Abstract
We have performed a systematic structure-function analysis of Saccharomyces cerevisiae TAF25, an evolutionarily conserved, single-copy essential gene which encodes the 206-amino-acid TAF25p protein. TAF25p is an integral subunit of both the 15-subunit general transcription factor TFIID and the multisubunit, chromatin-acetylating transcriptional coactivator SAGA. We used hydroxylamine mutagenesis, targeted deletion, alanine-scanning mutagenesis, high-copy suppression methods, and two-hybrid screening to dissect TAF25. Temperature-sensitive mutant strains generated were used for coimmunoprecipitation and transcription analyses to define the in vivo functions of TAF25p. The results of these analyses show that TAF25p is comprised of multiple mutable elements which contribute importantly to RNA polymerase II-mediated mRNA gene transcription.
mRNA gene transcription is mediated by RNA polymerase II working in concert with multiple general transcription factors (GTFs). The basal mRNA gene transcription machinery, as originally defined in vitro, is comprised of the GTFs TFIIA, TFIIB, TFIID, TFIIE, TFIIF, and TFIIH, which function with RNA polymerase II to promote preinitiation complex (PIC) formation in vitro and in vivo (see references 29 and 55 for reviews). GTFs contribute to PIC formation in multiple ways, either acting sequentially in a stepwise fashion (7) to form a PIC or acting as a single unit comprising the RNA polymerase II holoenzyme (35, 39). Although the composition of the basal transcription machinery and its possible modes of action have been fairly well characterized, the mechanisms of transcriptional activation are still poorly understood (44). Transactivator proteins have been shown to interact with a variety of targets, including the protein components of the RNA polymerase II transcription machinery as well as chromatin constituents and activities which modify chromatin. Among these putative transactivator targets are the GTF TFIID and the SAGA (Spt-Ada-Gcn5 acetylase) histone acetyltransferase complex. Both of these multisubunit complexes have been extensively studied in yeasts and metazoans (see references 1, 4, 18, and 66 for recent reviews).
Yeast TFIID is composed of 14 TATA box DNA binding protein-associated factors (TAFs) exhibiting molecular masses ranging from 150 to 17 kDa (61). Although the identifies of TFIID subunits are known, the exact stoichiometry of these multiple subunits within the complex is not. With the exception of TAF30p (30), all TFIID subunits are encoded by single-copy essential genes, and all display a high degree of sequence conservation among eukaryotes. One of these subunits, yeast TAF130p (also known as TAF145p) (58), and its metazoan counterparts (human and Drosophila TAF250p) contain intrinsic enzymatic activities that contribute to transcription (15, 16, 47, 49, 53, 54, 56).
Genetic and biochemical experiments have indicated that direct interactions between the activation domains (AD) of transcriptional activators and the subunits of TFIID play key roles in transactivation (11, 22, 23, 38, 40, 45, 62, 63, 70, 73, 74, 77). This coactivator function may be manifested at the molecular level by DNA-bound activators either stabilizing (recruiting) TFIID on the TATA box-core promoter (TATA-INR-DPE) (8, 42, 64, 65) of cis-linked genes or, perhaps, by (also) activating latent enzymatic activities residing within the subunits of TFIID itself. Regardless of the exact mechanisms through which transactivation occurs, only by a systematic molecular genetic dissection of the components comprising the GTFs, particularly TFIID, will this complex process be fully understood.
The yeast SAGA complex contains at least 14 subunits with molecular masses ranging from 430 to 17 kDa. The Gcn5p subunit of SAGA carries the catalytic activity capable of acetylating nucleosomal histones (24). Interestingly, five of the subunits of TFIID, TAF90p, TAF61(68)p, TAF60p, TAF25p, and TAF17p, are shared with SAGA (25). Except for TRA1, which encodes the largest subunit, none of the other known, non-TAF SAGA subunits are encoded by essential genes. Presumably, this genetic nonessentiality reflects the redundant nature of the function(s) of SAGA with other chromatin-modifying complexes (see references 36, 66, and 75 for recent reviews). Like TFIID, SAGA has been shown to play a key role in gene induction and transcriptional regulation. Recent in vivo (13, 26, 41, 69) and in vitro (46, 52, 72) studies have demonstrated that SAGA specifically associates with target genes early in the transcriptional activation process. The association of these chromatin-modifying complexes is thought to be a direct effect of specific, high-affinity AD-SAGA interactions. Complex formation between AD and SAGA is mutationally sensitive and can be readily observed in vitro (33, 72). Clearly, a thorough and detailed understanding of the mechanisms of gene activation will also require extensive dissection of the components comprising SAGA.
Certain TAFp-TAFp interactions are understood at the molecular level. The cocrystal structures of two TAFp-TAFp (core) complexes have been solved by X-ray crystallography (5, 76). From this and other work it is now quite clear that a highly conserved protein-protein interaction motif, the histone fold (HF), is found in many interacting proteins (68), and TAFps are no exception. Indeed, Burley and Roeder proposed a key role for histone-like TAFps in mediating TFIID functions some years ago (9). The HF motif mediates protein interactions both within TFIID (i.e., between TAF61p-TAF48p, TAF60p-TAF17p, TAF25p-TAF47p, and TAF25p-TAF65p) and within SAGA, where HF-mediated interactions between TAF25p and Spt7p as well as TAF68p and Ada1p have been identified and characterized (18, 19, 20). Dissecting the molecular rules defining these protein-protein interactions will prove crucial to understanding the roles that these multisubunit transcription factors play in regulated mRNA gene transcription.
TAF25p, an HF-containing protein, provides a unique insight into the study of the mechanisms of transcriptional regulation because this TAF is an integral subunit of both TFIID and SAGA. We originally cloned TAF25 and characterized the encoded protein, TAF25p, due to its presence in our TFIID preparations (37, 57). It has been shown that TAF25p plays a key role in mediating transcription both in vitro (37) and in vivo (43, 60). However, since TAF25p is resident in both TFIID and SAGA, it was not possible to unambiguously determine which TAF25p-containing complex was responsible for the observed transcription effects in the aforementioned studies. In order to address this and other gaps in our understanding of TAF25p function, we initiated a systematic analysis of this protein, including a detailed analysis of the structure-function relationships of TAF25. In this report, we describe our efforts to dissect TAF25p into functional domains through genetic and biochemical experimentation.
MATERIALS AND METHODS
Yeast and bacterial strains.
The parental yeast strains YEK16 (MATa leu2-3,112 ura3-52 his3-Δ200 trp1-Δ901 suc2-Δ9 ade2-101 lys2-801 taf25Δ1::TRP1 [pRS416-HA3-TAF25 WT]) and yJK800 (MATα leu2Δ0 ura3Δ0 his3Δ1 taf25D::KAN [pRS416-HA3-TAF25]) were transformed with HIS3-marked CEN-ARS plasmids (pRS413) carrying TAF25 genes with the various mutations (Fig. 1). The plasmids were then exchanged by plating the resulting pseudodiploid strains on 5-fluoroorotic acid (5-FOA) to select for those which had lost URA3-marked plasmids (pRS416-HA3-TAF25 WT) (6). Other strains used in this study include YEK25.75 (60), YEK20 (60), and YSLS74 (MATα/MATa leu2Δ0/leu2Δ0 ura3Δ0/ura3Δ0 his3Δ1/his3Δ1 lys2Δ0/LYS2 met16Δ0/MET15 taf25Δ::KAN), from which yJK800 was derived by sporulation and dissection. Additional information on strains and plasmids is available upon request. Escherichia coli cells were grown in Luria-Bertani media supplemented with ampicillin. Yeast cells were cultured in liquid or on solid defined media (minimal defined [SD] or complete defined [SC]) or rich media (yeast extract-peptone-dextrose [YPD] supplemented with adenine as needed [YPAD]) formulated as described previously (27).
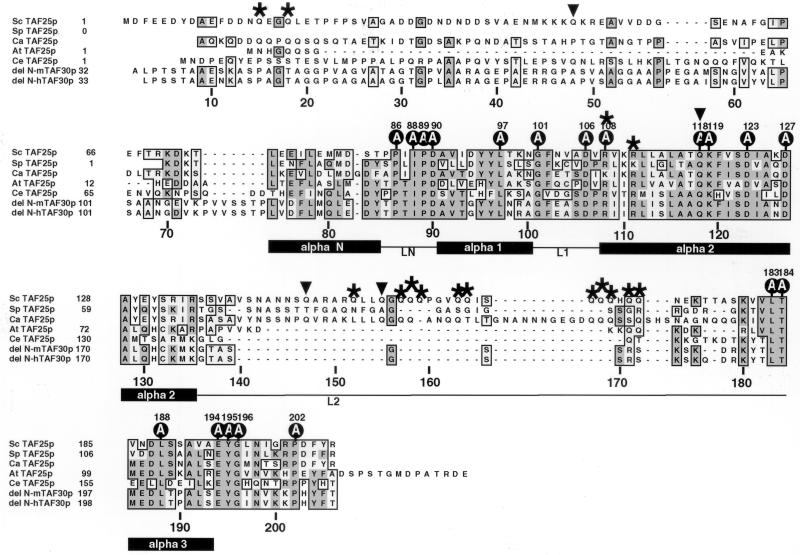
FIG. 1.
Alignment of TAF25p orthologs from various eukaryotes. Sequences were aligned with S. cerevisiae (Sc) TAF25p; the numbering shown below the aligned sequences refers to the amino acid sequence of this protein. Schizosaccharomyces pombe (Sp), C. albicans (Ca), Arabidopsis thaliana (At), Caenorhabditis elegans (Ce), and N-terminally truncated human and mouse (del N-m or del N-h) TAF25p orthologs were used for Clustal W alignment with MacVector software (version 6.5.3). These sequences are numbered on the left; the translational start site of the Ca ortholog has not been determined. The sequence of Ca TAF25p was obtained from the Stanford DNA Sequencing and Technology Center website at http://www-sequence.stanford.edu/group/candida; all other sequences were obtained from GenBank. Asterisks indicate arginine and glutamine residues (R108, R111, Q15, Q18, Q152, Q157, Q158, Q159, Q163, Q164, Q167, Q168, Q169, Q171, and Q172) which, when the cognate encoding codons were mutated with hydroxylamine to termination codons, produced C-terminally truncated TAF25 proteins conferring lethality. The locations of the four Q residues (Q48, Q118, Q147, and Q155) not affected by hydroxylamine are indicated by the filled inverted triangles. Residues individually mutated to alanine in the background of mini-TAF25 are indicated by the circles labeled A (Sc TAF25p residues P86A, I88A, P89A, D90A, L97A, G101A, D106A, R108A, Q118A, K119A, D123A, D127A, L183A, T184A, L188A, E194A, Y195A, G196A, and P202A). Shown below the sequence alignments is the putative HF consensus structure of four alpha helices separated by three linker regions (alpha N, LN, alpha 1, L1, alpha 2, L2, and alpha 3), as proposed by Gangloff et al. (18).
Plasmids.
TAF25 plasmids were constructed by standard techniques. In all cases, plasmid-based TAF25p expression was driven by the normal TAF25 regulatory sequences. The 2μm-based plasmids (pRS426) carrying ADA1, TAF47, and TAF65 were constructed by standard techniques. The 2μm vectors containing SPT7, TAF25, and TAF40 were kind gifts of Steve Buratowski and Fred Winston.
Molecular biological methods.
DNA manipulation, purification, analysis, RNA purification and hybridization, and yeast transformations were all performed as described previously (3, 12, 37, 60). Whole-cell extract (WCE) preparation, antibody preparation, immunoblotting, and immunoprecipitation were performed as detailed previously (61). Immunoblots were quantitated using a Fluor-S MultiImager (Bio-Rad). Yeast two-hybrid screening was performed using both Clontech yeast strain L40 (according to manufacturer protocols) and yeast strain PJ69-4A as detailed previously (34). TAF25 “bait” molecules used in the screening fused the TAF25p-encoding open reading frame (ORF) to either LexA or Gal4 DNA binding domains (DBD).
RESULTS
Inactivation of TAF25pG101E by a temperature shift dramatically reduces polymerase II-mediated gene transcription in vivo without complete disruption of TFIID or SAGA.
We performed hydroxylamine mutagenesis of TAF25 with the goal of generating temperature-sensitive mutant alleles of the gene which might prove useful for the characterization of TAF25p functions. We were successful in this endeavor and obtained several such mutant alleles, which clustered around TAF25 sequences encoding amino acids 101 to 111 (Fig. 1). In an earlier work, Sanders et al. (60) used a particular temperature-conditional mutant from this collection, strain YEK25.75, which expressed a form of TAF25p bearing a single mutation resulting in a G→E amino acid substitution at position 101 (TAF25pG101E). When YEK25.75 cells were shifted from a permissive (22°C) to a nonpermissive (37°C) temperature, high-level RNA polymerase II-mediated mRNA gene transcription was reduced ≥60% within 30 min (60). One can readily envision two limit cases to explain this transcriptional phenotype. On one hand, TFIID and/or SAGA complexes could be substantially or completely disrupted upon the temperature shift. Indeed, this is the situation for all HF motif-containing TAFs described in the literature (2, 25, 48, 50, 51, 52, 59, 60). Alternatively, a second possibility is that TAF25p is inactivated in situ within TFIID and/or SAGA complexes, and this inactivation disrupts critical, positively acting protein-protein contacts made between TAF25p and some yet-to-be-defined component(s) of the RNA polymerase II transcription machinery. The second of these two possibilities is mechanistically more interesting and experimentally more useful. Prior to embarking upon a detailed analysis of TAF25, we wanted to convince ourselves that we were not simply studying the effects of TFIID and/or SAGA complex dissociation. We therefore conducted an experiment which would allow us to distinguish between these two limit cases.
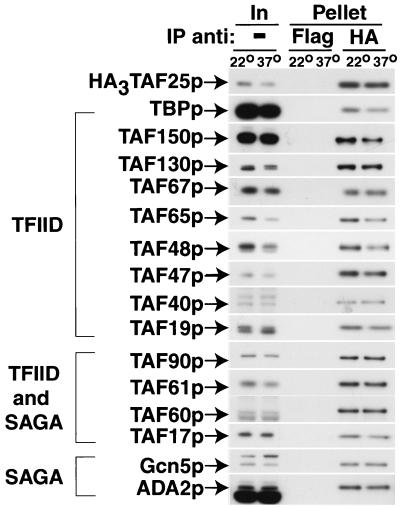
To probe TFIID and SAGA complex integrity, we performed a coimmunoprecipitation analysis of TAF25p-containing complexes. A 22°C-grown culture of YEK25.75 cells expressing a three-hemagglutinin (HA3)-tagged TAF25p mutant protein was split; we left half at 22°C and shifted the other half to 37°C. After 30 min, WCEs were generated, immunoprecipitates were prepared (antibody excess) using either control anti-FLAG monoclonal antibody (MAb) or anti-HA MAb, and the precipitated proteins were analyzed by immunoblotting. All components analyzed were present in both 22°C and 37°C WCEs (Fig. 2, In). Consistent with the results of a previous study (60), the amount of each of the subunits analyzed was slightly reduced in the 37°C WCE (10 to 25%), but there were still large amounts of intact TFIID and SAGA in these cells (Fig. 2, Pellet, HA). Thus, the primary cause of the transcription decrement in this mutant cell strain is not complete complex disruption of TFIID and SAGA. The transcription defect could be caused either by an inactivation of TAF25p in situ within both complexes or by the slight reduction of TFIID and SAGA levels. Regardless, encouraged by these data, which clearly demonstrated that TAF25p and mRNA gene transcription in vivo could be inactivated without the induction of complete disruption of TFIID and SAGA, we embarked upon a detailed genetic dissection of TAF25p structure-function relationships.
FIG. 2.
Neither TFIID nor SAGA complexes are significantly disrupted in YEK25.75 cells shifted from permissive to nonpermissive growth conditions. Logarithmically growing YEK25.75 cells expressing HA3-tagged TAF25p were cultured at 22°C in YPAD media. Cells were collected by centrifugation in two equal portions, and pelleted cells were resuspended in an equivalent volume of YPAD media either at 22°C or prewarmed to 37°C. Both the 22°C and the 37°C cultures were shaken at these temperatures for 30 min. WCEs were prepared, and immunoprecipitations (IP) were performed with either anti-FLAG or anti-HA MAb. Fractions of the Input (In, 2%) and the precipitate (Pellet, 4%) were analyzed by immunoblotting with the indicated antibody. Twenty-five percent less of the 22°C precipitate was analyzed to adjust for the ∼25% decrease in the amount of HA3-TAF25p in the 37°C WCE. −, no IP.
Amino acid sequences C terminal to glutamine 172 are required to produce functional TAF25p.
Although useful, the collection of TAF25 mutants described above gave us little insight into the overall structure-function relationships of the molecule. Indeed, one unlikely, yet plausible, explanation for not finding mutations throughout TAF25p was that only the residues of the protein centered around amino acids 101 to 111 contributed to function. To test this idea, we returned to using hydroxylamine mutagenesis but screened for mutations that were lethal. Of ∼30,000 colonies screened, 30 failed to grow on 5-FOA-containing plates. The relevant HIS3-marked TAF25-containing mutagenized plasmids were separately recovered from these strains, passaged through E. coli, and rescreened by plasmid shuffling as described above. Twenty-five continued to confer lethality, and the DNA sequences of the TAF25 genes were determined. Twenty-three carried nonsense mutations either in TAF25p glutamine residues (mutating CAA or CAG to UAA or UAG; 21 of 25) or arginine residues (CGA to UGA; 2 of 25). Of the 17 possible Q residue targets, only 4 were not identified in this screen, while both of the CGA-encoded R residues were mutated (Fig. 1). The other two mutations of this collection both mutated the initiator methionine residue. The Q157, Q159, Q171, and R111 mutants were isolated five, three, two, and two times, respectively,while all other Q or R mutants were isolated once, suggesting that the screen was relatively complete. These results indicated that sequences throughout the length of the 618-bp TAF25 ORF might be important for function and, more particularly, that the residues C terminal to Q172 could not be deleted without compromising TAF25p function. This portion of TAF25p (residues 172 to 206) defines a region of the molecule which is highly conserved.
Core residues of TAF25p comprising evolutionarily conserved sequences are sufficient to support growth.
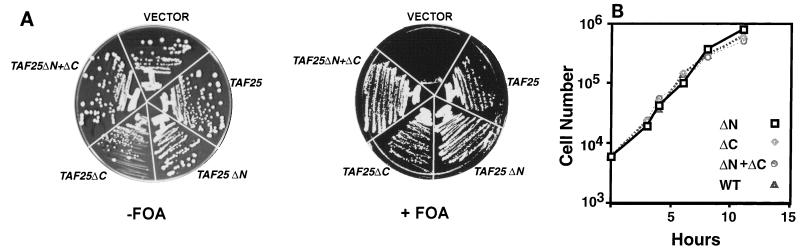
Alignment of TAF25p orthologs from two other yeasts, plants, worms, and mammals with Saccharomyces cerevisiae TAF25p showed, as noted previously, that only a portion of this protein is conserved among organisms (5, 21, 37, 51). Notably, two segments of S. cerevisiae TAF25p (amino acids 2 to 73 and amino acids 142 to 179) were not conserved (Fig. 1). In order to examine whether nonconserved N-terminal and C-terminal domains of TAF25p were critical for function, we generated deletion derivatives of TAF25 which removed the N-terminal portion (amino acids 2 to 74; termed TAF25ΔN), the C-terminal portion (amino acids 140 to 179; termed TAF25ΔC), or both nonconserved portions (TAF25ΔN+ΔC). Vector alone, the wild-type gene, and the three deletion forms of TAF25 were introduced into the yeast, and plasmid shuffling was used to test if these forms of TAF25 supported growth. None of the deletion mutant alleles of TAF25 exhibited strong dominant-negative effects on growth (Fig. 3A, −FOA). After shuffling, even the double-deletion form of TAF25 (TAF25ΔN+ΔC), here referred to as core TAF25 or mini-TAF25, supported wild-type levels of growth. Growth curves for these various strains in liquid media at 30°C were indistinguishable (Fig. 3B). Clearly, under these conditions (and in SD media; data not shown), the nonconserved portions of TAF25p were dispensable for normal growth properties.
FIG. 3.
Nonconserved TAF25p sequences are dispensable for yeast cell growth. (A) Shown is a photograph of two plates upon which the indicated yeast strains were inoculated. The plate on the right contained 5-FOA, which selects for cells which have lost the URA3-marked pRS416-TAF25WT plasmid, consequently revealing the chromosomal taf25Δ1::TRP1 allele and testing for the ability of the HIS3-marked pRS413 vector (pRS413) or wild-type TAF25 (pRS413-WT) and deletion alleles of TAF25 (pRS413-TAF25ΔN, pRS413-TAF25ΔC, and pRS413-TAF25ΔN+ΔC) to support growth; these constructs are labeled VECTOR, TAF25, TAF25ΔN, TAF25ΔC, and TAF25ΔN+ΔC, respectively. Plates were incubated at 30°C for 3 days (left) or 6 days (right) before being imaged. (B) Growth curves for various TAF25 deletion alleles at 30°C. Overnight cultures of the indicated deletion alleles of TAF25 were diluted in appropriate liquid media to an equal concentration, as determined by cell counts using a hemocytometer. The cultures were then grown at 30°C. At various time points, cell counts were performed. WT, wild type.
Residues in the conserved core of TAF25p are important for TAF25p function in vivo.
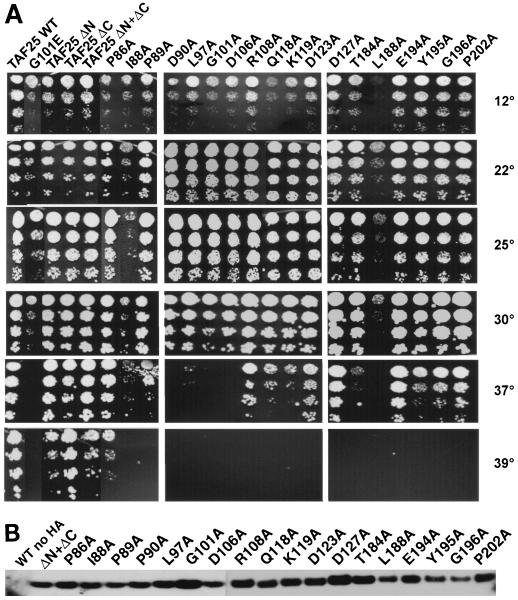
Although the experiments above proved useful, we decided to take a more systematic mutational approach toward elucidating the structure-function relationships of TAF25 for two reasons. First, we wanted to assess whether the amino acids within the conserved 96-amino-acid core (i.e., mini-TAF25p) really were important for TAF25p function. If so, then these sequences should be mutationally sensitive. Second, and more importantly, we wanted to prepare a panel of TAF25 genetic reagents that we could use to attempt to define domains of the protein that uniquely contribute to TFIID-specific and SAGA-specific functionalities. To accomplish these goals, we performed alanine-scanning mutagenesis of mini-TAF25. The triplets encoding 19 highly conserved amino acids within the core domain of TAF25p were separately mutated to encode alanine (Fig. 1, residues marked with the circles labeled “A”). All of these mutants were constructed with the double-deletion mini-TAF25 gene backbone and cloned in pRS413 as HA3-tagged proteins. Each construct was sequenced, including ∼200 bp of 5′ and 3′ sequences. The Ala mutants were introduced into yeast strain YEK16 to generate a family of TAF25 pseudodiploid strains; all mutants were recessive (data not shown).
Following plasmid shuffling, the ability of the Ala mutants to support growth was assessed. With the exception of mutant L183A, which was lethal, all the mutants supported growth at room temperature (∼22°C), although to various extents. With the exception of P86A, none of the mutants could grow at 39°C, while the control strain carrying mini-TAF25p could (Fig. 4A). Wild-type, TAF25ΔN, TAF25ΔC, TAF25ΔN+ΔC, and G101E strains were included as controls. The lethality of strain L183A could be due either to inactivity of that mutant form of TAF25p or to protein instability; this mutant protein was undetectable on Western blots (data not shown). Strains expressing the I88A and L188A mutant forms of TAF25p grew particularly slowly at extreme temperatures (12, 22, 37, and 39°C) (Fig. 4A). Control strain G101E displayed both low- and high-temperature-sensitive growth (Fig. 4A). Residue I88 (potential loop region) and residue L188 (alpha helix three) are 100% conserved among all TAF25p othologs and map to the putative HF region of TAF25p (Fig. 1) (18). In addition to the marked growth deficiencies noted above, more subtle defects were also apparent. Mutants D90A, G101A, D106A, Q118A, and K119A all exhibited a mild cold-sensitive growth phenotype; these patterns of conditional growth were observed in two genetic backgrounds (S288C and W303) (data not shown). Protein stability does not appear to explain the variable growth properties, as steady-state levels of the Ala mutant proteins are approximately the same (Fig. 4B). Collectively, these results indicated that the residues encoded by the conserved core of TAF25 are mutationally sensitive and indeed are important for the function(s) of the molecule.
FIG. 4.
Phenotypes of yeast strains expressing wild-type TAF25 and mutant alleles of TAF25. (A) TAF25 haploid yeast strains carrying the indicated wild-type (WT) or mutant TAF25 alleles were plated on YPAD media in order to test for growth at different temperatures. Serial fivefold dilutions of cells were plated. The 12, 22, 25, 30, 37, and 39°C plates were photographed after 330, 66, 66, 46, 66, or 66 h of incubation, respectively. (B) Steady-state levels of TAF25p produced from various alleles of TAF25. TAF25 haploid shuffled yeast cells carrying the indicated HA3-tagged mutant forms of mini-TAF25 were grown in selective media to mid-log phase, and equivalent amounts of cells (optical density at 600 nm, ∼5) were harvested by centrifugation and frozen at −80oC. Protein extracts were prepared from the frozen cells and fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and HA3-TAF25p content was measured by immunoblotting using peroxidase-conjugated anti-HA MAb (Roche).
In order to gain further insight into TAF25p function, the mutant collection was subjected to a battery of growth tests in an attempt to sort the members of the family into functional subsets. We tested the Ala mutant family for growth on various media either lacking certain nutrients (methionine or inositol) or containing alternate carbon sources (glycerol, galactose, maltose, or sucrose), nonspecific growth inhibitors (formamide, sorbitol, caffeine, or ethanol), or inhibitors of DNA or RNA synthesis (hydroxyurea, thiolutin, or azauracil). The use of selective media for growth tests often identifies defects in components of the transcriptional apparatus (28, 29). Moreover, mutations in certain genes encoding SAGA subunits have been reported to result in differential growth properties in some of these tests (67). The results of these screens are summarized in Table 1. The different members of the Ala mutant family behave in a distinct fashion in these disparate growth tests. Mutants I88A, L188A, and G101A as well as the non-Ala mutant G101E showed enhanced growth defects on media containing nonspecific growth inhibitors (caffeine, formamide, or ethanol). I88A also exhibited little or no growth on media lacking inositol or containing glycerol or maltose as a carbon source. Other strains (Q118A, K119A, and D90A) showed slight growth defects on both ethanol and formamide (Table 1). All of these residues are located in different portions of the canonical HF motif. In aggregate, these data suggested that functionally distinct domains of TAF25p exist and that these domains contribute differentially to specific transcriptional functions of the molecule.
TABLE 1.
Phenotypic analyses of the TAF25 mutant familya
| Strain | Growth on:
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MAL | GAL | GLYC | SUC | SORB | −MET | −INO | AZA-U | CAFF | THIO | FORM | ETH | HYD-U | |
| Wild type | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| G101E | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | + | + | + | + | ++ |
| TAF25ΔN | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| TAF25ΔC | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| TAF25ΔN + ΔC | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| P86A | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| I88A | ± | + | ± | ++ | + | + | ± | + | ± | + | + | ± | + |
| P89A | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| D90A | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | ++ | ++ | +++ |
| L97A | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| G101A | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | ++ | +++ | ± | ± | +++ |
| D106A | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| R108A | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| Q118A | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | ++ | ++ | +++ |
| K119A | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | ++ | ++ | +++ |
| D123A | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| D127A | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| T184A | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| L188A | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | + | ++ | − | + | ++ |
| E194A | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| Y195A | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| G196A | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| P202A | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
Haploid yeast strains carrying the indicated TAF25 alleles were plated on various solid media supplemented with different compounds, deficient in a certain growth component, or containing inhibitors in order to test for growth. Serial fivefold dilutions of cells were plated. Growth was assessed qualitatively compared to that of the the isogenic wild-type strain either 90 h (in the presence of maltose [MAL], galactose [GAL], glycerol [GLYC], sucrose [SUC], or sorbitol [SORB]; in the absence of methionine [−MET] or inositol [−INO]; or in the presence of azauracil [AZA-U], caffeine [CAFF], or thiolutin [THIO]) or 100 h (in the presence of formamide [FORM], ethanol [ETH], or hydroxyurea [HYD-U]) after inoculation. Growth ranged from little growth (±) to wild-type growth (+++). −, no growth.
Two hybrid screens identify a TFIID subunit, a SAGA subunit, and transacting factors as TAF25p-interacting partners.
To identify potential TAF25p-interacting proteins, we performed three sets of two-hybrid screens (17) using either Gal4 DBD-TAF25p or LexA DBD-TAF25p bait molecules in two different yeast strains and with two different yeast genomic DNA Gal4p-AD fusion libraries; we tested a total of ∼1.12 × 107 yeast colonies. The DNA sequences of 209 clones which met appropriate selective criteria were determined. Several of the hits in this assay are notable. First, both TFIID (TAF47) and SAGA (SPT7) subunit-encoding genes were identified. This result might be expected, since TAF25p is an integral subunit of both complexes. Interestingly, TAF25p-TAF47p and TAF25p-Spt7p protein-protein interactions have been observed recently by Gangloff et al. (19), who used directed two-hybrid screens with DBD-TAF25p and TAF47p-AD and Spt7p-AD fusions to demonstrate interactions between these HF-containing proteins. These results corroborate the nondirected two-hybrid interactions reported here. A second feature of these analyses is that genes encoding several known or putative transcriptional regulators (ASK10, UME6, YPR115W, TAO3, RIM15, RLM1, ECM22, RAP1, and YAP6) were identified. Finally, genes encoding components of RNA processing proteins were also identified (PRP22, CLP1, and PRP40). The specificity and physiological significance of these last two classes of genes remain to be authenticated.
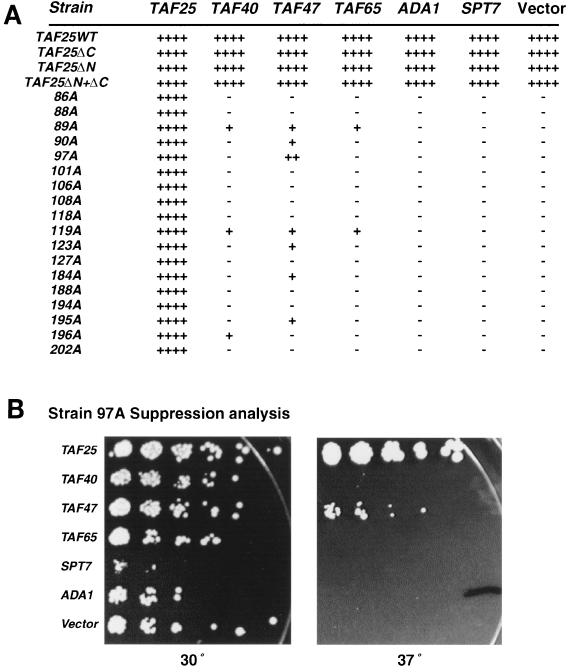
Overexpression of selected TFIID and SAGA subunits weakly suppresses the temperature-sensitive growth of distinct TAF25 Ala mutants.
We tested the effects of overexpression of the TFIID and SAGA subunits which interact with TAF25p (TAF47p, TAF65p, and Spt7p) (19) upon the ability of the TAF25 Ala mutant family to grow at nonpermissive temperatures. In addition, we tested overexpression of TAF40p, as it was identified as a putative TAF25p interactor by others (71). As expected, TAF25 but not a vector suppressed the temperature-sensitive growth of all temperature-sensitive strains (Fig. 5). We used ADA1 as a specificity control, since it encodes an HF motif SAGA-specific subunit but does not physically interact with TAF25p (19). Overexpression of TAF25 had no effect on the growth of any of the strains. Indeed, this was the situation with all genes examined, with the exception of SPT7. Overexpression of this gene reproducibly induced a slow-growth phenotype in all strains, even under permissive growth conditions (Fig. 5 and data not shown). Cooverexpression of TAF25 reversed, in a dose-dependent fashion, the dominant-negative growth effect of SPT7 overexpression (data not shown). The molecular mechanism of this phenomenon is unknown at present. Unfortunately, however, this dominant-negative effect of SPT7 prevented us from using the overexpression-suppression approach to full effect to map TFIID-specific and SAGA-specific domains within TAF25p. Overexpression of TAF47, TAF40, and TAF65 weakly suppressed the temperature-sensitive phenotype of particular members (P89A, D90A, L97A, K119A, D123A, T184A, Y195A, and G196A) of the TAF25 Ala mutant family. Although suppression in most cases was weak, it was reproducibly above the suppression levels conferred by the ADA1 control.
FIG. 5.
Suppression of temperature-sensitive growth patterns of yeast TAF25 mutants by overexpression of genes encoding TFIID and SAGA subunits. (A) Yeast strains bearing alleles of TAF25 were transformed with 2μm URA3-marked plasmids carrying the indicated TFIID or SAGA subunit-encoding genes. Serial fivefold dilutions of cells were plated on SC media and incubated at either a permissive (30°C) or a restrictive (37 to 39°C) temperature. Growth at the restrictive temperature (37, 38, or 39°C, as appropriate) was scored qualitatively in comparison to that of the pRS426-TAF25 positive control and is represented as + to ++++ (strains carrying 2μm TAF25); −, no growth. Note that all strains carrying 2μm SPT7 displayed a slow-growth phenotype at all temperatures examined compared to the same strains carrying the empty 2μm vector. (B) Suppression analysis. Strain 97A was transformed with the indicated plasmids, and serial fivefold dilutions were plated on selective SC media at either 30 or 37°C for 90 h and then imaged.
TAF25 Ala mutant family members display differential effects upon mRNA gene transcription in vivo.
The two-hybrid data, along with the data of Table 1 and Fig. 5, strongly suggested that TAF25p contributed substantially to RNA polymerase II-catalyzed mRNA gene transcription. Such results are consistent with previously published observations (37, 43, 60). Given these results, we believed it important to examine mRNA gene transcription in the Ala mutant collection and therefore tested both total and gene-specific mRNA syntheses.
The results of these transcription analyses are presented in Table 2. The numbers refer to the ratio of the number of transcripts detected from cultures grown at restrictive temperature 39°C for 1 h to the value for the control culture grown at 22°C. These data represent the averages of three independent analyses performed using three independent sets of yeast cultures subjected to the temperature shift regimen outlined above. Values deemed different (decreased by ∼2-fold or increased by ∼50%) from those for control strain TAF25ΔN+ΔC are indicated by bold type in Table 2. From these data it can be seen that total poly(A)+ mRNA synthesis was dramatically reduced (down 78%) when control strain G101E was shifted to the nonpermissive temperature, consistent with previous results (60). Total poly(A)+ mRNA synthesis was also decreased substantially in several of the Ala mutant family members, particularly mutant I88A and, to a lesser extent, mutants G101A, D123A, and L188A, upon temperature shift. These data are consistent with the idea that TAF25p function is integral to high-level mRNA gene transcription (43, 60).
TABLE 2.
Transcription analyses of the TAF25 mutant familya
| Strain | Total poly (A)+ mRNA | SAGA
|
TFIID
|
TAF25p
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| YHB1 | PHO84 | YJL012C | YDL124W | PMT4 | WSC2 | TRX1 | GIC2 | YOR248W | FAS2 | ||
| Wild type | 124 | 123 | 394 | 101 | 233 | 82 | 90 | 44 | 92 | 52 | 96 |
| G101E | 22 | 22 | 240 | 87 | 160 | 37 | 36 | 54 | 86 | 26 | 37 |
| TAF25ΔN | 87 | 26 | 334 | 47 | 168 | 40 | 64 | 45 | 82 | 34 | 31 |
| TAF25ΔC | 98 | 42 | 422 | 64 | 136 | 55 | 72 | 45 | 63 | 63 | 37 |
| TAF25ΔN+ΔC | 80 | 62 | 324 | 61 | 235 | 85 | 77 | 56 | 59 | 107 | 57 |
| 86A | 77 | 37 | 228 | 71 | 153 | 50 | 43 | 36 | 76 | 26 | 55 |
| 88A | 35 | 53 | 89 | 51 | 285 | 41 | 30 | 50 | 75 | 33 | 56 |
| 89A | 79 | 45 | 166 | 59 | 102 | 62 | 38 | 42 | 53 | 38 | 40 |
| 90A | 78 | 28 | 315 | 60 | 265 | 64 | 38 | 48 | 78 | 37 | 37 |
| 97A | 70 | 52 | 358 | 70 | 277 | 56 | 32 | 31 | 54 | 57 | 64 |
| 101A | 52 | 50 | 139 | 56 | 331 | 26 | 2 | 48 | 52 | 24 | 32 |
| 106A | 71 | 36 | 166 | 71 | 97 | 36 | 31 | 46 | 46 | 37 | 29 |
| 108A | 81 | 44 | 270 | 59 | 224 | 46 | 60 | 47 | 47 | 45 | 28 |
| 118A | 78 | 39 | 205 | 64 | 121 | 52 | 60 | 35 | 55 | 46 | 35 |
| 119A | 63 | 43 | 220 | 50 | 267 | 45 | 25 | 40 | 35 | 32 | 32 |
| 123A | 52 | 32 | 286 | 56 | 135 | 41 | 26 | 47 | 17 | 38 | 37 |
| 127A | 62 | 59 | 363 | 66 | 197 | 48 | 74 | 41 | 63 | 35 | 26 |
| 184A | 83 | 56 | 236 | 68 | 161 | 62 | 43 | 40 | 59 | 41 | 32 |
| 188A | 54 | 21 | 342 | 67 | 60 | 48 | 3 | 22 | 24 | 9 | 12 |
| 194A | 81 | 56 | 124 | 68 | 275 | 65 | 53 | 46 | 52 | 34 | 36 |
| 195A | 59 | 21 | 435 | 51 | 97 | 24 | 27 | 27 | 48 | 30 | 27 |
| 196A | 72 | 30 | 281 | 77 | 145 | 34 | 34 | 32 | 63 | 57 | 49 |
| 202A | 83 | 43 | 235 | 61 | 124 | 38 | 67 | 41 | 55 | 43 | 34 |
All analyses were performed three times using three independent cultures of the indicated yeast strains. RNA was either directly blotted and probed for total poly(A)+ mRNA by hybridization with 32P-labeled oligo(dT)20 or fractionated by denaturing agarose gel electrophoresis, blotted, and detected with the indicated 32P-labeled, PCR-generated gene-specific hybridization probes as detailed in Materials and Methods. TAF25ΔN+ΔC was the relevant control strain. Values that were at least 50% increased or 2-fold decreased relative to those for this control strain are in bold type; see the text for details. The values shown represent the means for three independent analyses; standard errors were <5% the indicated values for the total poly(A)+ mRNA analyses or <20% the indicated values for the RNA blotting experiments. The three classes of genes used for the transcription analyses were chosen based upon the data of Lee et al. (43). For TFIID-dependent genes PMT4, WSC2, and TRX1, transcription was reduced in a temperative-sensitive taf145(130) strain after 45 min at a nonpermissive temperature 3.2-, 4.8-, and 5.3- fold (mRNA half-lives of 10, 11, and 18 min), respectively. For the SAGA-dependent genes YHB1, PHO84, YDL124W, and YJL012C, transcription was reduced in a gcn5 null strain 4.1-, 20.1-, 3.5-, and 3.0- fold (half-lives of 6, 7, 14, and 14 min), respectively. For the TAF25-dependent genes GIC2, YOR248W, and FAS2 transcription was reduced 4.2-, 4.1-, and 3.1- fold (half-lives of 36, 13 and 13).
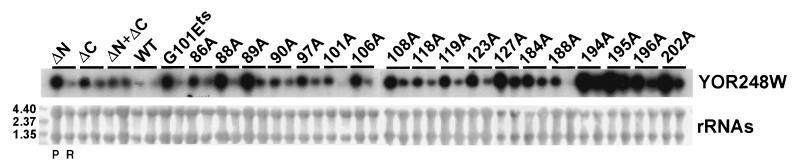
To more rigorously test the hypothesis that TAF25p function is required for mRNA synthesis, we examined specific mRNA gene transcription in this mutant family by RNA blotting. Three classes of genes were chosen for hybridization probes based upon the recently published gene array transcription data of Lee and colleagues (see Table 3 in reference 43 and website web.wi.mit.edu/young/pub/expressionanalysis.html). The first comprised genes whose transcription was reported to be uniquely but generally dependent upon TFIID (PMT4, WSC2, and TRX1). The second was composed of genes whose transcription was reported to uniquely depend upon SAGA (YHB1, PHO84, YDL124W, and YJL012C). The third represented genes whose transcription was reduced in an undefined temperature-sensitive taf25 strain at a nonpermissive temperature after 45 min (GIC2, YOR248W, and FAS2). An example of the results of these RNA blotting analyses is presented in Fig. 6, where YOR248W mRNA levels were examined; overall data are summarized in Table 2. These analyses were performed in triplicate, and the values shown in Table 2 represent the averages of three independent experiments. Analysis of the data indicates that the TAF25 mutant family displays a variety of gene-specific defects in transcription which often vary dramatically from allele to allele and transcript to transcript.
FIG. 6.
Transcriptional profiles of yeast strains expressing wild-type and mutant alleles of TAF25. Specific mRNA gene transcription was measured using denaturing agarose gel electrophoresis, blotting, and hybridization with a 32P-labeled gene-specific probe derived from YOR248W. Prior to hybridization, the blot was stained with 0.01% (wt/vol) methylene blue to detect rRNAs (the loading control) and 4.40-, 2.37-, and 1.35-kb RNA size markers (bottom panel). YOR248W mRNA content was detected by autoradiography and phosphorimaging (see Table 2). RNA samples were loaded in pairs—RNA from cells grown at a permissive (P) temperature of 22°C or a restrictive (R) temperature of 39°C (left and right lanes, respectively, under each bar)—for each of the strains shown. Phosphorimaging counts for the samples at 22 and 39°C, respectively, were as follows: ΔN (164,949 and 56,082), ΔC (151,639 and 103,114), ΔN+ΔC (123,804 and 115,840), wild type (WT) (48,114 and 21,494), temperature-sensitive G101E (G101Ets) (146,270 and 31,543), 86A (126,730 and 38,422), 88A (148,997 and 42,889), 89A (178,012 and 80,655), 90A (98,835 and 36,172), 97A (123,489 and 69,778), 101A (55,756 and 15,984), 106A (130,987 and 50,679), 108A (157,686 and 71,411), 118A (91,983 and 50,313), 119A (106,343 and 40,123), 123A (108,767 and 45,670), 127A (177,102 and 67,832), 184A (114,814 and 44,283), 188A (68,941 and 6,099), 194A (304,320 and 120,734), 195A (290,872 and 106,468), 196A (134,893 and 72,202), and 202A (218,445 and 84,486).
DISCUSSION
In this report we describe our efforts to systematically define the structure-function relationships of TAF25, a single-copy essential gene which encodes an HF motif-containing subunit shared by TFIID and SAGA. We have made a number of important observations regarding TAF25p function.
Inactivation of TAF25pG101E dramatically reduces polymerase II-mediated gene transcription in vivo without complete disruption of TFIID or SAGA.
It has been hypothesized that there are basically two types of TAFps with regard to transcriptional dependencies. One class is TAFps which induce a relatively large drop in total poly(A)+ mRNA synthesis when functionally inactivated. This group includes the HF motif-containing TAFps: TAF17p, TAF25p, TAF40p, TAF60p, and TAF61p. At least for TAF17p, this large drop in high-level poly(A)+ mRNA gene transcription translates to a correspondingly large drop in gene-specific mRNA gene transcription (2). The other group of TAFps is comprised of those that do not contain HF motifs. When these TAFps are inactivated, they fail to induce a large decrease in high-level total poly(A)+ mRNA synthesis; they include TAF150p, TAF130/145p, TAF90p, and TAF67p. One possible explanation for the differences in transcriptional output upon inactivation is that when the HF motif-containing TAFps are inactivated, the TFIID (and/or SAGA) complex is rapidly and totally disrupted, resulting in TAFp degradation (2, 25, 48, 50, 51, 52, 59, 60). This degradation could be due to the fact that the HF motif-containing TAFps fulfill key structural roles within TFIID and SAGA. Although the ability to induce a rapid loss of TFIID is a useful phenomenon for studying TFIID biology, mutants which induce rapid complex dissociation are less useful for biochemical and genetic experimentation aimed at elucidating the molecular mechanisms of action of these multisubunit complexes. Therefore, we were not as excited about molecularly dissecting TAF25 if the only mechanism by which mutation could inactivate its in vivo function(s) were TFIID and/or SAGA disruption. We consequently carefully analyzed the effects of a temperature shift upon complex integrity with a temperature-sensitive allele of TAF25 (60).
We used cells carrying the taf25G101E allele, a mutation which causes both temperature-sensitive growth and also dramatically reduces RNA polymerase II-mediated mRNA transcription in vivo (60), for SAGA and TFIID coimmunoprecipitation studies. It appears that the primary defect causing the precipitous drop in transcription is not complete complex dissociation per se (Fig. 2) (60), as transcription is decreased to a greater extent than complex dissociation at 30 min postshift. This observation argues that TAF25pG101E is inactivated in situ within TFIID and/or SAGA and that this inactivation is responsible for the primary in vivo decrement in transcription. If correct, then the data further suggest that TAF25p makes multiple, positively acting protein-protein interactions in one or both of these multisubunit assemblies which contribute critically to ongoing high-level RNA polymerase II transcription. Indeed, our own two-hybrid screens and the recent work of Gangloff et al. (19) support this hypothesis. TAF25p interacts specifically, through its HF motif, with the complementary HF domains of TAF47p, TAF65p, and Spt7p, all subunits of either TFIID or SAGA. These observations all add to the understanding of TAF25p. More importantly, though, these data encouraged us to analyze the structure-function relationships of this molecule in detail.
Less than half of the TAF25 ORF is required to produce a mini-TAF25p capable of supporting yeast cell growth.
To gain greater insight into TAF25p, we performed an additional analysis of the hydroxylamine-generated mutant family which had produced yeast strain YEK25.75 (60). We searched for hydroxylamine-induced mutations conferring lethality. Such mutations were readily identified and mapped to sequences distributed throughout the TAF25 ORF. These data indicated that nearly the entire length of TAF25 was mutable (Fig. 1); hence, sequences throughout TAF25p might contribute to its function.
The idea that the entire length of TAF25p contributed to its function was tested directly in two complementary ways: first, by sequence alignments coupled with deletion mutagenesis, and second, by site-directed alanine-scanning mutagenesis. Our deletion mutagenesis studies have shown that only the 96 amino acids which comprise evolutionarily conserved portions of TAF25p are required to support yeast cell growth. Indeed, this conserved region supports wild-type levels of growth (Fig. 3B). However, deletion of the nonconserved residues, amino acids 2 to 73 and amino acids 142 to 179, is not without effect. Strains expressing the ΔN and ΔN+ΔC deletion forms of TAF25p display different total poly(A)+ mRNA levels and exhibit different patterns of specific transcript levels compared to the wild type (Table 2). These results suggest that the nonconserved N-terminal portion of TAF25p could play some role in the function of either TFIID or SAGA or both.
TAF25p interacts with transcription factors as well as with TFIID and SAGA constituents.
One of the popular models for TAF function, originally formulated by Chen and colleagues (10) and Goodrich and colleagues (23), posits that TAFps function as coactivators or receptors which specifically and positively interact with the activation domains of DNA-bound transactivator proteins. Such interactions are thought to stabilize (recruit) TFIID on the cis-linked promoter, thereby stimulating PIC formation and thus transcription. This model is supported by an extensive body of experimental data derived from in vitro and in vivo studies performed with metazoans. Whether such a model is applicable to S. cerevisiae is unclear.
In an attempt to address the question of whether TAF25p might interact directly with transactivator proteins, we performed two-hybrid screens using either LexA-TAF25p or Gal4p-TAF25p as DBD baits. A number of putative TAF25p-interacting proteins were identified, and among these were TAF47p and Spt7p, two proteins which are integral subunits of TFIID and SAGA, respectively. We also identified genes encoding a number of known transcription factors—ASK10, UME6, YPR115W, TAO3, RIM5, RLM1, ECM22, RAP1, and YAP6—suggesting that these transcription factors could operate via an AD-TAFp interaction-coactivator mechanism. An intriguing additional class of hits observed in these screens was the group of genes (proteins) involved in RNA processing (PRP22, CLP1, and PRP40). RNA processing can be coupled to transcription through both TFIID and the C-terminal domain of RNA polymerase II (14, 31). It would be interesting if TAF25p within TFIID (or SAGA?) contributed specific contact surfaces for these RNA processing enzymes or proteins which would help localize these proteins to active genes. It will be important to follow up on these putative TAF25p-interacting proteins to determine if they are physiologically relevant and, if so, to determine those which make direct versus indirect contacts with TAF25p. It will also be informative to elucidate in which context, TFIID, SAGA, or both, such contacts are made.
Distinct TAF25p residues contribute differentially to gene-specific transcription.
We created a family of alanine-scanning mutants of TAF25 to generate reagents which we could use to assess the contributions of individual amino acid residues within TAF25p to the function of the molecule. Of the 19 amino acids targeted, we found that only the L183A mutant was lethal. The 19 amino acids subjected to mutation represent 18 of the amino acids which are 100% conserved (excluding A128) among the seven orthologs aligned (Fig. 1). Moreover, these 19 amino acids represent 54% of the invariant amino acid residues conserved in six of the seven orthologs compared. Some but not all of the targeted residues map to structured regions of the putative histone fold domain of TAF25p (Fig. 1). The residues chosen for mutagenesis, which span the length of mini-TAF25p, comprise a representative sampling of potential protein-protein interaction surfaces of the molecule.
Members of this family display distinct phenotypes of cold sensitivity, temperature sensitivity, and differential susceptibility to inhibitors. We reasoned that at least some of these phenotypes were caused by defects in transcription. We therefore performed a series of direct tests of transcription with the mutant cells. We analyzed both total poly(A)+ mRNA synthesis and gene-specific mRNA production in cells grown under permissive conditions and 60 min after a shift to the nonpermissive temperature. Others have carried out gene array studies which assessed the effect of yeast TAF protein inactivation upon mRNA gene transcription (32, 43). In these studies, single temperature-sensitive alleles of the genes in question were used, although it was stated that in some instances, another temperature-sensitive allele scored similarly in the assays (43). The actual nucleotide or amino acid lesions in the cognate genes or proteins were not reported.
We had two goals in generating our family of TAF25 Ala mutants. The first was to prepare a group of mutants which might allow us to identify domains of TAF25p which primarily contributed to TFIID function, to SAGA function, or to functions in both multisubunit complexes. The second was to use this panel of reagents to determine if any single temperature-sensitive mutant allele of TAF25 could describe the entirety of the transcriptional functions if TAF25p, as implied by Lee and colleagues (43). Our success in achieving these two goals was mixed, at least in part because a finite number of transcripts could be queried using RNA blotting methods rather than gene array technology. However, a number of conclusions can be drawn from our transcription experiments (Table 2). First, analysis of total poly(A)+ mRNA synthesis clearly indicated that conditional cell growth does not relate readily to overall RNA polymerase II transcription. Second, depending upon which strain one chooses as a baseline (wild type, TAF25ΔN, TAF25ΔC, or TAF25ΔN+ΔC), the actual percent decrease in mRNA synthesis upon a temperature shift can vary. Since all mutants were constructed with a TAF25ΔN+ΔC background, any values considered different (decreased by ∼2-fold or increased by ∼50%) from the value for control strain TAF25ΔN+ΔC are shown in bold type (note that mutant G101E was present in the full-length TAF25 ORF background).
Our TAF25 mutant collection exhibited a rather broad range of total mRNA synthesis phenotypes. Mutants I88A, G101A, D123A, and L188A all displayed decreased total mRNA synthesis levels (as did the G101E control) (60); mutant I88A displaying the largest decrease in total mRNA synthesis. The values for the other mutants were all near those for TAF25ΔN+ΔC. The fact that high-level total poly(A)+ mRNA values do not reveal the whole picture is underscored when the transcription of distinct mRNAs in the mutants is analyzed. There are clear and large allele-specific variations in gene-specific mRNA transcription among the members of the collection. For the 10 genes studied, only the transcription of YJL012C appears totally unaffected by mutation of TAF25, while for all other genes at least three different alleles affect transcription. In aggregate, these assays therefore clearly argue that no individual TAF25 mutant allele can describe the totality of the transcriptional repertoire of the protein.
The genes that we chose for our RNA blotting experiments were selected because they had been implicated (43) as being dependent upon TFIID (PMT4, WSC2, and TRX1) or SAGA (YHB1, PHO84, YJL012C, and YDL124W) or uniquely dependent upon TAF25p function (GIC2, YOR248W, and FAS2). We reasoned that if these TFIID and SAGA dependencies were correct, then this panel of genes should allow us to assign residues within TAF25p as contributing to shared and/or complex-specific functions of TFIID and SAGA. The three putative TAF25p-dependent genes, GIC2, YOR248W, and FAS2, were chosen to serve as controls, as transcription of these genes was reported to depend directly upon TAF25p function, displaying 4.2-, 4.1-, and 3.1-fold decreases (with excellent confidence) (43) when TAF25p was inactivated by a temperature shift. Analysis of these three genes also provided a crucial test of the assumption made in the gene array analyses that one temperature-sensitive TAF allele equals all transcriptional functions.
With the proviso that a limited sampling of genes has been analyzed, our data argue that it is problematic to use a single temperature-sensitive TAF mutant to define the totality of transcriptional functions of these molecules, at least for TAF25. Of the three putative TAF25p-dependent genes that we have analyzed in detail, only one behaved as would be predicted for a bona fide TAFp-dependent gene. YOR248W transcription in cells expressing all but the L97A and G196A TAF25 alleles was reduced at the nonpermissive temperature (range, 2.3- to 11.9-fold). In contrast, when we examined GIC2 and FAS2 expression in the Ala mutant family, even though two mutant alleles did show a decrease in transcription upon a temperature shift, transcription was essentially unaffected for all other alleles. For GIC2, 3 of 18 mutants displayed decreased transcription at the nonpermissive temperature, while for FAS2, only 5 of 18 mutants showed a decrease in transcription. The data obtained with YOR248W rule out the possibility that none of these alleles is compromised sufficiently to display a decrease in specific gene transcription upon a temperature shift.
The situation becomes even more clouded when the SAGA-dependent and TFIID-dependent genes are considered. Transcription of the TRX1 and YJL012C genes is essentially unaffected by TAF25 mutation, while transcription of the other five genes tested displays unique patterns of TAF25 Ala mutant dependencies. It is thus clear from a consideration of all of our data that TAF25p does contribute importantly to high-level, ongoing mRNA gene transcription. However, at this juncture, it is not yet feasible to ascribe complex-specific functions to discrete domains or residues of the protein. Such mechanistic details will require further intensive investigation.
Conclusions.
The studies described in this report argue that TAF25p contributes importantly to gene-specific transcription. Despite the fact that we have yet to totally analyze TAF25p, our experiments have dissected this critical, shared TFIID and SAGA subunit. We have also shown that at least for TAF25p, one can observe a very large decrease in transcription without a corresponding loss of TFIID or SAGA complex integrity. Finally, we have shown that distinct temperature-sensitive alleles of TAF25 display differential and unique patterns of gene-specific transcription in vivo. Our future studies will be aimed at characterizing the novel TAF25p protein-protein interactions described here as well as utilizing our collection of mutants for additional and more detailed functional tests of the molecular mechanisms by which TAF25p functions in RNA polymerase II-catalyzed mRNA gene transcription within the context of TFIID and SAGA.
ACKNOWLEDGMENTS
First, we thank all of our laboratory colleagues for freely sharing reagents, strains, and most of all their constructive criticism throughout the course of this work. Their input has made this a much better study. Second, we acknowledge Erin Baldwin's contributions to the overexpression complementation studies presented here. Third, we thank Steve Buratowski and Fred Winston for kindly supplying yeast expression plasmids for our experiments. Finally, we thank Laszlo Tora, Doris Kirschner, Gael Gangloff, and Irwin Davidson for freely sharing their ideas and information regarding TAF25 and histone folds with us.
This work was supported by NIH grant GM52461. J.K. received partial support from NIH training grant T32 CA09385. Sequencing of Candida albicans was accomplished with the support of the NIDR and the Burroughs Wellcome Fund.
REFERENCES
- 1.Albright S R, Tjian R. TAFs revisited: more data reveal new twists and confirm old ideas. Gene. 2000;242:1–13. doi: 10.1016/s0378-1119(99)00495-3. [DOI] [PubMed] [Google Scholar]
- 2.Apone L M, Virbasius C A, Holstege F C, Wang J, Young R A, Green M R. Broad, but not universal, transcriptional requirement for yTAF17, a histone H3-like TAF present in TFIID and SAGA. Mol Cell. 1998;2:653–661. doi: 10.1016/s1097-2765(00)80163-x. [DOI] [PubMed] [Google Scholar]
- 3.Bai Y, Perez G M, Beechem J M, Weil P A. Structure-function analysis of TAF130: identification and characterization of a high-affinity TATA-binding protein interaction domain in the N terminus of yeast TAF(II)130. Mol Cell Biol. 1997;17:3081–3093. doi: 10.1128/mcb.17.6.3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bell B, Tora L. Regulation of gene expression by multiple forms of TFIID and other novel TAFII-containing complexes. Exp Cell Res. 1999;246:11–19. doi: 10.1006/excr.1998.4294. [DOI] [PubMed] [Google Scholar]
- 5.Birck C, Poch O, Romier C, Ruff M, Mengus G, Lavigne A C, Davidson I, Moras D. Human TAF(II)28 and TAF(II)18 interact through a histone fold encoded by atypical evolutionary conserved motifs also found in the SPT3 family. Cell. 1998;94:239–249. doi: 10.1016/s0092-8674(00)81423-3. [DOI] [PubMed] [Google Scholar]
- 6.Boeke J D, Trueheart J, Natsoulis G, Fink G R. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- 7.Buratowski S, Hahn S, Guarente L, Sharp P A. Five intermediate complexes in transcription initiation by RNA polymerase II. Cell. 1989;56:549–561. doi: 10.1016/0092-8674(89)90578-3. [DOI] [PubMed] [Google Scholar]
- 8.Burke T W, Kadonaga J T. Drosophila TFIID binds to a conserved downstream basal promoter element that is present in many TATA-box-deficient promoters. Genes Dev. 1996;10:711–724. doi: 10.1101/gad.10.6.711. [DOI] [PubMed] [Google Scholar]
- 9.Burley S K, Roeder R G. Biochemistry and structural biology of transcription factor IID (TFIID) Annu Rev Biochem. 1996;65:769–799. doi: 10.1146/annurev.bi.65.070196.004005. [DOI] [PubMed] [Google Scholar]
- 10.Chen J L, Attardi L D, Verrijzer C P, Yokomori K, Tjian R. Assembly of recombinant TFIID reveals differential coactivator requirements for distinct transcriptional activators. Cell. 1994;79:93–105. doi: 10.1016/0092-8674(94)90403-0. [DOI] [PubMed] [Google Scholar]
- 11.Chiang C M, Roeder R G. Cloning of an intrinsic human TFIID subunit that interacts with multiple transcriptional activators. Science. 1995;267:531–536. doi: 10.1126/science.7824954. [DOI] [PubMed] [Google Scholar]
- 12.Choder M. A general topoisomerase I-dependent transcriptional repression in the stationary phase in yeast. Genes Dev. 1991;5:2315–2326. doi: 10.1101/gad.5.12a.2315. [DOI] [PubMed] [Google Scholar]
- 13.Cosma M P, Tanaka T, Nasmyth K. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally-regulated promoter. Cell. 1999;97:299–311. doi: 10.1016/s0092-8674(00)80740-0. [DOI] [PubMed] [Google Scholar]
- 14.Dantonel J C, Murthy K G, Manley J L, Tora L. Transcription factor TFIID recruits factor CPSF for formation of the 3′ end of mRNA. Nature. 1997;389:399–402. doi: 10.1038/38763. [DOI] [PubMed] [Google Scholar]
- 15.Dikstein R, Ruppert S, Tjian R. TAF250 is a bipartite protein kinase that phosphorylates the basal transcription factor RAP74. Cell. 1996;84:781–790. doi: 10.1016/s0092-8674(00)81055-7. [DOI] [PubMed] [Google Scholar]
- 16.Dunphy E L, Johnson T, Auerbach S S, Wang E H. Requirement for TAF(II)250 acetyltransferase activity in cell cycle progression. Mol Cell Biol. 2000;20:1134–1139. doi: 10.1128/mcb.20.4.1134-1139.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 18.Gangloff Y G, Romier C, Thualt S, Werten S, Davidson I. The histone fold is a key structural motif of transcription factor TFIID. Trends Biochem Sci. 2001;26:250–257. doi: 10.1016/s0968-0004(00)01741-2. [DOI] [PubMed] [Google Scholar]
- 19.Gangloff Y G, Sanders S L, Romier C, Kirschner D, Weil P A, Tora L, Davidson I. Histone folds mediate selective heterodimerization of yeast TAF(II)25 with TFIID components yTAF(II)47 and yTAF(II)65 and with SAGA component ySPT7. Mol Cell Biol. 2001;21:1841–1853. doi: 10.1128/MCB.21.5.1841-1853.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gangloff Y G, Werten S, Romier C, Carre L, Poch O, Moras D, Davidson I. The human TFIID components TAF(II)135 and TAF(II)20 and the yeast SAGA components Ada1 and TAF(II)68 heterodimerize to form histone-like pairs. Mol Cell Biol. 2000;20:340–351. doi: 10.1128/mcb.20.1.340-351.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Georgieva S, Kirschner D B, Jagla T, Nabirochkina E, Hanke S, Schenkel H, de Lorenzo C, Sinha P, Jagla K, Mechler B, Tora L. Two novel Drosophila TAF(II)s have homology with human TAF(II)30 and are differentially regulated during development. Mol Cell Biol. 2000;20:1639–1648. doi: 10.1128/mcb.20.5.1639-1648.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gill G, Pascal E, Tseng Z H, Tjian R. A glutamine-rich hydrophobic patch in transcription factor Sp1 contacts the dTAF110 component of the Drosophila TFIID complex and mediates transcriptional activation. Proc Natl Acad Sci USA. 1994;91:192–196. doi: 10.1073/pnas.91.1.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goodrich J A, Hoey T, Thut C J, Admon A, Tjian R. Drosophila TAF40 interacts with both a VP16 activation domain and the basal transcription factor TFIIB. Cell. 1993;75:519–530. doi: 10.1016/0092-8674(93)90386-5. [DOI] [PubMed] [Google Scholar]
- 24.Grant P A, Duggan L, Cote J, Roberts S M, Brownell J E, Candau R, Ohba R, Owen-Hughes T, Allis C D, Winston F, Berger S L, Workman J L. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- 25.Grant P A, Schieltz D, Pray-Grant M G, Steger D J, Reese J C, Yates J R, Workman J L. A subset of TAF(II)s are integral components of the SAGA complex required for nucleosome acetylation and transcriptional stimulation. Cell. 1998;94:45–53. doi: 10.1016/s0092-8674(00)81220-9. [DOI] [PubMed] [Google Scholar]
- 26.Gregory P D, Schmid A, Zavari M, Munsterkotter M, Horz W. Chromatin remodelling at the PHO8 promoter requires SWI-SNF and SAGA at a step subsequent to activator binding. EMBO J. 1999;18:6407–6414. doi: 10.1093/emboj/18.22.6407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guthrie C, Fink G R, editors. Guide to yeast genetics and molecular biology. New York, N.Y: Academic Press, Inc.; 1991. [Google Scholar]
- 28.Hampsey M. A review of phenotypes in Saccharomyces cerevisiae. Yeast. 1997;13:1099–1133. doi: 10.1002/(SICI)1097-0061(19970930)13:12<1099::AID-YEA177>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 29.Hampsey M. Molecular genetics of the RNA polymerase II general transcriptional machinery. Microbiol Mol Biol Rev. 1998;62:465–503. doi: 10.1128/mmbr.62.2.465-503.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Henry N L, Campbell A M, Feaver W J, Poon D, Weil P A, Kornberg R D. TFIIF-TAF-RNA polymerase II connection. Genes Dev. 1994;8:2868–2878. doi: 10.1101/gad.8.23.2868. [DOI] [PubMed] [Google Scholar]
- 31.Hirose Y, Manley J L. RNA polymerase II and the integration of nuclear events. Genes Dev. 2000;14:1415–1429. [PubMed] [Google Scholar]
- 32.Holstege F C, Jennings E G, Wyrick J J, Lee T I, Hengartner C J, Green M R, Golub T R, Lander E S, Young R A. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 33.Ikeda K, Steger D J, Eberharter A, Workman J L. Activation domain-specific and general transcription stimulation by native histone acetyltransferase complexes. Mol Cell Biol. 1999;19:855–863. doi: 10.1128/mcb.19.1.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.James P, Halladay J, Craig E A. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim Y J, Bjorklund S, Li Y, Sayre M H, Kornberg R D. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell. 1994;77:599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- 36.Kingston R E, Narlikar G J. ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev. 1999;13:2339–2352. doi: 10.1101/gad.13.18.2339. [DOI] [PubMed] [Google Scholar]
- 37.Klebanow E R, Poon D, Zhou S, Weil P A. Isolation and characterization of TAF25, an essential yeast gene that encodes an RNA polymerase II-specific TATA-binding protein-associated factor. J Biol Chem. 1996;271:13706–13715. doi: 10.1074/jbc.271.23.13706. [DOI] [PubMed] [Google Scholar]
- 38.Klemm R D, Goodrich J A, Zhou S, Tjian R. Molecular cloning and expression of the 32-kDa subunit of human TFIID reveals interactions with VP16 and TFIIB that mediate transcriptional activation. Proc Natl Acad Sci USA. 1995;92:5788–5792. doi: 10.1073/pnas.92.13.5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koleske A J, Young R A. An RNA polymerase II holoenzyme responsive to activators. Nature. 1994;368:466–469. doi: 10.1038/368466a0. [DOI] [PubMed] [Google Scholar]
- 40.Kotani T, Banno K, Ikura M, Hinnebusch A G, Nakatani Y, Kawaichi M, Kokubo T. A role of transcriptional activators as antirepressors for the autoinhibitory activity of TATA box binding of transcription factor IID. Proc Natl Acad Sci USA. 2000;97:7178–7183. doi: 10.1073/pnas.120074297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krebs J E, Kuo M H, Allis C D, Peterson C L. Cell cycle-regulated histone acetylation required for expression of the yeast HO gene. Genes Dev. 1999;13:1412–1421. doi: 10.1101/gad.13.11.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kutach A K, Kadonaga J T. The downstream promoter element DPE appears to be as widely used as the TATA box in Drosophila core promoters. Mol Cell Biol. 2000;20:4754–4764. doi: 10.1128/mcb.20.13.4754-4764.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee T I, Causton H C, Holstege F C, Shen W C, Hannett N, Jennings E G, Winston F, Green M R, Young R A. Redundant roles for the TFIID and SAGA complexes in global transcription. Nature. 2000;405:701–704. doi: 10.1038/35015104. [DOI] [PubMed] [Google Scholar]
- 44.Lemon B, Tjian R. Orchestrated response: a symphony of transcription factors for gene control. Genes Dev. 2000;14:2551–2569. doi: 10.1101/gad.831000. [DOI] [PubMed] [Google Scholar]
- 45.Macpherson N, Measday V, Moore L, Andrews B. A yeast taf17 mutant requires the Swi6 transcriptional activator for viability and shows defects in cell cycle-regulated transcription. Genetics. 2000;154:1561–1576. doi: 10.1093/genetics/154.4.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Massari M E, Grant P A, Pray-Grant M G, Berger S L, Workman J L, Murre C. A conserved motif present in a class of helix-loop-helix proteins activates transcription by direct recruitment of the SAGA complex. Mol Cell. 1999;4:63–73. doi: 10.1016/s1097-2765(00)80188-4. [DOI] [PubMed] [Google Scholar]
- 47.Matangkasombut O, Buratowski R M, Swilling N W, Buratowski S. Bromodomain factor 1 corresponds to a missing piece of yeast TFIID. Genes Dev. 2000;14:951–962. [PMC free article] [PubMed] [Google Scholar]
- 48.Michel B, Kamarnitsky P, Buratowski S. Histone-like TAFs are essential for transcription in vivo. Mol Cell. 1998;2:663–673. doi: 10.1016/s1097-2765(00)80164-1. [DOI] [PubMed] [Google Scholar]
- 49.Mizzen C A, Yang X J, Kokubo T, Brownell J E, Bannister A J, Owen-Hughes T, Workman J, Wang L, Berger S L, Kouzarides T, Nakatani Y, Allis C D. The TAF(II)250 subunit of TFIID has histone acetyltransferase activity. Cell. 1996;87:1261–1270. doi: 10.1016/s0092-8674(00)81821-8. [DOI] [PubMed] [Google Scholar]
- 50.Moqtaderi Z, Keaveney M, Struhl K. The histone H3-like TAF is broadly required for transcription in yeast. Mol Cell. 1998;2:675–682. doi: 10.1016/s1097-2765(00)80165-3. [DOI] [PubMed] [Google Scholar]
- 51.Moqtaderi Z, Yale J D, Struhl K, Buratowski S. Yeast homologues of higher eukaryotic TFIID subunits. Proc Natl Acad Sci USA. 1996;93:14654–14658. doi: 10.1073/pnas.93.25.14654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Natarajan K, Jackson B M, Rhee E, Hinnebusch A G. yTAF61 has a general role in RNA polymerase II transcription and is required by Gcn4p to recruit the SAGA coactivator complex. Mol Cell. 1998;2:683–692. doi: 10.1016/s1097-2765(00)80166-5. [DOI] [PubMed] [Google Scholar]
- 53.O'Brien T, Tjian R. Functional analysis of the human TAF250 N-terminal kinase domain. Mol Cell. 1998;1:905–911. doi: 10.1016/s1097-2765(00)80089-1. [DOI] [PubMed] [Google Scholar]
- 54.O'Brien T, Tjian R. Different functional domains of TAF250 modulate expression of distinct subsets of mammalian genes. Proc Natl Acad Sci USA. 2000;97:2456–2461. doi: 10.1073/pnas.97.6.2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Orphanides G, Lagrange T, Reinberg D. The general transcription factors of RNA polymerase II. Genes Dev. 1996;10:2657–2683. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- 56.Pham A D, Sauer F. Ubiquitin-activating/conjugating activity of TAF250, a mediator of activation of gene expression in Drosophila. Science. 2000;289:2357–2360. doi: 10.1126/science.289.5488.2357. [DOI] [PubMed] [Google Scholar]
- 57.Poon D, Bai Y, Campbell A M, Bjorklund S, Kim Y J, Zhou S, Kornberg R D, Weil P A. Identification and characterization of a TFIID-like multiprotein complex from Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1995;92:8224–8228. doi: 10.1073/pnas.92.18.8224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reese J C, Apone L, Walker S S, Griffin L A, Green M R. Yeast TAFs in a multisubunit complex required for activated transcription. Nature. 1994;371:523–527. doi: 10.1038/371523a0. [DOI] [PubMed] [Google Scholar]
- 59.Reese J C, Zhang Z, Kurpad H. Identification of a yeast transcription factor IID subunit, TSG2/TAF48. J Biol Chem. 2000;275:17391–17395. doi: 10.1074/jbc.M001635200. [DOI] [PubMed] [Google Scholar]
- 60.Sanders S L, Klebanow E R, Weil P A. TAF25p, a non-histone-like subunit of TFIID and SAGA complexes, is essential for total mRNA gene transcription in vivo. J Biol Chem. 1999;274:18847–18850. doi: 10.1074/jbc.274.27.18847. [DOI] [PubMed] [Google Scholar]
- 61.Sanders S L, Weil P A. Identification of two novel TAF subunits of the yeast Saccharomyces cerevisiae TFIID complex. J Biol Chem. 2000;275:13895–13900. doi: 10.1074/jbc.275.18.13895. [DOI] [PubMed] [Google Scholar]
- 62.Sauer F, Hansen S K, Tjian R. DNA template and activator-coactivator requirements for transcriptional synergism by Drosophila bicoid. Science. 1995;270:1825–1828. doi: 10.1126/science.270.5243.1825. [DOI] [PubMed] [Google Scholar]
- 63.Schock F, Sauer F, Jackle H, Purnell B A. Drosophila head segmentation factor buttonhead interacts with the same TATA box-binding protein-associated factors and in vivo DNA targets as human Sp1 but executes a different biological program. Proc Natl Acad Sci USA. 1999;96:5061–5065. doi: 10.1073/pnas.96.9.5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shen W C, Green M R. Yeast TAF(II)145 functions as a core promoter selectivity factor, not a general coactivator. Cell. 1997;90:615–624. doi: 10.1016/s0092-8674(00)80523-1. [DOI] [PubMed] [Google Scholar]
- 65.Smale S T, Baltimore D. The “initiator”as a transcription control element. Cell. 1989;57:103–113. doi: 10.1016/0092-8674(89)90176-1. [DOI] [PubMed] [Google Scholar]
- 66.Sterner D E, Berger S L. Acetylation of histones and transcription-related factors. Microbiol Mol Biol Rev. 2000;64:435–459. doi: 10.1128/mmbr.64.2.435-459.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sterner D E, Grant P A, Roberts S M, Duggan L J, Belotserkovskaya R, Pacella L A, Winston F, Workman J L, Berger S L. Functional organization of the yeast SAGA complex: distinct components involved in structural integrity, nucleosome acetylation, and TATA-binding protein interaction. Mol Cell Biol. 1999;19:86–98. doi: 10.1128/mcb.19.1.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sullivan S A, Aravind L, Makalowska I, Baxevanis A D, Landsman D. The histone database: a comprehensive WWW resource for histones and histone fold-containing proteins. Nucleic Acids Res. 2000;28:320–322. doi: 10.1093/nar/28.1.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Syntichaki P, Topalidou I, Thireos G. The Gcn5 bromodomain coordinates nucleosome remodelling. Nature. 2000;404:414–417. doi: 10.1038/35006136. [DOI] [PubMed] [Google Scholar]
- 70.Thut C J, Chen J L, Klemm R, Tjian R. p53 transcriptional activation mediated by coactivators TAF40 and TAF60. Science. 1995;267:100–104. doi: 10.1126/science.7809597. [DOI] [PubMed] [Google Scholar]
- 71.Uetz P, Biot L, Cagney G, Mansfield T A, Judson R S, Knight J R, Lockshon D, Narayan V, Strinivasan M, Pachart P, Qureshi-Emill A, Li Y, Godwin B, Conover D, Kalbflelsch T, Vijayadamodar G, Yang M, Johnston M, Fields S, Rothberg J M. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 72.Utley R T, Ikeda K, Grant P A, Cote J, Steger D J, Eberharter A, John S, Workman J L. Transcriptional activators direct histone acetyltransferase complexes to nucleosomes. Nature. 1998;394:498–502. doi: 10.1038/28886. [DOI] [PubMed] [Google Scholar]
- 73.Weissman J D, Howcroft, D., T K, Singer S. TAF(II)250-independent transcription can be conferred on a TAF(II)250-dependent basal promoter by upstream activators. J Biol Chem. 2000;275:10160–10167. doi: 10.1074/jbc.275.14.10160. [DOI] [PubMed] [Google Scholar]
- 74.Wolstein O, Silkov A, Revach M, Dikstein R. Specific interaction of TAF105 with OCA-B is involved in activation of octamer-dependent transcription. J Biol Chem. 2000;275:16459–16465. doi: 10.1074/jbc.275.22.16459. [DOI] [PubMed] [Google Scholar]
- 75.Workman J L, Kingston R E. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu Rev Biochem. 1998;67:545–579. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]
- 76.Xie X, Kokubo T, Cohen S L, Mirza U A, Hoffmann A, Chait B T, Roeder R G, Nakatani Y, Burley S K. Structural similarity between TAFs and the heterotetrameric core of the histone octamer. Nature. 1996;380:316–322. doi: 10.1038/380316a0. [DOI] [PubMed] [Google Scholar]
- 77.Zhou J, Zwicker J, Szymanski P, Levine M, Tjian R. TAFII mutations disrupt Dorsal activation in the Drosophila embryo. Proc Natl Acad Sci USA. 1998;95:13483–13488. doi: 10.1073/pnas.95.23.13483. [DOI] [PMC free article] [PubMed] [Google Scholar]