Summary
The low-density lipoprotein (LDL) receptor-related protein 2 (LRP2 or megalin) is representative of the phylogenetically conserved subfamily of giant LDL receptor-related proteins, which function in endocytosis and are implicated in diseases of the kidney and brain. Here we report high-resolution cryo-electron microscopy structures of LRP2 isolated from mouse kidney, at extracellular and endosomal pH. The structures reveal LRP2 to be a molecular machine that adopts a conformation for ligand binding at the cell surface and for ligand shedding in the endosome. LRP2 forms a homodimer, the conformational transformation of which is governed by pH-sensitive sites at both homodimer and intra-protomer interfaces. A subset of LRP2 deleterious missense variants in humans appears to impair homodimer assembly. These observations lay the foundation for further understanding the function and mechanism of LDL receptors and implicate homodimerization as a conserved feature of the LRP receptor subfamily.
Keywords: LRP2, megalin, LRP1, CD91, LDL, proteinuria, endocytosis, recycling, pH-sensitive, cryo-EM
Introduction
LRP2/megalin is the largest member of the LDL receptor (LDLR) family and a representative of the giant LDLR-related protein (LRP) subfamily that also includes LRP1 and LRP1b. All members of this subfamily adopt an architecture that is conserved from worms to humans1–5 (Figure S1). LRP2 was originally identified in kidney as the autoantigen gp330 in an animal model of autoimmune disease6–8, and subsequently named megalin due to its large size9,10. LRP2 is expressed in many absorptive epithelia, particularly the apical membrane of the proximal tubules of the kidney, but also epithelia of the inner ear, thyroid, parathyroid, and alveoli11–14. LRP2 is also expressed in the central nervous system (CNS), including in neurons15, glia16, and the choroid plexus17,18, and plays signaling roles key to CNS development19–24.
Functional analysis has shown that LRP2 is an endocytic receptor that binds and internalizes many ligands including protease-protease inhibitor complexes25,26, vitamin-binding protein complexes27,28, hormones29, lipocalins30,31, and lipoproteins26,32–35. LRP2 recycles from endosomes to the cell membrane following clathrin-mediated endocytosis, capturing and releasing ligands dozens of times during its lifetime8,36–39 (Figure 1A).
Figure 1. Biochemical characterization of endogenous LRP2 reveals a stable dimer.
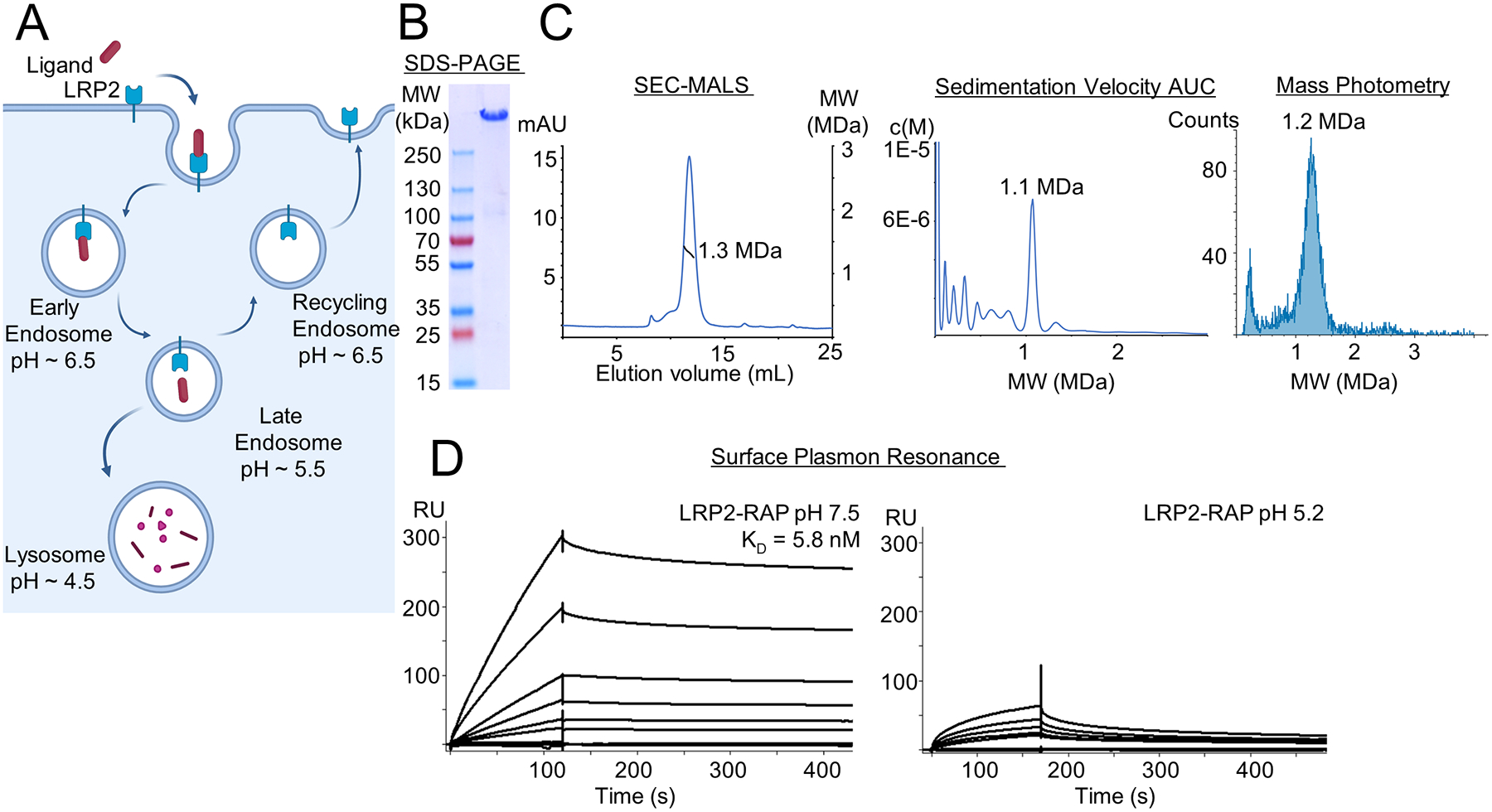
(A) Schematic representation of ligand binding by LRP2 and of LRP2 receptor recycling.
(B) SDS-PAGE reducing, denaturing gel of the purified LRP2 sample following SEC showing a single major species at ~600kD
(C) Biophysical characterization of LRP2 mass by SEC-MALS (left), Sedimentation velocity AUC (middle), and Mass Photometry (right). The three experiments demonstrate a dimeric form of purified LRP2 with a MW ~1.1 to 1.3 MDa. The fit of the MALS data is indicated as a black line on the SEC chromatogram.
(D) SPR characterization of RAP binding to purified LRP2 at pH 7.5 (left) and pH 5.2 (right) using injections of LRP2 varying from 0.82–200 nM over a surface of immobilized RAP. SPR sensorgrams for pH 7.5 could be fit to determine KD of 5.8 nM, but sensorgrams for pH 5.2 could not be fit. Qualitatively, binding is substantially reduced at pH 5.2.
LRP2 recovers low molecular weight (LMW) proteins that escape glomerular filtration barriers, a crucial component of normal kidney function. LMW proteinuria can lead to tubulointerstitial damage, and genome-wide association studies (GWAS) link LRP2 variants with chronic kidney disease40–45. Loss of function in LRP2 in humans leads to Donnai-Barrow syndrome (DBS, OMIM #222448), a Mendelian genetic disorder characterized by LMW proteinuria and CNS disorders including sensorineural hearing loss, myopia, hypertelorism, agenesis/hypoplasia of the corpus callosum, and forebrain malformations46. Thus, DBS implicates a role for LRP2 as a multi-ligand endocytic receptor essential for capturing ligands in both the kidney and brain.
Alongside LRP1, LRP2 is implicated in the capture of proteins associated with the pathogenesis of Alzheimer’s disease (AD) and other tauopathies. LRP1 regulates tau uptake and spread for both normal and pathogenic forms of tau47. LRP2 and LRP1 recognize a similar complement of ligands26,48,49, including apoE4 that modulates tau uptake and strongly associates with AD50,51. A role for LRP2 in AD pathogenesis was further suggested by polymorphisms in LRP2 associated with susceptibility to AD52,53 and studies showing that Aβ40 and Aβ42 peptides are cleared across the blood-brain barrier by LRP254,55. Similarly, clusterin/apoJ associates with AD in GWAS56,57 and is cleared across the blood-brain barrier by LRP254. These studies demonstrate the importance of LRP-mediated endocytosis of specific ligands implicated in common diseases of the brain.
LRP2 is a receptor for over 75 putative ligands1, and a complex domain structure enables this breadth of function. LDLR family members combine three types of modules in their ectodomains: epidermal growth factor (EGF)-like repeats (E), YWTD motif-containing β-propellers (P), and LDL receptor type A repeats (L)48,49. LRP2 is a single pass transmembrane protein comprising 8 β-propeller domains (P1-P8), 17 EGF-like domains (E1-E17), and 36 LDL receptor type A repeats (L1-L36) grouped in four separate clusters (R1-R4)9,10. Ligand-binding L repeats contain a conserved Ca2+-coordination site consisting of 4 acidic side chains and 2 backbone carbonyls (Table S1)58. Selected EGF-like domains contain a conserved Ca2+-binding (cb) motif implicated in ligand binding (cbE6, cbE11, cbE12, and cbE15) (Table S2)59–61. The β-propellers are typified by YWTD repeats62,63. Binding site mapping and structural studies have shown that the L repeats are primarily responsible for ligand binding to both LRP1 and LRP264–77, although the EGF-like domains and β-propellers have also been implicated in ligand binding78–92.
Additional insights into LRP receptor function can be inferred from biochemical analyses of LDLR. LDLR has 7 ligand-binding L repeats followed by a region with homology to the EGF precursor (EGFP), containing EGF domains flanking a β-propeller93. The EGFP region has been implicated in ligand release at endosomal pH by competing for ligand binding94,95. Receptors lacking the EGFP region bind LDL or β-VLDL but fail to release them in the endosome95,96. Thus, the LDLR ectodomain consists of one ligand-binding-and-release segment. The LRP receptors, by contrast, each have four clusters of L repeats, all abutted by a C-terminal region with EGFP homology, suggesting the possibility of four ligand-binding/release regions.
Knowledge of the structural biology of the LDLR family primarily stems from the crystal structure of the human LDLR ectodomain at endosomal pH97. The structure reveals LDLR in an inhibited conformation with the L4-L5 repeats binding the β-propeller in a characteristic motif expected to block ligand interaction: a lysine from the β-propeller hydrogen bonds with aspartates at a Ca2+-coordination site in the L repeat. This motif provides the basis for a proposed mechanism for ligand release and receptor recycling, whereby the β-propeller acts as an intramolecular ligand at endosomal pH to displace ligands captured at the cell surface97. Receptor-associated Protein (RAP), an ER chaperone that competes for ligand binding with all LDLR family members26,98–106, has since been observed to bind L repeats via the same motif66,69,107. Indeed, numerous structures have now confirmed the importance of this binding motif, including LDLR L3-L4:RAPd366, LDLR L4:β2-glycoprotein I domain V (B2GPI-DV)108, LRP1 L17:apoE(130–149)68, and LDLR L2:VZV G protein and LDLR L3:VZV G protein109. In contrast to this clearly observed motif in LDLR at endosomal pH, structures of LDLR at neutral pH, such as that between the ectodomain of LDLR and PCSK980, have been limited by disorder of the L repeats which extend away from the β-propeller and could not be modeled. Thus, while structural data from LDLR family protein domains has identified important functional motifs, it has not unveiled the mechanisms responsible for the family’s pH-sensitive structural transformations.
Here, we report high-resolution cryo-EM structures of endogenously purified LRP2 at endosomal and extracellular pH. In the context of a homodimer, LRP2 executes substantial structural transitions as it navigates endosomal compartments. These structural transitions reveal pH-sensitive homodimer interfaces, demonstrate new modes of ligand-binding regulation, and provide a mechanistic understanding of deleterious mutations in LRP2 that result in kidney and brain diseases.
Results
Biochemical characterization of endogenous LRP2 reveals a stable dimer
A prior study produced LRP1 ectodomain in a recombinant mammalian expression system, and EM analysis showed a “beads on a string” structure110. Building on landmark endogenous purifications of LRP1 and LRP2 instead25,106,111–114, we purified endogenous LRP2 at pH 7.5 from the apical membrane of the mouse kidney proximal tubule. Mass spectrometry analysis demonstrated the purity of our LRP2 sample (Table S3) as did SDS-PAGE gel (Figure 1B). Mass photometry, SEC/MALS, and analytical ultracentrifugation demonstrated a size of ~1.1–1.3 MDa for LRP2 (Figure 1C), suggesting that the endogenous assembly of LRP2 is dimeric. Our findings are consistent with Farquhar’s reports that LRP2 multimerizes in the rough ER during biosynthesis115,116. In surface plasmon resonance (SPR) experiments our LRP2 species bound RAP with high affinity at pH 7.5, but with substantially reduced affinity at pH 5.2, consistent with established biology (Figure 1D)39,110,117 and demonstrating that our purified LRP2 is an active protein receptor.
Cryo-EM analysis of LRP2
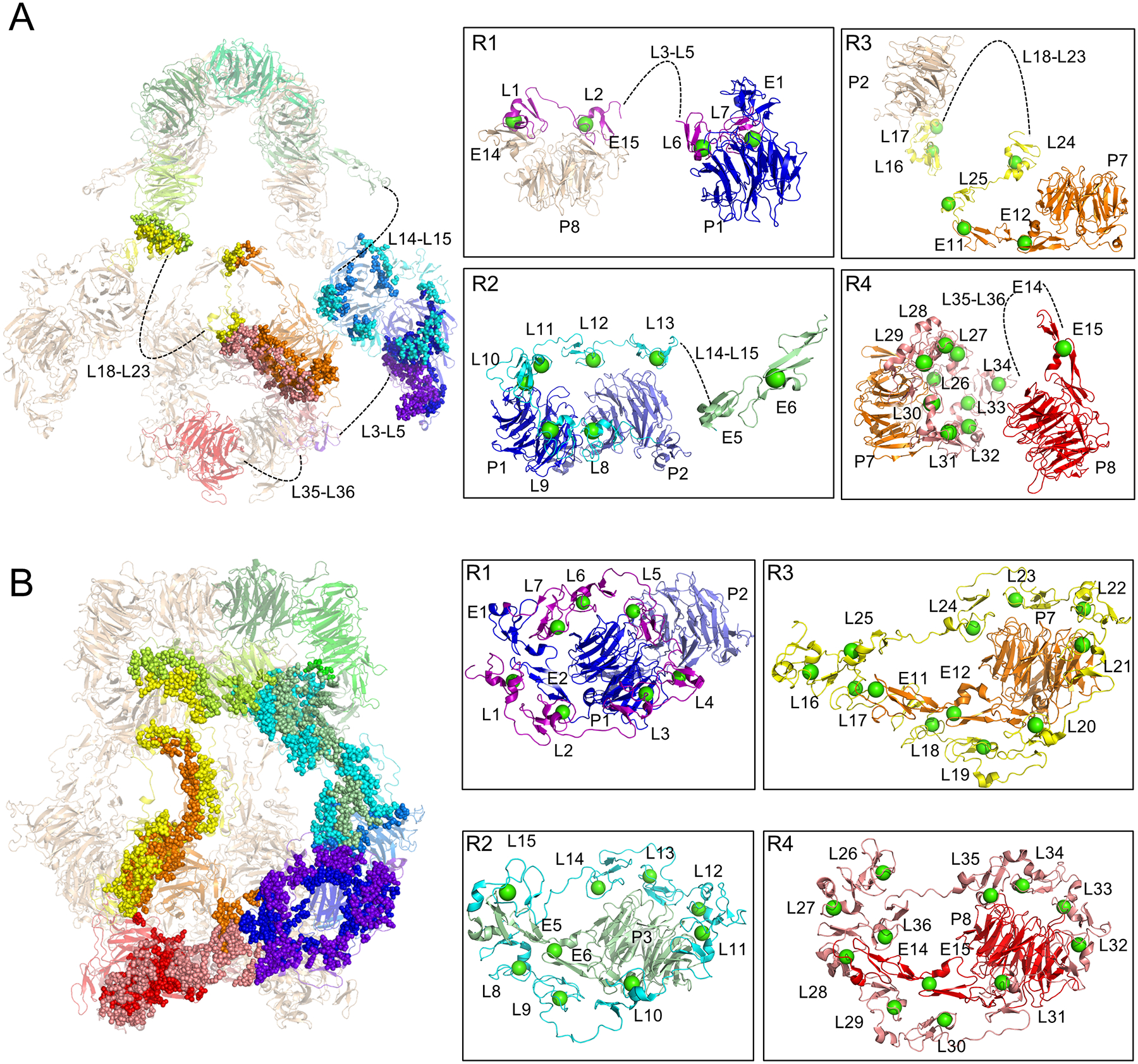
Endogenous preparations of LRP2 at pH 7.5 and pH 5.2 were frozen in vitreous ice, imaged with a Titan Krios microscope, and data were processed in Cryosparc, Relion, and CryoDRGN (Figure S2A–N, S3A–N)118–121 (see STAR Methods). The processed datasets revealed a homodimer with two-fold symmetry (Figure S2B–D, S3B–D) and yielded structures of LRP2 at resolutions of 2.83 Å for pH 7.5 (Figure S2H) and 2.97 Å for pH 5.2 (Figure S3H). Flexibility of the assembly was more significant at pH 7.5 where loops of low-resolution density extend out from a higher resolution core (Figure S2E, Movie S1, S2). As a result, at extracellular pH, portions of each cluster of ligand-binding repeats were left unmodeled because they extended rope-like out into the solvent. In contrast, nearly the entire Cα trace for the LRP2 ectodomain could be modeled for the more compact endosomal pH structure. The contrast in the overall flexibility of the two structures is consistent with a biological model where L repeats are free at extracellular pH but bound to EGFP domains at endosomal pH.
Architecture of the LRP2 homodimer at extracellular and endosomal pH
The resolved assembly at extracellular pH has dimensions of approximately 270 Å × 265 Å × 160 Å and displays two-fold symmetry (Figure 2A). Each protomer in the homodimer has a hairpin morphology and can be categorized into major regions in terms of the organization of the clusters of L repeats and EGFP domains: two “legs” including an ascending leg of L repeat clusters I and II (R1-R2) and β-propellers 1 and 2 of each protomer (P1-P2), a central “canopy” where the tandemly repeated EGFP domains of each protomer including β-propellers 3–6 (P3-P6) abut one another face to face at the apex of the structure to provide the major homodimer interface of the assembly (Figure 2B), and lastly a “descending leg” where L repeat clusters III and IV (R3-R4) and β-propellers 7 and 8 (P7-P8) of both protomers descend like a column toward the membrane insertion, at a distance of about 25 Å apart (Figure 2A). The N-terminus of one protomer forms a homodimer interface near the C-terminus of the second protomer. Notably, portions of each L repeat cluster are disordered as they extend rope-like through solvent (Figure 2B, 2E).
Figure 2. Architecture of the LRP2 homodimer at extracellular and endosomal pH.
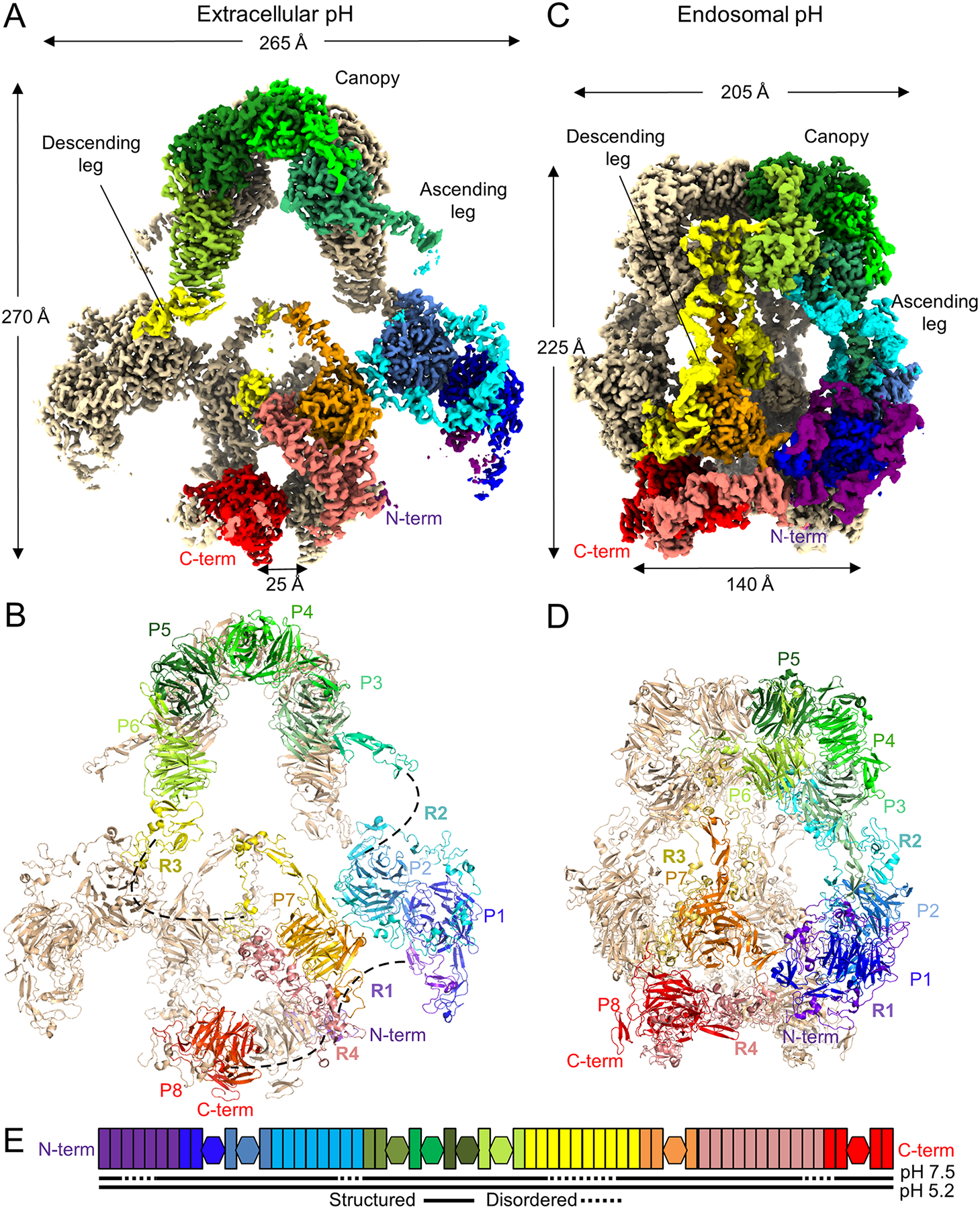
(A) Cryo-EM map of LRP2 at pH 7.5. One protomer is colored in rainbow, from the N-terminus (purple) to the C-terminus (red); the other is in wheat. Distinctive regions are labelled, and dimensions are shown. Coloring and labeling is consistent throughout the figure. Distance between the transmembrane insertions is labeled.
(B) Cryo-EM model of LRP2 at pH 7.5. β-propellers (P) and groups of L repeats (R) are labeled. Unmodeled portions of flexible ligand-binding regions in the structure are dashed lines.
(C) Cryo-EM map of LRP2 at pH 5.2.
(D) Cryo-EM model of LRP2 at pH 5.2. Labeling is as for panel B
(E) Domain diagram as in Figure S1, now colored in rainbow from N-terminus to C-terminus. Below the diagram, structured disordered domains are indicated by solid and dotted lines for pH 7.5 and pH 5.2 respectively.
See also Figures S2, S3, and Tables S1, S2, S4, and Movies S1, S2, S3
At endosomal pH, substantial conformational change renders the overall assembly more compact, with dimensions now 225 Å × 205 Å × 160 Å (Figure 2C). Despite the conserved hairpin contour to the overall structure, most intra-protomer and all homodimer interfaces are altered in the assembly by transition to endosomal pH (Figure S4A–B, Movie S3), and Cα RMSD between the two structures is 51 Å. All clusters of L repeats are now ordered and packed against β-propellers (Figure 2D, 2E), the buried surface area within the canopy at the apex of the structure is substantially reduced (Figure 2D), and the transmembrane insertion points have become widely spaced at >140 Å apart between protomers (Figure 2C).
A major consequence of these structural transformations is a shift from predominantly homodimer interface contacts at extracellular pH to predominantly intra-protomer contacts at endosomal pH (Figure S4A–B). This redistribution of buried area reflects a transition of LRP2 from a ligand-receptive state at extracellular pH to a ligand-shedding state at endosomal pH. The homodimer assembly maintains the ligand-binding R clusters in largely solvent-exposed orientations at extracellular pH (Figure 2B), and a reduction in homodimer contacts at endosomal pH makes available additional surfaces in the protomers where R clusters can pack to exclude solvent from ligand-binding sites (Figure 2D).
pH-specific homodimer interfaces regulate ligand binding
Homodimer interfaces are distinct between the two structures at pH 7.5 and pH 5.2 (Figure S4A–B). These pH-specific interfaces regulate ligand binding to LRP2. At extracellular pH, orientations of certain L repeats are fixed and intramolecular ligands are immobilized. At endosomal pH, the homodimer interfaces help prevent ligand binding by limiting ligand access to the center of the LRP2 assembly and by burying selected EGF-like domains.
Homodimer interfaces bury a total of 6472 Å2 of surface area at extracellular pH (Figure 3A). This homodimer assembly provides a scaffold that enables a portion of every R cluster to maintain a fully solvent-exposed loop that is receptive to ligand at extracellular pH (Figure 2B). Homodimer interfaces can also explicitly orient individual L repeats. At one interface, L17 of R3 in one protomer packs against P2 of the second protomer, such that the Ca2+-coordinating ligand-binding site is fixed facing outward and available to solvent, maintaining it in a ligand-receptive state (Figure 3B). L25 of R3 and L32 of R4 are similarly fixed in ligand-receptive states through homodimer contacts.
Figure 3. pH-specific homodimer interfaces regulate ligand-binding.
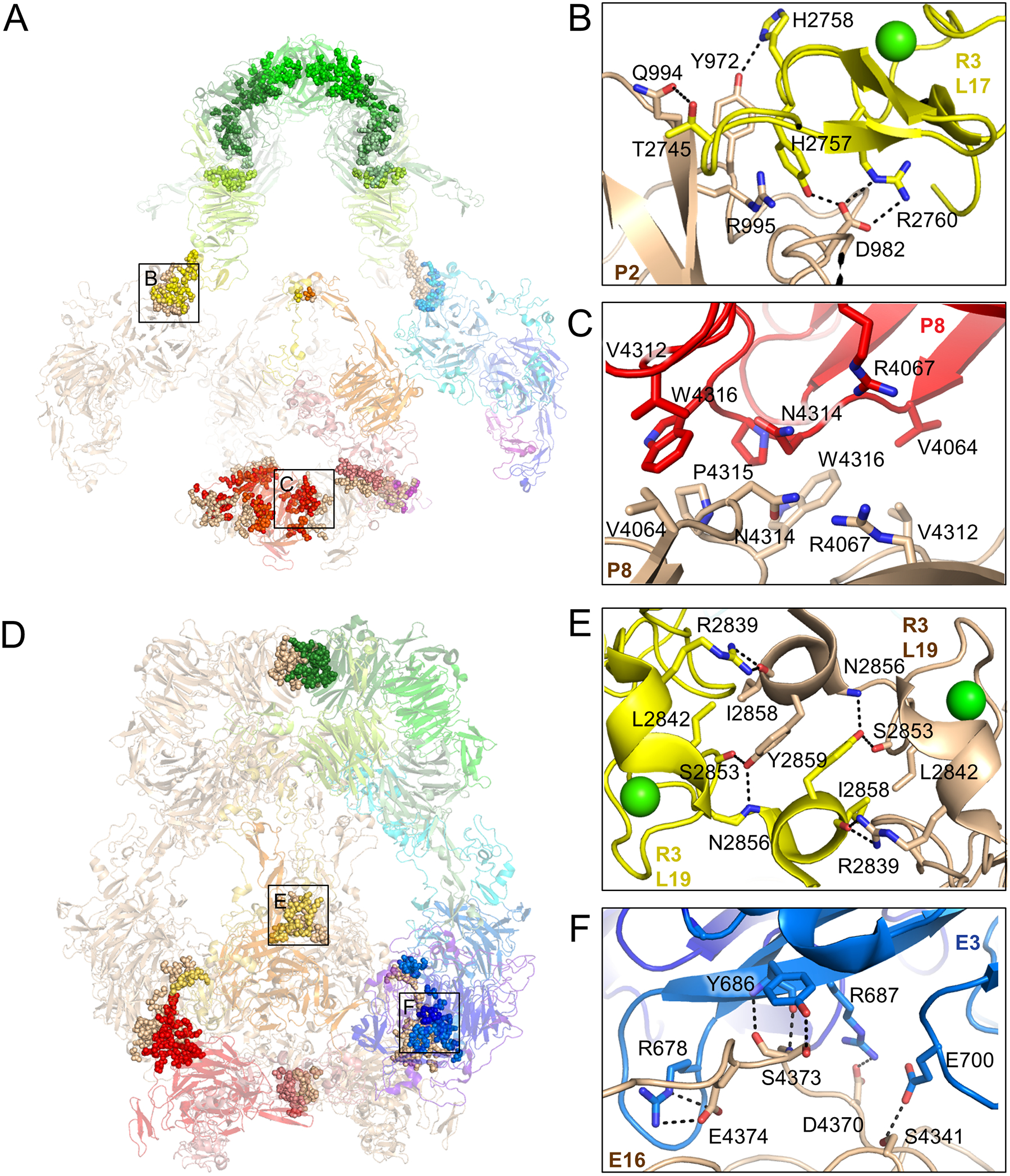
(A) Residues involved in homodimer contacts between protomers at pH 7.5 are rendered as spheres superimposed on a semi-transparent cartoon depiction of the LRP2 structure (left panel). Residue level interactions are shown at right for the corresponding labeled boxed regions. Identities of interacting domains are labeled.
(B) L17 of R3 is stabilized with its ligand-binding face facing solvent by virtue of polar contacts with P2 of the second protomer.
(C) R4067, which functions as an intramolecular ligand at pH 5.2, is buried in a hydrophobic interface between the P8 domains of the two protomers at pH 7.5, preventing it from competing with ligand at ligand-binding repeats.
(D) Depiction and labeling of LRP2 at pH 5.2 according to panel A.
(E) L19 from R3 of the two protomers engage in symmetric contacts that tether the protomers together at the center of the assembly, excluding solvent and making ligand-binding surfaces inaccessible.
(F) E3 of the first protomer packs against E16 of the second protomer in the homodimer, tethering the protomers together near the membrane insertion points.
See also Figure S4
Much of the homodimer interface at extracellular pH occurs between propellers in the canopy (2485 Å2) and at a symmetric P8-P8 interface (828 Å2). These interfaces serve to fix intramolecular ligands at significant distances from their cognate L repeats, making them incapable of displacing ligand at extracellular pH. There are 28 intramolecular ligands in LRP2 that engage L repeats at endosomal pH, but at extracellular pH they are separated by a median distance of 65 Å from their cognate repeats. In certain cases, intramolecular ligands are fully buried at homodimer interfaces, such as R4067 which packs inside the largely hydrophobic P8:P8 interface (Figure 3C).
At endosomal pH the homodimer interface is reduced to 3396 Å2, a change largely driven by the canopy region where now only 722 Å2 is buried (Figure 3D, Movie S3). The remaining homodimer interfaces now serve to negatively regulate ligand-binding. Eight L repeats are without an intra-molecular ligand at endosomal pH, yet most of these are still inaccessible to ligand-binding due to packing coordinated by the homodimer assembly. L19 of R3 engages in symmetric contacts across the homodimer interface to closely tether the two protomers together at the center of the assembly (Figure 3E). L15 and L18 lack intramolecular ligands at endosomal pH, but by virtue of the close approach of the protomers mediated by the symmetric R3 homodimer contacts, they remain inaccessible to ligand in an enclosed space near the center of the assembly. The close approach of E3 from one protomer with E16 of the second protomer that enables main chain to main chain hydrogen bonding also tethers the assembly together near its membrane insertions (Figure 3F).
pH-sensitive interfaces define the morphology of the canopy in the homodimer assembly
Local resolution analysis reveals that the canopy is the most rigid region in the homodimer at extracellular pH, suggesting that it plays a key role in the stability of the overall protein assembly (Figure S2E). Examination of the density in the canopy revealed two symmetrically related pairs of putative Ca2+-coordination sites that serve to facilitate contacts across the interface (Figure 4A), with the first pair of putative Ca2+ ions coordinated by D2257 and the second pair by D1621.
Figure 4. pH-sensitive interfaces define the morphology of the canopy in the homodimer assembly.
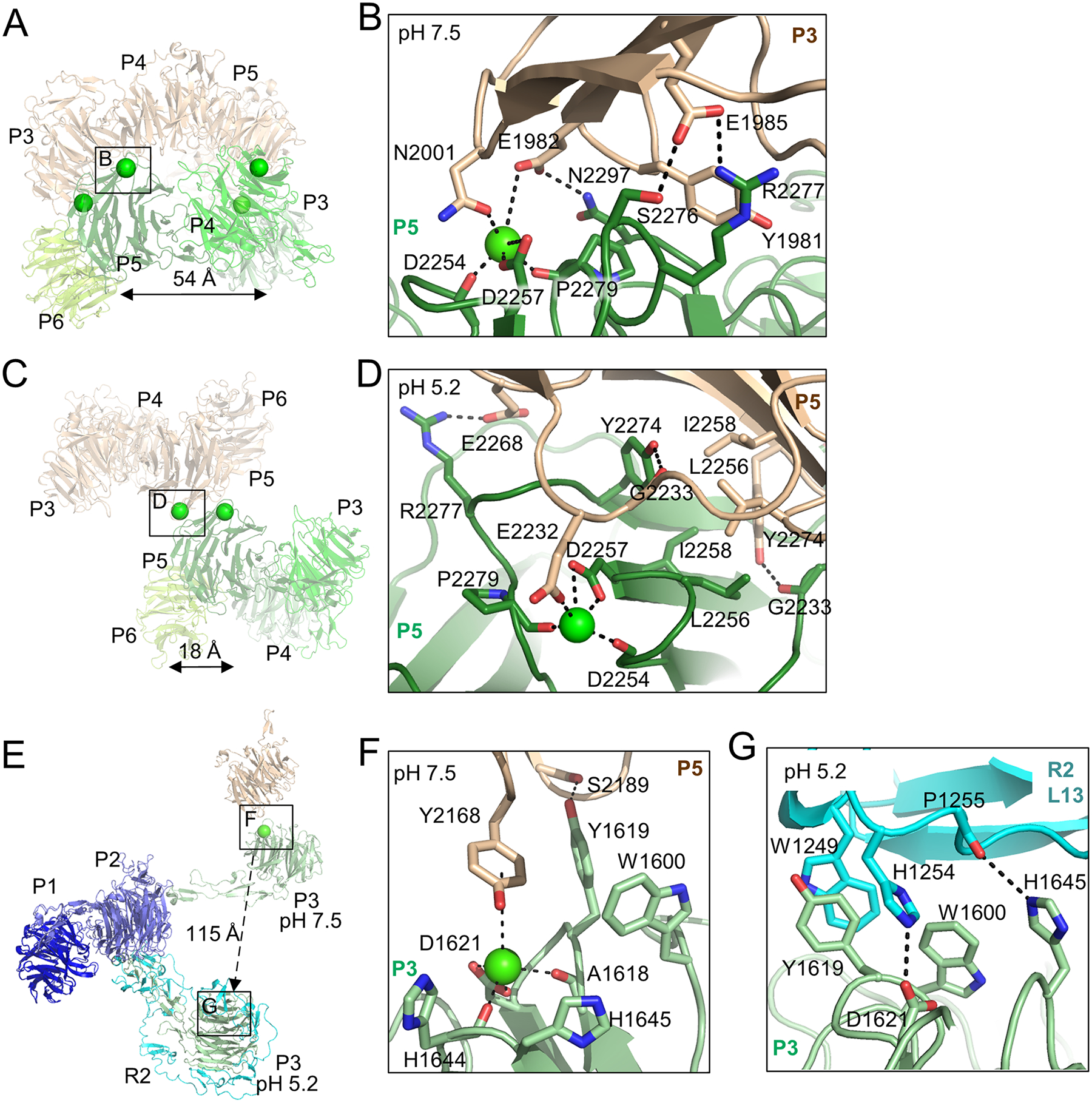
(A) Depiction of the canopy as a semi-transparent cartoon at pH 7.5. The two pairs of symmetrically related putative Ca2+ ions coordinated in the canopy are depicted by spheres. The putative Ca2+ ion coordinated by conserved D2257 is boxed. The distance between the two putative Ca2+ ions coordinated by D2257 is indicated.
(B) Residue-level interactions observed around the D2257-coordinated putative Ca2+ ion at pH 7.5.
(C) Depiction of the canopy at pH 5.2. One pair of putative Ca2+ ions has dissociated due to the reduced pH, but the putative Ca2+ ions coordinated by D2257 persist, one of which is boxed.
(D) Residue-level interactions observed around the D2257-coordinated putative Ca2+ ion at pH 5.2.
(E) P1 from the LRP2 structures at pH 7.5 and pH 5.2 have been superimposed with RMSD 1.3 Å. The P1-P3 regions from both structures are depicted based on this superimposition as cartoons. Relative to the fixed position of P1, P3 translates 115Å following pH reduction from 7.5 to 5.2, and the putative Ca2+ ion coordinated by conserved D1621 dissociates, enabling interaction of P3 with ligand-binding repeats from R2. The regions that include conserved D1621 are boxed on the P3 domains at both pH’s.
(F) Residue-level interactions observed around the D1621-coordinated putative Ca2+ ion at pH 7.5. Y2168 engages in a π-cation interaction with the putative Ca2+ ion.
(G) Residue-level interactions observed around D1621 at pH 5.2 following dissociation of the putative Ca2+ ion. D1621 now forms an intra-protomer salt bridge with H1254.
See also Figure S5
The first pair is coordinated by the side chain of D2257 at extracellular pH, and additional coordination is contributed by the backbone carbonyls of D2254 and P2279 and the side chains of N2001 and E1982 from the second protomer (Figure 4B). This putative Ca2+-coordination site orients the protomers to engage in multiple van der Waals contacts across the homodimer interface, including between P2279 and Y1981 (Figure 4B). At endosomal pH (Figure 4C), coordination by the D2257 side chain and carbonyls from P2279 and D2254 persists, but the homodimer interface changes, the putative Ca2+ ion pair translates from 54 Å to 18 Å apart (Figure 4C), and now the E2232 side chain from the second protomer participates in coordination (Figure 4D). Notably, D2257 is conserved across species in both LRP1 and LRP2 (Figure S5A, C, D).
The second pair of putative Ca2+ ions is present at extracellular pH but not at endosomal pH (Figure 4E). The side chain of D1621 coordinates at extracellular pH along with the backbone carbonyls from H1644 and A1618, with Y2168 from the second protomer coordinating via a π-cation interaction (Figure 4F). At endosomal pH, the pair dissociates and P3 from the canopy translates 115 Å relative to P1-P2 to engage in numerous intra-protomer contacts with R2 (Figure 4E, Movie S3). D1621 now forms a salt bridge with H1254 from the same protomer, and H1645 hydrogen bonds with a carbonyl from P1255 from the same protomer (Figure 4G). Like D2257, D1621 and H1645 are conserved across species in LRP1 and LRP2 (Figure S5A, B, D).
Thus, the pH-specific homodimer interfaces that help regulate ligand-binding (Figure 3) are undergirded by pH-sensitive homodimer contacts (Figure 4) mediated by highly conserved residues (Figure S5). These pH-sensitive homodimer contacts help define the morphology of the canopy region, such that 1763 Å2 of surface area is released from the canopy’s homodimer interface through the transition from extracellular to endosomal pH. This liberated surface area on the canopy’s β-propellers can now bind L repeats to displace ligand.
pH-dependent intra-protomer contacts govern ligand binding-site availability
The reduction in buried area at homodimer interfaces at endosomal pH enables an increase in intra-protomer interfaces. Buried area at intra-protomer interfaces has an inverse relationship to pH, with 8,168 Å2 of the ligand-binding clusters (R1-R4) buried against each protomer at extracellular pH and 18,258 Å2 buried at endosomal pH. Thus, LRP2 converts from a ligand-receptive state primarily characterized by homodimer interfaces at the cell surface (Figure 3A, D) to a ligand-shedding state characterized by intra-protomer interfaces (Figure 5A, B) in the endosomal compartment.
Figure 5. pH-dependent intra-protomer contacts govern ligand binding site availability.
(A) Residues involved in intra-protomer contacts at pH 7.5 are rendered as spheres superimposed on a semi-transparent cartoon depiction of the LRP2 structure (left panel). Flexible ligand-binding repeats that could not be modeled are indicated with dashed lines and labeled. Residue level interactions are shown at right in boxes for each of the four ligand-binding domains. Ca2+ ions in the L repeats are rendered as green spheres. Individual ligand-binding repeats within the larger domains are labeled. Domains from one protomer are colored in rainbow, and from the other protomer in wheat.
(B) Residues involved in intra-protomer contacts at pH 5.2 are rendered as spheres superimposed on a transparent cartoon depiction of the structure (left panel).
See also Figure S4
For each cluster of ligand-binding repeats (R1-R4), the transition to endosomal pH induces a thematic transformation: L repeats that were primarily either fully solvent-exposed or oriented with their Ca2+-coordinating site facing solvent are now packed with their Ca2+-coordinating site facing an EGFP domain (Figure 5B). R1 that was primarily solvent-exposed packs against P1, R2 releases from P1-P2 to pack against P3, R3 that was primarily solvent-exposed now packs against P3 and P7, and R4 that was primarily oriented with the Ca2+-coordinating sites of its L repeats fixed out towards solvent by P7 now packs against P8 (Figure 5A, B). The motif of an intramolecular ligand from an EGFP domain binding at the Ca2+-coordinating site of a L repeat to displace bound ligand that was first observed in LDLR97 is here recapitulated 28 times.
Published structures have not defined how the ligand-binding EGF-like domains enter a ligand-shedding state at endosomal pH. Our structures show that the EGF-like repeats also undergo a stereotypical change, whereby the transition to endosomal pH moves them from solvent-exposed positions (Figure 5A) to being encased on either side by adjacent L repeats: E1-E2 by R1, E5-cbE6 by R2, cbE11-cbE12 by R3, and E14-cbE15 by R4 (Figure 5B). The encasement of the EGF-like domains is facilitated by the fact that the EGF-like domains are themselves donors of intramolecular ligands to the very L repeats surrounding them. 11 of the 28 intramolecular ligands in LRP2 at endosomal pH are contributed by EGF-like domains, and these intramolecular ligands reside entirely within the EGF-like domain pairs (E1-E2, E5-cbE6, cbE11-cbE12, and E14-cbE15) (Table S2). In one example, cbE15 binds L29 and L31 via K4027 and R4048 respectively. In the process of inhibiting ligand binding in these L repeats, the EGF-like domain has at the same time brought the L repeats into such close proximity that they now concurrently inhibit its own ligand-binding (Figure 5B). The intra-protomer interactions between the L repeats and EGF-like domains are thus mutually inhibitory. Ligand-binding faces in the β-propellers are similarly buried when their intramolecular ligands bring L repeats into close apposition.
Notably, biochemical characterization of isolated groups of L repeats have shown similar ligand-binding affinities for different groups of L repeats64,65,75–77. However, our structural data show that each ligand-binding cluster has specific structural regulation at both extracellular and endosomal pH. At extracellular pH, one cluster is primarily flexible and open to solvent (R3), another is primarily fixed with its ligand-binding sites facing solvent (R4), and the other two clusters are a mix of flexible L repeats, L repeats fixed with ligand-binding sites facing solvent, and L repeats buried against EGFP domains bound with the classical inhibitory motif (R1 and R2). This variation in the disposition of L repeats across R1-R4 at extracellular and endosomal pH suggests a complex equilibrium where ligand-binding kinetics could vary substantially across different regions of the assembly and different groups of L repeats may be highly specific for certain ligands by virtue of their diverse contacts within the homodimer assembly.
pH-sensitive interfaces are essential for LRP2 function
We evaluated all known human mutations in LRP2 leading to loss-of-function to help identify critical features of the protein assembly. Over 40 mutations in LRP2 in humans leading to DBS have been published, but most involve frameshifts and truncations of the coded protein46,122. Of the published missense variants associated with human disease, many have plausible interpretations from sequence alone, such as production of unpaired cysteines (p.C286F)123, or disruption of secondary structure in β-propellers (p.R1578P)124.
A subset of LRP2 missense variants highlight the importance of pH-sensitive interfaces in the homodimer. A p.R3192Q mutation in human (p.R3194Q in mouse) completely ablates LRP2 expression and albumin endocytosis in the kidney proximal tubule in immunohistochemical experiments123, resulting in DBS. At extracellular pH, this arginine is not located at an intra-protomer or homodimer interface, but at endosomal pH, it serves as the intramolecular ligand for L20, where it docks with W2883, D2886, and D2890 of the ligand-binding repeat (Figure 6A). Mutation to glutamine ablates the π-cation interaction with tryptophan and breaks the salt bridges with aspartate residues. By thus inhibiting the formation of intra-protomer contacts at endosomal pH, the p.R3192Q mutation may impair either the biosynthetic trafficking of the LRP2 assembly or its recycling. Several reported missense variants of uncertain significance in LRP2 would inhibit the formation of similar intra-protomer contacts, including p.R1412W (ClinVar #1366634), p.H3096Q (ClinVar #1401841), p.R3554Q (ClinVar #1050333), and p.R4065Q (ClinVar #1438033). Mutation of the conserved D2256 residue (D2257 in mouse) that disturbs a putative Ca2+-coordination site within a pH-sensitive homodimer interface occurs in the compound heterozygote p.D2256Y/G2849A (Figure 6A). This genotype was reported in a patient with a syndrome potentially consistent with DBS, characterized by microcephaly, cerebellar abnormalities, and bilateral hearing loss125. D2256 is present at a symmetric homodimer interface at endosomal pH (Figure 4C, Figure 6A), suggesting that the p.D2256Y mutation may act in a dominant negative fashion.
Figure 6. pH-sensitive interfaces are essential for LRP2 function.
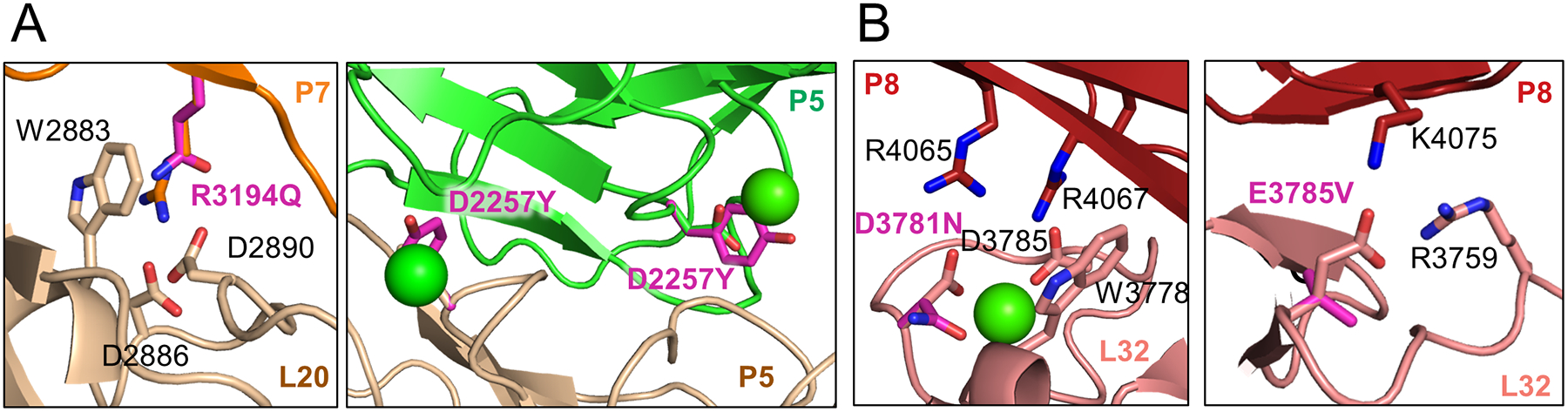
(A) Representation of the residues surrounding human mutations in LRP2 at pH 5.2 at pH-sensitive sites associated with syndromic phenotypes. The mutations disturb both intra-protomer (left panel) and homodimer (right panel) interfaces. The p.R3194Q mutation ablates a π-cation interaction with W2883 as well as salt bridges with D2886 and D2890. The p.D2257Y mutation eliminates the ability to coordinate a putative Ca2+ ion at a pH-sensitive interface within the canopy of the homodimer.
(B) Representation of the residues surrounding human mutations in LRP2 at pH 5.2 at pH-insensitive sites associated with non-syndromic phenotypes. The p.D3781N mutation weakens coordination of the Ca2+ ion and breaks a salt-bridge with R4065. The p.E3785V mutation breaks a salt bridge with R3759.
In contrast to mutations associated with syndromic phenotypes that affect pH-sensitive interfaces, mutations at pH-insensitive sites are associated with non-syndromic phenotypes including developmental delay and differences in sex development (DSD). These milder phenotypes are associated with missense variants that affect intra-protomer interactions without perturbing pH-sensitive interfaces. The p.D3779N mutation (p.D3781N in mouse) is associated with a delay in speech development and mild intellectual disability126. Our structure reveals that this mutation ablates an intra-protomer salt bridge and weakens the Ca2+-coordination site of the L repeat but leaves the adjacent pH-sensitive inhibitory motif intact (Figure 6B). The p.E3763V mutation (p.E3765V in mouse), associated with DSD and identified from a cell line bank from individuals with gonadal abnormalities, ablates a salt bridge in the linker region between L31 and L32 (Figure 6B). Thus, whereas LRP2 mutations affecting the pH-sensitive interfaces can lead to full loss of function, missense mutations apart from those sites could cause partial loss of function. Presumably, p.D3779N and p.E3763V impair the binding of a certain subset of ligands to LRP2 while nonetheless leaving LRP2 functional for endocytosis and recycling.
Discussion
We propose that the LRP2 homodimer assembly regulates extracellular ligand-binding specificity, enables efficient delivery and dissociation of ligand in the endosomal compartment, and facilitates recycling to the cell surface, a process it repeats every 10 to 30 minutes36,37,39. At extracellular pH, the homodimer interfaces fix intramolecular ligands distant from their cognate L repeats and maintain loops of L repeats in solvent-exposed, ligand-receptive orientations (Figure 2B, 3A, 7). The homodimer assembly also imposes specific structural regulation on each cluster of L repeats (Figure 5A). At endosomal pH, in contrast, a reduction in homodimer interfaces enables additional intra-protomer interfaces to form, allowing the open loops of L repeats to condense into a more compact assembly (Figure 2D, 5B, 7), and the remaining homodimer interfaces help exclude ligand from the center of the assembly by tethering together the two protomers (Figure 3D–F). The homodimer assembly coordinates all these diverse structural transitions, a feat difficult to conceive in an isolated monomer (Movie S3).
Figure 7. A homodimer coordinates the pH-sensitive LRP2 molecular machine.
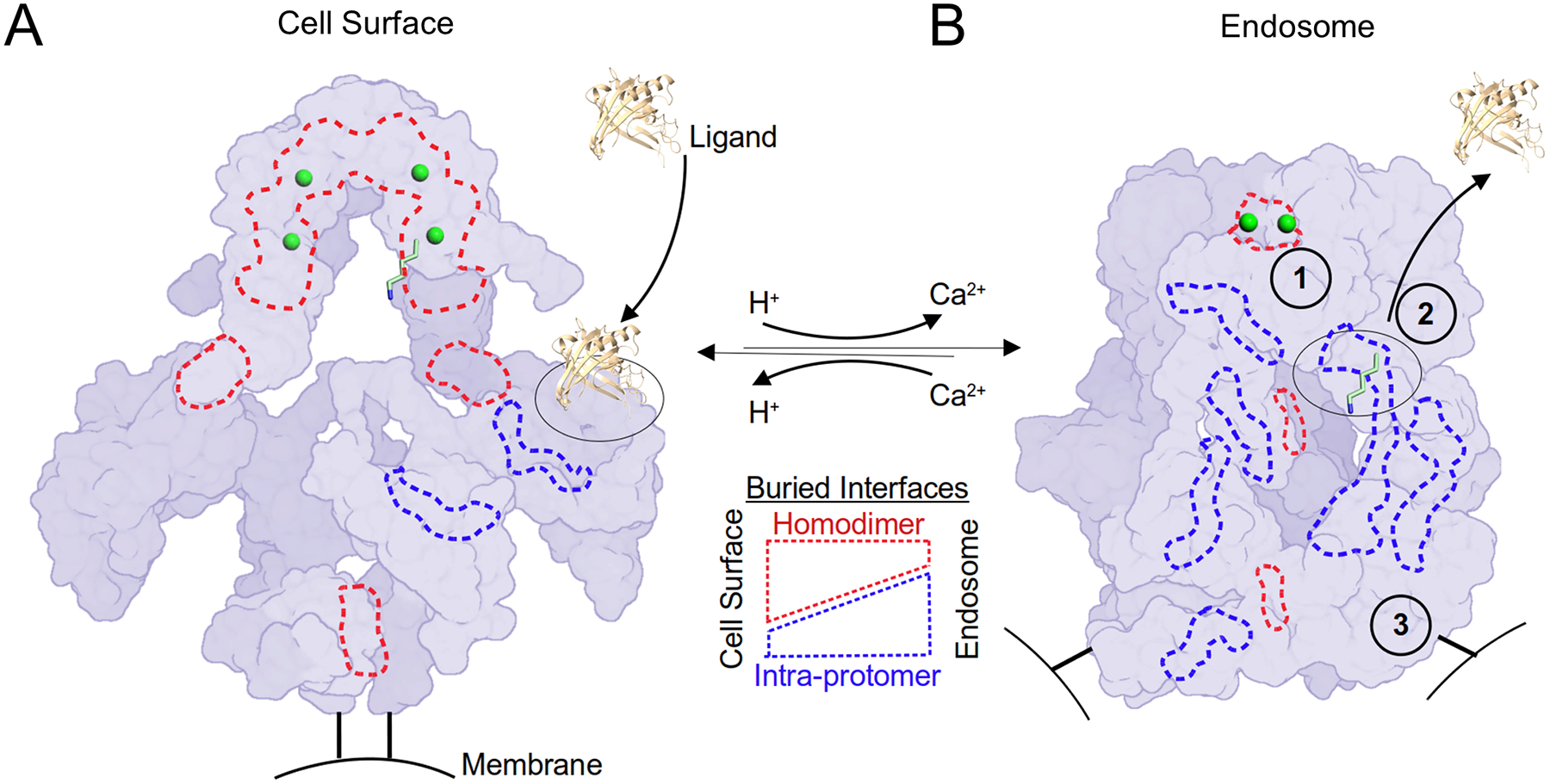
LRP2 is rendered as a schematic surface at extracellular and endosomal pH in the left and right panels respectively. Protomers are distinguished by two shades of purple. The C-termini of the extracellular domains that are directed towards the membrane insertions are rendered as black bars. Putative Ca2+ ions within the canopy are rendered as green spheres. K1430 is rendered in sticks with 6-fold magnification as one example of the 28 intramolecular ligands, and its binding partner, L8, is outlined with a dashed black oval. An extracellular ligand of ~20kDa rendered to scale is represented as a cartoon (PDB ID: 5JR8). Selected examples of buried surface area at the homodimer interfaces are outlined in dashed red lines, and intra-protomer interfaces are outlined in dashed blue lines. (A) At the cell surface, homodimer interfaces predominate. Hypothetical ligand binding to L8 is depicted. At the cell surface, the homodimer acts as a scaffold to keep L repeats open for ligand interactions and maintain distance between intramolecular ligands and their cognate L repeats. (B) In the endosomal compartment, 1) a pair of Ca2+ ions is released from the canopy and the homodimer interface unlocks to enable intramolecular ligands to interact with their cognate L repeats. 2) An intramolecular ligand displaces the extracellular ligand. As a result of these structural transitions, intra-protomer interfaces now predominate at endosomal pH. 3) The homodimer reorganization separates the membrane insertion points by >140 Å, which may serve as a compartment-specific trafficking signal.
See also Movie S3
pH-sensitive homodimer interfaces are crucial to drive the molecular machinery of the LRP2 homodimer that enables the transformation of the assembly from ligand-receptive at extracellular pH to ligand-shedding at endosomal pH (Figure 4, 7). Deleterious missense variants in humans affecting pH-sensitive sites in the homodimer lead to LRP2 loss-of-function and DBS (Figure 6A), emphasizing the physiologic importance of the homodimer assembly to LRP2 function in humans.
In addition to coordinating ligand binding and release, the homodimer assembly may also help regulate LRP2 trafficking. Our structures highlight that the homodimer also serves a role in positioning the cytoplasmic domains of LRP2. At extracellular pH, the membrane insertion points in the homodimer are adjacent but at endosomal pH are spaced >140 Å apart (Figure 2A, C). This repositioning of the cytoplasmic domains within the homodimer may alter how LRP2 binds adaptor proteins and could serve as a trafficking signal (Figure 7).
The LRP2 molecular machine is conceivably a prototype for the LRP subfamily as a whole. LRP1 may adopt a similar quaternary structure, given its nearly identical domain organization and conservation of the critical residues D1621 and D2257 involved in pH-sensitive homodimer interface contacts (Figure S5A–D). Indeed, dimers of LRP1 have been reported in neuronal cells127, and instances of neuronal calcium signaling have been posited to require LRP1 dimerization128. Notably, the tandem array of four β-propeller domains in LRP2 between the 2nd and 3rd clusters of L repeats is retained in LRP 4/5/6 as well as in LRP1 (Figure S1). Given that these β-propellers provide the primary contribution to the homodimer interface at extracellular pH in LRP2, this may point to a physiologic role for homodimerization in these LRPs as well.
In conclusion, we describe a novel pH-sensitive homodimer assembly in endogenously purified LRP2 that governs ligand binding and release through an equilibrium of homodimer and intra-protomer interfaces (Figure 7). This molecular machine provides a template for further understanding the structural biology of the LRP subfamily. LRP2 is critical for kidney tubular function by carrying out endocytosis of LMW proteins to create a non-toxic protein-free urine, and it is implicated in the pathogenesis of neurological disorders including Alzheimer’s Disease. Here, we have offered the beginnings of an understanding of its complex structural biology.
Limitations of the Study
While our purification of LRP2 from an endogenous source has made tractable the structural analysis of the giant LRP receptors for the first time, it has also limited out ability to pursue functional experiments to probe features observed in our structures. Heterologous expression systems of LRP proteins have not yet been able to reproduce their intricate native assembly. Thus, structure-function studies of apo LRP2 would require a gene-edited CRISPR mouse line, such that the mutated LRP2 could again be purified endogenously. A ligand-receptor structure, however, would enable probing the structure by mutations in ligand, and experiments to this end are underway.
The quality of the cryo-EM density at extracellular pH did not enable the LRP2 atomic model to include L3-L5, L14-L15, L18-L23, and L35-L36-E14. At both extracellular and endosomal pH, the cryo-EM density did not enable the full characterization of the extensive N-glycosylation networks on LRP2, some which appear to extend across intra-protomer and homodimer interfaces. At both extracellular and endosomal pH, the cytoplasmic domains were disordered, preventing any structural analysis.
STAR Methods
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Lawrence Shapiro (lss8@columbia.edu).
Materials availability
Endogenous LRP2 generated in this study will be shared upon request
Data and code availability
Cryo-EM maps and fitted coordinates of LRP2 at pH 7.5 and pH 5.2 have been deposited with accession codes, 8EM4 and 8EM7 respectively. This paper does not report original code. Particle stacks for LRP2 at pH 7.5 and pH 5.2 can be shared upon request. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
Endogenous LRP2 was purified from mice without regard to their sex, age, or genetic background. Each separate endogenous preparation was conducted on heterogeneous groups of mice that were no longer needed for their primary experiments and had been otherwise designated for euthanasia.
METHOD DETAILS
Protein Purification
All experiments were performed according to guidelines established by the Columbia University Institutional Animal Care and Use Committee. Following euthanasia, mouse kidneys were dissected, frozen on dry ice, and then stored at −80°C until use. At time of use, kidneys were thawed on wet ice then minced with a razor blade, and Dounce homogenized, 200mg at a time, in a 7mL chamber on ice for 10 strokes with a loose pestle followed by 10 strokes with a tight pestle. Homogenization buffer consisted of 25 mM Tris pH 7.5 (Corning, #46–030-CM), 150 mM NaCl (Fischer Scientific, #S671), 0.5 mM PMSF (Alexis, #270–184-G005), 0.5% BSA wt/vol (Gemini, #700–100P), 4% wt/vol mannitol (Sigma, #M4125), 2.5% wt/vol sucrose (Fischer Scientific, #S25590), 10 mM CaCl2 (Sigma, #C3881), 5% glycerol (Sigma, #G5516), 1ug/mL aprotinin (Sigma, 10236624001), 1ug/mL pepstatin A (Sigma, #P5318), and 5ug/mL leupeptin (Sigma, #L2884). 10 mM CaCl2 was included to achieve Ca2+-precipitation of the basolateral membrane129. All subsequent steps of differential centrifugation were conducted at 4°C. Following homogenization, sample was spun at 1000g × 10min to pellet cell debris on a Beckman Allegra 6KR centrifuge (Beckman Coulter Inc, NJ, USA). Supernatant was set aside on ice while cell debris was homogenized again as above, and re-spun at 1000g × 10min. Supernatant from the first two spins was then pooled and spun at 10,000g × 10min on a Sorvall RC-5B centrifuge (Thermo Fischer Scientific, MA, USA). Supernatant was again saved, and then spun at 100,000g × 60 min to pellet membrane in a Beckman L7 Ultracentrifuge. Membrane was now washed by resuspending with 1.5M NaCl, 25 mM Tris pH 7.5, 4% wt/vol mannitol and 2.5% wt/vol sucrose, and then three times with 25 mM Tris pH 7.5, 4% wt/vol mannitol, 2.5% wt/vol sucrose, spinning at 100,000g × 60min after each wash. Resuspension was accomplished by pipetting with a 5mL pipette, followed by aspirating through a 20-gauge needle until the solution was homogenous. 10g of mouse kidney yields approximately 100mg of washed membrane by this method. The presence of LRP2 in washed membrane was confirmed by Western blot with anti-LRP2 antibody (Abcam, #ab76969). After the final wash, membrane was solubilized in 1.6% β-octylglucoside (OG) (Research Products International, #N02007), 4% wt/vol mannitol, 2.5% wt/vol sucrose, 25 mM Tris pH 7.5, along with 0.5 mM PMSF and 0.2% Protease Inhibitor cocktail (Sigma, #P8340) and spun at 100,000g × 60min on a Beckman Ultracentrifuge. The supernatant was then filtered through a 0.22μM filter and loaded into a 150mL SuperLoop (Cytiva, #18102385). The exterior of the SuperLoop was packed with ice, and then the sample was loaded at 4mL/min onto six 5mL HiTrap Q HP columns (Cytiva, #17115301) mounted in series at room temperature on an AKTApurifier system equilibrated in 1% OG, 4% wt/vol mannitol, 75 mM sucrose, 25 mM Tris pH 7.5. Following the loading of sample, the HiTrap columns were detached, placed on ice, and then separately eluted with a continuous gradient from 0->1M NaCl in 1% OG, 225 mM mannitol, 75 mM sucrose, 25 mM Tris pH 7.5, with LRP2 primarily eluting from the final three of six columns in the series between 100 and 300 mM NaCl. LRP2-containing fractions were pooled and concentrated to 0.5mL using Amicon ultra centrifugal filter units (Millipore #UFC210024). The concentrated LRP2 sample was then injected over a Superose 6 10/300 (Cytiva, #17517201) size-exclusion column equilibrated in 150 mM NaCl, 0.8% OG, 25 mM Tris pH 7.5, 3 mM CaCl2. LRP2-containing fractions were concentrated to 2mg/ml and frozen at −80°C prior to further use. 100mg of washed membrane yields approximately 0.1mg of purified protein through this method. The sample tolerated free-thaw cycles without precipitation. A pGEX-KG-RAP construct was a generous gift from Joachim Herz’s lab, and purification was conducted as previously described98.
Nanoflow Liquid Chromatography coupled to Tandem Mass Spectrometry (nano-LC-MS/MS):
Proteins were separated by SDS- PAGE and digested in-gel with Trypsin (Mass Spectrometry Grade, Promega). Contaminants were removed from resulting tryptic peptides with in-house optimized, Empora C18 High Performance Extraction Disks (3M, MN) as previously described130. The eluted peptide solutions were partially dried under vacuum and then analyzed by nano-LC-MS/MS using a Thermo Easy nLC 1000 system coupled online to a Q Exactive HF with a NanoFlex source (Thermo Fisher Scientific, Waltham, MA) equipped with a self-packed 75 μm × 20-cm reverse phase column (ReproSil-Pur C18, 3M, Dr. Maisch GmbH, Germany) for peptide separation. Analytical column temperature was maintained at 50 °C by a column oven (Sonation GmBH, Germany). Peptides were eluted with a 3–50% acetonitrile gradient over 60 min at a flow rate of 250 nL/min. The mass spectrometer was operated in DDA mode with survey scans acquired at a resolution of 120,000 (at m/z 200) over a scan range of 300–1750 m/z. Up to 15 of the most abundant precursors from the survey scan were selected with an isolation window of 1.6 Th for fragmentation by higher-energy collisional dissociation with normalized collision energy (NCE) of 27. The maximum injection time for the survey and MS/MS scans was 60 ms and the ion target value (AGC) for both scan modes was set to 3e6. Raw mass spectrometry files were uploaded to Proteome Discoverer 1.4 and the resulting mgf files searched against Uniprot mouse database (mouse [Mus musculus] protein database; Uniprot; Reviewed, 16,950 entries, [12202017]) using Mascot (Matrix Science, London,UK; version 2.7.0; www.matrixscience.com)131; the search parameters were as follows: (i) up to two missed tryptic cleavage sites were allowed; (ii) precursor ion mass tolerance = 10 ppm; (iii) fragment ion mass tolerance = 0.5Da; and (iv) variable protein modifications were allowed for methionine oxidation, deamidation of asparagine and glutamines, and protein N-terminal acetylation. MudPit scoring was typically applied using significance threshold score p<0.01. Mascot search result was imported into Scaffold (Proteome Software, Inc., Portland, OR; version 4.11.0) to further analyze tandem mass spectrometry (MS/MS) based protein and peptide identifications (expressed as Exclusive Spectrum Counts). Decoy proteins were added to the search to allow for the calculation of false discovery rates (FDR). MudPit scoring was typically applied using significance threshold for protein at p<0.01. Decoy database search was always activated and, in general, LS-MS/MS analysis of a gel lane with p<0.01, protein false discovery rate averaged <1%. Mass spectra were also subjected to label-free quantitation by using MaxQuant proteomics data analysis workflow (version 1.5.5.1) with the Andromeda search engine132,133.
Size-exclusion chromatography / Multi-angle light scattering
1 μM LRP2 was run, in a buffer of TRIS 25 mM, NaCl 150 mM, CaCl2 2 mM, 0.8 % w/v octyl glycoside, pH 7.5, and elution buffer was the same, with a flow 0.5 mL/min at room temperature. 100 μL was injected using an AKTA FPLC system with a Superose 6 Increase 10/300 GL column. (https://www.cytivalifesciences.com) Light scattering data were collected using a Wyatt Dawn Heleos II detector, RI data was collected using a Wyatt Optilab T-rEX detector (Wyatt Technology, Santa Barbara, CA, USA), and UV data at 280 nm using the AKTA system. Molecular weight of the peaks was obtained from light scattering and RI data using ASTRA 6.1 software. (Wyatt Technology, Santa Barbara, CA, USA)
Analytical Ultracentrifugation Sedimentation Velocity
1 μM LRP2 in a buffer of TRIS 25 mM, NaCl 150 mM, CaCl2 2 mM, 0.8 % w/v octyl glycoside, pH 7.5 was run in a Beckman Optima XL-I/A analytical ultracentrifuge, (Beckman-Coulter, Palo Alto CA, USA) using interference detection at 660 nm. Two-channel cells with 12 mm path length and sapphire windows were used, the temperature was 25° C and the speed was 35000 rpm. Buffer density and viscosity and protein v-bar were calculated using the program SednTerp (Alliance Protein Laboratories, Corte Cancion, Thousand Oaks, CA, USA) (https://bitc.sr.unh.edu/index.php/Primary_Reference). Measuring scans were taken every 60 seconds, and the data were analyzed using Sedfit software (https://sedfitsedphat.nibib.nih.gov/software/default.aspx)134.
Mass Photometry
VWR microscope coverslips 22×50 mm No 1.5 (VWR, #48393–195) were cleaned sequentially with distilled Milli-Q water and isopropanol and then dried overnight in a fume hood. For each acquisition, 10uL of LRP2 at 10nM diluted in 25 mM Tris pH 7.5, 150 mM NaCl, 3 mM CaCl2, 0.8% OG, was applied to the coverslip and placed in the chamber of a Refeyn OneMP photometer (Refeyn Ltd, Oxford, UK)135. Following autofocus stabilization, movies of 180s duration were recorded. Data acquisition was conducted with AcquireMP (Refeyn, Ltd, v1.2.3) and processed and analyzed with Discover MP (Refeyn Ltd, v1.2.3).
Surface Plasmon Resonance
SPR assays for mouse LRP2 binding to RPA were performed using a Biacore T200 biosensor, equipped with a Series S CM4 chip at 25°C. RPA was immobilized over a single flow cell in HBS pH 7.4, supplemented with 0.005% (v/v) Tween-20, using amine-coupling chemistry, at approximately 2000 RU. An empty flow cell which has been activated and blocked but not immobilized served as a reference control. Full length LRP2 binding was tested in two buffer conditions, one of which was 25 mM TRIS-HCl pH 7.5, 150 mM NaCl, 3 mM CaCl2 and 0.8% (v/v) n-Octyl-β-D-Glucopyranoside, representing the extracellular pH, and the second 10 mM sodium acetate pH 5.2, 150 mM NaCl, 3 mM CaCl2 and 0.8% (v/v) n-Octyl-β-D-Glucopyranoside, representing the endosomal pH. LRP2 was prepared in each running buffer at six concentrations using a three-fold dilution series ranging from 0.82–200 nM. Samples were tested in order of increasing protein concentration. In experiments at the different pHs, blank buffer cycles were performed by injecting each running buffer instead of the analyte, after two LRP2 injections. The association and dissociation rates were each monitored for 120s and 300s respectively, at 50μL/min. The data was processed and fit to a 1:1 interaction model using Scrubber 2.0, with ±0.1 error in the calculated fit (BioLogic Software)136.
Cryo-EM Grid Preparation and Data Collection
3μL of LRP2 at 2mg/ml in 150 mM NaCl, 0.8% OG, 25 mM Tris pH 7.5, 3 mM CaCl2 was applied to glow-discharged 300 mesh UltraAuFoil R 1.2/1.3 gold grids (Electron Microscopy Sciences, #Q350AR13A) and blotted for 2.0s in a FEI Vitrobot Mark IV (Thermo Fischer) using a blotting force of 0 and wait time of 0 at 20°C and 100% humidity. Grids were plunge frozen in liquid ethane and then transferred to liquid nitrogen for storage.
Data for LRP2 at pH 7.5 were collected using the Leginon Software137 at the Simons Electron Microscopy Center of the New York Structural Biology Center on a 300keV FEI Titan Krios microscope with energy filter. A Gatan K3 direct detection camera was used in counting mode. 20,522 micrographs were collected at a pixel size of 1.083 using a dose of 54.87 e/Å over 50 frames with a defocus range of −0.5 to −1.5μm.
Data for LRP2 at pH 5.2 were collected using the Leginon Software137 at the Cryo-Electron Microscopy Center at the Vagelos College of Physicians and Surgeons at Columbia University using a 300keV FEI Titan Krios microscope with energy filter with a Gatan K3 direct detection camera in counting mode. 12,794 micrographs were collected at a pixel size of 0.83 using a dose of 58.06 e/Å over 50 frames with a defocus range of −0.5 to −1.5μm.
For both datasets, each micrograph was corrected for beam-induced movement using MotionCor2138 and estimation of contrast transfer function was conducted with ctffind4139.
Cryo-EM Data Processing
Particles from the pH 7.5 dataset were automatically picked in Cryosparc118 (Figure S2A) using a box size of 648, yielding 11,411,108 picks. Following bin8 extraction, 2D classes from these cryo-EM images revealed a twofold symmetric assembly (Figure S2B). An ab initio model on particles culled from these 2D classes in Cryosparc at ~12Å resolution confirmed the two-fold symmetry (Figure S2C). Density for a small micelle distal to the final C-terminal EGFP regions indicates the positioning of the transmembrane single-pass alpha helices of the LRP2 homodimer (Figure S2C). A cloud of density potentially representing multiple conformations of the cytosolic domains is visible in the 2D classes (Figure S2B) but is not appreciated in the 3D reconstruction. Iterative heterogeneous refinement on the full set of automatically picked particles using the ab initio model as a template provided a final dataset of 492,737 particles. Re-extraction of these particles with a box size of 512 followed by homogeneous refinement of these particles using the ab initio model as reference yielded identical structures for refinements conducted under C1 or C2 symmetry constraints (Figure S2D), although local resolution analysis conducted in Cryosparc and visualized with ChimeraX revealed significant flexibility in the assembly (Figure S2E). 3D classification of the dataset did not reveal any classes with apparent domain reorganizations (Figure 2F), and all particles were exported to Relion for CTF per-particle refinement and Bayesian polishing119,120. Continuous heterogenous refinement in Cryodrgn121 identified a subset of homogenous particles that was used for local refinement. Picked particles were downsampled from a box size of 512 to 120, and then analyzed in cryodrgn using train_vae on a 1024×1024 architecture with 3 layers for 50 iterations. Volumes derived from the principal component analysis of this training enabled an evaluation of the range of variation of the structures (Movie S1). Masks for local refinement were designed to enclose β-propellers and their adjacent L repeats and EGF-like domains, and these refinements yielded resolutions ranging from 2.9 to 3.3 Å (Figure S2G)140. A global cryo-EM density map was produced with nonuniform refinement in Cryosparc refined under C2 symmetry constraints141, with an overall resolution of 2.83 Å (Figure S2H) and no evidence of preferential orientation in the dataset (Figure S2I). A parallel procedure for LRP2 at pH 5.2 yielded 209,932 particles with an overall resolution of 2.97 Å (Figure S3A–I). Given less particles in the overall dataset at pH 5.2 compared to pH 7.5, this dataset was not further curated with continuous heterogeneous refinement as it was at pH 7.5. Post-processing with DeepEMhancer142 provided the maps into which models were built.
Model Building, Refinement and Validation
Using homology modeling based on the previously determined structure of LDLR (PDB ID: 1N7D), we built a model encompassing residues 28 to 4413 of LRP2 accounting for nearly the entirety of the extracellular domain at both pH 7.5 and pH 5.2. After establishing a Cα trace with Coot143, the model was extensively refined with Isolde to reduce clash score and correct Ramachandran and rotamer outliers144. Real-space refinement in Phenix145 corrected Cβ deviations and identified bond angle outliers. Bond angle outliers were manually corrected in Coot, and the model was then passed back into Phenix for final validation.
At pH 7.5, the map quality for the regions encompassing the solvent-exposed domains L3–5, L14–15, L18–23, and L35–36 was inadequate for an atomic model, as was a loop region encompassing residues 298–304. The extensive motion that these regions experience in the assembly explains the limitations of the map (Movie S1). Due to reduced flexibility of the assembly at pH 5.2 (Movie S2), the quality of the cryo-EM density was sufficient to identify orientations and Cα traces for all domains with the exception of the loop region encompassing residues 257–264. For regions of lower resolution in the maps where the domain orientation was clear but not all side chains could be modeled, side chains are rendered as alanines in the models. Models were refined with all side chains in place, and only after refinement was complete were side chains not fitting density pruned to alanines, with minimal effect on clash score. Density for numerous O-and N-glycosylations were present in the maps at both pH’s, but only the orientations of the first one or two glycans of each N-glycosylation could be modeled from the density. Mass spectrometry has previously predicted many sites of glycosylation in LRP2146–148, and evidence suggests they are important for ligand-binding147–149.
For both structures, Ca2+ was modeled at the canonical Ca2+ coordination site for all of the modeled L repeats and in the four calcium-binding (cb) EGF-like domains (E6, E11, E12, E15). Multiple previously unpredicted sites of putative Ca2+ coordination could be modeled within the β-propellers, six at pH 7.5 and four at pH 5.2. In addition, nine unpredicted putative Ca2+ per protomer could be modeled at pH 7.5 at a recurrent motif in the L repeats; all nine of these Ca2+ were dissociated at pH 5.2.
Validation of the structural models was conducted in Phenix150,151 and Molprobity152, using a composite of unsharpened maps from the local refinements. The DAQ score was used for residue-wise local quality estimation153. FSC model-map correlations ranged from 3.0 to 3.4Å for global and local portions of both models (Figure S2J and S3J), and CC-per-residue and DAQ-per-residue evaluations demonstrate good quality throughout both models with the best fit in the propeller regions (Figure S2K–M and S3K–M). Alphafold 2 was used to help further confirm the register of the polypeptide strands in the β-propellers (Figure S2N, S3N), with Cα RMSD between Alphafold predictions and the model ranging from 0.56 to 1.47 Å for the individual β-propeller domains. Global CC (mask) at pH 7.5 was 0.80 and at pH 5.2 was 0.69 (Table S4). Pymol (The PyMOL Molecular Graphics System, Version 2.0 Schrӧdinger, LLC) and UCSF ChimeraX were used to produce figures154.
Quantitation and Statistical Analysis
Sequence alignments were conducted using SnapGene (http:/www.snapgene.com), analysis of sequence conservation across the structures was conducted using Consurf155–158, analysis of SEC-MALS data was conducted with ASTRA 6.1 software. (Wyatt Technology, Santa Barbara, CA, USA), analysis of AUC data was done with Sedfit software https://sedfitsedphat.nibib.nih.gov/software/default.aspx)134, analysis of mass photometry data was accomplished with Discover MP (Refeyn Ltd, Oxford, UK)135, SPR data was analyzed with Scrubber 2.0 (BioLogic Software)136, cryo-EM processing was conducted with Cryosparc118 and Relion119,120, interface contacts and buried surface area were calculated with ChimeraX154, prediction of β-propeller structure was conducted with Alphafold 2159.
Supplementary Material
Table S1: Alignment of ligand-binding repeats (L) in LRP2 and LDLR
The four groups of L repeats in LRP2 are boxed at left. Regions of beta-sheet are boxed at 0.75 pt weight, and regions of alpha-helix are boxed at 1.5 pt weight. Arrows above the alignment identify Ca2+-coordinating residues: solid arrows indicating involvement of the residue’s side chain, and open arrows indicating involvement of the residue’s main chain carbonyl. Intramolecular ligands in L25 and L36 are in bold and underlined.
Related to Figure 1, Figure 2, Figure S1
Table S2: Alignment of EGF-like domains (E) in LRP2 and LDLR
EGF-like pairs are boxed at left. Regions of beta-sheet are boxed in the alignment. Residues from calcium-binding (cb) EGF-like domains whose side chain coordinates a Ca2+ ion are underlined and italicized. Intramolecular ligands from the EGF-like pairs are in bold and underlined.
Related to Figure 1, Figure 2, Figure S1
Table S3: Mass spectrometry of endogenously purified LRP2 from mouse kidney
Proteins identified in SDS-PAGE purified protein with apparent molecular weight 600kDa: tandem mass spectrometry (MS/MS) based protein and peptide identifications are expressed as exclusive spectrum count of the peptide spectral matches. Exclusive peptide count, identified protein percent sequence coverage, and the protein abundance index (emPAI factor that defines the ratio of the observed to observable peptides) are also listed.
Related to Figure 1
Table S4: Validations of the cryo-EM 3D reconstructions of LRP2 at pH 5.2 and pH 7.5
Validations were conducted in Phenix of the molecular models of LRP2 against a composite of the unsharpened locally refined maps.
Related to Figure 2
Movie S2: Flexibility of the LRP2 assembly at pH 5.2
Continuous heterogeneous classification of the LRP2 dataset bin4 at pH 5.2 was conducted in Cryodrgn121. The movie represents a principal component analysis of the dataset in a series of 10 volumes. In comparison to the LRP2 dataset at pH 7.5, movement within the assembly is of lesser magnitude and more evenly distributed across the structure.
Movie S1: Flexibility of the LRP2 assembly at pH 7.5
Continuous heterogeneous classification of the LRP2 dataset bin4 at pH 7.5 was conducted in Cryodrgn121. The movie represents a principal component analysis of the dataset in a series of 10 volumes. The lateral portions of the map that include R1, P1-P2, and R2 skew 15–17 Å relative to one another. The center portion of the map translates out of the plane established by the membrane insertion points by 10–15 Å. In relation to these changes, the canopy exhibits minimal movement.
Movie S3: The pH-dependent structural transition of LRP2
The LRP2 assembly is represented as a surface with one protomer in blue and the other in salmon. The structures at pH 7.5 and pH 5.2 are morphed into another using ChimeraX154. L3-L5, L14-L15, L18-L23, and L35-L36-E14 were not modeled in the atomic model at pH 7.5 since their Cα traces could not be well-defined due to the flexibility of the assembly. However, for the purposes of this morphing representation, they have been empirically docked into the low-resolution cryo-EM density corresponding to their positions at pH 7.5. All domains in the model undergo significant translations and re-orientations, all homodimer interfaces are pH-specific, and a decrease of homodimer interface at pH 5.2 enables increased intra-protomer contacts as L repeats pack against EGF-like domains and β-propellers.
Figure S1. Related to Figure 2; Domain organization of the LDLR family
Members of the LDLR family are schematically represented with their constituent domains, identified in the key. Domain organization is highly conserved between LRP1 and LRP2, the largest members of the family. Mouse LRP2 and human LRP2 have identical domain organization.
Figure S2. Related to Figure 2, Table S4, Movie S1; Cryo-EM data processing and validation for the LRP2 structure at pH 7.5
(A) The processing pipeline. (B) 2D Classification (C) Ab initio model (D) Homogeneous refinements with C1 and C2 symmetry constraints yield identical structures. (E) Local resolution (F) 3D classification did not demonstrate any substantial variation in domain organizations across the dataset. (G) Local refinement (H) Non-homogeneous refinement FSC (I) Viewing Direction Distribution (J) Map-Model FSC for the full model and locally refined areas. (K) CC per-residue with the average CC mask for the structure indicated. (L) DAQ per-residue (M) Model colored by DAQ score. (N) P domains superimposed on AF2 predictions.
Figure S4. Related to Figure 3, Figure 5, Movie S3; Contact Maps for the LRP2 structures at pH 5.2 and pH 7.5
(A) The domains of the two protomers are listed along the two short sides of the triangle, and intersecting squares are colored depending on whether contact is made between domains at pH 5.2, pH 7.5, or at both pH’s.
(B) Intra-protomer and homodimer interactions represented on domain diagrams. The red and blue lines represent domain-domain interactions at pH 5.2 and pH 7.5, respectively.
Figure S3. Related to Figure 2, Table S4, Movie S2; Cryo-EM data processing and validation for the LRP2 structure at pH 5.2
(A) The processing pipeline. (B) 2D Classification (C) Ab initio model (D) Homogeneous refinements with C1 and C2 symmetry constraints are identical. (E) Local resolution (F) 3D classification did not demonstrate any substantial variation in domain organizations across the dataset. (G) Local refinement (H) Non-homogeneous refinement FSC (I) Viewing Direction Distribution (J) Map-Model FSC for the full model and locally refined areas. (K) CC per-residue with the average CC mask for the structure indicated. (L) DAQ per-residue (M) Model colored by DAQ score. (N) P domains superimposed on AF2 predictions.
Figure S5. Related to Figure 4; Evolutionary conservation of residues at the putative Ca2+-coordinating sites in the canopy of LRP2
(A) Looking down upon the canopy of the homodimer at pH 7.5 in the direction of the cell membrane, the cleft between the two protomers of the homodimer is visible. Alignments of LRP1 and LRP2 sequences were run through the Consurf server to assess conservation. Magenta coloring of the cartoon depiction indicates high sequence conservation while teal coloring indicates high sequence variability. The majority of the loop regions connecting the β-strands are variable, with the exception of residues responsible for putative calcium coordination. Residue level detail is shown for the boxed regions. (B) D1621, H1645 are highly conserved at the putative Ca2+-coordination site despite residing in a loop region. (C) D2257 and S2276 are the only highly conserved residues in the loop region surrounding the putative Ca2+-coordination site. (D) Alignment of selected LRP1 and LRP2 sequences to show conservation of D1621, H1645, and D2257.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| PMSF | Alexis | Cat# 270–184-G005 |
| BSA | Gemini | Cat# 700–100P |
| Mannitol | Millipore Sigma | Cat# M4125 |
| Tris 1M, pH 7.5 | Corning | Cat# 46–030-CM |
| Sodium Chloride | Fischer Scientific | Cat# S671 |
| Sucrose | Fischer | Cat# S25590B |
| Glycerol | Millipore Sigma | Cat# G5516 |
| Pepstatin A | Millipore Sigma | Cat# P5318 |
| Leupeptin | Millipore Sigma | Cat# L2884 |
| β-octylglucoside | RPI | Cat# N02007 |
| Protease inhibitor cocktail | Millipore Sigma | Cat# P8340 |
| Anti-LRP2 antibody | Abcam | Cat# ab76969 |
| Aprotinin | Millipore Sigma | Cat# 10236624001 |
| Sodium acetate | Millipore Sigma | Cat #S7545 |
| Tween-20 | Millipore Sigma | Cat #P7949 |
| HEPES | Millipore Sigma | Cat #H3375 |
| Calcium chloride | Millipore Sigma | Cat# C3881 |
| Amine coupling kit | Cytiva | Cat# BR100050 |
| Critical commercial assays | ||
| SuperLoop | Cytiva | Cat# 18102385 |
| HiTrap Q HP | Cytiva | Cat# 17115301 |
| Amicron ultra centrifugal filter units | Millipore Sigma | Cat# UFC210024 |
| Series S Sensor Chip CM4 | Cytiva | Cat #BR100534 |
| Superose 6 30/300 | Cytiva | Cat# 17517201 |
| Recombinant DNA | ||
| pGEX-KG-RAP | Joachim Herz Lab | N/A |
| Deposited data | ||
| Cryo-EM structure of LRP2 at pH 7.5 | This paper | PDB: 8EM4 |
| Cryo-EM map of LRP2 at pH 7.5 | This paper | EMDB: EMD-28233 |
| Cryo-EM map of local refinement of P1-P2 domains of LRP2 at pH 7.5 | This paper | EMDB: EMD-28242 |
| Cryo-EM map of local refinement of P3-P6 domains of LRP2 at pH 7.5 | This paper | EMDB: EMD-28243 |
| Cryo-EM map of local refinement of P7 domain of LRP2 at pH 7.5 | This paper | EMDB: EMD-28250 |
| Cryo-EM map of local refinement of R4 domain of LRP2 at pH 7.5 | This paper | EMDB: EMD-28251 |
| Cryo-EM map of local refinement of P8 domain of LRP2 at pH 7.5 | This paper | EMDB: EMD-28252 |
| Cryo-EM structure of LRP2 at pH 5.2 | This paper | PDB: 8EM7 |
| Cryo-EM map of LRP2 at pH 5.2 | This paper | EMDB: EMD-28241 |
| Cryo-EM map of local refinement of P1-P2 domains of LRP2 at pH 5.2 | This paper | EMDB: EMD-28253 |
| Cryo-EM map of local refinement of P1-P2 domains of LRP2 at pH 5.2 | This paper | EMDB: EMD-28258 |
| Cryo-EM map of local refinement of P1-P2 domains of LRP2 at pH 5.2 | This paper | EMDB: EMD-28260 |
| Cryo-EM map of local refinement of P1-P2 domains of LRP2 at pH 5.2 | This paper | EMDB: EMD-28261 |
| Cryo-EM map of local refinement of P1-P2 domains of LRP2 at pH 5.2 | This paper | EMBD: EMD-28265 |
| Software and algorithms | ||
| Coot | Emsley et al.143,160 | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot |
| cryoSPARC | Punjani et al.118 | https://cryosparc.com |
| Leginon | Suloway et al.137 | https://sbgrid.org/software/titles/leginon |
| Molprobity | Davis et al.152 | http://molprobity.biochem.duke.edu |
| Phenix | Adams et al.161 | https://www.phenix-online.org |
| The PyMOL Molecular Graphics System, Version 2.0 | Schrödinger, LLC | https://pymol.org/2/support.html#page-top |
| RELION | Scheres, S.H.119 | https://www3.mrc-lmb.cam.ac.uk/relion/index.php/Main_Page |
| DeepEMhancer | Sanchez-Garcia et al.142 | https://github.com/rsanchezgarc/deepEMhancer |
| Mascot | Perkins et al.131 | http://www.matrixscience.com |
| MaxQuant | Cox et al.132 | https://www.maxquant.org |
| UCSF Chimera | Pettersen et al.162 | https://www.cgl.ucsf.edu/chimera/ |
| UCSF Chimera X | Pettersen et al.154 | https://www.cgl.ucsf.edu/chimerax/ |
| Refeyn One Acquire MP and Discover MP | Young et al.135 | http://www.refeyn.com |
| Python v3.8.3 | Python | https://www.python.org/ |
| ASTRA 6.1 | Wyatt Technology | www.wyatt.com |
| SednTerp | Laue, T.M.163 | https://bitc.sr.unh.edu/index.php/Primary_Reference |
| Sedfit | Schuck et al.134 | https://sedfitsedphat.nibib.nih.gov/software/default.aspx |
| Scrubber 2.0 | Sigmundsson et al136 | http://www.biologic.com.au/scrubber.html |
| SnapGene | Insightful Science | http://www.snapgene.com |
| BioRender | BioRender | http://BioRender.com |
| Consurf | Ashkenazy et al.155–158 | https://consurf.tau.ac.il/ |
| Alphafold 2 | Jumper et al.159 | http://github.com/deepmind/alphafold |
Acknowledgements
Some of this work was performed at the Columbia University Cryo-Electron Microscopy Center as well as at the Simons Electron Microscopy Center and National Resource for Automated Molecular Microscopy located at the New York Structural Biology Center, supported by grants from the Simons Foundation (SF349247), NYSTAR, and the NIH National Institute of General Medical Sciences (GM103310) with additional support from Agouron Institute (F00316) and NIH (OD019994). The authors are grateful for the support of R. Grassucci, Z. Zhang, and L. Urnavicius for help collecting cryo-EM data at the Columbia University Cryo-Electron Microscopy Center. We thank E. Eng for technical support at the New York Structural Biology Center. We thank F. Vallese for assistance with mass photometry data collection. Figure 1A and Figure 7 were created with BioRender.com. The bacterial expression construct for RAP was a generous gift from the Joachim Herz lab. A.B. was supported by R01-DK124667, TL1-TR001875, and T32-DK108741. Z.A., S.Y.R, and E.Y.C. were supported by U54-DK104309. H.E., T.A.N., Jonathan B., and L.S. acknowledge support from NIH NIDDK R01-DK124667. T.A.N. acknowledges support from 1S10RR027990 from NIH NCRR. A.W.P.F. acknowledges support from NIH NINDS U01NS110438 and U54NS110435.
Footnotes
Inclusion and Diversity
We support inclusive, diverse, and equitable conduct of research.
Declaration of Interests
The authors declare no competing interests.
REFERENCES
- 1.Nielsen R, Christensen EI, and Birn H (2016). Megalin and cubilin in proximal tubule protein reabsorption: from experimental models to human disease. Kidney Int 89, 58–67. 10.1016/j.kint.2015.11.007. [DOI] [PubMed] [Google Scholar]
- 2.Christensen EI, and Nielsen R (2007). Role of megalin and cubilin in renal physiology and pathophysiology. Rev Physiol Biochem Pharmacol 158, 1–22. 10.1007/112_0604. [DOI] [PubMed] [Google Scholar]
- 3.Gonias SL, and Campana WM (2014). LDL receptor-related protein-1: a regulator of inflammation in atherosclerosis, cancer, and injury to the nervous system. Am J Pathol 184, 18–27. 10.1016/j.ajpath.2013.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Consortium, C.e.S. (1998). Genome sequence of the nematode C. elegans: a platform for investigating biology. Science 282, 2012–2018. 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- 5.Yochem J, and Greenwald I (1993). A gene for a low density lipoprotein receptor-related protein in the nematode Caenorhabditis elegans. Proc Natl Acad Sci U S A 90, 4572–4576. 10.1073/pnas.90.10.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kerjaschki D, and Farquhar MG (1982). The pathogenic antigen of Heymann nephritis is a membrane glycoprotein of the renal proximal tubule brush border. Proc Natl Acad Sci U S A 79, 5557–5561. 10.1073/pnas.79.18.5557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raychowdhury R, Niles JL, McCluskey RT, and Smith JA (1989). Autoimmune target in Heymann nephritis is a glycoprotein with homology to the LDL receptor. Science 244, 1163–1165. 10.1126/science.2786251. [DOI] [PubMed] [Google Scholar]
- 8.Kerjaschki D, and Farquhar MG (1983). Immunocytochemical localization of the Heymann nephritis antigen (GP330) in glomerular epithelial cells of normal Lewis rats. J Exp Med 157, 667–686. 10.1084/jem.157.2.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saito A, Pietromonaco S, Loo AK, and Farquhar MG (1994). Complete cloning and sequencing of rat gp330/”megalin,” a distinctive member of the low density lipoprotein receptor gene family. Proc Natl Acad Sci U S A 91, 9725–9729. 10.1073/pnas.91.21.9725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hjalm G, Murray E, Crumley G, Harazim W, Lundgren S, Onyango I, Ek B, Larsson M, Juhlin C, Hellman P, et al. (1996). Cloning and sequencing of human gp330, a Ca(2+)-binding receptor with potential intracellular signaling properties. Eur J Biochem 239, 132–137. 10.1111/j.1432-1033.1996.0132u.x. [DOI] [PubMed] [Google Scholar]
- 11.Lundgren S, Carling T, Hjalm G, Juhlin C, Rastad J, Pihlgren U, Rask L, Akerstrom G, and Hellman P (1997). Tissue distribution of human gp330/megalin, a putative Ca(2+)-sensing protein. J Histochem Cytochem 45, 383–392. 10.1177/002215549704500306. [DOI] [PubMed] [Google Scholar]
- 12.Zheng G, Bachinsky DR, Stamenkovic I, Strickland DK, Brown D, Andres G, and McCluskey RT (1994). Organ distribution in rats of two members of the low-density lipoprotein receptor gene family, gp330 and LRP/alpha 2MR, and the receptor-associated protein (RAP). J Histochem Cytochem 42, 531–542. 10.1177/42.4.7510321. [DOI] [PubMed] [Google Scholar]
- 13.Kounnas MZ, Haudenschild CC, Strickland DK, and Argraves WS (1994). Immunological localization of glycoprotein 330, low density lipoprotein receptor related protein and 39 kDa receptor associated protein in embryonic mouse tissues. In Vivo 8, 343–351. [PubMed] [Google Scholar]
- 14.Assmann KJ, Lange WP, Tangelder MM, and Koene RA (1986). The organ distribution of gp-330 (Heymann antigen) and gp-90 in the mouse and the rat. Virchows Arch A Pathol Anat Histopathol 408, 541–553. 10.1007/BF00705307. [DOI] [PubMed] [Google Scholar]
- 15.LaFerla FM, Troncoso JC, Strickland DK, Kawas CH, and Jay G (1997). Neuronal cell death in Alzheimer’s disease correlates with apoE uptake and intracellular Abeta stabilization. J Clin Invest 100, 310–320. 10.1172/JCI119536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bento-Abreu A, Velasco A, Polo-Hernandez E, Perez-Reyes PL, Tabernero A, and Medina JM (2008). Megalin is a receptor for albumin in astrocytes and is required for the synthesis of the neurotrophic factor oleic acid. J Neurochem 106, 1149–1159. 10.1111/j.1471-4159.2008.05462.x. [DOI] [PubMed] [Google Scholar]
- 17.Carro E, Spuch C, Trejo JL, Antequera D, and Torres-Aleman I (2005). Choroid plexus megalin is involved in neuroprotection by serum insulin-like growth factor I. J Neurosci 25, 10884–10893. 10.1523/JNEUROSCI.2909-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chun JT, Wang L, Pasinetti GM, Finch CE, and Zlokovic BV (1999). Glycoprotein 330/megalin (LRP-2) has low prevalence as mRNA and protein in brain microvessels and choroid plexus. Exp Neurol 157, 194–201. 10.1006/exnr.1999.7052. [DOI] [PubMed] [Google Scholar]
- 19.Alvira-Botero X, Perez-Gonzalez R, Spuch C, Vargas T, Antequera D, Garzon M, Bermejo-Pareja F, and Carro E (2010). Megalin interacts with APP and the intracellular adapter protein FE65 in neurons. Mol Cell Neurosci 45, 306–315. 10.1016/j.mcn.2010.07.005. [DOI] [PubMed] [Google Scholar]
- 20.Christ A, Christa A, Kur E, Lioubinski O, Bachmann S, Willnow TE, and Hammes A (2012). LRP2 is an auxiliary SHH receptor required to condition the forebrain ventral midline for inductive signals. Dev Cell 22, 268–278. 10.1016/j.devcel.2011.11.023. [DOI] [PubMed] [Google Scholar]
- 21.Gajera CR, Emich H, Lioubinski O, Christ A, Beckervordersandforth-Bonk R, Yoshikawa K, Bachmann S, Christensen EI, Gotz M, Kempermann G, et al. (2010). LRP2 in ependymal cells regulates BMP signaling in the adult neurogenic niche. J Cell Sci 123, 1922–1930. 10.1242/jcs.065912. [DOI] [PubMed] [Google Scholar]
- 22.Gil SY, Youn BS, Byun K, Huang H, Namkoong C, Jang PG, Lee JY, Jo YH, Kang GM, Kim HK, et al. (2013). Clusterin and LRP2 are critical components of the hypothalamic feeding regulatory pathway. Nat Commun 4, 1862. 10.1038/ncomms2896. [DOI] [PubMed] [Google Scholar]
- 23.Willnow TE, Hilpert J, Armstrong SA, Rohlmann A, Hammer RE, Burns DK, and Herz J (1996). Defective forebrain development in mice lacking gp330/megalin. Proc Natl Acad Sci U S A 93, 8460–8464. 10.1073/pnas.93.16.8460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Willnow TE, and Christ A (2017). Endocytic receptor LRP2/megalin-of holoprosencephaly and renal Fanconi syndrome. Pflugers Arch 469, 907–916. 10.1007/s00424-017-1992-0. [DOI] [PubMed] [Google Scholar]
- 25.Moestrup SK, Nielsen S, Andreasen P, Jorgensen KE, Nykjaer A, Roigaard H, Gliemann J, and Christensen EI (1993). Epithelial glycoprotein-330 mediates endocytosis of plasminogen activator-plasminogen activator inhibitor type-1 complexes. J Biol Chem 268, 16564–16570. [PubMed] [Google Scholar]
- 26.Willnow TE, Goldstein JL, Orth K, Brown MS, and Herz J (1992). Low density lipoprotein receptor-related protein and gp330 bind similar ligands, including plasminogen activator-inhibitor complexes and lactoferrin, an inhibitor of chylomicron remnant clearance. J Biol Chem 267, 26172–26180. [PubMed] [Google Scholar]
- 27.Christensen EI, Moskaug JO, Vorum H, Jacobsen C, Gundersen TE, Nykjaer A, Blomhoff R, Willnow TE, and Moestrup SK (1999). Evidence for an essential role of megalin in transepithelial transport of retinol. J Am Soc Nephrol 10, 685–695. 10.1681/ASN.V104685. [DOI] [PubMed] [Google Scholar]
- 28.Nykjaer A, Dragun D, Walther D, Vorum H, Jacobsen C, Herz J, Melsen F, Christensen EI, and Willnow TE (1999). An endocytic pathway essential for renal uptake and activation of the steroid 25-(OH) vitamin D3. Cell 96, 507–515. 10.1016/s0092-8674(00)80655-8. [DOI] [PubMed] [Google Scholar]
- 29.Orlando RA, Rader K, Authier F, Yamazaki H, Posner BI, Bergeron JJ, and Farquhar MG (1998). Megalin is an endocytic receptor for insulin. J Am Soc Nephrol 9, 1759–1766. 10.1681/ASN.V9101759. [DOI] [PubMed] [Google Scholar]
- 30.Hvidberg V, Jacobsen C, Strong RK, Cowland JB, Moestrup SK, and Borregaard N (2005). The endocytic receptor megalin binds the iron transporting neutrophil-gelatinase-associated lipocalin with high affinity and mediates its cellular uptake. FEBS Lett 579, 773–777. 10.1016/j.febslet.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 31.Barasch J, Hollmen M, Deng R, Hod EA, Rupert PB, Abergel RJ, Allred BE, Xu K, Darrah SF, Tekabe Y, et al. (2016). Disposal of iron by a mutant form of lipocalin 2. Nat Commun 7, 12973. 10.1038/ncomms12973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Faber K, Hvidberg V, Moestrup SK, Dahlback B, and Nielsen LB (2006). Megalin is a receptor for apolipoprotein M, and kidney-specific megalin-deficiency confers urinary excretion of apolipoprotein M. Mol Endocrinol 20, 212–218. 10.1210/me.2005-0209. [DOI] [PubMed] [Google Scholar]
- 33.Moestrup SK, Schousboe I, Jacobsen C, Leheste JR, Christensen EI, and Willnow TE (1998). beta2-glycoprotein-I (apolipoprotein H) and beta2-glycoprotein-I-phospholipid complex harbor a recognition site for the endocytic receptor megalin. J Clin Invest 102, 902–909. 10.1172/JCI3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kounnas MZ, Loukinova EB, Stefansson S, Harmony JA, Brewer BH, Strickland DK, and Argraves WS (1995). Identification of glycoprotein 330 as an endocytic receptor for apolipoprotein J/clusterin. J Biol Chem 270, 13070–13075. 10.1074/jbc.270.22.13070. [DOI] [PubMed] [Google Scholar]
- 35.Stefansson S, Chappell DA, Argraves KM, Strickland DK, and Argraves WS (1995). Glycoprotein 330/low density lipoprotein receptor-related protein-2 mediates endocytosis of low density lipoproteins via interaction with apolipoprotein B100. J Biol Chem 270, 19417–19421. 10.1074/jbc.270.33.19417. [DOI] [PubMed] [Google Scholar]
- 36.Perez Bay AE, Schreiner R, Benedicto I, Paz Marzolo M, Banfelder J, Weinstein AM, and Rodriguez-Boulan EJ (2016). The fast-recycling receptor Megalin defines the apical recycling pathway of epithelial cells. Nat Commun 7, 11550. 10.1038/ncomms11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shah M, Baterina OY Jr., Taupin V, and Farquhar MG (2013). ARH directs megalin to the endocytic recycling compartment to regulate its proteolysis and gene expression. J Cell Biol 202, 113–127. 10.1083/jcb.201211110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kerjaschki D, Miettinen A, and Farquhar MG (1987). Initial events in the formation of immune deposits in passive Heymann nephritis. gp330-anti-gp330 immune complexes form in epithelial coated pits and rapidly become attached to the glomerular basement membrane. J Exp Med 166, 109–128. 10.1084/jem.166.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Czekay RP, Orlando RA, Woodward L, Lundstrom M, and Farquhar MG (1997). Endocytic trafficking of megalin/RAP complexes: dissociation of the complexes in late endosomes. Mol Biol Cell 8, 517–532. 10.1091/mbc.8.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chambers JC, Zhang W, Lord GM, van der Harst P, Lawlor DA, Sehmi JS, Gale DP, Wass MN, Ahmadi KR, Bakker SJ, et al. (2010). Genetic loci influencing kidney function and chronic kidney disease. Nat Genet 42, 373–375. 10.1038/ng.566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chasman DI, Fuchsberger C, Pattaro C, Teumer A, Boger CA, Endlich K, Olden M, Chen MH, Tin A, Taliun D, et al. (2012). Integration of genome-wide association studies with biological knowledge identifies six novel genes related to kidney function. Hum Mol Genet 21, 5329–5343. 10.1093/hmg/dds369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee J, Lee Y, Park B, Won S, Han JS, and Heo NJ (2018). Genome-wide association analysis identifies multiple loci associated with kidney disease-related traits in Korean populations. PLoS One 13, e0194044. 10.1371/journal.pone.0194044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parsa A, Fuchsberger C, Kottgen A, O’Seaghdha CM, Pattaro C, de Andrade M, Chasman DI, Teumer A, Endlich K, Olden M, et al. (2013). Common variants in Mendelian kidney disease genes and their association with renal function. J Am Soc Nephrol 24, 2105–2117. 10.1681/ASN.2012100983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pattaro C, Teumer A, Gorski M, Chu AY, Li M, Mijatovic V, Garnaas M, Tin A, Sorice R, Li Y, et al. (2016). Genetic associations at 53 loci highlight cell types and biological pathways relevant for kidney function. Nat Commun 7, 10023. 10.1038/ncomms10023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Elsakka EGE, Mokhtar MM, Hegazy M, Ismail A, and Doghish AS (2022). Megalin, a multi-ligand endocytic receptor, and its participation in renal function and diseases: A review. Life Sci 308, 120923. 10.1016/j.lfs.2022.120923. [DOI] [PubMed] [Google Scholar]
- 46.Kantarci S, Al-Gazali L, Hill RS, Donnai D, Black GC, Bieth E, Chassaing N, Lacombe D, Devriendt K, Teebi A, et al. (2007). Mutations in LRP2, which encodes the multiligand receptor megalin, cause Donnai-Barrow and facio-oculo-acoustico-renal syndromes. Nat Genet 39, 957–959. 10.1038/ng2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rauch JN, Luna G, Guzman E, Audouard M, Challis C, Sibih YE, Leshuk C, Hernandez I, Wegmann S, Hyman BT, et al. (2020). LRP1 is a master regulator of tau uptake and spread. Nature 580, 381–385. 10.1038/s41586-020-2156-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Christensen EI, and Birn H (2002). Megalin and cubilin: multifunctional endocytic receptors. Nat Rev Mol Cell Biol 3, 256–266. 10.1038/nrm778. [DOI] [PubMed] [Google Scholar]
- 49.Lillis AP, Van Duyn LB, Murphy-Ullrich JE, and Strickland DK (2008). LDL receptor-related protein 1: unique tissue-specific functions revealed by selective gene knockout studies. Physiol Rev 88, 887–918. 10.1152/physrev.00033.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cooper JM, Lathuiliere A, Migliorini M, Arai AL, Wani MM, Dujardin S, Muratoglu SC, Hyman BT, and Strickland DK (2021). Regulation of tau internalization, degradation, and seeding by LRP1 reveals multiple pathways for tau catabolism. J Biol Chem 296, 100715. 10.1016/j.jbc.2021.100715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ridler C (2018). APOE4 transforms the transcriptome. Nat Rev Neurol 14, 450–451. 10.1038/s41582-018-0034-7. [DOI] [PubMed] [Google Scholar]
- 52.Vargas T, Bullido MJ, Martinez-Garcia A, Antequera D, Clarimon J, Rosich-Estrago M, Martin-Requero A, Mateo I, Rodriguez-Rodriguez E, Vilella-Cuadrada E, et al. (2010). A megalin polymorphism associated with promoter activity and Alzheimer’s disease risk. Am J Med Genet B Neuropsychiatr Genet 153B, 895–902. 10.1002/ajmg.b.31056. [DOI] [PubMed] [Google Scholar]
- 53.Wang LL, Pan XL, Wang Y, Tang HD, Deng YL, Ren RJ, Xu W, Ma JF, Wang G, and Chen SD (2011). A single nucleotide polymorphism in LRP2 is associated with susceptibility to Alzheimer’s disease in the Chinese population. Clin Chim Acta 412, 268–270. 10.1016/j.cca.2010.10.015. [DOI] [PubMed] [Google Scholar]
- 54.Bell RD, Sagare AP, Friedman AE, Bedi GS, Holtzman DM, Deane R, and Zlokovic BV (2007). Transport pathways for clearance of human Alzheimer’s amyloid beta-peptide and apolipoproteins E and J in the mouse central nervous system. J Cereb Blood Flow Metab 27, 909–918. 10.1038/sj.jcbfm.9600419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zlokovic BV, Martel CL, Matsubara E, McComb JG, Zheng G, McCluskey RT, Frangione B, and Ghiso J (1996). Glycoprotein 330/megalin: probable role in receptor-mediated transport of apolipoprotein J alone and in a complex with Alzheimer disease amyloid beta at the blood-brain and blood-cerebrospinal fluid barriers. Proc Natl Acad Sci U S A 93, 4229–4234. 10.1073/pnas.93.9.4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Williams A, et al. (2009). Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat Genet 41, 1088–1093. 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lambert JC, Heath S, Even G, Campion D, Sleegers K, Hiltunen M, Combarros O, Zelenika D, Bullido MJ, Tavernier B, et al. (2009). Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer’s disease. Nat Genet 41, 1094–1099. 10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- 58.Fass D, Blacklow S, Kim PS, and Berger JM (1997). Molecular basis of familial hypercholesterolaemia from structure of LDL receptor module. Nature 388, 691–693. 10.1038/41798. [DOI] [PubMed] [Google Scholar]
- 59.Downing AK, Knott V, Werner JM, Cardy CM, Campbell ID, and Handford PA (1996). Solution structure of a pair of calcium-binding epidermal growth factor-like domains: implications for the Marfan syndrome and other genetic disorders. Cell 85, 597–605. 10.1016/s0092-8674(00)81259-3. [DOI] [PubMed] [Google Scholar]
- 60.Fehon RG, Kooh PJ, Rebay I, Regan CL, Xu T, Muskavitch MA, and Artavanis-Tsakonas S (1990). Molecular interactions between the protein products of the neurogenic loci Notch and Delta, two EGF-homologous genes in Drosophila. Cell 61, 523–534. 10.1016/0092-8674(90)90534-l. [DOI] [PubMed] [Google Scholar]
- 61.Rao Z, Handford P, Mayhew M, Knott V, Brownlee GG, and Stuart D (1995). The structure of a Ca(2+)-binding epidermal growth factor-like domain: its role in protein-protein interactions. Cell 82, 131–141. 10.1016/0092-8674(95)90059-4. [DOI] [PubMed] [Google Scholar]
- 62.Springer TA (1998). An extracellular beta-propeller module predicted in lipoprotein and scavenger receptors, tyrosine kinases, epidermal growth factor precursor, and extracellular matrix components. J Mol Biol 283, 837–862. 10.1006/jmbi.1998.2115. [DOI] [PubMed] [Google Scholar]
- 63.Jeon H, Meng W, Takagi J, Eck MJ, Springer TA, and Blacklow SC (2001). Implications for familial hypercholesterolemia from the structure of the LDL receptor YWTD-EGF domain pair. Nat Struct Biol 8, 499–504. 10.1038/88556. [DOI] [PubMed] [Google Scholar]
- 64.Croy JE, Brandon T, and Komives EA (2004). Two apolipoprotein E mimetic peptides, ApoE(130–149) and ApoE(141–155)2, bind to LRP1. Biochemistry 43, 7328–7335. 10.1021/bi036208p. [DOI] [PubMed] [Google Scholar]
- 65.Croy JE, Shin WD, Knauer MF, Knauer DJ, and Komives EA (2003). All three LDL receptor homology regions of the LDL receptor-related protein bind multiple ligands. Biochemistry 42, 13049–13057. 10.1021/bi034752s. [DOI] [PubMed] [Google Scholar]
- 66.Fisher C, Beglova N, and Blacklow SC (2006). Structure of an LDLR-RAP complex reveals a general mode for ligand recognition by lipoprotein receptors. Mol Cell 22, 277–283. 10.1016/j.molcel.2006.02.021. [DOI] [PubMed] [Google Scholar]
- 67.Guttman M, Prieto JH, Croy JE, and Komives EA (2010). Decoding of lipoprotein-receptor interactions: properties of ligand binding modules governing interactions with apolipoprotein E. Biochemistry 49, 1207–1216. 10.1021/bi9017208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Guttman M, Prieto JH, Handel TM, Domaille PJ, and Komives EA (2010). Structure of the minimal interface between ApoE and LRP. J Mol Biol 398, 306–319. 10.1016/j.jmb.2010.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jensen GA, Andersen OM, Bonvin AM, Bjerrum-Bohr I, Etzerodt M, Thogersen HC, O’Shea C, Poulsen FM, and Kragelund BB (2006). Binding site structure of one LRP-RAP complex: implications for a common ligand-receptor binding motif. J Mol Biol 362, 700–716. 10.1016/j.jmb.2006.07.013. [DOI] [PubMed] [Google Scholar]
- 70.Lee CJ, De Biasio A, and Beglova N (2010). Mode of interaction between beta2GPI and lipoprotein receptors suggests mutually exclusive binding of beta2GPI to the receptors and anionic phospholipids. Structure 18, 366–376. 10.1016/j.str.2009.12.013. [DOI] [PubMed] [Google Scholar]
- 71.Orlando RA, Exner M, Czekay RP, Yamazaki H, Saito A, Ullrich R, Kerjaschki D, and Farquhar MG (1997). Identification of the second cluster of ligand-binding repeats in megalin as a site for receptor-ligand interactions. Proc Natl Acad Sci U S A 94, 2368–2373. 10.1073/pnas.94.6.2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wolf CA, Dancea F, Shi M, Bade-Noskova V, Ruterjans H, Kerjaschki D, and Lucke C (2007). Solution structure of the twelfth cysteine-rich ligand-binding repeat in rat megalin. Journal of biomolecular NMR 37, 321–328. 10.1007/s10858-006-9129-3. [DOI] [PubMed] [Google Scholar]
- 73.Dagil R, O’Shea C, Nykjaer A, Bonvin AM, and Kragelund BB (2013). Gentamicin binds to the megalin receptor as a competitive inhibitor using the common ligand binding motif of complement type repeats: insight from the nmr structure of the 10th complement type repeat domain alone and in complex with gentamicin. The Journal of biological chemistry 288, 4424–4435. 10.1074/jbc.M112.434159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Horn IR, van den Berg BM, van der Meijden PZ, Pannekoek H, and van Zonneveld AJ (1997). Molecular analysis of ligand binding to the second cluster of complement-type repeats of the low density lipoprotein receptor-related protein. Evidence for an allosteric component in receptor-associated protein-mediated inhibition of ligand binding. J Biol Chem 272, 13608–13613. 10.1074/jbc.272.21.13608. [DOI] [PubMed] [Google Scholar]
- 75.Meijer AB, Rohlena J, van der Zwaan C, van Zonneveld AJ, Boertjes RC, Lenting PJ, and Mertens K (2007). Functional duplication of ligand-binding domains within low-density lipoprotein receptor-related protein for interaction with receptor associated protein, alpha2-macroglobulin, factor IXa and factor VIII. Biochim Biophys Acta 1774, 714–722. 10.1016/j.bbapap.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 76.Neels JG, van Den Berg BM, Lookene A, Olivecrona G, Pannekoek H, and van Zonneveld AJ (1999). The second and fourth cluster of class A cysteine-rich repeats of the low density lipoprotein receptor-related protein share ligand-binding properties. J Biol Chem 274, 31305–31311. 10.1074/jbc.274.44.31305. [DOI] [PubMed] [Google Scholar]
- 77.Obermoeller-McCormick LM, Li Y, Osaka H, FitzGerald DJ, Schwartz AL, and Bu G (2001). Dissection of receptor folding and ligand-binding property with functional minireceptors of LDL receptor-related protein. J Cell Sci 114, 899–908. 10.1242/jcs.114.5.899. [DOI] [PubMed] [Google Scholar]
- 78.Bottomley MJ, Cirillo A, Orsatti L, Ruggeri L, Fisher TS, Santoro JC, Cummings RT, Cubbon RM, Lo Surdo P, Calzetta A, et al. (2009). Structural and biochemical characterization of the wild type PCSK9-EGF(AB) complex and natural familial hypercholesterolemia mutants. J Biol Chem 284, 1313–1323. 10.1074/jbc.M808363200. [DOI] [PubMed] [Google Scholar]
- 79.Kwon HJ, Lagace TA, McNutt MC, Horton JD, and Deisenhofer J (2008). Molecular basis for LDL receptor recognition by PCSK9. Proc Natl Acad Sci U S A 105, 1820–1825. 10.1073/pnas.0712064105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lo Surdo P, Bottomley MJ, Calzetta A, Settembre EC, Cirillo A, Pandit S, Ni YG, Hubbard B, Sitlani A, and Carfi A (2011). Mechanistic implications for LDL receptor degradation from the PCSK9/LDLR structure at neutral pH. EMBO Rep 12, 1300–1305. 10.1038/embor.2011.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang DW, Lagace TA, Garuti R, Zhao Z, McDonald M, Horton JD, Cohen JC, and Hobbs HH (2007). Binding of proprotein convertase subtilisin/kexin type 9 to epidermal growth factor-like repeat A of low density lipoprotein receptor decreases receptor recycling and increases degradation. J Biol Chem 282, 18602–18612. 10.1074/jbc.M702027200. [DOI] [PubMed] [Google Scholar]
- 82.Esser V, Limbird LE, Brown MS, Goldstein JL, and Russell DW (1988). Mutational analysis of the ligand binding domain of the low density lipoprotein receptor. J Biol Chem 263, 13282–13290. [PubMed] [Google Scholar]
- 83.Huang S, Henry L, Ho YK, Pownall HJ, and Rudenko G (2010). Mechanism of LDL binding and release probed by structure-based mutagenesis of the LDL receptor. J Lipid Res 51, 297–308. 10.1194/jlr.M000422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zong Y, Zhang B, Gu S, Lee K, Zhou J, Yao G, Figueiredo D, Perry K, Mei L, and Jin R (2012). Structural basis of agrin-LRP4-MuSK signaling. Genes Dev 26, 247–258. 10.1101/gad.180885.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kim J, Han W, Park T, Kim EJ, Bang I, Lee HS, Jeong Y, Roh K, Kim J, Kim JS, et al. (2020). Sclerostin inhibits Wnt signaling through tandem interaction with two LRP6 ectodomains. Nat Commun 11, 5357. 10.1038/s41467-020-19155-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ren Q, Chen J, and Liu Y (2021). LRP5 and LRP6 in Wnt Signaling: Similarity and Divergence. Front Cell Dev Biol 9, 670960. 10.3389/fcell.2021.670960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tamai K, Semenov M, Kato Y, Spokony R, Liu C, Katsuyama Y, Hess F, Saint-Jeannet JP, and He X (2000). LDL-receptor-related proteins in Wnt signal transduction. Nature 407, 530–535. 10.1038/35035117. [DOI] [PubMed] [Google Scholar]
- 88.Pinson KI, Brennan J, Monkley S, Avery BJ, and Skarnes WC (2000). An LDL-receptor-related protein mediates Wnt signalling in mice. Nature 407, 535–538. 10.1038/35035124. [DOI] [PubMed] [Google Scholar]
- 89.Chen S, Bubeck D, MacDonald BT, Liang WX, Mao JH, Malinauskas T, Llorca O, Aricescu AR, Siebold C, He X, and Jones EY (2011). Structural and functional studies of LRP6 ectodomain reveal a platform for Wnt signaling. Dev Cell 21, 848–861. 10.1016/j.devcel.2011.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cheng Z, Biechele T, Wei Z, Morrone S, Moon RT, Wang L, and Xu W (2011). Crystal structures of the extracellular domain of LRP6 and its complex with DKK1. Nat Struct Mol Biol 18, 1204–1210. 10.1038/nsmb.2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Matoba K, Mihara E, Tamura-Kawakami K, Miyazaki N, Maeda S, Hirai H, Thompson S, Iwasaki K, and Takagi J (2017). Conformational Freedom of the LRP6 Ectodomain Is Regulated by N-glycosylation and the Binding of the Wnt Antagonist Dkk1. Cell Rep 18, 32–40. 10.1016/j.celrep.2016.12.017. [DOI] [PubMed] [Google Scholar]
- 92.Ahn VE, Chu ML, Choi HJ, Tran D, Abo A, and Weis WI (2011). Structural basis of Wnt signaling inhibition by Dickkopf binding to LRP5/6. Dev Cell 21, 862–873. 10.1016/j.devcel.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jeon H, and Blacklow SC (2005). Structure and physiologic function of the low-density lipoprotein receptor. Annu Rev Biochem 74, 535–562. 10.1146/annurev.biochem.74.082803.133354. [DOI] [PubMed] [Google Scholar]
- 94.Beglova N, Jeon H, Fisher C, and Blacklow SC (2004). Cooperation between fixed and low pH-inducible interfaces controls lipoprotein release by the LDL receptor. Mol Cell 16, 281–292. 10.1016/j.molcel.2004.09.038. [DOI] [PubMed] [Google Scholar]
- 95.Davis CG, Goldstein JL, Sudhof TC, Anderson RG, Russell DW, and Brown MS (1987). Acid-dependent ligand dissociation and recycling of LDL receptor mediated by growth factor homology region. Nature 326, 760–765. 10.1038/326760a0. [DOI] [PubMed] [Google Scholar]
- 96.Boswell EJ, Jeon H, Blacklow SC, and Downing AK (2004). Global defects in the expression and function of the low density lipoprotein receptor (LDLR) associated with two familial hypercholesterolemia mutations resulting in misfolding of the LDLR epidermal growth factor-AB pair. J Biol Chem 279, 30611–30621. 10.1074/jbc.M401412200. [DOI] [PubMed] [Google Scholar]
- 97.Rudenko G, Henry L, Henderson K, Ichtchenko K, Brown MS, Goldstein JL, and Deisenhofer J (2002). Structure of the LDL receptor extracellular domain at endosomal pH. Science 298, 2353–2358. 10.1126/science.1078124. [DOI] [PubMed] [Google Scholar]
- 98.Herz J, Goldstein JL, Strickland DK, Ho YK, and Brown MS (1991). 39-kDa protein modulates binding of ligands to low density lipoprotein receptor-related protein/alpha 2-macroglobulin receptor. J Biol Chem 266, 21232–21238. [PubMed] [Google Scholar]
- 99.Kounnas MZ, Moir RD, Rebeck GW, Bush AI, Argraves WS, Tanzi RE, Hyman BT, and Strickland DK (1995). LDL receptor-related protein, a multifunctional ApoE receptor, binds secreted beta-amyloid precursor protein and mediates its degradation. Cell 82, 331–340. 10.1016/0092-8674(95)90320-8. [DOI] [PubMed] [Google Scholar]
- 100.Narita M, Bu G, Herz J, and Schwartz AL (1995). Two receptor systems are involved in the plasma clearance of tissue-type plasminogen activator (t-PA) in vivo. J Clin Invest 96, 1164–1168. 10.1172/JCI118105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Willnow TE, Orth K, and Herz J (1994). Molecular dissection of ligand binding sites on the low density lipoprotein receptor-related protein. J Biol Chem 269, 15827–15832. [PubMed] [Google Scholar]
- 102.Willnow TE, Rohlmann A, Horton J, Otani H, Braun JR, Hammer RE, and Herz J (1996). RAP, a specialized chaperone, prevents ligand-induced ER retention and degradation of LDL receptor-related endocytic receptors. EMBO J 15, 2632–2639. [PMC free article] [PubMed] [Google Scholar]
- 103.Bu G, Geuze HJ, Strous GJ, and Schwartz AL (1995). 39 kDa receptor-associated protein is an ER resident protein and molecular chaperone for LDL receptor-related protein. EMBO J 14, 2269–2280. 10.1002/j.1460-2075.1995.tb07221.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mokuno H, Brady S, Kotite L, Herz J, and Havel RJ (1994). Effect of the 39-kDa receptor-associated protein on the hepatic uptake and endocytosis of chylomicron remnants and low density lipoproteins in the rat. J Biol Chem 269, 13238–13243. [PubMed] [Google Scholar]
- 105.Orlando RA, and Farquhar MG (1994). Functional domains of the receptor-associated protein (RAP). Proc Natl Acad Sci U S A 91, 3161–3165. 10.1073/pnas.91.8.3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ashcom JD, Tiller SE, Dickerson K, Cravens JL, Argraves WS, and Strickland DK (1990). The human alpha 2-macroglobulin receptor: identification of a 420-kD cell surface glycoprotein specific for the activated conformation of alpha 2-macroglobulin. J Cell Biol 110, 1041–1048. 10.1083/jcb.110.4.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Migliorini MM, Behre EH, Brew S, Ingham KC, and Strickland DK (2003). Allosteric modulation of ligand binding to low density lipoprotein receptor-related protein by the receptor-associated protein requires critical lysine residues within its carboxyl-terminal domain. J Biol Chem 278, 17986–17992. 10.1074/jbc.M212592200. [DOI] [PubMed] [Google Scholar]
- 108.Beglov D, Lee CJ, De Biasio A, Kozakov D, Brenke R, Vajda S, and Beglova N (2009). Structural insights into recognition of beta2-glycoprotein I by the lipoprotein receptors. Proteins 77, 940–949. 10.1002/prot.22519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Nikolic J, Belot L, Raux H, Legrand P, Gaudin Y, and A AA (2018). Structural basis for the recognition of LDL-receptor family members by VSV glycoprotein. Nat Commun 9, 1029. 10.1038/s41467-018-03432-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.De Nardis C, Lossl P, van den Biggelaar M, Madoori PK, Leloup N, Mertens K, Heck AJ, and Gros P (2017). Recombinant Expression of the Full-length Ectodomain of LDL Receptor-related Protein 1 (LRP1) Unravels pH-dependent Conformational Changes and the Stoichiometry of Binding with Receptor-associated Protein (RAP). J Biol Chem 292, 912–924. 10.1074/jbc.M116.758862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Jensen PH, Moestrup SK, and Gliemann J (1989). Purification of the human placental alpha 2-macroglobulin receptor. FEBS Lett 255, 275–280. 10.1016/0014-5793(89)81105-6. [DOI] [PubMed] [Google Scholar]
- 112.Kanalas JJ, and Makker SP (1990). Isolation of a 330-kDa glycoprotein from human kidney similar to the Heymann nephritis autoantigen (gp330). J Am Soc Nephrol 1, 792–798. 10.1681/ASN.V15792. [DOI] [PubMed] [Google Scholar]
- 113.Moestrup SK, and Gliemann J (1989). Purification of the rat hepatic alpha 2-macroglobulin receptor as an approximately 440-kDa single chain protein. J Biol Chem 264, 15574–15577. [PubMed] [Google Scholar]
- 114.Orlando RA, Kerjaschki D, Kurihara H, Biemesderfer D, and Farquhar MG (1992). gp330 associates with a 44-kDa protein in the rat kidney to form the Heymann nephritis antigenic complex. Proc Natl Acad Sci U S A 89, 6698–6702. 10.1073/pnas.89.15.6698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Biemesderfer D, Dekan G, Aronson PS, and Farquhar MG (1993). Biosynthesis of the gp330/44-kDa Heymann nephritis antigenic complex: assembly takes place in the ER. Am J Physiol 264, F1011–1020. 10.1152/ajprenal.1993.264.6.F1011. [DOI] [PubMed] [Google Scholar]
- 116.Lundstrom M, Orlando RA, Saedi MS, Woodward L, Kurihara H, and Farquhar MG (1993). Immunocytochemical and biochemical characterization of the Heymann nephritis antigenic complex in rat L2 yolk sac cells. Am J Pathol 143, 1423–1435. [PMC free article] [PubMed] [Google Scholar]
- 117.Williams SE, Ashcom JD, Argraves WS, and Strickland DK (1992). A novel mechanism for controlling the activity of alpha 2-macroglobulin receptor/low density lipoprotein receptor-related protein. Multiple regulatory sites for 39-kDa receptor-associated protein. J Biol Chem 267, 9035–9040. [PubMed] [Google Scholar]
- 118.Punjani A, Rubinstein JL, Fleet DJ, and Brubaker MA (2017). cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods 14, 290–296. 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- 119.Scheres SH (2012). RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol 180, 519–530. 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Scheres SH (2012). A Bayesian view on cryo-EM structure determination. J Mol Biol 415, 406–418. 10.1016/j.jmb.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zhong ED, Bepler T, Berger B, and Davis JH (2021). CryoDRGN: reconstruction of heterogeneous cryo-EM structures using neural networks. Nat Methods 18, 176–185. 10.1038/s41592-020-01049-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Pober BR, Longoni M, and Noonan KM (2009). A review of Donnai-Barrow and facio-oculo-acoustico-renal (DB/FOAR) syndrome: clinical features and differential diagnosis. Birth Defects Res A Clin Mol Teratol 85, 76–81. 10.1002/bdra.20534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Charlton JR, Tan W, Daouk G, Teot L, Rosen S, Bennett KM, Cwiek A, Nam S, Emma F, Jouret F, et al. (2020). Beyond the tubule: pathological variants of LRP2, encoding the megalin receptor, result in glomerular loss and early progressive chronic kidney disease. Am J Physiol Renal Physiol 319, F988–F999. 10.1152/ajprenal.00295.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Carss KJ, Arno G, Erwood M, Stephens J, Sanchis-Juan A, Hull S, Megy K, Grozeva D, Dewhurst E, Malka S, et al. (2017). Comprehensive Rare Variant Analysis via Whole-Genome Sequencing to Determine the Molecular Pathology of Inherited Retinal Disease. Am J Hum Genet 100, 75–90. 10.1016/j.ajhg.2016.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Benson KA, White M, Allen NM, Byrne S, Carton R, Comerford E, Costello D, Doherty C, Dunleavey B, El-Naggar H, et al. (2020). A comparison of genomic diagnostics in adults and children with epilepsy and comorbid intellectual disability. Eur J Hum Genet 28, 1066–1077. 10.1038/s41431-020-0610-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Vasli N, Ahmed I, Mittal K, Ohadi M, Mikhailov A, Rafiq MA, Bhatti A, Carter MT, Andrade DM, Ayub M, et al. (2016). Identification of a homozygous missense mutation in LRP2 and a hemizygous missense mutation in TSPYL2 in a family with mild intellectual disability. Psychiatr Genet 26, 66–73. 10.1097/YPG.0000000000000114. [DOI] [PubMed] [Google Scholar]
- 127.Makarova A, Bercury KK, Adams KW, Joyner D, Deng M, Spoelgen R, Koker M, Strickland DK, and Hyman BT (2008). The LDL receptor-related protein can form homo-dimers in neuronal cells. Neurosci Lett 442, 91–95. 10.1016/j.neulet.2008.06.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Bacskai BJ, Xia MQ, Strickland DK, Rebeck GW, and Hyman BT (2000). The endocytic receptor protein LRP also mediates neuronal calcium signaling via N-methyl-D-aspartate receptors. Proc Natl Acad Sci U S A 97, 11551–11556. 10.1073/pnas.200238297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Schmitz J, Preiser H, Maestracci D, Ghosh BK, Cerda JJ, and Crane RK (1973). Purification of the human intestinal brush border membrane. Biochim Biophys Acta 323, 98–112. 10.1016/0005-2736(73)90434-3. [DOI] [PubMed] [Google Scholar]
- 130.Rappsilber J, Mann M, and Ishihama Y (2007). Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc 2, 1896–1906. 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- 131.Perkins DN, Pappin DJ, Creasy DM, and Cottrell JS (1999). Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567. . [DOI] [PubMed] [Google Scholar]
- 132.Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, and Mann M (2011). Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res 10, 1794–1805. 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- 133.Tyanova S, Temu T, and Cox J (2016). The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat Protoc 11, 2301–2319. 10.1038/nprot.2016.136. [DOI] [PubMed] [Google Scholar]
- 134.Schuck P (2000). Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys J 78, 1606–1619. 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Young G, Hundt N, Cole D, Fineberg A, Andrecka J, Tyler A, Olerinyova A, Ansari A, Marklund EG, Collier MP, et al. (2018). Quantitative mass imaging of single biological macromolecules. Science 360, 423–427. 10.1126/science.aar5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Sigmundsson K, Masson G, Rice R, Beauchemin N, and Obrink B (2002). Determination of active concentrations and association and dissociation rate constants of interacting biomolecules: an analytical solution to the theory for kinetic and mass transport limitations in biosensor technology and its experimental verification. Biochemistry 41, 8263–8276. 10.1021/bi020099h. [DOI] [PubMed] [Google Scholar]
- 137.Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, Stagg S, Potter CS, and Carragher B (2005). Automated molecular microscopy: the new Leginon system. J Struct Biol 151, 41–60. 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 138.Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, and Agard DA (2017). MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 14, 331–332. 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Rohou A, and Grigorieff N (2015). CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol 192, 216–221. 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Punjani A, and Fleet DJ (2021). 3D variability analysis: Resolving continuous flexibility and discrete heterogeneity from single particle cryo-EM. J Struct Biol 213, 107702. 10.1016/j.jsb.2021.107702. [DOI] [PubMed] [Google Scholar]
- 141.Punjani A, Zhang H, and Fleet DJ (2020). Non-uniform refinement: adaptive regularization improves single-particle cryo-EM reconstruction. Nat Methods 17, 1214–1221. 10.1038/s41592-020-00990-8. [DOI] [PubMed] [Google Scholar]
- 142.Sanchez-Garcia R, Gomez-Blanco J, Cuervo A, Carazo JM, Sorzano COS, and Vargas J (2021). DeepEMhancer: a deep learning solution for cryo-EM volume post-processing. Commun Biol 4, 874. 10.1038/s42003-021-02399-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Emsley P, and Cowtan K (2004). Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60, 2126–2132. 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 144.Croll TI (2018). ISOLDE: a physically realistic environment for model building into low-resolution electron-density maps. Acta Crystallogr D Struct Biol 74, 519–530. 10.1107/S2059798318002425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Afonine PV, Poon BK, Read RJ, Sobolev OV, Terwilliger TC, Urzhumtsev A, and Adams PD (2018). Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr D Struct Biol 74, 531–544. 10.1107/S2059798318006551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Morelle W, Haslam SM, Ziak M, Roth J, Morris HR, and Dell A (2000). Characterization of the N-linked oligosaccharides of megalin (gp330) from rat kidney. Glycobiology 10, 295–304. 10.1093/glycob/10.3.295. [DOI] [PubMed] [Google Scholar]
- 147.Tian E, Wang S, Zhang L, Zhang Y, Malicdan MC, Mao Y, Christoffersen C, Tabak LA, Schjoldager KT, and Ten Hagen KG (2019). Galnt11 regulates kidney function by glycosylating the endocytosis receptor megalin to modulate ligand binding. Proc Natl Acad Sci U S A 116, 25196–25202. 10.1073/pnas.1909573116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Wang S, Mao Y, Narimatsu Y, Ye Z, Tian W, Goth CK, Lira-Navarrete E, Pedersen NB, Benito-Vicente A, Martin C, et al. (2018). Site-specific O-glycosylation of members of the low-density lipoprotein receptor superfamily enhances ligand interactions. J Biol Chem 293, 7408–7422. 10.1074/jbc.M117.817981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Hirano M, Totani K, Fukuda T, Gu J, and Suzuki A (2017). N-Glycoform-dependent interactions of megalin with its ligands. Biochim Biophys Acta Gen Subj 1861, 3106–3118. 10.1016/j.bbagen.2016.10.015. [DOI] [PubMed] [Google Scholar]
- 150.Liebschner D, Afonine PV, Baker ML, Bunkoczi G, Chen VB, Croll TI, Hintze B, Hung LW, Jain S, McCoy AJ, et al. (2019). Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr D Struct Biol 75, 861–877. 10.1107/S2059798319011471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Afonine PV, Klaholz BP, Moriarty NW, Poon BK, Sobolev OV, Terwilliger TC, Adams PD, and Urzhumtsev A (2018). New tools for the analysis and validation of cryo-EM maps and atomic models. Acta Crystallogr D Struct Biol 74, 814–840. 10.1107/S2059798318009324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Davis IW, Murray LW, Richardson JS, and Richardson DC (2004). MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res 32, W615–619. 10.1093/nar/gkh398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Terashi G, Wang X, Maddhuri Venkata Subramaniya SR, Tesmer JJG, and Kihara D (2022). Residue-wise local quality estimation for protein models from cryo-EM maps. Nat Methods 19, 1116–1125. 10.1038/s41592-022-01574-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Pettersen EF, Goddard TD, Huang CC, Meng EC, Couch GS, Croll TI, Morris JH, and Ferrin TE (2021). UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci 30, 70–82. 10.1002/pro.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Ashkenazy H, Abadi S, Martz E, Chay O, Mayrose I, Pupko T, and Ben-Tal N (2016). ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res 44, W344–350. 10.1093/nar/gkw408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Ashkenazy H, Erez E, Martz E, Pupko T, and Ben-Tal N (2010). ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res 38, W529–533. 10.1093/nar/gkq399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Glaser F, Pupko T, Paz I, Bell RE, Bechor-Shental D, Martz E, and Ben-Tal N (2003). ConSurf: identification of functional regions in proteins by surface-mapping of phylogenetic information. Bioinformatics 19, 163–164. 10.1093/bioinformatics/19.1.163. [DOI] [PubMed] [Google Scholar]
- 158.Landau M, Mayrose I, Rosenberg Y, Glaser F, Martz E, Pupko T, and Ben-Tal N (2005). ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res 33, W299–302. 10.1093/nar/gki370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Zidek A, Potapenko A, et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589. 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Emsley P, Lohkamp B, Scott WG, and Cowtan K (2010). Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66, 486–501. 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Adams PD, Gopal K, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Pai RK, Read RJ, Romo TD, et al. (2004). Recent developments in the PHENIX software for automated crystallographic structure determination. J Synchrotron Radiat 11, 53–55. 10.1107/s0909049503024130. [DOI] [PubMed] [Google Scholar]
- 162.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, and Ferrin TE (2004). UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 25, 1605–1612. 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 163.Laue TM (1997). Advances in sedimentation velocity analysis. Biophys J 72, 395–396. 10.1016/S0006-3495(97)78679-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: Alignment of ligand-binding repeats (L) in LRP2 and LDLR
The four groups of L repeats in LRP2 are boxed at left. Regions of beta-sheet are boxed at 0.75 pt weight, and regions of alpha-helix are boxed at 1.5 pt weight. Arrows above the alignment identify Ca2+-coordinating residues: solid arrows indicating involvement of the residue’s side chain, and open arrows indicating involvement of the residue’s main chain carbonyl. Intramolecular ligands in L25 and L36 are in bold and underlined.
Related to Figure 1, Figure 2, Figure S1
Table S2: Alignment of EGF-like domains (E) in LRP2 and LDLR
EGF-like pairs are boxed at left. Regions of beta-sheet are boxed in the alignment. Residues from calcium-binding (cb) EGF-like domains whose side chain coordinates a Ca2+ ion are underlined and italicized. Intramolecular ligands from the EGF-like pairs are in bold and underlined.
Related to Figure 1, Figure 2, Figure S1
Table S3: Mass spectrometry of endogenously purified LRP2 from mouse kidney
Proteins identified in SDS-PAGE purified protein with apparent molecular weight 600kDa: tandem mass spectrometry (MS/MS) based protein and peptide identifications are expressed as exclusive spectrum count of the peptide spectral matches. Exclusive peptide count, identified protein percent sequence coverage, and the protein abundance index (emPAI factor that defines the ratio of the observed to observable peptides) are also listed.
Related to Figure 1
Table S4: Validations of the cryo-EM 3D reconstructions of LRP2 at pH 5.2 and pH 7.5
Validations were conducted in Phenix of the molecular models of LRP2 against a composite of the unsharpened locally refined maps.
Related to Figure 2
Movie S2: Flexibility of the LRP2 assembly at pH 5.2
Continuous heterogeneous classification of the LRP2 dataset bin4 at pH 5.2 was conducted in Cryodrgn121. The movie represents a principal component analysis of the dataset in a series of 10 volumes. In comparison to the LRP2 dataset at pH 7.5, movement within the assembly is of lesser magnitude and more evenly distributed across the structure.
Movie S1: Flexibility of the LRP2 assembly at pH 7.5
Continuous heterogeneous classification of the LRP2 dataset bin4 at pH 7.5 was conducted in Cryodrgn121. The movie represents a principal component analysis of the dataset in a series of 10 volumes. The lateral portions of the map that include R1, P1-P2, and R2 skew 15–17 Å relative to one another. The center portion of the map translates out of the plane established by the membrane insertion points by 10–15 Å. In relation to these changes, the canopy exhibits minimal movement.
Movie S3: The pH-dependent structural transition of LRP2
The LRP2 assembly is represented as a surface with one protomer in blue and the other in salmon. The structures at pH 7.5 and pH 5.2 are morphed into another using ChimeraX154. L3-L5, L14-L15, L18-L23, and L35-L36-E14 were not modeled in the atomic model at pH 7.5 since their Cα traces could not be well-defined due to the flexibility of the assembly. However, for the purposes of this morphing representation, they have been empirically docked into the low-resolution cryo-EM density corresponding to their positions at pH 7.5. All domains in the model undergo significant translations and re-orientations, all homodimer interfaces are pH-specific, and a decrease of homodimer interface at pH 5.2 enables increased intra-protomer contacts as L repeats pack against EGF-like domains and β-propellers.
Figure S1. Related to Figure 2; Domain organization of the LDLR family
Members of the LDLR family are schematically represented with their constituent domains, identified in the key. Domain organization is highly conserved between LRP1 and LRP2, the largest members of the family. Mouse LRP2 and human LRP2 have identical domain organization.
Figure S2. Related to Figure 2, Table S4, Movie S1; Cryo-EM data processing and validation for the LRP2 structure at pH 7.5
(A) The processing pipeline. (B) 2D Classification (C) Ab initio model (D) Homogeneous refinements with C1 and C2 symmetry constraints yield identical structures. (E) Local resolution (F) 3D classification did not demonstrate any substantial variation in domain organizations across the dataset. (G) Local refinement (H) Non-homogeneous refinement FSC (I) Viewing Direction Distribution (J) Map-Model FSC for the full model and locally refined areas. (K) CC per-residue with the average CC mask for the structure indicated. (L) DAQ per-residue (M) Model colored by DAQ score. (N) P domains superimposed on AF2 predictions.
Figure S4. Related to Figure 3, Figure 5, Movie S3; Contact Maps for the LRP2 structures at pH 5.2 and pH 7.5
(A) The domains of the two protomers are listed along the two short sides of the triangle, and intersecting squares are colored depending on whether contact is made between domains at pH 5.2, pH 7.5, or at both pH’s.
(B) Intra-protomer and homodimer interactions represented on domain diagrams. The red and blue lines represent domain-domain interactions at pH 5.2 and pH 7.5, respectively.
Figure S3. Related to Figure 2, Table S4, Movie S2; Cryo-EM data processing and validation for the LRP2 structure at pH 5.2
(A) The processing pipeline. (B) 2D Classification (C) Ab initio model (D) Homogeneous refinements with C1 and C2 symmetry constraints are identical. (E) Local resolution (F) 3D classification did not demonstrate any substantial variation in domain organizations across the dataset. (G) Local refinement (H) Non-homogeneous refinement FSC (I) Viewing Direction Distribution (J) Map-Model FSC for the full model and locally refined areas. (K) CC per-residue with the average CC mask for the structure indicated. (L) DAQ per-residue (M) Model colored by DAQ score. (N) P domains superimposed on AF2 predictions.
Figure S5. Related to Figure 4; Evolutionary conservation of residues at the putative Ca2+-coordinating sites in the canopy of LRP2
(A) Looking down upon the canopy of the homodimer at pH 7.5 in the direction of the cell membrane, the cleft between the two protomers of the homodimer is visible. Alignments of LRP1 and LRP2 sequences were run through the Consurf server to assess conservation. Magenta coloring of the cartoon depiction indicates high sequence conservation while teal coloring indicates high sequence variability. The majority of the loop regions connecting the β-strands are variable, with the exception of residues responsible for putative calcium coordination. Residue level detail is shown for the boxed regions. (B) D1621, H1645 are highly conserved at the putative Ca2+-coordination site despite residing in a loop region. (C) D2257 and S2276 are the only highly conserved residues in the loop region surrounding the putative Ca2+-coordination site. (D) Alignment of selected LRP1 and LRP2 sequences to show conservation of D1621, H1645, and D2257.
Data Availability Statement
Cryo-EM maps and fitted coordinates of LRP2 at pH 7.5 and pH 5.2 have been deposited with accession codes, 8EM4 and 8EM7 respectively. This paper does not report original code. Particle stacks for LRP2 at pH 7.5 and pH 5.2 can be shared upon request. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


