Abstract
The class II transactivator (CIITA) is induced by gamma interferon (IFN-γ) and activates major histocompatibility complex class II; however, this report shows it suppresses other genes. An N-terminal 36 amino acids of CIITA mediates suppression of the collagen α2(I) promoter via binding to CREB-binding protein (CBP). Reconstitution of cells with CBP reverts this suppression. IFN-γ is known to inhibit collagen gene expression; to test if CIITA mediates this gene suppression, a mutant cell line defective in CIITA induction but not in the activation of STAT1/JAK/IRF-1 is studied. IFN-γ suppression of the collagen promoter and the endogenous gene is observed in the wild-type control but not in the mutant line. Suppression is restored when CIITA is introduced. Other targets of CIITA-mediated promoter suppression include interleukin 4, thymidine kinase, and cyclin D1.
The class II transactivator (CIITA) is a master regulator of major histocompatibility complex class II (MHC-II), Ii, and DM genes (8, 9, 10, 55, 56). CIITA was initially identified by complementation of an HLA-DR− mutant B-cell line, RJ2.2.5 (55), which was created by mutagenesis followed by negative immunoselection for MHC-II-defective cells (1). CIITA is important for the constitutive expression of MHC-II in B cells and dendritic cells and the cytokine induction of these genes in a variety of other cell types. Gamma interferon (IFN-γ) is a prime example of a cytokine which induces CIITA and subsequently MHC-II expression.
CIITA is a transcriptional coactivator that does not bind DNA (55) but does interact with RFX (consisting of RFX5-RFXANK-RFXAP), CREB, and NF-Y (NF-YA/B/C) (65), all of which directly bind to the X and Y element of the MHC-II promoters. Specifically, CIITA interacts with both RFX5 and RFXANK and NF-YB and NF-YC but not with NF-YA. In addition, CIITA also interacts with the histone acetyltransferase (HAT) CREB-binding protein (CBP) (20, 30), which can acetylate histone at lysine residues to allow gene activation (13, 19, 31, 57, 59, 62). Thus, CIITA appears to be a focal point of interaction for both DNA-binding proteins specific for the MHC-II promoters and for HATs to allow chromatin opening.
CIITA expression is strongly induced by IFN-γ (56). Two of its promoters, promoter III and promoter IV, are responsible for IFN-γ inducibility (39, 42, 43). Promoter III contains a proximal sequence that is responsible for the constitutive expression of CIITA in B cells and a distal sequence containing STAT1-responsive element which confers IFN-γ responsiveness. Promoter IV contains the major IFN-responsive sequence regulated by both STAT1 and IRF-1.
An invaluable tool to study IFN-γ pathways has been the use of mutant cell lines. Mutant lines which are defective in the IFN-γ induction of MHC-II genes but not defective in the induction of other IFN-γ functions have been useful in elucidating this pathway (9, 37). The G3A cell line was derived from the commonly used IFN-γ-responsive parental cell line 2fTGH (10). IFN-γ treatment of G3A cells causes the normal induction of most responsive genes but causes a selective failure to induce MHC-II, Ii, and DM. This is caused by a defect in the IFN induction of CIITA and places CIITA downstream of the JAK/STAT pathway.
In addition to its well-known function in inducing gene expression, IFN-γ also suppresses the expression of a number of genes. There are several examples of IFN-γ-suppressible promoters or genes, and these can be loosely grouped as the following: genes important for cell proliferation and cell differentiation, such as cyclin (51) and c-myc and N-myc (59); certain cytokine genes expressed by the TH2 subpopulation, such as interleukin 4 (IL-4) (15) and IL-10 (17); and genes coding for matrix proteins, such as the collagen (26, 29, 58) and proteoglycan (16, 50) genes. The mechanism by which IFN-γ causes gene suppression is not well known, and the present study shows that one mechanism involves the CIITA molecule.
The rationale for this study stems from a previous report, where we and others detected interactions of CIITA with a number of DNA-binding proteins that bind the MHC-II promoters (65). This prompted us to determine if CIITA can selectively induce the expression of MHC-II but simultaneously sequester DNA-binding proteins from their other gene targets. The squelching of transcription factors has been proposed before (4, 21, 34, 43); however, there are few good physiologic examples of squelching. In this report, we determined that CIITA can squelch the expression of NF-Y-dependent promoters. However, this was not reversed by the addition of NF-Y, and the CIITA domain that is required for this effect does not match the CIITA–NF-Y interaction site. Instead, the region within CIITA that is required for squelching maps to a 36-amino-acid (aa) region which binds the CBP. Significantly, squelching was observed under biologic conditions where the effects of endogenous CIITA on endogenous collagen gene expression were examined. These experiments show that this small peptide domain is a potent inhibitor of CBP function and elucidate a possible mechanism by which IFN-γ suppresses gene expression.
MATERIALS AND METHODS
Cell cultures.
NIH 3T3 cells were maintained in Dulbecco's modified Eagle medium (DMEM) (Sigma) supplemented with 10% Colorado calf serum (Colorado Serum Company). 2fTGH and G3A cells were cultured in DMEM (Sigma) supplemented with 10% fetal bovine serum (FBS) as previously described (10). Cells were grown in an incubator at 37°C and 5% CO2.
Plasmid constructs.
The PCR-amplified collagen α2(I) promoter from −357 to +55 bp (48) was introduced into pGL3-CAT basic and pGL3-luciferase basic vectors (Promega) at the EcoRV cloning site. The orientation was checked by restriction digestion and direct DNA sequencing. pGL3-DRA-CAT and pGL3-DRA-Luc reporter constructs bear 300 bp of the DRA promoter (10, 11). IL-4–chloramphenicol acetyltransferase (CAT) (32) and TK-CAT (7) were kindly provided by M. Li-Weber, German Cancer Research Center, Heidelberg, Germany. CD1-Luc was a generous gift from Albert Baldwin's Lab, UNC Lineberger Cancer Center (24). For mutagenesis studies FlagCIITA (11) was used as a parent template, and stop codons were introduced at residue position 335 to generate FlagCIITA(1-335) using the QuickChange mutagenesis protocol (Stratagene). FlagCIITA(1-297), FlagCIITA(1-251), FlagCIITA(1-215), FlagCIITA(1-177), FlagCIITA(1-139), FlagCIITA(1-94), and FlagCIITA(1-58) were constructed using FlagCIITA as a template together with a common upper-strand primer, GACCCAAAGCTTGGTACCGAG, that is contained in pcDNA3 and a series of lower-strand primers that distribute along the human CIITA sequences at the designated positions. CIITA(334-1130) was generated by introducing a BglI site into FlagCIITA using the Stratagene QuickChange protocol and inserting the BglII-EcoRI fragment into the BglII-EcoRI-digested pcDNA3HisC expression vector (Invitrogen). CIITA(59-94)Δ was generated by overlapping PCR by standard procedure using pcDNA3.1FlagCIITA as template with the following primers: (i) 5′ GGCGTGTACGGTGGGAGGTC (on pcDNA3.1), (ii) 5′ CCAGTTCCGCGATATTGGCATATCCAGCCAGGTCCTACTGGTC, (iii) 5′ GACCAGATGGACCTGGCTGGATATGCCAATATCGCGGAACTGG, and (iv) 5′ TGGTGGGGACAAACTGGATGGG. The PCR products of primers i and ii and primers iii and iv were combined and subjected to a second-round PCR using primers i and iv. The fused PCR product was reintroduced into pcDNA3.1FlagCIITA at NheI and Bsu36I sites. FlagNF-YA, FlagNF-YB, and FlagNF-YC have been previously described (65) and consist of a full-length gene coding for an individual NF-Y subunit tagged at the 5′ end with the Flag sequence. CMV5-CBP was kindly provided by R. H. Goodman, Vollum Institute, Oregon Health Science University (13). All plasmids were purified using a Qiagen column (Qiagen).
Transient transfection and CAT and/or luciferase assay.
Ninety thousand to 1.2 × 105 NIH 3T3 cells or 1.5 × 105 2fTGH or G3A cells were plated in 6-well plates (Fisher) and cultured for 18 h before transfection. One microgram or various amounts (as described in the text or in the figure legends) of reporter plasmid was cotransfected with 1.0 μg or various amounts (as described in the text or in the figure legends) of expression vector or its empty vector. Transfected cells were incubated with or without 500 U of IFN-γ/ml. Cells were harvested 48 h later and assayed for CAT and/or luciferase activity (Promega) following standard procedures (10, 43) with the same amount of protein as determined by the Bradford protein assay (Bio-Rad).
Immunoprecipitation and Western blot analyses of in vivo protein-protein interaction.
COS7 cells were cultured in a 37°C incubator with 5% CO2 in DMEM (Sigma) supplemented with 10% FBS. Cells were plated at 9 × 105 cells/100-mm-diameter dish and allowed to grow for 18 h. Cells were cotransfected with 3 μg of each plasmid as described in the figure legend for each experiment using Fugene 6 (Boehringer Mannheim) and following the manufacturer's instructions. After 30 to 40 h of culture, cells were washed twice with 1× phosphate-buffered saline and lysed with 1.5 ml of cold radioimmunoprecipitation assay (RIPA) buffer (49) (0.1% sodium dodecyl sulfate [SDS], 1% NP-40, 1% deoxycholate, 150 mM NaCl, 2 mM EDTA, 0.01 M sodium phosphate [pH 7.2], and 50 mM NaF) supplemented with a tablet of Complete protease inhibitor cocktail (Boehringer Mannheim) per 50 ml of solution. Immunoprecipitation and Western blotting were performed following standard procedures (47, 49, 54). Blots were detected by ECL (Pierce) using Kodak X-OMAT film. Detailed information is available upon request.
Total RNA isolation and RT-PCR.
Total RNA was extracted from the culture cells with the RNeasy mini-total RNA isolation kit (Qiagen) according to the manufacturer's instruction. The primers used for reverse transcriptase (RT)-PCR are as follows: for human collagen α2(I), 5′ GACTCAGCCACCCAGAGTGG and 5′ TGGTCAGCACCACCGATGTCC, which gave a 440-bp band; for human glyceraldehyde-3-phosphate dehydrogenase (GAPDH), 5′ GAGGGGCCATCCACAGTCTTC and 5′ CAAAAGGGTCATCATCTCTGC, which gave a 228-bp band. RT-PCR was performed using Access RT-PCR System (Promega) according to the manufacturer's instructions. One microgram of total RNA was used for each reaction and was subjected to 20 to 30 cycles of amplification. The linear phase of the amplification process was first determined for each primer set and then was used in all the experiments. Ten microliters of RT-PCR product was subjected to 1.5% agarose gel electrophoresis and stained with ethidium bromide for visualization and digital recording. One and a half micrograms of total RNA was reverse transcribed with Moloney murine leukemia virus reverse transcriptase (Gibco-BRL) in 50 μl of reaction mixture, and 3 μl of each reaction mixture was subjected to PCR amplification in the presence of 5 μCi of [α-32P]dCTP using primers 5′ CCTTACTGGTGCCAAGGGTGCTG and 5′ CCAGGGAATCCAATGTTGCCA, which yield a 577-bp product. As a positive control, samples of cDNA were also amplified in parallel with a pair of human GAPDH-specific primers, which yield a 220-bp product. Ten or 20 μl of the PCR mixtures were fractionated on a nondenatured polyacrylamide gel, visualized by autoradiography with Kodak X-ray film, and quantitated with NIH Image software (National Institutes of Health).
RESULTS
CIITA inhibits the transcriptional activation of the collagen promoter which contains an NF-Y binding site.
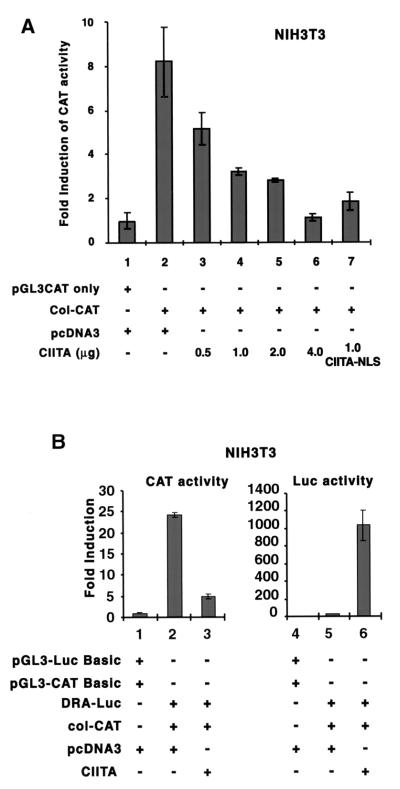
A previous report shows that CIITA directly binds to the NF-Y protein (65). One hypothesis is that CIITA may serve as a scaffolding protein that binds NF-Y and enriches its local concentration at the site of an MHC-II promoter. Accordingly, less NF-Y would be available for the transcription of other promoters. To examine this possibility, we determined if CIITA can inhibit promoters containing an NF-Y binding site. Two of the classical promoters which originally defined the NF-Y/CBF cognate binding site are those of MHC-II and collagen α2(I) (5, 18, 35, 36). To determine if CIITA can inhibit the activation of the collagen promoter, we cotransfected NIH 3T3 cells with a collagen α2(I) promoter CAT reporter construct (col-CAT) and increasing amounts of a CIITA expression plasmid (Fig. 1A) (14). CIITA inhibited col-CAT transcription activity in a dose-dependent manner (lanes 3 to 6) until the activity was extinguished. The process of inhibition could take place in the nucleus, as CIITA linked to a strong nuclear localization sequence (NLS) from simian virus 40 produced efficient suppression of col-CAT activity (compare lanes 7 and 4). This CIITA-NLS construct is expressed almost exclusively in the nucleus (data not shown). In the same cell where the suppression of col-CAT activity was observed (Fig. 1B, left panel), CIITA activated DRA-promoter-driven luciferase reporter expression (Fig. 1B, right panel). Thus, CIITA has dual effects: the suppression of a collagen promoter and the activation of an MHC-II promoter.
FIG. 1.
CIITA inhibits the collagen promoter in a dose-dependent manner. NIH 3T3 cells (1.1 × 105) were plated in a 6-well dish 18 h before transfection. (A) Collagen α2(I) CAT reporter gene (col-CAT) or an empty vector, pGL3-CAT, was transiently cotransfected with various amounts of CIITA or with an empty vector, pcDNA3. Cells were harvested 48 h later, and extracts were assayed for activity. A representative of three experiments is shown. The CAT activities were normalized based on the activity detected in the pGL3-CAT basic group. Error bars represent means ± standard errors. CIITA-NLS is the CIITA molecule with a simian virus 40 NLS at its C-terminal end. (B) The same cells were cotransfected with CIITA, col-CAT, and DRA-Luc reporter constructs. col-CAT monitors the effect of CIITA on the collagen promoter, and DRA-luc monitors the effect of CIITA on the DRA promoter. The panel on the left shows CAT activity, while the panel on the right shows luciferase activity. Experiments were repeated three times.
The inhibition of the collagen promoter by CIITA is not reversed by the addition of NF-Y proteins.
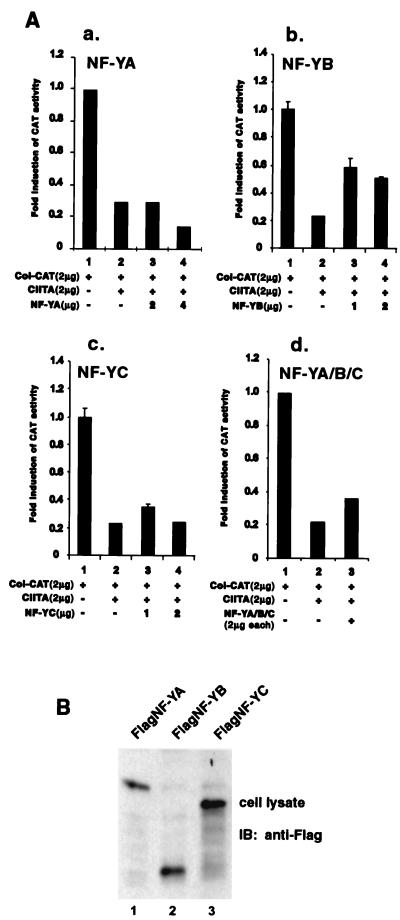
As described earlier, one of the premises for this study is the assumption that CIITA interacts with NF-Y proteins, thereby reducing their availability for the transcription of other promoters. If this were the case, the introduction of NF-Y expression vectors should overcome this sequestration. The collagen promoter activity was reduced by the presence of CIITA as described earlier (compare lanes 1 and 2 in Fig. 2A, panels a, b, c, and d). The addition of NF-YA had no positive effect on the collagen promoter (Fig. 2A, panel a, compare lane 2 to lanes 3 and 4). The addition of NF-YB resulted in a modest restoration of promoter activity (Fig. 2A, panel b, compare lane 2 to lanes 3 and 4), and the addition of NF-YC had less effect (Fig. 2A, panel c, compare lane 2 to lanes 3 and 4). The addition of all three NF-Y subunits together also had a minor effect (Fig. 2A, panel d). In all these experiments, the optimal results with the optimal quantities of NF-Y subunits are shown. This suggests that the direct sequestering of NF-Y is unlikely the primary, or only, mechanism for CIITA-mediated gene suppression. To verify that NF-Y proteins were expressed, lysates were isolated from cells that were separately transfected with NF-YA, NF-YB, or NF-YC. A Western analysis was performed, and it showed the robust expression of these proteins (Fig. 2B).
FIG. 2.
The addition of NF-Y subunits does not reverse CIITA-mediated suppression of the collagen α2(I) promoter. (A) The four panels show that the addition of exogenous NF-YA, NF-YB, NF-YC, or a combination of the three had little to modest effects on CIITA-mediated suppression of the collagen promoter. The NF-Y subunits, alone or in combination, were examined for their capacity to reverse CIITA-mediated suppression of the collagen promoter. All the experiments were repeated three times. (B) The expression of NF-YA, NF-YB, and NF-YC in COS7 cell transfectants was confirmed by immunoblotting (IB) using anti-Flag M5 antibody.
Inhibition of collagen α2(I) promoter activity by CIITA requires the N-terminal 59 to 94 aa of CIITA.
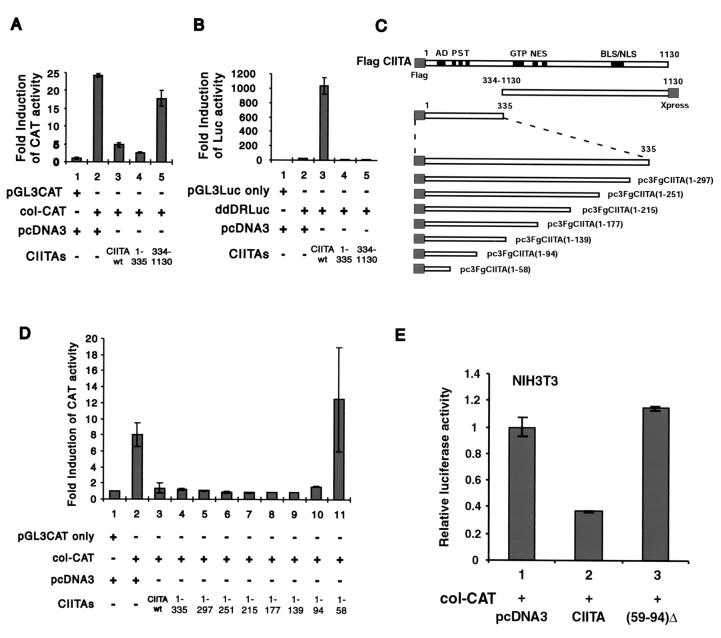
To elucidate the possible mechanism by which CIITA mediates gene suppression, efforts were undertaken to define the domain(s) within CIITA that is responsible for the suppressive activity. Two critical constructs were tested. One constitutes the N-terminal one-third of CIITA [designated Flag CIITA(1-335)], and the other represents the C-terminal two-thirds of CIITA [designated CIITA(334-1130)]. Each mutant, wild-type CIITA, or empty vector pcDNA3 was cotransfected with col-CAT into target cells. As shown in Fig. 3A, the collagen promoter was suppressed by Flag CIITA(1-335) as efficiently as full-length CIITA (lanes 2 to 4). In contrast, CIITA(334-1130) had little effect on collagen promoter activation. Again, full-length CIITA vigorously activated an MHC-II promoter (Fig. 3B, lane 3), but neither the N terminus nor the C terminus alone resulted in MHC-II promoter activation. This is expected on the basis of previous studies (11).
FIG. 3.
The N-terminal 59 to 94 aa of CIITA is responsible for the inhibition of collagen α2(I) promoter activities. (A) The collagen α2(I) CAT reporter gene or its empty vector, pGL3-CAT, was transiently cotransfected with the N terminus or C terminus of CIITA [designated Flag CIITA(1-335) and CIITA(334-1130), respectively] or its empty vector, pcDNA3, into NIH 3T3 cells as described in the legend to Fig. 1. A portion of the cell lysate was subjected to CAT assay for collagen α2(I) promoter activity. (B) DRA-Luc reporter gene or its empty vector, pGL3-Luc, was transfected into the same cells as those described for panel A. Luciferase (Luc) activity was used to show DRA promoter activity. CAT (A) or luciferase (B) activities were normalized based on the activity of the pGL3-CAT group (A) or the pGL3-Luc group (B), respectively. (C) A series of CIITA deletion constructs are shown. AD, acidic domain; PST, proline-, serine-, and threonine-rich domain; GTP, GTP binding site; NES, nuclear export sequence; BLS/NLS, nuclear localization sequence. (D) The experiment depicted was performed in a manner identical to that of the experiment depicted in panel A. The collagen α2(I) CAT reporter gene or its empty vector, pGL3-CAT, was transiently cotransfected with CIITA, its empty vector control, pcDNA3, or a series of C terminus deletion mutants, as depicted in the panel C. The results are expressed as relative fold induction of CAT activity over the negative control pGL3-CAT basic vector. Error bars represent means ± standard errors. Data shown have been reproduced three times. (E) The collagen α2(I) CAT reporter gene was transiently cotransfected with empty vector, wild-type (wt) CIITA, or CIITA(59-94)Δ.
To map the region within the N-terminal 335 aa of CIITA that is required for this suppressive activity, a series of CIITA mutants representing nested deletions of approximately 30 aa each were constructed (Fig. 3C). These mutants, wild-type CIITA, or empty vector pcDNA3 were cotransfected with the col-CAT reporter gene (Fig. 3D). Full-length CIITA suppressed most of the collagen promoter activity (lane 3), as did Flag CIITA(1-335) (lane 4). Further deletions of CIITA up to Flag CIITA(1-94) still caused a similar degree of suppression; however, the suppressive function was no longer observed with Flag CIITA(1-58), which contains the final N-terminal 58 aa. All of these deletion constructs were nonfunctional for the MHC-II promoter, as expected (data not shown).
To more definitively show that residues 59 to 94 are important for gene suppression, an internal deletion construct that lacks these residues was constructed (Fig 3E). This construct, CIITA(59-94)Δ, did not inhibit the collagen α2(I) promoter, while wild-type CIITA did (compare lanes 2 and 3). Taken together, the results shown in Fig. 3 strongly support the importance of residues 59 to 94 in suppressing the collagen promoter. It is noteworthy that a recent study shows that residues 518 to 642 of CIITA associate with NF-YB, while residues 218 to 335 associate with NF-YC (65). These residues lie outside of the region (residues 59 to 94) defined here as being important for gene suppression. This supports our earlier conclusion that the squelching of NF-Y is not likely the primary mechanism by which CIITA causes gene suppression.
CIITA residues 59 to 94 contain sequences that interact with the CBP.
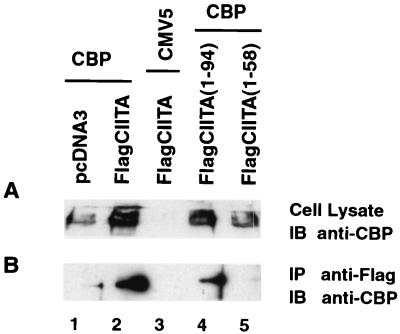
CBP interacts with CIITA and enhances the expression of MHC-II genes. Although fine mapping of the CIITA domain which interacts with CBP has not been performed, the N-terminal portion of the molecule is known to be important for this interaction (20, 30). Therefore, we tested if the inhibition of collagen α2(I) promoter activity by CIITA is mediated by the physical association of CIITA with CBP. According to this model, CIITA would squelch CBP such that the protein is unavailable for the activation of other CBP-dependent promoters. Experiments were performed to determine whether CBP interacts with the CIITA sequence that mediates gene suppression. COS7 cells were cotransfected with full-length or truncated forms of CIITA linked to the Flag epitope along with a CBP expression vector or empty vector control. The lysates were immunoprecipitated with anti-Flag antibodies to bring down CIITA or its truncated variants. The immunoprecipitates were analyzed for the presence of CBP by anti-CBP Western blotting. A fraction of the lysate was also tested for the expression of CBP. As shown in Fig. 4, CBP was coprecipitated with full-length CIITA (compare lanes 1 and 2 of Fig. 4B) and Flag CIITA(1-94) but not Flag CIITA(1-58). These results indicate that the CIITA domain important for interaction with CBP corresponds to the domain important for the inhibition of collagen α2(I) transcriptional activity.
FIG. 4.
CBP interacts with CIITA at its N terminus. (A) COS7 cells (9 × 105) were cotransfected with 3 μg of FlagCIITA or FlagCIITA(1-58) or FlagCIITA(1-94) or its corresponding control vector, pcDNA3, and 3 μg of CMV5-CBP or its empty vector, CMV5, as indicated. Cells were lysed with 1.5 ml of RIPA buffer 36 h after transfection, and 25 μl of the samples was subjected to SDS–10% polyacrylamide gel electrophoresis resolution. The expression of CBP was analyzed by immunoblotting (IB) using the rabbit anti-CBP antibody. (B) The cell lysate described above was immunoprecipitated (IP) with anti-Flag M2-agarose at 4°C overnight. The association of CBP with wild-type FlagCIITA (lane 2) and mutant FlagCIITA(1-94) was revealed by immunoblotting using the rabbit anti-CBP antibody.
The addition of exogenous CBP rescues collagen α2(I) promoter from the suppressive effects of CIITA.
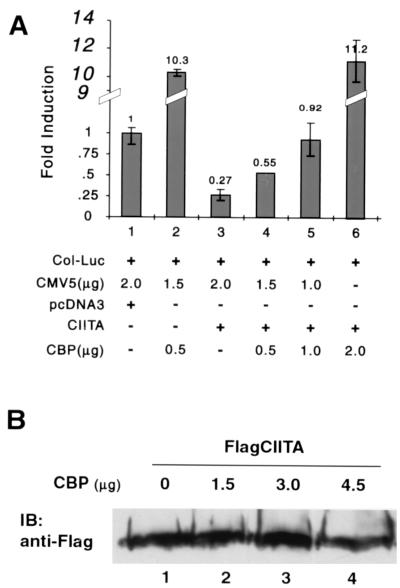
To investigate the biological significance of these physical interactions and to determine if CBP is the target of CIITA-mediated gene suppression, increasing doses of a CBP expression vector were transfected into cells. The col-Luc (collagen promoter, luciferase reporter) plasmid and the CIITA expression vector were also cotransfected into these cells. The assumption is that if CIITA sequesters CBP, then the addition of exogenous CBP should overcome the suppressive function of CIITA. As shown in Fig. 5A, CBP alone activated the collagen α2(I) promoter activity in comparison to the empty vector control (lanes 1 and 2). The addition of CIITA significantly suppressed collagen promoter activity (compare lanes 1 and 3). This suppression was maintained in the presence of low levels of exogenously expressed CBP (compare lane 2 to 4 or 5); however, it was released by the addition of a higher quantity (2 μg) of CBP (lane 6). This indicates that additional CBP reverses the CIITA inhibition of collagen α2(I) transcriptional activity, and CBP is likely the target of CIITA. Together with the mapping data shown in Fig. 4, these experiments strongly support the hypothesis that CIITA is squelching CBP molecules, resulting in gene suppression. To exclude the trivial explanation that CBP overexpression downregulates the expression of exogenous CIITA, equal amounts of CIITA were cotransfected with various amounts of CBP. As shown in Fig. 5B, the expression level of CIITA was not altered by the absence or presence of various amounts of CBP (compare lane 1 with 2, 3, or 4).
FIG. 5.
CBP relieves the inhibition of collagen α2(I) promoter activities by CIITA. (A) Increasing amounts of CBP were cotransfected with CIITA and/or carrier DNA, as depicted in the figure. The influence of CBP on CIITA-mediated suppression of the collagen promoter was measured by comparing the collagen luciferase reporter gene activity in the absence (lane 3) or presence (lane 4 to 6) of CBP. The quantity of CBP used to transfect the cells are depicted in the figure. These experiments were repeated three times. (B) COS7 cells (9 × 105) were cotransfected with 3 μg of FlagCIITA and various amounts of CBP (1.5, 3.0, and 4.5 μg for lanes 2, 3, and 4, respectively) or its empty control vector, CMV5 (lane 1). CMV5 empty vector was also added to lanes 1 to 3 such that the total amount of plasmid transfected is the same in all of the samples. Cells were lysed with 1.5 ml of RIPA buffer 36 h after transfection, and 25 μl of the samples was subjected to SDS–10% polyacrylamide gel electrophoresis resolution. The expression of Flag CIITA was analyzed by immunoblotting (IB) using anti-Flag antibody.
IFN-γ inhibition of promoter activity is associated with the presence of CIITA.
IFN-γ induces as well as suppresses gene expression (6, 33). CIITA mediates the IFN-γ activation of MHC-II genes, while this report finds that CIITA can suppress gene expression. Therefore, it was of interest to determine whether CIITA may be an underlying mediator of IFN-γ-induced gene suppression in cells that express CIITA. Interestingly, the collagen α2(I) gene studied here is a well-known target of IFN-γ-mediated suppression (22, 28, 46).
To test the physiologic relevance of CIITA-mediated suppression of the collagen α2(I) gene, we used a mutant cell line that is selectively defective in CIITA expression. Normally, IFN-γ induces the expression of CIITA in the parental fibrosarcoma 2fTGH cell line, which then leads to surface MHC-II expression (37). G3A is an MHC-II-negative mutant derived from 2fTGH, which was immunoselected for the lack of MHC-II antigen expression (37). All other known components of an IFN-γ response are normal in this cell line, including the functional IFN-γ receptor and members of the JAK/STAT1 pathway. The IFN-γ induction of IRF-1, guanylate binding protein, and β2-microglobulin is also normal (37). These results suggest that the defect in this cell line is specific to MHC-II gene control. However, NF-Y and RFX proteins are all found to be normal in G3A cells, indicating that the defect is not in these DNA-binding transcription factors (10). When CIITA was examined, its induction by IFN-γ was found to be defective. Semiquantitative RT-PCR shows that the CIITA level is approximately one-fiftieth that of 2fTGH (10). Defective CIITA induction by IFN-γ in this line is likely the basis for the lack of MHC-II induction, as the reintroduction of CIITA into this line restores MHC-II expression and causes MHC-II promoters to assume an open configuration (61).
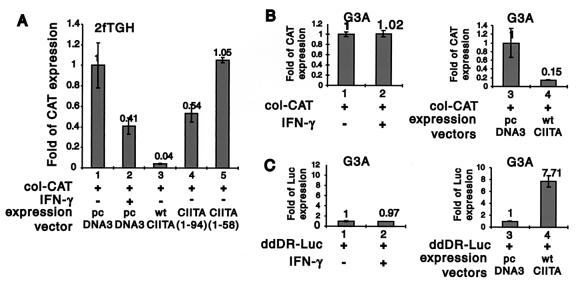
A comparison of responses in 2fTGH versus G3A cells provides a direct analysis of the physiologic role of CIITA upon IFN-γ treatment. First, to determine the effect of IFN-γ-induced expression of endogenous CIITA on the transcriptional activity of col-CAT, this reporter construct was transiently transfected into 2fTGH in the presence or absence of IFN-γ. IFN-γ repressed col-CAT activity (Fig. 6A, lanes 1 and 2), albeit to a lesser extent than that of exogenously expressed CIITA (lane 3). This may be due to the low level of CIITA that is induced by IFN-γ in these cells compared to the higher level derived from transfected CIITA. Mutant CIITA(1-94) also downregulated col-CAT activity, but to a lesser extent than wild-type CIITA, while CIITA(1-58) had no effect.
FIG. 6.
The use of 2fTGH and G3A cells to delineate the physiologic importance of CIITA in IFN-γ-mediated gene suppression. (A) The experiment was performed as described in the legend Fig. 3, except 2fTGH cells were treated with 500 U of IFN-γ/ml (lanes 1 and 2) 24 h after transfection. The inhibition of col-CAT activity by IFN-γ treatment in these cells is shown. This suppression is observed with CIITA or CIITA(1-94) but not with CIITA(1-58). (B) G3A cells were used in place of 2fTGH cells, and the experiment was performed as for panel A. col-CAT activity was not inhibited by IFN-γ treatment in G3A cells (lanes 1 and 2). The inclusion of exogenously expressed CIITA inhibited col-CAT activity (lanes 3 and 4). (C) The DRA-Luc control shows that the addition of IFN-γ to the same culture of G3A cells did not result in MHC-II promoter activation (compare lanes 1 and 2). In contrast, the addition of CIITA resulted in the induction of the MHC-II promoter. These experiments were reproduced three times. Luc, luciferase; wt, wild type.
To determine if CIITA suppresses gene expression during an IFN-γ response in a more physiologic system, an analysis was performed in the CIITA-defective mutant cell line G3A. In contrast to the 2fTGH cell line (Fig. 6B), treatment of these cells with IFN-γ (lanes 1 and 2) did not result in suppression of col-CAT expression. The introduction of CIITA restored IFN-γ suppression of col-CAT activity in these cells (lanes 3 and 4). This strongly supports the contention that CIITA mediates IFN-γ suppression of promoter activity. As a control, Fig. 6C shows that the addition of IFN-γ to the same culture of G3A cells did not result in MHC-II promoter activation (compare lanes 1 and 2). In contrast, the addition of CIITA resulted in the induction of the MHC-II promoter. These studies show that endogenous CIITA has dual activity, both as an inducer of MHC-II promoters and as an inhibitor of the collagen α2(I) promoter.
CIITA inhibits endogenous collagen α2(I) expression.
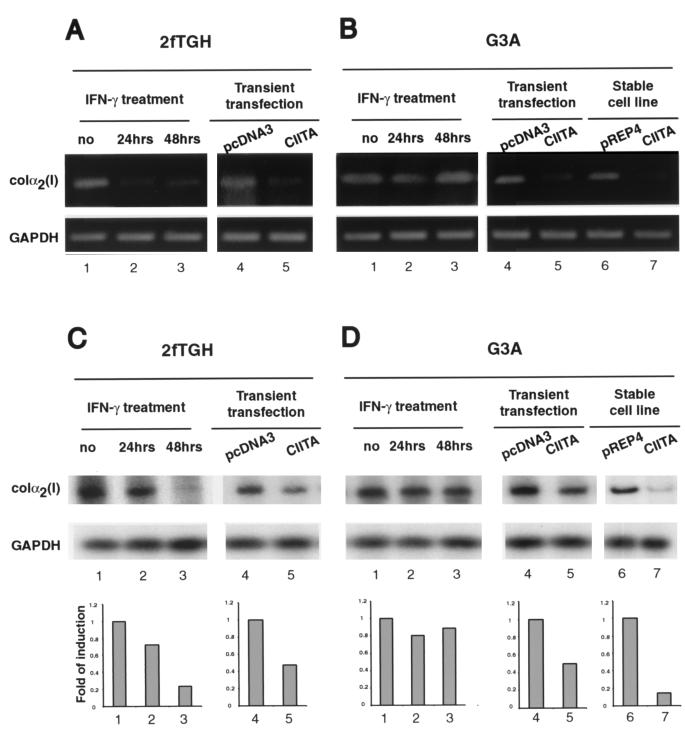
To further determine if endogenously expressed CIITA affects the expression of endogenous collagen α2(I) gene expression, 2fTGH and G3A cells were treated with IFN-γ for 24 or 48 h. As described above, the former has intact CIITA expression, while G3A has a defect in the IFN-γ induction of CIITA; thus, differences in these two lines in response to IFN-γ can be attributed to endogenous CIITA. RT-PCR was performed to examine endogenous collagen α2(I) transcript levels. The linear phase of amplification for each gene was preestablished and used in all the experiments to avoid artifacts associated with RT-PCR (see Materials and Methods). In addition, these experiments were repeated three times, and they produced consistent findings. In 2fTGH cells, IFN-γ suppressed collagen α2(I) without any effect on the housekeeping gene of GAPDH (Fig. 7A, compare lane 1 with lanes 2 and 3). The transient transfection of a CIITA expression construct also significantly suppressed endogenous collagen α2(I) gene expression in the absence of IFN-γ treatment.
FIG. 7.
IFN-γ treatment, or introduction of CIITA, reduced the expression of endogenous collagen α2(I) transcripts in 2fTGH cells. (A) Total RNA was isolated from 2fTGH cells with or without IFN-γ treatment for 24 (lane 2) or 48 h (lane 3), or cells were transiently transfected with empty vector (lane 4) or CIITA (lane 5). Total RNAs were subjected to amplification by RT-PCR with primers specific for the collagen α2(I) gene. GAPDH was used as a control. (B) The experiment depicted in lanes 1 to 5 is the same as that depicted in panel A, except that G3A cells were used. Additionally, both transient (lanes 4 and 5) and stably transfected (lanes 6 and 7) cells were analyzed. (C and D) Total RNAs (1.5 μg), prepared in a manner similar to that described for panels A and B, were reverse transcribed, and an aliquot of each sample was subjected to amplification by hot-PCR with the collagen-specific primers in the presence of [α-32P]dCTP. PCR samples were electrophoresed in nondenatured polyacrylamide gel, autoradiographed, and quantitated using the NIH Image software (lower graphs). GAPDH was used as a control (second row of the upper panel).
A comparative study with the G3A cells shows that IFN-γ failed to inhibit the expression of endogenous collagen α2(I) in this mutant line (Fig. 7B, compare lane 1 with lanes 2 and 3). To further specify the role of CIITA in this inhibitory pathway, G3A cells were transiently or stably transfected with CIITA, and the endogenous collagen α2(I) mRNA level was examined. The appropriate control plasmid for the transient transfectant is pcDNA3, while that for the stably transfected cell line is pREP4. Exogenously introduced CIITA inhibited the expression of endogenous collagen α2(I) transcript (Fig. 7B, compare lanes 4 to 5 and 6 to 7). Together, these experiments show that CIITA suppresses the expression of the endogenous collagen α2(I) gene.
For the quantitative measurement of the changes depicted in Fig. 7A and B, a new experiment similar to the one described in the legend to Fig. 7A and B was performed. Total RNA from each sample was reverse transcribed and subsequently amplified by PCR in the presence of [α-32P]dCTP. The findings, shown in Fig. 7C and D, confirm the qualitative results from Fig. 7A and B. These results were quantitated and plotted against the data from GAPDH-specific PCR-amplified product shown in the lower panel of Fig. 7C and D.
Treatment of 2fTGH with IFN-γ caused a reduction of the endogenous collagen α2(I) transcript, although in this experiment the reduction is less obvious at the 24-h time point but is apparent at the 48-h time point. The transient introduction of CIITA into cells which have not been treated with IFN-γ caused a 50% drop in collagen α2(I) transcript. This is found for both 2fTGH and G3A cells (compare lane 4 to 5 in Fig. 7C and D). This modest effect is likely attributed to the fact that transient transfection can only introduce the transfected gene in a portion of the cells. However, a G3A line which has been stably transfected with CIITA shows a dramatic reduction of collagen α2(I) transcript (Fig. 7D, lanes 6 and 7).
Additional promoters known to be inhibited by IFN-γ are also inhibited by CIITA.
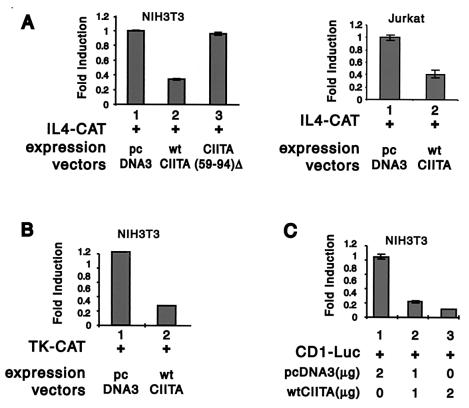
In addition to genes coding for matrix proteins such as collagen and fibronectin, IFN-γ is known to suppress other cellular processes. The most notable examples are the suppression of IL-4 production in TH2 cells (23) and the inhibition of cell growth (2, 45). To determine whether CIITA mediates suppression of genes which participate in alternate cellular processes, transcriptional activity of the IL-4 promoter was assessed using the IL-4 CAT reporter construct (Fig. 8A), the TK promoter using the TK-CAT construct (Fig. 8B), and the cyclin D1 promoter using CD1-Luc (Fig. 8C). All promoters were tested as described for the collagen promoter, except IL-4 CAT was tested in both the NIH 3T3 fibroblastic line and the Jurkat T-cell line. The latter was used because IL-4 is produced by cells of T-cell origin. Transcriptional activities of all these promoters were suppressed by CIITA. To further investigate if residues 59 to 94 of CIITA are important for this suppression, NIH 3T3 cells were transfected with the IL-4 CAT reporter gene along with wild-type CIITA or CIITA(59-94)Δ, which lacks the CBP interaction domain. As shown in Fig. 8A, CIITA(59-94)Δ did not inhibit IL-4 promoter transcriptional activity, while wild-type CIITA did (compare lanes 2 and 3). This demonstrates the importance of residues 59 to 94 in suppressing IL-4 promoter activity.
FIG. 8.
CIITA suppresses other promoters. The effect of CIITA on the IL-4 promoter (A), the TK promoter (B), and the cyclin D1 promoter (C) are shown. The IL-4 CAT reporter gene was transiently cotransfected with an empty vector (panel A, left-hand graph, lane 1), wild-type (wt) CIITA (same graph, lane 2), or CIITA(59-94)Δ (same graph, lane 3). The other two reporter constructs were only cotransfected with either CIITA or an empty vector.
DISCUSSION
CIITA is a master regulator of MHC-II gene expression through its interaction with DNA-binding transcription factors that bind the X and Y elements of MHC-II promoters. Specifically, distinct or overlapping domains of CIITA are required for interactions with NF-YB–NF-YC, RFX5-RFXANK and CREB (65). These are, respectively, proteins that recognize the Y, X1, and X2 elements of a MHC-II promoter. Based on these interactions, we propose that CIITA may serve as a scaffold protein, very much in the same vein as scaffold proteins which interact with members of the mitogen-activated protein kinase pathways to achieve specific cellular effects and not others (65). The original intent of this work was to determine if CIITA shares a unique property of other signaling scaffold proteins, e.g., the sequestration of mitogen-activated protein kinase members for signaling in one cellular pathway, thus reducing their availability for other pathways. In the case of transcription, squelching is thought to serve the same purpose by titrating general transcription factors which are limiting (44). We tested the possibility that CIITA may sequester first-tier DNA-binding proteins and reduce the availability of these proteins for the initiation of other transcripts. The initial finding (Fig. 1) shows that CIITA simultaneously enhances the transcription of an MHC-II promoter and reduces the transcription of a collagen α2(I) promoter which contains a canonical NF-Y binding site. However, two further tests show that the sequestration of NF-Y by CIITA is unlikely the primary mechanism by which CIITA causes gene suppression. First, the addition of NF-Y only modestly reverts gene repression. Second, the minimal domain within CIITA that is required for this repression is distinct from its interaction site with NF-Y (65). In contrast, these same tests show that CBP is the target because the addition of CBP rescues the collagen α2(I) promoter from CIITA-mediated repression. Furthermore, the minimal domain required for CIITA-mediated gene repression and for interaction with CBP is contained within the same 36 residues. In composite, these results strongly support the model where CIITA mediates gene repression via the sequestration of CBP while enhancing MHC-II transcription by interaction with CBP and the transcription factors that activate MHC-II promoters. This is of relevance to the control of a number of genes, including both immune and nonimmune genes. During the second review of this paper, a recent report found that CIITA sequesters p300 to cause the suppression of the IL-4 promoter (52). Thus, the squelching of CBP and p300 may be a common mechanism by which CIITA mediates gene suppression.
One question that arises from the present study is why CIITA cannot be recruited to the collagen promoter through NF-Y sites and activate its gene expression. Instead, it suppresses collagen gene expression. Indeed, the work from our lab and other laboratories shows that CIITA physically interacts with NF-Y (25, 65) through a domain that is distinct from the CBP-associative domain identified here (65). There are several possible explanations. First, although the NF-Y site exists on nearly 30% of promoters, the combinatorial influence of NF-Y with adjacent and distal elements is likely to influence how CIITA affects these promoters (60). As a matter of fact, the transactivation of MHC-II promoters by CIITA is dependent on the stereospecific alignment of the X-Y box binding protein, RFX, CREB, and NF-Y (40, 65). The collagen promoter does not contain adjacent promoter elements found in MHC-II, and this is likely the reason why CIITA does not activate the collagen gene. Otherwise, CIITA should have more global effects on gene expression. Second, although in vivo chromatin immunoprecipitation indirectly shows that CIITA interacts with NF-Y on the MHC-II promoter (3, 38), evidence is lacking that this occurs on the collagen promoter. It is possible that without the appropriate juxtaposed promoter elements this interaction between CIITA and NF-Y may not be stabilized to occur in cells.
An examination of CIITA-mediated gene repression reveals interesting physiologic relevance of this finding. The collagen promoter which was used as the prototype promoter to study CIITA-mediated gene repression turns out to be a primary target of IFN-γ-mediated gene repression (26, 27, 64). This led to our hypothesis that CIITA may be a mediator of the well-documented gene repression by IFN-γ. A comparison of the wild-type 2fTGH line and its CIITA-defective variant, G3A, shows that the lack of CIITA is associated with the lack of IFN-γ-mediated gene suppression. Direct evidence that CIITA is involved in IFN-γ-mediated gene repression was obtained when CIITA reproduced the effects of IFN-γ in 2fTGH cells and further caused promoter and endogenous gene repression in G3A cells to a level similar to that of 2fTGH. Thus, CIITA constitutes a novel pathway which contributes to IFN-γ-mediated gene suppression. An examination of three types of promoters that are known targets of IFN-γ repression shows that all can be repressed by CIITA. The three types of promoters include those for (i) matrix proteins, with collagen as a prototype target; (ii) cytokines produced by the T helper subset, TH2, exemplified by IL-4; and (iii) cell cycle genes important for cell cycle progression, such as the thymidine kinase and cyclin D1 genes. However, it is unlikely that CIITA is the only molecule that mediates IFN-γ repression, since not all IFN-γ-responsive cells produce CIITA. Other mechanisms in addition to CIITA must also exert their effects to cause gene repression upon IFN-γ treatment. Nonetheless, among cell types that express CIITA, CIITA should constitute an important mechanism by which gene repression is mediated.
The repression of gene products required for cell cycle progression by IFN-γ has been well documented in the literature (2, 12, 45, 51, 63), stemming from the early observations that IFN-γ treatment leads to cell cycle arrest. During our studies of CIITA, we have noted that long-term transfectants expressing ectopic CIITA were difficult to obtain, and the rare clones which did grow expressed a low level of CIITA. These observations are consistent with the notion that CIITA can repress certain genes important for cell cycle progression, thus reducing their proliferation and growth.
The repression of IL-4 by CIITA has been observed by another group (23), and that study was performed with mice where CIITA was introduced as a transgene into all cells. The authors noted a decrease in IL-4 synthesis, which was not observed in a mouse lacking MHC-II antigen expression. The authors detected CIITA expression by RT-PCR in a T-cell-enriched preparation and concluded that CIITA expression in the T-cell fraction leads to decreased IL-4 production. During the review of this paper, this group showed that the binding of p300 by CIITA may be responsible for this suppression.
The repression of CBP function is particularly interesting and of practical relevance because CBP is a critical HAT important for the accessibility of a large group of promoters (13, 41, 57, 59). Its role in transcriptional regulation explains much of promoter accessibility and chromatin structure. Its quantity is limiting and thus is a likely target of squelching by CIITA. There is some information that CIITA may interact with other members of the histone acetylase family, including the p300 family. It will be of interest to assess whether CIITA also affects these other HAT members. From another practical vantage point, CIITA is very effective in inhibiting the function of CBP through protein-protein interaction. Considering that the region necessary for CIITA interaction with CBP is likely smaller than the 36 aa defined here, it is possible that peptido-mimetics approaches based on this interaction may be employed to block the function of CBP. This potentially has therapeutic benefits for cancer in general and for drug-related acute leukemia where a fusion of the MLL gene to CBP occurs (53).
In conclusion, this report has several novel findings. First, it shows that CIITA can mediate the IFN-γ suppression of genes. This represents a clever design by nature, where CIITA is used to mediate both the upregulation of crucial immune genes and the downregulation of genes that may be nonessential during an IFN-γ response. Gene suppression by IFN-γ has been observed by many, yet the mechanism is not well understood. Our study provides a molecular basis for this suppression. Second, the mechanism of this repression is through the squelching of CBP. Another report has shown the squelching of p300 by CIITA (52); thus, squelching represents a common mechanism for CIITA-mediated gene repression. Third, this study finely delineates the region of CIITA that interacts with and squelches CBP. This delineation provides an important new reagent: a small but potent molecule to inhibit CBP function. Fourth, this study took advantage of the G3A mutant cell line and revealed the suppression of the endogenous collagen gene by the endogenous level of CIITA that is induced by IFN-γ. In sum, this report should have significant impact on a number of fields, and it interjects CIITA into several important areas of research, including the study of IFN-γ repression, collagen gene regulation, CBP function, cancer therapy through the inhibition of CBP, and transcriptional squelching.
ACKNOWLEDGMENTS
We thank M. Li-Weber for IL-4–CAT and TK-CAT, Albert Baldwin for CD-Luc, and R. H. Goodman for CMV5-CBP.
This study was supported by grants from the National Institutes of Health (AI45580, AI41751, AI29565, and DK38108 to J.P.-Y.T.) and the National Multiple Sclerosis Society (RG7815 to J.P.-Y.T.).
REFERENCES
- 1.Accolla R S, Jotterand-Bellomo M, Scarpellino L, Maffei A, Carra G, Guardiola J. aIr-1, a newly found locus on mouse chromosome 16 encoding a trans-acting activator factor for MHC class II gene expression. J Exp Med. 1986;164:369–374. doi: 10.1084/jem.164.1.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asao H, Fu X Y. Interferon-gamma has dual potentials in inhibiting or promoting cell proliferation. J Biol Chem. 2000;275:867–874. doi: 10.1074/jbc.275.2.867. [DOI] [PubMed] [Google Scholar]
- 3.Beresford G W, Boss J M. CIITA coordinates multiple histone acetylation modifications at the HLA-DRA promoter. Nat Immunol. 2001;2:652–657. doi: 10.1038/89810. [DOI] [PubMed] [Google Scholar]
- 4.Berger S L, Cress W D, Cress A, Triezenberg S J, Guarent L. Selective inhibition of activated but not basal transcription by the acidic activation domain of VP16: evidence for transcriptional adaptors. Cell. 1990;64:1199–1208. doi: 10.1016/0092-8674(90)90684-7. [DOI] [PubMed] [Google Scholar]
- 5.Bi W, Wu L, Coustry F, de Crombrugghe B, Maity S N. DNA binding specificity of the CCAAT-binding factor CBF/NF-Y. J Biol Chem. 1997;272:26562–26572. doi: 10.1074/jbc.272.42.26562. [DOI] [PubMed] [Google Scholar]
- 6.Boehm U, Klamp T, Groot M, Howard J C. Cellular responses to interferon-gamma. Annu Rev Immunol. 1997;15:749–795. doi: 10.1146/annurev.immunol.15.1.749. [DOI] [PubMed] [Google Scholar]
- 7.Boshart M, Kluppel M, Schmidt A, Schutz G, Luckow B. Reporter constructs with low background activity utilizing the cat gene. Gene. 1992;110:129–130. doi: 10.1016/0378-1119(92)90456-y. [DOI] [PubMed] [Google Scholar]
- 8.Chang C-H, Guerder S, Hong S-C, van Ewijk W, Flavell R A. Mice lacking the MHC class II transactivator (CIITA) show tissue-specific impairment of MHC class II expression. Immunity. 1996;4:167–178. doi: 10.1016/s1074-7613(00)80681-0. [DOI] [PubMed] [Google Scholar]
- 9.Chang C H, Fontes J D, Peterlin M, Flavell R A. Class II transactivator (CIITA) is sufficient for the inducible expression of major histocompatibility complex class II genes. J Exp Med. 1994;180:1367–1374. doi: 10.1084/jem.180.4.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chin K-C, Mao C, Skinner C, Riley J L, Wright K L, Moreno C S, Stark G R, Boss M J, Ting J P-Y. Molecular analysis of G1B and G3A IFN gamma mutants reveals that defects in CIITA or RFX result in defective MHC-II and Ii gene induction. Immunity. 1994;1:687–697. doi: 10.1016/1074-7613(94)90039-6. [DOI] [PubMed] [Google Scholar]
- 11.Chin K-C, Li G X, Ting J P-Y. Importance of acidic, proline/serine/threonine-rich, and GTP-binding regions in the major histocompatibility complex class II transactivator: generation of transdominant-negative mutants. Proc Natl Acad Sci USA. 1997;94:2501–2506. doi: 10.1073/pnas.94.6.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chin Y E, Kitagawa M, Su W C, You Z H, Iwamoto Y, Fu X Y. Cell growth arrest and induction of cyclin-dependent kinase inhibitor p21 WAF1/CIP1 mediated by STAT1. Science. 1996;272:719–722. doi: 10.1126/science.272.5262.719. [DOI] [PubMed] [Google Scholar]
- 13.Chrivia J C, Kwok R P, Lamb N, Hagiwara M, Montminy M R, Goodman R H. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- 14.Coustry F, Maity S N, de Crombrugghe B. Studies on transcription activation by the multimeric CCAAT-binding factor CBF. J Biol Chem. 1995;270:468–475. doi: 10.1074/jbc.270.1.468. [DOI] [PubMed] [Google Scholar]
- 15.Dickensheets H L, Donnelly R P. Inhibition of IL-4-inducible gene expression in human monocytes by type I and type II interferons. J Leukoc Biol. 1999;65:307–312. doi: 10.1002/jlb.65.3.307. [DOI] [PubMed] [Google Scholar]
- 16.Dodge G R, Diaz A, Sanz-Rodriguez C, Reginato A M, Jimenez S A. Effects of interferon-gamma and tumor necrosis factor alpha on the expression of the genes encoding aggrecan, biglycan, and decorin core proteins in cultured human chondrocytes. Arthritis Rheum. 1998;41:274–283. doi: 10.1002/1529-0131(199802)41:2<274::AID-ART11>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 17.Donnelly R P, Freeman S L, Hayes M P. Inhibition of IL-10 expression by IFN-gamma up-regulates transcription of TNF-alpha in human monocytes. J Immunol. 1995;155:1420–1427. [PubMed] [Google Scholar]
- 18.Dorn A, Bollekens J, Staub A, Benoist C, Mathis D. A multiplicity of CCAAT box-binding proteins. Cell. 1987;50:863–872. doi: 10.1016/0092-8674(87)90513-7. [DOI] [PubMed] [Google Scholar]
- 19.Eckner R, Ewen M E, Newsome D, Gerdes M, DeCaprio J A, Lawrence J B, Livingston D M. Molecular cloning and functional analysis of the adenovirus E1A-associated 300-kD protein (p300) reveals a protein with properties of a transcriptional adaptor. Genes Dev. 1994;8:869–884. doi: 10.1101/gad.8.8.869. [DOI] [PubMed] [Google Scholar]
- 20.Fontes J D, Kanazawa S, Jean D, Peterlin B M. Interactions between the class II transactivator and CREB binding protein increase transcription of major histocompatibility complex class II genes. Mol Cell Biol. 1999;19:941–947. doi: 10.1128/mcb.19.1.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gill G, Ptashne M. Negative effect of the transcriptional activator GAL4. Nature. 1988;334:721–724. doi: 10.1038/334721a0. [DOI] [PubMed] [Google Scholar]
- 22.Goldring M B, Sandell L J, Stephenson M L, Krane S M. Immune interferon suppresses levels of procollagen mRNA and type II collagen synthesis in cultured human articular and costal chondrocytes. J Biol Chem. 1986;264:9049–9055. [PubMed] [Google Scholar]
- 23.Gourley T, Roys S, Lukacs N W, Kunkel S L, Flavell R A, Chang C H. A novel role for the major histocompatibility complex class II transactivator CIITA in the repression of IL-4 production. Immunity. 1999;10:377–386. doi: 10.1016/s1074-7613(00)80037-0. [DOI] [PubMed] [Google Scholar]
- 24.Guttridge D C, Albanese C, Reuther J Y, Pestell R G, Baldwin A S., Jr NF-κB controls cell growth and differentiation through transcriptional regulation of cyclin D1. Mol Cell Biol. 1999;19:5785–5799. doi: 10.1128/mcb.19.8.5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hake S B, Masternak K, Kammerbauer C, Janzen C, Reith W, Steimle V. CIITA leucine-rich repeats control nuclear localization, in vivo recruitment to the major histocompatibility complex (MHC) class II enhanceosome, and MHC class II gene transactivation. Mol Cell Biol. 2000;20:7716–7725. doi: 10.1128/mcb.20.20.7716-7725.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Higashi K, Kouba D J, Song Y J, Uitto J, Mauviel A. A proximal element within the human alpha 2(I) collagen (COL1A2) promoter, distinct from the tumor necrosis factor-alpha response element, mediates transcriptional repression by interferon-gamma. Matrix Biol. 1998;16:447–456. doi: 10.1016/s0945-053x(98)90016-6. [DOI] [PubMed] [Google Scholar]
- 27.Jaffe H A, Gao Z, Mori Y, Li L, Varga J. Selective inhibition of collagen gene expression in fibroblasts by an interferon-gamma transgene. Exp Lung Res. 1999;25:199–215. doi: 10.1080/019021499270268. [DOI] [PubMed] [Google Scholar]
- 28.Jimenez S A, Freundlich B, Rosenbloom J. Selective inhibition of human diploid fibroblast collagen synthesis by interferons. J Clin Investig. 1984;74:1112–1116. doi: 10.1172/JCI111480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kahari V M, Chen Y Q, Su M W, Ramirezand F, Uitto J. Tumor necrosis factor-alpha and interferon-gamma suppress the activation of human type I collagen gene expression by transforming growth factor-beta 1. Evidence for two distinct mechanisms of inhibition at the transcriptional and posttranscriptional levels. J Clin Investig. 1990;86:1489–1495. doi: 10.1172/JCI114866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kretsovali A. Involvement of CREB binding protein in expression of major histocompatibility complex class II genes via interaction with the class II transactivator. Mol Cell Biol. 1998;18:6777–6783. doi: 10.1128/mcb.18.11.6777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kwok R P, Lundblad J R, Chrivia J C, Richards J P, Bachinger H P, Brennan R G, Roberts S G, Green M R, Goodman R H. Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature. 1994;370:223–226. doi: 10.1038/370223a0. [DOI] [PubMed] [Google Scholar]
- 32.Li-Weber M, Davydov I V, Krafft H, Krammer P H. The role of NF-Y and IRF-2 in the regulation of human IL-4 gene expression. J Immunol. 1994;153:4122–4133. [PubMed] [Google Scholar]
- 33.Link H. The cytokine storm in multiple sclerosis. Mult Scler. 1998;4:12–15. doi: 10.1177/135245859800400104. [DOI] [PubMed] [Google Scholar]
- 34.Liu X, Berk A J. Reversal of in vitro p53 squelching by both TFIIB and TFIID. Mol Cell Biol. 1995;15:6474–6478. doi: 10.1128/mcb.15.11.6474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maity S N, Golumbek P T, Karsenty G, de Crombrugghe B. Selective activation of transcription by a novel CCAAT binding factor. Science. 1988;241:582–585. doi: 10.1126/science.3399893. [DOI] [PubMed] [Google Scholar]
- 36.Maity S N, de Crombrugghe B. Role of the CCAAT-binding protein CBF/NF-Y in transcription. Trends Biochem Sci. 1998;23:174–178. doi: 10.1016/s0968-0004(98)01201-8. [DOI] [PubMed] [Google Scholar]
- 37.Mao C, Davies D, Kerr I M, Stark G R. Mutant human cells defective in induction of major histocompatibility complex class II genes by interferon gamma. Proc Natl Acad Sci USA. 1993;90:2880–2884. doi: 10.1073/pnas.90.7.2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Masternak K, Muhlethaler-Mottet A, Villard J, Zufferey M, Steimle V, Reith W. CIITA is a transcriptional coactivator that is recruited to MHC class II promoters by multiple synergistic interactions with an enhanceosome complex. Genes Dev. 2000;14:1156–1166. [PMC free article] [PubMed] [Google Scholar]
- 39.Muhlethaler-Mottet A, Di Berardino W, Otten L A, Mach B. Activation of the MHC class II transactivator CIITA by interferon-gamma requires cooperative interaction between Stat1 and USF-1. Immunity. 1998;8:157–166. doi: 10.1016/s1074-7613(00)80468-9. [DOI] [PubMed] [Google Scholar]
- 40.Nekrep N, Jabrane-Ferrat N, Peterlin B M. Mutations in the bare lymphocyte syndrome define critical steps in the assembly of the regulatory factor X complex. Mol Cell Biol. 2000;20:4455–4464. doi: 10.1128/mcb.20.12.4455-4461.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Penton Ogryzko V V, Schiltz R L, Russanova V, Howard B H, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 42.Piskurich J F, Wang Y, Linhoff M W, White L C, Ting J P. Identification of distinct regions of 5′ flanking DNA that mediate constitutive, IFN-gamma, STAT1, and TGF-beta-regulated expression of the class II transactivator gene. J Immunol. 1998;160:233–240. [PubMed] [Google Scholar]
- 43.Piskurich J F, Linhoff M W, Wang Y, Ting J P-Y. Two distinct gamma interferon-inducible promoters of the major histocompatibility complex class II transactivator gene are differentially regulated by STAT1: interferon regulatory factor 1, and transforming growth factor beta. Mol Cell Biol. 1999;19:431–440. doi: 10.1128/mcb.19.1.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ptashne M, Gann A A. Activators and targets. Nature. 1990;346:329–331. doi: 10.1038/346329a0. [DOI] [PubMed] [Google Scholar]
- 45.Ramana C V, Grammatikakis N, Chernov M, Nguyen H, Goh K C, Williams B R, Stark G R. Regulation of c-myc expression by IFN-gamma through Stat1-dependent and -independent pathways. EMBO J. 2000;19:263–272. doi: 10.1093/emboj/19.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rosenbloom J, Feldman G, Freundlich B, Jimenez S A. Transcriptional control of human diploid fibroblast collagen synthesis by gamma-interferon. Biochem Biophys Res Commun. 1984;123:365–372. doi: 10.1016/0006-291x(84)90422-4. [DOI] [PubMed] [Google Scholar]
- 47.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 48.Schmidt A, Rossi P, de Crombrugghe B. Transcriptional control of the mouse alpha 2(I) collagen gene: functional deletion analysis of the promoter and evidence for cell-specific expression. Mol Cell Biol. 1986;6:347–354. doi: 10.1128/mcb.6.2.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sefton B M. Analysis of protein phosphorylation. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1999. pp. 18.2.1–18.2.8. [Google Scholar]
- 50.Sharma B, Iozzo R V. Transcriptional silencing of perlecan gene expression by interferon-gamma. J Biol Chem. 1998;273:4642–4646. doi: 10.1074/jbc.273.8.4642. [DOI] [PubMed] [Google Scholar]
- 51.Sibinga N E, Wang H, Perrella M A, Endege W O, Patterson C, Yoshizumi M, Haber E, Lee M E. Interferon-gamma-mediated inhibition of cyclin A gene transcription is independent of individual cis-acting elements in the cyclin A promoter. J Biol Chem. 1999;274:12139–12146. doi: 10.1074/jbc.274.17.12139. [DOI] [PubMed] [Google Scholar]
- 52.Sisk T J, Gourley T, Roys S, Chang C H. MHC class II transactivator inhibits IL-4 gene transcription by competing with NF-AT to bind the coactivator CREB binding protein (CBP)/p300. J Immunol. 2000;165:2511–2517. doi: 10.4049/jimmunol.165.5.2511. [DOI] [PubMed] [Google Scholar]
- 53.Sobulo O M, Borrow J, Tomek R, Reshmi S, Harden A, Schlegelberger B, Housman D, Doggett N A, Rowley J D, Zeleznik-Le N J. MLL is fused to CBP, a histone acetyltransferase, in therapy-related acute myeloid leukemia with a t(11;16)(q23;p13.3) Proc Natl Acad Sci USA. 1997;94:8732–8737. doi: 10.1073/pnas.94.16.8732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Springer T A. Analysis of protein. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1999. pp. 10.16.1–10.16.11. [Google Scholar]
- 55.Steimle V, Otten L A, Zufferey M, Mach B. Complementation cloning of an MHC class II transactivator mutated in hereditary MHC class II deficiency (or bare lymphocyte syndrome) Cell. 1993;75:135–146. [PubMed] [Google Scholar]
- 56.Steimle V, Siegrist C A, Mottet A, Lisowska-Grospierre B, Mach B. Regulation of MHC class II expression by interferon-gamma mediated by the transactivator gene CIITA. Science. 1994;265:106–109. doi: 10.1126/science.8016643. [DOI] [PubMed] [Google Scholar]
- 57.Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- 58.Varga J, Olsen A, Herhal J, Constantine G, Rosenbloom J, Jimenez S A. Interferon-gamma reverses the stimulation of collagen but not fibronectin gene expression by transforming growth factor-beta in normal human fibroblasts. Eur J Clin Investig. 1990;20:487–493. doi: 10.1111/j.1365-2362.1990.tb01890.x. [DOI] [PubMed] [Google Scholar]
- 59.Wade P A, Wolffe A P. Histone acetyltransferases in gene control. Curr Biol. 1997;7:R82–R84. doi: 10.1016/s0960-9822(06)00042-x. [DOI] [PubMed] [Google Scholar]
- 60.Wright K L, Vilen B J, Itoh-Lindstrom Y, Moore T L, Li G, Criscitiello M, Cogswell P, Clarke J B, Ting J P. CCAAT box binding protein NF-Y facilitates in vivo recruitment of upstream DNA-binding transcription factors. EMBO J. 1994;13:4042–4053. doi: 10.1002/j.1460-2075.1994.tb06721.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wright K L, Chin K C, Linhoff M W, Skinner C, Brown J A, Boss J M, Stark G, Ting J P. CIITA stimulation of transcription factor binding to major histocompatibility complex class II and associated promoter in vivo. Proc Natl Acad Sci USA. 1998;95:6267–6272. doi: 10.1073/pnas.95.11.6267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang X J, Ogryzko V V, Nishikawa J, Howard B H, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 63.Yarden A, Kimchi A. Tumor necrosis factor reduces c-myc expression and cooperates with interferon-gamma in HeLa cells. Science. 1986;234:1419–1421. doi: 10.1126/science.3097823. [DOI] [PubMed] [Google Scholar]
- 64.Yuan W, Yufit T, Li L, Mori Y, Chen S J, Varga J. Negative modulation of alpha1(I) procollagen gene expression in human skin fibroblasts: transcriptional inhibition by interferon-gamma. J Cell Physiol. 1999;179:97–108. doi: 10.1002/(SICI)1097-4652(199904)179:1<97::AID-JCP12>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 65.Zhu X Z, Linhoff M W, Li G, Chin K, Ting J P. Transcriptional scaffold: CIITA interacts with NF-Y, RFX and CREB to cause stereospecific regulation of the MHC-II promoter. Mol Cell Biol. 2000;20:6051–6064. doi: 10.1128/mcb.20.16.6051-6061.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]