Abstract
Human cytomegalovirus (HCMV) is a ubiquitous pathogen that indelibly shapes the natural killer (NK) cell repertoire. Using transcriptomic, epigenomic, and proteomic approaches to evaluate peripheral blood NK cells from healthy human volunteers, we find that prior HCMV infection promotes NK cells with a T cell-like gene profile, including the canonical markers CD3ε, CD5, and CD8β, as well as the T cell lineage-commitment transcription factor Bcl11b. While Bcl11b expression is upregulated during NK maturation from CD56bright to CD56dim, we find a Bcl11b-mediated signature at the protein level for FcεRIγ, PLZF, IL-2Rβ, CD3γ, CD3δ, and CD3ε in later-stage HCMV-induced NK cells. BCL11B is targeted by Notch signaling in T cell development, and culture of NK cells with Notch ligand increases cytoplasmic CD3ε expression. The Bcl11b-mediated gain of CD3ε, physically associated with CD16 signaling molecules Lck and CD247, in NK cells is correlated with increased antibody-dependent effector function, including against HCMV-infected cells, identifying a potential mechanism for their prevalence in HCMV-infected individuals and their prospective clinical use in antibody-based therapies.
Introduction
Human natural killer (NK) cells are CD3−CD56+ lymphocytes that provide a rapid cytokine and cytotoxic response to virus-infected cells and tumor cells. Dissimilarities between NK cells and T cells among stimulatory ligands and functional effector properties suggest that they arise from distinct lymphocyte lineages (1–3). Association of the gene-rearranged, antigen-specific T cell receptor (TCR) with the CD3 complex is critical for the development, functionality, and specificity of each T cell. In contrast to T cells, NK cells express an array of germline-encoded cell surface stimulatory and inhibitory receptors (4). Human NK cells universally express CD247 as a signaling adaptor for CD16 (5), but do not routinely express any of the other components of the CD3 complex. Fetal NK cells are a notable exception, as they have been observed to express CD3 proteins in their cytoplasm (6, 7). The same studies, however, failed to detect CD3+ NK cells in adult NK cells, suggesting that the subset is gradually lost with aging.
Human cytomegalovirus (HCMV) infects and establishes a persistent infection in the majority of humans worldwide (8). HCMV is the only virus known to shape the NK cell repertoire in humans, giving rise in some individuals to an “adaptive NK” population, characterized by the surface expression of the activating heterodimer CD94/NKG2C, CD57, and self-HLA specific inhibitory KIR receptors and by the poor expression of the signaling molecules FcεRIγ, SYK, and EAT-2 and the transcription factor PLZF (9–12). The population exhibits higher capacity for IFN-γ and CD107a response for certain effector functions, notably humoral anti-viral immunity (13–15).
The phenotypic definition of the adaptive NK cell population is unclear, however, with incomplete overlap of surface protein and transcription factor expression levels, and the apparent non-essential role of NKG2C (16, 17). Broadly defining the NK cell populations that can be observed at high frequencies in HCMV-seropositive donors as “HCMV-induced NK cells”, we demonstrate that a high percentage of HCMV-induced NK cell populations in healthy adults and umbilical cord blood transplantation recipients express CD3ε intracellularly, and that NK cells expressing CD3ε have features that largely overlap with adaptive NK cells, such as expression of NKG2C and lack of expression of FcεRIγ (18). Furthermore, among the NKG2C+FcεRIγ− adaptive NK cell population, cytoplasmic CD3ε+ NK cells exhibit higher antibody-dependent cytokine response.
Transcriptional and epigenetic profiling of NK cells at two opposing developmental stages, comprised of a heterogeneous and less mature NKG2A+ NK cell population and the terminally differentiated NKG2C+ NK cell population, not only confirms transcriptional upregulation of CD3E in the HCMV-associated population but also identifies upregulation of the transcription factor BCL11B, a critical regulator of T cell programming (19). We show how a common viral infection indelibly shapes the phenotypic and functional NK repertoire, via expansion of NK cells exhibiting a Bcl11b-guided T cell program, resulting in a cytoplasmic CD3ε+ population with enhanced antibody-dependent function.
Materials and Methods
Study subjects and clinical samples
Buffy coats were collected from 150 volunteer blood donors at the New York Blood Center. Because the samples were obtained anonymously, the Memorial Sloan Kettering Cancer Center (MSKCC) Institutional Review Board waived the need for additional research consent. Subjects receiving live fibroblast-adapted chimeric Towne and Toledo strain HCMV vaccines provided informed consent, as described previously (21, 22).
Cells
The human erythroleukemia cell line K562, human neuroblastoma cell line BE(2)N, T cell lymphoblast cell line MOLT-4, peripheral blood mononuclear cells (PBMC), and purified NK cells were cultured in RPMI-1640 medium containing 10% FCS (MSKCC). Human foreskin fibroblasts (HFFs) were cultured in MEM medium alpha (GIBCO/Invitrogen) containing 10% FCS. NK cells were enriched by negative selection from PBMCs (Stemcell) and labeled with CellTrace™ Violet (CTV, Invitrogen). Human natural killer cell lines NKL and NK-92 cells were cultured in RPMI-1640 medium containing 10% FCS and 200IU/ml IL-2 (Roche). CD16+NK-92MI cells were cultured in RPMI-1640 medium containing 10% FCS. The mouse bone marrow stroma cell lines OP9 and OP9-DL1 were cultured in MEM medium alpha containing 20% FCS. The 1×105 OP9 and OP9-DL1 cells were seeded one day before coculture with purified NK cells at an E:T ratio of 1:1 in the presence of 200IU/ml IL-2 in 24-well plates. Transwell-based cultures were used to study NK cell expansion with OP9 and OP9-DL1 cells. Purified NK cells were cultured with OP9 or OP9-DL1 cells in lower well in the presence of an upper transwell reactor (Greiner) containing 1×106 PBMC from the same donor to support NK cell survival and proliferation.
Mice
Tissue from human CD3ε transgenic mice was kindly provided by Dr. Nai-Kong Cheung (MSKCC). Human CD3ε F1 heterozygotes were generated by breeding B6.Cg-Tg(CD3E)600Cpt/J and wildtype C57BL/6 mice as previously described (30). All mice used in this study were housed and bred under specific pathogen-free conditions at MSKCC and handled in accordance with the guidelines of the Institutional Animal Care and Use Committee (IACUC).
Preparation of viral stocks and infection of fibroblasts
An HCMV variant with a repaired UL40-ORF was generated by markerless mutagenesis on the basis of TB40-BACKL7-SE-EGFP. For preparation of virus stocks, supernatants were collected at 5–7 days post-infection and after cellular debris was removed by centrifugation at 2,800 × g for 10 min. Fibroblasts were infected using a multiplicity of infection (MOI) of 5 PFU/fibroblast for 1 hour. Expression of HLA-A, -B, and -C on infected fibroblasts was evaluated at 48 hours post-infection. HCMV-infected cells were detected by HCMV-IEA staining, and the infection rates of cells used in this study were >98% (Figure S3C). To upregulate HLA class I expression, recombinant human IFN-γ (PeproTech) was added at 1,000 units/ml for 24 hours.
Functional studies
Cryopreserved PBMCs were cultured overnight in RPMI 1640 media containing 10% FCS and 200 IU/ml IL-2 (Roche) and 1ng/ml IL-15 (Miltenyi) prior to functional analysis. NK cell degranulation and IFN-γ production assay was performed as previously described (13). To evaluate NK cell activity, PBMC were cultured for 5 hours with target cells in 96-well plates at a final E:T ratio of 5:1. CD107a was added at the beginning of co-culture, and GolgiStop (BD) was added to the co-culture one hour later. To assess antibody-dependent cellular cytotoxicity (ADCC), humanized anti-GD2 monoclonal antibody 3F8 (provided by Dr. Nai-Kong Cheung, MSKCC) was used at a concentration of 1μg/ml with a 1:20 dilution of sera collected from HCMV seropositive and seronegative healthy individuals. After surface marker staining, cells were fixed and permeabilized with a FIX&PERM kit (GAS004, Thermo Fisher) prior to intracellular IFN-γ staining. Dead cells were excluded from analysis using a fixable Aqua dead cell staining kit (ThermoFisher). Data were collected with a LSRFortessa flow cytometer (BD Bioscience) and analyzed using FlowJo (TreeStar).
Immunoprecipitation and western blot
Protein complexes were prepared using the Dynabeads™ co-immunoprecipitation kit (ThermoFisher). Dynabeads (1mg) were conjugated with 5μg of antibody one day prior to experiments. Cells were washed twice with cold PBS and lysed with extraction buffer for 15mins on ice with protease inhibitors. Cell lysates were incubated with conjugated beads for 30 mins at 4 degree. Immunoprecipitated proteins were eluted using elution buffer, denatured by the addition of Laemmli sample buffer and heating, resolved by SDS-PAGE, and analyzed by immunoblotting. For input samples, cells were lysed in RIPA buffer for 15mins on ice with protease inhibitors and denatured by the addition of Laemmli sample buffer and heating prior to processing for western blot detection.
Antibodies
The following antibodies were used for flow cytometry: BD Bioscience: CD3 epsilon (UCHT1, SP34–2 and SK7), CD122 (Mik-β3), CD8a (RPA-T8), PLZF (RI7–809). CD107a (H4A3), IFN-γ (B27), KIR3DL1 (DX9), KIR2DL2/L3/S2 (CH-L), CD16 (3G8), CD57 (NK-1), HLA-DR (G46–6), HLA-C (DT9), and CD86 (FUN-1). Biolegend: CD2 (RPA-2.10), PD-1 (EH12.2H7), Bcl-2 (100), CD3 epsilon (SK7, OKT3, HIT3a and APA1/1), HLA-ABC (W6/32), NKp46 (9E2), IgG1 (MOPC-21), IgG2a (MOPC-173), CD5 (UCHT2), Lck (LCK-01), and Bcl11b (25E6). Beckman Coulter: CD56 (NKH-1), KIR2DL2/L3/S2 (GL183), KIR2DL1/S1 (EB6), NKp46 (BAB281), CD247 (TIA-2), CD69 (TP1.55.3). Miltenyi: NKG2A (REA110), NKG2C (REA205), PD-1(PD1.3.1.3), LAG-3 (REA351), CD58(TS2/9), Ki-67 (REA183), Bw4 (REA274), and TCRrd (11F2). R&D Systems: NKG2C (134591), KIR2DL1 (143211), and KIR2DL3 (180701). Millipore: FcεRI (FCABS400F), HCMV-IEA (8B1.2). ThemoFisher: LIR-1 (HP-F1), TCRab (WT13), and CD8b (SIDI8BEE). Abcam: CD3 gamma (EPR4517), CD3 delta (EP4426) and Bcl11b (25E6). Jackson Immunoresearch: Donkey anti-rabbit IgG (H+L). The following antibodies were used for western blot: Biolegend: CD3 epsilon (SK7), IgG1 (MOPC-21), β-actin (W16197A), CD247 (6B10.2), pLck (A18002D), Lck (LCK-01), CD2 (RPA-2.10), and CD16 (3G8); pCD247 (C415.9A, Santa Cruz Biotechnology), CD3 gamma (EPR4517, Abcam), and CD3 delta (EP4426, Abcam).
Transcriptome sequencing
RNA from cells suspended in Trizol was extracted using Arcturus Picopure RNA Isolation Kit according to manufacturer protocol (Applied Biosystems). RNA was precipitated with isopropanol and washed with 75% ethanol. Samples were resuspended in RNase-free water. After RiboGreen quantification and quality control by Agilent BioAnalyzer, 1.2–2ng total RNA with RNA integrity numbers ranging from 7.7 to 10 underwent amplification using the SMART-Seq v4 Ultra Low Input RNA Kit (Clonetech), with 12 cycles of amplification. Subsequently, 10ng of amplified cDNA was used to prepare libraries with the KAPA Hyper Prep Kit (Kapa Biosystems) using 8 cycles of PCR. Samples were barcoded and run on a HiSeq 4000 or HiSeq 2500 in High Output mode in a 50bp/50bp paired end run, using the HiSeq 3000/4000 SBS Kit or TruSeq SBS Kit v4 (Illumina). An average of 38 million paired reads were generated per sample and the percent of mRNA bases per sample ranged from 45% to 78%.
ATAC Sequencing
Profiling of chromatin was performed by ATAC-Seq. Briefly, frozen primary NK cells were thawed on ice, washed in cold PBS, and lysed. The transposition reaction was incubated at 42°C for 45 minutes. The DNA was cleaned with the MinElute PCR Purification Kit (QIAGEN) and material was amplified for 5 cycles. After evaluation by real-time PCR, 4–7 additional PCR cycles were done. The final product was cleaned by aMPure XP beads (Beckman Coulter) at a 1X ratio. Libraries were sequenced on a HiSeq 2500 in High Output mode and a HiSeq4000 in a 50bp/50bp paired end run, using the TruSeq SBS Kit v4 or HiSeq 3000/4000 SBS Kit (Illumina). An average of 57 million paired reads were generated per sample.
RNA-seq analysis
Paired-end reads were trimmed for adaptors and removed of low quality reads using Trimmomatic (v0.36). Transcript quantification was based on the hg38 UCSC Known Gene models and performed using the quasi-mapping-based mode of Salmon (v0.8.2), correcting for potential GC bias. Counts were summarized to the gene level using tximport (v1.8.0). For those samples that were sequenced across two runs, summarized reads determined by tximport were summed, and the means of average transcript length offsets calculated for each run was used for downstream differential analyses executed by DESeq2 (v1.22.2). Genes were considered differentially expressed (DE) if they showed a false discovery rate (FDR)-adjusted P-value < 0.05. Gene set analysis was performed with GOseq (v.1.26.0), using either DE genes higher in NKG2C+ NK cells or DE genes higher in NKG2A+ NK cells, and those showing an absolute log2 fold change > 1. Gene sets were retrieved from the MSigDB database (v.3.0). FDR-corrected P-values were calculated from P-values calculated by GOseq, and only gene ontologies passing a threshold of p < 0.05 were considered. Up to the top 10 gene sets ranked on p-value are shown. Heatmaps were generated using ComplexHeatmap (v1.99.7), using log2-transformed normalized counts calculated by the rlog function in DESeq2.
ATAC-seq analysis
Trimmed reads were mapped to the Homo sapiens genome (hg38 assembly) using Bowtie2 (v2.2.9). For peak calling, all positive-strand reads were shifted 4 bp downstream and all negative-strand reads were shifted 5 bp upstream to center the reads on the transposon binding event. Shifted, concordantly aligned paired mates were used for peak calling by MACS2 (v2.1.1.20160309) at a P-value of 0.01. Irreproducible discovery rate (IDR) calculations using scripts provided by the ENCODE project (https://www.encodeproject.org/software/idr/;v2.0.3) were performed on all pairs of replicates using an oracle peak list called from pooled replicates for each condition, keeping only reproducible peaks showing an IDR value of 0.05 or less. IDR-thresholded peak regions from each condition were then merged to generate the final peak atlas. Read counts for the peak atlas were generated using the summarizeOverlaps function from the Genomic Alignments package (v1.10.1). Differential analyses were executed with DESeq2 (v1.14.1) using the hg38 UCSC Known Gene models as reference annotations. Features were considered differentially accessible (DA) if they showed an FDR-adjusted P-value < 0.05. Peak assignment was done using ChipPeakAnno. Promoter regions were defined as peaks that overlapped a region that was +2 kb to −0.5 kb from the transcriptional start site (TSS). Intragenic (intronic and exonic) peaks were defined as any peak that overlapped with annotated intronic and exonic regions, respectively, based on the annotation database. Intergenic peaks were defined as any non-promoter or non-intragenic peaks and were assigned to the gene of the nearest TSS based on the distance from the start of the peak. Priority was given to transcripts that were canonical, based on the UCSC Known Canonical database. The final analysis resulted in 5,549 DA regions out of a total of 46,059, after removing regions identified in alternate loci scaffolds and the mitochondrial chromosome. For gene ontology analysis, BED files of DA regions were used as input for analysis by GREAT using the online web interface at http://bejerano.stanford.edu/great/public/html/index.php (v3.0.0). All analyses used default settings with the whole genome as background considering analyses from the MSigDB Canonical Pathway databases. Enriched pathways were filtered for those that showed an adjusted P-value < 0.05 for both binomial and hypergeometric P-values as calculated by GREAT, and a region fold enrichment of greater than 2.
Chromatin accessibility heatmaps and gene tracks
For peak-centered heatmaps, average sample counts normalized using size factors calculated by DESeq2 were plotted in 40-bp bins across a 2 kb window using ComplexHeatmap (v1.18.1). To improve visualization, binned counts greater than the 75th percentile + 3 × (interquartile range) were capped at that value. Gene tracks were generated by converting BAM files to bigWig files using bedtools2 (v.2.26.0) and UCSC’s bedGraphToBigWig (v.4) and visualized using the Gviz R package (v.1.18.2). All tracks show normalized tag counts on the y-axis, using size factors calculated by DESeq2.
Statistical analysis
Nonparametric Wilcoxon signed rank sum test was used to compare paired percentages from the same donor. Paired student T test was used to compare paired geometric mean fluorescence intensity (gMFI) from the same donor. The Friedman test and post hoc pairwise Wilcoxon test were performed for multi-group comparison. Results were considered significant at the two-sided P level of 0.05. * p<0.05; ** p<0.01; *** p<0.001.
Results
Transcriptomic and epigenomic profiling identifies an enrichment of T cell related molecules among HCMV-induced NKG2C+ NK cells
HCMV infection expands a CD56dimCD57+NKG2C+ mature NK cell population with preferential co-expression of a self-HLA-C-specific KIR2DL receptor in approximately 30–40% of exposed individuals (13,20). To better understand the transcriptional differences associated with the divergence between NK cell populations, we performed global transcriptional profiling via RNA-seq of the NKG2A+ population and the HCMV-associated NKG2C+ population from five HCMV-seropositive donors with varying frequencies of adaptive NK cells. As shown in Figure S1A, NKG2A and NKG2C define transcriptionally distinct populations, with approximately ~1700 genes differentially expressed across both cell populations.
The top differentially expressed genes are highlighted in Figure S1A. Pathway analysis revealed enrichment among NKG2C+ cells of genes that are involved in PD-1 signaling, translocation of ZAP-70 to immunological synapse, phosphorylation of CD3ζ (CD247) chains, generation of second messenger molecules, co-stimulation by the CD28 family, and TCR signaling (Fig. 1A). We further identified locations of all active or poised regulatory elements by measuring global changes in chromatin accessibility through the Assay for Transposase-Accessible Chromatin using sequencing (ATAC-seq) from a representative donor. We found that NKG2C+ NK cells have distinct chromatin accessibility profiles compared to their NKG2A+ counterparts (Fig. S1B), with notably increased accessibility in genes associated with biological processes involved in TCR receptor signaling, NK cell cytotoxicity, cytokine signaling, and leukocyte activation (Fig. S1C).
FIGURE 1.
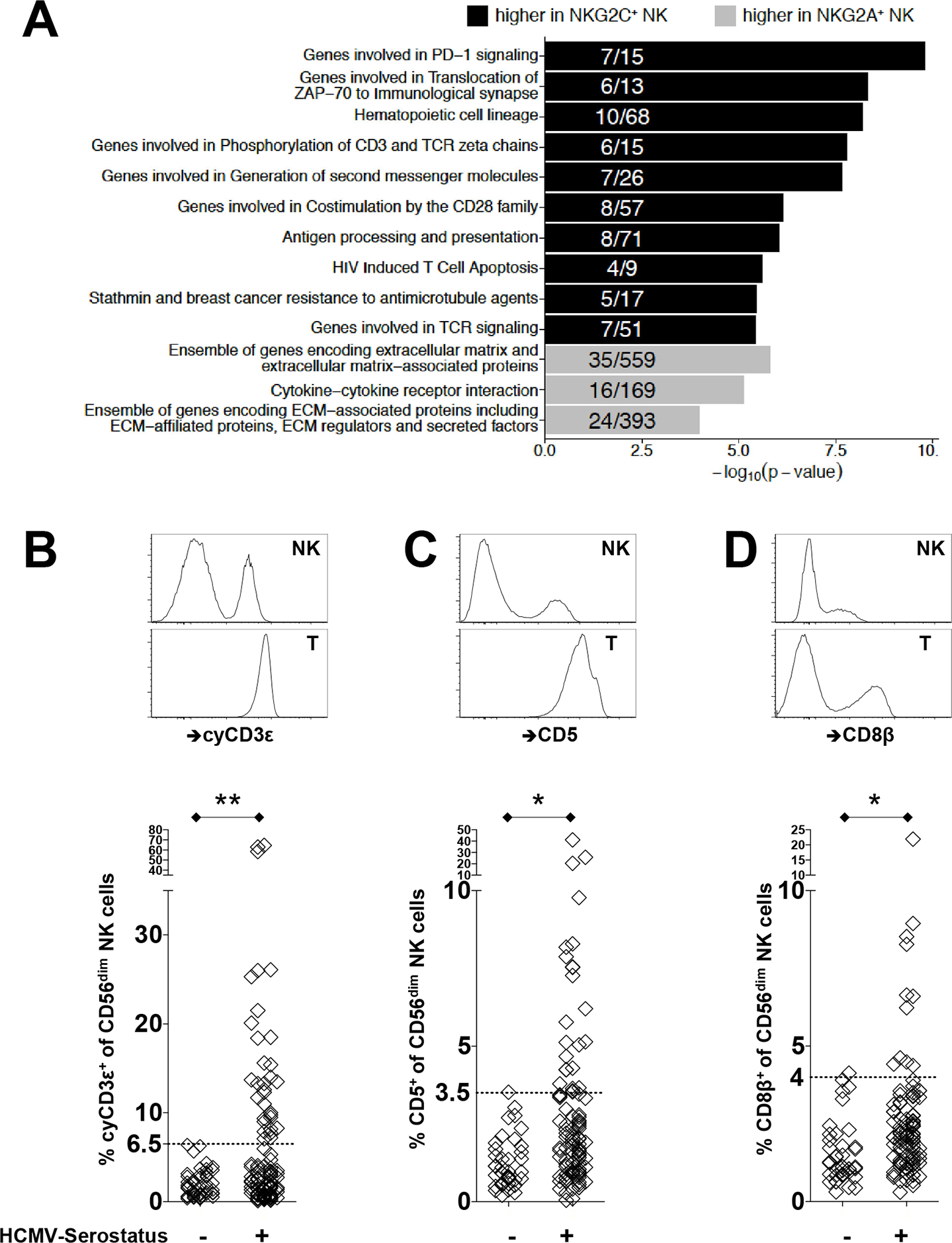
Transcriptomic and epigenomic profiling identifies an enrichment of T cell related molecules among HCMV-induced NKG2C+ NK cells. (A) Bar plots display −log10 (p-value) of gene set enrichment in genes higher in NKG2C+ (black) or higher in NKG2A+ (gray) NK cells. Depicted are the top pathways enriched among genes associated with higher expression within NKG2C+ or NKG2A+ NK cells. Fractions within bar plots indicate the number of differentially expressed (DE) genes found within the total gene set. (B) Representative flow cytometry plots to identify cyCD3ε+ NK cells from healthy donors are shown. CyCD3ε+ NK cells were defined as viable surfaceCD3ε(UCHT1)−CD56dimcytoplasmicCD3ε(SK7)+ lymphocytes. Intracellular staining of cyCD3ε is shown for NK cells and T cells from the same donor. Frequency of cyCD3ε+ NK cells among CD56dim NK cells from healthy HCMV seronegative (n=48) and seropositive (n=105) donors are shown below. Surface expression on NK cells and autologous T cells for (C) CD5 and (D) CD8β are shown with frequencies of CD5+ or CD8β+ NK cells among CD56dim NK cells from healthy HCMV seronegative (n=40) and seropositive (n=96) donors displayed below. The dotted horizontal line indicates the 6.5% threshold defining an expanded cyCD3ε+ cell population.
In order to more precisely identify the candidate molecules that contribute to differences between NKG2C+ and NKG2A+ NK cells, we focused on genes that were commonly regulated at both transcriptional and epigenetic levels. We noted that CD3E, which encodes CD3ε, a canonical subunit of the TCR-CD3 complex in T cells, was prominent among these genes, displaying increased gene expression in parallel with increased accessibility within its promoter region (Fig. S1D).
While antibody staining for CD3ε is specifically used as a negative surface marker for NK cells, routine protein detection by flow cytometry does not detect protein in the intracellular compartment. We therefore confirmed the presence of intracellular CD3ε protein in NK cells from healthy donors by flow cytometry. PBMCs were first stained with CD3ε (clone UCHT1) and CD56 on their surface, followed by intracellular staining of CD3ε (clone SK7). Cytoplasmic CD3ε (cyCD3ε) positive NK cells were defined as surfaceCD3ε(UCHT1)−CD56dimcytoplasmicCD3ε(SK7)+ lymphocytes. Using this strategy, we find in a number of tested individuals that CD3ε can readily be detected intracellularly among CD56dim NK cells but not on the cell surface, although at lower levels than in T cells from the same donors (Fig. 1B).
The increase of CD3E gene expression and locus accessibility in NKG2C+ NK cell suggests that intracellular CD3ε in NK cells may be related to previous HCMV infection. We therefore evaluated the presence of cyCD3ε+ NK cells among 153 healthy donors, segregated by HCMV serostatus. CyCD3ε+ NK cells were detected significantly more among HCMV seropositive donors compared to HCMV negative donors (6.83%±2.17 vs 1.99%±0.44 with 95% confidence limits) (Fig. 1B). Based on the highest percentage of cyCD3ε+ NK cells detected in HCMV-seronegative donors (range 0.12% to 6.42%), we designated individuals with at least 6.5% cyCD3ε+ NK cells within the CD56dim population as positive for (HCMV-induced) cyCD3ε+ NK cells. Using this cutoff, 32 out of 105 (30.5%) HCMV-seropositive donors exhibited cyCD3ε+ NK cells. In three donors, cyCD3ε positivity was strikingly found in more than 50% of CD56dim NK cells. Notably, cyCD3ε+ NK cells were not detected in immature, CD56bright NK cells (0.62%±0.79), irrespective of HCMV serostatus.
Transcriptome analysis further indicated that gene expression for the T cell related CD5 and CD8β molecules are also enriched in NKG2C+ NK cells. As for CD3ε, we detected significantly higher frequencies of CD5+ and CD8β+ NK cells from HCMV-seropositive donors, although these three molecules were not necessarily fully overlapped in a given NK cell. While expressed on the cell surface of NK cells, CD5 and CD8β are expressed to a lower extent than in autologous T cells (Fig. 1C and 1D).
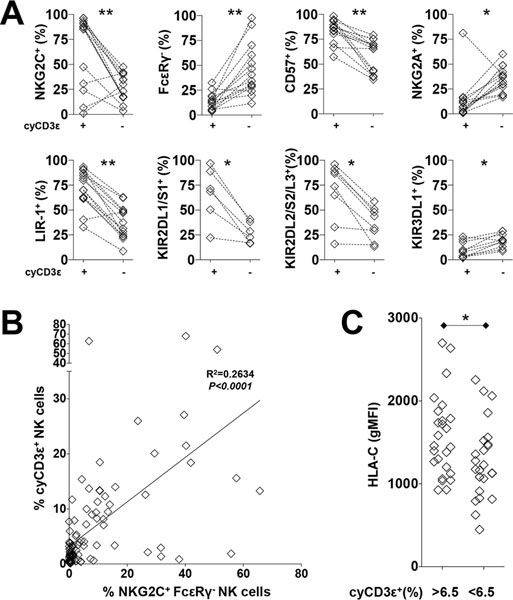
CyCD3ε+ NK cells are enriched for adaptive NK cell markers
In adults, cyCD3ε+ NK cells are significantly more likely than cyCD3ε− NK cells to co-express NKG2C, CD57, CD2, LIR-1, and Bcl-2 and less likely to express NKG2A and FcεRIγ (Fig. 2A and S1F). In addition, they have lower density of PLZF and NKp46 (Fig. S1F). Consistent with this observation, ATACseq analysis demonstrated that BCL2 and CD2 have increased accessibility within HCMV-induced NK cells (Fig. S1B). There is no difference in CD16 expression between cyCD3ε+ and cyCD3ε− cells; and resting cyCD3ε+ NK cells do not express PD-1 (Fig. S1F).
FIGURE 2.
CyCD3ε+ NK cells are enriched for adaptive NK cell markers. (A) Percentages of NKG2C+, FcεRIγ−, CD57+, NKG2A+, LIR-1+, KIR2DL1/S1+, KIR2DL2/S2/L3+, and KIR3DL1+ NK cells among cyCD3ε+ NK cells and cyCD3ε− NK cells are shown. Populations from the same donors are paired. The expression of KIR2DL1/S1 is evaluated on HLA-C2+ donors and KIR2DL2/S2/L3 is evaluated on HLA-C1+ donors. (B) Linear regression analysis between cyCD3ε+ NK cell frequency and NKG2C+FcεRIγ− NK cell frequency among the CD56dim NK cell population. (C) Donors were grouped based on cyCD3ε expression and geometric mean fluorescence intensity (gMFI) of HLA-C on their lymphocytes.
To determine if cyCD3ε+ NK cells fully comprise the adaptive NK cell population, commonly defined by NKG2C positivity and FcεRIγ deficiency (9, 18, 23), we evaluated NKG2C and FcεRIγ expression among cyCD3ε+ NK cells, finding that the overlap of the three molecules is highly variable between donors (Fig. S1G and S1H). While the majority of cyCD3ε+ NK cells (76.7%) express NKG2C, only 59.3% of cyCD3ε+ NK cells are negative for FcεRIγ. Indeed, there was only weak positive correlation between the cyCD3ε+ and the NKG2C+ FcεRIγ− adaptive NK phenotypes among CD56dim NK cells (r2=0.2634) (Fig. 2B). From this, we conclude that the cyCD3ε+ NK cell population does not fully recapitulate the previously described NKG2C+FcεRIγ− adaptive NK cell population, although there are many shared features.
HLA-C expression correlates with HCMV-induced cyCD3ε+ NK cells in healthy adults
HCMV-induced NKG2C+ NK cells preferentially express the self-HLA-C-specific inhibitory KIR2DL receptors, but not the HLA-Bw4 specific KIR3DL1 receptor (10). Similarly, compared to cyCD3ε− NK cells, cyCD3ε+ NK cells are more likely to express KIR2DL and less likely to express KIR3DL1 (Fig. 2A). We evaluated HLA-C and HLA-Bw4 expression on lymphocytes from 93 HCMV-seropositive donors. Compared to donors who do not have detectable HCMV-induced NK cells, donors with high frequencies of cyCD3ε+ NK cells demonstrate higher HLA-C expression levels as measured by gMFI on their lymphocytes (Fig. 2C). There is no difference in HLA-Bw4 expression between donors with or without cyCD3ε+ NK cells (Fig. S1I). The positive correlation of HLA-C and KIR2DL expression suggests that there is an advantage for HLA-C and KIR2DL interaction in generating and/or maintaining HCMV-induced NK cells.
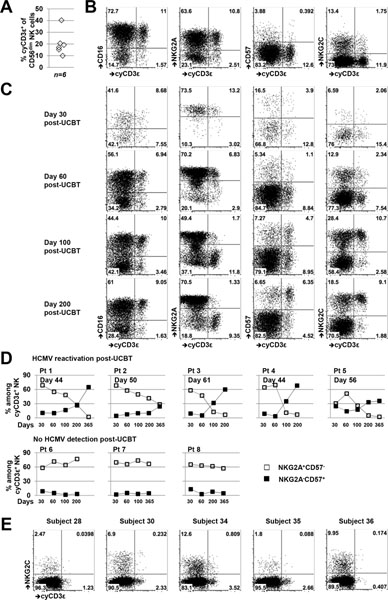
HCMV infection expands a population of mature cyCD3ε+ NK cells in UCB transplantation recipients
Fetal human NK cells frequently express cytoplasmic CD3ε (6, 7), although the protein’s function in this setting is unclear. Because the fetus is HCMV-naïve, umbilical cord blood does not contain the adaptive NK cell population found in HCMV-seropositive healthy adults, permitting characterization of cyCD3ε+ cells outside viral infection. In six umbilical cord blood samples, cyCD3ε+ NK cells were found with a mean frequency of 20.93% within the CD56dim NK cell population (range 9.94% to 40.3%) (Fig. 3A). These same cells co-express NKG2A, but lack CD57 expression (Fig. 3B), consistent with a less mature NK cell phenotype (24) and in contrast to the HCMV-induced cyCD3ε+ NK cells in adult healthy donors, which demonstrate a mature phenotype with NKG2C and CD57 expression (Fig. 2A).
FIGURE 3.
HCMV infection expands a population of mature cyCD3ε+ NK cells in UCB transplantation recipients, but fibroblast-adapted HCMV vaccines in healthy individuals do not. (A) Frequency of cyCD3ε+ NK cells among CD56dim umbilical cord blood NK cells are shown. (B) Representative flow cytometry demonstrating the CD16, NKG2A, CD57, and NKG2C expression among cyCD3ε+ cord blood NK cells. (C) Phenotype of total NK cells from a UCBT patient (patient 4) at indicated time points post transplantation. One representative staining of 8 UCBT patients is shown. (D) Expression of CD57 and NKG2A on cyCD3ε+ NK cells from UCBT patients with or without HCMV reactivation post-transplant. The percentages of NKG2A+CD57− (☐) or NKG2A−CD57+ (■) cells among cyCD3ε+ NK cells were evaluated. The day of HCMV reactivation post-UCBT is indicated. (E) Expression of NKG2C and cyCD3ε among CD56dim NK cells from 5 HCMV-vaccinated healthy donors at one-year post-vaccination.
Patients undergoing umbilical cord blood transplantation (UCBT) provide the unique opportunity to observe in a longitudinal fashion the development of the cyCD3ε+ NK cell population in the setting of HCMV reactivation. As the first lymphocyte population to emerge following UCBT, NK cells are nearly exclusively NKG2A+ (25). CyCD3ε+ NK cells can be detected at 30 days post-transplantation, and their NKG2A+ and CD57− phenotype resembles the cyCD3ε+ NK cells found in umbilical cord blood (Fig. 3C and 3D). In UCBT recipients who experience HCMV reactivation, however, the NKG2A+CD57−cyCD3ε+ population declines, and an NKG2A−CD57+cyCD3ε+ population, comprised mostly of NKG2C+ cells, emerges and expands (Fig. 3C–D and Fig. S2). In patients without HCMV reactivation, the cyCD3ε+ NK cell population remains NKG2A+CD57− (Fig. 3D). These data suggest strongly that HCMV infection is associated with the expansion of a mature cyCD3ε+ NK cell population.
HCMV vaccination capable of inducing a T cell response does not induce expansion of a cyCD3ε+ NK cell population
We examined whether the cyCD3ε+ NK cell population is induced in HCMV-seronegative individuals who seroconvert with an HCMV vaccine (22). In 36 individuals vaccinated with live fibroblast-adapted HCMV vaccines constructed as chimeras of Towne and Toledo strains (21), eight out of 11 subjects who seroconverted after vaccination also mounted weak CD8 T cell responses (22). We evaluated for HCMV-induced NK cells in five of these eight donors by assessing cyCD3ε, NKG2C, and FcεRIγ expression. Despite seroconverting and mounting a CD8 T cell response to the HCMV vaccine, the same individuals did not develop detectable HCMV-induced NK cells, by any phenotypic definition (Fig. 3E and S3A), instead exhibiting NK phenotypes similar to HCMV-seronegative donors (Fig. S3B). These data suggest that the immunogenic proteins and molecular conditions sufficient to elicit a T and B cell response to an HCMV vaccine are insufficient for generating an HCMV-induced cyCD3ε+ NK cell population.
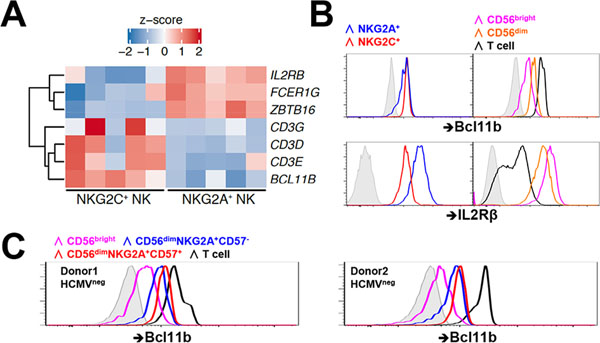
Bcl11b, a regulator of T-cell programming including CD3E, is upregulated at the transition from CD56bright to CD56dim in NK cell development
Transcriptome comparison between the NKG2A+ and the HCMV-associated NKG2C+ NK cell populations reveals increased transcription in the NKG2C+ cells of BCL11B. BCL11B encodes a transcription factor critical for activating a T cell specification gene regulatory network necessary for T lineage commitment, culminating in the silencing of some genes (ZBTB16, NFIL3, POU2AF1, IL2RB, KIT, ITGA2B, FCERIG, and TYROBP) and upregulation of others (GBP4, CD3E, CD3D, and CD3G) (26). Increased activity of BCL11B in HCMV-induced NKG2C+ NK cells is supported by the higher transcription of CD3E, CD3D, and CD3G and lower transcription of IL2RB, FCER1G, and ZBTB16 (Fig. 4A). Moreover, increased chromatin accessibility at the BCL11B locus among NKG2C+ NK cells is evident compared to NKG2A+ NK cells (Fig. S1E), further signifying that BCL11B expression among NK cells is epigenetically regulated. Flow cytometric assessment of BCL11B and IL2Rβ in primary NK cells parallels the transcriptional differences, with higher BCL11B and lower IL2Rβ measured in NKG2C+ cells (Fig. 4B), although neither to the extent found in autologous T cells. Notably, BCL11B appears to have two expression patterns in the NKG2A+ population, low and high, an observation explained by the CD56bright and CD56dim composition of the NKG2A population, where BCL11B expression is escalated in the more mature CD56dim NK compartment (Fig. 4B). Interestingly, the increase in BCL11B that occurs at the developmental transition marked by the phenotypic change from CD56bright to CD56dim can be observed even in HCMV-seronegative individuals. In fact, within the CD56dimNKG2A+ population, further maturation to CD57+ is associated with even higher expression of BCL11B (Fig. 4C). Thus, while it is commonly thought of as a T cell lineage commitment factor, BCL11B may play an important role throughout human NK cell development, independent of HCMV exposure.
FIGURE 4.
Bcl11b upregulation occurs in early NK development. (A) Heatmaps with differential expression of selected genes from NKG2A+ and NKG2C+ NK cells. Shown are z-scores of log2 normalized counts. (B) Bcl11b and IL2Rβ protein expression in NKG2A+ (blue), NKG2C+(red) NK cells. Bcl11b and IL2Rβ protein expression among CD56bright NK cells (magenta), CD56dim NK cells (orange), and T cells (black) is shown with isotype control (gray tint). One representative HCMV seropositive donor out of eight analyzed is shown. (C) Bcl11b protein expression among indicated lymphocyte populations from HCMV seronegative donors were shown.
Notch signaling promotes Bcl11b-regulated protein expression
Notch/Delta-like 1 (DL1) signaling induces cyCD3ε expression during human NK cell differentiation (27), a result consistent with reports that BCL11B is a downstream gene target of Notch signaling (19, 28). We tested whether Notch/DL1 signaling enhances expression of cyCD3ε+ NK cells from HCMV-seropositive individuals. Selecting eight donors who exhibit cyCD3ε+ NK cells, we cultured primary NK cells with parental OP9 cells or OP9 cells expressing the Notch ligand Delta-like 1 (OP9-DL1) for a week. Incubation with OP9-DL1 significantly enhanced CD3ε expression in cyCD3ε+ NK cells (Fig. S3C).
Notch ligands are highly enriched in the thymic microenvironment and, to a lesser extent, in bone marrow (29). To investigate cyCD3ε expression in NK cells from these tissues, we assessed human cyCD3ε expression among murine NK cells in a mouse transgenic for human CD3ε (huCD3ε) (30). We found that while few NK cells in the spleen express cytoplasmic huCD3ε (Fig. S3D), there is a notably higher frequency of cytoplasmic huCD3ε+ NK cells detected from the bone marrow. The highest frequency of cytoplasmic huCD3ε+ NK cells is observed in the thymus of the transgenic mouse. Collectively, these findings suggest that environments rich in Notch ligands enhance CD3ε expression among NK cells.
To determine whether Notch signaling upregulates BCL11B in human NK cells, we measured BCL11B protein levels of primary NK cells cultured with OP9 or OP9-DL1 cells. Co-incubation with OP9-DL1 enhanced expression of both BCL11B (Fig. S3E) and CD3ε among NK cells (Fig. S3C). Together, these data suggest a model in which Notch signaling increases BCL11B expression in NK cells, which then activates a T-specification pathway, leading to the upregulation of CD3ε expression.
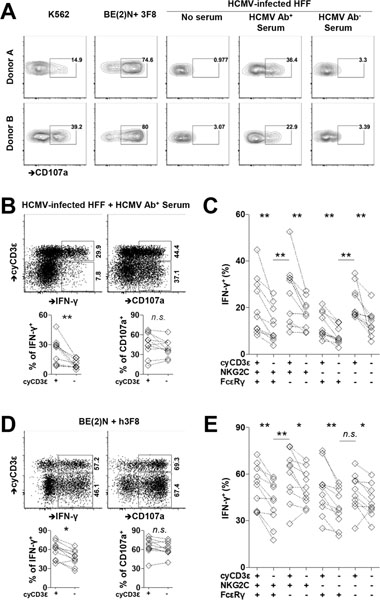
CyCD3ε+ NK cells are capable of enhanced antiviral humoral immunity against HCMV
We then sought to determine the functional consequences of T cell reprogramming of the HCMV-induced NK cell population. Having identified that cyCD3ε+ NK cells largely, but incompletely, overlap the previously described adaptive NK cell population found in HCMV-seropositive individuals, we evaluated the functional response of cyCD3ε+ NK cells from healthy donors to various stimuli. The cyCD3ε+ NK cell population degranulates readily to the MHC class I-negative and NK-sensitive K562 cells (Fig. 5A) and responds robustly in an antibody-dependent cellular cytotoxicity (ADCC) assay using the anti-GD2 monoclonal antibody 3F8 and the GD2+ neuroblastoma cell line BE(2)N (31). The cyCD3ε+ NK cells do not respond to fibroblasts infected in vitro with the HCMV strain TB40/E (Fig. 5A), despite the fact that infected fibroblasts express significantly lower levels of surface MHC class I proteins (Fig. S3F and S3G). The same cells, however, do degranulate robustly against infected cells in the presence of serum from HCMV-seropositive individuals.
FIGURE 5.
Enhanced activity of cyCD3ε+ NK cells in response to HCMV-infected cells in the presence of HCMV-specific antibody. PBMCs were collected from HCMV seropositive donors. (A) PBMCs were cultured with K562 cells, neuroblastoma cell line BE(2)N cells in the presence of 3F8, or HCMV infected fibroblasts with indicated sera. The surface CD107a expression on cyCD3ε+ NK cells from two donors is shown. (B) CD107a and IFN-γ responses in cyCD3ε+ and cyCD3ε− NK cells against HCMV-infected cells in the presence of HCMV-specific antibody. Responses of cyCD3ε+ or cyCD3ε− populations from the same individual are paired (n=10). The indicated percentages of positive cells in FACS plots were determined as percentage of cyCD3ε+ NK cells and cyCD3ε− NK cells. (C) IFN-γ responses of indicated NK cell subsets against HCMV-infected fibroblasts in the presence of HCMV-specific antibody. NK cell subsets were defined with the expression of cyCD3ε, NKG2C, and FcεRIγ among CD56dim NK cells. (D) and (E) CD107a and IFN-γ responses among indicated NK cell populations following co-culture with BE(2)N cells and the monoclonal antibody 3F8 are shown. The indicated percentages of positive cells in FACS plots were determined as percentage of cyCD3ε+ NK cells and cyCD3ε− NK cells.
We evaluated the activity of cyCD3ε+ NK cells from ten HCMV-seropositive individuals against K562 cells and HCMV-infected fibroblasts in the presence of serum from an HCMV-seropositive donor, comparing their activity to cyCD3ε− NK cells from the same individuals. CyCD3ε+ NK cells degranulate and produce IFN-γ in response to coincubation with K562 cells with similar frequency to cyCD3ε− NK cells (Fig. S3H). Similarly, cyCD3ε+ NK cells degranulate to HCMV-infected fibroblasts in the presence of HCMV antibodies with comparable frequency to cyCD3ε− NK cells (Fig. 5B). We observed, however, that cyCD3ε+ NK cells are significantly more likely to produce IFN-γ against infected fibroblasts with HCMV-specific antibodies compared to cyCD3ε− NK cells in all donors tested (23.8±12.1 vs 9.9±4.2, Fig. 5B).
Because cyCD3ε expression in adult NK cells frequently overlaps with NKG2C positivity and FcεRIγ deficiency (Fig. S1G), we evaluated cytokine response capacity of cell subsets expressing different combinations of cyCD3ε, NKG2C and FcεRIγ to ADCC stimulation to determine if heightened response could be narrowed to one specific phenotypic population. We found that cyCD3ε positivity is a marker of enhanced response among all subsets, including NKG2C+FcεRIγ+, NKG2C+FcεRIγ−, NKG2C−FcεRIγ+, and NKG2C−FcεRIγ− NK cells (Fig. 5C). Notably, the greatest IFN-γ response to ADCC occurs in the cyCD3ε+FcεRIγ− cell population, regardless of NKG2C expression. The escalated response suggests that losing FcεRIγ and acquiring CD3ε intrinsically promotes the response of HCMV-induced NK cells against infected cells in the presence of HCMV-specific antibodies (Fig. 5C).
To test whether the enhanced ADCC activity of cyCD3ε+ NK cells can be generalized to non-HCMV ADCC settings, we evaluated the activity of cyCD3ε+ NK cells against 3F8 antibody-coated BE(2)N neuroblastoma cells. We found similar patterns of responsiveness of the cyCD3ε+ and cyCD3ε− NK cell subsets as we did for HCMV-infected target cells (58.9±13.0 vs 46.7±11.6, Fig. 5D). When examining the cytokine response capacity of cell subsets expressing different combinations of cyCD3ε, NKG2C and FcεRIγ positivity to ADCC stimulation, we found that cyCD3ε+ NK cells are more responsive than cyCD3ε− NK cells, again with the cyCD3ε+NKG2C+FcεRIγ− cell population demonstrating the greatest IFN-γ response to ADCC stimulation (Fig. 5E). Our results suggest that cyCD3ε positivity in HCMV-induced NK cells enhances the cytokine response of NK cells in the setting of ADCC.
CD3ε complexes with CD16 signaling molecules in HCMV-induced NK cells with an enhanced expression of Lck and CD247
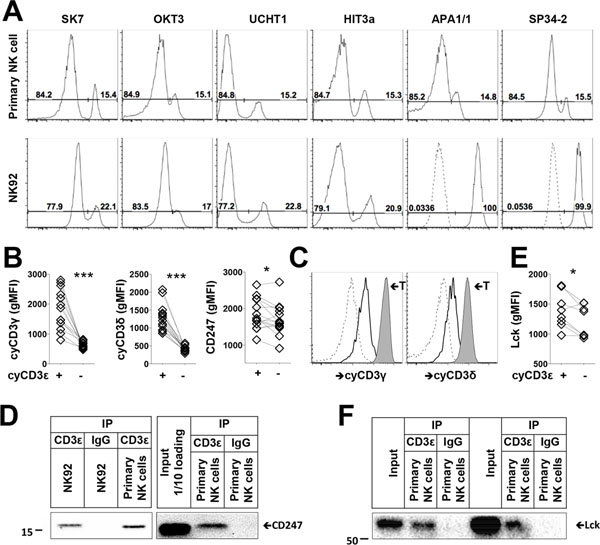
Seven different anti-CD3ε antibody clones stained the same percentage of cyCD3ε+ NK cells from healthy donors. Interestingly, the NK cell line NK92 displays a distinct cyCD3ε staining pattern with the antibody clones, whereby all of the cells are positive by staining with APA1/1 and SP34–2, which recognize the single CD3ε chain, but only 20% of the cells are positive by staining with SK7, OKT3, UCHT1, and HIT3a, which exclusively recognize complexed CD3ε (32) (Fig. 6A). The staining therefore suggests that CD3ε is complexed in all primary cyCD3ε+ NK cells but in only 20% of NK92 cells. Transcriptome analysis suggests that CD3G and CD3D expression are enriched in HCMV-induced NK cells (Fig. 4A). Accordingly, we confirmed by flow cytometry that cyCD3ε+ NK cells co-express cytoplasmic CD3γ and CD3δ (Fig. 6B), although all three CD3 chains are expressed at lower levels than in autologous T cells (Fig. 1B and 6C). Furthermore, we find that cyCD3ε+ NK cells express more CD247 than cyCD3ε− NK cells (Fig. 6B).
FIGURE 6.
CD3 subunits form complexes in HCMV-induced NK cells. (A) Detection of cyCD3ε+ NK cells using indicated CD3ε antibody clones among primary CD56dim NK cells and NK92 cells is shown. Isotype controls for APA1/1 and SP34–2 are indicated as dashed lines. (B) gMFIs of CD3γ, CD3δ, and CD247 on cyCD3ε+ and cyCD3ε− NK cells are shown. Populations from the same donors are paired (n=14). (C) Expression of cyCD3γ, and cyCD3δ are shown on cyCD3ε− NK cells (dashed), cyCD3ε+ NK cells (solid), and T cells (tinted) from the same donor. One representative donor out of fourteen analyzed is shown. (D) NK92 cells and purified NK cells from two cyCD3ε+ NK cell donors were lysed, immunoprecipitated with control IgG or anti-CD3ε antibody and blotted with anti-CD247. (E) gMFIs of Lck on cyCD3ε+ and cyCD3ε− NK cells are shown. Populations from the same donors are paired (n=8). (F) Purified NK cells from two cyCD3ε+ NK cell donors were lysed, and immunoprecipitated with control IgG or anti-CD3ε antibody for Lck detection.
To confirm that CD3ε forms a complex with other CD3 subunits, we immunoprecipitated CD3ε from the human T-lymphoblast cell line MOLT-4, and from the NK cell lines NKL and NK92. NK92 cells express 7-fold less cyCD3ε compared to MOLT-4 cells, which express cytoplasmic but not cell surface CD3 (7, Fig. S4A). NKL does not express CD3ε at all and serves as a negative control. As expected, CD3ε complexes with CD3γ, CD3δ, and CD247 in MOLT-4 cells. CD3ε complexes with CD3γ, but not CD3δ in NK92 cells, consistent with the finding that NK92 cells do not express CD3δ, as assessed by western blot (Fig. S4B). We further confirmed that CD3ε complexes with CD247 in primary cyCD3ε+ NK cells (Fig. 6D). These data indicate that the various CD3 subunits are present and can complex in this population of HCMV-induced NK cells.
CD16 mediates NK cell activation in a tyrosine-protein kinase-dependent manner principally via Lck (33, 35). As a signaling adaptor of CD16 in NK cells, CD247 is also a substrate of Lck (33, 34). Transcriptome analysis suggests that LCK expression is enriched in HCMV-induced NK cells (Fig. S4C), consistent with the findings that cyCD3ε+ NK cells express more Lck protein than cyCD3ε− NK cells (Fig. 6E). In T cells, CD3ε-Lck interaction promotes TCR signaling and CAR-T cell function (36, 37). To determine whether in NK cells Lck and CD3ε also directly associate, anti-Lck immunoblotting was performed using immunoprecipitates prepared from cyCD3ε+ primary NK cells (Fig. S4D), with NKL cells as negative controls (Fig. S4E). We find that, similar to T cells, CD3ε complexes with Lck in primary NK cells (Fig. 6F). To demonstrate that CD3+ NK cells have enhanced activity upon CD16 stimulation, we investigated the CD16+ NK92MI cells, of which 10% are CD3ε/γ complex positive. We observed an enhanced IFN-γ activity among these cyCD3ε+ cells upon stimulation with plate bound anti-CD16 antibody (Fig. S4F). Taken together, the higher expression of CD16 signaling-related molecules CD247 and Lck, the direct interaction of Lck with CD3ε, and the enhanced cytokine response to CD16 stimulation in cyCD3ε+ NK cells suggest that CD3ε contributes to CD16 proximal signaling in this HCMV-expanded NK population.
Discussion
We demonstrate that cyCD3ε+ NK cells can comprise up to 68% of all circulating CD56dim NK cells in healthy adults, that they bear a broader T cell signature regulated by the Notch-activated Bcl11b transcription factor, and that elevated frequency of the cells is associated with prior HCMV exposure.
In the HCMV-induced NK cell population, CD3ε is just one molecule of a wider T cell program, which includes the CD3 complex partners CD3δ and CD3γ, as well as CD5 and CD8β. In the same NK cells, we observe simultaneous downregulation of PLZF, FcεRIγ, and CD122, additional molecular features typical of T cell identity. A striking common characteristic among these molecules is their targeting by the zinc finger transcription factor Bcl11b (26), a critical regulator of T lineage commitment (28) and one whose gene transcription is also differentially upregulated in the HCMV-induced NK cell population. Bcl11b plays both activating and repressive roles in the T cell specification process, exerting its effects at different stages of T cell development (26). Among the genes silenced by Bcl11b are those distinctive to innate NK cells, ILCs, and innate-like T cells; notably, deletion of Bcl11b in mice results in a reprogramming of T cells at all developmental stages to adopt NK cell properties similar to conventional NK cells (19) Thus, a transcription factor responsible for directing T cell fate is absent in NK lineage cells at their earliest developmental stage, but emerges at the transition from CD56bright to CD56dim, a phenotypic juncture that marks significant functional distinctions: the immature cytokine-sensitive CD56bright stage with higher proliferative capacity and the more mature CD56dim stage with greater competence for degranulation (38). The continued increase in Bcl11b protein levels from CD57− to CD57+ in the CD56dim NK cell population indicates a role for Bcl11b in further reshaping the programming of these cells as they continue to mature. Indeed, the majority of the HCMV-induced NK cell population is CD57+, which may explain the striking presence of so many Bcl11b-associated changes.
T cells acquire NK cell properties and concomitantly lose T cell-associated gene expression upon Bcl11b deletion in mice, suggesting that NK cell gene programs require continual Bcl11b suppression (26). The discovery that human NK cells then express Bcl11b in the course of their maturation is therefore surprising. Transcriptome analyses of murine T cells reveal that Cd7, Cd160, Fcer1g, Gpr183, Il2rb, Sh2d1b1, Zbtb16, Tyrobp, Xcl1, and Kit are repressed by Bcl11b while Cd3d, Cd3g, Cd5, and Cd6 expression are dependent on Bcl11b (28), most of which we confirm in our human NK cell dataset, thereby verifying successful transcriptional activation of a T cell specification program in human NK cells. Provision of Notch signaling to NK cells enhances cyCD3ε expression, whether via culture with Notch ligand or via in vivo exposure to Notch ligand-rich tissue environments, suggesting a Notch/Bcl11b-driven T cell-related genetic program in NK cells. The exact binding sites and physical interaction partners of Bcl11b within human NK cells are of particular interest, given the implications for NK cell-specific selective gene activation and repression during maturation, and remain under investigation.
CyCD3ε+ NK cells exhibit higher cytokine response capacity with ADCC, particularly when the majority of cyCD3ε+ cells exhibit loss of FcεRIγ, but the exact role of the CD3ε molecule remains elusive. Canonical NK cells express FcεRIγ and CD247 as heterodimeric and homodimeric adaptor proteins for CD16 signaling (5). The number of immunoreceptor tyrosine-based activation motifs (ITAMs) is likely critical for the strength of CD16 signaling in human NK cells. Previous studies have proposed that loss of FcεRIγ, which expresses one ITAM each, may increase homodimerization of CD247 molecules, each of which expresses three ITAMs, thereby increasing CD16-mediated activation through the higher number of ITAMs (14). In addition, cyCD3ε+ NK cells express more of the CD16 signaling molecules Lck and CD247. We find that CD3ε complexes with CD247 and Lck, an observation that has not previously been made in NK cells, but which could stabilize and propagate CD16 signaling in NK cells. Although the functional association remains to be determined, their enhanced activity through CD16 is likely to depend on, in part, the availability and proximity of signaling molecules. We propose that altered and enhanced NK cell signaling that occurs through engagement of cytoplasmic CD3ε is a functional hallmark of a potent HCMV-induced NK cell population, comprising a potentially attractive population for adoptive cellular therapy.
Supplementary Material
Key points:
HCMV infection promotes expression of canonical T cell molecules in NK cells.
CD3-positive NK cells are functionally superior in ADCC response.
Acknowledgements:
We thank Dr. Marcel van den Brink for providing OP9 cells and Dr. Hongfen Guo for providing CD16+NK92MI cells.
Supported by NIH R01 AI125651, P30 CA008748 and U01 AI069197
Abbreviations used in this article:
- HCMV
Human cytomegalovirus
- NK
natural killer
- PBMC
Peripheral blood mononuclear cells
- UCBT
umbilical cord blood transplantation
- ADCC
antibody-dependent cellular cytotoxicity
- ITAM
immunoreceptor tyrosine-based activation motif
References
- 1.Spits H, Lanier LL, and Phillips JH. 1995. Development of human T and natural killer cells. Blood 85: 2654–2670. [PubMed] [Google Scholar]
- 2.Renoux VM, Zriwil A, Peitzsch C, Michaelsson J, Friberg D, Soneji S, and Sitnicka E. 2015. Identification of a Human Natural Killer Cell Lineage-Restricted Progenitor in Fetal and Adult Tissues. Immunity 43: 394–407. [DOI] [PubMed] [Google Scholar]
- 3.Sun JC, and Lanier LL. 2011. NK cell development, homeostasis and function: parallels with CD8(+) T cells. Nat Rev Immunol 11: 645–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Orr MT, and Lanier LL. 2010. Natural killer cell education and tolerance. Cell 142: 847–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lanier LL 2003. Natural killer cell receptor signaling. Curr Opin Immunol 15: 308–314. [DOI] [PubMed] [Google Scholar]
- 6.Phillips JH, Hori T, Nagler A, Bhat N, Spits H, and Lanier LL. 1992. Ontogeny of human natural killer (NK) cells: fetal NK cells mediate cytolytic function and express cytoplasmic CD3 epsilon,delta proteins. J Exp Med 175: 1055–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lanier LL, Chang C, Spits H, and Phillips JH. 1992. Expression of cytoplasmic CD3 epsilon proteins in activated human adult natural killer (NK) cells and CD3 gamma, delta, epsilon complexes in fetal NK cells. Implications for the relationship of NK and T lymphocytes. J Immunol 149: 1876–1880. [PubMed] [Google Scholar]
- 8.Cannon MJ, Schmid DS, and Hyde TB. 2010. Review of cytomegalovirus seroprevalence and demographic characteristics associated with infection. Rev Med Virol 20: 202–213. [DOI] [PubMed] [Google Scholar]
- 9.Guma M, Angulo A, Vilches C, Gomez-Lozano N, Malats N, and Lopez-Botet M. 2004. Imprint of human cytomegalovirus infection on the NK cell receptor repertoire. Blood 104: 3664–3671. [DOI] [PubMed] [Google Scholar]
- 10.Beziat V, Liu LL, Malmberg JA, Ivarsson MA, Sohlberg E, Bjorklund AT, Retiere C, Sverremark-Ekstrom E, Traherne J, Ljungman P, Schaffer M, Price DA, Trowsdale J, Michaelsson J, Ljunggren HG, and Malmberg KJ. 2013. NK cell responses to cytomegalovirus infection lead to stable imprints in the human KIR repertoire and involve activating KIRs. Blood 121: 2678–2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee J, Zhang T, Hwang I, Kim A, Nitschke L, Kim M, Scott JM, Kamimura Y, Lanier LL, and Kim S. 2015. Epigenetic modification and antibody-dependent expansion of memory-like NK cells in human cytomegalovirus-infected individuals. Immunity 42: 431–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schlums H, Cichocki F, Tesi B, Theorell J, Beziat V, Holmes TD, Han H, Chiang SC, Foley B, Mattsson K, Larsson S, Schaffer M, Malmberg KJ, Ljunggren HG, Miller JS, and Bryceson YT. 2015. Cytomegalovirus infection drives adaptive epigenetic diversification of NK cells with altered signaling and effector function. Immunity 42: 443–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu Z, Sinzger C, Frascaroli G, Reichel J, Bayer C, Wang L, Schirmbeck R, and Mertens T. 2013. Human cytomegalovirus-induced NKG2C(hi) CD57(hi) natural killer cells are effectors dependent on humoral antiviral immunity. J Virol 87: 7717–7725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang T, Scott JM, Hwang I, and Kim S. 2013. Cutting edge: antibody-dependent memory-like NK cells distinguished by FcRgamma deficiency. J Immunol 190: 1402–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Costa-Garcia M, Vera A, Moraru M, Vilches C, Lopez-Botet M, and Muntasell A. 2015. Antibody-mediated response of NKG2Cbright NK cells against human cytomegalovirus. J Immunol 194: 2715–2724. [DOI] [PubMed] [Google Scholar]
- 16.Della Chiesa M, Falco M, Bertaina A, Muccio L, Alicata C, Frassoni F, Locatelli F, Moretta L, and Moretta A. 2014. Human cytomegalovirus infection promotes rapid maturation of NK cells expressing activating killer Ig-like receptor in patients transplanted with NKG2C−/− umbilical cord blood. J Immunol 192: 1471–1479. [DOI] [PubMed] [Google Scholar]
- 17.Liu LL, Landskron J, Ask EH, Enqvist M, Sohlberg E, Traherne JA, Hammer Q, Goodridge JP, Larsson S, Jayaraman J, Oei VYS, Schaffer M, Tasken K, Ljunggren HG, Romagnani C, Trowsdale J, Malmberg KJ, and Beziat V. 2016. Critical Role of CD2 Co-stimulation in Adaptive Natural Killer Cell Responses Revealed in NKG2C-Deficient Humans. Cell Rep 15: 1088–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muntasell A, Pupuleku A, Cisneros E, Vera A, Moraru M, Vilches C, and Lopez-Botet M. 2016. Relationship of NKG2C Copy Number with the Distribution of Distinct Cytomegalovirus-Induced Adaptive NK Cell Subsets. J Immunol 196: 3818–3827. [DOI] [PubMed] [Google Scholar]
- 19.Li P, Burke S, Wang J, Chen X, Ortiz M, Lee SC, Lu D, Campos L, Goulding D, Ng BL, Dougan G, Huntly B, Gottgens B, Jenkins NA, Copeland NG, Colucci F, and Liu P. 2010. Reprogramming of T cells to natural killer-like cells upon Bcl11b deletion. Science 329: 85–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lopez-Verges S, Milush JM, Pandey S, York VA, Arakawa-Hoyt J, Pircher H, Norris PJ, Nixon DF, and Lanier LL. 2010. CD57 defines a functionally distinct population of mature NK cells in the human CD56dimCD16+ NK-cell subset. Blood 116: 3865–3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adler SP, Manganello AM, Lee R, McVoy MA, Nixon DE, Plotkin S, Mocarski E, Cox JH, Fast PE, Nesterenko PA, Murray SE, Hill AB, and Kemble G. 2016. A Phase 1 Study of 4 Live, Recombinant Human Cytomegalovirus Towne/Toledo Chimera Vaccines in Cytomegalovirus-Seronegative Men. J Infect Dis 214: 1341–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murray SE, Nesterenko PA, Vanarsdall AL, Munks MW, Smart SM, Veziroglu EM, Sagario LC, Lee R, Claas FHJ, Doxiadis IIN, McVoy MA, Adler SP, and Hill AB. 2017. Fibroblast-adapted human CMV vaccines elicit predominantly conventional CD8 T cell responses in humans. J Exp Med 214: 1889–1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hwang I, Zhang T, Scott JM, Kim AR, Lee T, Kakarla T, Kim A, Sunwoo JB, and Kim S. 2012. Identification of human NK cells that are deficient for signaling adaptor FcRgamma and specialized for antibody-dependent immune functions. Int Immunol 24: 793–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Strauss-Albee DM, Liang EC, Ranganath T, Aziz N, and Blish CA. 2017. The newborn human NK cell repertoire is phenotypically formed but functionally reduced. Cytometry B Clin Cytom 92: 33–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Della Chiesa M, Falco M, Podesta M, Locatelli F, Moretta L, Frassoni F, and Moretta A. 2012. Phenotypic and functional heterogeneity of human NK cells developing after umbilical cord blood transplantation: a role for human cytomegalovirus? Blood 119: 399–410. [DOI] [PubMed] [Google Scholar]
- 26.Longabaugh WJR, Zeng W, Zhang JA, Hosokawa H, Jansen CS, Li L, Romero-Wolf M, Liu P, Kueh HY, Mortazavi A, and Rothenberg EV. 2017. Bcl11b and combinatorial resolution of cell fate in the T-cell gene regulatory network. Proc Natl Acad Sci U S A 114: 5800–5807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Smedt M, Taghon T, Van de Walle I, De Smet G, Leclercq G, and Plum J. 2007. Notch signaling induces cytoplasmic CD3 epsilon expression in human differentiating NK cells. Blood 110: 2696–2703. [DOI] [PubMed] [Google Scholar]
- 28.Hosokawa H, Romero-Wolf M, Yui MA, Ungerback J, Quiloan MLG, Matsumoto M, Nakayama KI, Tanaka T, and Rothenberg EV. 2018. Bcl11b sets pro-T cell fate by site-specific cofactor recruitment and by repressing Id2 and Zbtb16. Nat Immunol 19: 1427–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tikhonova AN, Dolgalev I, Hu H, Sivaraj KK, Hoxha E, Cuesta-Dominguez A, Pinho S, Akhmetzyanova I, Gao J, Witkowski M, Guillamot M, Gutkin MC, Zhang Y, Marier C, Diefenbach C, Kousteni S, Heguy A, Zhong H, Fooksman DR, Butler JM, Economides A, Frenette PS, Adams RH, Satija R, Tsirigos A, and Aifantis I. 2019. The bone marrow microenvironment at single-cell resolution. Nature 569: 222–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang L, Hoseini SS, Xu H, Ponomarev V, and Cheung NK. 2019. Silencing Fc Domains in T cell-Engaging Bispecific Antibodies Improves T-cell Trafficking and Antitumor Potency. Cancer Immunol Res 7: 2013–2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tarek N, Le Luduec JB, Gallagher MM, Zheng J, Venstrom JM, Chamberlain E, Modak S, Heller G, Dupont B, Cheung NK, and Hsu KC. 2012. Unlicensed NK cells target neuroblastoma following anti-GD2 antibody treatment. J Clin Invest 122: 3260–3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salmeron A, Sanchez-Madrid F, Ursa MA, Fresno M, and Alarcon B. 1991. A conformational epitope expressed upon association of CD3-epsilon with either CD3-delta or CD3-gamma is the main target for recognition by anti-CD3 monoclonal antibodies. J Immunol 147: 3047–3052. [PubMed] [Google Scholar]
- 33.Salcedo TW, Kurosaki T, Kanakaraj P, Ravetch JV, and Perussia B. 1993. Physical and functional association of p56lck with Fc gamma RIIIA (CD16) in natural killer cells. J Exp Med 177: 1475–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lanier LL, Yu G, and Phillips JH. 1989. Co-association of CD3 zeta with a receptor (CD16) for IgG Fc on human natural killer cells. Nature 342: 803–805. [DOI] [PubMed] [Google Scholar]
- 35.Cone JC, Lu Y, Trevillyan JM, Bjorndahl JM, and Phillips CA. 1993. Association of the p56lck protein tyrosine kinase with the Fc gamma RIIIA/CD16 complex in human natural killer cells. Eur J Immunol 23: 2488–2497. [DOI] [PubMed] [Google Scholar]
- 36.Li L, Guo X, Shi X, Li C, Wu W, Yan C, Wang H, Li H, and Xu C. 2017. Ionic CD3-Lck interaction regulates the initiation of T-cell receptor signaling. Proc Natl Acad Sci U S A 114: E5891–E5899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hartl FA, Beck-Garcia E, Woessner NM, Flachsmann LJ, Cardenas RMV, Brandl SM, Taromi S, Fiala GJ, Morath A, Mishra P, Yousefi OS, Zimmermann J, Hoefflin N, Kohn M, Wohrl BM, Zeiser R, Schweimer K, Gunther S, Schamel WW, and Minguet S. 2020. Noncanonical binding of Lck to CD3epsilon promotes TCR signaling and CAR function. Nat Immunol 21: 902–913. [DOI] [PubMed] [Google Scholar]
- 38.Freud AG, Mundy-Bosse BL, Yu J, and Caligiuri MA. 2017. The Broad Spectrum of Human Natural Killer Cell Diversity. Immunity 47: 820–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.