Abstract
The highly abundant molecular chaperone Hsp90 functions with assistance from auxiliary factors, collectively referred to as Hsp90 cochaperones, and the Hsp70 system. Hsp104, a molecular chaperone required for stress tolerance and for maintenance of [psi+] prions in the budding yeast Saccharomyces cerevisiae, appears to collaborate only with the Hsp70 system. We now report that several cochaperones previously thought to be dedicated to Hsp90 are shared with Hsp104. We show that the Hsp90 cochaperones Sti1, Cpr7, and Cns1, which utilize tetratricopeptide repeat (TPR) domains to interact with a common surface on Hsp90, form complexes with Hsp104 in vivo and that Sti1 and Cpr7 interact with Hsp104 directly in vitro. The interaction is Hsp90 independent, as further emphasized by the fact that two distinct TPR domains of Sti1 are required for binding Hsp90 and Hsp104. In a striking parallel to the sequence requirements of Hsp90 for binding TPR proteins, binding of Sti1 to Hsp104 requires a related acidic sequence at the C-terminal tail of Hsp104. While Hsp90 efficiently sequesters the cochaperones during fermentative growth, respiratory conditions induce the interaction of a fraction of Hsp90 cochaperones with Hsp104. This suggests that cochaperone sharing may favor adaptation to altered metabolic conditions.
The molecular chaperone Hsp90 is a highly conserved and abundant protein that has been shown to be essential in both multicellular organisms and yeasts (6, 7, 36). In the budding yeast Saccharomyces cerevisiae, at least one of the two almost identical Hsp90 isoforms, Hsc82 and Hsp82, has to be maintained for viability (4).
Hsp90 fulfills its functions with a cohort of Hsp90 cochaperones, which are thought to modulate its substrate recognition and binding and its functions (reviewed in references 6, 7, 12, and 36). The Hsp90 cochaperones Sti1 (the yeast homolog of mammalian Hop), Cpr6 and Cpr7 (yeast cyclophilin-40), and Cns1 contain tetratricopeptide repeats (TPR) mediating binding to a common surface on Hsp90 encompassing a highly conserved C-terminal peptide sequence (see references 23 and 41 and references therein), and participate in complexes with many and perhaps most Hsp90 substrates. A subset of Hsp90 cochaperones, notably Cpr6 and Cpr7, Cdc37, and Sba1 (yeast homolog of mammalian p23), have chaperone activity on their own (5, 22, 26, 31, 37, 46), but it is not clear to what extent they interact directly with proteins other than molecular chaperones. Surprisingly, the Hsp90 cochaperones Sti1, Cpr6 and Cpr7, and Sba1 have proven to be dispensable for vegetative growth of S. cerevisiae under standard growth conditions (3, 14, 19, 20, 34, 45), although Δcpr7 cells display a slow-growth phenotype (14, 19), and Sti1 and Sba1 are essential during amino acid starvation (17).
Hsp104 is a molecular chaperone of the Hsp100/Clp family (for a review, see reference 42). While Hsp104 is dispensable for growth of S. cerevisiae at moderate temperatures, it is the key factor conferring stress-induced tolerance to extreme temperatures (28, 38) by promoting the resolubilization of protein aggregates (24, 35). Interestingly, through this function, Hsp104 controls the aggregation state of the Sup35-based [psi+] prion-like factor. Overexpression of Hsp104 results in the solubilization of these particles and cures yeast of these prions; deletion of the HSP104 gene has the same effect, perhaps because oversized particles get lost during mitosis (reviewed in reference 43). Thus, normal levels of Hsp104 are required for maintenance of the [psi+] (11) and other prions (33).
Hsp104 associates with the Hsp40-type molecular chaperone Ydj1 (24), a component of the Hsp70 chaperone system, with which Hsp104 also interacts genetically (39). Together these chaperones constitute a rescue team for aggregated proteins (24). Apart from substrate proteins, Ydj1 is the only known protein that interacts with Hsp104. In searching for proteins that interact with the TPR-containing Hsp90 cochaperones, we have discovered a novel chaperone network. Under certain circumstances, Hsp90 cochaperones associate with Hsp104.
MATERIALS AND METHODS
Plasmids.
Glutathione S-transferase (GST) and GST fused to Cpr7 were expressed constitutively in yeast from the glyceraldehyde-3-phosphate dehydrogenase promoter with plasmids p2U/GST-2 (45) and p2U/GST.Cpr7, respectively. The latter was constructed by inserting the PCR-generated Cpr7 open reading frame into p2U/GST-2. Plasmid pGEX/CPR7 was constructed similarly with vector pGEX-1 (Pharmacia). Plasmids pYesF/Cns1, pYesF/Hsp104, pYesF/Hsp104ΔC2, pYesF/Sti1, pYesF/Sti1(TPR1), and pYesF/Sti1(TPR2AB) contain the coding sequences for Cns1, Hsp104, Hsp104 lacking the C-terminal 8 amino acids, Sti1, Sti1 amino acids 1 to 200, and Sti1 amino acids 201 to 589 in the vector pYES/Flag, respectively. The last allows galactose-inducible expression of Flag-tagged proteins in yeast and was constructed by inserting the sequence GAGCTCAAAGCATGGACTACAAGGACGACGATGACAAGGGGATCC between the SacI and BamHI sites of plasmid pYES2.0 (Invitrogen).
For expression of His6-tagged proteins in Escherichia coli, Hsp104, Hsp104ΔC, and Sti1 coding sequences were inserted into plasmid pTrcHis C (Invitrogen) to yield constructs pTrcHis/Hsp104, pTrcHis/Hsp104ΔC2, and pTrcHis/Sti1, respectively; coding sequences for full-length Hsp82 and Hsp82 codons 1 to 704 were inserted into plasmids pET15b (Novagen) and pTrcHis B (Invitrogen) to yield vectors pET15b/His.Hsp82 and pTrcHisB/Hsp82ΔC, respectively.
Yeast strains and media.
As the wild-type strain we used RMY326 (MATa his3 leu2-3,112 trp1-1 ura3-52) except for experiments with GST.Cpr7, when it was BJ2168 (MATa leu2 pep4-3 prc1-407 prb1-1122 trp1 ura3-52). Strains HH1a-p2HG/Hsp82 and HH1a-p2HG/Hsp82(1-704) have been described (29). Yeast cells were either grown directly in YEP or, in the case of transformants with episomes, precultured in selective minimal medium and then switched to YEP for overnight incubation. Unless indicated, 2% glucose was used as the carbon source.
Antibodies and recombinant proteins.
The following primary antibodies were used: rabbit polyclonal antisera against Hsp104 (a gift from S. Lindquist and antiserum PA3-016 from Affinity BioReagents) and GST, and mouse monoclonal antibodies against the Flag (antibody M2 from Sigma) and His6 (antibody His-1 from Sigma) tags and against Sti1 (antibody Sti2; a gift from D. O. Toft). Recombinant proteins were expressed in E. coli and purified on glutathione-Sepharose (Pharmacia) or Ni-nitrilotriacetic acid-agarose (Qiagen) as directed by the manufacturer.
GST pull-down and immunoprecipitation experiments.
Transformants, induced with galactose by overnight incubation when appropriate, were grown at 30°C to low or high culture density corresponding to an optical density at 600 nm (OD600) of 0.5 and 20, respectively. Cells were washed once with TEG (25 mM Tris-HCl [pH 7.4], 15 mM EGTA, 10% glycerol, 1 mM dithiothreitol, yeast protease inhibitor cocktail [Sigma]) containing 150 mM NaCl. Cell pellets were then resuspended in a small volume of the same buffer and broken with glass beads by two 30-s pulses at maximum speed in a Mini-BeadBeater-8 (Biospec, Bartlesville, N.C.) at 4°C. After centrifugation at 15,000 rpm in a tabletop centrifuge at 4°C, the supernatant was quantitated and adjusted to 0.1% Triton X-100.
GST pull-down experiments with extracts were done as described (1). For immunoprecipitations, 3 mg of total cell extracts was incubated with the anti-Sti1 antibody for 2 h at 4°C with tumbling. Then protein G-Sepharose (Pharmacia) was added for an additional hour. In the case of Flag-tagged proteins, the immunoprecipitation was performed with the anti-Flag monoclonal antibody or by directly adding the M2 anti-Flag resin (Sigma).
Immunoprecipitation experiments with purified recombinant proteins were done at 4°C as follows. The anti-Sti1 antibody was bound to protein G-Sepharose in phosphate-buffered saline (PBS) for 90 min and washed several times with PBS containing 0.1% Triton X-100; 2 μg of purified Sti1 per sample was then added in PBS–0.1% Triton X-100 and tumbled for 2 h before addition of 2 μg of purified Hsp104, Hsp104ΔC, or Hsp82 and tumbling for an additional 2 h; following three washes with the same buffer, proteins were solubilized by boiling in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer. GST pull-down experiments with purified proteins were done similarly with 1 μg of protein; test proteins were added to glutathione-Sepharose-bound GST proteins.
Gel staining and immunoblot experiments.
Silver staining of SDS-PAGE gels was done according to published procedures (2, 44). For identification of new protein bands, SDS-PAGE gels were stained with Coomassie blue, and bands of interest were excised, digested in-gel with trypsin, and subjected to mass spectrometric analysis. Immunoblotting was done as described (1).
RESULTS
Hsp104 associates with Cpr7, Cns1, and Sti1.
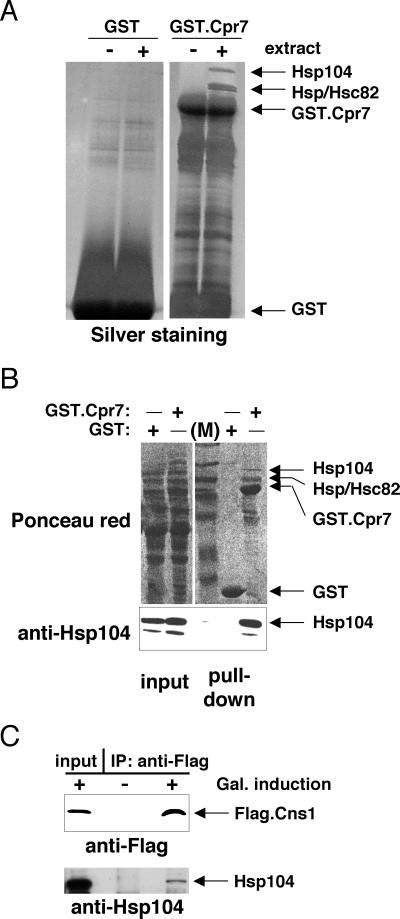
To identify new interaction partners of Cpr7 in particular and of Hsp90-interacting TPR proteins in general, we did a binding experiment with recombinant Cpr7. Cpr7, produced in bacteria as a GST fusion protein, was bound to glutathione beads and incubated with a whole-cell extract of a wild-type S. cerevisiae strain. The silver-stained gel displayed in Fig. 1A reveals three prominent proteins that are specifically retained by GST.Cpr7 and not by GST alone. As expected (18), the doublet above GST.Cpr7 corresponds to the two Hsp90 isoforms Hsp82 and Hsc82. The slower migrating third band was excised from a separate gel and subjected to sequence analysis by mass spectrometry. Several peptides identified this protein unambiguously as Hsp104 (data not shown).
FIG. 1.
Hsp104 forms complexes with Cpr7 and Cns1. (A) Recombinant GST.Cpr7 retains Hsp104 from a total yeast extract. GST or GST.Cpr7 purified from bacteria (4 μg) was bound to beads and incubated with 3 mg of yeast extract. Arrows on the right of the silver-stained SDS-PAGE gel point to identified bands. (B) GST.Cpr7 expressed in yeast pulls down Hsp104. The upper panel represents the Ponceau red-stained nitrocellulose filter of the anti-Hsp104 immunoblot in the lower panel. Lane M, molecular size marker proteins. (C) Flag-tagged Cns1 coprecipitates with Hsp104. Cells were grown either with glucose (−; no expression of Flag.Cns1) or in galactose (+; induced expression). Following immunoprecipitation (IP) with an anti- Flag antibody, Flag.Cns1 and endogenous Hsp104 were revealed by immunoblotting with anti-Flag and anti-Hsp104 antibodies, respectively. Input designates a small aliquot of the extract prior to immunoprecipitation.
To establish that Cpr7-Hsp104 complexes also exist in vivo, GST.Cpr7 was expressed in yeast. The GST pull-down experiment in Fig. 1B demonstrates that the same proteins copurify with GST.Cpr7 expressed in vivo. The specificity of the association of Hsp104 with GST.Cpr7 and not GST was confirmed by immunoblotting with a polyclonal anti-Hsp104 antiserum.
Cns1 is a TPR-containing protein that has been shown to interact with both Hsp90 and Cpr7 (15, 30). We expressed it with an N-terminal Flag epitope. As shown in Fig. 1C, an anti-Flag antibody specifically coprecipitates Hsp104 with the epitope-tagged Cns1.
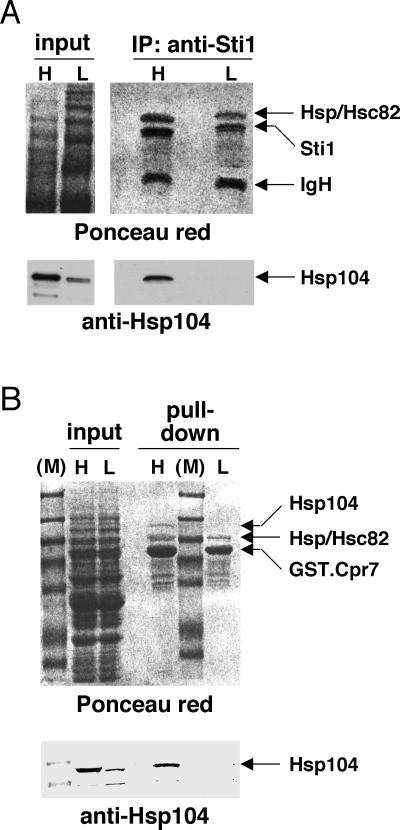
To facilitate the analysis of endogenous complexes, we switched to Sti1, an abundant TPR-containing Hsp90 cochaperone. Endogenous Hsp104 is coimmunoprecipitated with endogenous Sti1 (Fig. 2A; see also below). Although it is difficult to quantitate immunoprecipitation experiments, it is evident that only a fraction of both molecules copurify.
FIG. 2.
Hsp104 associates with endogenous Sti1 and exogenous GST.Cpr7 at high culture density. (A) Endogenous Sti1 is associated with endogenous Hsp104 at high (H) but not at low (L) cell culture density. Immunoprecipitation (IP) experiment with an antibody against Sti1 (anti-Sti1). IgH, antibody heavy-chain band. (B) GST.Cpr7 expressed in yeast is associated with Hsp104 at high density. GST pull-down experiment as in Fig. 1B. The upper panels represent the Ponceau red-stained nitrocellulose filters of the anti-Hsp104 immunoblots in the lower panels.
Interactions of Hsp104 with Sti1 and Cpr7 are regulated.
In the course of our experiments, we noticed that the abundance of Hsp104 complexes can be modulated. Figure 2 demonstrates that the association of endogenous Hsp104 with endogenous Sti1 and with GST.Cpr7 depends on cell culture density. The association is undetectable at low density (lanes L), whereas high-density culture conditions appear to induce it (lanes H). Although Hsp104 expression itself is increased as cells reach stationary phase, this increase is far too small to account for the marked difference in copurification (for example, compare lanes in the bottom panel of Fig. 2A).
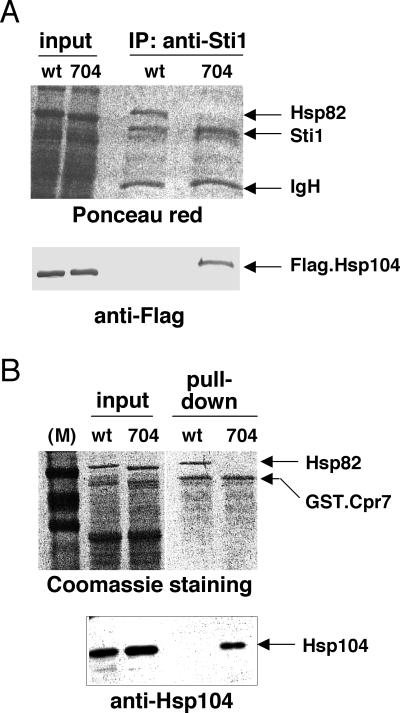
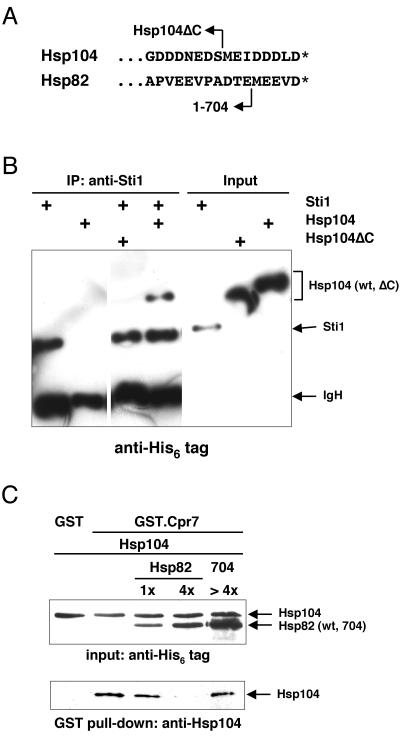
Since both Hsp90 and Hsp104 copurify with Sti1 and GST.Cpr7, we examined whether the association of Hsp104 with Sti1 and Cpr7 is influenced by Hsp90. The interaction of Hsp90 with Sti1 and Cpr7 depends on their TPR domains (9, 18, 27) and the C-terminal pentapeptide MEEVD of Hsp90 (41). We therefore took advantage of a truncation mutant lacking this pentapeptide, which we have previously shown to complement an hsp90 deletion strain without an obvious phenotype (29).
Our coprecipitation experiments with endogenous Sti1 and GST.Cpr7 expressed in a strain containing only a truncated version of Hsp82, lacking this pentapeptide (mutant 1-704), confirms the importance of the C-terminal pentapeptide for the Sti1-Hsp90 and Cpr7-Hsp90 interactions (Fig. 3, top panels). Remarkably, the immunoblots (Fig. 3, bottom panels) reveal that Hsp104 is associated with Sti1 and GST.Cpr7 even when these TPR proteins are unable to interact with Hsp90, as in the case of the 1-704 mutant (lanes 704). These data strongly argue that the interaction of Sti1 and Cpr7 with Hsp104 is not mediated by Hsp90. It is noteworthy that this experiment was performed with a low-density culture, and hence, Hsp104 was not expected to be pulled down by GST.Cpr7 in a strain with wild-type Hsp82. Yet even under those conditions Hsp104 is complexed with Sti1 and GST.Cpr7 when the interaction of Hsp90 with TPR proteins is weakened by truncation. This suggests that the binding to Hsp90 may be dominant and exclude Hsp104 unless some metabolic changes associated with high cell density elicit a switch of a fraction of Sti1 and Cpr7 molecules from Hsp90 to Hsp104.
FIG. 3.
Hsp90 and Hsp104 compete for interaction with Sti1 and Cpr7 in vivo. The association of Hsp104 with endogenous Sti1 (A) and GST.Cpr7 (B) is favored in a strain with an Hsp82 variant lacking the C-terminal pentapeptide that mediates interaction with TPR proteins. Extracts were prepared from cells grown to low density. wt and 704, strains HH1a-p2HG/Hsp82 and HH1a-p2HG/Hsp82(1-704), with wild-type Hsp82 and mutant Hsp82 lacking amino acids 705 to 709, respectively. In panel A, Hsp104 was expressed with a Flag epitope and revealed by immunoblotting with an anti-Flag antibody. Note that Flag.Hsp104 complements a Δhsp104 strain (data not shown). The upper panel in A represents the Ponceau red-stained nitrocellulose filter of the anti-Flag immunoblot in the lower panel.
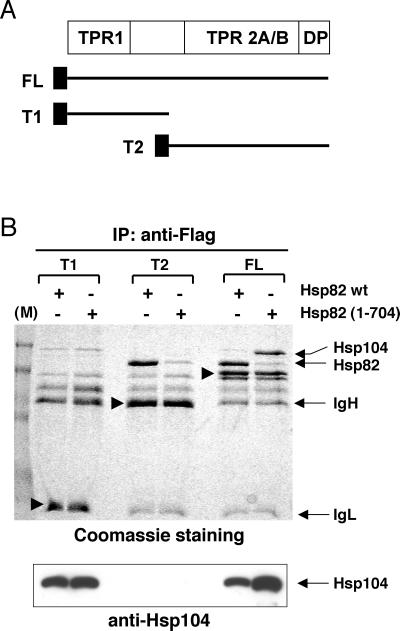
Interaction domains of Sti1.
TPR clusters 1 and 2 of the mammalian Sti1 homolog Hop have been shown to be required for differential binding to Hsp70 and Hsp90, respectively (9, 27, 41). To determine the Hsp104 binding domain of Sti1, we expressed the protein in two moieties (Fig. 4A) in yeast and performed immunoprecipitation experiments. As a control for this experiment, full-length Sti1 was also expressed with the Flag epitope. Figure 4B reveals that Hsp104 binds the N-terminal moiety encompassing TPR domain 1 (TPR1) and thus shares the interaction domain with Hsp70. Hsp90 binding requires the second TPR domain (TRP2A/B), and C-terminal truncation of Hsp90 [Hsp82(1-704)] all but abolishes this interaction. This truncation has no effect on the constitutive interaction of Hsp104 with the TPR1-containing moiety (lanes T1), whereas it favors the interaction with full-length Sti1 (lanes FL). These data prove that Sti1 uses different surfaces to interact with Hsp90 and Hsp104 and that its interaction with Hsp104 is not mediated by Hsp90.
FIG. 4.
Hsp104 and Hsp90 associate with different domains of Sti1. (A) Schematic representation of the domain structure of Sti1 and of the galactose-inducible Flag-tagged derivatives used in the experiment in panel B. TPR clusters and the DP domain are indicated. The latter comprises the C-terminal sequences DPEV and DPVM and contributes with TPR1 to Hsp70 binding (10). The Flag epitope is represented by the black boxes. (B) Hsp104 and Hsp90 binding to Sti1 requires TPR1 and TPR2, respectively. Hsp82 wt and Hsp82(1-704), strains with wild-type Hsp82 and the C-terminal truncation mutant lacking the last five amino acids, respectively. The black arrowheads indicate the bands corresponding to T1, T2, and FL. Note that cells were grown to high density to favor interaction between Sti1 and Hsp104. IgL, antibody light-chain band.
Interaction of Hsp104 with Sti1 is direct and dependent on its C-terminal tail.
We assessed the interaction between Hsp104 and the TPR protein Sti1 using recombinant protein purified from E. coli. Sti1 was mixed with Hsp104 and subjected to an immunoprecipitation experiment with the anti-Sti1 antibody (Fig. 5B). Hsp104 only coprecipitates with the anti-Sti1 antibody in the presence of Sti1, and thus, Sti1 and Hsp104 interact directly.
FIG. 5.
Interaction of Hsp104 with Sti1 and Cpr7 is direct. (A) Schematic representation of the very C-terminal sequences of Hsp104 and Hsp82 and their corresponding truncation mutants. ∗, C-terminal end. (B) In vitro interaction of purified Hsp104 and Sti1, dependent on C-terminal tail of Hsp104; 0.2 μg of input proteins was loaded for comparison. Note that there is unequal recognition of different His6-tagged proteins by the anti-His6 antibody. (C) Direct interaction of Hsp104 with Cpr7 fused to GST and competition by Hsp90 (Hsp82). The immunoblot of the upper panel represents aliquots of the input proteins prior to the GST pull-down. The Ponceau red-stained filter shows that equivalent amounts of GST.Cpr7 and GST were pulled down (data not shown).
The very C-terminal sequence of Hsp104 displays a striking sequence similarity to that of Hsp82, which is essential for the interaction with TPR proteins (Fig. 5A). We therefore produced a truncated version of Hsp104, denoted Hsp104ΔC, lacking the last 8 amino acids. Unlike the wild-type protein, Hsp104ΔC is defective for interaction with Sti1 both in vitro (Fig. 5B) and in vivo (data not shown).
Hsp104 and Hsp90 compete for direct interaction with Cpr7.
To extend the in vitro analysis to a protein that has only one TPR cluster, we mixed recombinant purified GST.Cpr7 with Hsp104 and performed a GST pull-down experiment (Fig. 5C). A fraction of Hsp104 copurifies with GST.Cpr7 but not with GST alone. Thus, this interaction is also direct. Moreover, purified Hsp82 efficiently competes with Hsp104 for binding to GST.Cpr7, whereas the C-terminally truncated Hsp82(1-704), which is unable to bind TPR proteins, does not. These results further support the notion that binding of Cpr7 to these two molecular chaperones is mutually exclusive.
Association of Sti1 with Hsp104 is induced by respiratory growth conditions.
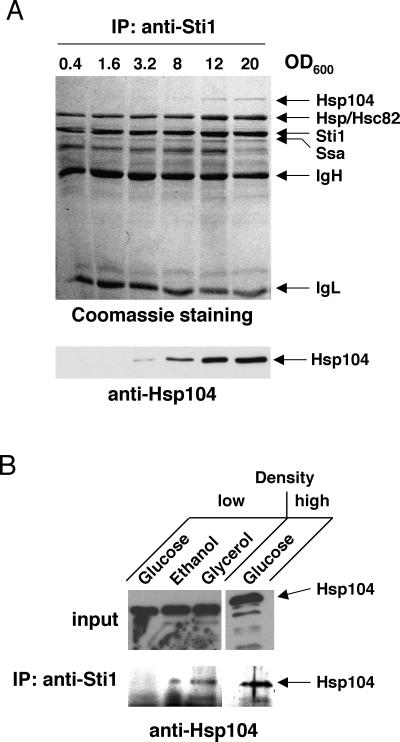
To begin to understand the nature of the metabolic conditions that promote the association of Hsp90 cochaperones with Hsp104, we examined the dependence on culture density and on carbon source more carefully. Extracts were made from cells grown to different densities. Endogenous protein complexes were immunoprecipitated with a monoclonal antibody against Sti1 and displayed by Coomassie blue staining of an SDS-PAGE gel and by immunoblotting with the anti-Hsp104 antiserum (Fig. 6A). Equal gel loading and Sti1 immunoprecipitation are apparent from the stained gel. Almost stoichiometric amounts of Hsp90 remain associated with Sti1 throughout exponential growth. In contrast, Sti1-associated Hsp104 only becomes detectable above a certain density and keeps increasing thereafter.
FIG. 6.
Respiratory growth conditions induce the association of Hsp104 with Sti1. (A) Increasing cell density coinciding with a switch to respiration induces the association. Immunoprecipitation (IP) experiment assessing coprecipitation of endogenous proteins with endogenous Sti1. Note that the total amounts of coprecipitated Sti1, Hsp90 (Hsc82/Hsp82), and Hsp70 (Ssa protein family) do not change significantly. OD600, optical density as a measure of cell density. The levels of Hsp104 in the input extracts can be seen in Fig. 2, where low and high culture densities correspond to an OD600 of 0.4 to 0.5 and 20, respectively. (B) Nonfermentable carbon sources (ethanol and glycerol) induce the association of Sti1 with Hsp104. A representative immunoblot experiment is shown.
This pattern suggested that the “switch” may be related to the diauxic shift of yeast cells progressing from a fermentative to a respiratory growth mode. Due to the progressive depletion of glucose prior to this shift, yeast cells switch from fermenting glucose as a carbon source to a respiratory utilization of its accumulated metabolite ethanol. Indeed, when cells are grown directly on nonfermentable carbon sources such as ethanol and glycerol, Hsp104 is associated with Sti1 even at low cell density (Fig. 6B), whereas the fermentable sugar galactose gives the same result as glucose (data not shown). Thus, in the presence of wild-type Hsp90, reduced concentrations of fermentable sugar and/or respiration, not high cell density per se, induce the association with Hsp104.
DISCUSSION
New chaperone connection.
Hsp90 cochaperones were thought to be dedicated to Hsp90. Among the few exceptions are the interactions of Sti1/Hop with Hsc70 and Hsp70 (9), of Cns1 with Cpr7 (15, 30), the binding of the mammalian Sba1 homolog p23 to some Hsp90 substrates (16, 21, 25), and the recruitment by CHIP of the protein degradation machinery to Hsp90 substrates (13). We were therefore surprised to discover that the molecular chaperone Hsp104 interacts with the Hsp90 cochaperones Sti1, Cpr7, and Cns1. Ydj1 was the only other known Hsp104 binding protein (24), but to our knowledge, Sti1 and Cpr7 are the first proteins for which a direct interaction with Hsp104 has been demonstrated.
Interestingly, related domains mediate the interaction of Sti1 with Hsp104 and Hsp90. Both types of interactions require a TPR domain of Sti1 and the very C-terminal peptide sequences of Hsp90 (9, 27, 41) and Hsp104. However, the two TPR clusters of Sti1 display marked differences in specificity. The N-terminal TPR domain TPR1 binds both Hsp70 and Hsp104, whereas Hsp90 binding is mediated by the second TPR cluster, TPR2A/B. The Hsp104 binding domain of Cpr7 has yet to be mapped, but its TPR domain may well be involved in this interaction.
It is important to point out that Hsp104 complexes, just like Hsp90 complexes, are heterogeneous, with any particular complex representing only a minor fraction at steady state. We have yet to investigate the stoichiometry, the relative proportions, and the dynamics of these novel chaperone complexes. With respect to the stoichiometry, data obtained by gel filtration are compatible with a 1:1 ratio of the components in Hsp104-Sti1 TPR1 complexes (data not shown).
There are no known interactions between the Hsp104 and Hsp90 chaperone machineries, and indeed, the complexes between Hsp90 cochaperones and Hsp104 are not mediated by Hsp90. Both Sti1 and Cpr7 interact with Hsp104 directly, and for Cpr7 we have even demonstrated that Hsp104 and Hsp90 binding is mutually exclusive. Similarly, the formation of Hsp104 complexes is favored in vivo when binding of TPR proteins to Hsp90 is impaired by truncating Hsp90. Preliminary data indicate that Cdc37 and Sba1 do not associate with Hsp104 (data not shown) suggesting that cochaperone sharing may be restricted to a subset of Hsp90 cochaperones. Thus, the two “core” chaperones share some regulatory and/or auxiliary components as part of distinct complexes with distinct functions.
Hsp104 cochaperone interactions are regulated.
The most striking finding about the new chaperone network is its regulation by growth conditions. When cells are grown with their favorite carbon source, glucose, Hsp90 cochaperones are not associated with Hsp104. The TPR proteins Sti1, Cpr7, and Cns1 interact with Hsp104 when cells are grown directly on nonfermentable carbon sources such as ethanol and glycerol, or once the fermentable carbon source is used up when cells have reached a certain density. Cells then begin to utilize nonfermentable metabolites. Hence, the association of Hsp90 cochaperones with Hsp104 correlates with respiratory growth and low (or no) concentrations of a fermentable sugar such as glucose and galactose. Remarkably, when the interaction between Hsp90 cochaperones and Hsp90 is reduced by truncating Hsp90, the interaction with Hsp104 occurs even in the presence of the fermentable sugar. This observation suggests that fermentable sugar and fermentation per se do not prevent the formation of Hsp104 complexes. If one assumes that the association with Hsp90 is the default, it is conceivable that respiration and/or the absence of a fermentable sugar inhibits the association with Hsp90 and, in turn, favors that to Hsp104. It is noteworthy that nitrogen starvation and classical types of stresses such as heat and high osmolarity do not induce the association of Hsp90 cochaperones with Hsp104 (data not shown). Current knowledge on glucose and galactose signaling (reviewed in reference 8), on the one hand, and on the regulation of molecular chaperone activity (see, for example, reference 32), on the other, is too limited to predict the “signaling pathway.” However, it will be interesting to explore this regulation as a paradigm of how extracellular or metabolic signals regulate chaperone networks and their activities.
What is the function?
The newly identified interactions could be important for Hsp104, the Hsp90 cochaperones, or both. Unfortunately, relatively little is known about the in vivo functions of Sti1, Cpr7, and Cns1. Therefore, we have explored potential effects on Hsp104 functions more extensively. We initially examined whether mutants carrying the double deletions Δhsp104 Δcpr7 and Δhsp104 Δsti1 display synthetic growth defects. However, with respect to growth at normal and moderately elevated temperatures and under both respiratory and fermentative conditions, these strains were indistinguishable from their parent strains (data not shown).
The potential role for the Hsp90 cochaperones in modulating Hsp104 function in protein disaggregation and refolding was also investigated (data not shown). We found that the Hsp104-dependent refolding of bacterial luciferase is not affected by respiratory growth conditions that promote the association of Hsp104 with Hsp90 cochaperones or by overexpression of the Sti1 TPR1 domain, which could be expected to block access of wild-type Sti1 and other TPR proteins to Hsp104, or by deletion of the STI1 gene. Moreover, the frequency of the Hsp104-modulated loss of the Sup35 prions underlying the [psi+] phenotype is not increased by overexpression of TPR1 or by growing cells on a nonfermentable carbon source. We have noticed a reduced competence to promote luciferase refolding for the C-terminally truncated Hsp104 (data not shown). Whether there is indeed a causal link between this reduced function and inability to bind TPR proteins remains to be established.
Therefore, the biological functions of the interaction of Hsp90 cochaperones with Hsp104 remain to be identified with more sophisticated assays and a larger array of different environmental and metabolic conditions. Indeed, Hsp104 has been shown to be required for tolerance to a large variety of stresses (40). It is also conceivable that the newly described interactions reveal a potential of Hsp104 to associate with TPR proteins other than the ones tested here. TPR proteins unrelated to the Hsp90 chaperone system are involved in a wide range of cellular processes. Since Hsp104-related proteins exist in many organisms, the newly discovered chaperone network is likely to be of importance beyond budding yeast.
ACKNOWLEDGMENTS
We are indebted to David Toft and Susan Lindquist for very generous gifts of antibodies. We thank Betty Craig, Susan Lindquist, and Peter Piper for plasmids. We are grateful to Rainer Warth for the construction of several plasmids, to Valentina Gburcik for GST, to the Swiss-2D service (Geneva) for mass spectrometric analysis, and to Peter Dudek for critical comments on the manuscript.
This work was supported by the Swiss National Science Foundation and the Canton de Genève.
REFERENCES
- 1.Abbas-Terki T, Donzé O, Picard D. The molecular chaperone Cdc37 is required for Ste11 function and pheromone-induced cell cycle arrest. FEBS Lett. 2000;467:111–116. doi: 10.1016/s0014-5793(00)01134-0. [DOI] [PubMed] [Google Scholar]
- 2.Blum H E, Beier H, Gross H J. Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis. 1987;8:93–99. [Google Scholar]
- 3.Bohen S P. Genetic and biochemical analysis of p23 and ansamycin antibiotics in the function of Hsp90-dependent signaling proteins. Mol Cell Biol. 1998;18:3330–3339. doi: 10.1128/mcb.18.6.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borkovich K A, Farrelly F W, Finkelstein D B, Taulien J, Lindquist S. Hsp82 is an essential protein that is required in higher concentrations for growth of cells at higher temperatures. Mol Cell Biol. 1989;9:3919–3930. doi: 10.1128/mcb.9.9.3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bose S, Weikl T, Bügl H, Buchner J. Chaperone function of Hsp90-associated proteins. Science. 1996;274:1715–1717. doi: 10.1126/science.274.5293.1715. [DOI] [PubMed] [Google Scholar]
- 6.Buchner J. Hsp90 & Co. — a holding for folding. Trends Biochem Sci. 1999;24:136–141. doi: 10.1016/s0968-0004(99)01373-0. [DOI] [PubMed] [Google Scholar]
- 7.Caplan A J. Hsp90's secrets unfold: new insights from structural and functional studies. Trends Cell Biol. 1999;9:262–268. doi: 10.1016/s0962-8924(99)01580-9. [DOI] [PubMed] [Google Scholar]
- 8.Carlson M. Glucose repression in yeast. Curr Opin Microbiol. 1999;2:202–207. doi: 10.1016/S1369-5274(99)80035-6. [DOI] [PubMed] [Google Scholar]
- 9.Chen S, Prapapanich V, Rimerman R A, Honoré B, Smith D F. Interactions of p60, a mediator of progesterone receptor assembly, with heat shock proteins Hsp90 and Hsp70. Mol Endocrinol. 1996;10:682–693. doi: 10.1210/mend.10.6.8776728. [DOI] [PubMed] [Google Scholar]
- 10.Chen S, Smith D F. Hop as an adaptor in the heat shock protein 70 (Hsp70) and Hsp90 chaperone machinery. J Biol Chem. 1998;273:35194–35200. doi: 10.1074/jbc.273.52.35194. [DOI] [PubMed] [Google Scholar]
- 11.Chernoff Y O, Lindquist S L, Ono B, Inge-Vechtomov S G, Liebman S W. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+] Science. 1995;268:880–884. doi: 10.1126/science.7754373. [DOI] [PubMed] [Google Scholar]
- 12.Cheung J, Smith D F. Molecular chaperone interactions with steroid receptors: an update. Mol Endocrinol. 2000;14:939–946. doi: 10.1210/mend.14.7.0489. [DOI] [PubMed] [Google Scholar]
- 13.Connell P, Ballinger C A, Jiang J, Wu Y, Thompson L J, Hohfeld J, Patterson C. The cochaperone CHIP regulates protein triage decisions mediated by heat-shock proteins. Nat Cell Biol. 2001;3:93–96. doi: 10.1038/35050618. [DOI] [PubMed] [Google Scholar]
- 14.Dolinski K, Muir S, Cardenas M, Heitman J. All cyclophilins and FK506 binding proteins are, individually and collectively, dispensable for viability in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1997;94:13093–13098. doi: 10.1073/pnas.94.24.13093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dolinski K J, Cardenas M E, Heitman J. CNS1 encodes an essential p60/Sti1 homolog in Saccharomyces cerevisiae that suppresses cyclophilin 40 mutations and interacts with Hsp90. Mol Cell Biol. 1998;18:7344–7352. doi: 10.1128/mcb.18.12.7344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Donzé O, Abbas-Terki T, Picard D. The Hsp90 chaperone complex is both a facilitator and a repressor of the dsRNA-dependent kinase PKR. EMBO J. 2001;20:3771–3780. doi: 10.1093/emboj/20.14.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Donzé O, Picard D. Hsp90 binds and regulates Gcn2, the ligand-inducible kinase of the α subunit of eukaryotic translation initiation factor 2. Mol Cell Biol. 1999;19:8422–8432. doi: 10.1128/mcb.19.12.8422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duina A A, Chang H-C J, Marsh J A, Lindquist S, Gaber R F. A cyclophilin function in Hsp90-dependent signal transduction. Science. 1996;274:1713–1715. doi: 10.1126/science.274.5293.1713. [DOI] [PubMed] [Google Scholar]
- 19.Duina A A, Marsh J A, Gaber R F. Identification of two CyP-40-like cyclophilins in Saccharomyces cerevisiae, one of which is required for normal growth. Yeast. 1996;12:943–952. doi: 10.1002/(sici)1097-0061(199608)12:10<943::aid-yea997>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 20.Fang Y, Fliss A E, Rao J, Caplan A J. SBA1 encodes a yeast hsp90 cochaperone that is homologous to vertebrate p23. Mol Cell Biol. 1998;18:3727–3734. doi: 10.1128/mcb.18.7.3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Freeman B C, Felts S J, Toft D O, Yamamoto K R. The p23 molecular chaperones act at a late step in intracellular receptor action to differentially affect ligand efficacies. Genes Dev. 2000;14:422–434. [PMC free article] [PubMed] [Google Scholar]
- 22.Freeman B C, Toft D O, Morimoto R I. Molecular chaperone machines: chaperone activities of the cyclophilin Cyp-40 and the steroid aporeceptor-associated protein p23. Science. 1996;274:1718–1720. doi: 10.1126/science.274.5293.1718. [DOI] [PubMed] [Google Scholar]
- 23.Frydman J, Höhfeld J. Chaperones get in touch: the Hip-Hop connection. Trends Biochem Sci. 1997;22:87–92. doi: 10.1016/s0968-0004(97)01005-0. [DOI] [PubMed] [Google Scholar]
- 24.Glover J R, Lindquist S. Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell. 1998;94:73–82. doi: 10.1016/s0092-8674(00)81223-4. [DOI] [PubMed] [Google Scholar]
- 25.Hu J, Toft D O, Seeger C. Hepadnavirus assembly and reverse transcription require a multicomponent chaperone complex which is incorporated into nucleocapsids. EMBO J. 1997;16:59–68. doi: 10.1093/emboj/16.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kimura Y, Rutherford S L, Miyata Y, Yahara I, Freeman B C, Yue L, Morimoto R I, Lindquist S. Cdc37 is a molecular chaperone with specific functions in signal transduction. Genes Dev. 1997;11:1775–1785. doi: 10.1101/gad.11.14.1775. [DOI] [PubMed] [Google Scholar]
- 27.Lässle M, Blatch G L, Kundra V, Takatori T, Zetter B R. Stress-inducible, murine protein mSTI1. J Biol Chem. 1997;272:1876–1884. doi: 10.1074/jbc.272.3.1876. [DOI] [PubMed] [Google Scholar]
- 28.Lindquist S, Kim G. Heat-shock protein 104 expression is sufficient for thermotolerance in yeast. Proc Natl Acad Sci USA. 1996;93:5301–5306. doi: 10.1073/pnas.93.11.5301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Louvion J-F, Warth R, Picard D. Two eukaryote-specific regions of Hsp82 are dispensable for its viability and signal transduction functions in yeast. Proc Natl Acad Sci USA. 1996;93:13937–13942. doi: 10.1073/pnas.93.24.13937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marsh J A, Kalton H M, Gaber R F. Cns1 is an essential protein associated with the hsp90 chaperone complex in Saccharomyces cerevisiae that can restore cyclophilin 40-dependent functions in cpr7Δ cells. Mol Cell Biol. 1998;18:7353–7359. doi: 10.1128/mcb.18.12.7353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mayr C, Richter K, Lilie H, Buchner J. Cpr6 and Cpr7, two closely related Hsp90-associated immunophilins from saccharomyces cerevisiae, differ in their functional properties. J Biol Chem. 2000;275:34140–34146. doi: 10.1074/jbc.M005251200. [DOI] [PubMed] [Google Scholar]
- 32.Morano K A, Thiele D J. The Sch9 protein kinase regulates Hsp90 chaperone complex signal transduction activity in vivo. EMBO J. 1999;18:5953–5962. doi: 10.1093/emboj/18.21.5953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moriyama H, Edskes H K, Wickner R B. [URE3] prion propagation in Saccharomyces cerevisiae: requirement for chaperone Hsp104 and curing by overexpressed chaperone Ydj1p. Mol Cell Biol. 2000;20:8916–8922. doi: 10.1128/mcb.20.23.8916-8922.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nicolet C M, Craig E A. Isolation and characterization of STI1, a stress-inducible gene from Saccharomyces cerevisiae. Mol Cell Biol. 1989;9:3638–3646. doi: 10.1128/mcb.9.9.3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parsell D A, Kowal A S, Singer M A, Lindquist S. Protein disaggregation mediated by heat-shock protein Hsp104. Nature. 1994;372:475–478. doi: 10.1038/372475a0. [DOI] [PubMed] [Google Scholar]
- 36.Pearl L H, Prodromou C. Structure and in vivo function of Hsp90. Curr Opin Struct Biol. 2000;10:46–51. doi: 10.1016/s0959-440x(99)00047-0. [DOI] [PubMed] [Google Scholar]
- 37.Pirkl F, Buchner J. Functional analysis of the Hsp90-associated human peptidyl prolyl cis/trans isomerases FKBP51, FKBP52 and Cyp40. J Mol Biol. 2001;308:795–806. doi: 10.1006/jmbi.2001.4595. [DOI] [PubMed] [Google Scholar]
- 38.Sanchez Y, Lindquist S L. HSP104 required for induced thermotolerance. Science. 1990;248:1112–1115. doi: 10.1126/science.2188365. [DOI] [PubMed] [Google Scholar]
- 39.Sanchez Y, Parsell D A, Taulien J, Vogel J L, Craig E A, Lindquist S. Genetic evidence for a functional relationship between Hsp104 and Hsp70. J Bacteriol. 1993;175:6484–6491. doi: 10.1128/jb.175.20.6484-6491.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sanchez Y, Taulien J, Borkovich K A, Lindquist S. Hsp104 is required for tolerance to many forms of stress. EMBO J. 1992;11:2357–2364. doi: 10.1002/j.1460-2075.1992.tb05295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scheufler C, Brinker A, Bourenkov G, Pegoraro S, Moroder L, Bartunik H, Hartl F U, Moarefi I. Structure of TPR domain-peptide complexes: critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine. Cell. 2000;101:199–210. doi: 10.1016/S0092-8674(00)80830-2. [DOI] [PubMed] [Google Scholar]
- 42.Schirmer E C, Glover J R, Singer M A, Lindquist S. HSP100/Clp proteins: a common mechanism explains diverse functions. Trends Biochem Sci. 1996;21:289–296. [PubMed] [Google Scholar]
- 43.Serio T R, Lindquist S L. Protein-only inheritance in yeast: something to get [PSI+]-ched about. Trends Cell Biol. 2000;10:98–105. doi: 10.1016/s0962-8924(99)01711-0. [DOI] [PubMed] [Google Scholar]
- 44.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 45.Warth R, Briand P-A, Picard D. Functional analysis of the yeast 40 kDa cyclophilin Cyp40 and its role for viability and steroid receptor regulation. Biol Chem. 1997;378:381–391. doi: 10.1515/bchm.1997.378.5.381. [DOI] [PubMed] [Google Scholar]
- 46.Weikl T, Abelmann K, Buchner J. An unstructured C-terminal region of the Hsp90 cochaperone p23 is important for its chaperone function. J Mol Biol. 1999;293:685–691. doi: 10.1006/jmbi.1999.3172. [DOI] [PubMed] [Google Scholar]