Summary
Type 1 diabetes (T1D) is an autoimmune disease targeting pancreatic islet beta cells that incorporates genetic and environmental factors1, including complex genetic elements2, patient exposures3, and the gut microbiome4. Viral infections5 and broader gut dysbioses6 have been identified as potential causes or contributing factors; however, human studies have not yet identified microbial compositional or functional triggers predictive of islet autoimmunity (IA) or T1D. We analyzed 10,913 metagenomes from 783 mostly white, non-hispanic children’s stool samples collected monthly from three months of age until the clinical end point (IA or T1D) in the TEDDY (The Environmental Determinants of Diabetes in the Young) study to characterize the natural history of the early gut microbiome in connection to IA, T1D diagnosis, and other common early life events such as antibiotic treatments and probiotics. Microbiomes of control children harbored more genes related to fermentation and short chain fatty acid (SCFA) biosynthesis, but these were not consistently associated with particular taxa across geographically diverse clinical centers, suggesting that microbial factors associated with T1D are taxonomically diffuse but functionally coherent. When investigating the broader establishment and development of the infant microbiome, both taxonomic and functional profiles were dynamic and highly individualized, dominated in the first year of life by one of three largely exclusive Bifidobacterium species (B. bifidum, B. breve, or B. longum) or by the phylum Proteobacteria. In particular, strain-specific carriage of human milk oligosaccharide utilization genes within a subset of Bifidobacterium longum was present specifically in breast-fed infants. These analyses of TEDDY gut metagenomes provide, to date, the largest and most detailed longitudinal functional profile of the developing gut microbiome in relation to IA, T1D, and other early childhood events. Together with existing evidence from human cohorts7,8 and T1D mouse model9, these data support the protective effects of SCFAs on early-onset human T1D.
Recent literature has linked multiple facets of gut health with T1D onset in humans and rodent models4,6,10. Altered intestinal microbiota in connection to T1D has been reported in Finnish7,8,11,12, German13, Italian14, Mexican15, U.S. (Colorado)16 and Turkish17 children. Common findings include increased Bacteroides species and deficiency of SCFA-producing bacteria7,8 in T1D or IA cases8,11,15,18. Corroborating these findings, an adult study found decreased SCFA-producing bacteria in adults with type 2 diabetes19. Additionally, increased intestinal permeability14 and decreased microbial diversity12 after IA but prior to T1D diagnosis have been reported. Studies using the nonobese diabetic (NOD) mouse model have elucidated immune mechanisms mediating the protective effects of SCFAs9 and the microbiome-linked sex bias in autoimmunity20. NOD mice fed specialized diets resulting in high bacterial release of SCFAs acetate and butyrate were almost completely protected from T1D9. A study in a streptozotocin-induced T1D mouse model demonstrated that bacterial products recognized in pancreatic lymph nodes contribute to pathogenesis21.
Even in the absence of immune perturbation, the first weeks, months, and years of life represent a unique human microbial environment that has only recently been detailed22,23. Infants have a markedly different gut microbial profile from adults, characterized by a distinct taxonomic profile, greater proportion of aerobic energy harvest metabolism, and more extreme dynamic change24. These differences gradually fade over the first several years of life, particularly in response to the introduction of solid food, and individual microbial developmental trajectories are influenced by environment, delivery mode, breast (versus formula) feeding, and antibiotics25–27. Most studies addressing gut microbiome development, both generally and in association with T1D, have used 16S rRNA gene analysis, leaving open the question of functional and strain-specific differences not easily detected by this technology that might contribute to disease pathogenesis12.
Bridging this gap is one goal of The Environmental Determinants of Diabetes in the Young (TEDDY) study, a prospective study aiming to identify environmental causes of T1D28. It includes six clinical research centers in the U.S. (Colorado, Georgia/Florida, Washington) and Europe (Finland, Germany, Sweden), which together have recruited several thousand newborns with a genetic predisposition for T1D or first-degree relative(s) with T1D. This has enabled the TEDDY study to collect a range of biospecimens, including monthly stool samples starting at three months of age, coupled with extensive clinical and personal data including diet, illnesses, medications, and other life experiences. To characterize microbial, environmental, genetic, immunological, and additional contributors to T1D development, the TEDDY study group further assembled nested case-control studies for IA (N = 418 case-control pairs) and T1D (N = 114)29. Case-control pairs were matched by clinical center, sex and family history of T1D, which are all known confounding factors for T1D susceptibility and microbiome composition.
Here, we assessed 783 children followed from three months to up to five years of age from six clinical centers in four countries (Finland, Germany, Sweden, U.S.) who either progressed to persistent IA or T1D or were matched as controls (Fig. 1a,b, Extended Data Table 1). Stool samples were collected, on average, monthly starting at three months of age and continuing until the clinical endpoint (IA or T1D). This study focused solely on analyzing metagenomic sequencing data (N = 10,903 samples, N = 783 subjects), while a companion manuscript by Stewart et al. interrogated corresponding 16S rRNA amplicon sequencing information.
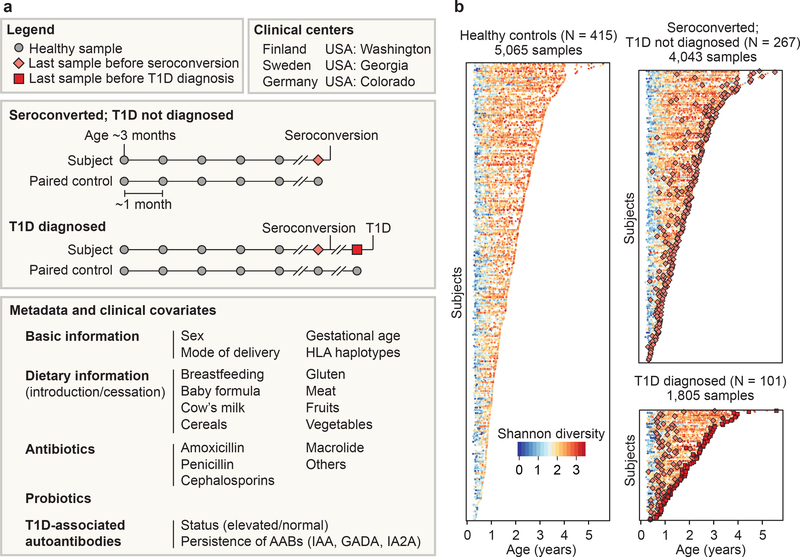
Figure 1: >10,000 longitudinal gut metagenomes from the TEDDY T1D cohort.
We analyzed 10,913 metagenomes collected longitudinally from 783 children (415 controls, 267 seroconverters, and 101 diagnosed with T1D) approximately monthly over the first five years of life. a, Subjects were recruited at six clinical centers. Primary endpoints were seroconversion (defined as persistent confirmed IA) and T1D diagnosis. Additional metadata analyzed for subjects and samples included breastfeeding status, birth mode, probiotics, antibiotics, formula feeding, and other dietary covariates. b, Overview of stool samples collected and microbiome development as summarized by Shannon alpha-diversity and stratified by endpoint. Median number of samples per individual N = 12 (healthy controls N = 10, seroconverters N = 13, T1D cases N = 16).
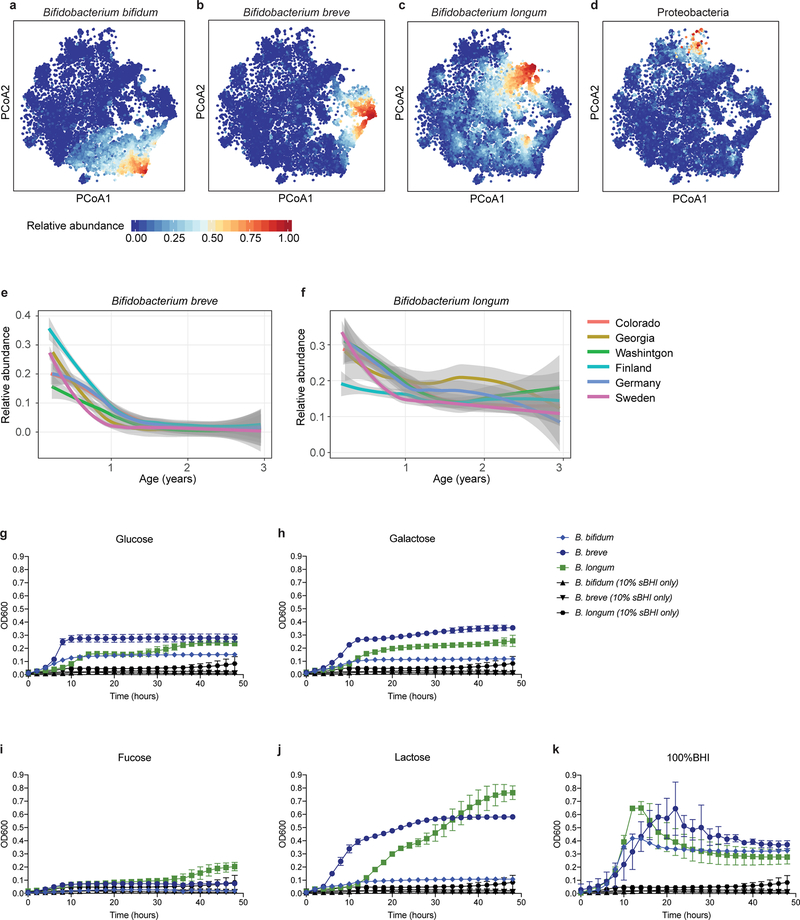
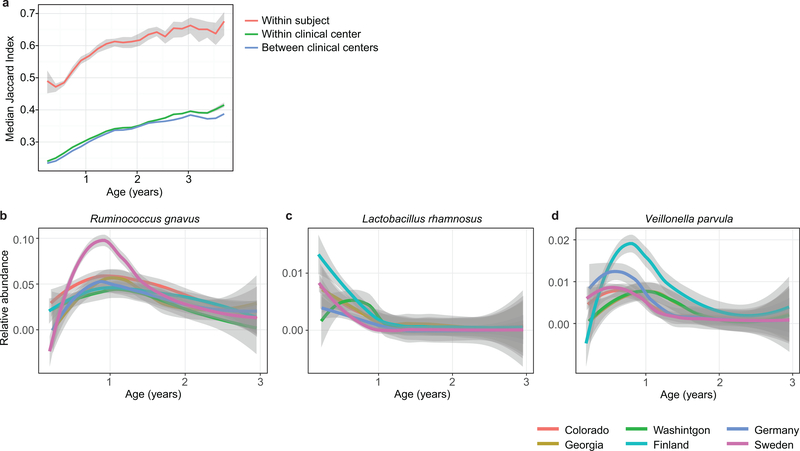
We first investigated the taxonomic composition of early gut metagenomes at the species level. Principal coordinate analysis (PCoA) ordination of Bray-Curtis beta diversities showed a strong longitudinal gradient and significant heterogeneity among the earliest samples (Fig. 2a, Extended Data Fig. 1a-k, Supplementary Note 1). Permutational analysis of variance (PERMANOVA) of Bray-Curtis beta diversities indicated that inter-subject differences explained 35% of microbial taxonomic variation (permutation test, p < 0.001, 1,000 permutations), followed by age at stool sampling at roughly 4% of variance (p < 0.001). We further analysed the data cross-sectionally to test for associations between taxonomic beta diversities and other collected metadata, finding that in addition to subject ID and age, geographic location and breastfeeding had strong and systematic effects on microbial community composition (Table S1, Extended Data Fig. 2a-d, Supplementary Note 1). To further investigate the stability and individuality of the microbial profiles, we compared intra- and inter-subject Bray-Curtis beta diversities. The gap between individual stability and similarity within or across clinical centers was largest at the beginning of the sampling period, indicating that the children had particularly dissimilar microbiotas during these early months (Fig. 2b, Supplementary Note 1). Finally, we tested microbial alpha-diversity (Shannon’s diversity index) of taxonomic profiles for associations with collected metadata, finding that breastfeeding cessation had the largest effect (ANOVA, partial η2 = 0.053) in the accrual of alpha diversity in early life (Table S2, Extended Data Fig. 3a-e, Supplementary Note 1).
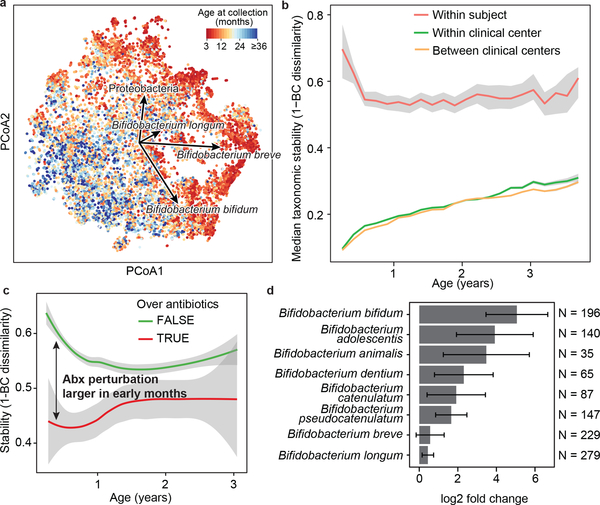
Figure 2: The early gut microbiome is characterized by early Bifidobacterium species heterogeneity and individualized accrual of taxa over time.
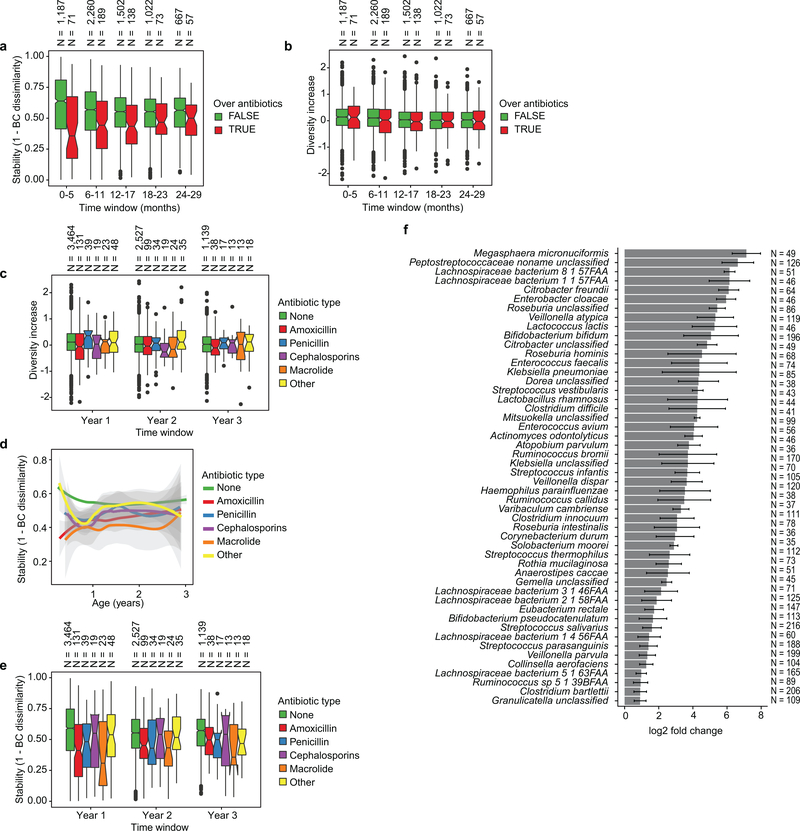
a, PCoA ordination of microbial beta diversities (N = 10,913 samples), measured by Bray-Curtis dissimilarity. Arrows show weighted averages of key taxonomic groups. b, Microbiota stability, measured by Bray-Curtis (BC) dissimilarity (N = 10,750 samples) in three-month time windows, over two-month increments, stratified in three groups: within subject, within clinical center, and between clinical centers. Lines show median per time window. Shaded area shows estimated 99% confidence interval. Gut microbial communities were highly individual. c, Influence of antibiotic courses on microbial stability, measured by Bray-Curtis dissimilarity over consecutive stool samples (<50 days apart) from the same individual during the first three years of life and stratified by whether antibiotics were given between the two samples (N = 654 observations with antibiotics, N = 6,734 observations without antibiotics). Curves show locally weighted scatterplot smoothing (LOESS) for the data per category. Shaded areas show permutation-based 95% confidence intervals for the fit. d, Decreases in the most common Bifidobacterium species in connection to oral antibiotic treatments. Fold change was measured between consecutive samples with an antibiotic course between them, given that the species in question was present in the first of the two samples. Sample size per species (N) indicates the number of sample pairs where the species in question was present in the sample preceding the antibiotic treatment. Bars show bootstrapped log2 fold change decrease (mean and standard deviation, N = 1,000 bootstrap samples).
We next investigated the effects of antibiotics on the early life microbiome. Oral antibiotic courses disrupted microbial stability, with a larger effect in the earliest comparisons (Fig. 2c, Extended Data Fig. 4a-f, Extended Data Table 2, Supplementary Note 2). Recent studies have found Bifidobacterium species to be especially vulnerable to antibiotics30,31, leading us to investigate how antibiotic perturbations influenced these common dominant members of the early gut. Comparing microbial relative abundances before and after antibiotics (assuming the given species was present in the preceding sample), we saw a drop in Bifidobacterium members B. bifidum, B. pseudocatenulatum, B. adolescentis, B. dentium, and B. catenulatum, whereas B. longum and B. breve did not systematically decline due to antibiotics (Fig. 2d), suggesting that certain Bifidobacterium species are particularly susceptible to out-competition by other community members after depletion by antibiotics. Given their dominance in the typical developing gut microbiota and finely-tuned balance of metabolic interactions with breast milk, this finding underscores the importance of approaching antibiotic prescriptions in early childhood with care, especially during breastfeeding.
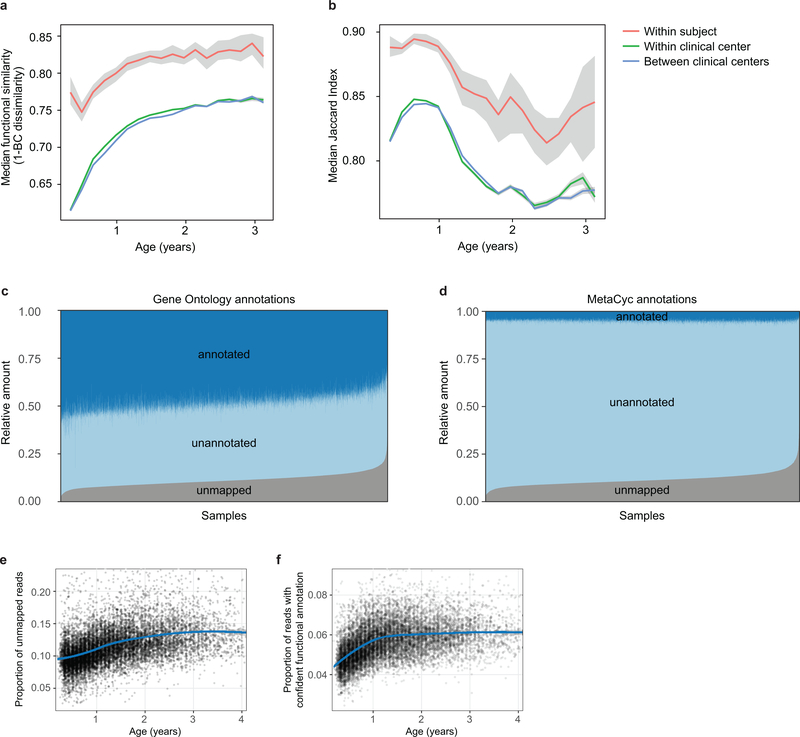
Accompanying our taxonomic profiling, functional profiling of these metagenomes suggested the development of a consistent microbial functional core during infancy, with a smaller subject-specific variable functional pool (Extended Data Fig. 5a,b, Supplementary Note 3). As in most microbial community studies32, microbial gene families of uncharacterized function made up a substantial fraction of these profiles, averaging roughly 50% based on Gene Ontology (GO)33 annotations (Extended Data Fig. 5c) and less than 10% based on more functionally-specific MetaCyc pathways (Extended Data Fig. 5d). We observed an increasing longitudinal trend in the proportion of unmapped reads (Extended Data Fig. 5e Pearson r = 0.318, p < 2.2e-16). However, within the reads mapping to either microbial pangenomes or known protein sequences (the proportion of which decreased with age), we saw an increase in the proportion of reads with MetaCyc annotation, mainly during the first year (Extended Data Fig. 5f, Pearson r = 0.391, p < 2.2e-16). This suggests that although the early life microbiome is relatively well-covered by current microbial reference genomes, less functional and biochemical characterization has been carried out on gene families within these microbes, which will thus particularly benefit from future work.
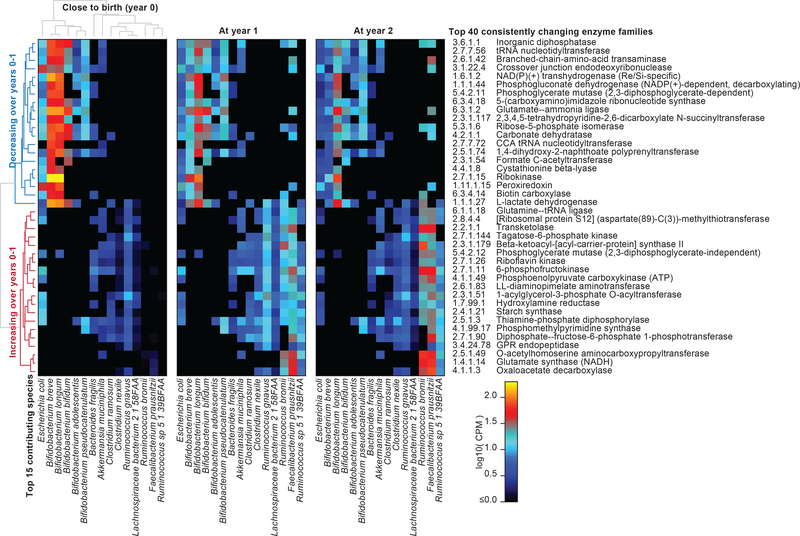
In addition to broadly-conserved and subject-specific functions, we identified a range of microbial metabolic enzymes that consistently increased or decreased in abundance over the first year of life, paralleling shifts in community structure and infant diet (Fig. 3, Supplementary Note 3, Table S3). For example, L-lactate dehydrogenase (1.1.1.27), an enzyme well-characterized in Bifidobacteria for its role in milk fermentation34, was among the most consistently declining enzymes over this period, notably coinciding with breastfeeding cessation in many infants (from 73% breastfed at month 3 to 28% at year 1). Conversely, transketolase (2.2.1.1), an enzyme implicated in fiber metabolism by Ruminococci35, was among the most consistently increasing enzymes, which also coincided with increased incorporation of solid food (a component of 53% of infants’ diets at month 3 versus 100% at year 1). Hence, these dramatic changes in community functional potential highlight the unique metabolic environment of the early infant gut and the subsequent transition to a more adult-like gut microbiome adapted to variable, fermentative energy sources.
Figure 3: Consistent changes in gut microbiome enzymatic content in early life.
We identified enzyme families (level-4 EC categories) that exhibited the most consistent within-subject changes in total community abundance between ages of three months and one year. The top 20 most consistent increases or decreases are presented and stratified according to their top 15 contributing species. Heatmap values reflect each species’ mean contribution to each enzyme over samples (N=733 at three months; 675 at 1 year; and 382 at 2 years). Values reflect units of “copies per million (CPM)” normalized over total read depth (including unmapped reads and reads mapped to gene families lacking EC annotation). Rows (enzymes) and columns (species) are clustered according to Spearman correlation at three months; subsequent years are ordered according to clustering at three months.
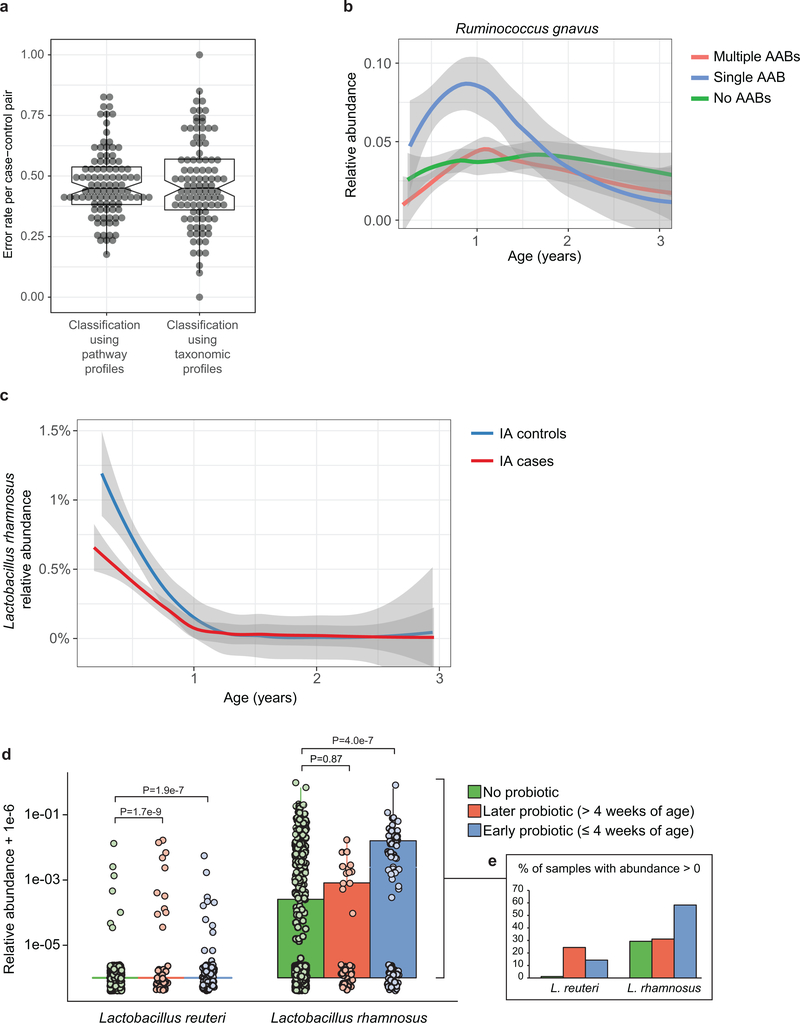
Combining taxonomic and functional profiles to test for differences between cases and controls, we used linear mixed effects modeling and identified a relatively small number of individual taxonomic and functional features associated with case-control outcome (Table S4), most with borderline statistical significance (FDR corrected q-values indicated below). We confirmed separation between cases and controls by random forest (RF) classifiers (Extended Data Fig. 6a, b, Supplementary Note 4). In the IA case-control cohort, healthy controls harbored higher levels Lactobacillus rhamnosus (q=0.055), supporting protection against IA by early probiotic supplementation36 (Extended Data Fig. 6c, d, Supplementary Note 5). IA controls also had more Bifidobacterium dentium (q=0.054), whereas cases had on average higher abundance of Streptococcus group mitis/oralis/pneumoniae species (q=0.11). In T1D case-control comparisons, controls had more Streptococcus thermophilus (q=0.078) and Lactococcus lactis (q=0.094) species, both common in dairy products, whereas cases harbored higher levels of such species as Bifidobacterium pseudocatenulatum (q=0.078), Roseburia hominis (q=0.11) and Alistipes shahii (q=0.14). Even though our modeling approach controlled for clinical centers (i.e. regional differences), we found additional but often weak associations with outcome in some clinical centers when tested separately (Table S4). Finnish IA cases had more Streptococcus group mitis/oralis/pneumoniae species (q=0.0008), IA controls from Colorado had more Streptococcus thermophilus (q=0.0059), and Swedish IA cases harbored more Bacteroides vulgatus (q=0.090).
Pathways with the highest statistical significance in case-control comparisons were related to bacterial fermentation (Table S4). Superpathway of fermentation (PWY4LZ-257) was increased in controls in the T1D cohort (q=0.019) and Finnish IA cohort (q=0.049). SCFAs - butyrate, acetate and propionate - are common by-products of bacterial fermentation, while butyrate and acetate protected NOD mice against T1D9. Consistently, we observed several bacterial pathways contributing to SCFA biosynthesis increased in healthy controls. Among pathways involved in butyrate production, superpathway of L-arginine, putrescine and 4-aminobutanoate degradation (ARGDEG-PWY) was increased in T1D controls cohort-wide (q=0.043), whereas acetyl coenzyme A fermentation to butanoate (PWY-5676) was more abundant in Finnish T1D controls (q=0.053). Acetylene degradation (P161-PWY), contributing to acetate production, was increased in T1D controls cohort-wide (q=0.14), and L-1,2-propanediol degradation (PWY-7013), involved in propionate biosynthesis, was higher in German T1D controls (q=0.019). These findings support existing evidence for protective effects of SCFAs from human T1D7,8 and T2D19 cohorts and NOD mouse model9.
As reflected by the community level analyses, human milk with its pro- and prebiotic functions is one of the main factors determining community composition of the infant gut microbiome. Subspecies B. longum infantis is a particularly versatile human milk oligosaccharide (HMO) degrader often found in stool samples collected during breastfeeding37. By following the families representing genes in the B. longum infantis HMO gene cluster38,39 in our data, we found that an additional 30 bacterial species carried at least one homolog with >50% sequence identity to one or more HMO utilization genes (Table S5). As expected, many Bifidobacteria carried multiple homologs, but surprisingly three Enterococcus species E. casseliflavus, E. faecalis and E. faecium also carried 7 or more homologs (Table S5).
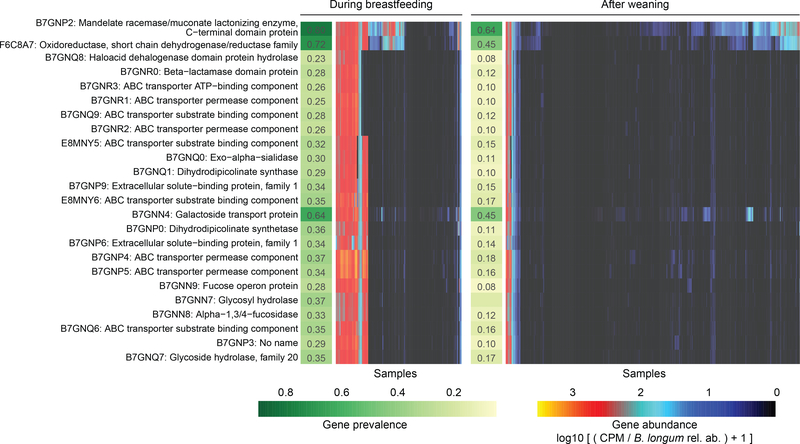
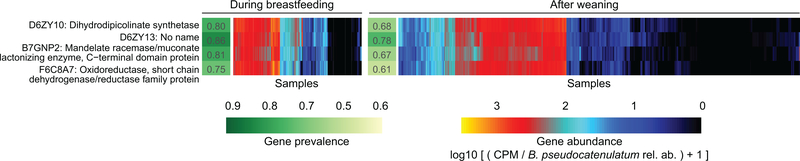
To identify strain-level adaptation similar to B. infantis, we further examined whether any of these genes showed contrasting prevalence between samples collected during breastfeeding and after weaning, given that the carrier species itself was present. Altogether, 41 gene families were observed more often during breastfeeding (Table S5, test of proportions, adjusted P < 0.001); the majority (37/41) were carried by B. longum (Fig. 4), and B. pseudocatenulatum harbored 4 such gene families (Extended Data Fig. 7, Table S5). In samples with B. longum, this implicated a clear strain shift after weaning, when fewer B. longum strains carried these genes (Fig. 4). In samples with B. pseudocatenulatum, four gene families showed a similar but less contrasting pattern (Extended Data Fig. 7). Overall, these observations identify new candidate species contributing to HMO processing or exploitation and link strain composition to specific driving molecular functions that potentially explain selective sweeps during microbiome development, in this case specifically related to breastfeeding.
Figure 4: Bifidobacterium longum strains are characterized by HMO gene content and stratified by breastfeeding status.
Gene families involved in HMO utilization and showing contrasting presence in B. longum genomes during breastfeeding (N = 1,584 samples) compared to after weaning (N = 3,705 samples). Abundance heatmap columns represent stool samples in which the relative abundance of species B. longum was >10 % (N = 5,289 samples). Rows and columns were ordered by hierarchical clustering using complete linkage method. As in Fig. 3, values reflect units of “copies per million (CPM)” and were further divided by B. longum relative abundance to obtain quantifications that are comparable between samples. UniRef90 identifiers and gene names or families are indicated on the left.
Despite ample sample size, scrutiny of the study design, and thorough statistical analyses, most of the taxonomic and functional signals we detected in case-control comparisons were modest in effect size and statistical significance. This could be due to multiple reasons - differences between T1D endotypes, temporally diffuse signals, geographic heterogeneity, lack of stool samples for the first two months of life - that should be considered in future investigations (Supplementary Note 6). Furthermore, the data used in these investigations was composed of samples from the genetically predisposed and mostly white, non-hispanic case-control groups designed into the TEDDY study. Results cannot be guaranteed to reflect the whole TEDDY cohort nor child populations in the respective countries.
Future targeted approaches to identify subject-specific connections between the gut microbiota and T1D pathogenesis may be beneficial, particularly given the apparent population-level heterogeneity revealed here. For example, laboratory experiments involving dietary factors that have been associated with T1D onset3 may reveal novel biochemically-specific signals mediated through the microbiome. Different endotypes of disease, such as differences in the first appearing autoantibody (IAA vs. GADA), number of appearing autoantibodies, time from seroconversion to T1D diagnosis, genetic host risk alleles and ethnic backgrounds may be characterized by distinct microbial configurations (Supplementary Note 6). Finally, microbiome components that were poorly measured in these data may also play critical roles: viruses, fungi, microbial transcription or small molecule biochemistry. By surveying these additional molecular activities both cross-sectionally and in more detailed longitudinal populations, this study lays the foundation to identify further gut microbial components predictive, protective, or potentially causal in T1D risk or pathogenesis.
Methods
Cohort and study design
The Environmental Determinants of Diabetes in the Young (TEDDY) is a prospective cohort study funded by the National Institutes of Health with the primary goal to identify environmental causes of type 1 diabetes (T1D). It includes six clinical research centers - three in the US: Colorado, Georgia/Florida, Washington and three in Europe: Finland, Germany, and Sweden. Detailed study design and methods have been previously published28,40,41. Written informed consents were obtained for all study participants from a parent or primary caretaker, separately, for genetic screening and participation in prospective follow-up. The TEDDY study was approved by local U.S. Institutional Review Boards and European Ethics Committee Boards in Colorado’s Colorado Multiple Institutional Review Board, Georgia’s Medical College of Georgia Human Assurance Committee (2004–2010), Georgia Health Sciences University Human Assurance Committee (2011–2012), Georgia Regents University Institutional Review Board (2013–2015), Augusta University Institutional Review Board (2015-present), Florida’s University of Florida Health Center Institutional Review Board, Washington state’s Washington State Institutional Review Board (2004–2012) and Western Institutional Review Board (2013-present), Finland’s Ethics Committee of the Hospital District of Southwest Finland, Germany’s Bayerischen Landesärztekammer (Bavarian Medical Association) Ethics Committee, Sweden’s Regional Ethics Board in Lund, Section 2 (2004–2012) and Lund University Committee for Continuing Ethical Review (2013-present). The study is monitored by External Advisory Board formed by the National Institutes of Health.
This analysis used stool samples and clinical metadata from two nested case-control studies (persistent, confirmed islet autoimmunity (IA) or T1D) using risk set sampling29. The data used here were collected as of May 31, 2012, a 1:1 match where one control per case of persistent confirmed IA or T1D were selected from the full TEDDY cohort. A control was a participant who had not developed persistent confirmed IA or T1D by the time the case to which it was matched had developed IA or T1D, within ±45 days of the event time. Matching factors were clinical center, sex and family history of T1D to control for differences in geographic area, genetic background and in sample/data handling between clinical centers. In all case-control comparisons, we removed all case-control pairs where the control later progressed to case status (i.e. they progressed to IA or T1D). Additionally, 17 subjects with missing breastfeeding information together with their matched pairs were excluded from the case-control comparisons to avoid confounding effects from unknown breastfeeding status.
The development of persistent confirmed IA was assessed every 3 months. Persistent autoimmunity was defined by the presence of confirmed islet autoantibody on two or more consecutive visits. Date of persistent autoimmunity was defined as the draw date of the first sample of the two consecutive samples which deemed the child persistent confirmed positive for a specific autoantibody (or any autoantibody). T1D was defined according to American Diabetes Association criteria for diagnosis42.
Stool samples were collected monthly starting at three months of age and continuing up until 48 months of age, then every three months until the age of 10 years and then biannually thereafter into the three plastic stool containers provided by the clinical center. Children who were antibody negative after 4 years of age were encouraged to submit 4 times a year even though after 4 years their visits schedule switched to biannual. Parents sent the stool containers at either ambient or +4°C temperature with guaranteed delivery within 24 hours in the appropriate shipping box to the NIDDK repository if living in the U.S. or their affiliated clinical center if living in Europe. The European clinical centers stored the stool samples and sent monthly bulk shipments of frozen stool to the NIDDK Repository. TEDDY Manual of Operations, including the stool sample collection protocol, can be accessed online at https://repository.niddk.nih.gov/static/studies/teddy/teddy_moop.pdf.
Metagenomic sequencing and initial bioinformatics
Samples were metagenomically sequenced as one library each multiplexed through Illumina HiSeq machines using using the 2×100 bp paired-end read protocol. Samples with limited DNA quantity and/or too few high quality reads were filtered out resulting in a discrepancy of sample frequencies between the metagenomic data and the 16S rRNA amplicon sequencing data analyzed in a companion paper [cite Stewart et al.]. Casava v1.8.2 (Illumina) output initial FASTQ files from the resulting data were processed using cutadapt v1.9dev2 for adapter removal, Trim Galore v0.2.8 (Babraham Bioinformatics) for removing low-quality bases and PRINSEQ v0.20.343 for sample demultiplexing. Bowtie2 v2.2.3 was used to map reads to the human genome for decontamination before subsequent analysis.
Taxonomic and functional profiling by MetaPhlAn and HUMAnN2
Taxonomic profiling of the metagenomic samples was performed using MetaPhlAn244 v2.6.0, which utilizes a library of clade-specific markers to provide pan-microbial (bacterial, archaeal, viral, and eukaryotic) quantification at the species level. MetaPhlAn2 was run using default settings.
Functional profiling was performed with HUMAnN245 v0.9.4. For an input metagenome, HUMAnN2 constructs a sample-specific reference database by concatenating and indexing the pangenomes of species detected in the sample by MetaPhlAn2 (pangenomes are pre-clustered, pre-annotated catalogs of open reading frames found across isolate genomes from a given species46). HUMAnN2 then maps sample reads against this database to quantify gene presence and abundance in a species-stratified manner, with unmapped reads further used in a translated search against UniRef9047 to include taxonomically-unclassified but functionally distinct gene family abundances. Finally, for community-total, species-stratified, and unclassified gene family abundance, HUMAnN2 reconstructs metabolic pathway abundance based on the subset of gene families annotated to metabolic reactions (based on reaction and pathway definitions from MetaCyc48). Enzyme [level-4 Enzyme Commision (EC) categories] abundances were further computed by summing the abundances of individual gene families annotated to each EC number based on UniRef90-EC annotations from UniProt49.
Phenotype and covariate analysis
This study includes extensive collection of clinical covariates covering several aspects of common and rare life events in early childhood from infancy through up to five years of age. In these analyses, we used information that is, according to literature, of high relevance in terms of the gut microbiome development. Information about mothers, pregnancy and birth was collected during the three-month clinic visit by questionnaire and included the mode of birth (vaginal birth vs. Caesarean section), gestational age, infant’s 5-minute Apgar score, information about maternal diabetes (T1D, T2D or gestational diabetes) and maternal insulin and medication use (antibiotics, ACE inhibitors, Metformin, Glyburide, antihypertensives) during pregnancy. Dietary information used in these analyses include the date of start (and end) for following dietary compounds: breastfeeding, baby formula, cow’s milk, gluten, cereals, meat, vegetables, fruits. The start of solid food (anything else than breast milk or cow’s milk) was also analyzed separately. T1D associated autoantibodies, IAA, GADA and IA2A, were analysed from serum samples collected at every clinic visit. In addition to the IA, defined as persistent confirmed autoantibody seropositivity, we analyzed the data in terms
of persistency of AABs and by counting cumulative frequency of AABs appeared. In TEDDY, all prescribed antibiotic courses are recorded. We further stratified these data by the type of antibiotic in five categories: amoxicillin, penicillin, cephalosporins, macrolide and other antibiotics. Information about probiotics covered the dates for starting and stopping probiotic supplementation, but not the specific types of probiotics used. Additionally, sex, information whether any first degree relatives in family had T1D and HLA haplotypes of the subjects were used in these analyses. Subjects screened from the general population were identified with high-risk alleles (89%) including: DRB1*04-DQA1*03-DQB1*03:02/DRB1*03-DQA1*05-DQB1*02:01 (DR3/4), DRB1*04-DQA1*03-DQB1*03:02/DRB1*04-DQA1*03-DQB1*03:02 (DR4/4), DRB1*04-DQA1*03-DQB1*03:02/DRB1*08-DQA1*04-DQB1*04:02 (DR4/8) and DRB1*03-DQA1*05-DQB1*02:01/DRB1*03-DQA1*05-DQB1*02:01 (DR3/3), plus six genotypes specific to first degree relatives28.
PCoA ordination was generated using t-distributed stochastic neighbor embedding (t-SNE) as implemented in Rtsne package in R with Bray-Curtis dissimilarity as the distance measure and perplexity (a free parameter) equal to 50. Statistical significance of the trends between early clusters and metadata were tested using mixed effect logistic regression and samples collected during the first year of life as follows. The target variable used was a binary indicator whether the relative abundance of the taxon of interest (three different Bifidobacterium species or phylum Proteobacteria) was greater than 0.5 (definition of the cluster). The age of sample collection, mode of delivery, clinical center, breastfeeding status (ongoing / stopped), solid food status (binary variable indicating whether solid food was introduced in the diet) and antibiotics status (binary variable indicating whether the subject received antibiotics during the last 30 days) were used as fixed effects and the subject ID was used as a random effect.
Associations between microbial feature abundances and metadata were determined using MaAsLin50. Briefly, this multivariate linear modeling system for microbial data selects from among a set of (potentially high-dimensional) covariates to associate with microbial taxon or pathway abundances. Mixed effects linear models using a variance-stabilizing arcsin square root transform on relative abundances are then used to determine the significance of putative associations from among this reduced set. Nominal p-values were adjusted using the Benjamini-Hochberg false discovery rate (FDR) method. Here, microbial features with corrected q < 0.25 were reported.
Associations between IA onset and microbial pathways were tested as described previously39. Briefly, pseudocount 26 was added to CPM values to stabilize the variation in lowly abundant and/or prevalent but highly variable categories. log2-transformed data were modeled using a mixed effect model (glmmPQL from the MASS package in R) with subject ID as a random effect and age of sample collection, mode of delivery, clinical center (for cohort-wide comparisons), breastfeeding status (ongoing / stopped), solid food status (binary variable indicating whether solid food was introduced in the diet), number of sequencing reads and IA case-control outcome as fixed effects. The nominal p-values for the IA case-control outcome fixed effect coefficient were adjusted using Benjamini-Hochberg FDR as above, and pathways with corrected q<0.25 were reported.
As previously described39, to associate microbial diversity with covariates while accounting for non-linear, age-dependent effects, we first fitted a sigmoid function (nls function in R) to account for the longitudinal trend. Residuals of this model were then used as inputs for a mixed-effect model (glmmPQL function in the MASS R package) with subject IDs as random effects to account for repeated measures in the data. Other factors were included in the model as fixed effects and their significance were evaluated using p-values reported by the model (Table S2).
Association between T1D case-control outcome and microbial alpha diversity in individual clinical centers was tested using a linear mixed effects model (glmmPQL function in MASS R package) on samples 730 days or less prior to T1D diagnosis. In the model, age at stool sample collection and T1D case-control outcome were used as fixed effects, and subject ID was used as a random effect.
Microbial variance explained by clinical and other covariates
Variance analysis was conducted using the adonis function in the vegan R package given a Bray-Curtis dissimilarity matrix of the taxonomic profiles and all TEDDY clinical metadata listed above. Briefly, adonis conducts multivariate analysis of variance (MANOVA) using the dissimilarity matrix (i.e. partitions the sums of squares) given the metadata as covariates. Statistical significance of the fit was assessed using permutation tests.
HMO gene homology
B. infantis HMO gene cluster homologs across multiple taxa were analyzed as follows. UniRef90 gene families corresponding to the protein sequences in B. infantis HMO gene cluster38 (protein sequences Blon_2331-Blon_2361 in NCBI protein sequence database) were identified by translated BLAST search against ChocoPhlAn pangenome collection46 utilized by HUMAnN2. Identified hits were further filtered by requiring >=50% alignment identity and >=80% mutual coverage. Combining this information with HUMAnN2 species-stratified UniRef90 gene family quantification enabled calling these genes present given that they had sufficient read coverage, here defined as log10(counts-per-million) > 0.1 in at least 50 samples collected during breastfeeding. Differential gene prevalence during breastfeeding was tested using the samples where the carrier species had >1% relative abundance. Testing was conducted using the test of equal or given proportions (prop.test() function in R) and by comparing the prevalence (proportion of the samples where the species in question harbored the gene according to the metagenomic data) of the gene in samples collected during breastfeeding with the samples collected after weaning. P-values were adjusted for multiple testing by Benjamini-Hochberg method (p.adjust function in R). All homologs together with their BLAST search metrics, prevalence in the metagenomic data and corresponding B. infantis HMO gene are reported in Table S5.
Bacterial growth assays
Bifidobacterium bifidum strain RJX-1201, Bifidobacterium breve RJX-1202 and Bifidobacterium longum RJX-1203 were streaked on brain heart infusion agar (BD) supplemented with 1% vitamin K/hemin solution (BD; sBHI), and incubated for 48 hours in a vinyl anaerobic chamber (Coy Laboratory Products) containing 5% CO2, 5%H2, and 90% N2 and maintained at 37 C. Cells were transferred to sBHI liquid medium (BHI broth, BD, supplemented as above) and grown for 24 hours in anaerobic conditions. Cultures were washed twice with PBS and OD600 measured using a BioTek PowerWave 340 plate reader. OD600 was normalized to 0.2 for all strains and 5 μl bacteria inoculum was added to a final volume of 200 μl containing 10% sBHI and 125 mM carbon source (glucose, fucose, galactose, or lactose) in a 96 well plate. OD600 was measured in the plate reader every hour for 48 hours with 5 seconds of medium shaking prior to each measurement. All the measurements were normalized to a medium-only blank. Experiment was repeated three times (n=3) in triplicate and one representative experiment is shown. Error bars are standard deviation of three technical replicates.
Data Availability
TEDDY Microbiome 16S and WGS data that support the findings of this are available in NCBI’s database of Genotypes and Phenotypes (dbGaP) with the primary accession code phs001443.v1, in accordance with the dbGaP controlled-access authorization process. Clinical metadata analyzed during the current study are available in the NIDDK Central Repository at https://www.niddkrepository.org/studies/teddy.
Code Availability
Code for Random Forest case-control comparisons and cohort wide MaAsLin association analyses in Table S4 has been made publicly available at https://github.com/tvatanen/broad_teddy_microbiome_analyses. Other analysis software including quality control, taxonomic, and functional profilers is publicly available and referenced as appropriate.
Extended Data
Extended Data Figure 1: Heterogeneity in early taxonomic profiles.
a-d, Relative abundances of taxonomic groups highlighted by weighted averages in Fig. 2a (arrows) shown separately (N = 10,913 samples). e,f, Average longitudinal abundance of B. breve (e) and B. longum (f) per clinical center (N = 10,194 samples). The curves show locally weighted scatterplot smoothing (LOESS) for the relative abundances and shaded area shows 95% confidence interval for each fit, as implemented in geom_smooth function in ggplot2 R package. g-k, Growth curves of human infant isolates of B. breve, B. bifidum and B. longum grown individually in low-nutrient medium (10% sBHI) supplemented with single carbon sources (glucose (g), galactose (h), fucose (i) and lactose (j) or grown in 100% sBHI (k). As a negative control, growth curves of each strain grown in 10% BHI without additional sugar are shown in black for each condition. Data are representative of three (N = 3) independent experiments and are presented as the mean (and SD) of triplicate assessments.
Extended Data Figure 2: Stability and regional differences of taxonomic profiles.
a, Stability of the microbiota, measured by Jaccard index (N = 10,750 samples) in three-month time windows, measured in two-month increments, stratified into three groups: within subject, within clinical center, and across clinical centers. The line shows the median per time window and shaded area shows its 99% confidence interval estimated using binomial distribution. Compare to Fig. 2b, which shows the same analysis measured by Bray-Curtis dissimilarity. b-d, Average longitudinal abundance of Ruminococcus gnavus (b), Lactobacillus rhamnosus (c) and Veillonella parvula (d) per clinical center (N = 10,194 samples). The curves show LOESS fit for the relative abundances as above.
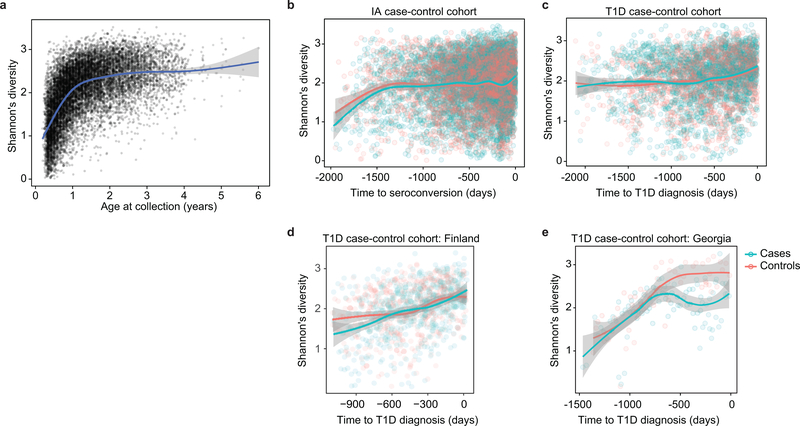
Extended Data Figure 3: Accrual of microbial alpha diversity.
a, Shannon’s diversity of the taxonomic profiles of the gut microbial communities (N = 10,913 samples) with respect to the age at the sample collection. The curve shows the generalized additive model (GAM) fit for the data and the shaded area shows 95% confidence interval the fit, as implemented in geom_smooth function in ggplot2 R package. b, Shannon’s diversity for the samples in IA case-control cohort (N = 7,051) with respect to time to the appearance of first autoantibody (seroconversion). The curves show LOESS fits for cases and controls separately, and the shaded area shows 95% confidence intervals for each fit. c, Shannon’s diversity for the samples in T1D case-control cohort (N = 3,309) with respect to time to T1D diagnosis. The curves and shaded areas are as in panel B. d, As panel (c), but only for data (N = 983 samples) for subjects in Finland. No difference between cases and controls. e, As panel (c), but only for data (N = 142 samples, N = 6 subjects) for subjects in Georgia, USA. Cases show a drop in alpha diversity prior to the T1D diagnosis (linear mixed effects model, p = 0.0033).
Extended Data Figure 4: Effects of antibiotics.
a, Influence of antibiotic courses on microbial stability, stratified in six-month time windows (x-axis). Stability was measured by Bray-Curtis (BC) dissimilarity over consecutive stool samples (<50 days apart) from the same individual between 3–29 months of age and stratified by whether antibiotics were given between the two samples. The box of the boxplot shows the interquartile range (IQR) of the data, and notch around the median (horizontal line in the box) show the approximation for 95% confidence interval (notch width equals 1.58 * IQR / sqrt(n), where n is number of samples per boxplot). Compare to Fig. 2c b., The influence of antibiotic courses on microbial diversity. Illustration shows boxplots of the increase (difference) in diversity between two consecutive stool samples (<50 days apart) stratified by antibiotic administration between the samples. Data shows no difference between the groups (antibiotics vs. no antibiotics). c, Influence of antibiotics courses on microbial diversity by antibiotic type; data on panel (b) stratified in one year time windows (x-axis) and antibiotic types (color of the boxplot). Data does not show consistent significant and differences between the antibiotic types. d,e, Influence of antibiotic courses on microbial stability by antibiotic type; data on Fig. 2c and Extended Data Fig. 3a stratified by antibiotic type. Panels show (d) LOESS fit for the relative abundances (shaded area shows 95% confidence interval for each fit, as implemented in geom_smooth function in ggplot2 R package) and (e) boxplot (as in previous panels) for the data per antibiotic type. Data does not show significant and consistent differences between the antibiotic types. No antibiotics, N = 7109; Amoxicillin, N = 268; Penicillin, N = 89; Cephalosporin, N = 51; Macrolide, N = 60; Other, N =99. f, Decreases in relative abundance of bacteria over antibiotic courses. Bacteria for which bootstrapped 95 % confidence interval of the fold change doesn’t overlap zero are shown. Fold change was measured between consecutive samples with an antibiotic course between them, given that the species in question was present in the first of the two samples. Sample size per species (N) indicate the number of sample pairs where the species in question was present in the sample preceding the antibiotic treatment. The bars show bootstrapped mean log2 fold change (decrease) and error bars show their standard deviations (N = 1,000 bootstrap samples).
Extended Data Figure 5: Dynamics of species-specific microbial functional potential during early gut development.
a,b, Stability of microbial pathways (N = 10,580 samples) measured by Bray-Curtis dissimilarity (a) and Jaccard Index (b) and stratified in three groups: within subject, within clinical center, and across clinical centers. While the baseline level of functional similarity is significantly greater than that of taxa (see Fig. 2b), functional states and development trajectories both retain a level of personalization as well. The stability of the functional profiles was evaluated in three-month time windows, over two-month increments. Lines show the median per time window and shaded area shows its 99% confidence interval estimated using binomial distribution. c,d, Proportion of metagenomic gene abundance with functional annotation through Gene Ontology (c) and MetaCyc (d) databases. The metagenomic reads was divided into following categories: reads that could be mapped to genes with functional assignment in the database in question (annotated), and reads with no annotation but alignment to species pangenomes or UniProt proteins (unannotated). The proportion of the unknown genes (unmapped) was estimated using the number of reads with unknown origin. e, The proportion of unmapped reads, reflecting the relative abundances of reads not mappable to any microbial pan-genomes in the available reference set or to UniProt. An increasing trend of unmapped reads with respect to the age at sample collection continued through approximately two years of age. f, The proportion of reads with confident functional annotation in MetaCyc within the genes that mapped to species pangenomes or UniProt proteins. The data again showed an increasing longitudinal trend, implicating a deficit of functional and biochemical annotations within microbes abundant during the first year of life.
Extended Data Figure 6: Error rates for RF classification between T1D cases and controls.
a, The gut microbiome functional (left) and taxonomic (right) profiles were classified between cases and controls using leave-one-out cross-validation (N = 3,366 samples), where one case-control pair was held-out in turn. Data shows error rates for classifying these held-out samples per fold (a data point per fold, N = 100 folds). This suggests weak but better-than-random classification between cases and controls. The box of the boxplot (overlaid on the data) shows the interquartile range (IQR) of the data, and notch around the median (horizontal line in the box) show the approximation for 95% confidence interval (notch width equals 1.58 * IQR / sqrt(n), where n is number of samples per boxplot). b, Average longitudinal abundance of Ruminococcus gnavus in Finland (N = 2,630 samples) stratified by the number of observed persistent AABs; no AABs (i.e. healthy control), a single AAB, or multiple (two or more) AABs. c, Average longitudinal abundance of Lactobacillus rhamnosus in IA cases and controls (N = 7,017 samples). L. rhamnosus is more abundant in controls (q = 0.055). The curves in (b) and (c) show LOESS fit per group and shaded areas shows 95% confidence interval for each fit, as implemented in geom_smooth function in ggplot2 R package. d, Abundance (left) and prevalence (right) of Lactobacillus reuteri and L. rhamnosus in the first stool sample of each individual (collected at approximately the age of three months) in association with early probiotic supplementation. “No probiotic” indicates no probiotics given prior to the first stool sample (N = 583); “later probiotic” refers to probiotics given later than the first four weeks but prior to the first stool sample (N = 45); “early probiotic” refers to probiotics given during the first four weeks of life (N = 84). N per clinical center are given in Extended Data Table 2. L. reuteri and L. rhamnosus were more abundant and prevalent in groups with probiotics supplementation. Visual jitter was added to make data equal to zero distinguishable and boxes show IQR of the data, when applicable. The shown p-values were obtained by applying Fisher’s Exact Test (two-sided) to presence/absence count data (counting samples where the species were present).
Extended Data Figure 7: Contrasting HMO utilization genes in B. pseudocatenulatum.
The gene families involved in HMO utilization and showing contrasting presence in B. pseudocatenulatum genomes during breastfeeding (N = 321 samples) compared to after weaning (N = 1,004 samples). Columns represent stool samples in which the relative abundance of species B. pseudocatenulatum relative abundance >10 % (N = 1,325 samples). Rows and columns were ordered by hierarchical clustering using complete linkage method. Compare to Fig. 4 which shows similar data for B. longum. UniRef90 identifiers and gene names or families are indicated on the left.
Extended Data Table 1. Summary of TEDDY microbiome cohort.
Data on subjects’ ethnic background was not systematically collected in European clinical centers but these study populations were predominantly white, non-hispanic. Reported antihypertensive drugs were Atenolol (N = 2), Bisoprolol (N = 1), Labetalol (N = 6), methyldopa (N = 1), Methyldopa + Methyldopate (N = 3), Metoprolol (N = 4), Nifedipine (N = 5). No use of angiotensin-converting enzyme (ACE) inhibitors was reported. Numbers indicate the number of subjects (N) if not specified otherwise.
| US, Colorado | US, Georgia | US, Washington | Finland | Germany | Sweden | |
|---|---|---|---|---|---|---|
| T1D cases (samples) | 14 (274) | 3 (89) | 8 (111) | 34 (553) | 13 (246) | 29 (532) |
| IA cases (samples) | 39 (689) | 17 (252) | 25 (368) | 70 (900) | 21 (292) | 95 (1,542) |
| Healthy controls (samples) | 61 (906) | 22 (250) | 36 (399) | 119 (1,273) | 40 (512) | 137 (1,725) |
| Sex | ||||||
| Male / Female | 61 / 53 | 19 / 23 | 51 / 18 | 117 / 106 | 30 / 44 | 152 / 109 |
| Ethnic background | ||||||
| White, non-hispanic | 86 (75.4%) | 41 (97.6%) | 56 (81.2%) | N/A | N/A | N/A |
| Mode of birth | ||||||
| Caesarean section | 41 (36.0%) | 22 (52.4%) | 25 (36.2%) | 42 (18.8%) | 23 (31.1%) | 46 (17.6%) |
| Probiotic supplementation | ||||||
| Probiotics during first 4 weeks | 0 | 2 (4.8%) | 0 | 67 (30.0%) | 7 (9.5%) | 14 (5.4%) |
| Probiotics during follow-up | 22 (19.3%) | 13 (31.0%) | 9 (13.0%) | 162 (72.6%) | 33 (44.6%) | 58 (22.2%) |
| Breastfeeding | ||||||
| Median duration (days) | 268 | 301 | 335 | 289 | 278 | 228 |
| duration, 25 percentile | 56 | 145 | 171 | 152 | 140 | 98 |
| duration, 75 percentile | 396 | 365 | 440 | 385 | 367 | 304 |
| Number of subjects never breastfed | 3 | 3 | 1 | 0 | 0 | 0 |
| Maternal characteristics | ||||||
| Maternal T1D | 7 (6.1%) | 0 | 3 (4.3%) | 14 (6.3%) | 18 (24.3%) | 7 (2.7%) |
| Maternal T2D | 2 (1.8%) | 0 | 0 | 0 | 0 | 0 |
| Gestational diabetes | 5 (4.4%) | 5 (11.9%) | 5 (7.2%) | 32 (14.3%) | 3 (4.1%) | 6 (2.3%) |
| Antibiotics during pregnancy | 21 (18.4%) | 10 (23.8%) | 5 (7.2%) | 40 (17.9%) | 13 (17.6%) | 29 (11.1%) |
| Metformin during pregnancy | 1 (0.9%) | 0 | 0 | 1 (0.4%) | 0 | 0 |
| Glyburide during pregnancy | 2 (1.8%) | 2 (4.8%) | 2 (2.9%) | 0 | 0 | 0 |
| Antihypertensives during pregnancy | 4 (3.5%) | 3 (7.1%) | 4 (5.8%) | 5 (2.2%) | 3 (4.1%) | 0 |
| Insulin during pregnancy | 9 (7.9%) | 0 | 3 (4.3%) | 23 (10.3%) | 19 (25.7%) | 8 (3.1%) |
Extended Data Table 2. Antibiotics and probiotics.
3,678 antibiotic prescriptions in TEDDY microbiome study population by clinical center (top). Early probiotic supplementation in TEDDY clinical centers (bottom). Probiotic use was stratified in three categories: probiotics during first 4 weeks of life (Early probiotic), probiotics prior to the first stool sample (roughly at three months) but not first 4 weeks (Later probiotic), and no probiotics prior to the first stool sample (No probiotic). Data for probiotics are presented as N (percentage). Abx = antibiotics.
| US, Colorado | US, Georgia | US, Washington | Finland | Germany | Sweden | |
|---|---|---|---|---|---|---|
| Subjects with abx prescriptions | 93 (81.6%) | 37 (88.1%) | 54 (78.3%) | 206 (92.4%) | 56 (75.7%) | 192 (73.6%) |
| Median number of abx per subject (25th and 75th percentile) | 2 (1–6) | 5 (2–9) | 2 (1–4) | 6 (3–11) | 2 (0–5) | 2 (0–4) |
| Number of abx by type (prescriptions per subject) | ||||||
| Amoxicillin | 242 (2.12) | 147 (3.50) | 104 (1.51) | 769 (3.45) | 45 (0.61) | 134 (0.51) |
| Cephalosporins | 87 (0.76) | 65 (1.55) | 31 (0.45) | 127 (0.57) | 51 (0.69) | 23 (0.09) |
| Macrolide | 54 (0.47) | 35 (0.83) | 47 (0.68) | 203 (0.91) | 33 (0.45) | 23 (0.09) |
| Penicillin | 6 (0.05) | 2 ( 0.05) | 3 (0.04) | 17 (0.08) | 13 (0.18) | 412 (1.58) |
| Other | 76 (0.67) | 80 (1.90) | 33 (0.48) | 521 (2.34) | 77 (1.04) | 154 (0.59) |
| Total | 465 (4.08) | 329 (7.83) | 218 (3.16) | 1,637 (7.34) | 219 (2.96) | 746 (2.86) |
| Probiotic use in early life | ||||||
| Early probiotic | 0 (0.0%) | 1 ( 2.9%) | 0 (0.0%) | 63 (30.7%) | 7 (10.0%) | 13 (5.6%) |
| Later probiotic | 1 (0.9%) | 1 ( 2.9%) | 2 (3.3%) | 16 (7.8%) | 8 (11.4%) | 17 (7.3%) |
| No probiotic | 109 (99.1%) | 32 (94.1%) | 59 (96.7%) | 126 (61.5%) | 55 (78.6%) | 202 (87.1%) |
Supplementary Material
Acknowledgements
This research was performed on behalf of the TEDDY Study Group, which is funded by U01 DK63829, U01 DK63861, U01 DK63821, U01 DK63865, U01 DK63863, U01 DK63836, U01 DK63790, UC4 DK63829, UC4 DK63861, UC4 DK63821, UC4 DK63865, UC4 DK63863, UC4 DK63836, UC4 DK95300, UC4 DK100238, UC4 DK106955, UC4 DK112243, UC4 DK117483, and Contract No. HHSN267200700014C from the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), National Institute of Allergy and Infectious Diseases (NIAID), National Institute of Child Health and Human Development (NICHD), National Institute of Environmental Health Sciences (NIEHS), Centers for Disease Control and Prevention (CDC), and JDRF. This work supported in part by the NIH/NCATS Clinical and Translational Science Awards to the University of Florida (UL1 TR000064) and the University of Colorado (UL1 TR001082). C.H. was supported by funding from JDRF (3-SRA-2016–141-Q-R) and NIDDK (U54DE023798, R24DK110499). H.V. and R.J.X. were supported by funding from JDRF (2-SRA-2016–247-S-B, 2-SRA-2018–548-S-B).
Footnotes
The authors declare no competing interests.
Supplementary Information
Supplementary Information is available in the online version of this paper.
References
- 1.Katsarou A et al. Type 1 diabetes mellitus. Nat Rev Dis Primers 3, 17016, doi: 10.1038/nrdp.2017.16 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Pociot F & Lernmark A Genetic risk factors for type 1 diabetes. Lancet 387, 2331–2339, doi: 10.1016/S0140-6736(16)30582-7 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Rewers M & Ludvigsson J Environmental risk factors for type 1 diabetes. Lancet 387, 2340–2348, doi: 10.1016/S0140-6736(16)30507-4 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Knip M & Siljander H The role of the intestinal microbiota in type 1 diabetes mellitus. Nat Rev Endocrinol 12, 154–167, doi: 10.1038/nrendo.2015.218 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Hober D & Sauter P Pathogenesis of type 1 diabetes mellitus: interplay between enterovirus and host. Nat Rev Endocrinol 6, 279–289, doi: 10.1038/nrendo.2010.27 (2010). [DOI] [PubMed] [Google Scholar]
- 6.Paun A, Yau C & Danska JS The Influence of the Microbiome on Type 1 Diabetes. J Immunol 198, 590–595, doi: 10.4049/jimmunol.1601519 (2017). [DOI] [PubMed] [Google Scholar]
- 7.de Goffau MC et al. Aberrant gut microbiota composition at the onset of type 1 diabetes in young children. Diabetologia 57, 1569–1577, doi: 10.1007/s00125-014-3274-0 (2014). [DOI] [PubMed] [Google Scholar]
- 8.de Goffau MC et al. Fecal microbiota composition differs between children with beta-cell autoimmunity and those without. Diabetes 62, 1238–1244, doi: 10.2337/db12-0526 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marino E et al. Gut microbial metabolites limit the frequency of autoimmune T cells and protect against type 1 diabetes. Nat Immunol 18, 552–562, doi: 10.1038/ni.3713 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Needell JC & Zipris D The Role of the Intestinal Microbiome in Type 1 Diabetes Pathogenesis. Curr Diab Rep 16, 89, doi: 10.1007/s11892-016-0781-z (2016). [DOI] [PubMed] [Google Scholar]
- 11.Davis-Richardson AG et al. Bacteroides dorei dominates gut microbiome prior to autoimmunity in Finnish children at high risk for type 1 diabetes. Front Microbiol 5, 678, doi: 10.3389/fmicb.2014.00678 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kostic AD et al. The dynamics of the human infant gut microbiome in development and in progression toward type 1 diabetes. Cell Host Microbe 17, 260–273, doi: 10.1016/j.chom.2015.01.001 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Endesfelder D et al. Compromised gut microbiota networks in children with anti-islet cell autoimmunity. Diabetes 63, 2006–2014, doi: 10.2337/db13-1676 (2014). [DOI] [PubMed] [Google Scholar]
- 14.Maffeis C et al. Association between intestinal permeability and faecal microbiota composition in Italian children with beta cell autoimmunity at risk for type 1 diabetes. Diabetes Metab Res Rev 32, 700–709, doi: 10.1002/dmrr.2790 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Mejia-Leon ME, Petrosino JF, Ajami NJ, Dominguez-Bello MG & de la Barca AM Fecal microbiota imbalance in Mexican children with type 1 diabetes. Sci Rep 4, 3814, doi: 10.1038/srep03814 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alkanani AK et al. Alterations in Intestinal Microbiota Correlate With Susceptibility to Type 1 Diabetes. Diabetes 64, 3510–3520, doi: 10.2337/db14-1847 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Soyucen E et al. Differences in the gut microbiota of healthy children and those with type 1 diabetes. Pediatr Int 56, 336–343, doi: 10.1111/ped.12243 (2014). [DOI] [PubMed] [Google Scholar]
- 18.Endesfelder D et al. Towards a functional hypothesis relating anti-islet cell autoimmunity to the dietary impact on microbial communities and butyrate production. Microbiome 4, 17, doi: 10.1186/s40168-016-0163-4 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao L et al. Gut bacteria selectively promoted by dietary fibers alleviate type 2 diabetes. Science 359, 1151–1156, doi: 10.1126/science.aao5774 (2018). [DOI] [PubMed] [Google Scholar]
- 20.Markle JG et al. Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science 339, 1084–1088, doi: 10.1126/science.1233521 (2013). [DOI] [PubMed] [Google Scholar]
- 21.Costa FR et al. Gut microbiota translocation to the pancreatic lymph nodes triggers NOD2 activation and contributes to T1D onset. J Exp Med 213, 1223–1239, doi: 10.1084/jem.20150744 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yatsunenko T et al. Human gut microbiome viewed across age and geography. Nature 486, 222–227, doi: 10.1038/nature11053 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Palmer C, Bik EM, DiGiulio DB, Relman DA & Brown PO Development of the human infant intestinal microbiota. PLoS Biol 5, e177, doi: 10.1371/journal.pbio.0050177 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koenig JE et al. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci U S A 108 Suppl 1, 4578–4585, doi: 10.1073/pnas.1000081107 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dominguez-Bello MG et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A 107, 11971–11975, doi: 10.1073/pnas.1002601107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Backhed F et al. Dynamics and Stabilization of the Human Gut Microbiome during the First Year of Life. Cell Host Microbe 17, 690–703, doi: 10.1016/j.chom.2015.04.004 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Yassour M et al. Natural history of the infant gut microbiome and impact of antibiotic treatment on bacterial strain diversity and stability. Sci Transl Med 8, 343ra381, doi: 10.1126/scitranslmed.aad0917 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hagopian WA et al. The Environmental Determinants of Diabetes in the Young (TEDDY): genetic criteria and international diabetes risk screening of 421 000 infants. Pediatr Diabetes 12, 733–743, doi: 10.1111/j.1399-5448.2011.00774.x (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee HS et al. Biomarker discovery study design for type 1 diabetes in The Environmental Determinants of Diabetes in the Young (TEDDY) study. Diabetes Metab Res Rev 30, 424–434, doi: 10.1002/dmrr.2510 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhernakova A et al. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science 352, 565–569, doi: 10.1126/science.aad3369 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Korpela K et al. Intestinal microbiome is related to lifetime antibiotic use in Finnish pre-school children. Nat Commun 7, 10410, doi: 10.1038/ncomms10410 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Joice R, Yasuda K, Shafquat A, Morgan XC & Huttenhower C Determining microbial products and identifying molecular targets in the human microbiome. Cell Metab 20, 731–741, doi: 10.1016/j.cmet.2014.10.003 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ashburner M et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25, 25–29, doi: 10.1038/75556 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O’Callaghan A & van Sinderen D Bifidobacteria and Their Role as Members of the Human Gut Microbiota. Front Microbiol 7, 925, doi: 10.3389/fmicb.2016.00925 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thurston B, Dawson KA & Strobel HJ Pentose utilization by the ruminal bacterium Ruminococcus albus. Appl Environ Microbiol 60, 1087–1092 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Uusitalo U et al. Association of Early Exposure of Probiotics and Islet Autoimmunity in the TEDDY Study. JAMA Pediatr 170, 20–28, doi: 10.1001/jamapediatrics.2015.2757 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Underwood MA, German JB, Lebrilla CB & Mills DA Bifidobacterium longum subspecies infantis: champion colonizer of the infant gut. Pediatr Res 77, 229–235, doi: 10.1038/pr.2014.156 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sela DA et al. The genome sequence of Bifidobacterium longum subsp. infantis reveals adaptations for milk utilization within the infant microbiome. Proc Natl Acad Sci U S A 105, 18964–18969, doi: 10.1073/pnas.0809584105 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vatanen T et al. Variation in Microbiome LPS Immunogenicity Contributes to Autoimmunity in Humans. Cell 165, 842–853, doi: 10.1016/j.cell.2016.04.007 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Teddy Study Group. The Environmental Determinants of Diabetes in the Young (TEDDY) study: study design. Pediatr Diabetes 8, 286–298, doi: 10.1111/j.1399-5448.2007.00269.x (2007). [DOI] [PubMed] [Google Scholar]
- 41.Teddy Study Group. The Environmental Determinants of Diabetes in the Young (TEDDY) Study. Ann N Y Acad Sci 1150, 1–13, doi: 10.1196/annals.1447.062 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.American Diabetes Association. (2) Classification and diagnosis of diabetes. Diabetes Care 38 Suppl, S8–S16, doi: 10.2337/dc15-S005 (2015). [DOI] [PubMed] [Google Scholar]
- 43.Schmieder R & Edwards R Quality control and preprocessing of metagenomic datasets. Bioinformatics 27, 863–864, doi: 10.1093/bioinformatics/btr026 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Truong DT et al. MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nat Methods 12, 902–903, doi: 10.1038/nmeth.3589 (2015). [DOI] [PubMed] [Google Scholar]
- 45.Abubucker S et al. Metabolic reconstruction for metagenomic data and its application to the human microbiome. PLoS Comput Biol 8, e1002358, doi: 10.1371/journal.pcbi.1002358 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang K et al. MetaRef: a pan-genomic database for comparative and community microbial genomics. Nucleic Acids Res 42, D617–624, doi: 10.1093/nar/gkt1078 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Suzek BE et al. UniRef clusters: a comprehensive and scalable alternative for improving sequence similarity searches. Bioinformatics 31, 926–932, doi: 10.1093/bioinformatics/btu739 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Caspi R et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res 44, D471–480, doi: 10.1093/nar/gkv1164 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.The UniProt Consortium. UniProt: the universal protein knowledgebase. Nucleic Acids Res 45, D158–D169, doi: 10.1093/nar/gkw1099 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morgan XC et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol 13, R79, doi: 10.1186/gb-2012-13-9-r79 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
TEDDY Microbiome 16S and WGS data that support the findings of this are available in NCBI’s database of Genotypes and Phenotypes (dbGaP) with the primary accession code phs001443.v1, in accordance with the dbGaP controlled-access authorization process. Clinical metadata analyzed during the current study are available in the NIDDK Central Repository at https://www.niddkrepository.org/studies/teddy.