Summary
The posttranslational modification lysine malonylation is found in many proteins, including histones. However, it remains unclear whether histone malonylation is regulated or functionally relevant. Here, we report that availability of malonyl-co-enzyme A (malonyl-CoA), an endogenous malonyl donor, affects lysine malonylation, and that the deacylase SIRT5 selectively reduces malonylation of histones. To determine if histone malonylation is enzymatically catalyzed, we knocked down each of the 22 lysine acetyltransferases (KATs) to test their malonyltransferase potential. KAT2A knockdown in particular reduced histone malonylation levels. By mass spectrometry, H2B_K5 was highly malonylated and regulated by SIRT5 in mouse brain and liver. Acetyl-CoA carboxylase (ACC), the malonyl-CoA producing enzyme, was partly localized in the nucleolus, and histone malonylation increased nucleolar area and ribosomal RNA expression. Levels of global lysine malonylation and ACC expression were higher in older mouse brains than younger mice. These experiments highlight the role of histone malonylation in ribosomal gene expression.
Subject areas: Biological sciences, Molecular biology, Omics, Proteomics
Graphical abstract
Highlights
-
•
Malonyl-CoA availability affects lysine malonylation
-
•
SIRT5 and KAT2A regulate histone malonylation
-
•
Histone malonylation increases ribosomal RNA expression
-
•
Lysine malonylation increases with age in mouse brain
Biological sciences; Molecular biology; Omics; Proteomics
Introduction
Lysine malonylation is a type of protein posttranslational modification.1,2 By adding a negatively charged malonyl group to the positively charged ε-amino group, malonylation increases the space occupied by the modified lysine residue, and alters the electrostatic force around the modified lysine residue. Similar to lysine acetylation, malonylation is reversible, and subject to the removal by SIRT5, a member of class III lysine deacetylases (also known as sirtuins). However, unlike other sirtuin family members, SIRT5 has very weak deacetylase activity. Instead, SIRT5 specifically removes acidic short-chain acyl groups, including malonyl-, succinyl-, and glutaryl-groups, from modified protein substrates.1,2,3 Although over a thousand malonylated proteins have been identified to date,4,5 their biological significance remains mostly unclear.
Like many other acyl-CoA species, malonyl-CoA, an intermediary metabolite in the de novo fatty acid synthesis pathway,6 is an endogenous provider of malonyl group for lysine malonylation. Acyl-CoA modifications can occur via enzymatic or non-enzymatic pathways. Owing to the reactive nature of acyl-CoA thioesters, acylations, including malonylation, are generally believed to be non-enzymatic in mitochondria which provide a basic environment that deprotonates the ε-amino group of lysine and make it more susceptible to nucleophilic attack toward acyl-CoA species.7
In our previous study, we identified 430 malonylated proteins in mouse livers by immuno-affinity enrichment and mass spectrometry (MS).4 These malonylated proteins were distributed across major subcellular compartments, including mitochondria, cytoplasm, and nucleus.4 Unlike mitochondria, the nucleus has a neutral pH,8 and therefore, lysine residues in nuclear proteins are less deprotonated. Meanwhile, malonyl-CoA appears to be less reactive than other acyl-CoA species with similar structures, such as succinyl-CoA and glutaryl-CoA.9 Therefore, within the nucleus, lysine malonylation may need enzymatic catalysis. Yet, no lysine malonyltransferase (KMaT) has so far been reported.
Histones are conserved nuclear proteins with abundant lysine and arginine residues. Using electrostatic attraction, DNA wraps around octamers of the four core histones (e.g., H2A, H2B, H3, and H4) to fit into the nucleus. Histones are subject to chemical modifications, including lysine acetylation, lysine methylation, and arginine methylation, which are some of the best characterized. These modifications play critical roles in regulating DNA replication and gene transcription.10 With the development of MS techniques in recent years, additional modifications, including lysine malonylation, have been identified in histones.1,2,11 However, the functional relevance and regulatory mechanism of histone malonylation are not well understood.
In the present study, we investigated the regulation of histone malonylation. First, we examined the effect of metabolism on lysine malonylation by changing malonyl-CoA levels. Second, we tested the possibility that a known lysine acetyltransferases (KATs) might be able to catalyze histone malonylation.12,13 Third, we tested the effect of histone malonylation on nucleolar expansion and ribosomal RNA expression. Finally, we examined malonylation levels in young and old mouse brains. Through this study, we found that histone malonylation is regulated by malonyl-CoA availability, SIRT5, and KAT2A. These observations support a role for histone malonylation in aging-associated epigenetic changes.
Results
Malonyl-CoA availability affects lysine malonylation
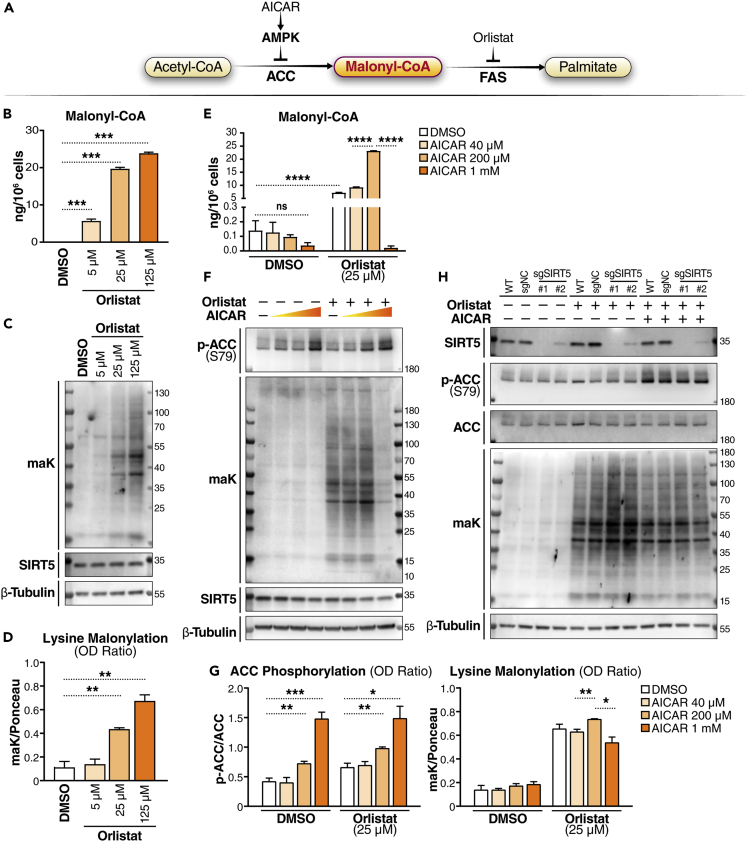
Malonyl-CoA thioester is an endogenous donor of malonyl group for lysine malonylation. Malonyl-CoA is a metabolic intermediate in the de novo fatty acid synthesis pathway, and is generated by acetyl-CoA carboxylase (ACC). Malonyl-CoA then serves as a substrate for the synthesis of median- and long-chain fatty acids through fatty acid synthase (FAS) (Figure 1A). To determine whether lysine malonylation is affected by malonyl-CoA levels, we used compounds that target the metabolism of malonyl-CoA. As a FAS inhibitor, orlistat, has been previously used to increase cellular malonyl-CoA levels.14,15 To verify the effect of orlistat treatment, we measured malonyl-CoA levels in K562 cells after orlistat treatment for 24 h using mass spectrometry. The results showed that malonyl-CoA levels were drastically increased (32– to 136-fold) by orlistat treatment in a dose-dependent manner (Figure 1B). By contrast, acetyl-CoA level was increased but not positively correlated with the dose of orlistat. With the increase of orlistat from 5 μM to 125 μM, acetyl-CoA level showed a trend of decrease (Figure S1A). Using western blotting and an antibody that recognizes malonylated lysine residues,2 we examined lysine malonylation in these cells. The results showed that lysine malonylation was barely detectable without orlistat treatment, and orlistat treatment increased the levels of lysine malonylation in a dose-dependent manner (Figures 1C and 1D), which is consistent with the change of malonyl-CoA levels. Next, we tested the effect of ACC inhibition with AICAR. This drug activates AMPK, an inhibitory upstream kinase of ACC through phosphorylation at Ser79.16,17,18 Our results showed that orlistat treatment drastically increased malonyl-CoA levels, and that AICAR treatment at 1 mM significantly decreased malonyl-CoA level (Figure 1E). Of interest, we also observed that, in orlistat-treated cells, malonyl-CoA level was significantly higher on AICAR treatment at 200 μM than AICAR treatment at 40 μM (Figure 1E). By contrast, acetyl-CoA levels were not significantly different between 40 μM and 200 μM AICAR treatments (Figure S1B). Western blotting results showed that orlistat treatment significantly increased lysine malonylation, whereas AICAR treatment at 1 mM reduced the orlistat-induced increase in lysine malonylation in K562 cells (Figures 1F and 1G). In consistent with malonyl-CoA levels, lysine malonylation level was higher upon AICAR treatment at 200 μM than AICAR treatment at 40 μM in orlistat-treated cells (Figures 1F and 1G). These results suggest that lysine malonylation levels are in line with malonyl-CoA levels upon orlistat and AICAR treatment at various doses.
Figure 1.
Malonyl-CoA availability affects lysine malonylation
(A) Schematic of the fatty acid synthesis pathway and compounds affecting malonyl-CoA level. AMPK, AMP-activated protein kinase; ACC, acetyl-CoA carboxylase; FAS, fatty acid synthase.
(B) Malonyl-CoA levels in K562 cells treated with orlistat at different doses as indicated for 24 h were measured with LC-MS. [13C3]-malonyl-CoA was added to each sample as a standard for quantification. Quantified levels are normalized to cell count in each sample. N = 3 per treatment. Values are shown as mean ± SEM. ∗∗∗p < 0.001 using unpaired Student’s t test.
(C) Malonyl-CoA levels in K562 cells treated with orlistat and AICAR at different doses as indicated for 24 h were measured with LC-MS. [13C3]-malonyl-CoA was added to each sample as a standard for quantification. Quantified levels are normalized to cell count in each sample. N = 3 per treatment. Values are shown as mean ± SEM. ns (not significant) ≧ 0.05, and ∗∗∗∗p < 10−4 using unpaired Student’s t test.
(D) Western blot (WB) of lysine malonylation (maK) in K562 cells at 24 h after orlistat treatment at the indicated concentrations.
(E) Quantification of WBs by calculating the OD ratio of maK to Ponceau S staining. N = 3. Values are shown as mean ± SEM. ∗∗p < 0.01 using unpaired Student’s t test.
(F) WB of maK in K562 cells at 24 h after drug treatment. Orlistat: 25 μM; AICAR: 40 μM, 200 μM, or 1 mM.
(G) Quantification of WBs by calculating the OD ratio of phosphorylated ACC (at Serine 79) to total ACC, and OD ratio of maK to Ponceau S staining. N = 3. Values are shown as mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001 using unpaired Student’s t test.
(H) WB of maK in K562 (wildtype) cells, and K562-dCas9-KRAB cells expressing sgNC, sgSIRT5#1 or sgSIRT5#2 at 24 h after drug treatment. Orlistat: 25 μM; AICAR: 1 mM.
SIRT5 is a prominent deacylase that removes malonyl from lysine residues. To determine the effect of SIRT5, we modified K562 cells by knocking down SIRT5 using a CRISPRi system19 with two different guide RNAs (gRNAs) targeting the SIRT5 promoter. Lysine malonylation was barely detected in both control (wildtype (WT) and sgNC (expressing scramble gRNA as negative control)) and both SIRT5 knockdown (KD) K562 cells without orlistat treatment (Figure 1H). After orlistat treatment at 25 μM for 24 h, lysine malonylation was induced, and as expected, lysine malonylation levels were higher in SIRT5 KD cells than controls (Figure 1H). However, following inhibition of ACC activity with AICAR, lysine malonylation levels in SIRT5 KD cells were no longer higher than in control cells (Figure 1H). This result suggests that SIRT5 lowers the hypermalonylation induced by the blockage of malonyl-CoA consumption (FAS inhibition), and the demalonylation function of SIRT5 becomes less effective when malonyl-CoA biogenesis is also reduced in K562 cells. Overall, these results indicate that lysine malonylation is affected by SIRT5 and malonyl-CoA availability.
SIRT5 and lysine acetyltransferases regulate histone malonylation
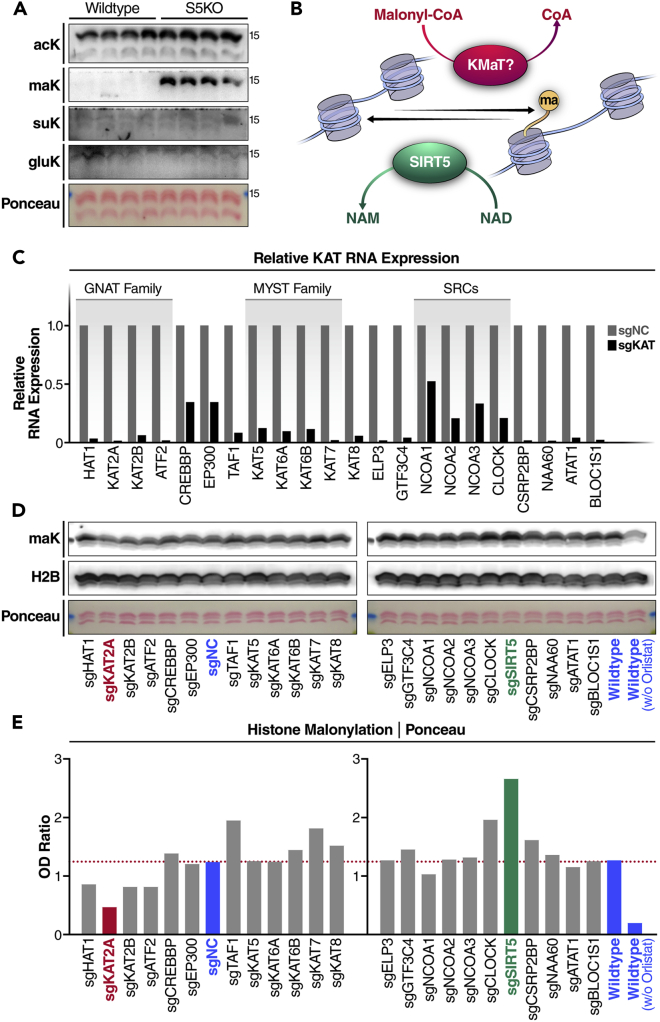
Next, we tested whether histone proteins are malonylated. Histones are basic nuclear proteins with abundant lysine residues that are subject to multiple acylations.20 We extracted histones from WT and SIRT5 knockout (S5KO) mouse liver, and examined their acylation levels using pan-lysine-acylation antibodies. Western blotting result showed that lysine acetylation was abundant in WT and S5KO mouse histones (Figure 2A). In agreement with the known low deacetylase activity of SIRT5,1 acetylation levels were not different in WT and S5KO histone samples (Figure 2A). Because SIRT5 functions mainly as a lysine demalonylase, desuccinylase, and deglutarylase,1,2,3,21 we probed lysine malonylation, succinylation, and glutarylation in the histone samples. Western blotting showed that lysine malonylation was barely detected in WT mouse liver histone samples, but was remarkably higher in S5KO mouse livers (Figure 2A). However, lysine succinylation and glutarylation levels were very low in histones, and no significant differences were observed in WT and S5KO mouse histone samples (Figure 2A). These results suggest that histones are more susceptible to malonylation than succinylation or glutarylation in the absence of SIRT5.
Figure 2.
SIRT5 and lysine acetyltransferases regulate histone malonylation
(A) Western blot (WB) of acetylation, malonylation, succinylation, and glutarylation in histone extracts derived from wildtype and SIRT5 knockout (S5KO) mouse livers.
(B) Schematic of the regulation of histone malonylation. KMaT, lysine malonyl-transferase.
(C) Lysine acetyltransferase (KAT) knockdown K562 cell lines were constructed using a CRISPR-dCsa9-KRAB system via lentiviral infection. Knockdown efficiencies were examined by qPCR.
(D) WB examination of malonylation on histones extracted from KAT knockdown K562 cells treated with 25 μM orlistat for 24 h WT cells without lentiviral infection, or with scramble lentiviral infection (sgNC), as control cell lines (shown in blue).
(E) Quantification of the optical densities (OD) of malonylation blots normalized to Ponceau S staining.
We next wanted to know if histone malonylation might be regulated by a lysine malonyltransferase (KMaT) (Figure 2B). Lysine acetyltransferases (KATs) transfer acetyl groups from acetyl-CoA to lysine residues. In the human proteome, 22 putative KATs have been identified so far. With some exceptions, KATs are mainly cataloged into three families (i.e., GCN5-related N-acetyltransferases (GNAT), the p300/CREB-binding protein (p300/CBP), and the MOZ, Ybf2, Sas2, and Tip60 (MYST) family).22 Importantly, KATs are not exclusive acetyltransferases; some also catalyze non-acetyl lysine acylations.12
Using a CRISPRi system,19 we knocked down each of the 22 KATs in K562 cells, and tested their malonyl-transferring potential. For each KAT gene, we used two different gRNAs with the highest predicted targeting potentials, and then chose the one with higher knockdown efficiency in K562-expressing dCas9-KRAB (Krüppel-associated box) cells to evaluate histone malonylation (Table S1). RNA expression levels of different KAT genes on CRISPRi KD were reduced by 47–99%, and 18 of 22 KAT genes had a knockdown efficiency >80% (Figure 2C). These sgKAT-expressing K562 cells were then treated with 25 μM orlistat for 24 h before histone extraction. Western blotting using a pan-malonyl lysine antibody detected lysine malonylation in histones in different sgRNA-expressing K562 cells, and, as predicted, malonylation levels were much lower in K562 cells without orlistat treatment (Figure 2D, right most lane). WT (not expressing gRNA) and sgNC K562 cells had comparative levels of histone malonylation (Figures 2D and 2E). As expected, histone malonylation level in SIRT5 KD cells (shown in green) was higher than in WT or sgNC cells (Figures 2D and 2E). Western blotting results also showed that some KAT KD cells (e.g., sgTAF1, sgKAT7, and sgCLOCK) had higher, whereas some KAT KD cells (e.g., sgHAT1, sgKAT2A, sgKAT2B, sgATF2) had lower histone malonylation levels than WT or sgNC cells (Figures 2D and 2E). Of particular interest, among all KAT KD K562 cells, the knockdown of KAT2A (shown in red) caused the highest level of reduction (by > 60%) in histone malonylation compared to WT or sgNC cells (Figures 2D and 2E), which suggests KAT2A as a robust regulator of histone malonylation.
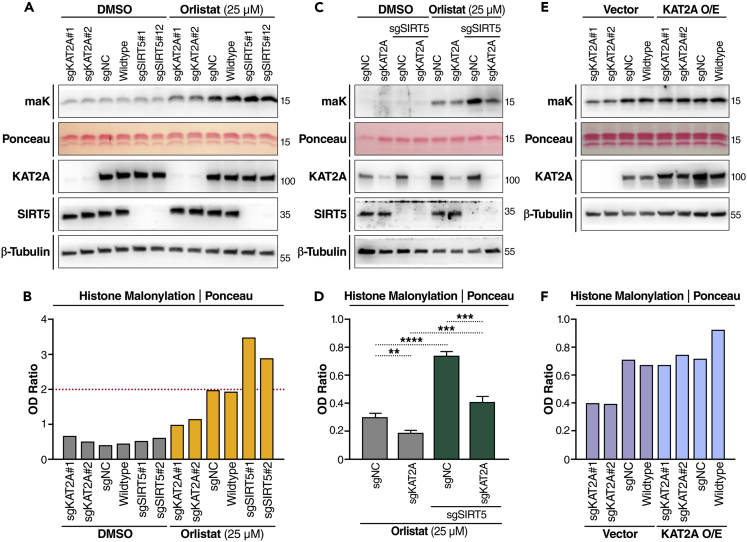
To confirm the observation that KAT2A KD reduces histone malonylation in the CRISPRi screening experiment, we examined histone malonylation levels in two KAT2A KD K562 cell lines that express two different KAT2A-targeting sgRNAs. As shown in Figures 3A and 3B, after orlistat treatment, two different SIRT5 KD K562 cell lines both had 50–100% higher levels of histone malonylation, whereas two different KAT2A KD cell lines had lower levels (decreased by ∼50%). These results confirmed that knockdown of KAT2A reduces orlistat-induced histone malonylation. Then we made a SIRT5 and KAT2A double knockdown K562 cell line and examined its histone malonylation levels. After orlistat treatment, knockdown of SIRT5 alone significantly increased histone malonylation, whereas knockdown of KAT2A significantly reduced histone malonylation in SIRT5 KD cells (Figures 3C and 3D), indicating that histone malonylation is reversible and regulated by both SIRT5 and KAT2A. Next, we examined if forced expression of KAT2A would increase histone malonylation. Histone malonylation levels in KAT2A KD K562 cells were about half of the level in WT and sgNC control cells, and the overexpression of KAT2A rescued the reduction in histone malonylation levels in KAT2A KD cells (Figures 3E and 3F). However, KAT2A overexpression did not further increase histone malonylation levels in WT or sgNC K562 cells (Figures 3E and 3F), suggesting the sufficiency of endogenous KAT2A for the overall histone malonylation in K562 cells. These results confirmed that KAT2A regulates histone malonylation, and contributes to the increased histone malonylation induced by orlistat treatment.
Figure 3.
KAT2A regulates histone malonylation
(A) Western blot (WB) of malonylation in histone extracts. sgKAT2A (two different sgRNAs), sgNC, WT, and sgSIRT5 (two sgRNAs) K562 cell lines were treated with 25 μM orlistat or DMSO for 24 h.
(B) Optical density (OD) quantification of histone malonylation normalized to Ponceau S staining in (A).
(C) WB of malonylation in KAT2A single knockdown and SIRT5;KAT2A double knockdown K562 cells.
(D) OD quantification of histone malonylation normalized to Ponceau S staining. n = 4 per cell type. Error bar: mean ± SEM. ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 10−4 using unpaired Student’s t test.
(E) WB of malonylation in histone extracts. Empty vector or KAT2A O/E vector transfected into the corresponding K562 cells for 48 h. Cells were treated with 25 μM orlistat for the last 24 h.
(F) OD quantification of histone malonylation normalized to Ponceau S staining in (E).
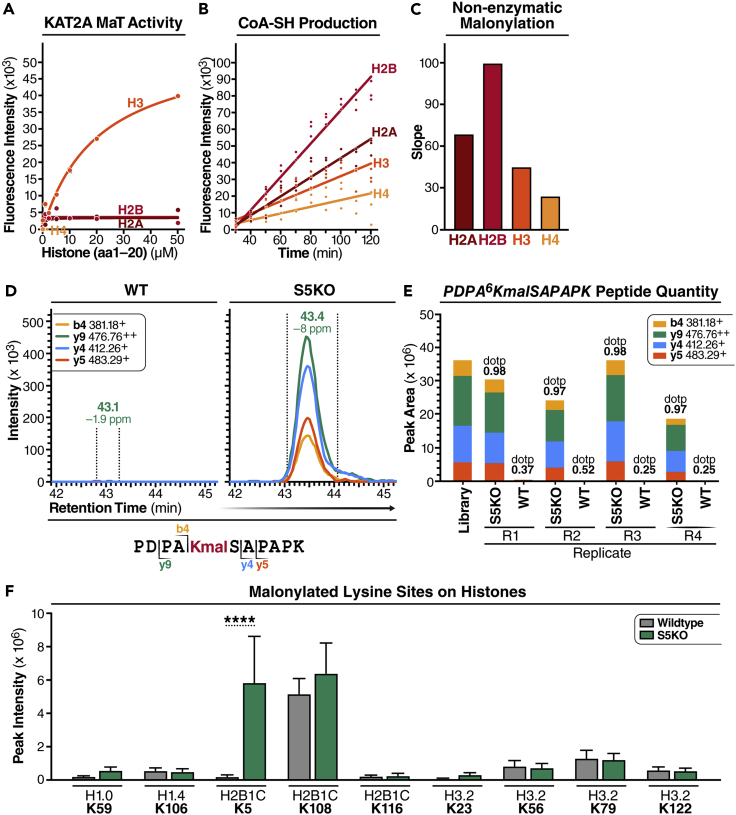
Given that KAT2A regulates histone malonylation, we next wanted to know which histone protein is regulated by KAT2A. To study the malonyltransferase activity of KAT2A, we used an in vitro KMaT activity assay. Because the N-terminal tails of histone proteins are flexible regions with abundant lysine residues, we synthesized the N-terminal (amino acids (a.a.) 1–20) peptides of the four core human histones (i.e., H2A, H2B, H3, and H4 (Table S3)), and incubated each with recombinant human KAT2A protein (catalytic domain, a.a. 497–663) in the presence of malonyl-CoA. The in vitro malonylation reactions were incubated at 37°C for 20 min with titration of histone peptides. Malonylation activity was evaluated by measuring the production of free co-enzyme A (CoA-SH), which was detected by the fluorogenic probe CPM (7-diethylamino-3-(4′-maleimidylphenyl)-4-methylcoumarin) and spectrophotometry. The KMaT activity results showed that KAT2A selectively catalyzed the malonylation of H3 (a.a. 1–20), whereas the malonylation of H2A, H2B, and H4 N-terminal peptides by KAT2A was barely detectable (Figure 4A). To evaluate whether histone malonylation can also be formed non-enzymatically, we incubated N-terminal histone peptides with malonyl-CoA in the absence of KAT2A and examined the production of CoA-SH over time. Among the four N-terminal peptides of core histones, H2B had the highest and H4 had the lowest levels of non-enzymatic malonylation (Figures 4B and 4C). These results indicate that the malonylation of histone peptides are partially enzymatic. Particularly, H3 had the highest level of enzymatic malonylation catalyzed by KAT2A, and H2B had the highest level of non-enzymatic malonylation.
Figure 4.
Histone H2B is malonylated at K5 in SIRT5 KO mouse brain and liver
(A) In vitro lysine malonyltransferase (KMaT) activity assay of the catalytic domains of KAT2A using synthesized histone peptides (20 amino acids at N terminus) as substrates at given concentrations in the presence of 50 μM malonyl-CoA. The reactions were stopped after incubation at 37°C for 20 min. KMaT activity was evaluated by measuring CoA-SH production using CPM at Ex = 390 nm/Em = 460 nm.
(B and C) Non-enzymatic malonylation was examined by incubating synthesized histone peptides with 50 μM malonyl-CoA, and measuring the production of CoA-SH using CPM over time at Ex = 390 nm/Em = 460 nm (B). The slope was calculated to reflect the velocity of non-enzymatic of malonylation (C).
(D) Extracted ion chromatograms of PDPA6KmalSAPAPK (m/z 589.81, z = 2+) from mouse histone H2B type 2-B (Q64525) obtained by data-independent acquisition, that indicates that malonylation of K5 is increased in S5KO mouse brain tissues compared to wildtype (WT).
(E) Quantification using the peak areas of four transitions of PDPA6KmalSAPAPK (m/z 589.81, z = 2+) in four biological replicates of S5KO and WT mouse brain tissues, respectively.
(F) Malonylated lysine sites on histones were previously identified in WT and S5KO mouse livers (Nishida, et al., Mol Cell. 2015). n = 5. Error bar: mean ± SD. ∗∗∗∗p < 10−4 using Sidak multiple comparisons test following two-way ANOVA.
To identify the lysine sites in histones that are actually malonylated in vivo, we examined the malonylation proteome in mouse tissues. We used MS and data-independent acquisitions23,24 to profile the malonylation proteome in mouse brain after immune-affinity enrichment of malonylated peptides. H2B_K5 was highly malonylated in SIRT5 KO (S5KO) mouse brains, but not detectable in WT mouse brains (Log2(S5KO vs WT) = 9.21, with pvalue <0.0001) (Figures 4D and 4E), suggesting that H2B_K5 is highly malonylated and regulated by SIRT5. To confirm this observation, we went back to our previous study, in which we probed malonylated proteins in WT and S5KO mouse liver tissue lysates by MS.4 Re-analysis of the dataset identified nine malonylated lysine sites in histones in mouse liver (Figure 4F). Like mouse brain, liver showed H2B_K5 malonylation levels also higher in S5KO vs. WT mouse livers (Figure 4F). These results suggest that H2B_K5 malonylation is regulated by SIRT5.
Histone malonylation increases ribosomal RNA expression
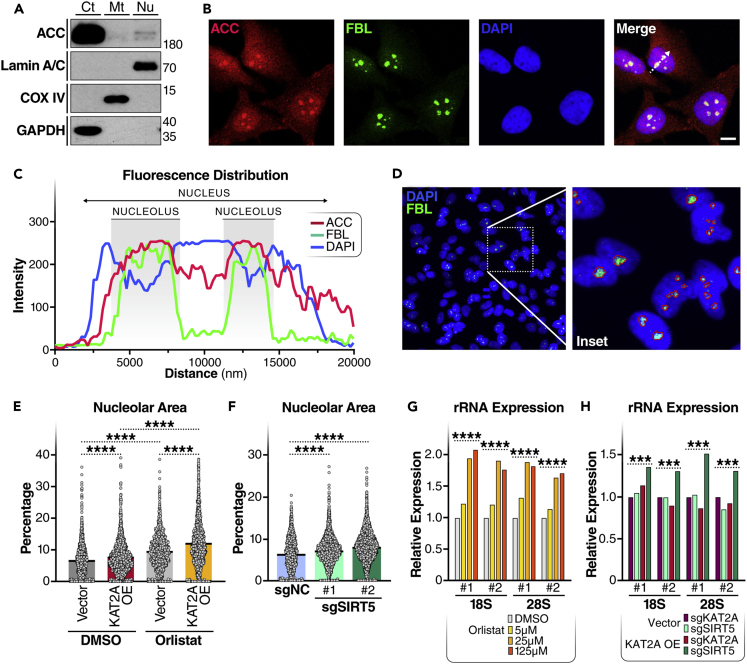
Next, we tested a possible biological role for histone malonylation. Because lysine malonylation is affected by local availability of malonyl-CoA, the localization of malonyl-CoA-producing enzyme, ACC, should inform the biological function of malonylation. Two isoforms of the ACC protein (i.e., ACC1 and ACC2) are expressed in mammals. ACC1 mainly localizes to the cytoplasm, and ACC2 localizes to both cytoplasm and the outer membrane of mitochondria.25 To examine the localization of ACC1, we used western blotting to examine ACC expression in subcellular fractions of HEK293T cells. Although ACC was expressed highest in the cytosolic fraction, a low level of ACC was detected in the nuclear fraction (Figure 5A), suggesting the nucleus as an unconventional subcellular compartment for ACC1. Nuclear localization of ACC1 is also reported in the human protein atlas (HPA) database (proteinatlas.org), which shows that ACC1 (also known as ACACA) was identified in nucleoli (Figure S2). To confirm this result, we performed confocal imaging after immunofluorescence staining of ACC and fibrillarin (FBL), a nucleolar marker, in HeLa cells. By fluorescence distribution analysis, we observed that ACC was detected in the nuclei, including nucleoli (where FBL was stained), in HeLa cells (Figures 5B and 5C). Hence, ACC1 may function as a local malonyl-CoA provider, thereby increasing malonyl-CoA availability and histone malonylation in nucleoli.
Figure 5.
Lysine malonylation increases ribosomal RNA expression
(A) WB with subcellular fractions of HEK293T cells. Ct, cytosol; Mt, mitochondria; Nu, nucleus.
(B) Representative image of immunofluorescence staining and confocal imaging of ACC (red), fibrillarin (FBL, green), and DAPI (blue) in HeLa cells. Scale bar: 10 μm. White dotted arrow area in the overlay (Merge) image was sampled for fluorescence distribution analysis in (C).
(C) The distribution of fluorescence signals of ACC (red), FBL (green), and DAPI (blue) along the dotted arrow in the overlay image in (B) was plotted to show the relative localizations of ACC, FBL, and DAPI.
(D) Representative image of immunofluorescence staining and wide-field imaging of FBL (green) and DAPI (blue) in HeLa cells using a 40x objective. Inset shows nucleolar areas identified (circled with red lines) using a machine learning algorithm.
(E) Nucleolar percentage area (ratio of nucleolar area to nuclear area) was quantified in HeLa cells transfected with empty vector (Vec) or KAT2A expressing plasmid (KAT2A OE) 24 h before DMSO or orlistat treatment (25 μM) for another 24 h. N = 1201–2265. ∗∗∗∗p < 10−4 using unpaired Student’s t test.
(F) Nucleolar percentage area was quantified in HEK293T cells expressing MeCP2-KRAB-dCas9 with scramble guild RNA (sgNC) or two different gRNAs targeting SIRT5 promoter (sgSIRT5#1 and #2). N = 1352–3054. ∗∗∗∗p < 10−4 using unpaired Student’s t test.
(G) qPCR examination of ribosomal RNA levels in HeLa cells after DMSO or orlistat treatment for 24 h. rRNA expression levels were normalized to β-actin expression level. ∗∗∗∗p < 10−4 using Sidak multiple comparisons test following two-way ANOVA.
(H) qPCR examination of ribosomal RNA levels in sgKAT2A, sgSIRT5 K562 cells upon vec or KAT2A overexpression (O/E). rRNA expression levels were normalized to β-actin expression level. ∗∗∗p < 0.001 using Sidak multiple comparisons test following two-way ANOVA.
To investigate if histone malonylation is affected by malonyl-CoA availability specifically in ribosomal DNA (rDNA) locus, we treated K562 cells with orlistat for 24 h, and performed cleavage under targets and release using nuclease (CUT&RUN) with anti-malonyl lysine antibody and normal rabbit IgG as negative control. DNA enrichment was assayed by qPCR using sets of primer pairs spanning the entire human rDNA repeat26 (Figure S3A). The results showed that the enrichment of the upstream region of rDNA promoter (H42) for malonylation was significant higher in orlistat treated cells compared with DMSO-trated cells (Figure S3B). Although not statistically significant, the trend holds for other regions of rDNA locus (Figure S3B). These results suggest that orlistat treatment increases histone malonylation at rDNA locus.
The malonyl group is bulkier than the acetyl group, and malonylation changes the positive charge at the lysine side chain to a negative charge might play a role in chromatin compaction in nucleoli which might be reflected in nucleolar size. To test this hypothesis, we measured nucleolar size using immunofluorescence staining and a machine-learning algorithm to quantify nucleolar area (as a percentage of nuclear area) in HeLa cells (Figure 5D). Cells treated with 25 μM orlistat for 24 h had larger nucleolar percentage areas than DMSO-treated cells (Figure 5E). KAT2A overexpression increased nucleolar percentage area in both DMSO or orlistat treated HeLa cells (Figure 5E). In regard that KAT2A is a well-characterized histone acetyltransferase, it is possible that histone acetylation, in addition to malonylation, contributes to the enlarged nucleolar area on KAT2A overexpression. Next, we quantified nucleolar areas in SIRT5 KD HEK293T cells. The nucleolar percentage areas were significantly higher in SIRT5 KD cells than in control cells (Figure 5F). These results support the notion that inducing histone malonylation increases nucleolar area.
Because ribosomal RNAs (rRNAs) are transcribed and produced in the nucleoli, we next examined rRNA expression levels by qPCR in these cells. Orlistat treatment increased the expression levels of both 18S and 28S rRNAs in a dose-dependent manner in HeLa cells (Figure 5G). Similarly, we examined rRNA expression levels in K562 cells treated with 25 μM orlistat for 24 h. Overexpressing KAT2A increased the expression levels of both 18S and 28S rRNAs in SIRT5 KD cells, but not in KAT2A KD cells (Figure 5H), suggesting that the KD of SIRT5 is required for the induction of rRNA expression on KAT2A overexpression. These results indicate that lysine malonylation increases rRNA expression.
Lysine malonylation increases with age in mouse brain
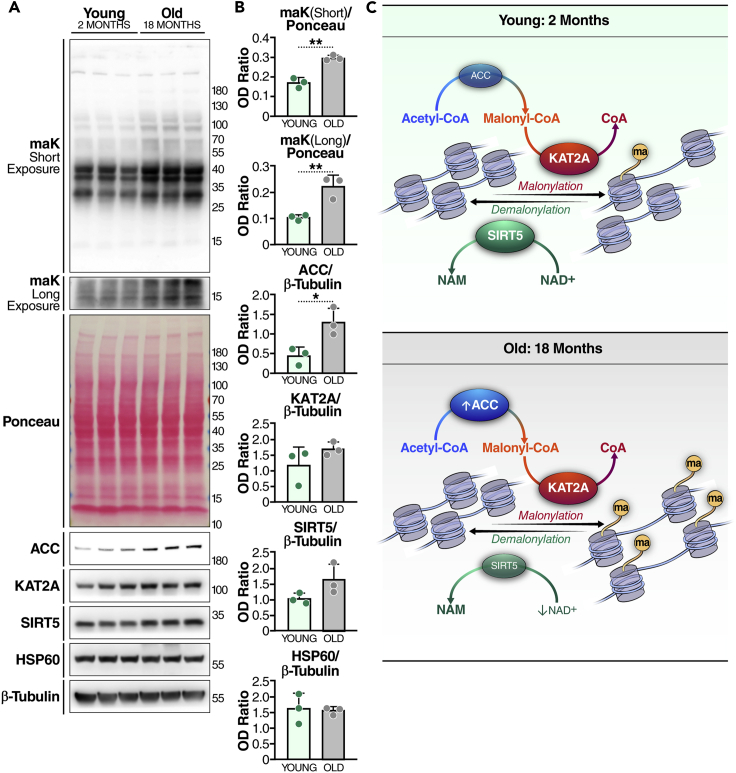
In light of the observations and adverse effect of nucleolar expansion in both premature and physiological aging,27,28 we tested whether lysine malonylation could be involved in the aging process. We extracted protein lysates from young (2-month-old) and older (18-month-old) mouse brains, and examined lysine malonylation levels by western blotting. Global malonylation levels, including malonylation of proteins around 15 kDa, where histones are located (shown on long exposure in Figure 6A), were higher in older than in young mice (Figures 6A and 6B), suggesting that levels of lysine malonylation increase with aging. Expression levels of SIRT5 and KAT2A were not different in young and older mice (Figures 6A and 6B), but the levels of ACC, the enzyme that generates malonyl-CoA, were higher in brains of older mice (Figures 6A and 6B), suggesting that older mice may have a higher level of fatty acid synthesis in the brain that contributes to the increase in lysine malonylation. To confirm these observations, we also performed nuclear isolation using whole brain tissues from young (3–6 months old) and middle-aged (12–14-month-old) mice. Western blotting results confirmed that lysine malonylation levels around 15 kDa, where core histone proteins reside, were significantly higher in the nuclear protein samples extracted from elder mouse brains than in younger ones (Figures S4A and S4B). Although not statistically significant, ACC levels in the nucleus tend to be higher in elder mouse brains than in younger ones as well (Figures S4A and S4B). These results suggest that lysine malonylation and ACC levels both increase with age in the nucleus in mouse brain.
Figure 6.
Histone malonylation as a function of age
(A) Western blot of protein samples prepared from young (2 months old) and middle-aged (18 months old) female C57BL/6 mouse brains.
(B) Quantification of the ODs of blots. n = 3 per group. Error bar: mean ± SD, ∗p < 0.05, ∗∗p < 0.01 with unpaired Student’s t test.
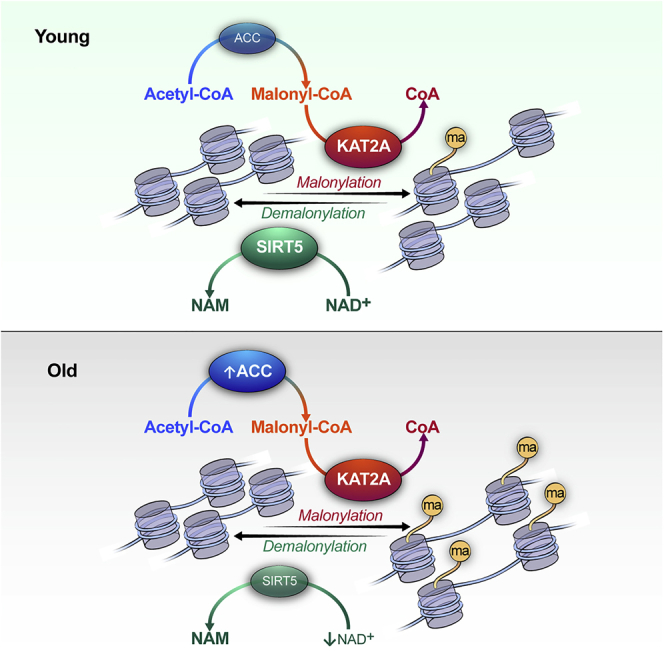
(C) Schematic of histone malonylation regulated by malonyl-CoA level, KAT2A, and SIRT5 emerges as a mechanistic link connecting metabolism, gene expression, and aging.
We next examined ribosomal RNA expression levels in RNA samples extracted from mouse brains at different ages. Real-time qPCR results showed that 18S rRNA expression levels in the brain were significantly higher in old (12–21 months old) than in young (6 months old) male mice (Figure S5A). Although not statistically significant, in female mice, 18S rRNA levels tend to be higher in elder (12–14 months old) than in younger (3–6 months old) mouse brains (Figure S5A). Similarly, 28S rRNA levels tend to be higher in old mouse brains than in young ones in both genders (Figure S5B). Linear regression analysis results confirmed that 18S rRNA level in the brain is positively correlated with age in male mice, and this trend holds for female mice (Figure S5C). Although not statistically significant, 28S rRNA level in the brain also tends to be positively correlated with age (Figure S5D). These results suggest that rRNA expression levels increase with age, which is at least partially because of increased histone malonylation, in mouse brain.
As a sirtuin family member, the deacylase activity of SIRT5 requires the availability of co-substrate NAD+.29 NAD levels decline with age,30,31 thereby restraining the activities of sirtuins, including SIRT5, at older age. Although the western blotting showed that the protein level of SIRT5 was not significantly different in young and older mouse brains (Figures 6A, 6B, S4A, and S4B), a decreased SIRT5 activity might still be a contributing factor to the increased level of lysine malonylation in old mice. Overall, these results suggest a possible association between histone malonylation and aging (Figure 6C).
Discussion
In the present study, we showed that lysine malonylation is affected by malonyl-CoA availability and found that histone malonylation is regulated by SIRT5 and KAT2A. Importantly, we observed that rRNA expression is subject to the regulation by lysine malonylation, and that global malonylation increases with age in mouse brain. Hence, this study reveals a possible role for histone malonylation in epigenetic regulations.
Malonyl-CoA thioester is a moderately reactive metabolic intermediate in de novo fatty acid synthesis pathway that provides malonyl group to modify free lysine residues. Genetic ablation of malony-CoA consuming enzymes, including MCD and FAS, globally increases lysine malonylation,5,14 whereas knocking out malonyl-CoA synthetase, ACSF3, reduces lysine malonylation in mitochondria.32 Thus, lysine malonylation appears to be tightly linked to malonyl-CoA metabolism. We confirmed the importance of malonyl-CoA availability in regulating lysine malonylation through pharmacological manipulation of the malony-CoA metabolic pathway. The basal level of lysine malonylation is barely detectable, even upon SIRT5 KD, in K562 cells, possibly because of constant proliferation and lack of malonylation accumulation in cultured cells. However, as shown here and elsewhere,15 lysine malonylation can be highly induced by orlistat (FAS inhibitor) treatment in cultured cells, and knockdown of SIRT5 further increased the level of orlistat-induced global lysine malonylation. On the other hand, orlistat-induced malonylation can be reduced by inhibiting ACC activity by activating AMPK using AICAR at a high dosage (1 mM). These observations suggest lysine malonylation as a function of malonyl-CoA metabolism.
SIRT5 is the main deacylase targeting malonyl, succinyl, and glutaryl groups for removal.1,2,3 In the present study, we found that levels of histone malonylation are dramatically increased in SIRT5 KO mouse liver, confirming that SIRT5 is a histone demalonylase. However, no difference in the overall histone succinylation or glutarylation was detected on SIRT5 KO, possibly because of low levels of succinylation and glutarylation in histones. This result is not entirely surprising because succinyl-CoA is a TCA cycle intermediate, and glutaryl-CoA is an intermediate in amino acid metabolism, and both metabolites are generated in mitochondria rather than in the nucleus. However, malonyl-CoA is a fatty acid building block generated in cytoplasm and, possibly, in nucleus (regarding the nuclear localization of ACC), which allows it to modify histones. By MS, we further determined that histone H2B was highly malonylated at K5 in both brain and liver tissues in SIRT5 KO mice. Although H2B_K108 was also identified to be highly malonylated in mouse liver, it appears that the malonyl group on H2B_K108 was not subject to the removal by SIRT5, possibly because of the lack of accessibility of H2B_K108 for SIRT5. Hence, these results suggest that H2B_K5 malonylation is selectively regulated by SIRT5.
With the discovery of a growing list of lysine acylations, KATs have been reported to possess an expanding repertoire of acyltransferase activities.12 Besides lysine acetylation, EP300 catalyzes propionylation, butyrylation, crotonylation, and β-hydroxybutyrylation in histones.33,34,35,36 Structural studies suggest that EP300 contains a deep aliphatic pocket within its active site that accommodates a wide range of acyl-CoA species.35 However, during the screening of KAT KD K562 cells for histone malonyltransferase, we observed no difference in histone malonylation on EP300 knockdown. This discrepancy might be because of several reasons: (1) Relatively low knockdown efficiency (65%) of EP300 in K562 cells, (2) high selectivity of EP300-regulated histone malonylation on the chromatin, or (3) redundancy of malonyltransferase activity between EP300 and other KATs, such as CBP. By contrast, we observed that KD of KAT2A strongly decreased histone malonylation. This observation is in line with recent findings that KAT2A regulates short-chain acidic acylations, including lysine succinylation and glutarylation, in histones.37,38 However, unlike EP300, KAT2A does not appear to have a deep aliphatic pocket in its active site to accommodate longer-chain acyl-CoA species. For this reason, the acyltransferase activity of KAT2A rapidly decreases with the increase of the length of the acyl group among acetyl-, propionyl-, and butyryl-CoA species.39 It is unclear if KAT2A undergoes structural rearrangement to accommodate short-chain acidic acyl-CoA species. Hence, further structural studies will be needed to resolve the molecular mechanism.
Compartmentalized acyl-CoA metabolism is important for chromatin regulation.40 Studies have shown that histone acylations, including acetylation, crotonylation, and succinylation, are regulated by the corresponding acyl-CoA-producing enzymes localized in the nucleus.37,41,42,43,44,45 In the present study, we found that, in addition to the canonical cytoplasmic localization, ACC was present in nucleus, and partly localized in nucleoli. Consequently, we observed that increasing malonylation (by orlistat treatment, KAT2A overexpression, or knocking down of SIRT5) increased nucleolar area and rRNA expression. Recent studies have shown that nucleolar enlargement is associated with aging.27,28 In the present study, we observed that lysine malonylation and ACC expression levels were higher in older mouse brains. Hence, increased histone malonylation might be a contributing factor that leads to aging-associated nucleolar expansion. Future profiling of histone malonylation at a genome-wide scale should help provide a broader view of aging-associated epigenetic changes.
To summarize, we demonstrated that malonylation is affected by malonyl-CoA availability, and regulated by SIRT5 and KAT2A. Of interest, we also found that inducing histone malonylation increases nucleolar size and rRNA expression. Finally, our finding of an age-related increase in malonlylation and the previously reported link between nucleolar size and aging suggest a possible role for histone malonylation in the aging process.
Limitations of the study
In this study, we identified KAT2A as a regulator of histone malonylation, but we would not rule out the possibility that some other KATs may also regulate histone malonylation at a lower scale, or in a more genomic region-specific manner, that was not detected by western blotting on CRISPRi in K562 cells. Moreover, the in vitro malonyltransferase activity assay of KAT2A was performed using recombinant histone peptides, the malonylation sites in which were not necessarily the same as in vivo. Future study needs to determine the lysine sites targeted by KAT2A for malonylation, and to profile the distribution of histone malonylation across the genome, to fully understand its biological functions.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit monoclonal anti-β-tubulin | Cell Signaling Technology | Cat. #2128, RRID:AB_823664 |
| Rabbit monoclonal anti-SIRT5 | Cell Signaling Technology | Cat. #8782, RRID:AB_2716763 |
| Rabbit monoclonal anti-acetyl-lysine | Cell Signaling Technology | Cat. #9814, RRID:AB_10544700 |
| Rabbit monoclonal anti-malonyl-lysine | PTM Biolabs | Cat. #PTM-901 |
| Rabbit monoclonal anti-succinyl-lysine | PTM Biolabs | Cat. #PTM-401 |
| Rabbit monoclonal anti-glutaryl-lysine | PTM Biolabs | Cat. #PTM-1151 |
| Rabbit monoclonal anti-ACC | Cell Signaling Technology | Cat. #3676, RRID:AB_2219397 |
| Rabbit polyclonal anti-phospho-ACC (Ser79) | Cell Signaling Technology | Cat. #3661, RRID:AB_330337 |
| Rabbit monoclonal anti-H2B | Cell Signaling Technology | Cat. #12364, RRID:AB_2714167 |
| Rabbit monoclonal anti-KAT2A | Cell Signaling Technology | Cat. #3305, RRID:AB_2128281 |
| Mouse monoclonal anti-HSP60 | Abcam | Cat. #ab59457, RRID:AB_2121285 |
| Rabbit polyclonal anti-Lamin A/C | Cell Signaling Technology | Cat. #2032, RRID:AB_2136278 |
| Rabbit monoclonal anti-COX IV | Abcam | Cat. #ab16056 |
| Rabbit monoclonal anti-GAPDH | Cell Signaling Technology | Cat. #5174, RRID:AB_10622025 |
| Mouse monoclonal anti-Fibrillarin | Abcam | Cat. #ab4566, RRID:AB_304523 |
| Goat anti-rabbit IgG, Alexa Fluor 555 | Invitrogen | Cat. #A-21428 |
| Goat anti-mouse IgG, Alexa Fluor 488 | Invitrogen | Cat. #A-11001 |
| Rabbit monoclonal anti-Tri-Methyl-Histone H3 (Lys4) (C42D8) | Cell Signaling Technology | Cat. #9751, RRID:AB_2616028 |
| Rabbit (DA1E) isotype control mAb IgG | Cell Signaling Technology | Cat. #66362, RRID:AB_2924329 |
| Chemicals, peptides, and recombinant proteins | ||
| Orlistat | Selleck | Cat. #1629 |
| AICAR | Selleck | Cat. #S1802 |
| Trichostatin A | Cayman Chem | Cat. #89730 |
| Nicotinamide | Sigma-Aldrich | Cat. #72340 |
| Recombinant KAT domain of human KAT2A | Enzo | Cat. #BML-SE272 |
| Human N-terminal (a.a. 1–20) histone peptides | This study, see Table S3 | N/A |
| 7-diethylamino-3-(4′-maleimidylphenyl)-4-methylcoumarin (CPM) | Invitrogen | Cat. #D346 |
| Critical commercial assays | ||
| Lipofectamine 3000 | Invitrogen | Cat. #L3000 |
| Histone extraction kit | EpiGentek | Cat. #OP-0006 |
| nuclear extraction kit | Epigentek | Cat. #OP-0002 |
| CUT&RUN assay kit | Cell Signaling Technology | Cat. #86652 |
| Deposited data | ||
| Raw data and complete mouse brain malonylation proteome MS datasets | This paper | MassIVE ID number: MSV000089318; ProteomeXchange ID: PXD033458http://massive.ucsd.edu/ProteoSAFe/status.jsp?task=a23d5ddef1b54e89b858221ce57863ab |
| Raw data and complete mouse liver malonylation proteome MS datasets | Nishida et al., 20154 | MassIVE ftp://MSV000079116:a@massive.ucsd.edu. |
| Experimental models: Cell lines | ||
| Human: K562 | ATCC | CRL-3343 |
| Human: K562-dCas9-KRAB | Gilbert et al., 201419 | N/A |
| Human: HEK293T | ATCC | CRL-3216 |
| Human: HeLa | ATCC | CCL-2 |
| Experimental models: Organisms/strains | ||
| Mouse: B6;129-Sirt5tm1Fwa/J | The Jackson Laboratory | Strain #:012757; RRID:IMSR_JAX:012757 |
| Mouse: C57BL/6J | The Jackson Laboratory | Strain #:000664; RRID:IMSR_JAX:000664 |
| Oligonucleotides | ||
| Primers for qPCR | This study, see Table S2 | N/A |
| Recombinant DNA | ||
| pEBB-Flag-GCN5 | Addgene | RRID:Addgene_74784 |
| pU6-sgRNA-EF1Alpha-puro-T2A-BFP | Addgene | RRID:Addgene_60955 |
| pCMV-VSV-G envelope vector | Cell Biolabs | Cat. # RV-110 |
| pCMV-dR8.91 packaging vector | Creative Biogen | Cat. #OVT2971 |
| Software and algorithms | ||
| GraphPad Prism 6 | GraphPad | https://graphpad.com/ |
| ImageJ | Schindelin et al., 201252 | https://imagej.nih.gov/ij/ |
| U-NET | Ronneberger et al., 201548 | https://lmb.informatik.uni-freiburg.de/resources/opensource/unet/ |
| Skyline-daily (version 21.1.1.223) | MacLean et al., 201051 | https://skyline.ms |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Eric Verdin (EVerdin@buckinstitute.org).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Mice
SIRT5 knockout (KO) mice (B6;129-Sirt5tm1Fwa/J, https://www.jax.org/strain/012757), and wild-type C57BL/6J were purchased from the Jackson Laboratory. Brain tissues harvested from 18-monthold female SIRT5 KO and wild-type littermates were used for malonylation proteomics analysis. Brain tissues harvested from male and female wild-type C57BL/6J mice at various ages were used for western blotting and qPCR analyses. All animal procedures were approved by Institutional Animal Care and Use Committee at Gladstone Institutes; University of California, San Francisco; and Buck Institute for Research on Aging. Mice were housed (12-h light/dark cycle, 22°C) and given unrestricted access to food and water.
Cell culture
K562 cells were cultured in RPMI1640 medium, and HEK293T and HeLa cells were cultured in Dulbecco’s modified Eagle medium (DMEM), with 10% fetal bovine serum (FBS) and 1% penicillin–streptomycin. The cells were grown at 37°C in a humidified incubator containing 5% CO2. Orlistat (Selleck, Cat. #1629) and AICAR (Selleck, Cat. #S1802) were dissolved in DMSO and diluted to specific concentrations for drug treatment. For transient overexpression of KAT2A, K562 or HeLa cells were transfected with pEBB-Flag-GCN5 (Addgene, Plasmid #74784) or an empty vector using Lipofectamine 3000 (Invitrogen, Cat. #L3000), according to manufacturer’s instructions 48 h before fixation or harvest.
Method details
Construction of CRISPRi K562 cells
K562 cells stably expressing a dCas9-KRAB fusion protein were kindly provided by Jonathan S. Weissman.19 pU6-sgRNA-EF1Alpha-puro-T2A-BFP (Addgene, Plasmid #60955) was used to express sgKAT and sgNC sgRNAs; and pU6-sgRNA-EF1Alpha-BSR-T2A-BFP (puromycin-resistance gene replaced by blasticidin S-resistance gene) was used to express sgSIRT5 sgRNAs. The generation of stably gene knockdown K562 cells was performed using a CRISPRi system.19 Briefly, double-stranded sgRNA oligos (Table S1) were synthesized and inserted into backbone plasmids using BstXI and BlpI restriction sites. Lentivirus expressing sgRNAs were packaged and produced using HEK293T cells with pCMV-VSV-G envelope vector (Cell Biolabs, Cat. # RV-110) and pCMV-dR8.91 packaging vector (Creative Biogen, Cat. #OVT2971). K562-dCas9-KRAB cells stably expressing sgRNAs were generated upon lentiviral infection, and selected using corresponding antibiotics and fluorescence-activated cell sorting of BFP-positive cells.
Malonyl-CoA measurement
K562 cells were harvested, and rinsed briefly with 1 mL saline solution (0.9% NaCl in water) before snap freezing in liquid nitrogen. Cell numbers were counted for quantification. 0.5 ml 80% MeOH/water was added to each sample on dry ice. Geno/Grinder was used to lyase cells (1500 rpm, 1 min). 10 μl 50 ng/μL [13C3]-malonyl-CoA was added to each sample placed on ice. Vortex for 1 min before centrifugation at 20,000 g for 10 minat 4°C. Supernatant was dried using speed vacuum concentrator. Metabolite extract was dissolved in 30 μl 50 mM ammonium acetate and 8 μl was injected for LC-MS analysis. For acyl-CoA analysis, a reversed phase liquid chromatography method employing a Luna C18 column (100 × 2.0 mm i.d., 3 μm; Phenomenex) was used.46 LC was coupled with Q Exactive mass spectrometer (Thermo Scientific) for metabolite separation and subsequent detection. The Q Exactive mass spectrometer was equipped with a HESI probe, and it was operated in the positive ion mode with a full-scan range of 150–1000 (m/z). LC-MS data were analyzed using Sieve 2.0 (Thermo Scientific), and the integrated area under metabolite peak was used to compare the relative abundance of each metabolite in different samples in the same batch. Retention times for each metabolite are based on in-house library and determined by comparing to standard compounds when available, m/z and characteristic fragments.
Reverse transcription and qPCR
RNA was extracted from HeLa and K562 cells using RNA STAT-60 (Fisher Scientific, Cat. #NC9489785). Reverse transcription was performed using a First-Strand cDNA Synthesis kit (New England Biolabs, Cat. #E6560) and random primers, according to manufacturer’s instructions. RNA expression levels were quantified by qPCR with a SYBR Green reagent (Thermo Fisher Scientific, Cat. #K0223). Primers for qPCR are shown in Table S2.
Western blotting
Cells and tissues are lysed and homogenized with RIPA buffer (Thermo Fisher Scientific, Cat. #89900) containing protease and phosphatase inhibitor cocktail (Thermo Fisher Scientific, Cat. # PI78447), 5 M trichostatin A (Cayman Chem, Cat. #89730), and 5 mM nicotinamide (Sigma-Aldrich, Cat. #72340). Histones were extracted from cells and tissues using a total histone extraction kit (EpiGentek, Cat. #OP-0006). Before SDS-PAGE, protein samples were heated at 70°C for 5 min. Antibodies used are listed as follows: anti-β-tubulin (Cell Signaling Technology, Cat. #2128), anti-SIRT5 (Cell Signaling Technology, Cat. #8782), anti-acetyl-lysine (Cell Signaling Technology, Cat. #9814), anti-malonyl-lysine (PTM Biolabs, Cat. #PTM-901), anti-succinyl-lysine (PTM Biolabs, Cat. # PTM-401), anti-glutaryl-lysine (PTM Biolabs, Cat. #PTM-1151), anti-ACC (Cell Signaling Technology, Cat. #3676), anti-phospho-ACC (Ser79) (Cell Signaling Technology, Cat. #3661), anti-H2B (Cell Signaling Technology, Cat. #12364), anti-KAT2A (Cell Signaling Technology, Cat. #3305), anti-HSP60 (Abcam, Cat. #ab59457), anti-Lamin A/C (Cell Signaling Technology, Cat. #2032), anti-COX IV (Abcam, Cat. #ab16056), and anti-GAPDH (Cell Signaling Technology, Cat. #5174). Ponceau S solution (Sigma-Aldrich, Cat. #P7170) was used for protein staining.
Subcellular fractionation and nuclear isolation
Subcellular fractionation was performed as described.47 Briefly, HEK293T cells were gently homogenized in hypotonic buffer on ice with a dounce homogenizer till the vast majority of cells were ruptured, and the nuclei were blue stained by Trypan blue. Cells were centrifuged at 600 g for 3 minat 4°C to obtain a pellet (nuclear fraction). The supernatant was further centrifuged at 15,000 g for 10 minat 4°C to pellet mitochondria, and the supernatant was collected as the cytosolic fraction. For western blotting, equal protein amounts of each fraction were assessed. Nuclear isolation from mouse brain tissues were performed using a nuclear extraction kit (Epigentek, Cat. #OP-0002) according to manufacturer’s instructions. Briefly, mouse brain tissues were cut in half, and half brains were pulvorized in liquid nitrogen-chilled grinder before lysed in 1 mL diluted NE1 on ice. Single cells/nuclei in buffer NE1 (contains PIC and DTT) were collected using 40 μm strainer. Cytoplasmic fraction was collected as supernatant in buffer NE1, and nuclear fraction in the pellet was collected and lysed in buffer NE2. For western blotting, samples from equal proportions of brain tissues were assessed.
CUT&RUN
Histone malonylation levels at rDNA regions in K562 cells were examined using a Cleavage under targets and release using nuclease (CUT&RUN) assay kit (Cell Signaling Technology, Cat. # 86652) according to manufacturer’s instructions. Briefly, K562 cells treated with DMSO or 25 μM orlistat for 24 h were harvested at room temperature to minimize stress on the cells. 50,000 cells were collected for each reaction, and an additional 50,000 (same amount) cells for the input sample. Reactions for the positive control Tri-Methyl-Histone H3 (Lys4) (C42D8) Rabbit (Cell Signaling Technology, Cat. mAb #9751) and the negative control Rabbit (DA1E) mAb IgG XP® Isotype Control (Cell Signaling Technology, Cat. #66362) were included in addition to anti-malonyl-lysine (PTM Biolabs, Cat. #PTM-901). Cells were mildly fixed (0.1% formaldehyde) at room temperature for 2 min, and crosslinking was stopped in 125 mM glycine. After binding of Concanavalin A beads and primary antibody, and binding of pAG-MNase enzyme, DNA was digested and diffused. DNA was then purified using DNA spin columns (Cell Signaling Technology, Cat. # 14209). 2 μl of purified DNA was used for qPCR-mediated quantification of rDNA regions. Primer pairs were designed based on a previous study,26 and are provided in Table S2.
Immunofluorescence staining and microscopy
HeLa cells were seeded on to slide chambers (Waston Bio Lab, Cat. #192–008), and treated with DMSO or orlistat for 24 h before fixation with 4% paraformaldehyde (PFA) for 15 minat room temperature. Cells were permeabilized with PBS containing 1% BSA and 0.01% Triton X-100 for 15 min, and blocked with PBS containing 1% BSA, 5% goat serum (same source as the secondary antibodies) and 0.1% Tween 20 for 1 hat room temperature. Cells were incubated with primary antibodies at 4°C overnight. Primary antibodies used are anti-ACC (Cell Signaling Technology, Cat. #3676, 1:200 dilution) and anti-FBL (Abcam, Cat. #ab4566, 1:500 dilution). Cells were incubated with secondary antibodies at room temperature for 1 h away from light. Secondary antibodies were goat anti-rabbit IgG, Alexa Fluor 555 (Invitrogen, Cat. #A-21428, 1:500 dilution) and goat anti-mouse IgG, Alexa Fluor 488 (Invitrogen, Cat. #A-11001, 1:500 dilution). Nuclei were stained with DAPI (Sigma-Aldrich, Cat. #D9542) before mounting with ProLong antifade mountant (Invitrogen, Cat. #P10144).
For confocal microscopy, Zeiss LSM 780 was used with 63x oil immersion objective at Ex. (nm)/Detection (nm) = 405/410–508 (DAPI), 488/490–570 (Alexa Fluor 488), and 561/566–697 (Alexa Fluor 555). Fluorescence intensity distribution was generated using Zeiss Zen 3.4 (blue edition). Forwide-field microscopy, Keyence BZ-X800 was used with a 40x objective. 20 images were taken for each genotype and treatment group. Nucleolar percentage area was quantified using a machine-learning algorithm, where we used U-NET for image segmentation to automatically detect DAPI-stained nuclei and fibrillarin-stained nucleoli and then quantified nucleoli and nuclei area.48 Several U-NET models were trained, including HeLa nuclei with four images, HeLa nucleoli with four images, HEK293T nuclei with four images, and HEK293T nucleoli with eight images. Each training session utilized data augmentation, including rotation, scaling, and brightness variation, and was performed for 1000 epochs. After training, U-NET was used to predict the nuclei and nucleoli area for each cell type. Each detected region was then scanned using a flood fill algorithm to quantify area in pixels.
In-vitro malonyltransferase activity assay
The malonyltransferase activities of recombinant KAT domain of human KAT2A (Enzo, Cat. #BML-SE272) were examined against synthesized human N-terminal (a.a. 1–20) histone peptides (Table S3). The in-vitro malonylation reaction was performed in an assay buffer containing 100 mM HEPES (pH 7.5), 100 mM NaCl, and 0.01% Triton X-100. The enzymatic reactions were initiated by adding malonyl-CoA (to 50 μM) to assay mixture containing 40 nM KAT2A, and incubated for 20 minat 37 °C. Non-enzymatic controls took place in the assay buffer without KAT peptide. Mixtures were allowed to equilibrate for 5 minat room temperature before adding malonyl-CoA and moving to 37 °C. Upon completion, for each the 50-μl mixture, the reaction was stopped with 50 μl of cold isopropanol. Then, 100 μl of 25 μM CPM (7-diethylamino-3-(4′-maleimidylphenyl)-4-methylcoumarin, Invitrogen, Cat. #D346) in assay buffer were added (200 μl total volume). After incubation for 15 minat room temperature, plates were read in a fluorescence microplate reader (ClLARIOStar; BMG Labtech) at Ex. (nm)/Em. (nm) = 390/460 and a gain setting of 938.
Mass spectrometry
Identification of malonylation sites in histones in mouse liver tissues was performed as reported.4 Briefly, livers were collected from five WT and five SIRT5 KO male mice at 10 months of age. Anti-malonyl-lysine antibodies (Cell Signaling Technology, H4597 and BL13640) were used for immunoaffinity enrichment of malonylated peptides. All samples used for MS1 filtering experiments were analyzed by reverse-phase LC-ESI-MS/MS with an Eksigent UltraPlus nano-LC 2D HPLC system connected to a quadruple time-of-flight TripleTOF 6600 mass spectrometer (MS) (SCIEX) in direct injection mode. Datasets were analyzed and searched using Mascot server version 2.3.02 (Matrix Sciences) and ProteinPilot (AB SCIEX 4.5beta) with the Paragon algorithm (4.5, 1656). All data files were searched using the SwissProt 2013_01 database with a total of 538,849 sequences but restricted to Mus musculus (16,580 protein sequences).
To identify malonylation sites in histones, four 18-month-old WT and four SIRT5 KO female mice were fasted for 24 h and refed for 6 h before sacrifice. Frozen brains were immersed in lysis buffer containing 8 M urea, 200 mM triethylammonium bicarbonate (TEAB), pH 8.5, 75 mM sodium chloride, 1 μM trichostatin A, 3 mM nicotinamide, and 1x protease/phosphatase inhibitor cocktail (Thermo Fisher Scientific, Waltham, MA), homogenized for 2 cycles with a Bead Beater TissueLyser II (Qiagen, Germantown, MD) at 30 Hz for 3 min each, and sonicated for 10 min. Lysates were clarified by spinning at 14,000 x g for 10 min at 4°C, and the supernatant containing the soluble proteins was collected. Protein concentrations were determined using a bicinchoninic acid protein assay (Thermo Fisher Scientific), and subsequently 5 mg of protein from each sample were brought to an equal volume using a solution of 8 M urea in 50 mM TEAB, pH 8. Proteins were reduced using 20 mM dithiothreitol in 50 mM TEAB for 30 min at 37°C, and after cooling to room temperature, alkylated with 40 mM iodoacetamide for 30 min at room temperature in the dark. Samples were diluted four-fold with 50 mM TEAB, pH 8, and proteins were digested overnight with a solution of sequencing-grade trypsin (Promega, San Luis Obispo, CA) in 50 mM TEAB at a 1:50 (wt:wt) enzyme:protein ratio at 37°C. This reaction was quenched with 1% formic acid (FA), and the sample was clarified by centrifugation at 2,000 x g for 10 minat room temperature. Clarified peptide samples were desalted with Oasis 10-mg Sorbent Cartridges (Waters, Milford, MA). 100 μg of each peptide elution were aliquoted for analysis of protein-level changes, after which all desalted samples were vacuum dried. The 100 μg of whole lysate were re-suspended in 0.2% FA in water at a final concentration of 1 μg/μL and stored for MS analysis. The remaining 4.9 mg of digests were re-suspended in 1.4 mL of immunoaffinity purification buffer (Cell Signaling Technology, Danvers, MA) containing 50 mM 4-morpholinepropanesulfonic acid (MOPS)/sodium hydroxide, pH 7.2, 10 mM disodium phosphate, and 50 mM sodium chloride for enrichment of posttranslational modifications. Peptides were enriched for malonylation with anti-malonyl antibody conjugated to agarose beads from the Malonyl-Lysine Motif Kit (Cell Signaling Technology, Danvers, MA). This process was performed according to the manufacturer protocol; however, each sample was incubated in half the recommended volume of washed beads. Peptides were eluted from the antibody-bead conjugates with 0.15% trifluoroacetic acid in water and were desalted using C18 stagetips made in-house. Samples were vacuum dried and re-suspended in 0.2% FA in water. Finally, indexed retention time standard peptides (iRT; Biognosys, Schlieren, Switzerland)49 were spiked in the samples according to manufacturer’s instructions. Samples were analyzed in data-independent acquisition (DIA) mode on a Dionex UltiMate 3000 system coupled to an Orbitrap Eclipse Tribrid MS (both from Thermo Fisher Scientific, San Jose, CA). The solvent system consisted of 2% ACN, 0.1% FA in water (solvent A) and 98% ACN, 0.1% FA in water (solvent B). Peptides enriched in posttranslational modifications (4 μL) were loaded onto an Acclaim PepMap 100C18 trap column (0.1 x 20 mm, 5 μm particle size; Thermo Fisher Scientific) over 5 minat 5 μL/min with 100% solvent A. Peptides were eluted on an Acclaim PepMap 100C18 analytical column (75 μm x 50 cm, 3 μm particle size; Thermo Fisher Scientific) at 0.3 μL/min using the following gradient of solvent B: linear from 2–20% in 125 min, linear from 20–32% in 40 min, and up to 80% in 1 min, with a total gradient length of 210 min. For the DIA analysis, full MS spectra were collected at 120,000 resolution (AGC target: 3e6 ions, maximum injection time: 60 ms, 350–1,650 m/z), and MS2 spectra at 30,000 resolution (AGC target: 3e6 ions, maximum injection time: Auto, NCE: 27, fixed first mass 200 m/z). The isolation scheme consisted in 26 variable windows covering the 350–1,650 m/z range with a window overlap of 1 m/z (Table S4).50
A spectral library was generated in Spectronaut (version 14.10.201222.47784; Biognosys, Schlieren, Switzerland) from the DIA acquisitions using a Mus musculus UniProtKB-TrEML database (58,430 entries, 01/31/2018). BGS settings were set as defaults settings, except for the Pulsar search for which four missed cleavages were allowed and lysine malonylation was added as variable modification. The final library contained 13,304 modified peptides and 3,112 protein groups (Table S5). The library was further imported into Skyline-daily (version 21.1.1.223).51 DIA data extraction for the peptide PDPAKSAPAPK (H2B2B_MOUSE) was performed using the following parameters: up to 10 of the most intense product ions (monocharged y- and b-type ions) from ion 2 to last ion-1 per precursor ion were extracted. Resolving power was set to 30,000 at 400 m/z, and all matching scans were including. Finally, chromatographic peaks were investigated to manually adjust peak integration boundaries and remove interfered transitions (four final transitions). Signal at the peptide level was obtained by summing the corresponding transition peak areas. Statistical analysis was performed in Skyline using default settings.
Quantification and statistical analysis
GraphPad Prism 6 (GraphPad Software, San Diego, CA) was used for statistical analyses. Unpaired two-tailed Student’s t-test was used when two groups of data were compared. Sidak multiple comparisons test following two-way ANOVA was used when malonylation levels between WT and S5KO groups on a certain lysine site were compared, and when rRNA expression levels between WT and S5KO groups were compared. p values less than 0.05 were considered statistically significant. Optical densities of western blots were quantified using ImageJ software.52
Acknowledgments
We thank Dr. Xueshu Xie for discussion and suggestions. We thank Dr. Marius Walter for assistance with CRISPRi K562 cell construction; Dr. Herbert Kasler for fluorescence-activated cell sorting; Ms. Lei Wei for assistance with sample processing for mass spectrometry, Dr. Wenjuan He for providing mouse tissue samples; Ms. Rebeccah Riley and Mr. Ryan Kwok for mouse colony maintenance; Mr. John Carroll for graphic design; Mr. Gary Howard for language editing. This study was supported by NIH grant R24DK085610 (Eric Verdin and Birgit Schilling), Glenn Foundation for Medical Research (Ran Zhang), and NCRR shared instrumentation grants 1S10 OD016281 (TripleTOF 6600, Buck Institute) and 1S10 OD028654 (Orbitrap Eclipse Tribrid, PI: Birgit Schilling).
Author contributions
R.Z., B.S., and E.V. designed the experiments and analyzed the data. J.B., J.R., and B.S. performed malonylation mass spectrometry analysis. G.S. and X.L. performed malonyl-CoA measurement using mass spectrometry. O.B. made sgSIRT5 HEK293T cell lines. I.H. and M.S.K. performed nucleolar area quantification analysis. R.Z. performed the rest of the experiments. R.Z. and E.V. wrote the manuscript. All authors discussed the results and commented on the manuscript.
Declaration of interests
Eric Verdin is a scientific co-founder of Napa Therapeutics and serves on the scientific advisory board of Seneque.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: February 13, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.106193.
Supplemental information
Data and code availability
-
•
Raw data and complete MS datasets have been uploaded to the Center for Computational Mass Spectrometry, to the MassIVE repository at UCSD, and can be downloaded using the following link: http://massive.ucsd.edu/ProteoSAFe/status.jsp?task=a23d5ddef1b54e89b858221ce57863ab (MassIVE ID number: MSV000089318; ProteomeXchange ID: PXD033458).
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Du J., Zhou Y., Su X., Yu J.J., Khan S., Jiang H., Kim J., Woo J., Kim J.H., Choi B.H., et al. Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science. 2011;334:806–809. doi: 10.1126/science.1207861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peng C., Lu Z., Xie Z., Cheng Z., Chen Y., Tan M., Luo H., Zhang Y., He W., Yang K., et al. The first identification of lysine malonylation substrates and its regulatory enzyme. Mol. Cell. Proteomics. 2011;10 doi: 10.1074/mcp.M111.012658. M111.012658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tan M., Peng C., Anderson K.A., Chhoy P., Xie Z., Dai L., Park J., Chen Y., Huang H., Zhang Y., et al. Lysine glutarylation is a protein posttranslational modification regulated by SIRT5. Cell Metabol. 2014;19:605–617. doi: 10.1016/j.cmet.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nishida Y., Rardin M.J., Carrico C., He W., Sahu A.K., Gut P., Najjar R., Fitch M., Hellerstein M., Gibson B.W., Verdin E. SIRT5 regulates both cytosolic and mitochondrial protein malonylation with glycolysis as a major target. Mol. Cell. 2015;59:321–332. doi: 10.1016/j.molcel.2015.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Colak G., Pougovkina O., Dai L., Tan M., Te Brinke H., Huang H., Cheng Z., Park J., Wan X., Liu X., et al. Proteomic and biochemical studies of lysine malonylation suggest its malonic aciduria-associated regulatory role in mitochondrial function and fatty acid oxidation. Mol. Cell. Proteomics. 2015;14:3056–3071. doi: 10.1074/mcp.M115.048850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saggerson D. Malonyl-CoA, a key signaling molecule in mammalian cells. Annu. Rev. Nutr. 2008;28:253–272. doi: 10.1146/annurev.nutr.28.061807.155434. [DOI] [PubMed] [Google Scholar]
- 7.Wagner G.R., Payne R.M. Widespread and enzyme-independent Nepsilon-acetylation and Nepsilon-succinylation of proteins in the chemical conditions of the mitochondrial matrix. J. Biol. Chem. 2013;288:29036–29045. doi: 10.1074/jbc.M113.486753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Casey J.R., Grinstein S., Orlowski J. Sensors and regulators of intracellular pH. Nat. Rev. Mol. Cell Biol. 2010;11:50–61. doi: 10.1038/nrm2820. [DOI] [PubMed] [Google Scholar]
- 9.Wagner G.R., Bhatt D.P., O'Connell T.M., Thompson J.W., Dubois L.G., Backos D.S., Yang H., Mitchell G.A., Ilkayeva O.R., Stevens R.D., et al. A class of reactive acyl-CoA species reveals the non-enzymatic origins of protein acylation. Cell Metabol. 2017;25:823–837.e8. doi: 10.1016/j.cmet.2017.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stillman B. Histone modifications: insights into their influence on gene expression. Cell. 2018;175:6–9. doi: 10.1016/j.cell.2018.08.032. [DOI] [PubMed] [Google Scholar]
- 11.Dai Z., Ramesh V., Locasale J.W. The evolving metabolic landscape of chromatin biology and epigenetics. Nat. Rev. Genet. 2020;21:737–753. doi: 10.1038/s41576-020-0270-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sabari B.R., Zhang D., Allis C.D., Zhao Y. Metabolic regulation of gene expression through histone acylations. Nat. Rev. Mol. Cell Biol. 2017;18:90–101. doi: 10.1038/nrm.2016.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Simithy J., Sidoli S., Yuan Z.F., Coradin M., Bhanu N.V., Marchione D.M., Klein B.J., Bazilevsky G.A., McCullough C.E., Magin R.S., et al. Characterization of histone acylations links chromatin modifications with metabolism. Nat. Commun. 2017;8:1141. doi: 10.1038/s41467-017-01384-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bruning U., Morales-Rodriguez F., Kalucka J., Goveia J., Taverna F., Queiroz K.C.S., Dubois C., Cantelmo A.R., Chen R., Loroch S., et al. Impairment of angiogenesis by fatty acid synthase inhibition involves mTOR malonylation. Cell Metabol. 2018;28:866–880.e15. doi: 10.1016/j.cmet.2018.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kulkarni R.A., Worth A.J., Zengeya T.T., Shrimp J.H., Garlick J.M., Roberts A.M., Montgomery D.C., Sourbier C., Gibbs B.K., Mesaros C., et al. Discovering targets of non-enzymatic acylation by thioester reactivity profiling. Cell Chem. Biol. 2017;24:231–242. doi: 10.1016/j.chembiol.2017.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Munday M.R., Campbell D.G., Carling D., Hardie D.G. Identification by amino acid sequencing of three major regulatory phosphorylation sites on rat acetyl-CoA carboxylase. Eur. J. Biochem. 1988;175:331–338. doi: 10.1111/j.1432-1033.1988.tb14201.x. [DOI] [PubMed] [Google Scholar]
- 17.Davies S.P., Carling D., Hardie D.G. Tissue distribution of the AMP-activated protein kinase, and lack of activation by cyclic-AMP-dependent protein kinase, studied using a specific and sensitive peptide assay. Eur. J. Biochem. 1989;186:123–128. doi: 10.1111/j.1432-1033.1989.tb15185.x. [DOI] [PubMed] [Google Scholar]
- 18.Sullivan J.E., Brocklehurst K.J., Marley A.E., Carey F., Carling D., Beri R.K. Inhibition of lipolysis and lipogenesis in isolated rat adipocytes with AICAR, a cell-permeable activator of AMP-activated protein kinase. FEBS Lett. 1994;353:33–36. doi: 10.1016/0014-5793(94)01006-4. [DOI] [PubMed] [Google Scholar]
- 19.Gilbert L.A., Horlbeck M.A., Adamson B., Villalta J.E., Chen Y., Whitehead E.H., Guimaraes C., Panning B., Ploegh H.L., Bassik M.C., et al. Genome-scale CRISPR-mediated control of gene repression and activation. Cell. 2014;159:647–661. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang H., Sabari B.R., Garcia B.A., Allis C.D., Zhao Y. SnapShot: histone modifications. Cell. 2014;159:458–458.e1. doi: 10.1016/j.cell.2014.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Z., Tan M., Xie Z., Dai L., Chen Y., Zhao Y. Identification of lysine succinylation as a new post-translational modification. Nat. Chem. Biol. 2011;7:58–63. doi: 10.1038/nchembio.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ali I., Conrad R.J., Verdin E., Ott M. Lysine acetylation goes global: from epigenetics to metabolism and Therapeutics. Chem. Rev. 2018;118:1216–1252. doi: 10.1021/acs.chemrev.7b00181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gillet L.C., Navarro P., Tate S., Röst H., Selevsek N., Reiter L., Bonner R., Aebersold R. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Mol. Cell. Proteomics. 2012;11 doi: 10.1074/mcp.O111.016717. O111.016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Collins B.C., Hunter C.L., Liu Y., Schilling B., Rosenberger G., Bader S.L., Chan D.W., Gibson B.W., Gingras A.C., Held J.M., et al. Multi-laboratory assessment of reproducibility, qualitative and quantitative performance of SWATH-mass spectrometry. Nat. Commun. 2017;8:291. doi: 10.1038/s41467-017-00249-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tong L., Harwood H.J., Jr. Acetyl-coenzyme A carboxylases: versatile targets for drug discovery. J. Cell. Biochem. 2006;99:1476–1488. doi: 10.1002/jcb.21077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grandori C., Gomez-Roman N., Felton-Edkins Z.A., Ngouenet C., Galloway D.A., Eisenman R.N., White R.J. c-Myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I. Nat. Cell Biol. 2005;7:311–318. doi: 10.1038/ncb1224. [DOI] [PubMed] [Google Scholar]
- 27.Tiku V., Jain C., Raz Y., Nakamura S., Heestand B., Liu W., Späth M., Suchiman H.E.D., Müller R.U., Slagboom P.E., et al. Small nucleoli are a cellular hallmark of longevity. Nat. Commun. 2017;8 doi: 10.1038/ncomms16083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buchwalter A., Hetzer M.W. Nucleolar expansion and elevated protein translation in premature aging. Nat. Commun. 2017;8:328. doi: 10.1038/s41467-017-00322-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Houtkooper R.H., Cantó C., Wanders R.J., Auwerx J. The secret life of NAD+: an old metabolite controlling new metabolic signaling pathways. Endocr. Rev. 2010;31:194–223. doi: 10.1210/er.2009-0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu X.H., Lu M., Lee B.Y., Ugurbil K., Chen W. In vivo NAD assay reveals the intracellular NAD contents and redox state in healthy human brain and their age dependences. Proc. Natl. Acad. Sci. USA. 2015;112:2876–2881. doi: 10.1073/pnas.1417921112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lautrup S., Sinclair D.A., Mattson M.P., Fang E.F. NAD(+) in brain aging and neurodegenerative disorders. Cell Metabol. 2019;30:630–655. doi: 10.1016/j.cmet.2019.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bowman C.E., Rodriguez S., Selen Alpergin E.S., Acoba M.G., Zhao L., Hartung T., Claypool S.M., Watkins P.A., Wolfgang M.J. The mammalian malonyl-CoA synthetase ACSF3 is required for mitochondrial protein malonylation and metabolic efficiency. Cell Chem. Biol. 2017;24:673–684.e4. doi: 10.1016/j.chembiol.2017.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen Y., Sprung R., Tang Y., Ball H., Sangras B., Kim S.C., Falck J.R., Peng J., Gu W., Zhao Y. Lysine propionylation and butyrylation are novel post-translational modifications in histones. Mol. Cell. Proteomics. 2007;6:812–819. doi: 10.1074/mcp.M700021-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sabari B.R., Tang Z., Huang H., Yong-Gonzalez V., Molina H., Kong H.E., Dai L., Shimada M., Cross J.R., Zhao Y., et al. Intracellular crotonyl-CoA stimulates transcription through p300-catalyzed histone crotonylation. Mol. Cell. 2015;58:203–215. doi: 10.1016/j.molcel.2015.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaczmarska Z., Ortega E., Goudarzi A., Huang H., Kim S., Márquez J.A., Zhao Y., Khochbin S., Panne D. Structure of p300 in complex with acyl-CoA variants. Nat. Chem. Biol. 2017;13:21–29. doi: 10.1038/nchembio.2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang H., Zhang D., Weng Y., Delaney K., Tang Z., Yan C., Qi S., Peng C., Cole P.A., Roeder R.G., Zhao Y. The regulatory enzymes and protein substrates for the lysine beta-hydroxybutyrylation pathway. Sci. Adv. 2021;7 doi: 10.1126/sciadv.abe2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Y., Guo Y.R., Liu K., Yin Z., Liu R., Xia Y., Tan L., Yang P., Lee J.H., Li X.J., et al. KAT2A coupled with the alpha-KGDH complex acts as a histone H3 succinyltransferase. Nature. 2017;552:273–277. doi: 10.1038/nature25003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bao X., Liu Z., Zhang W., Gladysz K., Fung Y.M.E., Tian G., Xiong Y., Wong J.W.H., Yuen K.W.Y., Li X.D. Glutarylation of histone H4 lysine 91 regulates chromatin dynamics. Mol. Cell. 2019;76:660–675.e9. doi: 10.1016/j.molcel.2019.08.018. [DOI] [PubMed] [Google Scholar]
- 39.Ringel A.E., Wolberger C. Structural basis for acyl-group discrimination by human Gcn5L2. Acta Crystallogr. D Struct. Biol. 2016;72:841–848. doi: 10.1107/S2059798316007907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trefely S., Lovell C.D., Snyder N.W., Wellen K.E. Compartmentalised acyl-CoA metabolism and roles in chromatin regulation. Mol. Metabol. 2020;38 doi: 10.1016/j.molmet.2020.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu X., Si W., He L., Yang J., Peng Y., Ren J., Liu X., Jin T., Yu H., Zhang Z., et al. The existence of a nonclassical TCA cycle in the nucleus that wires the metabolic-epigenetic circuitry. Signal Transduct. Targeted Ther. 2021;6:375. doi: 10.1038/s41392-021-00774-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nagaraj R., Sharpley M.S., Chi F., Braas D., Zhou Y., Kim R., Clark A.T., Banerjee U. Nuclear localization of mitochondrial TCA cycle enzymes as a critical step in mammalian zygotic genome activation. Cell. 2017;168:210–223.e11. doi: 10.1016/j.cell.2016.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sutendra G., Kinnaird A., Dromparis P., Paulin R., Stenson T.H., Haromy A., Hashimoto K., Zhang N., Flaim E., Michelakis E.D. A nuclear pyruvate dehydrogenase complex is important for the generation of acetyl-CoA and histone acetylation. Cell. 2014;158:84–97. doi: 10.1016/j.cell.2014.04.046. [DOI] [PubMed] [Google Scholar]
- 44.Liu S., Yu H., Liu Y., Liu X., Zhang Y., Bu C., Yuan S., Chen Z., Xie G., Li W., et al. Chromodomain protein CDYL acts as a crotonyl-CoA hydratase to regulate histone crotonylation and spermatogenesis. Mol. Cell. 2017;67:853–866.e5. doi: 10.1016/j.molcel.2017.07.011. [DOI] [PubMed] [Google Scholar]
- 45.Wellen K.E., Hatzivassiliou G., Sachdeva U.M., Bui T.V., Cross J.R., Thompson C.B. ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 2009;324:1076–1080. doi: 10.1126/science.1164097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu X., Sadhukhan S., Sun S., Wagner G.R., Hirschey M.D., Qi L., Lin H., Locasale J.W. High-resolution metabolomics with acyl-CoA profiling reveals widespread remodeling in response to diet. Mol. Cell. Proteomics. 2015;14:1489–1500. doi: 10.1074/mcp.M114.044859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Park J., Chen Y., Tishkoff D.X., Peng C., Tan M., Dai L., Xie Z., Zhang Y., Zwaans B.M.M., Skinner M.E., et al. SIRT5-mediated lysine desuccinylation impacts diverse metabolic pathways. Mol. Cell. 2013;50:919–930. doi: 10.1016/j.molcel.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ronneberger O., Fischer P., Brox T. Springer International Publishing; 2015. U-net: Convolutional Networks for Biomedical Image Segmentation. Held in Cham; pp. 234–241. [Google Scholar]
- 49.Escher C., Reiter L., MacLean B., Ossola R., Herzog F., Chilton J., MacCoss M.J., Rinner O. Using iRT, a normalized retention time for more targeted measurement of peptides. Proteomics. 2012;12:1111–1121. doi: 10.1002/pmic.201100463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bruderer R., Bernhardt O.M., Gandhi T., Xuan Y., Sondermann J., Schmidt M., Gomez-Varela D., Reiter L. Optimization of experimental parameters in data-independent mass spectrometry significantly increases depth and reproducibility of results. Mol. Cell. Proteomics. 2017;16:2296–2309. doi: 10.1074/mcp.RA117.000314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.MacLean B., Tomazela D.M., Shulman N., Chambers M., Finney G.L., Frewen B., Kern R., Tabb D.L., Liebler D.C., MacCoss M.J. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26:966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Raw data and complete MS datasets have been uploaded to the Center for Computational Mass Spectrometry, to the MassIVE repository at UCSD, and can be downloaded using the following link: http://massive.ucsd.edu/ProteoSAFe/status.jsp?task=a23d5ddef1b54e89b858221ce57863ab (MassIVE ID number: MSV000089318; ProteomeXchange ID: PXD033458).
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.