replying to: T. Spisak et al. Nature 10.1038/s41586-023-05745-x (2023)
In our previous study1, we documented the effect of sample size on the reproducibility of brain-wide association studies (BWAS) that aim to cross-sectionally relate individual differences in human brain structure (cortical thickness) or function (resting-state functional connectivity (RSFC)) to cognitive or mental health phenotypes. Applying univariate and multivariate methods (for example, support vector regression (SVR)) to three large-scale neuroimaging datasets (total n ≈ 50,000), we found that overall BWAS reproducibility was low for n < 1,000, due to smaller than expected effect sizes. When samples and true effects are small, sampling variability, and/or overfitting can generate ‘statistically significant’ associations that are likely to be reported due to publication bias, but are not reproducible2–5, and we therefore suggested that BWAS should build on recent precedents6,7 and continue to aim for samples in the thousands. In the accompanying Comment, Spisak et al.8 agree that larger BWAS are better5,9, but argue that “multivariate BWAS effects in high-quality datasets can be replicable with substantially smaller sample sizes in some cases” (n = 75–500); this suggestion is made on the basis of analyses of a selected subset of multivariate cognition/RSFC associations with larger effect sizes, using their preferred method (ridge regression with partial correlations) in a demographically more homogeneous, single-site/scanner sample (Human Connectome Project (HCP), n = 1,200, aged 22–35 years).
There is no disagreement that a minority of BWAS effects can replicate in smaller samples, as shown with our original methods1). Using the exact methodology (including cross-validation) and code of Spisak et al.8 to repeat 64 multivariate BWAS in the 21-site, larger and more diverse Adolescent Brain Cognitive Development Study (ABCD, n = 11,874, aged 9–11 years), we found that 31% replicated at n = 1,000, dropping to 14% at n = 500 and none at n = 75. Contrary to the claims of Spisak et al.8, replication failure was the outcome in most cases when applied to this larger, more diverse dataset. Basing general BWAS sample size recommendations on the largest effects has at least two fundamental flaws: (1) failing to detect other true effects (for example, reducing the sample size from n = 1,000 to n = 500 leads to a 55% false-negative rate), therefore restricting BWAS scope, and (2) inflation of reported effects3,10–12. Thus, regardless of the method, associations based on small samples can remain distorted and lack generalizability until confirmed in large, diverse, independent samples.
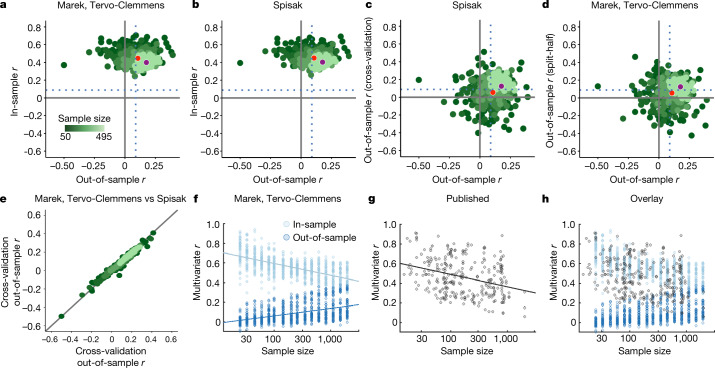
We always test for BWAS replication with null models (using permutation tests) of out-of-sample estimates to ensure that our reported reproducibility is unaffected by in-sample overfitting. Nonetheless, Spisak et al.8 argue against plotting inflated in-sample estimates1,10 on the y axis, and out-of-sample values on the x axis, as we did (Fig. 1a). Instead, they propose plotting cross-validated associations from an initial, discovery sample (Fig. 1b (y axis)) against split-half out-of-sample associations (x axis). However, cross-validation—just like split-half validation—estimates out-of-sample, and not in-sample, effect sizes13. The in-sample associations1,10 for the method of Spisak et al.8 (Fig. 1b), that is, from data in the sample used to develop the model, show the same degree of overfitting (Fig. 1a versus Fig. 1b). The plot of Spisak et al.8 (Fig. 1c) simply adds an additional out-of-sample test (cross-validation before split half), and therefore demonstrates the close correspondence between two different methods for out-of-sample effect estimation14. Analogously, we can replace the cross-validation step in the code of Spisak et al.8 with split-half validation (our original out-of-sample test), obtaining split-half effects in the first half of the sample, and then comparing them to the split-half estimates from the full sample (Fig. 1d). The strong correspondences between cross-validation followed by split-half (Spisak et al. method8; Fig. 1c) and repeated split-half validation (Fig. 1d) are guaranteed by plotting out-of-sample estimates (from the same dataset) against one another. Here, plotting cross-validated discovery sample estimates on the y axis (Fig. 1c,d) provides no additional information beyond the x axis out-of-sample values. The critically important out-of-sample predictions, required for reporting multivariate results1, generated using the method of Spisak et al.8 and our method are nearly identical (Fig. 1e).
Fig. 1. In-sample versus out-of-sample effect estimates in multivariate BWAS.
a–e, Methods comparison between our previous study1 (split-half) and Spisak et al.8 (cross-validation followed by split-half). ‘Marek, Tervo-Clemmens’ and ‘Spisak’ refer to the methodolgies described in ref. 1 and ref. 8, respectively. For a–e, HCP 1200 Release (full correlation) data were used to predict age-adjusted total cognitive ability. Analysis code and visualizations (x,y scaling; colours) are the same as in Spisak et al.8. The x axes in a–e always display the split-half out-of-sample effect estimates from the second (replication) half of the data (correlation between true scores and predicted scores; as in Spisak et al.8 and in our previous study1; Supplementary Methods). a, In-sample (training correlation; y axis) as a function of out-of-sample associations (plot convention in our previous study1). b, Matched comparison of the true in-sample association (training correlations, mean across folds; y axis) in the method proposed by Spisak et al.8. c, The proposed correction by Spisak et al.8 that inserts an additional cross-validation step to evaluate the first half of data, which by definition makes this an out-of-sample association (y axis). d, Replacing the cross-validation step from Spisak et al.8 with a split-half validation provides a different (compared with c) out-of-sample association of the first half of the total data (that is, each of the first stage split halves is one-quarter of the total data; y axis). The appropriate and direct comparison of in-sample associations between Spisak et al.8 and our previous study1 is comparing b to a, rather than c to a. The Spisak et al. method8 (cross-validation followed by split-half validation) does not reduce in-sample overfitting (b) but, instead, adds an additional out-of-sample evaluation (c), which is nearly identical to split-half validation twice in a row (d), and makes it clear why the out-of-sample performance of these two methods is likewise nearly identical. e, Correspondence between out-of-sample associations (to the left-out half) from the additional cross-validation step proposed by Spisak et al.8 (mean across folds; y axis) and the original split-half validation from our previous study1 (x axis). The identity line is shown in black. f, In-sample (r; light blue) and out-of-sample (r; dark blue) associations as a function of sample size. Data are from figure 4a–d of ref. 1. g, Published literature review of multivariate r (y axis) as a function of sample size (data from ref. 15) displayed with permission. For f and g, best fit lines are displayed in log10 space. h, Overlap of f and g.
As Spisak et al.8 highlight, cross-validation of some type is considered to be standard practice10, and yet the distribution of out-of-sample associations (Fig. 1f (dark blue)) does not match published multivariate BWAS results (Fig. 1g), which have largely ranged from r = 0.25 to 0.9, decreasing with increasing sample size10,15,16. Instead, published effects more closely follow the distribution of in-sample associations (Fig. 1h). This observation suggests that, in addition to small samples, structural problems in academic research (for example, non-representative samples, publication bias, misuse of cross-validation and unintended overfitting) have contributed to the publication of inflated effects12,17,18. A recent biomarker challenge5 showed that cross-validation results continued to improve with the amount of time researchers spent with the data, and the models with the best cross-validation results performed worse on never-seen held-back data. Thus, cross-validation alone has proven to be insufficient and must be combined with the increased generalizability of large, diverse datasets and independent out-of-sample evaluation in new, never before seen data5,10.
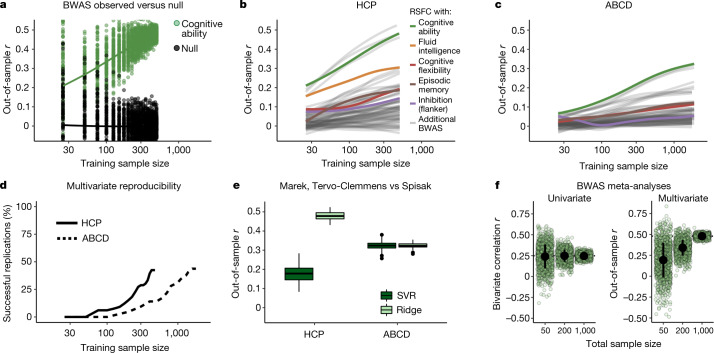
The use of additional cross-validation in the discovery sample by Spisak et al.8 does not affect out-of-sample prediction accuracies (Fig. 1e). However, by using partial correlations and ridge regression on HCP data, they were able to generate higher out-of-sample prediction accuracies than our original results in ABCD (Fig. 2a). The five variables they selected are strongly correlated19 cognitive measures from the NIH Toolbox (mean r = 0.37; compare with the correlation strength for height versus weight r = 0.44)20 and age (not a complex behavioural phenotype), unrepresentative of BWAS as a whole (Fig. 2b (colour versus grey lines)). As the HCP is the relatively smallest and most homogeneous dataset, we applied the exact method and code of Spisak et al.8 to the ABCD data (Fig. 2c and Supplementary Table 2). At n = 1,000 (training; n = 2,000 total), only 31% of BWAS (44% RSFC, 19% cortical thickness) were replicable (Fig. 2d; defined as in Spisak et al.8; Supplementary Information). Expanding BWAS scope beyond broad cognitive abilities towards complex mental health outcomes therefore requires n > 1,000 (Fig. 2b–d). The absolute largest BWAS (cognitive ability: RSFC, green) reached replicability only using n = 400 (n = 200 train; n = 200 test) with an approximate 40% decrease in out-of-sample prediction accuracies from HCP to ABCD (Fig. 2e (lighter green, left versus right)). The methods of Spisak et al.8 and our previous study1 returned equivalent out-of-sample reproducibility for this BWAS (cognitive ability: RSFC) in the larger, more diverse ABCD data (Fig. 2e (right, dark versus light green)). Thus, the smaller sample sizes (Fig. 2b,c) that are required for out-of-sample reproducibility (Fig. 2e) reported by Spisak et al.8 in the HCP data did not generalize to the larger ABCD dataset. See also our previous study1 for a broader discussion of convergent evidence across HCP and ABCD datasets.
Fig. 2. BWAS reproducibility, scope and prediction accuracy using the method of Spisak et al.
a, Example bootstrapped BWAS of total cognitive ability (green) and null distribution (black) (y axis), as a function of sample size (x axis) from the suggested method of Spisak et al.8 (RSFC by partial correlation; prediction by ridge regression) in the HCP dataset (n = 1,200, 1 site, 1 scanner, 60 min RSFC/participant, 76% white). Sample sizes were log10-transformed for visualization. b, Out-of-sample correlation (between true scores and predicted scores) from ridge regression (y axis; code from Spisak et al.8) as a function of training sample size (x axis, log10 scaling) for 33 cognitive and mental health phenotypes (Supplementary Information) in the HCP dataset. Each line displays a smoothed fit estimate (through penalized splines in general additive models) for a brain (RSFC (partial correlations, as proposed by Spisak et al.8), cortical thickness) phenotype pair (66 total) that has 100 bootstrapped iterations from sample sizes of 25 to 500 (inclusive) in increments of 25 (20 total bins). Sample sizes were log10-transformed (for visualization) before general additive model fitting. c, The same as in b, but in the ABCD dataset (n = 11,874, 21 sites, 3 scanner manufacturers, 20 min RSFC/participant, 56% white) using 32 cognitive and mental health phenotypes at sample sizes of 25, 50, 75 and from 100 to 1,900 (inclusive) in increments of 100 (22 total bins). d, The percentage of brain–phenotype pairs (BWAS) from b and c with significant replication on the basis of the method of Spisak et al.8 (Supplementary Information). e, Comparison of our original method in our previous study1 and the method proposed by Spisak et al.8 at the full split-half sample size of HCP (left) and ABCD (right). Out-of-sample correlations (RSFC with total cognitive ability, y axis) for the method used in our previous study1 (dark green; RSFC by correlation, PCA, SVR) and by Spisak et al.8 (light green; RSFC by partial correlation, ridge regression). Repeating the method proposed by Spisak et al.8 in ABCD (right) and comparing this to the method used in our previous study1 results in a very similar out-of-sample r. f, Simulated individual studies (light green circles; n = 1,000 per sample size) and meta-analytic estimates (black dot, ±1 s.d.) using the method of Spisak et al.8 (partial correlations in the HCP dataset) for the largest univariate association (left; y axis, bivariate correlation) and multivariate association (right; y axis, out-of-sample correlation) for total cognitive ability versus RSFC, as a function of total sample size (x axis; bivariate correlation for sample sizes of 50, 200 and 1,000, and multivariate sum of train and test samples, each 25, 100 and 500). For univariate approaches, studies of any sample size, when appropriately aggregated to a large total sample size, can correctly estimate the true effect size. However, for multivariate approaches, even when aggregating across 1,000 independent studies, studies with a small sample size produce prediction accuracies that are downwardly biased relative to large sample studies, highlighting the need for large samples in multivariate analyses.
Notably, the objections of Spisak et al.8 raise additional reasons to stop the use of smaller samples in BWAS that were not highlighted in our original article. Multivariate BWAS prediction accuracies—absent overfitting—are systematically suppressed in smaller samples5,9,21, as prediction accuracy scales with increasing sample size1,9. Thus, the claim that “cross-validated discovery effect-size estimates are unbiased” does not account for out-of-dataset generalizability and downward bias. In principle, if unintended overfitting and publication bias could be fully eliminated, meta-analyses of small-sample univariate BWAS would return the correct association strengths (Fig. 2f (left)). However, meta-analyses of small multivariate BWAS would always be downwardly biased (Fig. 2f (right)). If we are interested in maximizing prediction accuracy, essential for clinical implementation of BWAS22, large samples and advancements in imaging and phenotypic measurements1 are necessary.
Repeatedly subsampling the same dataset, as Spisak et al.8 and we have done, overestimates reproducibility compared with testing on a truly new, diverse dataset. Just as in genomics23, BWAS generalization failures have been highlighted5,24. For example, BWAS models trained on white Americans transferred poorly to African Americans and vice versa (within dataset)24. Historically, BWAS samples have lacked diversity, neglecting marginalized and under-represented minorities25. Large studies with more diverse samples and data aggregation efforts can improve BWAS generalizability and reduce scientific biases contributing to massive health inequities26,27.
Spisak et al.8 worry that “[r]equiring sample sizes that are larger than necessary for the discovery of new effects could stifle innovation”. We appreciate the concern that rarer populations may never be investigated with BWAS. Yet, there are many non-BWAS brain–behaviour study designs (fMRI ≠ BWAS) focused on within-patient effects, repeated-sampling and signal-to-noise-ratio improvements that have proven fruitful down to n = 1 (ref. 28). By contrast, the strength of multivariate BWAS lies in leveraging large cross-sectional samples to investigate population-level questions. Sample size requirements should be based on expected effect sizes and real-world impact, and not resource availability. Through large-scale collaboration and clear standards on data sharing, GWAS has reached sample sizes in the millions29–31, pushing genomics towards new horizons. Similarly, BWAS analyses of the future will not be limited to statistical replication of the same few strongest effects in small homogeneous populations, but also have broad scope, maximum prediction accuracy and excellent generalizability.
Reporting summary
Further information on experimental design is available in the Nature Portfolio Reporting Summary linked to this Article.
Online content
Any methods, additional references, Nature Portfolio reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at 10.1038/s41586-023-05746-w.
Supplementary information
Supplementary Methods, Supplementary Tables 1 and 2 and Supplementary References.
Acknowledgements
Data used in the preparation of this Article were, in part, obtained from the Adolescent Brain Cognitive Development (ABCD) Study (https://abcdstudy.org), held in the NIMH Data Archive (NDA). This is a multisite, longitudinal study designed to recruit more than 10,000 children aged 9–10 years and follow them over 10 years into early adulthood. The ABCD Study is supported by the National Institutes of Health and additional federal partners under award numbers U01DA041022, U01DA041028, U01DA041048, U01DA041089, U01DA041106, U01DA041117, U01DA041120, U01DA041134, U01DA041148, U01DA041156, U01DA041174, U24DA041123, U24DA041147, U01DA041093 and U01DA041025. A full list of supporters is available online (https://abcdstudy.org/federal-partners.html). A listing of participating sites and a complete listing of the study investigators is available online (https://abcdstudy.org/scientists/workgroups/). ABCD consortium investigators designed and implemented the study and/or provided data but did not necessarily participate in the analysis or the writing of this report. This Article reflects the views of the authors and may not reflect the opinions or views of the NIH or ABCD consortium investigators. Data were provided, in part, by the HCP, WU-Minn Consortium (U54 MH091657) funded by the 16 NIH Institutes and Centers that support the NIH Blueprint for Neuroscience Research; and by the McDonnell Center for Systems Neuroscience at Washington University. This work used the storage and computational resources provided by the Masonic Institute for the Developing Brain (MIDB), the Neuroimaging Genomics Data Resource (NGDR) and the Minnesota Supercomputing Institute (MSI). The NGDR is supported by the University of Minnesota Informatics Institute through the MnDRIVE initiative in coordination with the College of Liberal Arts, Medical School and College of Education and Human Development at the University of Minnesota. This work used the storage and computational resources provided by the Daenerys Neuroimaging Community Computing Resource (NCCR). The Daenerys NCCR is supported by the McDonnell Center for Systems Neuroscience at Washington University, the Intellectual and Developmental Disabilities Research Center (IDDRC; P50 HD103525) at Washington University School of Medicine and the Institute of Clinical and Translational Sciences (ICTS; UL1 TR002345) at Washington University School of Medicine. This work was supported by NIH grants MH121518 (to S.M.), NS090978 (to B.P.K.), MH129616 (to T.O.L.), 1RF1MH120025-01A1 (to W.K.T), MH080243 (to B.L.), MH067924 (to B.L.), DA041148 (to D.A.F.), DA04112 (to D.A.F.), MH115357 (to D.A.F.), MH096773 (to D.A.F. and N.U.F.D.), MH122066 (to D.A.F. and N.U.F.D.), MH121276 (to D.A.F. and N.U.F.D.), MH124567 (to D.A.F. and N.U.F.D.), NS088590 (to N.U.F.D.), and the Andrew Mellon Predoctoral Fellowship (to B.T.-C.), the Staunton Farm Foundation (to B.L.), the Lynne and Andrew Redleaf Foundation (to D.A.F.) and the Kiwanis Neuroscience Research Foundation (to N.U.F.D.).
Author contributions
Conception: B.T.-C., S.M., D.A.F. and N.U.F.D. Design: B.T.-C., S.M., R.J.C., D.A.F. and N.U.F.D. Data acquisition, analysis and interpretation: B.T.-C., S.M., R.J.C., A.V.N., B.P.K., W.K.T., T.E.N., B.T.T.Y., D.A.F. and N.U.F.D. Manuscript writing and revising: B.T.-C., S.M., R.J.C., A.V.N., B.P.K., T.O.L., W.K.T., T.E.N., B.T.T.Y., D.M.B., B.L., D.A.F. and N.U.F.D. We note that the reply author list differs from the original paper in number and in order to accurately reflect its more focused scope compared with the original work.
Data availability
Participant-level data from all datasets (ABCD and HCP) are openly available pursuant to individual, consortium-level data access rules. The ABCD data repository grows and changes over time (https://nda.nih.gov/abcd). The ABCD data used in this report came from ABCD collection 3165 and the Annual Release 2.0 (10.15154/1503209). Data were provided, in part, by the HCP, WU-Minn Consortium (principal investigators: D. Van Essen and K. Ugurbil; 1U54MH091657) funded by the 16 NIH Institutes and Centers that support the NIH Blueprint for Neuroscience Research; and by the McDonnell Center for Systems Neuroscience at Washington University. Some data used in the present study are available for download from the HCP (www.humanconnectome.org). Users must agree to data use terms for the HCP before being allowed access to the data and ConnectomeDB, details are provided online (https://www.humanconnectome.org/study/hcp-young-adult/data-use-terms).
Code availability
Manuscript analysis code specific to this study is available at GitHub (https://gitlab.com/DosenbachGreene/bwas_response). Code for processing ABCD data is provided at GitHub (https://github.com/DCAN-Labs/abcd-hcp-pipeline). MRI data analysis code is provided at GitHub (https://github.com/ABCD-STUDY/nda-abcd-collection-3165). FIRMM software is available online (https://firmm.readthedocs.io/en/latest/release_notes/). The ABCD Study used v.3.0.14.
Competing interests
D.A.F. and N.U.F.D. have a financial interest in Turing Medical and may financially benefit if the company is successful in marketing FIRMM motion monitoring software products. A.N.V., D.A.F. and N.U.F.D. may receive royalty income based on FIRMM technology developed at Washington University School of Medicine and Oregon Health and Sciences University and licensed to Turing Medical. D.A.F. and N.U.F.D. are co-founders of Turing Medical. These potential conflicts of interest have been reviewed and are managed by Washington University School of Medicine, Oregon Health and Sciences University and the University of Minnesota.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Brenden Tervo-Clemmens, Scott Marek
These authors jointly supervised this work: Damien A. Fair, Nico U. F. Dosenbach
Contributor Information
Brenden Tervo-Clemmens, Email: btervo-clemmens@mgh.harvard.edu.
Scott Marek, Email: smarek@wustl.edu.
Damien A. Fair, Email: faird@umn.edu
Nico U. F. Dosenbach, Email: ndosenbach@wustl.edu
Supplementary information
The online version contains supplementary material available at 10.1038/s41586-023-05746-w.
References
- 1.Marek S, et al. Reproducible brain-wide association studies require thousands of individuals. Nature. 2022;603:654–660. doi: 10.1038/s41586-022-04492-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schönbrodt, F. D. & Perugini, M. At what sample size do correlations stabilize? J. Res. Pers.47, 609–612 (2013).
- 3.Button KS, et al. Confidence and precision increase with high statistical power. Nat. Rev. Neurosci. 2013;14:585–586. doi: 10.1038/nrn3475-c4. [DOI] [PubMed] [Google Scholar]
- 4.Varoquaux G. Cross-validation failure: small sample sizes lead to large error bars. Neuroimage. 2018;180:68–77. doi: 10.1016/j.neuroimage.2017.06.061. [DOI] [PubMed] [Google Scholar]
- 5.Traut N, et al. Insights from an autism imaging biomarker challenge: promises and threats to biomarker discovery. Neuroimage. 2022;255:119171. doi: 10.1016/j.neuroimage.2022.119171. [DOI] [PubMed] [Google Scholar]
- 6.Casey BJ, et al. The Adolescent Brain Cognitive Development (ABCD) study: imaging acquisition across 21 sites. Dev. Cogn. Neurosci. 2018;32:43–54. doi: 10.1016/j.dcn.2018.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Littlejohns TJ, et al. The UK Biobank imaging enhancement of 100,000 participants: rationale, data collection, management and future directions. Nat. Commun. 2020;11:2624. doi: 10.1038/s41467-020-15948-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spisak, T., Bingel, U. & Wager, T. D. Multivariate BWAS can be replicable with moderate sample sizes. Nature10.1038/s41586-023-05745-x (2023). [DOI] [PMC free article] [PubMed]
- 9.Schulz, M.-A., Bzdok, D., Haufe, S., Haynes, J.-D. & Ritter, K. Performance reserves in brain-imaging-based phenotype prediction. Preprint at 10.1101/2022.02.23.481601 (2022). [DOI] [PMC free article] [PubMed]
- 10.Poldrack RA, Huckins G, Varoquaux G. Establishment of best practices for evidence for prediction: a review. JAMA Psychiatry. 2020;77:534–540. doi: 10.1001/jamapsychiatry.2019.3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Poldrack RA. The costs of reproducibility. Neuron. 2019;101:11–14. doi: 10.1016/j.neuron.2018.11.030. [DOI] [PubMed] [Google Scholar]
- 12.Button KS, et al. Power failure: why small sample size undermines the reliability of neuroscience. Nat. Rev. Neurosci. 2013;14:365–376. doi: 10.1038/nrn3475. [DOI] [PubMed] [Google Scholar]
- 13.Kohavi, R. A study of cross-validation and bootstrap for accuracy estimation and model selection. In Proc. IJCAI 95 (ed. Mellish, C. S.) 1137–1143 (Morgan Kaufman, 1995).
- 14.Scheinost D, et al. Ten simple rules for predictive modeling of individual differences in neuroimaging. Neuroimage. 2019;193:35–45. doi: 10.1016/j.neuroimage.2019.02.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sui J, Jiang R, Bustillo J, Calhoun V. Neuroimaging-based individualized prediction of cognition and behavior for mental disorders and health: methods and promises. Biol. Psychiatry. 2020;88:818–828. doi: 10.1016/j.biopsych.2020.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Woo C-W, Chang LJ, Lindquist MA, Wager TD. Building better biomarkers: brain models in translational neuroimaging. Nat. Neurosci. 2017;20:365–377. doi: 10.1038/nn.4478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ioannidis JPA. Why most discovered true associations are inflated. Epidemiology. 2008;19:640–648. doi: 10.1097/EDE.0b013e31818131e7. [DOI] [PubMed] [Google Scholar]
- 18.Pulini AA, Kerr WT, Loo SK, Lenartowicz A. Classification accuracy of neuroimaging biomarkers in attention-deficit/hyperactivity disorder: effects of sample size and circular analysis. Biol. Psychiatry Cogn. Neurosci. Neuroimaging. 2019;4:108–120. doi: 10.1016/j.bpsc.2018.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Funder, D. C. & Ozer, D. J. Evaluating effect size in psychological research: sense and nonsense. Adv. Methods Pract. Psychol. Sci.2, 156–168 (2019).
- 20.Meyer, G. J. et al. Psychological testing and psychological assessment: a review of evidence and issues. Am. Psychol.56, 128–165 (2001). [PubMed]
- 21.He T, et al. Deep neural networks and kernel regression achieve comparable accuracies for functional connectivity prediction of behavior and demographics. Neuroimage. 2020;206:116276. doi: 10.1016/j.neuroimage.2019.116276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leptak C, et al. What evidence do we need for biomarker qualification? Sci. Transl. Med. 2017;9:eaal4599. doi: 10.1126/scitranslmed.aal4599. [DOI] [PubMed] [Google Scholar]
- 23.Weissbrod O, et al. Leveraging fine-mapping and multipopulation training data to improve cross-population polygenic risk scores. Nat. Genet. 2022;54:450–458. doi: 10.1038/s41588-022-01036-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li J, et al. Cross-ethnicity/race generalization failure of behavioral prediction from resting-state functional connectivity. Sci. Adv. 2022;8:eabj1812. doi: 10.1126/sciadv.abj1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Henrich J, Heine SJ, Norenzayan A. The weirdest people in the world? Behav. Brain Sci. 2010;33:61–83. doi: 10.1017/S0140525X0999152X. [DOI] [PubMed] [Google Scholar]
- 26.Bailey ZD, et al. Structural racism and health inequities in the USA: evidence and interventions. Lancet. 2017;389:1453–1463. doi: 10.1016/S0140-6736(17)30569-X. [DOI] [PubMed] [Google Scholar]
- 27.Martin AR, et al. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat. Genet. 2019;51:584–591. doi: 10.1038/s41588-019-0379-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gratton C, Nelson SM, Gordon EM. Brain-behavior correlations: two paths toward reliability. Neuron. 2022;110:1446–1449. doi: 10.1016/j.neuron.2022.04.018. [DOI] [PubMed] [Google Scholar]
- 29.Levey, D. F et al. Bi-ancestral depression GWAS in the Million Veteran Program and meta-analysis in >1.2 million individuals highlight new therapeutic directions. Nature24, 954–963 (2021). [DOI] [PMC free article] [PubMed]
- 30.Muggleton N, et al. The association between gambling and financial, social and health outcomes in big financial data. Nat. Hum. Behav. 2021;5:319–326. doi: 10.1038/s41562-020-01045-w. [DOI] [PubMed] [Google Scholar]
- 31.Yengo L, et al. A saturated map of common genetic variants associated with human height. Nature. 2022;610:704–712. doi: 10.1038/s41586-022-05275-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Methods, Supplementary Tables 1 and 2 and Supplementary References.
Data Availability Statement
Participant-level data from all datasets (ABCD and HCP) are openly available pursuant to individual, consortium-level data access rules. The ABCD data repository grows and changes over time (https://nda.nih.gov/abcd). The ABCD data used in this report came from ABCD collection 3165 and the Annual Release 2.0 (10.15154/1503209). Data were provided, in part, by the HCP, WU-Minn Consortium (principal investigators: D. Van Essen and K. Ugurbil; 1U54MH091657) funded by the 16 NIH Institutes and Centers that support the NIH Blueprint for Neuroscience Research; and by the McDonnell Center for Systems Neuroscience at Washington University. Some data used in the present study are available for download from the HCP (www.humanconnectome.org). Users must agree to data use terms for the HCP before being allowed access to the data and ConnectomeDB, details are provided online (https://www.humanconnectome.org/study/hcp-young-adult/data-use-terms).
Manuscript analysis code specific to this study is available at GitHub (https://gitlab.com/DosenbachGreene/bwas_response). Code for processing ABCD data is provided at GitHub (https://github.com/DCAN-Labs/abcd-hcp-pipeline). MRI data analysis code is provided at GitHub (https://github.com/ABCD-STUDY/nda-abcd-collection-3165). FIRMM software is available online (https://firmm.readthedocs.io/en/latest/release_notes/). The ABCD Study used v.3.0.14.