Abstract
Interaction scaffolds that selectively recognize disordered protein strongly shape protein interactomes. An important scaffold of this type that contributes to transcription is the TFIIS N-terminal domain (TND). The TND is a five-helical bundle that has no known enzymatic activity, but instead selectively reads intrinsically disordered sequences of other proteins. Here, we review the structural and functional properties of TNDs and their cognate disordered ligands known as TND-interacting motifs (TIMs). TNDs or TIMs are found in prominent members of the transcription machinery, including TFIIS, super elongation complex, SWI/SNF, Mediator, IWS1, SPT6, PP1-PNUTS phosphatase, elongin, H3K36me3 readers, the transcription factor MYC, and others. We also review how the TND interactome contributes to the regulation of transcription. Because the TND is the most significantly enriched fold among transcription elongation regulators, TND- and TIM-driven interactions have widespread roles in the regulation of many transcriptional processes.
Keywords: intrinsically disordered proteins, molecular scaffolds, structural biology, transcription
Introduction
The emergence of diverse functional roles for intrinsically disordered regions (IDRs) has highlighted their enormous regulatory potential [1–5]. One important route by which IDRs exert distinct functions is their ability to mediate selective assembly with folded interaction platforms on their binding partners [2]. To date, a number of structurally conserved interaction scaffolds that selectively ‘read’ IDRs have been identified. This includes TFIIS N-terminal domains (TNDs) [6, 7], WD40 repeat (WDR) domains [8, 9], Src Homology 2 (SH2) [10], Src Homology 3 (SH3) [11], PDZ [12, 13], WW [14, 15], and many other domains. These domains represent ‘landing pads’ that bind disordered short linear motifs (SLiMs) through structurally conserved mechanisms [16].
IDRs are particularly enriched among human transcription and chromatin regulators [2]. Compared with other protein scaffolds that selectively bind IDRs, TNDs are found with remarkable selectively in proteins that regulate gene expression (Figure 1A) [6]. As a result of this selective enrichment, TNDs are uniquely poised to aid IDR-mediated assembly of the transcriptional machinery. Here, we summarize the structural features and interaction modes maintained by this protein domain family, as well as the roles they exert in transcription.
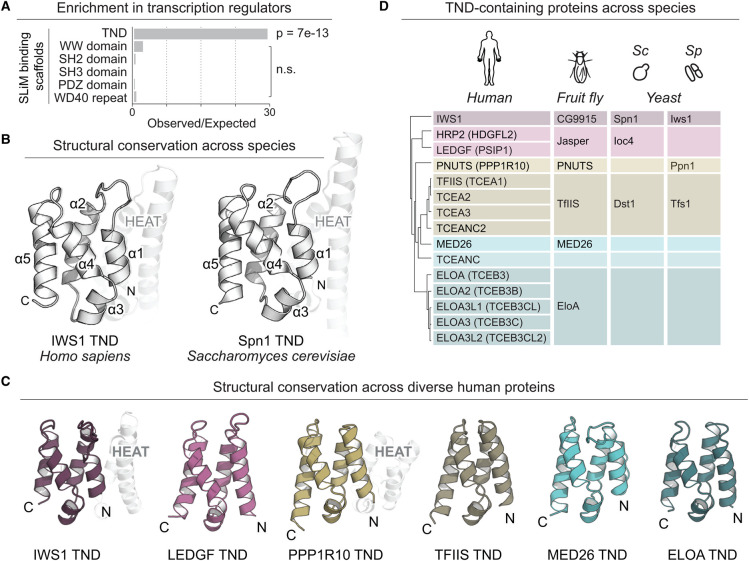
Figure 1. The TFIIS N-terminal domain (TND) is a structurally conserved fold enriched among transcription regulators.
(A) Enrichment of diverse SLiM interaction scaffolds among transcription regulators associated with GO term GO:0006351. (B) Characteristic right-handed five-helix bundle (α1–α5) of TNDs. The N-terminal HEAT subdomain is highlighted (PDB accessions 3O8Z and 6ZV1). (C) Example structures of TNDs in human transcription regulators (PDB accessions 6ZV1, 6ZV0, 6ZV2, 6ZUY, 6ZUZ and 6ZV3). (D) Human, fruit fly, and yeast proteins that contain a TND. Abbreviations: Sc, Saccharomyces cerevisiae; Sp, Schizosaccharomyces pombe.
TNDs are conserved binding scaffolds for disordered TND-interacting motifs
Conservation and structural properties
The TND is conserved from humans to yeast (Figure 1B–D), with notable expansion and diversification in multicellular organisms. While the human proteome harbors at least 15 TND-containing factors, only four TND-containing proteins are currently annotated in the budding or fission yeast proteomes (Figure 1D) [17, 18]. Structurally, the TND fold is a right-handed bundle of five helices, reminiscent of a pair of HEAT repeats (Figure 1B,C) [6, 19–21]. In several human as well as yeast proteins, the TND is immediately preceded by an N-terminal HEAT subdomain (Figure 1B,C) [6, 21, 22].
Such helical repeat domains are frequently found to be protein scaffolds utilized in large protein–protein complexes, for example, ATP-dependent chromatin remodelers [23–27] or protein kinases implicated in DNA repair [28, 29]. Their high structural stability relies on tight packing of helix-turn-helix motifs into a supercoiled arrangement. Four- to five-helix bundles like the TND are the minimal viable helical repeat domains enabling variability of interaction surfaces while retaining stability [30, 31]. While overall sequence conservation of TNDs is low (27% identity and 47% similarity for 71 annotated PROSITE PS51319 domains), the fold-stabilizing core residues are more invariant (41% identity and 67% similarity) [32]. In the human proteome, the strongest conservation is observed for buried hydrophobic residues in the domain core, while side chains from poorly conserved residues are solvent exposed [6] and therefore accessible to confer interaction specificity. Because there is currently no known catalytic activity associated with the TND fold, these domains are thought to act primarily as interaction platforms [6, 33].
The TND selectively reads short, disordered TND-interacting motifs
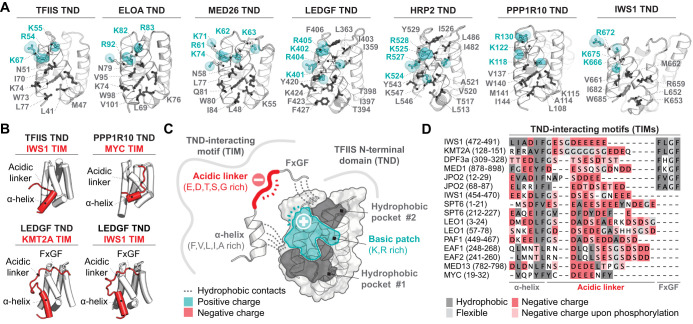
Detailed structural analysis of TND-mediated protein complexes revealed core elements of the binding interfaces that are structurally conserved across TND-containing factors and across species [6, 7, 21, 22, 34]. Each TND coordinates its interactome by two relatively shallow binding pockets (each representing ∼80 Å2) that accommodate bulky hydrophobic residues, as well as a positively charged surface patch containing 3–5 positively charged residues positioned between pockets (Figure 2A). This recurrent configuration gives rise to a characteristic charge patterning of the TND surface that accommodates a corresponding pattern of hydrophobic and negatively charged residues on TND-interacting motifs (TIMs). Correspondingly, TIMs engage TNDs through distinct motif features: an obligate α-helix and acidic linker sequence, and an optional FxGF motif (Figure 2B). While the α-helix anchors the TIM through conserved phenylalanine, valine, leucine, and isoleucine side-chains in the first hydrophobic binding pocket created by α3, α4, and α5 of TNDs, the FxGF motif occupies the second, more shallow binding pocket via its two phenylalanines (Figure 2C). The FxGF motif plays a unique role in the recognition of TNDs found in H3K36me3 readers LEDGF and HRP2, where it significantly contributes to TIM binding. However, many TIMs lack the FxGF motif and only engage the first pocket through the short α-helix (Figure 2D). The selectivity of each TND towards the TIMs lies in differences in the configurations of these hydrophobic pockets and charge patterns, particularly their compatibility with aliphatic residues and charge patterns on the TIM amino acid sequence.
Figure 2. TNDs are selective binding scaffolds for disordered TND-interacting motifs (TIMs).
(A) Comparison of human TNDs and interaction interfaces that each TND utilizes for TIM recognition. Residues forming the hydrophobic pockets (dark gray) and residues forming the positively charged basic patch (teal) are highlighted and labeled. (B) Examples of TIMs (red) in complexes with their cognate TND scaffolds (white). (C) Summary of important features that govern TND:TIM interactions. (D) Multiple sequence alignment of TIMs found in different human proteins.
In addition to aliphatic contacts from the α-helix and FxGF motifs, the flexible acidic linker also contributes to overall affinity. This variable linker spans 8–16 amino acid residues in length and is enriched in glutamate and aspartate residues that recognize the basic patch on TNDs. The acidic linker is also often enriched in glycine residues that confer flexibility, as well as serine and threonine residues that are often also negatively charged due to post-translational phosphorylation (Figure 2D) [6, 7, 33]. In particular, the acidic linkers of IWS1, KMT2A (also known as MLL1), JPO2 and other TIMs are phosphorylated by casein kinase 2 (CK2) [7]. TIM phosphorylation enhances affinity towards TND-containing binding partners, thereby enabling switching between low- and high-affinity states of these interactions [6, 7]. However, many questions regarding the regulatory roles of PTMs in the TND interactome remain open. For example: Is CK2 unique, or are there other kinases that phosphorylate TIMs? What structural features confer selectivity of kinases for distinct TIMs? Is TIM phosphorylation constitutive or regulated? If it is regulated, what phosphatases remove these post-translational marks? How does TIM phosphorylation contribute to different stages of transcription? Phosphorylation of the acidic linker represents a primary avenue for regulation of these interactions, therefore answering these exciting questions would expand our mechanistic understanding of how regulated TND:TIM interactions contribute to transcription.
The TND governs assembly of higher-order structures
Several proteins possess more than a single TIM. For example, SPT6, LEO1, JPO2 and CDC7-ASK contain each two such motifs and IWS1 harbors three distinct TIMs in series [6, 7]. Therefore, these proteins have the capacity to regulate higher-order complex assemblies by engaging multiple TND-containing factors through their TIMs at the same time. Currently, the best characterized example of multiprotein complex assembly through these surfaces is human IWS1. IWS1 contains three TIMs, but also harbors its own TND and simultaneously engages these surfaces to bring together four other transcription regulators through TND:TIM interactions [6]. Importantly, regulation of higher-order structures of these factors can also be enhanced through multimerization of TND-containing factors. For example, dimerization of LEDGF is stabilized by TND domain swapping and additional electrostatic ‘stapling’ of the negatively charged α helix formed in the IDR C-terminal to the TND [35]. Importantly, the TIM interaction sites on the TNDs remain structurally unperturbed by domain swapping [35]. Such an arrangement has the potential to aid assembly of higher-order structures.
Transcriptional roles of TND across proteins and species
The TFIIS TND links transcription regulators to RNAP2
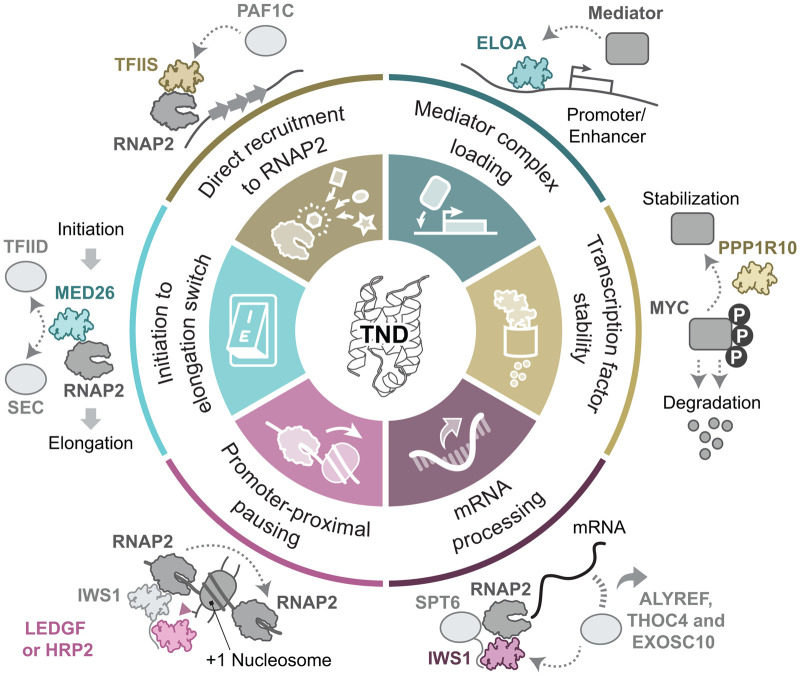
The transcription elongation factor TFIIS increases the overall transcription rate of RNAP2 by rescuing backtracked polymerases [36, 37]. TFIIS is well conserved from human to yeast, however; homologs are also found in archaea and in some viral genomes [20, 38]. Despite the eponymous naming of the TND from TFIIS (where it is also known as domain I, or LW domain), the mechanisms by which this domain contributes to TFIIS-dependent regulation of transcription remained unclear for many years, in part because the TND is not required for backtrack rescue in vitro [39]. Mutational analysis of the human TFIIS TND linked this domain with the nuclear localization of TFIIS [40]. Additionally, the TFIIS TND was implicated in early transcription events distinct from TFIIS's role in elongation, in particular, for efficient formation of RNAP2 preinitiation complexes and promoter recruitment [41]. Even though CryoEM revealed the structure of TFIIS bound to other transcription elongation complexes and RNAP2 [42–45], the TFIIS TND remained a dynamic component of these complexes and contacts to other transcription regulators mediated by this domain remained hidden. Similar to other members of the TND family, the TFIIS TND acts as an interaction scaffold for disordered TIMs, including motifs in transcriptional regulators IWS1, LEO1, PAF1 and others [6]. The TFIIS TND directly links these factors to RNAP2, thereby mediating their proximity to the transcriptional machinery, which is expected to influence their functional roles (Figure 3).
Figure 3. TND-mediated interactions govern many transcriptional and co-transcriptional processes.
TND-containing proteins and mechanisms by which their TNDs contribute to transcription.
The TND in IWS1 links transcription and mRNA processing machinery
IWS1 (Interacts with SPT6) is a transcription elongation regulator conserved from human to yeast, where the ortholog is known as Spn1. Both human IWS1 and yeast Spn1 harbor a TND, which recognizes a disordered TIM in the histone chaperone SPT6 (SUPT6H or Spt6) [6, 22]. While other TNDs recognize multiple TIMs with comparable affinity, the IWS1 TND has >200-fold higher affinity to SPT6 TIM than other measured interactions, and is thus the most stable complex supported by the IWS1 TND identified so far [6]. In mammalian cells, the IWS1:SPT6 complex was implicated in elongation-coupled placement of H3K36me3, a signature of active transcription written by the histone methyltransferase SETD2 [46]. In yeast, the ortholog Spn1 influences methylation of both H3K36 and H3K4 across the genome and acts as a histone chaperone at highly expressed genes [46, 47]. Interestingly, Spn1 is not required for the interaction of Spt6 with RNAP2, but rather plays a role in optimal Spt6 recruitment to chromatin [47]. In human cells, IWS1 localizes at actively transcribed genes, with peak occupancy close to the transcription start site [6] and, together with SPT6 and RNAP2, recruits mRNA processing factors including ALYREF/THOC4 and EXOSC10 to ensure proper mRNA maturation and export (Figure 3) [48].
A recent structure of the yeast RNAP2 elongation complex revealed that the Spn1 TND preceded by HEAT subdomain is recruited to RNAP2 through association with the Spt5 NGN and KOW2 domains using an interface distinct from the one needed for association with TIMs, leaving the TIM binding site open for interaction with Spt6 [49]. Importantly, the IWS1-TND:SPT6-TIM interaction interface in the context of fully assembled elongation complexes and RNAP2 is structurally similar to the binary complex resolved by protein NMR or crystallography [6, 21, 22, 49], confirming that the binary TND:TIM interactions exist in the context of larger assembled complexes.
As the only structured domain of IWS1, the TND is localized in the middle of the IWS1 sequence and is surrounded by IDRs. The disordered region N-terminal to the IWS1 TND harbors a series of three unique TIMs that selectively interact with different TND-containing factors: While TNDs from TFIIS and ELOA compete for the TIM1 of IWS1, TIM2 is recognized by the PP1-PNUTS phosphatase TND, and the TNDs of H3K36me3 readers LEDGF and HRP2 compete for TIM3 [6]. Additionally, the IWS1 TND independently associates with SPT6 via its TIM [6]. Therefore, IWS1 acts as a central factor that coordinates many elongation and RNA processing factors.
The TND in mediator subunit MED26 enables molecular switching between initiation and elongation
The multiprotein Mediator complex is conserved in eukaryotes [50], where it serves as a scaffold for the assembly of a functional preinitiation complex and as a bridge communicating information from gene-specific regulatory proteins to the basal RNAP2 transcription machinery. The TND within this complex is found in an N-terminal portion of a metazoan-specific subunit MED26 [51], hence the utilization of the Mediator TND is not structurally conserved to yeast. In human cells, MED26 is recruited to the Mediator complex through an interaction between its C-terminal domain and the MED4/7 subunits, which leaves the TND accessible for interactions with accessory proteins [50]. A single binding site on the MED26 TND is employed in two distinct contexts: At distinct moments, the TND mediates interactions either by recruiting super-elongation complex containing ELL/EAF family members through their disordered TIMs, or by associating with TFIID and elongation complexes [6, 51, 52]. Mutation of the interaction site on the MED26 TND does not affect Mediator-dependent binding of TFIID to the promoter, and hence the MED26 TND is not exclusively responsible for Mediator's interaction with TFIID. However this mutation does prevent Mediator from recruiting RNAP2 elongation factors [51]. Therefore, the MED26 TND was proposed to participate in molecular signaling activity that instructs RNAP2 to transition from initiation into productive elongation (Figure 3).
TNDs in H3K36me3 readers LEDGF (PSIP1) and HRP2 (HDGFL2) mediate regulation of chromatin structure
Both LEDGF and HRP2 are chromatin readers that each contain a TND and Pro-Trp-Trp-Pro (PWWP) domain that recognizes H3K36me2/3 methylated histone tails [53, 54]. The TNDs of both proteins directly interact with TIMs in transcription regulators, including the KMT2A histone methyltransferase [55, 56], IWS1 [6, 33] or JPO2 [33]. LEDGF also directly interacts with TIMs in MED1 [7] and CDC7-ASK [7]. As described in further detail below, the HRP2 TND additionally binds a TIM in DPF3a [57]. All currently known interaction partners of LEDGF and HRP2 TNDs possess the FxGF portion on their TIMs, where it is essential for their interaction. This finding suggests the FxGF portion may be generally required for interaction with these TNDs.
Although LEDGF and HRP2 share some functional redundancy, the shared and unique roles of these two proteins remain to be fully understood. Both proteins influence RNAP2 transcription elongation by functioning as histone chaperones [58]. In differentiated myoblasts, these chromatin readers are required for efficient transcription elongation genome-wide [58], where they functionally substitute for loss of histone chaperone activity by the FACT complex at the +1 nucleosome. Moreover, transcription elongation defects similar to genetic depletion of HRP2 and LEDGF were observed upon mutation of the IWS1 TIM that selectively recognizes HRP2 and LEDGF TNDs [6]. Affected genes similarly displayed increased RNAP2 pausing near the +1 nucleosome, suggesting that the contributions of these H3K36me3 readers towards pause release near the +1 nucleosome is governed in part through their interaction with IWS1 (Figure 3).
The HRP2 TND also contributes to regulation of chromatin structure through its interactions with a TIM in DPF3a, a subunit of SWI/SNF chromatin remodeling complexes [57]. This activity of HRP2 is dependent on the H3K36me3 mark and is regulated by phosphorylation of DPF3a, which enhances interaction with the HRP2 TND. Importantly, HRP2:DPF3a activity is essential for myogenesis and muscle regeneration in vivo. Its ability to recruit SWI/SNF ATPase activity [57] suggests that LEDGF and HRP2 TNDs may have pleiotropic molecular functions.
The ELOA TND acts as mediator loading platform
Elongin is an RNAP2-associated complex that is conserved to nematodes. While the C-terminus of ELOA (Elongin A) enables interaction with other subunits of the Elongin complex, the N-terminal region of ELOA harbors a TND that is accessible for other factors and complexes [59, 60]. In vivo, ELOA regulates RNAP2 promoter proximal pausing [61] and acts as a substrate recognition subunit of a Cullin-RING E3 ubiquitin ligase that targets stalled RNAP2 and promotes RNAP2 polyubiquitination and proteasomal degradation [62].
Interestingly, the ubiquitination activity of ELOA is independent of its elongation regulatory activity in vivo [63]. Even though the TND of isolated ELOA is dispensable for transcriptional activation in vitro [59], it directly interacts with TIMs conserved in IWS1, PAF1, LEO1, MED13 and other transcription elongation regulators [6]. Importantly, the ELOA TND directly links this protein to purified Mediator and facilitates recruitment of Mediator complex to promoters of stress response genes (Figure 3) [64], highlighting the role of this domain as a linker between different transcription regulatory machines.
The PP1-PNUTS phosphatase TND regulates protein stability and transcription rate
PP1-PNUTS serine/threonine phosphatase is a negative regulator of RNAP2 elongation rate [65] that also plays a role in transition between transcription stages and recycling transcriptional machinery [65–67], control of chromatin structure [68, 69], cell cycle progression [70–72] and many other cellular processes. The TND is located in the PPP1R10 subunit, also known as PNUTS, p99, FB19, or CAT53. PPP1R10 is a scaffold protein mediating the formation of the phosphatase [69]. The PPP1R10 directly interacts with TIMs in transcription elongation regulators including IWS1, SPT6 and PAF1 [6], as well as the transcription factor MYC [34]. While the exact regulatory roles exerted by association of PPP1R10 with the transcription elongation factors IWS1, SPT6, or PAF1 remains unclear, the function of MYC and PPP1R10 have been evaluated due to the prominent role of MYC in cancer. MYC and PP1-PNUTS phosphatase interact across multiple cell types and co-occupy MYC target gene promoters [73]. Disruption of PP1 activity results in MYC hyperphosphorylation, which compromises its ability to bind to chromatin and leads to reduction in MYC levels due to proteasomal degradation (Figure 3) [73]. Interestingly, PPP1R10 and MYC are co-amplified in breast cancer cells [73], suggesting that elevated PP1-PNUTS expression may confer a growth advantage by increasing MYC protein stability.
In addition to interactions mediated through the TND:TIM module, an N-terminal fragment of PPP1R10 containing the TND interacts directly with WDR82 and TOX4 [69]. While the PPP1R10 interaction with WDR82 prevents transcription–replication conflicts by promoting RNAP2 degradation [74], the association with TOX4 restricts pause release in early elongation and promotes late elongation [75], both via regulation of the phospho-state of the RNAP2 CTD. The exact mechanism of association WDR82 and TOX4 with PPP1R10 remains uncertain, however, the PPP1R10 TND may act as an interaction platform supporting these processes in a manner similar to its interaction with MYC.
The TND in disease and as a therapeutic target
TNDs and TIMs are present in proteins that represent the core of transcriptional machinery, and the factors that harbor them are generally essential. Like other pan-essential proteins, TND- and TIM-containing factors are also infrequently associated with disease-related mutations, suggesting a degree of protection from mutation and underscoring their importance as regulators of basic cellular functions. However, there are many instances when the endogenous activities of these proteins are hijacked in disease settings described below.
Viral mimicry
Due to their short length and simple interaction modes, short linear motifs like TIMs are often hijacked by viruses [76, 77]. Indeed, LEDGF and HRP2 have generated considerable interest because the TNDs of these H3K36me2/3 readers are hijacked by lentiviral integrases [78, 79]. These readers act as molecular tethers for viral pre-integration complexes, which biases viral integration into the bodies of actively transcribed genes in the host chromatin. Interestingly, HIV-1 integrase has higher affinity for the LEDGF TND compared with the HRP2 TND, and hence HIV-1 primarily uses LEDGF as an integration cofactor. However, HRP2 is also sufficient to guide site selection for viral integration in the absence of LEDGF [78]. Small-molecule antivirals known as LEDGINs that target the HIV-1 integrase and disrupt its interaction with the TND were successfully developed and currently serve as an important research tool with potential future clinical application [80].
Deregulation in cancer
As mentioned above, the PPP1R10 phosphatase subunit regulates MYC phosphorylation and stability by directly interacting with the TIM in MYC. Indeed, PP1-PNUTS expression is amplified in several cancer settings, including breast [73] and prostate cancers [81], where PPP1R10 protein levels are predictor of poor prognosis. Therefore, the PPP1R10:MYC interaction represents an interesting potential target in these cancer settings. Additionally, the chromatin tethering role of HRP2 and LEDGF are hijacked in acute leukemia, as their TNDs directly interact with oncogenic KMT2A fusions [55, 56, 82]. Interestingly, both of these TNDs have similar affinities to the TIMs in KMT2A fusions, however, while LEDGF is crucial for leukemic transformation [55, 83, 84], HRP2 is not required [56]. Separately, the interaction between LEDGF and JPO2 is a potential therapeutic target in medulloblastoma, due to its ability to promote AKT signaling [85]. HRP2 is also frequently overexpressed in human hepatocellular carcinoma tissues, where the HRP2:IWS1 complex promotes cell growth by enhancing expression of key oncogenes [86].
Small-molecule targeting of the TND
Targeting of specific TND:TIM complexes may be beneficial in diverse disease settings. However, successful protein–protein inhibitors frequently target deep grooves or pockets rather than shallow surfaces like TIM binding sites on TNDs. Additionally, given the close resemblance of all TND:TIM protein complexes, designing small molecules to selectively target a single TND:TIM surface represents a challenge. Nevertheless, targeting the disease-related activities supported by TND:TIM modules may be achieved by degradation of full-length proteins using PROTACs [87–89] designed for recognition of different parts of these proteins, or by design of small covalent molecules selectively recognizing disordered TIMs, similar to those that were recently developed for targeting MYC [90].
Concluding remarks
The conservation, diversification, and widespread utilization of TNDs underscores the functional importance of this ancient scaffold for assembly of the transcriptional machinery. As a result, addressing how the interactomes of individual TNDs are regulated to coordinate the transcription machinery represents a promising research direction. More generally, a key frontier for molecular biology is to decipher the interactions between disordered sequences and folded protein domains. The identification of TNDs as selective interaction platforms for disordered TIMs highlights one avenue by which disordered protein can influence cellular activities with high specificity by engaging in selective, well-defined interactions. However, many more motifs have been predicted to engage a variety of folded domains [91]. For this reason, identifying the underlying logic and grammar for these many interactions, as well as their influence on subnuclear organization, remain important goals.
Perspectives
TFIIS N-terminal domains (TNDs) are conserved and have diverse functional roles in many prominent regulators of transcription.
TNDs mediate specific interactions with intrinsically disordered motifs called TIMs found in other transcription regulators. Interactions between TNDs and TIMs guide the organization of the transcription machinery.
Identifying the functional grammar and spatial organization of TND:TIM interactions represents an important future direction. Additionally, TND:TIM contacts may enable structural characterization of higher-order assemblies mediated by these interaction modules.
Acknowledgements
We thank members of the Hodges lab (BCM) and the Laboratory of Structural Biology (IOCB) for helpful feedback during manuscript preparation.
Abbreviations
- CTD
C-terminal domain
- HEAT
Helical scaffold, acronym named for Huntingtin elongation factor 3 (EF3), protein phosphatase 2A (PP2A), and TOR1
- IDR
Intrinsically disordered region
- PDZ
Domain, acronym named for Post synaptic density protein (PSD95), Drosophila disc large tumor suppressor (Dlg1), and Zonula occludens-1 protein (zo-1)
- PWWP
proline — tryptophan — tryptophan — proline motif containing domain
- RNAP2
RNA polymerase 2
- SH2
Src Homology 2 domain
- SH3
Src Homology 2 domain
- SLiM
Short linear motif
- TIM
TND-interacting motif
- TND
TFIIS N-terminal domain
- WD40
tryptophan-aspartic acid dipeptide containing 40 amino acid long structural motif
- WDR
WD40 repeat
- WW
tryptophan-tryptophan containing structural motif
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
This work was supported by grants from the NIH (R35GM137996 to H.C.H.), the Cancer Prevention and Research Institute of Texas (RR170036 to H.C.H.), GACR grant (22-03028S to V.V.) and Chemical Biology for Drugging Undruggable Targets (ChemBioDrug) grant CZ.02.1.01/0.0/0.0/16_019/0000729 (V.V.).
Author Contributions
K.C.: Conceptualization, Writing — Original Draft, Writing — Review and Editing, Visualization; V.V.: Writing — Review and Editing, Supervision, Funding acquisition; H.C.H.: Writing — Review and Editing, Visualization, Supervision, Funding acquisition.
References
- 1.Babu, M.M. (2016) The contribution of intrinsically disordered regions to protein function, cellular complexity, and human disease. Biochem. Soc. Trans. 44, 1185–1200 10.1042/BST20160172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cermakova, K. and Hodges, H.C. (2023) Interaction modules that impart specificity to disordered protein Trends Biochem. Sci. (in press) [Google Scholar]
- 3.Musselman, C.A. and Kutateladze, T.G. (2021) Characterization of functional disordered regions within chromatin-associated proteins. iScience 24, 102070 10.1016/j.isci.2021.102070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sabari, B.R., Dall'Agnese, A., Boija, A., Klein, I.A., Coffey, E.L., Shrinivas, K.et al. (2018) Coactivator condensation at super-enhancers links phase separation and gene control. Science 361, eaar3958 10.1126/science.aar3958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cubuk, J., Alston, J.J., Incicco, J.J., Singh, S., Stuchell-Brereton, M.D., Ward, M.D.et al. (2021) The SARS-CoV-2 nucleocapsid protein is dynamic, disordered, and phase separates with RNA. Nat. Commun. 12, 1936 10.1038/s41467-021-21953-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cermakova, K., Demeulemeester, J., Lux, V., Nedomova, M., Goldman, S.R., Smith, E.A.et al. (2021) A ubiquitous disordered protein interaction module orchestrates transcription elongation. Science 374, 1113–1121 10.1126/science.abe2913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sharma, S., Cermáková, K., De Rijck, J., Demeulemeester, J., Fábry, M., El Ashkar, S.et al. (2018) Affinity switching of the LEDGF/p75 IBD interactome is governed by kinase-dependent phosphorylation. Proc. Natl Acad. Sci. U.S.A. 115, E7053–E7062 10.1073/pnas.1803909115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kruusvee, V., Lyst, M.J., Taylor, C., Tarnauskaite, Ž, Bird, A.P. and Cook, A.G. (2017) Structure of the MeCP2-TBLR1 complex reveals a molecular basis for Rett syndrome and related disorders. Proc. Natl Acad. Sci. U.S.A. 114, E3243–E3250 10.1073/pnas.1700731114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Justin, N., Zhang, Y., Tarricone, C., Martin, S.R., Chen, S., Underwood, E.et al. (2016) Structural basis of oncogenic histone H3K27M inhibition of human polycomb repressive complex 2. Nat. Commun. 7, 11316 10.1038/ncomms11316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marasco, M., Berteotti, A., Weyershaeuser, J., Thorausch, N., Sikorska, J., Krausze, J.et al. (2020) Molecular mechanism of SHP2 activation by PD-1 stimulation. Sci. Adv. 6, eaay4458 10.1126/sciadv.aay4458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gemperle, J., Hexnerová, R., Lepšík, M., Tesina, P., Dibus, M., Novotný, M.et al. (2017) Structural characterization of CAS SH3 domain selectivity and regulation reveals new CAS interaction partners. Sci. Rep. 7, 8057 10.1038/s41598-017-08303-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Genera, M., Quioc-Salomon, B., Nourisson, A., Colcombet-Cazenave, B., Haouz, A., Mechaly, A.et al. (2021) Molecular basis of the interaction of the human tyrosine phosphatase PTPN3 with the hepatitis B virus core protein. Sci. Rep. 11, 944 10.1038/s41598-020-79580-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kristal Kaan HY, Chan SW, Tan SKJ, Guo F, Lim CJ, Hong W, et al. (2017) Crystal structure of TAZ-TEAD complex reveals a distinct interaction mode from that of YAP-TEAD complex. Sci. Rep. 7:2035. 10.1038/s41598-017-02219-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vargas, R.E., Duong, V.T., Han, H., Ta, A.P., Chen, Y., Zhao, S.et al. (2020) Elucidation of WW domain ligand binding specificities in the hippo pathway reveals STXBP 4 as YAP inhibitor. EMBO J. 39, e102406 10.15252/embj.2019102406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kanelis, V., Rotin, D. and Forman-Kay, J.D. (2001) Solution structure of a Nedd4 WW domain-ENaC peptide complex. Nat. Struct. Biol. 8, 407–412 10.1038/87562 [DOI] [PubMed] [Google Scholar]
- 16.Kumar, M., Gouw, M., Michael, S., Sámano-Sánchez, H., Pancsa, R., Glavina, J.et al. (2020) ELM-the eukaryotic linear motif resource in 2020. Nucleic Acids Res. 48, D296–D306 10.1093/nar/gkz1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paysan-Lafosse, T., Blum, M., Chuguransky, S., Grego, T., Pinto, B.L., Salazar, G.A.et al. (2022) Interpro in 2022. Nucleic Acids Res. 51, D418–D427 10.1093/nar/gkac993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Benjamin, B., Sanchez, A.M., Garg, A., Schwer, B. and Shuman, S. (2021) Structure-function analysis of fission yeast cleavage and polyadenylation factor (CPF) subunit Ppn1 and its interactions with Dis2 and Swd22. PLoS Genet. 17, e1009452 10.1371/journal.pgen.1009452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cherepanov, P., Sun, Z.Y.J., Rahman, S., Maertens, G., Wagner, G. and Engelman, A. (2005) Solution structure of the HIV-1 integrase-binding domain in LEDGF/p75. Nat. Struct. Mol. Biol. 12, 526–532 10.1038/nsmb937 [DOI] [PubMed] [Google Scholar]
- 20.Booth, V., Koth, C.M., Edwards, A.M. and Arrowsmith, C.H. (2000) Structure of a conserved domain common to the transcription factors TFIIS, Elongin A, and CRSP70. J. Biol. Chem. 275, 31266–31268 10.1074/jbc.M002595200 [DOI] [PubMed] [Google Scholar]
- 21.Diebold, M.L., Koch, M., Loeliger, E., Cura, V., Winston, F., Cavarelli, J.et al. (2010) The structure of an Iws1/Spt6 complex reveals an interaction domain conserved in TFIIS, Elongin A and Med26. EMBO J. 29, 3979–3991 10.1038/emboj.2010.272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McDonald, S.M., Close, D., Xin, H., Formosa, T. and Hill, C.P. (2010) Structure and biological importance of the Spn1-Spt6 interaction, and its regulatory role in nucleosome binding. Mol. Cell 40, 725–735 10.1016/j.molcel.2010.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nodelman, I.M., Das, S., Faustino, A.M., Fried, S.D., Bowman, G.D. and Armache, J.P. (2022) Nucleosome recognition and DNA distortion by the Chd1 remodeler in a nucleotide-free state. Nat. Struct. Mol. Biol. 29, 121–129 10.1038/s41594-021-00719-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Han, Y., Reyes, A.A., Malik, S. and He, Y. (2020) Cryo-EM structure of SWI/SNF complex bound to a nucleosome. Nature 579, 452–455 10.1038/s41586-020-2087-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ye, Y., Wu, H., Chen, K., Clapier, C.R., Verma, N., Zhang, W.et al. (2019) Structure of the RSC complex bound to the nucleosome. Science 366, 838–843 10.1126/science.aay0033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patel, A.B., Moore, C.M., Greber, B.J., Luo, J., Zukin, S., Ranish, J.et al. (2019) Architecture of the chromatin remodeler RSC and insights into its nucleosome engagement. eLife 8, e54449 10.7554/eLife.54449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mashtalir, N., Suzuki, H., Farrell, D.P., Sankar, A., Luo, J., Filipovski, M.et al. (2020) A structural model of the endogenous human BAF complex informs disease mechanisms. Cell 183, 802–817.e24 10.1016/j.cell.2020.09.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sibanda, B.L., Chirgadze, D.Y., Ascher, D.B. and Blundell, T.L. (2017) DNA-PKcs structure suggests an allosteric mechanism modulating DNA double-strand break repair. Science 355, 520–524 10.1126/science.aak9654 [DOI] [PubMed] [Google Scholar]
- 29.Sharif, H., Li, Y., Dong, Y., Dong, L., Wang, W.L., Mao, Y.et al. (2017) Cryo-EM structure of the DNA-PK holoenzyme. Proc. Natl Acad. Sci. U.S.A. 114, 7367–7372 10.1073/pnas.1707386114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Groves, M.R. and Barford, D. (1999) Topological characteristics of helical repeat proteins. Curr. Opin. Struct. Biol. 9, 383–389 10.1016/S0959-440X(99)80052-9 [DOI] [PubMed] [Google Scholar]
- 31.Yoshimura, S.H. and Hirano, T. (2016) HEAT repeats - versatile arrays of amphiphilic helices working in crowded environments? J. Cell Sci. 129, 3963–3970 10.1242/jcs.185710 [DOI] [PubMed] [Google Scholar]
- 32.Stothard, P. (2000) The sequence manipulation suite: JavaScript programs for analyzing and formatting protein and DNA sequences. Biotechniques 28, 1102 10.2144/00286ir01 [DOI] [PubMed] [Google Scholar]
- 33.Tesina, P., Cermáková, K., Horejší, M., Procházková, K., Fábry, M., Sharma, S.et al. (2015) Multiple cellular proteins interact with LEDGF/p75 through a conserved unstructured consensus motif. Nat. Commun. 6, 7968 10.1038/ncomms8968 [DOI] [PubMed] [Google Scholar]
- 34.Wei, Y., Redel, C., Ahlner, A., Lemak, A., Johansson-Åkhe, I., Houliston, S.et al. (2022) The MYC oncoprotein directly interacts with its chromatin cofactor PNUTS to recruit PP1 phosphatase. Nucleic Acids Res. 50, 3505–3522 10.1093/nar/gkac138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lux, V., Brouns, T., Čermáková, K., Srb, P., Fábry, M., Mádlíková, M.et al. (2020) Molecular mechanism of LEDGF/p75 dimerization. Structure 28, 1288–1299.e7 10.1016/j.str.2020.08.012 [DOI] [PubMed] [Google Scholar]
- 36.Izban, M.G. and Luse, D.S. (1992) The RNA polymerase II ternary complex cleaves the nascent transcript in a 3′ → 5′ direction in the presence of elongation factor SII. Genes Dev. 6, 1342–1356 10.1101/gad.6.7.1342 [DOI] [PubMed] [Google Scholar]
- 37.Farnung, L., Ochmann, M., Garg, G., Vos, S.M. and Cramer, P. (2022) Structure of a backtracked hexasomal intermediate of nucleosome transcription. Mol. Cell 82, 3126–3134.e7 10.1016/j.molcel.2022.06.027 [DOI] [PubMed] [Google Scholar]
- 38.Langer, D. and Zillig, W. (1993) Putative tflls gene of sulfolobus acidocaldarius encoding an archaeal transcription elongation factor is situated directly downstream of the gene for a small subunit of DNA-dependent RNA polymerase. Nucleic Acids Res. 21, 2251 10.1093/nar/21.9.2251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakanishi, T., Shimoaraiso, M., Kubo, T. and Natori, S. (1995) Structure-function relationship of yeast S-II in terms of stimulation of RNA polymerase II, arrest relief, and suppression of 6-azauracil sensitivity. J. Biol. Chem. 270, 8991–8995 10.1074/jbc.270.15.8991 [DOI] [PubMed] [Google Scholar]
- 40.Ling, Y., Smith, A.J. and Morgan, G.T. (2006) A sequence motif conserved in diverse nuclear proteins identifies a protein interaction domain utilised for nuclear targeting by human TFIIS. Nucleic Acids Res. 34, 2219–2229 10.1093/nar/gkl239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim, B., Nesvizhskii, A.I., Rani, P.G., Hahn, S., Aebersold, R. and Ranish, J.A. (2007) The transcription elongation factor TFIIS is a component of RNA polymerase II preinitiation complexes. Proc. Natl Acad. Sci. U.S.A. 104, 16068–16073 10.1073/pnas.0704573104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Filipovski, M., Soffers, J.H.M., Vos, S.M. and Farnung, L. (2022) Structural basis of nucleosome retention during transcription elongation. Science 376, 1313–1316 10.1126/science.abo3851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu, Y., Bernecky, C., Lee, C.T., Maier, K.C., Schwalb, B., Tegunov, D.et al. (2017) Architecture of the RNA polymerase II-Paf1C-TFIIS transcription elongation complex. Nat. Commun. 8, 15741 10.1038/ncomms15741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vos, S.M., Farnung, L., Boehning, M., Wigge, C., Linden, A., Urlaub, H.et al. (2018) Structure of activated transcription complex Pol II–DSIF–PAF–SPT6. Nature 560, 607–612 10.1038/s41586-018-0440-4 [DOI] [PubMed] [Google Scholar]
- 45.Ehara, H., Kujirai, T., Fujino, Y., Shirouzu, M., Kurumizaka, H. and Sekine, S. (2019) Structural insight into nucleosome transcription by RNA polymerase II with elongation factors. Science 363, 744–747 10.1126/science.aav8912 [DOI] [PubMed] [Google Scholar]
- 46.Yoh, S.M., Lucas, J.S. and Jones, K.A. (2008) The Iws1:Spt6:CTD complex controls cotranscriptional mRNA biosynthesis and HYPB/Setd2-mediated histone H3K36 methylation. Genes Dev. 22, 3422–3434 10.1101/gad.1720008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reim, N.I., Chuang, J., Jain, D., Alver, B.H., Park, P.J. and Winston, F. (2020) The conserved elongation factor Spn1 is required for normal transcription, histone modifications, and splicing in saccharomyces cerevisiae. Nucleic Acids Res. 48, 10241–10258 10.1093/nar/gkaa745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yoh, S.M., Cho, H., Pickle, L., Evans, R.M. and Jones, K.A. (2007) The Spt6 SH2 domain binds Ser2-P RNAPII to direct Iws1-dependent mRNA splicing and export. Genes Dev. 21, 160–174 10.1101/gad.1503107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ehara, H., Kujirai, T., Shirouzu, M., Kurumizaka, H. and Sekine, S.I. (2022) Structural basis of nucleosome disassembly and reassembly by RNAPII elongation complex with FACT. Science 377, eabp9466 10.1126/science.abp9466 [DOI] [PubMed] [Google Scholar]
- 50.Tsai, K.L., Tomomori-Sato, C., Sato, S., Conaway, R.C., Conaway, J.W. and Asturias, F.J. (2014) Subunit architecture and functional modular rearrangements of the transcriptional mediator complex. Cell 157, 1430–1444 10.1016/j.cell.2014.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Takahashi, H., Parmely, T.J., Sato, S., Tomomori-Sato, C., Banks, C.A.S., Kong, S.E.et al. (2011) Human mediator subunit MED26 functions as a docking site for transcription elongation factors. Cell 146, 92–104 10.1016/j.cell.2011.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lens, Z., Cantrelle, F.X., Peruzzini, R., Hanoulle, X., Dewitte, F., Ferreira, E.et al. (2017) Solution structure of the N-terminal domain of mediator subunit MED26 and molecular characterization of Its interaction with EAF1 and TAF7. J. Mol. Biol. 429, 3043–3055 10.1016/j.jmb.2017.09.001 [DOI] [PubMed] [Google Scholar]
- 53.Van Nuland, R., Van Schaik, F.M.A., Simonis, M., Van Heesch, S., Cuppen, E., Boelens, R.et al. (2013) Nucleosomal DNA binding drives the recognition of H3K36-methylated nucleosomes by the PSIP1-PWWP domain. Epigenetics Chromatin 6, 12 10.1186/1756-8935-6-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang, H., Farnung, L., Dienemann, C. and Cramer, P. (2020) Structure of H3K36-methylated nucleosome–PWWP complex reveals multivalent cross-gyre binding. Nat. Struct. Mol. Biol. 27, 8–13 10.1038/s41594-019-0345-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Čermaková, K., Tesina, P., Demeulemeester, J., El Ashkar, S., Méreau, H., Schwaller, J.et al. (2014) Validation and structural characterization of the LEDGF/p75-MLL interface as a new target for the treatment of MLL-dependent leukemia. Cancer Res. 74, 5139–5151 10.1158/0008-5472.CAN-13-3602 [DOI] [PubMed] [Google Scholar]
- 56.Van Belle, S., Ashkar, S.E., Čermáková, K., Matthijssens, F., Goossens, S., Canella, A.et al. (2021) Unlike its paralog LEDGF/p75, HRP-2 is dispensable for MLL- R leukemogenesis but important for leukemic cell survival. Cells 10, 192 10.3390/cells10010192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhu, X., Lan, B., Yi, X., He, C., Dang, L., Zhou, X.et al. (2020) HRP2-DPF3a-BAF complex coordinates histone modification and chromatin remodeling to regulate myogenic gene transcription. Nucleic Acids Res. 48, 6563–6582 10.1093/nar/gkaa441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.LeRoy, G., Oksuz, O., Descostes, N., Aoi, Y., Ganai, R.A., Kara, H.O.et al. (2019) LEDGF and HDGF2 relieve the nucleosome-induced barrier to transcription in differentiated cells. Sci. Adv. 5, eaay3068 10.1126/sciadv.aay3068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Aso, T., Haque, D., Barstead, R.J., Conaway, R.C. and Conaway, J.W. (1996) The inducible elongin A elongation activation domain: Structure, function and interaction with the elongin BC complex. EMBO J. 15, 5557–5566 10.1002/j.1460-2075.1996.tb00940.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yasukawa, T., Kamura, T., Kitajima, S., Conaway, R.C., Conaway, J.W. and Aso, T. (2008) Mammalian Elongin A complex mediates DNA-damage-induced ubiquitylation and degradation of Rpb1. EMBO J. 27, 3256–3266 10.1038/emboj.2008.249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang, Y., Hou, L., Behfar Ardehali, M., Kingston, R.E. and Dynlacht, B.D. (2021) Elongin A regulates transcription in vivo through enhanced RNA polymerase processivity. J. Biol. Chem. 296, 100170 10.1074/jbc.RA120.015876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Weems, J.C., Slaughter, B.D., Unruh, J.R., Hall, S.M., McLaird, M.B., Gilmore, J.M.et al. (2015) Assembly of the Elongin A ubiquitin ligase is regulated by genotoxic and other stresses. J. Biol. Chem. 290, 15030–15041 10.1074/jbc.M114.632794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yasukawa, T., Bhatt, S., Takeuchi, T., Kawauchi, J., Takahashi, H., Tsutsui, A.et al. (2012) Transcriptional elongation factor elongin A regulates retinoic acid-induced gene expression during neuronal differentiation. Cell Rep. 2, 1129–1136 10.1016/j.celrep.2012.09.031 [DOI] [PubMed] [Google Scholar]
- 64.He, Y., Sato, S., Tomomori-Sato, C., Chen, S., Goode, Z.H., Conaway, J.W.et al. (2021) Elongin functions as a loading factor for mediator at ATF6α-regulated ER stress response genes. Proc. Natl Acad. Sci. U.S.A. 118, e2108751118 10.1073/pnas.2108751118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cortazar, M.A., Sheridan, R.M., Erickson, B., Fong, N., Glover-Cutter, K., Brannan, K.et al. (2019) Control of RNA Pol II speed by PNUTS-PP1 and Spt5 dephosphorylation facilitates termination by a “sitting duck torpedo” mechanism. Mol. Cell 76, 896–908.e4 10.1016/j.molcel.2019.09.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kecman, T., Kuś, K., Heo, D.H., Duckett, K., Birot, A., Liberatori, S.et al. (2018) Elongation/Termination factor exchange mediated by PP1 phosphatase orchestrates transcription termination. Cell Rep. 25, 259–269.e5 10.1016/j.celrep.2018.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Parua, P.K., Booth, G.T., Sansó, M., Benjamin, B., Tanny, J.C., Lis, J.T.et al. (2018) A Cdk9-PP1 switch regulates the elongation-termination transition of RNA polymerase II. Nature 558, 460–464 10.1038/s41586-018-0214-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Landsverk, H.B., Kirkhus, M., Bollen, M., Küntziger, T. and Collas, P. (2005) PNUTS enhances in vitro chromosome decondensation in a PP1-dependent manner. Biochem. J. 390, 709–717 10.1042/BJ20050678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lee, J.H., You, J., Dobrota, E. and Skalnik, D.G. (2010) Identification and characterization of a novel human PP1 phosphatase complex. J. Biol. Chem. 285, 24466–24476 10.1074/jbc.M110.109801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.De Castro, I.J., Budzak, J., Di Giacinto, M.L., Ligammari, L., Gokhan, E., Spanos, C.et al. (2017) Repo-Man/PP1 regulates heterochromatin formation in interphase. Nat. Commun. 8, 14048 10.1038/ncomms14048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Grallert, A., Boke, E., Hagting, A., Hodgson, B., Connolly, Y., Griffiths, J.R.et al. (2015) A PP1-PP2A phosphatase relay controls mitotic progression. Nature 517, 94–98 10.1038/nature14019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wu, J.Q., Guo, J.Y., Tang, W., Yang, C.S., Freel, C.D., Chen, C.et al. (2009) PP1-mediated dephosphorylation of phosphoproteins at mitotic exit is controlled by inhibitor-1 and PP1 phosphorylation. Nat. Cell Biol. 11, 644–651 10.1038/ncb1871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dingar, D., Tu, W.B., Resetca, D., Lourenco, C., Tamachi, A., De Melo, J.et al. (2018) MYC dephosphorylation by the PP1/PNUTS phosphatase complex regulates chromatin binding and protein stability. Nat. Commun. 9, 3502 10.1038/s41467-018-05660-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Landsverk, H.B., Sandquist, L.E., Bay, L.T.E., Steurer, B., Campsteijn, C., Landsverk, O.J.B.et al. (2020) WDR82/PNUTS-PP1 prevents transcription-replication conflicts by promoting RNA polymerase II degradation on chromatin. Cell Rep. 33, 108469 10.1016/j.celrep.2020.108469 [DOI] [PubMed] [Google Scholar]
- 75.Liu, Z., Wu, A., Wu, Z., Wang, T., Pan, Y., Li, B.et al. (2022) TOX4 facilitates promoter-proximal pausing and C-terminal domain dephosphorylation of RNA polymerase II in human cells. Commun. Biol. 5, 300 10.1038/s42003-022-03214-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Elkhaligy, H., Balbin, C.A. and Siltberg-Liberles, J. (2022) Comparative analysis of structural features in SLiMs from eukaryotes, bacteria, and viruses with importance for host-pathogen interactions. Pathogens 11, 583 10.3390/pathogens11050583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hraber, P., O'Maille, P.E., Silberfarb, A., Davis-Anderson, K., Generous, N., McMahon, B.H.et al. (2020) Resources to discover and Use short linear motifs in viral proteins. Trends Biotechnol. 38, 113–127 10.1016/j.tibtech.2019.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schrijvers, R., Vets, S., De Rijck, J., Malani, N., Bushman, F.D., Debyser, Z.et al. (2012) HRP-2 determines HIV-1 integration site selection in LEDGF/p75 depleted cells. Retrovirology 9, 84 10.1186/1742-4690-9-84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Busschots, K., Vercammen, J., Emiliani, S., Benarous, R., Engelborghs, Y., Christ, F.et al. (2005) The interaction of LEDGF/p75 with integrase is lentivirus-specific and promotes DNA binding. J. Biol. Chem. 280, 17841–7 10.1074/jbc.M411681200 [DOI] [PubMed] [Google Scholar]
- 80.Christ, F., Voet, A., Marchand, A., Nicolet, S., Desimmie, B.A., Marchand, D.et al. (2010) Rational design of small-molecule inhibitors of the LEDGF/p75-integrase interaction and HIV replication. Nat. Chem. Biol. 6, 442–448 10.1038/nchembio.370 [DOI] [PubMed] [Google Scholar]
- 81.Marx, A., Luebke, A.M., Clauditz, T.S., Steurer, S., Fraune, C., Hube-Magg, C.et al. (2020) Upregulation of phosphatase 1 nuclear-targeting subunit (PNUTS) Is an independent predictor of poor prognosis in prostate cancer. Dis. Markers 2020, 7050146 10.1155/2020/7050146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cermakova, K., Weydert, C., Christ, F., De Rijck, J. and Debyser, Z. (2016) Lessons learned: HIV points the way towards precision treatment of mixed-lineage leukemia. Trends Pharmacol. Sci. 37, 660–671 10.1016/j.tips.2016.05.005 [DOI] [PubMed] [Google Scholar]
- 83.Yokoyama, A. and Cleary, M.L. (2008) Menin critically links MLL proteins with LEDGF on cancer-associated target genes. Cancer Cell 14, 36–46 10.1016/j.ccr.2008.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Méreau, H., De Rijck, J., Čermáková, K., Kutz, A., Juge, S., Demeulemeester, J.et al. (2013) Impairing MLL-fusion gene-mediated transformation by dissecting critical interactions with the lens epithelium-derived growth factor (LEDGF/p75). Leukemia 27, 1245–1253 10.1038/leu.2013.10 [DOI] [PubMed] [Google Scholar]
- 85.Chan, T.S.Y., Hawkins, C., Krieger, J.R., McGlade, C.J. and Huang, A. (2016) JPO2/CDCA7L and LEDGF/p75 are novel mediators of pi3k/akt signaling and aggressive phenotypes in medulloblastoma. Cancer Res. 76, 2802–2812 10.1158/0008-5472.CAN-15-2194 [DOI] [PubMed] [Google Scholar]
- 86.Gao, K., Xu, C., Jin, X., Wumaier, R., Ma, J., Peng, J.et al. (2015) HDGF-related protein-2 (HRP-2) acts as an oncogene to promote cell growth in hepatocellular carcinoma. Biochem. Biophys. Res. Commun. 458, 849–855 10.1016/j.bbrc.2015.02.042 [DOI] [PubMed] [Google Scholar]
- 87.Békés, M., Langley, D.R. and Crews, C.M. (2022) PROTAC targeted protein degraders: the past is prologue. Nat. Rev. Drug Discov. 21, 181–200 10.1038/s41573-021-00371-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Samarasinghe, K.T.G. and Crews, C.M. (2021) Targeted protein degradation: A promise for undruggable proteins. Cell Chem. Biol. 28, 934–951 10.1016/j.chembiol.2021.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cermakova, K. and Courtney Hodges, H. (2018) Next-generation drugs and probes for chromatin biology: From targeted protein degradation to phase separation. Molecules 23, 1958 10.3390/molecules23081958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Boike, L., Cioffi, A.G., Majewski, F.C., Co, J., Henning, N.J., Jones, M.D.et al. (2021) Discovery of a functional covalent ligand targeting an intrinsically disordered cysteine within MYC. Cell Chem. Biol. 28, 4–13.e17 10.1016/j.chembiol.2020.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tompa, P., Davey, N.E., Gibson, T.J. and Babu, M.M. (2014) A million peptide motifs for the molecular biologist. Mol. Cell 55, 161–169 10.1016/j.molcel.2014.05.032 [DOI] [PubMed] [Google Scholar]