Objectives:
Latent infection by HIV hinders viral eradication despite effective antiretroviral treatment (ART). Among proposed contributors to viral latency are cellular small RNAs that have also been proposed to shuttle between cells in extracellular vesicles. Thus, we profiled extracellular vesicle small RNAs during different infection phases to understand the potential relationship between these extracellular vesicle associated small RNAs and viral infection.
Design:
A well characterized simian immunodeficiency virus (SIV)/macaque model of HIV was used to profile extracellular vesicle enriched blood plasma fractions harvested during preinfection, acute infection, latent infection/ART treatment, and rebound after ART interruption.
Methods:
Measurement of extracellular vesicle concentration, size distribution, and morphology was complemented with qPCR array for small RNA expression, followed by individual qPCR validations. Iodixanol density gradients were used to separate extracellular vesicle subtypes and virions.
Results:
Plasma extracellular vesicle particle counts correlated with viral load and peaked during acute infection. However, SIV gag RNA detection showed that virions did not fully explain this peak. Extracellular vesicle microRNAs miR-181a, miR-342–3p, and miR-29a decreased with SIV infection and remained downregulated in latency. Interestingly, small nuclear RNA U6 had a tight association with viral load peak.
Conclusion:
This study is the first to monitor how extracellular vesicle concentration and extracellular vesicle small RNA expression change dynamically in acute viral infection, latency, and rebound in a carefully controlled animal model. These changes may also reveal regulatory roles in retroviral infection and latency.
Keywords: antiretroviral therapy, ectosomes, exosomes, HIV, microvesicles, miRNA, retrovirus, simian immunodeficiency virus, small nuclear RNA, small RNA, U6, viral rebound
Introduction
Although HIV can be well controlled virologically with antiretroviral treatment (ART), chronic inflammation and other mechanisms lead to early aging disorders and serious non-AIDS events (SNAEs) including the HIV-associated neurocognitive disorders (HAND) [1,2]. Eradication of HIV reservoirs also cannot yet be achieved routinely [3,4]. New biomarkers of SNAEs, inflammation, and responses to treatments and eradication approaches would be useful. Animal models of HIV facilitate understanding of disease and biomarker discovery. These models allow control of ART dosing, compliance, disease monitoring, and other factors and also access to normally inaccessible compartments like the brain. Pig-tailed macaques (Macaca nemestrina) and rhesus macaques (Macaca mulatta) are well characterized SIV models of HIV disease, with some species-specific differences [5–7].
Extracellular vesicles are nanosized membranous vesicles released from most cells. Extracellular vesicles have roles in viral pathogenesis [8–10] and, much like retroviruses, shuttle molecules between cells, including microRNAs (miRNAs) [11,12] and small nuclear RNAs (snRNAs) [13–15]. We and others have reported correlations of miRNA expression with HIV infection, virus replication, and central nervous system (CNS) diseases using HIV-infected CD4+ cell lines [16,17], total plasma [18–21], and extracellular vesicles [22–26]. However, our previous work identified relatively few robust miRNA differences in total plasma [18], and a preinfection time point was required to establish informative changes in abundance. This may be because a large proportion of RNA in the blood is found in non-extracellular vesicle fractions [11,27] that represent a relatively constant baseline of nonspecific release from dead or dying cells. We reason that extracellular vesicle release might better reflect the state of living cells and responses to infection events, but dynamic changes of extracellular vesicles attributes including miRNAs have not yet been fully characterized at different stages of retroviral infection and treatment.
We thus characterized extracellular vesicles, extracellular vesicles miRNA, and U6 snRNA in longitudinal samples from pig-tailed and rhesus macaque models [28–31], comparing preinfection with, acute (viremic peak), latent (ART-suppressed), and rebound [ART-treatment interruption (ATI)] phases. We report that average particle counts correlate with infection, while levels of several extracellular vesicle miRNAs are altered even during latent infection. Remarkably, U6 snRNA is highly upregulated during infection, even in extracellular vesicles purified by density gradient.
Materials and methods
Sample collection
All samples were from archives of studies approved by the Johns Hopkins University Institutional Animal Care and Use Committee and conducted in accordance with the Weatherall Report, the Guide for the Care and Use of Laboratory Animals, and the USDA Animal Welfare Act. For initial studies and verification of extracellular vesicle separations, plasma samples were obtained from pigtailed macaques that were not infected (n = 2) or dual-inoculated with SIV swarm B670 and clone SIV/17E-Fr and untreated (n = 3) or treated then treatment interruption (“rebound,” n = 3); (Supplemental Table 1). Longitudinal verification samples were from two cohorts of six pigtailed macaques dual-inoculated as above [28,29] and treated with ART (consisting of once daily subcutaneous 2.5 mg/kg dolutegravir, 20 mg/kg tenofovir, and 30 mg/kg emtricitabine) boosted or not with maraviroc, and a cohort of six rhesus macaques infected with SIVmac251 [30,31] and treated with ART (Supplemental Table 2). For pigtails, ART started at 12 days postinoculation (dpi), and for rhesus, at 14 dpi. Time points were preinfection (two draws), acute infection (7, 14 dpi), latent infection (ART-suppressed) (86, 154 dpi), rebound [12 days ART treatment interruption (postrelease (dpr)], and necropsy. Additional pigtailed samples (uninfected, n = 3 and acute infected 7 dpi, n = 3) were used for density separations (Supplemental Table 3).
Separation of plasma extracellular vesicle enriched and protein-enriched fractions
Plasma was thawed on ice and centrifuged twice at 2500 × g (15 min, 4°C) to deplete residual platelets and debris [32,33]. 0.1 (Samples listed in Supplemental Table 2), 0.5 (Supplemental Table 1), or 8 (Supplemental Table 3) ml of platelet-depleted plasma (PDP) was separated by size-exclusion chromatography (SEC) with qEVsingle/70 nm, qEVoriginal/70 nm, or qEV10/70 nm columns (Izon Science). PBS was used to elute fractions of 0.2 ml (qEVsingle/70 nm), 0.5 ml (qEVoriginal/70 nm), or 5 ml (qEV10/70 nm). Extracellular vesicle enriched fractions (F6–8, qEVsingle/70 nm; F7–9, qEVoriginal/70 nm; and F1–4, qEV10/70 nm) were pooled and concentrated by 100 kilodalton (kDa) MWCO concentrators (Thermo Fisher 88503, 88524, 88532). Pooled fractions 10–12 and 13–15 from qEV original columns were collected and concentrated as protein-enriched fractions. All fractions were stored at –80°C.
Iodixanol gradient separation
Iodixanol gradients (Optiprep, Sigma-Aldrich D1556) were made by layering 2.5 ml each of 18, 14, 10, 6% iodixanol (from 60% iodixanol at 1.320 ± 0.001 g/ml) diluted in PBS. extracellular vesicle enriched fractions in 1.6 ml PBS were loaded onto the gradient. After ultracentrifugation at 200 000 × g (60 min, 4°C, TH-641 rotor, 13.2 ml thinwall polypropylene tubes, acceleration/deceleration 9), 12 fractions (0.96 ml/each) were collected from the top. Fraction densities were measured by absorbance at 340 nm. Fractions were diluted in 5 ml PBS and washed by ultracentrifugation at 200 000 × g (60 min, 4°C, AH-650 rotor, 5 ml Beckman Ultra-Clear tubes, acceleration/deceleration 9). Pellets were pipetted up and down 10 times and vortexed for 5 s in 120 μl PBS. Tubes were placed on ice (20 min) followed by another round of pipetting/vortexing as above. Resuspensions were stored at –80°C.
Nanoflow cytometry
Concentration and size of extracellular vesicle and protein-enriched fractions were measured for 1 min by side-scatter using NFCM (NanoFCM) calibrated for concentration and size with 200 nm polystyrene beads and silica nanospheres, respectively (NanoFCM).
Transmission electron microscopy
Extracellular vesicle preparations (10 μl) were adsorbed to glow-discharged 400 mesh ultra-thin carbon-coated grids (EMS CF400-CU-UL) for 2 min followed by three rinses in TBS and staining in 1% uranyl acetate with 0.05 Tylose. After aspiration and drying, grids were observed with a Philips CM120 instrument at 80 kV. Images were captured by XR80 charge-coupled device (8 megapixel; AMT Imaging, Woburn, Massachusetts, USA).
Western blotting
Extracellular vesicle containing fractions were lysed in 1X RIPA. Protein concentrations were determined by microBCA protein assay (Thermo Fisher, 23235). Equivalent protein amounts (extracellular vesicles and proteins) were separated on 4–15% stain-free precast SDS-PAGE gradient gels (Bio-Rad 5678083) under nonreducing conditions and transferred onto PVDF membranes (Sigma Aldrich IPVH00005). After 1 h blocking (5% nonfat milk, Bio-Rad 170–6404) at room temperature (RT), membranes were incubated with antibodies against CD63 (1 : 1000, BD Biosciences 556019), CD81 (1 : 500, Santa Cruz Biotechnology sc23962), calnexin (1 : 2000, Abcam ab22595), GM130 (1 : 1000, Abcam, ab76154), albumin (1 : 1000, Abcam ab28405), AGO2 (1 : 500, Sigma-Aldrich SAB4200085), ApoB100 (1 : 1000, Academy Bio-Medical 20A-G1b), ApoA1 (1 : 1000, Academy Bio-Medical 11A-G2b), and ApoC1 (1 : 1000, Academy Bio-Medical 31A-G1b) overnight at 4°C. Membranes were washed three times for 8 min in PBST with shaking, then incubated with HRP-conjugated secondary mouse antirabbit IgG or mouse IgG kappa binding protein antibodies (1 : 10 000, Santa Cruz Biotechnology sc-2357 and sc-516102) at RT for 1 h. After a PBST wash, membranes were incubated with SuperSignal West Pico PLUS chemiluminescent substrate (Thermo Fisher 34580) and visualized by iBright (Thermo Fisher, Waltham, Massachusetts, USA).
Total RNA extraction
RNA was extracted by miRNeasy Serum/Plasma Kit (Qiagen 217184) per manufacturer's instructions, with 4 × 108 copies of cel-miR-39 miRNA mimic (Qiagen 339390) spiked into to the sample after addition of lysis buffer.
SIV Gag RNA quantification by RT-qPCR/ddPCR
Viral RNA was measured by quantitative reverse transcription-PCR (qPCR) or digital droplet PCR (ddPCR) as described [34,35]. RNA from 140 μl of plasma was isolated by QIAamp Viral RNA Minikit (Qiagen 1020953). qPCR of SIV gag RNA was by QuantiTect Virus kit (Qiagen 211011) or ddPCR using One-Step RT ddPCR Adv kit (Bio-Rad 1864022). Copy numbers were calculated with a regression curve from control transcript standards and normalization to the volume of extracted plasma. Primers/probes for SIV gag RNA were: SIV21 forward 5’-GTCTGCGTCATCTGGTGCATTC-3’; SIV22 reverse 5‘’-CACTAGGTGTCTCTGCACTATCTGTTTTG-3’; SIV23, 5’ FAM/3’-Black hole-labeled probe 5′-CTTCCTCAGTGTGTTTCACTTTCTCTTCTG-3 (Integrated DNA Technologies).
SIV p27 ELISA
SIV p27 Gag was quantified by ELISA (ZeptoMetrix 0801169) per manufacturer's instructions.
miRNA profiling by custom TaqMan OpenArray Panel
A custom 112-assay TaqMan OpenArray MicroRNA panel (Thermo Fisher 4471121) was designed with miRNA assays chosen based on previous investigations of infectious and inflammatory diseases and identity of human and Macaca mulatta (mml-) miRNAs. Stem-loop primer reverse transcription and preamplification were done with manufacturer's reagents as described [18], but with 16 cycles of preamplification. qPCR was performed by QuantStudio 12K. Data were collected using SDS software, and quantification cycle (Cq) values were extracted with EXPRESSION SUITE v1.0.4 (Thermo Fisher Scientific). miRNAs with amplification score more than 1.1 and detected in more than 90% of samples were included. Cq values were normalized by quantiles.
Individual quantitative PCR assays
Individual qPCR assays (Thermo Fisher 4366596, 4440038) were performed as described [18] for U6 snRNA (Assay ID 001973), miRs-181a (000480), 342–3p (002260), 29a (0002112), 16 (000391), 192-5p (000491), 193b (002367),126 (000450), 21 (000397), and let-7b (002619). Cq values were adjusted to the mean Cq of cel-miR-39 spike-in.
Statistical analysis
Statistical significance of differences in extracellular vesicle/particle concentration, particle/protein ratio, and miRNA level between two groups were assessed by two-tailed Student's t-test for overall ranking. Correlations between average extracellular vesicle/particle count and viral RNA were evaluated by Pearson's correlation coefficient (r). Receiver operating characteristic (ROC) analyses were done in SPSS Statistics (bi-negative exponential model).
Results
Plasma size exclusion chromatography fractionation
Macaque plasma was separated by SEC into early, middle, and late pooled fractions (F7–9, F10–12, and F13–15) and characterized per MISEV recommendations [32]. Despite similar particle concentrations (Figure S1A), F7–9 had significantly less protein versus other fractions, consistent with relatively pure EVs. Transmission electron microscopy (TEM) showed cup-shaped oval/round particles in F7–9 but non-extracellular vesicle aggregates in others (Figure S1B). Western blot for extracellular vesicle-enriched membrane markers CD63 and CD8; argonaute protein 2 (AGO2, which may be present at low levels in extracellular vesicles but is mostly outside extracellular vesicles in plasma); cellular markers GM130 and calnexin; and albumin and lipoproteins ApoB100, ApoA1, and ApoC1 (Figure S1C and D). We thus defined F7–9 as extracellular vesicles, and F10–12 and F13–15 as proteins. Individual qPCR assays confirmed the abundance of several miRNAs, including miRs-126, 21, and 16, in non-extracellular vesicle fractions such as low-density lipoprotein particles (LDL), high-density lipoprotein particles (HDL), and an albumin-rich protein fractions (ALB) (Figure S1E).
Particle counts and sizes in uninfected, infected, and rebound groups
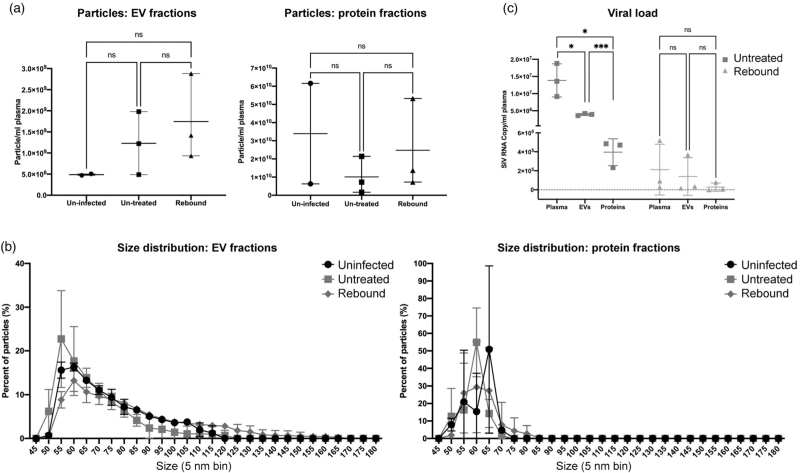
Particle counts and size profiles of extracellular vesicle and protein fractions were characterized from a small number of uninfected (n = 2), infected and untreated (44–49 dpi, n = 3), and rebound (infected/ART treatment interrupted) (Supplemental Table 1) subjects. Five of six infected samples from untreated and rebound groups had greater particle counts than uninfected in extracellular vesicle (though not statistically different due to the small sample size used) but not protein fractions (Fig. 1a). The extracellular vesicle fractions of infected, untreated animals had smaller average particle sizes, but following treatment interruption, there were more extracellular vesicles with diameter more than 120 nm (Fig. 1b). As the majority of particles had diameters less than 100 nm, intact virions (100–130 nm) could not fully explain these differences.
Fig. 1.
Characterization of plasma extracellular vesicle and protein-enriched fractions from SIV-uninfected and infected nonhuman primate models.
(a) Particle concentrations of EV (left) and protein (right) enriched fractions of uninfected, untreated, and rebound animals were measured by HSFCM. Particle concentration for each group was normalized by plasma input (per 1 ml). (b) Size distributions of EV (left) and protein (right) fractions were measured by HSFCM and calculated as particles in a specific size bin versus total detected particles in each sample (percentage). (c) Viral load (GAG RNA qPCR) as copy number per ml plasma for unfractionated plasma, EVs, and proteins. Data are mean ± SD. ns, no significant difference (P > 0.05), ∗P ≤ 0.05, ∗∗∗P ≤ 0.001 by two-tailed Welch's t-test.
Distribution of an simian immunodeficiency virus gag RNA amplicon
Approximately one-third of total plasma viral gag RNA signal was recovered in extracellular vesicle fractions (Fig. 1c): one order of magnitude greater than recovery from protein fractions. During rebound, most plasma gag RNA (mean 2.1 × 105 copies/ml) was recovered in extracellular vesicles (mean 1.4 × 105 copies/ml): two orders of magnitude greater than recovery from protein (mean 2.81 × 103 copies/ml).
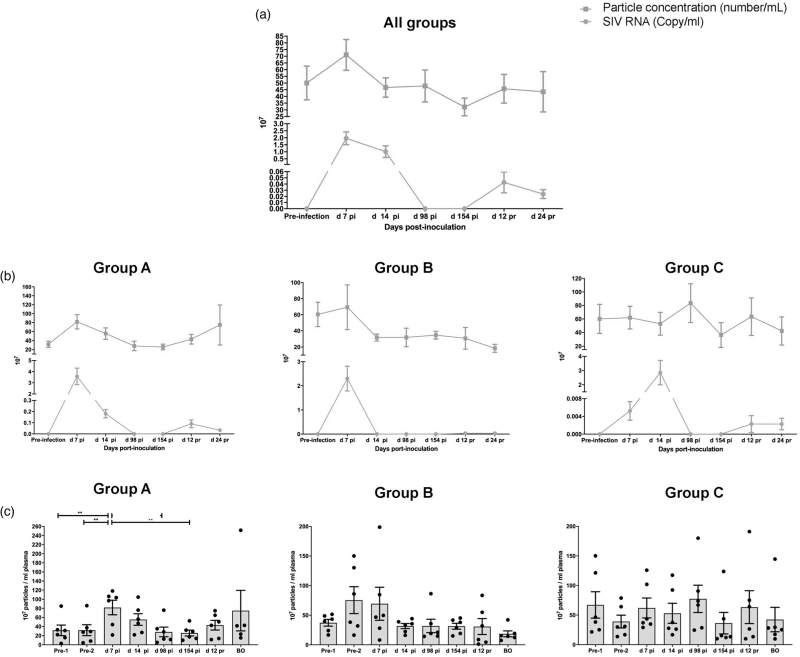
Longitudinal particle and viral RNA concentrations in three HIV/simian immunodeficiency virus models
We next examined species, SIV strain, and ART regimens with three cohorts of NHP (n = 6 each, Supplemental Table 2; viral load, Figure S2). Group A was pigtailed macaques receiving ART at 12 dpi. For Group B, ART was augmented with CCR5 inhibitor Maraviroc. Group C was rhesus macaques treated with ART at 14 dpi. Combining all participants, average particle concentration in extracellular vesicle fractions was positively correlated with average plasma SIV RNA throughout infection phases (R = 0.8916, P < 0.05; Fig. 2a). However, the correlation of particle concentration and viral RNA was significant only in Group A (R = 0.88, P < 0.05; Fig. 2b), nearing nominal statistical significance in Group B (R = 0.71, P = 0.07; Fig. 2b) but not in Group C (R = -0.12, P = 0.79; Fig. 2b; see also Fig. 2c for individual particle counts). Average viral loads at 7dpi were highest for Group A, followed by B and C (Fig. 2b). Virions, which comprise only a small percentage of total particles, do not explain these differences. Size distribution in extracellular vesicle fractions was similar between groups and time points, with the majority of detected particles less than 100 nm in all groups (Figure S3).
Fig. 2.
Plasma particle concentration and viral load in longitudinal samples from nonhuman primate (NHP) models.
Plasma EV particle concentration measured by HSFCM and SIV GAG RNA copy number (qPCR) of 18 NHPs (a) and NHPs separated by groups based on species and treatment regimen (b). Particle concentration and viral RNA copy number for each group was normalized by plasma input (per 1 ml). (c) Particle concentrations of EV-enriched fractions at different infection phases were measured by HSFCM. Particle concentration for each group was normalized by plasma input (per 1 ml). Data are mean ± SD. ∗P ≤ 0.05, ∗∗P ≤ 0.01, ∗∗∗P ≤ 0.001, ∗∗∗∗P ≤ 0.0001 by two-tailed Welch's t-test.
Plasma extracellular vesicle sRNA profiles during simian immunodeficiency vurus infection
Because of the infection-associated differences in particle counts in the extracellular vesicle fractions, and as our previous work focused on total plasma miRNAs, we next measured the abundance of extracellular vesicle associated small RNAs (sRNAs, mostly miRNAs) by custom microarray. Fifty-six features satisfied inclusion criteria. Significantly differentially abundant sRNAs with fold difference in abundance of more than 1.5 in acute infection, latent infection (ART-suppressed), and rebound (after treatment interruption) vs. preinfection are shown in Figure S4A. Most sRNA differences were found in Group A, with the largest acute and rebound viral loads. Similarly, more sRNAs were dysregulated during acute infection than in other phases (Figure S4A). Several sRNAs were consistently dysregulated across two or more groups. During acute infection, these included miR-29a and miR-145 (Groups A,B) and miR-342–3p and U6 (all groups). During ART treatment, there were no consistent sRNA changes compared with preinfection. During viral rebound, miR-192 (A,B) and miR-146b and miR-342–3p (A,C) were partly consistent. Thus, the only consistent changes across groups were of miR-342–3p and U6 during acute infection.
Extracellular vesicle miRNA dysregulation: individual quantitative PCR validation
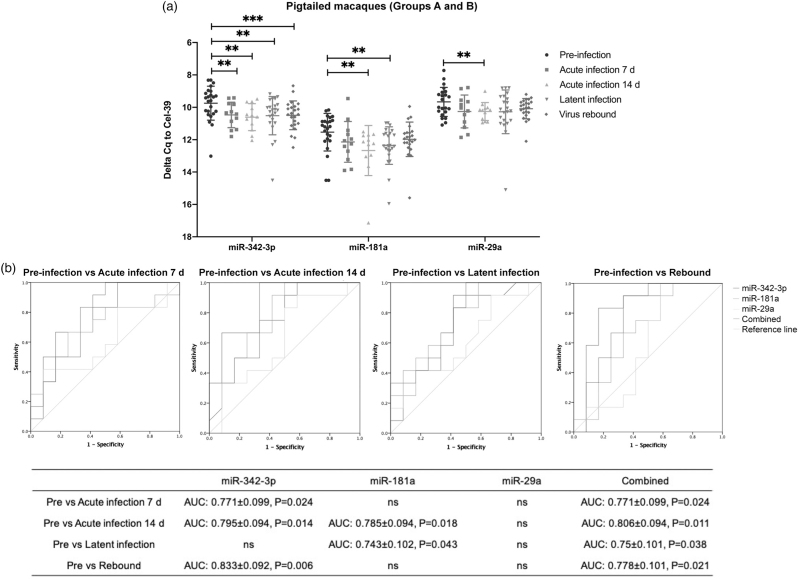
Individual qPCR assays confirmed downregulation of miRs-342–3p, 181a, and 29a after SIV infection in pigtails (A,B, Supplemental Table 2) (Fig. 3a). miRs-342–3p and 181a were not only dysregulated in acute infection, but also remained downregulated during latent and rebound phases. To assess association with phase, receiver operating characteristic (ROC) curves were generated (Fig. 3b). miR-342–3p alone had greater area-under-the-curve (AUC, 0.83 ± 0.10) in discriminating preinfection from rebound, while a combination of three miRNAs had greater AUC in other comparisons: 0.77 ± 0.10 (acute 7 dpi), 0.81 ± 0.10 (acute 14 dpi), 0.75 ± 0.10 (latency). We also examined how these miRNAs changed dynamically in the 12 participants individually (Figure S5A). Despite large inter-participant variation, miRs-342–3p, 181a, and 29a were downregulated after SIV infection in at least half of the participants. In contrast, for rhesus macaques (Figure S5B, Group C), only miR-29a was differentially abundant between acute infection (14 dpi) and latent infection, but this could be largely attributed to one outlier (Figure S5C).
Fig. 3.
EV miRNA validation in pigtailed macaques.
(a) qPCR validation for miR-342–3p, miR-181a, and miR-29a in pigtailed macaques (n = 12) at different infection phases. Delta Cq values were normalized to the spike in cel-miR-39 control. Data are mean ± SD. ∗P ≤ 0.05, ∗∗P ≤ 0.01, ∗∗∗P ≤ 0.001, ∗∗∗∗P ≤ 0.0001 by two-tailed Welch's t-test. (b) Receiver operating characteristic (ROC) curves for the levels of three individual EV miRNAs and a combined three-miRNA panel to differentiate preinfection from acute infection (7 and 14 dpi), latency, and rebound.
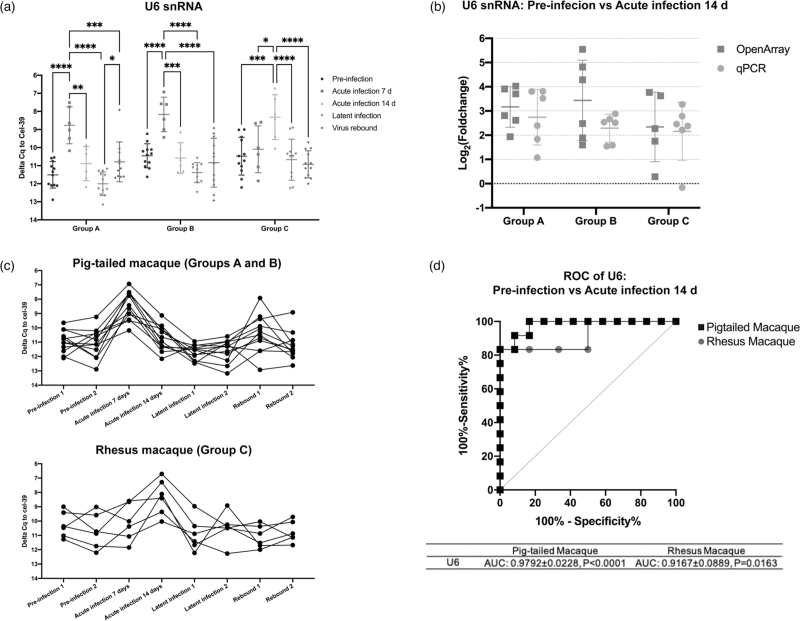
U6 snRNA correlates with plasma viral RNA peak in pigtailed and rhesus macaques
U6 is a commonly used reference in miRNA qPCR assays, especially for examination of cellular and tissue RNA. Extracellular vesicle associated U6 snRNA increased in both pigtailed and rhesus macaques during peak viral load in acute infection (7 dpi, pigtails, 14 dpi, rhesus, Fig. 4a; viral load, Figure S2), with log2 fold changes versus preinfection of 2–3 on average (Fig. 4b). Tracking the abundance of extracellular vesicle U6 snRNA across infection (Fig. 4c), changes in extracellular vesicle U6 levels were highly consistent. Association with infection was also supported by ROC curves (Fig. 4d).
Fig. 4.
EV U6 snRNA level is tightly associated with the acute infection phase.
(a) Individual qPCR validation of U6 snRNA in Groups A, B, and C at different infection phases. ∗P ≤ 0.05, ∗∗P ≤ 0.01, ∗∗∗P ≤ 0.001, ∗∗∗∗P ≤ 0.0001 by two-tailed Welch's t-test. (b) Log2 (fold change) of EV U6 snRNA in acute infection compared to preinfection from OpenArray and individual qPCR analyses. (c) U6 snRNA levels in pigtailed and rhesus macaques in longitudinal samples. (a)–(c) Delta Cq values were normalized to cel-miR-39 control. Data are presented as mean ± SD. (d) Receiver operating characteristic (ROC) curves for the levels of EV U6 to differentiate preinfection from acute infection in pigtailed and rhesus macaques.
U6 snRNA in gradient-separated extracellular vesicles
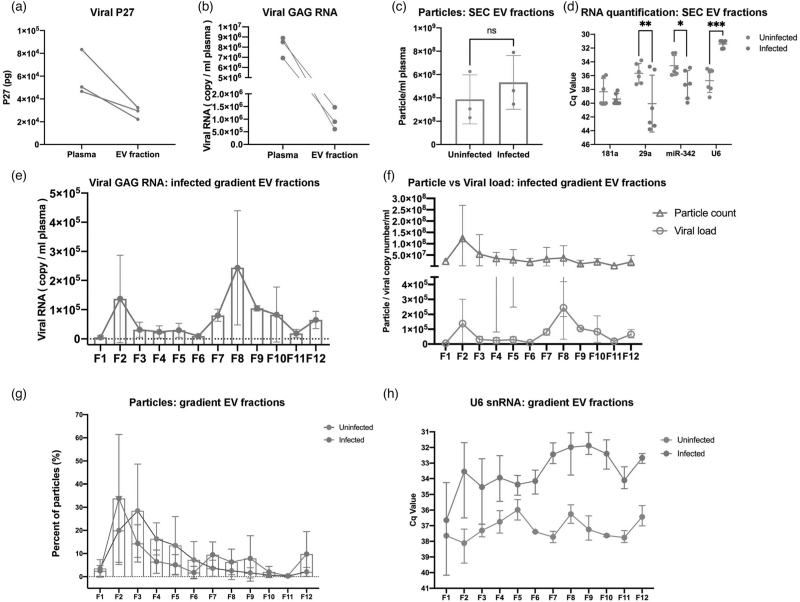
Increased U6 in extracellular vesicle fractions during infection could be due to packaging into extracellular vesicles, virions, or both. We thus further fractionated extracellular vesicles from uninfected and acute infected samples (Supplemental Table 3) into lighter extracellular vesicle fractions and denser virus populations using differential gradient ultracentrifugation with iodixanol. For acute infected samples, SIV p27 Gag protein (Fig. 5a) and RNA (Fig. 5b) were determined for input plasma and extracellular vesicle enriched SEC fractions. Consistent with the findings in Fig. 1, not all viral protein and RNA was recovered in the extracellular vesicle fractions, likely because some material is lost to the column matrix. Also as before, total particles by NFCM tended to be greater in infection (Fig. 5c). qPCR confirmed that miRs-342–3p and 29a were less abundant, while U6 snRNA was more abundant, in acute infection samples, although miR-181a abundance was similar (Fig. 5d).
Fig. 5.
EV and EV small RNA characterization in iodixanol density fractions.
SIV P27 GAG protein (a) measured by ELISA and GAG RNA and (b) by qPCR in raw plasma and EV-enriched SEC fraction from SIV acutely infected pigtailed macaque plasma (n = 3). (c) Particle concentrations of EV-enriched SEC fractions of uninfected (n = 3) and acutely infected (n = 3) pigtailed macaques as measured by HSFCM. Particle concentration for each group was normalized by plasma input (per 1 ml). ns, no significant difference (P > 0.05) by two-tailed t-test. (d) qPCR validation for miRNAs and U6 snRNA in EV-enriched SEC fractions from uninfected and acutely infected plasma. ∗P ≤ 0.05, ∗∗P ≤ 0.01, ∗∗∗P ≤ 0.001, ∗∗∗∗P ≤ 0.0001 by two-tailed t-test. (e) GAG RNA level detected by qPCR in 12 EV fractions separated by iodixanol density gradient in acutely infected plasma (n = 3). (f) EV particle concentration and SIV GAG RNA copy number were plotted for 12 EV iodixanol fractions of acutely infected plasma (n = 3). Particle concentration and viral RNA copy number for each group was normalized by plasma input (per 1 ml). (g) Particle number distribution of 12 iodixanol fractions from uninfected (n = 3) and acutely infected plasma (n = 3). Particle concentration for each fraction was measured by HSFCM and calculated as particles in each fraction versus total particles recovered from 12 fractions (percentage). (h) The level of U6 in 12 iodixanol fractions from uninfected (n = 3) and acute infected plasma (n = 3). (a)–(h) Data are mean ± SD.
Following gradient separation, the density distribution of 12 collected fractions was similar for uninfected and acute infected samples, ranging from 0.961 to 1.187 g/ml (Figure S6A). Particles with extracellular vesicle morphology were found by TEM in all fractions, although it was difficult to identify virions (Figure S6B). Transmembrane protein CD9 was detected in F2–9 (acute infected), and F2–4 (uninfected; Figure S6C). The pattern of CD9 distribution for the acute infected sample suggests successful separation of a light extracellular vesicle fraction (∼F2–4) and a heavy extracellular vesicle and/or virion fraction (∼F7–9). Gag RNA was detected in most fractions with a peak around F8, consistent with presence of virions, while Gag protein was below the limit of detection (Fig. 5e and data not shown). There was no significant correlation between particle concentration and viral RNA (R = 0.3501, P = 0.2645; Fig. 5f). Particle recovery was greatest in F2 and F7-F9 (acute infected), but in F3 for uninfected, with few particles in F7–9 (Fig. 5g). U6 snRNA was more abundant in all infected fractions except for F1, with the highest average abundance in the heavier F7–10 (Fig. 5h). miRs-342–3p and 29a, while detected, were highly variable (Figure S6B).
Discussion
As the gold standard to measure the latent HIV reservoir, quantitative virus outgrowth assays (qVOAs) are laborious, expensive, and require a large amount of blood samples [36]. We posited that the presence of latent infection could potentially be verified using extracellular vesicles released from live cells, which are easily accessible in blood and whose contents, including host RNAs, may reflect infection status. Here, for the first time, we examined extracellular vesicle concentration and small RNA contents in longitudinal samples from several SIV models, finding that extracellular vesicle concentration, miRNA, and U6 snRNA levels are linked to SIV infection. Several extracellular vesicle miRNAs that were differentially regulated during acute-phase infection remained downregulated even during the latency phase, when viral RNA was undetectable. These miRNAs could potentially signal the presence of a latent infection and could conceivably have regulatory roles. Interestingly, density separation of extracellular vesicle subtypes revealed strong enrichment of U6 snRNA in all extracellular vesicle subtypes in acutely infected vs. uninfected plasma.
To be sure, overlapping contents and physical properties make EVs and virions (or “host EVs” and “viral EVs”) nontrivial to separate [37–40]. Although density gradients are a standard method for virion/extracellular vesicle separation [41–45], most methods have been optimized for cell culture medium [42–44] or large quantities of virions spiked into plasma [45]. However, our extracellular vesicle and extracellular vesicle exRNA results are likely not skewed by the presence of virions. First, retrovirions are less abundant than extracellular vesicles in biofluids, even in uncontrolled viral replication. We observed a significantly lower viral GAG RNA copy number than nanoparticle number in extracellular vesicle enriched SEC fractions, even during viral peak. Both GAG RNA and GAG antigen levels were significantly lower in extracellular vesicle enriched SEC fractions vs. unfractioned plasma. In addition, gradient-separated extracellular vesicles did not contain detectable GAG antigen but did have GAG RNAs. Second, GAG RNA was also detected in protein fractions, with particles of much smaller diameter than virions. Finally, viral RNA fragments could also be incorporated into host extracellular vesicles [46,47]. Thus, increased particles in infected plasmas are not explained solely by retrovirions.
Previous studies reported increased plasma extracellular vesicle concentration in HIV-infected patients compared with uninfected controls [23,24,48], albeit with no differences between ART-naive and ART-suppressed patients [23]. In some cases, extracellular vesicles were quantitated by acetylcholinesterase (AChE) [48], which may not be specific for EVs [49]. Moreover, separating extracellular vesicles from, for example, lipoproteins is difficult using ultracentrifugation or polymer precipitation [32]. Here, we used SEC, a good option for obtaining relatively pure extracellular vesicles [50,51]. We also further separated extracellular vesicle subtypes and virions by gradient, revealing extracellular vesicle and extracellular vesicle small RNA distributions. Although we cannot rule out all co-isolates, and thus refer to “EV-enriched fractions,” our results support more extracellular vesicle production during SIV infection. Even so, changes appeared to vary with species and individual subject, and more work is needed to understand mechanisms behind this and previous findings on release of extracellular vesicles after retroviral infection [52–54].
Where do our miRNA findings fit into the existing literature? We previously reported that total plasma miRNA profiles change during acute SIV infection [18] and that extracellular vesicle borne miRNA miR-186–5p is downregulated in cervicovaginal lavage (CVL) and regulates HIV replication in macrophages [55]. Here, we identified several miRNAs that were dysregulated during latent and rebound infections, but only very few compared with the acute infection phase, consistent with a study comparing productive and latent infection in CD4+ T-cell models [16]. Some identified miRNAs were previously reported in HIV infection or inflammation, such as miR-29a [56–59] and miR-181a [16,60,61], while miR-342–3p was not. However, unlike particle concentration, which varied with viral load, extracellular vesicle miRNAs remained downregulated after infection even when viral RNA was undetectable. Of note, host miRNA changes are probably not specific to HIV infection. For example, miR-29a, identified by us and other groups, was previously observed to be downregulated in tuberculosis [62] and hepatitis C virus (HCV) [57] infection. Extracellular vesicle miRNA may thus be useful to indicate persistent infection generally but may not be a good indicator for specific disease monitoring.
Our findings on U6 were unexpected and potentially informative. U6 snRNA is one of the most highly conserved RNAs across species and is thus commonly used as an internal control gene in, for example, miRNA qPCR assays [63–66] despite some individual and disease-related differences [67–71]. Although a few studies revealed upregulation of extracellular vesicle U6 in cancer and smoking [72–74], such changes have not to our knowledge been reported in infectious diseases. Our data suggest that U6 is not an ideal reference RNA, at least in retroviral infection. Interestingly, U6 snRNA is also one of the host cellular RNAs reported to be specifically packaged into retroviral particles [75–77], for example, in murine leukemia virus (MLV) infection [75,78]. Retroviral packaging of U6 may be independent of viral full-length RNA transcripts [77,79] but affected by the nucleocapsid domain of the Gag polyprotein [79]. However, standard virus purifications [76,77,79] also co-isolate extracellular vesicles. How U6 is packaged into extracellular vesicles is unclear, as is the possible function of this RNA in the cell or in the extracellular vesicle during retroviral infection.
In summary, our rigorous EV separation and characterization strategy showed that plasma extracellular vesicles and several extracellular vesicle sRNAs change consistently during the course of SIV infection across several related models. Our findings raise new questions about the distribution of host RNAs released in ‘host’ EVs compared with ‘hijacked’ extracellular vesicles (retrovirions) and sound a cautionary note about the use of U6 as a reference RNA, at least in retroviral infection. We are now applying these methods to studies of human plasma to understand if extracellular vesicle RNAs will provide value in monitoring HIV latent infection.
Acknowledgements
The authors thank members of the Witwer Laboratory for discussions and support. They are particularly grateful to members of the Retrovirus Laboratory for access to samples from the animal models and for helpful suggestions. They also thank Angela M. Zivkovic and Joanne K. Agus, University of California, Davis, for providing non-extracellular vesicle plasma fractions. Electron microscopy images were acquired in the Johns Hopkins University School of Medicine Institute for Basic Biomedical Sciences Microscope Facility. The authors would like to thank ViiV Pharmaceuticals for their generous donation of dolutegravir and maraviroc, and Gilead Pharmaceuticals for their generous donations of tenofovir and emtricitabine. This work was supported in part by the U.S. National Institutes of Health, National Institute on Drug Abuse (NIDA, DA040385, and DA047807 to K.W.W.), and by two pilot grants awarded to Y.H. through the Johns Hopkins NIMH Center (supported by MH075673) and the Johns Hopkins University Center for AIDS Research (supported by AI094189). The Witwer laboratory is also supported in part by NCI/Common Fund CA241694, NIAID AI144997, NIMH MH118164, and the Richman Family Precision Medicine Center of Excellence in Alzheimer's Disease. Samples used in this study were derived in part from research supported by U42OD013117 (Johns Hopkins, NIH-supported pigtailed macaque breeding colony), NINDS NS089482 (to Joseph L. Mankowski), and NIMH MH070306 (to Janice E. Clements).
Conflicts of interest
There are no conflicts of interest.
Supplementary Material
Supplementary Material
Footnotes
Supplemental digital content is available for this article.
References
- 1.Tedaldi EM, Minniti NL, Fischer T. HIV-associated neurocognitive disorders: the relationship of Hiv infection with physical and social comorbidities. Biomed Res Int 2015; 2015:641913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smail RC, Brew BJ. HIV-associated neurocognitive disorder. In: Aminoff MJ, Boller F, Swaab R (editors), Handbook of clinical neurology. Elsevier B.V; 2018. pp. 75–97. [DOI] [PubMed] [Google Scholar]
- 3.Marsh Sung J, M. Margolis D. HIV persistence on antiretroviral therapy and barriers to a cure. In: Zhang L, Lewin SR (editors), Advances in experimental medicine and biology. New York: Springer New York LLC; 2018. pp. 165–185. [DOI] [PubMed] [Google Scholar]
- 4.Siliciano RF, Greene WC. HIV latency. Cold Spring Harb Perspect Med 2011; 1:a007096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beck SE, Kelly KM, Queen SE, Adams RJ, Zink MC, Tarwater PM, et al. Macaque species susceptibility to simian immunodeficiency virus: increased incidence of SIV central nervous system disease in pigtailed macaques versus rhesus macaques. J Neurovirol 2015; 21:148–158. [DOI] [PubMed] [Google Scholar]
- 6.Batten CJ, De Rose R, Wilson KM, Agy MB, Chea S, Stratov I, et al. Comparative evaluation of simian, simian-human, and human immunodeficiency virus infections in the pigtail macaque (Macaca nemestrina) model. AIDS Res Hum Retroviruses 2006; 22:580–588. [DOI] [PubMed] [Google Scholar]
- 7.Zink MC, Amedee AM, Mankowski JL, Craig L, Didier P, Carter DL, et al. Pathogenesis of SIV encephalitis: selection and replication of neurovirulent SIV. Am J Pathol 1997; 151:793–803. [PMC free article] [PubMed] [Google Scholar]
- 8.Stenovec M, Lasič E, Dominkuš PP, Bobnar ST, Zorec R, Lenassi M, et al. Slow release of HIV-1 protein Nef from vesicle-like structures is inhibited by cytosolic calcium elevation in single human microglia. Mol Neurobiol 2019; 56:102–118. [DOI] [PubMed] [Google Scholar]
- 9.Booth AM, Fang Y, Fallon JK, Yang JM, Hildreth JEK, Gould SJ, et al. Exosomes and HIV Gag bud from endosome-like domains of the T cell plasma membrane. J Cell Biol 2006; 172:923–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Madison MN, Okeoma CM. Exosomes: implications in HIV-1 pathogenesis. Viruses 2015; 7:4093–4118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Turchinovich A, Weiz L, Langheinz A, Burwinkel B. Characterization of extracellular circulating microRNA. Nucleic Acids Res 2011; 39:7223–7233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 2007; 9:654–659. [DOI] [PubMed] [Google Scholar]
- 13.Huang Y, Cheng L, Turchinovich A, Mahairaki V, Troncoso JC, Pletniková O, et al. Influence of species and processing parameters on recovery and content of brain tissue-derived extracellular vesicles. J Extracell Vesicles 2020; 9:1785746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nolte-’t Hoen EN, Buermans HP, Waasdorp M, Stoorvogel W, Wauben MH, t Hoen PA. Deep sequencing of RNA from immune cell-derived vesicles uncovers the selective incorporation of small noncoding RNA biotypes with potential regulatory functions. Nucleic Acids Res 2012; 40:9272–9285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tosar JP, Gambaro F, Sanguinetti J, Bonilla B, Witwer KW, Cayota A. Assessment of small RNA sorting into different extracellular fractions revealed by high-throughput sequencing of breast cell lines. Nucleic Acids Res 2015; 43:5601–5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu X, Yang J, Wu H, Yang Z, Jin C, Wang J, et al. High-throughput sequencing identifies HIV-1-replication- and latency-related miRNAs in CD4+ T cell lines. Arch Virol 2017; 162:1933–1942. [DOI] [PubMed] [Google Scholar]
- 17.Sun G, Li H, Wu X, Covarrubias M, Scherer L, Meinking K, et al. Interplay between HIV-1 infection and host microRNAs. Nucleic Acids Res 2012; 40:2181–2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Witwer KW, Sarbanes SL, Liu J, Clements JE. A plasma microRNA signature of acute lentiviral infection: biomarkers of central nervous system disease. AIDS 2011; 25:2057–2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cárdenas-Bedoya J, Marquez-Pedroza J, Morán-Moguel MC, Escoto-Delgadillo M, Vázquez-Valls E, González-Enríquez GV, et al. Microrna-296-5p is differentially expressed in individuals with and without hiv-1 infection. Genet Mol Biol 2020; 43:1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Biswas S, Haleyurgirisetty M, Lee S, Hewlett I, Devadas K. Development and validation of plasma miRNA biomarker signature panel for the detection of early HIV-1 infection. EBioMedicine 2019; 43:307–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Asahchop EL, Akinwumi SM, Branton WG, Fujiwara E, Gill MJ, Power C. Plasma microRNA profiling predicts HIV-associated neurocognitive disorder. AIDS 2016; 30:2021–2031. [DOI] [PubMed] [Google Scholar]
- 22.O’Meara T, Kong Y, Chiarella J, Price RW, Chaudhury R, Liu X, et al. Exosomal MicroRNAs associate with neuropsychological performance in individuals with HIV infection on antiretroviral therapy. J Acquir Immune Defic Syndr 2019; 82:514–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hubert A, Subra C, Jenabian MA, Labrecque PFT, Tremblay C, Laffont B, et al. Elevated abundance, size, and MicroRNA content of plasma extracellular vesicles in viremic HIV-1+ patients: correlations with known markers of disease progression. J Acquir Immune Defic Syndr 2015; 70:219–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chettimada S, Lorenz DR, Misra V, Wolinsky SM, Gabuzda D. Small RNA sequencing of extracellular vesicles identifies circulating miRNAs related to inflammation and oxidative stress in HIV patients. BMC Immunol 2020; 21:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ruiz-de-León MJ, Jiménez-Sousa MA, Moreno S, García M, Gutiérrez-Rivas M, León A, et al. Lower expression of plasma-derived exosome miR-21 levels in HIV-1 elite controllers with decreasing CD4 T cell count. J Microbiol Immunol Infect 2019; 52:667–671. [DOI] [PubMed] [Google Scholar]
- 26.Marques de Menezes EG, Jang K, George AF, Nyegaard M, Neidleman J, Inglis HC, et al. Seminal plasma-derived extracellular-vesicle fractions from HIV-infected men exhibit unique MicroRNA signatures and induce a proinflammatory response in cells isolated from the female reproductive tract. J Virol 2020; 94:e00525–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A 2011; 108:5003–5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beck SE, Queen SE, Metcalf Pate KA, Mangus LM, Abreu CM, Gama L, et al. An SIV/macaque model targeted to study HIV-associated neurocognitive disorders. J Neurovirol 2018; 24:204–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zink MC, Suryanarayana K, Mankowski JL, Shen A, Piatak M, Jr, Spelman JP, et al. High viral load in the cerebrospinal fluid and brain correlates with severity of simian immunodeficiency virus encephalitis. J Virol 1999; 73:10480–10488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abreu CM, Veenhuis RT, Avalos CR, Graham S, Parrilla DR, Ferreira EA, et al. Myeloid and CD4 T cells comprise the latent reservoir in antiretroviral therapy-suppressed SIVmac251-infected macaques. MBio 2019; 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Monceaux V, Viollet L, Petit F, Cumont MC, Kaufmann GR, Aubertin AM, et al. CD4+ CCR5+ T-cell dynamics during simian immunodeficiency virus infection of Chinese rhesus macaques. J Virol 2007; 81:13865–13875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Théry C, Witwer KW, Aikawa E, Alcaraz MJ, Anderson JD, Andriantsitohaina R, et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles 2018; 7:1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muth DC, Powell BH, Zhao Z, Witwer KW. miRNAs in platelet-poor blood plasma and purified RNA are highly stable: a confirmatory study. BMC Res Notes 2018; 11:273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shen A, Zink MC, Mankowski JL, Chadwick K, Margolick JB, Carruth LM, et al. Resting CD4 + T lymphocytes but not thymocytes provide a latent viral reservoir in a simian immunodeficiency virus- Macaca nemestrina model of human immunodeficiency virus type 1-infected patients on highly active antiretroviral therapy. J Virol 2003; 77:4938–4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abreu CM, Veenhuis RT, Avalos CR, Graham S, Queen SE, Shirk EN, et al. Infectious virus persists in CD4 + T cells and macrophages in antiretroviral therapy-suppressed simian immunodeficiency virus-infected macaques. J Virol 2019; 93:e00065–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stone M, Rosenbloom DIS, Bacchetti P, Deng X, Dimapasoc M, Keating S, et al. Assessing the suitability of next-generation viral outgrowth assays to measure human immunodeficiency virus 1 latent reservoir size. J Infect Dis 2020; 224:1209–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hoen EN, Cremer T, Gallo RC, Margolis LB. Extracellular vesicles and viruses: are they close relatives?. Proc Natl Acad Sci U S A 2016; 113:9155–9161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McNamara RP, Dittmer DP. Modern techniques for the isolation of extracellular vesicles and viruses. J Neuroimmune Pharmacol 2020; 15:459–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ott DE. Cellular proteins detected in HIV-1. Rev Med Virol 2008; 18:159–175. [DOI] [PubMed] [Google Scholar]
- 40.Orentas RJ, Hildreth JEK. Association of host cell surface adhesion receptors and other membrane proteins with HIV and SIV. AIDS Res Hum Retroviruses 1993; 9:1157–1165. [DOI] [PubMed] [Google Scholar]
- 41.Konadu KA, Chu J, Huang MB, Amancha PK, Armstrong W, Powell MD, et al. Association of cytokines with exosomes in the plasma of HIV-1–seropositive individuals. J Infect Dis 2015; 211:1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.R C, J D, D B, AM T, C G. Discrimination between exosomes and HIV-1: purification of both vesicles from cell-free supernatants. J Immunol Methods 2008; 338:21–30. [DOI] [PubMed] [Google Scholar]
- 43.Barclay RA, Khatkar P, Mensah G, DeMarino C, Chu JSC, Lepene B, et al. An omics approach to extracellular vesicles from HIV-1 infected cells. Cells 2019; 8:787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martin-Jaular L, Nevo N, Schessner JP, Tkach M, Jouve M, Dingli F, et al. Unbiased proteomic profiling of host cell extracellular vesicle composition and dynamics upon HIV-1 infection. EMBO J 2021; 40:e105492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vaillancourt M, Hubert A, Subra C, Boucher J, Bazié WW, Vitry J, et al. Velocity gradient separation reveals a new extracellular vesicle population enriched in miR-155 and mitochondrial DNA. Pathogens (Basel, Switzerland) 2021; 10:526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Demarino C, Pleet ML, Cowen M, Barclay RA, Akpamagbo Y, Erickson J, et al. Antiretroviral drugs alter the content of extracellular vesicles from HIV-1-infected cells. Sci Rep 2018; 8:1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barclay RA, Schwab A, Demarino C, Akpamagbo Y, Lepene B, Kassaye S, et al. Exosomes from uninfected cells activate transcription of latent HIV-1. J Biol Chem 2017; 292:11682–11701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bazié WW, Boucher J, Vitry J, Goyer B, Routy JP, Tremblay C, et al. Plasma extracellular vesicle subtypes may be useful as potential biomarkers of immune activation in people with HIV. Pathog Immun 2021; 6:1–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liao Z, Jaular LM, Soueidi E, Jouve M, Muth DC, Schøyen TH, et al. Acetylcholinesterase is not a generic marker of extracellular vesicles. J Extracell Vesicles 2019; 8:1628592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Böing AN, van der Pol E, Grootemaat AE, Coumans FAW, Sturk A, Nieuwland R. Single-step isolation of extracellular vesicles by size-exclusion chromatography. J Extracell Vesicles 2014; 3.doi10.3402/jev.v3.23430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Karimi N, Cvjetkovic A, Jang SC, Crescitelli R, Hosseinpour Feizi MA, Nieuwland R, et al. Detailed analysis of the plasma extracellular vesicle proteome after separation from lipoproteins. Cell Mol Life Sci 2018; 75:2873–2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lenassi M, Cagney G, Liao M, Vaupotič T, Bartholomeeusen K, Cheng Y, et al. HIV Nef is secreted in exosomes and triggers apoptosis in bystander CD4+ T cells. Traffic 2010; 11:110–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Subra C. Dendritic cells pulsed with HIV-1 release exosomes that promote apoptosis in Cd4+ T lymphocytes. J Clin Cell Immunol 2013; 04:1. [Google Scholar]
- 54.Lee JH, Schierer S, Blume K, Dindorf J, Wittki S, Xiang W, et al. HIV-Nef and ADAM17-containing plasma extracellular vesicles induce and correlate with immune pathogenesis in chronic HIV infection. EBioMedicine 2016; 6:103–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhao Z, Muth DC, Mulka K, Liao Z, Powell BH, Hancock GV, et al. miRNA profiling of primate cervicovaginal lavage and extracellular vesicles reveals miR-186-5p as a potential antiretroviral factor in macrophages. FEBS Open Bio 2020; 10:2021–2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Monteleone K, Selvaggi C, Cacciotti G, Falasca F, Mezzaroma I, D’Ettorre G, et al. MicroRNA-29 family expression and its relation to antiviral immune response and viro-immunological markers in HIV-1-infected patients. BMC Infect Dis 2015; 15:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Moghoofei M, Najafipour S, Mostafaei S, Tavakoli A, Bokharaei-Salim F, Ghorbani S, et al. MicroRNAs profiling in HIV, HCV, and HIV/HCV co-infected patients. Curr HIV Res 2021; 19:27–34. [DOI] [PubMed] [Google Scholar]
- 58.Hu G, Yao H, Chaudhuri AD, Duan M, Yelamanchili SV, Wen H, et al. Exosome-mediated shuttling of microRNA-29 regulates HIV Tat and morphine-mediated neuronal dysfunction. Cell Death Dis 2012; 3:e381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu J, Sisk JM, Gama L, Clements JE, Witwer KW. Tristetraprolin expression and microRNA-mediated regulation during simian immunodeficiency virus infection of the central nervous system. Mol Brain 2013; 6:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhu J, Yao K, Guo J, Shi H, Ma L, Wang Q, et al. miR-181a and miR-150 regulate dendritic cell immune inflammatory responses and cardiomyocyte apoptosis via targeting JAK1-STAT1/c-Fos pathway. J Cell Mol Med 2017; 21:2884–2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hutchison ER, Kawamoto EM, Taub DD, Lal A, Abdelmohsen K, Zhang Y, et al. Evidence for miR-181 involvement in neuroinflammatory responses of astrocytes. Glia 2013; 61:1018–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ndzi EN, Nkenfou CN, Mekue LM, Zentilin L, Tamgue O, Pefura EWY, et al. MicroRNA hsa-miR-29a-3p is a plasma biomarker for the differential diagnosis and monitoring of tuberculosis. Tuberculosis 2019; 114:69–76. [DOI] [PubMed] [Google Scholar]
- 63.Oh JG, Lee P, Gordon RE, Sahoo S, Kho C, Jeong D. Analysis of extracellular vesicle miRNA profiles in heart failure. J Cell Mol Med 2020; 24:7214–7227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ng EKO, Chong WWS, Jin H, Lam EKY, Shin VY, Yu J, et al. Differential expression of microRNAs in plasma of patients with colorectal cancer: a potential marker for colorectal cancer screening. Gut 2009; 58:1375–1381. [DOI] [PubMed] [Google Scholar]
- 65.Wang H, Zhang P, Chen W, Feng D, Jia Y, Xie L. Serum microRNA signatures identified by Solexa sequencing predict sepsis patients’ mortality: a prospective observational study. PLoS One 2012; 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ji F, Yang B, Peng X, Ding H, You H, Tien P. Circulating microRNAs in hepatitis B virus-infected patients. J Viral Hepat 2011; 18:e242–e251. [DOI] [PubMed] [Google Scholar]
- 67.Ding X, Ding J, Ning J, Yi F, Chen J, Zhao D, et al. Circulating microRNA-122 as a potential biomarker for liver injury. Mol Med Rep 2012; 5:1428–1432. [DOI] [PubMed] [Google Scholar]
- 68.Qi R, Weiland M, Gao XH, Zhou L, Mi QS. Identification of endogenous normalizers for serum MicroRNAs by microarray profiling: U6 small nuclear RNA is not a reliable normalizer. Hepatology 2012; 55:1640–1642. [DOI] [PubMed] [Google Scholar]
- 69.Gouin K, Peck K, Antes T, Johnson JL, Li C, Vaturi SD, et al. A comprehensive method for identification of suitable reference genes in extracellular vesicles. J Extracell Vesicles 2017; 6:1347019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Benz F, Roderburg C, Cardenas DV, Vucur M, Gautheron J, Koch A, et al. U6 is unsuitable for normalization of serum miRNA levels in patients with sepsis or liver fibrosis. Exp Mol Med 2013; 45:e42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sohel MH. Extracellular/circulating microRNAs: release mechanisms, functions and challenges. Achiev Life Sci 2016; 10:175–186. [Google Scholar]
- 72.Chiam K, Wang T, Watson DI, Mayne GC, Irvine TS, Bright T, et al. Circulating serum exosomal miRNAs as potential biomarkers for esophageal adenocarcinoma. J Gastrointest Surg 2015; 19:1208–1215. [DOI] [PubMed] [Google Scholar]
- 73.Puigdelloses M, González-Huárriz M, García-Moure M, Martínez-Vélez N, Esparragosa Vázquez I, Bruna J, et al. RNU6-1 in circulating exosomes differentiates GBM from nonneoplastic brain lesions and PCNSL but not from brain metastases. Neuro-Oncology Adv 2020; 2:vdaa010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Badrnya S, Baumgartner R, Assinger A. Smoking alters circulating plasma microvesicle pattern and microRNA signatures. Thromb Haemost 2014; 112:128–136. [DOI] [PubMed] [Google Scholar]
- 75.Eckwahl MJ, Telesnitsky A, Wolin SL. Host RNA packaging by retroviruses: a newly synthesized story. MBio 2016; 7:e02025–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Houzet L, Paillart JC, Smagulova F, Maurel S, Morichaud Z, Marquet R, et al. HIV controls the selective packaging of genomic, spliced viral and cellular RNAs into virions through different mechanisms. Nucleic Acids Res 2007; 35:2695–2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tian C, Wang T, Zhang W, Yu XF. Virion packaging determinants and reverse transcription of SRP RNA in HIV-1 particles. Nucleic Acids Res 2007; 35:7288–7302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gwizdek C, Ossareh-Nazari B, Brownawell AM, Doglio A, Bertrand E, Macara IG, et al. Exportin-5 mediates nuclear export of minihelix-containing RNAs. J Biol Chem 2003; 278:5505–5508. [DOI] [PubMed] [Google Scholar]
- 79.Didierlaurent L, Racine PJ, Houzet L, Chamontin C, Berkhout B, Mougel M. Role of HIV-1 RNA and protein determinants for the selective packaging of spliced and unspliced viral RNA and host U6 and 7SL RNA in virus particles. Nucleic Acids Res 2011; 39:8915–8927. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.