Abstract
Recent technical advances have enabled unbiased transcriptomic and epigenetic analysis of each cell, known as “single-cell analysis”. Single-cell analysis has a variety of technical approaches to investigate the state of each cell, including mRNA levels (transcriptome), the immune repertoire (immune repertoire analysis), cell surface proteins (surface proteome analysis), chromatin accessibility (epigenome), and accordance with genome variants (eQTLs; expression quantitative trait loci). As an effective tool for investigating robust immune responses in coronavirus disease 2019 (COVID-19), many researchers performed single-cell analysis to capture the diverse, unbiased immune cell activation and differentiation. Despite challenges elucidating the complicated immune microenvironments of chronic inflammatory diseases using existing experimental methods, it is now possible to capture the simultaneous immune features of different cell types across inflamed tissues using various single-cell tools. In this review, we introduce patient-based and experimental mouse model research utilizing single-cell analyses in the field of chronic inflammatory diseases, as well as multi-organ atlas targeting immune cells.
Keywords: autoimmune disease, chronic inflammatory disease, coronavirus disease 2019, mouse model, multi-organ atlas, single-cell genomics
INTRODUCTION
Unbiased profiling of single cells from inflamed status allows understanding of immune networks between heterogeneous cell types. Although single-cell profiling technology, such as flow cytometry, has been used to evaluate the protein expression of a few dozen antigens, recent technical advances have enabled unbiased transcriptomic and epigenetic analysis of each cell. In this context, the human cell atlas, the most comprehensive reference map of the molecular state of cells, was established based on high-throughput single-cell profiling (Regev et al., 2017). Immunologists have actively used single-cell genomics studies to rapidly and comprehensively reveal the pathogenesis of coronavirus disease 2019 (COVID-19) in the recent pandemic era (Lee et al., 2020; Stephenson et al., 2021; Unterman et al., 2022). In this review, we introduce recent progress in understanding inflammatory diseases by applying single-cell genomics technologies and propose new directions for future studies.
CURRENT SINGLE-CELL TOOLS FOR IMMUNOLOGICAL STUDIES
Single-cell analysis offers a variety of windows by which to observe the state of each cell, including cell surface proteins, intracellular proteins, mRNA levels, DNA methylation, genome sequence, chromatin accessibility, and histone modifications (Stuart et al., 2019). Exploring the immune system, these integrative tools can be applied to measure multiple immune cell characteristics not only in the transcriptome, but also in the genome, methylome, and proteome.
Single-cell immune repertoire analysis
V(D)J recombination constructs highly diverse repertoires of antigen receptors in T cells and B cells, which is important for the development of adaptive immunity (Schatz and Ji, 2011). Compared to the early T cell receptor (TCR) analysis methods, which can capture partial sequences, the advent of NGS (next-generation sequencing) and single-cell analysis allowed the capture of paired TCRαβ chain information at single-cell resolution with transcriptome data (Pai and Satpathy, 2021). With the development of immune profiling, TCR sequencing has been applied to study the immunological responses of COVID-19 patients, showing that a diverse TCR repertoire was absent in patients with severe conditions (Zhang et al., 2020). In line with prior studies, robust TCR clonal expansion and less B cell receptor (BCR) clonal expansion was found in peripheral blood mononuclear cell (PBMC) samples from asymptomatic COVID-19 patients compared to moderate and severe patients (Zhao et al., 2021). Using LIBRA-seq to map the antigen specificity of B cells in high-throughput at single-cell levels (Setliff et al., 2019), a transition from immunoglobulin M (IgM) to IgG was observed in severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)-specific and cross-reactive B cells in response to BNT162b2 vaccination, suggesting an improvement in SARS-CoV-2 antibody specificities and cross-reactivity after vaccination (Kramer et al., 2022). Apart from COVID-19 studies, Piper et al. (2022) reported that CD4+ GM-CSF+ IFN-γ- T cells possess a distinct TCR repertoire compared to other CD4+ clusters, unraveling their heterogeneity in gastrointestinal tract GVHD (graft-versus-host disease) (Piper et al., 2022). The number of studies investigating TCR or BCR repertoires in human subjects is growing rapidly, but the diversity of TCR or BCR sequences is still considerably larger than the scale of current commercial single-cell genomics platforms.
Single-cell surface proteome analysis
Immunophenotyping with flow cytometry (fluorescence-activated cell sorting [FACS]) has been the principal method of identifying immune cell subsets by staining surface markers or intracellular cytokines (Perfetto et al., 2004). Using oligonucleotide-labeled antibodies, cellular indexing of transcriptomes and epitopes by sequencing (CITE-seq) can identify protein markers with unbiased transcriptome profiling at the single-cell level. CITE-seq can detect separate pools based on the CD8a fluorescence of flow cytometry, indicating its applicability in immunophenotyping using single-cell transcriptomics (Stoeckius et al., 2017). Considering that CITE-seq has no optical limitations, future proteomics analyses will be able to include higher dimensions than FACS.
Single-cell epigenome analysis
Post-translational modification of chromatin, such as DNA and histone methylation, is dynamic and changes during the development and differentiation of cells. These modifications, which are connected to chromatin accessibility, reflect how certain genomic regions function in regulating gene expression (Turner, 2007). First applied to the single-cell platform by Buenrostro et al. (2015) and Cusanovich et al. (2015), single-cell assay for transposase-accessible chromatin sequencing (ATAC-seq) has become a well-liked and straightforward method for profiling chromatin accessibility spanning thousands of individual cells (Minnoye et al., 2021). Global chromatin remodeling has been observed in the epigenomic profiles of SARS-CoV-2 patients using single-cell ATAC-seq, which promotes the activation and differentiation of monocytes and SARS-CoV-specific effector and memory CD8+ T cells (You et al., 2021). Extensive analysis of chromatin accessibility and gene expression has identified a unique TH1-like pathogenic TH17 program shared by pTH17 cells and TH1 cells and discovered BACH2 as a novel regulator of TH17 pathogenicity (Thakore et al., 2022). Nevertheless, single-cell ATAC-seq requires nucleus extraction before generating libraries, and combining TCR and/or BCR immune repertoires can be challenging.
Single-cell expression quantitative trait loci (eQTLs)
Single-cell eQTL overcame the pre-existing limitations of eQTL with bulk RNA sequencing (RNA-seq) which represents average gene expression in selective cell types or tissues. By connecting the single-cell level of gene expression with disease-associated genomic variant information provided by GWASs (genome-wide association studies), researchers can better investigate eQTL effects based on the dynamic states of individual cells. Yazar et al. (2022) discovered 990 trans-eQTL effects outside the MHC locus and identified 305 loci contributing to autoimmune disease based on transcriptomic changes in specific cell types. To better depict the plasticity and diversity of memory T cells, Nathan et al. (2022) focused on continuous transcriptional landscapes when dissecting eQTL effects at single-cell resolution. They demonstrated that continuous multimodally defined cell states were responsible for most of the cis-eQTL effects, indicating the importance of cell-state context in understanding eQTL pathogenicity. Context-specific gene regulation was identified and related to immune disease-associated variants in CD4+ T cells in accordance with prior studies (Soskic et al., 2022).
COVID-19 AS A MODEL DISEASE
After a new coronavirus known as SARS-CoV-2 appeared in the Chinese city of Wuhan in 2019, COVID-19 rapidly spread worldwide and presented a severe danger to global public health (Hu et al., 2021). As of September 18, 2022, more than 637 million confirmed COVID-19 cases and 6.62 million deaths have been reported worldwide (https://ourworldindata.org/). Single-cell analysis has been an effective tool for investigating robust immune responses in COVID-19, capturing diverse, unbiased immune cell activation and differentiation. Since Spring 2020, there have been more than 200 publications on single-cell analysis of COVID-19 (Liu et al., 2022a). COVID-19 serves as a disease model that enables the improvement and development of single-cell analytic methods and technologies.
Based on a large set of data from patients, researchers have constructed a dynamic immune cell atlas of COVID-19. Large-scale single-cell RNA-seq of PBMCs, bronchoalveolar lavage fluid, and sputum in 196 COVID-19 patients revealed upregulated transcriptomic levels in interferon-stimulated genes (ISGs) of SARS-CoV-2-infected epithelial and immune cell types (Ren et al., 2021). Levels of inflammatory genes, such as S100A9 and S100A8, are also increased in the monocytes and megakaryocytes of patients, contributing to “cytokine storms”. Expansion of nonclassical monocytes, megakaryocyte-committed progenitors, and effector CD8+ T cells have been identified by single-cell multi-omics analysis, including immune profiling and CITE-seq (Stephenson et al., 2021).
Approximately 20% of COVID-19 patients have an exacerbation of severe diseases (Wu and McGoogan, 2020), followed by a hyperinflammatory response and immune dysfunction. Our group performed single-cell RNA-seq in patients with mild COVID-19, severe COVID-19, or severe influenza (Lee et al., 2020), finding distinctive upregulation of tumor necrosis factor (TNF)/interleukin (IL)-1β-driven inflammation and the type I interferon (IFN) response in PBMCs from severe COVID-19 patients compared to others, indicating a significant role in hyperinflammation in severe COVID-19. In severe COVID-19 patients with acute respiratory distress syndrome, the transition of plasmablasts to neutrophils with type I IFN-driven inflammatory signatures and human leukocyte antigen (HLA) class II downregulation of monocytes were observed by cellular trajectory analysis (Wilk et al., 2020). Single-nucleus RNA-seq of the lungs of patients who died from COVID-19 has shown dense infiltration of activated macrophages, transition of alveolar type 1 cells to type 2 cells, and CTHRC1+ pathological fibroblasts, exacerbating pulmonary fibrosis in COVID-19 (Melms et al., 2021).
COVID-19 has been the subject of many immunological studies, but the main issue is preparation for the next pandemic. It is essential to maintain a system of effective research on treating critically ill patients with inflammatory diseases whose vital signs are affected by unknown infectious diseases.
LARGE-SCALE COHORT-BASED APPROACHES TO CHRONIC INFLAMMATORY DISEASES
The intricate immune microenvironments of chronic inflammatory diseases have been difficult to study with existing methods, which are insufficient to distinguish the heterogenic transcriptional changes in myriad types of cells that participate in inflammation. However, with the advent of high-resolution technologies, unbiased single-cell analysis-based approaches can capture the simultaneous immune features of different cell types across inflamed tissues. In Table 1, we summarize the studies that have used single-cell genomics to investigate the pathogenesis of chronic inflammatory diseases (Table 1).
Table 1.
Current single-cell analysis-based studies on chronic inflammatory diseases
| Inflammatory disease | Species | Tissue | Single-cell analysis techniques | Reference |
|---|---|---|---|---|
| Ankylosing spondylitis | Human | PBMC | Transcriptome, epigenome | (Xu et al., 2021) |
| Human | PBMC | Transcriptome, surface proteome analysis | (Alber et al., 2022) | |
| Human | PBMC, SFMC | Transcriptome, immune repertoire analysis | (Simone et al., 2021) | |
| Inflammatory bowel diseases | Human | Intestinal epithelium, lamina propria from terminal ileum resections | Transcriptome | (Jaeger et al., 2021) |
| Human | Lamina propria from ileum, PBMC | Transcriptome | (Martin et al., 2019) | |
| Human | Rectum, PBMC | Immune repertoire analysis | (Boland et al., 2020) | |
| Human | Colonic biopsies | Transcriptome (both single cell and spatial) | (Garrido-Trigo et al., 2022) | |
| Multiple sclerosis | Human | Cerebrospinal fluid, PBMC | Transcriptome | (Schafflick et al., 2020) |
| Human | Post mortem brain tissue, PBMC | Transcriptome (both single cell and spatial), epigenome | (Kaufmann et al., 2021) | |
| Human | Post mortem brain tissue | Transcriptome | (Absinta et al., 2021) | |
| Mouse | CD11b+ myeloid cell from spleen | Transcriptome | (Lu et al., 2020) | |
| Mouse | MOG-specific CD4+ T cells | Transcriptome | (Krienke et al., 2021) | |
| Mouse | Memory-phenotype CD4+ T cells |
Transcriptome, immune repertoire analysis | (Cho et al., 2022) | |
| Chronic inflammatory skin diseases | Human | Skin biopsies | Transcriptome, epigenome | (Liu et al., 2022b) |
| Human | Skin biopsies | Transcriptome | (Nakamizo et al., 2021) | |
| Human | Skin biopsies | Transcriptome | (Kim et al., 2021) | |
| Human | Skin biopsies | Transcriptome (single cell and spatial) | (Schäbitz et al., 2022) | |
| Rheumatoid arthritis | Human | Synovial biopsies | Transcriptome | (Zhang et al., 2019) |
| Human | Synovial biopsies, PBMC | Transcriptome | (Wu et al., 2021) | |
| Human | Synovial fluid, PBMC | Immune repertoire analysis | (Argyriou et al., 2022) | |
| Human | Synovial biopsies | Transcriptome | (Edalat et al., 2022) | |
| Mouse | CD4+ T cells from lymph node, spleen | Immune repertoire analysis | (Ashouri et al., 2022) | |
| Systemic lupus erythematosus | Human | PBMC | Transcriptome | (Nehar-Belaid et al., 2020) |
| Human | PBMC | Transcriptome | (Guo et al., 2022) | |
| Human | PBMC | Transcriptome, eQTLs | (Perez et al., 2022) | |
| Mouse | B cells from spleen | Transcriptome, immune repertoire analysis | (Zheng et al., 2022) | |
| Primary Sjögren’s syndrome | Human | PBMC | Immune repertoire analysis | (Hou et al., 2022) |
| Human | PBMC | Transcriptome | (Hong et al., 2021) | |
| Human | Labial minor salivary gland biopsies | Transcriptome (both single cell and spatial) | (Nayar et al., 2022) | |
| Mouse | Submandibular gland | Transcriptome | (Horeth et al., 2021) | |
| Type 1 diabetes mellitus | Human | Pancreatic islet, PBMC | Transcriptome, epigenome | (Chiou et al., 2021) |
| Human | Pancreatic islet | Transcriptome | (Fasolino et al., 2022) | |
| Mouse | Pancreatic islet | Transcriptome | (Zakharov et al., 2020) |
PBMC, peripheral blood mononuclear cell; SFMC, synovial fluid mononuclear cell; MOG, myelin oligodendrocyte glycoprotein; eQTLs, expression quantitative trait loci.
Ankylosing spondylitis (AS)
AS is a chronic inflammatory disease featuring inflammation at the spine and sacroiliac joints with bone remodeling and ankylosis (Mauro et al., 2021). Applying single-cell RNA-seq and ATAC-seq on PBMCs from AS patients, NFKB expression and TNF signaling pathways have been shown to be upregulated in CD8+ T cells with abnormal accessibility of FOS, JUN, and JUNB (Xu et al., 2021). This finding suggests the role of effector CD8+ T cells in inflammatory cytokine secretion, including TNF-α. Single-cell CITE-seq has shown that TNFSF10 and IL-18Rα are overexpressed by CD16+ monocytes and cytotoxicity-related genes are upregulated in effector memory CD8+ T cells and NK cells (Alber et al., 2022). In immunological studies, Th17 cells have primarily been studied in terms of their pathological role in AS, but recent single-cell transcriptome or epigenome studies suggest major roles of other immune cell populations, including CD8+ T cells. Interactions between specific immune cell subpopulations need to be revealed to explain different features of inflammation in AS.
Inflammatory bowel diseases (IBDs)
IBD is characterized by chronic inflammatory processes of the digestive tract and comprises Crohn’s disease (CD) and ulcerative colitis (UC). CD is characterized by patchy transmural inflammation occurring at the distal parts of the small intestine (up to 80%) and colon (up to 30%), whereas UC is associated with persistent superficial inflammation in the colon (Neurath, 2019). Jaeger et al. (2021) observed an increase in activated TH17 cells compared to other T cell subsets in CD patients’ intraepithelial lymphocytes and lamina propria. GIMATS module, consisting of IgG plasma cells, inflammatory mononuclear phagocytes, activated T cells, and stromal cells was defined in a subgroup of large ileal CD cohorts by mapping the ileal lamina propria using single-cell analysis (Martin et al., 2019). Moreover, GIMATS module scores are increased in CD patients with anti-TNF resistance. In UC, pro-inflammatory resident memory CD8+ T cells have increased expression of EOMES, which is known as the master regulator of CD8+ effector and memory T cells (Boland et al., 2020).
Multiple sclerosis (MS)
MS is a chronic inflammatory neurological disease that causes demyelination and neurodegeneration. Various MS-associated features, including responses to antigens and IL-17-producing capacities, are widely recognized to be related to diverse innate and adaptive immune cells (Attfield et al., 2022). For example, cytotoxic-like helper T cells, follicular helper T cells (TFH), and B lineage cells are increased in the CSF of MS patients, supporting the efficacy of B-cell-depleting therapies (Schafflick et al., 2020). Because CNS (central nervous system)-resident inflammatory immune cells are mainly infiltrated from peripheral blood, Kaufmann et al. (2021) performed single-cell RNA-seq and CITE-seq on PBMCs from MS patients with a relapsing-remitting disease course (RRMS). They discovered unique CNS-homing CD4+ T cells that promote white matter demyelination and are situated in the cortical brain, causing progression to disability in RRMS. To profile the ongoing inflammatory environment of demyelinated white matter lesions, single-cell nucleus sequencing with magnetic resonance imaging identified ‘microglia inflamed in MS’ (MIMS), with complement component 1q (C1q) as a significant mediator of MIMS (Absinta et al., 2021).
Chronic inflammatory skin diseases
Chronic inflammatory skin lesions commonly occur in two immune diseases: atopic dermatitis (AD) and psoriasis vulgaris (PV). Different from allergic features of TH2-related AD, PV are defined by the key pathological mediator IL-23 and IL-17 secreted by TH17 and TH1 cells (Armstrong and Read, 2020). Performing single-cell RNA-seq and CITE-seq on the sorted CD45+ cells from AD and PV skin biopsies, transcriptomic differences in resident memory T cells (Trm) from AD and PV were discovered in accordance with previous findings (Liu et al., 2022b). Nakamizo et al. (2021) analyzed myeloid lineages in the skin of PS patients and identified a novel subtype of dendritic cells and its role, which is IL-1β and IL-23A producing CD14+ type 3 dendritic cells, promoting simultaneous inflammations in PS. Semimature dendritic cells and type 17 T cells expressing IL-23A and IL36G have been found in unbiasedly harvested samples of PS skin biopsies (Kim et al., 2021).
Rheumatoid arthritis (RA)
One of the most common autoimmune diseases worldwide, RA is a chronic inflammatory joint disease characterized by the infiltration of the synovial membrane of joints by immune cells, leading to bone erosion and cartilage degradation (Aletaha and Smolen, 2018; Ryu et al., 2019). Using specific immune-related cell types sorted from the synovial tissues of patients with RA and osteoarthritis, Zhang et al. (2019) performed a comprehensive analysis combining single-cell RNA-seq, FACS, mass cytometry, and bulk RNA-seq. They observed increased THY1+ HLA-DRAhi fibroblasts, IL1B+ inflammatory monocytes, PDCD1+ peripheral helper T cells (TPH), TFH, and ITGAX+ TBX21+ autoimmune-associated B cells in RA patients (Zhang et al., 2019). Because RA can be categorized by the presence of anticitrullinated-peptide antibodies (ACPAs), Wu et al. (2021) studied the differences in the immunopathogenesis of ACPA+ and ACPA- RA and discovered decreased HLA-DRB5+ B cells and upregulated CCL13, CCL18, and MMP3 expression by myeloid cells in ACPA- RA patients. Based on these discoveries, they reclassified RA subtypes as ACPA- with inflammatory myeloid cells and ACPA+ with lymphoid cells according to the key cell types that contributed to the disease. Similar to the prior study, single-cell RNA-seq of the synovial CD4+ T cells from both ACPA+ and ACPA- RA patients identified clonal expansion of GPR56+ LAG3+ CXCL13high TPH in ACPA+ RA, which was confirmed by flow cytometry (Argyriou et al., 2022). However, RA has not yet been reported to be caused by a single pathogenic subset of immune cells. A larger cohort including RA patients with heterogenous immunological features is needed to depict diverse clinical aspects.
Systemic lupus erythematosus (SLE)
SLE is a systemic autoimmune disease across the skin, joints, CNS, and kidneys. Antinuclear antibody production and the preponderance in childbearing-age females are well-known clinical characteristics of SLE (Kaul et al., 2016). Nehar-Belaid et al. (2020) discovered high expression of ISGs in most immune cell subtypes among the PBMCs of 33 young SLE patients. They also discovered associations between ISG-expressing cell populations and high disease activity in adult cohorts. Because dysfunctional Treg cells mainly contribute to various autoimmune diseases (Dominguez-Villar and Hafler, 2018), upregulated exhaustion-related gene expression, which is similar to ISG, has been identified in CCR7low CD74hi Treg subgroups from SLE patients (Guo et al., 2022). Perez et al. (2022) profiled PBMCs in a large SLE cohort and showed elevated ISG expression in monocytes, decline in naïve CD4+ T cells, and expanded repertoire-restricted cytotoxic GZMH+ CD8+ T cells. They also performed single-cell eQTL and discovered that elevated IFN levels can modify the genetic effects of cis-eQTLs, highlighting the pathological role of IFN in the SLE microenvironment.
Primary Sjögren’s syndrome (pSS)
pSS is a systemic autoimmune disease with dryness of mouth and eyes, fatigue, and joint pains, which are caused by infiltration of the exocrine glands by lymphocytes (Mariette and Criswell, 2018). Analyzing the PBMCs of pSS patients by single-cell immune profiling, Hou et al. (2022) found differentially expressed genes in multiple immune cell subtypes but did not capture any significant changes in the TCR repertoire. Expansion of CD4+ cytotoxic lymphocytes has been identified with chemokine receptor gene expression and upregulated type 1 IFN-related genes across immune cell types in the PBMCs of pSS patients (Hong et al., 2021). Nayar et al. (2022) performed single-cell RNA-seq and spatial transcriptomics to better understand the pathological role of tertiary lymphoid structures (TLS) in the minor salivary glands of pSS patients. They found that immunofibroblasts and stromal cells expressing ACKR3 and CD55 are key components of TLS formation in salivary glands, supporting immune cell migration and survivability.
Type 1 diabetes mellitus (T1DM)
T1DM is a chronic autoimmune disease with loss of the pancreatic islet β-cells leading to insulin deficiency and hyperglycemia (Katsarou et al., 2017). By performing single nuclei ATAC-seq to identify candidate cis-regulatory elements (cCREs), T1DM variants were discovered to be broadly enriched in T cell cCREs (Chiou et al., 2021).
ANIMAL MODEL-BASED APPROACHES TO CHRONIC INFLAMMATORY DISEASES
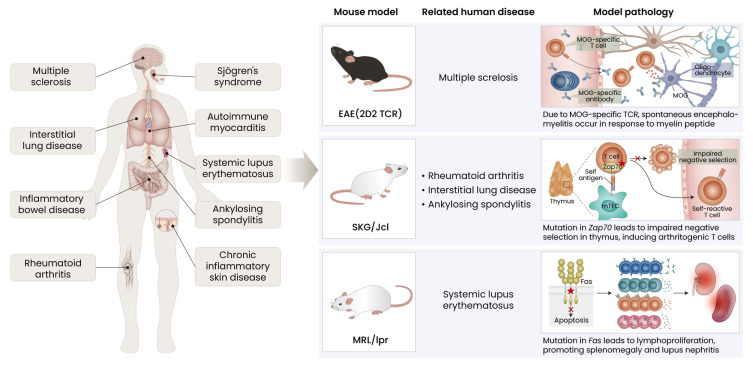
Despite its significance in clinical applications, using patient samples to study autoimmune and chronic inflammatory diseases has limitations. Firstly, unlike patients with infectious diseases, which begin with exposure to pathogens, it is difficult to know the starting point of chronic inflammatory diseases. Secondly, patients have genetically diverse backgrounds and studies have limitations in using cross-sectional samples. Thirdly, with successive developments in clinical approaches to autoimmune diseases, it has been difficult to observe certain cases that progress in severity without treatment. Animal model-based approaches have the advantages of carefully controlled studies, and it is easier to configure immune mechanisms by using certain genetically engineered mice. Because there are various animal models designed to exhibit immune responses similar to humans (Jeon and Choi, 2016), we are going to introduce widely used experimental models in the field of chronic inflammatory and autoimmune diseases (Fig. 1).
Fig. 1. Animal model-based approaches to chronic inflammatory diseases in human patients.
EAE, experimental autoimmune encephalomyelitis; TCR, T cell receptor; MOG, myelin oligodendrocyte glycoprotein; mTEC, medullary thymic epithelial cell.
Experimental autoimmune encephalomyelitis (EAE)
EAE is a commonly used mouse model for MS. However, the model is induced by myelin antigens with adjuvants, which is not similar to MS pathology of non-specified antigens (Constantinescu et al., 2011). Producing a conditionally Stat3-deleted mouse in the EAE model to discover the role of Stat3 in MS lesion-infiltrating monocytes, Lu et al. (2020) showed downregulated expression of genes involved in the innate immune response, antigen presentation and processing, cytokine, and inflammatory responses in the CD11b+ cells of Stat3-deleted mice. Krienke et al. (2021) applied nanoparticle-formulated 1 methylpseudouridine-modified messenger RNA (m1 Ψ mRNA) in the EAE model to induce immune tolerance by CD11c+ dendritic cells as a vaccine. Surprisingly, they observed a significant surge in effector Treg cells followed by upregulated expression of immunosuppressive genes, whereas the populations of inflammatory TH1, TH17, and TH1/17 cells were reduced compared to control groups.
SKG mouse model
With a recessive point mutation of ZAP-70 on the BALB/C genetic background, the SKG/Jcl mouse features spontaneous development of CD4+ T cell-mediated chronic autoimmune diseases, including RA (Sakaguchi et al., 2003). The SKG mouse model has also been widely used to study other types of chronic inflammatory diseases, including AS and interstitial lung disease (Jeong et al., 2017; Shiomi et al., 2014). Ashouri et al. (2022) established a Nur77-eGFP reporter SKG mouse (SKGNur mouse) to identify antigen-reactive T cells. With single-cell RNA-seq and immune profiling, they identified a cluster of naïve CD4+ T cells with upregulated TCR signaling (T.N4Nr4a1). T.N4Nr4a1 cells from the SKGNur mouse model have an increased number of especially early activated subgroups (Egr module) with a biased TCR variable beta gene repertoire, defining arthritogenic naïve CD4+ T cells.
MRL/LPR mouse model
MRL/LPR mice have defects in Fas expression, which interfere with negative selection during thymic development of T cells, triggering the escape of autoreactive T cells. Due to this, MRL/LPR mice are prone to lymphadenopathy and systemic autoimmune disease—not only SLE, but also other autoimmune diseases (Watanabe-Fukunaga et al., 1992). Zheng et al. (2022) generated Ezh2 conditional knockout in CD19+ cells from MRL/LPR mice and performed single-cell RNA-seq and BCR seq. They discovered downregulation of XBP1 in B cells, which led to decreased B cell development, autoantibody production, and improved glomerulonephritis.
Non-obese diabetic (NOD) mouse model
NOD/ShiLtJ mice have a genetic susceptibility related to the Ag7 MHCII allele and multiple non-MHCII genes affecting T-cell function, which leads to the destruction of insulin-producing β cells in islets by lymphocyte infiltration (De Riva et al., 2013). The NOD mouse is widely used not only for studying T1DM, but also Sjögren’s syndrome (Park et al., 2015). Defining the longitudinal stage of T1DM, memory CD4+ and cytotoxic CD8 T+ cells with proinflammatory-polarizing islet macrophages have been observed in the pancreatic islets of NOD mice (Zakharov et al., 2020).
MULTI-ORGAN ATLAS OF INFLAMMATION
Represented by RA and SLE, some chronic inflammatory diseases feature symptoms related to systemic inflammation in multiple organs. Approximately 25% of patients with autoimmune diseases develop additional autoimmune diseases (Cojocaru et al., 2010), indicating the need for a comprehensive multi-organ and multi-timepoint atlas of immune responses for chronic inflammatory diseases.
To distinguish tissue-specific features of immune cells, Dominguez Conde et al. (2022) performed single-cell RNA-seq and immune profiling on immune cells from up to 16 organs from healthy donors. They developed CellTypist, an extensive reference database of immune cell types and annotated 15 major populations and 43 minor subtypes of T cells, B cells, ILCs, and mononuclear phagocytes. In the context of T cells, they discovered resident memory T cells (TRM) that produce IFN-γ/IL-17A, though with tissue restriction of clonal expansion. Apart from immune cells, but related to our topic, Korsunsky et al. (2022) analyzed tissue-resident fibroblasts in four different chronic inflammatory diseases (RA, inflammatory bowel disease, interstitial lung disease, and Sjögren’s syndrome) and identified two distinct fibroblasts across all tissues: CXCL10+ CCL19+ immune-interacting and SPARC+ COL3A1+ vascular-interacting fibroblasts, which expanded under inflammatory conditions. Multi-organ single-cell transcriptomic atlas studies have been performed by Tabula Muris Consortium et al. (2018), Tabula Sapiens Consortium et al. (2022), and Gökcen Eraslan et al. (2022) that also feature immune cells and immune-related cells, such as fibroblasts.
However, few studies are available to show the application of single-cell analysis in a mouse and human multi-organ atlas targeting immune cells. Our group is currently constructing one of the single-cell level atlases of a chronic arthritis model.
CONCLUDING REMARKS
Single-cell genomics data associated with immunological diseases is primarily focused on uncovering rare or unique cell types, which are defined by transcriptome. The next step will be to determine their pathological roles in inflammatory diseases. Though genetically engineered animal models may provide confirmation of the pathological roles of newly discovered cell types, cohort studies including relevant patient specimens will provide real-world evidence to develop new therapeutic strategies. Larger size, longitudinal sampling from initial diagnosis, and multi-organ sampling would be required to establish a human atlas of chronic immune diseases. Integrating transcriptome, TCR/BCR, epigenetic, and proteomic data will be increasingly important to develop answers to immunological questions in inflammatory diseases.
ACKNOWLEDGMENTS
This work was supported by a grant from the National Research Foundation of Korea (2022R1A4A3034038, 2022M3A9D3016848, and NRF-2022R1C1C1012634).
Footnotes
AUTHOR CONTRIBUTIONS
S.J. and J.S.L. conceived idea, wrote the manuscript, and secured funding.
CONFLICT OF INTEREST
The authors have no potential conflicts of interest to disclose.
REFERENCES
- Absinta M., Maric D., Gharagozloo M., Garton T., Smith M.D., Jin J., Fitzgerald K.C., Song A., Liu P., Lin J.P., et al. A lymphocyte-microglia-astrocyte axis in chronic active multiple sclerosis. Nature. 2021;597:709–714. doi: 10.1038/s41586-021-03892-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alber S., Kumar S., Liu J., Huang Z.M., Paez D., Hong J., Chang H.W., Bhutani T., Gensler L.S., Liao W. Single cell transcriptome and surface epitope analysis of ankylosing spondylitis facilitates disease classification by machine learning. Front. Immunol. 2022;13:838636. doi: 10.3389/fimmu.2022.838636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aletaha D., Smolen J.S. Diagnosis and management of rheumatoid arthritis: a review. JAMA. 2018;320:1360–1372. doi: 10.1001/jama.2018.13103. [DOI] [PubMed] [Google Scholar]
- Argyriou A., Wadsworth M.H., 2nd, Lendvai A., 2nd, Christensen S.M., 2nd, Hensvold A.H., 2nd, Gerstner C., 2nd, van Vollenhoven A., 2nd, Kravarik K., 2nd, Winkler A., 2nd, Malmstrom V., 2nd, et al. Single cell sequencing identifies clonally expanded synovial CD4(+) T(PH) cells expressing GPR56 in rheumatoid arthritis. Nat. Commun. 2022;13:4046. doi: 10.1038/s41467-022-31519-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong A.W., Read C. Pathophysiology, clinical presentation, and treatment of psoriasis: a review. JAMA. 2020;323:1945–1960. doi: 10.1001/jama.2020.4006. [DOI] [PubMed] [Google Scholar]
- Ashouri J., McCarthy E., Yu S., Perlmutter N., Lin C., DeRisi J., Ye C.J., Weiss A. Naïve arthritogenic SKG T cells have a defect in anergy and a repertoire pruned by superantigen. BioRxiv. 2022 doi: 10.1101/2022.01.13.476250. https://doi.org/10.1101/2022.01.13.476250. [DOI] [Google Scholar]
- Attfield K.E., Jensen L.T., Kaufmann M., Friese M.A., Fugger L. The immunology of multiple sclerosis. Nat. Rev. Immunol. 2022;22:734–750. doi: 10.1038/s41577-022-00718-z. [DOI] [PubMed] [Google Scholar]
- Boland B.S., He Z., Tsai M.S., Olvera J.G., Omilusik K.D., Duong H.G., Kim E.S., Limary A.E., Jin W., Milner J.J., et al. Heterogeneity and clonal relationships of adaptive immune cells in ulcerative colitis revealed by single-cell analyses. Sci. Immunol. 2020;5:eabb4432. doi: 10.1126/sciimmunol.abb4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro J.D., Wu B., Litzenburger U.M., Ruff D., Gonzales M.L., Snyder M.P., Chang H.Y., Greenleaf W.J. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature. 2015;523:486–490. doi: 10.1038/nature14590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou J., Geusz R.J., Okino M.L., Han J.Y., Miller M., Melton R., Beebe E., Benaglio P., Huang S., Korgaonkar K., et al. Interpreting type 1 diabetes risk with genetics and single-cell epigenomics. Nature. 2021;594:398–402. doi: 10.1038/s41586-021-03552-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho M.Z., Lee H.G., Yoon J.W., Kim G.R., Koo J.H., Taneja R., Edelson B.T., Lee Y.J., Choi J.M. Bystander memory-phenotype conventional CD4+T cells exacerbating autoimmune neuroinflammation. BioRxiv. 2022 doi: 10.1101/2022.06.17.496529. https://doi.org/10.1101/2022.06.17.496529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cojocaru M., Cojocaru I.M., Silosi I. Multiple autoimmune syndrome. Maedica (Bucur.) 2010;5:132–134. doi: 10.1007/s40619-018-0394-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantinescu C.S., Farooqi N., O'Brien K., Gran B. Experimental autoimmune encephalomyelitis (EAE) as a model for multiple sclerosis (MS) Br. J. Pharmacol. 2011;164:1079–1106. doi: 10.1111/j.1476-5381.2011.01302.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusanovich D.A., Daza R., Adey A., Pliner H.A., Christiansen L., Gunderson K.L., Steemers F.J., Trapnell C., Shendure J. Multiplex single cell profiling of chromatin accessibility by combinatorial cellular indexing. Science. 2015;348:910–914. doi: 10.1126/science.aab1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Riva A., Varley M.C., Bluck L.J., Cooke A., Deery M.J., Busch R. Accelerated turnover of MHC class II molecules in nonobese diabetic mice is developmentally and environmentally regulated in vivo and dispensable for autoimmunity. J. Immunol. 2013;190:5961–5971. doi: 10.4049/jimmunol.1300551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez Conde C., Xu C., Jarvis L.B., Rainbow D.B., Wells S.B., Gomes T., Howlett S.K., Suchanek O., Polanski K., King H.W., et al. Cross-tissue immune cell analysis reveals tissue-specific features in humans. Science. 2022;376:eabl5197. doi: 10.1101/2021.04.28.441762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Villar M., Hafler D.A. Regulatory T cells in autoimmune disease. Nat. Immunol. 2018;19:665–673. doi: 10.1038/s41590-018-0120-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edalat S.G., Gerber R., Houtman M., Kuret T., Ižanc N., Micheroli R., Burki K., Burja B., Pauli C., Rotar Ž., et al. A comprehensive single-cell atlas of freshly dissociated human synovium in inflammatory arthritis with an optimized dissociation protocol for prospective fresh synovial biopsy collection. BioRxiv. 2022 doi: 10.1101/2022.06.01.493823. https://doi.org/10.1101/2022.06.01.493823. [DOI] [Google Scholar]
- Eraslan G., Drokhlyansky E., Anand S., Fiskin E., Subramanian A., Slyper M., Wang J., Van Wittenberghe N., Rouhana J.M., Waldman J., et al. Single-nucleus cross-tissue molecular reference maps toward understanding disease gene function. Science. 2022;376:eabl4290. doi: 10.1126/science.abl4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasolino M., Schwartz G.W., Patil A.R., Mongia A., Golson M.L., Wang Y.J., Morgan A., Liu C., Schug J., Liu J., et al. Single-cell multi-omics analysis of human pancreatic islets reveals novel cellular states in type 1 diabetes. Nat. Metab. 2022;4:284–299. doi: 10.1038/s42255-022-00531-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido-Trigo A., Corraliza A.M., Veny M., Dotti I., Melon-Ardanaz E., Rill A., Crowell H.L., Corbí Á., Gudiño V., Esteller M., et al. Macrophage and neutrophil heterogeneity at single-cell spatial resolution in inflammatory bowel disease. BioRxiv. 2022 doi: 10.1101/2022.11.28.518139. https://doi.org/10.1101/2022.11.28.518139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C., Liu Q., Zong D., Zhang W., Zuo Z., Yu Q., Sha Q., Zhu L., Gao X., Fang J., et al. Single-cell transcriptome profiling and chromatin accessibility reveal an exhausted regulatory CD4+ T cell subset in systemic lupus erythematosus. Cell Rep. 2022;41:111606. doi: 10.1016/j.celrep.2022.111606. [DOI] [PubMed] [Google Scholar]
- Hong X., Meng S., Tang D., Wang T., Ding L., Yu H., Li H., Liu D., Dai Y., Yang M. Single-cell RNA sequencing reveals the expansion of cytotoxic CD4+ T lymphocytes and a landscape of immune cells in primary Sjögren's syndrome. Front. Immunol. 2021;11:594658. doi: 10.3389/fimmu.2020.594658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horeth E., Oyelakin A., Song E.C., Che M., Bard J., Min S., Kiripolsky J., Kramer J.M., Sinha S., Romano R.A. Transcriptomic and single-cell analysis reveals regulatory networks and cellular heterogeneity in mouse primary Sjögren's syndrome salivary glands. Front. Immunol. 2021;12:729040. doi: 10.3389/fimmu.2021.729040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou X., Hong X., Ou M., Meng S., Wang T., Liao S., He J., Yu H., Liu L., Yin L., et al. Analysis of gene expression and TCR/B cell receptor profiling of immune cells in primary Sjögren's syndrome by single-cell sequencing. J. Immunol. 2022;209:238–249. doi: 10.4049/jimmunol.2100803. [DOI] [PubMed] [Google Scholar]
- Hu B., Guo H., Zhou P., Shi Z.L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2021;19:141–154. doi: 10.1038/s41579-020-00459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger N., Gamini R., Cella M., Schettini J.L., Bugatti M., Zhao S., Rosadini C.V., Esaulova E., Di Luccia B., Kinnett B., et al. Single-cell analyses of Crohn's disease tissues reveal intestinal intraepithelial T cells heterogeneity and altered subset distributions. Nat. Commun. 2021;12:1921. doi: 10.1038/s41467-021-22164-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon Y.H., Choi Y.S. Follicular helper T (Tfh) cells in autoimmune diseases and allograft rejection. Immune Netw. 2016;16:219–232. doi: 10.4110/in.2016.16.4.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong H., Bae E.K., Kim H., Eun Y.H., Kim I.Y., Kim H., Lee J., Jeon C.H., Koh E.M., Cha H.S. Estrogen attenuates the spondyloarthritis manifestations of the SKG arthritis model. Arthritis Res. Ther. 2017;19:198. doi: 10.1186/s13075-017-1407-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsarou A., Gudbjornsdottir S., Rawshani A., Dabelea D., Bonifacio E., Anderson B.J., Jacobsen L.M., Schatz D.A., Lernmark A. Type 1 diabetes mellitus. Nat. Rev. Dis. Primers. 2017;3:17016. doi: 10.1038/nrdp.2017.16. [DOI] [PubMed] [Google Scholar]
- Kaufmann M., Evans H., Schaupp A.L., Engler J.B., Kaur G., Willing A., Kursawe N., Schubert C., Attfield K.E., Fugger L., et al. Identifying CNS-colonizing T cells as potential therapeutic targets to prevent progression of multiple sclerosis. Med (N. Y.) 2021;2:296–312.e8. doi: 10.1016/j.medj.2021.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaul A., Gordon C., Crow M.K., Touma Z., Urowitz M.B., van Vollenhoven R., Ruiz-Irastorza G., Hughes G. Systemic lupus erythematosus. Nat. Rev. Dis. Primers. 2016;2:16039. doi: 10.1038/nrdp.2016.39. [DOI] [PubMed] [Google Scholar]
- Kim J., Lee J., Kim H.J., Kameyama N., Nazarian R., Der E., Cohen S., Guttman-Yassky E., Putterman C., Krueger J.G. Single-cell transcriptomics applied to emigrating cells from psoriasis elucidate pathogenic versus regulatory immune cell subsets. J. Allergy Clin. Immunol. 2021;148:1281–1292. doi: 10.1016/j.jaci.2021.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korsunsky I., Wei K., Pohin M., Kim E.Y., Barone F., Major T., Taylor E., Ravindran R., Kemble S., Watts G.F.M., et al. Cross-tissue, single-cell stromal atlas identifies shared pathological fibroblast phenotypes in four chronic inflammatory diseases. Med (N. Y.) 2022;3:481–518.e14. doi: 10.1016/j.medj.2022.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer K.J., Wilfong E.M., Voss K., Barone S.M., Shiakolas A.R., Raju N., Roe C.E., Suryadevara N., Walker L.M., Wall S.C., et al. Single-cell profiling of the antigen-specific response to BNT162b2 SARS-CoV-2 RNA vaccine. Nat. Commun. 2022;13:3466. doi: 10.1038/s41467-022-31142-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krienke C., Kolb L., Diken E., Streuber M., Kirchhoff S., Bukur T., Akilli-Ozturk O., Kranz L.M., Berger H., Petschenka J., et al. A noninflammatory mRNA vaccine for treatment of experimental autoimmune encephalomyelitis. Science. 2021;371:145–153. doi: 10.1126/science.aay3638. [DOI] [PubMed] [Google Scholar]
- Lee J.S., Park S., Jeong H.W., Ahn J.Y., Choi S.J., Lee H., Choi B., Nam S.K., Sa M., Kwon J.S., et al. Immunophenotyping of COVID-19 and influenza highlights the role of type I interferons in development of severe COVID-19. Sci. Immunol. 2020;5:eabd1554. doi: 10.1126/sciimmunol.abd1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Jia J., Dai Y., Chen W., Pei G., Yan Q., Zhao Z. Delineating COVID-19 immunological features using single-cell RNA sequencing. Innovation (Camb.) 2022a;3:100289. doi: 10.1016/j.xinn.2022.100289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Wang H., Taylor M., Cook C., Martinez-Berdeja A., North J.P., Harirchian P., Hailer A.A., Zhao Z., Ghadially R., et al. Classification of human chronic inflammatory skin disease based on single-cell immune profiling. Sci. Immunol. 2022b;7:eabl9165. doi: 10.1126/sciimmunol.abl9165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H.C., Kim S., Steelman A.J., Tracy K., Zhou B., Michaud D., Hillhouse A.E., Konganti K., Li J. STAT3 signaling in myeloid cells promotes pathogenic myelin-specific T cell differentiation and autoimmune demyelination. Proc. Natl. Acad. Sci. U. S. A. 2020;117:5430–5441. doi: 10.1073/pnas.1913997117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariette X., Criswell L.A. Primary Sjogren's syndrome. N. Engl. J. Med. 2018;378:931–939. doi: 10.1056/NEJMcp1702514. [DOI] [PubMed] [Google Scholar]
- Martin J.C., Chang C., Boschetti G., Ungaro R., Giri M., Grout J.A., Gettler K., Chuang L.S., Nayar S., Greenstein A.J., et al. Single-cell analysis of Crohn's disease lesions identifies a pathogenic cellular module associated with resistance to anti-TNF therapy. Cell. 2019;178:1493–1508.e20. doi: 10.1016/j.cell.2019.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauro D., Thomas R., Guggino G., Lories R., Brown M.A., Ciccia F. Ankylosing spondylitis: an autoimmune or autoinflammatory disease? Nat. Rev. Rheumatol. 2021;17:387–404. doi: 10.1038/s41584-021-00625-y. [DOI] [PubMed] [Google Scholar]
- Melms J.C., Biermann J., Huang H., Wang Y., Nair A., Tagore S., Katsyv I., Rendeiro A.F., Amin A.D., Schapiro D., et al. A molecular single-cell lung atlas of lethal COVID-19. Nature. 2021;595:114–119. doi: 10.1038/s41586-021-03569-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minnoye L., Marinov G.K., Krausgruber T., Pan L., Marand A.P., Secchia S., Greenleaf W.J., Furlong E.E.M., Zhao K., Schmitz R.J., et al. Chromatin accessibility profiling methods. Nat. Rev. Methods Primers. 2021;1:10. doi: 10.1038/s43586-020-00008-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamizo S., Dutertre C.A., Khalilnezhad A., Zhang X.M., Lim S., Lum J., Koh G., Foong C., Yong P.J.A., Tan K.J., et al. Single-cell analysis of human skin identifies CD14+ type 3 dendritic cells co-producing IL1B and IL23A in psoriasis. J. Exp. Med. 2021;218:e20202345. doi: 10.1084/jem.20202345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan A., Asgari S., Ishigaki K., Valencia C., Amariuta T., Luo Y., Beynor J.I., Baglaenko Y., Suliman S., Price A.L., et al. Single-cell eQTL models reveal dynamic T cell state dependence of disease loci. Nature. 2022;606:120–128. doi: 10.1038/s41586-022-04713-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayar S., Turner J.D., Asam S., Fennell E., Pugh M., Colfrancesco S., Berardicurti O., Smith C.G., Flint J., Teodosio A., et al. A cellular and spatial map of salivary glands at single cell resolution reveals the functional basis of tertiary lymphoid structure formation in Sjogren's syndrome. BioRxiv. 2022 doi: 10.1101/2022.11.03.514908. https://doi.org/10.1101/2022.11.03.514908. [DOI] [Google Scholar]
- Nehar-Belaid D., Hong S., Marches R., Chen G., Bolisetty M., Baisch J., Walters L., Punaro M., Rossi R.J., Chung C.H., et al. Mapping systemic lupus erythematosus heterogeneity at the single-cell level. Nat. Immunol. 2020;21:1094–1106. doi: 10.1038/s41590-020-0743-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neurath M.F. Targeting immune cell circuits and trafficking in inflammatory bowel disease. Nat. Immunol. 2019;20:970–979. doi: 10.1038/s41590-019-0415-0. [DOI] [PubMed] [Google Scholar]
- Pai J.A., Satpathy A.T. High-throughput and single-cell T cell receptor sequencing technologies. Nat. Methods. 2021;18:881–892. doi: 10.1038/s41592-021-01201-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y.S., Gauna A.E., Cha S. Mouse models of primary Sjogren's syndrome. Curr. Pharm. Des. 2015;21:2350–2364. doi: 10.2174/1381612821666150316120024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez R.K., Gordon M.G., Subramaniam M., Kim M.C., Hartoularos G.C., Targ S., Sun Y., Ogorodnikov A., Bueno R., Lu A., et al. Single-cell RNA-seq reveals cell type-specific molecular and genetic associations to lupus. Science. 2022;376:eabf1970. doi: 10.1126/science.abf1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perfetto S.P., Chattopadhyay P.K., Roederer M. Seventeen-colour flow cytometry: unravelling the immune system. Nat. Rev. Immunol. 2004;4:648–655. doi: 10.1038/nri1416. [DOI] [PubMed] [Google Scholar]
- Piper C., Hainstock E., Yin-Yuan C., Chen Y., Khatun A., Kasmani M.Y., Evans J., Miller J.A., Gorski J., Cui W., et al. Single-cell immune profiling reveals a developmentally distinct CD4+ GM-CSF+ T-cell lineage that induces GI tract GVHD. Blood Adv. 2022;6:2791–2804. doi: 10.1182/bloodadvances.2021006084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regev A., Teichmann S.A., Lander E.S., Amit I., Benoist C., Birney E., Bodenmiller B., Campbell P., Carninci P., Clatworthy M., et al. The human cell atlas. Elife. 2017;6:e27041. doi: 10.7554/eLife.27041.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X., Wen W., Fan X., Hou W., Su B., Cai P., Li J., Liu Y., Tang F., Zhang F., et al. COVID-19 immune features revealed by a large-scale single-cell transcriptome atlas. Cell. 2021;184:1895–1913.e19. doi: 10.1016/j.cell.2021.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu H., Kim J., Kim D., Lee J.E., Chung Y. Cellular and molecular links between autoimmunity and lipid metabolism. Mol. Cells. 2019;42:747–754. doi: 10.14348/molcells.2019.0196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi N., Takahashi T., Hata H., Nomura T., Tagami T., Yamazaki S., Sakihama T., Matsutani T., Negishi I., Nakatsuru S., et al. Altered thymic T-cell selection due to a mutation of the ZAP-70 gene causes autoimmune arthritis in mice. Nature. 2003;426:454–460. doi: 10.1038/nature02119. [DOI] [PubMed] [Google Scholar]
- Schäbitz A., Hillig C., Mubarak M., Jargosch M., Farnoud A., Scala E., Kurzen N., Pilz A.C., Bhalla N., Thomas J., et al. Spatial transcriptomics landscape of lesions from non-communicable inflammatory skin diseases. Nat. Commun. 2022;13:7729. doi: 10.1038/s41467-022-35319-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafflick D., Xu C.A., Hartlehnert M., Cole M., Schulte-Mecklenbeck A., Lautwein T., Wolbert J., Heming M., Meuth S.G., Kuhlmann T., et al. Integrated single cell analysis of blood and cerebrospinal fluid leukocytes in multiple sclerosis. Nat. Commun. 2020;11:247. doi: 10.1038/s41467-019-14118-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz D.G., Ji Y. Recombination centres and the orchestration of V(D)J recombination. Nat. Rev. Immunol. 2011;11:251–263. doi: 10.1038/nri2941. [DOI] [PubMed] [Google Scholar]
- Setliff I., Shiakolas A.R., Pilewski K.A., Murji A.A., Mapengo R.E., Janowska K., Richardson S., Oosthuysen C., Raju N., Ronsard L., et al. High-throughput mapping of B cell receptor sequences to antigen specificity. Cell. 2019;179:1636–1646.e15. doi: 10.1016/j.cell.2019.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiomi A., Usui T., Ishikawa Y., Shimizu M., Murakami K., Mimori T. GM-CSF but not IL-17 is critical for the development of severe interstitial lung disease in SKG mice. J. Immunol. 2014;193:849–859. doi: 10.4049/jimmunol.1303255. [DOI] [PubMed] [Google Scholar]
- Simone D., Penkava F., Ridley A., Sansom S., Al-Mossawi M.H., Bowness P. Single cell analysis of spondyloarthritis regulatory T cells identifies distinct synovial gene expression patterns and clonal fates. Commun. Biol. 2021;4:1395. doi: 10.1038/s42003-021-02931-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soskic B., Cano-Gamez E., Smyth D.J., Ambridge K., Ke Z., Matte J.C., Bossini-Castillo L., Kaplanis J., Ramirez-Navarro L., Lorenc A., et al. Immune disease risk variants regulate gene expression dynamics during CD4(+) T cell activation. Nat. Genet. 2022;54:817–826. doi: 10.1038/s41588-022-01066-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson E., Reynolds G., Botting R.A., Calero-Nieto F.J., Morgan M.D., Tuong Z.K., Bach K., Sungnak W., Worlock K.B., Yoshida M., et al. Single-cell multi-omics analysis of the immune response in COVID-19. Nat. Med. 2021;27:904–916. doi: 10.1038/s41591-021-01329-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeckius M., Hafemeister C., Stephenson W., Houck-Loomis B., Chattopadhyay P.K., Swerdlow H., Satija R., Smibert P. Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods. 2017;14:865–868. doi: 10.1038/nmeth.4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., 3rd, Hao Y., 3rd, Stoeckius M., 3rd, Smibert P., 3rd, Satija R., 3rd Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabula Muris Consortium, Overall coordination, Logistical coordination, Organ collection and processing, Library preparation and sequencing, Computational data analysis, Cell type annotation, Writing group, Supplemental text writing group, and Principal investigators, author. Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature. 2018;562:367–372. doi: 10.1038/s41586-018-0590-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones R.C., Karkanias J., Krasnow M.A., Pisco A.O., Quake S.R., Salzman J., Yosef N., Bulthaup B., Brown P., et al. Tabula Sapiens Consortium*, author. The Tabula Sapiens: a multiple-organ, single-cell transcriptomic atlas of humans. Science. 2022;376:eabl4896. doi: 10.1126/science.abl4896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakore P.I., Schnell A., Zhao M., Huang L., Hou Y., Christian E., Zaghouani S., Wang C., Singh V., Ma S., et al. The chromatin landscape of Th17 cells reveals mechanisms of diversification of regulatory and pro-inflammatory states. BioRxiv. 2022 doi: 10.1101/2022.02.26.482041. https://doi.org/10.1101/2022.02.26.482041. [DOI] [Google Scholar]
- Turner B.M. Defining an epigenetic code. Nat. Cell Biol. 2007;9:2–6. doi: 10.1038/ncb0107-2. [DOI] [PubMed] [Google Scholar]
- Unterman A., Sumida T.S., Nouri N., Yan X., Zhao A.Y., Gasque V., Schupp J.C., Asashima H., Liu Y., Cosme C., Jr., et al. Single-cell multi-omics reveals dyssynchrony of the innate and adaptive immune system in progressive COVID-19. Nat. Commun. 2022;13:440. doi: 10.1038/s41467-021-27716-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe-Fukunaga R., Brannan C.I., Copeland N.G., Jenkins N.A., Nagata S. Lymphoproliferation disorder in mice explained by defects in Fas antigen that mediates apoptosis. Nature. 1992;356:314–317. doi: 10.1038/356314a0. [DOI] [PubMed] [Google Scholar]
- Wilk A.J., Rustagi A., Zhao N.Q., Roque J., Martinez-Colon G.J., McKechnie J.L., Ivison G.T., Ranganath T., Vergara R., Hollis T., et al. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat. Med. 2020;26:1070–1076. doi: 10.1038/s41591-020-0944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Liu Y., Jin S., Wang M., Jiao Y., Yang B., Lu X., Ji X., Fei Y., Yang H., et al. Single-cell sequencing of immune cells from anticitrullinated peptide antibody positive and negative rheumatoid arthritis. Nat. Commun. 2021;12:4977. doi: 10.1038/s41467-021-25246-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z., McGoogan J.M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) OUTBREAK in China: summary of a report of 72 314 cases from the Chinese Center for Disease Control and Prevention. JAMA. 2020;323:1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- Xu H., Yu H., Liu L., Wu H., Zhang C., Cai W., Hong X., Liu D., Tang D., Dai Y. Integrative single-cell RNA-seq and ATAC-seq analysis of peripheral mononuclear cells in patients with ankylosing spondylitis. Front. Immunol. 2021;12:760381. doi: 10.3389/fimmu.2021.760381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazar S., Alquicira-Hernandez J., Wing K., Senabouth A., Gordon M.G., Andersen S., Lu Q., Rowson A., Taylor T.R.P., Clarke L., et al. Single-cell eQTL mapping identifies cell type-specific genetic control of autoimmune disease. Science. 2022;376:eabf3041. doi: 10.1126/science.abf3041. [DOI] [PubMed] [Google Scholar]
- You M., Chen L., Zhang D., Zhao P., Chen Z., Qin E.Q., Gao Y., Davis M.M., Yang P. Single-cell epigenomic landscape of peripheral immune cells reveals establishment of trained immunity in individuals convalescing from COVID-19. Nat. Cell Biol. 2021;23:620–630. doi: 10.1038/s41556-021-00690-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakharov P.N., Hu H., Wan X., Unanue E.R. Single-cell RNA sequencing of murine islets shows high cellular complexity at all stages of autoimmune diabetes. J. Exp. Med. 2020;217:e20192362. doi: 10.1084/jem.20192362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Wei K., Slowikowski K., Fonseka C.Y., Rao D.A., Kelly S., Goodman S.M., Tabechian D., Hughes L.B., Salomon-Escoto K., et al. Defining inflammatory cell states in rheumatoid arthritis joint synovial tissues by integrating single-cell transcriptomics and mass cytometry. Nat. Immunol. 2019;20:928–942. doi: 10.1038/s41590-019-0378-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.Y., Wang X.M., Xing X., Xu Z., Zhang C., Song J.W., Fan X., Xia P., Fu J.L., Wang S.Y., et al. Single-cell landscape of immunological responses in patients with COVID-19. Nat. Immunol. 2020;21:1107–1118. doi: 10.1038/s41590-020-0762-x. [DOI] [PubMed] [Google Scholar]
- Zhao X.N., You Y., Cui X.M., Gao H.X., Wang G.L., Zhang S.B., Yao L., Duan L.J., Zhu K.L., Wang Y.L., et al. Single-cell immune profiling reveals distinct immune response in asymptomatic COVID-19 patients. Signal Transduct. Target. Ther. 2021;6:342. doi: 10.1038/s41392-021-00753-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X., Dozmorov M.G., Strohlein C.E., Bastacky S., Sawalha A.H. Ezh2 knockout in B cells impairs plasmablast differentiation and ameliorates lupus-like disease in MRL/lpr mice. BioRxiv. 2022 doi: 10.1101/2022.07.21.500990. https://doi.org/10.1101/2022.07.21.500990. [DOI] [PMC free article] [PubMed] [Google Scholar]