Abstract
In response to viral infections, the mammalian innate immune system induces the production of the second messenger 2’-5’ oligoadenylate (2–5A) to activate latent ribonuclease L (RNase L) that restricts viral replication and promotes apoptosis. A subset of rotaviruses and coronaviruses encode 2’,5’-phosphodiesterase enzymes that hydrolyze 2–5A, thereby inhibiting RNase L activation. We report the crystal structure of the 2’,5’-phosphodiesterase domain of group A rotavirus protein VP3 at 1.39 Å resolution. The structure exhibits a 2H phosphoesterase fold and reveals conserved active site residues, providing insights into the mechanism of 2–5A degradation in viral evasion of host innate immunity.
Keywords: phosphodiesterase, 2–5A, 2H phosphoesterase, RNase L, oligoadenylate synthase, rotavirus, innate immunity, immune evasion
Introduction
The mammalian innate immune system detects viral infections through pattern recognition receptors and initiates an immune response mediated predominantly by type I interferons (IFNs).1 IFN-mediated cell signaling subsequently results in the transcriptional upregulation of a number of interferon-response genes encoding antiviral restriction factors that block viral propagation by diverse mechanisms.2,3 A potent antiviral restriction mechanism involves the oligoadenylate synthase (OAS)–ribonuclease (RNase) L pathway.4 OAS genes are induced in response to IFN stimulation.5,6 Upon binding and activation by viral double-stranded RNAs (dsRNA), catalytically active OAS isoforms convert ATP into 5’-triphosphorylated, 2’-5’ linked adenylate oligomers (2–5A).7 Trimeric and longer species of 2–5A bind to the inactive monomeric form of the pseudokinase-ribonuclease RNase L to induce dimerization of the enzyme and its catalytic activation.8,9 Activated RNase L exerts its antiviral effect by cleaving single-stranded RNA regions in viral RNAs, as well as by promoting apoptosis in infected cells through degradation of cellular mRNAs and rRNAs.10 Thus, OAS enzymes function as pattern recognition receptors of viral dsRNA, while 2–5A is a conserved second messenger of viral infection whose only known biological function is to activate RNase L.3,7
In the host-pathogen arms race, viruses have evolved diverse mechanisms to evade the innate immune response of the host, including direct inhibition of OAS-RNase L pathway. The genomes of coronaviruses and rotaviruses encode 2’,5’-phosphodiesterase (PDE) enzymes that degrade 2–5A and prevent its cellular accumulation, thereby blocking RNase L activation. Mouse hepatitis virus (MHV), an enveloped (+)RNA virus of the coronavirus family, expresses a cytoplasmic 30-kDa 2’,5’-phosphodiesterase protein ns2 that functions as an IFN antagonist and is required for the induction of hepatitis.11–13 Group A rotavirus (RVA), a member of the rotavirus family of segmented dsRNA viruses that cause severe infant diarrhea, encodes the capping enzyme VP3 that contains a C-terminal phosphodiesterase domain (Figure 1A).14 Both the VP3 PDE domain and ns2 have been shown to cleave 2-5A in vitro and in vivo, thereby antagonizing RNaseL activation while enhancing viral growth and pathogenesis.14,15 The proteins share 18% sequence identity (Figure 1B) and are functionally interchangeable, as the VP3 PDE domain is able to rescue the replication of ns2 mutant MHV in cells expressing RNase L.14 MHV ns2 and the C-terminal domain of RVA VP3 are members of the 2H phosphoesterase family and contain two characteristic His-X-Ser/Thr motifs in their catalytic sites. Both proteins are structurally homologous to A-kinase-anchoring protein-7 (AKAP7), a 2H phosphoesterase domain-containing protein that mediates signaling within cAMP microdomains by anchoring multiple signaling proteins.16 AKAP7 exhibits 2’,5’-phosphodiesterase activity and can complement an inactive ns2 gene in a MHV mutant to restore infectivity of the virus.17
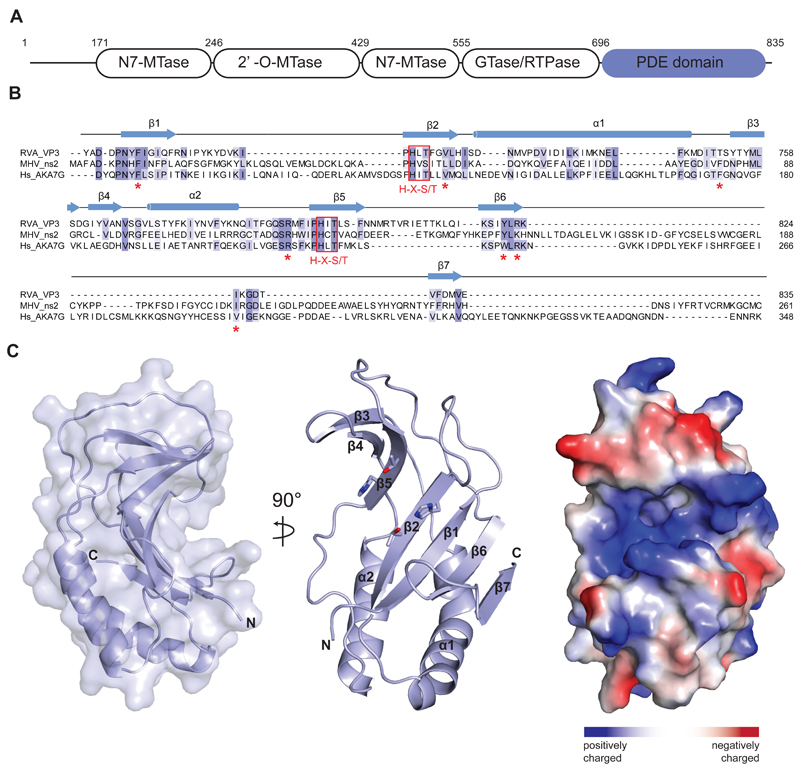
Figure 1.
(A) Domain architecture of group A rotavirus VP3 protein (UniProt ID A2T3S1). The protein consists of an N7-methyltransferase (N7-MTase), 2’-O-methyltransferase (2’-O-MTase) and guanylyltransferase/RNA-triphosphatase (GTase/RTPase) domains, followed by a C-terminal phosphodiesterase (PDE) domain spanning residues 696–835. (B) Multiple sequence alignment for 2H phosphoesterase superfamily members displaying 2’,5’-phosphodiesterase activity. Primary sequences of RVA VP3 (Uniprot ID A2T3S1), MHV ns2 (UniProt ID P19738) and human AKAP7 (Hs_AKA7G, UniProt ID Q9P0M2) proteins were aligned using MAFFT34 with the E-INS-I strategy. Conserved catalytic His-X-Ser/Thr motifs (red boxes) and active site residues (red asterisks) are highlighted. Secondary structure elements of RVA VP3 are indicated above the sequence. (C) Crystal structure of the VP3 PDE domain. Left and middle: Two perpendicular views shown in cartoon representation. Histidine and threonine sidechains of the two His-X-Thr catalytic motifs are shown as sticks. Right: Surface representation colored according to electrostatic surface potential.
Despite their essential role in antagonizing the OAS-RNase L innate immune response pathway, there is little structural information available for viral 2’,5’-phosphodiesterase enzymes. Here we report the structure of the PDE domain of the RVA VP3 protein (UniProt entry A2T3S1, residues 696–835) at a resolution of 1.39 Å, highlighting its evolutionary relationship with cellular phosphodiesterases and providing insights into the catalytic mechanism of 2–5A cleavage.
Materials and Methods
Protein purification and crystallization
The DNA sequence encoding the PDE domain of the minor core protein VP3 from group A rotavirus (residues 696–835) was synthesized by GeneArt (Life Technologies) and subcloned into bacterial expression vector 1B (obtained from the UC Berkeley MacroLab, sequence available from Addgene, www.addgene.org) by ligation-independent cloning (LIC). The VP3 PDE domain was expressed in Escherichia coli BL21 (DE3) Rosetta 2 (Novagen) cells as a fusion protein containing an N-terminal hexahistidine (His6) affinity tag, the polypeptide sequence of the maltose binding protein (MBP), and the tobacco etch virus (TEV) protease cleavage site. Cells were grown to an optical density OD600 of 0.8 and protein expression was induced by addition of 0.2 mM isopropyl-β-D-thiogalactopyranoside (IPTG) for 16 h at 18°C. Cells were lysed in 20 mM Tris pH 8.0, 500 mM NaCl and 5 mM imidazole pH 8.0 using a homogenizer (Avestin). The lysate was clarified by centrifugation at 20,000 g for 30 min and applied to a 5 ml Ni-NTA Superflow cartridge (Qiagen). The cartridge was washed with lysis buffer and bound protein was eluted using lysis buffer supplemented with 250 mM imidazole. Eluted protein was dialyzed against 20 mM Tris pH 8.0, 150 mM NaCl and 10 % glycerol overnight at 4 °C in the presence of His6-tagged TEV protease to cleave the His6-MBP affinity tag. Dialyzed protein was re-applied to a 5 ml Ni-NTA Superflow cartridge to remove TEV protease and the affinity tag. The flow through was concentrated and applied to a Superdex 75 16/600 column (GE Healthcare), eluting in 20 mM Tris pH 8.0, 150 mM NaCl. The purified protein was concentrated to 15 mg ml-1 using a 10,000 MWCO centrifugal concentrator (Merck).
Crystallization and structure determination
Crystals of the VP3 PDE domain were grown at 20 °C using the hanging drop vapor diffusion method. The drop was composed of equal volumes (1 μl + 1 μl) of purified protein and reservoir solution (0.1 M Tris-acetate pH 7.0, 0.2 M KSCN and 22 % PEG monomethyl ether 2000). For cryoprotection, crystals were briefly transferred into 0.1 M Tris-acetate pH 7.0, 0.2 M KSCN, 25 % PEG monomethyl ether 2000, 15 % ethylene glycol, and flash-cooled in liquid nitrogen. Native X-ray diffraction data were collected at beam line X06DA (PXIII) of the Swiss Light Source (Paul Scherrer Institute, Villigen, Switzerland) to a resolution of 1.39 Å. The crystals belonged to space group P21 and contained four molecules in the asymmetric unit. Phases were obtained from a single-wavelength anomalous diffraction (SAD) experiment using the anomalous diffraction properties of endogenous sulfur atoms. 15 data sets were collected by exposing different parts of the same crystal, rotating the crystal through 360° in each data set and changing the kappa angle between datasets in 5 degree increments to ensure data completeness and redundancy. Data were processed using XDS18 and scaled using SCALA.19 Sulfur sites were located using Phenix.Hyss20 and phases calculated by the SAD phasing routine implemented in Phenix.Phaser21, resulting in a readily interpretable electron density map. The initial atomic model was built automatically using ARP/wARP22 and completed by manual building in COOT.23 The atomic model was refined against the high-resolution native data using PHENIX.refine.24 A single molecule of PEG was initially placed in the asymmetric unit using PHENIX.LigandFit25 and adjusted manually. The final model contains four copies of the VP3 PDE domain (residues 696–835), 597 water molecules, one PEG molecule and two K+ ions. Atomic coordinates and structure factors have been deposited in the Protein Data Bank (PDB) under accession code 5AF2.
Results and Discusssion
To elucidate the mechanism of 2–5A degradation by viral 2’, 5’-phosphodiesterases involved in immune evasion, we determined a high resolution structure of the C-terminal PDE domain of the RVA VP3 protein (residues 696–835). The structure was solved by a single-wavelength anomalous diffraction experiment and refined at a resolution of 1.39 Å with an Rfree of 19.3 % (Table 1). The asymmetric unit of the crystal contained four copies of the VP3 PDE domain, although the protein is monomeric in solution as determined by size exclusion chromatography. The structure reveals a compact domain of approximate dimensions of 40 Å × 30 Å × 20 Å. The domain adopts a concave α-β fold typical of the LigT-like clade of the 2H-phosphoesterase superfamily26, consisting of 2 α-helices and 7 β-sheets (Figure 1B, C). The concave surface of the fold features a positively charged groove that reaches approximately 10 Å into the core of the protein (Figure 1C). The base of the groove is formed by the antiparallel strands β2 and β5. Each of the two strands harbors one of the catalytic His-X-Thr motifs (His718 and Thr720 in β2, His797 and Thr799 in β5), a characteristic feature of 2H phosphoesterases that form the catalytic site of the PDE domain (Figures 1B and 2B). The catalytic groove is flanked by two lobes. The lower lobe consists of 4 antiparallel β-sheets (β1, β2, β6, β7) capped by 2 α-helices (α1, α2) showing similarities to a Rossmann fold, a structural motif known to bind nucleotides27, whereas the upper lobe contains three antiparallel β-strands (β3-5).
Table 1. Data collection and refinement statistics.
| Dataset | Native | Sulfur SAD |
|---|---|---|
| X-ray source | SLS X06DA (PXIII) | SLS X06DA (PXIII) |
| Space group | P21 | P21 |
| Cell dimensions | ||
| a, b, c (Å) | 61.3, 75.0, 63.2 | 61.3, 75.1, 63.0 |
| α, β, γ (°) | 90, 112.8, 90 | 90, 112.7, 90 |
| Wavelength (Å) | 1.00000 | 2.06640 |
| Resolution (Å)* | 45.12–1.39 (1.43–1.39) | 45.96–2.14 (2.19–2.14) |
| Rsym (%)* | 7.2 (62.22) | 6.2 (13.5) |
| CC1/2* | 0.999 (0.891) | 1.000 (0.974) |
| I/σI* | 20.1 (3.7) | 52.67 (6.72) |
| Observations* | 1409763 (85738) | 2019684 (7779) |
| Unique reflections* | 105693 (7685) | 53486 (2350) |
| Multiplicity* | 13.3 (11.7) | 37.8 (3.3) |
| Completeness (%)* | 99.7 (98.4) | 92.7 (55.3) |
| Refinement | ||
| Resolution (Å) | 45.12–1.39 | |
| No. reflections | 105679 | |
| Rwork / Rfree | 0.171 / 0.193 | |
| No. atoms | ||
| Protein | 4692 | |
| Ligands | 72 | |
| Water | 597 | |
| B-factors | ||
| mean | 21.8 | |
| Protein | 20.6 | |
| Ligands | 30.8 | |
| Water | 29.5 | |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.006 | |
| Bond angles (°) | 1.12 | |
| Ramachandran plot | ||
| % favored | 97.9 | |
| % allowed | 2.1 | |
| % outliers | 0.0 |
Values in parentheses denote highest resolution shell
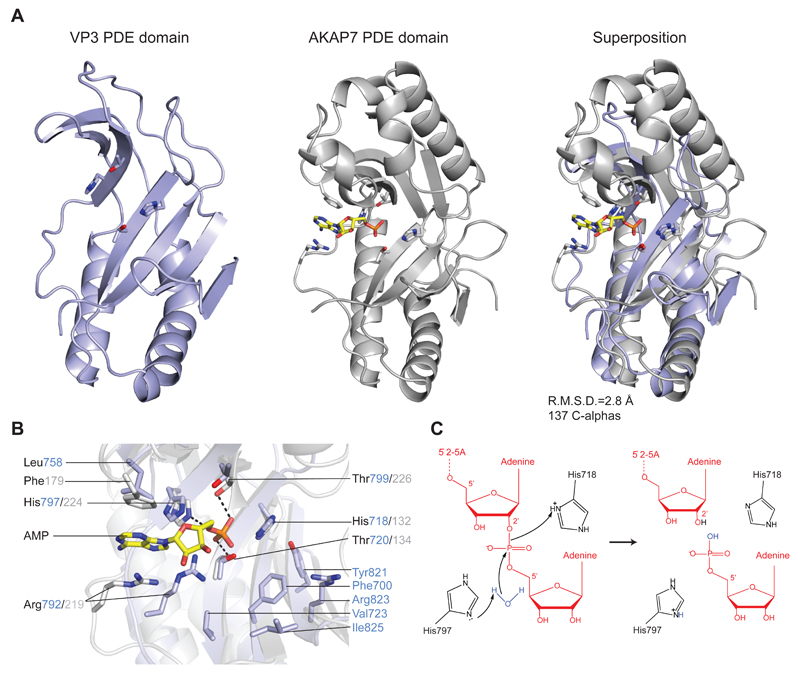
Figure 2.
(A) Comparison of RVA VP3 (light blue) and human AKAP7 (grey, PDB ID 2FVK) phosphodiesterase domains. The structures were superimposed using the DALI server and are shown in identical orientations. The catalytic His and Thr residues as well as bound AMP ligand in AKAP7 are shown in stick representation. The AMP molecule is colored yellow. (B) Zoomed-in view of the overlaid catalytic centers of the PDE domains of VP3 and AKAP7. Catalytic and substrate binding residues are shown as sticks and colored as in (A). Hydrogen bond interactions are depicted as dashed lines. (C) Proposed model for the 2’,5’-phosphodiesterase catalytic mechanism of VP3. The sidechain of His797 acts as a general base to abstract a proton from a water molecule, promoting the nucleophilic attack on the phosphate group. His718 acts as a general acid to donate a proton to the 2’-oxygen of the leaving group. The 2’-5’-oligoadenylate substrate is depicted in red.
We compared the VP3 PDE domain with the structures of other 2H phosphoesterase enzymes using the DALI server.28 Superposition with the phosphoesterase domain of AKAP7 29 (PDB ID 2VFK) reveals substantial structural homology, with a root mean square deviation of 2.8 Å over 137 Cα atoms (Figure 2A). AKAP7 adopts a globular α-β architecture consisting of four α-helices and eight β-sheets.29 In contrast to AKAP7 and a predicted structural model of the VP3 PDE domain14, the crystal structure of the VP3 PDE domain lacks two α-helical segments and one β-strand in the upper lobe. The DALI search additionally identified human 3’-5’ exoribonuclease USB130 (C16orf57, PDB ID 4H7W), 2’-5’ RNA ligases from Pyrococcus horikoshii31 (PDB ID 1VDX) and Thermus thermophilus32 (PDB ID 1IUH) and the 1’-2’-cyclic nucleotide 2’-phosphodiesterase from Arabidopsis thaliana33 (PDB ID 1JH7) as close structural homologs, all of which contain the two additional α-helices in their upper lobes. The observed differences in secondary and tertiary structures are likely in part due to the diversity of substrate specificities across the 2H phosphoesterase superfamily. Nevertheless, the overall structural similarity underscores a clear evolutionary relationship of VP3 to vertebrate 2H phosphoesterase domain-containing proteins including AKAP7, USB1, as well as CGI-18 and LENG9.26 This supports the hypothesis that a vertebrate host phosphodiesterase served as the ancestral precursor of 2’,5’-phosphodiesterases harbored by rotaviruses and coronaviruses.17
The C-terminal domain of VP3 has 2’,5’-phosphodiesterase activity that cleaves 5’-AMP from the 2’-termini of 2-5A molecules.14 To obtain structural insights into its catalytic mechanism of 2–5A hydrolysis, we collected diffraction data from VP3 PDE domain crystals soaked with 5’-AMP as well as crystals grown in the presence of 50 mM 5’-AMP. In neither case were we able to observe electron density corresponding to the bound cleavage product. Closer inspection of the crystal packing interfaces revealed that in all four copies in the asymmetric unit, the catalytic site of the VP3 PDE domain is blocked by the insertion of a loop (Ser759-Asp760) from a symmetry-related molecule, likely explaining the lack of ligand binding in the structure. Comparison of the active sites of AKAP7 and the VP3 PDE domain reveals that the catalytic His-X-Thr motifs are found in equivalent positions, suggesting that the catalytic mechanism of substrate cleavage is conserved in the two proteins. We therefore modeled the binding of the ligand in the active site of the VP3 PDE domain based on the superposition with the structure of AKAP7 in complex with 5’-AMP (PDB ID 2FVK). In AKAP7, both active site threonines donate hydrogen bonds to the phosphate oxygen atoms of the AMP product, while His224 of the second catalytic motif (equivalent to His797 in VP3) forms an additional hydrogen bond to the bridging ester oxygen (Figure 2B). A similar binding mode is expected in VP3, with the phosphate group being hydrogen-bonded to Thr720 and Thr799. The adenine moiety of the AMP ligand is sandwiched between the side chains of Arg219 and Phe179 in AKAP7, the two residues interacting with the base via cation-π and π-π stacking interactions, respectively.29 While the conserved Arg792 likely fulfills the same function in VP3, the phenylalanine position is occupied by Leu758, suggesting that the precise mode of product binding might differ slightly between VP3 and AKAP7. Assuming that the bound AMP represents the product cleaved off from the 2’ terminus of the 2–5A substrate, the adenosine residue upstream of the scissile phosphate would likely bind in the vicinity of amino acid residues Phe700, Val723, Tyr821, Arg823 and Ile825, in a conserved pocket adjacent to the catalytic site (Figures 1B, C and 2B). Hydrolysis of phosphodiesters by 2H phosphoesterases involves a nucleophilic attack on the scissile phosphate. The product of the reaction, however, varies depending on the attacking group and the type of ribose linkage in the substrate. We propose a catalytic mechanism of 2-5A hydrolysis for the VP3 PDE domain reminiscent of the catalytic mechanisms predicted for T. thermophilus 2’-5’ ligase32 and A. thaliana 1’-2’-cyclic nucleotide 2’-phosphodiesterase.33 In VP3, the unprotonated form of His797 would act as a general base, deprotonating a water molecule for in-line nucleophilic attack on the scissile phosphate (Figure 2C). In turn, His718 acts as a general acid, facilitating the cleavage reaction by protonating the 2’-oxygen atom of the leaving group. During the reaction, the scissile phosphate is anchored by hydrogen bonds to Thr720 and Thr799, while the buildup of negative charge on the pentacovalent reaction intermediate might be stabilized by the overall positive surface electrostatic potential of the active site cleft.
In summary, our crystal structure of the C-terminal phosphodiesterase domain of rotavirus VP3 confirms its 2H phosphoesterase fold and pinpoints the location of conserved active site residues, providing clues about the molecular mechanisms of 2–5A recognition and hydrolysis. The global structural homology between the VP3 PDE domain, AKAP7 as well as other 2H phosphoesterase proteins, together with conservation of principal active site residues, suggest that viral 2’,5’-phosphodiesterases such as RVA VP3 and MHV ns2 have emerged by acquisition of an ancestral host 2’,5’-phosphodiesterase whose structural architecture was subsequently shaped by evolutionary pressures of the virus-host conflict.
Acknowledgements
The authors thank V. Olieric and the staff at beamline X06DA of the Swiss Light Source for assistance with X-ray diffraction data collection and phasing. They thank B. Blattman and C. Stutz-Ducommun from the University of Zurich Protein Crystallization Center for initial crystallization screening. They are grateful to members of the Jinek laboratory for critical reading of the manuscript. This work was supported by the ERC Starting Grant ANTIVIRNA (Grant. no. 337284).
Footnotes
The authors declare no conflict of interest.
References
- 1.Sen GC. Viruses and interferons. Annual review of microbiology. 2001;55:255–281. doi: 10.1146/annurev.micro.55.1.255. [DOI] [PubMed] [Google Scholar]
- 2.Stark GR, Kerr IM, Williams BR, Silverman RH, Schreiber RD. How cells respond to interferons. Annual review of biochemistry. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 3.Sadler AJ, Williams BR. Interferon-inducible antiviral effectors. Nature reviews Immunology. 2008;8(7):559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Player MR, Torrence PF. The 2-5A system: modulation of viral and cellular processes through acceleration of RNA degradation. Pharmacology & therapeutics. 1998;78(2):55–113. doi: 10.1016/S0163-7258(97)00167-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chebath J, Benech P, Revel M, Vigneron M. Constitutive expression of (2'-5') oligo A synthetase confers resistance to picornavirus infection. Nature. 1987;330(6148):587–588. doi: 10.1038/330587a0. [DOI] [PubMed] [Google Scholar]
- 6.Zhou A, Hassel BA, Silverman RH. Expression cloning of 2-5A-dependent RNAase: a uniquely regulated mediator of interferon action. Cell. 1993;72(5):753–765. doi: 10.1016/0092-8674(93)90403-d. [DOI] [PubMed] [Google Scholar]
- 7.Hartmann R, Justesen J, Sarkar SN, Sen GC, Yee VC. Crystal structure of the 2'-specific and double-stranded RNA-activated interferon-induced antiviral protein 2'-5'-oligoadenylate synthetase. Molecular cell. 2003;12(5):1173–1185. doi: 10.1016/s1097-2765(03)00433-7. [DOI] [PubMed] [Google Scholar]
- 8.Han Y, Whitney G, Donovan J, Korennykh A. Innate immune messenger 2-5A tethers human RNase L into active high-order complexes. Cell reports. 2012;2(4):902–913. doi: 10.1016/j.celrep.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 9.Huang H, Zeqiraj E, Dong B, Jha BK, Duffy NM, Orlicky S, Thevakumaran N, Talukdar M, Pillon MC, Ceccarelli DF, Wan LC, et al. Dimeric structure of pseudokinase RNase L bound to 2-5A reveals a basis for interferon-induced antiviral activity. Molecular cell. 2014;53(2):221–234. doi: 10.1016/j.molcel.2013.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Silverman RH. Viral encounters with 2',5'-oligoadenylate synthetase and RNase L during the interferon antiviral response. Journal of virology. 2007;81(23):12720–12729. doi: 10.1128/JVI.01471-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schwarz B, Routledge E, Siddell SG. Murine coronavirus nonstructural protein ns2 is not essential for virus replication in transformed cells. Journal of virology. 1990;64(10):4784–4791. doi: 10.1128/jvi.64.10.4784-4791.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bredenbeek PJ, Noten AF, Horzinek MC, Spaan WJ. Identification and stability of a 30-kDa nonstructural protein encoded by mRNA 2 of mouse hepatitis virus in infected cells. Virology. 1990;175(1):303–306. doi: 10.1016/0042-6822(90)90212-A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zoltick PW, Leibowitz JL, Oleszak EL, Weiss SR. Mouse hepatitis virus ORF 2a is expressed in the cytosol of infected mouse fibroblasts. Virology. 1990;174(2):605–607. doi: 10.1016/0042-6822(90)90114-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang R, Jha BK, Ogden KM, Dong B, Zhao L, Elliott R, Patton JT, Silverman RH, Weiss SR. Homologous 2',5'-phosphodiesterases from disparate RNA viruses antagonize antiviral innate immunity. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(32):13114–13119. doi: 10.1073/pnas.1306917110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao L, Jha BK, Wu A, Elliott R, Ziebuhr J, Gorbalenya AE, Silverman RH, Weiss SR. Antagonism of the interferon-induced OAS-RNase L pathway by murine coronavirus ns2 protein is required for virus replication and liver pathology. Cell host & microbe. 2012;11(6):607–616. doi: 10.1016/j.chom.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wong W, Scott JD. AKAP signalling complexes: focal points in space and time. Nature reviews Molecular cell biology. 2004;5(12):959–970. doi: 10.1038/nrm1527. [DOI] [PubMed] [Google Scholar]
- 17.Gusho E, Zhang R, Jha BK, Thornbrough JM, Dong B, Gaughan C, Elliott R, Weiss SR, Silverman RH. Murine AKAP7 has a 2',5'-phosphodiesterase domain that can complement an inactive murine coronavirus ns2 gene. mBio. 2014;5(4):e01312–01314. doi: 10.1128/mBio.01312-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kabsch W. Xds. Acta crystallographica Section D, Biological crystallography. 2010;66(Pt 2):125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Evans PR. An introduction to data reduction: space-group determination, scaling and intensity statistics. Acta crystallographica Section D, Biological crystallography. 2011;67(Pt 4):282–292. doi: 10.1107/S090744491003982X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zwart PH, Afonine PV, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, McKee E, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, et al. Automated structure solution with the PHENIX suite. Methods in molecular biology. 2008;426:419–435. doi: 10.1007/978-1-60327-058-8_28. [DOI] [PubMed] [Google Scholar]
- 21.Read RJ, McCoy AJ. Using SAD data in Phaser. Acta crystallographica Section D, Biological crystallography. 2011;67(Pt 4):338–344. doi: 10.1107/S0907444910051371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Langer G, Cohen SX, Lamzin VS, Perrakis A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nature protocols. 2008;3(7):1171–1179. doi: 10.1038/nprot.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta crystallographica Section D, Biological crystallography. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 24.Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, Adams PD. Towards automated crystallographic structure refinement with phenix.refine. Acta crystallographica Section D, Biological crystallography. 2012;68(Pt 4):352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Terwilliger TC, Adams PD, Moriarty NW, Cohn JD. Ligand identification using electron-density map correlations. Acta crystallographica Section D, Biological crystallography. 2007;63(Pt 1):101–107. doi: 10.1107/S0907444906046233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mazumder R, Iyer LM, Vasudevan S, Aravind L. Detection of novel members, structure-function analysis and evolutionary classification of the 2H phosphoesterase superfamily. Nucleic acids research. 2002;30(23):5229–5243. doi: 10.1093/nar/gkf645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rao ST, Rossmann MG. Comparison of super-secondary structures in proteins. Journal of molecular biology. 1973;76(2):241–256. doi: 10.1016/0022-2836(73)90388-4. [DOI] [PubMed] [Google Scholar]
- 28.Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic acids research. 2010;38(Web Server issue):W545–549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gold MG, Smith FD, Scott JD, Barford D. AKAP18 contains a phosphoesterase domain that binds AMP. Journal of molecular biology. 2008;375(5):1329–1343. doi: 10.1016/j.jmb.2007.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hilcenko C, Simpson PJ, Finch AJ, Bowler FR, Churcher MJ, Jin L, Packman LC, Shlien A, Campbell P, Kirwan M, Dokal I, et al. Aberrant 3' oligoadenylation of spliceosomal U6 small nuclear RNA in poikiloderma with neutropenia. Blood. 2013;121(6):1028–1038. doi: 10.1182/blood-2012-10-461491. [DOI] [PubMed] [Google Scholar]
- 31.Rehse PH, Tahirov TH. Structure of a putative 2'-5' RNA ligase from Pyrococcus horikoshii. Acta crystallographica Section D, Biological crystallography. 2005;61(Pt 9):1207–1212. doi: 10.1107/S0907444905017841. [DOI] [PubMed] [Google Scholar]
- 32.Kato M, Shirouzu M, Terada T, Yamaguchi H, Murayama K, Sakai H, Kuramitsu S, Yokoyama S. Crystal structure of the 2'-5' RNA ligase from Thermus thermophilus HB8. Journal of molecular biology. 2003;329(5):903–911. doi: 10.1016/s0022-2836(03)00448-0. [DOI] [PubMed] [Google Scholar]
- 33.Hofmann A, Grella M, Botos I, Filipowicz W, Wlodawer A. Crystal structures of the semireduced and inhibitor-bound forms of cyclic nucleotide phosphodiesterase from Arabidopsis thaliana. The Journal of biological chemistry. 2002;277(2):1419–1425. doi: 10.1074/jbc.M107889200. [DOI] [PubMed] [Google Scholar]
- 34.Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic acids research. 2002;30(14):3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]