Abstract
Resistance to radiation therapy (RT) remains an obstacle in HPV-negative head and neck squamous cell carcinomas (HNSCCs)—even with a combined RT-immunotherapy approach. Jak-Stat proteins have long been studied for both their immune regulatory role in the host immune response as well as their cancer cell signaling role in shaping the tumor microenvironment (TME). Here, we identify STAT1 as a mediator of radioresistance in HPV-negative preclinical mouse models of HNSCC, by which knockout of STAT1 in the cancer cell (STAT1 KO)—but not in the host—resulted in decreased tumor growth alongside increased immune activation. We show that RT increases STAT1/pSTAT1 expression, which may act as a marker of radioresistance. Whereas RT increased JAK-STAT and interferon (IFN) signaling, transcriptomic analysis revealed that STAT1 KO in the cancer cell resulted in decreased expression of IFN-associated genes of resistance. In vitro experiments showed that STAT1 KO increased T cell chemoattraction and decreased baseline growth. These results indicate that STAT1 may serve a tumor-promoting role in the cancer cell and will inform biomarker development and treatment regimens for HNSCC incorporating RT.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00262-021-03059-3.
Keywords: HNSCC, STAT1, Radiation therapy, Interferon signaling, Tumor microenvironment, Cancer immunology
Introduction
The expression and activation of various signal transducer and activator of transcription (STAT) proteins are crucial in shaping the tumor microenvironment (TME) [1]. STAT proteins respond to engagement of cytokine receptors in the TME, and upon phosphorylation from receptor-associated Janus kinases (JAKs) translocate to the nucleus to promote specific genes [1] resulting in an array of pro-tumor or anti-tumor autocrine and paracrine signals [2]. While there are many redundancies in their transcriptional targets, STAT proteins are still known to have distinct roles [3]. While STAT3 has been implicated in immunosuppression and cancer cell survival, STAT1 has been typically reported to drive anti-tumor responses [2]. However, STAT1 has also been shown to act as a tumor promoter, both in its ability to promote cancer cell growth as well as influence immunosuppressive immune populations [4]. As JAK-STAT signaling is involved in nearly all immune regulation, [2] understanding how STAT1 may be acting in cancer growth and progression is crucial given its controversial role.
Here, we investigate the role of STAT1 signaling in the setting of head and neck squamous cell carcinoma (HNSCC), a cancer that bears an immunologically cold TME, [5] responding poorly even to radiation therapy (RT) and immunotherapies in attempts to spark an inflammatory response and attract tumor-infiltrating lymphocytes [6, 7]. STAT1 has been widely studied in context of the host immune response in relation to invigorating the CD8 T cell response, [1, 8] but the expression of STAT1 specifically in the cancer cell does not necessarily yield the same anti-tumor effect [4]. In HNSCC cell lines, we observed that RT increased the phosphorylation of STAT1 and increased CXCL9 and CXCL10 secretion. We further observed that irradiated cell lines induce T cell proliferation and activation. We, therefore, hypothesized that activation of STAT1 on the cancer cells induces an inflammatory response leading to T cell recruitment, activation, and proliferation. Contrary to our hypothesis, we show instead that knockout of STAT1 on the cancer cell reduces tumor growth and is associated with increased activation of T effector populations when combined with RT in two mouse models of HNSCC. Additionally, the absence of cancer cell STAT1 signaling resulted in a reduced transcriptional signature of interferon-stimulated genes associated with resistance to radiation therapy. These data reveal a distinct role for STAT1 in HNSCC tumor cells, indicating that the pro- or anti-tumor effect of STAT1 signaling depends on compartment and context in the TME.
Results
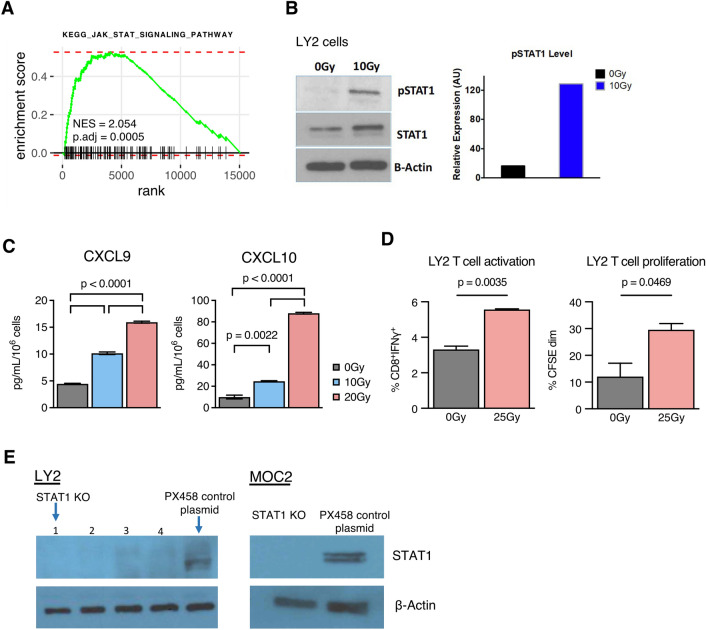
RT induces STAT1 phosphorylation, CXCL9/10 release by cancer cells, and activation and proliferation of effector T cells
We have previously demonstrated that RT synergizes with anti-PD-L1 to reduce tumor growth in HNSCC tumor models [9]. To understand how radiation’s cytotoxic effect on the cancer cell induces an immune response within the TME, we performed RNA-Seq on LY2 HNSCC tumors with and without RT. We observed significant induction of KEGG JAK-STAT signaling pathway [10] (Fig. 1a), which ranked highly among several other significantly enriched KEGG pathways (Fig. S1). From previously reported [11] analysis of this dataset of Hallmark pathways, [12] we also observed significant increases in STAT family-related and inflammation-related pathways, including Hallmark IFNγ response and Hallmark IFNα response pathways, which both involve STAT proteins [2].
Fig. 1.
RT induces STAT1 phosphorylation, CXCL9/10 release by cancer cells, and activation and proliferation of effector T cells a Enrichment curve for KEGG_JAK_STAT_SIGNALING_PATHWAY from RNA-Seq expression data in LY2 tumors harvested 3 days post-RT (10 Gy) compared to 0 Gy controls. b Western blot quantification of STAT1, pSTAT1, and β-Actin in LY2 cancer cell line 24 h post-RT (10 Gy) versus 0 Gy controls. c CXCL9 and CXCL10 concentration in conditioned media harvested from irradiated LY2 cells 3 days post-RT (10 Gy) versus 0 Gy controls. d Characterization of CD8 T cells co-cultured with irradiated LY2 cells in the presence of IL-2: proportion of CD8 T cells expressing IFNγ and proportion showing dim CFSE signal. Comparisons shown for LY2 cells irradiated with 25 Gy dose prior to co-culture versus 0 Gy controls. e Western blot quantifications of STAT1 KO cell line clones and corresponding PX458 control cell lines
Type I and II interferons, including IFNα and IFNγ, have been established to signal through STAT1 [3]. Given the increase in transcriptomic pathways concerning JAK-STAT and these interferons, we measured expression of STAT1 from LY2 cells following RT. Western blot analysis showed increases in both total and phosphorylated STAT1 after exposure to RT (Fig. 1b). Evaluating conditioned media from LY2 cells, we observed significant increases in CXCL9 and CXCL10 in response to increasing doses of RT (Fig. 1c). Analysis of mRNA expression in HNSCC tumors from The Cancer Genome Atlas (TCGA) showed that CXCL9 and CXCL10 correlate more significantly with STAT1 than with any other STAT family proteins (Fig. S2A–D). CXCL9 and CXCL10 are potent T cell chemoattractants that respond to IFNγ [13], which signals through STATs including STAT1 [2]. These data suggest an association between RT-induced STAT1 and cancer cell secretion of CXCL9/10.
To assess how these RT-induced changes in the cancer cell might influence an immune effect, we co-cultured irradiated LY2 cells with CD8 T cells and observed significant upregulation of IFNγ expression (Fig. 1d). We also observed a significant increase in CD8+ T cell proliferation as determined by CFSE staining (Fig. 1d). Together, these data indicated that RT induces STAT1 expression and phosphorylation and CXCL9/10 release, promoting the activation and proliferation of effector T cells. Given these findings, we used CRISPR-Cas9 to develop isogenic cancer cell lines from LY2 and MOC2 with STAT1 knockout (STAT1 KO) or PX458 control plasmid (Fig. 1e) [14].
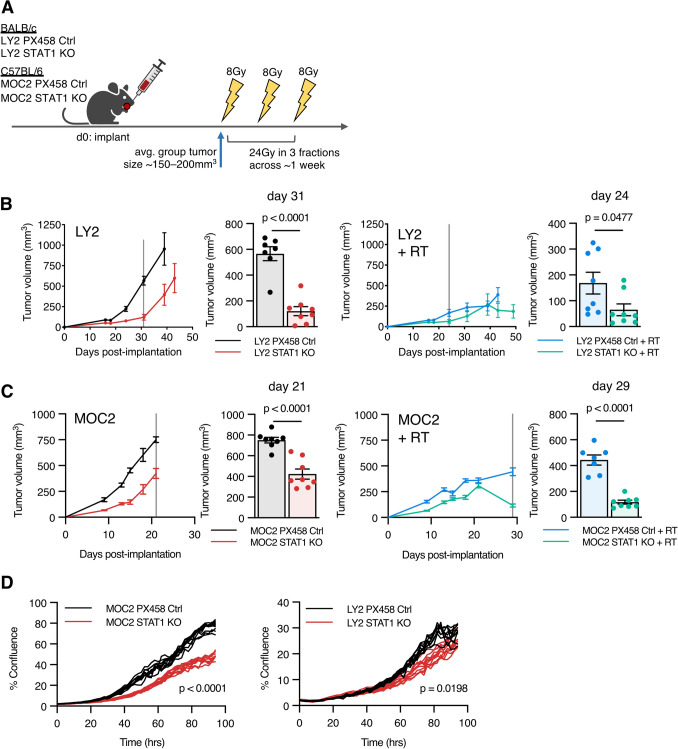
Loss of STAT1 in the cancer cell has an anti-tumoral and anti-proliferative effect
Given the observed activation of STAT1 in response to RT and its association with increased T cell chemoattractants CXCL9 and CXCL10, we sought to determine the functional implications of cancer cell STAT1 loss in vivo. The use of isogenic cell lines with STAT1 CRISPR knockout allowed us to explore the intrinsic effect of STAT1 signaling on the cancer cell, while leaving STAT1 intact in the host immune system. LY2 STAT1 KO or MOC2 STAT1 KO cell lines and appropriate controls were implanted into the buccal mucosa, and tumors were treated with a hypofractionated RT dose of 24 Gy in 3 fractions (Fig. 2a). We observed that loss of STAT1 unexpectedly resulted in reduced tumor growth, with or without RT in both tumor models (Fig. 2b, c). Using an in vitro cell growth assay, we found that loss of STAT1 resulted in significant cancer cell growth reduction (Fig. 2d).
Fig. 2.
Loss of STAT1 in the cancer cell has an anti-tumoral and anti-proliferative effect a Experimental design for murine orthotopic tumor models treated with hypofractioned RT. LY2 and MOC2 tumors were established in BALB/c and C57BL/6 mice, respectively. Administration of RT (24 Gy in 3 fractions) over approximately 1 week began when tumors from respective groups were approximately 150–200 mm3, given on days 8, 12, and 15 for MOC2 PX458, days 15, 18, and 21 for MOC2 STAT1 KO, days 24, 28, and 32 for LY2 PX458, and days 32, 36, and 40 for LY2 STAT1 KO. b Tumor growth curves for LY2 STAT1 KO versus LY2 PX458 Ctrl with or without RT; replicate values shown as bar chart for all mice alive on day indicated by vertical line; unpaired two-tailed t test. c Tumor growth curves for MOC2 STAT1 KO versus MOC2 PX458 Ctrl with or without RT; replicate values shown as bar chart for all mice alive on day indicated by vertical line; unpaired two-tailed t test. d Percent confluence for indicated cancer cell lines; unpaired two-tailed t test calculated at final time point
Reduced cancer cell growth in vitro suggests that a cancer-cell-intrinsic mechanism contributes toward reduced tumor growth in vivo, but this hypothesis still allows for the contribution of effect from an altered host immune response. This notion is supported by the observation that LY2 STAT1 KO tumors still have significantly reduced tumor growth, even with a smaller in vitro growth reduction. Thus, we hypothesized that presence of STAT1 in the cancer cell may contribute toward tumor growth and impart resistance to RT through an immune-mediated mechanism.
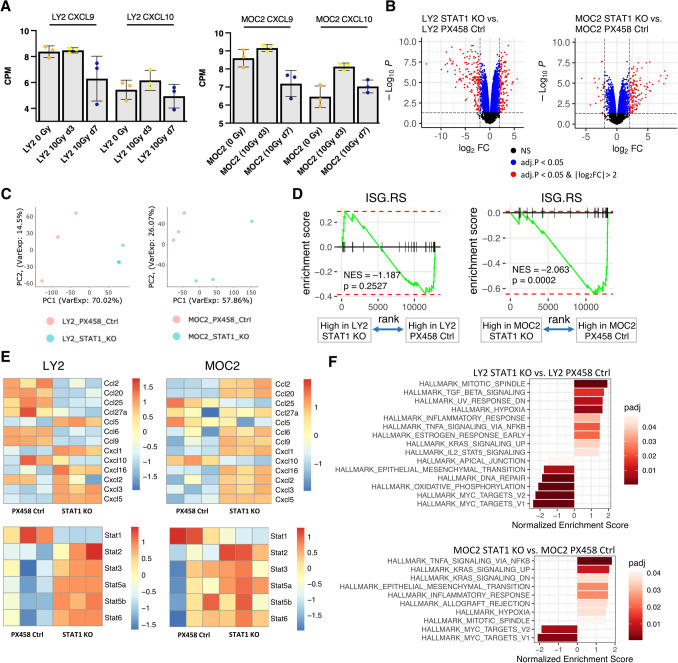
RT-induced CXCL9/10 are transiently expressed, and STAT1 KO alters the cancer-cell expressed chemokine profile
Reduced tumor growth with the loss of cancer cell STAT1 disputed our initial hypothesis of STAT1 being indicative of a robust immune response associated with T cell chemokines CXCL9 and CXCL10, so we analyzed these chemokines at a later time point (7 days post-RT). Transcriptomic analysis of tumors from LY2 and MOC2 tumor-bearing mice did not suggest that expression of CXCL9 or CXCL10 was any greater at later time point 7 days post-RT compared to 0 Gy controls (Fig. 3a), despite our initial in vitro experiments showing a significant increase 3 days post-RT (Fig. 1c). We hypothesized that cancer cell STAT1 had better prognostic value as a measure of resistance to RT, as supported by previous studies in applying RT to a human xenograft oral squamous cell carcinoma. [15, 16] To explore potential immune mechanisms by which cancer cell STAT1 enhances tumor cell growth, we analyzed STAT1 KO cell lines by RNA-Seq. Both cell lines with STAT1 KO showed significant differential expression compared to control cell lines (Fig. 3b). Dimension reduction using principal component analysis (PCA) showed distinct phenotypes in LY2 and MOC2 cell lines with STAT1 KO compared to control cell lines (Fig. 3c).
Fig. 3.
RT-induced CXCL9/10 are transiently expressed, and STAT1 KO alters the cancer-cell expressed chemokine profile a Transcriptome expression of CXCL9 and CXCL10 from tumors of LY2 and MOC2 tumor-bearing mice. b Volcano plots showing differential expression in STAT1 KO versus PX458 control cell lines. Positive log2 fold change indicates greater expression in STAT1 KO groups. Y-axis indicates the negative logarithm of BH-adjusted p-values. c Principal component analysis of STAT1 KO versus PX458 control cell lines. d Enrichment curves for interferon stimulated genes of resistance from cell line transcriptome expression, signature derived from Benci et al.[17] e Heatmaps for detected CC chemokines, CXC chemokines, CC ligands, and STAT family gene expression in STAT1 KO and PX458 control cell lines. f Waterfall plots depicting all significant Hallmark pathways for indicated comparisons for cell line transcriptome expression. Darker red colors indicate smaller p-adjusted value
Since STAT1 enables effects on gene expression downstream of type I and II interferons [2], we sought to determine how altered interferon signaling might be associated with the reduced tumor growth seen in the STAT1 KO tumors. Benci et al. demonstrated that STAT1 orchestrates a mechanism of resistance through alteration of cancer cell interferon signaling in two mouse models of melanoma, [17, 18] and this resistance gene signature (that they refer to as ISG.RS) was validated in a model to predict clinical outcomes in melanoma patients. [17] We applied this human signature to manually curated predicted murine orthologous equivalents (Fig. S3) to test the pathway in our knockout cell lines. Using gene set enrichment analysis, MOC2 STAT1 KO cell line showed a significant decrease in this resistance signature compared to PX458 control, with a similar trend for LY2 STAT1 KO (Fig. 3d).
Both cell lines showed, as expected, a significant reduction in STAT1 expression in the STAT1 KO CRISPR cell lines (Fig. 3e). CXCL9 expression was not detected, but its paralogous gene, CXCL10, showed decreased expression with the loss of STAT1 (Fig. 3e), consistent with the hypothesis that STAT1 and CXCL9/10 expression is highly correlated in these models (Fig. 1b, c, Fig. S2). LY2 STAT1 KO cell line, and to a lesser extent MOC2 STAT1 KO, showed compensatory increases in other STAT1 proteins (Fig. 3e).
Pathway analysis showed reductions in Hallmark MYC targets (Fig. 3f). Activation of the MYC pathway has been associated with tumor progression in multiple cancers including head and neck cancer [19]. Additionally, both STAT1 KO cell lines showed significantly increased Hallmark TNFα signaling via NFκB (Fig. 3f). In agreement with upregulation of this pathway, loss of STAT1 in both cell lines associated with significantly increased CXCL1, CXCL3, and CXCL5 expression, with CCL5 significantly increased in LY2 STAT1 KO and CXCL2 significantly increased in MOC2 KO. (Fig. 3e). CCL25 was significantly decreased in both cell lines with the loss of STAT1 (Fig. 3e), which in conjunction with its receptor CCR9 can suppress apoptosis and lead to cancer cell proliferation and metastasis [20]. Collectively, RNA-Seq analysis suggests that cancer cell STAT1 KO results in decreased IFN-associated resistance, increased TNFα signaling pathway, and increases in immune recruitment by CC and CXC chemokines.
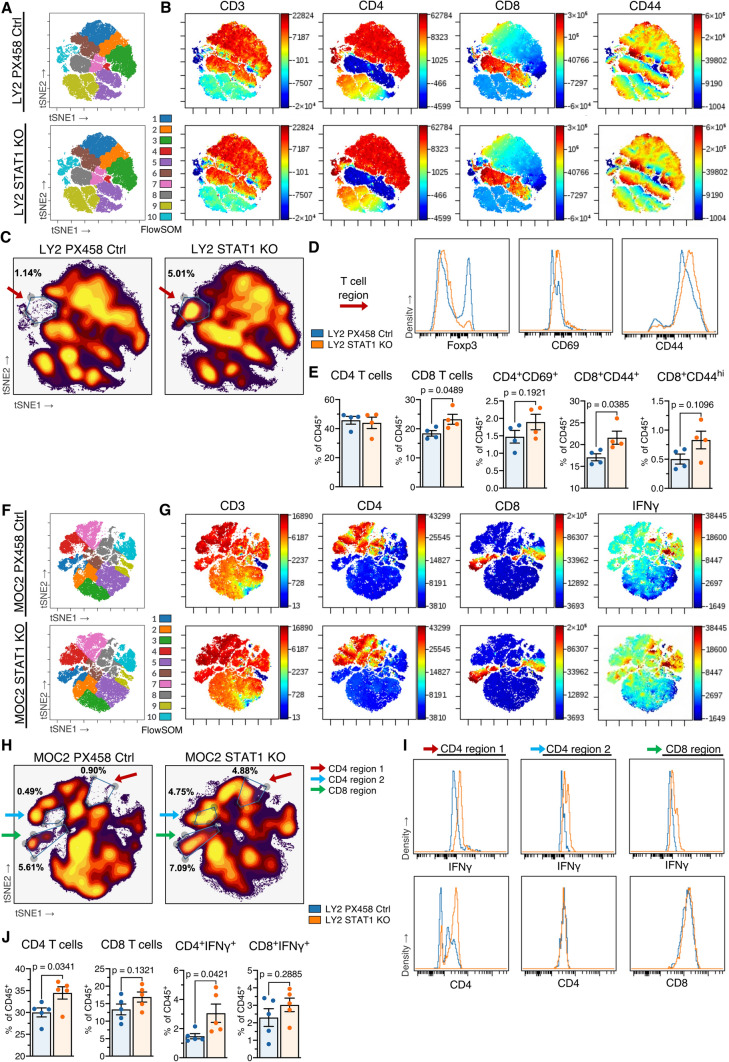
STAT1 KO tumor-bearing mice exhibit increased immune activation
We hypothesized that this altered JAK-STAT signaling and a reduced signature of interferon resistance genes would affect the communication between cancer cell and host, whereby loss of STAT1 on the cancer cell would result in an improved host immune response, considering the observed tumor growth reduction (Fig. 2b, c). Flow cytometric analysis followed by high-dimensional clustering was performed on CD45+ leukocytes in tumor-draining lymph nodes from LY2 and MOC2 tumor-bearing mice after the same 3 × 8 Gy hypofractionated RT regimen was administered. FlowSOM cluster 10 from the LY2 model (Fig. 4a) appeared to demonstrate a T cell phenotype based on expression of CD3, CD4, CD8, and CD44 (Fig. 4b). A region of these nodal T cells showed an increase in proportion in mice bearing LY2 STAT1 KO tumors compared to control tumors, encircled in a density plot (Fig. 4c). These nodal T cells showed higher expression of CD69 and CD44 in LY2 STAT1 KO tumor-bearing mice, suggesting a more activated T cell phenotype, and also showed lower expression of Foxp3, indicating decreased frequency of immunosuppressive Treg populations (Fig. 4d). Conventional gating techniques (Fig. S4A, B) confirmed that lymph nodes in the STAT1 KO group showed a significant increase in CD8 T cells and CD44-expressing CD8 T cells (Fig. 4e).
Fig. 4.
STAT1 KO tumor-bearing mice exhibit increased immune activation a tSNE plots for nodal CD45 + leukocytes from mice bearing LY2 STAT1 KO or LY2 PX458 control tumors, colored by FlowSOM predicted populations. b Expression across the nodal leukocyte landscape for CD3, CD4, CD8, and CD44. c Density plot of nodal leukocytes, with gate drawn around indicated T cell region and percent in the gate out of total CD45 + leukocytes. d Histograms depicting Foxp3, CD69, and CD44 expression of the T cell region. e Characterization of nodal CD4 and CD8 T cells as percent of CD45+ leukocytes. Unpaired two-tailed t test. f tSNE plots for nodal live CD45 + leukocytes from mice bearing MOC2 STAT1 KO or MOC2 PX458 control tumors, colored by FlowSOM predicted populations. g Expression across the nodal leukocyte landscape for CD3, CD4, CD8, and IFNγ. h Density plot of nodal leukocytes, with gates drawn around indicated CD4 and CD8 T cell regions and percent in each gate out of total live CD45 + leukocytes. i Histograms depicting IFNγ and CD4 expression of CD4 region 1, CD4 region 2, and CD8 region. j Characterization of nodal CD4 and CD8 T cells as percent of CD45+ leukocytes. Unpaired two-tailed t test
Mice bearing MOC2 tumors also showed an altered T cell phenotype in tumor-draining lymph nodes. FlowSOM clusters 1, 4, and 7 (Fig. 4f) showed increased frequency when STAT1 was knocked out in cancer cells, demonstrating T cell phenotypes marked by CD3, CD4, and CD8 expression (Fig. 4g). IFNγ expression was highly prevalent across many of these T cell regions for mice bearing MOC2 STAT1 KO tumors (Fig. 4g). Gating on regions of CD4 and CD8 T cells that were increased in the STAT1 KO model (Fig. 4h), we observe that these CD4+ and CD8+ cells show higher expression of IFNγ (Fig. 4i). Likewise, conventional gating techniques corroborate these results, showing significant increases in CD4 T cells and IFNγ-expressing CD4 T cells, with similar trends for CD8 T cells (Fig. 4J). Taken together, these results showed—in two HNSCC models—that STAT1 KO on the cancer cell results in expansion of activated CD4 and CD8 T cells.
STAT1 KO cell lines attract T cells and show no altered invasive properties
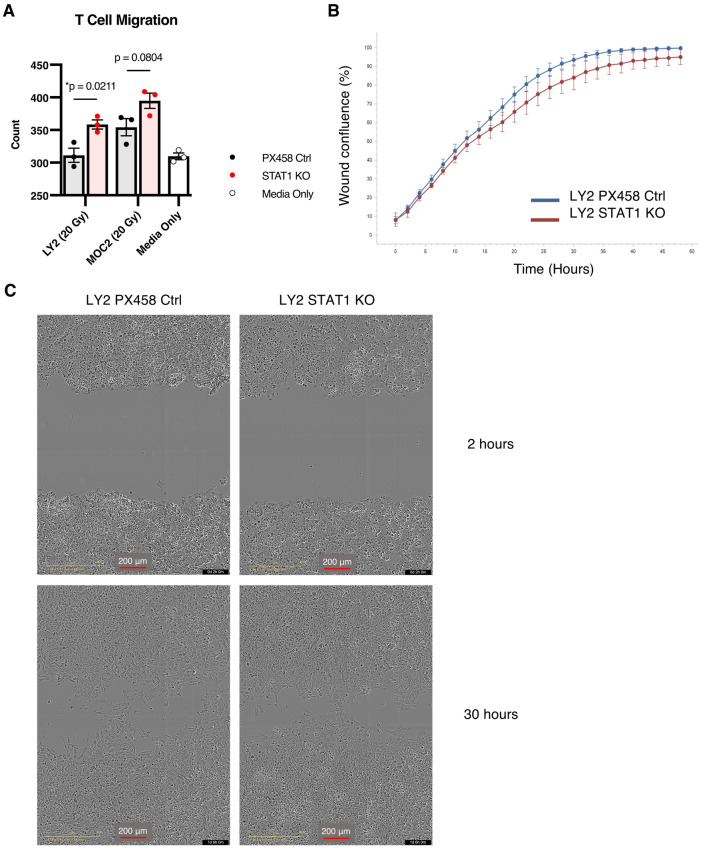
Given the improved T cell response observed in both tumor models in vivo, we hypothesized that STAT1 KO cancer cells may be promoting T cell activity via release of various chemoattractants. We tested this hypothesis in vitro using a Boyden chamber assay with conditioned media from the STAT1 KO cancer cells placed in the bottom chamber serving as a chemoattractant for T cells in the top chamber. RT was administered to cells prior to media collection to mimic treatment used in our in vivo models. After 4 h we visualized an increased number of T cells migrating out of the top chamber in STAT1 KO cell lines compared to PX458 control cell lines and control wells (Fig. 5a), indicating that a difference in conditioned media was responsible for increased T cell migration. These results suggest that the altered chemokine profile secreted from STAT1 KO cancer cells (Fig. 3e) is—at least in part—responsible for increased T cell attraction, and the assay design using conditioned media segregates this mechanism from other cancer-cell-intrinsic mechanisms that might be at play.
Fig. 5.
STAT1 KO cell lines attract T cells and show no altered invasive properties a Count of migrating T cells through boyden chamber after 4 h toward conditioned media harvested from STAT1 KO or PX458 control cell lines compared with normal media. Unpaired two-tailed t-test. b Incucyte results of scratch wound assay showing wound confluence over a 48 h period comparing invasion of LY2 STAT1 KO cell line versus LY2 PX458 control cell line. (n = 8 wells per group). c Representative images of LY2 STAT1 KO and LY2 PX458 control cell lines subjected to scratch wound assay at 2 h and 30 h time points
LY2 STAT1 KO cell line showed decreased Hallmark epithelial-mesenchymal transition pathway compared to controls (Fig. 3f), so we aimed to evaluate whether the loss of STAT1 would impact the cancer cell’s invasive abilities based on this cancer-cell-intrinsic property. Scratch wound assay to assess invasion, however, revealed no significant difference between STAT1 KO and control cell line (Fig. 5b, c).
Discussion
STAT1 plays a major role in the TME, impacting the transcription of thousands of genes through type I and II IFN signaling, [2] and Jak-Stat proteins as a whole sit downstream of over 50 cytokines [3]. While a number of cancer-cell-intrinsic anti-tumoral effects may occur downstream [1, 2], STAT1 may act as a tumor-promoter through its influence on the immune response, including upregulation of PD-L1 expression, promotion of immunosuppressive myeloid cells such as MDSCs or TAMs, and release of immunosuppressive cytokines [4]. In this study, we show that knockout of STAT1 in the cancer cell is sufficient to activate T cells in the tumor-draining lymph nodes, while maintaining function of STAT1 in the host.
IFNγ secretion has been shown to be a key driver of the PD-1/PD-L1 axis in HNSCC [21]. An IFNγ-related gene signature (including STAT1, CXCL9, CXCL10) predicts a T cell-inflamed TME responsive to PD-1 blockade [22], and patients with higher PD-L1 expression show better response to PD-1 blockade [23]. One might suggest, therefore, that STAT1 and IFN signaling provide a powerful stimulus to T cell-mediated immunity, while the simultaneous induction of PD-L1 in adaptive resistance can be effectively managed by targeting PD-1/PD-L1. However, upregulation of PD-L1 is only one of multiple mechanisms by which STAT1 drives therapeutic resistance, and most HNSCC patients are failing PD-1-targeted therapies in clinical trials [24]. Benci et al.[18] documents a mechanism by which interferon signaling in conjunction with STAT1 is responsible for immune suppression independent of PD-L1. They comment that interferon signaling and associated genes may correlate with favorable responses when conventional treatments are combined with agents targeting the PD-1/PD-L1 axis. This commentary provides powerful insight: STAT1 may appear protective in some analyses by virtue of its relationship with T cell infiltration or response to PD-1/PD-L1 blockade, but there remains additional immunosuppressive potential by other means that may contradict this protective role, instead providing evidence of STAT1 as a tumor-promoter in the cancer cell. This idea warrants careful interpretation of biomarker expression in tumor types that might vary in immune infiltration and composition.
In our initial hypothesis, we had observed that an increase in STAT1 expression was associated with response to RT, where STAT1/pSTAT1 initiate T cell chemotaxis and activation through CXCL9/CXCL10. However, these data do not exclude the possibility that STAT1 is also mediating negative feedback to provide resistance to this initial response. In one study, a human tumor xenograft selected for radioresistance by serial passage (nu61) demonstrated increased expression of IFN-related genes, of which STAT1α and STAT1β isoforms were the most highly expressed compared to its parental cell line [15]. This line was selected using 5 Gy in vivo irradiation [15], but our data suggest that STAT1 may contribute toward radioresistance even at higher doses of 8 Gy.
STAT1 may have differing prognostic values depending on the compartment. While STAT1 on the cancer cell has been previously demonstrated to contribute to the development of intrinsic radioresistance [15], how that affects the host tumor response and its contribution to TME radioresistance have been overlooked. Previously, STAT1 has been demonstrated to act as a tumor suppressor by promoting NK cell and T cell cytotoxicity [4]. This tumor-suppressing function may be more relevant in the host immune response rather than the cancer cell, as STAT1-deficient mice exhibit accelerated tumor growth in B4B8 and LY2 tumor models [25]. Furthermore, a study showed in mouse melanoma models that type I IFN signaling in the host is crucial for T cell priming by DCs, and knockout of either the type I IFN receptor IFNAR1 or STAT1 in the host is sufficient to prevent this recognition of the tumor [8]. Our data show that knockout of STAT1 on the cancer cell does not inhibit this process in the host immune response, as mice bearing cancer cell STAT1 KO tumors showed increased T cell activation. This role of STAT1 in the host for generating T cell immunity may explain, in part, why STAT1 expression in tumor samples may appear as a protective biomarker [22], even though it may be a major component of acquired radioresistance and other cell survival mechanisms.
In conclusion, this study highlights how cancer cell STAT1 interacts with the immune tumor microenvironment to mediate radioresistance. Translation to therapeutics remains a difficult challenge, as Jak-Stat inhibitors such as Ruxolitinib target multiple STAT proteins, which not infrequently have opposing functions, and thus their applications remain mostly limited toward inflammatory diseases and a limited set of cancers [26]. The implications of our study in regard to biomarker development are promising, as increased clarity in expression analysis by compartment become more available by single-cell sequencing, and as incorporation of large panels of genes improve prognostic accuracy, reducing the noise that a gene such as STAT1 might contribute in certain cases where opposing function by compartment may exist. STATs will remain an important focus in the design of treatments, as any interventions that target particular cytokines or immune checkpoint molecules and impact cytokine availability necessarily have a STAT-altering component, impacting JAK-STAT pathways [3]. STAT1, existing in a role as a major mediator of the inflammatory and immune response, is of particular importance to designing treatment strategies incorporating RT and immunotherapy.
Methods
Cell lines, cell culture, and animal tumor models
LY2 and MOC2 squamous cell carcinoma murine cell lines were used for in vivo and in vitro studies, and tumor models were established orthotopically in the buccal, all of which are previously described [11]. BALB/c mice (Charles River, Wilmington MA) were used for the LY2 tumor model with 1 million cells implanted, and C57BL/6 mice (The Jackson Laboratory, Bar Harbor, Maine, USA) were used for the MOC2 tumor model with 100,000 cells implanted. For CRISPR knockouts, LY2 or MOC2 cells were transfected with STAT1 plasmids or PX458 control plasmid using PEI transfection agent at 1 mg/mL concentration (Sigma, St. Louis, MO, USA). STAT1 plasmids were obtained from the Functional Genomics University of Colorado Cancer Center Shared Resource. pSpCas9(BB)-2A-GFP (PX458) was a gift from Feng Zhang (Addgene plasmid #48,138; http://n2t.net/addgene:48138; RRID:Addgene_48138) Single cells with positive GFP expression were sorted using a MoFlo XDP cell sorter (Backman Coulter, Indianapolis, IN, USA) and resulting populations were validated for loss of STAT1 by Western blot. Mouse experiments were approved and mice showing signs of morbidity according to the guidelines set by the Institutional Animal Care and Use Committee (IACUC) were sacrificed immediately.
Irradiation
RT was performed on mouse buccal tumors as previously described. [11] Cells were irradiated with the same parameters at a dose rate of 2.0 Gy/min.
Statistical analysis
Using GraphPad Prism version 9.1.0 for Mac (GraphPad Software, San Diego, CA, USA), one-way ANOVA followed by Tukey’s multiple comparisons test was used for all comparisons with three or more groups, and unpaired two-tailed t-test was used for all comparisons between two groups. Other tests used were described in figure legends or methods.
Figures
Figure 2a was created using BioRender.com.
Western blot
Cell lysate preparation, gel electrophoresis, and blocking and staining protocols were performed as previously described.[27] Membranes were stained with primary antibody overnight at 4 °C for approximately 18 h. Anti-Stat1 and anti-pStat1 antibodies were purchased from Cell Signaling Technology, Danvers, MA, USA and anti-mouse horseradish peroxidase-conjugated secondary antibody was purchased from Sigma (St. Louis, MO, USA).
Multiplex cytokine detection
A murine U-Plex array (Meso Scale Diagnostics, Rockville, MD, USA) was used to detect CXCL9 and CXCL10 in conditioned media according to kit instructions.
T cell co-culture
Tumor cells were irradiated with 25 Gy dose and subsequently incubated for 48 h. Purified T cells (Stemcell Technologies, Vancouver, BC, Canada) were introduced to irradiated or control tumor cells at a 10:1 ratio (T cell: Tumor cell). Mixed T cells and tumor cells were co-cultured for 24 h in the presence of IL-2. After 24 h, T cells were harvested and processed for flow cytometry analyzed for IFNγ production. To determine T cell proliferation, the same experiment was repeated, but in the presence of CFSE-labeled T cells. CFSE label was analyzed by flow cytometry.
Incucyte assay
Cells were grown to 90% confluence in appropriate media in Incucyte ImageLock 96-well plates (Essen BioScience, Inc., Ann Arbor, MI, USA). Wells were scratched and subsequently rinsed carefully twice with PBS. Plates were imaged on a S3 Incucyte machine provided by the University of Colorado Cancer Center Cell Technologies Shared Resource using the Scratch Wound Module, and results were analyzed using the Scratch Wound analysis module in Incucyte software.
Boyden chamber assay
50,000 T cells were isolated from splenocytes of C57BL/6 mice with a CD3+ T cell isolation kit (Stemcell Technologies, Vancouver, BC, Canada) and were loaded into the top chamber on top of matrigel membrane matrix (Corning Inc., Corning, NY, USA), and conditioned media from STAT1 KO or PX458 control cells collected 72 h post- 20 Gy irradiation were loaded into the bottom chamber. After 4 h, chambers were removed and cells in the process of migration were fixed in the bottom chamber with 4% paraformaldehyde and stained with 0.1% crystal violet. Chambers were visualized with a microscope and T cells were counted in each chamber for comparison among groups. Control chambers contained non-conditioned normal LY2 or MOC2 media as appropriate.
TCGA analysis
The TCGA HNSCC Firehose legacy set was used for analysis in Supplemental Fig. 2. Scatterplots and correlation statistics were generated in the web app in cBioportal.[28] https://www.cbioportal.org/.
Rna-Seq
Cell lines for RNA-Seq were harvested from growing cultures using an RNA miniprep kit (Zymo research, Irvine CA). Sequencing and library prep were performed by The Genomics and Microarray Shared Resource at University of Colorado Denver Cancer Center. mRNA was used for library prep and sequencing was performed on an Illumina NovaSEQ 6000 with 2 × 150 paired end reads at a depth of 40 million PE reads per sample. Illumina adapters and the first 12 base pairs of each read were trimmed using BBDuk (BBMap – Bushnell B. – sourceforge.net/projects/bbmap/), and reads < 50 bp post-trimming were discarded. Reads were aligned and quantified using STAR (2.6.0a) [29] against the Ensembl mouse transcriptome (mg38.p6 genome (release 96)). Lowly expressing genes were removed if mean raw count < 1 or mean counts per million (CPM) < 1 for the dataset. Reads were normalized to CPM using the edgeR R package [30]. Differential expression was calculated using the voom function in the limma R package [31]. Analysis of RNA-Seq from mouse tumors (Fig. 1a, Fig. S1) was performed using data previously sequenced and described [11].
Volcano plots were generated using the EnhancedVolcano R package [32]. Gene set enrichment analysis was performed using the full ranked list of genes by log2 fold change for the indicated comparisons using the fgsea R package [33]. The p-value using the fgsea package was reported in Fig. 3d for ISG.RS pathways, and the BH-adjusted p-value for multiple comparisons was reported for Fig. 1a according to fgsea output from surveying all KEGG pathways.
Flow cytometry
Tumor-draining lymph nodes were harvested and processed for flow cytometric analysis as previously described.11 For analysis of immune cells, the following conjugated antibodies were used for the LY2 STAT1 KO versus control experiment: PercP/Cyanine5.5-CD3e (145-2C11, BD Biosciences), Super Bright 436-CD4 (GK1.5, Invitrogen), Brilliant Blue 515-CD8 (53–6.7, BD Biosciences), Brilliant Violet 650-CD11c (N418, Biolegend), PE/Cyanine5-CD44 (IM7, Biolegend), PerCP-CD45 (30-F11, BD Biosciences), APC-CD62L (MEL-14, Biolegend), Brilliant Violet 480-CD80 (16-10A1, BD Biosciences), Brilliant Violet 785-CD69 (H1.2F3, Biolegend), Alexa Fluor 532-Foxp3 (FJK-16 s, Invitrogen), PE/Cyanine7-IFNγ (XMG1.2, Biolegend), APC/eF780-Ki-67 (SolA15, Invitrogen), Brilliant Ultraviolet 615-CD274 (MIH5, BD Biosciences), BV750 TNF-α(Clone: MP6-XT22 BD Biosciences). For MOC2 STAT1 KO versus control experiment: PercP/Cy5.5-CD3 (145-2C11, BD Biosciences), Super Bright 436-CD4 (GK1.5, Invitrogen), Brilliant Blue 515-CD8 (53–6.7, BD Biosciences), Brilliant Ultraviolet-CD11b (M1/70, BD Biosciences), PE/Cyanine5-CD11c (N418, Biolegend), Super Bright 436-CD44 (IM7, Invitrogen), PerCP-CD45 (30-F11, BD Biosciences), APC-CD62L (MEL-14, Biolegend), PerCP/Cyanine5.5-CD80 (16-10A1, Biolegend), PE/Dazzle594-CD103 (2E7, Biolegend), Alexa Fluor 532-Foxp3 (FJK-16 s, Invitrogen), APC/Cyanine7-IFNγ (XMG1.2, Invitrogen), Brilliant Ultraviolet 395-Ki-67 (B56, BD Biosciences), Brilliant Violet 605-Ly-6C (AL-21, BD Biosciences), Brilliant Violet 480-CD274 (MIH5, BD Biosciences). Samples were analyzed on an Aurora spectral cytometer (Cytek Biosciences, Fremont, CA, USA) through The University of Colorado Cancer Center Flow Cytometry Shared Resource. FlowJo software (version 10.7.2) was used for initial selection of singlet leukocytes and exclusion of debris. CD45+ singlets from experimental group replicate for LY2 were concatenated or CD45+ live singlets from experimental group replicate for MOC2 were concatenated and used for input to Cytobank software [34] using all markers for clustering except CD45 and live dead aqua markers, and viSNE analysis was performed after transforming all parameters with asinh function. Populations were visualized with FlowSOM within Cytobank. Select populations were manually gated in FlowJo (Fig. 4e, j) to confirm observations from clustering analysis (Fig. 4a–d, f–i). Representative plots are shown in Supplemental Fig. 4.
Supplementary Information
Below is the link to the electronic supplementary material.
Authors’ contributions
SDK and MWK designed the study and all experiments. MWK, TEB, SBu, and DN performed mouse studies. BVC performed irradiation. MWK, SBh, SC, DN, and AO performed in vitro experiments. AO performed cell proliferation assays and western blotting. MWK, LBD, and JG performed flow cytometry and analysis. MP and MWK performed scratch wound assays. Sbu and MWK performed boyden chamber assays and analysis. All authors advised throughout the course of the study. SDK and MWK wrote the manuscript. All authors discussed, revised, and approved the final manuscript.
Funding
Dr. Sana D. Karam is funded by the NIDCR/NCI (R01 DE028529-01, R01 DE028282-01, 1 P50 CA261605-01) and receives preclinical funding from Roche and clinical trial funding from AstraZeneca, both unrelated to this research. This study was also partly supported by the National Institutes of Health P30CA046934 Cancer Center Support Grant, including the Bioinformatics and Biostatistics Shared Resource, the Cancer Center Flow Cytometry Shared Resource, the Genomics and Microarray Shared Resource, and the Cell Technologies Shared Resource at the University of Colorado. This study was also partly supported by the Cancer League of Colorado.
Declarations
Conflicts of interest
The authors declare no potential conflicts of interest.
Data Availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Code availability
Available from the corresponding author upon reasonable request.
Consent for publication
All listed authors have contributed significantly to the research described and have read and approved this manuscript for submission to Cancer Immunology, Immunotherapy.
Footnotes
Précis
HNSCCs have responded poorly to current RT-immunotherapy combinations, and mechanisms of resistance remain elusive. Here, we provide compelling data for cancer cell-specific STAT1 directing resistance in immune -dependent and -independent behavior.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Yu H, Pardoll D, Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nat Rev Cancer. 2009;9:798–809. doi: 10.1038/nrc2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Owen KL, Brockwell NK, Parker BS. JAK-STAT signaling: a double-edged sword of immune regulation and cancer progression. Cancers (Basel) 2019;11:2002. doi: 10.3390/cancers11122002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Villarino AV, Kanno Y, O'Shea JJ. Mechanisms and consequences of Jak-STAT signaling in the immune system. Nat Immunol. 2017;18:374–384. doi: 10.1038/ni.3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meissl K, Macho-Maschler S, Muller M, Strobl B. The good and the bad faces of STAT1 in solid tumours. Cytokine. 2017;89:12–20. doi: 10.1016/j.cyto.2015.11.011. [DOI] [PubMed] [Google Scholar]
- 5.Mandal R, et al. The head and neck cancer immune landscape and its immunotherapeutic implications. JCI Insight. 2016;1:e89829. doi: 10.1172/jci.insight.89829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Annals of Oncology (2019) 30 (suppl_5): v851-v934. 10.1093/annonc/mdz394.
- 7.Cohen EE, Ferris RL, Psyrri A, et al. Primary results of the phase III JAVELIN head & neck 100 trial: Avelumab plus chemoradiotherapy (CRT) followed by avelumab maintenance vs CRT in patients with locally advanced squamous cell carcinoma of the head and neck (LA SCCHN) ESMO Virtual Congress. 2020;31:S658. [Google Scholar]
- 8.Fuertes MB, et al. Host type I IFN signals are required for antitumor CD8+ T cell responses through CD8{alpha}+ dendritic cells. J Exp Med. 2011;208:2005–2016. doi: 10.1084/jem.20101159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oweida A, et al. Resistance to radiotherapy and PD-L1 blockade is mediated by TIM-3 upregulation and regulatory T-cell infiltration. Clin Cancer Res. 2018;24:5368–5380. doi: 10.1158/1078-0432.CCR-18-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Knitz MW, et al. Targeting resistance to radiation-immunotherapy in cold HNSCCs by modulating the Treg-dendritic cell axis. J Immunother Cancer. 2021;9:e001955. doi: 10.1136/jitc-2020-001955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liberzon A, et al. The molecular signatures database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xanthou G, Duchesnes CE, Williams TJ, Pease JE. CCR3 functional responses are regulated by both CXCR3 and its ligands CXCL9, CXCL10 and CXCL11. Eur J Immunol. 2003;33:2241–2250. doi: 10.1002/eji.200323787. [DOI] [PubMed] [Google Scholar]
- 14.Ran FA, et al. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khodarev NN, et al. STAT1 is overexpressed in tumors selected for radioresistance and confers protection from radiation in transduced sensitive cells. Proc Natl Acad Sci U S A. 2004;101:1714–1719. doi: 10.1073/pnas.0308102100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khodarev NN, et al. Signal transducer and activator of transcription 1 regulates both cytotoxic and prosurvival functions in tumor cells. Cancer Res. 2007;67:9214–9220. doi: 10.1158/0008-5472.CAN-07-1019. [DOI] [PubMed] [Google Scholar]
- 17.Benci JL, et al. Opposing functions of interferon coordinate adaptive and innate immune responses to cancer immune checkpoint blockade. Cell. 2019;178(933–948):e914. doi: 10.1016/j.cell.2019.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Benci JL, et al. Tumor interferon signaling regulates a multigenic resistance program to immune checkpoint blockade. Cell. 2016;167(1540–1554):e1512. doi: 10.1016/j.cell.2016.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bhattacharya N, Roy A, Roy B, Roychoudhury S, Panda CK. MYC gene amplification reveals clinical association with head and neck squamous cell carcinoma in Indian patients. J Oral Pathol Med. 2009;38:759–763. doi: 10.1111/j.1600-0714.2009.00781.x. [DOI] [PubMed] [Google Scholar]
- 20.Xu B, et al. CCR9 and CCL25: a review of their roles in tumor promotion. J Cell Physiol. 2020;235:9121–9132. doi: 10.1002/jcp.29782. [DOI] [PubMed] [Google Scholar]
- 21.Chen S, et al. Mechanisms regulating PD-L1 expression on tumor and immune cells. J Immunother Cancer. 2019;7:305. doi: 10.1186/s40425-019-0770-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ayers M, et al. IFN-gamma-related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest. 2017;127:2930–2940. doi: 10.1172/JCI91190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cohen EEW, et al. Pembrolizumab versus methotrexate, docetaxel, or cetuximab for recurrent or metastatic head-and-neck squamous cell carcinoma (KEYNOTE-040): a randomised, open-label, phase 3 study. Lancet. 2019;393:156–167. doi: 10.1016/S0140-6736(18)31999-8. [DOI] [PubMed] [Google Scholar]
- 24.Nanda R, et al. Pembrolizumab in patients with advanced triple-negative breast cancer: phase Ib KEYNOTE-012 study. J Clin Oncol. 2016;34:2460–2467. doi: 10.1200/JCO.2015.64.8931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ryan N, et al. STAT1 inhibits T-cell exhaustion and myeloid derived suppressor cell accumulation to promote antitumor immune responses in head and neck squamous cell carcinoma. Int J Cancer. 2020;146:1717–1729. doi: 10.1002/ijc.32781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hosseini A, et al. Janus kinase inhibitors: a therapeutic strategy for cancer and autoimmune diseases. J Cell Physiol. 2020;235:5903–5924. doi: 10.1002/jcp.29593. [DOI] [PubMed] [Google Scholar]
- 27.Bhatia S, et al. Inhibition of EphB4-Ephrin-B2 signaling enhances response to cetuximab-radiation therapy in head and neck cancers. Clin Cancer Res. 2018;24:4539–4550. doi: 10.1158/1078-0432.CCR-18-0327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cerami E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47–e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blighe, K., Rana, S. & Lewis, M. (2021). EnhancedVolcano: Publication-ready volcano plots with enhanced colouring and labeling. R package version 1.10.0, https://github.com/kevinblighe/EnhancedVolcano.
- 33.Korotkevich, G. et al. (2021) An algorithm for fast preranked gene set enrichment analysis using cumulative statistic calculation. BioRxiv, 060012.
- 34.Kotecha N, Krutzik PO, Irish JM. Web-based analysis and publication of flow cytometry experiments. Curr Protoc Cytom. 2010 doi: 10.1002/0471142956.cy1017s53. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.