Significance
Mucosal immunity is a key component of protection against many pathogens. Robust and effective mucosal immune responses are generally induced following infection with a replication-competent pathogen at a mucosal surface. Several studies have attempted to develop viral vector-based enteric mucosal vaccines; however, the most advanced of these are still in clinical development. Here, we successfully induced systemic and mucosal antibody responses against both rotavirus and norovirus following inoculation of recombinant rotaviruses expressing the human norovirus capsid proteins. These responses are likely to function as correlates of protection. Live-attenuated rotavirus vaccines have already proven safe and effective worldwide. These findings confirm the potential utility of using rotaviruses as a dual enteric vaccine platform for other important human enteric pathogens.
Keywords: mucosal vaccination, enteric viral vector, rotavirus, norovirus, enteric pathogens
Abstract
Rotaviruses (RVs) preferentially replicate in the small intestine and frequently cause severe diarrheal disease, and the following enteric infection generally induces variable levels of protective systemic and mucosal immune responses in humans and other animals. Rhesus rotavirus (RRV) is a simian RV that was previously used as a human RV vaccine and has been extensively studied in mice. Although RRV replicates poorly in the suckling mouse intestine, infection induces a robust and protective antibody response. The recent availability of plasmid only-based RV reverse genetics systems has enabled the generation of recombinant RVs expressing foreign proteins. However, recombinant RVs have not yet been experimentally tested as potential vaccine vectors to immunize against other gastrointestinal pathogens in vivo. This is a newly available opportunity because several live-attenuated RV vaccines are already widely administered to infants and young children worldwide. To explore the feasibility of using RV as a dual vaccine vector, we rescued replication-competent recombinant RRVs harboring bicistronic gene segment 7 that encodes the native RV nonstructural protein 3 (NSP3) protein and a human norovirus (HuNoV) VP1 protein or P domain from the predominant genotype GII.4. The rescued viruses expressed HuNoV VP1 or P protein in infected cells in vitro and elicited systemic and local antibody responses to HuNoV and RRV following oral infection of suckling mice. Serum IgG and fecal IgA from infected suckling mice bound to and neutralized both RRV and HuNoV. These findings have encouraging practical implications for the design of RV-based next-generation multivalent enteric vaccines to target HuNoV and other human enteric pathogens.
Mucosal immunity plays a critical role in protecting against many pathogens in the respiratory and intestinal tracts. Live virus infections generally trigger more robust and effective mucosal immune response than oral administration of inactivated viruses or target protein antigens because they are self-amplifying and can more effectively elicit cellular as well as humoral immunity (1–4). Several studies have attempted to utilize recombinant viruses as vaccine vectors to induce an immune response against enteric pathogens (5–8); however, the most advanced of such enteric vaccine vectors are still in early stages of clinical development.
Rotaviruses (RVs), the leading cause of acute gastroenteritis in infants, are a promising candidate for enteric vaccine vectors for several reasons. A) RV preferentially replicates in the small intestine, distinguishing it from several other enteric viruses that can also infect systemically or the colon. B) RV infection is acute, and the virus does not integrate into the host genome. C) RV is highly immunogenic and induces both systemic and mucosal immune responses in infected animals and humans (9, 10). D) Several live-attenuated human RV vaccines have been shown to be both safe and effective to use in very young children [e.g., RotaTeq (Merck) and Rotarix (GlaxoSmithKline)]. Other effective live-attenuated RV vaccines [Rotasiil, Rotavac, Lanzhou lamb rotavirus vaccine (LLR), and Rotavin-M1] are also licensed for use globally or primarily in their country of origin (11). E) Following substantial public health efforts, RV vaccines are now widely available in many low- and middle-income countries, as well as the more developed countries, and hence the administration of RV-based vaccines that included other heterologous antigens could potentially be piggybacked onto current RV immunization programs used globally. F) The RV double-stranded RNA (dsRNA) genome is segmented in nature, permitting easy genetic manipulation. G) With the insertion of heterologous antigens, RV replication can become attenuated in vitro (12, 13).
Since a plasmid-based reverse genetics system was established in 2017, several studies have reported the generation of recombinant RVs that express fluorescent and bioluminescent reporter proteins (GFP, RFP, luciferase, etc.) and exogenous nucleotide sequences [e.g., endoribonuclease Csy4 target sequence and sequences encoding the receptor binding domain of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike protein] in vitro (12–22). To facilitate the assessment and development of RVs as potential enteric vaccine vectors, the capacity of recombinant RVs to induce an enteric immune response against other gastrointestinal (GI) pathogens needs to be evaluated in well-characterized preclinical small animal models. Rhesus rotavirus (RRV) is a prototype laboratory strain of simian RV that efficiently replicates in vitro (23, 24). Although RRV does not replicate well in a murine model (25–27), it does induce both systemic and mucosal immune responses in infected mice (28). In addition, RRV itself and RRV-based RV vaccine candidates have previously been shown to be a highly immunogenic and protective in several human vaccine trials and were, for a time, licensed for use in children in the United States (29, 30).
Human norovirus (HuNoV) is a major cause of acute gastroenteritis in both young children and adults. Although B cells and human intestinal organoids support HuNoV replication (31, 32), there is not yet a widely available robust cell culture system for efficient HuNoV cultivation, which has impeded both the assessment of HuNoV immunity and vaccine development. The HuNoV virion consists of major capsid protein VP1 and minor capsid protein VP2 surrounding a positive-sense RNA genome (33–35). Exogenously expressed VP1 can form virus-like particles (VLPs) that are structurally and antigenically similar to HuNoV virions (36–38), and the parenteral administration of such VLPs provides some level of protective immunity to HuNoV in adults (39–41). Moreover, expression of the protruding or P domain of VP1 that bears the major antigenic sites of HuNoV can yield subunit “P particles” that can also induce immune responses (42, 43). Here, we demonstrate the induction of both systemic and mucosal antibody responses against HuNoV in suckling mice using recombinant RRVs expressing HuNoV VP1 or P domain. Our data suggest that recombinant RVs represent a potentially effective small-intestine–targeted vaccination platform to express exogenous genes in the human intestine and to protect people from other enteric pathogens such as HuNoV as well as RV.
Results
Generation of a Recombinant RV Expressing HuNoV VP1.
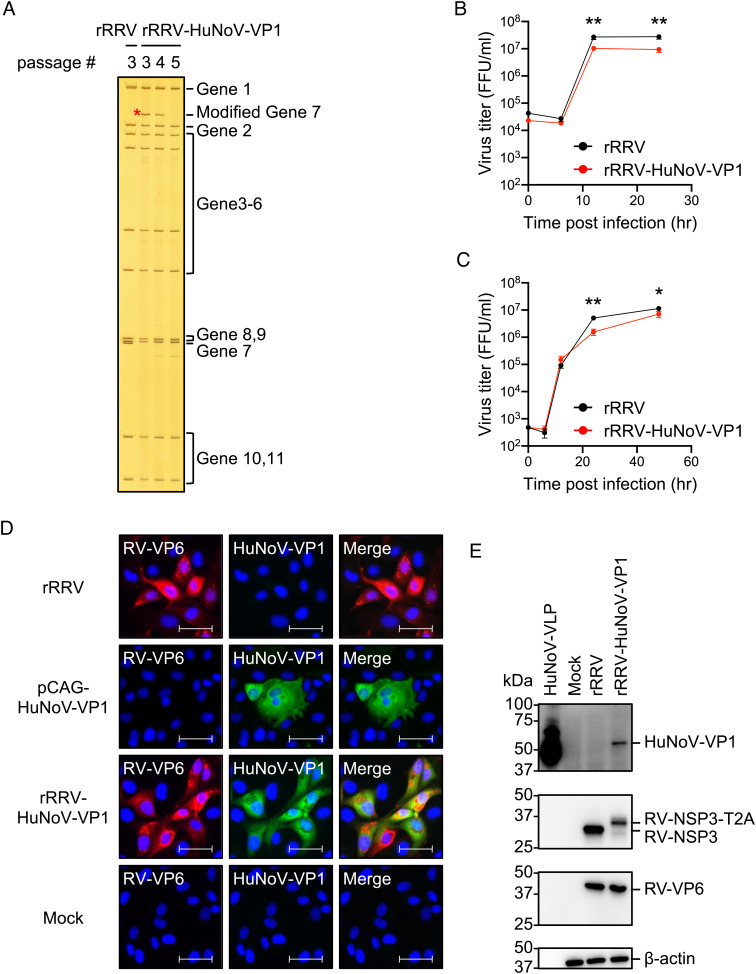
To express HuNoV VP1 from RV, we first constructed a plasmid with a T7 promoter driving the expression of RV nonstructural protein 3 (NSP3), a “self-cleavage” peptide derived from Thosea asigna virus 2A (T2A), and the coding sequence of major capsid protein VP1 of GII.4 HuNoV Sydney strain into the RV gene segment 7 (SI Appendix, Fig. S1A). We confirmed the expression of the expected proteins from this construct design by immunostaining (SI Appendix, Fig. S1B) and western blot (SI Appendix, Fig. S1C) following transfection of this plasmid into baby hamster kidney cells expressing T7 RNA polymerase (BHK-T7 cells). A faint signal for the fusion protein (RV-NSP3-T2A-HuNoV-VP1) was also detected by both the HuNoV VP1- and RV NSP3-specific sera in the western blot, but the cleavage mediated by T2A to release NSP3 (36 kDa) and VP1 (~58 kDa) appeared quite efficient (SI Appendix, Fig. S1C). Using an improved RV reverse genetics system previously described (12), we rescued a recombinant RRV harboring the HuNoV VP1 gene (rRRV-HuNoV-VP1). To confirm that the rRRV-HuNoV-VP1 actually harbored the modified gene segment 7, we extracted viral dsRNA from virions of rRRV-HuNoV-VP1 and compared the electrophoretic pattern of the dsRNA with that from wild-type recombinant RRV (rRRV). RNA polyacrylamide gel electrophoresis (RNA-PAGE) analysis showed that rRRV-HuNoV-VP1 passage 3 stock has an extra band between gene segment 1 (3,302 bp) and gene segment 2 (2,708 bp), which corresponds in size to the modified gene segment 7 size (2,752 bp). Of note, the rRRV-HuNoV-VP1 lost this extra band between gene segments 1 and 2 during serial passage (Fig. 1A). This finding suggests that the modified gene segment 7 was incorporated into progeny virion, but that the resulting recombinant RV was not genetically stable during multiple passages in cell culture. Because of the limited genetic stability of rRRV-HuNoV-VP1, we exclusively used passage 3 stocks in all the subsequent in vitro and in vivo experiments described below. The growth kinetics of rRRV-HuNoV-VP1 in MA104 cells showed that the virus replicated efficiently at multiplicity of infection (MOI) of 1 and 0.01 focus forming unit (FFU)/cell although the recombinant HuNoV-VP1 expressing virus titer was slightly lower than that of wild-type rRRV (Fig. 1 B and C). We next demonstrated that rRRV-HuNoV-VP1 expressed HuNoV VP1 in RV-infected cells by immunostaining (Fig. 1D). The immunostaining data showed that 98.9% (453 out of 458) of RV NSP3 positive cells expressed HuNoV VP1. We also determined that the molecular size of the expressed HuNoV VP1 corresponded to HuNoV VLP by western blot assay (Fig. 1E). Taken together, these data demonstrate that rRRV-HuNoV-VP1 harbors the modified RV gene segment 7 during viral replication and produces HuNoV VP1 in infected cells as designed.
Fig. 1.
In vitro characterization of rRRV-HuNoV-VP1. (A) Electrophoretic analysis of viral dsRNA. Viral dsRNAs were purified from rRRV and different passages of rRRV-HuNoV-VP1, separated by RNA-PAGE, and visualized by silver staining. The modified gene segment 7 is indicated by a red asterisk. (B and C) Growth kinetics of rRRV and rRRV-HuNoV-VP1 in MA104 cells. MA104 cells were infected with either rRRV or rRRV-HuNoV-VP1 at an MOI of (B) 1 FFU/cell or (C) 0.01 FFU/cell. The samples were frozen at each indicated time point and the virus titers were determined by a focus forming unit (FFU) assay. Representative data of two independent experiments with triplicate samples are shown as mean with SD. Statistical analysis was performed by Student’s t test for each time point. Statistical significance is indicated as *P < 0.05, **P < 0.01. (D) Immunostaining analysis of protein expression by rRRV-HuNoV-VP1 in MA104 cells. MA104 cells were infected with the rRRV or rRRV-HuNoV-VP1 at MOI of 1 FFU/cell and fixed at 24 h postinfection, or the cells were transfected with pCAG-HuNoV-VP1 plasmid and fixed at 3 d posttransfection. The cells were stained with antibodies specific to RV-VP6 (red), HuNoV-VP1 (green), and DAPI (blue). Representative data of two independent experiments are shown. (Scale bar, 50 μm.) (E) Western blotting analysis of protein expression by rRRV-HuNoV-VP1 in MA104 cells. MA104 cells were infected with rRRV or rRRV-HuNoV-VP1 at an MOI of 4 FFU/cell and harvested at 12 h postinfection. The cells were lysed with Laemmli buffer and resolved by sodium dodecyl-sulfate polyacrylamide gel electrophoresis (SDS-PAGE). Protein expression of HuNoV-VP1, RV-NSP3, RV-VP6, and β-actin was detected by the specific antibodies. The numbers indicate the molecular weights (in kDa) of components of the protein ladder. Representative data of two independent experiments are shown.
Effect of HuNoV VP1 Gene Insertion on RV Replication and Pathogenesis In Vivo.
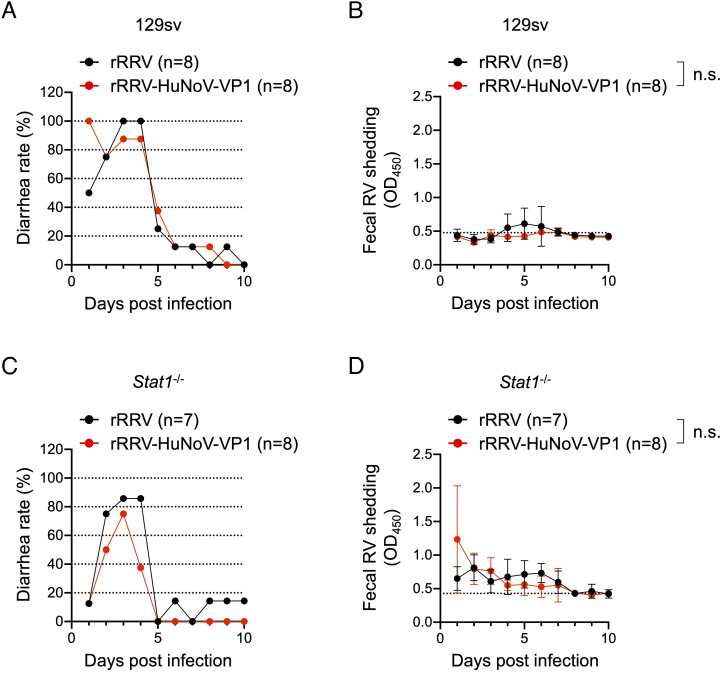
Although RRV does not replicate robustly in the small intestine of neonatal mice, it causes diarrhea in pups at early time points following inoculation (26, 44). To assess the effect of HuNoV VP1 insertion on RRV replication in vivo, we inoculated immunocompetent 5-d-old 129sv pups with 3.9 × 105 FFUs of rRRV-HuNoV-VP1 or the parental rRRV as a control and compared the diarrhea rate and fecal RV shedding over 10 d. Like rRRV, rRRV-HuNoV-VP1 caused diarrhea at early times postinoculation in 129sv pups, but neither virus was detectable by enzyme-linked immunosorbent assay (ELISA) in the stools of the 129sv pups (Fig. 2 A and B). Previous studies demonstrated that RRV replicates better in immunodeficient Stat1−/− pups than in immunocompetent pups, presumably because STAT1 serves as a vital mediator in the interferon-induced anti-RV signaling cascade (26, 27, 45). To test replication of rRRV-HuNoV-VP1 in suckling mice under less restrictive growth conditions, we examined diarrheal rates and fecal RV shedding in 5-d-old Stat1−/− pups. Both rRRV and rRRV-HuNoV-VP1 caused diarrhea from 1 to 4 d postinfection (Fig. 2C), which was less than that of 129sv mice. These data suggest that the innate immune response may play a role in RRV-induced diarrhea. We detected modest fecal RV shedding by rRRV and rRRV-HuNoV-VP1 from 1 to 7 d post-inoculation in the Stat1−/− pups (Fig. 2D). The amount of fecal RV shedding by rRRV-HuNoV-VP1 was not significantly different from that by rRRV (Fig. 2D). These data suggest that administration of rRRV-HuNoV-VP1 was associated with diarrhea to a similar extent as rRRV in both 129sv and Stat1−/− mice and that both viruses were shed in stools significantly more in the Stat1−/− pups following oral inoculation with high titer stock (3.9 × 105 FFU/pups).
Fig. 2.
In vivo characterization of rRRV-HuNoV-VP1. (A and C) Diarrhea rate by rRRV and rRRV-HuNoV VP1 in (A) 129sv or (C) Stat1−/− mice. Five-day-old 129sv pups were orally inoculated with rRRV (n = 8) or rRRV-HuNoV-VP1 (n = 8) (3.9 × 105 FFU/pup), and 5-d-old Stat1−/− pups were orally inoculated with rRRV (n = 7) or rRRV-HuNoV-VP1 (n = 8) (3.9 × 105 FFU/pup). Diarrhea was monitored daily following gentle abdominal palpation until 10 d postinfection. Pups were regarded as positive for diarrhea if liquid or unformed stools were observed. The daily percentage of diarrhea is shown. (B and D) Fecal shedding curve in (B) 129sv or (D) Stat1−/− mice after rRRV or rRRV-HuNoV-VP1 inoculation. Fecal RV antigens in stools collected from 1 to 10 d postinfection were measured by ELISA. Data are shown as mean scores of net optical density at 450 nm (OD450) with SD. The dotted lines show background signals measured by stools from uninfected pups. Statistical analysis was performed with two-way ANOVA. Statistical significance is indicated as n.s.: not significant.
Induction of Serum IgG Responses in 129sv and Stat1−/− Suckling Mice Following rRRV-HuNoV-VP1 Infection.
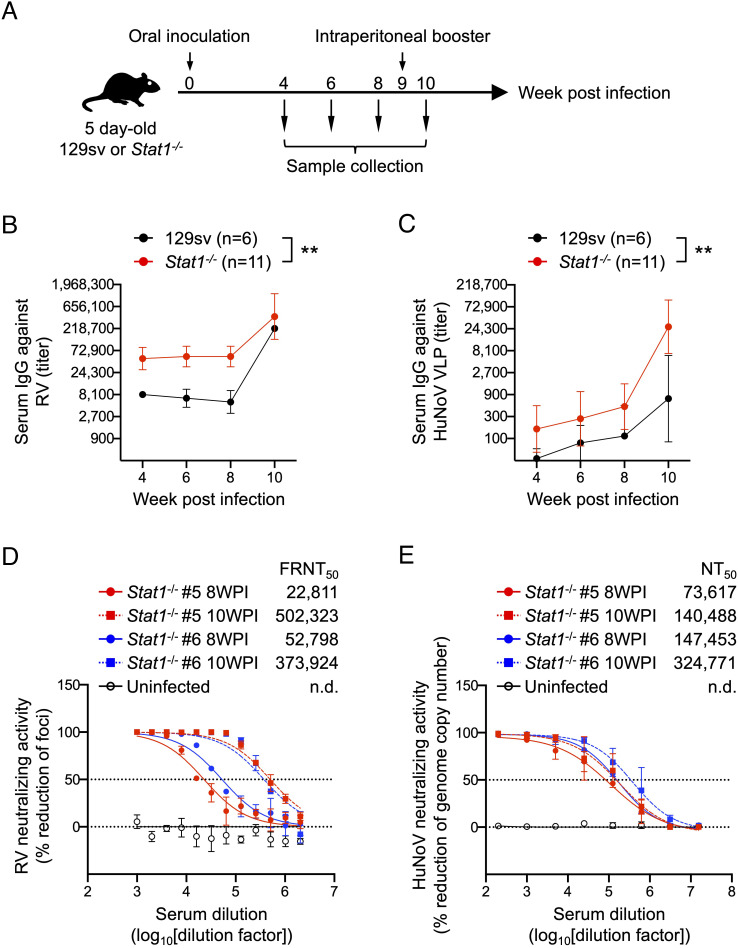
Next, we examined the ability of rRRV-HuNoV-VP1 to induce a systemic antibody response against HuNoV VP1 in the suckling mice. We orally inoculated 5-d-old 129sv and Stat1−/− pups with 3.9 × 105 FFUs of rRRV-HuNoV-VP1 or the wild-type rRRV and collected blood and stool specimens at 4-, 6-, and 8-wk postinoculation (WPI). In the case the primary infection with rRRV-HuNoV-VP1 induced no or a weak immune response, we also provided a parenteral intraperitoneal (IP) booster immunization with the same RV dose (3.9 × 105 FFU/pup) at 9 WPI and collected blood and stool specimens 1 wk later (10 WPI) (Fig. 3A). To examine whether the mouse sera contained antibodies against HuNoV VP1, we first established a new immunostaining assay. We expressed either HuNoV VP1 or RV VP6, a broadly conserved RV antigen, in BHK-T7 cells, and stained for these proteins with the mouse sera collected at 4 WPI. Consistent with previous studies (28), sera from 129sv pups infected with either rRRV-HuNoV VP1 or rRRV group reacted robustly to the BHK-T7 cell expressed RV VP6. However, we did not observe a clear HuNoV VP1 staining signal with these sera (SI Appendix, Fig. S2 A and B). The sera from an uninfected 129sv mouse served as a negative control and, as expected, failed to react with either RV VP6 or HuNoV VP1 (SI Appendix, Fig. S2C). Interestingly, we found that sera from nine of the eleven Stat1−/− mice inoculated with rRRV-HuNoV-VP1 recognized both RV VP6 and HuNoV VP1 (SI Appendix, Fig. S3A). As expected, the sera from Stat1−/− mice infected with rRRV only reacted with RV VP6, while the sera from an uninfected Stat1−/− mouse did not react to either RV VP6 or HuNoV VP1 (SI Appendix, Fig. S3 B and C). These data suggest that the sera from rRRV-HuNoV-VP1-infected Stat1−/− mice reacted specifically to HuNoV VP1.
Fig. 3.
Systemic antibody responses against RV and HuNoV VLPs in 129sv and Stat1−/− mouse following rRRV-HuNoV-VP1 infection. (A) Schematic presentation of the infection and immunization experiments. Five-day-old 129sv (n = 6) or Stat1−/− (n = 11) pups were orally inoculated with rRRV-HuNoV-VP1 (3.9 × 105 FFU/pup) at 0 WPI and intraperitoneally boostered with the virus (3.9 × 105 FFU/pup) at 9 WPI. Blood and stool samples were collected at 4, 6, 8, and 10 WPI. (B and C) Temporal dynamics of serum IgG titers against RV and HuNoV VLP. Mouse sera were serially diluted and tested in ELISA for (B) anti-RV IgG or (C) anti-HuNoV VLP. The IgG titer was defined as the highest serum dilution at which the OD score is higher than an uninfected mouse serum. Data are shown as mean with SD. Statistical analysis was performed by two-way ANOVA. Statistical significance is indicated as **P < 0.01. (D) Serum neutralization activities against RV. Mouse sera were serially diluted, mixed with RRV, and incubated at 37 °C for 1 h. The virus titer in the mixture of the serum and RRV was determined using MA104 cells in an FFU assay. Data are shown as mean scores of the percentage reduction of the focus number with SD. n.d.: not determined. (E) Serum neutralization activities against HuNoV. Mouse sera were serially diluted, mixed with human HuNoV GII.4 Sydney [P16] strain, and incubated at 37 °C for 1 h. The virus titer was determined using HIO monolayers and quantitative real-time RT-PCR 24 h after inoculation. Data are shown as mean scores of the percentage reduction of the number of genomic copies with SD. n.d.: not determined.
To further characterize the serum immunoglobulin G (IgG) antibody responses induced in the 129sv and Stat1−/− mice following rRRV-HuNoV-VP1 infection, we determined the ELISA binding titers of anti-RV and anti-HuNoV VLP IgG at different time points postinoculation. Consistent with the immunostaining data, all sera from both 129sv and Stat1−/− mice infected with rRRV-HuNoV-VP1 reacted to RV as early as 4 WPI, and the serum anti-RV IgG titers increased further by 10 WPI following the IP boost (Fig. 3B and SI Appendix, Table S1). Remarkably, three of six 129sv mice also demonstrated seroconversion to HuNoV VLP at 6 WPI, showing a titer of 1:114 by 8 WPI (Fig. 3C). The IP booster seroconverted two more mice that were negative at 8 WPI and enhanced the titer to 1:736 at 10 WPI (Fig. 3C and SI Appendix, Table S1). Sera from all Stat1−/− mice showed higher serum anti-HuNoV VLP IgG titers than seen in the 129sv mice at all time points (1:494 at 8 WPI and 1:26,852 at 10 WPI) (Fig. 3C and SI Appendix, Table S1). Based on these findings, we concluded that sera from 129sv and Stat1−/− infected with rRRV-HuNoV-VP1 contain antibodies against both RV and HuNoV VLP and that a parenteral IP boost enhanced the immune response to RV and HuNoV VP1.
Neutralization of RV and HuNoV by Sera from rRRV-HuNoV-VP1-Infected Stat1−/− Mice.
We further evaluated the neutralizing activity of serum antibodies against RV and HuNoV. Because Stat1−/− mice #5 and #6 showed the highest anti-HuNoV VLP binding titers among the 11 mice tested (SI Appendix, Table S1), and because of limitations on our ability to carry out multiple HuNoV neutralization assays, we focused neutralization testing on specimens from these two mice (hereafter called sera #5 and #6) from 8 and 10 WPI. Not surprisingly, both sera #5 and #6 neutralized RV. Serum #5 had a 50% focus reduction neutralization titer (FRNT50) of 1:22,811 on 8 WPI and 1:502,323 on 10 WPI, while serum #6 showed a FRNT50 of 1:52,798 on 8 WPI and 1:373,924 on 10 WPI (Fig. 3D). Human HuNoV recognizes the host histo-blood group antigen (HBGA) molecules through the P2 subdomain in the P domain of the HuNoV VP1 protein (46–48). The expression of the P domain can form a self-assembling subunit P particle that exposes the P2 subdomain on its surface and binds to HBGA in the same manner as VLPs (42, 43, 49). As an alternative to a traditional restriction of replication-based neutralization assay, we first assessed whether sera #5 and #6 were capable of blocking P particle binding to HBGA, the cell surface receptor for HuNoV (43). Serum #5 inhibited P particle binding to HBGA at a 50% blocking titer (BT50) of 1:12.5 on 8 WPI and 1:50 on 10 WPI, while serum #6 inhibited binding at a BT50 of 1:6.25 on 10 WPI (Table 1). Furthermore, treatment of HuNoV with sera #5 or #6 prior to infection reduced the HuNoV genome copy number in a human intestinal organoid culture-based neutralization assay (50). Serum #5 had a 50% neutralization titer (NT50) of 1:73,617 on 8 WPI and 1:140,488 on 10 WPI, while serum #6 had a NT50 of 1:147,453 on 8 WPI and 1:324,771 on 10 WPI (Fig. 3E). Collectively, these data support the conclusions that rRRV-HuNoV-VP1 induced antibody responses against both RV and HuNoV in Stat1−/− suckling mice and both postimmunization sera developed the capacity to effectively neutralize HuNoV in two different neutralization assays.
Table 1.
Blocking of HuNoV P particle binding to HBGA by sera from Stat1−/− mice infected with rRRV-HuNoV-VP1
| Sample |
50% Blocking titer (BT50) of HuNoV P particle binding to HBGA |
|---|---|
| Stat1−/− #5 8WPI | 12.5 |
| Stat1−/− #5 10WPI | 50 |
| Stat1−/− #6 8WPI | <12.5 |
| Stat1−/− #6 10WPI | 6.25 |
| Uninfected | <3.125 |
Induction of Fecal IgA Responses in 129sv and Stat1−/− Mice Following rRRV-HuNoV-VP1 Infection.
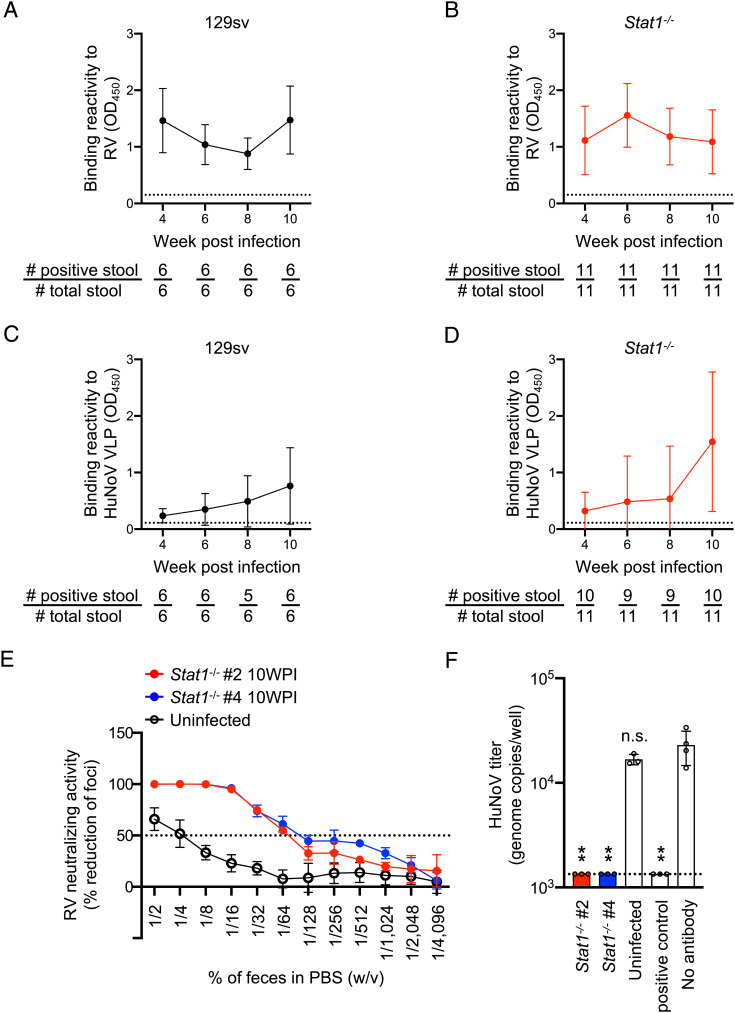
To investigate whether rRRV-HuNoV-VP1 is capable of inducing a mucosal immune response in orally infected mice, we quantified the local enteric immunoglobulin A (IgA) responses to RV and HuNoV in stool specimens collected from the 129sv and Stat1−/− immunized mice. Consistent with previous studies (28, 44, 51, 52), we detected RV-specific IgA in the fecal specimens from both the 129sv and Stat1−/− immunized mice (Fig. 4 A and B). Of note, we found that the postinfection fecal samples also contained HuNoV VLP-specific fecal IgA in the 129sv and Stat1−/− mice (Fig. 4 C and D). We also stained for HuNoV VP1 expressed in BHK-T7 cells with the fecal samples from Stat1−/− mice followed by a secondary antibody specific to the α chain of murine IgA. The BHK-T7 immunostaining also clearly demonstrated that the fecal samples contained HuNoV VP1 specific IgA (SI Appendix, Fig. S5A). Secretory IgA in the intestine has been frequently proposed to be a key component for the protection against enteric pathogens (53). Thus, we tested the ability of the fecal samples to neutralize both RV and HuNoV. We prepared fecal supernatants from Stat1−/− mouse #2 and #4 (hereafter called fecal samples #2 and #4). These mice showed the highest ELISA values against HuNoV VLP at 10 WPI (Fig. 4D and SI Appendix, Table S2). Fecal samples #2 and #4 demonstrated FRNT50 against RV at a dilution of 1/64% (w/v). A control stool suspension from uninfected mice showed a very low background reactivity at lower dilutions of 1/2 and 1/4% (w/v) of feces in phosphate-buffered saline (PBS) (Fig. 4E). Of note, fecal samples #2 and #4 also decreased the titer of HuNoV to the neutralization threshold in our human intestinal organoid assay using 1% (w/v) fecal suspensions (Fig. 4F). These data suggest that rRRV-HuNoV-VP1 induced a functional enteric immune response against both RV and HuNoV in suckling mice.
Fig. 4.
Mucosal antibody responses against RV and HuNoV VLPs in 129sv and Stat1−/− mouse following rRRV-HuNoV-VP1 infection. (A and B) Temporal dynamics of fecal IgA responses to RV. The amount of fecal IgA against RV in (A) 1% (w/v) of fecal suspension from 129sv or (B) 0.1% (w/v) of fecal suspension from Stat1−/− was determined by ELISA. Data are shown as the mean score of the net OD450 with SD. The dotted lines show the limit of detection by stools from uninfected pups. A stool with a higher OD score than the dotted line was considered positive. The number of positive stools and total stools are shown below the graphs. (C and D) Temporal dynamics of fecal IgA responses to HuNoV VLP. The amount of fecal IgA against HuNoV VLP in 10% (w/v) of fecal suspension from (C) 129sv or (D) Stat1−/− pups were determined by ELISA. Data are shown as the mean score of net OD450 with SD. The dotted lines show background signals by stools from uninfected pups. A stool with a higher OD score than the dotted line was considered positive. The number of positive stools and total stools are shown below the graphs. (E) Fecal neutralization activities against RV. Mouse stool suspensions were serially diluted, mixed with RRV, and incubated at 37 °C for 1 h. The virus titer in the mixture of stool and RRV was determined using MA104 cells in an FFU assay. Data are shown as mean scores of the percentage focus reduction of the focus number with SD. (F) HuNoV genome copy numbers in HIOs following infection with HuNoV pretreated with fecal specimens. Mouse stool suspensions [1% (w/v)] were mixed with HuNoV GII.4 Sydney [P16] strain, incubated at 37 °C for 1 h, and inoculated to differentiated HIO monolayers. The virus titer in HIO was determined by quantitative real-time RT-PCR 24 h after inoculation. Data are shown as mean scores of the HuNoV genome copy numbers. Statistical analysis was performed by one-way ANOVA test with Dunnett's post-test. Statistical significance is indicated as n.s.: not significant, **P < 0.01.
Genetic Stability and Antigenicity of a Recombinant RRV Expressing HuNoV P Protein in Suckling Mice.
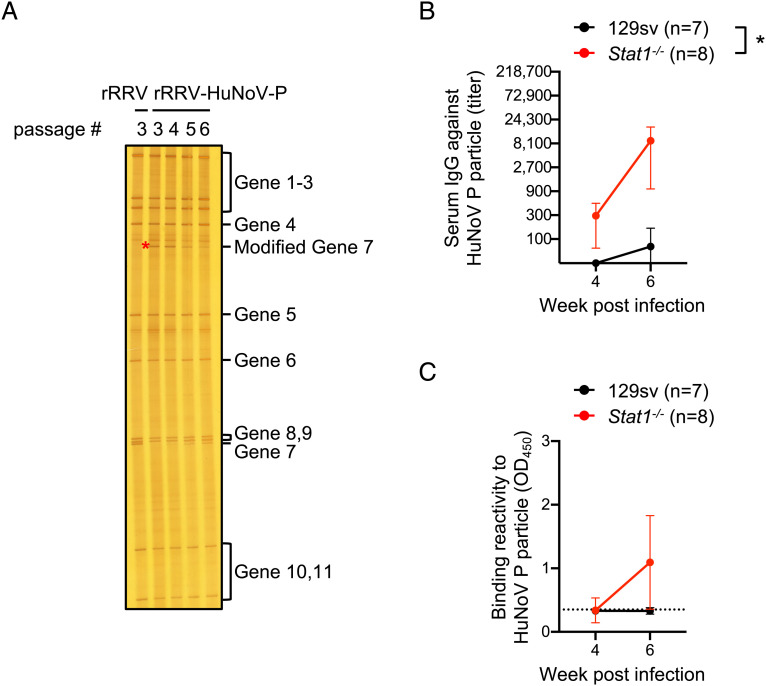
Despite its ability to clearly induce anti-HuNoV VLP antibody responses in immunized mice, rRRV-HuNoV-VP1 lost the HuNoV VP1 gene (1,623 bp) by passage 5 (Fig. 1A). Following the submission of this manuscript, Philip and Patton published a paper demonstrating that the HuNoV P domain (~960 bp) was stably maintained for five passages in RV gene segment 7 by rescuing a recombinant simian RV SA11 strain harboring HuNoV P domain (54). No data on immunogenicity (either systemic or local) was provided in these studies. To test the genetic stability and antigenicity of HuNoV P domain in the RRV genetic backbone, we generated a recombinant RRV harboring the P domain of GII.4 HuNoV VA387 strain whose P particle has been well characterized in terms of structural property and antigenicity (rRRV-HuNoV-P) (SI Appendix, Fig. S6 A and B) (49). On RNA-PAGE analysis, we observed that rRRV-HuNoV-P maintained the modified RV gene segment 7 (2,089 bp) over passages 3 to 6; however, the virus started to show a band corresponding in size to wild-type RV gene segment 7 initially in passage 4 but still present in passage 6 (Fig. 5A). Next, we examined the immunogenicity of rRRV-HuNoV-P in 129sv and Stat1−/− mice following oral inoculation and IP boost immunization with passage 3 stocks (SI Appendix, Fig. S7A). All tested serum and fecal specimens from 129sv and Stat1−/− mice reacted to RV (SI Appendix, Fig. S7 B and C and Tables S3 and S4). Among tested serum and fecal specimens from 129sv, only sera collected after the IP boost reacted to HuNoV P particles (Fig. 5 B and C). As expected, based on prior studies by us and others, serum and fecal specimens from Stat1−/− mice reacted more strongly to HuNoV P particles (Fig. 5 B and C). Notably, selected serum samples from Stat−/− mice blocked HuNoV P particles binding to HBGA, showing enhanced BT50 after the IP boost (Table 2 and SI Appendix, Figs. S3 and S4) which served as a surrogate assay for neutralizing activity. Taken together, these data strongly suggest that rRRV-HuNoV-P demonstrated enhanced genetic stability of inserted HuNoV gene without losing HuNoV antigenicity in immunized mice.
Fig. 5.
Genome stability and antigenicity of a recombinant RRV expressing HuNoV P protein. (A) Electrophoretic analysis of viral dsRNA. Viral dsRNAs were purified from rRRV and different passages of rRRV-HuNoV-P, separated by RNA-PAGE, and visualized by silver staining. The modified gene segment 7 is indicated by a red asterisk. (B) Temporal dynamics of serum IgG titer against HuNoV P particle. Mouse sera were serially diluted and tested in ELISA for anti-HuNoV P particle. The IgG titer was defined as the highest serum dilution at which the OD score is higher than an uninfected mouse serum. Data are shown as mean with SD. Statistical analysis was performed by two-way ANOVA. Statistical significance is indicated as *P < 0.05. (C) Temporal dynamics of fecal IgA responses to HuNoV P particle. The amount of fecal IgA against HuNoV P particle in 10% (w/v) of fecal suspension from 129sv or Stat1−/− pups were determined by ELISA. Data are shown as the mean scores of net OD450 with SD. The dotted lines show background signals by stools from uninfected pups.
Table 2.
Blocking of HuNoV P particle binding to HBGA by sera from Stat1−/− mice infected with rRRV-HuNoV-P
| Sample |
50% Blocking titer (BT50) of HuNoV P particle binding to HBGA |
|---|---|
| Stat1−/− #12 4WPI | 50 |
| Stat1−/− #12 6WPI | 200 |
| Stat1−/− #14 4WPI | <12.5 |
| Stat1−/− #14 6WPI | 200 |
| Stat1−/− #15 4WPI | <12.5 |
| Stat1−/− #15 6WPI | 800 |
| Stat1−/− #17 4WPI | <12.5 |
| Stat1−/− #17 6WPI | 1,600 |
| Uninfected | <12.5 |
Discussion
In this study, we generated recombinant RVs harboring the gene encoding HuNoV capsid protein VP1 and P domain (Figs. 1A and 5A and SI Appendix, Figs. S1A and S6A). The viruses expressed the HuNoV VP1 and P protein in infected cells (Fig. 1 D and E and SI Appendix, Fig. S6B) and induced both systemic and mucosal immune responses to both RV and HuNoV following oral administration of the recombinant RV to naïve suckling mice (Figs. 3 A–E, 4 A–F, and 5 B and C and Tables 1 and 2 and SI Appendix, Figs. S2 A–C, S3 A–C, S4 A and B, S5 A and B, and S7 A–C and Tables S1–S4). Our data provide proof of principle that RV can be engineered as a “dual-use” enteric vaccine vector (i.e., a candidate live viral vaccine for combined RV and HuNoV immunization). We selected the RRV infection mouse model for these initial studies for several reasons. RRV has excellent in vitro replication characteristics as an expression vector. Insertion of exogenous genes into the RV genome often leads to a reduction in virus titer. Thus, it is preferable to use a RV that replicates to a high titer as a vaccine expression vector. In addition, inoculation of a high titer stock was likely more favorable for the enhanced expression of the exogenous protein target (in this case HuNoV VP1 and P protein) in vivo. By virtue of a rhesus rotavirus RRV strain that grows to high titer in cell culture, rRRV-HuNoV-VP1 and rRRV-HuNoV-P could be readily tested at a high dose in this initial proof of concept study. The majority of currently licensed RV vaccines are based either on human RVs or reassortants between human and bovine RVs (11). It seems logical, if a vectored two-in-one RV-based enteric vaccine strategy is further pursued, to focus on modifying one of the already existing safe and effective licensed RV vaccines to express a second target antigen for use in humans as the most straightforward and practical path forward.
Of interest, RRV has already been extensively studied in both humans and animal models to better understand humoral and cellular immune responses to RV (26–28, 44, 45, 55–59). In fact, RRV represents the genetic backbone of a highly immunogenic human RV vaccine in multiple human clinical trials, but had issues related to safety in post-licensure surveillance which precluded its further use as a vaccine (29, 30). Prior studies of RRV infection in the suckling mouse model, including work by our group and others, documented enhanced RRV replication in mice deficient in components of innate immune signaling (26, 27, 45, 60–62). For the reasons considered above, we chose to study both immunocompetent 129sv and immunodeficient Stat1-/- suckling mice. Of note, in a paper by Vancott et al. (45), the antibody response against RV in Stat1−/− mice was noted to be significantly higher than in 129sv mice following enteric RV infection. Given the reduced replication capacity of RRV in immunocompetent mice, we took advantage of the ability to enhance replication and immunogenicity in this only minimally permissive model system by using Stat1−/− mice. As anticipated, rRRV-HuNoV-VP1 and rRRV-HuNoV-P demonstrated enhanced replication in Stat1−/− mice as assessed by the level of RV fecal shedding (Fig. 2 B and D) and by the induction of increased systemic and local intestinal origin antibody responses to RV and HuNoV VLP or P particle (Figs. 3 B and C, 4 A–D, and 5 B and C). These findings strongly suggest that there is a correlation between viral replication in vivo and the level of systemic and local antibodies in our study. Although Stat1−/− mice are more permissive to RRV infection, the lack of STAT1 results in a defect in the interferon signaling pathway which could raise a concern that the immune response in Stat1−/− mice may not reflect a natural immune response to rRRV-HuNoV-VP1 or rRRV-HuNoV-P in an immunocompetent animal model. Fortuitously, we did not see a blunted humoral adaptive immune response to either RV or HuNoV in Stat1−/− mice. Since young immunocompetent RV naïve rats have been shown to actually much more efficiently support RRV infection (59) and gnotobiotic piglets have been shown to have a protective immune response to RV following immunization with RRV (63), we expect that follow-up immunization studies in the neonatal rat and gnotobiotic piglet models will provide further information regarding the immune response to a vectored RV-based dual antigen vaccine constructs under more natural viral replication conditions.
To develop a HuNoV vaccine, several HuNoV VLPs have been assessed in clinical trials (39–41). In addition, some viral vectors expressing HuNoV VLPs have been used to induce a mucosal response against HuNoV VLPs in mice or humans (i.e., Venezuelan equine encephalitis virus replicon, vesicular stomatitis virus, and adenovirus) (5–8). Among these viruses, the adenovirus type 5-based HuNoV vaccine is the only one to have been studied in phase I human clinical trials (8, 40). In that model, development of immunity to the adenovirus type 5 expression vector itself would not add significant protection against a second important pediatric enteric infection and so RV immunity would need to be addressed by a separate vaccine.
By taking advantage of the natural intestinal tissue tropism of RV, we developed an RV-based viral vector against HuNoV that potentially could induce humoral immunity to two separate highly important pediatric enteric pathogens. rRRV-HuNoV-VP1 and rRRV-HuNoV-P induced antibody responses not only in the systemic circulation but also in the GI tract where RVs and HuNoVs preferentially replicate (Figs. 3C, 4 C and D, and 5 B and C). Importantly, we observed the induction of neutralizing antibodies in both sera and stool specimens in rRRV-HuNoV-VP1 inoculated Stat1−/− mice despite the fact that the suckling mice are, at best, only semipermissive for RRV replication (Figs. 3E and 4F). Furthermore, we confirmed that the rRRV-HuNoV-VP1 or rRRV-HuNoV-P-infected Stat1−/− mouse sera blocked HuNoV P particle binding to HBGA (Tables 1 and 2). Because of technical capacity limitations on our ability to carry out multiple HuNoV neutralization assays, we tested HuNoV neutralizing activity with only two serum samples (#5 and #6) and two stool specimens (#2 and #4) that had previously shown the highest score in the ELISA. Future studies should examine whether the virus induces neutralizing antibodies in larger numbers of suckling mice and whether the neutralization response to the HuNoV VP1 or P protein in Stat1−/− mice (or possibly immunocompetent suckling rats) is similar or identical to that generated in humans after natural HuNoV infection.
Although rRRV-HuNoV-VP1 induced immune responses against HuNoV VLP, the HuNoV VP1 sequence (1,623 bp) was not stably maintained in the recombinant RRV (Fig. 1A and SI Appendix, Fig. S1A). Previous studies clearly demonstrated an association between the size of an exogenous gene and its genomic stability during RV replication. Recombinant RVs stably retained foreign genes during multiple rounds of replication when the gene size was less than 1 kbp [e.g., eGFP, ZsGreen, AsRed2, and mCherry (693 to 720 bp), UnaG (420 bp), and nanoluciferase (516 bp)] (12–17, 19, 64). On the other hand, recombinant simian SA11 strain with the gene for SARS-CoV-2 spike protein S1 fragment (2,055 bp) lost a part of the spike protein gene following three serial passages in cell culture (21). Based on the study by Philip and Patton (54), we expected that rRRV-HuNoV-P would stably retain the gene for HuNoV P domain (960 bp). Indeed, the virus did not lose the modified RV gene segment 7 as quickly as rRRV-HuNoV-VP1 (Figs. 1A and 5A). However, we did observe a band corresponding in size to the wild-type RV gene segment 7 after passage 4 (Fig. 5A), which suggests that, in the case of RRV, the virus did not stably retain the HuNoV P domain during multiple passages. The more precise length of time and passage of genetic stability of an exogenous gene in RV expression vectors needs to be resolved more completely in further studies if such recombinant viruses are to be used for actual use of an RV-based enteric vaccine vector in humans. A potential solution to increase the genetic stability of the exogenous gene even further is to include a larger sequence of 3′ untranslated region (UTR) of RV gene (64).
If these initial HuNoV immunization proof of concept studies prove successful, the RV-based dual vaccine vector system could be expanded to other small enteric immune target antigens. Potential candidates might include astrovirus capsid protein and Escherichia coli heat-labile enterotoxin produced by enterotoxigenic E. coli (65, 66). The current findings provide an initial demonstration of the feasibility of developing a new approach to inducing mucosal immune responses to multiple enteric pathogens using a single enteric virus as simultaneously a live RV vaccine and a viral vaccine vector. The data presented here potentially have broad practical implications for the design of RV-based multivalent mucosal vaccine candidates against other common viral, bacterial, and parasitic enteric pathogens.
Materials and Methods
The rescue plasmids (pT7-RRV-NSP3-HuNoV-VP1 and pT7-RRV-NSP3-HuNoV-P) were constructed by replacing the GFP gene in pT7-RRV-NSP3-GFP constructed previously (12) with those encoding the HuNoV VP1 (GII.4 Sydney strain) or the HuNoV P domain (GII.4 VA387 strain). Recombinant viruses (rRRV-HuNoV-VP1 and rRRV-HuNoV-P) were rescued using an optimized reverse genetics protocol (12). HuNoV VP1 or P protein expression was analyzed by western blotting and immunostaining in MA104 cells. To check serum and mucosal antibody responses to RV, HuNoV VLP, or HuNoV P particle in vivo, 5-d-old 129sv or Stat1−/− mice were orally inoculated with rRRV-HuNoV-VP1 or rRRV-HuNoV-P and intraperitoneally boosted with the same viruses. Serum IgG and fecal IgA responses to RV, HuNoV VLP, and HuNoV P particles were determined by ELISA using RV infected cell lysate, HuNoV VLP (GII.4 MD145 strain) (67), or HuNoV P particles (GII.4 VA387 strain) (42) as antigens. Blocking activity in selected serum specimens of the binding of HuNoV P particles (GII.4 VA387 strain) to HBGA was quantified (43). Neutralizing activities in selected serum and fecal specimens to HuNoV were evaluated by determining the reduction of HuNoV (GII.4 Sydney strain) genome copy number in human intestinal organoids (HIOs) as described (50). More detailed descriptions of the materials and methods, including plasmid constructions, reverse genetics protocols, mouse experimental protocols, immunofluorescent staining and western blotting procedures, ELISAs used to detect serum IgG and fecal IgA to RV and HuNoV, HIOs culture conditions, and RV and HuNoV neutralizing assays are provided in SI Appendix.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We thank members of the Greenberg and Ding lab for the helpful discussion, Linda Jacob for her secretarial work, and Taufeeq Ahmed for his technical assistance. We thank Dr. Philippe H. Jaïs [Eukarÿs société par actions simplifiée (SAS)] for sharing the C3P3-G1 plasmid, Dr. Susana López (Universidad Nacional Autónoma de México) for sharing the rhesus rotavirus (RRV) rescue plasmid, Dr. Sean Tucker (Vaxart Inc.) for providing the plasmid encoding HuNoV VP1, and Dr. Didier Poncet (Université Paris-Saclay) for sharing rabbit antiserum to RV NSP3. The findings and conclusions in this article are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention. This work is supported by Stanford Maternal and Child Health Research Institute Postdoctoral Support FY2020 (T.K.), Division of Intramural Research, National Institute of Allergy and Infectious Diseases (NIAID) (K.Y.G.), R01 AI150796 (S.D.), R01 AI125249, U19 AI116484 NIH grants, and a Veterans Affairs (VA) Merit Grant (GRH0022) (H.B.G.).
Author contributions
S.D. and H.B.G. designed research; T.K., L.S.-T., N.F., V.P.C., and M.T. performed research; T.K., N.F., V.P.C., and M.T. analyzed data; V.P.C., M.T., X.J., K.Y.G., and J.V. contributed new reagents/analytic tools; and T.K., S.D., and H.B.G. wrote the paper.
Competing interests
The authors have organizational affiliations to disclose: Greenberg currently consults for the following companies: Pfizer, Vaxart, FluGen, and Aridis.
Footnotes
This article is a PNAS Direct Submission.
Contributor Information
Siyuan Ding, Email: siyuan.ding@wustl.edu.
Harry B. Greenberg, Email: harry.greenberg@stanford.edu.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Ball J. M., et al. , Recombinant Norwalk virus-like particles given orally to volunteers: Phase I study. Gastroenterology 117, 40–48 (1999). [DOI] [PubMed] [Google Scholar]
- 2.Hird T. R., Grassly N. C., Systematic review of mucosal immunity induced by oral and inactivated poliovirus vaccines against virus shedding following oral poliovirus challenge. PLoS Pathog. 8, e1002599 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mohn K. G., Smith I., Sjursen H., Cox R. J., Immune responses after live attenuated influenza vaccination. Hum. Vaccin. Immunother. 14, 571–578 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Minor P. D., Live attenuated vaccines: Historical successes and current challenges. Virology 479–480, 379–392 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Baric R. S., et al. , Expression and self-assembly of norwalk virus capsid protein from venezuelan equine encephalitis virus replicons. J. Virol. 76, 3023–3030 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harrington P. R., Lindesmith L., Yount B., Moe C. L., Baric R. S., Binding of Norwalk virus-like particles to ABH histo-blood group antigens is blocked by antisera from infected human volunteers or experimentally vaccinated mice. J. Virol. 76, 12335–12343 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma Y., Li J., Vesicular stomatitis virus as a vector to deliver virus-like particles of human norovirus: A new vaccine candidate against an important noncultivable virus. J. Virol. 85, 2942–2952 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim L., et al. , Safety and immunogenicity of an oral tablet norovirus vaccine, a phase I randomized, placebo-controlled trial. JCI Insight 3, e121077 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Desselberger U., Huppertz H. I., Immune responses to rotavirus infection and vaccination and associated correlates of protection. J. Infect. Dis. 203, 188–195 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Angel J., Franco M. A., Greenberg H. B., Rotavirus immune responses and correlates of protection. Curr. Opin. Virol. 2, 419–425 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Glass R. I., Tate J. E., Jiang B., Parashar U., The rotavirus vaccine story: From discovery to the eventual control of rotavirus disease. J. Infect. Dis. 224, S331–S342 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sánchez-Tacuba L., et al. , An optimized reverse genetics system suitable for efficient recovery of simian, human, and murine-like rotaviruses. J. Virol. 94, e01294-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu Y., et al. , A recombinant murine-like rotavirus with nano-luciferase expression reveals tissue tropism, replication dynamics, and virus transmission. Front. Immunol. 13, 911024 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kanai Y., et al. , Entirely plasmid-based reverse genetics system for rotaviruses. Proc. Natl. Acad. Sci. U.S.A. 114, 2349–2354 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Komoto S., et al. , Generation of recombinant rotaviruses expressing fluorescent proteins by using an optimized reverse genetics system. J. Virol. 92, e00588-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kanai Y., et al. , Development of stable rotavirus reporter expression systems. J. Virol. 93, e01774-18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Philip A. A., et al. , Generation of recombinant rotavirus expressing NSP3-UnaG fusion protein by a simplified reverse genetics system. J. Virol. 93, e01616-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang-Graham A. L., et al. , Rotavirus calcium dysregulation manifests as dynamic calcium signaling in the cytoplasm and endoplasmic reticulum. Sci. Rep. 9, 10822 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Philip A. A., Patton J. T., Expression of separate heterologous proteins from the rotavirus NSP3 genome segment using a translational 2A stop-restart element. J. Virol. 94, e00959-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Papa G., et al. , CRISPR-Csy4-mediated editing of rotavirus double-stranded RNA genome. Cell Rep. 32, 108205 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Philip A. A., Patton J. T., Rotavirus as an expression platform of domains of the SARS-CoV-2 spike protein. Vaccines (Basel) 9, 449 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Diebold O., et al. , Using species a rotavirus reverse genetics to engineer chimeric viruses expressing SARS-CoV-2 spike epitopes. J. Virol. 96, e0048822 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stuker G., Oshiro L. S., Schmidt N. J., Antigenic comparisons of two new rotaviruses from rhesus monkeys. J. Clin. Microbiol. 11, 202–203 (1980). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kitamoto N., Ramig R. F., Matson D. O., Estes M. K., Comparative growth of different rotavirus strains in differentiated cells (MA104, HepG2, and CaCo-2). Virology 184, 729–737 (1991). [DOI] [PubMed] [Google Scholar]
- 25.Fenaux M., Cuadras M. A., Feng N., Jaimes M., Greenberg H. B., Extraintestinal spread and replication of a homologous EC rotavirus strain and a heterologous rhesus rotavirus in BALB/c mice. J. Virol. 80, 5219–5232 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feng N., et al. , Role of interferon in homologous and heterologous rotavirus infection in the intestines and extraintestinal organs of suckling mice. J. Virol. 82, 7578–7590 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feng N., Yasukawa L. L., Sen A., Greenberg H. B., Permissive replication of homologous murine rotavirus in the mouse intestine is primarily regulated by VP4 and NSP1. J. Virol. 87, 8307–8316 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ishida S. I., Feng N., Gilbert J. M., Tang B., Greenberg H. B., Immune responses to individual rotavirus proteins following heterologous and homologous rotavirus infection in mice. J. Infect. Dis. 175, 1317–1323 (1997). [DOI] [PubMed] [Google Scholar]
- 29.Midthun K., Kapikian A. Z., Rotavirus vaccines: An overview. Clin. Microbiol. Rev. 9, 423–434 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Widdowson M. A., Bresee J. S., Gentsch J. R., Glass R. I., Rotavirus disease and its prevention. Curr. Opin. Gastroenterol. 21, 26–31 (2005). [PubMed] [Google Scholar]
- 31.Jones M. K., et al. , Enteric bacteria promote human and mouse norovirus infection of B cells. Science 346, 755–759 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ettayebi K., et al. , Replication of human noroviruses in stem cell-derived human enteroids. Science 353, 1387–1393 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Greenberg H. B., et al. , Proteins of Norwalk virus. J. Virol. 37, 994–999 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xi J. N., Graham D. Y., Wang K. N., Estes M. K., Norwalk virus genome cloning and characterization. Science 250, 1580–1583 (1990). [DOI] [PubMed] [Google Scholar]
- 35.Glass P. J., et al. , Norwalk virus open reading frame 3 encodes a minor structural protein. J. Virol. 74, 6581–6591 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiang X., Wang M., Graham D. Y., Estes M. K., Expression, self-assembly, and antigenicity of the Norwalk virus capsid protein. J. Virol. 66, 6527–6532 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prasad B. V., Rothnagel R., Jiang X., Estes M. K., Three-dimensional structure of baculovirus-expressed Norwalk virus capsids. J. Virol. 68, 5117–5125 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prasad B. V., et al. , X-ray crystallographic structure of the Norwalk virus capsid. Science 286, 287–290 (1999). [DOI] [PubMed] [Google Scholar]
- 39.Zhang M., Fu M., Hu Q., Advances in human norovirus vaccine research. Vaccines (Basel) 9, 732 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tan M., Norovirus vaccines: Current clinical development and challenges. Pathogens 10, 1641 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vesikari T., et al. , Immunogenicity of a bivalent virus-like particle norovirus vaccine in children from 1 to 8 years of age: A phase 2 randomized, double-blind study. Vaccine 40, 3588–3596 (2022). [DOI] [PubMed] [Google Scholar]
- 42.Tan M., Jiang X., The P domain of norovirus capsid protein forms a subviral particle that binds to histo-blood group antigen receptors. J. Virol. 79, 14017–14030 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tan M., et al. , Noroviral P particle: Structure, function and applications in virus-host interaction. Virology 382, 115–123 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Feng N., Burns J. W., Bracy L., Greenberg H. B., Comparison of mucosal and systemic humoral immune responses and subsequent protection in mice orally inoculated with a homologous or a heterologous rotavirus. J. Virol. 68, 7766–7773 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vancott J. L., McNeal M. M., Choi A. H., Ward R. L., The role of interferons in rotavirus infections and protection. J. Interferon Cytokine Res. 23, 163–170 (2003). [DOI] [PubMed] [Google Scholar]
- 46.Bu W., et al. , Structural basis for the receptor binding specificity of Norwalk virus. J. Virol. 82, 5340–5347 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Choi J. M., Hutson A. M., Estes M. K., Prasad B. V., Atomic resolution structural characterization of recognition of histo-blood group antigens by Norwalk virus. Proc. Natl. Acad. Sci. U.S.A. 105, 9175–9180 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tenge V. R., et al. , Glycan recognition in human norovirus infections. Viruses 13, 2066 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tan M., Jiang X., Norovirus P particle: A subviral nanoparticle for vaccine development against norovirus, rotavirus and influenza virus. Nanomedicine (Lond.) 7, 889–897 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Costantini V., et al. , Human norovirus replication in human intestinal enteroids as model to evaluate virus inactivation. Emerg. Infect. Dis. 24, 1453–1464 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Merchant A. A., Groene W. S., Cheng E. H., Shaw R. D., Murine intestinal antibody response to heterologous rotavirus infection. J. Clin. Microbiol. 29, 1693–1701 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Khoury C. A., Brown K. A., Kim J. E., Offit P. A., Rotavirus-specific intestinal immune response in mice assessed by enzyme-linked immunospot assay and intestinal fragment culture. Clin. Diagn. Lab. Immunol. 1, 722–728 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pietrzak B., Tomela K., Olejnik-Schmidt A., Mackiewicz A., Schmidt M., Secretory IgA in intestinal mucosal secretions as an adaptive barrier against microbial cells. Int. J. Mol. Sci. 21, 9254 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Philip A. A., Patton J. T., Generation of recombinant rotaviruses expressing human norovirus capsid proteins. J. Virol. 96, e0126222 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kapikian A. Z., et al. , Rotavirus: The major etiologic agent of severe infantile diarrhea may be controllable by a “Jennerian” approach to vaccination. J. Infect. Dis. 153, 815–822 (1986). [DOI] [PubMed] [Google Scholar]
- 56.Vesikari T., Kapikian A. Z., Delem A., Zissis G., A comparative trial of rhesus monkey (RRV-1) and bovine (RIT 4237) oral rotavirus vaccines in young children. J. Infect. Dis. 153, 832–839 (1986). [DOI] [PubMed] [Google Scholar]
- 57.Christy C., et al. , Safety and immunogenicity of live attenuated rhesus monkey rotavirus vaccine. J. Infect. Dis. 154, 1045–1047 (1986). [DOI] [PubMed] [Google Scholar]
- 58.Ciarlet M., Estes M. K., Conner M. E., Simian rhesus rotavirus is a unique heterologous (non-lapine) rotavirus strain capable of productive replication and horizontal transmission in rabbits. J. Gen. Virol. 81, 1237–1249 (2000). [DOI] [PubMed] [Google Scholar]
- 59.Ciarlet M., Conner M. E., Finegold M. J., Estes M. K., Group A rotavirus infection and age-dependent diarrheal disease in rats: A new animal model to study the pathophysiology of rotavirus infection. J. Virol. 76, 41–57 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Broquet A. H., Hirata Y., McAllister C. S., Kagnoff M. F., RIG-I/MDA5/MAVS are required to signal a protective IFN response in rotavirus-infected intestinal epithelium. J. Immunol. 186, 1618–1626 (2011). [DOI] [PubMed] [Google Scholar]
- 61.Sen A., et al. , Innate immune response to homologous rotavirus infection in the small intestinal villous epithelium at single-cell resolution. Proc. Natl. Acad. Sci. U.S.A. 109, 20667–20672 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lin J. D., et al. , Distinct roles of type I and type III interferons in intestinal immunity to homologous and heterologous rotavirus infections. PLoS Pathog. 12, e1005600 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gaul S. K., Simpson T. F., Woode G. N., Fulton R. W., Antigenic relationships among some animal rotaviruses: Virus neutralization in vitro and cross-protection in piglets. J. Clin. Microbiol. 16, 495–503 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Desselberger U., What are the limits of the packaging capacity for genomic RNA in the cores of rotaviruses and of other members of the Reoviridae? Virus Res. 276, 197822 (2020). [DOI] [PubMed] [Google Scholar]
- 65.Ykema M., Tao Y. J., Structural Insights into the human astrovirus capsid. Viruses 13, 821 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Croxen M. A., Finlay B. B., Molecular mechanisms of Escherichia coli pathogenicity. Nat. Rev. Microbiol. 8, 26–38 (2010). [DOI] [PubMed] [Google Scholar]
- 67.Bok K., et al. , Chimpanzees as an animal model for human norovirus infection and vaccine development. Proc. Natl. Acad. Sci. U.S.A. 108, 325–330 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
All study data are included in the article and/or SI Appendix.