Abstract
The E2F transcription factor controls the cell cycle-dependent expression of many S-phase-specific genes. Transcriptional repression of these genes in G0 and at the beginning of G1 by the retinoblasma protein Rb is crucial for the proper control of cell proliferation. Rb has been proposed to function, at least in part, through the recruitment of histone deacetylases. However, recent results indicate that other chromatin-modifying enzymes are likely to be involved. Here, we show that Rb also interacts with a histone methyltransferase, which specifically methylates K9 of histone H3. The results of coimmunoprecipitation experiments of endogenous or transfected proteins indicate that this histone methyltransferase is the recently described heterochromatin-associated protein Suv39H1. Interestingly, phosphorylation of Rb in vitro as well as in vivo abolished the Rb-Suv39H1 interaction. We also found that Suv39H1 and Rb cooperate to repress E2F activity and that Suv39H1 could be recruited to E2F1 through its interaction with Rb. Taken together, these data indicate that Suv39H1 is involved in transcriptional repression by Rb and suggest an unexpected link between E2F regulation and heterochromatin.
The retinoblastoma protein Rb is a key regulator of mammalian cell proliferation. In its active hypophosphorylated form, it prevents the cell from progressing to the S phase (22). This block must be relieved to allow cells to progress into the S phase. During a normal cell cycle, Rb is inactivated at the end of G1 through the concerted phosphorylation by cyclin D- and cyclin E-dependent kinase complexes (40). The gene encoding the retinoblastoma protein is subjected to inactivating mutations in a great variety of human tumors. In addition, viral transforming proteins such as the adenovirus E1A protein inhibit Rb functions through a direct physical interaction. The mechanisms by which Rb controls cell proliferation have been extensively studied in the past few years.
One of the major protein targets of Rb is the E2F transcription factor (34). E2F binding sites are present within the promoters of many genes whose products are required for S-phase progression. The E2F transcription factor binds to these sites as a heterodimer between a so-called E2F protein and a DRTF1 polypeptide (DP) protein (26). So far, six E2F proteins (E2F1 to E2F6) and two DP proteins have been described. At the end of G1 and the beginning of S phase, E2F-DP heterodimers (free E2F) activate transcription of their target genes through a transcriptional activation domain present within the E2F protein. The only exception is E2F6 (33, 49), which does not harbor any activation domain but rather represses transcription. At G0 and at the beginning of G1, proteins from the Rb family (called pocket proteins) bind directly to the activation domain of the E2F protein. Rb itself interacts with E2F1, E2F2, and E2F3, whereas the two related proteins, p107 and p130, target E2F4 and E2F5 (22).
Through their interaction with E2F, proteins of the Rb family are recruited to E2F sites. This binding leads to transcriptional repression of E2F-regulated genes through a transcriptional repression domain present within the pocket protein (12, 55). Many bits of evidence indicate that transcriptional repression by pocket proteins is crucial for the proper control of cell proliferation. First, E2F sites play mainly a repressive role on transcription (22). Second, inactivation of pocket protein function, either by phosphorylation, mutation, or viral transforming proteins, results in the loss of transcriptional repression properties (12, 44). Finally, a basal unrepressed level of transcription of E2F-regulated genes can be sufficient in some instances to induce cell transformation (16, 23).
Transcriptional repression by pocket proteins is mediated through their conserved domain, which is called the pocket (11). This domain of Rb is a hot spot of mutations in cancer. Recently, transcriptional repression by Rb has been shown to correlate with the ability of Rb to interact with proteins containing the so-called LXCXE motif (14). This motif has been first described as the Rb interaction site of viral transforming proteins such as E1A. Since then, a very large number of cellular proteins that use this motif to interact with Rb have been described (19). Consistent with the presumably important role of transcriptional repression by Rb, the domain responsible for the interaction with LXCXE-containing proteins is required for Rb to induce permanent cell cycle arrest (9, 14).
Several different molecular mechanisms for transcriptional repression by Rb have been proposed (21, 54). However, recent reports indicate that, at least in some instances, it involves recruitment of histone deacetylases (HDs) (6, 30, 31). Indeed, Rb has been shown to interact directly with the histone deacetylase HDAC1, through an LXCXE-like motif present within this protein (31). Furthermore, transcriptional repression of some E2F target genes can be relieved by HD inhibitors (30). Consistent with this model, histones on E2F-regulated promoters evolve during G1 from a hypoacetylated state to a hyperacetylated state (47).
Acetylation is a common posttranslational modification of nucleosomal histones which has long been known to correlate with activation of transcription (53). Histone acetylation status is controlled through a balance between histone acetyltransferase (HATs) and HDs. HATs are generally transcriptional activators, whereas HDs are often transcriptional repressors. They are believed to be recruited to specific promoters through physical interaction with coactivators and corepressors (24). Little is known about the molecular consequences of histone acetylation. A likely model is that acetylation of histones induces a decompaction of chromatin structure, thereby allowing a greater accessibility of transcription factors to DNA. An alternative hypothesis, called the histone code hypothesis, was recently proposed (45). This hypothesis relies on the fact that histone N-terminal tails are extensively modified, not only by acetylation but also by phosphorylation and methylation (45). According to the histone code hypothesis, a precise combination of modifications would lead to a specific consequence for chromatin function. Consistent with this, although histone acetylation is generally linked with transcriptional activation, acetylation of K12 of histone H4 in Saccharomyces cerevisiae is important for the formation of compact silenced heterochromatin (5).
If the histone code hypothesis is correct, then the other histone modifications could be as important as acetylation for chromatin function. Indeed, phosphorylation of histone H3 or the histone variant H2AX is likely to play a major role in cell cycle control (10, 29) and DNA repair (42), respectively. The discovery of the first histone methyltransferase (HMT) has also recently renewed our interest in histone methylation (41). This HMT was previously known as the human homologue of the Drosophila Su(Var)3.9 protein (1). The Su(Var)3.9 protein and its homologue in Schizosaccharomyces pombe Clr4 have been cloned as proteins involved in centromer function and pericentric heterochromatin silencing (3, 50). Suv39H1 also localizes at heterochromatin foci and centromers (1). Suv39H1 and the closely related Suv39H2 protein methylate specifically K9 from histone H3 (39, 41). Recent results suggest that methylation of K9 from histone H3 induces the formation of a high-affinity binding site on chromatin for proteins of the heterochromatin protein 1 (HP1) family (4, 25). These proteins are present in organisms from yeasts (Swi6 in S. pombe) to humans (HP1α, -β, and -γ) and like the members of the Suv39H1 family, they localize at heterochromatin foci and they are involved in pericentric heterochromatin silencing (15). The results of these studies indicate that histone H3 methylation very likely plays a major role in chromatin structure and function.
Importantly, the various posttranslational modifications of nucleosomal histones occur dependently on each other. For example, phosphorylation of S10 of histone H3 increases the efficiency of K14 acetylation (10, 29). Similarly, methylation of K9 interferes with phosphorylation of S10 (41). Furthermore, we recently described a physical interaction between the HAT CBP and an HMT (51). These data led us to test whether other histone-modifying enzymes could also be involved in Rb-mediated transcriptional repression. Indeed, it has been shown recently that in vitro, transcriptional repression by Rb requires nucleosomes but not HDs (43). Here, we found that Rb interacts through its growth-regulating domain with an HMT that we identified as Suv39H1. Furthermore, through this interaction, Suv39H1 could be targeted to E2F1. In transient-transfection experiments, the E2F1-Rb-Suv39H1 ternary complex repressed transcription. Taken together, these results indicate that Suv39H1 could be important for transcriptional repression by Rb. To our knowledge, it is the first demonstration of an HMT functioning as a promoter-specific transcriptional regulator. Finally, our findings suggest the existence of a functional link between E2F-regulated genes and heterochromatin.
MATERIALS AND METHODS
Plasmids.
pCMV NeoBam Rb 379-928 (pCMV Rb), pCMV NeoBam E2F1, pGEX2TK-Rb 379-928, pGEX2TK-Rb 379-928 706 C-F (Rb Mut) and empty vectors, E2F-luciferase and pCMV lacZ reporter vectors were described previously (31). Vectors allowing the expression of cyclin D, cyclin E, cdk4, and cdk2 were kind gifts from A. Harel-Bellan. Gal4-luc and TK-luc reporter vectors were kind gifts from H. G. Stunnenberg and H. Richard-Foy, respectively. pCMVGT, pHKGT, pHKG E2F1 380-437 (pHKG E2F1-AD), pGEX2TKp-E2F1 359-437 (E2F1 AD), pGEX-p53, pGEX-MyoD, and pGEX-pCAF were kind gifts from T. Kouzarides. pSG5 Rb was a kind gift from W. G. Kaelin. Myc-Suv39H1 expression vector was a kind gift from T. Jenuwein. The vector expressing GAL4-Suv39H1 fusion protein was made by inserting myc-Suv39H1 cDNA in frame in the pCMVGT expression vector. Suv39H1 1-332 was constructed by PCR and fully sequenced. pCMV2N3T Suv39H1 and pCMV 2N3T Suv39H1 1-332 were constructed by inserting the corresponding fragment into the pCMV 2N3T empty vector. They express the corresponding protein with N-terminal nuclear localization signal and hemagglutinin (HA) tags. Details of constructions are available upon request.
Cell culture and transfection.
U2OS, SAOS2, and HeLa cells were maintained in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum. Jurkat cells were maintained in RPMI medium supplemented with 10% fetal calf serum. U2OS and SAOS2 cells were transfected by the calcium phosphate coprecipitation procedure, whereas HeLa cells were transfected using Fugene Reagent (Roche Diagnostics), according to the manufacturer's instructions. For coimmunoprecipitation experiments, transfections were performed using 2 × 106 cells in 10-cm-diameter dishes. For reporter activity assays, transfections were performed using 4 × 105 cells in six-well plates. The amount of cytomegalovirus (CMV) promoter in the transfection was kept constant using empty vectors. Cells were harvested 24 or 48 h after transfection. For luciferase assays, pCMV lacZ was included in each experiment as a control for transfection efficiency. Luciferase and β-galactosidase activities were measured using Promega and Tropix kits, respectively, according to the manufacturer's instructions.
Immunoprecipitations and GST pulldown experiments.
Immunoprecipitations were performed by the method of Nicolas et al. (37). For glutathione S-transferase GST-pulldown experiments, Jurkat cell nuclear extracts or whole-cell extracts from transfected cells (prepared by the method of Nicolas et al. [37]) were diluted using IPH buffer (51) and subjected to a preclearing step with glutathione beads. Beads containing the various GST fusion proteins (prepared as described previously [37]) and with the amount of fusion protein standardized by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by Coomassie blue staining) were added to the precleared nuclear extracts, and reaction mixtures were incubated for 1 h at 4°C on a rotating wheel. For peptide competition experiments, beads were preincubated with peptides (simian virus 40 [SV40] T-antigen peptide, NEENLFCSEEMPSSDD; irrelevant peptide, GKEKSKEPRDPDQLYC) for 1 h at 4°C. After extensive washing, bound proteins were analyzed by Western blotting or were assayed for HMT activity. For phosphorylation experiments, GST-Rb beads were incubated for 1 h at 37°C in phosphorylation buffer (25 mM Tris [pH 7.5], 0.1 mM NaOV, 0.1 mM EGTA, 10 mM MgCl2, 0.04 mM dithiothreitol, 0.1 μM ZnCl2, 0.1 mM ATP) with purified baculovirus-expressed cyclin E-cdk2 (kind gift from B. Ducommun) (2). After extensive washing, beads were used in GST pulldown experiments.
HMT assays.
Beads from GST pulldown experiments or immunoprecipitations were resuspended in 30 μl of IPH buffer supplemented with 0.8 μM of S-adenosyl [methyl-3H]methionine (Amersham) and either 2 μg of histones (prepared from duck erythrocytes as described previously [51]) or 30 μM histone H3-derived peptides. The sequences of peptides were as follows: ARTKQTARKSTGGKAPRKQLATKA for H3 wt(1–24); the same sequence for K4Mut, K9Mut, and K14Mut, except that K4, K9, or K14 was replaced by an alanine; and ARTKQTARKSTGGKAPR for H3 wt(1–17). Methylation was then quantified using the filter binding assay as described previously (8).
Western blots and antibodies.
Western blots were performed using standard procedures. We used the following antibodies: 9E10 (Roche Diagnostics) as an anti-myc antibody (to detect myc-Suv39H1), KH95 (Santa Cruz) as an anti-E2F1 antibody, either C15 or C15G (Santa Cruz) as an anti-Rb antibody for immunoprecipitations, and either XZ55 or G3-245 (both from Pharmingen) for Western blots. Other antibodies are indicated in figure legends. To produce the anti-Suv39H1 antibody, a rabbit was immunized with two peptides derived from Suv39H1 (amino acids 67 to 82 and 101 to 115). Immune serum efficiently immunoprecipitated transfected myc-tagged Suv39H1 (data not shown).
RESULTS
Rb interacts with an HMT.
Recent reports have emphasized the notion that various chromatin modifications work in an interdependent manner (10, 18, 29, 36, 41, 48, 52, 56). Since Rb can repress E2F activity through HDs (6, 30, 31), we tested the possible involvement of other histone-modifying enzymes in Rb function.
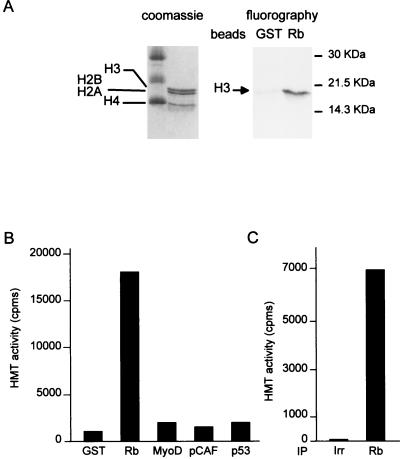
In order to test whether Rb interacts with an HMT activity, we incubated Jurkat cell nuclear extracts with beads containing either bacterially produced GST-Rb fusion protein or control GST. After extensive washing, bound proteins were assayed for HMT activity by adding S-adenosyl [methyl-3H]methionine and purified histones. Transfer of the methyl group to histones was then analyzed by SDS-PAGE. Fluorography results (Fig. 1A) indicated that Rb could physically recruit a histone H3-specific HMT from cell extracts. In control experiments, GST-Rb fusion protein did not show any HMT activity by itself (data not shown).
FIG. 1.
Physical interaction between Rb and an HMT. (A) Glutathione-agarose beads containing 2 μg of recombinant bacterially expressed GST Rb 379–928 (Rb) or control GST (GST) were incubated with 200 μl of Jurkat cell nuclear extracts. After extensive washing, beads were subjected to an HMT assay using 2 μg of purified histones. Histones were then separated by SDS-PAGE (18% polyacrylamide) and were detected by Coomassie blue staining or fluorography. (B) Beads containing the indicated GST fusion proteins were incubated with 25 μl of Jurkat cell nuclear extracts and, after extensive washing, were subjected to an HMT assay using the histone H3 peptide [wt(1–24)] as a substrate (final concentration, 30 μM). Methylation was quantified using the filter binding assay. (C) Jurkat cell nuclear extracts (150 μl) were subjected to immunoprecipitation (IP) using 5 μg of either a control anti-HA antibody (Irr) (Santa Cruz) or an anti-Rb antibody. Immunoprecipitates were then assayed for HMT activity as described above for panel B.
We also used a more quantitative assay, in which we directly spotted the reaction product on P81 chromatographic Whatman paper, and we performed the classical filter binding assay (8). Using this assay, we were able to show that a peptide derived from the first 24 amino acids from the histone H3 was efficiently methylated (Fig. 1B), indicating that the histone H3 N-terminal tail is the target of methylation. In addition, we found that the ability to recruit an HMT from cell extracts is specific to Rb, since neither recombinant MyoD, pCAF, nor p53 significantly interacted with any HMT activity.
These experiments relied on the use of large amounts of recombinant Rb protein. We thus tested whether the endogenous Rb protein was also complexed with an HMT. We found that immunoprecipitation of endogenous Rb from Jurkat cell nuclear extracts led to the coimmunoprecipitation of a high level of HMT activity (Fig. 1C), which was specific since it was not seen using an irrelevant antibody. Taken together, these results indicate that Rb is physically associated with a histone H3-specific HMT in living cells.
The ability to interact with an HMT correlates with the growth inhibitory properties of Rb.
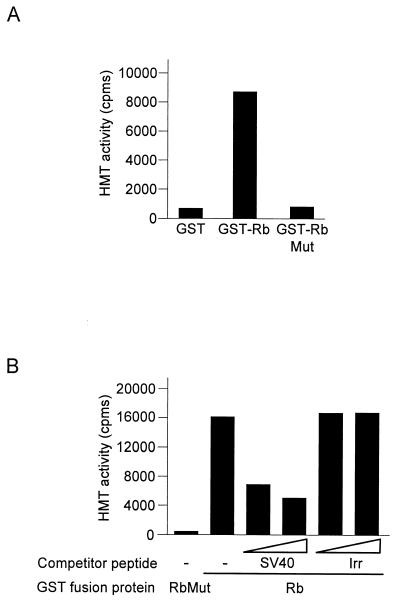
The retinoblastoma susceptibility gene is a hot spot of mutations in cancer, leading to the expression of an inactive protein. We tested whether a tumor-derived mutation abolishes the ability of Rb to interact with an HMT. As already shown (Fig. 1B), incubation of GST-Rb with Jurkat cell nuclear extracts led to the recruitment of a robust HMT activity (Fig. 2A). In a similar experiment, a tumor-derived point mutant of Rb was not able to recruit any significant HMT activity from cell extracts (GST-RbMut). This mutation resides in the so-called pocket domain of Rb, which is targeted by viral transforming proteins such as the SV40 T antigen. This domain is responsible for Rb binding to E2F and to proteins containing an LXCXE motif, a sequence found in many Rb-binding proteins of viral or cellular origin (22). To test which binding site of Rb was involved in the interaction, we performed peptide competition experiments (Fig. 2B). We found that preincubation of Rb beads with an LXCXE-containing peptide derived from the SV40 T antigen led to a specific decrease in the Rb-associated HMT, whereas an irrelevant peptide had no effect. This result suggests that the cellular HMT interacts with Rb through an LXCXE motif. Interestingly, Rb represses transcription and induces a permanent cell cycle arrest through the binding to LXCXE-containing proteins (9, 14). Thus, these data suggest that the ability to associate with an HMT could be important for transcriptional repression and growth suppression by Rb. Furthermore, the Rb-HMT interaction could be an important target of S-phase-inducing viral transforming proteins, such as the SV40 T antigen.
FIG. 2.
Binding of the HMT correlates with Rb antiproliferative activity. (A) Beads containing bacterially expressed GST Rb 379–928 (GST-Rb), GST Rb 379–928 706C-F (GST-Rb Mut), or control GST were incubated with 25 μl of Jurkat cell nuclear extracts and, after extensive washing, were subjected to an HMT assay using the histone H3 peptide [wt(1–24)] as a substrate (final concentration, 30 μM). Methylation was quantified using the filter binding assay. (B) Beads containing either fusion protein GST-Rb or GST-RbMut were used in pulldown reactions as described above for panel A, except that, prior to the addition of Jurkat cell nuclear extracts, beads were incubated with or without (−) 5 or 10 μg (amount indicated by the height of the white triangle) of an SV40 T-antigen-derived peptide or an irrelevant peptide (Irr), as indicated. Bound HMT activity was measured as described in the legend to Fig. 1B.
The Rb-associated HMT is the heterochromatin-associated Suv39H1 protein.
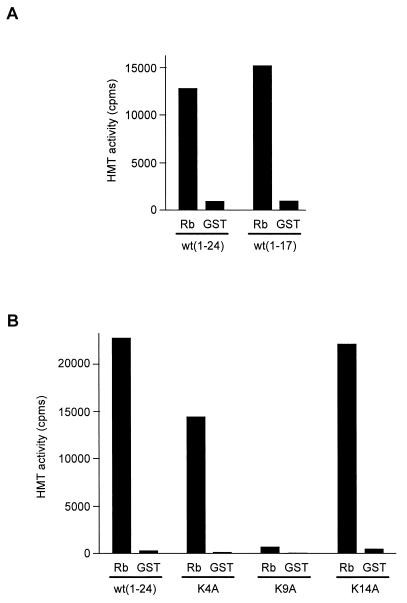
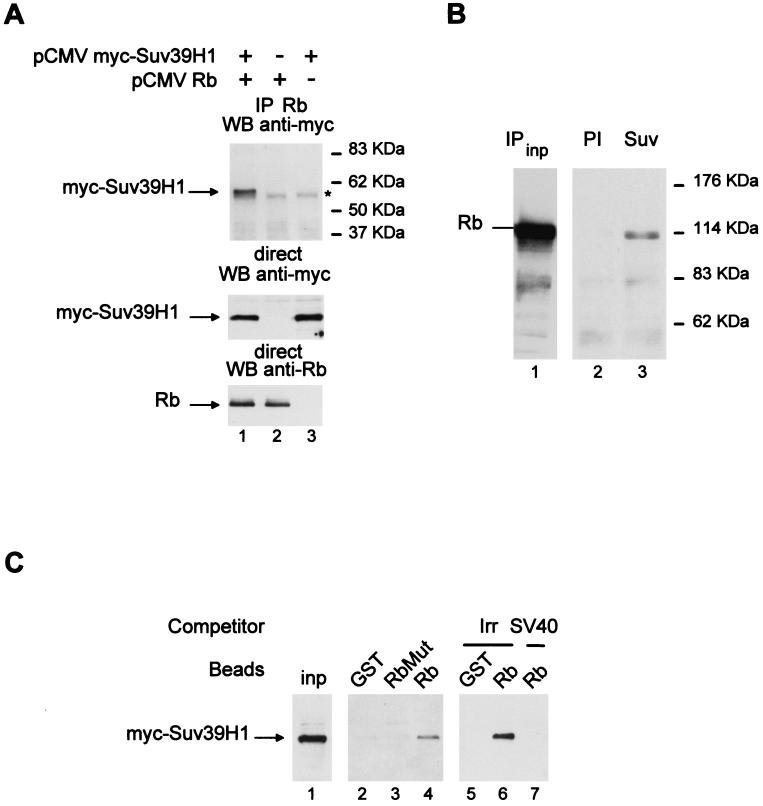
As a first step towards the identification of the Rb-associated HMT, we determined its substrate specificity. As already shown in Fig. 1B, a peptide containing the first 24 amino acids from histone H3 [wt(1–24)] was efficiently methylated by the Rb-interacting enzyme (Fig. 3A). By using a shorter peptide, we were able to demonstrate that the main methylation events occurred within the first 17 amino acids of histone H3 [wt(1–17)] (Fig. 3A). We then individually mutated each of the three lysines present within this peptide. When peptides mutated either at K4 or at K14 were used as substrates, no significant difference in the methylation efficiency could be detected (Fig. 3B). In contrast, a peptide mutated at K9 could not be methylated at all, indicating that K9 is the major methylation site of the Rb-interacting enzyme. This strict substrate specificity is reminiscent of the newly described Suv39H1 HMT (41). Suv39H1 is a mammalian homologue of Drosophila Su(Var)3.9, which is involved in pericentric heterochromatin formation (50). We thus tested whether Suv39H1 could interact with Rb. Immunoprecipitation of Rb from transfected-cell extracts led to the coimmunoprecipitation of transfected Suv39H1 (Fig. 4A, top gel, lane 1). This interaction was specific, since it was not detected in the absence of exogenous Rb (lane 3) or in the absence of transfected Suv39H1 (lane 2). Taken together, these data indicate that Rb interacts with Suv39H1 in transfected cells.
FIG. 3.
The Rb-associated HMT specifically methylates K9 from histone H3. (A) Beads containing either GST-Rb or control GST fusion protein were incubated with 25 μl of Jurkat cell nuclear extracts and were assayed for bound HMT activity using peptides containing either the first 24 amino acids [wt(1–24)] or the first 17 amino acids [wt(1–17)] of histone H3. (B) Beads containing either GST-Rb or control GST fusion protein were incubated with 25 μl of Jurkat cell nuclear extracts and were assayed for bound HMT activity using either the wild-type H3 peptide [wt(1–24)] or the same peptide with the indicated lysine mutated.
FIG. 4.
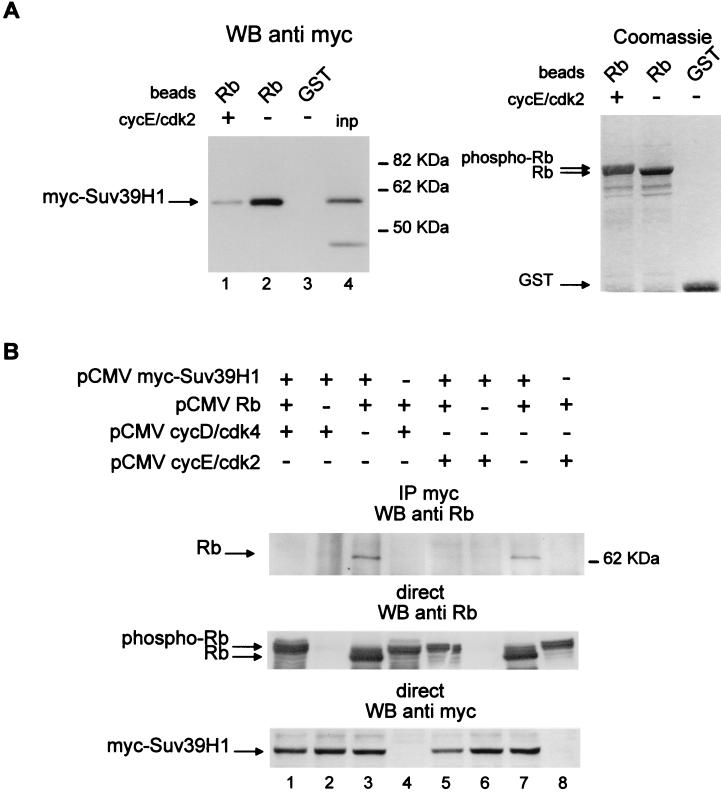
Suv39H1 physically associates with Rb. (A) U2OS cells were transfected with 10 μg of pCMV myc-Suv39H1 and/or pCMV-Rb 379–928 as indicated. Total cell extracts were then immunoprecipitated using an anti-Rb antibody, and immunoprecipitates (IP) were tested for the presence of myc-Suv39H1 by Western blotting (WB) (top gel). The position of the immunoglobulin heavy chain of the immunoprecipitating antibody is indicated by an asterisk. The expression levels of transfected Rb or myc-Suv39H1 are shown in the lower gels. (B) Jurkat cell nuclear extracts (200 μl) were immunoprecipitated with the indicated antibody (anti-Suv39H1 [Suv] or preimmune serum [PI]), and immunoprecipitates were tested for the presence of endogenous Rb by Western blotting using the G3-245 antibody (Pharmingen). In lane 1, 2 μl of Jurkat nuclear extracts was directly loaded as input (inp). (C) Beads containing 0.2 μg of GST, GST-Rb Mut, or GST-Rb fusion protein (middle gel) or 2 μg of GST-Rb (Rb) or control GST (GST) fusion protein (right gel) were incubated with whole extracts (80 μl) from cells transfected with 2.7 μg of myc-Suv39H1 expression vector. Competitor peptides (10 μg) were added where indicated (right gel). After extensive washing, the amount of myc-Suv39H1 pulled down was tested by Western blotting. In lane 1, whole-cell extracts (13 μl) were directly loaded as input.
The latter experiment was performed by overexpressing proteins. To test whether Rb and Suv39H1 produced at physiological levels were also physically associated in living cells, we performed coimmunoprecipitation experiments from Jurkat cell nuclear extracts.
Immunoprecipitation of endogenous Suv39H1 (Fig. 4B) led to the coimmunoprecipitation of endogenous Rb, whereas the control immunoprecipitation performed with the preimmune serum did not. These results indicate that endogenous Rb and Suv39H1 are physically present within the same complex.
The interaction between Rb and the cellular HMT is dependent upon Rb pocket integrity and is competed away by an LXCXE-containing peptide (Fig. 2B). We thus tested whether Rb interacts with Suv39H1 using the same domain. When incubated with whole-cell extracts, GST-Rb beads were able to recruit specifically transfected myc-Suv39H1 (Fig. 4C, lanes 4 and 6), whereas beads harboring the Rb point mutant RbMut were not (lane 3). Furthermore, this interaction is abolished in the presence of the LXCXE-containing SV40 peptide (compare lanes 6 and 7). Thus, Suv39H1 interacts with Rb through an LXCXE motif. Taken together, these data suggest that Suv39H1 is likely to be the Rb-associated HMT. Strikingly, Suv39H1 does not contain any sequence resembling the LXCXE motif, suggesting that it interacts with Rb indirectly (see Discussion).
The Rb-Suv39H1 interaction is abolished by Rb phosphorylation.
The activity of the retinoblastoma protein is controlled by its phosphorylation by cyclin D- and cyclin E-dependent kinases (40). Consequently, protein-protein interactions critical for the growth inhibitory functions of Rb are abolished upon Rb phosphorylation. We thus tested the effect of Rb phosphorylation on its ability to interact with Suv39H1. As already shown (Fig. 4C), incubation of transfected-cell extracts with GST-Rb beads led to the specific recruitment of exogenous myc-Suv39H1 (Fig. 5A, left gel, compare lane 2 and lane 3). When GST-Rb beads phosphorylated in vitro by purified recombinant cyclin E-cdk2 complex were used, we found that the amount of Suv39H1 retained on the beads was largely reduced (compare lanes 1 and 2), although the amount of recombinant Rb protein was similar (right gel). Thus, in vitro phosphorylation of Rb by cyclin E-cdk2 reduces its ability to interact with Suv39H1.
FIG. 5.
Phosphorylation of Rb abolishes its interaction with Suv39H1. (A) Beads containing GST-Rb or GST proteins were phosphorylated in vitro using purified cyclin E-cdk2 kinase complex. The extent of Rb phosphorylation was assessed by Coomassie blue staining. Note the slight change in the migration velocity of GST-Rb upon cyclin E-cdk2 treatment. These beads were used in GST pulldown experiments with whole extracts from cells transfected with the myc-Suv39H1 expression vector, as described in the legend to Fig. 4C (left gel). Bound proteins were detected by Western blotting (WB) using the anti-myc antibody. In lane 4, whole-cell extracts (13 μl) were loaded directly as input (inp). (B) U2OS cells were transfected as described in the legend to Fig. 3B with 10 μg of the indicated expression vectors. Total cell extracts were immunoprecipitated with the anti-myc antibody, and immunoprecipitates (IP) were tested for the presence of Rb by Western blotting (WB) (top gel). In the middle and bottom gels, expression levels of transfected Rb and myc-Suv39H1 are shown. Exogenous Rb migrates at about 60 kDa because the N-terminal part of the protein was deleted (see Materials and Methods). Note that addition of cyclin-cdk expression vectors led to a shift in the migration of transfected Rb (phosphorylated Rb [phospho-Rb] in the middle gel).
To test whether the same was true in living cells, we transfected U2OS cells with Rb and Suv39H1 expression vectors in the presence or absence of exogenous cyclin-cdk's. As expected, in the absence of exogenous kinase, immunoprecipitation of myc-Suv39H1 led to the coimmunoprecipitation of exogenous Rb (Fig. 5B, top gel, lanes 3 and 7). In the presence of exogenous kinases, exogenous Rb was efficiently phosphorylated, as indicated by the shift in its migration (middle gel, compare lanes 3 and 7 to lanes 1, 4, 5, and 8). Phosphorylated Rb was not significantly coimmunoprecipitated with myc-Suv39H1 (lanes 1 and 5), although Rb and Suv39H1 were expressed at high levels (middle and bottom gels, lanes 1 and 5). Thus, phosphorylation of Rb by cyclin-cdk's abolishes its ability to interact with Suv39H1 in living cells.
Suv39H1 functions as a transcriptional corepressor of the E2F transcription factor.
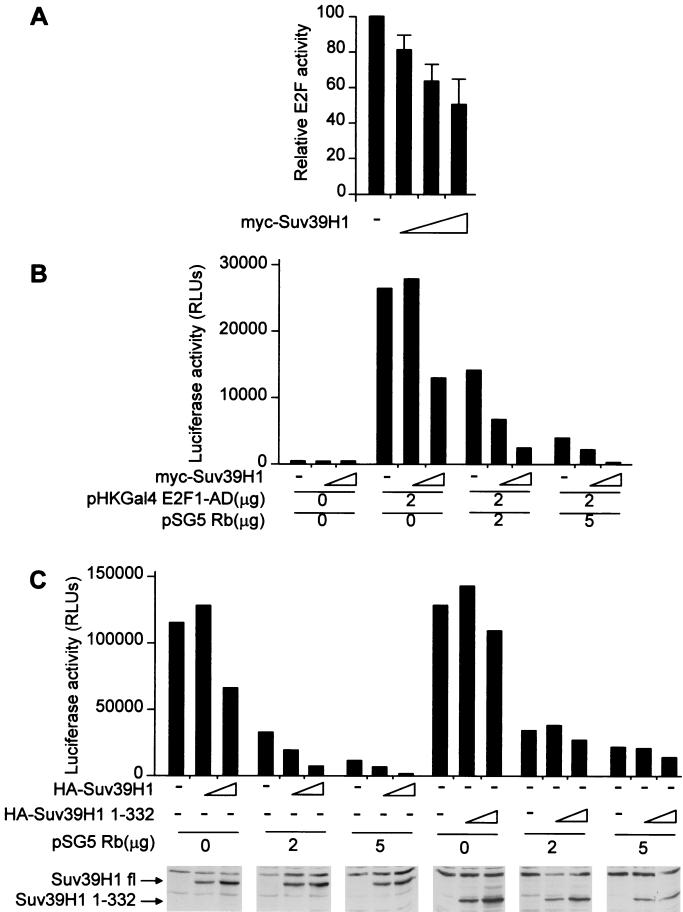
One of the major targets of Rb is the E2F transcription factor (22). To test whether Suv39H1 could be involved in transcriptional regulation by E2F, we transfected HeLa cells, which express low levels of endogenous Suv39H1 (1), with a reporter construct in which the luciferase-encoding gene is cloned downstream of E2F sites (Fig. 6A). Increasing amounts of Suv39H1 expression vector led to a dose-dependent repression of this reporter vector, indicating that Suv39H1 repressed E2F activity (measured as the ratio between the activity of the E2F-TK luciferase reporter vector to the activity of the empty thymidine kinase [TK] luciferase reporter vector).
FIG. 6.
Suv39H1 represses E2F activity. (A) HeLa cells were transiently transfected with 2 μg of E2F-TK luc or control TK-luc reporter vectors, 20 ng of pCMV NeoBam, and 100 ng of pCMV lacZ to monitor transfection efficiency, and with increasing amounts of myc-Suv39H1 expression vector (0, 0.5, 1, and 2 μg [amount indicated by the height of the white triangle]). Luciferase and β-galactosidase activities were measured 48 h later. E2F activity (normalized to that of empty reporter construct) was calculated relative to 100% in the absence (−) of exogenous Suv39H1. The means of four independent experiments are shown. (B) HeLa cells were transiently transfected with 2 μg of GAL4-luc reporter construct, with the indicated amount of SV40 promoter-driven Gal4 E2F1-AD (pHKGal4 E2F1-AD) and/or Rb (pSG5 Rb) expression vectors and with either 0, 1, or 2 μg of pCMV myc-Suv39H1 expression vector. Luciferase activity (in relative light units [RLU]) was measured 48 h later. The result of a typical experiment is shown. Note that transcriptional repression by Rb is more efficient in the presence than in the absence (−) of exogenous Suv39H1. (C) HeLa cells were transiently transfected with 2 μg of GAL4-luc reporter construct, 2 μg of pHKGal4 E2F1-AD, the indicated amount of pSG5 Rb, and various amounts (0, 1, or 2 μg) of either pCMV 2N3T Suv39H1 (HA-Suv39H1) or pCMV 2N3T Suv39H1 1–332 (HA-Suv39H1 1–332), as indicated. Luciferase activity was measured 48 h later. The result of a typical experiment is shown. In the lower panels, the expression levels of HA-tagged Suv39H1 fl or Suv39H1 1–332 were assayed by anti-HA Western blotting.
Since Suv39H1 interacts with Rb (see above), we wondered whether Suv39H1 could cooperate with Rb for transcriptional repression. In order to test this hypothesis, we transfected HeLa cells with a GAL4 luciferase reporter vector and an expression vector for Gal4-E2F1AD (E2F1 activation domain) fusion protein (Fig. 6B). Increasing doses of an expression vector for myc-tagged Suv39H1 did not have any effect in the absence of Gal4-E2F1 and induced a slight repression in its presence (two fold at most). In contrast, in the presence of exogenous Rb, Suv39H1 led to an important decrease in Gal4-E2F1 activity (up to 10-fold repression in the presence of 5 μg of pSG5 Rb). This decrease is unlikely to be due to changes in the expression of the transfected proteins, since both Rb and Gal4-E2F1 were expressed from the SV40 promoter, which was not affected by overexpressed Suv39H1 (data not shown). Furthermore, the expression of exogenous Suv39H1 was slightly lower in the presence of Rb than in its absence (data not shown). Similarly, the efficiency of transcriptional repression by Rb increased in the presence of exogenous Suv39H1. For example in Fig. 6B, for 2 μg of Rb expression vector, the efficiency went from less than twofold in the absence of exogenous Suv39H1 up to sixfold with 2 μg of Suv39H1 expression vector. Taken together, these data indicate that Rb and Suv39H1 cooperate to repress E2F activity and support Suv39H1 acting as a corepressor of E2F.
In order to test whether the methyltransferase activity of Suv39H1 is involved in this corepressor activity, we constructed an expression vector for either Suv39H1 fl (Suv39H1) or Suv39H1 1-332, in which some amino acids critical for its HMT activity have been deleted (41), both tagged with HA epitopes and nuclear localization signals. As expected, we found that, like myc-Suv39H1 in Fig. 6B, HA-Suv39H1 was able to cooperate with Rb to repress E2F activity (Fig. 6C). The version of Suv39H1 with a deletion (Suv39H1 1-332) had hardly any effect on E2F activity, either in the absence or presence of Rb. Since this mutant was well expressed (Fig. 6C) and bound Rb as efficiently as the wild type in GST pulldown assays (data not shown), this result suggests that the HMT activity of Suv39H1 is required for its ability to cooperate with Rb.
Rb targets Suv39H1 to the E2F1 activation domain.
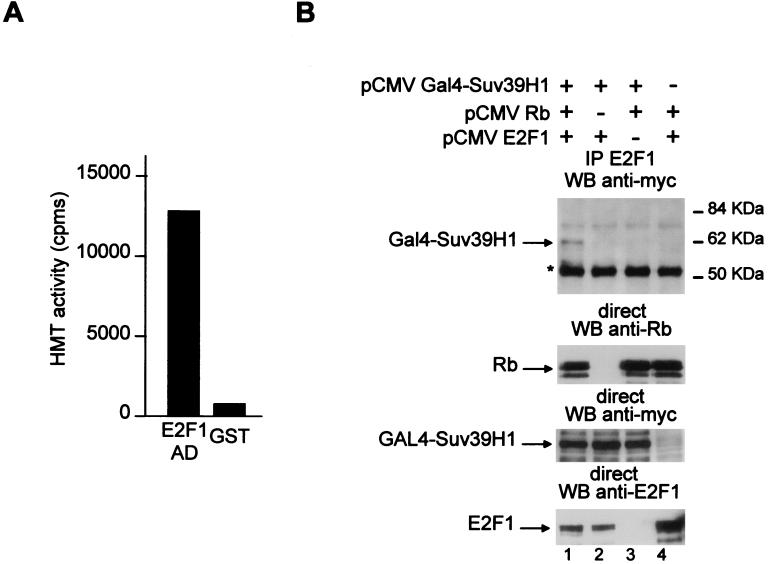
We then hypothesized that Suv39H1 could be recruited to E2F-regulated promoters through its physical interaction with Rb. To test this possibility, we first investigated whether E2F1 could interact with a cellular HMT. We produced beads harboring recombinant bacterially produced fusion proteins in which the activation domain of E2F1 is fused to GST. When these beads were used in GST pulldown experiments, as in Fig. 1B, we found that they efficiently recruited HMT activity from cell extracts (Fig. 7A). Thus, the E2F1 activation domain, which binds Rb, interacts with a cellular HMT.
FIG. 7.
Ternary complex formation between E2F1, Rb, and Suv39H1. (A) Beads containing 10 μg of either GST-E2F1 359–437 (E2F1 AD) or control GST fusion proteins were incubated with 200 μl of Jurkat cell nuclear extract, and bound proteins were assayed for HMT activity. (B) U2OS cells were transfected as described in the legend to Fig. 3B with 5 to 15 μg of the indicated expression vectors. Note that the Gal4-Suv39H1 fusion protein is tagged with the myc tag. Total cell extracts were immunoprecipitated with the anti-E2F1 antibody, and immunoprecipitates were tested for the presence of Gal4-Suv39H1 by Western blotting (WB) (top gel). The position of the immunoglobulin heavy chain of the immunoprecipitating antibody is indicated by an asterisk. In the lower gels, expression levels of transfected Rb, Gal4-Suv39H1, and E2F1 are shown.
We then intended to test whether this last result was due to the formation of a ternary complex containing E2F1, Rb, and Suv39H1. To that end, we transfected U2OS cells with expression vectors encoding E2F1, Rb, and GAL4-myc-Suv39H1 fusion protein. We used this larger protein version rather than myc-Suv39H1, because the latter protein could not be adequately detected in the immunoprecipitates due to its comigration with the strong band of the heavy chain of the immunoprecipitating anti-E2F1 antibody (Fig. 7B, top gel). In the presence of all three expression vectors, immunoprecipitation of E2F1 led to the coimmunoprecipitation of GAL4-myc-Suv39H1 (Fig. 7B, top gel, lane 1). This coimmunoprecipitation was specific, since it was not seen in the absence of exogenous E2F1 (lane 3) or GAL4-myc-suv39H1 (lane 4). Interestingly, this coimmunoprecipitation was also dependent upon the presence of exogenous Rb (lane 2), although GAL4-myc-Suv39H1 and E2F1 expressions were similar (lower panels). Taken together, these results indicate that Rb can recruit Suv39H1 to the E2F1 protein through physical interactions with both proteins.
Suv39H1 represses transcription once recruited on a promoter.
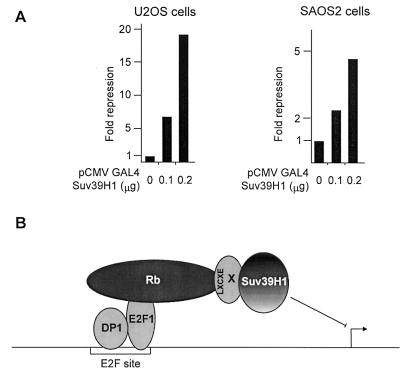
We therefore tested the effect on transcription of Suv39H1 recruitment to a heterologous promoter. U2OS cells were transfected with a reporter vector in which the luciferase gene is cloned downstream of GAL4 sites. Increasing amounts of an expression vector for Suv39H1 fused to the Gal4 DNA binding domain led to a dose-dependent repression of luciferase activity (Fig. 8A, left panel), which required the Gal4 DNA binding domain (data not shown). Furthermore, it had no effect on the same reporter vector without GAL4 sites (data not shown). These data indicate that the HMT Suv39H1 represses transcription when targeted to a promoter, as already shown by others in a different cell type (17). Similar repression, albeit less efficient, was also observed in Rb-negative SAOS2 cells (right panel), indicating that transcriptional repression by Suv39H1 is not a mere consequence of binding to Rb.
FIG. 8.
Transcriptional repression through artificial recruitment of Suv39H1. (A) U2OS cells or Rb-negative SAOS2 cells were transiently transfected with 2 μg of the GAL4-luciferase reporter construct and the indicated doses of pCMVGal4-Suv39H1. Fold repression by Gal4-Suv39H1 is calculated relative to luciferase activity in the absence of Gal4-Suv39H1 expression vector. (B) Model for transcriptional repression of E2F-regulated promoters by Suv39H1. Rb recruits Suv39H1 through an indirect interaction (protein X, which contains an LXCXE motif) to E2F1 and E2F-regulated promoters. Once on the promoter, Suv39H1 represses transcription.
DISCUSSION
In this paper, we show that Rb physically interacts with an HMT, which is likely to be Suv39H1. This interaction could be important for the function of Rb, since (i) it is dependent upon the integrity of the pocket domain of Rb (Fig. 2 and 4), (ii) it is competed away by peptides derived from viral transforming proteins (Fig. 2 and 4), and (iii) it is lost upon phosphorylation of Rb by cyclin-dependent kinases (Fig. 5). Through this physical interaction, Rb could recruit Suv39H1 to the E2F transcription factor (Fig. 7), leading to the repression of E2F-regulated promoters (Fig. 6). Taken together, our results suggest a model in which transcriptional repression of E2F-regulated promoters could involve the recruitment of the HMT Suv39H1 (Fig. 8B). Consistent with this model, Rb interacts with Suv39H1 through its transcriptional repression domain (LXCXE-dependent) (Fig. 4C) (14). It has to be noted, however, that we did not succeed in directly demonstrating the recruitment of Suv39H1 to E2F-regulated promoters by chromatin immunoprecipitations.
Many other proteins, including HDs, have been proposed to be involved in the transcriptional repression by Rb (6, 30, 31). Thus, it is important to understand to what extent Suv39H1 is important for transcriptional repression by Rb. Transcriptional repression by Rb in vitro requires the DNA template to be assembled in nucleosomes (43). However, HD inhibitors have no effect on this repression (43). Thus, these results indicate that Rb represses transcription through a mechanism involving nucleosomes but independent of the acetylation status of histones. Our results suggest that histone methylation could be involved in this in vitro repression. What about in vivo? No specific inhibitors of HMTs are available so far. Furthermore, we did not manage to inhibit Suv39H1 expression by using antisense RNA or double-stranded RNA (data not shown). However, most importantly, in mouse embryo fibroblasts derived from Suv39H1 and Suv39H2−/− mice, cyclin E expression is deregulated (T. Kouzarides, personal communication). Since cyclin E is regulated through E2F sites, this experiment strongly suggests that Suv39H1 is important for Rb function. Consistent with that, overexpression of Suv39H1 in 3T3 cells induces a slight decrease in the percentage of cells that enter S phase (17).
The interaction of Suv39H1 and Rb is competed away by an LXCXE-containing peptide. However, analysis of the primary structure of human Suv39H1 does not show any sequence resembling a known LXCXE motif (data not shown). Although at this stage we cannot rule out the possibility of direct contacts between Rb and Suv39H1, this result suggests that the interaction between Suv39H1 and Rb is indirect and is mediated through an LXCXE-containing protein.
Suv39H1 is thought to be involved in heterochromatin formation. Indeed, it localizes at heterochromatic foci in mammalian interphase cells (1, 15, 32). Furthermore, Drosophila and S. pombe homologues of Suv39H1 have been cloned in genetic screens for proteins involved in silencing through heterochromatin (3, 15, 50). The fact that Suv39H1 physically interacts with Rb suggests a link between Rb and heterochromatin. Transcriptional repression by Rb could thus involve the formation on E2F-regulated genes of a heterochromatin-like structure. Such a mechanism has already been proposed for transcriptional repression by the repressor KAP1 (also called TIF1β) (38). Through their interaction with Suv39H1, E2F-regulated genes could be relocalized within the cell nuclei to a heterochromatic compartment. A similar silencing through relocalization has already been described for transcriptional repression by the differentiation-associated protein Ikaros (7). Indeed, subnuclear localization appears to be an important feature of transcriptional regulation (13).
To our knowledge, our results are the first example of an HMT being involved in transcriptional regulation by sequence-specific transcription factors. What could be the molecular basis for this repressing effect? The methyltransferase activity of Clr4, the S. pombe homologue of Suv39H1, is required for silencing through heterochromatin formation (4). Suv39H1 specifically methylates K9 from histone H3. Recent results indicate that the histone H3 methylated on K9 is a binding site for proteins from the HP1 family (4, 25). According to these studies, HP1 would recognize methylated histone H3 through its chromo-domain. HP1β, a member of the HP1 family, also interacts with Suv39H1 (1). Thus, the presence of Suv39H1 on a promoter could induce the formation of a high-affinity binding site for proteins of the HP1 family, resulting both from the methylation of histone H3 (4, 25) and from the physical interaction with Suv39H1 itself (1). Transcriptional repression would then result from HP1 recruitment, consistent with previous observations (27, 28, 38).
Another important question raised by our results deals with the relationship between HDs and HMTs. Indeed, Rb has been shown to repress transcription through the recruitment of HDs, including the histone deacetylase HDAC1 (6, 30, 31). Our results suggest that it also involves the HMT Suv39H1. Furthermore, silencing through heterochromatin involves Suv39H1 homologues (3, 50), and histones within heterochromatin are largely hypoacetylated (20). Finally, we found that transcriptional repression by Suv39H1 of a heterologous promoter requires HDs (our unpublished results). What could be the basis of this cooperation? A likely possibility invokes the existence of a physical interaction between Suv39H1 and HDs. Alternatively, since both enzymes modify the same substrate, it is tempting to speculate that one modification might affect the efficiency of the other. Indeed, such influences between various histone posttranslational modifications have already been documented (10, 29, 41). Thus, methylation of histone H3 could favor its deacetylation, or conversely, methylation of deacetylated histones could be more efficient. Consistent with this explanation, acetylation of K9 of histone H3 blocks methylation by Suv39H1 (41). Thus, deacetylation of K9 by HDs could be required for histone H3 methylation on K9. Such a mechanism would be consistent with the observation that localization of HP1 proteins, which is dependent upon histone H3 K9 methylation, is slowly lost upon inhibition of HDs (46). Also, recent results indicate that, in S. pombe, K9 methylation by Clr4 is dependent upon the activity of the histone deacetylase Clr3 (35).
ACKNOWLEDGMENTS
L. Vandel and E. Nicolas contributed equally to this work.
We thank T. Jenuwein, A. Harel-Bellan, H. Richard-Foy, T. Kouzarides, and B. Ducommun for materials, and we thank M. Grigoriev, H. Richard-Foy, and C. Monod for critical reading of the manuscript.
This work was supported in part by grants from the Ligue Nationale Contre le Cancer as an Equipe Labellisée and from the French Ministry as an Action Concertée Incitative. E.N. and L.V. are recipients of a scholarship from the French ministry and a fellowship from the Association de Recherche sur le Cancer, respectively. R.F. is supported by the Comité du Val d'Oise de la Ligue contre le Cancer.
REFERENCES
- 1.Aagaard L, Laible G, Selenko P, Schmid M, Dorn R, Schotta G, Kuhfittig S, Wolf A, Lebersorger A, Singh P B, Reuter G, Jenuwein T. Functional mammalian homologues of the Drosophila PEV-modifier Su(var)3-9 encode centromere-associated proteins which complex with the heterochromatin component M31. EMBO J. 1999;18:1923–1938. doi: 10.1093/emboj/18.7.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ait-Si-Ali S, Ramirez S, Barre F X, Dkhissi F, Magnaghi-Jaulin L, Girault J A, Robin P, Knibiehler M, Pritchard L L, Ducommun B, Trouche D, Harel-Bellan A. Histone acetyltransferase activity of CBP is controlled by cycle-dependent kinases and oncoprotein E1A. Nature. 1998;396:184–186. doi: 10.1038/24190. [DOI] [PubMed] [Google Scholar]
- 3.Allshire R C, Nimmo E R, Ekwall K, Javerzat J P, Cranston G. Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev. 1995;9:218–233. doi: 10.1101/gad.9.2.218. [DOI] [PubMed] [Google Scholar]
- 4.Bannister A J, Zegerman P, Partridge J F, Miska E A, Thomas J O, Allshire R C, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- 5.Braunstein M, Sobel R E, Allis C D, Turner B M, Broach J R. Efficient transcriptional silencing in Saccharomyces cerevisiae requires a heterochromatin histone acetylation pattern. Mol Cell Biol. 1996;16:4349–4356. doi: 10.1128/mcb.16.8.4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brehm A, Miska E A, McCance D J, Reid J L, Bannister A J, Kouzarides T. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature. 1998;391:597–601. doi: 10.1038/35404. [DOI] [PubMed] [Google Scholar]
- 7.Brown K E, Guest S S, Smale S T, Hahm K, Merkenschlager M, Fisher A G. Association of transcriptionally silent genes with Ikaros complexes at centromeric heterochromatin. Cell. 1997;91:845–854. doi: 10.1016/s0092-8674(00)80472-9. [DOI] [PubMed] [Google Scholar]
- 8.Brownell J E, Allis C D. An activity gel assay detects a single, catalytically active histone acetyltransferase subunit in Tetrahymena macronuclei. Proc Natl Acad Sci USA. 1995;92:6364–6368. doi: 10.1073/pnas.92.14.6364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen T-T, Wang J Y. Establishment of irreversible growth arrest in myogenic differentiation requires the RB LXCXE-binding function. Mol Cell Biol. 2000;20:5571–5580. doi: 10.1128/mcb.20.15.5571-5580.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheung P, Tanner K G, Cheung W L, Sassoni-Corsi P, Denu J M, Allis C D. Synergistic coupling of histone H3 phosphorylation and acetylation in response to epidermal growth factor stimulation. Mol Cell. 2000;5:905–915. doi: 10.1016/s1097-2765(00)80256-7. [DOI] [PubMed] [Google Scholar]
- 11.Chow K N, Dean D C. Domains A and B in the Rb pocket interact to form a transcriptional repressor motif. Mol Cell Biol. 1996;16:4862–4868. doi: 10.1128/mcb.16.9.4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chow K N, Starostik P, Dean D C. The Rb family contains a conserved cyclin-dependent-kinase-regulated transcriptional repressor motif. Mol Cell Biol. 1996;16:7173–7181. doi: 10.1128/mcb.16.12.7173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cockell M, Gasser S M. Nuclear compartments and gene regulation. Curr Opin Genet Dev. 1999;9:199–205. doi: 10.1016/S0959-437X(99)80030-6. [DOI] [PubMed] [Google Scholar]
- 14.Dahiya A, Gavin M R, Luo R X, Dean D C. Role of the LXCXE binding site in Rb function. Mol Cell Biol. 2000;20:6799–6805. doi: 10.1128/mcb.20.18.6799-6805.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eissenberg J C, Elgin S C. The HP1 protein family: getting a grip on chromatin. Curr Opin Genet Dev. 2000;10:204–210. doi: 10.1016/s0959-437x(00)00058-7. [DOI] [PubMed] [Google Scholar]
- 16.Field S J, Tsai F Y, Kuo F, Zubiaga A M, Kaelin W G, Jr, Livingston D M, Orkin S H, Greenberg M E. E2F-1 functions in mice to promote apoptosis and suppress proliferation. Cell. 1996;85:549–561. doi: 10.1016/s0092-8674(00)81255-6. [DOI] [PubMed] [Google Scholar]
- 17.Firestein R, Cui X, Huie P, Cleary M L. Set domain-dependent regulation of transcriptional silencing and growth control by SUV39H1, a mammalian ortholog of Drosophila Su(var)3-9. Mol Cell Biol. 2000;20:4900–4909. doi: 10.1128/mcb.20.13.4900-4909.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fuks F, Burgers W A, Brehm A, Hughes-Davies L, Kouzarides T. DNA methyltransferase Dnmt1 associates with histone deacetylase activity. Nat Genet. 2000;24:88–91. doi: 10.1038/71750. [DOI] [PubMed] [Google Scholar]
- 19.Grana X, Garriga J, Mayol X. Role of the retinoblastoma protein family, pRB, p107 and p130 in the negative control of cell growth. Oncogene. 1998;17:3365–3383. doi: 10.1038/sj.onc.1202575. [DOI] [PubMed] [Google Scholar]
- 20.Grunstein M. Yeast heterochromatin: regulation of its assembly and inheritance by histones. Cell. 1998;93:325–328. doi: 10.1016/s0092-8674(00)81160-5. [DOI] [PubMed] [Google Scholar]
- 21.Hagemeier C, Cook A, Kouzarides T. The retinoblastoma protein binds E2F residues required for activation in vivo and TBP binding in vitro. Nucleic Acids Res. 1993;21:4998–5004. doi: 10.1093/nar/21.22.4998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harbour J W, Dean D C. Rb function in cell-cycle regulation and apoptosis. Nat Cell Biol. 2000;2:E65–E67. doi: 10.1038/35008695. [DOI] [PubMed] [Google Scholar]
- 23.Krek W, Xu G, Livingston D M. Cyclin A-kinase regulation of E2F-1 DNA binding function underlies suppression of an S phase checkpoint. Cell. 1995;83:1149–1158. doi: 10.1016/0092-8674(95)90141-8. [DOI] [PubMed] [Google Scholar]
- 24.Kuo M H, Allis C D. Roles of histone acetyltransferases and deacetylases in gene regulation. Bioessays. 1998;20:615–626. doi: 10.1002/(SICI)1521-1878(199808)20:8<615::AID-BIES4>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 25.Lachner M, O'Carrol D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 26.La Thangue N B. E2F and the molecular mechanisms of early cell-cycle control. Biochem Soc Trans. 1996;24:54–59. doi: 10.1042/bst0240054. [DOI] [PubMed] [Google Scholar]
- 27.Le Douarin B, Nielsen A L, Garnier J M, Ichinose H, Jeanmougin F, Losson R, Chambon P. A possible involvement of TIF1 alpha and TIF1 beta in the epigenetic control of transcription by nuclear receptors. EMBO J. 1996;15:6701–6715. [PMC free article] [PubMed] [Google Scholar]
- 28.Lehming N, Le Saux A, Schuller J, Ptashne M. Chromatin components as part of a putative transcriptional repressing complex. Proc Natl Acad Sci USA. 1998;95:7322–7326. doi: 10.1073/pnas.95.13.7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lo W-S, Trievel R C, Rojas J R, Duggan L, Hsu J-Y, Allis C D, Marmostein R, Berger S L. Phosphorylation of serine 10 in histone H3 is functionally linked in vitro and in vivo to Gcn5-mediated acetylation at lysine 14. Mol Cell. 2000;5:917–926. doi: 10.1016/s1097-2765(00)80257-9. [DOI] [PubMed] [Google Scholar]
- 30.Luo R X, Postigo A A, Dean D C. Rb interacts with histone deacetylase to repress transcription. Cell. 1998;92:463–473. doi: 10.1016/s0092-8674(00)80940-x. [DOI] [PubMed] [Google Scholar]
- 31.Magnaghi-Jaulin L, Groisman R, Naguibneva I, Robin P, Lorain S, Le Villain J P, Troalen F, Trouche D, Harel-Bellan A. Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature. 1998;391:601–605. doi: 10.1038/35410. [DOI] [PubMed] [Google Scholar]
- 32.Minc E, Courvalin J C, Buendia B. HP1 gamma associates with euchromatin and heterochromatin in mammalian nuclei and chromosomes. Cytogenet Cell Genet. 2000;90:279–284. doi: 10.1159/000056789. [DOI] [PubMed] [Google Scholar]
- 33.Morkel M, Wenkel J, Bannister A J, Kouzarides T, Hagemeier C. An E2F-like repressor of transcription. Nature. 1997;390:567–568. doi: 10.1038/37507. [DOI] [PubMed] [Google Scholar]
- 34.Muller H, Helin K. The E2F transcription factors: key regulators of cell proliferation. Biochim Biophys Acta. 2000;1470:M1–M12. doi: 10.1016/s0304-419x(99)00030-x. [DOI] [PubMed] [Google Scholar]
- 35.Nakayama J, Rice J C, Strahl B D, Allis C D, Grewal S I. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- 36.Ng H H, Bird A. DNA methylation and chromatin modification. Curr Opin Genet Dev. 1999;9:158–163. doi: 10.1016/s0959-437x(99)80024-0. [DOI] [PubMed] [Google Scholar]
- 37.Nicolas E, Morales V, Magnaghi-Jaulin L, Harel-Bellan A, Richard-Foy H, Trouche D. RbAp48 belongs to the histone deacetylase complex that associates with the retinoblastoma protein. J Biol Chem. 2000;275:9797–9804. doi: 10.1074/jbc.275.13.9797. [DOI] [PubMed] [Google Scholar]
- 38.Nielsen A L, Ortiz J A, You J, Oulad-Abdelghani M, Khechumian R, Gansmuller A, Chambon P, Losson R. Interaction with members of the heterochromatin protein 1 (HP1) family and histone deacetylation are differentially involved in transcriptional silencing by members of the TIF1 family. EMBO J. 1999;18:6385–6895. doi: 10.1093/emboj/18.22.6385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O'Carroll D, Scherthan H, Peters A H, Opravil S, Haynes A R, Laible G, Rea S, Schmid M, Lebersorger A, Jerratsch M, Sattler L, Mattei M G, Denny P, Brown S D, Schweizer D, Jenuwein T. Isolation and characterization of suv39h2, a second histone H3 methyltransferase gene that displays testis-specific expression. Mol Cell Biol. 2000;20:9423–9433. doi: 10.1128/mcb.20.24.9423-9433.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Planassilva M D, Weinberg R A. The restriction point and control of cell proliferation. Curr Opin Cell Biol. 1997;9:768–772. doi: 10.1016/s0955-0674(97)80076-2. [DOI] [PubMed] [Google Scholar]
- 41.Rea S, Eisenhaber F, O'Carroll D, Strahl B D, Sun Z W, Schmid M, Opravil S, Mechtler K, Ponting C P, Allis C D, Jenuwein T. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- 42.Rogakou E P, Pilch D R, Orr A H, Ivanova V S, Bonner W M. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 43.Ross J F, Näär A, Cam H, Gregory R, Dynlacht B D. Active repression and E2F inhibition by pRB are biochemically distinguishable. Genes Dev. 2001;15:392–397. doi: 10.1101/gad.858501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Starostik P, Chow K N, Dean D C. Transcriptional repression and growth suppression by the p107 pocket protein. Mol Cell Biol. 1996;16:3606–3614. doi: 10.1128/mcb.16.7.3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Strahl B D, Allis C D. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 46.Taddei A, Maison C, Roche D, Almouzni G. Reversible disruption of pericentric heterochromatin and centromere function by inhibiting deacetylases. Nat Cell Biol. 2001;3:114–120. doi: 10.1038/35055010. [DOI] [PubMed] [Google Scholar]
- 47.Takahashi Y, Rayman J B, Dynlacht B D. Analysis of promoter binding by the E2F and pRB families in vivo: distinct E2F proteins mediate activation and repression. Genes Dev. 2000;14:804–816. [PMC free article] [PubMed] [Google Scholar]
- 48.Tong J K, Hassig C A, Schnitzler G R, Kingston R E, Schreiber S L. Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature. 1998;395:917–921. doi: 10.1038/27699. [DOI] [PubMed] [Google Scholar]
- 49.Trimarchi J M, Fairchild B, Verona R, Moberg K, Andon N, Lees J A. E2F-6, a member of the E2F family that can behave as a transcriptional repressor. Proc Natl Acad Sci USA. 1998;95:2850–2855. doi: 10.1073/pnas.95.6.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tschiersch B, Hofmann A, Krauss V, Dorn R, Korge G, Reuter G. The protein encoded by the Drosophila position-effect variegation suppressor gene Su(var)3-9 combines domains of antagonistic regulators of homeotic gene complexes. EMBO J. 1994;13:3822–3831. doi: 10.1002/j.1460-2075.1994.tb06693.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vandel L, Trouche D. Physical association between the histone acetyl transferase CBP and a histone methyl transferase. EMBO Rep. 2001;2:21–26. doi: 10.1093/embo-reports/kve002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wade P A, Jones P L, Vermaak D, Wolffe A P. A multiple subunit Mi-2 histone deacetylase from Xenopus laevis cofractionates with an associated Snf2 superfamily ATPase. Curr Biol. 1998;8:843–846. doi: 10.1016/s0960-9822(98)70328-8. [DOI] [PubMed] [Google Scholar]
- 53.Wade P A, Pruss D, Wolffe A P. Histone acetylation: chromatin in action. Trends Biochem Sci. 1997;22:128–132. doi: 10.1016/s0968-0004(97)01016-5. [DOI] [PubMed] [Google Scholar]
- 54.Weintraub S J, Chow K N, Luo R X, Zhang S H, He S, Dean D C. Mechanism of active transcriptional repression by the retinoblastoma protein. Nature. 1995;375:812–815. doi: 10.1038/375812a0. [DOI] [PubMed] [Google Scholar]
- 55.Weintraub S J, Prater C A, Dean D C. Retinoblastoma protein switches the E2F site from positive to negative element. Nature. 1992;358:259–261. doi: 10.1038/358259a0. [DOI] [PubMed] [Google Scholar]
- 56.Zhang Y, LeRoy G, Seelig H P, Lane W S, Reinberg D. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell. 1998;95:279–289. doi: 10.1016/s0092-8674(00)81758-4. [DOI] [PubMed] [Google Scholar]