Abstract
CITE-seq, a single-cell multi-omics technology that measures RNA and protein expression simultaneously in single cells, has been widely applied in biomedical research, especially in immune related disorders and other diseases such as influenza and COVID-19. Despite the proliferation of CITE-seq, it is still costly to generate such data. Although data integration can increase information content, this raises computational challenges. First, combining multiple datasets is prone to batch effects that need to be addressed. Secondly, it is difficult to combine multiple CITE-seq datasets because the protein panels in different datasets may only partially overlap. Integrating multiple CITE-seq and single-cell RNA-seq (scRNA-seq) datasets is important because this allows the utilization of as many data as possible to uncover cell population heterogeneity. To overcome these challenges, we present sciPENN, a multi-use deep learning approach that supports CITE-seq and scRNA-seq data integration, protein expression prediction for scRNA-seq, protein expression imputation for CITE-seq, quantification of prediction and imputation uncertainty, and cell type label transfer from CITE-seq to scRNA-seq. Comprehensive evaluations spanning multiple datasets demonstrate that sciPENN outperforms other current state-of-the-art methods.
Keywords: single-cell multi-omics, CITE-seq, single-cell RNA-seq, protein prediction, deep learning
Introduction
The proliferation of single-cell multi-omics profiling in biological research has advanced our understanding of cell heterogeneity and subpopulations 1, 2. In particular, the increasing availability of the CITE-seq protocol has greatly contributed to these advances. CITE-seq allows simultaneous profiling of RNA gene expression along with a panel of cell surface proteins 3, 4. It is well-known that proteins are much more abundant than RNA and are functionally directly involved in cell signaling and cell-cell interactions 5, 6. CITE-seq holds the potential to uncover cellular heterogeneity that is missed by single-modality single-cell RNA-seq (scRNA-seq).
Despite the promises of CITE-seq multi-modality expression profiling, technical difficulties persist. CITE-seq data remain expensive to generate relative to scRNA-seq data. One potential solution is to learn the relationship between RNA and proteins, borrowing information from a large reference dataset, and then make protein predictions for the scRNA-seq data. Seurat 4 7 and totalVI 8 have been introduced to fulfill this function, but both face limitations. TotalVI and especially Seurat 4 are computationally expensive. Furthermore, investigating more complex scenarios where multiple CITE-seq datasets whose protein panels do not completely overlap are consolidated give researchers the ability to increase the number of cells. However, Seurat 4 does not have the capability to do so. While totalVI can do it in theory, this problem has not been explored.
To address these challenges, we developed sciPENN (single cell imputation Protein Embedding Neural Network), a deep learning framework for predicting and imputing protein expression, quantifying uncertainty, integrating datasets in a low-dimension embedding, and merging multiple CITE-seq datasets together. sciPENN can integrate multiple CITE-seq datasets, even when their protein panels do not totally overlap using a censored loss approach. sciPENN’s strengths lie in its capacity to provide more robust and accurate results than totalVI and Seurat 4, while also being highly scalable and computationally efficient. Through comprehensive evaluations, we demonstrate that sciPENN performs dramatically faster than its peers. As the scale of multi-modality datasets continues to grow, computational methods that are both accurate and efficient are of great importance for scaling their applications in practice.
Results
Overview of sciPENN and evaluation strategies
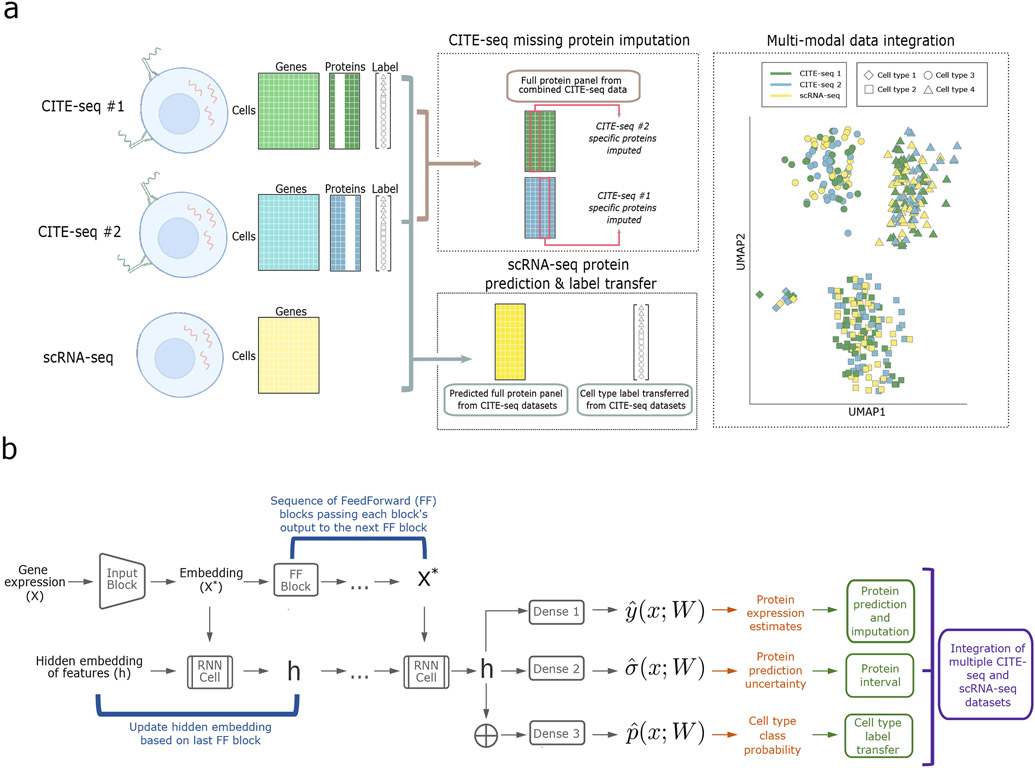
The model architecture of sciPENN is depicted in Fig. 1. The overall goal of sciPENN is to learn from one or more CITE-seq reference datasets. If the CITE-seq references do not completely overlap, sciPENN can impute the missing proteins for each reference dataset. After learning from CITE-seq references, sciPENN can predict all proteins for a scRNA-seq query dataset and integrate multiple datasets together in a common embedding space. Our model estimates mean protein expression, quantifies estimation uncertainty, and optionally transfers cell type labels from the CITE-seq reference to the query data. The structure of the model consists of blocks, sequences of layers that are used repeatedly throughout the model.
Fig. 1 ∣. Overview of sciPENN.
a, sciPENN is a flexible method which supports completion of multiple CITE-seq references (by imputing missing proteins for each reference) as well as protein expression prediction in an scRNA-seq test set, all in one framework. Simultaneously, sciPENN can transfer cell type labels from a training set to a test set, and can also integrate cells from the multiple datasets into a common latent space. b, sciPENN’s model architecture is comprised by an input block, followed by a sequence of FeedForward blocks interleaved with updates to an internally maintained hidden state updated via an RNN cell. The final hidden state is passed through three dense layers to compute protein predictions, protein prediction bounds, and cell type class probability vectors.
sciPENN can be used to predict protein expression in an external scRNA-seq dataset using a training CITE-seq dataset. sciPENN can also integrate multiple CITE-seq datasets. More specifically, an investigator may wish to jointly analyze multiple CITE-seq datasets, whose protein panels are not identical. sciPENN can integrate these datasets and impute expression for the proteins missing in each dataset. We train our model jointly on the multiple CITE-seq datasets by using a censored loss function approach in which the loss is only computed for measured proteins and censored for unmeasured proteins for a given cell. The trained model can be used to then impute missing proteins for each CITE-seq dataset and also predict protein expression for external scRNA-seq datasets.
We compared sciPENN to totalVI 8 and Seurat 4 7 for multi-modality integration, protein expression prediction and imputation, uncertainty quantification, and cell type label transfer. We have multiple goals in our analyses. First, we wish to demonstrate that sciPENN can both make predictions on external scRNA-seq datasets accurately and effectively integrate multiple CITE-seq datasets. Furthermore, we aim to demonstrate that sciPENN consistently performs well across diverse settings, even when the single-cell protocols vary substantially between datasets, and can recover expression trends in specific protein biomarkers of interest.
Seurat PBMC to MALT prediction
In our first analysis, we used a dataset of 161,764 human peripheral blood mononuclear cells (PBMCs) reported in the Seurat 4 paper 7, which we refer to as the PBMC dataset. This dataset includes 224 proteins. For the test set, we used the Mucosa-Associated Lymphoid Tissue (MALT) dataset, which contains 8,412 cells generated by 10x Genomics. Among the 17 proteins in the MALT dataset, 10 overlapped with the proteins in the PBMC data. We held out the protein expression for the MALT data and evaluated how well each method can recover the protein expression. Among proteins sequenced in both datasets, average protein expression was over four times higher in the MALT dataset than in the PBMC dataset, highlighting inherent differences between these two datasets.
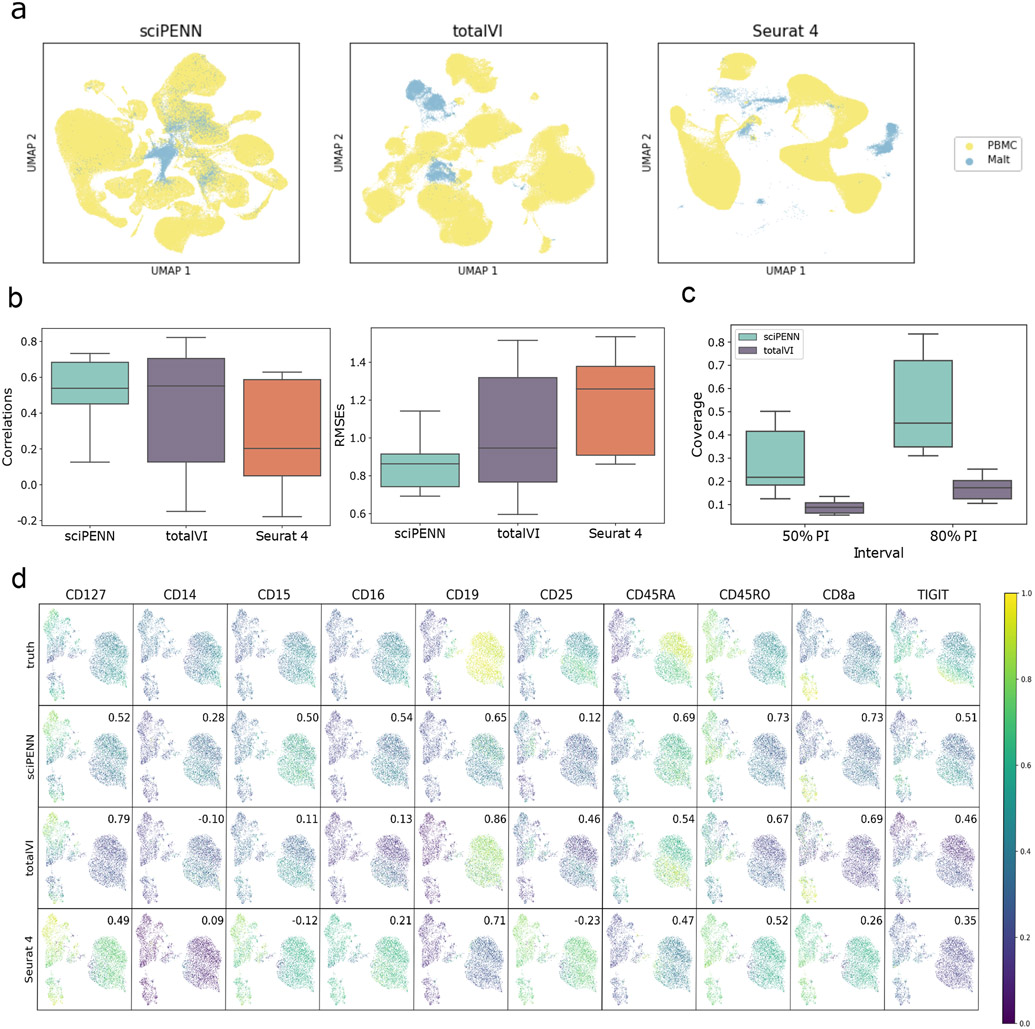
We analyzed these data using each of the three approaches. First, we embedded the PBMC CITE-seq reference and MALT RNA query data together into a single latent space using each method (Fig. 2a). Owing to the substantial differences between the PBMC and MALT query data, sciPENN, totalVI, and Seurat 4 all struggled to fully mix the two datasets together in the latent embedding space even with the internal batch correction strategies incorporated into all three methods. However, sciPENN did the best at integrating the two datasets and achieved partial mixing in its latent embedding.
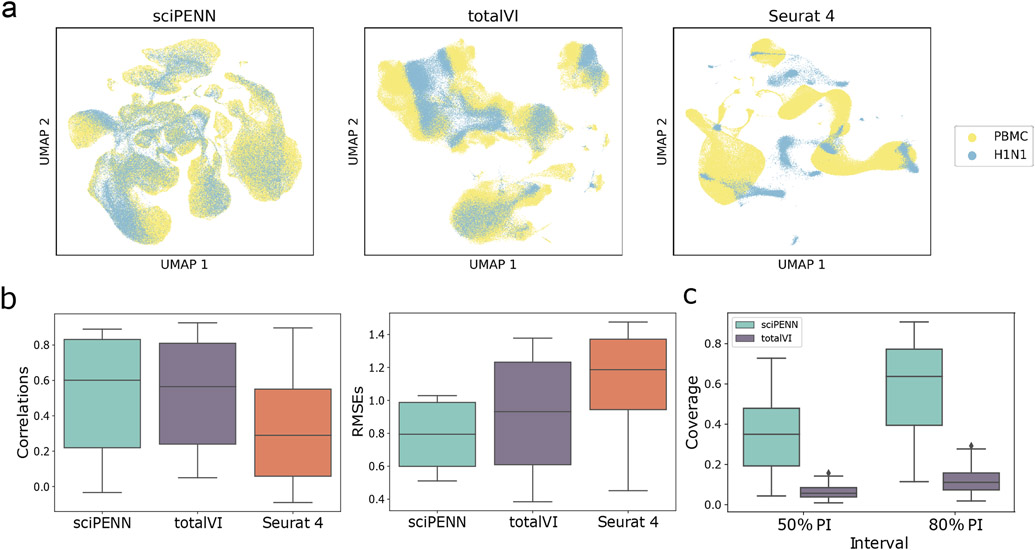
Fig. 2 ∣. Protein expression prediction in the MALT dataset using the Seurat 4 PBMC dataset as reference.
a, UMAP Embeddings visualizing the integrated hidden representation of the data, for each method. Each cell is colored according to the dataset from which it was sequenced. b, Box plots that display the correlation (left) and the RMSE (right) between each MALT protein’s predicted and true values for each method. The lower and upper hinges correspond to the first and third quartiles, and the center refers to the median value. The upper (lower) whiskers extend from the hinge to the largest (smallest) value no further (at most) than 1.5 × interquartile range from the hinge. Results are based on the analysis of 8,412 cells in the MALT dataset and 161,764 cells in the Seurat 4 PBMC dataset. c, Box plots that visualize the empirical test coverage of nominal 50% and 80% PIs per protein computed with sciPENN and totalVI. The lower and upper hinges correspond to the first and third quartiles, and the center refers to the median value. The upper (lower) whiskers extend from the hinge to the largest (smallest) value no further (at most) than 1.5 × interquartile range from the hinge. Results are based on the analysis of 8,412 cells in the MALT dataset and 161,764 cells in the Seurat 4 PBMC dataset. d, Feature plots for every MALT protein. The scatterplot is a UMAP representation of the true protein counts for the MALT data. In each feature plot, we color each cell in the scatterplot according to the intensity of its relative value for the specified protein. In the first row, we use the true values to guide the feature plot color mapping. In the subsequent rows, we color each cell according to the protein’s predicted expression, as predicted by sciPENN, totalVI, and Seurat 4. The number in the top right in each plot is the correlation between the gold standard (true) protein expression counts and the predicted counts.
Next, we examined the protein expression prediction accuracy of each method. We quantified prediction accuracy both by computing Pearson correlation and the root mean squared error (RMSE) between the predicted and observed protein expression, where the RMSE for each protein was calculated in the z-score standardized feature space. Fig. 2b shows that sciPENN achieved the highest protein prediction accuracy among all proteins, as quantified by both correlations and RMSEs.
We further evaluated the coverage probabilities of sciPENN and totalVI’s prediction intervals. We could not include Seurat 4 in this comparison as it does not quantify protein expression prediction uncertainty. Fig. 2c shows that for both the nominal 50% and 80% prediction intervals (PIs), sciPENN’s PIs have much better coverage than totalVI’s PIs. sciPENN’s 50% and 80% PIs have 22.1% and 44.6% median empirical coverage, while totalVI’s median coverages were only 9.8% and 18.3%, respectively.
Lastly, we examined feature plots for individual proteins (Fig. 2d). Again, sciPENN performs the best overall. For example, for CD8a, the cells are embedded into three clusters roughly and CD8a is expressed much more highly in the bottom left cluster than the other clusters when using the true protein expression. sciPENN recovered this trend for the test data, predicting much higher expression in the bottom cluster than the other clusters. totalVI incorrectly predicted moderately high expression in the upper right cluster. Seurat 4 struggled the most and predicted moderately high expression in all three clusters. We observed similar patterns for other proteins. For example, CD45RO is expressed in both left clusters but not the right cluster. sciPENN recovered this trend, but totalVI underestimated expression in the bottom left cluster. Seurat 4 again failed to distinguish all three clusters. However, totalVI performed well in some scenarios. For example, it outperformed sciPENN for CD19.
Monocyte to monocyte prediction
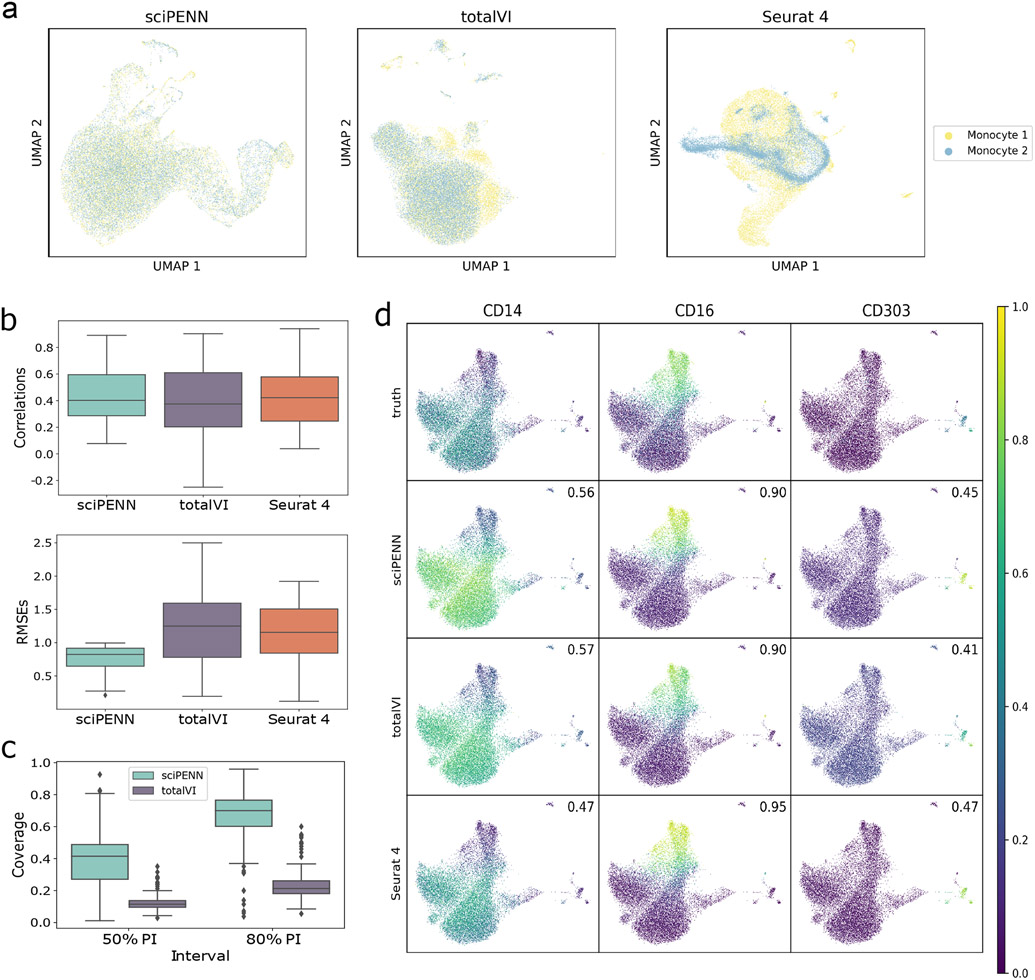
In this next evaluation, we consider a more even-handed balance between the query and reference sets. We used a human blood monocyte and dendritic cell CITE-seq dataset, referred to as the Monocyte dataset, which we generated. Monocytes play distinct, but poorly defined, roles in human cardiovascular disease 9. Human circulating monocytes can be divided into three subsets based on surface protein markers, classical (CD14++/CD16−), intermediate (CD14++/CD16+), and non-classical “patrolling” (CD14dim/CD16++) subpopulations. Clinical cardiovascular disease outcomes are directly associated with levels of circulating monocytes, specifically with higher proportions of classical and intermediate subsets 10-15. To better understand the role of monocyte subpopulations in homeostasis and disease, we generated a CITE-seq dataset that consists of 37,212 cells and 283 proteins obtained from 8 samples in 4 subjects. To create a reference and query dataset, we allocated 4 samples to the reference and the other 4 samples to the query. We held out true expression for the test set to see how well each method can recover it. Fig. 3a shows that sciPENN achieved complete mixing of the two datasets in its embedding. totalVI achieved nearly complete mixing as well, with only minor non-overlapping of the two datasets. Seurat 4 did not mix the two datasets as well as the other methods, but the two datasets still overlapped substantially with considerable mixing.
Fig. 3 ∣. Protein expression prediction in the Monocyte dataset.
This figure illustrates the prediction of proteins in the Monocyte test data (samples RPM215A, RPM215B, RPM218A, RPM218B) using the Monocyte training data (samples RPM211A, RPM211B, RPM232A, RPM232B) as reference. a, UMAP embeddings visualizing the integrated hidden representation of the data, for each method. Each cell is colored according to the dataset from which it was sequenced. “Monocyte 1” represents the training data, “Monocyte 2” represents the test data. b, Box plots that display the correlation (left) and the RMSE (right) between each Monocyte protein’s predicted and true values for each method. The lower and upper hinges correspond to the first and third quartiles, and the center refers to the median value. The upper (lower) whiskers extend from the hinge to the largest (smallest) value no further (at most) than 1.5 × interquartile range from the hinge. Results are based on the analysis of 37,112 cells in the monocyte study (19,516 cells in training and 17,596 cells in testing). c. Box plots that visualize the empirical test coverage of nominal 50% and 80% PIs per protein computed with sciPENN and totalVI. The lower and upper hinges correspond to the first and third quartiles, and the center refers to the median value. The upper (lower) whiskers extend from the hinge to the largest (smallest) value no further (at most) than 1.5 × interquartile range from the hinge. Results are based on the analysis of 37,112 cells in the monocyte study (19,516 cells in training and 17,596 cells in testing). d, Feature plots for selected proteins CD14, CD16, and CD303. The scatterplot is a UMAP representation of the true protein counts for the Monocyte data. In each feature plot, we color each cell in the scatterplot according to the intensity of its relative value for the specified protein. In the first row, we use the true values to guide the feature plot color mapping. In the subsequent rows, we color each cell according to the protein’s predicted expression, as predicted by sciPENN, totalVI, and Seurat 4. The number in the top right of each plot is the correlation between the gold standard (true) protein expression counts and the predicted counts.
Next, we examined the correlations and RMSEs between predictions and truth for each protein (Fig. 3b). On the correlation scale, all three methods performed highly effectively in this analysis. sciPENN was the leader when considering RMSE as the metric of interest, likely because its estimates not only were correlated with the truth but also were close to it resulting in an overall lower error. We also repeated the random split of training and testing samples 10 times and found the degree of randomness for the prediction is small (Extended Data Fig. 1a).
Additionally, we evaluated both sciPENN and totalVI’s empirical test coverage probabilities (Fig. 3c). sciPENN performed reasonably well: its 50% PI achieved a 41.9% median coverage probability across all proteins, while its 80% PI achieved 71.7% median coverage. totalVI struggled to quantify uncertainty: its median coverage probabilities were only 16.7% and 21.2%, respectively, which are well below the nominal coverage rates.
Lastly, we examined feature plots for proteins CD14, CD16, and CD303 (Fig. 3d). These three proteins are of special interest because CD14 is a marker for classical monocytes and CD16 is a marker for nonclassical monocytes 16, while CD303 is a marker for dendritic cells 17. All three methods performed relatively well for all three proteins, exhibiting similar correlations with the truth and recovering the main trends observed in the true expression data.
PBMC to PBMC prediction
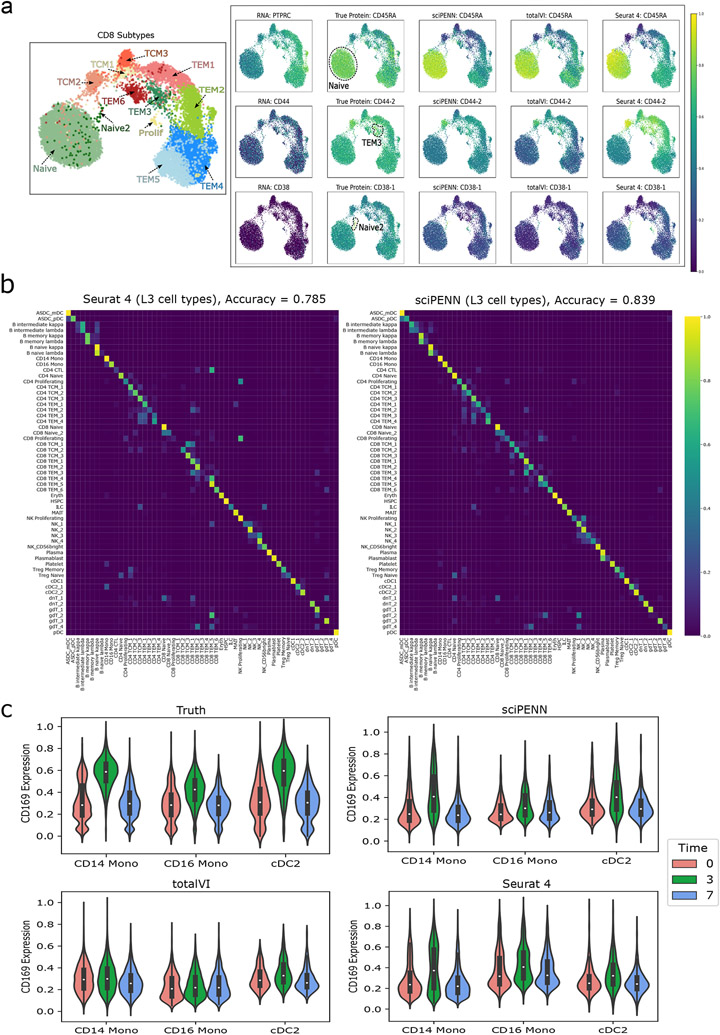
For this evaluation, we randomly split the full PBMC data into a training half and a test half. First, we consider sciPENN’s ability to recover marker protein trends (Fig. 4a). We chose three proteins: CD45RA, CD44-2, and CD38-1, representing protein markers for CD8 subtypes identified in the Seurat 4 paper 7. CD8 T cells are mediators of adaptive immunity and they respond adaptively to the type of encountered pathogens 18. It is important to characterize CD8 T cell subpopulations and understand how different factors, e.g., tissue site, type of pathogens and stimuli, influence T cell persistence and function. For each protein, we first checked the expression dynamics of its encoding RNA gene (PTPRC, CD44, and CD38, respectively) and verified that the encoding RNA gene alone is not enough to identify CD8 cell subtypes. We then examined the true protein expression across CD8 subtypes for each protein to see which cell subtypes express each protein highly. Lastly, we examined the magnitude of predicted expression across CD8 subtypes to see how well each prediction method recovers the truth and can be used to identify marker proteins. We examined that CD45RA is an apparent marker for CD8 Naïve, CD44-2 is an apparent marker for CD8 TEM3 and for CD8 TCM2 to a lesser extent, and CD38-1 is an apparent marker for CD8 Naïve 2. sciPENN’s protein predictions accurately recovered these trends, allowing the investigator to detect which cell subtypes a protein is expressed highly in using sciPENN predictions only. totalVI and Seurat 4 also performed well, albeit marginally worse. Seurat 4 underestimated the expression of CD44-2 in CD8 TEM3, and totalVI underestimated the expression of CD38-1 in CD8 Naïve 2. We also repeated the random split of training and testing samples 10 times and found the degree of randomness for the prediction is small (Extended Data Fig. 1b).
Fig. 4 ∣. Protein expression prediction and cell type label transfer in the PBMC dataset.
This figure illustrates the prediction of proteins and cell type label transfer in the PBMC test data (donors P2, P5, P6, P8) using the PBMC training data (donors P1, P3, P4, P7) as reference. a, UMAP plot on the left shows the CD8 cell subtypes reported in the Seurat 4 paper. UMAP plots on the right demonstrate the necessity of protein data to identify cell subpopulations by comparing UMAP colored by the true protein to the UMAP colored by the protein’s encoding RNA gene. Additional UMAPs colored by sciPENN, totalVI and Seurat 4 protein predictions demonstrate the utility of protein predictions for recovering these subpopulation behaviors when true protein data are missing, and sciPENN’s utility compared to other methods for most consistently recovering such trends. b, Confusion matrices which demonstrate the cell type prediction accuracy of sciPENN and Seurat 4 for each true cell type. Rows represent true cell type and columns represent predicted cell type. The raw matrix is first computed, and then normalized by each row’s sum, i.e., by the number of cells of each type. Element i, j of the numeric matrix can be thought of as the proportions of cells of type i which were classified as type j. c, Violin plots visualizing the CD169 protein’s feature values immediately before reception of a VSV-vectored HIV vaccine (Time = 0), 3 days after administration of the vaccine (Time = 3), and 7 days after administration (Time = 7). We examine the true CD169 expression with respect to Time, as well as sciPENN predicted, totalVI predicted, and Seurat 4 predicted CD169 expression with respect to time.
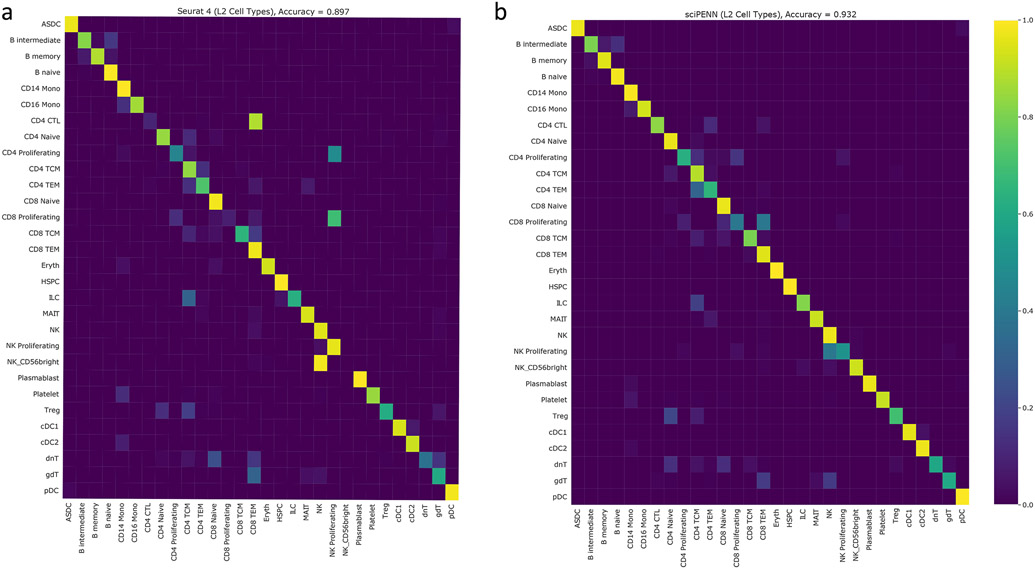
Next, we evaluated sciPENN and Seurat 4’s abilities to transfer cell type labels from a CITE-seq reference to a scRNA-seq test set (Fig. 4b). We omitted totalVI since it is not designed for cell type label transfer. The PBMC dataset has three resolutions of cell type labels provided by the Seurat 4 paper: L1 (8 types), L2 (30 types), and L3 (57 types). We evaluated the performance using L3 labels in the main text as this represents the most challenging task due to the close relatedness of the 57 cell types. Fig. 4b shows a row-normalized confusion matrix, where rows represent true cell types and columns represent predicted cell types. Overall, sciPENN outperformed Seurat 4 for predicting cell type labels, in spite of using the labels originally assigned using Seurat 4. sciPENN achieved 83.9% accuracy, while Seurat 4 achieved only 78.5% accuracy. The confusion matrices suggest this performance gap arises because sciPENN correctly classified certain cell subtypes significantly better than Seurat 4. We also evaluated the performance of sciPENN using L2 labels and the results are shown in Extended Data Fig. 2.
Lastly, we evaluated sciPENN’s ability to recover protein expression trends triggered by stimuli. Donors in the PBMC dataset were administered a VSV-vectored HIV vaccine. Expression of cells were profiled from patients immediately before, 3 days after, and then 7 days after the vaccine. In the Seurat 4 paper, Hao et al. 7 reported that CD169 protein showed a clear response to the vaccine in CD14 Monocytes, CD16 Monocytes, and cDC2 cells. In all three cell types, CD169 expression spiked 3 days after the vaccine was received when patients were experiencing their immune response to the vaccine, and then returned their pre-vaccine baseline after 7 days once the immune response ended. This suggests that CD169 is a biomarker for immune response to the vaccine. Identifying biomarkers like CD169 can be of great importance to understanding diseases and corresponding vaccine development.
We visualized CD169’s expression in CD14 monocytes, CD16 monocytes, and cDC2 cells at each of the three time points (Fig. 4c). sciPENN recovered the CD169’s response to the vaccine, while totalVI struggled with this, and Seurat 4 did a reasonable job. For sciPENN, a clear spike in the predicted expression of CD169 is observed at Time 3 for all three cell types. In totalVI, the spike in CD169 is observed for cDC2, but it appears to be small in CD14 Monocytes and nearly non-existent in CD16 monocytes. In Seurat 4, the spike in CD169 is clear in CD14, but less so in the other two cell types. To assess this quantitatively, we tested whether the mean CD169 expression is not the same across the three times within each method using the Kruskal Wallis test and sciPENN had the highest −log10(p-value) for all three cell types. For CD14, sciPENN’s −log10(p-value) was greater than 100 while totalVI’s was 87 and Seurat 4’s was also greater than 100. For CD16, sciPENN’s metric was 27.4 while totalVI’s was just 2.34 and Seurat 4’s was 50.8. Finally, for cDC2, sciPENN achieved a metric of 27.4, while totalVI was 17.7 and Seurat 4 was 19.8. Results from this analysis indicate that sciPENN can help identify stimulus biomarkers like this vaccine immune response biomarker.
PBMC to H1N1 prediction
In this evaluation, we consider a situation where the query set is moderately different from the reference set. Specifically, we reused the Seurat 4 PBMC dataset as the reference, but used a new H1N1 influenza dataset 19 as the query. The H1N1 dataset includes CITE-seq data of 53,201 cells and 87 proteins from PBMCs in healthy donors, which was used to investigate the response of these donors to influenza vaccination. Since the H1N1 dataset also contains PBMCs, the Seurat 4 PBMC dataset is a natural reference to use to recover the held-out protein expression of the H1N1 data. 59 of the proteins in the H1N1 and the Seurat 4 PBMC datasets overlapped. Fig. 5a shows that sciPENN and totalVI were both mixed these relatively different datasets in the embedding space reasonably well. On the other hand, Seurat 4 was not effective in mixing these dataset batches. Additionally, sciPENN and totalVI predicted protein expression effectively than Seurat 4, as measured by correlation and RMSE between the predicted and true protein expression (Fig. 5b). sciPENN and totalVI had near-identical performance when using correlation as the metric. sciPENN leads in prediction accuracy when considering RMSE, but the gap between the three methods is not substantial.
Fig. 5 ∣. Protein expression prediction in the H1N1 dataset using the Seurat 4 PBMC dataset as reference.
a, UMAP Embeddings visualizing the integrated hidden representation of the data, for each method. Each cell is colored according to the dataset from which it was sequenced. b, Box plots that display the correlation (left) and the RMSE (right) between each H1N1 protein’s predicted and true values for each method. The lower and upper hinges correspond to the first and third quartiles, and the center refers to the median value. The upper (lower) whiskers extend from the hinge to the largest (smallest) value no further (at most) than 1.5 × interquartile range from the hinge. Results are based on the analysis of 53,201 cells in the H1N1 dataset and 161,764 cells in the Seurat 4 PBMC dataset. c, Box plots that visualize the empirical test coverage of nominal 50% and 80% PIs per protein computed with sciPENN and totalVI. The lower and upper hinges correspond to the first and third quartiles, and the center refers to the median value. The upper (lower) whiskers extend from the hinge to the largest (smallest) value no further (at most) than 1.5 × interquartile range from the hinge. Results are based on the analysis of 53,201 cells in the H1N1 dataset and 161,764 cells in the Seurat 4 PBMC dataset.
sciPENN quantified uncertainty prediction for its protein predictions much more effectively than totalVI in this analysis (Fig. 5c). sciPENN’s 50% nominal PI achieved 34.9% median coverage probability, and its 80% nominal PI also achieved 63.7% median coverage. By contrast, the median coverage probabilities for totalVI are only 5.69% and 11.1%, respectively.
Integration of COVID datasets
In the last evaluation, we consider a more complex problem of integration in which we combine multiple CITE-seq datasets as reference. Since different CITE-seq datasets may have different protein panels, some proteins are sequenced in only some of the CITE-seq datasets we wish to combine. Our challenge is to fill in the unmeasured proteins for each CITE-seq dataset. To evaluate this scenario, we consider two CITE-seq datasets generated from a mix of healthy people and patients infected with COVID-19. The first dataset consists of 647,366 cells and 192 proteins generated by the Haniffa Lab 20, and the second dataset consists of 240,627 cells and 192 proteins generated by the Sanger Institute 21. 110 proteins overlapped between these two datasets. Being able to effectively integrate and impute protein expression for these datasets are of great clinical interest, as the Coronavirus pandemic has massively disrupted societies around the world, increasing interest in understanding this coronavirus.
To set up our experiment, we identified the 110 proteins shared between the datasets, and dropped all other proteins. To mimic the situation of merging two CITE-seq datasets with partially overlapping protein panels, we randomly partitioned the 110 proteins into three groups of equal size: proteins only present in Haniffa, proteins only in Sanger, and proteins present in both. For each of the two datasets, we set aside the true protein expression for the proteins designated as missing. For the Sanger data we set aside the expression data for proteins designated as present only in Haniffa (the Haniffa proteins). Likewise, for the Haniffa data we set aside the expression data for proteins designated as present only in Sanger (the Sanger proteins).
We then took our two partially overlapping CITE-seq datasets and trained both sciPENN and totalVI to integrate the datasets and impute the missing protein expression for each dataset, where the imputation used RNA expression only and the protein expression levels were not included in the imputation. We did not include Seurat 4 in this evaluation because it is only able to map a reference CITE-seq dataset to a query RNA dataset. This integration experiment was quite challenging for both sciPENN and totalVI due to the large number of cells. However, sciPENN was able to integrate the two datasets into a common embedding efficiently, mixing the two datasets well (Fig. 6a). totalVI struggled considerably, failing to mix the two datasets.
Fig. 6 ∣. Integration of two COVID CITE-seq datasets.
a, UMAP Embeddings visualizing the integrated hidden representation of the data, for each method. Each cell is colored according to the dataset from which it was sequenced. b, Box plots that plot the correlation (left) and the RMSE (right) between each imputed protein’s predicted and true values for each method. Note that the box plots for Haniffa involves the proteins that were missing from Haniffa and imputed, and likewise the box plots for Sanger involves the proteins that were missing from Sanger and were imputed. The lower and upper hinges correspond to the first and third quartiles, and the center refers to the median value. The upper (lower) whiskers extend from the hinge to the largest (smallest) value no further (at most) than 1.5 × interquartile range from the hinge. Results are based on the analysis of 647,366 cells in the Haniffa data and 240,627 cells in the Sanger data. c, Feature plots for selected proteins CD7, TCR_Va7.2, CD123, and HLA-DR. The first two are proteins that were imputed into the Haniffa dataset, the second two are proteins that were imputed into the Sanger dataset. The scatterplot is a UMAP representation of the true protein expression for the missing protein data. One UMAP representation is computed for the missing proteins in the Haniffa data, and another UMAP representation is computed for the missing proteins in the Sanger data. In each feature plot, we color each cell in the scatterplot according to the intensity of its relative value for the specified protein. In the first row, we use the true values to guide the feature plot color mapping. In the subsequent rows, we color each cell according to the protein’s predicted expression, as predicted by sciPENN and totalVI. The number in the top right is the correlation between the gold standard (true) protein expression counts and the predicted counts.
Next, we examined protein imputation accuracy. Fig. 6b shows that imputing the Sanger proteins in the Haniffa data was a difficult task for both methods because the sequencing depth in the Haniffa data is only ~50% of the Sanger data, making RNA expression in the Haniffa data less predictive for protein expression. Despite this, sciPENN clearly outperformed totalVI in both correlation and RMSE with the truth. On the other hand, imputing the Haniffa proteins into the Sanger data was a much easier problem for both methods. sciPENN outperformed totalVI on both the correlation and RMSE metrics (Fig. 6b), but totalVI still made useful imputations.
We also examined feature plots for a few selected proteins (Fig. 6c). The first two proteins, CD7 and TCR_Va7.2, were Sanger proteins imputed into the Hannifa dataset. sciPENN and totalVI performed decently well at imputing CD7, with sciPENN leading totalVI. In TCR_Va7.2, both methods struggled and totalVI failed to predict the protein, a reflection of how difficult imputing into the Haniffa dataset is. The latter two proteins, CD123 and HLA-DR, were Haniffa proteins imputed into the Sanger dataset. Both methods did much better with sciPENN leading totalVI, but only by modest margins. This better performance is a reflection of the lower difficulty at imputing protein expression in the Sanger data.
Finally, we examined the ability of sciPENN to predict protein expression in the PBMC and the H1N1 RNA-seq data. We did not compare with totalVI because its loss function rapidly decayed to NaN. For the proteins predicted in each test dataset, we categorized them into three categories: only present in Haniffa, only present in Sanger, and present in both. As shown in Extended Data Fig. 3, the common proteins are more accurately predicted than the unique proteins, which is expected because larger sample size in the training set yields better predictions. These results underscore the importance of combining multiple CITE-seq datasets for protein expression prediction.
Discussion
We have developed sciPENN, a deep learning model that can predict and impute protein expression, integrate multiple CITE-seq datasets, and quantify prediction and imputation uncertainty. We accomplish this by designing both the internal network structure, as well as the loss function and optimization strategy of sciPENN in order to maximize its protein prediction and imputation accuracy. The network is built as a stack of dense, batchnorm, PReLU, dropout layer blocks which help the model learn progressively finer latent cell representations. These design choices enabled sciPENN to perform well for supervised protein prediction.
Across the three supervised analyses we considered, sciPENN consistently integrated the reference CITE-seq dataset with the query dataset in the latent embedding the best when compared to totalVI and Seurat 4. sciPENN also consistently had the highest protein prediction accuracy both by the correlation and RMSE metrics. This high protein prediction accuracy allows sciPENN to recover protein expression patterns accurately.
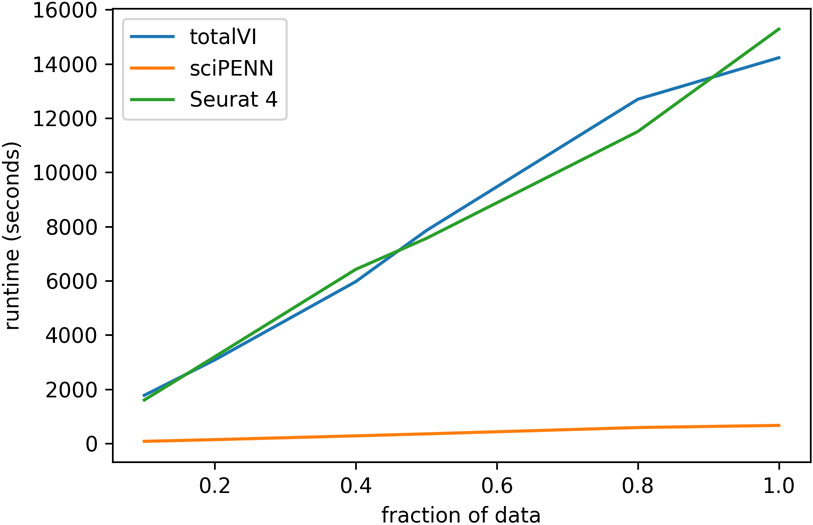
One challenge in CITE-seq analysis is the integration of multiple CITE-seq datasets. Such integration is not trivial because the protein panels for different CITE-seq datasets usually have some non-overlap, which prevents simple concatenation. To circumvent this, we introduced a censored loss function scheme for sciPENN, where a protein loss is masked and does not contribute to backpropagation whenever it is missing from a cell. This allows sciPENN to learn from multiple CITE-seq datasets with partially non-overlapping protein panels, impute the missing proteins of each constituent CITE-seq dataset, and even predict protein expression in external scRNA-seq datasets after learning from the partially overlapping CITE-seq datasets, a task that was not achievable by totalVI and Seurat 4. Additionally, sciPENN is an order of magnitude faster than totalVI and Seurat 4 (Extended Data Fig. 4), which makes it a desirable tool for integrative CITE-seq and scRNA-seq data analysis.
Methods
The sciPENN workflow (Fig. 1) involves four main steps: preprocessing, training, imputation, and prediction. Below we describe each of these steps in detail.
Step 1: preprocessing
Suppose there are k CITE-seq datasets that we wish to integrate with a possibly query scRNA-seq data for which we wish to predict proteins. Let the ith CITE-seq dataset of ni cells be represented by an ni × gi RNA array Xi and ni × pi protein array Yi. Additionally, let the query scRNA-seq dataset of nq cells be denoted by an nq × gq RNA-array Xq. For each CITE-seq dataset and the query scRNA-seq dataset, a cell is removed if the number of expressed RNA genes is less than 200, and a gene is removed if the number of cells expressing the gene is less than 30.
Next, we normalize expression values for both RNA genes and proteins. In the first step, cell level normalization is performed in which expression for a given gene in each cell is divided by the total gene expression across all genes in the cell, multiplied by the median total expression for that gene across all cells in that specific dataset, and then transformed to a natural log scale. We also do this cell level normalization for the protein modality of each CITE-seq dataset. In the second step, we find the set of RNA genes that are available in every dataset (all CITE-seq datasets, and the query dataset If one exists). We then proceed by finding highly variable RNA genes (HVGs) among them. HVGs are selected based on the log-normalized counts using the approach introduced by Stuart and Butler 22 and implemented in the “pp.highly_variable_genes” function with “batch_key” parameter in the SCANPY Python package (version >=1.4) 23, where each dataset is treated as a batch. In the last major step of preprocessing, we z-score normalize features in the dataset by batch for both RNA genes and proteins.
After the last major step of preprocessing, we do a few final operations before wrapping up preprocessing. First, we merge the protein data together across multiple CITE-seq datasets. When proteins are not available in a cell, we fill the missing protein values of this cell with 0. The merged protein dataset Ytrain is of dimension ntrain × p, where ntrain = Σini and p is the number of proteins available in the union of all available proteins. If we have a query dataset, we create a corresponding test set by splitting the full gene array Xall into a training RNA-array Xtrain of dimension ntrain × g and the now-normalized query array Xq of dimension nq × g. If no query dataset exists, then Xtrain can be taken as Xall.
Step 2: Training the network
In the next step we perform minibatch gradient descent to train the model. We obtain input gene expression vectors for the minibatch cells from Xtrain, pass the inputs through the network, and then use these outputs, along with the corresponding true protein expression data for the minibatch from the protein array Ytrain to compute the loss function. The gradients are computed using reverse mode automatic differentiation and used to update the weights of the network.
To help manage overfitting and optimize model performance, we use an early stopping strategy with learning rate decay to fit the model. Precisely speaking, we set aside a prespecified, randomly selected fraction f of our available training cells to use as a validation set, and then leave the remaining 1 – f fraction of cells for training. For each epoch, we loop over the training cells, grabbing random minibatches of these cells, computing the loss and the gradients of the loss with respect to model weights, and update the network weights using the Adam optimizer 24 before proceeding to the next randomly selected minibatch. Once we have looped over all training cells, we then check the validation loss. We loop over the validation dataset, grabbing minibatches of cells and updating the running validation minibatch loss, but not using these cells to compute gradients. Once we have looped over the validation dataset, we record the validation minibatch loss for the epoch. After computing the minibatch validation loss, we check learning rate decay and early stopping conditions. For details, please see Supplementary Note 1.
Step 3: Imputation of protein expression in CITE-seq data
In the application of sciPENN, the user may want to integrate multiple CITE-seq datasets with protein panels that only partially overlap. The proteins which are not measured in the specific CITE-seq dataset from which the cell is sequenced are missing, so they are arbitrarily filled with 0s as a placeholder when creating the merged protein array Ytrain that spans all CITE-seq datasets. Once sciPENN has been trained, the user can opt to impute the missing proteins for each cell. The main focus of imputation is to fill the missing values of Ytrain with predicted expression values, but in addition we will also store quantile estimates and optionally transferred cell type labels as well. Let Qtrain,qi be an ntrain × p array storing the estimates of quantile qi, and Yj, Qj,qi, and Xj represent row j of Ytrain, Qtrain,qi, and Xtrain, respectively.
sciPENN passed Xi (j = 1 to ntrain) as input to obtain corresponding estimates , , of the protein mean, quantiles, and predicted cell type class probabilities of cell , respectively, as described in the “Model architecture” section. Since we have true cell type labels for the training data, we discard . will be an array of all the quantile estimates of shape p × k where k is the number of quantiles. Simply loop over the columns from s = 1 to k and set Qj,qs equal to the sth column of , where qs denotes the quantile represented by column s of . To update Ytrain, we want to fill in predictions only for proteins that are missing. To do so, let bj be a vector of length p whose sth element equals 1 if and only if the sth protein is sequenced for cell j. Then we update Yj as follows:
where “·” represents the dot-product operator. We perform these updates for each individual cell in the training data.
Step 4: Prediction of protein expression in scRNA-seq data
The last step a user may consider is predicting protein expression in scRNA-seq, which is distinct from imputation described in Step 3. Let Xq be the ntest × 1000 test set RNA gene expression array after we selected the top 1000 HVGs. Similar to the imputation process, let Qtest,qi be an ntest array storing the estimates of quantile qi, and Ytest store the protein predictions, and C be an ntest length vector to store predicted cell type labels. Let Yj, Qj,qi, and Xj represent row j of Ytest, Qtest,qi, and Xtest, respectively.
Take Xj (j = 1 to ntest), and pass it as input to sciPENN, and obtain corresponding estimates , , . is used to update Qj,qi, for i = 1, 2, … , ntest, as described in the imputation section. Unlike with imputation where we only needed protein mean estimates for missing proteins, we want to store predictions for all proteins for test set prediction. For this reason, we simply set Yj equal to to update Ytest. To store the predicted cell type label for cell j, we set the jth element of C equal to .
Model architecture
Suppose we have a (merged) CITE-seq RNA-array Xtrain of shape ntrain × g and a corresponding (merged) protein-array Ytrain of shape ntrain × p with some missing proteins that we wish to impute. Suppose further that we wish to estimate k quantiles for each corresponding protein prediction to quantify uncertainty. Here, is an estimate of a protein’s mean expression for a cell with gene expression vector x. is a vector estimate of the k prediction quantiles for the protein’s expression, and is the estimate of quantile qi. Lastly, is a vector of predicted cell type class probabilities. S(x; W) is our neural network,
parametrized by weights W. The network structure is best described using the concept of blocks: sequences of elementary layers that are stacked together in a standard way and used as smaller parts for building a more complex model. The two key blocks used by the network are an Input Block and a FeedForward block. The Input Block is described first.
| Input Block |
|---|
| Receive as input: gene expression vector x x ← BatchNorm(x; W1) x ← Dropout(x) x ← Dense(x; W2) x ← BatchNorm(x;W3) x ← PReLU (x; W4) x ← Dropout(x) return x |
The FeedForward Block is described next. This block receives an embedding as input and will only runs BatchNorm and Dropout after passing the embedding through a dense layer. Otherwise, this block is similar to the Input Block.
| FeedForward Block |
|---|
| Receive as input: embedding vector x x ← Dense(x; W1) x ← BatchNorm(x; W2) x ← PReLU(x; W3) x ← Dropout(x) return x |
With these blocks introduced, we can now discuss the construction of the network S(x, W). First, the gene expression x is passed into an Input Block, which computes an embedding from the gene expression data. Then, we pass this embedding to a sequence of FeedFoward Blocks. After we compute the output of each FeedForward Block, we pass this output to a recurrent cell, which maintains a hidden embedding of features that it updates using the FeedForward block’s output. Note that the hidden embedding is initialized as vector of 0s. Once the hidden embedding is updated, we pass the FeedForward Block’s output to the next FeedForward Block and repeat the process. After we obtain the final updated RNN hidden state from the last FeedForward Block, we use it as the final embedding for visualization of the data integration. We further use this hidden embedding to compute estimates , , . We do this by passing the hidden embedding through three dense layers (one for each of the three estimated quantities). The entire computation graph is described in Fig. 1. In our computation graph, the symbol ⨁ represents the “detach” operation, which satisfies the following condition:
Essentially, the detach operation treats the output of any function of weights as a constant with respect to the weights, so that all operations downstream of the detached function output will not contribute to gradient updates of the function’s weights. In this context, if h is the hidden embedding after the last update from the RNN Cell and g(x; W) is a function which encapsulates all of the layers used to map the input to this embedding, then the detach operation is used in the
Loss function for minibatch gradient descent
We are interested not only in predicting protein expression, but also in quantifying the uncertainty of our prediction using interval estimation. To that end, we will want to estimate not only the mean expected protein expression given the RNA expression profile of the cell (), but also a vector of quantiles which can be used to construct prediction intervals. To train the model to estimate these quantities, we need a loss function to minimize. For the remainder of this section, we will suppress the notational dependence of , , on input genes x and weights W. Define and . Let Q = {q1, q2, … , qk} be the set of quantiles we wish to estimate. Then we want to estimate and for q ∈ Q such that we minimize the following objective
Since we wish to predict cell type assignment probability, we also need a loss function for cell type classification. A natural choice is the categorical cross-entropy function, which is simply the log-probability of the true class. Let the true class for the cell be denoted by ct and the random variable which represents a cell’s class be denoted by C, then the loss is
The total loss for a cell is then as follows:
Recall that for any given cell, only a subset of the proteins may have been measured since we allow for the merging of multiple CITE-seq datasets whose protein panels do not totally overlap. Accordingly, we must handle the loss and gradient computation with care, since not all of the p predicted proteins will necessarily have true sequenced expression values for us to compute losses with for any given cell. The missing proteins for a cell were filled with artificial 0 values when merging the CITE-seq protein arrays, but these 0s are simply placeholders with no biological significance.
To handle the missing proteins, we dynamically compute the loss function of a cell only over sequenced proteins, and this set of sequenced proteins is permitted to vary from cell to cell in a minibatch to accommodate minibatches with cells from different datasets. When computing our loss function for backpropagation for a cell, we average the protein-specific losses only of proteins sequenced for the cell. Specifically, let Lij be the total loss for protein j in cell i. Define the set Pi such that j ∈ Pi if and only if protein j is expressed in cell i. The loss for cell i is computed as follows:
This can be thought of as a “censored loss” approach in which the contribution of a protein to the total loss is censored if the protein is not sequenced for that cell. For a minibatch of cells, we simply average these cell losses across the minibatch of cells to obtain a single minibatch loss and then update the network weights, just as we would do for any typical application of minibatch gradient descent. The key idea here is that the cell-specific loss varies functionally from cell to cell due to protein censoring. As a consequence, each protein contributes to the overall minibatch gradient only through cells for which the protein was sequenced in the panel.
CITE-seq data generation in the Monocyte study
4 mL of blood was drawn into sodium heparin tubes and processed immediately in the Clinical Research Center at Columbia University Irving Medical Center. PBMCs were isolated by Ficoll-paque (GE Healthcare: 17-5442-02) density gradient centrifugation from four human subjects. Cells were then incubated with Human TruStain FcX (BioLegend: 422302) for 10 minutes at room temperature. Subsequently, samples were simultaneously stained with a pre-titrated pool of TotalSeq-A antibodies from BioLegend (99787) and fluorescent antibodies (CD14-AF488, CD16-PE-Cy7, HLA-DR-APC-eFluor 780, and Lineage markers) for 30 minutes at 4°C then washed 3 times in staining buffer (2% FBS, 5mM EDTA, 20mM HEPES, 100mM sodium pyruvate). Cells were then incubated with Sytox Blue viability die. Monocytes and monocytes/dendritic cells were sorted on a BD FACSAriaII for 10x genomics and sequencing analysis.
Extended Data
Extended Data Fig. 1. Evaluation of the robustness of sciPENN by randomly splitting training and testing data.
a, Boxplot for correlations between predicted and observed protein expression based on a random split of the Monocyte data into training and testing. The lower and upper hinges correspond to the first and third quartiles, and the center refers to the median value. The upper (lower) whiskers extend from the hinge to the largest (smallest) value no further (at most) than 1.5 × interquartile range from the hinge. Displayed are results for 10 random splits. The number of cells in the analysis is 37,122. b, Boxplot for correlations between predicted and observed protein expression based on a random split of the PBMC data into training and testing. The lower and upper hinges correspond to the first and third quartiles, and the center refers to the median value. The upper (lower) whiskers extend from the hinge to the largest (smallest) value no further (at most) than 1.5 × interquartile range from the hinge. Displayed are results for 10 random splits. The number of cells in the analysis is 161,764.
Extended Data Fig. 2. Cell type label transfer accuracy for L2 labels of Seurat 4 PBMC data.
In this figure, we demonstrate the accuracy of cell type label transfer for both Seurat 4 and sciPENN using the L2 labels of the Seurat 4 PBMC data. The color intensity of cell type (i, j) in the confusion matrix reflects the proportion of cells of type i that were misclassified as cells of type j. a, Confusion matrix of Seurat 4. b, Confusion matrix of sciPENN.
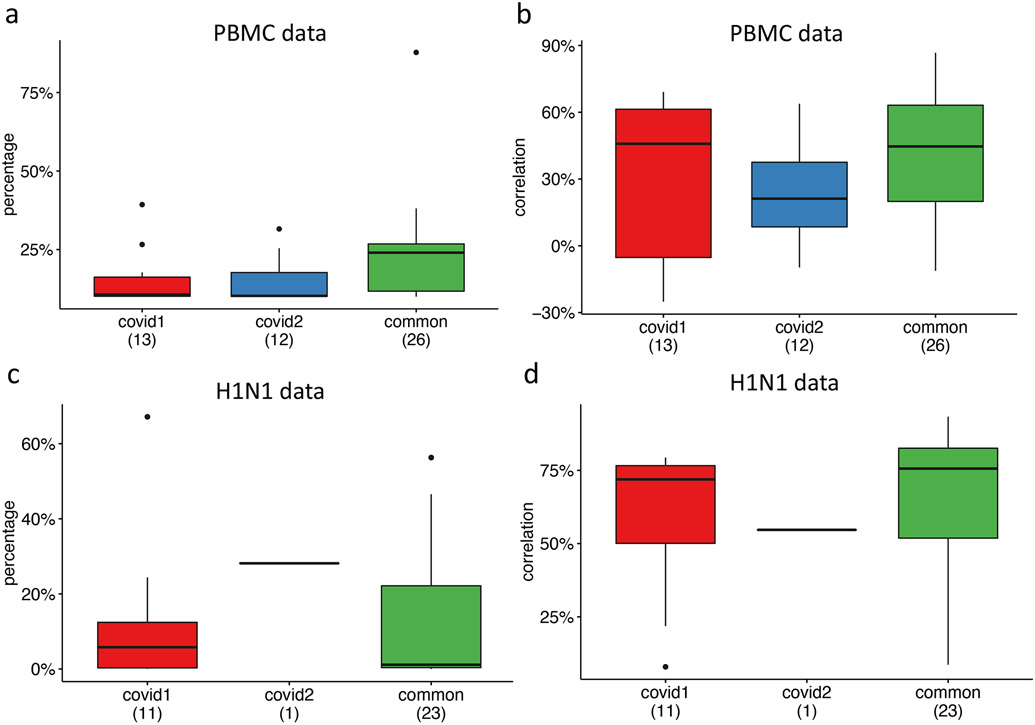
Extended Data Fig. 3. Protein expression prediction in the PBMC and the H1N1 datasets using the combined Haniffa (covid1, 647,366 cells) and Sanger (covid2, 240,627 cells) CITE-seq datasets as reference.
a, Percentage of cells whose protein expression is correctly predicted within the of 1st and 3rd quantiles of the prediction interval in the PBMC data. The lower and upper hinges correspond to the first and third quartiles, and the center refers to the median value. The upper (lower) whiskers extend from the hinge to the largest (smallest) value no further (at most) than 1.5 × interquartile range from the hinge. The number of cells in the PBMC data is 161,764. b, Correlation between predicted and observed protein expression in the PBMC data. The lower and upper hinges correspond to the first and third quartiles, and the center refers to the median value. The upper (lower) whiskers extend from the hinge to the largest (smallest) value no further (at most) than 1.5 × interquartile range from the hinge. The number of cells in the PBMC data is 161,764. c, Percentage of cells whose protein expression is correctly predicted within the of 1st and 3rd quantiles of the prediction interval in the H1N1 data. The lower and upper hinges correspond to the first and third quartiles, and the center refers to the median value. The upper (lower) whiskers extend from the hinge to the largest (smallest) value no further (at most) than 1.5 × interquartile range from the hinge. The number of cells in the H1N1 data is 53,201. d, Correlation between predicted and observed protein expression in the H1N1 data. The lower and upper hinges correspond to the first and third quartiles, and the center refers to the median value. The upper (lower) whiskers extend from the hinge to the largest (smallest) value no further (at most) than 1.5 × interquartile range from the hinge. The number of cells in the H1N1 data is 53,201.
Extended Data Fig. 4. Runtime comparison of methods.
In this figure, we demonstrate the speed of sciPENN relative to other methods. Specifically, we use the Seurat 4 PBMC data as reference (161,746 cells) and the H1N1 data (53,201 cells) as query. Given a fraction f, we train each method using (100 * f)% of the Seurat 4 PBMC data and predict on (100 * f)% of the query data and record how long this process takes. We perform this for various fractions f for each method and plot the results.
Supplementary Material
Acknowledgments
This work was supported by the following grants: R01GM125301 (to M.L.), R01EY030192 (to M.L.), R01EY031209 (to M.L.), R01HL113147 (to M.L. and M.P.R.), and R01HL150359 (to M.L. and M.P.R.). This study was conceived of and led by M.L.. J.L. designed the model and algorithm, implemented the sciPENN software, and led data analysis with input from M.L., A.S., M.L., A.B., and M.P.R.. A.S. contributed to comparison with totalVI and Seurat 4. A.B. and M.P.R. generated the human monocyte data and provided input on the Monocyte data analysis. J.L. and M.L. wrote the paper with feedback from all coauthors.
Footnotes
Competing Interests
The authors declare no competing interests.
Data Availability
We analyzed multiple published CITE-seq datasets throughout the evaluations. These data are available as follows (accession numbers provided where possible): 1) Mucosa-Associated Lymphoid Tissue (MALT) dataset (https://www.10xgenomics.com/resources/datasets/10-k-cells-from-a-malt-tumor-gene-expression-and-cell-surface-protein-3-standard-3-0-0); 2) Seurat 4 human peripheral blood mononuclear cells (PBMCs) (GEO: GSE164378); 3) H1N1 influenza PBMC dataset (https://doi.org/10.35092/yhjc.c.4753772, ref. 25 ); 4) human monocyte dataset (https://upenn.box.com/s/64c9fsex50g1bhv67893cpdg9c5jqjzo). All participants provided written informed consent under Columbia University IRB protocol AAAR5004. 5) Haniffa COVID Dataset (https://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-10026/); 6) Sanger COVID Dataset (https://covid19.cog.sanger.ac.uk/submissions/release2/vento_pbmc_processed.h5ad); Details of these datasets can be found from Details of these datasets were described in Supplementary Table 1.
Code Availability
An open-source implementation of the sciPENN algorithm is available at the online data warehouse: https://github.com/jlakkis/sciPENN. The codes are available via Zenodo at https://doi.org/10.5281/zenodo.6944521 (ref. 26). All analyses conducted in this paper can be reproduced using this repository at the online data warehouse: https://github.com/jlakkis/sciPENN_codes. The codes are available via Zenodo at https://doi.org/10.5281/zenodo.6944525 (ref. 27).
References
- 1.Chappell L, Russell AJC & Voet T Single-Cell (Multi)omics Technologies. Annu Rev Genomics Hum Genet 19, 15–41 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Stuart T & Satija R Integrative single-cell analysis. Nat Rev Genet 20, 257–272 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Stoeckius M et al. Simultaneous epitope and transcriptome measurement in single cells. Nat Methods 14, 865–868 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peterson VM et al. Multiplexed quantification of proteins and transcripts in single cells. Nat Biotechnol 35, 936–939 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Berridge MJ Unlocking the secrets of cell signaling. Annu Rev Physiol 67, 1–21 (2005). [DOI] [PubMed] [Google Scholar]
- 6.Davis DM Intercellular transfer of cell-surface proteins is common and can affect many stages of an immune response. Nat Rev Immunol 7, 238–243 (2007). [DOI] [PubMed] [Google Scholar]
- 7.Hao Y et al. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587 e3529 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gayoso A et al. Joint probabilistic modeling of single-cell multi-omic data with totalVI. Nat Methods 18, 272–282 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghattas A, Griffiths HR, Devitt A, Lip GY & Shantsila E Monocytes in coronary artery disease and atherosclerosis: where are we now? J Am Coll Cardiol 62, 1541–1551 (2013). [DOI] [PubMed] [Google Scholar]
- 10.Horne BD et al. Which white blood cell subtypes predict increased cardiovascular risk? J Am Coll Cardiol 45, 1638–1643 (2005). [DOI] [PubMed] [Google Scholar]
- 11.Berg KE et al. Elevated CD14++CD16- monocytes predict cardiovascular events. Circ Cardiovasc Genet 5, 122–131 (2012). [DOI] [PubMed] [Google Scholar]
- 12.Zhou X et al. The Kinetics of Circulating Monocyte Subsets and Monocyte-Platelet Aggregates in the Acute Phase of ST-Elevation Myocardial Infarction: Associations with 2-Year Cardiovascular Events. Medicine (Baltimore) 95, e3466 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rogacev KS et al. CD14++CD16+ monocytes independently predict cardiovascular events: a cohort study of 951 patients referred for elective coronary angiography. J Am Coll Cardiol 60, 1512–1520 (2012). [DOI] [PubMed] [Google Scholar]
- 14.Rogacev KS et al. CD14++CD16+ monocytes and cardiovascular outcome in patients with chronic kidney disease. Eur Heart J 32, 84–92 (2011). [DOI] [PubMed] [Google Scholar]
- 15.Cappellari R et al. Shift of monocyte subsets along their continuum predicts cardiovascular outcomes. Atherosclerosis 266, 95–102 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Kapellos TS et al. Human Monocyte Subsets and Phenotypes in Major Chronic Inflammatory Diseases. Front Immunol 10, 2035 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ziegler-Heitbrock L et al. Nomenclature of monocytes and dendritic cells in blood. Blood 116, e74–80 (2010). [DOI] [PubMed] [Google Scholar]
- 18.Kok L, Masopust D & Schumacher TN The precursors of CD8(+) tissue resident memory T cells: from lymphoid organs to infected tissues. Nat Rev Immunol (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kotliarov Y et al. Broad immune activation underlies shared set point signatures for vaccine responsiveness in healthy individuals and disease activity in patients with lupus. Nat Med 26, 618–629 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stephenson E et al. Single-cell multi-omics analysis of the immune response in COVID-19. Nat Med 27, 904–916 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chan Zuckerberg Initiative Single-Cell COVID-19 Consortia Single cell profiling of COVID-19 patients: an international data resource from multiple tissues. medRxiv; doi: 10.1101/2020.11.20.20227355 (2020). [DOI] [Google Scholar]
- 22.Stuart T et al. Comprehensive integration of single-cell data. Cell 177, 1888–1902. e1821 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wolf FA, Angerer P & Theis FJ SCANPY: large-scale single-cell gene expression data analysis. Genome biology 19, 15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kingma DP & Ba J Adam: a method for stochastic optimization. Preprint at arXiv:1412.6980 (2014). [Google Scholar]
- 25.Kotliarov Y et al. Broad immune activation underlies shared set point signatures for vaccine responsiveness in healthy individuals and disease activity in patients with lupus [Data set]. Nature Medicine 26, 618–629 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lakkis J et al. A multi-use deep learning method for CITE-seq and single-cell RNA-seq data integration with cell surface protein prediction and imputation [Software codes]. Zenodo, 10.5281/zenodo.6944521 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lakkis J et al. A multi-use deep learning method for CITE-seq and single-cell RNA-seq data integration with cell surface protein prediction and imputation [Analysis codes]. Zenodo, 10.5281/zenodo.6944525 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
We analyzed multiple published CITE-seq datasets throughout the evaluations. These data are available as follows (accession numbers provided where possible): 1) Mucosa-Associated Lymphoid Tissue (MALT) dataset (https://www.10xgenomics.com/resources/datasets/10-k-cells-from-a-malt-tumor-gene-expression-and-cell-surface-protein-3-standard-3-0-0); 2) Seurat 4 human peripheral blood mononuclear cells (PBMCs) (GEO: GSE164378); 3) H1N1 influenza PBMC dataset (https://doi.org/10.35092/yhjc.c.4753772, ref. 25 ); 4) human monocyte dataset (https://upenn.box.com/s/64c9fsex50g1bhv67893cpdg9c5jqjzo). All participants provided written informed consent under Columbia University IRB protocol AAAR5004. 5) Haniffa COVID Dataset (https://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-10026/); 6) Sanger COVID Dataset (https://covid19.cog.sanger.ac.uk/submissions/release2/vento_pbmc_processed.h5ad); Details of these datasets can be found from Details of these datasets were described in Supplementary Table 1.
An open-source implementation of the sciPENN algorithm is available at the online data warehouse: https://github.com/jlakkis/sciPENN. The codes are available via Zenodo at https://doi.org/10.5281/zenodo.6944521 (ref. 26). All analyses conducted in this paper can be reproduced using this repository at the online data warehouse: https://github.com/jlakkis/sciPENN_codes. The codes are available via Zenodo at https://doi.org/10.5281/zenodo.6944525 (ref. 27).