Abstract
CA150 represses RNA polymerase II (RNAPII) transcription by inhibiting the elongation of transcripts. The FF repeat domains of CA150 bind directly to the phosphorylated carboxyl-terminal domain of the largest subunit of RNAPII. We determined that this interaction is required for efficient CA150-mediated repression of transcription from the α4-integrin promoter. Additional functional determinants, namely, the WW1 and WW2 domains of CA150, were also required for efficient repression. A protein that interacted directly with CA150 WW1 and WW2 was identified as the splicing-transcription factor SF1. Previous studies have demonstrated a role for SF1 in transcription repression, and we found that binding of the CA150 WW1 and WW2 domains to SF1 correlated exactly with the functional contribution of these domains for repression. The binding specificity of the CA150 WW domains was found to be unique in comparison to known classes of WW domains. Furthermore, the CA150 binding site, within the carboxyl-terminal half of SF1, contains a novel type of proline-rich motif that may be recognized by the CA150 WW1 and WW2 domains. These results support a model for the recruitment of CA150 to repress transcription elongation. In this model, CA150 binds to the phosphorylated CTD of elongating RNAPII and SF1 targets the nascent transcript.
A complex array of general transcription factors, DNA binding activators and repressors, and a multitude of coregulators mediate transcription in eukaryotes (32, 48, 49). Regulation of the frequency of transcription initiation is a well-documented means of controlling gene expression (32, 48, 64), but many genes are also controlled by modulation of the ability of RNA polymerase II (RNAPII) to elongate transcripts (11, 15–17, 37, 53, 82, 93). Although the mechanisms controlling RNAPII elongation efficiency are not completely understood, it is clear that the interplay of multiple protein factors regulates RNAPII elongation efficiency (21, 70, 75). Some of these elongation factors have been identified, and they belong to two classes: positive transcription elongation factors (P-TEFs), such as positive elongation factor b (P-TEFb) (38, 47, 67, 68), and negative transcription elongation factors (N-TEFs), such as DRB sensitivity-inducing factor (DSIF) and negative elongation factor (NELF) (28, 84, 90). In addition to trans-acting elongation factors, nucleic acid sequences in the template and transcript can modulate elongation (11, 44, 82). A topic central to elongation control is the role of the carboxyl-terminal domain (CTD) of the largest subunit of RNAPII (25). The CTD contains 52 repeats of a 7-amino-acid sequence with the consensus YSPTSPS and is the substrate for several kinases, including P-TEFb (25, 68, 99). Phosphorylation of the CTD occurs during a transitional step of the transcription cycle, the switch from the initiation phase to the elongation phase. Hypophosphorylated RNAPII (designated RNAPIIA) is preferentially recruited to a promoter and initiates transcription. Subsequently, the RNAPII becomes hyperphosphorylated (RNAPIIO) as it clears the promoter. The CTD acts as a platform that recruits regulatory factors to RNAPII transcription complexes, and the phosphorylation of the CTD serves as a switch to regulate this recruitment. Regulatory initiation factors, such as the Mediator complex, bind to the hypophosphorylated CTD (48, 60). CTD phosphorylation causes the release of these initiation factors from the CTD and allows elongation factors to bind (65, 87). The phosphorylated CTD (phospho-CTD) can also recruit pre-mRNA processing proteins, which include factors involved in 5′-end cap formation (e.g., capping enzyme) and splicing (e.g., the U1 small nuclear ribonucleoprotein snRNP-associated protein Prp40) (10, 22, 27, 30, 34, 35, 39, 54, 56, 58, 59, 62, 76, 94). The CTD also functions in 3′-end formation events (34, 55). Thus, the CTD of RNAPII serves as a control center for regulating transcript initiation, elongation, and processing. There are compelling reports that these processes are functionally coupled in cells and extracts (4, 23, 24, 29, 96).
The human transcription factor CA150 is a negative regulator of RNAPII transcription elongation (80, 81). Overexpression of CA150 in human cells inhibits transcript elongation in a promoter-specific manner, affecting the human immunodeficiency virus long terminal repeat and the human α4-integrin promoter while having no influence on several other viral promoters (such as simian virus 40 and cytomegalovirus promoters) (80). The specificity of the repression is dictated by core promoter elements, mainly the TATA box (80), which has also been observed for Tat-mediated activation of elongation (15, 50, 63). Although the mechanism of CA150-mediated repression is not known, an insight into this process was the discovery that CA150 binds directly to the phospho-CTD of RNAPII (18). The primary sequence of CA150 contains several types of domains that characteristically function to mediate protein-protein interactions. Within the carboxyl-terminal half of CA150, there are six repeats of a recently identified sequence element termed the FF repeat motif, so called because of flanking conserved phenylalanine residues (7, 18). The FF repeats are protein interaction modules, about 50 amino acids in length, which have a predicted α-helical structure. It was shown that the CA150 FF repeats are responsible for binding to the phospho-CTD of RNAPII (18). This finding led us to hypothesize that CA150 may target RNAPII elongation complexes by binding to the phosphorylated CTD.
The amino-terminal half of CA150 contains three WW domains. These domains are versatile protein interaction modules, about 35 amino acids in length, that form a stable triple-stranded, antiparallel, β-sheet structure (77–79, 95). These compact WW domains interact with specific types of short proline-rich polypeptide sequences. Four classes of proline-rich ligands have been identified for WW domains: PPXY, PPLP, PR, and phospho-SP/phospho-TP (9, 78). Individual WW domains generally recognize one type of ligand, and the determinants of WW domain specificity are only now becoming understood (51, 83, 95). The ligands of the CA150 WW domains and their function in transcription were not known before the findings presented here.
In this study, we assessed the function of the protein interaction domains of CA150 in transcription. First, we tested whether the CA150 FF repeats, which bind to the phosphorylated CTD of RNAPII, were required for CA150-mediated repression of transcription. Deletion of the FF repeats caused a loss of function in repression, suggesting that interaction with the phospho-CTD was a key component in the pathway of CA150-mediated repression. Additionally, the amino-terminal half of CA150, which contains the WW domains, was necessary for efficient repression. We determined that WW1 and WW2 were important for CA150-mediated repression while WW3 was not required. This result led us to hypothesize that the CA150 WW1 and WW2 domains may bind to factors necessary for repression. To identify this protein(s), we used a binding assay to purify a protein from HeLa cells that interacted specifically with the CA150 WW1 and WW2 domains. This protein, splicing factor 1 (SF1), was originally identified as a constitutive splicing factor (40, 43); however, it has also been shown to repress transcription (97, 98). We have mapped the CA150 binding site in SF1 to a small proline-rich motif present in the carboxyl-terminus. This sequence may represent a new class of WW ligand. Finally, we compared the ligand binding specificity of CA150 WW domains to that of other classes of WW domains and ligands. The implications of these findings in relation to transcription regulation and pre-mRNA splicing are discussed.
MATERIALS AND METHODS
Plasmids.
All glutathione S-transferase (GST) fusion constructs were made in the pGEX2TK vector (Amersham-Phamacia) by cloning PCR products into the BamHI and EcoRI sites. PCR cloning was done with Pfu-Turbo (Stratagene) or Bio-X-Act (Denville Scientific) DNA polymerases. The pGEX2TK-N-CA150 construct contains amino acids 235 to 631 of CA150 fused to GST. pGEX2TK-WW1 contains CA150 amino acids 129 to 169, pGEX2TK-WW2 contains amino acids 427 to 467, and pGEX2TK-WW3 contains amino acids 526 to 566. CA150 expression constructs were cloned by inserting CA150 PCR products with BglII ends into the BamHI site of the mammalian expression vector pEFBOST7, which contains an amino-terminal T7 epitope tag. pEFBOST7 CA150 was described previously (80). SF1 constructs were subcloned from the SF1-Bo isoform, provided by Angela Kramer (Université de Genève, Geneva, Switzerland), by inserting SF1 PCR products into the BamHI and XbaI sites of the pcDNA3.1HisC mammalian expression vector (Invitrogen). This vector contains both His6 and T7 epitope tags at the amino terminus. The SF1-Bo isoform is identical to HeLa SF1 (SF1-HL1 and HL2) except for 42 amino acids at the carboxyl terminus (2, 42). LacZ-SF1(aa420–500) containing amino acids 420 to 500 of SF1-Bo and LacZ-SF1(aa461–500) containing amino acids 461 to 500 of SF1 were cloned by insertion of SF1 PCR fragments with KpnI ends into pcDNA3.1HisBLacZ (Invitrogen). The α4-integrin promoter reporter gene p(−300)Alpha-4CAT and the transfection efficiency control reporter pTK-LUC were described previously (80). The pcDNA3.1HisC-LacZ transfection efficiency control plasmid was purchased from Invitrogen.
Site-directed mutagenesis.
Site-directed mutagenesis of CA150 WW domains was performed using the Quickchange (Stratagene) method as specified by the manufacturer. For the mutation of the WW domains in CA150, the following constructs were generated by site-directed mutagenesis: pEFBOST7-CA150 WW1mt had amino acids Y148 Y149 Y150 changed to AAA, pEFBOST7-CA150 WW2mt had amino acids Y446 Y447 Y448 changed to AAA, pEFBOST7-CA150 WW3 had amino acids F545 F546 Y547 changed to AAA, and pEFBOST7-CA150 WW1mt+WW2mt had amino acids Y148 Y149 Y150 changed to AAA and amino acids Y446 Y447 Y448 changed to AAA. The GST expression vector pGEXTK-N-CA150 was used as a template to create WW2 or WW3 mutants in the N-CA150 far-Western probe. pGEX2TK-N-CA150 WW2mt was created by mutating amino acids Y446 Y447 Y448 to AAA. pGEX2TK-N-CA150 WW3mt was created by changing amino acids F545 F546 Y547 to AAA.
GST fusion protein purification.
GST fusion proteins were purified from Escherichia coli BL21 (Stratagene) grown to an optical density at 600 nm of 0.6 and then induced for 2 h using 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Cells were lysed with lysozyme in 1× phosphate-buffered saline (PBS) and sonication three times for 30 s at 10 W. Triton X-100 was added to a final concentration of 1%. Cell debris was removed by centrifugation at 10,000 × g for 30 min. A 1-ml bed volume of glutathione-agarose beads (Sigma) was added to the supernatant and allowed to bind for 1 h at 4°C. The beads were washed three times with 1× PBS containing 1 M NaCl. Purified proteins were eluted from the beads in 50 mM Tris-HCl (pH 8.0) containing 10 mM glutathione. Protein concentrations were quantified by the Bradford assay (Bio-Rad).
Immunoprecipitations.
GST and CA150 antibodies were antigen affinity purified from the same rabbit polyclonal serum (raised against a GST-CA150 fusion protein) by the method of Harlow and Lane (33). A 50-μl volume of HeLa cell nuclear extract was diluted to 200 μl (final volume) with IP buffer (20 mM HEPES [pH 7.9], 150 mM KCl, 20% glycerol, 1 mM dithiothreitol, 1% Triton- X-100, 0.5% NP-40, 0.2 mM EDTA). Then 15 μg of each antibody was added to the diluted nuclear extract, and the mixture was incubated with end-over-end rotation at 4°C for 4 h. Immune complexes were collected with 500 to 750 μg of magnetic protein A beads (BioMag protein A beads; Perceptive Biosciences) and a magnet. The pellets were washed four times with 1 ml of IP buffer by rotating for 5 min at 4°C. The pellets were then separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels (12.5% polyacrylamide) and analyzed by Western blotting with anti-CA150 antibody and by far-Western blotting with the N-CA150 probe.
Western and far-Western blotting.
To prepare the membranes for use in the Western and far-Western assays, protein samples were separated by SDS-PAGE and transferred to Immobilon-P membranes (Millipore). Western detection was performed using standard techniques. The monoclonal anti-T7 antibody (Novagen) or anti-HisG (Invitrogen) was used to detect T7 or His6 epitope-tagged proteins, respectively. CA150 was detected using rabbit polyclonal antibodies. Enhanced chemiluminescence (Amersham-Pharmacia) was used for all Western blot detections.
Far-Western assays were performed using radioactive probes consisting of GST fusions proteins produced and purified for E. coli. These proteins possess a phosphorylation site in the linker between the GST moiety and the CA150 fragment, which permitted labeling with heart kinase (Sigma) and [γ-32P]ATP. Probes were labeled and purified by centrifugation-chromatography through Sephadex G-50 (Amersham-Pharmacia). The specific activity of the probes was quantified with a scintillation counter. Portions (50 μg) of each probe were used in a final volume of 50 ml of PBSTM (PBS with 0.1% Tween 20 and 5% nonfat dry milk) at 500,000 cpm/ml. This amount gives a final concentration of probe at 33 nM for the GST-WW1, GST-WW2, and GST-WW3 proteins. The concentration of the N-CA150 probe was 16 nM. Target proteins were separated by SDS-PAGE and blotted to Immobilon-P membranes. The membranes were blocked for at least 1 h in PBSTM, and probe was added. Binding was allowed to occur for 4 h at 4°C on a rotating shaker. The blots were then washed three times at 25°C with 100 ml of PBSTM for 10 min per wash. The blot was dried and visualized using a PhosphorImager (Molecular Dynamics).
Far-Western assays of 293T whole-cell lysates (WCL) were prepared from cells transiently transfected with 2 μg of SF1 or lacZ expression vectors using Lipofectamine. The cells were grown for 48 h, washed with PBS, resuspended in 150 μl of PBS, and lysed by three cycles of freezing and thawing. Then 20-μl volumes of the WCL were analyzed by far-Western blotting as described above.
The peptide probes used to characterize the CA150 WW domain binding specificity are as follows: SmB, biotin-PPGMRPPPPGMRRGPPPPGMRPPRP; CDC25, biotin-SGSGEQPLphospho-TPVTDL; Ld10, biotin-SGSGAPPTPPPLPP; WBP1, biotin-SGSGGTPPPPYTVG; and P3, biotin-GVSVRGRGAAPPPPPVPRGRGVGP. These probes were detected using streptavidin-horseradish peroxidase and enhanced chemiluminescence (Amersham-Pharmacia). The SmB peptide is from the SmB subunit common to the U snRNP complexes. Ld10 peptide is from the Limb deformity 10 gene. The P3 peptide is from the Sam68 protein. The FBP11 and FBP21 GST fusions contain the two WW domains from each respective protein as described previously (19). The FBP30 GST fusion contains WW domain A and was first described by Bedford et al. (6, 9). The YAP WW domain was described by Bedford et al. (5). The PIN1 WW domain was provided by Gerhard Niederfellner.
Transfection of 293T cells.
The human cell line 293T was used in all cell culture experiments. Transfections were performed using Lipofectamine as specified by the manufacturer (Gibco BRL). Cells were grown on BioCoat poly-d-lysine six-well plates (Beckton-Dickinson) for transfection. Liposomes were formed and added to the cells for 5 h. For functional assays, 1.5 μg of the p(−300)Alpha-4CAT was used as a reporter construct along with either 10 ng of pHSV-TK-Luc or pcDNA-LacZ as transfection efficiency controls. The amounts of CA150 expression constructs were carefully controlled by using equimolar amounts of the constructs in each transfection, with the remaining mass of DNA transfected always balanced with carrier tRNA. Portions (2 μg) of pEFBOST7-CA150 expression plasmids or pEFBOST7 vector control were cotransfected with the reporter plasmids. Following transfection, the cells were grown for 48 h and then harvested for enzyme assays. The cells were washed twice in PBS and then resuspended in 150 μl of 50 mM Tris-HCl(pH 8.0). WCL were prepared by three cycles of freezing and thawing rapidly from −80 to 37°C. Cellular debris was removed by centrifuging at 20,000 × g for 5 min at 4°C. The supernatant was then removed and stored as the WCL.
Reporter gene assays.
Chloramphenicol acetyltransferase (CAT) and luciferase assays were performed as described previously (80). β-Galactosidase activity was measured using chlorophenol rad-β-d-galactopyranoside (CPRG) substrate. A 10-μg portion of WCL was mixed with CPRG reagent and incubated for 7 min at 25°C. Reactions were stopped by addition of 0.5 M Na2CO3. The product was measured at a wavelength of 574 nm. CAT reporter gene activity was measured in at least three independent samples for each CA150 test construct. CAT activities were measured and corrected for transfection efficiency by dividing the CAT activity by the luciferase or β-galactosidase activity. The percent inhibition was determined by first calculating the fold inhibition of corrected CAT activity of wild-type CA150 in relation to the vector control, which was on average 5-to 10-fold inhibition. The fold inhibition by wild-type CA150 was set as 100% inhibition. The percent inhibition of the CA150 mutation and deletion test constructs was then calculated relative to that of the wild-type CA150.
Chromatography and microsequencing.
All chromatography was performed using a Pharmacia fast protein liquid chromatography system. Protease inhibitor Complete tablets (Boehringer-Mannheim) were used in all buffers. Fractions were assayed for CIP80 by far-Western analysis with the N-CA150 probe. A phosphocellulose P11 (Whatmann) column with a 50-ml bed volume was equilibrated in HEK100 (HEPES [pH 7.9], 10 mM EDTA, 100 mM KCl). Then 30 ml of HeLa nuclear extract was applied to the column, and bound proteins were eluted by sequential washes with the following gradient: HEK250, HEK500, HEK750, and HEK1000. The 500 mM KCl fraction contained the CIP80 peak. The fraction was dialyzed into HEK100 and applied to a 5-ml-bed-volume HighS column (Bio-Rad). Proteins were eluted with a linear gradient from 100 to 1,000 mM KCl, Coomassie stained, and assayed by far-Western blotting with the N-CA150 probe. A 300-μl volume of the CIP80 peak fraction was precipitated using 2 volumes of acetone and then separated by SDS-PAGE. The 80-kDa band corresponding to CIP80, visualized by Coomassie staining, was excised from the gel and used for microsequencing. Protein microsequencing analysis was performed by John Leszyk (Protein Microsequencing Laboratory, University of Massachusetts Medical School, Shrewsbury, Mass.). A tryptic digest of CIP80 was analyzed by matrix-assisted laser desorption mass spectrometry (MALDI-MS). Nine peptides were identified and sequenced, all of which belong to the protein SF1 (and also designated ZFM1, mBBP, and ZFP162) (2, 41, 42, 88). Peptide sequences were confirmed by mass spectrometry and post-source decay fragmentation analysis.
RESULTS
The phospho-CTD binding sites of CA150 are required for repression of transcription.
We performed a structure-function analysis of several of the protein interaction domains of CA150 to determine their role in CA150-mediated repression. The functional assay that we used involves a CAT reporter gene, whose expression is controlled by the human α4-integrin promoter (80). The α4-integrin promoter is controlled by both DNA binding activators and repressors and is regulated during hematopoeitic development and differentiation (3, 26, 45, 46, 72). Previously we demonstrated that CA150 inhibits the α4-integrin promoter in a dose-dependent manner and that core promoter elements, containing the TATA box, were required for mediating this effect. We had also previously shown that the FF repeats of CA150 mediate binding to the phosphorylated CTD of RNAPII (18). To assess the function of the FF repeats, a CA150 construct with a deletion of all six repeats, CA150(1–663), was created and tested for repression of α4-integrin (Fig. 1). CA150(1–663) exhibited a 70% loss in repression activity compared to full-length CA150. Thus, the FF repeats are necessary for efficient CA150-mediated repression. These results are consistent with the view that the interaction of CA150 with the phospho-CTD of elongating RNAPII is required for efficient repression. It was not clear, however, whether CTD binding is sufficient for this activity. For instance, the repression by CA150 could conceivably be the result of competitive binding between CA150 FF repeats and other elongation factors for binding to the phospho-CTD. To test this possibility, a construct, CA150(590–1098), that contains all six FF repeats but is lacking the N-terminal half of the protein was assayed for repression. CA150(590–1098) had greatly diminished activity relative to full-length CA150, exhibiting an 80% loss of repression (Fig. 1). This result demonstrates that the FF repeats are not sufficient for repression. The expression of the variant CA150 proteins was confirmed by Western blot analysis (Fig. 1). These CA150 proteins all contain the nuclear localization signal and were shown to be localized in the nucleoplasm, as assayed by immunofluorescence using an antibody against the amino-terminal T7 epitope tag of each CA150 construct (data not shown). We have also determined that the FF repeats were necessary for direct binding to phospho-CTD; full-length CA150 bound to phospho-CTD, while CA150(1–663) did not (A. C. Goldstrohm, S. Carty, A. Greenleaf, and M. Garcia-Blanco, unpublished data). Moreover, CA150(590–1098) was shown to coimmunoprecipitate RNAPII as well as full-length CA150 did (18). Therefore, the FF repeats appear to be necessary and sufficient for phospho-CTD binding whereas they are necessary but not sufficient for efficient repression of transcription.
FIG. 1.
The carboxyl-terminal and amino-terminal halves of CA150, containing FF repeats and WW domains, respectively, are required for repression of the α4-integrin promoter. CA150 test constructs were overexpressed in 293T cells with an α4-integrin promoter driving the expression of the CAT reporter gene to assay CA150-mediated repression. CA150 constructs shown on the left indicate the domains contained in each protein. CA150 WT is the wild-type 1,098-amino-acid protein. CA150(1–663) and CA150(590–1098) contain amino acids 1 to 663 and 590 to 1098, respectively. CA150 protein domain abbreviations: PP, polyproline-rich region; WW, WW domain; QA, glutamine-alanine repeats; NLS, nuclear localization signal; FF, FF repeats. The percent inhibition of CAT activity was calculated as described in Materials and Methods. The activity of each construct was assayed at least three times independently, and the error bars represent the standard deviation. The right panel is a representative Western blot, against the amino-terminal T7 epitope tag of each protein, showing the expression of each construct.
The WW domains of CA150 are required for efficient repression.
The loss of repression by CA150(590–1098) (Fig. 1) demonstrated that important determinants of CA150 function exist in the amino-terminal half of the protein. We hypothesized that CA150 may inhibit transcription by binding to the RNAPII CTD via the FF repeats and recruiting a repressor(s) to the transcription complex through the protein interaction domains in its amino-terminal half. The three amino-terminal WW domains of CA150 were strong candidates for such a function (CA150 WW domains are shown in Fig. 2A). We created point mutations in or deletions of each WW domain of CA150 and tested these constructs for repression activity. Since the results for mutations and deletions are nearly identical, we present only the data for the point mutations of CA150 WW domains in Fig. 2B. The highly conserved central aromatic amino acids of each WW domain were mutated to alanine (WW1 and WW2, YYY to AAA; WW3, FFY to AAA [Fig. 2]). These three aromatic amino acids are part of the proline binding pocket of WW domains (36, 83, 95). It has been shown that mutation of these residues does not destabilize the domain; therefore it is likely that the mutated WW domains were folded correctly (51). The mutated CA150 constructs were then tested for repression of the α4-integrin promoter. Mutation of WW1 (CA150 WW1mt) and WW2 (CA150 WW2mt) reproducibly resulted in 35 and 38% loss of activity, respectively, while mutation of WW3 had no effect on repression (Fig. 2B). Although the effect of disrupting WW1 and WW2 was modest, the results were highly reproducible. These data suggest that WW1 and WW2 contribute to CA150-mediated repression. We also created a double mutation of both WW1 and WW2 (CA150 WW1mt+WW2mt), which caused a 63% loss of activity relative to wild type, further supporting the requirement of WW1 and WW2 for repression (Fig. 2B). To evaluate a possible ancillary role for WW3, we disrupted WW1 and WW3 in combination and WW2 and WW3 in combination. These combinations behaved as the single WW1 and WW2 disruptions, suggesting that WW3 did not play a supportive role in the repression activity. CA150 protein expression levels from each construct were essentially identical as determined by Western blotting against the amino-terminal T7 epitope tag of each construct (Fig. 2B). In addition, we found that replacement of the CA150 WW1 domain with a functional WW domain from another protein (YAP) (52, 89), which has a ligand specificity distinct from that of CA150 WW1 and WW2 (for instance, see Fig. 5), also resulted in a loss of CA150 activity, identical to deletion or mutation of CA150 WW1 domain (data not shown). Therefore, the WW1 and WW2 domains of CA150 are specifically required for the repression activity. These results indicate that factors that interact with WW1 and WW2 are important for CA150-mediated repression.
FIG. 2.
The WW1 and WW2 domains of CA150 are required for efficient repression of the α4-integrin promoter. (A) Sequence comparison of CA150 WW domains WW1, WW2, and WW3. Alignments were constructed using the Clustal alignment function of the Mac Vector 6.5.3 program. A bracket highlights the central aromatic residues of the domains, YYY and FFY, which constitute part of the proline recognition site. These are the amino acids that were mutated in the CA150 WW domain mutant constructs in panel B and Fig. 3A. (B) Mutation of WW1 or WW2 reduced CA150-mediated repression of the α4-integrin promoter relative to wild type. A double-mutant CA150, containing mutations in both WW1 and WW2 (CA150 WW1mt+WW2mt), resulted in an even greater loss of repression activity. The percent inhibition of CAT activity is represented in the graph at the right. The standard error is also indicated. The site-directed mutagenesis of each domain is described in Materials and Methods. Diagrams of each CA150 test construct are depicted. Abbreviations are the same as those used in Fig. 1. YYY-AAA indicates the mutations of tyrosine to alanine in the central aromatic residues of WW1 and WW2. FFY-AAA indicates mutations of phenylalanine and tyrosine to alanine in the central aromatic residues of WW3. The bottom panel shows a representative anti-T7 Western blot demonstrating equal levels of expression for each CA150 protein.
FIG. 5.
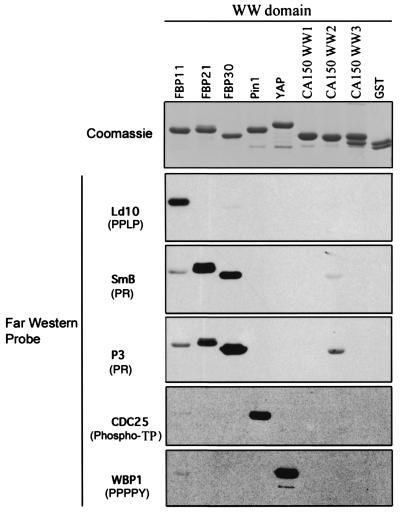
The ligand binding specificity of recombinant CA150 WW domains WW1, WW2, and WW3 was compared to that of WW domains with previously determined specificity. The identity of each WW domain is indicated at the top of the figure. Ligand binding specificities of the WW domains are as follows. The WW domain from the FBP11 protein specifically recognizes the PPLP motif from the ld10 protein. The FBP21 and FBP30 WW domains have a distinct binding preference for proline-arginine motifs (PR), such as those found in the SmB snRNP protein (designated SmB) and Sam68 (designated P3). The Pin1 WW domain binds specifically to motifs containing phosphoserine or phosphothreonine followed by a proline residue (phospho-SP/phospho-TP), such as that found in CDC25. Finally, the YAP WW domain interacts exclusively with a proline-rich sequence followed by tyrosine (PPPPY), which is found in WBP1. The top panel is a Coomassie-stained SDS-PAGE gel containing equal amounts of each WW domain. Purified GST protein served as a negative control. Peptides corresponding to the known classes of proline-rich ligand were used as probes in far-Western assays, shown in the bottom five panels. These probes are described in Materials and Methods. The identity of the proline-rich probes is indicated on the left, with the class of proline-rich motif indicated in parentheses. This analysis shows that CA150 WW domains did not bind strongly with the known classes of proline-rich ligands, indicating that they may recognize a novel proline-rich motif.
A protein of 80 kDa interacts with CA150 WW domains.
Given the important role played by WW1 and WW2 in CA150-mediated repression, we sought to identify factors that interact with these domains. We chose to use a far-Western (protein interaction) blotting approach to identify CA150-interacting proteins (CIPs). WW domains are particularly amenable to this approach because they can autonomously fold into their native structure and their ligands are small proline-rich motifs that can easily renature. This type of analysis has been extensively used to characterize other WW domains and their ligands (9, 20). We created a GST fusion protein, designated N-CA150, containing the WW2 and WW3 domains, to be used as a probe for detecting CIPs (Fig. 3A). HeLa cell nuclear extract (NE) was separated by SDS-PAGE, blotted to a membrane and renatured, and probed with nanomolar concentrations of the N-CA150 or negative-control GST probes (Fig. 3B). As expected, GST did not interact with any proteins in NE. N-CA150 detected a strong interaction with a CIP of 80 kDa, tentatively named CIP80 (Fig. 3B). Several weaker CIPs were also observed, some of which varied among extract preparations. We chose to analyze CIP80 because it was consistently the strongest interaction observed, both by far-Western assay and coimmunoprecipitation analysis (see below).
FIG. 3.
A CA150-interacting protein of 80 kDa (CIP80), which interacts directly with CA150, is the splicing-transcription factor SF1. (A) Diagram showing the recombinant CA150 proteins used as probes in the far-Western analysis. Abbreviations are the same as those used in Fig. 1. N-CA150, WW2mt, and WW3mt contain amino acids 235 to 631 of CA150, except that the WW2mt probe had the central aromatic residues of WW2 domain mutated from YYY to AAA and WW3mt had the corresponding residues mutated from FFY to AAA. These mutations ablate the recognition of proline-containing ligands by the WW domains. (B) Far-Western analysis of HeLa nuclear extract with the N-CA150 probe, as indicated at the top of the figure, detected a CA150-interacting protein with an apparent mass of 80 kDa, designated CIP80. The negative-control far-Western blot with a GST probe did not bind to nuclear proteins, as expected. (C) Far-Western analysis of HeLa nuclear extract showed that mutation of the WW2 domain ablates binding to CIP80 while mutation of the WW3 domain has no effect. CA150 probes are indicated at the top, and CIP80 is indicated on the left. (D) Coimmunoprecipitation of CIP80 with CA150 from HeLa nuclear extract. CA150 was immunoprecipitated using antigen affinity-purified rabbit polyclonal antibodies, as indicated at the top. The immunoprecipitates were analyzed by Western blotting with anti-CA150 antibodies (top panel) and by far-Western blotting using the N-CA150 probe (bottom panel), as labeled on the left. (E) Purification scheme for isolating CIP80. The CIP80 protein was excised from an SDS-PAGE gel and microsequenced by mass spectrometry. CIP80 corresponds to the previously identified SF1 protein. (F) Fractions from HighS chromatography, developed with a linear gradient of 100 to 500 mM KCl, were analyzed by far-Western blotting with the N-CA150 probe. Molecular weight markers are indicated on the right of the figure in thousands. CIP80 is labeled on the left. The peak CIP80-containing fraction from the phosphocellulose step was included in the analysis and labeled Input. A 110-kDa protein that also interacts with the N-CA150 probe, albeit weakly, copurified with CIP80.
To examine the specificity of the interaction of N-CA150 with CIP80, we mutated the central aromatic residues of the WW2 or WW3 domain to alanine (N-CA150 WW2mt and N-CA150 WW3mt, respectively) and used these proteins as probes in far-Western assays of HeLa NE (Fig. 3A). This is the same mutation (YYY to AAA) that caused loss of repression activity when introduced into the WW2 domain in the full-length CA150 (Fig. 2). The WW2mt protein was not capable of binding to CIP80, while the WW3mt protein bound as well as the wild type did (Fig. 3C). Therefore, WW2, but not WW3, is specifically required for interaction with CIP80. Furthermore, in Fig. 4 (see below) we demonstrate that an individual WW domain can recognize CIP80; both WW1 and WW2 can bind to CIP80. Hence, WW1 and WW2 domains of CA150 are necessary and sufficient for interaction with CIP80.
FIG. 4.
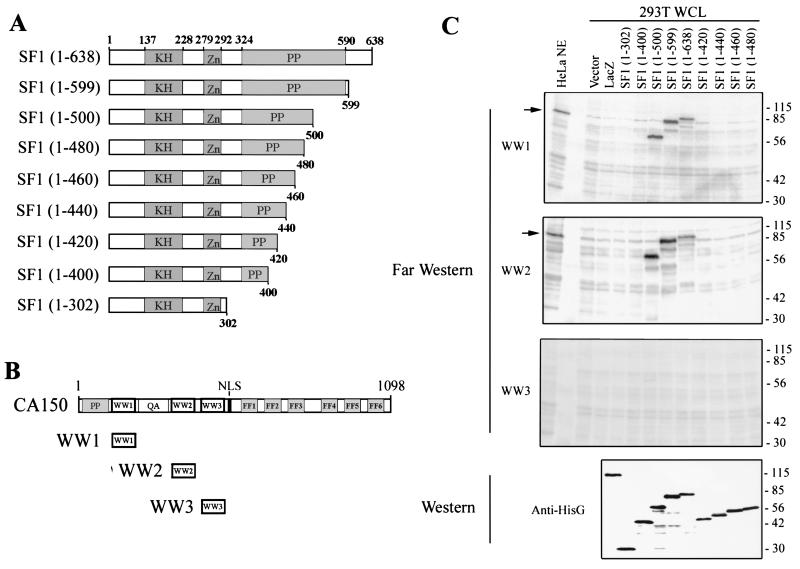
Full-length SF1(1–638) was used to create a deletion series of the carboxyl-terminal half of the protein. The SF1 amino acids contained in each construct are indicated in parentheses. KH is the hnRNP K homology domain, and Zn represents the Zn knuckle motif. PP indicates the proline-rich region. (B) The CA150 WW1, WW2, and WW3 domain probes, represented in this diagram, were used in far-Western analysis. (C) Far-Western analysis of WCL from 293T cells overexpressing the SF1 constructs indicated at the top. HeLa NE was included as a positive control, and endogenous HeLa SF1 is indicated by an arrow on the left. 293T WCL transfected with empty vector or the lacZ expression vector were included as negative controls. Far-Western blotting of blots of these WCLs with each WW domain from CA150, as labeled on the left, detected protein-protein interactions with the overexpressed SF1 constructs. The bottom panel, labeled Western, is an anti-HisG Western blot of the overexpressed proteins that contained an amino-terminal His tag. Protein molecular weight markers are labeled on the right in thousands.
We sought to confirm the interaction between CA150 and CIP80 by determining if the proteins interact in nuclear extracts from HeLa cells. To test this, we used antigen affinity-purified antibodies to selectively immunoprecipitate CA150 from HeLa NE. This antibody is specific since it recognizes only CA150 in a Western blot of NE (81). Anti-GST antibodies were used as a negative control for nonspecific interactions in the immunoprecipitation. The immunoprecipitate pellets were separated by SDS-PAGE and analyzed by Western blotting for CA150 and by far-Western blotting using the N-CA150 probe to detect CIP80. Comparison of the input and the supernatant fractions revealed that only a small fraction of CIP80 was immunoprecipitated with anti-CA150 antibodies (data not shown). Nonetheless, this analysis revealed that CIP80 is associated with CA150 in NE (Fig. 3D) and provides an independent means of demonstrating that CA150 and CIP80 can interact. The coimmunoprecipitation of CIP80 with CA150 is specific, since other abundant nuclear proteins such as TATA binding protein and proliferating-cell nuclear antigen did not coimmunoprecipitate (reference 18 and data not shown). We conclude that CA150 and CIP80 exist in a preformed complex in NE.
CIP80 is the splicing and transcription factor SF1.
To determine the identity of CIP80, we purified it and obtained microsequence data. The purification protocol is described below and is outlined in Fig. 3E. HeLa nuclear extract containing CIP80 was separated by phosphocellulose chromatography, and fractions were assayed for the presence of CIP80 by far-Western blotting. CIP80 peak fractions were pooled and applied to a HighS column (Fig. 3F). The HighS peak fraction of CIP80 was then separated by SDS-PAGE, the 80-kDa band corresponding to CIP80 was excised from the gel, and peptides were generated and microsequenced using mass spectrometry. This analysis revealed that CIP80 is the previously identified pre-mRNA splicing-transcription factor SF1 (2, 12, 13, 40, 42, 69). Multiple isoforms of SF1 have been identified (see Discussion). The major isoform of SF1 in HeLa cells contains 638 amino acids and has a mobility of approximately 80 kDa on SDS-PAGE (2, 42, 69). In addition to its role in pre-mRNA splicing, SF1 is a transcription repressor, thus providing a possible explanation for its interaction with CA150 (97, 98).
The primary structure of SF1 contains an amino-terminal KH domain and a zinc knuckle, both of which are necessary for its relatively nonspecific RNA binding activity (2, 13, 14). The carboxyl-terminal half of SF1 is proline rich, with many polyproline motifs. We conjectured that this proline-rich region was the likely binding site for CA150, given the propensity of WW domains to recognize proline-rich ligands. We mapped the binding site for CA150 in SF1 by creating a deletion series of the proline-rich region of SF1 (Fig. 4A). These SF1 constructs were transfected and expressed in 293T cells, and WCL were prepared, separated by SDS-PAGE, and transferred to membranes. As negative controls, we included WCL from cells transfected with empty vector or a lacZ expression construct. All of the SF1 constructs contained an amino-terminal HisG epitope tag that allowed a comparison of relative expression levels by Western blot analysis with an anti-HisG antibody (Fig. 4C); each protein was expressed at essentially the same level. We then performed far-Western analysis on these membranes using probes composed of each individual WW domain from CA150 (WW1, WW2, or WW3) (Fig. 4B). By this approach, we determined that the CA150 WW1 and WW2 domains bind to the same region of SF1. The results are shown in Fig. 4C. The CA150 WW1 and WW2 probes bound to SF1 (1–638), SF1 (1–599), and SF1 (1–500) but not to the other SF1 deletion constructs. The WW3 domain did not bind to any proteins in this assay (Fig. 4C). It should be noted that a band migrating as expected for endogenous 293T SF1 binds WW1 and WW2 but not WW3. Several conclusions can be drawn from this experiment. First, we confirm that SF1 is CIP80 by demonstrating that CA150 probes bound to the recombinant SF1 expressed from our cDNA clone. CA150 WW1 and WW2 probes also bound to affinity-purified SF1 (data not shown). Second, this analysis allowed us to determine that an individual WW domain was sufficient for binding to SF1. The WW1 and WW2 domains of CA150 interacted with SF1, while the WW3 domain did not, in agreement with our mutational analysis in the context of the N-CA150 probe. It is noteworthy that sequence comparison of the three WW domains demonstrates that WW1 and WW2 are most similar to each other whereas WW3 is the most divergent of the three (Fig. 2A). Third, far-Western analysis of the SF1 deletion series demonstrates that the carboxyl-terminal 138 amino acids of SF1 were fully dispensable for interaction with the WW domains and that amino acids within residues 1 to 500 were necessary. Loss of binding to SF1(1–480) suggests that amino acids near this region were involved in CA150 WW binding. In addition, WW1 and WW2 appeared to bind to the same region of SF1 because both interacted with SF1(1–500) but not SF1(1–480). Finally, we note that the binding of WW1 and WW2 to SF1 directly correlated with the requirement of these domains for CA150-mediated transcription repression. In additional experiments, we have attempted to directly enhance CA150-mediated repression of the α4-integrin promoter by overexpression of both CA150 and SF1. This approach did not significantly enhance CA150 repression (data not shown), a result that is not all that surprising given that SF1 is an abundant nuclear protein and therefore may not be limiting. Future experiments that develop and utilize SF1-dependent assays will be required to directly test the function of the CA150-SF1 interaction and to assess the function of the RNA binding activity of SF1 in CA150-mediated repression.
A proline-rich sequence in SF1 is sufficient for binding to CA150 WW domains.
To characterize the ligand binding specificity of CA150 WW domains, we used the far-Western assay with recombinant WW1, WW2, and WW3 proteins and tested their ability to interact with prototypic peptides of the known classes of ligands (Fig. 5) (8, 9, 78, 79). As controls, we included WW domains with documented ligand specificity. Equal amounts of each purified GST-WW domain were separated by SDS-PAGE, transferred to membranes, and probed with the peptide ligands (Fig. 5). The WW2 domain of CA150 bound weakly to the PR motif-containing ligands; however, this binding was considerably weaker than that of FBP21 or FBP30, which exhibit high-affinity, specific binding to proline-arginine motifs (PR) (Fig. 5) (9). Thus, the PR motif is a poor ligand for WW2. The CA150 WW1 and WW3 domains did not bind any of these peptide ligands. In addition, some WW domains, including that of the Ess1 protein, can bind to the phospho-CTD of RNAPII (YpSPTpSPS ligand, where “p” indicates phosporylation of serine). The three WW domains of CA150 were tested for binding to phospho-CTD, and they did not bind (Goldstrohm et al., unpublished). These results indicate that the CA150 WW domains display a ligand binding specificity distinct from those of previously classified WW domains.
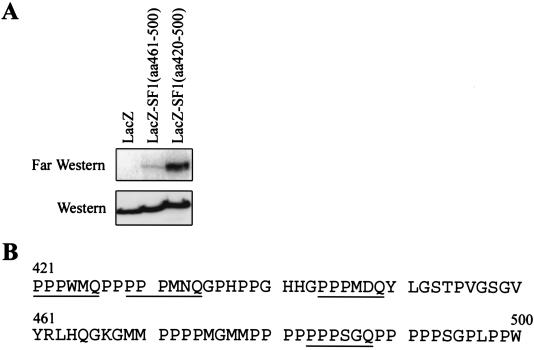
With the knowledge that CA150 WW domains did not recognize the known classes of ligands, we wished to further characterize the binding site, within the proline-rich region of SF1, for the CA150 WW domains and perhaps gain insight into their ligand specificity. To achieve this, we fused portions of the proline-rich region of SF1 onto the amino terminus of the lacZ gene and expressed these constructs in 293T cells. Based on the deletion analysis in Fig. 4C and the fact that WW domains usually recognize short proline-rich sequences, it was likely that the CA150 binding site was located between amino acids 400 and 500 of SF1. Far-Western analysis with the WW2 domain of CA150 determined that amino acids 420 to 500 of SF1 were sufficient for strong binding to WW2 (Fig. 6). SF1 amino acids 461 to 500 bound weakly to WW2, while the lacZ control did not interact (Fig. 6A). Based on these results and the SF1 deletion analysis (Fig. 4), we conclude that amino acids 420 to 500 of SF1 are necessary and sufficient for the binding of CA150 to SF1. Sequence analysis of SF1 amino acids 420 to 500 revealed multiple proline-rich motifs that may be recognized by CA150 WW1 and WW2 domains; moreover, this region did not contain sequences that match the known classes of WW domain ligands (Fig. 6B). Multiple motifs with the consensus PPPxxQ (where x is a variable amino acid) are found in this portion of SF1 and may constitute the WW domain ligands. We note that four of these motifs are contained in SF1 amino acids 420 to 500 whereas only one motif was present in amino acids 461 to 500 (Fig. 6B). The strong binding of WW2 to amino acids 420 to 500 and the weaker binding to amino acids 461 to 500 can be explained by this difference in the number of PPPxxQ motifs. These results show that CA150 WW domains possess distinct specificity for proline-rich ligands. The CA150-SF1 binding data suggest that the WW1 and WW2 domains may bind to a new type of ligand, the PPPXQ motif. As a final point, the multiple PPPXXQ motifs found in SF1 could allow both CA150 WW1 and WW2 domains to interact simultaneously with SF1, thereby strengthening the CA150-SF1 interaction.
FIG. 6.
Amino acids 420 to 500 of SF1 are sufficient for binding to the WW2 domain of CA150. (A) The SF1 amino acids 461 to 500 or 420 to 500 were fused to the lacZ gene and expressed in 293T cells. WCL were prepared from these cells or control lacZ-transfected cells and subjected to far-Western analysis with the a WW2 domain probe, shown in the top panel. The expression constructs are indicated at the top of the figure. Anti-T7 Western blot analysis of the WCL demonstrates equivalent expression of each protein. (B) The CA150 binding site in SF1, composed of amino acids 420 to 500. The PPPxxQ motifs, which are the probable ligands of the WW1 and WW2 domains of CA150, are underlined.
DISCUSSION
CA150 is a transcription repressor that inhibits the elongation efficiency of RNAPII in vivo (80) and in vitro (A. C. Goldstrohm and M. Garcia-Blanco, unpublished data). Previous work demonstrated that CA150 FF repeats interact directly and specifically with the phospho-CTD of RNAPII (18), and here we show that these repeats of CA150 are required for maximal repression. Taken together, these observations suggest that CA150 is targeted to the elongating RNAPII via the FF repeats-phospho CTD interaction. Once targeted to elongating RNAPII, CA150 can repress transcript elongation by one of several mechanisms. CA150 could potentially displace P-TEFs from the phospho-CTD; however, the fact that the FF repeats of CA150 are sufficient for phospho-CTD binding but not for repression in vivo (this study) or in vitro (Goldstrohm and Garcia-Blanco, unpublished) makes this possibility unlikely. CA150 could modify the enzymatic activity of RNAPII, as has been shown for the transcript cleavage-inducing factor TFIIS, which stimulates elongation by facilitating the release of the polymerase from an arrested state (86). Alternatively, CA150 could modify the activity of P-TEFs that also interact with RNAPII. We have observed that CA150 associates with multiple positive elongation factors including P-TEFb, Tat-SF1, and TFIIF (Goldstrohm and Garcia-Blanco, unpublished). CA150 may inhibit transcription by suppressing the action of these positive factors, or, conversely, these factors may associate with and counteract CA150. Finally, CA150 could mediate the recruitment of other effectors (N-TEFs) to the elongation complex. This possibility led us to look for other proteins that interacted with CA150.
The amino-terminal half of CA150 contains three WW domains, two of which, WW1 and WW2, are important for repression of transcription. We posited that these two domains could be important in recruiting other proteins that could collaborate with CA150. We identified a factor, SF1, which physically interacts with these WW domains. We found that the requirement of WW1 and WW2 domains for CA150-mediated repression correlated precisely with the binding specificity of WW1 and WW2 for SF1. These observations lead to two non-mutually exclusive suggestions about the role of SF1 on CA150 repression: CA150 recruits SF1 to act as an N-TEF, and/or CA150 uses SF1 to target the elongating RNAPII. It must be noted that we have not formally proven that the interaction with SF1 is required for CA150 function and thus we must also consider that the observed interaction is a surrogate interaction.
Could SF1 be an N-TEF?
SF1 has indeed been shown to repress activation of transcription by certain chimeric transcription activators (97, 98). SF1 also represses transcription from a promoter when tethered to a Gal4 DNA binding domain and targeted to a promoter containing Gal4 binding sites (97). Taken together, these results suggest that recruitment of SF1 to a transcription complex, whether by a DNA binding factor or by CA150, can result in repression. The CA150 binding site resides in the proline-rich carboxyl terminus of SF1. Currently, the function of the proline-rich region is not known, and it has been reported to be dispensable for SF1 splicing activity (31, 69). Zhang and Childs found that a portion of this region of SF1 could bind to the activation domain of certain transcription factors, suggesting that the proline-rich region may function in transcription (97). At least 10 alternatively spliced SF1 isoforms have been identified, which vary in the proline-rich region and carboxyl terminus (2, 42; A. Kramer, personal communication). Several isoforms lack portions of the CA150 binding site (SF1 amino acids 420 to 500). So far we have determined that at least one of these isoforms, ZFM1-ΔE12/E13 (42), interacts very weakly with CA150 in the far-Western assay, demonstrating that the CA150-SF1 interaction is isoform specific (data not shown). The ZFM1-ΔE12/E13 isoform diverges from the major HeLa isoforms, SF1-HL1, SF1-HL2, and SF1-Bo, at amino acid 469, which resides within the CA150 binding site (42). Also, based on our binding analysis we predict that the SF1 isoform ZFM1-B3 would not bind to CA150, since it possesses an entirely different C -terminus beginning at residue 448 (42). In conclusion, the isoform-specific CA150-SF1 interaction advocates a possible mechanism for regulating CA150 activity depending on the repertoire of SF1 isoforms present in a cell. As an additional consideration, we note that two other mammalian proteins that possess WW domains have been shown to interact with SF1. The FBP11 protein, an ortholog of the U1-associated snRNP protein Prp40, and the U2 snRNP-associated protein FBP21 both bind to SF1. The binding site in SF1 for these proteins has not been mapped. However, because the WW domains from FBP11 and FBP21 exhibit different ligand specificities in comparison to CA150 (see Fig. 5), we expect that they do not compete with CA150 for binding to SF1.
Could SF1 target CA150 to elongating transcripts?
SF1 binds RNA via its KH and Zn knuckle domains with rather poor sequence- specificity (2, 13). Could this RNA binding activity be a clue to its role in CA150-mediated repression? We propose that CA150 and SF1 together may target elongating RNAPII complexes by recognition of the two distinguishing characteristics of an RNAPII elongation complex: the phosphorylated CTD and the nascent RNA transcript. This idea is consistent with the pathway used by other elongation factors, which function by binding to the transcribing polymerase and the transcript. One example is the yeast Nrd1-Nab3 complex, which binds to the phospho-CTD of RNAPII (via Nrd1) and recognizes a sequence in the nascent RNA (both Nrd1 and Nab3 bind to RNA), leading to termination of transcription (22). One noteworthy difference between Nrd1 and CA150 repression mechanisms is that Nrd1 requires a specific cis-acting RNA sequence, designated U6R∗, whereas we have not detected a transcript sequence specificity for CA150 activity. Another negative regulator of RNAPII elongation that may follow a similar pathway to that of CA150 is the multisubunit DSIF-NELF complexes. DSIF-NELF is responsible for inhibition of early elongation events, and this effect is reversed by P-TEFb (28, 85, 90). DSIF-NELF associates with RNAPII, mediated by the Spt5 subunit of DSIF, which binds directly to the large subunit of RNAPII (91). However, unlike CA150, DSIF-NELF binds to and inhibits unphosphorylated RNAPII (85, 90). The RD subunit of NELF has similarity to Nrd1 in that it contains a single RNA recognition motif that very probably binds to RNA, thus making it probable that DSIF-NELF contacts RNAPII and the nascent transcript to inhibit elongation (28, 90). Finally, prokaryotic RNAP is also regulated by elongation factors that bind the polymerase and the transcript. The transcription termination factor Rho interacts with the NusG protein to mediate termination. Rho also has an RNA binding domain that recognizes the nascent transcript, while NusG binds directly to the polymerase (71). In addition, the phage lambda N protein, an antitermination factor, affects elongation through a similar pathway. N is a positive elongation factor that binds to a cis-acting element in the nascent transcript, nut, and mediates antitermination in conjunction with cellular factors, including the NusA protein that binds directly to the elongating RNAP (57). Therefore, dual interactions with the polymerase and the nascent transcript may be a common mechanism used by elongation factors to identify their targets and carry out their function.
Does the CA150-SF1 interaction play a role in pre-mRNA splicing?
Multiple studies have also demonstrated a role for SF1 in pre-mRNA splicing (1, 12, 13, 31, 40, 41, 69, 73, 74). The apparent dichotomy of function of SF1 in splicing and transcription can be viewed in two ways. SF1 may play independent roles in transcription and splicing, or it may participate in coupling the two processes (see below). The transcription assays used in our studies did not include introns; therefore, the transcription repression that we observe is not likely to be a splicing-related process. However, accumulating evidence suggests a role for CA150 in pre-mRNA splicing. Neubauer et al. discovered that purified spliceosomes contain CA150 (61). Likewise, we have observed that the snRNP Sm proteins coimmunoprecipitate with CA150, suggesting that CA150 can associate with snRNPs (Goldstrohm and Garcia-Blanco, unpublished). CA150 also coimmunoprecipitates with Tat-SF1 (Goldstrohm and Garcia-Blanco, unpublished), which, in addition to its ability to affect elongation, has been shown to be the mammalian ortholog of the splicing factor CUS2 and to associate with the splicing factor SF3a (66, 92). An interesting possibility is that the CA150-SF1 complex could function to coordinate the rates of transcript elongation and pre-mRNA processing. This idea is highly speculative, and future work is required to test it.
ACKNOWLEDGMENTS
We thank Eric J. Wagner, Rob Brazas, and Arno Greenleaf for helpful discussions; Angela Kramer (University of Geneva) for providing SF1 clones; and John Lezsyk (University of Massachusetts) for microsequencing expertise. We also thank Angela Kramer, Arno Greenleaf (Duke University), and Sherry Carty (Duke University) for sharing unpublished results.
REFERENCES
- 1.Abovich N, Rosbash M. Cross-intron bridging interactions in the yeast commitment complex are conserved in mammals. Cell. 1997;89:403–412. doi: 10.1016/s0092-8674(00)80221-4. [DOI] [PubMed] [Google Scholar]
- 2.Arning S, Gruter P, Bilbe G, Kramer A. Mammalian splicing factor SF1 is encoded by variant cDNAs and binds to RNA. RNA. 1996;2:794–810. [PMC free article] [PubMed] [Google Scholar]
- 3.Audet J F, Masson J Y, Rosen G D, Salesse C, Guerin S L. Multiple regulatory elements control the basal promoter activity of the human alpha 4 integrin gene. DNA Cell Biol. 1994;13:1071–1085. doi: 10.1089/dna.1994.13.1071. [DOI] [PubMed] [Google Scholar]
- 4.Bauren G, Wieslander L. Splicing of Balbiani ring 1 gene pre-mRNA occurs simultaneously with transcription. Cell. 1994;76:183–192. doi: 10.1016/0092-8674(94)90182-1. [DOI] [PubMed] [Google Scholar]
- 5.Bedford M T, Chan D C, Leder P. FBP WW domains and the Abl SH3 domain bind to a specific class of proline-rich ligands. EMBO J. 1997;16:2376–2383. doi: 10.1093/emboj/16.9.2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bedford M T, Frankel A, Yaffe M B, Clarke S, Leder P, Richard S. Arginine methylation inhibits the binding of proline-rich ligands to Src homology 3, but not WW, domains. J Biol Chem. 2000;275:16030–16036. doi: 10.1074/jbc.M909368199. [DOI] [PubMed] [Google Scholar]
- 7.Bedford M T, Leder P. The FF domain: a novel motif that often accompanies WW domains. Trends Biochem Sci. 1999;24:264–265. doi: 10.1016/s0968-0004(99)01417-6. [DOI] [PubMed] [Google Scholar]
- 8.Bedford M T, Reed R, Leder P. WW domain-mediated interactions reveal a spliceosome-associated protein that binds a third class of proline-rich motif: the proline glycine and methionine-rich motif. Proc Natl Acad Sci USA. 1998;95:10602–10607. doi: 10.1073/pnas.95.18.10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bedford M T, Sarbassova D, Xu J, Leder P, Yaffe M B. A novel pro-Arg motif recognized by WW domains. J Biol Chem. 2000;275:10359–10369. doi: 10.1074/jbc.275.14.10359. [DOI] [PubMed] [Google Scholar]
- 10.Bentley D. Coupling RNA polymerase II transcription with pre-mRNA processing. Curr Opin Cell Biol. 1999;11:347–351. doi: 10.1016/S0955-0674(99)80048-9. [DOI] [PubMed] [Google Scholar]
- 11.Bentley D L. Regulation of transcriptional elongation by RNA polymerase II. Curr Opin Genet Dev. 1995;5:210–216. doi: 10.1016/0959-437x(95)80010-7. [DOI] [PubMed] [Google Scholar]
- 12.Berglund J A, Abovich N, Rosbash M. A cooperative interaction between U2AF65 and mBBP/SF1 facilitates branchpoint region recognition. Genes Dev. 1998;12:858–867. doi: 10.1101/gad.12.6.858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berglund J A, Chua K, Abovich N, Reed R, Rosbash M. The splicing factor BBP interacts specifically with the pre-mRNA branchpoint sequence UACUAAC. Cell. 1997;89:781–787. doi: 10.1016/s0092-8674(00)80261-5. [DOI] [PubMed] [Google Scholar]
- 14.Berglund J A, Fleming M L, Rosbash M. The KH domain of the branchpoint sequence binding protein determines specificity for the pre-mRNA branchpoint sequence. RNA. 1998;4:998–1006. doi: 10.1017/s1355838298980499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berkhout B, Jeang K T. Functional roles for the TATA promoter and enhancers in basal and Tat- induced expression of the human immunodeficiency virus type 1 long terminal repeat. J Virol. 1992;66:139–149. doi: 10.1128/jvi.66.1.139-149.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blau J, Xiao H, McCracken S, O'Hare P, Greenblatt J, Bentley D. Three functional classes of transcriptional activation domain. Mol Cell Biol. 1996;16:2044–2055. doi: 10.1128/mcb.16.5.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brown S A, Weirich C S, Newton E M, Kingston R E. Transcriptional activation domains stimulate initiation and elongation at different times and via different residues. EMBO J. 1998;17:3146–3154. doi: 10.1093/emboj/17.11.3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carty S M, Goldstrohm A C, Sune C, Garcia-Blanco M A, Greenleaf A L. Protein-interaction modules that organize nuclear function: FF domains of CA150 bind the phosphoCTD of RNA polymerase II. Proc Natl Acad Sci USA. 2000;97:9015–9020. doi: 10.1073/pnas.160266597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chan D C, Bedford M T, Leder P. Formin binding proteins bear WWP/WW domains that bind proline-rich peptides and functionally resemble SH3 domains. EMBO J. 1996;15:1045–1054. [PMC free article] [PubMed] [Google Scholar]
- 20.Chang A, Cheang S, Espanel X, Sudol M. Rsp5 WW domains interact directly with the carboxyl-terminal domain of RNA polymerase II. J Biol Chem. 2000;275:20562–20571. doi: 10.1074/jbc.M002479200. [DOI] [PubMed] [Google Scholar]
- 21.Conaway J W, Conaway R C. Transcription elongation and human disease. Annu Rev Biochem. 1999;68:301–319. doi: 10.1146/annurev.biochem.68.1.301. [DOI] [PubMed] [Google Scholar]
- 22.Conrad N K, Wilson S M, Steinmetz E J, Patturajan M, Brow D A, Swanson M S, Corden J L. A yeast heterogeneous nuclear ribonucleoprotein complex associated with RNA polymerase II. Genetics. 2000;154:557–571. doi: 10.1093/genetics/154.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cramer P, Caceres J F, Cazalla D, Kadener S, Muro A F, Baralle F E, Kornblihtt A R. Coupling of transcription with alternative splicing: RNA pol II promoters modulate SF2/ASF and 9G8 effects on an exonic splicing enhancer. Mol Cell. 1999;4:251–258. doi: 10.1016/s1097-2765(00)80372-x. [DOI] [PubMed] [Google Scholar]
- 24.Cramer P, Pesce C G, Baralle F E, Kornblihtt A R. Functional association between promoter structure and transcript alternative splicing. Proc Natl Acad Sci USA. 1997;94:11456–11460. doi: 10.1073/pnas.94.21.11456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dahmus M E. Reversible phosphorylation of the C-terminal domain of RNA polymerase II. J Biol Chem. 1996;271:19009–19012. doi: 10.1074/jbc.271.32.19009. [DOI] [PubMed] [Google Scholar]
- 26.De Meirsman C, Schollen E, Jaspers M, Ongena K, Matthijs G, Marynen P, Cassiman J J. Cloning and characterization of the promoter region of the murine alpha- 4 integrin subunit. DNA Cell Biol. 1994;13:743–754. doi: 10.1089/dna.1994.13.743. [DOI] [PubMed] [Google Scholar]
- 27.Du L, Warren S L. A functional interaction between the carboxy-terminal domain of RNA polymerase II and pre-mRNA splicing. J Cell Biol. 1997;136:5–18. doi: 10.1083/jcb.136.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garber M E, Jones K A. HIV-1 Tat: coping with negative elongation factors. Curr Opin Immunol. 1999;11:460–465. doi: 10.1016/S0952-7915(99)80077-6. [DOI] [PubMed] [Google Scholar]
- 29.Ghosh S, Garcia-Blanco M A. Coupled in vitro synthesis and splicing of RNA polymerase II transcripts. RNA. 2000;6:1325–1334. doi: 10.1017/s1355838200992537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Greenleaf A L. Positive patches and negative noodles: linking RNA processing to transcription? Trends Biochem Sci. 1993;18:117–119. doi: 10.1016/0968-0004(93)90016-g. [DOI] [PubMed] [Google Scholar]
- 31.Guth S, Valcarcel J. Kinetic role for mammalian SF1/BBP in spliceosome assembly and function after polypyrimidine tract recognition by U2AF. J Biol Chem. 2000;275:38059–38066. doi: 10.1074/jbc.M001483200. [DOI] [PubMed] [Google Scholar]
- 32.Hampsey M. Molecular genetics of the RNA polymerase II general transcriptional machinery. Microbiol Mol Biol Rev. 1998;62:465–503. doi: 10.1128/mmbr.62.2.465-503.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harlow E, Lane D. Using antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1998. [Google Scholar]
- 34.Hirose Y, Manley J L. RNA polymerase II and the integration of nuclear events. Genes Dev. 2000;14:1415–1429. [PubMed] [Google Scholar]
- 35.Hirose Y, Tacke R, Manley J L. Phosphorylated RNA polymerase II stimulates pre-mRNA splicing. Genes Dev. 1999;13:1234–1239. doi: 10.1101/gad.13.10.1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang X, Poy F, Zhang R, Joachimiak A, Sudol M, Eck M J. Structure of a WW domain containing fragment of dystrophin in complex with beta-dystroglycan. Nat Struct Biol. 2000;7:634–638. doi: 10.1038/77923. [DOI] [PubMed] [Google Scholar]
- 37.Isel C, Karn J. Direct evidence that HIV-1 Tat stimulates RNA polymerase II carboxyl-terminal domain hyperphosphorylation during transcriptional elongation. J Mol Biol. 1999;290:929–941. doi: 10.1006/jmbi.1999.2933. [DOI] [PubMed] [Google Scholar]
- 38.Kanazawa S, Okamoto T, Peterlin B M. Tat competes with CIITA for the binding to P-TEFb and blocks the expression of MHC class II genes in HIV infection. Immunity. 2000;12:61–70. doi: 10.1016/s1074-7613(00)80159-4. [DOI] [PubMed] [Google Scholar]
- 39.Kim E, Du L, Bregman D B, Warren S L. Splicing factors associate with hyperphosphorylated RNA polymerase II in the absence of pre-mRNA. J Cell Biol. 1997;136:19–28. doi: 10.1083/jcb.136.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kramer A. Purification of splicing factor SF1, a heat-stable protein that functions in the assembly of a presplicing complex. Mol Cell Biol. 1992;12:4545–4552. doi: 10.1128/mcb.12.10.4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kramer A. The structure and function of proteins involved in mammalian pre-mRNA splicing. Annu Rev Biochem. 1996;65:367–409. doi: 10.1146/annurev.bi.65.070196.002055. [DOI] [PubMed] [Google Scholar]
- 42.Kramer A, Quentin M, Mulhauser F. Diverse modes of alternative splicing of human splicing factor SF1 deduced from the exon-intron structure of the gene. Gene. 1998;211:29–37. doi: 10.1016/s0378-1119(98)00058-4. [DOI] [PubMed] [Google Scholar]
- 43.Kramer A, Utans U. Three protein factors (SF1, SF3 and U2AF) function in pre-splicing complex formation in addition to snRNPs. EMBO J. 1991;10:1503–1509. doi: 10.1002/j.1460-2075.1991.tb07670.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Landick R. RNA polymerase slides home: pause and termination site recognition. Cell. 1997;88:741–744. doi: 10.1016/s0092-8674(00)81919-4. [DOI] [PubMed] [Google Scholar]
- 45.Larouche K, Leclerc S, Giasson M, Guerin S L. Multiple nuclear regulatory proteins bind a single cis-acting promoter element to control basal transcription of the human alpha 4 integrin gene in corneal epithelial cells. DNA Cell Biol. 1996;15:779–792. doi: 10.1089/dna.1996.15.779. [DOI] [PubMed] [Google Scholar]
- 46.Larouche N, Larouche K, Beliveau A, Leclerc S, Salesse C, Pelletier G, Guerin S L. Transcriptional regulation of the alpha 4 integrin subunit gene in the metastatic spread of uveal melanoma. Anticancer Res. 1998;18:3539–3547. [PubMed] [Google Scholar]
- 47.Lee D K, Duan H O, Chang C. Androgen receptor interacts with the positive elongation factor p-tefb and enhances the efficiency of transcriptional elongation. J Biol Chem. 2001;276:9978–9984. doi: 10.1074/jbc.M002285200. [DOI] [PubMed] [Google Scholar]
- 48.Lee T L, Young R A. Transcription of eukaryotic protein-coding genes. Annu Rev Genet. 2000;34:77–137. doi: 10.1146/annurev.genet.34.1.77. [DOI] [PubMed] [Google Scholar]
- 49.Lemon B, Tjian R. Orchestrated response: a symphony of transcription factors for gene control. Genes Dev. 2000;14:2551–2569. doi: 10.1101/gad.831000. [DOI] [PubMed] [Google Scholar]
- 50.Lu X, Welsh T M, Peterlin B M. The human immunodeficiency virus type 1 long terminal repeat specifies two different transcription complexes, only one of which is regulated by Tat. J Virol. 1993;67:1752–1760. doi: 10.1128/jvi.67.4.1752-1760.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Macias M J, Gervais V, Civera C, Oschkinat H. Structural analysis of WW domains and design of a WW prototype. Nat Struct Biol. 2000;7:375–379. doi: 10.1038/75144. [DOI] [PubMed] [Google Scholar]
- 52.Macias M J, Hyvonen M, Baraldi E, Schultz J, Sudol M, Saraste M, Oschkinat H. Structure of the WW domain of a kinase-associated protein complexed with a proline-rich peptide. Nature. 1996;382:646–649. doi: 10.1038/382646a0. [DOI] [PubMed] [Google Scholar]
- 53.Marciniak R A, Sharp P A. HIV-1 Tat protein promotes formation of more-processive elongation complexes. EMBO J. 1991;10:4189–4196. doi: 10.1002/j.1460-2075.1991.tb04997.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McCracken S, Fong N, Yankulov K, Ballantyne S, Pan G, Greenblatt J, Patterson S D, Wickens M, Bentley D L. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature. 1997;385:357–361. doi: 10.1038/385357a0. [DOI] [PubMed] [Google Scholar]
- 55.Minvielle-Sebastia L, Keller W. mRNA polyadenylation and its coupling to other RNA processing reactions and to transcription. Curr Opin Cell Biol. 1999;11:352–357. doi: 10.1016/S0955-0674(99)80049-0. [DOI] [PubMed] [Google Scholar]
- 56.Misteli T, Spector D L. RNA polymerase II targets pre-mRNA splicing factors to transcription sites in vivo. Mol Cell. 1999;3:697–705. doi: 10.1016/s1097-2765(01)80002-2. [DOI] [PubMed] [Google Scholar]
- 57.Mogridge J, Mah T, Greenblatt J. A protein-RNA interaction network facilitates the template-independent cooperative assembly on RNA polymerase of a stable antitermination complex containing the lamda N protein. Genes Dev. 1995;9:2831–2845. doi: 10.1101/gad.9.22.2831. [DOI] [PubMed] [Google Scholar]
- 58.Morris D P, Greenleaf A L. The splicing factor, Prp40, binds the phosphorylated carboxyl-terminal domain of RNA polymerase II. J Biol Chem. 2000;275:39935–39943. doi: 10.1074/jbc.M004118200. [DOI] [PubMed] [Google Scholar]
- 59.Mortillaro M J, Blencowe B J, Wei X, Nakayasu H, Du L, Warren S L, Sharp P A, Berezney R. A hyperphosphorylated form of the large subunit of RNA polymerase II is associated with splicing complexes and the nuclear matrix. Proc Natl Acad Sci USA. 1996;93:8253–8257. doi: 10.1073/pnas.93.16.8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Myers L C, Kornberg R D. Mediator of transcriptional regulation. Annu Rev Biochem. 2000;69:729–749. doi: 10.1146/annurev.biochem.69.1.729. [DOI] [PubMed] [Google Scholar]
- 61.Neubauer G, King A, Rappsilber J, Calvio C, Watson M, Ajuh P, Sleeman J, Lamond A, Mann M. Mass spectrometry and EST-database searching allows characterization of the multi-protein spliceosome complex. Nat Genet. 1998;20:46–50. doi: 10.1038/1700. [DOI] [PubMed] [Google Scholar]
- 62.Neugebauer K M, Roth M B. Transcription units as RNA processing units. Genes Dev. 1997;11:3279–3285. doi: 10.1101/gad.11.24.3279. [DOI] [PubMed] [Google Scholar]
- 63.Olsen H S, Rosen C A. Contribution of the TATA motif to Tat-mediated transcriptional activation of human immunodeficiency virus gene expression. J Virol. 1992;66:5594–5597. doi: 10.1128/jvi.66.9.5594-5597.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Orphanides G, Lagrange T, Reinberg D. The general transcription factors of RNA polymerase II. Genes Dev. 1996;10:2657–2683. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- 65.Otero G, Fellows J, Li Y, de Bizemont T, Dirac A M, Gustafsson C M, Erdjument-Bromage H, Tempst P, Svejstrup J Q. Elongator, a multisubunit component of a novel RNA polymerase II holoenzyme for transcriptional elongation. Mol Cell. 1999;3:109–118. doi: 10.1016/s1097-2765(00)80179-3. [DOI] [PubMed] [Google Scholar]
- 66.Perriman R, Ares M., Jr ATP can be dispensable for prespliceosome formation in yeast. Genes Dev. 2000;14:97–107. [PMC free article] [PubMed] [Google Scholar]
- 67.Ping Y H, Rana T M. Tat-associated kinase (P-TEFb): a component of transcription preinitiation and elongation complexes. J Biol Chem. 1999;274:7399–7404. doi: 10.1074/jbc.274.11.7399. [DOI] [PubMed] [Google Scholar]
- 68.Price D H. P-TEFb, a cyclin-dependent kinase controlling elongation by RNA polymerase II. Mol Cell Biol. 2000;20:2629–2634. doi: 10.1128/mcb.20.8.2629-2634.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rain J C, Rafi Z, Rhani Z, Legrain P, Kramer A. Conservation of functional domains involved in RNA binding and protein- protein interactions in human and Saccharomyces cerevisiae pre-mRNA splicing factor SF1. RNA. 1998;4:551–565. doi: 10.1017/s1355838298980335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Reines D, Conaway R C, Conaway J W. Mechanism and regulation of transcriptional elongation by RNA polymerase II. Curr Opin Cell Biol. 1999;11:342–346. doi: 10.1016/S0955-0674(99)80047-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Richardson J P. Structural organization of transcription termination factor Rho. J Biol Chem. 1996;271:1251–1254. doi: 10.1074/jbc.271.3.1251. [DOI] [PubMed] [Google Scholar]
- 72.Rosen G D, Barks J L, Iademarco M F, Fisher R J, Dean D C. An intricate arrangement of binding sites for the Ets family of transcription factors regulates activity of the alpha 4 integrin gene promoter. J Biol Chem. 1994;269:15652–15660. [PubMed] [Google Scholar]
- 73.Rutz B, Seraphin B. A dual role for BBP/ScSF1 in nuclear pre-mRNA retention and splicing. EMBO J. 2000;19:1873–1886. doi: 10.1093/emboj/19.8.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rutz B, Seraphin B. Transient interaction of BBP/ScSF1 and Mud2 with the splicing machinery affects the kinetics of spliceosome assembly. RNA. 1999;5:819–831. doi: 10.1017/s1355838299982286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shilatifard A, Conaway J W, Conaway R C. Mechanism and regulation of transcriptional elongation and termination by RNA polymerase II. Curr Opin Genet Dev. 1997;7:199–204. doi: 10.1016/s0959-437x(97)80129-3. [DOI] [PubMed] [Google Scholar]
- 76.Steinmetz E J. Pre-mRNA processing and the CTD of RNA polymerase II: the tail that wags the dog? Cell. 1997;89:491–494. doi: 10.1016/s0092-8674(00)80230-5. [DOI] [PubMed] [Google Scholar]
- 77.Sudol M. Structure and function of the WW domain. Prog Biophys Mol Biol. 1996;65:113–132. doi: 10.1016/s0079-6107(96)00008-9. [DOI] [PubMed] [Google Scholar]
- 78.Sudol M, Hunter T. New wrinkles for an old domain. Cell. 2000;103:1001–1004. doi: 10.1016/s0092-8674(00)00203-8. [DOI] [PubMed] [Google Scholar]
- 79.Sudol M, Sliwa K, Russo T. Functions of WW domains in the nucleus. FEBS Lett. 2001;490:190–195. doi: 10.1016/s0014-5793(01)02122-6. [DOI] [PubMed] [Google Scholar]
- 80.Suñé C, Garcia-Blanco M A. Transcriptional cofactor CA150 regulates RNA polymerase II elongation in a TATA-box-dependent manner. Mol Cell Biol. 1999;19:4719–4728. doi: 10.1128/mcb.19.7.4719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Suñé C, Hayashi T, Liu Y, Lane W S, Young R A, Garcia-Blanco M A. CA150, a nuclear protein associated with the RNA polymerase II holoenzyme, is involved in Tat-activated human immunodeficiency virus type 1 transcription. Mol Cell Biol. 1997;17:6029–6039. doi: 10.1128/mcb.17.10.6029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Uptain S M, Kane C M, Chamberlin M J. Basic mechanisms of transcript elongation and its regulation. Annu Rev Biochem. 1997;66:117–172. doi: 10.1146/annurev.biochem.66.1.117. [DOI] [PubMed] [Google Scholar]
- 83.Verdecia M A, Bowman M E, Lu K P, Hunter T, Noel J P. Structural basis for phosphoserine-proline recognition by group IV WW domains. Nat Struct Biol. 2000;7:639–643. doi: 10.1038/77929. [DOI] [PubMed] [Google Scholar]
- 84.Wada T, Orphanides G, Hasegawa J, Kim D K, Shima D, Yamaguchi Y, Fukuda A, Hisatake K, Oh S, Reinberg D, Handa H. FACT relieves DSIF/NELF-mediated inhibition of transcriptional elongation and reveals functional differences between P-TEFb and TFIIH. Mol Cell. 2000;5:1067–1072. doi: 10.1016/s1097-2765(00)80272-5. [DOI] [PubMed] [Google Scholar]
- 85.Wada T, Takagi T, Yamaguchi Y, Watanabe D, Handa H. Evidence that P-TEFb alleviates the negative effect of DSIF on RNA polymerase II-dependent transcription in vitro. EMBO J. 1998;17:7395–7403. doi: 10.1093/emboj/17.24.7395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wind M, Reines D. Transcription elongation factor SII. Bioessays. 2000;22:327–336. doi: 10.1002/(SICI)1521-1878(200004)22:4<327::AID-BIES3>3.0.CO;2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wittschieben B O, Otero G, de Bizemont T, Fellows J, Erdjument-Bromage H, Ohba R, Li Y, Allis C D, Tempst P, Svejstrup J Q. A novel histone acetyltransferase is an integral subunit of elongating RNA polymerase II holoenzyme. Mol Cell. 1999;4:123–128. doi: 10.1016/s1097-2765(00)80194-x. [DOI] [PubMed] [Google Scholar]
- 88.Wrehlke C, Wiedemeyer W R, Schmitt-Wrede H P, Mincheva A, Lichter P, Wunderlich F. Genomic organization of mouse gene zfp 162. DNA Cell Biol. 1999;18:419–428. doi: 10.1089/104454999315303. [DOI] [PubMed] [Google Scholar]
- 89.Yagi R, Chen L F, Shigesada K, Murakami Y, Ito Y. A WW domain-containing yes-associated protein (YAP) is a novel transcriptional co-activator. EMBO J. 1999;18:2551–2562. doi: 10.1093/emboj/18.9.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yamaguchi Y, Takagi T, Wada T, Yano K, Furuya A, Sugimoto S, Hasegawa J, Handa H. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell. 1999;97:41–51. doi: 10.1016/s0092-8674(00)80713-8. [DOI] [PubMed] [Google Scholar]
- 91.Yamaguchi Y, Wada T, Watanabe D, Takagi T, Hasegawa J, Handa H. Structure and function of the human transcription elongation factor DSIF. J Biol Chem. 1999;274:8085–8092. doi: 10.1074/jbc.274.12.8085. [DOI] [PubMed] [Google Scholar]
- 92.Yan D, Perriman R, Igel H, Howe K J, Neville M, Ares M., Jr CUS2, a yeast homolog of human Tat-SF1, rescues function of misfolded U2 through an unusual RNA recognition motif. Mol Cell Biol. 1998;18:5000–5009. doi: 10.1128/mcb.18.9.5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yankulov K, Blau J, Purton T, Roberts S, Bentley D L. Transcriptional elongation by RNA polymerase II is stimulated by transactivators. Cell. 1994;77:749–759. doi: 10.1016/0092-8674(94)90058-2. [DOI] [PubMed] [Google Scholar]
- 94.Yuryev A, Patturajan M, Litingtung Y, Joshi R V, Gentile C, Gebara M, Corden J L. The C-terminal domain of the largest subunit of RNA polymerase II interacts with a novel set of serine/arginine-rich proteins. Proc Natl Acad Sci USA. 1996;93:6975–6980. doi: 10.1073/pnas.93.14.6975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zarrinpar A, Lim W A. Converging on proline: the mechanism of WW domain peptide recognition. Nat Struct Biol. 2000;7:611–613. doi: 10.1038/77891. [DOI] [PubMed] [Google Scholar]
- 96.Zeng C, Berget S M. Participation of the C-terminal domain of RNA polymerase II in exon definition during pre-mRNA splicing. Mol Cell Biol. 2000;20:8290–8301. doi: 10.1128/mcb.20.21.8290-8301.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhang D, Childs G. Human ZFM1 protein is a transcriptional repressor that interacts with the transcription activation domain of stage-specific activator protein. J Biol Chem. 1998;273:6868–6877. doi: 10.1074/jbc.273.12.6868. [DOI] [PubMed] [Google Scholar]
- 98.Zhang D, Paley A J, Childs G. The transcriptional repressor ZFM1 interacts with and modulates the ability of EWS to activate transcription. J Biol Chem. 1998;273:18086–18091. doi: 10.1074/jbc.273.29.18086. [DOI] [PubMed] [Google Scholar]
- 99.Zhou M, Halanski M A, Radonovich M F, Kashanchi F, Peng J, Price D H, Brady J N. Tat modifies the activity of CDK9 to phosphorylate serine 5 of the RNA polymerase II carboxyl-terminal domain during human immunodeficiency virus type 1 transcription. Mol Cell Biol. 2000;20:5077–5086. doi: 10.1128/mcb.20.14.5077-5086.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]