Abstract
The guide RNA-binding protein gBP21 has been characterized as a mitochondrial RNA/RNA annealing factor. The protein co-immunoprecipitates with RNA editing ribonucleoprotein complexes, which suggests that gBP21 contributes its annealing activity to the RNA editing machinery. In support of this view, gBP21 was found to accelerate the hybridization of cognate guide (g)RNA/pre-edited mRNA pairs. Here we analyze the mechanism of the gBP21-mediated RNA annealing reaction. Three possible modes of action are considered: chaperone function, matchmaker function and product stabilization. We conclude that gBP21 works as a matchmaker by binding to gRNAs as one of the two RNA annealing reactants. Three lines of evidence substantiate this model. First, gBP21 and gRNAs form a thermodynamically and kinetically stable complex in a 1 + 1 stoichiometry. Secondly, gRNA-bound gBP21 stabilizes single-stranded RNA, which can be considered the transition state in the annealing reaction. Thirdly, gBP21 has a low affinity for double-stranded RNAs, suggesting the release of the annealed reaction product after the hybridization step. In the process, up to six ionic bonds are formed between gBP21 and a gRNA, which decreases the net negative charge of the RNA. As a consequence, the electrostatic repulsion between the two annealing reactants is reduced favoring the hybridization reaction.
INTRODUCTION
RNA editing in kinetoplastid organisms is a processing reaction whereby the sequence of precursor mRNAs (pre-mRNAs) is post-transcriptionally modified (reviewed in 1). As a result, non-translatable primary transcripts are converted into translatable mRNAs, which makes RNA editing an essential reaction pathway for the expression of mitochondrial genes. The generated sequence alterations are insertions and deletions of exclusively uridylate residues. RNA editing is catalyzed by a high molecular mass ribonucleoprotein (RNP) complex that consists of three enzymatic core activities: a guide RNA (gRNA)-dependent endonuclease, a terminal uridylyltransferase and an RNA ligase (2–5). As trans-acting components the process involves small, metabolically stable mitochondrial transcripts, known as gRNAs (6). gRNAs hybridize to pre-edited mRNAs and dictate the insertion and deletion of the U nucleotides by anti-parallel base pairing. Chemical cross-linking experiments suggest that both termini of a gRNA are base paired to a pre-edited mRNA. This defines the editing reaction center between two, or possibly three, helical RNA elements (7). As RNA editing has a 3′ to 5′ processivity (with respect to the pre-mRNA molecule) and requires multiple gRNAs for the processing of one pre-mRNA, the reaction center must undergo dynamic changes. As a consequence, factors binding the RNA reactants as well as proteins promoting and resolving RNA/RNA interactions are probably important components in the process. Candidate polypeptides, which possibly contribute some of the described activities to the editing machinery, are the proteins mHel61p (8), REAP1 (9), TBRGG1 (10), RBP16 (11) and gBP21 (12). mHel61p is a mitochondrial DEAD box protein that has been shown to affect exclusively edited mitochondrial transcripts. The protein, probably acting as an RNA helicase, was suggested to be involved in unwinding gRNAs from fully edited mRNAs. TBRGG1 and RBP16 have poly(U) binding activity and thus might interact with the non-encoded 3′ oligo(U) extensions of gRNAs. REAP1 is an editing complex-associated polypeptide that has been demonstrated to preferentially bind to pre-edited mRNAs (13). gBP21 has recently been shown to have RNA/RNA annealing activity (14). Although the protein is not essential for RNA editing (15), its annealing activity is probably important for the editing machinery. Several lines of evidence support such a scenario: (i) cognate gRNA/pre-mRNA pairs have been found to be good annealing substrates for the protein (14); (ii) a polyclonal anti-gBP21 antiserum inhibits both RNA editing and RNA annealing in vitro (14,15) and (iii) a monoclonal antibody against gBP21 was found to immunoprecipitate deletional as well as insertional RNA editing activities (16).
In the present study, we address several questions concerning the molecular and thermodynamic basis of the RNA annealing activity of gBP21. We consider two principal modes of action: first, the protein might act as a positively charged catalyst in order to reduce the charge repulsion between the two polyanionic RNA reactants (17). This should increase the RNA/RNA collision frequency and consequently should kinetically favor the formation of the gRNA/pre-mRNA hybrid (17,18). Secondly, gBP21 might modulate the structure of one or both RNA reaction partners (19). This could be realized in three ways. (i) gBP21 could act as chaperone protein. Non-specific binding to one or both RNA reactants would induce conformational changes in order to resolve kinetically trapped RNA conformations. As a result, the concentration of RNAs that are competent to undergo a successful annealing event would increase and the protein could dissociate from the RNP complex. (ii) gBP21 could function as a matchmaker polypeptide. In a specific binding reaction an annealing-competent RNA conformation could be stabilized, gBP21 would remain bound to the RNA until the two RNA annealing partners met, at which time gBP21 would dissociate from the double-stranded reaction product (19–21). (iii) gBP21 could thermodynamically stabilize the hybridized RNA/RNA reaction product. In this case, the protein would not affect the substrate association kinetics (22).
Our data show that gBP21 performs its RNA annealing activity by acting as a matchmaker protein. This is deduced from the following results. gBP21 forms a kinetically and thermodynamically stable complex with gRNAs. In its gRNA-bound form, the protein is capable of stabilizing single-stranded (ss)RNA conformations in a sequence-non-specific manner. Furthermore, gBP21 has a low affinity for double-stranded (ds)RNA, which could trigger the release of the annealed RNA product. The matchmaker function is assisted by positive surface charges on the protein, which compensate negative charges of the gRNA.
MATERIALS AND METHODS
Expression of recombinant gBP21 and RNA synthesis
Recombinant gBP21 was expressed and purified essentially as described (12). Guide RNAs gA6-14, gND7-506 and gA6-48 were synthesized by run-off in vitro transcription using standard procedures (23). A dsRNA of 55 bp with 5′-overhangs of 5 and 9 nt was created from two complementary ssRNAs. Hybridization was achieved by heat denaturation followed by slowly cooling to room temperature (1°C/min). The dsRNA product was purified on a non-denaturing 8% (w/v) polyacrylamide gel.
RNP stoichiometry determination
Recombinant gBP21, radioactively labeled gRNA gA6-14 and preformed gA6-14/gBP21 complexes were analyzed on linear 5–35% (v/v) glycerol gradients in 6 mM HEPES/KOH pH 7.5, 50 mM KCl, 2.1 mM MgCl2, 0.1 mM Na2EDTA and 0.5 mM dithiothreitol (DTT). Centrifugation was carried out in a Beckman SW41 rotor at 35 000 r.p.m. for 36 h at 4°C. Gradients were fractionated and assayed for the presence of protein by SDS–PAGE and for radioactively labeled gRNA by scintillation counting. Apparent sedimentation coefficients were determined by comparison to five marker proteins: bovine pancreas chymotrypsinogen α [(2.5 Svedberg units (S)], ovalbumin (3.5 S), BSA (4.3 S), pig heart lactate dehydrogenase (6.93 S) and rabbit skeletal muscle aldolase (7.35 S) (24). The apparent S-value of the complex was correlated with the Perrin factor (F) following the equation F = Mr (1 – Vxδ)/(6 π η rH S NA). Next, F was used to calculate the apparent axial ratios for the gA6-14/gBP21 complex (assuming different complex stoichiometries) according to F = (1 – p2)1/2/(p2/3 ln{[1+(1 – p2)1/2]/p}) (25). This equation is valid for a prolate ellipsoid, where p = short axis/long axis. Mr is the molecular mass (gBP21, 21 000 g mol–1; gRNA, 27 000 g mol–1; gA6-14/gBP21 complex, varies depending on the stoichiometry), Vx is the partial specific volume [0.73 ml/g for protein (25), 0.56 ml/g for RNA (26), 0.63 ml/g for the gBP21/gRNA complex (27)], δ is the density of the liquid (1.02 g ml–1 for all three molecules), rH is the hydrated radius of the equivalent sphere and S is the observed Svedberg constant (see above). The viscosity of the liquid in the gradient, η, was set to 1 g m–1 s–1 for all molecules, as the calibration of the gradient with the standard proteins includes the frictional characteristics of the gradient. The hydrated radius rH was estimated from rH = [3Mr(Vx + Vlδx)/4πNA]1/3 (25). Mr is the molecular mass, NA is Avogadro’s number, δx is the hydration (w/w) of the macromolecule [0.35 for protein; 0.60 for RNA (25) and 0.50 for the RNP complex] and Vl is the partial specific volume of the liquid (0.98 ml g–1).
Adsorption filtration
Filter binding assays were performed essentially as described by Köller et al. (12). Unless stated otherwise, all binding experiments were performed with 100 nM gBP21 in 6 mM HEPES/KOH pH 7.5, 50 mM KCl, 2.1 mM MgCl2, 0.1 mM Na2EDTA, 0.5 mM DTT and 750 nM BSA containing ∼0.3% (v/v) glycerol from the protein storage buffer. Experimental data were fitted to the Langmuir adsorption isotherm [AB] = n[B]/(Kd + [B]) where AB is the complex, B is the ligand and n is the number of binding sites. Identical results were obtained when the Scatchard equation was used. In order to obtain different pH values in the binding reaction, 6 mM HEPES pH 7.5 was substituted by 1 mM HEPES plus 5 mM of the following buffers: MES–KOH (final pH 5.8–7.1), MOPS–KOH (final pH 6.6–7.5), HEPES–KOH (final pH 6.6–7.9) and Tris–HCl (final pH 7.1–8.8).
The number of ion bonds (m) was determined using equation: m φ = –∂logKA/∂log[M+] where the empirical factor φ is, on average, 0.85 for RNA and KA is Kd–1 (28). The energetic contribution of the ionic bonds was calculated from ΔGOBS = ΔG0 + ΔGION (ΔGOBS is the observed free energy, ΔGION is the ionic contribution and ΔG0 is the non-ionic contribution to the free energy). ΔG values were calculated from ΔG = –RT lnKA where R is the gas constant and T is the absolute temperature. For ΔGOBS, the measured KA was used. For ΔG0 a KA extrapolated to a monovalent ion concentration of 1 M was used (28).
Competition experiments contained a fixed concentration (15 nM) of radioactively labeled gRNA gA6-14 and varying amounts of competitor RNA. The data were fitted to [AB] = (n[A]/Kd,A)/{1 + ([A]/Kd,A) + ([C]/Kd,C)} where AB is the complex, A is the labeled ligand, C is the competitor, Kd,A is the Kd between A and B, Kd,C is the Kd between A and C and n is the number of binding sites (29).
Optical biosensor measurements
Kinetic measurements of the complex formation between gRNAs and gBP21 were analyzed in real time using a resonant mirror system (Affinity Sensors, Cambridge, UK). Recombinant gBP21 was covalently attached to the aminosilane surface of an IAsys micro-cuvette (Affinity Sensors) using the bifunctional cross-linker bis-(sulfosuccinimidyl) suberate. Remaining activated sites were blocked with BSA. To obtain equilibrium binding constants, the equilibrium resonant angle shifts were plotted as a function of the gRNA concentration and fitted to the Langmuir adsorption isotherm or the Scatchard equation. First order association rate constants (k1) were obtained by exponential fits of the signal intensities as a function of time. Second order association rate constants (k2) were determined by plotting k1 as a function of the gRNA concentration. The slope of the resulting line equals k2. Dissociation rate constants kdiss were obtained from kdiss = Kd × k2. The half-life of the complex (t1/2) equals ln2/kdiss.
In vitro selection (SELEX)
The starting DNA library and the amplification primers are described by Homann and Göringer (30). The initial RNA transcription was performed with T7 RNA polymerase using standard procedures yielding an estimated pool no. 1 complexity of 1.6 × 1015 unique RNA sequences. All transcription reactions were carried out in the presence of [α-32P]NTP to radioactively label the RNA. gBP21-bound RNA was isolated by filter binding using nitrocellulose/cellulose acetate mixed bed filters. To minimize the selection of aptamers against the filter material, pre-selections were carried out by filtering all pool RNAs through two filter disks. The percentage of gBP21-bound RNA was calculated by scintillation counting and bound RNA was extracted with 20 mM Tris–HCl pH 7.5, 4 M guanidinium thiocyanate. To select for high affinity aptamers, initial protein and RNA concentrations were chosen at least 5-fold below the known Kd for gRNAs (12) (RNA 1.5 nM, gBP21 0.95 µM). During the first five SELEX rounds the selection stringency was increased stepwise by decreasing the amount of gBP21 (from 400 to 2.4 µg), decreasing the amount of RNA (from 850 µg to 500 ng) and increasing the reaction volume from 20 to 80 ml. Conditions were kept constant after round no. 5 (RNA 0.22 nM, gBP21 1.4 nM). Selected RNAs were reverse transcribed using MuLV-RT for 25 min at 42°C followed by a 5 min incubation at 48°C. Products were purified and PCR amplified. DNA templates of SELEX round no. 10 were cloned into pUC18 and sequenced by dideoxy terminator sequencing. Sequence analysis and RNA secondary structure analysis was done using the GCG software package (Wisconsin Package v.9.1, Genetics Computer Group, Madison, WI). Control sequences were randomized either unbiased (25% probability for each base) or with the same bias as the selected aptamer pool no. 10 (8% G, 24% A, 28% T and 40% C). All randomizations were executed using the AWK desktop calculator on Unix.
Individual aptamers were analyzed for their affinity to gBP21 resulting in Kd values of 3 ± 1 nM. The Kd of pool no. 10 RNA was 3.5 nM while the Kd for pool no. 1 RNA was 10 nM. Thus, during the selection the average affinity for gBP21 increased by a factor of three with no significant affinity differences between individual aptamers. The specificity of the selected aptamers for the gRNA binding site on gBP21 was determined in a competition assay (see above). From pool no. 1 to pool no. 10 specificity increased by a factor of seven. Most aptamers competed radioactively labeled gRNA with a Kd <20 nM.
RNA gel electrophoresis
Analytical denaturing gel electrophoresis was performed in 10% (w/v) polyacrylamide [acrylamide:bis-(acrylamide) 19:1] slab gels containing 8 M urea and buffer TBE pH 8.3 (89 mM Tris–borate pH 8.3, 2 mM Na2EDTA). Non-denaturing gels contained 8% (w/v) polyacrylamide [acrylamide:bis-(acrylamide) 19:1], 0.5× TBE pH 8.3 and were run at 4°C and 11 V/cm for 5 h. RNA mobilities within the native gels were analyzed based on the Lumpkin–Zimm equation (31). The RNAs were treated as prolate ellipsoids and axial ratios were estimated by calibration to the known geometries of tRNA and oligo(C)75 (32,33).
RESULTS
The gBP21/gRNA complex is kinetically stable and has a 1 + 1 stoichiometry
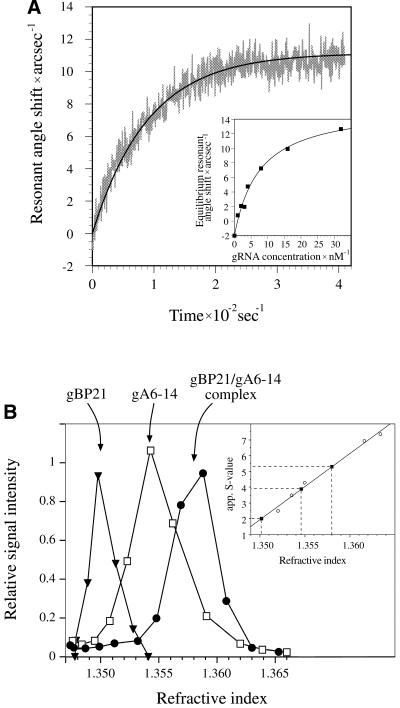
In order to function as an RNA annealing factor, gBP21 has to recognize and stably bind one or both RNA reactants. The resulting RNP complex should have defined characteristics such as (i) a thermodynamic stability representing a high affinity interaction, (ii) a defined protein/RNA stoichiometry, and (iii) a half-life of the RNP complex sufficient to execute its function. Previous results have established that gBP21 is capable of forming RNP complexes with gRNAs (12,34). gBP21/gRNA complexes are characterized by Kd values of ≤10 nM (12) and thus meet the required high affinity characteristics. In contrast, gBP21 does not directly interact with pre-mRNA molecules, the other RNA substrate in the annealing reaction (14). In order to measure the kinetic stability of the gBP21/gRNA complex we performed time-dependent binding experiments using a resonant mirror detection system. Recombinant gBP21 was covalently immobilized on an aminosilane surface and the association with different gRNA ligands was monitored in real time (Fig. 1A). From the binding curves we derived first order association rate constants (k1), which were converted to second order association rate constants (k2) by plotting k1 as a function of the gRNA concentration. Dissociation rate constants (kdiss) were determined as outlined in the Materials and Methods. Table 1 summarizes the resulting data for three different gRNA ligands. The derived kdiss values are on average 0.018 s–1, which correlates with an average half-life (t1/2) for the different gBP21/gRNA complexes of 43 s.
Figure 1.
Characterization of the gBP21/gRNA complex. (A) Determination of the kinetic and thermodynamic stability of the gBP21/gRNA complex using an optical biosensor (Affinity Sensors). gBP21 was covalently coupled to the surface of a micro-cuvette and the progress of the interaction with added gRNA gA6-14 was recorded in real time. The plot shows the change in the resonant angle shift as a function of time. A monoexponential curve fit of the data points allowed the determination of first order association rate constants. (Insert) Equilibrium signals were plotted as a function of the gRNA concentration and curve fitted. The obtained equilibrium dissociation constants are identical to the results obtained by adsorption filtration (Kd = 8 ± 2 nM) (12). The kinetic data were used to calculate the half-lives of three gBP21/gRNA complexes (see Table 1). (B) Determination of apparent sedimentation coefficients for gBP21, radioactively labeled gRNA (gA6-14) and the gA6-14/gBP21 complex. The three molecules were sedimented in linear glycerol gradients and plotted as a function of the refractive index of the gradient fractions. In comparison to the sedimentation behavior of standard proteins (insert) the following apparent S-values were derived: gBP21, 2.0 S; gRNA gA6-14, 3.9 S; gA6-14/gBP21 complex, 5.3 S. The apparent S-value of the complex was used to calculate the axial ratio of the gBP21/gRNA complex (see Materials and Methods), in agreement with a 1 + 1 stoichiometry.
Table 1. Kinetic and thermodynamic characteristics of three different gBP21/gRNA complexesa.
| gRNA | k2 (106 M–1 s–1)b | Kd (nM)b | kdiss (s–1) | t1/2 (s) |
|---|---|---|---|---|
| gA6-14 | 1.6 ± 0.2 | 8 | 0.013 | 53 |
| gA6-48 | 2.8 ± 0.9 | 5 | 0.014 | 50 |
| gND7-506 | 4.5 ± 1.8 | 6 | 0.027 | 26 |
aSecond order binding rate constants (k2), dissociation constants (Kd), dissociation rate constants (kdiss) and half-lives (t1/2). Errors are uncertainties of the mean.
bThe values are derived from resonant mirror experiments.
To determine the stoichiometry of the gBP21/gRNA complex, we performed isokinetic density gradient centrifugations. Recombinant gBP21, a radioactively labeled gRNA (gA6-14) and preformed gA6-14/gBP21 complexes were sedimented in linear glycerol gradients (Fig. 1B). In comparison with molecules of known S-values, we derived an apparent S-value for the gBP21/gRNA complex of 5.3 S. As outlined in the Materials and Methods, this sedimentation coefficient was used to calculate axial ratios for the gBP21/gRNA complex assuming different protein/RNA stoichiometries. For the 1 + 1 RNP complex we derived an axial ratio of 1:1.8. Any higher order stoichiometry [(gBP21)2/RNA or (RNA)2/gBP21 and higher] resulted in very asymmetric (greater than 1:6) and thus unrealistic axial ratios. Therefore, the gBP21/gRNA complex probably has a 1 + 1 stoichiometry.
gRNA-bound gBP21 stabilizes ssRNA
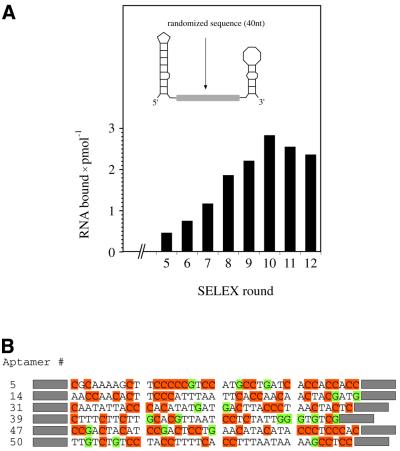
Boundary experiments and RNase protection data have shown that the primary RNA binding site for gBP21 is an irregular hairpin structure, such as the stem–loop, that can form near the 3′ end of gRNAs (14,23,35). Müller et al. (14) further demonstrated that an RNA hairpin is sufficient for the high affinity interaction with gBP21. However, the nucleotides that base pair to the pre-mRNA during the annealing reaction are located at the 5′ end of a gRNA. This sequence stretch, known as the anchor sequence, has been shown to be involved in a stem–loop structure of low thermodynamic stability (23,35). It must be presented in a single-stranded conformation in order to be available for the gRNA/pre-mRNA interaction. This suggests that gBP21 has a second RNA binding domain adjacent to the 3′ hairpin, which provides a presentation platform for the gRNA anchor. To test whether gRNA-bound gBP21 has an affinity for ssRNA, we performed an in vitro selection (SELEX) experiment using a combinatorial RNA library. The SELEX technology has been used before to determine the RNA recognition elements of different proteins (36). To account for the hairpin region as the primary gRNA binding site, we used a structurally biased combinatorial RNA library. It consists of a randomized region of 40 nt flanked by primer binding sites, which are able to form hairpin structures (Fig. 2A, insert). Twelve SELEX rounds were performed. The amount of gBP21-bound RNA increased from round no. 5 to a maximum in round no. 10 (Fig. 2A). PCR products of round no. 10 were cloned and 45 clones were sequenced. Representative sequences are shown in Figure 2B. They are characterized by a high C nucleotide content (39 ± 8%) and a low abundance of G nucleotides (8 ± 4%). A and T nucleotides are present at equal ratios (A, 25 ± 7%; T, 28 ± 7%). Predictions of minimal free energy structures for the various aptamers suggest a single-stranded conformation for the 40 nt sequence stretch with an average Gibbs free energy of –68 ± 2 kJ mol–1. This differs significantly from the calculated ΔG values of unbiased randomized sequences of identical length (–85 ± 3 kJ mol–1).
Figure 2.
Selection of RNA aptamers to recombinant gBP21. (A) (Insert) Schematic representation of the starting RNA pool (pool no. 1), which consists of a central 40 nt randomized sequence stretch flanked by primer binding sites that are predicted to fold into irregular hairpin structures. Either of the two hairpins can function as a binding site for gBP21 (14,35). Bar graph of the amount of bound RNA during 12 SELEX rounds. Maximal binding was achieved in round no. 10, which was cloned and further analyzed. (B) DNA sequences of six representative aptamers. C nucleotides are highlighted in orange and G nucleotides in green to emphasize the nucleotide bias of the selected aptamers. Primer binding sites are shown as gray boxes.
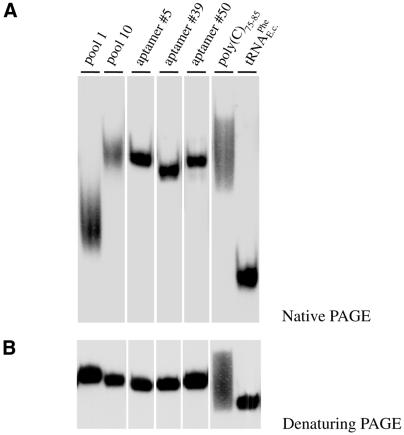
To experimentally verify the high proportion of single strand within the selected aptamers, we examined the electrophoretic mobility of three representative RNAs (aptamers no. 5, 39 and 50; Fig. 2B) in non-denaturing polyacrylamide gels (Fig. 3A). Native PAGE has successfully been applied to monitor the global architecture of RNAs in many systems (37). As controls we used a compactly folded tRNA, SELEX pool no. 1 and pool no. 10 and single-stranded oligo(C)75–85. Pool no. 1 RNA shows an electrophoretic mobility between that of tRNA and oligo(C). In contrast, pool no. 10 RNA, as well as the three aptamers, shows a reduced electrophoretic mobility similar to the migration of oligo(C). As no electrophoretic difference was detected in urea containing polyacrylamide gels (Fig. 3B), the above described reduced migration is indicative of a high degree of single strand within the aptamers. To gain a more quantitative measurement we calculated the overall geometries for the three aptamers, assuming the general structure of a stretched ellipsoid. The axial ratios ranged between 1:7 and 1:10 in support of a high amount of single strand within the three RNAs.
Figure 3.
Gel electrophoretic analysis of gBP21 aptamers. (A) Autoradiogram of a gel electrophoretic separation of three representative radioactively labeled RNA aptamers (aptamers no. 5, 39 and 50; for primary sequences see Fig. 2B) in a non-denaturing polyacrylamide gel. For comparison, RNAs from pool no. 1, pool no. 10, oligo(C)75–85 and tRNAPhe from E.coli were run at the same time. The reduced electrophoretic mobility of the three aptamers and of pool no. 10 is indicative of a high degree of single strand within their structures. (B) The same RNA samples as in (A) but separated in a denaturing polyacrylamide gel. As expected, the three aptamers and the two RNA pools show identical electrophoretic mobilities.
gBP21 has a low affinity for dsRNA
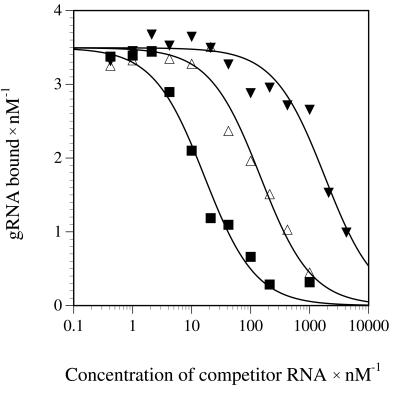
In order to test whether the partially double-stranded gRNA/pre-mRNA annealing product is stabilized by gBP21, we determined the affinity of the protein for dsRNA. This was done in a competition experiment where a radioactively labeled gBP21-bound gRNA was competed out of the RNP complex by increasing amounts of dsRNA. As controls, we measured competition curves with gRNAs and tRNA molecules. Figure 4 shows a representative result. Curve fitting of the data allowed the calculation of Kd values to the gRNA binding site: 800 nM for dsRNA, 100 nM for tRNA and 8 nM for gRNA. Thus, the Kd for the gBP21/dsRNA complex is two orders of magnitude above the Kd for the gBP21/gRNA complex. This corresponds to a difference of 12 kJ mol–1 in the complex stability (ΔΔG) and demonstrates a significant discriminative range between the two RNA ligands. The fact that some single-stranded oligonucleotide homopolymers can function as effective competitors has been shown previously (34,38,39).
Figure 4.
gBP21 has a low affinity for dsRNA. Radiolabeled gRNA gA6-14 was bound to gBP21 and incubated with increasing amounts of a 55 bp dsRNA as a competitor RNA. The amount of bound gRNA was determined by filter binding and plotted as a function of the competitor concentration. The data were fitted as outlined in the Materials and Methods to derive a Kd value of 800 nM (filled triangles). Control competition experiments were performed with the homologous competitor gA6-14 (filled squares, Kd = 8 nM) and tRNAPhe as a non-homologous RNA (open triangles, Kd = 100 nM).
Up to six ion bonds stabilize the gBP21/gRNA complex
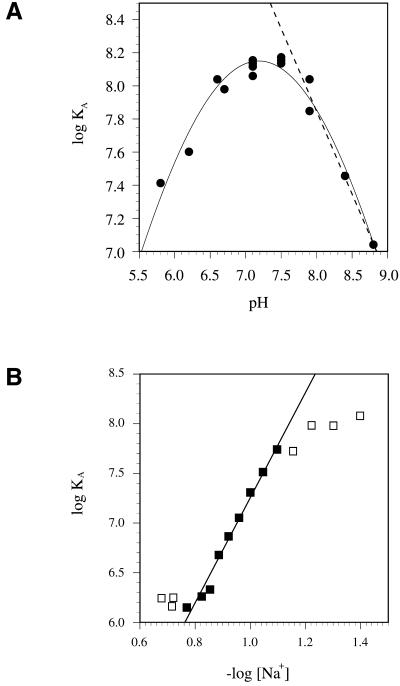
The amino acid composition of gBP21 makes it probable that charge/charge interactions contribute to the mechanism of the annealing reaction. The protein has 18 arginines, 10 lysines, 10 histidines and a pI of 9.7 (12). It is predicted to carry five positive surface charges at neutral pH. This should be reflected in the pH dependence of the stability of the gBP21/gRNA complex. We measured the dependence of the logKA as a function of the pH by adsorption filtration. As expected from the interaction of two oppositely charged macromolecules, this plot shows a bell-shaped curve (Fig. 5A). The affinity maximum is between pH 7.0 and 7.5. The slope of the pH dependence (δlogKA/δpH) in the alkaline range lies between –1.0 and –1.8, corresponding to one to two proton titratable groups that influence the interaction (40). Half of this slope is reached at pH 7.5, representing the upper limit of the pKa values of these groups. This value approaches the pKa of histidine, which is in the range of 5.1–7.2 (41). Thus, one to two titratable ionic bonds, probably between histidines and phosphate groups, contribute to the pH dependence of the gBP21/gRNA interaction.
Figure 5.
Electrostatic contributions to the gBP21/gRNA interaction. (A) pH dependence of the binding affinity. The logKA was determined by adsorption filtration and plotted as function of the pH. Linear regression of the data points in the descending region of the curve according to deHaseth et al. (40) allows the conclusion that one to two proton titratable ionic bonds are involved in the gBP21/gRNA interaction. (B) Dependence of the binding affinity on the monovalent ion concentration. The logKA was determined by adsorption filtration and plotted as function of –log[Na+]. A linear least square fit of the data points between 70 and 150 mM [Na+] has a slope (δlogKA/δ – log[Na+]) of 5.8. Using the semi-empirical equation of Record et al. (28), involvement of up to six ionic bonds in the interaction is suggested.
To determine the total number of ionic bonds that contribute to the formation of the gBP21/gRNA complex, we titrated the ionic groups by increasing the concentration of monovalent ions in the binding reaction (28). The interaction affinities were again determined in a filter binding assay. The relation between logKA and –log[Na+] is shown as a linear function between 70 and 150 mM Na+ (Fig. 5B). The slope (δlogKA/δ –log[Na+]) is 5.3, corresponding to an upper limit of 6.2 ionic bonds. Thus, gBP21 reduces the negative charge of the gRNA molecule in the interaction and the association is stabilized by up to six ionic bonds, one to two of which are probably mediated by histidine residues. The absence of a pronounced curvature in the plot of Figure 5B (between 70 and 150 mM Na+) indicates that a possible anionic contribution to the binding reaction is negligible. The free energy contribution of the six ionic bonds was calculated to be –35 kJ mol–1, which is ∼75% of the total free energy of the interaction (–46 kJ mol–1).
DISCUSSION
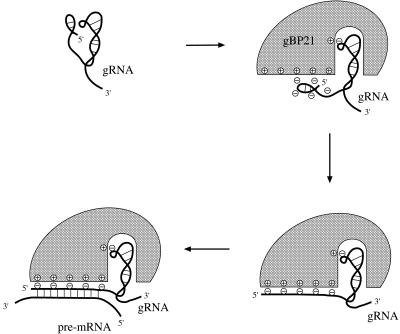
In this study we analyzed the interaction between gBP21 and gRNAs to gain insight into the mechanism by which the protein promotes the annealing of base complementary RNAs. We would like to propose the first pathway for the gBP21-mediated RNA annealing reaction based on the described results and published material. The protein acts on the basis of two mechanistic principles. First, it functions as a matchmaker protein by binding to one of the RNA reactants and converting it into an annealing-active conformation. Secondly, gBP21 decreases the electrostatic repulsion between the two RNA annealing substrates, thereby favoring the formation of the gRNA/pre-mRNA hybrid. Figure 6 illustrates the essential stages of the process.
Figure 6.
Matchmaker model of the gBP21-mediated RNA/RNA annealing reaction. Schematic representation of the basic steps of the gBP21-mediated annealing reaction. First, a kinetically and thermodynamically stable 1 + 1 gBP21/gRNA complex is formed. One binding determinant is the binding site for the 3′ hairpin of gRNAs (23,35), which has been shown to be sufficient for the high affinity interaction (14). Secondly, the 5′ end of the gRNA is unfolded in order to present the anchor region for the annealing reaction. The identified six ionic bonds might participate in this step in order to reduce the negative surface charge of the gRNA. This should increase the collision frequency with the pre-mRNA. gBP21 stays bound to the structurally altered gRNA, which is characteristic of a matchmaker-type protein. Thirdly, the anchor sequences of the two RNAs hybridize, forming a partially dsRNA product, which probably triggers the dissociation of gBP21 from the complex.
The initial step of the reaction is the formation of a thermodynamically and kinetically stable (gRNA)1/(gBP21)1 complex. For this, the protein relies on two gRNA binding determinants: a binding pocket for the 3′ hairpin (14,23,35) and a presentation platform for the anchor sequence at the 5′ end of gRNAs. During or shortly after the formation of the gBP21/gRNA complex, the 5′ end of the gRNA is unfolded to present the anchor sequence in a single-stranded conformation. This reaction step is consistent with a matchmaker as well as a chaperone function and can be considered the transition state in the annealing pathway. The energy input for the unfolding of the gRNAs 5′ end is very small as the anchor sequence is involved in a hairpin structure of low thermodynamic stability (23,35). The results from the SELEX experiment confirm that gBP21 is capable of stabilizing ssRNA sequences in a sequence-independent context. All selected RNA aptamers are characterized by a strong nucleotide bias (8% G, 25% A, 29% U and 39% C). They probably fold into structures with a great deal of single strand and are of low thermodynamic stability. Thus, the aptamer sequences resemble, from a structural point of view, the 5′ ends of natural gRNAs. The correlation between nucleotide bias and low energy structures can be made plausible if the base pairing potential of the 4 nt is considered: G nucleotides have the highest potential through the formation of hydrogen bonds with C and U nucleotides. That means that RNAs with a low G content form, on average, lower energy structures. The low G content of the aptamers is compensated by a high C content. As C nucleotides form canonical base pairs only with G residues, a combination of a high C and a low G content leads to unpaired bases and weak secondary structures.
One way of providing sequence-independent RNA/protein binding is a charge/charge interaction between the polyanionic nucleic acid and cationic side groups of the protein. We identified an upper value of six ionic bonds that contribute to the formation of the gBP21/gRNA complex. Although the exact positions of the six positive charges on the surface of gBP21 have not been identified, three arginine-rich regions of the protein are potential candidates (12). R-rich domains have been shown to confer binding of ssRNA ligands (42). An additional result of the formation of ionic bonds between gRNAs and gBP21 is a reduced net negative charge of the bound gRNA. This should disfavor the electrostatic repulsion of the two RNA annealing partners and thus favor the formation of the double-stranded gRNA/pre-mRNA product.
As stated above, the unfolding of the gRNA 5′ end is in line with a matchmaker as well as a chaperone function. However, the average dissociation rate constant of the gBP21/gRNA complex is 0.018 s–1. This is equivalent to a half-life of 43 s. The value is similar to other well-characterized RNA/protein interactions (43,44) and indicates that gBP21 is still complexed to the gRNA when the two annealing substrates meet. Thus, gBP21 probably acts as a matchmaker protein. It remains bound to the gRNA even after the RNA refolding step. In contrast, a chaperone protein would dissociate from the gBP21/gRNA complex in response to the change in RNA structure (20,21).
After the formation of the gRNA/pre-mRNA double strand, our data suggest that gBP21 dissociates from the annealed RNA/RNA product to bind free gRNA. This is deduced from the fact that the affinity of gBP21 for dsRNA is very low. With a ΔG of –46 kJ mol–1 the protein stabilizes gRNA, as one of the two RNA reactants, significantly more than the final gRNA/pre-mRNA annealing product (ΔG = –35 kJ mol–1). Thus, it is unlikely that gBP21 mediates the annealing of complementary RNAs through a mechanism that stabilizes the double-stranded reaction product. In addition, two further features may contribute to the dissociation of the protein: as the editing reaction proceeds, the gRNA/pre-mRNA double strand becomes gradually larger. Along with this extension of the gRNA/pre-mRNA hybrid, the gRNA 3′ hairpin progressively unfolds in order to provide its template function for the editing machinery. As a consequence gBP21 will, at some point in the reaction pathway, lose one of its RNA binding sites and might dissociate from the complex.
The dissociation of a matchmaker protein from the annealed product is, in most cases, energized by the hydrolysis of ATP (21). At present, there are no data that would indicate an ATP dependence for the gBP21-mediated annealing reaction. However, at least one matchmaker has been identified that does not require ATP hydrolysis for dissociation: the σ70 subunit of Escherichia coli RNA polymerase. The protein works as a matchmaker in guiding the holoenzyme to the promoter (21). The dissociation from the promoter is driven by the local melting of the DNA double helix, i.e. conformational changes of the bound product (21,45,46). Similarly, gBP21 could use conformational changes of the bound nucleic acid to dissociate. As in this case a double helix is formed and not melted, gBP21 would use the base pairing energy to dissociate. The low affinity for dsRNA supports this hypothesis.
The rate limiting step of the gBP21-mediated annealing reaction must be the contact between the gBP21/gRNA complex and the pre-mRNA for the following reason: the on-rate of the gBP21/gRNA interaction is ∼3 × 106 M–1 s–1, whereas the stimulation of the RNA/RNA annealing reaction has a second order rate constant of 3 × 105 M–1 s–1. As the gBP21/gRNA complex is sufficiently stable to wait for the second reaction step to occur, the second step must be rate limiting, i.e. the association between the pre-mRNA and the gBP21/gRNA complex. A contributing factor for the high rate of the first reaction step might be the 3′ oligo(U) extension of gRNAs. This part of the RNA seems to speed up the interaction to gBP21 by a factor of eight (data not shown), which could be a consequence of the structural flexibility of the oligo(U) tail (23,35). The increase in binding rates between macromolecules by flexible elements has been described previously (47).
Together, our data provide a first model for the role of gBP21 in the annealing of gRNAs to cognate pre-mRNAs. We suggest a mechanism that involves high affinity binding of gBP21 to gRNAs and stabilization of a bound gRNA in an annealing-competent conformation. The gBP21/gRNA complex involves up to six ionic bonds and the protein probably dissociates from the complex after the formation of the partially double-stranded gRNA/pre-mRNA annealing product. Thus, we suggest that gBP21 acts as a matchmaker-type annealing factor, which reduces the electrostatic repulsion of the two interacting RNA reactants.
Acknowledgments
ACKNOWLEDGEMENTS
We thank J. Kaiser for his help in the sequence randomization experiments. M. Homann is thanked for advice performing the SELEX experiment and A. S. Paul, A. Fuß and A. Best for critically reading the manuscript. This work was supported in part by the Deutsche Forschungsgemeinschaft (DFG) and the Human Frontier Science Program (HFSP) to H.U.G.
REFERENCES
- 1.Estévez A.M. and Simpson,L. (1999) Uridine insertion/deletion RNA editing in trypanosome mitochondria—a review. Gene, 240, 247–260. [DOI] [PubMed] [Google Scholar]
- 2.Pollard V.W., Harris,M.E. and Hajduk,S.L. (1992) Native mRNA editing complexes from Trypanosoma brucei mitochondria. EMBO J., 11, 4429–4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Correll R.A., Read,L.K., Riley,G.R., Nelissery,J.K., Allen,T.E., Kable,M.L., Wachal,M.D., Seiwert,S.D., Myler,P.J. and Stuart,K. (1996) Complexes from Trypanosoma brucei that exhibit deletion editing and other editing-associated properties. Mol. Cell. Biol., 16, 1410–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cruz-Reyes J. and Sollner-Webb,B. (1996) Trypanosome U-deletional RNA editing involves guide RNA-directed endonuclease cleavage, terminal U exonuclease and RNA ligase activities. Proc. Natl Acad. Sci. USA, 93, 8901–8906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rusché L.N., Cruz-Reyes,J., Piller,K.J. and Sollner-Webb,B. (1997) Purification of a functional enzymatic editing complex from Trypanosoma brucei mitochondria. EMBO J., 16, 4069–4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blum B., Bakalara,N. and Simpson,L. (1990) A model for RNA editing in kinetoplastid mitochondria: ‘guide’ RNA molecules transcribed from maxicircle DNA provide the edited information. Cell, 60, 189–198. [DOI] [PubMed] [Google Scholar]
- 7.Leung S.S. and Koslowsky,D.J. (2001) RNA editing in Trypanosoma brucei: characterization of gRNA U-tail interactions with partially edited mRNA substrates. Nucleic Acids Res., 29, 703–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Missel A., Souza,A.E., Nörskau,G. and Göringer,H.U. (1997) Disruption of a gene encoding a novel mitochondrial DEAD-box protein in Trypanosoma brucei affects edited mRNAs. Mol. Cell. Biol., 17, 4895–4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Madison-Antennucci S., Sabatini,R.S., Pollard,V.W. and Hajduk,S.L. (1998) Kinetoplastid RNA-editing-associated protein 1 (REAP-1): a novel editing complex protein with repetitive domains. EMBO J., 17, 6368–6376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vanhamne L., Perez-Morga,D., Marchal,C., Speijer,D., Lambert,L., Geuskens,M., Alexandre,S., Ismaili,N., Göringer,H.U., Benne,R. and Pays,E. (1998) Trypanosoma brucei TBRGG1, a mitochondrial oligo(U)-binding protein that co-localizes with an in vitro RNA editing activity. J. Biol. Chem., 273, 21825–21833. [DOI] [PubMed] [Google Scholar]
- 11.Hayman M.L. and Read,L.K. (1999) Trypanosoma brucei RBP16 is a mitochondrial Y-box family protein with guide RNA binding activity. J. Biol. Chem., 274, 12067–12074. [DOI] [PubMed] [Google Scholar]
- 12.Köller J., Müller,U.F., Schmid,B., Missel,A., Kruft,V., Stuart,K. and Göringer,H.U. (1997) Trypanosoma brucei gBP21. An arginine-rich mitochondrial protein that binds to guide RNA with high affinity. J. Biol. Chem., 272, 3749–3757. [DOI] [PubMed] [Google Scholar]
- 13.Madison-Antenucci S. and Hajduk,S. (2001) RNA editing-associated protein 1 is an RNA binding protein with specificity for preedited mRNA. Mol. Cell, 7, 879–886. [DOI] [PubMed] [Google Scholar]
- 14.Müller U.F., Lambert,L. and Göringer,H.U. (2001) Annealing of RNA editing substrates facilitated by guide RNA-binding protein gBP21. EMBO J., 20, 1394–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lambert L., Müller,U.F., Souza,A.E. and Göringer,H.U. (1999) The involvement of gRNA-binding protein gBP21 in RNA editing—an in vitro and in vivo analysis. Nucleic Acids Res., 27, 1429–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Allen T.E., Heidmann,S., Reed,R., Myler,P.J., Göringer,H.U. and Stuart,K.D. (1998) Association of guide RNA binding protein gBP21 with active RNA editing complexes in Trypanosoma brucei.Mol. Cell. Biol., 18, 6014–6022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pontius B.W. and Berg,P. (1991) Rapid renaturation of complementary DNA strands mediated by cationic detergents: a role for high-probability binding domains in enhancing the kinetics of molecular assmbly processes. Proc. Natl Acad. Sci. USA, 88, 8237–8241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nedbal W., Homann,M. and Sczakiel,G. (1997) The association of complementary ribonucleic acids can be strongly increased without lowering Arrhenius activation energies or significantly altering structures. Biochemistry, 36, 13552–13557. [DOI] [PubMed] [Google Scholar]
- 19.Herschlag D. (1995) RNA chaperones and the RNA folding problem. J. Biol. Chem., 270, 20871–20874. [DOI] [PubMed] [Google Scholar]
- 20.Racker E. (1992) Chaperones and matchmakers: inhibitors and stimulators of protein phosphorylation. Curr. Top. Cell. Reg., 33, 127–143. [DOI] [PubMed] [Google Scholar]
- 21.Sancar A. and Hearst,J.E. (1993) Molecular matchmakers. Science, 259, 1415–1420. [DOI] [PubMed] [Google Scholar]
- 22.Eguchi Y. and Tomizawa,J.-I. (1990) Complex formed by complementary RNA stem–loops and its stabilisation by a protein: function of colE1 rom protein. Cell, 60, 199–209. [DOI] [PubMed] [Google Scholar]
- 23.Schmid B., Riley,G.R., Stuart,K. and Göringer,H.U. (1995) The secondary structure of guide RNA molecules from Trypanosoma brucei. Nucleic Acids Res., 23, 3093–3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sober H.A. (1968) Handbook of Biochemistry. The Chemical Rubber Co., Cleveland, OH.
- 25.Cantor C.R. and Schimmel,P.R. (1980) Biophysical Chemistry. W. H. Freeman and Co., San Francisco, CA.
- 26.Allen S.H. and Wong,K.-P. (1978) The hydrodynamic and spectroscopic properties of 16 S RNA from Escherichia coli ribosome in reconstitution buffer. J. Biol. Chem., 253, 8759–8766. [PubMed] [Google Scholar]
- 27.Niewenhuysen P. and Clauwaert,J. (1981) Physicochemical characterization of ribosomal particles from the eucaryote artemia.J. Biol. Chem., 256, 9626–9632. [PubMed] [Google Scholar]
- 28.Record M.T., Jr, Lohman,M.L. and deHaseth,P. (1976) Ion effects on ligand-nucleic acid interactions. J. Mol. Biol., 107, 145–158. [DOI] [PubMed] [Google Scholar]
- 29.Klotz I.M., Triwush,H. and Walker,F.M. (1948) The binding of organic ions by proteins. Competition phenomena and denaturation effects. J. Am. Chem. Soc., 70, 2935–2941. [DOI] [PubMed] [Google Scholar]
- 30.Homann M. and Göringer,H.U. (1999) Combinatorial selection of high affinity RNA ligands to live African trypanosomes. Nucleic Acids Res., 27, 2006–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lumpkin O.J. and Zimm,B.H. (1982) Mobility of DNA in gel electrophoresis. Biopolymers, 21, 2315–2316. [DOI] [PubMed] [Google Scholar]
- 32.Holbrook S.R., Sussman,J.L., Warrant,R.W. and Kim,S.-H. (1978) Crystal structure of yeast phenylalanine transfer RNA. J. Mol. Biol., 123, 631–660. [DOI] [PubMed] [Google Scholar]
- 33.Broido M.S. and Kearns,D.R. (1982) 1H NMR evidence for a left-handed helical structure of poly(ribocytodylic acid) in neutral solution. J. Am. Chem. Soc., 104, 5207–5216. [Google Scholar]
- 34.Köller J., Nörskau,G., Paul,A.S., Stuart,K. and Göringer,H.U. (1994) Different Trypanosoma brucei guide RNA molecules associate with an identical complement of mitochondrial proteins in vitro. Nucleic Acids Res., 22, 1988–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hermann T., Schmid,B., Heumann,H. and Göringer,H.U. (1997) A three-dimensional working model for a guide RNA from Trypanosoma brucei.Nucleic Acids Res., 25, 2311–2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hirao I., Spingola,M., Peabody,D. and Ellington,A.D. (1999) The limits of specificity: an experimental analysis with RNA aptamers to MS2 coat protein variants. Mol. Diversity, 4, 75–89. [DOI] [PubMed] [Google Scholar]
- 37.Heilman-Miller S.L., Thirumalai,D. and Woodson,S.A. (2001) Role of counterion condensation in folding of the Tetrahymena ribozyme. I. Equilibrium stabilization by cations. J. Mol. Biol., 306, 1157–1166. [DOI] [PubMed] [Google Scholar]
- 38.Leegwater P., Speijer,D. and Benne,R. (1995) Identification by UV cross-linking of oligo(U)-binding proteins in mitochondria of the insect trypanosomatid Crithidia fasciculata. Eur. J. Biochem., 227, 780–786. [DOI] [PubMed] [Google Scholar]
- 39.Blom D., van den Burg,J., Breek,C.K., Speijer,D., Muijsers,A.O. and Benne,R. (2001) Cloning and characterization of two guide RNA-binding proteins from mitochondria of Crithidia fasciculata: gBP27, a novel protein and gBP29, the orthologue of Trypanosoma brucei gBP21. Nucleic Acids Res., 29, 2950–2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.deHaseth P.L., Lohman,T.M. and Record,M.T.,Jr (1977) Nonspecific interaction of lac repressor with DNA: an association reaction driven by counterion release. J. Mol. Biol., 107, 145–158. [DOI] [PubMed] [Google Scholar]
- 41.Antosiewicz J. and McCammon,J.A. (1996) The determinants of pKas in proteins. Biochemistry, 35, 7819–7833. [DOI] [PubMed] [Google Scholar]
- 42.Chen C.-X., Cho,D.-S.C., Wang,Q., Lai,F., Carter,K.C. and Nishikura,K. (2000) A third member of the RNA-specific adenosine deaminase gene family, ADAR3, contains both single- and double-stranded RNA binding domains. RNA, 6, 755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carey J. and Uhlenbeck,O.C. (1983) Kinetic and thermodynamic characterization of the R17 coat protein-ribonucleic acid interaction. Biochemistry, 22, 2610–2615. [DOI] [PubMed] [Google Scholar]
- 44.Mougel M., Ehresmann,B. and Ehresmann C. (1986) Binding of Escherichia coli ribosomal protein S8 to 16S rRNA: kinetic and thermodynamic characterization. Biochemistry, 25, 4803–4821. [DOI] [PubMed] [Google Scholar]
- 45.Travers A.A. and Burgess,R.R. (1969) Cyclic re-use of the RNA polymerase sigma factor. Nature, 222, 537–540. [DOI] [PubMed] [Google Scholar]
- 46.Gralla J.D. (1991) Transcriptional control-lessons from an E.coli promoter data base. Cell, 66, 415–418. [DOI] [PubMed] [Google Scholar]
- 47.Pontius B.W. (1993) Close encounters: why unstructured, polymeric domains can increase rates of specific macromolecular association. Trends Biochem. Sci., 18, 181–186. [DOI] [PubMed] [Google Scholar]