Roach et al. 10.1073/pnas.0502272102. |
Supporting Text
Supporting Data Set 1
Supporting Figure 4
Supporting Figure 5
Supporting Figure 6
Supporting Figure 7
Supporting Data Set 1
Dataset 1. Information about sequences discussed in this paper.
Fig. 4.
Multiple alignment used to compute sequence distances.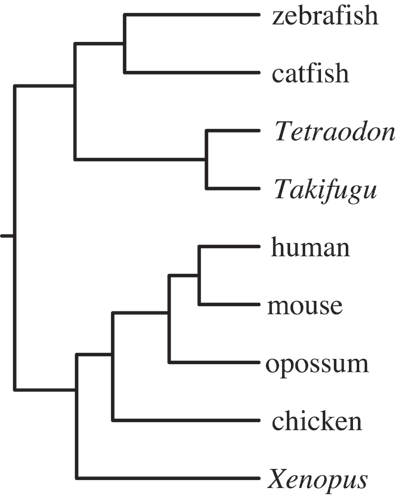
Fig. 5.
Molecular tree of Toll-like receptor 3 (TLR3). The topology of the tree is preserved; branch lengths are equalized. The molecular evolution of the TLR3 family recapitulates the evolution of species (subject to the caveat that the rooting of the bony fish clade is not absolutely established in the literature).
Fig. 6.
Molecular tree of the family containing TLR1. The topology of the tree is preserved; branch lengths are equalized to better display branch labels. The molecular evolution of the TLR2 subfamily recapitulates the evolution of species. The molecular evolution of the TLR14 subfamily and the TLR1 subfamily is more complex. The TLR14 subfamily appears to have been lost in amniotes but expanded in amphibians. The TLR1 subfamily probably expanded independently in different lineages. In particular, eutherian TLR1 and TLR6 arose from a duplication of TLR1L, which remained single copy in marsupials.
Fig. 7.
Multidimensional scaling of the molecular distances between the TIR domains of metazoan Toll-like receptors. Large type indicates a subfamily of vertebrate TLRs. The distance between most clusters compared with the distances within clusters is so great that portraying this information as a molecular tree could be misleading. All of the insect TLRs group together, so this representation does not support a hypothesis that Drosophila TLR9 is more closely related to vertebrate TLRs than it is to other insect TLRs.