Abstract
A potential strategy to alleviate the aggregation of intrinsically disordered proteins (IDPs) is to maintain the native functional state of the protein by small molecule binding. However, the targeting of the native state of IDPs by small molecules has been challenging due to their heterogeneous conformational ensembles. To tackle this challenge, we applied a high-throughput chemical microarray surface plasmon resonance imaging screen to detect the binding between small molecules and monomeric full-length Tau, a protein linked with the onset of a range of Tauopathies. The screen identified a novel set of drug-like fragment and lead-like compounds that bound to Tau. We verified that the majority of these hit compounds reduced the aggregation of different Tau constructs in vitro and in N2a cells. These results demonstrate that Tau is a viable receptor of drug-like small molecules. The drug discovery approach that we present can be applied to other IDPs linked to other misfolding diseases such as Alzheimer’s and Parkinson’s diseases.
Keywords: Alzheimer’s Disease, drug discovery, high throughput screening, inhibitor, protein aggregation, Tau, Taupathies, thearapeutic
Introduction
Tauopathies [1] represent a subset of neurodegenerative disorders, including Alzheimer’s disease (AD) [2] and frontotemporal dementias (FTD) [1], which are linked to the accumulation of aberrant protein deposits known as neurofibrillary tangles (NFTs), which are formed by microtubule-associated protein Tau (MAPT) in patients’ brains. NFTs consist of paired helical filaments (PHFs) of Tau generated by the aggregation of monomeric Tau in neurons. Tau is an intrinsically disordered protein (IDP) with six main isoforms in the human CNS [3], the longest of which is of 441 residues. While no Tau mutations have been linked yet to the early onset of AD, familial mutations in the Tau gene on chromosome 17 (FTDP-17) have been identified that cause early-onset forms of FTD with Parkinsonism [4]. Under normal conditions Tau is known to stabilize microtubules within cells. In AD, however, Tau is hyperphosphorylated, which reduces its affinity towards microtubules [5]. This lack of binding to microtubules could be one of the reasons why Tau aggregates abnormally, initially in small oligomers and later in PHFs [6].
A potential strategy to alleviate the aggregation of proteins and their consequent toxicity linked to disease is to use small molecules to inhibit the aggregation process itself. This approach has been pursued for Tau [7–11] by applying high-throughput screens to detect the formation of protein fibrils. Some of these screening efforts resulted in Tau aggregation inhibitors, which have been shown to be neuroprotective [12–15]. However, a majority of hit compounds derived from such screens have been shown to inhibit fibril formation through nonspecific or reactive mechanisms [11; 12; 16; 17], a situation that impedes the application of traditional medicinal chemistry approaches to further optimize these compounds. Because NFT deposits have been reported to be less toxic compared to their oligomeric precursors [18–21], the detection and elimination of small oligomeric species should be the principal goal of new screening efforts [22].
To reduce potential toxic oligomer and fibril formation, a possible approach is to target the early phases of the aggregation process. This goal may be accomplished through small molecules that bind to the monomeric proteins and thereby reduce their misfolding and aggregation through stabilizing and promoting the functional native states of the proteins. This type of approach has been successful in the development of a small-molecule therapeutic for the globular folded protein transthyretin [23], whose misfolding is linked to systemic amyloidosis and related disorders [24]. This strategy could also work for inhibiting Tau aggregation, for instance because compounds could bind to and stabilize a specific conformational state of the monomeric ensemble of Tau, known as the paperclip conformation [25], a preferred global folding of Tau which brings the N-terminal and C-terminal domains into the vicinity of the repeat domain, and thus protects against pathological aggregation. The stabilization of the monomeric form by small molecules should reduce the rates of all the monomer-dependent processes during aggregation [26]. Therefore, delaying or stopping the initial steps of oligomer formation this way should lead to the reduction or elimination of toxic Tau oligomers. Such a therapeutic approach should be able to slow down the progression of Tauophathies and could also prove to be preventive. However, the structural heterogeneity of the conformational ensemble of Tau and other IDPs presents a major challenge [27]. The feasibility of binding of drug-like small molecules specifically to the monomeric forms of Tau and other IDPs has not yet been definitively established.
In this study, we pursue an alternative approach to those reported previously for identifying small molecule inhibitors of Tau [7–11]. First, we apply a high-throughput chemical microarray surface plasmon resonance imaging screen (HT-CM-SPR) [28; 34–36], which has the ability to detect the interaction between immobilized small molecules and monomeric full length Tau (hTau2N4Rwt), to probe whether the binding of fragments and/or lead-like compounds to Tau is feasible. In contrast to more common SPR approaches, in which the protein target itself is immobilized on the sensor surface, this “reverse SPR” scheme exploits the advantages of having large collections of compounds immobilized on chemical microarrays (Fig. 1). Because the surface chemistry applied for the chemical microarray allows controlling the orientation and density of the compounds precisely, the technique is applicable to the screening of a variety of biomolecules (including structured and intrinsically disorder proteins, and antibodies) with a variety of structures. This universal screening platform has been a powerful tool for the identification of small molecule binders to proteins providing valuable starting points for hit-to-lead optimization in various drug discovery projects [28; 34–36]. Next, we identified novel small molecule binders capable of modulating Tau aggregation in vitro and in N2a cells. Our overall goal is to identify drug-like small molecules that bind to monomeric Tau and can reduce its aggregation. By specifically targeting the monomeric state of Tau, we anticipate that it will be possible to identify small molecules that modulate Tau aggregation at the earliest phases of its fibrillization pathway.
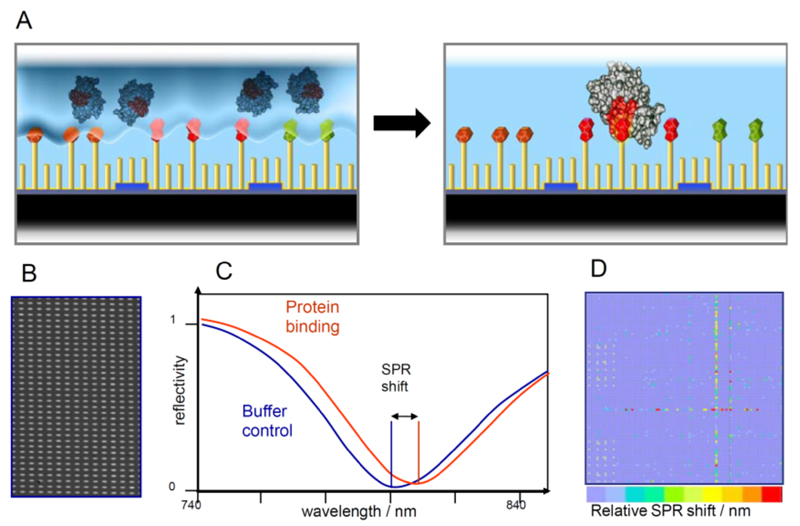
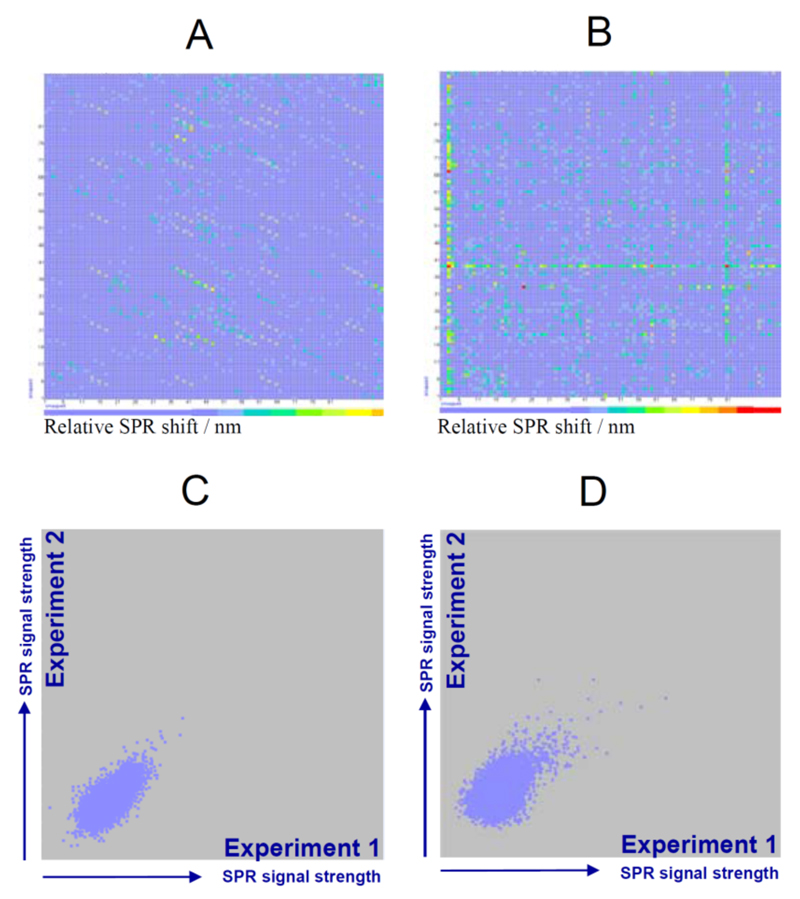
Fig. (1). The HT-CM-SPR scheme.
(A) The protein analyte is allowed to float over the array surface under controlled conditions to allow binding events to happen. SPR Imaging enables the detection of binding events: (B) Close-up of a grey scale picture obtained by CCD camera imaging of chemical microarray. (C) Grey scale analysis resulted in the parallel detection of 9,216 SPR minima per microarray exhibiting a shift in the resonance wavelength upon protein binding to the immobilized compounds. (D) Generic example of a color coded visualization of one array experiment.
Materials and Methods
Expression and purification of hTau2N4Rwt, Tau3RD and Tau4RDΔK280
The human Tau constructs were expressed in pNG2 vector, a derivative of pET-3a (Merck-Novagen, Darmstadt) in E.coli strain BL21(DE3) (Merck-Novagen, Darmstadt). The expressed protein was purified from bacterial extract by making use of the heat stability of Tau protein. The cell pellet was resuspended in the boiling-extraction buffer (50 mM MES, 500 mM NaCl, 1 mM MgCl2, 1 mM EGTA, 5 mM DTT, pH 6.8) complemented with protease inhibitor coctail (1 mM PMSF, 1 mM EDTA, 1 mM EGTA, 1 mM benzamidin, 1 µg/ml leupeptin, 1 µg/mL aprotinin, and 1 µg/ml pepstatin). The cells were disrupted with a French pressure cell and subsequently boiled for 20 min. The soluble extract was isolated by centrifugation, the supernatant was dialyzed against the cation exchange chromatography buffer A (20 mM MES, 50 mM NaCl, 1 mM EGTA, 1 mM MgCl2, 2 mM DTT, 0.1 mM PMSF, pH 6.8) and loaded on cation exchange SP-Sepharose column (GE Healthcare). The protein was eluted by a linear gradient of cation exchange chromatography buffer B (20 mM MES, 1 M NaCl, 1 mM EGTA, 1 mM MgCl2, 2 mM DTT, 0.1 mM PMSF, pH 6.8). The short Tau constructs representing Tau repeat domains Tau3RD (3 repeats; (M)Q244-S372 lacking V275-S305 => 99 residues) or Tau4RDDK280 (4 repeats; (M)Q244-E372 lacking K on position 280 => 129 residues) were subsequently concentrated and rebuffered on Amicon Ultra-15 device (Millipore, Bedford, MA).
The full length Tau isoform (hTau2N4Rwt) was in the first step similar purified on cation exchange SP-Sepharose column and in the next step the protein was loaded on size exclusion column Superdex G200 (GE Healthcare) and eluted with PBS Buffer (pH 7.4; 1 mM DTT), to remove Tau breakdown products.
Protein concentration was determined by the Bradford assay. The purity of the proteins was analyzed by SDS-PAGE (10% or 17%) and the monomeric state of hTau2N4Rwt was assessed by analysis of gel filtration column G200 fractions on SDS PAGE in non-reducing conditions.
Chemical Microarrays
The construction of such arrays and their use for primary screening in drug discovery was already described elsewhere [28; 29]. Briefly, chemical microarrays with footprints of 8.2 × 12.4 cm comprise 9,216 sensor fields each and were constructed by photolithographic methods. Microstructuring of glass plates and metal deposition techniques allowed generating arrays of sensor fields with diameters of 400µm covered with a thin gold film of defined thickness. The latter enables the optical read-out of the chips by SPR imaging and also facilitated a chip surface modification based on binary self-assembled monolayers (SAM) exhibiting maleimide moieties (anchors) at a controlled density. All 110,000 library compounds, the synthesis and QC of which was described before [30], were coupled to a flexible, long, hydrophilic thiol-linker, called “chemtag”, the structure of which was defined previously [30]. Upon pintool spotting, the linker-compounds constructs were allowed to react covalently with the maleimide moieties on the array surface to yield a surface architecture comprising the glass/gold/SAM surface/covalently attached chemtag and the immobilized ligands. The surface architecture resulting from coupling the chemtag-compounds conjugate to the chemically modified chips surface was also described previously elsewhere [28; 29]. The optimized surface chemistry was designed to be resistant to unspecific protein binding exhibiting only marginal background in the SPR screening experiments.
SPR Imaging of Chemical Microarrays
The microarrays were read out by SPR using Graffinity’s in-house developed SPR Imager®. The optical set-up in the instrument allowed illuminating the entire chip area with parallel light of defined incidence angle and wavelength via a high refractive index prism in a Kretschmann configuration. Reflection images of the chip were recorded by means of a cooled, low-noise CCD camera. While keeping the incidence angle of the incoming beam fixed, the wavelength was varied over a certain range covering the SPR resonance conditions for the given chip/prism combination. Recorded array images (close up in Fig. 1C) were deconvoluted by automatic spot finding routines and grey scale analysis. Plotting the reflectivity of the individual sensor areas versus the applied wavelength yielded 9,216 SPR minima (Fig. 1D) resulting from the excitation of surface plasmons. Binding of analytes (e.g. folded or unfolded proteins) to the immobilized library compounds altered the optical resonance conditions for the corresponding sensor fields. Such differences to analyte-free buffer were picked up by monitoring (red) shifts of the wavelength dependent SPR minima with time. Additionally, bulk refractive index changes upon buffer exchange were taken into account by control spots distributed across the array. Typical incubation times of analytes on the arrays ranged from 15 minutes to 3 hours during which scans were recorded repeatedly. SPR shifts were plotted in color coded plots (Fig. 1D). Software routines guided the hit selection process on the array level by fitting Gauss-like functions to the SPR signal distribution and suggesting hit thresholds per array allowing to remove non-hits. A great deal of knowledge about the interaction of library components tethered on the chip with other proteins is has been gathered from prior screens. Therefore, compounds showing promiscuous interactions (frequent binders identified to have hit > 50% of screened targets) were excluded from initial hits to extract compounds with possibly higher target specificity.
SPR Screening of hTau2N4Rwt
For the screening set-up, the binding behavior of the intrinsically disorder protein hTau2N4Rwt was evaluated on a set of 96 control ligands of defined physicochemical properties in order to optimize parameters such as buffer composition, surface density of ligands and protein concentration. A minimum of unspecific binding, low background and acceptable signal/noise ratio was obtained at a final screening concentration of 400 nM hTau2N4Rwt in 50 mM Hepes at pH 7.4, with 50 mM NaCl, 1 mM DTT and 0.01% Tween.
Measurement of absorption spectra of hit compounds
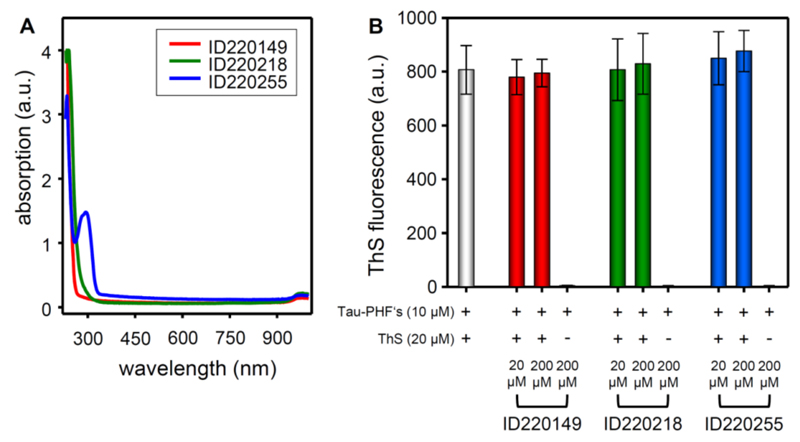
To examine possible optical interference by the compounds with the fluorescence of ThS absorption scans were performed. 200 µM compound in PBS-buffer plus 2% DMSO were scanned in a 384 well plate (flat bottom transparent, Greiner, Frickenhausen) in the range of 230 to 1000 nm using a Safire spectrometer (Tecan, Männerdorf, Switzerland). For three selected compounds the results are shown in Fig. 7A.
Fig. (7).
(A) Absorption properties of selected compounds. 200 µM of the compound (in PBS + 2% DMSO) were sanned over a wavelength range of 230 to 1000 nm and plotted against the absorption value. (B) Staining of pre-formed Tau fibrils with Thioflavin S (ThS) in combination with selected compounds or with compound alone. The y-axis represents the amount of aggregated Tau protein monitored by ThS-fluorescence.
Thioflavin-S in vitro assays of Tau aggregation and inhibition
ThS-based assays of Tau-aggregation [9; 31; 32] were also performed in different buffer systems in order to optimize aggregation properties [33] of hTau2N4Rwt (50 µM) or Tau4RDΔK280 (10 µM) in 20 mM BES-buffer (pH 7.4, 25 mM NaCl) plus heparin3000 (12.5 µM) – in case of Tau4RDDK280 without heparin – and DTT (1 mM) were heated up at 98 °C for 20 min. After cooling down to room temperature a protease-inhibitor-mix (1 mM benzamidin, 1 µg/ml leupeptin, 1 µg/mL aprotinin, and 1 µg/ml pepstatin) and the compound to be tested (300 µM for hTau2N4Rwt, 50 µM in case of Tau4RDΔK280; molar ratio of compound to protein = 6:1 for hTau2N4Rwt and 5:1 for Tau4RDΔK280) were added and the sample was incubated for 10 days at 37°C, with addition of 1 mM DTT every day. The samples contained DMSO at a final concentration of 2% resulting from the solubilization of the compounds. Before testing the sample was diluted to 10 µM Tau concentration in PBS buffer, and 20 µM ThS was added (molar ratio of protein to ThS = 1:2) and incubated for 30 min at room temperature. The measurement was performed at excitation and emission wavelengths of 440 nm and 521 nm, respectively.
Construct Tau3RD (3R repeat domain of Tau, 10 µM) incubated in 50 mM NaAc (pH 6.8) plus heparin3000 (2.5 µM) and 60 µM compound (molar ratio of compound to protein = 6:1) were mixed and the sample incubated over night (16 h) at 37 °C. Before testing 20 µM of Thioflavin-S was added to the sample (ratio of protein to ThS = 1:2) and incubated for 30 min at room temperature. The measurement was performed at excitation wavelength of 440 nm and emission wavelength of 521 nm.
For three selected compounds the results are shown in Fig. 3A and Table 4.
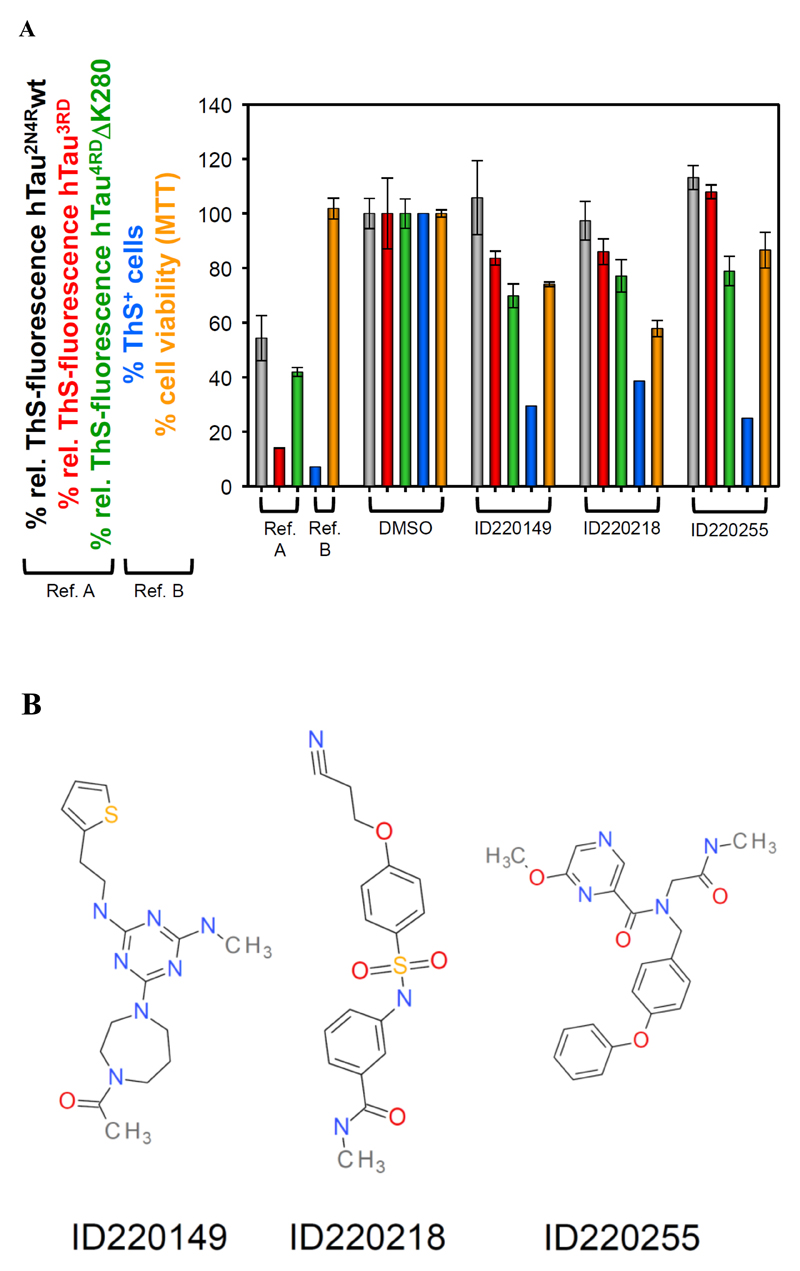
Fig. (3).
(A) Thioflavin S fluorescence as indicator for the extent of Tau aggregation after incubation with compounds. Bar colors: Grey hTau2N4Rwt; red Tau3RD; green Tau4RDDK280; blue ThS+ N2a cells (expressing Tau4RDDK280 with aggregates); orange viability of N2a cells (MTT assay). Reference results are shown for earlier studies with different classes of compounds: in vitro studies with bb14 [40],, cellular studies with BSc3094 [41]. The in vitro ThS-signal of the soluble Tau protein was subtracted from all samples. (B) Examples of chemical structures of Tau binding compounds identified by HT-CM-SPR screen.
Table 4. Summary of the effects of selected synthesized hits on Tau aggregation in vitro and in N2a cells.
(Column 5 – 7) The relative ThS-fluorescence was used to indicate the degree of tau fibril formation after compound treatment. The fluorescence of the compound untreated control (“DMSO - Dox”) was normalized to 100% (note: 2% DMSO were added to the controls to ensure comparable conditions to the compound treated samples). (Column 8) The amount of ThS+ cells (cells with tau aggregates stained by ThS and quantified by FACS) in the induced compound-untreated sample (“DMSO + Dox”) was normalized to 100%. (Column 9) The cell viability (monitored by MTT-assay) of the uninduced compound-untreated sample (entry #1, right column) was normalized to 100%. MTT-values reflect the viability of the N2a cells in the presence of the compounds without expression of tau protein. (Column 10) Concentrations at which synthesized hits were soluble and no compound aggregates were detected by DLS.
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
|---|---|---|---|---|---|---|---|---|---|
| Hit ID | Hit Series | MW | Affinity HT-CM-SPRa | hTau2N4Rwtb | Tau3RD c | Tau 4RD ΔK280d | Tau4RD ΔK280 N2a cellse | viability N2a cells (MTT) | solubility (µM) |
| bb14[34] | 452.0 | 54.3 ± 8.3 | 14.0 ± 0.1 | 42.0 ± 1.6 | |||||
| BSc3094[35] | 380.4 | 7.2 | 101.9 ± 3.8 | ||||||
| DMSO (- Dox) | 100 ± 5.5 | 100 ± 13.0 | 100 ± 5.4 | 0.1 | 100 ± 1.3 | ||||
| DMSO (+ Dox) | 100 | ||||||||
| ID220149 | 7 | 375.6 | medium | 105.8±13.6 | 83.6± 2.6 | 69.8 ± 4.3 | 29.6 | 74.1 ± 0.8 | 300.0 |
| ID220218 | 3 | 359.4 | weak | 97.4± 7.1 | 86.0± 4.7 | 77.1 ± 5.9 | 38.6 | 57.8 ± 3.0 | 300.0 |
| ID220255 | 12 | 406.4 | weak | 113.2 ± 4.4 | 107.9± 2.5 | 78.9 ± 5.4 | 25.0 | 86.6 ±6.5 | 150.0 |
aRelative affinity of immobilized hits to hTau2N4Rwt detected by SPR in the HT-CM-SPR. Percentage of aggregated hTau2N4Rwtb, Tau3RD c and Tau4RDΔK280d detected by ThS fluorescence in the presence of hit compound compared to control (100%).
Test of displacement of ThS from Tau fibrils by compounds
10 µM Tau3RD in 50 mM NH4Ac with 2.5 µM heparin3000 was incubated for 3 days at 37°C. After incubation the Tau aggregates were stained with (a) 20 µM ThS plus 20 µM compound, (b) 20 µM ThS plus 200 µM compound, or (c) 200 µM compound alone. After incubation for 1h at room temperature the fluorescence was measured at 440 nm/521 nm (ex/em). For three selected compounds the results are shown in Fig. 7B and Table 4.
Inhibition of Tau aggregation in N2a cells measured by FACS
Cell suspensions of inducible N2aTau4RDΔK280 cells were distributed on 24-well pates (1 ml per well) and 0.0005% (12 µM) Thioflavin-S was added to each sample. For induction of Tau expression 1 µg/ml doxycyclin was added, except for the uninduced negative control. 60 µM compounds were added, except for the untreated positive control (all compounds were tested at 60 µM concentration). An additional assay was performed with some selected compounds in the concentration range of 0 to 60 µM (Fig. 6B-D; Table 4). The cells were incubated for 4 days at 37 °C. Floating and adherent cells were combined, pelleted (5 min, 295 g) and washed once with PBS. Cells were then counted in a BD FACSCantoTMII Flow Cytometry System by the fluorescent signal intensity in the FITC channel (Excitation: 495 nm, Emission: 519 nm).
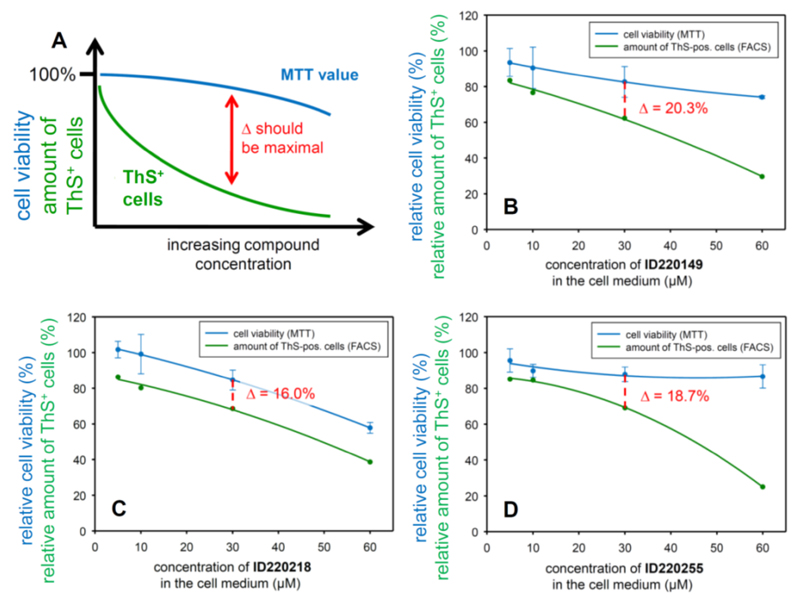
Fig. (6). Comparison of N2a cell viability (MTT assay) with the amount of ThS+ cells (FACS) at increasing compound concentrations.
Promising compounds should show a relatively high stable cell viability value over a broad concentration range whereas the amount of ThS+ cells should drop already at low compound concentrations (A). By this criterion, compound ID220149 (B) is the best inhibitor in this series wereas ID220218 (C) and ID220255 (D) show less effectivity and drug-like performance. Note: The MTT value in the untreated sample (without compound) is defined as 100% cell viability. The amount of ThS+ cells in the Tau-induced control is normalized to 100%.
Cell viability in the presence of compounds
The cell viability of N2a cells in the presence of compounds was monitored by an MTT-assay. Uninduced N2a cells were incubated in the presence of 60 µM compound (all compounds) or at several compound concentrations (0 to 60 µM; Fig. 6B-D; Table 4) in the cell culture medium over 4 days at 37°C. The MTT-assay was performed according to the manufacturer's instructions (Roche, Cell Proliferation Kit, Cat.No. 11465007001).
Dynamic Light Scattering (DLS) analyzes of synthesized hits
Representative size distributions of the 70 synthesized hits in DMSO, PBS and 1 mM DTT at pH 7.4 at 300 mM, 150 mM, 75 mM, 37.5 mM, 18.75 mM and 9.375 mM concentrations were recorded at 25 ºC on DynaPro Plate reader (Wyatt) and analyzed using Dynamics software from the same vendor from 300 µM in 0.5% DMSO. Buffers and samples were filtered using a 22 micron filter prior to analyses. A 45 µl sample of the compound solution was added to the well of a 384 well plate (in duplicate).
Resynthesis of Library Compounds
In general synthesis of compounds for biochemical testing was done using a linker replacement strategy described in Neumann et al. [28]. In all cases, the linker present in the original screening hits was replaced by a methyl group.
Results
Identification of small molecule binders of full length Tau (hTau2N4Rwt) by HT-CM-SPR
Monomeric hTau2N4Rwt (Fig. S1) was screened against an in-house library of small molecules at Graffinity Pharmaceuticals (Graffinity library at the time of screening: 110,000 drug-like compounds, which include 23,000 fragment-like compounds with a molecular weight (MW) less than 300Da) immobilized on microarrays using HT-CM-SPR.
To identify optimal screening conditions for HT-CM-SPR of full-length Tau (hTau2N4Rwt), the binding behavior of hTau2N4Rwt to a set of 96 immobilized control compounds (reference array) with diverse physicochemical and pharmacological properties (frequent binders, target class specific binders, positive and negative controls) was evaluated with various buffer compositions, surface densities of compounds and protein concentrations. Overall, these experiments resulted in identifying a screening buffer containing 50 mM Hepes at pH 7.4, 50 mM NaCl, 1 mM DTT, 0.01% Tween, and the use of an optimized surface density of immobilized ligands. Screening was done at room temperature by incubating the chemical microarrays with 400 nM hTau2N4Rwt for 3 hours. To probe if hTau2N4Rwt had a tendency to aggregate over time under the screening conditions applied, hTau2N4Rwt samples were incubated for 3 hours in the screening buffer and samples from it were taken in every hour. Potentially formed pellets from these samples were resuspended and analyzed using SDS PAGE. Precipitation was not observed and monomeric hTau2N4Rwt was detected throughout the time course of a typical array experiment of 3 hours. By using the final screening conditions minimal nonspecific binding to hTau2N4Rwt, a low background and acceptable signal/noise ratio were observed. All these observations suggest that the detected SPR signal in the screen reflects the interactions between monomeric hTau2N4Rwt and immobilized compounds.
Monomeric hTau2N4Rwt was screened against the Graffinity library using the in-house developed Graffinity SPR Imagers, which allowed recording the SPR minima of four chemical arrays in parallel with 9,216 sensor fields each. For manual analysis of the array data, SPR signals were visualized in coloured two-dimensional fingerprints with the colour representing the intensity of SPR shift upon incubation with protein. To demonstrate this process, Fig. 2 shows representative examples of hTau2N4Rwt SPR fingerprints in an array view. For fragment libraries compounds were spotted in triplicates, hence hits for the fragment library appear as triplicate spots on a diagonal (Fig. 2A). In the case of lead-like libraries derived from combinatorial synthesis approaches, each row and column consists of a common molecular moiety in combination with 96 other diverse analouges (Fig. 2B), thus hit series identified for hTau2N4Rwt appear as line pattern (Fig. 2B). The plots on the right in figure 2A and B compare the SPR signals of two independent repetition experiments (different operator, reader, date, same hTau2N4Rwt batch). Multiplicate SPR signals were averaged for each immobilized array compound (e.g. fragments signals: 2 experiments x 3 spots = hexaplicate signals). A filtering step removed potential hits for which the standard deviation of the SPR signal was higher than 0.8 nm (see Fig. 1C). The threshold for selection of Tau-binding compounds was defined as the SPR minimum that was separated from the background noise by at least 1 nm and for which the reproducibility criteria was satisfied. In addition, immobilized compounds that had low purity before spotting to the surface of the array and had low saturation per spot (< 80%) were disregarded.
Fig. (2). SPR Screening Results.
Example of colour coded SPR signals obtained by chemical microarray screening of hTau2N4Rwt under optimized conditions against: (A) a fragment library of 3,000 fragments (out of 25,000 fragments present in the small molecule library screened) spotted in triplicates, and (B) a lead-like library combining (96×96 =9,216) motifs. Reproducibility of screening experiments for chemical microarray with (C) fragments and for (D) lead-like compounds immobilized.
Generally, the overall data quality, signal strength and hit rate were lower compared to previously screened globular proteins. For previous screens of globular proteins of similar molecular weight compared to hTau2N4Rwt, hit signals were clearly separated from the background and signal to noise ratios of > 10 were regularly obtained, while for hTau2N4Rwt the array signals were rather small with S/N ratios ranging between 1.5-2.0.
Analysis of the screening results by computational methods and by visual inspection of the array data identified 834 immobilized compounds as hits, which conveys a hit rate of approximately 0.76% for the screen. Hits were distributed over various compound classes and a large number of structurally independent singletons were also identified. Out of this initial set of 834 screening hits, we selected 118 representative hits based on parameters such as physicochemical properties, molecular weight, SPR signal strength, structural diversity, chemical tractability and overall drug-like attractiveness. None of these hit compounds have been reported previously to bind to Tau or affect the aggregation of the protein. These hits were arranged into ten different molecular scaffold classes based on their common substructures and prevalent physicochemical features using cheminformatic analyses. 41 structurally unrelated lead-like and 26 fragment hits were also identified (Table 1). The different molecular scaffold classes had a high degree of structural diversity with a high number of negatively charged compounds. The ranges of some molecular properties (such as MW, clogP, H-bond donors and acceptors, ring count) for the above mentioned 118 representative array-linked hits are summarized in Tables 2 and 3.
Table 1.
Summary of hit molecular scaffolds identified by the HT-CM-SPR of hTau2N4Rwt.
| Series | Series Eponym | Number of hits | Affinity in HT-CM-SPRa |
|---|---|---|---|
| 1 | phenoxy-acetamides | 10 | weak |
| 2 | benzimidazoles | 6 | weak |
| 3 | sulfonamides | 6 | weak |
| 4 | ureas | 5 | weak |
| 5 | carboxylic acids | 7 | medium |
| 6 | tetrazoles | 3 | weak |
| 7 | triazines | 9 | medium |
| 8 | mesyl-phenyl-oxo-butyramides | 4 | weak |
| 9 | cyano-benzamides | 4 | weak |
| 10 | piperidinyl-methylfuran-carboxamides | 4 | weak |
| 11 | structurally unrelated fragments | 19 | weak |
| 12 | structurally unrelated lead-like compounds | 41 | weak-medium |
Average relative affinity of immobilized hits to hTau2N4Rwt detected by SPR in the HT-CM-SPR
Table 2.
Molecular weight distribution of hits identified by the HT-CM-SPR of hTau2N4Rwt.
| Molecular Weigh Distribution | Number of hits |
|---|---|
| fragments < 275 Da | 13 |
| 275 Da < fragments < 300 Da | 13 |
| 300 Da < lead-like < 350 Da | 37 |
| 300 Da < lead-like < 400 Da | 31 |
| 400 Da < lead-like | 24 |
Table 3.
Ranges of selected physicochemical properties for the 118 hits identified by the HTCM-SPR of full length hTau2N4Rwt as well as for the 70 resynthesized compounds.
| 118 linked array hits |
70 compounds resynthesized |
|||||||
|---|---|---|---|---|---|---|---|---|
| Min | Mean | Median | Max | Min | Mean | Median | Max | |
| MW | 209.2 | 350.8 | 344.9 | 539.6 | 223.2 | 346.8 | 344.4 | 528.5 |
| ClogP | -2.32 | 1.76 | 2.07 | 4.21 | -0.87 | 1.69 | 1.89 | 4.21 |
| #ACCEPTORS | 2 | 4.3 | 4 | 9 | 2 | 3.9 | 4 | 6 |
| #DONORS | 1 | 2.9 | 3 | 8 | 1 | 2.6 | 3 | 6 |
| #ROTBONDS | 2 | 6.1 | 6 | 12 | 2 | 5.6 | 6 | 11 |
| #RINGCOUNT | 1 | 2.7 | 3 | 4 | 1 | 2.7 | 3 | 4 |
| HAC | 15 | 24.9 | 25 | 38 | 16 | 24.8 | 25 | 38 |
| TPSA | 35.6 | 95.7 | 89.8 | 200.9 | 35.6 | 87.5 | 81.3 | 157.9 |
For immobilization to the chip surface all library compounds were tethered to the chip via a linker group R. In order to calculate the ClogP, H-bond acceptor count, H-bond donor count, the count of rotatable bonds and topological polar surface area for the 118 array hits, this R-group was virtually replaced by a carbon. For the calculation of the molecular weight (MW) and Heavy Atom Count (HAC) the R group was replaced by hydrogen. In case of the 70 resynthesized compounds the linker group R at the attachment point at the compound was synthetically replaced by other groups such as methyl groups, which are included in the calculation of all properties. The following software was used: Hivol for the calculation of the counts, Biobyte for the calculation of ClogP and Openbabel v. 2.3.1 for the calculation of total polar surface area (TPSA).
SPR signal intensities were categorized into weak, medium and strong, which reflect the relative binding affinities of the immobilized compounds to the protein. Overall, most hits were detected to bind weakly to the protein (approximately Kd > 100 µM), with only 19 hits binding with medium affinity (approximately Kd < 100 µM). Many of the hits that bound with medium affinity were charged, 7 of the hits have a total net charge of -1 while 4 have a total net charge of +1.
Hits were selected for synthesis on the basis of their binding ability to hTau2N4Rwt and on the basis of covering the full chemical space of the identified hits from the screen. 70 hits were synthesized devoid of the chemtag linker moiety, which was used to tether the compound to the SPR chip and replaced with appropriate atoms/groups such as methyl groups. The properties of these selected compounds can also be found in Table 3.
Tau binding compounds can affect the aggregation of Tau in vitro
Our next interest was to elucidate whether the variety of small molecules, which we identified to bind to hTau2N4Rwt, could modulate its aggregation. To explore whether the synthesized hits can influence the aggregation propensity of Tau in vitro, we ran hTau2N4Rwt, Tau3RD (three-repeat-domain; see Materials and Methods) (with heparin), and Tau4RDΔK280 (four-repeat-domain construct with pro-aggregant deletion of K280 [37]; see Materials and Methods) (without heparin) aggregation assays in the presence of hit compounds (molar ratio of protein to compound 1:6 for hTau2N4Rwt, Tau3RD and 1:5 for Tau4RDΔK280) using Thioflavin S (ThS) to detect fibril formation of the proteins.
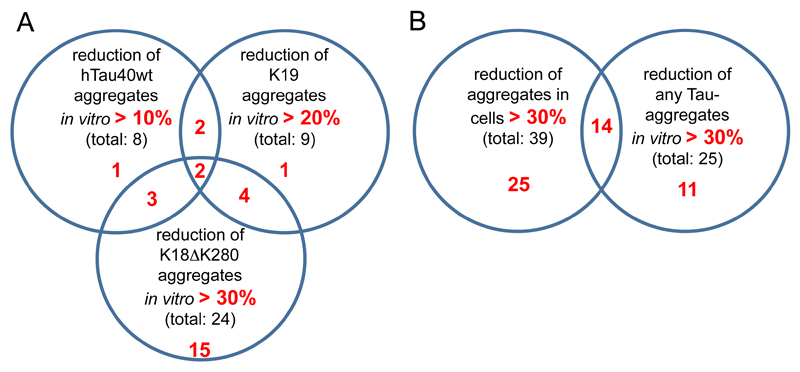
A number of the synthesized hits were able to inhibit fibril formation of Tau4RDΔK280 (example illustrated by electron microscopy in Figure S2), Tau3RD and hTau2N4Rwt. In the case of hTau2N4Rwt aggregation assays, only about 15% of the synthesized hits had some inhibitory effect on the aggregation of hTau2N4Rwt (reduction of aggregation by ~10-14% compared to control), while control compound bb14 inhibited aggregation by ~46%. In the case of the Tau3RD aggregation assays, over 20% of the synthesized hits had a significant effect in reducing the aggregation of Tau3RD (up to ~36% compared to control), while control compound bb14 inhibited aggregation by ~84%. Lastly, in the case of the Tau4RDΔK280 aggregation assays, over 35% of the synthesized hits had a significant effect in reducing the aggregation of Tau4RDΔK280 (up to ~54% compared to control), while reference compound bb14 inhibited aggregation by ~58%. In summary, the synthesized hits reduced fibril formation of the tested Tau constructs by different extents (Fig. 4). 6 compounds significantly reduced aggregation of Tau3RD and of Tau4RDΔK280, 5 compounds significantly reduced aggregation of hTau2N4Rwt and of Tau4RDΔK280, and 4 compounds significantly reduced aggregation of hTau2N4Rwt and of Tau3RD. Only 2 compounds had a significant effect in reducing the aggregation of all Tau constructs tested in vitro.
Fig. (4).
Graphical representation of the effect of hit compounds on the aggregation of different Tau constructs in vitro and in N2a cells. No significant correlation was observed in the ability of hit compounds to reduce aggregation of the different Tau constructs (A) in vitro and (B) Tau4RD∆K280 in N2a cells. Nevertheless, several hit compounds had significant aggregation inhibition effect in both cellular and the in vitro aggregation assays.
A few of the compounds promoted a strong increase in the ThS-fluorescence signal, up to ~8-25 fold compared to the control. This increase does not reflect a higher extent of Tau aggregation (as ascertained by pelleting and SDS-PAGE), but rather an interference between the compound and ThS which changes the fluorescence properties (note that, depending on isoform and conditions, ~40-80% of the protein aggregates, so that any aggregation-promoting compound could at best enhance the extent of aggregation by up to ~2.5-fold). Analogous non-linear increases in ThS fluorescence have been observed in other conditions, for example during Tau aggregation in presence of arachidonic acid near the critical micellar concentration[38; 39]. We therefore conclude that these compounds are not Tau aggregation promoters.
Specific selected examples of compounds, ID220149, ID220218 and ID220255 (see Fig. 3B), which reduced Tau fibril formation in ThS aggregation assays are shown in Fig. 3A and Table 4, representing three distinct compound series from among the Tau binders identified in the HT-CM-SPR screen. The relative ThS fluorescence in the ThS aggregation assays was measured as degree of Tau fibril formation, which was already confirmed by other methods (filter- and pelleting assay [10]). For comparison, the Tau aggregation inhibitor bb14 (from the rhodanine class of compounds described previously [40]) was tested as well as reference value.
A majority of Tau binding compounds affect the aggregation of Tau4RDΔK280 in regulatable N2a cells
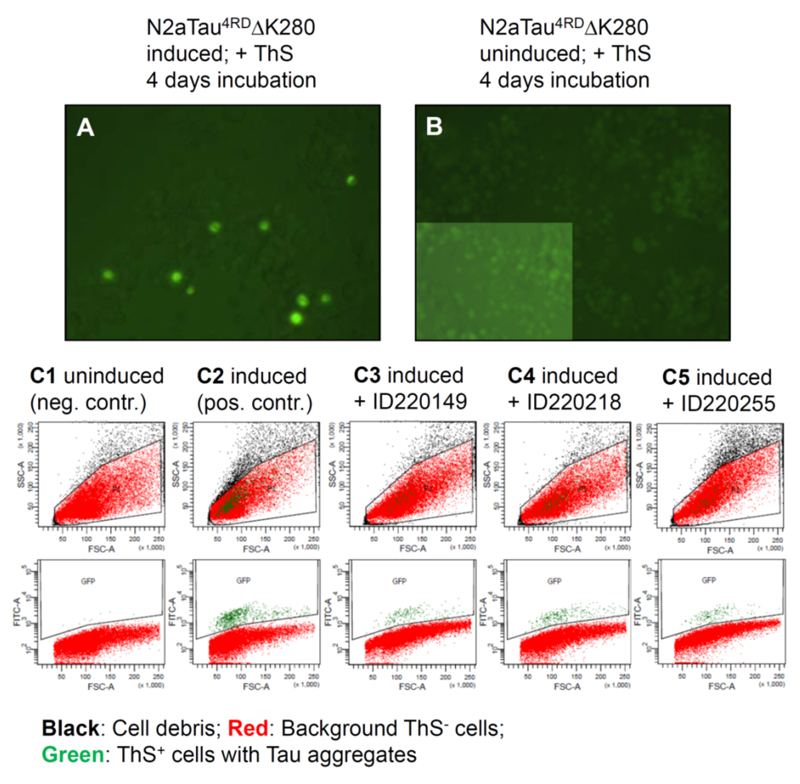
Next, we tested the synthesized hits for their ability to inhibit Tau aggregation in an inducible N2a cell model, which expresses Tau construct Tau4RDΔK280. N2a cells with Tau4RDΔK280 aggregates were stained by ThS and visualized by fluorescence microscopy (Fig. 5A). This allows discrimination of cells with aggregated Tau from cells with soluble Tau (Fig. 5A, background) or cells without Tau expression (Fig. 5B, highlighted inset demonstrates the presence of cells). Fig. 5 (C1-C5) shows examples of the quantification of Tau aggregation in uninduced N2a cells (column C1), induced cells (column C2) and 60 µM compound treated cells (column C3 – C5) by FACS analysis. In parallel the cell viability was measured at the same compound concentrations by MTT assay. The extent of Tau4RDΔK280 aggregation and cell viability in the compound-free control is normalized to 100% (results summarized in Table 4). Notably, at least two thirds of the hits had a substantial effect in reducing Tau4RDΔK280 aggregation. The most potent half dozen compounds were between 70-84% compared to control, among which were compounds, ID220149, ID220218 and ID220255 shown in Fig. 3 and Table 4, representing three distinct compound series from among the Tau binders identified in the HT-CM-SPR screen. As a reference for aggregation in cells, the Tau aggregation inhibitor BSc3094 (from the phenylthiazolyl-hydrazide class [41]) was tested as well, which reduced aggregation of Tau4RDΔK280 by 93%.
Fig. (5). N2a cells with Tau aggregates stained by Thioflavin S.
Fluorescence microscopy shows a representative section of the N2a cell population after 4 days of Tau expression. Cells bearing higher Tau aggregates appear bright green (A) whereas cells with soluble or oligomeric Tau show only background staining (A, green background), similar to cells without any Tau expression (B). (C1 – C5) shows examples of cell distributions by FACS counting after 4 days of incubation, without Tau expression (C1, negative control), with Tau expression (C2, positive control), and with Tau expression in the presence of Tau-aggregation inhibitors ID220149 (C3), ID220218 (C4) and ID220255 (C5). The cell signals were plotted with regard to cell morphology (upper panels: FSC (forward scatter indicating the particle size) vs. SSC (side scatter indicating the particle granularity), and with regard to the amount of cells with Tau aggregates (lower panels: FSC vs. fluorescence intensity measured in the FITC channel). Cell debris would appear as small particles (small FSC values) with high density (high SSC values). Note that the green ThS+ cells are well separated from the cell debris population shown in black. The reduction of aggregate-bearing cells by the inhibitor is apparent from the lower panels.
The rhodanine derivative bb14 showed in the cellular assays an increasing fluorescence staining of the whole cell population in the FITC channel when excitated at 495 nm (Fig. S3C). Because of this phenomenon we used compound BSc3094 as reference for the cell experiments (FACS and MTT-assay) where this effect does not occur (Fig. S3D). None of the novel compounds showed such a fluorescence-shifting effect in the cellular system (for example see Fig. 5C3 – C5).
Selected hit compounds inhibit Tau aggregation in a dose dependent manner in N2a cells inducible to over-express Tau4RDΔK280
We selected three representative synthesized hits, ID220149, ID220218 and ID220255, due to their superior ability to reduce aggregation of the different constructs of Tau in vitro and in N2a cells and for their CNS drug-like physico-chemical properties, and tested them for their ability to inhibit Tau aggregation and cell viability in the inducible N2aTau4RDΔK280 cell line [37] at increasing concentrations (0, 5, 10, 30 and 60 µM). The aggregate load in the N2a cells was quantified via ThS+ cells counted by FACS and the cell viability was measured by MTT-assay (Fig. 6).
An optimal aggregation inhibitor compound should have low cytotoxicity (high MTT value) over a wide concentration range, with a decrease in cell viability only at high concentrations. By contrast, the amount of ThS+ cells should decrease with increasing compound concentrations, indicating that the compound inhibits Tau aggregation effectively in a dose responsive manner. In this latter case the separation between the MTT- and ThS+ cell-curves should become maximal (Fig. 6A).
Overall, all three hits tested in N2a cells showed a dose-dependent inhibition of Tau4RDΔK280 aggregation. Compound ID220149 (Fig. 6B) causes a moderate decrease in cell viability at higher concentrations tested but shows a relatively strong decrease in the amount of ThS+ cells at lower tested concentrations. Since the aim is to maximize cell viability and minimize the amount of ThS+ cells; one can best judge the quality of these parameters at the concentration of 30 µM. In this respect ID220149 showed the best dose-response effect on the N2a cells among the presented compounds. Treatment with ID220218 (Fig. 6C) illustrates a case where the curves of the MTT-value and ThS+ cells decrease in a parallel manner. This suggests that the reduction of ThS+ cells could be caused by the increasing cytotoxicity at higher compound concentrations. By contrast, ID220255 (Fig. 6D) shows a stable value of cell viability over the entire concentration range but a reduction of the amount of ThS+ cells only at high compound concentration. This could be explained by some metabolic instability of the compound or an insufficient ability to pass the cell membrane.
Cell active Tau aggregation inhibitors do not compete with ThS for binding to Tau fibrils
To examine possible optical interference by the compounds with the fluorescence of ThS (excitation and emission of ThS that is bound to aggregated Tau), we measured absorption scans in the range of 230 to 1000 nm (Fig. 7A). Almost no absorption by the hits was observed, especially at the wavelengths of 440 nm and 521 nm, which suggest that the compounds do not interfere with the fluorescence of ThS.
It is plausible that some of our Tau binding compounds may bind to Tau fibrils. Thus, they could compete with ThS for binding to Tau fibrils, which could potentially lead to the dislocation of ThS from Tau aggregates, leading to a reduced fluorescence signal and therefore cause false-negative results. (Note that ThS bound to Tau fibrils containing beta structure leads to a change in its fluorescence emission spectrum when excited at 440 nm. The increase at 521 nm serves as an indicator of Tau aggregation.) To test this, we performed co-staining experiments of Tau3RD fibrils with ThS and compounds ID220149, IS220218 and ID220255 at molar ratios of 1:1 and 1:10 (ThS:compound). If some of these compounds were able to displace ThS from the protein one would expect a reduced fluorescence signal compared with the control of ThS alone (Fig. 7B). However, compared with the compound-untreated control (Fig. 7B, entry #1) there is no significant difference in the fluorescence signal when treating with ThS plus compound or with ThS alone. This indicates that the compounds are not able to displace ThS from Tau fibrils.
Characterization of the self-aggregation ability of synthesized hits by DLS
The mechanism of protein aggregation modulation by a compound may occur by the self-assembly of the compound into a multimer/oligomer particle and consequent interactions with monomeric or oligomeric forms of the aggregating protein [16; 17]. This is a probable mechanism of many compounds inhibiting amyloid formation for a variety of proteins [16]. To investigate this possibility, the oligomerization properties of all 70 synthesized hits were investigated by measuring the hydrodynamic radii of the compounds by dynamic light scattering (DLS) under similar sample conditions to those used in the aggregation assays (Table 4, column 10). These DLS experiments showed, for example, that compounds that inhibited Tau aggregation in the N2a cells in a dose dependent manner (ID220149, ID220218, ID220255) are soluble and do not form detectable compound aggregates in the assayed conditions. These results suggest that these synthesized hits function in the aggregation assays as monomers.
Discussion
We have pursued an SPR array screening approach to identify novel fragment and lead-like small molecules capable of binding hTau2N4Rwt and elucidated the ability of the binders to modulate the aggregation of Tau3RD and Tau4RDΔK280 in vitro and in N2a cells. Our experiments resulted in the identification of dozens of novel compounds that inhibit the aggregation of distinct Tau constructs in vitro and in cells. These results strongly provide initial support for our drug discovery approach to develop small molecules that can bind to Tau in its intrinsically disordered state, and protect it from aggregating.
A prominent challenge is the identification of small molecule binders of intrinsically disordered proteins, because these proteins lack stable tertiary structure. Moreover, an additional complication is represented by the limited availability of high-throughput screening techniques with the ability to detect direct binding of small molecules to label-free and solution-free biomolecule receptors. In fact, there is no other high-throughput screening technique available yet, other than HT-CM-SPR, which has these capabilities and is capable of performing under near physiological conditions and concentrations of Tau. A further advantage of this optical label-free technique for our studies is its high sensitivity to detect weak intermolecular interactions. This results from the fact that high molecular weight protein analytes, such as hTau2N4Rwt, induce large changes in refractive index when binding to the immobilized compounds, and hence effectively increase the sensitivity and limit of detection of the method (Fig. 1B-D).
Compounds identified in the HT-CM-SPR screen that interact with hTau2N4Rwt may be binding to distinct locations and regions of Tau. Further studies will be required to established whether those compounds that bind to the four-repeat region of Tau, which is responsible for formation of the core structure of Tau fibrils [42], are most likely to have a role in modulating Tau aggregation. Also, compounds that bind to other parts of Tau, where they may be involved in stabilizing transient tertiary interactions that support the overall “paperclip” structure of Tau [25], may also affect the aggregation propensity of Tau. Stabilization of the Tau paperclip structure could reduce misfolding of Tau, for example, by protecting the domain of the repeats from intermolecular interactions [25]. Likewise, these compounds may simply support the native disordered ensemble of Tau and protect it from misfolding. Therefore, the locations of the binding sites of the compounds can have a pronounced effect on Tau structural stability and misfolding. This idea is supported by the data that some binders reduced the aggregation of distinct constructs of Tau to different to different extent, while others did not show any observable effect.
It was not clear whether the weak binding of the compounds to hTau2N4Rwt in the HT-CM-SPR assay would have enough impact to interfere with the protein-protein interaction involved in the aggregation process. For example, a weak-binding compound may be displaced by another Tau monomer, by cofactors such as heparin or other polyanions. Nevertheless, the Tau binding hits were able to reduce fibril formation of Tau constructs to different extents. They reduced the fibril formation of Tau4RDΔK280 the most, then of Tau3RD and the least of hTau2N4Rwt. The full-length isoform hTau2N4Rwt (2N4R, 441 amino acid residues) is approximately 4 times larger than the 3-repeat construct Tau3RD (99 residues). Given that Tau is a largely unstructured protein, accessible to solvent along the whole chain, compounds could possibly bind to several sites along the chain. In this case, the compound concentration, which might affect Tau aggregation in the repeat domain, could be effectively diluted several-fold when comparing the repeat domain to full-length Tau. The aggregation of hTau2N4Rwt in vitro is very slow and has to be catalysed by heparin. Heparin binds to monomeric Tau, which likely neutralizes the charge in the basic repeat region of Tau [33] thereby inducing structural changes in the protein [43] that enhances a structural ensemble favoring nucleation [44]. The N- and C-terminal domains of hTau2N4Rwt slow down aggregation because they protect the four repeat region [25], so that it requires incubation times up to 10 days and 5-fold higher protein concentrations than that required for Tau3RD and Tau4RDΔK280 aggregation assays, in order to obtain sufficient fibrillization. Heparin cannot be reduced appreciably because this would lead to even longer aggregation times. Such long incubation times at 37°C may influence hit compound stability and activity in these assays. Aggregation assays with Tau3RD construct are much faster (12 h at 37°C) in the presence of heparin [32]. In both assays, hit compounds had a significant effect in reducing fibril formation or promoting more fibril formation, however the reducing effects were not as substantial as observed in Tau4RDΔK280 aggregation assays. We speculate that one of the reasons for the small aggregation modulation effects of the hits in the hTau2N4Rwt and Tau3RD assays is that heparin competes with hit compounds for binding to hTau2N4Rwt. NMR studies demonstrated that heparin can bind to monomeric full length Tau along its whole sequence with an affinity as high as low μM at some sites [43]. (The binding of negatively charged heparin to Tau may be non-specific and driven mostly by salt bridge formation.) Furthermore, these NMR experiments [43] and recent single molecule FRET measurements [44] showed that the interaction between heparin and Tau induces substantial structural changes throughout the protein, for example, β-strand structure in certain regions in the four repeat region and α-helical structure in some regions outside of the four repeat region [43], while it releases long-range intramolecular interactions and compacts the microtubule binding region. Such structural changes in Tau also likely limit the ability of hit compounds to interact with monomeric Tau.
In case of the Tau construct Tau4RDΔK280, which contains one of the FTDP-17 deletion mutations, the aggregation assay can be performed in the absence of heparin because of its high propensity to aggregate [38; 45]. These results allow one to exclude artificial perturbations to Tau structure due to heparin. Indeed, in this assay two thirds of our Tau binding compounds had a significant effect in modulating Tau4RDΔK280 aggregation and notably 35% of them had a substantial effect. As a result of the use of heparin and the different Tau constructs, it is not surprising that no correlation in the degree of aggregation reduction by the hits was observed, although several hit compounds had significant aggregation inhibition effect in all in vitro and cellular assays (Table 4).
In N2a cells, the hit compounds, in general, had a more extensive ability to reduce fibril formation of Tau4RDΔK280, compared to in vitro experiments. One reason for this may be the fact that there is less Tau4RDΔK280 in the cells and thus less amount of compound is needed to have a functional effect. No significant correlation in the ability of hit compounds to reduce aggregation of Tau4RDΔK280 in N2a cells compared to in vitro was observed, however, several hit compounds had significant aggregation inhibition effect in both cellular and the in vitro assay (Table 4 and Fig. 4). We further confirmed our findings by demonstrating that the three selected compounds (Table 4) reduced the aggregation of Tau4RDΔK280 in Na2 cells in a dose dependent manner. These hit compounds are likely to act via single compound monomer species, because DLS experiments did not detect self-aggregated compound species at the assayed conditions. The concentration of the compound that can modulate aggregation is likely to depend on the relative ratio of protein and compound, therefore, the necessary compound concentration for in vivo aggregation modulation is determined by the absolute level of soluble monomeric Tau in neurons, which may be as low as 20 nM [22]. The functional effect of our compounds in reducing Tau aggregation is comparable to other reported optimized aggregation inhibitors. For example, the rhodanine based inhibitor bb14 [40] was less than 2 times more potent in reducing Tau4RDΔK280 aggregation in vitro compared to our best hits, while the phenylthiazolyl-hydrazide based inhibitor BSc3094 [41] was about 2-3 more effective in reducing aggregation in N2a cells compared to our best hits. Independently of their inhibitory capacity, the reported hit compounds (Fig. 3 and Table 4) are promising starting points for hit optimization efforts, because these compound have no reactive fragments and generally have drug-like physicochemical properties.
The use of absorption and fluorescence assays to monitor different physiological effect of compounds on cellular system (e.g. MTT-Assay, LDH-Assay) could be restricted by the optical properties of the investigated compound (see e.g. absorption spectra of methylene blue and azur A in [46]). In these cases ThS is not usable as an indicator of overall aggregation and has to be replaced by other methods (e.g. quantitative electron microscopy [47], filter trap [12; 48] or pelleting assays [10; 11]. However, the compounds used in the present work have no absorption in the region critical for ThS fluorescence and therefore do not interfere with the fluorescence-based methods.
Overall, the results suggest that hit compounds exert their functional effect by binding to Tau. Hit compounds may reduce the likelihood of Tau self-interactions and may also affect monomer to oligomeric/aggregated Tau interactions and thereby reduce secondary nucleation and aggregate growth. Indeed, it has been suggested that the rate-limiting step in the fibrillization pathway of Tau may be dimer formation, and filament elongation happens by addition of Tau monomers to nascent filament ends [26]. We can not exclude the possibility that hit compounds may also have an affinity toward oligomeric Tau and exert a functional effect that way a case that requires further investigation. Moreover, further studies will be needed to establish whether the compounds that we have presented here bind specifically to Tau or also to other proteins. Our approach of targeting the initial step of the fibrillization pathway is aimed at finding compounds that reduce the generation of toxic Tau oligomers and protect against Tau toxicity. Such therapeutic approach should be able to slow down progression of the disease as well as provide preventative effect.
Parallel to our own efforts reported in this study, fluorescently labeled monomeric Aβ40 was screened by Chen and co-workers [49] using small molecules immobilized on microarrays. This screen resulted in identifying small molecule binders to Aβ40, which were found to promote the aggregation of Aβ40 and reduced the toxicity of Aβ40 in PC12 cells. Although the study did not find Aβ40 aggregation inhibitors, it supports one of the conclusions of this study that the presented screening approach can be extended to other IDPs.
Conclusion
We applied a high-throughput chemical microarray surface plasmon resonance imaging screen (HT-CM-SPR) to identify small molecule binders to monomeric hTau2N4Rwt. We identfied a diverse set of novel fragment and lead-like small molecules capable of binding Tau, some of which had the ability to inhibit the aggregation of full length Tau, three-repeat Tau construct Tau3RD, and pro-aggregant mutant four-repeat Tau construct Tau4RDΔK280 in vitro and in N2a cells. The discovery of these small-molecule binders to Tau demonstrates that despite its heterogeneous conformational ensemble and a lack of stable tertiary structure, it can be a viable receptor of small drug-like molecules, a finding that may be valid for other IDPs and is in line with our and other recent reports[49; 50]. Some of the small-molecule binders to Tau exerted a functional effect by reducing the aggregation propensity of Tau (Table 4). Several of the identified novel aggregation inhibitors (Table 4) are drug-like small molecules that are suitable starting scaffolds for hit to lead optimization and for efficacy studies in relevant in vivo models of AD. Overall, these results support the potential and practical feasibility of the therapeutic strategy to target the early phases of the aggregation pathway of IDPs by a small molecule to reduce initial or secondary nucleation events, thereby eliminating the formation of potential toxic oligomers. Such a strategy for Tauophaties could be of therapeutic value in both disease modifying and preventive contexts. The presented drug discovery paradigm has general applicability because it can be applied to any IDPs linked to misfolding diseases such as AD, Parkinson’s Disease, FTDP and others.
Supplementary Material
Fig. (S1). HTau2N4Rwt stock preparation, used as starting material fort the screen, showing that the protein was monomeric. 10% SDS-PAGE of hTau2N4Rwt stock prepared in PBS buffer at pH 7.4 with 1 mM DTT at 1 mg/ml (22 µM) concentration. HTau2N4Rwt appears as a single band by Coomassie Blue staining. The purity of the protein is > 95%.
Fig. (S2). Electron micrographs of Tau4RDΔK280 fibrils after 10 days of incubation. A and B – without compound; C – in the presence of anti-aggregation compound (10 µM Tau protein + 60 µM bb14[34] after 12 hours incubation at 37 °C).
Fig. (S3). Comparison of tau expressing N2a cells when treated with anti-aggregation compounds. N2a cells expressing the tau construct K18ΔK280 were incubated with the rhodanine derivative bb14 [34] and the phenyl-thiazolyl-hydrazide derivative BSc3094 [35] (15 µM concentration in the medium) for 4 days and the amount of ThS-positive cells was analyzed by FACS. The induced cells without compounds shows an amount of ThS-positive cells (located in the “GFP+”-field) which decreases in the presence of BSc3094 (compare B and D). In the samples treated with bb14 we observed a general upward shift of the whole cell population in the FITC channel. This observation suggests an artificial fluorescence effect of this compound in our cellular system. Because of this we used only the BSc3094 compound as reference for our cellular assays (FACS and MTT).
Acknowledgements
We thank the Wellcome Trust (UK), Medical Research Council (UK), Elan Pharmaceuticals (USA), the Canadian Institutes of Health Research (Canada) and the Alzheimer Society of Ontario (Canada), and Hungarian Brain Research Program (KTIA_NAP_13-2014-0009) for funding.
Footnotes
Author contributions
G. T. conceived the study and managed the overall execution of the study. E. M., L. M., M. P., G. T. designed screen and follow up experiments. T. N. optimized and performed high-throughput screen. D. S. synthesized hit compounds. M. P. performed all in vitro aggregation and cell based experiments. K. C. performed all DLS experiments. G. T., M. P., E. M., L. M., C. M. D., E. M. M., P. H., M. V., D. S., T. N. analyzed, interpreted the data and wrote the paper.
Competing financial interests
We have read the journal's policy and make the following statements on conflict of interests: some of the authors were employees and/or shareholders of Elan Pharmaceuticals as designated by Elan affiliation. G.T. was supported in part by a grant from Elan Pharmaceuticals. This does not alter our adherence to all the CAR policies on sharing data and materials.
References
- [1].Lee VM, Goedert M, Trojanowski JQ. Neurodegenerative tauopathies. Annu Rev Neurosci. 2001;24:1121–59. doi: 10.1146/annurev.neuro.24.1.1121. [DOI] [PubMed] [Google Scholar]
- [2].Goedert M, Spillantini MG. A century of Alzheimer's disease. Science. 2006;314(5800):777–81. doi: 10.1126/science.1132814. [DOI] [PubMed] [Google Scholar]
- [3].Goedert M, Spillantini MG, Jakes R, Rutherford D, Crowther RA. Multiple isoforms of human microtubule-associated protein tau: sequences and localization in neurofibrillary tangles of Alzheimer's disease. Neuron. 1989;3(4):519–26. doi: 10.1016/0896-6273(89)90210-9. [DOI] [PubMed] [Google Scholar]
- [4].Wszolek ZK, Slowinski J, Golan M, Dickson DW. Frontotemporal dementia and parkinsonism linked to chromosome 17. Folia neuropathologica / Association of Polish Neuropathologists and Medical Research Centre, Polish Academy of Sciences. 2005;43(4):258–70. [PubMed] [Google Scholar]
- [5].Busciglio J, Lorenzo A, Yeh J, Yankner BA. beta-amyloid fibrils induce tau phosphorylation and loss of microtubule binding. Neuron. 1995;14(4):879–88. doi: 10.1016/0896-6273(95)90232-5. [DOI] [PubMed] [Google Scholar]
- [6].Lippens G, Amniai L, Wieruszeski JM, Sillen A, Leroy A, Landrieu I. Towards understanding the phosphorylation code of tau. Biochem Soc T. 2012;40(4):698–703. doi: 10.1042/BST20120050. [DOI] [PubMed] [Google Scholar]
- [7].Crowe A, Ballatore C, Hyde E, Trojanowski JQ, Lee VM. High throughput screening for small molecule inhibitors of heparin-induced tau fibril formation. Biochem Biophys Res Commun. 2007;358(1):1–6. doi: 10.1016/j.bbrc.2007.03.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Crowe A, Huang W, Ballatore C, Johnson RL, Hogan AM, Huang R, et al. Identification of aminothienopyridazine inhibitors of tau assembly by quantitative high-throughput screening. Biochemistry. 2009;48(32):7732–45. doi: 10.1021/bi9006435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Pickhardt M, Gazova Z, von Bergen M, Khlistunova I, Wang Y, Hascher A, et al. Anthraquinones inhibit tau aggregation and dissolve Alzheimer's paired helical filaments in vitro and in cells. J Bio Chem. 2005;280(5):3628–35. doi: 10.1074/jbc.M410984200. [DOI] [PubMed] [Google Scholar]
- [10].Pickhardt M, von Bergen M, Gazova Z, Hascher A, Biernat J, Mandelkow EM, et al. Screening for inhibitors of tau polymerization. Curr Alzheimer Res. 2005;2(2):219–26. doi: 10.2174/1567205053585891. [DOI] [PubMed] [Google Scholar]
- [11].Taniguchi S, Suzuki N, Masuda M, Hisanaga S, Iwatsubo T, Goedert M, et al. Inhibition of heparin-induced tau filament formation by phenothiazines, polyphenols, and porphyrins. J Biol Chem. 2005;280(9):7614–23. doi: 10.1074/jbc.M408714200. [DOI] [PubMed] [Google Scholar]
- [12].Akoury E, Pickhardt M, Gajda M, Biernat J, Mandelkow E, Zweckstetter M. Mechanistic basis of phenothiazine-driven inhibition of Tau aggregation. Angew Chem. 2013;52(12):3511–5. doi: 10.1002/anie.201208290. [DOI] [PubMed] [Google Scholar]
- [13].Messing L, Decker JM, Joseph M, Mandelkow E, Mandelkow EM. Cascade of tau toxicity in inducible hippocampal brain slices and prevention by aggregation inhibitors. Neurobiol Aging. 2013;34(5):1343–54. doi: 10.1016/j.neurobiolaging.2012.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Fatouros C, Pir GJ, Biernat J, Koushika SP, Mandelkow E, Mandelkow EM, et al. Inhibition of tau aggregation in a novel Caenorhabditis elegans model of tauopathy mitigates proteotoxicity. Hum Mol Gen. 2012;21(16):3587–603. doi: 10.1093/hmg/dds190. [DOI] [PubMed] [Google Scholar]
- [15].Congdon EE, Figueroa YH, Wang L, Toneva G, Chang E, Kuret J, et al. Inhibition of tau polymerization with a cyanine dye in two distinct model systems. J Biol Chem. 2009;284(31):20830–9. doi: 10.1074/jbc.M109.016089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Feng BY, Toyama BH, Wille H, Colby DW, Collins SR, May BC, et al. Small-molecule aggregates inhibit amyloid polymerization. Nat Chem Biol. 2008;4(3):197–9. doi: 10.1038/nchembio.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Lendel C, Bertoncini CW, Cremades N, Waudby CA, Vendruscolo M, Dobson CM, et al. On the mechanism of nonspecific inhibitors of protein aggregation: dissecting the interactions of alpha-synuclein with Congo red and lacmoid. Biochemistry. 2009;48(35):8322–34. doi: 10.1021/bi901285x. [DOI] [PubMed] [Google Scholar]
- [18].Wittmann CW, Wszolek MF, Shulman JM, Salvaterra PM, Lewis J, Hutton M, et al. Tauopathy in Drosophila: neurodegeneration without neurofibrillary tangles. Science. 2001;293(5530):711–4. doi: 10.1126/science.1062382. [DOI] [PubMed] [Google Scholar]
- [19].Brunden KR, Trojanowski JQ, Lee VM. Evidence that non-fibrillar tau causes pathology linked to neurodegeneration and behavioral impairments. J Alzheimer's Dis. 2008;14(4):393–9. doi: 10.3233/jad-2008-14406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Sydow A, Van der Jeugd A, Zheng F, Ahmed T, Balschun D, Petrova O, et al. Reversibility of Tau-related cognitive defects in a regulatable FTD mouse model. J Mol Neurosci. 2011;45(3):432–7. doi: 10.1007/s12031-011-9604-5. [DOI] [PubMed] [Google Scholar]
- [21].Sydow A, Van der Jeugd A, Zheng F, Ahmed T, Balschun D, Petrova O, et al. Tau-induced defects in synaptic plasticity, learning, and memory are reversible in transgenic mice after switching off the toxic Tau mutant. J Mol Neurosci. 2011;31(7):2511–25. doi: 10.1523/JNEUROSCI.5245-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Brunden KR, Trojanowski JQ, Lee VM. Advances in tau-focused drug discovery for Alzheimer's disease and related tauopathies. Nat Rev Drug Discov. 2009;8(10):783–93. doi: 10.1038/nrd2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Bulawa CE, Connelly S, Devit M, Wang L, Weigel C, Fleming JA, et al. Tafamidis, a potent and selective transthyretin kinetic stabilizer that inhibits the amyloid cascade. Proc Natl Acad Sci USA. 2012;109(24):9629–34. doi: 10.1073/pnas.1121005109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Benson MD. Familial amyloidotic polyneuropathy. Trends Neurosci. 1989;12(3):88–92. doi: 10.1016/0166-2236(89)90162-8. [DOI] [PubMed] [Google Scholar]
- [25].Jeganathan S, von Bergen M, Brutlach H, Steinhoff HJ, Mandelkow E. Global hairpin folding of tau in solution. Biochemistry. 2006;45(7):2283–93. doi: 10.1021/bi0521543. [DOI] [PubMed] [Google Scholar]
- [26].Congdon EE, Kim S, Bonchak J, Songrug T, Matzavinos A, Kuret J. Nucleation-dependent tau filament formation: the importance of dimerization and an estimation of elementary rate constants. J Biol Chem. 2008;283(20):13806–16. doi: 10.1074/jbc.M800247200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Metallo SJ. Intrinsically disordered proteins are potential drug targets. Curr Opin Chem Biol. 2010;14(4):481–8. doi: 10.1016/j.cbpa.2010.06.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Neumann T, Junker HD, Schmidt K, Sekul R. SPR-based fragment screening: advantages and applications. Curr Topics Med Chem. 2007;7(16):1630–42. doi: 10.2174/156802607782341073. [DOI] [PubMed] [Google Scholar]
- [29].Neumann T, Sekul R. SPR Screening of Chemical Microarrays for Fragment Based Discovery. In: Mayr LM, Cooper MA, editors. Label-free Technologies for Drug Discovery. Wiley-Blackwell; 2011. [Google Scholar]
- [30].Maier S, Frank M, Rau H, Lewandrowski P, Uhrig R, Keil O, et al. Synthesis and Quality Control of Thiol Tagged Compound Libraries for Chemical Microarrays. QSAR & Combinatorial Science. 2006;25:1047–54. [Google Scholar]
- [31].Friedhoff P, Schneider A, Mandelkow EM, Mandelkow E. Rapid assembly of Alzheimer-like paired helical filaments from microtubule-associated protein tau monitored by fluorescence in solution. Biochemistry. 1998;37(28):10223–30. doi: 10.1021/bi980537d. [DOI] [PubMed] [Google Scholar]
- [32].Friedhoff P, von Bergen M, Mandelkow EM, Davies P, Mandelkow E. A nucleated assembly mechanism of Alzheimer paired helical filaments. Proc Natl Acad Sci USA. 1998;95(26):15712–7. doi: 10.1073/pnas.95.26.15712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Jeganathan S, von Bergen M, Mandelkow EM, Mandelkow E. The natively unfolded character of tau and its aggregation to Alzheimer-like paired helical filaments. Biochemistry. 2008;47(40):10526–39. doi: 10.1021/bi800783d. [DOI] [PubMed] [Google Scholar]
- [34].Neumann T, Junker HD, Keil O, Burkert K, Ottleben H, Gamer J, et al. Discovery of Thrombin Fragments from Chemical Microarray Screening. Letters in Drug Design & Discovery. 2005;2:563–66. [Google Scholar]
- [35].Dickopf S, Frank M, Junker HD, Maier S, Metz G, Ottleben H, et al. Custom chemical microarray production and affinity fingerprinting for the S1 pocket of factor VIIa. Anal Biochem. 2004;335(1):50–7. doi: 10.1016/j.ab.2004.08.033. [DOI] [PubMed] [Google Scholar]
- [36].Heim-Riether A, Taylor SJ, Liang S, Gao DA, Xiong Z, Michael August E, et al. Improving potency and selectivity of a new class of non-Zn-chelating MMP-13 inhibitors. Bioorg Med Chem Lett. 2009;19(18):5321–4. doi: 10.1016/j.bmcl.2009.07.151. [DOI] [PubMed] [Google Scholar]
- [37].Khlistunova I, Biernat J, Wang Y, Pickhardt M, von Bergen M, Gazova Z, et al. Inducible expression of Tau repeat domain in cell models of tauopathy: aggregation is toxic to cells but can be reversed by inhibitor drugs. J Biol Chem. 2006;281(2):1205–14. doi: 10.1074/jbc.M507753200. [DOI] [PubMed] [Google Scholar]
- [38].Barghorn S, Mandelkow E. Toward a unified scheme for the aggregation of tau into Alzheimer paired helical filaments. Biochemistry. 2002;41(50):14885–96. doi: 10.1021/bi026469j. [DOI] [PubMed] [Google Scholar]
- [39].King ME, Ahuja V, Binder LI, Kuret J. Ligand-dependent tau filament formation: implications for Alzheimer's disease progression. Biochemistry. 1999;38(45):14851–9. doi: 10.1021/bi9911839. [DOI] [PubMed] [Google Scholar]
- [40].Bulic B, Pickhardt M, Khlistunova I, Biernat J, Mandelkow EM, Mandelkow E, et al. Rhodanine-based tau aggregation inhibitors in cell models of tauopathy. Angew Chem. 2007;46(48):9215–9. doi: 10.1002/anie.200704051. [DOI] [PubMed] [Google Scholar]
- [41].Pickhardt M, Larbig G, Khlistunova I, Coksezen A, Meyer B, Mandelkow EM, et al. Phenylthiazolyl-hydrazide and its derivatives are potent inhibitors of tau aggregation and toxicity in vitro and in cells. Biochemistry. 2007;46(35):10016–23. doi: 10.1021/bi700878g. [DOI] [PubMed] [Google Scholar]
- [42].von Bergen M, Barghorn S, Biernat J, Mandelkow EM, Mandelkow E. Tau aggregation is driven by a transition from random coil to beta sheet structure. Arch Biochem Biophys. 2005;1739(2–3):158–66. doi: 10.1016/j.bbadis.2004.09.010. [DOI] [PubMed] [Google Scholar]
- [43].Sibille N, Sillen A, Leroy A, Wieruszeski JM, Mulloy B, Landrieu I, et al. Structural impact of heparin binding to full-length Tau as studied by NMR spectroscopy. Biochemistry. 2006;45(41):12560–72. doi: 10.1021/bi060964o. [DOI] [PubMed] [Google Scholar]
- [44].Elbaum-Garfinkle S, Rhoades E. Identification of an aggregation-prone structure of tau. J Am Chem Soc. 2012;134(40):16607–13. doi: 10.1021/ja305206m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].von Bergen M, Barghorn S, Li L, Marx A, Biernat J, Mandelkow EM, et al. Mutations of tau protein in frontotemporal dementia promote aggregation of paired helical filaments by enhancing local beta-structure. J Biol Chem. 2001;276(51):48165–74. doi: 10.1074/jbc.M105196200. [DOI] [PubMed] [Google Scholar]
- [46].Bulic B, Pickhardt M, Mandelkow E. Progress and developments in tau aggregation inhibitors for Alzheimer disease. J Med Chem. 2013;56(11):4135–55. doi: 10.1021/jm3017317. [DOI] [PubMed] [Google Scholar]
- [47].Congdon EE, Necula M, Blackstone RD, Kuret J. Potency of a tau fibrillization inhibitor is influenced by its aggregation state. Arch Biochem Biophys. 2007;465(1):127–35. doi: 10.1016/j.abb.2007.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Chang E, Kim S, Yin H, Nagaraja HN, Kuret J. Pathogenic missense MAPT mutations differentially modulate tau aggregation propensity at nucleation and extension steps. J Neurochem. 2008;107(4):1113–23. doi: 10.1111/j.1471-4159.2008.05692.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Chen J, Armstrong AH, Koehler AN, Hecht MH. Small molecule microarrays enable the discovery of compounds that bind the Alzheimer's Abeta peptide and reduce its cytotoxicity. J Am Chem Soc. 2010;132(47):17015–22. doi: 10.1021/ja107552s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Zhu M, De Simone A, Schenk D, Toth G, Dobson CM, Vendruscolo M. Identification of small-molecule binding pockets in the soluble monomeric form of the Abeta42 peptide. J Chem Phys. 2013;139(3):035101. doi: 10.1063/1.4811831. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.