Abstract
Retroviral integration, like all forms of DNA transposition, proceeds through a series of DNA cutting and joining reactions. During transposition, the 3′ ends of linear transposon or donor DNA are joined to the 5′ phosphates of a double-stranded cut in target DNA. Single-end transposition must be avoided in vivo because such aberrant DNA products would be unstable and the transposon would therefore risk being lost from the cell. To avoid suicidal single-end integration, transposons link the activity of their transposase protein to the combined functionalities of both donor DNA ends. Although previous work suggested that this critical coupling between transposase activity and DNA ends occurred before the initial hydrolysis step of retroviral integration, work in the related Tn10 and V(D)J recombination systems had shown that end coupling regulated transposase activity after the initial hydrolysis step of DNA transposition. Here, we show that integrase efficiently hydrolyzed just the wild-type end of two different single-end mutants of human immunodeficiency virus type 1 in vivo, which, in contrast to previous results, proves that two functional DNA ends are not required to activate integrase's initial hydrolysis activity. Furthermore, despite containing bound protein at their processed DNA ends, these mutant viruses did not efficiently integrate their singly cleaved wild-type end into target DNA in vitro. By comparing our results to those of related DNA recombination systems, we propose the universal model that end coupling regulates transposase activity after the first chemical step of DNA transposition.
Transposition is a specialized type of DNA recombination that results in the movement of transposon or donor DNA from a preexisting genomic location to a new DNA target site. Transposition can be nonreplicative, wherein the transposon leaves its old location for the new site, or replicative, when a copy of the donor DNA is retained at the original genomic position. Examples of nonreplicative and replicative prokaryotic transposons are Tn10 and bacteriophage Mu, respectively. V(D)J recombination and retroviral integration represent nonreplicative and replicative eukaryotic transposition systems, respectively. Retroviruses in this sense are packaged RNA intermediates of replicative DNA transposition.
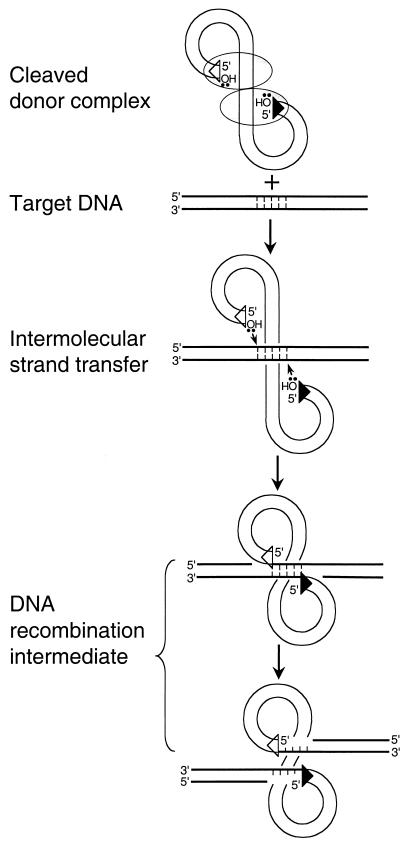
Certain mechanistic features group these seemingly disparate elements into the same class of DNA recombination. Most notably, transposition proceeds through a common DNA recombination intermediate. This intermediate is formed by transposase-mediated intermolecular strand transfer of donor DNA 3′ ends to the 5′-phosphates of a double-stranded staggered cut in target DNA (Fig. 1). In the intermediate, the 5′ ends of donor DNA remain unjoined to the target. Subsequent DNA repair or replication enzymes fill in and ligate the single-strand gaps, yielding the sequence duplication of the target DNA cut flanking the newly integrated element.
FIG. 1.
Formation of the transpositional DNA recombination intermediate. Transposase protein and linear donor DNA interact in a higher-order nucleoprotein complex called the cleaved donor complex or type 1 transpososome (reviewed in reference 8). Although a dimer of transposase is shown engaging both DNA ends, the precise stoichiometry of transposase to donor DNA is undefined for a number of transposition systems. The 3′-oxygens at the ends of donor DNA are the nucleophiles that cut target DNA (bold lines) during intermolecular strand transfer. Although a 5-bp target DNA cut is shown, the size of this cut varies among transposons. The triangles at the ends of donor DNA represent sequences important for transposase binding and activity. For simplicity, transposase was omitted from the later reaction steps.
Transposing through a higher-order nucleoprotein complex with a multimer of transposase engaging both donor DNA ends as illustrated in Fig. 1 helps ensure the proper formation of the recombination intermediate. A precise spacing separates the two phosphodiesters that are cut during intermolecular strand transfer. Thus, the higher-order nature of the cleaved donor complex helps position donor 3′-OHs for nucleophilic attack. End synapsis also helps to orchestrate transposase activity such that the frequency of unwanted strand transfer events, for example, the integration of just one end of donor DNA into target, is reduced. Integrating just one donor end would be nonproductive because the resulting branched Y-mer recombination intermediate would likely resolve into its starting donor and target DNA components during DNA repair. Since productive transposition requires the integration of both donor DNA ends into both strands of target DNA, the hypothesis is that transposase activity is at some level coupled to the functionalities of both donor DNA ends. This is referred to here as “end coupling.”
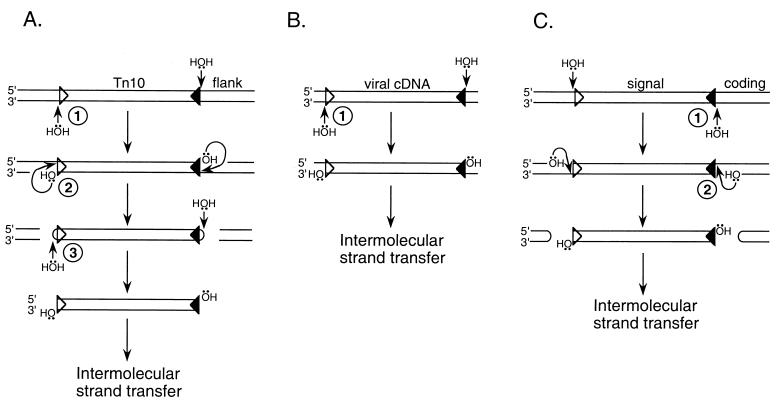
Although intermolecular strand transfer yields a common recombination intermediate, different transposons use different reaction pathways to generate their precursor 3′ ends. This is due to different transposon lifestyles. For example, Tn10 is a nonreplicative transposon that breaks free from its preexisting genomic location prior to intermolecular strand transfer (21). To accomplish double-strand breaks, Tn10 transposase catalyzes two different chemical reactions, first hydrolysis and then interstrand DNA strand transfer (Fig. 2A). Transposase then catalyzes a third step, hydrolysis, that both resolves donor DNA hairpins and generates 3′-OHs for intermolecular strand transfer (Fig. 2A) (24). In contrast, retroviral cDNA is synthesized by reverse transcription as a linear blunt-ended molecule. Thus, retroviruses do not break preexisting bonds in both DNA strands, and because of this retroviral integrase catalyzes just a single hydrolysis step, referred to as 3′ processing, to generate the cleaved donor complex (Fig. 2B). V(D)J recombination represents a third transpositional lifestyle. Similar to Tn10 transposase, RAG1/2 recombinase yields double-strand breaks via an initial hydrolysis and then interstrand transfer (Fig. 2C). Due to the polarity of the initial cut, however, the excised donor DNA is competent for intermolecular strand transfer without an additional hydrolysis step; the flanking DNA contains the hairpinned ends in this case (Fig. 2C). Clearly, different transposons use quite different strategies to generate their 3′-OHs for intermolecular strand transfer (Fig. 2).
FIG. 2.
DNA transposition systems. (A) Tn10. Tn10 transposase catalyzes three distinct chemical reactions to form the precursor 3′-OHs for intermolecular strand transfer: step 1, site-specific hydrolysis of transposon 3′ ends; step 2, interstrand transfer; and step 3, hairpin resolution (24). (B) Retroviral DNA integration. In an initial processing step (step 1), integrase hydrolyzes a specific site near each 3′ end of donor DNA. The resulting processed cDNA is the substrate for intermolecular strand transfer (18). (C) V(D)J recombination. Following the initial hydrolysis step (step 1), RAG1/2 recombinase catalyzes interstrand transfer, forming hairpinned coding ends (step 2). Although the physiological role of V(D)J recombination is the eventual joining of these coding DNA ends, the excised signal DNA can undergo intermolecular strand transfer in vitro (1, 22). For simplicity, donor DNAs are drawn as linear molecules. Whereas the triangles in panel A represent the outside ends of IS10 (20, 23), those in panels B and C represent viral DNA attachment (att) sites (5) and RSSs (17), respectively.
The point at which end coupling regulates transposase activity has been analyzed in different transposition systems. The cis-acting elements important for V(D)J recombination, called recombination signal sequences (RSSs) (Fig. 2C), are comprised of conserved heptamer and nonamer sequences separated by a variable 12- or 23-bp spacer (reviewed in reference 17). Since recombination normally occurs between 12-bp-containing and 23-bp-containing RSSs, end coupling in V(D)J recombination is also known as the 12/23 rule (17). Whereas DNA substrates containing a pair of nonfunctional 12-by-12 or 23-by-23 RSSs supported RAG1/2 hydrolysis at each RSS, only a physiologically relevant 12-by-23 substrate supported efficient interstrand transfer at both RSSs (40). Similarly, Tn10 transposase asymmetrically cleaved just the wild-type end of transposition-defective mutants carrying a mutation at one end of donor DNA (20). Thus, for both Tn10 and V(D)J, transposase apparently sensed the combined functionalities of both DNA ends after the initial hydrolysis step. In contrast, it was reported previously that integrase was unable to cleave either end of Moloney murine leukemia virus (MoMLV) carrying a mutation at just one DNA end (30). Based on this, it was proposed that integrase had to interact with two functional DNA ends before it could be activated to cleave either end in vivo (30). Consistent with this interpretation, neither the wild-type nor mutant viral end supported functional protein binding as detected by Mu-mediated PCR (MM-PCR) footprinting (39).
Overall, it appeared that end coupling functioned after the initial hydrolysis step of Tn10 transposition and V(D)J recombination but prior to the first chemical step of retroviral integration. We hypothesized that this might be the case for one of two different reasons. Since retroviral integrase catalyzes fewer overall steps than does either Tn10 transposase or RAG1/2 recombinase (Fig. 2), coupling integrase activity to the functionalities of both ends prior to hydrolysis might be necessary in this case to suppress unwanted strand transfer events. On the other hand, the retroviral model was based on a limited number of donor end mutants, highlighted by a single replication-defective virus (30, 39), and analyses of large sets of Tn10 (23) and IS903 (12) mutants identified different classes based on the severity of the transpositional defect. Because of this, we decided to analyze a larger set of mutants to determine if end coupling indeed functioned prior to the 3′-processing step of retroviral integration. Since we recently described 24 different donor end mutants of human immunodeficiency virus type 1 (HIV-1) that displayed a variety of replication phenotypes (4), we analyzed a subset of these for levels of (i) integrase 3′-processing activity at each donor DNA end, (ii) protein binding to each DNA end, and (iii) intermolecular strand transfer activity. Our results show that integrase can efficiently process just one end of HIV-1 in vivo and that the resulting viral preintegration complexes (PICs), despite containing bound protein at the processed DNA end, do not efficiently integrate this singly cleaved end in vitro. Thus, we conclude that, as in related DNA recombination systems, end coupling does not regulate retroviral integrase activity until after the first chemical step of DNA transposition.
MATERIALS AND METHODS
Plasmids and viruses.
Plasmids carrying the wild-type NL4-3 strain of HIV-1, as well as donor DNA and integrase mutant derivatives, were previously described (4, 15). Viruses were prepared by transfecting 293T cells in the presence of calcium phosphate as previously described (11). For cotransfection, the ratio of two different plasmids was varied while keeping the total amount of DNA constant.
Preparation of HIV-1 PICs.
PICs were prepared from acutely infected CD4-positive T-lymphoid cells as previously described (11). In brief, C8166 cells (1.2 × 108) infected for 7 h with 80 ml of DNase I-treated virus were lysed in 4 ml of buffer K (20 mM HEPES [pH 7.6], 150 mM KCl, 5 mM MgCl2, 1 mM dithiothreitol, 20 μg of aprotinin/ml) containing 0.025% (wt/vol) digitonin. The resulting cytoplasmic extract was treated with RNase A (0.1 mg/ml) for 30 min at room temperature.
Analysis of 3′-processing activity.
To analyze levels of integrase 3′-processing activity, HIV-1 cDNA purified from cytoplasmic extract (1 ml) by deproteinization was reacted with 20 U each of HaeIII and HindIII as previously described (10). The digested DNA was fractionated through DNA sequencing gels as previously described (10). After electrophoresis, DNA was transferred to a Duralon-UV membrane (Stratagene, La Jolla, Calif.) using a Genie electrophoretic blotter (Idea Scientific, Minneapolis, Minn.) as described previously (10).
The structures of the U3 and U5 ends of donor DNA were analyzed by indirect end labeling using strand-specific riboprobes (10, 28, 32). For detecting the 3′ end of minus-strand cDNA, a U3-specific riboprobe was generated using T7 RNA polymerase and BamHI-digested pMM104 plasmid DNA (28) as previously described (10). Whereas the 3′ end of the U5 plus strand was detected using a riboprobe generated from HindIII-cut pMM105 (28) using T7 RNA polymerase, the 5′ end of the nonprocessed U3 plus strand was detected using RNA generated from EcoRV-digested pMM104 and T3 RNA polymerase (10). Membranes were hybridized, washed, and processed for autoradiography as previously described (10). 3′-processing activity, expressed as the percentages of the full-length U3 minus strand and U5 plus strand converted into their −2 reaction products, was quantified by densitometry (IS-1000 Digital Imaging System; Alpha Innotech Corp., San Leandro, Calif.).
In vitro integration assays.
To analyze DNA strand transfer activity, cytoplasmic extract (0.25 ml) was reacted with linearized φX174 target DNA (750 ng) at 37°C for 45 min. Following deproteinization and ethanol precipitation, the DNA was fractionated through agarose and transferred to a GeneScreen Plus membrane (NEN Life Science Products Inc., Boston, Mass.) as previously described (11). HIV-1 cDNA was detected using a U3-specific riboprobe as previously described (4, 9–11). Integration activity was quantified by either a PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.) or a densitometer. Wild-type activity was calculated as the percentage of the 9.7-kb cDNA substrate converted into the linear 15.1-kb integration product, and mutant activity was expressed as the percentage of wild-type activity. Integration activities represent averages of at least three independent infections.
MM-PCR footprinting and Western blotting.
To detect protein-DNA complex formation at the ends of donor DNA, cytoplasmic extract (1.8 ml) was purified through 10-ml, 10 to 50% (wt/vol) Nycodenz gradients as previously described (11). Following centrifugation and fractionation, HIV-1 PICs (0.25 ml) were reacted with an equal volume of preassembled Mu transpososomes as previously described (11). Following deproteinization and ethanol precipitation, the reaction products were subjected to two rounds of PCR as described previously (11). For detecting the U3 end of donor DNA, HIV-1-specific primers AE330 (11) and AE347 (4) were used in the first and second PCRs, respectively, as previously described (10, 11). HIV-1 primers AE529 (11) and AE609 (4) were used in the first and second PCR rounds, respectively, to analyze the U5 end. PCR products fractionated through DNA sequencing gels were detected by autoradiography as described previously (11).
Gradient-purified HIV-1 PICs were subjected to Western blotting using an anti-integrase monoclonal antibody as previously described (10, 11).
RESULTS
Experimental strategy.
To analyze the relationship between end coupling and retroviral integrase activity, we required a recombination system that supported efficient integration of both ends of donor DNA during intermolecular strand transfer. After reverse transcription, integrase and retroviral cDNA form part of a large nucleoprotein PIC, and PICs isolated from infected cells can integrate their endogenous cDNA into an added target DNA in vitro (6, 16, 18). Integrase's 3′-processing and DNA strand transfer activities, however, can be analyzed using a variety of experimental setups (reviewed in reference 5). For example, recombinant integrase protein purified following its expression in bacteria can process and integrate synthetic oligonucleotide substrates that model the ends of retroviral cDNA. A limitation of such simplified integration assays, however, is that purified integrase preferentially integrates just a single end of donor DNA. Although the frequency of authentic two-ended HIV-1 integration can be increased by changing the source of the integrase protein, the structure of the DNA substrate, and/or reaction conditions (7, 19), these optimized systems fall short of recapitulating the frequency with which PICs promote the coupled integration of both ends of endogenous cDNA (5). Because of this, we studied PICs isolated from HIV-1-infected cells.
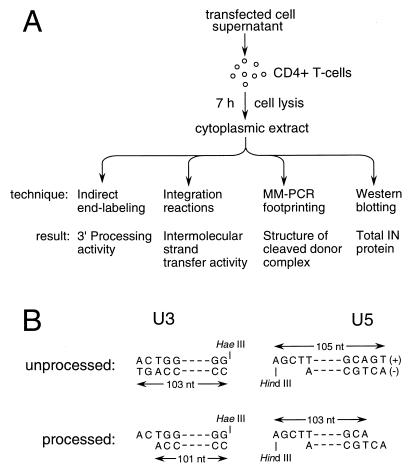
PICs prepared from cytoplasmic extracts were treated in a variety of ways to determine levels of integrase catalytic activities, as well as protein-DNA complex formation at each cDNA end (Fig. 3A). To measure levels of integrase 3′ processing at the U3 and U5 ends of donor DNA, deproteinized PICs were analyzed by indirect end labeling. Native PICs were reacted with φX174 target DNA in in vitro integration reactions to measure levels of intermolecular strand transfer activity (Fig. 3A). Since PICs were either deproteinized or reacted with target DNA immediately after their preparation, in vivo levels of 3′-processing activity could be compared to in vitro levels of DNA strand transfer activity. To detect protein-DNA interactions, PICs purified by Nycodenz gradient centrifugation were analyzed by MM-PCR footprinting. A portion of the purified PICs were analyzed for total integrase protein by Western blotting (Fig. 3A).
FIG. 3.
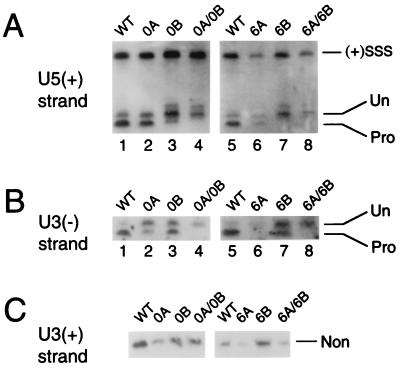
Experimental strategy. (A) Infection and downstream manipulations. CD4-positive T cells were lysed 7 h postinfection. Cytoplasmic extracts, which contain PICs in their native form, were processed using the indicated techniques to measure levels of integrase catalytic activities and PIC-associated protein. (B) Strategy for determining 3′ processing of the U3 and U5 ends of HIV-1 DNA. The sequences of the unprocessed and processed plus- and minus-strand termini are shown. Deproteinized PICs were digested with HaeIII and HindIII, which cut HIV-1 approximately 100 bp from the U3 and U5 ends, respectively. Membranes were probed with strand-specific riboprobes following denaturing polyacrylamide gel electrophoresis and electroblotting. Whereas the unprocessed and processed U3 minus strand is 103 and 101 bases, respectively, the unprocessed and processed U5 plus strand is 105 and 103 bases, respectively. nt, nucleotides.
We used two different experimental approaches, each of which was previously used in related transposition systems, to address the role of end coupling during HIV-1 integration. Whereas one approach analyzed the kinetics of 3′ processing at each end of wild-type DNA (2, 13, 20), the other relied on the analysis of mutants carrying changes in the end regions of donor DNA known to be important for transposition (20, 30, 36). To begin with, we analyzed the kinetics of donor end processing.
Asymmetric processing of HIV-1 cDNA in vivo.
During 3′ processing, integrase removes two nucleotides from the U3 minus and U5 plus strands of blunt-ended HIV-1 (Fig. 3B), and as mentioned above, indirect end labeling (18, 32, 33) was used to detect the loss of these nucleotides from each 3′ end. For this, HIV-1 cDNA was cleaved with HaeIII and HindIII (10, 28, 32). Considering the U3 end, HaeIII cleavage yields a 103-base minus-strand terminus (Fig. 3B). If integrase removed two nucleotides, however, this HaeIII product would be 101 bases (Fig. 3B). U3 donor end processing was quantified as the fraction of the 101-base product over the total U3 minus strand that was synthesized by reverse transcription. The level of U5 end processing was similarly calculated (Fig. 3B).
Tissue culture infections were initiated by mixing cell-free viral supernatant with susceptible CD4-positive T cells (Fig. 3A). Under these conditions, HIV-1 cDNA synthesis peaked 7 to 8 h postinfection (11; data not shown). DNA was prepared from cells 3, 4, 5, 6, 7, and 8 h postinfection. About 50% of each end of donor DNA was cleaved at the early time point, and the extent of 3′ processing at each end increased roughly in parallel until maximum cleavage, which ranged from 70 to 90%, was reached 7 to 8 h postinfection (data not shown). Thus, we conclude that both HIV-1 ends were synchronously processed by integrase under these assay conditions. The U3 and U5 ends of HIV-1 were also similarly processed at various times postinfection using a different infection technique that mixed virus producer cells with uninfected target cells (28). Since these kinetic approaches revealed that the ends of wild-type HIV-1 were synchronously processed by integrase, we next turned our attention to the analysis of donor end mutant viruses.
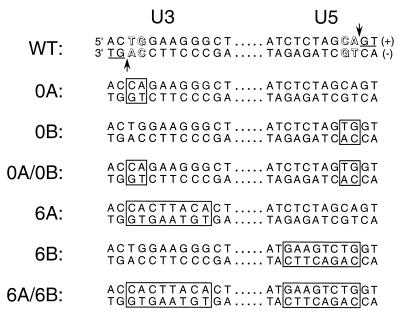
Previous genetic analyses revealed that only relatively small regions of HIV-1 DNA, consisting of about 10 bp at the U3 and U5 ends of unintegrated cDNA, are important for integration in vivo (4, 27). Furthermore, out of these regions, the TG/AC base pairs near the very ends of DNA play the most critical roles (Fig. 4). Integrase processes each end of donor DNA adjacent to the conserved adenine of this motif (Fig. 3B and 4) and then uses the exposed adenine 3′-OHs to cut target DNA during intermolecular strand transfer. Because the TG/AC base pairs play critical roles in integrase binding and catalysis, we analyzed two sets of viruses mutated at these sites.
FIG. 4.
Donor DNA mutant viruses. The sequences of the wild-type (WT) U3 and U5 DNA attachment (att) sites are shown. These sequences are important for integrase catalytic activity and HIV-1 replication (4, 5, 27). Highlighted within the att sites are the TG/AC base pairs that are conserved among all retroviruses (outlined), the phosphodiester bonds in the U3 minus and U5 plus strands that are cleaved by integrase during 3′ processing (vertical arrows), and the GT dinucleotides that are removed as a result of 3′ processing (underlined). The bases that differed from wild type are boxed in the indicated mutant viruses.
Each set consisted of a U3 mutant, a U5 mutant, and a double U3-U5 mutant. Mutant 0A carried the 2-bp substitution of CA/GT for TG/AC at the U3 end of donor DNA, 0B contained the analogous U5 substitution, and 0A-0B combined both of these changes (Fig. 4). The second set of mutants was analogous to the first, with the addition that the mutations extended inward to include a total of 8 bp at each DNA end (Fig. 4). We previously showed that, whereas 0A and 0B displayed slight growth delays in vivo compared to wild-type HIV-1, 6A and 6B grew about 8 days and 2 weeks delayed, respectively. In contrast, both double-end mutant viruses were completely replication defective (4).
Levels of integrase 3′-processing activity at each HIV-1 end were determined at the peak of wild-type cDNA synthesis (7 h postinfection). As mentioned above, integrase efficiently processed each wild-type end under these conditions: in repeated experiments, levels of U3 and U5 processing ranged from 72 to 90% and 77 to 90%, respectively (Fig. 5A and B, lanes 1 and 5; Table 1). In contrast, neither end of replication-defective 0A and 0B supported a detectable level of 3′ processing (Fig. 5A and B, lanes 4; Table 1). On the other hand, both ends of 0A and 0B were cleaved, but in each case integrase processed the intact DNA end more efficiently than the mutant end. Whereas processing of the intact U5 end in 0A was reduced about 1.4-fold below that of the wild type, processing of the mutant U3 end was down 2- to 2.5-fold in repeated experiments (Fig. 5A and B, lanes 2; Table 1). For 0B, processing of the mutant U5 end was down three- to fourfold below that of the wild type, but cutting at the intact U3 end was reduced only about twofold (Fig. 5A and B, lanes 3; Table 1).
FIG. 5.
3′ Processing of wild-type (WT) and mutant HIV-1. (A) U5 end processing of wild type and the indicated mutant viruses. (+)SSS, DNA cleavage product of the plus-strand strong-stop intermediate of reverse transcription; Un, the unprocessed 105-base strand; Pro, the processed 103-base product (Fig. 3B). (B) U3 end processing. Un, the unprocessed 103-base strand; Pro, the processed 101-base product (Fig. 3B). (C) The nonprocessed (Non) 103-base U3 plus strand (Fig. 3B). The size of each DNA strand in the panels was confirmed by comparing migration distances to those of an M13 DNA sequencing ladder (10, 33).
TABLE 1.
3′ processing of wild-type and mutant donor DNA endsa
| Virusb | Expt 1
|
Expt 2
|
||
|---|---|---|---|---|
| U3 | U5 | U3 | U5 | |
| Wild type | 86 (100) | 77 (100) | 90 (100) | 90 (100) |
| 0A (U3) | 34 (40) | 62 (81) | 44 (49) | 66 (73) |
| 0B (U5) | 51 (59) | 18 (23) | 49 (54) | 30 (33) |
| 0A-0B | NDc | ND | ND | ND |
| Wild type | 89 (100) | 78 (100) | 72 (100) | 78 (100) |
| 6A (U3) | NDd | 42 (54) | NDd | 44 (56) |
| 6B (U5) | 48 (54) | ND | 41 (57) | ND |
| 6A-6B | ND | ND | ND | ND |
Activities for two independent experiments are shown. Numbers in parentheses are percentages of wild-type activity in each experiment.
The mutated donor DNA end is indicated in parentheses. Wild type is listed twice because the 0A-0B and 6A-6B virus sets were analyzed in separate experiments.
ND, not detected (<3% activity).
The level of detection for 6A was <9%.
Asymmetric processing by integrase was more dramatic with the 6A-6B set of viruses. As was the case for replication-defective 0A-0B, neither the U3 nor the U5 end of 6A-6B supported a detectable level of 3′ processing (Fig. 5A and B, lanes 8; Table 1). The mutant U5 ends of 6B and 6A-6B were indistinguishable: in both cases, levels of integrase 3′-processing activity were undetectable (Fig. 5A, lanes 7 and 8). However, integrase's ability to cleave the intact U3 end of mutant 6B was reduced only about twofold below that of the wild type (Fig. 5B, lanes 5 and 7; Table 1). This pattern of asymmetric processing was also observed with 6A. That is, whereas processing of the mutant U3 end was undetectable, the intact U5 end was processed about half as efficiently as was wild type (Fig. 5A and B, lanes 6; Table 1). In this case, however, our ability to detect 3′ processing of the mutant DNA end was reduced about threefold below that of 6B (Table 1), probably because 6A yields about half as much cDNA as does wild type in infected cells (4) (Fig. 5A, lanes 5 and 6). To further probe the structure of the mutant end in 6A, nonprocessed U3 plus strands (Fig. 3B) were analyzed by indirect end labeling (Fig. 5C). The results of this experiment revealed (i) that 6A-6B also yielded less cDNA than did wild type (Fig. 5A, lanes 5 and 8; Fig. 5C) and (ii) that, more importantly, the mutant U3 ends of 6A and 6A-6B were full length and thus were not subject to degradation by cellular enzymes (Fig. 5B, lanes 6 and 8; Fig. 5C).
Intermolecular strand transfer activities of wild-type and mutant PICs.
Having determined that each single-end mutant supported some level of integrase 3′-processing activity, we next assayed the intermolecular strand transfer activity of the different viruses. For this, wild-type and mutant PICs were reacted with linearized φX174 target DNA in in vitro integration reactions (Fig. 3A). Since endogenous HIV-1 cDNA is 9.7 kb and φX174 is 5.4 kb, intermolecular strand transfer of both donor DNA ends yields a linear 15.1-kb integration product (Fig. 6) containing single-strand gaps at the donor-target DNA junctions (Fig. 1) (18).
FIG. 6.
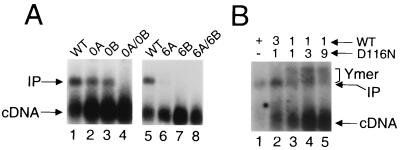
Intermolecular strand transfer activities of wild-type (WT) and mutant PICs. (A) Integration reactions contained PICs that were derived from either wild type (lanes 1 and 5) or the indicated mutant virus. cDNA, 9.7-kb HIV-1 DNA substrate; IP, 15.1-kb linear integration product. (B) Reaction mixtures contained either wild type PICs (lane 1) or those derived from viruses using the indicated molar ratio of wild type and D116N mutant plasmid DNA. Ymer, integration products that migrated more slowly than the standard linear 15.1-kb product. Other labeling is the same as for panel A.
Wild-type HIV-1 integrated between 30 and 60% of its cDNA substrate in repeated experiments (Fig. 6A, lanes 1 and 5; Fig. 6B, lane 1). As expected from the results in Fig. 5, neither 0A-0B nor 6A-6B supported a detectable level (<2% of wild type) of DNA strand transfer activity (Fig. 6A, lanes 4 and 8). Whereas mutant 0A yielded about 12% as much integration product as did wild type (lane 2), 0B supported about 18% activity (lane 3) in repeated experiments. Mutant 6B did not display a detectable level of intermolecular strand transfer activity (lane 7), but 6A was about 4% as active as was wild type (lane 6).
Since integrase efficiently processed the wild-type U3 end of 6B without detectably cleaving the mutant U5 end (Fig. 5), we reasoned that 6B PICs might integrate just their singly cleaved U3 end into φX174 target DNA in vitro. Overexposing the autoradiogram in Fig. 6A, however, did not reveal evidence for any novel DNA recombination product. What might we expect for the product of single-end strand transfer? Similar to the 15.1-kb linear product of normal two-ended integration, integration of just one end of HIV-1 into φX174 would also yield a 15.1-kb product. In this case, however, the product would be a population of branched Y-mers. Although both the linear and Y-mer products have the same mass, Y-mers should migrate more slowly in agarose due to their branched structures. Although we did not detect any novel DNA recombination products, we nonetheless attempted to construct a control for single-end integration. For this, we modeled a mixing experiment after results from the Mu transposition field.
Purified retroviral integrase protein preferentially integrates just a single end of recombinant viral DNA in in vitro integration reactions. In contrast, a tetramer of MuA transposase efficiently integrates two synapsed transposon ends in in vitro transposition reactions (25, 29). Single-end integration of Mu DNA, however, can be enhanced by using a mixture of wild-type and transposition-defective mutant proteins that carry amino acid substitutions in the active site of MuA (3). Whereas the level of single-end transposition product relative to that of the normal double-end product was increased by increasing the level of active-site mutant protein in the reaction mixture, mixtures of two different active-site mutants were inactive (3). Based on these results, we constructed a series of phenotypically mixed HIV-1 PICs by varying the ratio of two different plasmids in cotransfections. Whereas one plasmid encoded wild type, the other expressed an integration-defective mutant carrying the Asp116→Asn substitution in the active site of HIV-1 integrase (14, 15). Unlike MuA, the number of integrase protomers in the active-integrase multimer is unknown. However, HIV-1 contains on the order of 50 to 100 integrase molecules per virion (5), and since integrase is cleaved from a Gag-Pol polyprotein precursor during virus assembly and the D116N active-site mutant does not suffer any adverse defects in the HIV-1 life cycle prior to the integration step (11, 15), we reasoned that wild-type and D116N integrase protomers would randomly assort during virus assembly and the subsequent steps of reverse transcription and PIC formation in infected cells.
As predicted from previous analyses of Mu transposition, PICs containing a phenotypic mixture of wild-type and D116N integrase yielded two DNA recombination products, the normal linear product and a novel population of products displaying a lower electrophoretic mobility (Fig. 6B). We note that the formation of both products was dependent on (i) the presence of target DNA in the integration reactions (data not shown) and (ii) both the wild-type and active-site mutant proteins, as PICs containing either D116N alone (11) or a 1:1 mixture of two different integrase active-site mutants did not form a detectable level of either DNA product (data not shown). Also as predicted from the Mu field, the relative level of the novel integration product increased as the proportion of D116N integrase was increased in the phenotypic mixture (Fig. 6B, lanes 2 to 4). At the 9:1 ratio of D116N to wild type, the novel recombination product predominated, although the overall level of integration activity was reduced (Fig. 6B, lane 5). Based on the parallel behaviors of phenotypically mixed MuA tetramers and HIV-1 PICs in in vitro integration reactions, we tentatively identify the more slowly migrating DNA as products of single-end integration. We therefore speculate that the absence of strand transfer with mutant 6B was not due to our inability to detect the integration of single HIV-1 ends.
Single-end mutants display asymmetric protein-DNA structures.
Despite 6A and 6B processing nearly 50% of their wild-type end, the results of the previous section indicated that neither 6A nor 6B could efficiently integrate its singly cleaved end in in vitro integration reactions. To further investigate this apparent block to intermolecular strand transfer, we next analyzed the protein-DNA structure at each end of HIV-1 donor DNA using MM-PCR footprinting.
MM-PCR is an in vitro footprinting technique that uses the intermolecular strand transfer activity of MuA transposase to cleave DNA and probe the structure of protein-DNA complexes (38). Proteins important for PIC function protect several hundred base pairs near each end of retroviral cDNA from Mu transposition (11, 38). In addition to these footprinted regions, sequences close to each cDNA end form hot spots for Mu transposition (4, 11, 38). This bipartite nucleoprotein structure comprised of footprinted and transpositional enhanced regions at each cDNA end defines the retroviral intasome. Various analyses of wild-type and mutant MoMLV and HIV-1 PICs have established a central role for the intasome nucleoprotein complex in the integration of endogenous retroviral cDNA (9–11, 38, 39).
As with all DNA footprinting techniques, the frequency and distribution of Mu transposition into native PICs (Fig. 7, even-numbered lanes) were compared to those of deproteinized controls (odd-numbered lanes). Wild type displayed the expected patterns of transposition protection and enhancement at each cDNA end (Fig. 7, compare lanes 2 to lanes 1). In contrast, each mutant PIC for the most part lacked strong footprinted regions (Fig. 7). Because of this, we focused our comparison of wild-type and mutant protein-DNA structures on the transpositional enhanced regions. Wild-type PICs that display relatively high levels of intermolecular strand transfer activity in in vitro integration reactions display more intensely footprinted regions than do less active PICs (11; H. Chen and A. Engelman, unpublished observations). We therefore speculate that the virtual absence of footprinted regions from the different mutant PICs reflected their relatively weak or undetectable levels of DNA strand transfer activity (Fig. 6A).
FIG. 7.
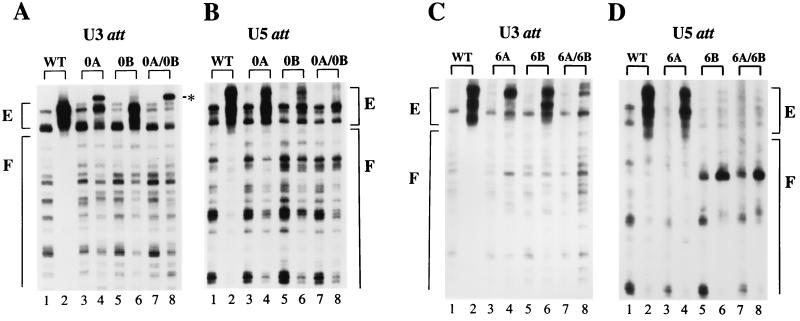
Wild-type (WT) and mutant protein-DNA complex formation. (A and C) The U3 ends of the indicated viruses. Odd-numbered lanes contained deproteinized cDNA; even-numbered lanes contain native PICs. E, regions of transpositional enhancements; F, wild-type footprinted regions; ∗, the novel Mu insertion at the U3 ends of native 0A and 0A-0B. (B and D) U5 end structures. Other labeling is the same as for panels A and C. Although the wild-type U3 and U5 footprints extend for about 250 bp (11), only about 80 bp of each DNA end is shown. Due to the polarity of Mu transposition, only the termini of the nonprocessed U3 plus and U5 minus strands can be visualized by MM-PCR (11, 38). att, attachment.
Consistent with the lack of detectable 3′-processing and DNA strand transfer activities, neither the U3 nor the U5 end of 0A-0B supported the wild-type pattern of transpositional enhancements (Fig. 7A and B, compare lanes 7 and 8 to lanes 1 and 2). A novel warm spot for Mu insertion, however, was detected near the very end of U3 (Fig. 7A, lane 8, indicated by ∗). Since wild-type PICs did not support Mu transposition at this position (Fig. 7A, lane 2) and the U3 end of 0A-0B was not detectably cleaved by integrase (Fig. 5B, lane 4), we interpret this band as indicative of defective end structure. The mutant end of 0A also showed this novel insertion, along with some evidence for the normal transpositional enhancements (Fig. 7A, lanes 3 and 4). The wild-type enhancement pattern, however, was more evident at the intact U5 end of 0A (Fig. 7B, lanes 3 and 4). In 0B, the wild-type pattern of transpositional enhancements was more evident at the intact U3 end than it was at the mutant U5 end (Fig. 7A and B, lanes 5 and 6).
Similar to the results of 3′ processing, asymmetric protein-DNA complexes were more evident with the 6A-6B set of viruses. Like 0A-0B, neither end of 6A-6B supported the wild-type pattern of transpositional enhancements (Fig. 7C and D, lanes 7 and 8). Transpositional enhanced regions were more evident at the intact U5 end of 6A than at the mutant U3 end (Fig. 7C and D, lanes 3 and 4). More dramatically, the wild-type pattern of enhancements was detected at the intact U3 end of 6B but was absent from the defective U5 end (Fig. 7C and D, lanes 5 and 6). Gradient-purified wild-type and mutant PICs contained levels of integrase protein that were indistinguishable by Western blotting (data not shown), suggesting that the diminished transpositional enhancements observed at certain mutant cDNA ends were not due to gross perturbations in the overall level of PIC-associated integrase protein.
DISCUSSION
To succeed in life, transposons must integrate both of their 3′ ends into both strands of a DNA target. Single-end integration must be suppressed in vivo because DNA repair would tend to detach the transposon from the target, which would likely result in the loss of the transposon from the cell. A convenient way to avoid this predicament is to link transposase activity to the combined functionalities of both donor DNA ends. That is, if a situation arose wherein both DNA ends could not be integrated, then neither end would get integrated. Although previous work suggested that this type of end coupling functioned early on in the retroviral integration pathway, the results presented here show that end coupling does not regulate integrase activity until relatively late in HIV-1 integration, after the chemical step of 3′ processing.
End coupling and HIV-1 integrase activity.
Previous work with MoMLV suggested that a pair of functional ends had to interact in vivo before integrase could be activated to cleave either end (30, 39). In that case, a mutation at just one end of donor DNA blocked processing of both the wild-type and mutant DNA ends. In contrast, our results with HIV-1 mutants 6A and 6B show that a wild-type end can be efficiently cleaved by integrase under conditions where processing of the other mutant end is undetectable (Fig. 5 and Table 1). Thus, we conclude that two functional DNA ends are not required to activate HIV-1 integrase's 3′-processing activity in vivo.
Although the reason(s) for the different results is unclear, we speculate that it lies with the mutations that were analyzed in the two studies. Previous work with prokaryotic transposons provided the basis for this interpretation. DNA end mutants of Tn10 and IS903 can be grouped into different categories based on the severity of the transpositional defect (12, 23). Approximately 16 bp of donor DNA forms the primary cis-acting determinants for Tn10 and IS903 transposition (12, 23). Whereas single-end mutations located roughly 6 to 13 bp from donor termini inhibited Tn10 and IS903 transposition as much as 500- and 5 × 108-fold, respectively, changes within the terminal 3 bp inhibited recombination at most 11-fold (12, 23). Based on these results, it was proposed that the more deleterious changes identified sequences important for transposase binding to DNA and that the less deleterious changes disrupted transposition after binding and transposase-mediated synapsis of donor DNA ends (12, 20, 23). Indeed, less defective single-end mutants of Tn10 display asymmetric processing of the intact end of donor DNA (20). Based on this, we propose that the previously analyzed MoMLV U3 mutant is analogous to the more defective class of Tn10 and IS903 mutants and that it is likely that this single-end mutation prohibited an essential interaction between integrase and DNA that precluded end synapsis of the U3 and U5 ends of MoMLV. This interpretation is consistent with the result that neither the mutant nor the wild-type end supported a detectable level of protein binding (39). In contrast, the 6A and 6B mutations studied here inhibited HIV-1 integration after an initial integrase-DNA binding step. Integration in these cases was blocked between the chemical steps of 3′ processing (Fig. 5) and DNA strand transfer (Fig. 6). Since neither 6A nor 6B appeared to integrate its singly cleaved wild-type end in vitro, we conclude that integrase's DNA strand transfer activity as assayed in the context of HIV-1 PICs is dependent upon functional end coupling. The requirement for end coupling before or at the DNA strand transfer step ensures proper integration of both viral DNA ends in vivo.
End synapsis and the cleaved donor intasome nucleoprotein complex.
Recombinant HIV-1 integrase preferentially integrates just a single end of donor DNA during intermolecular strand transfer. Whereas oligonucleotide U3 or U5 substrates containing substitutions of the conserved TG/AC did not support detectable levels of 3′-processing activity (26, 35, 37), cleavage of the analogous mutant ends in 0A and 0B was reduced only 2.5- and 4-fold, respectively, compared to that for wild type (Table 1). Thus, the intact wild-type ends of 0A and 0B largely overrode the negative impact of these mutations on integrase's 3′-processing activity. Based on this, we speculate that 3′ processing of 0A and 0B occurred in the context of a higher-order nucleoprotein complex that minimally contained integrase, U3, and U5 and by extension that the U3 and U5 ends of HIV-1 are normally synapsed prior to 3′ processing in infected cells. This interpretation is consistent with our finding that the U3 and U5 ends of wild-type HIV-1 were synchronously processed by integrase in vivo. Since the mutant U5 end of 6B did not support a detectable level of integrase processing (Fig. 5A) or Mu transpositional enhancements (Fig. 7D), it is unclear whether processing of the wild-type U3 end occurred in the context of a higher-order integrase-DNA complex or whether asymmetric processing of this virus occurred in the absence of end synapsis. Since the initial hydrolysis step of V(D)J recombination can occur in the absence of end synapsis (42), it seems plausible that U3 end processing in mutant 6B may have occurred without synapsis.
Although 6A and 6B were highly defective, we note that they each supported HIV-1 replication in vivo (4). In each case, we speculate that virus replication required functional restoration of the mutant DNA end. For example, the virus that grew out of 6B-infected cells reverted the G/C base pair at the mutant U5 terminus to the wild-type A/T (4) (Fig. 4). Thus, it seems likely that the original mutant ends of 6A and 6B were processed at some level by integrase, but in each case this level was below the detection limit of our indirect end-labeling assay (Table 1). We also note that 6A PICs supported about 4% of the wild-type level of intermolecular strand transfer activity (Fig. 6A). Although the level of protein binding to the mutant U3 end of 6A was reduced in comparison to that for the analogous wild-type end, MM-PCR footprinting revealed that this mutant end nonetheless supported the wild-type pattern of Mu transpositional enhancements (Fig. 7C). Based on these observations, we conclude that only those PICs that supported Mu enhancements at both ends of donor DNA were competent for intermolecular strand transfer (Fig. 6 and 7). We therefore propose the following refined definition for the retroviral intasome as the PIC-associated nucleoprotein complex that supports Mu transpositional enhancements at both ends of donor DNA in MM-PCR footprinting assays and catalyzes two-ended intermolecular strand transfer activity in in vitro integration assays. Using this operational definition, the retroviral intasome is analogous to the type 1 transpososome or cleaved donor nucleoprotein complex of Mu transposition (8) (Fig. 1).
Similarity to related DNA recombination systems.
Results of previous studies showed that end coupling regulated Tn10 transposase (20) and RAG1/2 recombinase (40) activity after the initial hydrolysis step of DNA transposition. Since we found that HIV-1 mutants 6A and 6B were apparently blocked in integration between the chemical steps of 3′ processing and DNA strand transfer (Fig. 5 and 6), it is tempting to speculate that end coupling functions to regulate the DNA strand transfer activity of a variety of transposase and integrase proteins. One apparent exception to this model comes from the Mu field. Mu transposition is analogous to retroviral integration in that MuA transposase catalyzes two chemical steps, an initial hydrolysis and then intermolecular strand transfer of processed 3′ ends, to form the transpositional DNA recombination intermediate (8). It was previously shown that a single base change at one terminus of Mu donor DNA blocked hydrolysis of both Mu ends in vitro (36). By analogy to other prokaryotic transposons, single-end mutations in this region of Mu DNA would not be expected to inhibit hydrolysis of both ends (12, 20, 23). Including the MuA accessory protein MuB along with target DNA in in vitro transposition reactions, however, overcame the block and permitted Mu hydrolysis at both donor DNA ends (36). Since MuB and target DNA promote the formation of the precleaved stable synaptic complex or type 0 Mu transpososome (8, 29), these results are consistent with the notion that the single base pair mutation in Mu DNA inhibited end synapsis and transpososome assembly. Thus, in the absence of MuB and target, this single-end Mu mutant behaved like the highly defective class of Tn10, IS903, and MoMLV single-end mutants.
The efficiency with which MuA promotes coupled integration in vitro has led to precise localization of the key players in transposition. That is, the MuA monomer bound at one Mu DNA terminus catalyzes hydrolysis and intermolecular strand transfer of the other Mu end, and vice versa (31, 34, 41). This arrangement helps ensure the integration of both ends of donor DNA during transposition. Although the presence of two functional donor DNA ends is not essential to activate the initial hydrolysis activity of either Tn10 transposase (20), MuA transposase (36), RAG1/2 recombinase (40), or HIV-1 integrase (Fig. 5 and Table 1), a pair of properly cleaved ends was required for efficient strand transfer of Mu DNA in vitro (41). Thus, a pair of functional donor DNA ends is apparently required to activate efficient strand transfer activity of MuA transposase, RAG1/2 recombinase (40), and HIV-1 integrase (Fig. 6). Future experiments are planned to further dissect the functionality of the HIV-1 intasome following its isolation from infected cells. The results of these experiments not only are expected to add to our understanding of DNA recombination in general but also should aid the development of antiviral drugs targeted against integration, an essential step in the HIV-1 life cycle.
ACKNOWLEDGMENTS
We thank M. Mizuuchi for purified MuA protein and A. Limón, R. Lu, and Å. Öhagen for critical review of the manuscript.
This work was supported by NIH grant AI39394, by the G. Harold and Lelia Y. Mathers Foundation, and by the Friends of Dana-Farber Cancer Institute.
REFERENCES
- 1.Agrawal A, Eastman Q M, Schatz D G. Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature. 1998;394:744–751. doi: 10.1038/29457. [DOI] [PubMed] [Google Scholar]
- 2.Bainton R, Gamas P, Craig N L. Tn7 transposition in vitro proceeds through an excised transposon intermediate generated by staggered breaks in DNA. Cell. 1991;65:805–816. doi: 10.1016/0092-8674(91)90388-f. [DOI] [PubMed] [Google Scholar]
- 3.Baker T A, Kremenstova E, Luo L. Complete transposition requires four active monomers in the Mu transposase tetramer. Genes Dev. 1994;8:2416–2428. doi: 10.1101/gad.8.20.2416. [DOI] [PubMed] [Google Scholar]
- 4.Brown H E V, Chen H, Engelman A. Structure-based mutagenesis of the human immunodeficiency virus DNA attachment site: effects on integration and cDNA synthesis. J Virol. 1999;73:9011–9020. doi: 10.1128/jvi.73.11.9011-9020.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown P O. Integration. In: Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 161–203. [PubMed] [Google Scholar]
- 6.Brown P O, Bowerman B, Varmus H E, Bishop J M. Correct integration of retroviral DNA in vitro. Cell. 1987;49:347–356. doi: 10.1016/0092-8674(87)90287-x. [DOI] [PubMed] [Google Scholar]
- 7.Carteau S, Batson S C, Poljak L, Mouscadet J-F, Rocquigny H, Darlix J-L, Roques B P, Kas E, Auclair C. Human immunodeficiency virus type 1 nucleocapsid protein specifically stimulates Mg2+-dependent DNA integration in vitro. J Virol. 1997;71:6225–6229. doi: 10.1128/jvi.71.8.6225-6229.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chaconas G, Lavoie B D, Watson M A. DNA transposition: jumping gene machine, some assembly required. Curr Biol. 1996;6:817–820. doi: 10.1016/s0960-9822(02)00603-6. [DOI] [PubMed] [Google Scholar]
- 9.Chen H, Engelman A. The barrier-to-autointegration protein is a host factor for HIV type 1 integration. Proc Natl Acad Sci USA. 1998;95:15270–15274. doi: 10.1073/pnas.95.26.15270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen H, Engelman A. Characterization of a replication-defective human immunodeficiency virus type 1 att site mutant that is blocked after the 3′ processing step of retroviral integration. J Virol. 2000;74:8188–8193. doi: 10.1128/jvi.74.17.8188-8193.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen H, Wei S-Q, Engelman A. Multiple integrase functions are required to form the native structure of the human immunodeficiency virus type 1 intasome. J Biol Chem. 1999;274:17358–17364. doi: 10.1074/jbc.274.24.17358. [DOI] [PubMed] [Google Scholar]
- 12.Derbyshire K M, Hwang L, Grindley N D F. Genetic analysis of the interaction of the insertion sequence IS903 transposase with its terminal inverted repeats. Proc Natl Acad Sci USA. 1987;84:8049–8053. doi: 10.1073/pnas.84.22.8049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eastman Q M, Schatz D G. Nicking is asynchronous and stimulated by synapsis in 12/23 rule-regulated V(D)J cleavage. Nucleic Acids Res. 1997;25:4370–4378. doi: 10.1093/nar/25.21.4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Engelman A, Craigie R. Identification of conserved amino acid residues critical for human immunodeficiency virus type 1 integrase function in vitro. J Virol. 1992;66:6361–6369. doi: 10.1128/jvi.66.11.6361-6369.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Engelman A, Englund G, Orenstein J M, Martin M A, Craigie R. Multiple effects of mutations in human immunodeficiency virus type 1 integrase on viral replication. J Virol. 1995;69:2729–2736. doi: 10.1128/jvi.69.5.2729-2736.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Farnet C M, Haseltine W A. Integration of human immunodeficiency virus type 1 DNA in vitro. Proc Natl Acad Sci USA. 1990;87:4164–4168. doi: 10.1073/pnas.87.11.4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fugmann S D, Lee A I, Shockett P E, Villey I J, Schatz D G. The RAG proteins and V(D)J recombination: complexes, ends, and transposition. Annu Rev Immunol. 2000;18:495–527. doi: 10.1146/annurev.immunol.18.1.495. [DOI] [PubMed] [Google Scholar]
- 18.Fujiwara T, Mizuuchi K. Retroviral DNA integration: structure of an integration intermediate. Cell. 1988;54:497–504. doi: 10.1016/0092-8674(88)90071-2. [DOI] [PubMed] [Google Scholar]
- 19.Goodzari G, Im G-J, Brackmann K, Grandgenett D P. Concerted integration of retrovirus-like DNA by human immunodeficiency virus type 1 integrase. J Virol. 1995;69:6090–6097. doi: 10.1128/jvi.69.10.6090-6097.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haniford D, Kleckner N. Tn10 transposition in vivo: temporal separation of cleavages at the two transposon ends and roles of terminal basepairs subsequent to interaction of ends. EMBO J. 1994;13:3401–3411. doi: 10.1002/j.1460-2075.1994.tb06643.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haniford D B, Benjamin H W, Kleckner N. Kinetic and structural analysis of a cleaved donor intermediate and a strand transfer intermediate in Tn10 transposition. Cell. 1991;11:171–179. doi: 10.1016/0092-8674(91)90218-n. [DOI] [PubMed] [Google Scholar]
- 22.Hiom K, Melek M, Gellert M. DNA transposition by the RAG1 and RAG2 proteins: a possible source of oncogenic translocations. Cell. 1998;94:463–470. doi: 10.1016/s0092-8674(00)81587-1. [DOI] [PubMed] [Google Scholar]
- 23.Huisman O, Errada P R, Signon L, Kleckner N. Mutational analysis of IS10's outside end. EMBO J. 1989;8:2101–2109. doi: 10.1002/j.1460-2075.1989.tb03619.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kennedy A K, Guhathakurta A, Kleckner N, Haniford D B. Tn10 transposition via a DNA hairpin intermediate. Cell. 1998;95:125–134. doi: 10.1016/s0092-8674(00)81788-2. [DOI] [PubMed] [Google Scholar]
- 25.Lavoie B D, Chan B S, Allison R G, Chaconas G. Structural aspects of a higher order nucleoprotein complex: induction of an altered DNA structure at the Mu-host junction of the type 1 transpososome. EMBO J. 1991;10:3051–3059. doi: 10.1002/j.1460-2075.1991.tb07856.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leavitt A D, Rose R B, Varmus H E. Both substrate and target oligonucleotide sequences affect in vitro integration mediated by the human immunodeficiency virus type 1 integrase protein produced in Saccharomyces cerevisiae. J Virol. 1992;66:2359–2368. doi: 10.1128/jvi.66.4.2359-2368.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Masuda T, Kuroda M J, Harada S. Specific and independent recognition of U3 and U5 att sites by human immunodeficiency virus type 1 integrase in vivo. J Virol. 1998;72:8396–8402. doi: 10.1128/jvi.72.10.8396-8402.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller M D, Farnet C M, Bushman F D. Human immunodeficiency type 1 preintegration complexes: studies of organization and function. J Virol. 1997;71:5382–5390. doi: 10.1128/jvi.71.7.5382-5390.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mizuuchi M, Baker T A, Mizuuchi K. Assembly of the active form of the transposase-Mu DNA complex: a critical control point in Mu transposition. Cell. 1992;70:303–311. doi: 10.1016/0092-8674(92)90104-k. [DOI] [PubMed] [Google Scholar]
- 30.Murphy J E, Goff S P. A mutation at one end of Moloney murine leukemia virus DNA blocks cleavage at both ends by the viral integrase in vivo. J Virol. 1992;66:5092–5095. doi: 10.1128/jvi.66.8.5092-5095.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Namgoong S-Y, Harshey R M. The same two monomers within a MuA tetramer provide the DDE domains for the strand cleavage and strand transfer steps of transposition. EMBO J. 1998;17:3775–3785. doi: 10.1093/emboj/17.13.3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pauza C D. Two bases are deleted from the termini of HIV-1 linear DNA during integrative recombination. Virology. 1990;179:886–889. doi: 10.1016/0042-6822(90)90161-j. [DOI] [PubMed] [Google Scholar]
- 33.Roth M J, Schwartzberg P L, Goff S P. Structure of the termini of DNA intermediates in the integration of retroviral DNA: dependence on IN function and terminal DNA sequence. Cell. 1989;58:47–54. doi: 10.1016/0092-8674(89)90401-7. [DOI] [PubMed] [Google Scholar]
- 34.Savilhati H, Mizuuchi K. Mu transpositional recombination: donor DNA cleavage and strand transfer in trans by the Mu transposase. Cell. 1996;85:271–280. doi: 10.1016/s0092-8674(00)81103-4. [DOI] [PubMed] [Google Scholar]
- 35.Sherman P A, Dickson M L, Fyfe J A. Human immunodeficiency virus type 1 integration protein: DNA sequence requirements for cleavage and joining reaction. J Virol. 1992;66:3593–3601. doi: 10.1128/jvi.66.6.3593-3601.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Surette M G, Harkness T, Chaconas G. Stimulation of the Mu A protein-mediated strand cleavage reaction by the Mu B protein, and the requirement of DNA nicking for stable type 1 transpososome formation. J Biol Chem. 1991;266:3118–3124. [PubMed] [Google Scholar]
- 37.Vink C, van Gent D C, Elgersma Y, Plasterk R H A. Human immunodeficiency virus integrase protein requires a subterminal position of its viral DNA recognition sequence for efficient cleavage. J Virol. 1991;65:4636–4644. doi: 10.1128/jvi.65.9.4636-4644.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wei S-Q, Mizuuchi K, Craigie R. A large nucleoprotein assembly at the ends of the viral DNA mediates retroviral DNA integration. EMBO J. 1997;16:7511–7520. doi: 10.1093/emboj/16.24.7511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wei S-Q, Mizuuchi K, Craigie R. Footprints on the viral DNA ends in Moloney murine leukemia virus preintegration complexes reflect a specific association with integrase. Proc Natl Acad Sci USA. 1998;95:10535–10540. doi: 10.1073/pnas.95.18.10535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.West R B, Lieber M R. The RAG-HMG1 complex enforces the 12/23 rule of V(D)J recombination specifically at the double-hairpin formation step. Mol Cell Biol. 1998;18:6408–6415. doi: 10.1128/mcb.18.11.6408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Williams T L, Jackson E L, Carritte A, Baker T A. Organization and dynamics of the Mu transpososome: recombination by communication between two active sites. Genes Dev. 1999;13:2725–2737. doi: 10.1101/gad.13.20.2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu K, Lieber M R. The nicking step in V(D)J recombination is independent of synapsis: implications for the immune repertoire. Mol Cell Biol. 2000;20:7914–7921. doi: 10.1128/mcb.20.21.7914-7921.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]