Abstract
Cuproptosis was a copper-dependent and unique kind of cell death that was separate from existing other forms of cell death. The last decade has witnessed a considerable increase in investigations of programmed cell death, and whether copper induced cell death was an independent form of cell death has long been argued until mechanism of cuproptosis has been revealed. After that, increasing number of researchers attempted to identify the relationship between cuproptosis and the process of cancer. Thus, in this review, we systematically detailed the systemic and cellular metabolic processes of copper and the copper-related tumor signaling pathways. Moreover, we not only focus on the discovery process of cuproptosis and its mechanism, but also outline the association between cuproptosis and cancers. Finally, we further highlight the possible therapeutic direction of employing copper ion ionophores with cuproptosis-inducing functions in combination with small molecule drugs for targeted therapy to treat specific cancers.
Keywords: Cuproptosis, Copper, Cancer, Targeted therapy, Immunotherapy, Drug resistance, Metabolism
Background
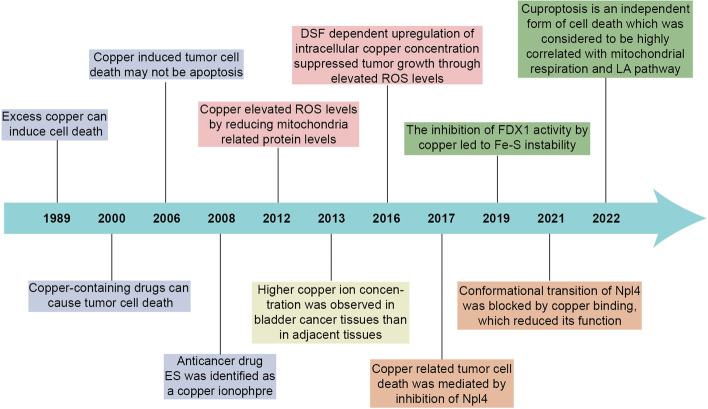
In the recent years, cuproptosis, a novel form of regulated cell death which is copper dependent has been identified [1, 2], may be implicated in the process of various cancers. Copper is a trace element in the human body and has been strongly associated with various signaling pathways and tumor-related biological behaviors [3]. Moreover, excess copper can lead to cell death, and for a long time the mechanisms and specific forms of copper-induced cell death have remained unclear. Until early this year, it has been suggested by a recent study that cuproptosis is an independent form of cell death, which was considered to be highly correlated with mitochondrial respiration and lipoic acid(LA) pathway [4]. We briefly summarize some of the findings on copper-induced cell death that have driven progress in the field (Fig. 1).
Fig. 1.
Timeline illustrating the discovery of cuproptosis. The historical events contributing to the discovery of cuproptopsis and oncological research advances of copper associated cell death are depicted in the timeline
A considerable number of researchers focusing on the pivotal relationship between cuproptosis and cancers. On the one hand, cancer has multiple types, with sufficient multi-omics data. On the other hand, cuproptosis is highly related to cellular metabolism, and certain cancer types usually exhibits high aerobic respiration levels. Some tumor types such as melanoma, breast cancer and leukemia [5, 6], some cancers with tumor stem cells [7, 8] and some drug-resistant tumors exhibit a high mitochondrial metabolic state [9–13]. Tumor cells treated with certain antitumor drugs such as proteasome inhibitors(PI) have also been found to exhibit higher mitochondrial metabolism [14, 15]. A growing number of researchers focusing on the vital link between cuproptosis and cancer process through bioinformatic analysis. Some studies have focused on the relationship between expression levels of cuproptosis key genes (CKGs), genes identified and validated in the previous studies of Tsvetkov et al., and tumor prognosis. To avoid the effects of gene interactions, some investigators have constructed Cuproptosis-related signatures by cuproptosis related genes (CRGs) to identify the association of Cuproptosis with cancer. Copper ionophores played a major contribution in the discovery of cuproptosis, and have been considered for possible use in antitumor therapy in the past [16, 17]. However, their specific mechanisms and applicable populations have not been fully analyzed. With the discovery of the cuproptosis, the interactions between these drugs, copper and the mitochondria are becoming clear, which makes the antitumor clinical application of these drugs possible. This review focusing on discovery of the mechanism of cuproptosis and the pivotal relationship between cuproptosis and cancers. We aimed to provide possible directions for future studies related to cuproptosis and cancers.
Systemic and cellular copper homeostasis
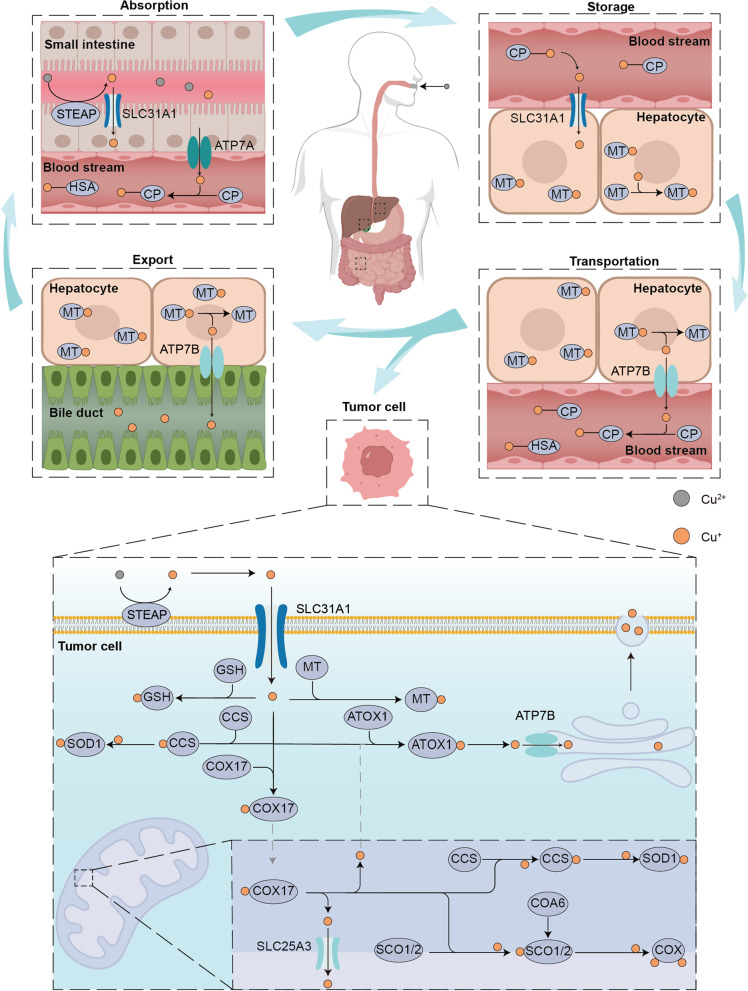
Copper, a kind of indispensable transition metal, has two sides for cell. On the one hand, it served as co-factor for many enzymes by donating or receipting electronics [3], on the other hand, the accumulation of copper can lead to a series of cellular metabolic dysfunctions and eventually cell death [18]. People mainly obtain copper from food, out of which organ meats and shellfish tend to be the richest food sources of copper, and the current recommended intake of copper for adults should be 0.8–2.4 mg/day to maintain systemic copper homeostasis [19] (Fig. 2). Copper uptake occurs mainly through the small intestine, the small intestine epithelium took up copper ions via copper transporter 1 (CTR1) or called solute carrier family 31 member 1(SLC31A1), the transporter encoding by slc31a1 on the cell surface. The copper was transported to another side of epithelium through copper chaperone antioxidant 1 copper chaperone (ATOX1) and exported into the bloodstream through the action of ATPase copper transporting alpha (ATP7A) [20]. Copper ions are transported in the blood by binding to proteins rather than being free. About 75% of copper ions are bound to ceruloplasmin (CP) in the non-exchangeable form, about 25% of copper ions are bound to human serum albumin (HSA) in the exchangeable form and about 0.2% of copper ions are bound to Histidine [21, 22]. Copper ions were then transported through portal system to the liver [23], which is the main organ for copper repository and also the main organ for copper excretion in the body. The copper storage function was believed to be mediated by metallothionein1/2 (MT1/2), two thiol-rich proteins, which bind copper ions in a pH-dependent manner through their cystine residues, however, their specific ability to bind and transfer copper is still unknown. Through the function of ATPase copper transporting beta (ATP7B), excess copper is excreted into the bile and leaves the body [24].
Fig. 2.
Schematic of systemic and cellular copper metabolism. The body absorbs copper mostly through the small intestine, where it is then transported by blood to the liver for excretion into the bile. In tumor cells, interactions between several proteins maintain copper homeostasis. The entry and departure of copper ions into and out of the cell are controlled by the copper ion transporters SLC31A1 and ATP7B, whereas the transit of copper ions through the outer and inner mitochondrial membranes is controlled by COX17 and SLC25A3, respectively. Copper ions entering the cytoplasm and mitochondrial intermembrane space bind to GSH and MT or form copper-containing molecular chaperones such as SOD1 which is crucial for proper function of copper. To sustain normal cellular functions, COA6, SCO1 and SCO2 work together to mediate the transfer of copper to COX in the mitochondrial intermembrane space
The role of copper in tumor processes has also been of interest to researchers. A significant increase in serum copper ion levels in tumor patients compared to normal patients has been observed in studies of lung cancer [25], prostate cancer [26], breast cancer [27], carcinoma of gallbladder [28], stomach cancer [29] and thyroid cancers [30]. Higher levels of copper ions were also observed in the gallbladder tissue of patients with gallbladder cancer [28]. Further, among lung cancer patients, those with worse clinical stage had higher serum copper ion concentrations and higher serum copper ion concentrations were also associated with worse clinical prognosis. The normal function of copper ions in cancer cells is dependent on the regulation of copper homeostasis and the interaction of different types of proteins (Fig. 2). The first group is proteins related to copper transport across the membrane. Consistent with small intestinal epithelial cells, the uptake of copper ions by tumor cells also requires the involvement of CTR1, and the elevated and decreased expression levels of SLC31A1 directly affect intracellular copper ion levels [31]. CTR1 mainly transports monovalent copper ions. After being transported to the cell surface in the blood, divalent copper ions are reduced to monovalent copper ions catalyzed by steap proteins [32], which are bound and maintained in the reduced state by two His-Met-Asp clusters at the nitrogen terminus of CTR [33], and thus transported into the cell. Cu was transported from the intermembrane space of mitochondria across the inner membrane into the mitochondrial matrix by the transmembrane transport protein solute carrier family 25 member 3 (SLC25A3) [34]. However how copper ions enter the intermembrane space of mitochondria through the outer membrane is still unknown. ATPases, including ATP7A and ATP7B, associated with the extracellular excretion of copper, export Cu ions bound to metal binding sites in the presence of ATP [35]. These proteins that mediate the transmembrane transportion of copper ions regulate their intracellular distribution. The second group is proteins that bind and store copper ions. MT and glutathione(GSH) served as naturally intracellular copper ion chelators, binding copper and thus preventing it from causing cell damage [36]. The third group is copper ion chaperones [37], interaction of which ensures proper copper cellular function. Cytoplasmic copper ion chaperones ATOX1 bind Cu(I) via two cysteine residues and transport it to the metal binding sites of ATP7B for further exportation. Copper chaperone for superoxide dismutase(CCS) directly interacted with and transported copper ions to superoxide dismutase 1(SOD1) [38]. With the involvement of O2, CCS can accelerate the disulfide formation of SOD1, which is essential for the correct spatial conformation and the enzyme activity [39]. SOD1 plays a role in catalyzing the generation of H2O2 from superoxide radicals and plays a key role in maintaining intracellular reactive oxygen species(ROS) homeostasis, and the inactivation of which can lead to the onset of cell death [40]. In addition to this there are a series of intra-mitochondrial copper ion chaperones that play an important role in the function of cytochrome c oxidase (COX), an important component of oxidative phosphorylation. These chaperones are involved in the composition and function of COX by storing or delivering copper ions [41]. Cytochrome c oxidase copper chaperone(COX17) carried copper ions from the cytoplasm into the intermembrane space of mitochondria [42], and further delivered copper ions to the cysteine residues of SCO1 with which it formed disulfide bonds [43]. Cytochrome c oxidase assembly factor 6(COA6) served as a thiol-disulfide oxidoreductase to reduce the formation of disulfide bonds between cysteine residues in synthesis of cytochrome c oxidase1/2 (SCO1/2) and substances other than copper [44], thus allowing copper binding [45]. The deficiency of COA6 lead to disorder of respiratory complex IV biogenesis [46]. In the effect of COA6, SCO1 and SCO2 transferred the copper ions obtained from COX17 to COX and participate in cytochrome c oxidase assembly [47]. Moreover, SCO1 and SCO2 were also involved in the regulation of cellular copper homeostasis, and the absence of both decreases cellular copper ion levels [48]. The maintenance of intracellular homeostasis of copper is dependent on the interaction of these four types proteins, and dysregulation of copper homeostasis will lead to disruption of cellular metabolism and even cell death.
Cuproptosis and cancer signaling pathways
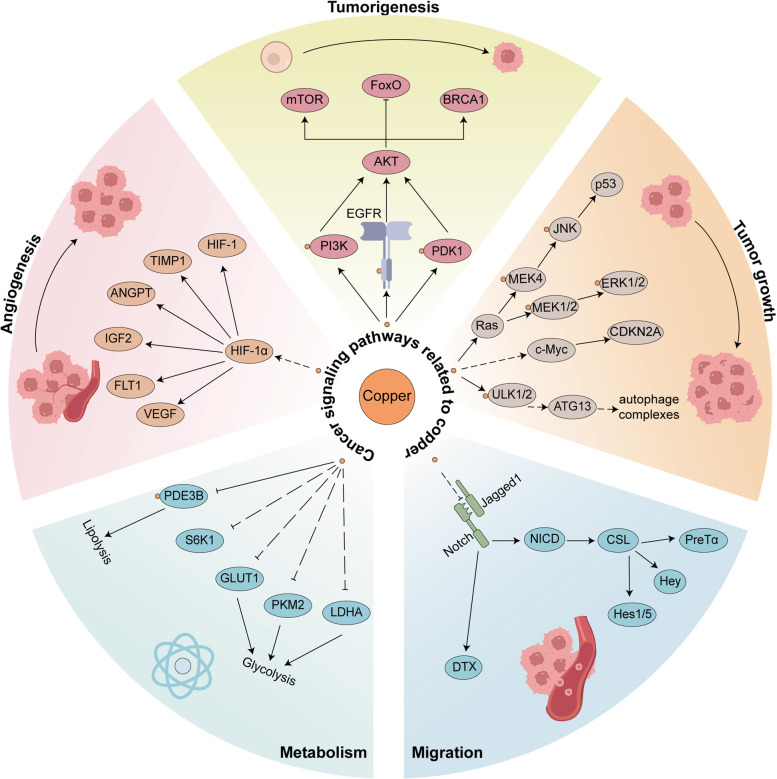
Copper is thought to be directly related to multiple signaling pathways in tumor cells, by binding and activating key molecules in multiple signaling pathways (Fig. 3). Copper were considered to play a critical role in receptor tyrosine kinase-related signaling pathways, which can bind and phosphorylate receptor tyrosine kinase(RTK) with no ligand binding and further lead to RKT activation. Activated RTK subsequently lead to phosphorylation of downstream extracellular regulated protein kinases(ERK) and agammaglobulinaemia tyrosine kinase(ATK), ultimately lead to cell migration and proliferation [49]. Copper ions are also thought to cause downstream activation by acting on different molecules of the phosphoinositide-3-kinase (PI3K)-AKT signaling pathway. On the one hand, Copper can directly activate the PI3K which leads to downstream AKT activation [50]. On the other hand copper bind to the histidine117 and histidine203 sites of pyruvate dehydrogenase kinase 1(PDK1) which lead to activation of AKT [51]. The AKT activation triggered by copper can further catalyze the phosphorylation and subcellular redistribution of forkhead box O1a(FoxO1a) and forkhead box O4(FoxO4), which promoted the cancer cell proliferation and tumor growth [52]. Activation of mitogen-activated protein kinase(MAPK) signaling pathway is also dependent on the presence of copper ions [53], Copper can directly bind to mitogen-activated proteinkinase kinase 1(MEK1) to promote the phosphorylation of ERK1/2 and further activate the downstream c-Jun N-terminal kinase(JNK) to regulate tumor growth [54].
Fig. 3.
Copper and cancer singnaling pathways. Copper is strongly associated with the process of cancer and impacts them in a direct or indirect ways. Copper directly binds or activates EGFR, PDK1 or PI3K to promotes tumorigenesis. Copper also influences MAPK and autophagic pathways or indirectly changes c-Myc stability to influence tumor growth. Copper ions indirectly promotes HIFα or indirectly inhibits the Notch pathway ligand Jagged1 thus promoting vascular neoplastic migration. In addition, copper can also regulate PDE3B or S6K1 and thus modulates tumor metabolism
The autophagy pathway can recycle metabolic waste from tumor cells to ensure their energy needs or allow them to escape apoptosis, ultimately leading to proliferation of tumor cells. Copper directly binds Unc-51 Like autophagy activating kinase(ULK) and acts as its regulator to promote phosphorylation and activation of autophagy related 13(ATG13), resulting in the formation of the autophagic complex and ultimately tumor growth [55, 56]. Consistent with the MAPK signaling pathway, autophagic pathway can also promote cancer cell survival and is directly affected by copper ions. In B-Raf proto-oncogene(BRAF)-driven lung adenocarcinoma cells, loss of CTR1 leads to a decrease in copper ion concentration that is directly associated with reduced function of MEK1/2 and ULK1/2, key kinases in both signaling pathways [57]. In addition to interacting directly with important proteins in the pathway, copper ions can also affect them indirectly to regulate the biological behavior of cancers. The Notch pathway is often considered as a tumor suppressor, which is extensively involved in the development of malignant tumours [58]. Copper ions promote shedding of the notch ligand Jagged1 on cell surface and promote tumor cell migration [59]. Numerous reports have revealed that copper ions have a close relationship with tumor angiogenesis [60], which was dependent on the interaction between copper and hypoxia inducible factor 1 subunit alpha(HIF-1α)-related signaling pathways. Copper mediates HIF-1α binding to the critical motifs of target gene promoters through a CCS-dependent manner thereby upregulating the expression of affected genes such as hypoxia inducible factor 1(HIF-1) [61, 62]. Even under normoxic conditions, copper can directly increase the stability of HIF-1α, which in turn promotes the expression of target genes such as vascular endothelial growth factor(VEGF), leading to tumor angiogenesis [63]. Previous research has also emphasized the significance of copper in inflammation promoting effects by interacting with NFκB pathways [64]. Inflammatory cytokines promote elevated intracellular copper levels and lead to X-linked inhibitor of apoptosis(XIAP) activation, which promotes NFκB activation and tumorigenesis. A strong relationship between copper and lipolysis pathways has been reported in the previous research. The Wnt signaling pathway maintains the renewal balance of human cells, and activation of whose genes exerts a pro-tumor effect in tumor cells [65]. A much-debated question is whether copper up or down regulated the level of C-myc. The previous study has found that increasing intracellular copper ion concentration through disulfiram(DSF) leads to a decrease in the expression of β-catenin and C-myc, two important molecules of the Wnt pathway, thereby inhibiting tumor growth [66]. However, a more recent research argued that the Copper elevates C-myc stability by promoting phosphorylation at its threonine 58 and serine 62 sites [67]. The level of tumor cell metabolism also has an impact on the biological behavior of tumors, and copper ions can regulate tumor metabolism through interactions with related molecules within the lipid or sugar metabolic pathways. Copper has been shown to regulate lipolysis through interaction with cysteine residues of phosphodiesterase 3B(PDE3B), the phosphodiesterase that degrades cAMP [68]. Moreover, Cu is also thought to inhibit the expression of S6K1 and its downstream glycolysis-related molecules, including GLUT1, PKM2 and LDHA, thereby suppressing tumor growth [69]. Collectively, copper plays a direct or indirect role in cancer signaling pathways and cancer properties, which further emphasizes its importance in cancers.
From copper induced cell death to cuproptosis
During the past few years, the link between copper and programmed cell death has long been at the center of much attention and the mechanism of copper induced cell death has long been researched. Copper has been known to cause cell death in the 1980s [70], however, the exact mechanism has not been elucidated. Copper ionophores, a lipid-soluble molecule that binds copper ions reversibly, played an important role in the discovery of cuproptosis, and may be involved in clinical treatment as antitumor agents [71]. Copper ionophores may transport copper ions through the plasma membrane or mitochondrial membrane structure of a cell. DSF, a drug that has been used to treat alcohol dependence, also functions as a copper ionophore and is thought to cause cell death [72], in the same way that elesclomol(ES), another copper ionophores, was also believed to have the ability to kill cells [73]. In studies on copper ionophores ES and DSF, many researchers have investigated the mechanism by which these copper ionophores cause cell death, suggesting that such cell death was caused by copper rather than copper ionophores, however, the exact mechanism was not indicated [74]. ROS is mainly derived from intracellular redox reactions in which mitochondria play a key role [75], which is also consistent with the correlation between copper induced cell death and mitochondrial metabolism. Among the studies on ES-induced cell death, researchers generally agree that the cell death caused by ES is mediated by elevated levels of ROS due to various mitochondrial-related factors. A study in 2012 on melanoma cell lines concluded that ES transported copper and led to reduced levels of mitochondria related proteins, thereby lead to increase of ROS and further inhibition of tumor cell proliferation [74]. In a 2013 study conducted in Human leukemia K562 cells, copper ions were suggested to be able to oxidize ascorbic acid and react with H2O2 to produce more damaging ROS after entering cells via ES transport [76]. It was suggested in the study on ES in 2015 that ES-Cu may have multiple roles, including blocking cells in the G1 phase, damaging DNA and affecting mitochondrial membrane potential [16]. A 2016 study on AT-rich interaction domain 1A(ARID1A) concluded that ES can act on the mitochondrial respiratory chain and lead to increased levels of intracellular reactive oxygen species and cell death through a ROS-mediated mechanism [77]. Other studies on tumor targeting by copper ionophores have proposed the same mechanism as previously described [78, 79]. However, in 2019 Tsvetkov et al. found that Hi-Mito condition (a rise in mitochondrial respiration, which can be induced by replacing Glucose with Galactose) leads to a resistance to PI but together with a higher vulnerability to ES, the copper ionophore. Ferredoxin 1(FDX1) was identified as the gene most associated with ES sensitivity, which was directly bound by ES-Cu and lead to inhibition of the iron-sulfur cluster (Fe-S cluster) formation function [14]. In 2021, studies in glioblastoma stem like cells revealed that ES-Cu can act directly on the mitochondrial membrane and lead to changes in the mitochondrial membrane potential, with low concentrations of ES leading to hyperpolarization and high concentrations of ES leading to depolarization, and this effect on the mitochondrial membrane potential can be inhibited by tetrathiomolybdate(TTM) [7]. In common with ES, DSF can also transport copper into the cell, and copper is thought to act on the mitochondrial respiratory chain, leading to elevated levels of ROS [17]. In addition to causing elevated levels of ROS, the interaction of DSF-Cu with Npl4 was also thought to be closely associated with copper-induced cell death. Copper is thought to inhibit the ubiquitinated protein degradation function of p97 by interacting with Npl4, possibly leading to its aggregation [80, 81] or directly binding and inhibiting its conformational transition [82], ultimately leading to cell death. In summary, in past studies on Cu-induced cell death, most researchers have attributed this cell death to the action of copper on the mitochondria resulting in the production of ROS. However, in studies on the mechanism of action of ES, the cytotoxic effect caused by ES-Cu was not eliminated by using 5 mM ROS inhibitor N-Acetylcysteine (NAC), and the cytotoxic effect was only partially eliminated by 10 mM NAC, so ROS-mediated cell death may not be the main mode of cell death caused by copper. The exact form of cell death caused by copper ions has been controversial, and in the past researchers have mostly regarded it as apoptosis [83–86], autophage [87, 88] or ferroptosis [89, 90], which can be proved by the evidence of cell viability assay, western blotting, flow cytometry and immunofluorescence staining. Until March 2022, Tsvetkov et al. identified the mechanism of copper induced cell death, which was named cuproptosis, researches on the mechanism of copper induced cell death have reached a new milestone. None of the inhibitors for apoptosis, necroptosis, ROS induced cell death or ferroptosis but the copper chelator can rescue cells from copper induced cell death [4], and the expression level of cleaved caspases were not increased. In other words, among the aforementioned evidence, the evidence of cell viability assay and western blotting have been disproved, while the others remain to be further verified.
The cell death triggered by ES decreased significantly by reducing copper binding function and completely disappeared after removal of this function. ES that does not carry copper ions cannot independently cause cell death [14]. Except for ES and DSF, other ionophores can also lead to same cellular effects. GSH was a natural intracellular chelator of copper ions, reduction of which can also lead to an increase in intracellular copper concentrations and ultimately lead to the cell death [4]. Moreover, in a WD-related model, it was found that breakdown of copper homeostasis through downregulation of ATP7B could also lead to the development of cell death [91].In summary, cell death caused by copper ionophores is a type of cell death that is triggered by copper independently of the known modes of cell death.
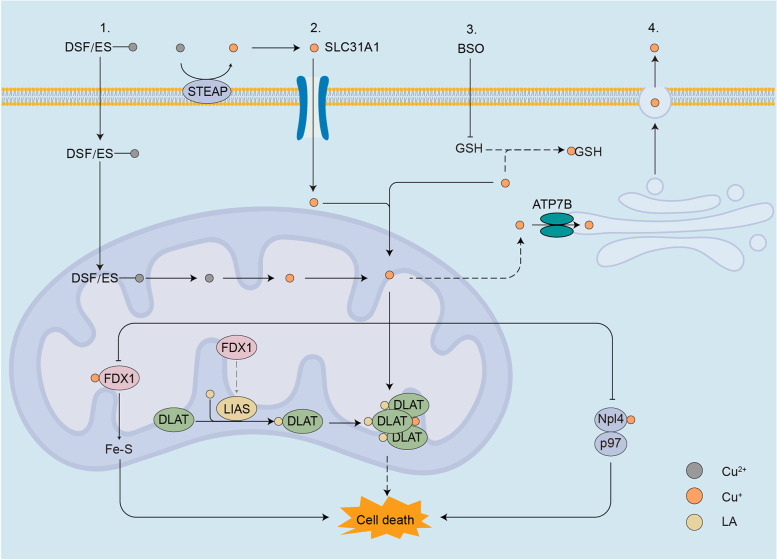
Cuproptosis was thought to be interact with components of the TCA cycle in mitochondria and involves a conserved post-transcriptional protein modification pathway, lipoylation [4], the mechanism of which displayed in the form of a schematic (Fig. 4). To identify the exact component of mitochondrial respiration that interacted with copper, Oligomycin (ATPase inhibitor), FCCP (uncoupler) and antimycin A/rotenone (electron transport chain inhibitor) has been used to measure the oxygen consumption rate (OCR). The OCR showed significantly reduced spare capacity of respiration rather than basal respiration or ATP-linked respiration, which proved that copper directly interact with the TCA cycle rather than neither the electron transport chain (ETC) complex nor the ATP production component [4]. Through genome wide knockout screens, metabolism screens and individual gene knockout studies, CKGscrucial for cuproptosis has been discovered and validated (Table 1), knockout of seven of which could lead to the rescue of copper ionophores [4]. These seven genes can be divided into three groups, FDX1, LA pathway-related genes (LIAS and LIPT1) and genes encoding components of pyruvate dehydrogenase complex (PDC) which play a crucial role in mitochondrial respiration (DLAT, DLD, PDHA1 and PDHB), all of which were correlated with LA pathway. FDX1 was considered may act as the upstream of LA pathway, and under the regulation of FDX1, LIAS linked lipoyl moiety to DLAT, which was essential for the function of mitochondrial PDC. Cu(I) directly bound with lipoyl moiety of lipoylated DLAT through disulfide bond and further lead to DLAT oligomerization and further proteotoxic stress and finally result in cell death [4].
Fig. 4.
Schematic of cuproptosis mechanism. Cuproptosis can be triggered by elevating intracellular free copper ion concentration in four ways involved in the absorption, export and storage of copper: (1) treatment with copper ionophores, which shuttle copper into the cell directly, such as ES and DSF; (2) overexpression of SLC31A1, the copper permease specific for reduced copper ion; (3) inhibition of glutathione (GSH) synthesis through BSO, without which free copper ion was released; (4) knockdown of ATP7B, decreasing copper export. Excessive Cu(I) binds to lipoyled DLAT and further leads to DLAT oligomerization, which, together with copper-induced reduction of Fe-S stability or inactivation of Npl4-p97, can lead to the onset of copper-induced cell death. ES elesclomol, DSF disulfiram, BSO L-Buthionine-sulfoximine
Table 1.
Functions and clinical values of cuproptosis validated genes
| Gene | Full name | Subcellular locations | Functions | Role in cuproptosis | Clinical values | Ref |
|---|---|---|---|---|---|---|
| FDX1 | Ferredoxin 1 | Mitochondrion matrix | Fe-S cluster biosynthesis ruduce Cu(II) to Cu(I) synthesis of various steroid hormones electron transport intermediate for mitochondrial cytochromes P450 | act as the upstream of LA pathway ruduce Cu(II) to Cu(I) | FDX1 high expression correlaed with better prognosis in many cancers | [14, 91–99] |
| LIAS | Lipoic Acid Synthetase | Mitochondrion | converting the octanoylated domains into lipoylated derivatives | Regulated by FDX1, and involved in lipoylation pathway | LIAS high expression led to worse OS and FP, and correlated with more advanced lung cancer staging | [100, 101] |
| LIPT1 | Lipoyltransferase 1 | Mitochondrion | Catalyzes the transfer of the lipoyl group from lipoyl-AMP to the specific lysine residue of lipoyl domains of lipoate-dependent enzymes | Regulated by FDX1, and involved in lipoylation of DLAT | LIPT1 high expression led to better prognosis | [102–104] |
| DLD | Dihydrolipoamide Dehydrogenase | mitochondrion and nucleus | E3 component of the pyruvate dehydrogenase complex | not montioned | NA | [105, 106] |
| DLAT | Dihydrolipoamide S-Acetyltransferase | Mitochondrion matrix | component of pyruvate dehydrogenase complex, mediate the conversion of pyruvate to acetyl-CoA | under the influence of copper, lipoylated DLAT oligomerization lead to cell death | NA | [107] |
| PDHA1 | Pyruvate Dehydrogenase E1 Subunit Alpha 1 | Mitochondrion matrix | component of pyruvate dehydrogenase complex, mediate the conversion of pyruvate to acetyl-CoA | not montioned | PDHA1 high expression led to worse OS and FP in LUAD and low expression correlaed with HCC | [108] |
| PDHB | Pyruvate Dehydrogenase E1 Subunit Beta | Mitochondrion matrix | component of pyruvate dehydrogenase complex, mediate the conversion of pyruvate to acetyl-CoA | not montioned | NA | NA |
| MTF1 | Metal Regulatory Transcription Factor 1 | Nucleus and Cytoplasm | Zinc-dependent transcriptional regulator for metal ions adaption | knock out lead to sensitive of cuproptosis | NA | [109] |
| GLS | Glutaminase | Mitochondrion. Cytoplasm and cytosol | Catalyzes the catabolism of glutamine | knock out lead to sensitive of cuproptosis | GLS high expression led to worse OS | [110, 111] |
| CDKN2A | Cyclin Dependent Kinase Inhibitor 2A | Nucleus and Cytosol | inducing cell cycle arrest in G1 and G2 phases | knock out lead to sensitive of cuproptosis | CDKN2A higher expression led to risk in LUAD but protective function in BRCA | [112–114] |
| SLC31A1 | Solute Carrier Family 31 Member 1 | Cell membrane | High-affinity, saturable copper transporter involved in dietary copper uptake | overactivaiton lead to intracellular copper accumulation | SLC31A1 alleles were associated with a worse prognosis in LUAD and BRCA | [112–120] |
| ATP7A | ATPase Copper Transporting Alpha | Cell membrane, trans-Golgi network membrane, plasma membrane | ATP-driven copper ion pump | knock out lead to intracellular copper accumulation | NA | [119] |
| ATP7B | ATPase Copper Transporting Beta | Cell membrane, trans-Golgi network and membrane | ATP-driven copper ion pump | knock out lead to intracellular copper accumulation | ATP7B alleles were associated with a decreased risk of LUAD | [99, 118] |
Although Tsvetkov et al. used concise cell lines and mouse models to elaborate part of the mechanism of copper action in cuproptosis, there are still some unanswered questions about cuproptosis that need further study. To begin with, the characteristic manifestations of cuproptosis have not been described. On the one hand, the cellular morphological changes of cuproptosis have not been described, whether cuproptosis occurs with characteristic or sequential morphological manifestations, and on the other hand, the characteristic changes that occur at the molecular or cellular level after the induction of cuproptosis have not been identified, thus lacking effective means to assess whether cuproptosis has occurred [4]. Furthermore, the downstream pathways of DLAT oligomers, which play an important role in cuproptosis, have not been described. In previous studies, only the toxic effects of DLAT oligomerization were described and it was assumed that this DLAT oligomerization leads to proteotoxic stress and ultimately to cell death. However, the direct mechanism between DLAT oligomerization and cell death has not been determined [4]. In addition, in studies on copper ionophores, either ES-Cu or DSF-Cu has been suggested to lead to increased proteotoxic stress in other ways besides DLAT oligomerization. For example, ES-Cu can lead to decreased stability of Fe-S through FDX1 [14] and DSF-Cu can affect the cellular ubiquitination degradation pathway by interaction with Npl4 [80–82], further leading to increased proteotoxic stress and ultimately leading to cell death. However, whether there are interactions between these events leading to increased cellular proteotoxic stress remains to be investigated. Finally, the function of FDX1 has not been sufficiently mentioned, FDX1 is thought to play a central role in the process of cuproptosis and was considered to act as an upstream of the LA pathway, but whether its function is achieved through direct interaction with LIAS or the action of some other proteins needs further investigation [4].
Validated cuproptosis key genes: functions and clinical values
Ten genes were identified by Tsvetkov et al. through whole genome knockout, deletion of which could lead to an altered risk of cuproptosis. While the analysis about the copper homeostasis suggested that SLC31A1, ATP7A and ATP7B could also affect cuproptosis by regulating the intracellular copper ion concentration [4]. Therefore these 13 genes might play an important role in the mechanism of cuproptosis and were considered as CKGs. The function and subcellular location of these CKGs are listed in Table 1. These CKGs have important cellular roles and are often closely related to energy metabolism and metal homeostasis processes. As the concept of cuproptosis was introduced, a considerable number of researchers focused on the role played by these CKGs in different tumors, indicating the expression levels and clinical significance of CKGs in different cancers.
FDX1 encodes Ferredoxin1, the iron–sulfur protein that was involved in multiple redox reactions. FDX1 served as monooxygenase of cytochrome P450 and was involved in steroidogenesis [92]. Also, it has proved that FDX1 was crucial for the iron–sulfur cluster biogenesis [93, 94]. In addition, FDX1 was also considered can reduce Copper ion from divalent to monovalent and exhibited stronger cytotoxic properties and cellular function [14]. In the process of cuproptosis, FDX1 played a central role which regulated the process of protein lipoylation as the upstream of LA pathway [4]. Compared to paired normal tissues, FDX1 expression levels were upregulated in glioblastoma (GBM) and female genital tumors, and downregulated in solid tumors like lung adenocarcinoma (LUAD) and hepatocellular carcinoma (HCC) [95]. High levels of FDX1 were considered to be associated with poor prognosis in patients with head and neck squamous cell carcinoma (HNSC) and low grade glioma (LGG). In contrast, higher FDX1 expression lead to better prognosis in patients with cervical squamous cell carcinoma(CESC) and clear cell renal cell carcinoma (KIRC) [95–97]. In KIRC and colorectal cancer (CRC),patients with lower expression level of FDX1 was condisered to suffer more advanced and metastatic cancers together with poor overall survival(OS) and disease free survival(DFS) [98, 121].
Lipoic acid is a crucial substance which is required for enzymes involved in oxidative decarboxylation of mitochondrial metabolism intermediates [122]. Protein expressed by LIPT1, LIAS all belong to the LA pathway, which mediated the post-transcriptional lipoic modification of proteins like PDC [122], was both crucial for cell normal cellular activity and cuproptosis. LIAS synthesized lipoic acid by introducing two sulfhydryl groups at the C6 and C8 sites of the octanoic acid moiety, deficiency of which led to neonatal epilepsy, disorder in mitochondrial energetic metabolism and elevated glycine [100]. Similarly, LIAS expression levels tend to correlate with prognosis in different patients, high LIAS expression in lung cancer was suggested to have a poor prognosis, while in KIRC and ovarian cancer it was considered to be associated with a better prognosis [101]. LIPT1catalyzed the transfer of a lipoyl group to the lysine residue of the target enzymes [102], deficiency of which will lead to Leigh disease together with a deficiency of PDC [103].In addition, cancer cells growth and invasion were prevented by knocking down the expression of the LIPT1 gene [104].
DLAT, DLD, PDHA1 and PDHB served as three vital subunits of PDC [123], which is the important component of the mitochondrial aerobic respiration process [124] and played a key role in the cuproptosis process [4], and the expression levels of which were also believed to be related to tumor prognosis. PDHA1 and PDHB form the α1 and β subunits of the PDC E1 component, respectively, and together form pyruvate dehydrogenase mediating pyruvate decarboxylation [125]. High levels of PDHA1 were thought to be associated with a better prognosis for lung cancer patients and can be used as a biomarker for the tumor microenvironment [108]. DLAT form the E2 component of PDC that acts as a dihydrolipoamide acetyltransferase catalyzing the biosynthesis of acetyl coenzyme A(acetyl-CoA) [107]. Lipoylated DLAT played an important role in the process of cuproptosis, where its oligomerization in the presence of copper leads to proteotoxic stress and ultimately to cell death. While DLD participated in the formation of the E3 component of PDC as dihydrolipoamide dehydrogenase catalyzes the formation of NADH [105, 106]. PDC generates acetyl-CoA and NADPH through cooperative interactions between three subunits, which is the control step for the mitochondrial oxidative phosphorylation [126], and its phosphorylation mediated by PDC kinase leads to inactivation [127].
CDKN2A, GLS and MTF1 have also been demonstrated that related to the cell cuproptosis sensitivity [4]. GLS mainly catalyzes the catabolism of glutamine, which convert glutamine into glutamate, and is also involved in the maintenance of glutamate homeostasis. Dysfunction of GLS can lead to glutamine overload affecting the physiology and structure of the central nervous system [110], and its hyperactivity can lead to glutamate overload also leading to neurodevelopmental delays [111]. MTF1 served as a transcriptional regulator of cellular adaptation to heavy metals, which activates the transcription of copper binding protein MT, by binding to the metal response element in the promoter of MT [109]. GLS and MTF-1 may influence the sensitivity of cells to cuproptosis by affecting the intracellular levels of the copper ion-binding substances GSH and MT. CDKN2A was suggested to interact with and inhibit cyclin‑dependent kinase inducing cell cycle arrest in G1 phases. In the past, studies have focused on the tumor suppressor gene role of CDKN2A, and mutations of which led to loss of growth control in breast cancer(BRCA) [112], HNSC [113] and ovarian cancer cells [114], but its role in cuproptosis remains to be further investigated. CDKN2A was suggested to be highly expressed in BRCA and LUAD, and in addition, high CDKN2A expression was thought to correlate with immune cell infiltration levels [116, 117].
Dysregulation of aforementioned copper homeostasis maintainer SLC31A1, ATP7A and ATP7B also leads to disruption of cellular functions [3]. As previously described, SLC31A1 mediates copper entry into cells while ATP7A and ATP7B mediate copper exit from cells, all functioning as copper carriers that closely related to the shuttle of copper [115, 118, 119]. High expression of SLC31A1 has been considered to be associated with poor clinical outcome of BRCA patients by two independent researches [120, 128]. Researchers also found that the microsatellites of SLC31A1 and ATP7B were associated with lung cancer risk, suggesting that the expression levels of copper homeostasis-related genes may also influence the process of lung cancer development [99]. Previous research on these genes has uncovered the ways in which they may influence or be influenced by cuproptosis, as well as the potential significance of their involvement in the connection between cuproptosis and cancers. However, further studies are needed to explore and validate these functions.
Cuproptosis related genes and cancer prognosis
There is a growing interest in cuproptosis since demonstration and most investigators made exploration on the relationship between cuproptosis and cancers. Researchers have used online databases to analyze some of the numerous genes that may play an important role in the link between cuproptosis and cancer, and based on which to predict the cancer features and prognosis of patients. As shown in Table 2, prognostic signatures constructed by CRGs and their clinical values have been listed.
Table 2.
Association of cuproptosis related genes and cancers
| Ref | database | cancer | risk factor | protective factor | microenviroment condition | Clinical value |
|---|---|---|---|---|---|---|
| [129] | TCGA | LUAD | DLAT, DLD, DLST and PDHA1 | DBT and LIPT1 | high risk related to immune cell infiltration reduction and less activity in HLA, type I and II IFN response | high risk led to worse OS together with higher mortality and more advanced T and N stage |
| [130] | TCGA, GSE68465 | LUAD | DLAT | CDKN2A, PDHA1and MTF1 | high risk led to less immune cell infiltration | higher risk score corelated with worse OS, more advanced pathological stage, positive lymph nodes and more severe tumor status |
| [131] | TCGA, GTEx, NODE, ICGC | HCC | ATP7A, LIPT1, DLAT, MTF1, GLS, and CDKN2A | none | high risk related to immunosuppressive microenviroment, immune cell infiltration reduction and macrophage polarization inhibition | high risk score correlated with worse survival |
| [132] | TCGA | liver cancer | DLAT | ATP7A, GLS | higher risk score correlated with worse immune status and higher expression of immune check point genes | higher risk correlated with worse OS and clinicopathological features |
| [133] | TCGA, GSE76427 | HCC | MANEA, PGM2, PTTG1 | CGNL1, ALAS1 | higher risk score correlated with actived CD4 + T | higher risk score correlated with higher mortality and worse OS |
| [134] | TCGA, ICGC, TISCHGSE64041, GSE14520/GPL3921, GSE76427, GSE104580, GSE109211, and GSE25097 | HCC | CAT, SLC27A, EHHADH, ALDH5A1 | none | higher risk score correlated with protumor immune infiltration and higher expression of immun check point genes | higher risk score correlated with worse OS, PFS and clinicopathological features |
| [135] | TCGA, GSE76427 | HCC | PBK, MMP1, GNAZ, GPC1 | AKR1D1 | higher risk score correlated with protumor immune infiltration | higher risk score correlated with worse OS |
| [136] | TCGA, GSE14520 | HCC | KIF2C, PTTG1, CENPM, CDC20, SFN | CYP2C9, CFHR3 | higher risk score correlated with less immune cell infiltration and higher expression of immun check point genes | higher risk score correlated with worse OS |
| [137] | TCGA, GTEx | CC | ATP7A, DLAT, GCSH | DBT, FDX1, LIPT1, PDHA1 | higher risk score correlated with less immune cell infiltration | higher risk score correlated with worse OS and more advanced clinical stage |
| [138] | TCGA | USEC | CDKN2A, GLS, and LIPT1 | none | not mentioned | higher risk correlated with worse OS, PFS and DFS |
| [139] | TCGA, GSE22138 | UVM | LIPT1, DLD, PDHA1, CDKN2A | LIAS, PDHB, MTF1, GLS | no significant difference in the total immune cell infiltration | higher risk correlated with more advanced clinical stage |
| [140] | TCGA, GTEx, CGGA | LGG | C21orf62, DRAXIN, ITPRID2, MAP3K1, and MOXD1 | none | higher risk score correlated with higher TME scores and lower CSC index, also with higher expression of immun check point genes | higher risk correlated with worse OS and more cases of death |
| [141] | TCGA, GTEx, CGGA311, CGGA668, GSE108474, GSE13041, GSE16011, GSE43289, GSE43378, GSE4412, GSE4412, GSE68838, and GSE83300 | giloma | H19, CYTOR, IGFBP2 and CHI3L1 | KLRC2 and C5orf38 | higher risk score correlated with higher immune infiltration and less immune function | higher risk correlated with more advanced clinical stage |
| [142] | TCGA, CGG, AGSE84465 | giloma | SLC31A1, NFE2L2, MT1M, MT1H, MAP1LC3A, FDX1, COX19, ARF1, AOC1 | LIAS, CYP1A1, ATP7B | higher risk score correlated with higher immune infiltration and less immune function | higher risk correlated with more advanced clinical stage |
| [143] | TCGA, CGGA | giloma | FDX1, DLD, MTF1 | DLAT, CDKN2A | higher risk score correlated with higher protumor immune infiltration | higher risk score correlated with worse OS |
| [144] | TCGA, GSE17538, GSE29623, GSE39582 | CRC | CDKN2A, GLS | DLAT | higher risk score correlated with less immune cell infiltration and more stormal cells | higher risk correlated with more advanced clinical stage and worse OS |
| [145] | TCGA, GSE61304 | BC | PGK1, PRDX1, MAL2, and SURF4 | RPL14, PSME1 | higher risk score correlated with higher immune activation | higher risk correlated with worse OS and PFS |
| [146] | TCGA, GSE58812, GSE135565 and GSE65194 | TNBC | ATP7A | LIPT1 | higher risk score correlated with less immune cell infiltration and lower TME score | higher risk correlated with worse OS and more advanced clinical stage |
| [147] | TCGA, GSE168410, GSE20685 and GSE20711 | BC | DLAT, SNX3, TTC3, PHF20, RTN4, SURF4, SDC1, KDELR2, BAMBI, ANXA5, MARVELD | RBP1, TPT1, MDK, RPLP1, ETV6 | higher risk score correlated with more active immune function and higher immune check point genes expression | higher risk correlated with worse prognosis |
| [148] | TCGA, GSE40435 and GSE53757 | KIRC | CDKN2A | FDX1, DLAT | not mentioned | higher risk correlated with worse OS and PFS |
| [149] | TCGA, E-MTAB-1980, ICGC, GSE64052 | KIRC | FDX1, CDC42BPG, C11orf52, GNG7, PAQR5, ENAM, WDR72, SDR42E1, BSPRY and KDF1 | TMEM214, CCM2 and P3H4 | higher risk score correlated with higher expression of immune check point genes | higher risk correlated with worse OS and DFS |
| [150] | TCGA, GSE12606, GSE53000, and GSE53757 | KIRC | none | ENAM, WDR72, CLDN10, HMGCS2, CYFIP2, QRFPR | higher risk score correlated with more immune cells infiltration | higher risk correlated with worse clinicopathological features |
| [151] | TCGA | HNSCC | PRKN, MT1E, CXCL8, COX11, COX5A, COX19, ACLY | CYP2D6, ABCB1, CCL5, LOXL1, CDKN2A, BCL2, DAPK2 | higher risk score correlated with less immune cell infiltration and immune function | higher risk correlated with worse OS and clinicopathological features |
| [152] | TCGA | HNSCC | not mentioned | not mentioned | higher risk score correlated with higher expression of immune check point genes, less immune function and lower TME score | higher risk correlated with worse OS, PFS, DFS and clinicopathological features |
| [153] | TCGA, GSE41613 and GSE65858 | HNSCC | PRELID2, ANP32B, MRPL47, and CCDC59 | CDKN2A, WDR90, NLRX1 and KCNK6 | higher risk score correlated with higher expression of immune check point genes | higher risk correlated with worse OS |
| [154] | TCGA, GSE41613 and GSE42743 | HNSCC | not mentioned | not mentioned | higher risk score correlated with higher expression of immune check point genes | higher risk correlated with worse OS and more advanced clinical stage |
| [155] | TCGA | ESCA | SLC25A5, SLC23A2, PDHX, ATP7A, and COX7B | PHID2 | higher risk score correlated with more immune cells infiltration | higher risk correlated with worse prognosis |
| [156] | TCGA, ICGC | PAAD | KRAS, TP53, BRCA1, BRCA2, DLAT | CDKN2A, SMAD4, LIAS, LIPT1, DLD, PDHA1, MTF1, GLS | higher risk score correlated with low TME scores and expression of immune check point genes | higher risk correlated with worse OS |
| [157] | TCGA, GTEx | PAAD | DLAT | LIPT, LIAS | higher risk score correlated with more immune cells infiltration | higher risk correlated with worse OS |
| [158] | TCGA, ICGC, GTEX, IMvigor210 | PAAD | DLAT, TIMMDC1, GSS | NDUFB2, LIAS, NDUFA8, ISCA2 | higher risk score correlated with more immune supression | higher risk correlated with less survival probablity |
| [159] | TCGA, GSE30219, GSE31210 and GSE37745 | LUAD | LINC00205, LINC00592 and AL162632.3S | AC026355.2, LINC02848 and ZNF571AS1 | higher risk score correlated with greater potential for immune escape and suppressed immune function | higher risk score correlated with worse OS, PFS and higher mortality |
| [160] | TCGA, GSE130740 | LUAD | AL031667.3, AL606489.1 and MIR31HG | AC008764.2, AL022323.1, ELN-AS1 and LINC00578 | higher risk score correlated with lower score of most pathways related to immune and less immune cells infiltration | higher risk correlated with worse OS |
| [161] | TCGA | HCC | miR-767-5p, miR-5003-3p, miR-137-3p, miR-760, miR-548f-3p, miR-3171, miR-3189-3p, miR-3620-3p, miR-3911, miR-4652-3p, miR-504-3p, miR-892a, miR-548aq-5p | miR-67645p | higher risk score correlated with higher expression of immuno check point genes | higher risk correlated with worse OS |
| [132] | TCGA | liver cancer | POLH-AS, AL117336.2, MKLN1-AS, AC005479.2, AL928654.1, AL031985.3 | none | higher risk score correlated with worse immune status and higher expression of immune check point genes | higher risk correlated with worse OS, more advanced clinical stage |
| [162] | TCGA, GTEx | HCC | AC138904.1, DEPDC1-AS1, GIHCG, AC145343.1 | AC099329.2, DNMBP-AS1 | higher risk score correlated with actived CD4 + T and higher expression of immune check ponit genes | higher risk correlated with worse OS |
| [163] | TCGA | PHC | MIR210HG, AC099850.3, AL031985.3, C012073.1, MKLN1-AS, KDM4A-AS1 and PLBD1-AS1 | none | risk score correlated with less immunce cell infiltration | higher risk correlated with worse OS |
| [164] | TCGA, GTEx | CC | AL354733.3 and AC009902.2 | AL441992.1, LINC01305, AL354833.2, CNNM3-DT and SCAT2 | risk score correlated with less immunce cell infiltration and down regulation of immune functions and less Immune checkpoint gene expression | higher risk correlated with worse OS and PFS and worse clinicalpathological features |
| [165] | TCGA, GSE16088 | OS | AL645608.6, AL591767.1 and UNC5B-AS1 | CARD8-AS1, AC098487.1 and AC005041.3 | higher risk score correlated with less immune infiltration and immune function | higher risk correlated with worse OS |
| [166] | TARGET | OS | AL033384.2 | AL031775.1, AC110995.1 and LINC00565 | higher risk score correlated with higher infiltration of immunosuupressive cell and less immune effective cell infiltration and immune function | higher risk correlated with worse OS |
| [167] | TCGA | SKCM | none | VIM-AS1, AC012443.2, MALINC1, AL354696.2 and HSD11B1-AS1 | higher risk score correlated with less immune infiltration, downregulation of immune function and immune check point gene expression | higher risk correlated with worse OS |
| [168] | TCGA | CM | AC009495.1 | LINC01150, EBLN3P, MIR100HG, WAC − AS1, LINC00339 and USP30 − AS1 | higher risk score correlated with less immune infiltration and less immune check point genes expression | higher risk correlated with worse OS |
| [169] | TCGA, GTEx, CGGA | LGG | CRNDE, FAM181A-AS1 | HAR1A | higher risk score correlated with higher immune cells infiltration, TME scores and immune check point genes expression | higher risk correlated with worse clinicalpathological features, lower survival rate and worse clinical outcone |
| [170] | TCGA | CRC | AP003119.3, RNF216P1, AC156455.1, AL360270.1, AC139720.2, AC092614.1, LRP4-AS1, AP003555.1, AL513550.1 and AL512306.2 | AC073896.3, LINC00511, AC026979.4, AC103703.1, LRP1-AS | higher risk score correlated with supressed immune function | higher risk correlated with worse OS and PFS |
| [171] | TCGA | CRC | AP001619.1, AC020917.2, AC002066.1, LINC01252, AC010789.2, LINC02542, AL356804.1 and ZFHX2-AS1 | AC008752.2 and AC012313.5 | higher risk score correlated with supressed immune function, less immune cell infiltration and less expression of immune check point genes | higher risk correlated with worse OS and clinical outcome |
| [172] | TCGA | CRC | LINC00861, AC090517.2, AC01233.5, AL513550.1, AC026979.4, AC064836.3, PRKAR1BAS2, LINC02175, ZNF775AS1, AL161729.4 | AC073896.3 | higher risk score correlated with less iimmune cells infiltration | higher risk correlated with worse clinical outcome |
| [173] | TCGA, GSE42743 | OSCC | C6orf99, AC010894.2, AC099850.4, RPL23AP7 | AC090587.2, AL513190.1, AC098484.2 | higher risk score correlated with lower TME score | higher risk correlated with worse prognosis and higher mortality rate |
| [174] | TCGA | BC | AC079922.2, ZNF197-AS1, AC002398.1, AL451085.3, LINC02446 | GORAB-AS1, AL589765.4, AC005696.4, CYTOR, YTHDF3-AS1, AC008771.1 | higher risk score correlated with inactive immune function and less expression of immune check point genes | higher risk correlated with worse OS |
| [175] | TCGA, E-MTAB-1980 | KIRC | FOXD2-AS1, NUP153-AS1, LINC02154 | SUCLG2-AS1, LINC00271 | higher risk score correlated with less iimmune cells infiltration and higher TME score | higher risk correlated with worse OS and DFS |
| [176] | TCGA | KIRC | LINC01605, AGAP2-AS1, FOXD2-AS1, LINC02195 | none | higher risk score correlated with more immune check point genes expression | higher risk correlated with worse OS and clinicopathological features |
| [177] | TCGA | KIRC | HHLA3, H110-AS1, PICSAR, SNHG15, LINC00471, LINC02154, MINCR | LINC02027, SNHG8, EIF1B-AS1 | higher risk score correlated with higher TME score and immune check point genes expression | higher risk correlated with worse OS |
| [152] | TCGA | HNSCC | AL359397.1, AC098679.1, AC008014.1, C6orf99, LINC01106 | LINC00278, AC106820.3, AC007406.3, AC106820.5, AC022182.2, AP006621.4, TTTY14, AP006545.1, AC067930.3, DCST1 − AS1, AC022167.3, AC022098.1, AC020907.1 | higher risk correlaed with CD4 memory resting T cells infiltration | higher risk correlated with shorter survival |
| [178] | TCGA | HNSCC | AL132800.1, AC 079,160.1, AL157888.1, SNHG16 | AC021148.2, AC090587.1, AC011462.4, GRHL3-AS1 | higher risk score correlated with low immune escape | higher risk correlated with worse OS and more advanced clinical stage |
| [179] | TCGA | HNSCC | AC004943.2, TTN-AS1, AL132800.1, WDFY3-AS2 | AC090587.1, AL136419.3, AC012313.5, AC106820.5, AL162458.1, CDKN2A-DT | higher risk score correlated with lower TME score and more immune dysfunction | higher risk correlated with worse OS and clinicopathological features |
| [180] | TCGA | STAD | LINC01094, AC022182.1, AC011747.1, LINC02476, AC090809.1, AC084781.2, SENCR | AC005014.2, AC010422.4 | higher risk score correlated with higher expression of immune check point genes | higher risk correlated with worse prognosis |
| [181] | TCGA | STAD | LINC01150, SNAP25-AS1, HAGLR | LINC00571 | higher risk score correlated with higher immune function and expression of immune check point geens | higher risk correlated with worse OS and clinicopathological features |
| [182] | TCGA | BLCA | AC080023.1 | AC010168.2, AC018653.3 | higher risk score correlated with higher immune functions and immune cells infiltration | higher risk correlated with worse OS and clinicopathological features |
CKGs have related cellular functions that interact with each other, complicating the assessment of tumor prognosis by any of these genes. CKGs have different roles among different tumors. DLD was considered as a risk factor in studies of different tumors such as uveal melanoma(UVM), glioma, and LUAD [129, 139, 143, 183]. In contrast, LIAS was analyzed as a protective factor in studies of UVM, glioma, and pancreatic adenocarcinoma(PAAD) [139, 142, 156–158]. In studies, DLAT was analyzed as a PAAD risk factor [156–158] and conversely was considered to be associated with a protective effect in glioma [142, 143]. Even in different studies on the same tumor, different investigators considered FDX1 and PDHA1 as risk or protective factor respectively [129, 130, 148, 149]. A potential association among CDKN2A, MTF1 and GLS and patient prognosis has been noted in different studies, which were believed had a consistent effect on the prognosis of patients [130, 131, 139, 156]. In one of the researches on glioma, SLC31A1 was considered a risk factor, while ATP7B was considered a protective factor [142], which further suggested that there may be a link between the copper homeostasis and cancer. Many investigators have focused on the expression levels of CKGs in different tumors and analyzed the relationship between CKGs and tumor prognosis either alone or by constructing signatures, however, the lack of biological knowledge about cuproptosis prevents direct analysis of the role of these genes in the link between cuproptosis and tumorigenesis. In addition to the expression levels and clinical significance of CKG among different tumors, the investigators also focused on the profiles of these CKGs in SNV, CNV, methylation, pathway cross-talk and so on through the pan-cancer analysis, and these genes were found to exhibit different properties in different tumors [184].
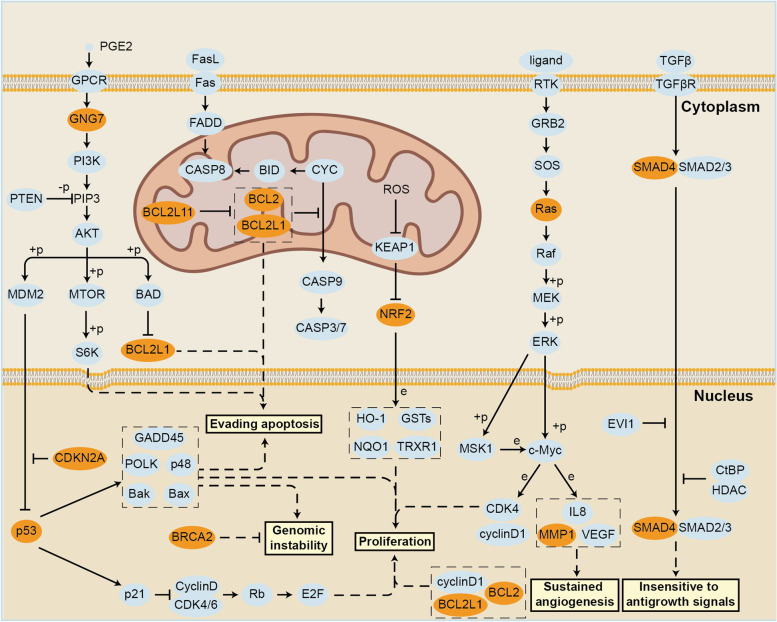
CRGs are located in several important tumor-related signaling pathways and play a critical role (Fig. 5), the exploration of which could provide possible guidance for future studies. Although so many CRGs were involved in the construction of the risk model, only a few genes were involved in the construction of different signatures, which may play a more important role. Dihydrolipoamide Branched Chain Transacylase E2(DBT) catalyzed α-keto acid to acyl-CoA, which is thought to be associated with protective effects in both LUAD and cervical cancer(CC) patients [159, 164]. Cytochrome c oxidase assembly factor 19(COX19) was involved in SCO1-dependent signaling essential for copper homeostasis [185], which was thought to be a risk factor in both gliomas and low-grade gliomas [140, 142]. Pituitary tumor-transforming gene 1 protein(PTTG1), a key regulator of the p53/TP53 pathway and DNA repair, has been considered as a risk factor in studies of HCC by different investigators [133, 135]. Surfeit 4(SURF4) is a regulator of lipoprotein export, and its high expression leads to poor prognosis of patients with BRCA [145, 147]. In addition, Enamelin(ENAM) and WD repeat domain 72(WDR72) were considered to be both associated with enamel construction in previous studies [186] and as risk or protective factors in studies on KIRC, respectively [149, 150]. MKLN1-AS and AL031985.3 were both considered risk factors for patients with hepatocellular carcinoma [136, 163], FOXD2-AS1 and LINC02154 were both considered risk factors for KIRC patients [175–177], AL513550.1 was suggested as a risk factor for CRC by different investigators [121, 172], and AL132800.1 was thought to be a risk factor for HNSCC [178, 179]. C6orf99 was regarded as a risk factor for patients with OSCC and HNSCC, two tumors with similar pathological types [152, 173]. In contrast, upregulation of AC090587.1 and AC012313 was identified to be associated with a better prognosis in HNSCC patients [178, 179], while AC073896.3 was used as a protective factor in the prognostic analysis of CRC patients [121, 172]. Certain lncRNAs were also hinted that may serve as protective or risk factors in different prognostic signatures of the same or different tumors, respectively. AC026979.4 was considered as a risk or protective factor among the different prognostic signatures of CRC patients [121, 172], and upregulation of LINC01150 was associated with better or worse prognosis in the prognostic analyses of STAD patients and CM patients [168, 181]. These possible relationships need to be verified by deeper mechanistic studies and multi-omics level analysis.
Fig. 5.
Cuproptosis related genes and cancer signaling pathways. Cuproptosis related genes occupy an important position in the tumor signaling pathways which have significant relevance to various processes of cancers, including proliferation, genomic instability, evading apoptosis, sustained angiogenesis and insensitive to antigrowth signals. The molecules linked together to accomplish the function by creating the complex, while the molecules framed by the dotted line cooperate to achieve the function. Essential genes in CRG-related cancer pathways and associated cancer processes are listed, CRGs are marked in orange
Except for the models constructed by survival analysis described above, there are also cuproptosis related signatures constructed by other methods, which was demonstrated that can also predict clinical prognosis for patients with bladder cancer and KIRC [187, 188]. As a programmed cell death, several studies have demonstrated that cuproptosis may interact with necroptosis or ferroptosis, and constructed risk models by co-opting cuproptosis-related genes with necroptosis- or ferroptosis-related genes, respectively. In a study on the prognostic risk of LGG patients, cuproptosis-related genes were analyzed by cox regression together with necroptosis-related genes, and the five genes that contributed most to the model construction were selected to construct the prognostic signature, however, these 5 genes were all considered to be necroptosis related in previous studies [189]. In the study on CRC, investigators selected cuproptosis-related genes together with ferroptosis-related genes to construct a prognostic signature, however, the contribution of each gene to the model was not described [190].
In addition to patient prognosis, the researchers have also focused on the correlation between CRGs and other cancer features like the tumor microenvironment. Less immune activation, lower levels of immune cells infiltration or worse TME scores were prevalent in patients of high-risk groups except for patients suffering gliomas [140, 141, 143, 169]. Moreover, some researchers have also focused on the expression levels of immune checkpoint genes(ICGs) such as PD-1, which are thought to associated with tumor immunosuppression and immunotherapy [191]. In most studies, ICGs were consistently highly expressed in the high-risk score group, however, the expression levels of were lower in the high-risk group of CC, BRCA and skin cutaneous melanoma(SKCM) [164, 167, 168, 174]. Most investigators performed functional enrichment analysis of CRGs based on GO and KEGG analysis, which are often highly correlated with metabolism and immune.
Although a growing number of researchers aim to construct cuproptosis-related gene signatures to predict tumor prognosis, only a few of them have been biologically validated in cell lines or clinical samples, detected by qRT-PCR, WB or IHC. Genes cited as risk factors tend to be expressed at higher levels in tumor cell lines and in patient tumor samples. However, the expression levels of risk factors in tumor cell lines and clinical tumor tissues in CRC showed inverse levels between the two [171], which may be due to the distinct tumor microenvironment of tumor cells from cell lines and clinical samples. Future studies on the cuproptosis should take note of the impact of this difference in the tumor microenviroment.
As one of the emerging therapeutic approaches, immunotherapy is often used among patients with advanced stage cancer or metastases [192]. Previous research has established that copper can modulate the expression of PD-L1, the important immune checkpoint gene crucial for immunotherapy [193], whereas the relationship between the CRGs and the efficacy of immunotherapy has been of great interest to researchers. However, most of these researchers predict the efficacy indirectly by the expression level of immune checkpoint genes or TIDE scores, only in some studies, investigators used the immunotherapy cohorts from the public databases to directly evaluate the prognosis of patients under immunotherapy. In most studies validated using immunotherapy cohorts, the low-risk group had a better prognosis for immunotherapy [152, 156, 158, 173, 194], except for those on glioma [140, 141]. Moreover, in the study on UVM, it has been found that immunotherapy was ineffective in both populations [139]. Due to the lack of biological evidence, it remains unclear what role these cuproptosis-related genes might play in the immunotherapeutic process of patients.
Some researchers have also focused on the predictive effect of cuproptosis related genetic signatures on chemotherapy efficacy. It is noteworthy that bortezomib, one of the PIs, was found to have better efficacy in different risk groups of cancers [165, 168], which further indicated the relationship between PIs and cuproptosis. However, these studies on drug sensitivity were not verified by preclinical experimental studies.
Cuproptosis and potential cancer treatment
The discovery of the mechanism of cuproptosis provides a direction for future drug research, and copper ionophores, or called cuproptosis related drugs, that can induce the cuproptosis may have some application prospects in the future treatment of cancers [195]. ES and DSF can induce cell death by translocating copper ions into cells and mitochondria, further leading to DLAT oligomerization, reduced Fe-S stability and interaction with Npl4.
Some antimicrobial drugs also serve as copper ionophores, which inhibit the growth of microorganisms by elevating intracellular copper ion concentration. Zinc pyrithione mediated copper influx can inhibit the growth of yeast [196], 4-Br-A23187 and Dimethyldithiocarbamate can increase cell copper level and play an antibacterial role [197, 198]. Lipographic copper containing complexes with bis (thiosemicarbazone) ligands can also increase the concentration of copper ions in cancer cells and host cells of chlamydial [199, 200]. Moreover, derivatives of quinolines are also considered to have the role of copper ionophores, and their modification can change their properties for better performance [201–203]. Derivatives obtained by modification of simple compounds like 3-Hydroxyflavone [204] as well as more complex copper ionophores such as Hydrophilic Temperature-Sensitive Liposome [205] and the copper ionophore designed based on salicylaldehyde isonicotinoyl hydrazone [206] can also elevate intracellular copper levels.
Among these drugs that elevate cellular copper ion concentration, DSF and ES have received the most attention and were subjected to clinical trials. In current most clinical trials on ES and DSF, both were not found to be clinically beneficial in unselected populations but the safety of both was evaluated comprehensively (Table 3). DSF, an FDA-approved drug for the treatment of alcohol dependence, is recommended at an average dose of 125 to 500 mg per day and is well tolerated by patients [17]. ES was first developed as an anti-tumor drug, although it has not shown good effect in a past clinical trial [207], its safety has also been proven. Nanomedicines combining copper ions with copper ionophores are currently being extensively studied, and the targeting of tumors makes it possible to achieve more precise tumor killing through cuproptosis [208, 209].
Table 3.
Clinical trials for copper ionophores
| NCT Number | Status | Phases | Enrollment | Conditions | Drugs | Result | Ref |
|---|---|---|---|---|---|---|---|
| NCT00742911 | Completed | Phase 1 | 21 | solid tumors, hepatic metastases | DSF, Copper Gluconate | Disulfiram 250 mg daily was well tolerated and no objective remission was observed | [210] |
| NCT01907165 | Completed | Early Phase 1 | 21 | Glioblastoma | Temozolomide, DSF, Copper gluconate | Disulfiram combined with TMZ therapy has an acceptable safety profile and can improve PFS | [211] |
| NCT00256230 | Completed | Phase 1, Phase 2 | 7 | Stage IV Melanoma | DSF | / | Unpublished |
| NCT02770378 | Completed | Phase 1, Phase 2 | 10 | Glioblastoma | Temozolomide, Aprepitant, Minocycline, DSF, Celecoxib, Sertraline, Captopril, Itraconazole, Ritonavir, Auranofin | Nine drug combinations, including DSF, can be applied safely with careful monitoring | [212] |
| NCT03714555 | Completed | Phase 2 | 1 | Metastatic Pancreatic Cancer |
nab-paclitaxel /gemcitabine Protocol Plus DSF/Copper Gluconate FOLFIRINOX regimen Plus DSF/Copper Gluconate Single-agent gemcitabine regimen Plus DSF/Copper Gluconate |
/ | Unpublished |
| NCT02101008 | Completed | Phase 2 | 12 | Melanoma | DSF and chelated zinc | / | Unpublished |
| NCT03034135 | Completed | Phase 2 | 23 | Recurrent Glioblastoma | DSF/Copper, Temozolomide (TMZ) | DSF combined with TMZ treatment is well tolerated but has limited effect on unselected populations | [213] |
| NCT02678975 | Completed | Phase 2, Phase 3 | 88 | Glioma, Glioblastoma | DSF, Copper, Alkylating Agents | DSF combined with Alkylating treatment has limited risk profile | [214] |
| NCT00312819 | Completed | Phase 2, Phase 3 | 60 | Non-small Cell Lung Cancer | chemotherapy ± DSF | DSF in combination with cisplatin and vincristine is well tolerated and may prolong patient survival | [215] |
| NCT01118741 | Completed | / | 19 | Prostate Cancer | DSF | DSF treatment is tolerated but has no clinical benefit | [216] |
| NCT00571116 | Terminated | Phase 1 | 9 | Metastatic Melanoma | DSF, Arsenic trioxide | / | Unpublished |
| NCT02963051 | Terminated | Phase 1 | 9 | Prostate Cancer | Copper, DSF, Copper gluconate | No significant PSA decline with imaging response was observed | [217] |
| NCT03151772 | Terminated | Early Phase 1 | 3 | Glioblastoma | DSF, Metformin | / | Unpublished |
| NCT00808418 | Completed | Phase 1 | 34 | Prostate Cancer | ES, Docetaxel | / | Unpublished |
| NCT00088114 | Completed | Phase 1 | 50 | Neoplasms | ES, paclitaxel | / | Unpublished |
| NCT00827203 | Suspended | Phase 1 | 30 | Metastatic Solid Tumors | ES | ES treatment in combination with paclitaxel was well tolerated, with a toxicity profile consistent with that of paclitaxel alone | [218] |
| NCT00084214 | Completed | Phase 1, Phase 2 | 103 | Melanoma | ES, Paclitaxel | ES combined with paclitaxel treatment doubled median PFS with an acceptable toxicity profile | [219] |
| NCT00088088 | Completed | Phase 1, Phase 2 | 86 | Stage IIIB/IV Non-Small Cell Lung Cancer | Paclitaxel, Carboplatin, ES | / | Unpublished |
| NCT00888615 | Completed | Phase 2 | 58 | Primary and recurrent fallopian tube cancer, primary and recurrent ovarian cancer, primary and recurrent primary peritoneal cancer | ES, Paclitaxel | ES combined with paclitaxel was well tolerated, but its response rate was inadequate | [220] |
| NCT00087997 | Completed | Phase 2 | 80 | Soft Tissue Sarcoma | ES | ES enhances the efficacy of taxane through the action of HSP70 | [221] |
| NCT00522834 | Terminated | Phase 3 | 630 | Melanoma | ES, Paclitaxel | ES combined with paclitaxel did not significantly improve PFS, and combination therapy improved PFS in patients with normal serum LDH levels | [207] |
There is a significant correlation between cuproptosis caused by copper ionophores and the level of mitochondrial metabolism, which should be considered comprehensively during future studies of exploring possible drugs that rely on cuproptosis for cancers treatment. Certain tumors inherently exhibit higher levels of mitochondrial metabolism, such as melanoma, breast cancer and leukemia [5]. Some cancer stem cell-like cells among cancers like glioblastoma [7] and cholangiocarcinoma [8] also exhibit higher levels of aerobic respiration. some drug-resistant tumors exhibit a high mitochondrial metabolic state [9–13]. Higher levels of mitochondrial respiration were demonstrated in certain drug-resistant tumors treated with chemotherapy by cisplatin [9] or 5-fluorouracil [11] or target therapy by anti-EGFR [12] or anti-BCL-2 [13], which were also thought to be associated with drug resistance. Therefore, copper ionophores may be used in combination with small molecule targeting agents that act on EGFR or BCL-2 to achieve better clinical outcomes, which needs further clinical trials. Tumor cells treated with certain antitumor drugs such as PI have also been found to promote the transformation of tumor cells to a high mitochondrial metabolic state [14, 15], combination with which may lead to better results in cuproptosis related therapy. In conclusion, copper ionophores may be more effective in tumors with higher levels of mitochondrial metabolism, and these patients may be the potential beneficiaries of future cuproptosis-inducing therapy, and treatment by other drugs inducing high mitochondrial respiratory state of the tumor in combination with copper ionophores is also a potential therapeutic direction. In the phase III clinical trial of ES, there was no statistical difference in efficacy between the experimental and control groups. However, among patients with low serum LDH levels the effect of ES differed between the two groups [207]. In future practical clinical use, serum LDH levels may be used as an indicator of whether or not to treat with cuproptosis related drugs and to determine the likely efficacy of these drugs. Moreover, Researchers are also focused on investigating new copper ionophores as drugs to target tumors [222]. Copper ionophores have different physicochemical properties and may be used in cancers with different characteristics. In addition to copper ionophores, copper complexes can also increase intracellular copper ion concentrations leading to cancer cell death [84], may also be used in the future as a cuproptosis-related treatment. Also noteworthy in the development of copper ionophores for clinical therapeutic use is the fact that relatively small changes in their structure can cause changes in properties and functions, like the different derivatives of bis(N4-methylthiosemicarbazone) [223, 224] or quinolines [225, 226]. In conclusion, copper ionophores can be used in combination with targeted therapeutic agents such as TKI and PI, which should be used in tumors with a high mitochondrial metabolic status, and LDH may be used as a predictor and prognostic indicator to guide treatment before and after drug administration, respectively. Further work is required to determine the viability of using cuproptosis-related therapy in certain patients with specific cancers.
Conclusion and future perspectives
Copper is essential for cell life, yet its excess has also been found to cause cell death [37]. As the concept of cuproptosis was introduced, a large number of publications in the form of research highlight described it [1, 2]. Some researcher also focused on copper ionophores which played an important role in the discovery of cuproptosis and reviewed them [71]. However, there is still no review systematically addressing this field.
The discovery of the copper death concept relied on the study of copper ionophores that have antitumor effects. Among the studies on copper ionophores, it is commonly observed that these ionophores cause cell death by importing copper [74]. Yet, the mechanism of copper-induced cell death [7, 14, 82] and the exact manner in which it occurs [72, 73, 89, 90] has been controversial in the past until the convincing mechanism of cuproptosis was proposed in 2022 [4]. Cuproptosis was described that showed close relationship with mitochondrial respiration level and LA pathway. However, the mechanism of cuproptosis remains to be further explored. On the one hand, the specific pathways of action of key factors such as FDX1 remain unexplored. On the other hand, certain mechanism through which cuproptosis is inhibited among normal cells have not been clearly described. Furthermore, the relationship between several possible mechanisms thought to contribute to copper induced cell death in past studies also need to be further clarified [14, 81, 82]. Finally, characteristic changes in cells undergoing cuproptosis at the cellular morphological and molecular levels have also not been described.
With the introduction of the concept of cuproptosis, an increasing number of researchers are intent on exploring its relationship with tumors. Studies on the relationship have covered most common cancer types and analyzed the links between CRGs and various aspects of tumor characteristics. However, these studies have only indirectly demonstrated the links between cuproptosis and cancer due to the insufficient biological evidence and experimental validation, whether these genes play a direct role in the relationship between cuproptosis and tumors or receive indirect effects from both is still unknown. Several genes have been identified by different investigators in these studies to construct signatures that may play a more significant role in the association between cuproptosis and cancers. Further studies, which examine the relationship between these repeatedly mentioned CRGs and cuproptosis, will need to be undertaken.
Past studies on copper have also provided a sufficient basis for future studies on the specific mechanisms of cuproptosis and its relationship with tumors. A variety of drugs have been identified in past studies to elevate intracellular copper ion concentrations including the aforementioned copper ionophores, copper ion complexes, and many copper chelators that block the elevation of copper ion concentrations such as triethylenetetramine dihydrochloride [227], 8-hydroxyquinoline [228], penicillamine [229], methanobactin [230], Bathocuproine disulphonate [231] and choline tetrathiomolybdate [232]. Through these substances, the intracellular concentration of copper ions can be regulated to induce or suppress the occurrence of cuproptosis. Researchers have been always focusing on their clinical antitumor effects [207], and their safety has been verified by sufficient clinical trials. In future cuproptosis-inducing therapies, patient populations that may be benefited should be preemptively evaluated, as certain cancers with higher levels of mitochondrial metabolism [5], certain cancers with tumor stem cell-like cells [7, 8], and certain drug-resistant cancers may have better efficacy. Proteasome inhibitors [14] can induce a high mitochondrial metabolic state in cells, with which the combination of copper ionophores for tumor therapy is also a possible future application direction. On the one hand, the concept of cuproptosis has brought the current investigations on the mechanism of copper-induced cell death to a new milestone, and there is sufficient basement for the future mechanistic studies on cuproptosis. On the other hand, there has been a large amount of exploration on the indirect relationship between cuproptosis and tumors, which provides guidance for future studies on the direct relationship of both. In summary, it has been shown that cuproptosis is an independent and novel cell death pattern. However, the specific mechanism by which cuproptosis occurred and the links between cuproptosis and cancer still needs further study.
Abbreviations
- Acetyl-CoA
Acetyl coenzyme A
- ARID1A
AT-rich interaction domain 1A
- ATG13
Autophagy related 13
- ATK
Agammaglobulinaemia tyrosine kinase
- ATOX1
Antioxidant 1 copper chaperone
- ATP7A
ATPase copper taransporting alpha
- ATP7B
ATPase copper transporting beta
- BRAF
B-Raf proto-oncogene
- BRCA
Breast cancer
- CC
Cervical cancer
- CCS
Copper chaperone for superoxide dismutase
- CDKN2A
Cyclin dependent kinase inhibitor 2A
- CESC
Cervical squamous cell carcinoma
- CKGs
Cuproptosis key genes
- COA6
Cytochrome c oxidase assembly factor 6
- COX
Cytochrome c oxidase
- COX19
Cytochrome c oxidase assembly factor 19
- CP
Ceruloplasmin
- CRC
Colorectal cancer
- CRGs
Cuproptosis related genes
- CTR1
Copper transporter 1
- DBT
Dihydrolipoamide branched chain transacylase E2
- DFS
Disease free survival
- DLAT
Dihydrolipoamide s-acetyltransferase
- DLD
Dihydrolipoamide dehydrogenase
- DSF
Disulfiram
- ENAM
Enamelin
- ERK
Extracellular regulated protein kinases
- ES
Elesclomol
- ETC
Electron transport chain
- FDX1
Ferredoxin 1
- Fe-S cluster
Iron-sulfur cluster
- FoxO1a
Forkhead box O1a
- FoxO4
Forkhead box O4
- GBM
Glioblastoma
- GLS
Glutaminase
- GSH
Glutathione
- HCC
Hepatocellular carcinoma
- HIF-1
Hypoxia inducible factor 1
- HIF-1α
Hypoxia inducible factor 1 subunit alpha
- HNSC
Head and neck squamous cell carcinoma
- ICGs
Immune checkpoint genes
- JNK
C-Jun N-terminal kinase
- KIRC
Clear cell renal cell carcinoma
- LA
Lipoic acid
- LGG
Low grade glioma
- LIAS
Lipoic acid synthetase
- LIPT1
Lipoyltransferase 1
- LUAD
Lung adenocarcinoma
- MAPK
Mitogen-activated protein kinase
- MEK1
Mitogen-activated proteinkinase kinase 1
- MT1/2
Metallothionein1/2
- MTF1
Metal regulatory transcription factor 1
- NAC
N-Acetylcysteine
- OS
Overall survival
- PAAD
Pancreatic adenocarcinoma
- PDC
Pyruvate dehydrogenase complex
- PDE3B
Phosphodiesterase 3B
- PDHA1
Pyruvate dehydrogenase e1 subunit alpha 1
- PDHB
Pyruvate dehydrogenase e1 subunit beta
- PI
Proteasome inhibitors
- PI3K
Phosphoinositide-3-kinase
- PTTG1
Pituitary tumor-transforming gene 1 protein
- ROS
Reactive oxygen species
- RTK
Receptor tyrosine kinase
- SCO1/2
Cytochrome c oxidase1/2
- SKCM
Skin cutaneous melanoma
- SLC25A3
Solute carrier family 25 member 3
- SLC31A1
Solute carrier family 31 member 1
- SOD1
Superoxide dismutase 1
- SURF4
Surfeit 4
- TTM
Tetrathiomolybdate
- ULK
Unc-51 like autophagy activating kinase
- UVM
Uveal melanoma
- VEGF
Vascular endothelial growth factor
- WDR72
WD repeat domain 72
- XIAP
X-linked inhibitor of apoptosis
Authors’ contributions
Y.G. and J.H. conceived the projects and revised the manuscript. J.X. and Y.Y. wrote the manuscript and summarized the table and prepared figures. All authors reviewed the manuscript and approved the final manuscript.
Funding
This work was supported by the National Key R&D Program of China (2021YFC2501900), National Natural Science Foundation of China (82122053, 82188102), the Beijing Municipal Science & Technology Commission (Z191100006619118), R&D Program of Beijing Municipal Education Commission(KJZD20191002302), CAMS Innovation Fund for Medical Sciences (CIFMS) (2021-I2M-1–012, 2021-I2M-1–067), Non-profit Central Research Institute Fund of Chinese Academy of Medical Sciences (2021-PT310-001, 2021-RC310-020), Key-Area Research and Development Program of Guangdong Province (2021B0101420005), Aiyou Foundation (KY201701), Shenzhen High-level Hospital Construction Fund, and Sanming Project of Medicine in Shenzhen, Shenzhen Science and Technology Program (ZDSYS20220606101604009).
Availability of data and materials
Not applicable.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
This manuscript has been read and approved by all authors and is not under consideration for publication elsewhere.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Jiaming Xie and Yannan Yang contributed equally to this work.
Contributor Information
Yibo Gao, Email: gaoyibo@cicams.ac.cn.
Jie He, Email: hejie@cicams.ac.cn.
References
- 1.Wang Y, Zhang L, Zhou F. Cuproptosis: a new form of programmed cell death. Cell Mol Immunol China. 2022;19:867–868. doi: 10.1038/s41423-022-00866-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tang D, Chen X, Kroemer G. Cuproptosis: a copper-triggered modality of mitochondrial cell death. Cell Res England. 2022;32:417–418. doi: 10.1038/s41422-022-00653-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Festa RA, Thiele DJ. Copper: an essential metal in biology. Curr Biol. 2011;21:R877–883. doi: 10.1016/j.cub.2011.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsvetkov P, Coy S, Petrova B, Dreishpoon M, Verma A, Abdusamad M, et al. Copper induces cell death by targeting lipoylated TCA cycle proteins. Science. 2022;375:1254–1261. doi: 10.1126/science.abf0529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Porporato PE, Filigheddu N, Pedro JMB-S, Kroemer G, Galluzzi L. Mitochondrial metabolism and cancer. Cell Res. 2018;28:265–80. doi: 10.1038/cr.2017.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sriskanthadevan S, Jeyaraju DV, Chung TE, Prabha S, Xu W, Skrtic M, et al. AML cells have low spare reserve capacity in their respiratory chain that renders them susceptible to oxidative metabolic stress. Blood. 2015;125:2120–2130. doi: 10.1182/blood-2014-08-594408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buccarelli M, D’Alessandris QG, Matarrese P, Mollinari C, Signore M, Cappannini A, et al. Elesclomol-induced increase of mitochondrial reactive oxygen species impairs glioblastoma stem-like cell survival and tumor growth. J Exp Clin Cancer Res. 2021;40:228. doi: 10.1186/s13046-021-02031-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raggi C, Taddei ML, Sacco E, Navari N, Correnti M, Piombanti B, et al. Mitochondrial oxidative metabolism contributes to a cancer stem cell phenotype in cholangiocarcinoma. J Hepatol Netherlands. 2021;74:1373–1385. doi: 10.1016/j.jhep.2020.12.031. [DOI] [PubMed] [Google Scholar]
- 9.Cruz-Bermúdez A, Laza-Briviesca R, Vicente-Blanco RJ, García-Grande A, Coronado MJ, Laine-Menéndez S, et al. Cisplatin resistance involves a metabolic reprogramming through ROS and PGC-1α in NSCLC which can be overcome by OXPHOS inhibition. Free Radic Biol Med United States. 2019;135:167–181. doi: 10.1016/j.freeradbiomed.2019.03.009. [DOI] [PubMed] [Google Scholar]
- 10.Matassa DS, Amoroso MR, Lu H, Avolio R, Arzeni D, Procaccini C, et al. Oxidative metabolism drives inflammation-induced platinum resistance in human ovarian cancer. Cell Death Differ. 2016;23:1542–1554. doi: 10.1038/cdd.2016.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Denise C, Paoli P, Calvani M, Taddei ML, Giannoni E, Kopetz S, et al. 5-fluorouracil resistant colon cancer cells are addicted to OXPHOS to survive and enhance stem-like traits. Oncotarget. 2015;6:41706–41721. doi: 10.18632/oncotarget.5991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Rosa V, Iommelli F, Monti M, Fonti R, Votta G, Stoppelli MP, et al. Reversal of Warburg Effect and Reactivation of Oxidative Phosphorylation by Differential Inhibition of EGFR Signaling Pathways in Non-Small Cell Lung Cancer. Clin Cancer Res United States. 2015;21:5110–5120. doi: 10.1158/1078-0432.CCR-15-0375. [DOI] [PubMed] [Google Scholar]
- 13.Lagadinou ED, Sach A, Callahan K, Rossi RM, Neering SJ, Minhajuddin M, et al. BCL-2 inhibition targets oxidative phosphorylation and selectively eradicates quiescent human leukemia stem cells. Cell Stem Cell. 2013;12:329–341. doi: 10.1016/j.stem.2012.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsvetkov P, Detappe A, Cai K, Keys HR, Brune Z, Ying W, et al. Mitochondrial metabolism promotes adaptation to proteotoxic stress. Nat Chem Biol. 2019;15:681–689. doi: 10.1038/s41589-019-0291-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luengo A, Gui DY, Vander Heiden MG. Targeting Metabolism for Cancer Therapy. Cell Chem Biol. 2017;24:1161–1180. doi: 10.1016/j.chembiol.2017.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hasinoff BB, Wu X, Yadav AA, Patel D, Zhang H, Wang D-S, et al. Cellular mechanisms of the cytotoxicity of the anticancer drug elesclomol and its complex with Cu(II) Biochem Pharmacol England. 2015;93:266–276. doi: 10.1016/j.bcp.2014.12.008. [DOI] [PubMed] [Google Scholar]
- 17.Jiao Y, Hannafon BN, Ding WQ. Disulfiram’s Anticancer Activity: Evidence and Mechanisms. Anticancer Agents Med Chem. 2016;16:1378–1384. doi: 10.2174/1871520615666160504095040. [DOI] [PubMed] [Google Scholar]
- 18.Cobine PA, Moore SA, Leary SC. Getting out what you put in: copper in mitochondria and its impacts on human disease. Biochim Biophys Acta Mol Cell Res. 2021;1868:118867. doi: 10.1016/j.bbamcr.2020.118867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bost M, Houdart S, Oberli M, Kalonji E, Huneau J-F, Margaritis I. Dietary copper and human health: current evidence and unresolved issues. J Trace Elem Med Biol. Germany. 2016;35:107–115. doi: 10.1016/j.jtemb.2016.02.006. [DOI] [PubMed] [Google Scholar]
- 20.Lönnerdal B. Intestinal regulation of copper homeostasis: a developmental perspective. Am J Clin Nutr. United States. 2008;88:846S–S850. doi: 10.1093/ajcn/88.3.846S. [DOI] [PubMed] [Google Scholar]
- 21.Kirsipuu T, Zadorožnaja A, Smirnova J, Friedemann M, Plitz T, Tõugu V, et al. Copper(II)-binding equilibria in human blood. Sci Rep. 2020;10:5686. doi: 10.1038/s41598-020-62560-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cabrera A, Alonzo E, Sauble E, Chu YL, Nguyen D, Linder MC, et al. Copper binding components of blood plasma and organs, and their responses to influx of large doses of (65)Cu, in the mouse. Biometals. 2008;21:525–543. doi: 10.1007/s10534-008-9139-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lutsenko S. Dynamic and cell-specific transport networks for intracellular copper ions. J Cell Sci. 2021;134:jcs240523. doi: 10.1242/jcs.240523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hernandez S, Tsuchiya Y, García-Ruiz JP, Lalioti V, Nielsen S, Cassio D, et al. ATP7B copper-regulated traffic and association with the tight junctions: copper excretion into the bile. Gastroenterology. United States. 2008;134:1215–1223. doi: 10.1053/j.gastro.2008.01.043. [DOI] [PubMed] [Google Scholar]
- 25.Wang W, Wang X, Luo J, Chen X, Ma K, He H, et al. Serum Copper Level and the Copper-to-Zinc Ratio Could Be Useful in the Prediction of Lung Cancer and Its Prognosis: A Case-Control Study in Northeast China. Nutr Cancer. United States. 2021;73:1908–1915. doi: 10.1080/01635581.2020.1817957. [DOI] [PubMed] [Google Scholar]
- 26.Saleh SAK, Adly HM, Abdelkhaliq AA, Nassir AM. Serum Levels of Selenium, Zinc, Copper, Manganese, and Iron in Prostate Cancer Patients. Curr Urol. 2020;14:44–49. doi: 10.1159/000499261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pavithra V, Sathisha TG, Kasturi K, Mallika DS, Amos SJ, Ragunatha S. Serum levels of metal ions in female patients with breast cancer. J Clin Diagn Res. 2015;9:BC25–c27. doi: 10.7860/JCDR/2015/11627.5476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Basu S, Singh MK, Singh TB, Bhartiya SK, Singh SP, Shukla VK. Heavy and trace metals in carcinoma of the gallbladder. World J Surg. United States. 2013;37:2641–2646. doi: 10.1007/s00268-013-2164-9. [DOI] [PubMed] [Google Scholar]
- 29.Yaman M, Kaya G, Yekeler H. Distribution of trace metal concentrations in paired cancerous and non-cancerous human stomach tissues. World J Gastroenterol. 2007;13:612–618. doi: 10.3748/wjg.v13.i4.612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kosova F, Cetin B, Akinci M, Aslan S, Seki A, Pirhan Y, et al. Serum copper levels in benign and malignant thyroid diseases. Bratisl Lek Listy. Slovakia. 2012;113:718–720. doi: 10.4149/bll_2012_162. [DOI] [PubMed] [Google Scholar]
- 31.Yu Z, Zhou R, Zhao Y, Pan Y, Liang H, Zhang JS, et al. Blockage of SLC31A1-dependent copper absorption increases pancreatic cancer cell autophagy to resist cell death. Cell Prolif. 2019;52:e12568. doi: 10.1111/cpr.12568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ohgami RS, Campagna DR, McDonald A, Fleming MD. The Steap proteins are metalloreductases. Blood. 2006;108:1388–1394. doi: 10.1182/blood-2006-02-003681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kar S, Sen S, Maji S, Saraf D, Ruturaj, Paul R, et al. Copper(II) import and reduction are dependent on His-Met clusters in the extracellular amino terminus of human copper transporter-1. J Biol Chem. 2022;298:101631. doi: 10.1016/j.jbc.2022.101631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu X, Boulet A, Buckley KM, Phillips CB, Gammon MG, Oldfather LE, et al. Mitochondrial copper and phosphate transporter specificity was defined early in the evolution of eukaryotes. Elife. 2021;10:e64690. doi: 10.7554/eLife.64690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.La Fontaine S, Ackland ML, Mercer JFB. Mammalian copper-transporting P-type ATPases, ATP7A and ATP7B: emerging roles. Int J Biochem Cell Biol. 2010;42:206–209. doi: 10.1016/j.biocel.2009.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Rensburg MJ, van Rooy M, Bester MJ, Serem JC, Venter C, Oberholzer HM. Oxidative and haemostatic effects of copper, manganese and mercury, alone and in combination at physiologically relevant levels: an ex vivo study. Hum Exp Toxicol. England. 2019;38:419–433. doi: 10.1177/0960327118818236. [DOI] [PubMed] [Google Scholar]
- 37.Kim BE, Nevitt T, Thiele DJ. Mechanisms for copper acquisition, distribution and regulation. Nat Chem Biol. 2008;4:176–185. doi: 10.1038/nchembio.72. [DOI] [PubMed] [Google Scholar]
- 38.Casareno RL, Waggoner D, Gitlin JD. The copper chaperone CCS directly interacts with copper/zinc superoxide dismutase. J Biol Chem. United States. 1998;273:23625–23628. doi: 10.1074/jbc.273.37.23625. [DOI] [PubMed] [Google Scholar]
- 39.Furukawa Y, Torres AS, O’Halloran TV. Oxygen-induced maturation of SOD1: a key role for disulfide formation by the copper chaperone CCS. EMBO J. 2004;23:2872–2881. doi: 10.1038/sj.emboj.7600276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dong X, Zhang Z, Zhao J, Lei J, Chen Y, Li X, et al. The rational design of specific SOD1 inhibitors via copper coordination and their application in ROS signaling research. Chem Sci. 2016;7:6251–6262. doi: 10.1039/C6SC01272H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dodani SC, Leary SC, Cobine PA, Winge DR, Chang CJ. A targetable fluorescent sensor reveals that copper-deficient SCO1 and SCO2 patient cells prioritize mitochondrial copper homeostasis. J Am Chem Soc. 2011;133:8606–8616. doi: 10.1021/ja2004158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Punter FA, Adams DL, Glerum DM. Characterization and localization of human COX17, a gene involved in mitochondrial copper transport. Hum Genet. Germany. 2000;107:69–74. doi: 10.1007/s004390000339. [DOI] [PubMed] [Google Scholar]
- 43.Banci L, Bertini I, Ciofi-Baffoni S, Hadjiloi T, Martinelli M, Palumaa P. Mitochondrial copper(I) transfer from Cox17 to Sco1 is coupled to electron transfer. Proc Natl Acad Sci U S A. 2008;105:6803–6808. doi: 10.1073/pnas.0800019105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pacheu-Grau D, Wasilewski M, Oeljeklaus S, Gibhardt CS, Aich A, Chudenkova M, et al. COA6 Facilitates Cytochrome c Oxidase Biogenesis as Thiol-reductase for Copper Metallochaperones in Mitochondria. J Mol Biol. 2020;432:2067–2079. doi: 10.1016/j.jmb.2020.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Soma S, Morgada MN, Naik MT, Boulet A, Roesler AA, Dziuba N, et al. COA6 Is Structurally Tuned to Function as a Thiol-Disulfide Oxidoreductase in Copper Delivery to Mitochondrial Cytochrome c Oxidase. Cell Rep. 2019;29:4114–4126.e5. doi: 10.1016/j.celrep.2019.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ghosh A, Trivedi PP, Timbalia SA, Griffin AT, Rahn JJ, Chan SSL, et al. Copper supplementation restores cytochrome c oxidase assembly defect in a mitochondrial disease model of COA6 deficiency. Hum Mol Genet. 2014;23:3596–3606. doi: 10.1093/hmg/ddu069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Horng Y-C, Leary SC, Cobine PA, Young FBJ, George GN, Shoubridge EA, et al. Human Sco1 and Sco2 function as copper-binding proteins. J Biol Chem. United States. 2005;280:34113–34122. doi: 10.1074/jbc.M506801200. [DOI] [PubMed] [Google Scholar]
- 48.Leary SC, Cobine PA, Kaufman BA, Guercin G-H, Mattman A, Palaty J, et al. The human cytochrome c oxidase assembly factors SCO1 and SCO2 have regulatory roles in the maintenance of cellular copper homeostasis. Cell Metab. United States. 2007;5:9–20. doi: 10.1016/j.cmet.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 49.He F, Chang C, Liu B, Li Z, Li H, Cai N, et al. Copper (II) Ions Activate Ligand-Independent Receptor Tyrosine Kinase (RTK) Signaling Pathway. Biomed Res Int. 2019;2019:4158415. doi: 10.1155/2019/4158415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ostrakhovitch EA, Lordnejad MR, Schliess F, Sies H, Klotz L-O. Copper ions strongly activate the phosphoinositide-3-kinase/Akt pathway independent of the generation of reactive oxygen species. Arch Biochem Biophys. United States. 2002;397:232–239. doi: 10.1006/abbi.2001.2559. [DOI] [PubMed] [Google Scholar]
- 51.Guo J, Cheng J, Zheng N, Zhang X, Dai X, Zhang L, et al. Copper Promotes Tumorigenesis by Activating the PDK1-AKT Oncogenic Pathway in a Copper Transporter 1 Dependent Manner. Adv Sci (Weinh) 2021;8:e2004303. doi: 10.1002/advs.202004303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Walter PL, Kampkötter A, Eckers A, Barthel A, Schmoll D, Sies H, et al. Modulation of FoxO signaling in human hepatoma cells by exposure to copper or zinc ions. Arch Biochem Biophys. United States. 2006;454:107–113. doi: 10.1016/j.abb.2006.08.016. [DOI] [PubMed] [Google Scholar]
- 53.Turski ML, Brady DC, Kim HJ, Kim B-E, Nose Y, Counter CM, et al. A novel role for copper in Ras/mitogen-activated protein kinase signaling. Mol Cell Biol. 2012;32:1284–1295. doi: 10.1128/MCB.05722-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Baldari S, Di Rocco G, Heffern MC, Su TA, Chang CJ, Toietta G. Effects of Copper Chelation on BRAF(V600E) Positive Colon Carcinoma Cells. Cancers (Basel) 2019;11:659. doi: 10.3390/cancers11050659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tsang T, Posimo JM, Gudiel AA, Cicchini M, Feldser DM, Brady DC. Copper is an essential regulator of the autophagic kinases ULK1/2 to drive lung adenocarcinoma. Nat Cell Biol. 2020;22:412–424. doi: 10.1038/s41556-020-0481-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Polishchuk EV, Merolla A, Lichtmannegger J, Romano A, Indrieri A, Ilyechova EY, et al. Activation of Autophagy, Observed in Liver Tissues From Patients With Wilson Disease and From ATP7B-Deficient Animals, Protects Hepatocytes From Copper-Induced Apoptosis. Gastroenterology. United States. 2019;156:1173–1189.e5. doi: 10.1053/j.gastro.2018.11.032. [DOI] [PubMed] [Google Scholar]
- 57.Tsang T, Gu X, Davis CI, Posimo JM, Miller ZA, Brady DC. BRAFV600E-Driven Lung Adenocarcinoma Requires Copper to Sustain Autophagic Signaling and Processing. Mol Cancer Res. 2022;20:1096–1107. doi: 10.1158/1541-7786.MCR-21-0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nowell CS, Radtke F. Notch as a tumour suppressor. Nat Rev Cancer. England. 2017;17:145–159. doi: 10.1038/nrc.2016.145. [DOI] [PubMed] [Google Scholar]
- 59.Parr-Sturgess CA, Tinker CL, Hart CA, Brown MD, Clarke NW, Parkin ET. Copper modulates zinc metalloproteinase-dependent ectodomain shedding of key signaling and adhesion proteins and promotes the invasion of prostate cancer epithelial cells. Mol Cancer Res. United States. 2012;10:1282–1293. doi: 10.1158/1541-7786.MCR-12-0312. [DOI] [PubMed] [Google Scholar]
- 60.Onuma T, Mizutani T, Fujita Y, Yamada S, Yoshida Y. Copper content in ascitic fluid is associated with angiogenesis and progression in ovarian cancer. J Trace Elem Med Biol. Germany. 2021;68:126865. doi: 10.1016/j.jtemb.2021.126865. [DOI] [PubMed] [Google Scholar]
- 61.Wu Z, Zhang W, Kang YJ. Copper affects the binding of HIF-1α to the critical motifs of its target genes. Metallomics England. 2019;11:429–438. doi: 10.1039/C8MT00280K. [DOI] [PubMed] [Google Scholar]
- 62.Feng W, Ye F, Xue W, Zhou Z, Kang YJ. Copper regulation of hypoxia-inducible factor-1 activity. Mol Pharmacol. 2009;75:174–182. doi: 10.1124/mol.108.051516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Martin F, Linden T, Katschinski DM, Oehme F, Flamme I, Mukhopadhyay CK, et al. Copper-dependent activation of hypoxia-inducible factor (HIF)-1: implications for ceruloplasmin regulation. Blood United States. 2005;105:4613–4619. doi: 10.1182/blood-2004-10-3980. [DOI] [PubMed] [Google Scholar]
- 64.Liao Y, Zhao J, Bulek K, Tang F, Chen X, Cai G, et al. Inflammation mobilizes copper metabolism to promote colon tumorigenesis via an IL-17-STEAP4-XIAP axis. Nat Commun. 2020;11:900. doi: 10.1038/s41467-020-14698-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nusse R, Clevers H. Wnt/β-Catenin Signaling, Disease, and Emerging Therapeutic Modalities. Cell United States. 2017;169:985–999. doi: 10.1016/j.cell.2017.05.016. [DOI] [PubMed] [Google Scholar]
- 66.Wang L, Chai X, Wan R, Zhang H, Zhou C, Xiang L, et al. Disulfiram Chelated With Copper Inhibits the Growth of Gastric Cancer Cells by Modulating Stress Response and Wnt/β-catenin Signaling. Front Oncol. 2020;10:595718. doi: 10.3389/fonc.2020.595718. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 67.Chen GS, Chen SY, Liu ST, Hsieh CC, Lee SP, Huang SM. Stabilization of the c-Myc Protein via the Modulation of Threonine 58 and Serine 62 Phosphorylation by the Disulfiram/Copper Complex in Oral Cancer Cells. Int J Mol Sci. 2022;23:9137. doi: 10.3390/ijms23169137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Krishnamoorthy L, Cotruvo JAJ, Chan J, Kaluarachchi H, Muchenditsi A, Pendyala VS, et al. Copper regulates cyclic-AMP-dependent lipolysis. Nat Chem Biol. 2016;12:586–592. doi: 10.1038/nchembio.2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Du C, Guan X, Liu Y, Xu Z, Du X, Li B, et al. Disulfiram/copper induces antitumor activity against gastric cancer cells in vitro and in vivo by inhibiting S6K1 and c-Myc. Cancer Chemother Pharmacol Germany. 2022;89:451–458. doi: 10.1007/s00280-022-04398-3. [DOI] [PubMed] [Google Scholar]
- 70.Halliwell B, Gutteridge JM. Oxygen toxicity, oxygen radicals, transition metals and disease. Biochem J. 1984;219:1–14. doi: 10.1042/bj2190001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Oliveri V. Selective targeting of cancer cells by copper ionophores: an overview. Front Mol Biosci. 2022;9:841814. doi: 10.3389/fmolb.2022.841814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cen D, Brayton D, Shahandeh B, Meyskens FL, Farmer PJ. Disulfiram facilitates intracellular Cu uptake and induces apoptosis in human melanoma cells. J Med Chem. 2004;47:6914–6920. doi: 10.1021/jm049568z. [DOI] [PubMed] [Google Scholar]
- 73.Kirshner JR, He S, Balasubramanyam V, Kepros J, Yang CY, Zhang M, et al. Elesclomol induces cancer cell apoptosis through oxidative stress. Mol Cancer Ther. 2008;7:2319–2327. doi: 10.1158/1535-7163.MCT-08-0298. [DOI] [PubMed] [Google Scholar]
- 74.Nagai M, Vo NH, Shin Ogawa L, Chimmanamada D, Inoue T, Chu J, et al. The oncology drug elesclomol selectively transports copper to the mitochondria to induce oxidative stress in cancer cells. Free Radic Biol Med. 2012;52:2142–2150. doi: 10.1016/j.freeradbiomed.2012.03.017. [DOI] [PubMed] [Google Scholar]
- 75.Sabharwal SS, Schumacker PT. Mitochondrial ROS in cancer: initiators, amplifiers or an Achilles’ heel? Nat Rev Cancer. 2014;14:709–721. doi: 10.1038/nrc3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yadav AA, Patel D, Wu X, Hasinoff BB. Molecular mechanisms of the biological activity of the anticancer drug elesclomol and its complexes with Cu(II), Ni(II) and Pt(II) J Inorg Biochem. United States. 2013;126:1–6. doi: 10.1016/j.jinorgbio.2013.04.013. [DOI] [PubMed] [Google Scholar]
- 77.Kwan S-Y, Cheng X, Tsang YTM, Choi JS, Kwan SY, Izaguirre DI, et al. Loss of ARID1A expression leads to sensitivity to ROS-inducing agent elesclomol in gynecologic cancer cells. Oncotarget. 2016;7:56933–56943. doi: 10.18632/oncotarget.10921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rogolino D, Cavazzoni A, Gatti A, Tegoni M, Pelosi G, Verdolino V, et al. Anti-proliferative effects of copper(II) complexes with hydroxyquinoline-thiosemicarbazone ligands. Eur J Med Chem. France. 2017;128:140–153. doi: 10.1016/j.ejmech.2017.01.031. [DOI] [PubMed] [Google Scholar]
- 79.Stefani C, Al-Eisawi Z, Jansson PJ, Kalinowski DS, Richardson DR. Identification of differential anti-neoplastic activity of copper bis(thiosemicarbazones) that is mediated by intracellular reactive oxygen species generation and lysosomal membrane permeabilization. J Inorg Biochem. United States. 2015;152:20–37. doi: 10.1016/j.jinorgbio.2015.08.010. [DOI] [PubMed] [Google Scholar]
- 80.Skrott Z, Mistrik M, Andersen KK, Friis S, Majera D, Gursky J, et al. Alcohol-abuse drug disulfiram targets cancer via p97 segregase adaptor NPL4. Nature. 2017;552:194–199. doi: 10.1038/nature25016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Skrott Z, Majera D, Gursky J, Buchtova T, Hajduch M, Mistrik M, et al. Disulfiram’s anti-cancer activity reflects targeting NPL4, not inhibition of aldehyde dehydrogenase. Oncogene. England. 2019;38:6711–6722. doi: 10.1038/s41388-019-0915-2. [DOI] [PubMed] [Google Scholar]
- 82.Pan M, Zheng Q, Yu Y, Ai H, Xie Y, Zeng X, et al. Seesaw conformations of Npl4 in the human p97 complex and the inhibitory mechanism of a disulfiram derivative. Nat Commun. 2021;12:121. doi: 10.1038/s41467-020-20359-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lei W, Xu J, Ya Y, Zhang J, Hou X, Zhai Q, et al. Disulfiram-copper activates chloride currents and induces apoptosis with tyrosine kinase in prostate cancer cells. Asia Pac J Clin Oncol. 2022;18:e46–55. doi: 10.1111/ajco.13551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li DD, Yagüe E, Wang LY, Dai LL, Yang ZB, Zhi S, et al. Novel Copper Complexes That Inhibit the Proteasome and Trigger Apoptosis in Triple-Negative Breast Cancer Cells. ACS Med Chem Lett. 2019;10:1328–1335. doi: 10.1021/acsmedchemlett.9b00284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nurmamat M, Yan H, Wang R, Zhao H, Li Y, Wang X, et al. Novel Copper(II) Complex with a 4-Acylpyrazolone Derivative and Coligand Induce Apoptosis in Liver Cancer Cells. ACS Med Chem Lett. 2021;12:467–476. doi: 10.1021/acsmedchemlett.0c00680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Xia Y, Liu X, Zhang L, Zhang J, Li C, Zhang N, et al. A new Schiff base coordinated copper(II) compound induces apoptosis and inhibits tumor growth in gastric cancer. Cancer Cell Int. 2019;19:81. doi: 10.1186/s12935-019-0801-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lee ZY, Leong CH, Lim KUL, Wong CCS, Pongtheerawan P, Arikrishnan SA, et al. Induction of Apoptosis and Autophagy by Ternary Copper Complex Towards Breast Cancer Cells. Anticancer Agents Med Chem. Netherlands. 2022;22:1159–1170. doi: 10.2174/1871520621666210726132543. [DOI] [PubMed] [Google Scholar]
- 88.Gul NS, Khan TM, Chen M, Huang KB, Hou C, Choudhary MI, et al. New copper complexes inducing bimodal death through apoptosis and autophagy in A549 cancer cells. J Inorg Biochem. United States. 2020;213:111260. doi: 10.1016/j.jinorgbio.2020.111260. [DOI] [PubMed] [Google Scholar]
- 89.Gao W, Huang Z, Duan J, Nice EC, Lin J, Huang C. Elesclomol induces copper-dependent ferroptosis in colorectal cancer cells via degradation of ATP7A. Mol Oncol. 2021;15:3527–3544. doi: 10.1002/1878-0261.13079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ren X, Li Y, Zhou Y, Hu W, Yang C, Jing Q, et al. Overcoming the compensatory elevation of NRF2 renders hepatocellular carcinoma cells more vulnerable to disulfiram/copper-induced ferroptosis. Redox Biol. 2021;46:102122. doi: 10.1016/j.redox.2021.102122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Polishchuk EV, Concilli M, Iacobacci S, Chesi G, Pastore N, Piccolo P, et al. Wilson disease protein ATP7B utilizes lysosomal exocytosis to maintain copper homeostasis. Dev Cell. 2014;29:686–700. doi: 10.1016/j.devcel.2014.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sheftel AD, Stehling O, Pierik AJ, Elsässer HP, Mühlenhoff U, Webert H, et al. Humans possess two mitochondrial ferredoxins, Fdx1 and Fdx2, with distinct roles in steroidogenesis, heme, and Fe/S cluster biosynthesis. Proc Natl Acad Sci U S A. 2010;107:11775–11780. doi: 10.1073/pnas.1004250107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shi Y, Ghosh M, Kovtunovych G, Crooks DR, Rouault TA. Both human ferredoxins 1 and 2 and ferredoxin reductase are important for iron-sulfur cluster biogenesis. Biochim Biophys Acta. 2012;1823:484–492. doi: 10.1016/j.bbamcr.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cai K, Tonelli M, Frederick RO, Markley JL. Human Mitochondrial Ferredoxin 1 (FDX1) and Ferredoxin 2 (FDX2) Both Bind Cysteine Desulfurase and Donate Electrons for Iron-Sulfur Cluster Biosynthesis. Biochemistry. 2017;56:487–499. doi: 10.1021/acs.biochem.6b00447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhang C, Zeng Y, Guo X, Shen H, Zhang J, Wang K, et al. Pan-cancer analyses confirmed the cuproptosis-related gene FDX1 as an immunotherapy predictor and prognostic biomarker. Front Genet. 2022;13:923737. doi: 10.3389/fgene.2022.923737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Xiao C, Yang L, Jin L, Lin W, Zhang F, Huang S, et al. Prognostic and immunological role of cuproptosis-related protein FDX1 in pan-cancer. Front Genet. 2022;13:962028. doi: 10.3389/fgene.2022.962028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yang L, Zhang Y, Wang Y, Jiang P, Liu F, Feng N. Ferredoxin 1 is a cuproptosis-key gene responsible for tumor immunity and drug sensitivity: a pan-cancer analysis. Front Pharmacol. 2022;13:938134. doi: 10.3389/fphar.2022.938134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Huang X, Wang T, Ye J, Feng H, Zhang X, Ma X, et al. FDX1 expression predicts favourable prognosis in clear cell renal cell carcinoma identified by bioinformatics and tissue microarray analysis. Front Genet. 2022;13:994741. Available from: https://app.dimensions.ai/details/publication/pub.1151070456. [Cited 2022 Sep 24]. [DOI] [PMC free article] [PubMed]
- 99.Yun Y, Wang Y, Yang E, Jing X. Cuproptosis-Related Gene - SLC31A1, FDX1 and ATP7B - Polymorphisms are Associated with Risk of Lung Cancer. Pharmgenomics Pers Med. 2022;15:733–742. doi: 10.2147/PGPM.S372824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mayr JA, Zimmermann FA, Fauth C, Bergheim C, Meierhofer D, Radmayr D, et al. Lipoic acid synthetase deficiency causes neonatal-onset epilepsy, defective mitochondrial energy metabolism, and glycine elevation. Am J Hum Genet. 2011;89:792–797. doi: 10.1016/j.ajhg.2011.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cai Y, He Q, Liu W, Liang Q, Peng B, Li J, et al. Comprehensive analysis of the potential cuproptosis-related biomarker LIAS that regulates prognosis and immunotherapy of pan-cancers. Front Oncol. 2022;12:952129. doi: 10.3389/fonc.2022.952129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ni M, Solmonson A, Pan C, Yang C, Li D, Notzon A, et al. Functional Assessment of Lipoyltransferase-1 Deficiency in Cells, Mice, and Humans. Cell Rep. 2019;27:1376–1386.e6. doi: 10.1016/j.celrep.2019.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Soreze Y, Boutron A, Habarou F, Barnerias C, Nonnenmacher L, Delpech H, et al. Mutations in human lipoyltransferase gene LIPT1 cause a Leigh disease with secondary deficiency for pyruvate and alpha-ketoglutarate dehydrogenase. Orphanet J Rare Dis. 2013;8:192. doi: 10.1186/1750-1172-8-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yan C, Niu Y, Ma L, Tian L, Ma J. System analysis based on the cuproptosis-related genes identifies LIPT1 as a novel therapy target for liver hepatocellular carcinoma. J Transl Med. 2022;20:452. doi: 10.1186/s12967-022-03630-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Brautigam CA, Wynn RM, Chuang JL, Machius M, Tomchick DR, Chuang DT. Structural insight into interactions between dihydrolipoamide dehydrogenase (E3) and E3 binding protein of human pyruvate dehydrogenase complex. Structure. 2006;14:611–621. doi: 10.1016/j.str.2006.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Park Y-H, Patel MS. Characterization of interactions of dihydrolipoamide dehydrogenase with its binding protein in the human pyruvate dehydrogenase complex. Biochem Biophys Res Commun. 2010;395:416–419. doi: 10.1016/j.bbrc.2010.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Casteel J, Miernyk JA, Thelen JJ. Mapping the lipoylation site of Arabidopsis thaliana plastidial dihydrolipoamide S-acetyltransferase using mass spectrometry and site-directed mutagenesis. Plant Physiol Biochem. France. 2011;49:1355–1361. doi: 10.1016/j.plaphy.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 108.Deng L, Jiang A, Zeng H, Peng X, Song L. Comprehensive analyses of PDHA1 that serves as a predictive biomarker for immunotherapy response in cancer. Front Pharmacol. 2022;13:947372. doi: 10.3389/fphar.2022.947372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Brugnera E, Georgiev O, Radtke F, Heuchel R, Baker E, Sutherland GR, et al. Cloning, chromosomal mapping and characterization of the human metal-regulatory transcription factor MTF-1. Nucleic Acids Res. 1994;22:3167–3173. doi: 10.1093/nar/22.15.3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rumping L, Büttner B, Maier O, Rehmann H, Lequin M, Schlump J-U, et al. Identification of a Loss-of-Function Mutation in the Context of Glutaminase Deficiency and Neonatal Epileptic Encephalopathy. JAMA Neurol. 2019;76:342–350. doi: 10.1001/jamaneurol.2018.2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Rumping L, Tessadori F, Pouwels PJW, Vringer E, Wijnen JP, Bhogal AA, et al. GLS hyperactivity causes glutamate excess, infantile cataract and profound developmental delay. Hum Mol Genet. England. 2019;28:96–104. doi: 10.1093/hmg/ddy330. [DOI] [PubMed] [Google Scholar]
- 112.Agarwal P, Sandey M, DeInnocentes P, Bird RC. Tumor suppressor gene p16/INK4A/CDKN2A-dependent regulation into and out of the cell cycle in a spontaneous canine model of breast cancer. J Cell Biochem. United States. 2013;114:1355–1363. doi: 10.1002/jcb.24476. [DOI] [PubMed] [Google Scholar]
- 113.Riese U, Dahse R, Fiedler W, Theuer C, Koscielny S, Ernst G, et al. Tumor suppressor gene p16 (CDKN2A) mutation status and promoter inactivation in head and neck cancer. Int J Mol Med. Greece. 1999;4:61–65. doi: 10.3892/ijmm.4.1.61. [DOI] [PubMed] [Google Scholar]
- 114.Bian Z, Yu Y, Yang T, Quan C, Sun W, Fu S. Effect of tumor suppressor gene cyclin-dependent kinase inhibitor 2A wild-type and A148T mutant on the cell cycle of human ovarian cancer cells. Oncol Lett. 2014;7:1229–1232. doi: 10.3892/ol.2014.1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wu G, Peng H, Tang M, Yang M, Wang J, Hu Y, et al. ZNF711 down-regulation promotes CISPLATIN resistance in epithelial ovarian cancer via interacting with JHDM2A and suppressing SLC31A1 expression. EBioMedicine. 2021;71:103558. doi: 10.1016/j.ebiom.2021.103558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chen Z, Guo Y, Zhao D, Zou Q, Yu F, Zhang L, et al. Comprehensive Analysis Revealed that CDKN2A is a Biomarker for Immune Infiltrates in Multiple Cancers. Front Cell Dev Biol. 2021;9:808208. doi: 10.3389/fcell.2021.808208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Cheng T, Wu Y, Liu Z, Yu Y, Sun S, Guo M, et al. CDKN2A-mediated molecular subtypes characterize the hallmarks of tumor microenvironment and guide precision medicine in triple-negative breast cancer. Front Immunol. 2022;13:970950. doi: 10.3389/fimmu.2022.970950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Mangala LS, Zuzel V, Schmandt R, Leshane ES, Halder JB, Armaiz-Pena GN, et al. Therapeutic Targeting of ATP7B in Ovarian Carcinoma. Clin Cancer Res. 2009;15:3770–3780. doi: 10.1158/1078-0432.CCR-08-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Samimi G, Safaei R, Katano K, Holzer AK, Rochdi M, Tomioka M, et al. Increased expression of the copper efflux transporter ATP7A mediates resistance to cisplatin, carboplatin, and oxaliplatin in ovarian cancer cells. Clin Cancer Res. United States. 2004;10:4661–4669. doi: 10.1158/1078-0432.CCR-04-0137. [DOI] [PubMed] [Google Scholar]
- 120.Li X, Ma Z, Mei L. Cuproptosis-related gene SLC31A1 is a potential predictor for diagnosis, prognosis and therapeutic response of breast cancer. Am J Cancer Res. 2022;12:3561–3580. [PMC free article] [PubMed] [Google Scholar]
- 121.Wang L, Cao Y, Guo W, Xu J. High expression of cuproptosis-related gene FDX1 in relation to good prognosis and immune cells infiltration in colon adenocarcinoma (COAD) Germany: J Cancer Res Clin Oncol; 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Solmonson A, DeBerardinis RJ. Lipoic acid metabolism and mitochondrial redox regulation. J Biol Chem. 2018;293:7522–7530. doi: 10.1074/jbc.TM117.000259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Patel MS, Nemeria NS, Furey W, Jordan F. The pyruvate dehydrogenase complexes: structure-based function and regulation. J Biol Chem. 2014;289:16615–16623. doi: 10.1074/jbc.R114.563148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sheu KF, Hu CW, Utter MF. Pyruvate dehydrogenase complex activity in normal and deficient fibroblasts. J Clin Invest. 1981;67:1463–1471. doi: 10.1172/JCI110176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Li D, Wang C, Ma P, Yu Q, Gu M, Dong L, et al. PGC1α promotes cholangiocarcinoma metastasis by upregulating PDHA1 and MPC1 expression to reverse the Warburg effect. Cell Death Dis. 2018;9:466. doi: 10.1038/s41419-018-0494-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Randle PJ, Priestman DA, Mistry S, Halsall A. Mechanisms modifying glucose oxidation in diabetes mellitus. Diabetologia. Germany. 1994;37 Suppl 2:S155–161. doi: 10.1007/BF00400839. [DOI] [PubMed] [Google Scholar]
- 127.Sugden MC, Holness MJ. Interactive regulation of the pyruvate dehydrogenase complex and the carnitine palmitoyltransferase system. FASEB J. United States. 1994;8:54–61. doi: 10.1096/fasebj.8.1.8299890. [DOI] [PubMed] [Google Scholar]
- 128.Li L, Li L, Sun Q. High expression of cuproptosis-related SLC31A1 gene in relation to unfavorable outcome and deregulated immune cell infiltration in breast cancer: an analysis based on public databases. BMC Bioinformatics. 2022;23:350. doi: 10.1186/s12859-022-04894-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zhang H, Shi Y, Yi Q, Wang C, Xia Q, Zhang Y, et al. A novel defined cuproptosis-related gene signature for predicting the prognosis of lung adenocarcinoma. Front Genet. 2022;13:975185. Available from: https://app.dimensions.ai/details/publication/pub.1150233359. [Cited 2022 Sep 23]. [DOI] [PMC free article] [PubMed]
- 130.Hu Q, Wang R, Ma H, Zhang Z, Xue Q. Cuproptosis predicts the risk and clinical outcomes of lung adenocarcinoma. Front Oncol. 2022;12:922332. doi: 10.3389/fonc.2022.922332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Fu J, Wang S, Li Z, Qin W, Tong Q, Liu C, et al. Comprehensive multiomics analysis of cuproptosis-related gene characteristics in hepatocellular carcinoma. Front Genet. Switzerland. 2022;13:942387. doi: 10.3389/fgene.2022.942387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Liu Y, Liu Y, Ye S, Feng H, Ma L. Development and validation of cuproptosis-related gene signature in the prognostic prediction of liver cancer. Front Oncol. 2022;12:985484. Available from: https://app.dimensions.ai/details/publication/pub.1150184726. [Cited 2022 Sep 23]. [DOI] [PMC free article] [PubMed]
- 133.Wang Y, Zhang Y, Wang L, Zhang N, Xu W, Zhou J, et al. Development and experimental verification of a prognosis model for cuproptosis-related subtypes in HCC. Hepatol Int. United States. 2022;16:1435–1447. doi: 10.1007/s12072-022-10381-0. [DOI] [PubMed] [Google Scholar]
- 134.Zhang Z, Zeng X, Wu Y, Liu Y, Zhang X, Song Z. Cuproptosis-Related Risk Score Predicts Prognosis and Characterizes the Tumor Microenvironment in Hepatocellular Carcinoma. Front Immunol. 2022;13:925618. doi: 10.3389/fimmu.2022.925618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Xiao J, Liu Z, Wang J, Zhang S, Zhang Y. Identification of cuprotosis-mediated subtypes, the development of a prognosis model, and influence immune microenvironment in hepatocellular carcinoma. Front Oncol. 2022;12:941211. Available from: https://app.dimensions.ai/details/publication/pub.1150597479. [Cited 2022 Sep 24]. [DOI] [PMC free article] [PubMed]
- 136.Liu Z, Qi Y, Wang H, Zhang Q, Wu Z, Wu W. Risk model of hepatocellular carcinoma based on cuproptosis-related genes. Front Genet. 2022;13:1000652. Available from: https://app.dimensions.ai/details/publication/pub.1151029313. [Cited 2022 Sep 23]. [DOI] [PMC free article] [PubMed]
- 137.Lei L, Tan L, Sui L. A novel cuproptosis-related gene signature for predicting prognosis in cervical cancer. Front Genet. 2022;13:957744. doi: 10.3389/fgene.2022.957744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Chen Y. Identification and validation of cuproptosis-related prognostic signature and associated regulatory axis in uterine corpus endometrial carcinoma. Front Genet. 2022;13:912037. doi: 10.3389/fgene.2022.912037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Chen Y, Chen X, Wang X. Identification of a prognostic model using cuproptosis-related genes in uveal melanoma. Front Cell Dev Biol. 2022;10:973073. doi: 10.3389/fcell.2022.973073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Bao J-H, Lu W-C, Duan H, Ye Y-Q, Li J-B, Liao W-T, et al. Identification of a novel cuproptosis-related gene signature and integrative analyses in patients with lower-grade gliomas. Front Immunol. 2022;13:933973. doi: 10.3389/fimmu.2022.933973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Wang W, Lu Z, Wang M, Liu Z, Wu B, Yang C, et al. The cuproptosis-related signature associated with the tumor environment and prognosis of patients with glioma. Front Immunol 2022;13:998236. Available from: https://app.dimensions.ai/details/publication/pub.1150594801. [Cited 2022 Sep 23]. [DOI] [PMC free article] [PubMed]
- 142.Chen B, Zhou X, Yang L, Zhou H, Meng M, Zhang L, et al. A Cuproptosis Activation Scoring model predicts neoplasm-immunity interactions and personalized treatments in glioma. Comput Biol Med. United States. 2022;148:105924. doi: 10.1016/j.compbiomed.2022.105924. [DOI] [PubMed] [Google Scholar]
- 143.Ye Z, Zhang S, Cai J, Ye L, Gao L, Wang Y, et al. Development and validation of cuproptosis-associated prognostic signatures in WHO 2/3 glioma. Front Oncol. 2022;12:967159. doi: 10.3389/fonc.2022.967159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Du Y, Lin Y, Wang B, Li Y, Xu D, Gan L, et al. Cuproptosis patterns and tumor immune infiltration characterization in colorectal cancer. Front Genet. 2022;13:976007. Available from: https://app.dimensions.ai/details/publication/pub.1150956563. [Cited 2022 Sep 23]. [DOI] [PMC free article] [PubMed]
- 145.Li Z, Zhang H, Wang X, Wang Q, Xue J, Shi Y, et al. Identification of cuproptosis-related subtypes, characterization of tumor microenvironment infiltration, and development of a prognosis model in breast cancer. Front Immunol. 2022;13:996836. doi: 10.3389/fimmu.2022.996836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Sha S, Si L, Wu X, Chen Y, Xiong H, Xu Y, et al. Prognostic analysis of cuproptosis-related gene in triple-negative breast cancer. Front Immunol. 2022;13:922780. doi: 10.3389/fimmu.2022.922780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Guan X, Lu N, Zhang J. Construction of a prognostic model related to copper dependence in breast cancer by single-cell sequencing analysis. Front Genet. 2022;13:949852. Available from: https://app.dimensions.ai/details/publication/pub.1150424732. [Cited 2022 Sep 23]. [DOI] [PMC free article] [PubMed]
- 148.Bian Z, Fan R, Xie L. A Novel Cuproptosis-Related Prognostic Gene Signature and Validation of Differential Expression in Clear Cell Renal Cell Carcinoma. Genes (Basel) 2022;13:851. doi: 10.3390/genes13050851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Yuan H, Qin X, Wang J, Yang Q, Fan Y, Xu D. The cuproptosis-associated 13 gene signature as a robust predictor for outcome and response to immune- and targeted-therapies in clear cell renal cell carcinoma. Front Immunol. 2022;13:971142. Available from: https://app.dimensions.ai/details/publication/pub.1150759817. [Cited 2022 Sep 23]. [DOI] [PMC free article] [PubMed]
- 150.Ji Z-H, Ren W-Z, Wang H-Q, Gao W, Yuan B. Molecular Subtyping Based on Cuproptosis-Related Genes and Characterization of Tumor Microenvironment Infiltration in Kidney Renal Clear Cell Carcinoma. Front Oncol. 2022;12:919083. doi: 10.3389/fonc.2022.919083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Zhang S, Zhang L, Lu H, Yao Y, Liu X, Hou J. A cuproptosis and copper metabolism-related gene prognostic index for head and neck squamous cell carcinoma. Front Oncol. 2022;12:955336. doi: 10.3389/fonc.2022.955336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Ding Q, Chen X, Hong W, Wang L, Liu W, Cai S, et al. The prognostic role of cuproptosis in head and neck squamous cell carcinoma patients: a comprehensive analysis. Dis Markers. 2022;2022:9996946. doi: 10.1155/2022/9996946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Huang J, Xu Z, Yuan Z, Cheng L, Zhou C, Shen Y. Identification of cuproptosis-related subtypes and characterization of the tumor microenvironment landscape in head and neck squamous cell carcinoma. J Clin Lab Anal. 2022;36:e24638. doi: 10.1002/jcla.24638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Tang S, Zhao L, Wu XB, Wang Z, Cai LY, Pan D, et al. Identification of a Novel Cuproptosis-Related Gene Signature for Prognostic Implication in Head and Neck Squamous Carcinomas. Cancers (Basel) 2022;14:3986. doi: 10.3390/cancers14163986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Jiang R, Huan Y, Li Y, Gao X, Sun Q, Zhang F, et al. Transcriptional and genetic alterations of cuproptosis-related genes correlated to malignancy and immune-infiltrate of esophageal carcinoma. Cell Death Discov. 2022;8:370. doi: 10.1038/s41420-022-01164-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Huang X, Zhou S, Tóth J, Hajdu A. Cuproptosis-related gene index: a predictor for pancreatic cancer prognosis, immunotherapy efficacy, and chemosensitivity. Front Immunol. 2022;13:978865. doi: 10.3389/fimmu.2022.978865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Xu Y, Li H, Lan A, Wu Q, Tang Z, Shu D, et al. Cuprotosis-Related Genes: Predicting Prognosis and Immunotherapy Sensitivity in Pancreatic Cancer Patients. J Oncol. 2022;2022:2363043. Available from: https://app.dimensions.ai/details/publication/pub.1150896293. [Cited 2022 Sep 23]. [DOI] [PMC free article] [PubMed]
- 158.Liu T, Liu Q, Wang Y, Yang R, Tian F. Cuproptosis scoring model predicts overall survival and assists in immunotherapeutic decision making in pancreatic carcinoma. Front Genet. 2022;13:938488. doi: 10.3389/fgene.2022.938488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Wang F, Lin H, Su Q, Li C. Cuproptosis-related lncRNA predict prognosis and immune response of lung adenocarcinoma. World J Surg Oncol. 2022;20:275. Available from: https://app.dimensions.ai/details/publication/pub.1150679459. [Cited 2022 Sep 23]. [DOI] [PMC free article] [PubMed]
- 160.Mo X, Hu D, Yang P, Li Y, Bashir S, Nai A, et al. A novel cuproptosis-related prognostic lncRNA signature and lncRNA MIR31HG/miR-193a-3p/TNFRSF21 regulatory axis in lung adenocarcinoma. Front Oncol. 2022;12:927706. doi: 10.3389/fonc.2022.927706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Jin Z, Wang M, Meng Y, Chen D, Xu Y, Jiang X, et al. Prognostic Implication of a Cuproptosis-Related miRNA Signature in Hepatocellular Carcinoma. J Healthc Eng. England. 2022;2022:4694323. doi: 10.1155/2022/4694323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Zhang G, Sun J, Zhang X. A novel Cuproptosis-related LncRNA signature to predict prognosis in hepatocellular carcinoma. Sci Rep. 2022;12:11325. Available from: https://app.dimensions.ai/details/publication/pub.1149234217. [Cited 2022 Sep 23]. [DOI] [PMC free article] [PubMed]
- 163.Luo L, Hu X, Huang A, Liu X, Wang L, Du T, et al. A Noval Established Cuproptosis-Associated LncRNA Signature for Prognosis Prediction in Primary Hepatic Carcinoma. Evid Based Complement Alternat Med. 2022;2022:1–15. Available from: https://app.dimensions.ai/details/publication/pub.1151038751. [Cited 2022 Sep 23]. [DOI] [PMC free article] [PubMed]
- 164.Liu X, Zhou L, Gao M, Dong S, Hu Y, Hu C. Signature of seven cuproptosis-related lncRNAs as a novel biomarker to predict prognosis and therapeutic response in cervical cancer. Front Genet. 2022;13:989646. Available from: https://app.dimensions.ai/details/publication/pub.1151150251. [Cited 2022 Sep 23]. [DOI] [PMC free article] [PubMed]
- 165.Yang M, Zheng H, Xu K, Yuan Q, Aihaiti Y, Cai Y, et al. A novel signature to guide osteosarcoma prognosis and immune microenvironment: Cuproptosis-related lncRNA. Front Immunol. 2022;13:919231. doi: 10.3389/fimmu.2022.919231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Ni S, Hong J, Li W, Ye M, Li J. Construction of a cuproptosis-related lncRNA signature for predicting prognosis and immune landscape in osteosarcoma patients. Cancer Med United States. 2022. [DOI] [PMC free article] [PubMed]
- 167.Yang X, Wang X, Sun X, Xiao M, Fan L, Su Y, et al. Construction of five cuproptosis-related lncRNA signature for predicting prognosis and immune activity in skin cutaneous melanoma. Front Genet. 2022;13:972899. Available from: https://app.dimensions.ai/details/publication/pub.1150819206. [Cited 2022 Sep 23]. [DOI] [PMC free article] [PubMed]
- 168.Zhou Y, Shu Q, Fu Z, Wang C, Gu J, Li J, et al. A novel risk model based on cuproptosis-related lncRNAs predicted prognosis and indicated immune microenvironment landscape of patients with cutaneous melanoma. Front Genet. 2022;13:959456. doi: 10.3389/fgene.2022.959456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Yan X, Wang N, Dong J, Wang F, Zhang J, Hu X, et al. A cuproptosis-related lncRNAs signature for prognosis, chemotherapy, and immune checkpoint blockade therapy of low-grade glioma. Front Mol Biosci. 2022;9:966843. doi: 10.3389/fmolb.2022.966843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Wang Y, Huang X, Chen S, Jiang H, Rao H, Lu L, et al. In Silico Identification and Validation of Cuproptosis-Related LncRNA Signature as a Novel Prognostic Model and Immune Function Analysis in Colon Adenocarcinoma. Curr Oncol Switzerland. 2022;29:6573–6593. doi: 10.3390/curroncol29090517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Xu M, Mu J, Wang J, Zhou Q, Wang J. Construction and validation of a cuproptosis-related lncRNA signature as a novel and robust prognostic model for colon adenocarcinoma. Front Oncol. 2022;12:961213. doi: 10.3389/fonc.2022.961213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Hou D, Tan J-N, Zhou S-N, Yang X, Zhang Z-H, Zhong G-Y, et al. A novel prognostic signature based on cuproptosis-related lncRNA mining in colorectal cancer. Front Genet. 2022;13:969845. doi: 10.3389/fgene.2022.969845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Li X, Zhou W, Zhu C, Liu J, Ming Z, Ma C, et al. Multi-omics analysis reveals prognostic and therapeutic value of cuproptosis-related lncRNAs in oral squamous cell carcinoma. Front Genet. 2022;13:984911. doi: 10.3389/fgene.2022.984911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Jiang Z-R, Yang L-H, Jin L-Z, Yi L-M, Bing P-P, Zhou J, et al. Identification of novel cuproptosis-related lncRNA signatures to predict the prognosis and immune microenvironment of breast cancer patients. Front Oncol. 2022;12:988680. Available from: https://app.dimensions.ai/details/publication/pub.1151151046. [Cited 2022 Sep 23]. [DOI] [PMC free article] [PubMed]
- 175.Huili Y, Nie S, Zhang L, Yao A, Liu J, Wang Y, et al. Cuproptosis-related lncRNA: prediction of prognosis and subtype determination in clear cell renal cell carcinoma. Front Genet. 2022;13:958547. doi: 10.3389/fgene.2022.958547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Xin S, Mao J, Cui K, Li Q, Chen L, Li Q, et al. A cuproptosis-related lncRNA signature identified prognosis and tumour immune microenvironment in kidney renal clear cell carcinoma. Front Mol Biosci. 2022;9:974722. Available from: https://app.dimensions.ai/details/publication/pub.1150989957. [Cited 2022 Sep 23]. [DOI] [PMC free article] [PubMed]
- 177.Xu S, Liu D, Chang T, Wen X, Ma S, Sun G, et al. Cuproptosis-Associated lncRNA Establishes New Prognostic Profile and Predicts Immunotherapy Response in Clear Cell Renal Cell Carcinoma. Front Genet. 2022;13:938259. Available from: https://app.dimensions.ai/details/publication/pub.1149495871. [Cited 2022 Sep 23]. [DOI] [PMC free article] [PubMed]
- 178.Yang L, Yu J, Tao L, Huang H, Gao Y, Yao J, et al. Cuproptosis-Related lncRNAs are Biomarkers of Prognosis and Immune Microenvironment in Head and Neck Squamous Cell Carcinoma. Front Genet. 2022;13:947551. Available from: https://app.dimensions.ai/details/publication/pub.1149682363. [Cited 2022 Sep 23]. [DOI] [PMC free article] [PubMed]
- 179.Li YJ, Li HY, Zhang Q, Wei SL. The prognostic value and immune landscape of a cuproptosis-related lncRNA signature in head and neck squamous cell carcinoma. Front Genet. 2022;13:942785. doi: 10.3389/fgene.2022.942785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Yu W, Huo H, You Z, Lu R, Yao T, Huang J. Identification of cuproptosis-associated IncRNAs signature and establishment of a novel nomogram for prognosis of stomach adenocarcinoma. Front Genet. 2022;13:982888. Available from: https://app.dimensions.ai/details/publication/pub.1150882142. [Cited 2022 Sep 23]. [DOI] [PMC free article] [PubMed]
- 181.Feng A, He L, Chen T, Xu M. A novel cuproptosis-related lncRNA nomogram to improve the prognosis prediction of gastric cancer. Front Oncol. 2022;12:957966. doi: 10.3389/fonc.2022.957966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Zhang Y, Li X, Li X, Zhao Y, Zhou T, Jiang X, et al. Comprehensive analysis of cuproptosis-related long noncoding RNA immune infiltration and prediction of prognosis in patients with bladder cancer. Front Genet. 2022;13:990326. Available from: https://app.dimensions.ai/details/publication/pub.1150990568. [Cited 2022 Sep 23]. [DOI] [PMC free article] [PubMed]
- 183.Ouyang Z, Zhang H, Lin W, Su J, Wang X. Bioinformatic profiling identifies the glutaminase to be a potential novel cuproptosis-related biomarker for glioma. Front Cell Dev Biol. 2022;10:982439. Available from: https://app.dimensions.ai/details/publication/pub.1150881518. [Cited 2022 Sep 23]. [DOI] [PMC free article] [PubMed]
- 184.Liu H. Pan-cancer profiles of the cuproptosis gene set. Am J Cancer Res. 2022;12:4074–81. Available from: https://app.dimensions.ai/details/publication/pub.1148392888. [Cited 2022 Sep 23]. [PMC free article] [PubMed]
- 185.Leary SC, Cobine PA, Nishimura T, Verdijk RM, de Krijger R, de Coo R, et al. COX19 mediates the transduction of a mitochondrial redox signal from SCO1 that regulates ATP7A-mediated cellular copper efflux. Mol Biol Cell. 2013;24:683–691. doi: 10.1091/mbc.e12-09-0705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Katsura KA, Horst JA, Chandra D, Le TQ, Nakano Y, Zhang Y, et al. WDR72 models of structure and function: a stage-specific regulator of enamel mineralization. Matrix Biol. 2014;38:48–58. doi: 10.1016/j.matbio.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Song Q, Zhou R, Shu F, Fu W. Cuproptosis scoring system to predict the clinical outcome and immune response in bladder cancer. Front Immunol. 2022;13:958368. doi: 10.3389/fimmu.2022.958368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Jin L, Mei W, Liu X, Sun X, Xin S, Zhou Z, et al. Identification of cuproptosis -related subtypes, the development of a prognosis model, and characterization of tumor microenvironment infiltration in prostate cancer. Front Immunol. 2022;13:974034. Available from: https://app.dimensions.ai/details/publication/pub.1151152566. [Cited 2022 Sep 23]. [DOI] [PMC free article] [PubMed]
- 189.Miao Y, Liu J, Liu X, Yuan Q, Li H, Zhang Y, et al. Machine learning identification of cuproptosis and necroptosis-associated molecular subtypes to aid in prognosis assessment and immunotherapy response prediction in low-grade glioma. Front Genet. 2022;13:951239. Available from: https://app.dimensions.ai/details/publication/pub.1150929742. [Cited 2022 Sep 23]. [DOI] [PMC free article] [PubMed]
- 190.Li Y, Wang RY, Deng YJ, Wu SH, Sun X, Mu H. Molecular characteristics, clinical significance, and cancer immune interactions of cuproptosis and ferroptosis-associated genes in colorectal cancer. Front Oncol. 2022;12:975859. doi: 10.3389/fonc.2022.975859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Ayati N, Lee ST, Zakavi SR, Cheng M, Lau WFE, Parakh S, et al. Response Evaluation and Survival Prediction After PD-1 Immunotherapy in Patients with Non-Small Cell Lung Cancer: Comparison of Assessment Methods. J Nucl Med United States. 2021;62:926–933. doi: 10.2967/jnumed.120.254508. [DOI] [PubMed] [Google Scholar]
- 192.Constantinidou A, Alifieris C, Trafalis DT. Targeting Programmed Cell Death -1 (PD-1) and Ligand (PD-L1): a new era in cancer active immunotherapy. Pharmacol Ther England. 2019;194:84–106. doi: 10.1016/j.pharmthera.2018.09.008. [DOI] [PubMed] [Google Scholar]
- 193.Voli F, Valli E, Lerra L, Kimpton K, Saletta F, Giorgi FM, et al. Intratumoral Copper Modulates PD-L1 Expression and Influences Tumor Immune Evasion. Cancer Res. United States. 2020;80:4129–4144. doi: 10.1158/0008-5472.CAN-20-0471. [DOI] [PubMed] [Google Scholar]
- 194.Yuan H, Qin X, Wang J, Yang Q, Fan Y, Xu D. The cuproptosis-associated 13 gene signature as a robust predictor for outcome and response to immune- and targeted-therapies in clear cell renal cell carcinoma. Front Immunol. 2022;13:971142. doi: 10.3389/fimmu.2022.971142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Oliveri V. Biomedical applications of copper ionophores. Coord Chem Rev. 2020;422:213474. Available from: https://www.sciencedirect.com/science/article/pii/S0010854520303349
- 196.Reeder NL, Kaplan J, Xu J, Youngquist RS, Wallace J, Hu P, et al. Zinc pyrithione inhibits yeast growth through copper influx and inactivation of iron-sulfur proteins. Antimicrob Agents Chemother. 2011;55:5753–5760. doi: 10.1128/AAC.00724-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Senges CHR, Warmuth HL, Vázquez-Hernández M, Uzun HD, Sagurna L, Dietze P, et al. Effects of 4-Br-A23187 on Bacillus subtilis cells and unilamellar vesicles reveal it to be a potent copper ionophore. Proteomics. Germany. 2022;22:e2200061. doi: 10.1002/pmic.202200061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Menghani SV, Rivera A, Neubert M, Hagerty JR, Lewis L, Galgiani JN, et al. Demonstration of N, N-Dimethyldithiocarbamate as a Copper-Dependent Antibiotic against Multiple Upper Respiratory Tract Pathogens. Microbiol Spectr. United States. 2021;9:e0077821. doi: 10.1128/Spectrum.00778-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Marsh JW, Djoko KY, McEwan AG, Huston WM. Copper(II)-bis(thiosemicarbazonato) complexes as anti-chlamydial agents. Pathog Dis United States. 2017;75:7. [DOI] [PubMed]
- 200.Cater MA, Pearson HB, Wolyniec K, Klaver P, Bilandzic M, Paterson BM, et al. Increasing intracellular bioavailable copper selectively targets prostate cancer cells. ACS Chem Biol United States. 2013;8:1621–1631. doi: 10.1021/cb400198p. [DOI] [PubMed] [Google Scholar]
- 201.Oliveri V. New Glycoconjugates for the Treatment of Diseases Related to Metal Dyshomeostasis. ChemistryOpen Germany. 2015;4:792–795. doi: 10.1002/open.201500155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Tardito S, Barilli A, Bassanetti I, Tegoni M, Bussolati O, Franchi-Gazzola R, et al. Copper-dependent cytotoxicity of 8-hydroxyquinoline derivatives correlates with their hydrophobicity and does not require caspase activation. J Med Chem. 2012;55:10448–10459. doi: 10.1021/jm301053a. [DOI] [PubMed] [Google Scholar]
- 203.Oliveri V, Lanza V, Milardi D, Viale M, Maric I, Sgarlata C, et al. Amino- and chloro-8-hydroxyquinolines and their copper complexes as proteasome inhibitors and antiproliferative agents. Metallomics. England. 2017;9:1439–1446. doi: 10.1039/C7MT00156H. [DOI] [PubMed] [Google Scholar]
- 204.Dai F, Yan W-J, Du Y-T, Bao X-Z, Li X-Z, Zhou B. Structural basis, chemical driving forces and biological implications of flavones as Cu(II) ionophores. Free Radic Biol Med. United States. 2017;108:554–563. doi: 10.1016/j.freeradbiomed.2017.04.023. [DOI] [PubMed] [Google Scholar]
- 205.Gaál A, Garay TM, Horváth I, Máthé D, Szöllősi D, Veres DS, et al. Development and In Vivo Application of a Water-Soluble Anticancer Copper Ionophore System Using a Temperature-Sensitive Liposome Formulation. Pharmaceutics. 2020;12:466. doi: 10.3390/pharmaceutics12050466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Ji Y, Dai F, Zhou B. Designing salicylaldehyde isonicotinoyl hydrazones as Cu(II) ionophores with tunable chelation and release of copper for hitting redox Achilles heel of cancer cells. Free Radic Biol Med. United States. 2018;129:215–226. doi: 10.1016/j.freeradbiomed.2018.09.017. [DOI] [PubMed] [Google Scholar]
- 207.O’Day SJ, Eggermont AM, Chiarion-Sileni V, Kefford R, Grob JJ, Mortier L, et al. Final results of phase III SYMMETRY study: randomized, double-blind trial of elesclomol plus paclitaxel versus paclitaxel alone as treatment for chemotherapy-naive patients with advanced melanoma. J Clin Oncol. 2013;31:1211–1218. doi: 10.1200/JCO.2012.44.5585. [DOI] [PubMed] [Google Scholar]
- 208.Shi H, Suo Y, Zhang Z, Liu R, Liu H, Cheng Z. Copper(II)-disulfiram loaded melanin-dots for cancer theranostics. Nanomedicine. United States. 2021;32:102340. doi: 10.1016/j.nano.2020.102340. [DOI] [PubMed] [Google Scholar]
- 209.Meng X, Shi Y, Chen Z, Song L, Zhao M, Zou L, et al. Extending Hypochlorite Sensing from Cells to Elesclomol-Treated Tumors in Vivo by Using a Near-Infrared Dual-Phosphorescent Nanoprobe. ACS Appl Mater Interfaces. United States. 2018;10:35838–35846. doi: 10.1021/acsami.8b14717. [DOI] [PubMed] [Google Scholar]
- 210.Kelley KC, Grossman KF, Brittain-Blankenship M, Thorne KM, Akerley WL, Terrazas MC, et al. A Phase 1 dose-escalation study of disulfiram and copper gluconate in patients with advanced solid tumors involving the liver using S-glutathionylation as a biomarker. BMC Cancer. 2021;21:510. doi: 10.1186/s12885-021-08242-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Huang J, Campian JL, Gujar AD, Tran DD, Lockhart AC, DeWees TA, et al. A phase I study to repurpose disulfiram in combination with temozolomide to treat newly diagnosed glioblastoma after chemoradiotherapy. J Neurooncol. United States. 2016;128:259–266. doi: 10.1007/s11060-016-2104-2. [DOI] [PubMed] [Google Scholar]
- 212.Halatsch M-E, Kast RE, Karpel-Massler G, Mayer B, Zolk O, Schmitz B, et al. A phase Ib/IIa trial of 9 repurposed drugs combined with temozolomide for the treatment of recurrent glioblastoma: CUSP9v3. Neurooncol Adv. 2021;3:vdab075. doi: 10.1093/noajnl/vdab075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Huang J, Chaudhary R, Cohen AL, Fink K, Goldlust S, Boockvar J, et al. A multicenter phase II study of temozolomide plus disulfiram and copper for recurrent temozolomide-resistant glioblastoma. J Neurooncol. United States. 2019;142:537–544. doi: 10.1007/s11060-019-03125-y. [DOI] [PubMed] [Google Scholar]
- 214.Jakola AS, Werlenius K, Mudaisi M, Hylin S, Kinhult S, Bartek JJ, et al. Disulfiram repurposing combined with nutritional copper supplement as add-on to chemotherapy in recurrent glioblastoma (DIRECT): Study protocol for a randomized controlled trial. F1000Res. 2018;7:1797. doi: 10.12688/f1000research.16786.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Nechushtan H, Hamamreh Y, Nidal S, Gotfried M, Baron A, Shalev YI, et al. A phase IIb trial assessing the addition of disulfiram to chemotherapy for the treatment of metastatic non-small cell lung cancer. Oncologist. 2015;20:366–367. doi: 10.1634/theoncologist.2014-0424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Schweizer MT, Lin J, Blackford A, Bardia A, King S, Armstrong AJ, et al. Pharmacodynamic study of disulfiram in men with non-metastatic recurrent prostate cancer. Prostate Cancer Prostatic Dis. 2013;16:357–361. doi: 10.1038/pcan.2013.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Zhang T, Kephart J, Bronson E, Anand M, Daly C, Spasojevic I, et al. Prospective clinical trial of disulfiram plus copper in men with metastatic castration-resistant prostate cancer. Prostate. United States. 2022;82:858–866. doi: 10.1002/pros.24329. [DOI] [PubMed] [Google Scholar]
- 218.Berkenblit A, Eder JPJ, Ryan DP, Seiden MV, Tatsuta N, Sherman ML, et al. Phase I clinical trial of STA-4783 in combination with paclitaxel in patients with refractory solid tumors. Clin Cancer Res. United States. 2007;13:584–590. doi: 10.1158/1078-0432.CCR-06-0964. [DOI] [PubMed] [Google Scholar]
- 219.O’Day S, Gonzalez R, Lawson D, Weber R, Hutchins L, Anderson C, et al. Phase II, randomized, controlled, double-blinded trial of weekly elesclomol plus paclitaxel versus paclitaxel alone for stage IV metastatic melanoma. J Clin Oncol. 2009;27:5452–5458. doi: 10.1200/JCO.2008.17.1579. [DOI] [PubMed] [Google Scholar]
- 220.Monk BJ, Kauderer JT, Moxley KM, Bonebrake AJ, Dewdney SB, Secord AA, et al. A phase II evaluation of elesclomol sodium and weekly paclitaxel in the treatment of recurrent or persistent platinum-resistant ovarian, fallopian tube or primary peritoneal cancer: An NRG oncology/gynecologic oncology group study. Gynecol Oncol. 2018;151:422–427. doi: 10.1016/j.ygyno.2018.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Gehrmann M. Drug evaluation: STA-4783–enhancing taxane efficacy by induction of Hsp70. Curr Opin Investig Drugs. England. 2006;7:574–580. [PubMed] [Google Scholar]
- 222.Asleh K, Negri GL, Spencer Miko SE, Colborne S, Hughes CS, Wang XQ, et al. Proteomic analysis of archival breast cancer clinical specimens identifies biological subtypes with distinct survival outcomes. Nat Commun. 2022;13:896. doi: 10.1038/s41467-022-28524-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Donnelly PS, Liddell JR, Lim S, Paterson BM, Cater MA, Savva MS, et al. An impaired mitochondrial electron transport chain increases retention of the hypoxia imaging agent diacetylbis(4-methylthiosemicarbazonato)copperII. Proc Natl Acad Sci U S A. United States. 2012;109:47–52. doi: 10.1073/pnas.1116227108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Munakata M, Kodama H, Fujisawa C, Hiroki T, Kimura K, Watanabe M, et al. Copper-trafficking efficacy of copper-pyruvaldehyde bis(N4- methylthiosemicarbazone) on the macular mouse, an animal model of Menkes disease. Pediatr Res. United States. 2012;72:270–276. doi: 10.1038/pr.2012.85. [DOI] [PubMed] [Google Scholar]
- 225.Summers KL, Dolgova NV, Gagnon KB, Sopasis GJ, James AK, Lai B, et al. PBT2 acts through a different mechanism of action than other 8-hydroxyquinolines: an X-ray fluorescence imaging study. Metallomics. England. 2020;12:1979–1994. doi: 10.1039/d0mt00222d. [DOI] [PubMed] [Google Scholar]
- 226.Jiang H, Taggart JE, Zhang X, Benbrook DM, Lind SE, Ding W-Q. Nitroxoline (8-hydroxy-5-nitroquinoline) is more a potent anti-cancer agent than clioquinol (5-chloro-7-iodo-8-quinoline) Cancer Lett. Ireland. 2011;312:11–17. doi: 10.1016/j.canlet.2011.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Castoldi F, Hyvönen MT, Durand S, Aprahamian F, Sauvat A, Malik SA, et al. Chemical activation of SAT1 corrects diet-induced metabolic syndrome. Cell Death Differ. 2020;27:2904–2920. doi: 10.1038/s41418-020-0550-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Vetrik M, Mattova J, Mackova H, Kucka J, Pouckova P, Kukackova O, et al. Biopolymer strategy for the treatment of Wilson’s disease. J Control Release. Netherlands. 2018;273:131–138. doi: 10.1016/j.jconrel.2018.01.026. [DOI] [PubMed] [Google Scholar]
- 229.Mao L, Huang C-H, Shao J, Qin L, Xu D, Shao B, et al. An unexpected antioxidant and redox activity for the classic copper-chelating drug penicillamine. Free Radic Biol Med. United States. 2020;147:150–158. doi: 10.1016/j.freeradbiomed.2019.12.013. [DOI] [PubMed] [Google Scholar]
- 230.Kenney GE, Rosenzweig AC. Chemistry and biology of the copper chelator methanobactin. ACS Chem Biol. 2012;7:260–268. doi: 10.1021/cb2003913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Lenartowicz M, Moos T, Ogórek M, Jensen TG, Møller LB. Metal-Dependent Regulation of ATP7A and ATP7B in Fibroblast Cultures. Front Mol Neurosci. 2016;9:68. doi: 10.3389/fnmol.2016.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Lee K, Briehl MM, Mazar AP, Batinic-Haberle I, Reboucas JS, Glinsmann-Gibson B, et al. The copper chelator ATN-224 induces peroxynitrite-dependent cell death in hematological malignancies. Free Radic Biol Med. 2013;60:157–167. doi: 10.1016/j.freeradbiomed.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.