Abstract
Deletion of the Saccharomyces cerevisiae TOP3 gene, encoding Top3p, leads to a slow-growth phenotype characterized by an accumulation of cells with a late S/G2 content of DNA (S. Gangloff, J. P. McDonald, C. Bendixen, L. Arthur, and R. Rothstein, Mol. Cell. Biol. 14:8391–8398, 1994). We have investigated the function of TOP3 during cell cycle progression and the molecular basis for the cell cycle delay seen in top3Δ strains. We show that top3Δ mutants exhibit a RAD24-dependent delay in the G2 phase, suggesting a possible role for Top3p in the resolution of abnormal DNA structures or DNA damage arising during S phase. Consistent with this notion, top3Δ strains are sensitive to killing by a variety of DNA-damaging agents, including UV light and the alkylating agent methyl methanesulfonate, and are partially defective in the intra-S-phase checkpoint that slows the rate of S-phase progression following exposure to DNA-damaging agents. This S-phase checkpoint defect is associated with a defect in phosphorylation of Rad53p, indicating that, in the absence of Top3p, the efficiency of sensing the existence of DNA damage or signaling to the Rad53 kinase is impaired. Consistent with a role for Top3p specifically during S phase, top3Δ mutants are sensitive to the replication inhibitor hydroxyurea, expression of the TOP3 mRNA is activated in late G1 phase, and DNA damage checkpoints operating outside of S phase are unaffected by deletion of TOP3. All of these phenotypic consequences of loss of Top3p function are at least partially suppressed by deletion of SGS1, the yeast homologue of the human Bloom's and Werner's syndrome genes. These data implicate Top3p and, by inference, Sgs1p in an S-phase-specific role in the cellular response to DNA damage. A model proposing a role for these proteins in S phase is presented.
DNA topoisomerases play several important roles in DNA metabolism (61, 62, 67). These enzymes catalyze the interconversion of topological isomers of DNA and are required for the resolution of torsional stress in DNA and for the unlinking of topologically intertwined molecules. Budding yeasts express three topoisomerases, designated topoisomerases I, II, and III, all of which are highly conserved in mammalian cells. The role of yeast topoisomerase III (Top3p) in DNA metabolism has remained enigmatic, partly because this enzyme possesses only a very weak DNA relaxation activity on negatively supercoiled DNA and is therefore thought unlikely to participate in the maintenance of DNA supercoiling homeostasis (25, 61). It has been suggested that Top3p performs a nonessential role in the segregation of newly replicated DNA in Saccharomyces cerevisiae, although there is no direct evidence to support this proposal (61). Strains of S. cerevisiae that lack Top3p (top3Δ) show hyperrecombination in repetitive sequences and a severe slow-growth phenotype, which is due to an accumulation of cells in the late S/G2 phase of the cell cycle (15). In the fission yeast Schizosaccharomyces pombe, the top3+ gene is essential for viability, and in that organism there is direct evidence that top3Δ mutants display abnormal nuclear division (17, 38). The bacterial Top3p enzyme shows catalytic properties in vitro very similar to those of its eukaryotic counterparts and has been implicated in the unlinking of DNA strands during DNA replication to permit nascent chain elongation and the separation of daughter molecules (21). There are at least two homologues of the TOP3 gene product in vertebrates (18, 24), one of which (Topo IIIα) has been shown to be essential for embryonic development in mice (30). Drosophila Topo IIIβ has been shown to efficiently relax hypernegatively supercoiled DNA (68), but the physiological role of this activity remains unclear.
One feature of the phenotypic effects of loss of Top3p function that has aroused considerable interest is the ability of the slow-growth and hyperrecombination phenotypes of S. cerevisiae top3Δ mutants to be suppressed by mutation of SGS1 (15). This genetic interaction is conserved in S. pombe, in which loss of the RecQ helicase, Rqh1, suppresses deletion of top3+ (17, 38). The SGS1 gene encodes the sole member of the RecQ subfamily of DExH box-containing helicases in budding yeast, and deletion of SGS1 leads to hyperrecombination (15, 63) and defects in chromosome segregation and sporulation (40, 64). Of the five known homologues of SGS1 that exist in human cells, three are linked to disease conditions; mutations in the BLM gene give rise to the cancer-prone condition Bloom's syndrome (9); mutations in WRN lead to the premature-ageing condition Werner's syndrome (72); and mutations in RECQ4 give rise to Rothmund-Thomson syndrome (27), which is associated with cancer predisposition as well as skin and skeletal abnormalities (58). Mutation of any one of the SGS1, BLM, WRN, and RECQ4 genes leads to genomic instability and an abnormally high rate of genetic recombination events and/or chromosomal rearrangements (reviewed in references 5 and 23).
Cells respond to DNA damage or to an inhibition of DNA replication by delaying cell cycle progression in order to permit time to resolve the resulting abnormal DNA structures. These cell cycle checkpoint pathways are highly conserved in different eukaryotic species and act to preserve both genome integrity and cell viability (32, 36, 43, 47, 50, 65). The genetic and biochemical composition of a particular checkpoint pathway is determined, at least in part, by the point in the cell cycle at which the perturbation of DNA structure or function occurs. Entry into S phase is inhibited if cells incur DNA damage in G1, such as through UV irradiation, and this G1/S DNA damage checkpoint is dependent, among others, upon the RAD9, RAD17, RAD24, MEC1, MEC3, DDC1, and RAD53 (also called MEC2, SAD1, or SPK1) genes in S. cerevisiae (2, 33, 34, 52–55). These genes, in conjunction with PDS1, are also required for the G2/M checkpoint, which acts to inhibit entry into anaphase when DNA damage has not been repaired (66, 71). An additional DNA damage checkpoint that has been identified in budding yeast, termed the intra-S checkpoint, functions to slow the rate of progression through S phase in the presence of DNA damage (45). The majority of the G1/S and G2/M checkpoint genes play a minor role, at least in budding yeast, in the intra-S checkpoint (46, 47). Two exceptions to this are MEC1 and RAD53, which are essential genes and are required for checkpoint pathways that operate in all phases of the cell cycle (2, 48, 52, 66). In addition to the “dedicated” checkpoint genes that play a role in S phase, there are certain gene products that function in S phase as part of the DNA replication machinery while also performing some as-yet-unidentified role in checkpoint surveillance of the genome. These include Pri1p (the catalytic subunit of DNA primase) and Rfc5p, which act in the intra-S checkpoint (39, 56), and replication protein A (RPA), which is a target for Mec1p in the cellular response to both DNA replication blockade and DNA damage (4).
In addition to subdividing checkpoints on the basis of the phase of the cell cycle in which they act, it is possible to categorize individual checkpoint proteins on the basis of whether they function in a DNA damage or abnormal structure “sensory” role, as signal transducers, or as targets of the signaling cascade (37). Thus, loss of a DNA damage sensor, such as Rad9p or Rad24p, causes failure to transduce the signal required for arrest of the cell cycle (26). Part of the signal transduction cascade that is absent in this class of mutants is the MEC1-dependent phosphorylation of Rad53p (7, 32, 37, 57, 65).
We have investigated the function of Top3p in S. cerevisiae. We show that deletion of TOP3 causes sensitivity to a variety of DNA-damaging agents. While top3Δ mutants show proficiency in checkpoint responses to DNA perturbations occurring outside of S phase, they fail to adequately delay S-phase transit in the presence of DNA damage. This is the first indication that a eukaryotic topoisomersase is required for protection of cells against DNA-damaging agents. We present a revised version of the checkpoint response cascade that links the processing of DNA structural abnormalities to DNA repair.
MATERIALS AND METHODS
Yeast strains.
All experiments were performed in the YP1 strain background except where stated. The following yeast strains were used: YP1 background, RKC1a (MATa leu2Δ his4-R ura3-52 lys2 ade2-101 top3::LEU2), JMK22d (MATa leu2Δ his4-R ura3-52 lys2 ade2-101 sgs1::LYS2), JMK253 (MATa leu2Δ his4-R ura3-52 lys2 ade2-101 rad24::LYS2), JMK469 (MATa leu2Δ his4-R ura3-52 lys2 ade2-101 rad24::LYS2 top3::LEU2), RKC1c (MATa leu2Δ his4-R ura3-52 lys2 ade2-101 sgs1::LYS2 top3::LEU2), and RKC 1d (MATa leu2Δ his4-R ura3-52 lys2 ade2-101); A364a background, RKC 31a (MATa ura3 his3 trp1 leu2), RKC 31b (MATa ura3 his3 trp1 leu2 top3::G418R), AG1 (MATα ura3 his3 trp1 leu2 sgs1::G418R), JMK245 (MATa ura3 his3 trp1 leu2 rad24::G418R), DLY264 (MATa ura3 his3 trp1 leu2 mec2-1::URA3 [44]), and DLY 285 (MATa ura3 his3 trp1 leu2 mec1-1::HIS3 [44]).
In the YP1 background, TOP3 was disrupted using the NotI-digested plasmid pWJ258, kindly provided by R. Rothstein. This disruption contains the LEU2 gene. Transformants were checked by Southern blot analysis and by PCR using primers flanking the TOP3 gene and within LEU2. In both the A364a and the CG378 strains, TOP3 was disrupted using pRKC20. In this plasmid, the full-length open reading frame of TOP3 incorporating 5′ BamHI and 3′ NotI sites was cloned into pT7. Blue (Invitrogen), and the coding sequence was disrupted by insertion of the KanMx:4 module (59) between the HpaI and BstXI sites. The product of a BamHI and NotI digest was used for the transformation, and the resulting transformants were analyzed by PCR as described above but using internal KanMx primers. The phenotype of top3Δ was also confirmed genetically in all cases by the ability of an sgs1Δ mutation to rescue the slow growth of the top3Δ strains (15).
Flow cytometric analyses.
Cells were grown in yeast extract-peptone-dextrose (YPD) medium, collected by centrifugation, and resuspended in 70% ethanol. The cells were then washed in 50 mM sodium citrate, pH 7.0, and resuspended in the same buffer containing 0.25 mg of RNase A/ml. The cells were sonicated and then incubated at 50°C for 60 min. Proteinase K was added to a final concentration of 1 mg/ml, and the cells were incubated at the same temperature for a further 60 min. The cells were then allowed to cool, and propidium iodide was added to a concentration of 8 μg/ml. Samples were analyzed using a Becton Dickinson FACScan machine incorporating LYSIS2 software. We confirmed that the peak shifting seen in flow cytometric analyses was a reflection of chromosomal DNA synthesis. The shift in the flow cytometric histograms from the G1 to the G2/M position was inhibited by α-factor, which induces a G1 arrest, and by hydroxyurea (HU), which inhibits DNA synthesis.
Cell cycle blockade and irradiation procedures.
All experiments were performed in YPD medium. For cell cycle blockade experiments, cultures were grown to early log phase (optical density, at 600 nm, 0.3 or 0.4). G1 arrest was induced by adding α-factor to a final concentration of 20 μg/ml, and G2/M arrest was achieved by adding nocodazole to a final concentration of 20 μg/ml. After the appropriate time intervals, the cells were checked microscopically and by flow cytometry to confirm the appropriate arrest phenotype. The method for UV irradiation (254 nm) has been described previously (1). Cell cycle delay during S phase was analyzed as described by Paulovich and Hartwell (45). Irradiation of nocodazole-arrested cells was done with 80 J of UV light/m2 before the cells were washed and returned to drug-free medium for 90 min.
Determination of population doubling time and viability.
The strains were grown overnight in YPD medium, diluted, and grown for several hours at 30°C. At time zero and at various times after dilution, the number of cells was determined with a Coulter Counter following brief sonication of the culture. To determine viability, a specific number of cells (usually 200 to 800) were spread on YPD agar, and the resulting colonies were counted after 3 days of incubation at 30°C.
Survival curves. (i) MMS.
Cells were grown as described above before addition of methyl methanesulfonate (MMS) to concentrations of 0.005 to 0.03% for 60 min. The MMS was then inactivated by the addition of sodium thiosulfate to 5%, and the percent survival was determined in relation to untreated controls.
(ii) UV irradiation.
Cells were grown to log phase, diluted at different concentrations in distilled H2O, and spread on YPD agar. The plates were irradiated at the indicated doses with 254-nm-wavelength UV light, and 72 h later surviving colonies were counted. The percent survival was compared to that of an unirradiated control.
(iii) HU.
Early-log-phase liquid cultures of the appropriate strains grown in minimal medium were exposed to 0.2 M HU for various times. The percent survival compared to that at time zero was determined by plating different dilutions of the culture on YPD agar and counting the resulting colonies 72 h later.
Northern blot analysis.
Total cellular RNA was isolated using an RNeasy Mini kit (Qiagen). RNA was separated on glyoxal gels, transferred to nylon membranes (Hybond-N), hybridized, and probed as described previously (1). Northern blots were quantitated (PhosphorImager; Molecular Dynamics Ltd.), and the values were normalized to an ACT1 loading control.
Preparation of yeast extracts and Western blot analysis.
Total cellular protein extracts were prepared using a trichloroacetic acid extraction technique (10) and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using 6.5% acrylamide gels prepared at an 80:1 acrylamide-bisacrylamide ratio. A rabbit polyclonal antiserum to Rad53p (NLO16 [7]) was used at a final dilution of 1:10,000 in 1% fat-free milk in phosphate-buffered saline containing 0.02% Tween 20, with a primary incubation period of 12 h. Horseradish peroxidase-linked secondary antibody (Sigma) was used at 1:10,000 with a 60-min incubation period. Chemiluminescent detection was performed with an ECL kit (Amersham).
Chromosome preparation and electrophoretic separation.
Intact yeast chromosomal-DNA samples were prepared from log-phase or arrested cells, as described previously (35). Briefly, samples representing approximately 107 cells were applied to a 1% agarose gel slab, and the chromosomes were separated using a Bio-Rad contour-clamped homogenous electric field apparatus. Separation was achieved in 24 h, with initial and final switching times of 60 and 90 s, respectively, at 200 V. The running buffer used was 0.5× Tris-borate-EDTA, which was maintained at a temperature of 11°C by rapid recirculation while the gel was run at an ambient temperature of 4°C. The DNA was then transferred to nylon membranes, and chromosome IV was detected with an HO gene probe, using standard protocols.
RESULTS
top3Δ cells demonstrate a cell cycle delay at G2/M.
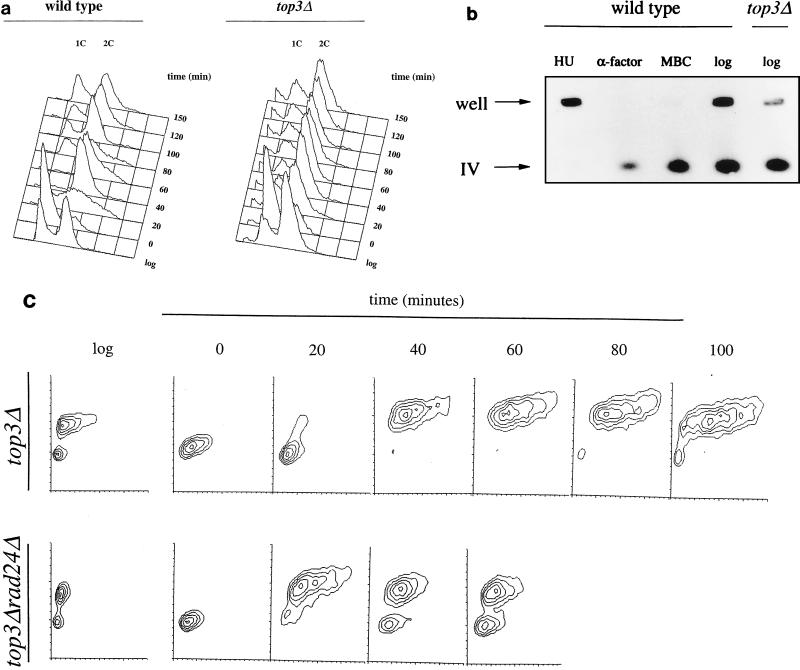
The cell cycle transit time of S. cerevisiae top3Δ mutants is approximately double that of isogenic wild-type strains (60). Previous microscopic studies of asynchronous top3Δ cultures revealed an accumulation of large-budded cells (≈70% of the total population) containing a single nucleus close to the neck of the mother cell (15), suggesting a delay in the cell cycle in the late S/G2 phase. In order to characterize this arrest phenotype in more detail, we performed flow cytometric analyses of wild-type and top3Δ cells at timed intervals following release from G1 cell cycle arrest induced by the α-factor mating pheromone. Figure 1a shows that following release from this arrest, top3Δ cells progressed through S phase at a rate similar to that of wild-type cells but then delayed in the cell cycle at a point where they had a 2C content of DNA. Consistent with previous studies (15), we calculated that the extended cell cycle time in top3Δ strains was fully accounted for by this late S/G2 delay. Analysis of cell size using contour plots indicated that top3Δ cells showed a marked and progressive increase in size during the late S/G2 delay period, indicating that while cell division was blocked in top3Δ mutants, cellular growth (increase in mass) continued (Fig. 1c). It should be noted that the top3Δ cells emerged from an α-factor arrest about 10 min earlier than did wild-type cells, as assessed microscopically by the appearance of small-budded cells (data not shown). This probably reflects the larger size of the G1-arrested top3Δ cells, which would influence the rate at which cells pass the cell size restriction point before Start (49). Synchrony experiments (not shown) confirmed previous results (15) showing that deletion of SGS1 in a top3Δ background completely corrected the cell cycle defect associated with mutation of TOP3.
FIG. 1.
top3Δ mutants show a G2/M-phase delay. (a) Log-phase cultures of wild-type (RKC1d) and top3Δ (RKC1a) cells were arrested in G1 with α-factor for 150 min, washed twice, and then released into fresh, warmed medium. Flow cytometric analysis was performed at timed intervals. Peaks representing cells with a 1C and a 2C content of DNA are indicated. (b) Intact chromosomal DNA from log-phase cultures of wild-type and top3Δ strains or wild-type cells arrested with HU, α-factor, or MBC was separated by pulsed-field gel electrophoresis. After transfer to a nylon membrane, the DNA was probed with the HO gene, which reveals chromosome IV. The positions of the wells and of chromosome IV that had migrated into the gel are indicated. (c) Flow cytometric analysis in the form of contour plots (with the DNA content on the vertical axis and forward light scatter, indicative of cell size, on the horizontal axis) of a top3 mutant and a top3 rad24 double mutant showing asynchronous log-phase cultures (log) in each case, α-factor-arrested cells (time zero), and cells at various times after release from α-factor arrest.
It is not possible to distinguish between late S-phase and G2/M cells by using flow cytometry, and therefore we considered the possibility that a residual level of unreplicated DNA exists in top3Δ cells that is sufficient to induce a late S-phase delay. To determine whether the late S/G2-arrested cells had completed DNA replication, intact chromosomes of log-phase cells were separated by pulsed field gel electrophoresis (Fig. 1b). In this assay, only fully replicated DNA enters the gel, whereas incompletely replicated DNA is retarded in the wells (20). As controls, wild-type cells were treated with HU (which blocks in S phase), α-factor (which blocks in G1), or methyl benzimidazol-Zyl-carbamate (MBC) (which blocks in M). As expected, DNA from HU-treated cells failed to enter the gel, whereas the majority of the fully replicated DNA in α-factor- or MBC-treated cells entered the gel. In common with the findings for wild-type cells, the majority of DNA from log-phase top3Δ cells entered the gel (70.0% ± 5.1% of total DNA for wild-type cells versus 82.3% ± 4.2% for top3Δ cells). The latter figure was consistently greater than the percentage of both large-budded cells and cells with a 2C DNA content. We conclude that top3Δ cells are able to complete bulk DNA replication and arrest at a point in the cell cycle after S phase but prior to the onset of anaphase.
The progressive increase in cell size when top3Δ mutants attain a 2C content of DNA (Fig. 1c) would be consistent with activation of the G2/M DNA damage checkpoint. To analyze this, we deleted separately the RAD24 and RAD9 DNA damage checkpoint genes in a top3Δ background and determined the cell cycle distribution of the resulting double mutants. An asynchronous population of rad24Δ top3Δ double mutant cells showed a marked reduction in the proportion of cells with a 2C content of DNA compared to an equivalent population of top3Δ cells (Fig. 1c). The proportions of cells gated for a 2C content of DNA were 61% (top3Δ) and 40.9% (rad24Δ top3Δ). The double-mutant cells with a 2C content of DNA were of a more uniform size than those of the top3Δ single mutant, also indicating that the double-mutant cells were progressing more rapidly through G2/M (Fig. 1c). Similar results were seen with a rad9 top3 double mutant (data not shown). Analysis of cell size and morphology by microscopy, coupled with DAPI (4′,6′-diamidino-2-phenylindole) staining of nuclear DNA, confirmed that following release from α-factor arrest, top3 rad24 double mutants were smaller and of a more uniform size than top3 mutant cells (data not shown). Taken together, these data suggest that the cell cycle delay in top3Δ cells is dependent on RAD24 and RAD9 and, by inference, on the G2/M DNA damage checkpoint. Consistent with this suggestion, the cell synchronization experiment shown in Fig. 1c confirmed that deletion of RAD24 in a top3Δ background had the effect of shortening the G2/M phase. Thus, at 60 min following release from α-factor arrest, the rad24Δ top3Δ cells had begun to progress into the next cell cycle, whereas the top3Δ cells were still largely held at G2/M even at the 100-min time point. These results were confirmed in two independent strain backgrounds.
Given that deletion of RAD24 had the effect of shortening the period of G2 arrest in top3Δ cells, we analyzed the effects that this had on growth rate and viability. We considered the possibility that the G2/M checkpoint arrest that occurs in every cell cycle in top3Δ mutants could be important for their continued survival. Consistent with the fluorescence-activated cell sorter data presented above, top3 rad24 double mutants showed a shorter doubling time than top3Δ mutants (120 min for top3 rad24 cells compared to 210 min for top3 cells). However, this more rapid proliferation did not appear to adversely affect survival, since top3 rad24 double mutants and top3 single mutants showed comparable levels of overall cell viability (approximately 25%).
top3Δ cells are sensitive to DNA-damaging agents.
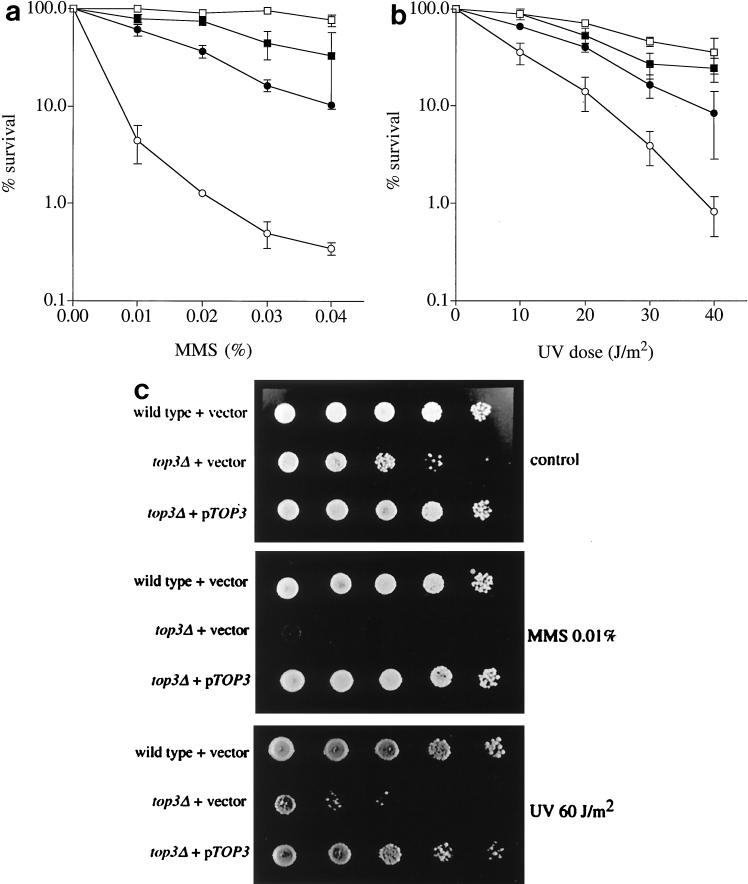
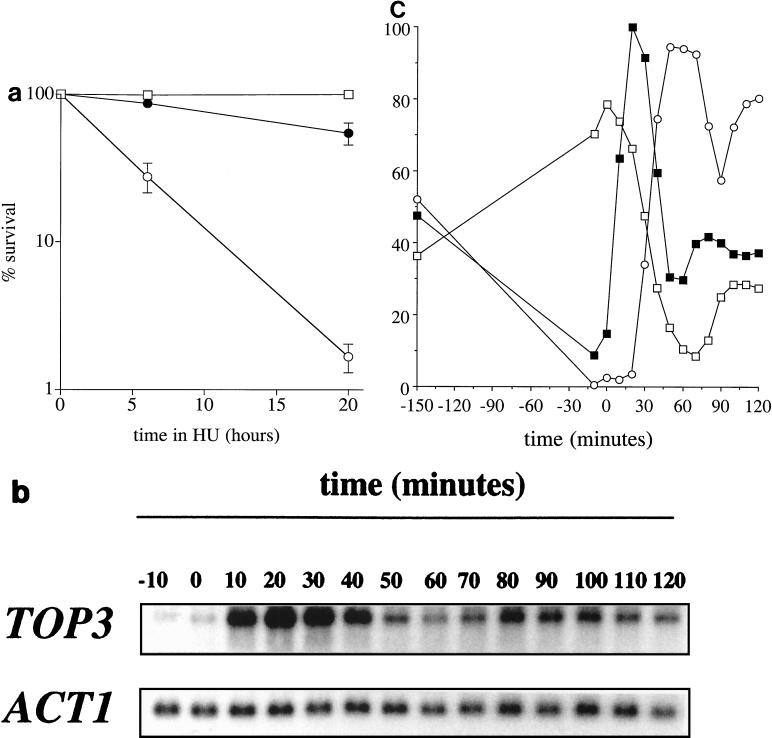
One explanation for the above-mentioned results is that top3Δ cells accumulate abnormal DNA structures in either S phase or G2/M as a consequence of a defect in the processing and/or repair of DNA structural abnormalities. We therefore tested whether top3Δ mutants are sensitive to killing by DNA-damaging agents. top3Δ cells were found to be sensitive to the DNA-damaging agents MMS and UV light (Fig. 2a and b) and, to a lesser extent (approximately 1.5-fold), to γ-irradiation (data not shown). These results were confirmed in three independent strain backgrounds. The sensitivity of top3Δ strains to DNA-damaging agents was substantially suppressed by deletion of SGS1 (Fig. 2a and b) or by ectopic expression of a wild-type TOP3 gene (Fig. 2c).
FIG. 2.
top3Δ cells are sensitive to DNA-damaging agents. (a and b) Strains RKC1d (wild type; □), JMK22d (sgs1; ▪), RKC1c (sgs1 top3; ●), and RKC1a (top3; ○) were grown to early log phase in YPD medium and then exposed to either MMS (a) or UV (b), and percent survival was determined. The means and standard errors of three independent experiments are shown. (c) Tenfold serial dilutions of exponentially growing yeast cultures from wild-type cells harboring an empty vector, a top3Δ strain harboring an empty vector, or a top3Δ strain harboring plasmid-encoded TOP3 (pTOP3), as indicated on the left. The cells on the upper plate were untreated, those on the middle plate were treated with 0.01% MMS, and those on the lower plate were irradiated with 60 J of UV light/m2, as indicated on the right.
top3Δ cells are defective in the intra-S checkpoint but not in the G1/S or G2/M DNA damage checkpoints.
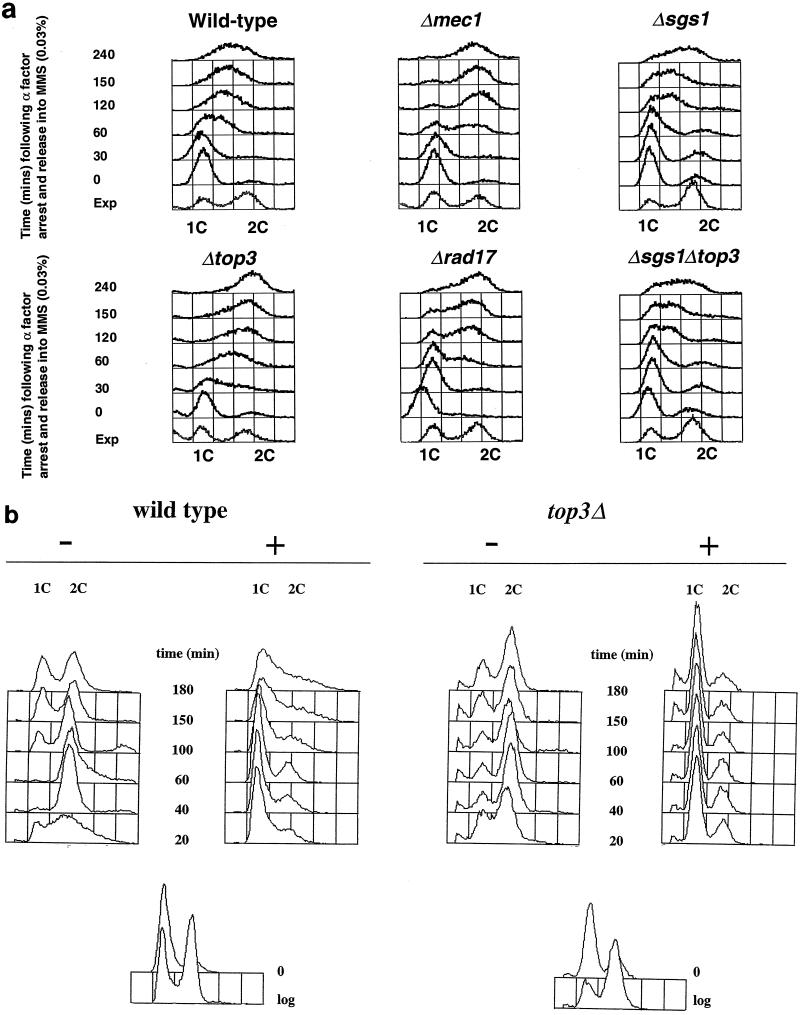
The intra-S checkpoint acts to slow the rate of DNA synthesis when DNA is damaged during S phase, and it is dependent upon the products of a number of genes, including MEC1, RAD53, RFC-5, PRI-1, RAD24, and RAD17 (45, 65). The data in Fig. 3a show that when top3Δ cells were released from an α-factor-induced G1 arrest into medium containing 0.03% MMS, most cells achieved a 2C content of DNA after 120 to 150 min, whereas wild-type cells treated similarly progressed through S phase much more slowly and still had not attained a 2C content of DNA by 240 min. Indeed, wild-type cells had still not fully completed DNA replication by 360 min (data not shown). Control experiments showed that wild-type cells and top3Δ mutants each progressed through S phase in approximately 60 min in the absence of MMS (data not shown). To analyze this apparent intra-S checkpoint defect in top3Δ mutants further, the rate of S-phase progression in the presence of MMS in top3 cells was compared directly to the previously reported partial defect of rad17Δ mutants and the complete defect of mec1 mutants in arresting S phase in the presence of MMS (45). As shown in Fig. 3a, the magnitude of the intra-S checkpoint defect in top3Δ cells was comparable to and consistently a little more severe than that observed in a rad17Δ mutant (and in rad24Δ cells [not shown]) but less severe than that seen in a mec1 mutant. In contrast, both sgs1Δ and sgs1Δ top3Δ mutants behaved essentially as wild type, indicating that deletion of SGS1 in a top3Δ background restores a largely functional intra-S checkpoint.
FIG. 3.
top3Δ cells are defective in the intra-S checkpoint but proficient in the G1/S DNA damage checkpoint. (a) To assess the intra-S checkpoint, A364a strains (as indicated above each panel) were grown in YPD medium to early log phase and then arrested in G1 with α-factor for 150 min. The cells were then washed twice with fresh warmed medium (time zero) and released into medium containing 0.03% MMS for the times indicated on the left. The DNA content was assessed by flow cytometry, and the peaks representing cells with a 1C or a 2C content of DNA are indicated. The sample denoted Exp represents a control log-phase cell population to show the positions of the 1C and 2C peaks. (b) To assess the G1/S checkpoint, cells were arrested as above and then irradiated with 80 J of UV light/m2 (+) or mock treated (−) before being washed twice and released into fresh medium. Similar results were obtained from four independent experiments.
In contrast to the above results, cell cycle checkpoint responses to DNA damage occurring outside of S phase were apparently unaffected by deletion of TOP3. Figure 3b shows that when α-factor-arrested top3Δ cells were UV irradiated and then released into fresh medium, they showed a marked delay in the rate of progression through the G1/S phase transition compared to nonirradiated controls. The cell cycle delay seen in top3Δ strains after UV irradiation in G1 was dependent upon functional RAD24 (data not shown). Hence, we conclude that the G1/S DNA damage checkpoint is intact in top3Δ mutants. The data presented in Fig. 1 indicate that top3Δ cells delay transiently at a G2/M checkpoint in the absence of exogenously added DNA-damaging agents. Consistent with this, when top3Δ cells were arrested in G2/M with nocodazole and then UV irradiated, the extents of subsequent delay in the cell cycle following removal of nocodazole were comparable in wild-type and top3Δ cells (data not shown). This confirms that the G2/M DNA damage checkpoint is intact in top3Δ mutants.
Rad53p phosphorylation is defective in top3Δ mutants following DNA damage in S phase in top3Δ cells.
Checkpoint proteins can be categorized on the basis of whether they function as sensors of abnormal DNA structures, signal transducers, or targets of the signaling apparatus (37, 65). Loss of sensory function might be expected to result in an absence of all downstream responses following DNA damage or replication inhibition, including a failure to phosphorylate Rad53p.
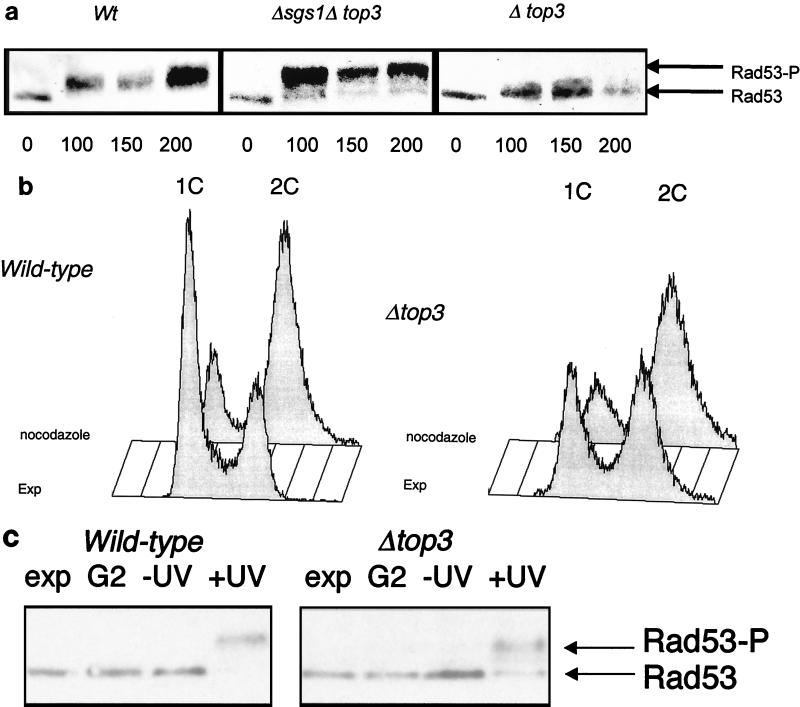
In order to position Top3p function in relation to this broad outline of the checkpoint pathway, we studied Rad53 phosphorylation in wild-type and top3Δ cells following release from G1 arrest into medium containing MMS. Figure 4a shows that phosphorylation of Rad53p (as indicated by the appearance of slower-migrating species on SDS-PAGE) was significantly impaired in top3Δ cells following exposure to MMS. Indeed, the phosphorylation of Rad53p in top3Δ cells was quantitatively and qualitatively different from that seen in wild-type cells. In MMS-treated wild-type cells, all of the Rad53p was fully phosphorylated within 100 min, as evidenced by the Rad53 species running with a substantially slower migration than the unphosphorylated Rad53p extracted from untreated cells. In contrast, in top3Δ cells, only a small fraction (approximately 25%) of the total Rad53p was phosphorylated, and the degree to which this modified Rad53 had a retarded migration was far less marked that that seen in wild-type cells. These data indicate that Top3p acts at an early step in the checkpoint cascade, upstream of Rad53p, and that in the absence of Top3p there is a diminution in signal transduction. Consistent with the observed restoration of the intra-S checkpoint by deletion of SGS1 in a top3Δ background (Fig. 3), an sgs1 top3 double mutant showed an apparently normal degree of Rad53p phosphorylation following MMS treatment in S phase (Fig. 4a).
FIG. 4.
Rad53 phosphorylation in top3Δ mutants. (a) Wild-type (Wt), top3, and top3 sgs1 cells were grown in YPD medium to early log phase and then arrested in G1 with α-factor for 150 min. The cells were then released into YPD medium containing 0.03% MMS. Protein extracts derived from cells removed at timed intervals (as indicated in minutes below the lanes) were then separated by SDS-PAGE, transferred to nylon, and immunoblotted with an anti-Rad53 antibody. A representative Western blot for extracts from wild-type, top3 sgs1, and top3 cells is shown. The positions of unphosphorylated Rad53 and phosphorylated Rad53 (Rad53-P) are shown on the right. (b) Flow cytometric analysis to confirm the arrest of wild-type (left) and top3 (right) strains after exposure to nocodazole for 90 min. A population of exponentially growing cells (EXP) is shown in each case for comparison. Peaks representing cells with a 1C and a 2C DNA content are indicated. (c) Phosphorylation of Rad53 in wild-type and top3 strains (as indicated above the lanes) following UV irradiation in G2/M. Lanes: Exp, unirradiated exponentially growing culture; G2, nocodazole-arrested culture; −UV, cells released from nocodazole arrest, mock irradiated, and incubated for 90 min; +UV, cells released from nocodazole arrest, UV irradiated, and incubated for 90 min. The positions of the nonphosphorylated and phosphorylated bands of Rad53 protein, detected by Western blotting as for panel a, are indicated on the right.
To confirm that top3Δ mutants did not show a general defect in signaling during a check point-mediated cell cycle arrest, we studied Rad53 phosphorylation during a G2/M checkpoint arrest induced after UV irradiation of nocadazole-treated cells (Fig. 4b). Figure 4c shows that comparable levels of Rad53p phosphorylation were seen in wild-type and top3Δ strains following irradiation, confirming that signaling to Rad53 in the G2/M DNA damage checkpoint is intact in top3Δ strains.
Genetic interactions between top3 and other checkpoint-deficient mutants.
The MEC1 and RAD53 genes encode essential proteins involved in the signal transduction pathway that is activated during S phase by inhibition of DNA replication and during all phases of the cell cycle by DNA damage (2, 52, 66). Combination of a top3Δ mutation in the A364a strain with alleles of MEC1 (mec1-1) and RAD53 (mec2-1) resulted in synthetic lethality (either microcolonies that could not be propagated or no visible colony) after sporulation of the appropriate heterozygous diploids (in each case, at least 40 tetrads were dissected). In an effort to characterize genetic interactions between top3Δ and other mutations that disable potential targets of the Mec1/Rad53 signal transduction pathway, we examined the effect of combining deletion of TOP3 with conditional mutations in RFA2, which encodes the 34-kDa subunit of the heterotrimeric single-stranded DNA binding protein RPA, and PRI1, which encodes the large subunit of DNA primase. Rfa2p has roles in both DNA replication and DNA repair in budding yeast. Rfa2p is phosphorylated in a MEC1-dependent manner in response to DNA replication block or DNA damage (4). Combination of top3Δ with either of two independent rfa2 alleles (rfa2-1 and rfa2-2) resulted in synthetic lethality, either at the permissive temperature for top3Δ rfa2-1 double mutants (25°C; n = 20 tetrads dissected) or at the semipermissive temperature for top3Δ rfa2-2 double mutants (30°C; n = 20). Similarly, combination of a top3Δ mutation with the pri1-M4 mutation, which itself leads to a defect in S-phase checkpoint responses to DNA damage (39), resulted in synthetic lethality at the permissive temperature for the pri1-M4 strain (25°C; n = 20). One interpretation of these data is that deletion of TOP3 is lethal in combination with any mutation that perturbs DNA replication. However, this proved not to be the case, since a combination of top3 with pol2-12 (42) produced viable spore colonies for the double mutant (dissection of 40 tetrads).
Further evidence for an S-phase role for Top3p.
Thus far, our data implicate Top3p in an S-phase-specific role in response to DNA perturbations. To gain further evidence to substantiate this suggestion, we studied whether top3Δ strains are sensitive to the ribonucleotide reductase inhibitor HU, which inhibits DNA replication, and whether the pattern of TOP3 gene expression was indicative of a role in S phase. Figure 5a shows that top3Δ mutants are highly sensitive to killing by HU compared to wild-type control cells and that this sensitivity is substantially suppressed by deletion of SGS1. To assess cell cycle regulation of TOP3 gene expression, wild-type cells were arrested in G1 with α-factor and then released into fresh medium. RNA samples were prepared at timed intervals for Northern blot analysis, and the cell cycle position was assessed in parallel by analysis of both the budding index and DNA content by flow cytometry. In the representative experiment shown in Fig. 5b, synchrony was maintained until the middle of the second cycle. The level of TOP3 mRNA peaked 20 min after release from α-factor arrest at a level 11-fold higher than that in the arrested cells (Fig. 5c). This time point coincided with the onset of the decline of G1 cells, as measured by flow cytometry, but was prior to the appearance of budded cells. In the second cycle, the peak in TOP3 mRNA levels was coincident with the rise in the proportion of G1 cells. We estimated the size of the TOP3 transcript to be ≈2.5 kb. This pattern of transcription was confirmed in a second strain background, and the 2.5-kb transcript was undetectable in RNA derived from top3Δ cells (data not shown). Thus, it appears that TOP3 transcripts are few in early G1, appear abruptly around Start, and then decline during late S/G2.
FIG. 5.
Further evidence for an S-phase-specific role for Top3p. (a) top3Δ mutants are sensitive to HU. Wild-type (□), top3 (○), and top3 sgs1 (●) strains (as indicated in Fig. 2) were exposed to 0.2 M HU for the times indicated, and percent survival was determined. The means and standard errors of three independent experiments are shown. (b and c) Cell cycle regulation of TOP3 mRNA. Wild-type cells (RKC1d) were grown to early log phase, arrested in G1 as described in the legend to Fig. 4, and then released into fresh warmed medium. The cells were removed at timed intervals for RNA extraction, measurement of the budding index, and flow cytometric analysis of the DNA content. (b) Levels of the TOP3 and ACT1 transcripts at timed intervals prior to and following release from G1 arrest. (c) The percentage of budded cells (○) and cells gated with a G1 content of DNA (□) plotted relative to TOP3 transcript levels (▪) quantified from the data in panel a. The maximal transcript level is given an arbitrary value of 100%. Note that cells lose synchrony at about the midpoint of the second cycle.
DISCUSSION
We have shown that S. cerevisiae top3Δ mutants are sensitive to DNA-damaging agents and HU and have an abrogated intra-S checkpoint response to DNA damage. These defects can be rescued by deletion of the SGS1 gene. In contrast, top3Δ mutants are proficient in DNA damage checkpoint responses that operate in the G1 or G2 phase of the cell cycle. We have also provided evidence that Top3p lies upstream of the Mec1-Rad53-dependent signal transduction cascade in the cellular response to DNA structural perturbations occurring within S phase.
top3Δ strains are defective in cell cycle progression in the absence of exogenous DNA-damaging agents.
Previous work has shown that the extended doubling time of top3Δ strains is a result of an accumulation of cells in the late S/G2 phase of the cell cycle (15). We have shown here that these cells have completed bulk DNA replication and are arrested at the G2/M DNA damage checkpoint. The most economical explanation for these findings is that some form of abnormal DNA structure and/or DNA lesion is generated during the process of DNA replication and that while this is not sufficient to prevent completion of DNA synthesis, it nevertheless is recognized by the G2/M DNA damage checkpoint machinery as abnormal. Although top3Δ strains delay at the G2/M checkpoint for an extended period, when this checkpoint is disabled by deletion of RAD24, the already-reduced viability of top3Δ strains is not further decreased. Indeed, the doubling time of the double mutant is shortened compared to that of top3Δ mutants. This might indicate that a substantial fraction of the DNA lesions that lead to induction of the G2/M checkpoint arrest in top3Δ mutants are either irreparable, at least in G2/M, or can be tolerated in subsequent cell cycles.
top3Δ strains are sensitive to several classes of DNA-damaging agents and replication inhibitors.
Mutations in a wide variety of genes encoding DNA repair enzymes or checkpoint proteins confer sensitivity to DNA-damaging agents. For example, mutation of nucleotide excision repair genes leads to sensitivity to UV light, while mutation of recombinational repair genes confers sensitivity primarily to ionizing radiation and MMS. top3Δ mutants are unusual in being sensitive to MMS and UV light, but not markedly to γ rays, suggesting that the Top3 protein probably does not play a dedicated role in one of the major pathways for the repair of specific DNA lesions but instead operates more generally in the cellular response to DNA damage. To our knowledge, this represents the first evidence in eukaryotes that a topoisomerase can protect cells from the cytotoxic effects of DNA-damaging agents. Given the intact nature of DNA damage checkpoint responses occurring in G1 and G2, but a failure to adequately invoke the intra-S DNA damage checkpoint, it would appear that DNA damage arising during S phase presents the most (or possibly only) serious challenge to top3Δ mutants. A major goal for the future is to identify the abnormal DNA structures that might occur during progression through S phase in these mutants. It is known that aberrant replicative structures resembling recombination intermediates or late Cairns-type structures can be observed on two-dimensional gels following drug-mediated inactivation of topoisomerase I or II in budding yeast (29). However, we have shown that DNA samples derived from asynchronous cultures of wild-type and top3Δ strains exhibit no consistent differences on two-dimensional gels (our unpublished data). We conclude that any putative abnormal replication intermediates that might arise in top3Δ strains either fall outside the group of structures detectable by this method or are accumulated at levels below the detection limit. We have shown that top3Δ strains are highly sensitive to the ribonucleotide reductase inhibitor HU. This sensitivity is suppressed by deletion of SGS1. Some of the genes required for protection against HU, such as MEC1 and RAD53, are required to prevent mitosis from occurring during arrest in S phase (loss of the so-called S/M checkpoint) (7, 32, 37, 57, 65). Our recent work indicates that HU-treated top3Δ cells do not obviously enter mitosis directly from an early S-phase arrest, as evidenced by the fact that there is no progressive elongation of the mitotic spindle. Nevertheless, during exposure to HU, top3Δ strains do show a progressive increase in the percentage of cells displaying aberrant nuclear DNA staining, including cells with marked DNA fragmentation (unpublished data). Further work will be required to characterize the terminal phenotype of HU-treated top3 mutants.
TOP3 is a putative new member of the SCB box-containing family of genes.
The proposal that Top3p has a role in S phase is supported by our data showing that TOP3 transcript levels are cell cycle regulated, arising in G1 and declining in late S/G2. A genome-wide transcript analysis also indicated that the TOP3 mRNA is induced in G1 (6). A number of genes are transcribed exclusively in the late G1 phase or at the G1/S boundary, including the G1 cyclins and certain genes required for DNA synthesis (reviewed in reference 51). These late-G1-activated genes can be classified into two groups on the basis of cis-acting sequences found within their promoter sequences. The first group of genes includes the DNA metabolism genes (e.g., RFA1-3, POL1-3, and DBF4) and the CLB5 and CLB6 cyclin genes, and the promoter regions of these genes contain an element similar to the Mlu1 cell cycle box (MCB element). The second group of genes, including CLN1, CLN2, and HCS26, contain a promoter motif, termed the SCB element, which acts as a late-G1-specific upstream activating sequence and binds the Swi4-Swi6 complex (44). Following S phase, transcription of both of these groups of genes is down-regulated. In this context, we have identified a potential SCB element in the 5′ flanking region of TOP3 (CGCGAAA, at positions −130 to −124 from the ATG start codon), suggesting that TOP3 is a new member of the group of genes regulated by the Swi4-Swi6 complex. While this finding would be consistent with the G1 activation of TOP3 gene expression, it should be noted that the minimal promoter region and the positions of any potential transcription start sites in the TOP3 gene have yet to be characterized.
How does loss of Top3p activity result in loss of checkpoint proficiency?
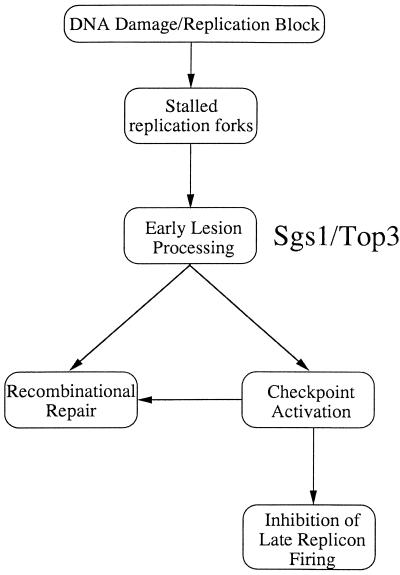
The TOP3 gene shows genetic interactions with SGS1, and biochemical analyses have shown that the products of these genes physically associate (3, 15). Moreover, we and others have shown that topoisomerase IIIα and BLM also physically interact in human cells, confirming that the association between the topoisomerase III and RecQ helicase enzymes is highly conserved (22, 69). It is not unreasonable to assume, therefore, that Sgs1p and Top3p act in concert while performing many, if not all, of their cellular functions. However, in contrast to top3Δ mutants, cells lacking Sgs1p show only a modest growth defect, are only slightly more sensitive to DNA-damaging agents than are wild-type cells, and are reported to have only a minor intra-S checkpoint defect (see the discussion below). How can the above-mentioned findings be incorporated into a model that explains the role of Top3p during normal DNA replication or when replication is perturbed by DNA damage? Following DNA damage, replication forks can stall, and we suggest that Sgs1p-Top3p is involved in the processing of the resulting abnormal DNA structures or lesions (Fig. 6). The suggestion that the enzymatic activity of Top3p is required for function is consistent with the finding that the sensitivity of top3Δ mutants to DNA-damaging agents cannot be corrected by expression of Top3p which has been mutated at its catalytic active site (our unpublished data). The Sgs1p-Top3p complex could serve two functions, which are not mutually exclusive, in the cellular response to S-phase perturbation. First, it could act in the generation of DNA structures that are a necessary intermediate in the activation of the checkpoint cascade. Second, it could prepare the damaged DNA for the DNA repair machinery. Specifically, we propose that during S phase this repair could exploit the availability of the genetic information on the intact sister chromatid and therefore proceed via the Rad52-dependent recombinational-repair pathway. Murray et al. (41) have suggested a similar role for fission yeast rqh1+ in preparing DNA lesions at blocked replication forks for the recombinational repair machinery. Consistent with this proposal, we have shown recently that Sgs1p interacts with the Rad51 recombinase (70). This model would also be consistent with the proposed role of the RecQ protein, the Escherichia coli homologue of Sgs1p, which in concert with RecA can initiate homologous recombination and disrupt joint molecules formed by aberrant recombination (19). In further support of this general concept are the observations that recombination intermediates (Holliday junctions) can be detected in yeast during S phase and that perturbation of replication leads to an elevation in their frequency (73).
FIG. 6.
Model for the role of Top3p in mediating S-phase checkpoint responses. We propose that the Sgs1-Top3 complex is involved in the early processing of aberrant structures arising at blocked or stalled replication forks. The complex may play at least two roles: preparation of lesions for repair by the recombinational repair pathway and activation of the checkpoint that leads to an inhibition of cell cycle progression. In the absence of Top3, there is a failure to adequately integrate the cellular response to the disturbance of replication.
Deletion of SGS1 at least partially suppresses all of the studied phenotypes in top3Δ mutants, including the intra-S checkpoint defect. One explanation for this could be that the phenotype of top3Δ mutants relates primarily to a deregulation of Sgs1p enzymatic activity. Such deregulated activity could interfere, either directly or indirectly, with the checkpoint machinery. Alternatively, deletion of SGS1 could permit the utilization of a redundant pathway that could lead to activation of the S-phase checkpoint. It has been suggested previously that a key role for checkpoint proteins is to process certain DNA structural abnormalities in readiness for their repair by dedicated repair proteins (reviewed in reference 65). If Top3p were to participate in such a role, one implication would be that some degree of lesion processing by the Sgs1p-Top3p complex is required in order for a robust S-phase checkpoint response to be invoked. This putative role would likely require the catalytic activity of Top3p to resolve structures created by the Sgs1 helicase. Evidence in support of the concept that some processing of lesions is required to invoke certain checkpoint responses comes from the finding that in DNA repair-deficient rad14 mutants, the UV-induced G1/S checkpoint is not RAD9 dependent, as it is in wild-type cells (53). Whether Top3 performs roles independent of Sgs1 will require additional studies. This suggestion is not unreasonable, however, given that sgs1 top3 mutants grow more slowly and are more UV/MMS sensitive than are sgs1 mutants.
Recent data indicate that deletion of SGS1 leads to a partial defect in the intra-S checkpoint, which is not associated with an alteration in phosphorylation of Rad53p after DNA damage (12, 13). However, in combination with deletion of RAD24, loss of Sgs1 function leads to some attenuation in the extent of Rad53 modification. Further, Sgs1p and Rad53p have been shown to colocalize in S-phase-specific foci (12). Our results are consistent with those of Frei and Gasser (12) in that we have shown that Top3p acts upstream of Rad53 in the S-phase response to DNA damage. However, our data show that top3 mutants have a much more severe S-phase checkpoint defect than do sgs1 mutants and, moreover, that deletion of SGS1 has the effect of strongly suppressing the defects in top3 mutants. Indeed, at least in the strain background that we analyzed, any effects of an sgs1 mutation alone or the combination of an sgs1 and a top3 mutation on intra-S checkpoint proficiency and Rad53 phosphorylation were not obvious. A second DNA helicase in S. cerevisiae, Srs2p, is required for normal activation of Rad53p during S phase, and srs2Δ strains show a defect in the intra-S checkpoint (31). In combination with deletion of SGS1 or TOP3, srs2Δ strains show very low viability, which is apparently associated with an accumulation of aberrant genetic recombination structures (8, 16, 28). Interestingly, recent data indicate that defects in a recently identified protein, Tof1p, which was first identified through interactions with topoisomerase I, have phenotypic consequences similar to those reported here for top3Δ strains. (11). These similarities include sensitivity to DNA-damaging agents and HU and an S-phase-specific defect in DNA damage checkpoint signaling to Rad53p. Further work will be required to assess whether Top1p is functionally connected with Sgs1-Top3p
In summary, we have shown that functional topoisomerase III is required for the normal response of S. cerevisiae cells to DNA-damaging agents. A recent report indicated that expression of a truncated form of human topoisomerase IIIα can partially reverse certain phenotypes associated with ataxia telangiectasia cells, which are defective in the response to DNA damage (14). Since the expression of hTOP3α was shown to correct both radio-resistant DNA synthesis (analogous to the intra-S checkpoint in yeast) and hyperrecombination, it is possible that the general model described here is also applicable in certain circumstances to higher eukaryotes.
ACKNOWLEDGMENTS
We thank R. Rothstein, J. Wang, T. Weinert, C. Santocanale, and M. Foiani for providing yeast strains, plasmids, and antibodies. We also thank C. Norbury and L. Wu for helpful comments on the manuscript and J. Pepper for preparation of the manuscript.
Funding was provided by the Imperial Cancer Research Fund and the Medical Research Council. R.K.C. was a Medical Research Council Clinical Training Fellow.
R.K.C., J.M.K., and T.J.O. contributed equally to the work.
REFERENCES
- 1.Aboussekhra A, Vialard J E, Morrison D E, de la Torre-Ruiz M-A, Cernakova L, Fabre F, Lowndes N F. A novel role for the budding yeast RAD9 checkpoint gene in DNA damage dependent transcription. EMBO J. 1996;15:3912–3922. [PMC free article] [PubMed] [Google Scholar]
- 2.Allen J B, Zhou Z, Siede W, Friedberg E C, Elledge S J. The SAD1/RAD53 protein kinase controls multiple checkpoints and DNA damage-induced transcription in yeast. Genes Dev. 1994;8:2401–2428. doi: 10.1101/gad.8.20.2401. [DOI] [PubMed] [Google Scholar]
- 3.Bennett R J, Noirot-Gros M F, Wang J C. Interaction between yeast sgs1 helicase and DNA topoisomerase III. J Biol Chem. 2000;275:26898–26905. doi: 10.1074/jbc.M003137200. [DOI] [PubMed] [Google Scholar]
- 4.Brush G S, Morrow D M, Hieter P, Kelly T J. The ATM homologue MEC1 is required for phosphorylation of replication protein A in yeast. Proc Natl Acad Sci USA. 1996;93:15075–15080. doi: 10.1073/pnas.93.26.15075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chakraverty R K, Hickson I D. Defending genome integrity during DNA replication: a proposed role for RecQ family helicase. Bioessays. 1999;21:286–294. doi: 10.1002/(SICI)1521-1878(199904)21:4<286::AID-BIES4>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 6.Cho R J, Campbell M J, Winzeler E A, Steinmetz L, Conway A, Wodicka L, Wolfsberg T G, Gabrielian A E, Landsman D, Lockhart D J, Davis R W. A genome-wide transcription analysis of the mitotic cell cycle. Mol Cell. 1998;2:65–73. doi: 10.1016/s1097-2765(00)80114-8. [DOI] [PubMed] [Google Scholar]
- 7.de la Torre-Ruiz M-A, Green C M, Lowndes N F. RAD9 and RAD24 define two additive, interacting branches of the DNA damage checkpoint pathway in budding yeast normally required for Rad53 modification and activation. EMBO J. 1998;9:2687–2698. doi: 10.1093/emboj/17.9.2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duno M, Thomsen B, Westergaard O, Krejci L, Bendixen C. Genetic analysis of the Saccharomyces cerevisiae Sgs1 helicase defines an essential function for the Sgs1-Top3 complex in the absence of SRS2 or TOP1. Mol Gen Genet. 2000;264:89–97. doi: 10.1007/s004380000286. [DOI] [PubMed] [Google Scholar]
- 9.Ellis N A, Groden J, Ye T-Z, Straughen J, Lennon D J, Ciocci S, Proytcheva M, German J. The Bloom's syndrome gene product is homologous to RecQ helicases. Cell. 1995;83:655–666. doi: 10.1016/0092-8674(95)90105-1. [DOI] [PubMed] [Google Scholar]
- 10.Foiani M, Marini F, Gamba D, Lucchini G, Plevani P. The B subunit of the DNA polymerase alpha-primase complex in Saccharomyces cerevisiae executes an essential function at the initial stages of DNA replication. Mol Cell Biol. 1994;14:923–933. doi: 10.1128/mcb.14.2.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Foss E J. Tof1p regulates DNA damage responses during S phase in Saccharomyces cerevisiae. Genetics. 2001;157:567–577. doi: 10.1093/genetics/157.2.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frei C, Gasser S M. The yeast Sgs1p helicase acts upstream of Rad53p in the DNA replication checkpoint and colocalizes with Rad53p in S-phase-specific foci. Genes Dev. 2000;14:81–96. [PMC free article] [PubMed] [Google Scholar]
- 13.Frei C, Gasser S M. RecQ-like helicases: the DNA replication checkpoint connection. J Cell Sci. 2000;113:2641–2646. doi: 10.1242/jcs.113.15.2641. [DOI] [PubMed] [Google Scholar]
- 14.Fritz E, Elsea S H, Patel P I, Meyn S. Overexpression of a truncated human topoisomerase III partially corrects multiple aspects of the ataxia telangiectasia phenotype. Proc Natl Acad Sci USA. 1997;94:4538–4542. doi: 10.1073/pnas.94.9.4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gangloff S, McDonald J P, Bendixen C, Arthur L, Rothstein R. The yeast type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog: a potential eukaryotic reverse gyrase. Mol Cell Biol. 1994;14:8391–8398. doi: 10.1128/mcb.14.12.8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gangloff S, Soustelle C, Fabre F. Homologous recombination is responsible for cell death in the absence of the Sgs1 and Srs2 helicases. Nat Genet. 2000;25:192–194. doi: 10.1038/76055. [DOI] [PubMed] [Google Scholar]
- 17.Goodwin A, Wang S-W, Toda T, Norbury C, Hickson I D. Topoisomerase III is essential for accurate nuclear division in Schizosaccharomyces pombe. Nucleic Acids Res. 1999;27:4050–4058. doi: 10.1093/nar/27.20.4050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanai R, Caron P R, Wang J C. Human TOP3: a single-copy gene encoding DNA Top3p. Proc Natl Acad Sci USA. 1996;93:3653–3657. doi: 10.1073/pnas.93.8.3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harmon F G, Kowalczykowski S C. RecQ helicase, in concert with RecA and SSB proteins, initiates and disrupts DNA recombination. Genes Dev. 1998;12:1134–1144. doi: 10.1101/gad.12.8.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hennessy K M, Lee A, Chen E, Botstein D. A group of interacting yeast DNA replication genes. Genes Dev. 1991;5:958–969. doi: 10.1101/gad.5.6.958. [DOI] [PubMed] [Google Scholar]
- 21.Hiasa H, Marians K J. Top3p, but not Topoisomerase I, can support nascent chain elongation during theta-type DNA replication. J Biol Chem. 1994;269:32655–32659. [PubMed] [Google Scholar]
- 22.Johnson F B, Lombard D B, Neff N F, Mastrangelo M A, Dewolf W, Ellis N A, Marciniak R A, Yin Y, Jaenisch R, Guarente L. Association of the Bloom syndrome protein with topoisomerase IIIalpha in somatic and meiotic cells. Cancer Res. 2000;60:1162–1167. [PubMed] [Google Scholar]
- 23.Karow J K, Wu L, Hickson I D. RecQ family helicases: roles in cancer and aging. Curr Opin Genet Dev. 2000;10:32–38. doi: 10.1016/s0959-437x(99)00039-8. [DOI] [PubMed] [Google Scholar]
- 24.Kawasaki K, Minoshima S, Nakato E, Shibuya K, Shintani A, Schmeits J L, Wang J, Shimizu N. One-megabase sequence analysis of the human Immunoglobulin λ gene locus. Genome Res. 1997;7:250–261. doi: 10.1101/gr.7.3.250. [DOI] [PubMed] [Google Scholar]
- 25.Kim R A, Wang J C. Identification of the yeast TOP3 gene product as a single strand-specific DNA topoisomerase. J Biol Chem. 1992;267:17178–17185. [PubMed] [Google Scholar]
- 26.Kiser G L, Weinert T A. Distinct roles of yeast MEC and RAD checkpoint genes in transcriptional induction after DNA damage and implications for function. Mol Biol Cell. 1996;7:703–718. doi: 10.1091/mbc.7.5.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kitao S, Shimamoto A, Goto M, Miller R W, Smithson W A, Lindor N M, Furuichi Y. Mutations in RECQL4 cause a subset of cases of Rothmund-Thomson syndrome. Nat Genet. 1999;22:82–84. doi: 10.1038/8788. [DOI] [PubMed] [Google Scholar]
- 28.Lee S K, Johnson R E, Yu S L, Prakash L, Prakash S. Requirement of yeast SGS1 and SRS2 genes for replication and transcription. Science. 1999;286:2339–2342. doi: 10.1126/science.286.5448.2339. [DOI] [PubMed] [Google Scholar]
- 29.Levac P, Moss T. Inactivation of topoisomerase I or II may lead to recombination or aberrant replication on both SV40 and yeast 2μm DNA. Chromosoma. 1996;105:250–260. doi: 10.1007/BF02528774. [DOI] [PubMed] [Google Scholar]
- 30.Li W, Wang J C. Mammalian DNA Top3pα is essential in early embryogenesis. Proc Natl Acad Sci USA. 1998;95:1010–1013. doi: 10.1073/pnas.95.3.1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liberi G, Chiolo I, Pellicioli A, Lopes M, Plevani P, Muzi-Falconi M, Foiani M. Srs2 DNA helicase is involved in checkpoint response and its regulation requires a functional Mec1-dependent pathway and cdk1 activity. EMBO J. 2000;19:5027–5038. doi: 10.1093/emboj/19.18.5027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Longhese M P, Foiani M, Muzi-Falconi M, Lucchini G, Plevani P. DNA damage checkpoint in budding yeast. EMBO J. 1998;17:5525–5528. doi: 10.1093/emboj/17.19.5525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Longhese M P, Fraschini R, Plevani P, Lucchini G. Yeast pip3/mec3 mutants fail to delay entry into S-phase and to slow replication in response to DNA damage, and they define a functional link between Mec3 and DNA primase. Mol Cell Biol. 1996;16:3235–3244. doi: 10.1128/mcb.16.7.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Longhese M P, Paciotti V, Fraschini R, Zaccarini R, Plevani P, Lucchini G. The novel DNA damage checkpoint protein Ddc1p is phosphorylated periodically during the cell cycle and in response to DNA damage in budding yeast. EMBO J. 1997;16:5216–5226. doi: 10.1093/emboj/16.17.5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Louis E J, Haber J E. The Subtelomeric Y′ repeat family in Saccharomyces cerevisiae: an experimental system for repeated sequence evolution. Genetics. 1990;124:533–545. doi: 10.1093/genetics/124.3.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lowndes N F, Murguia J R. Sensing and responding to DNA damage. Curr Opin Genet Dev. 2000;10:17–25. doi: 10.1016/s0959-437x(99)00050-7. [DOI] [PubMed] [Google Scholar]
- 37.Lydall D, Weinert T. From DNA damage to cell cycle arrest and suicide: a budding yeast perspective. Curr Opin Genet Dev. 1996;5:12–16. doi: 10.1016/s0959-437x(96)90003-9. [DOI] [PubMed] [Google Scholar]
- 38.Maftahi M, Han C S, Langston L D, Hope J C, Zigouras N, Freyer G A. The top3+ gene is essential in Schizoccharomyces pombe and the lethality associated with its loss is caused by Rad12 helicase activity. Nucleic Acids Res. 1999;27:4715–4724. doi: 10.1093/nar/27.24.4715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marini F, Pellicioli A, Paciotti V, Lucchini G, Plevani P, Stern D F, Foiani M. A role for DNA primase in coupling DNA replication to DNA damage response. EMBO J. 1997;16:639–650. doi: 10.1093/emboj/16.3.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miyajima A, Seki M, Onoda F, Shiratori M, Odagiri N, Ohta K, Kikuchi Y, Ohno Y, Enomoto T. Sgs1 helicase activity is required for mitotic but apparently not for meiotic functions. Mol Cell Biol. 2000;20:6399–6409. doi: 10.1128/mcb.20.17.6399-6409.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murray J M, Lindsay H D, Munday C A, Carr A M. Role of Schizosaccharomyces pombe RecQ homolog, recombination, and checkpoint genes in UV damage tolerance. Mol Cell Biol. 1997;17:6868–6875. doi: 10.1128/mcb.17.12.6868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Navas T A, Sanchez Y, Elledge S J. RAD9 and DNA polymerase ɛ form parallel sensory branches for transducing the DNA damage checkpoint in Saccharomyces cerevisiae. Genes Dev. 1996;10:2632–2643. doi: 10.1101/gad.10.20.2632. [DOI] [PubMed] [Google Scholar]
- 43.Norbury C, Hickson I D. Cellular responses to DNA damage. Annu Rev Pharmacol Toxicol. 2001;41:367–401. doi: 10.1146/annurev.pharmtox.41.1.367. [DOI] [PubMed] [Google Scholar]
- 44.Ogas J, Andrews B J, Herskowitz I. Transcriptional activation of the CLN1, CLN2, and a putative new G1 cyclin (HCS26) by Swi4, a positive regulator of G1-specific transcription. Cell. 1991;66:1015–1026. doi: 10.1016/0092-8674(91)90445-5. [DOI] [PubMed] [Google Scholar]
- 45.Paulovich A G, Hartwell L H. A checkpoint regulates the rate of progression through S-phase in S. cerevisiae in response to DNA damage. Cell. 1995;82:841–847. doi: 10.1016/0092-8674(95)90481-6. [DOI] [PubMed] [Google Scholar]
- 46.Paulovich A G, Margulies R U, Garvik B M, Hartwell L H. RAD9, RAD17 and RAD24 are required for S-phase regulation in Saccharomyces cerevisiae in response to DNA damage. Genetics. 1997;145:45–62. doi: 10.1093/genetics/145.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paulovich A G, Toczyski D P, Hartwell L H. When checkpoints fail. Cell. 1997;88:315–321. doi: 10.1016/s0092-8674(00)81870-x. [DOI] [PubMed] [Google Scholar]
- 48.Pellicioli A, Lucca C, Liberi G, Marini F, Lopes M, Plevani P, Romano A, Di Fiore P P, Foiani M. Activation of Rad53 kinase in response to DNA damage and its effect in modulating phosphorylation of the lagging strand DNA polymerase. EMBO J. 1999;18:6561–6572. doi: 10.1093/emboj/18.22.6561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pringle J R, Hartwell L H. The Saccharomyces cerevisiae cell cycle. In: Strathern J N, Jones E W, Broach J R, editors. The molecular biology of the yeast Saccharomyces. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1981. pp. 97–142. [Google Scholar]
- 50.Rhind N, Russell P. Chk1 and cds1: linchpins of the DNA damage and replication checkpoint pathways. J Cell Sci. 2000;113:3889–3896. doi: 10.1242/jcs.113.22.3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schwob E, Nasmyth K. Cell cycle control of DNA replication in Saccharomyces cerevisiae. In: Blow J J, editor. Eukaryotic DNA replication. Oxford, United Kingdom: Oxford University Press; 1996. pp. 165–196. [Google Scholar]
- 52.Siede W, Allen J B, Elledge S J, Friedberg E C. The Saccharomyces cerevisiae MEC1 gene, which encodes a homolog of the human ATM gene, is required for G1 arrest following irradiation treatment. J Bacteriol. 1996;178:5841–5843. doi: 10.1128/jb.178.19.5841-5843.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Siede W, Friedberg A S, Dianova I, Friedberg E C. Characterisation of G1 checkpoint control in the yeast Saccharomyces cerevisiae following exposure to DNA damaging agents. Genetics. 1994;138:271–281. doi: 10.1093/genetics/138.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Siede W, Friedberg A S, Friedberg E C. RAD9-dependent G1 arrest defines a second checkpoint for damaged DNA in the cell cycle of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1993;90:7985–7989. doi: 10.1073/pnas.90.17.7985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Siede W, Nusspaumer G, Portillo V, Rodriguez R, Friedberg E C. Cloning and characterisation of RAD17: a gene controlling cell cycle responses to DNA damage in Saccharomyces cerevisiae. Nucleic Acids Res. 1996;24:1669–1675. doi: 10.1093/nar/24.9.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sugimoto K, Ando S, Shimomura T, Matsumoto K. Rfc5, a replication factor C component, is required for regulation of Rad53 protein kinase in the yeast checkpoint pathway. Mol Cell Biol. 1997;17:5905–5914. doi: 10.1128/mcb.17.10.5905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sun Z, Fay D S, Marini F, Foiani M, Stern D F. Spk1/Rad53 is regulated by Mec-1 dependent protein phosphorylation in DNA replication and damage checkpoint pathways. Genes Dev. 1996;10:395–406. doi: 10.1101/gad.10.4.395. [DOI] [PubMed] [Google Scholar]
- 58.Vennos E M, James W D. Rothmund-Thomson syndrome. Dermatol Clin. 1995;13:143–150. [PubMed] [Google Scholar]
- 59.Wach A, Brachat A, Pohlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 60.Wallis J W, Chrebet G, Brodsky G, Rolfe M, Rothstein R. A hyper-recombination mutation in S. cerevisiae identifies a novel eukaryotic topoisomerase. Cell. 1989;58:409–419. doi: 10.1016/0092-8674(89)90855-6. [DOI] [PubMed] [Google Scholar]
- 61.Wang J C. DNA topoisomerases. Annu Rev Biochem. 1996;65:639–692. doi: 10.1146/annurev.bi.65.070196.003223. [DOI] [PubMed] [Google Scholar]
- 62.Watt P M, Hickson I D. Structure and function of type II topoisomerases. Biochem J. 1994;303:681–695. doi: 10.1042/bj3030681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Watt P M, Hickson I D, Borts R H, Louis E J. Sgs1, a homologue of the Bloom's and Werner's Syndrome genes, is required for maintenance of genome stability in Saccharomyces cerevisiae. Genetics. 1996;144:935–945. doi: 10.1093/genetics/144.3.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Watt P M, Louis E J, Borts R H, Hickson I D. Sgs1: a homolog of E. coli RecQ that interacts with topoisomerase II in vivo and is required for faithful chromosome segregation. Cell. 1995;81:253–260. doi: 10.1016/0092-8674(95)90335-6. [DOI] [PubMed] [Google Scholar]
- 65.Weinert T. DNA damage checkpoints update: getting molecular. Curr Opin Genet Dev. 1998;8:185–193. doi: 10.1016/s0959-437x(98)80140-8. [DOI] [PubMed] [Google Scholar]
- 66.Weinert T A, Kiser G L, Hartwell L H. Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair. Genes Dev. 1994;8:652–655. doi: 10.1101/gad.8.6.652. [DOI] [PubMed] [Google Scholar]
- 67.Wigley D B. Structure and mechanism of DNA topoisomerases. Annu Rev Biophys Biomol Struct. 1995;24:185–208. doi: 10.1146/annurev.bb.24.060195.001153. [DOI] [PubMed] [Google Scholar]
- 68.Wilson T M, Chen A D, Hsieh T. Cloning and characterization of Drosophila topoisomerase IIIbeta. Relaxation of hypernegatively supercoiled DNA. J Biol Chem. 2000;275:1533–1540. doi: 10.1074/jbc.275.3.1533. [DOI] [PubMed] [Google Scholar]
- 69.Wu L, Davies S, North P S, Goulaouic H, Riou J-F, Turley H, Gatter K C, Hickson I D. The Bloom's syndrome gene product interacts with topoisomerase III. J Biol Chem. 2000;275:9636–9644. doi: 10.1074/jbc.275.13.9636. [DOI] [PubMed] [Google Scholar]
- 70.Wu L, Davies S L, Levitt N C, Hickson I D. Potential role for the BLM helicase in recombinational repair via a conserved interaction with Rad51. J Biol Chem. 2001;276:19375–19381. doi: 10.1074/jbc.M009471200. [DOI] [PubMed] [Google Scholar]
- 71.Yamamoto A, Guacci V, Koshland D. Pds1p is required for faithful execution of anaphase in the yeast, Saccharomyces cerevisiae. J Cell Biol. 1996;133:85–97. doi: 10.1083/jcb.133.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yu C-E, Oshima J, Fu Y-H, Wijsman E M, Hisama F, Alisch R, Mathews S, Nakura T, Miki T, Ouais S, Martin G M, Mulligan J, Schellenberg G D. Positional cloning of the Werner's syndrome gene. Science. 1996;272:258–262. doi: 10.1126/science.272.5259.258. [DOI] [PubMed] [Google Scholar]
- 73.Zhou H, Rothstein R. Holliday junctions accumulate in DNA replication mutants via a RecA homolog-independent mechanism. Cell. 1997;90:87–96. doi: 10.1016/s0092-8674(00)80316-5. [DOI] [PubMed] [Google Scholar]