Abstract
Position-specific integration of the retroviruslike element Ty3 near the transcription initiation sites of tRNA genes requires transcription factors IIIB and IIIC (TFIIIB and TFIIIC). Using a genetic screen, we isolated a mutant with a truncated 95-kDa subunit of TFIIIC (TFIIIC95) that reduced the apparent retrotransposition of Ty3 into a plasmid-borne target site between two divergently transcribed tRNA genes. Although TFIIIC95 is conserved and essential, no defect in growth or transcription of tRNAs was detected in the mutant. Steps of the Ty3 life cycle, such as protein expression, proteolytic processing, viruslike particle formation, and reverse transcription, were not affected by the mutation. However, Ty3 integration into a divergent tDNA target occurred exclusively in one orientation in the mutant strain. Investigation of this orientation bias showed that TFIIIC95 and Ty3 integrase interacted in two-hybrid and glutathione S-transferase pulldown assays and that interaction with the mutant TFIIIC95 protein was attenuated. The orientation bias observed here suggests that even for wild-type Ty3, the protein complexes associated with the long terminal repeats are not equivalent in vivo.
Genomic sequence analysis has shown that retroelements account for a significant proportion of the genomes of plants, animals, and microbes. Among various host organisms, the budding yeast Saccharomyces cerevisiae offers one of the most genetically tractable model systems for studying these elements. Currently, five retrotransposons, Ty1 through Ty5, have been identified in S. cerevisiae. Of these retroelements, Ty1, Ty2, Ty4, and Ty5 belong to the copialike family whereas Ty3 belongs to the gypsylike family. Although Ty3 is limited to an intracellular life cycle, It has many similarities to retroviruses, including a number of steps in its life cycle (34). Retrotransposons and retroviruses rely on proteins encoded in the element or virus and in host cells. Full-length Ty3 DNA is approximately 5.4 kb in length (10). It encodes Gag3p and Gag3-Pol3p polyproteins, which are processed into mature proteins in the context of the Ty3 viruslike particle by the Ty3 protease. Gag3p is processed into major structural proteins, capsid (CA) and nucleocapsid. Gag3p-Pol3p is processed into catalytic proteins PR, reverse transcriptase and integrase (IN) (17). The life cycle requires host factors such as transcription and translation machinery and unknown factors involved in such processes as assembly, uncoating, nuclear import, and target site recognition.
Several host factors affect the efficiency or position of retroelement integration. For retroviral integration, in vitro experiments showed enhancement of integration by Ini1 (22), HMG1 (1), and HMG I(Y) (14) and reduction of autointegration by barrier to autointegration factor (29). For yeast retrotransposons, in vivo experiments have identified chromatin-associated proteins that enhance the efficiency of integration into promoter regions of RNA polymerase II (pol II)-transcribed genes or heterochromatic DNA or affect the general efficiency of integration. For example, Ubc2 and CAF-I affect the integration preference of Ty1 (20). Ty5 targets heterochromatic DNA through contacts mediated by Sir proteins (42). Most genomic Ty1 and Ty2 elements are found within 750 bp of the 5′ end of tRNA genes (24), and this targeting presumably involves host proteins.
Ty3 is distinguished from other retroelements by its extreme integration specificity. It integrates within a few nucleotides of the transcription initiation sites of pol III genes on plasmids (9) and of chromosomal tRNA genes (24). There is no sequence similarity among pol III transcription initiation sites, suggesting that the structure of the transcription initiation complex, rather than a consensus DNA sequence, is responsible for the specificity of integration (9). The tRNA and U6 gene transcription preinitiation complexes are composed of transcription factors IIIC and IIIB (TFIIIC and TFIIIB). TFIIIC binds to promoter elements, box B and box A, and recruits the initiation factor TFIIIB, which binds upstream of the initiation site. In the case of the tRNA and U6 genes, TFIIIC is required for transcription in vivo, but in defined in vitro systems TFIIIB can mediate TATA box-dependent transcription in the absence of TFIIIC (39). Similar to what is observed for transcription, only TFIIIB is required for integration upstream of the U6 gene in vitro (40). However, in vitro, chromatographic fractions containing TFIIIB and TFIIIC are required for position-specific integration of Ty3 upstream of a tRNA gene (27). As is the case for transcription in vivo, a point mutation in box B that abolishes TFIIIC binding abrogates the activity of a tRNA or U6 in Ty3-targeting assays (9). Although the in vitro studies suggest that direct contacts must occur between TFIIIB and the preintegration complex (PIC), they did not address whether the in vivo role of TFIIIC in transposition is indirect, that is via loading TFIIIB, or direct, through interactions with the PIC.
In this study, we identified a mutation that caused truncation of the 95-kDa subunit of TFIIIC (TFIIIC95) and severely reduced the recovery of Ty3 integrants in an in vivo assay. The mutant strain had no detectable defect in growth or transcription of tRNAs. Although intermediates of the Ty3 life cycle were not altered, Ty3 elements integrated between a pair of divergent tRNA genes in the mutant strain showed orientation bias. Furthermore, interaction observed between Ty3 IN and TFIIIC95 was attenuated for the mutant protein, suggesting that TFIIIC95 participates directly in docking the PIC. These results provide the first genetic evidence directly linking the targeting of a Ty element to a specific component of the pol III transcription initiation complex. The orientation bias observed here offers insights into the asymmetry of a retroelement PIC.
MATERIALS AND METHODS
Yeast strains and plasmids.
The strains and plasmids used in this work are listed in Tables 1 and 2. Media and standard techniques for yeast were as previously described (35). The haploid yeast strain yMA1322 used to generate mutants was derived from YPH500 (36). First, YPH500 was transformed with the LYS2 gene excised from pDP6 (32), resulting in the lysine prototrophic strain yMA1241. An ochre allele of the LYS2 gene was generated by PCR amplification of the wild-type LYS2 gene present on the pDP6 plasmid template, with primers 489 and 485 (sequences of the oligonucleotides used in this study are shown in Table 3). This amplification converts Tyr31 to an ochre codon. The PCR product was transformed into yMA1241, and transformants were plated onto α-aminoadipate medium to select for lysine auxotrophs. Multiple lysine auxotrophs were transformed with the pTIT plasmid (L. Yieh, unpublished work), which carries the SUP2bo suppressor tDNA, and SUP2bo-dependent lysine prototrophs were selected. One of the strains carrying a suppressible lys2o allele was designated yMA1322. A LEU2 gene excised with BamHI and NarI from plasmid YEp351 was transformed into yMA1322 to obtain leucine-prototrophic strain yMA1342, the reference strain which is referred to as wild type in this study.
TABLE 1.
Yeast strains used in this study
| Strain | Relevant genotype | Source or reference |
|---|---|---|
| YPH500 | matα ade2-101o lys2-801am his3-Δ200 trp1-Δ63 leu2-Δ1 ura3-52 | 36 |
| yMA1235 | LEU2 derivative of YPH500 | This work |
| yMA1241 | LYS2 derivative of YPH500 | This work |
| yMA1322 | lys2o derivative of YPH500 | This work |
| yMA1342 | LEU2 derivative of yMA1322 | This work |
| yMA1343 | tfc1::mTn, yMA1322 (isolated from the screen) | This work |
| yMA1344 | tfc1::mTn, yMA1322 (reconstructed mutant) | This work |
| yMA1356 | rad52Δ derivative of yMA1322 | This work |
| yMA1357 | rad52Δ derivative of yMA1343 | This work |
TABLE 2.
Plasmids used in this study
| Plasmid | Description | Source or reference |
|---|---|---|
| pTM45 | GAL1-10UAS::Ty3 CEN TRP1 | 31 |
| pCH2bo19V | sup2bo tDNAValCEN HIS3 | 25 |
| pEH2b19V | sup2b tDNAVal 2μm HIS3 | 25 |
| pTIT | pCH2bo19V with a Ty3 insertion | L. Yieh, unpublished |
| pDLC348 | GAL1-10UAS::Ty3-Neo, CEN, URA3 | 9 |
| pDLC356 | SUP2b tDNA 2μm HIS3 | 9 |
| pDLC374 | sup2+A tDNA 2μm HIS3 | 9 |
| pDP6 | LYS2 plasmid | 32 |
| pRS316 | CEN, URA3 vector | 36 |
| pMA1715 | GAL1-10UAS::Ty3-H CEN URA3 | This work |
| pTFC1 | TFC1 CEN URA3 (the same as YCpCS7) | 11 |
| pTFC1::mTn3 | pTFC1 with mTn3 insertion | This work |
| pACT55 | Gal4 AD fused to TFIIIC55 | 30 |
| pACT95 | Gal4 AD fused to TFIIIC95 | 30 |
| pAS95 | GAL4 BD fused to TFIIIC95 | 30 |
| pAS95ΔC | Gal4 BD fused to TFIIIC95 (1–510 aa) | This work |
| pAS95 (C) | Gal4 BD fused to TFIIIC95 (511–649 aa) | This work |
| pACT IN | Gal4 AD fused to Ty3 IN | J. A. Claypool, unpublished |
| pAS IN | Gal4 BD fused to Ty3 IN | J. A. Claypool, unpublished |
| pAS IN (A) | Gal4 BD fused to Ty3 IN (1–61 aa) | S. L. Dildine, unpublished |
| pAS IN (B) | Gal4 BD fused to Ty3 IN (62–304 aa) | S. L. Dildine, unpublished |
| pAS IN (C) | Gal4 BD fused to Ty3 IN (305–536 aa) | S. L. Dildine, unpublished |
| pAS IN (AB) | Gal4 BD fused to Ty3 IN (1–304 aa) | S. L. Dildine, unpublished |
| pAS IN (BC) | Gal4 BD fused to Ty3 IN (62–536 aa) | S. L. Dildine, unpublished |
| pGST τ95 | GST fused to TFIIIC95 in pGEX2T (Pharmacia) | This work |
| pGST τ95ΔC | GST fused to TFIIIC95 (1–510 aa) | This work |
| pGST IN (A) | GST fused to IN (1–61 aa) | This work |
| pJK788 | pT7::IN in pYES2 (Invitrogen) | Kirchner, unpublished |
| pT7 IN (A) | pT7::IN (1–61 aa) in pCRII-Topo (Invitrogen) | This work |
| pT7 IN (N) | pT7::IN (1–150 aa) in pCRII-Topo vector | This work |
| pT7 IN (AB) | pT7::IN (1–304 aa) in pCRII-Topo vector | This work |
TABLE 3.
Oligonucleotides used in this study
| Oligonucleotide | Sequence | Application |
|---|---|---|
| 489 | 5′-CCACAACAAGAACCTTAAACGAAACAAGC-3′ | To generate the lys2o allele by PCR |
| 485 | 5′-AAACCAAGATGAAGCTGCCA-3′ | To generate the lys2o allele by PCR |
| 470 | 5′-CGATGTTCCAGATTACGCTGGATCCCGAAAGAATTATTCCCTAGAAG-3′ | To create pAS IN-BC from pAS IN |
| 471 | 5′-CTCGAACTATCAGAGACCTTCTGAACGTTCATAAAACACATATG-3′ | To create pAS IN-A from pAS IN |
| 472 | 5′-CGATGTTCCAGATTACGCTGGATCCCCATTTGAAATTGATTTAGG-3′ | To create pAS IN-C from pAS IN |
| 473 | 5′-CTACTAGAACACTTGGAAAATCATGAACGTTCATAAAACACATATG-3′ | To create pAS IN-AB from pAS IN |
| 762 | 5′-GATCGGACTACTAGCAGCTG-3′ | PCR amplification of IN fragments |
| 763 | 5′-CTCAGAAGGTCTCTGATAGTTCG-3′ | PCR amplification of IN-A |
| 764 | 5′-CTCATGGTTGTAATAGTCCATG-3′ | PCR amplification of IN-N |
| 765 | 5′-CTCATCCAAGTGTTCTAGTAGGTG-3′ | PCR amplification of IN-AB |
| 637 | 5′-GAAGGAGAGGACGCTGTCTGTCGAAGGTAAGGAACGGACGAGAGAAGGGAGAG-3′ | Anchor bubble primer for vectorette PCR |
| 638 | 5′-GACTCTCCCTTCTCGAATCGTAACCGTTCGTACGAGAATCGATGTCCTCTCCTTC-3′ | Anchor bubble primer for vectorette PCR |
| 639 | 5′-CGAATCGTAACCGTTCGTACGAGAATCGCT-3′ | Primer for vectorette PCR |
| 642 | 5′-GTAAAACGACGGGATCCCCCTTAACGTGAG-3′ | Primer for vectorette PCR |
| 12 | 5′-GATTTCGTAGGTTACCTGATAAATTACAG-3′ | Primer extension of SUP2b pre-tRNA |
| 83 | 5′-TGGTAGCGCCGCTGCGTTTCGATCCGAGG-3′ | Probe for northern blot of tRNA
|
Strain yMA1343 (25-41A mutant) was identified in the genetic screen as having a reduced frequency of transposition-dependent activation of the suppressor tRNA, sup2bo. The pMA1922 (pTFC1::mTn) plasmid was generated by the plasmid gap rescue method (16) from yMA1343. Briefly, pTFC1 (yCpCS7) DNA (11) was cleaved with AatII and HpaI to generate a gap within the TFC1 coding region flanking the mTn3 insertion site. This gapped plasmid was transformed into yMA1343, and Ura+ transformants carrying plasmids repaired by gene conversion from the TFC1 locus were selected. Plasmids (pTFC1::mTn) from Ura+ colonies were rescued by transforming DNA from those strains into Escherichia coli HB101. The ApaI-SacII fragment containing part of the TFC1 gene with an mTn3 insertion was excised from pTFC1::mTn plasmid and transformed into the yMA1322 strain, and Leu+ transformants were selected to obtain yMA1344. DNA isolated from yMA1344 was analyzed by Southern blotting using a TFC1-specific probe to confirm the TFC1 disruption in this strain.
To minimize homologous recombination, which generates background in the helper-donor transposition assay, the RAD52 gene was deleted using a knockout construct. The pBJC302 plasmid (12) cleaved with PvuII was transformed into the wild-type (yMA1322) or tfc1 strain, and transformants were selected on synthetic complete medium lacking uracil (SC-Ura). Subsequent loss of the URA3 gene from these cells was selected on 5-fluoroorotic acid medium. Gene disruption was confirmed by Southern and PCR analysis.
Yeast two-hybrid plasmids for Ty3 IN were constructed by cloning a BamHI fragment with Ty3 IN sequence into pMA424 (Gal4 BD fusion) and pGAD2F (Gal4 AD fusion) vectors. Constructs for Ty3 IN domains amino-terminally fused to Gal4 BD were generated by single-stranded mutagenesis (28) from pAS IN by using oligonucleotides 471 (pAS IN-A), 472 (pAS IN-C), 473 (pAS IN-AB), and 470 (pAS IN-BC) and from pAS IN-BC by using oligonucleotide 473 (pAS IN-B). Plasmids pAS95, pACT95, and pACT55 were described previously (30). pAS95ΔC was constructed by cloning the Ncol fragment of TFC1 from pAS95 into the NcoI site of pAS1-CYH2. The same NcoI fragment was excised from pAS95 and the backbone was religated to yield pAS95(C).
To generate glutathione S-transferase (GST) fusion constructs, the pGEX2T vector (Pharmacia) was linearized with BamHI and blunt ends were created by filling in with Klenow polymerase. The SmaI-SalI and NcoI-SalI fragments of TFC1 from pAS95 were treated with Klenow polymerase to generate blunt ends and cloned into the pGEX2T vector described above to create pGSTτ95 and pGSTτ95ΔC, respectively. The BamHI fragment containing the IN-A domain from pAS IN-A was treated with Klenow and cloned into the same vector to create pGST IN-A.
To generate templates for in vitro transcription and translation reactions, fragments of IN were amplified by PCR from pJK788 (J. Kirchner, unpublished work) using primers 762 and 763 (A), 762 and 764 (N), and 762 and 765 (AB). Each PCR product was cloned into the pCRII-Topo vector (Invitrogen). Plasmids containing IN fragments downstream of the T7 promoter were identified by restriction and sequence analyses.
Yeast mutagenesis.
Shuttle mutagenesis was performed as previously described (6). Briefly, DNA was prepared from library pools separately mutagenized with mini-Tn3::lacZ/LEU2 (kindly provided by M. Snyder, Yale University), cleaved with NotI, and transformed into strain yMA1322 carrying pTM45 and pPK689 (pCH2bo19V) by the lithium acetate procedure (21). Genomic loci disrupted by mTn3 insertion in each mutant of interest were amplified by vectorette PCR (http://genome-www.stanford.edu/group/botlab/protocols/vectorette.html) and identified by sequence analysis. Briefly, total yeast DNA prepared from the mutant strain was cleaved with RsaI and ligated with preannealed anchor bubble primers (primer 637 and 638). One-tenth of the reaction mixture was used as the input for amplification of the genomic disruption using primers 639 and 642. PCR mixtures were prepared as previously described (31). Following an initial incubation at 95°C for 2.5 min, 35 cycles of PCR consisting of 20 s at 92°C, 30 s at 67°C, and 2.5 min at 72°C were performed. PCR products were separated by electrophoresis, and each product was excised. DNA was extracted by using the QiaexII kit (Qiagen) and sequenced by using primer 642 and a Thermosequenase kit (Amersham). Each sequence generated was compared to complete S. cerevisiae genomic DNA using the Blastn search of the Saccharomyces Genome Database (http://genome-www.stanford.edu /Saccharomyces/).
Ty3 retrotransposition assay.
A target-specific genetic assay (25) was slightly modified to screen mutants for the Ty3 transposition phenotype. Leu+ mutant transformants or wild-type strain yMA1342, carrying pTM45 and pPK689, were patched onto SC-His-Trp-Leu (synthetic dropout medium with glucose). After incubation for 24 h at 30°C, each plate was replica plated to SC(Gal)-His-Trp-Leu (galactose as the carbon source) for induction of Ty3 expression. After 48 h of growth at 30°C on this medium, yeast cells expressing Ty3 were replica plated to minimal medium (with glucose and supplemented with uracil) for detection of retrotransposition events. The latter plates were incubated at 30°C for 5 days, and the number of papillae within each patch was compared. The cells on SC-His-Trp-Leu plates after 1 day at 30°C were replica plated to minimal plates with glucose and uracil as the negative control.
Immunoblot analysis.
Whole-cell extracts (WCEs) were prepared from 10-ml cultures as described previously (31). Then 20-μg portions of WCEs were fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to nitrocellulose membranes (Hybond ECL; Amersham), and incubated with rabbit polyclonal antibodies to CA and IN. Secondary antibodies to rabbit immunoglobulin G (IgG) were detected by the ECL system.
Southern blot analysis.
RNA-free total yeast DNA (1 μg) was digested with EcoRI, separated by electrophoresis, transferred to a nylon membrane (Duralon UV; Stratagene), and immobilized by UV cross-linking in a Stratalinker 1800 (Stratagene). Hybridization was performed with 32P-labeled internal BglII fragment of Ty3.
Northern blot and primer extension analysis.
Yeast cells grown for 6 h with 2% raffinose or 2% galactose were harvested. Total RNA was extracted, separated in an 8% polyacrylamide–8.3 M urea gel by electrophoresis, transferred to a GeneScreen Plus membrane (Stratagene), and probed with 32P-labeled oligonucleotide specific for mature tRNA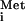 . Primer extension analysis was performed as previously described (26).
. Primer extension analysis was performed as previously described (26).
BR500 transcription extract preparations and in vitro transcription assays.
Yeast transcription extracts were prepared from 12 liters of stationary-phase cultures as previously described (23). Briefly, cell pellets were washed, resuspended, and lysed with glass beads in a bead-beater chamber. Centrifugation of lysates for 1 h at 100,000 × g yielded S100 supernatant. S100 extract was fractionated by (NH4)2SO4 precipitation. The pellet precipitated by 35 to 70% ammonium sulfate was resuspended and loaded on to a BioRex70 ion-exchange column. Elution with 500 mM NaCl from this column yielded BR500 extracts.
In vitro transcriptions were performed with 30 μg of BR500 as previously described (23). Each reaction mixture contained 110 mM NaCl, 8 mM MgCl2, 20 mM HEPES (pH 7.8), 250 μM each ATP, CTP, and UTP, 15 μM GTP, 10 μCi of [α-32P]GTP (16.7 Ci/mmol), 200 ng of template tRNA gene containing plasmid, and 800 ng of pIBI20 as nonspecific DNA, in a total volume of 40 μl. The reaction mixtures were incubated for 30 min at 30°C, and the reactions were terminated by the addition of 160 μl of stop solution (27 mM EDTA, 0.27% SDS, 0.33 μg of salmon sperm DNA per μl, 1.33 M LiCl). The transcription products were separated by electrophoresis on an 8% polyacrylamide gel and visualized by autoradiography.
Yeast two-hybrid assay.
Yeast two-hybrid filter assays were performed as previously described (37). Two-hybrid constructs were transformed into the SF526 strain, and the filter assay for β-galactosidase activity was conducted on at least three different transformants.
GST pulldown assay.
GST pulldown assays were performed as previously described (37). Expression of fusion proteins was induced by isopropyl-β-d-thiogalactopyranoside (IPTG) at a final concentration of 500 μM for 3 h at 37°C in E. coli HB101, and fusion proteins were purified by batch binding to glutathione-Sepharose beads. Ty3 IN or its domains were labeled with [35S]methionine in a coupled transcription-translation system (Promega) in vitro. Labeled protein was incubated with GST fusion proteins or GST alone bound to the beads equilibriated with 0.3% bovine serum albumin fraction V (Sigma) in buffer C (20 mM HEPES [pH 7.8], 100 mM NaCl, 20% glycerol, 1 mM EDTA, 1 mM dithiothreitol). After incubation for 30 min at room temperature with gentle agitation, the beads were washed three times with buffer C. The proteins retained on the beads were eluted by boiling in sample buffer and were separated by SDS-PAGE (10% polyacrylamide). Each gel was fixed in 40% methanol and 10% acetic acid, soaked in Amplify (Amersham), dried, and exposed to film.
Recovery of plasmids with Ty3-Neo insertions.
To recover plasmids with Ty3 insertions, pEH2b19V target plasmid together with pTM45 (helper) and pDLC348 (donor) plasmids were transformed into rad52Δ versions of the wild-type (yMA1356) and tfc1 (yMA1357) strains. The transposition assay was performed as previously described (9). Transposition was induced by growing transformants for 4 to 6 days at 30°C on SC-Trp-Ura-His, containing galactose as the carbon source. These cells were patched to yeast extract-peptone-dextrose (YPD) containing 700 μg of G418 per liter to allow loss of Ty3 plasmids and to enrich for plasmids with Ty3 insertions. G418-resistant cells that had lost the URA3-marked donor plasmid were selected on medium containing 5-fluoroorotic acid, and colonies that retained the target plasmid were identified on SC-His medium containing G418. DNA was isolated from these cells, and plasmids containing Ty3-N insertions were recovered by transformation of E. coli and selecting for kanamycin and ampicillin resistance. Plasmid DNA was isolated from a single E. coli transformant per galactose-induced colony to ensure that the Ty3-N insertions were independent. The position and the orientation of Ty3-N insertions were determined by sequence analysis.
RESULTS
Genetic screen to identify yeast strains with the Ty3 retrotransposition phenotype.
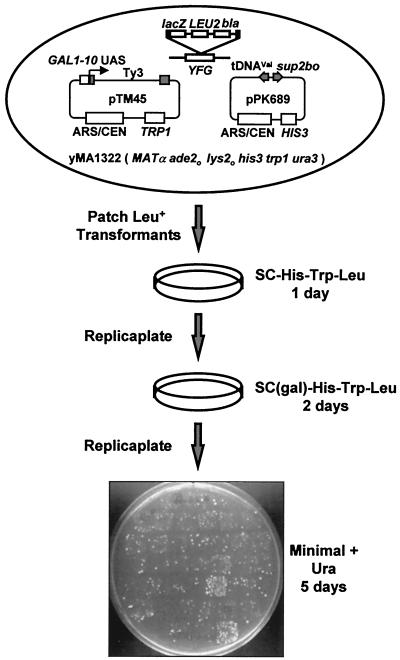
Insertional mutagenesis (6) coupled with a genetic assay (25) was used to identify host factors that affect retrotransposition of Ty3 (Fig. 1). In the assay, Ty3 transcription is induced by growth on galactose and Ty3 integration is detected using a plasmid-borne tDNA target. The target tRNAVal gene is positioned so that it interferes with expression of a neighboring, divergent ochre suppressor tRNATyr gene, sup2bo. In addition, the latter is inactivated by a tract of pyrimidines on the nontranscribed strand in the transcription initiation region. Ty3 position-specific integration into this target both alleviates the interference between the divergent genes and changes the sequence composition upstream of the suppressor, thereby activating its expression. Haploid yeast mutants were generated as described in Materials and Methods. Leu+ mutants and wild-type strain yMA1342, carrying pTM45 with a galactose-inducible Ty3 element and pPK689 with the divergent tDNA target, were patched onto SC-His-Trp-Leu plates and replica plated to SC(Gal)-His-Trp-Leu for induction of Ty3 expression. These patches were replica plated to minimal medium supplemented with uracil. Cells that had undergone transposition and activated the suppressor expression grew in the absence of adenine and lysine and were identified as papillae within each patch. The relative frequency of retrotransposition was determined by comparison between the number of papillae arising from mutant and wild-type patches (Fig. 1). A total of 27,000 mutants containing mTn3 insertions generated from various pools of the insertion library were screened for the Ty3 phenotype. Details of the screen with a complete list of the genes isolated will be described in a separate paper.
FIG. 1.
Outline of the genetic assay for Ty3 retrotransposition. Leu+ transformants resulting from mutagenesis were patched onto SC-His-Trp-Leu medium and replicaplated onto galactose-containing medium to induce the expression of Ty3. These cultures were replica plated onto minimal medium plates supplemented with uracil to select for colonies in which suppressor activation by Ty3 insertion into the target plasmid allowed growth in medium lacking adenine and lysine.
Truncation of the TFC1 gene reduces Ty3 transposition.
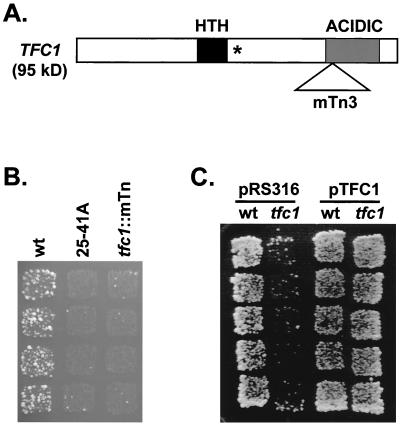
One mutant, 25-41A, exhibited an 11-fold decrease in transposition (data not shown) by a quantitative version of the divergent tRNA gene target assay. To identify the host mutation responsible for the Ty3 phenotype, vectorette PCR was used as described in Materials and Methods to amplify part of the mTn3 insertion and flanking genomic DNA. The sequence of the resulting PCR product was determined and compared to the Saccharomyces Genome Database. This search identified TFC1, which encodes an essential 95-kDa subunit of TFIIIC (TFIIIC95). This subunit has a helix-turn-helix motif and an acidic C-terminal domain (38). TFIIIC95 binds to tDNA (15) and is centrally located within TFIIIC over the box A promoter element (5). Sequence analysis indicated that mTn3 insertion at nucleotide position 1571 of TFC1 coding region would lead to expression of a truncated protein with the N-terminal 526 amino acids instead of the 649-residue full-length protein (Fig. 2A).
FIG. 2.
Truncation of TFC1 reduces detectable insertions into a divergent tRNA gene target plasmid. (A) Schematic of the TFC1 coding region and its motifs. The TFC1 coding region (open box) contains a helix-turn-helix (HTH) DNA-binding motif (black box) closely followed by a putative nuclear localization signal (asterisk). The C-terminal acidic domain (gray box) is truncated by mTn3 insertion (open triangle). (B) Retrotransposition assay of wild-type and mutant strains. The mutant strain (25-41A) isolated from the genetic screen and the reconstructed mutant (tfc1::mTn) exhibit severely reduced retrotransposition compared to the wild-type strain (wt). The retrotransposition assay was performed as described above. Each column represents four independent transformants assayed for indicated strain as described in the legend of Fig. 1. (C) Extrachromosomal expression of TFC1 restored retrotransposition to wild-type frequencies in the mutant strain. Wild-type (wt) and mutant (tfc1) strains were transformed with either control plasmid (pRS316) or TFC1 expression plasmid (pTFC1), and retrotransposition was assayed as described above using appropriate selective media. Each column represents five independent transformants.
To test whether the mTn3 insertion into the TFC1 gene was solely responsible for reduced Ty3 retrotransposition phenotype, the TFC1::mTn allele was recovered from the 25-41A mutant strain by the plasmid gap repair method (16) using a gapped plasmid-borne TFC1 gene. The mutant allele was then used to disrupt TFC1 in the wild-type strain by homologous recombination. Southern analysis with a TFC1-specific probe confirmed that this gene was disrupted in the reengineered strain as in the original mutant (data not shown). The resulting strain showed the same severely reduced transposition phenotype as the 25-41A mutant (Fig. 2B). Next, the effect of the wild-type TFC1 gene on the transposition frequency in the mutant strain was tested. The wild-type and mutant strains were transformed with either a control plasmid, pRS316, or a low-copy plasmid carrying TFC1, pTFC1, and retrotransposition was tested for multiple transformants. The mutant transformants with pRS316 showed severely reduced retrotransposition, but those with pTFC1 exhibited wild-type level of retrotransposition (Fig. 2C). These results showed that the reduced-transposition phenotype was caused by truncation of TFC1 gene and that the mutant allele was recessive since the mutant phenotype was rescued by a plasmid-borne wild-type gene.
Truncation of TFC1 has no detectable effect on growth or on pol III transcription.
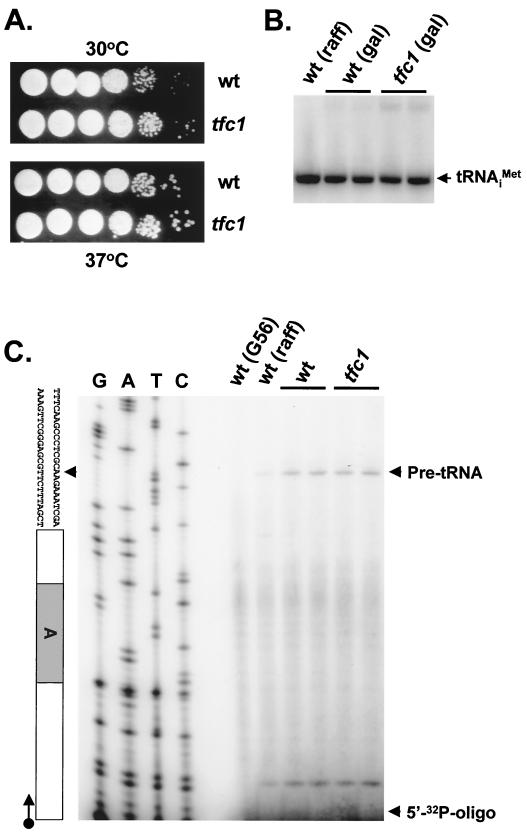
Although TFC1 is essential (38), the mutant with a truncated allele is viable. To test if this mutation causes a growth defect and thus might indirectly affect the transposition frequency, serial dilutions of wild-type and mutant cultures in mid-log phase were spotted onto YPD medium and incubated at 30°C for 2 days or at 37°C for 3 days. The mutant strain grew as well as the wild-type strain at both temperatures (Fig. 3A). Growth curves of liquid cultures also showed no significant difference in growth rate between the two strains (data not shown). These results indicated that even at 37°C, the mutant strain has no growth-limiting defect in pol III transcription.
FIG. 3.
Truncation of TFC1 does not have detectable effects on growth at 37°C or on the levels of tRNA test species. (A) Serial dilutions of wild-type (wt) and mutant (tfc1) cultures were spotted onto YPD medium and incubated at 30°C for 2 days (top) or at 37°C for 3 days (bottom). (B) Northern blot analysis of tRNAMet. Yeast strains were grown in appropriate synthetic medium with raffinose (raff) or galactose (gal) as the carbon source. Total yeast RNA extracted from each culture was used for Northern blot analysis with 32P-labeled oligonucleotide specific for mature tRNA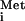 . (C) Autoradiograph of the primer extension reaction. Total yeast RNA was used as the template, and 32P-labeled SUP2b-specific oligonucleotide was used as the primer for each extension reaction. A DNA-sequencing ladder was generated from the pDLC356 plasmid using the same oligonucleotide. A strain with a C56G mutation in the box B promoter element (G56) was used as a negative control. The transcription initiation site, the primer extension product from pre-tRNA, and the 5′-end-labeled free oligonucleotide are indicated by arrowheads.
. (C) Autoradiograph of the primer extension reaction. Total yeast RNA was used as the template, and 32P-labeled SUP2b-specific oligonucleotide was used as the primer for each extension reaction. A DNA-sequencing ladder was generated from the pDLC356 plasmid using the same oligonucleotide. A strain with a C56G mutation in the box B promoter element (G56) was used as a negative control. The transcription initiation site, the primer extension product from pre-tRNA, and the 5′-end-labeled free oligonucleotide are indicated by arrowheads.
To test more specifically whether pol III transcripts were limiting in the mutant strain, cells carrying a plasmid with the SUP2b tRNA gene (pDLC356) (9) were grown with raffinose or galactose as the carbon source and total RNA was prepared. Northern blot analysis (Fig. 3B) showed no significant difference in mature tRNAMet levels between samples from the mutant and the wild type. To determine the accuracy of pol III transcription initiation in the mutant, reverse primer extension analysis was performed on the same RNA samples with an oligonucleotide primer specific for the SUP2b pre-tRNA intron (Fig. 3C). Comparison of results from mutant and wild-type samples showed identical sizes and amounts of the primer extension products, indicating that neither the transcription initiation site nor the amount of pre-tRNA was dramatically affected in the mutant strain. In addition, no dramatic difference in transcription activity was detected by in vitro transcription experiments using pol III transcription extracts prepared from wild-type and mutant strains (data not shown). To determine if Ty3 insertions into the divergent target could activate the expression of the suppressor tRNA gene, a target plasmid containing a Ty3 insertion was transformed into the tfc1 strain. Transformants grew readily on media requiring expression of the sup2bo gene. These results showed that the decrease in Ty3 retrotransposition was not due to a dramatic decrease in pol III transcription activity.
Truncation of TFC1 does not affect Ty3 intermediates.
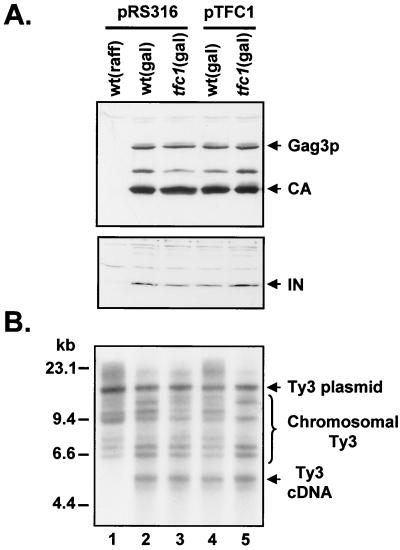
To find how truncation of TFC1 affects Ty3 transposition, the amounts of Ty3 CA, IN, and DNA intermediates in wild-type and mutant cells were compared. For these assays, wild-type and mutant strains carrying pTM45 and either pRS316 or pTFC1 were grown to early log-phase (absorbance at 600 nm [A600], 0.2 to 0.4) in synthetic medium with raffinose as the carbon source. Expression of Ty3 was induced by addition of galactose to a final concentration of 2%. After 6 h, the cells were harvested and proteins or DNA was isolated. Immunoblot analysis with anti-Ty3 CA and anti-Ty3 IN antibodies showed no significant difference in the amounts or processing of CA or IN protein between the wild-type or the mutant extracts with and without pTFC1 (Fig. 4A, lanes 2 to 5). Results of immunoblot analysis of VLPs prepared from these strains were consistent with these results (data not shown).
FIG. 4.
Truncation of TFC1 has no significant effect on the early steps of the Ty3 life cycle (A) Immunoblots with antibodies to Ty3 CA or IN proteins were performed on yeast extracts prepared from cultures grown in media containing raffinose or galactose as the carbon source. Each strain contained control plasmid (pRS316) or TFC1 expression plasmid (pTFC1) in addition to pTM45. Antibodies to CA recognize mature CA protein (26 kDa) and precursor Gag3p (38 kDa). Antibodies to IN recognize mature IN of 61 kDa. (B) Total yeast DNA was digested with EcoRI, and Southern blot analysis was performed with 32P-labeled, Ty3-specific probe, which hybridizes to full-length cDNA of 5.4 kbp as well as to Ty3 donor plasmid and chromosomal Ty3 elements (brace).
After particle assembly and protein maturation, Ty3 DNA is reverse transcribed from the genomic RNA template. Total DNA was extracted from wild-type and mutant cultures, and Southern blot analysis was performed using a radiolabeled Ty3-specific probe. Quantitative analysis of the blot using PhosphorImager and ImageQuant software (Molecular Dynamics) showed that similar amounts of full-length cDNA were present in both wild-type and mutant strains irrespective of the presence of pTFC1 (Fig. 4B).
Ty3 integration is qualitatively different in the tfc1 mutant.
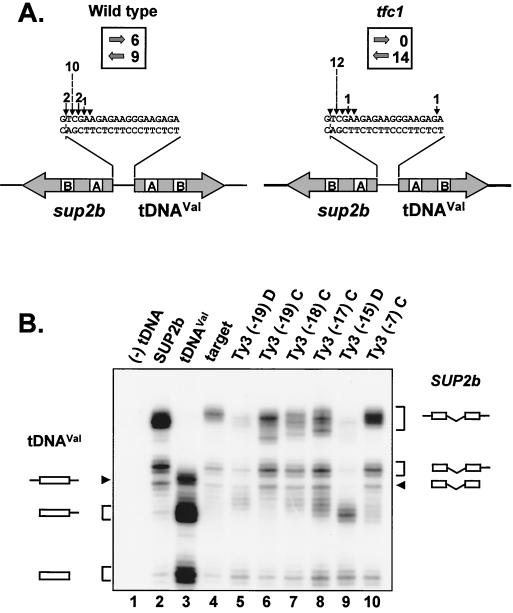
The previous finding that TFIIIC is essential for in vitro integration of Ty3 at a tRNA gene (27), coupled with the results described above, suggested that either the efficiency or the position of Ty3 integration was defective in the tfc1 mutant. To address the possibility that Ty3 integration was altered in the mutant strain so that integration events failed to occur in positions that activated sup2bo, Ty3 insertions in vivo were selected independent of the sup2bo expression. To facilitate the recovery of insertions into target plasmids, the ochre anticodon of sup2bo was changed to the wild-type anticodon (sup2b) and the target plasmid was converted into a high-copy plasmid. Wild-type and tfc1 mutant strains were transformed with pTM45; the modified target, pEH2b19V; and pDLC348, carrying a galactose-inducible, Neo-marked Ty3 (Ty3-N). Ty3-N insertions into the target plasmid were selected in yeast and plasmid DNA was recovered in bacteria as described in Materials and Methods. The Ty3 insertion sites in 15 plasmids recovered from the wild-type strain and 14 recovered from the tfc1 strain were determined by sequencing. All the Ty3-N insertions from the wild-type strain and 13 of 14 insertions from mutant strains were within the 5-bp window expected to activate sup2bo expression (Fig. 5A). A total of 12 of the 14 insertions recovered from the mutant and 10 of the 15 insertions from the wild type occurred at position −19. These data confirm the previous finding that tDNAVal is the target for the vast majority of insertions, although the spacing of the two genes results in Ty3 insertions occurring much closer to the 5′ end of sup2b. In addition, the characteristic 5-bp duplication of flanking sequence was observed for all 10 insertions for which the sequence at both Ty3-plasmid junctions was determined. Thus, the tfc1 strain showed no significant alteration in integration specificity.
FIG. 5.
(A) Orientation of Ty3 integration into a target plasmid is severely skewed in the mutant strain. A target plasmid with a nonsuppressor sup2b allele and a Neo-marked Ty3 were used to collect plasmids containing insertions. These were analyzed as described in Materials and Methods. The position of the Ty3-Neo insertions (the strand transfer site distal to the tDNAVal target) into the divergent tDNA target is indicated by the arrowheads, and the distribution of events is indicated by the height of the bars. The orientation of Ty3-Neo insertions is indicated above each figure by block arrows, and the numbers of events analyzed are indicated by the corresponding numbers. (B) Orientation of Ty3 affects the sup2b in vitro transcription. In vitro transcription reactions using BR500 extracts were performed as described in Materials and Methods. The reaction with no tDNA is indicated as (−) tDNA (lane 1). SUP2b (lane 2) and tDNAVal (lane 3) were used as positive control templates for each tRNA species. The tDNA templates used are indicated; the numbers in parentheses indicate the distance, in base pairs, of the integrated Ty3 sequence from the 5′ end of the coding region of tDNAVal. D and C refer to Ty3 insertions divergent (opposite) to and corresponding to (the same as) the orientation of tDNAVal, respectively. Diagrams to the left and right of the blot indicate RNA species deduced from control reactions and previously identified processed intermediates.
Inspection of Ty3-N insertions in tfc1 as a set showed that they were distinct from insertions in the wild-type strain in their orientation. Whereas 9 of 15 insertions in the wild-type strain occurred so that the transcriptional orientation of Ty3-N was opposing that of the tRNAVal target gene, all 14 insertions in the tfc1 mutant were in that orientation (Fig. 5A). A chi-square test of the mutant indicates that this distribution is not random (P < 0.005). Lack of recovery of Ty3-N in the same transcriptional orientation as the tDNAVal was not due to inability to select for Ty3-N in that orientation in the mutant background, since plasmids with Ty3-N insertions recovered from wild-type cells and transformed into mutant cells readily conferred resistance to G418 (data not shown). These data indicated that the orientation of Ty3 integration into the target plasmid was biased in the mutant strain.
The 11-fold reduction in the recovery of Ty3 insertions into the divergent target in the mutant strain was significantly greater than the 40 to 50% reduction predicted for loss of insertions in the same orientation as the target tDNA. We first tested whether Ty3 insertions in the two orientations affect sup2bo expression differently and thus contribute to the bias in detection of Ty3 integration. BR500 extracts from the wild-type strain were used to transcribe the tRNA genes on target plasmids with Ty3 integrated at different positions and in each orientation (Fig. 5B). Plasmid templates carrying either SUP2b (Fig. 5B, lane 2) or tDNAVal (lane 3) alone were included to identify the various RNA species generated. As expected, SUP2bo pre-tRNA was transcribed inefficiently from the divergent tRNA gene target plasmid compared to SUP2b alone (lane 4 compared to lane 2). Target plasmids with Ty3 integrated, at positions −19, −18, −17, and −7 relative to the 5′ end of the mature tRNA, in the same transcriptional orientation as the tDNAVal yielded slightly more unprocessed and processed RNA species than did the target plasmid alone (compare lanes 6, 7, 8, 10, and 4). Surprisingly, target plasmids with Ty3 at positions −19 and −15 and in the opposite orientation generated lower levels of pre-tRNAs and processed species than did the the target alone (compare lanes 5, 9, and 4). Thus, only Ty3 insertions in the same orientation as tDNAVal activated transcription. Similar results were obtained using extracts from the mutant strain (data not shown). If Ty3 insertion in vivo affected sup2bo expression similarly to what was observed in vitro, insertions in one orientation might not have been detected in the mutant or wild-type strains by the suppressor activation assay. To test this hypothesis, cells with independent Ty3 insertions into pPK689 in the wild-type strain were selected as described for Fig. 1. Of 30 insertions, 27 were found to be in the same orientation as the target tDNAVal by Southern blot analysis (data not shown). This suggested that in the wild-type background, where insertions of Ty3-N in both orientations were observed, Ty3 insertion activated sup2bo only when inserted in the same transcriptional orientation as tDNAVal. Thus, although insertions probably occurred in the tfc1 strain for Ty3, as they did for Ty3-N, they would not have been in the orientation that activated sup2bo.
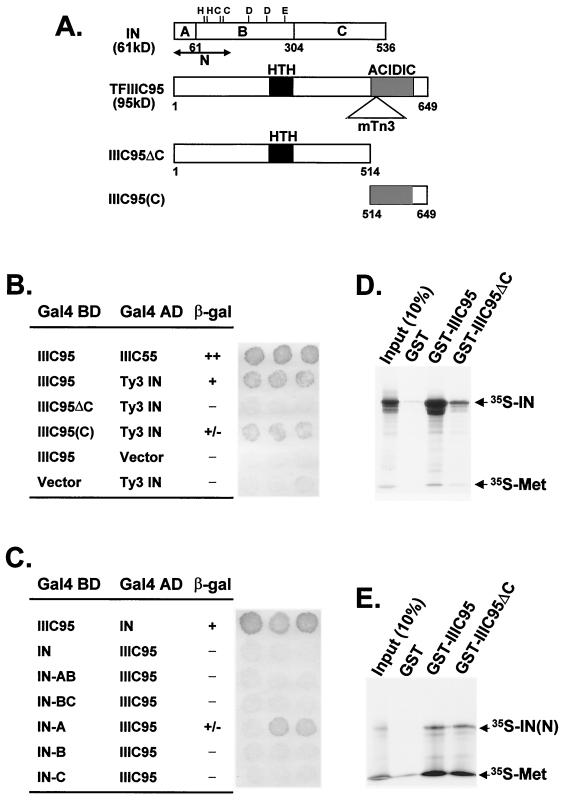
TFIIIC95 interacts with the amino-terminal domain of IN.
The fact that pol III transcription did not appear to be altered in the tfc1 mutant suggested that the orientation bias was not due to an indirect effect on the number of initiation complexes. Rather, it suggested that specific contacts between the Ty3 PIC and the preinitiation complex might be lacking in the mutant. The possibility that TFIIIC95 and Ty3 IN might interact directly was therefore investigated. As shown in Fig. 6B, TFIIIC95 fused to the DNA-binding domain of Gal4 (Gal4 BD; amino acids 1 to 147), when tested with Ty3 IN fused to the activation domain of Gal4 (Gal4 AD; amino acids 768 to 881) gave a robust signal in a two-hybrid assay. This signal was abrogated when the C-terminal region was deleted from TFIIIC95 (TFIIIC95ΔC), suggesting that this region is important for interaction with Ty3 IN. In addition, the C-terminal domain of TFIIIC95 (TFIIIC95C) gave a weak but significant signal when tested against IN. Full-length IN or IN-AB (amino acids 1 to 304) fused to the Gal4 BD did not show interaction with TFIIIC95 fused to Gal4 AD. However, equivalent interactions are not always observed in the two-hybrid assay in each of the two possible expression contexts (4). Testing of individual domains showed that the IN-A domain (amino acids 1 to 61), but not the other domains, interacted with TFIIIC95 (Fig. 6C).
FIG. 6.
TFIIIC95 interacts with Ty3 IN. (A) Domains of Ty3 IN and TFIIIC95 used for protein-protein interaction. IN domains cloned into two-hybrid vectors include A (amino acids 1 to 61), B (amino acids 62 to 304), and C (amino acids 305 to 536). The labels for TFIIIC95 are the same as in Fig. 2A. The numbers below represent the amino acid residues of TFIIIC95. (B) Filter assay for the yeast two-hybrid interaction. The Gal4 DNA-binding domain (BD) or the Gal4 activation domain (AD) was fused to full-length TFIIIC95, portions of TFIIIC 95, or Ty3 IN as indicated in the table. At least three independent transformants were tested for β-galactosidase (β-gal) activity for each pair of constructs. Positive and negative results are indicated by + and −, respectively, and weakly positive results are indicated by +/−. The interaction between IIIC95 and IIIC55 has been observed previously (30), and these transformants were used as a positive control. (C) The amino-terminal domain of Ty3 IN (IN-A) interacts with TFIIIC95 in the two-hybrid assay. The labels are as in panel B. (D) TFIIIC95 physically interacts with Ty3 IN in the GST pulldown assay. GST fusion proteins of full-length TFIIIC95, truncated protein (TFIIIC95ΔC), or GST bound to glutathione-Sepharose beads were incubated with 35S-labeled Ty3 IN. After repeated washing, proteins that remained bound to the beads were eluted and separated by SDS-PAGE. Labeled IN was visualized by autoradiography. Ten percent of the labeled protein used for incubation is shown for comparison, and full-length IN is indicated by an arrow. (E) IN-N (amino acids 1 to 150) physically interacts with TFIIIC95 in the GST pulldown assay. The in vitro 35S-labeled IN(N) fragment, used as the probe, is indicated by an arrow.
To further investigate the IN-TFIIIC95 interaction, GST fusions with full-length and truncated TFIIIC95 were expressed in E. coli and purified on glutathione-Sepharose beads. These GST-TFCIII95 fusion proteins were tested for interaction with IN labeled with [35S]methionine produced in a coupled transcription-translation system (Promega) (Fig. 6D). In accordance with two-hybrid results, full-length IN interacted strongly with GST-TFIIIC95 and truncated TFIIIC retained significantly less 35S-IN (compare the two right lanes). Moreover, in a GST pulldown assay, 35S-labeled TFIIIC95 showed weak interaction with GST–IN-A (data not shown). Because the IN-A contains few methionine residues, it does not incorporate [35S]methionine at a significant level. Therefore, IN-N (amino acids 1 to 150) was tested for interaction with GST-TFIIIC95. This experiment showed that the IN-N domain was sufficient to interact with TFIIIC95, but no differential interaction was observed between IN-N and GST-TFIIIC95 or GST-TFIIIC95ΔC (Fig. 6E). These data suggested that the N-terminal domain of IN was minimally required to interact with TFIIIC95 but that a larger fragment, possibly full-length IN, was required to distinguish between the full-length TFIIIC95 and TFIIIC95ΔC. The fact that the C terminus of TFIIIC95 was important for optimal protein-protein interaction with IN argued that both TFIIIC95 and IN are involved in mediating Ty3 integration orientation.
DISCUSSION
For both transcription and transposition in vivo, the box B element, which is bound by TFIIIC, is required by TATA-containing genes and TATA-less genes. However, similar to pol III, in vitro Ty3 requires TFIIIC and TFIIIB for interaction at TATA-less tRNA genes but only TFIIIB at a TATA-containing gene (40). This would seem to suggest that the role of TFIIIC in transcription and transposition is indirect, that is, to load TFIIIB. However, subunits of pol III do interact directly with TFIIIC (13, 19), raising the possibility that TFIIIC contributes directly to recruitment of pol III and potentially Ty3 as well. In this work, we describe the recovery from a large-scale screen of a novel, viable mutant that has a truncated subunit of TFIIIC. In the mutant background, Ty3 transposition is position specific but occurs in only one orientation in the context of a synthetic divergent tRNA gene target. Investigation of the basis of this effect indicated interactions between the nonconserved, N-terminal domain of Ty3 IN and the acidic C-terminal domain of TFIIIC95. While these data did not demonstrate that a direct contact between the Ty3 PIC and TFIIIC is required for specificity, they did provide the first evidence that both TFIIIC and IN could interact directly during Ty3 integration. In addition, they show the potential of the two Ty3 ends for dramatically different integration activity.
The acidic C-terminal domain of TFIIIC95 is not essential.
Studies of the yeast and human TFIIIC transcription factors have shown that aspects of structure and function are conserved. Both complexes are comprised of multiple subunits: yeast TFIIIC (yTFIIIC) contains six subunits, and human TFIIIC (hTFIIIC) has at least nine polypeptides (39). The human homologue of yTFIIIC95 is a 63-kDa protein (hTFIIIC63) (19) containing helix-loop-helix and C-terminal acidic domains. Yeast TFIIIC95 contacts the internal promoter box A element (5) and interacts with the 55-kDa subunit of TFIIIC (30). Interestingly, hTFIII63 shows physical interactions with hTFIIIC90 and hTFIIIB90 (Brf), as well as with a subunit of pol III (19). In yeast, a point mutation in TFC1 affects TFIIIB complex formation (S. Jourdain et al., unpublished work). This observation also suggests interaction of TFIIIC95 with TFIIIB. Although the C-terminal acidic region is conserved between the human and yeast proteins, its function is not known. Presumably it is not required for DNA binding or pol III interaction under normal growth conditions, since neither the deletion mutant isolated in this screen nor a strain expressing TFIIIC95 with β-galactosidase fused at the C-terminal end showed obvious growth defects (11). This domain is also not essential for interaction of TFIIIC95 with TFIIIC55 (S. Jourdain, unpublished work). However, the genetic and biochemical experiments reported here suggest that the TFIIIC95 C-terminal region could be a protein-protein interaction domain.
The amino-terminal domain of Ty3 IN interacts with TFIIIC.
Two-hybrid and GST pulldown assays indicated that the Ty3 IN amino-terminal domain is likely to mediate at least some of the contact between the PIC and TFIIIC. Retroelement IN proteins can be divided into amino-terminal, core, and carboxyl-terminal domains. The N-terminal domain contains a Zn2+-binding HHCC motif but is otherwise not well conserved (2). This domain is required for multimerization of IN (41) and strand transfer but not for disintegration, the reverse of strand transfer, in vitro (7). The Ty3 IN amino-terminal sequence is almost 100 amino acids longer than that of human immunodeficiency virus IN and also contains a zinc-binding motif (17). The present study showed that the first 61 amino acids of Ty3 IN can interact weakly with TFIIIC95 and that the interaction is enhanced by the presence of the HHCC domain (data not shown and Fig. 6E). The amino-terminal domain also contains several patches of charged residues. Ty3 IN mutants with substitutions of alanine for basic amino acids at positions 53 and 54 and with substitutions of alanine for basic amino acids from positions 62 and 63 fall to transpose but are only slightly reduced for replicated cDNA (33), suggesting a defect at a late step in the life cycle. It will be of interest to determine whether these mutants display orientation bias in integration.
Model for Ty3 position-specific integration.
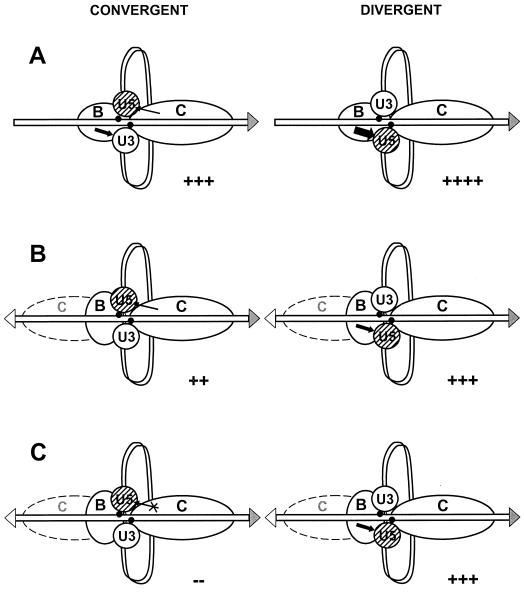
Based on previous in vivo results showing that the TFIIIC binding site is required for Ty3 integration at TATA-containing and TATA-less targets (9) and on in vitro results with TATA-containing genes showing that TFIIIB is sufficient for Ty3 integration, a model was proposed that TFIIIC was required for integration as the TFIIIB loading factor but was not directly involved in contacts with the PIC. Results from the present in vivo study have contributed to significant revision and extension of this model for Ty3 position specific integration. This revised model (Fig. 7) has four features. (i) TFIIIB is a major determinant of Ty3 targeting and can target integration in either orientation (Fig. 7A). This feature is based on the observation that TFIIIB is sufficient to target in vitro integration in both orientations at the TATA-containing SNR6 gene. No integration is observed for TFIIIC alone (40). (ii) The Ty3 PIC is asymmetric. The existence of a Ty3 tDNA target makes it possible to define orientation for Ty3 insertions, and because Ty3 inserts with dramatic orientation bias relative to the tDNAVal target gene, we know that the Ty3 PIC itself cannot be symmetric. A completely symmetric Ty3 PIC could not display insertion bias at any insertion site. Although the synthetic, divergent tDNA target showed the asymmetric behavior of the Ty3 PIC in a particularly dramatic pattern, previous observations are also consistent with asymmetric behavior of the Ty3 PIC at genomic targets. In a study of 91 independent insertions into the genome, examination of seven pairs of independent integrations, each at the same genomic tRNA gene, showed an overall distribution of orientations similar to the distribution of orientations of preexisting genomic insertions, but each individual pair of insertions at a particular target occurred in the same orientation. The probability that both pair members at each site would be in the same orientation is quite low (8). This result would be consistent with differential interaction of at least one end of the cDNA in the PIC with the target complex but also suggests that individual tRNA genes might differ with respect to features that affect orientation, such as strand sequence or TFIIIC occupancy. Figure 7 shows how preferential interactions between TFIIIB and U5, coupled with interactions between TFIIIC and U5 but not U3, could lead to insertions in either orientation at a genomic tDNA (Fig. 7A) or at the divergent target in the wild-type background (Fig. 7B) but might lead to biased integration at the divergent target in the tfc1 mutant background (Fig. 7C). (A detailed explanation of the diagram is given in the legend.) Although preferential interactions between the factors and the ends of the Ty3 PIC would be consistent with our observations, the specific interactions shown in Fig. 7 are for illustrative purposes; there are no data that demonstrate particular preferential interactions between U3 and U5 or specific protein domains exposed at these ends and TFIIIB or TFIIIC. (iii) TFIIIC is probably associated with at least some targets during integration. It was shown in the present study that TFIIIC95 interacts with IN. The distribution of Ty3-N insertions for the divergent target is broader in the wild type than in the tfc1 strains, indicating that an interaction between IN and TFIIIC95 could influence integration site selection in vivo. Interestingly, the pattern of integrations observed in this study in the presence and absence of the C-terminal domain of TFIIIC95 is similar to the pattern of in vitro integration at a SNR6 target in the presence and absence, respectively, of TFIIIC (L. Yieh et al., unpublished data). (iv) Contact between Ty3 IN and the TFIIIC95 C-terminal domain, while not essential, either facilitates integration in the same transcriptional orientation as the target tDNA or impedes it in the opposite orientation (Fig. 7, band C). The synthetic target is neither transcribed nor used as a transposition target as efficiently as a wild-type tRNA gene (25). Among other possibilities, this could result from attenuated TFIIIB function caused by steric interference with proper TFIIIB binding upstream of the target tRNAVal gene by TFIIIC or TFIIIB bound to the divergent sup2bo gene. With loss of the C-terminal domain of TFIIIC95, Ty3 insertions in the same transcriptional orientation as tDNAVal in the divergent target did not occur; however, insertions were still observed in this orientation at chromosomal tRNA gene targets (M. Aye et al., unpublished data). These observations argue that some feature of the divergent tDNA target, such as attenuated TFIIIB function, causes increased dependence on TFIIIC (Fig. 7B) for Ty3 insertions in the same orientation as the target tDNAVal and that this dependence is associated with the TFIIIC95 C-terminal domain. Ty3 IN mutations that disrupt the interaction between IN and TFIIIC95 could be used to directly test this aspect of the model since they would be predicted to bias insertion in a similar way to the TFIIIC95 truncation.
FIG. 7.
Model of Ty3 integration at an isolated tRNA gene and at tDNAVal (divergent target). (A) Integration into an isolated tRNA gene target. Hypothetical preferential interaction of TFIIIB with the U5 end and interaction of TFIIIC with the U5 end are shown. (B) Integration into the divergent tDNA target in the wild-type background. This diagram shows one possible scenario—that interference by TFIIIC bound to sup2bo with TFIIIB binding to the tDNAVal in the divergent target attenuates the TFIIIB interaction (particularly the weaker interaction at U3), thereby enhancing the dependence on the TFIIIC interaction with U5. (C) Integration into the divergent tDNA target in the tfc1 mutant. The diagram illustrates one possible scenario, i.e., that loss of the TFIIIC contact in the tfc1 mutant results in absolute dependence on the remaining TFIIIB interaction and commensurate bias in the orientation of insertions. Contacts leading to convergent insertions (same direction of transcription as the target tDNA) are shown on the left, and those leading to insertions with a divergent direction of transcription relative to the tDNAVal target are shown on the right. Target DNA is shown as an open bar, and Ty3 DNA is shown as a ribbon. Black dots indicate sites of the strand transfer reaction. TFIIIB and TFIIIC are shown as ellipses labeled B and C, respectively. A second TFIIIC bound to sup2bo in panels B and C is shown as a dashed ellipse. IN bound to the U3 and U5 ends of the DNA is shown as open and hatched balls, respectively. Interactions between DNA or protein domains at U3 and U5 and TFIIIC or TFIIIB are shown as line or block arrows correlating with the extent of the interaction. Loss of interaction in the tfc1 mutant is indicated by the X. The relative frequency of insertions is shown at the bottom of each figure. The direction of transcription of the isolated tRNA gene (A) and tRNAVal genes (B and C) is shown by shaded arrowheads. The direction of transcription of sup2bo (B and C) is shown by the open arrowhead.
One novel aspect of the Ty3 integration model introduced here is that the ends of the element, in spite of containing perfect inverted repeats for IN recognition, clearly have different integration activities and are likely to reflect this in their interactions with TFIIIB and TFIIIC. The key observation of orientation bias was possible only because a large set of Ty3 integrations can be observed at a specific target, a situation which does not occur for retroviruses. Orientation specificity has also been observed in Tn7, a bacterial transposon with a defined target, (3). While difficult to elucidate in retroviruses, orientation has clear implications for the effect of proviral enhancers and promoters on flanking genes (18) and is a relatively unexplored aspect of the retrovirus-host interaction.
Although Ty3 is similar to retroviruses in organization and proteins encoded, it has a high degree of position specificity. Ty1 to Ty4 are each associated with tRNA genes; however, this is the first report of direct contact between a member of the PIC and a pol III transcription factor. The contribution of this contact for integration may vary for each target, and our ability to use a targeted insertion assay was crucial for distinguishing different effects of the tfc1 mutation at different loci. These studies suggest that retroelements with targeting properties might be also be useful as reporters of protein occupancy at the loci where they insert. In cases where members of a gene family may be differentially associated with protein factors, these elements could offer insights into whether the distribution into different states reflects some kinetic parameter of the population as a whole or the differential behavior of some subset of genes or cells where transposition occurred.
ACKNOWLEDGMENTS
We thank M. Snyder for providing the mTn3::lacZ/LEU2 library and C. Friddle for providing the vectorette PCR protocol. We also thank E. Chen for assistance with the mutant screen, M. H. Nymark-McMahon and L. Yieh for assistance with VLP and BR500 preparations, J. Steffan for assistance with GST pulldown assays, T. Menees for helpful discussions, and A. Sentenac for critical reading of the manuscript.
This work was supported by Public Health Service grant GM33281 to S.B.S. and by the Synthesis and Structure of Biological Macromolecules training grant GM07311-24 (M.A.).
REFERENCES
- 1.Aiyar A, Hindmarsh P, Skalka A M, Leis J. Concerted integration of linear retroviral DNA by the avian sarcoma virus integrase in vitro: dependence on both long terminal repeat termini. J Virol. 1996;70:3571–3580. doi: 10.1128/jvi.70.6.3571-3580.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrake M D, Skalka A M. Retroviral Integrase, putting the pieces together. J Biol Chem. 1996;271:19633–19636. doi: 10.1074/jbc.271.33.19633. [DOI] [PubMed] [Google Scholar]
- 3.Arciszewska L K, Drake D, Craig N L. Transposon Tn7. cis-acting sequences in transposition and transposition immunity. J Mol Biol. 1989;207:35–52. doi: 10.1016/0022-2836(89)90439-7. [DOI] [PubMed] [Google Scholar]
- 4.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. 4, section 20.1. New York, N.Y: Greene Publishing Associates/Wiley-Interscience; 1999. [Google Scholar]
- 5.Bartholomew B, Kassavetis G A, Braun B R, Geiduschek E P. The subunit structure of Saccharomyces cerevisiae transcription factor IIIC probed with a novel photocrosslinking reagent. EMBO J. 1990;9:2197–2205. doi: 10.1002/j.1460-2075.1990.tb07389.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burns N, Grimwade B, Ross-Macdonald P B, Choi E-Y, Finberg K, Roeder G S, Snyder M. Large-scale analysis of gene expression, protein localization, and gene disruption in Saccharomyces cerevisiae. Genes Dev. 1994;8:1087–1105. doi: 10.1101/gad.8.9.1087. [DOI] [PubMed] [Google Scholar]
- 7.Cannon P M, Wilson W, Byles E, Kingsman S M, Kingsman A J. Human immunodeficiency virus type 1 integrase: effect on viral replication of mutations at highly conserved residues. J Virol. 1994;68:4768–4775. doi: 10.1128/jvi.68.8.4768-4775.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chalker D L, Sandmeyer S B. Transfer RNA genes are genomic targets for de novo transposition of the yeast retrotransposon Ty3. Genetics. 1990;126:837–850. doi: 10.1093/genetics/126.4.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chalker D L, Sandmeyer S B. Ty3 integrates within the region of RNA polymerase III transcription initiation. Genes Dev. 1992;6:117–128. doi: 10.1101/gad.6.1.117. [DOI] [PubMed] [Google Scholar]
- 10.Clark D J, Bilanchone V W, Haywood L J, Dildine S L, Sandmeyer S B. A yeast sigma composite element, Ty3, has properties of a retrotransposon. J Biol Chem. 1988;263:1413–1423. [PubMed] [Google Scholar]
- 11.Conesa C, Swanson R N, Schultz P, Oudet P, Sentenac A. On the subunit composition, stoichiometry, and phosphorylation of the yeast transcription factor TFIIIC/tau. J Biol Chem. 1993;268:18047–18052. [PubMed] [Google Scholar]
- 12.Curcio M J, Garfinkel D J. Heterogeneous functional Ty1 elements are abundant in the Saccharomyces cerevisiae genome. Genetics. 1994;136:1245–1259. doi: 10.1093/genetics/136.4.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dumay H, Rubbi L, Sentenac A, Marck C. Interaction between yeast RNA polymerase III and transcription factor TFIIIC via ABC10alpha and tau131 subunits. J BiolChem. 1999;274:33462–33468. doi: 10.1074/jbc.274.47.33462. [DOI] [PubMed] [Google Scholar]
- 14.Farnet C M, Bushman F D. HIV-1 cDNA integration: requirement of HMG I(Y) protein for function of preintegration complexes in vitro. Cell. 1997;88:483–492. doi: 10.1016/s0092-8674(00)81888-7. [DOI] [PubMed] [Google Scholar]
- 15.Gabrielsen O S, Marzouki N, Ruet A, Sentenac A, Fromogeot P. Two polypeptide chains in yeast transcription factor τ interact with DNA. J Biol Chem. 1989;264:7505–7511. [PubMed] [Google Scholar]
- 16.Guthrie C, Fink G R. Guide to yeast genetics and molecular biology. San Diego, Calif: Academic Press, Inc.; 1991. [Google Scholar]
- 17.Hansen L J, Chalker D L, Sandmeyer S B. Ty3, a yeast retrotransposon associated with tRNA genes, has homology to animal retroviruses. MolCellBiol. 1988;8:5245–5256. doi: 10.1128/mcb.8.12.5245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hayward W S, Neel B G, Astrin S M. Activation of a cellular oncogene by promoter insertion in ALV-induced lymphoid leukosis. Nature. 1981;290:475–480. doi: 10.1038/290475a0. [DOI] [PubMed] [Google Scholar]
- 19.Hsieh Y, Wang Z, Kovelman R, Roeder R G. Cloning and characterization of two evolutionarily conserved subunits (TFIIIC102 and TFIIIC63) of human TFIIIC and their involvement in functional interactions with TFIIIB and RNA polymerase III. MolCellBiol. 1999;19:4944–4952. doi: 10.1128/mcb.19.7.4944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang H, Hong J Y, Burck C L, Liebman S W. Host genes that affect the target-site distribution of the yeast retrotransposon Ty1. Genetics. 1999;151:1393–1407. doi: 10.1093/genetics/151.4.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kalpana G V, Marmon S, Wang W, Crabtree G R, Goff S P. Binding and stimulation of HIV-1 integrase by a human homolog of yeast transcription factor SNF5. Science. 1994;266:2002–2006. doi: 10.1126/science.7801128. [DOI] [PubMed] [Google Scholar]
- 23.Kassavetis G A, Riggs D L, Negri R, Nguyen L H, Geiduschek E P. Transcription factor IIIB generates extended DNA interactions in RNA polymerase III transcription complexes on tRNA genes. MolCellBiol. 1989;9:2551–2566. doi: 10.1128/mcb.9.6.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim J M, Vanguri S, Boeke J D, Gabriel A, Voytas D F. Transposable elements and genome organization: a comprehensive survey of retrotransposons revealed by the complete Saccharomyces cerevisiae genome sequence. Genome Res. 1998;8:464–478. doi: 10.1101/gr.8.5.464. [DOI] [PubMed] [Google Scholar]
- 25.Kinsey P, Sandmeyer S. Ty3 transposes in mating populations of yeast: a novel transposition assay for Ty3. Genetics. 1995;139:81–94. doi: 10.1093/genetics/139.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kinsey P T, Sandmeyer S B. Adjacent pol II and pol III promoters: transcription of the yeast retrotransposon Ty3 and a target tRNA gene. Nucleic Acids Res. 1991;19:1317–1324. doi: 10.1093/nar/19.6.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kirchner J, Connolly C M, Sandmeyer S B. Requirement of RNA polymerase III transcription factors for in vitro position-specific integration of a retroviruslike element. Science. 1995;267:1488–1491. doi: 10.1126/science.7878467. [DOI] [PubMed] [Google Scholar]
- 28.Kunkel T A. Rapid and efficient site-specific mutagenesis without phenotypic selection. ProcNatlAcadSciUSA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee M S, Craigie R. A previously unidentified host protein protects retroviral DNA from autointegration. ProcNatlAcadSciUSA. 1998;95:1528–1533. doi: 10.1073/pnas.95.4.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Manaud N, Arrebola R, Buffin-Meyer B, Lefebvre O, Voss H, Riva M, Conesa C, Sentenac A. A chimeric subunit of yeast transcription factor IIIC forms a subcomplex with tau95. MolCellBiol. 1998;18:3191–3200. doi: 10.1128/mcb.18.6.3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Menees T M, Sandmeyer S B. Transposition of the yeast retroviruslike element Ty3 is dependent on the cell cycle. MolCellBiol. 1994;14:8229–8240. doi: 10.1128/mcb.14.12.8229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morris M E, Jinks-Robertson S. Nucleotide sequence of the LYS2 gene of Saccharomyces cerevisiae: homology to Bacillus brevis tyrocidine synthetase. Gene. 1991;98:141–145. doi: 10.1016/0378-1119(91)90117-t. [DOI] [PubMed] [Google Scholar]
- 33.Nymark-McMahon M H, Sandmeyer S B. Mutations in nonconserved domains of Ty3 integrase affect multiple stages of the Ty3 life cycle. J Virol. 1999;73:453–465. doi: 10.1128/jvi.73.1.453-465.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sandmeyer, S. B., M. Aye, and T. M. Menees. Ty3: a position-specific, cypsylike element in Saccharomyces cerevisiae. In N. L. Craig, R. Craigie, M. Gellert, and A. M. Lambowitz (ed.), Mobile DNA II, in press. ASM Press, Washington, D.C.
- 35.Sherman F, Fink G R, Hicks J B. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1986. [Google Scholar]
- 36.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficent manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Steffan J S, Keys D A, Dodd J A, Nomura M. The role of TBP in rDNA transcription by RNA polymerase I in Saccharomyces cerevisiae: TBP is required for upstream activation factor-dependent recruitment of core factor. Genes Dev. 1996;10:2551–2563. doi: 10.1101/gad.10.20.2551. [DOI] [PubMed] [Google Scholar]
- 38.Swanson R N, Conesa C, Lefebvre O, Carles C, Ruet A, Quemeneur E, Gagnon J, Sentenac A. Isolation of TFC1, a gene encoding one of two DNA-binding subunits of yeast transcription factor tau (TFIIIC) ProcNatlAcadSciUSA. 1991;88:4887–4891. doi: 10.1073/pnas.88.11.4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.White R J. RNA polymerase III transcription. Berlin, Germany: Springer-Verlag KG and R. G. Landes; 1998. [Google Scholar]
- 40.Yieh L, Kassavetis G, Geiduschek E P, Sandmeyer S B. The Brf and TBP subunits of the RNA polymerase III transcription factor IIIB mediate position-specific integration of the gypsy-like element, Ty3. J Biol Chem. 2000;275:29767–29771. doi: 10.1074/jbc.M003149200. [DOI] [PubMed] [Google Scholar]
- 41.Zheng R, Jenkins T M, Craigie R. Zinc folds the N-terminal domain of HIV-1 integrase, promotes multimerization, and enhances catalytic activity. ProcNatlAcadSciUSA. 1996;93:13659–13664. doi: 10.1073/pnas.93.24.13659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhu Y, Zou S, Wright D A, Voytas D F. Tagging chromatin with retrotransposons: target specificity of the Saccharomyces Ty5 retrotransposon changes with the chromosomal localization of Sir3p and Sir4p. Genes Dev. 1999;13:2738–2749. doi: 10.1101/gad.13.20.2738. [DOI] [PMC free article] [PubMed] [Google Scholar]