Abstract
Hedgehog (Hh) signaling is evolutionarily conserved and plays an instructional role in embryonic morphogenesis, organogenesis in various animals, and the central nervous system organization. Multiple feedback mechanisms dynamically regulate this pathway in a spatiotemporal and context-dependent manner to confer differential patterns in cell fate determination. Hh signaling is complex due to canonical and non-canonical mechanisms coordinating cell–cell communication. In addition, studies have demonstrated a regulatory framework of Hh signaling and shown that cholesterol is vital for Hh ligand biogenesis, signal generation, and transduction from the cell surface to intracellular space. Studies have shown the importance of a specific cholesterol pool, termed accessible cholesterol, which serves as a second messenger, conveying signals between smoothened (Smo) and patched 1 (Ptch1) across the plasma and ciliary membranes. Remarkably, recent high-resolution structural and molecular studies shed new light on the interplay between Hh signaling and cholesterol in membrane biology. These studies elucidated novel mechanistic insight into the release and dispersal of cholesterol-anchored Hh and the basis of Hh recognition by Ptch1. Additionally, the putative model of Smo activation by cholesterol binding and/or modification and Ptch1 antagonization of Smo has been explicated. However, the coupling mechanism of Hh signaling and cholesterol offered a new regulatory principle in cell biology: how effector molecules of the Hh signal network react to and remodel cholesterol accessibility in the membrane and selectively activate Hh signaling proteins thereof. Recognizing the biological importance of cholesterol in Hh signaling activation and transduction opens the door for translational research to develop novel therapeutic strategies. This review looks in-depth at canonical and non-canonical Hh signaling and the distinct proposed model of cholesterol-mediated regulation of Hh signaling components, facilitating a more sophisticated understanding of the Hh signal network and cholesterol biology.
Keywords: Hedgehog, Cholesterol, Smo, Ptch1, Plasma membrane, Canonical signaling, Non-canonical signaling
Introduction
Hedgehog (Hh) signaling is one of the intricate signal transduction pathways that plays an instructional role during embryonic development, stem cell biology, and tissue homeostasis [1–4]. Dysregulation of Hh signaling resulted in developmental defects, such as holoprosencephaly, microencephaly, cyclopia, congenital syndromes, and other developmental malformations [5, 6]. Nevertheless, embryogenesis and tumorigenesis share common characteristics, such as synchronized mechanisms of proliferation, differentiation, and migration [7]. Therefore, modulation of the Hh signaling components and disruption of the regulatory trafficking network of the Hh pathway are also related to tumorigenesis and facilitate the aggressive phenotype of various human cancers [8–10]. Hh signaling is a unique regulatory pathway that controls subcellular milieus and offers targets for translational research from basic biology to clinical application [11, 12]. Notably, the versatility of the conserved Hh pathway in cell fate determination emphasizes its role in multiple signaling contexts throughout an organism’s life span from embryonic development to postnatal physiology and pathophysiology [4, 13]. Moreover, Hh signals are transmitted in an autocrine, paracrine, and juxtracrine manner and coordinate diverse functions within a target field [14, 15].
Several lines of evidence support that Hh signaling consists of canonical and non-canonical mechanisms that coordinate cell–cell communication in multiple aspects [16, 17]. Canonical Hh signaling occurs when secreted Hh ligand binds to cell surface receptor patched1 (Ptch1), leading to inactivation of Ptch1, subsequently releasing Ptch1-mediated suppression of smoothened (Smo) [16]. The free Smo translocates into the ciliary membrane and transactivates glioma-associated oncogene (Gli) to the nucleus, ultimately triggering Gli-dependent activation of the downstream targets [16, 18]. In non-canonical Hh signaling, the components signal outside the Hh–Ptch–Smo–Gli paradigm and perform a vital role in activating Gli transcription [19–21]. However, in the past few years, several investigations have elucidated that Hh can also signal through a Gli-independent mechanism or Gli can be directly activated without receptors Smo or Ptch1, also referred to as a non-canonical signaling pathway [21, 22]. Studies have also indicated a regulatory framework of Hh signaling, in which cholesterol plays a critical role in generating and transducing Hh signals from the cell surface to the intracellular space [23–25]. Defects in cholesterol biosynthesis and its depletion significantly affect the activity of Hh signaling components, leading to attenuation of the signaling [25].
Cholesterol is a polycyclic amphipathic molecule that serves as a building material for cellular membranes and plays an essential structural role in maintaining the fluidity of eukaryotic cell membranes and various molecular signaling pathways [26]. Cholesterol is highly enriched in two specialized areas termed lipid rafts and caveolae [27, 28]. Several receptors are localized to these cholesterol-rich microdomains and function as “signaling gateways” into the cell [26, 29]. Both receptors Smo and Ptch1 are situated in caveolin-1-enriched/raft microdomains. It was suggested that depletion of plasmalemmal cholesterol, affects the Hh receptor complex distribution in the cholesterol-rich microdomains and affects Hh signaling [30]. In response to Hh gradient signals, the high-level Hh signal transmission requires Smo oligomerization/higher-order clustering in lipid rafts [31]. The organization of cholesterol in the ciliary membrane is also needed to control the activity of cilia-localized signaling proteins including, Smo and Ptch1 [32]. In addition, the study showed that sterol depletion via defects in cholesterol biosynthesis influences the activity of Smo, and response to the Hh signal is compromised [33]. Hence, cell–cell communication is coordinated by coupling cholesterol and Hh signaling components in multiple ways. Understanding the mechanistic roles of cholesterol in Hh signaling activation and transduction is essential to develop novel therapeutic strategies and open new avenues for translational research.
Hh acts as a morphogen, and the ability to signal both short and long distances depends on the degree of processing, post-translational modification, and the accessory regulatory factors which control its diffusion [11, 34]. Hh ligands that initiate signaling by covalently anchoring with a palmitoyl moiety at the N-terminus and cholesterol molecule at the C-terminus coordinate the functional signaling activity [35]. The mechanisms governing the processing, secretion, delivery to target cells, and signal transduction of Hh are fascinating due to the hydrophobic nature of Hh ligands [36, 37]. The many regulatory proteins of the Hh cascade, such as cell-surface receptors, membrane-anchored proteins, cell-adhesion molecules, and co-receptors, also require cholesterol in target cells to receive Hh signals [38, 39]. Smo functions as a core receptor of Hh signaling, and its activity is regulated by cholesterol concentration or accessibility in the ciliary and plasma membrane [32, 40]. Smo is activated by both cholesterol binding and modification, which help reinforce and sustain the Hh signal [41]. Hence, cholesterol emerged as the endogenous ligand for Smo and acts by binding and/or covalently linked to Smo for its activation and functions [40]. Another membrane receptor of Hh signaling, Ptch1, is also implicated in the cholesterol efflux and modulates the intracellular cholesterol concentration [42]. Furthermore, the local concentration of cholesterol affects the trafficking of both Smo and Ptch1 and plays a distinctive role as a second messenger that supports the functional interaction between Ptch1 and Smo to activate and trigger Hh signaling [24, 43]. Intriguingly, Hh signaling involves two cholesterol-dependent aspects: In signal-producing cells, dispatched (Disp) control the release of cholesterol-modified Hh. In contrast, in Hh-responding cells, Ptch1 regulates Smo activation by transporting cholesterol from cells [44, 45]. In this way, both receptors Disp and Ptch1 require cholesterol for Hh signal transfer and activation. Specifically, Disp and Ptch1 function as cation-powered transporters, and distinct cation gradients power cholesterol transport at a different point in Hh signaling across the plasma membrane [46]. Hence, cholesterol biosynthesis, accessible cholesterol in the membrane, and cholesterol modification of the Hh cascade components drive the duration and dynamics of Hh signaling.
Multiple feedback mechanisms dynamically synchronize Hh signaling in a spatiotemporal and context-dependent manner that confers differential patterns in cell fate determination [47, 48]. In the past decade, numerous researchers examined the potential role of cholesterol in Hh signaling and its regulatory phenomenon using biochemical, genetic, structure- and physiology-based approaches. Interestingly, each new finding sparked another question mark to answer the underlying mechanism that makes this pathway complex and fascinating. These findings provided significant outputs to comprehend the mechanistic view and underlying concept of regulation. Therefore, this article summarizes the depth of the Hh signaling network, current research findings, and the distinct proposed model of cholesterol-mediated regulation of Hh signal transduction. We first emphasize the detail of Hh pathway mediators in signal transduction at the cellular level. Then, we explain the current model of intracellular Hh signaling as a canonical and non-canonical Hh/Gli cascade. Finally, we focus on linking Hh signaling and cholesterol and describing their cross-regulatory mechanism(s). This review will cover the following topics: (1) activation of Smo by cholesterol, (2) cholesterol modification of Hh ligand, (3) mechanism of efficient secretion of cholesterol-anchored Hh, (4) the role of cholesterol in the directive of Hh morphogen gradient, and association with trafficking regulators, (5) involvement of Ptch in cholesterol transport, (6) the concept of ciliary cholesterol in Hh signaling, (7) the connection between cholesterol, lipid rafts, and Hh signaling, (8) developmental impact of cholesterol-modified Hh signaling, (9) conception of cholesterol-free and cholesterol-modified Hh secretions and their actions, and (10) cation gradients’ influence on cholesterol transport in the Hh signaling. Collectively, this review aims to provide the cumulative evidence supporting Hh signaling association with cholesterol to promote further inquiry into this area.
Hedgehog signal transduction and regulatory elements of Hh signaling cascade: at a glance
The Hh signaling mechanism is evolutionarily conserved from flies to humans [49] and considered a pivotal regulator of multiple fundamental processes, including cell fate determination, proliferation, differentiation, tissue polarity, patterning, morphogenesis, regeneration, and repair in adults by regulating the various progenitor cells [4, 34, 50]. The Hh gene was initially identified by Christiane Nüsslein-Volhard and Eric F. Weischaus in the late 1970s as a result of genetic analysis of the segmentation of fruit fly Drosophila melanogaster. In 1995, they were awarded the Nobel Prize for studying genetic mutations in Drosophila embryogenesis [51]. The origin of the name of the Hh gene is the ensuing appearance of a continuous lawn of denticles and hair-like bristles reminiscent of hedgehog spines projecting from larvae cuticle containing null allele of hh [11]. In vertebrates, the Hh family comprises three Hh-related genes sonic hedgehog (Shh), desert hedgehog (Dhh), and Indian hedgehog (Ihh), which share high sequence homology and act as initial ligands to trigger Hh signaling [1, 52]. All three Hh ligands are similarly processed and secreted from responding cells and activate Hh signaling in target cells [1, 34]. The critical physiological role of Hh involves its function as a morphogen, a mitogen, a survival factor, and even a guidance factor to induce the distinct cell fates [53–55]. Shh is mainly engaged in nervous system cell-type specification and limb patterning, while Ihh is marked in skeletal development, particularly endochondral ossification, and interestingly, Dhh is restricted to the gonads, particularly in granulosa cells of ovaries and sertoli cells of the testis [56–60].
In the first step of the Hh cascade, the Hh transcript is translated into ~ 45 kDa pro-protein consisting of an amino-terminal (N-terminal) signal sequence, a Hedge domain, followed by a carboxy-terminal (C-terminal) Hog domain [35, 61]. After eliminating the signal sequence, Hh polypeptides are moved into the endoplasmic reticulum (ER), and the Golgi apparatus undergoes an autocatalytic process. The N-terminal half of Hh is covalently anchored with cholesterol through an intramolecular proteolytic reaction that concurrently cleaves off the inactive C-terminal polypeptide [61, 62]. Subsequently, the palmitoyl group is transferred to the extreme N-terminus cysteine of Hh using membrane-bound O-acyltransferase Skinny Hh in flies and Hh acyltransferase (HHAT) in vertebrates [63, 64]. A distinctive N-terminus signal sequence is also eliminated during trafficking to the plasma membrane [35]. The processed/mature Hh is ~ 19 kDa protein carrying two lipid moieties that exhibits all biological activities in an autocrine-, paracrine-, or juxtracrine-dependent signal transmission [35, 65]. The mature Hh is secreted as a monomer, multimer, or component of lipoprotein particles, exosomes, or cytonemes [66–69]. The generation and release of extracellular mature Hh are facilitated by a membrane-bound protein named Disp [44]. In the extracellular space, signal peptide-CUB-EGF domain-containing protein), a chaperone family protein, binds to mature Hh and directs its trafficking with the aid of other membrane-anchored proteins (Scube) and Hh co-receptors [39, 70, 71]. Generally, the switch between active and inactive Hh signaling depends on the rapid translocation of the regulatory component of Hh signaling to the cilium [24].
Next, Hh reaches the surface of target cells, and the signals are transmitted into cells by two primary membrane receptors, Ptch and Smo [47]. Ptch is a 12-span transmembrane protein that comprises a sterol-sensing domain (SSD). It shows structural similarity to a member of the resistance-nodulation and cell division (RND) family of bacterial transporters. In the absence of Hh ligand, it remains in the ciliary membrane [45, 72, 73]. The mammalian genome has two Ptch genes called Ptch1 and Ptch2, and both share structural similarities and overlapping functions. [74, 75]. Studies also suggested the distinct functional properties of Ptch2 isoforms compared with Ptch1 [76]. The negative regulation of Hh is governed by Hedgehog-interacting protein (Hip) that competes with Ptch1 on Hh binding [77]. When cells encounter processed Hh, Ptch1 binds to Hh and forms a complex that translocate from primary cilia to the plasma membrane [78]. In Drosophila, the Hh–Ptch complex involves the adhesion molecules Interference hedgehog (Ihog) and Brother of ihog (Boi) [79, 80], whereas in vertebrates, the complex contains CAM-related/downregulated by oncogene (Cdo), Brother of Cdo (Boc), and growth arrest-specific-1(Gas1) [38, 81, 82]. These regulatory proteins located in the plasma membrane, including filopodial and distinctive localization, account for the variations in temporal and/or spatial Hh signal transmission. Next, Hh–Ptch undergoes endocytosis with the involvement of membrane-remodeling GTPase dynamin and the HECT-domain ubiquitin E3 ligases Smurf1/2; then, both Hh and Ptch are degraded in the lysosome, abolishing Ptch-mediated inhibition of Smo [83–87]. Smo is a seven-transmembrane protein, a Frizzled class (class-F) related to the G-protein coupled receptor (GPCR) superfamily. It comprises three functional domains, the N-terminal cysteine-rich domain (CRD), hepta-helical transmembrane domain (TMD), and a long cytosolic tail [88–90]. The long C-terminal domain (C-tail) is indispensable for Hh-dependent signal transduction. Importantly, Smo activation involves two regulatory steps: Smo translocation from an intracellular vesicle to the cell surface and its subsequent post-translational modification [91–93]. Also, Smo activation includes a conformational switch, leading to Smo accumulation on the cell surface and Hh signal transduction [94, 95]. After clearance of Ptch from the ciliary membrane, Smo is translocated from the plasma membrane to the cilium [96]. Subsequently, Smo is phosphorylated at particular serine residues (carboxyl intracellular region) and interacts with β-arrestin-2 [93, 97]. β-arrestin 1 or β-arrestin 2 plays an essential role in intracellular localization of Smo to primary cilia and is required for the activity and stability of Smo, thereby regulating Gli activation [97]. In addition, Smo phosphorylation is the critical step of Hh signal transduction, and known protein kinases include protein kinase A (PKA), casein kinase 1α (CK1-α), or CK1-ϒ and GPCR kinase known as Gprk2 or Grk2 [98, 99].
Ultimately, activation of the Hh signal potentially induces the activation and translocation of transcription factor cubitus interruptus (Ci) in Drosophila, and Gli family members in vertebrates into the nucleus, leading to specific target genes expression [11, 53]. Gli is a Krüppel-like transcription factor comprising zinc-finger DNA-binding domains with dual activity [100, 101]. The three genes in this family known as Gli1, Gli2, and Gli3 appear to have similar DNA binding specificities [102, 103]. All Gli members contain the carboxyl terminus activator domain, while Gli2 and Gli3 contain an N-terminus transcriptional repressor domain [104]. Their post-translational proteolytic processing balances the activator and repressor forms of Gli proteins [105, 106]. Gli1 has diverged evolutionally as a full-length transcriptional activator, whereas Gli2 and Gli3 can serve as positive Gli2A and Gli3A activators, or negative Gli2R and Gli3R repressors [107, 108]. The presence of Gli2 regulates the conversion of Gli3A to Gli3R; therefore, Gli2 functions as a transcriptional activator, and Gli3 is mainly a repressor [104]. Surprisingly, in response to the Hh signal, Gli3 directly binds to the Gli1 promoter and stimulates Gli1 transcription [109].
Generally, in the absence of an extracellular Hh ligand, Ptch1 is enriched in primary cilia and blocks Smo activity [110]. This repression is mediated via two known mechanisms, either disruption of Smo localization or catalytic suppression of Smo [87, 110]. In vertebrates, full-length Gli is held in a microtubule-associated protein complex containing kinesin protein (Kif7), Suppressor of fused (Sufu), and kinases including, CK1, Gsk3β, and PKA which facilitates Gli phosphorylation; while in Drosophila, the regulatory proteins, such as Costal2 (Cos2), the kinase fused (Fu), Sufu and kinases controls Ci processing and activation [16, 111–113]. In addition, in the absence of Hh, the increased calcium (Ca2+) level in the cytoplasm and cilium induces ciliary cAMP and PKA activity [114]. Consequently, kinases, including PKA, glycogen synthase kinase3 β (Gsk3β), and CK, serve as negative regulators of the Hh pathway by phosphorylating Ci/Gli [19, 115, 116]. Phosphorylated Gli is recognized by the Skp-cullin-F-box (SCF) ubiquitin ligase complex, which usually is composed of culin-3 and E3-type ubiquitin ligase β-transducin repeat-containing protein (β-TrCP) [117]. The SCF complex generates the proteolytic cleavage of the full-length form of Gli into a truncated repressor form that ultimately represses subsets of the Hh target gene [106]. On the other hand, in the presence of extracellular Hh ligand, Ptch1 moves out of the cilia and forms a Ptch–Hh complex, leading to internalization [78]. Next, Smo is dynamically trafficked to both the plasma membrane and cilium and monitors Gli processing and its action [96]. Simultaneously, Hh stimulation reduces ciliary cAMP through a G protein-independent mechanism that needs extracellular Ca2+ entry, PKA activity, and stabilization of full-length Gli1 protein [114, 118]. Thus, the full-length active form of Gli1 is translocated into the nucleus, binds to DNA target sites, and directs various target genes' expression regulating multiple cellular processes [47, 105].
Hh signaling is also negatively regulated by the scaffold protein Suppressor of fused (Sufu), which acts on Gli [119–121]. In the absence of the Hh signal, Sufu directly binds to Gli and stabilizes it; thus, Gli is retained in the cytosol and undergoes degradation [121, 122]. Sufu-mediated attenuation of Gli nuclear translocation thereby significantly prevents Hh pathway activation. With ligand activation, Smo accumulates in the primary cilium, and the suppression of Gli activity by Sufu is diminished by hyperphosphorylation of Sufu [123, 124]. However, the mechanism of Sufu action based on its phosphorylation status is complicated. Previous study identified that Gsk3β specifically binds and phosphorylates Sufu, which leads to positive regulation of Hh signaling [125]. Another study reported that Sufu is phosphorylated at Ser-342 and Ser-346 through Gsk3β and PKA, respectively. Dual phosphorylation stabilizes Sufu against Hh signaling-induced degradation [126]. Sufu–Gli complex trafficking occurs in a Smo-dependent manner that allows Gli to be hyperphosphorylated and dissociated from Sufu [116]. The Sufu–Gli complex dissociation attenuates Gli full-length processing and shifts the processing to the activated form Gli-A [124]. Thus, these reports suggest Sufu acts by sequestering Gli activators in the cytosol or facilitating the production of the Gli truncated repressor form [126]. However, the underlying specific mechanisms through which Smo interacts with Sufu as well as how Sufu inactivates Gli is elusive.
Multiple feedback regulatory mechanisms dynamically control Hh signal activation and its signal transduction. Hh signal amplification and signal attenuation confer various cell fates in target cells. Hh-binding proteins, such as Ptch and Hip, are induced by Hh signaling and sequester the Hh ligand [127]. Moreover, Hh co-receptors Boc/Cdo expression is negatively regulated by Hh signaling. However, this complex Boc/Cdo does not participate in Ptch-mediated feedback regulation but in Ptch-mediated Hh reception [81]. Importantly, several kinases, including PKA, protein kinase C (PKC), CK1, mitogen-activated protein kinase kinase (Mek1), Gsk3, GPCR kinase (Grk) and dual-specificity Yak1-related kinase (DYRK1) modulate the effector molecules of the Hh cascade and control Hh signal activation or its inhibition [98, 99, 116, 117, 128]. Thus, Hh signaling is a spatial, temporal, and cell-contextual regulatory network of intracellular signaling mediators, including membrane receptors, several binding proteins, protein kinases, and effector molecules that determine the signaling kinetics and responsiveness to the signal (Fig. 1).
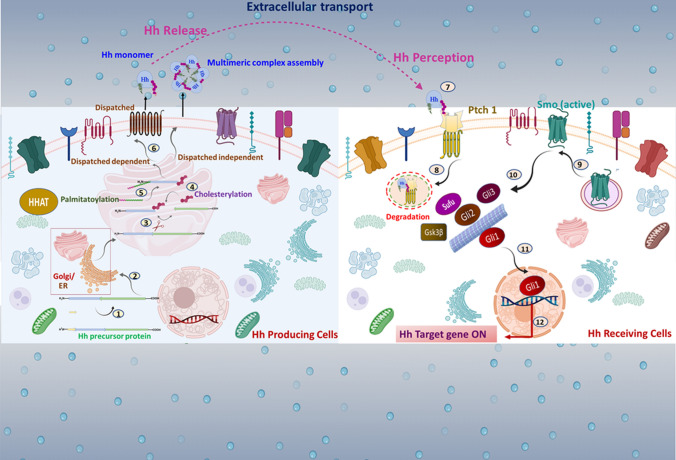
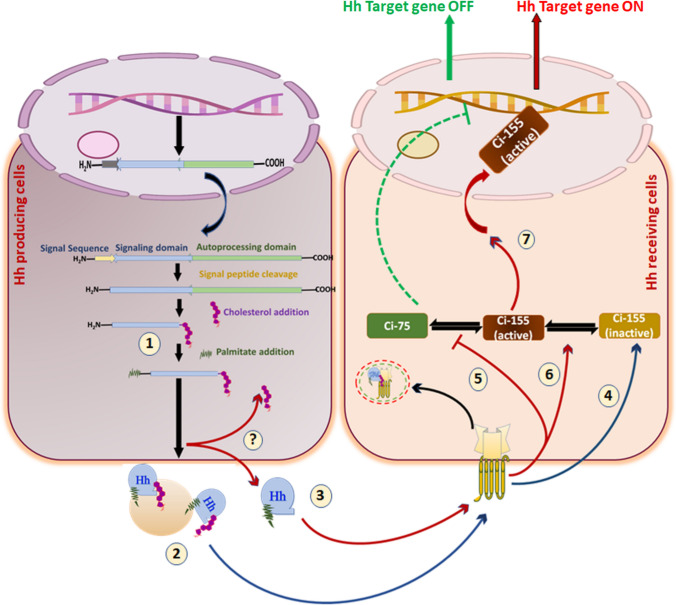
Fig. 1.
Insight on molecular events involved in Hh signal transmission: in Hh-producing cells, the Hh transcript is translated as a ~ 45 kDa pro-protein consisting of a signal sequence at the N-terminal domain followed by the C-terminal domain. Multiple steps are implicated in Hh signal transmission: 1. The signal sequence is removed. 2. Hh polypeptides are moved into the ER and the Golgi apparatus (A rectangular magnified box represents it). 3. Hh pro-protein undergoes autocatalytic processing 4. The N-terminal half of Hh is covalently anchored with cholesterol via cholesterylation. 5. A palmitoyl group is transferred to Hh’s extreme N-terminus cysteine using HHAT. 6. In the extracellular space, the mature Hh is secreted distinctly as a Disp-dependent monomer or a Disp-independent multimer. 7. Hh reaches the surface of target cells. 8. In Hh receiving cells, Hh complexes with Ptch1, leading to internalization. 9. Smo is released from Ptch1-mediated suppression, and free Smo translocates into the membrane 10. Activation of Smo regulates Gli family proteins, such as Gli1, Gli2, and Gli3, with the help of multiple protein kinases. 11. An active form of Gli, such as Gli1, trans-locates into the nucleus. 12. Gli triggers activation of downstream targets
The intracellular Hh signal transduction network: canonical and non-canonical Hh/Gli signaling mechanism
To date, numerous findings have highlighted the prominence of molecular crosstalk between Hh signaling and other signaling cascades that regulate signal strength and influence the specificity of molecular and cellular phenotypes. The tight regulation of Hh signaling ensures the graded responses to Hh in responsive cells in a temporal and spatial context. Hh signaling occurs by canonical and non-canonical mechanisms [16, 22, 129]. The current canonical Hh signaling paradigm involves the ligand-dependent interaction or receptor-induced signaling ruled by the binding of mature Hh ligand to its receptor Ptch1 and its co-receptors Boc/Cdo, Gas, etc. In response, Smo becomes activated and accumulates at the membranes and initiates a downstream signaling cascade, involving Gli1 processing and transactivation [130]. Activation of Gli promotes its nuclear translocation and induction of target genes, which include a self-amplifying loop of Gli itself, leading to cellular response (Fig. 2). In non-canonical Hh signaling, the response to the Hh signal diverges from this paradigm, independent of transcriptional changes mediated by the Gli family of transcription factors [131]. According to current studies, three scenarios of non-canonical Hh signaling have been described: (1) type I Smo-independent or Ptch1-dependent mechanism that regulates cellular proliferation and survival; (2) type II Smo-dependent or Gli-independent mechanism can modulate Ca2+ signaling, and cytoskeleton rearrangement for metabolic rewiring; and (3) type III consists of all other regulatory mechanisms of Gli-family member activation, which are independent of upstream Ptch1–Smo interaction [16, 17, 131] (Fig. 3).
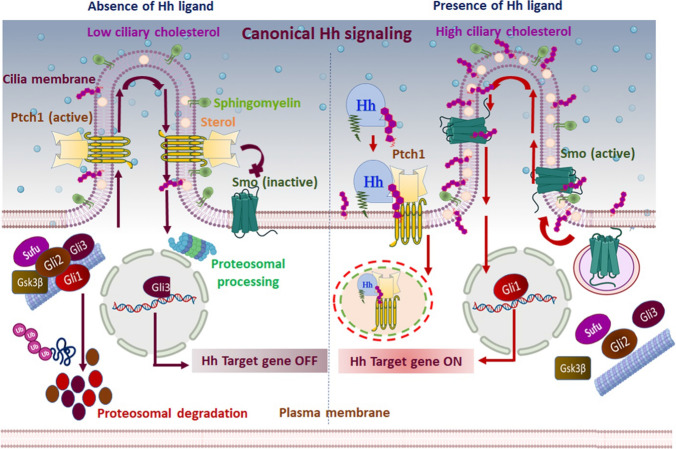
Fig. 2.
Canonical Hh signaling mechanism: the canonical/classical Hh signaling is a ligand-dependent interaction regulated by a bifunctional transcription factor that can activate or repress the transcription of target genes based on nuclear translocation of suppressor/truncated or activator/full-length forms. In the absence of Hh ligand (left panel), Ptch1 accumulates on the primary cilium and inhibits the translocation and functional activation of Smo. Subsequently, Sufu restrains Gli activity, and Ptch1 facilitates the activation of protein kinases (CK1, PKA, and Gsk3β) that induce the phosphorylation of Gli family members. Complete degradation of Gli1, Gli2, and partial cleavage of Gli3 ensue by ubiquitination. The partial cleavage of Gli3 generates the truncated Gli3 that trans-locates to the nucleus and acts as a transcriptional repressor for Hh target genes. Furthermore, Ptch1 inhibits Smo activity by reducing the accessible cholesterol in the ciliary membrane. Without Smo activity, the Gli proteins undergo proteasomal degradation and turn off Hh signal transmission. In the presence of the Hh signal (right panel), Hh binding to Ptch1 inhibits the function of Ptch1 and induces its clearance from the primary cilium via lysosomal degradation of the Hh–Ptch1 complex. Consequentially, inhibition of Smo is lifted, and it relocates to the primary cilium. Activated Smo transmits the Hh signal across the membrane by antagonizing Sufu and protein kinases, ultimately preventing degradation of Gli proteins. Activated Gli protein is translocated into the nucleus and acts as a transcriptional activator for Hh target genes. In addition, the inactivation of Ptch1 raises the accessible cholesterol level in the ciliary membrane, allowing Smo to adopt an active conformation and induce activated Gli. Active Gli trans-locates into the nucleus and ultimately turns on the Hh signal transmission
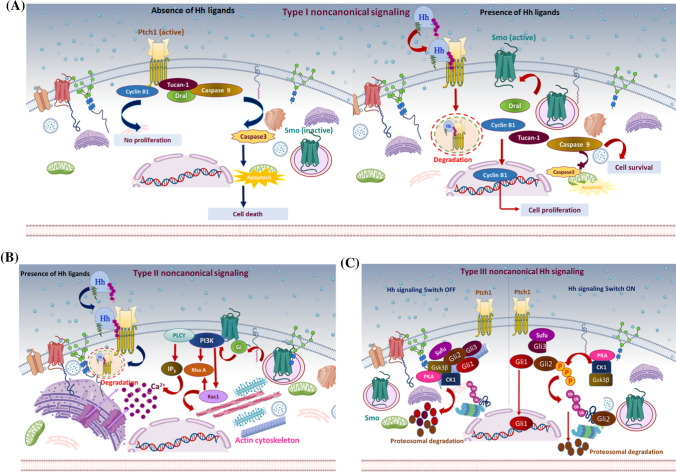
Fig. 3.
Non-canonical hedgehog signaling mechanism: different proposed models of non-canonical Hh signaling regulation. a Type I non-canonical Hh signaling: the C-terminal motif of Ptch1 interacts with cyclin B1 and a proapoptotic complex, including caspase 9, Tucan-1, and Dral. In the absence of the Hh signal (left panel), Ptch1 sequesters cyclin B1 and inhibits its nuclear translocation, inhibiting cell proliferation/survival. Also, cleavage of the C-terminal domain of Ptch1 by caspase 3 exposes the proapoptotic domain and promotes nucleation and activation of caspase 3, eventually leading to apoptosis. In the presence of Hh ligand (right panel), Hh binds Ptch1, and the interaction of Ptch1 with cyclin B1 or the proapoptotic assembly is disrupted, leading to increased cellular proliferation/survival. b Type II non-canonical Hh signaling: In the presence of Hh ligand, Ptch1 becomes degraded, releasing activated Smo to regulate the actin cytoskeleton through a small GTPase in a context-dependent manner. In some cell types, activated Smo releases Ca+2 from the ER via Gi and PLCϒ-dependent generation of IP3 and IP3 channel opening. c Type III non-canonical Hh signaling: In the absence of an Hh signal (left panel), protein kinases PKA, CK1, and Gsk3β phosphorylate Gli family members. This induces ubiquitin-mediated degradation of Gli1 and Gli2. Meanwhile, Gli3 is converted into its truncated repressor form and is translocated into the nucleus. Truncated Gli3 is released from Sufu and blocks Gli1-mediated activation of the target genes. In the presence of the Hh signal (right panel), Gli is released from phosphorylation-mediated proteasomal degradation. Subsequently, Gli1 localizes to the nucleus and activates Hh responsive genes
Type-I non-canonical Hh signaling relies on Ptch1 and is distinctive from Smo inhibition. Hh acts as a survival factor and Ptch1 as a dependence receptor where it stimulates apoptosis in the absence of Hh ligand. Its proapoptotic activity is obstructed in the presence of Hh ligand [132, 133]. Ptch1 contains extracellular domains that bind to Hh ligand, whereas its intracellular C-terminal domain modulates Ptch1 activity but is not vital for canonical Hh pathway regulation [127]. Therefore, C-terminal fragments serve as an alternative signal transducer that directly translocates to the nucleus and modulates the transcriptional activity of Gli1 [134]. Additionally, the C-terminal motif of Ptch1 is cleaved by caspase, particularly at a conserved aspartic acid residue (Asp1392) to expose the proapoptotic domain [134]. In the absence of Hh, Ptch1 interacts with the adaptor protein DRAL known as FHL2, which recruits CARD-domain-containing proteins named TUCAN or NALP1 and apical caspase-9, which triggers caspase-9 activation. Therefore, Ptch1 acts as the anchor protein for this caspase-activating complex and induces apoptosis via caspase 9 [135]. A C-terminal-truncated Ptch1 mutant resulted in Ptch1-mediated cell death irrespective of the presence of a Hh signal, and a mutation in the cleavage site of Ptch1 significantly abolished the cell-death efficiency; these findings point to the importance of the C-terminal domain in regulating Ptch1 activity [134, 135]. Furthermore, Ptch1-dependent non-canonical signaling involves interaction with cyclin B1, leading to localization of M-phase promoting factor (MPF) to regulate cell cycle progression at the G2/M checkpoint [136]. Phosphorylation-dependent changes in the nuclear import rate and export of cyclin-B1-Cdk1 and their regulator Cdc25C control the onset of mitosis [137]. Ptch1 serves as a tumor suppressor by restricting G1–S phase and G2–M phase and is considered the “gatekeeper” of cell cycle progression [138]. Hh ligand binding disrupts the interaction between Ptch1 and cyclin B1, allowing nuclear translocation of cyclin B1, resulting in enhanced mitosis and increased cellular proliferation and survival. On the contrary, in the absence of the Hh signal, the Ptch1–cyclin B1 interaction inhibits cellular proliferation by sequestering cyclin B1 in the cytosol [136, 137]. Remarkably, Hh-mediated conformational changes in Ptch1 increase its affinity towards GRK2. Consequently, Ptch1 releases cyclin B1, which localizes to the nucleus and activates genetic programming supporting cell proliferation and survival [138, 139]. These mechanistic insights constitute the Smo-independent regulation of Hh signaling via Ptch1.
Type-II non-canonical Hh signaling involves Gli1-independent activities of Smo through modulation of Ca2+ flux, leading to regulation of the actin cytoskeleton through the Rho family of small GTPase, i.e., RhoA and Rac1. This mechanism promotes actin stress fiber formation, endothelial tubulogenesis, migration, axon turning, cell polarity, and dendritic spine formation in a Smo-dependent manner [140, 141]. Regulation is context-dependent, as Smo interacts with Gli protein and PI3K in fibroblasts, and Tiam1 or Src and Fyn, an Src kinase family (SFK) member, in neurons [140, 142]. Smo releases Ca2+ spikes from the ER in spinal neurons through the Gβϒ subunit upon Gli activation [143]. In addition, activated Gli promotes activation of phospholipase C-ϒ (PLC-ϒ) to generate inositol3-phosphate (IP3) and, subsequently, to open an IP3-dependent channel [16, 143]. Interestingly, a recent study showed the importance of primary cilia in non-canonical Hh signaling. The ciliary-dependent non-canonical Hh signaling stimulates α-tubulin acetylation via Smo, leading to post-translational microtubule regulation and modulation of cell behavior [144].
Type-III non-canonical Hh signaling consists of all other signaling mechanisms that independently activate Gli without Smo or Ptch1 activation [10]. This type of mechanism usually occurs through post-translational modification, including phosphorylation, sumoylation, acetylation, and O-GlcNAcylation of Gli-family members. Phosphorylation induces activator or repressor forms of Gli; protein kinases, such as PKA, DYRK, ULK3, S6K1, AMPK, MEKK1, and Hck promote activation, while CK1 and Gsk3β can promote either activation or suppression [116, 128, 145–149]. Ubiquitination promotes proteasomal degradation or generation of the nonfunctional truncated form of Gli that interferes with Gli protein activity. Numerous E3 ligase complexes, such as Cul1/β-TrCP and E3 ubiquitin-protein ligase Itchy homolog (Itch), catalyze the ubiquitination of the Gli protein [150, 151]. Interestingly, Cul1/β-TrCP-mediated ubiquitination occurs on the inactive form of Gli, resulting in complete protein degradation, while Itch is implicated in the degradation of activated Gli. SUMOylation is a nuclear localization signal that stabilizes Gli1 via competing with ubiquitination. Small ubiquitin-related modifier (SUMO) as Pias1 ligase reduces Gli1 ubiquitination, whereas sentrin-specific peptidase 1 (SENP1)-mediated deSUMOylation attenuates Gli activity [151, 152]. Moreover, crosstalk with other molecular signaling pathways induces the transcriptional activation of Gli [131].
Activation of Smo by cholesterol
Vertebrate Hh signal transduction requires cellular sterol binding to membrane protein Smo, which controls Smo activity by controlling its access to cholesterol [40]. Genetic defects in cholesterol biosynthesis and sterol depletion affect the activity of Smo and are implicated in the attenuation of Hh signal response [33]. Notably, cholesterol has emerged as a candidate endogenous activator of Smo and is essential for Hh signaling [40]. The crystal structure of the Smo CRD domain complexed with sterol demonstrated that cholesterol can itself bind and directly activate Smo; however, cholesterol has a much greater affinity for full-length Smo [40]. Structural insights revealed that cholesterol occupies a central position, interacting with all three Smo domains, i.e., CRD, linker domain, and TMD domains. It adopts an extended conformation with its tetracyclic sterol ring that facilitates binding to a shallow groove of extracellular CRD [89]. The CRD hydrophobic residues are predominantly lined on the sterol-binding site of Smo, and these residues stabilize the flat α-face of cholesterol. Mutations in several amino residues (Leu108, Trp109, Pro164, and Phe166) of Smo are projected to attenuate Smo binding to cholesterol and impair Hh signaling [89]. Moreover, a high-resolution structural analysis revealed that sterols occupy the site on SmoCRD in a head-to-tail orientation. SmoCRD distinguishes sterols from other lipids using both shape and amphipathic properties that emphasize the exquisite sterol specificity of Smo [40]. However, a class of metabolites produced by cholesterol oxidation and several different sterols can bind to Smo and stimulate a dramatic CRD conformational change of the binding site, which is adequate to activate Smo. Smo–sterol interaction is crucial for allosteric Smo activation (ligand-induced conformational changes) and distinctive among CRD-containing receptors. However, the activity of Hh signaling hinges on a balance between active and inactive forms of the wild-type Smo receptor. In contrast, oncogenic Smo has a higher intrinsic ability for aberrant activation of this pathway [153]. Additionally, Smo activity is restricted by changes in cholesterol abundance as well as the accessibility of a specific pool of cholesterol [43]. An acute increase in plasma membrane cholesterol can initiate signaling from the cell surface; hence cholesterol acts as a ligand for Smo [89] [154].
Biochemical studies revealed that covalent cholesterol modification termed cholesterylation is a post-translational modification of Smo on the Asp95 (D95) residue through an ester bond formed between the 3β-OH of cholesterol and the side chain –COOH [41]. Reports suggested the Smo (D95N) mutation abolished cholesterylation and decreased Hh-stimulated ciliary localization and native Hh signal transmission. Similarly, homozygous SmoD99N/D99N (the equivalent residue in mouse) knock-in caused embryonic lethality with severe cardiac defects in mice and phenocopied the Smo−/− mice. Intriguingly, this cholesterylation process is abrogated by Ptch receptor but stimulated by Shh ligand. Interestingly, the cholesterylation of Smo is unlikely to modify cholesterol binding to Hh due to lack of autocatalysis, results in the Smo size being unchanged after modification [41].
Following crystal structure determination and biochemical studies on cholesterol and Smo, Xiao et al. [41] concluded that cholesterol initially binds to the SmoCRD. After this interaction, esterification occurs between the 3β-OH of cholesterol and the side chain-COOH of the SmoD95 residue. However, both cholesterol binding and modification are able to activate Smo and downstream signaling. Indeed, cholesterylation of Smo helps reinforce and sustain Hh signaling.
Cholesterol modification of Hh ligand
Hh proteins are vital secreted protein ligands/morphogens that coordinate cell–cell communication by activating the Hh signaling components. Post-translational modification of Hh proteins is required for their activity and native signal transduction [25, 155, 156]. Hh ligand is produced as an approximately 45 kDa precursor protein and then translocated to the ER to remove its signal peptide. An autocatalytic cleavage reaction of the Hh protein results in the production of a 19 kDa N-terminal fragment (N-Hh) and 25 kDa C-terminal fragment (C-Hh) [157]. C-Hh was observed as the catalytic domain, and N-Hh is dispensable for signal transmission. Structurally, Hh proteins are composed of a Hedge domain-containing signaling sequence (SS) at the N-terminus and a Hog domain-containing sterol-recognition region (SSR) and Hint (Hedgehog/intein) module at the C-terminus. The Hint domain accounts for the autocatalytic activity of self-cleavage between C-Hh and N-Hh [61]. After autocatalytic cleavage of the peptide bond between the two domains of Hh, the N-Hh fragment undergoes lipid modification. A cholesteryl residue is bound at the C-terminus of N-Hh by autocatalytic conjugation [62], and a palmitoyl residue is linked at the N-terminus by the palmityl transferase Skinny hedgehog [63]. This cholesterylation process is a prerequisite for efficient secretion of N-Hh and for its signal activity [25]. Biochemical analysis elucidated that the cholesterol modification on Hh proteins involves a two-step auto-processing reaction. First, the thiol group of cysteine makes a nucleophilic attack on the carbonyl group of the preceding residue, glycine, replacing the peptide bond with a thioester bond. Subsequently, the 3β-OH of cholesterol is attached to the same carbon of the thioester intermediate, resulting in an ester-associated adduct to N-Hh and free C-Hh [158] [62] (Fig. 4).
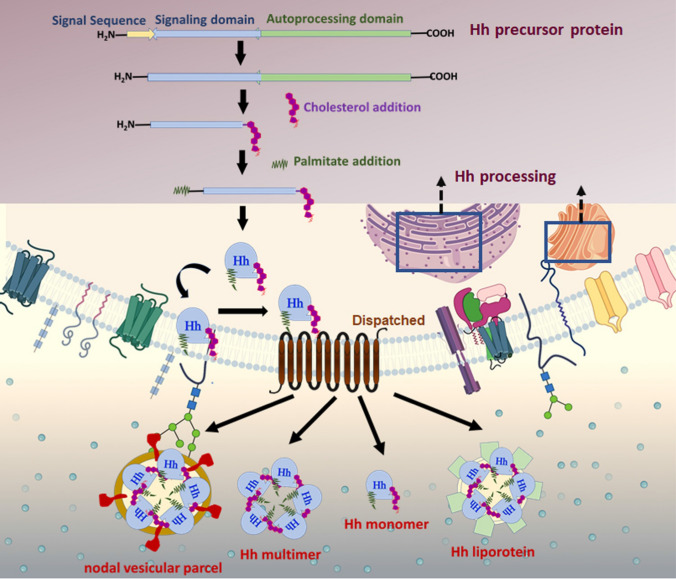
Fig. 4.
Hh protein biogenesis and secretion: Hh is produced as a precursor protein ~ 45 kDa, transported to the ER, and post-translationally modified to generate a biologically active Hh protein. Hh pre-protein comprises a signal sequence, an N-terminal domain, and a C-terminal domain. Upon entrance into the secretory pathway, the N-terminal signal sequence is cleaved. Next, an autocatalytic reaction removes the C-terminal domain of Hh and catalyzes an intramolecular cholesteryl transfer reaction at the C-terminal. The final modification is completed by adding a palmitic acid moiety on a cysteine residue near the N-terminus by the acyltransferase skinny hedgehog. The dually modified Hh is secreted and released by the multi-pass transmembrane protein Disp 1 into the extracellular compartment. Disp facilitates the assembly of soluble, high molecular weight multimeric complexes. Hh protein is delivered to responsive cells via cytonemes and released from the plasma membrane via multiple possible mechanisms as a soluble multimer, a nodal vesicular parcel, or within lipoprotein particles or exovesicles
The N-Hh is responsible for all Hh signaling activities. The C-Hh is involved in the auto-processing reaction and is responsible for the peptide bond cleavage that enables cholesterol transfer [159]. Notably, the crystal structure of a Hh auto-processing domain shows structural homology with self-splicing proteins [159]. The structure of C-Hh is specific, characterized by a conserved amino acid sequence, present in the different homologs of Hh (Shh, Ihh, and Dhh). These amino acid sequences are implicated explicitly in forming active sites and generating the thioester intermediate during cholesterylation. His329, Thr326, and Cys400 amino acids are essential for thioester formation, and Asp303 (replaced by histidine in some Hh homologues) forms a hydrophobic pocket activating the cholesterol hydroxyl; 63 C-terminal amino acids are required for cholesterol attachment [160]. A cholesterol analogue showed that the 3β position of the hydroxy group of cholesterol is essential for the successful cleavage and attachment of sterol. Replacement of the –OH group with ketone, ester, or thiol resulted in the abolition of auto-processing, whereas the adjunct of non-essential functionalities, such as addition or branching of the aliphatic side chain, did not cause noticeable changes in Hh processing. Furthermore, the addition of an –OH group to the side chain or ring (positions 4 and 19) of cholesterol significantly decreased its auto-processing efficiency [159] [160].
Mechanism of efficient secretion of the cholesterol- modified Hh ligand
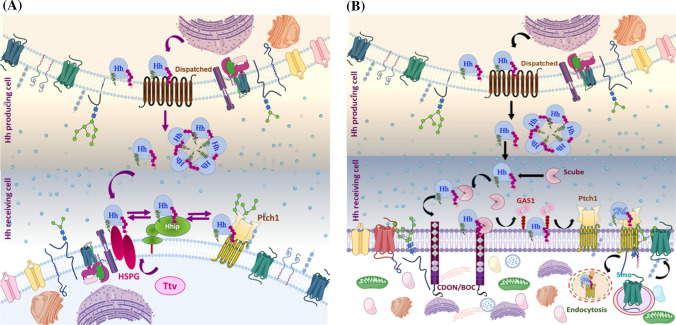
Hh ligand is strongly hydrophobic, firmly tethering it to the plasma membrane of expressing cells. Due to its hydrophobic nature, how Hh exits cells and reaches distal target cells is intriguing. Genetic analysis revealed that Disp, a multi-spanning membrane protein containing a sterol-sensing domain, which shares homology with RND bacterial efflux pumps, is required for Hh release in both flies as well as vertebrates [44, 161] (Fig. 5). Additionally, long-range Hh signal transmission requires the Scube family of secreted glycoproteins (Scube1, Scube2, Scube3) [71, 162]. In Disp mutant cells, only juxtracrine Hh signaling is activated due to inadequate release of lipid-modified Hh. Therefore, precisely, Disp facilitates lipid-modified Hh secretion [67, 163]. Hh binds physically to Disp through its cholesterol moiety and participates in releasing Hh from cells. Scube proteins are involved in Hh biosynthesis, secretion, and release via cholesterol or palmitate moiety [44, 70, 164]. Both proteins are implicated in Hh function outside the signal-producing cells. Scube protein contains CUB domains (C1r/C1s, Uegf, and Bmp1), which act as a regulator of proteolytic activity [71]. Cholesterol-modified Hh requires Disp and Scube proteins for successful interactions, efficient secretion, and signal diffusion [164]. Tukachinsky et al. [164] reported that DispA and Scube2 cooperate during human sonic hedgehog (hShh) secretion. During secretion, the unique cholesterol modification on hSHH participates in two distinct and synergistic binding events. The study showed that DispA transfers hShh to Scube2 via a hand-off mechanism that relies on structural recognition of cholesterol. This mechanism prevents the precipitation of Hh ligand and ensures the cholesterol anchor of Hh and never contacts the aqueous environment directly and thus retains its soluble state. Initially, modified Hh ligand bind to DispA in a cholesterol-dependent manner, and from the heterologous protein (Hh-Disp), the Hh is transferred to Scube [164]. There are two Scube-dependent proposed models for Hh solubility and dispersal. One possibility is Scube2 participates in chaperoning Shh and helps it to form a soluble multimeric species (cholesterol anchors are shielded from the aqueous atmosphere by the interaction between Shh monomers) [164]. In another model, Scube proteins act only transiently during Shh release. Such protein–protein interaction is expected to be dynamic, to allow Hh spreading and formation of soluble Shh species [71].
Fig. 5.
Possible scheme for the release and uptake of cholesterol-modified Hh ligand: lipidated Hh associates with the plasma membrane (lipid-raft domain) and binds with the sterol-sensing domain of DispA, and this interaction is essential for Hh release. a Disp and Ttv mediate Hh secretion in Drosophila: once Disp-dependent Hh is released, it moves from the secretory cell to the responding cell by interacting with a Ttv-dependent membrane-tethered proteoglycan HSPG. Further, Ptch1 and Hip limit Hh diffusion by sequestering the Ptch–Hh complex. b Disp and Scube mediate Hh secretion in Vertebrates: From DispA, Hh is transferred to the Scube2 via a hand-off mechanism. Scube binds cholesterol-anchored Hh, producing a highly active and soluble morphogen. Co-receptors, such as CDON/BOC and Gas1, cooperate to move Hh from Scube2 to Ptch1. Briefly, CDON/BOC recruits Scube-bound Hh to the cell surface and facilitates Hh transfer to Gas1, which catalyzes Hh transfer to receptor Ptch1 in responding cells. The binding of Hh to Ptch1 promotes endocytosis of the Ptch–Hh complex, which facilities Smo translocation
Notably, the generation of the active form of Hh from the lipidated form involves multiple steps. Suggested mechanisms consist of movement by lipoprotein particle [68], transport by cytonemes (cellular extensions) [69], Scube2-mediated extraction [70, 164], Hh ectodomain shedding from the cell surface [165, 166], and finally, ADAM (A disintegrin and metalloproteases) sheddases-mediated conversion of cell surface-tethered multimer Hh into truncated morphogen clusters [165, 167]. Intriguingly, Scube releases the cholesterol-modified Hh but blocks its direct signaling through Ptch by obscuring its unprocessed N-terminus. Previous studies have clarified that Hh is solubilized by proteolytic processing called shedding, which involves removing N-terminal acetylated peptides from Hh, which thereby exposes the Ptch binding site of the solubilized clusters and promotes Shh release [71, 166]. Indeed, Scube2 functions as protease enhancers and augments the N- and C-terminal Hh shedding in the producing cells. Scube2-mediated Hh solubilization is further enhanced by activated shedding. Hence, sheddases and Scube2 cooperatively increase the soluble bioactive Hh pool [71].
Palmitoylation of Hh (attachment of palmitic acid to the conserved α-amino group of the N-terminal cysteine of Hh) is also indispensable for Hh biological activity, efficient delivery of secreted Hh to its receptor Ptch1, and deactivation of Ptch1, which activates downstream Hh signaling [168, 169]. Palmitoylation of Hh occurs in the ER with the help of Hedgehog acyltransferase (HHAT), a membrane-bound O-acyl transferase (MBOAT) that encourages the transfer of palmitoyl-CoA from the cytosolic to the luminal side of the ER membrane [169]. Therefore, the mechanism by which Hh distinguishes Ptch is a critical aspect. Recently, cryo-EM structures of human Ptch-1 alone and complex with native Shh provided atomic level understanding into recognizing Shh by Ptch1 to regulate Hh signaling. Structural analysis revealed that palmitoylated Shh inserts into a cavity between the two homologous ECD domains of Ptch1 and dominates the Ptch1-Shh interface; co-receptors or another Ptch1 bind to Shh at a discrete interface [168].
Recently, Wierbowski et al. [39] elaborated the precise molecular pathway that shuttles the Shh morphogen from signal-producing cells to target cells. Scube-Shh signaling requires several coreceptors, including transmembrane proteins CDON (cell-adhesion-molecule related/down-regulated by oncogene) and BOC (bioregional Cdon-binding protein), and the unrelated GPI-anchored protein Gas1 (growth-arrest-specific 1), which recruit Scube-Shh to responding cells and transfer Shh to Ptch1 [170]. Gas1 is an evolutionarily conserved, vertebrate-specific positive regulator of Shh signaling. Elimination of Gas1 expression resulted in the loss of function, i.e., Shh dose-dependent loss of cell identities, which is consistent with diminished Shh signaling. In contrast, ectopic Gas1 expression resulted in Shh-dependent promotion of cells fates [171, 172]. Shh reception by responding cells involves both CDON/BOC recruitment as well as Gas1-mediated lipid transfer. The Scube–Shh complex is recruited to the cell surface as a ternary complex with CDON/BOC. Afterward, Gas1 interacts with Shh in a lipid-dependent manner and disengages Scube. Overall, CDON/BOC recruits Scube–Shh to cells, and Gas1 catalyzes Shh transfer to Ptch1, initiating downstream signaling [39]. Importantly, Shh reception by responding cells occurs in two steps, including a recruitment step and a lipid transfer step. In the absence of co-receptors, direct handover of Shh lipids from Scube2 to Ptch1 occurs with low affinity because Scube2 blocks the palmitate-dependent interaction between Shh and Ptch1. Thus, in the absence of co-receptors, Ptch1 engages with Scube2-Shh via a pseudo-active site on Shh, which reduces Shh activity. In contrast, with the involvement of coreceptors, the Shh-N and Ptch1 interaction has higher potency [39].
Cholesterol-dependent regulation of the Hh morphogen gradient: Trafficking regulators
Hh activity depends on the degree of its processing, modification and the accessory regulatory factors that control its diffusion [173]. Different post-translational modifications modulate the Hh activity. The imperative role of cholesterol in Hh function raises the question of how does modified Hh execute both short- and long-range Hh biological activity? Initial studies have demonstrated that tout-velu (Ttv) is essential for diffusion of lipid-modified Hh in Drosophila [174], and the processed form of Hh appears to show all morphogenic functions for local and distant-range signaling [175]. Strikingly, cholesterol-modified Hh, but not un-modified Hh depend on Ttv’s target cells to move across the field. The Ttv gene encodes a glycosaimoglycan (GAG) transferase enzyme, homologous to the vertebrate exostoses (EXT1), which is needed for the biosynthesis of heparan sulfate proteoglycans (HSPG). The interaction of cholesterol-modified Hh with HSPG suggested that proteoglycans are obligatory for the diffusion of cholesterol-modified Hh [176]. In addition, another protein Disp has been reported in the release of a fully functional Hh protein from Hh-producing cells [177]. In Drosophila, Disp-deficient cells displayed cholesterol-bound Hh, but cholesterol-free Hh accumulated in signal-producing cells and was not released to the target field. However, Hh synthesis and lipid modifications were unaltered in Disp-deficient cells. Hh signaling under Disp deficiency is limited to cells adjacent to the producing cells, and long-range signaling is abrogated due to the retainment of cholesterol-modified Hh [44]. Thus, Disp functions to release cholesterol-anchored Hh and facilitate intracellular trafficking of the Hh signal. Likewise, cholesterol is needed to restrict Hh diffusion and determine its range of action via Ttv and Disp activities [44]. Moreover, Gallet et al. [175] suggested that cholesterol modification is needed for Hh assembly into lipopolysaccharides (LPSs), and its apical sorting is dependent on Disp. Additionally, LPS apical movement involves a Ttv-dependent proteoglycan for the adjacent anterior cell to perceive the Hh signal, while basolateral Hh localization is Rho-dependent.
Hh presentation from a distinct cellular membrane compartment was suggested to account for the functional diversity of Hh signaling, and accordingly, it permits the Hh-receiving cells to differentially respond to the Hh input [175]. Importantly, unmodified Hh does not require Disp for its secretion. Non-cholesterol anchored Hh, which includes a transmembrane domain or glycosylphosphatidylinositol that transmits the signal to immediate adjacent cells even in the presence of Disp in the signal-producing cell [25]. Thus, Disp is involved in the generation of a signaling aggregate, as well as intracellular trafficking of only cholesterol-anchored Hh in the secretory pathway, or displacement of cholesterol-modified Hh from the cell membrane after reaching the surface of the cell [25, 44, 164]. Contrary to this, a study has shown that cholesterol binding to Hh is essential for its long-range function, whereas unmodified Hh involves only short-range action [178]. In agreement with this, Zeng et al. [179] elucidated that cholesterol-modified Hh has the proficiency to oligomerize while un-modified Hh did not oligomerize, suggesting the cholesterol moiety mediates Hh multimerization needed for long-range signal transmission. The cumulative gradient of Hh is a composite of pools secreted by distinct routes, either apical or basolateral. A cellular summation of these composites is obligatory for proper signaling to understand the absolute value of the extracellular morphogen gradient and determine the requisite response [180]. Two antagonist models have been anticipated: Ayers et al. [180] speculated that apical Hh pool is responsible for a dramatic change in long-range low-threshold target genes activation, whereas the basolateral pool of Hh regulates short-range activity. The study also implied that the glypican Dally and hydrolase Notum regulate apical Hh levels and long-range activity hydrolase [180]. On the contrary, Callejo et al. [163] proposed that the basolateral route for Hh release to form the long-range morphogenetic gradient is mediated by Disp in Drosophila wing disk epithelium. Disp regulates the vascular trafficking required for basolateral release of Hh, Dlp (glypican Dally-like protein), and Ihog (Ig-like and FNNIII domain protein Interference) [163]. Specifically, Hh undergoes endocytosis in producing cells, Dlp and Disp interact with Hh and mediate its release and intracellular trafficking, whereas Ihog attaches extracellular Hh to the basolateral epithelium. Secreted Hh in the form of extracellular vesicles/exovesicles is derived from the endocytic compartment and is released into the extracellular space. In particular, the secretion of Hh exovesicles occurs via two possible pathways: multivesicular body (MVB)-mediated secretion or microvesicle blebbing/shedding; both mechanisms are dependent on endosomal sorting complexes required for transport (ESCRT) proteins [181, 182].
Furthermore, Simon et al. [67] summarized that different apicobasal polarity models of the Hh gradient emerged based on the Hh distribution (apical or basolateral) in the wing imaginal disc epithelium. The proposed “recycling model of Hh secretory trafficking” includes Hh secretion, autocrine internalization, apical to basolateral transport, and ligand representation. Remarkably, internalization of the apical pool of Hh occurs with the help of dynamin and Rab family members, i.e., Rab4, Rab5, and Rab5. After apical internalization, Hh follows the MVB pathway for basolateral secretion and long-range travel associated with cytonemes. In contrast, another study reported that the internalized apical Hh is re-secreted apically via Vps4 (vacuolar protein sorting-associated protein 4) and ESCRT machinery that formed Hh exovesicles via blebbing of the apical plasma membrane in Hh gradient formation [67].
Previous studies have revealed that Ptch also plays a pivotal role in shaping the gradient, i.e., limiting the range of Hh action [183]. Mechanistically, Ptch binds and serves to sequester free Hh and prevent its further movement that reflects the unique self-limiting mechanism of Ptch, by which Hh impedes its range of action. A feedback mechanism whereby Hh upregulates Ptch and thereby limits its own movement serves to balance short and long-range Hh signaling, depending on the developmental context [183, 184]. Indeed, cholesterol-modified and unmodified Hh have a similar binding affinity for Ptch [25]. However, Ptch needs the cholesterol moiety for effective sequestration of Hh [178], and surprisingly, a mutation in the SSD domain of Ptch does not impact the internalization/sequestration of Hh. This finding suggests that the enhanced sequestration of cholesterol-anchored Hh by Ptch is possible through other cholesterol-dependent processes such as lipid raft association [25, 27].
In vertebrates, the morphogen gradient is furthermore refined by Hip, a membrane glycoprotein that interacts with all three mammalian Hh ligands (Ihh, Shh, and Dhh), establishes a negative feedback loop, and modulates Hh downstream signaling [77]. Hip-expressing cells are situated next to Hh-producing cells, and studies have shown that ectopic Hh signaling induces Hip expression, whereas its expression was lost in Hh mutant cells. Moreover, up-regulation of Hh signaling was evident in Hip1-mutant cells, further corroborating the role of Hip1 as a negative regulator of Hh signaling [48, 77]. Cholesterol modified Hh interacts with Hh signaling components, such as Ttv, Disp, Dlp, Ihog, Ptch, and Hip, to fine-tune the Hh morphogen gradient for signal diffusion and transmission. Overall, these studies suggest that multiple layers of regulation are involved in Hh concentration gradient formation.
Involvement of Ptch in cholesterol transport
Ptch is projected to serve as a pump that can change the concentration of a small molecule implicated in Smo regulation. This activity of Ptch requires the suppression of Smo levels in the plasma membrane [42, 87]. Additionally, Ptch acts as a lipoprotein receptor and modulates intracellular lipid homeostasis in a Hh-independent manner [185]. Further, the cryo-EM structure of human Ptch1 with a modified Hh ligand uncovered the structural basis of sterol recognition by Ptch1. Ten sterol molecules surrounded the membrane-embedded part of Ptch1 located at the inner and outer leaflet of the lipid bilayer portion of the protein. This structure may explain how sterol translocation occurs across the lipid bilayer through Ptch1 and other homologous transporters [186].
Apart from Ptch-dependent Smo regulation, Ptch mediates the cholesterol efflux from cells and modulates the intracellular cholesterol concentration. However, exogenous exposure of the N-terminal domain of unmodified murine Shh protein (ShhN) triggers Hh signaling in mouse fibroblast cells. Remarkably, exposure to cholesterol led to an enrichment of Smo at the cell surface compared to cells treated with ShhN protein or Smo agonist. In contrast, inhibiting cholesterol synthesis prevented the Smo accumulation on the plasma membrane by ShhN protein [42]. This suggests that intracellular cholesterol concentration might be pivotal for Smo enrichment in the plasma membrane [42]. The characteristic feature of the Ptch1 structure is a hydrophobic conduit with sterol-like contents required for cholesterol transport and Smo suppression. Zhang et al. [187] stated that Ptch1 expression reduces the cholesterol action in the inner leaflet of the plasma membrane but is promptly restored by Hh stimulation. Therefore, Ptch1 facilitates Hh-reversible reduction of cholesterol activity as well as regulates enrichment of Smo activity in the plasma membrane by controlling cholesterol availability [42, 187]. A recent study suggested that cholesterol access in the cellular membrane regulates Hh signaling, whereby the Hh ligand increases the cholesterol accessibility in the membrane by inactivating Ptch1. Counterintuitively, the trapping of this accessible cholesterol attenuates Hh signal transmission across the membrane. Further, the emerging evidence revealed that plasma membrane cholesterol is sorted into the accessible and inaccessible pool of cholesterol. Accessible cholesterol is defined as the thermodynamically distinctive fraction of total cholesterol suitable for Smo regulation and Hh signal transmission [43]. In summary, Ptch is implicated in cholesterol transport from the plasma membrane’s inner and outer leaflets to the membrane of targeted cells or acceptor proteins [42]. It is also associated with acquiring cholesterol from a donor such as Smo and transporting it to the membrane, thereby regulating the accessibility of cholesterol in the plasma membrane [43].
The concept of ciliary cholesterol in the regulation of Hh signaling
Hh migrates towards target cells from the site of synthesis and synchronizes cellular outcomes depending on the local concentration of Hh. However, accessible cholesterol is responsible for regulating both Hh signal transmission and cholesterol biosynthesis and governs it by spatial segregation [43]. Another mechanism that maintains homeostasis between cholesterol synthesis and Hh signaling is Ptch1-mediated Smo regulation by ciliary cholesterol [32]. Cilia are small hair-like protuberances outside the eukaryotic cells with a distinct protein, and lipid composition compared to the plasma membrane and decode various signals [188]. A recent study deciphered that extracellular ligand, such as Shh, Smo agonist (SAG) or cholesterol depleting agent Methyl-β-cyclodextrin (MβCD), modifies ciliary cholesterol to control the activity of ciliary-localized proteins, which in turn controls Hh signaling. Enzymes in the cholesterol biosynthesis pathway positively regulate Hh signaling, and cellular sphingomyelin suppresses the Hh pathway by sequestering the cholesterol [32]. Ptch1 is prominently localized in a punctate pattern along the membrane of the cilia and ciliary pocket (membrane invagination around the base of the cilium) [32, 43]. The ciliary-membrane-anchored Ptch1 inhibits Smo by inhibiting its accumulation within cilia. However, when Shh is bound to Ptch1, Shh induces changes in the localization of Ptch1 and Smo, and Ptch1 leaves the cilia, inducing Smo and signaling [78]. The abundance of Ptch1 at the ciliary membrane and the cholesterol pumping activity of Ptch1 are both responsible for depleting accessible cholesterol from the ciliary membrane [32]. The direction of cholesterol transport by Ptch1 could be inward from the ciliary membrane to a sterol transport protein or membrane compartment in the cytosol, or outward from the ciliary membrane to an extracellular acceptor [32].
Another observation suggested that the primary cilia of mammalian cultured cells constituted a higher concentration of total sphingomyelin than the plasma membrane. A higher abundance of sphingomyelin in the ciliary membrane reduces the accessible cholesterol and modulates Hh signaling. Interestingly, plasma membranes were found to maintain a constant level of sphingomyelin/cholesterol complexes over wide-ranging cholesterol concentrations using biochemical analysis with ostreolysin A (OlyA). OlyA binds only membranes containing both sphingomyelin and cholesterol [189]. In addition, two models for ligand-mediated changes in cholesterol accessibility of the ciliary membrane were proposed. One observation suggested that in response to Shh, the level of accessible cholesterol in the ciliary membrane increases, raising the total cholesterol in cilia; the second model suggested Shh switches sphingomyelin-sequestered cholesterol to accessible cholesterol, regulating Hh signal transmission.
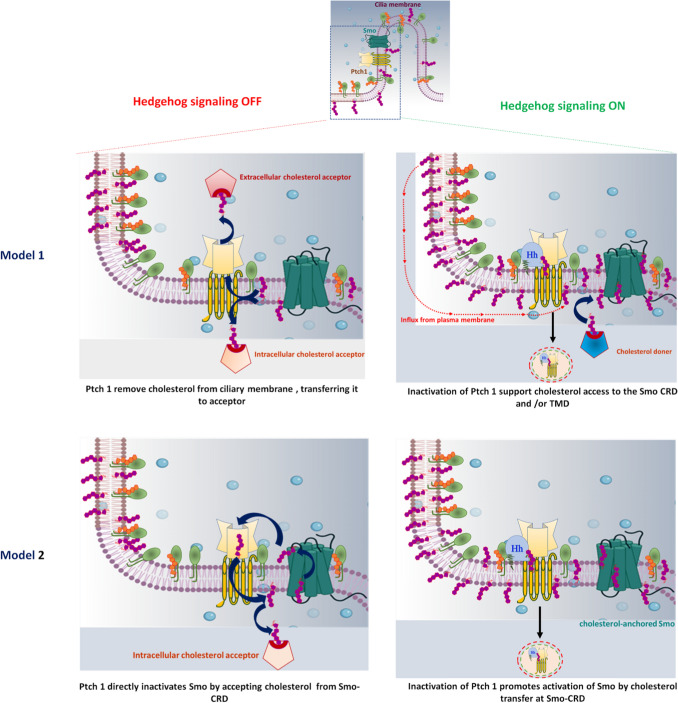
To maintain an equilibrium of accessible cholesterol between the ciliary and plasma membranes, an active transport system is required (Fig. 6). Several models are considered for Ptch1-mediated Smo regulation in the ciliary membrane. According to the 'pump-leak' model, the accessible cholesterol level drops lower than the threshold level expected to activate Smo due to the pumping action of Ptch1 at the ciliary membrane. Ptch1 utilizes its energy-driven transporter function to remove cholesterol above the threshold level from the ciliary membrane to either an extracellular or intracellular acceptor. Further, the continuous leaking of cholesterol is opposed by the Shh-driven suppression of Ptch1, allowing an influx of cholesterol back into the ciliary membrane, the subcellular location for Smo signaling [32, 43]. The recent studies on Niemann-Pick type C (NPC1) proteins examined how cholesterol interacts with the N-terminal domain of NPC1 [190] via a ‘direct inactivation mechanism.’ In this model, the extracellular domain of Ptch1 accepts cholesterol directly from the Smo-CRD and transports it to the membrane, which directly inactivates Smo [43]. Moreover, recent reports anticipated that Ptch1 increases ciliary sphingomyelin or promotes sphingomyelin–cholesterol interaction and enhances the expulsion of accessible cholesterol in ciliary exovesicles [32, 43].
Fig. 6.
Variations in accessible cholesterol affect the function of Ptch1 and Smo at the ciliary membrane: A cilium is shown at the top, with a rectangle representing a ciliary pocket magnified below. a In model 1: Ptch1 utilizes its energy-driven transporter function to remove cholesterol from the ciliary membrane, transporting it to an extracellular or intracellular acceptor. By this function, the accessible cholesterol level falls below the threshold needed for Smo activation, and the level of sphingomyelin increases at the ciliary membrane (left panel). In the presence of Hh ligand, inactivated Ptch1 allows an influx of cholesterol into the ciliary membrane thus accessible ciliary cholesterol increases and activates Smo (right panel). b In model 2: The extracellular domain of Ptch1 directly accepts cholesterol from Smo-CRD, transferring it to the membrane, and directly inactivates Smo (left panel). In the presence of Hh ligand, inactivation of Ptch1 increases accessible ciliary cholesterol and promotes cholesterol interaction with Smo-CRD, resulting in activation of Smo (right panel)
Next, trans-bilayer distribution of cholesterol in Ptch-Smo regulation is also an important aspect and raises a concern about how cholesterol gets access to Smo between the two leaflets. Zheng et al. clarified that overexpression of Ptch1 throughout the plasma membrane significantly reduces the activity of cholesterol in the inner leaflet of the plasma membrane. By contrast, Ptch1 inactivation leads to increased abundance of inner leaflet cholesterol [187] and allows access to Smo’s seven-transmembrane site for binding cholesterol [191, 192]. Though the CRD of Smo receives cholesterol from the outer leaflet, Shh-mediated inactivation of Ptch1 increases outer leaflet accessible cholesterol at the ciliary membrane [32, 43]. However, studies indicate the inactivation of Ptch1 causes an overall rise in accessible cholesterol levels in both inner and outer leaflets of the membrane. The possible explanation is the ‘flip-flop mechanism’, i.e., rapid trans-bilayer movement of cholesterol between the two leaflets of plasma membrane results in alterations in cholesterol activity in one leaflet, which is likely to be reflected in another leaflet of the membrane [43].
The connection between cholesterol, lipid rafts, and Hh signaling
As described earlier, Hh signaling contains a distinguishing signal reception system, including two membrane proteins, the Ptch receptor and transducer Smo. Both play a crucial role in maintaining Hh signal transmission, both short and long-range, and require accessible cholesterol. The functional relevance of lipid rafts in Hh signaling and the underlying mechanism by which Smo regulation occurs on the plasma membrane is also an exciting part of Hh signaling. Previous studies have revealed that the cholesterol/sphingomyelin-rich domain, known as lipid rafts in lipid bilayers, is involved in regulating the Hh signaling cascade [31]. Lipid rafts (membrane rafts) are a specific membrane structure that makes a liquid-ordered phase and acts as an oligomerization platform and a hub for signal transduction proteins through protein–protein or protein–lipid interactions [29, 193]. Ptch and Smo are localized in caveolin-enriched microdomains termed caveolae [30]. Caveolae are invaginated microdomains of the plasma membrane that are rich in cholesterol, sphingomyelin, and intrinsic membrane proteins called caveolins such as caveolin-1 and -2. Caveolae are associated with endocytosis, cholesterol trafficking, sequestration of various lipid-modified signaling proteins, and a signaling center for multiple pathways [28, 194, 195]. Indeed, caveolin-1 directly binds to cholesterol or cholesterol-modified proteins involved in trafficking. The idea is that Ptch and Smo form a complex early on, probably in the Golgi, and are trafficked intact to lipid-enriched microdomains [30]. Notably, Ptch1 interacts with caveolin-1 in cholesterol-rich microdomain; however, Smo interacts only with Ptch1. Caveolin-1 associates with numerous proteins through an intracellular region termed the caveolin-1 scaffolding domain, and proteins that bind caveolin-1, including Ptch, contain a particular caveolin-binding sequence motif. The local cholesterol concentration impacts the trafficking of both Ptch1 and caveolin-1 in the caveolin-enriched microdomain because depletion of plasmalemmal cholesterol reduces the amount of both proteins in the lipid rafts [30]. Sequestration of Ptch in these lipid rafts might be a selective method for the cell to discriminate other signaling components and encourage Ptch–Smo interaction. Shi et al. [31] revealed that the N-terminal extracellular domain and transmembrane domain of Smo make oligomers/higher-order clusters in the plasma membrane lipid rafts in response to the Hh signal and promote Smo activity. Thus, the oligomerization of the Smo C-terminal tail is critical for high-level Hh signal transduction [31]. Overall, these observations indicate that lipid rafts play a vital function in Hh signal transmission either in the secretion of Hh morphogen or interaction of Hh with Ptch, leading to accumulation of Smo.
Developmental impact of the cholesterol-modified Hh signal
As described, Hh is involved in developmental patterning in various systems, including the neural tube, limbs, and somites [4]. The vertebrate Hh signal originating from the ventral midline of the neural tube plays a pivotal function in the dorsoventral patterning of the brain. In addition, the signal appears to represent early patterning activity along the proximo-distal axis of the developing eyes [196].
Auto-processing in the biogenesis of Hh is an essential aspect of Hh patterning functions and developmental consequences [61, 65]. The autoproteolytic activity of the carboxy-terminal domain generates an amino-terminal Hh product, which accounts for all signaling activity. The N-Hh product also covalently links cholesterol, increasing the hydrophobic character of the signaling and, in turn, regulating its distribution [158, 197]. The truncated unprocessed N-Hh led to embryonic mis-patterning, altered spatial and subcellular distribution, illuminating the importance of auto-processing in Hh signaling regulation [158]. Furthermore, abolishment of the cholesterol modification of Smo resulted in severe developmental defects such as embryonic lethality with severe cardiac defects, highlighting the importance of cholesterol in Hh signaling [198]. Another study demonstrated that expression of unmodified Hh (a form that lacks cholesterol) causes a gain-of-function phenotype with aberrant signaling in Drosophila [44]. Hence, cholesterol appears to be implicated in limiting the diffusion of modified-Hh (cholesterol anchor). Intriguingly, on cholesterol modification seems to have the opposite effect in the mammalian limb. Cholesterol-modified Shh showed long-range activity directly over a few hundred microns (up to 30 cell diameters). Although unmodified Shh retained similar polarizing/biological activity, signaling was posteriorly restricted [178]. These data suggest that unmodified Shh, like modified Shh, generates a high level of posterior signaling activity that specifies the most posterior digits (digits 5 and 4). However, unlike modified Shh, unmodified Shh signaling did not extend the anterior regions of the limb field (digits 3 and 2) [57, 178]. Overall, cholesterol modification seems to be critical for long-range patterning [178]. Jeong and McMohan [25] resolved this inconsistency between fly and mouse data by providing possible explanations of differences between the tissues under investigation. In the fly (ectoderm and wing disc), Hh signaling occurs within a continuous sheet of the epithelium; in contrast, in mice (limb bud), the signaling ensues across the mesenchyme. The gradient area established in mice is more extensive than the fly. Also, distinct methods were applied to express modified and unmodified Hh in both experimental models [25]. Taken together, cholesterol modification on Hh ligands plays a pivotal role in regulating the Hh signal range and calibrating the morphogen gradient. As such, cholesterol participates in coordinating various organ development patterning, including cell fate specification and tissue homeostasis. Nevertheless, cholesterol is not required for the signaling activity of Hh protein.
Cholesterol-free and cholesterol-modified Hh secretions and actions
The Hh gene family encodes secreted ligands (morphogens), which are covalently modified with palmitate and cholesterol moieties that are conserved from flies to humans [199]. These modifications are critical for correct patterning and growth during development and tissue homeostasis. Mechanistically, the palmitoyl adduct is essential for Hh secretion, and cholesterol affects the spread of Hh and regulates the Hh signaling range and its action within tissues [158, 200]. However, the exact function of the cholesterol modification is controversial because reports in Drosophila have suggested that it either increases or decreases the Hh range [200]. One study proposed that cholesterol is required for long-range Hh transport and two other reports suggested that the cholesterol moiety limits the range of the Hh protein [201–203]. Moreover, several lines of evidence have indicated that a cholesterol-free form of Hh is also produced and secreted in both flies and cultured human cells. Diverse research groups are working hand in hand to investigate the modified-Hh mechanism in long- and short-range action in the drosophila wing disc [204] (Fig. 7). The Eaton research group deciphered that morphogen function requires lipophorin, which bears lipid-linked morphogen on its surface and serves as a vehicle. In brief, Hh boards lipophorin, and Hh-Lpp (lipoprotein–lipophorein complex) move across tissues that allow long-range signaling activity. Intriguingly, lipoprotein particles mediate Hh intracellular transportation and are also endocytosed together with Hh [68, 205]. Additionally, a study revealed that megalin, an endocytic receptor, is a new regulatory component of Hh signaling and exhibits a crucial role in interaction with the lipoprotein-associated form of Hh and internalization of Hh [206, 207]. Remarkably, the complement of proteins and lipid present on lipoprotein particles control the Hh activity, as lipid in lipophorin suppresses Hh signaling in the absence of Hh ligand [205]. Palm et al. reported that Hh protein could be secreted in both lipoparticle-associated and non-associated forms with complementary and synergic functions [206]. The study revealed that secretion of lipoprotein associated Hh requires either a palmitate or cholesterol moiety; these two moieties are sufficient to promote Hh association with lipoprotein. The proposed model of anchored-Hh secretion and actions suggested that processed/mature Hh (modified with lipid moiety) is secreted in an Lpp-dependent manner. The Hh–Lpp complex stabilizes inactive full-length Ci (Cubitus interruptus, a transcription factor of Hh signaling), which alone is insufficient to activate Hh target genes. Surprisingly, a putative unknown esterase activity also generates a cholesterol-free pool of Hh that is secreted in an Lpp-free manner. The Lpp free Hh reduces the amount of cleaved Ci and promotes the switch from inactive to active full-length Ci. Thus, Lpp-free Hh acts synergistically with Lpp-bound Hh to trigger Hh target gene expression [204, 206]. However, further investigation is necessary to endorse this model and analyze how these two forms of Hh bind to Ptch and stimulate precise and differential Hh responses.
Fig. 7.
Proposed model of cholesterol-free and cholesterol-bound Hh secretion in Drosophila: Cholesterol-bound and free forms of Hh are both secreted and have distinct and synergic functions. 1. Hh-producing cells synthesize lipid-anchored Hh protein. 2. Processed Hh protein can be secreted as an Lpp-bound form. An unknown esterase mechanism removes cholesterol and generates a cholesterol-free pool of Hh. 3. Cholesterol-free Hh can be secreted without Lpp. 4. Lpp-associated Hh stabilizes inactive full-length Ci-155. 5. Lpp-free Hh reduces the amount of cleaved Ci-75. 6. Lpp-free Hh also promotes the conversion of inactive Ci-155 into active full-length Ci-155. 7. Lpp-free Hh functions in synergy with Lpp-associated Hh to ultimately activate Hh target gene expression
Cation gradients influence cholesterol transport in Hh signaling
The mechanism of the Hh response is distinctive among the other known signaling pathways due to its dual function in ligand reception and intracellular signal transmission between secreted cells and target cells. Ptch acts as an ion-driven transporter and has sequence homology with prokaryotic RND receptors [87, 187]. The energy source behind Ptch1’s function in Ptch-Smo communication is still in question. According to one proposed model, Ptch1 is an ion-driven transporter that regulates the availability of endogenous lipid-anchored Smo in the ciliary membrane, where substantial Hh signaling regulation takes place. Another leading model, based on the resemblance between Ptch1 and RND, suggested that a proton-motive force drives the export of substances. However, direct evidence that Ptch1 has intrinsic transporter activity or that a proton gradient exists in the metazoan plasma membrane remain to be explored. Conventional approaches for measuring Hh pathway activity use downstream transcriptional effector readouts or alterations in the ciliary Hh pathway components, which are both long-term and indirect. Recent findings demonstrated that the Ptch1 effects on Smo are unexpectedly dynamic and can occur without cilia-specific proteins or metabolites. It was suggested that Ptch1 utilizes a transmembrane cation gradient to block activation of Smo by cholesterol. Moreover, another RND protein Disp1 was proposed as a cation gradient transporter that uses the transmembrane Na+ gradient to catalyze cholesterol-modified Shh secretion [46].
Conclusion and future insights
Due to rapid progress in unraveling the Hh pathway over the past years, this review aims to provide comprehensive details about Hh signal transduction and its association with cholesterol. We covered the current structural, biochemical, and molecular studies that have interrogated how cholesterol regulates the Hh signal, its downstream targets, auto-regulation, crosstalk with other signaling elements, and feedback control. Past decades demonstrated the functional involvement of cholesterol in Hh ligand biogenesis and receptor and coreceptor activation. Recent studies have focused on accessible cholesterol and Hh traffickings, such as the biochemical mechanism of efficient secretion of cholesterol-anchored Hh and the co-receptors that shuttle the Hh morphogen from producing to responding cells. Our review summarizes critical lipid-dependent signaling and transport pathways, distinct cation gradients that mobilize cholesterol for Hh pathway transmission, and spatial, temporal, and cell-contextual Hh/Gli cascade regulation. It also describes the concept of ‘cholesterol modification of Hh family proteins’ where cholesterol serves as the master regulator of the Hh pathway. Several researchers tried to investigate the mode of action of Hh transportation using different co-receptors and regulatory proteins, such as Disp, Scube, and Gas1. This review, for the first time, covers all possible aspects of the cholesterol mediated Hh signaling. It offers a comprehensive overview from basic concepts to advanced understanding of Hh signaling and novel mechanistic insights. We also summarized the involvement of Hh cascade regulatory proteins in Hh signaling and their association with cholesterol (Table 1).
Table 1.
Functional involvement of Hh signaling regulatory proteins in Hh pathway and their association with cholesterol
| Hh signaling proteins | Functional contribution | |
|---|---|---|
| Involvement in Hh signaling | Association with cholesterol | |
| Hedgehog (Hh) |
Ligands of Hh signaling serves as a morphogen, mitogen, survival factor, and guidance [11, 47, 53, 54] Hh family comprises three Hh-related genes sonic hedgehog (Shh), desert hedgehog (Dhh), and Indian hedgehog (Ihh) [86] |
Various roles of cholesterol in Hh protein biogenesis and signal transmission [155] During Hh auto-processing, the N-terminal half of Hh is covalently anchored with cholesterol [61, 160] |
| Cholesterol-modification of Hh is required for Hh activity and native signal transduction [62, 156] | ||
| Smoothened (Smo) |
Smo encodes a serpentine protein, a member of the G-protein coupled receptor (GPCR) superfamily that is implicated as a core receptor of Hh signaling [88, 90] Smo activation involves Smo translocation from intracellular vesicle to the cell surface and its post-translational modification [91, 96] Hh-mediated Smo activity is regulated by inducing a conformational switch [94] The long C-terminal domain (C-tail) of Smo is indispensable for Hh-dependent signal transduction [95] |
Cholesterol serves as an endogenous ligand for Smo [40, 154] Cholesterol binding is required for allosteric Smo activation [40, 154] Cholesterol modification of Smo is needed to reinforce and sustain the Hh signaling [41] |
| Smo activity is regulated by cholesterol concentration or accessibility in the ciliary and plasma membrane [32, 40, 192] | ||
| Local concentration of cholesterol affects the trafficking of Smo [32] | ||
| Patched 1 (Ptch1) | A twelve-span transmembrane receptor of Hh signaling [73] |
Cation-powered Ptch1transporter uses distinct cation gradients power for cholesterol transport [46] Ptch1 is implicated in cholesterol efflux [187] Ptch1 modulates the intracellular cholesterol concentration [187] |
|
Ptch1 involves in the suppression of the Smo enrichment in the plasma membrane [87] The sterol-sensing domain of Ptch1 appears to control Smo activity through vesicular trafficking [45] | ||
|
Extracellular domains of Ptch1 bind to Hh ligand [127] Ptch1 acts as a dependence receptor [135]: Intracellular C-terminal domain modulates Ptch1 activity and has pro-apoptotic activity [134, 135] Acts as tumor-suppressor gene and restricts cell cycle progression [136, 138] |
Local concentration of cholesterol affects the trafficking of Ptch1 Ptch1 regulates the Smo activation by transporting cholesterol from cells [24] |
|
| Ptch1 acts as a lipoprotein receptor and modulates intracellular lipid homeostasis in a Hh-independent manner [185] | ||
| Dispatched (Disp) |
Twelve-transmembrane membrane receptor[44] In signal-producing cells, Disp controls the release of fully functional Hh protein [37, 177] |
Disp is a cation-powered transporter that requires cholesterol for Hh signal transfer [46, 177] Hh binds physically to Disp through cholesterol moiety [164] Disp facilitates efficient secretion of the cholesterol modified Hh ligand [44, 164] Disp regulates long-range cholesterol modified Hh signal transmission [163] |
|
Cell-adhesion-molecule related/down-regulated by oncogene (CDON) Bioregional Cdon-binding protein (BOC) |
CDON and BOC co-receptors recruit Scube-Hh into the responding cells as a ternary complex with CDON/BOC [38] | |
| Growth-arrest-specific (Gas1) |
Gas1 co-receptor catalyzes Hh transfer to Ptch receptor [170] Gas 1 acts as a positive regulator of the Hh signaling [172] Gas1 extends the range of Hh action by assisting its signaling [171] |
Gas1 interacts with Hh in a lipid-dependent manner and dissociates its interaction with Scube [39] |
| Suppressor of fused (Sufu) | Sufu, a scaffold protein, acts as a negative regulator of the Hh signaling acting on Gli [120, 121] | |
| Signal peptide-CUB-EGF domain-containing protein (Scube) |
Scube is secreted glycoprotein, and Scube family of proteins are Scube1, Scube2, Scube3, and its activity is required for Hh signal transduction [70, 162] Scube activity is essential for efficient Hh secretion and long-range Hh signal transmission [71, 162, 164] Scube acts as a chaperone and contributes to safeguarding Hh by facilitating a soluble multimeric Hh complex [70, 164] |
Scube facilitates the release of dually lipid-modified Hh signal in soluble form [70] A heterologous protein complex, i.e., cholesterol anchored Hh-Disp, is transferred to the Scube from Disp in extracellular space for efficient cholesterol-modified Hh secretion [164] |
| Glioma-associated oncogene (Gli) |
Gli is a Krüppel-like transcription factor comprising zinc-finger DNA-binding domains with dual activity [100, 101] Gli family comprises three genes Gli1, Gli2, and Gli3 appear to have similar DNA binding specificities [102, 103] The post-translational proteolytic processing of Gli proteins generates the activator and repressor form of Gli, such as GliA and GliR [105, 107] In response to the Hh signal, Gli-family protein directly regulates Gli1 activation and stimulates numerous Hh target gene [109] |
|
| Protein Kinases, such as PKA, DYRK, ULK3, S6K1, AMPK, MEKK1, and Hck |
Regulates phosphorylation of Gli-family members and encourages activation of the Hh pathway [116, 146–150] PKA is also involved in Smo phosphorylation [98] |
|
| CK1 and Gsk3β |
Represented dual role thus it can promote or activate Hh pathway in a context-dependent manner [145] CK1 induces the Smo phosphorylation that is the critical step of Hh signal transduction [98] Gli phosphorylation by CK1 positively regulates Hh pathway activity [145] Gsk3β induces the phosphorylation of Sufu and Gli [17, 19, 125] |
|
|
E3 ligase complexes, such as Cul1/β-TrCP and E3 ubiquitin-protein ligase Itchy homolog (Itch) |
Catalyzes ubiquitination to Gli protein and influences Gli protein activity [150, 151] | |
| Hedgehog-interacting protein (Hip) |
Hip is a membrane glycoprotein that interacts with Hh ligands and modulates the Hh downstream signaling [77] Hip sequestrates the Hh ligand, hence acts as a negative regulator of the Hh signaling pathway [48, 77] |
|
Understanding the cholesterol modification of Hh signaling suggested the therapeutic potential for Hh pathway-related disorders by targeting several Hh signaling downstream effector molecules. Remarkably, the interplay of Hh family proteins (Ptch1, Smo, Disp, and Scube) and cholesterol in Hh signaling is much better understood, but multiple paradoxes remain to be explored. Notably, the Hh family protein involvement with other signaling pathways, the effective mechanism(s) for short- and long-range Hh transmission, and the role of the ciliary, plasma membrane, Golgi, and ER in Hh trafficking have elicited numerous fascinating questions. In addition, questions remain unanswered on about how Ptch1 functions as a receptor and transporter, how cholesterol-binding mediates Smo activation, and how the cholesterol metabolic pathway(s) contributes to the Hh pathway. Intriguingly, the advancement of science and technology opens up new research avenues to resolve the functional complexity of the Hh pathway.
Acknowledgements
The authors thank Dr. Jessica Mercer for editing this manuscript.
Author contributions
JBK conceived the idea, researched the data, wrote the main body of the manuscript, and prepared figures. SR and SKB critically reviewed and finalized the manuscript. All authors have approved the final version of the manuscript.
Funding
The authors of this review article are supported, in parts, by the grants from the National Institutes of Health and National Cancer Institute, USA, Grants (RO1 CA247763, R21 CA238953, PO1 CA217798) and Department of Defence (DoD), USA, Grants Prostate Cancer Research Program DoD W81XWH-21-10340 (JBK), DoD W81XWH-18-1-0308 (SKB).
Availability of data and materials
The data sets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Declarations
Conflict of interest
SKB is one of the co-founders of Sanguine Diagnostics and Therapeutics, Inc. The other authors declare no competing interests.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Surinder K. Batra, Email: sbatra@unmc.edu
Satyanarayana Rachagani, Email: srachagani@unmc.edu.
References
- 1.Ingham PW, McMahon AP. Hedgehog signaling in animal development: paradigms and principles. Genes Dev. 2001;15(23):3059–3087. doi: 10.1101/gad.938601. [DOI] [PubMed] [Google Scholar]
- 2.Jia J, Jiang J. Decoding the Hedgehog signal in animal development. Cell Mol Life Sci. 2006;63(11):1249–1265. doi: 10.1007/s00018-005-5519-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pyczek J, et al. Hedgehog signaling activation induces stem cell proliferation and hormone release in the adult pituitary gland. Sci Rep. 2016 doi: 10.1038/srep24928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Petrova R, Joyner AL. Roles for Hedgehog signaling in adult organ homeostasis and repair. Development (Cambridge) 2014;141(18):3445–3457. doi: 10.1242/dev.083691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belloni E, et al. Identification of sonic hedgehog as a candidate gene responsible for holoprosencephaly. Nat Genet. 1996;14(3):353–356. doi: 10.1038/ng1196-353. [DOI] [PubMed] [Google Scholar]
- 6.Nieuwenhuis E, Hui CC. Hedgehog signaling and congenital malformations. Clin Genet. 2005;67(3):193–208. doi: 10.1111/j.1399-0004.2004.00360.x. [DOI] [PubMed] [Google Scholar]
- 7.Ma Y, Zhang P, Wang F, Yang J, Yang Z, Qin H. The relationship between early embryo development and tumourigenesis. J Cell Mol Med. 2010;14(12):2697–2701. doi: 10.1111/j.1582-4934.2010.01191.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peacock CD, et al. Hedgehog signaling maintains a tumor stem cell compartment in multiple myeloma. Proc Natl Acad Sci USA. 2007;104(10):4048–4053. doi: 10.1073/pnas.0611682104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fan YH, et al. Aberrant hedgehog signaling is responsible for the highly invasive behavior of a subpopulation of hepatoma cells. Oncogene. 2016;35(1):116–124. doi: 10.1038/onc.2015.67. [DOI] [PubMed] [Google Scholar]
- 10.Doheny D, Manore SG, Wong GL, Lo HW. Hedgehog signaling and truncated GLI1 in cancer. Cells. 2020 doi: 10.3390/cells9092114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Varjosalo M, Taipale J. Hedgehog: functions and mechanisms. Genes Dev. 2008;22(18):2454–2472. doi: 10.1101/gad.1693608. [DOI] [PubMed] [Google Scholar]
- 12.Yao E, Chuang PT. Hedgehog signaling: from basic research to clinical applications. J Formosan Med Assoc. 2015;114(7):569–576. doi: 10.1016/j.jfma.2015.01.005. [DOI] [PubMed] [Google Scholar]
- 13.Briscoe J, Thérond PP. The mechanisms of Hedgehog signalling and its roles in development and disease. Nat Rev Mol Cell Biol. 2013;14(7):418–431. doi: 10.1038/nrm3598. [DOI] [PubMed] [Google Scholar]
- 14.Harris LG, Samant RS, Shevde LA. Hedgehog signaling: networking to nurture a promalignant tumor microenvironment. Mol Cancer Res. 2011;9(9):1165–1174. doi: 10.1158/1541-7786.MCR-11-0175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaushal JB, Popli P, Sankhwar P, Shukla V, Dwivedi A. Sonic hedgehog protects endometrial hyperplasial cells against oxidative stress via suppressing mitochondrial fission protein dynamin-like GTPase (Drp1) Free Radic Biol Med. 2018;129:582–599. doi: 10.1016/J.FREERADBIOMED.2018.10.427. [DOI] [PubMed] [Google Scholar]
- 16.Robbins DJ, Fei DL, Riobo NA. The hedgehog signal transduction network. Sci Signal. 2012 doi: 10.1126/scisignal.2002906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carballo GB, Honorato JR, De Lopes GPF, Spohr TCLDSE. A highlight on sonic hedgehog pathway. Cell Commun Signal. 2018 doi: 10.1186/s12964-018-0220-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Milenkovic L, Scott MP, Rohatgi R. Lateral transport of smoothened from the plasma membrane to the membrane of the cilium. J Cell Biol. 2009;187(3):365–374. doi: 10.1083/JCB.200907126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaushal JB, et al. The regulation of Hh/Gli1 signaling cascade involves Gsk3β- mediated mechanism in estrogen-derived endometrial hyperplasia. Sci Rep. 2017;7(1):1–16. doi: 10.1038/s41598-017-06370-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaushal JB, et al. Repurposing niclosamide for targeting pancreatic cancer by inhibiting Hh/Gli non-canonical axis of Gsk3β. Cancers. 2021;13(13):3105. doi: 10.3390/CANCERS13133105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jenkins D. Hedgehog signalling: emerging evidence for non-canonical pathways. Cell Signal. 2009;21(7):1023–1034. doi: 10.1016/J.CELLSIG.2009.01.033. [DOI] [PubMed] [Google Scholar]
- 22.Brennan D, Chen X, Cheng L, Mahoney M, Riobo NA. Noncanonical Hedgehog signaling. Vitam Horm. 2012;88:55–72. doi: 10.1016/B978-0-12-394622-5.00003-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ingham PW. Hedgehog signalling: How cholesterol modulates the signal. Curr Biol. 2000;10(5):R180–R183. doi: 10.1016/S0960-9822(00)00346-8. [DOI] [PubMed] [Google Scholar]
- 24.Ao H, Bao-Liang S. The interplay of patched, Smoothened and cholesterol in Hedgehog signaling. Curr Opin Cell Biol. 2019;61:31–38. doi: 10.1016/J.CEB.2019.06.008. [DOI] [PubMed] [Google Scholar]
- 25.Jeong J, McMahon AP. Cholesterol modification of Hedgehog family proteins. J Clin Investig. 2002;110(5):591–596. doi: 10.1172/jci16506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fantini J, Barrantes FJ. How cholesterol interacts with membrane proteins: an exploration of cholesterol-binding sites including CRAC, CARC, and tilted domains. Front Physiol. 2013;4:31. doi: 10.3389/FPHYS.2013.00031/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kai S, Robert E. Cholesterol, lipid rafts, and disease. J Clin Investig. 2002;110(5):597–603. doi: 10.1172/JCI16390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parton RG. Caveolae and caveolins. Curr Opin Cell Biol. 1996;8(4):542–548. doi: 10.1016/S0955-0674(96)80033-0. [DOI] [PubMed] [Google Scholar]
- 29.Simons K, Toomre D. Lipid rafts and signal transduction. Nat Rev Mol Cell Biol. 2000;1(1):31–39. doi: 10.1038/35036052. [DOI] [PubMed] [Google Scholar]
- 30.Karpen HE, Bukowski JT, Hughes T, Gratton JP, Sessa WC, Gailani MR. The sonic Hedgehog receptor patched associates with caveolin-1 in cholesterol-rich microdomains of the plasma membrane. J Biol Chem. 2001;276(22):19503–19511. doi: 10.1074/jbc.M010832200. [DOI] [PubMed] [Google Scholar]
- 31.Shi D, et al. Smoothened oligomerization/higher order clustering in lipid rafts is essential for high hedgehog activity transduction. J Biol Chem. 2013;288(18):12605–12614. doi: 10.1074/jbc.M112.399477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kinnebrew M, et al. Cholesterol accessibility at the ciliary membrane controls hedgehog signaling. Elife. 2019 doi: 10.7554/eLife.50051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cooper MK, et al. A defective response to Hedgehog signaling in disorders of cholesterol biosynthesis. Nat Genet. 2003;33(4):508–513. doi: 10.1038/ng1134. [DOI] [PubMed] [Google Scholar]
- 34.Petrov K, Wierbowski BM, Salic A. Sending and receiving hedgehog signals. Annu Rev Cell Dev Biol. 2017;33:145–168. doi: 10.1146/annurev-cellbio-100616-060847. [DOI] [PubMed] [Google Scholar]
- 35.Bürglin TR. The Hedgehog protein family. Genome Biol. 2008;9(11):241. doi: 10.1186/gb-2008-9-11-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lum L, Beachy PA. The hedgehog response network: sensors, switches, and routers. Science. 2004;304(5678):1755–1759. doi: 10.1126/science.1098020. [DOI] [PubMed] [Google Scholar]
- 37.Qi X, Li X. Mechanistic insights into the generation and transduction of Hedgehog signaling. Trends Biochem Sci. 2020;45(5):397–410. doi: 10.1016/j.tibs.2020.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Allen BL, et al. Overlapping roles and collective requirement for the coreceptors GAS1, CDO, and BOC in SHH pathway function. Dev Cell. 2011;20(6):775–787. doi: 10.1016/j.devcel.2011.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wierbowski BM, Petrov K, Aravena L, Gu G, Xu Y, Salic A. Hedgehog pathway activation requires coreceptor-catalyzed, lipid-dependent relay of the sonic Hedgehog ligand. Dev Cell. 2020;55(4):450–467.e8. doi: 10.1016/j.devcel.2020.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang P, et al. Cellular cholesterol directly activates smoothened in Hedgehog signaling. Cell. 2016;166(5):1176–1187.e14. doi: 10.1016/j.cell.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xiao X, et al. Cholesterol modification of smoothened is required for Hedgehog signaling. Mol Cell. 2017;66(1):154–162.e10. doi: 10.1016/j.molcel.2017.02.015. [DOI] [PubMed] [Google Scholar]
- 42.Bidet M, et al. The hedgehog receptor patched is involved in cholesterol transport. PLoS ONE. 2011;6(9):e23834. doi: 10.1371/journal.pone.0023834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Radhakrishnan A, Rohatgi R, Siebold C. Cholesterol access in cellular membranes controls Hedgehog signaling. Nat Chem Biol. 2020;16(12):1303–1313. doi: 10.1038/s41589-020-00678-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Burke R, et al. Dispatched, a novel sterol-sensing domain protein dedicated to the release of cholesterol-modified Hedgehog from signaling cells. Cell. 1999;99(7):803–815. doi: 10.1016/S0092-8674(00)81677-3. [DOI] [PubMed] [Google Scholar]
- 45.Martín V, Carrillo G, Torroja C, Guerrero I. The sterol-sensing domain of patched protein seems to control smoothened activity through patched vesicular trafficking. Curr Biol. 2001;11(8):601–607. doi: 10.1016/S0960-9822(01)00178-6. [DOI] [PubMed] [Google Scholar]
- 46.Kostadin P, Wierbowski BM, Jingjing L, Adrian S. Distinct cation gradients power cholesterol transport at different key points in the hedgehog signaling pathway. Dev Cell. 2020;55(3):314–327. doi: 10.1016/J.DEVCEL.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hooper JE, Scott MP. Communicating with hedgehogs. Nat Rev Mol Cell Biol. 2005;6(4):306–317. doi: 10.1038/nrm1622. [DOI] [PubMed] [Google Scholar]
- 48.Chuang PT, Kawcak T, McMahon AP. Feedback control of mammalian Hedgehog signaling by the Hedgehog-binding protein, Hip1, modulates Fgf signaling during branching morphogenesis of the lung. Genes Dev. 2003;17(3):342–347. doi: 10.1101/gad.1026303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goodrich LV, Johnson RL, Milenkovic L, McMahon JA, Scott MP. Conservation of the hedgehog/patched signaling pathway from flies to mice: Induction of a mouse patched gene by Hedgehog. Genes Dev. 1996;10(3):301–312. doi: 10.1101/gad.10.3.301. [DOI] [PubMed] [Google Scholar]
- 50.Jia Y, Wang Y, Xie J. The Hedgehog pathway: role in cell differentiation, polarity and proliferation. Arch Toxicol. 2015;89(2):179–191. doi: 10.1007/s00204-014-1433-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nüsslein-volhard C, Wieschaus E. Mutations affecting segment number and polarity in drosophila. Nature. 1980;287(5785):795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- 52.Pereira J, et al. Evolutionary genomics and adaptive evolution of the Hedgehog gene family (Shh, Ihh and Dhh) in vertebrates. PLoS ONE. 2014;9(12):e74132. doi: 10.1371/journal.pone.0074132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Torroja C, Gorfinkiel N, Guerrero I. Mechanisms of Hedgehog gradient formation and interpretation. J Neurobiol. 2005;64(4):334–356. doi: 10.1002/neu.20168. [DOI] [PubMed] [Google Scholar]
- 54.Cai C, Thorne J, Grabel L. Hedgehog serves as a mitogen and survival factor during embryonic stem cell neurogenesis. Stem Cells. 2008;26(5):1097–1108. doi: 10.1634/stemcells.2007-0684. [DOI] [PubMed] [Google Scholar]
- 55.Falkenstein KN, Vokes SA. Transcriptional regulation of graded hedgehog signaling. Semin Cell Dev Biol. 2014;33:73–80. doi: 10.1016/j.semcdb.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dessaud E, McMahon AP, Briscoe J. Pattern formation in the vertebrate neural tube: a sonic Hedgehog morphogen-regulated transcriptional network. Development. 2008;135(15):2489–2503. doi: 10.1242/dev.009324. [DOI] [PubMed] [Google Scholar]
- 57.Zhu J, Nakamura E, Nguyen MT, Bao X, Akiyama H, Mackem S. Uncoupling Sonic hedgehog control of pattern and expansion of the developing limb bud. Dev Cell. 2008;14(4):624–632. doi: 10.1016/J.DEVCEL.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wijgerde M, Ooms M, Hoogerbrugge JW, Grootegoed JA. Hedgehog signaling in mouse ovary: Indian Hedgehog and desert Hedgehog from granulosa cells induce target gene expression in developing theca cells. Endocrinology. 2005;146(8):3558–3566. doi: 10.1210/en.2005-0311. [DOI] [PubMed] [Google Scholar]
- 59.Hung-Chang Yao H, Whoriskey W, Capel B. Desert Hedgehog/patched 1 signaling specifies fetal Leydig cell fate in testis organogenesis. Genes Dev. 2002;16(11):1433–1440. doi: 10.1101/gad.981202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bitgood MJ, Shen L, McMahon AP. Sertoli cell signaling by desert Hedgehog regulates the male germline. Curr Biol. 1996;6(3):298–304. doi: 10.1016/S0960-9822(02)00480-3. [DOI] [PubMed] [Google Scholar]
- 61.Lee JJ, Ekker SC, Von Kessler DP, Porter JA, Sun BI, Beachy PA. Autoproteolysis in Hedgehog protein biogenesis. Science (80-) 1994;266(5190):1528–1537. doi: 10.1126/science.7985023. [DOI] [PubMed] [Google Scholar]
- 62.Porter JA, Young KE, Beachy PA. Cholesterol modification of Hedgehog signaling proteins in animal development. Science (80-) 1996;274(5285):255–259. doi: 10.1126/science.274.5285.255. [DOI] [PubMed] [Google Scholar]
- 63.Chamoun Z, et al. Skinny Hedgehog, an acyltransferase required for palmitoylation and activity of the Hedgehog signal. Science (80-) 2001;293(5537):2080–2084. doi: 10.1126/science.1064437. [DOI] [PubMed] [Google Scholar]
- 64.Buglino JA, Resh MD. Hhat is a palmitoylacyltransferase with specificity for N-palmitoylation of Sonic Hedgehog. J Biol Chem. 2008;283(32):22076–22088. doi: 10.1074/jbc.M803901200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Porter JA, et al. The product of hedgehog autoproteolytic cleavage active in local and long-range signalling. Nature. 1995;374(6520):363–366. doi: 10.1038/374363a0. [DOI] [PubMed] [Google Scholar]
- 66.Ryan KE, Chiang C. Hedgehog secretion and signal transduction in vertebrates. J Biol Chem. 2012;287(22):17905–17913. doi: 10.1074/jbc.R112.356006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Simon E, Aguirre-Tamaral A, Aguilar G, Guerrero I. Perspectives on intra- and intercellular trafficking of Hedgehog for tissue patterning. J Dev Biol. 2016 doi: 10.3390/jdb4040034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Paná D, Sprong H, Marois E, Thiele C, Eaton S (2005) Lipoprotein particles are required for Hedgehog and wingless signaling. 2005. [Online]. www.nature.com/nature. Accessed 27 Apr 2021 [DOI] [PubMed]
- 69.Bischoff M, et al. Cytonemes are required for the establishment of a normal Hedgehog morphogen gradient in Drosophila epithelia. Nat Cell Biol. 2013;15(11):1269–1281. doi: 10.1038/ncb2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Creanga A, Glenn TD, Mann RK, Saunders AM, Talbot WS, Beachy PA. Scube/You activity mediates release of dually lipid-modified Hedgehog signal in soluble form. Genes Dev. 2012;26(12):1312–1325. doi: 10.1101/gad.191866.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jakobs P, et al. Scube2 enhances proteolytic Shh processing from the surface of Shh-producing cells. J Cell Sci. 2014;127(8):1726–1737. doi: 10.1242/jcs.137695. [DOI] [PubMed] [Google Scholar]
- 72.Stone DM, et al. The tumour-suppressor gene patched encodes a candidate receptor for Sonic hedgehog. Nature. 1996;384(6605):129–134. doi: 10.1038/384129a0. [DOI] [PubMed] [Google Scholar]
- 73.Marigo V, Davey RA, Zuo Y, Cunningham JM, Tabin CJ. Biochemical evidence that patched is the hedgehog receptor. Nature. 1996;384(6605):176–179. doi: 10.1038/384176a0. [DOI] [PubMed] [Google Scholar]
- 74.Carpenter D, et al. Characterization of two patched receptors for the vertebrate hedgehog protein family. Proc Natl Acad Sci USA. 1998;95(23):13630–13634. doi: 10.1073/pnas.95.23.13630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhulyn O, Nieuwenhuis E, Liu YC, Angers S, Chung Hui C. Ptch2 shares overlapping functions with Ptch1 in Smo regulation and limb development. Dev Biol. 2015;397(2):191–202. doi: 10.1016/J.YDBIO.2014.10.023. [DOI] [PubMed] [Google Scholar]
- 76.Rahnama F, Toftgård R, Zaphiropoulos PG. Distinct roles of PTCH2 splice variants in Hedgehog signalling. Biochem J. 2004;378(2):325–334. doi: 10.1042/BJ20031200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chuang PT, McMahon AP. Vertebrate hedgehog signalling modulated by induction of a hedgehog- binding protein. Nature. 1999;397(6720):617–621. doi: 10.1038/17611. [DOI] [PubMed] [Google Scholar]
- 78.Rohatgi R, Milenkovic L, Scott MP. Patched1 regulates hedgehog signaling at the primary cilium. Science (80-) 2007;317(5836):372–376. doi: 10.1126/science.1139740. [DOI] [PubMed] [Google Scholar]
- 79.Yao S, Lum L, Beachy P. The Ihog cell-surface proteins bind hedgehog and mediate pathway activation. Cell. 2006;125(2):343–357. doi: 10.1016/j.cell.2006.02.040. [DOI] [PubMed] [Google Scholar]
- 80.Yan D, Wu Y, Yang Y, Belenkaya TY, Tang X, Lin X. The cell-surface proteins Dally-like and Ihog differentially regulate Hedgehog signaling strength and range during development. Development. 2010;137(12):2033–2044. doi: 10.1242/dev.045740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tenzen T, Allen BL, Cole F, Kang JS, Krauss RS, McMahon AP. The cell surface membrane proteins Cdo and Boc are components and targets of the hedgehog signaling pathway and feedback network in mice. Dev Cell. 2006;10(5):647–656. doi: 10.1016/j.devcel.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 82.Kavran JM, Ward MD, Oladosu OO, Mulepati S, Leahy DJ. All mammalian hedgehog proteins interact with cell adhesion molecule, down-regulated by oncogenes (CDO) and brother of CDO (BOC) in a conserved manner. J Biol Chem. 2010;285(32):24584–24590. doi: 10.1074/jbc.M110.131680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Incardona JP, Lee JH, Robertson CP, Enga K, Kapur RP, Roelink H. Receptor-mediated endocytosis of soluble and membrane-tethered sonic hedgehog by patched-1. Proc Natl Acad Sci USA. 2000;97(22):12044–12049. doi: 10.1073/pnas.220251997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ferguson SM, De Camilli P. Dynamin, a membrane-remodelling GTPase. Nat Rev Mol Cell Biol. 2012;13(2):75–88. doi: 10.1038/nrm3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Huang S, et al. Activation of Smurf E3 ligase promoted by smoothened regulates hedgehog signaling through targeting patched turnover. PLoS Biol. 2013 doi: 10.1371/journal.pbio.1001721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li S, Li S, Wang B, Jiang J. Hedgehog reciprocally controls trafficking of Smo and Ptc through the Smurf family of E3 ubiquitin ligases. Sci Signal. 2018 doi: 10.1126/scisignal.aan8660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Taipale J, Cooper MK, Maiti T, Beachy PA. Patched acts catalytically to suppress the activity of smoothened. Nature. 2002;418(6900):892–897. doi: 10.1038/nature00989. [DOI] [PubMed] [Google Scholar]
- 88.Van den Heuvel M, Ingham PW. Smoothened encodes a receptor-like serpentine protein required for hedgehog signalling. Nature. 1996;382(6591):547–551. doi: 10.1038/382547a0. [DOI] [PubMed] [Google Scholar]
- 89.Byrne EFX, et al. Structural basis of Smoothened regulation by its extracellular domains. Nature. 2016;535(7613):517–522. doi: 10.1038/nature18934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nachtergaele S, et al. Structure and function of the Smoothened extracellular domain in vertebrate Hedgehog signaling. Elife. 2013 doi: 10.7554/eLife.01340.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang Y, Zhou Z, Walsh CT, McMahon AP. Selective translocation of intracellular smoothened to the primary cilium in response to Hedgehog pathway modulation. Proc Natl Acad Sci USA. 2009;106(8):2623–2628. doi: 10.1073/pnas.0812110106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Apionishev S, Katanayeva NM, Marks SA, Kalderon D, Tomlinson A. Drosophila smoothened phosphorylation sites essential for Hedgehog signal transduction. Nat Cell Biol. 2005;7(1):86–92. doi: 10.1038/ncb1210. [DOI] [PubMed] [Google Scholar]
- 93.Li S, et al. Regulation of smoothened phosphorylation and high-level hedgehog signaling activity by a plasma membrane associated kinase. PLoS Biol. 2016 doi: 10.1371/journal.pbio.1002481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhao Y, Tong C, Jiang J. Hedgehog regulates smoothened activity by inducing a conformational switch. Nature. 2007;450(7167):252–258. doi: 10.1038/nature06225. [DOI] [PubMed] [Google Scholar]
- 95.Fan J, Liu Y, Jia J. Hh-induced Smoothened conformational switch is mediated by differential phosphorylation at its C-terminal tail in a dose- and position-dependent manner. Dev Biol. 2012;366(2):172–184. doi: 10.1016/j.ydbio.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Corbit KC, Aanstad P, Singla V, Norman AR, Stainier DYR, Reiter JF. Vertebrate Smoothened functions at the primary cilium. Nature. 2005;437(7061):1018–1021. doi: 10.1038/nature04117. [DOI] [PubMed] [Google Scholar]
- 97.Chen W, et al. Activity-dependent internalization of smoothened mediated by β-Arrestin 2 and GRK2. Science (80-) 2004;306(5705):2257–2260. doi: 10.1126/science.1104135. [DOI] [PubMed] [Google Scholar]
- 98.Jia J, Tong C, Wang B, Luo L, Jiang J. Hedgehog signalling activity of smoothened requires phosphorylation by protein kinase a and casein kinase I. Nature. 2004;432(7020):1045–1050. doi: 10.1038/nature03179. [DOI] [PubMed] [Google Scholar]
- 99.Chen Y, et al. Sonic hedgehog dependent phosphorylation by CK1α and GRK2 is required for ciliary accumulation and activation of smoothened. PLoS Biol. 2011;9(6):e1001083. doi: 10.1371/journal.pbio.1001083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.The Krüppel-like zinc finger transcription factor, GLI-similar 1, is regulated by hypoxia-inducible factors via non-canonical mechanisms— PubMed. https://pubmed.ncbi.nlm.nih.gov/24383088/. Accessed 9 June 2021 [PubMed]
- 101.Walterhouse D, et al. gli, a zinc finger transcription factor and oncogene, is expressed during normal mouse development. Dev Dyn. 1993;196(2):91–102. doi: 10.1002/aja.1001960203. [DOI] [PubMed] [Google Scholar]
- 102.Ruiz I Altaba A. Gli proteins and hedgehog signaling: development and cancer. Trends Genet. 1999;15(10):418–425. doi: 10.1016/S0168-9525(99)01840-5. [DOI] [PubMed] [Google Scholar]
- 103.Hui CC, Slusarski D, Platt KA, Holmgren R, Joyner AL. Expression of three mouse homologs of the drosophila segment polarity gene cubitus interruptus, Gli, Gli-2, and Gli-3, in ectoderm-and mesoderm-derived tissues suggests multiple roles during postimplantation development. Dev Biol. 1994;162(2):402–413. doi: 10.1006/dbio.1994.1097. [DOI] [PubMed] [Google Scholar]
- 104.Sasaki H, Nishizaki Y, Hui CC, Nakafuku M, Kondoh H. Regulation of Gli2 and Gli3 activities by an amino-terminal repression domain: Implication of Gli2 and Gli3 as primary mediators of Shh signaling. Development. 1999;126(17):3915–3924. doi: 10.1242/dev.126.17.3915. [DOI] [PubMed] [Google Scholar]
- 105.Aza-Blanc P, Ramírez-Weber FA, Laget MP, Schwartz C, Kornberg TB. Proteolysis that is inhibited by hedgehog targets cubitus interruptus protein to the nucleus and converts it to a repressor. Cell. 1997;89(7):1043–1053. doi: 10.1016/S0092-8674(00)80292-5. [DOI] [PubMed] [Google Scholar]
- 106.Pan Y, Bai CB, Joyner AL, Wang B. Sonic hedgehog signaling regulates Gli2 transcriptional activity by suppressing its processing and degradation. Mol Cell Biol. 2006;26(9):3365–3377. doi: 10.1128/mcb.26.9.3365-3377.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Müller B, Basler K. The repressor and activator forms of Cubitus interruptus control Hedgehog target genes through common generic Gli-binding sites. Development. 2000;127(14):2999–3007. doi: 10.1242/dev.127.14.2999. [DOI] [PubMed] [Google Scholar]
- 108.Aza-Blanc P, Lin HY, Ruiz I Altaba A, Kornberg TB. Expression of the vertebrate Gli proteins in Drosophila reveals a distribution of activator and repressor activities. Development. 2000;127(19):4293–4301. doi: 10.1242/dev.127.19.4293. [DOI] [PubMed] [Google Scholar]
- 109.Dai P, Akimaru H, Tanaka Y, Maekawa T, Nakafuku M, Ishii S. Sonic hedgehog-induced activation of the Gli1 promoter is mediated by GLI3. J Biol Chem. 1999;274(12):8143–8152. doi: 10.1074/jbc.274.12.8143. [DOI] [PubMed] [Google Scholar]
- 110.Goetz SC, Anderson KV. The primary cilium: a signalling centre during vertebrate development. Nat Rev Genet. 2010;11(5):331–344. doi: 10.1038/nrg2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Cheung HOL, et al. The kinesin protein Kif7 is a critical regulator of Gli transcription factors in mammalian Hedgehog signaling. Sci Signal. 2009;2(76):ra29. doi: 10.1126/scisignal.2000405. [DOI] [PubMed] [Google Scholar]
- 112.Robbins DJ, Nybakken KE, Kobayashi R, Sisson JC, Bishop JM, Thérond PP. Hedgehog elicits signal transduction by means of a large complex containing the kinesin-related protein costal2. Cell. 1997;90(2):225–234. doi: 10.1016/S0092-8674(00)80331-1. [DOI] [PubMed] [Google Scholar]
- 113.Jia J, Tong C, Jiang J. Smoothened transduces Hedgehog signal by physically interacting with Costal2/Fused complex through its C-terminal tail. Genes Dev. 2003;17(21):2709–2720. doi: 10.1101/gad.1136603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Asaoka Y. Phosphorylation of Gli by cAMP-dependent protein kinase. In: Litwack G, editor. Vitamins and hormones. Academic Press Inc; 2012. pp. 293–307. [DOI] [PubMed] [Google Scholar]
- 115.Price MA, Kalderon D. Proteolysis of the Hedgehog signaling effector Cubitus interruptus requires phosphorylation by glycogen synthase kinase 3 and casein kinase 1. Cell. 2002;108(6):823–835. doi: 10.1016/S0092-8674(02)00664-5. [DOI] [PubMed] [Google Scholar]
- 116.Chen Y, Jiang J. Decoding the phosphorylation code in Hedgehog signal transduction. Cell Res. 2013;23(2):186–200. doi: 10.1038/cr.2013.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Tempé D, Casas M, Karaz S, Blanchet-Tournier M-F, Concordet J-P. Multisite protein kinase A and glycogen synthase kinase 3β phosphorylation leads to Gli3 ubiquitination by SCF βTrCP. Mol Cell Biol. 2006;26(11):4316–4326. doi: 10.1128/mcb.02183-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Moore BS, et al. Cilia have high cAMP levels that are inhibited by Sonic Hedgehog-regulated calcium dynamics. Proc Natl Acad Sci USA. 2016;113(46):13069–13074. doi: 10.1073/pnas.1602393113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ding Q, et al. Mouse suppressor of fused is a negative regulator of Sonic hedgehog signaling and alters the subcellular distribution of Gli1. Curr Biol. 1999;9(19):1119–1122. doi: 10.1016/S0960-9822(99)80482-5. [DOI] [PubMed] [Google Scholar]
- 120.Barnfield PC, Zhang X, Thanabalasingham V, Yoshida M, Hui CC. Negative regulation of Gli1 and Gli2 activator function by suppressor of fused through multiple mechanisms. Differentiation. 2005;73(8):397–405. doi: 10.1111/j.1432-0436.2005.00042.x. [DOI] [PubMed] [Google Scholar]
- 121.Huang D, Wang Y, Tang J, Luo S. Molecular mechanisms of suppressor of fused in regulating the hedgehog signalling pathway. Oncol Lett. 2018;15(5):6077–6086. doi: 10.3892/ol.2018.8142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Chen M-H, et al. Cilium-independent regulation of Gli protein function by Sufu in Hedgehog signaling is evolutionarily conserved. Genes Dev. 2009;23(16):1910–1928. doi: 10.1101/gad.1794109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Tukachinsky H, Lopez LV, Salic A. A mechanism for vertebrate Hedgehog signaling: recruitment to cilia and dissociation of SuFu-Gli protein complexes. J Cell Biol. 2010;191(2):415–428. doi: 10.1083/jcb.201004108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Humke EW, Dorn KV, Milenkovic L, Scott MP, Rohatgi R. The output of Hedgehog signaling is controlled by the dynamic association between suppressor of fused and the Gli proteins. Genes Dev. 2010;24(7):670–682. doi: 10.1101/gad.1902910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Takenaka K, Kise Y, Miki H. GSK3β positively regulates Hedgehog signaling through Sufu in mammalian cells. Biochem Biophys Res Commun. 2007;353(2):501–508. doi: 10.1016/j.bbrc.2006.12.058. [DOI] [PubMed] [Google Scholar]
- 126.Chen Y, Yue S, Xie L, Pu XH, Jin T, Cheng SY. Dual phosphorylation of suppressor of fused (Sufu) by PKA and GSK3β regulates its stability and localization in the primary cilium. J Biol Chem. 2011;286(15):13502–13511. doi: 10.1074/JBC.M110.217604/ATTACHMENT/AC941FE3-42DD-406A-8FC6-FF97D76DA5D6/MMC1.PDF. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Beachy PA, Hymowitz SG, Lazarus RA, Leahy DJ, Siebold C. Interactions between Hedgehog proteins and their binding partners come into view. Genes Dev. 2010;24(18):2001–2012. doi: 10.1101/gad.1951710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Mao J, et al. Regulation of Gli1 transcriptional activity in the nucleus by Dyrk1. J Biol Chem. 2002;277(38):35156–35161. doi: 10.1074/jbc.M206743200. [DOI] [PubMed] [Google Scholar]
- 129.Teperino R, Aberger F, Esterbauer H, Riobo N, Pospisilik JA. Canonical and non-canonical hedgehog signalling and the control of metabolism. Semin Cell Dev Biol. 2014;33:81–92. doi: 10.1016/j.semcdb.2014.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Huangfu D, Anderson KV. Signaling from Smo to Ci/Gli: conservation and divergence of Hedgehog pathways from Drosophila to vertebrates. Development. 2006;133(1):3–14. doi: 10.1242/dev.02169. [DOI] [PubMed] [Google Scholar]
- 131.Dagan J. Hedgehog signalling: emerging evidence for non-canonical pathways. Cell Signal. 2009;21(7):1023–1034. doi: 10.1016/J.CELLSIG.2009.01.033. [DOI] [PubMed] [Google Scholar]
- 132.Thibert C, Teillet MA, Lapointe F, Mazelin L, Le Douarin NM, Mehlen P. Inhibition of neuroepithelial patched-induced apoptosis by sonic hedgehog. Science (80-) 2003;301(5634):843–846. doi: 10.1126/science.1085405. [DOI] [PubMed] [Google Scholar]
- 133.Bredesen DE, Mehlen P, Rabizadeh S. Apoptosis and dependence receptors: a molecular basis for cellular addiction. Physiol Rev. 2004;84(2):411–430. doi: 10.1152/physrev.00027.2003. [DOI] [PubMed] [Google Scholar]
- 134.Kagawa H, et al. A novel signaling pathway mediated by the nuclear targeting of c-terminal fragments of mammalian patched 1. PLoS ONE. 2011 doi: 10.1371/journal.pone.0018638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Mille F, et al. The Patched dependence receptor triggers apoptosis through a DRAL-caspase-9 complex. Nat Cell Biol. 2009;11(6):739–746. doi: 10.1038/ncb1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Barnes EA, Kong M, Ollendorff V, Donoghue DJ. Patched1 interacts with cyclin B1 to regulate cell cycle progression. EMBO J. 2001;20(9):2214–2223. doi: 10.1093/emboj/20.9.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Takizawa CG, Morgan DO. Control of mitosis by changes in the subcellular location of cyclin-B1-Cdk1 and Cdc25C. Curr Opin Cell Biol. 2000;12(6):658–665. doi: 10.1016/S0955-0674(00)00149-6. [DOI] [PubMed] [Google Scholar]
- 138.Adolphe C, Hetherington R, Ellis T, Wainwright B. Patched1 functions as a gatekeeper by promoting cell cycle progression. Cancer Res. 2006;66(4):2081–2088. doi: 10.1158/0008-5472.CAN-05-2146. [DOI] [PubMed] [Google Scholar]
- 139.Jiang X, Yang P, Ma L, Lefkowitz RJ. Kinase activity-independent regulation of cyclin pathway by GRK2 is essential for zebrafish early development. Proc Natl Acad Sci USA. 2009;106(25):10183–10188. doi: 10.1073/pnas.0812105106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Polizio AH, Chinchilla P, Chen X, Kim S, Manning DR, Riobo NA. Heterotrimeric Gi proteins link hedgehog signaling to activation of Rho small GTPases to promote fibroblast migration. J Biol Chem. 2011;286(22):19589–19596. doi: 10.1074/jbc.M110.197111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Renault MA, et al. Sonic hedgehog induces angiogenesis via Rho kinase-dependent signaling in endothelial cells. J Mol Cell Cardiol. 2010;49(3):490–498. doi: 10.1016/j.yjmcc.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Yam PT, Bastien S, Langlois D, Morin S, Dé Ric Charron F. Article sonic hedgehog guides axons through a noncanonical, src-family-kinase-dependent signaling pathway. Neuron. 2009;62(3):349–362. doi: 10.1016/j.neuron.2009.03.022. [DOI] [PubMed] [Google Scholar]
- 143.Belgacem YH, Borodinsky LN. Sonic hedgehog signaling is decoded by calcium spike activity in the developing spinal cord. Proc Natl Acad Sci USA. 2011;108(11):4482–4487. doi: 10.1073/pnas.1018217108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Lee H, Ko HW. Ciliary smoothened-mediated noncanonical hedgehog signaling promotes tubulin acetylation. Biochem Biophys Res Commun. 2016;480(4):574–579. doi: 10.1016/j.bbrc.2016.10.093. [DOI] [PubMed] [Google Scholar]
- 145.Shi Q, Li S, Li S, Jiang A, Chen Y, Jiang J. Hedgehog-induced phosphorylation by CK1 sustains the activity of Ci/Gli activator. Proc Natl Acad Sci. 2014;111(52):E5651–E5660. doi: 10.1073/PNAS.1416652111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Maloverjan A, Piirsoo M, Michelson P, Kogerman P, Østerlund T. Identification of a novel serine/threonine kinase ULK3 as a positive regulator of Hedgehog pathway. Exp Cell Res. 2010;316(4):627–637. doi: 10.1016/J.YEXCR.2009.10.018. [DOI] [PubMed] [Google Scholar]
- 147.Wang Y, et al. The crosstalk of mTOR/S6K1 and hedgehog pathways. Cancer Cell. 2012;21(3):374–387. doi: 10.1016/J.CCR.2011.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Antonucci L, et al. Mitogen-activated kinase kinase kinase 1 inhibits hedgehog signaling and medulloblastoma growth through GLI1 phosphorylation. Int J Oncol. 2019;54(2):505–514. doi: 10.3892/IJO.2018.4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Shi X, Zhan X, Wu J. A positive feedback loop between Gli1 and tyrosine kinase Hck amplifies shh signaling activities in medulloblastoma. Oncogenesis. 2015;4(11):e176–e176. doi: 10.1038/oncsis.2015.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Rui Z, et al. Dual degradation signals destruct GLI1: AMPK inhibits GLI1 through β-TrCP-mediated proteasome degradation. Oncotarget. 2017;8(30):49869–49881. doi: 10.18632/ONCOTARGET.17769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Di Marcotullio L, et al. Numb activates the E3 ligase Itch to control Gli1 function through a novel degradation signal. Oncogene. 2011;30(1):65–76. doi: 10.1038/onc.2010.394. [DOI] [PubMed] [Google Scholar]
- 152.Liu H, Yan S, Ding J, Yu T-T, Cheng SY. DeSUMOylation of Gli1 by SENP1 attenuates sonic hedgehog signaling. Mol Cell Biol. 2017 doi: 10.1128/MCB.00579-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Talpale J, et al. Effects of oncogenic mutations in smoothened and patched can be reversed by cyclopamine. Nature. 2000;406(6799):1005–1009. doi: 10.1038/35023008. [DOI] [PubMed] [Google Scholar]
- 154.Luchetti G, Sircar R, Kong JH, Nachtergaele S, Sagner A Cholesterol activates the G-protein coupled receptor smoothened to promote hedgehog signaling recommended citation. 10.7554/eLife.20304.001 [DOI] [PMC free article] [PubMed]
- 155.Beachy PA, et al. Multiple roles of cholesterol in hedgehog protein biogenesis and signaling. Cold Spring Harb Symp Quant Biol. 1997;62:191–204. doi: 10.1101/sqb.1997.062.01.025. [DOI] [PubMed] [Google Scholar]
- 156.Banavali NK. The mechanism of cholesterol modification of hedgehog ligand. J Comput Chem. 2020;41(6):520–527. doi: 10.1002/jcc.26097. [DOI] [PubMed] [Google Scholar]
- 157.Chen X, et al. Processing and turnover of the hedgehog protein in the endoplasmic reticulum. J Cell Biol. 2011;192(5):825–838. doi: 10.1083/jcb.201008090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Porter JA, et al. Hedgehog patterning activity: role of a lipophilic modification mediated by the carboxy-terminal autoprocessing domain. Cell. 1996;86(1):21–34. doi: 10.1016/S0092-8674(00)80074-4. [DOI] [PubMed] [Google Scholar]
- 159.Tanaka Hall TM, Porter JA, Young KE, Koonin EV, Beachy PA, Leahy DJ. Crystal structure of a hedgehog autoprocessing domain: homology between hedgehog and self-splicing proteins. Cell. 1997;91(1):85–97. doi: 10.1016/S0092-8674(01)80011-8. [DOI] [PubMed] [Google Scholar]
- 160.Ciepla P, Magee AI, Tate EW. Cholesterylation: a tail of hedgehog. Biochem Soc Trans. 2015;43(2):262–267. doi: 10.1042/BST20150032. [DOI] [PubMed] [Google Scholar]
- 161.Amanai K, Jiang J. Distinct roles of central missing and dispatched in sending the hedgehog signal. Development. 2001;128(24):5119–5127. doi: 10.1242/dev.128.24.5119. [DOI] [PubMed] [Google Scholar]
- 162.Johnson JLFA, et al. Scube activity is necessary for Hedgehog signal transduction in vivo. Dev Biol. 2012;368(2):193–202. doi: 10.1016/j.ydbio.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 163.Callejo A, et al. Dispatched mediates Hedgehog basolateral release to form the long-range morphogenetic gradient in the Drosophila wing disk epithelium. Proc Natl Acad Sci USA. 2011;108(31):12591–12598. doi: 10.1073/pnas.1106881108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Tukachinsky H, Kuzmickas RP, Jao CY, Liu J, Salic A. Dispatched and scube mediate the efficient secretion of the cholesterol-modified hedgehog ligand. Cell Rep. 2012;2(2):308–320. doi: 10.1016/j.celrep.2012.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Ohlig S, et al. Sonic hedgehog shedding results in functional activation of the solubilized protein. Dev Cell. 2011;20(6):764–774. doi: 10.1016/j.devcel.2011.05.010. [DOI] [PubMed] [Google Scholar]
- 166.Ohlig S, et al. An emerging role of Sonic hedgehog shedding as a modulator of heparan sulfate interactions. J Biol Chem. 2012;287(52):43708–43719. doi: 10.1074/jbc.M112.356667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Dierker T, Dreler R, Petersen A, Bordych C, Grobe K. Heparan sulfate-modulated, metalloprotease-mediated sonic hedgehog release from producing cells. J Biol Chem. 2009;284(12):8013–8022. doi: 10.1074/jbc.M806838200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Qi X, Schmiege P, Coutavas E, Wang J, Li X. Structures of human Patched and its complex with native palmitoylated sonic hedgehog. Nature. 2018;560(7716):128–132. doi: 10.1038/s41586-018-0308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Resh MD. Palmitoylation of hedgehog proteins by hedgehog acyltransferase: roles in signalling and disease. Open Biol. 2021;11(3):200414. doi: 10.1098/rsob.200414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Izzi L, et al. Boc and gas1 each form distinct shh receptor complexes with ptch1 and are required for shh-mediated cell proliferation. Dev Cell. 2011;20(6):788–801. doi: 10.1016/j.devcel.2011.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Martinelli DC, Fan CM. Gas1 extends the range of hedgehog action by facilitating its signaling. Genes Dev. 2007;21(10):1231–1243. doi: 10.1101/gad.1546307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Allen BL, Tenzen T, McMahon AP. The hedgehog-binding proteins Gas1 and Cdo cooperate to positively regulate Shh signaling during mouse development. Genes Dev. 2007;21(10):1244–1257. doi: 10.1101/gad.1543607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Farzan SF, Singh S, Schilling NS, Robbins DJ. The adventures of sonic hedgehog in development and repair. III. Hedgehog processing and biological activity. Am J Physiol Gastrointest Liver Physiol. 2008;294(4):G844. doi: 10.1152/ajpgi.00564.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Bellaiche Y, The I, Perrimon N. Tout-velu is a Drosophila homologue of the putative tumour suppressor EXT-1 and is needed for Hh diffusion. Nature. 1998;394(6688):85–88. doi: 10.1038/27932. [DOI] [PubMed] [Google Scholar]
- 175.Gallet A, Rodriguez R, Ruel L, Therond PP. Cholesterol modification of Hedgehog is required for trafficking and movement, revealing an asymmetric cellular response to Hedgehog. Dev Cell. 2003;4(2):191–204. doi: 10.1016/S1534-5807(03)00031-5. [DOI] [PubMed] [Google Scholar]
- 176.The I, Bellaiche Y, Perrimon N. Hedgehog movement is regulated through tout velu-dependent synthesis of a heparan sulfate proteoglycan. Mol Cell. 1999;4(4):633–639. doi: 10.1016/S1097-2765(00)80214-2. [DOI] [PubMed] [Google Scholar]
- 177.Ma Y, et al. Hedgehog-mediated patterning of the mammalian embryo requires transporter-like function of dispatched. Cell. 2002;111(1):63–75. doi: 10.1016/S0092-8674(02)00977-7. [DOI] [PubMed] [Google Scholar]
- 178.Lewis PM, et al. Cholesterol modification of sonic hedgehog is required for long-range signaling activity and effective modulation of signaling by Ptc1. Cell. 2001;105(5):599–612. doi: 10.1016/S0092-8674(01)00369-5. [DOI] [PubMed] [Google Scholar]
- 179.Zeng X, Goetz JA, Suber LM, Scott WJ, Schreiner CM, Robbins DJ. A freely diffusible form of sonic hedgehog mediates long-range signalling. Nature. 2001;411(6838):716–720. doi: 10.1038/35079648. [DOI] [PubMed] [Google Scholar]
- 180.Ayers KL, Gallet A, Staccini-Lavenant L, Thérond PP. The long-range activity of hedgehog is regulated in the apical extracellular space by the glypican dally and the hydrolase notum. Dev Cell. 2010;18(4):605–620. doi: 10.1016/j.devcel.2010.02.015. [DOI] [PubMed] [Google Scholar]
- 181.Parchure A, Vyas N, Ferguson C, Parton RG, Mayor S. Oligomerization and endocytosis of Hedgehog is necessary for its efficient exovesicular secretion. Mol Biol Cell. 2015;26(25):4700–4717. doi: 10.1091/mbc.E15-09-0671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Vyas N, et al. Vertebrate Hedgehog is secreted on two types of extracellular vesicles with different signaling properties. Sci Rep. 2014;4(1):1–12. doi: 10.1038/srep07357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Torroja C, Gorfinkiel N, Guerrero I. Patched controls the Hedgehog gradient by endocytosis in a dynamin-dependent manner, but this internalization does not play a major role in signal transduction. Development. 2004;131(10):2395–2408. doi: 10.1242/DEV.01102. [DOI] [PubMed] [Google Scholar]
- 184.Adolphe C, Junker JP, Lyubimova A, van Oudenaarden A, Wainwright B. Patched receptors sense, interpret, and establish an epidermal hedgehog signaling gradient. J Investig Dermatol. 2017;137(1):179–186. doi: 10.1016/J.JID.2016.06.632. [DOI] [PubMed] [Google Scholar]
- 185.Callejo A, Culi J, Guerrero I. Patched, the receptor of hedgehog, is a lipoprotein receptor. Proc Natl Acad Sci USA. 2008;105(3):912–917. doi: 10.1073/pnas.0705603105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Qi C, Di Minin G, Vercellino I, Wutz A, Korkhov VM. Structural basis of sterol recognition by human hedgehog receptor PTCH1. Sci Adv. 2019;5(9):eaaw6490. doi: 10.1126/sciadv.aaw6490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Zhang Y, et al. Structural basis for cholesterol transport-like activity of the hedgehog receptor patched. Cell. 2018;175(5):1352–1364.e14. doi: 10.1016/j.cell.2018.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Nachury MV, Mick DU. Establishing and regulating the composition of cilia for signal transduction. Nat Rev Mol Cell Biol. 2019;20(7):389–405. doi: 10.1038/s41580-019-0116-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Endapally S, Frias D, Grzemska M, Gay A, Tomchick DR, Radhakrishnan A. Molecular discrimination between two conformations of sphingomyelin in plasma membranes. Cell. 2019;176(5):1040–1053.e17. doi: 10.1016/j.cell.2018.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Winkler MBL, et al. Structural insight into eukaryotic sterol transport through niemann-pick type C proteins. Cell. 2019;179(2):485–497.e18. doi: 10.1016/j.cell.2019.08.038. [DOI] [PubMed] [Google Scholar]
- 191.Huang P, et al. Structural basis of smoothened activation in hedgehog signaling. Cell. 2018;174(2):312–324.e16. doi: 10.1016/j.cell.2018.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Deshpande I, et al. Smoothened stimulation by membrane sterols drives Hedgehog pathway activity. Nature. 2019;571(7764):284–288. doi: 10.1038/s41586-019-1355-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Yao Y et al The differential protein and lipid compositions of noncaveolar lipid microdomains and caveolae. nature.com. [Online]. https://www.nature.com/articles/cr200927. Accessed 15 May 2021 [DOI] [PubMed]
- 194.Murata M, Peränen J, Schreiner R, Wieland F, Kurzchalia TV, Simons K. VIP21/caveolin is a cholesterol-binding protein. Proc Natl Acad Sci USA. 1995;92(22):10339–10343. doi: 10.1073/pnas.92.22.10339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Zheng YZ, et al. Differential impact of caveolae and caveolin-1 scaffolds on the membrane raft proteome. Mol Cell Proteom. 2011 doi: 10.1074/mcp.M110.007146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Ekker SC, et al. Patterning activities of vertebrate hedgehog proteins in the developing eye and brain. Curr Biol. 1995;5(8):944–955. doi: 10.1016/S0960-9822(95)00185-0. [DOI] [PubMed] [Google Scholar]
- 197.Fietz MJ, Jacinto A, Taylor AM, Alexandre C, Ingham PW. Secretion of the amino-terminal fragment of the Hedgehog protein is necessary and sufficient for hedgehog signalling in Drosophila. Curr Biol. 1995;5(6):643–650. doi: 10.1016/S0960-9822(95)00129-1. [DOI] [PubMed] [Google Scholar]
- 198.Hu A, et al. Cholesterylation of smoothened is a calcium-accelerated autoreaction involving an intramolecular ester intermediate. Cell Res. 2022;2022:1–14. doi: 10.1038/s41422-022-00622-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Manikowski D, Ehring K, Gude F, Jakobs P, Froese J, Grobe K. Hedgehog lipids: promotors of alternative morphogen release and signaling? BioEssays. 2021;43(11):2100133. doi: 10.1002/BIES.202100133. [DOI] [PubMed] [Google Scholar]
- 200.Wendler F, Franch-Marro X, Vincent JP. How does cholesterol affect the way hedgehog works? Development. 2006;133(16):3055–3061. doi: 10.1242/dev.02472. [DOI] [PubMed] [Google Scholar]
- 201.Gallet A, Ruel L, Staccini-Lavenant L, Thérond PP. Cholesterol modification is necessary for controlled planar long-range activity of hedgehog in Drosophila epithelia. Development. 2006;133(3):407–418. doi: 10.1242/DEV.02212. [DOI] [PubMed] [Google Scholar]
- 202.Callejo A, Torroja C, Quijada L, Guerrero I. Hedgehog lipid modifications are required for Hedgehog stabilization in the extracellular matrix. Development. 2006;133(3):471–483. doi: 10.1242/DEV.02217. [DOI] [PubMed] [Google Scholar]
- 203.Dawber RJ, Hebbes S, Herpers B, Docquier F, Van Den Heuvel M. Differential range and activity of various forms of the Hedgehog protein. BMC Dev Biol. 2005 doi: 10.1186/1471-213X-5-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Ducuing A, Querenet M. Cholesterol-free and cholesterol-bound hedgehog: two sparring-partners working hand in hand in the Drosophila wing disc? Fly (Austin) 2013;7(4):23554573. doi: 10.4161/fly.25655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Eugster C, Panáková D, Mahmoud A, Eaton S. Lipoprotein-heparan sulfate interactions in the Hh pathway. Dev Cell. 2007;13(1):57–71. doi: 10.1016/j.devcel.2007.04.019. [DOI] [PubMed] [Google Scholar]
- 206.Palm W, Swierczynska MM, Kumari V, Ehrhart-Bornstein M, Bornstein SR, Eaton S. Secretion and signaling activities of lipoprotein-associated hedgehog and non-sterol-modified hedgehog in flies and mammals. PLoS Biol. 2013;11(3):1001505. doi: 10.1371/journal.pbio.1001505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.McCarthy RA, et al. Megalin functions as an endocytic sonic hedgehog receptor. J Biol Chem. 2002;277(28):25660–25667. doi: 10.1074/jbc.M201933200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data sets used and/or analyzed during the current study are available from the corresponding author on reasonable request.