Abstract
Monitoring protein biomarker levels in the cerebrospinal fluid (CSF) can help assess injury severity and outcome after traumatic brain injury (TBI). Determining injury-induced changes in the proteome of brain extracellular fluid (bECF) can more closely reflect changes in the brain parenchyma, but bECF is not routinely available. The aim of this pilot study was to compare time-dependent changes of S100 calcium-binding protein B (S100B), neuron-specific enolase (NSE), total Tau, and phosphorylated Tau (p-Tau) levels in matching CSF and bECF samples collected at 1, 3, and 5 days post-injury from severe TBI patients (n = 7; GCS 3–8) using microcapillary-based western analysis. We found that time-dependent changes in CSF and bECF levels were most pronounced for S100B and NSE, but there was substantial patient-to-patient variability. Importantly, the temporal pattern of biomarker changes in CSF and bECF samples showed similar trends. We also detected two different immunoreactive forms of S100B in both CSF and bECF samples, but the contribution of the different immunoreactive forms to total immunoreactivity varied from patient to patient and time point to time point. Our study is limited, but it illustrates the value of both quantitative and qualitative analysis of protein biomarkers and the importance of serial sampling for biofluid analysis after severe TBI.
Keywords: biomarker, cMD, CSF, protein, temporal, traumatic brain injury
Introduction
Cerebrospinal fluid (CSF) is uniquely qualified for protein biomarker analysis after traumatic brain Injury (TBI) because of its proximity to the brain (for reviews, see previous works1–3). Cerebral microdialysis (cMD)4 has greatly contributed to the better understanding of changes in intracranial metabolism after TBI by enabling continuous sampling and analysis of brain extracellular fluid (bECF) for changes in lactate, pyruvate, and glucose levels.5–9,10 The bECF proteome can closely reflect cerebral tissue-level changes—at least at the sampling site11—and elevated lactate-to-pyruvate ratio was found to be associated with a specific bECF proteome consisting of cytoarchitectural and mitochondrial proteins as well as a unique peptide with a mass/charge 4733.5, a candidate protein marker of metabolic crisis in TBI patients.12 Additional analyses have identified changes in the bECF proteome after both experimental and clinical TBI.13 Studies have shown that high initial Tau levels are indicative of poor outcome,14 and that elevated total Tau and beta-amyloid levels correlate with injury severity in focal and/or mixed types of TBI14,15 and identified the inflammatory response after TBI.16 However, cMD is not widely performed and the qualitative, quantitative, and temporal relationships between protein biomarker levels in the CSF versus bECF thus are currently poorly understood.1,2
The protein biomarkers S100 calcium-binding protein B (S100B), neuron-specific enolase (NSE), Tau, and phosphorylated Tau (p-Tau) have been extensively studied in TBI,17,18 and their elevated serum and CSF levels have been shown to indicate the extent of neuronal, glial, and axonal damage as well as correlate with injury severity and outcome.19–24 In this pilot study, we used microcapillary electrophoresis-coupled western analysis (WES) to determine the qualitative, quantitative, and temporal relationships between CSF and bECF levels of S100B, NSE, Tau, and p-Tau. WES is a highly sensitive proteomic platform that requires very low sample volume (microliters), and, like traditional westerns, it can separate immunoreactive proteins by molecular weight.
Methods
Patients and clinical parameters
Patients in our study were a subset of a larger population from a prospective observational study performed at the North Carolina Central University at Karolinska University Hospital (Stockholm, Sweden) under ethical approval #2009/1112-31/3 by Stockholm County branch of the Central Ethical Review Board, now called the Swedish Ethical Review Authority (Table 1). Study details, including inclusion and exclusion criteria, patient management, and sample acquisition, are as described in detail earlier.25 For this study, we selected patients who had matching CSF and bECF samples at three acute post-injury time points (days 1, 3, and 5 as detailed in Table 2).
Table 1.
Patient Demographics of the Study Cohort
| Patient ID | 6 | 7 | 10 | 17 | 11 | 13 | 14 |
|---|---|---|---|---|---|---|---|
| Sex | M | M | M | M | M | M | M |
| Age, years | 22 | 23 | 25 | 36 | 42 | 59 | 62 |
| GCS | 7 | 8 | 8 | 7 | 3 | 7 | 3 |
| ISS | 29 | 16 | 25 | 26 | 38 | 25 | 25 |
| AIS | 4 | 4 | 5 | 4 | 5 | 5 | 5 |
| Pupil responsiveness | 1 | 0 | 0 | 0 | 1 | 1 | 0 |
| Outcome (GOS)a | 3 | 4 | 5 | 4 | 3 | 3 | 1 |
Outcome was determined 6 months after the injury by a neurorehabilitation board-certified physician (P.H.G.); GOS categories: 1) dead, 2) persistent vegetative state, 3) severe disability, 4) moderate disability, and 5) low disability.
M, male; GCS, Glasgow Coma Scale; ISS, Injury Severity Score; AIS, Abbreviated Injury Scale; GOS, Glasgow Outcome Scale.
Table 2.
An Overview of Samples Analyzed by WES
| Time points | Day 1 | Day 3 | Day 5 |
|---|---|---|---|
| Original collection time points (h) to be combined | 6, 12, 18, 24 h | 54, 60, 66, 72 h | 102, 108, 114, 120 h |
| Patient nos. | |||
| 6 | CSF; bECF | CSF; bECF | CSF; bECF |
| 7 | CSF; bECF | CSF; bECF | CSF; bECF |
| 10 | CSF; bECF | CSF; X | CSF; X |
| 11 | CSF; bECF | CSF; bECF | CSF; bECF |
| 13 | CSF; bECF | X; X | CSF; bECF |
| 14 | CSF; bECF | CSF; bECF | X; X |
| 17 | CSF; bECF | CSF; bECF | CSF; bECF |
Note: “X” refers to missing samples.
WES, western analysis; CSF, cerebrospinal fluid; bECF, brain extracellular fluid.
Biosamples
Cerebrospinal fluid
CSF was collected using a catheter (conventional ventricular drain) placed in the ventricle and connected to a pump (Liquoguard®) collecting CSF at a rate of 2 mL/h, as long as intracranial pressure was >2 mm Hg. CSF was collected every 6 h, centrifuged, and the supernatant was transferred into collection tubes and stored in a −70°C freezer.
Brain extracellular fluid
cMD was performed as part of the clinical routine at the Neurointensive Care Unit of the Department of Neurosurgery at the Karolinska Hospital to monitor brain metabolism.7–9,26–28 A 0.6-mm-wide microdialysis catheter with a 10-mm dialysis membrane at its tip (100-kDa cutoff) was surgically introduced into the brain tissue of interest (in the border zone close to the injury). A pump perfused the interior of the catheter with a perfusion fluid, which equilibrated with the interstitial tissue surrounding the catheter. Equilibration occurred by diffusion of chemicals over the dialysis membrane. Using a perfusion flow of 0.3 μL/min, the recovery of glucose, lactate, pyruvate, and glutamate in the dialysate was ∼70% of the concentration in the interstitial fluid.29 Samples were continuously collected into microvials analyzed at bedside by a CMA 600 microdialysis analyzer every hour for changes in glucose, pyruvate, lactate, glycerol, and glutamate. In the same area, a similar catheter with a 100-kDa cutoff was introduced to collect proteins. The perfusion fluid was the same as for the 20-kDa catheter, but samples were collected every sixth hour and frozen at −70°C. The final collection tubes contained a protease and phosphatase inhibitor cocktail.30,31
Because of the low protein concentrations of bECF samples, we needed to combine four consecutive collections (e.g., 6, 12, 18, and 24 h) to be able to assay them using WES (see Table 2). To match the bECF samples, we also pooled equal volumes of CSF samples collected at time points matching the bECF collections. The final, combined bECF and CSF samples represent three post-injury time points: days 1, 3, and 5 (Table 2).
Protein analysis
CSF and bECF samples were analyzed by using WES (Simple Western microcapillary-based Western; ProteinSimple, Santa Clara, CA). Samples were diluted with 5X Fluorescent Master Mix (400 mM of dithiothreitol and 5X Sample Buffer; Prod # SM-W004; ProteinSimple), making 0.48 mg/mL as the final protein concentration for CSF. Samples and standard ladders were denatured at 70°C for 20 min, then set on ice for 10 min. Primary antibody dilutions were optimized for CSF and bECF samples using antigen-antibody binding titration before the assay (Supplementary Table S1).
The WES performs protein separation, blocking, incubation with the primary and horseradish peroxidase (HRP)-conjugated secondary antibodies, washing steps, and signal detection automatically.32 Samples, along with the diluted primary antibodies, HRP-conjugated secondary antibody, detection reagents, and wash buffers, were loaded onto the Simple Western assay plates according to the company's protocol. Plates were centrifuged at 2500 rpm for 5 min at room temperature, then the 25-slot microcapillary cartridge and plates were placed in the WES platform for size selection to be completed (3 h). Immunodetection was performed using the WES's default setting for the 12- to 230-kDA size-based assay. Chemiluminescent signal intensities were acquired by using the company's Compass software. Intensities were normalized to a signal-to-noise ratio >10. Relative abundance of each protein was then calculated as the area under the curve (AUC) for each of the detected peaks .
Results
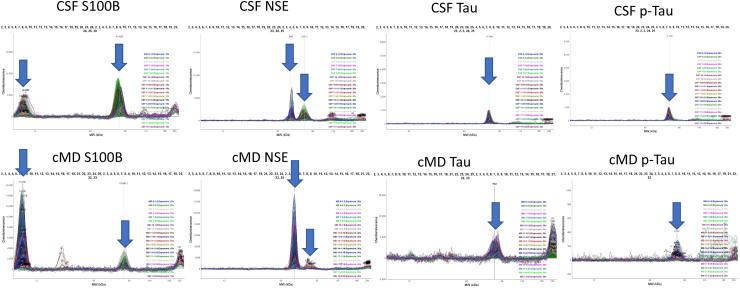
In addition to the expected ∼10-kDa S100B immunoreactivity, we detected a second S100B peak at ∼60 kDa in every CSF and bECF sample, but the contribution of the two immunoreactivities differed between CSF and bECF samples (Fig. 1 and Table 3), with most patients showing CSF: 60 kDa >10 kDa vs. bECF 10 kDa >60 kDa. A few bECF samples contained a third, very small S100B immunoreactive peak at ∼20 kDa. Similarly, for NSE, in addition to the expected ∼50-kDa immunoreactive peak, there was a second immunoreactive peak at ∼60 kDa detected in all CSF samples, but it was barely detectable in bECF samples. Again, the ratio between the different immunoreactive forms varied from patient to patient (Fig 1 and Table 3). We detected the expected ∼55-kDa Tau and p-Tau immunoreactivities in CSF as well as in bECF samples, but in bECF samples there was a second Tau and p-Tau peak at >200 kDa, likely representing large Tau and p-Tau proteins likely aggregated in vitro.
FIG. 1.
Distribution of immunoreactivities of S100B, NSE, Tau, and p-Tau in CSF and bECF samples. Blue arrows point to the immunoreactive peaks that were quantified. bECF, brain extracellular fluid; CSF, cerebrospinal fluid; NSE, neuron-specific enolase; p-Tau, phosphorylated Tau; S100B, S100 calcium-binding protein B.
Table 3.
Percentage Distribution of Different Immunoreactive Forms of S100B and NSE in Different Biofluid Compartments
| Marker |
S100B |
NSE |
||||||
|---|---|---|---|---|---|---|---|---|
| Biofluid |
CSF |
bECF |
CSF |
bECF |
||||
| MW (kDa) | 10 kDa | 60 kDa | 10 kDa | 60 kDa | 50 kDa | 60 kDa | 50 kDa | 60 kDa |
| Patients | ||||||||
| PT 6 | 32.6 | 67.3 | N/A | 100 | 68.6 | 31.3 | 100 | N/A |
| PT 7 | 56.3 | 43.6 | 76.1 | 23.8 | 66.9 | 33.01 | 86.7 | 13.2 |
| PT 10 | 46.3 | 53.6 | N/A | N/A | 38.2 | 61.7 | N/A | N/A |
| PT 11 | 12.6 | 87.32 | 63.8 | 36.1 | 21.3 | 78.6 | 55.9 | 44.08 |
| PT 13 | 23.3 | 76.6 | 100 | N/A | 49.6 | 50.3 | 100 | N/A |
| PT 14 | 17.1 | 82.8 | 89.6 | 10.3 | 13.2 | 86.7 | 78.8 | 21.1 |
| PT 17 | N/A | 100 | 89.4 | 10.5 | 69.6 | 30.3 | 90.3 | 9.6 |
S100B, S100 calcium-binding protein B; NSE, neuron-specific enolase; CSF, cerebrospinal fluid; bECF, brain extracellular fluid; MW, molecular weight; N/A, not applicable.
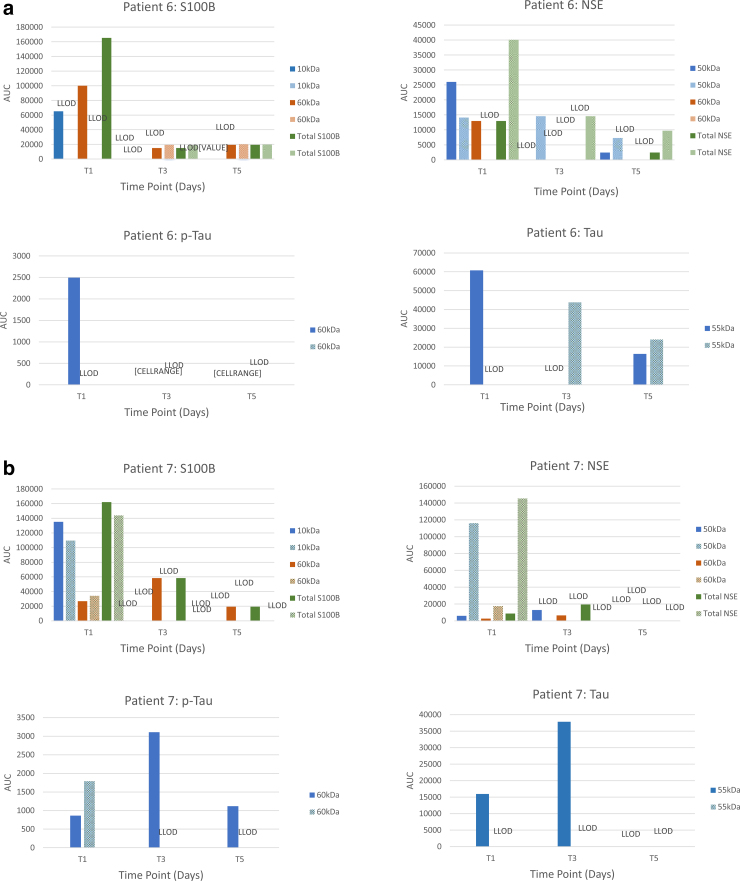
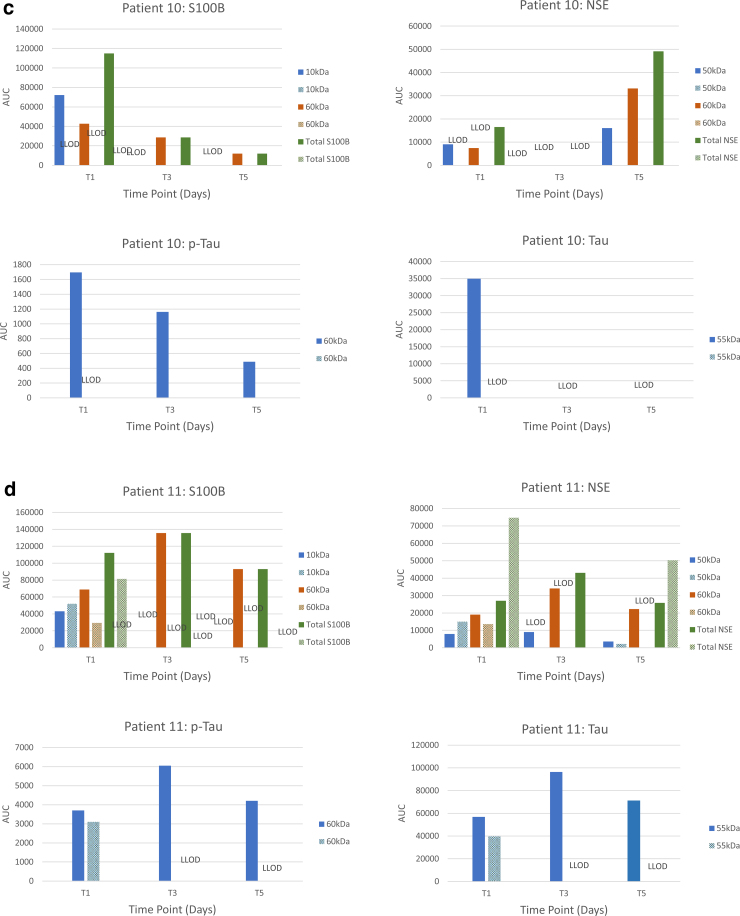
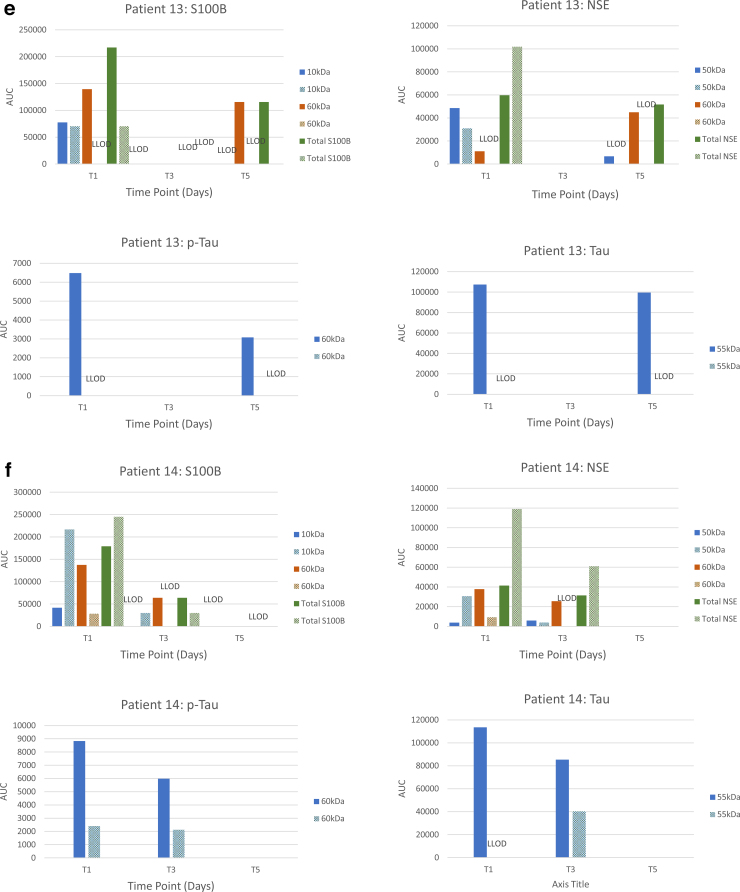
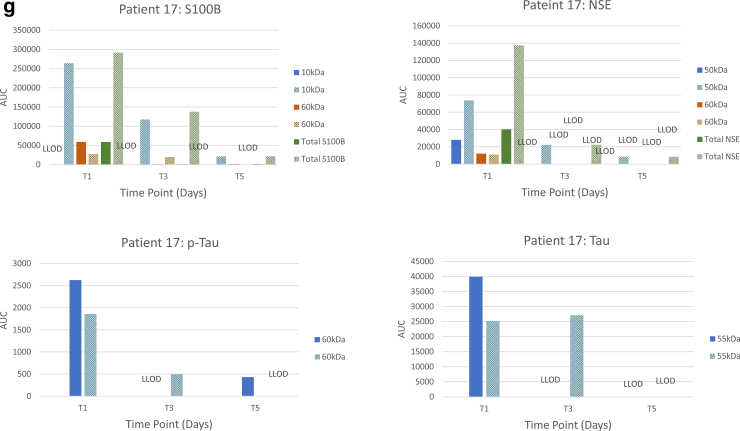
Semiquantitative analysis of immunoreactivities (area under the peak) showed substantial variability in biomarker levels between patients, biomarkers and their immunoreactive forms, and time points (Fig. 2A–G). But, the overall pattern of time-dependent changes showed similar trends in matching CSF and bECF samples, such that the relative concentrations of S100B and NSE immunoreactivities in both biofluids decreased over time. Highest relative concentrations of both proteins were detected at the earliest time point (T1 or day 1) and were substantially lower at T5. It should be noted that contributions of the different S100B and NSE immunoreactive forms to total immunoreactivity varied from patient to patient, time point to time point, and compartment to compartment. Tau and p-Tau levels showed similar trends, but in some patients their levels remained elevated at T5.
FIG. 2.
Time-dependent changes in the protein biomarker values of matching CSF and bECF samples in the individual patients (A, patient 6; B, patient 7, C, patient 10; D, patient 11; E, patient 13; F, patient 14; G, patient 17). Pay attention to the scales. The scales reflect the relative abundance of proteins and vary substantially between proteins and biosamples. AUC, area under the curve; bECF, brain extracellular fluid; CSF, cerebrospinal fluid; LLOD, lower limit of detection.
Discussion
The goal of this prospective longitudinal pilot study was to understand the relationship between protein biomarker levels measured in matching CSF and bECF samples collected from severe TBI patients during the acute stage of injury. TBI-induced changes in CSF levels of protein biomarkers have been extensively studied, although most studies have used single and varying post-injury time points (for reviews, see past works3,33). Therefore, this is the first study that has coanalyzed matching, serially collected CSF and bECF samples.
Consistent with the earlier report that analyzed some of the same CSF samples using a different analytical platform,25 we found that CSF levels of S100B and NSE decreased over time. We also detected a similar temporal pattern in matching bECF samples. S100B is one of the best-characterized protein markers in TBI (for review, see past works18,34), and its very short half-life (∼0.5 h)35 makes it ideal as a marker of de novo release. Serum S100B levels have been established as part of the Scandinavian TBI management guidelines.36–38
The two biofluids CSF and bECF represent distinct intracranial environments that can differently allow and/or promote multimerization of S100B and/or secondary modifications to NSE. We detected both monomeric and multimeric (hexamer) forms of S100B protein in both CSF and bECF samples, but the ratio between the monomeric and multimeric forms varied between patients and post-injury time points. Monomeric S100B is a ∼10-kDa protein, but it forms multimers, dimers, hexamers, and even amyloids in metal ion-dependent manner.39,40 Extracellular S100B proteins are related to the group of proteins alarmins, also called damage-associated molecular patterns, that coordinate adaptive cellular stress response to tissue damage.39,40 Multimeric S100B, including hexamers, can bind to receptor for advanced glycation end product (RAGE) and Toll-like receptor-4 and activate the inflammatory response to central nervous system injury.41–50
Since its invention, cMD has identified changes in brain metabolism post-TBI.4,5,9,12,14,15,51,52 Studies have also reported injury-induced changes in the bECF proteome,53–57 but there have not been any studies (to our knowledge) that directly compared matching bECF and CSF samples for injury-induced changes in protein biomarker levels. It should be noted that cMD technology has known issues that can affect the outcome of protein analysis of bECF samples, such as the non-specific binding of proteins to the catheter.52,58–60
There are important technical issues that can be responsible for the detected S100B and NSE immunoreactive forms. The main issue, as in all antibody-based analysis, is the specificity of the antibodies. We have tested the antibodies for specificity before using them in WES, but cross-reactivity can still occur.32,61–64 Though all samples were treated identically after collection, protease inhibitors could not be added to the microdialysis vials during collection, which could have affected direct comparisons between compartments. However, CSF and bECF samples were continuously collected, prepared, and assayed under identical conditions by trained professionals; therefore, intersample variability should be negligible.
Elevated CSF levels of Tau proteins have been found in CSF65 as well as in bECF15 (for review, see a previous work66). We also found elevated Tau and p-Tau levels in both CSF and bECF samples. Tau and p-Tau levels in both biofluids showed a similar temporal pattern to S100B and NSE, but the rate of decrease over time appeared to be slower in the CSF. These changes can be interpreted in several ways, including a potentially long half-life of these axoskeletal proteins in the extracellular environment. The exact half-life of these protein biomarkers that are released from the intracellular environment is still not well known.67 Further, the extracellular environment can be altered by the severity and type of injury that can selectively activate extracellular proteases as part of the secondary injury process.68–70 In addition, Tau is an intrinsically disordered protein with a high propensity for self-aggregation,71 as also indicated by the presence of >200-kDa aggregates in our study.
Limitations
This is a pilot study with limited numbers of patients who were all males. Technical issues include the use of standard CSF perfusion fluid that was used to perfuse the catheters, which may be suboptimal for protein recovery.72 Because of various issues, including patient safety, we were unable to collect sufficient quantities of bECF samples at all time points that would have enabled higher (e.g., daily or even higher) temporal resolution.
Conclusion
Our pilot study focused on the acute stage of TBI in a clinically heterogeneous population. However, we found that the temporal pattern of changes in the CSF and bECF levels of four well-established neural injury markers were generally similar, suggesting that CSF levels of protein biomarkers can reflect intraparenchymal changes after TBI. The apparent multimeric S100B form detected in CSF and bECF samples may indicate other functions for S100B in the intracranial environment of the injured brain, for example, involvement in the inflammatory response after TBI upon binding to receptor RAGE.42,43,73 Given the biological significance of RAGE signaling in injury repair,43 these analyses need to be repeated on a larger scale using different analytical platforms. Our study is small, but it illustrates the value of both quantitative and qualitative analysis of protein biomarkers in serially sampled biofluids, especially CSF after TBI. It also demonstrates some of the challenges protein biomarker studies face vis-à-vis a complex, dynamically changing condition such as severe TBI.
Supplementary Material
Acknowledgments
We express our thanks and gratitude to the patients and their families for participating in this study. We thank the nurses of the Neurointensive Care Unit of the Karolinska Hospital for their help in collecting the clinical samples. E.P.T. acknowledges funding support from Strategic Research Area Neuroscience (StratNeuro), The Erling-Persson Family Foundation, and Region Stockholm (Clinical Research Appointment). The funders had no role in the design or conduct of this research. The views and opinions here are those of the authors and do not represent the view of USUHS, DoD, or the Karolinska Institutet.
Abbreviations Used
- AIS
Abbreviated Injury Scale
- AUC
area under the curve
- bECF
brain extracellular fluid
- cMD
cerebral microdialysis
- CSF
cerebrospinal fluid
- GCS
Glasgow Coma Scale
- HRP
horseradish peroxidase
- ISS
Injury Severity Score
- NSE
neuron-specific enolase
- p-Tau
phosphorylated Tau
- RAGE
receptor for advanced glycation end product
- S100B
S100 calcium-binding protein B
- TBI
traumatic brain injury
- WES
western analysis
Authors' Contributions
E.P.T. and B.-M.B. designed and supervised the clinical study. D.V.A. led the protein biomarker study, wrote the study report, and wrote the manuscript. I-H.L., A.K., J.M., and R.B. performed the biomarker analyses. R.A. and M.E. analyzed the data. I-H.L. created the graphs. P.H.G. performed the neurobehavioral assessments. M.R., M.S., and D.W.N. reviewed the data, and all authors reviewed the manuscript. A.K. performed the final editing.
Funding Information
The study was funded by NINDS (award no.: RO3NS087350).
Author Disclosure Statement
No competing financial interests exist.
Supplementary Material
Cite this article as: Lin IH, Kamnaksh A, Aniceto R, et al. Time-dependent changes in the biofluid levels of neural injury markers in severe TBI patients; cerebrospinal fluid and cerebral microdialysates: a longitudinal prospective pilot study. Neurotrauma Reports 2023:4(1):107–117. doi: 10.1089/neur.2022.0076.
References
- 1. Saatman KE, Duhaime AC, Bullock R, et al. . Classification of traumatic brain injury for targeted therapies. J Neurotrauma 2008;25(7):719–738; doi: 10.1089/neu.2008.0586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Agoston DV, Shutes-David A, Peskind ER. Biofluid biomarkers of traumatic brain injury. Brain Inj 2017;31(9):1195–1203; doi: 10.1080/02699052.2017.1357836 [DOI] [PubMed] [Google Scholar]
- 3. Santacruz CA, Vincent JL, Bader A, et al. . Association of cerebrospinal fluid protein biomarkers with outcomes in patients with traumatic and non-traumatic acute brain injury: systematic review of the literature. Crit Care 2021;25(1):278; doi: 10.1186/s13054-021-03698-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ungerstedt U. Microdialysis—principles and applications for studies in animals and man. J Intern Med 1991;230(4):365–373; doi: 10.1111/j.1365-2796.1991.tb00459.x [DOI] [PubMed] [Google Scholar]
- 5. Hutchinson PJ, Jalloh I, Helmy A, et al. . Consensus statement from the 2014 International Microdialysis Forum. Intensive Care Med 2015;41(9):1517–1528; doi: 10.1007/s00134-015-3930-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hillered L, Persson L, Nilsson P, et al. . Continuous monitoring of cerebral metabolism in traumatic brain injury: a focus on cerebral microdialysis. Curr Opin Crit Care 2006;12(2):112–118; doi: 10.1097/01.ccx.0000216576.11439.df [DOI] [PubMed] [Google Scholar]
- 7. Nelson DW, Bellander BM, Maccallum RM, et al. . Cerebral microdialysis of patients with severe traumatic brain injury exhibits highly individualistic patterns as visualized by cluster analysis with self-organizing maps. Crit Care Med 2004;32(12):2428–2436; doi: 10.1097/01.ccm.0000147688.08813.9c [DOI] [PubMed] [Google Scholar]
- 8. Nelson DW, Thornquist B, MacCallum RM, et al. . Analyses of cerebral microdialysis in patients with traumatic brain injury: relations to intracranial pressure, cerebral perfusion pressure and catheter placement. BMC Med 2011;9:21; doi: 10.1186/1741-7015-9-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bellander BM, Cantais E, Enblad P, et al. . Consensus meeting on microdialysis in neurointensive care. Intensive Care Med 2004;30(12):2166–169; doi: 10.1007/s00134-004-2461-8 [DOI] [PubMed] [Google Scholar]
- 10. Stocchetti N, Le Roux P, Vespa P, et al. . Clinical review: neuromonitoring—an update. Crit Care 2013;17(1):201; doi: 10.1186/cc11513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Oddo M, Hutchinson PJ. Understanding and monitoring brain injury: the role of cerebral microdialysis. Intensive Care Med 2018;44(11):1945–1948; doi: 10.1007/s00134-017-5031-6 [DOI] [PubMed] [Google Scholar]
- 12. Lakshmanan R, Loo JA, Drake T, et al. . Metabolic crisis after traumatic brain injury is associated with a novel microdialysis proteome. Neurocrit Care 2010;12(3):324–336; doi: 10.1007/s12028-010-9342-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hanrieder J, Wetterhall M, Enblad P, et al. . Temporally resolved differential proteomic analysis of human ventricular CSF for monitoring traumatic brain injury biomarker candidates. J Neurosci Methods 2009;177(2):469–478; doi: 10.1016/j.jneumeth.2008.10.038 [DOI] [PubMed] [Google Scholar]
- 14. Magnoni S, Esparza TJ, Conte V, et al. . Tau elevations in the brain extracellular space correlate with reduced amyloid-beta levels and predict adverse clinical outcomes after severe traumatic brain injury. Brain 2012;135(Pt 4):1268–1280; doi: 10.1093/brain/awr286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Marklund N, Blennow K, Zetterberg H, et al. . Monitoring of brain interstitial total tau and beta amyloid proteins by microdialysis in patients with traumatic brain injury. J Neurosurg 2009;110(6):1227–1237; doi: 10.3171/2008.9.jns08584 [DOI] [PubMed] [Google Scholar]
- 16. Clausen F, Marklund N, Hillered L. Acute inflammatory biomarker responses to diffuse traumatic brain injury in the rat monitored by a novel microdialysis technique. J Neurotrauma 2019;36(2):201–211; doi: 10.1089/neu.2018.5636 [DOI] [PubMed] [Google Scholar]
- 17. Wang KK, Yang Z, Zhu T, et al. . An update on diagnostic and prognostic biomarkers for traumatic brain injury. Expert Rev Mol Diagn 2018;18(2):165–180; doi: 10.1080/14737159.2018.1428089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Amoo M, Henry J, O'Halloran PJ, et al. . S100B, GFAP, UCH-L1 and NSE as predictors of abnormalities on CT imaging following mild traumatic brain injury: a systematic review and meta-analysis of diagnostic test accuracy. Neurosurg Rev 2022;45(2):1171–1193; doi: 10.1007/s10143-021-01678-z.Epub2021Oct28 [DOI] [PubMed] [Google Scholar]
- 19. Bohmer AE, Oses JP, Schmidt AP, et al. Neuron-specific enolase, S100B, and glial fibrillary acidic protein levels as outcome predictors in patients with severe traumatic brain injury. Neurosurgery 2011;68(6):1624–1630; discussion, 1630–1631; doi: 10.1227/NEU.0b013e318214a81f [DOI] [PubMed] [Google Scholar]
- 20. Berger RP, Adelson PD, Richichi R, et al. . Serum biomarkers after traumatic and hypoxemic brain injuries: insight into the biochemical response of the pediatric brain to inflicted brain injury. Dev Neurosci 2006;28(4–5):327–335; doi: 10.1159/000094158 [DOI] [PubMed] [Google Scholar]
- 21. Cheng F, Yuan Q, Yang J, et al. . The prognostic value of serum neuron-specific enolase in traumatic brain injury: systematic review and meta-analysis. PLoS One 2014;9(9):e106680; doi: 10.1371/journal.pone.0106680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Paczkowska E, Gołąb-Janowska M, Bajer-Czajkowska A, et al. . Increased circulating endothelial progenitor cells in patients with haemorrhagic and ischaemic stroke: the role of endothelin-1. J Neurol Sci 2013;325(1–2):90–99; doi: 10.1016/j.jns.2012.12.005 [DOI] [PubMed] [Google Scholar]
- 23. Yates D. Traumatic brain injury: serum levels of GFAP and S100B predict outcomes in TBI. Nat Rev Neurol 2011;7(2):63; doi: 10.1038/nrneurol.2010.207 [DOI] [PubMed] [Google Scholar]
- 24. Morochovic R, Racz O, Kitka M, et al. . Serum S100B protein in early management of patients after mild traumatic brain injury. Eur J Neurol 2009;16(10):1112–1117; doi: 10.1111/j.1468-1331.2009.02653.x [DOI] [PubMed] [Google Scholar]
- 25. Lindblad C, Nelson DW, Zeiler FA, et al. . Influence of blood-brain barrier integrity on brain protein biomarker clearance in severe traumatic brain injury: a longitudinal prospective study. J Neurotrauma 2020;37(12):1381–1391; doi: 10.1089/neu.2019.6741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Raboel PH, Bartek J Jr, Andresen M, et al. . Intracranial pressure monitoring: invasive versus non-invasive methods—a review. Crit Care Res Pract 2012;2012:950393; doi: 10.1155/2012/950393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rostami E, Bellander BM. Monitoring of glucose in brain, adipose tissue, and peripheral blood in patients with traumatic brain injury: a microdialysis study. J Diabetes Sci Technol 2011;5(3):596–604; doi: 10.1177/193229681100500314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Thelin EP, Nelson DW, Ghatan PH, et al. . Microdialysis monitoring of CSF parameters in severe traumatic brain injury patients: a novel approach. Front Neurol 2014;5:159; doi: 10.3389/fneur.2014.00159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hutchinson PJ, O'Connell MT, Al-Rawi PG, et al. . Clinical cerebral microdialysis: a methodological study. J Neurosurg 2000;93(1):37–43; doi: 10.3171/jns.2000.93.1.0037 [DOI] [PubMed] [Google Scholar]
- 30. Gyorgy AB, Walker J, Wingo D, et al. . Reverse phase protein microarray technology in traumatic brain injury. J Neurosci Methods 2010;192(1):96–101; doi: 10.1016/j.jneumeth.2010.07.029 [DOI] [PubMed] [Google Scholar]
- 31. Kwon SK, Kovesdi E, Gyorgy AB, et al. . Stress and traumatic brain injury: a behavioral, proteomics, and histological study. Front Neurol 2011;2:12; doi: 10.3389/fneur.2011.00012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Harris VM. Protein detection by Simple Western™ analysis. Methods Mol Biol 2015;1312:465–468; doi: 10.1007/978-1-4939-2694-7_47 [DOI] [PubMed] [Google Scholar]
- 33. Teunissen CE, Verheul C, Willemse EAJ. The use of cerebrospinal fluid in biomarker studies. Handb Clin Neurol 2017;146:3–20; doi: 10.1016/b978-0-12-804279-3.00001-0 [DOI] [PubMed] [Google Scholar]
- 34. Thelin EP, Nelson DW, Bellander BM. A review of the clinical utility of serum S100B protein levels in the assessment of traumatic brain injury. Acta Neurochir (Wien) 2017;159(2):209–225; doi: 10.1007/s00701-016-3046-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jönsson H, Johnsson P, Höglund P, et al. . Elimination of S100B and renal function after cardiac surgery. J Cardiothorac Vasc Anesth 2000;14(6):698–701; doi: 10.1053/jcan.2000.18444 [DOI] [PubMed] [Google Scholar]
- 36. Ingebrigtsen T, Romner B, Kock-Jensen C. Scandinavian guidelines for initial management of minimal, mild, and moderate head injuries. The Scandinavian Neurotrauma Committee. J Trauma 2000;48(4):760–766; doi: 10.1097/00005373-200004000-00029 [DOI] [PubMed] [Google Scholar]
- 37. Astrand R, Unden J, Romner B. Clinical use of the calcium-binding S100B protein. Methods Mol Biol 2013;963:373–384; doi: 10.1007/978-1-62703-230-8_23 [DOI] [PubMed] [Google Scholar]
- 38. Undén J, Astrand R, Waterloo K, et al. . Clinical significance of serum S100B levels in neurointensive care. Neurocrit Care 2007;6(2):94–99; doi: 10.1007/s12028-007-0005-0 [DOI] [PubMed] [Google Scholar]
- 39. Baudier J, Deloulme JC, Shaw GS. The Zn(2+) and Ca(2+)-binding S100B and S100A1 proteins: beyond the myths. Biol Rev Camb Philos Soc 2020;95(3):738–758; doi: 10.1111/brv.12585 [DOI] [PubMed] [Google Scholar]
- 40. Michetti F, D'Ambrosi N, Toesca A, et al. . The S100B story: from biomarker to active factor in neural injury. J Neurochem 2019;148(2):168–187; doi: 10.1111/jnc.14574 [DOI] [PubMed] [Google Scholar]
- 41. Thulin E, Kesvatera T, Linse S. Molecular determinants of S100B oligomer formation. PLoS One 2011;6(3):e14768; doi: 10.1371/journal.pone.0014768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Moysa A, Steczkiewicz K, Niedzialek D, et al. A model of full-length RAGE in complex with S100B. Structure 2021;29(9):989–1002.e6; doi: 10.1016/j.str.2021.04.002 [DOI] [PubMed] [Google Scholar]
- 43. Bianchi R, Kastrisianaki E, Giambanco I, et al. . S100B protein stimulates microglia migration via RAGE-dependent up-regulation of chemokine expression and release. J Biol Chem 2011;286(9):7214–7226; doi: 10.1074/jbc.M110.169342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ostendorp T, Leclerc E, Galichet A, et al. . Structural and functional insights into RAGE activation by multimeric S100B. EMBO J 2007;26(16):3868–3878; doi: 10.1038/sj.emboj.7601805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Donato R, Sorci G, Riuzzi F, et al. . S100B's double life: intracellular regulator and extracellular signal. Biochim Biophys Acta 2009;1793(6):1008–1022; doi: 10.1016/j.bbamcr.2008.11.009 [DOI] [PubMed] [Google Scholar]
- 46. Fritz G, Botelho HM, Morozova-Roche LA, et al. . Natural and amyloid self-assembly of S100 proteins: structural basis of functional diversity. FEBS J 2010;277(22):4578–4590; doi: 10.1111/j.1742-4658.2010.07887.x [DOI] [PubMed] [Google Scholar]
- 47. Heizmann CW, Fritz G, Schäfer BW. S100 proteins: structure, functions and pathology. Front Biosci 2002;7:d1356–d1368; doi: 10.2741/A846 [DOI] [PubMed] [Google Scholar]
- 48. Leclerc E, Fritz G, Vetter SW, et al. . Binding of S100 proteins to RAGE: an update. Biochim Biophys Acta 2009;1793(6):993–1007; doi: 10.1016/j.bbamcr.2008.11.016 [DOI] [PubMed] [Google Scholar]
- 49. Dumurgier J, Sabia S, Zetterberg H, et al. ; Alzheimer's Disease Neuroimaging Initiative. A pragmatic, data-driven method to determine cutoffs for CSF biomarkers of Alzheimer disease based on validation against PET imaging. Neurology 2022;99(7):e669–e678; doi: 10.1212/wnl.0000000000200735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sorci G, Bianchi R, Riuzzi F, et al. . S100B protein, a damage-associated molecular pattern protein in the brain and heart, and beyond. Cardiovasc Psychiatry Neurol 2010;2010:656481; doi: 10.1155/2010/656481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ungerstedt U, Rostami E. Microdialysis in neurointensive care. Curr Pharm Des 2004;10(18):2145–2152; doi: 10.2174/1381612043384105 [DOI] [PubMed] [Google Scholar]
- 52. Hillered L, Dahlin AP, Clausen F, et al. . Cerebral microdialysis for protein biomarker monitoring in the neurointensive care setting—a technical approach. Front Neurol 2014;5:245; doi: 10.3389/fneur.2014.00245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Willie JT, Lim MM, Bennett RE, et al. . Controlled cortical impact traumatic brain injury acutely disrupts wakefulness and extracellular orexin dynamics as determined by intracerebral microdialysis in mice. J Neurotrauma 2012;29(10):1908–1921; doi: 10.1089/neu.2012.2404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wijman CA, Smirnakis SM, Vespa P, et al. . Research and technology in neurocritical care. Neurocrit Care 2012;16(1):42–54; doi: 10.1007/s12028-011-9609-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Schwetye KE, Cirrito JR, Esparza TJ, et al. . Traumatic brain injury reduces soluble extracellular amyloid-beta in mice: a methodologically novel combined microdialysis-controlled cortical impact study. Neurobiol Dis 2010;40(3):555–564; doi: 10.1016/j.nbd.2010.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Vespa PM. Multimodality monitoring and telemonitoring in neurocritical care: from microdialysis to robotic telepresence. Curr Opin Crit Care 2005;11(2):133–138; doi: 10.1097/01.ccx.0000155353.01489.58 [DOI] [PubMed] [Google Scholar]
- 57. Hillered L, Vespa PM, Hovda DA. Translational neurochemical research in acute human brain injury: the current status and potential future for cerebral microdialysis. J Neurotrauma 2005;22(1):3–41; doi: 10.1089/neu.2005.22.3 [DOI] [PubMed] [Google Scholar]
- 58. Dahlin AP, Hjort K, Hillered L, et al. . Multiplexed quantification of proteins adsorbed to surface-modified and non-modified microdialysis membranes. Anal Bioanal Chem 2012;402(6):2057–2067; doi: 10.1007/s00216-011-5614-y [DOI] [PubMed] [Google Scholar]
- 59. Dahlin AP, Purins K, Clausen F, et al. . Refined microdialysis method for protein biomarker sampling in acute brain injury in the neurointensive care setting. Anal Chem 2014;86(17):8671–8679; doi: 10.1021/ac501880u [DOI] [PubMed] [Google Scholar]
- 60. Dahlin AP, Wetterhall M, Caldwell KD, et al. . Methodological aspects on microdialysis protein sampling and quantification in biological fluids: an in vitro study on human ventricular CSF. Anal Chem 2010;82(11):4376–4385; doi: 10.1021/ac1007706 [DOI] [PubMed] [Google Scholar]
- 61. Algenäs C, Agaton C, Fagerberg L, et al. . Antibody performance in western blot applications is context-dependent. Biotechnol J 2014;9(3):435–445; doi: 10.1002/biot.201300341 [DOI] [PubMed] [Google Scholar]
- 62. Pillai-Kastoori L, Heaton S, Shiflett SD, et al. . Antibody validation for Western blot: by the user, for the user. J Biol Chem 2020;295(4):926–939; doi: 10.1074/jbc.RA119.010472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Signore M, Manganelli V, Hodge A. Antibody validation by western blotting. Methods Mol Biol 2017;1606:51–70; doi: 10.1007/978-1-4939-6990-6_4 [DOI] [PubMed] [Google Scholar]
- 64. Signore M, Reeder KA. Antibody validation by Western blotting. Methods Mol Biol 2012;823:139–155; doi: 10.1007/978-1-60327-216-2_10 [DOI] [PubMed] [Google Scholar]
- 65. Ost M, Nylen K, Csajbok L, et al. . Initial CSF total tau correlates with 1-year outcome in patients with traumatic brain injury. Neurology 2006;67(9):1600–1604; doi: 10.1212/01.wnl.0000242732.06714.0f [DOI] [PubMed] [Google Scholar]
- 66. Tsitsopoulos PP, Marklund N. Amyloid-β peptides and tau protein as biomarkers in cerebrospinal and interstitial fluid following traumatic brain injury: a review of experimental and clinical studies. Front Neurol 2013;4:79; doi: 10.3389/fneur.2013.00079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Thelin EP, Zeiler FA, Ercole A, et al. . Serial sampling of serum protein biomarkers for monitoring human traumatic brain injury dynamics: a systematic review. Front Neurol 2017;8:300; doi: 10.3389/fneur.2017.00300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Abdul-Muneer PM, Pfister BJ, Haorah J, et al. . Role of matrix metalloproteinases in the pathogenesis of traumatic brain injury. Mol Neurobiol 2016;53(9):6106–6123; doi: 10.1007/s12035-015-9520-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. George N, Geller HM. Extracellular matrix and traumatic brain injury. J Neurosci Res 2018;96(4):573–588; doi: 10.1002/jnr.24151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Zhang H, Adwanikar H, Werb Z, et al. . Matrix metalloproteinases and neurotrauma: evolving roles in injury and reparative processes. Neuroscientist 2010;16(2):156–170; doi: 10.1177/1073858409355830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Sabbagh JJ, Dickey CA. The metamorphic nature of the tau protein: dynamic flexibility comes at a cost. Front Neurosci 2016;10:3; doi: 10.3389/fnins.2016.00003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Giorgi-Coll S, Thelin EP, Lindblad C, et al. . Dextran 500 improves recovery of inflammatory markers: an in vitro microdialysis study. J Neurotrauma 2020;37(1):106–114; doi: 10.1089/neu.2019.6513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Balança B, Desmurs L, Grelier J, et al. . DAMPs and RAGE pathophysiology at the acute phase of brain injury: an overview. Int J Mol Sci 2021;22(5):2439; doi: 10.3390/ijms22052439 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.