Abstract
Poly(ADP-ribose) polymerases (PARPs) are abundant nuclear proteins that synthesize ADP ribose polymers (pADPr) and catalyze the addition of (p)ADPr to target biomolecules. PARP1, the most abundant and well-studied PARP, is a multifunctional enzyme that participates in numerous critical cellular processes. A considerable amount of PARP research has focused on PARP1’s role in DNA damage. However, an increasing body of evidence outlines more routine roles for PARP and PARylation in nearly every step of RNA biogenesis and metabolism. PARP1’s involvement in these RNA processes is pleiotropic and has been ascribed to PARP1’s unique flexible domain structures. PARP1 domains are modular self-arranged enabling it to recognize structurally diverse substrates and to act simultaneously through multiple discrete mechanisms. These mechanisms include direct PARP1-protein binding, PARP1-nucleic acid binding, covalent PARylation of target molecules, covalent autoPARylation and induction of noncovalent interactions with PAR molecules. A combination of these mechanisms has been implicated in PARP1’s context-specific regulation of RNA biogenesis and metabolism. We examine the mechanisms of PARP1 regulation in transcription initiation, elongation and termination, co-transcriptional splicing, RNA export, and post-transcriptional RNA processing. Finally, we consider promising new investigative avenues for PARP1 involvement in these processes with an emphasis on PARP1 regulation of subcellular condensates.
Graphical abstract:
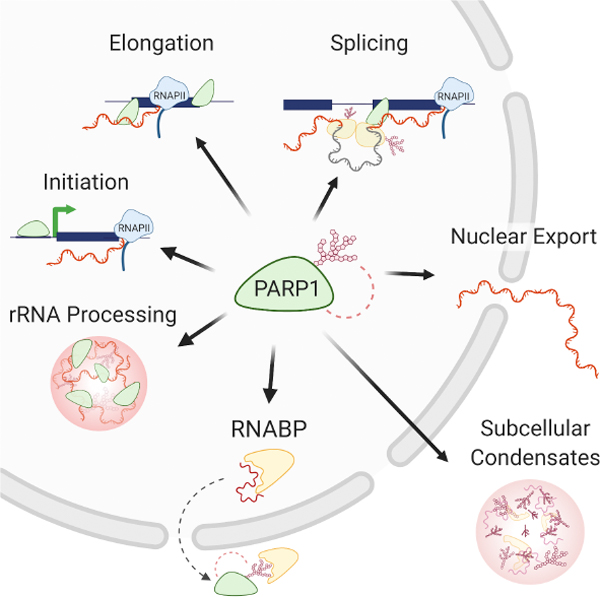
PARP1 plays functional roles in the mRNA life cycle: from biogenesis (transcription, splicing and polyadenylation) to modifications (m6A), stability, export, and ribosome assembly (translation). This regulation occurs either through its direct binding to its binding partners (DNA, RNA, protein) and/or through PARylation or PAR recognition by target molecules.
1. INTRODUCTION
The genome is stored in the nucleus as chromatin, which is composed of DNA and proteins. The basic unit of chromatin is the nucleosome, which consists of roughly 146 bp DNA wound around an octamer of histone proteins. The structural organization of repeated nucleosomes allows for orderly compaction of vast genomic material within the confines of the nucleus and helps to protect an organism’s genetic information from damage. However, the tight compaction of chromatin must be alleviated for nuclear processes, which require direct access to DNA such as transcription, replication and DNA repair. Thus, factors that regulate the structure of chromatin must encompass mechanisms for both deliberate, targeted alterations to chromatin structure in response to cell signaling and to respond rapidly to environmental cues and DNA damage. Poly (ADP-ribose) polymerase (PARP) is one such family of multifunctional enzymes that are able to mediate these complex changes to chromatin structure.
PARP1 belongs to a group of proteins also known as ADP-ribosyl transferases, composed of 18 members in humans (Barkauskaite, Jankevicius, & Ahel, 2015). PARP1, the founding member of this group of proteins, is often called the guardian of the genome. PARP1 has well described roles in responding to DNA damage, either by loosening chromatin at specific loci to promote DNA repair, or in the case of extensive damage, promoting cell death for the maintenance of genome integrity. More recently it has been understood that in the absence of DNA damage, PARP1 also plays an integral role in regulating gene expression. Given PARP1’s influence over chromatin structure, it is logical to envision PARP’s role in the opening of chromatin to enable transcriptional initiation. However, over the last decade, it is becoming increasingly clear that PARP1’s influence on gene expression is not limited to initiation. Indeed, PARP1 is now known to be involved in many steps of the RNA life cycle from biogenesis to splicing to post-transcriptional processing. We focus on the association of PARP with RNA and novel roles of PARP in RNA biogenesis and metabolism.
2. DOMAIN FUNCTIONS OF PARP
Aside from histones, the most abundant nuclear proteins are the PARP family of enzymes (Virag & Szabo, 2002). PARPs catalyze the transfer of an ADP-ribose (ADPr) moiety from the oxidized form of nicotinamide adenine dinucleotide (NAD+) to a target molecule. This deposition can be mono, branched or linear and depends on the PARP family member involved, as well as the cellular context. Only a few of the PARPs (PARP1, 2, 5A, 5B) are capable of true polymerization (PARylation) while the majority, transfer only a single ADPr (Barkauskaite et al., 2015; Leung, 2014; Vyas & Chang, 2014) (MARylation) onto target molecules. For many years it was thought that ADPr transfer (MARylation and PARylation) occurred only on proteins via ester linkage at 7 amino acid residues (asparagine, aspartic acid, glutamic acid, arginine, lysine, serine and cysteine) (Bonfiglio, Colby, & Matic, 2017; Daniels, Ong, & Leung, 2017; Palazzo et al., 2018; Vyas & Chang, 2014). However, recent studies now show MARylation and PARylation occurring on DNA (Munnur, Bartlett, et al., 2019; Munnur, Somers, et al., 2019; Talhaoui et al., 2016), RNA (Munnur, Bartlett, et al., 2019), antibiotics (reviewed in Palazzo, Mikoc, and Ahel (2017)) and other small molecules (Kirby & Cohen, 2019). This covalent modification is degraded primarily by poly (ADP-ribose) glycohydrolase (PARG). The cellular levels of ADPrs (mono and poly), are maintained by locally altering enzyme kinetics between PARP and PARG activity.
PARP activity has been demonstrated in all types of eukaryotic cells with the exception of yeast (Ame, Spenlehauer, & de Murcia, 2004). Identification of the 18 members of the human PARP family has been either via homology and/or direct observation of PARylation activity (Ame et al., 2004). While each PARP family member contains the PARylation signature, they vary in the presence and composition of additional domains, hinting at distinct functional roles for the PARP proteins. The large diversity of PARPs in humans confounds studies that attempt to elucidate the function of PARPs. More so, though human PARPs have different domains, the catalytic domain is highly conserved, making catalytic inhibitors selective to individual PARPs impossible. The Drosophila genome on the other hand, which encodes only PARP (PARP1) and a Tankyrase, has become an ideal model for studying the function of PARP1.
PARP1, the most abundant and highly studied of the PARPs, is responsible for approximately 90% of PARylation activity (Burkle, Diefenbach, Brabeck, & Beneke, 2005). PARP1 consists of three major functional domains: 1) an N-terminal nucleic acid binding domain containing three zinc fingers (Zn1, Zn2, Zn3), 2) a BRCT-containing automodification domain, and 3) a C-terminal domain responsible for both protein interactions via a WGR subdomain as well as PARylation via the catalytic domain (Figure 1A). Within this catalytic domain are two subdomains: an ADP-ribosyltransferase subdomain that contains the active site and a fold that is conserved in all PARP family members and a helical domain (HD) conserved in the DNA damage-dependent PARPs 1, 2, and 3 (Hottiger, Hassa, Luscher, Schuler, & Koch-Nolte, 2010). Following DNA damage, PARP1 is activated, forms long, branched PAR chains, which are transferred onto its automodification domain, as well as target molecules. PARylation of PARP1 occurs at several residues outside of the BRCT region (Gagne et al., 2005; Gagne et al., 2012), causing a conformational shift in PARP1’s structure, which results in its release from the damaged DNA substrate, and subsequent inactivation (Steffen, McCauley, & Pascal, 2016). Once released from the damaged DNA region, PARP1 aids in the shuttling and recycling of its associated proteins (Ke, Zhang, Lv, Zeng, & Ba, 2019).
Figure 1. PARP1 Domain Structure.
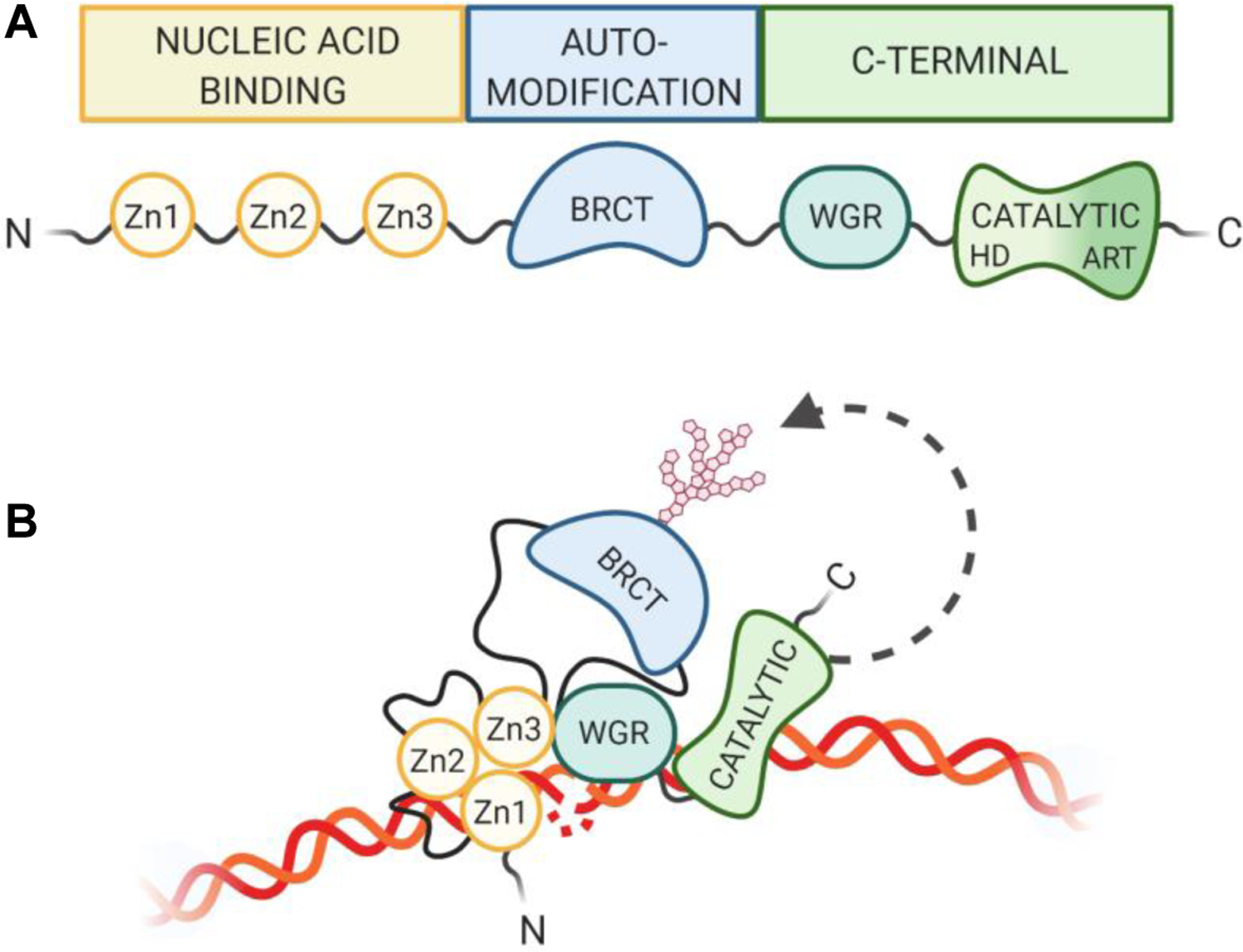
(A) PARP1 is composed of 3 main regions, consisting of 6 independently folded domains connected by flexible linker regions. The N-terminal nucleic acid binding region contains three zinc finger domains (Zn1, Zn2, Zn3). The BRCT-containing automodification domain lies in the PARP1 interior. The C-terminal region contains an additional nucleic acid binding motif (WGR) as well as the catalytic domain, which is made up of an alpha helical subdomain and the highly conserved PAR signature subdomain. (B) PARP1 adopts a collapsed conformation for variable substrate recognition during activation (Langelier, Zandarashvili, Aguiar, Black, & Pascal, 2018; Rudolph, Mahadevan, Dyer, & Luger, 2018).
Despite decades of investigation into the mechanism of PARP1 activation and automodification, there remains conflict as to whether PARP1 binds its targets as a monomer or dimer (Alemasova & Lavrik, 2019). PARP1 likely acts in both manners, depending on its target molecule. Especially, as PARP1 is known to interact with several substrates at once, possibly using different domains or combinations of domains. Thus, depending on the context (binding partners), PARP1 could be binding either as a monomer or a dimer. Furthermore, the activating partners of PARP1 vary, encompassing damaged/structured DNA (Lonskaya et al., 2005; Potaman, Shlyakhtenko, Oussatcheva, Lyubchenko, & Soldatenkov, 2005), nuclear proteins (M. Y. Kim, Mauro, Gevry, Lis, & Kraus, 2004; Kraus, 2008; Kraus & Lis, 2003) and post-translational modifications (Cohen-Armon et al., 2007), which all could change the mode of PARP1 binding in non-active and activated circumstances.
Within the three main domains discussed above, PARP1 has six subdomains, which fold independently. These subdomains are separated by flexible linker regions, allowing for modular organization of domain-specific interactions with a particular substrate (Eustermann et al., 2015). This dynamic organization thus allows the multifunctionality of the PARP1 protein. For example, the Zn1, Zn3, WGR and catalytic domains converge on double stranded DNA breaks (Langelier, Planck, Roy, & Pascal, 2012; Langelier, Servent, Rogers, & Pascal, 2008) while the Zn1, Zn2 domains cooperate to respond to nicked DNA (Eustermann et al., 2015) (Figure 1B). PARP1 can also be activated through C-terminal interactions with proteins, such as H4 (Thomas et al., 2019).
Newer studies have identified PARPs in RNA-binding (Guetg, Scheifele, Rosenthal, Hottiger, & Santoro, 2012; E. A. Matveeva, Al-Tinawi, Rouchka, & Fondufe-Mittendorf, 2019; E.A. Matveeva, Mathbout, & Fondufe-Mittendorf, 2019; Melikishvili, Matveeva, & Fondufe-Mittendorf, 2017). Since it is now widely accepted that RNA is an integral component of chromatin (reviewed in Li and Fu (2019)) with functional and regulatory roles, the finding of PARP1 binding to both RNA and DNA is not surprising. While these studies have shown that PARP1 binds to RNA, the questions of which domains are involved in PARP1-RNA binding and whether RNA binding is able to activate PARylation are still controversial (D. S. Kim et al., 2019; Nakamoto, Rudolph, Wuttke, & Luger, 2019). Full length PARP1 binds to both DNA and RNA, with a preference for DNA. Additionally, Melikishvili, Chariker, Rouchka, and Fondufe-Mittendorf (2017) showed that deletion of the known DNA-binding domain Zn1, Zn2, converted PARP1 from a DNA binding to an RNA binding protein, suggesting a role for Zn3 in RNA binding (Huambachano, Herrera, Rancourt, & Satoh, 2011). However, PARP1 does not have the typical RNA recognition motif (RRM), thus it is not known what regions PARP1 uses to bind RNA. In an attempt to define the RNA binding domain, Melikishvili, Chariker, et al. (2017), using several deletion mutants of PARP1 domains, showed that all the domains of PARP1 were still capable of binding RNA, suggesting that these domains all contribute to PARP1-RNA binding. Specifically, it suggests that perhaps PARP1 utilizes different domain combinations to recognize RNA structures, as seen in its ability to distinguish between SS and DS DNA breaks (Langelier et al., 2012; Langelier et al., 2008). Collectively these studies suggest that while all domains are capable of binding RNA, Zn3 and the remaining C-terminal end of PARP1 are the most important for RNA interactions.
Our results showing that PARP1’s Zn3 is important for its in-vitro, is consistent with other studies that use deletion mutants to detail PARP1’s RNA binding characteristics (Desroches & Denault, 2019; Guetg et al., 2012; Huambachano et al., 2011). However, despite the importance of Zn3, in its absence, PARP1 is still able to bind RNA (Melikishvili, Chariker, et al., 2017), suggesting that other regions of PARP1 also might be contributing to its RNA binding function. Indeed, the WGR domain (Huambachano et al., 2011) and the BRCT domain (Desroches & Denault, 2019) of PARP1 have also been implicated in RNA binding. Other PARP family members have been shown to shown to bind RNA using different domains. For instance, PARP7, PARP12 and PARP13 bind RNA through CCCH ZnFs while PARP10 and PARP14 have RNA recognition motifs (RRMs) (Bock, Todorova, & Chang, 2015). PARP1 and PARP2 do not have any of the above domains yet they bind RNA. PARP2 lacks the N-terminal ZnFs and the BRCT domain found in PARP1, but shares high homology in its C-terminal end, which contains both WGR and CAT domains. Leger et al (Leger, Bar, Savic, Santoro, & Hottiger, 2014) showed that PARP2 binds rRNA intermediates with its SAF-A/B Acinus and PIAS (SAP) motif located in its N-terminal domain. Interestingly, while we show that deletion of PARP1’s N-terminal ZnFs shifts PARP1 from a DNA binder to an RNA binder, Leger et al’s deletion of PARP2 N-terminal SAP domain completely abolishes its ability to bind rRNA intermediates even though the homologous C-terminal region remains intact. It is possible that, while the differences in PARP1 and PARP2 C-terminal regions are few, they are significant for RNA binding. These results suggest that multiple domains could be important in PARP1-RNA binding, and more detailed studies will be needed to comprehensively determine the PARP1-RNA binding domains and their contributions.
Recent studies have identified several RNA substrates that activate PARP1 or PARP2 during cellular processes (Chen, Kassab, Dantzer, & Yu, 2018; Huambachano et al., 2011; D. S. Kim et al., 2019). However, activation of PARP1 by RNA was recently disputed in an in vitro study (Nakamoto et al., 2019). It is possible that RNA activation of PARylation could be dependent on the particular PARP isoform, since several other PARPs (PARP10, PARP11, PARP15, TRPT1) were recently shown to MARylate phosphorylated ends of RNA (Munnur, Somers, et al., 2019). On the other hand, since PARP1 can act as a scaffold between RNA and effector proteins (Desroches & Denault, 2019), it is possible that activation is through these effector proteins. More detailed studies will be needed to unravel the exact mechanisms and functional outcomes of PARP-RNA interactions in vivo.
3. THE ROLE OF PARP1 IN TRANSCRIPTIONAL REGULATION
The levels of a transcript are regulated at several critical steps in RNA biogenesis and stability of the transcript, both of which cumulate in determining the amount of protein eventually generated. RNA biogenesis starts with transcriptional initiation, followed by elongation, splicing, processing, and eventually export of the mature mRNA to the cytoplasm for translation. PARP1 plays a role in almost all of these steps.
3.1. PARP1’s role in transcription initiation
For transcription initiation to occur, RNA polymerase and transcription factors must access promoter DNA sequences present in chromatin. Thus, for transcription to take place, the chromatin structure needs to be remodeled to become accessible to these regulatory proteins. PARP regulates transcription initiation by triggering and maintaining changes in chromatin structure as well as through direct recruitment of transcription factors.
3.1.1. Regulation of local chromatin decondensation
PARP1 binds near the promoters of transcriptionally active genes, where it outcompetes the repressive histone H1 to drive gene expression (Krishnakumar et al., 2008; Krishnakumar & Kraus, 2010b). In response to certain developmental and environmental cues, PARP1 PARylates histones opening the chromatin structure to allow binding of the transcriptional machinery (Martinez-Zamudio & Ha, 2012; Schiewer & Knudsen, 2014) (Figure 2A). In addition, PARP1 PARylates the lysine demethylase 5B (KDM5B) inhibiting KDM5B’s ability to bind to chromatin, preventing demethylation of H3K4me3, with consequences in gene expression maintenance (Krishnakumar & Kraus, 2010b). In support of PARP1’s role in gene activation, PARP1 PARylates DNMT1, inhibiting its DNA methylation activity (Althaus, 2005; Caiafa, Guastafierro, & Zampieri, 2009; Ciccarone et al., 2012). This interplay of PARP1 and DNA methylation was seen in a genome-wide analysis showing that PARP1 preferentially localizes at hypomethylated regions with active histone marks (Nalabothula et al., 2015). However, how PARP1 is recruited to specific regions to drive gene expression is still unknown.
Figure 2. PARP in Transcription Initiation.
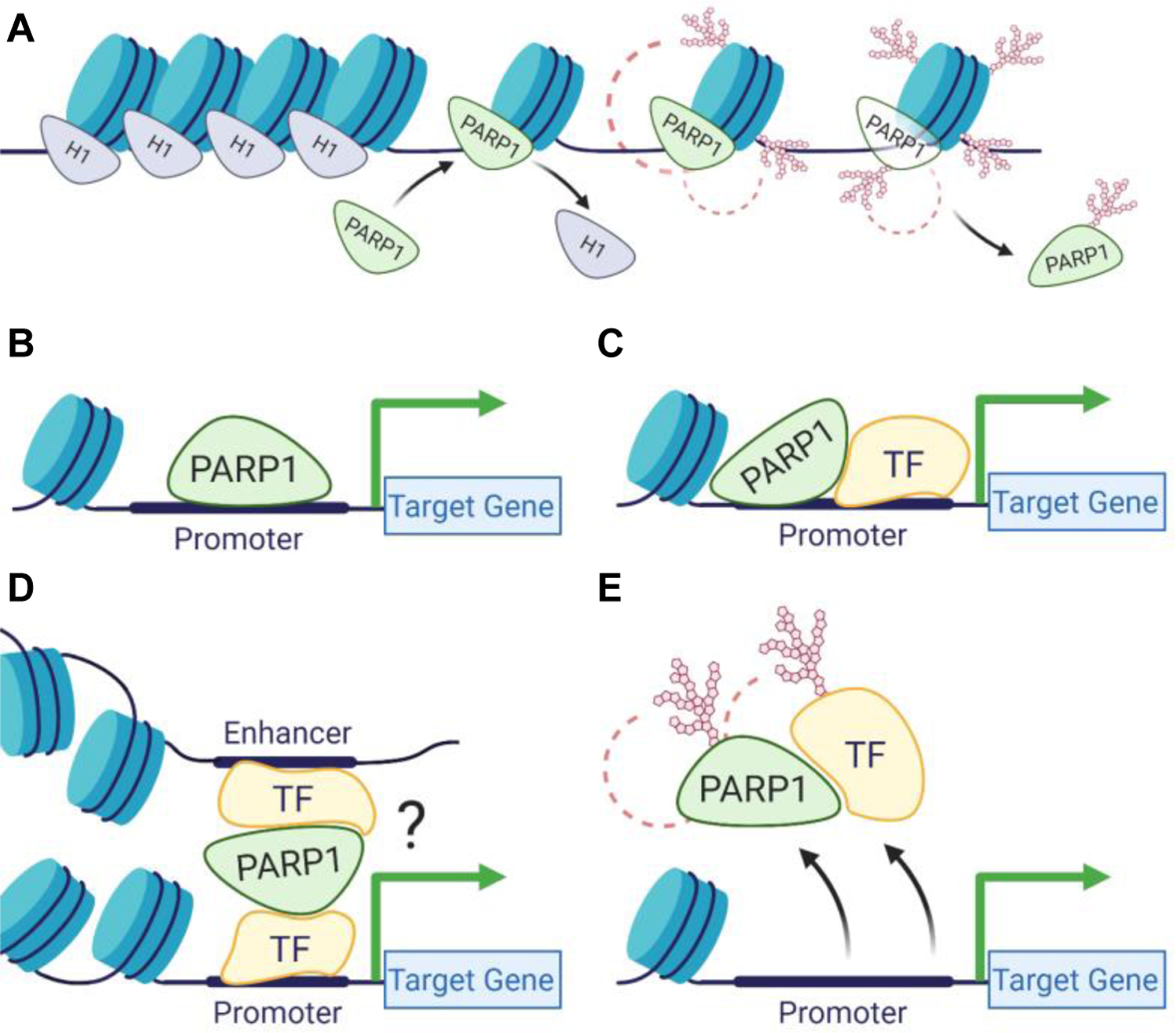
PARP regulates transcription initiation by (A) promoting local chromatin loosening at active genes; (B) acting as a canonical transcription factor through direct binding of promoter sequences; (C) acting as a co-factor for other transcription factors; (D) influencing long range chromatin structure; and (E) altering the DNA binding capabilities of transcription factors.
A recent study (Hau et al., 2017) shows that at the start of differentiation of neural progenitor cells to neurons, pre-B cell leukemia homeobox 1 and myeloid-like eco-integration site transcription factors cooperate to recruit PARP1 to loosen the chromatin structure. Interestingly, this recruitment of PARP1 results in PARylation and removal of the repressive H1 to drive expression of genes critical for neuronal cell fate maintenance. It therefore seems as if PARP1 acts in a context-specific manner. Thus, understanding whether PARP1 acts by itself or in association with other factors is critical in understanding its function in gene regulation. Some studies have demonstrated that nuclear factors important in transcription allosterically regulate PARP1 activity. These factors include non-histone chromatin associated proteins (such as HPF1 (Suskiewicz et al., 2020), HMGM1 (Masaoka et al., 2012), YB-1 (Alemasova & Lavrik, 2019), canonical and variant histone proteins (Kozlowski et al., 2018; Nusinow et al., 2007), as well as modified histones such as phosphorylated H2Av (Cohen-Armon et al., 2007; Kotova, Jarnik, & Tulin, 2010; Thomas et al., 2019). These results suggest that PARP1 mediates a context-specific activation of gene expression. However, is it is still not known which PARP1 interacting partners or which of its domains regulate its activation to drive specific transcription initiation programs.
3.1.2. H4-dependent activation of PARP1
Transcription factors such as YY1 (Doetsch, Gluch, Poznanovic, Bode, & Vidakovic, 2012; Oei & Shi, 2001a) and histones have been reported to stimulate PARP1 activity during transcriptional activation. It seems as if this activation by different factors occurs with the help of different PARP1 domains. For instance, PARP1 activation by transcription factor YY1 involves the BRCT domain, whereas its C-terminal is important for its activation by the N-terminal tail of H4 (Pinnola, Naumova, Shah, & Tulin, 2007; Thomas et al., 2016). Therefore, activation of PARP1 by transcription factors gives proper local, spatio-temporal control critical for selective PARP1 transcriptional regulation. In the case of activation by H4, which is present across the genome, specification of when and where PARylation occurs is likely regulated by dynamic structural composition of surrounding nucleosomes. For example, PARP1 can be localized to promoter regions by a preference for binding nucleosomes containing histone variants such as H2Av and macroH2A1 (Ciccarone, Zampieri, & Caiafa, 2017), which gives its constituent nucleosome a larger PARP1 interacting surface. And the H2A subunits in nearby nucleosomes then act as a brake, preventing the spread of PARylation to nearby nucleosomes, thus setting up chromatin domains (Ciccarone et al., 2017). Nearby histone PTMs such as H2AK5Ac can release the H2A inhibition and help position PARP1 in an active conformation by increasing its interaction with H4. Further, it was shown that post-translational modifications on H2Av, H2A and H4 also can promote H4 ability to activate PARP1 (reviewed in Kotova et al. (2010); Thomas et al. (2016)).
Activation of PARP1 can be long and continuous as in the case of H4-mediated PARP1 activation during transcription. Such prolonged activation ensures a sustained open chromatin state required for transcriptional activity. On the other hand, the recognition of DNA damage by the PARP1-DBD initiates a highly activated but brief state of PARP1 activity specific to the transient nature of DNA repair (Thomas et al., 2019). Furthermore, cooperation between the C-terminal domain with the DBD during H4 dependent activation directs PARP1 to promoter regions by ‘walking’ (scanning) along the DNA (Rudolph et al., 2018; Thomas et al., 2019). PARP1 therefore appears to act as a tightly regulated switch, opening up chromatin to initiate transcription in response to multiple environmental and cellular cues.
3.1.3. PARP1 regulates 3D chromosomal structure
PARP1’s influence on chromatin structure is not limited to local activity on histones. A recent flurry of papers show that PARP1 also regulates large scale 3D organization of the genome through interaction with CCCTC-binding factor (CTCF), the genome’s 3D master regulator (Corces & Corces, 2016). PARP1 co-localizes with CTCF at specific sites (Nalabothula et al., 2015). Additionally, CTCF directly induces PARP1’s PARylation activity in the absence of DNA damage, suggestive of an interaction between these two proteins in vivo (Yu et al., 2004). Both proteins seem to help each other’s activities, as PARP1 stabilizes the binding of CTCF to DNA (Lupey-Green et al., 2018), and in so doing, CTCF stimulates PARP1 activity (Guastafierro et al., 2008). Both activities then cooperatively open up the chromatin structure for gene expression at specific gene target. On the other hand, PARP1 PARylates CTCF controlling the transcription of imprinted genes and ribosomal DNA (Yu et al., 2004; Zlatanova & Caiafa, 2009). PARylation of CTCF would facilitate the regulation of intra-chromosomal interactions, suggesting an involvement of PARP1 in regulating chromatin loop formation. Such interactions of PARP1 and CTCF would help to maintain or mediate changes at boundaries of long-range chromatin states (Lupey-Green et al., 2018; Ong & Corces, 2014; Yu et al., 2004) (Pugacheva et al., 2020).
3.1.4. Co-regulatory function of PARP1
PARP1 can act as a classical transcription factor, binding directly to promoters of cTnT (K. Huang et al., 2004), HTLV-I (Zhang, Hildebrandt, Simbulan-Rosenthal, & Anderson, 2002) and Reg (Akiyama et al., 2001) to regulate transcription (Figure 2B). PARP1 can also interact directly with transcription factors to enhance, inhibit or alter their DNA binding ability (Krishnakumar & Kraus, 2010a; Lin, Tang, Zhu, Shu, & Han, 2011). In doing so, it acts as a co-activator or co-repressor depending on its binding partner (Figure 2C–E). For instance, PARP1 co-activates B-MYB (Cervellera & Sala, 2000; Kraus & Lis, 2003; Musa, Aynaud, Mirabeau, Delattre, & Grunewald, 2017), and together they activate AP-2-mediated transcription (Erener, Hesse, Kostadinova, & Hottiger, 2012). PARP1 also co-activates NF-κB transcription factor, contributing to the transcription activation of a subset of NF-κB target genes (Erener, Petrilli, et al., 2012). Other studies show that PARP1 is not only recruited, but in a sequence-specific manner it co-binds with other transcription factors to drive particular gene expression patterns. Several examples include its sequence-specific co-binding with TEF1 at MCAT1 elements to regulate muscle-specific gene expression (Hu et al., 2013; Morales et al., 2014). PARP1 also interacts with transcription factors YY1 (Oei & Shi, 2001b), E47 (Dear et al., 1997), and p53 (Fischbach et al., 2018; Wiman, 2013). Thus, it is possible that PARP1 is specifically recruited to regions close to transcription factor binding sites to regulate gene expression, or that PARP1 acts as a scaffold, recruiting and PARylating transcription factors (Fig. 2E) to effect specific gene expression. More studies will be needed to tease out PARP1’s co-regulatory activity in a context-dependent manner. Though earlier studies have shown PARP1 involvement in chromatin remodeling, transcription activation and repression, a consensus among these functions remains poorly understood. PARP1 spatial localization on the human genome landscape would provide extremely valuable insights for understanding how PARP1 coordinates maintenance of chromatin architecture with gene expression regulatory networks.
3.2. PARP1 and its role in regulation of RNA polymerase II elongation
Aside from its role in structurally enabling loading of RNAPII onto DNA to initiate RNA synthesis, PARP1 also appears to be intimately involved in pausing RNAPII during elongation (Gibson et al., 2016; E. A. Matveeva et al., 2019; Vispe, Yung, Ritchot, Serizawa, & Satoh, 2000). Early RNAPII pausing is an important transcriptional checkpoint needed to ensure proper recruitment of splicing factors prior to elongation. If this goes awry, it results in defects in gene expression levels and propagation (Akhtar et al., 2019). The early elongation complex is under the control of pausing factors DSIF/NELF (DRB sensitivity-inducing factor and negative elongation factor), which stall RNAPII by binding nascent RNA. Release factors must then be recruited to disengage NELF from RNAPII before entry into productive elongation is permitted. Failure to release NELF results in termination of synthesis. Therefore, factors that promote the release of RNAPII from NELF should determine the rate of RNA synthesis (Adelman & Lis, 2012). Indeed, rapid pause and release is associated with higher levels of gene expression (Gressel, Schwalb, & Cramer, 2019; Vihervaara et al., 2017). PARP1 has been implicated in regulation of RNAPII pausing (Ji & Tulin, 2009, 2013; E. A. Matveeva et al., 2019; E. A. Matveeva et al., 2016; Petesch & Lis, 2012b). Through a clever analog sensitive PARP approach, Gibson et al. (2016) identified NELF components as targets of PARP1 activity in mammalian cells. PARylation of NELF-E by PARP1 inhibits its RNA binding, causing NELF to release RNAPII pausing, thereby promoting, productive elongation. Interestingly, PARylation of NELF-E is dependent on phosphorylation, thus PARP1 and CDK9 (a member of the P-TEFb) kinase colocalize at low levels of RNAPII pausing. Depletion of PARP1 resulted in increased RNAPII pausing (Gibson et al., 2016). Additionally, PARP1 blocked transcription (pause RNAPII) itself by binding pre-mRNA, and only after PARP1 becomes automodified is this block released (Vispe et al., 2000). Together these data present a picture where PARP1 and NELF-E bind to nascent pre-mRNA at the site of RNAPII pausing. After PARP1 activation, RNAPII is released from pausing, which unclasps NELF-E and PARP1’s grip on the pre-mRNA allowing RNAPII to continue with elongation (Figure 3).
Figure 3. PARP1’s role in transcription elongation.
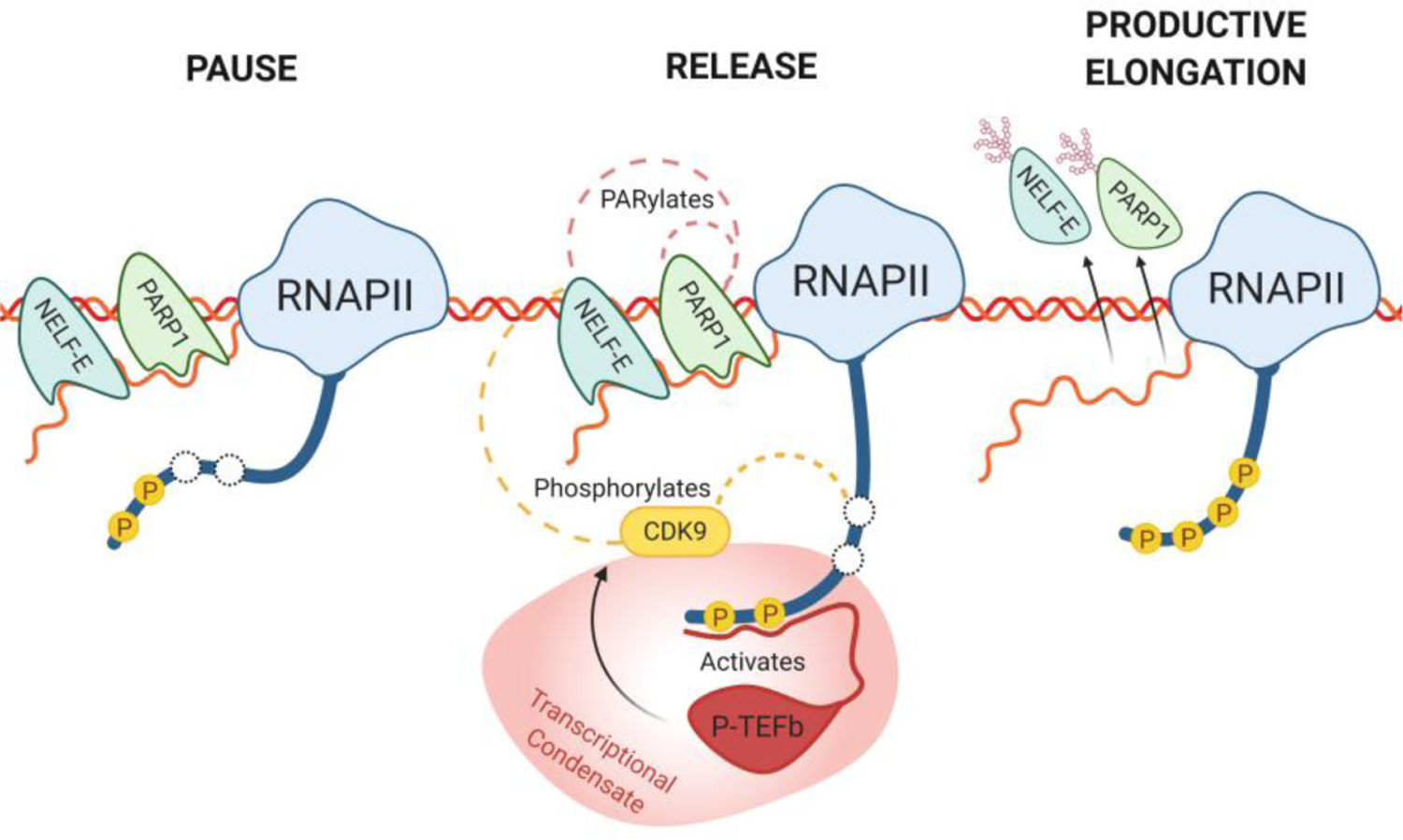
During RNAPII pausing, a chromatin associated PARP1 assists NELF by binding to the nascent pre-mRNA. Partial phosphorylation of RNAPII CTD recruits the low complexity region of P-TEFb, activating CDK9 (reviewed in Thomas et al. (2019)). To release RNAPII, CDK9 then hyperphosphorylates RNAPII’s CTD and also phosphorylates NELF-E. Phosphorylation of NELF-E then instigates PARylation of NELF-E by PARP1, which is also autoPARylated. PARylation of NELF-E and PARP1 inhibit their RNA-binding, releasing both proteins from the nascent pre-mRNA, and allowing the RNAPII complex to enter productive elongation. Splicing condensate is shown as the reddish structure.
3.3. PARP1’s regulation of co-transcriptional alternative splicing
Once RNAPII transitions from the paused state at a promoter to productive elongation, its progression through the gene body faces other barriers. This is supported by frequent RNAPII pausing that occurs throughout transcription (Kwak, Fuda, Core, & Lis, 2013; Mayer et al., 2015). Intragenic transcriptional pausing generates opportunities for the regulation of RNA processing—specifically, splicing. Recent findings suggest that alternative splicing (AS) is mediated through epigenetic processes. As evidenced, specific histone modifications, DNA methylation, and nucleosome positioning have all been implicated in splice choice selection (Allemand, Batsche, & Muchardt, 2008; Aslanzadeh, Huang, Sanguinetti, & Beggs, 2018; de la Mata et al., 2011; Gelfman & Ast, 2013; Gelfman, Cohen, Yearim, & Ast, 2013; Haque & Oberdoerffer, 2014; Luco et al., 2010; Nieto Moreno, Giono, Cambindo Botto, Munoz, & Kornblihtt, 2015; Schor, Gomez Acuna, & Kornblihtt, 2013; Shayevitch, Askayo, Keydar, & Ast, 2018; Xu, Zhao, Olson, Prabhakara, & Zhou, 2018). Thus proteins that modulate chromatin structure, such as PARP1, are likely to regulate AS. Supporting this idea is the recent finding that PARP1 is an AS regulator (E. A. Matveeva et al., 2019; E. Matveeva et al., 2016). PARP1’s new role in AS has led to a proposed model whereby PARP1 is implicated in a network of interactions between RNA and chromatin to regulate AS.
3.3.1. PARP1 chromatin association in alternative splicing
PARP1 binds nucleosomes at target exon/intron boundaries and PARP1 knockdown (KD) or inhibition of its PARylation activity results in specific AS patterns (Figure 4). Interestingly, the AS patterns provoked by PARP1 KD, differ from those due to PARylation-inhibition. These results suggest that PARP1’s physical association with chromatin structure directly affects splicing decisions, while PARP1 PARylation may regulate splicing indirectly through the PARylation of splicing factors (Ji & Tulin, 2009, 2013) and/or through the PARylation of histones (Karch, Langelier, Pascal, & Garcia, 2017; Yang et al., 2020).
Figure 4. PARP1 and Co-Transcriptional Alternative Splicing.
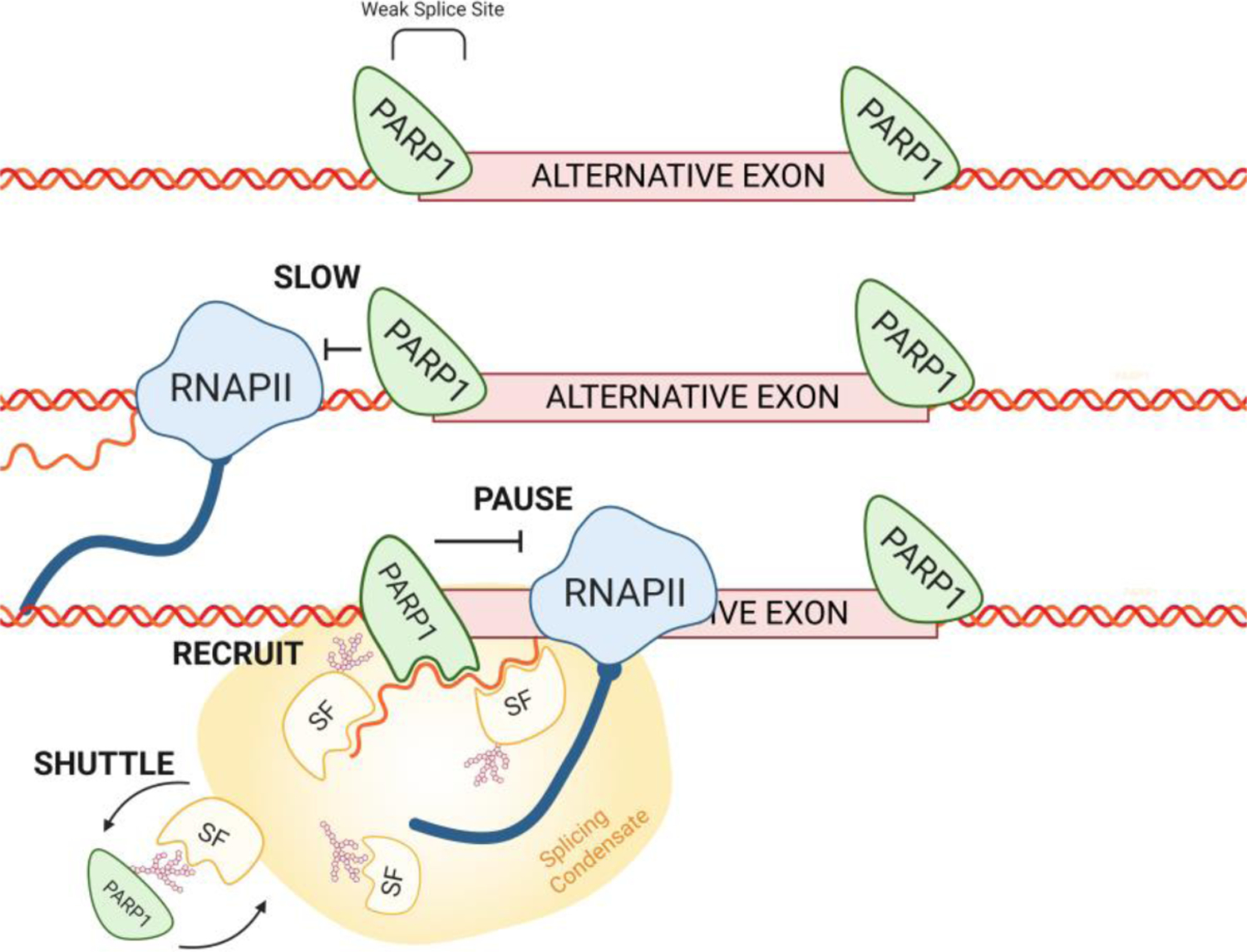
PARP1 associates with chromatin at intron/exon boundaries. PARP1 can influence splicing decisions by 1) controlling RNAPII elongation kinetics and 2) once RNAPII passes, PARP1-chromatin reassembles and can bind to the nascent mRNA, where it controls the recruitment and RNA-binding capacities of splicing factors at splice sites. Splicing condensate is shown as the yellowish structure.
Two non-mutually exclusive models have been proposed to explain how chromatin structure regulates AS. In the kinetic model, the chromatin structure regulates the speed of RNAPII elongation to influence splicing outcomes (de la Mata et al., 2003; Hsin & Manley, 2012; Ip et al., 2011; Jonkers & Lis, 2015; Lenasi & Barboric, 2010; Saldi, Fong, & Bentley, 2018) and in the adaptor/recruitment model, chromatin or its associated factors recruit splicing factors, bridging chromatin to nascent mRNAs (Jabre et al., 2019) to direct/orchestrate splicing choices. Recent studies suggest that PARP1 could act in both models. In support of the kinetic model, PARP1-chromatin structure creates a barrier for RNAPII elongation within genes (E. A. Matveeva et al., 2019). These results are consistent with previous studies showing that the PARP1-chromatin complex creates a chromatin structure that pauses RNAPII at the heat-shock-gene promoters (Petesch & Lis, 2012b). In support of the adapter/recruitment model, PARP1 binds pre-mRNA (E. Matveeva et al., 2016; Melikishvili, Chariker, et al., 2017) and recruits splicing factor 3B1 (SF3B1), a U2 spliceosomal member (E. Matveeva et al., 2016), possibly bridging the spliceosomal complex to RNA. Thus, PARP1 appears to be a major player in AS regulation (Figure 4).
While these studies have suggested PARP1-chromatin in AS, it is interesting that PARP1 could also directly recruit or modify elongation factors to regulate RNAPII elongation. Thus, a kinetic understanding of PARP1-mediated recruitment/assembly of elongation factors may also be key in delineating its role in regulating the rate of RNAPII elongation and eventually, splicing. RNAPII elongation at promoters occurs sequentially—recruitment of the DSIF-NELF, followed by recruitment of the Integrator complex. RNAPII elongation at promoters occurs sequentially, i.e., recruitment of the DSIF-NELF is followed by recruitment of the Integrator complex. For pause-release, this recruitment then aids in recruitment of the positive elongation factor (pTEFb), followed by the super elongation complex (SEC) (Bartholomeeusen, Xiang, Fujinaga, & Peterlin, 2012; Luo, Lin, Guest, et al., 2012; Luo, Lin, & Shilatifard, 2012) and together, the pTEFb-SEC complex then acts as the central elongation scaffold (N. He, Pezda, & Zhou, 2006). PARP1 seems to play roles in recruitment/assembly of this central scaffold. NELF (Petesch & Lis, 2012a) and ELL (a member of the SEC complex) (Ardehali et al., 2009) were shown to regulate poised RNAPII transcription at the heat-shock promoter through PARP1’s stabilization of ELL to form a stable elongation complex (J. Y. Huang et al., 2006). A kinetic understanding of PARP1-mediated recruitment/assembly of elongation factors may be key in delineating its role in the rate of RNAPII elongation and splicing.
In summary, determining the factors regulating the rate limiting steps in transcriptional control is fundamentally important for understanding the mechanisms that govern transcription. While studies in unicellular organisms have pointed to ‘initiation’ as the rate-limiting step in transcription, a large body of work in metazoans indicates that the transition to productive transcriptional elongation also constitutes a critical step. Despite these advances, much remains unknown about how the PARP1-chromatin complex mechanistically regulates and coordinates RNAPII elongation, RNA, and splicing factors to regulate co-transcriptional splicing.
3.3.2. PARP1 regulating the activity of RNA binding proteins in splicing
PARP1’s role in splicing is not limited to transcriptional kinetics. A large body of evidence suggests that PARP1 can also impact splicing through modifying the functioning of RNABP. The recruitment model of alternative splicing posits that the types of splicing factors present on pre-mRNA during processing primarily inform splicing choices. Three major types of RNABP are involved in splicing: small nuclear ribonucleoproteins (snRNP), SR proteins (SRP) and heterogeneous ribonucleoproteins (hnRNP).
PARP-1 can functionally alter splicing factor activity through several ways: through 1) protein-protein interactions, 2) PARylation of these splicing factors, and 3) non-covalent interactions of these proteins with pADPr via PAR binding motifs (PBMs) (Figure 5). Several proteomics approaches have identified multiple splicing factors and RNA processing factors as part of the PARP1 interactome (Isabelle et al., 2010) PARylated by PARP1 (Jungmichel et al., 2013; Zhen & Yu, 2018) or PAR-binding proteins (Teloni & Altmeyer, 2016). Expectedly, many of these RNABPs are differentially targeted by PARP1 in a cell-type specific manner, consistent the idea of cell-type specific differential alternative splicing (Zhen & Yu, 2018).
Figure 5. PARylation and Modes of PARP1 Interactions.
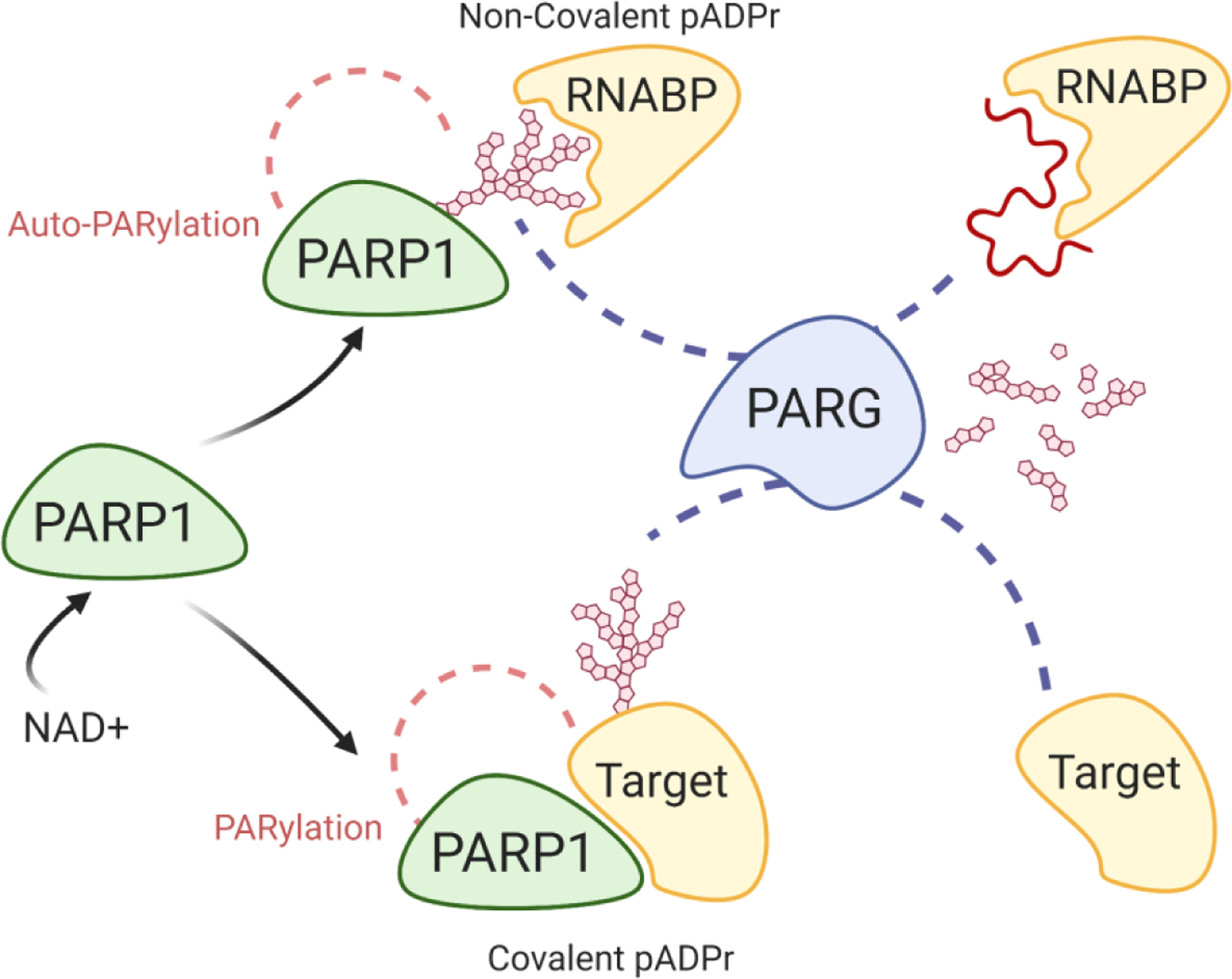
Upon activation, PARP1 can influence protein functions through either covalent PARylation of target proteins or through non-covalent interactions after autoPARylation. PAR associates with RNA binding domains where it competes for binding with target RNA. PARG then degrades PAR to mono ADP-ribose, which can then be further digested by ADP-ribosyl hydrolase 3 or TARG (not shown).
Currently, PARP1 regulation of core spliceosomal snRNP is limited to its direct interaction with U2. PARP1 stably binds to the SF3B1 subunit of the U2 RNP complex, recruiting U2 to PARP1-nucleosomes near splice sites. This recruitment is through direct PARP1-SF3B1 interaction, rather than on PARylation, as knockdown of PARP1, and not PARylation inhibition, diminishes SF3B1 association with PARP1-nucleosomes (E. A. Matveeva et al., 2016). In addition to direct recruitment by PARP1, splicing factors are activated through PARylation (Gibson et al., 2016; Isabelle et al., 2010). This PARylation of RNABPs can also alter RNABP function through inhibiting their ability to bind RNA/DNA as seen with NELF-E and PARP1 itself (Gibson et al., 2016; Zlatanova & Caiafa, 2009). Furthermore, non-covalent binding of the PAR moiety has been observed at many SRPs and hnRNPs. These proteins use a PAR-binding motif to bind PAR. The conformation of PAR mimics nucleic-acid moieties leading to competition with RNA for binding. For instance, hnRNP stably binds pADPr through a conserved PBM (Gagne et al., 2005). The PAR molecule then competes with the binding of RNA at positively charged R-rich, RRM and RGG motifs. As such, changes in the relative concentrations of PAR and RNA can therefore alter RNA-protein complexes (Krishnakumar & Kraus, 2010a) and affect alternative splicing decisions (Ji & Tulin, 2013). This was demonstrated in Drosophila, showing that the splicing factors Squid and hrp38, when bound to PAR, alter splicing choices through inhibition of their RNA binding. Indeed, PAR binding inhibited splicing of the Hsrω-RC pre-mRNA intron, while it promoted splicing of the Ddc pre-mRNA intron (Ji & Tulin, 2009). These findings align with those examining PARP1 kinetic regulation of splicing and underscore the importance of epigenetic context in understanding PARP influence on alternative splicing both in terms of transcriptional kinetics as well as in recruitment of splicing factors. In summary these results suggest that splicing decisions are made by a host of factors where competition and local abundance likely play a factor. Complicating the activities of PAR, reading of these PAR signals by PAR-binding proteins (Thomas et al., 2019) constitutes a novel and interesting aspect of PAR biology that is still very much in its infancy.
Heterogenous ribonucleoproteins (hnRNPs) and splicing regulatory proteins (SRPs) regulate many other aspects of the mRNA life cycle including transcriptional initiation and elongation, nuclear export, mRNA stability, all contributing to the translational efficiency of mRNA to proteins (Geuens, Bouhy, & Timmerman, 2016; Gittings et al., 2019). As a result, their functions can change based on their subcellular localization. Therefore, control of the intracellular transport and localization of these proteins is critical to the fate of the cell. Due to their transient nature, post-translational modifications are well suited for spatiotemporal regulation of proteins and are commonly involved in transport of RNABP. PARylation of hnRNP A1 regulates its translocation to the cytoplasm. However, a majority of what is known of PARP1’s role in RNABP subcellular localization involves non-covalent interactions with pADPr. PARylation of these RNABPs can influence their shuttling to different regions of the cell with functional consequences. A clear example is seen with the PAR-dependent shuttling of TARG1 from nucleolar to nucleoplasm during DNA damage (Butepage et al., 2018). It will be interesting to determine the many RBPs whose localization is modulated by PAR.
3.4. RNA poly-A regulation and RNA export by PARP1
The final process in generation of mature mRNAs consists of endonucleolytic cleavage by a polyadenylation factor and the synthesis of the poly(A) tail by poly (A) polymerases (PAP). Polyadenylation is a highly regulated process that completes the maturation of nearly all eukaryotic mRNAs controlling their turnover and nuclear export. During thermal stress, PARP1 binds and PARylates PAP, resulting in its dissociation from RNA, and decreased global polyadenylation (Di Giammartino, Shi, & Manley, 2013). This decreased polyadenylation would then impact RNA stability/turnover and export, resulting in decreased protein translation efficiency.
Eukaryotic gene expression requires export of protein-coding mRNAs and non-coding RNA molecules from the nucleus to the cytoplasm. The major classes of cellular RNAs are exported as RNA–protein complexes, comprising RBPs and receptor proteins that interact with the nuclear pore complexes (Bjork & Wieslander, 2017). PARP1 has been implicated in mRNA export mediated by the chromosome maintenance region 1 receptor (reviewed in Ke et al. (2019)). Aside from the receptors, other complexes mediate mRNA export. One such complex is the THO/TREX complex, which is required for nuclear export of mRNA and is coupled to transcriptional elongation (Heath, Viphakone, & Wilson, 2016). Subunits of the THO/TREX complex, specifically, the THO subunits 1, 2, 5 and 6, were identified by mass spectrometry as part of the PARP1 interactome (Isabelle et al., 2010; Miyazaki et al., 2014). THOC4 is highly PARylated in all breast cancer cells (Zhen & Yu, 2018). PARylation of these subunits stimulated their interaction between key subunits of the THO/TREX complex. A complete elucidation of the composition, function and interactions of PARP1 with the THO/TREX complex will provide the framework for understanding the molecular basis of its role not only in mRNA export but in how it might impact diseases due to defects in mRNA export.
3.5. PARP1’s role in mRNA stability
We recently showed a role of PARP1 in mRNA stability (E.A. Matveeva et al., 2019). These results were in line with the previous finding that PARP1 KD resulted in alternative spliced isoforms whose stability was also impacted. Additionally, KD of PARP1 increased transcription of genes involved in NMD (Melikishvili, Chariker, et al., 2017), possibly linking PARP1 to the stability of these misspliced isoforms. In fact, NMD pathway controls the quality of mRNAs, degrading unproductive spliced or aberrant transcripts in several organisms including C. elegans (Mitrovich & Anderson, 2005), S. cerevisiae (F. He et al., 2003; Sayani, Janis, Lee, Toesca, & Chanfreau, 2008) and Drosophila (Hansen et al., 2009). Thus, if PARP1 is also important in this pathway, it might survey the quality of mRNAs produced before they are exported out of the nucleus. In support of PARP1’s role in mRNA stability, PARylation of HuR, a protein that binds to AREs, increases mRNA stability (Ke et al., 2017). While this idea that PARP1 would regulate mRNA stability and decay is exciting, the challenge that lies ahead is to determine the mechanism by which it does so. It will also be worthwhile to determine and tease out direct vs. indirect roles of PARP1 in mRNA stability.
3.6. The role of PARP1 in ribosome biogenesis and translation
The nucleolus is a non-membrane bound compartment of the nucleus (Nunez Villacis et al., 2018) and serves as the site for ribosome biogenesis including ribosome assembly and rRNA synthesis. Proper ribosome biogenesis leads to proper protein translation. An impressive body of evidence has accumulated in recent years supporting the idea that dysregulation of ribosome biogenesis causes diseases now known as ribosomopathies (Kampen, Sulima, Vereecke, & De Keersmaecker, 2020). Defects in factors that regulate ribosome biogenesis may lead to development of these diseases. PARP1 plays both a direct and an indirect role in proper functioning of the ribosome. PARP1 contains a nucleolar localization signal within its N-terminal domain (Meder, Boeglin, de Murcia, & Schreiber, 2005), and in the absence of genotoxic stress about 40% of PARP1 locates to the nucleolus, suggesting a critical role for PARP1 in ribosomal biogenesis (Mortusewicz, Ame, Schreiber, & Leonhardt, 2007; Rancourt & Satoh, 2009). In Drosophila, PARP1 is essential in functional nucleoli by stabilizing the general nucleolar architecture through formation of complexes with nucleolar proteins such as fibrillarin and nucleophosmin (Meder et al., 2005). Additionally, PARylation of TIP5, a component of the nucleolar-remodeling complex, promotes silencing of rDNA chromatin during replication. This finding uncovered a mechanism by which PARP1 ensures that silent rDNA regions are properly inherited (Guetg et al., 2012; Isabelle et al., 2010). PARylation is also involved in all the major stages of ribosomal biogenesis, including pre-rRNA processing, post-transcriptional modification and pre-ribosome assembly. It was posited that it does so by directing nucleolar proteins within proximity of pre-rRNA (Boamah, Kotova, Garabedian, Jarnik, & Tulin, 2012). PARP1’s regulation of nucleolar proteins has been identified in mammalian cells and this regulation occurs via distinct mechanisms. PARP1 PARylation is required for localization of XRCC1 to the nucleolus while PARP1 regulation of WRN localization is independent of its enzymatic activity and instead is dependent upon interactions with automodified PARP1 (Veith et al., 2019). Meanwhile, PARP1 recruitment of DDX21 to the nucleolus operates in two steps. First, automodified PARP1 interacts non-covalently with the RNA binding domain of DDX21. This close interaction with DDX21 allows PARP1 to covalently PARylate DDX21 N-terminal, leading to nucleolar localization (D. S. Kim et al., 2019). Interestingly, unlike Drosophila, mammalian PARP1 deficient cells are not altered in rRNA transcription efficiency (Meder et al., 2005), possibly because of redundancy with the many other PARPs.
ADP-ribosylation could also disrupt ribosome assembly by interfering with the interaction between ribosomal proteins and rRNA. One ribosomal subunit RPS8 was found to be extensively ADP-ribosylated in MDA-MB468 cells (Zlatanova & Caiafa, 2009). The ADP-ribosylated isoform then binds to 28S rRNA (part of the 60S subunit) through an inter-subunit bridge called eB11 (M. Y. Kim et al., 2004). Disruption of this linkage could lead to weaker binding between subunits and disrupted ribosomal assembly. Finally, it has been shown that many ribosomal proteins are also PARylated, and some have been identified as part of the PARP1 interactome, possibly impacting protein translation (Isabelle et al., 2010). These studies are consistent with the finding that several ribosomal proteins are part of the PARP1 interactome (E.A. Matveeva et al., 2019). While several components of the ribosomal network have been shown to be PARylated, a comprehensive catalog of PARylated ribosomal proteins, as well as steps in RNA biogenesis impacted remains unknown. Since PARP1 plays a significant role in RNA biogenesis, future studies will be worthwhile to tease out the steps in which PARP1/PARylation is critical. This is of great interest as disruption of ribosome biogenesis at any of the steps can promote cell cycle arrest, senescence or apoptosis which have been linked to disease pathogenesis.
4. PARP1 AND REGULATION OF SUBCELLULAR COMPARTMENTALIZATION
It is becoming clear that PARP1 is highly involved in the formation and composition of many membraneless sub compartments in the cell, including the nucleolus (D. S. Kim et al., 2019), Cajal bodies (Kotova et al., 2010), histone locus bodies (Duronio & Marzluff, 2017; Kotova et al., 2010), and stress granules (Duan et al., 2019) many of which are responsible for processing, transport and recycling/turnover of ribonucleoprotein (RNP) complexes (Anko et al., 2012; Muller-McNicoll & Neugebauer, 2013) (Figure 6). RNP granules can be formed through classic liquid-liquid phase separation, nucleation with a polymer, and through the bridging of long polymers (Peng & Weber, 2019). Post-translational modifications often regulate their assembly and biomolecular makeup. Recent analysis of pADPr-interacting proteins and PARylated proteins show enrichment for proteins containing low complexity regions, which facilitate phase separation of subcellular condensates (Gibson et al., 2016; Isabelle et al., 2010). Additionally, either altering the abundance of PARP1, PARG or inhibition of PARylation would disrupt the spatiotemporal dynamics of these condensates (Leung, 2020).
Figure 6. PARP-1 Regulation of Subcellular Condensates.
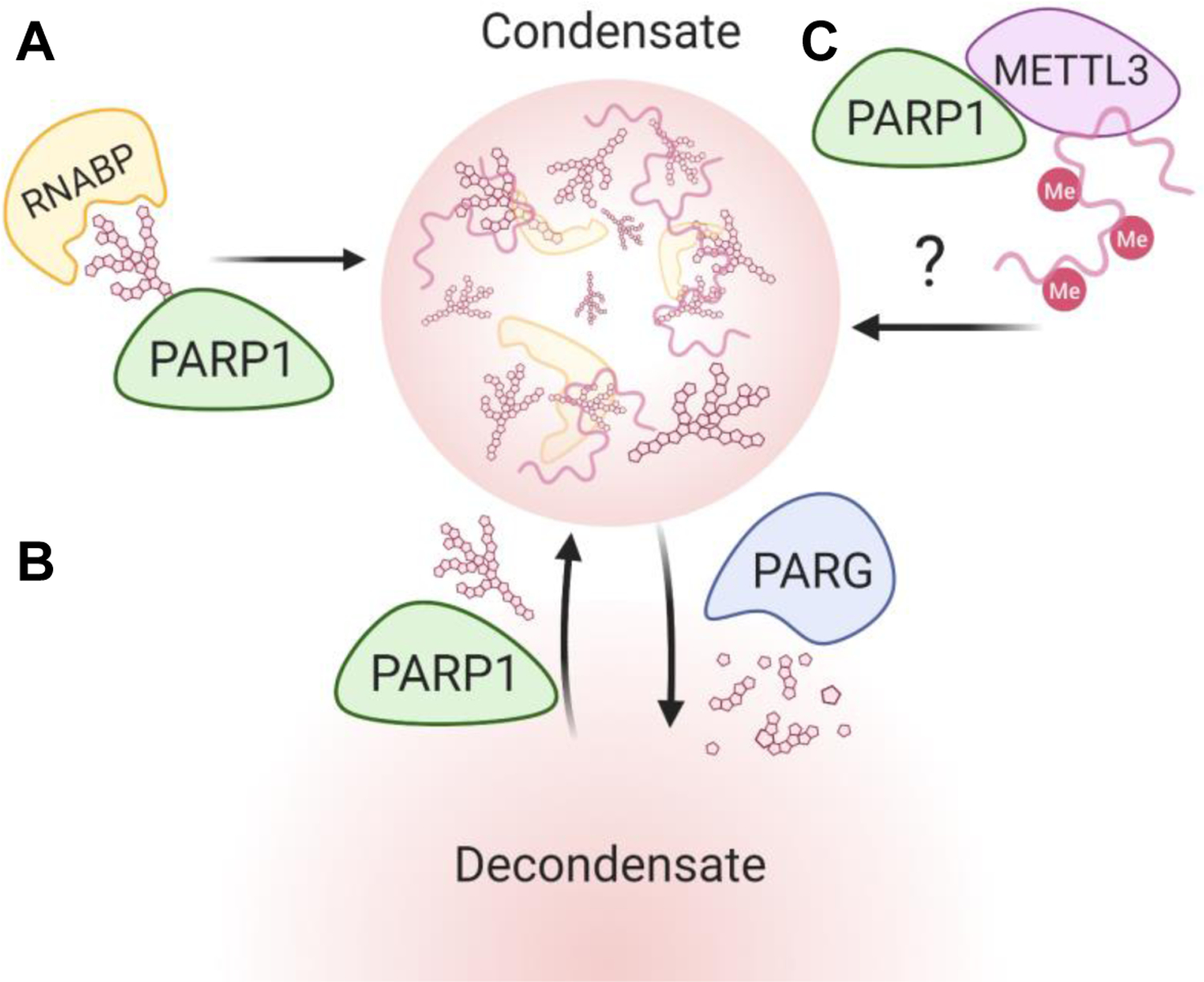
(A) PARP1 influences condensate composition through protein shuttling. (B) PAR levels can regulate spatiotemporal dynamics of condensates. (C) PARP1 recruitment of METTL3 may enhance phase separation through hypermethylation of constituent RNA.
4. 1. Cajal bodies
are transient nuclear condensates where the biogenesis and turnover of transcriptional and splicing RNP complexes occur, including RNA polymerase. PARP1 activation and automodification is required to shuttle proteins to Cajal bodies via interaction with pARDr via PBMs (Fig. 6) (Kotova et al., 2010; Thomas et al., 2019). PARylation also controls Cajal body formation dynamics, promotes assembly of Cajal bodies, and cleavage of pADPr by PARG is required for disassembly (Thomas et al., 2019). Therefore, a careful balance between PARylation and de-PARylation is needed for proper biogenesis and turnover of transcriptional RNP complexes.
4. 2. Stress granules.
RNA binding proteins can often form RNP granules through liquid-liquid phase separation (LLPS). Stress granules are RNP granules that form in response to cellular stress and are involved in the pathology of many neurodegenerative diseases. Duan et al. (2019) found that PARylation levels critically regulate formation of stress granules containing RNABPs (hnRNP A1 and TDP-43) involved in neurodegenerative disease. Additionally, PARP has two effects on hnRNP A1—covalent modification of hnRNP A1 controls nucleocytoplasmic translocation, whereas interaction of hnRNP A1 PBM with pADPr controls its association with stress granules and promotes demixing with TDP-43.
Other RNA binding proteins that drive some disease pathogenesis also mediate the formation of stress granules. For instance, FUS is an RNA binding protein that facilitates cotranscriptional splicing (Mastrocola, Kim, Trinh, Rodenkirch, & Tibbetts, 2013; Mortusewicz et al., 2007; Naumann et al., 2018). FUS is necessary for correct splicing of developmental genes in frogs (Dichmann & Harland, 2012) and its depletion leads to accumulation of phosphorylated RNAPII at transcriptional start sites, suggesting it is also necessary for proper transcriptional elongation (Schwartz et al., 2012). Interestingly, FUS binds to pADPr in response to DNA damage (Singatulina et al., 2019) promoting condensate formation of concentrated DNA damage needed to promote efficient repair. Just as with Cajal bodies, the dynamics of these FUS condensates are regulated by pADPr levels. As DNA is repaired, PARylation activity returns to basal levels allowing PARG activity to effectively reduce pADPr leading to dissociation of the complex. Given the involvement of FUS in splicing and RNAPII elongation it is possible that FUS and PARP1 cooperate to regulate transcriptional and splicing condensates during RNA synthesis. Additionally, PARylation of RNAPII CTD cooperates with its phosphorylation, to cause the CTD to disassociate from transcriptional initiation condensates into neighboring condensates containing elongation and splicing factors (reviewed in Maita and Nakagawa (2020)). These data, combined with the knowledge that PARylation already regulates condensates that impact RNAPII activity, suggest that PARPs play a role in transcriptional condensates.
5. RNA METHYLATION ENHANCED LIQUID-LIQUID PHASE SEPARATION
Recently, Ries et al. (2019) demonstrated that m6A modification on RNA enhances phase separation, and subsequently affects a series of cellular biological processes. Interestingly, a recent study demonstrated that PARP1 helps to produce RNA with m6A RNA modifications at sites of DNA damage likely through recruitment of the m6A methyltransferase, METTL3. This accumulation of poly-m6As on RNAs is required for transcriptional re-initiation at the DNA damage site (Xiang et al., 2017). These studies thus provide another new area where PARP1 plays a significant role. More open questions arise: 1) Does this enhanced PARP1-mediated m6A RNA modification at damaged DNA sites occur only during DNA damage or are there other environmental signals that can drive this as well? 2) Are there other proteins that aid in mediating such activities of PARP1 at these sites? 3) Is this activity of PARP due to PARP itself or PARylation solely? Given that addition of m6A occurs co-transcriptionally and is frequently transient in nature, it is possible that PARP1 works together with the m6A writer complex METTL3/14 to fine-tune nuclear droplet formation. The increased interest in recent years has shown that PARP1 is biologically relevant in nearly every stage of RNA lifecycle, including importance of subcellular condensates.
Conclusion
PARP1 has been implicated in many cellular processes such as DNA repair, transcription and RNA biogenesis. We focused on how PARP1 is involved in RNA biogenesis and metabolism. Our recent findings show PARP1 binding to RNA (Melikishvili, Chariker, et al., 2017), which now adds PARP1 to the growing list of proteins that bind both DNA and RNA to affect gene regulation (Hendrickson, Kelley, Tenen, Bernstein, & Rinn, 2016; Hudson & Ortlund, 2014). These studies suggest that PARP1 is involved in a more intertwined gene regulatory network than has been previously appreciated. Indeed, splicing is tightly integrated with gene transcription (Neugebauer, 2002; Shukla & Oberdoerffer, 2012), and splicing has also been shown to control gene expression via Nonsense-mediated (Glisovic, Bachorik, Yong, & Dreyfuss, 2008) or spliceosome-mediated (Haimovich et al., 2013) decay pathways. PARP1 has now been shown to be involved in regulating several steps of RNA biogenesis, such as RNA metabolism (Gibson et al., 2016), mRNA metabolism, protein synthesis (Caruso et al., 2018) and co-transcriptional splicing (E. A. Matveeva et al., 2019; E. Matveeva et al., 2016). We also recently observed a role for PARP1 in mRNA stability and decay (E.A. Matveeva et al., 2019). Thus, we believe PARP1 acts as a genome ‘surveyor’ with multiple roles for directing RNA and DNA in gene regulation. However, more studies will need to be carried in out teasing out direct vs. indirect effects of PARP1 in these processes. We also assume that these pleiotropic effects of PARP1 on RNA biogenesis are context dependent. Thus, studies dissecting the temporal-spatial context-specificity of PARP1 and its activity will greatly improve our understanding of this pleiotropic protein in the many steps of RNA biogenesis.
Acknowledgments
We would like to thank the members of the Fondue-Mittendorf laboratory for critical reading of the manuscript. We would also like to thank the Markey Cancer Center’s Research Communications Office for manuscript editing (P30 CA177558).
Funding Information
Research reported in this publication was supported by the National Institutes of Environmental Health grant R01 ES024478 (YFN-M), National Science Foundation grant MCB 1517986 (YFN-M), and GRF 1839289 (RE). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIEHS or NSF.
References
- Adelman K, & Lis JT (2012). Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat Rev Genet, 13(10), 720–731. doi: 10.1038/nrg3293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhtar J, Kreim N, Marini F, Mohana G, Brune D, Binder H, & Roignant JY (2019). Promoter-proximal pausing mediated by the exon junction complex regulates splicing. Nat Commun, 10(1), 521. doi: 10.1038/s41467-019-08381-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama T, Takasawa S, Nata K, Kobayashi S, Abe M, Shervani NJ, … Okamoto H (2001). Activation of Reg gene, a gene for insulin-producing beta -cell regeneration: poly(ADP-ribose) polymerase binds Reg promoter and regulates the transcription by autopoly(ADP-ribosyl)ation. Proc Natl Acad Sci U S A, 98(1), 48–53. doi: 10.1073/pnas.240458597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alemasova EE, & Lavrik OI (2019). Poly(ADP-ribosyl)ation by PARP1: reaction mechanism and regulatory proteins. Nucleic Acids Res, 47(8), 3811–3827. doi: 10.1093/nar/gkz120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allemand E, Batsche E, & Muchardt C (2008). Splicing, transcription, and chromatin: a menage a trois. Curr Opin Genet Dev, 18(2), 145–151. doi: 10.1016/j.gde.2008.01.006 [DOI] [PubMed] [Google Scholar]
- Althaus FR (2005). Poly(ADP-ribose): a co-regulator of DNA methylation? Oncogene, 24(1), 11–12. doi: 10.1038/sj.onc.1208382 [DOI] [PubMed] [Google Scholar]
- Ame JC, Spenlehauer C, & de Murcia G (2004). The PARP superfamily. Bioessays, 26(8), 882–893. doi: 10.1002/bies.20085 [DOI] [PubMed] [Google Scholar]
- Anko ML, Muller-McNicoll M, Brandl H, Curk T, Gorup C, Henry I, … Neugebauer KM (2012). The RNA-binding landscapes of two SR proteins reveal unique functions and binding to diverse RNA classes. Genome Biol, 13(3), R17. doi: 10.1186/gb-2012-13-3-r17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardehali MB, Yao J, Adelman K, Fuda NJ, Petesch SJ, Webb WW, & Lis JT (2009). Spt6 enhances the elongation rate of RNA polymerase II in vivo. EMBO J, 28(8), 1067–1077. doi: 10.1038/emboj.2009.56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslanzadeh V, Huang Y, Sanguinetti G, & Beggs JD (2018). Transcription rate strongly affects splicing fidelity and cotranscriptionality in budding yeast. Genome Res, 28(2), 203–213. doi: 10.1101/gr.225615.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkauskaite E, Jankevicius G, & Ahel I (2015). Structures and Mechanisms of Enzymes Employed in the Synthesis and Degradation of PARP-Dependent Protein ADP-Ribosylation. Mol Cell, 58(6), 935–946. doi: 10.1016/j.molcel.2015.05.007 [DOI] [PubMed] [Google Scholar]
- Bartholomeeusen K, Xiang Y, Fujinaga K, & Peterlin BM (2012). Bromodomain and extra-terminal (BET) bromodomain inhibition activate transcription via transient release of positive transcription elongation factor b (P-TEFb) from 7SK small nuclear ribonucleoprotein. J Biol Chem, 287(43), 36609–36616. doi: 10.1074/jbc.M112.410746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjork P, & Wieslander L (2017). Integration of mRNP formation and export. Cell Mol Life Sci, 74(16), 2875–2897. doi: 10.1007/s00018-017-2503-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boamah EK, Kotova E, Garabedian M, Jarnik M, & Tulin AV (2012). Poly(ADP-Ribose) polymerase 1 (PARP-1) regulates ribosomal biogenesis in Drosophila nucleoli. PLoS Genet, 8(1), e1002442. doi: 10.1371/journal.pgen.1002442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock FJ, Todorova TT, & Chang P (2015). RNA Regulation by Poly(ADP-Ribose) Polymerases. Mol Cell, 58(6), 959–969. doi: 10.1016/j.molcel.2015.01.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonfiglio JJ, Colby T, & Matic I (2017). Mass spectrometry for serine ADP-ribosylation? Think o-glycosylation! Nucleic Acids Res, 45(11), 6259–6264. doi: 10.1093/nar/gkx446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkle A, Diefenbach J, Brabeck C, & Beneke S (2005). Ageing and PARP. Pharmacol Res, 52(1), 93–99. doi: 10.1016/j.phrs.2005.02.008 [DOI] [PubMed] [Google Scholar]
- Butepage M, Preisinger C, von Kriegsheim A, Scheufen A, Lausberg E, Li J, … Luscher B (2018). Nucleolar-nucleoplasmic shuttling of TARG1 and its control by DNA damage-induced poly-ADP-ribosylation and by nucleolar transcription. Sci Rep, 8(1), 6748. doi: 10.1038/s41598-018-25137-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caiafa P, Guastafierro T, & Zampieri M (2009). Epigenetics: poly(ADP-ribosyl)ation of PARP-1 regulates genomic methylation patterns. FASEB J, 23(3), 672–678. doi: 10.1096/fj.08-123265 [DOI] [PubMed] [Google Scholar]
- Caruso LB, Martin KA, Lauretti E, Hulse M, Siciliano M, Lupey-Green LN, … Tempera I (2018). Poly(ADP-ribose) Polymerase 1, PARP1, modifies EZH2 and inhibits EZH2 histone methyltransferase activity after DNA damage. Oncotarget, 9(12), 10585–10605. doi: 10.18632/oncotarget.24291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervellera MN, & Sala A (2000). Poly(ADP-ribose) polymerase is a B-MYB coactivator. J Biol Chem, 275(14), 10692–10696. doi: 10.1074/jbc.275.14.10692 [DOI] [PubMed] [Google Scholar]
- Chen Q, Kassab MA, Dantzer F, & Yu X (2018). PARP2 mediates branched poly ADP-ribosylation in response to DNA damage. Nat Commun, 9(1), 3233. doi: 10.1038/s41467-018-05588-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccarone F, Klinger FG, Catizone A, Calabrese R, Zampieri M, Bacalini MG, … Caiafa P (2012). Poly(ADP-ribosyl)ation acts in the DNA demethylation of mouse primordial germ cells also with DNA damage-independent roles. PLoS One, 7(10), e46927. doi: 10.1371/journal.pone.0046927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccarone F, Zampieri M, & Caiafa P (2017). PARP1 orchestrates epigenetic events setting up chromatin domains. Semin Cell Dev Biol, 63, 123–134. doi: 10.1016/j.semcdb.2016.11.010 [DOI] [PubMed] [Google Scholar]
- Cohen-Armon M, Visochek L, Rozensal D, Kalal A, Geistrikh I, Klein R, … Seger R (2007). DNA-independent PARP-1 activation by phosphorylated ERK2 increases Elk1 activity: a link to histone acetylation. Mol Cell, 25(2), 297–308. doi: 10.1016/j.molcel.2006.12.012 [DOI] [PubMed] [Google Scholar]
- Corces MR, & Corces VG (2016). The three-dimensional cancer genome. Curr Opin Genet Dev, 36, 1–7. doi: 10.1016/j.gde.2016.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels CM, Ong SE, & Leung AKL (2017). ADP-Ribosylated Peptide Enrichment and Site Identification: The Phosphodiesterase-Based Method. Methods Mol Biol, 1608, 79–93. doi: 10.1007/978-1-4939-6993-7_7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Mata M, Alonso CR, Kadener S, Fededa JP, Blaustein M, Pelisch F, … Kornblihtt AR (2003). A slow RNA polymerase II affects alternative splicing in vivo. Mol Cell, 12(2), 525–532. [DOI] [PubMed] [Google Scholar]
- de la Mata M, Munoz MJ, Allo M, Fededa JP, Schor IE, & Kornblihtt AR (2011). RNA Polymerase II Elongation at the Crossroads of Transcription and Alternative Splicing. Genet Res Int, 2011, 309865. doi: 10.4061/2011/309865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dear TN, Hainzl T, Follo M, Nehls M, Wilmore H, Matena K, & Boehm T (1997). Identification of interaction partners for the basic-helix-loop-helix protein E47. Oncogene, 14(8), 891–898. doi: 10.1038/sj.onc.1200912 [DOI] [PubMed] [Google Scholar]
- Desroches A, & Denault JB (2019). Caspase-7 uses RNA to enhance proteolysis of poly(ADP-ribose) polymerase 1 and other RNA-binding proteins. Proc Natl Acad Sci U S A, 116(43), 21521–21528. doi: 10.1073/pnas.1909283116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giammartino DC, Shi Y, & Manley JL (2013). PARP1 represses PAP and inhibits polyadenylation during heat shock. Mol Cell, 49(1), 7–17. doi: 10.1016/j.molcel.2012.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dichmann DS, & Harland RM (2012). fus/TLS orchestrates splicing of developmental regulators during gastrulation. Genes Dev, 26(12), 1351–1363. doi: 10.1101/gad.187278.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doetsch M, Gluch A, Poznanovic G, Bode J, & Vidakovic M (2012). YY1-binding sites provide central switch functions in the PARP-1 gene expression network. PLoS One, 7(8), e44125. doi: 10.1371/journal.pone.0044125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Y, Du A, Gu J, Duan G, Wang C, Gui X, … Fang Y (2019). PARylation regulates stress granule dynamics, phase separation, and neurotoxicity of disease-related RNA-binding proteins. Cell Res, 29(3), 233–247. doi: 10.1038/s41422-019-0141-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duronio RJ, & Marzluff WF (2017). Coordinating cell cycle-regulated histone gene expression through assembly and function of the Histone Locus Body. RNA Biol, 14(6), 726–738. doi: 10.1080/15476286.2016.1265198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erener S, Hesse M, Kostadinova R, & Hottiger MO (2012). Poly(ADP-ribose)polymerase-1 (PARP1) controls adipogenic gene expression and adipocyte function. Mol Endocrinol, 26(1), 79–86. doi: 10.1210/me.2011-1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erener S, Petrilli V, Kassner I, Minotti R, Castillo R, Santoro R, … Hottiger MO (2012). Inflammasome-activated caspase 7 cleaves PARP1 to enhance the expression of a subset of NF-kappaB target genes. Mol Cell, 46(2), 200–211. doi: 10.1016/j.molcel.2012.02.016 [DOI] [PubMed] [Google Scholar]
- Eustermann S, Wu WF, Langelier MF, Yang JC, Easton LE, Riccio AA, … Neuhaus D (2015). Structural Basis of Detection and Signaling of DNA Single-Strand Breaks by Human PARP-1. Mol Cell, 60(5), 742–754. doi: 10.1016/j.molcel.2015.10.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischbach A, Kruger A, Hampp S, Assmann G, Rank L, Hufnagel M, … Mangerich A (2018). The C-terminal domain of p53 orchestrates the interplay between non-covalent and covalent poly(ADP-ribosyl)ation of p53 by PARP1. Nucleic Acids Res, 46(2), 804–822. doi: 10.1093/nar/gkx1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagne JP, Bonicalzi ME, Gagne P, Ouellet ME, Hendzel MJ, & Poirier GG (2005). Poly(ADP-ribose) glycohydrolase is a component of the FMRP-associated messenger ribonucleoparticles. Biochem J, 392(Pt 3), 499–509. doi: 10.1042/BJ20050792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagne JP, Pic E, Isabelle M, Krietsch J, Ethier C, Paquet E, … Poirier GG (2012). Quantitative proteomics profiling of the poly(ADP-ribose)-related response to genotoxic stress. Nucleic Acids Res, 40(16), 7788–7805. doi: 10.1093/nar/gks486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelfman S, & Ast G (2013). When epigenetics meets alternative splicing: the roles of DNA methylation and GC architecture. Epigenomics, 5(4), 351–353. doi: 10.2217/epi.13.32 [DOI] [PubMed] [Google Scholar]
- Gelfman S, Cohen N, Yearim A, & Ast G (2013). DNA-methylation effect on cotranscriptional splicing is dependent on GC architecture of the exon-intron structure. Genome Res, 23(5), 789–799. doi: 10.1101/gr.143503.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geuens T, Bouhy D, & Timmerman V (2016). The hnRNP family: insights into their role in health and disease. Hum Genet, 135(8), 851–867. doi: 10.1007/s00439-016-1683-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson BA, Zhang Y, Jiang H, Hussey KM, Shrimp JH, Lin H, … Kraus WL (2016). Chemical genetic discovery of PARP targets reveals a role for PARP-1 in transcription elongation. Science, 353(6294), 45–50. doi: 10.1126/science.aaf7865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gittings LM, Foti SC, Benson BC, Gami-Patel P, Isaacs AM, & Lashley T (2019). Heterogeneous nuclear ribonucleoproteins R and Q accumulate in pathological inclusions in FTLD-FUS. Acta Neuropathol Commun, 7(1), 18. doi: 10.1186/s40478-019-0673-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glisovic T, Bachorik JL, Yong J, & Dreyfuss G (2008). RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett, 582(14), 1977–1986. doi: 10.1016/j.febslet.2008.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gressel S, Schwalb B, & Cramer P (2019). The pause-initiation limit restricts transcription activation in human cells. Nat Commun, 10(1), 3603. doi: 10.1038/s41467-019-11536-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guastafierro T, Cecchinelli B, Zampieri M, Reale A, Riggio G, Sthandier O, … Caiafa P (2008). CCCTC-binding factor activates PARP-1 affecting DNA methylation machinery. J Biol Chem, 283(32), 21873–21880. doi: 10.1074/jbc.M801170200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guetg C, Scheifele F, Rosenthal F, Hottiger MO, & Santoro R (2012). Inheritance of silent rDNA chromatin is mediated by PARP1 via noncoding RNA. Mol Cell, 45(6), 790–800. doi: 10.1016/j.molcel.2012.01.024 [DOI] [PubMed] [Google Scholar]
- Haimovich G, Medina DA, Causse SZ, Garber M, Millan-Zambrano G, Barkai O, … Choder M (2013). Gene expression is circular: factors for mRNA degradation also foster mRNA synthesis. Cell, 153(5), 1000–1011. doi: 10.1016/j.cell.2013.05.012 [DOI] [PubMed] [Google Scholar]
- Hansen KD, Lareau LF, Blanchette M, Green RE, Meng Q, Rehwinkel J, … Brenner SE (2009). Genome-wide identification of alternative splice forms down-regulated by nonsense-mediated mRNA decay in Drosophila. PLoS Genet, 5(6), e1000525. doi: 10.1371/journal.pgen.1000525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haque N, & Oberdoerffer S (2014). Chromatin and splicing. Methods Mol Biol, 1126, 97–113. doi: 10.1007/978-1-62703-980-2_7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hau AC, Grebbin BM, Agoston Z, Anders-Maurer M, Muller T, Gross A, … Schulte D (2017). MEIS homeodomain proteins facilitate PARP1/ARTD1-mediated eviction of histone H1. J Cell Biol, 216(9), 2715–2729. doi: 10.1083/jcb.201701154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F, Li X, Spatrick P, Casillo R, Dong S, & Jacobson A (2003). Genome-wide analysis of mRNAs regulated by the nonsense-mediated and 5’ to 3’ mRNA decay pathways in yeast. Mol Cell, 12(6), 1439–1452. [DOI] [PubMed] [Google Scholar]
- He N, Pezda AC, & Zhou Q (2006). Modulation of a P-TEFb functional equilibrium for the global control of cell growth and differentiation. Mol Cell Biol, 26(19), 7068–7076. doi: 10.1128/MCB.00778-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath CG, Viphakone N, & Wilson SA (2016). The role of TREX in gene expression and disease. Biochem J, 473(19), 2911–2935. doi: 10.1042/BCJ20160010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson DG, Kelley DR, Tenen D, Bernstein B, & Rinn JL (2016). Widespread RNA binding by chromatin-associated proteins. Genome Biol, 17, 28. doi: 10.1186/s13059-016-0878-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hottiger MO, Hassa PO, Luscher B, Schuler H, & Koch-Nolte F (2010). Toward a unified nomenclature for mammalian ADP-ribosyltransferases. Trends Biochem Sci, 35(4), 208–219. doi: 10.1016/j.tibs.2009.12.003 [DOI] [PubMed] [Google Scholar]
- Hsin JP, & Manley JL (2012). The RNA polymerase II CTD coordinates transcription and RNA processing. Genes Dev, 26(19), 2119–2137. doi: 10.1101/gad.200303.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B, Wu Z, Hergert P, Henke CA, Bitterman PB, & Phan SH (2013). Regulation of myofibroblast differentiation by poly(ADP-ribose) polymerase 1. Am J Pathol, 182(1), 71–83. doi: 10.1016/j.ajpath.2012.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huambachano O, Herrera F, Rancourt A, & Satoh MS (2011). Double-stranded DNA binding domain of poly(ADP-ribose) polymerase-1 and molecular insight into the regulation of its activity. J Biol Chem, 286(9), 7149–7160. doi: 10.1074/jbc.M110.175190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang JY, Chen WH, Chang YL, Wang HT, Chuang WT, & Lee SC (2006). Modulation of nucleosome-binding activity of FACT by poly(ADP-ribosyl)ation. Nucleic Acids Res, 34(8), 2398–2407. doi: 10.1093/nar/gkl241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K, Tidyman WE, Le KU, Kirsten E, Kun E, & Ordahl CP (2004). Analysis of nucleotide sequence-dependent DNA binding of poly(ADP-ribose) polymerase in a purified system. Biochemistry, 43(1), 217–223. doi: 10.1021/bi0301800 [DOI] [PubMed] [Google Scholar]
- Hudson WH, & Ortlund EA (2014). The structure, function and evolution of proteins that bind DNA and RNA. Nat Rev Mol Cell Biol, 15(11), 749–760. doi: 10.1038/nrm3884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip JY, Schmidt D, Pan Q, Ramani AK, Fraser AG, Odom DT, & Blencowe BJ (2011). Global impact of RNA polymerase II elongation inhibition on alternative splicing regulation. Genome Res, 21(3), 390–401. doi: 10.1101/gr.111070.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isabelle M, Moreel X, Gagne JP, Rouleau M, Ethier C, Gagne P, … Poirier GG (2010). Investigation of PARP-1, PARP-2, and PARG interactomes by affinity-purification mass spectrometry. Proteome Sci, 8, 22. doi: 10.1186/1477-5956-8-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabre I, Reddy ASN, Kalyna M, Chaudhary S, Khokhar W, Byrne LJ, … Syed NH (2019). Does co-transcriptional regulation of alternative splicing mediate plant stress responses? Nucleic Acids Res, 47(6), 2716–2726. doi: 10.1093/nar/gkz121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y, & Tulin AV (2009). Poly(ADP-ribosyl)ation of heterogeneous nuclear ribonucleoproteins modulates splicing. Nucleic Acids Res, 37(11), 3501–3513. doi: 10.1093/nar/gkp218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y, & Tulin AV (2013). Post-transcriptional regulation by poly(ADP-ribosyl)ation of the RNA-binding proteins. Int J Mol Sci, 14(8), 16168–16183. doi: 10.3390/ijms140816168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonkers I, & Lis JT (2015). Getting up to speed with transcription elongation by RNA polymerase II. Nat Rev Mol Cell Biol, 16(3), 167–177. doi: 10.1038/nrm3953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungmichel S, Rosenthal F, Altmeyer M, Lukas J, Hottiger MO, & Nielsen ML (2013). Proteome-wide identification of poly(ADP-Ribosyl)ation targets in different genotoxic stress responses. Mol Cell, 52(2), 272–285. doi: 10.1016/j.molcel.2013.08.026 [DOI] [PubMed] [Google Scholar]
- Kampen KR, Sulima SO, Vereecke S, & De Keersmaecker K (2020). Hallmarks of ribosomopathies. Nucleic Acids Res, 48(3), 1013–1028. doi: 10.1093/nar/gkz637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karch KR, Langelier MF, Pascal JM, & Garcia BA (2017). The nucleosomal surface is the main target of histone ADP-ribosylation in response to DNA damage. Mol Biosyst, 13(12), 2660–2671. doi: 10.1039/c7mb00498b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke Y, Han Y, Guo X, Wen J, Wang K, Jiang X, … Zeng X (2017). PARP1 promotes gene expression at the post-transcriptiona level by modulating the RNA-binding protein HuR. Nat Commun, 8, 14632. doi: 10.1038/ncomms14632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke Y, Zhang J, Lv X, Zeng X, & Ba X (2019). Novel insights into PARPs in gene expression: regulation of RNA metabolism. Cell Mol Life Sci, 76(17), 3283–3299. doi: 10.1007/s00018-019-03120-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DS, Camacho CV, Nagari A, Malladi VS, Challa S, & Kraus WL (2019). Activation of PARP-1 by snoRNAs Controls Ribosome Biogenesis and Cell Growth via the RNA Helicase DDX21. Mol Cell, 75(6), 1270–1285 e1214. doi: 10.1016/j.molcel.2019.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MY, Mauro S, Gevry N, Lis JT, & Kraus WL (2004). NAD+-dependent modulation of chromatin structure and transcription by nucleosome binding properties of PARP-1. Cell, 119(6), 803–814. doi: 10.1016/j.cell.2004.11.002 [DOI] [PubMed] [Google Scholar]
- Kirby IT, & Cohen MS (2019). Small-Molecule Inhibitors of PARPs: From Tools for Investigating ADP-Ribosylation to Therapeutics. Curr Top Microbiol Immunol, 420, 211–231. doi: 10.1007/82_2018_137 [DOI] [PubMed] [Google Scholar]
- Kotova E, Jarnik M, & Tulin AV (2010). Uncoupling of the transactivation and transrepression functions of PARP1 protein. Proc Natl Acad Sci U S A, 107(14), 6406–6411. doi: 10.1073/pnas.0914152107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlowski M, Corujo D, Hothorn M, Guberovic I, Mandemaker IK, Blessing C, … Ladurner AG (2018). MacroH2A histone variants limit chromatin plasticity through two distinct mechanisms. EMBO Rep, 19(10). doi: 10.15252/embr.201744445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus WL (2008). Transcriptional control by PARP-1: chromatin modulation, enhancer-binding, coregulation, and insulation. Curr Opin Cell Biol, 20(3), 294–302. doi: 10.1016/j.ceb.2008.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus WL, & Lis JT (2003). PARP goes transcription. Cell, 113(6), 677–683. [DOI] [PubMed] [Google Scholar]
- Krishnakumar R, Gamble MJ, Frizzell KM, Berrocal JG, Kininis M, & Kraus WL (2008). Reciprocal binding of PARP-1 and histone H1 at promoters specifies transcriptional outcomes. Science, 319(5864), 819–821. doi: 10.1126/science.1149250 [DOI] [PubMed] [Google Scholar]
- Krishnakumar R, & Kraus WL (2010a). The PARP side of the nucleus: molecular actions, physiological outcomes, and clinical targets. Mol Cell, 39(1), 8–24. doi: 10.1016/j.molcel.2010.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnakumar R, & Kraus WL (2010b). PARP-1 regulates chromatin structure and transcription through a KDM5B-dependent pathway. Mol Cell, 39(5), 736–749. doi: 10.1016/j.molcel.2010.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak H, Fuda NJ, Core LJ, & Lis JT (2013). Precise maps of RNA polymerase reveal how promoters direct initiation and pausing. Science, 339(6122), 950–953. doi: 10.1126/science.1229386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langelier MF, Planck JL, Roy S, & Pascal JM (2012). Structural basis for DNA damage-dependent poly(ADP-ribosyl)ation by human PARP-1. Science, 336(6082), 728–732. doi: 10.1126/science.1216338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langelier MF, Servent KM, Rogers EE, & Pascal JM (2008). A third zinc-binding domain of human poly(ADP-ribose) polymerase-1 coordinates DNA-dependent enzyme activation. J Biol Chem, 283(7), 4105–4114. doi: 10.1074/jbc.M708558200 [DOI] [PubMed] [Google Scholar]
- Langelier MF, Zandarashvili L, Aguiar PM, Black BE, & Pascal JM (2018). NAD(+) analog reveals PARP-1 substrate-blocking mechanism and allosteric communication from catalytic center to DNA-binding domains. Nat Commun, 9(1), 844. doi: 10.1038/s41467-018-03234-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leger K, Bar D, Savic N, Santoro R, & Hottiger MO (2014). ARTD2 activity is stimulated by RNA. Nucleic Acids Res, 42(8), 5072–5082. doi: 10.1093/nar/gku131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenasi T, & Barboric M (2010). P-TEFb stimulates transcription elongation and pre-mRNA splicing through multilateral mechanisms. RNA Biol, 7(2), 145–150. [DOI] [PubMed] [Google Scholar]
- Leung AK (2014). Poly(ADP-ribose): an organizer of cellular architecture. J Cell Biol, 205(5), 613–619. doi: 10.1083/jcb.201402114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung AK (2020). Poly(ADP-ribose): A Dynamic Trigger for Biomolecular Condensate Formation. Trends in Cell Biology. doi:DOI: 10.1016/j.tcb.2020.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Xiao, & Fu Xiang-Dong. (2019). Chromatin-associated RNAs as facilitators of functional genomic interactions. Nature Reviews Genetics, 20(9), 503–519. doi: 10.1038/s41576-019-0135-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Tang X, Zhu Y, Shu T, & Han X (2011). Identification of PARP-1 as one of the transcription factors binding to the repressor element in the promoter region of COX-2. Arch Biochem Biophys, 505(1), 123–129. doi: 10.1016/j.abb.2010.09.016 [DOI] [PubMed] [Google Scholar]
- Lonskaya I, Potaman VN, Shlyakhtenko LS, Oussatcheva EA, Lyubchenko YL, & Soldatenkov VA (2005). Regulation of poly(ADP-ribose) polymerase-1 by DNA structure-specific binding. J Biol Chem, 280(17), 17076–17083. doi: 10.1074/jbc.M413483200 [DOI] [PubMed] [Google Scholar]
- Luco RF, Pan Q, Tominaga K, Blencowe BJ, Pereira-Smith OM, & Misteli T (2010). Regulation of alternative splicing by histone modifications. Science, 327(5968), 996–1000. doi: 10.1126/science.1184208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Z, Lin C, Guest E, Garrett AS, Mohaghegh N, Swanson S, … Shilatifard A (2012). The super elongation complex family of RNA polymerase II elongation factors: gene target specificity and transcriptional output. Mol Cell Biol, 32(13), 2608–2617. doi: 10.1128/MCB.00182-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Z, Lin C, & Shilatifard A (2012). The super elongation complex (SEC) family in transcriptional control. Nat Rev Mol Cell Biol, 13(9), 543–547. doi: 10.1038/nrm3417 [DOI] [PubMed] [Google Scholar]
- Lupey-Green LN, Caruso LB, Madzo J, Martin KA, Tan Y, Hulse M, & Tempera I (2018). PARP1 Stabilizes CTCF Binding and Chromatin Structure To Maintain Epstein-Barr Virus Latency Type. J Virol, 92(18). doi: 10.1128/JVI.00755-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maita H, & Nakagawa S (2020). What is the switch for coupling transcription and splicing? RNA Polymerase II C-terminal domain phosphorylation, phase separation and beyond. Wiley Interdiscip Rev RNA, 11(1), e1574. doi: 10.1002/wrna.1574 [DOI] [PubMed] [Google Scholar]
- Martinez-Zamudio R, & Ha HC (2012). Histone ADP-ribosylation facilitates gene transcription by directly remodeling nucleosomes. Mol Cell Biol, 32(13), 2490–2502. doi: 10.1128/MCB.06667-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masaoka A, Gassman NR, Kedar PS, Prasad R, Hou EW, Horton JK, … Wilson SH (2012). HMGN1 protein regulates poly(ADP-ribose) polymerase-1 (PARP-1) self-PARylation in mouse fibroblasts. J Biol Chem, 287(33), 27648–27658. doi: 10.1074/jbc.M112.370759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastrocola AS, Kim SH, Trinh AT, Rodenkirch LA, & Tibbetts RS (2013). The RNA-binding protein fused in sarcoma (FUS) functions downstream of poly(ADP-ribose) polymerase (PARP) in response to DNA damage. J Biol Chem, 288(34), 24731–24741. doi: 10.1074/jbc.M113.497974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matveeva EA, Al-Tinawi QMH, Rouchka EC, & Fondufe-Mittendorf YN (2019). Coupling of PARP1-mediated chromatin structural changes to transcriptional RNA polymerase II elongation and cotranscriptional splicing. Epigenetics Chromatin, 12(1), 15. doi: 10.1186/s13072-019-0261-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matveeva E, Maiorano J, Zhang Q, Eteleeb AM, Convertini P, Chen J, … Fondufe-Mittendorf YN (2016). Involvement of PARP1 in the regulation of alternative splicing. Cell Discov, 2, 15046. doi: 10.1038/celldisc.2015.46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matveeva EA, Maiorano J, Zhang Q, Eteleeb AM, Convertini P, Chen J, … Fondufe-Mittendorf YN (2016). Involvement of PARP1 in the regulation of alternative splicing. Cell Discov, 2, 15046. doi: 10.1038/celldisc.2015.46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matveeva EA, Mathbout LF, & Fondufe-Mittendorf YN (2019). PARP1 is a versatile factor in the regulation of mRNA stability and decay. Sci Rep, 9(1), 3722. doi: 10.1038/s41598-019-39969-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer A, di Iulio J, Maleri S, Eser U, Vierstra J, Reynolds A, … Churchman LS (2015). Native elongating transcript sequencing reveals human transcriptional activity at nucleotide resolution. Cell, 161(3), 541–554. doi: 10.1016/j.cell.2015.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meder VS, Boeglin M, de Murcia G, & Schreiber V (2005). PARP-1 and PARP-2 interact with nucleophosmin/B23 and accumulate in transcriptionally active nucleoli. J Cell Sci, 118(Pt 1), 211–222. doi: 10.1242/jcs.01606 [DOI] [PubMed] [Google Scholar]
- Melikishvili M, Chariker JH, Rouchka EC, & Fondufe-Mittendorf YN (2017). Transcriptome-wide identification of the RNA-binding landscape of the chromatin-associated protein PARP1 reveals functions in RNA biogenesis. Cell Discov, 3, 17043. doi: 10.1038/celldisc.2017.43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melikishvili M, Matveeva E, & Fondufe-Mittendorf Y (2017). Erratum to: Methodology to Identify Poly-ADP-Ribose Polymerase 1 (PARP1)-mRNA Targets by PAR-CLiP. Methods Mol Biol, 1608, E1. doi: 10.1007/978-1-4939-6993-7_30 [DOI] [PubMed] [Google Scholar]
- Mitrovich QM, & Anderson P (2005). mRNA surveillance of expressed pseudogenes in C. elegans. Curr Biol, 15(10), 963–967. doi: 10.1016/j.cub.2005.04.055 [DOI] [PubMed] [Google Scholar]
- Miyazaki T, Kim YS, Yoon J, Wang H, Suzuki T, & Morse HC 3rd. (2014). The 3’−5’ DNA exonuclease TREX1 directly interacts with poly(ADP-ribose) polymerase-1 (PARP1) during the DNA damage response. J Biol Chem, 289(47), 32548–32558. doi: 10.1074/jbc.M114.547331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales J, Li L, Fattah FJ, Dong Y, Bey EA, Patel M, … Boothman DA (2014). Review of poly (ADP-ribose) polymerase (PARP) mechanisms of action and rationale for targeting in cancer and other diseases. Crit Rev Eukaryot Gene Expr, 24(1), 15–28. doi: 10.1615/critreveukaryotgeneexpr.2013006875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortusewicz O, Ame JC, Schreiber V, & Leonhardt H (2007). Feedback-regulated poly(ADP-ribosyl)ation by PARP-1 is required for rapid response to DNA damage in living cells. Nucleic Acids Res, 35(22), 7665–7675. doi: 10.1093/nar/gkm933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller-McNicoll M, & Neugebauer KM (2013). How cells get the message: dynamic assembly and function of mRNA-protein complexes. Nat Rev Genet, 14(4), 275–287. doi: 10.1038/nrg3434 [DOI] [PubMed] [Google Scholar]
- Munnur D, Bartlett E, Mikolcevic P, Kirby IT, Matthias Rack JG, Mikoc A, … Ahel I (2019). Reversible ADP-ribosylation of RNA. Nucleic Acids Res, 47(11), 5658–5669. doi: 10.1093/nar/gkz305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munnur D, Somers J, Skalka G, Weston R, Jukes-Jones R, Bhogadia M, … Malewicz M (2019). NR4A Nuclear Receptors Target Poly-ADP-Ribosylated DNA-PKcs Protein to Promote DNA Repair. Cell Rep, 26(8), 2028–2036 e2026. doi: 10.1016/j.celrep.2019.01.083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musa J, Aynaud MM, Mirabeau O, Delattre O, & Grunewald TG (2017). MYBL2 (B-Myb): a central regulator of cell proliferation, cell survival and differentiation involved in tumorigenesis. Cell Death Dis, 8(6), e2895. doi: 10.1038/cddis.2017.244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamoto MY, Rudolph J, Wuttke DS, & Luger K (2019). Nonspecific Binding of RNA to PARP1 and PARP2 Does Not Lead to Catalytic Activation. Biochemistry, 58(51), 5107–5111. doi: 10.1021/acs.biochem.9b00986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalabothula N, Al-Jumaily T, Eteleeb AM, Flight RM, Xiaorong S, Moseley H, … Fondufe-Mittendorf YN (2015). Genome-Wide Profiling of PARP1 Reveals an Interplay with Gene Regulatory Regions and DNA Methylation. PLoS One, 10(8), e0135410. doi: 10.1371/journal.pone.0135410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumann M, Pal A, Goswami A, Lojewski X, Japtok J, Vehlow A, … Hermann A (2018). Impaired DNA damage response signaling by FUS-NLS mutations leads to neurodegeneration and FUS aggregate formation. Nat Commun, 9(1), 335. doi: 10.1038/s41467-017-02299-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neugebauer KM (2002). On the importance of being co-transcriptional. J Cell Sci, 115(Pt 20), 3865–3871. [DOI] [PubMed] [Google Scholar]
- Nieto Moreno N, Giono LE, Cambindo Botto AE, Munoz MJ, & Kornblihtt AR (2015). Chromatin, DNA structure and alternative splicing. FEBS Lett, 589(22), 3370–3378. doi: 10.1016/j.febslet.2015.08.002 [DOI] [PubMed] [Google Scholar]
- Nunez Villacis L, Wong MS, Ferguson LL, Hein N, George AJ, & Hannan KM (2018). New Roles for the Nucleolus in Health and Disease. Bioessays, 40(5), e1700233. doi: 10.1002/bies.201700233 [DOI] [PubMed] [Google Scholar]
- Nusinow DA, Hernandez-Munoz I, Fazzio TG, Shah GM, Kraus WL, & Panning B (2007). Poly(ADP-ribose) polymerase 1 is inhibited by a histone H2A variant, MacroH2A, and contributes to silencing of the inactive X chromosome. J Biol Chem, 282(17), 12851–12859. doi: 10.1074/jbc.M610502200 [DOI] [PubMed] [Google Scholar]
- Oei SL, & Shi Y (2001a). Poly(ADP-ribosyl)ation of transcription factor Yin Yang 1 under conditions of DNA damage. Biochem Biophys Res Commun, 285(1), 27–31. doi: 10.1006/bbrc.2001.5115 [DOI] [PubMed] [Google Scholar]
- Oei SL, & Shi Y (2001b). Transcription factor Yin Yang 1 stimulates poly(ADP-ribosyl)ation and DNA repair. Biochem Biophys Res Commun, 284(2), 450–454. doi: 10.1006/bbrc.2001.4985 [DOI] [PubMed] [Google Scholar]
- Ong CT, & Corces VG (2014). CTCF: an architectural protein bridging genome topology and function. Nat Rev Genet, 15(4), 234–246. doi: 10.1038/nrg3663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palazzo L, Leidecker O, Prokhorova E, Dauben H, Matic I, & Ahel I (2018). Serine is the major residue for ADP-ribosylation upon DNA damage. elife, 7. doi: 10.7554/eLife.34334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palazzo L, Mikoc A, & Ahel I (2017). ADP-ribosylation: new facets of an ancient modification. FEBS J, 284(18), 2932–2946. doi: 10.1111/febs.14078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng A, & Weber SC (2019). Evidence for and against Liquid-Liquid Phase Separation in the Nucleus. Noncoding RNA, 5(4). doi: 10.3390/ncrna5040050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petesch SJ, & Lis JT (2012a). Activator-induced spread of poly(ADP-ribose) polymerase promotes nucleosome loss at Hsp70. Mol Cell, 45(1), 64–74. doi: 10.1016/j.molcel.2011.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petesch SJ, & Lis JT (2012b). Overcoming the nucleosome barrier during transcript elongation. Trends Genet, 28(6), 285–294. doi: 10.1016/j.tig.2012.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinnola A, Naumova N, Shah M, & Tulin AV (2007). Nucleosomal core histones mediate dynamic regulation of poly(ADP-ribose) polymerase 1 protein binding to chromatin and induction of its enzymatic activity. J Biol Chem, 282(44), 32511–32519. doi: 10.1074/jbc.M705989200 [DOI] [PubMed] [Google Scholar]
- Potaman VN, Shlyakhtenko LS, Oussatcheva EA, Lyubchenko YL, & Soldatenkov VA (2005). Specific binding of poly(ADP-ribose) polymerase-1 to cruciform hairpins. J Mol Biol, 348(3), 609–615. doi: 10.1016/j.jmb.2005.03.010 [DOI] [PubMed] [Google Scholar]
- Pugacheva EM, Kubo N, Loukinov D, Tajmul M, Kang S, Kovalchuk AL, … Lobanenkov VV (2020). CTCF mediates chromatin looping via N-terminal domain-dependent cohesin retention. Proc Natl Acad Sci U S A, 117(4), 2020–2031. doi: 10.1073/pnas.1911708117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rancourt A, & Satoh MS (2009). Delocalization of nucleolar poly(ADP-ribose) polymerase-1 to the nucleoplasm and its novel link to cellular sensitivity to DNA damage. DNA Repair (Amst), 8(3), 286–297. doi: 10.1016/j.dnarep.2008.11.018 [DOI] [PubMed] [Google Scholar]
- Ries RJ, Zaccara S, Klein P, Olarerin-George A, Namkoong S, Pickering BF, … Jaffrey SR (2019). m(6)A enhances the phase separation potential of mRNA. Nature, 571(7765), 424–428. doi: 10.1038/s41586-019-1374-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph J, Mahadevan J, Dyer P, & Luger K (2018). Poly(ADP-ribose) polymerase 1 searches DNA via a ‘monkey bar’ mechanism. elife, 7. doi: 10.7554/eLife.37818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldi T, Fong N, & Bentley DL (2018). Transcription elongation rate affects nascent histone pre-mRNA folding and 3’ end processing. Genes Dev, 32(3–4), 297–308. doi: 10.1101/gad.310896.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayani S, Janis M, Lee CY, Toesca I, & Chanfreau GF (2008). Widespread impact of nonsense-mediated mRNA decay on the yeast intronome. Mol Cell, 31(3), 360–370. doi: 10.1016/j.molcel.2008.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiewer MJ, & Knudsen KE (2014). Transcriptional roles of PARP1 in cancer. Mol Cancer Res, 12(8), 1069–1080. doi: 10.1158/1541-7786.MCR-13-0672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schor IE, Gomez Acuna LI, & Kornblihtt AR (2013). Coupling between transcription and alternative splicing. Cancer Treat Res, 158, 1–24. doi: 10.1007/978-3-642-31659-3_1 [DOI] [PubMed] [Google Scholar]
- Schwartz JC, Ebmeier CC, Podell ER, Heimiller J, Taatjes DJ, & Cech TR (2012). FUS binds the CTD of RNA polymerase II and regulates its phosphorylation at Ser2. Genes Dev, 26(24), 2690–2695. doi: 10.1101/gad.204602.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shayevitch R, Askayo D, Keydar I, & Ast G (2018). The importance of DNA methylation of exons on alternative splicing. RNA, 24(10), 1351–1362. doi: 10.1261/rna.064865.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla S, & Oberdoerffer S (2012). Co-transcriptional regulation of alternative pre-mRNA splicing. Biochim Biophys Acta, 1819(7), 673–683. doi: 10.1016/j.bbagrm.2012.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singatulina AS, Hamon L, Sukhanova MV, Desforges B, Joshi V, Bouhss A, … Pastre D (2019). PARP-1 Activation Directs FUS to DNA Damage Sites to Form PARG-Reversible Compartments Enriched in Damaged DNA. Cell Rep, 27(6), 1809–1821 e1805. doi: 10.1016/j.celrep.2019.04.031 [DOI] [PubMed] [Google Scholar]
- Steffen JD, McCauley MM, & Pascal JM (2016). Fluorescent sensors of PARP-1 structural dynamics and allosteric regulation in response to DNA damage. Nucleic Acids Res, 44(20), 9771–9783. doi: 10.1093/nar/gkw710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suskiewicz MJ, Zobel F, Ogden TEH, Fontana P, Ariza A, Yang JC, … Ahel I (2020). HPF1 completes the PARP active site for DNA damage-induced ADP-ribosylation. Nature. doi: 10.1038/s41586-020-2013-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talhaoui I, Lebedeva NA, Zarkovic G, Saint-Pierre C, Kutuzov MM, Sukhanova MV, … Ishchenko AA (2016). Poly(ADP-ribose) polymerases covalently modify strand break termini in DNA fragments in vitro. Nucleic Acids Res, 44(19), 9279–9295. doi: 10.1093/nar/gkw675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teloni F, & Altmeyer M (2016). Readers of poly(ADP-ribose): designed to be fit for purpose. Nucleic Acids Res, 44(3), 993–1006. doi: 10.1093/nar/gkv1383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C, Ji Y, Lodhi N, Kotova E, Pinnola AD, Golovine K, … Tulin AV (2016). Non-NAD-Like poly(ADP-Ribose) Polymerase-1 Inhibitors effectively Eliminate Cancer in vivo. EBioMedicine, 13, 90–98. doi: 10.1016/j.ebiom.2016.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C, Ji Y, Wu C, Datz H, Boyle C, MacLeod B, … Tulin AV (2019). Hit and run versus long-term activation of PARP-1 by its different domains fine-tunes nuclear processes. Proc Natl Acad Sci U S A, 116(20), 9941–9946. doi: 10.1073/pnas.1901183116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veith S, Schink A, Engbrecht M, Mack M, Rank L, Rossatti P, … Mangerich A (2019). PARP1 regulates DNA damage-induced nucleolar-nucleoplasmic shuttling of WRN and XRCC1 in a toxicant and protein-specific manner. Sci Rep, 9(1), 10075. doi: 10.1038/s41598-019-46358-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vihervaara A, Mahat DB, Guertin MJ, Chu T, Danko CG, Lis JT, & Sistonen L (2017). Transcriptional response to stress is pre-wired by promoter and enhancer architecture. Nat Commun, 8(1), 255. doi: 10.1038/s41467-017-00151-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virag L, & Szabo C (2002). The therapeutic potential of poly(ADP-ribose) polymerase inhibitors. Pharmacol Rev, 54(3), 375–429. [DOI] [PubMed] [Google Scholar]
- Vispe S, Yung TM, Ritchot J, Serizawa H, & Satoh MS (2000). A cellular defense pathway regulating transcription through poly(ADP-ribosyl)ation in response to DNA damage. Proc Natl Acad Sci U S A, 97(18), 9886–9891. doi: 10.1073/pnas.170280397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyas S, & Chang P (2014). New PARP targets for cancer therapy. Nat Rev Cancer, 14(7), 502–509. doi: 10.1038/nrc3748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiman KG (2013). p53 talks to PARP: the increasing complexity of p53-induced cell death. Cell Death Differ, 20(11), 1438–1439. doi: 10.1038/cdd.2013.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Laurent B, Hsu CH, Nachtergaele S, Lu Z, Sheng W, … Shi Y (2017). RNA m(6)A methylation regulates the ultraviolet-induced DNA damage response. Nature, 543(7646), 573–576. doi: 10.1038/nature21671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Zhao W, Olson SD, Prabhakara KS, & Zhou X (2018). Alternative splicing links histone modifications to stem cell fate decision. Genome Biol, 19(1), 133. doi: 10.1186/s13059-018-1512-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G, Chen Y, Wu J, Chen SH, Liu X, Singh AK, & Yu X (2020). Poly(ADP-ribosyl)ation mediates early phase histone eviction at DNA lesions. Nucleic Acids Res doi: 10.1093/nar/gkaa022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Ginjala V, Pant V, Chernukhin I, Whitehead J, Docquier F, … Ohlsson R (2004). Poly(ADP-ribosyl)ation regulates CTCF-dependent chromatin insulation. Nat Genet, 36(10), 1105–1110. doi: 10.1038/ng1426 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Hildebrandt EF, Simbulan-Rosenthal CM, & Anderson MG (2002). Sequence-specific binding of poly(ADP-ribose) polymerase-1 to the human T cell leukemia virus type-I tax responsive element. Virology, 296(1), 107–116. doi: 10.1006/viro.2002.1385 [DOI] [PubMed] [Google Scholar]
- Zhen Y, & Yu Y (2018). Proteomic Analysis of the Downstream Signaling Network of PARP1. Biochemistry, 57(4), 429–440. doi: 10.1021/acs.biochem.7b01022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlatanova J, & Caiafa P (2009). CTCF and its protein partners: divide and rule? J Cell Sci, 122(Pt 9), 1275–1284. doi: 10.1242/jcs.039990 [DOI] [PubMed] [Google Scholar]


