Abstract
In cell models, changes in the ‘accessible’ pool of plasma membrane (PM) cholesterol are linked with the regulation of endoplasmic reticulum sterol synthesis and metabolism by the Aster family of nonvesicular transporters; however, the relevance of such nonvesicular transport mechanisms for lipid homeostasis in vivo has not been defined. Here we reveal two physiological contexts that generate accessible PM cholesterol and engage the Aster pathway in the liver: fasting and reverse cholesterol transport. During fasting, adipose-tissue-derived fatty acids activate hepatocyte sphingomyelinase to liberate sequestered PM cholesterol. Aster-dependent cholesterol transport during fasting facilitates cholesteryl ester formation, cholesterol movement into bile and very low-density lipoprotein production. During reverse cholesterol transport, high-density lipoprotein delivers excess cholesterol to the hepatocyte PM through scavenger receptor class B member 1. Loss of hepatic Asters impairs cholesterol movement into feces, raises plasma cholesterol levels and causes cholesterol accumulation in peripheral tissues. These results reveal fundamental mechanisms by which Aster cholesterol flux contributes to hepatic and systemic lipid homeostasis.
The PM contains up to 90% of the total cellular cholesterol1. Das and colleagues have stratified PM cholesterol into three discrete pools: an accessible pool that is available for transport to the endoplasmic reticulum (ER) to signal cellular cholesterol sufficiency; a sphingomyelin (SM)-sequestered pool that can be rendered accessible by SM hydrolysis; and an essential pool that is sequestered by phospholipids and proteins and cannot be liberated without reducing cell viability2. Much of the cellular machinery important for responding to changes in PM cholesterol resides in the ER, including the SCAP complex for sterol regulatory-element binding protein (SREBP) processing and enzymes for cholesterol modification (acyl-coA cholesterol acyltransferase and cholesterol hydroxylases)3-5. It therefore makes intuitive sense that rapid communication of changes in accessible PM cholesterol to the ER would be critical for membrane homeostasis. Indeed, adding cholesterol to the PM in cultured cells suppresses SREBP cleavage within 1 h (ref. 3). The mechanisms underlying rapid sterol movement between the PM and ER have only recently begun to be understood.
The Aster family of proteins (Aster-A, Aster-B and Aster-C; encoded by Gramd1a, Gramd1b and Gramd1c) mediate nonvesicular transport of cholesterol from the PM to the ER6. Asters are single-pass ER membrane proteins that localize to contact sites between the ER and the PM in cells loaded with high-density lipoprotein (HDL) or cholesterol–cyclodextrin complexes. Notably, it is the accessible PM cholesterol fraction that seems to be recognized specifically by Asters in mouse, hamster and human cell lines6-9. The accessible pool of cholesterol in the PM is detectable when the cholesterol concentration exceeds ~35 mol% of total PM lipids, approximately the same concentration that triggers the recruitment of Aster proteins to the PM. Loss of Aster function in cells expands the accessible PM cholesterol pool at the expense of the ER, triggering activation of the SREBP-2 pathway6-8,10.
Mice lacking Aster-B have low cholesterol ester (CE) stores in the adrenal cortex and reduced corticosteroid production due to impaired movement of HDL-derived cholesterol from the PM to the ER6; however, the importance of nonvesicular transport pathways in other contexts is unknown. In particular, the functional relevance of Aster-mediated cholesterol transport in the liver—an organ central to systemic metabolism—remains to be established. Furthermore, the physiological factors that generate accessible cholesterol in the PM and promote its removal by Asters have not been elucidated11. We hypothesized that fasting and reverse cholesterol transport (RCT) might be physiological contexts that require Aster-mediated cholesterol transport from PM to the ER in hepatocytes.
Hepatic cholesterol biosynthesis is suppressed by fasting in mammals12,13. Nevertheless, CEs accumulate in the liver during fasting to sustain very low-density lipoprotein (VLDL) output and bile acids continue to be produced from cholesterol in preparation for the next meal; however, the source of cholesterol for VLDL output and bile acid synthesis during nutrient deprivation is unclear. RCT involves the movement of cholesterol from peripheral tissues to hepatocytes via HDL for excretion into feces14. Liver HDL cholesterol uptake is mediated by scavenger receptor class B member 1 (SR-BI)15. Activation of the nuclear hormone receptor FXR promotes RCT by inducing SR-BI16; however, the pathways that move HDL cholesterol from the PM to the ER of hepatocytes downstream of SR-BI are unknown.
Here we show that nonvesicular sterol transport by Aster proteins plays key roles in both hepatocellular cholesterol redistribution during fasting and RCT. We demonstrate that hepatocyte CE production during fasting is sustained through liberation of PM cholesterol by sphingomyelinase (SMase) and subsequent Aster-dependent cholesterol transport to the ER. Expression of Aster-C, the dominant family member in hepatocytes, is coordinately regulated along with SR-BI by FXR. Loss of Aster expression in the liver impairs HDL cholesterol movement into feces, raises HDL cholesterol levels in the plasma and causes cholesterol accumulation in peripheral tissues. Our findings show that Aster proteins in the liver are required to maintain cholesterol homeostasis and underscore the importance of nonvesicular sterol transport in hepatic lipid metabolism.
Results
Asters transport accessible PM cholesterol in hepatocytes
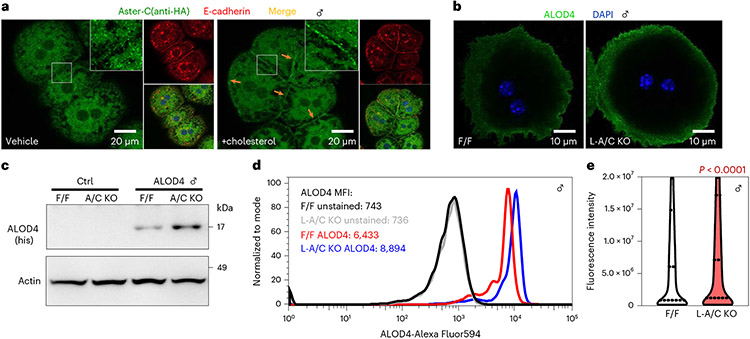
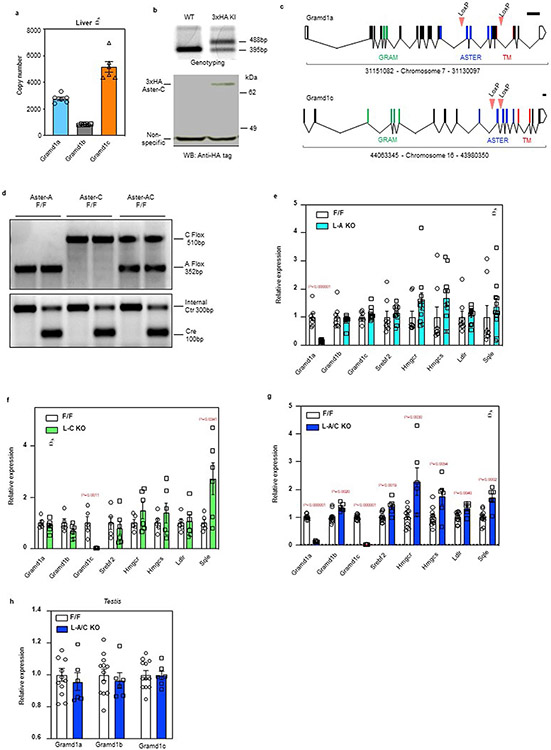
Transcripts encoding Aster-A and Aster-C are highly expressed in the liver of mice fed a standard chow diet, whereas transcripts encoding Aster-B are less abundant (Extended Data Fig. 1a). To visualize the localization of endogenous Aster-C in hepatocytes, we inserted a 3×HA tag into the Gramd1c locus of mice by CRISPR-Cas9 editing (Extended Data Fig. 1b). Immunofluorescence microscopy with an HA antibody revealed that Aster-C was distributed throughout the ER of hepatocytes at baseline and was recruited to the PM by cholesterol loading (Fig. 1a). The observation that PM cholesterol levels regulate the movement of Aster-C in hepatocytes prompted us to further explore Aster function in that cell type.
Fig. 1 ∣. Asters transport accessible PM cholesterol in hepatocytes.
a, Localization of Aster-C (green) in primary hepatocytes from 3xHA-Aster-C knock-in mice. Red, E-cadherin (PM marker). b, ALOD4 staining (green) of PM accessible cholesterol in primary hepatocytes from F/F control and L-A/C KO mice. Blue, DAPI nuclear stain. Data in a and b are representative of three independent samples. DAPI, 4,6-diamidino-2-phenylindole. c, Immunoblot analysis of ALOD4 binding in hepatocytes from b. Actin was used as a loading control. d, Flow cytometry plots of ALOD4 staining in hepatocytes from b. MFI, mean fluorescence intensity. e, Violin plots of cellular fluorescence intensity quantified in hepatocytes from b. The 25th, 50th and 75th percentiles are shown as dashed lines. P values were determined by two-sided Student’s t-test.
To study Aster function in the liver, we generated mice with hepatocyte-specific deletion of either Aster-A (L-A knockout (KO)), Aster-C (L-C KO) or both (L-A/C KO) (Extended Data Fig. 1c,d). Aster-A expression was ~70% lower in the livers of L-A KO and L-A/C KO mice (Extended Data Fig. 1e,g). The residual Aster-A expression was likely due to the presence of Aster-A in natural killer cells, B cells and endothelial cells, based on publicly available single-cell sequencing datasets17. Aster-C expression was undetectable in the livers of L-C KO and L-A/C KO mice (Extended Data Fig. 1f,g). Aster-A and -C expression was unchanged in the testes of L-A/C KO mice, consistent with liver-specific gene ablation (Extended Data Fig. 1h). The liver messenger RNA levels of SREBP-2 target genes were modestly elevated in the Aster-A and Aster-C single KO mice (Extended Data Fig. 1e,f); however, L-A/C KO mice exhibited a striking induction of SREBP-2 target genes, indicating that Asters-A and Asters-C compensate for each other in the liver when one is inactivated (Extended Data Fig. 1g). We therefore focused subsequent experiments on L-A/C KO mice due to the lack of Aster-B and the apparent redundancy of Aster-A and Aster-C in hepatocytes.
In cultured cells, a failure to transport cholesterol efficiently from the PM to the ER expands the accessible cholesterol pool8-10. We assessed the accessible cholesterol pool in primary hepatocytes with ALOD4, a probe that binds accessible cholesterol18. Confocal microscopy revealed more ALOD4 staining on the PM of L-A/C KO than in control hepatocytes (Fig. 1b). Western blotting and flow cytometry for ALOD4 binding further confirmed that loss of Aster function expands the accessible cholesterol pool in hepatocytes (Fig. 1c,d). Quantification of fluorescence intensity from ~30,000 cells confirmed enhanced ALOD4 binding in the absence of Aster (Fig. 1e; P < 0.0001). Thus, Aster-mediated cholesterol removal from the PM of hepatocytes determines the size of the accessible PM cholesterol pool.
Asters facilitate hepatocyte PM to ER cholesterol transport
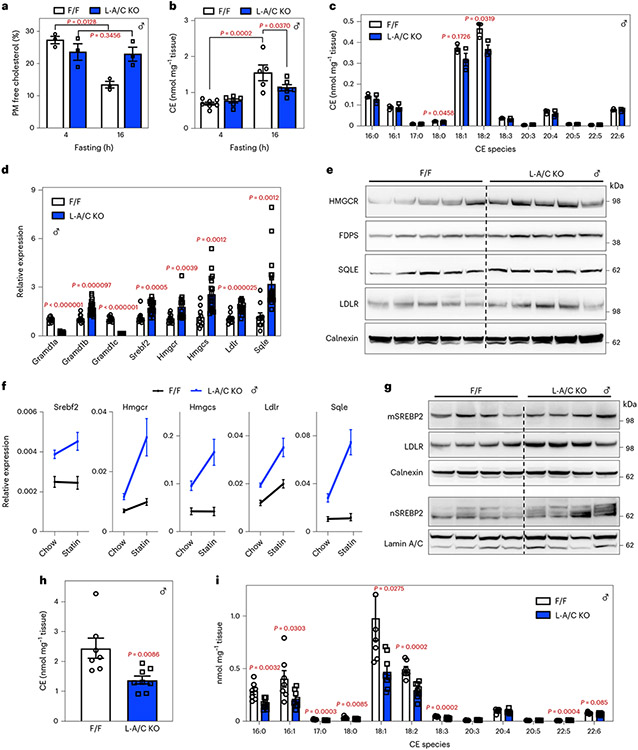
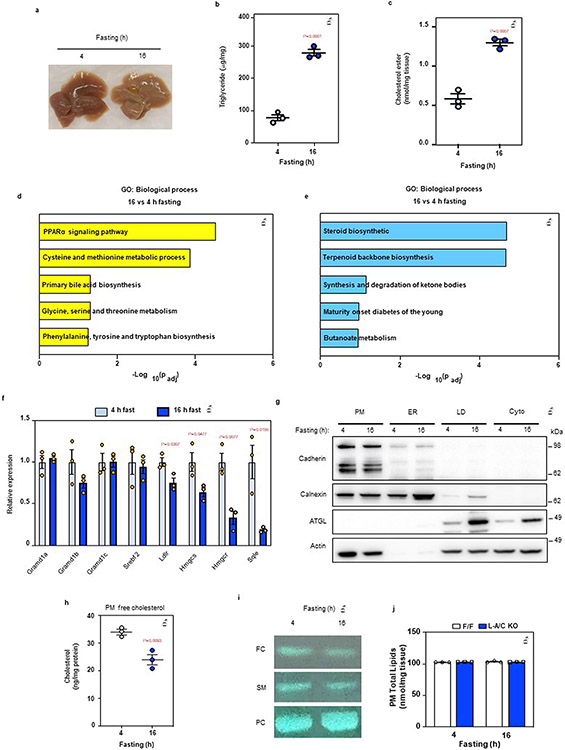
To study changes in liver cholesterol distribution during fasting, we compared wild-type (WT) mice that had undergone prolonged fasting (16 h) to those that had undergone a short (4 h) fast. Prolonged fasting promoted liver triglyceride (TG) and CE accumulation compared to a short fast (Extended Data Fig. 2a-c). Pathway analysis after liver RNA sequencing (RNA-seq) showed that PPARα signaling was the most robustly induced pathway after a 16-h fast (Extended Data Fig. 2d), whereas the SREBP-2 pathway was the most repressed (Extended Data Fig. 2e). qPCR analysis confirmed suppression of SREBP-2 target genes during prolonged fasting (Extended Data Fig. 2f), despite liver CE accumulation (Extended Data Fig. 2c). Notably, purification of PMs from WT mice (Extended Data Fig. 2g) showed that PM free cholesterol was reduced after a 16-h fast, coinciding with liver CE accumulation and suppression of the SREBP-2 pathway (Extended Data Fig. 2h,i).
Floxed control mice had lower PM free cholesterol after a 16-h fast compared to a 4-h fast and this depletion was blocked in L-A/C KO mice (Fig. 2a and Extended Data Fig. 2j), strongly implying that Asters mediate the movement of hepatocyte cholesterol from the PM to the ER in response to fasting. Furthermore, liver CE accumulation in control mice was greater after a 16-h fast than after a 4-h fast (Fig. 2b). Fasting-induced liver CE accumulation was lower in L-A/C KO mice (Fig. 2b). Of note, the most abundant CE species (18:2 CE) was significantly lower in L-A/C KO mice (Fig. 2c). At the same time, L-A/C KO mice had higher mRNA and protein levels for SREBP-2 target genes after a 16-h fast (Fig. 2d,e). These findings suggested that induction of the SREBP-2 pathway was likely compensating for impaired PM–ER cholesterol transport in L-A/C KO mice during fasting.
Fig. 2 ∣. Asters facilitate hepatocyte PM to ER cholesterol transport during fasting.
a, PM free cholesterol from F/F and L-A/C KO mice after 4 or 16 h fasting (n = 3 and 3). b, Total hepatic CEs from F/F and L-A/C KO mice after 4 and 16 h fasting (n = 7, 6, 5 and 6). c, Individual hepatic CE species (nmol mg−1 liver) from F/F and L-A/C KO mice after 16 h fasting (n = 3 and 3). d, Hepatic mRNA expression of SREBP-2 pathway targets from livers of F/F and L-A/C KO mice after 16 h fasting (n = 10 and 18). e, Immunoblot analysis of indicated proteins from livers of F/F and L-A/C KO mice after 16 h fasting (n = 5 and 5). Calnexin was used as a sample processing control. f, Hepatic mRNA expression of SREBP-2 pathway targets from livers of F/F and L-A/C KO mice after chow or statin diet feeding for 5 d (n = 10 and 18 for chow diet and n = 7 and 8 for statin diet). g, Immunoblot analysis of membrane (top) and nuclear (bottom) SREBP-2 protein levels from livers of F/F and L-A/C KO mice after statin diet feeding (n = 4 and 4). Calnexin (top) and Lamin A/C (bottom) were used as sample processing controls. h, Total hepatic CEs from livers of F/F and L-A/C KO mice after statin diet feeding (n = 7 and 8). i, Individual hepatic CE species (nmol mg−1 liver) from livers of F/F and L-A/C KO mice after statin diet feeding (n = 7 and 8). All data are presented as mean ± s.e.m. P values were determined by two-way analysis of variance (ANOVA) with Sidak’s correction for multiple comparisons (a,b), two-sided Student’s t-test (h) or two-sided Student’s t-test with Benjamini, Krieger and Yekutieli correction for multiple comparisons (c,d,i). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
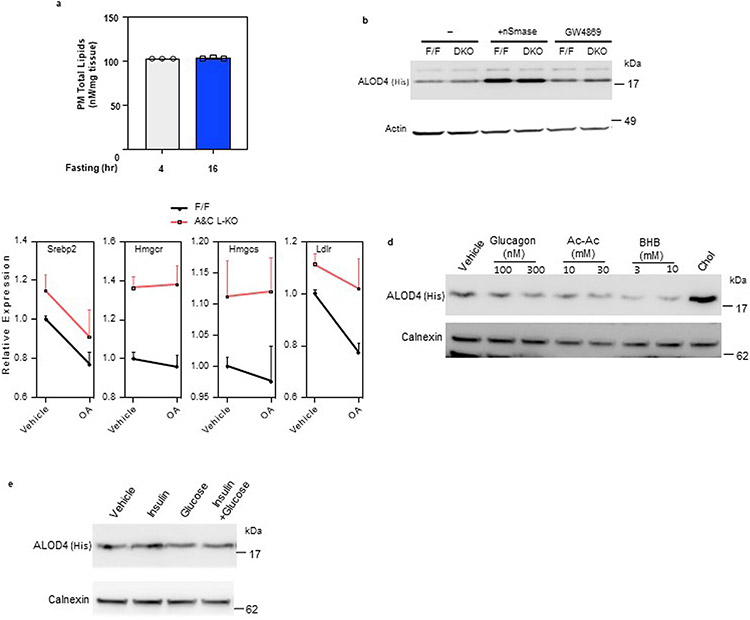
To bypass potential compensatory effects of enhanced cholesterol synthesis, we treated control and L-A/C KO mice with statins (that block endogenous cholesterol synthesis by inhibiting HMGCR). The mRNA expression of SREBP-2 pathway targets was higher in livers of L-A/C KO mice fed a statin diet than in mice fed a chow diet (Fig. 2f). Additionally, nuclear fractions from livers of L-A/C KO fed a statin diet had higher levels of mature SREBP-2 than in controls, despite comparable levels of the membrane-bound precursor (Fig. 2g). Total CE levels and levels of most major CE species, were dramatically lower in the livers of L-A/C KO mice fed the statin diet after a 16-h fast (Fig. 2h,i). We conclude from these studies that loss of Aster-A and Aster-C in hepatocytes impairs the formation fasting-induced CE stores, which is preferentially derived from PM cholesterol. In the absence of Asters, hepatocytes compensate by upregulating de novo cholesterol synthesis.
Adipose-derived fatty acids free sequestered PM cholesterol
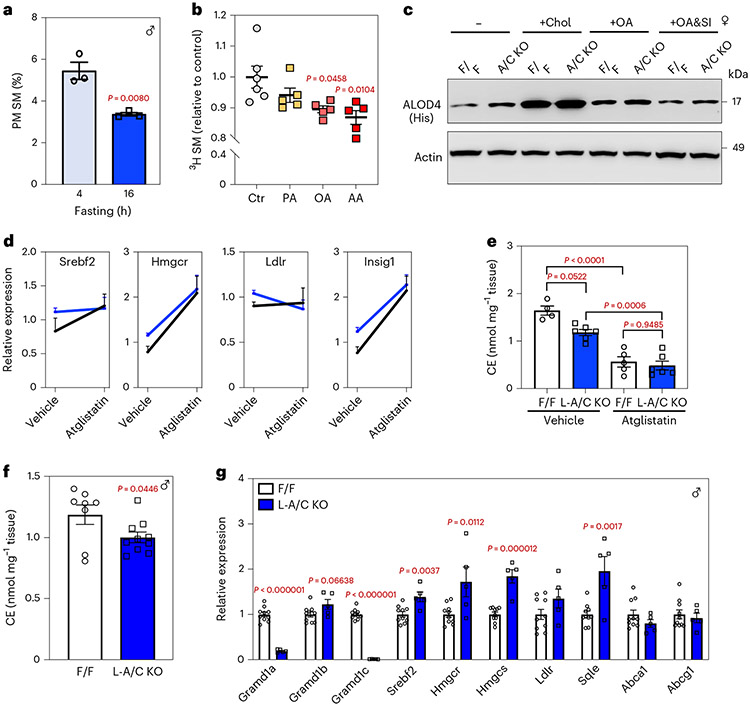
We next sought to determine how fasting promotes Aster-mediated cholesterol removal from the PM. Lipidomic analysis of PMs isolated from mouse liver showed that prolonged fasting reduced SM abundance (Fig. 3a and Extended Data Fig. 3a). Fasting promotes intracellular TG hydrolysis in adipose tissue to supply the liver with fatty acids. We found that BSA-conjugated oleic acid (OA) or arachidonic acid, but not palmitic acid, treatment promoted the breakdown of [3H]choline-labeled SM in cultured hepatocytes (Fig. 3b), consistent with earlier work19. We therefore hypothesized that fatty acids released from adipose tissue during fasting might promote the hydrolysis of SM in hepatocyte PMs to render the SM-sequestered cholesterol pool ‘accessible’ for Aster-mediated transport to the ER.
Fig. 3 ∣. Adipose-derived fatty acids liberate sequestered PM cholesterol during fasting.
a, Lipidomic analysis of PM SM from mice that had been fasted for 4 or 16 h (n = 3 and 3). b, [3H]choline-labeled SM in HepG2 cells after incubation with 300 μM BSA-conjugated PA, OA or AA for 1 h (n = 6, 5, 5 and 5). c, Immunoblot analysis of ALOD4 binding in F/F and L-A/C KO primary hepatocytes after treatment with vehicle, MβCD-cholesterol (35 μM), OA (30 μM) and OA (30 μM) plus GW4869 (10 μM) for 1 h. Actin was used as a loading control. d, Expression levels of the indicated genes in livers of mice that had been administered with vehicle or Atglistatin (200 μmol kg−1) for 16 h (n = 3, 4 for vehicle and n = 5 and 5 for Atglistatin). e, Total hepatic CEs in mice from d (n = 4 and 6 for vehicle and n = 5 and 6 for Atglistatin). f, Total hepatic CEs from F/F and L-A/C KO mice after acute cold exposure (n = 8 and 10). g, Hepatic mRNA expression of SREBP-2 pathway targets from livers of F/F control and L-A/C KO mice after 5 h cold exposure (n = 10 and 5). All data are presented as mean ± s.e.m. P values were determined by two-sided Student’s t-test (a,f), two-sided Student’s t-test with Benjamini, Krieger and Yekutieli correction for multiple comparisons (g), one-way ANOVA with Tukey’s post-test (b) or two-way ANOVA with Sidak’s correction for multiple comparisons (e). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
We first tested whether free fatty acids could generate accessible cholesterol in PM of primary hepatocytes, as assessed by ALOD4 binding. At baseline, L-A/C KO primary hepatocytes had more accessible PM cholesterol than control primary hepatocytes, consistent with our previous experiments (Fig. 3c; lanes 1 and 2). Treatment of hepatocytes with mβCD-cholesterol served as a positive control for elevated accessible cholesterol and increased ALOD4 binding (Fig. 3c lanes 3 and 4). Oleic acid treatment increased accessible cholesterol in both control and L-A/C KO primary hepatocytes (Fig. 3c; lanes 5 and 6). Treatment with an inhibitor of neutral SMases (GW4869) blocked the increase in accessible cholesterol induced by OA (Fig. 3c; lanes 7 and 8, and Extended Data Fig. 3b). L-A/C KO primary hepatocytes had higher ALOD4 binding than in controls under all conditions, consistent with an impaired ability to remove accessible cholesterol from the PM (Fig. 3c). OA treatment also suppressed SREBP-2 target genes in control primary hepatocytes, consistent with increased cholesterol transport to ER; this suppression was blunted in L-A/C KO hepatocytes (Extended Data Fig. 3c).
We next treated both control and L-A/C KO mice with Atglistatin (an inhibitor of adipose TG lipase) and then fasted the mice for 16 h to promote hepatic PM–ER cholesterol movement. Expression of SREBP-2 pathway target genes was enhanced by Atglistatin, suggesting that inhibition of fatty acid flux from adipose tissue retards cholesterol movement to the ER of hepatocytes (Fig. 3d). This increased expression of SREBP-2 pathway targets by Atglistatin was blunted in the livers of L-A/C KO mice (Fig. 3d). Total liver CEs were lower in Atglistatin-treated control mice than in vehicle-treated control mice (Fig. 3e). Vehicle-treated L-A/C KO mice had lower CEs after fasting compared to floxed control mice (Fig. 3e). Atglistatin treatment did not further decrease liver CEs in the L-A/C KO mice compared to Atglistatin-treated control mice (Fig. 3e). These data show that the magnitude of the decrease in CE accumulation in response to Atglistatin treatment is smaller in L-A/C KO mice compared to floxed controls.
In addition to promoting fatty acid release from adipose tissue, fasting increases glucagon and ketones and decreases insulin and glucose in circulation. We therefore assessed the effects of glucagon, ketones (acetoacetate and hydroxybutyrate), insulin and glucose on PM accessible cholesterol levels, as assessed by ALOD4 binding, in primary hepatocytes. In contrast to fatty acids, glucagon or ketone body treatment of primary hepatocytes slightly decreased PM accessible cholesterol (Extended Data Fig. 3d). Furthermore, ALOD4 binding to primary hepatocytes was unchanged by insulin and glucose (Extended Data Fig. 3e). These results show that, of the fasting-induced molecules tested, fatty acids are unique in their ability to acutely generate accessible cholesterol in PM.
To further examine the role of adipose-derived fatty acids in hepatic cholesterol movement in vivo, we exposed mice to a 5-h cold challenge, which acutely stimulates adipose-tissue lipolysis20. We found the CE accumulation in the livers of L-A/C KO mice was lower (Fig. 3f) and SREBP-2 pathway activity was higher (Fig. 3g), compared to floxed mice in response to cold. These results provide further evidence of an important role for adipose-derived fatty acids in hepatocyte PM cholesterol movement. Collectively, these data indicate that adipose-tissue-derived fatty acids increase the accessible cholesterol pool in hepatocyte PMs and that this effect is mediated by neutral SMase release of sphingomyelin-sequestered cholesterol.
Fasting induces Smpd3 to regulate cholesterol metabolism
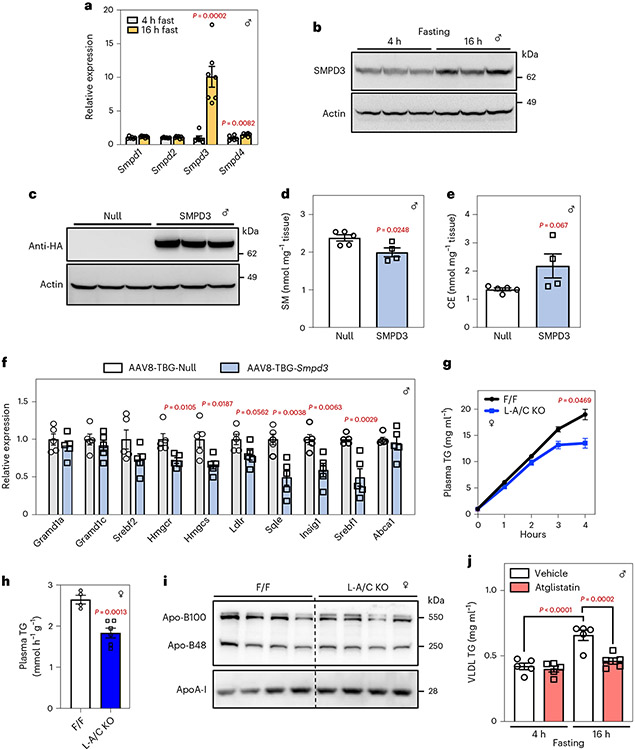
There are four SMase enzymes (encoded by Smpd1–4) in liver17. Smpd1 encodes a lysosomal acidic SMase, while Smpd2–4 encode neutral SMases21. The mRNA and protein levels of Smpd3 were increased in the livers after a 16-h fast (Fig. 4a,b), consistent with physiological regulation of SMase expression. We expressed Smpd3 in hepatocytes of WT mice with an intravenous injection of an AAV8-3xHA-Smpd3 vector driven by the liver-specific thyroxine binding globulin (TBG) promotor (Fig. 4c). Smpd3 expression decreased hepatic SM levels, increased liver CE and suppressed SREBP-2 pathway target gene expression (Fig. 4d-f). Thus, fasting induces liver Smpd3 expression and the increased SM activity promotes cholesterol movement to the ER.
Fig. 4 ∣. Fasting induces Smpd3 expression to regulate cholesterol metabolism and VLDL production.
a, Hepatic mRNA expression of Smpd1, Smpd2, Smpd3 and Smpd4 in mice fasted for 4 h or 16 h (n = 6 and 7). b, Immunoblot analysis of hepatic SMPD3/nSMase2 in the livers of mice fasted for 4 h or 16 h (n = 3 and 3). Actin was used as a loading control. c, Immunoblot analysis of HA-tagged SMPD3/nSMase2 in the livers of mice transduced with AAV8-TBG-Ctr or AAV8-TBG-Smpd3 for 3 d (n = 3 and 3). Actin was used as a loading control. d,e, Hepatic SM (d) and CEs (e) in mice 3 d after i.v. injection of AAV8-TBG-Ctr or AAV8-TBG-Smpd3 (n = 5 and 4). f, Hepatic mRNA expression of indicated genes from WT mice transduced with AAV8-TBG-Ctr or AAV8-TBG-Smpd3 for 3 d (n = 5 and 5). g, VLDL-TG secretion in F/F control and L-A/C KO mice (n = 4 and 6). h. The rate of VLDL-TG production from F/F control and L-A/C KO mice (n = 4 and 6). i, Apo-B and ApoA-I protein levels in plasma of F/F and L-A/C KO mice (n = 4 and 4). j, TGs in isolated VLDL particles from mice treated with vehicle or Atglistatin (200 μmol kg−1) for 16 h (n = 5). All data are presented as mean ± s.e.m. P values were determined by two-sided Student’s t-test (d,e,h), two-sided Student’s t-test with Benjamini, Krieger and Yekutieli correction for multiple comparisons (a,f), or two-way ANOVA with Sidak’s correction for multiple comparisons (g,j). *P < 0.05, **P < 0.01, ***P < 0.001.
Nonvesicular sterol transfer supports fasting VLDL secretion
To determine the impact of PM–ER cholesterol transport during fasting on VLDL secretion, we injected mice with the lipase inhibitor, Poloxamer-407 after a 16-h fast and measured the appearance of TG in plasma. L-A/C KO mice had impaired liver VLDL-TG secretion after a prolonged fast (Fig. 4g,h), suggesting that impaired PM–ER cholesterol movement blunts hepatic lipid output during fasting. Plasma apolipoprotein B100 also tended to be lower in the plasma of L-A/C KO after a 16-h fast (without a change in Apo-B48), consistent with reduced VLDL output. Conversely, plasma ApoA-I levels were higher in L-A/C KO mice (Fig. 4i). Reducing free fatty acid delivery to the liver by Atglistatin treatment of WT mice also impaired fasting-induced VLDL secretion (Fig. 4j). These data raise the possibility that Asters move cholesterol to the ER of hepatocytes to support CE formation and VLDL output during fasting.
Aster-C is a direct target gene of FXR in mouse liver
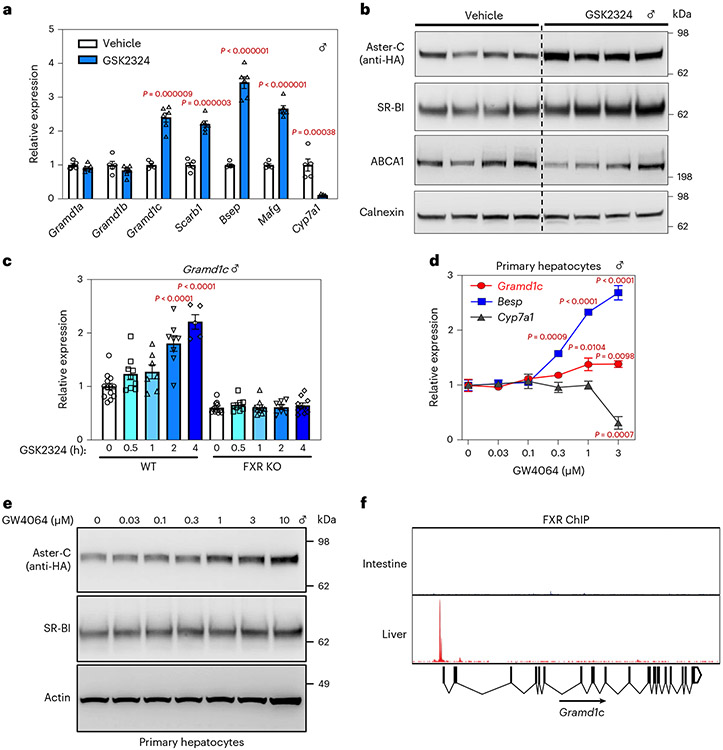
Earlier studies showed that the gene encoding Aster-B is regulated by the nuclear receptor LXR in macrophages6. We observed that Aster-C and SR-BI were induced at the mRNA and protein levels in the liver of 3×HA-Aster-C knock-in mice in response to the FXR agonist GSK2324 (Fig. 5a,b). This response was abolished in FXR KO mice (Fig. 5c). The FXR agonist GW4046 also increased Aster-C expression in a dose-dependent manner in primary mouse hepatocytes (Fig. 5d,e). Furthermore, chromatin immunoprecipitation-sequencing (ChIP-seq) data supported FXR binding to the promoter region of Aster-C in the liver but not intestine (Fig. 5f)22. These data identified the gene encoding Aster-C as a direct transcriptional target of FXR in liver and suggested that Aster-C may operate downstream of SR-BI to move HDL cholesterol to the ER.
Fig. 5 ∣. Aster-C is a direct target gene of FXR in mouse liver.
a, Hepatic mRNA expression of Asters and FXR target genes after vehicle or GSK2324 treatment (n = 5 and 6). b, Immunoblot analysis of hepatic Aster-C (HA tag), SR-BI and ABCA1 from a. Calnexin was used as a sample processing control (n = 4 and 4). c, Hepatic mRNA expression of Aster-C from WT or FXR KO mice after vehicle or GSK2324 treatment (n = 13, 9, 7, 8, 5, 11, 8, 9, 7 and 10). d, qPCR analysis of indicated genes in response to GW4064 treatment for 16 h in primary hepatocytes (n = 3 for each time point). e, Immunoblot analysis of Aster-C (HA tag) and SR-BI in response to GW4064 treatment for 16 h in primary hepatocytes. Actin was used as a loading control for SRB1 and a sample processing control for HA-Aster-C. f, ChIP assay of FXR recruitment to the Aster-C promoter in the intestine or liver. All data are presented as mean ± s.e.m. P values were determined by two-sided Student’s t-test with Benjamini, Krieger and Yekutieli correction for multiple comparisons (a) or one-way ANOVA with Dunnett’s correction for multiple comparisons (c,d). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Asters transfer lipoprotein cholesterol to the ER in livers
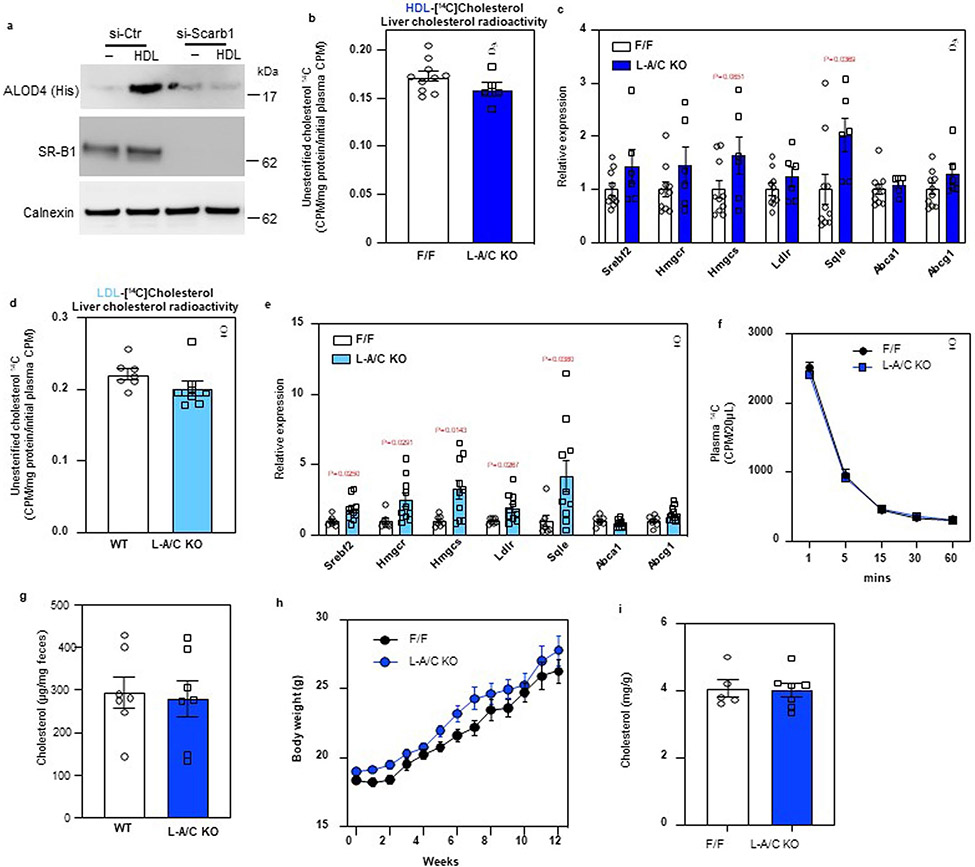
RCT involves the movement of HDL cholesterol from peripheral tissues to hepatocytes via the HDL receptor, SR-BI. HDL predominantly carries cholesterol in the form of CEs; however, HDL loading of cells for 1 h increased PM accessible cholesterol, as indicated by higher ALOD4 staining (Extended Data Fig. 4a). This increase in ALOD4 binding was abolished by knocking down Scarb1 (Extended Data Fig. 4a). As ALOD4 recognizes unesterified accessible cholesterol and not CEs, these results demonstrate that SR-BI loads cholesterol from HDL into the PM where it becomes part of the accessible cholesterol pool, presumably by a currently unidentified cholesterol esterase.
To determine whether hepatic Asters participate in RCT, we conducted tracer studies with HDL containing [14C]cholesterol23. Control and L-A/C KO mice were fasted for 16 h and then given an intravenous (i.v.) injection of the HDL (Fig. 6a). The rate of HDL clearance from the circulation was similar in the different groups of mice (Fig. 6b). Total 14C counts and 14C-labeled free cholesterol were modestly lower in livers of L-A/C KO mice than in controls (Fig. 6c and Extended Data Fig. 4b); however, 14C-labeled CE accumulation was dramatically lower (Fig. 6d). L-A/C KO livers had a higher proportion of 14C in free cholesterol and a lower proportion of 14C in CE compared to controls, consistent with impaired cholesterol movement from PM to ER (Fig. 6e). Furthermore, the expression of SREBP-2 pathway target genes was higher in livers of L-A/C KO mice 1 h after administration of the radiolabeled HDL (Extended Data Fig. 4c). These data indicate that Aster proteins function downstream of SR-BI to transport HDL-derived cholesterol from the hepatocyte PM to the ER.
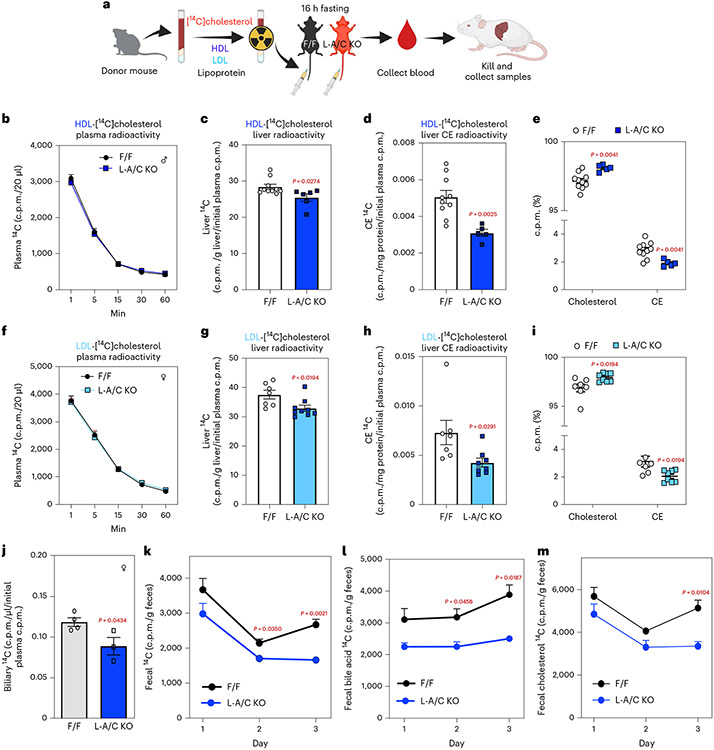
Fig. 6 ∣. Hepatic Aster deficiency impairs transport of lipoprotein-derived cholesterol and RCT.
a, Experiment schematic of the [14C]cholesterol-labeled lipoprotein tracing studies. HDL/LDL particles were freshly isolated from donor mice by density-gradient ultracentrifugation. F/F control and L-A/C KO mice received intravenous injections of [14C]-HDL or -LDL and blood was collected at the indicated timepoints for up to 1 h. After 1 h, mice were killed for tissue collection and analysis. Liver total [14C] radioactivity was normalized per gram of liver and expressed relative to radioactivity in plasma 1 min after lipoprotein injection (which we defined as 100% of radioactivity administered). [14C] radioactivity in liver CEs was normalized per mg protein and expressed relative to radioactivity in plasma 1 min after lipoprotein injection. b, Rate of [14C]-HDL clearance from circulation, as indicated by radioactive counts in plasma at 1, 5, 15, 30 and 60 min after lipoprotein administration (n = 9 and 6). c, Total [14C] counts in livers (n = 10 and 6). d, [14C] counts in liver CE of mice from c. e, The ratio of radioactivity in free cholesterol or CE to total radioactivity in cholesterol from c (n = 10 and 5). f, Rate of [14C]-LDL clearance from circulation, as indicated by radioactive counts in plasma at 1, 5, 15, 30 and 60 min after lipoprotein administration (n = 7 and 9). g, Total [14C] counts in livers (n = 7 and 9). h, [14C] counts in liver CE of mice from g. i, The ratio of radioactivity in free cholesterol or CE to total radioactivity in cholesterol from g (n = 7 and 8). j, Total biliary [14C] counts from F/F and L-A/C KO mice 3 d after [14C]-HDL administration (n = 4 and 3). Total biliary [14C] counts are normalized per μL bile and expressed relative to radioactivity in plasma 1 min after [14C]-HDL administration. k, Total [14C] counts in feces of mice over 3 d after [14C]-HDL administration (n = 6 and 5). l, [14C] counts in the fecal bile acids from k (n = 6 and 5). m, [14C] counts in fecal cholesterol from k (n = 6 and 5). All data are presented as mean ± s.e.m. P values were determined by two-sided Student’s t-test (c–e,g–h,j) or two-way ANOVA with Sidak’s correction for multiple comparisons (k–m). *P < 0.05, **P < 0.01.
Low-density lipoprotein (LDL) is internalized by clathrin-mediated endocytosis following binding to LDL receptors on the hepatocyte cell surface—a pathway distinct from SR-BI-mediated HDL uptake24. From the surface of the late endosome/lysosome, most of the LDL cholesterol is transported to the PM before moving to the ER2,25. Aster proteins have been shown to contribute to post-lysosomal movement of LDL cholesterol in cultured cells8,9, but whether this pathway operates in vivo is unknown11. To address this, we isolated LDL from donor mice, labeled it with [14C]cholesterol and administered it i.v. to recipient mice. The rate of [14C]LDL clearance was similar in control and L-A/C KO mice (Fig. 6f). Total liver 14C counts were modestly lower in L-A/C KO mice compared to controls and free [14C]cholesterol tended to be lower (Fig. 6g and Extended Data Fig. 4d); however, 14C-labeled CE accumulation was substantially lower in livers of L-A/C KO mice (Fig. 6h). Consistent with impaired LDL cholesterol movement to the ER, SREBP-2 pathway target genes were higher in livers of L-A/C KO mice 1 h after administration of the radiolabeled LDL (Extended Data Fig. 4e). The proportion of 14C in free cholesterol was higher in L-A/C KO livers, whereas the proportion of 14C in CE was lower (Fig. 6i). Thus, Aster proteins mediate PM–ER movement of both HDL- and LDL-derived cholesterol in the liver.
Hepatic Aster deficiency impairs RCT
Once internalized into hepatocytes, HDL-derived cholesterol is secreted into bile in the form of free cholesterol and bile acids as part of RCT26. To assess the contribution of Aster-mediated cholesterol transport to bile and subsequent excretion, we injected [14C]cholesterol-labeled HDL into mice and collected feces over 3 d. The rate of [14C]HDL clearance was similar in control and L-A/C KO mice (Extended Data Fig. 4f); however, total 14C counts were lower in gallbladders of L-A/C KO mice than in controls (Fig. 6j). Also, L-A/C KO mice had lower fecal 14C counts (Fig. 6k) and lower radiolabel abundance in fecal bile acids and free cholesterol than in controls (Fig. 6l,m). Collectively, these data demonstrate that Asters mediate a key step in RCT by moving HDL cholesterol through hepatocytes and into the gut for excretion.
To determine whether the total mass of cholesterol leaving the body is different between L-A/C KO mice compared to floxed controls, we fed mice a cholesterol-free diet and collected feces over 48 h before assessing fecal cholesterol output. Fecal cholesterol concentrations were similar between L-A/C KO mice and controls (Extended Data Fig. 4g), suggesting that higher endogenous cholesterol synthesis in the livers of the KO mice and subsequent secretion into bile may compensate for the impaired flux from lipoproteins into bile, as determined by the radiolabeled tracer studies.
Asters are required for hepatic dietary cholesterol storage
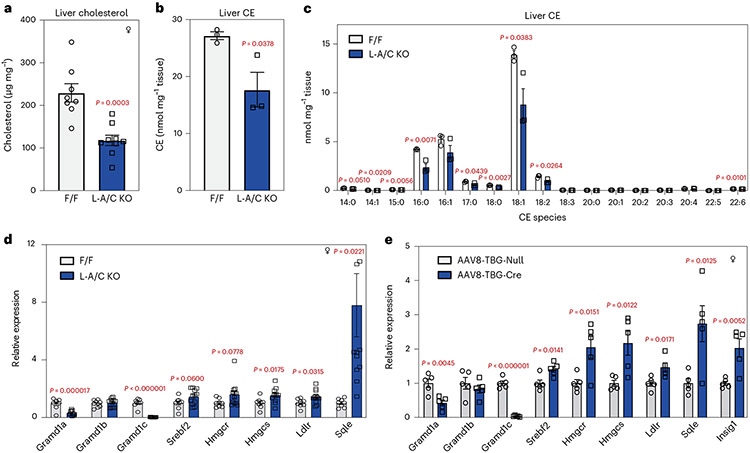
To further explore the role of Asters in PM–ER cholesterol transport in the liver, we challenged mice with a western diet (WD), which suppresses endogenous cholesterol synthesis and results in the delivery of large amounts of cholesterol to the basolateral surface of hepatocytes. Body weight gain was comparable between controls and L-A/C KO mice over 12 weeks of WD feeding (Extended Data Fig. 4h); however, total cholesterol, CE and most major CE species were dramatically lower in the livers of L-A/C KO mice than in controls (Fig. 7a-c). Hepatic SREBP-2 target gene expression was also higher in L-A/C KO mice fed the WD (Fig. 7d). We also acutely deleted Asters in hepatocytes by administering an intravenous injection of an AAV8-TBG-Cre vector to Aster-A/C double-floxed mice. The SREBP-2 pathway was markedly upregulated after acute deletion of Aster-A and Aster-C in the liver after 4 weeks of WD feeding (Fig. 7e), consistent with the phenotype observed in the developmental knockout line. These data show that Asters play a key role in the movement of dietary cholesterol into the hepatocyte for storage as CE.
Fig. 7 ∣. Asters are required for storage of dietary cholesterol as CEs in hepatocytes.
a, Total liver cholesterol in F/F and L-A/C KO mice after 12 weeks of WD feeding (n = 8 and 9). b, Total liver CEs from a (n = 3 and 3). c, Individual hepatic CE species (nmol mg−1 liver) from a (n = 3 and 3). d, Expression levels of the indicated genes from a (n = 8 and 12). e, Expression levels of the indicated genes in livers from Aster-A and Aster-C F/F mice transduced with AAV8-TBG-Cre compared to control AAV8-TBG-null for 2 weeks. Mice were then fed with WD for 4 weeks (n = 5 and 5). All data are presented as mean ± s.e.m. P values were determined by two-sided Student’s t-test (a,b), or two-sided Student’s t-test with Benjamini, Krieger and Yekutieli correction for multiple comparisons (c–e). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Hepatic Aster deficiency disrupts systemic sterol balance
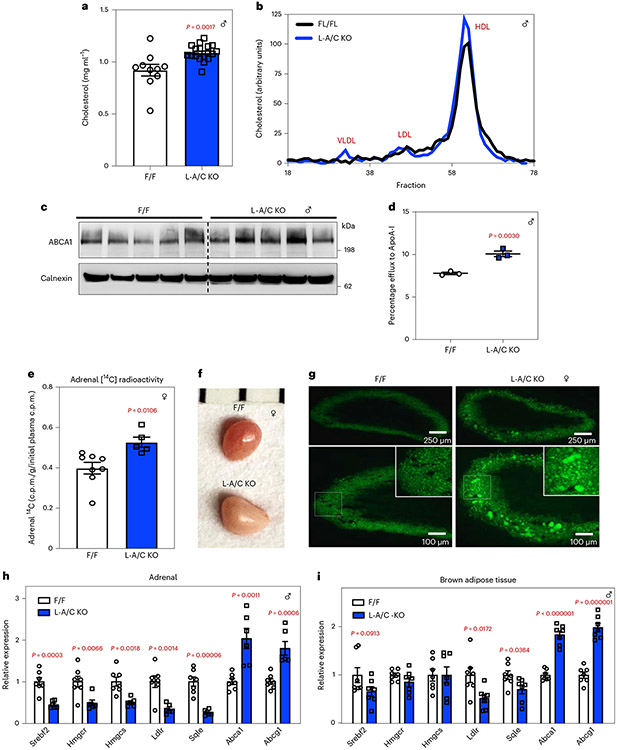
The liver plays a central role in determining plasma lipid levels. Total plasma cholesterol levels were higher in L-A/C KO mice than in the controls (Fig. 8a). The higher plasma cholesterol levels were mainly due to increased amounts of HDL cholesterol (Fig. 8b). HDL is produced primarily in hepatocytes and enterocytes through lipidation of apolipoprotein AI (ApoA-I) by PM-localized ABCA1 (refs. 27-30). ABCA1 protein levels were higher in the livers of L-A/C KO mice than in control mice (Fig. 8c). Consistent with this finding, cholesterol efflux to ApoA-I was higher in primary hepatocytes from L-A/C KO mice than in primary hepatocytes from control mice (Fig. 8d). These observations suggest that, in the absence of the Aster nonvesicular transport pathway, HDL cholesterol accumulated in hepatocyte PMs is effluxed to ApoA-I rather than trafficked to the ER.
Fig. 8 ∣. Hepatic Aster deficiency disrupts systemic cholesterol homeostasis.
a, Plasma total cholesterol from 8-week-old F/F control and L-A/C KO mice fed a chow diet (n = 10 and 18). b, Cholesterol content of plasma lipoproteins fractionated by FPLC. c, Immunoblot analysis of hepatic ABCA1. Calnexin was used as a sample processing control (n = 5 and 5). d, Cholesterol efflux to ApoA-I by control and L-A/C KO primary hepatocytes (n = 3 and 3). e, Total adrenal [14C] radioactivity 3 d after [14C]-HDL administration (n = 8 and 5). Total adrenal [14C] counts are normalized per gram tissue and expressed relative to radioactivity in plasma 1 min after [14C]-HDL administration. f, Gross appearance of adrenal glands from representative 8-week-old F/F control and L-A/C KO mice (1 mm scale). g, Histological sections of the adrenal cortex stained with BODIPY from f. Data are representative of three independent samples. h,i, mRNA expression levels of the indicated genes in adrenal glands (h; n = 7 and 6) and brown adipose tissue (i, n = 7 and 7) from F/F control and L-A/C KO mice. All data are presented as mean ± s.e.m. P values were determined by two-sided Student’s t-test (a,d,e) or two-sided Student’s t-test with Benjamini, Krieger and Yekutieli correction for multiple comparisons (h,i). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, *****P < 0.00001.
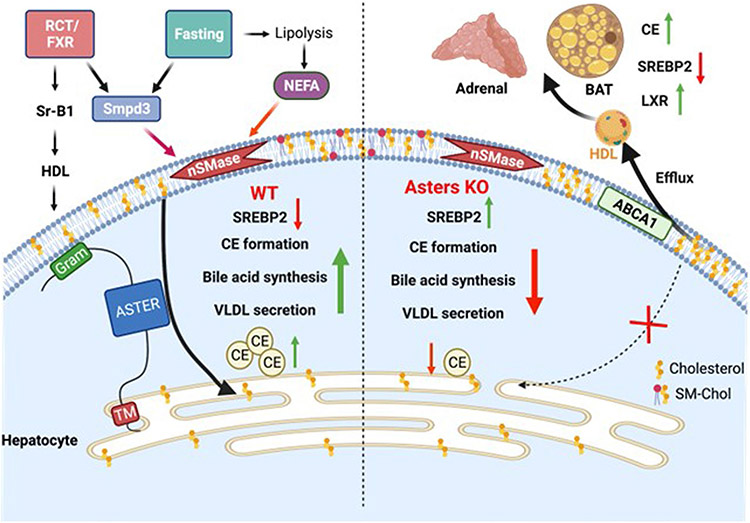
We further investigated the effects of loss of Aster function in the liver on cholesterol homeostasis in peripheral tissues. Rodent adrenal glands rely on HDL cholesterol for steroidogenesis31. Loss of hepatic Aster expression resulted in greater accumulation of 14C counts in adrenal glands after intravenous administration of [14C]cholesterol-labeled HDL for 3 d (Fig. 8e). The adrenal glands of L-A/C KO mice were paler by visual inspection, indicating an enrichment of neutral lipids (Fig. 8f). Indeed, BODIPY staining revealed a marked increase in neutral lipid stores in the adrenal cortex of L-A/C KO mice (Fig. 8g). Moreover, SREBP-2 pathway genes were suppressed in the adrenals of L-A/C KO mice, whereas LXR target genes (Abca1 and Abcg1) were increased (Fig. 8h). SREBP-2 suppression and LXR induction was also observed in brown adipose tissue, suggesting increased cholesterol delivery to multiple peripheral tissues in the absence of hepatic Aster expression (Fig. 8i). Whole-body cholesterol content was similar between L-A/C KO and F/F mice (Extended Data Fig. 4i). These data suggest that impaired flux from lipoproteins into bile in L-A/C KO mice is balanced by higher plasma cholesterol levels (due to enhanced PM cholesterol efflux to HDL), higher cholesterol in peripheral tissues and higher endogenous cholesterol synthesis in the liver to result in unchanged whole-body cholesterol content compared to F/F mice. Therefore, impaired Aster-mediated PM–ER cholesterol transport in the liver alters systemic cholesterol distribution (Extended Data Fig. 5).
Discussion
The three Aster family members (Aster-A, Aster-B and Aster-C) are expressed in a tissue-specific manner. Loss of Aster-B impairs CE formation and steroid hormone synthesis in the adrenal gland6, but the role of these proteins in systemic lipid metabolism is unknown. The concept of accessible PM cholesterol is important for contextualizing the function of Asters in hepatic physiology. Accessible cholesterol is rapidly transported to the ER and maintains an equilibrium with the ER cholesterol pool2, but the role that accessible cholesterol plays in physiology—and the endogenous signals that promote release of sequestered cholesterol in vivo—have not been defined. Here we show that Aster proteins move accessible cholesterol to the ER in two physiological contexts—fasting and RCT (Extended Data Fig. 5). We showed that fatty acids activate SMase to liberate sequestered cholesterol in the hepatocyte PM during fasting and that Aster proteins move the augmented pool of accessible cholesterol to the ER to support CE production. Asters are also an important component of the RCT pathway owing to their function in moving HDL-derived cholesterol within hepatocytes. Loss of Aster function decreases the appearance of HDL-derived cholesterol in the feces, raises plasma cholesterol levels and causes peripheral cholesterol accumulation.
The role that adipose TG lipolysis plays in liver TG accumulation and the secretion of VLDL TGs during fasting is well established, but little is known about how adipose tissue nutrient stores affect liver cholesterol homeostasis. Our experiments define a pathway whereby fatty acids from adipose tissue increase Aster-mediated cholesterol movement in the liver by promoting hydrolysis of SM in liver PM. Impaired fatty acid flux from adipose tissue or loss of Aster function in the liver reduces VLDL output and bile acid synthesis. Our work reveals how hepatic intracellular cholesterol movement is regulated by signals derived from adipose tissue to control systemic cholesterol homeostasis.
Our data also show that liver PM cholesterol pools are dynamic and responsive to nutritional cues and energy supply. Fasting requires mammals to utilize lipid metabolites from pre-existing stores. It has long been appreciated that fasting promotes release of fatty acids from adipose tissue, TG accumulation in the liver and ultimately hepatic VLDL secretion to promote redistribution of lipid nutrients to peripheral tissues. CEs also accumulate in the liver during fasting, which is dependent on cholesterol delivery to the ER. Given that the energetically costly SREBP-2 cholesterol biosynthetic pathway is suppressed during fasting32, the source of cholesterol for fasting-induced CE synthesis has been unclear. We showed that fatty acids released by adipose TG lipolysis activate liver neutral SMase to liberate SM-sequestered cholesterol in the PM of hepatocytes, thereby augmenting the accessible cholesterol pool. Asters transport the accessible cholesterol to the ER. Therefore, the SM-sequestered cholesterol pool plays a physiological role in providing cholesterol to the ER of hepatocytes during periods of nutrient deprivation. Redistribution of cholesterol from the PM to the ER of hepatocytes promotes CE formation, VLDL output, bile acid synthesis and suppression of the SREBP-2 pathway during fasting. The concept of a SM-cholesterol/cholesterol switch represents a new regulatory mechanism that might be broadly relevant to other physiological or pathological conditions involving cholesterol transport, including metabolism, cancer and infection. It is possible that, in addition to fatty acids, other endogenous signals affect PM accessible cholesterol by altering intracellular cholesterol transport pathways or changing phospholipid composition of the PM. Determining these signals may uncover further roles for accessible cholesterol movement in different cell types.
The RCT pathway from peripheral cells to the liver has been studied extensively. Similarly, the importance of transporters for moving sterols and bile acids into bile is well documented33,34; however, a notable gap in our understanding of RCT is the path that cholesterol follows between SR-BI uptake at the PM and excretion in bile. The hepatocyte is a polarized cell with basolateral (sinusoidal) and apical (canalicular) PM. ABCG5/8 mediates biliary cholesterol secretion at the canalicular PM33,34. A potential hypothesis would be that basolateral PM cholesterol equilibrates across the entire PM surface before being secreted by ABCG5/8; however, we found that mice with impaired nonvesicular PM–ER cholesterol transport have reduced movement of HDL-derived cholesterol into bile and accumulate cholesterol at the PM. Our results suggest that SR-BI binding of HDL generates accessible PM cholesterol and that a substantial portion of this cholesterol is transported by Asters to the ER before appearing at the apical surface of hepatocytes for efflux into the bile. It is noteworthy that humans carry most of their plasma cholesterol in LDL. Our experiments show that Asters transport LDL-derived cholesterol, in addition to HDL-derived cholesterol, in the liver. Therefore, while the relative proportions of LDL and HDL are different in mice and humans, Aster-mediated cholesterol transport downstream of both lipoprotein classes is likely conserved.
FXR agonists offer considerable therapeutic promise for patients with non-alcoholic fatty liver disease (NAFLD)/non-alcoholic steatohepatitis (NASH). Current literature suggests that FXR agonists act to lower intestinal lipid absorption and hepatic lipogenesis35. But, despite cholesterol accumulation being a key feature of the diseases, the contribution of intracellular cholesterol transport pathways to NAFLD/NASH is still poorly understood. Our finding that Aster-C is an FXR target in the liver makes it conceivable that Aster-mediated cholesterol movement contributes to some of the effects of FXR agonists on NAFLD/NASH. Further studies are required to fully examine the contribution of hepatic Asters to fatty liver diseases.
Systemic consequences of impaired liver PM–ER cholesterol movement after loss of Aster function in the liver include elevated plasma HDL cholesterol levels (due to enhanced liver cholesterol efflux) and increased cholesterol delivery to peripheral tissues (for example, adrenal gland and brown adipose tissue). Overall, these findings have uncovered key functions for nonvesicular sterol transport in mammalian lipid physiology.
Methods
Mice
Conditional KO alleles for Aster-A and Aster-C (Gramd1a and Gramd1c) were generated on a C57BL/6N background using a TALENs strategy by inserting LoxP sites into the locations outlined in Extended Data Fig. 1c. AsterF/F mice were crossed with albumin-Cre transgenic mice from The Jackson Laboratory. Cre-negative, littermate floxed controls from albumin-Cre breeding pairs was combined and analyzed as a single control group. The 3xHA-Aster-C knock-in mice were generated on a C57BL/6N background by inserting a 3xHA tag into the first exon of Aster-C (Gramd1c) gene using a CRISPR/Cas9 strategy. crRNA (GCGGGAGGGTGCGGCGATGG) and tracrRNA (IDT, 1073189) were complexed with Cas9 protein (NEB, M0646T) and homology directed repair templates contained the 3xHA sequence (Supplementary Table 2) flanked by ~50-bp homology arms. Microinjections of CRISPR constructs was conducted as UC San Diego Diabetes Research Center Transgenic, CRISPR Mutagenesis and Knockout Mouse Core. Fxr-knockout mice were previously described36. All mice were housed in a temperature-controlled room (22 °C with humidity approximately 50–65%) with a 12-h light–dark cycle and under pathogen-free conditions. Mice were fed a chow diet (PicoLab Rodent Diet 20, 5053) or WD (Research Diets, D12079B) unless otherwise stated. For statin diet experiments, mice were fed a chow diet containing 0.008% Atorvastatin (Research Diets, D18062105i) and provided with drinking water containing 0.8% simvastatin sodium salt (Millipore Sigma, 5670215MG). For AAV8 infections, AAV8-TBG-Cre (plasmid 107787) or AAV8-TBG-null (plasmid 105536) was purchased from Addgene. The 8–10-week-old male AsterF/F mice were injected via the tail vein with 5 × 1010 genome copies per mouse. AAV8-TBG-HA-Smpd3 was packaged at the Vector Core of University of Michigan. The 8-week-old WT mice (C57BL/6J) from The Jackson Laboratory were injected via the tail vein with 1 × 1011 genome copies per mouse. For FXR agonist experiments, mice were administered with either vehicle or GSK2324 via intraperitoneal injection once daily at 30 mg kg−1 body weight for 3 d. Mice were fasted for 4 h before being killed to collect plasma and tissues37,38. For ATGL inhibitor experiments, mice were administered with either vehicle or Atglistatin (Millipore Sigma, SML1075) via intraperitoneal injection at 200 μmol kg−1 body weight20. Animals were fasted for 16 h before being killed. For the acute cold exposure experiment, mice were fasted for 1 h at room temperature. Mice were then singly housed at approximately 4 °C in pre-chilled cages without food or bedding for an additional 5 h before being killed. For assessment of fecal cholesterol output, F/F and L-A/C KO mice were fed a cholesterol-free diet (Research Diets, D02031501Ni) for 2 d, during which time feces was collected and processed for lipid analysis as outlined below. Experiments were performed in male and female mice. Age-matched 8–15-week-old mice were used for experiments when sufficiently large cohorts became available, which is why mice with different sexes were used in different experiments. All animal experiments were approved by the UCLA Animal Research Committee (protocols 1999-131 and 2003-166).
Primary hepatocytes
Primary hepatocytes were isolated from floxed control and L-A/C KO or 3xHA-Aster-C knock-in mice. The mice were anesthetized with 200 μl ketamine/xylazine solution by intraperitoneal injection before a cannula was inserted into the vena cava. The liver was perfused through the portal vein with 50 ml of solution A (Hanks’s solution with 1 mM EGTA, 20 mM HEPES and 1× pen/strep), followed by 50 ml of solution B (William Medium E with 1× GlutaMax, 20 mM HEPES, 1× pen/strep and 50 μg ml−1 liberase) at a speed of 5 ml min−1. The liver was placed in a dish, extraneous tissue was removed and the liver was dismantled using a forceps to release hepatocytes into the medium. Using a 25-ml pipette, hepatocytes were filtered through a 70-μm cell strainer into a fresh 50-ml conical tube. Cells were centrifuged at 50 g (400 r.p.m.) at 4 °C for 3 min to gently pellet cells. Cells were gently centrifuged an additional two times to wash. Cells were plated in warmed Washing/Plating medium (William Medium E medium with 1× GlutaMax, 20 mM HEPES, 1× pen/strep, 1 μM dexamethasone, 100 nM insulin and 5% FBS) at a density of 5 × 105 cells per well in collagen-coated six-well plates. Cells were allowed to settle for 3–4 h in Washing/Plating medium before experiments were conducted as described below. To determine Aster-C localization, hepatocytes from 3xHA-Aster-C knock-in mice were plated as described above. Hepatocytes were then either loaded with 200 μM MβCD-cholesterol in maintenance medium (William Medium E medium with 5% BSA, 0.1 μM dexamethasone and 100 nM insulin) or medium alone for 1 h before being washed three times with PBS and fixed with 4% PFA for 15 min. After fixation, cells were washed three times with PBS. Cells were incubated with anti-HA (Cell Signaling Technology, 3724S, diluted 1:1,000) or anti-E-cadherin (BD Biosciences, 610181, diluted 1:1,000) antibodies overnight before incubation with fluorescently labeled secondary antibodies (goat anti-mouse IgG H&L Alexa Fluor 555 Invitrogen A21424 or goat anti-rabbit IgG H&L Alexa Fluor 488 Invitrogen A11034; each diluted 1:1,000) for 1 h at room temperature. Cells were mounted on slides with ProLong Diamond Antifade Mountant with DAPI (Invitrogen, P36962). Images were acquired using an Inverted Leica TCS-SP8-SMD Confocal Microscope. For the GW4064 treatment, hepatocytes were treated with or without GW4064 in maintenance medium for 16 h for qPCR analysis. For OA experiments, primary hepatocytes that had been cultured in maintenance medium with Ro 48-8071 (1 μM) overnight were treated with or without OA (30 μM) for 6 h.
Western blot
Whole cell and tissues were lysed using RIPA buffer (Boston BioProducts, BP-115) supplemented with protease inhibitor (Thermo Scientific Pierce, A32965) and phosphatase inhibitor (Thermo Scientific Pierce, A32957) cocktail tablets. For determination of SREBP-2 membrane and nuclear isoforms, tissues were homogenized in lysis buffer (50 mM Tris-HCl, pH 7.8, 1 mM EDTA and protease inhibitors) at 1,000 r.p.m. 30 times on an overhead stirrer (Caframo, BDC2002). Nuclei was pelleted by centrifuging at 1,000 g for 5 min at 4 °C. Nuclear pellets were resuspended in nuclear extraction buffer (20 mM HEPES/KOH, pH 7.6; 2.5% (v/v) glycerol; 1.5 mM MgCl2; 0.42 M NaCl; 1 mM EDTA; 1 mM EGTA and protease inhibitors) and the suspension was rotated at 4 °C for at least 1 h. The nuclear suspension was centrifuged at 12,500g at 4 °C for 15 min and the supernatant was designated as the nuclear extract. Membranes were pelleted by centrifuging the post-nuclear supernatant at 100,000 g for 1 h at 4 °C. Membrane pellets were resuspended in RIPA buffer using a 1-ml syringe with a 23-G needle. Membrane pellets were then centrifuged at 12,500 g at 4 °C for 15 min and the supernatant was designated the Membrane Fraction. Protein concentrations were measured using the Bicinchoninic Acid Assay Protein Assay kit (Thermo Scientific Pierce, PI23223/4) and western blots were performed. To measure plasma apolipoproteins, plasma was diluted 20-fold with water and loading buffer. Equal amounts of protein were loaded onto NuPAGE 4–12% gels (Invitrogen, NP0321) for ApoA-I and 4% sodium dodecyl sulfate polyacrylamide gels for Apo-B. Western blot band density of target proteins was quantified with ImageJ and expressed relative to the density of loading controls or sample preparation controls, as indicated. Western blot quantification data can be found in Extended Data Fig. 6. Details regarding antibodies used are listed in Supplementary Table 1.
Gene expression analysis
RNA was extracted using TRIzol reagent (Invitrogen, 15596018) for both qPCR and total RNA-seq. Complementary DNA was generated by reverse transcription and quantified using iTaq Universal SYBR Green Supermix (Bio-Rad, 1725125) and the QuantStudio 6 Flex 384-well qPCR system (Applied Biosystems). Each gene of interest was normalized to 36b4. qPCR primers can be found in Supplementary Table 2. For RNA-seq, library preparation and sequencing were performed at GENEWIZ. Adaptor and quality trimming of raw FASTQ files was performed using Trimmomatic39. FastQC was used to analyze FASTQ files before and after trimming (Babraham Bioinformatics). Trimmed FASTQ files were aligned to GRCm38/mm10 using STAR40. HTSeq-count was used to extract gene counts and differential gene expression analysis was performed using DESeq2 (refs. 41, 42). Pathway analysis was performed using Enrichr43. To measure Aster-A, Aster-B and Aster-C copy numbers in the liver, RNA was extracted from liver as described above. cDNA was generated by reverse transcription and quantified relative to Aster-A, Aster-B or Aster-C plasmids that were diluted to give a standard curve in which Asters were present at 300,000 copies, 30,000 copies, 3,000 copies, 300 copies and 30 copies.
Tissue, feces and plasma lipid measurements
Tissues were disrupted in homogenization buffer (10 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA and protease inhibitors) and lipids were extracted from 0.5–1 mg liver protein using the Folch method44. Fecal samples were pulverized using a mortar and pestle and lipids were extracted from 200 mg feces using the Folch method. Liver and fecal cholesterol were measured by colorimetric assay (WAKO, NC9138103). Plasma total cholesterol (WAKO, NC9138103), TG (Sekisui, 36-100-4169) and non-esterified fatty acids (WAKO, 991-34891) were measured using colorimetric assays. To resolve lipoprotein classes, plasma samples were injected into a Superose 6 10/300 (GE Healthcare Life Sciences) column attached to ÄKTA Pure FPLC unit (GE Healthcare) and fractions were collected for measurement of cholesterol by colorimetric assay (WAKO, NC9138103).
Lipidomic analysis
Liver tissue (50–100 mg) was collected in a 2-ml homogenizer tube pre-loaded with 2.8-mm ceramic beads (Omni 19-628). PBS (0.75 ml) was added to the tube and the sample was homogenized in an Omni Bead Ruptor Elite (three cycles of 10 s at 5 m s−1 with a 10-s dwell time). Homogenate containing 2–6 mg of original tissue was transferred to a glass tube for extraction. A modified Bligh and Dyer extraction was carried out on all samples45. Before biphasic extraction, an internal standard mixture consisting of 70 lipid standards across 17 subclasses was added to each sample (AB Sciex, 5040156; Avanti, 330827, 330830, 330828 and 791642). Following two successive extractions, pooled organic layers were dried down in a Thermo SpeedVac SPD300DDA using ramp setting 4 at 35 °C for 45 min with a total run time of 90 min. Lipid samples were resuspended in 1:1 methanol/dichloromethane with 10 mM ammonium acetate and transferred to robovials (Thermo, 10800107) for analysis. Samples were analyzed on the Sciex 5500 with DMS device (Lipidyzer Platform) with an expanded targeted acquisition list consisting of 1,450 lipid species across 17 subclasses (or the original acquisition list of 1,100 lipids across 13 subclasses). A Differential Mobility Device on Lipidyzer was tuned with EquiSPLASH LIPIDOMIX (Avanti, 330731). Data analysis was performed on an in-house data analysis platform comparable to the Lipidyzer Workflow Manager46. Instrument method including settings, tuning protocol and MRM list is available from Su et. al.46 Quantitative values were normalized to mg of tissue.
BODIPY staining
Adrenal glands were dissected carefully and the surrounding fat tissue was removed. The glands were embedded in O.C.T. compound (Tissue-Tek, 4583), placed on dry ice for 30 min, then moved to −80 °C. Tissue was sectioned (12-mm thick) using a Microm HM 505 E cryostat and sections were placed on glass microscope slides. Before staining, sections were washed three times with PBS and fixed with 4% PFA for 30 min. After washing, sections were stained with 1 mg ml−1 BODIPY 493/503 (Thermo Fisher, D3922) for 30 min and washed with PBS. Sections were then mounted on slides with ProLong Diamond Antifade Mountant with DAPI (Invitrogen, P36962) and images were captured on a Zeiss Axioskop 2 Plus microscope with Zeiss ZEN software.
Plasma membrane isolation
Liver PMs were isolated as described previously47. In brief, fresh tissues (150–200 mg) were homogenized in TES buffer (250 mM sucrose, 10 mM Tris, pH 7.4, and 0.5 mM EDTA) at 4 °C. The homogenate was then centrifuged at 17,000 g with a SS-34 rotor for 15 min to separate pellet A and supernatant A. The top lipid layer was collected as lipid droplet fraction. Pellet A was washed by being resuspended in TES buffer and centrifuged at 17,000 g for 20 min and resuspended in TES buffer and gently layered on top of 1.12 M sucrose buffer cushion in ultracentrifuge tubes. The samples were then centrifuged at 105,000 g with a SW55Ti rotor for 20 min to separate pellet B, interface B and supernatant B. Interface B was collected, resuspended in TES buffer and centrifuged at 60,000 g for 9 min in an Optima MAX-XP Ultracentrifuge to obtain pellet C. Pellet C was then washed by being resuspended in TES buffer and centrifuged at 60,000g for 9 min to obtain the PM fraction. The supernatant A was centrifuged at 390,000 g for 75 min to separate pellet D and supernatant D. Pellet D was washed by being resuspended in TES buffer and centrifuged at 390,000 g for 60 min and was collected as the ER fraction. Supernatant D was collected as the cytosol fraction. PM free cholesterol was measured by Amplex Red Cholesterol Assay kit (Fisher Scientific, A12216). PM SM was measured by mass spectrometry as described above. PM cholesterol or SM was normalized to total PM lipids as measured by mass spectrometry. For assessment of PM lipids by thin-layer chromatography (TLC), lipids were extracted from isolated PMs (1 mg protein) using the Folch method. Lipid extracts were dried under nitrogen gas, resuspended in chloroform, spotted on silica plates and separated using the solvent systems chloroform:methanol:acetic acid:water (50:30:12:4) and heptane:isopropyl ether:acetic acid (60:40:4). The plates were sprayed with 8-anilino-1-naphthalenesulfonic acid and bands were visualized with a Transilluminator FBTI-88 (Fisher Scientific).
Purification of ALOD4
pALOD4 was a gift from A. Radhakrishnan (Addgene, plasmids 111026)18 and was purified as previously described48. Briefly, plasmids encoding specified proteins were expressed in BL21 (DE3) pLysS Escherichia coli (Invitrogen). Cell pellets were lysed by sonication in lysis buffer containing 50 mM NaH2PO4, pH 7.0, 300 mM NaCl, 1 mg ml−1 lysozyme, 1 mM dithiothreitol, 2 mM phenylmethylsulfonyl fluoride and protease inhibitor cocktail tablet (Themo Scientific). The protein was bound to HisPur Ni-NTA Agarose resin (Thermo Scientific), washed twice with 50 mM imidazole, eluted with 300 mM imidazole and subjected to size exclusion chromatography on Superdex 200. Fractions containing specified protein were concentrated to 1 mg ml−1 and stored at −80 °C with glycerol until use. For ALOD4 labeling, ALOD4 was conjugated to Alexa Fluor 594 followed by affinity chromatography using HisPur Ni-NTA Agarose resin as above. Samples were dialyzed to remove imidazole before determination of labeling efficiency on Nanodrop (Thermo Fisher).
ALOD4 staining
Primary hepatocytes were washed with DPBS containing 0.2% BSA three times. The ALOD4 working solution (20 μg ml−1 in DPBS + 0.2% BSA) was passed through a 0.45-μm filter before addition to cells at 4 °C in the dark for 2 h. For confocal imaging, cells were washed three times with DPBS before being fixed with 3% PFA for 15 min at room temperature. After fixation, cells were washed with DPBS three times. Cells were incubated with an anti-His antibody (27E8, Cell Signaling, 2366S; diluted 1:1,000) overnight before incubation with fluorescently labeled secondary antibodies for 1 h at room temperature (goat anti-mouse IgG H&L, Alexa Fluor 488, Invitrogen, A11001; diluted 1:1,000). Cells were mounted on slides with ProLong Diamond Antifade Mountant with DAPI (Invitrogen, P36962). Images were acquired using an Inverted Leica TCS-SP8-SMD Confocal Microscope. For western blot analysis, cells were lysed with RIPA buffer before being probed with anti-His (27E8, Cell Signaling, 2366S; diluted 1:1,000) or anti-actin antibodies (Millipore Sigma, A2066; diluted 1:10,000).
The method for the ALOD4 signal quantification in cells has been previously described49. Briefly, images were taken at the UCLA Molecular Screening Shared Resource core facility on a Molecular Devices ImageXpress confocal imaging system using a ×20 objective (Nikon Plan Fluor; NA 0.3). Integrated fluorescence intensity profiles were exported and analyzed in R with the ggplot2 package or in GraphPad Prism 9. For flow cytometry analysis, primary hepatocytes were stained with ALOD4-Alexa Fluor 594 and Fixable Viability Dye eFluor450 (eBioscience, to exclude dead cells) before fixation with PFA. After washing, cells were resuspended in FACS buffer (2% FCS and 2 mM EDTA in DPBS) and analyzed on a flow cytometer (LSR-II). ALOD4 levels of eFluor450-negative, live cells were analyzed by FlowJo v.10.
Isolation and radiolabeling of HDL and LDL
LDL (density 1.019–1.063 g ml−1) and HDL (density 1.07–1.21 g ml−1) was isolated from WT mice by density-gradient ultracentrifugation before being extensively dialyzed (5 mM Tris (pH 7.4), 50 mM NaCL and 5 mM EDTA) and labeled with [14C]-cholesterol, as described previously23,50. Briefly, [14C]-cholesterol (20 μCi) was dried under nitrogen and sonicated in 1 ml phosphate-buffered saline for 30 min at 37 °C. The suspension was incubated with LDL or HDL (3 mg total protein content) in the presence of thimerosal (3 mM; Sigma, T5125), butylated hydroxytoluene (11.24 mM; Sigma, Phr1117), 6-hydroxy-2,5,7,8-tetramethylchroman-2-carboxylic acid (3.33 mM; Sigma, 238813), protease inhibitors (Thermo Scientific Pierce, A32965) and 3 ml lipoprotein-deficient serum (LPDS; density >1.21 g ml−1). LPDS was heated at 60 °C for 10 min to inactivate lecithin:cholesterol acyltransferase activity before use50,51. The mixture was layered with nitrogen gas in a glass tube and incubated at 37 °C for 24 h with gentle shaking (300 r.p.m.) before re-isolation of the radiolabeled lipoproteins by density-gradient ultracentrifugation. The lipoproteins were dialyzed for 24 h including three buffer changes at 4 °C before determination of protein concentration by bicinchoninic acid assay. Radiolabeled lipoproteins were stored at 4 °C under nitrogen and used within 1 week.
Determination of hepatic HDL and LDL uptake in mice
Radiolabeled HDL or LDL was injected into mice via the tail vein. Blood was collected at 1, 5, 10, 15, 30 and 60 min after injection. The radioactivity in plasma at 1 min after injection was be defined as 100% of injected radiolabel. For acute lipoprotein uptake studies, mice were killed 60 min after the initial injection for collection of the liver and gallbladder bile. For assessment of fecal bile acid and cholesterol output, mice were killed 3 d after lipoprotein injection. Radioactivity was determined in plasma at each time point and in terminal gallbladder bile by liquid scintillation counting. Lipids were extracted from the whole liver by the method of Folch. Cholesterol and CE were separated by TLC on silica plates using the solvent system heptane: isopropyl ether: acetic acid (60:40:4). The radioactivity in each lipid fraction was determined by liquid scintillation counting after scraping the relevant bands. Fecal samples (100 mg) were separated into aqueous (containing bile salts) and lipid fractions by the method of Folch before determination of the radioactivity in each fraction. Liver total [14C] radioactivity was normalized per gram of liver and expressed relative to radioactivity in plasma 1 min after lipoprotein injection (which we defined as 100% of radioactivity administered). [14C] radioactivity in liver CEs was normalized per mg protein and expressed relative to radioactivity in plasma 1 min after lipoprotein injection. Total biliary [14C] counts are normalized per μl bile and expressed relative to radioactivity in plasma 1 min after lipoprotein injection. Radioactivity in 14C-cholesterol-HDL and 14C-cholesterol-LDL experiments is expressed as c.p.m.
SM degradation in cultured cells
Cellular SM degradation in response to OA, AA or PA treatment of HepG2 cells was assessed as described previously with minor modifications52. Briefly, HepG2 cells were plated in six-well plates at a density of 1 × 106 cells per well and incubated with 1 μCi ml−1 [methyl-3H]choline chloride (PerkinElmer, NET109250UC) for 72 h. After incubation, cells were washed three times with Hank’s Balanced Salt Solution before being treated with 300 μM BSA-conjugated oleate, arachidonate or palmitate in plain EMEM for 60 min. Cells were then washed and collected in homogenization buffer (10 mM Tris (pH 7.4), 150 mM NaCl and 1 mM EDTA). The protein content of the samples was determined by BCA assay and lipids were extracted from 60 μg protein by the Folch method. Lipids were dried under nitrogen gas and resuspended in 100 μl of assay buffer consisting of 100 mM Tris-HCL (pH 7.4), 6 mM MgCI2, 0.1% Triton X-100, 10 nmol ml−1 18:1 phosphatidylserine (Avanti, 840035 C) and 1 U ml−1 SMase from Bacillus cereus (Avanti, S9396). Reactions were incubated at 37 °C for 2 h. Reactions were terminated by addition of 1.5 ml chloroform:methanol (2:l). Water (200 μl) was added to complete the Folch extraction. SMase treatment releases [3H]-phosphorylcholine from SM, which can be recovered in the aqueous phase of a Folch extraction. Radioactivity in [3H]-SM was measured by drying and counting the aqueous phase and radioactivity in [3H]-PC was measured by drying and counting the organic phase. Radioactivity in SM was normalized to radioactivity in PC.
VLDL isolation
Plasma (150 μl) was overlayed with 150 μl of 0.15 M NaCl solution with density d = 1.006 kg l−1. The samples were then centrifuged at 627,000 g in an Optima MAX-XP Ultracentrifuge for 100 min at 18 °C. After centrifugation, the top layer was collected as the VLDL fraction (~100 μl).
VLDL secretion assays
F/F and L-A/C KO mice were fasted overnight before receiving an intraperitoneal injection of Poloxamer-407 in sterile PBS (1 g kg−1). Blood was collected at the time of injection and every subsequent hour for 4 h. Plasma TG concentrations were assessed by enzymatic assay (Sekisui, 36-100-4169). The VLDL production rate was calculated from the slope of the line of plasma TG concentrations relative to time for each individual mouse.
Cholesterol efflux assay
The cholesterol efflux protocol was adapted and modified from previous studies53. Briefly, primary hepatocytes were plated on collagen-coated six-well plates and incubated with maintenance medium (William Medium E medium with 5% BSA, 0.1 uM dexamethasone and 100 nM insulin) containing [3H]-cholesterol (3 μCi per well) and 2 μg ml−1 acyl-coA cholesterol acyltransferase inhibitor (Sandoz) for 16 h at 37 °C. After 16 h, cells were rinsed three times with PBS containing 2 mg ml−1 BSA and incubated with 1.5 ml of maintenance medium containing ApoA-I (20 μg ml−1) for 4 h at 37 °C. Medium was collected from each well into 2-ml Eppendorf tubes and centrifuged at 1,500 g for 10 min at room temperature to pellet floating cells. The supernatant was carefully removed and 1.2 ml supernatant was used to determine radioactivity by liquid scintillation counting in 5 ml scintillation buffer. Radioactive counts were normalized to protein content as determined by bicinchoninic acid assay.
siRNA knockdown
Immortalized human aortic endothelial cells were plated in MCDB-131 complete medium (VEC Technologies, WOFBS) containing 10% FBS at a density of 1 × 106 cells per well of a six-well plate for 24 h. The cells were washed once before being transfected with 100 nM control siRNA (Thermo Fisher, 4390843) or Scarb1 siRNA (Thermo Fisher, s2648) using Lipofectamine 3000 (Thermo Fisher, L3000008) in OptiMEM for 5 h. Cells were switched to MCDB-131 complete medium with 10% FBS for 24 h before being depleted overnight in MCDB-131 containing 1% LPDS, simvastatin (5 mM) and mevalonate (50 mM). Depleted cells were either loaded with HDL (400 μg ml−1) or maintained in LPDS for 1 h before ALOD4 staining and collection for western blotting as described above.
Determination of whole-body cholesterol content
Whole-body cholesterol content of F/F and L-A/C KO mice was determined as described previously54. Briefly, each mouse was killed and placed in a 250 ml beaker containing 10 g KOH and 150 ml ethanol. The beaker was covered in aluminum foil and left at room temperature for 1 week, stirring contents once per day. The beaker was then placed on a boiling water bath and stirred until the volume of ethanol was reduced to approximately 50 ml. The contents were filtered through gauze into a volumetric flask. The beaker was rinsed three times some ethanol and the remaining contents were filtered into the flask. The total volume of the of flask was brought up to 100 ml with ethanol. Lipids were extracted from 2 ml flask contents and the cholesterol content was measured by enzymatic assay (WAKO, NC9138103).
Experimental replicates and quantification
Replicates are described in the figure legends. For cellular assays, n corresponds to the number of biological replicates. For animal experiments, n corresponds to the number of mice used per genotype or condition. All data are presented as mean ± s.e.m. as described in the figure legends.
Statistical analysis
All data are presented as mean ± s.e.m. and analyzed using Prism (GraphPad v.9). A two-sided Student’s t-test was used for single variable comparison between two groups. Multiple comparisons were corrected for using the Benjamini, Krieger and Yekutieli method where appropriate. A two-way ANOVA followed by Sidak’s post-test was used to examine interactions between multiple variables. A one-way ANOVA with Tukey’s or Dunnett’s post-test was used to compare a single independent variable between multiple groups. P < 0.05 was considered statistically significant. Exact P values are indicated in the figures.
Extended Data
Extended Data Fig. 1 ∣. Generation of Aster-C 3xHA KI and Asters hepatocyte specific KO mice.
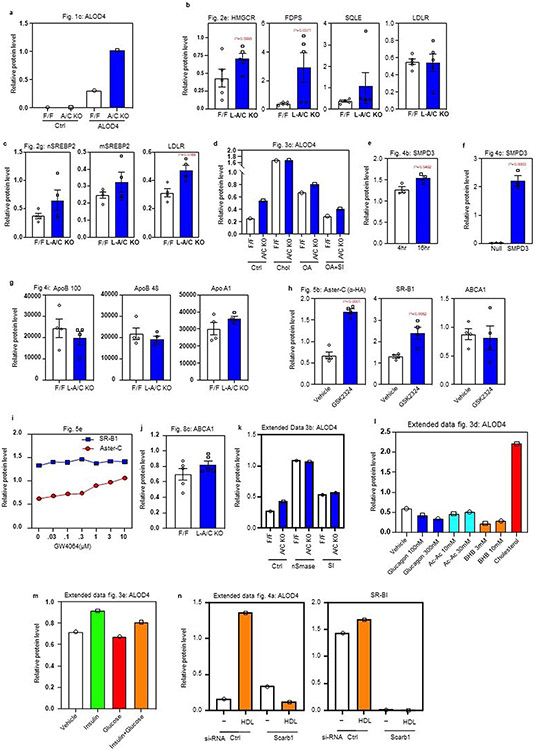
a, Quantitative PCR of Asters expression in mouse liver (n = 6). b, Evaluation of 3xHA-Aster-C KI mice. Genotyping results (upper) and anti-HA tag immunoblot (bottom) for 3xHA-Aster-C KI mice. The genotyping result is representative of at least 50 similar results. The anti-HA-Aster-C western blot was repeated independently in Fig. 5e. c, Strategy for generating Gramd1a (Aster-A) and Gramd1c (Aster-C) Flox/Flox (F/F) mice. Coding exons are depicted in black. Exons that correspond to the GRAM domain, ASTER domain, and transmembrane (TM) domain are depicted in green, blue, and red respectively. Scale bar represents 1 kb. d, Genotyping results for L-A KO, L-C KO and L-A/C KO mice. The genotyping results are representative of at least 100 similar results per condition. *****(e, f and g) Expression levels of the indicated genes in liver from F/F control and L-A KO mice (e, n = 8/11); L-C KO mice (f, n = 5/6); L-A/C KO mice (g, n = 12/6). h, Gramd1a, Gramd1b and Gramd1c expression level in the testis of F/F control and L-A/C KO mice (n = 12/6). All data are presented as mean ± SEM. P values were determined by two-sided Student’s t-test with Benjamini, Krieger and Yekutieli correction for multiple comparisons (e, f, g and h).
Extended Data Fig. 2 ∣. Fasting stimulates hepatic PM–ER cholesterol transport.
a, Gross appearance of livers from mice fasted for 4- or 16-h. b, Hepatic triglycerides in mice fasted for 4- or 16-h (n = 3/3). c, Hepatic CE in mice fasted for 4- or 16-h (n = 3/3). d, Significantly upregulated pathways in the livers of mice fasted for 16-h compared to 4-h according to pathway analysis of RNA Sequencing data. e, Significantly downregulated pathways in the livers of mice fasted for 16-h compared to 4-h according to pathway analysis of RNA Sequencing data. f, Hepatic mRNA expression of SREBP-2 pathway targets from the livers of mice fasted for 4- or 16-h (n = 3/3). g, Quality control of plasma membrane isolation from the mouse liver. Cadherin: PM maker; Calnexin: ER maker; ATGL: lipid droplet (LD) maker; Actin: cytoskeleton maker. This analysis was completed once as the organelle isolation method has been previously validated (further method details are in the Methods section). h, Free cholesterol analysis from purified PMs of wild-type mice (n = 3/3). i, TLC analysis of free cholesterol (FC), sphingomyelin (SM) and phosphatidylcholine (PC) from the livers of mice fasted for either 4 or 16 h. j, PM total lipids as measured by mass-spec from livers of F/F control and L-A/C KO after 4- and 16-h fasting (n = 3/3). All data are presented as mean ± SEM. P values were determined by two-sided Student’s t-test (b, c and h), or two-sided Student’s t-test with Benjamini, Krieger and Yekutieli correction (f).
Extended Data Fig. 3 ∣. Aster-mediated cholesterol transport determines the size of the accessible PM cholesterol pool.
a, PM total lipids from mouse liver after 4- and 16-h fasting as determined by mass spec (n = 3/3). b, Immunoblot analysis of ALOD4 binding in F/F control and L-A/C KO primary hepatocytes after treatment with vehicle, nSmase (100 mU/ml) and GW4869 (10 μM) for 1-h. Actin was used as a loading control. c, Expression levels of the indicated genes in primary hepatocyte that had been cultured in Maintenance medium and Ro 48-8071 (1μM) overnight before being treated with or without oleic acid (30μM) for 6-h (n = 3). d, Immunoblot analysis of ALOD4 binding in primary hepatocytes after treatment with vehicle or indicated concentration of glucagon, acetoacetate (Ac-Ac), beta-hydroxybutyrate (BHB) or MβCD-cholesterol (35 μM) for 1-h. Calnexin was used as a loading control. e, Immunoblot analysis of ALOD4 binding in primary hepatocytes after treatment with vehicle or insulin (300 nM), glucose (50 mM) and both for 1-h. Calnexin was used as a loading control. All data are presented as mean ± SEM.
Extended Data Fig. 4 ∣. Asters transport lipoprotein-derived cholesterol from the PM to ER in hepatocytes.
a. Immunoblot analysis of ALOD4 binding in control or Sr-b1 knockdown HAECs after treatment with vehicle or HDL (400 ug/ml) for 1-h. Calnexin was used as a sample processing control. b, [14C] counts in liver unesterified cholesterol of mice from Fig. 6c (n = 10/6). c, mRNA expression levels of the indicated genes in the livers of mice from Fig. 6c (n = 10/6). d, [14C] counts in unesterified cholesterol in the livers of mice from Fig. 6g (n = 7/9). e, mRNA expression levels of the indicated genes in the livers of mice from Fig. 6g (n = 7/9). f, The rate of [14C] lipoprotein clearance from the circulation (n = 6/5), related to Figs. 6j-6m. Data are represented as mean ± SEM with individual animals noted as dots. *p < 0.05. g. Fecal cholesterol analysis of F/F and L-A/C KO mice fed a cholesterol-free diet for 48 hours (n = 7/7). h, Growth curves for F/F and L-A/C KO mice fed a Western diet from 8 weeks of age (n = 8/12). Masses are shown as mean ± SEM. i, Whole-body cholesterol content from F/F control and L-A/C KO mice on chow diet at 15 weeks old (n = 5/7). All data are presented as mean ± SEM. P values were determined by two-sided Student’s t-test (b, d, g and i), two-sided Student’s t-test with Benjamini, Krieger and Yekutieli correction (c and e), or two-way ANOVA with Sidak’s correction for multiple comparisons (h).
Extended Data Fig. 5 ∣. Model for the role of hepatic Asters in liver and systemic cholesterol homeostasis.
In normal physiology (left side of schematic), fasting stimulates fatty acid release from adipose tissue to promote hepatic sphingomyelinase activity, which liberates sequestered cholesterol in the hepatocyte PM. Aster proteins recognize this newly accessible cholesterol and transport it to the ER for CE formation, suppression of SREBP-2 pathway targets, bile acid synthesis and VLDL production. Hepatic Asters are also induced by FXR and function in the RCT pathway by moving HDL-derived cholesterol (and LDL-derived cholesterol) within hepatocytes. Loss of hepatic Aster function (right side of the schematic) impairs CE formation and VLDL output during fasting. Loss of Asters in the liver also decreases the appearance of HDL-derived cholesterol in bile and feces, raises plasma cholesterol levels (due to enhanced liver cholesterol efflux), and causes peripheral cholesterol accumulation (for example, adrenal gland, brown adipose tissue).
Extended Data Fig. 6 ∣. Western blot quantifications.
a, Western blot quantifications of ALOD4 binding from Fig. 1c. Normalized to loading control Actin (n = 1). b, Western blot quantifications of indicated proteins from Fig. 1c. Normalized to sample processing control calnexin (n = 5). c, Western blot quantifications of indicated proteins from Fig. 2g. Normalized to sample processing controls calnexin or lamin a/c (n = 4). d, Western blot quantifications of ALOD4 binding from Fig. 3c. Normalized to loading control actin (n = 1). e and f, Western blot quantifications of SMPD3 from Fig. 4b and Fig. 4c. Normalized to loading control actin (n = 3). g, Western blot quantifications of indicated proteins from Fig. 4i. Equal amounts of protein was loaded for each line (n = 4). h, Western blot quantifications of indicated proteins from Fig. 5b. Normalized to sample processing control calnexin (n = 4). i, Western blot quantifications of indicated proteins from Fig. 5e. Normalized to actin which served as a loading control for SRB1 and a sample processing control for Aster-C (n = 1). j, Western blot quantifications of ABCA1 from Fig. 8c. Normalized to sample processing control calnexin (n = 5). k, l and m, Western blot quantifications of ALOD4 binding from extended data Figs. 3b, 3d and 3e. Normalized to loading control actin or calnexin (n = 1). n, Western blot quantifications of ALOD4 binding and Sr-B1 from Fig. 4a. Normalized to calnexin which served as a loading control for ALOD4 and a sample processing control for SRB1 (n = 1). Data are presented as mean ± SEM. P values were determined by two-sided Student’s t-test (b, c, e, f and h).
Supplementary Material
Acknowledgements
We thank all members of the Tontonoz, Tarling-Vallim, Edwards, Villanueva, Young and Bensinger laboratories at UCLA for useful advice and discussions and for sharing reagents and resources. We thank K. Williams, G. Su and staff at UCLA Lipidomics core for the lipidomics analysis. Confocal microscopy was performed at the California NanoSystems Institute of Advanced Light Microscopy/Spectroscopy Facility. We thank the Vector Core of the University of Michigan for AAV packaging. We thank J. Smothers and A. Radhakrishnan for the helpful suggestions about the ALOD4 protein purification. This work was supported by NIH grant R01 DK126779 and Fondation Leducq Transatlantic Network of Excellence (19CVD04). X.X. was supported by AHA Postdoctoral Fellowship (18POST34030388). J.P.K. is supported by AHA Postdoctoral Fellowship (903306). A.F. was funded by Ermenegildo Zegna Founder’s Scholarship (2017) and by American Diabetes Association postdoctoral fellowship (1-19-PDF-043-RA). Y.G. is supported by Damon Runyon Cancer Research Foundation and Mark Foundation postdoctoral fellowship (DRG2424-21). R.T.N. was supported by a T32GM008042 grant to the UCLA-Caltech Medical Scientist Training Program. A.N. was supported by the NIDDK of the National Institutes of Health under Award Number T32DK007180.
Footnotes
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Competing interests
The authors declare no competing interests.
Extended data is available for this paper at https://doi.org/10.1038/s42255-022-00722-6.
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s42255-022-00722-6.
Data availability
The RNA-seq dataset generated for this paper has been deposited to the National Center for Biotechnology Information and is available at accession no. GSE206278. For RNA-seq analysis, trimmed FASTQ files were aligned to GRCm38/mm10 using STAR40. Source data are provided with this paper.
References
- 1.Lange Y, Swaisgood MH, Ramos BV & Steck TL Plasma membranes contain half the phospholipid and 90% of the cholesterol and sphingomyelin in cultured human fibroblasts. J. Biol. Chem 264, 3786–3793 (1989). [PubMed] [Google Scholar]
- 2.Das A, Brown MS, Anderson DD, Goldstein JL & Radhakrishnan A Three pools of plasma membrane cholesterol and their relation to cholesterol homeostasis. eLife 3, e02882 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Radhakrishnan A, Goldstein JL, McDonald JG & Brown MS Switch-like control of SREBP-2 transport triggered by small changes in ER cholesterol: a delicate balance. Cell Metab. 8, 512–521 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson RA et al. Identification of a form of acyl-CoA: cholesterol acyltransferase specific to liver and intestine in nonhuman primates. J. Biol. Chem 273, 26747–26754 (1998). [DOI] [PubMed] [Google Scholar]
- 5.Pikuleva IA Cholesterol-metabolizing cytochromes P450. Drug Metab. Dispos 34, 513–P520 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Sandhu J et al. Aster proteins facilitate nonvesicular plasma membrane to ER cholesterol transport in mammalian cells. Cell 175, 514–529 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Naito T et al. Movement of accessible plasma membrane cholesterol by GRAMD1 lipid transfer protein complex. eLife 8, e51401 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xiao X et al. Selective Aster inhibitors distinguish vesicular and nonvesicular sterol transport mechanisms. Proc. Natl Acad. Sci. USA 118, e2024149118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trinh MN et al. Interplay between Asters/GRAMD1s and phosphatidylserine in intermembrane transport of LDL cholesterol. Proc. Natl Acad. Sci. USA 119, e2120411119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferrari A et al. Aster proteins regulate the accessible cholesterol pool in the plasma membrane. Mol. Cell. Biol 40, e00255–20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kennelly JP & Tontonoz P Cholesterol transport to the endoplasmic reticulum. Cold Spring Harb. Perspect. Biol 10.1101/cshperspect.a041263 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edwards PA, Muroya H & Gould RG In vivo demonstration of the circadian rhythm of cholesterol biosynthesis inthe liver and intestine of the rat. J. Lipid Res 13, 396–401 (1972). [PubMed] [Google Scholar]
- 13.Horton JD, Bashmakov Y, Shimomura I & Shimano H Regulation of sterol regulatory element binding proteins in livers of fasted and refed mice. Proc. Natl Acad. Sci. USA 95, 5987–5992 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tall A An overview of reverse cholesterol transport. Atherosclerosis 109, 337 (1994). [Google Scholar]
- 15.Acton S et al. Identification of scavenger receptor SR-BI as a high-density lipoprotein receptor. Science 271, 518–520 (1996). [DOI] [PubMed] [Google Scholar]
- 16.Zhang Y et al. Identification of novel pathways that control farnesoid X receptor-mediated hypocholesterolemia. J. Biol. Chem 285, 3035–3043 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schaum N et al. Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature 562, 367–372 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gay A, Rye D & Radhakrishnan A Switch-like responses of two cholesterol sensors do not require protein oligomerization in membranes. Biophys. J 108, 1459–1469 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Worgall TS, Johnson RA, Seo T, Gierens H & Deckelbaum RJ Unsaturated fatty acid-mediated decreases in sterol regulatory element-mediated gene transcription are linked to cellular sphingolipid metabolism. J. Biol. Chem 277, 3878–3885 (2002). [DOI] [PubMed] [Google Scholar]
- 20.Simcox J et al. Global analysis of plasma lipids identifies liver-derived acylcarnitines as a fuel source for brown fat thermogenesis. Cell Metab. 26, 509–522 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Insausti-Urkia N, Solsona-Vilarrasa E, Garcia-Ruiz C & Fernandez-Checa JC Sphingomyelinases and liver disease. Biomolecules 10, 1497 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thomas AM et al. Genome-wide tissue-specific farnesoid X receptor binding in mouse liver and intestine. Hepatology 51, 1410–1419 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Redgrave TG, Roberts DCK & West CE Separation of plasma lipoproteins by density-gradient ultracentrifugation. Anal. Biochem 65, 42–49 (1975). [DOI] [PubMed] [Google Scholar]
- 24.Brown MS & Goldstein JL A receptor-mediated pathway for cholesterol homeostasis. Science 232, 34–47 (1986). [DOI] [PubMed] [Google Scholar]
- 25.Tabas I, Rosoff WJ & Boykow GC Acyl coenzyme A: cholesterol acyl transferase in macrophages utilizes a cellular pool of cholesterol oxidase-accessible cholesterol as substrate. J. Biol. Chem 263, 1266–1272 (1988). [PubMed] [Google Scholar]
- 26.Ji Y et al. Hepatic scavenger receptor BI promotes rapid clearance of high-density lipoprotein free cholesterol and its transport into bile. J. Biol. Chem 274, 33398–33402 (1999). [DOI] [PubMed] [Google Scholar]
- 27.Feng Y et al. Hepatocyte-specific ABCA1 transfer increases HDL cholesterol but impairs HDL function and accelerates atherosclerosis. Cardiovasc. Res 88, 376–385 (2010). [DOI] [PubMed] [Google Scholar]
- 28.Singaraja RR et al. Human ABCA1 BAC transgenic mice show increased high-density lipoprotein cholesterol and ApoAI-dependent efflux stimulated by an internal promoter containing liver X receptor response elements in intron 1. J. Biol. Chem 276, 33969–33979 (2001). [DOI] [PubMed] [Google Scholar]
- 29.Basso F et al. Role of the hepatic ABCA1 transporter in modulating intrahepatic cholesterol and plasma HDL cholesterol concentrations. J. Lipid Res 44, 296–302 (2003). [DOI] [PubMed] [Google Scholar]
- 30.Timmins JM et al. Targeted inactivation of hepatic Abca1 causes profound hypoalphalipoproteinemia and kidney hypercatabolism of apoA-I. J. Clin. Invest 115, 1333–1342 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rigotti A et al. A targeted mutation in the murine gene encoding the high-density lipoprotein (HDL) receptor scavenger receptor class B type I reveals its key role in HDL metabolism. Proc. Natl Acad. Sci. USA 94, 12610–12615 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu X-Y et al. Feeding induces cholesterol biosynthesis via the mTORC1–USP20–HMGCR axis. Nature 588, 479–484 (2020). [DOI] [PubMed] [Google Scholar]
- 33.Yu L et al. Expression of ABCG5 and ABCG8 is required for regulation of biliary cholesterol secretion. J. Biol. Chem 280, 8742–8747 (2005). [DOI] [PubMed] [Google Scholar]
- 34.Kosters A et al. The mechanism of ABCG5/ABCG8 in biliary cholesterol secretion in mice 1. J. Lipid Res 47, 1959–1966 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clifford BL et al. FXR activation protects against NAFLD via bile-acid-dependent reductions in lipid absorption. Cell Metab. 33, 1671–1684 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sinal CJ et al. Targeted disruption of the nuclear receptor FXR/BAR impairs bile acid and lipid homeostasis. Cell 102, 731–744 (2000). [DOI] [PubMed] [Google Scholar]
- 37.de Aguiar Vallim TQ et al. MAFG is a transcriptional repressor of bile acid synthesis and metabolism. Cell Metab. 21, 298–311 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tarling EJ et al. RNA-binding protein ZFP36L1 maintains posttranscriptional regulation of bile acid metabolism. J. Clin. Invest 127, 3741–3754 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bolger AM, Lohse M & Usadel B Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dobin A et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Anders S, Pyl PT & Huber W HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Love MI, Huber W & Anders S Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen EY et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14, 128 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Folch J, Lees M & Stanley GHS A simple method for the isolation and purification of total lipids from animal tissues. J. Biol. Chem 226, 497–509 (1957). [PubMed] [Google Scholar]
- 45.Hsieh W-Y et al. Toll-like receptors induce signal-specific reprogramming of the macrophage lipidome. Cell Metab. 32, 128–143 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Su B et al. A DMS shotgun lipidomics workflow application to facilitate high-throughput, comprehensive lipidomics. J. Am. Soc. Mass Spectrom 32, 2655–2663 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lyu K et al. A membrane-bound diacylglycerol species induces PKCϵ-mediated hepatic insulin resistance. Cell Metab. 32, 654–664 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Endapally S, Infante RE & Radhakrishnan A Monitoring and modulating intracellular cholesterol trafficking using ALOD4, a cholesterol-binding protein. Methods Mol. Biol 1949, 153–163 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou QD et al. Interferon-mediated reprogramming of membrane cholesterol to evade bacterial toxins. Nat. Immunol 21, 746–755 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Robichaud JC, van der Veen JN, Yao Z, Trigatti B & Vance DE Hepatic uptake and metabolism of phosphatidylcholine associated with high-density lipoproteins. Biochim. Biophys. Acta 1790, 538–551 (2009). [DOI] [PubMed] [Google Scholar]
- 51.Terpstra AH, Nicolosi RJ & Herbert PN In vitro incorporation of radiolabeled cholesteryl esters into high and low-density lipoproteins. J. Lipid Res 30, 1663–1671 (1989). [PubMed] [Google Scholar]
- 52.Jayadev S, Linardic CM & Hannun YA Identification of arachidonic acid as a mediator of sphingomyelin hydrolysis in response to tumor necrosis factor α. J. Biol. Chem 269, 5757–5763 (1994). [PubMed] [Google Scholar]
- 53.Low H, Hoang A & Sviridov D Cholesterol efflux assay. J. Vis. Exp 10.3791/3810 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zabalawi M et al. Inflammation and skin cholesterol in LDLr−/−, apoA-I−/− mice: link between cholesterol homeostasis and self-tolerance? J. Lipid Res 48, 52–65 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq dataset generated for this paper has been deposited to the National Center for Biotechnology Information and is available at accession no. GSE206278. For RNA-seq analysis, trimmed FASTQ files were aligned to GRCm38/mm10 using STAR40. Source data are provided with this paper.