Abstract
Yeast TAF90p is a component of at least two transcription regulatory complexes, the general transcription factor TFIID and the Spt-Ada-Gcn5 histone acetyltransferase complex (SAGA). Broad transcription defects have been observed in mutants of other TAFIIs shared by TFIID and SAGA but not in the only two TAF90 mutants isolated to date. Given that the numbers of mutants analyzed thus far are small, we isolated and characterized 11 temperature-sensitive mutants of TAF90 and analyzed their effects on transcription and integrity of the TFIID and SAGA complexes. We found that the mutants displayed a variety of allele-specific defects in their ability to support transcription and maintain the structure of the TFIID and SAGA complexes. Sequencing of the alleles revealed that all have mutations corresponding to the C terminus of the protein, with most clustering within the conserved WD40 repeats; thus, the C terminus of TAF90p is required for its incorporation into TFIID and function in SAGA. Significantly, inactivation of one allele, taf90-20, caused the dramatic reduction in the levels of total mRNA and most specific transcripts analyzed. Analysis of the structure and/or activity of both TAF90p-containing complexes revealed that this allele is the most disruptive of all. Our analysis defines the requirement for the WD40 repeats in preserving TFIID and SAGA function, demonstrates that the defects associated with distinct mutations in TAF90 vary considerably, and indicates that TAF90 can be classified as a gene required for the transcription of a large number of genes.
TFIID is one of several general initiation factors required for the reconstitution of RNA polymerase II (Pol II) transcription in vitro. In yeast, it is composed of the TATA-binding protein (TBP) and at least 14 associated factors collectively referred to as TAFIIs (23, 30, 34, 41, 42, 43, 46, 53). Individual TAFII polypeptides contain distinct structural motifs and functional domains that are likely to be responsible for their specialized functions. For example, several functional domains have been identified in hTAFII250, which include histone acetyltransferase (HAT), kinase, and ubiquitin-conjugating domains and bromodomains (8, 21, 32, 40). Many TAFIIs contain another specialized motif, the histone-like fold (11, 12, 13, 19, 36, 59). These include TAF60p, TAF17p, TAF68/61p, TAF40p, TAF19p, TAF47p, TAF25p, and TAF48p; for simplicity, only the yeast TAFIIs are mentioned, although their eukaryotic homologues also contain this motif (for reviews, see references 13, 19, and 36).
Perhaps the least understood motif found in TAFIIs is the WD40 repeats of TAF90/hTAFII100/dTAFII80 (9, 10, 42, 51, 52). The function of this domain is not known, but it is speculated to be involved in protein-protein interactions, as has been proposed for other WD40-containing proteins (49). Despite the highly conserved nature of this region of the protein, it has been reported that the WD40 repeat domain is not necessary for incorporation of hTAFII100 into TFIID (9). Moreover, the Schizosaccharomyces pombe homologue of TAF90, taf72+, cannot complement the function of Saccharomyces cerevisiae TAF90, but a chimera encoding the N terminus of TAF90p and the C terminus of Taf72p can do so (60), indicating that the N terminus of TAF90p provides the species specificity. Therefore, all of the available evidence suggests that the N terminus of TAF90p, and those of its homologues, is most important for its incorporation into TFIID. That is not to say that the C terminus is dispensable, as the only temperature-sensitive mutants characterized to date have mutations located within the C terminus (2).
There is an ongoing debate on what fraction of the genome is dependent upon certain TAFII genes for its expression and on what accounts for the differences observed between laboratories and species (1, 17, 18). Mutation or depletion of certain TAFII subunits results in restricted transcription defects (2, 20, 26, 33, 53), whereas mutation of other subunits reduces the transcription of many genes (3, 25, 31, 35, 37, 43, 45). The identification of certain TAFIIs besides TFIID in nuclear complexes (15, 22, 29, 39, 55) provided a possible explanation for the differences observed among TAFII genes. For example, genome-wide studies demonstrated that TAF145 and GCN5 perform some redundant functions in transcription, as a double taf145 gcn5 mutant shows defects in the transcription of a larger fraction of the genome than for the sum of the individual mutations (26). These observations suggest that certain TAFIIs are responsible for the transcription of a large number of genes because they are shared by TFIID and the Spt-Ada-Gcn5 HAT complex (SAGA). Nonetheless, an inconsistency in this theory is that mutation of TAF90, encoding a subunit of TFIID and SAGA, affects only a small number of genes (2, 26). Of the TAFIIs shared between TFIID and SAGA, TAF90p is the only one whose loss of function does not broadly affect transcription. However, the number of mutants characterized thus far has been small, and different mutant alleles might be expected to have diverse consequences for the structures of TFIID and SAGA. Another factor that complicates the interpretation of the results obtained using various TAFII mutants is that the effects of these mutations on TFIID and SAGA integrity have not been thoroughly examined.
A comprehensive analysis of TAF90 was performed in an attempt to understand its role in the function of TFIID and SAGA. Eleven temperature-sensitive mutants were isolated and were characterized in regard to their effects on transcription and on the structures of TFIID and SAGA. We found that all of the mutants contained amino acid substitutions within the conserved C terminus of the protein, particularly within the WD40 repeats. Mutations within this region caused defects in the ability of TAF90p to interact with TFIID and SAGA. While 10 of the 11 alleles displayed a variety of weak and selective transcription phenotypes, a single allele of TAF90 was identified that caused a rapid reduction in the mRNA levels of a large number of genes. Our data indicate that the breadth and severity of the transcription defects of each mutant correlated strongly with their effects on TFIID structure but less so with their effects on SAGA activity. The allele-specific defects we report here illustrate the point that multiple alleles need to be examined to fully understand the role of TAFIIs in transcription.
MATERIALS AND METHODS
Yeast strains and genetic manipulations.
The strains used in this study were as follows: YJR224 (matα Δtaf90::TRP1 [pRS316-TAF90] ade2-101 his3Δ200 leu2-3,112 ura3-52 trp1-901 lys2-801 suc2-Δ9), YJR226 (same as YJR224 except containing pRS313-HA-TAF90 or pRS313-HA-taf90-x in place of pRS316-TAF90, where x is the allele number), YJR240 (mata Δtaf90::HIS3 [pRS316-TAF90] ade2-101 his3Δ200 leu2-Δ1 ura3-Δ99 lys2-801), YJR241 (same as YJR240 except containing pRS415-HA-TAF90 or pRS415-HA-taf90-x), YJR500 (mata/α taf90::TRP1/TRP1 [pRS316-TAF90] his4-917δ/ his4-917δ lys2-17R2/lys2-17R2 leu2/leu2 ura3-52/ura3-52), YJR501 (mata taf90::TRP1 [pRS316-TAF90] his4-917δ lys2-17R2 leu2 ura3-52), and YJR530 (same as YJR501 except containing pRS415-HA-TAF90-x).
Mutants were created by hydroxylamine mutagenesis of pRS313-HA-TAF90 in vitro for 60, 90, and 120 min at 70°C. After purification of the plasmid, each pool was independently transformed into YJR224 and plated onto histidine dropout plates. The transformants were subsequently replica plated onto 5-fluoro-orotic acid (48) and placed at 24, 30, and 37°C. Temperature-sensitive mutants were identified based on growth at 24°C but inviability at 37°C. The mutants were verified, and the coding region was sequenced to identify the base substitutions. The mutants were transferred into pRS415 by gap repair, and the resulting plasmids were used to transfer the mutations into additional genetic backgrounds (YJR241 and YJR530). For the SPT analysis, strain FY632 was transformed with a taf90::TRP1 DNA fragment generated by PCR (4), and the resulting transformants were selected for on synthetic complete medium without tryptophan. Individual colonies were screened by PCR and by Southern blotting to confirm the disruption of TAF90. YJR500 cells were transformed with pRS316-TAF90, and the transformants were sporulated on solid medium for 4 days. Tetrads were dissected, and genotypes of the resulting spores were identified by replica plating onto selective media and mating type testing. The resulting strain, YJR501, was transformed with pRS415 containing TAF90 or its mutant derivatives. Next, transformants were selected for twice on 5-fluoro-orotic acid plates, and the temperature-sensitive mutants were confirmed by monitoring growth at 24 and 37°C.
Analysis of RNA.
To measure poly(A)+ mRNA levels, total RNA was prepared by the acid-phenol extraction method, and 10 μg was applied to charged nylon using a dot blot manifold. The membrane was blocked and hybridized with a 32P-labeled dT20 oligonucleotide (Pharmacia) as described previously (54). For Northern blotting (44) and S1 nuclease protection (7), total RNA was prepared by the glass bead disruption procedure (2, 53). For the genes RPS5, TRX1, CLB2, CLN2, SUC2, HUG1, and scR1, gel-purified PCR-generated probes were used. ADH2 RNA was detected using an oligonucleotide probe specific for a region nonhomologous to ADH1.
Whole-cell extracts and immunoprecipitation (IP).
Yeast cells were grown in 2% peptone–1% yeast extract–2% dextrose (YPD) supplemented with 20 μg of adenine sulfate per ml (YAPD) (500 ml) to an optical density at 600 nm (OD600) of approximately 1.0 to 1.2. An equal volume of 24°C YPD (for permissive-temperature extracts) or 50°C YPD (for nonpermissive-temperature extracts) was added, and the cells were grown for an additional 3 h at 24 or 37°C, respectively. Whole-cell extracts were prepared as described previously (42, 58). One milliliter of whole-cell extract was adjusted to a total protein concentration of 1.5 mg/ml with 0.2 M buffer T (same as buffer B described by Reese et al. [42] but with 0.2 M potassium acetate), and the insoluble material was removed by centrifugation for 10 min at 14,000 rpm in a Microfuge. Two microliters of anti-TAF90p or anti-TAF145p polyclonal antibody (53) was added, and the mixture was incubated at 4°C for about 4 h. The tubes were centrifuged for 10 min at 14,000 rpm, and the supernatant was transferred to a new tube. Thirty microliters of a 50% slurry of protein A beads (Repligen) in 0.2 M buffer T was added, and the tubes were incubated overnight at 4°C with end-over-end mixing. The protein A beads were collected by low-speed centrifugation (500 rpm, 4°C, 5 min), and the supernatant was carefully removed. The beads were washed four times with 0.9 ml of ice-cold wash buffer (same as 0.2 M buffer T but containing 0.1% NP-40). After the final wash, the supernatant was carefully removed and the proteins were eluted by heating in 1.5× sodium dodecyl sulfate (SDS) loading buffer. Samples were fractionated on 8% (14- by 10-cm) and 12.5% (8- by 10-cm) SDS-polyacrylamide gels and transferred to nitrocellulose. For the inputs, 30 μg of extract was loaded onto similar gels.
Purification of SAGA.
Six 500-ml cultures of yeast were grown at room temperature in YAPD until an OD600 of 1 to 2 was reached and then were diluted with an equal volume of prewarmed YAPD (50°C). SAGA was purified essentially as described by Grant et al. (14). Extracts containing an equal quantity of total protein (440 mg in about 40 to 43 ml) were mixed overnight at 4°C with 7.5 ml of Ni-nitrilotriacetic acid (Ni-NTA) agarose (Qiagen). The slurry was poured into a 1.5- by 10-cm column (Kontes) at 4°C, washed with wash buffer (20 mM imidazole [pH 7.0], 100 mM NaCl, 0.1% Tween 20, 10% glycerol, 2 μg of pepstatin A per ml, 2 μg of leupeptin per ml, 5 μg of aprotinin per ml, and 1 mM phenylmethylsulfonyl fluoride [PMSF]), and eluted in same buffer containing 300 mM imidazole (pH 7.0). The Ni-NTA column eluate was loaded onto a Mono Q HR 5/5 column (Pharmacia), and the column was washed with 1 column volume of 100 mM Q-buffer (50 mM Tris-HCl [pH 8.0], 100 mM NaCl, 0.1% Tween 20, 10% glycerol, 2 μg of pepstatin A per ml, 2 μg of leupeptin per ml, 5 μg of aprotinin per ml, and 1 mM PMSF) and eluted using a linear gradient from 100 to 500 mM NaCl through a volume of 25 ml. The SAGA-containing fractions were pooled (about 3 ml) and concentrated to a final volume of about 400 μl using a Centricon YM-30 filtration unit (Millipore). A 250-μl portion was loaded onto a Superose 6 HR 10/30 column equilibrated with 40 mM HEPES (pH 7.5)–0.5 M NaCl–0.1% Tween 20–10% glycerol–1 mM PMSF and eluted at a flow rate of 0.2 ml/min using a Biologic HR system (Bio-Rad). Fractions (0.5 ml) were collected and frozen at −80°C after the addition of 50 μg of insulin.
RESULTS
Isolation and characterization of TAF90 mutants.
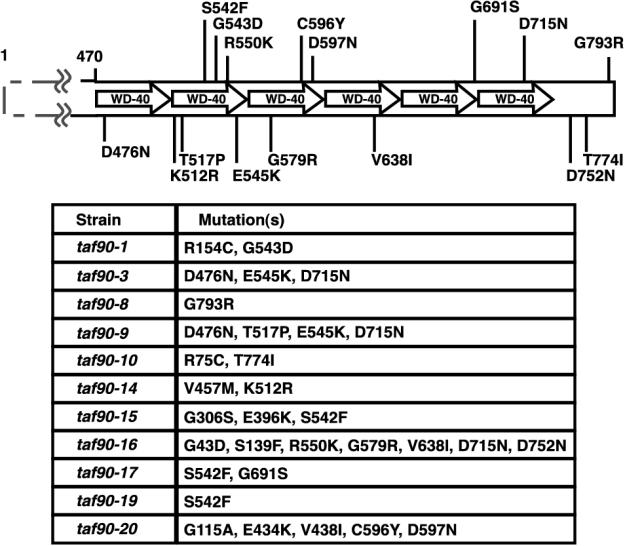
To study the function of TAF90, a screen for temperature-sensitive mutants was performed. Eleven mutants that failed to support growth at 37°C were isolated, and their mutations were identified by DNA sequencing (Fig. 1). All but two mutants (taf90-8 and -19) have multiple amino acid substitutions. Strikingly, every mutant has at least one amino acid change within the C terminus, and 9 of the 11 mutants have substitutions within a WD40 repeat. Interestingly, 8 out of the 11 mutants have an amino acid substitution within the second WD40 repeat. Several of the mutants have mutations that lie close to the locations of those in two previously identified TAF90 mutants, referred to as taf90ts2-1 (S702D, G793R) and taf90ts3-1 (G711E, G712S) by Apone et al. (2). In fact, taf90-8 shares the exact amino acid substitution (G793R) as one of the two mutations contained in taf90ts2-1. In no case did we isolate a mutant that contained changes exclusively within the N terminus of the protein. While we have not delineated the contributions of each of the substitutions in the mutants with multiple mutations, clearly the data as a whole suggest that mutations in the C terminus largely are responsible for the loss of function of these mutants.
FIG. 1.
Isolation of TAF90 mutants. A table of amino acid substitutions and the locations of those in the C terminus relative to the putative WD40 repeats are shown.
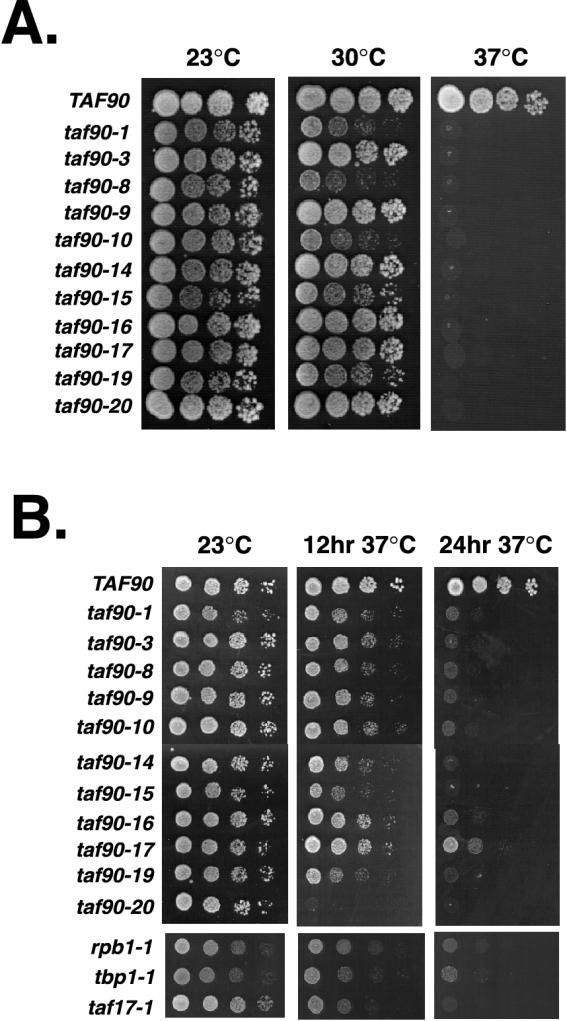
The growth phenotypes of each allele were examined at 23, 30, and 37°C (Fig. 2A). As expected, none of the mutants grew at 37°C, consistent with their isolation as temperature-sensitive mutants. Differences in growth rates were observed among the mutants at 23, 30, and 34°C (not shown), suggesting a range of severity among the alleles. The taf90-1, taf90-8, and taf90-10 mutants displayed a slight reduction in growth rate at 23°C, which was more prevalent at 30°C (Fig. 2A). In addition, a number of mutants grew significantly slower than wild-type cells at 23°C in liquid medium, such as the taf90-1, taf90-3, and taf90-19 mutants, despite showing comparatively milder defects on solid medium at the same temperature (not shown).
FIG. 2.
Characterization of growth phenotypes. (A) Growth phenotypes on rich medium. Cultures were grown in rich medium to an OD600 of 1.0, and 2 μl of 10-fold serial dilutions was spotted onto agar plates. Plates were grown for 3 days at the temperatures indicated. (B) Recovery of TAF90 mutants from exposure to the restrictive temperature. Cultures were grown in liquid rich medium to an OD600 of 1.0, and then 2 μl of 10-fold serial dilutions was spotted onto solid rich medium. Plates were grown either for 3 days at 23°C or for 12 or 24 h at 37°C and then for 3 days at 23°C. Since the cells were plated onto agar plates at room temperature, the exact time that the cells were exposed to a temperature of 37°C is not known.
It has been proposed that certain weak temperature-sensitive (leaky) alleles of TAFII mutants fail to display true loss-of-function phenotypes because they have residual activity at 37°C, despite their apparent lack of growth in liquid culture (25, 31). For example, mutants with certain temperature-sensitive alleles of TAF40 that display restricted transcription phenotypes do not lose viability even after 2 days at the nonpermissive temperature, whereas a mutant with another allele that displays broader transcription defects loses viability during that time (25). These observations necessitated that we analyze the ability of each mutant to recover from a transient exposure to the nonpermissive temperature, so as to judge the efficacy of the mutations in destroying TAF90 function. In addition to the TAF90 mutants, we analyzed three well-characterized temperature-sensitive mutants considered to be tight based on the rapid disruption of transcription that results from their transfer to 37°C. These mutants are those carrying the rpb1-1 allele of the gene encoding the largest subunit of Pol II (20, 38), the tbpts-1 allele of the gene encoding TBP (7), and the taf17-L68P,E148G allele of the TBP-associated factor 17 (31). Serial dilutions of each culture were spotted onto rich medium and either were grown solely at the permissive temperature or were exposed to the restrictive temperature for 12 or 24 h before being returned to the permissive temperature to resume growth. The results indicate that most of the TAF90 mutants showed a loss of viability similar to that of the rpb1-1, tbpts-1, and taf17-L68P,E148G mutants (Fig. 1B). The noted exceptions are the taf90-20 and taf90-17 alleles. The taf90-20 mutant failed to recover from a 12-h exposure to 37°C and was by far the most severely affected among all the general transcription mutants examined here. This mutant was even more strongly affected than the rpb1-1 and tbpts-1 strains. On the other hand, the taf90-17 strain recovered even after a 24-h incubation at 37°C. Therefore, with the exception of the taf90-17 mutant, all of the TAF90 mutants described here were at least as defective in their recovery from an exposure to the restrictive temperature as the rpb1-1, tbpts-1, and taf17-L68P,E148G mutants. This analysis uncovered differences in the growth phenotypes of the taf90-17 and taf90-19 mutants: the taf90-19 allele is more deleterious to cell viability. Moreover, the taf90-17 mutant grows well at 34°C, whereas the taf90-19 mutant grows very poorly at this temperature (not shown). Therefore, based upon these two growth phenotypes, the additional amino acid substitution, G691S, contained in the taf90-17 mutant is somewhat compensatory for the defects caused by the S542F mutation found in both mutants.
Mutations in certain SAGA subunits can suppress Ty insertions (SPT phenotype) and cause inositol and galactose auxotrophies (16, 56); therefore, we screened each mutant for these phenotypes. The TAF90 mutants were reconstructed in a strain containing a Ty insertion at the HIS4 locus, his4-914 (56, 57), and plated onto medium lacking histidine. Whereas a strain containing a mutation in spt3 grew on medium lacking histidine, indicating suppression, none of the TAF90 mutants showed this phenotype (not shown). In addition, none of the mutants displayed obvious galactose or inositol auxotrophies under conditions that would support growth (not shown).
Allele-specific defects in Pol II-mediated transcription.
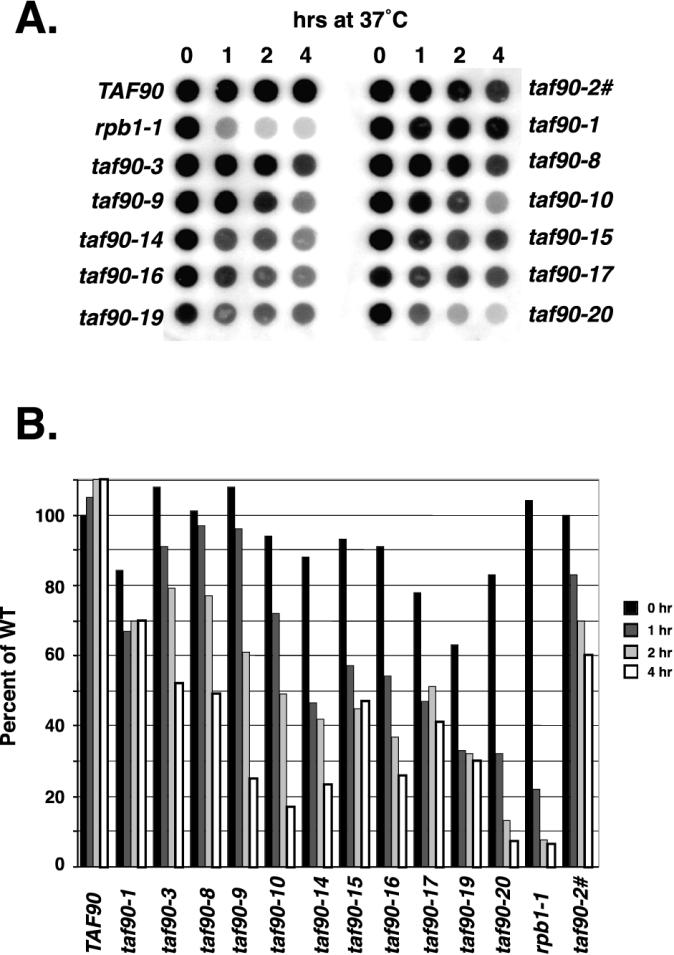
To determine the effects of TAF90 mutations on Pol II-mediated transcription, changes in mRNA levels were detected by probing a blot of total RNA with radiolabeled oligo(dT). This method has been used repeatedly to analyze transcription defects in TAFII mutants (3, 25, 31, 46, 54). From the data presented in Fig. 3, it appears that shifting each mutant to 37°C had diverse consequences on the levels of total poly(A)+ RNA. Based upon the magnitude and kinetics of RNA loss, the mutants can be roughly grouped into classes. The first category of mutants exhibited a reduction in mRNA at the permissive temperature, a small drop at the 1-h time point, and then maintenance of that level to 4 h. Examples of this class are the taf90-1, taf90-15, taf90-17, and taf90-19 mutants. The next class showed slight, gradual reductions for the first 2 h and then a sharp decline at 4 h, such as the taf90-8 mutant. Most mutants represent the third type, which exhibited a steady decrease immediately after the temperature shift. Examples of this class are the taf90-3, taf90-10, taf90-14, and taf90-16 mutants and the previously described taf90ts2-1 mutant (2, 54). Even within this group the kinetics and magnitude of RNA loss varied considerably, from the weaker phenotypes exhibited by the taf90-3 and taf90ts2-1 mutants to the stronger phenotype of the taf90-14 mutant. Finally, there is one mutant, taf90-20, which is clearly unique. Shifting this mutant to 37°C resulted in a rapid, and significant, reduction in poly(A)+ RNA levels (Fig. 3). Significantly, the kinetics and magnitude of mRNA loss observed in the taf90-20 mutant were very similar to those of the Pol II mutant, rpb1-1. Specifically, the mRNA levels in the taf90-20 mutant were reduced to 32% of the wild-type level by 1 h and to 7% by 4 h, while the quantities in the rpb1-1 strain declined to 20 and 6%, respectively. In summary, it appears that a variety of transcription defects were observed, ranging from weakly affected, as in the case of taf90-1 and taf90-3, to very severe, as seen in the taf90-20 mutant.
FIG. 3.
Poly(A)+ RNA levels. (A) Cultures were grown in YPAD to an OD600 of approximately 0.3 and then shifted to 37°C. Aliquots were removed prior to the shift (0 h) and after 1, 2, and 4 h at 37°C, and RNA was isolated by acid-phenol extraction. Ten micrograms of total RNA was spotted onto nitrocellulose, and poly(A)+ RNA was detected by probing with a 32P-labeled dT20 probe. (B) Quantification. Spots were analyzed using a phosphorimager. The percentage of wild-type (WT) levels was determined by comparing the counts in each spot with those in the wild-type spot from 0 h. The taf90-2 strain (#) is a previously described noncogenic temperature-sensitive strain used as a control (2). The relative levels of mRNA in the mutants versus the wild type varied between 5 and 15% between experiments, but most points varied by less than 10%.
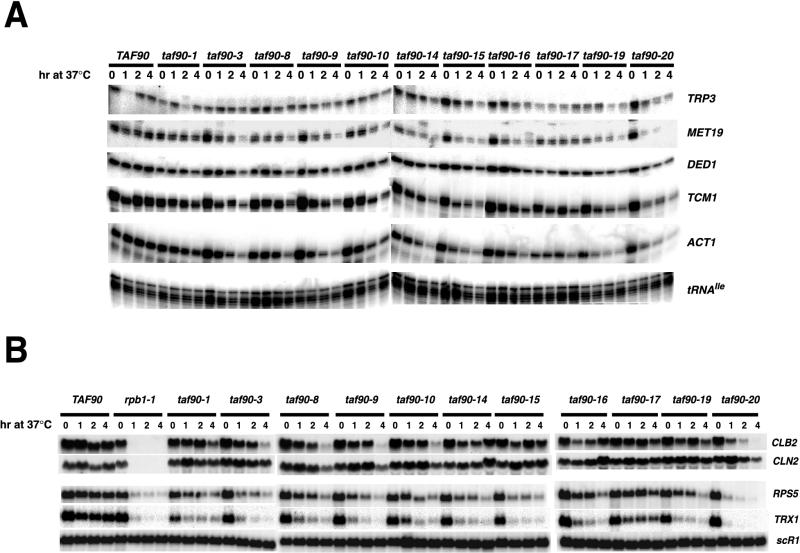
Next, we wanted to determine if the changes in total poly(A)+ RNA described above reflect the loss of a few very abundant RNAs or a reduction in the expression of many different RNAs. If the latter is true, we expect that the kinetics and magnitude of the reductions of specific transcripts will match those observed in the poly(A)+ blots. Genes that have been routinely used to characterize other TAFII mutants were selected (2, 3, 25, 35, 36, 37, 43, 46, 53). The steady-state levels of TRP3, ACT1, DED1, TCM1, and MET19 RNAs were examined by S1 nuclease protection, and the transcripts of CLB2, CLN2, RPS5, and TRX1 were detected by Northern blotting. Shifting cells to the restrictive temperature significantly affected the transcription of TRP3, MET19, TCM1, and ACT1 in nearly every mutant (Fig. 4). The RNA levels of these genes showed a gradual decrease over time, and the severity of the defects varied among the mutants, similar to what was observed in the analysis of poly(A)+ RNA quantities. For example, shifting the taf90-1 mutant to 37°C only weakly affected the levels of total poly(A)+ RNA up to 4 h, and similarly, nearly wild-type levels of mRNA for each of these individual genes were observed over the same time period (Fig. 4). Moreover, the taf90-17 mutant displayed reduced total mRNA levels at 23°C, which were not strongly reduced further by the temperature shift, and likewise it displayed a reproducible reduction in TRP3, MET19, DED1, and ACT1 RNA levels at 23°C that was not exacerbated by the temperature shift. Shifting the taf90-20 mutant to 37°C caused rapid and dramatic losses of TRP3, MET19, TCM1, and ACT1 mRNAs by 1 h, similar to what was observed for the levels of poly(A)+ RNA. Therefore, the losses of poly(A)+ RNA observed in each mutant closely correlated with the magnitude and kinetics of the losses of the individual transcripts examined here as well. In contrast to the case for these other genes, DED1 transcription was only weakly affected in all mutants, including the taf90-20 strain. Therefore, even with the strongest allele, some genes are not strongly affected by the inactivation of TAF90.
FIG. 4.
Transcription analysis of specific messages. Cultures were grown, treated, and processed as described in the legend to Fig. 3. (A) S1 analysis of specific messages. Ten micrograms of total RNA was analyzed by S1 nuclease protection (7). tRNAIle, a Pol III transcript, was used as an RNA control. (B) Northern blotting. Fifteen micrograms of total RNA was fractionated on formaldehyde-containing gels and transferred to a membrane, and the specific transcripts were detected by hybridization with 32P-labeled probes corresponding to the coding regions of CLB2, CLN2, TRX1, and RPS5. The Pol III-transcribed scR1 was used as a loading control.
The messages for TRX1, RPS5, CLN2, and CLB2 were analyzed because they are particularly sensitive to mutations in TAF145 and other TAFIIs (3, 20, 25, 47, 54; J. C. Reese, unpublished data). The results presented in Fig. 4B indicate that the inactivation of most alleles, with the exception of taf90-17, strongly affected the transcription of RPS5, and all mutants displayed significant reductions in TRX1 mRNA (Fig. 4B). Once again, inactivation of taf90-20 had the most deleterious effect on the levels of TRX1 and RPS5 RNAs; their transcription is nearly abolished by 1 h at the restrictive temperature. This is the only allele that exhibited a reduction in these transcripts similar to that for the rpb1-1 mutant. Even mutants such as taf90-1, taf90-3, and taf90-8, which displayed weaker reductions in total poly(A)+ RNA (Fig. 3) and in the specific transcripts analyzed in Fig. 4A at up to 4 h, exhibited more rapid and larger reductions in the expression of these two genes. Thus, it appears that the expression of RPS5 and TRX1 is especially sensitive to TAF90 mutations and is not as prone to allelic variations.
Cyclin expression is abolished in certain TAFII mutants, and in some cases this may explain the cell cycle phenotypes observed in those mutants (20, 26, 28, 54). Our preliminary analysis revealed that only the taf90-9 and taf90-20 mutants displayed a convincing arrest in G2/M at the restrictive temperature, whereas only very weak arrest phenotypes were observed in other mutants (J. C. Reese, A. K. Fisher, and T. A. Albright-Frey, unpublished data). The expression of the cyclins CLN2 and CLB2 was analyzed following a temperature shift, and with the exception of taf90-20, it appears that the alleles had a weak effect or no effect on CLB2 and CLN2 transcription (Fig. 4B). Most mutants, such as taf90-10, showed a decrease in transcription only at the 4-h time point, a time when the decrease in transcription may result from decreased cell vitality.
Activated transcription in TAF90 mutants.
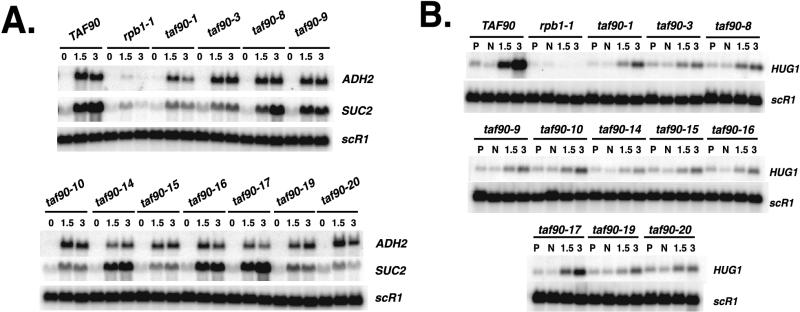
The genes analyzed thus far were actively transcribed at the moment the mutants were inactivated. We next examined the activation of genes when the mutants were exposed to the restrictive temperature. We first examined the derepression of catabolite-repressed genes, specifically, ADH2 and SUC2. It has been reported that the depletion of TAF90p leads to a decrease in ADH2 gene expression and that TFIID interacts with Ard1p, the specific activator of this gene. (24). SUC2 was chosen for analysis because we have found that its derepression is severely impaired in certain TAFII mutants (Reese, unpublished data). The wild-type strain and each of the mutants were grown to log phase in high-dextrose medium, shifted to 37°C, and then switched to prewarmed low-dextrose medium for 1.5 and 3 h. Although none of these mutants displayed abated ADH2 transcription, mRNA levels were reduced in the taf90-1, taf90-14, and taf90-17 mutants (Fig. 5A). Several alleles exhibited wild-type levels of ADH2 RNA, reaffirming the allele-specific nature of these mutations. In addition, we found that the expression of ADH2 is only weakly affected in the taf90-20 mutant, which displayed the strongest transcription phenotype. We cannot account for the differences between our results and those of Komarnitsky et al. (24) with certainty. The previous study utilized a copper-inducible shutoff strategy that required significantly longer incubation times under the restrictive conditions, which may have resulted in secondary effects that adversely affected ADH2 expression.
FIG. 5.
Induced transcription. (A) Derepression of SUC2 and ADH2. Cultures of YJR241-x were grown initially in YPAD at 23°C. Following the removal of an aliquot from each culture, cells were centrifuged, resuspended in YPA containing a low concentration of dextrose (0.05%) prewarmed to 37°C, and grown at the restrictive temperature. RNA was isolated from aliquots that were removed after 1.5 and 3 h at 37°C in low-dextrose medium. Transcripts were detected by Northern blotting. scR1 was used as a loading control. (B) Analysis of a DNA damage-induced gene. Cultures of YJR241-x were grown in rich medium and shifted to the nonpermissive temperature. Aliquots were removed (permissive [lanes P]) prior to the temperature shift. After 1 h at 37°C, MMS was added to 0.02%. Aliquots were taken at 1.5 and 3 h after treatment with MMS. A separate culture was maintained at 37°C but left untreated (nonpermissive [lanes N]) for 4 h. HUG1 RNA was analyzed by Northern blotting. scR1 was used as a loading control.
Several TAF90 mutants showed large reductions in the levels of SUC2 mRNA (Fig. 5A), including the taf90-1 and taf90-3 mutants, which we classify as having weaker transcription phenotypes. Interestingly, the taf90-14 and taf90-17 mutations appear to have no effect of SUC2 transcription, whereas they were two that had a stronger effect on ADH2 expression. Our analysis of these two catabolite-repressed genes highlights the allele- and gene-specific nature of the transcription defects in each mutant and clearly demonstrates the compensatory nature of the G691S substitution in the taf90-17 mutant. The activation of SUC2 is not strongly affected by the taf90-17 allele, whereas it is drastically reduced by the taf90-19 allele.
Depletion of TAF90p or inactivation of a temperature-sensitive allele strongly reduces the expression of DNA damage-induced genes (27). We therefore examined the expression of a representative of this class of genes, HUG1, in the mutants. Cells were shifted to 37°C and treated for 1.5 and 3 h with the DNA-damaging agent methyl methanesulfonate (MMS). A separate untreated aliquot of cells was maintained at 37°C for 3 h to detect changes in the uninduced level of transcription (Fig. 5B). The results presented in Fig. 5B indicate that inactivation of every allele affected the DNA damage-induced expression of HUG1. Expression was significantly reduced even in the taf90-1, taf90-3, and taf90-17 mutants (although to a lesser extent), which display the most selective transcription defects of all of the mutants described here. In agreement with our previous analysis of the TAFII dependence of DNA damage-inducible genes (27), these alleles appeared to specifically affect the derepression of this gene, as the uninduced level of RNA was not significantly reduced even after 3 h at 37°C (Fig. 5).
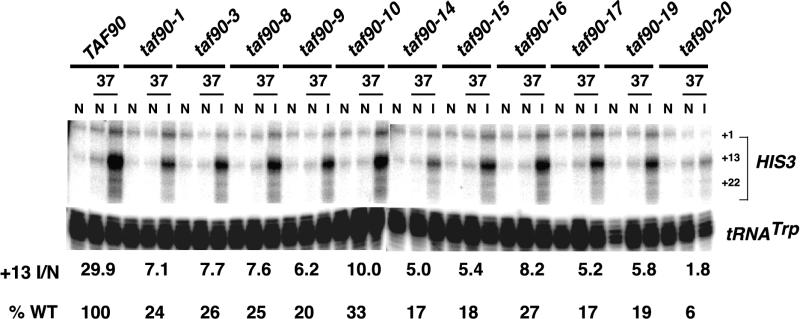
Some mutants of TAFIIs shared by TFIID and SAGA affect Gcn4p-activated transcription (3, 35, 37); therefore, we examined the expression of the Gcn4p-responsive gene HIS3 in the mutants. HIS3 contains several transcription start sites; a nonconsensus TATA element (TC) is responsible for constitutive transcription from position +1, while a consensus TATA element (TR) is responsible for transcription from +13 (5). Gcn4p preferentially activates transcription from the +13 start site (5, 6). The cells were grown for 3 h at 37°C in complete medium (noninducing conditions) or in medium lacking histidine for 1.5 h at 37°C, followed by another 1.5 h in the presence of 30 mM 3-aminotriazole (inducing conditions). Comparing the levels of HIS3 mRNA when the cells are grown under noninduced conditions at 23°C (permissive) versus 37°C (nonpermissive) gives an indication of the effect of each mutant on the basal transcription level. In all but one mutant, shifting the cells to 37°C for 3 h did not affect the uninduced levels of the +1 or +13 transcripts from HIS3 (Fig. 6). A reduction in the +1 transcript was observed in the taf90-20 mutant in this and other experiments. In contrast, the level of starvation-induced transcription from the +13 transcript was strongly reduced for every allele. The magnitudes of induction varied between 33 and 6% of the wild-type level, but most were reduced to approximately 20%. Not surprisingly, the taf90-20 mutant displayed the largest reduction in induced transcription, to 6% of wild-type levels. Interestingly, unlike what was observed in the numerous examples presented above, the taf90-19 and taf90-17 alleles were equally defective for the activation of HIS3 by Gcn4p. Thus, the compensatory nature of the G691S substitution is not universal and displays some gene specificity. These results demonstrate that TAF90 is essential for the activation of HIS3 by Gcn4, similar to the case for other TAFIIs found in both TFIID and SAGA.
FIG. 6.
Effects of TAF90 mutations on Gcn4p-mediated transcription. Cells were grown under noninducing conditions in synthetic complete medium at 24°C (lanes N) and at 37°C for 3 h (lanes N, 37). A separate culture was grown in synthetic complete medium without histidine, shifted to 37°C for 1.5 h, and incubated for another 1.5 h in the presence of 30 mM 3-aminotriazole (lanes I, 37). Analysis of HIS3 was performed by S1 nuclease protection. The induction level (+13 I/N) is the ratio of the +13 transcript from cells grown under inducing conditions at 37°C to that from cells grown under noninducing conditions at 37°C. The reduced tRNA signal in the taf90-19 sample is a gel-loading artifact and is not reproducible. WT, wild type.
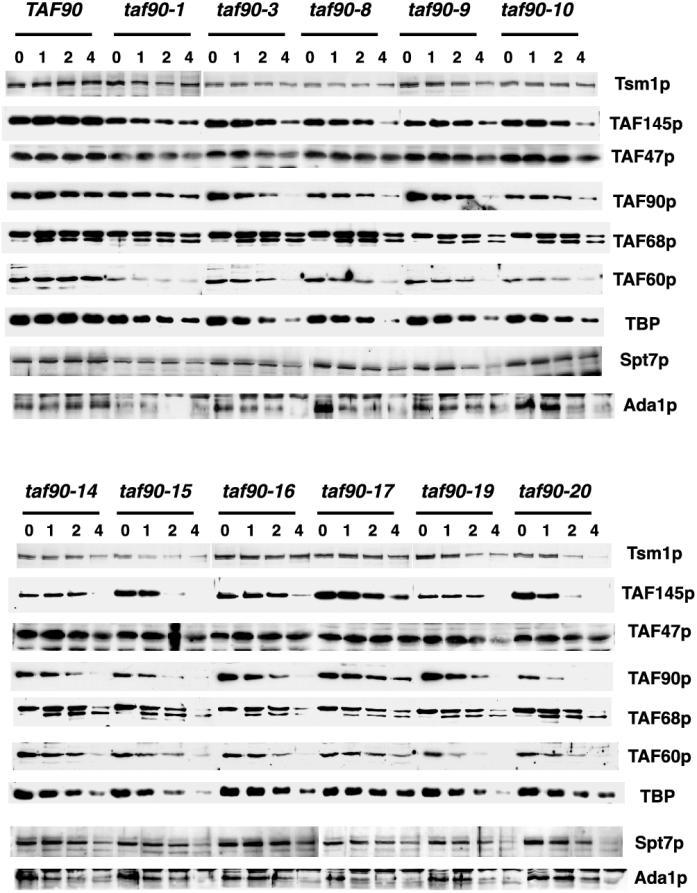
Inactivation of TAF90 mutants reduces TFIID and SAGA subunit levels.
Inactivation of certain TAFII temperature-sensitive alleles, or depletion of its protein, results in the degradation of multiple TFIID and SAGA subunits (25, 31, 43, 46, 53). Codegradation of TAFIIs is a reasonable indicator of the disruption of complex structure; therefore, we monitored the levels of TAFIIs, TBP, and two SAGA components after shifting the cells to 37°C. In many mutants, the levels of TFIID subunits were not strongly reduced until 4 h at 37°C (Fig. 7). The clearest exception was the taf90-20 mutant, which showed a steady decrease in TAF90p, TAF145p, TAF60p, and, to a lesser degree, TBP immediately upon the temperature shift. Thus, by this criterion the taf90-20 allele is the most detrimental to the structure of the TFIID complex. Overall, the degree to which inactivation of these mutants reduced TAFII levels correlated with their growth and transcriptional defects. The taf90-20 and taf90-17 mutants had the strongest and weakest effects on the levels of TFIID subunits, respectively, and likewise they had the most and least severe growth and transcriptional defects. Additionally, the taf90-19 mutant displayed stronger growth and transcriptional defects than the taf90-17 mutant (Fig. 2, 4, and 5) and likewise displayed more drastic reductions in TAFIIs. Finally, the taf90-1 mutant, which showed significant growth defects at the permissive temperature (Fig. 2A and data not shown), had measurable reductions in TFIID and SAGA subunits, even when grown at 23°C. This is most obvious for TAF145p, TAF60p, TAF47p, TAF17p (see below), and Ada1p. The steady-state levels of two SAGA-specific components, Ada1 and Spt7, were also examined. Inactivation of each allele had very different consequences for these two proteins. Whereas Spt7p remained relatively stable in most mutants until the 4-h time point, the levels of Ada1p were significantly reduced earlier in the temperature shift. This is particularly apparent in the taf90-8, taf90-14, taf90-17, and taf90-19 mutants, which showed noticeable reductions in Ada1p by 1 h. Interestingly, the taf90-20 mutant, which had the largest reductions in TFIID subunits, exhibited relatively minor reductions in Spt7p and Ada1p up until the 4-h time point. Therefore, in regard to the degradation of these two SAGA subunits, this allele appears to be no more destructive than the other mutations.
FIG. 7.
Steady-state levels of TFIID and SAGA subunits following a temperature shift. Wild-type and mutant strains were grown at 24°C in YPAD to an OD600 of approximately 0.4, and after removal of an aliquot for the t = 0 time point, they were transferred to a 37°C shaking water bath. Aliquots of culture were removed after 1, 2, and 4 h at the nonpermissive temperature. Extracts prepared from these cells (20 μg of protein) were fractionated by SDS-polyacrylamide gel electrophoresis and transferred to nitrocellulose, and the specific proteins were detected by immunoblotting.
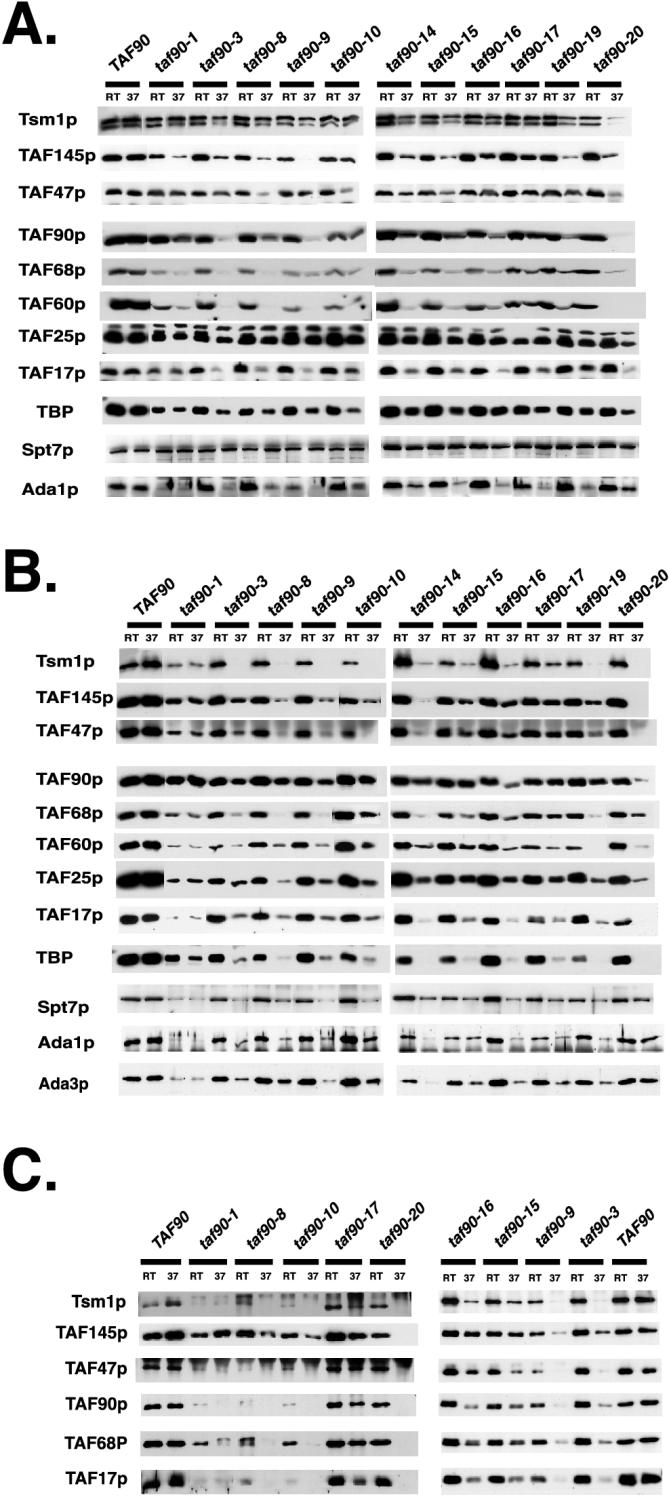
Mutations in TAF90 affect its ability to associate with TFIID and SAGA.
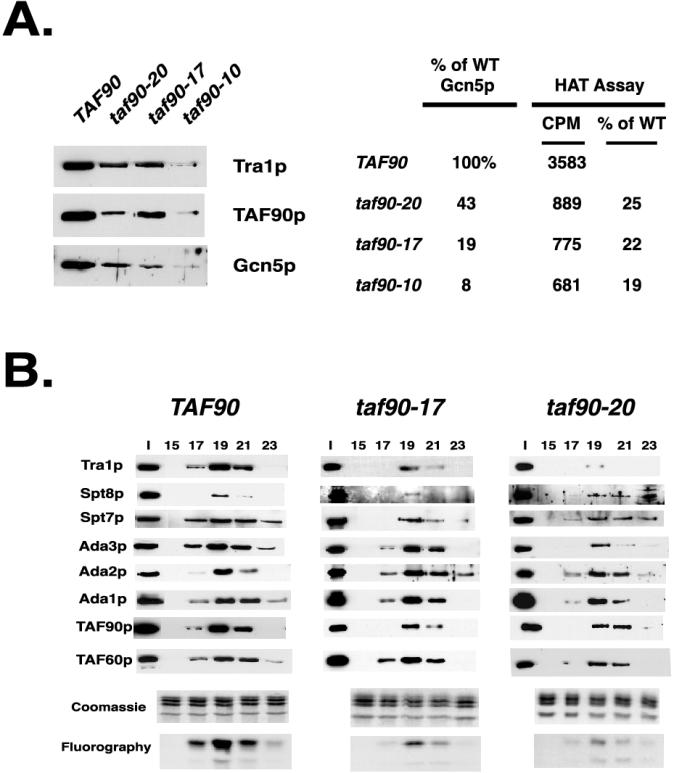
Next, we examined the ability of each mutant to bind to TFIID and SAGA in extracts prepared from cells grown at the permissive temperature and after 3 h of growth at 37°C by co-IP using anti-TAF90p antisera. The ability of each mutant to incorporate into TFIID was judged by the IP efficiencies of Tsm1p, TAF145p, and TAF47p, since they are not subunits of SAGA (15). Likewise, immunoblotting for Spt7p, Ada3p, and Ada1p monitored their association with the SAGA complex. Unfortunately, the antibodies to Ada3p were not of sufficient quality to detect the levels of these proteins in whole-cell extracts; nonetheless, they could be used to analyze the immunopurified material, and these results are presented. The loads and IP material are presented in Fig. 8A and B, respectively. The quantity of TAF90p immunoprecipitated from each sample largely correlated with its relative abundance in the extracts (input samples) and in many cases was not significantly different for samples from cells grown at 23 and 37°C (Fig. 8B). Only the samples from the taf90-16 and taf90-20 mutants showed significantly reduced amounts of TAF90p in the IPs. Virtually every mutant displayed a temperature-dependent defect in its ability to interact with most TFIID subunits examined here. The two exceptions were the taf90-1 mutant, which showed a temperature-independent reduction in its ability to interact with other TFIID subunits, and the taf90-17 mutant, which showed only minor reductions in extracts from cells grown at 37°C (Fig. 8B). The taf90-20 mutant showed the strongest defects. However, since the levels of TAFIIs were severely reduced in the extracts of this mutant, we can be certain only that the taf90-20 allele is extremely disruptive to the TFIID complex. Small amounts of TAF90p, TAF60p, TAF68p, and TAF25p was detected in the IP material from the extracts of this mutant grown at 37°C, which may be attributed to TAFIIs contained in the SAGA complex (see below). This assay also highlights differences between the taf90-17 and taf90-19 alleles. In agreement with what was observed for these two alleles in regard to their effects on the steady-state levels of TAFIIs (Fig. 7), the binding of TFIID subunits to the taf90-19p mutant protein was reduced to a greater degree than that to the taf90-17p mutant protein.
FIG. 8.
Co-IP studies with extracts of TAF90 mutants. Whole-cell extracts were prepared from cells grown at room temperature (approximately 24°C) or at 37°C for 3 h. Complexes containing TAF90p were immunoprecipitated from cell extracts using antibodies raised against TAF90p or TAF145p. Cell extracts and IPs were analyzed by Western blotting using the antibodies indicated. The input material (A) and complexes immunoprecipitated using antibody raised against TAF90p (B) or TAF145p (C) are shown.
Interestingly, while the levels of Tsm1p in the 37°C extracts were not significantly reduced in each of the mutants (Fig. 8A), very strong reductions in its ability to co-IP with the TAF90p mutants were observed (Fig. 8B). Moreover, the taf90-3, taf90-16, and taf90-19 mutants appeared to be more strongly affected in their ability to co-IP Tsm1p versus either TAF145p or TAF47p, suggesting that these mutants somewhat selectively affect the interaction of Tsm1p with TFIID. The ability of TBP to co-IP with TAF90p was severely compromised in all but the taf90-1 mutant when the cells were grown at 37°C (Fig. 8B). Therefore, the majority of the TBP in these extracts (Fig. 8A) likely represents free TBP or TBP distributed among other complexes. In a number of cases, the reduction of TBP in the IP material was disproportionately lower than that of TAF145p; therefore, the TAF145p-TBP interaction alone is not sufficient to maintain TBP in the TFIID complex. A similar result was also found in the analysis of a TAF68/61 mutant (43).
The presence of Spt7p, Ada1p, and Ada3p in the immunoprecipitates was examined to ascertain each mutant's ability to incorporate into the SAGA complex. Most mutants, such as taf90-3, taf90-8, taf90-9, taf90-10, taf90-14, taf90-16, taf90-17, and taf90-19, displayed temperature-dependent interactions with Spt7p, Ada1p, and Ada3p, while the taf90-1 mutant displayed a temperature-independent reduction in the IP efficiency of these three SAGA subunits. Thus, most of the mutants described here are defective for interacting with SAGA in a manner similar to that for TFIID. An interesting exception is the taf90-20 mutant. This allele is the most disruptive to TFIID, but the quantities of these three SAGA subunits detected in the IPs were not significantly reduced. Therefore, the mutations contained in this allele do not affect its ability to interact with at least some SAGA components, and it is significantly more competent to bind to SAGA than most of the other mutants described here. Comparing the results for the taf90-17 and taf90-19 alleles revealed differences in the ability of these two mutants to interact with SAGA versus TFIID components. Whereas the additional amino acid substitution contained in the taf90-17 mutant protein stabilized its interaction with multiple TFIID subunits (see above), it did not confer the same effect on SAGA components: the amounts of Spt7p, Ada1p, and Ada3p contained in the IPs from extracts of the taf90-17 and taf90-19 mutants were similar (Fig. 8B). Thus, it appears that the additional amino acid substitution is compensatory for TFIID but not for SAGA interactions. This hypothesis is consistent with the results indicating that both mutants are equally defective for HIS3 activation (Fig. 6), a process heavily dependent upon SAGA function (for reviews, see references 16 and 56).
While the results of Fig. 7 suggest otherwise, it is a formal possibility that these mutations leave TFIID denuded of TAF90p but otherwise intact after a temperature shift. The assay shown in Fig. 8B measures the ability of TAF90p to interact with TFIID rather than the integrity of the complex; therefore, we repeated the IPs using antiserum to TAF145p (Fig. 8C). Overall, the results obtained using anti-TAF145p antibodies to co-IP TAFIIs confirmed those in Fig. 8B; that is, the quantities of TAFIIs coimmunoprecipitated from extracts of mutants grown at 37°C were significantly reduced. The noted difference is that less Tsm1p, TAF68p, and TAF47p were detected in the anti-TAF145 IPs from the extracts of the taf90-1 and taf90-10 mutants grown at the permissive temperature than in the anti-TAF90p IPs (compare Fig. 8B and C). In addition, no TFIID subunits were detected in the anti-TAF145p IPs from the 37°C extracts of the taf90-20 mutant, even in overexposed blots (not shown); therefore, the small quantities of TAF90p, TAF68p, TAF60p, and TAF25p detected in the TAF90p IPs (Fig. 8B) likely originated from the SAGA complex. In conclusion, our results show that each mutation has different effects on TFIID structure, with taf90-1 and taf90-17 being the least severe and taf90-20 being the most. In addition, comparison of the abilities of the taf90-17p, taf90-19p, and taf90-20p mutant proteins to copurify with TFIID and SAGA components suggests that TAF90p interacts with these two complexes differently.
TAF90 mutations reduce SAGA abundance and activity.
The results described in Fig. 8 indicate that inactivation of most TAF90p mutants adversely affects SAGA subunit levels and/or their ability to interact with the complex. Nonetheless, these are indirect assays for SAGA complex activity, and the ability to co-IP SAGA subunits, as is observed in the taf90-20 mutant, does not necessarily indicate that the mutant complex is functional. The preparation and fractionation of extracts from each of these 11 TAF90 mutants to analyze SAGA, at least in duplicate, would be a significant undertaking. Therefore, three mutants were selected to more carefully determine their effect on the ability of the SAGA complex to acetylate nucleosomes. The taf90-20 mutant was chosen since it completely disrupts TFIID structure and yet appears not to strongly affect its ability to copurify with some SAGA components (Fig. 8B). Thus, it is important to determine if SAGA, or a subcomplex, containing the mutant protein is active. The taf90-17 mutant was chosen because it has the weakest effect on TFIID structure and transcription yet displayed measurable defects in its ability to bind SAGA subunits. Finally, taf90-10 was chosen as a representative of a mutant that displayed defects in both complexes and exhibited a moderate transcription phenotype.
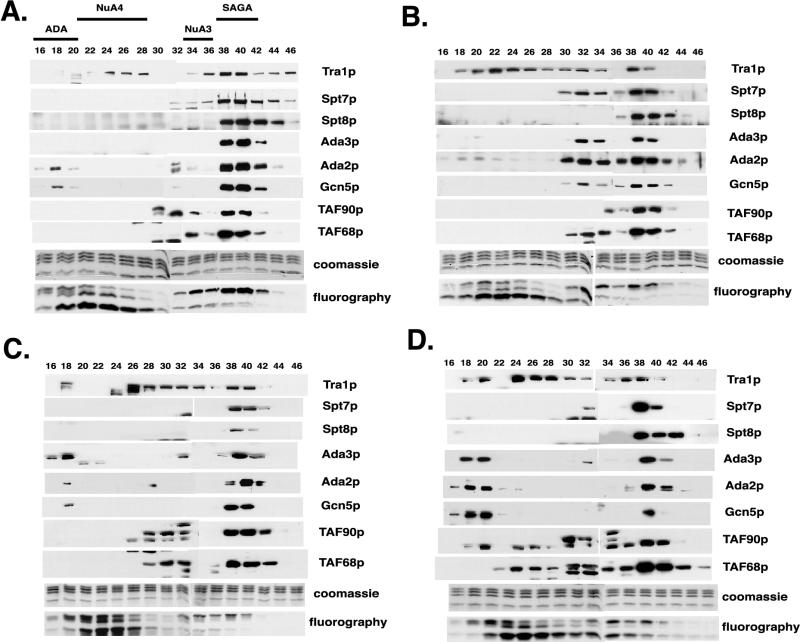
Extracts were prepared from cells grown at 37°C for 3 h, and HAT complexes were purified by sequential chromatography over Ni-NTA affinity and Mono Q ion-exchange columns (14). Aliquots of alternating Mono Q fractions were used in HAT assays and analyzed by Western blotting. Each of the four distinct HAT complexes (14) retained on Ni-NTA-agarose were resolved in the sample from wild-type cells, and their locations in the elution profile are indicated above Fig. 9A. Their identities were assigned by Western blotting for unique subunits (see below and data not shown) and by histone substrate specificity. In each of the mutants, four HAT activities were also resolved, but the amount contained in the fractions where SAGA elutes (fractions 38 to 42) was significantly reduced (Fig. 9B to D). In contrast, the activities of the TAF90p-independent NuA4, ADA, and NuA3 complexes were comparable to those of the wild-type sample and were within the normal variation observed between different preparations (Fig. 9 and data not shown). Next, we examined the elution profiles of individual SAGA components by Western blotting. The Mono Q elution profile of the HAT complexes retained on Ni-agarose from wild-type extracts reveals that all of the SAGA subunits examined coeluted predominantly with their HAT activity in fractions 38 to 42, with the exception of those found in the ADA and NuA4 complexes (Fig. 9A). Western blotting of the fractions from each of the mutants revealed that significant amounts of most subunits were also in earlier fractions (Fig. 9B to D), indicating a breakdown of the mutant SAGA complexes. This is most obvious in the fractionation profile for the taf90-20 mutant (Fig. 9B). Specifically, Tra1p broadly eluted in fractions 18 to 40, and significant amounts of Spt7p, Ada3p, Ada2p, Gcn5p, and TAF68p were detected in fractions 30 to 34 (Fig. 9B). It is not know if this represents a stable subcomplex of SAGA or free subunits that coincidentally coelute in these fractions. In addition to the appearance of breakdown products, the amounts of SAGA components eluting in fractions 38 to 42 were somewhat reduced in the taf90-17 (Fig. 9C) and taf90-20 (Fig. 9B) samples and were drastically reduced in the taf90-8 (Fig. 9D) samples (for a more accurate estimation, see Fig. 10).
FIG. 9.
Purification of HAT complexes from TAF90 mutants. Extracts prepared from mutants grown at 37°C for 3 h were subjected to chromatographic analysis as described in Materials and Methods. Aliquots of the Mono Q column fractions were analyzed by Western blotting and HAT assays using nucleosomes as substrates. Elution profiles for the wild type (A) and the taf90-20 (B), taf90-17 (C), and taf90-10 (D) mutants are shown. Note that the blots presented in panel D were deliberately exposed longer than those in panels A to C to detect the small amounts of SAGA subunits eluting in fractions 38 to 42.
FIG. 10.
Analysis of mutant SAGA complexes. (A) SAGA has reduced HAT activity in TAF90 mutants. Fractions containing SAGA (fractions 38 to 42 from Fig. 9) were pooled and analyzed by Western blotting and HAT assays using oligonucleosomes as substrates. The relative levels of Gcn5p were determined from appropriately exposed blots using NIH Image software and are expressed as the percentage of the wild-type (WT) level. HAT activity was compared to that of SAGA isolated from wild-type cells, which was set at 100%. (B) Size exclusion chromatography. The pooled SAGA-containing fractions from the Mono Q chromatography (fractions 38 to 42 from Fig. 9) were concentrated by filtration and fractionated on a Superose 6 column. Each fraction was analyzed by Western blotting and HAT assays using nucleosomes as a substrate. The void volume corresponds to fraction 14. No HAT activity or SAGA components were detected in later-eluting fractions, and thus only fractions 15 to 23 are shown.
The reduced HAT activity observed in each TAF90 mutant may be due to a reduction in the amount of SAGA, an altered complex with decreased catalytic activity, or both. Therefore, we directly compared the SAGA levels and activity in the peak Mono Q column fractions. Equal amounts (volumes) of the pooled fractions were used to measure the HAT activity using oligonucleosomes as substrates (Fig. 10A, left panel), and an aliquot was withdrawn and subjected to immunoblotting following the HAT assay (Fig. 10A, right panel). The results show that the levels of Gcn5p in the Mono Q fractions were reduced about 2.5- and 5-fold in the taf90-20 and taf90-17 mutants, respectively, whereas the level was reduced approximately 10-fold in the taf90-10 mutant. The level of HAT activity in the taf90-17 samples was roughly proportional to the levels of Gcn5p, but the taf90-20 fractions contained an additional reduction in activity that cannot be accounted for by reduced SAGA levels (Fig. 10A, right panel). For example, this mutant has 43% of wild-type Gcn5p levels but 25% of wild-type HAT activity (Fig. 10A). Thus, even though the SAGA complex from the taf90-20 mutant is largely intact, its mutations reduce the activity of the complex. Higher HAT activity was detected in the taf90-10 fractions compared to the level of Gcn5p. This is likely due to the contamination of these fractions with quantities of the NuA3 complex, which elutes near the SAGA complex (14) (Fig. 9A).
The integrity and activity of the mutant SAGA complexes from the taf90-17 and taf90-20 strains were analyzed by gel filtration chromatography. Unfortunately, the quantities of the SAGA complex from the taf90-10 mutant were not sufficient for this analysis. The results presented in Fig. 10B show that the wild-type and mutant SAGA complexes eluted from the Superose 6 column with the expected mass of approximately 2 MDa (14, 15). While this analysis cannot rule out the possibility of a subtle change in structure or composition, it does suggest that the size of the mutant complexes is not significantly different from that of the wild type. In addition, we did not detect the presence of additional complexes eluting in later fractions by either HAT assays or Western blotting for SAGA subunits (data not shown). It is important to note that as observed in the Mono Q fractions, the nucleosomal HAT activity was significantly reduced (Fig. 10B). The HAT activities, measured in liquid assays, for the taf90-20 and taf90-17 mutants were approximately 19 and 15% of that for the wild type, respectively (not shown). We observed a slightly larger difference in HAT activities between the mutants and the wild-type complexes eluting from the sizing column versus that observed in the Mono Q fractions. This may be attributed to a further reduction in activity in the mutants during the processing of the samples for size exclusion chromatography or their separation from contaminating NuA3 complex. From these results, it appears that mutations in TAF90, like for TAF68/61 (15), disturb the function of the SAGA complex by reducing the total amount of the SAGA complex present in the cell and impairing its ability to acetylate nucleosomes.
DISCUSSION
TAF90 mutants display allele-specific and broad transcription defects.
The relatively thorough characterization of a large number of mutants has allowed us to make a number of observations regarding TAF90 function. The first significant conclusion is that allele-specific defects in transcription can be observed among different mutants of the same gene, despite the fact that all alleles seem tight based upon their temperature-sensitive growth phenotypes. There were a number of clear examples where the expression of a gene was strongly reduced in one or more mutants but not in others. This argues that results obtained with any single temperature-sensitive allele may not reveal all of the functions of this gene. While we have largely concentrated our discussion on cases where the differences between alleles are very obvious, even the subtle differences in the magnitude and kinetics of RNA loss observed among all of the alleles described here could have consequences for the classification of certain genes as being TAF90 dependent when a cutoff is applied. By no means does our study diminish the impact of the genome-wide studies performed to date. However, it already has been recognized that analyzing the genome-wide expression profile of any given mutant under a single growth condition does not constitute a comprehensive analysis of its function (for a review, see reference 61), and now it is clear that multiple alleles of the same gene may be necessary when conditional mutants are used.
The second important observation is that mutations in TAF90 that cause a broad transcription defect can be isolated, as seen in mutants of other TAFIIs that are shared by TFIID and SAGA. Mutants of TAF68/61, TAF60, TAF25, and TAF17 that satisfy this criterion had been described previously (3, 31, 37, 43, 46), but until this study such a mutant of TAF90 had not been. The thermoinactivation of this mutant results in the rapid reduction in total poly(A)+ RNA and in most of the specific messages analyzed here. Significantly, the magnitude and timing of these reductions were strikingly similar to those observed in the rpb1-1 mutant, which is considered the standard for judging primary transcription defects (20, 26). This phenotype is unlikely to result from a reduction in only a small number of abundant messages, because we found that many specific messages, some of which are of low to medium abundance, were reduced likewise. Therefore, mutations in TAF90 that abolish transcription of many yeast genes can be isolated. We recognize that the analysis performed on this mutant thus far cannot accurately determine the exact fraction of the genome that is affected. Nonetheless, it is clear that this mutation does have much stronger effects on the expression of more genes than any of the other TAF90 mutations described here or elsewhere. Equally clear is that inactivation of even the strongest mutant described here did not significantly affect the expression of all genes examined here, thus reinforcing the notion that TAFIIs are unlikely to be obligate transcription factors for the entire genome.
The cause of the broad transcription defects observed in certain TAFII mutants is slowly being revealed. It has been proposed that redundancy between TFIID and SAGA explains this phenotype (26). While this explanation may account for an increase in the total number of genes affected in these mutants, it cannot solely account for the differences between these mutants and mutants of other TAFIIs found only in TFIID. The first evidence for this is that mutations in TAF40 or depletion of TAF48p, two TAFIIs not in SAGA, cause reduced expression of many genes (25, 43). The second is the results presented here. Many of the TAF90 mutants described here show significant reductions in their ability to interact with the SAGA components Ada3p, Ada1p, and Spt7p in co-IP experiments, yet they do not display the broad transcription phenotypes exhibited by the taf90-20 mutant. Moreover, we found that the levels and activities of SAGA complex isolated from the taf90-17, taf90-10, and taf90-8 (not shown) mutants were reduced at least as much as those of the taf90-20 mutant. In light of these results, redundancy between SAGA and TFIID is at best an important contributing factor to the broad transcription defects observed in certain TAFII mutants.
A reoccurring phenotype exhibited by most TAFII mutants that show reduced expression of a large number of genes is that shifting them to the restrictive temperature results in dramatic reductions in the steady-state levels of most TFIID subunits (25, 31, 43, 46). This also is true of the taf90-20 allele characterized here. It is striking how well the magnitudes of the transcription defects described here match the consequences of each mutant for the structure of TFIID. The TAF90 mutants that display the most dramatic reductions in TAFII levels when shifted to 37°C, and are the least competent to bind to TFIID in the co-IP studies, are also those that show the strongest transcription phenotypes. This is particularly apparent when comparing the weakest and strongest classes of mutants, taf90-1 and taf90-17 versus taf90-20, respectively. Therefore, our study argues that it is the rapid loss of many TFIID subunits that, when combined, results in broad transcription defects. This hypothesis is supported by a recent genome-wide analysis of transcription that showed that the combined effect of the loss of function of each individual TFIID subunit can account for the expression of greater than 70% of the genome (26). What now needs to be debated is what type of mutant, or depletion system, is most appropriate to assign functions to TAF90. Depletion strategies and mutants such as taf90-20 are prone to secondary effects. It is likely that a comprehensive analysis of multiple alleles will be necessary.
The C terminus of TAF90p is required for TFIID and SAGA function.
This study has established the importance of the WD40 repeats and the C terminus in maintaining the integrity of the TFIID and SAGA complexes. This result would not have been predicted from previous analysis of hTAF100, which suggested that the WD40 repeats are dispensable for its incorporation into TFIID or other TAF-containing complexes (9). Does this mean that human TFIID and yeast TFIID are different? It is more likely that these results are explained by the incorporation of the truncated hTAF100 into partial TFIID complexes or alternative TAF-containing complexes that had not been identified at the time of that study. We demonstrate that the C terminus of TAF90p is important for its association with TFIID and SAGA, but we cannot say that it is sufficient or that the N terminus does not make important contributions to this function as well. It is likely that regions across the entire length of the protein are required for TFIID and SAGA function. Interestingly, inactivation of nearly every mutant severely compromises its ability to co-IP with Tsm1p, TAF17p, and TBP, even though it retained some ability to copurify with other TAFIIs. This was even true for some of the alleles that have weaker effects on the co-IP of other TFIID subunits, such as TAF47p and TAF145p. This suggests that the interaction of Tsm1, TAF17p, and TBP is strictly dependent upon the integrity of the C terminus of TAF90p. This agrees very well with another analysis of hTAFII100, which demonstrated that it directly interacts with hTAFII31 (TAF17p homologue) and TBP (52). It is not known if Tsm1p, or its homologues, binds to the WD40 repeats of TAF90p.
Our results also indicate that TAF90 is required for SAGA abundance and activity, as the complexes isolated from the taf90-8 (not shown), taf90-10, taf90-17, and taf90-20 mutants are significantly reduced in abundance and/or activity. Even though we did not purify SAGA from every mutant, essentially every allele showed impaired interactions with some SAGA subunits in co-IP studies, in particular Ada1p. Since Ada1 is essential for the structure and function of the SAGA complex (50), we can predict with a high degree of certainty that the activity of SAGA is compromised to some degree in each of the mutants described here. This prediction is supported by the inability of all mutants to support wild-type levels of starvation-induced transcription of HIS3, which is particularly sensitive to mutations in SAGA subunits. The C terminus of TAF90p is required for both SAGA and TFIID function, but it is likely that it binds to these two complexes differently. The first line of evidence for this is that the mutations contained in the taf90-20 allele are the most disruptive to the integrity of TFIID and the least disruptive to that of SAGA. The second is the phenotypes of the taf90-17 and taf90-19 alleles. It is clear that the taf90-19 allele has stronger effects on cell viability, on the integrity of the TFIID complex, and on transcription. However, both alleles are equally disruptive to the SAGA complex as judged by the co-IP studies (Fig. 8B) and are equally defective for the activation of HIS3 (Fig. 6). Interestingly, both alleles contain the same S542F mutation, but taf90-17 has an additional amino acid substitution that is compensatory in nature in regard to its interaction with TFIID but not SAGA. Molecular modeling of the WD40 repeat region of TAF90p, based on the known structure of another WD40 repeat protein, predicts that these mutations are on the same surface of the protein (not shown). Whereas we did not isolate either a TFIID- or a SAGA-specific mutant in this screen, scoring for a temperature-sensitive phenotype may not be selective enough for the identification of such a mutant. Our discovery and analysis of the taf90-17, taf90-19, and taf90-20 mutants suggest that isolating such a mutant is possible. Alternative screens that are more selective, such as an SPT phenotype for SAGA-specific mutants, may result in the identification of these types of mutants, which will further our understanding of the role of TAF90 in these two important transcription regulatory complexes.
ACKNOWLEDGMENTS
We are especially gratefully to Patrick Grant and Jerry Workman for advice on SAGA purification and antibodies. We thank Michael Green, Fred Winston, Tony Weil, Shelley Berger, Steve Buratowski, Rick Young, and Kevin Struhl for providing strains used in our studies and members of the Reese lab and the Penn State gene regulation group for advice and comments on this work.
This research was supported by funds provided by the National Institutes of Health (grant GM58672) to J.C.R.
REFERENCES
- 1.Albright S R, Tjian R. TAFs revisited: more data reveal new twists and confirm old ideas. Gene. 2000;242:1–13. doi: 10.1016/s0378-1119(99)00495-3. [DOI] [PubMed] [Google Scholar]
- 2.Apone L M, Virbasius C M, Reese J C, Green M R. Yeast TAF(II)90 is required for cell-cycle progression through G2/M but not for general transcription activation. Genes Dev. 1996;10:2368–2380. doi: 10.1101/gad.10.18.2368. [DOI] [PubMed] [Google Scholar]
- 3.Apone L M, Virbasius C A, Holstege F C, Wang J, Young R A, Green M R. Broad, but not universal, transcriptional requirement for yTAFII17, a histone H3-like TAFII present in TFIID and SAGA. Mol Cell. 1998;2:653–661. doi: 10.1016/s1097-2765(00)80163-x. [DOI] [PubMed] [Google Scholar]
- 4.Brachmann C B, Davies A, Cost G J, Caputo E, Li J, Heiter P, Boeke J D. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 5.Chen W, Struhl K. Saturation mutagenesis of a yeast his3 TATA element: genetic evidence for a specific TATA-binding protein. Proc Natl Acad Sci USA. 1988;85:2691–2695. doi: 10.1073/pnas.85.8.2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collart M A, Struhl K. CDC39, an essential nuclear protein that negatively regulates transcription and differentially affects the constitutive and inducible HIS3 promoters. EMBO J. 1993;12:177–186. doi: 10.1002/j.1460-2075.1993.tb05643.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cormack B P, Struhl K. The TATA-binding protein is required for transcription by all three nuclear RNA polymerases in yeast cells. Cell. 1992;69:685–696. doi: 10.1016/0092-8674(92)90232-2. [DOI] [PubMed] [Google Scholar]
- 8.Dikstein R, Ruppert S, Tjian R. TAFII250 is a bipartite protein kinase that phosphorylates the base transcription factor RAP74. Cell. 1996;84:781–790. doi: 10.1016/s0092-8674(00)81055-7. [DOI] [PubMed] [Google Scholar]
- 9.Dubrovskaya V, Lavigne A C, Davidson I, Acker J, Staub A, Tora L. Distinct domains of hTAFII100 are required for functional interaction with transcription factor TFIIF beta (RAP30) and incorporation into the TFIID complex. EMBO J. 1996;15:3702–3712. [PMC free article] [PubMed] [Google Scholar]
- 10.Dynlacht B D, Weinzierl R O, Admon A, Tjian R. The dTAFII80 subunit of Drosophila TFIID contains beta-transducin repeats. Nature. 1993;363:176–179. doi: 10.1038/363176a0. [DOI] [PubMed] [Google Scholar]
- 11.Gangloff Y G, Werten S, Romier C, Carre L, Poch O, Moras D, Davidson I. The human TFIID components TAF(II)135 and TAF(II)20 and the yeast SAGA components ADA1 and TAF(II)68 heterodimerize to form histone-like pairs. Mol Cell Biol. 2000;20:340–351. doi: 10.1128/mcb.20.1.340-351.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gangloff Y G, Sanders S L, Romier C, Kirschner D, Weil P A, Tora L, Davidson I. Histone folds mediate selective heterodimerization of yeast TAF(II)25 with TFIID components yTAF(II)47 and yTAF(II)65 and with SAGA component ySPT7. Mol Cell Biol. 2001;21:1841–1853. doi: 10.1128/MCB.21.5.1841-1853.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gangloff Y, Romier C, Thuault S, Werten S, Davidson I. The histone fold is a key structural motif of transcription factor TFIID. Trends Biochem Sci. 2001;26:250–257. doi: 10.1016/s0968-0004(00)01741-2. [DOI] [PubMed] [Google Scholar]
- 14.Grant P A, Duggan L, Cote J, Roberts S M, Brownell J E, Candau R, Ohba R, Owen-Hughes T, Allis C D, Winston F, Berger S L, Workman J L. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- 15.Grant P A, Schieltz D, Pray-Grant M G, Steger D J, Reese J C, Yates J R, 3rd, Workman J L. A subset of TAF(II)s are integral components of the SAGA complex required for nucleosome acetylation and transcriptional stimulation. Cell. 1998;94:45–53. doi: 10.1016/s0092-8674(00)81220-9. [DOI] [PubMed] [Google Scholar]
- 16.Grant P A, Sterner D E, Duggan L J, Workman J L, Berger S L. The SAGA unfolds: convergence of transcription regulators in chromatin-modifying complexes. Trends Cell Biol. 1998;8:193–197. doi: 10.1016/s0962-8924(98)01263-x. [DOI] [PubMed] [Google Scholar]
- 17.Green M R. TBP-associated factors (TAFIIs): multiple, selective transcriptional mediators in common complexes. Trends Biochem Sci. 2000;25:59–63. doi: 10.1016/s0968-0004(99)01527-3. [DOI] [PubMed] [Google Scholar]
- 18.Hahn S. The role of TAFs in RNA polymerase II transcription. Cell. 1998;95:579–582. doi: 10.1016/s0092-8674(00)81625-6. [DOI] [PubMed] [Google Scholar]
- 19.Hoffmann A, Chiang C M, Oelgeschlager T, Xie X, Burley S K, Nakatani Y, Roeder R G. A histone octamer-like structure within TFIID. Nature. 1996;380:356–359. doi: 10.1038/380356a0. [DOI] [PubMed] [Google Scholar]
- 20.Holstege F C, Jennings E G, Wyrick J J, Lee T I, Hengartner C J, Green M R, Golub T R, Lander E S, Young R A. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 21.Jacobson R H, Ladurner A G, King D S, Tjian R. Structure and function of human TAFII250 double bromodomain module. Science. 2000;288:1422–1425. doi: 10.1126/science.288.5470.1422. [DOI] [PubMed] [Google Scholar]
- 22.John S, Howe L, Tafrov S T, Grant P A, Sternglanz R, Workman J L. The something about silencing protein, Sas3, is the catalytic subunit of NuA3, a yTAF(II)30-containing HAT complex that interacts with the Spt16 subunit of the yeast CP (Cdc68/Pob3)-FACT complex. Genes Dev. 2000;14:1196–1208. [PMC free article] [PubMed] [Google Scholar]
- 23.Klebanow E R, Poon D, Zhou S, Weil P A. Isolation and characterization of TAF25, an essential yeast gene that encodes an RNA polymerase II-specific TATA-binding protein-associated factor. J Biol Chem. 1996;271:13706–13715. doi: 10.1074/jbc.271.23.13706. [DOI] [PubMed] [Google Scholar]
- 24.Komarnitsky P B, Klebanow E R, Weil P A, Denis C L. ADR1-mediated transcriptional activation requires the presence of an intact TFIID complex. Mol Cell Biol. 1998;18:5861–5867. doi: 10.1128/mcb.18.10.5861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Komarnitsky P B, Michel B, Buratowski S. TFIID-specific yeast TAF40 is essential for the majority of RNA polymerase II-mediated transcription in vivo. Genes Dev. 1999;13:2484–2489. doi: 10.1101/gad.13.19.2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee T I, Causton H C, Holstege F C, Shen W C, Hannett N, Jennings E G, Winston F, Green M R, Young R A. Redundant roles for the TFIID and SAGA complexes in global transcription. Nature. 2000;405:701–704. doi: 10.1038/35015104. [DOI] [PubMed] [Google Scholar]
- 27.Li B, Reese J C. Derepression of DNA damage-regulated genes requires yeast TAF(II)s. EMBO J. 2000;19:4091–4100. doi: 10.1093/emboj/19.15.4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Macpherson N, Measday V, Moore L, Andrews B. A yeast taf17 mutant requires the Swi6 transcriptional activator for viability and shows defects in cell cycle-regulated transcription. Genetics. 2000;154:1561–1576. doi: 10.1093/genetics/154.4.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martinez E, Kundu T K, Fu J, Roeder R G. A human SPT3-TAFII31-GCN5-L acetylase complex distinct from transcription factor IID. J Biol Chem. 1998;273:23781–23785. doi: 10.1074/jbc.273.37.23781. [DOI] [PubMed] [Google Scholar]
- 30.Matangkasombut O, Buratowski R M, Swilling N W, Buratowski S. Bromodomain factor 1 corresponds to a missing piece of yeast TFIID. Genes Dev. 2000;14:951–962. [PMC free article] [PubMed] [Google Scholar]
- 31.Michel B, Komarnitsky P, Buratowski S. Histone-like TAFs are essential for transcription in vivo. Mol Cell. 1998;2:663–673. doi: 10.1016/s1097-2765(00)80164-1. [DOI] [PubMed] [Google Scholar]
- 32.Mizzen C A, Yang X J, Kokubo T, Brownell J E, Bannister A J, Owen-Hughes T, Workman J, Wang L, Berger S L, Kouzarides T, Nakatani Y, Allis C D. The TAF(II)250 subunit of TFIID has histone acetyltransferase activity. Cell. 1996;87:1261–1270. doi: 10.1016/s0092-8674(00)81821-8. [DOI] [PubMed] [Google Scholar]
- 33.Moqtaderi Z, Bai Y, Poon D, Weil P A, Struhl K. TBP-associated factors are not generally required for transcriptional activation in yeast. Nature. 1996;383:188–191. doi: 10.1038/383188a0. [DOI] [PubMed] [Google Scholar]
- 34.Moqtaderi Z, Yale J D, Struhl K, Buratowski S. Yeast homologues of higher eukaryotic TFIID subunits. Proc Natl Acad Sci USA. 1996;93:14654–14658. doi: 10.1073/pnas.93.25.14654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moqtaderi Z, Keaveney M, Struhl K. The histone H3-like TAF is broadly required for transcription in yeast. Mol Cell. 1998;2:675–682. doi: 10.1016/s1097-2765(00)80165-3. [DOI] [PubMed] [Google Scholar]
- 36.Nakatani Y, Bagby S, Ikura M. The histone folds in transcription factor TFIID. J Biol Chem. 1996;271:6575–6578. doi: 10.1074/jbc.271.12.6575. [DOI] [PubMed] [Google Scholar]
- 37.Natarajan K, Jackson B M, Rhee E, Hinnebusch A G. yTAFII61 has a general role in RNA polymerase II transcription and is required by Gcn4p to recruit the SAGA coactivator complex. Mol Cell. 1998;2:683–692. doi: 10.1016/s1097-2765(00)80166-5. [DOI] [PubMed] [Google Scholar]
- 38.Nonet M, Scafe C, Sexton J, Young R. Eukaryotic RNA polymerase conditional mutant that rapidly ceases mRNA synthesis. Mol Cell Biol. 1987;7:1602–1611. doi: 10.1128/mcb.7.5.1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ogryzko V V, Kotani T, Zhang X, Schlitz R L, Howard T, Yang X J, Howard B H, Qin J, Nakatani Y. Histone-like TAFs within the PCAF histone acetylase complex. Cell. 1998;94:35–44. doi: 10.1016/s0092-8674(00)81219-2. [DOI] [PubMed] [Google Scholar]
- 40.Pham A-D, Sauer F. Ubiquitin-activating/conjugating activity of TAFII250, a mediator of activation of gene expression in Drosophila. Science. 2000;289:2357–2360. doi: 10.1126/science.289.5488.2357. [DOI] [PubMed] [Google Scholar]
- 41.Poon D, Bai Y, Campbell A M, Bjorklund S, Kim Y J, Zhou S, Kornberg R D, Weil P A. Identification and characterization of a TFIID-like multiprotein complex from Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1995;92:8224–8228. doi: 10.1073/pnas.92.18.8224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reese J C, Apone L, Walker S S, Griffin L A, Green M R. Yeast TAFIIs in a multisubunit complex required for activated transcription. Nature. 1994;371:523–527. doi: 10.1038/371523a0. [DOI] [PubMed] [Google Scholar]
- 43.Reese J C, Zhang Z, Kurpad H. Identification of a yeast transcription factor IID subunit, TSG2/TAF48. J Biol Chem. 2000;275:17391–17398. doi: 10.1074/jbc.M001635200. [DOI] [PubMed] [Google Scholar]
- 44.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 45.Sanders S L, Klebanow E R, Weil P A. TAF25p, a non-histone-like subunit of TFIID and SAGA complexes, is essential for total mRNA gene transcription in vivo. J Biol Chem. 1999;274:18847–18850. doi: 10.1074/jbc.274.27.18847. [DOI] [PubMed] [Google Scholar]
- 46.Sanders S L, Weil P A. Identification of two novel TAF subunits of the yeast Saccharomyces cerevisiae TFIID complex. J Biol Chem. 2000;275:13895–13900. doi: 10.1074/jbc.275.18.13895. [DOI] [PubMed] [Google Scholar]
- 47.Shen W C, Green M R. Yeast TAF(II)145 functions as a core promoter selectivity factor, not a general coactivator. Cell. 1997;90:615–624. doi: 10.1016/s0092-8674(00)80523-1. [DOI] [PubMed] [Google Scholar]
- 48.Sikorski R S, Boeke J D. In vitro mutagenesis and plasmid shuffling: from cloned gene to mutant yeast. Methods Enzymol. 1991;194:302–318. doi: 10.1016/0076-6879(91)94023-6. [DOI] [PubMed] [Google Scholar]
- 49.Smith T F, Gaitatzes C, Saxena K, Neer E J. The WD repeat: a common architecture for diverse functions. Trends Biochem Sci. 1999;24:181–185. doi: 10.1016/s0968-0004(99)01384-5. [DOI] [PubMed] [Google Scholar]
- 50.Sterner D E, Grant P A, Roberts S M, Duggan L J, Belotserkovskaya R, Pacella L A, Winston F, Workman J L, Berger S L. Functional organization of the yeast SAGA complex: distinct components involved in structural integrity, nucleosome acetylation, and TATA-binding protein interaction. Mol Cell Biol. 1999;19:86–98. doi: 10.1128/mcb.19.1.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tanese N, Saluja D, Vassallo M F, Chen J L, Admon A. Molecular cloning and analysis of two subunits of the human TFIID complex: hTAFII130 and hTAFII100. Proc Natl Acad Sci USA. 1996;93:13611–13616. doi: 10.1073/pnas.93.24.13611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tao Y, Guermah M, Martinez E, Oelgeschlager T, Hasegawa S, Takada R, Yamamoto T, Horikoshi M, Roeder R G. Specific interactions and potential functions of human TAFII100. J Biol Chem. 1997;272:6714–6721. doi: 10.1074/jbc.272.10.6714. [DOI] [PubMed] [Google Scholar]
- 53.Walker S S, Reese J C, Apone L M, Green M R. Transcription activation in cells lacking TAFIIs. Nature. 1996;383:185–188. doi: 10.1038/383185a0. [DOI] [PubMed] [Google Scholar]
- 54.Walker S S, Shen W C, Reese J C, Apone L M, Green M R. Yeast TAF(II)145 required for transcription of G1/S cyclin genes and regulated by the cellular growth state. Cell. 1997;90:607–614. doi: 10.1016/s0092-8674(00)80522-x. [DOI] [PubMed] [Google Scholar]
- 55.Wieczorek E, Brand M, Jacq X, Tora L. Function of TAF(II)-containing complex without TBP in transcription by RNA polymerase II. Nature. 1998;393:187–191. doi: 10.1038/30283. [DOI] [PubMed] [Google Scholar]
- 56.Winston F. Analysis of SPT genes: a genetic approach toward analysis of TFIID, histone, and other transcription factors in yeast. In: McKnight S L, Yamamoto K R, editors. Transcriptional regulation. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 1271–1293. [Google Scholar]
- 57.Winston F, Chaleff D T, Valent B, Fink G R. Mutations affecting Ty-mediated expression of the HIS4 gene of Saccharomyces cerevisiae. Genetics. 1984;107:179–197. doi: 10.1093/genetics/107.2.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wootner M, Wade P A, Bonner J, Jaehning J A. Transcriptional activation in an improved whole-cell extract from Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:4555–4560. doi: 10.1128/mcb.11.9.4555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xie X, Kokubo T, Cohen S L, Mirza U A, Hoffmann A, Chait B T, Roeder R G, Nakatani Y, Burley S K. Structural similarity between TAFs and the heterotetrameric core of the histone octamer. Nature. 1996;380:316–322. doi: 10.1038/380316a0. [DOI] [PubMed] [Google Scholar]
- 60.Yamamoto T, Poon D, Weil P A, Horikoshi M. Molecular genetic elucidation of the tripartite structure of the Schizosaccharomyces pombe 72 kDa TFIID subunit which contains a WD40 structural motif. Genes Cells. 1997;2:245–254. doi: 10.1046/j.1365-2443.1997.1180316.x. [DOI] [PubMed] [Google Scholar]
- 61.Young R A. Biomedical discovery with DNA arrays. Cell. 2000;102:9–15. doi: 10.1016/s0092-8674(00)00005-2. [DOI] [PubMed] [Google Scholar]