Introduction
DRG neurons provide connectivity between peripheral targets and the spinal cord. These neurons show significant heterogeneity in relation to their morphology, functional properties, growth factor-dependence and transcriptional profile [1,4,64]. This reflects the highly specialised nature of these neurons subserving distinct sensory modalities, including temperature, pain, itch, touch and proprioception. Recent single cell RNA-seq studies have provided a means to classify these neurons and have identified multiple DRG neuron sub-groups [39,68]. Pathologies of DRG neurons—for instance in the form of acquired or inherited peripheral neuropathies—have a significant impact on human health as a consequence of sensory loss and neuropathic pain [13] and are destined to become more common with an ageing population and increased prevalence of Type II diabetes.
A wide variety of injuries applied to sensory neurons, whether traumatic or metabolic, result in marked alterations in transcription of protein-coding genes [14,35,54]. Such changes can have either beneficial or maladaptive outcomes; including increased expression of regeneration-associated genes [10,52] and altered expression of ion channels resulting in enhanced DRG neuronal excitability and neuropathic pain. The DRG has therefore become a model system to study the transcriptional changes following injury. This focus on RNAs encoding proteins is understandable, given their obvious link with function. However, there are other types of RNA and one of these, long non-coding RNA (LncRNA) has been relatively neglected and little studied in the context of sensory neurones.
LncRNAs are usually multi-exonic transcripts of more than 200 base pairs which, can modulate gene expression through cis and trans signalling, and have important functional effects [6,32,59,71,78]. The mechanisms by which LncRNAs may alter gene expression are very diverse, including: complementary binding of antisense LncRNAs, transcriptional interference at promoter sites, altered chromatin structure, competing for miRNA binding, and binding to transcription factors [27,45,83].
Ion channels are key determinants of the excitability and hence the functional properties of sensory neurons. Antisense LncRNAs have previously been identified to the voltage-gated potassium channel, KCNA2 [88] and the voltage-gated sodium channel, SCN9A [33]. In the former case induction of the antisense LncRNA following nerve injury was shown to result in reduced expression of KCNA2 (which acts as an excitability break), leading to sensory neuronal hyper-excitability and the development of neuropathic pain. These are selected examples of functionally relevant LncRNAs that illustrate the important role they can play. However, there has to date been no comprehensive analysis of LncRNA expression within the DRG partly because LncRNAs are typically expressed at low levels and are known to vary by species-, tissue- and developmental stage [32,69]. Our aim was to use high coverage RNA-seq combined with a dedicated bioinformatics platform to identify as comprehensively as possible LncRNAs expressed in DRG. We compared rat and two different mouse strains (which show differing degrees of mechanical hypersensitivity following nerve injury [68]). We wished to determine if LncRNAs were expressed in a cell type-specific manner and also to assess the effect of nerve injury on LncRNA expression. In addition we also assessed LncRNA expression in human IPSC-derived sensory neurons.
Material and Methods
18 supplementary spreadsheets are available at http://doi.org/10.6084/m9.figshare.6508205, 17 tables, 11 figures and supplementary methods are in Supplementary Digital Content.
Animals: Welfare, Tissue and sample collection
Rat
All procedures on rats were carried out in accordance with UK home office regulations and in line with the Animals Scientific Procedures Act 1986 at a licensed facility at King’s College London. Animals were group housed in temperature and humidity controlled rooms where food and water was available ad libitum, with a 12 hour light dark cycle. The welfare of all animals was continually assessed throughout all procedures. In total 24 rats were used.
Rats were humanely culled. L5 DRG tissue from male Wistar rats was collected 21 days after the SNT surgery, placed into sterile tubes, frozen on dry ice and stored at -80. Each sample comprises 3 pooled animals and we had 4 samples of each condition (SNT vs sham).
Mouse
All procedures in mice were carried out in McGill University, Montreal, Canada, were approved by the McGill University Animal Care Committee and are fully consistent with Canadian Council on Animal Care guidelines. All mice strains were procured from Jackson Laboratories (Bar Harbor, ME, USA) at 4-8 weeks of age. All animals of the same sex were group housed in a vivarium at ≈21°C in standard shoebox cages, 2–4 per cage, with access to food (Harlan Teklad 8604) and tap water ad libitum. Average weights were 20.2 (SD=2.89) for BALB/c mice and 21.4 (SD=3.48) for B10.D2 mice (N=12 per strain, for each strain 6 SNI – 6 Sham stratified for sex). All operations were performed on adult mice. Brain and DRG tissue has also been dissected from 3 wild type mice (C57/bl6) and was used to determine relative expression of mRNA using qPCR (see below). In total 27 mice were used.
The tip of the iliac bone, “the first articular process more than 1mm rostral to the iliac crest” [62] was used as the landmark for identifying L5 DRG in all samples. L3 and L4 DRGs were dissected from all mice 28 days after peripheral nerve injury. Each sample represents 1 animal and consists of both L3 and L4 DRG. 12 mice BALB/c mice and 12 B10.D2 mice, stratified for condition and sex were used. All dissections were performed on dry ice and RNase Decontamination Solution was used in order to prevent RNA degradation. Tissue was placed into sterile Eppendorf tubes and initially stored on dry ice. For long-term storage samples were stored in a -80C freezer.
IPS derived human neurons
Human fibroblast derived IPSC were generated as described previously [12]. Neural differentiation was performed using [8] protocol with modifications.
Animal models of pain
Mouse spared nerve injury (SNI)
Surgical procedure for SNI followed a published protocol developed for rat [16] and adapted for mice [65]. Under general anaesthesia (isoflurane and oxygen) the common peroneal and the sural branch of the sciatic nerve were cut and the tibial branch spared. For sham surgery, the same surgical and anaesthetization procedures were followed, but the nerve branches were simply exposed and not damaged We assessed mechanical hypersensitivity after SNI surgery on the ipsilateral mouse paw.
Rat spinal nerve transection (SNT)
The left L5 spinal nerve was ligated and transected and the L4 and L6 branches were left intact. In sham animals the spinal nerve was exposed but not ligated.
Behavioural tests
Behaviour was carried out in a specially allocated room in the animal facility unit at McGill University, performed at a consistent time of day and by the same experimenter. Mice habituated to the vivarium for at least one week before testing. Mechanical pain-related hypersensitivity in mice was assessed using von Frey filaments and the up-down method of Dixon [11] to determine the 50% withdrawal threshold. Mice were first acclimatised to behaviour equipment and baseline behaviour performed 3 times and an average calculated prior to surgery. Baseline paw withdrawal threshold was 1.27g (SD=0.22) for BALB/c strain and 1.36g (SD=0.23) for B10.D2 strain (N=12). Mice were assigned into sham or SNI group randomly and post injury mechanical sensitivity tested at day 1, 7, 14, 21 and 28 (N = 12 per strain stratified for sex and condition, 6 SNI – 6 Sham mice per strain). Assuming an effect of 30% and an SD=20% we need an N=6 to achieve power=80 at an a=0.05 two-sided one way ANOVA. Mice from both strains and surgery groups were tested on the same day. The experimenter was not informed about the condition (injury vs sham) of animals but could not be blinded because of the coat colour of the different strains.
RNA isolation and library preparation
RNA was extracted using a hybrid method of phenol extraction (TriPure, Roche) and combined with column purification (High Pure RNA tissue Kit, Roche) [15]. DRG samples were first homogenised in Tripure using a handheld homogeniser (Cole-Palmer). For IPS cells Tripure was added directly to the well following removal of media. The concentration of RNA in the samples was measured using a nanodrop. Total RNA was provided to the sequencing centre, and the ribodepleted fraction was selected for further sequencing. In rat this was the poly-adenylated fraction. It was then converted to cDNA using the strand-specific deoxy-UTP strand-marking protocol (dUTP).
Sequencing and mapping
All samples were sequenced at the Oxford Genomics Centre. Sequencing was performed using the Illumina HiSeq4000 paired-end protocol with 100bp reads for mouse DRG, 75bp for human IPSC/neurons and Illumina HiSeq2000 – 100bp reads for rat DRG.
DRG from 24 mice (12 per strain stratified for sex and condition) was sent for sequencing. During library preparation 2 samples (Sample 72 BALB/c SNI Male and Sample 68 B10.D2 SNI Female) were accidentally mixed together and destroyed. From the 22 samples sent for sequencing 2 were excluded (Sample 59 B10.D2 SHAM Male and Sample 66 BALB/c SNI Female) due to having more ambiguously mapped reads, lower percentage of mapped reads and higher Cook’s distance than all the other samples.
Mapping to the genome was done using STAR aligner [19]. Reads were mapped on the mm10 mouse genome, rn6 rat genome and Hg38 human genome, all downloaded from ENSEMBL. Conditions and strains were multiplexed in lanes and library batches. Lanes were merged as BAM files after mapping [40].
DE and counting features
DE analysis was done using DESeq2 [43] using default settings. Significance cut-off in all cases was FDR adjusted p.value < 0.05. Counting of features was done using HTSeq [3] and the intersection not empty strategy to resolve ambiguously counted reads.
All visualisations used regularised log2 transformed counts [43]. PCA was always performed on regularized log transformed counts using the top 10000 genes in mouse and human, 5000 genes in rat ranked by their standard deviation. Hierarchical clustering was done on regularised log2 counts of the whole gene set, using euclidean distances and complete linkage.
GO enrichment for DE genes was carried in R using topGO and GSEA [2,50]. Enrichment of neuron sub-type specific genes was calculated with the fisher exact test and enrichment of BP in network modules was calculated using hyper geometric distribution.
Identification of novel LncRNAs
We used a customised reference-based transcript assembly pipeline that requires a reference genome and gene set annotations. Workflow similar [7,23,30] with modifications to produce annotations at the gene level. Doing this we get a non-redundant annotation of unique genes of LncRNAs suitable for count based DE analysis [3,43]. The concept of islands of expression (I.o.E) is described in [23]. Coverage cut-off has been set as in [7].
Only properly paired and uniquely mapped reads were selected. We selected splicing junctions covered with > 2 reads and with lengths > 20 and < 100000. We discarded all reads overlapping annotated gene models. We then used the remaining subset of RNA-seq reads to identify I.o.E outside known gene models using a coverage window approach. Gene models were extended by 1000bp in each direction to ensure that elongated UTRs or not yet annotated exons would not be considered putative novel genes. We selected continuous regions above the coverage threshold of more than a read-mate length to ensure that overlapping read-mates would not artificially increase coverage. For I.o.E length >= 100 and depth > 2. I.o.E were identified using the function “BAM_to_IOE”. I.o.E were collapsed and clustered as co-overlapping features connected by splicing junctions. A connectivity matrix was created holding all interconnecting I.o.E. In each cluster consensus introns were calculated by the relative frequency of each discrete segment of a set of splicing junctions. We then subtracted the genomic intervals of these consensus introns from the genomic intervals of the grouped (I.o.E) to reconstruct full length putative LncRNAs. For novel I.o.E with no overlapping splicing junctions we first selected only the intersect of the respective genomic regions across all samples. Then mono-exonic putative LncRNAs were kept for further analysis only if the length normalised coverage had Pr (>|Z|) < 0.1. Coverage across I.o.E was fed into a smoothed z-score signal processing algorithm. Z-score thresholding was used to identify introns not identified by the aligner and sudden coverage drops indicating end of transcription activity [5]. Rolling coverage was calculated over a smoothing window of 31bp, the minimum coverage drop threshold was set to 5 and the minimum intron length to 20bp. We only kept novel intronic genes if they were supported by evidence of novel splicing junction and did not contain retained introns.
We included putative LncRNAs in this novel annotation only if they were present in all replicates of a biological condition or strain. Annotations were exported in the Gene Transfer Format (GTF). Subsequently we filtered out transcripts with length < 200 bp and we used CPAT [74] to assess coding potential. An average expression cut-off threshold similar to [57] of > 0.5 fpkm for at least one condition was applied to novel LncRNAs carried out for downstream analysis. The pipeline was scripted in R using bioconductor [22] packages and custom scripts. All iterative processes were executed in parallel to optimise run times using parallel and BiocParallel [49,57]. More details in Supplementary Methods. All scripts of the workflow are available in github: http://github.com/gbaskozos/Scripts_LncRNAs.
Transcription Start Sites mapping to mm10
TSS data was downloaded from FANTOM 5 database [21,41]. We downloaded TSS data that has been classified as “True TSS” by the “TSS classifier”. The UCSC Lift Over tool [47] has been used to translate genomic coordinates from the mm9 genome to the mm10. 51% of the true TSS were unambiguously mapped to mm10.
Tissue specificity
Tissue specificity was calculated using the tau metric [79]:
Gene co-expression network
WGCNA [24] was used to create a weighted gene co-expression network. Analysis was carried out only in mouse as it requires an n > 15. Weighted bi-correlation was used as a robust correlation metric. An unsigned network was constructed using only genes that had > 10 counts in 25% of samples. Then the top 25% of genes ranked according to their median absolute deviations was used for the analysis. Scale free topology was achieved with a soft threshold = 5. Modules detected with hierarchical clustering and dynamic tree cut, minimum module size = 30, cut height = 0.995 and deep split = 2. Merged threshold was 0.2. Module eigengenes were calculated as the first principal component of each module. Module membership of LncRNAs was calculated as the absolute (unsigned) bi-correlation with the module eigengenes. The hyper-geometric distribution and the Fisher Exact test were used to identify the top GO enriched (p.value < 0.05) terms for each module.
Primer Design
Primers for the detection of LncRNA and reference gene expression were designed using Primer-BLAST (https://www.ncbi.nlm.nih.gov/tools/primer-blast/). Primers were designed not to overlap any other annotated gene. Thus primers designed for antisense LncRNAs were not able to detect regions complementary to sense gene exons. Primer efficiency and specificity were validated before experimental use.
Reverse transcription PCR
For qPCR analysis RNA samples were converted into cDNA using Evoscript Universal cDNA Master kit (Roche) and by following the manufacturer’s instructions. This kit uses random primers. For LncRNA2754, the primers designed also detected a putative UTR region and therefore a strand specific RT reaction was used. Strand specific RT primers (see table 18) were used for LncRNA2754 and for HPRT1 (final concentration 0.5µM) and 200ng of RNA was used for each reaction. Strand specific reverse translation into cDNA synthesis was carried out using Transcriptor reverse transcriptase (Roche) and dNTPs (Promega).
Quantitative Real Time PCR (qPCR)
For qPCR analysis, cDNA (5ng) and primer pairs (1μM, See table 18) were mixed with LightCycler 480 SYBR Green Master (Roche) in a 1:1 ratio and added to white 384 well plates (Roche). Plates were run on a 45-cycle protocol using the LC 480 II system (Roche). Gene expression for each mouse target primer was normalized against the reference gene HPRT1 and the relative expression (delta CT) calculated. For human IPS cells transcript expression was normalised against the average CT of GAPDH and HPRT1. For each target transcript expression is shown relative to a control group (e.g. Sham). Significance was calculated using ANOVA with a design of ~ sex + condition for each mouse strain and ~ condition for human IPSC averaging over all cell lines. N= 10 per strain (6 Sham 4 SNI for BALB/c, 5 Sham - 5 SNI for B10.D2, for strand specific qPCR B10.D2 strain N =8, 5 Sham – 3 SNI. qPCR was also used to assess relative mRNA expression in brain vs DRG, N=3. Significance was calculated using one-way ANOVA.
In-situ hybridisation
In situ hybridization (ISH) was performed as previously shown [15]. Once cut, sections were air-dried onto superfrost slides in the cryostat for 0.5 hours and then stored in the -80C. ISH was carried out using the RNAScope 2.5 RED chromogenic assay kit and by following the manufacturer’s instructions (Advanced Cell Diagnostics). Briefly, slides containing tissue sections were removed from the -80, allowed to equilibrate to RT and re-hydrated in PBS. Pre-treatment required a hydrogen peroxide step at RT; followed by an antigen retrieval step and protease treatment in a hybridization oven at 40C. Slides were then incubated with the target or control probes at 40C for 2 hours. For HAGLR mRNA the probes were designed to target position 1153-2443 of NR_110445.1. Following probe incubation, slides were subjected to 6 rounds of amplification and the probe signal was developed via a reaction with fast red. To combine with immunohistochemistry (IHC), tissue sections were then washed with PBS-Tx (0.3%) and treated with either the isolectin B4 (IB4) conjugated to biotin (Sigma, 1:100) or primary antibodies against NF200 (mouse anti-NF200, Sigma, 1:250) and CGRP (rabbit anti-CGRP, Peninsula Labs, 1:1000) overnight at room temperature. IB4 and primary antibodies were diluted in PBS-Tx (0.3%). Slidesd were then washed with PBS-Tx (0.3%) and then incubated with the appropriate secondary antibody (anti-mouse Pacific Blue or ant-rabbit Alexa 488, Thermo Fisher Scientific) or a Pacific blue streptavidin (Thermo Fisher Scientific) at a concentration of 1:500 for 3-4 hours at room temperature. Slides were then washed, mounted using Vectashield and imaged using a confocal microscope (Zeiss).
Results
Novel LncRNAs expressed in rodent DRG
To identify unknown transcribed regions outside known gene models (i.e. representation of RNA transcripts produced by a gene) and then group them into transcriptional units representing putative novel LncRNAs on the gene level, we performed RNA-sequencing (RNA-Seq) of DRG tissue harvested from animal models of nerve injury versus sham controls. We profiled novel LncRNAs alongside known annotated protein-coding genes and LncRNAs from the ENSEMBL genome database [81]. To obtain computational predictions of novel LncRNAs we used a reference-based customised pipeline [46] that relies on a reference genome and gene set annotations. It uses the output of the STAR [19] aligner and the quality of predictions is dependent on the aligner accuracy, the quality of the reference genome and the completeness of the gene set annotation. A coverage threshold is used [7] to identify non-annotated continuously transcribed regions [23], i.e. Islands of Expression (I.o.E), and then clustered together and trimmed these regions using de-novo identified splicing junctions (SJ) from mapping the RNA-seq reads to the organism’s genome. We also applied a signal processing, smoothed z-score thresholding algorithm to further identify coverage drops (putative introns and transcription ends) and peaks (putative exons). To identify non-annotated I.o.E and to differentiate them from untranslated regions (UTRs) or not-yet-annotated exons belonging to known gene models we filtered out reads overlapping ENSEMBL and RefSeq annotations as well as genomic regions of 1000 bp from the 5’ and 3’ end of known gene models. Doing so we have excluded any predictions overlapping with a region of 1000bp from both ends of annotated gene models. The mean length of 3’ UTRs is 424bp and 524bp in rodents and Human respectively. For the 5’ UTRs mean lengths are 127bp in rodents and 146bp in Human [56]. We should note that we used both major annotation consortia so an incomplete gene annotation (i.e. missing exons, UTRs, isoforms) in just one of the annotated gene sets would not influence results. Given the fact that we discarded all predictions overlapping annotated genes we could not also identify extra-coding RNAs as these overlap protein-coding genes on the sense strand [63].
32.6% of read pairs on average were overlapping regions outside gene models (as we used paired-end data the units of evidence for gene expression are always pairs of reads). This finding is consistent with [23].
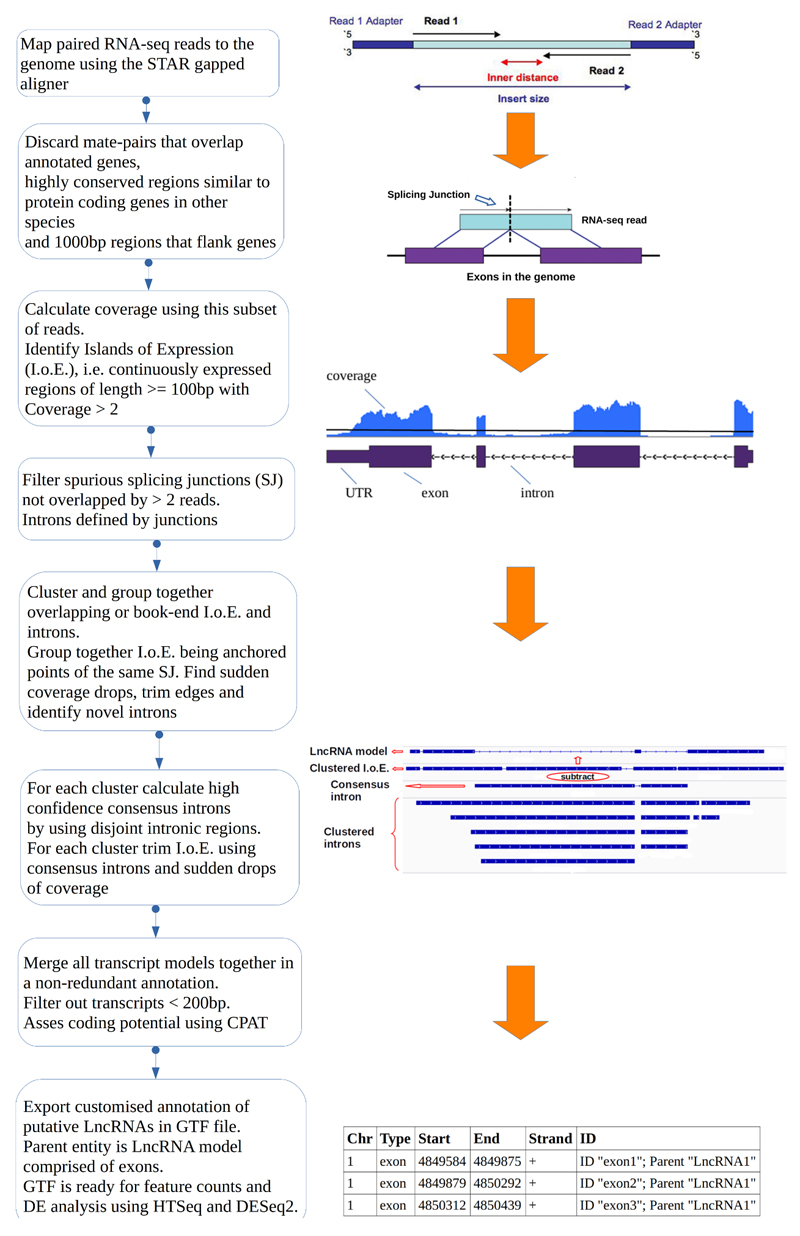
To acquire a complete representation of the non-annotated transcribed regions we intentionally applied a low coverage threshold (> 2 sequence coverage for the region) for at least the length of an RNA-seq read (>=100bp). The cut-off threshold is similar to the one applied in [23]. Then we clustered and trimmed these regions using splicing information from novel SJs identified by at least two RNA-seq read-pairs and a smoothed z-score over a rolling window to identify coverage drops. To predict and analyse LncRNAs on the gene level, we merged together all identified transcripts from individual samples to create a unified set of non-redundant, novel, putative LncRNAs in the form of a gene transfer format (GTF) file. The bioinformatic workflow is illustrated in figure 1, for more details see methods.
Figure 1.
Overview of the computational pipeline used for the identification of novel LncRNAs using RNA-seq data. See Materials and Methods - Identification of novel LncRNAs and Supplementary Methods – Identification of novel LncRNAs.
We then calculated the coding potential and performed transcriptional profiling in order to identify Differentially Expressed (DE) novel and known gene models between animal models of peripheral neuropathy and control (sham surgery) samples. To carry out DE analysis without overestimating fold changes for lowly expressed transcripts we used the analysis software DESeq2 [43] and filtered DE results according to expression levels and consistency. Significance cut-off for DE was adjusted p.value < 0.05 in all cases. We filtered out novel LncRNAs which were not expressed in at least all replicates of a condition or strain or were below an average expression threshold of > 0.5 fpkm in at least one condition. We particularly focused on antisense LncRNAs, i.e. overlapping exonic regions of the gene on the opposite strand, and intergenic LncRNAs, lying on the intergenic space between known gene models. All expression data including antisense and intergenic LncRNAs are available in http://www.painnetworks.org [55], all RNA-seq data (raw data and the whole gene set i.e. novel LncRNAs and annotated genes) are available in Gene Expression Omnibus (GEO) (GSE107182, GSE107180, GSE107181). Supplemental spreadsheets of the complete dataset are available at http://doi.org/10.6084/m9.figshare.6508205.
LncRNAs are known to have relatively poor conservation between species, so we aimed to compare one rat (Wistar) and two mouse strains (BALB/c and B10.D2 strains). RNA-seq quality was good for both experiments (supplementary tables 1-4). Quality was assessed based on: the percentage of uniquely mapped reads (89% for mouse and 72.51% for rat), the number of properly and concordantly paired reads, the average Phred score for read quality (32.2 for mouse and 34.3 for rat), the base calling at the extremities of reads (0.08 for mouse and 0 for rat low Phred scores at the ends of reads), the median of the insert between paired read-mates (192.1 for mouse and 153.4 for rat), and the GC content of reads for all sequencing lanes and samples (48.2% for mouse and 51.4% for rat). Two mouse samples were excluded as they had much lower mapped reads (Sample 59 73.3%), a very high percentage of unmapped reads as they were too short (Sample 59 18.1%), much more reads mapped to multiple loci (Sample 66 and Sample 59) and higher Cook’s distance (supplementary figure 1). Uniquely mapped read pairs were used for downstream analysis. Raw and processed gene expression data are available in Gene Expression Omnibus (GEO).
In total we had on average 64 million uniquely aligned pairs of reads per sample for mouse and 41 million pairs of reads for rat, more than enough both for identification of de-novo LncRNAs and then to ask if they were DE in injury versus sham conditions [20].
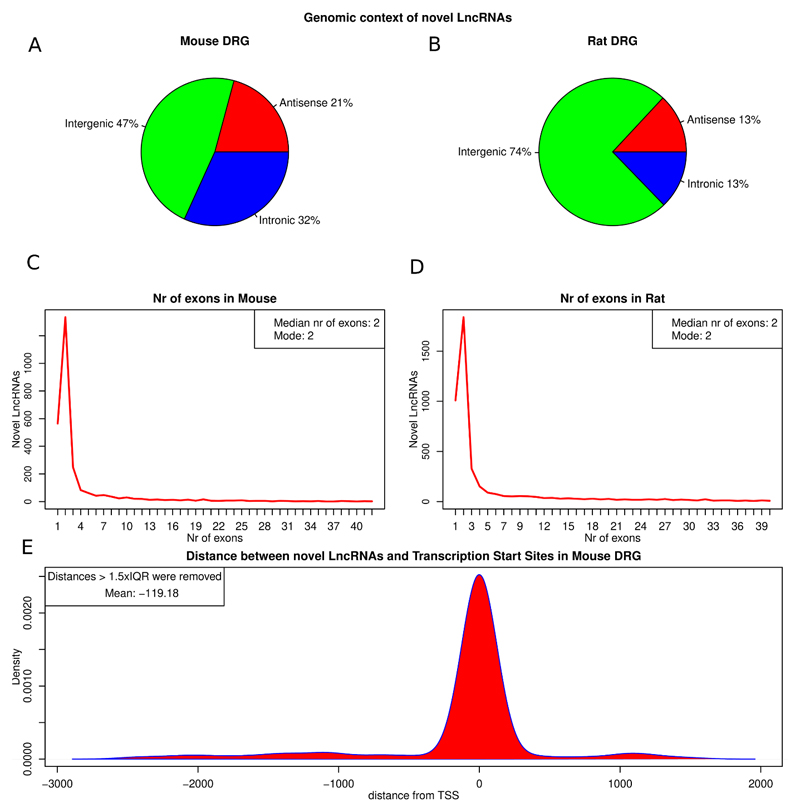
Using this approach we initially identified and reconstructed thousands of non-annotated transcribed loci in mouse and rat DRG with length of more than 200 bp. Then we evaluated whether these novel transcripts were protein coding or non-coding. After coding potential calculation using the coding potential calculator CPAT [74] we obtained 6657 long consistently expressed transcripts classified as non-coding in rat DRG and 4729 in mouse DRG. 4315 of these passed the expression threshold in Rat and 2693 in mouse and were retained for downstream analysis. These long novel transcripts with no coding potential were considered putative novel LncRNAs. The majority of novel LncRNAs were intergenic in both species, with 21%, and 13% antisense (overlapping exonic regions of protein coding genes on the opposite strand) in mouse and rat DRG respectively (figure 2A, B). Most novel LncRNAs were multi-exonic, with a distribution of exons heavily skewed towards bi-exonic transcripts (figure 2C, D). This exon distribution is very similar to GENCODE findings [28].
Figure 2.
Attributes of novel LncRNAs identified in mouse and rat.
A, B: Classification of novel LncRNAs according to genomic context in mouse (A) and rat (B). C, D: Exon distribution of novel LncRNAs in mouse (C) and rat (D). E: Kernel density of distances between novel LncRNAs and 5’ CAGE TSS. Distance is measured in genomic bases. Outlying distances > 1.5 * IQR of the distribution were removed (All data in supplementary figure 2). Median distance of TSS is 0bp of the predicted LncRNA transcript.
The usage of ribo-depleted RNA for the library preparation allowed us to completely sample and interrogate the non-coding transcriptome and led to a relatively high proportion of intronic non-coding transcripts being identified [30].
In order to increase confidence and to gather more evidence regarding the completeness and expression of predicted novel LncRNAs we examined the relationship between their genomic loci and annotated transcription start sites (TSS). To do this we used 5’ CAGE (cap analysis gene expression) TSS data [41] which is available in the mouse. CAGE is a technique for high-throughput identification of sequence tags corresponding to 5' ends of RNA at the cap sites and the identification of the transcription start sites [66]. As TSS data was not available for the mouse reference genome mm10 we translated and mapped coordinates from the mm9 genome to mm10. We then calculated the kernel density of the distance between the TSS that were mapped to mouse genome mm10 (51% of the TSS available for mm9) and a region within 100 bp of the 5’ end of the putative LncRNA similarly to [28]. For both known and novel LncRNAs the kernel density was highest at 0 but with more spread for the novel LncRNAs (figure 2E, supplementary Figure 2). 33.7% of the antisense and intergenic LncRNAs had a predicted TSS on their 5’ end on the same strand of the genome. When we removed the outlying distant TSS—i.e. more than 1.5 times the inter quantile range (IQR) of the distribution—the mean distance of TSS and novel LncRNAs was 119 bp upstream of the predicted transcript. In a GENCODE study [28], 15% of identified LncRNAs in human had a TSS on their 5’ end. These results highlight how close the 5’ end of novel LncRNAs was to experimentally determined TSS. Due to the fact that only a fraction of TSS were mapped to the current mouse genome, this data is likely to be an underestimate. These findings are in concordance with those of [18]. We also note that a significant fraction of novel LncRNAs are either incomplete, not yet annotated extended UTRs or that LncRNAs are indeed independent transcription units arising from TSS that are not yet annotated, possibly due to their low expression level.
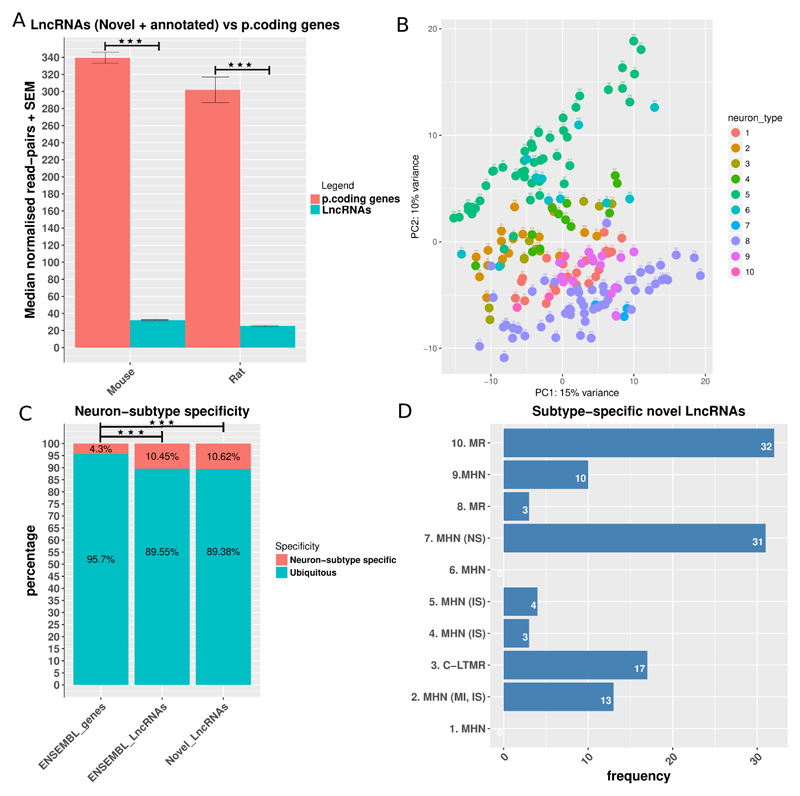
In both species we found that LncRNAs were significantly and consistently expressed at a lower level than protein-coding genes (figure 3A). This ratio of about 10-fold lower expression of LncRNAs to protein coding genes is similar to previous studies [71,78].
Figure 3.
Features of LncRNA’s expression.
A: Median read counts of LncRNAs (novel and ENSEMBL annotated) vs protein coding genes in mouse and rat. Data is presented as mean plus SEM. Significance was assessed with the Mann–Whitney U test (MWW). B: PCA plot of the expression of novel LncRNAs in different DRG neuron-sub types. Neuron subtypes are as follows: 1. MHN, 2. MHN (MI, IS), 3. C-LTMR, 4. MHN (IS), 5. MHN (IS), 6. MHN, 7. MHN (NS), 8. MR, 9. MHN, 10. MR. Axis represent PCs and show percentage of original data’s variance explained by the respective PC (PCA plot of ENSEMBL genes in supplementary figure 3A). C: Neuron subtype specificity (tau > 0.8, average log 2 counts > 3 for at least one neuron sub-type). Enrichment was assessed with the Fisher's Exact Test for Count Data. Kernel density of the Tau specificity metric in supplementary figure 3B. D: Distribution of neuron sub-type specific novel LncRNAs in different neuron subtypes. MHN: mechanoheat receptors, MI: mechanically insensitive, MS: mechanically sensitive, IS: itch-sensitive, C-LTMR: c-fiber low-threshold mechanoreceptors, MN: mechanical nociceptor, MR: mechanoreceptor. P < 0.5 *, p < 0.01 **, P < 0.001 ***.
Nociceptors are a major component of DRG and a number of pain genes are selectively expressed by these neurons. We therefore studied the genomic context of identified LncRNAs and in particular whether they were antisense or in close genomic proximity with known pain genes downloaded from the Pain Genes Database [36]. The Pain Genes Database catalogues genes that have been shown to have an impact on pain-related behaviour in rodents based on transgenic knockout experiments. Out of the 449 genes in the database, we have found 13 novel LncRNAs antisense to pain genes in mouse and 19 in rat (supplementary tables 5 – 6). 23 intergenic LncRNAs had a pain gene as their closest genomic neighbour in mouse and 57 in rat (supplementary tables 7 – 8). Ion channels are key determinants of sensory neuron excitability. We identified LncRNAs antisense to voltage gated sodium channels, potassium channels, calcium channels, chloride channels and TRP channels, within mouse and rat DRG, as shown in supplementary tables 9 and 10.
In general we had modest syntenic conservation (i.e. in equivalent genomic positions) between species. We used synteny portal [38] to retrieve synteny blocks conserved between human (GRCh38.88), mouse (mm10) and rat (rn5) with a resolution of 150000bp. We lifted genomic coordinates from rn5 to rn6 genome and found in total 912 conserved synteny blocks in human and mouse and 443 uniquely mapped in the current rat genome. 800 (18.5%) novel LncRNAs in rat and 1271 (47%) in mouse were in these syntenic blocks conserved between the three species. Moreover 649 LncRNAs in mouse and 782 in rat were in 200 common syntenic blocks between the two species (Supplementary data available at http://doi.org/10.6084/m9.figshare.6508205 - S. Data 1). 509 LncRNAs in rat and 397 in mouse were antisense of the same orthologous genes in mouse and rat (S. Data 2-3).
Different types of DRG neurons selectively express LncRNAs
DRG neurons are heterogenous both in terms of physiology and molecular profile; in order to identify whether the expression of LncRNA may be DRG subtype-specific we re-analysed a published dataset of single cell RNA sequencing data [39] derived from C57BL/6 mouse DRG neurons for expression of the novel LncRNAs which we had identified. The authors had previously generated 10 different DRG neuron subtypes from their analysis of 197 neurons. We found that we could effectively identify transcriptome- and size-based neuronal types based on the selective expression of both ENSEMBL annotated genes and our novel LncRNAs.
Principal component analyses (PCA) of the expression set of novel LncRNAs showed that samples belonging to most of the sensory neuron sub-types were clustered together (Figure 3B), but with a higher spread than for annotated genes (supplementary figure 3A), suggesting that these can be important transcriptional units the expression of which is highly related to the different subtypes of DRG neurons.
We then used the Tau tissue specificity metric [79] to identify highly DRG subtype-specific LncRNAs expressed in mouse DRG. The density plot of the Tau index distribution (supplementary figure 3B and S. Data 4) suggested that the Tau > 0.8 was an appropriate threshold for separating neuron sub-type specific from ubiquitous annotated genes and novel LncRNAs. To consider a gene or LncRNA neuron subtype specific we also imposed a cut-off mean expression threshold of > 3 log2 normalised read pairs across all samples of a neuronal subtype. 87 novel LncRNAs, 119 annotated LncRNAs and 597 protein coding genes were neuron sub-type specific. This reveals a significant (Fisher Exact test p.value < 0.001) enrichment of novel and annotated LncRNAs amongst the neuron sub type specific transcripts (figure 3C). This quantification of the DRG subtype specificity confirms that LncRNAs’ expression pattern is more tissue specific than protein coding genes’. In studying the different DRG subtypes (as defined by Li et al., 2016) we noted that there was an uneven distribution of subtype specific LncRNAs, figure 3D.
IPSC - derived sensory neurons differentially express known and novel LncRNAs compared to their respective IPS cells
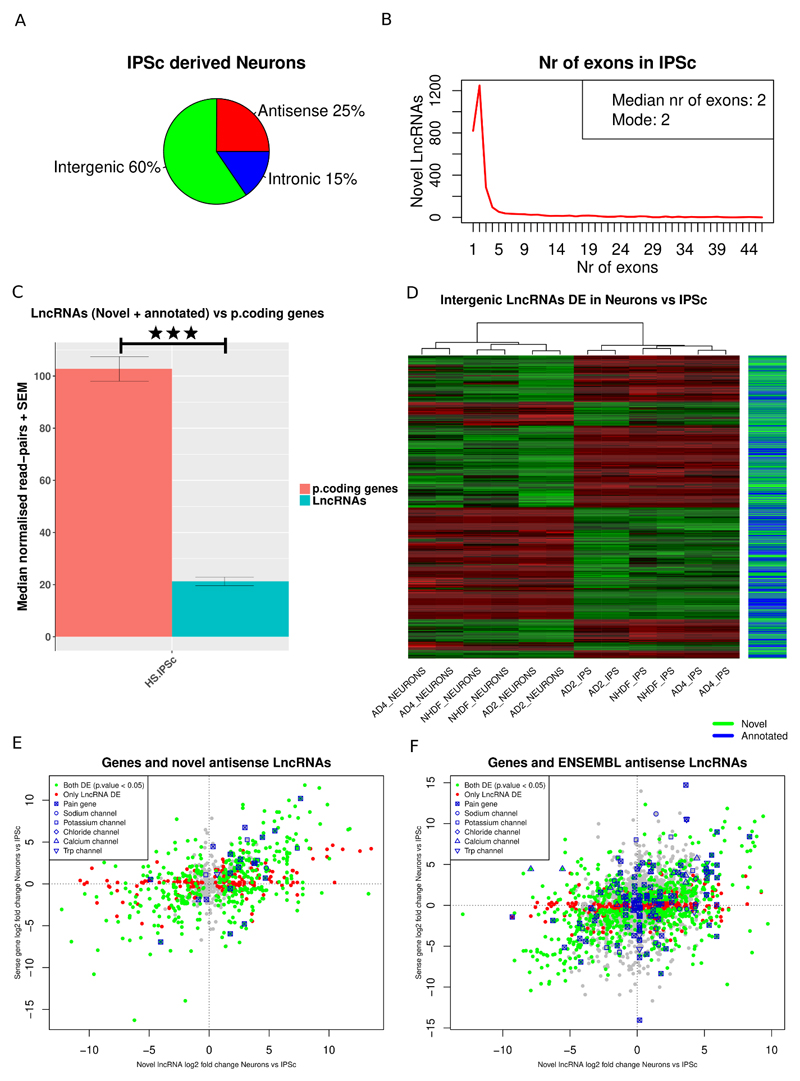
We assessed LncRNA expression in human IPSC derived sensory neurons generated from healthy individuals. We followed a differentiation protocol known to produce neurons with a gene expression profile and functional characteristics that are very similar to rodent sensory neurons [8,12,82]. Virtually all the resulting neurons express the sensory neuron marker Brn3a, project extensively arborized neurites, a subset can be myelinated and exhibit mature electrophysiological characteristics of sensory afferents [8,75]. At least 84% of the ion channel genes known to be expressed in human DRG were shown to be expressed by sensory neurons generated using this protocol [82]. We compared gene expression in IPSC-derived sensory neurons to expression in IPSC parent lines in order to focus on LncRNAs enriched in differentiated sensory neurons (GEO GSE107181, S. Data 5-7). We first interrogated genes annotated in the ENSEMBL GRCh38.88 gene set and we also identified and profiled 2948 novel LncRNAs in human IPSC and IPSC-derived sensory neurons, most of them were intergenic (Figure 4A) and bi-exonic (Figure 4B). Again we found that LncRNAs were significantly lower expressed than protein coding RNAs (Figure 4C). IPSC-derived sensory neurons from three different cell lines (identified as AD2, AD4 and NHDF) were very similar to each other, and considerably different to the IPSC parent lines (supplementary figure 4A). 42% of all expressed genes were DE in IPSC-derived sensory neurons vs IPSCs, which is a remarkable transcriptional change. A total of 6103 annotated LncRNAs (ENSEMBL GRCh38.88) were consistently expressed in at least all samples of either IPSCs or IPSC-derived sensory neurons. A total of 1830 (30%), of them were significantly DE in all three cell lines between IPSC-derived sensory neurons and IPSCs; the majority of these were intergenic (47%). 77% of the expressed novel LncRNAs were significantly DE between IPSC-derived sensory neurons and IPS cells. All three cell lines had in common 371 novel LncRNAs significantly DE.
Figure 4.
Expression patterns of LncRNAs in Human IPSC-derived neurons.
A: Classification of novel LncRNAs according to genomic context. B: Exon distribution of novel LncRNAs. C: Median read counts of LncRNAs (novel and ENSEMBL annotated) vs protein coding genes in human IPSC-derived neurons. Data is presented as mean plus SEM. Significance was assessed with the Mann–Whitney U test (MWW). D: Heatmap of novel and annotated intergenic LncRNAs DE between Human IPSC-derived neurons and IPSC. E, F: Expression plot of novel antisense LncRNAs (E) and annotated ENSEMBL antisense LncRNAs (F) vs the sense protein coding gene. LncRNAs antisense to DE genes Kcnj6 and Trpm3 were DE with opposite log2 fold changes to the DE sense gene. P < 0.5 *, p < 0.01 **, P < 0.001 ***.
In both annotated and novel intergenic LncRNAs, distance was modestly but significantly anti-correlated with expression correlation with their closest genomic neighbour (supplementary figure 4B, C). 2743 intergenic LncRNAs (novel and annotated) were significantly DE between IPSC derived Neurons and IPSC (Figure 4D). 80 annotated and 16 novel LncRNAs were antisense to pain genes (S. Data 8) and 15 annotated and 18 novel LncRNAs had a pain gene as its closest genomic neighbour (S. Data 9). 8 of the LncRNAs antisense to pain genes were significantly DE between IPSC-derived sensory neurons and IPSCs with a significantly DE gene on the opposite strand and anti-correlated expression changes, 1 of them was novel (supplementary table 11). IPSC derived sensory neurons expressed LncRNAs (including both annotated and novel LncRNAs) antisense to TRP, voltage gated sodium, potassium, chloride and calcium ion channels; in some cases these showed the opposite expression changes relative to the sense gene on the opposite strand (Figure 4E,F).
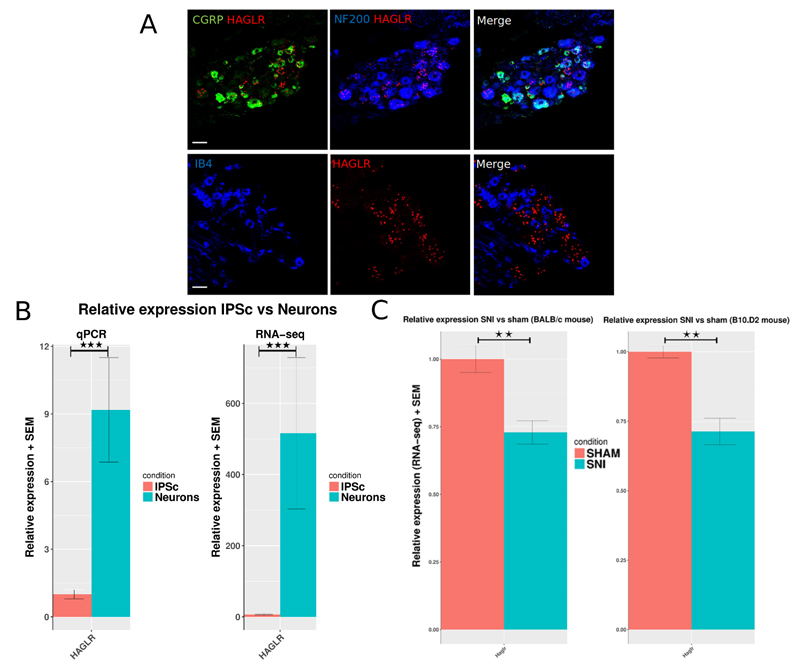
One example of the antisense LncRNAs, the HOXD Cluster Antisense RNA 1 (HAGLR) was further investigated because it was very highly up-regulated in neurons vs IPSC and HoxD genes have been implicated in nociceptor specification [26]. HAGLR was ranked first amongst the significantly DE annotated antisense LncRNAs by its log2 fold change (8.74) and base mean counts (261.5). Differential expression was validated by qPCR (figure 5, supplementary table 12). In-situ hybridization also revealed that it was expressed by mouse DRG cells in vivo and it was found to be significantly down-regulated in mouse DRG following nerve injury (see below as well as figure 5).
Figure 5.
A: In-situ hybridisation for Haglr LncRNA (mouse ortholog of human HAGLR) shows expression in mouse WT L4 DRG. The ISH signal was developed using a fast red reaction. From right to left: Representative images of mouse DRG sections stained for Haglr (red) and NF200 (blue), CGRP (green), and IB4 (blue). Scale bar 50µm. B: Quantification of expression change of HAGLR in human IPSC vs IPSC-derived neurons. Relative expression assessed by qPCR of HAGLR LncRNA. C: RNA-seq determined relative expression in SNI vs Sham BALB/c and B10.D2 mice DRG. Data is presented as mean plus SEM. P < 0.5 *, p < 0.01 **, P < 0.001 ***.
We also observed similar extent of syntenic conservation between human, mouse and rat in novel LncRNAs identified in human IPSC and IPSC-derived sensory neurons. 1312 (44.5%) were in syntenically conserved blocks between the three species. 459 LncRNAs in human IPSC and 495 in mouse were in 126 common syntenic blocks between the two species (S. Data 10) and 522 were antisense orthologous genes in human and mouse (S. Data 11).
Human DRG eQTLs (expression quantitative trait loci) have recently been identified and associated with pain phenotypes [53]. We interrogated this dataset for overlaps with novel and annotated LncRNAs expressed in IPSC-derived sensory neurons as this may provide an underlying mechanism through which a SNP at this site may impact on gene expression. We only considered overlaps valid if they were found on exons of the respective transcript. We found that 4 annotated LncRNAs (3 antisense, 1 intergenic) which had DRG eQTLS were expressed in IPSC-derived sensory neurons, and also that 5 novel LncRNAs were overlapping DRG eQTLS (table 1).
Table 1. Annotated and novel LncRNAs overlapped by eQTLs.
Reported p.values are FDR adjusted.
| ENSEMBL annotated LncRNAs | ||||||||
| Intergenic LncRNA | ||||||||
| LncRNA ID | LncRNA symbol | eQTL | Close/Sense Gene symbol/ID | Log2 FoldChange LncRNA | LncRNA p.value | Log2 FoldChange gene | Gene p.value | |
| ENSG00000253641 | LINCR-0001 | rs11782819 | ENSG00000272505 | -0.029 | 0.97 | 0.10 | 0.9 | |
| Antisense LncRNAs | ||||||||
| ENSG00000268516 | LOC105372482 | rs260461 | ZNF544 | 1.36 | < 0.001 | -0.5 | 0.01 | |
| ENSG00000247809 | NR2F2-AS1 | rs1437588 | NR2F2 | 2.93 | 0.04 | 10.02 | < 0.001 | |
| ENSG00000234456 | MAGI2-AS3 | rs7802883 | MAGI2 | 4.78 | < 0.001 | -0.045 | 0.79 | |
| Novel LncRNAs | ||||||||
| Intergenic LncRNAs | ||||||||
| LncRNA1562 | 6:30528975-30529861(-) | rs2534823 | GNL1 | 6.95 | < 0.001 | 0.3 | 0.001 | |
| Antisense LncRNAs | ||||||||
| LncRNA5059 | 4:173079010-173163269(-) | rs4419455 | ENSG00000241652 | -3.5 | <0.01 | NA | NA | |
| LncRNA5322 | 6:29808275-30003297(+) | rs9260408 | ZNRD1ASP | 1.5 | < 0.001 | -0. | 0.2 | |
| LncRNA1444 | 6:32183289-32184686(+) | rs1800624 | AGER | -0.23 | 0.7 | -1.86 | < 0.001 | |
| LncRNA5453 | 6:32646756-32699597(-) | rs17205373 | HLA-DQA1 | 1.01 | 0.21 | 1.6 | 0.2 | |
Expression profiling of LncRNAs following nerve injury
We then assessed differential expression of LncRNAs within DRG following nerve injury both in rat and mouse. We used the spared nerve injury (SNI) model in mice and the spinal nerve transection (SNT) model in rats. We used two different mouse strains: BALB/c and B10.D2 which have previously been shown to develop different levels of mechanical hypersensitivity following nerve injury [68].
Transcriptional changes of LncRNAs in Rat DRG
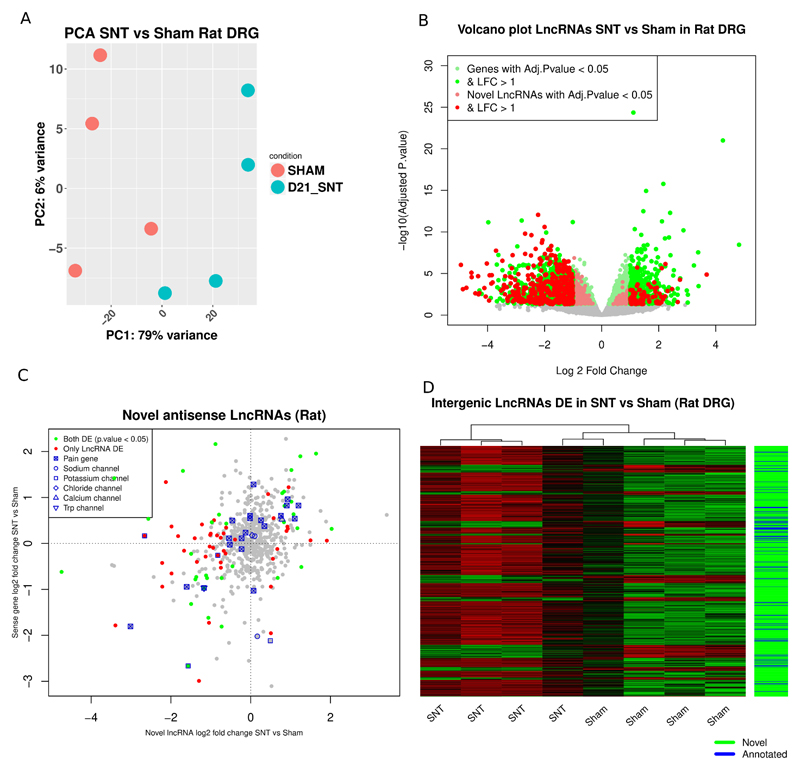
We confirmed that the expression patterns of the top 5000 ENSEMBL annotated genes and novel LncRNAs ranked by their standard deviation (SD) could separate samples according to condition (figure 6A). We also observed a significant transcriptional response after nerve injury in rat DRG which amounts to 25.5% (4215 ENSEMBL annotated genes + novel LncRNAs) of all expressed genes (figure 6B, S. Data 12). There was a significant enrichment of Biological Process (BP) Gene Ontology (GO) terms related to ion channel transport, signal transduction and response to mechanical stimulus in the ENSEMBL annotated DE genes (supplementary figure 5). Novel LncRNAs were a substantial component of the DE genes following peripheral nerve injury (highlighted in figure 6B).
Figure 6.
DRG transcriptional response in rat following peripheral neuropathy.
A: PCA plot of samples based on the regularised log2 transformed counts of novel LncRNAs and ENSEMBL genes (1st 5000 genes ranked by their standard deviation) in rat DRG. Axis represent PCs and show percentage of original data’s variance explained by the respective PC. B: Volcano plot of the whole gene set (ENSEMBL annotated genes and novel LncRNAs). X-axis Log2(Fold Change), y-axis -log10(FDR adjusted p.value). Significantly DE ENSEMBL annotated genes and novel LncRNAs are highlighted. C: Expression plot of novel antisense LncRNAs vs the sense protein coding gene. D: Heatmap of novel and annotated intergenic LncRNAs DE between SNT and Sham operated animals.
541 out of the 3169 annotated LncRNAs in rat (ENSEMBL Rnor_6.0.92) were expressed in DRG. Out of these 82 (16%) were significantly DE. From the 4316 putative novel LncRNAs 708 (16.4%) were significantly DE in rat (PainNetworks, GEO GSE107180). Out of the 629 novel antisense LncRNAs 253 (40%) had an anti-correlated expression pattern to the sense gene following spinal nerve transection (S. Data 13). In total 31 reach significance with a significant DE gene on the opposite strand. 9 of them were significantly DE on the opposite strand of a significantly DE pain gene with opposite log fold changes (supplementary table 13). There were novel LncRNAs antisense of pain genes, voltage gated potassium and sodium channels (Figure 6C).
654 intergenic LncRNAs were found to be significantly DE in SNT vs sham, 575 of them were novel (Figure 6D, S. Data 14). 5 of them (4 novel and one annotated) were adjacent to and highly correlated with pain genes (supplementary table 14). Most intergenic LncRNAs were positively correlated with their closest genomic neighbour. Also the closer they were to their closest neighbour the stronger the correlation (supplementary figure 6), both for annotated and novel LncRNAs.
Transcriptional changes of LncRNAs in Mouse DRG
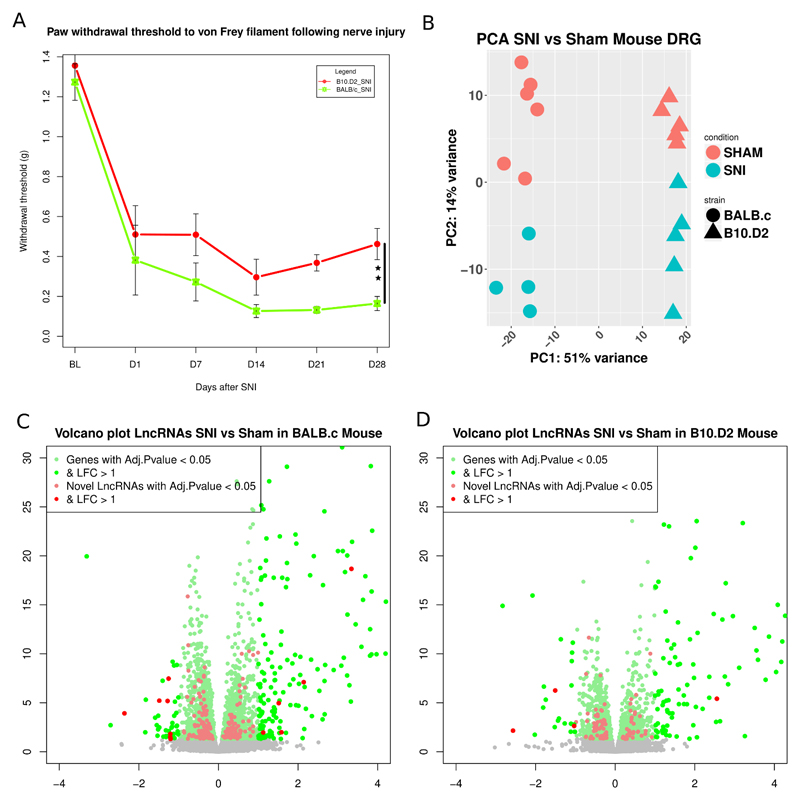
Withdrawal thresholds to Von Frey filament stimulation confirmed previous findings that the B10.D2 demonstrate less mechanical pain related hypersensitivity following SNI than BALB/c [68] (figure 7A). There were no significant behavioural differences between male and female mice for either strain. Principal components analysis of the expression of the top 10000 ENSEMBL annotated genes and novel LncRNAs (ranked by SD) showed that they were able to optimally separate mouse samples according to sex, strain and within them, condition (figure 7B).
Figure 7.
DRG transcriptional response in mouse following peripheral neuropathy.
A: Hindpaw withdrawal thresholds to von Frey filament stimulation + SEM in grams. We calculated the area over the curve (AOC) for each strain and obtained the percentage of maximum induced hypersensitivity. Two way ANOVA showed a significant effect of surgery and a significant interaction of strain:surgery (p= 0.001). One-way ANOVA between SNI groups on D28 showed significant difference in % of maximum hypersensitivity (p=0.002). Black bar shows comparison between SNI groups, p < 0.01 : **. N=12 per strain, N=6 per SNI group. B: PCA plot of samples based on the expression of novel LncRNAs and ENSEMBL genes (1st 10000 genes ranked by their standard deviation) in mouse DRG. Axis represent PCs and show percentage of original data’s variance explained by the respective PC. C, D: Volcano plots of the whole gene set for BALB/c strain (C) and B10.D2 strain (D). X-axis Log2(Fold Change), y-axis -log10(FDR adjusted p.value). Significantly DE ENSEMBL annotated genes and novel LncRNAs are highlighted.
Volcano plots show the range of transcriptional changes of ENSEMBL annotated genes and novel LncRNAs in mouse after peripheral nerve injury (figure 7C, D, S. Data 15). Novel LncRNAs were a substantial component of the DE genes following the SNI pain model (highlighted in figure 7C, D)
Regarding the BALB/c mouse strain, which showed significantly more pain-related hypersensitivity from day 7 onwards than the B10.D2 strain, we found significantly more DE ENSEMBL annotated genes and novel LncRNAs, 2750 compared to 1441. In comparison to rat we found less DE genes in the mouse following nerve injury. SNT is a more severe injury model than SNI (in which less sensory neurons are axotomised and the injury is more distal) and so this may reflect model severity rather than species differences.
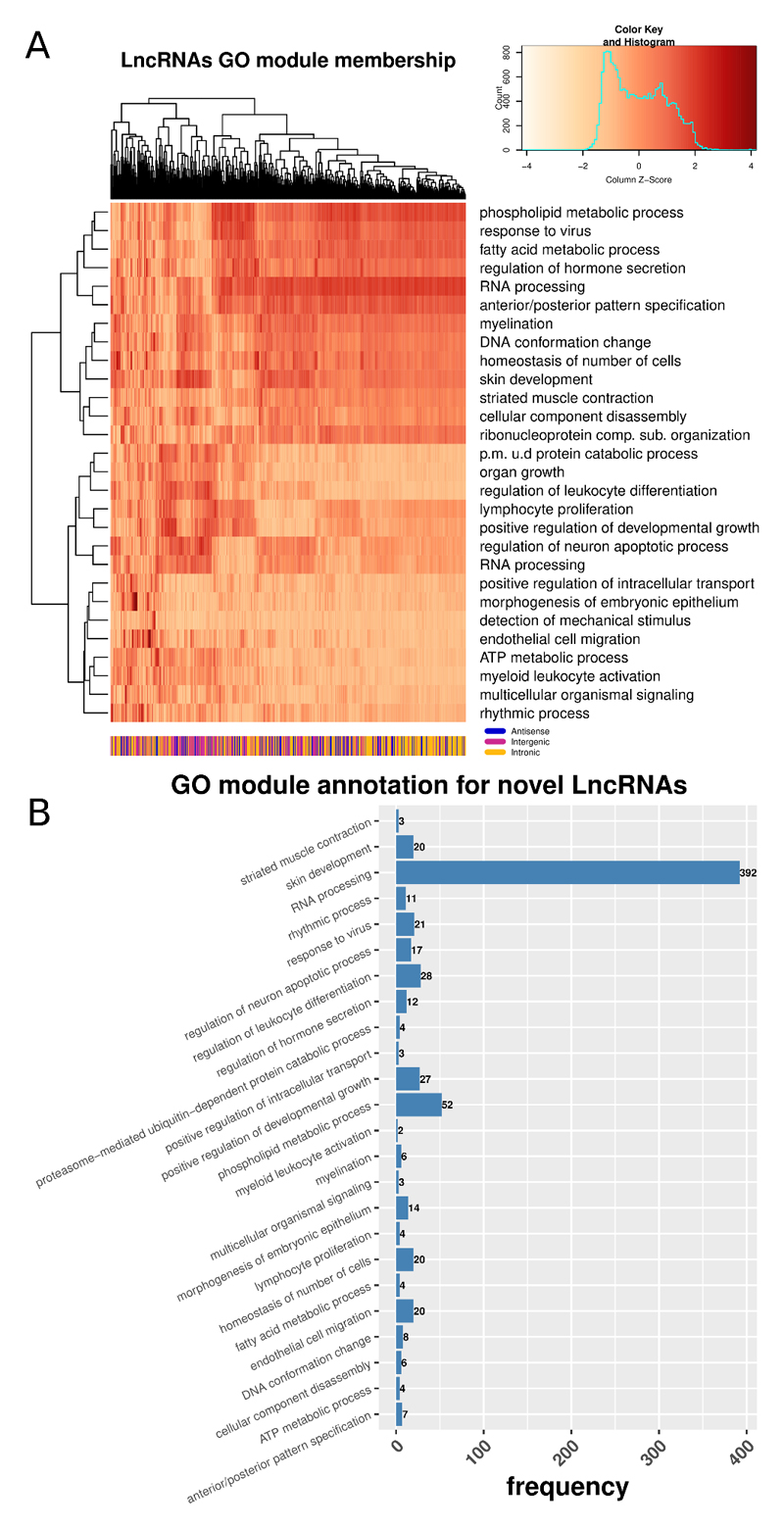
In those ENSEMBL annotated genes that were DE we found significant GO enrichment in terms related to the nervous system, regulation of excitability, signal transduction and response to injury (supplementary figure 7). As novel LncRNAs were not assigned with GO terms we used their expression profiling context, in an unbiased way, to gather more insights regarding their possible functional importance. Under the assumption that they may regulate gene expression either in-cis or in-trans we further studied them in the context of modules of closely co-expressed genes and associated them with enriched GO BP terms. We first created a weighted gene co-expression network (WGCNA) [37] and then identified modules of highly co-expressed ENSEMBL annotated genes and novel LncRNAs (scale independence and dendrogram of merged/um-merged modules in supplementary figure 8). The absolute weighted bi-correlation across all samples (n = 20) was used to construct the network. An n > 15 is needed to robustly calculate expression correlations. We also calculated the representative gene (i.e. eigengene) for each module, defined as the first principal component of the expression of all member genes. We then quantified the novel LncRNAs module membership by calculating the robust weighted bi-correlation of the LncRNAs with these eigengenes. Next we performed a GO enrichment analysis and annotated each module with its top enriched BP term, based on an over-representation analysis of the annotated genes. The strength of module membership for novel LncRNAs (Figure 8A) shows highly correlated modules of LncRNAs associated with RNA-processing and some related to the nervous system, signalling and regeneration. The vast majority of novel LncRNAs were in the module associated with RNA-processing (Figure 8B).
Figure 8.
Network analysis of annotated genes and novel LncRNAs.
A: Heatmap of module membership for novel LncRNAs. Module membership quantified with absolute bi-correlation. Colours represent z-values of absolute bi-correlation. B: Distribution of novel LncRNAs in modules enriched with GO terms of Biological Process (BP).
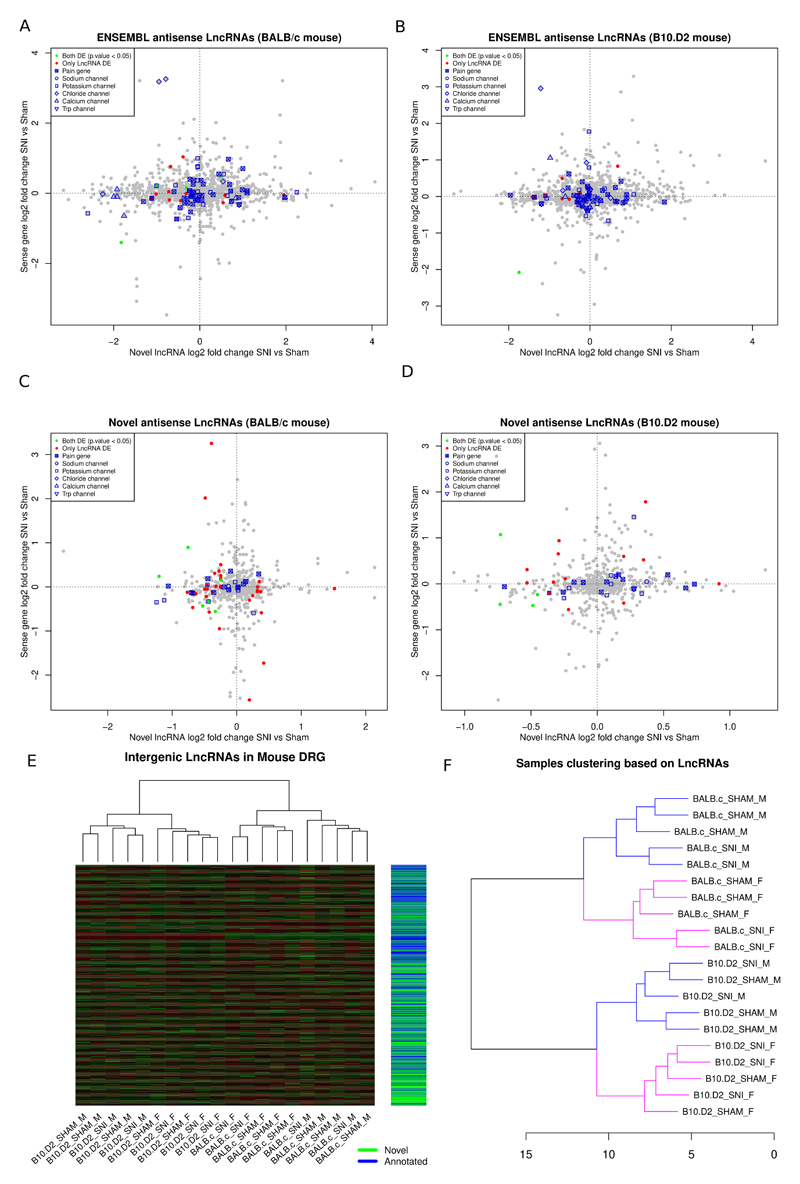
Out of the 7990 annotated ENSEMBL (GRCm38.87) LncRNAs, 2406 were expressed in mouse DRG. A total of 296 LncRNAs were significantly DE in BALB/c strain SNI vs. sham, 193 of them were novel. In B10.D2 strain 146 LncRNAs, 97 of which were novel, were significantly DE (PainNetworks and GEO GSE107180). Although the absolute numbers are much smaller in comparison to annotated genes, percentages of significantly dysregulated transcripts are similar. Most of them were intergenic (S. Data 16) and antisense (S. Data 17).
40% of the 1776 annotated antisense LncRNAs in mouse had an anticorrelated expression pattern with their sense gene (Figure 9A and B). 44.8% of the novel antisense LncRNAs had opposite fold changes in comparison with their protein coding gene on the opposite strand (Figure 9C and D). Some demonstrated the opposite expression pattern to their sense gene following nerve injury (Figure 9 A-D, supplementary table 15). The significantly DE LncRNAs antisense to sodium and potassium channels are in supplementary table 16. The majority of LncRNAs antisense to potassium channels were down-regulated after nerve injury and between IPSC-derived neurons and IPSC. On the other hand, although the LncRNA antisense to Scn9a was down-regulated after nerve injury, all other LncRNAs antisense to sodium channels were up-regulated in IPSC-derived neurons vs IPSC. In some cases these showed an opposite expression pattern to their sense gene.
Figure 9.
Expression patterns of LncRNAs in mouse DRG.
A, B: Expression plot of annotated ENSEMBL antisense LncRNAs in BALB/c mouse (A) and B10.D2 mouse (B). C, D: Novel antisense LncRNAs vs the sense protein coding gene in BALB/c (C) and B10.D2 (D) strains. Novel LncRNAs antisense to DE genes Inpp1, Epyc, Kcnmb1, Nefl, Nalcn, Nbea, Ttc39aos1, Cgref1, Zyg11b are significantly DE. E: Heatmap of novel and annotated intergenic LncRNAs DE between SNT and Sham operated animals. F: Hierarchical samples’ clustering based on the expression of ENSEMBL annotated and novel LncRNAs in mouse. Counts were transformed using the regularized log2 transformation, euclidean distance was used as a dissimilarity measure and complete linkage was used for clustering. Male samples are in blue and female in pink colour.
In total 2365 intergenic LncRNAs were expressed in mouse DRG, 1282 of them were novel and 126 were significantly DE in mouse SNI vs Sham (Figure 9E). Similarly to rat and human, intergenic LncRNAs showed positive correlation with their adjacent gene (supplementary figure 9). 23 novel and 18 known intergenic LncRNAs were adjacent to pain genes in mouse. 2 of these novel intergenic LncRNAs were significantly DE and highly correlated with their adjacent pain gene (supplementary table 17). In some cases multiple novel LncRNAs related to coding genes in the same pain-related signalling pathway could be identified and were DE in mouse and rat. For instance two intergenic LncRNAs upstream of the opioid receptor genes, Oprl1 and Oprd1, were significantly DE and highly correlated with their adjacent significantly DE gene in both species. Oprl1 and Oprd1 form heterodimers and appear in the same network of highly correlated genes [55].
We also performed unsupervised clustering of samples based on the expression of all novel and annotated LncRNAs. We observed a pattern of highly sex-specific expression changes within strains and then, within sex, we had separation according to condition (Figure 9F). This indicated that LncRNAs are dysregulated following peripheral nerve injury in a sex and strain dependent manner.
We selected and validated 7 representative novel LncRNAs based on the expression strength, the significance and size of the effect (log2 fold change) and the genomic context. These novel LncRNAs were all found to be significantly DE in mouse DRG following nerve injury and amongst these are antisense and intergenic, table 2 (primers in supplementary table 18, primer binding in supplementary figure 10).
Table 2. Representative qPCR validated novel LncRNAs.
Base Mean column holds average library size normalised counts across sham samples.
| BALB/c mouse | B10.D2 mouse | |||||
|---|---|---|---|---|---|---|
| BaseMean | Log2 FoldChange | FDR adjusted p.value | Log2 FoldChange | FDR adjusted p.value | Genomic context | |
| LncRNA2754 | 194.6 | -0.53 | 0.002 | -0.49 | 0.006 | Antisense of Nefl (Neurofilament Protein, Light Chain) |
| LncRNA1528 | 112.6 | -0.5 | 0.007 | -0.45 | 0.02 | Antisense of Htra1 (High-Temperature Requirement A Serine Peptidase 1) |
| LncRNA1779 | 615.0 | -0.42 | 0.001 | -0.44 | < 0.001 | Intergenic upstream to Scn4b (Sodium Voltage-Gated Channel Beta Subunit 4) |
| LncRNA1291 | 79.96 | -0.07 | 0.96 | -2.57 | 0.007 | Intergenic downstream to Lrrc4 (Leucine Rich Repeat Containing 4 – related to axon guidance) |
| LncRNA4834 | 413.3 | 0.77 | < 0.001 | 0.29 | 0.04 | Intergenic upstream to Cdc7 (Cell Division Cycle 7) |
| LncRNA4714 | 445.4 | -0.77 | < 0.001 | -0.66 | < 0.001 | Intergenic upstream to Oprd1 (Opioid Receptor Delta 1) |
| LncRNA561 | 76.8 | 0.56 | 0.007 | -0.05 | 0.91 | Intergenic |
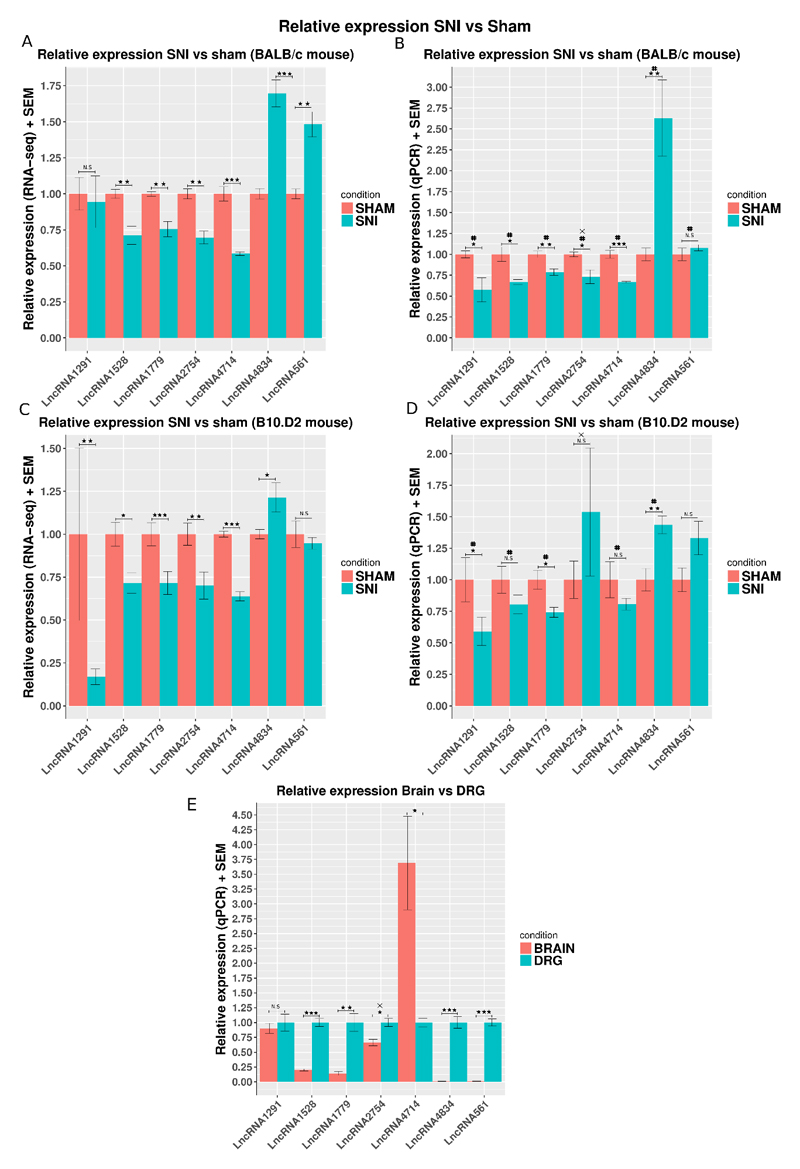
QPCR confirmed the expression of these novel LncRNAs in mouse DRG and validated their dysregulation following nerve injury, Figure 10. With the exception of 1 LncRNA these all demonstrated higher expression in DRG compared to brain (figure 10E). This comprehensive study [48] showed that upstream of genes where these LncRNAs lie there were no previously unannotated lengthened 3' UTRs. With the exception of LncRNA4714, upstream of Oprd1, where the multi-exonic transcript we have identified is much longer than any predicted elongated UTR. When studying the changes in LncRNA expression evoked by SNI in B10.D2 and BALB/c mouse strains there was in general good agreement between RNA-seq and qPCR findings. We also found that in all cases, where RNA-sequencing showed significant dysregulation, qPCR confirmed the result (Figure 10).
Figure 10.
Relative expression of 7 novel LncRNAs in SNI vs Sham mouse DRG assessed by qPCR and RNA-seq.
A, C: Relative expression assessed by RNA-seq. RNA-seq counts were normalised by the sham average and the effective library size using DESeq2. Significance as obtained by DESeq2 using the following GLM ~ sex + strain*condition for the whole gene set of ENSEMBL annotated genes and novel LncRNAs, N = 10 per strain (6 Sham – 4 SNI BALB/c, 5 Sham – 5 SNI B10.D2). B, D: Relative expression assessed by qPCR. Expression was normalized against the average expression in Sham. Significance was obtained using a one-way ANOVA and the linear model ~ sex + condition, N= 10 per strain.E: Relative expression of 7 novel LncRNAs in Brain vs DRG. Expression was measured by qPCR using the delta delta CT method. Expression was normalized against the average expression in brain. Data is presented as mean plus SEM. P < 0.5 *, p < 0.01 **, P < 0.001 ***, same direction of change between RNA-seq and qPCR #. X indicates strand specific RT-PCR, N = 8 for B10.D2 (5 Sham, 3 SNI).
Discussion
We used high-depth RNA sequencing and a dedicated bioinformatic pipeline to identify thousands of known and putative novel LncRNAs expressed in mouse and rat DRG and Human IPSC-derived sensory neurons. Many of these LncRNAs were antisense or adjacent to known pain and ion channel genes. A significant proportion demonstrated selective expression in DRG neuron subtypes. Novel LncRNAs were DE following peripheral nerve injury in a species- and strain-dependent manner, including novel antisense LncRNAs with opposite transcriptional changes to their sense genes and intergenic LncRNAs highly correlated with their adjacent gene. Thus, LncRNAs that have been a relatively unexplored part of the DRG transcriptome demonstrate remarkable complexity in terms of their relationship to genes known to impact on sensory function, DRG cell type-specific expression and the transcriptional response to nerve injury.
LncRNAs have been shown to be: smaller than protein-coding genes, spliced, bi-exonic, transcribed as independent transcription units, expressed at a lower level than protein coding genes, and show highly spatially constrained correlation with their antisense and adjacent genes [18,71,83]. These characteristics are highly consistent with the antisense and intergenic LncRNAs that we identified in this study as being expressed in DRG.
No previous attempts have comprehensively determined LncRNA expression within the DRG. Previous studies have undertaken a candidate gene approach [33,88], while others have used a high-throughput approach such as RNA-seq but have only interrogated the expression of previously annotated LncRNAs [25,31,77]. As we have shown, this strategy will miss thousands of unannotated and potentially important LncRNAs. In fact, 8% of genes within the Pain Genes Database in the mouse, 16% in the rat and 7% in human IPSC were found to have an antisense or neighbouring intergenic LncRNA. Furthermore LncRNAs antisense to ion channels were not limited to KCNA2 and SCN9A, but could also be identified to other sodium, potassium, calcium and also TRP channels, all of which have key roles in modulating sensory neuron function. As with all computational predictions, this study has the limitation of the possible inclusion of false positives in our list of novel LncRNAs however we have validated the expression of a number of novel LncRNAs using the independent technique of qPCR.
It is known that LncRNAs can be highly tissue- and cell type-specific [58,70]. DRG cells are heterogeneous in terms of their physiology, anatomy and connectivity; recent single-cell analysis shows how gene expression relates to such specialised features, but also reveals even greater complexity in DRG subtypes based on molecular profiling [39,72]. We showed that both novel and annotated LncRNAs demonstrate selective expression within sensory neuron subtypes. As such, identification of LncRNAs may help in the molecular classification of DRG cells. Moreover, both novel and annotated LncRNAs were significantly enriched in neuron-subtype specific genes versus protein coding genes. The functional specialisation of different neuron subtypes will principally arise due to the neuron subtype-dependent expression of protein-coding genes however such expression may be sculpted by LncRNAs.
We have also defined LncRNAs expressed in human IPSC-derived sensory neurons. Advantages of studying LncRNA expression in this model (rather than, e.g., cadaveric human DRG) is the ready source of high-quality RNA from a pure sensory neuronal population (with very few contaminating Schwann cells) and the ability to compare expression to the parent IPSC line. Disadvantages include the fact that although sub-populations of sensory neurons exist in these cultures, these are not as diverse as native DRG cells and are less mature given the lack of target interactions.
We identified almost 2000 previously annotated and over 2000 novel putative LncRNAs, which were DE when comparing IPSC-derived sensory neurons and parent IPSC lines. These demonstrated many characteristics of the LncRNAs expressed in rodent DRG. 29% of genes in the Pain Genes Database had a potentially relevant relationship to a novel or annotated LncRNA either antisense LncRNAs on the opposite strand or adjacent to an intergenic LncRNA. Interestingly, some of these LncRNAs may have a role in shaping the expression of the sense pain gene during differentiation, as both novel and annotated, DE antisense LncRNAs demonstrated anti-correlated expression changes to their DE sense pain gene. We also identified human LncRNAs which were antisense to voltage-gated ion channel genes. It has been shown that many LncRNAs overlap Genome-Wide Association Studies (GWAS) traits and LncRNAs overlapping trait-associated single nucleotide polymorphisms (SNPs) are specifically expressed in cell types relevant to the traits [29]. The identification of human LncRNAs expressed in IPSC-derived sensory neurons may therefore enable investigation of the genetic basis of chronic pain states in humans, especially under conditions of neuropathic pain wherein maladaptive plasticity in the DRG is an important pathophysiological driver. A recent study has mapped eQTLs in human DRG [53], and a number of SNPs that impacted upon gene expression were found to be within annotated LncRNAs. In our data 9 LncRNAs expressed in IPSC-derived sensory neurons were overlapped by eQTLs. 3 of the novel and 3 of the known were also DE in IPSC-derived neurons vs IPSC. DRG eQTLs identified by [53] were found among hits in numerous GWAS. Such studies highlight the utility in identifying LncRNAs, in this case by providing explanatory power as to how a SNP linked to complex disorders may impact on gene expression and phenotype. As more tissue becomes available for RNA-seq it will also be possible to extend LncRNA discovery to post-mortem human DRG [61].
Nerve injury is known to elicit remarkable transcriptional changes within DRG, which has deleterious consequences such as neuropathic pain, but in certain contexts can also be adaptive; e.g. by promoting nerve repair [52]. We investigated the expression of LncRNAs in Wistar rat following SNT and in two different mouse strains following SNI. The BALB/c strain was previously shown to develop greater levels of mechanical hypersensitivity following SNI versus the B10.D2 strain [68]. Hundreds of LncRNAs were DE in both rat and mouse following nerve injury. We found that more protein-coding genes, annotated LncRNAs and novel LncRNAs were DE in the high pain BALB/c strain compared to the low pain B10.D2 strain. Most of the variance of the LncRNAs expression in our dataset was between strains and not conditions; namely, LncRNAs demonstrated strain-dependent expression plasticity after nerve injury. 27% of LncRNAs were significantly DE between mouse strains compared to 12.2% for ENSEMBL protein coding genes. Considering the rapidly increasing literature on sex differences in pain processing [49,67] that there were sex-dependent effects on LncRNA expression.
Weighted gene co-expression network analysis revealed that novel LncRNAs in the mouse DRG were in network modules related to RNA-processing and some of them in modules related to myelination, development and regeneration of the nervous system, immune response and signalling. This could indicate that these LncRNAs function together with genes acting as transcriptional regulators associated with post-transcriptional modifications. This enrichment is different from that observed in the ENSEMBL annotated gene set, where terms related to axon guidance, ion channel transport, regulation of synapse assembly and of neuron apoptotic process, nervous system development and memory were significantly enriched. These findings are similar to known biological processes enriched after nerve injury [85,86] and in pain genes [42].
Enrichment in biological processes of splicing, mRNA processing and polyadenylation (i.e. parent, child and related terms to RNA-processing) have however been reported [84,85]. Our finding that the expression of novel LncRNAs changes together with annotated genes regulating RNA-processing is consistent with known mechanisms by which LncRNAs alter gene expression (discussed below).
LncRNAs can regulate expression in cis and in trans [34], but the genomic context of LncRNAs is important for both antisense transcripts regulating the gene on the sense strand, or intergenic LncRNAs. Intergenic and antisense LncRNAs, which tend to lie in genomic regions populated with genes [32], have correlated expression patterns to their adjacent genes and may regulate gene expression (Figure 4; [59]). We found that the shorter the distance the stronger the correlation in all species, both for known and annotated LncRNAs. However, the network analysis we carried out allowed us to identify in an unbiased way modules of highly correlated genes and LncRNAs across all samples. Thus these LncRNAs are putative both in-cis and in-trans regulators. Regarding the relation of LncRNAs with their genomic context we found that more than 45% of antisense LncRNAs had anti-correlated - opposite expression changes in respect to the sense gene, while only 10% of intergenic LncRNAs had negative correlation to the expression of their closest genomic neighbour. 12 pairs of antisense LncRNA/sense gene with opposite LFCs reached significance between pain models and control animals. These antisense LncRNAs fit into the paradigm of Kcna2 antisense [88] which silences the gene on the opposite strand. LncRNAs are also known for regulating clusters of imprinted genes or close genes like the Hoxd cluster [44,80]. One of these LncRNAs, HAGLR, was the most up-regulated and stronger expressed LncRNA in Human IPSC-derived neurons and was also found by in-situ hybridization to be expressed in mouse DRG and significantly down-regulated in both mouse strains after nerve injury (supplementary table 12).
We used qPCR to validate the expression of a number of novel LncRNAs in close genomic relationship to protein coding genes of relevance to sensory function: A novel LncRNA antisense of Nefl gene implicated in the Charcot-Marie-Tooth (CMT) disease [87], and an intergenic LncRNA close to and highly correlated with the pain gene Oprd1 were found DE and validated with qPCR. We also describe a novel intergenic LncRNA in close proximity to the Lrrc4 gene that relates to axon guidance and synapse organisation [17,76] and finally, a significantly DE intergenic LncRNA was validated close to the voltage gated sodium channel subunit Scn4b. The majority of LncRNAs validated by qPCR were found to be more highly expressed in DRG than brain. LncRNAs are putative therapeutic agents that could regulate the expression of target genes related to disease [73]. Although application of such therapeutics to pain would need to overcome the hurdle of delivering therapeutics to DRG cell bodies.
In summary, we have provided a resource, in which we have defined LncRNA expression within DRG across species. We show that marked changes in LncRNA expression occur following nerve injury and during sensory neuron differentiation. LncRNAs expression is DRG subtype-specific, and there is often highly spatially-constrained correlation/anti-correlation with their antisense and adjacent genes.
Supplementary Material
Acknowledgements
The project was supported by a strategic award from the Wellcome Trust (Ref. 102645); DLB is a Wellcome senior clinical scientist (202747/Z/16/Z). A.A.-M., S.B.M., D.L.H.B., J.L., and C.O. were part of the Europain Collaboration, which has received support from the Innovative Medicines Initiative Joint Undertaking, under grant agreement no 115007, resources of which are composed of financial contribution from the European Union’s Seventh Framework Programme (FP7/2007-2013) and EFPIA companies’ in kind contribution. The research leading to these results has also received support from the Innovative Medicines Initiative Joint Undertaking under grant agreement no. 115439, resources of which are composed of financial contribution from the European Union’s Seventh Framework Programme (FP7/2007-2013) and EFPIA companies’ in kind contribution. DLB is a member of the DOLORisk consortium funded by the European Commission Horizon 2020 (ID633491). JSM and DLB received a collaborative funding award from the Oxford-McGill-Zurich partnership.
Footnotes
Data availability
Data is in GSE107182 super series which consists of GSE107180 (rodents DRG) and GSE107181 (human IPSC). Splicing junctions, DE data analysis results and GTF files with annotations used are included as downloadable supplementary files of the GEO series GSE107180 and GSE107181. Supplemental data spreadsheets are available at http://doi.org/10.6084/m9.figshare.6508205.
IGV genome browser tracks for novel LncRNAs are publicly available.
Mouse: https://storage.googleapis.com/lncrnatracks/Mouse_DRG_LncRNAs/Novel_lncRNAs_mouse_DRG.gtf (from IGV version 2.4x)
gs://lncrnatracks/Mouse_DRG_LncRNAs/Novel_lncRNAs_mouse_DRG.gtf (until IGV version 2.3x)
Rat: https://storage.googleapis.com/lncrnatracks/Rat_DRG_LncRNAs/Novel_lncRNAs_rat_DRG.gtf (from IGV version 2.4x)
gs://lncrnatracks/Rat_DRG_LncRNAs/Novel_lncRNAs_rat_DRG.gtf (until IGV version 2.3x)
Human IPSC: https://storage.googleapis.com/lncrnatracks/Human_IPS_LncRNAs/Novel_lncRNAs_IPS.gtf (from IGV version 2.4x)
gs://lncrnatracks/Human_IPS_LncRNAs/Novel_lncRNAs_IPS.gtf (until IGV version 2.3x)
Processed RNA-seq mouse DRG samples for IGV genome browser are publicly available at: https://storage.googleapis.com/lncrnatracks/Mouse_DRG_BAM/Sample50_BALB.c_SHAM_M.sorted.bam
https://storage.googleapis.com/lncrnatracks/Mouse_DRG_BAM/Sample55_BALB.c_SHAM_F.sorted.bam
https://storage.googleapis.com/lncrnatracks/Mouse_DRG_BAM/Sample58_B10.D2_SHAM_M.sorted.bam
https://storage.googleapis.com/lncrnatracks/Mouse_DRG_BAM/Sample77_B10.D2_SHAM_F.sorted.bam
https://storage.googleapis.com/lncrnatracks/Mouse_DRG_BAM/Sample65_BALB.c_SNI_F.sorted.bam
https://storage.googleapis.com/lncrnatracks/Mouse_DRG_BAM/Sample70_BALB.c_SNI_M.sorted.bam
https://storage.googleapis.com/lncrnatracks/Mouse_DRG_BAM/Sample78_B10.D2_SNI_F.sorted.bam
https://storage.googleapis.com/lncrnatracks/Mouse_DRG_BAM/Sample90_B10.D2_SNI_M.sorted.bam
All data has been integrated to PainNetworks. In http://www.painnetworks.org the user can examine a gene (or set of genes) of interest alongside known interaction partners on the protein level. This information is displayed by the resource in the form of a network. Moreover the user can access all expression data (log2 fold change and FDR adjusted p.values) and download these in the form of spreadsheets. A tutorial on how to use painnetworks can be accessed following this link http://www.painnetworks.org/tutorials/RefMan.pdf.
All intergenic and antisense LncRNAs profiling data is accessible in PainNetworks (http://www.painnetworks.org) → ExpressionData → Mouse centric / Rat centric / Human centric. Experiment names are GB-BALBC-LNCRNAS for BALB/c mouse, GB-SNI-B10D2-LNCRNAS for B10.D2 mouse, GB-RAT-LNCRNAS for rat and IPSC_HS_AD2-LNCRNAS for IPSC-derived neurons. Naming is as follows: Closest {gene or sense gene}_LNCRNA_{IG or nothing}_chr:start-end(strand). Examples:
ENSMUSG00000000093_LNCRNA_IG:11:85830666-85831495(+) is the intergenic LncRNA with coordinates 11:85830666-85831495(+) close to the ENSMUSG00000000093 gene.
ENSMUSG00000000094_LNCRNA:11:85897018-85900613(-) is the antisense LncRNA with coordinates 11:85897018-85900613(-) on the opposite strand of ENSMUSG00000000094 gene.
Author contributions
GB designed bioinformatic workflow and performed data analysis. JGL integrated data into the painnetworks database. AAM, JMD, JSA, TT performed surgery, behavioural analysis and sample preparation. JMD performed qPCR validation. LM and AJC generated IPSC derived sensory neurons and extracted RNA. DLHB, JSM, SBM and CO conceived of and managed this study. GB, DLHB, JSM, CO and SBM drafted the manuscript which was reviewed and approved by all authors.
Conflict of interest: The authors declare no conflicts of interest
Bibliography
- [1].Abraira VE, Ginty DD. The sensory neurons of touch. Neuron. 2013;79:618–639. doi: 10.1016/j.neuron.2013.07.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Alexa A, Rahnenfuhrer J. topGO: Enrichment analysis for Gene Ontology. 2010.
- [3].Anders S, Pyl PT, Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Basbaum AI, Bautista DM, Scherrer G, Julius D. Cellular and molecular mechanisms of pain. Cell. 2009;139:267–284. doi: 10.1016/j.cell.2009.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].van Brakel J-P. algorithm - Peak signal detection in realtime timeseries data. Stack Overflow; 2014. [Accessed 1 May 2018]. Available: https://stackoverflow.com/questions/22583391/peak-signal-detection-in-realtime-timeseries-data/22640362. [Google Scholar]
- [6].Brown JB, Boley N, Eisman R, May GE, Stoiber MH, Duff MO, Booth BW, Wen J, Park S, Suzuki AM, Wan KH, et al. Diversity and dynamics of the Drosophila transcriptome. Nature. 2014 doi: 10.1038/nature12962. advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Chambers SM, Qi Y, Mica Y, Lee G, Zhang X-J, Niu L, Bilsland J, Cao L, Stevens E, Whiting P, Shi S-H, et al. Combined small-molecule inhibition accelerates developmental timing and converts human pluripotent stem cells into nociceptors. Nat Biotechnol. 2012;30:715–720. doi: 10.1038/nbt.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Chambers SM, Qi Y, Mica Y, Lee G, Zhang X-J, Niu L, Bilsland J, Cao L, Stevens E, Whiting P, Shi S-H, et al. Combined small-molecule inhibition accelerates developmental timing and converts human pluripotent stem cells into nociceptors. Nat Biotechnol. 2012;30:715–720. doi: 10.1038/nbt.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Chandran V, Coppola G, Nawabi H, Omura T, Versano R, Huebner EA, Zhang A, Costigan M, Yekkirala A, Barrett L, Blesch A, et al. A Systems-Level Analysis of the Peripheral Nerve Intrinsic Axonal Growth Program. Neuron. 2016;89:956–970. doi: 10.1016/j.neuron.2016.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Chaplan SR, Bach FW, Pogrel JW, Chung JM, Yaksh TL. Quantitative assessment of tactile allodynia in the rat paw. J Neurosci Methods. 1994;53:55–63. doi: 10.1016/0165-0270(94)90144-9. [DOI] [PubMed] [Google Scholar]
- [12].Clark AJ, Kaller MS, Galino J, Willison HJ, Rinaldi S, Bennett DLH. Co-cultures with stem cell-derived human sensory neurons reveal regulators of peripheral myelination. Brain J Neurol. 2017;140:898–913. doi: 10.1093/brain/awx012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Colloca L, Ludman T, Bouhassira D, Baron R, Dickenson AH, Yarnitsky D, Freeman R, Truini A, Attal N, Finnerup NB, Eccleston C, et al. Neuropathic pain. Nat Rev Dis Primer. 2017;3:17002. doi: 10.1038/nrdp.2017.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Costigan M, Befort K, Karchewski L, Griffin RS, D’Urso D, Allchorne A, Sitarski J, Mannion JW, Pratt RE, Woolf CJ. Replicate high-density rat genome oligonucleotide microarrays reveal hundreds of regulated genes in the dorsal root ganglion after peripheral nerve injury. BMC Neurosci. 2002;3:16. doi: 10.1186/1471-2202-3-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Dawes JM, Weir GA, Middleton SJ, Patel R, Chisholm KI, Pettingill P, Peck LJ, Sheridan J, Shakir A, Jacobson L, Gutierrez-Mecinas M, et al. Immune or Genetic-Mediated Disruption of CASPR2 Causes Pain Hypersensitivity Due to Enhanced Primary Afferent Excitability. Neuron. 2018;97:806–822.e10. doi: 10.1016/j.neuron.2018.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Decosterd I, Woolf CJ. Spared nerve injury: an animal model of persistent peripheral neuropathic pain. Pain. 2000;87:149–158. doi: 10.1016/S0304-3959(00)00276-1. [DOI] [PubMed] [Google Scholar]
- [17].DeNardo LA, de Wit J, Otto-Hitt S, Ghosh A. NGL-2 regulates input-specific synapse development in CA1 pyramidal neurons. Neuron. 2012;76:762–775. doi: 10.1016/j.neuron.2012.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, Lagarde J, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].ENCODE consortium. ENCODE - Guidelines and Best Practices for RNA-Seq. 2016 Available: https://www.encodeproject.org/documents/cede0cbe-d324-4ce7-ace4-f0c3eddf5972/@@download/attachment/ENCODE%20Best%20Practices%20for%20RNA_v2.pdf.
- [21].FANTOM5 CAGE profiles of human and mouse samples. [Accessed 12 Oct 2017];Scientific Data. doi: 10.1038/sdata.2017.112. n.d. Available: https://www.nature.com/articles/sdata2017112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Gentleman RC, Carey VJ, Bates DM, et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Gerstein MB, Rozowsky J, Yan K-K, Wang D, Cheng C, Brown JB, Davis CA, Hillier L, Sisu C, Li JJ, Pei B, et al. Comparative analysis of the transcriptome across distant species. Nature. 2014;512:445–448. doi: 10.1038/nature13424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Ghazalpour A, Doss S, Zhang B, Wang S, Plaisier C, Castellanos R, Brozell A, Schadt EE, Drake TA, Lusis AJ, Horvath S. Integrating Genetic and Network Analysis to Characterize Genes Related to Mouse Weight. PLOS Genet. 2006;2:e130. doi: 10.1371/journal.pgen.0020130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Gong L, Wu J, Zhou S, Wang Y, Qin J, Yu B, Gu X, Yao C. Global analysis of transcriptome in dorsal root ganglia following peripheral nerve injury in rats. Biochem Biophys Res Commun. 2016;478:206–212. doi: 10.1016/j.bbrc.2016.07.067. [DOI] [PubMed] [Google Scholar]
- [26].Guo T, Mandai K, Condie BG, Wickramasinghe SR, Capecchi MR, Ginty DD. An evolving NGF-Hoxd1 signaling pathway mediates development of divergent neural circuits in vertebrates. Nat Neurosci. 2011;14:31–36. doi: 10.1038/nn.2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Han TW, Jan LY. Making antisense of pain. Nat Neurosci. 2013;16:986–987. doi: 10.1038/nn.3475. [DOI] [PubMed] [Google Scholar]
- [28].Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, Barnes I, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Hon C-C, Ramilowski JA, Harshbarger J, Bertin N, Rackham OJL, Gough J, Denisenko E, Schmeier S, Poulsen TM, Severin J, Lizio M, et al. An atlas of human long non-coding RNAs with accurate 5’ ends. Nature. 2017;543:199–204. doi: 10.1038/nature21374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Ilott NE, Ponting CP. Predicting long non-coding RNAs using RNA sequencing. Methods San Diego Calif. 2013;63:50–59. doi: 10.1016/j.ymeth.2013.03.019. [DOI] [PubMed] [Google Scholar]
- [31].Jiang B-C, Sun W-X, He L-N, Cao D-L, Zhang Z-J, Gao Y-J. Identification of lncRNA expression profile in the spinal cord of mice following spinal nerve ligation-induced neuropathic pain. Mol Pain. 2015;11:43. doi: 10.1186/s12990-015-0047-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Kapusta A, Feschotte C. Volatile evolution of long noncoding RNA repertoires: mechanisms and biological implications. Trends Genet. 2014;30:439–452. doi: 10.1016/j.tig.2014.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Koenig J, Werdehausen R, Linley JE, Habib AM, Vernon J, Lolignier S, Eijkelkamp N, Zhao J, Okorokov AL, Woods CG, Wood JN, et al. Regulation of Nav1.7: A Conserved SCN9A Natural Antisense Transcript Expressed in Dorsal Root Ganglia. PloS One. 2015;10:e0128830. doi: 10.1371/journal.pone.0128830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Kornienko AE, Guenzl PM, Barlow DP, Pauler FM. Gene regulation by the act of long non-coding RNA transcription. BMC Biol. 2013;11:59. doi: 10.1186/1741-7007-11-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].LaCroix-Fralish ML, Austin J-S, Zheng FY, Levitin DJ, Mogil JS. Patterns of pain: meta-analysis of microarray studies of pain. Pain. 2011;152:1888–1898. doi: 10.1016/j.pain.2011.04.014. [DOI] [PubMed] [Google Scholar]
- [36].Lacroix-Fralish ML, Ledoux JB, Mogil JS. The Pain Genes Database: An interactive web browser of pain-related transgenic knockout studies. Pain. 2007;131:3.e1–4. doi: 10.1016/j.pain.2007.04.041. [DOI] [PubMed] [Google Scholar]
- [37].Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Lee J, Hong W, Cho M, Sim M, Lee D, Ko Y, Kim J. Synteny Portal: a web-based application portal for synteny block analysis. Nucleic Acids Res. 2016;44:W35–W40. doi: 10.1093/nar/gkw310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Li C-L, Li K-C, Wu D, Chen Y, Luo H, Zhao J-R, Wang S-S, Sun M-M, Lu Y-J, Zhong Y-Q, Hu X-Y, et al. Somatosensory neuron types identified by high-coverage single-cell RNA-sequencing and functional heterogeneity. Cell Res. 2016;26:83–102. doi: 10.1038/cr.2015.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinforma Oxf Engl. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Lizio M, Harshbarger J, Shimoji H, Severin J, Kasukawa T, Sahin S, Abugessaisa I, Fukuda S, Hori F, Ishikawa-Kato S, Mungall CJ, et al. Gateways to the FANTOM5 promoter level mammalian expression atlas. Genome Biol. 2015;16:22. doi: 10.1186/s13059-014-0560-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Lötsch J, Doehring A, Mogil JS, Arndt T, Geisslinger G, Ultsch A. Functional genomics of pain in analgesic drug development and therapy. Pharmacol Ther. 2013;139:60–70. doi: 10.1016/j.pharmthera.2013.04.004. [DOI] [PubMed] [Google Scholar]
- [43].Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Maamar H, Cabili MN, Rinn J, Raj A. linc-HOXA1 is a noncoding RNA that represses Hoxa1 transcription in cis. Genes Dev. 2013;27:1260–1271. doi: 10.1101/gad.217018.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Marques AC, Ponting CP. Intergenic lncRNAs and the evolution of gene expression. Curr Opin Genet Dev. 2014;27:48–53. doi: 10.1016/j.gde.2014.03.009. [DOI] [PubMed] [Google Scholar]
- [46].Martin JA, Wang Z. Next-generation transcriptome assembly. Nat Rev Genet. 2011;12:671–682. doi: 10.1038/nrg3068. [DOI] [PubMed] [Google Scholar]
- [47].Meyer LR, Zweig AS, Hinrichs AS, Karolchik D, Kuhn RM, Wong M, Sloan CA, Rosenbloom KR, Roe G, Rhead B, Raney BJ, et al. The UCSC Genome Browser database: extensions and updates 2013. Nucleic Acids Res. 2012;41:D64–D69. doi: 10.1093/nar/gks1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Miura P, Shenker S, Andreu-Agullo C, Westholm JO, Lai EC. Widespread and extensive lengthening of 3′ UTRs in the mammalian brain. Genome Res. 2013;23:812–825. doi: 10.1101/gr.146886.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Mogil JS. Sex differences in pain and pain inhibition: multiple explanations of a controversial phenomenon. Nat Rev Neurosci. 2012;13:859–866. doi: 10.1038/nrn3360. [DOI] [PubMed] [Google Scholar]
- [50].Morgan M, Falcon S, Gentleman R. GSEABase: Gene set enrichment data structures and methods. 2017. [Google Scholar]
- [51].Morgan M, Obenchain V, Lang M, Thompson R. BiocParallel: Bioconductor facilities for parallel evaluation. 2017 Available: https://github.com/Bioconductor/BiocParallel. [Google Scholar]
- [52].O’Donovan KJ. Intrinsic Axonal Growth and the Drive for Regeneration. Front Neurosci. 2016;10:486. doi: 10.3389/fnins.2016.00486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Parisien M, Khoury S, Chabot-Doré A-J, Sotocinal SG, Slade GD, Smith SB, Fillingim RB, Ohrbach R, Greenspan JD, Maixner W, Mogil JS, et al. Effect of Human Genetic Variability on Gene Expression in Dorsal Root Ganglia and Association with Pain Phenotypes. Cell Rep. 2017;19:1940–1952. doi: 10.1016/j.celrep.2017.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Perkins JR, Antunes-Martins A, Calvo M, Grist J, Rust W, Schmid R, Hildebrandt T, Kohl M, Orengo C, McMahon SB, Bennett DL. A comparison of RNA-seq and exon arrays for whole genome transcription profiling of the L5 spinal nerve transection model of neuropathic pain in the rat. Mol Pain. 2014;10:7. doi: 10.1186/1744-8069-10-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Perkins JR, Lees J, Antunes-Martins A, Diboun I, McMahon SB, Bennett DLH, Orengo C. PainNetworks: a web-based resource for the visualisation of pain-related genes in the context of their network associations. Pain. 2013;154:2586.e1–12. doi: 10.1016/j.pain.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Pesole G, Liuni S, Grillo G, Licciulli F, Mignone F, Gissi C, Saccone C. UTRdb and UTRsite: specialized databases of sequences and functional elements of 5′ and 3′ untranslated regions of eukaryotic mRNAs. Update 2002. Nucleic Acids Res. 2002;30:335–340. doi: 10.1093/nar/30.1.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Petryszak R, Keays M, Tang YA, Fonseca NA, Barrera E, Burdett T, Füllgrabe A, Fuentes AM-P, Jupp S, Koskinen S, Mannion O, et al. Expression Atlas update—an integrated database of gene and protein expression in humans, animals and plants. Nucleic Acids Res. 2016;44:D746–D752. doi: 10.1093/nar/gkv1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Ponjavic J, Ponting CP, Lunter G. Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Res. 2007;17:556–565. doi: 10.1101/gr.6036807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- [60].R Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2017. Available: https://www.R-project.org/ [Google Scholar]
- [61].Ray P, Torck A, Quigley L, Wangzhou A, Neiman M, Rao C, Lam T, Kim J-Y, Kim TH, Zhang MQ, Dussor G, et al. Comparative transcriptome profiling of the human and mouse dorsal root ganglia: an RNA-seq-based resource for pain and sensory neuroscience research. Pain. 2018 doi: 10.1097/j.pain.0000000000001217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Rigaud M, Gemes G, Barabas M-E, Chernoff DI, Abram SE, Stucky CL, Hogan QH. Species and strain differences in rodent sciatic nerve anatomy: implications for studies of neuropathic pain. Pain. 2008;136:188–201. doi: 10.1016/j.pain.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Savell KE, Gallus NVN, Simon RC, Brown JA, Revanna JS, Osborn MK, Song EY, O’Malley JJ, Stackhouse CT, Norvil A, Gowher H, et al. Extra-coding RNAs regulate neuronal DNA methylation dynamics. Nat Commun. 2016;7:12091. doi: 10.1038/ncomms12091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Willis William D., Jr . Sensory Mechanisms of the Spinal Cord. Vol. 1. Springer; [Accessed 13 Jul 2017]. n.d. p. Available: http://www.springer.com/cn/book/9780306480331. [Google Scholar]
- [65].Shields SD, Eckert WA, Basbaum AI. Spared nerve injury model of neuropathic pain in the mouse: a behavioral and anatomic analysis. J Pain Off J Am Pain Soc. 2003;4:465–470. doi: 10.1067/s1526-5900(03)00781-8. [DOI] [PubMed] [Google Scholar]
- [66].Shiraki T, Kondo S, Katayama S, Waki K, Kasukawa T, Kawaji H, Kodzius R, Watahiki A, Nakamura M, Arakawa T, Fukuda S, et al. Cap analysis gene expression for high-throughput analysis of transcriptional starting point and identification of promoter usage. Proc Natl Acad Sci. 2003;100:15776–15781. doi: 10.1073/pnas.2136655100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Sorge RE, Mapplebeck JCS, Rosen S, Beggs S, Taves S, Alexander JK, Martin LJ, Austin J-S, Sotocinal SG, Chen D, Yang M, et al. Different immune cells mediate mechanical pain hypersensitivity in male and female mice. Nat Neurosci. 2015;18:1081–1083. doi: 10.1038/nn.4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Sorge RE, Trang T, Dorfman R, Smith SB, Beggs S, Ritchie J, Austin J-S, Zaykin DV, Meulen HV, Costigan M, Herbert TA, et al. Genetically determined P2X7 receptor pore formation regulates variability in chronic pain sensitivity. Nat Med. 2012;18:595–599. doi: 10.1038/nm.2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Tsoi LC, Iyer MK, Stuart PE, Swindell WR, Gudjonsson JE, Tejasvi T, Sarkar MK, Li B, Ding J, Voorhees JJ, Kang HM, et al. Analysis of long non-coding RNAs highlights tissue-specific expression patterns and epigenetic profiles in normal and psoriatic skin. Genome Biol. 2015;16:24. doi: 10.1186/s13059-014-0570-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Tsoi LC, Iyer MK, Stuart PE, Swindell WR, Gudjonsson JE, Tejasvi T, Sarkar MK, Li B, Ding J, Voorhees JJ, Kang HM, et al. Analysis of long non-coding RNAs highlights tissue-specific expression patterns and epigenetic profiles in normal and psoriatic skin. Genome Biol. 2015;16:24. doi: 10.1186/s13059-014-0570-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Ulitsky I, Bartel DP. lincRNAs: Genomics, Evolution, and Mechanisms. Cell. 2013;154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Usoskin D, Furlan A, Islam S, Abdo H, Lönnerberg P, Lou D, Hjerling-Leffler J, Haeggström J, Kharchenko O, Kharchenko PV, Linnarsson S, et al. Unbiased classification of sensory neuron types by large-scale single-cell RNA sequencing. Nat Neurosci. 2015;18:145–153. doi: 10.1038/nn.3881. [DOI] [PubMed] [Google Scholar]
- [73].Wahlestedt C. Targeting long non-coding RNA to therapeutically upregulate gene expression. Nat Rev Drug Discov. 2013;12:433–446. doi: 10.1038/nrd4018. [DOI] [PubMed] [Google Scholar]
- [74].Wang L, Park HJ, Dasari S, Wang S, Kocher J-P, Li W. CPAT: Coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucleic Acids Res. 2013;41:e74. doi: 10.1093/nar/gkt006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Weir GA, Middleton SJ, Clark AJ, Daniel T, Khovanov N, McMahon SB, Bennett DL. Using an engineered glutamate-gated chloride channel to silence sensory neurons and treat neuropathic pain at the source. Brain J Neurol. 2017;140:2570–2585. doi: 10.1093/brain/awx201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Wu M, Huang H, Chen Q, Li D, Zheng Z, Xiong W, Zhou Y, Li X, Zhou M, Lu J, Shen S, et al. Leucine-rich repeat C4 protein is involved in nervous tissue development and neurite outgrowth, and induction of glioma cell differentiation. Acta Biochim Biophys Sin. 2007;39:731–738. doi: 10.1111/j.1745-7270.2007.00338.x. [DOI] [PubMed] [Google Scholar]
- [77].Wu S, Marie Lutz B, Miao X, Liang L, Mo K, Chang Y-J, Du P, Soteropoulos P, Tian B, Kaufman AG, Bekker A, et al. Dorsal root ganglion transcriptome analysis following peripheral nerve injury in mice. Mol Pain. 2016;12 doi: 10.1177/1744806916629048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Xie C, Yuan J, Li H, Li M, Zhao G, Bu D, Zhu W, Wu W, Chen R, Zhao Y. NONCODEv4: exploring the world of long non-coding RNA genes. Nucleic Acids Res. 2014;42:D98–103. doi: 10.1093/nar/gkt1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Yanai I, Benjamin H, Shmoish M, Chalifa-Caspi V, Shklar M, Ophir R, Bar-Even A, Horn-Saban S, Safran M, Domany E, Lancet D, et al. Genome-wide midrange transcription profiles reveal expression level relationships in human tissue specification. Bioinforma Oxf Engl. 2005;21:650–659. doi: 10.1093/bioinformatics/bti042. [DOI] [PubMed] [Google Scholar]
- [80].Yarmishyn AA, Batagov AO, Tan JZ, Sundaram GM, Sampath P, Kuznetsov VA, Kurochkin IV. HOXD-AS1 is a novel lncRNA encoded in HOXD cluster and a marker of neuroblastoma progression revealed via integrative analysis of noncoding transcriptome. BMC Genomics. 2014;15(Suppl 9):S7. doi: 10.1186/1471-2164-15-S9-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Yates A, Akanni W, Amode MR, Barrell D, Billis K, Carvalho-Silva D, Cummins C, Clapham P, Fitzgerald S, Gil L, Girón CG, et al. Ensembl 2016. Nucleic Acids Res. 2016;44:D710–D716. doi: 10.1093/nar/gkv1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Young GT, Gutteridge A, Fox HDE, Wilbrey AL, Cao L, Cho LT, Brown AR, Benn CL, Kammonen LR, Friedman JH, Bictash M, et al. Characterizing human stem cellderived sensory neurons at the single-cell level reveals their ion channel expression and utility in pain research. Mol Ther J Am Soc Gene Ther. 2014;22:1530–1543. doi: 10.1038/mt.2014.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Young RS, Ponting CP. Identification and function of long non-coding RNAs. Essays Biochem. 2013;54:113–126. doi: 10.1042/bse0540113. [DOI] [PubMed] [Google Scholar]
- [84].Zhang Y, Wang J, Ji L-J, Li L, Wei M, Zhen S, Wen C-C. Identification of Key Gene Modules of Neuropathic Pain by Co-Expression Analysis. J Cell Biochem. 2017;118:4436–4443. doi: 10.1002/jcb.26098. [DOI] [PubMed] [Google Scholar]
- [85].Zhang Y, Zhan Y, Han N, Kou Y, Yin X, Zhang P. Analysis of temporal expression profiles after sciatic nerve injury by bioinformatic method. Sci Rep. 2017;7:9818. doi: 10.1038/s41598-017-10127-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Zhao H, Duan L-J, Sun Q-L, Gao Y-S, Yang Y-D, Tang X-S, Zhao D-Y, Xiong Y, Hu Z-G, Li C-H, Chen S-X, et al. Identification of Key Pathways and Genes in L4 Dorsal Root Ganglion (DRG) After Sciatic Nerve Injury via Microarray Analysis. J Investig Surg Off J Acad Surg Res. 2018:1–9. doi: 10.1080/08941939.2018.1452996. [DOI] [PubMed] [Google Scholar]
- [87].Zhao J, Brown K, Liem RKH. Abnormal neurofilament inclusions and segregations in dorsal root ganglia of a Charcot-Marie-Tooth type 2E mouse model. PloS One. 2017;12:e0180038. doi: 10.1371/journal.pone.0180038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Zhao X, Tang Z, Zhang H, Atianjoh FE, Zhao J-Y, Liang L, Wang W, Guan X, Kao S-C, Tiwari V, Gao Y-J, et al. A long noncoding RNA contributes to neuropathic pain by silencing Kcna2 in primary afferent neurons. Nat Neurosci. 2013;16:1024–1031. doi: 10.1038/nn.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.