Abstract
BACKGROUND
Ultrasound is indispensable to gestational age estimation and thus to quality obstetrical care, yet high equipment cost and the need for trained sonographers limit its use in low-resource settings.
METHODS
From September 2018 through June 2021, we recruited 4695 pregnant volunteers in North Carolina and Zambia and obtained blind ultrasound sweeps (cineloop videos) of the gravid abdomen alongside standard fetal biometry. We trained a neural network to estimate gestational age from the sweeps and, in three test data sets, assessed the performance of the artificial intelligence (AI) model and biometry against previously established gestational age.
RESULTS
In our main test set, the mean absolute error (MAE) (±SE) was 3.9±0.12 days for the model versus 4.7±0.15 days for biometry (difference, −0.8 days; 95% confidence interval [CI], −1.1 to −0.5; P<0.001). The results were similar in North Carolina (difference, −0.6 days; 95% CI, −0.9 to −0.2) and Zambia (−1.0 days; 95% CI, −1.5 to −0.5). Findings were supported in the test set of women who conceived by in vitro fertilization (MAE of 2.8±0.28 vs. 3.6±0.53 days for the model vs. biometry; difference, −0.8 days; 95% CI, −1.7 to 0.2) and in the set of women from whom sweeps were collected by untrained users with low-cost, battery-powered devices (MAE of 4.9±0.29 vs. 5.4±0.28 days for the model vs. biometry; difference, −0.6; 95% CI, −1.3 to 0.1).
CONCLUSIONS
When provided blindly obtained ultrasound sweeps of the gravid abdomen, our AI model estimated gestational age with accuracy similar to that of trained sonographers conducting standard fetal biometry. Model performance appears to extend to blind sweeps collected by untrained providers in Zambia using low-cost devices. (Funded by the Bill and Melinda Gates Foundation.)
Introduction
Accurate estimation of gestational age is fundamental to quality obstetrical care. Gestational age is established as early as feasible in pregnancy and then used to determine the timing of subsequent care.1 Providers use gestational age to interpret abnormalities of fetal growth, to plan referrals, and to decide if, or when, to intervene for fetal benefit. By convention, gestational age is expressed as the time elapsed since the start of the last menstrual period (LMP). Although easily solicited, self-reported LMP has long been recognized as problematic.2 Some women may be uncertain of the LMP date. Some (perhaps most3) will have a menstrual cycle that varies from the “normal” 28-day length with ovulation on day 14. It is therefore considered best practice to confirm gestational age dating with an ultrasound examination in early pregnancy.4 This is achieved by fetal biometry, the practice of measuring standard fetal structures and applying established formulas to estimate gestational age.
Although it is ubiquitous in industrialized regions, obstetrical ultrasound is used infrequently in low- and middle-income countries.5 Reasons for this disparity include the expense of traditional ultrasound machines, their requirement of reliable electrical power, the need for obstetrics-trained sonographers to obtain images, and the need for expert interpretation. However, two recent developments offer solutions to these obstacles. The first is the expanding availability of point-of-care ultrasound devices. There are now several manufacturers of battery-powered transducers that connect to a smartphone or tablet computer and cost considerably less than a traditional ultrasound machine.6,7 The second is rapid advancement in the field of computer vision. Deep-learning algorithms are increasingly capable of interpreting medical images, and these artificial intelligence (AI) models can be deployed on mobile devices.8,9
Methods
The Fetal Age Machine Learning Initiative (FAMLI) is an ongoing project that is developing technologies to expand obstetrical ultrasound access to low-income settings. Prospective data collection commenced in September 2018 at two sites in Chapel Hill, North Carolina, and in January 2019 at two sites in Lusaka, Zambia. For the present analysis, we enrolled women who were at least 18 years of age, had a confirmed singleton intrauterine pregnancy, and provided written informed consent. The study protocol and informed consent documents were approved by the University of North Carolina Institutional Review Board, the University of Zambia Biomedical Research Ethics Committee, and the Zambia National Health Research Authority prior to initiation.
SONOGRAPHY
The study employed certified, obstetrics-trained sonographers, each of whom was credentialled by the relevant authority in their respective country (i.e., the Health Professions Council of Zambia or the American Registry for Diagnostic Medical Sonography). Participants were recruited during prenatal care and completed a single study visit with no required follow-up; however, we did allow repeat study visits no more frequently than fortnightly. Evaluation was conducted with a commercial ultrasound machine (multiple makes and models; Table S1 in the Supplementary Appendix). We performed fetal biometry by crown rump length (if less than 14 weeks) or by biparietal diameter, head circumference, abdominal circumference, and femur length (if 14 weeks or greater). Each fetal structure was measured twice and the average taken.
During the same examination, we also collected a series of blind sweep cineloop videos. These were free-hand sweeps with a two-dimensional probe, approximately 10 seconds in length, across the gravid abdomen in multiple directions and using multiple probe configurations. Craniocaudal sweeps started at the pubis and ended at the level of the uterine fundus, with the probe indicator facing toward the maternal right either perpendicular (90°) or angled (15 and 45°) to the line of probe movement. Lateral sweeps were performed with the probe indicator facing superiorly, starting just above the pubis and sweeping from the left to the right lateral uterine borders. Each subsequent lateral sweep moved progressively cephalad until either the uterine fundus was reached or six sweeps were obtained. Complete sets of blind sweep videos were collected by the study sonographer on both the commercial ultrasound machine and a low-cost, battery-powered device (Butterfly iQ; Butterfly Networks Inc., Guilford, CT). In June 2020, we began collecting a third series of sweeps at the Zambia sites. These “novice blind sweeps” were obtained by a nurse midwife with no training in sonography and included three sweeps in the craniocaudal axis and three in the lateral axis with the low-cost probe (Video 1, available at evidence.nejm.org). Before obtaining the sweeps, the novice measured the participant’s symphysial-fundal height and set the depth parameter on the ultrasound device as follows: fundus not palpable, 11-cm depth; fundus palpable but less than 25 cm, 13-cm depth; and fundus 25 cm or greater, 15-cm depth.
Except for a small number of participants who had conceived by in vitro fertilization (IVF), the “ground-truth” gestational age (i.e., gestational age established by the best method available for that participant) was established by the first ultrasound received. Our approach differed somewhat by country according to prevailing care practices. At the North Carolina sites, women presented early in pregnancy, and gestational age was set according to the American College of Obstetricians and Gynecologists practice guidelines, which incorporate fetal biometry from the first scan and the reported LMP.4 At the Zambia sites, women presented later in pregnancy,10 and the LMP was less reliable.11 We thus assigned gestational age solely on the basis of the results of the first scan, an approach that antedates the FAMLI protocol.12,13
TRAINING, TUNING, AND TESTING DATA SETS
Participants with viable singleton pregnancies enrolled between September 2018 and June 2021 were included in this study (Fig. 1). We applied participant-level exclusions to women whose available medical records did not allow a ground-truth gestational age to be established. We applied visit-level exclusions to study scans that did not contain at least two blind sweep cineloops, were uninterpretable because of missing image metadata, or were conducted before 9 weeks of gestation (because they were too infrequent to allow model training). After applying exclusions, we apportioned the remaining data into five nonoverlapping groups of participants to develop the deep-learning model (training and tuning sets) and to evaluate its performance (three test sets).
Figure 1. Study Flow Chart.
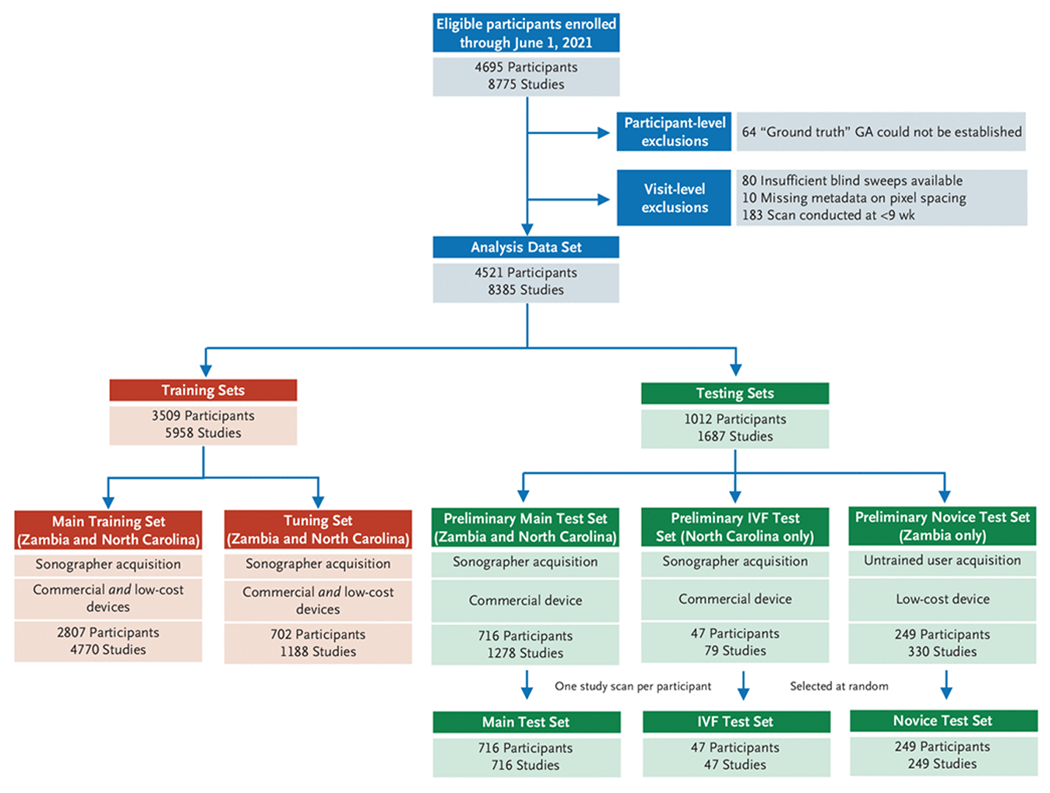
After applying participant and visit-level exclusions, we created 2 training sets to develop and tune the deep learning model and 3 test sets to assess its performance. To be eligible for inclusion in a test set a participant must have at least one study with both blind sweeps and sonographer-acquired biometry available and have their “ground truth” gestational age (GA) established by a prior scan or in vitro fertilization (IVF). The IVF test set comprises all participants who conceived by IVF. The novice test set comprises all participants in whom at least one study visit included sweeps collection by a novice user on a low-cost device. (There were 8 such novices; all were nurse midwives.) The main test set was selected at random from among all remaining eligible participants Some participants apportioned to the test sets had contributed more than one study scan; in such cases we selected a single study scan at random. The training sets comprise all participants who remain after creation of the test sets and were split randomly, by participant, in a 4:1 ratio, into a main training set and a tuning set.
The three test sets were created first. The IVF test set comprised women who conceived by IVF (and thus whose gestational age was known with certainty); all were enrolled in North Carolina. The novice test set contained women who contributed at least one study scan from the novice blind sweep protocol; all were enrolled in Zambia. Our primary assessments were made on an independent main test set, which was created as a simple random sample of 30% of eligible women who remained after creation of the other test sets. It included participants from both Zambia and North Carolina. After establishing the participant members of each test set, we ensured that each woman contributed only a single study scan to her respective test set through random selection (Fig. 1). Sensitivity analyses that include all participant study scans are presented in Tables S3 and S4.
To be included in a test set, a pregnancy had to be dated by either a prior ultrasound or IVF; this establishes the ground truth against which both the deep-learning model and biometry are measured. In Zambia, a single ultrasound provided by the FAMLI protocol may have been the only scan received. In North Carolina, a single ultrasound provided by the FAMLI protocol may have been conducted on the same day as the participant’s clinical dating ultrasound. In such cases without a prior gestational age benchmark, comparison of the model’s estimate with that of biometry is not possible. Thus, these women were included only in the data sets used for training. After creation of the three test sets, all remaining participants were randomly allocated in a 4:1 ratio into a main training set (80%) and a tuning set (20%).
TECHNICAL METHODS OF THE DEEP-LEARNING MODEL
Our deep-learning model received blind sweep cineloop videos as input and provided a gestational age estimate as output. Details of the model architecture and its constituent parts, including preprocessing steps, training procedures and parameters, and inference procedure are provided in Supplementary Appendix Section 1 and Fig. S1.
STATISTICAL ASSESSMENT OF DIAGNOSTIC ACCURACY
Predictive performance of both the model and the biometry was assessed by comparing each approach’s estimate with the previously established ground-truth gestational age. The absolute difference between these quantities was the absolute error of the prediction. We report the mean absolute error (MAE; ±SE), along with the root mean squared error of each approach. We used a paired t-test to assess the mean of the pairwise difference between the model absolute error and the biometry absolute error (|Model Error| − |Biometry Error|). Our null hypothesis was that the mean of this pairwise difference is zero; a negative mean of the pairwise difference whose 95% confidence interval (CI) does not include zero would indicate that the AI model meets our definition of statistical superiority to biometry dating.
We compared the model MAE with that of biometry in the overall test data sets and in subsets by geography (Zambia vs. North Carolina) and trimester (defined as 97 days or less, 98 to 195 days, or 196 days or more as dated by ground truth). We also plotted the empirical cumulative distribution function (CDF) for the absolute error produced by the model and the biometry. From the CDF, we compared the proportion of study scans in which the absolute error was less than 7 or 14 days for the model versus biometry, using the McNemar test. Wald-type 95% CIs for the difference in proportions were also computed. Finally, for the novice test set only, we present the diagnostic accuracy of the LMP reported at the first patient visit, because this is the relevant comparator for implementation of this technology in low-resource settings. No formal statistical analysis plan was made for this diagnostic study. The primary outcome is by default the model versus biometry in the main test set and IVF test set. No multiplicity adjustments for the secondary and exploratory end points were defined. Therefore, only point estimates and 95% CIs are provided. The CIs have not been adjusted for multiple comparisons and should not be used to infer definitive diagnostic accuracy.
Results
From September 2018 through June 2021, 4695 participants contributed 8775 ultrasound studies at the four research sites (Fig. 1). After applying participant- and visit-level exclusions, we created the three test sets as follows: 716 participants (360 from North Carolina and 356 from Zambia) formed the main test set, 47 participants (all from North Carolina) formed the IVF test set, and 249 participants (all from Zambia) formed the novice test set. As outlined earlier, participants were allowed to contribute only a single study scan (chosen at random from all the scans contributed by a single woman) to their respective test set. The 3509 participants who remained after creation of the test sets were randomly apportioned into the main training and tuning sets in a 4:1 ratio. Collectively, these women contributed 5958 study scans comprising 109,806 blind sweeps comprising 21,264,762 individual image frames for model training and tuning. Baseline characteristics of the women included in the combined training sets and the three test sets are presented in Table 1.
Table 1.
Characteristics of Participants in the Combined Training and Tuning Sets and in the Three Test Sets.*
| Training and Tuning Sets (n=3509) |
Main Test Set (n=716) |
IVF Test Set, North Carolina (n=47) | Novice Test Set, Zambia (n=249) | |||
|---|---|---|---|---|---|---|
| Characteristic† | North Carolina (n=854) | Zambia (n=2655) | North Carolina (n=360) | Zambia (n=356) | ||
| Age — yr | 30 (26 to 33) | 27 (23 to 32) | 30 (27 to 34) | 27 (24 to 31) | 37 (33 to 39) | 27 (23 to 30) |
|
| ||||||
| BMI‡ | 25.3 (22.1 to 29.2) | 24.5 (21.8 to 28.3) | 25.2 (22.0 to 29.5) | 24.6 (22.1 to 29.0) | 24.6 (22.7 to 28.1) | 24.2 (21.8 to 27.2) |
|
| ||||||
| <18.5 | 14 (1.6) | 97 (3.7) | 7 (1.9) | 11 (3.1) | 1 (2.1) | 7 (2.8) |
|
| ||||||
| 18.5 to 30 | 553 (64.8) | 1998 (75.3) | 234 (65.0) | 258 (72.5) | 34 (72.3) | 199 (79.9) |
|
| ||||||
| >30 | 163 (19.1) | 454 (17.1) | 74 (20.6) | 72 (20.2) | 7 (14.9) | 22 (8.9) |
|
| ||||||
| Parity | 2.0 (1.0 to 3.0) | 3.0 (2.0 to 4.0) | 2.0 (1.0 to 3.0) | 3.0 (2.0 to 4.0) | 2 (1 to 4) | 2 (2 to 4) |
|
| ||||||
| Gestational age at dating ultrasound — wk | 9.0 (8.0 to 11.6) | 26.3 (20.5 to 31.4) | 8.9 (7.7 to 11.0) | 23.9 (19.4 to 27.6) | NA§ | 23.7 (19.8 to 27.4) |
|
| ||||||
| No. of studies contributed | 1 (1 to 11) | 1 (1 to 7) | 1 (1 to 11) | 1 (1 to 5) | 1 (1 to 5) | 1 (1 to 5) |
|
| ||||||
| Chronic hypertension¶ | 30 (3.5) | 165 (6.2) | 16 (4.4) | 19 (5.3) | 0 (0) | 11 (4.4) |
|
| ||||||
| Diabetes‖ | 43 (5.0) | 17 (0.6) | 22 (6.1) | 1 (0.3) | 6 (12.8) | 3 (1.2) |
|
| ||||||
| HIV infection | 2 (0.2) | 719 (27.1) | 3 (0.8) | 137 (38.5) | 0 (0) | 38 (15.3) |
|
| ||||||
| Syphilis in pregnancy | 3 (0.4) | 78 (2.9) | 1 (0.3) | 22 (6.2) | 0 (0) | 7 (2.8) |
|
| ||||||
| Asthma** | 145 (17.0) | 49 (1.8) | 48 (13.3) | 11 (3.1) | 8 (17.0) | 3 (1.2) |
|
| ||||||
| Tuberculosis | 0 (0) | 80 (3.0) | 1 (0.3) | 13 (3.7) | 0 (0) | 2 (0.8) |
|
| ||||||
| Alcohol use during pregnancy | 196 (23.0) | 220 (8.3) | 89 (24.7) | 57 (16.0) | 13 (27.7) | 17 (6.8) |
|
| ||||||
| Tobacco use during pregnancy | 68 (8.0) | 6 (0.2) | 33 (9.2) | 4 (1.1) | 0 (0) | 0 (0) |
|
| ||||||
| Pregnancy conceived by IVF | 2 (0.2)†† | 0 (0) | 0 (0) | 0 (0) | 47 (100) | 0 (0) |
Data are presented as median (interquartile range) or n (%). HIV denotes human immunodeficiency virus, IVF in vitro fertilization.
Information on medical conditions was obtained from both patient interview and medical record review.
The body-mass index (BMI) is the weight in kilograms divided by the square of the height in meters. Missing height prevents BMI calculation of 5 in the IVF test set, 21 in the novice test set, 60 in the main test set, and 230 in the combined training sets.
NA denotes not applicable because gestational age was assigned by known fertilization date in the IVF test set.
Chronic hypertension is defined as ever having a diagnosis of hypertension outside of pregnancy or a new diagnosis in the current pregnancy prior to 20 weeks of gestation.
Diabetes includes both pregestational and gestational diabetes.
Asthma is defined as ever having received the diagnosis of asthma.
Two women who conceived by IVF had blind sweeps collected with the Butterfly iQ device but not a commercial ultrasound machine; hence, they were not able to be included in the IVF test set.
MODEL VERSUS BIOMETRY IN THE MAIN TEST SET AND IVF TEST SET
In the main test set, the deep-learning model outperformed biometry, with an overall MAE (±SE) of 3.9±0.12 days for the model versus 4.7±0.15 days for biometry (difference, −0.8 days; 95% CI, −1.1 to −0.5; P<0.001; Fig. 2 and Table 2). The observed difference manifested primarily in the third trimester, in which the mean of the pairwise difference in absolute error was −1.3 days (95% CI, −1.8 to −0.8; P<0.001). On the basis of the empirical CDF, the percentage of study scans that were correctly classified within 7 days was higher for the model than for biometry (86.0% vs. 77.0%; difference, 9.1 percentage points; 95% CI, 5.7 to 12.5 percentage points; P<0.001). The model similarly outperformed biometry using a 14-day classification window (98.9% vs. 96.9%; difference, 2.0 percentage points; 95% CI, 0.5 to 3.4 percentage points; P = 0.01). In a sensitivity analysis limiting the main test set to women whose pregnancy was dated by a first trimester ultrasound (322 from North Carolina and 31 from Zambia), the model performed favorably (Fig. S4 and Table S5), with an MAE (±SE) of 3.5±0.15 days for the model versus 4.0±0.20 days for biometry (difference, −0.5 days; 95% CI, −0.9 to −0.2).
Figure 2. Model Versus Biometry in the Main Test Set and IVF Test Set.
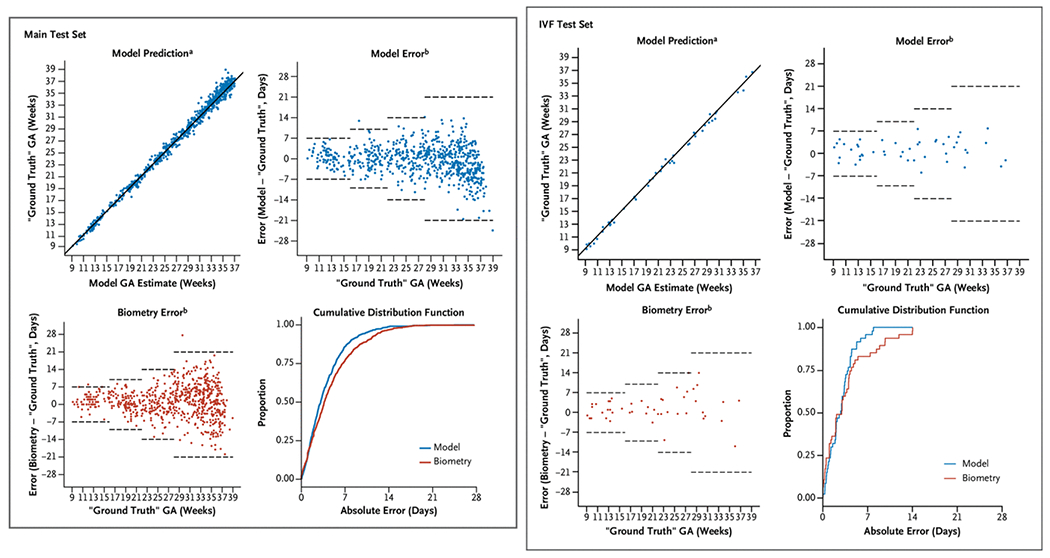
a solid line indicates y = x, b dashed horizontal lines represent expected error bounds of ultrasound biometry according to the American College of Obstetricians and Gynecologists.4 In Zambia, “ground truth” gestational age is defined by the first ultrasound. In North Carolina it is defined by an algorithm incorporating both the last menstrual period and the first ultrasound4 (main test set) or by the known fertilization date (IVF test set). GA denotes gestational age and IVF in vitro fertilization.
Table 2.
Gestational Age Estimation of Deep-Learning Model Compared with Trained Sonographer in the Main Test Set and In Vitro Fertilization (IVF) Test Set.*
| Main Test Set (n = 716)† |
IVF Test Set (n=47)‡ |
|||||
|---|---|---|---|---|---|---|
| Estimate | Model | Biometry | Difference (95% CI) | Model | Biometry | Difference (95% CI) |
| MAE (±SE) — d | 3.9±0.12 | 4.7±0.15 | −0.8 (−1.1 to −0.5) | 2.8±0.28 | 3.6±0.53 | −0.8 (−1.7 to 0.2) |
|
| ||||||
| RMSE (±SE) — d | 5.1±0.18 | 6.1±0.19 | −1.1 (−1.5 to −0.6) | 3.4±0.31 | 5.1±0.67 | −1.7 (−2.8 to −0.5) |
|
| ||||||
| Estimate of bias (±SE) — d§ | −0.7±0.19 | 1.3±0.22 | 1.3±0.46 | 1.7±0.70 | ||
|
| ||||||
| First trimester — d¶ | ||||||
|
| ||||||
| MAE (±SE) | 2.1±0.19 | 2.0±0.23 | 0.2 (−0.4 to 0.7) | 2.3±0.34 | 2.2±0.38 | |
|
| ||||||
| Estimate of bias (±SE) | 0.8±0.32 | 1.4±0.29 | 0.8±0.67 | 0.9±0.66 | ||
|
| ||||||
| Second trimester — d | ||||||
|
| ||||||
| MAE (±SE) | 3.1±0.16 | 3.3±0.17 | −0.2 (−0.5 to 0.1) | 2.7±0.45 | 3.4±0.84 | |
|
| ||||||
| Estimate of bias (±SE) | 0.2±0.25 | 0.7±0.26 | 1.3±0.69 | 2.0±1.04 | ||
|
| ||||||
| Third trimester — d | ||||||
|
| ||||||
| MAE (±SE) | 4.7±0.18 | 6.0±0.22 | −1.3 (−1.8 to −0.8) | 3.7±0.69 | 5.7±1.38 | |
|
| ||||||
| Estimate of bias (±SE) | −1.4±0.29 | 1.6±0.36 | 1.8±1.23 | 2.4±2.13 | ||
|
| ||||||
| Absolute error (±SE) — % | ||||||
|
| ||||||
| <7 d | 86.0±1.3 | 77.0±1.6 | 9.1 (5.7 to 12.5) | 95.7±2.9 | 83.0±5.5 | |
|
| ||||||
| <14 d | 98.9±0.4 | 96.9±0.6 | 2.0 (0.5 to 3.4) | 100.0 | 100.0 | |
|
| ||||||
| North Carolina — d | ||||||
|
| ||||||
| MAE (±SE) | 3.4±0.15 | 4.0±0.20 | −0.6 (−0.9 to −0.2) | |||
|
| ||||||
| Estimate of bias (±SE) | −0.3±0.23 | 0.7±0.28 | ||||
|
| ||||||
| Zambia — d | ||||||
|
| ||||||
| MAE (±SE) | 4.4±0.19 | 5.4±0.22 | −1.0 (−1.5 to −0.5) | |||
|
| ||||||
| Estimate of bias (±SE) | −1.0±0.30 | 1.9±0.35 | ||||
CI denotes confidence interval, MAE mean absolute error, and RMSE root mean square error
The main test set comprises a 30% random sample of participants who are dated by a prior ultrasound and who are not included in the IVF or novice test sets. Participants were enrolled in either North Carolina or Zambia. Blind sweeps and fetal biometry were collected by a sonographer on a commercial ultrasound machine
The IVF test set comprises studies conducted in women who conceived by in vitro fertilization. All participants were enrolled in North Carolina. Blind sweeps and fetal biometry were collected by a sonographer on a commercial ultrasound machine.
Estimate of bias is reported as the estimated mean of the error.
Trimesters are defined as 97 days or less, 98 to 195 days, or 196 days or more.
Among the 47 study scans in the IVF test set, the MAE (±SE) was 2.8±0.28 days for the model compared with 3.6±0.53 days for biometry (difference, −0.8 days; 95% CI, −1.7 to 0.2; P=0.10). As was observed in the main test set, the difference was most pronounced in the third trimester, in which the estimated mean of the pairwise difference in absolute error was −2.0 days. On the basis of the empirical CDF, the proportion of study scans that were classified correctly within 7 days was higher for the model than for biometry (95.7% vs. 83.0%). Owing to the small sample size in our IVF test set, we did not perform statistical tests on the difference by trimester or the difference in proportion. Both the model and biometry categorized 100% of cases correctly within 14 days ( Table 2).
MODEL VERSUS BIOMETRY AND LMP IN THE NOVICE TEST SET
The novice test set contains 249 sets of blind sweeps obtained on the Butterfly iQ battery-powered device by an untrained user (Fig. 3 and Table S2). A total of eight untrained users (all nurse midwives) participated. As described earlier, we compared model estimates with biometry obtained by a trained sonographer on a commercial ultrasound machine. We also compared the model estimates with the gestational age that would have been calculated had only the LMP been available (as is overwhelmingly the case in Zambia). In the novice test set, the model and biometry performed similarly, with an overall MAE (±SE) of 4.9±0.29 days for the model versus 5.4±0.28 days for biometry (difference, −0.6 days; 95% CI, −1.3 to 0.1). However, when compared with the LMP, the model was clearly superior, with an MAE of 4.9±0.29 days versus 17. 4±1.17 days for the LMP (difference, −12.7 days; 95% CI, −15.0 to −10.3). On the basis of the empirical CDF, the percentage of study scans that were correctly classified within 7 days was substantially higher for the model than for the LMP (75.1% vs. 40.1%; difference, 36.1 percentage points; 95% CI, 28.0 to 44.2 percentage points). The model similarly outperformed the LMP using a 14-day classification window (95.6% vs. 55.1%; difference, 40.5 percentage points; 95% CI, 33.9 to 47.1 percentage points).
Figure 3. Model Versus Biometry and LMP in the Novice Test Set.
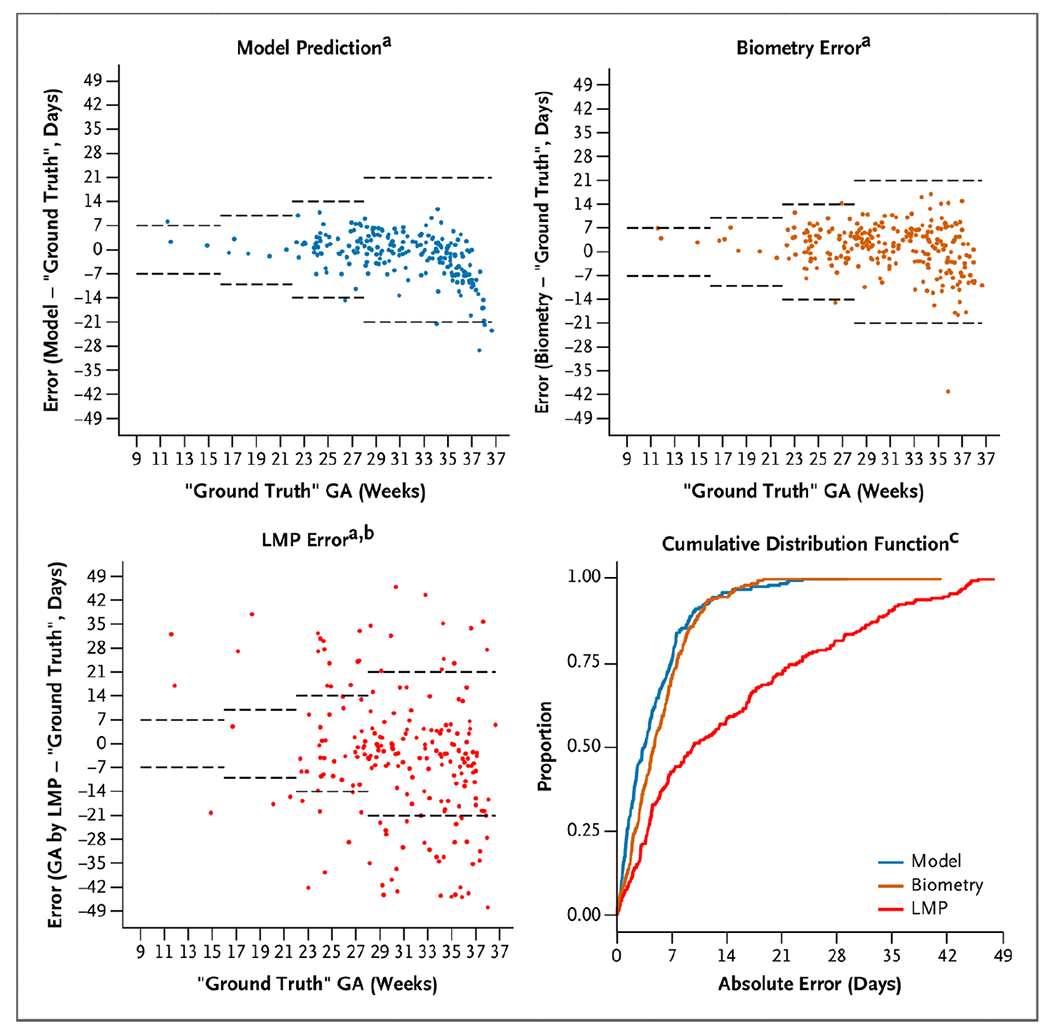
a dashed horizontal lines represent expected error bounds of ultrasound biometry according to the American College of Obstetricians and Gynecologists.4 b data missing from 22 participants who could not recall their LMP c 13 studies from GA by LMP excluded from the plot because the absolute error is truncated at 49 days In Zambia, “ground truth” gestational age is defined by the first ultrasound. GA denotes gestational age and LMP last menstrual period.
Discussion
Quality obstetrical care requires accurate knowledge of gestational age. We built a deep-learning model that can perform this critical assessment from blindly obtained ultrasound sweeps of the gravid abdomen. Expressed as the MAE or as the percentage of estimates that fall within 7 or 14 days of a previously defined ground-truth gestational age, the model performance is superior to that of a trained sonographer performing fetal biometry on the same day. Results were consistent across geographic sites and were supported in a test set of women who conceived by IVF (whose ground-truth gestational age was unequivocally established) and in a test set of women from whom the ultrasound blind sweeps were obtained by a novice provider using a low-cost, battery-powered device.
This research addresses a shortcoming in the delivery of obstetrical care in low- and middle-income countries. In the Lusaka public sector, which is typical of care systems across the sub-Saharan Africa and parts of Asia, few women have access to ultrasound pregnancy dating, and the median gestational age at presentation for antenatal care is 23 weeks (interquartile range, 19 to 26 weeks).10 This means that each year in the city of Lusaka, more than 100,000 pregnancies14 must be managed with an unacceptably low level of gestational age precision (Fig. 3).11,15 The availability of a resource-appropriate technology that could assign gestational age in the late second and third trimesters with reasonable accuracy could provide those caring for these expectant mothers a higher standard of care than is currently available.
This study collected thousands of images from each participant in the form of blind sweeps. Each cineloop frame in the sweep is itself a two-dimensional ultrasound image that is provided to the neural network during training. Although most of these frames would be considered clinically suboptimal views, the sheer number of them (more than 21 million) provides a comprehensive picture of the developing fetus from multiple angles across the spectrum of gestational age. Considered as individual images rather than participants, studies, or sweeps, our training set is two orders of magnitude larger than most of the prior high-profile applications of deep learning to medical imaging.16–19 As we have verified through manual review, it is rare for a blind sweep to contain the ideal image planes necessary for standard fetal biometry. Although the nature of the deep-learning algorithm is such that we do not know exactly which image features the model uses to make its predictions, it seems likely to be incorporating many facets of the available data to accomplish its task, rather than simply mimicking that which is acquired when a trained sonographer performs biometry. The very large sample of training images may also help explain the model’s excellent performance even though our Zambian training data include some studies from women who presented late for care and whose clinically established gestational age was therefore imprecisely estimated.
Strengths of this study include its prospective nature and bespoke blind sweep sonography procedures. We used several different makes and models of ultrasound scanners for data collection, a feature that likely bolsters the model’s generalizability. Although this technology seems primarily suited for low-resource settings, we included participants from North Carolina to increase the heterogeneity in our training sets. We see combining these disparate populations in model training as an overall strength of the research, but note that our design does not include a truly external validation cohort. Although we did not deliberately impose a lower gestational age limit on enrollment, our data set includes very few scans at less than 9 gestational weeks, and we thus are unable to make estimates below this threshold. Data were similarly sparse beyond 37 weeks (term gestation), and the model appears to systematically underestimate gestational age beyond this point in the novice test set. We note, however, that this limitation would only affect that minority of women who attend prenatal care but have no visits between 9 and 37 weeks. From our prior population-based study of 115,552 pregnancies in Lusaka, less than 1% of women would meet these criteria.10 Finally, we acknowledge that a blind sweep approach to obstetrical sonography would be a departure from current diagnostic practices and might be seen as a threat to diagnostic capacity building in low-resource settings. However, the technology has the capacity to fill a real gap in obstetrics care within currently available resources.
Our data show that AI can estimate gestational age from a series of blindly obtained ultrasound sweeps with accuracy similar to that of a trained sonographer conducting standard biometry. This performance appears to extend to sweeps collected by untrained providers in Zambia using low-cost ultrasound devices. Whether this technology can be successfully disseminated into extant health care systems in low-resource settings will require further study.
Supplementary Material
Acknowledgments
This work was funded by the Bill and Melinda Gates Foundation (Grants OPP1191684 and INV003266). Complementary resources were provided by the University of North Carolina School of Medicine and the National Institutes of Health (Grants T32 HD075731 and K01 TW010857 to Dr. Price, UL1 TR002489 to Dr. Kosorok, R01 AI157758 to Dr. Cole, K24AI120796 to Dr. Chi, and P30 AI50410 to Drs. Chi, Cole, and J. S. A. Stringer). Butterfly Network Inc. donated ultrasound probes for this research. The conclusions and opinions expressed in this article are those of the authors and do not necessarily reflect those of the Bill and Melinda Gates Foundation or the National Institutes of Health. The funders had no role in study design, data collection, analysis, preparation of the manuscript, or interpretation of results.
We thank our research participants in both Lusaka and Chapel Hill for providing data to this study. We thank Drs. Sindura Ganapathi, Rasa Izadnegahdar, Hilary Gammill, Arunan Skandarajah, Bryan Ranger, Anisha Gururaj, Alan Rosenbaum, David Stamilio, Xiaoning Jiang, and Sergey Feldman for support in conceptualizing and planning this work.
Footnotes
Disclosures
Disclosure forms provided by the authors are available with the full text of this article at evidence.nejm.org.
The data sets used for this research will be made available to other investigators upon request through a third-party data-sharing platform. Access is subject to specific terms around acceptable use and attribution.
References
- 1.World Health Organization. WHO Recommendations on Antenatal Care for a Positive Pregnancy Experience. 2016. (https://www.who.int/reproductivehealth/publications/maternal_perinatal_health/anc-positive-pregnancy-experience/en/). [PubMed]
- 2.Kramer MS, McLean FH, Boyd ME, Usher RH. The validity of gestational age estimation by menstrual dating in term, preterm, and postterm gestations. JAMA 1988;260:3306–3308. [PubMed] [Google Scholar]
- 3.Matsumoto S, Nogami Y, Ohkuri S. Statistical studies on menstruation: a criticism on the definition of normal menstruation. Gunma J Med Sci 1962;11:294–318. [Google Scholar]
- 4.Chiazze L Jr., et al. The length and variability of the human menstrual cycle. JAMA 1968;203:377–380. [PubMed] [Google Scholar]
- 5.Yadav H, Shah D, Sayed S, Horton S, Schroeder LF. Availability of essential diagnostics in ten low-income and middle-income countries:results from national health facility surveys. Lancet Glob Health 2021;9:e1553–e1560. DOI: 10.1016/S2214-109X(21)00442-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marsh-Feiley G, Eadie L, Wilson P. Telesonography in emergency medicine: A systematic review. PLoS One 2018;13:e0194840. DOI: 10.1371/journal.pone.0194840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Becker DM, Tafoya CA, Becker SL, Kruger GH, Tafoya MJ, Becker TK. The use of portable ultrasound devices in low- and middle-income countries: a systematic review of the literature. Trop Med Int Health 2016;21:294–311. DOI: 10.1111/tmi.12657. [DOI] [PubMed] [Google Scholar]
- 8.Carin L, Pencina MJ. On deep learning for medical image analysis. In: Livingston EH, Lewis RJ, eds. JAMA guide to statistics and methods. New York: McGraw-Hill Education, 2019;1192–1193. [DOI] [PubMed] [Google Scholar]
- 9.Esteva A, Robicquet A, Ramsundar B, et al. A guide to deep learning in healthcare. Nat Med 2019;25:24–29. DOI: 10.1038/s41591-018-0316-z. [DOI] [PubMed] [Google Scholar]
- 10.Chi BH, Vwalika B, Killam WP, et al. Implementation of the Zambia electronic perinatal record system for comprehensive prenatal and delivery care. Int J Gynaecol Obstet 2011;113:131–136. DOI: 10.1016/j.ijgo.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Price JT, Winston J, Vwalika B, et al. Quantifying bias between reported last menstrual period and ultrasonography estimates of gestational age in Lusaka, Zambia. Int J Gynaecol Obstet 2019;144: 9–15. DOI: 10.1002/ijgo.12686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Castillo MC, Fuseini NM, Rittenhouse K, et al. The Zambian Preterm Birth Prevention Study (ZAPPS): cohort characteristics at enrollment. Gates Open Res 2019;2:25. DOI: 10.12688/gatesopenres.12820.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Price JT, Vwalika B, Rittenhouse KJ, et al. Adverse birth outcomes and their clinical phenotypes in an urban Zambian cohort. Gates Open Res 2020;3:1533. DOI: 10.12688/gatesopenres.13046.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zambian Ministry of Health. Annual Health Statistics Report 2017-2019. October 2020. (https://www.moh.gov.zm/?wpfb_dl=159).
- 15.Vwalika B, Price JT, Rosenbaum A, Stringer JSA. Reducing the global burden of preterm births. Lancet Glob Health 2019;7:e415. DOI: 10.1016/S2214-109X(19)30060-9. [DOI] [PubMed] [Google Scholar]
- 16.Gulshan V, Peng L, Coram M, et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA 2016;316:2402–2410. DOI: 10.1001/jama.2016.17216. [DOI] [PubMed] [Google Scholar]
- 17.Milea D, Najjar RP, Zhubo J, et al. Artificial intelligence to detect papilledema from ocular fundus photographs. N Engl J Med 2020; 382:1687–1695. DOI: 10.1056/NEJMoa1917130. [DOI] [PubMed] [Google Scholar]
- 18.McKinney SM, Sieniek M, Godbole V, et al. International evaluation of an AI system for breast cancer screening. Nature 2020;577:89–94. DOI: 10.1038/s41586-019-1799-6. [DOI] [PubMed] [Google Scholar]
- 19.Esteva A, Kuprel B, Novoa RA, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017;542: 115–118. DOI: 10.1038/nature21056. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


