Abstract
Early-onset colorectal cancer (EOCRC), defined as a diagnosis under age 50, is an emerging public health burden. As many of these individuals fall outside of screening guidelines, the development of a minimally invasive, accurate screening modality for this population is warranted. We evaluated the FDA-approved blood-based biomarker methylated Septin9 (mSEPT9) test as screening tool for EOCRC. EOCRC plasma, healthy plasma, and serum-free conditioned media from cancer cell lines was collected. Cell-free DNA (cfDNA) was isolated and bisulfite converted for use in the assay. mSEPT9 and ACTB measured using Epi proColon® V2.0. EOCRC plasma was collected at Massachusetts General Hospital (2005–2019) and controls were collected at the National Institutes of Health and by ZenBio Inc. (prior to 2019). Twenty-seven EOCRC cases, 48 healthy controls <50 years old, and 39 healthy controls ≥50 years old were included in this study. mSEPT9 was detected more frequently in EOCRC cases (88.9%) compared to healthy controls age <50 (4.2%) and ≥50 (15.4%), respectively (p<0.001). The sensitivity, specificity, positive predictive value, and negative predictive values of the mSEPT9 assay to detect EOCRC was 90.8% (95% CI: 84.7–96.9%), 88.9% (95% CI: 77.0–100.0%), 96.3% (95% CI: 92.3–100.0%), and 75.0% (95% CI 60.0–90.0%), respectively, compared to all healthy controls. mSEPT9 cfDNA level was an independent predictor of survival (p=0.02). mSEPT9 is a sensitive and specific biomarker for EOCRC detection. These results suggest that mSEPT9 may be useful in the detection of EOCRC, providing a minimally invasive method for screening in this growing population of CRC patients.
Keywords: early-onset, colorectal cancer, early detection, biomarkers, methylated Septin9
INTRODUCTION
Incidence of early-onset colorectal cancer (EOCRC), defined as a colorectal cancer (CRC) diagnosis under the age of 50 years, has dramatically increased over the last several decades in the United States and globally (1). The risk factors contributing to the rising trends of EOCRC remain undefined, though several factors, such as increased obesity, dietary changes, and sedentary lifestyle, have been proposed (2–4). Despite the increasing incidence, unless there is a known genetic predisposition, most individuals with EOCRC are not screened until they are symptomatic. Though prominent gastroenterological societies have begun recommending endoscopic screening at age 45, the influx of new screen-eligible individuals will be difficult to manage given systemic constraints, workforce shortages, and the high cost of implementation (5,6). Therefore, creating solutions for this unforeseen issue needs to be prioritized.
Despite an increase in the incidence of CRC in individuals under the age of 50 years, as a proportion of all CRC cases, EOCRC is still small and widespread screening of this age group may not be the most feasible or cost-effective strategy. However, a tiered screening strategy with the use of a less-invasive approach like fecal immunochemical test (FIT), Cologuard® or a blood-based test has been proposed (6). The integration of a sensitive and specific blood-based assay may fill the EOCRC screening and detection gap. Plasma-based circulating biomarkers, such as cell-free DNA (cfDNA), are often used for the detection of somatic alterations in cancer and sensitive modalities for its detection have been recently developed (7–10). The use of cfDNA for cancer detection is promising, as tumor-derived cfDNA is abundant compared to normal circulating cfDNA and remains relatively stable during long-term storage (11–13). This has been established in lung, prostate, breast, and colorectal cancers (14–17). Further, the addition of blood-based biomarkers, such as methylated SEPT9 (mSEPT9), to FIT has demonstrated improved overall screening sensitivity (18–20).
mSEPT9 has displayed efficacy as a plasma-based circulating biomarker for the detection of CRC, as SEPT9 production is regulated by epigenetic events which have proven critical in the initiation and progression of cancer (21,22). Moreover, mSEPT9 can be easily and reliably detected in plasma collected from tumor-bearing individuals (23,24). Further, numerous clinical studies have demonstrated high sensitivity and specificity of mSEPT9 for the detection of CRC (23,25,26). A recent meta-analysis of published case-control studies evaluating the performance of mSEPT9 showed a pooled sensitivity of 74% (95% CI: 61–84%) and specificity of 84% (95% CI: 81–87%), comparing CRC to healthy individuals (27). These and other studies provided compelling evidence to grant U.S. Food and Drug Administration (FDA) approval for Epi proColon®, a commercially available mSEPT9 detection kit (28). Epi proColon® is the only FDA-approved blood-based screening tool for CRC, however its approval is limited to individuals age 50 and older who have refused colonoscopy or fecal-based screening methods (29,30). Therefore, in the current study, we seek to extend the population utility of Epi proColon®.
Due to the increasing trend in EOCRC and the significant burden on the healthcare system for CRC screening, a rapid, non-invasive modality to triage potential EOCRC cases is needed. However, no studies have evaluated the efficacy of mSEPT9 as a CRC screening modality in a younger population. In this study, we evaluated the efficacy of the commercially available mSEPT9 assay, Epi proColon® V2.0, for the detection of CRC in a retrospective case-control study of archived EOCRC plasma samples, compared to control plasma collected from healthy individuals <50 years and healthy controls ≥50 years. We hypothesized that mSEPT9 would be a sensitive and specific biomarker for EOCRC detection in this cohort, comparable to that reported for individuals ≥50 years old for which Epi proColon® is FDA-approved.
MATERIALS AND METHODS
Plasma collection, preparation, and patient information
Plasma from cases with an EOCRC diagnosis under age 50 and healthy (disease-free at time of blood collection) controls younger than age 50 at time of collection were used in the current study. All EOCRC plasma samples were treatment naïve and collected prior to surgery. The study protocol and use of biospecimens were reviewed and determined exempt by the National Institutes of Health (NIH) Institutional Review Board. Healthy donor blood was acquired from the NIH Clinical Center (CC) and a commercial vendor (ZenBio, Inc., Research Triangle, NC). Blood acquired through the NIH CC was collected into EDTA vacutainer tubes and transported Frederick National Laboratory for Cancer Research (FNLCR). Upon arrival, blood samples were spun for 10 minutes at 500 g. The top layer (plasma) was transferred and pooled into a 15 ml conical tube and spun at 2000 g for 10 minutes. Plasma was stored in 0.5 ml aliquots at −80⁰C until DNA extraction. Similarly, plasma procured from the commerical vendor was collected in EDTA vacutainer tubes and processed and stored according to the vendors specifications.
EOCRC plasma samples were collected at Massachusetts General Hospital between May 2005 and February 2019. Briefly, venous blood was collected by standard phlebotomy into EDTA vacutainer tubes and sent for processing. Upon arrival, samples were centrifuged at 1600 g for 10 minutes and plasma supernatent transferred to a 15 ml centrifuge tube for an additional centrifugation step for 10 minutes at 3000 g. The plasma was transferred to a fresh 15 ml tube, gently mixed, and stored in 1 ml aliquots. Aliquots were stored at −80⁰C at Massachusetts General Hospital until shipment on dry ice to FNLCR, where they were stored at −80⁰C upon arrival. All EOCRC cases inlcuded in the study had biopsy confirmed CRC. Controls were healthy (cancer-free) at the time of collection and acquired from the NIH (2018–2019) and the commerical vendor ZenBio Inc. (prior to 2019). Demographic, diagnostic, and prognostic information of EOCRC cases and demographic information of controls were collected. All samples were deidentified.
EOCRC sample collection was approved by the Massachusetts General Hospital Institutional Review Board (IRB 14–046) and the study was deemed exempt from NIH Institutional Review Board approval (IRB000294). Written informed consent from all participants was obtained at their respective collection sites. The study was conducted in accordance with the U.S. Common Rule.
Cell culture
CRC cell lines HCT-116 (obtained 2018), HT-29 (obtained 2018), and LoVo (obtained 2018), lung cancer cell line HOP-92 (obtained 2017), breast cancer cell line MCF7 (obtained 2021) and melanoma cell line RPMI-7951 (obtained 2021) were obtained through the NCI Cell Line Repository (Division of Cancer Treatment and Diagnosis (DCTD) Tumor Repository, NCI at Frederick, Frederick, MD). The prostate cancer cell line PC-3 were provided by Dr. Esta Sterneck (NCI-Frederick, obtained 2019). HCT-116, HT-29, LoVo, HOP-92, MCF7, RPMI-7951, and PC-3 cells were cultured in RPMI 1640 Medium supplemented with 10% FBS, 1% P/S, and 2mM L-Glutamine. All cells were incubated at 5% CO2 at 37⁰C. Cell lines were tested for mycoplasma contamination by PPLO culture under aerobic and anaerobic conditions and orcein staining of Fogh or by PCR-based assay. Cell lines obtained from the NCI DCTD Tumor Respository (HCT-116, HT-29, LoVo, HOP-92, MCF7, RPMI-7951) were authenticated using Applied Biosystems AmpFISTR® Identifiler with PCR amplification prior to cell line receipt. PC-3 cells were authenticated using CellCheck, (IDEXX BioAnalytics, Columbia, MO) a comprehensive cell line authentication service that utilizes STR-based DNA profiling and multiplex PCR to detect both contamination and misidentification of cell lines.
Serum-free conditioned media collection
Cell lines were thawed according to repository guidelines. Passage number between thawing and serum-free conditioned media (SFCM) collection was kept to a minimum. Cells were grown to 90% confluence in 75 mm2 flasks, washed with 3 ml 1xDPBS, and serum- and antibiotic-free media was added to the cells and incubated at 37⁰C overnight. SFCM was collected, centrifuged briefly to rid of cellular debris, and stored in 1 ml aliquots at −80⁰C until use.
Epi proColon® V2.0 assay kit
The Epi proColon® V2.0 plasma circulating cfDNA test kit protocol was performed according to the manufacturers’ protocol, however adapted to a smaller sample volume (1 ml), as demonstrated in Hitchins et al. 2019 (31). Briefly, 1 ml plasma and assay controls were thawed at room temperature for 30 minutes. Samples were transferred to a 15 ml conical tube and 1 ml Epi proColon® Lysis Binding Buffer added, briefly vortexed, and incubated at room temperature for 10 minutes. Following incubation, 25.7 μl magnetic beads and 714 μl molecular grade absolute ethanol was added to each sample, then mixed by inversion and rotated for 45 minutes to complete DNA binding. Upon completion, samples were incubated at 56⁰C for 10 minutes, washed with 500 μl Epi proColon® Wash Buffer A, and bound DNA eluted into 50 μl Epi proColon® Elution Buffer. Next, bisulfite conversion was performed by adding 75 μl Epi proColon® Bisulfite and 12.5 μl Epi proColon® Protection Buffer to the extracted DNA. Samples were briefly vortexed, spun down, and incubated at 80⁰C for 45 minutes. Immediately after incubation, samples were briefly spun down and 500 μl Epi proColon® Wash Buffer A and 10 μl Epi proColon® Magnetic Beads were added to complete DNA binding. Samples were briefly vortexed, centrifuged, and incubated at 23⁰C while shaking at 1000 rpm. The magnetic bead solutions were then centrifuged and placed in a magnetic rack to remove the remaining buffer. The bound beads were washed three times, first with 500 μl Epi proColon® Wash A Buffer, and subsequently with 400 μl and 200 μl Epi proColon® Wash B. After removing all wash buffer, the beads were dried at 23⁰C for 10 minutes and bisulfite-converted DNA (bisDNA) eluted into 17 μl of Epi proColon® Elution Buffer. Internal positive and negative controls were included in each batch (Epi proColon® Sensitive PCR Kit, Epigenomics, Inc.).
SFCM volumes of 1 ml, 500 μl, 250 μl, and 125 μl were used for volume titration of the Epi proColon® V2.0 kit. Samples were diluted with 1xDPBS to a volume of 1 ml then processed in the same manner as the plasma samples.
Quantitative PCR
Immediately following the isolation of bisDNA, the samples were randomized in batches and analyzed by quantitative polymerase chain reaction (qPCR) using the Epi proColon® Sensitive PCR Kit. A volume of 15 μl of PCR Master Mix was added to 15 μl of bisDNA and the plate was briefly centrifuged. All samples were run using an Applied Biosystems QuantStudio 5. Thermal cycle program conditions were as follows: (1) denaturation for 20 minutes at 94⁰C (40% ramp rate), (2) annealing and extension for 5 seconds at 62⁰C (80% ramp rate), 35 seconds at 55.5⁰C (80% ramp rate), and 30 seconds at 93⁰C (40% ramp rate) for 45 cycles, and (3) extension for 5 seconds at 40⁰C (80% ramp rate). A valid assay run had postive control mSEPT9 and ACTB thresholds less than cycle threshold (Ct)≤41.4 and Ct≤29.8, respectively, and negative control mSEPT9 and ACTB thresholds undetermined and Ct≤37.2, respectively. Patient plasma samples were considered positive if ACTB Ct≤32.1 and mSEPT9 Ct<45, negative if ACTB Ct≤32.1 and mSEPT9 undetermined, and invalid ACTB Ct>32.1.
Statistical analysis
As the protocol was adapted to 1 ml plasma (1/3 of the original protocol volume), a single real-time PCR reaction was performed in a single well for each sample. mSEPT9 positivity was determined using the 1/1 testing algorithm, whereby if the result for mSEPT9 and internal ACTB reached the specified threshold, then the sample was considered positive. If the assay and sample controls passed quality control, sample mSEPT9 levels were evaluated. If mSEPT9 was detected below a Ct of 45, the sample was determined positive. Each case and control was analyzed with a dichotomous (postive, negative) outcome and relative methylation was determined using the ΔΔCt method for DNA methylation, as described elsewhere (32,33). Receiver operating curves (ROC) were generated using qPCR Ct values. Statistical differences in relative methylation were determined by one-way analysis of variance (ANOVA) or Mann-Whitney U. A p-value less than 0.05 was considered statistically significant. Analyses were performed in GraphPad Prism 8 for Windows (GraphPad Software, Inc., San Diego, CA) and sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) were calculated in SAS 9.4 (SAS Institute, Cary, NC).
Data availability
The datasets generated and/or analyzed during the current study are not publicly available due to the sensitivity of the data but are available from the corresponding author upon reasonable request.
RESULTS
The study cohort included 34 EOCRC cases, 50 healthy controls <50 years old, and 40 healthy controls ≥50 years old. Of these, 10 samples were excluded due to failed tests. The final cohort for which complete data were obtained included 27 EOCRC cases, 48 healthy controls <50 years old, and 39 healthy controls ≥50 years old (114 total). EOCRC cases had a median age of 44 years (range 25.9–49), were 81% white and 59% male (Table 1). Healthy controls <50 years old had a median age of 44 (range 29–49), were 48% black and 65% male, while healthy controls ≥50 years old had a median age of 56 (range 50–77), were 54% white and 64% male. Majority of the EOCRC cases were rectal cancers (66.7%), late stage (62.9% stage III/IV), and had a family history of cancer (77.8%) (Table 2).
Table 1.
Demographics of healthy controls <50 years old, healthy controls ≥50 years old, and EOCRC cases.
| Variable | Healthy controls <50 years (N=48) | Healthy controls ≥50 years (N=39) | EOCRC (N=27) |
|---|---|---|---|
|
| |||
| Age (median, range) | 44 (29–49) | 56 (50–77) | 44 (25.9–49) |
|
| |||
| Race (n, %) | |||
|
| |||
| White | 9 (18.8) | 21 (53.8) | 22 (81.5) |
| Black | 23 (47.9) | 4 (10.3) | 2 (7.4) |
| Hispanic | 15 (31.3) | 7 (17.9) | 0 (0.0) |
| Asian | 1 (2.1) | 5 (12.8) | 1 (3.7) |
| Unknown | 0 (0.0) | 1 (2.6) | 2 (7.4) |
|
| |||
| Sex (n, %) | |||
| Female | 17 (35.4) | 14 (35.9) | 11 (40.7) |
| Male | 31 (64.6) | 25 (64.1) | 16 (59.3) |
|
| |||
| mSEPT9 assay (n, %) | |||
| Positive | 2 (4.2) | 6 (15.4) | 24 (88.9) |
| Negative | 46 (95.8) | 33 (84.6) | 3 (11.1) |
Table 2.
Clinical characteristics of EOCRC cases.
| Variable | EOCRC (N=27) |
|---|---|
|
| |
| Cancer site (n, %) | |
| Colon | 3 (11.1) |
| Rectum | 18 (66.7) |
| Unspecified colorectal site | 6 (22.2) |
|
| |
| Stage (n, %) | |
| I | 4 (14.8) |
| II | 1 (3.7) |
| III | 5 (18.5) |
| IV | 12 (44.4) |
| Unknown | 5 (18.5) |
|
| |
| Tumor grade (n, %) | |
| Low, low/intermediate | 11 (40.7) |
| Intermediate | 6 (22.2) |
| High | 7 (25.9) |
| Unknown | 3 (11.1) |
|
| |
| Survival status (n, %) | |
| Alive | 21 (77.8) |
| Deceased | 6 (22.2) |
|
| |
| Family history of cancer (n, %) | |
| Yes | 21 (77.8) |
| No | 5 (18.5) |
| Unknown | 1 (3.7) |
|
| |
| Family history of CRC (n, %) | |
| Yes | 9 (33.3) |
| No | 17 (63.0) |
| Unknown | 1 (3.7) |
|
| |
| History of IBD or chronic inflammation (n, %) | |
| Yes | 5 (18.5) |
| No | 22 (81.5) |
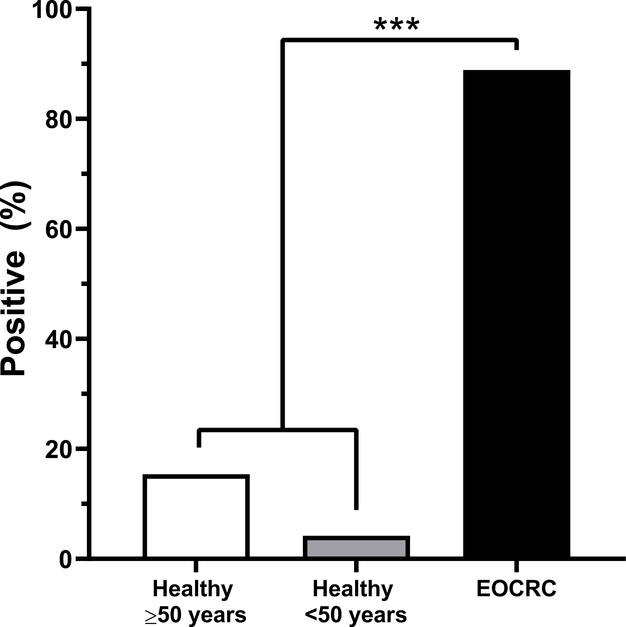
Abiding by the thresholds established in the Epi proColon® V2.0 kit, significantly more EOCRC samples were positive for mSEPT9 compared to healthy controls <50 years and healthy controls ≥50 years (p<0.001) (Figure 1). ACTB values were not statistically different between EOCRC cases and healthy controls (p=0.53). Specifically, 4.2% (2/48) of healthy controls <50 years old, 15.4% (6/39) of healthy controls ≥50 years old, and 88.9% (24/27) of EOCRC cases were positive for mSEPT9. Interestingly, no healthy samples under the age of 40 were mSEPT9 positive, and the highest percentage of mSEPT9 positive healthy controls were the in 50–55-year age group (21%) (Table 3).
Figure 1. EOCRC cases showed significantly higher mSEPT9 positivity than healthy controls.
Significantly more EOCRC cases (CRC ≤50 years) were mSEPT9 positive, compared to plasma from healthy controls <50 years old and ≥50 years old (p<0.001).
Table 3.
mSEPT9 status by demographics of EOCRC cases and healthy controls.
| Variable | Healthy controls <50 years (N=48) | Healthy controls ≥50 years (N=39) | EOCRC (N=27) |
|---|---|---|---|
|
| |||
| Age (n, %) | |||
| 26–29 | 0/1 (0.0) | - | 1/1 (100.0) |
| 30–34 | 0/1 (0.0) | - | 1/1 (100.0) |
| 35–39 | 0/4 (0.0) | - | 3/3 (100.0) |
| 40–44 | 1/20 (5.0) | - | 10/11 (90.9) |
| 45–49 | 1/23 (4.3) | - | 9/11 (81.8) |
| 50–54 | - | 3/17 (17.6) | - |
| 55–59 | - | 1/7 (14.3) | - |
| 60–64 | - | 1/8 (12.5) | - |
| 65–69 | - | 1/5 (20.0) | - |
| 70+ | - | 0/2 (0.0) | - |
|
| |||
| Race (n, %) | |||
| White | 1/9 (11.1) | 4/21 (19.0) | 19/22 (86.3) |
| Black | 1/23 (4.3) | 0/5 (0.0) | 2/2 (100.0) |
| Hispanic | 0/15 (0.0) | 2/7 (28.6) | 0/0 (0.0) |
| Asian | 0/1 (0.0) | 0/5 (0.0) | 1/1 (100.0) |
| Unknown | 0/0 (0.0) | 0/1 (0/0) | 2/2 (100.0) |
|
| |||
| Sex (n, %) | |||
| Female | 0/17 (0.0) | 2/14 (14.3) | 11/11 (100.0) |
| Male | 2/21 (9.5) | 4/25 (16.0) | 13/16 (81.3) |
|
| |||
| Cancer site (n, %) | |||
| Colon | - | - | 3/3 (100.0) |
| Rectum | - | - | 15/18 (83.3) |
| Unspecified colorectal site | - | - | 6/6 (100.0) |
|
| |||
| Stage (n, %) | |||
| I | - | - | 4/4 (100.0) |
| II | - | - | 1/1 (100.0) |
| III | - | - | 5/5 (100.0) |
| IV | - | - | 12/12 (100.0) |
| Unknown | - | - | 2/5 (40.0) |
|
| |||
| Tumor grade (n, %) | |||
| Low, low/intermediate | - | - | 10/11 (90.9) |
| Intermediate | - | - | 5/6 (83.3) |
| High | - | - | 6/7 (85.7) |
| Unknown | - | - | 3/3 (100.0) |
|
| |||
| Survival status, overall (n, %) | |||
| Alive | - | - | 18/21 (85.7) |
| Deceased | - | - | 6/6 (100.0) |
|
| |||
| Family history of cancer (n, %) | |||
| Yes | - | - | 19/21 (90.5) |
| No | - | - | 4/5 (80.0) |
| Unknown | - | - | 1/1 (100.0) |
|
| |||
| Family history of CRC (n, %) | |||
| Yes | - | - | 8/9 (88.9) |
| No | - | - | 15/17 (88.2) |
| Unknown | - | - | 1/1 (100.0) |
|
| |||
| History of IBD or chronic inflammation (n, %) | |||
| Yes | - | - | 5/5 (100.0) |
| No | - | - | 19/22 (86.4) |
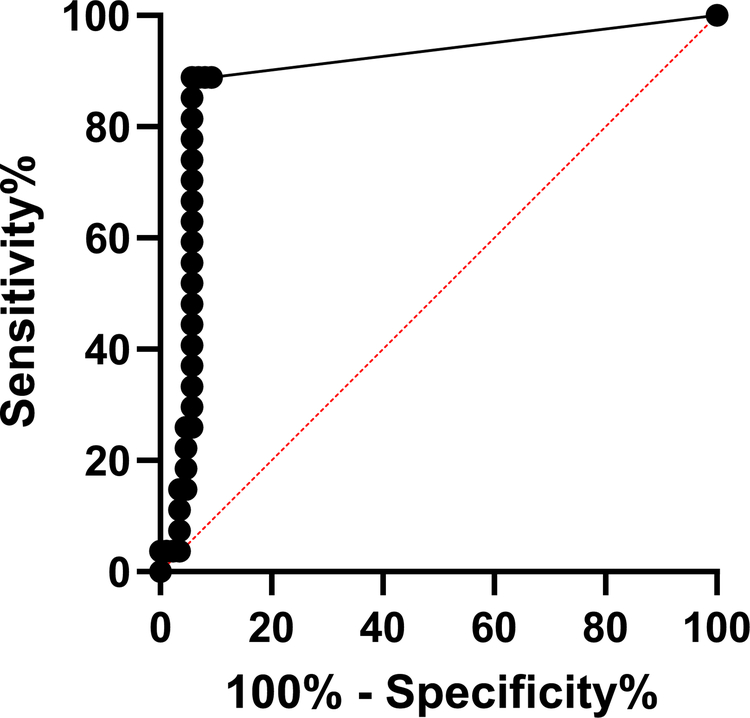
mSEPT9 was detected at similar frequency in EOCRC stages I-IV. Additional control and EOCRC demographics are reported in Table 3, as well as mSEPT9 positivity by EOCRC clinicopathological characteristics. The overall sensitivity (for EOCRC of all stages I-IV), specificity, PPV, and NPV of the mSEPT9 assay were calculated to be 90.8% (95% CI: 84.7–96.9%), 88.9% (95% CI: 77.0–100.0%), 96.3% (95% CI: 92.3–100.0%), and 75.0% (95% CI: 60.0–90.0%), respectively. ROC curves were generated to evaluate the performance of the assay in distinguishing CRC cases from non-CRC controls (healthy controls ≤50 and >50 years old, combined). Defining CRC cases as positive and non-CRC healthy controls as negative produced an area under the curve (AUC) of 0.89 (95% CI: 0.81–0.97, p<0.001), suggesting that the mSEPT9 assay can sensitively and specifically distinguish CRC from non-CRC (Figure 2).
Figure 2. Receiver operating curve (ROC) of mSEPT9 comparison between EOCRC cases and healthy controls.
The ROC was generated by comparing the mSEPT9 cycle threshold (Ct) values of EOCRC cases and all healthy controls. The AUC (0.89) was statistically significant (standard error=0.04, 95% CI=0.81–0.97, p<0.001).
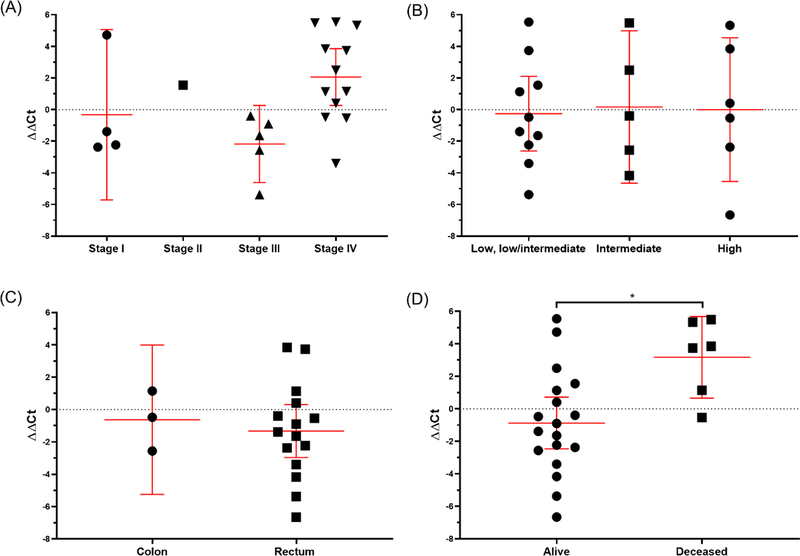
We next decided to quantitatively evaluate the positive EOCRC cases, normalizing sample Ct values to the within batch controls (ΔΔCt). No significant differences in ΔΔCt were noted between stages (p=0.06, p for trend=0.13), tumor grade (p=0.98, p for trend=0.88), or tumor site (p=0.65) (Figure 3A–C). However, we did observe a significant difference in patient outcome. EOCRC cases with a follow-up status of deceased had significantly greater levels of plasma mSEPT9 (ΔΔCt) compared to cases with a follow-up status of alive (p=0.02), suggesting mSEPT9 plasma levels are prognostic (Figure 3D). Overall, among positive EOCRC cases, level of plasma mSEPT9 was an independent predictor of overall survival.
Figure 3. Presence of mSEPT9 was significantly associated with EOCRC survival status.
Comparison of presence of mSEPT9, normalized by batch and ACTB, by (A) stage (p=0.06, p for trend=0.13), (B) tumor grade (p=0.98, p for trend=0.88), (C) tumor site (p=0.65), and (D) survival status (p=0.02).
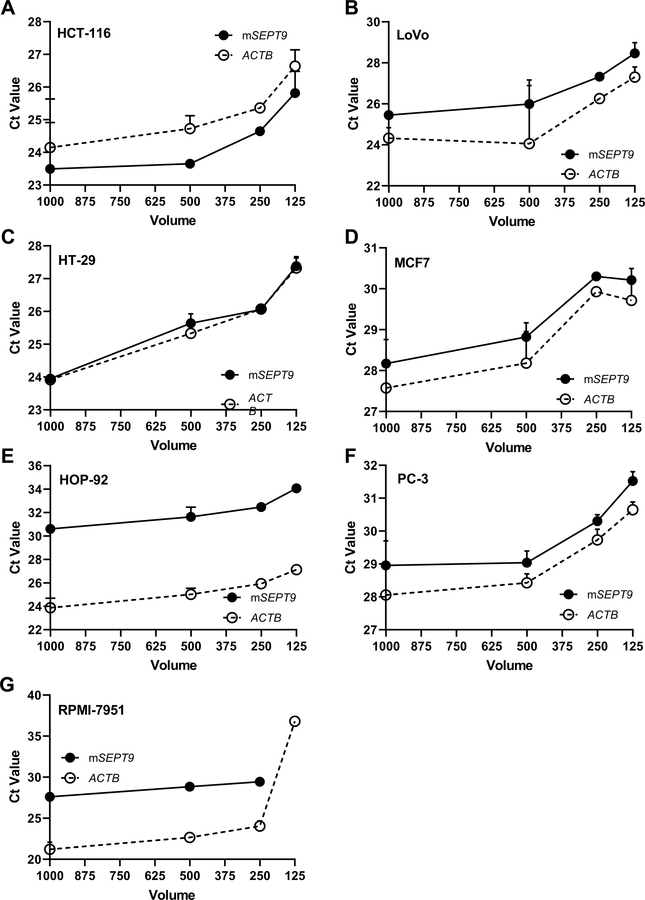
We and others have established the ability of the Epi proColon V2.0® kit to detect mSEPT9 in small volumes of plasma collected from individuals with CRC, however the production of mSEPT9 as a biomarker in additional cancer types remains unexplored. We evaluated conditioned media collected from cancer cell lines. We collected SFCM from cell culture (HCT-116, LoVo, HT-29, HOP-92, PC-3, MCF-7, RPMI-7951) for bisDNA conversion. mSEPT9 and ACTB was in the detectable range in the SFCM of most cell lines, down to a volume of 125 ul (Figure 4A–G). mSEPT9 was not detectable in 125 μl of melanoma RPMI-7951 SFCM, though ACTB was present (Figure 4G). Ct values for mSEPT9 and ACTB were within the same range for CRC, prostate, lung, breast, and melanoma cancer cell lines. For all cell lines, Ct values were similar between 500 μl and 1 ml of SFCM.
Figure 4. mSEPT9 was detectable in small volumes of serum-free conditioned media (SFCM) in CRC and non-CRC cell lines.
Volume titration of SFCM from CRC and non-CRC cell lines. mSEPT9 and ACTB detection was evaluated in 1000 μl, 500 μl, 250 μl, and 125 μl of SFCM in (A) HCT-116, (B) LoVo, (C) HT-29, (D) MCF7, (E) HOP-92, (F) PC-3, and (G) RPMI-7951.
DISCUSSION
In the current study, we found that plasma mSEPT9 was specific and sensitive for the detection of EOCRC. EOCRC cases were found more frequently positive for mSEPT9, compared to healthy controls <50 years and healthy controls ≥50 years. Furthermore, we were able to detect consistently and accurately mSEPT9 in samples using a small plasma volume (1 ml) and measurement in a single real-time PCR reaction (31). To our knowledge, this is the first evaluation of the utility of mSEPT9 as a screening modality in the early-onset population. Previous investigations of mSEPT9 among individuals of screening age (≥50 years old) and in individuals with Lynch syndrome, have demonstrated the potential of mSEPT9 as a sensitive and specific blood-based biomarker for CRC (24,26,31,32,34). Our study adds to this growing evidence base supporting the expansion of mSEPT9 as a biomarker for CRC detection in the population under 50 years of age.
Employing methylation for early detection of cancer can be challenging, as epigenetic markers accumulate along CpG islands with increasing age and over time (35–37). Some methylation changes associated with aging are predictable, such as methylation of ELOVL2, which is considered one of the most robust biomarkers associated with age, and methylation profiles differ between aging and cancer (38,39). Methylation of SEPT9 has not been described in aging profiles, suggesting its specificity to cancer. In addition to CRC, mSEPT9 has been associated with overall survival in head and neck squamous cell carcinoma, cholangiocarcinoma, lymph node status in bladder cancer, non-basal breast cancer, and lung cancer (40–44). Despite the prognostic implications, mSEPT9 has been moved forward as a diagnostic biomarker for CRC (45).
EOCRC is a rising public health problem in the United States and globally (46,47). However, the majority of these younger individuals fall outside of the current screening guidelines. Furthermore, initiating screening by colonoscopy or sigmoidoscopy at earlier ages would place additional burden on an already overwhelmed system (48). Alternative approaches that are more accessible and cost-effective, including blood-based screening, provides an opportunity to fill this notable screening gap. On a population scale, blood-based approaches, such as Epi proColon®, could be used to triage individuals under the age of 50, prior to receiving a colonoscopy.
A limitation to the current Epi proColon® assay is the evaluation of a single blood-based biomarker, though mSEPT9 demonstrated high sensitivity and specificity. Using a multiplexed platform could improve the diagnostic ability of mSEPT9. For example, an evaluation of KISS1R, SEPT9, and CSAD methylation in bladder cancer improved the AUC for predicting lymph node status (AUC=0.68–0.72), compared to KISS1R (AUC=0.67) SEPT9 (AUC=0.58), or CSAD (AUC=0.70) alone (42). Moreover, utilizing a multi-marker blood-based approach may afford an opportunity to simultaneously screen for multiple cancers, however this would require the identification of organ-specific gene or methylation signatures. Multigene or methylation panels may soon allow for this type of approach (49). An additional limitation of note is that EOCRC cases and controls were not collected simultaneously. Cases and controls were, however processed using almost identical protocols and stored without freeze/thaw at −80⁰C until used in the mSEPT9 assay. Therefore, we are confident that all caution was taken to handle the biospecimens the same despite different collection locations. Finally, it is important to note that the current study was limited in its sample size. Though EOCRC incidence is rising, availability of samples remains limited. Even though the EOCRC sample size was small, we were able to observe strong associations between mSEPT9 and different outcomes, which would only be strengthened with a larger study population.
Though the current study provided new evidence in support of the utility of mSEPT9 for the detection of EOCRC, our study had additional limitations to note. First, we conducted a case-control study using archival plasma samples with known cancer outcomes. Though incidence is increasing, EOCRC is infrequent and prospective collection is difficult. The use of archival biospecimens allowed for the current analysis extending mSEPT9 detection into EOCRC. We are confident that the results obtained in our archival cohort reflect what would be measured in a fresh collection, as several studies have demonstrated that circulating tumor DNA remains stable with long-term storage (50) and is concordant with tumor tissue profiles (13). The results of the current study provide support for the institution of a prospective EOCRC cohort (of greater than average risk) to thoroughly evaluate mSEPT9 as a screening tool using freshly collected plasma. Next, the number of available EOCRC cases was limited. The participant pool was restricted to CRC-confirmed individuals under the age of 50 who were treatment naïve, as the effect of chemotherapy and radiation on mSEPT9 is unknown. This limited the number of biospecimens available in the Massachusetts General Hospital biorepository. We additionally did not choose to extend our search for biospecimens to additional biorepositories or archival collections, in an attempt to limit the variability in sample collection and storage. Designing the case-control study in this manner limited the number of included samples. Despite the small sample size, we were able to observe significant differences in mSEPT9 detection between EOCRC cases and healthy controls, as well as significant associations with clinical characteristics. This is the first evaluation of mSEPT9 for the detection of CRC in this population, lending novelty to the analysis despite limited cases. We anticipate that expanding the study, or a subsequent study, to include more cases will strengthen our findings.
The strengths of the study are not without mention. We showed that mSEPT9 could be detected at high sensitivity and specificity in 1 ml of plasma from both EOCRC cases and healthy controls, and in cell line SFCM at increasingly small volumes, highlighting the feasibility of this assay in a clinical setting where limited biospecimens are available. It is possible that mSEPT9 will perform even better in optimal clinical screening settings. Further, we were able to quantitatively measure mSEPT9 in non-CRC cell lines, suggesting that mSEPT9 may have applicability as a biomarker in other cancer types, recent data suggests this to be the case for esophageal, gastric, and liver cancer (51,52). Future studies could focus on a pan-cancer evaluation of mSEPT9 combined with organ specific markers to distinguish the biomarker origin.
In conclusion, we demonstrated that mSEPT9 is a sensitive and specific biomarker for the detection of CRC among individuals age under 50 years. Due to the increasing public health concern of EOCRC, the development of non-invasive screening modalities is warranted. Current research suggests that the detection of mSEPT9 in plasma may help fill this gap. Additional studies are essential to develop and improve EOCRC screening modalities.
SIGNFICANCE.
mSEPT9 may be a novel biomarker for the detection of early-onset colorectal cancer, as it demonstrated high sensitivity and specificity in our study.
ACKNOWLEDGEMENTS
We would like to thank the NIH Clinical Center staff for providing healthy control samples and Travis Kerr for preparing and aliquoting plasma samples. Finally, we would like to thank the patients for participating in this research.
This study was funded in whole or in part with Federal funds from the Chris4Life Research Award from the Colorectal Cancer Alliance (R. Alvarez, M.P. Hitchins) and the ASCO Career Development Award (A. Parikh), the NIH Diabetes and Digestive and Kidney Diseases (K23DK103119) (M. Gala), and the National Institutes of Health, Department of Health and Human Services, under Contract No. 75N91019D00024 (Y. Song).
CONFLICTS OF INTEREST DISCLOSURE STATEMENT
Dr. Parikh has consulted for or received fees from Eli Lilly, Pfizer, Checkmate Pharm, DSMC Roche, Equity C2I Genomics, Research to Institution, PureTech, PMV, Plexxicon, Takeda, BMS, and Novartis. Dr. Gala has equity in New Amsterdam Genomics, Inc.
ABBREVIATIONS
- 95% CI
95% confidence interval
- ACTB
β-actin
- ANOVA
analysis of variance
- AUC
area under the curve
- bisDNA
bisulfite-converted DNA
- CC
Clinical Center
- cfDNA
cell-free DNA
- CRC
colorectal cancer
- Ct
cycle threshold
- EOCRC
early-onset colorectal cancer
- FBS
fetal bovine serum
- FDA
Food and Drug Administration
- FNLCR
Frederick National Laboratory for Cancer Research
- mSEPT9
methylated Septin9
- NPV
negative predictive value
- NIH
National Institutes of Healt
- P/S
penicillin and streptomycin
- PPV
positive predictive value
- ROC
receiver operating curve
- SFCM
serum-free conditioned media
Footnotes
All other authors declare that they have no conflicts of interest. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
DECLARATIONS
Ethics approval and consent to participate
The collection of samples included in this study was approved by the Massachusetts General Hospital Institutional Review Board (IRB 14-046) and the study was deemed exempt from NIH Institutional Review Board approval (IRB000294).
Consent for publication
Not applicable
REFERENCES
- 1.Siegel RL, Torre LA, Soerjomataram I, Hayes RB, Bray F, Weber TK, et al. Global patterns and trends in colorectal cancer incidence in young adults. Gut 2019;68:2179–85. [DOI] [PubMed] [Google Scholar]
- 2.Sanford NN, Giovannucci EL, Ahn C, Dee EC, Mahal BA. Obesity and younger versus older onset colorectal cancer in the United States, 1998–2017. J Gastrointest Oncol 2020;11(1):121–6 doi 10.21037/jgo.2019.12.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nguyen LH, Liu PH, Zheng X, Keum N, Zong X, Li X, et al. Sedentary Behaviors, TV Viewing Time, and Risk of Young-Onset Colorectal Cancer. JNCI Cancer Spectr 2018;2(4):pky073 doi 10.1093/jncics/pky073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zheng X, Hur J, Nguyen LH, Liu J, Song M, Wu K, et al. Comprehensive Assessment of Diet Quality and Risk of Precursors of Early-Onset Colorectal Cancer. J Natl Cancer Inst 2020. doi 10.1093/jnci/djaa164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peterse EFP, Meester RGS, Siegel RL, Chen JC, Dwyer A, Ahnen DJ, et al. The impact of the rising colorectal cancer incidence in young adults on the optimal age to start screening: Microsimulation analysis I to inform the American Cancer Society colorectal cancer screening guideline. Cancer 2018;124(14):2964–73 doi 10.1002/cncr.31543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ladabaum U, Mannalithara A, Meester RGS, Gupta S, Schoen RE. Cost-Effectiveness and National Effects of Initiating Colorectal Cancer Screening for Average-Risk Persons at Age 45 Years Instead of 50 Years. Gastroenterology 2019;157(1):137–48 doi 10.1053/j.gastro.2019.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen JD, Javed AA, Thoburn C, Wong F, Tie J, Gibbs P, et al. Combined circulating tumor DNA and protein biomarker-based liquid biopsy for the earlier detection of pancreatic cancers. Proceedings of the National Academies of Science USA 2017;114(38):10202–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haselmann V, Gebhardt C, Brechtel I, Duda A, Czerwinski C, Sucker A, et al. Liquid Profiling of Circulating Tumor DNA in Plasma of Melanoma Patients for Companion Diagnostics and Monitoring of BRAF Inhibitor Therapy. Clin Chem 2018;64(5):830–42 doi 10.1373/clinchem.2017.281543. [DOI] [PubMed] [Google Scholar]
- 9.Stroun M, Anker P, Maurice P, Lyautey J, Lederrey C, Beljanski M. Neoplastic Characteristics of the DNA Found in the Plasma of Cancer Patients. Oncology 1989;46(5):318–22 doi 10.1159/000226740. [DOI] [PubMed] [Google Scholar]
- 10.Volik S, Alcaide M, Morin RD, Collins C. Cell-free DNA (cfDNA): Clinical Significance and Utility in Cancer Shaped By Emerging Technologies. Mol Cancer Res 2016;14(10):898–908 doi 10.1158/1541-7786.MCR-16-0044. [DOI] [PubMed] [Google Scholar]
- 11.Shen SY, Singhania R, Fehringer G, Chakravarthy A, Roehrl MHA, Chadwick D, et al. Sensitive tumour detection and classification using plasma cell-free DNA methylomes. Nature 2018;563(7732):579–83 doi 10.1038/s41586-018-0703-0. [DOI] [PubMed] [Google Scholar]
- 12.Diaz LA, Jr., Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol 2014;32(6):579–86 doi 10.1200/JCO.2012.45.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nesic M, Bodker JS, Terp SK, Dybkaer K. Optimization of Preanalytical Variables for cfDNA Processing and Detection of ctDNA in Archival Plasma Samples. Biomed Res Int 2021;2021:5585148 doi 10.1155/2021/5585148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoon KA, Park S, Lee SH, Kim JH, Lee JS. Comparison of circulating plasma DNA levels between lung cancer patients and healthy controls. J Mol Diagn 2009;11(3):182–5 doi 10.2353/jmoldx.2009.080098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma L, Du J, Sui Y, Wang S. Clinical significance of plasma free DNA in patients with non-small cell lung cancer. J Int Med Res 2019;47(11):5593–600 doi 10.1177/0300060519872046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Papadopoulou E, Davilas E, Sotiriou V, Georgakopoulos E, Georgakopoulou S, Koliopanos A, et al. Cell-free DNA and RNA in plasma as a new molecular marker for prostate and breast cancer. Annals of the New York Academy of Sciences 2006;1075:235–43 doi 10.1196/annals.1368.032. [DOI] [PubMed] [Google Scholar]
- 17.Scholer LV, Reinert T, Orntoft MW, Kassentoft CG, Arnadottir SS, Vang S, et al. Clinical Implications of Monitoring Circulating Tumor DNA in Patients with Colorectal Cancer. Clin Cancer Res 2017;23(18):5437–45 doi 10.1158/1078-0432.CCR-17-0510. [DOI] [PubMed] [Google Scholar]
- 18.Jin P, Kang Q, Wang X, Yang L, Yu Y, Li N, et al. Performance of a second-generation methylated SEPT9 test in detecting colorectal neoplasm. J Gastroenterol Hepatol 2015;30(5):830–3 doi 10.1111/jgh.12855. [DOI] [PubMed] [Google Scholar]
- 19.Chen CH, Yan SL, Yang TH, Chen SF, Yeh YH, Ou JJ, et al. The Relationship between the Methylated Septin-9 DNA Blood Test and Stool Occult Blood Test for Diagnosing Colorectal Cancer in Taiwanese People. J Clin Lab Anal 2017;31(1) doi 10.1002/jcla.22013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Niedermaier T, Weigl K, Hoffmeister M, Brenner H. Fecal immunochemical tests in combination with blood tests for colorectal cancer and advanced adenoma detection-systematic review. United European Gastroenterol J 2018;6(1):13–21 doi 10.1177/2050640617737004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wasserkort R, Kalmar A, Valcz G, Spisak S, Krispin M, Toth K, et al. Aberrant septin 9 DNA methylation in colorectal cancer is restricted to a single CpG island. BMC Cancer 2013;13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okugawa Y, Grady WM, Goel A. Epigenetic Alterations in Colorectal Cancer: Emerging Biomarkers. Gastroenterology 2015;149(5):1204–25 e12 doi 10.1053/j.gastro.2015.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun J, Fei F, Zhang M, Li Y, Zhang X, Zhu S, et al. The role of (m)SEPT9 in screening, diagnosis, and recurrence monitoring of colorectal cancer. BMC Cancer 2019;19(1):450 doi 10.1186/s12885-019-5663-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Warren JD, Xiong W, Bunker AM, Vaughn CP, Furtado LV, Roberts WL, et al. Septin 9 methylated DNA is a sensitive and specific blood test for colorectal cancer. BMC Medicine 2011;14(9):133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Church TR, Wandell M, Lofton-Day C, Mongin SJ, Burger M, Payne SR, et al. Prospective evaluation of methylated SEPT9 in plasma for detection of asymptomatic colorectal cancer. Gut 2014;63(2):317–25 doi 10.1136/gutjnl-2012-304149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fu B, Yan P, Zhang S, Lu Y, Pan L, Tang W, et al. Cell-Free Circulating Methylated SEPT9 for Noninvasive Diagnosis and Monitoring of Colorectal Cancer. Dis Markers 2018;2018:6437104 doi 10.1155/2018/6437104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun G, Meng J, Duan H, Zhang D, Tang Y. Diagnostic Assessment of septin9 DNA Methylation for Colorectal Cancer Using Blood Detection: A Meta-Analysis. Pathol Oncol Res 2019;25(4):1525–34 doi 10.1007/s12253-018-0559-5. [DOI] [PubMed] [Google Scholar]
- 28.Lamb YN, Dhillon S. Epi proColon((R)) 2.0 CE: A Blood-Based Screening Test for Colorectal Cancer. Mol Diagn Ther 2017;21(2):225–32 doi 10.1007/s40291-017-0259-y. [DOI] [PubMed] [Google Scholar]
- 29.United States Preventive Services Task Force, Bibbins-Domingo K, Grossman DC, Curry SJ, Davidson KW, Epling JW Jr., et al. Screening for Colorectal Cancer: US Preventive Services Task Force Recommendation Statement. JAMA 2016;315(23):2564–75 doi 10.1001/jama.2016.5989. [DOI] [PubMed] [Google Scholar]
- 30.United States Preventive Services Task Force. Screening for Colorectal Cancer: An Evidence Update for the U.S Preventive Services Task Force. In: Services USDoHaH, editor2020. [Google Scholar]
- 31.Hitchins MP, Vogelaar IP, Brennan K, Haraldsdottir S, Zhou N, Martin B, et al. Methylated SEPTIN9 plasma test for colorectal cancer detection may be applicable to Lynch syndrome. BMJ Open Gastroenterol 2019;6(1):e000299 doi 10.1136/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Song L, Wang J, Wang H, Chen Y, Jia J, Guo S, et al. The quantitative profiling of blood mSEPT9 determines the detection performance on colorectal tumors. Epigenomics 2018;10(12):1569–83. [DOI] [PubMed] [Google Scholar]
- 33.Dietrich D, Hasinger O, Liebenberg V, Field JK, Kristiansen G, Soltermann A. DNA methlyation of the homeobox genes PITX2and SHOX2 predicts outcome in non-small-cell lung cancer patients. Diagnostic Molecular Pathology 2012;21(2):93–104. [DOI] [PubMed] [Google Scholar]
- 34.Xie L, Jiang X, Li Q, Sun Z, Quan W, Duan Y, et al. Diagnostic Value of Methylated Septin9 for Colorectal Cancer Detection. Front Oncol 2018;8:247 doi 10.3389/fonc.2018.00247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lau CE, Robinson O. DNA methylation age as a biomarker for cancer. Int J Cancer 2021. doi 10.1002/ijc.33451. [DOI] [PubMed] [Google Scholar]
- 36.Michalak EM, Burr ML, Bannister AJ, Dawson MA. The roles of DNA, RNA and histone methylation in ageing and cancer. Nat Rev Mol Cell Biol 2019;20(10):573–89 doi 10.1038/s41580-019-0143-1. [DOI] [PubMed] [Google Scholar]
- 37.Maugeri A, Barchitta M, Magnano San Lio R, Li Destri G, Agodi A, Basile G. Epigenetic Aging and Colorectal Cancer: State of the Art and Perspectives for Future Research. Int J Mol Sci 2020;22(1) doi 10.3390/ijms22010200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Y, Zhang J, Xiao X, Liu H, Wang F, Li S, et al. The identification of age-associated cancer markers by an integrative analysis of dynamic DNA methylation changes. Sci Rep 2016;6:22722 doi 10.1038/srep22722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Garagnani P, Bacalini MG, Pirazzini C, Gori D, Giuliani C, Mari D, et al. Methylation of ELOVL2 gene as a new epigenetic marker of age. Aging Cell 2012;11(6):1132–4. [DOI] [PubMed] [Google Scholar]
- 40.Schrock A, Leisse A, de Vos L, Gevensleben H, Droge F, Franzen A, et al. Free-Circulating Methylated DNA in Blood for Diagnosis, Staging, Prognosis, and Monitoring of Head and Neck Squamous Cell Carcinoma Patients: An Observational Prospective Cohort Study. Clin Chem 2017;63(7):1288–96 doi 10.1373/clinchem.2016.270207. [DOI] [PubMed] [Google Scholar]
- 41.Branchi V, Schaefer P, Semaan A, Kania A, Lingohr P, Kalff JC, et al. Promoter hypermethylation of SHOX2 and SEPT9 is a potential biomarker for minimally invasive diagnosis in adenocarcinomas of the biliary tract. Clin Epigenetics 2016;8:133 doi 10.1186/s13148-016-0299-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stubendorff B, Wilhelm K, Posselt K, Catto J, Hartmann A, Bertz S, et al. A three-gene methylation marker panel for the nodal metastatic risk assessment of muscle-invasive bladder cancer. J Cancer Res Clin Oncol 2019;145(4):811–20 doi 10.1007/s00432-018-02829-4. [DOI] [PubMed] [Google Scholar]
- 43.Matsui S, Kagara N, Mishima C, Naoi Y, Shimoda M, Shimomura A, et al. Methylation of the SEPT9_v2 promoter as a novel marker for the detection of circulating tumor DNA in breat cancer patients. Oncology Reports 2016;36(4):2225–35. [DOI] [PubMed] [Google Scholar]
- 44.Powrozek T, Krawczyk P, Kucharczyk T, Milanowski J. Septin 9 promoter region methylation in free circulating DNA-potential role in noninvasive diagnosis of lung cancer: preliminary report. Med Oncol 2014;31(4):917 doi 10.1007/s12032-014-0917-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.deVos T, Molnar B. Screening for Colorectal Cancer Based on the Promoter Methylation Status of the Septin 9 Gene in Plasma Cell Free DNA. Journal of Clinical Epigenetics 2017;03(01) doi 10.21767/2472-1158.100040. [DOI] [Google Scholar]
- 46.Loomans-Kropp HA, Umar A. Increasing Incidence of Colorectal Cancer in Young Adults. J Cancer Epidemiol 2019;2019:9841295 doi 10.1155/2019/9841295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dairi O, Anderson JC, Butterly LF. Why is colorectal cancer increasing in younger age groups in the United States? Expert Rev Gastroenterol Hepatol 2021:1–10 doi 10.1080/17474124.2021.1876561. [DOI] [PubMed] [Google Scholar]
- 48.Seeff LC, Richards TB, Shapiro JA, Nadel MR, Manninen DL, Given LS, et al. How many endoscopies are performed for colorectal cancer screening? Results from CDC’s survey of endoscopic capacity. Gastroenterology 2004;127(6):1670–7 doi 10.1053/j.gastro.2004.09.051. [DOI] [PubMed] [Google Scholar]
- 49.Aravanis AM, Lee M, Klausner RD. Next-Generation Sequencing of Circulating Tumor DNA for Early Cancer Detection. Cell 2017;168(4):571–4 doi 10.1016/j.cell.2017.01.030. [DOI] [PubMed] [Google Scholar]
- 50.Ward Gahlawat A, Lenhardt J, Witte T, Keitel D, Kaufhold A, Maass KK, et al. Evaluation of Storage Tubes for Combined Analysis of Circulating Nucleic Acids in Liquid Biopsies. Int J Mol Sci 2019;20(3) doi 10.3390/ijms20030704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li B, Huang H, Huang R, Zhang W, Zhou G, Wu Z, et al. SEPT9 Gene Methylation as a Noninvasive Marker for Hepatocellular Carcinoma. Dis Markers 2020;2020:6289063 doi 10.1155/2020/6289063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Song L, Chen Y, Gong Y, Wan J, Guo S, Liu H, et al. Opportunistic screening and survival prediction of digestive cancers by the combination of blood mSEPT9 with protein markers. Ther Adv Med Oncol 2020;12:1758835920962966 doi 10.1177/1758835920962966. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and/or analyzed during the current study are not publicly available due to the sensitivity of the data but are available from the corresponding author upon reasonable request.