Abstract
Downstream sequences influence activity of the rice tungro bacilliform virus (RTBV) promoter in protoplasts derived from cultured rice cells. We previously identified a DNA element located between positions +50 and +90 relative to the transcription start site to which rice nuclear proteins bind. In this study, using DNA UV crosslinking assays, we show that two rice nuclear proteins bind specifically to this DNA element. We demonstrate that the DNA element enhances RTBV promoter activity in a copy number-dependent manner when transferred to a position upstream of the promoter. In addition, using electrophoretic mobility shift assays, we show that at least two novel nuclear proteins from rice cell suspension cultures bind to a subregion (from +50 to +59) of the DNA element and that a protein from rice root, but not shoot, nuclear extracts interacts with a perfect palindromic sequence motif located within the sequence +45 to +59. Furthermore, a position-dependent GAGA motif, present in three copies within downstream promoter sequences from +1 to +50, is involved in the regulation of RTBV promoter activity.
INTRODUCTION
Sequences located downstream of the transcription start site of protein coding genes can affect gene expression. At the post-transcriptional level, processes such as pre-mRNA processing (1), transcript stability (2) and translation efficiency (3–6) can be affected. However, in many eukaryotic genes, downstream sequences have been shown to act transcriptionally. Such downstream promoter sequences have been found both in TATA-containing (7–10) as well as in TATA-less promoters (11–13). Many of these downstream promoter sequences are involved in basal transcription by providing sites of interaction for the TFIID complex (8,10,14–16).
A conserved downstream core promoter element (DPE), located ∼30 nt downstream of the transcription start site, is required for sequence-specific binding of TFIID to a subset of TATA-less promoters (11). The DPE consensus (A/G)G(A/T)CGTG (11,12) appears to be as common as the TATA box in Drosophila core promoters (17). Analysis of interactions of purified TFIID revealed that Drosophila TAFII60 and TAFII40 specifically interact with the DPE (12). These observations suggest that downstream promoter elements may function as part of the core promoter by increasing TFIID–promoter complex formation and/or stability through direct interactions with TAFIIs. In other cases, gene-specific factors binding to downstream sequences stimulate transcription (7). The complex regulatory region downstream of the transcription start site in human immunodeficiency virus type 1 (HIV-1) comprises both DNA and RNA elements (18,19) and interacts with proteins in crude nuclear extracts (20,21)
Relatively little is known about regulation of gene expression by downstream promoter sequences in plants, although an increasing number of plant genes transcribed by RNA polymerase II have been found to contain downstream sequences that act transcriptionally (22–26); in other cases, whether regulation is at the transcriptional or translational level has not been determined (27). Sequences both upstream and downstream of the transcription start site are required for expression of the Arabidopsis thaliana EF-1α gene in leaves (28) and full light-regulated expression of the A.thaliana ferredoxin gene (29–31), indicating a combinatorial manner of regulation of expression of these genes (32).
Downstream promoter sequences influencing gene expression have also been found in some plant viruses (33–36). In rice tungro bacilliform virus (RTBV), the downstream promoter sequence (dps) contains a position-dependent element close to the transcription start site and a position-independent DNA enhancer element located between +50 and +90, which interacts with a nuclear protein(s) (33). The dps is necessary for promoter activity in protoplasts (33,37,38). In transgenic plants it modulates mRNA production, most likely at the level of RNA polymerase processivity (34). Upstream RTBV promoter elements are also required for quantitative regulation and for tissue specificity (34,37–41).
In the present study we have further dissected the downstream promoter elements and tested their effects on transcription from the RTBV promoter in a transient expression system. We provide evidence that the DNA element from +50 to +90 stimulates RTBV promoter activity in a copy number-dependent manner. We show that a GAGAG motif present in three copies within the region +1 to +50 of the dps is important for promoter activity. Furthermore, using electrophoretic mobility shift assays (EMSA) and in vitro footprinting, we have identified cell type-specific proteins that interact with downstream promoter elements and mapped a cluster of binding sites for these nuclear proteins.
MATERIALS AND METHODS
Plasmid constructions
Standard cloning techniques were used for all plasmid constructions (42). Plasmid R-218 contains RTBV promoter sequences up to position –218, the full-length RTBV leader sequences and a chloramphenicol acetyltransferase (CAT) gene fused to RTBV ORF I (33,37).
Internally deleted DNA fragments covering sequences from –218 to the first 20 nt of the CAT ORF were generated by ligating different combinations of 5′- and 3′-truncated fragments amplified by PCR and these were then cloned into the XbaI–XhoI sites of R-218 to create internal deletions RΔ8–90, RΔ50–90 and RΔ33–50.
A series of plasmids containing one, two or four copies of the DNA element (from +50 to +90) in an upstream position was constructed by insertion of the required number of copies of the element, flanked by XbaI sites, into the XbaI sites of RΔ8–90 and RΔ50–90, respectively. The resulting plasmids were designated 1-, 2- and 4-dRΔ8–90 and 1-, 2- and 4-dRΔ50–90, respectively.
RL50 was generated by cloning the PCR amplified DNA fragment corresponding to sequence –218 to +50 of the RTBV promoter, with an XbaI site at the 5′-end and the sequence 5′-atcaccATGgagctcgagaaa-3′ (translation start codon in upper case, NcoI and XhoI sites underlined) at the 3′-end, into the XbaI–XhoI sites of the construct R-218.
dRL50 was created by cloning the NheI–XhoI fragment from RL50 into the corresponding sites of 1-dRΔ50–90.
Constructs dRL50Δ+9/+27, dRL50Δ+28/+32, dRL50Δ+34/+39 and dRL50Δ+44/+48 were generated by an overlapping extension PCR strategy with Pfu DNA polymerase (Stratagene) using RL50 as template. Fixed primers were a 5′-end primer (P5) corresponding to sequences –218 to –200 of the RTBV promoter and a 3′-end oligonucleotide primer (P3) covering the sequences from +210 to +230 (including an EcoRI site) on the antisense strand relative to the translation start codon in the CAT gene. Four pairs of oligonucleotides corresponding to 5′- and 3′-truncated fragments were used as internal primers. The resulting PCR products were digested with NheI and XhoI and inserted between the corresponding sites of dRL50.
To generate plasmids dRL50ins1 and dRL50ins2, two PCR amplified DNA fragments, one from –218 to +10, flanked by (CA)10 at the 3′-end, and the other covering sequences from +11 of the leader to +230 of the CAT gene relative to the translation start codon and harboring (CA)10 at the 5′-end, were used as templates to amplify a mutated DNA fragment using P5 and P3. These PCR products were digested with NheI and XhoI, followed by cloning into the NheI–XhoI sites of dRL50 to yield dRL50ins1 carrying (CA)10 and dRL50ins2 with (CA)30.
To obtain plasmid pBL150, a PCR amplified DNA fragment spanning positions +1 to +150 was cloned into the XbaI and EcoRI (filled-in) sites of pBluescript II KS(+). Two PCR fragments covering sequences from +1 to +90 or +150, flanked by an XbaI site at the 5′-end and a HindIII site at the 3′-end, were inserted between the XbaI and HindIII sites of M13mp18 or pGEM-1 to create plasmids mL90 and pGL150, respectively.
Restriction digestion and DNA sequencing confirmed all deletions and mutations. Plasmids were isolated from Escherichia coli (strain DH5α) using a Qiagen plasmid kit.
Nuclear protein extraction
Nuclear extracts from cell suspensions of Oryza sativa line Oc were prepared as described (33). Nuclear extracts from 2-week-old O.sativa plant shoots and roots were prepared as described (37).
Electrophoretic mobility shift assay
A PCR amplified DNA fragment covering sequences +1 to +90, digested with XbaI at the 5′-end, was labeled with [α-32P]dCTP using the Klenow fragment of DNA polymerase. The resulting probe was purified on a 5% native polyacrylamide gel. EMSAs were performed essentially as described (37). 15 000 c.p.m. of labeled DNA probe (∼0.5 ng DNA) and 15–25 µg nuclear extract proteins were used for each reaction.
Copper-phenanthroline footprinting analysis
A 5′-end-labeled DNA probe was generated by digesting construct pBL150 with XbaI and labeling the bottom strand with [α-32P]dCTP using the Klenow fragment of DNA polymerase, followed by a secondary digestion with EcoRV. This labeled DNA probe was purified on a 5% native polyacrylamide gel. A 3′-end-labeled DNA probe was prepared in the same way, using HindIII-digested pGL150, with secondary digestion by SmaI. Footprinting analysis was performed as described (37). Chemical sequencing reactions were carried out according to the Maxam–Gilbert method (42).
DNA UV crosslinking assays
Single-stranded mL90 DNA was prepared as described previously (42). A uniformly labeled DNA fragment [with thymidine residues substituted by bromodeoxyuridine (BrdU)] was prepared with [α-32P]dCTP as described (43) and then digested with XbaI and HindIII, followed by purification on a 5% native polyacrylamide gel. Binding reactions were performed as described for EMSA and included 1 × 105 c.p.m. of the uniformly labeled DNA probe. Following a 20 min incubation period, the reaction mixtures were irradiated with UV light (312 nm) for 30 min in a UV Stratalinker 1800 (Stratagene) at 4°C. Samples were then digested with 10 U each of DNase I and micrococcal nuclease in the presence of 10 mM CaCl2 for 30 min at 37°C. The resulting mixtures were resolved on a 14% SDS–polyacrylamide gel.
Protoplast preparation and transfection
Protoplasts from O.sativa line Oc were isolated from cell suspension cultures as described (44). Protoplasts were transfected by the polyethyleneglycol method as described (44,45). Routinely, 10 µg test plasmid DNA was used to transfect 0.6 × 106 protoplasts, and an internal control plasmid expressing β-glucuronidase (GUS) under control of the cauliflower mosaic virus 35S promoter was co-transfected in all experiments.
Transient expression assays
Protein extracts were prepared after overnight incubation of protoplasts (46,47) and were analyzed for CAT (CAT ELISA kit; Roche Diagnostics Ltd) and GUS activities (47,48). CAT activities were calculated relative to the internal control in all cases. Relative expression levels did not vary more than ±15% with the given constructs in this study. All constructs were tested in at least three independent experiments with at least three independent isolates of each plasmid.
RESULTS
Cell type-specific nuclear proteins interact with downstream promoter elements
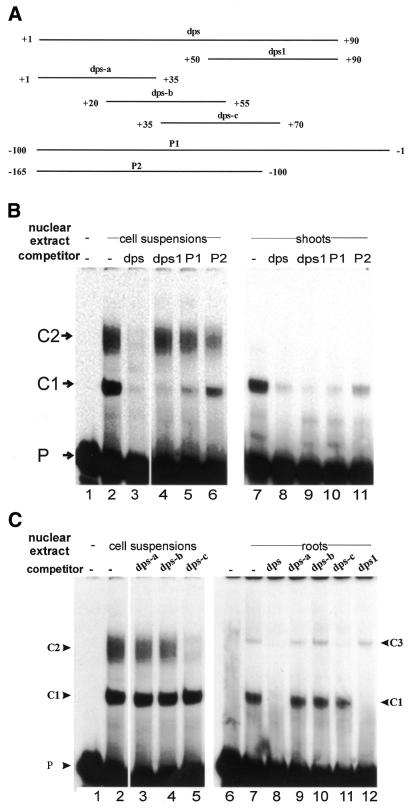
A nuclear protein prepared from rice cell suspensions interacts with a DNA element located in the region from +50 to +90 downstream of the RTBV transcription start site (here called dps1) forming a complex, C1 (33). To investigate whether additional proteins bind to other downstream sequences, we performed EMSA using greater amounts of nuclear extracts prepared from rice cell suspensions, shoots and roots. In addition to C1, which had been observed in all types of nuclear extracts (33) (Fig. 1B, lanes 2 and 7, and C, lane 7), DNA–protein complexes C2 and C3 were detected in cell suspension and in root nuclear extracts, respectively, using a radiolabeled dps fragment from +1 to +90 (Fig. 1B, lane 2, and C, lane 7).
Figure 1.
Analyses of the interactions between dps from +1 to +90 and proteins in different types of rice nuclear extracts. EMSAs were performed using a labeled DNA probe from +1 to +90. (A) DNA fragments (dps, P1 or P2) and double-stranded oligonucleotides having overlapping sequences between +1 and +90 (dps1 and dps-a to dps-c) were used as cold competitors at a 200-fold molar excess in EMSAs. (B) EMSAs were performed without (lane 1) or with nuclear extracts from cell suspensions or shoots in the absence (–) or presence of competitors (dps, dps1, P1 or P2). DNA–protein complexes (C1 and C2) are indicated. P is free DNA probe. (C) EMSAs were carried out without (lanes 1 and 6) or with cell suspension or root nuclear extracts in the absence (–) or presence of competitors (dps, dps1 or dps-a to dps-c). DNA–protein complexes are designated C1–C3.
To examine the specificity of these complexes, competitor DNA fragments (Fig. 1A) were used at a 200-fold molar excess. Unlabeled dps (+1 to +90) competed all complexes in all three types of nuclear extract (Fig. 1B and C), with complex C1, but not C2 and C3, being competed completely, confirming our previous results (33). However, complex C1 can also be competed by a DNA fragment spanning the promoter sequence from –100 to –1 (P1), and partially by sequences from –165 to –100 (P2). Fragment dps-c (+35 to +70; Fig. 1A), but not dps-a or dps-b, competed complexes C2 and C3 totally. We conclude that complexes C2 and C3 result from interactions of proteins with DNA sequences from +35 to +70. We refer to this region as dps2.
The formation of complex C1 requires the whole 40 bp of dps1
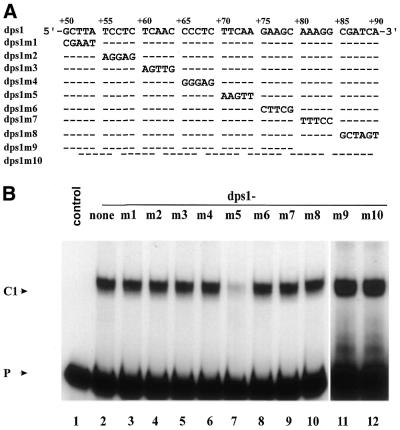
To determine the minimal sequence requirement for formation of complex C1, we used a series of double-stranded oligonucleotides with complementary mutations replacing stretches of 5 nt within the sequence from +50 to +90 and two shorter double-stranded oligonucleotides (+50 to +79 and +55 to +90) as competitors in EMSA (Fig. 2A). None of these competitors, except dps1m5 (+70 to +74 mutated) (Fig. 2B, lane 7), was able to compete complex C1 (Fig. 2B).
Figure 2.
Mutational analysis of the minimal binding site of complex C1 on dps1. (A) Wild-type dps1 and mutants were used as competitors at a 200-fold molar excess in EMSAs. Dashes indicate identity with the wild-type sequence. Only the mutated bases are indicated. (B) EMSA experiment with labeled DNA probe from +1 to +90 in the absence (lane 1) or presence (lanes 2–12) of nuclear extract from rice shoots. Competitors used are indicated at the top of the gel.
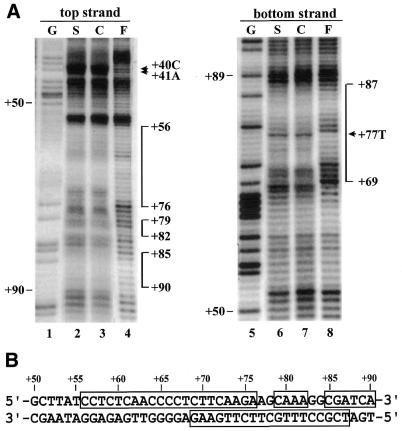
To further confirm these results, we performed copper-phenanthroline footprinting assays. EMSAs were carried out with nuclear extracts prepared from rice cell suspension cultures and shoots using a DNA probe from +1 to +150 labeled on either the top or bottom strand. Complex C1 was subjected to footprinting analysis. The results showed that footprinting patterns resulting from both shoot (S) and cell suspension (C) nuclear extracts are identical (Fig. 3A). The footprinted regions span sequences from +56 to +76, +79 to +82, and +85 to +90 on the top strand and from +69 to +87 on the bottom strand (Fig. 3A and B).
Figure 3.
Copper-phenanthroline footprinting analysis. (A) A DNA fragment covering +1 to +150 was 5′-end-labeled on either the top or bottom strand and incubated with rice shoot (S) or cell suspension (C) nuclear extracts. The mixture was resolved on a native polyacrylamide gel. DNA–protein complex C1 and free probe (F) were then digested in situ with 1,10-phenanthroline-copper ion. DNAs were eluted from the gel and resolved on a 6% polyacrylamide sequencing gel. A Maxam–Gilbert sequencing ladder (G) was run in parallel. The protected areas are depicted on the right of the gel by vertical bars. Numbers correspond to base pairs downstream of the transcription start site. The hypersensitive sites are indicated with arrowheads. (B) Nucleotide sequence of dps1 showing the areas protected in complex C1 (open boxes).
Taken together, these results suggest that formation of complex C1 requires the whole dps1 of 40 bp and that the CT-rich sequences are a major determinant of protein binding. The CA residues at positions +40 and +41 in the top strand and the T at position +77 in the bottom strand showed enhanced reactivities in the presence of proteins (Fig. 3A), probably indicating induced local conformational changes in the DNA upon protein binding.
The sequence required for formation of complex C2 is located in a subregion of dps1
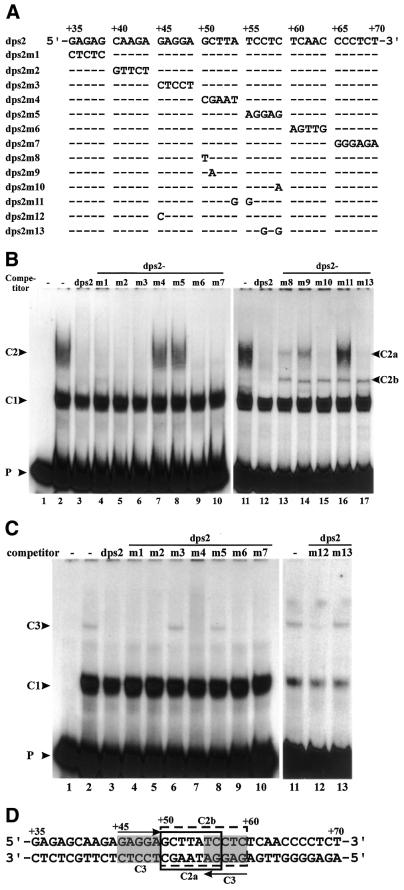
To localize the protein-binding sites of complex C2 precisely, complementary mutations were introduced in 5 nt units in the sequence +35 to +70 (Fig. 4A) (dps2m7 has 6 nt mutated). These mutated fragments were used as competitors in EMSA. Most of these competed for complex C2 as effectively as wild-type dps2 (Fig. 4B); only dps2m4 and dps2m5 failed to compete (Fig. 4B, lanes 7 and 8). This indicates that the sequence from +50 to +59 is required for protein binding. Replacing AT at +54 and +55 with GG (dps2m11) abolished competition. Mutations at positions +50 or +51 (dps2m8 and dps2m9) strongly reduced competition, while mutations at positions +57 and +59 (dps2m10 and dps2m13) had no effect. Interestingly, an additional complex migrating between complexes C2 and C1 appeared in the presence of all the latter set of mutant competitors (Fig. 4B, lanes 13–17), indicating that complex C2 contains more than one DNA-binding protein. To reflect this finding, C2 was renamed C2a and the intermediate complex C2b.
Figure 4.
Mutational analysis of the minimal binding sites of complexes C2 and C3 on dps2. (A) Wild-type dps2 and mutants used as competitors at a 200-fold molar excess in EMSAs. Dashes in the sequences indicate identity with the wild-type sequence. Only the mutated bases are indicated. (B) EMSAs were performed with labeled DNA probe from +1 to +90 in the absence (lane 1) or presence (lanes 2–17) of cell suspension nuclear extracts. Competitors used are indicated at the top of the gels. DNA–protein complexes are indicated. (C) EMSAs were performed as in (B) but with root nuclear extracts. (D) Nucleotide sequence of dps2. Open boxes with solid and dotted lines represent the binding sites of complexes C2a and C2b, respectively. Gray boxes indicate binding sites of complex C3. Arrows depict palindromic sequences.
In summary, these results indicate that the protein-binding site for formation of complex C2a is located within the sequence +50 to +56 (Fig. 4D), that formation of C2b requires sequences in the region +50 to +59 (Fig. 4D) and that these protein-binding sites overlap.
A nuclear protein from rice roots interacts with a palindromic sequence
DNA sequences involved in formation of complex C3 were similarly analyzed. The binding site could be localized to a palindromic sequence overlapping the site of complex C2 formation (Fig. 4C and D). C3 complex formation was abolished by mutation of either side of the GAGGA(N)5TCCTC palindrome from +45 to +59 (dps2m3 and m5) or by altering the CTC at positions +57 to +59 to GTG (dps2m13) (Fig. 4C). Complex formation was not affected by mutation of sequences separating the palindrome halves (dps2m4) and only barely affected by a change of the first G to C (dps2m12). In the sequence, the palindrome extends 2 nt further upstream and downstream, but mutation of these flanking nucleotides had no effect on C3 complex formation (dps2m2 and dps2m6, Fig. 4C, lanes 5 and 9). Figure 4D compiles an interpretation of the binding analyses of complexes C2a, C2b and C3.
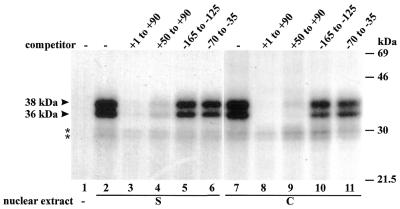
Two major proteins with apparent molecular masses of 36 and 38 kDa are specifically crosslinked to the dps
To further investigate DNA–protein interactions with the dps, we performed DNA UV crosslinking assays with a BrdU-substituted, uniformly radiolabeled DNA probe from +1 to +90 and nuclear extracts prepared from rice cell suspensions and shoots. Two major proteins with apparent sizes of 36 and 38 kDa were crosslinked to the labeled probe in both shoot (S) and cell suspension (C) nuclear extracts (Fig. 5). The proteins detected bound specifically, since binding of both of them could be competed totally by wild-type DNA (+1 to +90) and by a DNA fragment from +50 to +90, but not by upstream promoter sequences (from –165 to –125 and –70 to –35) (Fig. 5). The two weak bands (marked with asterisks) were not significantly competed by any of the competitors and are probably derived from the labeled DNA.
Figure 5.
Analysis of the DNA-binding proteins by DNA UV crosslinking. A radiolabeled DNA probe substituted with BrdU was incubated without (lane 1) or with nuclear extracts from rice shoots (S) or cell suspensions (C) in the absence (–) or presence of a 100-fold molar excess of the competitors indicated. UV crosslinked proteins were separated in a 14% SDS–polyacrylamide gel. The sizes of the marker proteins are indicated on the right. The arrowheads indicate the apparent molecular masses of the crosslinked proteins. Asterisks indicate incomplete digestion of the labeled DNA.
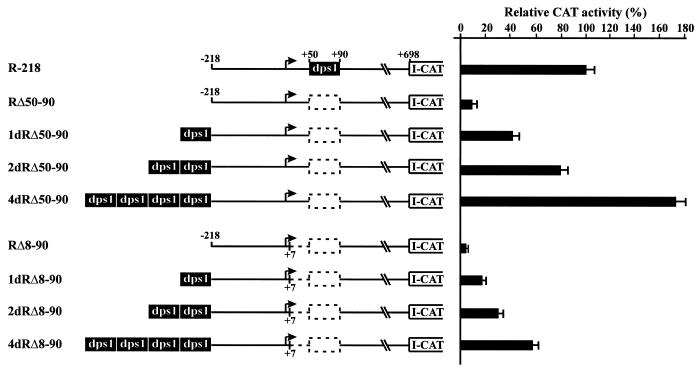
Dps1 enhances RTBV promoter activity in a copy number-dependent manner
We have previously shown that, in rice protoplasts, full RTBV promoter activity depends on the first 90 bp of dps in the context of upstream promoter sequences to position –681 (full length) or –218 (33). Within this dps region, dps1 (+50 to +90) was found to be position-independent, since it also functioned when transferred to an upstream position, although less efficiently than in its original position (33).
To further examine the functional contribution of dps1, we placed one, two or four copies of dps1 218 bp upstream of the transcription start site of the RTBV promoter, while simultaneously deleting it from its original position. The activity of these constructs was evaluated in transfected rice protoplasts. The activity of the construct 1-dRΔ50–90, which contains one upstream copy of dps1, was increased in comparison with the corresponding construct without the insertion (Fig. 6). Insertion of two copies of dps1 resulted in a significant enhancement of promoter activity, while four copies increased promoter activity to 171% compared to the wild-type construct R-218 (Fig. 6). These observations demonstrate that dps1 enhances promoter activity efficiently from an upstream position in a copy number-dependent manner, further confirming that dps1 contains a position-independent transcription regulatory element.
Figure 6.
dps1 enhances RTBV promoter activity from an upstream position. Constructs used for transfection of O.sativa protoplasts are shown on the left. The filled box indicates dps1 (+50 to +90). Dotted boxes depict deletions of dps1. The dotted line represents deletion of dps3 (+8 to +50). A bent arrow indicates the transcription start site. All constructs were tested in at least three independent transfections. For each construct the mean promoter activity is indicated as a percentage of the activity of the wild-type construct R-218 (set as 100%).
To test whether dps1 in an upstream position could also restore promoter activity when the complete dps region (from +8 to +90) was deleted in a downstream position, a similar series of constructs, but with additional deletion of the dps fragment from +8 to +90, was tested (Fig. 6). Promoter activity increased with the number of copies of dps1 inserted upstream, but was not completely restored (Fig. 6). Insertion of four copies of dps1 resulted in an 11-fold increase in promoter activity over construct RΔ8–90, but this level is still only 57% of that of the wild-type R-218.
These results indicate that the enhancement effect of one or more copies of dps1 is independent of the presence of the first 50 bp but that this region is required for full promoter activity. We refer to this region as dps3.
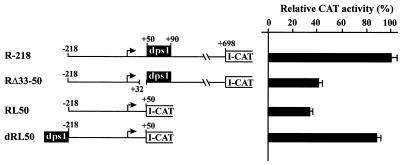
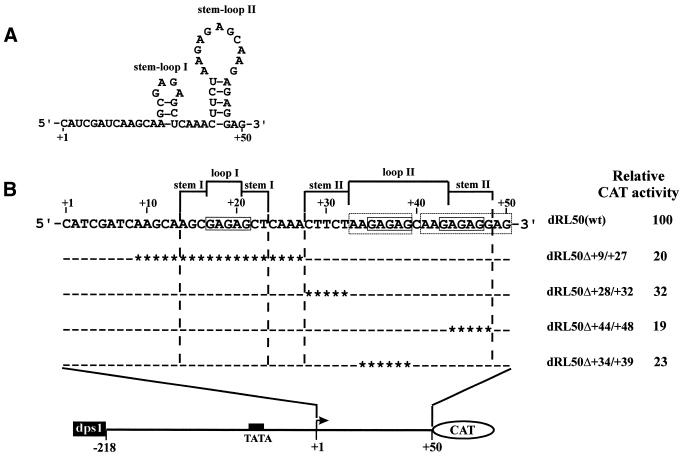
GAGAG motifs in dps3 contribute significantly to efficient promoter activity in vivo
Complete (+8 to +50) deletion of dps3 from plasmid RΔ50–90 (plasmid RΔ8–90) or partial (+33 to +50) deletion of dps3 from plasmid R-218 resulted in a 2.5- to 3-fold reduction in expression (Figs 6 and 7). Expression analysis with these constructs could be complicated by post-transcriptional contributions of the transcribed dps3 sequence to the complex expression strategy of RTBV. To reduce this complexity, we studied dps3 in the context of plasmid dRL50 (Fig. 7), which lacks large parts of the RTBV 35S RNA leader sequence, including dps1 and all sequences required for ribosome shunt, splicing and polyadenylation (34,49–52). dps1 was then inserted into an upstream position to increase the otherwise very low expression levels. Several deletions within dps3 were introduced into dRL50. They all reduced expression 3- to 5-fold. The dps3 region is characterized by a redundancy of GAGAG motifs and all deletions causing the most severe reductions remove one of these motifs (Fig. 8). The slightly less severe effect in dRL50d+28/+32 is caused by removal of another CT/GA-rich motif, which could also be involved in formation of a stem–loop structure at the RNA level (Fig. 8). GAGAG is a known binding site for transcription modulation factors (53), but we have not detected any direct DNA–protein interaction in the dps3 region.
Figure 7.
Deletion analysis of the RTBV leader sequences. Constructs consisting of the RTBV upstream promoter sequence (to position –218) and the RTBV leader sequence (either complete or truncated as indicated) are shown schematically. All constructs were tested in transfected O.sativa protoplasts in at least three independent experiments. For each construct the mean promoter activity is indicated as a percentage of the activity of the wild-type construct R-218 (set as 100%).
Figure 8.
GAGAG motifs contribute to RTBV promoter activity. (A) Nucleotide sequence and potential secondary structure of the dps3 transcript. (B) Schematic representation of the constructs tested in transfected O.sativa protoplasts showing the expanded dps3 region with deletions. A filled box indicates the TATA box. A bent arrow represents the transcription start site. Open boxes show the GAGAG motifs. Dashes in the sequences indicate identity with the wild-type sequence. Asterisks indicate deleted bases. Constructs and relative CAT activities are shown on the left. The relative CAT activity given for each construct is an average of at least three independent transient expression assays.
The spacing between the initiation site and the GAGAG motif is required for promoter activity
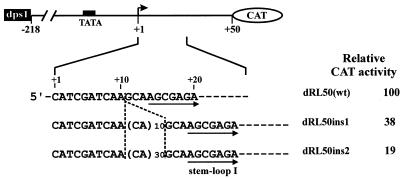
The fact that single deletions of any of the GAGAG motifs resulted in the same or an even greater reduction in expression as was previously found for removal of the whole dps3 (33) indicates that dps3 requires either a certain structure (possibly at the RNA level) or a certain spacing of several DNA elements to exert its function. This function can also be disturbed by alterations in the distances of the motifs from the transcription start site. Insertion of an unstructured sequence (CA)n between positions +10 and +11 reduced expression to 38% for n = 10 and 19% for n = 30 in RL50 derivatives (Fig. 9), with similar reductions in RL-218 derivatives (not shown).
Figure 9.
Effect of the spacing between the initiation site and the GAGAG motif on promoter activity. A schematic representation of the constructs tested in transfected O.sativa protoplasts is shown at the top, with the expanded sequence underneath indicating insertions between +10 and +11. Inserted nucleotides are shown in parentheses. Corresponding CAT activities are shown on the left. The results shown are the average of at least three independent transient expression assays.
DISCUSSION
Sequences downstream of the transcription start site can contain important determinants of gene expression efficiency. Such sequences, like all other regions of a gene, can contain motifs that attract enhancing or repressing proteins, i.e. classical enhancers or silencers. Their location close to the transcription start also allows a direct influence on formation of the basal transcription complex (11,12,14,15,17). Alternatively, sequences in the 5′-transcribed region can control RNA polymerase processivity (19,54,55), which is low at some promoters without specific activation.
The dps (+1 to +90) of the RTBV promoter contributes significantly to transcriptional activity. The region from +35 to +90 contains several different, independent, protein-binding regions. CT-rich sequences within dps1 (+50 to +90) form a complex (C1) with 36 and 38 kDa nuclear proteins from all rice cells tested. Similar sequences occur in the 5′-untranslated regions of several plant genes (22,23,26,56). CT-rich elements in the leader sequence of nuclear-encoded plastid proteins such as spinach PetE, PsaF and AtpC act transcriptionally rather than post-transcriptionally (22,23) and interact with nuclear proteins (23). In other cases, e.g. in the Arabidopsis biotin carboxylase or enoyl-acyl carrier protein reductase genes, the role of such CT-rich elements remains unknown (26,56).
Our results show that the DNA–protein complex C1 was competed by high amounts of an upstream DNA fragment from –100 to –1 (Fig. 1B, lanes 5 and 10), which itself binds proteins in an activator element (AE) from –70 to –35 (37). However, protein binding to AE was not competed by dps (unpublished observation). Since there is also no extended sequence homology between AE and dps1, we assume that the competition observed here is caused by interaction between dps1-binding proteins and proteins bound to the AE (our previous analysis indicates a number of proteins interacting with the DNA-contacting proteins at the AE; 37).
DNA–protein complex C2 forms around position +55 and comprises at least two DNA-binding proteins to form complexes C2a and C2b (Fig. 4B). Formation of complex C2 required flanking sequences at the 5′-end of the protein-binding site (Fig. 1B, lane 4). In contrast to complex C1, which was detected in all types of nuclear extracts tested, complex C2 was restricted to cell suspension nuclear extracts and possibly contributes to the differential effects of downstream sequences on RTBV promoter activity observed in transgenic rice plants (34) and transfected rice protoplasts (33 and this study).
A third, minor, protein complex (C3) is formed between root nuclear proteins and a palindromic [GAGGA(N5)TCCTC] sequence motif (dps2) overlapping the site of complex C2.
dps3 (+1 to +50) contains several GAGAG motifs, which could represent consensus binding sites for GAGA transcription factor (GAF). GAF is an essential protein in Drosophila, important for the transcriptional regulation of numerous genes (53). In plants, binding of a nuclear factor to the GAGA element located immediately downstream of the putative TATA box positively affects transcription of the gene gsa1 (57). Our results show that the GAGAG motifs within dps3 contribute significantly to efficient promoter activity. These effects might be position-dependent, since insertion of unstructured sequence between +10 and +11 resulted in a significant reduction in CAT activity (Fig. 9). Such insertions may also disturb sequence-specific interactions between basal transcription factors and downstream promoter sequences.
Studies in transgenic plants revealed the effects of dps1 and dps3 to be more complex (34). RTBV promoter constructs containing the dps region were active in more cell types in transgenic rice plants than those without (34). However, individual deletion of dps1 or dps3 resulted in an increase in expression (34), suggesting a negative effect. Nevertheless, dps1, dps2 and dps3 clearly function as positive elements in transfected rice cell suspension protoplasts. The upstream region of the RTBV promoter important for activity in the vascular system of plants (34,40) also associates with nuclear proteins from cell suspensions, but is rather an inhibitory element in protoplasts (38). The apparent contradiction in these findings might be due to differences in the nature of polymerase complexes formed at the RTBV promoter in different expression systems.
The presence of the RTBV polyadenylation signal at its native position, ∼220 nt downstream of the transcription start, results in the production of a short stop RNA in addition to the ‘full-length’ reporter-encoding RNA. Since the ratio of these two RNAs in transgenic plants varied with the promoter, it was suggested that RNA polymerase II complexes with different processivities associate with the RTBV initiation site (34). Low processivity would lead to increased pausing and transcription termination, i.e. production of short stop RNA, while high processivity would lead to a more efficient bypass and production of functional mRNA. In protoplasts, a very high proportion of the RNA is short stop RNA (52), suggesting that mainly low processivity complexes are present. This could be a general feature of the protoplast system or due to a lack of tissue-specific activators. Different processivities may depend on properties of the loaded polymerase complexes, as has been suggested for the HIV-1 promoter (58), or may result from incomplete activation by other promoter-associated factors. The differing effects of dps1/dps3 deletions in transgenic plant and protoplast systems may possibly be explained thus: in a system with (almost) exclusively low processivity complexes (protoplasts), any reduction in the number of such complexes will reduce expression, while in a system where high and low processivity complexes compete for promoter association (plants), a reduction in the latter may allow higher expression because of increased promoter clearing (58). According to this model, the dps elements would be involved in loading a basal, non-activated RNA polymerase complex to the promoter (59). Similar roles have been discussed for GA-rich and CT-rich downstream regions of the HIV-1 promoter (60). The sequence and location of the RTBV dps would be in keeping with such a scenario. Recruitment may involve direct contacts between the dps and TFIID, as described for other promoters with important elements just downstream of the transcription start site, or it may occur via chromatin effects of GAGA-associated factors. Thus, the dps region of RTBV and its associated factors may provide a useful model system to study transcriptional regulation at the level of initiation and early elongation in plants.
Acknowledgments
ACKNOWLEDGEMENTS
We especially thank Matthias Müller for his expert technical assistance. We greatly appreciate Drs Helen Rothnie, Patrick Matthias and Mikhail Pooggin for thorough critical reading of the manuscript. We also thank Peter Müller for synthesis of the oligonucleotides. The Friedrich Miescher Institute is part of the Novartis Research Foundation.
REFERENCES
- 1.Simpson G.G. and Filipowicz,W. (1996) Splicing of precursors to mRNA in higher plants: mechanism, regulation and sub-nuclear organisation of the spliceosomal machinery. Plant Mol. Biol., 32, 1–41. [DOI] [PubMed] [Google Scholar]
- 2.Abler M.L. and Green,P.J. (1996) Control of mRNA stability in higher plants. Plant Mol. Biol., 32, 63–78. [DOI] [PubMed] [Google Scholar]
- 3.Fütterer J. and Hohn,T. (1996) Translation in plants—rules and exceptions. Plant Mol. Biol., 32, 159–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gallie D.R. (1996) Translational control of cellular and viral mRNAs. Plant Mol. Biol., 32, 145–158. [DOI] [PubMed] [Google Scholar]
- 5.Gallie D.R. and Walbot,V. (1992) Identification of the motifs within the tobacco mosaic virus 5′-leader responsible for enhancing translation. Nucleic Acids Res., 20, 4631–4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stripecke R., Oliveira,C.C., McCarthy,J.E. and Hentze,M.W. (1994) Proteins binding to 5′ untranslated region sites: a general mechanism for translational regulation of mRNAs in human and yeast cells. Mol. Cell. Biol., 14, 5898–5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pelletier M.R., Hatada,E.N., Scholz,G. and Scheidereit,C. (1997) Efficient transcription of an immunoglobulin kappa promoter requires specific sequence elements overlapping with and downstream of the transcriptional start site. Nucleic Acids Res., 25, 3995–4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Emanuel P.A. and Gilmour,D.S. (1993) Transcription factor TFIID recognizes DNA sequences downstream of the TATA element in the Hsp70 heat shock gene. Proc. Natl Acad. Sci. USA, 90, 8449–8453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Purnell B.A., Emanuel,P.A. and Gilmour,D.S. (1994) TFIID sequence recognition of the initiator and sequences farther downstream in Drosophila class II genes. Genes Dev., 8, 830–842. [DOI] [PubMed] [Google Scholar]
- 10.Nakatani Y., Horikoshi,M., Brenner,M., Yamamoto,T., Besnard,F., Roeder,R.G. and Freese,E. (1990) A downstream initiation element required for efficient TATA box binding and in vitro function of TFIID. Nature, 348, 86–88. [DOI] [PubMed] [Google Scholar]
- 11.Burke T.W. and Kadonaga,J.T. (1996) Drosophila TFIID binds to a conserved downstream basal promoter element that is present in many TATA-box-deficient promoters. Genes Dev., 10, 711–724. [DOI] [PubMed] [Google Scholar]
- 12.Burke T.W. and Kadonaga,J.T. (1997) The downstream core promoter element, DPE, is conserved from Drosophila to humans and is recognized by TAFII60 of Drosophila. Genes Dev., 11, 3020–3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Minchiotti G., Contursi,C. and Di Nocera,P.P. (1997) Multiple downstream promoter modules regulate the transcription of the Drosophila melanogaster I, Doc and F elements. J. Mol. Biol., 267, 37–46. [DOI] [PubMed] [Google Scholar]
- 14.Knutson A., Castaño,E., Oelgeschläger,T., Roeder,R.G. and Westin,G. (2000) Downstream promoter sequences facilitate the formation of a specific transcription factor IID-promoter complex topology required for efficient transcription from the megalin/low density lipoprotein receptor-related protein 2 promoter. J. Biol. Chem., 275, 14190–14197. [DOI] [PubMed] [Google Scholar]
- 15.Lewis B.A., Kim,T.K. and Orkin,S.H. (2000) The downstream element in the human β-globin promoter: evidence of extended sequence-specific transcription factor IID contacts. Proc. Natl Acad. Sci. USA, 97, 7172–7177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Purnell B.A. and Gilmour,D.S. (1993) Contribution of sequences downstream of the TATA element to a protein-DNA complex containing the TATA-binding protein. Mol. Cell. Biol., 13, 2593–2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kutach A. and Kadonaga,J.T. (2000) The downstream promoter element DPE appears to be as widely used as the TATA box in Drosophila core promoter. Mol. Cell. Biol., 20, 4754–4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones K.A. (1993) Tat and the HIV-1 promoter. Curr. Opin. Cell Biol., 5, 461–468. [DOI] [PubMed] [Google Scholar]
- 19.Jones K.A. and Peterlin,B.M. (1994) Control of RNA initiation and elongation at the HIV-1 promoter. Annu. Rev. Biochem., 63, 717–743. [DOI] [PubMed] [Google Scholar]
- 20.Garcia J.A., Wu,F.K., Mitsuyasu,R. and Gaynor,R.B. (1987) Interactions of cellular proteins involved in the transcriptional regulation of the human immunodeficiency virus. EMBO J., 6, 3761–3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones K.A., Luciw,P.A. and Duchange,N. (1988) Structural arrangements of transcription control domains within the 5′-untranslated leader regions of the HIV-1 and HIV-2 promoters. Genes Dev., 2, 1101–1114. [DOI] [PubMed] [Google Scholar]
- 22.Bolle C., Herrmann,R.G. and Oelmuller,R. (1996) Different sequences for 5′-untranslated leaders of nuclear genes for plastid proteins affect the expression of the beta-glucuronidase gene. Plant Mol. Biol., 32, 861–868. [DOI] [PubMed] [Google Scholar]
- 23.Bolle C., Sopory,S., Lubberstedt,T., Herrmann,R.G. and Oelmuller,R. (1994) Segments encoding 5′-untranslated leaders of genes for thylakoid proteins contain cis-elements essential for transcription. Plant J., 6, 513–523. [DOI] [PubMed] [Google Scholar]
- 24.Helliwell C.A., Webster,C.I. and Gray,J.C. (1997) Light-regulated expression of the pea plastocyanin gene is mediated by elements within the transcribed region of the gene. Plant J., 12, 499–506. [DOI] [PubMed] [Google Scholar]
- 25.Curie C., Axelos,M., Bardet,C., Atanassova,R., Chaubet,N. and Lescure,B. (1993) Modular organization and development activity of an Arabidopsis thaliana EF-1 alpha gene promoter. Mol. Gen. Genet., 238, 428–436. [DOI] [PubMed] [Google Scholar]
- 26.de Boer G.J., Testerink,C., Pielage,G., Nijkamp,H.J. and Stuitje,A.R. (1999) Sequences surrounding the transcription initiation site of the Arabidopsis enoyl-acyl carrier protein reductase gene control seed expression in transgenic tobacco. Plant Mol. Biol., 39, 1197–1207. [DOI] [PubMed] [Google Scholar]
- 27.Rouster J., van Mechelen,J. and Cameron-Mills,V. (1998) The untranslated leader sequence of the barley lipoxygenase 1 (Lox1) gene confers embryo-specific expression. Plant J., 15, 435–440. [DOI] [PubMed] [Google Scholar]
- 28.Curie C. and McCormick,S. (1997) A strong inhibitor of gene expression in the 5′ untranslated region of the pollen-specific LAT59 gene of tomato. Plant Cell, 9, 2025–2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bovy A., Van den Berg,C., De Vrieze,G., Thompson,W.F., Weisbeek,P. and Smeekens,S. (1995) Light-regulated expression of the Arabidopsis thaliana ferredoxin gene requires sequences upstream and downstream of the transcription initiation site. Plant Mol. Biol., 27, 27–39. [DOI] [PubMed] [Google Scholar]
- 30.Caspar T. and Quail,P.H. (1993) Promoter and leader regions involved in the expression of the Arabidopsis ferredoxin A gene. Plant J., 3, 161–174. [DOI] [PubMed] [Google Scholar]
- 31.Dickey L.F., Gallo-Meagher,M. and Thompson,W.F. (1992) Light regulatory sequences are located within the 5′ portion of the Fed-1 message sequence. EMBO J., 11, 2311–2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Singh K.B. (1998) Transcriptional regulation in plants: the importance of combinatorial control. Plant Physiol., 118, 1111–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen G., Rothnie,H.M., He,X., Hohn,T. and Fütterer,J. (1996) Efficient transcription from the rice tungro bacilliform virus promoter requires elements downstream of the transcription start site. J. Virol., 70, 8411–8421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klöti A., Henrich,C., Bieri,S., He,X., Chen,G., Burkhardt,P.K., Wünn,J., Lucca,P., Hohn,T., Potrykus,I. and Fütterer,J. (1999) Upstream and downstream sequence elements determine the specificity of the rice tungro bacilliform virus promoter and influence RNA production after transcription initiation. Plant Mol. Biol., 40, 249–266. [DOI] [PubMed] [Google Scholar]
- 35.Hohn T., Corsten,S., Rieke,S., Müller,M. and Rothnie,H. (1996) Methylation of coding region alone inhibits gene expression in plant protoplasts. Proc. Natl Acad. Sci. USA, 93, 8334–8339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dey N. and Maiti,I.B. (1999) Structure and promoter/leader deletion analysis of mirabilis mosaic virus (MMV) full-length transcript promoter in transgenic plants. Plant Mol. Biol., 40, 771–782. [DOI] [PubMed] [Google Scholar]
- 37.He X., Hohn,T. and Fütterer,J. (2000) Transcriptional activation of the rice tungro bacilliform virus gene is critically dependent on an activator element located immediately upstream of the TATA box. J. Biol. Chem., 275, 11799–11808. [DOI] [PubMed] [Google Scholar]
- 38.He X., Fütterer,J. and Hohn,T. (2001) Sequence specific, methylation dependent and independent binding of rice nuclear proteins to a rice bacilliform virus vascular bundle expression element J. Biol. Chem., 276, 2644–2651. [DOI] [PubMed] [Google Scholar]
- 39.Yin Y. and Beachy,R. (1995) The regulatory regions of the rice tungro bacilliform virus promoter and interacting nuclear factors in rice (Oryza sativa L). Plant J., 7, 969–980. [DOI] [PubMed] [Google Scholar]
- 40.Yin Y., Chen,L. and Beachy,R. (1997) Promoter elements required for phloem-specific gene expression from the RTBV promoter in rice. Plant J., 12, 1179–1188. [DOI] [PubMed] [Google Scholar]
- 41.Yin Y., Zhu,Q., Da,S., Lamb,C. and Beachy,R. (1997) RF2a, a bZIP transcriptional activator of the phloem-specific rice tungro bacilliform virus promoter, functions in vascular development. EMBO J., 16, 5247–5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 43.Jackson S.P. (1993) Identification and characterization of eukaryotic transcription factors. In Hames,B.D. and Higgins,S.J. (eds), Gene Transcription. Oxford University Press, Oxford, UK, pp. 189–242.
- 44.Datta S.K., Peterhans,A., Datta,K. and Potrykus,I. (1990) Genetically engineered fertile Indica-rice recovered from protoplasts. Biotechnology, 8, 736–740. [Google Scholar]
- 45.Shillito R.D., Saul,M.W., Paszkowski,J., Müller,M. and Potrykus,I. (1985) High efficiency direct gene transfer to plants. Biotechnology, 3, 1099–1103. [Google Scholar]
- 46.Bonneville J.M., Sanfaçon,H., Fütterer,J. and Hohn,T. (1989) Posttranscriptional trans-activation in cauliflower mosaic virus. Cell, 59, 1135–1143. [DOI] [PubMed] [Google Scholar]
- 47.Fütterer J. and Hohn,T. (1991) Translation of a polycistronic mRNA in the presence of the cauliflower mosaic virus transactivator protein. EMBO J., 10, 3887–3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jefferson R.A., Kavanagh,T.A. and Bevan,M.W. (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J., 6, 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fütterer J., Potrykus,I., Bao,Y., Li,L., Burns,T.M., Hull,R. and Hohn,T. (1996) Position-dependent ATT initiation during plant pararetrovirus rice tungro bacilliform virus translation. J. Virol., 70, 2999–3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fütterer J., Potrykus,I., Valles,B.M., Dasgupta,I., Hull,R. and Hohn,T. (1994) Splicing in a plant pararetrovirus. Virology, 198, 663–670. [DOI] [PubMed] [Google Scholar]
- 51.Fütterer J., Rothnie,H.M., Hohn,T. and Potrykus,I. (1997) Rice tungro bacilliform virus open reading frames II and III are translated from polycistronic pregenomic RNA by leaky scanning. J. Virol., 71, 7984–7989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rothnie H.M., Chen,G., Fütterer,J. and Hohn,T. (2001) Polyadenylation in rice tungro bacilliform virus: cis-acting signals and regulation. J. Virol., 75, 4184–4194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wilkins R.C. and Lis,J.T. (1998) GAGA factor binding to DNA via a single trinucleotide sequence element. Nucleic Acids Res., 26, 2672–2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Krumm A., Hickey,L.B. and Groudine,M. (1995) Promoter-proximal pausing of RNA polymerase II defines a general rate-limiting step after transcription initiation. Genes Dev., 9, 559–572. [DOI] [PubMed] [Google Scholar]
- 55.Reeder T.C. and Hawley,D.K. (1996) Promoter proximal sequences modulate RNA polymerase II elongation by a novel mechanism. Cell, 87, 767–777. [DOI] [PubMed] [Google Scholar]
- 56.Bao X., Shorrosh,B.S. and Ohlrogge,J.B. (1997) Isolation and characterization of an Arabidopsis biotin carboxylase gene and its promoter. Plant Mol. Biol., 35, 539–550. [DOI] [PubMed] [Google Scholar]
- 57.Frustaci J.M., Sangwan,I. and O’Brian,M.R. (1995) gsa1 is a universal tetrapyrrole synthesis gene in soybean and is regulated by a GAGA element. J. Biol. Chem., 270, 7383–7393. [DOI] [PubMed] [Google Scholar]
- 58.Yankulov K., Blau,J., Purton,T., Roberts,S. and Bentley,D.L. (1994) Transcriptional elongation by RNA polymerase II is stimulated by transactivators. Cell, 77, 749–759. [DOI] [PubMed] [Google Scholar]
- 59.Sypes M.A. and Gilmour,D.S. (1994) Protein/DNA crosslinking of a TFIID complex reveals novel interactions downstream of the transcription start. Nucleic Acids Res., 22, 807–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.el Kharroubi A. and Martin,M.A. (1996) cis-acting sequences located downstream of the human immunodeficiency virus type 1 promoter affect its chromatin structure and transcriptional activity. Mol. Cell. Biol., 16, 2958–2966. [DOI] [PMC free article] [PubMed] [Google Scholar]