Summary
Salmonella invade non-phagocytic cells by inducing massive actin rearrangements, resulting in membrane ruffle formation and phagocytosis of the bacteria. This process is mediated by a cohort of effector proteins translocated into the host cell by type III secretion system 1, which is encoded by genes in the Salmonella Pathogenicity Island 1 regulon. This network is precisely regulated and must be induced outside of host cells. In vitro invasive Salmonella are prepared by growth in synthetic media although the details vary. Here we show that, culture condition affects the frequency of Salmonella Pathogenicity Island 1 induced bacteria and therefore invasion efficiency and also can affect the ability of Salmonella to adapt to its intracellular niche following invasion. Aerobically grown late-log bacteria were more invasive and this was associated with a greater frequency of Salmonella Pathogenicity Island 1 induced, motile bacteria as revealed by single-cell analysis of gene expression. Culture condition also affected the ability of Salmonella to adapt to the intracellular environment since it caused marked differences in intracellular replication. These findings show that induction of Salmonella Pathogenicity Island 1 under different pre-invasion growth conditions can impact the ability of Salmonella to interact with eukaryotic host cells.
Keywords: epithelial, flagella, invasion, motility, Salmonella-containing vacuole, transcriptome
Introduction
Type III secretion systems (T3SS) are important virulence determinants for many Gram-negative pathogens. These systems consist of a specialized secretion apparatus that translocates effector proteins from the bacterial cytoplasm into the host cell. Translocated T3SS effector proteins effectively allow the bacteria to “hijack” many essential intracellular processes. Salmonella enterica serovar Typhimurium (S. Typhimurium) is a facultative intracellular pathogen that causes gastroenteritis in humans and a systemic typhoid-like disease in susceptible mice. S. Typhimurium virulence is dependent on two T3SS (T3SS1 and T3SS2) that are used for invasion of non-phagocytic host cells and modification of the intracellular environment. T3SS1 translocates effector proteins required for invasion of non-phagocytic host cells and some post-invasion events [for review (McGhie et al., 2009)]. In contrast, T3SS2 is induced intracellularly and its effectors are translocated into the host cell across the membrane of the Salmonella-containing vacuole (SCV), a modified phagosome within which the bacteria survive and replicate (Steele-Mortimer, 2008). Most of the genes encoding T3SS1 and T3SS2 structural components as well as many regulatory factors and effectors are located on Salmonella pathogenicity islands (SPI) 1 and 2, respectively, although some effectors proteins are encoded on other pathogenicity islands or islets (Hansen-Wester & Hensel, 2001; Marcus et al., 2000).
Various environmental signals such as oxygen concentration, osmolarity and the bacterial growth state have been shown to influence the expression of SPI1 genes and the secretion of T3SS1 effectors (Ellermeier & Slauch, 2007). A number of in vitro culture conditions for obtaining SPI1-induced invasive S. Typhimurium have been described. In aerated culture, the invasive phenotype is induced during a brief period at the end of exponential growth, or late logarithmic phase (Song et al., 2004; Steele-Mortimer et al., 1999), although other studies have shown that oxygen limiting, or microaerophilic, conditions are required for optimal induction and that, under these conditions, invasiveness is induced during stationary phase (Bajaj et al., 1996; Behlau & Miller, 1993; Jones & Falkow, 1994; Lee & Falkow, 1990; Schiemann & Shope, 1991; Temme et al., 2008).
In this study we have grown S. Typhimurium under two distinct “invasion inducing” conditions and then analyzed gene expression and the ability of the bacteria to invade, and replicate within, cultured epithelial cells. Bacteria were grown in LB-Miller broth, either to late-logarithmic phase with aeration (aer-LL) (Steele-Mortimer et al., 1999) or to stationary phase in the absence of aeration under microaerophilic conditions (μaer-ST). Aer-LL bacteria were more invasive than μaer-ST bacteria and, although transcriptome analysis did not reveal significant variation in SPI1 expression, single cell analysis revealed differences in frequency of SPI1-duced bacteria, which were predominant in the aer-LL culture. Moreover, co-expression of flagella and SPI1 genes were associated with higher invasion levels. Finally, we found that intracellular replication could be impacted by pre-invasion growth conditions and this was associated with differences in intracellular expression of SPI1 genes. Altogether this study suggests that pre-invasion growth conditions used to induce invasive bacteria in vitro can be an influential factor in the outcome of Salmonella-host cell interactions.
Methods
Bacterial cultures and growth conditions
Salmonella enterica serovar Typhimurium SL1344 (Hoiseth & Stocker, 1981) was used in all experiments unless otherwise indicated. ΔSPI1::kan has been described previously (Drecktrah et al., 2006). The flagellar mutants fliC::Tn10, fljB::Mud-Cm and flgB::Tn10 and the fimAICDHF::kan mutant were obtained by P22 transduction of S. enterica serovar Typhimurium SL1344 with phages kindly provided by Ed Miao (Miao et al., 2006) and Andreas Bäumler (Weening et al., 2005). Bacteria were initially streaked on Luria-Bertani (LB)-Miller agar (10 g tryptone l−1, 5 g yeast extract l−1, 10 g NaCl l−1, pH 7.0) supplemented with streptomycin (100 μg ml−1), kanamycin (50 μg ml−1), chloramphenicol (30 μg ml−1), carbenicillin (50 μg ml−1) and/or tetracycline (10 μg ml−1). Fresh plates were streaked every week and stored at 4 °C. The μaer-ST bacteria were prepared by inoculating one colony into 3 ml of LB-Miller broth in a 17 × 100 mm, 14 ml polypropylene round bottom test tube (Becton Dickinson) with a loose cap, and then incubating at 37 °C without aeration (no shaking) for 18 h. Aer-LL bacteria were prepared by inoculating one colony into 2 ml of LB-Miller broth, in a 17 × 100 mm, 14 ml polypropylene round bottom test tube (Becton Dickinson) with a loose cap, and incubating at 37 °C with aeration (shaking at 225 rpm) for 16-18 h. Thereafter a 300 μl aliquot of this culture was immediately subcultured into 10 ml of LB-Miller broth, in a 150 ml glass Erlenmeyer flask with a loose cap, and incubated at 37 °C for 3.5 h with aeration (shaking at 225 rpm).
Measurement of bacterial growth
For measurement of bacterial growth, Salmonella was cultured as described then samples were taken every hour for optical density at 600 nm (OD600) readings in a SmartSpec 3000 spectrophotometer (Bio Rad) and diluted in sterile PBS and plated on LB-Miller agar for enumeration of viable bacteria.
Plasmid stability
Plasmid stabilities in culture, aerated and non-aerated as described, or in infected cells were confirmed by plating on LB Miller agar and by comparing the c.f.u. obtained in the presence (50 μg ml−1carbenicillin) or absence (no antibiotic) of selection.
Salmonella Affymetrix array
The Salmonella serovar Typhimurium SL1344 (NCTC 13347) genome was first analyzed by Integrated Genomics to remove genetic redundancy and provide annotation for intergenic and opposite strand coding regions using the sequence contigs from the Sanger Institute (www.sanger.ac.uk). The microarray is an 11 micron, 49-5241 format array containing 108,166 probe pairs or about 9,833 eleven probe pair probe sets specific to 4,712 open reading frames (ORFs) and 5,121 intergenic and opposite strand coding targets (Affymetrix).
Microarray experiments
For each condition (μaer-ST or aer-LL Salmonella) six biological replicates were prepared. Total RNA from each sample was stabilized with RNAProtect (QIAGEN). The concentrate was fragmented with a QIAshredder (QIAGEN) and total RNA was purified in 96-well format using the RNeasy 96 kit centrifugation system (QIAGEN) as described previously (Virtaneva et al., 2005). RNA samples were treated with DNA-free DNase I (Ambion) and sample quality was monitored on Agilent RNA Pico II LabChips (Agilent Technologies). Additionally, possible DNA contamination was tested by qPCR (see below). 10 μg of total RNA from each sample was reverse-transcribed to cDNA using 4.5 μg random hexamer primers (Invitrogen) and 30 U SuperScript III reverse transcriptase (Invitrogen). RNA was removed by treatment with 0.25 N NaOH for 30 min at 65 °C and then neutralized with 1 N HCl. cDNA was purified using a QIAQuick 96 PCR purification kit (QIAGEN), quantified and fragmented with 0.6 U DNase I (Roche). Enzo BioArray Terminal Labeling Kit with Biotin-ddUTP (EnzoLite Sciences) was used to label the 3′ termini of the fragmented cDNA products. Labeled and fragmented cDNA (2 μg) was hybridized to Salmonella GeneChips following a previously described procedure (Virtaneva et al., 2005).
Transcriptome analysis
Affymetrix GeneChip Operating Software (GCOS v1.4, http://www.affymetrix.com) was used to perform the preliminary analysis of the custom chips at the probe-set level. A pivot table with all samples was created including calls, call P-value and signal intensities for each gene. The pivot table was then imported into GeneSpring GX 7.3 (Agilent), where hierarchical clustering (condition tree) using a Pearson correlation similarity measure with average linkage was used to produce the dendrogram indicating biological replicates were grouping together (data not shown). The CEL files were imported into Partek® Genomics Suite™ and quantile normalized to produce a principal component analysis (PCA) plot as a secondary statistical check on biological replicates grouping together. A one-way ANOVA was run from this data set to produce a false discovery rate (FDR) report of false positive reduced P-values for each of the treatments. Once the data had passed all preliminary statistical tests, the biological replicates were combined. A custom Microsoft Excel worksheet was then used to combine correlated replicates of all test conditions and controls. Quality filters based upon combined calls and signal intensities are used in the worksheet in order to further evaluate individual gene comparisons. Present and marginal are treated as the same while absent calls are negatively weighted for the filters and dropped completely from quality summations. All individual genes passing the above filters and combined from all usable replicates have the ratios of test/control reported with associated probability of Student's t-test values. Significance analysis of microarrays (SAM) (Tusher et al., 2001) was also run within the Excel sheet and a column added with the results. An additional column was added for Partek's ANOVA P-values that pass the FDR.
Gene set enrichment analysis (GSEA)
GSEA was performed using gene sets obtained from KEGG (http://www.genome.jp/kegg/pathway.html) and ERGO (Integrated Genomics) (Kanehisa et al., 2008). These databases were curated to include the SPI1 and SPI2 regulon gene sets from previous microarray analysis (Eriksson et al., 2003; Hautefort et al., 2008). The enriched sets were performed using GSEA (Mootha et al., 2003; Subramanian et al., 2005; Subramanian et al., 2007) and SAM (Tusher et al., 2001) with 1,000 permutations and a FDR of 0.25 for the significance threshold.
qPCR Analysis
Briefly, RNA samples were obtained from separate experiments and verified on Agilent RNA Pico II LabChips. For qPCR the CellsDirect™ One-Step qRT-PCR kit (Invitrogen) was used according to manufacturer's instructions. Primers and probes for different genes were designed using Primer Express® v2.0 software for multiplex qPCR (Applied Biosystems) and 100% amplification efficiency and compatibility were verified with S. Typhimurium genomic DNA. Relative quantitation of the gene expression was determined with the comparative threshold (ΔΔCT) method (Applied Biosystems). ftsZ was chosen as an internal control because no significant variation in expression for this gene was observed under the conditions used [this study and (Drecktrah et al., 2006)]. In order to detect the correlation between microarray and qPCR fold changes, the Pearson product-moment correlation coefficient (r) was calculated (Microsoft Excel®).
Transcriptional fusions
A destabilized variant of green fluorescent protein, GFP[LVA], with a half-life of approximately 40 min in S. Typhimurium (data not shown) was used as a reporter of gene expression (Andersen et al., 1998). Promoters from the following six genes were selected to drive gfp[LVA] expression: invF, prgH, sopB, ssaG, pipB and fliC. PinvF was amplified with the oligonucleotides invF-Xho (5’ ccg ctc gag aag at gag gcg cca tgt 3’) and invF-Kpn (5’ cgg ggt acc aca acg gcc tgc tcg caa 3’) (engineered restriction sites are underlined). PprgH was amplified with prgH-Xho (5’ ccg ctc gag ttg taa atg ctc ttt att 3’) and prgH-Kpn (5’ cgg ggt acc aca aag agt gtt cgg cct 3’). PssaG was amplified with ssaH-Xho (5’ ccg ctc gag tgg tag ttt ggg act 3’) and ssaH-Kpn (5’ cgg ggt acc ata acc gtt agc gct 3’). These amplicons were digested with XhoI and KpnI and ligated into SalI/KpnI digested pGFP[LVA] (Clontech Laboratories). PsopB was amplified with the oligonucleotides PsigD-gfp-XbaF (5’tgc tct aga tgt tca agc atg gaa 3’) and PsigD-gfp-Kpn2R (5’cgt tgt atg gta cct ttt gta gg 3’). PpipB was amplified with PpipB-gfp-XbaF (5’ tgc tct aga cgc att tat tct ggt ata 3’) and PpipB-gfp-KpnR (5’ cgg ggt acc ggc cgc atg caa ata tct 3’). PfliC was amplified with PfliC-gfp-XbaF (5’ tgc tct aga cag tag tta agc gcg 3’) and PfliC-gfp-KpnR (5’ cgg ggt acc cag agc gga ctg gga 3’). These three amplicons were digested with XbaI and KpnI and ligated into the corresponding sites of pGFP[LVA].
The promoter-gfp[LVA]fusion cassettes were next inserted into the plasmid pMPM-A3ΔPlac, made by removing the lac promoter from pMPM-A3 (Mayer, 1995). Plac was removed by inverse PCR with the oligonucleotides, delta Plac-for (5’ cgg ggt acc agt cgg gaa acc tgt cgt 3’) and delta Plac-rev (5’ gga aca aaa gct ggg tac cgg g 3’). The resulting amplicon was digested with KpnI and self-ligated to give pMPM-A3ΔPlac. Promoter-gfp[LVA]fusion cassettes were excised from pGFP[LVA] by HindIII digestion and ligated into HindIII-digested pMPM-A3 ΔPlac. All fusions are in the opposite direction to the α-lacZ gene. The reporter genes are designated: pMPMA3ΔPlac PinvF-gfp[LVA]/R, pMPMA3ΔPlac PprgH-gfp[LVA]/R, pMPMA3ΔPlac PsopB-gfp[LVA]/R, pMPMA3ΔPlac PssaG-gfp[LVA]/R, pMPMA3ΔPlac PpipB-gfp[LVA]/R and pMPMA3ΔPlac PfliC-gfp[LVA]/R. To construct a negative control reporter fusion, a fragment containing gfp[LVA] without any promoter was removed from pGFP[LVA] by HindIII digestion and ligated into pMPM-A3ΔPlac to give pMPMA3ΔPlac null-gfp[LVA]/R. All fusion sequences were verified by DNA sequencing (NIAID genomics facility, Rocky Mountain Laboratories).
Flow cytometry
100 μl aliquots of bacterial cultures in a round-bottom 96-well plate (Falcon) were centrifuged at 2,250 × g for 6 min, washed with PBS and resuspended in freshly made 1% paraformaldehyde (PFA; w/v) in PBS. Bacteria were then washed twice in PBS and immunostained with an anti-Salmonella LPS mAb (Meridian Life Science) diluted (1:2,000) in PBS for 10 min. Bacteria were again washed twice with PBS, followed by incubation with a Cy5-conjugated donkey anti-mouse antibody (1:1,000 in PBS; Jackson ImmunoResearch Laboratories) for 10 min and two more PBS washes. For all measurements a BD™ LSR II flow cytometer (BD Bioscience) was used and results were analyzed using FlowJo software (Tree Star).
Electron Microscopy
1 ml of bacterial culture, containing approximately 6 × 109 or 1.5 × 109 c.f.u. of aer-LL or μaer-ST respectively, were centrifuged at 1,000 × g for 10 min, washed gently with 1 ml of PBS and resuspended in 500 μl of PBS. 10 μl droplets were then adsorbed for 5 min on carbon-coated formvar grids (Ted Pella). Excess fluids were removed from the grid surface by blotting with filter paper, and the sample was stained for 2 min with 2% ammonium molybdate in double distilled water. Excess stain was removed by blotting and the samples dried at room temperature prior to being viewed at 80 kV in a Hitachi 7500 transmission electron microscope (Hitachi High-Technologies). Images were acquired with a digital camera system (Advanced Microscopy Techniques) and processed with Adobe Photoshop v. 7 (Adobe Systems).
Immunofluorescence detection of flagella and type 1 fimbriae
Bacterial cultures were prepared using either WT Salmonella Typhimurium or a flgB::Tn10 mutant, both harboring the reporter plasmid pMPMA3ΔPlac PprgH-gfp[LVA]/R, or a fimAICDHF::kan mutant. Bacteria were collected by centrifugation at 1,000 × g for 10 min and resuspended carefully in PBS. PFA was then added (1% w/v final concentration) and the sample incubated for 10 min. Fixed bacteria were washed twice with PBS and stained with mouse anti-FliC mAb (1:100; BioLegend) and rabbit anti-Salmonella LPS antibodies (1:300; Difco) or rabbit polyclonal anti-FimA (1:100; provided by Andreas Bäumler) and mouse anti-Salmonella LPS mAb (1:2,000). Secondary antibodies were AlexaFluor 568-conjugated goat anti-mouse antibodies (1:1,000; Invitrogen) and Cy5-conjugated donkey anti-rabbit antibodies (1:1,000; Jackson ImmunoResearch Laboratories). All antibodies were diluted in 0.1% (w/v) saponin, 10% (w/v) normal goat serum in PBS (SS-PBS). To label DNA bacteria were incubated with 1 μg ml−1 Hoechst 33342 solution (Invitrogen). After gentle washing with PBS, bacteria were resuspended in PBS and mounted on glass slides using ProLong® Gold anti-fade reagent (Invitrogen). FliC-positive and FimA-positive Salmonella were enumerated visually using an upright fluorescence microscope (Nikon Eclipse E800).
Motility assays
Motility assays on semisolid agar were performed as described with some modifications (Kim & Surette, 2003). Briefly, 0.5 μl of overnight cultures were spotted in the middle of a swimming plate (LB-Miller, 0.25% agar) in duplicate and allowed to dry for 1 h at room temperature. Plates were incubated at 37 °C for 6 h and growth halo measured in mm.
Microscopy based bacterial motility measurements were performed using Salmonella harboring plasmid pFPV25.1 for the constitutive expression of gfpmut3 under the control of the rpsM promoter (Valdivia & Falkow, 1996). Salmonella motility in swimming plates was not affected by either the presence of this plasmid or the expression of GFP (Fig. S3). μaer-ST and aer-LL cultures were diluted 1:12 and 1:30, respectively, in PBS. 10 μl drops of bacterial suspension on a glass slide were covered with a coverslip, which was sealed with wax, and the samples imaged within 5 minutes. Images were collected using a Nikon Eclipse Ti Perfect Focus inverted microscope fitted with a Nipkow spinning disk, and a 60×, 1.4 NA oil immersion objective (Nikon). Image acquisition by a Photometrics CascadeII camera was controlled by Metamorph software (Molecular Devices). A time-lapse series of 26 sequential images (100 ms exposure) was taken over 5 sec (n=10 fields). To identify non-motile Salmonella, i.e. those that remained in the same location in images #1, 13 and 26, these three images were overlaid in ImageJ (Rasband, W.S., ImageJ, U. S. NIH, Bethesda, MD, USA, http://rsb.info.nih.gov/ij/, 1997-2008). To enumerate the total number of bacteria in the sample, Salmonella were fixed (1% PFA) and mounted before imaging (n=10 fields). The percentage motile bacteria was determined by subtracting from 100 the percentage of total non-motile bacteria.
Invasion and replication assays
These assays were done as previously described (Drecktrah et al., 2006; Steele-Mortimer et al., 1999; Steele-Mortimer, 2007). Briefly, HeLa cells (human cervical adenocarcinoma epithelial, ATCC CCL-2) were grown in growth media (GM), consisting of Eagle's medium (Mediatech) supplemented with 10% (v/v) heat-inactivated fetal bovine serum (Invitrogen), in a humidified 37 °C, 5% CO2 incubator. HeLa cells were seeded 24 h prior to infection, either in 24-well plates at 5 × 104 cells/well or in 6-well plates at 2.4 × 105 cells/well. Salmonella were collected by centrifugation at 5,900 × g for 2 min, resuspended in Hank's buffered saline solution (HBSS) and used immediately to infect epithelial cells for 10 min at 37 °C. Inoculum counts were checked by plating on LB agar plates. In some experiments, monolayers were centrifuged at 1,000 × g for 5 min immediately after addition of the bacteria and then incubated for a further 10 min at 37 °C. Extracellular bacteria were removed by aspiration and monolayers were washed twice with HBSS. Monolayers were then incubated at 37 °C for 20 min in GM after which time 50 μg gentamicin ml−1 was added for 30 min to kill extracellular bacteria. Subsequently GM with 10 μg gentamicin ml−1 was added for the remainder of the experiment. Monolayers were lysed in 1 ml of 1% Triton X-100 (v/v), 0.1% sodium dodecyl sulfate (w/v) in PBS and viable intracellular bacteria enumerated by plating on LB agar.
Immunofluorescence microscopy
HeLa cells were plated 24 h prior to infection on glass coverslips in 24-well plates (6 × 104 cells/well). To measure LAMP1 acquisition by the SCV, infected cells were fixed in 2.5% PFA (w/v) for 10 min at 37 °C, then permeabilized and blocked with SS-PBS for 20 min. All antibodies were diluted in SS-PBS. Primary antibodies were mouse anti-human Lysosomal Associated Membrane Protein 1 (LAMP1) (H4A3; 1:1,500; Developmental Studies Hybridoma Bank) and rabbit anti-Salmonella LPS (1:2,000; Difco). Secondary antibodies were AlexaFluor 488-conjugated goat anti-mouse IgG antibodies and AlexaFluor 568-conjugated goat anti-rabbit IgG (1:800; Invitrogen). After extensive washing in PBS, coverslips were mounted on glass slides using Mowiol (Sigma-Aldrich). LAMP1-positive SCVs were scored visually using a Nikon Eclipse E800 upright fluorescent microscope.
For quantification of intracellular GFP-positive bacteria, HeLa cells on glass coverslips were infected with Salmonella containing the reporter plasmid constructs. Cells were then fixed in 2.5% PFA and processed for indirect immunofluorescence microscopy with mouse mAb against Salmonella LPS (1:5,000; Meridian Life Science), followed by incubation with a Cy5-conjugated donkey anti-mouse antibody (1:800; Jackson ImmunoResearch Laboratories) to detect extracellular bacteria. Cells were then permeabilized and blocked with SS-PBS and total bacteria detected with rabbit anti-Salmonella LPS antibodies (1:2,000; Difco) followed by AlexaFluor 568-conjugated goat anti-rabbit antibodies (1:800; Invitrogen). Samples were mounted as above. The percentage of intracellular GFP-positive Salmonella was enumerated visually using a Nikon Eclipse E800 upright microscope. All values represent the means ± SD from at least three independent experiments.
QuantiGene detection of intracellular gene expression
Direct measurement of RNA was performed using the QuantiGene reagent system v. 2.0 (Panomics) according to the manufacturer's directions. Probe sets were custom designed by Panomics and detection efficiency was previously standardised with bacterial cultures from both growth conditions (data not shown). Briefly, HeLa cells in 6-well plates (2.2 × 105 cells/well) were infected at an m.o.i. similar to that used for invasion assays. Infected monolayers were lysed in 0.8 ml QuantiGene lysis buffer supplemented with 150 ng Proteinase K ml−1 (Panomics), and solubilised by incubation at 65 °C for 30 min. One-tenth of each sample (80 μl) was combined with blocking buffer and probe sets for each gene in a capture plate, followed by incubation at 55 °C for 16-20 h. The wells were then washed three times with wash buffer (0.1x SSC, 0.03 % lithium lauryl sulfate). Signal for the bound mRNA was developed by sequential hybridization with branched DNA and alkaline phosphatase-conjugated label probe (Panomics) at 55 °C for 1 h. After three washes with wash buffer the dioxetane substrate was added to the wells and incubated at 50 °C for 1 h. Luminescence was measured over 1000 ms using a Safire2 microplate reader (Tecan). Uninfected HeLa cells were used to determine the background signal and this value was subtracted from each sample. To normalize the expression of each gene, nusG was chosen as an internal control because no significant variation of expression for this gene was observed (Eriksson et al., 2003).
Results
Induction of invasive S. Typhimurium
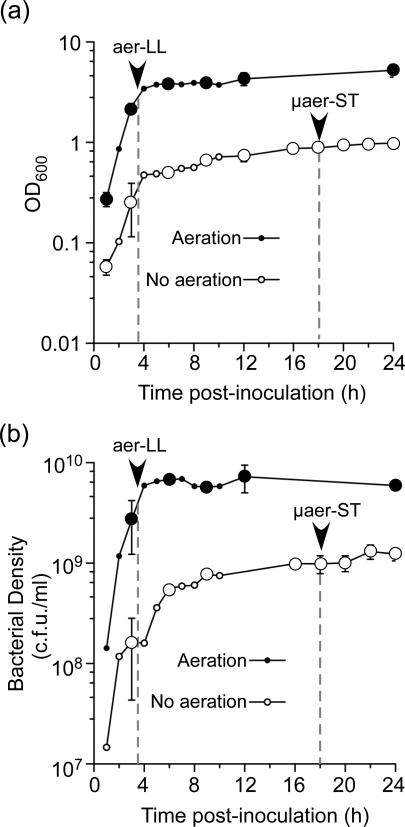
In order to induce SPI1 gene expression we grow Salmonella with aeration overnight and then dilute in fresh media and grow with aeration to late logarithmic phase. Originally we used LB broth containing 5 g l−1 NaCl (Steele-Mortimer et al., 1999) but have since found that increasing the NaCl concentration to 10 g l−1 (LB Miller) results in better induction of SPI1 [not shown and (Song et al., 2004)]. Other groups have shown that SPI1 can also be induced in stationary phase even in the absence of aeration. We hypothesized that SPI1 induction under different physiological conditions could affect the outcome of Salmonella-host cell interactions. In order to address this hypothesis we grew S. Typhimurium SL1344 in LB Miller broth either with aeration to late logarithmic phase (aer-LL) or without aeration to stationary phase (μaer-ST). As previously shown, growth curves (Fig. 1a) show that the bacteria grown with aeration reach stationary phase slightly earlier (approximately 4 h) than those grown without aeration and grow to higher cell densities (Song et al., 2004).
Figure 1.
S. Typhimurium growth curves. Bacteria were grown in LB-Miller broth at 37 °C, with (closed circles) or without (open circles) aeration. Bacterial growth was determined by OD600 (a) or culturable bacterial numbers (b). Arrowheads and dashed vertical lines indicate the time points at which aer-LL and μaer-ST bacteria are harvested from aerated and non-aerated cultures respectively. Larger symbols indicate n=3, smaller symbols n=2.
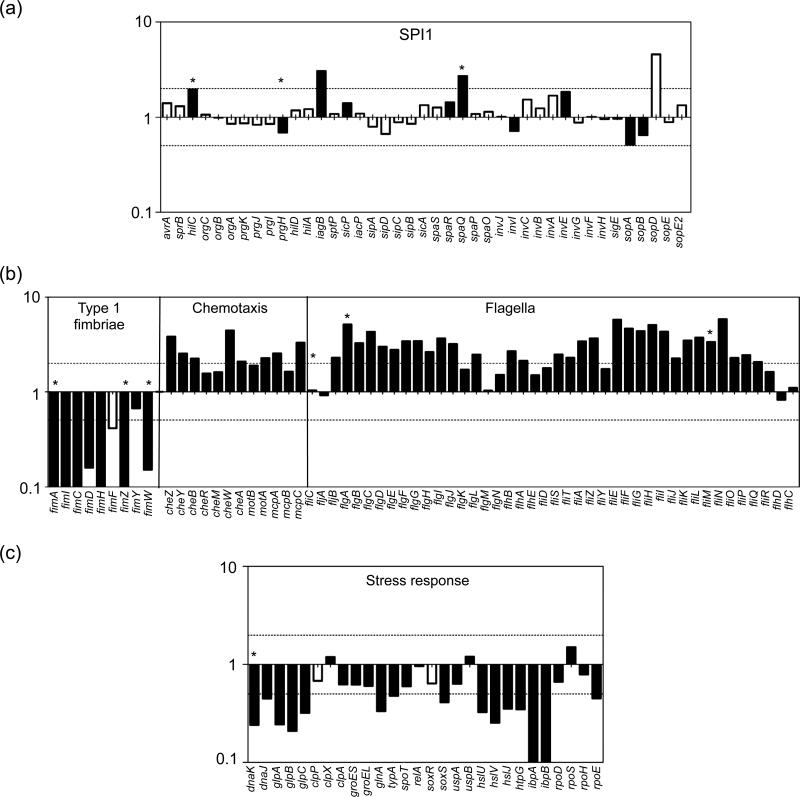
To compare these two cultures we first did a transcriptome analysis in order to confirm 1) that SPI1 genes are similarly expressed and 2) to detect differences in other gene groups. Using a custom made microarray 1009 genes were identified with statistically significant changes in the level of expression. Of these, 517 genes were expressed higher and 492 genes expressed lower in aer-LL bacteria compared to μaer-ST bacteria (Table S1). These data have been submitted to the NCBI GEO database (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE13343. To identify which functional groups of genes were differentially expressed in aer-LL and μaer-ST bacteria we used gene set enrichment analysis (GSEA), a computational method that determines whether defined sets of genes show statistically significant, concordant differences between two biological states (Subramanian et al., 2005; Tintle et al., 2008). As predicted we did not detect significant differences in the SPI1 regulon gene set, whereas flagellar-based motility and chemotaxis gene sets were highly activated in aer-LL bacteria compared to μaer-ST bacteria (GSEA rank 1 and 2, not shown). Comparison of individual genes in these functional groups (Fig. 2) confirmed that the majority of genes in the T3SS1 regulon were similarly expressed in aer-LL and μaer-ST bacteria. In contrast, genes involved in flagella biosynthesis (including flh, flg, fli and flj operons) and chemotaxis (che, mot and mcp) had higher expression levels in aer-LL bacteria and genes involved in type 1 fimbriae (fim) production and the stress response were more highly expressed in μaer-ST bacteria.
Figure 2.
Relative expression levels of selected genes in S. Typhimurium. A value of 1 indicates no detectable difference in expression between aer-LL and μaer-ST bacteria. Values >1 indicate higher expression in aer-LL bacteria and values <1 indicate higher expression in μaer-ST bacteria. Dotted lines indicate the two-fold threshold. (a) SPI1 regulon genes. (b) Motility associated genes. (c) Stress related genes. Black bars denote data that passed the P-value threshold. Asterisks indicate data confirmed by qPCR.
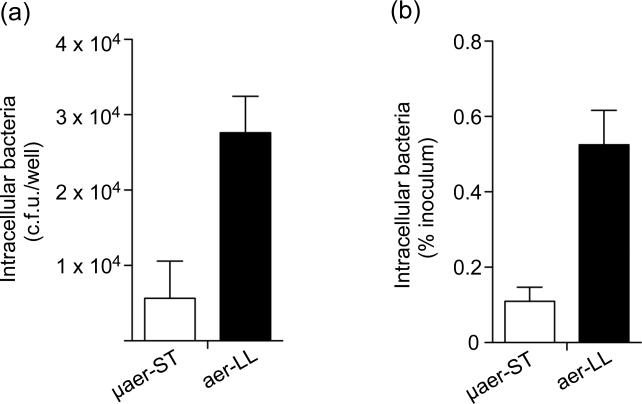
We next compared the invasiveness of aer-LL and μaer-ST bacteria using a gentamicin protection assay (Fig. 3). HeLa cells were infected with equivalent c.f.u. from each culture and viable intracellular bacteria were estimated at 1 h post-infection (p.i.). Cells infected with aer-LL bacteria contained four- to five-fold more Salmonella (E 2.8 × 104 per well) than those infected with μaer-ST bacteria (E 0.6 × 104 per well), which is within the same ranges as that reported under similar conditions previously (Song et al., 2004). Expression of these data as % of the initial inoculum, to account for any differences in inoculum size, showed invasion efficiency of 0.52%±0.09 for aer-LL bacteria and 0.11%±0.04 for μaer-ST bacteria. The apparently low invasion efficiency is a reflection of the invasion conditions since HeLa cells were incubated with a relatively high m.o.i. (E 50-60) for a short infection time (10 min). These conditions were selected in order to minimize the effect of environmental change on gene expression while still internalizing sufficient numbers of bacteria. Given that there are approximately 8 × 104 HeLa cells per well (data not shown), less than half of the cells are infected, even with the more invasive aer-LL bacteria, demonstrating that these infection conditions are well below saturation levels. Analysis by fluorescence microscopy showed that the distribution of bacteria within cells was not affected by growth condition and the majority (>70%) of infected cells contained less than 6 bacteria (Fig. S1). Altogether these data show that both growth conditions produce invasive bacteria but aer-LL bacteria are significantly more invasive than μaer-ST bacteria.
Figure 3. Invasion of epithelial cells by SPI1-induced bacteria.
To directly compare the invasiveness of aer-LL and μaer-ST S. Typhimurium, HeLa cells were infected for 10 min at 37 °C (m.o.i. = 50-60). At 1 h p.i. cells were lysed and intracellular bacteria enumerated by plating. Shown are the means ± SD from three independent experiments. (a) Invasion expressed as c.f.u. (b) Invasion expressed as % of inoculum. In both cases P<0.001, Student's t-test.
Single-cell analysis reveals differences in SPI1 gene expression
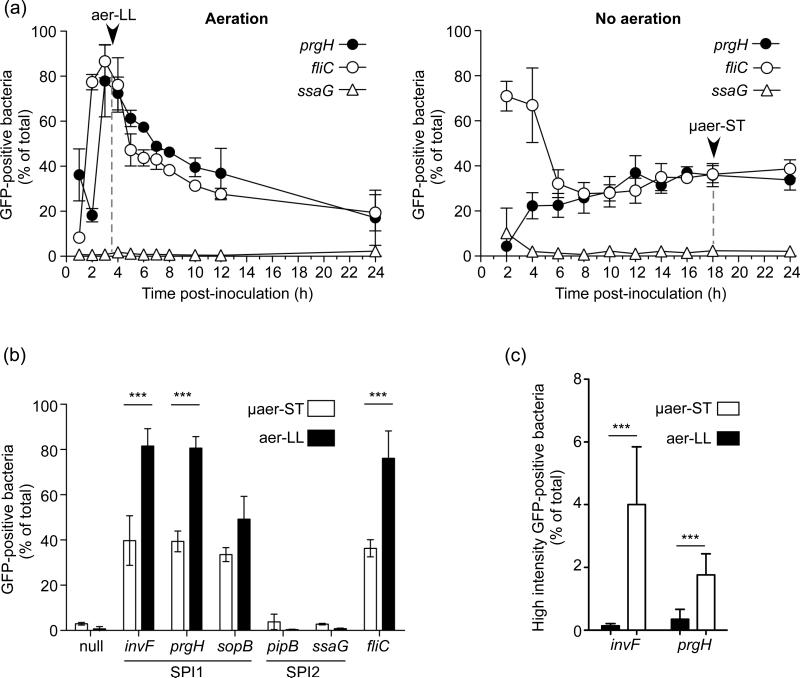
Previous studies have shown that, under SPI1-inducing conditions, only a fraction of the population are induced and that single cell experiments are required to characterize the population heterogeneity (Bumann, 2002; Hautefort et al., 2003; Temme et al., 2008). Here we used transcriptional fusions to a destabilized variant of GFP, GFP[LVA], to identify induced bacteria (Drecktrah et al., 2006). Promoters of three SPI1-regulon genes (invF, prgH and sopB) were compared to two SPI2-associated genes (ssaG and pipB) and a flagellin gene (fliC). Plasmid borne fusions were introduced into WT S. Typhimurium and the numbers of GFP-positive bacteria assessed by flow cytometry following growth under aerated and non-aerated conditions (Fig. 4a). Control experiments showed that under these conditions loss of the plasmid was negligible (Fig. S2). Initial experiments, in which PprgH activity was measured, confirmed that SPI1 induction peaks at approximately 3-4 h post inoculation in aerated culture with approximately 80% GFP-positive bacteria. In contrast, in non-aerated culture, GFP-positive bacteria never accounted for more than 40% of the total population. PfliC activity was very similar to PprgH in aerated culture but in non-aerated culture revealed very different activation levels at early time points. PfliC activity was initially high (71±7% GFP-positive bacteria), whereas very little PprgH activity could be detected (4±1% GFP-positive bacteria). Subsequently, PprgH activity increased and PfliC activity decreased to so that by 8 h there were similar numbers of GFP-positive bacteria, 26±7 and 28±1% respectively. Thereafter there were no significant changes up to 24 h. When plasmids containing a SPI2 gene transcriptional fusion (PssaG-gfp[LVA]) or GFP without a promoter (null-gfp[LVA]) were used, no significant numbers of GFP-positive bacteria were detected at any time point (Fig. 4 and data not shown). When aer-LL and μaer-ST S. Typhimurium, harvested from aerated and non-aerated cultures at 3.5 h and 18 h respectively, were directly compared for promoter activities for the three SPI1-regulon genes invF, prgH and sopB the aer-LL culture contained 82, 81 and 49% GFP-positive bacteria respectively compared to 40, 39 and 34% GFP-positive bacteria in the μaer-ST culture (Fig. 4b). Similarly the fliC promoter was induced in 76±12 and 36±4% of bacteria in aer-LL and μaer-ST cultures, respectively. Virtually no activity could be detected in either culture for the SPI2 promoters PpipB and PssaG or for the null construct. Altogether, single-cell analysis revealed a higher frequency of SPI1-induced bacteria in aer-LL compared to μaer-ST bacteria.
Figure 4.
Single-cell analysis of promoter activity for representative SPI1 and flagellin genes. S. Typhimurium harboring gfp[LVA] transcriptional fusions to selected promoters were analysed by flow cytometry. (a) Activity of promoters for cultures grown with (left panel) or without (right panel) aeration. Arrowheads indicate the time points at which aer-LL and μaer-ST bacteria are harvested. (b) Promoter activity in aer-LL and μaer-ST bacteria. Comparison of SPI1 regulon genes (prgH, invF and sopB), SPI2 regulon genes (pipB and ssaG), and the flagellin gene fliC. Promoterless gfp[LVA] (null) was included as a negative control. (c) Comparison of the frequencies of high intensity GFP (>104 relative fluorescent units) expressing bacteria in aer-LL and μaer-ST bacteria. Shown are means ± SD from at least three independent experiments. ***= P<0.001 Student's t-test.
To account for the differences between the transcriptome and single-cell data, levels of induction must be higher in SPI1-induced μaer-ST bacteria compared to SPI1-induced aer-LL bacteria. To investigate this possibility we compared the GFP intensity profiles in the flow cytometry data from the above experiments. This revealed that a small, but significant, population of μaer-ST Salmonella (2-4%) had a higher GFP intensity level (RFU > 104) than detected in any aer-LL bacteria (Fig. 4c). Thus, although fewer Salmonella are SPI1-induced in the μaer-ST compared to the aer-LL culture, a small percentage of “hyper–induced” SPI1 bacteria in μaer-ST cultures results in similar average expression levels of SPI1 genes as indicated by the transcriptome analysis.
Flagella are required for efficient Salmonella invasion
Salmonella achieve motility from 5-10 peritrichous, or randomly placed, filamentous flagellae that extend from the cell surface. S. Typhimurium has two loci encoding different flagellins, fliC and fljB, the expression of which is governed by a switch mechanism so that only one flagellin is expressed at a time [reviewed in (McQuiston et al., 2008)]. Our transcriptome data suggested that fljB was induced in aer-LL bacteria whereas no difference was detected in fliC expression (Fig. 2b). However, in our hands a fliC mutant was more affected in motility and invasion of HeLa cells compared to a fljB mutant (Fig. S3 and data not shown). Since comparative analysis does not exclude the possibility that fliC is induced in both conditions, similar to the SPI1-regulon genes, we used the GFP-based transcriptional fusion assay to see if the SPI1-induced bacteria are also flagellated. Salmonella bearing the PprgH-gfp[LVA] construct were scored for GFP fluorescence and/or flagella using an anti-FliC antibody. Using this method 48±5% of aer-LL and 23±3% of μaer-ST bacteria were GFP-positive. Of these SPI1-induced bacteria 75±8% of the aer-LL and 94±5% of the μaer-ST culture were also FliC-positive, whereas only one-third of the GFP-negative bacteria were FliC-positive. The physical presence of flagella on the surface of both μaer-ST and aer-LL bacteria was confirmed by electron microscopy (Fig. S4), which also revealed that fimbriae were only present on the surface of μaer-ST bacteria.
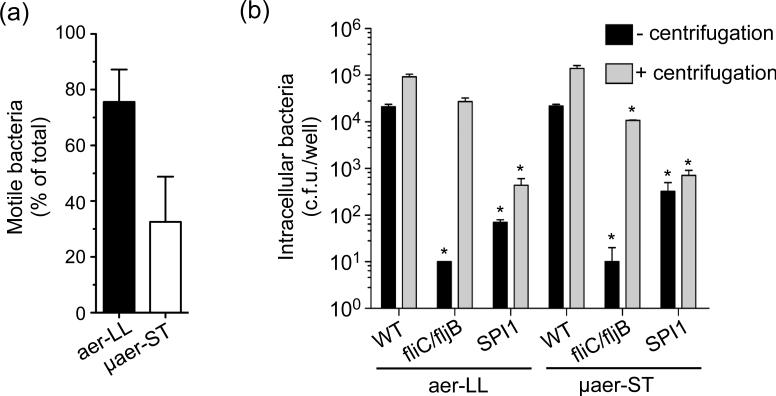
We hypothesized that the physical presence of flagella may not accurately reflect the functional competence of these structures i.e. motility. To compare the motility of aer-LL and μaer-ST Salmonella we developed a rapid microscopy-based assay, which allowed us to assess motility within minutes of harvesting the bacteria, using S. Typhimurium constitutively expressing GFP from the pFPV25.1 plasmid (Valdivia & Falkow, 1996). This assay revealed that motile bacteria predominate in the aer-LL (76±12%) compared to the μaer-ST (33±16%) culture (Fig. 5a) in agreement with the higher expression levels of motility and chemotaxis related genes.
Figure 5.
Flagella-based motility is required for optimal invasion of epithelial cells. (a) Bacterial motility was assessed by fluorescence microscopy using S. Typhimurium constitutively expressing GFP. Shown are means± SD from three independent experiments. P<0.05, Student's t-test. (b) Comparison of the role of flagellar-based motility in invasion. HeLa cells were infected with aer-LL or μaer-ST S. Typhimurium either with (grey bars) or without (black bars) centrifugation. Intracellular bacteria (1 h p.i.) were enumerated by plating. Strains were wild type S. Typhimurium (WT), fliC::Tn10 fljB::Mud-Cm mutant (fliC/fljB) and a ΔSPI1::kan mutant (SPI1). Shown are means ± SD from at least three independent experiments. *= P<0.05 Student's t-test (compared to WT without centrifugation).
The above experiments revealed a correlation between SPI1 expression and motility, with SPI1-induced motile bacteria predominating in the aer-LL culture. We next considered the contribution of flagellar-based motility in invasion of either μaer-ST or aer-LL S. Typhimurium into HeLa cells. To do this we compared the invasion efficiency of a non-motile S. Typhimurium flagellar mutant, fliC::Tn10 fljB::Mud-Cm, which is deficient for both flagellin proteins, to WT S. Typhimurium, as well as a noninvasive ΔSPI1 mutant (Drecktrah et al., 2006). The absence of flagella and motility in the flagellar mutant was confirmed by electron microscopy and soft agar motility assay (Fig. S3 and data not shown). To directly compare the requirement for motility in both cultures the inoculum size of μaer-ST bacteria was increased (5-fold higher m.o.i. compared to aer-LL) so that similar numbers of WT bacteria were internalized (Fig. 5b). In comparison to WT both the SPI1-deletion mutant, which lacks T3SS1 and is invasion defective, and the flagellar mutant had massive defects in the numbers of internalized bacteria (E3% and <1% of WT respectively) (Khoramian-Falsafi et al., 1990). We then confirmed that the invasion deficiency of the flagellar mutant could be overcome by centrifugation to increase contact with host cells (Fig. 5b), since this can compensate for the lack of motility but not the lack of SPI1 (van Asten et al., 2004). Altogether these data show that flagella-based motility is an important component in the initiation of contact with epithelial cells for both μaer-ST and aer-LL Salmonella.
Pre-invasion bacterial growth conditions can affect intracellular replication and intracellular virulence gene expression
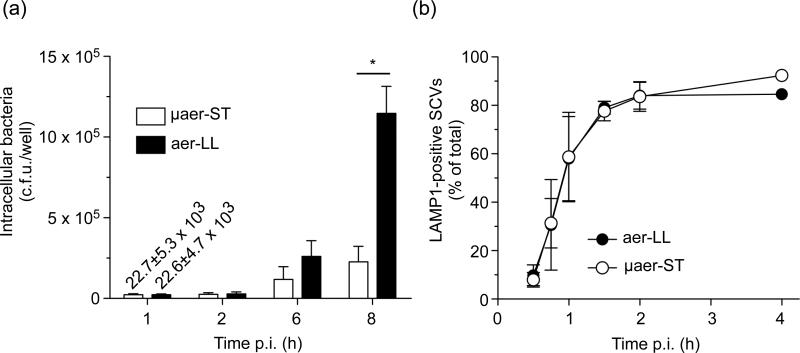
The ability of Salmonella to survive and replicate within host cells is a multifactorial process that can be affected by a variety of bacterial genes [for review see (Ibarra & Steele-Mortimer, 2009)]. Since aer-LL and μaer-ST Salmonella are grown under different conditions and are induced for different gene sets we next compared the ability of these bacteria to replicate inside HeLa cells. For these experiments we used the gentamicin protection assay, adjusting the inoculum size to ensure that there was <1% difference in the numbers of bacteria internalized and no significant difference in the intracellular distribution of bacteria at 1 h p.i. (Fig. S1). This is important since even small differences in the numbers of bacteria internalized, as opposed to the invasion efficiency, could be amplified into significant differences once replication is initiated. As shown in Fig. 6a, intracellular S. Typhimurium initiated intracellular replication by 6 h p.i. irrespective of the pre-invasion growth conditions, but by 8 h aer-LL bacteria showed a marked increase in replication compared to μaer-ST bacteria (51-fold increase vs 10-fold increase compared to 1 h).
Figure 6.
Intracellular replication is affected by pre-invasion growth conditions. (a) Infected HeLa cells were lysed for enumeration of intracellular S. Typhimurium or (b) fixed and processed for immunofluorescence using anti-LPS and anti-LAMP1 antibodies. To obtain comparable invasion cells were infected with an m.o.i. of ~50-60 for aer-LL and ~150-180 for μaer-ST. The number of intracellular aer-LL (black symbols) or μaer-ST (white symbols) S. Typhimurium with coincident LAMP1 staining cell was scored visually by fluorescence microscopy. Shown are means ± SD from three independent experiments. *= P<0.001, Bonferroni's post-hoc test.
The marked difference in intracellular replication of aer-LL and μaer-ST Salmonella suggested that pre-invasion growth conditions can affect the ability of Salmonella to adapt to the intracellular environment. To address this, we first asked whether there was any apparent defect in SCV biogenesis, by following the recruitment of the lysosomal membrane protein LAMP1, which occurs with well-established kinetics (Steele-Mortimer et al., 1999). However, no difference in LAMP1 acquisition could be detected (Fig. 6b) suggesting that the differences in intracellular replication cannot be attributed to overt changes in vacuole biogenesis.
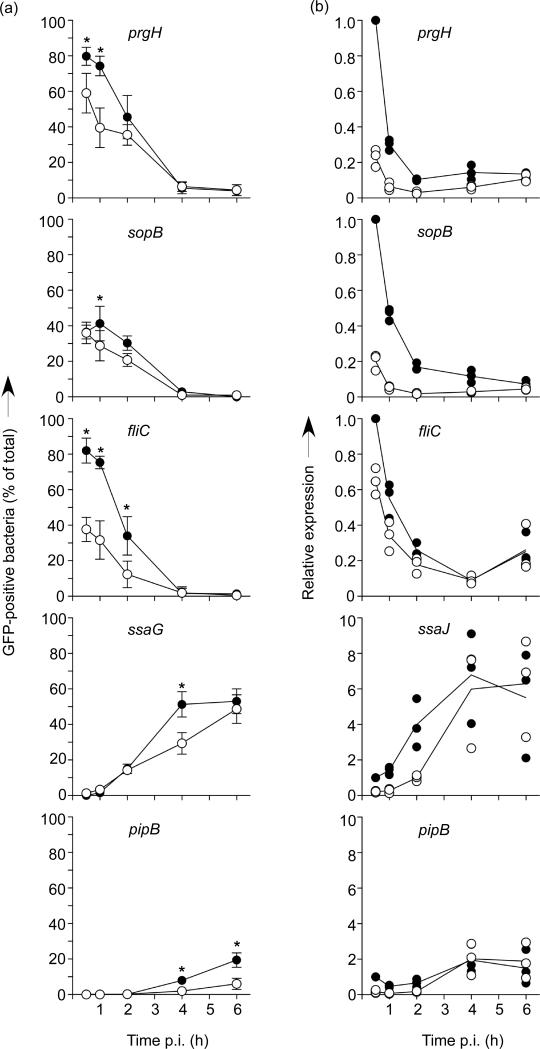
We next considered that intracellular virulence gene expression might be affected by the pre-invasion growth condition. Previous experiments have shown that SPI1 genes are rapidly down-regulated following internalization into host cells (Boddicker & Jones, 2004; Drecktrah et al., 2006; Knodler et al., 2009) so we used the GFP[LVA] transcriptional fusions to follow this process in individual bacteria following invasion of HeLa cells by aer-LL and μaer-ST bacteria. As expected the numbers of SPI1-induced GFP-positive bacteria (prgH and sopB) rapidly decreased after internalization and by 4 h p.i. virtually no GFP-positive bacteria could be detected. However, when aer-LL and μaer-ST bacteria harboring the PprgH-gfp[LVA] fusion were compared (Fig. 7a), we observed an apparent delay in down-regulation in aer-LL bacteria such that at 0.5 h p.i. more aer-LL bacteria were GFP-positive (80±5% compared to 59±11%). A similar, though more dramatic, trend was observed with the PfliC-gfp[LVA] fusion with 82±7% of aer-LL bacteria compared to 38±7% of μaer-ST bacteria being GFP-positive at 0.5 h. In contrast, we did not detect any difference in the down regulation of sopB, presumably because this gene is expressed at lower levels. When intracellular expression of the SPI2 regulon genes ssaG and pipB was compared, we observed induction in both cases, although there were far fewer GFP-positive bacteria when the PpipB-gfp[LVA] fusion was used, likely due to low expression levels of pipB. Comparison of numbers of GFP-positive bacteria again revealed a slight dependence on the pre-invasion growth method, suggesting that post-invasion induction of SPI2 genes is more efficient in aer-LL bacteria. However, this was less dramatic than that seen for the prgH and fliC promoters.
Figure 7.
Intracellular virulence gene expression can be influenced by pre-invasion growth conditions. (a) gfp[LVA] transcriptional fusions were used to assess promoter activity for prgH, sopB, ssaG, pipB and fliC following invasion of HeLa cells. GFP-positive “induced” bacteria were scored visually by fluorescence microscopy. Extracellular S. Typhimurium were stained with anti-LPS mAb before permeabilisation and excluded from analysis. Shown are means ± SD from at least three separate experiments. *= P<0.05, two-way ANOVA and Bonferroni's post-hoc analysis. (b) QuantiGene detection of prgH, sopB, ssaJ, pipB and fliC gene expression. The expression of each gene was normalized to nusG as an internal control and then compared to the value for aer-LL bacteria at 30 min. Each symbol represents the mean from one experiment in duplicate. The statistical means from three (prgH, sopB and fliC) or four (ssaJ and pipB) experiments are indicated by horizontal lines.
Limitations of the plasmid based GFP transcriptional fusion assay used here are that the presence of plasmid and/or the expression of GFP could affect invasion or the ability of the bacteria to respond to the intracellular environment [Ibarra, Knodler and Steele-Mortimer unpublished observations (Clark et al., 2009; Knodler et al., 2005)]. Therefore, we independently confirmed these results by assessing the relative levels of RNA obtained from intracellular WT S. Typhimurium (Canales et al., 2006; Flagella et al., 2006). This method uses oligonucleotide probe sets that bind a contiguous region of the target RNA. Unfortunately, technical limitations of probe design made it impossible to make ssaG probes. Instead we analyzed another SPI2 gene, ssaJ, which is in the same operon as ssaG (Lober et al., 2006). Probe sets were first tested on aer-LL and μaer-ST bacterial cultures, and gave similar results to quantitative PCR for the SPI1-regulon genes, prgH and sopB, the flagellin gene fliC (not shown). As expected no SPI2-regulon gene expression was detected in these cultures. In intracellular bacteria this method confirmed the results obtained with the transcriptional fusion assay for the prgH, sopB and fliC promoters but was unable to recapitulate any significant differences in the SPI2 gene induction (Fig.7b). Altogether, these gene expression analyses show that the increased multiplication efficiency of aer-LL S. Typhimurium inside epithelial cells is associated with higher expression of SPI1 genes at early times post invasion.
Discussion
Salmonella enterica serovars invade nonphagocytic host cells by inducing membrane ruffling on the host cell surface, a well-characterized process that is mediated by effector proteins translocated via T3SS1 (Patel & Galan, 2005). In vitro the SPI1 regulon, which encodes T3SS1, can be induced under several different conditions including growth with aeration to late log (aer-LL) and growth without aeration to stationary phase (μaer-ST). While the expression of SPI1 genes under similar conditions has been examined (Croinin & Dorman, 2007; Mangan et al., 2006; Song et al., 2004; Temme et al., 2008) the effect on pathogen-host interactions of SPI1 induction under different physiological backgrounds has not been addressed. In this study we studied aer-LL and μaer-ST S. Typhimurium cultures, and their ability to interact with cultured epithelial cells, in a comparative analysis using both population based and single cell methods. Our main findings are; (i) that pre-invasion conditions impact invasion, intracellular replication and post-invasion expression of SPI1 genes, (ii) that single cell analysis of SPI1 induction reveals differences in aer-LL and μaer-ST cultures not revealed by transcriptome analysis and (iii) that co-expression of SPI1 and flagella is associated with higher invasion levels. Here we found that pre-invasion growth conditions can significantly impact the invasiveness of S. Typhimurium as well as the ability of the bacteria to replicate intracellularly. In contrast a recent report that showed that, unlike for S. Typhi, growth conditions have no effect on the invasiveness of S. Typhimurium (Bishop et al., 2008). The differences in results are doubtless due in part to the methods or strains used, but also Bishop et al. did not include late logarithmic bacteria in their studies. Previously we have shown a very strong correlation between late logarithmic growth and invasiveness using a different Typhimurium strain (14028s) (Steele-Mortimer et al., 1999) and here we have used SL1344, however it is possible that other strains behave differently.
Our single cell analysis of gene expression revealed significant differences between aer-LL and μaer-ST S. Typhimurium in both the frequency of SPI1-induced bacteria and the maximum induction levels in agreement with previous findings (Passerat et al., 2009; Song et al., 2004) as well as in the expression of flagellar motility. Regulation of the SPI1 regulon is complex and subjected to the actions of a number of factors, which are still not well understood but themselves could be differentially regulated under the conditions described here. Transcriptional regulators necessary for optimal induction in aerobic conditions include HilA, HilC, EnvZ/OmpR and SirA/BarA (Song et al., 2004). Some may be important under both conditions, for example the positive regulator Fis, which under aerobic growth is expressed only in early logarithmic phase, is expressed throughout stationary phase in non-aerobic growth conditions (Croinin & Dorman, 2007). This sustained expression of Fis in non-aerobic conditions would explain why SPI1 is induced in μaer-ST bacteria. Another global regulator involved in SPI1 regulation is the integration host factor (IHF), the expression of which peaks in late-logarithmic and stationary phases and plays a positive role in the expression of SPI1 and flagella in the early late-log phase of growth in aerobic conditions (Mangan et al., 2006). This is consistent with our finding that in aer-LL bacteria, SPI1 and flagella genes are highly expressed and a model that proposes co-regulation of invasion and motility genes (Baxter & Jones, 2005; Kage et al., 2008; Saini et al., 2008). Nevertheless, other groups have found repression of motility under conditions of SPI1-induction and vice versa and the regulatory connection between motility and virulence remains unclear (Teplitski et al., 2003; Thijs et al., 2007). Regardless, our data clearly defines both SPI1 expression and flagellar motility as being required for optimal invasion by Salmonella.
We, and others, have previously shown that the mode of bacterial entry and SPI1-induction can affect the intracellular responses of Salmonella in phagocytic cells (Drecktrah et al., 2006; van der Velden et al., 2000). Here, however, we report the novel observation that SPI1-induction via different pre-invasion growth conditions can affect intracellular gene expression of Salmonella in non-phagocytic cells. Unlike phagocytic cells, non-phagocytic epithelial cells only internalize invasive, T3SS1-induced Salmonella. Accordingly, invasion should select specifically for SPI1-induced bacteria irrespective of the pre-invasion growth conditions and thus, since only SPI1-induced bacteria are internalized, these bacteria should react similarly to the intracellular environment. One caveat is that non-SPI1 induced “bystander” bacteria can be internalized into the ruffles stimulated by invasive SPI1-induced bacteria (Steele-Mortimer et al., 2002). In order to minimize this possibility, we did not centrifuge the bacteria onto the monolayer and after a brief 10 min invasion immediately removed non-internalized bacteria by washing. Under these conditions, we found that SPI1 genes are down-regulated following invasion in agreement with quantitative PCR showing rapidly decreasing sopB transcript levels in HeLa cells (Knodler et al., 2009) and studies showing the most SopB is translocated early (Patel et al., 2009). A recent study found that rather than being down-regulated as reported here, SPI1 is instead induced following invasion of epithelial cells (Hautefort et al., 2008). While it is difficult to reconcile these contradictions, we believe that they are most likely due to differences in methodology, including a centrifugation step to optimize internalization of bacteria, especially since this later step could increase the numbers of “bystander” non-SPI1 induced cells that are internalized. When we compared intracellular expression of SPI1 genes following internalization of either aer-LL or μaer-ST bacteria, the aer-LL bacteria exhibited sustained intracellular SPI1 expression that was accompanied by higher intracellular replication. While we have not shown a direct link between intracellular replication and sustained SPI1 expression in this study, effectors translocated by the SPI1-encoded T3SS1 can have significant post invasion roles, for example SopB mediates sustained Akt activation in infected epithelial cells (Steele-Mortimer et al., 2000) and also, though to a less obvious extent, SCV biogenesis (Hernandez et al., 2004). Also, despite rapid down-regulation of gene expression, the persistence of some T3SS1 effectors, such as SopB, inside host cells has been demonstrated (Drecktrah et al., 2005; Hernandez et al., 2004; Knodler et al., 2009). Overall, our data suggest that aer-LL bacteria are better able to adapt to the intracellular environment than μaer-ST bacteria. This could be due to something as simple as the difference in adapting from an oxygen rich environment (aer-LL) compared to an oxygen poor environment (μaer-ST), or to multiple combined factors.
Here we have defined pre-invasion bacterial growth status as a factor that can influence the outcome of Salmonella-host cell interactions in vitro. We conclude that, in the context of host-pathogen interactions, the physiological state of the bacteria should be considered, particularly when different growth conditions can be used for induction of an invasive phenotype.
Supplementary Material
Supplementary Material
Figure S1. Distribution of S. Typhimurium in infected HeLa cells. To obtain comparable invasion cells were infected with an m.o.i. of ~50-60 for aer-LL and ~150-180 for μaer-ST. Extracellular S. Typhimurium were stained with anti-LPS mAb before permeabilisation and excluded from analysis. After permeabilisation, all bacteria were stained with rabbit anti-LPS antibodies followed by AlexaFluor 568-conjugated secondary antibodies. The number of intracellular S. Typhimurium per infected cell was scored visually. Shown are means ± SD from three independent experiments (n =100).
Figure S2. Plasmids are retained in S. Typhimurium. Plasmids stabilities were determined by culturing bacteria in either μaer-ST (a) or aer-LL (b) conditions without antibiotic selection, thereafter bacteria were plated on LB-Miller agar with or without carbenicillin. Shown are means ± SD from three independent experiments.
Figure S3. Motility of S. Typhimurium strains. Flagellar function was determined by rate of spread, after 6 h incubation at 37 °C in semisolid agar. Strains were WT S. Typhimurium (WT), fliC::Tn10 (fliC), fljB::Mud-Cm mutant (fljB), fliC::Tn10 fljB::Mud-Cm mutant (fliC/fljB), WT S. Typhimurium bearing pFPV25.1 (pFPV25.1) or PprgH-gfp[LVA] (PprgH-gfp[LVA]). Shown are means ± SD, from three independent experiments.
Figure S4. Electron micrographs showing flagella (black arrows) on the surface of aer-LL and μaer-ST bacteria, but fimbriae (white arrows) only on the surface of μaer-ST bacteria. Scale bars are 500 nm (left panels) or 100 nm (right panels).
Table S1. Genome wide-expression changes for aer-LL compared to μaer-ST S. Typhimurium SL1344. Probe-set identification (ID) for S. Typhimurium SL1344 is shown and the corresponding S. Typhimurium LT2 gene or synonym is denoted where identified by BLAST analysis. Check marks show genes where both a 2-fold (aer-LL/μaer-ST fold Δ) change and P-value passing the false discovery rate at a significant level was obtained (2X and P-value Sig.).
Acknowledgements
We thank Seth Winfree, Patricia Fuentes, Ron Messer, Alissa Curda, Craig Martens, Greg Farneth, Damon Ellison and Luke Wicke for expert experimental and technical assistance. We are grateful to Ed Miao for kindly providing phages, Andreas Bäumler for providing phage stock and the anti-FimA antibody, the RML Genomics Unit for DNA sequencing analysis, members of our lab for their helpful discussions and criticism and to the reviewers for constructive suggestions. This research was supported by the Intramural Research Program (DIR) of the NIH, NIAID.
Abbreviations
- aer-LL
Salmonella grown with aeration to late logarithmic phase
- FDR
false discovery rate
- GM
growth medium
- p.i.
post-infection
- PFA
paraformaldehyde
- SCV
Salmonella-containing vacuole
- SPI
Salmonella Pathogenicity Island
- μaer-ST
Salmonella grown without aeration to stationary phase
- T3SS
Type III Secretion System
- WT
wild type
References
- Andersen JB, Sternberg C, Poulsen LK, Bjorn SP, Givskov M, Molin S. New unstable variants of green fluorescent protein for studies of transient gene expression in bacteria. Appl Environ Microbiol. 1998;64:2240–2246. doi: 10.1128/aem.64.6.2240-2246.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajaj V, Lucas RL, Hwang C, Lee CA. Co-ordinate regulation of Salmonella typhimurium invasion genes by environmental and regulatory factors is mediated by control of hilA expression. Mol Microbiol. 1996;22:703–714. doi: 10.1046/j.1365-2958.1996.d01-1718.x. [DOI] [PubMed] [Google Scholar]
- Baxter MA, Jones BD. The fimYZ genes regulate Salmonella enterica Serovar Typhimurium invasion in addition to type 1 fimbrial expression and bacterial motility. Infect Immun. 2005;73:1377–1385. doi: 10.1128/IAI.73.3.1377-1385.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behlau I, Miller SI. A PhoP-repressed gene promotes Salmonella typhimurium invasion of epithelial cells. J Bacteriol. 1993;175:4475–4484. doi: 10.1128/jb.175.14.4475-4484.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop A, House D, Perkins T, Baker S, Kingsley RA, Dougan G. Interaction of Salmonella enterica serovar Typhi with cultured epithelial cells: roles of surface structures in adhesion and invasion. Microbiology. 2008;154:1914–1926. doi: 10.1099/mic.0.2008/016998-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddicker JD, Jones BD. Lon protease activity causes down-regulation of Salmonella pathogenicity island 1 invasion gene expression after infection of epithelial cells. Infect Immun. 2004;72:2002–2013. doi: 10.1128/IAI.72.4.2002-2013.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bumann D. Examination of Salmonella gene expression in an infected mammalian host using the green fluorescent protein and two-colour flow cytometry. Mol Microbiol. 2002;43:1269–1283. doi: 10.1046/j.1365-2958.2002.02821.x. [DOI] [PubMed] [Google Scholar]
- Canales RD, Luo Y, Willey JC, Austermiller B, Barbacioru CC, Boysen C, Hunkapiller K, Jensen RV, Knight CR, other authors Evaluation of DNA microarray results with quantitative gene expression platforms. Nat Biotechnol. 2006;24:1115–1122. doi: 10.1038/nbt1236. [DOI] [PubMed] [Google Scholar]
- Clark L, Martinez-Argudo I, Humphrey TJ, Jepson MA. GFP plasmid-induced defects in Salmonella invasion depend on plasmid architecture, not protein expression. Microbiology. 2009;155:461–467. doi: 10.1099/mic.0.025700-0. [DOI] [PubMed] [Google Scholar]
- Croinin TO, Dorman CJ. Expression of the Fis protein is sustained in late-exponential- and stationary-phase cultures of Salmonella enterica serovar Typhimurium grown in the absence of aeration. Mol Microbiol. 2007;66:237–251. doi: 10.1111/j.1365-2958.2007.05916.x. [DOI] [PubMed] [Google Scholar]
- Drecktrah D, Knodler LA, Galbraith K, Steele-Mortimer O. The Salmonella SPI1 effector SopB stimulates nitric oxide production long after invasion. Cell Microbiol. 2005;7:105–113. doi: 10.1111/j.1462-5822.2004.00436.x. [DOI] [PubMed] [Google Scholar]
- Drecktrah D, Knodler LA, Ireland R, Steele-Mortimer O. The mechanism of Salmonella entry determines the vacuolar environment and intracellular gene expression. Traffic. 2006;7:39–51. doi: 10.1111/j.1600-0854.2005.00360.x. [DOI] [PubMed] [Google Scholar]
- Ellermeier JR, Slauch JM. Adaptation to the host environment: regulation of the SPI1 type III secretion system in Salmonella enterica serovar Typhimurium. Curr Opin Microbiol. 2007;10:24–29. doi: 10.1016/j.mib.2006.12.002. [DOI] [PubMed] [Google Scholar]
- Eriksson S, Lucchini S, Thompson A, Rhen M, Hinton JC. Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol Microbiol. 2003;47:103–118. doi: 10.1046/j.1365-2958.2003.03313.x. [DOI] [PubMed] [Google Scholar]
- Flagella M, Bui S, Zheng Z, Nguyen CT, Zhang A, Pastor L, Ma Y, Yang W, Crawford KL, other authors A multiplex branched DNA assay for parallel quantitative gene expression profiling. Anal Biochem. 2006;352:50–60. doi: 10.1016/j.ab.2006.02.013. [DOI] [PubMed] [Google Scholar]
- Hansen-Wester I, Hensel M. Salmonella pathogenicity islands encoding type III secretion systems. Microbes Infect. 2001;3:549–559. doi: 10.1016/s1286-4579(01)01411-3. [DOI] [PubMed] [Google Scholar]
- Hautefort I, Proenca MJ, Hinton JC. Single-copy green fluorescent protein gene fusions allow accurate measurement of Salmonella gene expression in vitro and during infection of mammalian cells. Appl Environ Microbiol. 2003;69:7480–7491. doi: 10.1128/AEM.69.12.7480-7491.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hautefort I, Thompson A, Eriksson-Ygberg S, Parker ML, Lucchini S, Danino V, Bongaerts RJ, Ahmad N, Rhen M, Hinton JC. During infection of epithelial cells Salmonella enterica serovar Typhimurium undergoes a time-dependent transcriptional adaptation that results in simultaneous expression of three type 3 secretion systems. Cell Microbiol. 2008;10:958–984. doi: 10.1111/j.1462-5822.2007.01099.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez LD, Hueffer K, Wenk MR, Galan JE. Salmonella modulates vesicular traffic by altering phosphoinositide metabolism. Science. 2004;304:1805–1807. doi: 10.1126/science.1098188. [DOI] [PubMed] [Google Scholar]
- Hoiseth SK, Stocker BA. Aromatic dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature. 1981;291:238–239. doi: 10.1038/291238a0. [DOI] [PubMed] [Google Scholar]
- Ibarra JA, Steele-Mortimer O. Salmonella--the ultimate insider. Salmonella virulence factors that modulate intracellular survival. Cell Microbiol. 2009;11:1579–1586. doi: 10.1111/j.1462-5822.2009.01368.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones BD, Falkow S. Identification and characterization of a Salmonella typhimurium oxygen-regulated gene required for bacterial internalization. Infect Immun. 1994;62:3745–3752. doi: 10.1128/iai.62.9.3745-3752.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kage H, Takaya A, Ohya M, Yamamoto T. Coordinated regulation of expression of Salmonella pathogenicity island 1 and flagellar type III secretion systems by ATP-dependent ClpXP protease. J Bacteriol. 2008;190:2470–2478. doi: 10.1128/JB.01385-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, other authors KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoramian-Falsafi T, Harayama S, Kutsukake K, Pechere JC. Effect of motility and chemotaxis on the invasion of Salmonella typhimurium into HeLa cells. Microb Pathog. 1990;9:47–53. doi: 10.1016/0882-4010(90)90039-s. [DOI] [PubMed] [Google Scholar]
- Kim W, Surette MG. Swarming populations of Salmonella represent a unique physiological state coupled to multiple mechanisms of antibiotic resistance. Biol Proced Online. 2003;5:189–196. doi: 10.1251/bpo61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knodler LA, Bestor A, Ma C, Hansen-Wester I, Hensel M, Vallance BA, Steele-Mortimer O. Cloning vectors and fluorescent proteins can significantly inhibit Salmonella enterica virulence in both epithelial cells and macrophages: implications for bacterial pathogenesis studies. Infect Immun. 2005;73:7027–7031. doi: 10.1128/IAI.73.10.7027-7031.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knodler LA, Winfree S, Drecktrah D, Ireland R, Steele-Mortimer O. Ubiquitination of the bacterial inositol phosphatase, SopB, regulates its biological activity at the plasma membrane. Cell Microbiol. 2009;11:1652–1670. doi: 10.1111/j.1462-5822.2009.01356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CA, Falkow S. The ability of Salmonella to enter mammalian cells is affected by bacterial growth state. Proc Natl Acad Sci U S A. 1990;87:4304–4308. doi: 10.1073/pnas.87.11.4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lober S, Jackel D, Kaiser N, Hensel M. Regulation of Salmonella pathogenicity island 2 genes by independent environmental signals. Int J Med Microbiol. 2006;296:435–447. doi: 10.1016/j.ijmm.2006.05.001. [DOI] [PubMed] [Google Scholar]
- Mangan MW, Lucchini S, Danino V, Croinin TO, Hinton JC, Dorman CJ. The integration host factor (IHF) integrates stationary-phase and virulence gene expression in Salmonella enterica serovar Typhimurium. Mol Microbiol. 2006;59:1831–1847. doi: 10.1111/j.1365-2958.2006.05062.x. [DOI] [PubMed] [Google Scholar]
- Marcus SL, Brumell JH, Pfeifer CG, Finlay BB. Salmonella pathogenicity islands: big virulence in small packages. Microbes Infect. 2000;2:145–156. doi: 10.1016/s1286-4579(00)00273-2. [DOI] [PubMed] [Google Scholar]
- Mayer MP. A new set of useful cloning and expression vectors derived from pBlueScript. Gene. 1995;163:41–46. doi: 10.1016/0378-1119(95)00389-n. [DOI] [PubMed] [Google Scholar]
- McGhie EJ, Brawn LC, Hume PJ, Humphreys D, Koronakis V. Salmonella takes control: effector-driven manipulation of the host. Curr Opin Microbiol. 2009;12:117–124. doi: 10.1016/j.mib.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuiston JR, Fields PI, Tauxe RV, Logsdon JM., Jr. Do Salmonella carry spare tyres? Trends Microbiol. 2008;16:142–148. doi: 10.1016/j.tim.2008.01.009. [DOI] [PubMed] [Google Scholar]
- Miao EA, Alpuche-Aranda CM, Dors M, Clark AE, Bader MW, Miller SI, Aderem A. Cytoplasmic flagellin activates caspase-1 and secretion of interleukin 1beta via Ipaf. Nat Immunol. 2006;7:569–575. doi: 10.1038/ni1344. [DOI] [PubMed] [Google Scholar]
- Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, other authors PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- Passerat J, Got P, Dukan S, Monfort P. Respective roles of culturable and viable-but-nonculturable cells in the heterogeneity of Salmonella enterica serovar typhimurium invasiveness. Appl Environ Microbiol. 2009;75:5179–5185. doi: 10.1128/AEM.00334-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel JC, Galan JE. Manipulation of the host actin cytoskeleton by Salmonella--all in the name of entry. Curr Opin Microbiol. 2005;8:10–15. doi: 10.1016/j.mib.2004.09.001. [DOI] [PubMed] [Google Scholar]
- Patel JC, Hueffer K, Lam TT, Galan JE. Diversification of a Salmonella virulence protein function by ubiquitin-dependent differential localization. Cell. 2009;137:283–294. doi: 10.1016/j.cell.2009.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini S, Brown JD, Aldridge PD, Rao CV. FliZ Is a posttranslational activator of FlhD4C2-dependent flagellar gene expression. J Bacteriol. 2008;190:4979–4988. doi: 10.1128/JB.01996-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiemann DA, Shope SR. Anaerobic growth of Salmonella typhimurium results in increased uptake by Henle 407 epithelial and mouse peritoneal cells in vitro and repression of a major outer membrane protein. Infect Immun. 1991;59:437–440. doi: 10.1128/iai.59.1.437-440.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song M, Kim HJ, Kim EY, Shin M, Lee HC, Hong Y, Rhee JH, Yoon H, Ryu S, other authors ppGpp-dependent stationary phase induction of genes on Salmonella pathogenicity island 1. J Biol Chem. 2004;279:34183–34190. doi: 10.1074/jbc.M313491200. [DOI] [PubMed] [Google Scholar]
- Steele-Mortimer O, Meresse S, Gorvel JP, Toh BH, Finlay BB. Biogenesis of Salmonella typhimurium-containing vacuoles in epithelial cells involves interactions with the early endocytic pathway. Cell Microbiol. 1999;1:33–49. doi: 10.1046/j.1462-5822.1999.00003.x. [DOI] [PubMed] [Google Scholar]
- Steele-Mortimer O, Knodler LA, Marcus SL, Scheid MP, Goh B, Pfeifer CG, Duronio V, Finlay BB. Activation of Akt/protein kinase B in epithelial cells by the Salmonella typhimurium effector sigD. J Biol Chem. 2000;275:37718–37724. doi: 10.1074/jbc.M008187200. [DOI] [PubMed] [Google Scholar]
- Steele-Mortimer O, Brumell JH, Knodler LA, Meresse S, Lopez A, Finlay BB. The invasion-associated type III secretion system of Salmonella enterica serovar Typhimurium is necessary for intracellular proliferation and vacuole biogenesis in epithelial cells. Cell Microbiol. 2002;4:43–54. doi: 10.1046/j.1462-5822.2002.00170.x. [DOI] [PubMed] [Google Scholar]
- Steele-Mortimer O. Infection of Epithelial Cells With Salmonella enterica. Methods Mol Biol. 2007;431:201–212. doi: 10.1007/978-1-60327-032-8_16. [DOI] [PubMed] [Google Scholar]
- Steele-Mortimer O. The Salmonella-containing vacuole-Moving with the times. Curr Opin Microbiol. 2008;11:38–45. doi: 10.1016/j.mib.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, other authors Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Kuehn H, Gould J, Tamayo P, Mesirov JP. GSEA-P: a desktop application for Gene Set Enrichment Analysis. Bioinformatics. 2007;23:3251–3253. doi: 10.1093/bioinformatics/btm369. [DOI] [PubMed] [Google Scholar]
- Temme K, Salis H, Tullman-Ercek D, Levskaya A, Hong SH, Voigt CA. Induction and relaxation dynamics of the regulatory network controlling the type III secretion system encoded within Salmonella pathogenicity island 1. J Mol Biol. 2008;377:47–61. doi: 10.1016/j.jmb.2007.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teplitski M, Goodier RI, Ahmer BM. Pathways leading from BarA/SirA to motility and virulence gene expression in Salmonella. J Bacteriol. 2003;185:7257–7265. doi: 10.1128/JB.185.24.7257-7265.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thijs IM, De Keersmaecker SC, Fadda A, Engelen K, Zhao H, McClelland M, Marchal K, Vanderleyden J. Delineation of the Salmonella enterica serovar Typhimurium HilA regulon through genome-wide location and transcript analysis. J Bacteriol. 2007;189:4587–4596. doi: 10.1128/JB.00178-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tintle NL, Best AA, DeJongh M, Van Bruggen D, Heffron F, Porwollik S, Taylor RC. Gene set analyses for interpreting microarray experiments on prokaryotic organisms. BMC Bioinformatics. 2008;9:469. doi: 10.1186/1471-2105-9-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdivia RH, Falkow S. Bacterial genetics by flow cytometry: rapid isolation of Salmonella typhimurium acid-inducible promoters by differential fluorescence induction. Mol Microbiol. 1996;22:367–378. doi: 10.1046/j.1365-2958.1996.00120.x. [DOI] [PubMed] [Google Scholar]
- van Asten FJ, Hendriks HG, Koninkx JF, van Dijk JE. Flagella-mediated bacterial motility accelerates but is not required for Salmonella serotype Enteritidis invasion of differentiated Caco-2 cells. Int J Med Microbiol. 2004;294:395–399. doi: 10.1016/j.ijmm.2004.07.012. [DOI] [PubMed] [Google Scholar]
- van der Velden AW, Lindgren SW, Worley MJ, Heffron F. Salmonella pathogenicity island 1-independent induction of apoptosis in infected macrophages by Salmonella enterica serotype typhimurium. Infect Immun. 2000;68:5702–5709. doi: 10.1128/iai.68.10.5702-5709.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virtaneva K, Porcella SF, Graham MR, Ireland RM, Johnson CA, Ricklefs SM, Babar I, Parkins LD, Romero RA, other authors Longitudinal analysis of the group A Streptococcus transcriptome in experimental pharyngitis in cynomolgus macaques. Proc Natl Acad Sci U S A. 2005;102:9014–9019. doi: 10.1073/pnas.0503671102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weening EH, Barker JD, Laarakker MC, Humphries AD, Tsolis RM, Baumler AJ. The Salmonella enterica serotype Typhimurium lpf, bcf, stb, stc, std, and sth fimbrial operons are required for intestinal persistence in mice. Infect Immun. 2005;73:3358–3366. doi: 10.1128/IAI.73.6.3358-3366.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material
Figure S1. Distribution of S. Typhimurium in infected HeLa cells. To obtain comparable invasion cells were infected with an m.o.i. of ~50-60 for aer-LL and ~150-180 for μaer-ST. Extracellular S. Typhimurium were stained with anti-LPS mAb before permeabilisation and excluded from analysis. After permeabilisation, all bacteria were stained with rabbit anti-LPS antibodies followed by AlexaFluor 568-conjugated secondary antibodies. The number of intracellular S. Typhimurium per infected cell was scored visually. Shown are means ± SD from three independent experiments (n =100).
Figure S2. Plasmids are retained in S. Typhimurium. Plasmids stabilities were determined by culturing bacteria in either μaer-ST (a) or aer-LL (b) conditions without antibiotic selection, thereafter bacteria were plated on LB-Miller agar with or without carbenicillin. Shown are means ± SD from three independent experiments.
Figure S3. Motility of S. Typhimurium strains. Flagellar function was determined by rate of spread, after 6 h incubation at 37 °C in semisolid agar. Strains were WT S. Typhimurium (WT), fliC::Tn10 (fliC), fljB::Mud-Cm mutant (fljB), fliC::Tn10 fljB::Mud-Cm mutant (fliC/fljB), WT S. Typhimurium bearing pFPV25.1 (pFPV25.1) or PprgH-gfp[LVA] (PprgH-gfp[LVA]). Shown are means ± SD, from three independent experiments.
Figure S4. Electron micrographs showing flagella (black arrows) on the surface of aer-LL and μaer-ST bacteria, but fimbriae (white arrows) only on the surface of μaer-ST bacteria. Scale bars are 500 nm (left panels) or 100 nm (right panels).
Table S1. Genome wide-expression changes for aer-LL compared to μaer-ST S. Typhimurium SL1344. Probe-set identification (ID) for S. Typhimurium SL1344 is shown and the corresponding S. Typhimurium LT2 gene or synonym is denoted where identified by BLAST analysis. Check marks show genes where both a 2-fold (aer-LL/μaer-ST fold Δ) change and P-value passing the false discovery rate at a significant level was obtained (2X and P-value Sig.).