Abstract
Introduction
The burden of metabolic (dysfunction) associated fatty liver disease (MAFLD) is rising mirrored by an increase in hepatocellular cancer (HCC). MAFLD and its sequelae are characterized by perturbations in lipid handling, inflammation, and mitochondrial damage. The profile of circulating lipid and small molecule metabolites with the development of HCC is poorly characterized in MAFLD and could be used in future studies as a biomarker for HCC.
Methods
We assessed the profile of 273 lipid and small molecule metabolites by ultra-performance liquid chromatography coupled to high-resolution mass spectrometry in serum from patients with MAFLD (n = 113) and MAFLD-associated HCC (n = 144) from six different centers. Regression models were used to identify a predictive model of HCC.
Results
Twenty lipid species and one metabolite, reflecting changes in mitochondrial function and sphingolipid metabolism, were associated with the presence of cancer on a background of MAFLD with high accuracy (AUC 0.789, 95% CI: 0.721–0.858), which was enhanced with the addition of cirrhosis to the model (AUC 0.855, 95% CI: 0.793–0.917). In particular, the presence of these metabolites was associated with cirrhosis in the MAFLD subgroup (p < 0.001). When considering the HCC cohort alone, the metabolic signature was an independent predictor of overall survival (HR 1.42, 95% CI: 1.09–1.83, p < 0.01).
Conclusion
These exploratory findings reveal a metabolic signature in serum which is capable of accurately detecting the presence of HCC on a background of MAFLD. This unique serum signature will be taken forward for further investigation of diagnostic performance as biomarker of early stage HCC in patients with MAFLD in the future.
Keywords: Metabolic (dysfunction) associated fatty liver disease, Hepatocellular cancer, Lipidomics, Metabolomics, Detection, Survival
Translational Relevance
Derangement of lipid metabolism is a hallmark of MAFLD pathogenesis; however, these changes have not been explored in MAFLD-associated HCC. We identified a circulating 21 metabolite signature that accurately predicts for the presence of HCC in patients with MAFLD. Once validated, this signature could be used as a screening tool for the identification of HCC in patients with MAFLD.
Introduction
Primary liver cancer is the fourth most common cause of cancer-related death worldwide, and unlike other cancer types, mortality rates continue to rise [1]. The vast majority of deaths are caused by hepatocellular cancer (HCC) associated with chronic liver disease (CLD). Currently, the commonest causes of CLD worldwide are hepatitis B virus, hepatitis C virus, and alcohol-related liver disease; however, metabolic (dysfunction) associated fatty liver disease (MAFLD)-related HCC incidence is rapidly rising in parallel with the global epidemic of obesity, type 2 diabetes, and the metabolic syndrome and is poised to overtake viral hepatitis as the lead cause of HCC worldwide [2]. In the USA, Europe, and Australasia, MAFLD currently accounts for approximately 8–14% of HCC cases, and though prevalence of MAFLD-related HCC is lower in Asian countries at present, a similar exponential trajectory is expected as rates of metabolic syndrome increase in Asia, in parallel with the dual burden of CLD from MAFLD and viral hepatitis [3].
MAFLD is characterized by excessive hepatic fat accumulation (>5%) histologically on liver biopsy, or by imaging, associated with overweight/obesity, type 2 diabetes, or metabolic dysregulation [4]. Metabolic steatohepatitis, part of the spectrum of MAFLD, is characterized by steatosis with concomitant inflammation and hepatocyte ballooning, and is associated with greater risk of adverse outcomes including cirrhosis and HCC [5]. MAFLD is an independent risk factor for the development of HCC with an estimated incidence of 0.44 per 1,000 persons/year; however, the relative risk of HCC increases to 5.29 per 1,000 persons/year for patients with evidence of steatohepatitis [6]. Although a significant number of MAFLD-associated HCC occur in the absence of cirrhosis [7], when cirrhosis is present the annual rate of HCC is estimated as high as 12.8% [8]. Despite the societal burden of MAFLD, radiologic surveillance for HCC in patients without cirrhosis is not recommended due to limitations visualizing the liver due to body habitus, limited cost-effectiveness, and the increased risk; nonliver-related mortality may mitigate surveillance-related survival benefits [9].
There is increasing evidence illustrating the relationship between inflammation, lipotoxicity, and lipid signaling in MAFLD. In particular, several lipid mediators such as diacylglycerols, oxysterols, free fatty acids, and ceramide (CER) are associated with lipotoxicity driving progression toward steatohepatitis [10]. While the focus of earlier studies was on the role of free fatty acid [11, 12], perturbations in lipid metabolism in MAFLD are increasingly being appreciated both within tumors and in the circulation [13, 14]. MAFLD is fundamentally a disease characterized by excess storage of toxic lipids and their metabolism, and while there have been a number of studies investigating the impact of changes in lipid profile on progression to steatohepatitis, there has been no study that considers changes in lipidomics in patients that develop HCC on a background of MAFLD. Using a cohort analysis approach, we undertook an unsupervised, exploratory metabolic phenotyping analysis (including both lipidomics and metabolomics) to determine whether MAFLD-associated HCC is associated with a distinct metabolic signature that could be used in future work as a biomarker of HCC. We also investigated if the presence of a distinct signature is prognostic for survival in patients with MAFLD-associated HCC.
Methods
Subjects
Patients were recruited from six different international centers; Imperial College NHS Healthcare Trust (ICHNT) (UK) (N = 155), Newcastle Upon Tyne NHS Foundation Trust (UK) (N = 61), Westmead Hospital, Sydney (Australia) (N = 28), Mt Sinai Hospital, New York, NY, USA (N = 7), and University of Bologna, Italy (N = 6). Serum samples from 113 patients with MAFLD and 144 patients with MAFLD-associated HCC were used in this study. MAFLD samples were collected from two different ICHNT sites: St Mary's Hospital (N = 76) and Hammersmith Hospital (N = 37). MAFLD was defined at each center by either MRI imaging findings or liver biopsy where the presence of >5% liver steatosis was taken as being diagnostic of MAFLD. In the St Mary's cohort, 36 patients were diagnosed with MAFLD on liver biopsy and the remainder by imaging. All MAFLD patients from Hammersmith Hospital were defined by imaging. Diurnal timing of sample collection was not observed or recorded. All patients with HCC were treatment naïve. The Institutional Review Board in each participating institution approved the study protocol (Sydney West Area Health Service, the University of Sydney, Human Research Ethics Committee [5522], Health Research Authority [17/NE/0127], Sheffield Human Research Ethics Committee [17/YH/0015], London Brent Research Ethics Committee [14/LO/0645]). Written informed consent was obtained from all patients recruited in this study in accordance with the Declaration of Helsinki and Good Clinical Practice guidelines.
Clinical variables were retrieved including patient demographics, body mass index (BMI), history of diabetes, hypertension, or hypercholesterolaemia. Fasting HBA1C, serum cholesterol, triglyceride, high-density lipoprotein (HDL), low-density lipoproteins, and baseline liver tests were identified, and Child-Pugh-Turcotte (CPT) score was determined. Metabolic syndrome was defined according to WHO 1999 criteria [15]. For patients with HCC, information regarding presence of cirrhosis, number of tumors, size of tumors, presence of portal vein thrombosis (PVT), and Barcelona Clinic Liver Cancer (BCLC) staging score was also collected.
Patient and Public Involvement
Patients and public were not involved in the design or conduct of the study.
Metabolic Profiling Methods
Samples (N = 257) were prepared and run by ultra-performance liquid chromatography mass spectrometry using previously published analytical and quality control (QC) procedures [16, 17]. Analysis was undertaken in a run-order designed to be orthogonal to key subject metadata with potential study outcome-related significance. For QC assessment and data preprocessing, a pooled QC sample was prepared by mixing equal parts of each study sample and a dilution series was created from the pooled QC sample (10 × 100%, 5 × 80%, 3 × 60%, 3 × 40%, 5 × 20%, 10 × 1%). Samples were subjected to reversed-phase chromatography (RPC) tailored for the separation of neutral and complex lipids and hydrophilic interaction liquid chromatography (HILIC) for the separation of small polar metabolites. In brief, 50 μL aliquots (RPC) and 25 μL (HILIC) were taken from each sample and the pooled QC and diluted 1:1 and 1:3 vol/vol with ultrapure water for RPC and HILIC analysis, respectively. Protein was removed by addition of organic solvent (diluted sample/isopropanol in 1:4 vol/vol ratio for lipid RPC profiling and diluted sample/acetonitrile in 1:3 vol/vol ratio for HILIC profiling). Mixtures of method-specific chemical standards were added (at dilution stage for HILIC and protein precipitation stage for RPC) in order to monitor data quality during acquisition. Analyses were performed on ACQUITY UPLC instruments (Waters Corp., Milford, MA, USA) coupled to Xevo G2-S TOF mass spectrometers (Waters Corp., Manchester, UK) via a Z-spray electrospray ionization source operating in either positive or negative ion modes to produce lipid-positive (lipid RPC+) and lipid-negative (lipid RPC−) and HILIC-positive (HILIC+) datasets. Sub-aliquots of a pooled QC sample were interleaved every 10 study samples throughout the analysis, and a set of dilution series samples were acquired at the start and end of the experiment. Raw data were converted to the mzML open source format, and signals below an absolute intensity threshold of 100 counts were removed using the MSConvert tool in ProteoWizard [18]. Targeted feature extraction was performed using peakPantheR, an R package for targeted integration of chemical signals from LC-MS datasets [19]. Using peakPantheR, for each LC-MS dataset, empirical retention time and theoretical m/z values from an in-house database of metabolite/lipid annotations were integrated, to yield complimentary datasets of known, pre-assigned metabolites. All datasets were preprocessed using the nPYc-Toolbox [20], including elimination of potential run-order effects and feature filtering. Only features measured with high analytical quality (relative standard deviation [RSD] in pooled QC <30%, dilution series Pearson correlation to dilution factor >0.7, RSD in study samples >1.1* RSD in pooled QC) were retained and put forward for biological analysis. After feature filtering, targeted extraction datasets contained the following number of variables: lipid RPC+: 174; lipid RPC−: 56; HILIC+: 43. Across all serum LC-MS datasets, 3 samples were missing or contained insufficient volume, an additional sample was excluded in RPC datasets, and 5 additional samples contained insufficient volume for HILIC, leaving 254 and 250 samples for data analysis in RPC and HILIC datasets, respectively. The analysis pipeline and any exclusions are illustrated in online supplementary Figure 1 (for all online suppl. material, see www.karger.com/doi/10.1159/000525911).
Signature Discovery Using Partial Least Squares Regression
Samples from Hammersmith Hospital (n = 81) were used as training dataset, whereas the remaining samples were randomly split into validation dataset 1 (n = 84) and validation dataset 2 (n = 83). We observed a moderate batch effect among samples collected from different institutes, which was expected from a multi-center metabolomics study. To obtain a list of stable metabolites across centers for downstream classification models, we performed three filtering steps of raw features: (1) Kolmogorov-Smirnov test was applied to exclude features with significantly different distributions; (2) moderated t test was applied to exclude features that are significantly different comparing cancer cases in training dataset and cancer cases in validation dataset 1, using “limma” package; (3) overlapping features, differentially expressed comparing MAFLD and MAFLD-associated HCC cases in both training dataset and validation dataset 1, were included, using “limma” package.
Partial least squares (PLS) regression was used to develop classification models to predict the presence or absence of HCC using the pre-filtered metabolites. A PLS model was initially fit in the training dataset using “pls” package. Root mean squared error of prediction (RMSEP), as a measure of cross-validation in the training set, was then plotted against the number of components included in each model. The number of components which produced the minimal RMSEP was selected for the final model.
The relative perturbations in the resulting metabolites, with optimal coefficients, were used to calculate a predictive index for each individual in the training dataset to demonstrate the presence or absence of HCC independent of MAFLD. The predictive index was termed the predictive metabolite vector (PMV). The PMV was then applied to two validation datasets using the “plot ROC” and “ROC” package. Confusion matrices were derived using the “caret” package and a nomogram developed using the “rms” package.
Alternative Signature Development Using Least Absolute Shrinkage and Selection Operator Regression
Least absolute shrinkage and selection operator (LASSO) was used to develop predictive model of HCC using the minimal metabolite vector. The panel of identified metabolites was included as an input in the LASSO regression to predict HCC in the same training set as used in the PLS model. After 10-fold cross-validation, the model which produced the least binomial deviance was selected as the final optimal model, which was then applied and tested in two validation sets.
Inter-Metabolite Correlations
A simple spectral cluster analysis was performed using the metabolic profiling data. First, a similarity measure was computed for each metabolite linking to all other metabolites using Pearson correlation coefficient in the entire cohort. The correlation matrix was then clustered using hierarchical clustering method. The heatmap was generated using the “heatmap.plus” package.
Exploration of Prognostic Metabolites
Given the increased risk of developing HCC on a background of cirrhosis, the relationship between PMV and cirrhosis was explored. We also explored if there was an association between PMV and liver function (CPT) in the HCC cohort and as it is expected that liver dysfunction and more advanced cancer would be associated with perturbations of lipid homeostasis. The PMV was used to perform subsequent continuous Cox regression and Kaplan-Meier analysis with known predictors of overall survival (OS) including the presence of PVT, maximal tumor diameter, CPT, and BCLC stage in the HCC cohort. “ggplot2” package was used for the analysis above. All statistical analyses were performed using R 3.5.2.
Results
The baseline clinical characteristics of the studied population groups are given in Table 1. The cancer population was significantly older (71.6 vs. 53.3 years, p < 0.001) and was more likely to be male (61.6 vs. 38.4%, p = 0.02). They also had a higher incidence of metabolic syndrome (92.0 vs. 29.6%, p < 0.001) and diabetes mellitus (73.0 vs. 27.0%, p < 0.01) compared to the MAFLD group. Total serum cholesterol (mean 4.9 mmol/L vs. 3.9 mmol/L, p < 0.001) and triglycerides (1.9 mmol/L vs. 1.4 mmol/L, p < 0.01) were higher in MAFLD cohort compared to the cancer cohort. No differences were observed in HBA1C levels. More patients in the cancer cohort had cirrhosis (87.0%) compared with the MAFLD cohort (13.0%) (p < 0.01). In both cohorts, underlying liver function was preserved (Child-Pugh A in MAFLD cohort 99.0% and 72.9% in HCC cohort). The median OS of the cancer group was 23.0 months (95% CI: 17.2–28.8).
Table 1.
Baseline characteristics of the study population
| Baseline characteristic | MAFLD, n (%) N = 113 (44.0) | MAFLD-HCC, n (%) N = 144 (56.0) | p value |
|---|---|---|---|
| Age (mean, IQR) | 53.3 (13) | 71.6 (10) | <0.001 |
| Sex | |||
| Male | 66 (58.4) | 107 (74.3) | 0.02 |
| Female | 47 (41.6) | 37 (25.7) | |
| BMI (mean, IQR) (N = 188) | 30.3 (6.1) | 31.2 (7.9) | 0.27 |
| Metabolic syndrome* (present) (N = 193) | 34 (33.7) | 81 (88.8) | <0.001 |
| Diabetes (present) (N = 243) | 40 (38.1) | 108 (78.3) | <0.001 |
| Hypertension (present) (N = 162) | 40 (38.8) | 48 (81.4) | <0.001 |
| Ethnicity** | |||
| White | 57 (50.4) | 110 (76.4) | |
| Asian | 18 (15.9) | 16 (11.1) | |
| Black | 13 (11.5) | 3 (2.1) | <0.001 |
| Middle Eastern | 4 (3.5) | 6 (4.2) | |
| Chinese | 3 (2.7) | 3 (2.1) | |
| Other | 18 (15.9) | 6 (4.2) | |
| HBA1C (mean, IQR) (N = 116)*** | 44.0 (7.0) | 48.6 (33.8) | 0.35 |
| Total cholesterol (mean, IQR) (N = 141)*** | 4.9 (1.3) | 3.9 (1.2) | <0.001 |
| Triglyceride levels (mean, IQR) (N = 140)*** | 1.9 (1.1) | 1.4 (0.8) | <0.01 |
| HDL levels (mean, IQR) (N = 118)*** | 1.2 (0.4) | 1.1 (0.4) | 0.47 |
| Cirrhosis | |||
| Absent | 89 (84.8) | 36 (25.2) | <0.001 |
| Present | 16 (15.2) | 107 (74.8) | |
| Child-Turcotte-Pugh class (N = 241) | |||
| A | 100 (99.0) | 99 (70.7) | |
| B | 1 (1.0) | 35 (25.0) | <0.001 |
| C | 6 (4.3) | ||
| BCLC (N = 127) | |||
| A | 37 (29.1) | ||
| B | 39 (30.7) | ||
| C | 39 (30.7) | ||
| D | 12 (9.4) | ||
| Maximum tumor diameter (N = 128) | |||
| ≤7 cm | 92 (71.9) | ||
| >7 cm | 36 (28.1) | ||
| PVT (N = 127) | |||
| Absent | 69 (54.3) | ||
| Present | 58 (45.7) | ||
| Metastases (N = 112)**** | |||
| Absent | 94 (83.4) | ||
| Present | 18 (16.0) | ||
| Treatments, n | |||
| 0 | 18 (12.8) | ||
| 1 | 105 (72.9) | ||
| 2 | 4 (2.8) | ||
| >3 | 7 (4.9) | ||
| OS | 23.0 (17.71–28.83) | ||
Metabolic syndrome defined according to WHO 1999 criteria. Data regarding metabolic syndrome were not available for Mt Sinai cohort.
Other as identified by patients (N = 24).
Data pertaining for HBAIC, cholesterol, triglycerides, and HDL levels not available for Newcastle, Mt Sinai, and Bologna cohort.
Presence of extrahepatic disease not available for Sydney cohort.
Identification of a Metabolic Signature Predictive of HCC in the Presence of MAFLD
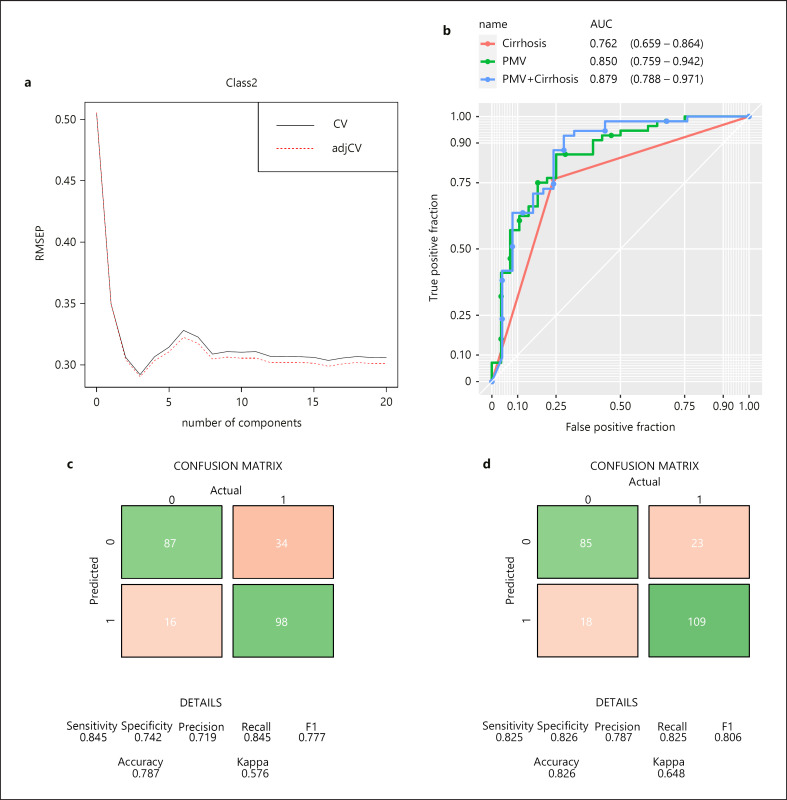
Metabolic phenotyping analysis was carried out that identified 230 lipid species belonging to 24 lipid classes and 43 small molecule metabolites. The primary aim of this study was to investigate whether a metabolic signature is present in serum that can accurately predict the presence of HCC on a background of MAFLD. First, we identified changes in 21 metabolites that were stable across the six different centers involved in this study. Using a PLS regression approach in the training set (Fig. 1a), we confirmed that relative changes in 21 metabolites accurately differentiated the presence of HCC on a background of MAFLD (area under curve [AUC] = 0.94, 95% CI: 0.885–0.993). The combined relative changes of these 21 metabolites were labeled the PMV and were derived for each individual in the study. The PMV was further tested in two validation sets, and consistent predictive power was exhibited (validation set 1: AUC = 0.85, 95% CI: 0.759–0.942; validation set 2: AUC = 0.72, 95% CI: 0.605–0.837; combined validation sets: AUC = 0.789, 95% CI: 0.721–0.858) (Fig. 1b; online suppl. Fig. A, B). In the combined validation cohorts, the positive predictive value of the PMV was 0.79 (95% CI: 0.70–0.86), and the negative predictive value was 0.86 (95% CI: 0.79–0.91). Demographic details across training and validation sets are given in online supplementary Table 1.
Fig. 1.
Building of predictive metabolite vector (PMV) to differentiate MAFLD from MAFLD-HCC. a Selection of optimal partial least squares regression (PLSR) model based on 21 metabolites. Root mean-squared error of prediction (RMSEP) (x-axis) is plotted against number of components (y-axis) after 10-fold cross-validation. Three components from PLSR are used to build PMV. Receiver operating characteristic (ROC) curves are plotted to demonstrate the predictive power of PMV and cirrhosis in the combined validation sets (b). Area under curve (AUC) values are shown under each plot. Confusion matrices summarizing the predicted HCC cases vs. actual HCC cases using models based on cirrhosis only (c) or PMV and cirrhosis (d) in the combined set.
The presence of cirrhosis per se is an established risk factor for the development of HCC [21]. Moreover, previous work has shown that liver dysfunction can influence metabolite profiling [22] and the relationship between CPT and PMV was explored. On multivariable analysis, both PMV and cirrhosis status were significantly associated with HCC (PMV: odds ratio [OR] = 7.84 [5.2–11.8], p < 0.001; cirrhosis: OR = 7.95 [5.32–11.9], p < 0.001), suggesting they act as independent predictors of cancer. No association was observed between CPT and PMV. We then explored whether the predictive ability of PMV in detecting HCC could be enhanced with the addition of cirrhosis. We illustrate a further improvement in the predictive model (validation set 1: AUC = 0.879 [95% CI: 0.788–0.971]; validation set 2: AUC = 0.841 [95% CI: 0.751–0.931]; combined validation sets: AUC = 0.855 [95% CI: 0.793–0.917]) (Fig. 1b; online suppl. Fig. 2A, B). A confusion matrix was generated, after accepting the minimum sensitivity as 0.8, to determine an optimal threshold of PMV that predicted the presence of HCC with high specificity. The PMV-integrated model gave a specificity of 0.826 (95% CI: 0.75–0.89), 11% more specific than a cirrhosis-only model (specificity: 0.742, 95% CI: 0.66–0.81) (Fig. 1c, d). We then derived a nomogram (online suppl. Fig. 3) for clinical application that combines individual patients' PMV and the presence or absence of cirrhosis as determinant for the presence of HCC. Of interest, PMV did not correlate with the BMI, presence of metabolic syndrome, or diabetes in the entire study cohort.
Metabolites Associated with HCC Are Inter-Correlated
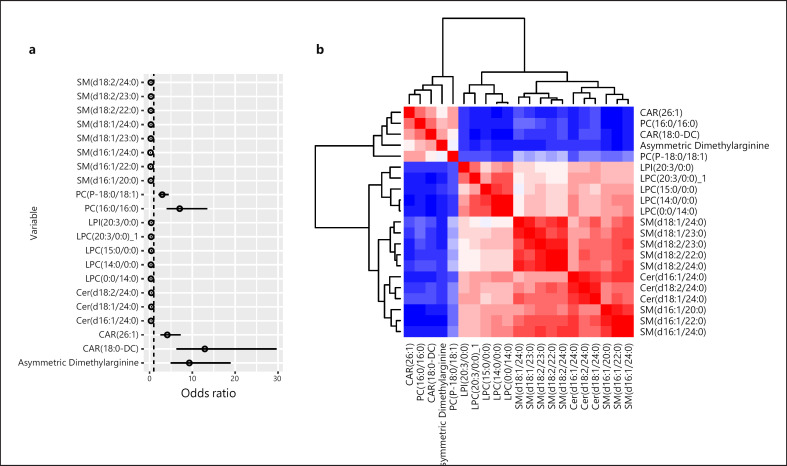
The majority of the metabolites in the PMV corresponded to lipid species, predominantly belonging to phosphatidylcholines (PCs), CERs, and sphingomyelins (SMs) lipid subclasses. Two carnitine (CAR) species and asymmetric dimethylarginine (ADMA) were also included (Table 2).
Table 2.
Lipid profile predictive of HCC on background of NAFLD
| Metabolite name | Coefficient |
|---|---|
| Asymmetrie dimethylarginine | 16.4728789 |
| CAR(18:0–DQ | 2.643794344 |
| CAR(26:1) | –1.115760821 |
| Cer(d16:1/24:0) | 6.636870471 |
| Cer(d18:2/24:0) | 4.846124 |
| Cer(d18:1/24:0) | 4.204558483 |
| LPC(0:0/14:0) | –4.033630203 |
| LPC(14:0/0:0) | –5.50639709 |
| LPC(15:0/0:0) | –7.825665552 |
| LPC(20:3/0:0)_1 | 1.310188498 |
| LPI(20:3/0:0) | –3.339869581 |
| PC(16:0/16:0) | 4.506361519 |
| PC(P-18:0/18:1) | 9.056127427 |
| SM(d16:1/20:0) | –3.502104041 |
| SM(d16:1/22:0) | –0.45509791 |
| SM(d16:1/24:0) | –1.614155581 |
| SM(d18:2/23:0) | –2.593395293 |
| SM(d18:2/24:0) | –4.584937325 |
| SM(d18:1/24:0) | –4.774057238 |
| SM(d18:2/22:0) | –5.331737388 |
| SM(d18:1/23:0) | –5.646288334 |
Coefficient of variance is given. PC, phosphocholine; Cer, ceramide; CAR, Fatty acyl carnitine; LPC, lysophosphocholine; SM, sphingomyelin; LPI, lysophosphatidylinositol.
When considering the OR for the presence of HCC, forest plot analysis illustrated that lipids belonging to PCs, CAR (26:1) and (18:0-DC), and ADMA were associated with the presence of HCC, whereas SM, CER lipid classes, lysophosphatidylcholine (LPC), and phosphatidylinositol were inversely associated with the presence of HCC (Fig. 2a). A correlation analysis of the 21 metabolites associated with the presence of HCC was conducted (Fig. 2b). A clear division was observed between the lipid subclasses such that CAR (26:1) and (18:0-DC), PC (16:0/16:0) and (P-18:0/18:1), and ADMA were tightly correlated, and a correlation was observed between the remaining lipid species illustrating an interplay between the lipids identified rather than individual lipid species driving pathogenicity.
Fig. 2.
Inter-correlations among lipid species making up the PMV. a Forest plot analysis illustrating the association between the 21 metabolites and HCC. b Heatmap demonstrating the inter-metabolite correlations within the entire cohort. Red color indicates high similarity between the two species, whereas blue indicates low similarity.
To simplify the PMV, LASSO regression was undertaken considering only the 21 metabolites that make up the PMV (online suppl. Fig. 4A, B). We identified a seven-lipid signature (online suppl. Table 2) which predicted the presence of HCC with high accuracy in both the training (AUC = 0.99, 95% CI: 0.998–1) and two validation sets (validation set 1: AUC = 0.92, 95% CI: 0.850–0.998; validation set 2: AUC = 0.79, 95% CI: 0.697–0.900) (online suppl. Fig. 4C–E).
The Metabolic Signature Is an Independent Predictor of Clinical Outcome in MAFLD-Associated HCC
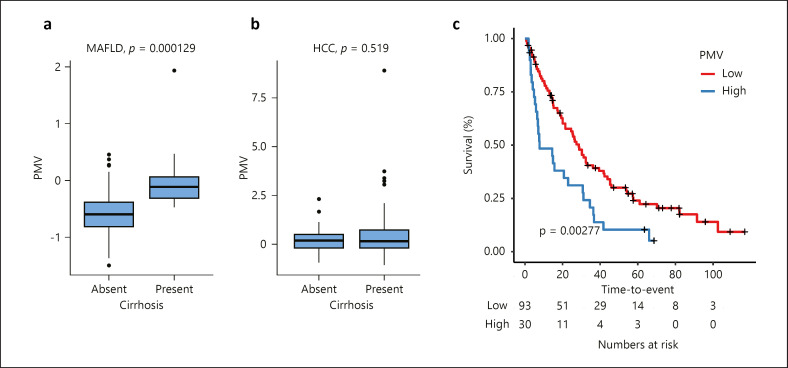
Considering the HCC cohort alone, we observed a strong relationship between PMV and BCLC stage such that PMV increases with BCLC C/D compared with A/B (p < 0.001). Kaplan-Meier analysis illustrated a significant relationship between PMV and OS, and PMV was significantly associated with poor OS in patients with MALFD-HCC (log-rank test; HR 1.97, 95% CI: 1.25–3.09, p < 0.01) (Fig. 3). We then considered the utility of the PMV when compared to known prognostic factors: BCLC stage, presence of PVT, CPT, and tumor size, using a Cox regression model (Table 3). PMV was associated with poor OS in the univariate model (HR 1.28, 95% CI: 1.06–1.55, p = 0.01) and remained an independent prognostic factor of OS in patients with HCC on a background of MAFLD (HR 1.42, 95% CI: 1.09–1.83, p < 0.01). As BMI is likely to decline with worsening disease stage, we explored changes in PMV within BCLC stage while considering changes in BMI; however, no significant association between PMV and BMI was observed (p = 0.279).
Fig. 3.
a–c Kaplan-Meier survival curve illustrating the association between the PMV and overall survival. Upper quartile is used as the cut-off to define PMV-high and PMV-low groups. p value was given by log-rank test.
Table 3.
Effects of PMV and common prognostic factors on overall survival in HCC cohort (N = 144)
| Predictor | Univariable models |
Multivariable models |
||
|---|---|---|---|---|
| hazard ratio (95% CI) | p | hazard ratio (95% CI) | p | |
| BCLC stage | 1.45 (1.18–1.78) | <0.001 | 1.18 (0.870–1.60) | 0.29 |
| Tumor size >7 cm | 1.60 (1.02–2.51) | 0.039 | 0.847 (0.457–1.57) | 0.59 |
| PVT present | 1.56 (1.05–2.3) | 0.028 | 1.57 (0.944–2.62) | 0.082 |
| CPT B/C vs. A | 1.54 (1.02–2.33) | 0.041 | 1.03 (0.542–1.96) | 0.93 |
| PMV | 1.28 (1.06–1.55) | 0.011 | 1.42 (1.09–1.86) | 0.0089 |
BCLC, Barcelona Clinic Liver Class; PVT, portal vein thrombosis; CPT, Child-Pugh Score; PMV, predictive metabolite vector.
Discussion
There are currently no screening programs for MAFLD-associated HCC in the absence of cirrhosis, and as a result, most patients will present with advanced stage disease [3, 8] where curative therapies are not an option. As 30% of the adult population worldwide has MAFLD [3], conventional screening with imaging is not feasible and there is pressing need for the development of novel methodologies of detecting HCC in this population group. As alterations in lipid metabolism are a central component in the pathogenesis and progression of MAFLD, we used ultra-performance liquid chromatography and mass spectrometry to assess whether specific changes in lipids occur with the presence of HCC.
We identified a unique metabolic signature in serum that accurately detects the presence of HCC on a background of MAFLD. We observed robust differences in 20 lipid species and the protein methylation by-product, ADMA. Collectively, this panel of 21 metabolites gave a high predictive accuracy of HCC. Importantly, the PMV was associated with the presence of cirrhosis in those patients with MAFLD, a precursor to HCC, and the combination of PMV and presence of cirrhosis significantly improved the predictive accuracy of the model. Multivariable modeling further demonstrated that in the subgroup of patients with HCC, PMV was an independent prognostic factor of OS. With regard to the lipid classes contributing to the PMV, we observed relative increase in acylcarnitines and arginine and perturbations in sphingolipids species. As illustrated, rather than a panel of independent bioactive lipids that predict the presence of HCC, there is an interplay between these lipid species such that there is a clear inter-relationship between CAR (26:1) and (18:0-DC), PCs (16:0/16:0) and (P-18:0/18:1), and ADMA, all of which are increased relative to the remaining lipid species.
Mitochondrial dysfunction secondary to lipotoxicity is central to the progression of MAFLD [23]. It is postulated that initially the increased lipid availability stimulates mitochondrial capacity to protect against MAFLD but eventually results in oxidative stress, release of pro-inflammatory cytokines, mitochondrial exhaustion, and development of NASH. Changes in mitochondrial function and fatty acid oxidation are reflected by changes in circulating acylcarnitine.
The carnitine shuttle system plays a central role in β-oxidation, the main process for energy production within the mitochondria. Consistent with our findings, long-chain acylcarnitines (C14 and above) have been shown to accumulate in both the serum and tissue in patients with NASH and HCC as these fatty acyl species rely on the carnitine shuttle to enter the mitochondria [24, 25, 26]. The accumulation of long-chain acylcarnitines is hypothesized to result from a reduction in carnitine palmitoyltransferase 1 (CPT2), an enzyme that controls the reaction rate of β-oxidation in the mitochondria. Expression of CPT2 has been shown to be reduced in MAFLD-associated HCC tissue compared to adjacent noncancer tissue both in vivo and in clinical samples [25, 27]. Enooku and colleagues [26] explored the relationship between serum acylcarnitine profiles in 241 patients with MAFLD, including 23 with MAFLD-associated HCC, and observed an increase in long-chain acylcarnitines with progression of fibrosis and HCC which is supported by an earlier study investigating lipid changes with MAFLD progression [22, 26]. Interestingly, Fujiwara and colleagues [27] postulated that acylcarnitines could be used as biomarker of HCC in the setting of steatohepatitis and reported a sequential increase in serum levels of acylcarnitines in 3 patients that paralleled the development of HCC, findings that are supported by our results.
We observed significant perturbations in the sphingolipids, notably a relative increase in PC and reduction in CERs, SMs, and LPC species in the HCC cohort relative to the MAFLD cohort. CERs play an important anti-proliferative and pro-apoptotic role in tissue homeostasis [28]. However, the enzymes that generate CER are often altered in cancer resulting in the disruption of the tumor suppressor effects of CER, and low CER levels have been shown to be associated with tumor progression and poor prognosis [29, 30]. An increase in CER is well documented during MAFLD development which is then subsequently reduced with the development of HCC, results that are corroborated by our findings [13, 31, 32]. Previous tissue studies have illustrated a reduction in the CER/SM ratio implicating an impaired sphingomyelinase activity, a pathway that has been shown to have anti-apoptotic and pro-survival roles in malignancy [33, 34]. However, CER levels can also be reduced through deacetylation and phosphorylation through other pathways lending further complexity to the role of CER in hepatocarcinogenesis [35]. Drugs that regulate CER-related enzymes are gaining interest as therapeutic targets by which CER levels can be increased resulting in autophagy and inhibition of tumor growth [30].
PC can be formed de novo via the Kennedy pathway which has been shown to be upregulated in numerous cancers, through over-expression of choline kinase-α (CHKA) [36, 37, 38]. Moreover, the metabolism of SM results in an increase in Kennedy pathway substrates per se [39]. We observed an increase in PC in the HCC cohort, suggesting upregulation of the Kennedy pathway driven by CHKA, an assumption that is confirmed by TCGA analysis which illustrates an increase in CHKA expression in HCC compared to normal liver (online suppl. Fig. 5). In addition to the increase in PC, we observed a decrease in LPC a reaction that is catalyzed by the enzyme LPC acyltransferase-1 that has been shown to be strongly expressed in HCC tissue samples [40] suggesting upregulation of Land's cycle through which fatty acids can further be incorporated into PC [41]. Our results are consistent with previous work illustrating significant reduction in LPC in serum of patients with HCC [42, 43, 44]. In a recent multi-omics study, Hall and colleagues [45] illustrated impaired β-oxidation and a significant increase in hepatic mono-unsaturated fatty acid-PC (36:1) associated with hepatocyte proliferation which was attributed to increased CHKA, findings which are consistent with our results particularly as we also observed an increase in the mono-unsaturated fatty acid-PC (−18:/18:1) in the presence of HCC. Gorden and colleagues [13], when investigating differences in MAFLD and NASH, also noted an increase in phosphocholine pool with cirrhosis which they attribute to increase cellular proliferation, marking a transition point from a relatively benign phenotype to a premalignant type.
The methylated arginine species ADMA is generated following cleavage of proteins that are post-translationally methylated at the arginine residues. ADMA regulates the amount of nitrous oxide produced in the body by competing with L-arginine for nitrous oxide synthase with elevated levels of ADMA associated with endothelial dysfunction. ADMA is metabolized by dimethylarginine dimethylaminohydrolase which is highly expressed within the liver, and it is well established that levels of ADMA increase with liver cirrhosis [46]. Two studies have reported elevated levels of ADMA in patients with MAFLD which is hypothesized to be associated with the cardiovascular consequences of the metabolic syndrome associated with this population group [47, 48, 49]. It could be postulated that the increase in ADMA observed in this study with HCC could be the result of progressive parenchymal damage from malignancy and not just a reflection of liver dysfunction [49].
Interestingly, even though we observed significant differences in triglycerides and cholesterol levels between the HCC and MAFLD cohorts, these are not included in the PMV. This suggests the changes in the lipid profile observed reflect metabolic reprogramming in HCC, a hallmark of cancer, which in turn has systemic effects including loss of weight and reduced appetite which may also contribute to the differences in lipidomics profiles observed. Correlation with tumor tissue would be of importance to further strengthen this hypothesis.
Another key consideration is that we have shown that there is an inter-relationship between individual lipids and no individual lipid exerts an effect on its own reflecting the known interplay of each sphingolipid species and is a more relevant biologic approach [39]. Acknowledging the limitation of analyzing multiple lipids in the clinical setting, we undertook LASSO regression to ascertain whether a smaller number of lipids could be used to detect the presence of HCC. A 7-lipid signature was developed that identified HCC with high accuracy which could be taken forward in further validation studies. Moving forward, it would be of importance to explore the PMV in a longitudinal study of patients with MAFLD in order to examine at what stage is there a metabolic switch and to delineate the role of the PMV as a screening tool for HCC in this population group. We observed a significant impact of the collection center on the lipids profiled. This is a recognized limitation of metabolic profiling when samples are collected using different protocols. By selecting for metabolites that were stable across sample collection sites (with comparable distributions), our approach to this challenge ensures that the resulting biomarkers are robust with respect to any procedural differences in sample collection. This approach may be overly stringent, and the use of more homogenous sample collection procedures may have resulted in additional metabolic insights. Future work would be strengthened by universal collection protocols to minimize the impact of sample handling on results. In addition, we noted a higher incidence of metabolic syndrome in those patients with HCC compared to the MAFLD group reflecting the higher incidence of hypertension and diabetes in those with HCC. A significant difference was observed in the age of the two cohorts with the MAFLD group being younger overall which may explain the differences in incidence of these comorbidities. There is a significant amount of missing data that may also impact on the differences in baseline demographic data. Similarly, differences in cholesterol and HBA1C may be attributable to weight loss and cachexia in the HCC subgroup, but should be interpreted with caution given the large amount of missing data.
In conclusion, through exploratory lipidomics and metabolomics, we have identified a unique metabolic signature that accurately discriminates the presence of HCC on a background of MAFLD. The PMV reflects alterations in β-oxidation and sphingolipid metabolism, the biology of which requires further elucidation. The PMV should be taken forward in prospective, longitudinal studies to validate its utility.
Statement of Ethics
Written informed consent was obtained from all patients recruited in this study in accordance with the Declaration of Helsinki and Good Clinical Practice (GCP) guidelines. The Institutional Review Board in each participating institution approved the study protocol (Sydney West Area Health Service, the University of Sydney, Human Research Ethics Committee [5522], Health Research Authority [17/NE/0127], Sheffield Human Research Ethics Committee [17/YH/0015], London Brent Research Ethics Committee [14/LO/0645]).
Conflict of Interest Statement
Augusto Villanueva is an Editorial Board Member of Liver Cancer. Rohini Sharma received lecture fees from Bayer Healthcare and Shingoi; consulting fees from EISAI, Roche, SIRTEX; and research funding (to institution) from Incyte, Boston Scientific, AAA, and Astex Pharmaceuticals. Haonan Lu, Jacob George, Mohammed Eslam, Luigi Bolondi, Helen L. Reeves, Misti McCain, Edward Chambers, Caroline Ward, Dewi Sartika, Caroline Sands, Lynn Maslen, Matthew R. Lewis, and Ramya Ramaswami have no conflicts of interest to disclose.
Funding Sources
This work was supported by the Medical Research Council and National Institute for Health Research (Grant No. MC_PC_12025). Infrastructure support was provided by the National Institute for Health Research (NIHR) Imperial Biomedical Research Centre (BRC) in the form of resource support. Imperial Experimental Cancer Medicines Centre, Cancer Research UK (C2536/A16584) to Rohini Sharma. Helen L. Reeves has received funding for staff and resources from European Community's Seventh Framework Programme (FP7/2001-2013) under grant agreement HEALTH-F2-2009-241762 for the project FLIP, Cancer Research UK (CR UK) Centre Grant C9380/A18084; Programme Grant C18342/A23390; and Accelerator Award C9380/A26813. These grants enabled sample and clinical data collection. Jacob George and Mohammed Eslam are supported by a NSW Cancer Institute Translational Cancer Research Centre Grant 15/TRC/1-01, Cancer Council NSW Grants APP1070076, APP1145008, and APP1069733. Funding was provided to support salaries and sample collection. Haonan Lu, Luigi Bolondi, Misti McCain, Edward Chambers, Caroline Ward, Augusto Villanueva, Dewi Sartika, Caroline Sands, Lynn Maslen, Matthew R. Lewis, and Ramya Ramaswami have no funding sources to declare.
Author Contributions
Haonan Lu: data analysis and manuscript preparation; Jacob George, Helen L. Reeves, and Misti McCain: resource provision, manuscript preparation, and review of final manuscript; Mohammed Eslam, Augusto Villanueva, Luigi Bolondi, and Edward Chambers: resource provision and review of final manuscript; Caroline Ward: sample collection, resource provision, and review of final manuscript; Dewi Sartika: data curation and review of final manuscript; Caroline Sands, Lynn Maslen, and Matthew R. Lewis: experimental design, study conduct, data analysis, and manuscript preparation; Ramya Ramaswami: concept, design, funding, and review of manuscript; Rohini Sharma: concept, funding, sample provision, study conduct, and data analysis.
Data Availability Statement
The data generated and analyzed in this study are included in this article and/or its online supplementary material. Further inquiries can be directed to the corresponding author.
Supplementary Material
Supplementary data
Supplementary data
Supplementary data
Acknowledgments
This research study was supported by the Imperial College NIHR Imperial Biomedical Research Centre and Imperial College Experimental Cancer Medicines Centre. The views expressed are those of the authors and not necessarily those of the NIHR or the Department of Health and Social Care.
Funding Statement
This work was supported by the Medical Research Council and National Institute for Health Research (Grant No. MC_PC_12025). Infrastructure support was provided by the National Institute for Health Research (NIHR) Imperial Biomedical Research Centre (BRC) in the form of resource support. Imperial Experimental Cancer Medicines Centre, Cancer Research UK (C2536/A16584) to Rohini Sharma. Helen L. Reeves has received funding for staff and resources from European Community's Seventh Framework Programme (FP7/2001-2013) under grant agreement HEALTH-F2-2009-241762 for the project FLIP, Cancer Research UK (CR UK) Centre Grant C9380/A18084; Programme Grant C18342/A23390; and Accelerator Award C9380/A26813. These grants enabled sample and clinical data collection. Jacob George and Mohammed Eslam are supported by a NSW Cancer Institute Translational Cancer Research Centre Grant 15/TRC/1-01, Cancer Council NSW Grants APP1070076, APP1145008, and APP1069733. Funding was provided to support salaries and sample collection. Haonan Lu, Luigi Bolondi, Misti McCain, Edward Chambers, Caroline Ward, Augusto Villanueva, Dewi Sartika, Caroline Sands, Lynn Maslen, Matthew R. Lewis, and Ramya Ramaswami have no funding sources to declare.
References
- 1.Ferlay J, Colombet M, Soerjomataram I, Mathers C, Parkin DM, Pineros M, et al. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int J Cancer. 2019;144((8)):1941–1953. doi: 10.1002/ijc.31937. [DOI] [PubMed] [Google Scholar]
- 2.Akinyemiju T, Abera S, Ahmed M, Alam N, Alemayohu MA, et al. Global Burden of Disease Liver Cancer Collaboration The burden of primary liver cancer and underlying etiologies from 1990 to 2015 at the Global, Regional, and National Level: Results from the Global Burden of Disease Study 2015. JAMA Oncol. 2017;3((12)):1683–1691. doi: 10.1001/jamaoncol.2017.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Younossi Z, Anstee QM, Marietti M, Hardy T, Henry L, Eslam M, et al. Global burden of NAFLD and NASH: trends, predictions, risk factors and prevention. Nat Rev Gastroenterol Hepatol. 2018;15((1)):11–20. doi: 10.1038/nrgastro.2017.109. [DOI] [PubMed] [Google Scholar]
- 4.Eslam M, Sanyal AJ, George J, International Consensus Panel MAFLD: a consensus-driven proposed nomenclature for metabolic associated fatty liver disease. Gastroenterology. 2020;158((7)):1999–2014.e1. doi: 10.1053/j.gastro.2019.11.312. [DOI] [PubMed] [Google Scholar]
- 5.Chalasani N, Younossi Z, Lavine JE, Charlton M, Cusi K, Rinella M, et al. The diagnosis and management of nonalcoholic fatty liver disease: practice guidance from the American Association for the Study of Liver Diseases. Hepatology. 2018;67((1)):328–357. doi: 10.1002/hep.29367. [DOI] [PubMed] [Google Scholar]
- 6.White DL, Kanwal F, El-Serag HB. Association between nonalcoholic fatty liver disease and risk for hepatocellular cancer, based on systematic review. Clin Gastroenterol Hepatol. 2012;10((12)):1342–59.e2. doi: 10.1016/j.cgh.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Piscaglia F, Svegliati-Baroni G, Barchetti A, Pecorelli A, Marinelli S, Tiribelli C, et al. Clinical patterns of hepatocellular carcinoma in nonalcoholic fatty liver disease: a multicenter prospective study. Hepatology. 2016;63((3)):827–838. doi: 10.1002/hep.28368. [DOI] [PubMed] [Google Scholar]
- 8.Dyson J, Jaques B, Chattopadyhay D, Lochan R, Graham J, Das D, et al. Hepatocellular cancer: the impact of obesity, type 2 diabetes and a multidisciplinary team. J Hepatol. 2014;60((1)):110–117. doi: 10.1016/j.jhep.2013.08.011. [DOI] [PubMed] [Google Scholar]
- 9.Singal AG, El-Serag HB. Rational HCC screening approaches for patients with NAFLD. J Hepatol. 2022;76((1)):195–201. doi: 10.1016/j.jhep.2021.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Malhi H, Gores GJ. Molecular mechanisms of lipotoxicity in nonalcoholic fatty liver disease. Semin Liver Dis. 2008;28((4)):360–369. doi: 10.1055/s-0028-1091980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feldstein AE, Werneburg NW, Canbay A, Guicciardi ME, Bronk SF, Rydzewski R, et al. Free fatty acids promote hepatic lipotoxicity by stimulating TNF-alpha expression via a lysosomal pathway. Hepatology. 2004;40((1)):185–194. doi: 10.1002/hep.20283. [DOI] [PubMed] [Google Scholar]
- 12.Malhi H, Bronk SF, Werneburg NW, Gores GJ. Free fatty acids induce JNK-dependent hepatocyte lipoapoptosis. J Biol Chem. 2006;281((17)):12093–101. doi: 10.1074/jbc.M510660200. [DOI] [PubMed] [Google Scholar]
- 13.Gorden DL, Myers DS, Ivanova PT, Fahy E, Maurya MR, Gupta S, et al. Biomarkers of NAFLD progression: a lipidomics approach to an epidemic. J Lipid Res. 2015;56((3)):722–736. doi: 10.1194/jlr.P056002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clarke JD, Novak P, Lake AD, Shipkova P, Aranibar N, Robertson D, et al. Characterization of hepatocellular carcinoma related genes and metabolites in human nonalcoholic fatty liver disease. Dig Dis Sci. 2014;59((2)):365–374. doi: 10.1007/s10620-013-2873-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alberti KGM, Zimmet P, Shaw J, IDF Epidemiology Task Force Consensus Group The metabolic syndrome: a new worldwide definition. Lancet. 2005;366((9491)):1059–1062. doi: 10.1016/S0140-6736(05)67402-8. [DOI] [PubMed] [Google Scholar]
- 16.Lewis MR, Pearce JTM, Spagou K, Green M, Dona AC, Yuen AHY, et al. Development and application of ultra-performance liquid chromatography-TOF MS for precision large scale urinary metabolic phenotyping. Anal Chem. 2016;88((18)):9004–9013. doi: 10.1021/acs.analchem.6b01481. [DOI] [PubMed] [Google Scholar]
- 17.Izzi-Engbeaya C, Comninos AN, Clarke SA, Jomard A, Yang L, Jones S, et al. The effects of kisspeptin on beta-cell function, serum metabolites and appetite in humans. Diabetes Obes Metab. 2018;20((12)):2800–2810. doi: 10.1111/dom.13460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chambers MC, Maclean B, Burke R, Amodei D, Ruderman DL, Neumann S, et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat Biotechnol. 2012;30((10)):918–920. doi: 10.1038/nbt.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wolfer AM, Turaga N. Phenomecentre/peakPantheR: Bioc 3.10 release version (v1.0.0) 2019.
- 20.Sands CJ, Wolfer AM, Correia GDS, Sadawi N, Ahmed A, Jimenez B, et al. The nPYc-Toolbox, a Python module for the pre-processing, quality-control and analysis of metabolic profiling datasets. Bioinformatics. 2019;35((24)):5359–5360. doi: 10.1093/bioinformatics/btz566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.European Association for the Study of the Liver, European Organisation for Research and Treatment of Cancer EASL-EORTC clinical practice guidelines: management of hepatocellular carcinoma. J Hepatol. 2012;56((4)):908–943. doi: 10.1016/j.jhep.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 22.Peng KY, Watt MJ, Rensen S, Greve JW, Huynh K, Jayawardana KS, et al. Mitochondrial dysfunction-related lipid changes occur in nonalcoholic fatty liver disease progression. J Lipid Res. 2018;59((10)):1977–1986. doi: 10.1194/jlr.M085613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marra F, Svegliati-Baroni G. Lipotoxicity and the gut-liver axis in NASH pathogenesis. J Hepatol. 2018;68((2)):280–295. doi: 10.1016/j.jhep.2017.11.014. [DOI] [PubMed] [Google Scholar]
- 24.Lu Y, Li N, Gao L, Xu YJ, Huang C, Yu K, et al. Acetylcarnitine is a candidate diagnostic and prognostic biomarker of hepatocellular carcinoma. Cancer Res. 2016;76((10)):2912–2920. doi: 10.1158/0008-5472.CAN-15-3199. [DOI] [PubMed] [Google Scholar]
- 25.Lu X, Zhang X, Zhang Y, Zhang K, Zhan C, Shi X, et al. Metabolic profiling analysis upon acylcarnitines in tissues of hepatocellular carcinoma revealed the inhibited carnitine shuttle system caused by the downregulated carnitine palmitoyltransferase 2. Mol Carcinog. 2019;58((5)):749–759. doi: 10.1002/mc.22967. [DOI] [PubMed] [Google Scholar]
- 26.Enooku K, Nakagawa H, Fujiwara N, Kondo M, Minami T, Hoshida Y, et al. Altered serum acylcarnitine profile is associated with the status of nonalcoholic fatty liver disease (NAFLD) and NAFLD-related hepatocellular carcinoma. Sci Rep. 2019;9((1)):10663. doi: 10.1038/s41598-019-47216-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fujiwara N, Nakagawa H, Enooku K, Kudo Y, Hayata Y, Nakatsuka T, et al. CPT2 downregulation adapts HCC to lipid-rich environment and promotes carcinogenesis via acylcarnitine accumulation in obesity. Gut. 2018;67((8)):1493–1504. doi: 10.1136/gutjnl-2017-315193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morad SAF, Cabot MC. Ceramide-orchestrated signalling in cancer cells. Nat Rev Cancer. 2013;13((1)):51–65. doi: 10.1038/nrc3398. [DOI] [PubMed] [Google Scholar]
- 29.Lin M, Liao W, Dong M, Zhu R, Xiao J, Sun T, et al. Exosomal neutral sphingomyelinase 1 suppresses hepatocellular carcinoma via decreasing the ratio of sphingomyelin/ceramide. FEBS J. 2018;285((20)):3835–3848. doi: 10.1111/febs.14635. [DOI] [PubMed] [Google Scholar]
- 30.Ogretmen B. Sphingolipid metabolism in cancer signalling and therapy. Nat Rev Cancer. 2018;18((1)):33–50. doi: 10.1038/nrc.2017.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krautbauer S, Meier EM, Rein-Fischboeck L, Pohl R, Weiss TS, Sigruener A, et al. Ceramide and polyunsaturated phospholipids are strongly reduced in human hepatocellular carcinoma. Biochim Biophys Acta. 2016;1861((11)):1767–1774. doi: 10.1016/j.bbalip.2016.08.014. [DOI] [PubMed] [Google Scholar]
- 32.Puri P, Wiest MM, Cheung O, Mirshahi F, Sargeant C, Min HK, et al. The plasma lipidomic signature of nonalcoholic steatohepatitis. Hepatology. 2009;50((6)):1827–1838. doi: 10.1002/hep.23229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee MJ, van Brocklyn JR, Thangada S, Liu CH, Hand AR, Menzeleev R, et al. Sphingosine-1-phosphate as a ligand for the G protein-coupled receptor EDG-1. Science. 1998;279((5356)):1552–1555. doi: 10.1126/science.279.5356.1552. [DOI] [PubMed] [Google Scholar]
- 34.Cuvillier O, Pirianov G, Kleuser B, Vanek PG, Coso OA, Gutkind JS, et al. Suppression of ceramide-mediated programmed cell death by sphingosine-1-phosphate. Nature. 1996;381((6585)):800–803. doi: 10.1038/381800a0. [DOI] [PubMed] [Google Scholar]
- 35.Simon J, Ouro A, Ala-Ibanibo L, Presa N, Delgado TC, Martinez-Chantar ML, et al. Sphingolipids in non-alcoholic fatty liver disease and hepatocellular carcinoma: ceramide turnover. Int J Mol Sci. 2019;21((1)):40. doi: 10.3390/ijms21010040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Celik O, Hascalik S, Sarac K, Meydanli MM, Alkan A, Mizrak B, et al. Magnetic resonance spectroscopy of premalignant and malignant endometrial disorders: a feasibility of in vivo study. Eur J Obstet Gynecol Reprod Biol. 2005;118((2)):241–245. doi: 10.1016/j.ejogrb.2004.06.030. [DOI] [PubMed] [Google Scholar]
- 37.de Molina AR, Sarmentero-Estrada J, Belda-Iniesta C, Taron M, de Molina VR, Cejas P, et al. Expression of choline kinase alpha to predict outcome in patients with early-stage non-small-cell lung cancer: a retrospective study. Lancet Oncol. 2007;8((10)):889–897. doi: 10.1016/S1470-2045(07)70279-6. [DOI] [PubMed] [Google Scholar]
- 38.Trousil S, Lee P, Pinato DJ, Ellis JK, Dina R, Aboagye EO, et al. Alterations of choline phospholipid metabolism in endometrial cancer are caused by choline kinase alpha overexpression and a hyperactivated deacylation pathway. Cancer Res. 2014;74((23)):6867–6877. doi: 10.1158/0008-5472.CAN-13-2409. [DOI] [PubMed] [Google Scholar]
- 39.Hannun YA, Obeid LM. Sphingolipids and their metabolism in physiology and disease. Nat Rev Mol Cell Biol. 2018;19((3)):175–191. doi: 10.1038/nrm.2017.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morita Y, Sakaguchi T, Ikegami K, Goto-Inoue N, Hayasaka T, Hang VT, et al. Lysophosphatidylcholine acyltransferase 1 altered phospholipid composition and regulated hepatoma progression. J Hepatol. 2013;59((2)):292–299. doi: 10.1016/j.jhep.2013.02.030. [DOI] [PubMed] [Google Scholar]
- 41.Shindou H, Shimizu T. Acyl-CoA:lysophospholipid acyltransferases. J Biol Chem. 2009;284((1)):1–5. doi: 10.1074/jbc.R800046200. [DOI] [PubMed] [Google Scholar]
- 42.Chen S, Kong H, Lu X, Li Y, Yin P, Zeng Z, et al. Pseudotargeted metabolomics method and its application in serum biomarker discovery for hepatocellular carcinoma based on ultra high-performance liquid chromatography/triple quadrupole mass spectrometry. Anal Chem. 2013;85((17)):8326–8333. doi: 10.1021/ac4016787. [DOI] [PubMed] [Google Scholar]
- 43.Chen S, Yin P, Zhao X, Xing W, Hu C, Zhou L, et al. Serum lipid profiling of patients with chronic hepatitis B, cirrhosis, and hepatocellular carcinoma by ultra fast LC/IT-TOF MS. Electrophoresis. 2013;34((19)):2848–2856. [PubMed] [Google Scholar]
- 44.Baniasadi H, Gowda GAN, Gu H, Zeng A, Zhuang S, Skill N, et al. Targeted metabolic profiling of hepatocellular carcinoma and hepatitis C using LC-MS/MS. Electrophoresis. 2013;34((19)):2910–2917. doi: 10.1002/elps.201300029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hall Z, Chiarugi D, Charidemou E, Leslie J, Scott E, Pellegrinet L, et al. Lipid remodelling in hepatocyte proliferation and hepatocellular carcinoma. Hepatology. 2021;73((3)):1028–1044. doi: 10.1002/hep.31391. [DOI] [PubMed] [Google Scholar]
- 46.Kimoto M, Whitley GSJ, Tsuji H, Ogawa T. Detection of NG, NG-dimethylarginine dimethylaminohydrolase in human tissues using a monoclonal antibody. J Biochem. 1995;117((2)):237–238. doi: 10.1093/jb/117.2.237. [DOI] [PubMed] [Google Scholar]
- 47.Boga S, Alkim H, Koksal AR, Bayram M, Ozguven MBY, Ergun M, et al. Increased plasma levels of asymmetric dimethylarginine in nonalcoholic fatty liver disease: relation with insulin resistance, inflammation, and liver histology. J Investig Med. 2015;63((7)):871–877. doi: 10.1097/JIM.0000000000000230. [DOI] [PubMed] [Google Scholar]
- 48.Kasumov T, Edmison JM, Dasarathy S, Bennett C, Lopez R, Kalhan SC, et al. Plasma levels of asymmetric dimethylarginine in patients with biopsy-proven nonalcoholic fatty liver disease. Metabolism. 2011;60((6)):776–781. doi: 10.1016/j.metabol.2010.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ferrigno A, Di Pasqua LG, Berardo C, Richelmi P, Vairetti M. Liver plays a central role in asymmetric dimethylarginine-mediated organ injury. World J Gastroenterol. 2015;21((17)):5131. doi: 10.3748/wjg.v21.i17.5131. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data
Supplementary data
Supplementary data
Data Availability Statement
The data generated and analyzed in this study are included in this article and/or its online supplementary material. Further inquiries can be directed to the corresponding author.