Abstract
Amyotrophic lateral sclerosis (ALS) is a fatal neurodegenerative disease. Its complex pathogenesis and phenotypic heterogeneity hinder therapeutic development and early diagnosis. Altered RNA metabolism is a recurrent pathophysiologic theme, including distinct microRNA (miRNA) profiles in ALS tissues. We profiled miRNAs in accessible biosamples, including skin fibroblasts and whole blood and compared them in age- and sex-matched healthy controls versus ALS participants with and without repeat expansions to chromosome 9 open reading frame 72 (C9orf72; C9-ALS and nonC9-ALS), the most frequent ALS mutation. We identified unique and shared profiles of differential miRNA (DmiRNA) levels in each C9-ALS and nonC9-ALS tissues versus controls. Fibroblast DmiRNAs were validated by quantitative real-time PCR and their target mRNAs by 5-bromouridine and 5-bromouridine-chase sequencing. We also performed pathway analysis to infer biological meaning, revealing anticipated, tissue-specific pathways and pathways previously linked to ALS, as well as novel pathways that could inform future research directions. Overall, we report a comprehensive study of a miRNA profile dataset from C9-ALS and nonC9-ALS participants across two accessible biosamples, providing evidence of dysregulated miRNAs in ALS and possible targets of interest. Distinct miRNA patterns in accessible tissues may also be leveraged to distinguish ALS participants from healthy controls for earlier diagnosis. Future directions may look at potential correlations of miRNA profiles with clinical parameters.
Introduction
Amyotrophic lateral sclerosis (ALS) is a progressive neurodegenerative disease affecting motor neurons in the spinal cord, brainstem and brain (1). Motor neuron loss results in skeletal muscle atrophy and weakness, ultimately leading to respiratory failure and death within 2–4 years of diagnosis (1). ALS is familial in ~15% of patients and sporadic in the remaining 85%. Mutation to chromosome 9 open reading frame 72 (C9orf72) is the most common among the approximate 40 genes associated with ALS (2). Only two drugs, riluzole (3) and edaravone (4), are approved by the US Food and Drug Administration for treating ALS, but they only slow disease progression modestly. Clinical trial evidence suggests that treatment may be more effective if initiated earlier, including with edaravone, methylcobalamin and combined taurursodiol-sodium phenylbutyrate (4–6). However, this idea remains unproven, further emphasizing the critical need for novel ALS therapeutic targets as well as biomarkers to facilitate earlier diagnosis.
Unfortunately, the complex molecular mechanisms underlying ALS hinder these goals (2). Epigenetics and RNA processing are strong undercurrents in ALS pathogenesis since inclusion bodies of the protein TAR DNA-binding protein 43 (TDP-43) are an almost universal pathologic finding in ALS (2). TDP-43 binds DNA and RNA in cells, regulating transcriptional repression, pre-mRNA splicing, mRNA translation and microRNAs (miRNAs) biogenesis and processing (7,8). miRNAs are approximately 22 nucleotide-long non-coding RNAs that negatively regulate gene expression by destabilizing mRNA, which modulates numerous physiological processes (9–12). miRNAs are highly expressed in the nervous system (13,14) and may play a role in ALS pathogenesis through altered RNA and protein metabolism, neuromuscular junction structure and function, neurogenesis and inflammation (15). Additionally, miRNAs are dysregulated in ALS mouse models (16) and human tissues (17,18) and may reflect disease state and progression (19).
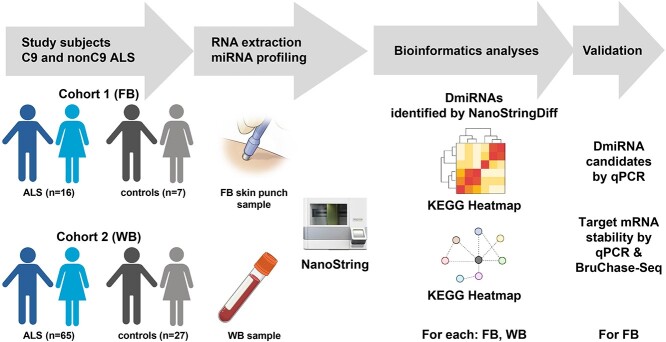
miRNA analysis in ALS can thus be leveraged for a dual purpose, providing both biological insight into disease mechanisms and serving as a diagnostic biomarker. The goal of the current investigation was to understand differences in miRNA levels across two tissues, skin fibroblasts and whole blood (WB), from C9orf72 positive (C9-ALS) and negative (nonC9-ALS) participants against age- and sex-matched controls (Fig. 1). Pathway analysis yielded biological insights that were conserved or unique across tissue types and genetic backgrounds associated with ALS and provided information for future investigations.
Figure 1.
Study design. miRNA levels in fibroblasts (FB) and WB were evaluated from two cohorts of C9-ALS and nonC9-ALS participants versus control samples using NanoString. Differential miRNAs (DmiRNAs) were identified by NanoStringDiff and compared across tissue type and in C9-ALS and nonC9-ALS versus controls. DmiRNAs were validated by qPCR and their predicted target mRNAs by BruChase-Seq. Biological meaning from DmiRNAs was inferred by pathway analysis, and random forest was applied to leverage DmiRNAs as biomarkers. KEGG, Kyoto Encyclopedia of Genes and Genomes. Generated in part using BioRender.com.
Results
Cohort
Fibroblasts were obtained and cultured from C9-ALS (n = 8), nonC9-ALS (n = 8) and controls without any neurological disorders (n = 7). WB samples were collected from a larger cohort, including15 C9-ALS, 50 nonC9-ALS and 27 control participants. Twelve out of 23 fibroblast samples were from the same subjects included in the WB samples. Demographics and clinical characteristics of the cohort, stratified by biosample, are outlined in Table 1.
Table 1.
Demographics and clinical characteristics of ALS participants and controls
| Fibroblasts | WB | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| NonC9-ALS | C9-ALS | Control | P-value | NonC9-ALS | C9-ALS | Control | P-value | ||
| N | 8 | 8 | 7 | 50 | 15 | 27 | |||
| Age | Years (mean ± SD) | 59.4 ± 6.8 | 58.2 ± 5.9 | 58.4 ± 7.2 | 0.71 | 62.9 ± 12.0 | 57.6 ± 5.7 | 61.6 ± 10.4 | 0.21 |
| Sex | Male | 5 | 4 | 4 | 0.88 | 25 | 6 | 15 | 0.63 |
| Female | 3 | 4 | 3 | 25 | 9 | 12 | |||
| Race | White | 8 | 8 | 7 | 49 | 14 | 23 | ||
| African American | . | . | . | . | . | 4 | |||
| Other/Not reported | . | . | . | 1 | 1 | . | |||
| Onset segment | Bulbar | 1 | 1 | . | 1.00 | 21 | 3 | . | 0.27 |
| Cervical | 2 | 2 | . | 13 | 5 | . | |||
| Lumbar | 5 | 5 | . | 16 | 7 | . | |||
| Initial El Escorial criteria | Definite | 11 | 2 | . | 0.42 | . | . | . | 0.59 |
| Probable | 15 | 8 | . | 6 | 5 | . | |||
| Probable, lab supported | 20 | 4 | . | 2 | 3 | . | |||
| Possible/suspected | 4 | 1 | . | . | . | . | |||
| Missing | . | . | . | . | . | . | |||
| Symptom duration | Days (mean ± SD) | 1268 ± 830 | 589 ± 53 | . | 0.04* | 1062 ± 930 | 579 ± 316 | . | 0.08 |
| ALS-FRS | Points (median + IQR) | 36.0 (34.8–40.0) | 36.0 (34.5–40.5) | . | 0.65 | 36.0 (33.0–41.0) | 35.0 (33.0–44.5) | . | 0.50 |
The significant differences among the groups (NonC9-ALS, C9-ALS and Control) were tested using one-way ANOVA for continuous variables and Chi-Square test for categorical variables with the significance cutoff of 0.05. * indicates P-value < 0.05. SD, Standard Deviation. ALS-FRS: Amyotrophic Lateral Sclerosis Functional Rating Scale. IQR, Inter-Quartile Range.
miRNA expression varies by tissue type
The two tissues were selected based on accessibility since we sought to leverage miRNA profiles as a potential diagnostic tool. Profiling indicated that fibroblasts and WB (all fractions including both plasma and blood cellular components) miRNAs had distinct non-overlapping miRNA expression profiles (Supplementary Material, Fig. S1A). Within each tissue type, group-specific samples (i.e. C9-ALS, nonC9-ALS, controls) only clustered with fibroblasts (Supplementary Material, Fig. S1B, left panel). There was a high degree of heterogeneity among the WB C9-ALS, nonC9-ALS and control groups (Supplementary Material, Fig. S1B, right panel).
C9-ALS and nonC9-ALS have differential miRNA levels in fibroblasts and WB
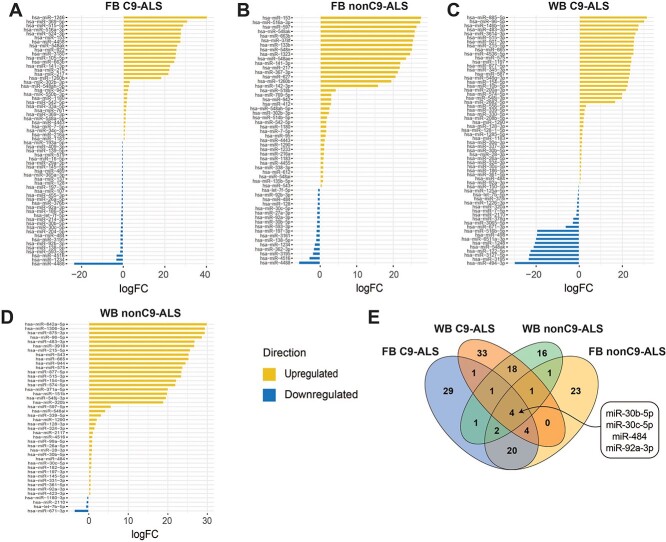
We identified differential miRNA (DmiRNA) levels in C9-ALS and nonC9-ALS versus controls using NanoStringDiff (20) (Supplementary Material, Tables S1 and S2). Raw count data were normalized with the positive and negative control probes as well as the reference probes, embedded in the NanoString nCount system, which showed consistent patterns across samples (Supplementary Material, Fig. S2). NanoStringDiff models count data using a generalized linear model of the negative binomial family and the likelihood ratio test, which has superior performance for identifying differentially expressed genes (20). We found a total of 62 fibroblast DmiRNAs (31 upregulated and 31 downregulated) in C9-ALS versus control samples (Fig. 2A). In nonC9-ALS fibroblasts, there were 55 DmiRNAs, including 38 increased and 17 decreased miRNAs versus control samples (Fig. 2B). WB also had slightly more DmiRNAs in C9-ALS than in nonC9-ALS versus control samples. C9-ALS WB had 62 DmiRNAs, of which 42 were increased and 20 were decreased in C9-ALS (Fig. 2C). Analysis of nonC9-ALS WB samples produced a total of 44 DmiRNAs, the majority of which (n = 40) were increased in nonC9-ALS, with only 4 decreased relative to controls (Fig. 2D).
Figure 2.
Fibroblast and WB DmiRNAs from C9-ALS and nonC9-ALS versus control participants. Fold-change (x-axis) of differential miRNAs (DmiRNAs; P < 0.05; y-axis) identified by NanoStringDiff. DmiRNAs that increased in ALS versus controls in yellow; DmiRNAs that decreased in ALS versus controls in blue. Plots for fibroblasts (FB) in (A) C9-ALS and (B) nonC9-ALS versus controls; plots for WB in (C) C9-ALS and (D) nonC9-ALS versus controls; yellow, upregulated in ALS versus controls; blue downregulated in ALS versus controls. (E) Venn diagram of the number of shared and unique DmiRNAs between fibroblasts and WB for C9-ALS and nonC9-ALS groups.
C9-ALS and nonC9-ALS have shared and unique DmiRNAs in fibroblasts and WB
C9-ALS and nonC9-ALS shared four DmiRNAs (miR-30b-5p, miR-30c-5p, miR-484, miR-92a-3p) across both fibroblasts (decreased miRNA levels) and WB (increased miRNA levels) (Fig. 2E, Supplementary Material, Tables S1 and S2). In fibroblasts, 30 DmiRNAs overlapped between C9-ALS and nonC9-ALS out of 87 total DmiRNAs (34%), which differed in ALS versus control samples. All shared fibroblast DmiRNAs had the same direction of change in C9-ALS and nonC9-ALS versus controls. In WB, 24 DmiRNAs overlapped between C9-ALS and nonC9-ALS out of 82 DmiRNAs (29%), which differed in ALS versus control samples. All shared WB DmiRNAs differed in ALS versus control in the same direction.
C9-ALS and nonC9-ALS share DmiRNA-regulated biological pathways in fibroblasts and WB
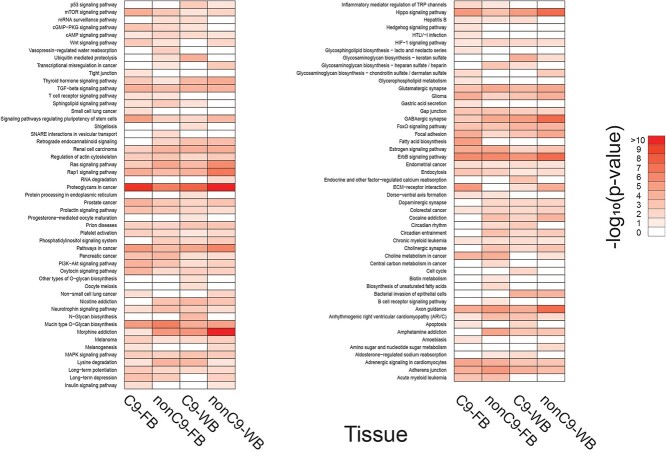
Functional enrichment analysis identified 102 overrepresented biological pathways from statistically significant DmiRNAs in fibroblasts and WB (Fig. 3). Of these, 44 enriched Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways were shared across all four groups (Fig. 3; Supplementary Material, Table S3). There were 54 KEGG pathways shared by C9-ALS and nonC9-ALS fibroblasts, including ‘proteoglycans in cancer’ and ‘ErbB signaling pathway’ as the top two most significant ones (Table 2). In WB, 56 KEGG pathways were shared by C9-ALS and nonC9-ALS, including ‘proteoglycans in cancer’, ‘morphine addiction’ and ‘GABAergic synapse’ (Table 3).
Figure 3.
KEGG pathway analysis of DmiRNAs. Heat-map of significantly enriched KEGG pathways identified for each of the DmiRNA datasets represented by a log10-based color and number index. FB, fibroblasts.
Table 2.
Top functions of differential miRNAs from fibroblasts
| Type | KEGG ID | Pathway description | #miRNAs | #genes | Adjusted P-value |
|---|---|---|---|---|---|
| C9-ALS | hsa05205 | Proteoglycans in cancer | 45 | 147 | 2.81E−10 |
| hsa04012 | ErbB signaling pathway | 44 | 71 | 1.18E−06 | |
| hsa04152 | AMPK signaling pathway | 45 | 99 | 1.30E−06 | |
| hsa04512 | ECM-receptor interaction | 38 | 57 | 5.98E−06 | |
| hsa04390 | Hippo signaling pathway | 46 | 109 | 6.24E−06 | |
| hsa00512 | Mucin type O-Glycan biosynthesis | 25 | 21 | 7.77E−06 | |
| hsa05200 | Pathways in cancer | 48 | 273 | 7.77E−06 | |
| hsa00061 | Fatty acid biosynthesis | 20 | 9 | 1.12E−05 | |
| hsa04550 | Signaling pathways regulating pluripotency of stem cells | 46 | 103 | 1.23E−05 | |
| hsa04261 | Adrenergic signaling in cardiomyocytes | 45 | 105 | 2.94E−05 | |
| hsa04915 | Estrogen signaling pathway | 43 | 71 | 5.11E−05 | |
| hsa04360 | Axon guidance | 45 | 95 | 5.11E−05 | |
| hsa04510 | Focal adhesion | 44 | 149 | 5.87E−05 | |
| hsa04015 | Rap1 signaling pathway | 46 | 151 | 5.87E−05 | |
| hsa04150 | mTOR signaling pathway | 44 | 51 | 6.26E−05 | |
| hsa05214 | Glioma | 44 | 49 | 6.26E−05 | |
| hsa04350 | TGF-beta signaling pathway | 41 | 59 | 7.58E−05 | |
| hsa04520 | Adherens junction | 44 | 59 | 7.58E−05 | |
| hsa05215 | Prostate cancer | 44 | 69 | 1.22E−04 | |
| hsa04151 | PI3K-Akt signaling pathway | 47 | 231 | 1.34E−04 | |
| nonC9-ALS | hsa00512 | Mucin type O-Glycan biosynthesis | 23 | 20 | 3.43E−07 |
| hsa05205 | Proteoglycans in cancer | 38 | 135 | 3.43E−07 | |
| hsa04012 | ErbB signaling pathway | 37 | 68 | 9.17E−07 | |
| hsa04360 | Axon guidance | 36 | 91 | 4.44E−06 | |
| hsa05032 | Morphine addiction | 36 | 65 | 5.11E−06 | |
| hsa04520 | Adherens junction | 39 | 58 | 6.67E−06 | |
| hsa05031 | Amphetamine addiction | 33 | 49 | 2.33E−05 | |
| hsa04152 | AMPK signaling pathway | 38 | 90 | 2.33E−05 | |
| hsa04014 | Ras signaling pathway | 41 | 153 | 2.79E−05 | |
| hsa04915 | Estrogen signaling pathway | 35 | 65 | 3.85E−05 | |
| hsa04727 | GABAergic synapse | 36 | 60 | 3.85E−05 | |
| hsa04724 | Glutamatergic synapse | 37 | 82 | 3.85E−05 | |
| hsa05231 | Choline metabolism in cancer | 38 | 76 | 7.47E−05 | |
| hsa04261 | Adrenergic signaling in cardiomyocytes | 37 | 99 | 9.34E−05 | |
| hsa00310 | Lysine degradation | 33 | 34 | 1.29E−04 | |
| hsa05211 | Renal cell carcinoma | 35 | 52 | 1.43E−04 | |
| hsa05212 | Pancreatic cancer | 35 | 49 | 2.08E−04 | |
| hsa04015 | Rap1 signaling pathway | 38 | 140 | 2.84E−04 | |
| hsa05200 | Pathways in cancer | 40 | 261 | 3.13E−04 | |
| hsa04350 | TGF-beta signaling pathway | 31 | 56 | 3.71E−04 |
Table 3.
Top functions of differential miRNAs from WB
| Type | KEGG ID | Pathway description | #miRNAs | #genes | Adjusted P-value |
|---|---|---|---|---|---|
| C9-ALS | hsa05205 | Proteoglycans in cancer | 57 | 157 | 1.74E−08 |
| hsa04012 | ErbB signaling pathway | 54 | 72 | 3.40E−06 | |
| hsa05032 | Morphine addiction | 54 | 72 | 3.40E−06 | |
| hsa04390 | Hippo signaling pathway | 53 | 113 | 7.86E−06 | |
| hsa04727 | GABAergic synapse | 51 | 67 | 9.30E−06 | |
| hsa05200 | Pathways in cancer | 61 | 286 | 2.39E−05 | |
| hsa04360 | Axon guidance | 56 | 97 | 6.49E−05 | |
| hsa00512 | Mucin type O-Glycan biosynthesis | 31 | 22 | 6.57E−05 | |
| hsa00533 | Glycosaminoglycan biosynthesis—keratan sulfate | 17 | 12 | 8.38E−05 | |
| hsa05100 | Bacterial invasion of epithelial cells | 51 | 63 | 8.95E−05 | |
| hsa05211 | Renal cell carcinoma | 48 | 54 | 9.14E−05 | |
| hsa04520 | Adherens junction | 51 | 60 | 9.14E−05 | |
| hsa04120 | Ubiquitin mediated proteolysis | 52 | 106 | 9.14E−05 | |
| hsa04015 | Rap1 signaling pathway | 57 | 157 | 9.85E−05 | |
| hsa04350 | TGF-beta signaling pathway | 48 | 62 | 1.28E−04 | |
| hsa04068 | FoxO signaling pathway | 53 | 104 | 1.40E−04 | |
| hsa05033 | Nicotine addiction | 46 | 31 | 2.13E−04 | |
| hsa00310 | Lysine degradation | 48 | 38 | 2.28E−04 | |
| hsa04919 | Thyroid hormone signaling pathway | 55 | 90 | 2.28E−04 | |
| hsa04724 | Glutamatergic synapse | 56 | 86 | 2.75E−04 | |
| nonC9-ALS | hsa05032 | Morphine addiction | 30 | 68 | 1.57E−11 |
| hsa05205 | Proteoglycans in cancer | 34 | 140 | 4.15E−11 | |
| hsa04727 | GABAergic synapse | 28 | 63 | 3.95E−08 | |
| hsa04360 | Axon guidance | 33 | 92 | 4.79E−08 | |
| hsa04390 | Hippo signaling pathway | 33 | 102 | 4.79E−08 | |
| hsa04012 | ErbB signaling pathway | 34 | 68 | 4.79E−08 | |
| hsa04015 | Rap1 signaling pathway | 36 | 147 | 1.87E−07 | |
| hsa05200 | Pathways in cancer | 37 | 255 | 6.01E−07 | |
| hsa04014 | Ras signaling pathway | 37 | 150 | 1.33E−06 | |
| hsa00512 | Mucin type O-Glycan biosynthesis | 20 | 20 | 1.39E−05 | |
| hsa04724 | Glutamatergic synapse | 33 | 77 | 3.45E−05 | |
| hsa04510 | Focal adhesion | 35 | 139 | 3.52E−05 | |
| hsa04919 | Thyroid hormone signaling pathway | 35 | 81 | 4.76E−05 | |
| hsa04512 | ECM-receptor interaction | 27 | 51 | 5.02E−05 | |
| hsa05211 | Renal cell carcinoma | 27 | 50 | 7.22E−05 | |
| hsa05214 | Glioma | 28 | 46 | 9.21E−05 | |
| hsa05030 | Cocaine addiction | 26 | 34 | 1.47E−04 | |
| hsa04350 | TGF-beta signaling pathway | 29 | 53 | 1.47E−04 | |
| hsa04068 | FoxO signaling pathway | 32 | 91 | 1.47E−04 | |
| hsa05100 | Bacterial invasion of epithelial cells | 30 | 55 | 1.49E−04 |
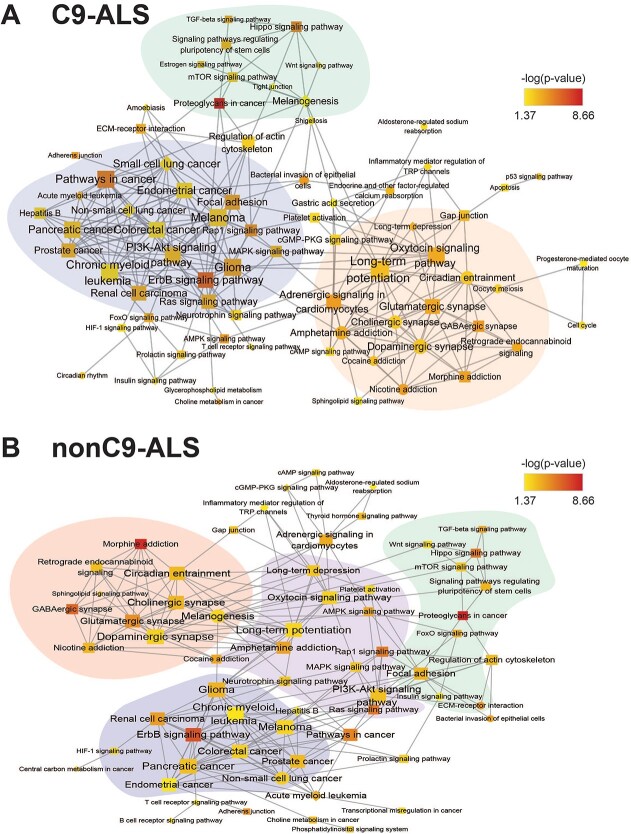
To identify the main themes of these enriched pathways, we built an association network using our in-house tool richR from the significant pathways in fibroblasts and WB. The generated network clusters enriched pathways with similar gene content. Highly interconnected subnetworks are highlighted by distinct colors (Fig. 4). In both C9-ALS (Fig. 4A) and nonC9-ALS (Fig. 4B) networks, subnetworks were centered around pathways related to neuronal functions (e.g. ‘glutamatergic synapse’, ‘dopaminergic synapse’, ‘long-term potentiation’), cancer or cell proliferation (e.g. ‘glioma’, ‘ErbB signaling pathway’) and metabolism and inflammation (e.g. ‘mTOR signaling pathway’).
Figure 4.
KEGG pathway association networks. Significantly enriched KEGG pathways were combined and visualized in a network for (A) C9-ALS and (B) nonC9-ALS samples. KEGG pathways are represented by nodes; shared gene content between pathways are represented by edges. Within the network, node shape indicates the tissue source of the enriched pathways: diamond, fibroblasts only; circle, WB only; square, both fibroblasts and WB. Node color is based on –log10 (P-value). Node size corresponds to the number of connections each node has. All networks were organized by the inverted self-organizing map layout with minimal manual node rearrangement for visibility. Highly inter-connected subnetworks were identified by Cytoscape MCODE and are highlighted by various colors. Single nodes, which are not connected to other nodes, were excluded from this network visualization.
Fibroblast DmiRNAs inversely correspond to target mRNA stability
miR-186-5p and miR-16-5p were underrepresented in C9-ALS fibroblasts, while miR-543 was overrepresented in nonC9-ALS fibroblasts by NanoString analysis (Fig. 2A and B). To validate these findings, we used quantitative real-time PCR (qPCR) to quantify miR-186-5p and miR-16-5p in C9-ALS and miR-543 in nonC9-ALS fibroblast RNA (Supplementary Material, Fig. S3). As anticipated, miR-186-5p levels were lower in C9-ALS versus control fibroblasts (P = 0.0236). Similarly, miR-16-5p transcripts were lower in C9-ALS versus control fibroblasts, although this only approached statistical significance (P = 0.0599). In nonC9-ALS fibroblasts, miR-543 was higher than in controls (P = 0.0423), whereas it did not statistically differ in C9-ALS versus control fibroblasts (P = 0.2773) as expected.
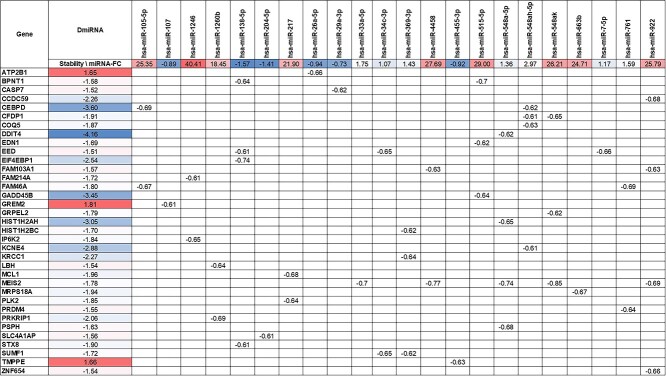
miRNAs negatively regulate their mRNA targets. We previously analyzed the stability of fibroblast RNA from C9-ALS and nonC9-ALS samples by Bru-seq and BruChase-seq, techniques that quantitatively measure mRNAs stability (21,22). We performed a correlation analysis between DmiRNA levels with their predicted mRNA target stability in fibroblasts corresponding to a subset of the published cohort (Fig. 5). Inverse Spearman correlations were observed for 22 DmiRNAs, including miR-1246 and miR-515-5p, and 35 mRNAs, including growth arrest and DNA-damage-inducible, beta (GADD45B) and inositol hexakisphosphate kinase 2 (IP6K2) (Fig. 5). We validated the stability of GADD45B mRNA, a predicted miR-515-5p target with the largest fold-change, and of IP6K2 mRNA, a predicted miR-1246 target with the third largest fold-change, in C9-ALS fibroblasts by qPCR (Fig. 6). miR-1246 was highly upregulated in C9-ALS fibroblasts, and, as anticipated, the stability of its mRNA IP6K2 target was significantly diminished versus controls (P = 0.0181). Similarly, miR-515-5p transcripts were increased in C9-ALS fibroblasts, which is reflected in a lower GADD45B mRNA stability relative to controls, although this only approached statistical significance (P = 0.0555).
Figure 5.
Identifying potential DmiRNA as regulators of mRNA stability in fibroblasts. miRNA fold-change (FC) for differential miRNAs (DmiRNAs; top row) in C9-ALS fibroblast. Predicted target mRNA (first column) stability values (mature/nascent; second column) are from our previously published study (21). Values in other table cells are the significant Pearson correlation coefficients between each pair of DmiRNA and its predicted mRNA targets; color represents the degree of differential expression in fold-changes (red up-regulation in ALS, blue down-regulation in ALS). Stability-FC and miRNA-FC were scaled independently.
Figure 6.
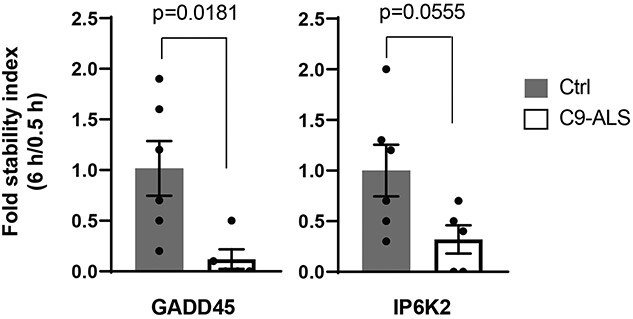
Validation of target mRNA in fibroblasts. Target mRNA in fibroblasts were validated by qPCR. Results were normalized to yWHAZ and presented as fold-change calculated by the 2-∆CT method for GADD45 [C9-ALS, n = 5; Control (Ctrl), n = 6], IP6K2 (C9-ALS, n = 5; Ctrl, n = 6). Transcript stability was determined as the ratio between transcript abundance at 6 h (pulse/chase) to 0.5 h (pulse) and compared with Ctrl. Experiment performed in duplicate; analysis by Student’s t-test; data represented as mean ± standard error of the mean.
Random forest analysis of DmiRNAs
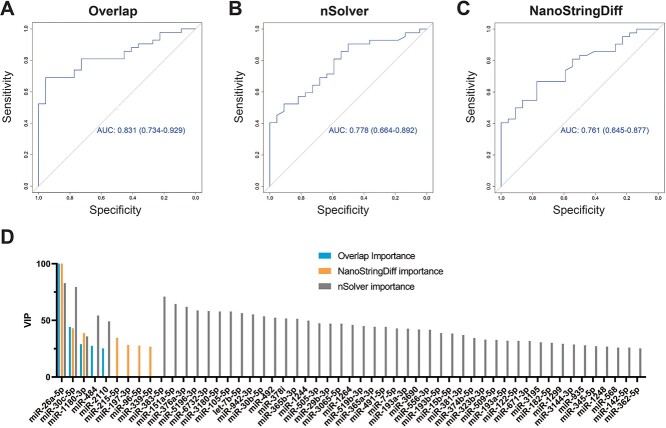
We next performed a random forest analysis on DmiRNAs from nonC9-ALS WB samples (n = 80) to determine whether miRNAs can classify nonC9-ALS participants from controls. There were too few C9-ALS samples (n = 16) for a random forest analysis. We ran the analysis in three formats, DmiRNAs identified by NanoStringDiff (n = 46), an additional DmiRNA set identified by a different analysis tool (n = 60), nSolver from NanoString, and the DmiRNAs (n = 11) overlapping between the two tools. Overlapping DmiRNAs produced a receiver operating characteristic (ROC) curve with the greatest area under the curve (AUC) of 0.831 [95% Confidence Interval (CI) 0.734–0.929; Fig. 7A], possibly because employing overlapping DmiRNAs minimized noise. The next best ROC had AUC 0.778 (95%CI 0.664–0.892), using nSolver (Fig. 7B). Last was the ROC generated using DmiRNAs identified by NanoStrongDiff (AUC 0.761, 95%CI 0.645–0.877; Fig. 7C). In all instances, miR-26a-5p emerged as the top candidate, which differentiated nonC9-ALS from control samples, followed by miR-30c-5p (Fig. 7D). This result suggests that the small subset of common DmiRNAs identified by the two tools have the most classifying power between ALS and control. Pathway analysis of genes regulated by miR-26a-5p identified enriched pathways, including ‘protein processing in endoplasmic reticulum’ (hsa04141, P = 6.09e−06), ‘hippo signaling pathway’ (hsa04390, P = 4.04e−05) and ‘biosynthesis of unsaturated fatty acids’ (hsa01040, P = 0.00046).
Figure 7.
Random forest analysis using overlapping DmiRNAs. ROC curves for random forest analysis of (A) overlap DmiRNAs (shared by NanoStringDiff and nSolver) with an AUC of 0.831 (95% CI 0.734–0.929), (B) nSolver AUC 0.778 (95%CI 0.664–0.892) and (C) NanoStringDiff AUC 0.761 (95%CI 0.645–0.877). (D) Variable importance in projection plot ranking importance of DmiRNAs to all three classifiers, overlap (blue), NanoStringDiff (orange) and nSolver (grey). miR-26a-5p was the top candidate, followed by miR-30c-5p, across all methods.
Discussion
Familial ALS comprises ~15% of all cases and the most common genetic mutation is a hexanucleotide repeat expansion in C9orf72 (2). The underlying disease etiology remains unknown in the remaining 85% of sporadic ALS patients. Cytoplasmic TDP-43 aggregates in motor neurons are an almost universal feature in ALS (2), although mutations to TAR DNA binding protein 1 (TARDBP) itself (gene encoding TDP-43) are uncommon. TDP-43 regulates miRNA processing by affecting the stability of or binding to Drosha or Dicer (7,8). Thus, altered RNA metabolism, including of miRNAs, may be generally disrupted in ALS. In the current study, we found that fibroblasts and WB from C9-ALS and nonC9-ALS patients exhibited a distinct DmiRNA profile versus controls. There were more DmiRNAs in C9-ALS (n = 114, total fibroblasts and WB) than in nonC9-ALS (n = 91) samples, possibly because of the comparative heterogeneity of nonC9-ALS samples. We also identified DmiRNAs by tissue type in fibroblasts (31 increased, 31 decreased) and WB (42 increased, 22 decreased) from C9-ALS and nonC9-ALS patients. Of these, four DmiRNAs were common to both tissues and groups (C9-ALS, nonC9-ALS) and may represent an ALS-specific panel of miR-30b-5p, miR-30c-5p, miR-484 and miR-92a-3p. Functional enrichment identified 102 biological pathways overall, of which 44 were shared across C9-ALS and nonC9-ALS in both tissues versus controls. Network analysis centered on pathways related to neuronal function, metabolism and cellular proliferation and structure. Finally, random forest of DmiRNAs produced an ROC curve with an AUC of 0.831 for differentiating ALS patients from control participants.
Previously, we profiled miRNA (18,23) and mRNA (23) in post mortem spinal cord tissue from sporadic ALS patients. We found that only miR-142-5p and miR-155-5p were upregulated in sporadic ALS spinal cord, but 88 miRNAs were downregulated, with miR-577 and miR-935 as the most suppressed in ALS (18). None of the spinal cord miRNAs overlapped with both fibroblast and WB miRNAs in this study. It is unclear whether differences in miRNAs between accessible biospecimens versus spinal cord tissue in ALS stems from disease pathology or from tissue-specific biomarkers independent of the disease process. Functional pathway analysis of spinal cord miRNAs highlighted cellular regulation and proliferation and immune response (18,23), which partially overlaps with fibroblast and WB miRNA pathways.
The role of miRNAs in ALS was first highlighted in mutant Superoxide Dismutase 1 (SOD1G93A) mice, which had upregulated skeletal muscle-specific miR-206 later in disease (24). Progression was slowed following miR-206 knockout. Several studies have corroborated overrepresentation of miR-206 in ALS muscle and plasma (17), but it is non-specific to ALS since it is also altered in muscular dystrophies (25) and other neurodegenerative diseases (26,27). This underscores the importance of including biosamples from similar yet distinct diseases to rule out miRNAs with significant overlap, which only a few studies have considered (28–30). Aside from miR-206, several other miRNAs are up- or downregulated in ALS, but with very little overlap across studies, which may be attributed to several factors. First, most studies recruited sporadic ALS patients at distinct stages of the disease, resulting in heterogeneous populations. Second, study sample sizes were small and likely unrepresentative of larger populations (17). Finally, studies profile different tissues. To overcome these limitations, we included both C9-ALS and nonC9-ALS from two tissues, fibroblasts and WB, from a relatively large total number of ALS samples (n = 81).
We report more DmiRNAs in C9-ALS than in nonC9-ALS versus controls with tissue-specific differences, although there was some overlap. This is aligned with other studies (31,32), which identified a 30-panel DmiRNA signature in familial ALS versus controls, but only 2 DmiRNAs in sporadic ALS versus controls, which the authors concluded could have arisen from greater heterogeneity of sporadic ALS. Another study of ALS muscle versus plasma found some but incomplete overlap in miRNA profiles (33), as we observed, indicating variation by tissue. Moving forward, it will be important to define the best tissue for analysis and stratify a sufficient sample size by genetic mutation to validate a strong ALS miRNA panel.
Currently, there is no sensitive molecular diagnostic test for ALS. With the goal of bridging this knowledge gap, we examined ALS-associated miRNA profiles in accessible WB, an immune cell-containing biofluid. Immune system dysfunction is a recurrent ALS theme (34). ALS participants exhibit distinct circulating immune cell populations versus healthy controls (35–38), advocating WB as a diagnostic medium for an ALS test. miRNAs are also attractive because they circulate widely in WB (39) and may represent a snapshot of disease status, including ALS (17). We performed random forest of all DmiRNAs from nonC9-ALS WB, which generated an ROC curve with an AUC of 0.832. The top candidate was miR-26a-5p, followed by miR-30c-5p, which most robustly differentiated ALS patients from control participants. Pathway analysis of miR-26a-5p-regulated genes yielded ‘protein processing in endoplasmic reticulum’, ‘hippo signaling pathway’ (see below) and ‘biosynthesis of unsaturated fatty acids’.
No study has identified an miRNA panel unique to nonC9-ALS. Our top two candidates, miR-26a-5p and miR-30c-5p, were identified in a study of 56 sporadic ALS WB samples (40) but were downregulated rather than upregulated. Other studies have noted miR-26a-5p downregulated in muscle (41) or upregulated in serum (42) in ALS. Therefore, there is a lack of consensus in the literature arising from the heterogeneity of sporadic ALS and small sample sizes. We did not evaluate miRNA changes longitudinally over the disease course, which may be an additional factor affecting consensus among results. Indeed, a pilot study suggests that miRNAs correlate with and change over time in relationship to clinical parameters (30). Furthermore, miRNAs have multiple targets, so there may be overlap in downstream biological pathways even if the overlap between DmiRNAs by studies is lacking.
Thus, after analyzing DmiRNAs, we next examined their target mRNAs to infer downstream biological pathways. miRNAs negatively regulate their targets by recruiting Argonaute proteins and assembling into an RNA-induced silencing complex, cleaving the target mRNA (43). We leveraged our Bru-seq and BruChase-seq dataset to correlate DmiRNAs to target mRNA stability (21,22). Bru-seq and BruChase-seq label nascent mRNA followed by a pulse with an orthogonal label after a time delay to assess stability (21). Less stable mRNAs decrease in level to a greater extent than more stable mRNAs over time. As anticipated, we found that C9-ALS fibroblast miRNA levels from this study correlated inversely with mRNA target stability from C9-ALS fibroblasts from our previously published study (21). Presumably, high-level miRNAs degraded their target mRNAs relatively rapidly, rendering them of lower stability, whereas low-level miRNAs degraded their tagets relatively more slowly, rendering them of higher stability. DmiRNA-to-mRNA correlations did not attain 100%, likely due to the presence of alternative mRNA-destabilizing or degrading pathways, such as decapping and base modifcations (44,45). We validated the DmiRNA-to-mRNA correlations by qPCR for two target mRNAs, GADD45B and IP6K2.
However, miRNAs have multiple targets, so we performed functional enrichment of all mRNA targets of significant DmiRNAs. We then clustered biologically enriched pathways by network analysis to identify the main themes. The subnetworks, along with the highly inter-connected nodes, provided interesting insight into ALS pathology. The largest inter-connected subnetwork in C9-ALS (shaded purple) contained several cancer KEGG pathways mostly shared by fibroblasts and WB, including ‘pathways in cancer’, ‘ErbB signaling pathway’ and ‘MAPK signaling pathway’. There were also cancer KEGGs unique to fibroblasts, such as ‘small cell lung cancer’. JAK–STAT signaling is central to ‘pathways in cancer’, which is overrepresented in C9-ALS samples. We previously found that JAK–STAT signaling is also prominent upon analysis of mRNA from sporadic ALS spinal cords (23). In an in vitro ALS model, we found that tofacitinib-mediated blocking of JAK–STAT signaling in natural killer cells inhibits their ability to attack motor neurons (46). Collectively, these results suggest that this pathway may serve as a therapeutic target in ALS, although further studies are warranted.
ErbBs are also of particular interest; they influence cell survival and proliferation and activate MAPK and PI3K-Akt signaling (47). ErbB is activated by its ligand neuregulin 1, which has lower expression in spinal cords from ALS patients and SOD1G93A mice, a model of familial ALS (48). Promoting neuregulin 1 expression in SOD1G93A animals slows disease progression in females. In humans, a Japanese family with familial ALS harbors mutant ERBB4, with a diminished capacity for neuregulin 1-mediated activation (49). MAPK signaling was also featured in this subnetwork (50); inhibiting p38 MAPK rescues retrograde axonal transport in SOD1G93A ALS mice (51), whereas blocking AMPKα prevents hydrogen peroxide-induced apoptosis of SOD1G93A embryonic neural stem cells (52). These examples illustrate how our pathway analysis can identify possible therapeutic ALS targets, e.g. ErbB, MAPK, which the literature shows can be pursued as potential therapies in pre-clinical studies with ALS mouse models.
There were two other highly inter-connected C9-ALS subnetworks with signaling pathways known to be dysfunctional in ALS. One subnetwork (shaded orange) was heavily centered on pathways in both fibroblasts and WB related to neuronal function and signaling, such as ‘glutamatergic synapse’, ‘GABAergic synapse’ and ‘long-term potentiation’. ‘Sphingolipid signaling pathway’ was also featured, which we have identified as a recurrent dysregulated pathway in sporadic ALS by metabolomics analysis of plasma (53,54). This pathway is also linked to familial ALS through mutations to SPTLC1, involved in sphingolipid synthesis (55,56). Additionally, the orange subnetwork contained some pathways that were WB predominant, e.g. ‘dopaminergic synapse’, ‘cholinergic synapse’. The second subnetwork (shaded green) contained pathways previously identified in ALS and linked to development, e.g. ‘Wnt signaling pathway’ (57) and ‘hippo signaling pathway’ (58), and is aligned with recent evidence of widespread neural network disruption in ALS (59).
Finally, when we examined the network constructed from nonC9-ALS KEGG pathways, similar clusters emerged as for C9-ALS. These spanned a cancer-predominant subnetwork (shaded blue) and a development subnetwork (shaded green). The orange C9-ALS subnetwork splits into two subnetworks in sporadic nonC9-ALS, one involving pathways related to neuronal function (shaded pink), which is highly interconnected with a second subnetwork encompassing various signaling pathways (shaded purple). Of pathways related to ALS, ‘PI3K-Akt signaling pathway’ (60) appeared in the nonC9-ALS network and clustered with MAPK. ‘FOXO signaling pathway’ (61) was linked with the neurodevelopment subnetwork in nonC9-ALS, rather than the cancer subnetwork in C9-ALS. Another notable difference in the nonC9-ALS network is greater integration of ‘regulation of actin cytoskeleton’ into the green development subnetwork, which bridges the development and cancer subnetworks in familial C9-ALS. Several ALS mutations have been identified in genes that regulate actin cytoskeleton (62), such as profilin 1 (PFN1), a regulator of actin polymerization (63), and ALS2, encoding the protein alsin, which has a RhoGEF domain for regulating Rho and actin dynamics (64). Although preliminary, these studies corroborate the overrepresented terms in our pathway analysis of miRNAs in ALS and may suggest therapeutic targets.
This study benefited from several strengths. First, the ability to access to two tissues. Second, ALS patients were stratified by the most prevalent C9orf72 alteration and examined DmiRNAs in the context of both familial and sporadic ALS. Third, DmiRNAs were identified by a multiplexed, untargeted approach, NanoString and rigorously analyzed by NanoStringDiff. This study also suffered limitations. Even with the large number of samples, the study still was not powered to detect sex differences, an important consideration in ALS, which is more prevalent in men (1). Moreover, familial ALS samples were limited to C9orf72 carriers, which comprise only half of familial cases. The qPCR validation of select DmiRNAs was done using a single reference rather than multiple references.
One major goal of this study was to seek ALS biomarkers in accessible tissue. Our top candidates, miR-26a-5p and miR-30c-5p, did not find overlap with all other ALS miRNA reports in the literature. The challenges faced for identifying a unifying miRNA biomarker panel in ALS are substantial. ALS is highly heterogeneous, with variation in clinical presentation (e.g. onset segment, speed of disease progression, disease stage), underlying genetic cause (with over 40 identified ALS mutations) and a possible environmental contribution (1,2). It is possible that this considerable extent of heterogeneity may defy determination of an ALS-specific miRNA panel. Nevertheless, our study underscores the importance of miRNAs for understanding ALS pathophysiology by providing miRNA pathway analysis of one of the largest ALS sample sizes to date, which uncovered both well-known, corroborating, pathways and less-known pathways of potential future interest.
Materials and Methods
Study participants and samples
All patients 18 years and older and able to communicate in English attending the University of Michigan Pranger Multidisciplinary ALS clinic were invited to submit samples to the University of Michigan ALS Patient Repository (UMAPR), an Institutional Review Board (IRB)-approved repository. Control participants were recruited separately. All participants provided written informed consent. For this study, participant-provided biospecimens were retrieved from UMAPR, which met the following criteria: diagnosis of ALS based on the Gold Coast criteria (65) and further classified by the initial and/or revised EI Escorial criteria (66). ALS participants were distributed between the sporadic ALS cohort (nonC9-ALS), by selecting patients without a family history of ALS and negative for C9orf72 expansion, and the familial ALS cohort (C9-ALS), by selecting patients with C9orf72 expansion determined by published methods (67). Age- and sex-matched healthy controls were then selected. Skin punch biopsy from the forearm was collected using standard protocols and participant-derived fibroblasts were isolated as previously reported (21). Fibroblasts were cultured in fibroblast media: Dulbecco's Modified Eagle Medium (DMEM) with 4.5 g/l D-glucose, with glutamine/without pyruvate (Gibco, Thermo Fisher Scientific, Grand Island, NY, USA), supplemented with 10% heat-inactivated fetal bovine serum (FBS) (Gibco, Thermo Fisher Scientific), 1X Glutamax-1 (Gibco, Thermo Fisher Scientific) and 1X MEM NEAA (Gibco, Thermo Fisher Scientific). Approximately 5 mL of WB was collected by standard venipuncture into PAXgene Blood RNA Tubes (cat # 762125, Qiagen, Germantown, MD, USA; for RNA isolation) and another 5 mL into EDTA tubes (plasma isolation by centrifugation); all tubes were stored at −80°C. The overall experimental outline is presented in Fig. 1.
RNA extraction and quality determination
Total RNA for miRNA profiling was isolated from fibroblasts using the miRNeasy Mini Kit (cat # 217004, Qiagen) and from WB with the PAXgene Blood miRNA Kit (cat # 63134, Qiagen), according to the manufacturer’s instructions. RNA was concentrated and purified using the Zymo RNA Clean & Concentrator-5 (cat # R1016, Zymo Research, Irvine ,CA, USA). RNA concentration was determined with a NanoDrop ND-1000 Spectrophotometer (NanoDrop Technologies, Thermo Fisher Scientific, Wilmington, DE, USA) and quality was assessed by the 260/280 and 260/230 nm ratios. RNA integrity was measured using the Agilent Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA).
miRNA expression profiling
Profiling was performed by the NanoString nCounter Human v2 (on fibroblasts) and v3 (on WB) miRNA Expression Panels (NanoString Technologies, Seattle, WA, USA), as previously described (68). Briefly, this assay detects 800 endogenous miRNAs, 5 reference transcripts, plus 6 positive and 6 negative controls. About 150 ng of total RNA per sample was used as input for the nCounter Human miRNA sample preparation and hybridized for 16 h at 65°C. Subsequently, the strip tubes were placed into the nCounter Prep Station for automated sample purification and subsequent reporter capture. Each sample was scanned for 555 fields of view on the nCounter Digital Analyzer and data were extracted using the nCounter RCC Collector (both NanoString). A quality control step identified and eliminated unrelated samples (outliers) using nSolver Analysis Software v3.1 (NanoString), according to manufacturer’s instruction. Principal component analysis visualized and examined the overall variation across samples. Next, data were analyzed with NanoStringDiff, an R package specifically designed for NanoString nCounter data (20). Data were normalized using positive controls, negative controls and reference genes (ACTB, B2M, GAPDH, RPL19, RPLP0) embedded in the nCounter system, and differential log fold-changes and multiple testing adjusted statistical significance q-values were obtained.
Pathway analysis
DIANA-miRPath v3.0 (69) was used to predict DmiRNA gene targets and characterize their biological functions and pathways using Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway terms. KEGG pathways with false discovery rate (FDR) < 0.05 were considered significantly enriched among the DmiRNAs from each tissue and genotype (C9-ALS or nonC9-ALS). To identify the overall theme of these enriched pathways, an association network for each tissue was generated from the gene-content overlap among the KEGG pathways using richR, our in-house analysis R package (https://github.com/hurlab/richR). The inter-relationship among the significant pathways within each tissue was visualized in Cytoscape (70) and highly-inter-connected pathway clusters were detected by MCODE (71), a Cytoscape application for network cluster analysis.
Fibroblast labeling and sequencing and mRNA stability
Fibroblast mRNA stability from controls (n = 3), nonC9-ALS (n = 4) and C9-ALS (n = 4) participants was determined by 5-bromouridine sequencing (Bru-seq) and 5-bromouridine-chase sequencing (BruChase-seq), as previously described (21). Briefly, fibroblasts were grown to confluency in duplicate in 150 mm petri dishes (Falcon/Corning, Corning, NY, USA) in fibroblast media: DMEM with 4.5 g/l D-glucose, with glutamine/without pyruvate (Gibco, Thermo Fisher) supplemented with 10% heat inactivated FBS (Gibco, Thermo Fisher), 1X Glutamax-1 (Gibco, Thermo Fisher) and 1X MEM NEAA (Gibco, Thermo Fisher), and 5-Bromouridine (2 mM; cat # 850187, Sigma-Aldrich, Burlington, MA, USA) was added to both petri dishes and incubated for 0.5 h at 37°C in 5% CO2 to label cells, which were either harvested (pulsed, first petri dish) or chased for 6 h with uridine (20 mM, second petri dish; cat # U3750, Sigma-Aldrich) at 37°C in 5% CO2. Cells were harvested in 3 mL of QIAzol (cat # 79306, Qiagen) and total RNA was isolated by phenol/chloroform extraction and resuspended in 100 μL of diethyl pyrocarbonate water. Bru-labeled RNA was immunoprecipitated with mouse anti-BrdU antibody (clone 3D4; cat # 555627, BD Pharmingen, Franklin Lakes, NJ, USA) conjugated to goat anti-mouse Dynabeads (cat # 11033, Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA). Strand-specific DNA libraries were prepared with the Illumina TruSeq Kit (Illumina, San Diego, CA, USA) and sequenced on an Illumina platform, as previously described (21). RNA stability was calculated as the ratio of transcript abundance at 6 h versus 0.5 h. We used a cutoff of ≥1.5-fold-change in stability of mRNA transcripts.
Quantitative real-time PCR
For miRNA, 30 ng of total RNA was reverse transcribed using the High-Capacity cDNA Reverse Transcription Kit (cat # 4368814) with Megaplex Primer Pools, Human Pools A v2.1 (cat # 4401009). cDNA was preamplified using TaqMan PreAmp Master Mix (cat # 4391128) and Megaplex PreAmp Primers, Human Pool A v2.1 (cat # 4399233). qPCR was performed with TaqMan Universal PCR Master Mix (cat # 4318157) and TaqMan microRNA Assays (cat # 4427975, all Thermo Fisher Scientific, Waltham, MA, USA). Each sample was run in triplicate. Transcripts levels were calculated by the ΔΔCT method and normalized to RNU48. The control group in each comparison was used as a calibrator (ΔΔCT = 0, 2^(−ΔΔCT) = 1). Amplification efficiency for each miRNA was assessed by technical duplicates method on a Bio-Rad Real-Time PCR System (Bio-Rad, Ann Arbor, MI, USA).
For mRNA stability, cDNA was generated using iScript Reverse Transcription Supermix (cat # 1708841, Bio-Rad) on the entire anti-Bru-immunoprecipitated pulse and pulse/chased RNA (Fibroblast labeling and sequencing and mRNA stability section). qPCR was performed on 2 μL of template with TaqMan Gene Expression Master Mix (cat # 4369016, Thermo Fisher Scientific) and TaqMan probes (Thermo Fisher Scientific) for GADD45B, IP6K2a and YWHAZ (normalization reference). The PCR program was 120 s at 50°C, 10 s at 95°C, 40x cycles (15 s at 95°C, 60 s at 60°C) on an Applied Biosystems StepOne Real-Time PCR system (Thermo Fisher Scientific). Transcript stability was determined as the ratio between transcript abundance at 6 h (pulse-chase/pulse) to 0.5 h (pulse).
Random forest
The Classification And REgression Training (CARET) package from R was used to build a random forest classification model (72). Ten-fold cross-validation was performed to construct and evaluate the model. The ROC curves were plotted for all folds’ prediction using the pROC package from R (73). Pathway analysis for miR-26a-5p was derived using mirPath (69). For DmiRNAs identified by nSolver, nCounter data by sample were normalized to the top 100 most highly expressed miRNAs across all samples and differential levels were examined by nSolver differential expression testing menu between groups. miRNA ratios between groups and statistical significance P-values were obtained, which were adjusted for multiple testing by the p.adjust function in R to calculate FDRs.
Statistical analysis
Raw data were analyzed using NanoStringDiff software to identify statistically significant DmiRNAs (P < 0.05) using normalized counts between sample groups. Data were expressed as mean ± standard error of the mean. Prism 5.01 (GraphPad, San Diego, CA, USA) was also used to analyze statistical significance (P < 0.05, two-tailed Student’s t-test).
Supplementary Material
Acknowledgements
We are grateful to the study participants from the University of Michigan Pranger ALS Multidisciplinary Clinic who generously provided biological samples. We also thank Crystal Pacut and Blake Swihart for study support.
Conflict of Interest statement. The authors declare that they have no competing interests related to the direct applications of this research.
Contributor Information
Junguk Hur, Department of Biomedical Sciences, University of North Dakota, Grand Forks, ND 58202, USA.
Ximena Paez-Colasante, Department of Neurology, University of Michigan, Ann Arbor, MI 48109, USA.
Claudia Figueroa-Romero, Department of Neurology, University of Michigan, Ann Arbor, MI 48109, USA; NeuroNetwork for Emerging Therapies, University of Michigan, Ann Arbor, MI 48109, USA.
Ting-wen Lo, Department of Chemical Engineering, University of Michigan, Ann Arbor, MI 48109, USA.
Sami J Barmada, Department of Neurology, University of Michigan, Ann Arbor, MI 48109, USA.
Michelle T Paulsen, Department of Radiation Oncology, University of Michigan, Ann Arbor, MI 48109, USA.
Mats Ljungman, Department of Radiation Oncology, University of Michigan, Ann Arbor, MI 48109, USA; Department of Internal Medicine, Division of Nephrology, University of Michigan, Ann Arbor, MI 48109, USA.
Fadhl M Alakwaa, Department of Neurology, University of Michigan, Ann Arbor, MI 48109, USA.
Masha G Savelieff, NeuroNetwork for Emerging Therapies, University of Michigan, Ann Arbor, MI 48109, USA.
Stephen A Goutman, Department of Neurology, University of Michigan, Ann Arbor, MI 48109, USA; NeuroNetwork for Emerging Therapies, University of Michigan, Ann Arbor, MI 48109, USA.
Eva L Feldman, Department of Neurology, University of Michigan, Ann Arbor, MI 48109, USA; NeuroNetwork for Emerging Therapies, University of Michigan, Ann Arbor, MI 48109, USA.
Funding
National ALS Registry/CDC/ATSDR (1R01TS000289; R01TS000327); National Institute of Neurological Disorders and Stroke (NINDS; R21NS102960, R01NS127188, R01NS120926); National Institute of Environmental Health Sciences (NIEHS; K23ES027221, R01ES030049); National Center for Advancing Translational Sciences at the NIH (UL1TR002240); ALS Association 20-IIA-532, Sinai Medical Staff Foundation Neuroscience Scholar Fund; Peter R. Clark Fund for ALS Research, Scott L. Pranger, University of Michigan; NeuroNetwork Therapeutic Discovery Fund; NeuroNetwork for Emerging Therapies, University of Michigan.
References
- 1. Goutman, S.A., Hardiman, O., Al-Chalabi, A., Chio, A., Savelieff, M.G., Kiernan, M.C. and Feldman, E.L. (2022) Recent advances in the diagnosis and prognosis of amyotrophic lateral sclerosis. Lancet Neurol., 21, 480–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Goutman, S.A., Hardiman, O., Al-Chalabi, A., Chio, A., Savelieff, M.G., Kiernan, M.C. and Feldman, E.L. (2022) Emerging insights into the complex genetics and pathophysiology of amyotrophic lateral sclerosis. Lancet Neurol., 21, 465–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bensimon, G., Lacomblez, L. and Meininger, V. (1994) A controlled trial of riluzole in amyotrophic lateral sclerosis. ALS/Riluzole Study Group. N. Engl. J. Med., 330, 585–591. [DOI] [PubMed] [Google Scholar]
- 4. Writing, G. and Edaravone, A.L.S.S.G. (2017) Safety and efficacy of edaravone in well defined patients with amyotrophic lateral sclerosis: a randomised, double-blind, placebo-controlled trial. Lancet Neurol., 16, 505–512. [DOI] [PubMed] [Google Scholar]
- 5. Kaji, R., Imai, T., Iwasaki, Y., Okamoto, K., Nakagawa, M., Ohashi, Y., Takase, T., Hanada, T., Shimizu, H., Tashiro, K. et al. (2019) Ultra-high-dose methylcobalamin in amyotrophic lateral sclerosis: a long-term phase II/III randomised controlled study. J. Neurol. Neurosurg. Psychiatry, 90, 451–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Paganoni, S., Macklin, E.A., Hendrix, S., Berry, J.D., Elliott, M.A., Maiser, S., Karam, C., Caress, J.B., Owegi, M.A., Quick, A. et al. (2020) Trial of Sodium Phenylbutyrate-Taurursodiol for Amyotrophic Lateral Sclerosis. N. Engl. J. Med., 383, 919–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kawahara, Y. and Mieda-Sato, A. (2012) TDP-43 promotes microRNA biogenesis as a component of the Drosha and Dicer complexes. Proc. Natl. Acad. Sci. U. S. A., 109, 3347–3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Di Carlo, V., Grossi, E., Laneve, P., Morlando, M., Dini Modigliani, S., Ballarino, M., Bozzoni, I. and Caffarelli, E. (2013) TDP-43 regulates the microprocessor complex activity during in vitro neuronal differentiation. Mol. Neurobiol., 48, 952–963. [DOI] [PubMed] [Google Scholar]
- 9. Lagos-Quintana, M., Rauhut, R., Lendeckel, W. and Tuschl, T. (2001) Identification of novel genes coding for small expressed RNAs. Science, 294, 853–858. [DOI] [PubMed] [Google Scholar]
- 10. Lau, N.C., Lim, L.P., Weinstein, E.G. and Bartel, D.P. (2001) An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science, 294, 858–862. [DOI] [PubMed] [Google Scholar]
- 11. Lee, R.C. and Ambros, V. (2001) An extensive class of small RNAs in Caenorhabditis elegans. Science, 294, 862–864. [DOI] [PubMed] [Google Scholar]
- 12. Selbach, M., Schwanhausser, B., Thierfelder, N., Fang, Z., Khanin, R. and Rajewsky, N. (2008) Widespread changes in protein synthesis induced by microRNAs. Nature, 455, 58–63. [DOI] [PubMed] [Google Scholar]
- 13. Nowak, J.S. and Michlewski, G. (2013) miRNAs in development and pathogenesis of the nervous system. Biochem. Soc. Trans., 41, 815–820. [DOI] [PubMed] [Google Scholar]
- 14. O'Carroll, D. and Schaefer, A. (2013) General principals of miRNA biogenesis and regulation in the brain. Neuropsychopharmacology, 38, 39–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rinchetti, P., Rizzuti, M., Faravelli, I. and Corti, S. (2018) MicroRNA Metabolism and Dysregulation in Amyotrophic Lateral Sclerosis. Mol. Neurobiol., 55, 2617–2630. [DOI] [PubMed] [Google Scholar]
- 16. Tsitkanou, S., Della Gatta, P.A. and Russell, A.P. (2016) Skeletal muscle satellite cells, mitochondria, and micrornas: their involvement in the pathogenesis of ALS. Front. Physiol., 7, 403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dardiotis, E., Aloizou, A.M., Siokas, V., Patrinos, G.P., Deretzi, G., Mitsias, P., Aschner, M. and Tsatsakis, A. (2018) The role of MicroRNAs in patients with amyotrophic lateral sclerosis. J. Mol. Neurosci., 66, 617–628. [DOI] [PubMed] [Google Scholar]
- 18. Figueroa-Romero, C., Hur, J., Lunn, J.S., Paez-Colasante, X., Bender, D.E., Yung, R., Sakowski, S.A. and Feldman, E.L. (2016) Expression of microRNAs in human post-mortem amyotrophic lateral sclerosis spinal cords provides insight into disease mechanisms. Mol. Cell. Neurosci., 71, 34–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Paez-Colasante, X., Figueroa-Romero, C., Sakowski, S.A., Goutman, S.A. and Feldman, E.L. (2015) Amyotrophic lateral sclerosis: mechanisms and therapeutics in the epigenomic era. Nat. Rev. Neurol., 11, 266–279. [DOI] [PubMed] [Google Scholar]
- 20. Wang, H., Horbinski, C., Wu, H., Liu, Y., Sheng, S., Liu, J., Weiss, H., Stromberg, A.J. and Wang, C. (2016) NanoStringDiff: a novel statistical method for differential expression analysis based on NanoString nCounter data. Nucleic Acids Res., 44, e151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tank, E.M., Figueroa-Romero, C., Hinder, L.M., Bedi, K., Archbold, H.C., Li, X., Weskamp, K., Safren, N., Paez-Colasante, X., Pacut, C. et al. (2018) Abnormal RNA stability in amyotrophic lateral sclerosis. Nat. Commun., 9, 2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Paulsen, M.T., Veloso, A., Prasad, J., Bedi, K., Ljungman, E.A., Tsan, Y.C., Chang, C.W., Tarrier, B., Washburn, J.G., Lyons, R. et al. (2013) Coordinated regulation of synthesis and stability of RNA during the acute TNF-induced proinflammatory response. Proc. Natl. Acad. Sci. U. S. A., 110, 2240–2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Figueroa-Romero, C., Hur, J., Bender, D.E., Delaney, C.E., Cataldo, M.D., Smith, A.L., Yung, R., Ruden, D.M., Callaghan, B.C. and Feldman, E.L. (2012) Identification of epigenetically altered genes in sporadic amyotrophic lateral sclerosis. PLoS One, 7, e52672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Williams, A.H., Valdez, G., Moresi, V., Qi, X., McAnally, J., Elliott, J.L., Bassel-Duby, R., Sanes, J.R. and Olson, E.N. (2009) MicroRNA-206 delays ALS progression and promotes regeneration of neuromuscular synapses in mice. Science, 326, 1549–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Coenen-Stass, A.M.L., Wood, M.J.A. and Roberts, T.C. (2017) Biomarker Potential of Extracellular miRNAs in Duchenne Muscular Dystrophy. Trends Mol. Med., 23, 989–1001. [DOI] [PubMed] [Google Scholar]
- 26. Xie, B., Liu, Z., Jiang, L., Liu, W., Song, M., Zhang, Q., Zhang, R., Cui, D., Wang, X. and Xu, S. (2017) Increased Serum miR-206 Level Predicts Conversion from Amnestic Mild Cognitive Impairment to Alzheimer's Disease: A 5-Year Follow-up Study. J. Alzheimers Dis., 55, 509–520. [DOI] [PubMed] [Google Scholar]
- 27. Grasso, M., Piscopo, P., Talarico, G., Ricci, L., Crestini, A., Tosto, G., Gasparini, M., Bruno, G., Denti, M.A. and Confaloni, A. (2019) Plasma microRNA profiling distinguishes patients with frontotemporal dementia from healthy subjects. Neurobiol. Aging, 84, 240 e241–240 e212. [DOI] [PubMed] [Google Scholar]
- 28. Russell, A.P., Wada, S., Vergani, L., Hock, M.B., Lamon, S., Léger, B., Ushida, T., Cartoni, R., Wadley, G.D., Hespel, P. et al. (2013) Disruption of skeletal muscle mitochondrial network genes and miRNAs in amyotrophic lateral sclerosis. Neurobiol. Dis., 49, 107–117. [DOI] [PubMed] [Google Scholar]
- 29. Waller, R., Goodall, E.F., Milo, M., Cooper-Knock, J., Da Costa, M., Hobson, E., Kazoka, M., Wollff, H., Heath, P.R., Shaw, P.J. et al. (2017) Serum miRNAs miR-206, 143-3p and 374b-5p as potential biomarkers for amyotrophic lateral sclerosis (ALS). Neurobiol. Aging, 55, 123–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Raheja, R., Regev, K., Healy, B.C., Mazzola, M.A., Beynon, V., Von Glehn, F., Paul, A., Diaz-Cruz, C., Gholipour, T., Glanz, B.I. et al. (2018) Correlating serum micrornas and clinical parameters in amyotrophic lateral sclerosis. Muscle Nerve, 58, 261–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Freischmidt, A., Muller, K., Zondler, L., Weydt, P., Volk, A.E., Bozic, A.L., Walter, M., Bonin, M., Mayer, B., von Arnim, C.A. et al. (2014) Serum microRNAs in patients with genetic amyotrophic lateral sclerosis and pre-manifest mutation carriers. Brain, 137, 2938–2950. [DOI] [PubMed] [Google Scholar]
- 32. Freischmidt, A., Muller, K., Zondler, L., Weydt, P., Mayer, B., von Arnim, C.A., Hubers, A., Dorst, J., Otto, M., Holzmann, K. et al. (2015) Serum microRNAs in sporadic amyotrophic lateral sclerosis. Neurobiol. Aging, 36, 2660.e2615–2660.e2620. [DOI] [PubMed] [Google Scholar]
- 33. de Andrade, H.M., de Albuquerque, M., Avansini, S.H., de S Rocha, C., Dogini, D.B., Nucci, A., Carvalho, B., Lopes-Cendes, I. and Franca, M.C., Jr. (2016) MicroRNAs-424 and 206 are potential prognostic markers in spinal onset amyotrophic lateral sclerosis. J. Neurol. Sci., 368, 19–24. [DOI] [PubMed] [Google Scholar]
- 34. Murdock, B.J., Bender, D.E., Segal, B.M. and Feldman, E.L. (2015) The dual roles of immunity in ALS: Injury overrides protection. Neurobiol. Dis., 77, 1–12. [DOI] [PubMed] [Google Scholar]
- 35. Murdock, B.J., Zhou, T., Kashlan, S.R., Little, R.J., Goutman, S.A. and Feldman, E.L. (2017) Correlation of Peripheral Immunity With Rapid Amyotrophic Lateral Sclerosis Progression. JAMA Neurol., 74, 1446–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Murdock, B.J., Bender, D.E., Kashlan, S.R., Figueroa-Romero, C., Backus, C., Callaghan, B.C., Goutman, S.A. and Feldman, E.L. (2016) Increased ratio of circulating neutrophils to monocytes in amyotrophic lateral sclerosis. Neurol. Neuroimmunol. Neuroinflamm., 3, e242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Murdock, B.J., Famie, J.P., Piecuch, C.E., Raue, K.D., Mendelson, F.E., Pieroni, C.H., Iniguez, S.D., Zhao, L., Goutman, S.A. and Feldman, E.L. (2021) NK cells associate with ALS in a sex- and age-dependent manner. JCI Insight, 6, e147129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Murdock, B.J., Goutman, S.A., Boss, J., Kim, S. and Feldman, E.L. (2021) Amyotrophic Lateral Sclerosis Survival Associates With Neutrophils in a Sex-specific Manner. Neurol. Neuroimmunol. Neuroinflamm., 8, e953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pogribny, I.P. (2018) MicroRNAs as biomarkers for clinical studies. Exp. Biol. Med., 243, 283–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Liguori, M., Nuzziello, N., Introna, A., Consiglio, A., Licciulli, F., D'Errico, E., Scarafino, A., Distaso, E. and Simone, I.L. (2018) Dysregulation of MicroRNAs and target genes networks in peripheral blood of patients with sporadic amyotrophic lateral sclerosis. Front. Mol. Neurosci., 11, 288–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kovanda, A., Leonardis, L., Zidar, J., Koritnik, B., Dolenc-Groselj, L., Ristic Kovacic, S., Curk, T. and Rogelj, B. (2018) Differential expression of microRNAs and other small RNAs in muscle tissue of patients with ALS and healthy age-matched controls. Sci. Rep., 8, 5609–5609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Taguchi, Y.H. and Wang, H. (2018) Exploring microRNA Biomarker for Amyotrophic Lateral Sclerosis. Int. J. Mol. Sci., 19, 1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kobayashi, H. and Tomari, Y. (2016) RISC assembly: Coordination between small RNAs and Argonaute proteins. Biochim. Biophys. Acta, 1859, 71–81. [DOI] [PubMed] [Google Scholar]
- 44. Houseley, J. and Tollervey, D. (2009) The many pathways of RNA degradation. Cell, 136, 763–776. [DOI] [PubMed] [Google Scholar]
- 45. Lee, Y., Choe, J., Park, O.H. and Kim, Y.K. (2020) Molecular Mechanisms Driving mRNA Degradation by m(6)A Modification. Trends Genet., 36, 177–188. [DOI] [PubMed] [Google Scholar]
- 46. Figueroa-Romero, C., Monteagudo, A., Murdock, B.J., Famie, J.P., Webber-Davis, I.F., Piecuch, C.E., Teener, S.J., Pacut, C., Goutman, S.A. and Feldman, E.L. (2022) Tofacitinib suppresses natural killer cells in vitro and in vivo: implications for amyotrophic lateral sclerosis. Front. Immunol., 13, 773288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mishra, R., Hanker, A.B. and Garrett, J.T. (2017) Genomic alterations of ERBB receptors in cancer: clinical implications. Oncotarget, 8, 114371–114392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Modol-Caballero, G., Garcia-Lareu, B., Verdes, S., Ariza, L., Sanchez-Brualla, I., Brocard, F., Bosch, A., Navarro, X. and Herrando-Grabulosa, M. (2020) Therapeutic Role of Neuregulin 1 Type III in SOD1-Linked Amyotrophic Lateral Sclerosis. Neurotherapeutics, 17, 1048–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Takahashi, Y., Fukuda, Y., Yoshimura, J., Toyoda, A., Kurppa, K., Moritoyo, H., Belzil, V.V., Dion, P.A., Higasa, K., Doi, K. et al. (2013) ERBB4 mutations that disrupt the neuregulin-ErbB4 pathway cause amyotrophic lateral sclerosis type 19. Am. J. Hum. Genet., 93, 900–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sahana, T.G. and Zhang, K. (2021) Mitogen-Activated Protein Kinase Pathway in Amyotrophic Lateral Sclerosis. Biomedicine, 9, 969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gibbs, K.L., Kalmar, B., Rhymes, E.R., Fellows, A.D., Ahmed, M., Whiting, P., Davies, C.H., Greensmith, L. and Schiavo, G. (2018) Inhibiting p38 MAPK alpha rescues axonal retrograde transport defects in a mouse model of ALS. Cell Death Dis., 9, 596–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sui, Y., Zhao, Z., Liu, R., Cai, B. and Fan, D. (2014) Adenosine monophosphate-activated protein kinase activation enhances embryonic neural stem cell apoptosis in a mouse model of amyotrophic lateral sclerosis. Neural Regen. Res., 9, 1770–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Goutman, S.A., Guo, K., Savelieff, M.G., Patterson, A., Sakowski, S.A., Habra, H., Karnovsky, A., Hur, J. and Feldman, E.L. (2022) Metabolomics identifies shared lipid pathways in independent amyotrophic lateral sclerosis cohorts. Brain. 10.1093/brain/awac025. Online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Goutman, S.A., Boss, J., Guo, K., Alakwaa, F.M., Patterson, A., Kim, S., Savelieff, M.G., Hur, J. and Feldman, E.L. (2020) Untargeted metabolomics yields insight into ALS disease mechanisms. J. Neurol. Neurosurg. Psychiatry, 91, 1329–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Mohassel, P., Donkervoort, S., Lone, M.A., Nalls, M., Gable, K., Gupta, S.D., Foley, A.R., Hu, Y., Saute, J.A.M., Moreira, A.L. et al. (2021) Childhood amyotrophic lateral sclerosis caused by excess sphingolipid synthesis. Nat. Med., 27, 1197–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Johnson, J.O., Chia, R., Miller, D.E., Li, R., Kumaran, R., Abramzon, Y., Alahmady, N., Renton, A.E., Topp, S.D., Gibbs, J.R. et al. (2021) Association of Variants in the SPTLC1 Gene With Juvenile Amyotrophic Lateral Sclerosis. JAMA Neurol., 78, 1236–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Vallée, A., Lecarpentier, Y., Guillevin, R. and Vallée, J.-N. (2018) Thermodynamics in neurodegenerative diseases: interplay between canonical WNT/Beta-catenin pathway-PPAR gamma, energy metabolism and circadian rhythms. NeuroMolecular Med., 20, 174–204. [DOI] [PubMed] [Google Scholar]
- 58. Wang, S.-P. and Wang, L.-H. (2016) Disease implication of hyper-Hippo signalling. Open Biol., 6, 160119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Dukic, S., McMackin, R., Buxo, T., Fasano, A., Chipika, R., Pinto-Grau, M., Costello, E., Schuster, C., Hammond, M., Heverin, M. et al. (2019) Patterned functional network disruption in amyotrophic lateral sclerosis. Hum. Brain Mapp., 40, 4827–4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Recabarren-Leiva, D. and Alarcón, M. (2018) New insights into the gene expression associated to amyotrophic lateral sclerosis. Life Sci., 193, 110–123. [DOI] [PubMed] [Google Scholar]
- 61. Zhang, T., Baldie, G., Periz, G. and Wang, J. (2014) RNA-processing protein TDP-43 regulates FOXO-dependent protein quality control in stress response. PLoS Genet., 10, e1004693–e1004693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Chia, R., Chio, A. and Traynor, B.J. (2018) Novel genes associated with amyotrophic lateral sclerosis: diagnostic and clinical implications. Lancet Neurol., 17, 94–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wu, C.H., Fallini, C., Ticozzi, N., Keagle, P.J., Sapp, P.C., Piotrowska, K., Lowe, P., Koppers, M., McKenna-Yasek, D., Baron, D.M. et al. (2012) Mutations in the profilin 1 gene cause familial amyotrophic lateral sclerosis. Nature, 488, 499–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Panzeri, C., De Palma, C., Martinuzzi, A., Daga, A., De Polo, G., Bresolin, N., Miller, C.C., Tudor, E.L., Clementi, E. and Bassi, M.T. (2006) The first ALS2 missense mutation associated with JPLS reveals new aspects of alsin biological function. Brain, 129, 1710–1719. [DOI] [PubMed] [Google Scholar]
- 65. Shefner, J.M., Al-Chalabi, A., Baker, M.R., Cui, L.Y., de Carvalho, M., Eisen, A., Grosskreutz, J., Hardiman, O., Henderson, R., Matamala, J.M. et al. (2020) A proposal for new diagnostic criteria for ALS. Clin. Neurophysiol., 131, 1975–1978. [DOI] [PubMed] [Google Scholar]
- 66. Brooks, B.R., Miller, R.G., Swash, M. and Munsat, T.L. (2000) El Escorial revisited: revised criteria for the diagnosis of amyotrophic lateral sclerosis. Amyotroph. Lateral Scler. Other Motor Neuron Disord., 1, 293–299. [DOI] [PubMed] [Google Scholar]
- 67. He, F., Jones, J.M., Figueroa-Romero, C., Zhang, D., Feldman, E.L., Goutman, S.A., Meisler, M.H., Callaghan, B.C. and Todd, P.K. (2016) Screening for novel hexanucleotide repeat expansions at ALS- and FTD-associated loci. Neurol. Genet., 2, e71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Valeri, N., Braconi, C., Gasparini, P., Murgia, C., Lampis, A., Paulus-Hock, V., Hart, J.R., Ueno, L., Grivennikov, S.I., Lovat, F. et al. (2014) MicroRNA-135b promotes cancer progression by acting as a downstream effector of oncogenic pathways in colon cancer. Cancer Cell, 25, 469–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Vlachos, I.S., Zagganas, K., Paraskevopoulou, M.D., Georgakilas, G., Karagkouni, D., Vergoulis, T., Dalamagas, T. and Hatzigeorgiou, A.G. (2015) DIANA-miRPath v3.0: deciphering microRNA function with experimental support. Nucleic Acids Res., 43, W460–W466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. McGregor, B.A., Eid, S., Rumora, A.E., Murdock, B., Guo, K., de Anda-Jauregui, G., Porter, J.E., Feldman, E.L. and Hur, J. (2018) Conserved transcriptional signatures in human and murine diabetic peripheral neuropathy. Sci. Rep., 8, 17678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Bader, G.D. and Hogue, C.W. (2003) An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics, 4, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kuhn, M. (2008) Building predictive models in R using the caret package. J. Stat. Softw., 28, 1–26.27774042 [Google Scholar]
- 73. Robin, X., Turck, N., Hainard, A., Tiberti, N., Lisacek, F., Sanchez, J.-C. and Müller, M. (2011) pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics, 12, 77–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.