Abstract
During a systematic analysis of conserved gene context in prokaryotic genomes, a previously undetected, complex, partially conserved neighborhood consisting of more than 20 genes was discovered in most Archaea (with the exception of Thermoplasma acidophilum and Halobacterium NRC-1) and some bacteria, including the hyperthermophiles Thermotoga maritima and Aquifex aeolicus. The gene composition and gene order in this neighborhood vary greatly between species, but all versions have a stable, conserved core that consists of five genes. One of the core genes encodes a predicted DNA helicase, often fused to a predicted HD-superfamily hydrolase, and another encodes a RecB family exonuclease; three core genes remain uncharacterized, but one of these might encode a nuclease of a new family. Two more genes that belong to this neighborhood and are present in most of the genomes in which the neighborhood was detected encode, respectively, a predicted HD-superfamily hydrolase (possibly a nuclease) of a distinct family and a predicted, novel DNA polymerase. Another characteristic feature of this neighborhood is the expansion of a superfamily of paralogous, uncharacterized proteins, which are encoded by at least 20–30% of the genes in the neighborhood. The functional features of the proteins encoded in this neighborhood suggest that they comprise a previously undetected DNA repair system, which, to our knowledge, is the first repair system largely specific for thermophiles to be identified. This hypothetical repair system might be functionally analogous to the bacterial–eukaryotic system of translesion, mutagenic repair whose central components are DNA polymerases of the UmuC-DinB-Rad30-Rev1 superfamily, which typically are missing in thermophiles.
INTRODUCTION
Most of the presently known archaeal species and many bacteria are thermophiles or hyperthermophiles whose optimal growth temperatures reach 115°C (1–3). The molecular basis of maintenance of genome stability in (hyper)thermophiles is, arguably, one of the most intriguing problems of modern biology (1,4). Thermophiles typically are resistant not only to high temperatures, but also to other damaging factors, such as ionizing and ultraviolet radiation and chemical agents; furthermore, spontaneous mutagenesis is accelerated at elevated temperatures (5–7). However, the genomic mutation rate in the thermophilic archaeon Sulfolobus acidocaldarius was shown to be about the same as in mesophiles (8). Thus, thermophiles must have highly efficient and probably specialized DNA repair systems. Experimental studies of repair in thermophiles so far have been scant, although several repair enzymes have been identified, including thermostable O-6-methylguanine-DNA methyltransferase (9), uracil-DNA-glycosylase (10–12), RecA/Rad51 family protein with unique DNase activity (13) and others reviewed in Grogan (4). Attempts to delineate repair systems of bacterial and particularly archaeal thermophiles from genome sequences by identification of homologs of well-characterized components of repair pathways from Escherichia coli and Saccharomyces cerevisiae have been only partially successful (4,14,15). This led to the hypothesis that a distinct DNA repair system might exist in thermophiles (4).
Currently, 12 complete genome sequences of thermophilic Archaea and two genome sequences of bacterial hyperthermophiles are available. Like prokaryotic genomes in general, the genomes in thermophiles show little conservation of gene order over long evolutionary distances (16). Nevertheless, comparative analysis of genomic context, i.e. organization of genes into partially conserved clusters that are likely to represent operons, has proved a powerful method for prediction of the functions of uncharacterized bacterial and archaeal genes (16–20). The central premise of genomic context analysis is that genes that belong to the same operon are almost certainly functionally connected. By inference, if a predicted operon contains one or more genes with a known function, functions can be predicted for other, uncharacterized members of the same operon, especially when context analysis is complemented by prediction of biochemical activity of the proteins in question by means of comparative sequence and structure analysis. Straightforward identification of conserved gene strings that are likely to represent operons is the principal approach that so far has been employed in genome context analysis (16,17,19). However, because of the extensive rearrangements of local gene order, even within operons, that is characteristic of prokaryotic evolution, this method is insufficient to extract all context information that potentially exists in bacterial and archaeal genomes. Several attempts have been made to identify partially conserved gene neighborhoods that may show little direct conservation of gene order, but consist of identical or substantially overlapping gene sets in different genomes. Gene neighborhoods typically are not present, in their entirety, in any single genome, but are held together by overlaps between partially conserved gene sets.
It has been noticed previously that orthologs of a relatively small fraction of bacterial and eukaryotic repair proteins are detectable in Archaea, although many proteins containing helicase, nuclease and DNA-binding domains were identified and, in principle, could be candidates for roles in repair (14,15). Thus, sequence analysis alone seems to be insufficient for confidently predicting archaeal repair systems (21). Recently, we utilized a combination of the analysis of conserved gene neighborhoods/gene fusions with sensitive sequence profile searches and structural comparisons to predict a novel prokaryotic DNA repair system that seems to be the counterpart of the eukaryotic Ku-dependent double strand break system (22). Here, by using a combination of gene neighborhood analysis and detailed sequence and structure analysis of protein domains, we predict another previously undetected DNA repair system in archaeal and bacterial genomes. To our knowledge, this is the first DNA repair system that appears to be largely confined to thermophiles in its phyletic distribution and could potentially fill a significant void in terms of archaeal DNA repair systems.
MATERIALS AND METHODS
Genome sequences, databases and sequence analysis
The genome sequences and the encoded protein sequences of the Archaea Archaeoglobus fulgidus (Aful) (23), Methanobacterium thermoautotrophicum (Mthe) (24), Methanococcus jannaschii (Mjan) (25), Pyrococcus horikoshii (Phor) (26), Pyrococcus abyssi (Paby) (R. Heilig, Genoscope; GenBank NC_000868), Thermoplasma volcanium (Euryarchaeota) (Tvol) and Aeropyrum pernix (Aper) (27) and Sulfolobus solfataricus (Ssol) (28) (Crenarchaeota), as well as the bacteria Thermotoga maritima (Tmar) (29), Aquifex aeolicus (Aaeo) (30), Bacillus halodurans (Bhal) (31), Mycobacterium tuberculosis (Mtub) (32), Streptococcus pyogenes (Spyo) (33) (bacteria) were retrieved from the Genomes division of the Entrez system (34). The preliminary genome sequence of the Euryarchaeon Pyrococcus furiosus was downloaded from http://comb5-156.umbi.umd.edu/genemate/pfu-info.html.
The non-redundant database of protein sequences at the National Center for Biotechnology Information (NIH, Bethesda) was iteratively searched using the PSI-BLAST program (35,36). The cut-off of E < 0.01 was normally employed for inclusion of sequences in the position-specific weight matrices. For detecting subtle sequence conservation, the PSI-BLAST search results were visually examined and sequences with greater E-values, but containing signature motifs of a given protein family, were included into profiles on a case by case basis (35–37). Nucleotide sequences translated in six reading frames were searched for protein sequence similarity using the TBLASTN program (35). Unfinished microbial genome sequences (http://www.ncbi.nlm.nih.gov/Microb_blast/unfinishedgenome.html) were searched using TBLASTN. Conserved domains in protein sequences were identified by searching the NCBI’s CD collection of domain-specific, position-dependent weight matrices using the Reverse PSI-BLAST program (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) and by searching the SMART collection of domain-specific hidden Markov models (38). Multiple alignments of protein sequences were constructed using the T-coffee program (39) and corrected on the basis of PSI-BLAST results. Protein secondary structure was predicted using the PHD program, with a multiple alignment submitted as the query (40). Protein sequence–structure threading was performed by using the hybrid fold-recognition method (41) and the 3D-PSSM method (42).
Phylogenetic analysis
Distance trees were constructed from multiple protein sequence alignments after excluding positions containing >70% gaps, by using the least-square method as implemented in the FITCH program of the PHYLIP package (43,44). Maximum likelihood trees were constructed using the ProtML program of the MOLPHY package, with the JTT-F model of amino acid substitutions, by optimizing the least-square trees with local rearrangements (45,46). Bootstrap analysis was performed for each maximum likelihood tree as implemented in MOLPHY using the resampling of estimated log-likelihoods (RELL) method (45–47).
Reconstruction of conserved gene neighborhoods
The procedure for reconstructing conserved gene neighborhoods will be described in detail elsewhere (I.B.Rogozin, K.S.Makarova and E.V.Koonin, unpublished data). Briefly, the following steps were implemented. The collection of clusters of orthologous groups (COGs) of proteins from complete genomes (48,49) was used as the source of information on orthologous relationships for detecting conserved gene pairs. A pair of genes from two COGs was considered ‘conserved’ if the corresponding genes were separated by none, one or two genes in at least three of the compared genomes. At the next step, overlapping gene pairs were joined in triplets, each of which was required to exist in at least one genome. Overlapping triplets were used to construct gene arrays by walk search in an oriented graph; a gene array may or may not be found in its entirety in any available genome (I.B.Rogozin, K.S.Makarova and E.V.Koonin, unpublished data). Finally, gene arrays that shared at least three COGs were clustered into neighborhoods by using a single-linkage clustering algorithm. All these steps were implemented in the program GENE_NEIGHBOR, which ran automatically without human control at intermediate stages (I.B.Rogozin, K.S.Makarova and E.V.Koonin, unpublished data; available upon request). The resulting gene neighborhoods were amended manually by adding genes that were located next to the genes of an automatically delineated neighborhood in at least one genome, but did not fit the criteria outlined above.
RESULTS AND DISCUSSION
Identification of a potential DNA repair system that is largely specific to thermophiles
An exhaustive search for conserved gene neighborhoods in the available complete bacterial and archaeal genomes (I.B.Rogozin, K.S.Makarova and E.V.Koonin, unpublished data) revealed only three large (more than 10 genes) neighborhoods that were represented predominantly in Archaea, along with a few bacterial species. Of these, the partially conserved superoperons that encode ribosomal proteins and subunits of the proton-transporting ATPase complex have been identified and analyzed in detail previously (16 and references therein). The third neighborhood, which is chiefly represented in Archaea and hyperthermophilic bacteria, is even larger, with genes that belong to over 20 COGs, but shows greater diversity than the ribosomal and ATPase neighborhoods, in terms of gene order. This neighborhood includes mostly genes without known or predicted functions. At the time of the genome comparison that resulted in the identification of this neighborhood, clear functional assignments were available for only two of its constituent genes. These genes encode a predicted DNA helicase and a predicted RecB family nuclease, which, by extension, could suggest a role in DNA repair for the entire neighborhood. Prompted by these observations, we undertook a detailed comparison of the potential operons comprising this neighborhood in different genomes and an in-depth analysis of the conserved domains that could be identified in the proteins encoded by uncharacterized genes.
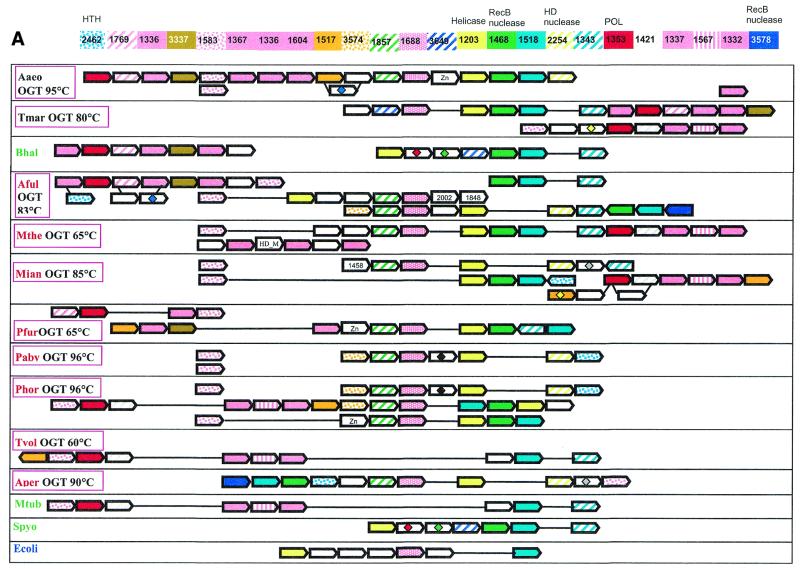
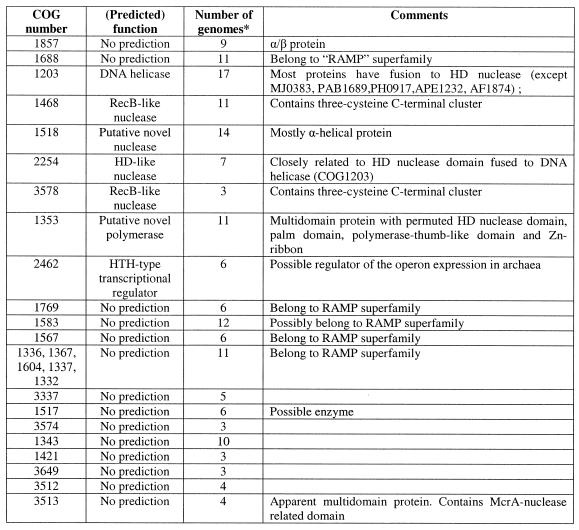
Diverse versions of this neighborhood were detected in all completely sequenced archaeal species, with the exception of Thermoplasma acidophilum (50) and Halobacterium sp. NRC-1 (51), both available genomes of bacterial hyperthermophiles, T.maritima and A.aeolicus, and some bacterial mesophiles, namely B.halodurans, M.tuberculosis and S.pyogenes. The corresponding genome region from the bacterial hyperthermophile A.aeolicus was chosen as a template to produce a template-anchored multiple alignment (16) of the analyzed neighborhood because it had the longest potential superoperon comprised of 18 genes (Fig. 1A). Although not a single gene is present in all genomes that have the analyzed neighborhood, a distinct group of five core genes that are conserved in the great majority of these genomes, often in the same order, was identified (Fig. 1A and Table 1). This conserved core of the putative new repair system shows the following predominant gene order: COG1857-COG1688-COG1203-COG1468-COG1518 (Fig. 1A). The sixth gene, which is not a part of this array, but is present within the neighborhood in most genomes, is COG1353, which typically is found in close proximity with one or more genes of COGs 1336, 1367, 1604, 1337 and 1332 (Fig. 1A).
Figure 1.
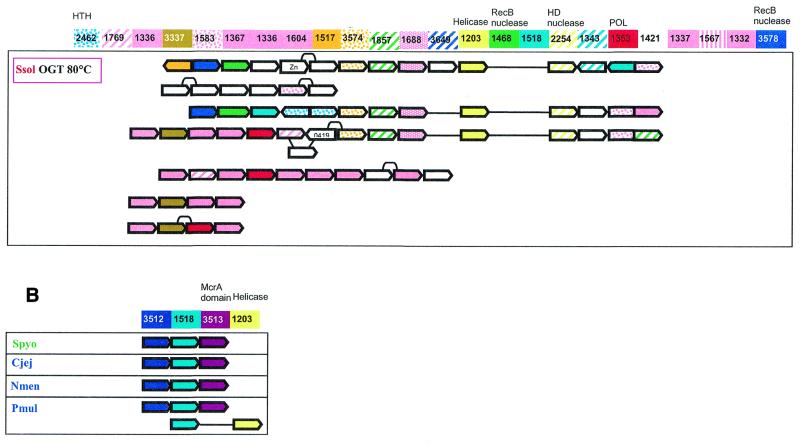
(Opposite) Organization of genes and potential operons in the genomic regions coding for protein components of the predicted novel DNA repair system. (A) The core (helicase-nuclease) and polymerase modules. Genes are shown not to scale; the direction of transcription is indicated by arrows. The multiple gene-by-gene alignment was produced by manually combining template-anchored genome alignments. For each column of the alignment, the corresponding COG number and predicted function is indicated. Generally, orthologous genes are shown by the same color and pattern. The exceptions are the RAMP proteins of COGs 1336, 1367, 1604, 1337 and 1332, which are all shown in pink. The remaining, more distant RAMPs (see text) are also shown in pink, with different patterns. Genes in each genome that are unique for this neighborhood are shown by white arrows; some of these unique genes belong to the following COGs: 2002, regulators of stationary/sporulation gene expression; AbrB, 1848, predicted nucleic acid-binding protein, contains PIN domain; 1458, uncharacterized protein, present only in Archaea and A.aeolicus; 0419, ATPase involved in DNA repair. Pairs of orthologous proteins that do not belong to COGs are marked by the same-colored diamonds. HTH, helix–turn–helix type transcriptional regulator; HD nuclease, HD conserved motif containing predicted nuclease conserved region; POL, novel predicted polymerase; HD_M, HD-hydrolase-domain-containing, apparently multidomain protein; Zn, Zn ribbon containing protein. The species abbreviations are as indicated in Materials and Methods. Species are color-coded as follows: Archaea, red; proteobacteria, blue; Gram-positive bacteria, green; other bacteria, black. The thermophilic species names are boxed and the optimal growth temperature (OGT) is indicated for each of them. Gene strings or individual genes shown on the figure are the following (from left to right for each genome): Aaeo, aq_387-aq_369, aq_173, aq_755; Tmar, TM1802-TM1791.1, TM1814-TM1807; Bhal, BH0327-BH0333, BH0336-BH0342; Aful, AF1869-AF1859, AF2436-AF2434, AF0072-AF0065, AF1870-AF1879; Mthe, MTH1091-MTH1078/1077, MTH328-MTH323; Mjan, MJ1234, MJ0380-MJ0386, MJ0375-MJ0379, MJ1666-MJ1665; Pfur, PF_1076764-PF_1077729-PF_1080337-PF_1081470, PF_1075331-PF_1074447-PF_1073960-PF_1072954-PF_1071624-PF_1070572-PF_1069932-PF_1067761-PF_1067252-PF_1066282-PF_1066279; Paby, PAB1064, PAB1613, PAB1685-PAB1691; Phor, PH0350, PH0921-PH0915, PH0161-PH0177, PH1252-PH1245; Tvol, TVN0114-TVN0105; Aper, APE1241-APE1228; Mtub, Rv2824c-Rv2816c; Spyo, Spy1567-Spy1561, Ecoli, YgcB-YgbT; Ssol, SSO1389-SSO1406, SSO1376-SSO1383, SSO1451-SSO1437, SSO1987-SSO2005, SSO1433-SSO1422, SSO1513-SSO1510, SSO1726-SSO1730. (B) A putative distinct bacterial operon centered on COG1518 and related to the predicted novel DNA repair system. The designations are as in (A). Gene strings: Spyo, Spy1048-Spy1046; Cjej, Cj1521c-Cj1523c; Nmen, NMA0629-NMA0631; Pmul, PM1125-PM1127, PM0311-PM0312.
Table 1. The genes comprising the predicted thermophile-specific DNA repair system.
*The number of completely sequenced genomes, in which the given COG is represented.
The core gene array includes those components of the putative repair system for which straightforward functional prediction was possible. All proteins of COG1203 contain a typical superfamily II helicase domain. In most of these proteins (with the exception of MJ0383, PAB1689, PH0917, APE1232 and AF1874), the helicase domain is fused to a predicted HD-nuclease domain (52). Fusion of helicase and nuclease domains is characteristic of many repair systems. For example, the bacterial RecB protein contains a fusion of a Superfamily I helicase and the eponymous nuclease, whereas the eukaryotic RAD1 protein is a Superfamily II helicase fused to a predicted ERCC4 family nuclease and the Werner syndrome protein displays a fusion of the 3′→5′ exonuclease and a SF-II helicase module (14,15,53). However, the specific combination of helicase and nuclease domains seen in the COG1203 proteins has not been described so far. Those species that have a stand-alone helicase in the core of the putative repair system (e.g. P.abyssi and A.pernix) possess either an extra copy of the fusion gene or a stand-alone predicted HD-nuclease (COG2254) that is typically adjacent to the core gene array (Fig. 1A).
The proteins of COG1468 belong to the RecB nuclease family and contain all the conserved catalytic residues characteristic of these nucleases. A distinctive feature of these proteins is the presence of a C-terminal module that contains three conserved cysteines and might mediate metal-dependent DNA binding. RecB protein, which is sporadically distributed in bacteria, contains helicase and nuclease domains and is a subunit of the RecBCD recombinase complex, one of the major systems of recombinational repair in E.coli (54–56). In addition to the RecB protein, RecB family nucleases are often fused to SF-I helicases of other subfamilies (e.g. in Synechocystis protein sll1582 to DNAI/HCS1-like helicase and in MTH472 from M.thermoautotrophicum to PCRA/UvrD-like helicase) (53).
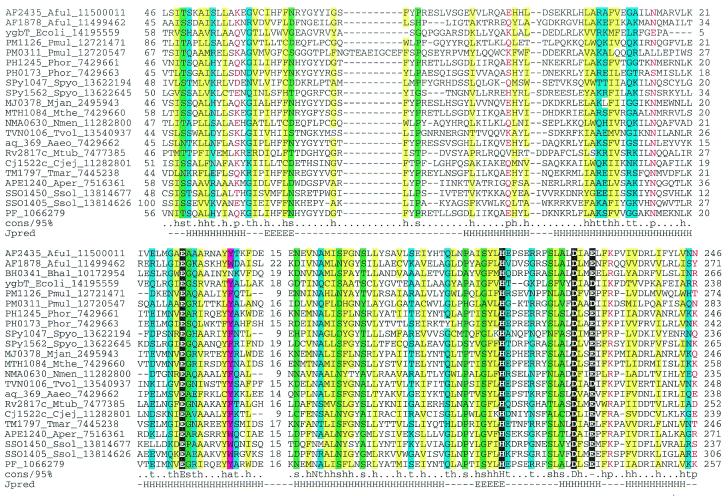
COG1518 consists of proteins that do not have detectable homologs with known functions. However, examination of the multiple alignment of these proteins revealed a pattern of conserved acidic residues (Fig. 2) that are often present in catalytic sites of various families of nucleases (53). Considering the strongly conserved association of the gene coding for this protein with the genes coding for two other nucleases (COG1203/2254 and COG1468) and a helicase (COG1203), we consider it likely that this protein is a previously undetected nuclease that functions within the putative novel DNA repair system. In contrast to most known nuclease families, which include α/β proteins, but similarly to the HD-superfamily (52), secondary structure prediction indicates that COG1518 proteins have an all-α structure that probably represents a novel nuclease fold. In addition to their position within the core of the putative repair system, genes for the COG1518 proteins were found in an alternative gene array that is conserved in several bacterial species (Fig. 1B). In particular, COG3513 consists of large, probably multidomain proteins that contain a diverged McrA/T4-Endo-VII nuclease domain (53).
Figure 2.
Multiple alignment of the predicted novel nuclease family (COG1518). The proteins are denoted by their systematic gene numbers, Gene Identification (GI) numbers from the GenBank database and abbreviated species names (see Materials and Methods for abbreviations). The positions of the first and the last residue of the aligned region in the corresponding protein are indicated for each sequence. The alignment coloring is based on the consensus shown underneath the alignment; b indicates a ‘big’ residue (E,K,R,I,L,M,F,Y,W), h indicates hydrophobic residues (A,C,F,I,L,M,V,W,Y), a indicates aromatic residues (F,Y,W), s indicates small residues (A,C,S,T,D,N,V,G,P), u indicates ‘tiny’ residues (G,A,S), p indicates polar residues (D,E,H,K,N,Q,R,S,T), c indicates charged residues (K,R,D,E,H), o indicates hydroxyl group containing residues (S,T), + indicates positively charged residues (R,K) and – indicates negatively charged residues (E,D). The secondary structure elements were predicted using the PHD program and a pre-constructed multiple alignment as the input and are shown above the alignment. H indicates α-helix and E indicates extended conformation (β-strand).
Two uncharacterized proteins that belong to the core of the putative repair system, COG1857 and COG1688, are predicted to possess α/β folds, as suggested by secondary structure prediction, but do not contain any conserved motifs with potential catalytic amino acids (data not shown). These proteins are homologous to the DevR and DevS gene products from Myxococcus xanthus, respectively. In M.xanthus, devRS is an autoregulated gene locus that is essential for fruiting body development, but this connection provides no clues as to the possible biochemical functions of the proteins in question (57). No further information was obtained on COG1857 despite extensive sequence searches. However, COG1688 turned out to be distantly related to several other COGs associated with this system as described below.
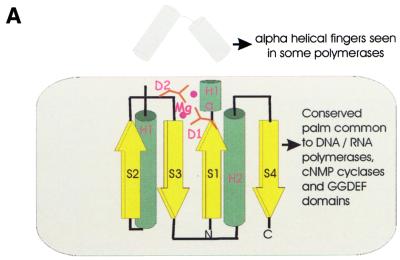
Another part of the analyzed gene neighborhood centers on the gene coding for multidomain proteins of COG1353. Many of the proteins in this COG contain an N-terminal domain that is a distinct version of an HD-superfamily hydrolase (52) with a circular permutation, in which the N-terminal metal-binding histidine is displaced to the extreme C-terminus of the HD domain (Fig. 3E). However, in some of these proteins, the HD domain is disrupted, whereas others, such as aq_357 from A.aeolicus, lack the HD domain altogether. The conserved C-terminus shared by almost all these proteins has three distinct domains (Fig. 4); the first of these is a distinct globular α + β domain that was not detected in any other proteins (Fig. 3D), whereas the second one is a Zn ribbon (Fig. 3C) that is seen in numerous contexts including nucleic acid interaction (58). The C-terminal domain of these proteins, which is present in a stand-alone form in SSO1429, is homologous to the catalytic domain of diverse DNA and RNA polymerases. Early sequence and structure comparisons showed that reverse transcriptases, viral RNA-dependent RNA polymerases and DNA polymerases of superfamilies A and B share a common catalytic core domain (59–61). This core domain was also detected in signal-transducing adenylyl cyclases and bacterial nucleotide cyclases typified by the GGDEF domain (62–64), some of which may possess diguanylate cyclase activity (65). The core palm-domain of these proteins contains a RNA recognition motif (RRM)-like fold with a β-α-β2-α-β topology; in nucleic acid polymerases, predominantly α-helical structures (the polymerase ‘fingers’) are inserted into the RRM-like domain upstream of helix-1 (Fig. 3A).
Figure 3.
(Opposite and above) The predicted novel DNA polymerase. (A) Topology of the conserved core of the polymerase-cyclase palm domain. The catalytic metal-coordinating residues and the variable inserted finger module in the polymerases are indicated. (B) Multiple alignment of different polymerase and cyclase domains. The structure-based sequence alignment was constructed using the proteins whose structures have been solved (PDB nos shown in brackets) and the core secondary structure elements were derived from this structural alignment. The novel predicted polymerases were first aligned using the T_coffee program and then aligned with the rest of the sequences using secondary structure prediction as a guide. The alignment consists of the following families of (predicted) polymerases and cyclases as indicated to the right of the aligned sequences: 1, B family DNA polymerases; 2, adenylate cyclases; 3, GGDEF family of (predicted) diguanylate cyclases; 4, predicted novel DNA polymerases; 5, RNA-dependent RNA polymerases (RDRP) of positive-strand RNA viruses; 6, reverse transcriptases (RT) of retroviruses and retroid elements. The shared secondary structure elements are indicated above the alignment and the catalytic residues are shown in reverse shading. The other designations are as in Figure 2. (C) Multiple alignment of the Zn ribbons seen in the predicted DNA polymerases. Note the disruption of the Zn-chelating residues in two of the proteins. The designations are as in Figure 2. (D) Multiple alignment of a putative polymerase-thumb-like domain shared by the COG1353 proteins. The designations are as in Figure 2. (E) Multiple alignment of the permuted HD hydrolase domain present at the extreme N-terminus of several members of COG1353. The designations are as in Figure 2.
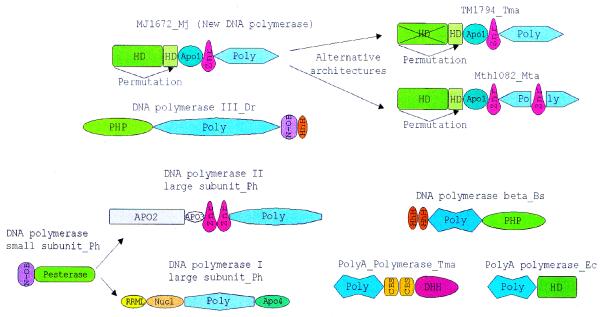
Figure 4.
The domain architecture of the predicted novel DNA polymerases compared with domain architectures of other nucleic acid polymerases that are associated with different phosphoesterase domains. The polymerase catalytic domains are abbreviated as ‘Poly’ and each distinct family of polymerases is shown by a different shape and shade. The other domain abbreviations are: HhH, helix–hairpin–helix domain; DHH, phosphoesterase domain with DHH motif; PHP, phosphoesterase domain shared by DNA polymerases and histidinol phosphosphatase; HD, phosphoesterase domain with the HD motif; Pesterase, calcineurin-like phosphoesterase domain; Znr, zinc ribbon domain; N-OB, nucleic acid binding OB-fold domain; Nucl, 3′→5′ nuclease domain; RRML, domain with RRM-like fold; CBS, cystathionine b synthase domain; Apo1-4, Archaeal-polymerase-specific domains 1–4.
PSI-BLAST searches with the C-terminal domains of the COG1353 proteins detected the GGDEF domains with statistically significant E-values (10–4–10–5 in 3rd–4th iterations). Furthermore, GGDEF domains and the putative polymerase domains of the COG1353 proteins shared an extended region of similarity beyond the core palm domain (62). Alignment-based secondary structure predictions (66) for these domains was compatible with the palm-domain structures of nucleic acid polymerases and nucleotide cyclases. Sequence–structure threading through the PDB database with both the combined fold recognition method (Z-score = 12.1) and 3D-PSSM method (E-value 0.02; E-values up to 0.8 are normally considered significant for threading through PDB database with this method) gave the adenylyl cyclases and DNA polymerases as the best hits. This strongly suggests that COG1353 proteins contain the same core fold as the palm domain of polymerases and cyclases.
A multiple alignment of the conserved portion of the C-terminal domain of COG1353 proteins with several families of polymerases, including family B DNA-directed DNA polymerases, reverse transcriptases, RNA-directed RNA polymerases of positive-strand RNA viruses and the two families of nucleotide cyclases, revealed the conservation of two distinct motifs (Fig. 3B) in this entire diverse array of proteins (Fig. 3A and B). These motifs contain the conserved negatively-charged residues (most often aspartates), which function as divalent metal ligands in the catalytic centers of these polymerases and are readily identifiable in the COG1353 proteins suggesting similar catalytic activities (Fig. 3A and B). Unlike the related GGDEF and adenylyl cyclase domains, the polymerase/cyclase-related domains of COG1353 proteins are never fused to the classic signaling domains such as PAS, GAF, HAMP or CACHE (64,67). Instead, they are fused to Zn ribbons, which are present in a variety of DNA polymerases, such as DNA polymerase ɛ of the eukaryotes and the euryarchaeal DNA polymerase II (58). In one of the COG1353 members, MTH1082, a Zn ribbon is inserted directly within the polymerase/cyclase domain after helix 1 (Fig. 4). The presence of the predicted HD-hydrolase domain fused to the N-terminus of this novel polymerase-related domain develops the theme of functional and, in most cases, physical association of various phosphohydrolase domain and nucleic acid polymerases (68) (Fig. 4). It has been hypothesized that these domains or subunits function as pyrophosphatases that cleave the pyrophosphate formed during nucleotide polymerization and thus drive forward the polymerase reaction (68). The same function is most likely for the HD-domain of the COG1353 proteins, although it is also possible that these predicted phosphoesterase domains function as uncharacterized nucleases in conjunction with the DNA polymerases. Taken together, these observations strongly suggest that the C-terminal domain of the COG1353 proteins is a previously undetected DNA polymerase distantly related to all of the above polymerase families. Given their degree of conservation and widespread presence in the Archaea, it appears most likely that this predicted DNA polymerase evolved from a common ancestor with other polymerases at an early stage of archaeal evolution. The GGDEF cyclases and adenylyl cyclase, which are more closely related to these predicted DNA polymerases, might have been derived from them and subsequently disseminated by horizontal gene transfer (HGT).
Genes of COGs 1336, 1367, 1604, 1337, 1332 and 1583 are always seen in close proximity to the predicted COG1353 polymerase, often in tandem (Fig. 1A). PSI-BLAST searches revealed relatively weak but, in many cases, statistically significant similarity between proteins from these COGs. For example, in a search starting with the sequence of the Rv2821c protein (COG1337), with the profile-inclusion cut-off set at E = 0.01, some members of COG1332 are detected in the second iteration, members of COG1604 and COG1336 appear in the fourth iteration and members of COG1367 in the fifth iteration. In the same search, a member of COG1567 (PH0166) appears above the cut-off in the fourth iteration. A reverse search started with the PH0166 sequence as the query detects all members of COG1567 and includes the first protein of COG1337 (MTH1080) on the fifth iteration. In the latter search, some proteins from other COGs represented in the neighborhood also appear, albeit with E-values that are below the cut-off. In particular, in the sixth iteration, proteins SSO1437 (COG1583) and AF0067 (COG1688) were detected with E-values of 0.13 and 0.29, respectively.
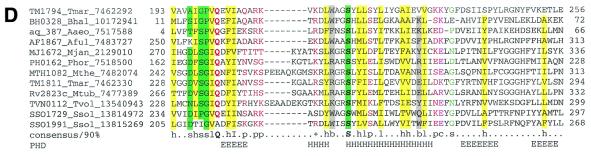
Proteins from COGs 1336, 1367, 1604, 1337 and 1332 produced a multiple alignment with many conserved positions and five common motifs (Fig. 5), which supports the notion that these COGs belong to the same protein family. The remaining COGs detected in BLAST searches failed to unequivocally align with the above five COGs, but shared at least some of the same conserved motifs and appeared to be compatible with the same fold as judged by similar patterns of predicted secondary structure elements (Fig. 5, alignments are available upon request). Thus, we believe that all these COGs comprise a previously undetected superfamily of repair-associated mysterious proteins (RAMPs). To identify all potential members of the RAMP superfamily and improve multiple alignments, we used all identified proteins as query sequences for exhaustive PSI-BLAST searches. This resulted in the identification of over 90 RAMPs, mostly in Archaea. All families of the RAMP superfamily share at least one C-terminal motif, which contains a glycine-rich loop (motif V, Fig. 5). Two other conserved motifs in the N-terminal portion of RAMPs show distinct structural features. Motif II also consists of a loop followed by an α-helix (Fig. 5). Motif I is a β-strand followed by a conserved glycine. However, none of these motifs was detectable in COG1583 members; this COG remains a provisional member of the RAMP superfamily (Fig. 5).
Figure 5.
The RAMP superfamily. The top part of the figure shows a multiple alignment of the major family of the RAMP superfamily. The designations are as in Figure 2. The bottom part shows a comparison of motifs derived from multiple alignments and secondary structure prediction for five families of RAMPs. Each family was aligned individually as described in Materials and Methods (alignments are available upon request). For each family, a 85% consensus was derived and secondary structure was predicted. The conserved motifs were aligned on the basis of PSI-BLAST alignments (when available; see Results), similarity of the conserved amino acid patterns and secondary structure prediction. Color coding and secondary structure element designations are as in Figure 2.
Genes coding for several other uncharacterized proteins tend to be associated with the core genes of the putative repair system. One of these (COG1343) shows a patchy distribution among bacteria and Archaea, and others are seen specifically in a subset of bacteria (e.g. COG3649 in T.maritima, B.halodurans and S.pyogenes) or Archaea (COG3574 in Pyrococci, A.fulgidus and S.solfataricus). COG3578, which is represented in A.fulgidus, S.solfataricus and A.pernix, includes additional members of the RecB nuclease family.
Several other proteins are typically encoded in the vicinity of the predicted new polymerase gene. Two of these (COG1517 and COG3574) are specific for Archaea and two others were detected in both Archaea and bacteria (COG3337 and COG1421). Members of COG1517 contain a distinct motif with a ‘hhDhoH’ signature and several other conserved polar amino acids, which could contribute to a potential catalytic center of an enzyme, perhaps yet another nuclease (alignment is available upon request). Members of the remaining polymerase-associated COGs are small proteins, for which no functional prediction is currently possible. At least one more functional prediction can be made on the basis of operon organization conservation. A gene for a predicted helix–turn–helix transcriptional regulator (COG2462) is located within the neighborhood in most archaeal genomes. This Archaea-specific protein is likely to regulate the expression of one or more of the operons in the analyzed neighborhood.
Although none of the genes in the analyzed neighborhood has been functionally characterized, the repertoire of predicted functions, which include a DNA helicase, several DNases and a polymerase (Table 1), strongly suggests that this neighborhood consists of genes together comprising a previously undetected DNA repair system. The uncharacterized proteins encoded within this neighborhood might function as accessory, DNA-binding or regulatory subunits of these repair complexes and perhaps as a sliding clamp for the predicted DNA polymerase. Of particular interest in this latter context are the RAMP proteins, which, given their remarkable proliferation, could form large hetero-oligomeric complexes.
A more specific functional role for the predicted repair system could be that of a functional equivalent of the mutagenic repair systems of bacteria and eukaryotes, which center around the translesion DNA polymerases of the UmuC-DinB-Rad30-Rev1 superfamily (69,70). A priori, such a system should be considered important in thermophiles, to counteract DNA damage caused by exposure to high temperatures. Among thermophilic organisms whose genomes have been completely or partially sequenced, only Sulfolobus encodes a predicted UmuC-DinB-Rad30-Rev1 superfamily polymerase (71). In particular, although this type of polymerase is present in most free-living bacteria, it is conspicuously missing in the hyperthermophiles T.maritima and A.aeolicus. In contrast, the only sequenced genome of a mesophilic archaeon, that of Halobacterium NRC-1, does have a DinB ortholog, but completely lacks the predicted new repair system. Thus, it seems plausible that the predicted novel repair system is the thermophilic counterpart of the mesophilic translesion repair system that also mediates adaptive mutagenesis (72). Technically, it is possible that the functional system encoded by the gene neighborhood described here is involved in RNA metabolism rather than in DNA repair. This, however, appears unlikely given that the most characteristic genes of this neighborhood encode a RecB family nuclease, which is specifically associated with repair systems (73) and a predicted polymerase fused to a phosphoesterase domain, an architecture typical of DNA polymerases (74).
Genomic diversity, modular organization, horizontal gene transfer and evolution of the predicted repair system
The predicted novel repair system shows conspicuous evolutionary plasticity. This becomes particularly obvious when genomes of closely related species, such as two Thermoplasmas, two Bacilli and three Pyrococci, are compared (Fig. 1A). Thermoplasma acidophilum does not encode any members of this system, whereas T.volcanium has what seems to be a rudimentary form, with only the predicted polymerase, the putative nuclease of COG1518, and several RAMPs and uncharacterized proteins. The disruption of the repair system probably started already in the common ancestor of the two Thermoplasma species, with T.acidophilum subsequently losing it completely. Bacillus halodurans has two large putative operons with genes for components of the predicted repair system, but B.subtilis has no counterpart to any of these genes. Among the three Pyrococcus species, P.abyssi resembles T.volcanium in having only remnants of the system, represented by the helicase, a stand-alone HD-nuclease and three RAMPs. Pyrococcus furiosus has a more complete system, which also includes the RecB family nuclease, the predicted polymerase and extra RAMPs. Finally, P.horikoshii has a complete, complex system, with triplication of the helicase and surrounding genes. Thus, substantial changes in this system tend to occur relatively rapidly, on a time-scale commensurate with the evolution of individual species. Gene loss is prominent among these evolutionary modifications, but amplification of parts of the system, and possibly acquisition of additional members through HGT (see below) also take place.
Cross-genome comparison of the repertoires and organization of the genes encoding components of the predicted novel repair system supports the notion that the core (helicase-nuclease) and the polymerase-RAMP modules of the superoperon have a degree of independence. The examples discussed above indicate that one of the modules can be independently lost. They also undergo rearrangement that includes reversal of the relative orientation of the modules in a superoperon (compare the gene organization in A.aeolicus and T.maritima in Figure 1A) and probable operon disruption (compare the gene organization in A.aeolicus and A.fulgidus). Furthermore, on some occasions, the modules have undergone independent amplification, such as the duplication of the polymerase module in T.maritima and triplication of the helicase-nuclease module in P.horikoshii.
The modular gene/operon organization of the predicted repair system probably also entails functional modularity. It seems likely that the stand-alone helicase-nuclease module that is present, for example, in P.abyssi and A.pernix (Fig. 1A) retains some limited functionality in repair, but probably functions on its own or in conjunction with a different polymerase. Conversely, in T.volcanium and M.tuberculosis, the predicted DNA polymerase might interact with a distinct helicase. Furthermore, the deletion of some of the predicted nucleases in P.abyssi, T.volcanium, B.halodurans, S.pyogenes and E.coli suggests a degree of redundancy among these enzymes. Such partial functional redundancy is typical of other repair pathways (75).
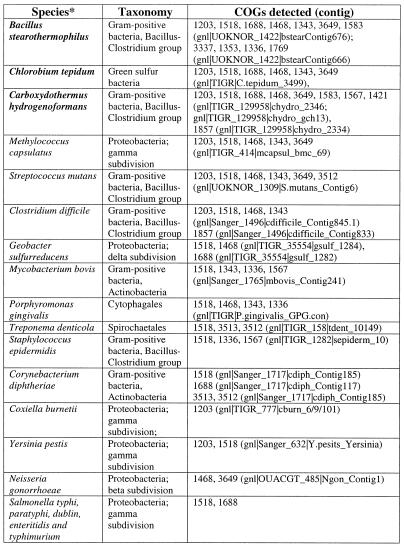
Overall, the number of genes coding for components of the predicted repair system varies to a great degree between genomes (Figs 1 and 6), with about 90 genes in S.solfataricus (>3% of all genes in this genome) and the minimal set of three genes in E.coli. The prevalence of this system in thermophiles is obvious. Bacillus halodurans is the only mesophile that has the principal genes of both the helicase-nuclease and the polymerase-RAMP modules and, even in this case, the system is less elaborate than it is in most thermophiles (Figs 1A and 6). Most mesophiles have no trace of this system, and several species, in which it is represented, have only remnants of one or both modules (Fig. 1A). Search of unfinished prokaryotic genomes detected homologs of different proteins from the new repair system, particularly of the helicase-nuclease module, in many diverse bacteria (Table 2). Again, the three thermophiles, for which large amounts of genome sequence were available, Chlorobium tepidum, Carboxydothermus hydrogenoformans and Bacillus stearothermophilus, showed a greater representation of this system than mesophiles (Table 2).
Figure 6.
Representation of the predicted novel repair system in different genomes. Pink rectangles, RAMP proteins; blue rectangles, other components of the system.
Table 2. Traces of the predicted new repair system in unfinished bacterial genomes.
*Thermophilic species are shown in bold.
The obvious plasticity of the new repair system raises the issue of a possible role of HGT in its evolution (as already alluded to above). The notion that HGT occurred more than once during evolution of this system is supported by notable conservation of certain gene arrays in phylogenetically distant genomes (Fig. 1A). The strongest case in point is the conservation of the gene order in the polymerase module between the archaeon A.fulgidus and the bacteria A.aeolicus and B.halodurans (Fig. 1A). These observations suggest that these gene cassettes (probable operons) disseminated via HGT as a single entity. In fact, in an early comparison, the apparent superoperon that comprises the predicted repair system in A.aeolicus has been noticed as the largest constellation of ‘archaeal’ genes in the genome of this hyperthermophilic bacterium, and its presence was one of the arguments supporting massive HGT between bacterial and archaeal hyperthermophiles (76).
To examine further the contribution of HGT to the evolution of the predicted new repair system, phylogenetic trees were constructed for the four genes that are most common in the conserved neighborhood (COGs 1203, 1518, 1468 and 1353). All four trees showed clear indications of multiple HGT events (Fig. 7). In particular, each tree supports HGT from Archaea to A.aeolicus, in agreement with the conservation of gene order between this bacterium and some Archaea (see above). The tree topologies suggest independent HGT events between Archaea and different bacterial groups as well as between different bacterial lineages. For example, in the tree for the putative novel nuclease (COG1518) and RecB-type nuclease (COG1468), the proteins from B.halodurans and B.stearothermophilus occupy very different positions instead of being adjacent as expected from the phylogeny of the corresponding species (Fig. 7A and C). The former belongs to a cluster of several bacterial species, which is located between two archaeal clusters, whereas the latter is part of another, smaller group of diverse bacterial species, which lies within one of the archaeal clusters (Fig. 7A and C). Furthermore, in the tree for COG1203 helicases, a third bacterial cluster, which combines proteins from proteobacteria, the Bacillus-Clostridium group of Gram-positive bacteria and a spirochete, joins the second archaeal cluster (Fig. 7B). Thus, the topology of this tree can be explained through three independent HGT events between Archaea and bacteria, followed by some additional HGT within the bacterial and possibly archaeal domains. Alternatively, it could be postulated that the existence of the third bacterial cluster, which is separated from the Archaea by a long branch, reflects vertical inheritance from the last common ancestor of Archaea and bacteria, with subsequent multiple gene losses resulting in the extant patchy phyletic distribution. Unlike the trees for the other three analyzed proteins, the tree for predicted polymerases has B.halodurans and B.stearothermophilus proteins in the same cluster (Fig. 7D); this emphasizes distinct evolutionary fates of different genes within the predicted new repair system.
Figure 7.
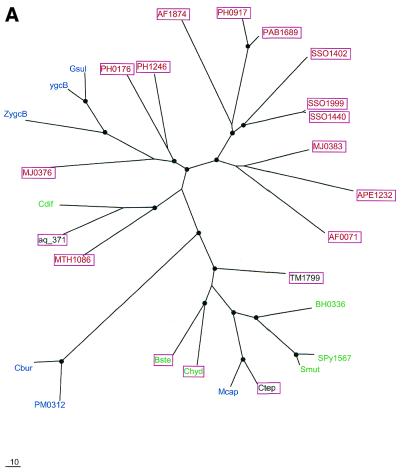
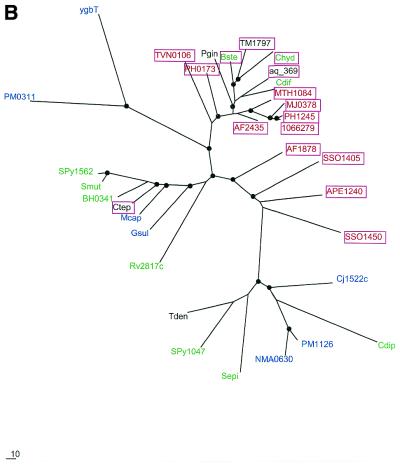
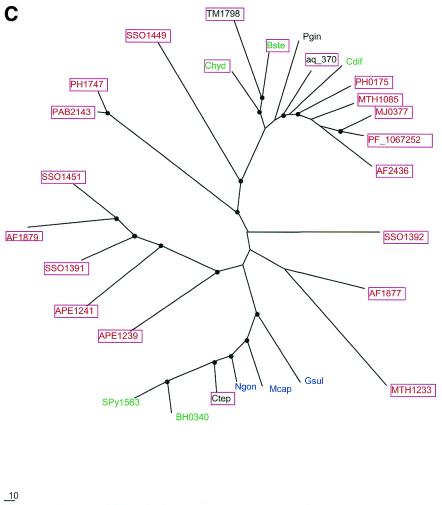
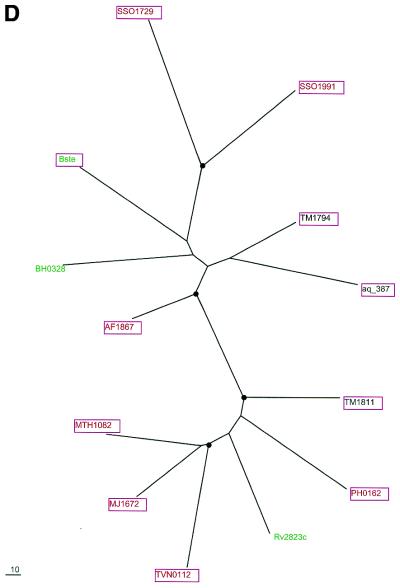
Phylogenetic trees for the most common components of the predicted novel repair system. (A) Putative novel nuclease (COG1518). (B) The helicase domain (COG1203). (C) The RecB family nuclease (COG1468). (D) The predicted novel polymerase (COG1353). Maximum likelihood trees constructed using the MOLPHY program are shown. Internal branches that were supported by bootstrap probability >70% are marked by black circles. In addition to the sequences from complete genomes, sequences that were identified by TBLASTN searches in the database of unfinished microbial genomes were used for phylogenetic analysis. Systematic gene names are used as branch designations except for sequences from unfinished genomes, which are designated using the corresponding species abbreviation. Archaeal genes are shown in red, genes from Gram-positive bacteria in green, proteobacterial genes in blue and genes from other bacteria in black. Genes from thermophiles are boxed.
The apparent multiple HGT and gene loss events preclude a definitive conclusion as to the origin of the predicted repair system described here. One scenario would posit that this system originally evolved in hyperthermophilic Archaea and subsequently was disseminated through the prokaryotic world via multiple HGTs. Under this scenario, many mesophilic bacteria acquired (parts of) this system from thermophiles and subsequently lost some of the acquired genes. An alternative possibility, which is best compatible with the hypothesis that the last universal common ancestor of modern life forms was a hyperthermophile (77,78), is that the core of this system already existed in this hypothetical ancestral organism, with numerous coordinated gene losses occurring in various lineages that became mesophilic. One such lineage is the eukaryotes whose common ancestor might have originally inherited this repair system. A variant of this hypothesis is that the helicase-nuclease and polymerase-RAMP modules evolved independently at a very early stage of evolution. Subsequently, they might have been brought together to form a single repair system in Archaea, and this system was acquired by some, primarily thermophilic bacteria via HGT. At a mechanistic level, the association of the predicted repair system with thermophily and the apparent near incompatibility of this system with the translesion repair pathway based on UmuC-DinB-Rad30-Rev1 superfamily polymerases remain mysterious and, hopefully, will be targets for future experimental studies.
CONCLUSIONS
A previously undetected DNA repair system that is largely specific for thermophiles was predicted through the use of a relatively permissive approach to gene context analysis, examination of partially conserved gene neighborhoods, which does not emphasize exact conservation of local gene order. The use of such an approach was important because of extreme evolutionary plasticity of the novel repair system. The evolution of this system appears to have involved frequent genomic rearrangements, modular and sporadic gene loss and multiple HGT events. Experimental validation of the predictions made here should include both demonstration of individual biochemical activities, particularly those of the predicted novel polymerase and nuclease, and of the RAMPs, and elucidation of the physiological role of the system as a whole. The latter type of experiments might shed light on the intriguing and unsolved question: how do thermophiles cope with the increased level of DNA damage that is inevitable in their natural habitats?
Acknowledgments
ACKNOWLEDGEMENTS
The release of unfinished genome sequences by The Institute of Genome Research, the Sanger Center, Oklahoma’s Advanced Center for Genome Technology and Washington University Genome Sequencing Center is gratefully acknowledged. K.M. is supported by the Microbial Genome Program, Office of Biological and Environmental Research, DOE (DE-FG02-98ER62583).
REFERENCES
- 1.Stetter K.O. (1996) Hyperthermophiles in the history of life. Ciba Found Symp., 202, 1–10. [PubMed] [Google Scholar]
- 2.Daniel R.M. and Cowan,D.A. (2000) Biomolecular stability and life at high temperatures. Cell. Mol. Life Sci., 57, 250–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nisbet E. (2000) The realms of Archaean life. Nature, 405, 625–626. [DOI] [PubMed] [Google Scholar]
- 4.Grogan D.W. (2000) The question of DNA repair in hyperthermophilic archaea. Trends Microbiol., 8, 180–185. [DOI] [PubMed] [Google Scholar]
- 5.Watrin L. and Prieur,D. (1996) UV and ethyl methanesulfonate effects in hyperthermophilic archaea and isolation of auxotrophic mutants of Pyrococcus strains. Curr. Microbiol., 33, 377–382. [DOI] [PubMed] [Google Scholar]
- 6.DiRuggiero J., Santangelo,N., Nackerdien,Z., Ravel,J. and Robb,F.T. (1997) Repair of extensive ionizing-radiation DNA damage at 95°C in the hyperthermophilic archaeon Pyrococcus furiosus. J. Bacteriol., 179, 4643–4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jacobs K.L. and Grogan,D.W. (1998) Spontaneous mutation in a thermoacidophilic archaeon: evaluation of genetic and physiological factors. Arch. Microbiol., 169, 81–83. [DOI] [PubMed] [Google Scholar]
- 8.Grogan D.W., Carver,G.T. and Drake,J.W. (2001) Genetic fidelity under harsh conditions: analysis of spontaneous mutation in the thermoacidophilic archaeon Sulfolobus acidocaldarius. Proc. Natl Acad. Sci. USA, 98, 7928–7933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Skorvaga M., Raven,N.D. and Margison,G.P. (1998) Thermostable archaeal O6-alkylguanine-DNA alkyltransferases. Proc. Natl Acad. Sci. USA, 95, 6711–6715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aravind L. and Koonin,E.V. (2000) The alpha/beta fold uracil DNA glycosylases: a common origin with diverse fates. Genome Biol., 1, Research0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sandigursky M. and Franklin,W.A. (1999) Thermostable uracil-DNA glycosylase from Thermotoga maritima a member of a novel class of DNA repair enzymes. Curr. Biol., 9, 531–534. [DOI] [PubMed] [Google Scholar]
- 12.Sandigursky M. and Franklin,W.A. (2000) Uracil-DNA glycosylase in the extreme thermophile Archaeoglobus fulgidus. J. Biol. Chem., 275, 19146–19149. [DOI] [PubMed] [Google Scholar]
- 13.Rashid N., Morikawa,M., Kanaya,S., Atomi,H. and Imanaka,T. (1999) A unique DNase activity shares the active site with ATPase activity of the RecA/Rad51 homologue (Pk-REC) from a hyperthermophilic archaeon. FEBS Lett., 445, 111–114. [DOI] [PubMed] [Google Scholar]
- 14.Aravind L., Walker,D.R. and Koonin,E.V. (1999) Conserved domains in DNA repair proteins and evolution of repair systems. Nucleic Acids Res., 27, 1223–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eisen J.A. and Hanawalt,P.C. (1999) A phylogenomic study of DNA repair genes, proteins, and processes. Mutat. Res., 435, 171–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wolf Y.I., Rogozin,I.B., Kondrashov,A.S. and Koonin,E.V. (2001) Genome alignment, evolution of prokaryotic genome organization, and prediction of gene function using genomic context. Genome Res., 11, 356–372. [DOI] [PubMed] [Google Scholar]
- 17.Overbeek R., Fonstein,M., D’Souza,M., Pusch,G.D. and Maltsev,N. (1999) The use of gene clusters to infer functional coupling. Proc. Natl Acad. Sci. USA, 96, 2896–2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Galperin M.Y. and Koonin,E.V. (2000) Who’s your neighbor? New computational approaches for functional genomics. Nat. Biotechnol., 18, 609–613. [DOI] [PubMed] [Google Scholar]
- 19.Huynen M., Snel,B., Lathe,W.,III and Bork,P. (2000) Predicting protein function by genomic context: quantitative evaluation and qualitative inferences. Genome Res., 10, 1204–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aravind L. (2000) Guilt by association: contextual information in genome analysis. Genome Res., 10, 1074–1077. [DOI] [PubMed] [Google Scholar]
- 21.DiRuggiero J., Brown,J.R., Bogert,A.P. and Robb,F.T. (1999) DNA repair systems in archaea: mementos from the last universal common ancestor? J. Mol. Evol., 49, 474–484. [DOI] [PubMed] [Google Scholar]
- 22.Aravind L. and Koonin,E.V. (2001) Prokaryotic homologs of the eukaryotic DNA-end-binding protein ku, novel domains in the ku protein and prediction of a prokaryotic double-strand break repair system. Genome Res., 11, 1365–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klenk H.P., Clayton,R.A., Tomb,J.F., White,O., Nelson,K.E., Ketchum,K.A., Dodson,R.J., Gwinn,M., Hickey,E.K., Peterson,J.D. et al. (1997) The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature, 390, 364–370. [DOI] [PubMed] [Google Scholar]
- 24.Smith D.R., Doucette-Stamm,L.A., Deloughery,C., Lee,H., Dubois,J., Aldredge,T., Bashirzadeh,R., Blakely,D., Cook,R., Gilbert,K. et al. (1997) Complete genome sequence of Methanobacterium thermoautotrophicum deltaH: functional analysis and comparative genomics. J. Bacteriol., 179, 7135–7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bult C.J., White,O., Olsen,G.J., Zhou,L., Fleischmann,R.D., Sutton,G.G., Blake,J.A., FitzGerald,L.M., Clayton,R.A., Gocayne,J.D. et al. (1996) Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science, 273, 1058–1073. [DOI] [PubMed] [Google Scholar]
- 26.Kawarabayasi Y., Sawada,M., Horikawa,H., Haikawa,Y., Hino,Y., Yamamoto,S., Sekine,M., Baba,S., Kosugi,H., Hosoyama,A. et al. (1998) Complete sequence and gene organization of the genome of a hyper-thermophilic archaebacterium, Pyrococcus horikoshii OT3. DNA Res., 5, 55–76. [DOI] [PubMed] [Google Scholar]
- 27.Kawarabayasi Y., Hino,Y., Horikawa,H., Yamazaki,S., Haikawa,Y., Jin-no,K., Takahashi,M., Sekine,M., Baba,S., Ankai,A. et al. (1999) Complete genome sequence of an aerobic hyper-thermophilic crenarchaeon, Aeropyrum pernix K1. DNA Res., 6, 83–101, 145–152. [DOI] [PubMed] [Google Scholar]
- 28.She Q., Singh,R.K., Confalonieri,F., Zivanovic,Y., Allard,G., Awayez,M.J., Chan-Weiher,C.C., Clausen,I.G., Curtis,B.A., De Moors,A. et al. (2001) The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc. Natl Acad. Sci. USA, 26, 7835–7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nelson K.E., Clayton,R.A., Gill,S.R., Gwinn,M.L., Dodson,R.J., Haft,D.H., Hickey,E.K., Peterson,J.D., Nelson,W.C., Ketchum,K.A. et al. (1999) Evidence for lateral gene transfer between Archaea and bacteria from genome sequence of Thermotoga maritima. Nature, 399, 323–329. [DOI] [PubMed] [Google Scholar]
- 30.Deckert G., Warren,P.V., Gaasterland,T., Young,W.G., Lenox,A.L., Graham,D.E., Overbeek,R., Snead,M.A., Keller,M., Aujay,M. et al. (1998) The complete genome of the hyperthermophilic bacterium Aquifex aeolicus. Nature, 392, 353–358. [DOI] [PubMed] [Google Scholar]
- 31.Takami H., Nakasone,K., Takaki,Y., Maeno,G., Sasaki,R., Masui,N., Fuji,F., Hirama,C., Nakamura,Y., Ogasawara,N. et al. (2000) Complete genome sequence of the alkaliphilic bacterium Bacillus halodurans and genomic sequence comparison with Bacillus subtilis. Nucleic Acids Res., 28, 4317–4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cole S.T., Brosch,R., Parkhill,J., Garnier,T., Churcher,C., Harris,D., Gordon,S.V., Eiglmeier,K., Gas,S., Barry,C.E.,III et al. (1998) Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature, 393, 537–544. [DOI] [PubMed] [Google Scholar]
- 33.Ferretti J.J., McShan,W.M., Ajdic,D., Savic,D.J., Savic,G., Lyon,K., Primeaux,C., Sezate,S., Suvorov,A.N., Kenton,S. et al. (2001) Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc. Natl Acad. Sci. USA, 98, 4658–4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tatusova T.A., Karsch-Mizrachi,I. and Ostell,J.A. (1999) Complete genomes in WWW Entrez: data representation and analysis. Bioinformatics, 15, 536–543. [DOI] [PubMed] [Google Scholar]
- 35.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Altschul S.F. and Koonin,E.V. (1998) Iterated profile searches with PSI-BLAST—a tool for discovery in protein databases. Trends Biochem. Sci., 23, 444–447. [DOI] [PubMed] [Google Scholar]
- 37.Aravind L. and Koonin,E.V. (1999) Gleaning non-trivial structural, functional and evolutionary information about proteins by iterative database searches. J. Mol. Biol., 287, 1023–1040. [DOI] [PubMed] [Google Scholar]
- 38.Schultz J., Copley,R.R., Doerks,T., Ponting,C.P. and Bork,P. (2000) SMART: a web-based tool for the study of genetically mobile domains. Nucleic Acids Res., 28, 231–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Notredame C., Higgins,D.G. and Heringa,J. (2000) T-Coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol., 302, 205–217. [DOI] [PubMed] [Google Scholar]
- 40.Rost B. and Sander,C. (1994) Combining evolutionary information and neural networks to predict protein secondary structure. Proteins, 19, 55–72. [DOI] [PubMed] [Google Scholar]
- 41.Fischer D. (2000) Hybrid fold recognition: combining sequence derived properties with evolutionary information. Pac. Symp. Biocomput., 119–130. [PubMed] [Google Scholar]
- 42.Kelley L.A., MacCallum,R.M. and Sternberg,M.J. (2000) Enhanced genome annotation using structural profiles in the program 3D-PSSM. J. Mol. Biol., 299, 499–520. [DOI] [PubMed] [Google Scholar]
- 43.Fitch W.M. and Margoliash,E. (1967) Construction of phylogenetic trees. Science, 155, 279–284. [DOI] [PubMed] [Google Scholar]
- 44.Felsenstein J. (1996) Inferring phylogenies from protein sequences by parsimony, distance, and likelihood methods. Methods Enzymol., 266, 418–427. [DOI] [PubMed] [Google Scholar]
- 45.Adachi J. and Hasegawa,M. (1992) MOLPHY: Programs for Molecular Phylogenetics. Institute of Statistical Mathematics, Tokyo, Japan. [Google Scholar]
- 46.Hasegawa M., Kishino,H. and Saitou,N. (1991) On the maximum likelihood method in molecular phylogenetics. J. Mol. Evol., 32, 443–445. [DOI] [PubMed] [Google Scholar]
- 47.Kishino H., Miyata,T. and Hasegawa,M. (1990) Maximum likelihood inference of protein phylogeny and the origin of chloroplasts. J. Mol. Evol., 31, 151–160. [Google Scholar]
- 48.Tatusov R.L., Koonin,E.V. and Lipman,D.J. (1997) A genomic perspective on protein families. Science, 278, 631–637. [DOI] [PubMed] [Google Scholar]
- 49.Tatusov R.L., Galperin,M.Y., Natale,D.A. and Koonin,E.V. (2000) The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res., 28, 33–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ruepp A., Graml,W., Santos-Martinez,M.L., Koretke,K.K., Volker,C., Mewes,H.W., Frishman,D., Stocker,S., Lupas,A.N. and Baumeister,W. (2000) The genome sequence of the thermoacidophilic scavenger Thermoplasma acidophilum. Nature, 407, 508–513. [DOI] [PubMed] [Google Scholar]
- 51.Ng W.V., Kennedy,S.P., Mahairas,G.G., Berquist,B., Pan,M., Shukla,H.D., Lasky,S.R., Baliga,N.S., Thorsson,V., Sbrogna,J. et al. (2000) Genome sequence of Halobacterium species NRC-1. Proc. Natl Acad. Sci. USA, 97, 12176–12181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aravind L. and Koonin,E.V. (1998) The HD domain defines a new superfamily of metal-dependent phosphohydrolases. Trends Biochem. Sci., 23, 469–472. [DOI] [PubMed] [Google Scholar]
- 53.Aravind L., Makarova,K.S. and Koonin,E.V. (2000) Holliday junction resolvases and related nucleases: identification of new families, phyletic distribution and evolutionary trajectories. Nucleic Acids Res., 28, 3417–3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kuzminov A. (1999) Recombinational repair of DNA damage in Escherichia coli and bacteriophage lambda. Microbiol. Mol. Biol. Rev., 63, 751–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang X.J. and Julin,D.A. (1999) Isolation and characterization of the C-terminal nuclease domain from the RecB protein of Escherichia coli. Nucleic Acids Res., 27, 4200–4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang J., Chen,R. and Julin,D.A. (2000) A single nuclease active site of the Escherichia coli RecBCD enzyme catalyzes single-stranded DNA degradation in both directions. J. Biol. Chem., 275, 507–513. [DOI] [PubMed] [Google Scholar]
- 57.Thony-Meyer L. and Kaiser,D. (1993) devRS, an autoregulated and essential genetic locus for fruiting body development in Myxococcus xanthus. J. Bacteriol., 175, 7450–7462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Aravind L. and Koonin,E.V. (1999) DNA-binding proteins and evolution of transcription regulation in the archaea. Nucleic Acids Res., 27, 4658–4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Poch O., Sauvaget,I., Delarue,M. and Tordo,N. (1989) Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J., 8, 3867–3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Delarue M., Poch,O., Tordo,N., Moras,D. and Argos,P. (1990) An attempt to unify the structure of polymerases. Protein Eng., 3, 461–467. [DOI] [PubMed] [Google Scholar]
- 61.Wang J., Sattar,A.K., Wang,C.C., Karam,J.D., Konigsberg,W.H. and Steitz,T.A. (1997) Crystal structure of a pol alpha family replication DNA polymerase from bacteriophage RB69. Cell, 89, 1087–1099. [DOI] [PubMed] [Google Scholar]
- 62.Pei J. and Grishin,N.V. (2001) GGDEF domain is homologous to adenylyl cyclase. Proteins, 42, 210–216. [DOI] [PubMed] [Google Scholar]
- 63.Murzin A.G. (1998) How far divergent evolution goes in proteins. Curr. Opin. Struct. Biol., 8, 380–387. [DOI] [PubMed] [Google Scholar]
- 64.Galperin M.Y., Natale,D.A., Aravind,L. and Koonin,E.V. (1999) A specialized version of the HD hydrolase domain implicated in signal transduction. J. Mol. Microbiol. Biotechnol., 1, 303–305. [PMC free article] [PubMed] [Google Scholar]
- 65.Tal R., Wong,H.C., Calhoon,R., Gelfand,D., Fear,A.L., Volman,G., Mayer,R., Ross,P., Amikam,D., Weinhouse,H. et al. (1998) Three cdg operons control cellular turnover of cyclic di-GMP in Acetobacter xylinum: genetic organization and occurrence of conserved domains in isoenzymes. J. Bacteriol., 180, 4416–4425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cuff J.A., Clamp,M.E., Siddiqui,A.S., Finlay,M. and Barton,G.J. (1998) JPred: a consensus secondary structure prediction server. Bioinformatics, 14, 892–893. [DOI] [PubMed] [Google Scholar]
- 67.Anantharaman V., Koonin,E.V. and Aravind,L. (2001) Regulatory potential, phyletic distribution and evolution of ancient, intracellular small-molecule-binding domains. J. Mol. Biol., 307, 1271–1292. [DOI] [PubMed] [Google Scholar]
- 68.Aravind L. and Koonin,E.V. (1998) Phosphoesterase domains associated with DNA polymerases of diverse origins. Nucleic Acids Res., 26, 3746–3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sutton M.D. and Walker,G.C. (2001) Managing DNA polymerases: Coordinating DNA replication, DNA repair, and DNA recombination. Proc. Natl Acad. Sci. USA, 98, 8342–8349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McDonald J.P., Tissier,A., Frank,E.G., Iwai,S., Hanaoka,F. and Woodgate,R. (2001) DNA polymerase iota and related rad30-like enzymes. Philos. Trans. R. Soc. Lond. B. Biol. Sci., 356, 53–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kulaeva O.I., Koonin,E.V., McDonald,J.P., Randall,S.K., Rabinovich,N., Connaughton,J.F., Levine,A.S. and Woodgate,R. (1996) Identification of a DinB/UmuC homolog in the archeon Sulfolobus solfataricus. Mutat. Res., 357, 245–253. [DOI] [PubMed] [Google Scholar]
- 72.McKenzie G.J., Lee,P.L., Lombardo,M., Hastings,P.J. and Rosenberg,S.M. (2001) SOS mutator DNA polymerase IV functions in adaptive mutation and not adaptive amplification. Mol. Cell, 7, 571–579. [DOI] [PubMed] [Google Scholar]
- 73.Jockovich M.E. and Myers,R.S. (2001) Nuclease activity is essential for RecBCD recombination in Escherichia coli. Mol. Microbiol., 41, 949–962. [DOI] [PubMed] [Google Scholar]
- 74.Aravind L. and Koonin,E.V. (1998) Phosphoesterase domains associated with DNA polymerases of diverse origins. Nucleic Acids Res., 26, 3746–3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Freedberg E.C. (1995) DNA Repair and Mutagenesis. ASM Press, Washington, DC. [Google Scholar]
- 76.Aravind L., Tatusov,R.L., Wolf,Y.I., Walker,D.R. and Koonin,E.V. (1998) Evidence for massive gene exchange between archaeal and bacterial hyperthermophiles. Trends Genet., 14, 442–444. [DOI] [PubMed] [Google Scholar]
- 77.Lazcano A. and Miller,S.L. (1996) The origin and early evolution of life: prebiotic chemistry, the pre-RNA world, and time. Cell, 85, 793–798. [DOI] [PubMed] [Google Scholar]
- 78.Di Giulio M. (2000) The universal ancestor lived in a thermophilic or hyperthermophilic environment. J. Theor. Biol., 203, 203–213. [DOI] [PubMed] [Google Scholar]