Abstract
Objectives:
The Environmental Determinants of Diabetes in the Young (TEDDY) study follows an HLA-risk selected birth cohort for celiac disease (CD) development using a uniform protocol. Children under investigation come from 6 different regions within Europe and the United States. Our aim was to identify regional differences in celiac disease autoimmunity (CDA) and CD cumulative incidence for children born between 2004 and 2010.
Methods:
Children (n=6,628) with DQ2.5 and/or DQ8.1 were enrolled prospectively from birth in Georgia, Washington, Colorado, Finland, Germany, and Sweden. Children underwent periodic study screening for tissue transglutaminase antibodies (tTGA) and then CD evaluation per clinical care. Population-specific estimates were calculated by weighting the study-specific cumulative incidence with the population-specific haplogenotype frequencies obtained from large stem cell registries from each site.
Results:
Individual haplogenotype risks for CDA and CD varied by region and affected the cumulative incidence within that region. The CD incidence by age 10 years was highest in Swedish children at 3%. Within the US, the incidence by age 10 in Colorado was 2.4%. In the HLA, sex, and family history-adjusted model, Colorado children had a 2.5-fold higher risk of CD compared to Washington. Likewise, Swedish children had a 1.4-fold and 1.8-fold higher risk of CD compared to Finland and Germany, respectively.
Conclusions:
There is high regional variability in cumulative incidence of CD which suggests differential environmental, genetic, and epigenetic influences even within the United States. The overall high incidence warrants a low threshold for screening and further research on region-specific CD triggers.
Keywords: Celiac disease, tissue transglutaminase, incidence
Introduction:
Determining the true incidence of celiac disease (CD) is not possible without nonbiased screening for the disease.[1] This is because many cases occur with neither a family history nor with classic symptoms.[2, 3] A recent meta-analysis of studies reporting population-based incidence estimates concluded that the pediatric incidence of biopsy-proven CD remains on the rise.[1] However, these estimates are highly dependent on the regional diagnostic practices. The prospective Diabetes Autoimmunity Study in the Young (DAISY) cohort study in Colorado estimated the 15-year cumulative incidence of biopsy-proven CD in the general pediatric population to be a remarkable 1.9% for children born between 1993 and 2004.[4] This estimate was an even higher 3.1% when accounting for those with high and persistent autoantibody levels who did not undergo a biopsy.
The Environmental Determinants of Diabetes in the Young (TEDDY) study prospectively follows children born between. 2004 and 2010 at genetic risk for both type 1 diabetes (T1D) and CD at six separate clinical sites in four countries. Children are longitudinally monitored for celiac disease autoimmunity with autoantibodies to tissue transglutaminase (tTGA) and the protocol is therefore designed for timely and thorough ascertainment of the development of persistent tTGA positivity or celiac disease autoimmunity (CDA) and subsequent CD. Individuals may have CDA without having CD if they have transient or fluctuating antibody levels, low antibody levels without biopsy evaluation, dietary modification influencing further evaluation, or potential celiac disease. The population contains various DQ2.5 and DQ8.1 combinations representing the highest risk HLA-DQ haplogenotypes for CD[5]. While prior publications have stratified celiac disease risk by HLA-DQ haplogenotype and country, incidence could not be extrapolated to the general population due to the enrichment of celiac disease permissive HLA-DQ haplogenotypes. In the present study, we identify regional differences in population-wide celiac disease cumulative incidence in the US and European countries using a highly unique cohort of children selected for close monitoring based on specific “high-risk” HLA-DQ haplogenotypes.
Methods:
Study Design and Population:
TEDDY examines environmental risk factors for the development of T1D and CD in individuals at high underlying genetic risk followed from birth to age 15 years at six clinical research centers: Colorado, Georgia, Washington, Finland, Germany, and Sweden, as described previously.[2, 5–7]
From September 2004 through February 2010, 424,788 newborns were screened for specific HLA haplogenotypes and 8,676 children were enrolled in TEDDY (Supplementary Figure 1). TEDDY eligible haplogenotypes include DQ2.5/DQ2.5, DQ2.5/DQ8.1, DQ8.1/DQ8.1 and DQ8.1/DQ4.2 (Supplementary Table 1). Participants were screened annually for tTGA starting at two years of age as described below. With the first tTGA positive result, all prior collected samples were then tested for tTGA to determine the earliest timepoint of autoimmunity. Sex and family history of CD and T1D were self-reported (Supplementary Table 2).
Ethical Considerations:
Informed consent was obtained for all studied children from a parent or legal guardian for HLA screening and participation in prospective follow-up. The study was approved at each site by its local IRB and was monitored by a National Institutes of Health External Evaluation Committee as described previously.[8]
Tissue Transglutaminase Autoantibody Measurements:
Tissue transglutaminase autoantibodies (tTGA-IgA and tTGA-IgG combined) were measured by radiobinding assays in two separate laboratories as previously described.[5] Blood samples were obtained and stored for TEDDY participants every three months until age 48 months, then at least every six months thereafter. Starting at the two year visit, tTGA was routinely tested and tested annually thereafter. If tTGA was positive at the annual screening, the child’s samples collected before this positive result were tested for tTGA to determine the timing of seroconversion.
Definitions of CD and CDA:
CDA was defined as positivity (>99th percentile of healthy control sera) in two consecutive tTGA tests at least three months apart. This was a primary study outcome. In seropositive children, CD was defined based on a duodenal biopsy with Marsh score ≥ 2.[9] The decision whether to biopsy was determined by the clinical gastroenterologist and was outside of the formal study protocol. When biopsy was not performed, individuals with an average tTGA of ≥100 units from two consecutive positive sera were considered to have CD for study purposes.[10] A post-hoc sensitivity analysis was performed including children with an average tTGA of ≥ 67.4 units from two consecutive positive sera in the outcome of CD. This lower cut-off was selected based on the findings of a receiver operating characteristic curve (data not shown).
Stem Cell Registries:
Stem Cell Donor Registries record the phased HLA-DQ genotypes of possible bone marrow transplant donors worldwide. The stem cell donor registries of the United States (National Marrow Donor Program), Finland (Finnish Stem Cell Registry), Germany (ZKRD - Zentrales Knochenmarkspender-Register Deutschland and Dusseldorf Blood Bank), and Sweden (Tobias Registry) were used to determine the major celiac risk-associated HLA-DQ haplogenotypes at each TEDDY site. For the United States, region-specific haplogenotype frequencies were used to weight the TEDDY proxy haplogenotype cumulative incidence. The HLA-DQ haplogenotypes (at 2-field resolution) included in the population risk estimates along with corresponding proxy haplogenotypes from TEDDY, are shown in Supplementary Table 3. For the purposes of describing registry haplotypes, any non-DQ2.5 and non-DQ8.1 haplotype are referred as “X” throughout this paper. Of note, TEDDY did not enroll individuals with the DQ2.5/X or DQ2.2/DQ7.5 haplogenotypes. Since these each have one DQA1*05 allele and one DQB1*02 allele, the TEDDY DQ2.5/8.1 incidence was used as a proxy for these haplogenotypes. Since the DQB1*02 has been shown to have a dose dependent effect, the DQ2.5/DQ2.2 frequency in the stem cell registry was included with the DQ2.5/2.5 frequency as these haplogenotypes confer similar risk.[11]
Statistical Methods:
In TEDDY, cumulative risk for CDA (or CD) was estimated from the Breslow cumulative baseline hazard estimates at the event times of each stratum by the TEDDY sites and HLA haplogenotypes. Risk of CDA (or CD) was compared between sites using hazard ratios from a Cox proportional hazard model adjusting for HLA, sex, and family history of CD.[5] In order to estimate the general population cumulative incidence in each region, the risk per clinical site and HLA haplogenotype by age was first multiplied by the corresponding HLA haplogenotype frequency obtained from the regional stem cell donor registries and then summed over a total of four TEDDY-defined haplogenotypes, as indicated in Supplementary Table 3. SAS version 9.4 (Cary, NC) was used for statistical analysis and R (4.0.5) package ggplot2 was used to generate figures.
Results:
Cohort Characteristics:
As of July 31, 2020, there were 6,628 HLA-typed eligible children carrying the DQ2.5, DQ8.1, or both haplotypes, who had undergone one or more tTGA tests and were thus included in the current analysis. The median follow-up was 11.5 years (interquartile range 10.0 to 13.1 years). Altogether, 580 children (9%) had a first-degree relative (FDR) with T1D and 317 (5%) reported a FDR with CD (Supplementary Table 2).
Of the 6,628 HLA-eligible screened children, 1,299 (20%) met the CDA outcome and 529 children (8%) met the study diagnostic criteria for CD based on biopsy or persistently high tTGA levels. The median age at CDA across all sites was 41 months (interquartile range 27–66 months). Symptom distribution in the TEDDY study has previously been reported and is notable for the fact that most children with CDA were asymptomatic.[2]
Cumulative Risks According to Site and Haplogenotype:
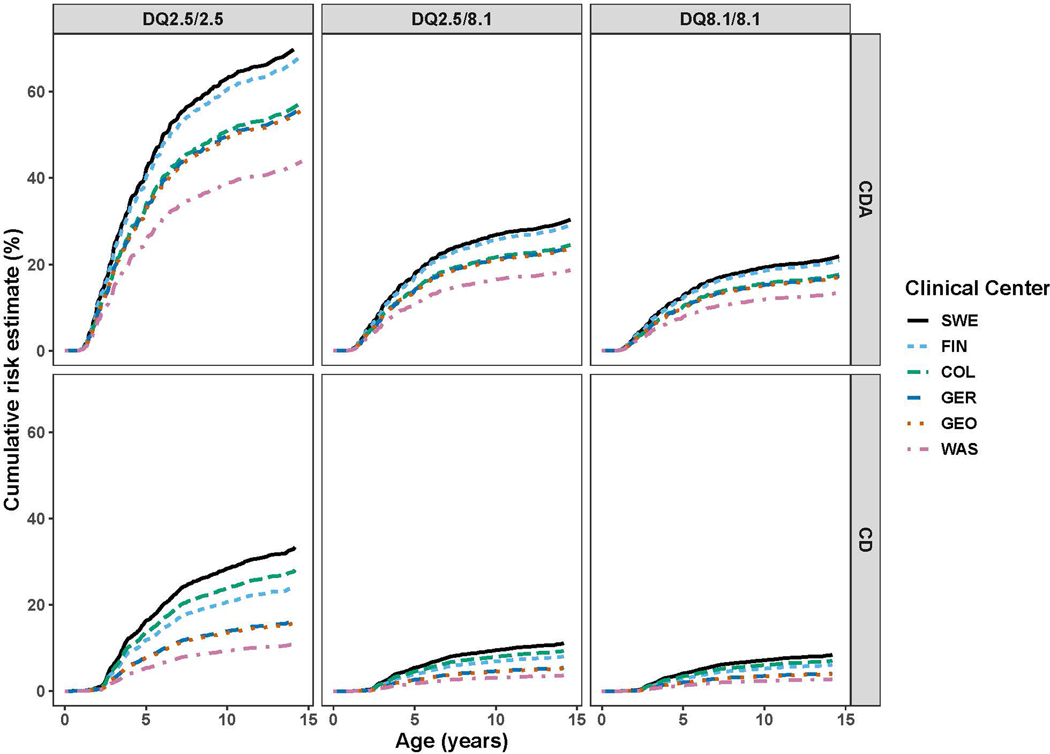
The observed regional cumulative risks for CDA and CD at each TEDDY site were compared according to the three highest risk HLA-DQ haplogenotypes (Figure 1). These observed cumulative risks are more completely detailed in Supplementary Table 4. Overall, Sweden continues to demonstrate the highest risk, with 63.1% and 28.3% of DQ2.5 subjects developing CDA and CD by age 10 respectively. This high rate of CD mirrors what was reported in earlier TEDDY publications.[5] Finland consistently had a higher incidence of CDA than Colorado, for example 60.4% vs. 50.9% for DQ2.5 subjects respectively, but had a lower incidence of CD when compared to Colorado (20.3% vs. 22.6% for DQ2.5 subjects respectively). CDA and CD risk varied substantially by haplogenotype and by clinical center, but the relative risk by region was preserved regardless of the haplogenotype. For example, the disease burden for each region remained highest in Sweden and lowest in Washington state for all haplogenotypes.
Figure 1: TEDDY Population Cumulative Risk of CDA and CD among DQ2.5/2.5, DQ2.5/8.1, DQ8.1/8.1 by site.
The cumulative risk of CDA and CD stratified by site and HLA-DQ haplogenotype. The y-axis shows the cumulative risk (%) and the x-axis shows the age (months). The figure legend is ordered from highest to lowest cumulative risk. The y-axis scales were adjusted for each panel based on the range of data.
Comparing Site-Specific Risk for CDA and CD:
Differences in site-specific CDA and CD risk are shown in Table 1. Adjusting for HLA, sex, and family history, children enrolled at the Colorado site were 2.5 times more likely (95%CI=1.7–3.5) to develop CD as those enrolled in Washington state. Finnish children were 0.70 times less likely (95%CI=0.55–0.89) than their Swedish counterparts to develop CD. Compared with the US sites, Swedish children were 1.3 times more likely to develop CDA (95%CI= 1.2–1.5) and 1.7 times more likely to develop CD (95%CI=1.4–2.0). Finnish children were 1.2 times as likely to develop CDA as those in the US (95%CI=1.1–1.4), but were not at higher risk of CD (HR=1.2 95%CI=0.91–1.5). Swedish children were at higher CD risk than those in Finland and Germany (HR=1.4 and HR=1.8; 95%CI=1.1–1.8 and 95%CI=1.2–2.8, respectively).
Table 1:
The Environmental Determinants of Diabetes in the Young (TEDDY) Hazard Ratios for Celiac Disease Autoimmunity and Celiac Disease
| Site | Celiac Disease Autoimmunity | Celiac Disease | ||
|---|---|---|---|---|
| Hazard Ratio (95% CI) | P-value | Hazard Ratio (95% CI) | P-value | |
| Washington b | 1.0 | 1.0 | ||
| Georgia | 1.3(1.0–1.6) | 0.05 | 1.4(0.92–2.3) | 0.11 |
| Colorado | 1.3(1.0–1.6) | 0.02 | 2.5(1.7–3.5) | <0.001 |
| Finland | 1.4(1.2–1.8) | <0.001 | 1.9(1.3–2.8) | <0.001 |
| Germany | 1.3(0.99–1.7) | 0.06 | 1.6(0.93–2.6) | 0.09 |
| Sweden | 1.5(1.3–1.9) | <0.001 | 2.8(2.0–3.9) | <0.001 |
Model adjusted for HLA haplogenotype, sex, and family history of CD
Washington was selected as the reference site as it had the lowest incidence of celiac disease autoimmunity and celiac disease.
Stem Cell Donor Registries for Haplogenotypes Frequencies:
According to stem cell donor registry data, DQ2.5/X and DQ8.1/X were the most common HLA-DQ haplogenotypes in the general population (Supplementary Table 5). Sweden had the highest frequency of celiac-permissive HLA-DQ haplogenotypes (41.6%) and Georgia the lowest (34.4%). It should be noted that, for the DQ2.5/DQ2.5, DQ2.5/DQ8.1, and DQ8.1/8.1 haplogenotypes which were directly detected in TEDDY, the Donor Registry frequencies in Supplementary Table 5 roughly match the frequencies observed in the general population newborns screened in TEDDY, especially the pattern of more frequent risk haplotypes in Sweden and to a lesser extent Colorado.[12]
Site-Specific Population Cumulative Incidence Adjusted by Regional Frequencies of Haplogenotypes:
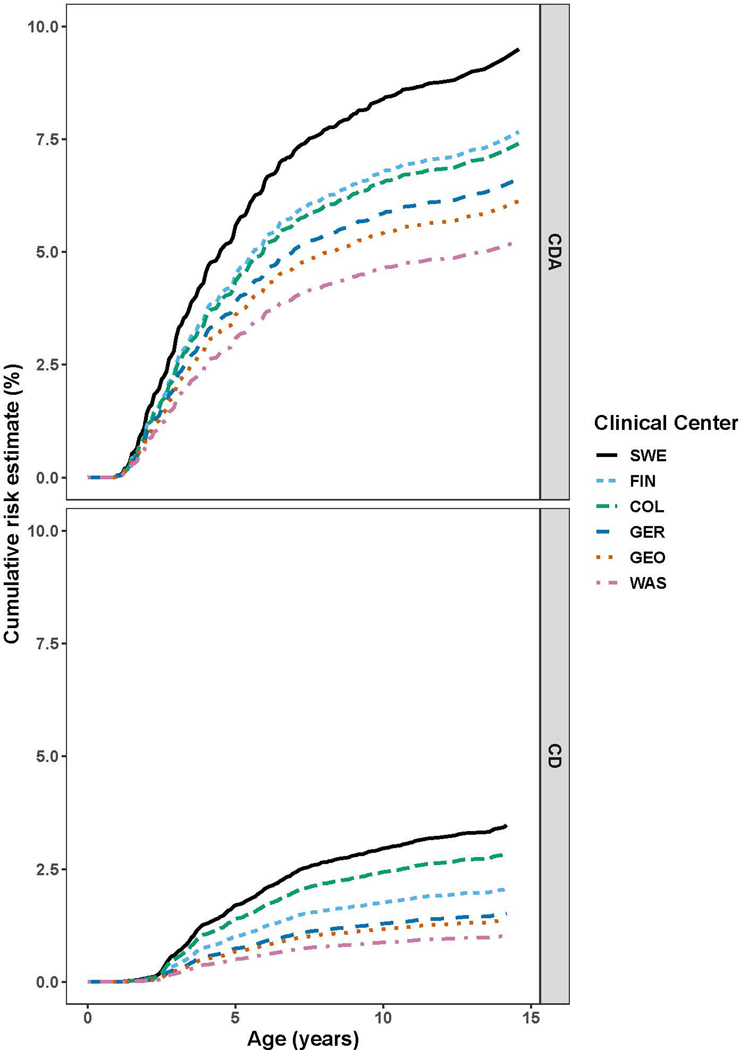
While DQ2.5 homozygosity conferred the highest risk of CDA and CD, DQ2.5 heterozygotes contributed most to the cumulative incidence. In this cohort born between 2004 and 2010, the 10-year cumulative incidence was highest in Sweden (CDA 8.4%, CD 3.0%). Within the US, Colorado had the highest cumulative incidence for both endpoints (CDA 6.5%; CD 2.4%) (Figure 2 and Table 2).
Figure 2: Estimated General Population Cumulative Incidence of CDA and CD by Site.
The cumulative incidence of CDA and CD stratified by TEDDY site. Observed incidences were weighted with the HLA-DQ haplogenotype frequencies from regional stem cell donor registries to obtain regional estimates.
Table 2:
Estimated General Population Cumulative Incidence of Celiac Disease Autoimmunity (CDA) and Celiac Disease (CD) by Site
| CDA | COLORADO, % | GEORGIA, % | WASHINGTON, % | FINLAND, % | GERMANY, % | SWEDEN, % |
|---|---|---|---|---|---|---|
| 5 years age | 4.4 (4.0–4.8)a | 3.6 (3.2–4.1) | 3.1 (2.8–3.4) | 4.5 (4.1–5.0) | 3.9 (3.3–4.5) | 5.6 (5.2–6.0) |
| 10 years age | 6.5 (6.0–7.2) | 5.4 (4.8–6.1) | 4.6 (4.2–5.2) | 6.8 (6.2–7.4) | 5.9 (5.0–6.8) | 8.4 (7.8–9.0) |
| CD | COLORADO | GEORGIA | WASHINGTON | FINLAND | GERMANY | SWEDEN |
| 5 years age | 1.4 (1.2–1.6) | 0.7 (0.5–0.8) | 0.5 (0.4–0.6) | 1.0 (0.9–1.2) | 0.7 (0.6–1.0) | 1.7 (1.5–1.9) |
| 10 years age | 2.4 (2.1–2.8) | 1.2 (0.9–1.5) | 0.9 (0.7–1.1) | 1.8 (1.5–2.1) | 1.3 (1.0–1.7) | 3.0 (2.7–3.3) |
Brackets represent the 95% confidence interval
Sensitivity Analyses with Lower Non-Biopsy tTGA threshold for CD
A post-hoc sensitivity analysis using a lower tTGA cutoff to define CD serologically (based on ROC data) was performed in order to reduce bias in site differences for biopsy referral and to increase the sensitivity of our definition of CD for incidence estimation. The original study strict serologic definition of CD of tTGA ≥100 avoids an over-diagnosis of CD, but at the risk of missing some potential cases that did not receive biopsy. When the tTGA cutoff to define CD was lowered to average two-visit tTGA≥ 67.4, more children met study serologic criteria for CD. Even with this lower cutoff, the differences in the risk of CD between clinical sites and countries were still observed with statistical significance (data not shown). This indicates that the regional differences in CD incidence could not be solely attributed to detection biases posed by differential biopsy rates.
Discussion:
In this study, the general population cumulative incidence of CD by age 10 is highly variable, ranging between 0.9% (Washington) to 2.4% (Colorado) within the US, and as high as 3% (Sweden). The rates of CDA are even higher indicating that not all who develop autoimmunity have progressed to CD. These differences remain after adjusting for key factors including HLA haplogenotypes, sex, and family history. Our findings overall are consistent with a recent meta-analysis of world-wide incidence data that suggested that CD incidence continues to rise and is highest amongst children, but is region dependent.[1] Our study specifically finds disparities in cumulative incidence even within the US participating sites and confirms dissimilarities between the US and Europe through a prospective and uniform screening program.[5, 13, 14]
Notably, each specific HLA-DQ haplogenotype was associated with consistently lower disease burden in Washington state versus Colorado, and similarly with lower disease burden in Germany versus Sweden and Finland, regardless of the specific HLA haplogenotype. This indicates that any specific HLA haplogenotype risk does not convey the same risk for disease from region to region. Finland consistently had a higher cumulative incidence of CDA, but not CD when compared to Colorado, suggesting that Finland may have higher rates of low level or transient autoimmunity or differing environmental factors that ameliorate progression to CD such as higher rates of probiotic supplementation in the first year of life.[15]
Multiple environmental factors likely account for the marked differences in autoimmunity amongst regions, such as diet, chemical exposures, vaccination patterns, gastrointestinal infections, or more likely an interaction of all of these exposures. There are also likely non-HLA and epigenetic influences that moderate the development of CDA and CD. The effect of some of these factors on CDA and CD has been well described in other studies[16–21] although the field still lacks a full understanding of the interplay of environment upon the development of CDA and CD. Other publications from the TEDDY cohort have highlighted differences in gluten intake[22], Rotavirus vaccination rates[23], early life gastrointestinal infections and season of birth[23, 24] at the respective sites. For example, Sweden, the site with the highest CD and CDA incidence, has previously been observed to have the lowest rotavirus vaccination rates and the highest median gluten intake among the TEDDY sites. Capturing environmental, genetic, and epigenetic exposures such as detailed dietary intake as well as detailed genetic identification of the stool virome and microbiome remains a primary focus of TEDDY. Future prospective studies isolating these modifiable factors should also be planned to assess causal pathways and plan for preventive strategies.
There were several limitations to this study. The self-reported nature of family history does pose the potential for information bias, but it would not have been feasible to have all family members of study participants screened as well. Since the decision to proceed to biopsy is outside of the study protocol, differences in biopsy rates between sites may introduce bias when estimating the incidence of CD. The inclusion of subjects with only very high tTGA levels helps capture additional CD cases but could be too stringent. However, when a lower threshold was used for the TEDDY serologic criteria for CD to account for this, the differences in regional risk were preserved. Furthermore, similar regional differences were seen for the outcome of CDA, which is not affected by these differential biopsy rates. However, it cannot be excluded that the observed regional differences resulted from random variation or differences in regional follow up and testing. Because initial screening for tTGA in the US is done by a lab that only measures the IgA isotype (as opposed to in Europe which measures both IgA and IgG isotypes), it is possible that a very small number of individuals in the US would be missed due to IgA deficiency. By using the less stringent definition of CDA and the more stringent definition of CD, we provide a range for estimated cumulative incidence for which the actual value of CD incidence likely falls within.[4, 25]
TEDDY only enrolled four of the described at-risk HLA-DQ haplogenotypes and, therefore, several assumptions were necessary in order to estimate the general population cumulative incidence. First, since TEDDY did not enroll DQ2.5/X subjects from the general population, this study used DQ2.5/DQ8.1 risk as a proxy for the calculation of the DQ2.5/X estimated cumulative incidence. While DQ8.1 is a risk haplotype on its own, it does not add to DQ2.5 risk when present in a DQ2.5/DQ8.1 haplogenotype.[4] Since DQ4.2 is not known to affect CD risk, the present study also uses DQ8.1/DQ4.2 risk as a proxy for DQ8.1/X risk. Another minor caveat to this estimate is that the additional risk incurred by having DQ2.2 alone was not included in the risk assessment, except when inherited with DQ7.5 or DQ2.5.[26–28] When considering DQ2.2/DQ7.5, the celiac literature indicates that the DQA1*05-DQB1*02 heterodimer confers similar disease risk in trans as in cis, and was therefore accounted for accordingly.[30] Participation in stem cell registries may pose selection bias. However, the HLA-DQ frequency data from newborn HLA screening from TEDDY and other birth cohort studies mirrors the data from the stem cell registries suggesting that these registries are representative.[29] Finally, some stem cell registries excluded participants based on a medical history of autoimmune diseases like CD and some children with CDA may have started a gluten-free diet before diagnostic confirmation of CD. However, these additional limitations would each only result in a further slight underestimation of the general population CD cumulative incidence. In contrast, our higher estimates are consistent with the prior DAISY cohort in Colorado including adolescents born prior to 2004. This study’s current estimate at 10 years of 2.4% in Colorado is similar to DAISY’s estimated cumulative incidence of 2.1–3.6% by 10 years.[4] Taken together, these findings indicate that pediatric CD incidence is currently extremely high in some regions, and may continue to manifest in rising rates of CD in adults. [1, 30] It also emphasizes the notion that CD remains under-ascertained in the general population and that current screening recommendations based on high-risk characteristics or symptoms may leave many cases unidentified.
In conclusion, there is a high cumulative incidence of CD that varies from country to country and even from state to state. From a policy standpoint, this informs future screening practices and supports efforts towards mass screening, at least in some areas. In the clinical setting, this points to the importance for clinicians to have a low threshold for CD screening in the appropriate clinical setting. The ongoing high incidence of CD in the pediatric population suggests that there are still active environmental and non-HLA drivers of disease at least in some regions. These differences cannot be explained simply by the proportion of HLA-DQ haplogenotype risk found within the various regional background populations, sex, or family history. Ongoing analyses of environmental, genetic, and epigenetic exposures in the TEDDY birth cohort will aid in the understanding of these marked differences in regional CDA and CD. Future analyses will focus on the interplay of these potential mediators and moderators in the development of CDA and CD.
Supplementary Material
WHAT IS KNOWN:
Celiac disease is common and reported incidence differs by study and by region.
Birth cohort studies suggest non-genetic factors may explain the observed differences.
WHAT IS NEW HERE:
There is a high rate of new celiac disease cases up to age 10 suggesting that there are still active drivers of disease.
Regional variation of celiac disease exists even within the USA.
Celiac disease was common in all studied regions and clinicians should have a low threshold to screen in the appropriate clinical scenario.
Acknowledgements:
Data Obtained from Stem Cell Registries including: Dusseldorf Blood Bank (Dr. Gesine Kogler), the ZKRD- The German National Bone Marrow Donor Registry (Dr. Hans-Peter Eberhard), The Finnish Stem Cell Registry (Matti Korhonen), Martin Maiers (National Marrow Donor Program / Be The Match Registry), and the Swedish National Cord Blood Bank and Tobias Registry (Dr. Anders).
Financial Support:
The TEDDY Study is funded by U01 DK63829, U01 DK63861, U01 DK63821, U01 DK63865, U01 DK63863, U01 DK63836, U01 DK63790, UC4 DK63829, UC4 DK63861, UC4 DK63821, UC4 DK63865, UC4 DK63863, UC4 DK63836, UC4 DK95300, UC4 DK100238, UC4 DK106955, UC4 DK112243, UC4 DK117483, U01 DK124166, U01 DK128847, and Contract No. HHSN267200700014C from the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), National Institute of Allergy and Infectious Diseases (NIAID), Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD), National Institute of Environmental Health Sciences (NIEHS), Centers for Disease Control and Prevention (CDC), and JDRF. This work is supported in part by the NIH/NCATS Clinical and Translational Science Awards to the University of Florida (UL1 TR000064) and the University of Colorado (UL1 TR002535). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Abbreviations:
- CDA
celiac disease autoimmunity
- CD
celiac disease
- DAISY
Diabetes Autoimmunity Study in the Young
- HLA
Human Leukocyte Antigen
- tTGA
tissue transglutaminase autoantibodies
- TEDDY
The Environmental Determinants of Diabetes in the Young
- T1D
type 1 diabetes
- US
United States
Footnotes
Guarantor of the article: Edwin Liu
Potential Competing Interests: There are no conflicts of interest
Previous Presentations: Preliminary data was presented as a poster presentation at DDW 2018 and is submitted for ICDS 2022.
RESOURCES
- 1.King JA, et al. , Incidence of Celiac Disease Is Increasing Over Time: A Systematic Review and Meta-analysis. Am J Gastroenterol, 2020. 115(4): p. 507–525. [DOI] [PubMed] [Google Scholar]
- 2.Agardh D, et al. , Clinical features of celiac disease: a prospective birth cohort. Pediatrics, 2015. 135(4): p. 627–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stahl MG, et al. , Mass Screening for Celiac Disease: The Autoimmunity Screening for Kids Study. Am J Gastroenterol, 2021. 116(1): p. 180–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu E, et al. , High Incidence of Celiac Disease in a Long-term Study of Adolescents With Susceptibility Genotypes. Gastroenterology, 2017. 152(6): p. 1329–1336 e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu E, et al. , Risk of pediatric celiac disease according to HLA haplotype and country. N Engl J Med, 2014. 371(1): p. 42–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lernmark B, et al. , Enrollment experiences in a pediatric longitudinal observational study: The Environmental Determinants of Diabetes in the Young (TEDDY) study. Contemp Clin Trials, 2011. 32(4): p. 517–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.The Environmental Determinants of Diabetes in the Young (TEDDY) study: study design. Pediatr Diabetes, 2007. 8(5): p. 286–98. [DOI] [PubMed] [Google Scholar]
- 8.Uusitalo U, et al. , Early Infant Diet and Islet Autoimmunity in the TEDDY Study. Diabetes Care, 2018. 41(3): p. 522–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oberhuber G, Histopathology of celiac disease. Biomed Pharmacother, 2000. 54(7): p. 368–72. [DOI] [PubMed] [Google Scholar]
- 10.Liu E, et al. , Fluctuating transglutaminase autoantibodies are related to histologic features of celiac disease. Clin Gastroenterol Hepatol, 2003. 1(5): p. 356–62. [DOI] [PubMed] [Google Scholar]
- 11.Lionetti E, et al. , Introduction of gluten, HLA status, and the risk of celiac disease in children. N Engl J Med, 2014. 371(14): p. 1295–303. [DOI] [PubMed] [Google Scholar]
- 12.Hagopian WA, et al. , The Environmental Determinants of Diabetes in the Young (TEDDY): genetic criteria and international diabetes risk screening of 421 000 infants. Pediatr Diabetes, 2011. 12(8): p. 733–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stordal K, et al. , Epidemiology of coeliac disease and comorbidity in Norwegian children. J Pediatr Gastroenterol Nutr, 2013. 57(4): p. 467–71. [DOI] [PubMed] [Google Scholar]
- 14.Olsson C, et al. , Regional variation in celiac disease risk within Sweden revealed by the nationwide prospective incidence register. Acta Paediatr, 2009. 98(2): p. 337–42. [DOI] [PubMed] [Google Scholar]
- 15.Uusitalo U, et al. , Early Probiotic Supplementation and the Risk of Celiac Disease in Children at Genetic Risk. Nutrients, 2019. 11(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Unalp-Arida A, et al. , Lower Prevalence of Celiac Disease and Gluten-Related Disorders in Persons Living in Southern vs Northern Latitudes of the United States. Gastroenterology, 2017. 152(8): p. 1922–1932.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kondrashova A, et al. , Lower economic status and inferior hygienic environment may protect against celiac disease. Ann Med, 2008. 40(3): p. 223–31. [DOI] [PubMed] [Google Scholar]
- 18.Aronsson CA, et al. , Age at gluten introduction and risk of celiac disease. Pediatrics, 2015. 135(2): p. 239–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sharma A, et al. , Identification of Non-HLA Genes Associated with Celiac Disease and Country-Specific Differences in a Large, International Pediatric Cohort. PLoS One, 2016. 11(3): p. e0152476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gaylord A, et al. , Persistent organic pollutant exposure and celiac disease: A pilot study. Environ Res, 2020. 186: p. 109439. [DOI] [PubMed] [Google Scholar]
- 21.Kuja-Halkola R, et al. , Heritability of non-HLA genetics in coeliac disease: a population-based study in 107 000 twins. Gut, 2016. 65(11): p. 1793–1798. [DOI] [PubMed] [Google Scholar]
- 22.Andrén Aronsson C, et al. , Association of Gluten Intake During the First 5 Years of Life With Incidence of Celiac Disease Autoimmunity and Celiac Disease Among Children at Increased Risk. Jama, 2019. 322(6): p. 514–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kemppainen KM, et al. , Factors That Increase Risk of Celiac Disease Autoimmunity After a Gastrointestinal Infection in Early Life. Clin Gastroenterol Hepatol, 2017. 15(5): p. 694–702.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lindfors K, et al. , Metagenomics of the faecal virome indicate a cumulative effect of enterovirus and gluten amount on the risk of coeliac disease autoimmunity in genetically at risk children: the TEDDY study. Gut, 2020. 69(8): p. 1416–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Almallouhi E, et al. , Increasing Incidence and Altered Presentation in a Population-based Study of Pediatric Celiac Disease in North America. J Pediatr Gastroenterol Nutr, 2017. 65(4): p. 432–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abadie V, et al. , Integration of genetic and immunological insights into a model of celiac disease pathogenesis. Annu Rev Immunol, 2011. 29: p. 493–525. [DOI] [PubMed] [Google Scholar]
- 27.Vader W, et al. , The HLA-DQ2 gene dose effect in celiac disease is directly related to the magnitude and breadth of gluten-specific T cell responses. Proc Natl Acad Sci U S A, 2003. 100(21): p. 12390–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pietzak MM, et al. , Stratifying risk for celiac disease in a large at-risk United States population by using HLA alleles. Clin Gastroenterol Hepatol, 2009. 7(9): p. 966–71. [DOI] [PubMed] [Google Scholar]
- 29.Liu E, et al. , High Incidence of Celiac Disease in a Long-term Study of Adolescents With Susceptibility Genotypes. Gastroenterology, 2017. 152(6): p. 1329–1336.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ludvigsson JF, et al. , Increasing incidence of celiac disease in a North American population. Am J Gastroenterol, 2013. 108(5): p. 818–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.