Abstract
Candida albicans undergoes a morphogenetic switch from budding yeast to hyphal growth form in response to a variety of stimuli and growth conditions. Multiple signaling pathways, including a Cph1-mediated mitogen-activated protein kinase pathway and an Efg1-mediated cyclic AMP/protein kinase A pathway, regulate the transition. Here we report the identification of a basic helix-loop-helix transcription factor of the Myc subfamily (Cph2) by its ability to promote pseudohyphal growth in Saccharomyces cerevisiae. Like sterol response element binding protein 1, Cph2 has a Tyr instead of a conserved Arg in the basic DNA binding region. Cph2 regulates hyphal development in C. albicans, as cph2/cph2 mutant strains show medium-specific impairment in hyphal development and in the induction of hypha-specific genes. However, many hypha-specific genes do not have potential Cph2 binding sites in their upstream regions. Interestingly, upstream sequences of all known hypha-specific genes are found to contain potential binding sites for Tec1, a regulator of hyphal development. Northern analysis shows that TEC1 transcription is highest in the medium in which cph2/cph2 displays a defect in hyphal development, and Cph2 is necessary for this transcriptional induction of TEC1. In vitro gel mobility shift experiments show that Cph2 directly binds to the two sterol regulatory element 1-like elements upstream of TEC1. Furthermore, the ectopic expression of TEC1 suppresses the defect of cph2/cph2 in hyphal development. Therefore, the function of Cph2 in hyphal transcription is mediated, in part, through Tec1. We further show that this function of Cph2 is independent of the Cph1- and Efg1-mediated pathways.
Candida albicans is one of the most frequently isolated fungal pathogens of humans. It is capable of causing superficial mucosal infections as well as life-threatening systemic infections in immunocompromised individuals. C. albicans can undergo reversible morphogenetic transitions among budding yeast, pseudohyphal, and hyphal growth forms. The pathogenicity of this fungus is linked to its capacity to switch among different growth forms (27).
A wide range of signals or culture conditions can trigger the yeast-to-hypha transition in C. albicans. These include serum, N-acetylglucosamine, proline, a temperature of 37°C, neutral pH, and microaerophilic conditions (8). Levels of expression of several genes have been shown to be associated with hyphal morphogenesis (hypha-specific genes), rather than with a specific hypha-inducing condition. Hypha-specific genes identified so far include ECE1, HWP1, HYR1, ALS3, RBT1, and RBT4 (2, 4, 7, 18, 43). Some of them, such as HWP1 (42), RBT1, and RBT4 (7), encode important virulence factors.
Molecular cloning and characterization of hyphal regulators or signaling pathways have been based largely on the strong conservation between C. albicans and other genetically tractable fungi. Cph1 is homologous to Saccharomyces cerevisiae Ste12, which encodes a transcription factor required for mating and filamentous growth (26). As in S. cerevisiae, Cph1 is regulated by a mitogen-activated protein (MAP) kinase cascade that includes Cst20, Hst7, and Cek1. Homozygous mutations in these genes of the MAP kinase pathway all display a medium-specific defect in hyphal development (9, 21, 24). Efg1, a basic helix-loop-helix (bHLH) protein similar to Phd1 of S. cerevisiae and StuA of Aspergillus nidulans, plays a major role in regulating hyphal development in C. albicans (44). efg1/efg1 null mutant strains are severely blocked in hyphal development under many conditions, including serum (27, 44). Efg1 may be regulated by the cyclic AMP/protein kinase A signaling pathway (6, 41). The Efg1-mediated pathway is distinct from that of Cph1 because cph1/cph1 efg1/efg1 double mutants are more defective than cph1/cph1 or efg1/efg1 single mutants in hyphal development under most conditions examined and are avirulent (27). Recently, a new member of the TEA/ATTS family of transcription factors, Tec1, has been shown to regulate hyphal development and virulence in C. albicans (40). TEA/ATTS family members AbaA in A. nidulans and Tec1 in S. cerevisiae are involved in the regulation of conidiophore formation and filamentous growth, respectively (1, 15, 34). Considering that C. albicans can respond to a large number of extracellular signals and growth conditions in controlling hyphal development, C. albicans cells are likely to utilize multiple signal transduction pathways to integrate these signals.
Here we report the identification of a novel hyphal regulator of C. albicans identified by using S. cerevisiae. The hyphal regulator is a bHLH protein of the Myc subfamily. It is important for hyphal development and the transcription of hypha-specific genes in a medium-specific manner. The functional relationship of the bHLH protein with Cph1, Efg1, and Tec1 in hyphal regulation is addressed.
MATERIALS AND METHODS
Media and C. albicans manipulation.
S. cerevisiae media were used for routine culturing of C. albicans, except that uridine instead of uracil was used for growing Ura− C. albicans strains. Several hypha-inducing media were used: Lee's medium (25) with either 2% glucose, 1% mannitol, or 2% succinic acid as the carbon source; yeast extract-peptone-dextrose (YPD)–serum; RPMI medium (40); synthetic succinate (SS) medium (0.0425% yeast nitrogen base without amino acids or ammonium sulfate [Difco], 0.125% ammonium sulfate, 2% succinic acid [pH 6.5]); and SSA medium (SS medium plus amino acids at the concentrations used for S. cerevisiae media). The lithium acetate method of Ito et al. (19) was used for C. albicans transformation, except that fewer cells were used (107 cells/ml), 1 M Tris (pH 7.4) (10 μl) was added to 50 μl of cells, more transforming DNA was used (∼2 μg), and heat shock was done for 22 min at 42°C. Photographs of cell and colony morphologies were obtained as described by Loeb et al. (28).
CPH2 cloning and plasmid construction.
Plasmids pHL17, pHL33, and pHL34 were isolated from a C. albicans genomic library based on their ability to promote invasive growth and cell elongation in diploid S. cerevisiae on SC-Ura–1 M sorbitol medium (26). Inserts from all three plasmids gave similar patterns of restriction digestion. pHL17 contained a 4-kb insert. Deletions from both directions narrowed down a region responsible for invasive-pseudohyphal growth. The DNA sequence of the region was determined. Half of the CPH2 coding sequence is not in the current C. albicans sequence database.
BES116-CPH2 (pHL541), a 3-kb KpnI-HindIII CPH2 fragment from pHL17, was subcloned into the HindIII-KpnI site of plasmid BES116, a C. albicans ADE2-integrating vector (13). BES116-PCK1p (BP1), a 1.4-kb NotI-BglII (blunt-ended) PCK1p fragment from plasmid CA01 (44), was subcloned into the NotI-EcoRV site of plasmid BES116. BP1-CPH1, a 2-kb CPH1 PCR fragment with HindIII at both ends, was subcloned into the HindIII site of plasmid BP1. The primers used for PCR were P303 (5′CCCAAGCTTGCCTAATACACTCTTTCGCC) and P304 (5′CCCAAGCTTACAAGTCCATAAACATAATGC). BP1-CPH2, a 1.1-kb KpnI-BspLU11I (blunt-ended) CPH2 fragment from plasmid BES116-CPH2, was subcloned into the KpnI-HindIII (blunt-ended) site of plasmid BP1. BES116-PCK1p-MluI (BP2), an MluI linker with ClaI ends, was subcloned into the ClaI site of plasmid BP1. The oligonucleotide annealed to make the linker was P 342 (5′ P-CGATGACGCGTCAT). BP2-TEC1, a 2.2-kb TEC1 PCR fragment with MluI at both ends, was subcloned into the MluI site of plasmid BP2. pGEX-2T-CPH2 C′, a 300-bp CPH2 PCR fragment with nucleotides corresponding to amino acids (aa) 190 to 290 of Cph2 and with BamHI at both ends, was subcloned into the BamHI site of plasmid pGEX-2T (Pharmacia Biotech). The primers used for PCR were P353 (5′CGGGATCCAACACCACTAAAAAACCGGCC) and P354 (5′CGGGATCCGCCTATGCAACTCAATATTG). All constructs were confirmed by DNA sequence analysis. The plasmids used in this study are listed in Table 1.
TABLE 1.
Plasmids used in this study
| Plasmid | Description | Reference or source |
|---|---|---|
| Library | C. albicans genomic library in an S. cerevisiae 2μm URA3 vector | 26 |
| pHL17 | Clone from the library with a 4-kb insertion carrying CPH2 | This study |
| pHL541 | CPH2 in BES116 | This study |
| BP1 | PCK1p in BES116 | This study |
| BP2 | PCK1p in BES116-MluI | This study |
| BP1-CPH2 | PCK1p-CPH2 in BES116 | This study |
| BP1-CPH1 | PCK1p-CPH1 in BES116 | This study |
| BP2-TEC1 | PCK1p-TEC1 in BES116 | This study |
| CAHUH | PCK1p-EFG1 | 44 |
| pGEX-2T-CPH2 C′ | GST-CPH2 C′ (aa 190–290) | This study |
C. albicans strain construction.
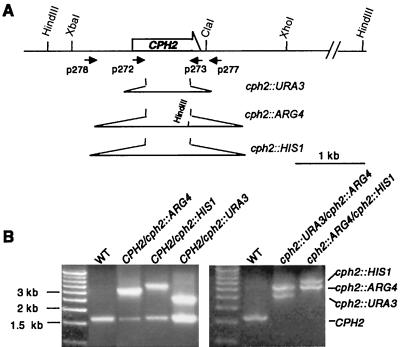
The C. albicans strains used in this study are listed in Table 2. CPH2 was deleted based on the method of Wilson et al. (46). Primers P273 (5′TTGATATATTCTGTAGCTTTGGTTAAAACACTAGCTTTGTTCAATTTAGATGCTGGTGTTTGTGGAATTGTGAGCGGATA) and P272 (5′GCATCTTTATATTCGTTTGATTTT GTTGATGCCGACGATTTCTTAGATTCGATATCTGGCGTTTTCCCAGTCACGACGTT) were used to amplify C. albicans HIS1, URA3, and ARG4 from plasmids pGEM-HIS1, pGEM-URA3, and pRS-ARG4ΔSpeI (46), respectively; the underlined sequences in the primers are the segments that annealed to the plasmids. Each primer also has 60 bp of sequence from the CPH2 coding region. The PCR products were used to transform C. albicans strain BWP17 (46), yielding cph2/CPH2 heterozygous strains HLY1909, HLY1906, and HLY1907 (Table 2). The replacement of one copy of the CPH2 genes by a selectable marker was detected by PCR with primers P277 (5′CCCATAACAGCAGCCATACATCCCAAC) and P278 (5′ATAACCAAGTGAAGGAAGAATACCC), which are located about 500 bp outside of the CPH2 coding region. P277 and P278 were also used to amplify DNA from cph2/CPH2 heterozygous strains to produce cph2 deletion fragments with 500-bp homologies on each end of the selectable markers. XcmI-digested genomic DNA from HLY1907 was used to generate cph2::ARG4. HpaI-digested genomic DNA from HLY1909 and HLY1906 was used for cph2::HIS1 and cph2::URA3, respectively. Both enzymes had a restriction site in CPH2, but not in the selectable markers, thus eliminating the PCR product from CPH2. cph2::URA3 and cph2::ARG4 PCR products were used to transform HLY1907 and HLY1909, respectively, generating cph2/cph2 homozygous deletion strains HLY1921 and HLY1927 (Table 2).
TABLE 2.
C. albicans strains used in this study
| Strain | Genotype | Reference or source |
|---|---|---|
| SC5314 | Wild type | 14 |
| BWP17 | ura3::1 imm434/ura3::1 imm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG | 46 |
| HLY1907 | cph2::ARG4/CPH2 ura3::1 imm434/ura3::1 imm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG | This study |
| HLY1909 | cph2::HIS1/CPH2 ura3::1 imm434/ura3::1 imm434 his1::hisG/hisI::hisG arg4::hisG/arg4::hisG | This study |
| HLY1906 | cph2::URA3/CPH2 ura3::1 imm434/ura3::1 imm434 his1::hisG/hisl::hisG arg4::hisG/arg4::hisG | This study |
| HLY1921 | cph2::ARG4/cph2::URA3 ura3::1 imm434/ura3::1 imm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG | This study |
| HLY1927 | cph2::ARG4/cph2::HIS1 ura3::1 imm434/ura3::1 imm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG | This study |
| HLY1928 | cph2::ARG4/cph2::HIS1 ura3::1 imm434/ura3::1 imm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG ADE2/ade2::URA3 | This study |
| HLY1929 | cph2::ARG4/cph2::HIS1 ura3::1 imm434/ura3::1 imm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG ADE2/ade2::CPH2-URA3 | This study |
| JKC19 | cph1::hisG/cph1::hisG-URA3-hisG ura3::1 imm434/ura3::1 imm434 | 26 |
| HLC52 | efg1::hisG/efg1::hisG-URA3-hisG ura3::1 imm434/ura3::1 imm434 | 44 |
| CAI4 | ura3::1 imm434/ura3::1 imm434 | 14 |
| HLY3119 | ura3::1 imm434/ura3::1 imm434 ADE2/ade2::PCK1p-CPH1-URA3 | This study |
| HLY3120 | ura3::1 imm434/ura3::1 imm434 ADE2/ade2::PCK1p-CPH2-URA3 | This study |
| HLY3125 | ura3::1 imm434/ura3::1 imm434 ADE2/ade2::PCK1p-EFG1-URA3 | This study |
| HLY3214 | ura3::1 imm434/ura3::1 imm434 ADE2/ade2::PCK1p-TEC1-URA3 | This study |
| HLY3183 | efg1::hisG/efg1::hisG ura3::1 imm434/ura3::1 imm434 ADE2/ade2::PCK1p-CPH2-URA3 | This study |
| HLY3139 | cph1::hisG/cph1::hisG ura3::1 imm434/ura3::1 imm434 ADE2/ade2::PCK1p-CPH2-URA3 | This study |
| HLY3220 | cph2::ARG4/cph2::HIS1 ura3::1 imm434/ura3::1 imm434 his1::hisG/his1::hisG arg4::hisG arg4::hisG ADE2/ade2::PCK1p-TEC1-URA3 | This study |
| HLY3156 | cph2::ARG4/cph2::HIS1 ura3::1 imm434/ura3::1 imm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG ADE2/ade2::PCK1p-CPH1-URA3 | This study |
| HLY3164 | cph2::ARG4/cph2::HIS1 ura3::1 imm434/ura3::1 imm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG efg1::URA3-PCK1p-EFG1 | This study |
Expression and purification of GST fusion proteins.
Glutathione S-transferase (GST) protein and a GST-Cph2 C′ (bHLH region) fusion protein were expressed from plasmid pGEX-2T transformed into Escherichia coli strain BL21 as described by Dooley et al. (11). Crude lysates (10 ml) were incubated with 3 ml of glutathione-agarose beads (Sigma) on a rotating wheel at 4°C for 4 h. The mixture was loaded onto a column, the flowthrough was collected, and the beads were washed five times with 4 column volumes (3 ml) of HEGN solution (50 mM HEPES [pH 7.6], 0.1 mM EDTA [pH 8.0], 10% glycerol, 0.1% Nonidet P-40) containing 0.1 M KCl, 1 mM dithiothreitol, 0.25 mM phenylmethylsulfonyl fluoride, 1 μg of leupeptin/ml, and 0.7 μg of pepstatin A/ml. The purified protein was eluted with 10 mM glutathione. The expression and purification of the proteins were verified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis followed by Coomassie blue staining. The most concentrated fractions were pooled and dialyzed (molecular weight cutoff, 12,000 to 14,000) in Z 0.1 solution (25 mM HEPES [pH 7.6], 2 mM MgCl2, 1 mM EDTA, 10% glycerol) at 4°C overnight.
Gel mobility shift experiments.
Gel mobility shift experiments were performed using 1 ng of labeled DNA probe and 0.33, 1, or 3.3 μg of GST-Cph2 C′ fusion protein in a 20-μl reaction mixture (39). For competition experiments, 200 ng of nonlabeled DNA was incubated with 1 μg of GST or GST-Cph2 C′ fusion protein before the addition of 1 ng of labeled DNA probe (45). Double-stranded DNA for probes or competitor were generated by annealing complementary oligonucleotides containing the wild-type −316 to −297 TEC1 upstream sequence as well as sequences with the first sterol regulatory element 1 (SRE-1) site or both SRE-1 sites mutated. All DNAs have 5′ GATC overhangs for polynucleotide kinase end labeling. See Fig. 8C for wild-type and mutant oligonucleotide probe sequences.
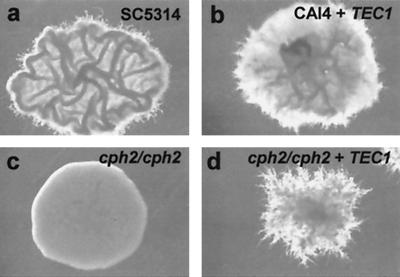
FIG. 8.
Ectopic TEC1 expression suppresses the cph2/cph2 defect in hyphal development. C. albicans wild-type strain (SC5314), wild type strain (CAI4) transformed with PCK1p-TEC1 (HLY3214), cph2/cph2 strain (HLY1928), and cph2/cph2 strain transformed with PCK1p-TEC1 (HLY3220) (a to d, respectively) were grown on SSA medium for 7 days at 37°C.
Northern analysis.
Methods for RNA isolation and Northern blot hybridization were as previously described (28). A ClaI-SalI ACT1 fragment from plasmid p1595/3 (10) was used as a probe for Northern analysis. A 1.5-kb CPH2 PCR product (primers P277 and P278) was used as a probe for Northern hybridization. PCR products of ECE1, HWP1, HYR1, and RBT4 were used for probing Northern blots. The images were scanned with a PhosphorImager (Molecular Dynamics) and quantified using ImageQuant (Molecular Dynamics) and Quantity One (Bio-Rad) software. The gene expression signal intensities were first normalized to those of actin before fold change values were calculated. The sizes of the mRNAs on the Northern blots correlated with the expected lengths based on information from the C. albicans genome database.
Sequence analysis.
The 800-bp upstream regions of hypha-specific genes and of 1,000 randomly chosen control genes were extracted from the Stanford Candida Genome Center (http://www-sequence.stanford.edu/group/candida/search.html). Weight matrices for SRE-1 and E-box motifs were entered into S. cerevisiae promoter database (SCPD) matrix search program (http://cgsigma.cshl.org/cgi-bin/jz/searchmatrix) to identify potential SRE-1-like or E-box motifs in the extracted upstream sequences. Motifs that were at least 80% identical to the matrices and contained the conserved core binding motif were considered potential SRE-1-like or E-box binding sites (see Table 3). The upstream sequences of ECE1, HWP1, HYR1 and ALS3 were also used to search for overrepresented motifs using the GibbsDNA program (23) (available at the SCPD site). Then, the new motifs were used to find potential matches to known transcription factor binding motifs using the SCPD and TFSearch (http://pdap1.trc.rwcp.or.jp/research/db/TFSEARCH.html). A Tec1 site (AbaA) was revealed in our search, and the string CATTCY was used to search for this site in all hypha-specific genes.
TABLE 3.
Computational search of potential Cph2 and Tec1 binding sitesa
| Gene | Cph2 sites
|
Tec1 sites (AbaA) | |
|---|---|---|---|
| SRE-1 | E-box | ||
| ECE1 | GTCACCTCAC (−724, −715) | TATCACCTGAT (−316, −306) | TCATTCT (−371, −365) |
| HWP1 | TATCACCTGTA (−222, −212) | TCATTCT (−153, −147) | |
| HYR1 | TCATTCT (−513, −519), TCATTCC (−270, −264), TCATTCT (−32, −26) | ||
| RBT4 | GTCATACCAC (−618, −609) | GCATTCT (−532, −526) | |
| SAP5 | TCATTCC (−116, −110), TCATTCC (−91, −85), TCATTCC (−66, −60) | ||
| ALS3 | TCATTCT (−808, −802) | ||
| Total no. of potential sites | 2 | 2 | 10 |
| Expected random occurrence | 0.356 × 6 = 2.14 | 0.235 × 6 = 1.41 | 0.728 × 6 = 4.37 |
SRE-1 and E-box weight matrices (see Fig. 6A) and the Tec1 consensus sequence were used to search for potential Cph2 and Tec1 binding sites in the upstream sequences of hypha-specific genes. The sequences and positions of putative binding sites found from the search are shown. The expected random occurrence of a motif in this set of hypha-specific genes is calculated by multiplying the frequency of its occurrence in the control set by the number of hypha-specific genes.
Nucleotide sequence accession number.
The GenBank accession number for the CPH2 nucleotide sequence is AF349507.
RESULTS
Cloning of a novel C. albicans bHLH gene that can promote pseudohyphal growth in S. cerevisiae.
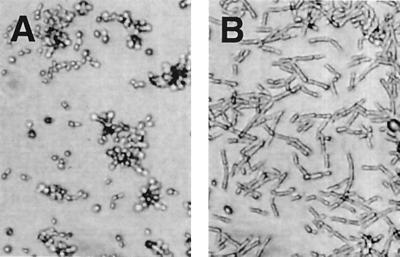
The absence of a sexual cycle and the diploid nature of the C. albicans genome prohibit the direct isolation of nonfilamentous mutants of C. albicans. Therefore, we chose to clone C. albicans genes that enabled S. cerevisiae cells to undergo filamentous growth on medium repressive for invasive and pseudohyphal growth. A C. albicans genomic library constructed in a high-copy-number S. cerevisiae vector (26) was transformed into diploid S. cerevisiae cells, and transformants were grown on SC-Ura medium. From 100,000 transformed colonies, about 200 transformed colonies were invasive and remained on the agar plates after the surface cells were washed off (Fig. 1A). Nine of the invasive colonies displayed elongated cell morphology (Fig. 1B). These nine clones represented two C. albicans genes, CPH1 and CPH2 (Candida pseudohyphal regulator).
FIG. 1.
Functional cloning of CPH2 in S. cerevisiae. Morphologies of S. cerevisiae cells remaining on SC-Ura–sorbitol plates after noninvasive colonies and surface cells of invasive colonies were washed away with water. (A) Round cells from an invasive colony. (B) Elongated cells from one of the positive colonies carrying CPH2.
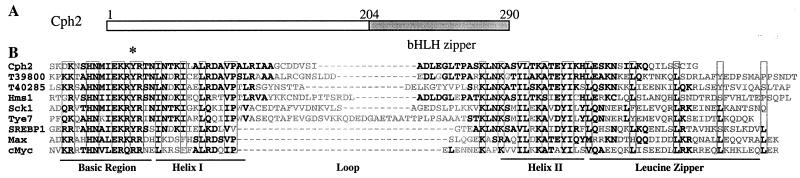
CPH2 encodes a protein with a bHLH domain in the Myc/Max subfamily (Fig. 2). The bHLH region of Cph2 is most similar to two Schizosaccharomyces pombe proteins of unknown function. It is also very similar to the S. cerevisiae bHLH proteins Hms1 and Tye7. HMS1 has been cloned as one of the multicopy suppressors of pseudohyphal growth in an ammonium permease mutant strain (30). Like that of CPH2, the overexpression of HMS1 enhances pseudohyphal growth in diploids. However, hms1/hms1 mutant strains have no detectable defects in pseudohyphal growth (30). The overexpression of TYE7 does not promote filamentous growth (unpublished data). The bHLH regions of Cph2, Hms1, and Tye7 share striking similarities with human SREBP1 (sterol response element binding protein 1), Max, and c-Myc proteins (Fig. 2). bHLH proteins of the Myc/Max subfamily bind to E-box motifs. Unlike Max and c-Myc proteins, SREBP1 binds to an E-box motif, as well as a non-E-box sequence, sterol regulatory element 1 (SRE-1) (20). The dual DNA binding specificity of SREBP1 is the result of an atypical Tyr residue in the conserved basic domain (20) (Fig. 2). Substitution of the atypical Tyr in the basic region with the Arg found in most bHLH proteins causes a restriction of only E-box binding. Cph2 has a Tyr residue at the position that correlates with the atypical Tyr in SREBP1 (Fig. 2) and is expected to have the same dual binding capacity as SREBP1. In addition, based on what is known about SREBPs, the Cph2 bHLH region predicts the formation of Cph2 homodimers (37).
FIG. 2.
CPH2 encodes a bHLH protein. (A) A schematic diagram illustrating the location of the bHLH domain in the deduced Cph2 protein. (B) Protein sequence alignment of the bHLH domain (aa 204 to 290) of Cph2 with two S. pombe proteins of unknown function (accession numbers T39800 and T40285 in the EMBO data library), S. cerevisiae Hms1 (30) and Tye7 (29), Kluyveromyces lactis Sck1 (KLSCK1 in the EMBO data library), and human SREBP1a (47), Max (5), and c-Myc (38). Darker (black) letters show identical residues or conserved changes between the Cph2 protein and other bHLH proteins. Conserved residues for the basic region, helix I, loop, helix II, and leucine zipper are indicated by boxes. The asterisk denotes Tyr residues of SREBPs and Cph2.
cph2/cph2 mutants show a medium-specific impairment in hyphal development.
To elucidate the function of CPH2 in C. albicans, we deleted both copies of CPH2 by homologous recombination as described by Wilson et al. (46). PCR fragments of C. albicans URA3, HIS1, or ARG4, flanked by 60 bp of sequence homologous to the CPH2 coding region, were used in a C. albicans transformation to delete the first copy of CPH2 (Fig. 3). To improve the efficiency of deleting the second CPH2 gene, a pair of outside primers from various cph2/CPH2 heterozygous strains obtained from the first round of transformation (Fig. 3) was used for PCR, generating cph2::URA3, cph2::HIS1, and cph2::ARG4 PCR fragments with 500 bp of sequence homologous to CPH2 flanking each selectable marker. These PCR fragments were then used for the second round of transformation.
FIG. 3.
Disruption of CPH2 in C. albicans. (A) Restriction map and disruption strategy for CPH2. Positions of PCR primers P272, P273, P278, and P277 are marked. The positions and relative lengths of the cph2 deletions are also marked. (B) Analysis of CPH2 deletions by PCR. PCRs with outside primers P278 and P277 for C. albicans strains SC5314 (wild type [WT]), HLY1907 (CPH2/cph2::ARG4), HLY1909 (CPH2/cph2::HIS1), HLY1906 (CPH2/cph2::URA3), HLY1921 (cph2::ARG4/cph2::URA3), and HLY1927 (cph2::HIS1/cph2::ARG4) are shown. The genomic DNA was treated with XcmI for HLY1901 and HpaI for HLY1909 and HLY1906 to reduce the amount of PCR product from the wild-type copy of CPH2. The deletion of one copy of CPH2 is based on the appearance of a larger PCR fragment (left panel). The deletion of the second copy is evident from the disappearance of the wild-type CPH2 PCR product and the existence of two larger deletion fragments (right panel).
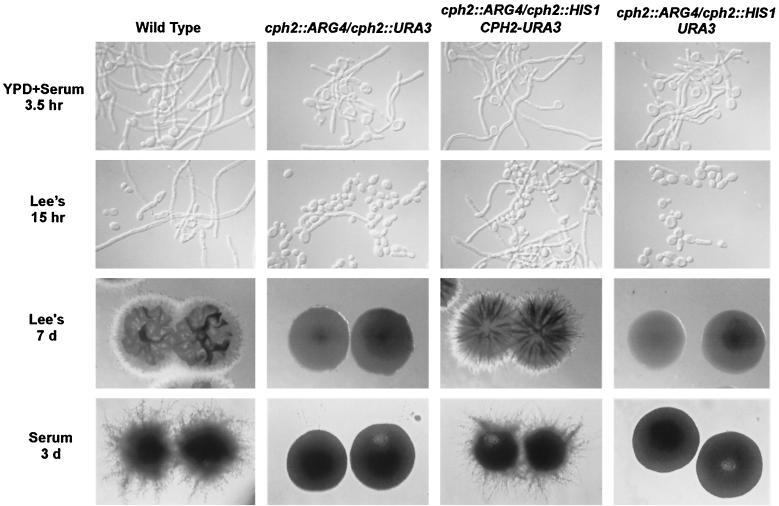
We examined the ability of cph2/cph2 strains to undergo hyphal development in several liquid hypha-inducing media. The homozygous mutant strains exhibited no discernible defect in germ tube or hyphal development in many liquid hypha-inducing media, including serum (Fig. 4, first row), N-acetylglucosamine, and proline (data not shown). However, the cph2/cph2 strains showed much less filamentation in Lee's medium (Fig. 4, second row). Changing the carbon source (mannitol, sucrose, or glucose) in Lee's medium did not seem to affect the level of filamentation of the cph2/cph2 strains. The fact that cph2/cph2 strains showed a defect in only certain hypha-inducing media suggested that Cph2 might be responsible for mediating medium-specific signals in hyphal development. The defect of cph2/cph2 in hyphal development was exacerbated on solid hypha-inducing media. The homozygous cph2/cph2 mutant strains exhibited a defect in hyphal colony formation on both serum-containing and solid Lee's media (Fig. 4). The defect in hyphal growth was directly linked to the CPH2 deletion, because reintroducing a wild-type CPH2 gene into cph2/cph2 strain HLY1927 rescued the defect in hyphal development, while the same cph2/cph2 strain with the vector alone was still defective in hyphal growth (Fig. 4).
FIG. 4.
Mutations in CPH2 suppress hyphal development. Cell and colony morphologies of wild-type and cph2/cph2 mutant strains after growth either in liquid (top two rows) or on solid (bottom two rows) Lee's or serum medium for the times (days [d]) indicated are shown. Strains: SC5314 (wild type), HLY1921 (cph2::ARG4/cph2::URA3), HLY1929 (cph2::ARG4/cph2::HIS1 ADE2/ade2::CPH2-URA3), and HLY1928 (cph2::ARG4/cph2::HIS1 ADE2/ade2::URA3).
cph2/cph2 strains are impaired in the induction of hypha-specific transcripts.
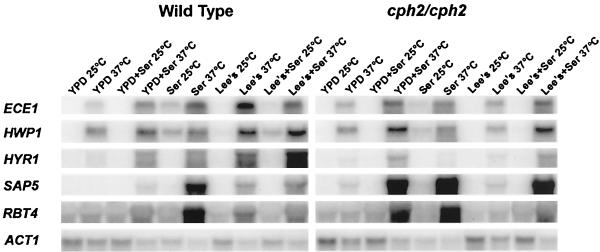
Since Cph2 encodes a Myc type of bHLH protein and cph2/cph2 mutants exhibit a medium-specific defect in hyphal development, we suspected that Cph2 might play a role in regulating the hyphal transcriptional program. We therefore examined the levels of expression of hypha-specific genes in the cph2/cph2 strains by Northern hybridization. The levels of ECE1, HWP1, HYR1, RBT4, and SAP5 expression in the cph2/cph2 strains were similar to those in the wild-type strains in YPD, YPD-serum, Lee's medium-serum, or serum alone but were reduced by about 20-fold in the cph2/cph2 strains in Lee's medium at 37°C (Fig. 5). The expression of ALS3 was similar to that of other hypha-specific genes (data not shown). Therefore, the cph2/cph2 strains exhibited a specific defect in the induction of hypha-specific genes in Lee's medium (Fig. 5, eighth lane from left) consistent with the medium-specific morphological defect shown in Fig. 4. Interestingly, we also observed that in some serum-containing media, such as YPD-serum, Cph2 appeared to have repressive activity for a group of hypha-specific genes, including RBT4 and SAP5 (Fig. 5, fourth lane from left).
FIG. 5.
Impairment in the transcription of hypha-specific genes in the cph2/cph2 mutant. Northern analysis of hypha-specific genes in wild-type (SC5314) and cph2/cph2 (HLY1921) strains is shown. Cells were diluted from overnight cultures into the media indicated and grown for 3 h in the conditions indicated, except for growth in Lee's medium for 6 h. Ser, serum.
Cph2 regulates TEC1 transcription.
Since Cph2 is responsible for the transcriptional induction of several hypha-specific genes in certain media, this activity of Cph2 may be mediated through the direct binding of Cph2 to SRE-1 or E-box motifs in the promoters of these hypha-specific genes. To address this possibility, we analyzed the upstream sequences of Cph2-regulated hypha-specific genes (Table 3). Surprisingly, the majority of the Cph2-regulated genes did not contain a potential Cph2 binding motif in their upstream regions. Of six genes, only ECE1 and RBT4 had a potential SRE-1-like site (Table 3). HWP1 and ECE1 were the only two genes found to have a potential E-box motif (Table 3). Moreover, many of these putative sites may not be actual Cph2 binding sites if one considers the possibility of random occurrences of these motifs in the genome (Table 3). In addition, SREBP1-regulated genes usually contain multiple binding sites in their promoters. Collectively, sequence analysis suggests that most of the hypha-specific genes may not be regulated directly by Cph2 but are regulated through a mediator(s).
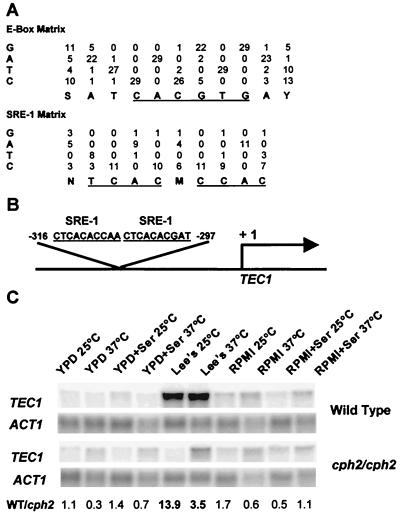
To identify potential Cph2-regulated mediators of hyphal development, we used the GibbsDNA sampling program (23) to analyze the 800-bp upstream sequences of four previously published hypha-specific genes: ECE1, HWP1, HYR1, and ALS3. Several motifs were found to be overrepresented in the upstream sequences of these four genes when compared to the occurrences in those of 1,000 randomly chosen C. albicans genes (unpublished data). In a search against known transcription factor binding sites, one of the motifs, TCATTCY, turned out to be very close to the binding site for A. nidulans AbaA and S. cerevisiae Tec1 (Table 3). AbaA binds to the sequence TTCATTCYTT (1), of which CATTCY is the core sequence for the TEA/ATTS family of transcription factors. The extracted motif is closer to the AbaA binding site than to other family members. The actual occurrence of the TCATTCY sequence in RBT4 and SAP5 upstream sequences was also much higher than what was expected to occur randomly (Table 3). The finding of Tec1 (AbaA) binding sites in hypha-specific genes was significant because Tec1 has recently been shown to regulate hyphal development in C. albicans (40). In addition, the TEC1 upstream region contains two highly conserved SRE-1-like sequences adjacent to each other (Fig. 6B). The proximity of the two SRE-1-like sites in the TEC1 upstream region is significant because two or more consecutive SRE-1 sites are a common feature for many SREBP1-regulated genes in mammalian cells (32). Therefore, we suspected that Cph2 may directly regulate the transcription of TEC1.
FIG. 6.
Cph2 regulates the transcription of TEC1. (A) Weight matrices for E-box (20) and SRE-1 (32) motifs were constructed from previously published data. Conserved core sequences are underlined. (B) Positions and sequences of two SRE-1-like motifs in the TEC1 upstream sequence. (C) Northern analysis of TEC1 in wild-type (SC5314) and cph2/cph2 (HLY1928) strains. Cells were grown in the conditions and media indicated for 3 h (YPD medium), 5 h (RPMI medium), and 6 h (Lee's medium with succinate). The fold change in TEC1 expression between wild-type (WT) and cph2/cph2 strains for each condition is indicated below each lane. Ser, serum.
Northern analysis of the TEC1 transcription level showed that TEC1 expression was medium dependent and was highest in Lee's medium (Fig. 6C). Interestingly, TEC1 expression was decreased in cph2/cph2 strains in Lee's medium (Fig. 6C) but was unchanged compared to that of the wild-type parent strains in other hypha-inducing media (Fig. 6C). Therefore, Cph2 was necessary for the transcriptional induction of TEC1 in Lee's medium. Additional regulators may control the basal expression of TEC1. The medium-dependent requirement of Cph2 for TEC1 transcriptional induction is consistent with the medium-specific impairment of cph2/cph2 in hyphal development.
Cph2 directly binds to the two SRE-1-like elements upstream of TEC1 as well as to an E-box sequence.
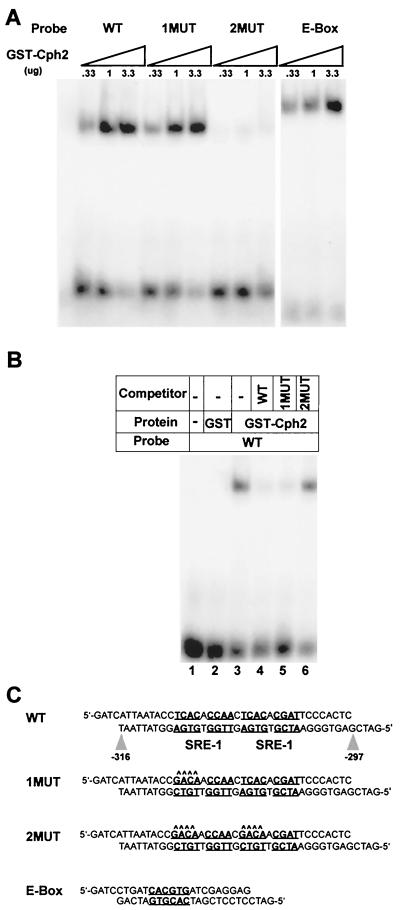
To address whether Cph2 binds directly to the SRE-1-like elements upstream of TEC1, we performed gel mobility shift experiments with a Cph2 recombinant protein. The Cph2 recombinant protein (including aa 190 to 290 of Cph2) contains all of the protein information necessary for DNA binding, protein dimerization, and transcriptional activation of this family of bHLH proteins (45). A DNA fragment corresponding to the two SRE-1-like elements from −316 to −297 of the TEC1 upstream sequence was used as a probe in the gel mobility shift experiments. The binding specificity was evaluated by gel mobility shift experiments using DNA probes for the TEC1 upstream sequence with mutations in one or both of the SRE-1-like elements (Fig. 7C). Mutating one of the two SRE-1-like elements did not abolish the binding of recombinant Cph2. However, mutating both of the SRE-1-like elements completely abolished the binding (Fig. 7A). Furthermore, competition with the same DNA mutated in both SRE-1 elements of the TEC1 upstream sequence failed to compete for Cph2 binding, while DNA with either one or both SRE-1-like elements intact efficiently competed with the probe for Cph2 binding (Fig. 7B). Therefore, the Cph2 recombinant protein bound specifically to the SRE-1-like elements (Fig. 7).
FIG. 7.
The bHLH region of Cph2 binds to the SRE-1 site in the TEC1 upstream sequence as well as an E-box sequence. (A) Analytical gel mobility shift analysis. Increasing concentrations of GST-Cph2 fusion protein were incubated with labeled DNA fragments containing SRE-1-like elements (wild type [WT]) and the corresponding DNAs with one (1MUT) or both (2MUT) SRE-1 sites mutated. Similarly, a labeled DNA probe corresponding to the E-box sequence was used in the titration experiments with recombinant GST-Cph2. (B) Competition binding reactions were carried out with the GST-Cph2 recombinant protein incubated first with unlabeled competitor DNA and then with a labeled SRE-1-containing (WT) probe (lanes 4 to 6). The labeled probe alone was loaded in lane 1. GST and the GST-Cph2 fusion protein were incubated with the probe in lanes 2 and 3, respectively. (C) DNA sequences of the probes and competitors used in the mobility shift analysis. The conserved SRE-1-like sites and E-box motifs are underlined and boldfaced. The mutated residues are indicated by carets.
To examine whether Cph2 is able to bind to an E-box sequence, we also performed a gel mobility shift experiment using the Cph2 recombinant protein with a DNA fragment that contains an E-box sequence (20). As shown in Fig. 7A, Cph2 bound to the E-box-containing DNA fragment as well as or even better than it bound to the SRE-1-like elements from TEC1. Therefore, Cph2 can bind to both SRE-1 elements and E-box sequences and has the same dual binding capacity as SREBP1.
Ectopic expression of TEC1 in cph2/cph2 strains suppresses the defect in hyphal development.
Although Cph2 is required for the transcriptional induction of TEC1 in Lee's medium and Cph2 binds directly to the two SRE-1-like elements upstream of TEC1, it is not clear whether the function of Cph2 in hyphal development is mediated through the regulation of TEC1 expression. To test this idea, we placed TEC1 under the control of the PCK1 promoter (44) and transformed the construct into wild-type and cph2/cph2 strains. To induce the expression of TEC1 from the PCK1 promoter, cells were grown on SSA medium. Wild-type cells produced wrinkled colonies with some filaments around the colonies after 7 days at 37°C (Fig. 8a). TEC1 overexpression in wild-type cells produced abundant and densely packed fine filaments in each colony (Fig. 8b). The cph2/cph2 strain produced completely smooth yeast colonies (Fig. 8c), and the ectopic expression of TEC1 in the cph2/cph2 strain suppressed the defect and produced colonies with fine filaments (Fig. 8d). However, the number of filaments was smaller than that seen with the wild-type strain with ectopic TEC1 expression (Fig. 8d versus Fig. 8b). TEC1 ectopic expression in wild-type and cph2/cph2 mutant strains showed similar phenotypes when the strains were grown on Lee's medium-succinate, but wild-type colonies were not as filamentous as those grown on SSA plates (data not shown). Our data suggest that TEC1 ectopic expression can suppress the cph2/cph2 defect in filamentation, a result which suggests that Cph2 regulates hyphal development through the regulation of TEC1 expression. However, phenotypic differences between wild-type cells and cph2/cph2 cells overexpressing TEC1 were observed (Fig. 8d versus Fig. 8b). This result indicates that Cph2 may have additional functions in regulating hyphal development that are independent of the regulation of TEC1 expression. Therefore, we suggest that Cph2 activity in regulating hypha-specific transcription is, in part, mediated through Tec1.
Overexpression of CPH2 promotes filamentation.
Since the overexpression of CPH2 in S. cerevisiae promoted pseudohyphal growth, we expected that the overexpression of CPH2 in C. albicans might also enhance hyphal formation. To test this idea, we placed CPH2 under the control of the PCK1 promoter (44) and transformed a PCK1p-CPH2 construct into a wild-type C. albicans strain. As shown in Fig. 9l, the CPH2 transcript (of the expected size of 0.9 kb) was readily detectable in strains carrying the PCK1p-CPH2 construct under inducing conditions for the PCK1 gene. However, CPH2 expression was not detected by Northern analysis in wild-type cells on Lee's medium with succinate as the carbon source at either 25 or 37°C, nor was it detected under several other hypha-inducing conditions examined, including YPD–10% serum and Lee's medium with mannitol as the carbon source (data not shown). The overexpression of CPH2 in C. albicans induced cell elongation under conditions usually favorable for yeast growth (Fig. 9c versus Fig. 9a). Therefore, Cph2 is a positive regulator of hyphal development in C. albicans.
FIG. 9.
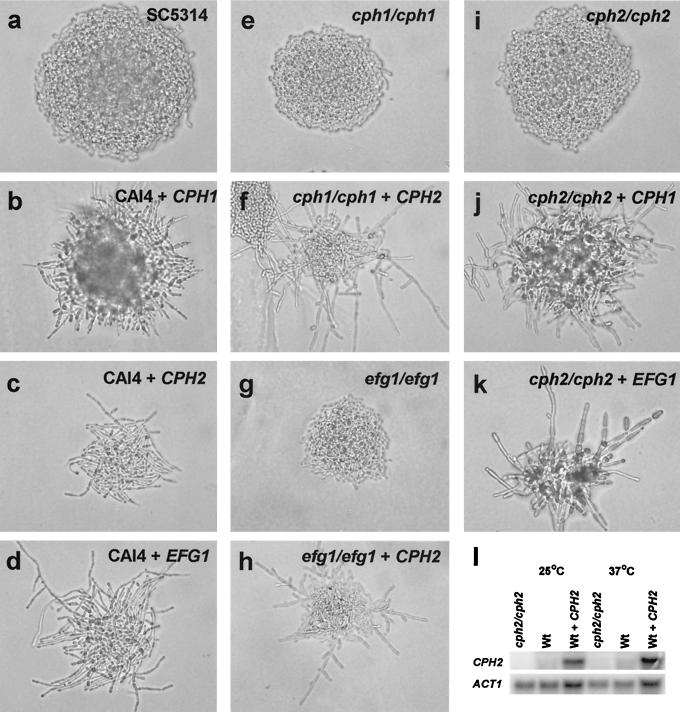
Functional relationship of Cph2 with Cph1 and Efg1 in C. albicans. Colony morphologies of various single and double mutant strains grown on SSA or SS media for 18 h are shown. (a to d) Colonies of a wild-type strain (SC5134) and a wild-type strain (CAI4) transformed with PCK1p-CPH1 (HLY3119), PCK1p-CPH2 (HLY3120), and PCK1p-EFG1 (HLY3125), respectively. (e and f) Colony morphologies of a cph1/cph1 strain (JKC19) and a cph1/cph1 strain transformed with PCK1p-CPH2 (HLY3138), respectively. (g and h) Colony morphologies of an efg1/efg1 strain (HLC52) and an efg1/efg1 strain transformed with PCK1p-CPH2 (HLY3183), respectively. (i to k) Colony morphologies of a cph2/cph2 strain (HLY1928) and a cph2/cph2 strain transformed with PCK1p-CPH1 (HLY3156) and PCK1p-EFG1 (HLY3164), respectively. Cells in panels g and h were grown on SS medium. Cells in all other panels were grown on SSA medium. (l) Northern blot analysis of CPH2 overexpression. A cph2/cph2 strain (HLY1928), a wild-type (Wt) strain (SC5314), and a wild-type strain transformed with PCK1p-CPH2 (HLY3120) were grown on Lee's medium with succinate as the carbon source at 25 or 37°C for 6 h before being harvested for Northern analysis.
Cph2 functions independently of the Cph1- and Efg1-mediated pathways in C. albicans.
To address whether Cph2 function is associated with the Cph1- or Efg1-mediated signaling pathway for hyphal development, we performed epistasis studies with C. albicans. The overexpression of CPH1, CPH2, or EFG1 from the PCK1 promoter (44) in wild-type cells on SSA medium promoted filamentation with elongated pseudohyphal cells (Fig. 9a to d). We then introduced several of these overexpression constructs into the appropriate C. albicans strains mutated in CPH1, CPH2, or EFG1 and compared the phenotypes of these strains with those of the original mutant strains. cph1/cph1 mutant strains were unable to undergo hyphal development on solid SSA medium (Fig. 9e). Overexpression of CPH2 in cph1/cph1 mutant strains could suppress the defect in hyphal formation (Fig. 9f). Similarly, efg1/efg1 single mutants were unable to undergo hyphal development on SS medium, and the defect was partially suppressed by introducing the PCK1p-CPH2 construct (Fig. 9g and h). cph2/cph2 mutant strains were defective in filamentation on SSA medium (Fig. 9i). The overexpression of either CPH1 or EFG1 from the PCK1 promoter suppressed the defect in hyphal development in cph2/cph2 strains on SSA medium (Fig. 9j and k). Based on the data that EFG1 overexpression could suppress filamentation in cph2/cph2 strains and, vice versa, that CPH2 overexpression could promote filamentation in efg1/efg1 strains, we suggest that Cph2 and Efg1 function in two different pathways. Similar reciprocal suppression was observed when CPH1 was expressed in cph2/cph2 strains and when CPH2 was expressed in cph1/cph1 strains, suggesting independent functions of Cph1 and Cph2 in regulating hyphal development.
DISCUSSION
In this study, we have identified a new Myc-type bHLH protein, Cph2, in C. albicans. Cph2 has a Tyr residue in the basic region at the position that correlates with the atypical Tyr in SREBP1 that is responsible for the dual DNA binding specificity of SREBP1. Like SREBP1, Cph2 also binds to both E-box motifs and non-E-box SRE-1-like elements. Therefore, Cph2 is the second naturally occurring bHLH protein with an atypical Tyr residue in the basic region that has been shown to bind to both classes of DNA motifs.
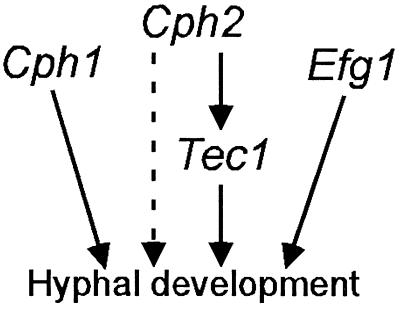
We have demonstrated that Cph2 plays an important role in hyphal development and in the induction of hypha-specific genes. Although Cph2 is necessary for the induction of all known hypha-specific genes examined, significant percentages of these genes do not have any potential Cph2 binding sites in their upstream sequences. Instead, potential Tec1 binding sites are present in the upstream regions of all known hypha-specific genes and even multiple times in some of them. Furthermore, TEC1 expression is induced particularly in the medium in which cph2/cph2 hyphal development is impaired. By gel mobility shift experiments with recombinant Cph2, we showed that Cph2 binds specifically to the two SRE-1-like elements upstream of TEC1. Therefore, Cph2 may regulate TEC1 transcription directly by binding to the two SRE-1 elements. Not only does Cph2 regulate TEC1 transcription but also the ectopic expression of TEC1 suppresses the defect in cph2/cph2 hyphal development. Together, our data suggest that the function of Cph2 in regulating hyphal development is likely mediated, in part, through Tec1 (Fig. 10). These data provide another example of a transcription factor cascade involved in regulating cellular differentiation in fungi. Similar transcription factor cascades have been shown for pseudohyphal growth in S. cerevisiae (31, 36) and conidiophore formation in A. nidulans (33). Interestingly, members of the TEA/ATTS family are the ones being regulated in the transcription factor cascades in all three fungi. However, the upstream regulators are different in each species. The functional relationship between Cph2 and Tec1 that we have discovered in C. albicans could not have been deduced by analogy to the regulation of pseudohyphal growth in S. cerevisiae or conidiophore formation in A. nidulans.
FIG. 10.
Proposed functional relationship between Cph2 and other regulators in hyphal development.
Our data do not exclude the possibility that Cph2 may also play a direct role in the transcriptional activation of some hypha-specific genes that contain SRE-1-like or E-box motifs in their upstream sequences (Table 3). The observation that TEC1 overexpression in cph2/cph2 cells does not generate the same level of hyphal growth as that seen in wild-type cells indicates that Cph2 may have other functions in addition to regulation of TEC1 expression. Because some of the hypha-specific genes contain SRE-1-like sequences or E-box motifs in their upstream sequences (Table 3) and we have shown that Cph2 binds to both types of DNA sequences, it is possible that Cph2 directly regulates the transcription of these genes. For mammalian cells, Max has been shown to act cooperatively with TEF-1, a mammalian member of the TEA/ATTS family, in regulating the expression of a cardiac α-myosin heavy-chain gene (16). The same cooperative transcriptional activation may exist between Cph2 and Tec1 in C. albicans. Interestingly, hypha-specific genes that have one SRE-1-like element or E-box motif in their upstream regions all have only one potential Tec1 binding site, while genes lacking potential Cph2 binding sites tend to have multiple potential Tec1 binding sites (Table 3). Therefore, the second group of genes may be induced by Tec1, which in turn is regulated by Cph2, while the first group of genes may be coordinately regulated by both Cph2 and Tec1 (Fig. 10).
How Cph2 activity is regulated during hyphal development in C. albicans is not clear. The Cph2 protein sequence predicts many potential phosphorylation sites for casein kinase II, protein kinase C, and cyclic AMP-dependent protein kinase. Therefore, phosphorylation may play a significant role in regulating its activity. Phosphorylation has been shown to lead to a change in the transcriptional activities of bHLH proteins, as in the case of Max, where the DNA binding activity of Max homodimers is inhibited by its phosphorylation by casein kinase II (3). The state of phosphorylation can also regulate the nuclear localization of bHLH proteins. For example, Pho4 is phosphorylated by the Pho80-Pho85 cyclin-dependent kinase complex and is subsequently exported to the cytoplasm when yeast cells are grown in phosphate-rich conditions (22, 35). Phosphorylation may also lead to a change in protein stability, as in the case of HLF in response to hypoxia (12). In some instances, the transcriptional activation of bHLH proteins is achieved by removal of their inhibitory interaction partners, which are modulated by phosphorylation (17). Determination of whether Cph2 is regulated by any of the known mechanisms requires further experiments.
We have data to suggest that Cph2 functions independently from the Cph1-mediated MAP kinase pathway and the Efg1-transmitted protein kinase A pathway. CPH2 overexpression is able to partially suppress the defect in hyphal development in efg1/efg1 mutant strains, and EFG1 overexpression overcomes the defect of filamentation in cph2/cph2 strains. Similarly, reciprocal suppression by CPH2 and CPH1 is observed in cph1/cph1 and cph2/cph2 strains, respectively. Furthermore, the medium-specific impairment in hyphal development displayed by each mutant also suggests that Cph1, Cph2, and Efg1 function in independent pathways. Therefore, different signaling pathways may respond to different growth and environmental signals in regulating filamentous growth (Fig. 10). Our data showing how Cph2 functions in hyphal development provide a clear molecular explanation for a mechanism by which C. albicans can integrate several different upstream signals into a common downstream output during hyphal development.
ACKNOWLEDGMENTS
The cloning of CPH2 was performed in the Fink Laboratory while H. Liu was a postdoctoral fellow. We thank Tim Osborne for helpful suggestions, discussions, and critical reading of the manuscript; the members of the Osborne and Dai Laboratories for advice and reagents for GST-Cph2 recombinant protein expression and purification and subsequent gel mobility shift experiments; Jiangye Chen for assistance with the cph2 deletion; and Gerald Fink, William Fonzi, and Joachim Ernst for strains and plasmids.
S. Lane is a predoctoral fellow supported by an NIH Carcinogenesis training grant, and S. Zhou is a predoctoral fellow supported by a training grant from the UC Systemwide Biotechnology Research and Education program. This work was supported by funds from Burroughs Wellcome and NIH (grant GM55155).
REFERENCES
- 1.Andrianopoulos A, Timberlake W E. The Aspergillus nidulans abaA gene encodes a transcriptional activator that acts as a genetic switch to control development. Mol Cell Biol. 1994;14:2503–2515. doi: 10.1128/mcb.14.4.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bailey D A, Feldmann P J, Bovey M, Gow N A, Brown A J. The Candida albicans HYR1 gene, which is activated in response to hyphal development, belongs to a gene family encoding yeast cell wall proteins. J Bacteriol. 1996;178:5353–5360. doi: 10.1128/jb.178.18.5353-5360.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berberich S J, Cole M D. Casein kinase II inhibits the DNA-binding activity of Max homodimers but not Myc/Max heterodimers. Genes Dev. 1992;6:166–176. doi: 10.1101/gad.6.2.166. [DOI] [PubMed] [Google Scholar]
- 4.Birse C E, Irwin M Y, Fonzi W A, Sypherd P S. Cloning and characterization of ECE1, a gene expressed in association with cell elongation of the dimorphic pathogen Candida albicans. Infect Immun. 1993;61:3648–3655. doi: 10.1128/iai.61.9.3648-3655.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blackwood E M, Eisenman R N. Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science. 1991;251:1211–1217. doi: 10.1126/science.2006410. [DOI] [PubMed] [Google Scholar]
- 6.Bockmuhl D P, Ernst J F. A Potential Phosphorylation Site for an A-Type Kinase in the Efg1 Regulator Protein Contributes to Hyphal Morphogenesis of Candida albicans. Genetics. 2001;157:1523–1530. doi: 10.1093/genetics/157.4.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Braun B R, Head W S, Wang M X, Johnson A D. Identification and characterization of TUP1-regulated genes in Candida albicans. Genetics. 2000;156:31–44. doi: 10.1093/genetics/156.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown A J, Gow N A. Regulatory networks controlling Candida albicans morphogenesis. Trends Microbiol. 1999;7:333–338. doi: 10.1016/s0966-842x(99)01556-5. [DOI] [PubMed] [Google Scholar]
- 9.Csank C, Schroppel K, Leberer E, Harcus D, Mohamed O, Meloche S, Thomas D Y, Whiteway M. Roles of the Candida albicans mitogen-activated protein kinase homolog, Cek1p, in hyphal development and systemic candidiasis. Infect Immun. 1998;66:2713–2721. doi: 10.1128/iai.66.6.2713-2721.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Delbruck S, Ernst J F. Morphogenesis-independent regulation of actin transcript levels in the pathogenic yeast Candida albicans. Mol Microbiol. 1993;10:859–866. doi: 10.1111/j.1365-2958.1993.tb00956.x. [DOI] [PubMed] [Google Scholar]
- 11.Dooley K A, Bennett M K, Osborne T F. A critical role for cAMP response element-binding protein (CREB) as a Co-activator in sterol-regulated transcription of 3-hydroxy-3-methylglutaryl coenzyme A synthase promoter. J Biol Chem. 1999;274:5285–5291. doi: 10.1074/jbc.274.9.5285. [DOI] [PubMed] [Google Scholar]
- 12.Ema M, Hirota K, Mimura J, Abe H, Yodoi J, Sogawa K, Poellinger L, Fujii-Kuriyama Y. Molecular mechanisms of transcription activation by HLF and HIF1alpha in response to hypoxia: their stabilization and redox signal-induced interaction with CBP/p300. EMBO J. 1999;18:1905–1914. doi: 10.1093/emboj/18.7.1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feng Q, Summers E, Guo B, Fink G. Ras signaling is required for serum-induced hyphal differentiation in Candida albicans. J Bacteriol. 1999;181:6339–6346. doi: 10.1128/jb.181.20.6339-6346.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fonzi W A, Irwin M Y. Isogenic strain construction and gene mapping in Candida albicans. Genetics. 1993;134:717–728. doi: 10.1093/genetics/134.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gavrias V, Andrianopoulos A, Gimeno C J, Timberlake W E. Saccharomyces cerevisiae TEC1 is required for pseudohyphal growth. Mol Microbiol. 1996;19:1255–1263. doi: 10.1111/j.1365-2958.1996.tb02470.x. [DOI] [PubMed] [Google Scholar]
- 16.Gupta M P, Amin C S, Gupta M, Hay N, Zak R. Transcription enhancer factor 1 interacts with a basic helix-loop-helix zipper protein, Max, for positive regulation of cardiac alpha-myosin heavy-chain gene expression. Mol Cell Biol. 1997;17:3924–3936. doi: 10.1128/mcb.17.7.3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hara E, Hall M, Peters G. Cdk2-dependent phosphorylation of Id2 modulates activity of E2A-related transcription factors. EMBO J. 1997;16:332–342. doi: 10.1093/emboj/16.2.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoyer L L, Payne T L, Bell M, Myers A M, Scherer S. Candida albicans ALS3 and insights into the nature of the ALS gene family. Curr Genet. 1998;33:451–459. doi: 10.1007/s002940050359. [DOI] [PubMed] [Google Scholar]
- 19.Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim J B, Spotts G D, Halvorsen Y D, Shih H M, Ellenberger T, Towle H C, Spiegelman B M. Dual DNA binding specificity of ADD1/SREBP1 controlled by a single amino acid in the basic helix-loop-helix domain. Mol Cell Biol. 1995;15:2582–2588. doi: 10.1128/mcb.15.5.2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kohler J R, Fink G R. Candida albicans strains heterozygous and homozygous for mutations in mitogen-activated protein kinase signaling components have defects in hyphal development. Proc Natl Acad Sci USA. 1996;93:13223–13228. doi: 10.1073/pnas.93.23.13223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Komeili A, O'Shea E K. Roles of phosphorylation sites in regulating activity of the transcription factor Pho4. Science. 1999;284:977–980. doi: 10.1126/science.284.5416.977. [DOI] [PubMed] [Google Scholar]
- 23.Lawrence C E, Altschul S F, Boguski M S, Liu J S, Neuwald A F, Wootton J C. Detecting subtle sequence signals: a Gibbs sampling strategy for multiple alignment. Science. 1993;262:208–214. doi: 10.1126/science.8211139. [DOI] [PubMed] [Google Scholar]
- 24.Leberer E, Harcus D, Broadbent I D, Clark K L, Dignard D, Ziegelbauer K, Schmidt A, Gow N A, Brown A J, Thomas D Y. Signal transduction through homologs of the Ste20p and Ste7p protein kinases can trigger hyphal formation in the pathogenic fungus Candida albicans. Proc Natl Acad Sci USA. 1996;93:13217–13222. doi: 10.1073/pnas.93.23.13217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee K L, Buckley H R, Campbell C C. An amino acid liquid synthetic medium for development of mycelial and yeast forms of Candida albicans. Sabouraudia. 1975;13:148–153. doi: 10.1080/00362177585190271. [DOI] [PubMed] [Google Scholar]
- 26.Liu H, Kohler J, Fink G R. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science. 1994;266:1723–1726. doi: 10.1126/science.7992058. [DOI] [PubMed] [Google Scholar]
- 27.Lo H J, Kohler J R, DiDomenico B, Loebenberg D, Cacciapuoti A, Fink G R. Nonfilamentous C. albicans mutants are avirulent. Cell. 1997;90:939–949. doi: 10.1016/s0092-8674(00)80358-x. [DOI] [PubMed] [Google Scholar]
- 28.Loeb J D, Sepulveda-Becerra M, Hazan I, Liu H. A G1 cyclin is necessary for maintenance of filamentous growth in Candida albicans. Mol Cell Biol. 1999;19:4019–4027. doi: 10.1128/mcb.19.6.4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lohning C, Ciriacy M. The TYE7 gene of Saccharomyces cerevisiae encodes a putative bHLH-LZ transcription factor required for Ty1-mediated gene expression. Yeast. 1994;10:1329–1339. doi: 10.1002/yea.320101010. [DOI] [PubMed] [Google Scholar]
- 30.Lorenz M C, Heitman J. Regulators of pseudohyphal differentiation in Saccharomyces cerevisiae identified through multicopy suppressor analysis in ammonium permease mutant strains. Genetics. 1998;150:1443–1457. doi: 10.1093/genetics/150.4.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Madhani H D, Fink G R. Combinatorial control required for the specificity of yeast MAPK signaling. Science. 1997;275:1314–1317. doi: 10.1126/science.275.5304.1314. [DOI] [PubMed] [Google Scholar]
- 32.Magana M M, Osborne T F. Two tandem binding sites for sterol regulatory element binding proteins are required for sterol regulation of fatty-acid synthase promoter. J Biol Chem. 1996;271:32689–32694. doi: 10.1074/jbc.271.51.32689. [DOI] [PubMed] [Google Scholar]
- 33.Mirabito P M, Adams T H, Timberlake W E. Interactions of three sequentially expressed genes control temporal and spatial specificity in Aspergillus development. Cell. 1989;57:859–868. doi: 10.1016/0092-8674(89)90800-3. [DOI] [PubMed] [Google Scholar]
- 34.Mosch H U, Fink G R. Dissection of filamentous growth by transposon mutagenesis in Saccharomyces cerevisiae. Genetics. 1997;145:671–684. doi: 10.1093/genetics/145.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O'Neill E M, Kaffman A, Jolly E R, O'Shea E K. Regulation of PHO4 nuclear localization by the PHO80-PHO85 cyclin-CDK complex. Science. 1996;271:209–212. doi: 10.1126/science.271.5246.209. [DOI] [PubMed] [Google Scholar]
- 36.Pan X, Heitman J. Sok2 regulates yeast pseudohyphal differentiation via a transcription factor cascade that regulates cell-cell adhesion. Mol Cell Biol. 2000;20:8364–8372. doi: 10.1128/mcb.20.22.8364-8372.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parraga A, Bellsolell L, Ferre-D'Amare A R, Burley S K. Co-crystal structure of sterol regulatory element binding protein 1a at 2.3 A resolution. Structure. 1998;6:661–672. doi: 10.1016/s0969-2126(98)00067-7. [DOI] [PubMed] [Google Scholar]
- 38.Prendergast G C, Lawe D, Ziff E B. Association of Myn, the murine homolog of Max, with c-Myc stimulates methylation-sensitive DNA binding and Ras cotransformation. Cell. 1991;65:395–407. doi: 10.1016/0092-8674(91)90457-a. [DOI] [PubMed] [Google Scholar]
- 39.Sanchez H B, Yieh L, Osborne T F. Cooperation by sterol regulatory element-binding protein and Sp1 in sterol regulation of low density lipoprotein receptor gene. J Biol Chem. 1995;270:1161–1169. doi: 10.1074/jbc.270.3.1161. [DOI] [PubMed] [Google Scholar]
- 40.Schweizer A, Rupp S, Taylor B N, Rollinghoff M, Schroppel K. The TEA/ATTS transcription factor CaTec1p regulates hyphal development and virulence in Candida albicans. Mol Microbiol. 2000;38:435–445. doi: 10.1046/j.1365-2958.2000.02132.x. [DOI] [PubMed] [Google Scholar]
- 41.Sonneborn A, Bockmuhl D P, Gerads M, Kurpanek K, Sanglard D, Ernst J F. Protein kinase A encoded by TPK2 regulates dimorphism of Candida albicans. Mol Microbiol. 2000;35:386–396. doi: 10.1046/j.1365-2958.2000.01705.x. [DOI] [PubMed] [Google Scholar]
- 42.Staab J F, Bradway S D, Fidel P L, Sundstrom P. Adhesive and mammalian transglutaminase substrate properties of Candida albicans Hwp1. Science. 1999;283:1535–1538. doi: 10.1126/science.283.5407.1535. [DOI] [PubMed] [Google Scholar]
- 43.Staab J F, Sundstrom P. Genetic organization and sequence analysis of the hypha-specific cell wall protein gene HWP1 of Candida albicans. Yeast. 1998;14:681–686. doi: 10.1002/(SICI)1097-0061(199805)14:7<681::AID-YEA256>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 44.Stoldt V R, Sonneborn A, Leuker C E, Ernst J F. Efg1p, an essential regulator of morphogenesis of the human pathogen Candida albicans, is a member of a conserved class of bHLH proteins regulating morphogenetic processes in fungi. EMBO J. 1997;16:1982–1991. doi: 10.1093/emboj/16.8.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vallett S M, Sanchez H B, Rosenfeld J M, Osborne T F. A direct role for sterol regulatory element binding protein in activation of 3-hydroxy-3-methylglutaryl coenzyme A reductase gene. J Biol Chem. 1996;271:12247–12253. doi: 10.1074/jbc.271.21.12247. [DOI] [PubMed] [Google Scholar]
- 46.Wilson R B, Davis D, Mitchell A P. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J Bacteriol. 1999;181:1868–1874. doi: 10.1128/jb.181.6.1868-1874.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yokoyama C, Wang X, Briggs M R, Admon A, Wu J, Hua X, Goldstein J L, Brown M S. SREBP-1, a basic-helix-loop-helix-leucine zipper protein that controls transcription of the low density lipoprotein receptor gene. Cell. 1993;75:187–197. [PubMed] [Google Scholar]