorena D, et al. Genetic analysis of over 1 million people identifies 535 new loci associated with blood pressure traits. Nat Genet 2018;50:1412–25. 10.1038/s41588-018-0205-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Richardson TG, Sanderson E, Palmer TM, et al. Evaluating the relationship between circulating lipoprotein lipids and apolipoproteins with risk of coronary heart disease: a multivariable Mendelian randomisation analysis. PLoS Med 2020;17:e1003062. 10.1371/journal.pmed.1003062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liu M, Jiang Y, Wedow R, et al. Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use. Nat Genet 2019;51:237–44. 10.1038/s41588-018-0307-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhong VW, Kuang A, Danning RD, et al. A genome-wide association study of bitter and sweet beverage consumption. Hum Mol Genet 2019;28:2449–57. 10.1093/hmg/ddz061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Said MA, van de Vegte YJ, Verweij N, et al. Associations of observational and genetically determined caffeine intake with coronary artery disease and diabetes mellitus. J Am Heart Assoc 2020;9:e016808. 10.1161/JAHA.120.016808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Klimentidis YC, Raichlen DA, Bea J, et al. Genome-Wide association study of habitual physical activity in over 377,000 UK Biobank participants identifies multiple variants including CADM2 and APOE. Int J Obes 2018;42:1161–76. 10.1038/s41366-018-0120-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. van de Vegte YJ, Said MA, Rienstra M, et al. Genome-Wide association studies and Mendelian randomization analyses for leisure sedentary behaviours. Nat Commun 2020;11:1770. 10.1038/s41467-020-15553-w [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
bmjmed-2022-000277supp001.pdf (526.3KB, pdf)
Data Availability Statement
Data are available in a public, open access repository. Data used are publicly available and the original sources are detailed in Table 1.