Abstract
The Polycomb group proteins are responsible for long-term repression of a number of genes in Drosophila melanogaster, including the homeotic genes of the bithorax complex. The Polycomb protein is thought to alter the chromatin structure of its target genes, but there has been little direct evidence for this model. In this study, the chromatin structure of the bithorax complex was probed with three separate assays for DNA accessibility: (i) activation of polymerase II (Pol II) transcription by Gal4, (ii) transcription by the bacteriophage T7 RNA polymerase (T7RNAP), and (iii) FLP-mediated site-specific recombination. All three processes are restricted or blocked in Polycomb-repressed segments. In contrast, control test sites outside of the bithorax complex permitted Gal4, T7RNAP, and FLP activities throughout the embryo. Several P insertions in the bithorax complex were tested, providing evidence that the Polycomb-induced effect is widespread over target genes. This accessibility effect is similar to that seen for SIR silencing in Saccharomyces cerevisiae. In contrast to SIR silencing, however, episomes excised from Polycomb-repressed chromosomal sites do not show an altered superhelix density.
The homeotic genes of Drosophila melanogaster are found in two gene clusters, the Antennapedia complex and the bithorax complex (BX-C). The complexes are large and contain many enhancers, which act over long distances to drive segment-specific expression of the homeotic genes (reviewed in reference 12). The BX-C is divided into at least nine regulatory domains, each of which is responsible for activating homeotic gene transcription in a given parasegment (parasegments 5 to 13 [PS5 to -13]). The domains are aligned on the chromosome in the linear order of the parasegments they affect.
In early embryogenesis, the expression domains of the homeotic genes are established by a group of short-lived regulators, products of the gap and pair-rule genes. These factors disappear at approximately 5 h into embryogenesis, and control is transferred to another group of factors, the Polycomb group (PcG). The PcG acts to maintain the proper segmental patterns of the target genes, “remembering” the pattern over many cell division cycles. In PcG mutants, the homeotic genes are misexpressed and are transcribed in all segments of the embryo (reviewed in references 38 and 45).
Recent biochemical findings suggest that PcG factors are found in large multiprotein complexes (34, 44). It is not clear how these complexes are targeted to DNA sites or how they maintain repression. However, several pieces of evidence suggest that transcriptional repression by the PcG might mimic the formation of heterochromatin. The Polycomb protein, the first PcG factor identified, shares a protein motif, the chromodomain, with the heterochromatin-associated factor HP-1 (37). Like heterochromatic regions, the BX-C appears underreplicated in the polytene chromosomes of the Drosophila salivary gland (24), a tissue in which the BX-C is transcriptionally inactive, and is thought to be repressed by the PcG. Numerous transgene insertions have been recovered in the BX-C, almost all of which appear to respond to PcG regulation (3, 30). Moreover, transgenes outside of the BX-C bearing Polycomb response elements (PREs) show variegated repression of neighboring reporter genes (9).
While it is clear from these results that the PcG is able to repress many enhancer-promoter combinations and to act over long distances, there is little direct evidence for chromatin modification by the PcG. Indeed, it has been suggested that the PcG might exert its repressive effect specifically on promoter regions or by inhibiting promoter-enhancer interactions (5, 39). It has also been postulated, based on in vitro data, that the PcG might affect chromatin structure indirectly by blocking the activity of other chromatin remodeling complexes (44), such as the brahma complex (36). The brahma gene has been identified as a member of the trithorax group of factors, which act as genetic antagonists to the PcG (reviewed in reference 22).
PcG-mediated repression shares similarities not only to heterochromatic position effects, but also to silencing mediated by the SIR complex of proteins of Saccharomyces cerevisiae. The SIR proteins appear to mediate long-range transcriptional repression over the target loci, and transgene insertions into these loci are invariably silenced (reviewed in reference 14). The SIR proteins themselves are thought to coat the silenced regions, and several pieces of experimental evidence suggest that SIR silencing is associated with an altered chromatin structure. This evidence includes reduced accessibility of the transcriptionally repressed DNA (17, 25, 47). There have been several previous attempts to identify similar PcG-dependent changes in DNA accessibility. Boivin and Dura (6) tested changes in DNA accessibility by using Escherichia coli dam DNA methyltransferase as a probe. Using transgenes containing a presumptive PRE from the polyhomeotic locus as their target DNA, they demonstrated ∼2-fold changes in the level of methylation in PcG mutant flies versus wild-type flies. However, Schloβherr et al. (42) examined endogenous sequences of the BX-C for restriction enzyme accessibility and failed to find any difference. Similarly, in a previous study from this laboratory, bacteriophage T7 RNA polymerase (T7RNAP) was used to probe DNA accessibility in the BX-C, and no sensitivity to the PcG was seen (29). These studies suggested that if an accessibility block is imposed by the PcG, it must be incomplete or selective.
In this study, we expand our analysis of DNA accessibility in the BX-C by using Gal4, T7RNAP, and FLP recombinase as probes. Each assay relies on in situ hybridization to fixed embryos, so that the products can be visualized cell by cell. The comparison of PcG-repressed and nonrepressed segments provides an internal control within each animal. By introducing Gal4, we tested for the ability of a foreign activator to elicit transcription from the fly's own polymerase II (Pol II) machinery under PcG-repressed conditions. Similarly, we compared the ability of a foreign polymerase, T7RNAP, to transcribe in PcG-repressed versus nonrepressed cells. T7RNAP has been used as a tool for recognizing altered chromatin states in yeast (10), trypanosome (33), and mammalian (19) systems. Although we had previously found no effect of PcG on T7RNAP, it seemed possible that the PcG might be more effective in blocking large protein complexes, such as the RNA Pol II transcription apparatus. Therefore, we created an enlarged version of T7RNAP, called “Goliath polymerase,” and compared it to the wild-type T7RNAP in its sensitivity to PcG modification of the DNA. We also tested the ability of the site-specific recombinase, FLP, to find and synapse its target sites and to recombine a circular episome out of the chromosome. We performed these assays with multiple P element insertions, spread throughout the BX-C, each of which contains target sites for Gal4, T7RNAP, and FLP. We found consistent effects by the PcG on all three proteins.
Our observations with the Gal4, T7RNAP, and FLP probes suggest that the DNA within our P insertions is somehow altered when the control region surrounding it is actively PcG repressed. In addition to reduced DNA accessibility, SIR silencing has been shown to correlate in vivo with an altered topology over the repressed DNA (4, 11). Changes in nucleosome density, nucleosome conformation, or the association of other DNA binding factors can alter the packaging of the DNA, and all have been shown to have measurable effects on DNA topology (27, 31, 41). Similarly, PRC1, a purified complex containing a subset of the PcG factors, has been shown to inhibit SWI/SNF remodeling of chromatin in vitro (44). The assay used involved visualization of a change in the topology of a nucleosomal template, because SWI/SNF removed negative supercoils. Thus, we might expect PcG-repressed DNA of the BX-C to have a greater negative superhelix density than nonrepressed DNA. In this study, we examined the supercoiling of PcG target DNA in vivo by a method very similar to that used with yeast to study SIR silencing. Circular episomes were produced from our FLP-inducible cassettes in order to trap the structure of the PcG-modified DNA. In contrast to SIR silencing, we do not see a difference between the topologies of PcG-repressed DNA and nonrepressed DNA.
MATERIALS AND METHODS
Plasmid constructions.
Each FRT in the construct illustrated in Fig. 1A is a 48-bp sequence derived from pJFS36 (43). The two tandem 49-bp T7 promoters were cloned by PCR from pT7–7 (kindly provided by Stan Tabor). The UAS sequence was derived from pUAST (8), and the LacZ reporter was derived from pCaSpeR-AUG-β-gal (51). The complete T7 promoter/UAS-LacZ cassette was subcloned into the pCasper-4 transformation vector and transformed into Drosophila to produce the control lines. The miniwhite reporter was removed from this construct and replaced with the homing fragment/rosy transformation marker cassette from the “homing pigeon” (3) to produce the P element of Fig. 1A. This P element was used to produce the five BX-C insertions shown in Fig. 2.
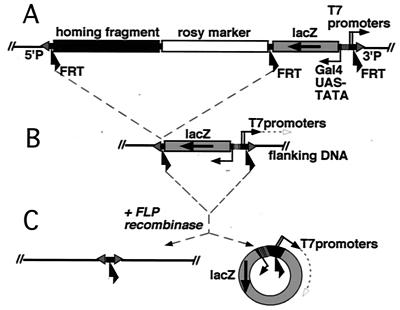
FIG. 1.
The P element construct serves as a target for Gal4, T7RNAP, and FLP. (A) Diagram of the P element. The initial P insertions contain two cassettes, each flanked by directly oriented FRT sequences. The left cassette contains the rosy transformation marker, and a “homing fragment,” which targets P elements to the BX-C chromosome region. The right cassette contains two tandem T7 promoters and a Gal4 activatable UAS-LacZ reporter. (B) Trimmed-down insertions, which lack the homing/rosy + cassette, were recovered as rosy− LacZ+ individuals following FLP induction in the germ line. The T7 promoters are poised to transcribe the genomic DNA flanking the 3′ P end. (C) Expression of FLP recombinase in a whole embryo results in excision of the T7 promoter/UAS-LacZ cassette from the chromosome, resulting in formation of a 4.9-kb circular episome in somatic cells. The T7 promoters are then poised to produce antisense copies of LacZ RNA.
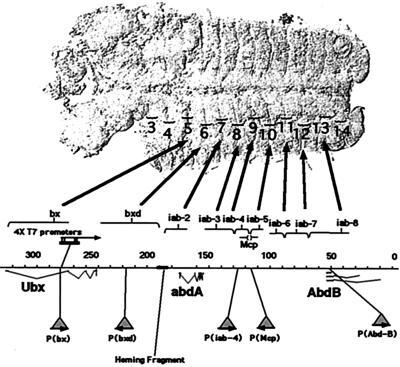
FIG. 2.
Five P element insertions were recovered within the BX-C. Transcription units are marked for the three homeotic genes, Ubx, abd-A, and Abd-B. Coordinates from the published sequence are shown (in kilobases). Above the map, control regions for each of the genes are denoted by brackets, and the large arrows point to the most anterior parasegments that they regulate in the fly embryo (displayed with the anterior portion to the left). The sites of insertion of the five P elements are indicated below the map as triangles. The location of the original 4× T7 promoters in the bx region (29) is indicated above the map as a black rectangle. Arrows denote the orientation of the T7 promoters.
The LacZ coding region in the Goliath polymerase fusion gene was recovered by PCR from the Adh-LacZ gene in PcaSpeR-AUG-β-gal (51) and subcloned into the EcoRI site of pAR3283 (13), which contains T7 gene 1 with a simian virus 40 (SV40) T-antigen nuclear localization signal (NLS) at its N terminus. This creates an in-frame fusion with the sequence N terminus–NLS–β-gal-T7RNAP–C terminus. This fusion was subcloned into the Drosophila transformation vector Pcasper-HS (50) and the Drosophila transformation vector pUAST (8). The wild-type UAS T7RNAP construct was engineered in a similar fashion by subcloning the NLS-T7RNAP gene from pAR3283 into pUAST. The mutant forms of T7RNAP, which included the promoter binding mutant (N748G) (40), the elongation mutant (thumb subdomain Δ3) (7), and the termination mutant (Δ271–2) (26), were subcloned behind the NLS in pAR3283 and subsequently subcloned into pUAST. Full details on the construction of all vectors are available upon request.
D. melanogaster stocks and crosses.
Germ line transformants were made as previously described (46). Locations of the new P insertions were identified by inverse PCR with primers in the P element ends (35). To produce trimmed-down insertions, flies with the initial P insertions were treated with FLP, and rosy− progeny were collected. These were screened by PCR for retention of the T7 promoter and UAS sequences; less than 1% of the rosy− candidates had the desired structure. All crosses were carried out at 25°C. Tests of LacZ patterns in PcG mutants utilized embryos homozygous for Pc3, PscII, and l(429b) and hemizygous for DfJA52, for testing zygotic loss of the Polycomb, Posterior Sex Combs, pleiohomeotic, and polyhomeotic genes, respectively. For extra sex combs, embryos lacking both maternal and zygotic product were generated by crossing esc10/esc2 males and females. To perform the FLP accessibility assays, females containing a heat shock-inducible source of FLP located on the X chromosome and a heat shock-inducible source of T7RNAP on the 2nd chromosome were crossed to males containing the accessibility constructs. The same source of FLP was used to produce circles for topology experiments in embryos and adults (Table 1). To generate circles from Polycomb mutant embryos, 70FLP4A, Pc3 /TM2 flies were crossed to either Pc3, p[Mcp] or Pc3, p[Abd-B] recombinant flies. Table 1 lists the transformant lines used in this paper.
TABLE 1.
Drosophila lines used in this paper
| Name in paper | Line name or genotype | Chromosome | Source or reference |
|---|---|---|---|
| p[bx] | pHFbx | III | This paper |
| p[bxd] Full length | pHF79A | III | This paper |
| p[iab-4] | pHFiab-4 | III | This paper |
| p[iab-4] Full length | pHF608B | III | This paper |
| p[Mcp] | pHFmcp | III | This paper |
| p[Abd-B] | pHFiab8 | III | This paper |
| X chromosome control | pCASFUBT X | I | This paper |
| 3rd chromosome control | pCASFUBT 85A | III | This paper |
| Ubiquitous Gal4 | 32B | III | 8 |
| 4× T7 promoters in bx | sbd, 2×8-1, e | III | 29 |
| CNS T7RNAP | 471I | II | 29 |
| CNS Goliath with 4× T7 promoters | pβT733E, sbd, 2×8-1, e | III | This paper |
| UAS T7RNAP | pUAST7 25A | II | This paper |
| UAS Goliath | pUASβT7 9A | III | This paper |
| Heat shock-inducible T7RNAP | 4710H | II | 29 |
| Heat shock-inducible FLP on X | y, w, p[hs70-FLP, ry+]122 | I | 48 |
| Heat shock-inducible FLP on III | 70FLP4A | III | 16 |
Whole-mount immunolocalization in Drosophila embryos.
Whole-mount immunolocalization of Goliath polymerase expression patterns was performed as previously described (21), with mouse anti-β-galactosidase monoclonal antibody (Promega) as the primary antibody and horseradish peroxidase-conjugated goat anti-mouse antibody (Bio-Rad) as the secondary antibody. Embryos were dissected as previously described (21) and mounted in Immu-mount (Shandon) for photography.
RNA in situ analysis of Drosophila embryos.
RNA in situ analysis was performed as previously described (15) with the following modifications. Hybridizations were carried out overnight at 45 to 50°C, and the alkaline phosphatase detection buffer was prepared with 2-amino-2-methyl-1-propanol (Sigma) instead of Tris-HCl. Digoxigenin-labeled RNA probes were prepared according to the protocol outlined for the Genius system (Boehringer Mannheim). The lacZ probe is complementary to 3 kb of LacZ coding sequence. The probe for transcription from p[bx], as well from the 4× T7 promoters, is from a 1.2-kb fragment, representing BX-C sequence coordinates 272,997 to 274,194 (sequence 89E of reference 28). A 1.4-kb fragment representing BX-C sequence 219,410 to 220,834 was used as a probe for transcription from p[bxd]. A 1.4-kb fragment representing BX-C sequence 125,812 to 127,224 was used as a probe for p[iab-4]. A 1.1-kb fragment representing BX-C sequence 113,733 to 114,900 was used as a probe for p[Mcp]. A 1.1-kb fragment representing BX-C sequence 48,720 to 49,853 was used as a probe for p[Abd-B]. The circle probe is complementary to 3 kb of the antisense strand of the LacZ coding sequence. To produce a probe for the promoters on the X chromosome, inverse PCR was performed as previously described (35) to capture ∼370 bp of the genomic sequences immediately adjacent to the X chromosome control insertion.
As reported previously (29), RNA products transcribed by T7RNAP were visible following a brief heat shock, but no such products were visible in the absence of a heat shock, despite constitutive expression of the polymerase. Transcription by Goliath polymerase similarly required a heat shock. All heat shocks to induce polymerase activity were carried out at 39°C in glass fly vials immersed in a circulating water bath for either 10 min (central nervous system [CNS] T7RNAP) or 60 min (ubiquitous Gal4-induced T7RNAP and CNS Goliath experiments). Embryos were fixed immediately after heat shock. Embryos were dissected and mounted as described above.
Analysis of supercoiled circles.
To prepare PcG-repressed DNA from adult flies, approximately 500 heads were severed by vortexing and sieved through a 710-μm-pore mesh. In a separate experiment, frozen flies were individually dissected, and their heads, thoraxes, and abdomens were collected separately. Whole adult flies were used to compare males and females containing the X chromosome insertion. To prepare genomic DNA from embryos, dechorionated embryos were ground in embryo grinding buffer (100 mM Tris [pH 8], 50 mM NaCl, 50 mM EDTA, 1% sodium dodecyl sulfate [SDS], 0.15 mM spermine, 0.5 mM spermidine). Adults were ground in fly grinding buffer (100 mM Tris [pH 9.1], 100 mM NaCl, 200 mM sucrose, 0.5% SDS). An equal volume of preheated phenol (saturated in 10 mM Tris [pH 8.0]– 1 mM EDTA [TE]) was immediately added, and samples were vortexed and then incubated at 65°C for >10 min. Chloroform was added 1:1 with the phenol, and samples were extracted. Samples were then phenol-chloroform extracted a second time and chloroform extracted once. Twenty micrograms of DNase-free RNase A was then added, and samples were incubated briefly at room temperature. DNA was precipitated in a mixture of 0.5 M NaCl and 12% polyethylene glycol (PEG) 6000 for 1 h at 4°C and collected by centrifugation. (Less of the DNA was nicked by PEG precipitation than by ethanol precipitation, although the distribution of topoisomers was the same with either method.) Pellets were washed in 70% ethanol, dried, and resuspended in TE. DNA samples were loaded into 1.5% SeaKem LE agarose gels, supplemented with 2 μg of chloroquine diphosphate (Sigma) per ml. Electrophoresis was performed in 40 mM Tris-acetate (pH 7.5)–1 mM EDTA (TAE) buffer, also supplemented with 2 μg of chloroquine per ml, at 1.6 V/cm, for about 36 h. Gels were capillary blotted to Magnacharge nylon membranes (Osmonics) and hybridized at 65°C against a random primed labeled probe representing the lacZ and SV40 sequences of the episomes. Images were analyzed with a Fujix PhosphorImager. Fujifilm Image Gauge V3.0 software was used for densitometric measurement of the areas under supercoiled DNA peaks used for computation of the average linking numbers.
RESULTS AND DISCUSSION
New P element insertions into the BX-C.
We constructed a P element with target sites for our three accessibility probes (Fig. 1). First, it carries binding sites for the yeast GAL4 transcriptional activator (GAL4-UAS) next to a minimal TATA-containing promoter driving a LacZ reporter. We can use LacZ to monitor Pol II-mediated transcription, with or without activation. Second, the P element carries two tandem 49-bp T7 promoters, oriented toward the outside of the P element. We can assay initiation and elongation by T7RNAP by looking for RNA copies of the genomic sequences flanking the 3′ end of the P element. Finally, the cassette with the T7 promoters and the lacZ gene is flanked by FRT recombination sites for the FLP enzyme. Excision of the cassette as a circular episome can be monitored indirectly by looking for T7RNAP transcription around the circle (Fig. 1C).
The initial challenge in probing the structure of PcG-repressed DNA is to introduce these P elements into a chromosomal region known to be PcG regulated. We have taken advantage of a fortuitous example of P element homing. A DNA fragment from the middle of the BX-C targets P elements preferentially to the region of the chromosome from which it originates (3). This “homing fragment” is included in our P element, along with the rosy+ marker for transformation, in a cassette that is also flanked by FRTs (Fig. 1A). Of 20 insertions generated by germ line transformation, 5 were within the BX-C. We were concerned that the rosy gene and/or the homing fragment could influence the structure of the neighboring DNA. Therefore, we used FLP-mediated recombination in the germ line to trim four of the five insertions down to 5.6-kb P elements containing only the T7 promoter/UAS-LacZ cassette (Fig. 1B). Most of our assays in this paper will focus on the four trimmed-down insertions (p[bx], p[iab-4], p[Mcp], and p[Abd-B]) (Fig. 2 and see below).
The new P element insertions are spaced fairly evenly throughout the homeotic gene cluster (Fig. 2). The most centromere-proximal insertion lies within the large intron of the Ubx gene, in the bx (PS5) control region. The most proximal base of the 8-bp target site repeat is position 274,398 on the sequence map of Martin et al. (28). A second insertion lies upstream of the Ubx gene, in the bxd (PS6) control region (at position 219,568). This insertion lies in the region of the bxd PRE, the best-defined PcG binding fragment (18). A third insertion lies within the iab-4 (PS9) control region, upstream of the abd-A gene (at position 125,800). A fourth insertion lies within the Mcp boundary region (32) between the iab-4 and iab-5 control regions (at 113,871). A fifth insertion (at 49,855) lies 658 bp downstream of the abd-B (class A) gene promoter.
In addition to the BX-C insertions, we generated lines with our accessibility test construct outside of the BX-C as non-PcG-regulated controls. These P elements do not contain the homing fragment and were identified by using the mini-white gene as a marker. Two of these control lines were used for the assays in this paper. One is located in the 18D region on the X chromosome and was mapped by inverse PCR (35) to the first exon of putative gene CG14200. The T7 promoters are oriented in the opposite direction to that of the putative transcript. A second control is located on the third chromosome and was not mapped. The LacZ reporters in both controls show no enhancer trap pattern during embryonic development (see below). Moreover, RNA in situ analysis of the X chromosome control with sense and antisense probes to the neighboring genomic DNA shows no transcription of the region from potential adjacent promoters during the developmental stages at which our accessibility assays were performed. (We did not test for expression during the larval, pupal, or adult stage.)
Gal4 activation of Pol II transcription is repressed by the PcG.
In the absence of Gal4, the UAS-LacZ reporter in each BX-C insertion responds to the neighboring enhancer sequences, giving rise to a LacZ expression pattern that is spatially restricted to particular segments (Fig. 3B to E). The controls show no LacZ expression in the absence of Gal4 (3rd chromosome control, Fig. 3A [X chromosome control not shown]). For each of the insertions in the BX-C, LacZ expression reflects segments in which the segmental control region surrounding the P insertion is active. The segments lacking LacZ expression are those in which the region of the insertion is repressed by the PcG (30). When Gal4 is expressed in control embryos from a ubiquitous source (line 32B), the LacZ gene is strongly activated in all segments (3rd chromosome control, Fig. 3F). In contrast, activation by Gal4 is segmentally restricted in the BX-C insertions (Fig. 3G to J). Although some activation occurs in PcG-repressed segments, a far greater number of cells show a high level of LacZ transcription in nonrepressed segments. Gal4 does not activate equally in all cells, even within the nonrepressed segments. Moreover, the Gal4-activated pattern is not an amplification of the nonactivated pattern. Gal4 appears to be interacting with the neighboring enhancer elements in a complicated fashion. However, in all cases, Gal4 activation is substantially blocked in PcG-repressed segments.
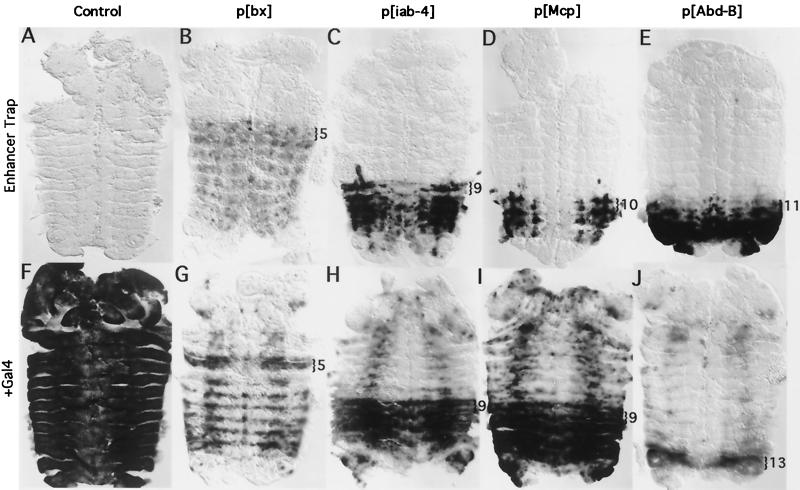
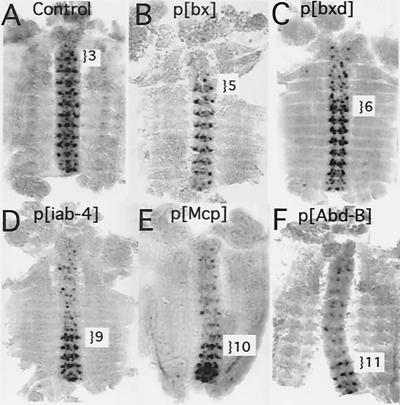
FIG. 3.
Gal4 is partially blocked by the PcG. (A to E) Enhancer trap patterns in the P insertion lines. RNA in situ hybridizations showing LacZ expression (in the absence of Gal4) in germ band retracted embryos. (A) A control embryo containing the UAS-LacZ cassette on the 3rd chromosome outside of the BX-C shows no LacZ expression. (B to E) Trimmed-down insertions p[bx], p[iab-4], p[Mcp], and p[Abd-B], respectively. The most anterior parasegment in which transcription occurs is marked with a bracket. (F to J) Gal4 activation of LacZ expression. (F) Gal4 strongly activates LacZ expression throughout the control embryo. (G to J) Gal4 activation in p[bx], p[iab-4], p[Mcp], and p[Abd-B], respectively. LacZ expression is much weaker or absent in PcG-repressed segments. The anterior-most parasegment in which strong activation of LacZ transcription occurs is marked with a bracket. All embryos shown in this and subsequent figures are dissected along the dorsal midline and are displayed as “pelts,” with the anterior oriented toward the top of the page.
The level of transcription from the Abd-B test site appears significantly lower in the presence of Gal4. This LacZ reporter lies ∼700 bp downstream of the RNA start site for Abd-B and is in the same orientation as the Abd-B transcript. We suspect that the nonactivated pattern (Fig. 3E) results from readthrough from the Abd-B promoter, creating an Abd-B-like pattern. Gal4 may bind to the UAS promoter in PS11 to -14, blocking readthrough from the Abd-B promoter. It may only succeed in activating high levels of transcription in PS13, in which the control region surrounding the insertion site is active and is not PcG repressed. In any case, the activated pattern better reflects the segmental control region (iab-8) in which the target is situated.
The p[Mcp] LacZ pattern begins in PS10 in nonactivated embryos, but starts in PS9 after Gal4 activation. This shift may be due to disruption of the Mcp boundary element between the iab-4 and iab-5 segmental domains (20). In fact, p[Mcp] adult flies show a weak Mcp phenotype (pigmented cuticle on the 4th abdominal tergite), which is significantly enhanced in flies expressing Gal4. None of the other lines show any phenotypic changes in the presence of Gal4.
Pattern changes in PcG mutants.
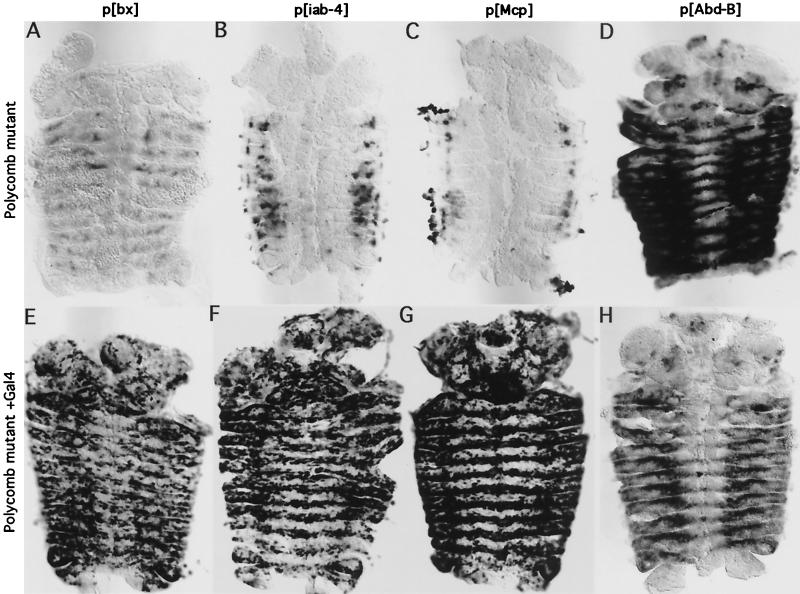
In Polycomb− (Pc−) mutant embryos, the homeotic genes are turned on inappropriately in anterior segments. Likewise, in Pc− embryos hosting our insertions, the LacZ pattern loses its segmental restriction. The Abd-B reporter shows strong expression throughout the thoracic and abdominal segments in a Polycomb mutant (Fig. 4D). Again, we suspect this reflects readthrough from the Abd-B promoter. However, in the bx, iab-4, and Mcp insertion lines, only a few cells in each segment turn on LacZ (Fig. 4A to C). In addition, the pattern is changed in posterior segments, with loss of expression in some cells. We also analyzed embryos with mutations specific for other PcG members, including extra sex combs, Posterior Sex Combs, polyhomeotic, and pleiohomeotic (see Materials and Methods for genotypes). All of the mutant embryos gave patterns similar to those of Pc− embryos (data not shown). We were initially surprised that our insertions did not show widespread transcription in PcG mutant backgrounds. Previously discovered BX-C enhancer traps had shown strong and widespread misexpression in PcG mutant embryos (30). Most of these enhancer traps were driven by the Ubx gene promoter (including 1.7 kb of 5′ flanking sequences), and all included the rosy gene as a reporter. The strong misexpression observed may have resulted from activation of enhancer sequences within the P elements. We believe the weak promoter in our new insertions is completely dependent upon the neighboring homeotic gene enhancers for activity and thus better reflects the effect of loss of PcG upon these homeotic gene control regions. We tested other P elements in the BX-C, which contain only LacZ driven from the P promoter (derived from “homing pigeons” of reference 3). These behave similarly to our new insertions, showing weak expression in Pc− embryos (not shown). These results show that the loss of the PcG does not, by itself, lead to widespread activation of the long-distance enhancers in the BX-C. Moreover, it shows that the PcG affects the activity of these enhancers not only in repressed segments, but also in transcriptionally active segments. However, the changes may be indirect, resulting from pleiotropic effects in the PcG mutants. The loss of the PcG should cause missexpression of genes other than the homeotics, some of which may be transcription factors capable of binding to the homeotic control regions.
FIG. 4.
Polycomb mutant embryos show a weak spread of LacZ expression, but allow Gal4 to activate LacZ transcription throughout the embryo. All panels show in situ hybridizations to LacZ RNA in germ band retracted embryos. (A to D) Enhancer trap patterns in Polycomb zygotic null embryos containing the p[bx], p[iab-4], p[Mcp], and p[Abd-B] insertions, respectively. (E to H) Gal4 activation of LacZ transcription in Polycomb zygotic null embryos.
Gal4 activation was also tested in Pc− embryos. In all four insertions tested (Fig. 4E to H), Gal4 strongly activated expression throughout the thoracic and abdominal segments. The level of activation drops off in the posterior-most segments (A6 to A8) for the iab-4 insertion for unknown reasons. The level of LacZ expression in Polycomb mutants with the Abd-B insertion is lower in the presence of Gal4 than in its absence, presumably due to competition between the UAS promoter and the upstream Abd-B gene promoter. In all of the BX-C insertions, Gal4 still does not activate transcription in all cells, as it does in a control (compare Fig. 4E to H to Fig. 3F). The neighboring BX-C sequences affect the Gal4 activation, resulting in a cell-specific pattern repeated in each segment, with the patterns differing for each LacZ insertion site. Moreover, the cell specificity of these patterns differs between the PcG mutant and wild-type state (compare Fig. 4E to H to Fig. 3G to J), even in posterior segments that are transcriptionally active in the presence of the PcG. Again, indirect effects of the PcG on the neighboring enhancers are an issue. However, the fact that Gal4 works more or less equally well in all segments in PcG mutants likely reflects a change in access or activity of Gal4 and, potentially, a change in access of the neighboring enhancer sequences to other trans-acting factors. In conjunction with the T7RNAP and FLP results reported below, this result supports the model that the PcG is responsible for long-range chromatin remodeling of these regions.
Transcription by T7RNAP is partially blocked by the PcG.
To test whether or not the PcG affects transcription by T7RNAP, we crossed the fly lines containing the new T7 promoter insertions to a source of T7RNAP that is constitutively expressed in cells of the CNS (29). Transcription by T7RNAP of the genomic sequences abutting the T7 promoter insertions was visualized by RNA in situ analysis of embryos (Fig. 5). As reported previously (29), brief treatment of the embryos at 37 to 39°C is required to see T7RNAP transcripts, perhaps due to increased transcript stability.
FIG. 5.
Transcription by T7RNAP is partially blocked by the PcG at multiple sites within the BX-C. RNA in situ hybridizations in germ band retracted embryos. All embryos contained a source of T7RNAP, which is expressed in the CNS. (A) The control is an embryo containing a P element with the T7 promoter/UAS-LacZ cassette inserted on the X chromosome. T7RNAP produces a transcript in the cells of the CNS from PS3 to -14, mirroring the expression pattern of the polymerase. (B to F) Transcription by T7RNAP in the BX-C insertions. In panels C and D, the initial full-length P elements were used, and in panels B, E, and F, the trimmed-down forms were used. The anterior-most parasegment in which the control region is active is indicated for each embryo. In the anterior segments repressed by the PcG, T7RNAP produces a transcript in only a subset of the polymerase-producing cells.
The CNS-T7RNAP efficiently transcribes the DNA neighboring the X chromosome control, resulting in a repeating pattern of RNA products in PS3 to -14 (Fig. 5A). This occurs despite the lack of endogenous transcription in this region. The pattern of the T7RNAP RNA products in this embryo mimics the pattern of expression of the polymerase, as visualized by anti-T7RNAP antibody (see Fig. 6C of reference 29). In contrast, in the BX-C insertions, a repeating pattern of transcription by T7RNAP is present only in the segments not blocked by PcG repression. In the more anterior segments (as well as the posterior-most segments) repressed by the PcG, only a subset of the polymerase-positive cells are marked by the T7RNAP transcription product. T7RNAP appears to be blocked in a random subset of cells, a pattern reminiscent of the variegated repression induced upon the miniwhite gene in transgenic constructs containing PREs (9). Since T7RNAP is capable of transcription without any Drosophila transcription factors, the block to transcription suggests that T7RNAP is unable to bind to its promoters and/or progress along PcG-repressed DNA.
Discordance with prior T7 assays.
A previous publication from this laboratory reported that T7RNAP was not affected by the PcG, as assayed by transcription from an insertion in the bx region containing four T7 promoters (29). Our repeats of that experiment verify the prior results. There are several differences between the present and previous experiments, which may explain the different outcomes. Each of our five new insertions contains only two T7 promoters, whereas most of the work in the previous study was performed on a single, four-promoter insertion. The probe used for RNA in situ analysis in the previous study included sequences immediately downstream of the promoters. In our study, the probe sequences begin ∼300 bp downstream of the transcription start site, since the polymerase must transcribe through FRT and 3′ P sequences before reaching the neighboring genomic DNA. The most significant difference may be the context of the T7 promoters. Although our new bx insertion line is located close to the position of the T7 promoter insertion described by McCall and Bender (29), it is possible that the ∼250 bp between the two insertions is enough to cause differences in the behaviors of the T7 promoters. It is also quite possible that the UAS-TATA Pol II promoter in our new insertions affects the neighboring T7 promoters. However, as is illustrated below, we do detect an accessibility effect on the simple T7 promoter target (29) by an enlarged version of T7RNAP, and so the PcG block cannot be solely an effect of a nearby Pol II promoter. We have also found that following prolonged expression of T7RNAP, no PcG block to T7RNAP transcription is apparent (see Fig. 6 and 7). Clearly the PcG block to T7 transcription is partial and is sensitive to assay conditions.
FIG. 6.
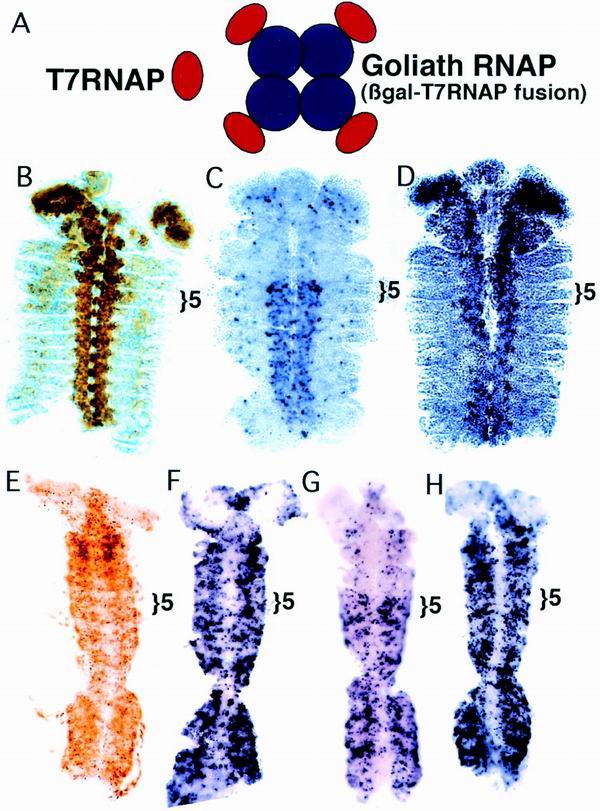
An enlarged polymerase is more sensitive to PcG repression. (A) Drawing comparing wild-type T7RNAP and Goliath polymerase. (B) Pattern of Goliath polymerase protein in the CNS Goliath line in a germ band retracted embryo. The location of Goliath polymerase was detected with a monoclonal antibody to β-galactosidase. PS5, the anterior limit of bx enhancer activity, is indicated with a bracket. (C) Transcription by Goliath polymerase on the 4× T7 promoters in the bx control region in a germ band retracted embryo. Transcription by Goliath is blocked anterior to PS5. (D) Transcription by Goliath polymerase in an extra sex combs mutant embryo (esc2/esc10, maternal and zygotic null). The repression in anterior segments is lost. (E to H) T7RNAP versus Goliath polymerase as shown by expression of the polymerases with the Gal4-UAS system. A ubiquitous Gal4 source (line 32B) was used to drive polymerase expression. All embryos are in the germ band extended stage. (E) Goliath polymerase protein pattern marked with an antibody to β-galactosidase. (F) Transcription by T7RNAP on the 4× T7 promoters in the bx control region. Transcription appears unaffected by the PcG. (G) Transcription by Goliath polymerase. Transcription is repressed by the PcG anterior to PS5. (H) Transcription by Goliath polymerase in a Polycomb mutant (zygotic null). Repression in the anterior segments is lost.
FIG. 7.
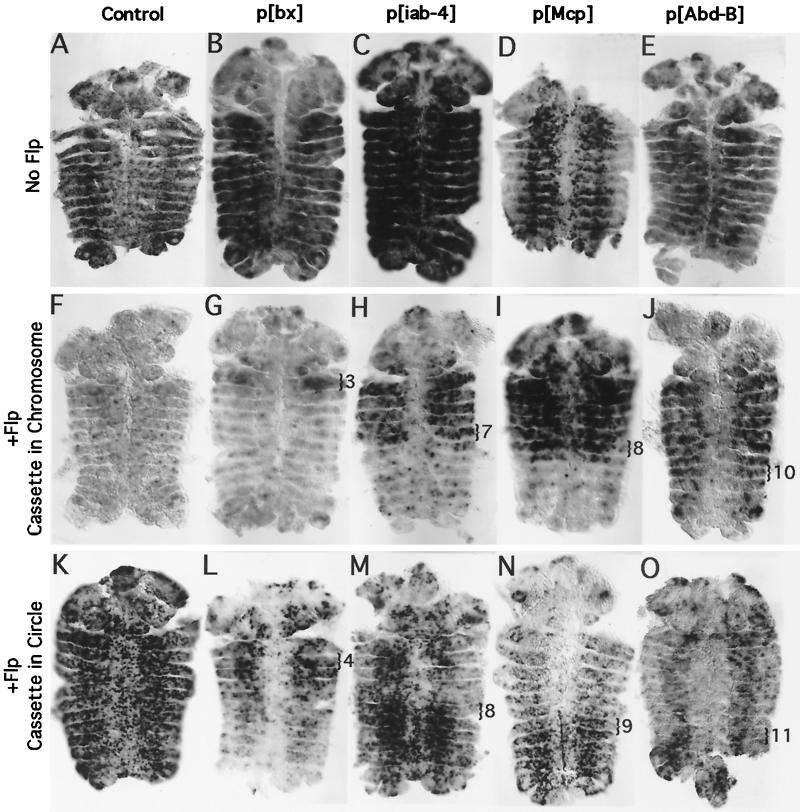
FLP recombinase is partially blocked by the PcG. RNA in situ hybridizations were used to distinguish T7 promoters in the chromosome or in FLP-induced circles in germ band retracted embryos. (A to E) Control embryos lacking FLP. RNA probes detect transcription of flanking genomic sequences from the T7 promoters in the P insertions. In the absence of FLP, all T7 promoter cassettes remain in the chromosome and are poised to transcribe the flanking genomic DNA. Under these assay conditions, no segmental bias in transcription by T7RNAP is apparent in any of the fly lines. (F to J) Transcription of chromosome sequences by T7RNAP marks cells in which FLP has failed to access its FRT target sites, which flank the T7 promoter/UAS-LacZ cassette. (F) FLP efficiently excises the cassette from the majority of the cells in a control line, in which the cassette is located on the X chromosome. (G to J) In the BX-C insertion lines, FLP fails to excise the cassette in many cells in PcG-repressed segments. The block to circle formation appears to occur one parasegment anterior to the block to Pol II transcription. The posterior-most parasegment in which chromosomal transcription is strong is marked with a bracket. (K to O) Transcription of antisense LacZ RNA by T7RNAP marks cells in which FLP has succeeded in producing a circular episome. (K) Circles are visible in most cells in the control embryo. (L to O) A greater number of circles are visible in non-PcG-repressed segments in the BX-C insertion lines. The anterior-most parasegment in which robust circle formation is visible is marked with a bracket.
An enlarged version of T7RNAP is more sensitive to PcG repression.
To test whether size matters to the PcG, we made T7RNAP larger by fusing it to β-galactosidase. This fusion protein forms a tight tetramer that is ∼860 kDa in size, over eight times the size of wild-type T7RNAP, but still substantially smaller than the Pol II holoenzyme. We call this fusion protein “Goliath polymerase” (Fig. 6A).
We introduced Goliath polymerase into flies and recovered an insertion on the 3rd chromosome, which expresses the polymerase constitutively in the CNS (Fig. 6B). We assayed the ability of Goliath polymerase to transcribe from the 4× T7 promoter insertion described by McCall and Bender (29), which is located in the bx control region (Fig. 2). Transcription by CNS Goliath polymerase was only visible in recombinant lines containing the source of Goliath polymerase and the T7 promoters on the same chromosome. Either two copies of the CNS Goliath polymerase source or homozygosity of the T7 promoter insertions was necessary to achieve high enough levels of transcription.
Transcription by Goliath polymerase appears to be blocked in most of the cells in PcG-repressed segments (Fig. 6C). This block is dependent upon the PcG, as demonstrated by repeating the experiment in an animal that lacks the Extra Sex Combs protein. In these PcG mutant embryos, Goliath polymerase transcribes equally well in all segments (Fig. 6D). We also tested CNS Goliath polymerase on the new p[bx] and p[Abd-B] insertions, again with both the source and the target homozygous. Goliath polymerase transcription from these promoters is weak, but tightly restricted to the appropriate segments of the CNS (data not shown).
In order to compare Goliath polymerase and normal T7RNAP more directly, we constructed fly lines that contain Gal4-inducible expression constructs of either T7RNAP or Goliath polymerase. This permitted us to express the polymerases in a variety of patterns and at comparable levels. Figure 6E shows the expression pattern of the polymerases when crossed to the 32B Gal4 driver, which is described as a ubiquitous Gal4 source (8). Under these conditions, as assayed from the 4× T7 promoter insertion, no block to transcription by T7RNAP is apparent (Fig. 6F). Transcription by Goliath polymerase, in contrast, is still strongly reduced in segments that are repressed by the PcG (Fig. 6G). The restriction disappears in an embryo lacking the Polycomb protein (Fig. 6H). We did not test the Gal4-inducible polymerases on any of the new lines, since these insertions contain Gal4 binding sites immediately upstream of the T7 promoters. Binding of Gal4 to these sites might influence the chromatin structure of the T7 promoters.
The segmental restriction of transcription by Goliath polymerase disappears in germ band retracted (stages 13 to 14) and older embryos (data not shown). However, Western analysis shows that a subset of Goliath polymerase (expressed under Gal4 induction) becomes clipped. Clipped forms are visible even in early embryos, but more clipped forms appear to accumulate in older embryos. Some of these are likely to be a functional polymerase, with only a small C-terminal fragment of β-galactosidase peptide remaining attached. Such a fusion would no longer be able to form tetramers and would be expected to behave like its wild-type counterpart. Wild-type T7RNAP, when expressed at high levels, is not inhibited by the PcG (Fig. 6F).
Although we favor the model that Goliath polymerase is more sensitive to PcG repression because of its size, it is possibly more sensitive because it is a less effective polymerase than native T7RNAP. Northern analysis of T7RNAP transcripts in embryos (0 to 24 h) shows a broad distribution of RNA products with an average transcript size of ∼1.5 kb (ranging up to ∼5 kb). Transcripts made by Goliath polymerase have a similar size distribution, but are ∼10× lower in abundance. It is possible, however, that a difference in size distribution of the transcripts is masked by a contribution of products of the clipped fusion protein.
To test the notion that crippled polymerases are differentially sensitive to PcG represion, we obtained three mutated T7 polymerases that were defective for promoter binding, elongation, and progression through pause sites, respectively (see Materials and Methods). All three forms were introduced into flies under Gal4 control. Based on in vitro comparisons, we expected the mutant forms to equal or exceed the activity of Goliath polymerase (data not shown). The T7RNAP defective in progression through pause sites worked equally well in all segments. Unfortunately, we could not detect transcription in flies by either the promoter binding or elongation mutant polymerases and were therefore unable to test their sensitivities to PcG repression.
FLP-mediated recombination is blocked by Polycomb.
The cassette containing the UAS-LacZ reporter and T7 promoters is flanked by FRTs oriented as direct repeats. Introduction of FLP recombinase into the somatic tissues of the embryo allows the excision of this cassette from the chromosome into a 4.9-kb circular episome (Fig. 1C). The T7 promoters on this episome are now poised to transcribe around the circle, producing antisense copies of LacZ. If FLP recombination is blocked, no episome is formed, and the T7 promoters remain in the chromosome, poised to transcribe the flanking genomic DNA. We therefore introduced both a source of FLP and a source of T7RNAP into our trimmed-down insertion lines, to assay for FLP activity cell by cell (Fig. 7). We performed cRNA in situ hybridizations with the flanking chromosomal DNA as probe to highlight cells in which FLP failed to excise the cassette (Fig. 7F to J) and a probe to antisense LacZ to highlight cells in which circles were present (Fig. 7K to O).
To perform the FLP accessibility experiment, we used both a heat shock-inducible source of FLP and a heat shock-inducible source of T7RNAP. Embryos were heat shocked for 1 h at 36.5°C to produce FLP and T7RNAP. They were then incubated for 1 h at room temperature to permit FLP activity. The embryos were then heat shocked a second time at 39°C for 1 h to allow T7RNAP products to accumulate. Under these conditions, no segmental bias is apparent in T7RNAP's ability to transcribe from its promoters, either in control embryos (X chromosome control, Fig. 7A) or in the BX-C test lines, which lack FLP (flanking probe, Fig. 7B to E). None of the lines showed any signal with the circle probe in the absence of FLP induction. In control embryos with FLP, circles are produced from the majority of cells, even though there is no endogenous transcription across the site of the control insertion (compare flanking probe, Fig. 7F, to circle probe, Fig. 7K). Analysis of the 3rd chromosome control with the circle probe showed that it behaved similarly (data not shown). There is no segmental bias in circle formation from control lines.
In contrast, FLP activity is blocked from the BX-C insertions in a segment-specific fashion. The effect is most obvious on the iab-4 and Mcp insertions. For both of these insertions, the chromosomal probe, which highlights cells in which FLP fails to work, selectively marks the anterior segments in which the PcG is expected to repress the DNA (Fig. 7H and I). The circle probe selectively marks the complementary posterior segments, in which the PcG is not expected to repress the DNA (Fig. 7M and N). Interestingly, the transitions in the staining patterns are apparent one parasegment ahead of the expected PcG restrictions (as determined by the LacZ enhancer trap patterns of Fig. 3). This suggests that a change in the chromatin structure may be graded with only partial repression of DNA regions at the edges of the PcG-repressed domain.
In the bx insertion, FLP restriction is less complete. Using the chromosomal probe to highlight cells in which FLP fails to work, PS3 is selectively highlighted (Fig. 7G). Again, this is one parasegment ahead of that expected; the bx enhancer drives expression of the LacZ reporter beginning in PS5. Many bx embryos show widespread FLP activity assayed by the circle probe (data not shown). However, the majority of the embryos show a clear bias with the circle probe (Fig. 7L), although this pattern is not strictly complementary to that of the chromosomal probe. PS4 and -5 are labeled most strongly, followed by PS3, followed by the abdominal parasegments. This does correspond with the levels of transcription from the LacZ reporter (Fig. 3B), which is higher in PS5 and -6 than it is in more posterior parasegments. However, the pattern of FLP activity is shifted one segment ahead of the LacZ pattern.
The Abd-B insertion also shows a reproducible bias in FLP activity, as assayed by the chromosomal probe; labeling is largely excluded from the posterior-most segments (PS11 to -13) (Fig. 7J). Many of the Abd-B insert embryos showed widespread FLP activity as assayed by the circle probe (data not shown). Some, however, showed a bias in circle formation in the posterior-most segments (PS11 to -13), as is illustrated in Fig. 7O. In general, FLP activity showed less segmental bias in the bx and Abd-B insertions than in the iab-4 and Mcp insertions. This is possibly because the former reside within actively transcribed regions of the homeotic genes, whereas the latter reside outside of the homeotic transcripts. The chromosomal probe shows a clearer segmental bias than the circle probe for all four insertion sites. An FLP target site lies immediately downstream of the T7 promoters. FLP binding alone may block transcription of the neighboring chromosomal sequences by T7RNAP. The circle probe, in contrast, should only produce a signal if FLP has both bound and recombined the DNA. If FLP binds the DNA in most of the nonrepressed cells but only produces a circle in a subset of these cells, we might observe a difference between the results with the chromosomal versus circle probes.
The fact that the FLP recombinase, a yeast protein with no role in transcription, is selectively blocked in a segment-specific manner from BX-C insertions, but not from control insertions, further suggests that the DNA structure of the BX-C is altered in PcG-repressed segments. This result is similar to that reported in reference 1, in which a strong inhibition of FLP recombination was observed at target sites located near centric heterochromatin.
The reduced accessibility of PcG-repressed DNA is not associated with a change in DNA supercoiling.
We compared the superhelix density of PcG-repressed and nonrepressed DNAs by using our FLP cassettes to produce extrachromosomal episomes. We crossed our accessibility test lines to a heat shock-inducible source of FLP recombinase, induced FLP activity, and then collected total genomic DNA, which should include newly excised circles (Fig. 1C). This DNA was subjected to gel electrophoresis in the presence of chloroquine, which resolves supercoiled circles into a ladder of distict bands. Adjacent rungs of the ladder differ by a single superhelical turn, and the topoisomers of the circles describe a Gaussian distribution. Chloroquine gels were Southern blotted and probed with the lacZ sequences contained within our FLP-inducible cassette (Fig. 8). By this technique, a reproducible change in linking number as small as 1 can be seen.
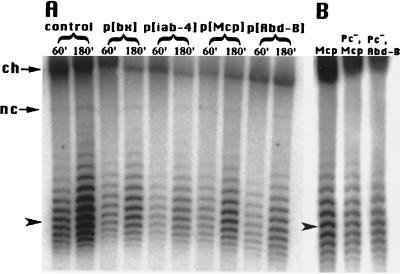
FIG. 8.
PcG repression is not associated with altered DNA supercoiling. Circular episomes produced from the FLP cassettes were subject to electrophoresis through agarose gels supplemented with 2 μg of chloroquine per ml. Under these conditions, more negatively supercoiled DNA has a faster mobility. The gels were Southern blotted and probed with lacZ sequence. (A) Total genomic DNA was prepared from adult fly heads, either immediately following a 1-h heat shock treatment to induce FLP activity (60′) or following 1 h of heat shock plus 2 h of recovery at room temperature (180′). All of the circles produced in the p[bx], p[iab-4], p[Mcp], and p[Abd-B] lines come from PcG-repressed DNA. The control is the 3rd chromosome insertion, which is not PcG repressed. The average linking number is approximately the same for all lanes. The locations of the unexcised DNA remaining in the chromosome (ch) and of nicked circles (nc) are marked with arrows. The average linking number for the sample in the first lane is indicated with an arrowhead (see Materials and Methods). (B) Total genomic DNA was prepared from whole embryos following 1 h of heat shock plus a half-hour of recovery at room temperature to induce FLP activity. This DNA was subjected to chloroquine gel and Southern blot analyses as described above. Circles produced from the p[Mcp] line with wild-type PcG activity are compared to circles produced from p[Mcp] and p[Abd-B] lines, which are null for the Polycomb protein. The loss of PcG repression does not alter the topology of the DNA in these regions.
We first asked whether PcG repression was maintained on newly excised circles. We examined the lacZ expression patterns in embryos with our insertions following FLP induction. LacZ expression patterns were similar before and after circle formation, but the patterns were weaker following FLP induction (data not shown). Likewise, similar patterns of lacZ were seen after Gal4 activation in the presence or absence of FLP. Again, the level of transcription was somewhat lower following circle formation. Thus, there is no evidence for loss of PcG repression due to circle formation; if anything, circles may become silenced in cells where the transgene was active in the chromosome. Ahmad and Golic (1) have shown that circles excised from heterochromatic locations become transcriptionally active. Their assay monitored changes in white gene expression in the adult eye, following FLP induction several days earlier, during larval or pupal development. Our results show that activation of the circles in embryos does not occur within the 3-h window of our experiments.
We then used chloroquine gel analysis to examine circles produced from whole embryos, to compare the topology of our BX-C insertions to control insertions. The greatest difference in the average linking number (ΔLK) between any pair of samples was less than 0.5 superhelical turns (data not shown). Thus, we do not believe there was a significant difference in the topology of circles collected from our control lines, as compared to any of our BX-C insertions. We expect the circles produced from the BX-C insertions were derived from a mixture of both PcG-repressed and nonrepressed segments. Moreover, we may have enriched for circles generated in nonrepressed segments, since FLP is partially inhibited by PcG repression (Fig. 7). In any case, this DNA is neither more negatively nor more positively supercoiled than the control.
In order to generate circles exclusively from PcG-repressed DNA, we induced FLP activity in adult flies and prepared DNA from fly heads, where the entire BX-C should be inactive. As a control, we also prepared circles from the 3rd chromosome insertion outside the BX-C (Fig. 8A). The greatest difference observed between samples produced by 1 h of FLP induction was a ΔLK of 0.7, between the control and the p[Mcp] line. The p[Mcp] line was less negatively supercoiled, which is a change opposite to the direction which we would expect, but we doubt this is a significant change. We do see a greater abundance of circles produced from control fly heads than from BX-C insertion fly heads (compare the first two lanes of Fig. 8A to the rest). This is in agreement with our observation that PcG repression partially inhibits FLP recombination in embryos (Fig. 7). However, FLP appears to overcome PcG repression with time, ultimately resulting in the recovery of many circles from PcG-repressed DNA. This is in agreement with results reported in yeast, where it was shown that the rate of excision by a recombinase is slower on silenced DNA, but that ultimately the same level of recombination is achieved on both silenced and nonsilenced DNA (2).
In yeast, SIR silencing and corresponding topology differences on episomes break down over time (4, 11). We examined circles collected immediately after 1 h of FLP induction, or after an additional 2-h room temperature incubation. Circles accumulate due to continued FLP activity over this time course. Circles may be slightly more positively supercoiled on average at the later time point, but again, the differences are small. The largest change occurred in the control line, with a ΔLK of 0.7. The p[bx] showed a ΔLK of 0.6, while the rest of the BX-C insertions showed changes smaller than 0.5 supercoil. (Fig. 8A).
We also compared circles prepared from different segments of the fly. Flies containing the 3rd chromosome control or the p[Mcp] or p[Abd-B] inserts were heat shocked to produce FLP circles, frozen, and then dissected into heads, thoraxes, and abdomens. Most or all of the DNA from the heads and thoraxes of the p[Mcp] and p[Abd-B] lines should be PcG repressed, while only a fraction of the cells in the abdomen should be PcG repressed. There was no significant difference in the supercoiling of circles between segments or between fly lines. (The greatest change was between the heads and abdomens of the control line [ΔLK of 0.5; data not shown].)
We also compared the topology of circles derived from wild-type embryos or Polycomb mutant embryos. We prepared recombinant fly lines containing a source of FLP on the same chromosome as a null allele for the Polycomb gene (Pc3). We also prepared recombinant 3rd chromosomes containing our BX-C insertions and the Pc3 allele. A cross between these lines produces a population of embryos in which only the Polycomb-null homozygotes will produce circles. We prepared circles from wild-type embryos with the p[Mcp] insertion and from Pc-null embryos with either the p[Mcp] or p[Abd-B] insertion. Note that segmental restriction of lacZ is lost in the absence of the PcG for both the p[Mcp] and p[Abd-B] insertions, but that lacZ is transcribed in fewer cells in Pc3, p[Mcp] embryos (Fig. 4C), whereas transcription is activated in many more cells in Pc3, p[Abd-B] embryos (Fig. 4D). Despite these changes, we see no difference in the topology of circles from wild-type or Pc mutant embryos (ΔLK less than 0.5; Fig. 8B). We also compared circles prepared from adult males versus adult females containing the X chromosome control insertions, to see if a change in DNA supercoiling might be associated with dosage compensation of the male X chromosome. Again, we could not detect any difference (data not shown).
In the experiments on SIR-silenced DNA in yeast, the linking number change observed was proportional to the size of the circle (4). A 4.3-kb circle derived from SIR-repressed DNA contained, on average, five more negative supercoils than nonrepressed DNA from the same location. Thus, a comparable modification on our 4.9-kb circles would be expected to create a linking number change of >5. It is possible that the structure of the DNA is perturbed upon circle formation in flies. However, there is no reason to believe that FLP-mediated recombination in flies should be any more disruptive to chromatin structure than it is in yeast. We heat shocked our fly lines to produce FLP. However, Cheng, et al. (11) analyzed circles formed at a variety of temperatures, as high as 37°C, and saw similar differences in topology at all temperatures. Moreover, we see inhibition of our accessibility probes following heat shock, suggesting that PcG repression is unaffected. The simplest interpretation is that supercoiling of DNA is not measurably altered in the presence or absence of the PcG.
Concluding remarks.
Our accessibility assays show that PcG repression blocks the activities of the Gal4 activator, the FLP recombinase, and two forms of T7RNAP. At least for FLP and T7RNAP, there should be no specific interactions between our probe proteins and any Drosophila proteins involved in transcription, and thus, the segment-specific differences in their activities should reflect differences in their ability to access the DNA. Our assays do not address the precise cause of the block. The inhibition of Gal4 could reflect a block to binding or to activation. The inhibition of the T7 polymerases could reflect a block to binding or to progression of the polymerase along the DNA template. The inhibition of FLP could reflect a block to binding or to recombination by the enzyme. Since all three enzymes are affected by the PcG, a block to binding for all three proteins seems the most simple explanation. Regardless of the exact nature of the block, it seems clear that there is a structural difference to PcG-repressed DNA. We note that the two control lines used in this paper were both transcriptionally silent in the absence of Gal4, but both allowed access to Gal4, T7RNAP, Goliath polymerase, and FLP throughout the embryo. If the controls represent a “typical” chromatin configuration, our results suggest that PcG-repressed DNA is less accessible than average. This suggests that the PcG does not simply prevent positive chromatin remodeling complexes from targeting the DNA.
The accessibility differences were seen at five different sites with the BX-C, which lie in very different contexts (one in a promoter, another in a boundary, a third in an intron, etc.). We note that our p[bx], p[bxd], p[iab-4], and p[Mcp] sites all lie in the vicinity of Polycomb binding regions, as defined by the cross-linking studies of Strutt et al. (49). However, our p[Abd-B] does not lie in a PRE by this criterion, or any other criterion. Since it is downstream of a promoter, we might expect it to be more accessible than other sites in the BX-C. p[Abd-B], however, behaves similarly to our other BX-C insertions. We note that homing does not target P insertions exclusively to suspected PcG binding sites (3). Moreover, PcG-mediated repression of lacZ transcription has been seen for many other insertions in the BX-C, distant from suspected sites of PcG binding (3, 30). The combined results of our five accessibility test sites suggest that the PcG induces a reduction in accessibility over large, contiguous stretches of DNA. It might do so directly by coating the chromosome or indirectly through modification of nucleosomes. The fact that T7RNAP appears more sensitive to PcG repression in our UAS-TATA-containing cassette than it was on simple insertions (via gene conversion) of the T7 promoter (29) may suggest that PcG repression may not function equivalently at all sites in the BX-C. Moreover, our results do not preclude the possibility that the large, multiprotein PcG complex plays multiple roles, perhaps mediating promoter-specific effects in addition to a more widespread chromatin effect. Our results with Goliath polymerase on the simple T7 promoter insertion, however, argue that the PcG must interact with sequences far from Pol II promoter regions.
The increased sensitivity of the Goliath polymerase suggests that the block created by the PcG may be more effective against larger molecules or complexes. Large activation complexes, like the SWI-SNF complex, are obvious candidates for Polycomb targets. Indeed, Shao et al. (44) demonstrated in vitro that the PRC1 complex, containing Polycomb, blocks chromatin remodeling by a mammalian SWI-SNF complex. Their assay measured changes in the supercoiling of a circular DNA template. SWI-SNF has been shown capable of creating an average ΔLK of ∼+0.32 per nucleosome in vitro (23). Similarly, SIR silencing has been shown to be associated with a ΔLK as large as ∼−0.36 per nucleosome, assuming 1 nucleosome per 216 bp of DNA (4). Our 4.9-kb circles are expected to accommodate ∼22 nucleosomes. If chromatin remodeling were limited to one or a few nucleosomes encompassing the promoter region of our constructs, we might not detect a change in superhelix density. It is clear, however, that widespread nucleosome remodeling, akin to that seen with the SIR complex in vivo or, indeed, with SWI-SNF in vitro, cannot account for the segment-specific accessibility differences of the DNA of the BX-C.
ACKNOWLEDGMENTS
We thank Rui Sousa and William McCallister for providing the T7RNAP mutants and Stan Tabor for help with in vitro analysis of T7RNAP variants. We also thank Steve Buratowski, Donald Morisato, and members of the W. Bender and D. Morisato laboratories for critical analysis and helpful discussions.
This work was supported by a grant from the National Institutes of Health to W.B.
REFERENCES
- 1.Ahmad K, Golic K G. Somatic reversion of chromosomal position effects in Drosophila melanogaster. Genetics. 1996;144:657–670. doi: 10.1093/genetics/144.2.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ansari A, Cheng T-H, Gartenberg M R. Isolation of selected chromatin fragments from yeast by site-specific recombination in vivo. Methods Companion Methods Enzymol. 1999;17:104–111. doi: 10.1006/meth.1998.0722. [DOI] [PubMed] [Google Scholar]
- 3.Bender W, Hudson A. P element homing to the Drosophila bithorax complex. Development. 2000;127:3981–3992. doi: 10.1242/dev.127.18.3981. [DOI] [PubMed] [Google Scholar]
- 4.Bi X, Broach J R. DNA in transcriptionally silent chromatin assumes a distinct topology that is sensitive to cell cycle progression. Mol Cell Biol. 1997;17:7077–7087. doi: 10.1128/mcb.17.12.7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bienz M, Müller J. Transcriptional silencing of homeotic genes in Drosophila. BioEssays. 1995;17:775–783. doi: 10.1002/bies.950170907. [DOI] [PubMed] [Google Scholar]
- 6.Boivin A, Dura J-M. In vivo chromatin accessibility correlates with gene silencing in Drosophila. Genetics. 1998;150:1539–1549. doi: 10.1093/genetics/150.4.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonner G, Lafer E M, Sousa R. The thumb subdomain of T7 RNA polymerase functions to stabilize the ternary complex during processive transcription. J Biol Chem. 1994;269:25129–25136. [PubMed] [Google Scholar]
- 8.Brand A H, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 9.Chan C-S, Rastelli L, Pirrotta V. A Polycomb response element in the Ubx gene that determines an epigenetically inherited state of repression. EMBO J. 1994;13:2553–2564. doi: 10.1002/j.1460-2075.1994.tb06545.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen W, Tabor S, Struhl K. Distinguishing between mechanisms of eukaryotic transcriptional activation with bacteriophage T7 RNA polymerase. Cell. 1987;50:1047–1055. doi: 10.1016/0092-8674(87)90171-1. [DOI] [PubMed] [Google Scholar]
- 11.Cheng T-H, Li Y-C, Gartenberg M R. Persistence of an alternate chromatin structure at silenced loci in the absence of silencers. Proc Natl Acad Sci. 1998;95:5521–5526. doi: 10.1073/pnas.95.10.5521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duncan I. The bithorax complex. Annu Rev Genet. 1987;21:285–319. doi: 10.1146/annurev.ge.21.120187.001441. [DOI] [PubMed] [Google Scholar]
- 13.Dunn J J, Krippl B, Bernstein K E, Westphal H, Studier F W. Targeting bacteriophage T7 RNA polymerase to the mammalian cell nucleus. Gene. 1988;68:259–266. doi: 10.1016/0378-1119(88)90028-5. [DOI] [PubMed] [Google Scholar]
- 14.Gartenberg M R. The Sir proteins of Saccharomyces cerevisiae: mediators of transcriptional silencing and much more. Curr Opin Microbiol. 2000;3:132–137. doi: 10.1016/s1369-5274(00)00064-3. [DOI] [PubMed] [Google Scholar]
- 15.Gavis E R, Lehmann R. Localization of nanos RNA controls embryonic polarity. Cell. 1992;71:301–313. doi: 10.1016/0092-8674(92)90358-j. [DOI] [PubMed] [Google Scholar]
- 16.Golic M M, Rong Y S, Petersen R B, Lindquist S L, Golic K G. Flp-mediated DNA mobilization to specific target sites in Drosophila chromosomes. Nucleic Acids Res. 1997;25:3665–3671. doi: 10.1093/nar/25.18.3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gottschling D E. Telomere proximal DNA in Saccharomyces cerevisiae is refractory to methyltransferase activity in vivo. Proc Natl Acad Sci USA. 1992;89:4062–4065. doi: 10.1073/pnas.89.9.4062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horard B, Tatout C, Poux S, Pirrotta V. Structure of a Polycomb response element and in vitro binding of Polycomb group complexes containing GAGA factor. Mol Cell Biol. 2000;20:3187–3197. doi: 10.1128/mcb.20.9.3187-3197.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jenuwein T, Forrester W C, Fernandez-Herrero L A, Laible G, Dull M, Grosschedl R. Extension of chromatin accessibility by nuclear matrix attachment regions. Nature. 1997;385:269–272. doi: 10.1038/385269a0. [DOI] [PubMed] [Google Scholar]
- 20.Karch F, Galloni M, Sipos L, Gausz J, Gyurkovics H, Schedl P. Mcp and Fab-7: molecular analysis of putative boundaries of cis-regulatory domains in the bithorax complex of Drosophila melanogaster. Nucleic Acids Res. 1994;22:3138–3146. doi: 10.1093/nar/22.15.3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karch F, Bender W, Weiffenbach B. abdA expression in Drosophila embryos. Genes Dev. 1990;4:1573–1587. doi: 10.1101/gad.4.9.1573. [DOI] [PubMed] [Google Scholar]
- 22.Kennison J A. Transcriptional activation of Drosophila homeotic genes from distant regulatory elements. Trends Genet. 1993;9:75–79. doi: 10.1016/0168-9525(93)90227-9. [DOI] [PubMed] [Google Scholar]
- 23.Kwon H, Imbalzano A N, Khavarl P A, Kingston R E, Green M R. Nucleosome disruption and enhancement of activator binding by a human SWI/SNF complex. Nature. 1994;370:477–481. doi: 10.1038/370477a0. [DOI] [PubMed] [Google Scholar]
- 24.Lamb M M, Laird C D. Three euchromatic DNA sequences under-replicated in polytene chromosomes of Drosophila are localized in constrictions and ectopic fibers. Chromosoma. 1987;95:227–235. doi: 10.1007/BF00294779. [DOI] [PubMed] [Google Scholar]
- 25.Loo S, Rine J. Silencers and domains of generalized repression. Science. 1994;264:1768–1771. doi: 10.1126/science.8209257. [DOI] [PubMed] [Google Scholar]
- 26.Lyakhov D L, He B, Zhang X, Studier F W, Dunn J J, McAllister W T. Mutant bacteriophage T7 RNA polymerases with altered termination properties. J Mol Biol. 1997;269:28–40. doi: 10.1006/jmbi.1997.1015. [DOI] [PubMed] [Google Scholar]
- 27.Lynch A S, Wang J C. Use of an inducible site-specific recombinase to probe the structure of protein-DNA complexes involved in F plasmid partition in Escherichia coli. J Mol Biol. 1994;236:679–684. doi: 10.1006/jmbi.1994.1179. [DOI] [PubMed] [Google Scholar]
- 28.Martin C H, Mayeda C A, Davis C A, Ericsson C L, Knafels J D, Mathog D R, Celniker S E, Lewis E B, Palazzolo M J. Complete sequence of the bithorax complex of Drosophila. Proc Natl Acad Sci USA. 1995;92:8398–8402. doi: 10.1073/pnas.92.18.8398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCall K, Bender W. Probes for chromatin accessibility in the Drosophila bithorax complex respond differently to Polycomb-mediated repression. EMBO J. 1996;15:569–580. [PMC free article] [PubMed] [Google Scholar]
- 30.McCall K, O'Connor M B, Bender W. Enhancer traps in the Drosophila Bithorax Complex mark parasegmental domains. Genetics. 1994;138:387–399. doi: 10.1093/genetics/138.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morales V, Richard-Foy H. Role of histone N-terminal tails and their acetylation in nucleosome dynamics. Mol Cell Biol. 2000;20:7230–7237. doi: 10.1128/mcb.20.19.7230-7237.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muller M, Hagstrom K, Gyurkovics H, Pirrotta V, Schedl P. The Mcp element from the Drosophila melanogaster bithorax complex mediates long-distance regulatory interactions. Genetics. 1999;153:1333–1356. doi: 10.1093/genetics/153.3.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Navarro M, Cross G A, Wirtz E. Trypanosoma brucei variant surface glycoprotein regulation involves coupled activation/inactivation and chromatin remodeling of expression sites. EMBO J. 1999;18:2265–2272. doi: 10.1093/emboj/18.8.2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ng J, Hart C M, Morgan K, Simon J A. ESC-E(Z) protein complex is distinct from other Polycomb group complexes and contains covalently modified ESC. Mol Cell Biol. 2000;20:3069–3078. doi: 10.1128/mcb.20.9.3069-3078.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ochman H, Gerber A S, Hartl D L. Genetic applications of an inverse polymerase chain reaction. Genetics. 1988;120:621–623. doi: 10.1093/genetics/120.3.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Papoulas O, Beek S J, Moseley S L, McCallum C M, Sarte M, Shearn A, Tamkun J W. The Drosophila trithorax group proteins BRM, ASH1, and ASH2 are subunits of distinct protein complexes. Development. 1998;125:3955–3966. doi: 10.1242/dev.125.20.3955. [DOI] [PubMed] [Google Scholar]
- 37.Paro R, Hogness D S. The Polycomb protein shares a homologous domain with a heterochromatin-associated protein of Drosophila. Proc Natl Acad Sci. 1991;88:263–267. doi: 10.1073/pnas.88.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pirrotta V. Polycombing the genome: PcG, trxG, and chromatin silencing. Cell. 1998;93:333–336. doi: 10.1016/s0092-8674(00)81162-9. [DOI] [PubMed] [Google Scholar]
- 39.Pirrotta V, Rastelli L. white gene expression, repressive chromatin domains and homeotic gene regulation in Drosophila. BioEssays. 1994;16:549–556. doi: 10.1002/bies.950160808. [DOI] [PubMed] [Google Scholar]
- 40.Raskin C A, Diaz G A, McAllister W T. T7 RNA polymerase mutants with altered promoter specificities. Proc Natl Acad Sci. 1993;90:3147–3151. doi: 10.1073/pnas.90.8.3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roth S Y, Shimizu M, Johnson L, Grunstein M, Simpson R T. Stable nucleosome positioning and complete repression by the α2 repressor are disrupted by amino-terminal mutations in histone H4. Genes Dev. 1992;6:411–425. doi: 10.1101/gad.6.3.411. [DOI] [PubMed] [Google Scholar]
- 42.Schloβherr J, Eggert H, Paro R, Cremer S, Jack R S. Gene inactivation in Drosophila mediated by the Polycomb gene product or by position-effect variegation does not involve major changes in the accessibility of the chromatin fibre. Mol Gen Genet. 1994;243:453–462. doi: 10.1007/BF00280476. [DOI] [PubMed] [Google Scholar]
- 43.Senecoff J F, Bruckner R C, Cox M M. The FLP recombinase of the yeast 2-micron plasmid: characterization of its recombination site. Proc Natl Acad Sci USA. 1985;82:7270–7274. doi: 10.1073/pnas.82.21.7270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shao Z, Raible F, Mollaaghababa R, Guyon J R, Wu C-T, Bender W, Kingston R E. Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell. 1999;98:37–46. doi: 10.1016/S0092-8674(00)80604-2. [DOI] [PubMed] [Google Scholar]
- 45.Simon J. Locking in stable states of gene expression: transcriptional control during Drosophila development. Curr Opin Cell Biol. 1995;7:376–385. doi: 10.1016/0955-0674(95)80093-x. [DOI] [PubMed] [Google Scholar]
- 46.Simon J, Peifer M, Bender W, O'Connor M. Regulatory elements of the bithorax complex that control expression along the anterior-posterior axis. EMBO J. 1990;9:3945–3956. doi: 10.1002/j.1460-2075.1990.tb07615.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Singh J, Klar A J S. Active genes in budding yeast display enhanced in vivo accessibility to foreign DNA methylases: a novel in vivo probe for chromatin structure of yeast. Genes Dev. 1992;6:186–196. doi: 10.1101/gad.6.2.186. [DOI] [PubMed] [Google Scholar]
- 48.Struhl C, Basler K. Organizing activity of wingless protein in Drosophila. Cell. 1993;72:527–540. doi: 10.1016/0092-8674(93)90072-x. [DOI] [PubMed] [Google Scholar]
- 49.Strutt H, Cavalli G, Paro R. Co-localization of Polycomb protein and GAGA factor on regulatory elements responsible for the maintenance of homeotic gene expression. EMBO J. 1997;16:3621–3632. doi: 10.1093/emboj/16.12.3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thummel C S, Pirrotta V. New PcaSpeR P element vectors. Drosophila Infect Serv. 1992;71:150. [Google Scholar]
- 51.Thummel C S, Boulet A M, Lipshitz H D. Vectors for Drosophila P-element-mediated transformation and tissue culture transfection. Gene. 1988;74:445–456. doi: 10.1016/0378-1119(88)90177-1. [DOI] [PubMed] [Google Scholar]