Abstract
Alzheimer’s disease (AD) is one of the most persistent and devastating neurodegenerative disorders of old age, and is characterized clinically by an insidious onset and a gradual, progressive deterioration of cognitive abilities, ranging from loss of memory to impairment of judgement and reasoning. Despite years of research, an effective cure is still not available. Autophagy is the cellular ‘garbage’ clearance system which plays fundamental roles in neurogenesis, neuronal development and activity, and brain health, including memory and learning. A selective sub-type of autophagy is mitophagy which recognizes and degrades damaged or superfluous mitochondria to maintain a healthy and necessary cellular mitochondrial pool. However, emerging evidence from animal models and human samples suggests an age-dependent reduction of autophagy and mitophagy, which are also compromised in AD. Upregulation of autophagy/mitophagy slows down memory loss and ameliorates clinical features in animal models of AD. In this review, we give an overview of autophagy and mitophagy and their link to the progression of AD. We also summarize approaches to upregulate autophagy/mitophagy. We hypothesize that age-dependent compromised autophagy/mitophagy is a cause of brain ageing and a risk factor for AD, while restoration of autophagy/mitophagy to more youthful levels could return the brain to health.
Keywords: Alzheimer’s disease, Autophagy, Mitophagy, Ageing
1. Autophagy: history, mechanism, and functions
Autophagy (from the Greek words “auto” meaning “self”, and “phagein” meaning “to eat”) is a cellular evolutionary conserved mechanism through which eukaryotic cells break down and recycle old, dysfunctional, or damaged subcellular components, such as defective organelles and misfolded protein aggregates [1], [2], [3]. Autophagy was first observed by Keith R. Porter and his student Thomas Ashford at the Rockefeller Institute in 1962, when they reported an increased number of lysosomes in rat liver cells after the addition of glucagon, and some displaced lysosomes towards the center of the cell containing other cell organelles such as mitochondria[4]. The term “autophagy” was coined by the Belgian biochemist Christian de Duve in 1963 based on his discovery of lysosome function[5]. In the 1990s studies identified several autophagy-related (ATG) genes in yeast that are highly conserved in mammals. In 2016 Professor Yoshinori Ohsumi was awarded the Nobel Prize in Physiology or Medicine for his discovery of the mechanisms behind autophagy (Table 1).
Table 1.
A summary of a list of autophagy and mitophagy inducers and their biomedical activities.
| Mitophagy inducers | Chemical formula | Mechanism | Main effects on AD models | Reference |
|---|---|---|---|---|
| 5-aminoimidazole-4-carboxamide riboside (AICAR) | CH14N4O5 | AMPK activator. | Increases spatial memory and improves motor function in young and old mice. | [1] |
| Berberine | C20H18NO4+ | Berberine induces autophagy through the class III PI3K/Beclin-1/Bcl-2 pathway. Additionally, berberine triggers the enhancement of lysosomal activity, which effectively degrades LC3-Ⅱ-positive autophagosomes. |
Promotes Aβ clearance and inhibits its production, improves learning capacity and memory retentions and attenuates the hyperphosphorylation of tau in 3 × Tg AD mice. | [2] |
| Carbamazepine | C15H12N2O | Carbamazepine stimulates autophagy by a mechanism dependent on the myo-inositol levels and AMPK activation. | Alleviates memory deficits and cerebral Aβ pathology in APP/PS1 mice | [3,4] |
| Cinnamic acid | C9H8O2 | Cinnamic acid activates the nuclear hormone receptor PPARα to transcriptionally upregulate TFEB and stimulates lysosomal biogenesis. | Stimulates lysosomal biogenesis, decreases Aβ plaque burden and improves memory and behavioral performance in 5XFAD mice. | [5] |
| Everolimus | C53H83NO14 | Allosteric mTORC1-specific inhibitor. | Reduces human APP/Aβ and human tau levels and improves cognitive function in 3 × Tg AD mice. | [6] |
| Gemfibrozil and Wy14643 | C15H22O3 and C14H14ClN3O2S | Both of them are PPARA activators and can activate autophagy through TFEB pathaway. | Reverse memory deficits and anxiety symptoms and reduce level of soluble and insoluble Aβ in APP-PSEN1ΔE9 mice. | [7] |
| Gypenoside XVII | C48H82O18 | Gypenoside XVII enhances lysosome biogenesis and autophagy flux through TFEB activation. | Restores the spatial learning and memory, decreases soluble and insoluble fraction of Aβ and prevents the formation of Aβ plaques in the hippocampus and cortex of APP/PS1 mice. | [7,8] |
| HEP14 | C25H34O5 | HEP14 induces PKC-dependent activation of TFEB. | Reduces Aβ plaque formation in APP/PS1 mice. | [9] |
| Latrepirdine | C21H25N3 | Latrepirdine enhances mTOR- and Atg5-dependent autophagy. | Improves memory, degrades p62 and reduces the accumulation of insoluble Aβ42 in TgCRND8 mice. | [10] |
| Lithium chloride | LiCl | Lithium chloride induces autophagy by inhibiting inositol monophosphatase. | Restores the long-term spatial memory deficit and reduce brain Aβ levels in APP/PS1 mice. | [11,12] |
| Melatonin | C13H16N2O2 | Melatonin exhibits a beneficial effect on mitochondria by reducing the oxygen consumption rate, oxygen flux and membrane potential and, therefore, suppressing ROS production. | Reduces Aβ generation and modulates and maintains tau phosphorylation | [13–15] |
| Metformin | C4H11N5 | Metformin triggers autophagy through AMPK activation and subsequent inhibition of mTOR. | Decreases tau phosphorylation in human tau transgenic mice. Decreases Aβ influx across the blood–brain barrier, improves memory impairment under diabetic context. Attenuates spatial memory deficit, hippocampal neuron loss, enhances neurogenesis, decreases Aβ plaque load and chronic inflammation in APP/PS1 mice. | [16,17,18] |
| Nicotinamide mononucleotide(NMN) | C11H15N2O8P | NMN is a NAD+ precursor and mitophagy activator. | Reduces Aβ load, decreases p-Tau levels, improves cognitive and memory functions, reduces neuroinflammation and promotes microglia phagocytic activity in APP/PS1, 3xTgAD, Aβ and Tau models of C.elegans. | [19] |
| Nicotinamide riboside (NR) | C11H15N2O5 | NR is a NAD+ precursor and mitophagy inducer. | Increases NAD+ levels, promotes neurogenesis, improves cognition and reduces p-Tau and Aβ load and aggregation in Aβ model of C. elegans, 3xTgAD and APP/PS1 mice, APP-SH-SY5Y cells. | [19,20] |
| Nicotinamide(NAM) | C6H6N2O | NAM could enhance acidification of intracellular acidic organelles. In NAM-treated 3xTgAD mice the LC3-II/LC3-I ratio is reduced. | Improves cognitive performance, reduces Aβ and p-Tau pathologies and improves mitochondrial dynamics in 3xTgAD mice. Increses SIRT1 expression, which mediates the stress resistance signaling in NAM-treated 3xTgAD mice. | [21,22] |
| Ouabain | C29H44O12 | Ouabain enhances activation of TFEB through inhibition of the mTOR pathway and induces downstream autophagy-lysosomal gene expression. | Reduces the accumulation of phosphorylated tau in tau transgenic flies as well as improves memory inTauP301L mice. | [23] |
| Rapamycin | C51H79NO13 | Inhibition of mTOR signaling. | Decreases intraneuronal accumulation of Aβ, decreases brain levels of pathogenic Aβ42 and attenuates age-dependent accumulation of phosphorylated and aggregated tau in 3 × Tg-AD and PDAPP mice. Prevents synaptic failure induced by Aβ oligomers. There are ongoing clinical trials in humans. | [24–26] |
| Resveratrol | C14H12O3 | Resveratrol activates autophagy through the mTOR-ULK1 pathway and SIRT1/AMPK signaling pathway. | Decreases brain Aβ levels.Modulates Aβ cleveage preventing its oligomerization. The effects of resveratrol in neurodegeneration are widely described in a review. | [27,28] |
| Simvastatin | C25H38O5 | Simvastatin can activates LBK1-AMPK-mTOR signalling pathway. | Efficiently reduces levels of Alzheimer’s Aβ in yeast and reduces cerebral Aβ42 and Aβ40 levels in the cerebrospinal fluid and brain homogenate. | [29,30] |
| Spermidine | C7H19N3 | Spermidine triggers autophagy mainly by promoting AMPK phosphorylation and mitophagy via PINK1/Parkin pathway. | Reduces neuroinflammation and soluble Aβ in APP/PS1 mice and prevents Tau fibrillization in rTg4510 mice. | [31,32] |
| Temsirolimus | C56H87NO16 | Allosteric mTORC1-specific inhibitor. | Induces autophagy, reduces both Aβ and p-Tau and improves motor functions in several AD mouse models, such as APP/PS1, P301S and Tg30. | [33–35] |
| Tomatidine | C27H45NO2 | Tomatidine maintains mitochondrial homeostasis by modulating mitochondrial biogenesis and PINK-1/DCT-1-dependent mitophagy. | Counteracts age-related deterioration of muscle function in C. elegans through the activation of cellular stress responses against metabolic and oxidative stress. | [36] |
| Trehalose | C12H22O11 | Trehalose regulates autophagy by inducing rapid and transient lysosomal enlargement and membrane permeabilization. | Inhibits Aβ generation in APP23 mice and in HAW and 20E2 cells. | [37,38] |
1. Kobilo, T. et al. AMPK agonist AICAR improves cognition and motor coordination in young and aged mice. Learn Mem21, 119–126 (2014).
2. Chen, Y. et al. Berberine mitigates cognitive decline in an Alzheimer’s disease mouse model by targeting both tau hyperphosphorylation and autophagic clearance. 121, 109,670 (2020).
3. Li, L. et al. Autophagy enhancer carbamazepine alleviates memory deficits and cerebral amyloid-β pathology in a mouse model of Alzheimer's disease. 10, 433–441 (2013).
4. Vasconcelos-Ferreira, A. et al. The autophagy-enhancing drug carbamazepine improves neuropathology and motor impairment in mouse models of Machado–Joseph disease. Neuropathology and Applied Neurobiology48, e12763 (2022).
5. Chandra, S., Roy, A., Jana, M. & Pahan, K.J.N.o.d. Cinnamic acid activates PPARα to stimulate Lysosomal biogenesis and lower Amyloid plaque pathology in an Alzheimer's disease mouse model. 124, 379–395 (2019).
6. Cassano, T. et al. Early intrathecal infusion of everolimus restores cognitive function and mood in a murine model of Alzheimer's disease. Exp Neurol311, 88–105 (2019).
7. Luo, R. et al. Activation of PPARA-mediated autophagy reduces Alzheimer disease-like pathology and cognitive decline in a murine model. 16, 52–69 (2020).
8. Meng, X. et al. Gypenoside XVII enhances lysosome biogenesis and autophagy flux and accelerates autophagic clearance of amyloid-β through TFEB activation. 52, 1135–1150 (2016).
9. Li, Y. et al. Protein kinase C controls lysosome biogenesis independently of mTORC1. 18, 1065–1077 (2016).
10. Steele, J.W. et al. Latrepirdine improves cognition and arrests progression of neuropathology in an Alzheimer's mouse model. 18, 889–897 (2013).
11. Pan, Y. et al. Cognitive benefits of lithium chloride in APP/PS1 mice are associated with enhanced brain clearance of β-amyloid. Brain, Behavior, and Immunity70, 36–47 (2018).
12. Sarkar, S. et al. Lithium induces autophagy by inhibiting inositol monophosphatase. The Journal of cell biology170, 1101–1111 (2005).
13. Li, X.-C., Wang, Z.-F., Zhang, J.-X., Wang, Q. & Wang, J.-Z.J.E.J.o.P. Effect of melatonin on calyculin A-induced tau hyperphosphorylation. 510, 25–30 (2005).
14. Fernández, A., Ordóñez, R., Reiter, R.J., González‐Gallego, J. & Mauriz, J.L.J.J.o.P.R. Melatonin and endoplasmic reticulum stress: relation to autophagy and apoptosis. 59, 292–307 (2015).
15. Luo, F. et al. Melatonin and autophagy in aging-related neurodegenerative diseases. 21, 7174 (2020).
16. Chen, F. et al. Antidiabetic drugs restore abnormal transport of amyloid-β across the blood–brain barrier and memory impairment in db/db mice. 101, 123–136 (2016).
17. Ou, Z. et al. Metformin treatment prevents amyloid plaque deposition and memory impairment in APP/PS1 mice. 69, 351–363 (2018).
18. Kickstein, E. et al. Biguanide metformin acts on tau phosphorylation via mTOR/protein phosphatase 2A (PP2A) signaling. Proceedings of the National Academy of Sciences of the United States of America107, 21830–21835 (2010).
19. Fang, E.F. et al. Mitophagy inhibits amyloid-β and tau pathology and reverses cognitive deficits in models of Alzheimer's disease. Nat Neurosci22, 401–412 (2019).
20. Sorrentino, V. et al. Enhancing mitochondrial proteostasis reduces amyloid-β proteotoxicity. Nature552, 187–193 (2017).
21. Liu, D. et al. Nicotinamide forestalls pathology and cognitive decline in Alzheimer mice: evidence for improved neuronal bioenergetics and autophagy procession. 34, 1564–1580 (2013).
22. Lautrup, S., Sinclair, D.A., Mattson, M.P. & Fang, E.F.J.C.m. NAD + in brain aging and neurodegenerative disorders. 30, 630–655 (2019).
23. Song, H.-L. et al. Ouabain activates transcription factor EB and exerts neuroprotection in models of Alzheimer's disease. 95, 13–24 (2019).
24. Ramirez, A., Pacheco, C., Aguayo, L. & Opazo, C.J.B.e.B.A.-M.B.o.D. Rapamycin protects against Aβ-induced synaptotoxicity by increasing presynaptic activity in hippocampal neurons. 1842, 1495–1501 (2014).
25. Caccamo, A., Majumder, S., Richardson, A., Strong, R. & Oddo, S. Molecular Interplay between Mammalian Target of Rapamycin (mTOR), Amyloid-β, and Tau: EFFECTS ON COGNITIVE IMPAIRMENTS*. Journal of Biological Chemistry285, 13107–13120 (2010).
26. Spilman, P. et al. Inhibition of mTOR by rapamycin abolishes cognitive deficits and reduces amyloid-beta levels in a mouse model of Alzheimer's disease. PLoS One5, e9979 (2010).
27. Ladiwala, A.R.A. et al. Resveratrol selectively remodels soluble oligomers and fibrils of amyloid Aβ into off-pathway conformers. 285, 24228–24237 (2010).
28. Rahman, M.H. et al. Resveratrol and Neuroprotection: Impact and Its Therapeutic Potential in Alzheimer's Disease. Frontiers in Pharmacology11 (2020).
29. Dhakal, S., Subhan, M., Fraser, J.M., Gardiner, K. & Macreadie, I.J.I.j.o.m.s. Simvastatin efficiently reduces levels of Alzheimer’s amyloid beta in yeast. 20, 3531 (2019).
30. Fassbender, K. et al. Simvastatin strongly reduces levels of Alzheimer's disease beta-amyloid peptides Abeta and Abeta40 in vitro and in vivo. Proceedings of the National Academy of Sciences98, 5856–5861 (2001).
31. Freitag, K. et al. The autophagy activator Spermidine reduces neuroinflammation and soluble amyloid beta in an Alzheimer’s disease mouse model. bioRxiv, 2021.2010.2028.466219 (2021).
32. Sandusky-Beltran, L.A. et al. Spermidine/spermine-N(1)-acetyltransferase ablation impacts tauopathy-induced polyamine stress response. Alzheimer's research & therapy11, 58–58 (2019).
33. Jiang, T. et al. Temsirolimus promotes autophagic clearance of amyloid-β and provides protective effects in cellular and animal models of Alzheimer's disease. Pharmacol Res81, 54–63 (2014).
34. Jiang, T. et al. Temsirolimus attenuates tauopathy in vitro and in vivo by targeting tau hyperphosphorylation and autophagic clearance. Neuropharmacology85, 121–130 (2014).
35. Frederick, C. et al. Rapamycin ester analog CCI-779/Temsirolimus alleviates tau pathology and improves motor deficit in mutant tau transgenic mice. J Alzheimers Dis44, 1145–1156 (2015).
36. Fang, E.F. et al. Tomatidine enhances lifespan and healthspan in C. elegans through mitophagy induction via the SKN-1/Nrf2 pathway. Sci Rep7, 46208 (2017).
37. Liu, Y., Wang, J., Hsiung, G.-Y.R. & Song, W.J.M.N. Trehalose inhibits Aβ generation and plaque formation in Alzheimer’s disease. 57, 3150–3157 (2020).
38. Rusmini, P. et al. Trehalose induces autophagy via lysosomal-mediated TFEB activation in models of motoneuron degeneration. Autophagy15, 631–651 (2019).
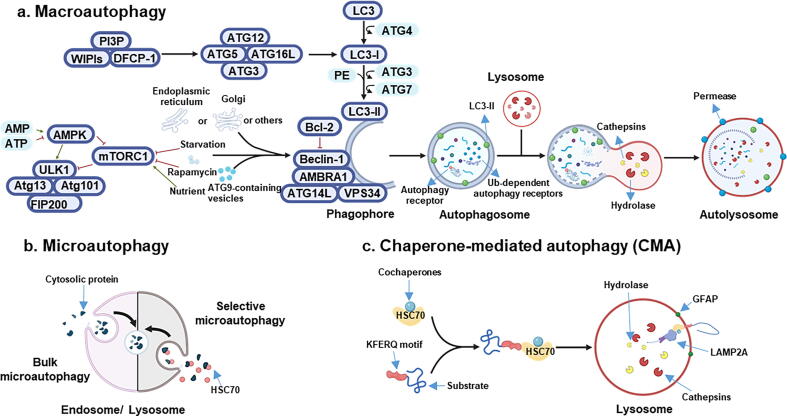
Three forms of autophagy have been identified based on the processes of cargo sequestration: macroautophagy, microautophagy, and chaperone-mediated autophagy (CMA)[3] (Fig. 1). While each is mechanistically different, all these pathways culminate with lysosomal degradation of the cargo. During macroautophagy (hereafter named autophagy), de novo synthesis of double-membraned vesicles occurs which promotes the formation of autophagosomes engulfing sub-cellular components. The proposed site for autophagosome formation is known as the phagophore assembly site (PAS). As the source of the isolation membrane, the PAS site has been described to mainly localize at the endoplasmic reticulum (ER) and additional membrane sources have been described, including Golgi complex, plasma membrane, mitochondria, ER-mitochondria contact sites, recycling endosomes[6]. Upon activation of autophagy, at this site, several ATG proteins are recruited[7]. These proteins play different and definite roles in all autophagy steps, including initiation, nucleation and elongation of the phagophore, sequestration of the cargo, delivery of the phagosome to a lysosome followed by their fusion, forming the autolysosome, and, lastly, degradation of the cargo by lysosomal hydrolases (Fig. 1A). Assembly of the core autophagic machinery normally begins with the inhibition of the mechanistic target of rapamycin (mTOR) and / or the activation of 5′AMP-activated protein kinase (AMPK)[2]. Depending on the trigger, autophagy can be in bulk or selective for specific cellular structures, such as aggregated proteins (aggrephagy), outside pathogens (xenophagy), damaged/superfluous mitochondria (mitophagy), lipid droplets (lipophagy) or others[8]. Details of the molecular mechanisms of many of these sub-type autophagic pathways and their linkages to health and disease have been reviewed by us and others[2], [9], and thus will not be repeated here.
Fig. 1.
Molecular machinery of autophagy. a) AMPK is the major energy-sensing kinase in the cell and responds to intracellular AMP/ATP ratio. A low energy state, associated with high AMP and a low ATP level, activates AMPK which in turn phosphorylates the TSC1/TSC2 complex by inhibiting the activity of mTORC1. In contrast, signals such as growth factors and nutrient abundance lead to inhibition of autophagy through mTORC1, which phosphorylates and associates with the induction complex ULK1/2-ATG13-ATG101-FIP2000 by preventing autophagic cascade. Under rapamycin treatment and starvation, mTORC1 dissociates from the induction complex, resulting in autophagy induction. This triggers phosphorylation of components of the class III PI3K (PI3KC3) complex I (including class III PI3K, VSP34, Beclin-1, ATG14, AMBRA1) which triggers nucleation of the phagophore. The source of membrane includes mitochondria, plasma membrane, Golgi apparatus, endoplasmic reticulum and ATG9-containg vesicles. The anti-apoptotic protein Bcl2 binds Beclin-1 and prevents its interaction with PI3KC3 and, in turn, autophagy. The PI3KC3 complex activates and recruits phosphatidylinositol-3-phosphate (PI3P) and its effector proteins WIPIs and DFCP1.This complex binds the ATG12-ATG5-ATG16L1 complex that promotes cleavage of LC3 by ATG4 to form LC3-I and its lipidated form with phosphatidylethanolamine (PE), which is integrated in the pre-autophagosomal and autophagosomal membrane, where LC3-II interacts with cargo receptors containing LC3-interacting motif (LIR). Finally, the mature autophagosome fuses with a lysosome, where acidic hydrolases and proteases, such as cathepsins, degrade the autophagic cargo. Lysosomal activity is mainly regulated by TFEB, whose nuclear translocation is promoted by a lower phosphorylation induced by AMPK activation. After degradation, component parts of the autophagic cargo are exported back into the cytoplasm through lysosomal permeases for use by the cell in biosynthetic processes or to produce energy. b) Microautophagy involves the direct uptake of the cargo through invagination of the lysosomal or late endosomal membrane. c) In chaperone-mediated autophagy (CMA), HSC70 together with co-chaperones binds KFERQ pentapeptide motif. Then, LAMP2A, a lysosomal membrane receptor, drives the degradation. The activity of this receptor is modulated by the glial fibrillary acidic protein (GFAP).
Autophagic receptors, such as p62, optineurin (OPTN), NBR1, nuclear dot protein 52 kDa (NDP52), NIX, BNIP3, bind to microtubule-associated protein light chain 3 (LC3) driving target sequestration, resulting in the formation of autophagosome. The mature autophagosome binds kinesins which promote the movement towards the lysosome [10]. Microautophagy allows the direct invagination of cellular components into the lysosome or into the late endosome either in bulk or in a selective way through the cytosolic chaperone heat shock cognate 71 kDa (HSC70) which then directly binds lipids on lysosome membrane [11] (Fig. 1B). While HSC70 is involved in all three different autophagy pathways [11], [12], its role is crucial in CMA [13]. Here, the substrates to be degraded are selectively recognized by HSC70 binding the pentapeptide KFERQ, which is essential to assign KFERQ-containing targeting proteins for lysosomal degradation [14]. The chaperone-substrate complex is recognized by the specific lysosomal membrane receptor lysosome-associated membrane protein type 2A (LAMP2A), which in turn translocates the complex inside the lysosome where the degradation takes place [13] (Fig. 1C).
Basal levels of autophagy are important for cellular homeostasis, including preservation of genomic and whole cellular integrity and function, stem cell turnover and suppression of age-related inflammation. Under stressful conditions, autophagy is considered an adaptive response that supports cellular survival [2]. Severe starvation induces autophagy to sustain cell viability via proteolysis of cellular compoments [7], [13]. It has been reported that 48 h starvation is sufficient to trigger degradation of up to 40 % proteins in the rat liver [15]. It has been noticed that there is a high level of autophagy in dying cells which resulted in the term ‘autophagic cell death’; mounting evidence suggests this is a misnomer, as increased autophagy during cellular death is a cellular compensatory response for survival [16]. In this review, we will focus mainly on the changes of macroautophagy and its sub-type mitophagy in the brain during ageing and in Alzheimer’s disease (AD).
2. Impaired autophagy and mitophagy in normal ageing brain
Compromised autophagy is a hallmark of ageing [2], [17], [18], [19]. Ageing is associated with the accumulation of misfolded proteins and senescent mitochondria, mainly due to failure of the cellular repair mechanisms [20]. Clearance of misfolded proteins is crucial for cell survival because protein misfolding impairs biological function and increases the tendency to form toxic aggregates [21], one of the characteristic signs of age-related neurodegenerative disorders [22]. In physiological conditions, misfolded proteins are initially removed by the ubiquitin-proteasomal system (UPS); however, big cargos, such as protein oligomers, aggregates, and damaged mitochondria, are too large to enter the proteasome of the UPS system, and are degraded by autophagic clearance. A coordinated activity between UPS and autophagy has been described to maintain homeostasis of functional protein levels. The ubiquitin tags of misfolded or aggregated proteins foster the deliver and the degradation by either the UPS or autophagy. Pharmaceutical or genetic approaches aiming to inhibit the UPS promote autophagy by increasing the expression of some autophagy genes, such as ATG5 and ATG7, upregulating of Beclin-1 and activating AMPK for instance. Similarly, compromised autophagy leads to the activation of the UPS, through upregulation of proteasomal subunit levels. Extensive descriptions of the crosstalk between UPS and autophagy are available elsewhere [23], [24].The role of autophagy is particularly important for long-lived post-mitotic cells such as neurons that are rarely replaced throughout the adult organism’s lifespan [22]. This protective mechanism decreases during ageing, which causes the accumulation of damaged structures both extra- and intra-lysosomally [2], [7].
A part of intracellular long-lived protein turnover is mediated by the Beclin-1-regulated autophagy pathway and levels of Beclin-1 correlate with autophagic activity [25], [26]. In post-mortem samples of human prefrontal cortices, this marker declined in an age-dependent manner. Both Beclin-1 mRNA and protein levels were 2-fold lower in the brain of older versus younger individuals [27]. The transcriptional downregulation of key autophagy-regulating genes such as Atg5 and Atg7 in older (>70 years old) compared to younger (<40 years old) human brain samples were found in a genome-wide study [28]. Another study compared the gene expression in hippocampal brain tissue, the key memory structure, of younger versus older men and women to explore the impact of sexual dimorphism and ageing on autophagy. In older women, they found reduced expression of PINK1 (a mitochondrial kinase crucial for mitophagy [29]), reduced HDAC6 (a deacetylase required for the maturation of autophagosomes and their fusion with lysosomes [30]), and also decreased expression of LC3 (a protein important for membrane elongation of phagophore [31]). An increased expression of Bcl-2 (the inhibitor of Beclin-1) and mTOR was seen in older men suggesting decreased autophagy activity [31].
Mitophagy is also a mitochondrial quality control mechanism crucial for the maintenance of homeostasis in neurons, and its impairment is associated with different neurodegenerative disorders, including AD, Parkinson’s disease (PD), Huntington's disease (HTT), and amyotrophic lateral sclerosis (ALS) [18], [32], [33], [34], [35], [36]. Upon stress, PINK1 and PARK2 cooperatively identify and label damaged mitochondria with phosphorylated poly-ubiquitin (p-S65-Ub) chains. This specific label is decoded by autophagy receptors that further facilitate mitophagy. p-S65-Ub is barely detectable in basal conditions but rapidly increases upon mitochondrial damage [37]. A strong correlation between p-S65-Ub levels and age was found in human post-mortem brain samples, specifically in the hippocampus, amygdala, and nucleus basalis of Meynerti, reflecting mitophagy dysfunction in the ageing human brain [38]. All these results suggest that the age-dependent decline in the expression of autophagy markers may contribute to the impairment of autophagy function and accumulation of misfolded proteins or damaged organelles during ageing in humans.
The study of ageing on brain autophagy in mammal models provides similar results. Moreover, the experimental manipulation of autophagy and mitophagy pathways has significantly influenced healthspan and lifespan in different model organisms [7]. Reductions in Beclin-1, VPS34, and Atg5 were observed in the hippocampi of 16-month-old mice compared to 3-month-olds, both in protein and mRNA levels [39]. Additionally, reduced autophagy activity was reflected by the decrease in LC3-II accumulation, the marker of autophagosomes, and by an increase in p62/ SQSTM1, a marker of autophagic flux [40], in neurons [39]. Similar age-related declines in autophagy markers (Beclin-1, Atg5-Atg12, LC3-II) together with increased protein levels of mTOR were found in whole mouse brain [41] and in rat hippocampi [42]. In addition, the reduction of Beclin-1 and increased LC3-II/LC3-I ratio were found in the hippocampi of aged cows which may indicate excessive accumulation of autophagosomes and impaired autophagosomal degradation [43]. Also, decreased mitophagy activity was observed in the brain of aged mice [44]. In vivo assessment of mitophagy in a transgenic mouse strain that expresses the fluorescent mitophagy reporter mt-Keima, has shown a decline of mitophagy activity by approximately 70 % in the hippocampal dentate gyrus in 21-month-old compared to 3-month-old mice [44].
Compromised autophagy during ageing is likely not a ‘bystander’ but a ‘culprit’ of brain deterioration and dysfunction, as autophagy maintenance/restoration delays ageing and impedes brain diseases. The effect of autophagy and mitophagy modulation on lifespan has been notably demonstrated by using several model organisms, such as the roundworm (Caenorhabditis elegans), fruit flies (Drosophila melanogaster), and mice (Mus musculus) [45], [46], [47], [48], [49], [50]. The upregulation of autophagy via mTOR inhibition doubled normal lifespan in nematodes [50], increased lifespan in flies [45], and prolonged mice lifespan by 14 % in females and 9 % in males [47].
Genetic induction of autophagy via enhancement of the levels of Atg8a, a member of the Atg8/LC3 protein family, in older fly brains extended fly lifespan by 56 % [49]. In addition, Atg8a mutant flies had a 53 % shorter lifespan in comparison to wild type; these Atg8a flies also developed a neurodegenerative phenotype, including accumulation of protein aggregates inside the neurons [49]. Atg5 is an autophagy protein important for autophagosome formation. Atg5 overexpression in mice enhanced autophagy and led to extensions of lifespan by 17.2 %. Additionally, these mice showed anti-ageing phenotypes, including improved motor function and leanness [48]. In view of the importance of Atg5, it is not surprising that Atg5 knockout resulted in neonatal lethality [51]. Forebrain-specific Atg7 deficient mice experienced age-related neurodegeneration with accumulation of ubiquitin and p62-positive protein aggregates, as well as the accumulation of p-Tau [52]. Facilitating the mitophagy pathway also has a beneficial effect on health and lifespan in different animal models. Overexpression of PINK1 and Parkin extended the healthspan and lifespan in flies [53], [54]. Similarly, overexpressing the mitochondrial fission protein dynamin-related protein 1 (Drp1) increased the lifespan in flies [55]. Extended lifespan was also demonstrated in nematodes after stimulation of mitophagy with tomatidine [46].
Collectively, the evidence indicates that autophagy decreases during ageing and that enhancing autophagy/mitophagy pathways may be an effective approach to delay ageing and extend health- and lifespan in diverse model organisms.
3. Impaired autophagy and mitophagy in AD
AD is on the rise, affecting approximately 55 million people worldwide, a figure estimated to triple by 2050 [56]. AD is the most common neurodegenerative disease of old age, characterized clinically by an insidious onset and a gradual, progressive deterioration of cognitive abilities [36]. Genetic factors, such as APP, presenilin 1 and 2 (PSEN1/PS1 and PSEN2/PS2) and APOE among others, have been identified as causative genes or risk factors for AD [57], [58]. The main disease-defining pathological features of AD are extracellular amyloid-beta (Aβ) aggregates (intracellular Aβ1-42 is also toxic to neurons) and intracellular neurofibrillary tangles of aberrantly hyper-phosphorylated microtubule-associated Tau (MAPT/p-Tau) [59]. Both these hallmarks lead to neurodegeneration associated with synaptic loss and neuronal dysfunction, or even neuronal death. While the Aβ hypothesis has been instrumental in directing our in-depth understanding of the aetiology and progression of AD [59], continued failure/slow movement in anti-AD drug development targeting on reducing Aβ plaques mandates to shift the focus on additional and alternate mechanisms such as to develop a multi-drug approach targeting both amyloid and tau [60], [61].
3.1. Autophagy and lysosomal dysfunction in AD
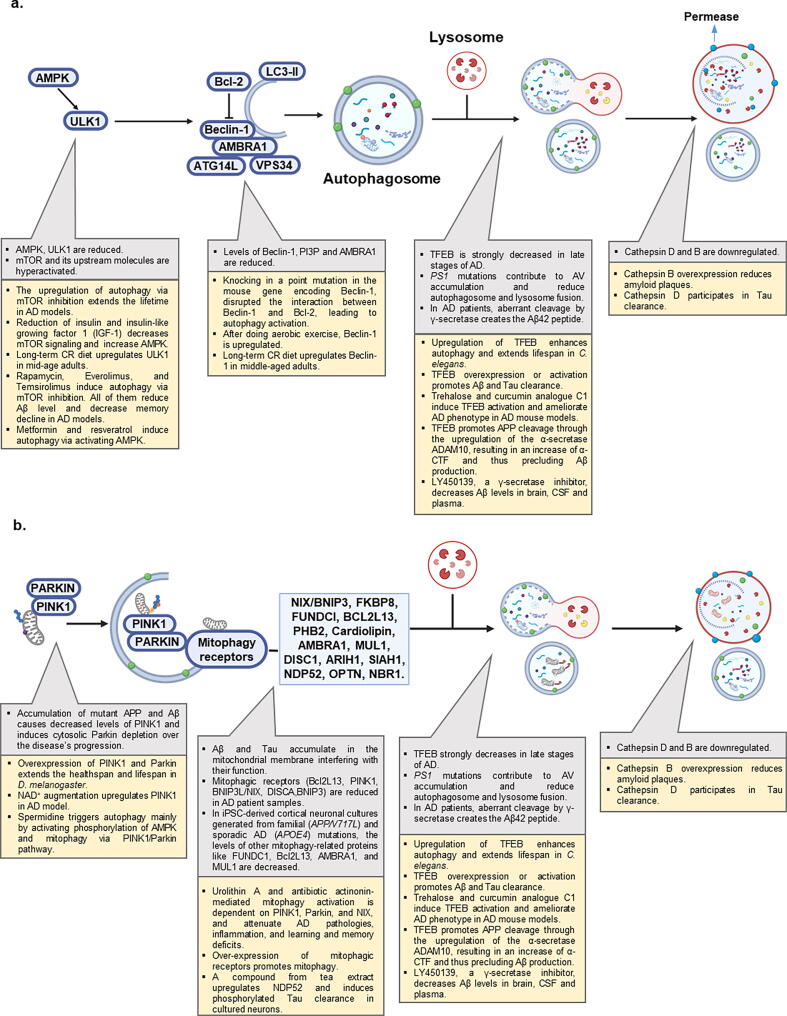
Autophagy is impaired in AD with compromised autophagy contributing to AD progression. Compelling evidence from hippocampi of AD patients showed hyperactivation of mTOR and its upstream molecules, suggesting an enduring autophagy inhibition [62]. Genetic ablation of mTOR in Tg2576 mice promoted autophagy and rescued memory deficits [63]. A summary on the known molecular mechanisms of impaired autophagy in AD is provided (Fig. 2A).
Fig. 2.
Autophagy and mitophagy in AD. a) Alterations of autophagic pathway in AD. b) Alterations of mitophagic pathway in AD. In each section strategies to induce and restore the pathway are reported.
One prominent feature of AD is neuronal loss accompanied by the presence of autophagic vacuoles (AVs) within neuronal processes, including in synaptic regions [64]. AVs have been observed in human AD brains and in transgenic mice expressing mutant P301L Tau [65] and are considered as late-stage autophagic remnants, suggesting an impaired autophagic pathway at the lysosomal degradation stage. Further, the expression of Beclin-1 declines in brains of AD patients compared with healthy controls [66]. In addition, the production of PI3P, which is mediated by Beclin-1/VSP34 complex, was down-regulated in brain tissue from AD patients [67]. AV accumulation appears at early stages of AD and before extracellular Aβ depositions, suggesting that macroautophagy activation is a very early response in the disease development [68]. Purified AVs were enriched with APP and its processing enzymes, such as γ-secretase (gamma secretase), being the first source of intracellular Aβ1-42 production and accumulation in neurons [69]. Indeed, in AD, neuronal maturation of autophagolysosomes and their retrograde transport are impeded, which makes lysosomal deficiency a prominent feature in normal ageing as well as in AD brains [70]. Disruption of lysosomal proteolysis in wild-type mice was shown to mimic the neuropathology of AD and exacerbate autophagic impairment and amyloidogenesis in AD mouse models [71].
γ-Secretase, working with β-secretase (beta secretase), cuts APP to produce Aβ1-42 peptide, the primary component of Aβ plaques. γ-secretase is a multi-subunit transmembrane protease complex, consisting of presenilin-1 (PS-1), PS-2, nicastrin, and APH-1. PS1 is a multi-pass membrane protein that serves as a catalytic site for γ-secretase and is involved in several cellular pathways, including synaptic plasticity, neurite growth, cell adhesion, and calcium homeostasis, among other functions [72]. Accentuated AVs in early-onset familiar AD, due to mutations of PS1, unveil a role of PS1 in lysosomal function [68], [73]. PS1 deletion prevents macroautophagic protein turnover by altering lysosome acidification caused by failed maturation of the proton trans-locating V0a1 subunit of the vacuolar (H+)-ATPase [74]. Moreover, PS1 deletion has been shown to reduce p62 levels, a protein cargo involved in Tau degradation, leading to impairment of Tau turnover [75]. In PS1 deficiency, MTORC1 is constitutively activated, which subsequently inhibits TFEB-mediated autophagy and lysosome biogenesis [76]. In addition, PS2 mutation was reported to alter autophagic flux by inhibiting autophagosome and lysosome fusion [77]. Lysosomal dysfunction has been also described in relation with the APP β-cleaved CTF (β-CTF) in Down syndrome, which normally further develops to AD [78]. Extensive studies in CA1 pyramidal neurons in the hippocampus of early and late-stage AD demonstrated that autophagy flux is upregulated in AD, but it is progressively impeded due to deficient substrate clearance, as reflected by autolysosomal accumulation of LC3-II and SQSTM1/p62 and expansion of autolysosomal size and total area [79].
Lysosomal dysfunction results in the accumulation of Aβ1-42 and aberrant Tau in AD [70]. A key protein involved in the biogenesis of lysosomes is transcription factor EB (TFEB). TFEB is normally sequestered in the cytoplasm by phosphorylation and, under specific stimuli, it translocates to the nucleus and binds to a specific DNA sequence called CLEAR (Coordinated Lysosomal Expression and Regulation) within the promoters of several lysosomal and autophagic genes, including WIPI1, PSEN2, LAMP1, CTSB, CTSD amongst others [80]. Approaches overexpressing and activating TFEB have been shown to possibly reverse AD pathology. A recent study reported variations of TFEB protein levels in a Braak-stage dependent manner [81]. Nuclear levels of TFEB strongly decreased in late stages of the disease, compensated by an increase in its cytoplasmatic levels, supporting the widely reported finding of lysosomal defects in AD [81]. This compensatory TFEB upregulation in the late stages of AD, due to increased autophagy induction in response to accumulated cellular Aβ and Tau proteins, may create favorable conditions for Aβ clearance, though insufficient to clear all the accumulated ‘garbage’. TFEB overexpression or activation promoted Aβ clearance through regulation of the autophagy-lysosome pathway (ALP), reduced Aβ-induced ROS production and cell apoptosis [82], [83], [84]. However, such approaches appear to be successful at early stages of AD, while they are insufficient in later ones, indicating the presence of more hampered autophagic machinery. Under basal conditions, TFEB-mediated ALP has also been shown to modulate Aβ turnover in neurons. TFEB promotes APP cleavage through the upregulation of the α-secretase ADAM10, resulting in an increase of α-CTF and thus precluding Aβ production [85]. Meanwhile, TFEB also augmented β-CTF levels possibly through altered proteasome-mediated catabolism [85]. Several studies reported a neurotoxic role for β-CTF [86] and thus upregulation of lysosomal pathway as a therapeutical strategy needs to be carefully evaluated. In addition to memory preservation, TFEB is likely involved in longevity. Downregulation of hlh-30 (the C. elegans orthologue of the mammalian TFEB) and daf-16 (the C. elegans orthologue of the mammalian FOXO) in C. elegans shortened lifespan in both wild type and long-lived daf-2 mutant [87], while its upregulation enhanced autophagy and extended lifespan [88].
Upregulation of TFEB in both microglia and astrocytes, cells that clear extracellular Aβ plaques via phagocytosis, accelerates their capacity to remove Aβ plaques [89], [90]. TFEB overexpression in different Tau mouse models of AD has been associated with higher clearance of NFTs through ALP without affecting normal Tau and improved neuronal survival and synaptic function [91], [92]. The mechanism by which TFEB rescued Tau aggregation is likely via the TFEB-PTEN-PI3K-Akt-mTOR pathway with an increased Tau degradation by cathepsin D [91], [92]; while the polymorphism of cathepsin D is an AD risk factor, its complete loss is associated with devastated neurodegenerative disorders [93], [94]. In response to trans-synaptic spreading of Tau [95], astroglial TFEB overexpression has been reported to enhance the lysosomal pathway and slow down Tau aggregation in some AD mouse models, such as rTg4510 and PS19 [96]. TFEB also regulates lysosomal exocytosis, which may have an unexplored role in cellular clearance in neurodegenerative diseases [97]. In this regard, TFEB is involved in Tau exocytosis. TFEB specifically recognized the microtubule-binding repeat (MTBR) - truncated Tau, but not the wild type one, and mediates its secretion through the lysosomal calcium channel TRPML1 [98]. Loss of TFEB induced a higher Tau accumulation [98]. To note, TFEB seems to be protective across different neurodegenerative diseases, as it eliminated aberrant aggregates in models of PD and HD, etc [99], [100]. Cumulatively, these data suggest that TFEB-dependent lysosomal/autophagic function is pivotal in the homeostasis of proteins (proteostasis) and neuroprotection.
3.2. Aβ and Tau-associated autophagic abnormalities
While the molecular interactions between Aβ, Tau, and autophagy are not fully understood, recent studies have uncovered the intertwined linkages among them. First, autophagy impairment induces Aβ production. Inducing a point mutation (F121A) in the mouse gene encoding Beclin-1, disrupted the interaction between Beclin-1 and Bcl-2, leading to autophagy activation [101]. AD mice carrying this mutation showed a dramatic reduction of amyloid plaques and a rescue of cognitive impairment [101]. Accordingly, Beclin-1 knock-out mice showed aggregation of both intercellular and extracellular Aβ compared to the control mice [66]. The benefits may be attributable to an induction of autophagy, which modulates both Aβ clearance and production. In addition to Beclin-1 knock out that is linked to enhanced AD pathology, conditional knockdown of Atg7 in an APP/PS1 mouse model of AD resulted in enhanced secretion of Aβ that contributes to the formation of extracellular plaques [102]. Indeed, autophagy has been implicated in secretion of undegraded integral membrane proteins via a secretory pathway from the endoplasmic reticulum (ER), to Golgi, and ultimately to the plasma membrane or through secretory lysosomes, a route through which APP is known to be transported and processed to Aβ [102]. Although autophagosomes may serve as sites for processing of APP to generate Aβ, how Aβ is then secreted following induction of autophagy is not known. However, in this context, it has been proposed that the increase in the accumulation of autophagosomes in AD promoted the release of Aβ into the extracellular space, facilitating the deposition of amyloid plaques [103]. Within the cell, impaired lysosomal function may give rise to accumulation of intracellular Aβ that participates in the cascade of events leading to neurodegeneration [102]. Promoting autophagic activity in AD models rescued neurodegeneration and inhibited the cognitive decline that correlated with an increase in clearance of Aβ [17], [104], [105]. For example, brain-specific upregulation of the anti-ageing hormone Klotho in APP/PS1 transgenic mice promoted autophagic clearance of Aβ and improved memory [106]. In humans, the heterozygous KLOTHO gene variant KL-vS which increases circulating levels of Klotho, abrogated APOE ε4-related phenotypes; carriers with APOE ε4/KL-vS heterozygosity had low AD risk [106].
APP’s cleaved product, APP-βCTF, also links to autophagy. AP2 (adaptor protein 2) is a specific LC3 cargo receptor that brings APP-βCTF directly to autophagosomes therefore leading to reduced Aβ1-42 prodution [107]. APP- βCTF removal via AP2 binding is also assisted by phosphatidylinositol binding clathrin assembly protein (PICALM), a known binding partner of AP2 involved in clathrin-mediated endocytosis [107]. Indeed, AD GWAS showed compelling evidence for association between AD and PICALM variants [108], supporting the idea that PICALM played an important role in APP-βCTF clearance and mutations in its locus link to a higher risk of AD [109]. PICALM expression is reduced in AD brains and its deficiency correlates with Aβ formation as well as accumulation of p-Tau [107], [110], [111]. Indeed, PICALM regulates both autophagosome formation and fusion with lysosomes via modulation of NSF attachment protein receptors (SNAREs) including VAMP2 and VAMP8 [111].
A physiological role for autophagy in Tau homeostasis is documented through p-Tau accumulation and neurodegeneration in mice with neuronal deletion of Atg7 [52]. A tight collaboration between the ubiquitin proteasome system (UPS) and autophagy, with p62 acting as a bridge, has been widely reviewed for Tau clearance [112]. Downregulation of p62, alone or together with other genetic factors, such as the APOE ε4 allele, caused changes in Tau conformation and solubility with a consequent failures in its removal [113]. Autophagy induction affects tauopathies. Tau mutant mice treated with rapamycin, an mTOR inhibitor, showed reduced Tau phosphorylation [114]. Conversely, TSC2-KO mice, which express an mTOR negative regulator in which mTOR is constitutively activated, displayed elevated Tau levels as well as Tau phosphorylation [114]. By using mTOR inhibitors more potent than rapamycin in patient iPSC-derived neuronal cells carrying a Tau mutation, Tau phosphorylation and insoluble Tau were reduced [115]. The autophagy receptor NDP52 recognized phosphorylated Tau in brains of AD mice models [116]. NDP52 upregulation by a compound from tea extract induced phosphorylated Tau clearance in cultured neurons [117]. Thus, impaired autophagy results in accumulated Aβ and Tau, which in turn block autophagy, initiating a ‘vicious cycle’ which may finally cause AD.
3.3. Mitophagy deficiency in AD
Neurons heavily depend on mitochondria to support their function, thus mitophagy plays an essential role in neuronal health and function via efficient degradation of damaged mitochondria. In neurons, clearance of autophagic substrates mainly occurs in the soma, where most of the lysosomes are located. Mitophagy is impaired in post-mortem human AD brain tissue as well as in AD iPSC-derived neurons and cross species AD animal models [17]. Initiation of the mitophagy machinery was impaired in postmortem brain tissue from AD patients; this was evidenced by reduced p-TBK1(Ser172) and p-ULK1(Ser555) in AD compared with cognitively normal control samples [17]. Studies in C. elegans suggest Aβ1-42 and Tau mutants inhibited PINK-1, DCT-1/NIX, or PDR-1/Parkin-dependent mitophagy pathways [17]. Defective mitophagy contributes to AD as restoration of neuronal and microglial mitophagy reduced insoluble Aβ1–42 and Aβ1–40, rescued memory deficits, and reduced the phosphorylation of Tau at several sites (Thr181, Ser202/205, Thr231, Ser262) in an AD mouse model [17]. Mitophagy stimulation in a human neuronal cell line increased the levels of a series of mitophagy-related proteins, including PINK1, Parkin, Beclin-1, Bcl2L13, AMBRA1, MUL1 and p-ULK1(Ser555) [17]. In addition, microglial mitophagy seems to be involved in eliminating extracellular Aβ plaques and reducing pro-inflammatory cytokines in AD [17], [118]. In neuroblastoma cells and C. elegans, expression of human wild-type Tau and frontotemporal dementia mutant Tau (P301L) inhibited mitophagy by increasing Tau-mediated sequestration of Parkin, which finally prevented its recruitment to damaged mitochondria without alterations in the mitochondrial membrane potential [119]. This was also shown in vivo in the C. elegans nervous system, wherein hTau expression reduced mitophagy while the mutant Tau completely inhibited mitophagy [119]. NH2-hTau fragments strongly associated with Parkin and Ubiquitin-C-terminal hydrolase L1 (UCHL-1) in cellular and animal AD models and in human AD brain tissue, leading to synaptic deterioration due to an improper mitochondrial clearance [120].
Defective mitophagy in AD, can happen at the initiation stage or lysosomal dysfunction stage [17], [121] (Fig. 2B), and may be caused by different mechanisms. The way Aβ enters into mitochondria seems to involve the translocase of the outer membrane (TOM) complex or the mitochondrial-associated endoplasmic reticulum (ER) membrane (MAM) [122], [123]. Both Aβ and p-Tau interacted and blocked voltage-dependent anion channel (VDAC) proteins, leading to mitochondrial dysfunction [124]. Accumulation of mutant APP and Aβ in the hippocampi of APP-transgenic mice caused defective biogenesis with reduced levels of PGC1α, NRF1, NRF2 and TFAM, increased levels of mitochondrial fission (Drp1 and Fis1), decreased levels of fusion (MFN1, MFN2 and Opa1), autophagy (ATG5 and LC3BI, LC3BII), mitophagy (PINK1 and TERT), synaptic (synaptophysin and PSD95) and dendritic (MAP2) proteins [125], [126]. Disrupted-in-schizophrenia-1 (DISC1), a recently discovered mitophagy receptor, has been shown to be reduced in AD human brain samples and in APP/PS1 mice [127]. DISC1′s overexpression protects synaptic plasticity from Aβ accumulation-induced toxicity through promoting mitophagy [127]. Additionally, mitochondrial imbalance activates mitochondrial unfolded protein response (UPRmt), a response which appears to be compromised in AD [128]. Pharmacologic and genetic restoration of UPRmt function reduced Aβ accumulation in both C. elegans and mouse models of AD [128].
Dysfunction of PINK1-Parkin-mediated mitophagy appears crucial in AD. Progressive Aβ accumulation significantly induced Parkin recruitment to depolarized mitochondria accompanied by cytosolic Parkin depletion over the disease’s progression, as confirmed in AD human samples [129]. In AD-derived patient fibroblasts, upregulating Parkin expression and enhancing autophagy restored mitochondrial membrane potential, reduced PINK1 expression, and hindered accumulation of damaged mitochondria [130]. Recently, Parkin has been shown to regulate PINK1 expression through PS1 [131]. Indeed, PS1, included in the catalytic core of the γ-secretase, led to the production of Aβ and its C-terminal counterpart AICD (APP intracellular domain), which in turn act on FOXO3, eventually upregulating PINK1 level [131]. In addition, AICD acts on other autophagy/mitophagy markers by upregulating LC3B and deregulating SQSTM1/p62 and TOMM20 together with MFN2 and Drp1 [131]. PINK1 also activates mitophagy in a Parkin-independent manner through some autophagy receptors, such as OPTN and NDP52 [132]. PINK-1 expression is significantly decreased in APP mouse models and AD human brains, supporting a correlation between Aβ spreading and PINK1. In addition, this led to an impairment in mitochondrial clearance. Approaches aimed at increasing PINK1 function both in vitro and in vivo promoted clearance of dysfunctional mitochondria and Aβ degradation, suppressed ROS production and improved synaptic plasticity [133]. In addition to turning up the PINK1/Parkin pathways, upregulation of other mitophagy pathways also inhibits AD. One example is MCL-1, a key anti-apoptotic protein but also a mitophagy receptor that interacts with LC3A to promote mitophagy; upregulation of the MCL-1-dependent mitophagy pathway inhibited memory loss in the APP/PS1 mice [134].
3.4. CMA and other autophagic pathways alterations in AD
Several pieces of literature show that both Aβ and Tau contain a KFERQ-like motif, suggesting they are eligible for degradation via CMA or microautophagy [135], [136]. APP contains a KFERQ motif in its C-terminus and, interestingly, its deletion did not affect its binding to HSC70, but it prevented the binding of either APP or its CTF with the lysosome for degradation [135]. Therefore, KFERQ deletion enhanced APP-βCTF accumulation and Tau phosphorylation, contributing to AD pathogenesis. Tau bears two KFERQ-motifs in its C‐terminal region, which are required for the HSC70 binding and LAMP2A-mediated lysosomal degradation [137]. Blocking of the internalization step occurred when the N-terminal was disrupted [137]. Inefficient translocation of the Tau fragments across the lysosomal membrane caused formation of Tau oligomers on the surface of lysosomes, which may act as precursors of aggregation and interfere with lysosomal functioning [138]. Recently, CMA has been shown to regulate a subset of the proteome prone to aggregation, and manipulation of its activity protected neurons from proteotoxicity in AD mice [136].
Non-canonical functions for several autophagy proteins in Aβ degradation have been documented. In physiological conditions, Aβ is engulfed and removed by microglia through LC3-associated endocytosis (LANDO), which is LC3+, Rab5+, and clathrin+ dependent. LANDO is significantly reduced in AD mice, and its restoration increased Aβ removal [139]. Microglial autophagic removal of Aβ is impaired in AD patients carrying TREM2 risk variants and in TREM2 -/- AD mice [140]. The key autophagy protein Beclin-1 (encoded by BECN1) plays a fundamental role in health and longevity [141]. Becn1+/- mice exhibited increased inflammation (higher IL-1β and IL-18) and Becn1+/- / APPS1 mice showed exacerbated microglial inflammation due to activation of the NLRP3-Caspase1-IL1β pathway [142].
Bcl-2 associated athanogene (BAG) proteins are a family of co-chaperones that interact with the ATPase domain of Hsp70 through a specific structural domain named BAG [143]. In humans this protein family consists of six members (BAG1-BAG6) all sharing the BAG domain with distinct protein domains (widely described in [144]). BAG proteins are involved in several biological process, including apoptosis, development, cytoskeleton organization and autophagy. Because of its ubiquitin-like domain, BAG1 drives the degradation of polyubiquitinated proteins via the proteosome, while BAG3 mediates lysosomal degradation, possibly also via the ubiquitin-binding protein p62/SQSTM1 [145]. Stressful stimuli, normal ageing and protein-prone neurological disease induced higher BAG3 expression associated with a functional switch from Hsp70-BAG1-mediated proteasomal degradation to Hsp70-BAG3-mediated selective autophagy [145]. BAG3-stimulated autophagy is interpretated as an adaptive response of the protein quality control system following proteosome impairment. BAG3-driven autophagy involves the formation of the “aggresome”, a perinuclear compartment with higher autophagic activity [146]. Indeed, HspB8 formed a stable complex with BAG3 in cells and the formation of this complex is essential for the degradation of a pathogenic form of huntingtin prone to aggregation [147].
A higher BAG3 expression has also been described in the astrocytes of post‐mortem human brain tissue from neurodegenerative disorders, such as AD, PD, and HD [148]. In line with the previous mentioned BAG1-BAG3 expression switch, inhibition of the proteasome in rat cortical neurons activated autophagy and reduced Tau levels [149]. There are also data showing BAG1 overexpression increased Tau levels, likely due to BAG1 facilitating the refolding of Tau and downregulating its degradation [150]. Genetic and pharmacological induction of BAG3, associated with proteasome inhibition, in rat primary neurons led to a significant reduction in Tau and p-Tau levels [151]. Recently, BAG3 has been defined as a “protector” against Tau accumulation [152]. Indeed, Tau preferentially accumulates in excitatory neurons, but not in inhibitory neurons, where BAG3 expression is higher. Downregulation of BAG3 in primary neurons enhanced Tau accumulation in inhibitory neurons. The balance between the proteasomal and lysosomal pathways may be also regulated by sAβPPα, a non-amyloidogenic trophic peptide produced by α-secretase cleavage of APP [153]. Indeed, under conditions of proteotoxic stress, sAβPPα increased proteasomal activity and decreased BAG3 expression in young cells. In aged cells, the irreversible changes of the proteasome machinery made sAβPPα unable to revert the protein turnover [153]. These data highlight roles for BAG3 in the mediation of Tau pathology, paving the way to consider it a drug target for drug development.
4. Approaches to upregulate autophagy: caloric restriction, exercise, and small compounds
Increasing evidence suggests that autophagy and mitophagy are significantly impaired during ageing and age-related disease such as AD [2]. Therapeutic intervention aiming to modulate these molecular pathways has the potential to mitigate the detrimental effect associated with ageing. Autophagy can be induced by multiple mechanisms, including lifestyle interventions such as caloric restriction [154] or exercise [155] and pharmacological interventions by synthetic drugs or natural compounds [156].
4.1. Lifestyle interventions
Caloric restriction (CR), reducing food intake by 25–50 % without malnutrition, is a non-genetic stimulator of autophagy that promotes lifespan and improves health in flies, worms, mice, and lemurs [157], [158]. In humans, short-term CR (from 1.5 − 3 days) increased autophagy markers in skeletal muscle samples [159], [160]. Likewise, long-term CR (6 ± 3 years) in middle-aged adults (58.7 ± 7.4 years) significantly upregulated many autophagy genes, including Beclin-1, ULK1, Atg12, LC3, and increased Beclin-1 and LC3 protein levels in the skeletal muscle. Additionally, CR decreases inflammation and increases serum cortisol resulting in enhanced cellular protein quality and a decrease in dysfunctional proteins and organelles [161]. Also, increased autophagy markers were observed after 4 consecutive days of a zero-calorie diet in healthy volunteers’ white blood cells suggesting that CR has a pleiotropic effect in multiple tissues [162].
The effect of CR on neuronal autophagy was investigated in animal models [163]. In mice, 24 h of food restriction induced autophagy in cortical neurons and Purkinje cells in the cerebellum [164]. In AD mouse models, short term CR enhanced autophagy but was insufficient to degrade brain Aβ [165], [166]. However, long-term CR for 68 weeks enhanced the autophagy and significantly decreased the Aβ plaques in the hippocampus, alongside an increased cerebral glucose metabolism and neuronal integrity resulting in an improved cognition of the AD mice model [166]. It was suggested that CR activates autophagy via two pathways: first, it reduces levels of insulin and insulin-like growth factor 1 (IGF-1), which decrease mTOR signaling. Second, it increases the AMP/ATP ratio which activates the nutrient-sensing kinase, AMPK [167], [168].
Similar to CR, physical exercise decreases the ATP/AMP ratio leading to AMPK activation [155]. In humans, physical activities upregulate the age-related decline in autophagy markers. After 8 weeks of aerobic exercise, the LC3-II/LC3-I ratio and the protein levels of Atg12, Atg16, and Beclin-1 were upregulated in peripheral blood mononuclear cells (PBMC) of older adults; similarly, autophagy inhibition markers such as phosphorylated ULK1 at serine 757 and p62/SQSTM1 levels were reduced in the same samples [169]. A similar effect on autophagy activation was observed in PBMC after 8 weeks of resistance exercise, which suggests that exercise has the potential to induce autophagy [170]. In addition to AD, the beneficial effect of exercise on autophagy was also observed in animal models of PD [171], [172], [173], suggesting broad benefits of exercise against common neurodegenerative diseases.
4.2. Pharmacological inducers
Different autophagy/mitophagy activators show translational potential against AD. The most studied pharmacological inducer of autophagy is the first mTOR inhibitor rapamycin. Oral intake of rapamycin has been shown to be effective in extending lifespan, slowing brain ageing, and protecting against neurodegeneration in various experimental models [47], [174], [175]. As a promising autophagy activator, the effect of rapamycin is already being tested in humans. Currently, ongoing clinical trials are aiming to determine the long-term efficacy of rapamycin in reducing clinical ageing measures in healthy older adults [176] and older adults with amnestic mild cognitive impairment and early-stage AD [177]. Chronic rapamycin treatment induces side effects, possibly due to its effect on inhibiting MTORC2. Therefore, specific MTORC1 inhibitors-rapamycin paralogues have been developed, such as everolimus and temsirolimus. Everolimus reduced human APP/Aβ and human tau levels and improved cognitive function in 3 × Tg AD mice [178]. Temsirolimus induced autophagy, reduced both Aβ and p-Tau and improved motor functions in several AD mouse models, such as APP/PS1, P301S and Tg30 [179], [180], [181].
mTOR-independent autophagy activators are normally designed to target the AMPK pathway. One of them is metformin, a biguanide anti-diabetic drug with neuroprotective properties [182]. Preclinical studies suggest a role for metformin in mitigating ageing [183]. Metformin increased the lifespan of C. elegans by 36 % [184]. In mice, metformin increased longevity and attenuated the deleterious effects of ageing, such as high cholesterol levels or the age-related rise of triglyceride and blood glucose levels [185], [186]. Treatment with metformin mimics some of the benefits of CR, such as prevention of the onset of metabolic syndrome and amelioration of physical performance [183]. Another AMPK activator AICAR (5-aminoimidazole-4-carboxamide ribonucleoside) has effectively delayed ageing by inducing autophagy/mitophagy with a beneficial effect on cognition and motor function [187].
Mitophagy is a sub-type of autophagy, molecules that induce mitophagy show similar benefits against AD laboratory models. Commonly studied mitophagy inducers are urolithin A (UA, a metabolite compound resulting from the transformation of ellagitannins bioactive polyphenols, found in some fruits, especially berries and nuts, by the gut bacteria) [188], antibacterial compound actinonin [32], polyamine spermidine [189], nicotinamide riboside (NR) and nicotinamide mononucleotide (NMN) which are precursors of nicotinamide adenine dinucleotide (NAD+) [190], as well as the natural compounds Kaempferol and Rhapontigenin [191].
Oral consumption of UA induced mitophagy in worms and mice resulting in extended lifespan and healthspan [192]. Additionally, promising results were shown in the mouse model of AD and Aβ and Tau C. elegans, where UA reversed memory impairment via activation of mitophagy [32]. In a first-in-human clinical trial, UA was administered over 4 weeks to healthy, sedentary elderly individuals. Results of this trial have demonstrated positive biological effects on mitochondrial health and, importantly, the compound showed a favourable safety and bioavailability profile [188]. Similarly, treatment with actinonin has shown positive effects in AD models. APP/PS1 mouse models were treated orally with actinonin for 2 months which restored hippocampal neuronal mitophagy, and ameliorated cognitive deficit and Aβ pathology [32]. In addition, actinonin induced robust neuronal mitophagy in AD C. elegans models [32]. Beneficial effects of both UA and AC are dependent on important mitophagy genes, such dct-1, pdr-1 and pink-1 as well as improvement of phagocytic activity of microglia [32].
Spermidine, a natural polyamine implicated in cell growth and survival, declines during ageing, and external supplementation of spermidine can extend the lifespan of worms, yeast, flies, mice, or human cells through enhanced autophagy [193], [194]. It has been shown that increased dietary spermidine inhibits memory loss in AD worms and improves locomotor capacity in a PD worm model both via the PINK1-PDR1- dependent mitophagy pathway [195]. Spermidine can trigger autophagy mainly by activating phosphorylation of AMPK [196] and mitophagy via PINK1/Parkin pathway [195]. Detailed molecular mechanisms of spermidine’s biological activity as well as its clinical potential are summarized in a recent review [197].
The NAD+ precursors NR and NMN induce mitophagy via different NAD+-dependent cell signaling pathways [18], [198]. NAD+ is a coenzyme that plays a pivotal role in energy metabolism and cell survival via cellular redox reactions, through which it becomes converted to NADH [18]. Reports have shown that an age-associated decline in NAD+ levels is associated with several age-related diseases, such as AD and PD, while the upregulation or replenishment of NAD+ metabolism with precursors might slow down certain aspects of ageing and forestall some age-related diseases [199]. It has been shown that NAD+ replenishment via NR supplementation restored the NAD+ profile and improved mitochondrial quality through mitophagy in both worm and fly models of Werner syndrome, a human premature ageing disease [200]. NAD+ repletion extended lifespan and delayed accelerated ageing in these model organisms [200]. Similarly, NAD+ depletion was found in models of accelerated ageing disease, such as ataxia-telangiectasia (AT) and xeroderma pigmentosum (XPA), which both exhibit neurodegeneration, and NAD+ augmentation had a beneficial effect on lifespan and healthspan [201], [202]. NAD+-enhanced mitophagy in AD mouse models reduced Aβ load, decreased p-Tau levels, improved cognitive and memory functions, reduced neuroinflammation and promoted microglia phagocytic activity [32], thus further supporting the role of impaired removal of mitochondria in AD aetiology. The beneficial effects of NAD+ supplementation are related to the activity of NAD+-consuming enzymes, such as sirtuins (SIRT1, SIRT3, SIRT6, SIRT7), cyclic ADP-ribose synthases CD38 and SARM1 (sterile alpha and TIR motif containing 1) and the DNA damage sensor PARPs (poly[ADP-ribose] polymerase) [190].
In view of the importance of mitophagy stimulation in treating AD, a lot of effort has been put into the identification of robust but low toxicity but potentially clinical applicable mitophagy inducers. While these mitophagy inducers are mainly identified using classic wet laboratory methods, modern techniques like AI have been used in drug development targeting on mitophagy induction. Kaempferol and Rhapontigenin were identified with the assistance of AI. Using unsupervised machine learning (involving vector representations of molecular structures, pharmacophore fingerprinting and conformer fingerprinting) and a cross-species approach, two potent mitophagy inducers, Kaempferol and Rhapontigenin, were identified from a total of 18 AI-high scored compounds. These compounds are able to induce mitophagy via transcriptionally upregulation of key mitophagy/autophagy proteins like PINK1 and ULK1. Intriguingly, these two compounds dramatically increased the survival and functionality of glutamatergic and cholinergic neurons, abrogated amyloid-β and tau pathologies, and improved 3xTg AD mice’s memory [191].
5. Outstanding questions and future perspectives
The importance of healthspan cannot be underestimated considering the potential impact of an increasingly ageing population worldwide [203], [204], [205]. The Lancet Commission on dementia prevention, intervention, and care identified a total of twelve potentially modifiable risk factors for dementia, which correlate with around 40 % of all cases of dementia worldwide [206]. Specific interventions may delay or even prevent the incidence of dementia and be protective for people even without a genetic risk. Many of these risk factors may also affect and impair autophagic and mitophagic pathways. The challenge will be to maintain youthful brain functions as ageing progresses, including through mitigation of mitochondrial damage and compromised autophagy [18], [207]. Thus, strategies to maintain autophagy or to restore autophagy to a ‘young’ level could provide an approach to prevent or slow down brain ageing and the occurrence of diseases. The genetic heterogeneity and etiological complexity of AD present a significant challenge in the search for a cure for AD [58], [208]. Mitochondria are linked to many pathways and events within the body, such as inflammation, senescence, stem cell exhaustion, and impaired phagocytosis of microglia and astrocytes, all of which could contribute to AD [32]. Thus, approaches that support mitophagy may provide additive or synergistic benefits, with targets inhibiting multiple AD-related pathways or events [209]. To note, turning up autophagy could be a double-edged sword. Normally, physiological level of autophagy is important on cell/tissue homeostasis and survival. In the conditions of aging and diseases, autophagy induction improves cell survival (e.g., in neurons) and healthy longevity; however, caution to be made that higher autophagy may facilitate cancer growth and resistance against chemotherapy [2], [210]. It urges the establishment of a guideline on how to safely manipulate autophagy to promote health and to limit side effects in different disease and health conditions.
Future studies in this emerging field could include a) creation of an autophagy and mitophagy “atlas” of the human brain covering both spatiotemporal and regional brain differences; b) investigation of activity levels and potential differential roles of autophagy and mitophagy in different brain cell types, such as excitatory neurons, inhibitory neurons, astrocytes, microglia, and oligodendrocytes; and c) investigation of the interactions between autophagy/mitophagy and other risks factors of the brain, such as impaired circadian rhythm and compromised glymphatic system. Furthermore, as the entorhinal cortex (EC) is the region where Tau pathology appears to originate, roles for compromised autophagy/mitophagy in the EC and in the spreading of Tau pathology is of paramount importance for the discovery of AD etiology and to secure new targets for anti-AD drug development [211]. Finally, AI has been increasingly used in the medical field, from assisting mechanistic investigation to translational applications; new AI approaches linked to large omics data from laboratory models and humans are needed to propel the development of strategies to maintain brain ageing in the healthy elderly and for treating AD and other neurodegenerative diseases.
Declaration of Competing Interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:E.F.F. has a CRADA arrangement with ChromaDex (USA) and a commercialization agreement with Molecule AG/VITADAO, and is consultant to Aladdin Healthcare Technologies (UK and Germany), the Vancouver Dementia Prevention Centre (Canada), Intellectual Labs (Norway), and MindRank AI (China).
Acknowledgments
The authors acknowledge the valuable work of the many investigators whose published articles they were unable to cite owing to space limitations. We thank Thale Patrick-Brown from the Fang lab who edited this paper. This project is supported by Cure Alzheimer’s Fund, HELSE SØR-ØST (#2020001, #2021021), the Research Council of Norway (#262175, #103553), Molecule AG/VITADAO (#282942), NordForsk Foundation (#119986), the National Natural Science Foundation of China (#81971327), Akershus University Hospital (#269901, #261973, #262960), the Civitan Norges Forskningsfond for Alzheimers sykdom (#281931), and the Rosa sløyfe/Norwegian Cancer Society & Norwegian Breast Cancer Society (#207819). The research leading to these results has received funding from the EEA/ Norway Grants 2014-2021 and the Technology Agency of the Czech Republic - project number TO01000215 (to M.V., K.V. and E.F.F.). MV and KV were supported by project nr. LX22NPO5107 (MEYS): Financed by EU – Next Generation EU. K.V. was supported by Czech Alzheimer foundation, scholarship for Female Scientists-Mothers. S.Q.Z. was funded by the China Scholarship Council [http:www.csc.edu.cn/]; The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Some figure elements were generated using BioRender via subscription through the Fang laboratory.
Contributor Information
Martin Vyhnalek, Email: martin.vyhnalek@lfmotol.cuni.cz.
Evandro F. Fang, Email: e.f.fang@medisin.uio.no.
References
- 1.Dikic I., Elazar Z. Mechanism and medical implications of mammalian autophagy. Nat Rev Mol Cell Biol. 2018;19:349–364. doi: 10.1038/s41580-018-0003-4. [DOI] [PubMed] [Google Scholar]
- 2.Aman Y., et al. Autophagy in healthy ageing and disease. Nat Aging. 2021;1:634–650. doi: 10.1038/s43587-021-00098-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klionsky D.J., et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2021;1–382 doi: 10.1080/15548627.2020.1797280. [DOI] [Google Scholar]
- 4.Ashford T.P., Porter K.R. Cytoplasmic components in hepatic cell lysosomes. J Cell Biol. 1962;12:198–202. doi: 10.1083/jcb.12.1.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klionsky D.J. Autophagy revisited: a conversation with Christian de Duve. Autophagy. 2008;4:740–743. doi: 10.4161/auto.6398. [DOI] [PubMed] [Google Scholar]
- 6.Rubinsztein D.C., Shpilka T., Elazar Z. Mechanisms of autophagosome biogenesis. Curr Biol. 2012;22:R29–R34. doi: 10.1016/j.cub.2011.11.034. [DOI] [PubMed] [Google Scholar]
- 7.Hansen M., Rubinsztein D.C., Walker D.W. Autophagy as a promoter of longevity: insights from model organisms. Nat Rev Mol Cell Biol. 2018;19:579–593. doi: 10.1038/s41580-018-0033-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gatica D., Lahiri V., Klionsky D.J. Cargo recognition and degradation by selective autophagy. Nat Cell Biol. 2018;20:233–242. doi: 10.1038/s41556-018-0037-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klionsky D.J., et al. Autophagy in major human diseases. Embo j. 2021;40:e108863. doi: 10.15252/embj.2021108863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Olsvik H.L., et al. FYCO1 Contains a C-terminally Extended, LC3A/B-preferring LC3-interacting region (LIR) motif required for efficient maturation of autophagosomes during basal autophagy. J Biol Chem. 2015;290:29361–29374. doi: 10.1074/jbc.M115.686915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sahu R., et al. Microautophagy of cytosolic proteins by late endosomes. Dev Cell. 2011;20:131–139. doi: 10.1016/j.devcel.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arndt V., et al. Chaperone-assisted selective autophagy is essential for muscle maintenance. Curr Biol. 2010;20:143–148. doi: 10.1016/j.cub.2009.11.022. [DOI] [PubMed] [Google Scholar]
- 13.Kaushik S., Cuervo A.M. The coming of age of chaperone-mediated autophagy. Nat Rev Mol Cell Biol. 2018;19:365–381. doi: 10.1038/s41580-018-0001-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dice J.F. Altered degradation of proteins microinjected into senescent human fibroblasts. J Biol Chem. 1982;257:14624–14627. [PubMed] [Google Scholar]
- 15.Pfeifer U., Strauss P. Autophagic vacuoles in heart muscle and liver. A comparative morphometric study including circadian variations in meal-fed rats. J Mol Cell Cardiol. 1981;13:37–49. doi: 10.1016/0022-2828(81)90227-3. [DOI] [PubMed] [Google Scholar]
- 16.Kroemer G., Levine B. Autophagic cell death: the story of a misnomer. Nat Rev Mol Cell Biol. 2008;9:1004–1010. doi: 10.1038/nrm2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fang E.F., et al. Mitophagy inhibits amyloid-β and tau pathology and reverses cognitive deficits in models of Alzheimer's disease. Nat Neurosci. 2019;22:401–412. doi: 10.1038/s41593-018-0332-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lautrup S., Sinclair D.A., Mattson M.P., Fang E.F. NAD(+) in brain aging and neurodegenerative disorders. Cell Metab. 2019;30:630–655. doi: 10.1016/j.cmet.2019.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Croteau D.L., Fang E.F., Nilsen H., Bohr V.A. NAD+ in DNA repair and mitochondrial maintenance. Cell Cycle. 2017;16:491–492. doi: 10.1080/15384101.2017.1285631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leidal A.M., Levine B., Debnath J. Autophagy and the cell biology of age-related disease. Nat Cell Biol. 2018;20:1338–1348. doi: 10.1038/s41556-018-0235-8. [DOI] [PubMed] [Google Scholar]
- 21.Hartl F.U. Protein Misfolding Diseases. Annu Rev Biochem. 2017;86:21–26. doi: 10.1146/annurev-biochem-061516-044518. [DOI] [PubMed] [Google Scholar]
- 22.Lou G., et al. Mitophagy and neuroprotection. Trends Mol Med. 2020;26:8–20. doi: 10.1016/j.molmed.2019.07.002. [DOI] [PubMed] [Google Scholar]
- 23.Kocaturk N.M., Gozuacik D. Crosstalk between mammalian autophagy and the ubiquitin-proteasome system. Front Cell Dev Biol. 2018;6:128. doi: 10.3389/fcell.2018.00128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Limanaqi F., et al. Promiscuous roles of autophagy and proteasome in neurodegenerative proteinopathies. Int J Mol Sci. 2020;21 doi: 10.3390/ijms21083028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hamacher-Brady A., Brady N.R., Gottlieb R.A. Enhancing macroautophagy protects against ischemia/reperfusion injury in cardiac myocytes. J Biol Chem. 2006;281:29776–29787. doi: 10.1074/jbc.M603783200. [DOI] [PubMed] [Google Scholar]
- 26.Kang R., Zeh H.J., Lotze M.T., Tang D. The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ. 2011;18:571–580. doi: 10.1038/cdd.2010.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shibata M., et al. Regulation of intracellular accumulation of mutant Huntingtin by Beclin 1. J Biol Chem. 2006;281:14474–14485. doi: 10.1074/jbc.M600364200. [DOI] [PubMed] [Google Scholar]
- 28.Lipinski M.M., et al. Genome-wide analysis reveals mechanisms modulating autophagy in normal brain aging and in Alzheimer's disease. Proc Natl Acad Sci U S A. 2010;107:14164–14169. doi: 10.1073/pnas.1009485107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fallaize D., Chin L.-S., Li L. Differential submitochondrial localization of PINK1 as a molecular switch for mediating distinct mitochondrial signaling pathways. Cell Signal. 2015;27:2543–2554. doi: 10.1016/j.cellsig.2015.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee J.Y., et al. HDAC6 controls autophagosome maturation essential for ubiquitin-selective quality-control autophagy. Embo j. 2010;29:969–980. doi: 10.1038/emboj.2009.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guebel D.V., Torres N.V. Sexual dimorphism and aging in the human hyppocampus: identification, validation, and impact of differentially expressed genes by factorial microarray and network analysis. Front Aging Neurosci. 2016;8 doi: 10.3389/fnagi.2016.00229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fang E.F., et al. Mitophagy inhibits amyloid-beta and tau pathology and reverses cognitive deficits in models of Alzheimer's disease. Nat Neurosci. 2019;22:401–412. doi: 10.1038/s41593-018-0332-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pickrell A.M., Youle R.J. The roles of PINK1, parkin, and mitochondrial fidelity in Parkinson's disease. Neuron. 2015;85:257–273. doi: 10.1016/j.neuron.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Franco-Iborra S., et al. Mutant HTT (huntingtin) impairs mitophagy in a cellular model of Huntington disease. Autophagy. 2021;17:672–689. doi: 10.1080/15548627.2020.1728096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Evans C.S., Holzbaur E.L.F. Autophagy and mitophagy in ALS. Neurobiol Dis. 2019;122:35–40. doi: 10.1016/j.nbd.2018.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kerr J.S., et al. Mitophagy and Alzheimer's disease: cellular and molecular mechanisms. Trends Neurosci. 2017;40:151–166. doi: 10.1016/j.tins.2017.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fiesel F.C., et al. (Patho-)physiological relevance of PINK1-dependent ubiquitin phosphorylation. EMBO Rep. 2015;16:1114–1130. doi: 10.15252/embr.201540514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hou X., et al. Age- and disease-dependent increase of the mitophagy marker phospho-ubiquitin in normal aging and Lewy body disease. Autophagy. 2018;14:1404–1418. doi: 10.1080/15548627.2018.1461294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Glatigny M., et al. Autophagy is required for memory formation and reverses age-related memory decline. Curr Biol. 2019;29:435–448.e438. doi: 10.1016/j.cub.2018.12.021. [DOI] [PubMed] [Google Scholar]
- 40.Bjørkøy G., et al. Monitoring autophagic degradation of p62/SQSTM1. Methods Enzymol. 2009;452:181–197. doi: 10.1016/s0076-6879(08)03612-4. [DOI] [PubMed] [Google Scholar]
- 41.Ott C., Konig J., Hohn A., Jung T., Grune T. Macroautophagy is impaired in old murine brain tissue as well as in senescent human fibroblasts. Redox Biol. 2016;10:266–273. doi: 10.1016/j.redox.2016.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu Y., et al. The alteration of autophagy and apoptosis in the hippocampus of rats with natural aging-dependent cognitive deficits. Behav Brain Res. 2017;334:155–162. doi: 10.1016/j.bbr.2017.07.003. [DOI] [PubMed] [Google Scholar]
- 43.De Biase D., et al. Amyloid precursor protein, lipofuscin accumulation and expression of autophagy markers in aged bovine brain. BMC Vet Res. 2017;13:102. doi: 10.1186/s12917-017-1028-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun N., et al. Measuring in vivo mitophagy. Mol Cell. 2015;60:685–696. doi: 10.1016/j.molcel.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bjedov I., et al. Mechanisms of life span extension by rapamycin in the fruit fly Drosophila melanogaster. Cell Metab. 2010;11:35–46. doi: 10.1016/j.cmet.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fang E.F., et al. Tomatidine enhances lifespan and healthspan in C. elegans through mitophagy induction via the SKN-1/Nrf2 pathway. Sci Rep. 2017;7:46208. doi: 10.1038/srep46208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Harrison D.E., et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460:392–395. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pyo J.O., et al. Overexpression of Atg5 in mice activates autophagy and extends lifespan. Nat Commun. 2013;4:2300. doi: 10.1038/ncomms3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Simonsen A., et al. Promoting basal levels of autophagy in the nervous system enhances longevity and oxidant resistance in adult Drosophila. Autophagy. 2008;4:176–184. doi: 10.4161/auto.5269. [DOI] [PubMed] [Google Scholar]
- 50.Vellai T. et al. Genetics: influence of TOR kinase on lifespan in C. elegans. Nature426, 620, doi:10.1038/426620a (2003). [DOI] [PubMed]
- 51.Kuma A., et al. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 52.Inoue K., et al. Macroautophagy deficiency mediates age-dependent neurodegeneration through a phospho-tau pathway. Mol Neurodegener. 2012;7:48. doi: 10.1186/1750-1326-7-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rana A., Rera M., Walker D.W. Parkin overexpression during aging reduces proteotoxicity, alters mitochondrial dynamics, and extends lifespan. Proc Natl Acad Sci. 2013;110:8638. doi: 10.1073/pnas.1216197110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Todd A.M., Staveley B.E. Expression of Pink1 with α-synuclein in the dopaminergic neurons of Drosophila leads to increases in both lifespan and healthspan. Genet Mol Res. 2012;11:1497–1502. doi: 10.4238/2012.May.21.6. [DOI] [PubMed] [Google Scholar]
- 55.Rana A., et al. Promoting Drp1-mediated mitochondrial fission in midlife prolongs healthy lifespan of Drosophila melanogaster. Nat Commun. 2017;8:448. doi: 10.1038/s41467-017-00525-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hou Y., Song H., Croteau D.L., Akbari M., Bohr V.A. Genome instability in Alzheimer disease. Mech Ageing Dev. 2017;161:83–94. doi: 10.1016/j.mad.2016.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Silva M.V.F., et al. Alzheimer’s disease: risk factors and potentially protective measures. J Biomed Sci. 2019;26:33. doi: 10.1186/s12929-019-0524-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Canter R.G., Penney J., Tsai L.H. The road to restoring neural circuits for the treatment of Alzheimer's disease. Nature. 2016;539:187–196. doi: 10.1038/nature20412. [DOI] [PubMed] [Google Scholar]
- 59.Selkoe D.J., Hardy J. The amyloid hypothesis of Alzheimer's disease at 25 years. EMBO Mol Med. 2016;8:595–608. doi: 10.15252/emmm.201606210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Prillaman M. Alzheimer's drug slows mental decline in trial - but is it a breakthrough? Nature. 2022;610:15–16. doi: 10.1038/d41586-022-03081-0. [DOI] [PubMed] [Google Scholar]
- 61.Becker R.E., Greig N.H., Giacobini E., Schneider L.S., Ferrucci L. A new roadmap for drug development for Alzheimer's disease. Nat Rev Drug Discov. 2014;13:156. doi: 10.1038/nrd3842-c2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sun Y.X., et al. Differential activation of mTOR complex 1 signaling in human brain with mild to severe Alzheimer's disease. J Alzheimers Dis. 2014;38:437–444. doi: 10.3233/jad-131124. [DOI] [PubMed] [Google Scholar]
- 63.Caccamo A., De Pinto V., Messina A., Branca C., Oddo S. Genetic reduction of mammalian target of rapamycin ameliorates Alzheimer's disease-like cognitive and pathological deficits by restoring hippocampal gene expression signature. J Neurosci. 2014;34:7988–7998. doi: 10.1523/jneurosci.0777-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nixon R.A., et al. Extensive involvement of autophagy in Alzheimer disease: an immuno-electron microscopy study. J Neuropathol Exp Neurol. 2005;64:113–122. doi: 10.1093/jnen/64.2.113. [DOI] [PubMed] [Google Scholar]
- 65.Lin W.L., Lewis J., Yen S.H., Hutton M., Dickson D.W. Ultrastructural neuronal pathology in transgenic mice expressing mutant (P301L) human tau. J Neurocytol. 2003;32:1091–1105. doi: 10.1023/b:Neur.0000021904.61387.95. [DOI] [PubMed] [Google Scholar]
- 66.Pickford F., et al. The autophagy-related protein beclin 1 shows reduced expression in early Alzheimer disease and regulates amyloid beta accumulation in mice. J Clin Invest. 2008;118:2190–2199. doi: 10.1172/jci33585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Morel E., et al. Phosphatidylinositol-3-phosphate regulates sorting and processing of amyloid precursor protein through the endosomal system. Nat Commun. 2013;4:2250. doi: 10.1038/ncomms3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yu W.H., et al. Macroautophagy–a novel Beta-amyloid peptide-generating pathway activated in Alzheimer's disease. J Cell Biol. 2005;171:87–98. doi: 10.1083/jcb.200505082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yu W.H., et al. Autophagic vacuoles are enriched in amyloid precursor protein-secretase activities: implications for β-amyloid peptide over-production and localization in Alzheimer’s disease. Int J Biochem Cell Biol. 2004;36:2531–2540. doi: 10.1016/j.biocel.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 70.Nixon R.A. Autophagy, amyloidogenesis and Alzheimer disease. J Cell Sci. 2007;120:4081–4091. doi: 10.1242/jcs.019265. [DOI] [PubMed] [Google Scholar]
- 71.Lee S., Sato Y., Nixon R.A. Lysosomal proteolysis inhibition selectively disrupts axonal transport of degradative organelles and causes an Alzheimer's-like axonal dystrophy. J Neurosci. 2011;31:7817–7830. doi: 10.1523/jneurosci.6412-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fraser PE. et al. Presenilin structure, function and role in Alzheimer disease. Biochim Biophys Acta (BBA) - Mol Basis Dis1502, 1-15, doi:https://doi.org/10.1016/S0925-4439(00)00028-4 (2000). [DOI] [PubMed]
- 73.Cataldo A.M., et al. Presenilin mutations in familial Alzheimer disease and transgenic mouse models accelerate neuronal lysosomal pathology. J Neuropathol Exp Neurol. 2004;63:821–830. doi: 10.1093/jnen/63.8.821. [DOI] [PubMed] [Google Scholar]
- 74.Lee J.H., et al. Lysosomal proteolysis and autophagy require presenilin 1 and are disrupted by Alzheimer-related PS1 mutations. Cell. 2010;141:1146–1158. doi: 10.1016/j.cell.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tung Y.-T., et al. Presenilin-1 regulates the expression of p62 to govern p62-dependent tau degradation. Mol Neurobiol. 2014;49:10–27. doi: 10.1007/s12035-013-8482-y. [DOI] [PubMed] [Google Scholar]
- 76.Reddy K., et al. Dysregulation of nutrient sensing and CLEARance in presenilin deficiency. Cell Rep. 2016;14:2166–2179. doi: 10.1016/j.celrep.2016.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fedeli C., Filadi R., Rossi A., Mammucari C., Pizzo P. PSEN2 (presenilin 2) mutants linked to familial Alzheimer disease impair autophagy by altering Ca(2+) homeostasis. Autophagy. 2019;15:2044–2062. doi: 10.1080/15548627.2019.1596489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jiang Y., et al. Lysosomal dysfunction in down syndrome is APP-dependent and Mediated by APP-βCTF (C99) J Neurosci. 2019;39:5255–5268. doi: 10.1523/jneurosci.0578-19.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bordi M., et al. Autophagy flux in CA1 neurons of Alzheimer hippocampus: Increased induction overburdens failing lysosomes to propel neuritic dystrophy. Autophagy. 2016;12:2467–2483. doi: 10.1080/15548627.2016.1239003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sardiello M., et al. A gene network regulating lysosomal biogenesis and function. Science. 2009;325:473–477. doi: 10.1126/science.1174447. [DOI] [PubMed] [Google Scholar]
- 81.Wang H., Wang R., Xu S., Lakshmana M.K. Transcription factor EB Is selectively reduced in the nuclear fractions of Alzheimer’s and amyotrophic lateral sclerosis brains. Neuroscience Journal. 2016;2016:4732837. doi: 10.1155/2016/4732837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang Y.D., Zhao J.J. TFEB participates in the aβ-induced pathogenesis of Alzheimer's disease by regulating the autophagy-lysosome pathway. DNA Cell Biol. 2015;34:661–668. doi: 10.1089/dna.2014.2738. [DOI] [PubMed] [Google Scholar]
- 83.Xiao Q., et al. Neuronal-targeted TFEB accelerates lysosomal degradation of APP, reducing Aβ generation and amyloid plaque pathogenesis. J Neurosci. 2015;35:12137–12151. doi: 10.1523/JNEUROSCI.0705-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Song J.-X., et al. A small molecule transcription factor EB activator ameliorates beta-amyloid precursor protein and Tau pathology in Alzheimer's disease models. Aging Cell. 2020;19:e13069. doi: 10.1111/acel.13069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yamamoto F., et al. TFEB-mediated enhancement of the autophagy-lysosomal pathway dually modulates the process of amyloid β-protein generation in neurons. Neuroscience. 2019;402:11–22. doi: 10.1016/j.neuroscience.2019.01.010. [DOI] [PubMed] [Google Scholar]
- 86.Lauritzen I., et al. Intraneuronal aggregation of the β-CTF fragment of APP (C99) induces Aβ-independent lysosomal-autophagic pathology. Acta Neuropathol. 2016;132:257–276. doi: 10.1007/s00401-016-1577-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lin X.-X., et al. DAF-16/FOXO and HLH-30/TFEB function as combinatorial transcription factors to promote stress resistance and longevity. Nat Commun. 2018;9:4400. doi: 10.1038/s41467-018-06624-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lapierre L.R., et al. The TFEB orthologue HLH-30 regulates autophagy and modulates longevity in Caenorhabditis elegans. Nat Commun. 2013;4:2267. doi: 10.1038/ncomms3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bao J., et al. Deacetylation of TFEB promotes fibrillar Aβ degradation by upregulating lysosomal biogenesis in microglia. Protein Cell. 2016;7:417–433. doi: 10.1007/s13238-016-0269-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Xiao Q., et al. Enhancing astrocytic lysosome biogenesis facilitates Aβ clearance and attenuates amyloid plaque pathogenesis. J Neurosci. 2014;34:9607–9620. doi: 10.1523/JNEUROSCI.3788-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Polito V.A., et al. Selective clearance of aberrant tau proteins and rescue of neurotoxicity by transcription factor EB. EMBO Mol Med. 2014;6:1142–1160. doi: 10.15252/emmm.201303671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang H, Wang R, Carrera I, Xu S, Lakshmana MK. TFEB Overexpression in the P301S model of tauopathy mitigates increased PHF1 levels and lipofuscin puncta and rescues memory deficits. eNeuro3, ENEURO.0042-0016.2016, doi:10.1523/ENEURO.0042-16.2016 (2016). [DOI] [PMC free article] [PubMed]
- 93.Schuur M., et al. Cathepsin D gene and the risk of Alzheimer's disease: a population-based study and meta-analysis. Neurobiol Aging. 2011;32:1607–1614. doi: 10.1016/j.neurobiolaging.2009.10.011. [DOI] [PubMed] [Google Scholar]
- 94.Steinfeld R., et al. Cathepsin D deficiency is associated with a human neurodegenerative disorder. Am J Hum Genet. 2006;78:988–998. doi: 10.1086/504159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wu J.W., et al. Neuronal activity enhances tau propagation and tau pathology in vivo. Nat Neurosci. 2016;19:1085–1092. doi: 10.1038/nn.4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Martini-Stoica H., et al. TFEB enhances astroglial uptake of extracellular tau species and reduces tau spreading. J Exp Med. 2018;215:2355–2377. doi: 10.1084/jem.20172158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Medina D.L., et al. Transcriptional activation of lysosomal exocytosis promotes cellular clearance. Dev Cell. 2011;21:421–430. doi: 10.1016/j.devcel.2011.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Xu Y., et al. TFEB regulates lysosomal exocytosis of tau and its loss of function exacerbates tau pathology and spreading. Mol Psychiatry. 2020 doi: 10.1038/s41380-020-0738-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Decressac M. et al. TFEB-mediated autophagy rescues midbrain dopamine neurons from α-synuclein toxicity. Proc Natl Acad Sci U S A110, E1817-1826, doi:10.1073/pnas.1305623110 (2013). [DOI] [PMC free article] [PubMed]
- 100.Tsunemi T. et al. PGC-1α Rescues Huntington’s disease proteotoxicity by preventing oxidative stress and promoting TFEB function. Sci Transl Med4, 142ra197-142ra197, doi:10.1126/scitranslmed.3003799 (2012). [DOI] [PMC free article] [PubMed]
- 101.Rocchi A. et al. A Becn1 mutation mediates hyperactive autophagic sequestration of amyloid oligomers and improved cognition in Alzheimer's disease. PLoS Genet13, e1006962, doi:10.1371/journal.pgen.1006962 (2017). [DOI] [PMC free article] [PubMed]
- 102.Nilsson P., et al. Aβ secretion and plaque formation depend on autophagy. Cell Rep. 2013;5:61–69. doi: 10.1016/j.celrep.2013.08.042. [DOI] [PubMed] [Google Scholar]
- 103.Nilsson P., Saido T.C. Dual roles for autophagy: degradation and secretion of Alzheimer's disease Aβ peptide. Bioessays. 2014;36:570–578. doi: 10.1002/bies.201400002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Vingtdeux V., et al. Novel synthetic small-molecule activators of AMPK as enhancers of autophagy and amyloid-β peptide degradation. Faseb j. 2011;25:219–231. doi: 10.1096/fj.10-167361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Schaeffer V., Goedert M. Stimulation of autophagy is neuroprotective in a mouse model of human tauopathy. Autophagy. 2012;8:1686–1687. doi: 10.4161/auto.21488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhao Y., et al. Klotho overexpression improves amyloid-β clearance and cognition in the APP/PS1 mouse model of Alzheimer's disease. Aging Cell. 2020;19:e13239. doi: 10.1111/acel.13239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tian Y, Chang JC, Fan EY, Flajolet M, Greengard P. Adaptor complex AP2/PICALM, through interaction with LC3, targets Alzheimer's APP-CTF for terminal degradation via autophagy. Proc Natl Acad Sci110, 17071-17076, doi:10.1073/pnas.1315110(2013). [DOI] [PMC free article] [PubMed]
- 108.Harold D., et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer's disease. Nat Genet. 2009;41:1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Jun G., et al. Meta-analysis confirms CR1, CLU, and PICALM as alzheimer disease risk loci and reveals interactions with APOE genotypes. Arch Neurol. 2010;67:1473–1484. doi: 10.1001/archneurol.2010.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ando K., et al. Clathrin adaptor CALM/PICALM is associated with neurofibrillary tangles and is cleaved in Alzheimer's brains. Acta Neuropathol. 2013;125:861–878. doi: 10.1007/s00401-013-1111-z. [DOI] [PubMed] [Google Scholar]
- 111.Moreau K., et al. PICALM modulates autophagy activity and tau accumulation. Nat Commun. 2014;5:4998. doi: 10.1038/ncomms5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lee M.J., Lee J.H., Rubinsztein D.C. Tau degradation: The ubiquitin–proteasome system versus the autophagy-lysosome system. Prog Neurobiol. 2013;105:49–59. doi: 10.1016/j.pneurobio.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 113.Ramesh Babu J., et al. Genetic inactivation of p62 leads to accumulation of hyperphosphorylated tau and neurodegeneration. J Neurochem. 2008;106:107–120. doi: 10.1111/j.1471-4159.2008.05340.x. [DOI] [PubMed] [Google Scholar]
- 114.Caccamo A., et al. mTOR regulates tau phosphorylation and degradation: implications for Alzheimer's disease and other tauopathies. Aging Cell. 2013;12:370–380. doi: 10.1111/acel.12057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Silva M.C., et al. Prolonged tau clearance and stress vulnerability rescue by pharmacological activation of autophagy in tauopathy neurons. Nat Commun. 2020;11:3258. doi: 10.1038/s41467-020-16984-1. <http://europepmc.org/abstract/MED/32591533 doi: 10.1038/s41467-020-16984-1 https://europepmc.org/articles/PMC7320012 https://europepmc.org/articles/PMC7320012?pdf=render> [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kim S., et al. NDP52 associates with phosphorylated tau in brains of an Alzheimer disease mouse model. Biochem Biophys Res Commun. 2014;454:196–201. doi: 10.1016/j.bbrc.2014.10.066. [DOI] [PubMed] [Google Scholar]
- 117.Chesser A.S., Ganeshan V., Yang J., Johnson G.V. Epigallocatechin-3-gallate enhances clearance of phosphorylated tau in primary neurons. Nutr Neurosci. 2016;19:21–31. doi: 10.1179/1476830515y.0000000038. [DOI] [PubMed] [Google Scholar]
- 118.Lautrup S., et al. Microglial mitophagy mitigates neuroinflammation in Alzheimer's disease. Neurochem Int. 2019;129 doi: 10.1016/j.neuint.2019.104469. [DOI] [PubMed] [Google Scholar]
- 119.Cummins N., Tweedie A., Zuryn S., Bertran-Gonzalez J., Götz J. Disease-associated tau impairs mitophagy by inhibiting Parkin translocation to mitochondria. EMBO J. 2019;38:e99360. doi: 10.15252/embj.201899360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Corsetti V., et al. NH2-truncated human tau induces deregulated mitophagy in neurons by aberrant recruitment of Parkin and UCHL-1: implications in Alzheimer's disease. Hum Mol Genet. 2015;24:3058–3081. doi: 10.1093/hmg/ddv059. [DOI] [PubMed] [Google Scholar]
- 121.Pareja-Cajiao M., et al. Age-related impairment of autophagy in cervical motor neurons. Exp Gerontol. 2021;144 doi: 10.1016/j.exger.2020.111193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Del Prete D., et al. Localization and processing of the amyloid-β protein precursor in mitochondria-associated membranes. J Alzheimers Dis. 2017;55:1549–1570. doi: 10.3233/JAD-160953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Cenini G., Rüb C., Bruderek M., Voos W. Amyloid β-peptides interfere with mitochondrial preprotein import competence by a coaggregation process. Mol Biol Cell. 2016;27:3257–3272. doi: 10.1091/mbc.E16-05-0313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Mao P., et al. Mitochondria-targeted catalase reduces abnormal APP processing, amyloid β production and BACE1 in a mouse model of Alzheimer's disease: implications for neuroprotection and lifespan extension. Hum Mol Genet. 2012;21:2973–2990. doi: 10.1093/hmg/dds128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Manczak M., Kandimalla R., Yin X., Reddy P.H. Hippocampal mutant APP and amyloid beta-induced cognitive decline, dendritic spine loss, defective autophagy, mitophagy and mitochondrial abnormalities in a mouse model of Alzheimer's disease. Hum Mol Genet. 2018;27:1332–1342. doi: 10.1093/hmg/ddy042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Reddy P.H., et al. Mutant APP and amyloid beta-induced defective autophagy, mitophagy, mitochondrial structural and functional changes and synaptic damage in hippocampal neurons from Alzheimer's disease. Hum Mol Genet. 2018;27:2502–2516. doi: 10.1093/hmg/ddy154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wang Z.T., et al. Disrupted-in-schizophrenia-1 protects synaptic plasticity in a transgenic mouse model of Alzheimer's disease as a mitophagy receptor. Aging Cell. 2019;18:e12860. doi: 10.1111/acel.12860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Sorrentino V., et al. Enhancing mitochondrial proteostasis reduces amyloid-β proteotoxicity. Nature. 2017;552:187–193. doi: 10.1038/nature25143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ye X., Sun X., Starovoytov V., Cai Q. Parkin-mediated mitophagy in mutant hAPP neurons and Alzheimer's disease patient brains. Hum Mol Genet. 2015;24:2938–2951. doi: 10.1093/hmg/ddv056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Martin-Maestro P., Gargini R., Perry G., Avila J., Garcia-Escudero V. PARK2 enhancement is able to compensate mitophagy alterations found in sporadic Alzheimer's disease. Hum Mol Genet. 2016;25:792–806. doi: 10.1093/hmg/ddv616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Checler F., Goiran T., Alves da Costa C. Presenilins at the crossroad of a functional interplay between PARK2/PARKIN and PINK1 to control mitophagy: Implication for neurodegenerative diseases. Autophagy. 2017;13:2004–2005. doi: 10.1080/15548627.2017.1363950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lazarou M., et al. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature. 2015;524:309–314. doi: 10.1038/nature14893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Du F., et al. PINK1 signalling rescues amyloid pathology and mitochondrial dysfunction in Alzheimer's disease. Brain. 2017;140:3233–3251. doi: 10.1093/brain/awx258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Cen X., et al. Pharmacological targeting of MCL-1 promotes mitophagy and improves disease pathologies in an Alzheimer’s disease mouse model. Nat Commun. 2020;11:5731. doi: 10.1038/s41467-020-19547-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Park J.-S., Kim D.-H., Yoon S.-Y. Regulation of amyloid precursor protein processing by its KFERQ motif. BMB Rep. 2016;49:337–342. doi: 10.5483/bmbrep.2016.49.6.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Bourdenx M., et al. Chaperone-mediated autophagy prevents collapse of the neuronal metastable proteome. Cell. 2021;184:2696–2714.e2625. doi: 10.1016/j.cell.2021.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Caballero B., et al. Interplay of pathogenic forms of human tau with different autophagic pathways. Aging Cell. 2018;17:e12692. doi: 10.1111/acel.12692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Wang Y., et al. Tau fragmentation, aggregation and clearance: the dual role of lysosomal processing. Hum Mol Genet. 2009;18:4153–4170. doi: 10.1093/hmg/ddp367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Heckmann B.L., et al. LC3-associated endocytosis facilitates β-amyloid clearance and mitigates neurodegeneration in murine Alzheimer's disease. Cell. 2019;178:536–551.e514. doi: 10.1016/j.cell.2019.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ulland T.K., et al. TREM2 maintains microglial metabolic fitness in Alzheimer's disease. Cell. 2017;170:649–663.e613. doi: 10.1016/j.cell.2017.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Fernández Á.F., et al. Disruption of the beclin 1-BCL2 autophagy regulatory complex promotes longevity in mice. Nature. 2018;558:136–140. doi: 10.1038/s41586-018-0162-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Houtman J., et al. Beclin1-driven autophagy modulates the inflammatory response of microglia via NLRP3. EMBO J. 2019;38 doi: 10.15252/embj.201899430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Takayama S., Xie Z., Reed J.C. An evolutionarily conserved family of Hsp70/Hsc70 molecular chaperone regulators. J Biol Chem. 1999;274:781–786. doi: 10.1074/jbc.274.2.781. [DOI] [PubMed] [Google Scholar]
- 144.Behl C. Breaking BAG: the co-chaperone BAG3 in health and disease. Trends Pharmacol Sci. 2016;37:672–688. doi: 10.1016/j.tips.2016.04.007. [DOI] [PubMed] [Google Scholar]
- 145.Gamerdinger M., et al. Protein quality control during aging involves recruitment of the macroautophagy pathway by BAG3. Embo j. 2009;28:889–901. doi: 10.1038/emboj.2009.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Johnston J.A., Ward C.L., Kopito R.R. Aggresomes: a cellular response to misfolded proteins. J Cell Biol. 1998;143:1883–1898. doi: 10.1083/jcb.143.7.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Carra S., Brunsting J.F., Lambert H., Landry J., Kampinga H.H. HspB8 participates in protein quality control by a non-chaperone-like mechanism that requires eIF2α phosphorylation*. J Biol Chem. 2009;284:5523–5532. doi: 10.1074/jbc.M807440200. [DOI] [PubMed] [Google Scholar]
- 148.Seidel K., et al. The HSPB8-BAG3 chaperone complex is upregulated in astrocytes in the human brain affected by protein aggregation diseases. Neuropathol Appl Neurobiol. 2012;38:39–53. doi: 10.1111/j.1365-2990.2011.01198.x. [DOI] [PubMed] [Google Scholar]
- 149.Krüger U., Wang Y., Kumar S., Mandelkow E.M. Autophagic degradation of tau in primary neurons and its enhancement by trehalose. Neurobiol Aging. 2012;33:2291–2305. doi: 10.1016/j.neurobiolaging.2011.11.009. [DOI] [PubMed] [Google Scholar]
- 150.Elliott E., Tsvetkov P., Ginzburg I. BAG-1 associates with Hsc70.Tau complex and regulates the proteasomal degradation of Tau protein. J Biol Chem. 2007;282:37276–37284. doi: 10.1074/jbc.M706379200. [DOI] [PubMed] [Google Scholar]
- 151.Lei Z., Brizzee C., Johnson G.V.W. BAG3 facilitates the clearance of endogenous tau in primary neurons. Neurobiol Aging. 2015;36:241–248. doi: 10.1016/j.neurobiolaging.2014.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Fu H., et al. A tau homeostasis signature is linked with the cellular and regional vulnerability of excitatory neurons to tau pathology. Nat Neurosci. 2019;22:47–56. doi: 10.1038/s41593-018-0298-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Renziehausen J., et al. The cleavage product of amyloid-β protein precursor sAβPPα modulates BAG3-dependent aggresome formation and enhances cellular proteasomal activity. J Alzheimers Dis. 2015;44:879–896. doi: 10.3233/jad-140600. [DOI] [PubMed] [Google Scholar]
- 154.Bagherniya M., Butler A.E., Barreto G.E., Sahebkar A. The effect of fasting or calorie restriction on autophagy induction: A review of the literature. Ageing Res Rev. 2018;47:183–197. doi: 10.1016/j.arr.2018.08.004. [DOI] [PubMed] [Google Scholar]
- 155.Andreotti D.Z., et al. Effects of physical exercise on autophagy and apoptosis in aged brain: human and animal studies. Front Nutr. 2020;7 doi: 10.3389/fnut.2020.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Partridge L., Fuentealba M., Kennedy B.K. The quest to slow ageing through drug discovery. Nat Rev Drug Discov. 2020;19:513–532. doi: 10.1038/s41573-020-0067-7. [DOI] [PubMed] [Google Scholar]
- 157.Donati A. et al. Age-related changes in the autophagic proteolysis of rat isolated liver cells: effects of antiaging dietary restrictions. J Gerontol A Biol Sci Med Sci56, B375-383, doi:10.1093/gerona/56.9.b375 (2001). [DOI] [PubMed]
- 158.Pifferi F. et al. Caloric restriction increases lifespan but affects brain integrity in grey mouse lemur primates. Commun Biol1, 30 (2018). <http://europepmc.org/abstract/MED/30271916 https://doi.org/10.1038/s42003-018-0024-8 https://europepmc.org/articles/PMC6123706 https://europepmc.org/articles/PMC6123706?pdf=render>. [DOI] [PMC free article] [PubMed]
- 159.Rittig N., et al. Anabolic effects of leucine-rich whey protein, carbohydrate, and soy protein with and without β-hydroxy-β-methylbutyrate (HMB) during fasting-induced catabolism: a human randomized crossover trial. Clin Nutr. 2017;36:697–705. doi: 10.1016/j.clnu.2016.05.004. [DOI] [PubMed] [Google Scholar]
- 160.Vendelbo M.H., et al. Fasting increases human skeletal muscle net phenylalanine release and this is associated with decreased mTOR signaling. PLoS One. 2014;9:e102031. doi: 10.1371/journal.pone.0102031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Yang L., et al. Long-term calorie restriction enhances cellular quality-control processes in human skeletal muscle. Cell Rep. 2016;14:422–428. doi: 10.1016/j.celrep.2015.12.042. [DOI] [PubMed] [Google Scholar]
- 162.Pietrocola F., et al. Metabolic effects of fasting on human and mouse blood in vivo. Autophagy. 2017;13:567–578. doi: 10.1080/15548627.2016.1271513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Escobar K.A., Cole N.H., Mermier C.M., VanDusseldorp T.A. Autophagy and aging: Maintaining the proteome through exercise and caloric restriction. Aging Cell. 2019;18:e12876. doi: 10.1111/acel.12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Alirezaei M., et al. Short-term fasting induces profound neuronal autophagy. Autophagy. 2010;6:702–710. doi: 10.4161/auto.6.6.12376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Chen X., Kondo K., Motoki K., Homma H., Okazawa H. Fasting activates macroautophagy in neurons of Alzheimer's disease mouse model but is insufficient to degrade amyloid-beta. Sci Rep. 2015;5:12115. doi: 10.1038/srep12115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Müller L., et al. Long-term caloric restriction attenuates β-amyloid neuropathology and is accompanied by autophagy in APPswe/PS1delta9 Mice. Nutrients. 2021;13:985. doi: 10.3390/nu13030985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Kim J., Kundu M., Viollet B., Guan K.-L. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Schwalm C., et al. Activation of autophagy in human skeletal muscle is dependent on exercise intensity and AMPK activation. FASEB J. 2015;29:3515–3526. doi: 10.1096/fj.14-267187. [DOI] [PubMed] [Google Scholar]
- 169.Mejías-Peña Y. et al. Effects of aerobic training on markers of autophagy in the elderly. Age (Dordr)38, 33-33, doi:10.1007/s11357-016-9897-y (2016). [DOI] [PMC free article] [PubMed]
- 170.Mejías-Peña Y., et al. Impact of resistance training on the autophagy-inflammation-apoptosis crosstalk in elderly subjects. Aging (Albany NY) 2017;9:408–418. doi: 10.18632/aging.101167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Almeida M.F., et al. Effects of mild running on substantia nigra during early neurodegeneration. J Sports Sci. 2018;36:1363–1370. doi: 10.1080/02640414.2017.1378494. [DOI] [PubMed] [Google Scholar]
- 172.Jang Y.C., et al. Association of exercise-induced autophagy upregulation and apoptosis suppression with neuroprotection against pharmacologically induced Parkinson's disease. J Exerc Nutrition Biochem. 2018;22:1–8. doi: 10.20463/jenb.2018.0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Zhao N., et al. The effects of treadmill exercise on autophagy in hippocampus of APP/PS1 transgenic mice. Neuroreport. 2018;29:819–825. doi: 10.1097/wnr.0000000000001038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Bové J., Martínez-Vicente M., Vila M. Fighting neurodegeneration with rapamycin: mechanistic insights. Nat Rev Neurosci. 2011;12:437–452. doi: 10.1038/nrn3068. [DOI] [PubMed] [Google Scholar]
- 175.Miller R.A., et al. Rapamycin-mediated lifespan increase in mice is dose and sex dependent and metabolically distinct from dietary restriction. Aging Cell. 2014;13:468–477. doi: 10.1111/acel.12194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Watson JP, 2020. (2020, January - 2023, December). Participatory Evaluation (of) Aging (With) Rapamycin (for) Longevity Study (PEARL). Identifier NCT04488601. [WWW Document]. URL https://clinicaltrials.gov/ct2/show/NCT04488601.
- 177.Seshadri SJ, Gonzales MJ, 2021. (2021, August – 2023, December). Rapamycin – Effects on Alzheimer’s and Cognitive Health (REACH). Identifier NCT04629495. [WWW Document]. URL https://clinicaltrials.gov/ct2/show/NCT04629495.
- 178.Cassano T., et al. Early intrathecal infusion of everolimus restores cognitive function and mood in a murine model of Alzheimer's disease. Exp Neurol. 2019;311:88–105. doi: 10.1016/j.expneurol.2018.09.011. [DOI] [PubMed] [Google Scholar]
- 179.Jiang T., et al. Temsirolimus promotes autophagic clearance of amyloid-β and provides protective effects in cellular and animal models of Alzheimer's disease. Pharmacol Res. 2014;81:54–63. doi: 10.1016/j.phrs.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 180.Jiang T., et al. Temsirolimus attenuates tauopathy in vitro and in vivo by targeting tau hyperphosphorylation and autophagic clearance. Neuropharmacology. 2014;85:121–130. doi: 10.1016/j.neuropharm.2014.05.032. [DOI] [PubMed] [Google Scholar]
- 181.Frederick C., et al. Rapamycin ester analog CCI-779/Temsirolimus alleviates tau pathology and improves motor deficit in mutant tau transgenic mice. J Alzheimers Dis. 2015;44:1145–1156. doi: 10.3233/jad-142097. [DOI] [PubMed] [Google Scholar]
- 182.Rotermund C, Machetanz G, Fitzgerald JC. The therapeutic potential of metformin in neurodegenerative diseases. Front Endocrinol (Lausanne)9, 400-400, doi:10.3389/fendo.2018.00400 (2018). [DOI] [PMC free article] [PubMed]
- 183.Anisimov V.N. Metformin: do we finally have an anti-aging drug? Cell Cycle. 2013;12:3483–3489. doi: 10.4161/cc.26928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Cabreiro F., et al. Metformin retards aging in C. elegans by altering microbial folate and methionine metabolism. Cell. 2013;153:228–239. doi: 10.1016/j.cell.2013.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Anisimov V.N., et al. Metformin slows down aging and extends life span of female SHR mice. Cell Cycle. 2008;7:2769–2773. doi: 10.4161/cc.7.17.6625. [DOI] [PubMed] [Google Scholar]
- 186.Martin-Montalvo A., et al. Metformin improves healthspan and lifespan in mice. Nat Commun. 2013;4:2192. doi: 10.1038/ncomms3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Kobilo T., et al. AMPK agonist AICAR improves cognition and motor coordination in young and aged mice. Learn Mem. 2014;21:119–126. doi: 10.1101/lm.033332.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Andreux P.A., et al. The mitophagy activator urolithin A is safe and induces a molecular signature of improved mitochondrial and cellular health in humans. Nature Metabolism. 2019;1:595–603. doi: 10.1038/s42255-019-0073-4. [DOI] [PubMed] [Google Scholar]
- 189.Qi Y, Qiu Q, Gu X, Tian Y, Zhang Y. ATM mediates spermidine-induced mitophagy via PINK1 and Parkin regulation in human fibroblasts. Sci Rep6, 24700-24700, doi:10.1038/srep24700 (2016). [DOI] [PMC free article] [PubMed]
- 190.Fang E.F. Mitophagy and NAD(+) inhibit Alzheimer disease. Autophagy. 2019;15:1112–1114. doi: 10.1080/15548627.2019.1596497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Xie C., et al. Amelioration of Alzheimer's disease pathology by mitophagy inducers identified via machine learning and a cross-species workflow. Nat Biomed Eng. 2022;6:76–93. doi: 10.1038/s41551-021-00819-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Ryu D., et al. Urolithin A induces mitophagy and prolongs lifespan in C. elegans and increases muscle function in rodents. Nat Med. 2016;22:879–888. doi: 10.1038/nm.4132. [DOI] [PubMed] [Google Scholar]
- 193.Eisenberg T., et al. Cardioprotection and lifespan extension by the natural polyamine spermidine. Nat Med. 2016;22:1428–1438. doi: 10.1038/nm.4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Eisenberg T., et al. Induction of autophagy by spermidine promotes longevity. Nat Cell Biol. 2009;11:1305–1314. doi: 10.1038/ncb1975. [DOI] [PubMed] [Google Scholar]
- 195.Yang X., et al. Spermidine inhibits neurodegeneration and delays aging via the PINK1-PDR1-dependent mitophagy pathway in C. elegans. Aging. 2020;12:16852–16866. doi: 10.18632/aging.103578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Fan J., et al. Spermidine coupled with exercise rescues skeletal muscle atrophy from D-gal-induced aging rats through enhanced autophagy and reduced apoptosis via AMPK-FOXO3a signal pathway. Oncotarget. 2017;8:17475–17490. doi: 10.18632/oncotarget.15728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Madeo F., Eisenberg T., Pietrocola F., Kroemer G. Spermidine in health and disease. Science. 2018;359 doi: 10.1126/science.aan2788. [DOI] [PubMed] [Google Scholar]
- 198.Xie C., Aman Y., Frank J., Donate-Lagartos M.J., Gudmundsrud R., Cechova K., et al. In: Autophagy in Health and Disease. Rothermel B., Diwan A., editors. Academic Press; 2021. Autophagic processes in early- and late-onset Alzheimer’s disease. [Google Scholar]
- 199.Fang E.F., et al. NAD+ in aging: molecular mechanisms and translational implications. Trends Mol Med. 2017 doi: 10.1016/j.molmed.2017.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Fang E.F., et al. NAD+ augmentation restores mitophagy and limits accelerated aging in Werner syndrome. Nat Commun. 2019;10:5284. doi: 10.1038/s41467-019-13172-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Fang E.F., et al. NAD(+) replenishment improves lifespan and healthspan in ataxia telangiectasia models via mitophagy and DNA repair. Cell Metab. 2016;24:566–581. doi: 10.1016/j.cmet.2016.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Fang E.F., et al. Defective mitophagy in XPA via PARP-1 hyperactivation and NAD(+)/SIRT1 reduction. Cell. 2014;157:882–896. doi: 10.1016/j.cell.2014.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Fang E.F., et al. A research agenda for ageing in China in the 21st century (2nd edition): Focusing on basic and translational research, long-term care, policy and social networks. Ageing Res Rev. 2020;64 doi: 10.1016/j.arr.2020.101174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Fang E.F., et al. A research agenda for aging in China in the 21st century. Ageing Res Rev. 2015;24:197–205. doi: 10.1016/j.arr.2015.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Cox LS. et al. Tackling immunosenescence to improve COVID-19 outcomes and vaccine response in older adults. Lancet Healthy Longev1, e55-e57, doi:10.1016/S2666-7568(20)30011-8 (2020). [DOI] [PMC free article] [PubMed]
- 206.Livingston G., et al. Dementia prevention, intervention, and care: 2020 report of the Lancet Commission. Lancet. 2020;396:413–446. doi: 10.1016/s0140-6736(20)30367-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Mattson M.P., Arumugam T.V. Hallmarks of brain aging: adaptive and pathological modification by metabolic states. Cell Metab. 2018;27:1176–1199. doi: 10.1016/j.cmet.2018.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Long J.M., Holtzman D.M. Alzheimer disease: an update on pathobiology and treatment strategies. Cell. 2019;179:312–339. doi: 10.1016/j.cell.2019.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Kingwell K. Turning up mitophagy in Alzheimer disease. Nat Rev Drug Discov. 2019 doi: 10.1038/d41573-019-00035-6. [DOI] [PubMed] [Google Scholar]
- 210.Shintani T., Klionsky D.J. Autophagy in health and disease: a double-edged sword. Science. 2004;306:990–995. doi: 10.1126/science.1099993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Kobro-Flatmoen A., et al. Re-emphasizing early Alzheimer's disease pathology starting in select entorhinal neurons, with a special focus on mitophagy. Ageing Res Rev. 2021;67 doi: 10.1016/j.arr.2021.101307. [DOI] [PubMed] [Google Scholar]