Abstract
Ovaries are central to development, fertility, and reproduction of women. A particularly interesting feature of ovaries is their accelerated aging compared to other tissues, leading to loss of function far before other organs senesce. The limited pool of ovarian follicles is generated before birth and once exhausted, menopause will inevitably commence around the age of 50 years marking the end of fertility. Yet, there are reports suggesting the presence of germline stem cells and neo-oogenesis in adult human ovaries. These observations have fueled a long debate, created experimental fertility treatments, and opened business opportunities. Our recent analysis of cell types in the ovarian cortex of women of fertile age could not find evidence of germline stem cells. Like before, our work has been met with critique suggesting methodological shortcomings. We agree that excellence starts with methods and welcome discussion on the pros and cons of different protocols. In this commentary, we discuss the recent re-interpretation of our work.
In 2020, we presented the first single-cell analysis of the adult human ovarian cortex.1 We subjected over 24 000 cells isolated from 7 healthy patients of reproductive age to single-cell RNA sequencing (scRNA-seq), and identified 6 main cell types: oocytes, granulosa cells, immune cells, endothelial cells, perivascular cells, and stromal cells. Our data is in agreement with independent single-cell datasets describing granulosa cells, immune cells, vascular system cells, and stroma/theca cells in adult human ovarian medulla,2 adult non-human primate ovaries,3 and adult mouse ovaries.4 None of these scRNA-seq studies has reported germline stem cells although these cells should be present in various species.5 Germline stem cells (also called oogonial stem cells [OSCs], egg precursor cells and non-oocyte germ cells) in adult human ovaries were first reported a decade ago together with a protocol for their antibody (Ab) based isolation by fluorescence- or magnetic-activated cell sorting (FACS or MACS) targeting an extracellular domain of DDX4.6,7 Following these protocols, we isolated such putative OSCs from human ovaries (DDX4 Ab+ cells) and carried out scRNA-seq and flow cytometry analysis, which revealed these cells to be perivascular cells.1 The findings of our scRNA-seq study are in agreement with many others who have not been able to validate human ovarian DDX4 Ab+ cells as OSCs,8-10 or simply have not found germline stem cells among thousands of analyzed cells.2-4 Considering the positive publication bias in research, we find it likely that many more groups have similarly failed to identify OSCs but have not reported it.
The age of women at first childbirth is increasing globally, and many will discover that their fertility is already declining by the time pregnancy is desired. The treatment of sub- and infertility in aging women is a growing challenge in reproductive medicine, and also presents a lucrative business opportunity. For example, the DDX4 Ab+ cells have already been used in experimental fertility treatments targeting women with low-quality oocytes.11 The autologous germline mitochondrial energy transfer (AUGMENT) technology by Ovascience promised to boost the quality of oocytes by injection of extra mitochondria extracted from “egg precursor cells” (a.k.a. OSCs).12 The first randomized controlled study assessing the AUGMENT treatment was discontinued because the protocol led to abnormal embryo development and significantly reduced blastocyst rates.13 In this triple-blinded, randomized controlled study, the OSCs were isolated by Ovascience following the protocols established by its founders.7,13
Recent criticism presented by Alberico et al regarding our study on cell types in adult human ovarian cortex suggests that we missed OSCs due to problems with (1) cell sorting, (2) numbers of cells, and (3) bioinformatic analyses.14 Further arguments presented relied on numbers of papers published on the topic, and on the importance of OSCs for fertility treatments. We agree that the clinical implications of stem cells generating functional new oocytes in humans are significant, and for that reason the existence of these cells must be robustly established, and their function and biological significance thoroughly characterized in pre-clinical settings before further clinical experiments are carried out.
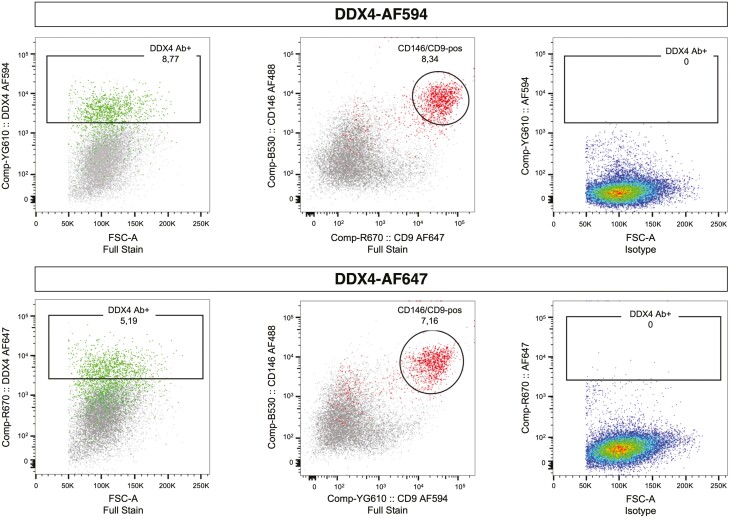
It was proposed that our data identifying DDX4 Ab+ cells as perivascular cells were a result of accidental sorting of vascular system cells due to autofluorescence from collagen and elastin in the red channel.14 Evidence for this was provided by experiments where a population of cells was sorted from cow ovaries based on autofluorescence. Then, the authors permeabilized the cells and showed them to be positive for cluster of differentiation 31 (CD31) and smooth muscle actin (SMA).14 Importantly, CD31 is a marker for endothelial cells, and we did not have significant numbers of endothelial cells in the DDX4 Ab+ population.1 In addition, collagen and elastin are not specific to perivascular cells in human ovaries.15 Finally, our antibody validation experiments that compared 3 DDX4 antibodies in parallel1 already showed that only the DDX4 Ab specifically recommended for OSC isolation6,7 separated a cell population, thereby contra-indicating a detection based on mere autofluorescence. Here, we set up new experiments to show that the DDX4 Ab+ population can be captured also with other fluorochrome-conjugates (Fig. 1). In addition, it is important to remember that the recommended DDX4 Ab labels perivascular cells in situ in ovarian cortex in addition to correctly labeling oocytes.1
Figure 1.
Isolation of DDX4 Ab+ cells with different fluorescent probes. DDX4 Ab+ cells were detected using secondary Ab labeled with either AF594 (excited by 561 nm laser, upper panel) or AF647 (excited by 640 nm laser, lower panel). DDX4-AF594 detects 8.8% of the cells as DDX4 Ab+ and allows for better separation of the 2 populations compared to DDX4-AF647 that detects 5.2% DDX4 Ab+ cells. Both labeling strategies mainly target perivascular cells identified as positive for CD146 and CD9. Green marks cells positive for DDX4 Ab, and red marks cells double positive for CD146/CD9. As a negative control, the isotype control Ab recommended by Abcam (ab171870) was used. Abbreviations: Ab, antibody; FSC, forward scatter.
A vivid discussion concerning sufficient numbers of cells for scRNA-seq experiments to uncover cell types is also on-going.14 The number of cells needed for identification of rare cell types can be estimated by statistical considerations. If an average percent yield of 1.7% DDX4 Ab+ cells in ovarian cortex is expected in FACS experiments, as reported by the authors6,7 (a number that the authors mathematically convert to 0.014% occurrence in the ovaries), then identification of this population by scRNA-seq would require in total 1800 ovarian cortical cells, assuming 6-10 clusters and aiming to find at least 20 OSCs (https://satijalab.org/howmanycells/). In each of our 4 samples, cells of >90% viability were prepped to the libraries with the aim to capture 8000 viable cells, and in agreement, the data output consisted of 5715 cells in unsorted gender reassignment patient (GRP) sample, 6445 cells in unsorted caesarean section (C-sec) sample, 5479 cells in DDX4 Ab+ sorted sample, and 6690 cells in DDX4 Ab-sorted sample. Hence, the cell numbers in our experiments far exceeded the recommended numbers needed to detect populations with even smaller expected frequencies.
Cell clusters that form the basis of cell type identification are defined by algorithms that bring cells together based on their transcriptomic similarity and difference to other cells present in the input data. Therefore, introducing a reference cluster of cells to the data can help identify cells that would not have otherwise formed a separate cluster. We integrated our adult ovary datasets with human fetal ovary data consisting of both ovarian somatic and germline cells.16 Robust co-clustering was found between fetal and adult ovarian somatic cells as well as oocytes, confirming the known ovarian biology where prenatally formed immature oocytes last into adulthood.1 No adult cells clustering together with fetal retinoic acid responsive or meiotic germ cells were found.1 In our set of 24 329 adult cells, 4 cells co-clustered with mitotic fetal germ cells. However, these cells did not express any markers of pluripotency, germline or oocytes.1
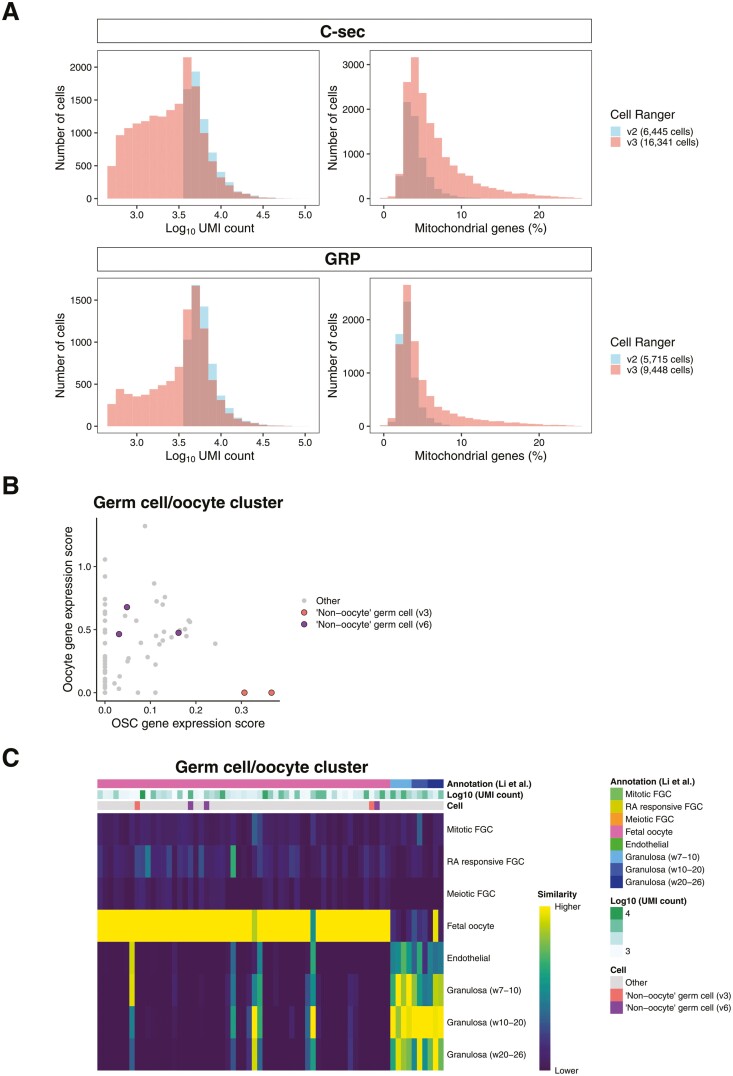
Additional concerns have been presented regarding our bioinformatic analyses, and a workflow optimization based on re-analyzing our data with newer versions of commonly used analysis pipelines and packages (Cell Ranger and Seurat) together with user-friendly visualization application provided by 10× Genomics (Loupe Browser) have been presented as the solution that enables discovery of OSCs in our data.14 Single-cell analysis technologies are rapidly evolving, allowing for more efficient identification of cellular identity and cell and molecule counting, enabling the re-evaluation of published datasets. The presented re-analysis focused on our unsorted ovarian cortical cells. When Cell Ranger v3 was applied, over 27 000 cells were discovered14 compared to the 12 160 found by us using Cell Ranger v2.1 Importantly, the cell suspension that we used for library preparation was calculated to contain 8000 viable cells for each sample based on MoxiZ automated cell counting, which was independently confirmed by the sequencing core facility by manual Bürker chamber counting prior to library preparation, making the discovery of over 27 000 cells in total from the 2 unsorted samples by changing bioinformatic pipeline surprising. Indeed, most of the new “cells” added by Cell Ranger v3 have low unique molecular identifier (UMI) counts and high mitochondrial gene expression, which is a known feature of v3 and indicative of non-viable cells, cellular debris, and empty droplets (https://kb.10xgenomics.com) (Fig. 2A).17 This re-analysis also increased the number of cells in the germ cell cluster from 18 to 62 cells, out of which 2 were now suggested as OSCs (“non-oocyte germ cells”). From here, the optimized pipeline presented by Alberico et al took the re-analysis to Cell Ranger v6 and replaced Seurat with Loupe Browser in the downstream analyses. This approach yielded again over 27 000 cells, and a germ cell cluster of 62 cells (or 65 cells by Seurat). This time, 3 of these cells were suggested as OSCs based on the co-expression of a combination of germline genes (DPPA3, IFITM3, TUBB8, and DDX4). Importantly, the 2 OSCs from v3 (identified based on expression scores of PRDM1, DPPA3, and DAZL) and the 3 from v6 are not the same (Fig. 2B). The v6 cells were annotated as oocytes in the v3 data, and vice versa. The 3 v6 OSCs express relatively high levels of oocyte markers (Fig. 2B) and when compared to fetal germline and somatic cells by SingleR analysis,18 they were all identified as oocytes (Fig. 2C). None of the cells in the germ cell cluster was annotated as mitotic, retinoic acid responsive, or meiotic germ cell by SingleR (Fig. 2C). Instead, some of the “new cells” in the germ cell cluster had more resemblance with granulosa cells than germline cells (Fig. 2C).
Figure 2.
Comparison of scRNA-seq output using different versions of Cell Ranger. (A) Histogram showing the UMI counts per cell (left graphs) and percentage of mitochondrial genes per cell (right graphs) in Cell Ranger v2 and v3 outputs. Use of Cell Ranger v3 increased the number of cells with low UMI counts and with a higher percentage of mitochondrial genes when compared to v2. Blue: sequencing output using Cell Ranger v2, red: sequencing output using Cell Ranger v3. C-sec, caesarean section; GRP, gender reassignment patient. (B) Scatter plot of the germ cell cluster cells showing the oocyte (FIGLA, OOSP2, GDF9, and ZP3) and oogonial stem cell (PRDM1, DPPA3, and DAZL) marker expression scores based on Cell Ranger v6. The 3 non-oocyte germ cells/OSCs identified with the “optimized workflow” using Cell Ranger v6 are marked with purple, and the 2 identified by the Cell Ranger v3 analysis are marked with pink. (C) Heatmap showing the similarity of the cells in the germ cell cluster (analyzed by Cell Ranger v6) to female fetal gonadal cells. The cell identity assigned by SingleR, UMI counts, and cell identity suggested by Alberico et al. are shown. Similarity scale indicates the transcriptional similarity of annotated cells ranging from low (dark blue) to high similarity (yellow) to fetal germ and somatic cells. Abbreviation: FGC, fetal germ cell; RA retinoic acid.
It has also been correctly noted that the perivascular cells that we isolated by FACS targeting DDX4 do not express DDX4 mRNA,14 as our in situ RNA-hybridization and scRNA-seq experiments previously showed.1 In conclusion, our data show that the recommended polyclonal rabbit Ab from Abcam targeting C terminus of DDX4 non-specifically binds to perivascular cells, in addition to labelling DDX4 in oocytes. This does not motivate the use of this Ab to isolate OSCs. We hope that these data would encourage all laboratories working with DDX4 Ab for germline stem cell isolation to employ stringent quality control parameters for characterizing and identifying putative OSCs.
We consider it essential for meaningful further development in the field that any germline stem cells, regardless of what they are called, are thoroughly characterized on single-cell level and by multiple technologies to be certain of their nature. This is equally applicable to “oocyte-like cells” that are reported to develop spontaneously or after elaborate differentiation protocols in cell cultures.6,19-25 Extensive work in mice has shown that generation of functional oocytes form regular embryonic stem cells is possible but requires closely mimicking the sequential steps of germline development in vivo.26 Even though mouse embryonic stem cells can be also directly programmed to oocyte-like cells, which can even be fertilized, the embryos do not develop to blastocysts due to aberrant nuclear maturation of the oocytes.27 These observations are critical, considering that the generation of viable offspring from DDX4 Ab+ cell derived oocytes in mice remains to be shown despite nearly 2 decades of research.6,21
To aid the community in taking ovarian research even further, we have prepared a user-friendly online tool where our data can be freely browsed (https://eovary.ki.se/ generated by shinyCell28). We hope that our comprehensive dataset will lead to more comparative studies between laboratories that can help reveal fascinating, yet undiscovered details of human ovarian biology.
Acknowledgments
We thank the clinicians helping us with patient recruitment, and the patients who gave ovarian tissue to our studies. The authors want to thank Petri Paavola for setting up the eovary server.
Contributor Information
Masahito Yoshihara, Department of Biosciences and Nutrition, Karolinska Institutet, Stockholm, Sweden; Institute for Advanced Academic Research, Chiba University, Chiba, Japan; Department of Artificial Intelligence Medicine, Graduate School of Medicine, Chiba University, Chiba, Japan.
Magdalena Wagner, Division of Obstetrics and Gynecology, Department of Clinical Science, Intervention and Technology, Karolinska Institutet and Karolinska University Hospital, Stockholm, Sweden.
Anastasios Damdimopoulos, Bioinformatics and Expression Analysis core facility, Department of Biosciences and Nutrition, Karolinska Institutet, Stockholm, Sweden.
Cheng Zhao, Division of Obstetrics and Gynecology, Department of Clinical Science, Intervention and Technology, Karolinska Institutet and Karolinska University Hospital, Stockholm, Sweden.
Sophie Petropoulos, Division of Obstetrics and Gynecology, Department of Clinical Science, Intervention and Technology, Karolinska Institutet and Karolinska University Hospital, Stockholm, Sweden; Centre de Recherche du Centre Hospitalier de l’Université de Montréal, Axe Immunopathologie, Montréal, Canada; Département de Médecine, Université de Montréal, Montréal, Canada.
Shintaro Katayama, Department of Biosciences and Nutrition, Karolinska Institutet, Stockholm, Sweden; Folkhälsan Research Center, Helsinki, Finland.
Juha Kere, Department of Biosciences and Nutrition, Karolinska Institutet, Stockholm, Sweden; Folkhälsan Research Center, Helsinki, Finland; Stem Cells and Metabolism Research Unit, University of Helsinki, Helsinki, Finland.
Fredrik Lanner, Division of Obstetrics and Gynecology, Department of Clinical Science, Intervention and Technology, Karolinska Institutet and Karolinska University Hospital, Stockholm, Sweden; Ming Wai Lau Center for Reparative Medicine, Stockholm node, Karolinska Institutet, Stockholm, Sweden.
Pauliina Damdimopoulou, Division of Obstetrics and Gynecology, Department of Clinical Science, Intervention and Technology, Karolinska Institutet and Karolinska University Hospital, Stockholm, Sweden.
Funding
This study received funding from Astellas Foundation for Research on Metabolic Disorders, Childhood Cancer Fund (PR2020-0096), Horizon 2020 innovation grant (ERIN, grant no. EU952516), Jane & Aatos Erkko Foundation, Japan Eye Bank Association, Japan Society for the Promotion of Science (JSPS) Overseas Research Fellowships, Scandinavia-Japan Sasakawa Foundation, Sigrid Jusélius Foundation, Swedish Research Council for Sustainable Development FORMAS (2020-01621), Svenska Sällskapet för Medicinsk Forskning (4-236-2107), The Canadian Institutes of Health Research (PJT-178082), and Swedish Research Council (2016-01919, 2020-02132). SP holds the Canada Research Chair in Functional Genomics of Reproduction and Development (950-233204).
Conflict of Interest
The authors declared no potential conflicts of interests.
Author Contributions
M.Y., P.D., F.L., M.W.: drafted the manuscript. M.Y., A.D.: carried out bioinformatic analyses. M.W.: carried out FACS experiments and analyses. Z.C.: created the e-ovary resource. All authors commented, read, and approved the final version of the commentary.
Ethics Statement
Human ovarian tissue from gender reassignment patients was collected in agreement with the Declaration of Helsinki and used in the flow cytometry experiments. The research was approved by the Swedish Ethical Review Authority #2015/798-31/2 and #2021-04563.
Data Availability
scRNA-seq data published in our original article1 were used and are available through EMBL-EBI under the accession code “E-MTAb−8381.” The scRNA-seq data re-analysis was performed using the scripts deposited for our original article1 and are available in github, “SingleCellOvary [https://github.com/wagmag/SingleCellOvary].”
References
- 1. Wagner M, Yoshihara M, Douagi I, et al. Single-cell analysis of human ovarian cortex identifies distinct cell populations but no oogonial stem cells. Nat Commun. 2020;11(1):1147. 10.1038/s41467-020-14936-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fan X, Bialecka M, Moustakas I, et al. Single-cell reconstruction of follicular remodeling in the human adult ovary. Nat Commun. 2019;10(1):3164. 10.1038/s41467-019-11036-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wang S, Zheng Y, Li J, et al. Single-cell transcriptomic atlas of primate ovarian aging. Cell. 2020;180(3):585-600.e19. 10.1016/j.cell.2020.01.009 [DOI] [PubMed] [Google Scholar]
- 4. Morris ME, Meinsohn MC, M C, et al. A single cell atlas of the cycling murine ovary. bioRxiv. 2022. 10.1101/2022.02.08.479522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Martin JJ, Woods DC, Tilly JL.. Implications and current limitations of oogenesis from female germline or oogonial stem cells in adult mammalian ovaries. Cells. 2019;8(2): 93. 10.3390/cells8020093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. White YA, Woods DC, Takai Y, et al. Oocyte formation by mitotically active germ cells purified from ovaries of reproductive-age women. Nat Med. 2012;18(3):413-421. 10.1038/nm.2669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Woods DC, Tilly JL.. Isolation, characterization and propagation of mitotically active germ cells from adult mouse and human ovaries [in Eng]. Nat Protocols. 2013;8(5):966-988. 10.1038/nprot.2013.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zarate-Garcia L, Lane SI, Merriman JA, Jones KT.. FACS-sorted putative oogonial stem cells from the ovary are neither DDX4-positive nor germ cells. Sci Rep. 2016;6:27991. 10.1038/srep27991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hernandez SF, Vahidi NA, Park S, et al. Characterization of extracellular DDX4- or Ddx4-positive ovarian cells. Nat Med. 2015;21(10):1114-1116. 10.1038/nm.3966 [DOI] [PubMed] [Google Scholar]
- 10. Zhang H, Panula S, Petropoulos S, et al. Adult human and mouse ovaries lack DDX4-expressing functional oogonial stem cells. Nat Med. 2015;21(10):1116-1118. 10.1038/nm.3775 [DOI] [PubMed] [Google Scholar]
- 11. Oktay K, Baltaci V, Sonmezer M, et al. Oogonial precursor cell-derived autologous mitochondria injection to improve outcomes in women with multiple IVF failures due to low oocyte quality: a clinical translation. Reprod Sci. 2015;22(12):1612-1617. 10.1177/1933719115612137 [DOI] [PubMed] [Google Scholar]
- 12. Woods DC, Tilly JL.. Autologous germline mitochondrial energy transfer (AUGMENT) in human assisted reproduction. Semin Reprod Med. 2015;33(6):410-421. 10.1055/s-0035-1567826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Labarta E, de Los Santos MJ, Herraiz S, et al. Autologous mitochondrial transfer as a complementary technique to intracytoplasmic sperm injection to improve embryo quality in patients undergoing in vitro fertilization-a randomized pilot study. Fertil Steril. 2019;111(1):86-96. 10.1016/j.fertnstert.2018.09.023 [DOI] [PubMed] [Google Scholar]
- 14. Alberico H, Fleischmann Z, Bobbitt T, et al. Workflow optimization for identification of female germline or oogonial stem cells in human ovarian cortex using single-cell RNA sequence analysis. Stem Cells. 2022;40(5):523-536. 10.1093/stmcls/sxac015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ouni E, Bouzin C, Dolmans MM, et al. Spatiotemporal changes in mechanical matrisome components of the human ovary from prepuberty to menopause. Hum Reprod. 2020;35(6):1391-1410. 10.1093/humrep/deaa100 [DOI] [PubMed] [Google Scholar]
- 16. Li L, Dong J, Yan L, et al. Single-cell RNA-seq analysis maps development of human germline cells and gonadal niche interactions. Cell Stem Cell. 2017;20(6):858-873.e4. 10.1016/j.stem.2017.03.007 [DOI] [PubMed] [Google Scholar]
- 17. Ilicic T, Kim JK, Kolodziejczyk AA, et al. Classification of low quality cells from single-cell RNA-seq data. Genome Biol. 2016;17:29. 10.1186/s13059-016-0888-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Aran D, Looney AP, Liu L, et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat Immunol. 2019;20(2):163-172. 10.1038/s41590-018-0276-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ding X, Liu G, Xu B, et al. Human GV oocytes generated by mitotically active germ cells obtained from follicular aspirates. Sci Rep. 2016;6:28218. 10.1038/srep28218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Silvestris E, Cafforio P, D’Oronzo S, et al. In vitro differentiation of human oocyte-like cells from oogonial stem cells: single-cell isolation and molecular characterization. Hum Reprod. 2018;33(3):464-473. 10.1093/humrep/dex377 [DOI] [PubMed] [Google Scholar]
- 21. Wu M, Lu Z, Zhu Q, et al. DDX04+ stem cells in the ovaries of postmenopausal women: existence and differentiation potential. Stem Cells. 2022;40(1):88-101. 10.1093/stmcls/sxab002 [DOI] [PubMed] [Google Scholar]
- 22. Bothun AM, Gao Y, Takai Y, et al. Quantitative proteomic profiling of the human ovary from early to mid-gestation reveals protein expression dynamics of oogenesis and folliculogenesis. Stem Cells Dev. 2018;27(11):723-735. 10.1089/scd.2018.0002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Clarkson YL, McLaughlin M, Waterfall M, et al. Initial characterisation of adult human ovarian cell populations isolated by DDX4 expression and aldehyde dehydrogenase activity. Sci Rep. 2018;8(1):6953. 10.1038/s41598-018-25116-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. MacDonald JA, Takai Y, Ishihara O, et al. Extracellular matrix signaling activates differentiation of adult ovary-derived oogonial stem cells in a species-specific manner. Fertil Steril. 2019;111(4):794-805. 10.1016/j.fertnstert.2018.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sequeira RC, Sittadjody S, Criswell T, et al. Enhanced method to select human oogonial stem cells for fertility research. Cell Tissue Res. 2021;386(1):145-156. 10.1007/s00441-021-03464-1 [DOI] [PubMed] [Google Scholar]
- 26. Hikabe O, Hamazaki N, Nagamatsu G, et al. Reconstitution in vitro of the entire cycle of the mouse female germ line. Nature. 2016;539(7628):299-303. 10.1038/nature20104 [DOI] [PubMed] [Google Scholar]
- 27. Hamazaki N, Kyogoku H, Araki H, et al. Reconstitution of the oocyte transcriptional network with transcription factors. Nature. 2021;589(7841):264-269. 10.1038/s41586-020-3027-9 [DOI] [PubMed] [Google Scholar]
- 28. Ouyang JF, Kamaraj US, Cao EY, et al. ShinyCell: simple and sharable visualisation of single-cell gene expression data. Bioinformatics 2021. 10.1093/bioinformatics/btab209 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
scRNA-seq data published in our original article1 were used and are available through EMBL-EBI under the accession code “E-MTAb−8381.” The scRNA-seq data re-analysis was performed using the scripts deposited for our original article1 and are available in github, “SingleCellOvary [https://github.com/wagmag/SingleCellOvary].”