SUMMARY
Organisms adapt to seasonal changes in photoperiod and temperature to survive; however, the mechanisms by which these signals are integrated in the brain to alter seasonal biology are poorly understood. We previously reported that EYES ABSENT (EYA) shows higher levels in cold temperature or short photoperiod and promotes winter physiology in Drosophila. Nevertheless, how EYA senses seasonal cues is unclear. Pigment-Dispersing Factor (PDF) is a neuropeptide important for regulating circadian output rhythms. Interestingly, PDF has also been shown to regulate seasonality, suggesting it may mediate the function of the circadian clock in modulating seasonal physiology. In this study, we investigated the role of EYA in mediating the function of PDF on seasonal biology. We observed that PDF abundance is lower in cold and short days as compared to warm and long days, opposite of what was previously observed for EYA. We observed that manipulating PDF signaling in eya+ fly brain neurons where EYA and PDF receptor are coexpressed modulates seasonal adaptations in daily activity rhythm and ovary development via EYA-dependent and independent mechanisms. At the molecular level, altering PDF signaling impacted EYA protein abundance. Specifically, we showed that protein kinase A (PKA), an effector of PDF signaling, phosphorylates EYA promoting its degradation, thus explaining the opposite responses of PDF and EYA abundance to changes in seasonal cues. In summary, our results support a model in which PDF signaling negatively modulates EYA levels to regulate seasonal physiology, linking the circadian clock to the modulation of seasonal adaptations.
eTOC Blurb
Organisms adapt to seasonal changes in photoperiod and temperature to survive. Hidalgo et al. uncover a mechanism by which the circadian clock conveys these environmental cues to modulate EYES ABSENT, a protein whose expression is sensitive to seasonal cues in insects, birds, and mammals.
Graphical Abstract

INTRODUCTION
As seasons change, organisms adapt to adverse environmental conditions to survive1,2. While some animals, like monarch butterflies3,4, migrate into more favorable environments, others undergo metabolic and physiological changes that allow them to prevail through the harsh winter5,6. These seasonal adaptations have been described in plants6,7 and animals8–11, and are triggered by changes in photoperiod12,13 (i.e. daylength) and temperature14. However, the molecular mechanisms that allow organisms to extract environmental information reflecting the calendar year is not well established. This is especially true in animals.
In several animals, endocrine signals drive seasonal adaptations. In mammals and birds, the increases in the hormone thyrotrophin (TSH) and the downstream T3 hormone, are key for photoperiodic summer responses15–18. In insects, reduction of insulin-like peptides (ILPs) and juvenile hormone (JH) are important for overwintering19–22. Although the hormonal components differ between species, studies have suggested that EYES ABSENT (EYA), a cotranscription factor with phosphatase activity, could be a key conserved molecular player within the seasonal timer, acting upstream of endocrine responses to modulate seasonal physiology. In the Japanese quail (Coturnix japonica), the expression of the eya homolog, Eya3, increases in the pars tuberalis of the pituitary gland upon transitioning into summer17. This phenomenon is also observed in soay sheep (Ovis aries), where EYA3 protein has also been proposed as an effector upstream TSH-β18,23. We recently described a functional role of EYA in the seasonal response of Drosophila melanogaster24. Specifically, we showed that eya expression in the insulin-producing cells (IPCs) within the pars intercerebralis (PI), the insect ortholog of the hypothalamus, is critical for reproductive dormancy, a seasonal adaptation characterized by a reduction in ovary size and egg production. Moreover, we reported that eya mRNA and EYA protein levels are sensitive to changes in photoperiod and temperature, increasing in simulated winter conditions24. Despite evidence from multiple organisms, the mechanism mediating changes in EYA expression in response to photoperiod and temperature has yet to be determined.
The Bünning hypothesis states that the circadian clock is required for appropriate photoperiodic measurement25,26. The core clock in Drosophila is a transcriptional translational feedback loop (TTFL) composed of the positive elements CLK-CYC, which promote the transcription of the negative elements period (per) and timeless (tim) as well as other clock-regulated genes27. Accumulation and subsequent nuclear translocation of PER-TIM then leads to the repression of CLK-CYC activity, which in turn shuts down the transcription of per and tim, preparing the clock for the next cycle. The proteasome mediates light-dependent and light-independent degradation of the negative elements, ensuring the proper functioning and entrainment of the TTFL. Bünning proposed that circadian oscillations are composed of light and dark-requiring phases26. In short days, light is only present in the light phase, while in long days, light is now present in both light and in part of the dark-requiring phase. This was later modified and presented as “coincidence model”28. This model then assumes that the elements of the circadian clock are required for photoperiodism and seasonal adaptations. Studies in several insects have been instrumental in establishing this relationship29–32. Recently, Wood et al. proposed a mechanism in mammals where Eya3 transcription is modulated by BMAL2-dependent chromatin remodeling in the pars tuberalis of the pituitary gland in sheep, highlighting the role of a circadian clock protein in regulating Eya3 expression33. In Drosophila however, the IPCs do not express a functional circadian clock34,35, although TIM expression in the fly brain is found to modulate EYA abundance24. This suggests that other unknown clock mechanisms must be involved in relaying environmental information to EYA-expressing cells to modulate EYA function in seasonal adaptations.
The circadian clock neuronal network provides extensive connections to and modulates the IPCs34–38. The clock network is composed of around 7 neuronal clusters: dorsal neurons-1 (DN1), dorsal neurons-2 (DN2), dorsal neurons-3 (DN3), lateral posterior neurons (LPN), dorsal lateral neurons (LNd), and the large and small ventral lateral neurons (l-LNvs and s-LNvs, respectively). Among clock network clusters, DN1 and s-LNvs are known to modulate the IPCs39–41. The s-LNvs secretes Pigment Dispersing Factor (PDF), a neuropeptide that enables the molecular synchronization of neuronal clusters within the clock network42,43, and is critical for the maintenance of anticipatory activity in light-dark cycles (LD) and the circadian rhythmicity in constant darkness (DD)44–47. Interestingly, the role of PDF in modulating seasonal adaptations has been examined in several organisms. In the blow fly and in the bean bug, ablation of the brain region containing the PDF-producing neurons prevents the discrimination between short and long photoperiod48,49. Moreover, ablation of Pdf in the brown-winged green bug impairs photoperiodic control of reproduction50. In Drosophila melanogaster, photoperiod sensitivity is lost in Pdf mutants51, whereas artificially increasing or decreasing the activity of Pdf-expressing cells inhibits or promotes reproductive dormancy, respectively52. More importantly, the role of PDF in reproductive dormancy appears to be mediated by direct modulation of the IPCs52.
Here, we explore the relationship between PDF and EYA in the context of seasonal biology. We provide evidence to support a mechanism by which PDF signaling regulates multiple aspects of seasonal physiology by conveying environmental cues to alter EYA levels in the fly brain via a PKA-dependent post-translational mechanism. We proposed that during summer-like conditions, PDF promotes EYA degradation by PKA-mediated phosphorylation, while a reduction in Pdf mRNA and PDF peptide in winter allows EYA accumulation, promoting winter adaptations.
RESULTS
PDF levels change with photoperiod and temperature
The circadian neuropeptide PDF is known to exhibit daily cycling in abundance in the fly brain and can be synchronized by light53. Furthermore, Pdf mRNA levels are sensitive to light intensity54. This suggests that PDF could be sensitive to environmental cues that change with seasons, namely photoperiod and temperature, and can subsequently convey these seasonal cues to alter EYA function in the fly brain. To test this hypothesis, we first examined the sensitivity of PDF abundance to key seasonal cues. We measured PDF levels in the dorsal terminals of the s-LNvs in flies subjected to different photoperiods and temperature conditions. First, we tested flies that were subjected to 7 days in simulated summer conditions, i.e. long photoperiod (LP, 16:8 LD) at 25°C, or simulated winter, i.e. short photoperiod (SP, 8:16 LD) at 10°C (Figure 1A–B). PDF levels showed daily rhythmicity in both conditions with peak phase at ZT0 (Rhythmicity LP25 p =5.4x10−3, Rhythmicity SP10 p = 3.19x10−7). Although we did not observe a difference in the amplitude of PDF cycling (p = 0.22), a reduction in the Midline Estimating Statistic Of Rhythm (MESOR) in simulated winter compared to simulated summer was observed, suggesting an overall reduction of PDF levels in winter-like conditions (MESOR LP25 = 2.41, MESOR SP10 = 1.26, p = 2.06x10−17; Figure 1C).
Figure 1. PDF levels are regulated by photoperiod and temperature.
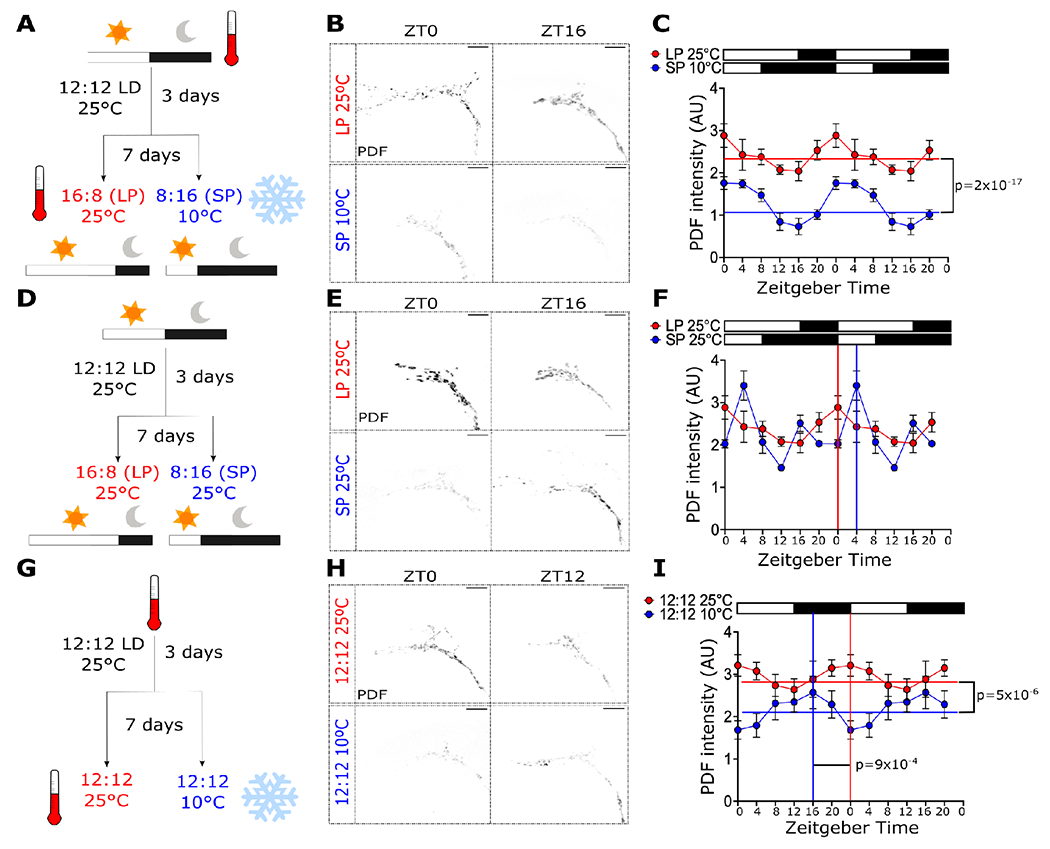
PDF levels in the s-LNv dorsal terminals were analyzed in flies entrained to (A) long-photoperiod (LP, 16:8 LD) at 25°C or short-photoperiod (SP, 8:16 LD) at 10°C, (D) LP or SP at 25°C, and (G) 12:12 LD cycles at 25°C or 10°C. Representative images for these conditions at ZT0 and ZT16 or ZT12 are shown in B, E and H, respectively. Double-plotted graphs of PDF signal for each condition are shown in C, F and I, respectively. White boxes represent lights-on, and black boxes represent lights-off. CircaCompare and RAIN was used to determine rhythmicity and phase for all datasets. MESOR (horizontal lines) and phase (vertical lines) are displayed in the graphs if statistically significant (p < 0.05), except for (F), where the phases detected between CircaCompare and RAIN were different. The phases shown in (F) were calculated using RAIN. Scale bars are 10 μm. Number of brains imaged were LP25: ZT0 n = 18, ZT4 n = 11, ZT8 n = 13, ZT12 n = 18, ZT16 = 9, ZT20 n = 14; SP10: ZT0 n = 9, ZT4 n = 8, ZT8 n = 14, ZT12 n = 14, ZT16 = 18, ZT20 n = 20; SP25 ZT0 n = 6, ZT4 n = 8, ZT8 n = 9, ZT12 n = 6, ZT16 =10, ZT20 n = 3; 12:12 25°C: ZT0 n = 14, ZT4 n = 14, ZT8 n = 15, ZT12 n = 19, ZT16 =10, ZT20 n = 15; 12:12 10°C: ZT0 n = 13, ZT4 n = 14, ZT8 n = 15, ZT12 n = 15, ZT16 =16, ZT20 n = 10.
To determine the effect of the photoperiod alone on PDF levels, the same experiment was conducted in flies entrained to either a long photoperiod or a short photoperiod but at constant temperature 25°C (Figure 1D–E). PDF abundance in both LP (p = 0.005) and SP (p = 0.034) were found to be rhythmic. When using CircaCompare55, no differences in the MESOR (p = 0.06), amplitude (p=0.9) or phase (p=0.72) were observed between these conditions (Figure 1F). Interestingly, the peak estimated for SP by CircaCompare was ZT1.5, far from the ZT4 peak based on visual inspection of the data. We therefore confirmed the phase of the peak using Rhythmicity Analysis Incorporating Nonparametric methods (RAIN)56. Compared to CircaCompare, RAIN is better suited for detection of peaks in skewed waveforms, as in the case for SP25°C (Figure 1F). While CircaCompare and RAIN were in agreement when computing the phases for all other datasets in Figure 1, RAIN determined the peak phase for SP25°C in Figure 1F to be ZT4 (p = 0.000015), matching that of visual inspection and indicating a 4-hr phase delay compared to LP25°C. To test the effect of temperature, flies were reared at 25°C or 10°C in 12:12 LD cycles (Figure 1G–H). We observed a reduction in the MESOR at 10°C compared to 25°C (MESOR 25°C = 2.95, MESOR 10°C = 2.16, p = 5x10−6; Figure 1I). Moreover, a change in the phase of the peak in PDF level was observed, with an estimated peak at ZT14 in 10°C and at ZT23 in 25°C (p = 9x10−4; Figure 1I).
As we observed low PDF levels in simulated winter, we hypothesized that a reduction in Pdf mRNA could be the cause. To test this hypothesis, we conducted fluorescent in situ hybridization (FISH) to detect Pdf transcripts in brains of flies expressing CD8::GFP in Pdf+ cells. We subjected these flies to either simulated summer or simulated winter. As Pdf mRNA expression does not show daily oscillation53, only one timepoint (ZT16) was assessed in this experiment. A clear signal, confined to CD8::GFP positive cells, was observed (Figure 2A). This signal was absent in brains from Pdf-null mutants (Pdf01), confirming the specificity of FISH library (Figure S1). A reduction in fluorescent signal was observed in the LNvs somas under simulated winter compared to flies reared in simulated summer (Figure 2B). Quantification of the signal from different brains show a consistent reduction in the normalized fluorescent intensity in the CD8::GFP+ cells consistent with lower levels of Pdf mRNA (t(13) = 3.267, p = 0.0061, Figure 2C).
Figure 2. Pdf mRNA is sensitive to seasonal cues.
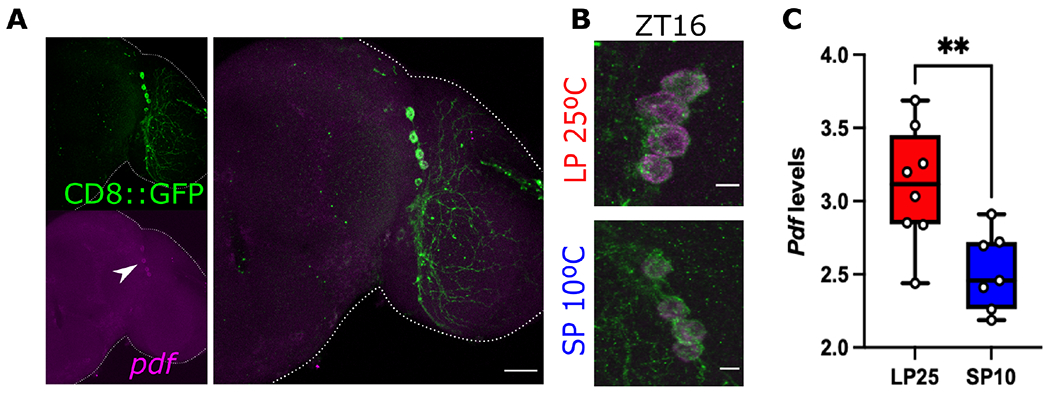
(A) Pdf mRNA (magenta) was detected using fluorescence in situ hybridization (FISH) in whole brains of flies entrained for 7 days to simulated-summer (LP at 25°C) or simulated-winter (SP at 10°C). CD8::GFP was expressed using the Pdf-gal4 driver to detect the PDF-producing LNvs. (B) Representative images of LNvs of flies entrained to LP 25°C (top panel) or SP 10°C (bottom panel). (C) Quantification of the normalized intensity of the Pdf signal at ZT16 is compared between conditions. Unpaired t-test, ** p <0.01. Number of brains imaged were LP25 n = 8, SP10 n = 7 brains. Scale bar in A is 25 μm and 5 μm in B. See also Figure S1 and Table S1.
Adaptation of daily locomotor activity rhythm to cold temperature and short photoperiod depends on PDF
PDF is a modulator of daily locomotor activity in Drosophila. Pdf01 mutants have previously been shown to exhibit an advancement in the evening peak of activity in LD and loss of rhythmicity under constant darkness46. As we observed a decrease in Pdf mRNA and PDF protein in simulated winter or when flies are exposed to lower temperature, we reasoned that changes in Pdf abundance should translate into changes in locomotor activity in response to photoperiodic and/or temperature changes, which has been shown to change over the calendar year in insects including flies. This would represent an effective readout of overall PDF abundance in flies. We therefore monitored daily activity in Pdf01 flies compared to Pdf01 mutants bearing a genomic rescue Pdf transgene (yw ; Pdf{WT} ; Pdf01, hereafter denoted as genomic rescue) in selected conditions. First, flies were monitored in 12:12 LD cycles but exposed to either 25°C or 10°C. As previously described46, Pdf01 flies display an advance in evening activity peak at 25°C consistent across the 5 days of monitoring. This is not observed in the genomic rescue, which has normal timing of evening peak immediately before the lights off transition at dusk (Figure 3A–B). In contrast, at 10°C, an advance in the evening peak was now observed in the genomic rescue flies and they behaved more similar to Pdf01 mutants (Figure 3C–D). Then, to test the effect of photoperiod, we monitored locomotor activity in short day conditions (8:16 LD) at 25°C in Pdf01 mutants and genomic rescue flies. Consistent with previous studies57, Pdf01 mutants showed a peak that is close to the lights-off transition under this condition (Figure 3E–F).
Figure 3. Locomotor adaptation to photoperiod and temperature in Pdf-null mutants.
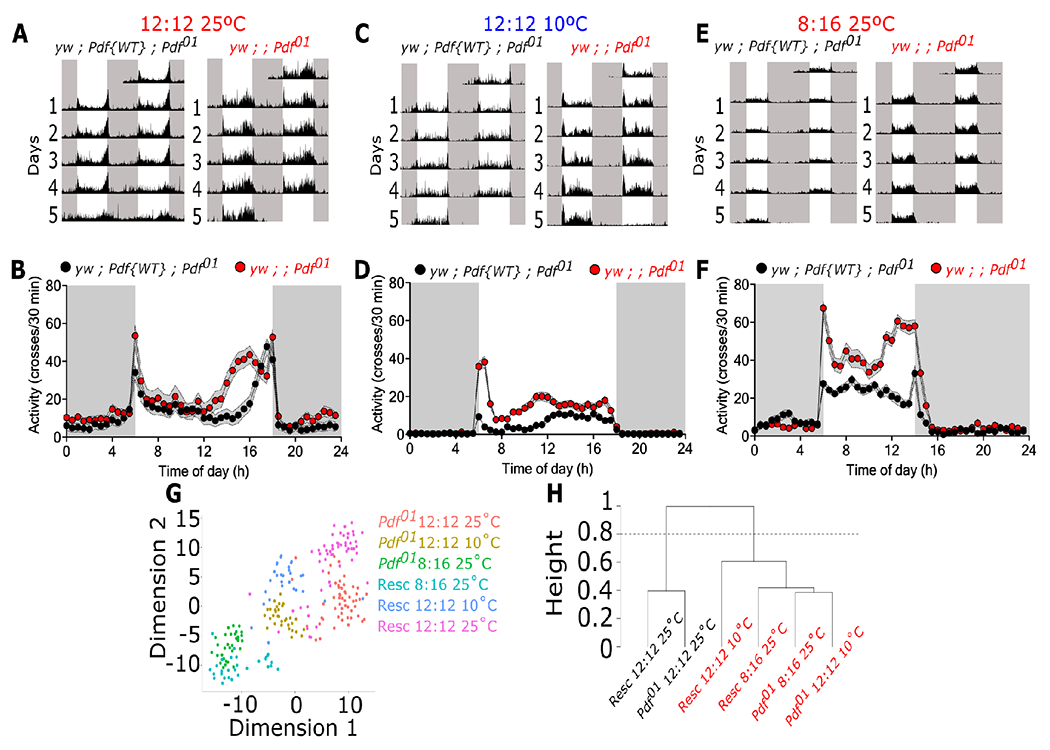
Locomotor activity was monitored in Pdf-null mutants (yw ; Pdf01) and in flies expressing a wild-type copy of Pdf (genomic rescue, yw ; Pdf{WT} ; Pdf01) subjected to (A-B) 12:12 LD at 25°C, (C-D) 12:12 LD at 10°C, or (E-F) 8:16 LD at 25°C. (G) Variance in locomotor activity under these environmental conditions as tSNE of the normalized locomotor activity for Rescue (Resc) and the mutant (Pdf01). Each dot represents an individual fly. (H) Hierarchical clustering of the normalized locomotor activity. Arbitrary cut of the tree at 0.8 (y-axis) generates two distinct groups denotated as black and red labels. White boxes represent lights-on while black boxes represent lights-off in panels A-F. Data were analyzed using a two-way ANOVA with Bonferroni’s multiple comparison test. Number of flies used were yw ; Pdf{WT} ; Pdf01 12:12 25°C n = 56, yw ; Pdf{WT} ; Pdf01 8:16 25°C n = 30, yw ; Pdf{WT} ; Pdf01 12:12 10°C n = 30, yw ; ; Pdf01 12:12 25°C n = 55, yw ; ; Pdf01 8:16 25°C n = 31, yw ; ; Pdf01 12:12 10°C n = 32.
To better visualize the overall similarities of fly activity profile between the different datasets, we conducted t-distributed Stochastic Neighbor Embedding (t-SNE) analysis on the observed locomotor activity rhythms using normalized averaged daily activity profiles over 5 days (see STAR Methods for details). Overall, three distinct groups can be defined by the environmental conditions tested. While data from the genomic rescue and the Pdf01 mutants at 25°C (magenta and red dots, respectively) cluster within two broad individual groups, data between the two genotypes at 10°C (blue and yellow dots) and 8:16 (green and cyan dots) are less differentiated (Figure 3G). To further define these clustering, we used an unsupervised hierarchical clustering analysis of the normalized averaged daily activity profiles per group (see STAR Methods for calculation) (Figure 3H). Two main branches are observed, one with rescue and mutant flies at 12:12 at 25°C (black labels) and the other one containing both mutants and rescue flies at 8:16 and 10°C (red labels). These suggest that at these environmental conditions, the structure of the daily locomotor activity in mutant and rescue flies are similar to each other than to the flies at 12:12 25°C. Overall, these data suggest that temperature and photoperiod differentially modulate the evening activity in Pdf01 mutants compared to control flies.
PDF modulates overall activity level through action in eya+ cells but its effect on timing of evening peak is not dependent on EYA
Upon release, PDF acts through activation of the G-coupled PDF receptor (PDFR)42. Thus, we reasoned that if PDF signaling acts through EYA to regulate changes in daily activity rhythms in response to photoperiodic and temperature changes, cells that are important in this process would coexpress eya and Pdfr. To explore this, we first used a single-cell transcriptomic (scRNA-seq) datasets available from the Fly Cell Atlas initiative58 to determine if Pdfr is coexpressed with eya in the fly brain. Using SCOPE, a visualization tool for large sequencing dataset59, we differentiated all cell/tissue types in the scRNA-seq dataset based on overall expression (Figure S2A), and also colored them specifically by Pdfr and eya expression (Figure S2B; green and red dots, respectively). Coexpression of both transcripts is observed in head tissue, gut, and testis cluster. Further examination showed that many of the cells expressing eya within the head cluster also express Pdfr (Figure S2B; insert).
To determine if PDF-dependent changes in daily activity rhythms in response to photoperiodic and temperature as observed in Figure 3 is mediated via EYA, we monitored locomotor activity under 12:12 LD cycles at 25°C in flies where Pdfr was knocked down in eya+ cells using an eya-Gal4 driver (Figure S3A)24. Interestingly, we did not observe a dramatic shift in the timing of the evening activity peak as in Pdf01 flies. This suggests that PDF signaling alters timing of evening peak through mechanisms other than EYA. We did observe, however, a decrease in the strength of the evening anticipation in eya > Pdfr-RNAi flies compared to controls indicated by reduced evening anticipation (Figure S3A–B). Notably the opposite is observed in flies driving a tethered version of PDF (t-PDF) using the same driver to activate PDF signaling (Figure S3C–D). This difference is even more pronounced when looking at the overall activity level of flies in constant darkness. Knocking down Pdfr in eya+ cells results in an overall reduction in total locomotion (H(2, 86) = 17.89, p = 0.0001; Figure 4A–B), while activating PDF signaling by expressing t-PDF in eya+ cells promotes an increase in activity (U = 333, p = 0.0384; Figure 4C–D).
Figure 4. PDF modulates overall locomotion through eya+ cells.
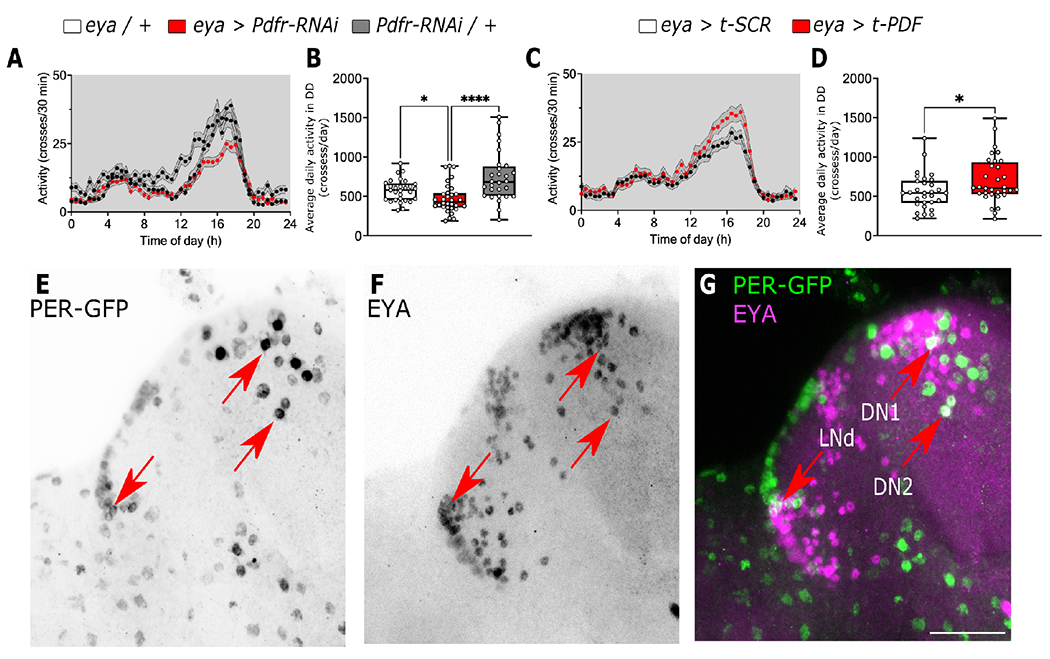
(A-B) Average locomotor activity of flies in constant darkness at 25°C where the PDF receptor was knocked down in eya+ cells (eya > Pdfr-RNAi red line and box) compared to controls (eya / + and Pdfr-RNAi / +, black and gray lines, respectively). (C-D) Average locomotor activity of flies where a membrane tethered version of PDF was expressed in eya+ cells (eya > t-PDF red line and box) compared to the expression of a scrambled version of the peptide (eya > t-SCR black line and box). (E-G) Confocal images of fly brain expressing endogenous GFP tagged PER (green) and stained against EYA (magenta). Red arrows points DN1, DN2 and LNd clock populations. Scale bar is 25 μm. Data in B were analyzed with Kruskal-Wallis test followed by Dunnett’s multiple comparison test, Mann-Whitney in D. Number of flies used were eya / + n = 29, eya > Pdfr-RNAi n = 32, Pdfr-RNAi / + n = 28, eya > t-SCR n = 31, eya > t-PDF n = 31, eya / + n = 30. See also Figures S2 and S3.
Previous studies have shown that PDF modulates evening anticipation by its action on non-Pdf-expressing clock cells within the clock network60. Thus, it is possible that the effect observed while knocking down Pdfr or expressing t-PDF in eya+ cells is due to the modulation of these neurons. To test if eya is expressed in clock neurons, we detected EYA using a monoclonal antibody (Figure 4F) in flies carrying a endogenous GFP-tagged version of PER61 to mark neurons in the clock network (Figure 4E). EYA was detected in some PER-expressing cells including DN1, LNd and DN2 neurons (Figure 4G) consistent with a role of these clusters in locomotor activity.
PDF modulates ovary size through action in eya+ insulin-producing cells
Expression of eya in the IPCs is critical for the regulation of seasonal reproductive dormancy24. To determine if PDF signaling works through EYA in IPCs to regulate seasonal phenotypes, we first tested if Pdfr is coexpressed with eya in these cells. Similar to a previous report62, we observed Pdfr+ cells widespread across the brain (Figure S2). A cluster of cells is observed in the PI where the IPCs are located (Figure 5A, inset). We used SCOPE to plot all the cells from the dilp2IPC SMART-seq dataset, generated from isolated IPCs by fluorescence-activated cell sorting58, and colored them by Pdfr and eya (Figure 5B; green and red dots). We observed eya expression in a small number of cells and some of them also express Pdfr (Figure 5B; yellow dots). We then detected EYA using a monoclonal antibody in Pdfr > CD8::GFP brains, focusing on this cluster. As previously described24, EYA expression is observed within these cells, consistent with its expression in the IPCs, and is within the Pdfr+ cells (Figure 5C, blue arrows). Thus, both transcriptomic and immunohistochemistry data support the notion that EYA and PDFR are coexpressed in the IPCs in fly brains.
Figure 5. PDF and EYA have opposite effects in the IPCs to modulate ovary size under summer-like conditions (16:8 LD at 25°C).
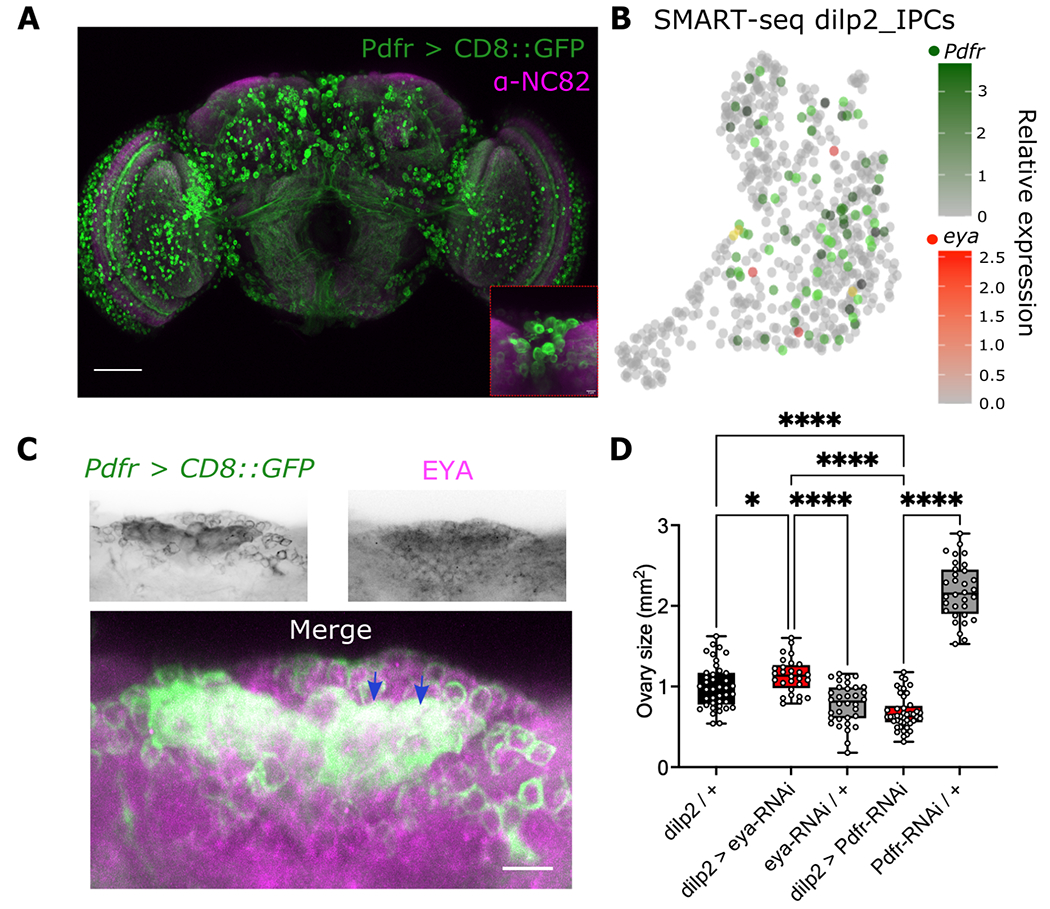
(A) Expression of CD8::GFP was driven in the Pdfr+ cells using a Pdfr-gal4 driver. Nc82 antibody was used as counterstaining. Expression of CD8::GFP is observed in several segments of the brain, including the pars intercerebralis, containing the IPCs (inset). Scale bar is 50μm. (B) SMART-seq dataset from dilp2+ isolated IPCs from Fly Cell Atlas53 as UMPA, colored by Pdfr (green) and eya (red) expression. Coexpression is shown as yellow. Color bar denotes the relationship between color intensity and relative expression levels. (C) EYA staining (upper right panel) in brains of flies expressing CD8::GFP in the Pdfr+ cells (Pdfr > CD8::GFP, upper left panel). Merged image is observed in the bottom panel. Scale is 5 μm. (D) Comparison of ovary size in mm2 while knocking down eya (dilp2 > eya-RNAi) or knocking down Pdfr (eya > Pdfr-RNAi) compared to genetic controls (black and grey boxes). Data was analyzed using a one-way ANOVA followed by Holm-šidák’s multiple comparison test. Number of flies used were dilp2 / + n = 40, dilp2 > eya-RNAi n = 28, eya-RNAi / + n = 34, dilp2 > Pdfr-RNAi n = 45, UAS-Pdfr-RNAi / + n = 32.
We showed that PDF levels decrease in winter-like conditions (Figures 1–2), while our previous study showed that EYA levels increase under similar conditions. This suggests opposite functions of PDF vs EYA. To validate the antagonistic relationship between PDF and EYA, we knocked down Pdfr or eya in the IPCs and examined ovary size in summer-like conditions. We hypothesized that, knocking down Pdfr in this condition, should render smaller ovaries and knocking down eya even in summer-like condition may produce bigger, more developed ovaries, as eya promotes winter physiology and reproductive dormancy. As expected, knocking down Pdfr generated a reduction in the ovary size in summer conditions compared to the parental controls, while the same manipulation for eya produced the opposite effect, suggesting opposite roles of EYA and PDF in ovary development in summer-like conditions (F(4, 173) = 177.8, p < 0.0001; Figure 5D). We did not perform a parallel experiment in winter-like conditions given we have previously shown that knocking down eya in winter-like conditions resulted in lower levels of reproductive dormancy24. Moreover, we do not anticipate further reduction of ovary size if we knock down Pdfr in winter-like conditions when ovaries are already undeveloped with no eggs. Taken together, these data suggest that PDF modulates ovary size through its action in eya expressing cells and PDF and EYA function antagonistically.
EYA protein but not mRNA is modulated by PDF
We next sought to determine if PDF could act upstream of EYA. First, we quantified eya mRNA in heads of Pdf01 and genomic rescue flies entrained in 12:12 LD cycles at 25°C. We observed no differences in eya abundance at all time points over the 24 hour cycle (F(1, 56) = 0.9281, p = 0.3395; Figure 6A). Nonetheless, statistical analysis of rhythmicity revealed that eya exhibits rhythmic expression in the genomic rescue flies (RAIN p = 0.0078), while it is not rhythmic in Pdf01 mutants (RAIN p = 0.833). Then, to determine if EYA protein level is different between the two genotypes, we assayed EYA abundance in protein extracts of heads from Pdf01 mutants or genomic rescue flies. Remarkably, we observed a dramatic increase in the overall amount of EYA protein in Pdf01 mutants as compared to the genomic rescue throughout the 24 hour cycle (F(1, 55) = 50.01, p < 0.0001; Figure 6B–C). Similar to our observation with eya mRNA, EYA protein showed daily rhythmic expression only in the genomic rescue (RAIN p = 0.0398), no robust daily rhythmicity was observed in Pdf01 mutants (RAIN p = 0.667).
Figure 6. PDF modulates EYA protein but not eya mRNA expression.
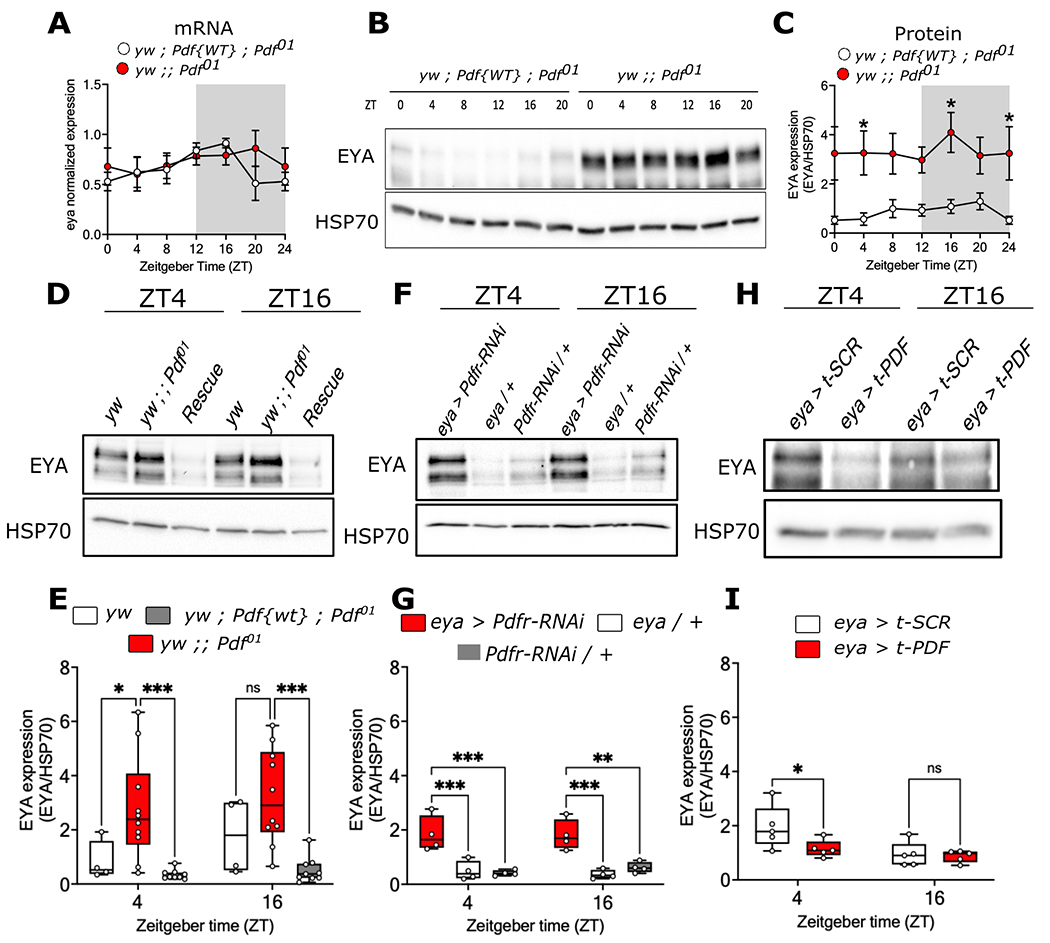
(A) eya mRNA daily oscillations in Pdf-null mutants (yw ; ; Pdf01 flies, red dots, RAIN p = 0.833) and in flies expressing a Pdf genomic rescue (yw ; Pdf{WT} ; Pdf01, white dots, RAIN p = 0.0078) in 12:12 LD cycles at 25°C. (B) Representative western blot and (C) quantification of EYA levels in whole-head lysates from Pdf null-mutant (red dots, RAIN p = 0.677) and in the genomic rescue (white dots, RAIN p = 0.039) at ZT0, 4, 8, 12, 16 and 20 in 12:12 LD 25°C cycles. (D) Representative western blot and (E) quantification of EYA levels from whole head lysates of yw (genetic control, white boxes), Pdf-null mutants (red boxes) and the genomic rescue (grey boxes) at ZT4 and ZT16 in 12:12 LD 25°C condition. (F) Representative western blot and (G) quantification of EYA levels in head lysates of flies where the PDF receptor was knocked down in eya+ cells (eya > Pdfr-RNAi, red boxes) and controls (eya / + and Pdfr-RNAi / +, white and grey boxes, respectively) at ZT4 and ZT16. (H) Representative western blot and (I) quantification of EYA levels in flies expressing a membrane-tethered version of PDF in eya+ cells (eya > t-PDF, red boxes) compared to a scrambled peptide as control (eya > t-SCR, white boxes) at ZT4 and ZT16. Data were analyzed using a two-way ANOVA followed by Tukey’s post hoc test in B and C, and šídák’s multiple comparison test in E, G and I. HSP70 was used for normalization in all cases. n > 3 biological replicates, each consisting of around 50 flies for each genotype.
To confirm these findings, we repeated the experiment using a control fly line in the same genetic background (yw) as the Pdf01 mutant, given that the genomic rescue might not fully recapitulate these control flies. An increase in EYA levels was still observed at ZT4 in Pdf01 mutants compared to control yw flies or the genomic rescue (F(2, 40) = 17.92, p < 0.0001; Figure 6D–E). To test if the effect of Pdf01 mutation in EYA protein levels is mediated by function of PDF signaling in eya+ cells, we knocked down Pdfr using the eya-Gal4 driver. Similarly to the Pdf01 mutant, an increase in the levels of EYA was observed in these flies (F(2, 18) = 30.64, p < 0.0001; Figure 6F–G). Conversely, expressing t-PDF to activate PDF signaling in the same eya+ cells triggered the opposite effect; flies expressing t-PDF showed lower EYA levels than flies expressing a scrambled (SCR) peptide (F(1, 16) = 8.268, p = 0.0110; Figure 6H–I). These results suggest that PDF signaling can modulate EYA protein levels.
PKA phosphorylates EYA to modulate its stability
Because PDF negatively affected EYA protein level with no changes in overall eya mRNA (Figure 6), we hypothesized that PDF modulates EYA stability through post-translational modification. PDFR activation leads to increased cAMP42,63 and subsequent activation of Protein kinase A (PKA) (Figure 7A). Interestingly, we found using pkaPS64, a prediction tool to find PKA target sites in a given protein, that EYA contains seven potential PKA phosphorylation sites: three of them in the C-terminal, one between the second Proline-Serine-Threonine (PST) rich domain and the EYA domain (ED1), and two within the ED1 domain. To test if PKA could phosphorylate EYA, we expressed EYA alone in Drosophila S2 cells or together with PKA-C1, the enzymatically active subunit of this kinase65. We observed slower migrating EYA isoforms when EYA and PKA-C1 are coexpressed (Figure 7B). When treated with lambda phosphatase (λPP), we observed a clear mobility shift to faster migrating isoforms indicating that EYA is indeed phosphorylated by PKA-C1 in S2 cells. This data combined with the observation of EYA accumulation upon knockdown of Pdfr (Figure 6) suggest that PKA potentially phosphorylates EYA to modulate its stability. To test this, we conducted a cycloheximide (CHX) chase experiment in S2 cells transfected with EYA alone or cotransfected with EYA and PKA-C1. CHX was added to S2 cells to inhibit translation of new proteins, thus enabling the measurement of EYA stability in the presence or absence of PKA-C1. Samples were collected at 0, 3, 6 and 9 hours after CHX application (Figure 7C). PKA expression increased EYA degradation rate, evidenced by a significant decrease in EYA levels after 3 hours of CHX addition as compared to the condition without PKA (F(1, 24) = 7.689, p = 0.0106; Figure 7C–D). Moreover, a mobility shift to higher molecular weight isoforms in EYA is clearly observed at 3, 6 and 9 hours post CHX in the presence of PKA-C1 coexpression (Figure 7C). Overall, our results suggest that PDF promotes PKA-dependent EYA phosphorylation and degradation.
Figure 7. PKA phosphorylates EYA and mediates its degradation.
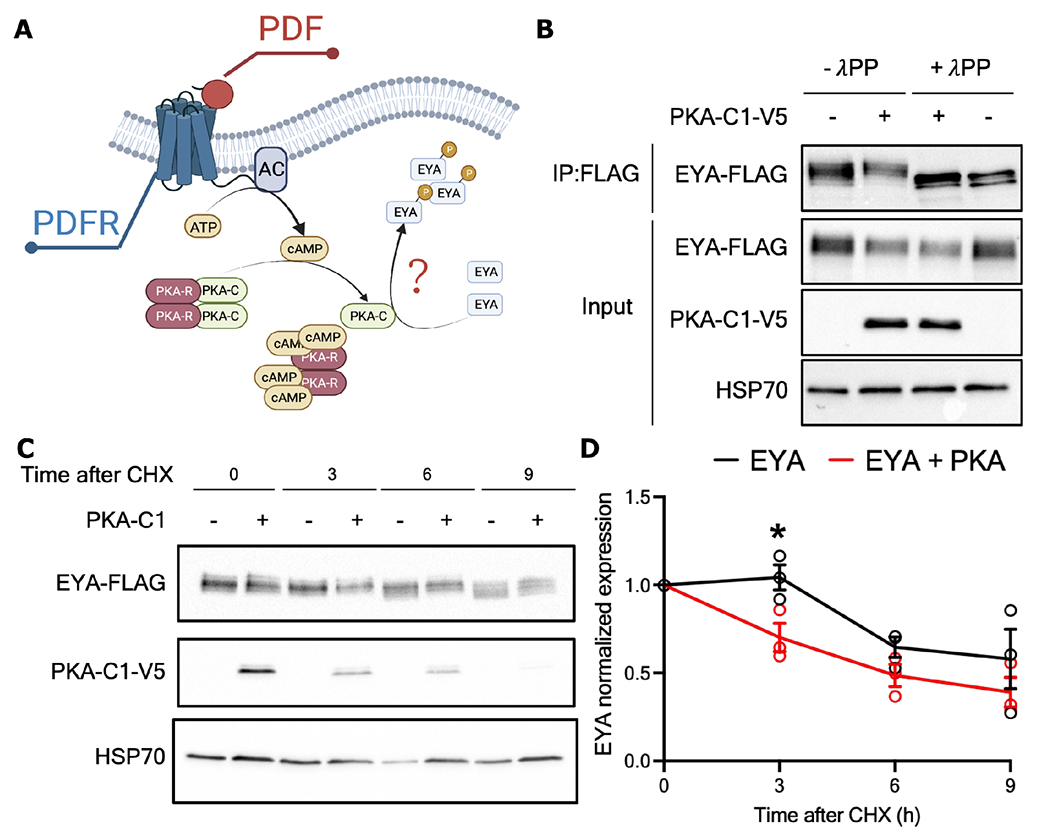
(A) PDF receptor triggers the activation of adenylate cyclase (AC) which produces cAMP. The binding of cAMP to the regulatory subunits of the protein kinase A (PKA-R) releases the catalytic subunits of the kinase (PKA-C) permitting their action. EYA has potential sites for PKA phosphorylation. (B) Drosophila S2 cells co-expressing EYA-FLAG and the active C1 domain of PKA tagged with V5 (PKA-C1-V5 + condition) or EYA alone. Samples were then subjected to phosphatase treatmet (+λPP) and compared to samples that were mock treated (−λPP). Immunoprecipitated EYA was detected using α-FLAG in the input. α-V5 and α-HSP70 were used to detect PKA-C1 and HSP70 (n>3, representative image is shown). (C) Representative western blot of a CHX chase assay in S2 cells co-expressing EYA-FLAG alone or with PKA-C1-V5. (D) Quantification showing the rates of EYA degradation in absence (black line) or presence (red line) of PKA-C1. Data analyzed with two-way ANOVA with Holm-šídák’s multiple comparison test. Each dot represents an independent experiment (n = 3, * p<0.05).
DISCUSSION
The mechanisms by which organisms sense photoperiod and temperature to adapt to seasons have been studied for many years, but the core molecular components involved in this process are still unclear. In this study, we showed that the circadian neuropeptide PDF can respond to environmental cues to modulate seasonal phenotypes via EYA-dependent and independent mechanisms. Our findings suggest that PDF level is responsive to both photoperiodic and temperature changes. We proposed that under summer days, high PDF levels promote EYA degradation via PKA-dependent phosphorylation. In contrast, under winter days, a reduction in PDF drives accumulation of EYA to induce EYA-dependent changes in seasonal phenotypes, such as reproductive dormancy and changes in overall activity levels.
While other studies have explored the differences in PDF levels with environmental conditions, the results have been ambiguous66,67. Here we showed that during short and cold days, PDF levels are greatly reduced compared to long and warm days, effect driven by a reduction in Pdf mRNA levels (Figure 1–2). This is consistent with a previous observation from our lab showing Pdf as one of the transcripts that is downregulated in head of Drosophila suzukii winter morphs68. Overall changes in PDF can be largely explained by temperature, however, it is not clear how temperature changes results in changes in Pdf levels. Several factors have been found to modulate Pdf/PDF expression. For instance, neuronal activity and expression of CLK repress Pdf transcription, possibly through Hormone Receptor 38 (HR38) and stripe (SR)39, a nuclear hormone receptor and a transcription factor, respectively. Pdf levels have also been shown to increase in response to high-intensity light through HR38 action54. In contrast, expression of VRILLE, a repressor in a secondary TTFL, promotes Pdf/PDF mRNA and protein accumulation in the sLNvs through a post-transcriptional mechanism40. Hence, it is possible that changes in core clock components are involved in the reduction in Pdf/PDF levels with cold temperatures. Interestingly, some core clock components are sensitive to temperature69–72. Alternative splicing of per and tim occurs at low temperature69–73. At 18°C, there is an increase in splicing of an intron located in the 3’ end of per, which leads to a phase advance in per levels, which ultimately drives a phase advance in the evening peak of locomotor activtiy70. We showed that at 10°C, control flies also display an advanced evening peak. This pattern resembled that of Pdf01 flies at 25°C, which show a similar evening peak advance (Figure 3). Also, Pdf01 evening peak at 12:12 LD at 25°C falls close to the lights off transition similar to what it would be on a short day as shown here and in previous studies57. Thus, it is possible that these changes rely on a per/Pdf dependent pathway. At similar temperatures, an intron retention event in tim generates a shorter version of tim named short and cold (tim-sc)72,73. Interestingly, overexpression of tim-sc using a tim-Gal4 driver generates akin but less pronounced changes in evening anticipation with flies initiating evening anticipation at around ZT7, similar to the time it starts in Pdf01 at 25°C or genomic rescue flies at 10°C72. We previously showed that TIM-SC protein is indeed expressed at 10°C and experiments in S2 cells showed that TIM-SC interacts with EYA to modulate its stability24. However, it is not known if this isoform has any impact on the function of the molecular clock, ultimately affecting PDF levels. Martin Anduaga et al.72 showed that TIM-SC has reduced binding to PER compared to TIM-L, the canonical TIM isoform expressed at 25°C. Thus, the presence of TIM-SC during cold temperatures could potentially reduce PER stability or its translocation to the nucleus, causing CLK-CYC to promote the Pdf reduction through HR38 and SR.
Considering our hypothesis that PDF signaling conveys photoperiodic and temperature cues to regulate seasonality via EYA and the previously identified role of EYA in winter physiology17,24,33, we expected that knocking down Pdfr or activating PDF signaling by overexpressing t-PDF in eya+ cells would alter seasonal adaptations. These manipulations rendered different locomotor and reproductive outputs. While we were unable to reproduce the phase advance in the evening peak observed in Pdf01 flies, we observed that knocking down Pdfr decreased overall activity akin to a winter-like quiescent state while expressing t-PDF in the same cells rendered the opposite effect (Figure 4). This suggest that PDF modulates evening phase and overall locomotion by EYA-independent and -dependent pathways, respectively. Also in accordance with our previous results, knocking down eya in the IPCs results in bigger ovaries even in long and warm days24. Notably, this is opposite to the phenotype observed when knocking down Pdfr, suggesting that PDF signaling may be antagonizing EYA function. This could be possible as the IPCs receive direct inputs from the s-LNvs to modulate seasonal reproduction. Activating PDF cells increases egg laying at 25°C74, and the same manipulation at 12°C inhibits reproductive dormancy52. In contrast, silencing the same neurons promotes this winter adaptation, likely through a direct effect of PDF on the IPCs. This is because Pdfr in the IPCs is sufficient to rescue dormancy levels in a Pdfr mutant52. Taken together, these data suggest that PDF functions in the IPCs to promote summer physiology, likely in an EYA-dependent manner.
We showed that cold temperature and short days trigger a reduction in Pdf/PDF mRNA and protein (Figure 1–2). Interestingly, this response is opposite of what we previously observed for EYA24, whose expression increase in winter, and the opposite role of PDF and EYA in seasonal adaptations. Thus, we reasoned that PDF may be upstream and convey environmental signals to modulate EYA. Consistent with this, EYA levels were observed to be increased in Pdf01 mutants, also suggesting a role of PDF in inhibiting EYA expression (Figure 6). Additionally, expression of t-PDF in eya+ cells was sufficient to decrease EYA levels, while an increase in EYA was observed when Pdfr was knocked down. This strongly suggests an inhibiting role of PDF on EYA. We proposed that the action of PDF signaling cascade would be involved in this. Consistent with this notion, we showed in S2 cells that the PKA-C1, the catalytic subunit of PKA and downstream target of PDF signaling, can modulate EYA levels by phosphorylation-dependent degradation (Figure 7). PKA-C1 is also constitutively expressed in the IPCs, thus a similar process is likely happening in vivo75. Nonetheless, although PDFR signals through a cAMP/PKA42,52, it is not the only receptor that uses this pathway to regulate seasonal biology. Indeed, dopamine and serotonin signaling in the IPCs, which also leverage cAMP/PKA, are required to promote dormancy76. Interestingly, this is opposite to what is observed for PDF signaling. Future studies to tease apart the contribution of PKA in the IPCs to regulate seasonal biology via EYA will require the identification and characterization of PKA-dependent EYA phosphorylation sites.
We previously showed that the interaction of TIM with EYA may be important for EYA stabilization. Interestingly, TIM is not expressed in the IPCs and EYA is not expressed in PDF-producing LNvs based on available data. These proteins seem to colocalize mainly in the optic lobes, potentially relaying seasonal cues to downstream neurons24. We therefore speculate TIM-EYA interaction in visual cells might be important to modulate the events in the IPCs; EYA-TIM interaction in the optic lobe could modulate the sLNvs and PDF levels downstream, consequently impacting EYA abundance and function in the IPCs to regulate seasonal physiology. Future work will be necessary to untangle the relationship between TIM and EYA in the context of the neuronal circuit.
Finally, a similar mechanism could be driving seasonal adaptations in other animals. Several studies have suggested a functional homology between the vasoactive intestinal peptide (VIP) and PDF in circadian rhythms and seasonal adaptations77,78. VIP is expressed by neurons located in the suprachiasmatic nucleus (SCN), the main hub for circadian timekeeping in the brain79. This peptide is important for circadian synchronization of the SCN individual clocks and activates the VPAC2 receptor, a class B G-protein coupled receptor that increases cAMP/PKA upon activation, similar to PDFR in Drosophila77. VIP is also indispensable for seasonal adaptations, as VIP knockout mice do not display behavioral or physiological adaptations while entrain to different photoperiods78. VIP is also expressed in the hypothalamus in birds and its levels are coupled to photoperiod and hormonal adaptations80,81. It would be interesting to explore possible modulations of EYA by VIP in the pars tuberalis and its effect on seasonal adaptations.
In summary, our study proposed a mechanism by which the circadian clock conveys photoperiod and temperature to clock outputs, to modulate seasonal adaptations. While we attribute the connection between the clock and the outputs to a role of a circadian neuropeptide, the molecular mechanism driving the modulation of the neuropeptide in response to environmental cues are still an open question. Furthermore, our study provides additional support for a role of EYA in seasonal adaptations and uncovers the importance of EYA post-translational modifications in the regulation of the seasonal biology.
STAR METHODS
Resource availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact Joanna C. Chiu (jcchiu@ucdavis.edu).
Materials Availability
All reagents used in this study are available from the lead contact without restriction.
Data and Code Availability
Raw data for all datasets used in this study, including microscopy, western blots, and locomotor activity data are available upon request to the lead contact. This paper also analyzes existing, publicly available data from single-cell RNA-sequencing58. These data are available at https://flycellatlas.org/scope.
Single-cell RNA-sequencing data was analyzed using SCOPE, available at https://scope.aertslab.org/#/FlyCellAtlas/*/welcome. ImageJ macros used to analyze microscopy data and R code used to analyze Drosophila locomotor activity data will be shared upon request to the lead contact.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental Model and Subject Details
Flies were reared on standard corn-yeast diet and kept at 25°C on a 12-hour light (L): 12-hour dark (D) cycle (12:12 LD). Fly stock were either obtained from Bloomington Drosophila Stock Center (BDSC), Vienna Drosophila Research Center (VDRC) or gifts from other labs described in the Key Resources Table.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-PDF (clone C7-C) | Developmental Studies Hybridoma Bank | RRID: AB_760350 and AB_2315084 |
| Mouse monoclonal anti-EYA (clone 10H6) | Developmental Studies Hybridoma Bank | RRID: AB_528232 |
| Mouse monoclonal anti-BRP (nc82) | Developmental Studies Hybridoma Bank | RRID: AB_2314866 |
| Chicken Polyclonal anti-GFP | Invitrogen | Cat#A10262; RRID: AB_2534023 |
| Sheep anti-mouse IgG-HRP | GE Healthcare | Cat#NA931; RRID: AB_772210 |
| Mouse monoclonal anti-V5 | Thermo Fisher Scientific | Car#R960-25; RRID: AB_2556564 |
| Mouse monoclonal anti-FLAG M2 | Sigma-Aldrich | Cat# F3165; RRID: AB_259529 |
| Goat Anti-mouse IgG Alexa647 | Cell Signaling | Cat#4410; RRID: AB_1904023 |
| Goat Anti-chicken IgG Alexa488 | Thermo Fisher Scientific | Cat#A-11039; RRID: AB_2534096 |
| Bacterial and virus strains | ||
| Biological samples | ||
| Chemicals, peptides, and recombinant proteins | ||
| Schneider’s Drosophila Medium | Thermo Fisher Scientific | Cat#21720-024 |
| Fetal Bovine Serum | VWR | Cat# 97068-085 |
| Penicillin/streptomycin | Thermo Fisher Scientific | Cat#15-140-148 |
| Trypsin/EDTA | Thermo Fisher Scientific | Cat#25300-062 |
| TRI regent | Sigma-Aldrich | Cat#T9424 |
| Aprotinin | Sigma-Aldrich | Cat#A1153 |
| Leupeptin | Sigma-Aldrich | Cat#L2884 |
| Pepstatin A | Sigma-Aldrich | Cat#P5318 |
| SIGMAFAST™ Protease Inhibitor Cocktail, EDTA-FREE | Sigma-Aldrich | Cat#S8830 |
| ANTI-FLAG M2 Affinity Gel | Sigma-Aldrich | Cat#A2220 |
| PhosSTOP | Sigma-Aldrich | Cat#4906845001 |
| Normal Goat Serum | Jackson Immunoresearch | Cat#005-000-121 |
| λ-Phosphatase | NEB | Cat#P0753S |
| SsoAdvanced SYBR green supermix | Bio-Rad | Cat#1725270 |
| PBS 10X Rnase free | Invitrogen | Cat#AM9624 |
| Nuclease-free water | Invitrogen | Cat#AM9932 |
| Acetic Acid | Fisher | Cat#A38S-500 |
| Sodium borohydride (99%) | Fisher | Cat#AC448481000 |
| SSC (20X) | Fisher | Cat#AM9763 |
| Hi-Di Formamide | Fisher | Cat#4311320 |
| Triton X-100 nuclease-free | Fisher | Cat#AC327371000 |
| Formamide (deionized) | Invitrogen | Cat#AM9342 |
| Denhard’s solution | Fisher | Cat#AAJ63135AD |
| tRNA from baker’s yeast | Roche | Cat#10109495001 |
| SDS 10% | Corning | Cat# 46-040-CI |
| UltraPure™ Salmon Sperm DNA Solution | Fisher | Cat# 15632011 |
| 4% PFA | EMS | Cat#157-4 |
| Rnase AWAY | Thermo Fisher Scientific | Cat#7002PK |
| Ethanol (200 Proof) | Fisher | Cat#BP2818500 |
| Critical commercial assays | ||
| Superscript IV | Thermo Fisher Scientific | Cat#18091050 |
| Effectene Transfection Reagents | QIAGEN | Cat#301425 |
| Pierce Coomassie Plus Assay Reagents | Thermo Fisher Scientific | Cat#1856210 |
| Deposited data | ||
| Experimental models: Cell lines | ||
| D. melanogaster: Cell line S2: S2-DRSC | Thermo Fisher Scientific | Cat#R69007 |
| Experimental models: Fly lines | ||
| D. melanogaster: w; Pdf-Gal4 | Park et al. 53 | N/A |
| D. melanogaster: w; UAS-mCD8::GFP | Bloomington Drosophila Stock Center | #32184 |
| D. melanogaster: yw; ; Pdf01 | Bloomington Drosophila Stock Center | #26654 |
| D. melanogaster: yw; Pdf-RescueB ; Pdf01 | Renn et al. 46 | N/A |
| D. melanogaster: yw | Bloomington Drosophila Stock Center | #1495 |
| D. melanogaster: w; ; eya-Gal4 | Bloomington Drosophila Stock Center | #49292 |
| D. melanogaster: w; UAS-Pdfr-RNAi | Bloomington Drosophila Stock Center | #38347 |
| D. melanogaster: w; UAS-t-PDF | Bloomington Drosophila Stock Center | #81111 |
| D. melanogaster: w; UAS-t-SCR | Bloomington Drosophila Stock Center | #81113 |
| D. melanogaster: w; UAS-eya-RNAi | Vienna Drosophila Resource Center | #108071 |
| D. melanogaster: w; per-AID-GFP | Chen et al.61 | #108071 |
| D. melanogaster: w; UAS-eya-RNAi | Vienna Drosophila Resource Center | #108071 |
| D. melanogaster: Pdfr-Gal4 , w | Bloomington Drosophila Stock Center | #33070 |
| D. melanogaster: w ; dilp2-Gal4 / Cyo | Bloomington Drosophila Stock Center | #37516 |
| Oligonucleotides | ||
| Primers for eya, see Methods details | This paper | N/A |
| Primers for cbp20, see Methods details | This paper | N/A |
| Library for Pdf detection, see Table S1 | Long et al.82 | N/A |
| Recombinant DNA | ||
| Plasmid: pMT-V5 | Thermo Fisher | Cat#V412020 |
| Plasmid: pMT-PKA-C1-V5 | This Paper | N/A |
| Plasmid: pAc-eya-3XFLAG-6XHis | Abrieux et al.24 | N/A |
| Software and algorithms | ||
| Fiji ImageJ (used for analysis of immunofluorescence microscopy images) | NIH Image | https://fiji.sc |
| GraphPad Prism 8 for Mac OS X | GraphPad Software | https://graphpad.com |
| Excel | Microsoft | Version 16.67 |
| R | R Core Team | Version 4.0.3 |
| RStudio | PBC | Version 2022.07.2 |
| CircaCompare | Parsons et al.55 | Release 0.1.1 |
| RAIN | Thaben & Westermark56 | Release 3.16 |
| Other | ||
Methods details
Drosophila whole-brain immunofluorescence
Newly emerged flies were collected and placed under 12:12 LD cycles at 25°C for 3 days before switching them to long photoperiod (16:8 LD at 25°C), short photoperiod (8:16 LD at 25°C), low temperature (12:12 LD at 10°C), simulated winter (8:16 LD at 10°C), simulated summer (16:8 LD at 25°C) or kept in equinox conditions (12:12 LD at 25°C) for 7 days. Flies were collected at ZT0, 4, 8, 12, 16 and 20 hours where ZT0 represents lights on time (Zeitgeber Time; ZT), fixed for 40 minutes in 4% PFA at room temperature and dissected immediately. Brains were re-fixed in 4% PFA for 20 minutes, washed 3 times in 0.2% PBST, 15 minutes per wash, and then incubated in blocking solution (5% NGS, 0.05% Sodium Azide in 0.2% PBST) for 30 minutes. All this process was conducted at room temperature. After that, brains were incubated with primary antibody for 2 days at 4°C. Brains were then washed 3 times in 0.2% PBST, 15 minutes per wash, and then incubated at room temperature with a secondary antibody tagged with a particular fluorophore. Finally, brains were mounted using ProLong Diamond Antifade Mountant (Life Technologies, Carlsbad, CA) and imaged using a Leica SP5 confocal microscope equipped with excitation diodes at 638 nm and OPSL exciting at 488. For the experiments determining PDF levels in the s-LNvs dorsal terminals, images were taken every 1 μm using a 40x oil objective and digital zoom. For images of the whole brain, a 20x objective was used instead. All images were analyzed using Fiji and PDF levels were assessed as the intensity in the s-LNVs dorsal terminal normalized by the background signal in the same dorsal region. This process was automatized in Fiji.
The dilution of the primary antibodies is as follow: α-PDF (1:200), α-EYA (1:200), α-brp (nc82; 1:50), and α-GFP (1:1,000). The dilution of the secondary antibodies is as follow: Alexa647 α-mouse IgG (1:1,000) and Alexa488 α-chicken IgG (1:1,000).
Fluorescence in situ hybridization (FISH)
FISH against Pdf was carried out as described in Long et al.82, with minor modifications. Flies were fixed in 4% PFA for 40 minutes and brains were dissected in cold 0.5% PBST (1X PBS, 0.5% Triton X-100) solution. Dissected brains were re-fixed in 2% PFA for 55 minutes, washed with 0.5% PBST and then dehydrated for storage with EtOH series at 30%, 50%, 70%, 100%, 100%, 100% for 10 min each. Next day, the tissue was rehydrated with inverse EtOH series 70%, 50%, 30% for 10 min each. Tissue was then washed with ice cold 1X PBS and re-fixed in 2% PFA for 55 min. After that, brains were permeated with 5% acetic acid for 5 min on ice and then washed with 0.5% PBST three times. Autofluorescence quenching was achieved by incubating the brains with 1% sodium borohydride in 3 consecutive washes for 10 min each. The tissue was primed with pre-hybridization solution (15% formamide, 2X SCC, and 0.1% Triton X-100) at 50°C for 2 hours and then hybridization was carried out in hybridization solution (10% Formamide, 2X SCC, 5X Denhard’s solution, 1 mg/mL Yeast tRNA, 100 μg/mL Salmon sperm DNA, 0.1% SDS) for 10 hours at 50°C followed by 10 hours at 37°C. Probes were tagged with CAL Fluor Red 635, purchased from Biosearch Technologies (Stellaris RNA FISH probes; Petaluma, CA) (Table S1). Tissue was washed with pre-hybridization solution 3 times, 10 min each, then with 30% formamide solution supplemented with 2X SCC and 0.06% Triton X-100 twice for 30 min each. After that, tissue was washed with 2X SCC solution with 0.06% Triton three times, 10 min each, followed by a 10 min wash with 1X PBS. Finally, brain was fixed with 2% PFA for 55 minutes, washed with 0.5% PBST and then mounted ProLong Diamond Antifade Mountant (Life Technologies).
Drosophila Activity Monitoring (DAM) system
Locomotor activity was measured using the DAM system (TriKinetics, MA)83. Flies were anesthetized under CO2 and loaded in individual glass tubes containing fly food (5% sucrose and 2% Bacto agar in distilled water). Tubes were loaded in monitors and placed inside the incubator at 25°C under 12:12 LD cycle. After 2 days of acclimation, locomotor activity was recorded for 5 days in LD conditions and released into constant darkness (DD) for 5 days. Only flies that survived until the last day of the experiments were considered for analysis. Average locomotor activity profiles were generated for LD and DD for the 5 days in both conditions and the total locomotor activity was quantified as the sum of all the locomotor events in that given period as beam breaks per 30-min bins. The evening anticipation was calculated as the difference between the normalized averaged activity in ZT9–11.5 and ZT5–7.5 to understand the strength/amplitude of the peak.
For clustering analysis, we transformed the raw Drosophila activity monitoring data into a 24-hour activity matrix that describes the normalized activity profile of each fly as follows. The average beam breaks (or activity counts) per 30 mins from ZT0 to ZT24 was obtained across 5 days to compute an average activity profile per fly composed of 48 values (1 value per 30-min bin). Each of those 48 values was then divided by the value of the bin with the highest activity count such that the bin with the highest value equals to 1. We chose to perform this normalization step to focus on the 24h activity profile without taking into account the differences in absolute activity level. These 48 values are now used as a descriptor for the flies’ activity profiles and analyzed using t-distributed Stochastic Neighbor Embedding (t-SNE). t-SNE was calculated using the “Rtsne” package84 on R, using perplexity 30, max iterations 1000 and 50 initial dimensions. Each dot represents the first two dimensions of the 48 values now reduced for each fly. t-SNE was used to mathematically transform high-dimensional data (48 values per fly) and project it into low-dimensional space85. The two dimensions on the t-SNE plot do not represent specific parameters. They hold no meaning on their own, unlike axes of conventional 2D graphs. The information gathered by this analysis is qualitative and is used to extrapolate the similarity of daily fly activity rhythms as visual clustering in a 2D space, i.e. data points that are closer to each other represent flies with more similar daily activity patterns.
Hierarchical clustering also used this 48 starting values per fly and the analysis was conducted using the “stat” package on R86. First, a distance matrix was created using “dist” function, using the “maximum” method. Then, clustering was conducted using “hclust” function using “ward.D” method. Finally, a dendrogram was generated using “as.dendrogram” function and plotted using the “plot” function from the “base” package.
Ovary size quantification
Virgin females were placed in simulated summer conditions (25°C, 16:8 LD) for 28 days and then dissected in 0.2% PBST (0.2% Triton X-100 in 1X PBS)24. Ovaries were imaged using a EVOS microscope (Life Technologies, CA) and ovary size was determined by measuring the surface area (mm2) using Fiji software.
Total RNA extraction and quantitative RT-PCR
Flies were collected at the indicated timepoints on dry ice and then heads were collected and stored at −80°C until sample processing. Around 20-50 μL of heads per condition were processed using TRI reagent (Sigma-Aldrich, St. Louis, MO), following manufacturer’s instructions. Total RNA was quantified and 1 μg was used as starting material for reverse transcription using the Superscript IV cDNA synthesis kit (Life Technologies) following manufacturer’s instruction. Quantitative PCR was performed using SsoAdvanced Universal SYBR green super mix (Bio Rad, Hercules, CA), with the following the program: initial 95°C for 30 seconds, then 40 cycles of 95°C for 5 seconds, followed by an annealing/extension phase at 60°C for 30 seconds. Primers to detect eya were 5’-GAGGCCTGGCTACAGATACG-3’ and 5’-AGTTGCGTGGAGGTTACCAG-3’, while cbp20 (5’-GTCTGATTCGTGTGGACTGG-3’ and 5’-CAACAGTTTGCCATAACCCC-3’) was used for normalization. Data were analyzed using the ΔΔCt method.
Protein extraction from heads and western blotting
Protein extraction from fly heads was conducted as previously described24,87–89. Flies were collected at the indicated timepoints and flash frozen using dry ice. Heads were then collected and ground on protein extraction buffer (0.1% Glycerol, 20mM Hepes pH7.5, 50mM KCl, 2mM EDTA, 1% Triton X-100, 0.40% NP 40, 1mM DTT, 10μg/mL Aprotinin, 5μg/mL Leupeptin, 1μ/mL Pepstatin A and 0.5mM PMSF). Protein samples were kept at −80°C in SDS loading buffer until electrophoresis in 8% SDS-PAGE gels. Transfer was conducted in a Trans-Blot semi-dry transfer system (Bio Rad) for 45 minutes to nitrocellulose membranes (0.45μm, Bio Rad). 5% blocking reagent (Bio Rad) in 0.05% TBST (0.05% Tween20 in 1XTBS) was used to block for 1 hour before antibody incubation overnight at 4°C. Anti-mouse IgG linked to HRP (Cytiva Life Sciences, Malborough, MA) was used as a secondary antibody at a dilution of 1:1,000 incubated for 1 hour. Membranes were incubated for 5 min with Clarity reagent (Bio Rad) and imaged on ChemiDoc MP Imaging system. Image analysis was performed using Image J. The dilution of the primary antibodies is as follow: μ-EYA (1:1,000), μ-HSP70 (1:7,000), μ-V5 (1:5,000), and μ-FLAG (1:7,000).
Drosophila S2 cell culture and transfection
Drosophila S2 cells were maintained at 22°C in Schneider’s Drosophila medium (Life Technologies) supplemented with 10% Fetal Bovine Serum (VWR, Radnor, PA). For both cycloheximide (CHX) chase and phosphatase experiments, on day one, 3 × 106 cells were transiently transfected with 0.8 μg pAc-eya-3XFLAG-6XHis and 0.2 μg pMT-PKA-C1-V5 or an empty pMT-V5 plasmid. PKA-C1 expression was induced 24 hours after transfection by addition of 500 μM CuSO4.
λ Phosphatase experiments
For phosphatase treatment, cells were treated with cycloheximide (CHX) for 3 hours to a final concentration of 10 μg/ml. Cells were harvested and proteins were extracted using EB2 (20 mM HEPES pH 7.5, 100 mM KCl, 5% glycerol, 5 mM EDTA, 1 mM DTT, 0.1% Triton X-100, 10 μg/ml Aprotinin, 5 μg/ml Leupeptin, 1 μg/ml Pepstatin A, 0.5 mM PMSF, 25 mM NaF)88 supplemented with 1X PhosSTOP (Roche, Indianapolis, IN).
Protein quantification was carried out using Coomassie reagent and the same amount of protein was aliquoted for PKA+ and PKA- conditions. α-FLAG immunoprecipitation was performed using anti-FLAG M2 agarose (Sigma) following protocol described in Lam et al.88. Beads were incubated with the protein lystes in and end-to-end rotator for 4 hours at 4°C. Immunocomplexes were then treated with Lambda phosphatase λ-PP or mock treated (New England Biolabs, Ipswich, MA) for 30 min at 30°C in a water bath, flicking the tube every 10 min. Finally, the beads were precipitated, and the proteins were obtained by adding 2x SDS sample buffer and then resolved in 8% SDS-PAGE.
Cycloheximide experiments
For CHX chase assay to measure protein degradation, CHX was added to the transfected cells, again at 24 hours after kinase induction to a final concentration of 10 μg/ml. S2 cells were harvested immediately, or at 3, 6, 9 hours after addition of CHX. Upon cell harvest, proteins were extracted using EB2 and resolved in 8% SDS-PAGE.
Quantification and statistical analysis
Data are represented as mean ± standard error of the mean (SEM) in XY graphs. For all other representations, whiskers and boxes were used, and the result of each biological replicate is represented in the graphs as a single dot. Circadian analysis was conducted using the R package CircaCompare55 or RAIN56 as indicated in each figure. All datasets were assessed for normality using a Shapiro-Wilk’s normality test to choose the appropriate parametric or non-parametric test. Tests used for each dataset are indicated in figure legends. Statistical differences reported throughout this work are ns p > 0.05, * p < 0.05, ** p < 0.01, *** p < 0.001 and **** p < 0.0001. The number of biological replicates is indicated in each figure or figure legend.
Supplementary Material
Highlights.
PDF level is sensitive to temperature and photoperiod.
PDF modulates seasonal biology through EYA-dependent and independent mechanisms.
EYA protein level is regulated by PDF signaling pathway.
EYA is phosphorylated by Protein Kinase A to promote its degradation.
ACKNOWLEDGEMENTS
We would like to thank all the members of the Chiu lab and Hamada lab for providing valuable comments in the development of the project. We also want to thank Dr. Yong Zhang for the PER-AID-GFP fly line and Dr. Pamela Ronald for access to the confocal microscope (Leica SP8 from NIH grant GM122968). S.H. is a Latin American Fellow in the Biomedical Sciences supported by the Pew Charitable Trusts. Research in the lab of JCC is supported by NIH R01 DK124068. Graphical abstract was generated on Biorender (license to UC Davis Chiu lab).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
The authors declare no competing interests.
INCLUSION AND DIVERSITY
We support inclusive, diverse, and equitable conduct of research.
REFERENCES
- 1.Stevenson TJ, Prendergast BJ, and Nelson RJ (2017). Mammalian Seasonal Rhythms: Behavior and Neuroendocrine Substrates. In Hormones, Brain and Behavior: Third edition (Academic Press; ), pp. 371–398. 10.1016/B978-0-12-803592-4.00013-4. [DOI] [Google Scholar]
- 2.Nelson R, (1990). Mechanisms Of Seasonal Cycles Of Behavior. Annu. Rev. Psychol 41, 81–108. 10.1146/annurev.psych.41.1.81. [DOI] [PubMed] [Google Scholar]
- 3.Alerstam T, and Bäckman J, (2018). Ecology of animal migration. Curr. Biol 28, R968–R972. 10.1016/J.CUB.2018.04.043. [DOI] [PubMed] [Google Scholar]
- 4.Chowdhury S, Fuller RA, Dingle H, Chapman JW, and Zalucki MP, (2021). Migration in butterflies: a global overview. Biol. Rev 96, 1462–1483. 10.1111/brv.12714. [DOI] [PubMed] [Google Scholar]
- 5.Ray S, Li M, Koch SP, Mueller S, Boehm-Sturm P, Wang H, Brecht M, and Naumann RK, (2020). Seasonal plasticity in the adult somatosensory cortex. Proc. Natl. Acad. Sci. U. S. A 117, 32136–32144. 10.1073/pnas.l922888117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu W, Feke A, Leung CC, Tarté DA, Yuan W, Vanderwall M, Sager G, Wu X, Schear A, Clark DA, et al. (2021). A metabolic daylength measurement system mediates winter photoperiodism in plants. Dev. Cell 56, 2501–2515.e5. 10.1016/j.devcel.2021.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cubas P, (2020). Plant Seasonal Growth: How Perennial Plants Sense That Winter Is Coming. Curr. Biol 30, R21–R23. 10.1016/j.cub.2019.11.044. [DOI] [PubMed] [Google Scholar]
- 8.Nelson RJ, (2004). Seasonal immune function and sickness responses. Trends Immunol. 25, 187–192. 10.1016/j.it.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 9.Webb AJS, Brain SAE, Wood R, Rinaldi S, and Turner MR, (2015). Seasonal variation in Guillain-Barré syndrome: a systematic review, meta-analysis and Oxfordshire cohort study. J. Neurol. Neurosurg. Psychiatry 86,1196–1201. 10.1136/jnnp-2014-309056. [DOI] [PubMed] [Google Scholar]
- 10.Tendler A, Bar A, Mendelsohn-Cohen N, Karin O, Kohanim YK, Maimon L, Milo T, Raz M, Mayo A, Tanay A, et al. (2021). Hormone seasonality in medical records suggests circannual endocrine circuits. Proc. Natl. Acad. Sci. U. S. A 118. 10.1073/pnas.2003926118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun L, Tang J, Liljenbäck H, Honkaniemi A, Virta J, Isojärvi J, Karjalainen T, Kantonen T, Nuutila P, Hietala J, et al. (2021). Seasonal variation in the brain μ-opioid receptor availability. J. Neurosci 41, 1265–1273. 10.1523/JNEUROSCI.2380-20.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hand SC, Denlinger DL, Podrabsky JE, and Roy R, (2016). Mechanisms of animal diapause: Recent developments from nematodes, crustaceans, insects, and fish. Am. J. Physiol. - Regul. Integr. Comp. Physiol 310, R1193–R1211. 10.1152/ajpregu.00250.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hazlerigg DG, and Wagner GC, (2006). Seasonal photoperiodism in vertebrates: from coincidence to amplitude. Trends Endocrinol. Metab 17, 83–91. 10.1016/j.tem.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 14.Anduaga AM, Nagy D, Costa R, and Kyriacou CP (2018). Diapause in Drosophila melanogaster- Photoperiodicity, cold tolerance and metabolites. J. Insect Physiol 105, 46–53. 10.1016/j.jinsphys.2018.01.003. [DOI] [PubMed] [Google Scholar]
- 15.Nakayama T, and Yoshimura T, (2018). Seasonal Rhythms: The Role of Thyrotropin and Thyroid Hormones. Thyroid 28, 4–10. 10.1089/thy.2017.0186. [DOI] [PubMed] [Google Scholar]
- 16.Ikegami K, Refetoff S, Van Cauter E, and Yoshimura T, (2019). Interconnection between circadian clocks and thyroid function. Nat. Rev. Endocrinol 15, 590–600. 10.1038/s41574-019-0237-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakao N, Ono H, Yamamura T, Anraku T, Takagi T, Higashi K, Yasuo S, Katou Y, Kageyama S, Uno Y, et al. (2008). Thyrotrophin in the pars tuberalis triggers photoperiodic response. Nature 452, 317–322. 10.1038/nature06738. [DOI] [PubMed] [Google Scholar]
- 18.Hazlerigg D, Lomet D, Lincoln G, and Dardente H, (2018). Neuroendocrine correlates of the critical day length response in the Soay sheep. J. Neuroendocrinol 30, el2631. 10.1111/jne.12631. [DOI] [PubMed] [Google Scholar]
- 19.Schiesari L, Kyriacou CP, and Costa R, (2011). The hormonal and circadian basis for insect photoperiodic timing. FEBS Lett. 585,1450–1460. 10.1016/j.febslet.2011.02.026. [DOI] [PubMed] [Google Scholar]
- 20.Xu WH, Lu YX, and Denlinger DL, (2012). Cross-talk between the fat body and brain regulates insect developmental arrest. Proc. Natl. Acad. Sci. U. S. A 109, 14687–14692. 10.1073/pnas.1212879109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Denlinger DL, Hahn DA, Merlin C, Holzapfel CM, and Bradshaw WE, (2017). Keeping time without a spine: What can the insect clock teach us about seasonal adaptation? Philos. Trans. R. Soc. B Biol. Sci 372. 10.1098/rstb.2016.0257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kurogi Y, Mizuno Y, Imura E, and Niwa R, (2021). Neuroendocrine Regulation of Reproductive Dormancy in the Fruit Fly Drosophila melanogaster: A Review of Juvenile Hormone-Dependent Regulation. Front. Ecol. Evol 9, 1–16. 10.3389/fevo.2021.715029. [DOI] [Google Scholar]
- 23.Dardente H, Wyse CA, Birnie MJ, Dupré SM, Loudon ASI, Lincoln GA, and Hazlerigg DG, (2010). A Molecular Switch for Photoperiod Responsiveness in Mammals. Curr. Biol 20, 2193–2198. 10.1016/j.cub.2010.10.048. [DOI] [PubMed] [Google Scholar]
- 24.Abrieux A, Xue Y, Cai Y, Lewald KM, Nguyen HN, Zhang Y, and Chiu JC, (2020). EYES ABSENT and TIMELESS integrate photoperiodic and temperature cues to regulate seasonal physiology in Drosophila. Proc. Natl. Acad. Sci 117,15293–15304. 10.1073/pnas.2004262117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bünning E, (1936). Die endogene Tagesrhythmik als Grundlage der photoperiodischen Reaktion. Berichte Dtsch. Bot. Gessellschaft [Google Scholar]
- 26.Bunning E, (1960). Circadian Rhythms and the Time Measurement in Photoperiodism. Cold Spring Harb. Symp. Quant. Biol 25, 249–256. 10.1101/SQB.1960.025.01.026. [DOI] [PubMed] [Google Scholar]
- 27.Patke A, Young MW, and Axelrod S, (2020). Molecular mechanisms and physiological importance of circadian rhythms. Nat. Rev. Mol. Cell Biol 21, 67–84. 10.1038/s41580-019-0179-2. [DOI] [PubMed] [Google Scholar]
- 28.Pittendrigh CS, and Minis DH, (1964). The Entrainment of Circadian Oscillations by Light and Their Role as Photoperiodic Clocks. Am. Nat 98, 261–294. [Google Scholar]
- 29.Ikeno T, Tanaka SI, Numata H, and Goto SG (2010). Photoperiodic diapause under the control of circadian clock genes in an insect. BMC Biol. 8, 116. 10.1186/1741-7007-8-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sandrelli F, Tauber E, Pegoraro M, Mazzotta G, Cisotto P, Landskron J, Stanewsky R, Piccin A, Rosato E, Zordan M, et al. (2007). A Molecular Basis for Natural Selection at the timeless Locus in Drosophila melanogaster. Science 316,1898–1900. 10.1126/science.1138426. [DOI] [PubMed] [Google Scholar]
- 31.Tauber E, Zordan M, Sandrelli F, Pegoraro M, Osterwalder N, Breda C, Daga A, Selmin A, Monger K, Benna C, et al. (2007). Natural Selection Favors a Newly Derived timeless Allele in Drosophila melanogaster. Science 316, 1895–1898. 10.1126/science.1138412. [DOI] [PubMed] [Google Scholar]
- 32.Pegoraro M, Gesto JS, Kyriacou CP, and Tauber E, (2014). Role for Circadian Clock Genes in Seasonal Timing: Testing the Bünning Hypothesis. PLoS Genet. 10. 10.1371/journal.pgen.1004603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wood SH, Hindle MM, Mizoro Y, Cheng Y, Saer BRCC, Miedzinska K, Christian HC, Begley N, McNeilly J, McNeilly AS, et al. (2020). Circadian clock mechanism driving mammalian photoperiodism. Nat. Commun 11, 4291. 10.1038/s41467-020-18061-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barber AF, Erion R, Holmes TC, and Sehgal A (2016). Circadian and feED1ng cues integrate to drive rhythms of physiology in Drosophila insulin-producing cells. Genes Dev. 30, 2596–2606. 10.1101/gad.288258.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cavanaugh DJ, Geratowski JD, Wooltorton JRA, Spaethling JM, Hector CE, Zheng X, Johnson EC, Eberwine JH, and Sehgal A (2014). Identification of a circadian output circuit for rest:activity rhythms in Drosophila. Cell 157, 689–701. 10.1016/j.cell.2014.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barber AF, Fong SY, Kolesnik A, Fetchko M, and Sehgal A, (2021). Drosophila clock cells use multiple mechanisms to transmit time-of-day signals in the brain. Proc. Natl. Acad. Sci. U. S. A 118, 1–8. 10.1073/pnas.2019826118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reinhard N, Bertolini E, Saito A, Sekiguchi M, Yoshii T, Rieger D, and Helfrich-Förster C, (2022). The lateral posterior clock neurons of Drosophila melanogaster express three neuropeptides and have multiple connections within the circadian clock network and beyond. J. Comp. Neurol 530, 1507–1529. 10.1002/cne.25294. [DOI] [PubMed] [Google Scholar]
- 38.Reinhard N, Schubert FK, Bertolini E, Hagedorn N, Manoli G, Sekiguchi M, Yoshii T, Rieger D, and Helfrich-Förster C, (2022). The Neuronal Circuit of the Dorsal Circadian Clock Neurons in Drosophila melanogaster. Front. Physiol 13, 1–29. 10.3389/fphys.2022.886432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mezan S, Feuz JD, Deplancke B, and Kadener S, (2016). PDF Signaling Is an Integral Part of the Drosophila Circadian Molecular Oscillator. Cell Rep. 17, 708–719. 10.1016/j.celrep.2016.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gunawardhana KL, and Hardin PE, (2017). VRILLE Controls PDF Neuropeptide Accumulation and Arborization Rhythms in Small Ventrolateral Neurons to Drive Rhythmic Behavior in Drosophila. Curr. Biol 27, 3442–3453e4. 10.1016/j.cub.2017.10.010. [DOI] [PubMed] [Google Scholar]
- 41.Taghert PH, and Shafer OT, (2006). Mechanisms of clock output in the Drosophila circadian pacemaker system. J. Biol. Rhythms 21, 445–457. 10.1177/0748730406293910. [DOI] [PubMed] [Google Scholar]
- 42.Shafer OT, Kim DJ, Dunbar-Yaffe R, Nikolaev VO, Lohse MJ, and Taghert PH, (2008). Widespread Receptivity to Neuropeptide PDF throughout the Neuronal Circadian Clock Network of Drosophila Revealed by Real-Time Cyclic AMP Imaging. Neuron 58, 223–237. 10.1016/j.neuron.2008.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin Y, Stormo GD, and Taghert PH, (2004). The neuropeptide pigment-dispersing factor coordinates pacemaker interactions in the Drosophila circadian system. J. Neurosci 24, 7951–7957. 10.1523/JNEUROSCI.2370-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Helfrich-Förster C, (1998). Robust circadian rhythmicity of Drosophila melanogaster requires the presence of lateral neurons: A brain-behavioral study of disconnected mutants. J. Comp. Physiol. - Sens. Neural Behav. Physiol 182, 435–453. 10.1007/s003590050192. [DOI] [PubMed] [Google Scholar]
- 45.Nitabach MN, Blau J, and Holmes TC, (2002). Electrical Silencing of Drosophila Pacemaker Neurons Stops the Free-Running Circadian Clock An important area of circadian rhythm research is the relationship between the function of the molecular clock in pacemaker neurons and the central physiological. Cell 109, 485–495. [DOI] [PubMed] [Google Scholar]
- 46.Renn SCP, Park JH, Rosbash M, Hall JC, and Taghert PH (1999). A pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila. Cell 99, 791–802. 10.1016/S0092-8674(00)81676-1. [DOI] [PubMed] [Google Scholar]
- 47.Seluzicki A, Flourakis M, Kula-Eversole E, Zhang L, Kilman V, and Allada R, (2014). Dual PDF Signaling Pathways Reset Clocks Via TIMELESS and Acutely Excite Target Neurons to Control Circadian Behavior. PLoS Biol. 12, 19–25. 10.1371/journal.pbio.1001810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shiga S, and Numata H, (2009). Roles of PER immunoreactive neurons in circadian rhythms and photoperiodism in the blow fly, Protophormia terraenovae. J. Exp. Biol 212, 867–877. 10.1242/jeb.027003. [DOI] [PubMed] [Google Scholar]
- 49.Ikeno T, Numata H, Goto SG, and Shiga S, (2014). Involvement of the brain region containing pigment-dispersing factor-immunoreactive neurons in the photoperiodic response of the bean bug, Riptortus pedestris. J. Exp. Biol 217, 453–462. 10.1242/jeb.091801. [DOI] [PubMed] [Google Scholar]
- 50.Hasebe M, Kotaki T, and Shiga S, (2022). Pigment-dispersing factor is involved in photoperiodic control of reproduction in the brown-winged green bug, Plautia stali. J. Insect Physiol 137, 104359. 10.1016/j.jinsphys.2022.104359. [DOI] [PubMed] [Google Scholar]
- 51.Ojima N, Hara Y, Ito H, and Yamamoto D, (2018). Genetic dissection of stress-induced reproductive arrest in Drosophila melanogaster females. PLoS Genet. 14, 1–15. 10.1371/journal.pgen.1007434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nagy D, Cusumano P, Andreatta G, Anduaga AM, Hermann-Luibl C, Reinhard N, Gesto J, Wegener C, Mazzotta G, Rosato E, et al. (2019). Peptidergic signaling from clock neurons regulates reproductive dormancy in Drosophila melanogaster. PLoS Genet. 15, 1–25. 10.1371/journal.pgen.1008158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Park JH, Helfrich-Förster C, Lee G, Liu L, Rosbash M, and Hall JC, (2000). Differential regulation of circadian pacemaker output by separate clock genes in Drosophila. Proc. Natl. Acad. Sci 97, 3608–3613. 10.1073/pnas.070036197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chatterjee A, Lamaze A, De J, Mena W, Chélot E, Martin B, Hardin P, Kadener S, Emery P, and Rouyer F, (2018). Reconfiguration of a Multi-oscillator Network by Light in the Drosophila Circadian Clock. Curr. Biol 28, 2007–2017.e4. 10.1016/j.cub.2018.04.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Parsons R, Parsons R, Garner N, Oster H, and Rawashdeh O, (2020). CircaCompare: a method to estimate and statistically support differences in mesor, amplitude and phase, between circadian rhythms. Bioinformatics 36, 1208–1212. 10.1093/bioinformatics/btz730. [DOI] [PubMed] [Google Scholar]
- 56.Thaben PF, and Westermark PO, (2014). Detecting Rhythms in Time Series with RAIN. J. Biol. Rhythms 29, 391–400. 10.1177/0748730414553029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yoshii T, Wülbeck C, Sehadova H, Veleri S, Bichler D, Stanewsky R, and Helfrich- Förster C, (2009). The neuropeptide pigment-dispersing factor adjusts period and phase of Drosophila’s clock. J. Neurosci 29, 2597–2610. 10.1523/JNEUROSCI.5439-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li H, Janssens J, de Waegeneer M, Kolluru SS, Davie K, Gardeux V, Saelens W, David FPA, Brbić M, Spanier K, et al. (2022). Fly Cell Atlas: A single-nucleus transcriptomic atlas of the adult fruit fly. Science 375. 10.1126/science.abk2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Davie K, Janssens J, Koldere D, De Waegeneer M, Pech U, Kreft Ł, Aibar S, Makhzami S, Christiaens V, Bravo González-Blas C, et al. (2018). A Single-Cell Transcriptome Atlas of the Aging Drosophila Brain. Cell 174, 982–998e20. 10.1016/j.cell.2018.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lear BC, Zhang L, and Allada R, (2009). The neuropeptide PDF acts directly on evening pacemaker neurons to regulate multiple features of circadian behavior. PLoS Biol. 7. 10.1371/journal.pbio.1000154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen W, Werdann M, and Zhang Y, (2018). The auxin-inducible degradation system enables conditional PERIOD protein depletion in the nervous system of Drosophila melanogaster. FEBS J. 285, 4378–4393. 10.1111/febs.l4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Im SH, and Taghert PH, (2010). PDF receptor expression reveals direct interactions between circadian oscillators in Drosophila. J. Comp. Neurol 518,1925–1945. 10.1002/cne.22311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hyun S, Lee Y, Hong ST, Bang S, Paik D, Kang J, Shin J, Lee J, Jeon K, Hwang S, et al. (2005). Drosophila GPCR han is a receptor for the circadian clock neuropeptide PDF. Neuron 48, 267–278. 10.1016/j.neuron.2005.08.025. [DOI] [PubMed] [Google Scholar]
- 64.Neuberger G, Schneider G, and Eisenhaber F, (2007). pkaPS: prediction of protein kinase A phosphorylation sites with the simplified kinase-substrate binding model. Biol. Direct 2. 10.1186/1745-6150-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Søberg K, and Skålhegg BS, (2018). The molecular basis for specificity at the level of the protein kinase a catalytic subunit. Front. Endocrinol 9, 538. 10.3389/FENDO.2018.00538/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Meiselman MR, Alpert MH, Cui X, Shea J, Gregg I, Gallio M, and Yapici N, (2022). Recovery from cold-induced reproductive dormancy is regulated by temperature-dependent AstC signaling. Curr. Biol 32, 1362–1375e8. 10.1016/j.cub.2022.01.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liao S, Broughton S, and Nässel DR, (2017). Behavioral senescence and aging-related changes in motor neurons and brain neuromodulator levels are ameliorated by lifespan-extending reproductive dormancy in Drosophila. Front. Cell. Neurosci 11, 1–20. 10.3389/fncel.2017.00111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shearer PW, West JD, Walton VM, Brown PH, Svetec N, and Chiu JC (2016). Seasonal cues induce phenotypic plasticity of Drosophila suzukii to enhance winter survival. BMC Ecol. 16, 11. 10.1186/sl2898-016-0070-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen WF, Low KH, Lim C, and Edery I, (2007). Thermosensitive splicing of a clock gene and seasonal adaptation. Cold Spring Harb. Symp. Quant. Biol 72, 599–606. 10.1101/sqb.2007.72.021. [DOI] [PubMed] [Google Scholar]
- 70.Majercak J, Sidote D, Hardin PE, and Edery I, (1999). How a circadian clock adapts to seasonal decreases in temperature and day length. Neuron 24, 219–230. 10.1016/S0896-6273(00)80834-X. [DOI] [PubMed] [Google Scholar]
- 71.Montelli S, Mazzotta G, Vanin S, Caccin L, Corrà S, De Pittà C, Boothroyd C, Green EW, Kyriacou CP, and Costa R, (2015). Period and timeless mRNA Splicing Profiles under Natural Conditions in Drosophila melanogaster. J. Biol. Rhythms 30, 217–227. 10.1177/0748730415583575. [DOI] [PubMed] [Google Scholar]
- 72.Anduaga AM, Evanta N, Patop IL, Bartok O, Weiss R, and Kadener S, (2019). Thermosensitive alternative splicing senses and mediates temperature adaptation in Drosophila. eLife 8, 1–31. 10.7554/eLife.44642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Foley LE, Ling J, Joshi R, Evantal N, Kadener S, and Emery P, (2019). Drosophila PSI controls circadian period and the phase of circadian behavior under temperature cycle via tim splicing. eLife 8, 1–28. 10.7554/eLife.50063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang C, Daubnerova I, Jang YH, Kondo S, Žitnan D, and Kim YJ, (2021). The neuropeptide allatostatin C from clock-associated DN1p neurons generates the circadian rhythm for oogenesis. Proc. Natl. Acad. Sci. U. S. A 118. 10.1073/pnas.2016878118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Machado Almeida P, Lago Solis B, Stickley L, Feidler A, and Nagoshi E, (2021). Neurofibromin 1 in mushroom body neurons mediates circadian wake drive through activating cAMP-PKA signaling. Nat. Commun 12, 1–17. 10.1038/s41467-021-26031-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Andreatta G, Kyriacou CP, Flatt T, and Costa R, (2018). Aminergic Signaling Controls Ovarian Dormancy in Drosophila. Sci. Rep 8, 1–14. 10.1038/s41598-018-20407-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Vosko AM, Schroeder A, Loh DH, and Colwell CS (2007). Vasoactive Intestinal Peptide and the Mammalian Circadian System. Gen. Comp. Endocrinol 152, 165–175. 10.1016/j.ygcen.2007.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lucassen EA, van Diepen HC, Houben T, Michel S, Colwell CS, and Meijer JH, (2012). Role of vasoactive intestinal peptide in seasonal encoding by the suprachiasmatic nucleus clock. Eur. J. Neurosci 35, 1466–1474. 10.1111/j.l460-9568.2012.08054.x. [DOI] [PubMed] [Google Scholar]
- 79.Aton SJ, Colwell CS, Harmar AJ, Waschek J, and Herzog ED, (2005). Vasoactive intestinal polypeptide mediates circadian rhythmicity and synchrony in mammalian clock neurons. Nat. Neurosci 8, 476–483. 10.1038/nnl419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yasuo S, Watanabe M, Tsukada A, Takagi T, ligo M, Shimada K, Ebihara S, and Yoshimura T, (2004). Photoinducible Phase-Specific Light Induction of Cry1 Gene in the Pars Tuberalis of Japanese Quail. Endocrinology 145, 1612–1616. 10.1210/en.2003-1285. [DOI] [PubMed] [Google Scholar]
- 81.el Halawani ME, Pitts GR, Sun S, Silsby JL, and Sivanandan V, (1996). Active immunization against vasoactive intestinal peptide prevents photo-induced prolactin secretion in turkeys. Gen. Comp. Endocrinol 104, 76–83. 10.1006/gcen.1996.0143. [DOI] [PubMed] [Google Scholar]
- 82.Long X, Colonell J, Wong AM, Singer RH, and Lionnet T, (2017). Quantitative mRNA imaging throughout the entire Drosophila brain. Nat. Methods 14, 703–706. 10.1038/nmeth.4309. [DOI] [PubMed] [Google Scholar]
- 83.Chiu JC, Low KH, Pike DH, Yildirim E, and Edery I, (2010). Assaying locomotor activity to study circadian rhythms and sleep parameters in Drosophila. J. Vis. Exp 10.3791/2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Krijthe JH, (2015). Rtsne: T-Distributed Stochastic Neighbor Embedding using Barnes-Hut Implementation.
- 85.Maaten L. van der, and Hinton G, (2008). Visualizing Data using t-SNE. J. Mach. Learn. Res 9, 2579–2605. [Google Scholar]
- 86.R Core Team (2020). R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing; ). [Google Scholar]
- 87.Chiu JC, Vanselow JT, Kramer A, and Edery I, (2008). The phospho-occupancy of an atypical SLIMB-binding site on PERIOD that is phosphorylated by DOUBLETIME controls the pace of the clock. Genes Dev. 22, 1758–1772. 10.1101/gad.1682708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lam VH, Li YH, Liu X, Murphy KA, Diehl JS, Kwok RS, and Chiu JC, (2018). CK1α collaborates with DOUBLETIME to regulate PERIOD function in the Drosophila circadian clock. J. Neurosci 38, 10631–10643. 10.1523/JNEUROSCI.0871-18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cai YD, Xue Y, Truong CC, Del Carmen-Li J, Ochoa C, Vanselow JT, Murphy KA, Li YH, Liu X, Kunimoto BL, et al. (2020). CK2 Inhibits TIMELESS Nuclear Export and Modulates CLOCK Transcriptional Activity to Regulate Circadian Rhythms. Curr. Biol 10.1016/j.cub.2020.10.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw data for all datasets used in this study, including microscopy, western blots, and locomotor activity data are available upon request to the lead contact. This paper also analyzes existing, publicly available data from single-cell RNA-sequencing58. These data are available at https://flycellatlas.org/scope.
Single-cell RNA-sequencing data was analyzed using SCOPE, available at https://scope.aertslab.org/#/FlyCellAtlas/*/welcome. ImageJ macros used to analyze microscopy data and R code used to analyze Drosophila locomotor activity data will be shared upon request to the lead contact.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


