Abstract
Chimeric antigen receptor (CAR) T-cell therapies exploit facile antibody-mediated targeting to elicit useful immune responses in patients. This work directly compares binding profiles of CAR and αβ T-cell receptors (TCR) with single cell and single molecule optical trap measurements against a shared ligand. DNA-tethered measurements of peptide-major histocompatibility complex (pMHC) ligand interaction in both CAR and TCR exhibit catch bonds with specific peptide agonist peaking at 25 and 14 pN, respectively. While a conformational transition is regularly seen in TCR–pMHC systems, that of CAR–pMHC systems is dissimilar, being infrequent, of lower magnitude, and irreversible. Slip bonds are observed with CD19-specific CAR T-cells and with a monoclonal antibody mapping to the MHC α2 helix but indifferent to the bound peptide. Collectively, these findings suggest that the CAR–pMHC interface underpins the CAR catch bond response to pMHC ligands in contradistinction to slip bonds for CARs targeting canonical ligands.
Graphical Abstract
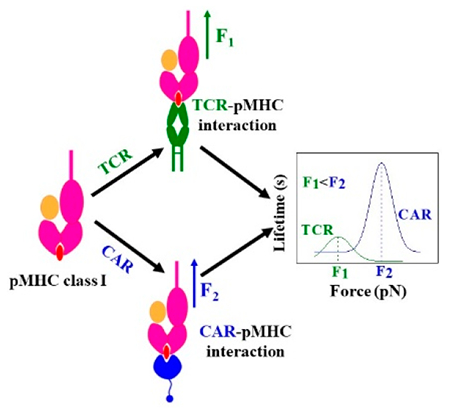
T-cells are crucial effectors and regulators of the adaptive immune system. Chimeric antigen receptor (CAR) T-cell therapies harness the response of conventional T-cells to fight disease.1–6 T-cells respond to nonself peptides (diseased, or virus-based) cradled within a major histocompatibility complex molecule (MHC) through force-mediated binding to an αβ heterodimer.7,8 Each T-cell displays tens of thousands of copies of a clone-specific αβ T-cell receptor (TCR) heterodimer noncovalently linked in an eight-member complex with the dimeric CD3 molecules (CD3εγ, CD3εδ, and CD3ζζ) which transduce the signal. (Here, all TCR designations refer to αβ TCRs unless indicated otherwise.) In contrast, CARs synthetically link an antigen-binding single chain variable fragment (scFv) domain derived from an antibody to the intracellular signaling domain of one such partnering molecule, CD3ζ. Subsequent-generation CARs incorporate additional signaling domains from costimulatory molecules including CD28 or 4-1BB.4,9 While αβ T-cells have been shown to harness physical bioforces to boost their sensitivity, the impact of force and mechanosensing on CAR T-cells has not been established.
Mechanical force was first implicated in T-cell triggering by Kim et al.7 where an optically trapped bead coated with peptide-bound MHC (pMHC) was used to apply an oscillating tangential mechanical force to a T-cell followed by continuous monitoring of intracellular calcium flux. Mechano-sensitivity of TCR was further confirmed by other groups using micropipette and biomembrane force probes (BFP),10,11 and the bond type was classified as a catch bond, a bond that strengthens with force. Catch bonds were observed in single molecule assays where the TCR–pMHC interaction was confined between a cover glass surface and a DNA-tethered bead, and the corresponding quaternary structural change of TCR was identified.12,13 Catch bonds were also observed in measurements using single DNA tethers to a single TCR on the T-cell surface.12 As few as two pMHC molecules with 10–20 pN shear force were optimal for triggering T-cells.14,15 Thus, single molecule profiles and cell activation studies performed at physical and chemical thresholds showed strong correlation between catch bond responses and acuity. The impact of mechanical force not only induced catch bond lifetimes but also elicited conformational change and molecular motor activation. Whether such mechanosensing contributes to CAR T-cell antigen interaction has not yet been established.
This study investigates CAR T-cell antigen recognition through similar optical trap force-based assays at the single cell and single molecule levels. We utilize two second-generation CARs with two different scFv that recognize Epstein–Barr virus LMP2 (CLGGLLTMV) and LMP1 (YLLEMLWRL) nonapeptides bound to the class I MHC complex HLA–A*0201 (LMP2/A*0201 and LMP1/A*0201),16–18 with CD28 and CD3ζ signaling domains (scFv-CD28-CD3ζ). We also employ a similar CD19 CAR which targets a B-cell antigen, CD19. In head-to-head experiments we compare three CARs to a TCR with the same LMP2 peptide specificity, all four expressed on CD8− Jurkat T-cells at similar levels. The measurements include (i) single molecule measurement on isolated single cells (SMSC) (Figure 1A) to probe individual bond force–lifetime profile (catch or slip), conformational transition (CT), and transition probability and (ii) single molecule (SM) (Figure 1B) experiments with purified LMP2 CAR antibody (LMP2 CAR antibody refers to the purified full-length monoclonal antibody used to generate the scFv ligand binding moiety incorporated into the LMP2 CAR which was assayed by SMSC) and control antibody–pMHC interactions measured directly with a pMHC-tethered bead against surface bound antibody. Understanding the mechanism aspect of CAR T-cell antigen interaction may help harness mechanosensing features used in native T-cells and improve future CAR T-cell therapies.
Figure 1.

Cartoon details of SMSC and SM assays. (A) SMSC is designed to measure the lifetime of a single αβ TCR/CAR-pMHC bond on the surface of a T-cell as a function of force. A 3500 bp DNA strand is connected to an optically trapped bead through a dig–antidig interaction. A single pMHC molecule which interacts with the TCR/CAR receptor is linked to the other end of the DNA strand by half of an antibody. The inset in (A) shows a photomicrograph (top view) of a T-cell interacting with a pMHC-coated bead during SMSC (yellow box). (B) SM assay to study the interaction between purified CAR/control antibody and pMHC on the cover glass surface. CAR/control antibody was bound to the PEG coverslip through disuccinimidyl suberate (DSS) and biotinylated protein G and streptavidin (SA). An antidigoxigenin-coated bead-3500 bp DNA–pMHC slurry was used to form tethers. A typical trap stiffness of 0.1–0.2 pN/nm was used for the experiment. Bead displacement from the trap center was measured until bond rupture. Force was calculated by multiplying ΔX (bead displacement from trap center) and trap stiffness. Cartoons are not to scale.
We sought to characterize the nature of the CAR–pMHC bonds with direct comparison to the TCR–pMHC bond at the single molecule level. TCR and CARs were produced and displayed on Jurkat76 cells as described19 (Figures S1–S4). The SMSC assay permits direct lifetime measurement of a single TCR/CAR-pMHC bond as a function of force by using optical tweezers to pull directly on a bead tethered via DNA linker to pMHC bound to a single receptor decorating the surface of a coverslip-affixed cell (Figure 1A).20,21 Note that pMHC is linked to the DNA strand by a half-antibody to ensure an individual interaction. Bond lifetime as a function of force is mapped along with conformational transition magnitude and probability. Representative SMSC traces of TCR and three CARs are shown in Figure 2A,B, Figure S5, and Figure S6, and the numbers of traces at different force ranges are given in Table S1. The TCR and two CARs (LMP2 and LMP1) exhibited catch bond profiles reaching maxima at 10–15 and 25 pN, respectively, with LMP2 CAR–LMP2/A*0201 and LMP1 CAR–LMP1/A*0201 exhibiting a peak bond lifetime (~28 and ~31 s, respectively) greater than that observed for TCR-LMP2/A*0201 (7 s) (Figure 2C). Similar catch bond behavior has been observed for TCR–pMHC conjugates by multiple groups.12,22 In striking contrast, the CD19 CAR–CD19 interaction formed a slip bond (Figure 2C) typical of antibody–antigen interactions23–25 with a peak lifetime of ~43 s. In control experiments with an irrelevant influenza M1 peptide (GILGFVFTL; a peptide different than the specific relevant LMP peptides), bound to the same MHC allele (M1/A*0201), stable tether formation to the same TCR or CAR T-cells (LMP2 and LMP1) was not observed (1/16, 1/18, and 1/21 attempts, respectively). Tether formation was not observed by using biotin–BSA as a control substrate with the CD 19 CAR system.
Figure 2.
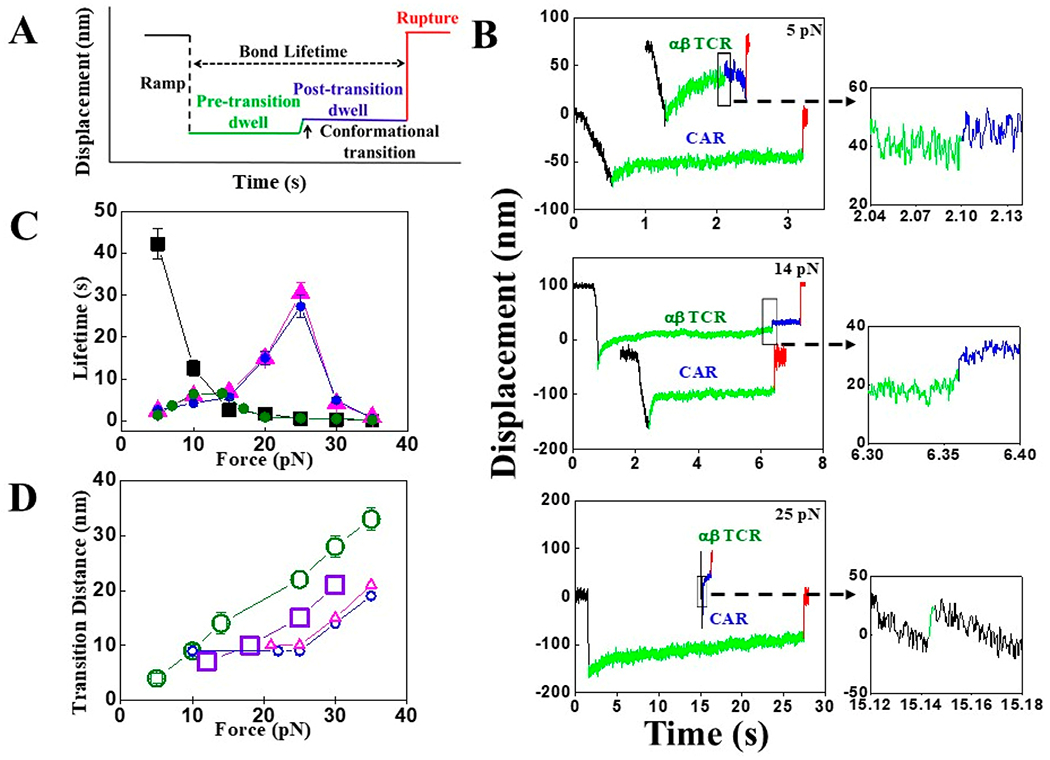
SMSC results. (A) Cartoon of a typical SMSC trace has four distinguishable regions: force loading “Ramp” is depicted as dotted black, “Pre-transition” as green, “Post-transition” as blue, and bond “Rupture” as red. “Bond Lifetime” and “Conformational transition” are shown by dotted black and solid black arrows, respectively. (B) Representative traces of single TCR and LMP2 CAR reacted with LMP2/A*0201 at three different forces: 5 pN (top), 14 pN (middle), and 25 pN (bottom). Conformational transition (indicated by black rectangles) for TCR appears in between “Ramp” and bond “Rupture” at 5 and 14 pN, whereas the transition is found in the “Ramp” of the third trace at 25 pN (bottom). (C) Bond lifetime plotted as a function of force for single TCR–LMP2/A*0201 (filled olive-green circles), LMP2 CAR–LMP2/A*0201 (filled blue circles), LMP1 CAR–LMP1/A*0201 (filled magenta triangles), and CD19 CAR-CD19 (filled black squares) interactions, indicating catch bond formation in all cases except the CD19 CAR. Error bars depict SEM. (D) Comparison of transition distance vs force plot for TCR (open olive-green circles), mouse TCR (open violet squares from Das et al.12), LMP2 CAR (five individual open blue circles), and LMP1 CAR (four individual open magenta triangles) interacting against their specific peptide. Conformational transitions are found in 29 (out of 37), 5 (out of 31), and 4 (out of 34) traces of TCR and LMP2 CAR and LMP1 CAR, respectively. Error bars depict SEM.
The SMSC assay reveals the magnitude and probability of the conformational transition typically seen in TCR–pMHC linkages. Conformational transitions are identified as sudden displacements of the bead toward the center of the trap within the displacement vs time trace. For TCR–pMHC systems, the transition is always seen in high-resolution single molecule measurements, even for irrelevant peptides bound to the same MHC as the cognate peptide.12,13 The magnitude of the transition distance also trends with potency of the agonist. While the transition formally can occur anywhere along the loading pathway, there are clues indicating that the location is within the constant (C) region of the TCR. Reagents such as H57 Fab stabilize the TCR–pMHC bond and also block the transition except at high force. Given that H57 binds an epitope on TCR–Cβ,26 the transition likely involves the TCR.12 In our studies performing measurements of TCR–LMP2/A*0201 conjugates, a similar conformational transition (28 nm at 30 pN) in TCR was observed with a high percentage of traces showing transitions (29 of 37 traces) (Figure 2D, Figure S7, and Table S2). Traces without a clear transition were typically those performed at lower force. We also observed conformational transitions in both LMP2 CAR–LMP2/A*0201 and LMP1 CAR–LMP1/A*0201 conjugates, but these transitions were of lower probability, 16% (5/31 traces) and 12% (4/34 traces), respectively, and of different magnitudes (19 and 21 nm at 35 pN) (Figure 2D, Figure 3, and Figure S5).
Figure 3.
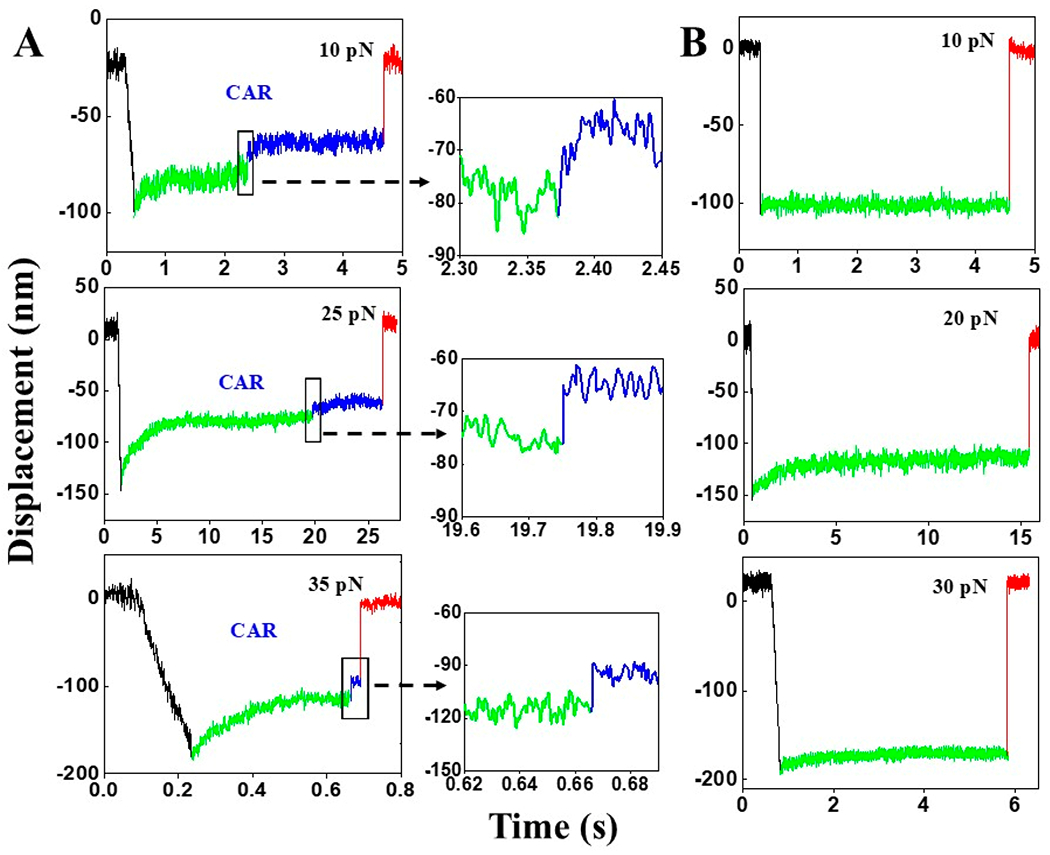
SMSC results of LMP2 CAR. (A) Representative SMSC traces (10, 25, and 35 pN) of the LMP2 CAR–LMP2/A*0201 interaction with clear conformational transition (indicated by black rectangles). 16% of the total traces contain such a transition. (B) Examples of the dominant population of traces (84%) which do not contain conformational transition.
To further probe this transition at high resolution, we prepared a single molecule (SM) assay with purified LMP2 CAR antibody bound to a PEG coverslip through biotin-tagged protein G. Tethered beads were formed through a 3500 bp strand of DNA linked to LMP2/A*0201 through a half-antibiotin (Figure 1B). Tethered beads were identified by eye and centered in the detection laser. Beads were pulled by translating the slide relative to a fixed trap, and bead positions were measured until bond rupture. Beads were calibrated for position sensing after the bond rupture. Figure 4A shows typical SM traces of the LMP2 CAR antibody–LMP2/A*0201 interaction at 10, 25, and 35 pN. LMP2 CAR antibody–LMP2/A*0201 exhibits a catch bond profile with a peak lifetime of ~21.4 s centered at 25 pN (Figure 4B), consistent with the SMSC results above (Figures 2C and 4B). Note that the “TCR-like” LMP2 CAR antibody binds to both the peptide and the HLA-A*0201 peptide binding groove (most likely both α1 and α2) (Figure 4C).27,28 Conformational transitions were also observed in traces that sustained 15–35 pN force; yet no transition was observed below 15 pN (Figure 4A and Table S2). Based on our SMSC and SM results, the transition appears to require a threshold force (Figure 4D). A shift in the transition distance as a function of force occurred around 25 pN, which was also seen in the SMSC assay on CAR–pMHC interactions (Figures 2D and 4D). The probability of observing the conformational transition was 53% overall (30/56 traces) (Figure 4D). While the LMP2 CAR–LMP2/A*0201 system produced visible tethers automatically in the field of view, control experiments using LMP2 CAR antibody–M1/A*0201 (irrelevant peptide) did not produce any tethers. Attempts to form tethers actively with this nonspecific peptide by trapping beads and rubbing them against the surface produced one tether out of 32 attempts.
Figure 4.
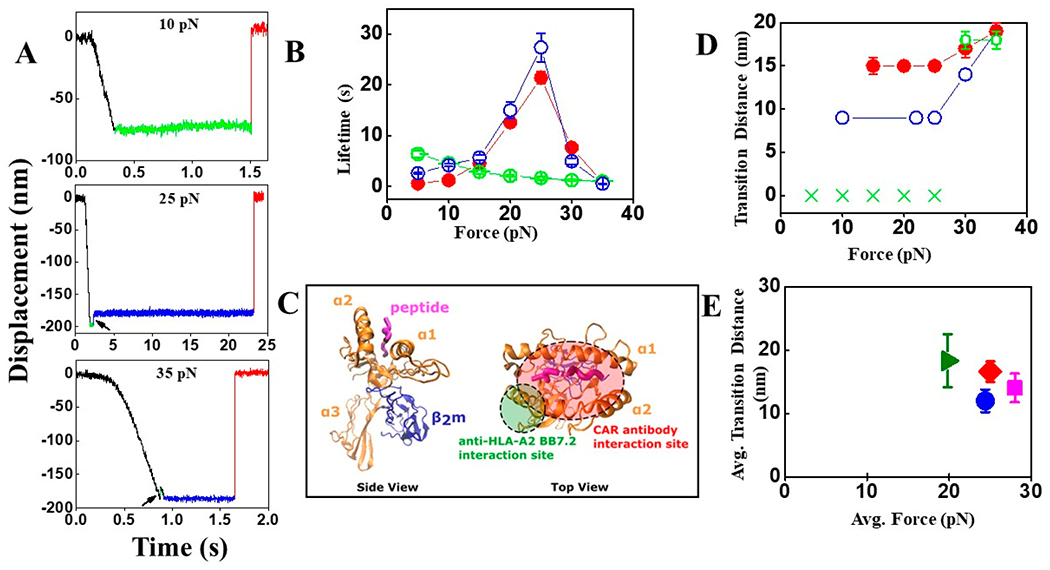
SM results of LMP2 CAR antibody/antihuman HLA-A2-LMP2/A*0201 interactions. (A) Representative SM traces of LMP2 CAR antibody–LMP2/A*0201 (10, 25, and 35 pN) interaction. Traces containing conformational transitions at 25 and 35 pN are indicated by black arrows. “Pre-transition” and “Post-transition” dwells are shown in green and blue colors, respectively. (B) Bond lifetimes of single LMP2 CAR antibody–LMP2/A*0201 (filled red circles) and antihuman HLA-A2 antibody (BB7.2)–LMP2/A*0201 (open green circles) are plotted as a function of force, showing catch and slip bond profiles, respectively. The open blue circles are SMSC results for the LMP2 CAR–LMP2/A*0201 interaction. Error bars depict SEM. (C) Probable interaction sites of CAR antibody (red ellipse) and antihuman HLA-A2 (BB7.2) (green circle) with HLA-A*0201. (D) Transition distance vs force plot for LMP2 CAR antibody–LMP2/A*0201 (filled red circles) and antihuman HLA-A2 antibody (BB7.2)–LMP2/A*0201 (open green circles and green crosses) interactions where transitions are found in 30 out of 56 and 10 out of 46 traces, respectively. The green crosses indicate no transition found. The open blue circles are SMSC results for the LMP2 CAR–LMP2/A*0201 interaction. Error bars depict SEM. (E) Plot of average transition distance vs average force for TCR (using SMSC; olive-green triangle), LMP2 CAR (using SMSC; blue circle), LMP1 CAR (using SMSC; magenta square), and CAR antibody (using SM; red rhombus). Error bars depict SEM.
We next sought to measure binding of an antibody recognizing an MHC region separate from the peptide binding pocket to compare to the LMP2 CAR antibody. Anti-HLA-A2 antibody BB7.2 primarily binds the α2-helix of HLA-A*0201, independently of the sequence of the presented peptide (Figure 4C).29 The affinity of TCR for pMHC complex is typically 1–100 μM.30 However, the two CARs (LMP2 and LMP1) exhibit higher affinities of 6.98 and 1.85 nM, respectively.16 The BB7.2 antibody is also expected to have an affinity of 14.9 nM.31 In our study, BB7.2 exhibited lower affinity than the LMP2 CAR antibody corroborates previous reports. The conformational transition was also observed at high forces (30 and 35 pN only) for BB7.2–LMP2/A*0201 (10/46 traces). The transition distances are similar to those observed previously for the LMP2 CAR antibody–LMP2/A*0201 pair (Figure 4D), suggesting the transition originates in the MHC domain. The slip bond seen here suggests that catch bond-like profile in the LMP2 CAR antibody–LMP2/A*0201 interaction is a feature of the antibody–peptide–MHC interface.
To further ascertain whether the transitions in the TCR and CAR systems have the same origin, we compared the average force vs average transition distance of TCR and CARs using SMSC and LMP2 CAR antibody using our SM assay with LMP2/A*0201 (Figure 4E). For TCR, average force and transition distances were 19.8 pN and 18.3 nm, respectively. For LMP2 CAR, LMP1 CAR, and LMP2 CAR antibody, average forces were higher, ranging from 25 to 28 pN, and displacements lower, ranging from 12 to 16 nm (Figure 4E). Thus, for each transition distance observed, a higher force was needed to achieve such a transition in the CAR system compared to TCR. The two systems showed different profiles of time to transition and frequency (Figure S8). While the load pathways in these systems have different compliances, this result along with difference in probability and temporal profile corroborates that the transitions are distinct. CAR appears to have a higher force window than TCR. We propose that lower probability conformational transition in pMHC, similar to that seen previously,32 may be the source of the transition in the CAR–pMHC response.
In summary, by employing SMSC and SM force spectroscopy assays, we directly compared receptor binding of TCR and CAR T-cells specific for the same pMHC. Surprisingly, LMP2 and LMP1 CAR–pMHC conjugate exhibited catch bonds, whereas the CD19 CAR–CD19 pair showed a slip bond. While typical TCR–pMHC catch bonds are centered at 10–20 pN,12 the CAR–pMHC catch bonds peaked at higher force, ~25 pN, and showed longer peak lifetimes of ~31 s. It is unclear how the higher force thresholds and longer lifetimes impact cell triggering, but such force and lifetime profiles may be advantageous in synthetic CAR systems to properly interface with T-cell signaling machinery.33
Both CAR (LMP2 and LMP1) and TCR systems show conformational transitions which were absent in the CD19 CAR–CD19 pair. However, the probability (16% and 12%) and magnitude (19 and 21 nm at 35 pN) for the CAR transition suggest the transition occurs at a different location than that seen in the TCR. Studies found that H57 Fab, which binds TCR-Cβ, not only blocks the transition but also typically blocks cell activation, suggesting that the conformational transition is located within C-domains of the TCR.7 Given that antibodies typically do not show transitions, it is likely that conformational change seen in the CAR–pMHC systems originates in the MHC domain. A similar conformational change was previously described in pMHC by using BFP, with less than 20% probability.32 This work included elegant controls where pMHC was conformationally closed by using cysteine bonds. The CAR–pMHC transition appears to have a threshold of 25 pN, below which only a constant displacement vs force was observed. Another indicator that the transition is different is that the one seen in the TCR (and pre-TCR) is reversible,13 yet no evidence of a reversible transition has been seen in the CAR–pMHC systems. A study comparing a wild-type γδ TCR to a chimera γδ/αβ chimeric TCR containing an identical ligand binding variable domain module shows a transition of high probability only for the chimera, which has the C-domains of αβ rather than those of γδ.34 This work further supports the C-domain as the origin of the conformational transition. We believe that transitioning and not just bond lifetime per se is important to drive signaling. Consistent with this notion, H57 typically blocks transitions in TCRs and associated T-cell activation while nevertheless substantially increasing bond lifetime.14,15 What is the purpose of an additional higher force, irreversible transition in pMHC? Perhaps it may redundantly participate in signaling by providing an additional opportunity for mechanotransduction involving coreceptors or, alternatively, impact antigen presenting cell function in ways yet to be elucidated.
When conventional T-cells activate, membrane-sequestered immunoreceptor tyrosine-based activation motif (ITAM) domains are released and phosphorylated to drive chemical signaling. It is thought that mechanical information is communicated through the TCR complex, leading to a destabilization/shift in the transmembrane domain complex organization. This is facilitated through a transition in the TCRα transmembrane domain,35 other TCR subunits, and vicinal lipids, which collectively dissociate TCR complex members. This process can be envisaged as a solid-to-fluid phase transition assisted by transport and motion of membrane-bound molecules that enhance or dampen signaling.36 Optical trap and traction force microscopy-based triggering near threshold limits showed coupling and activation of acto-myosin-based transport.14,37 Motility led to triggering when it was in a direction that would maintain the force within an optimal window of 10–15 pN where reversible transitioning occurs. The CAR–pMHC systems not only lacked this transition, but the few transitions seen in CAR–pMHC occurred above this optimal force threshold window for the TCR–pMHC interaction.
We have found similar catch bond profile and conformational transition in SM assay using LMP2 CAR antibody and LMP2/A*0201. Control experiments with BB7.2 corroborate the importance of the LMP2 CAR antibody and LMP2/A*0201 interaction pair for catch bond formation, as BB7.2 exhibits a slip bond response. As the LMP2 CAR antibody binds to the peptide and α1 and α2 helices of HLA-A*0201 while BB7.2 binds to the α2 domains of HLA-A*0201, we expect force-induced extension of the linker between the α1–α2 antigen presenting platform and α3 domains followed by separation of α1–α2 and α3–β2m domains, as observed earlier by using molecular dynamics simulation and single molecule biophysical approaches.32 The extension (13 nm at 15 pN) observed in silico32 is very close to our empirical measurements (15 nm at 15 pN). These studies suggest there are multiple locations for conformational change, including a high probability lower force (10–15 pN) conformational transition within the TCR, likely involving TCRβ, and a second lower probability but slightly higher force (20 pN) transition in pMHC.
Observed differences in bond lifetimes as well as transition distances, between CAR (SMSC) and CAR antibody (SM) (Figures 2C,D and 4B,D), are likely consequent to differences in load path connectivity and how the assays are executed, as discussed previously.34 Such differences may explain the lower frequency of conformational transition in SMSC than SM (16% and 12% vs 53%, respectively).
A recent molecular dynamics study suggests that force helps to organize the interface between a TCRαβ and its pMHC ligand.38 The peptide itself constitutes a “key” that with the CDR3α and CDR3β loops prominently serving to stabilize the dynamic bonding. Simulations involving the V module interface with constant domains present or absent suggest that TCRαβ interdomain mismatches are corrected with force application. In fact, the strongest pMHC binding was seen in the most compliant systems where only TCRαβ variable domains were present. Given that these CAR–pMHC systems exhibit a catch bond, it is likely that motions of MHC and the peptide underpin some of the interfacial strengthening and organization observed and that the CAR scFv somehow capitalizes on this phenomenon. Overall, through investigation of the biophysical features of LMP1 CAR, LMP2 CAR, and LMP2 CAR monoclonal antibody, all specific for their respective peptides cradled within MHC vs CD19 CAR and BB7.2 both lacking pMHC specificity, we have shown that the CAR–pMHC systems form catch bonds whereas the other systems exhibit the expected slip bonds of typical antibody–ligand interactions. It may be important that pMHC specific CARs can produce a catch bond response. That said, other CARs such as those CD19 systems work in the absence of catch bonds, presumably utilizing their high affinity scFv to bind non-MHC ligand and via cellular shear forces release CD3ζ ITAMs from the inner leaflet of the plasma membrane for subsequent tyrosine kinase-mediated phosphorylation and T-cell triggering. In this regard, the higher affinity of TCR γδ heterodimers for their natural ligands (being distinct from pMHC) and the inability to mechanosense,34 features shared in common with Fab and scFv fragments, suggest that γδ CARs may serve as facile receptors for immunotherapy.
Recent studies have identified important aspects of CAR T-cells. It has been reported that CAR T-cells form nonclassical immunological synapses, unlike the typical bulls-eye signature of central, peripheral, and distal supramolecular activation complexes that are observed between αβ T cells and their targets.39 Ligand-induced receptor dimerization was reported for CAR T-cells targeting soluble ligands.40,41 CAR signaling was found to be unique and different from that of TCRs and even distinct among various CAR constructs.9,42 Wu and coworkers compared bulk activation of TCR T-cells and CAR T-cells targeting LMP2/A*0201,19 revealing that although CAR binds the monomeric antigen due to its high affinity, it requires multimeric antigen for activation. CAR calcium flux reached and sustained a plateau, whereas for TCR it decreased after reaching a peak. Despite sustained calcium flux for CAR, its magnitude was much lower than TCR. Although CD8 coreceptor was recruited to the CAR synapse, its role in calcium flux was not significant, unlike for the TCR.19 Switchable dual receptor CAR-T cells (sdCAR-T) were found to be more effective than conventional CAR T-cells against solid tumors.43 Redirection and regulation of CAR T-cell activity were shown by using a bifunctional small molecule “switch”.44 Therefore, extensive investigations are required to unveil the mystery of CAR T-cells for the betterment of CAR T-cell therapy.
In conclusion, using single cell and single molecule optical trap measurements, we have identified several distinct features of TCR and CAR interactions with their specific ligands that can direct future single molecule studies to improve CAR T-cell therapies. Defining mechanobiology requirements for CAR-based therapies more broadly can serve to identify signaling features that will benefit the engineering and design of next-generation systems.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge all lab members of the Lang and Gascoigne groups for their help and support during the work. This work was supported by grants from the NIH (R01 AI136301 and P01 AI143565) to M.J.L. and by the Singapore Ministry of Health’s National Medical Research Council under its OFIRG19nov-0066 and by Ministry of Education under NUHSRO/2020/110/T1/SEED-MAR/06 to N.R.J.G.
Footnotes
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jpclett.1c02240.
Materials and methods, flow cytometry results (Figures S1–S4), SMSC traces for LMP1 CAR (Figure S5) and CD19 CAR (Figure S6) systems, conformational transition analysis (Figure S7), frequency of transition plots (Figure S8), and two tables indicating the number of SMSC and SM traces (Table S1) and conformational transition (Table S2) in TCR and CAR systems (PDF)
Complete contact information is available at: https://pubs.acs.org/10.1021/acs.jpclett.1c02240
The authors declare no competing financial interest.
Contributor Information
Debasis Banik, Department of Chemical and Biomolecular Engineering, Vanderbilt University, Nashville, Tennessee 37235, United States.
Maryam Hamidinia, Translational Immunology Research Program, Yong Loo Lin School of Medicine, Translational Cancer Research Program, Yong Loo Lin School of Medicine, and Department of Microbiology and Immunology, Yong Loo Lin School of Medicine, National University of Singapore, Singapore 117545, Singapore.
Joanna Brzostek, Translational Immunology Research Program, Yong Loo Lin School of Medicine, Translational Cancer Research Program, Yong Loo Lin School of Medicine, and Department of Microbiology and Immunology, Yong Loo Lin School of Medicine, National University of Singapore, Singapore 117545, Singapore.
Ling Wu, Translational Immunology Research Program, Yong Loo Lin School of Medicine, Translational Cancer Research Program, Yong Loo Lin School of Medicine, and Department of Microbiology and Immunology, Yong Loo Lin School of Medicine, National University of Singapore, Singapore 117545, Singapore.
Hannah M. Stephens, Department of Chemical and Biomolecular Engineering, Vanderbilt University, Nashville, Tennessee 37235, United States
Paul A. MacAry, Translational Immunology Research Program, Yong Loo Lin School of Medicine, Translational Cancer Research Program, Yong Loo Lin School of Medicine, and Department of Microbiology and Immunology, Yong Loo Lin School of Medicine, National University of Singapore, Singapore 117545, Singapore
Ellis L. Reinherz, Laboratory of Immunobiology, Dana-Farber Cancer Institute, Boston, Massachusetts 02115, United States; Department of Medical Oncology, Dana-Farber Cancer Institute and Department of Medicine, Harvard Medical School, Boston, Massachusetts 02115, United States
Nicholas R. J. Gascoigne, Translational Immunology Research Program, Yong Loo Lin School of Medicine, Translational Cancer Research Program, Yong Loo Lin School of Medicine, and Department of Microbiology and Immunology, Yong Loo Lin School of Medicine, National University of Singapore, Singapore 117545, Singapore
Matthew J. Lang, Department of Chemical and Biomolecular Engineering, Vanderbilt University, Nashville, Tennessee 37235, United States; Department of Molecular Physiology and Biophysics, Vanderbilt University School of Medicine, Nashville, Tennessee 37235, United States.
REFERENCES
- (1).Brown CE; Mackall CL CAR T Cell Therapy: Inroads to Response and Resistance. Nat. Rev. Immunol 2019, 19 (2), 73–74. [DOI] [PubMed] [Google Scholar]
- (2).Yip A; Webster RM The Market for Chimeric Antigen Receptor T Cell Therapies. Nat. Rev. Drug Discovery 2018, 17 (3), 161–162. [DOI] [PubMed] [Google Scholar]
- (3).Lindner SE; Johnson SM; Brown CE; Wang LD Chimeric Antigen Receptor Signaling: Functional Consequences and Design Implications. Sci. Adv 2020, 6 (21), eaaz3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).June CH; O’Connor RS; Kawalekar OU; Ghassemi S; Milone MC CAR T Cell Immunotherapy for Human Cancer. Science 2018, 359 (6382), 1361–1365. [DOI] [PubMed] [Google Scholar]
- (5).Li L; Yang Z; Chen X Recent Advances in Stimuli-Responsive Platforms for Cancer Immunotherapy. Acc. Chem. Res 2020, 53 (10), 2044–2054. [DOI] [PubMed] [Google Scholar]
- (6).Shi Y; Lammers T Combining Nanomedicine and Immunotherapy. Acc. Chem. Res 2019, 52 (6), 1543–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Kim ST; Takeuchi K; Sun Z-YJ; Touma M; Castro CE; Fahmy A; Lang MJ; Wagner G; Reinherz EL The αβ T Cell Receptor is an Anisotropic Mechanosensor. J. Biol. Chem 2009, 284 (45), 31028–31037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Kim ST; Shin Y; Brazin K; Mallis RJ; Sun Z-YJ; Wagner G; Lang MJ; Reinherz EL TCR Mechanobiology: Torques and Tunable Structures Linked to Early T Cell Signaling. Front. Immunol 2012, 3 (76), 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Wu L; Wei Q; Brzostek J; Gascoigne NRJ Signaling from T Cell Receptors (TCRs) and Chimeric Antigen Receptors (CARs) on T Cells. Cell. Mol. Immunol 2020, 17 (6), 600–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Li Y-C; Chen B-M; Wu P-C; Cheng T-L; Kao L-S; Tao M-H; Lieber A; Roffler SR Cutting Edge: Mechanical Forces Acting on T Cells Immobilized via the TCR Complex Can Trigger TCR Signaling. J. Immunol 2010, 184 (11), 5959–5963. [DOI] [PubMed] [Google Scholar]
- (11).Husson J; Chemin K; Bohineust A; Hivroz C; Henry N Force Generation upon T Cell Receptor Engagement. PLoS One 2011, 6 (5), e19680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Das DK; Feng Y; Mallis RJ; Li X; Keskin DB; Hussey RE; Brady SK; Wang J-H; Wagner G; Reinherz EL; Lang MJ; et al. Force-Dependent Transition in the T-Cell Receptor β-Subunit Allosterically Regulates Peptide Discrimination and pMHC Bond Lifetime. Proc. Natl. Acad. Sci. U. S. A 2015, 112 (5), 1517–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Das DK; Mallis RJ; Duke-Cohan JS; Hussey RE; Tetteh PW; Hilton M; Wagner G; Lang MJ; Reinherz EL Pre-T Cell Receptors (pre-TCRs) Leverage vβ Complementarity Determining Regions (CDRs) and Hydrophobic Patch in Mechanosensing Thymic Self-Ligands. J. Biol. Chem 2016, 291 (49), 25292–25305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Feng Y; Brazin KN; Kobayashi E; Mallis RJ; Reinherz EL; Lang MJ Mechanosensing Drives Acuity of αβ T-Cell Recognition. Proc. Natl. Acad. Sci. U. S. A 2017, 114 (39), E8204–E8213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Feng Y; Reinherz EL; Lang MJ αβ T Cell Receptor Mechanosensing Forces out Serial Engagement. Trends Immunol. 2018, 39 (8), 596–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Sim ACN; Too CT; Oo MZ; Lai J; Eio MY; Song Z; Srinivasan N; Tan DAL; Pang SW; Gan SU; et al. Defining the Expression Hierarchy of Latent T-Cell Epitopes in Epstein-Barr Virus Infection with TCR-Like Antibodies. Sci. Rep 2013, 3, 3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Lai J; Tan WJ; Too CT; Choo JAL; Wong LH; Mustafa FB; Srinivasan N; Lim APC; Zhong Y; Gascoigne NRJ; et al. Targeting Epstein-Barr Virus-Transformed B Lymphoblastoid Cells using Antibodies with T-Cell Receptor-Like Specificities. Blood 2016, 128 (10), 1396–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Lai J; Choo JAL; Tan WJ; Too CT; Oo MZ; Suter MA; Mustafa FB; Srinivasan N; Chan CEZ; Lim AGX; et al. TCR-Like Antibodies Mediate Complement and Antibody-Dependent Cellular Cytotoxicity against Epstein-Barr Virus-Transformed B Lymphoblastoid Cells Expressing Different HLA-A*02 Microvariants. Sci. Rep 2017, 7 (1), 9923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Wu L; Brzostek J; Sankaran S; Wei Q; Yap J; Tan TYY; Lai J; MacAry PA; Gascoigne NRJ Targeting CAR to the Peptide-MHC Complex Reveals Distinct Signaling Compared to That of TCR in a Jurkat T Cell Model. Cancers 2021, 13 (4), 867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Brau RR; Tarsa PB; Ferrer JM; Lee P; Lang MJ Interlaced Optical Force-Fluorescence Measurements for Single Molecule Biophysics. Biophys. J 2006, 91 (3), 1069–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Lang MJ; Fordyce PM; Engh AM; Neuman KC; Block SM Simultaneous, Coincident Optical Trapping and Single-Molecule Fluorescence. Nat. Methods 2004, 1 (2), 133–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Liu B; Chen W; Evavold BD; Zhu C Accumulation of Dynamic Catch Bonds between TCR and Agonist Peptide-MHC Triggers T Cell Signaling. Cell 2014, 157 (2), 357–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Schwesinger F; Ros R; Strunz T; Anselmetti D; Güntherodt H-J; Honegger A; Jermutus L; Tiefenauer L; Plückthun A Unbinding Forces of Single Antibody-Antigen Complexes Correlate with Their Thermal Dissociation Rates. Proc. Natl. Acad. Sci. U. S. A 2000, 97 (18), 9972–9977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Aubin-Tam M-E; Appleyard DC; Ferrari E; Garbin V; Fadiran OO; Kunkel J; Lang MJ Adhesion through Single Peptide Aptamers. J. Phys. Chem. A 2011, 115 (16), 3657–3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Li R; Ma C; Cai H; Chen W The CAR T-Cell Mechanoimmunology at a Glance. Adv. Sci 2020, 7 (24), 2002628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Gascoigne NRJ Transport and Secretion of Truncated T Cell Receptor β-Chain Occurs in the Absence of Association with CD3*. J. Biol. Chem 1990, 265 (16), 9296–9301. [PubMed] [Google Scholar]
- (27).Ataie N; Xiang J; Cheng N; Brea EJ; Lu W; Scheinberg DA; Liu C; Ng HL Structure of a TCR Mimic Antibody with Target Predicts Pharmacogenetics. J. Mol. Biol 2016, 428 (1), 194–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Mareeva T; Martinez-Hackert E; Sykulev Y How a T Cell Receptor-Like Antibody Recognizes Major Histocompatibility Complex-Bound Peptide. J. Biol. Chem 2008, 283 (43), 29053–29059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Hilton HG; Parham P Direct Binding to Antigen-Coated Beads Refines the Specificity and Cross-Reactivity of Four Monoclonal Antibodies that Recognize Polymorphic Epitopes of HLA Class I Molecules. Tissue Antigens 2013, 81 (4), 212–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).van der Merwe PA; Davis SJ Molecular Interactions Mediating T Cell Antigen Recognition. Annu. Rev. Immunol 2003, 21, 659–684. [DOI] [PubMed] [Google Scholar]
- (31).Inaguma Y; Akahori Y; Murayama Y; Shiraishi K; Tsuzuki-Iba S; A Endoh A; Tsujikawa J; Demachi-Okamura A; Hiramatsu K; Saji H; et al. Construction and Molecular Characterization of a T-Cell Receptor-Like Antibody and CAR-T Cells Specific for Minor Histocompatibility Antigen HA-1H. Gene Ther. 2014, 21 (6), 575–84. [DOI] [PubMed] [Google Scholar]
- (32).Wu P; Zhang T; Liu B; Fei P; Cui L; Qin R; Zhu H; Yao D; Martinez RJ; Hu W; et al. Mechano-Regulation of Peptide-MHC Class I Conformations Determines TCR Antigen Recognition. Mol. Cell 2019, 73 (5), 1015–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Brockman J; Salaita K Mechanical Proofreading: A General Mechanism to Enhance the Fidelity of Information Transfer Between Cells. Front. Phys 2019, 7, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Mallis RJ; Duke-Cohan JS; Das DK; Akitsua A; Luoma AM; Banik D; Stephens HM; Tetteh PW; Castro CD; Krahnke S; et al. Molecular Design of the γδ T Cell Receptor Ectodomain Encodes Biologically Fit Ligand Recognition in the Absence of Mechanosensing. Proc. Natl. Acad. Sci. U. S. A 2021, 118 (26), No. e2023050118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Brazin KN; Mallis RJ; Boeszoermenyi A; Feng Y; Yoshizawa A; Reche PA; Kaur P; Bi K; Hussey RE; Cohan JSD; et al. The T Cell Antigen Receptor α Transmembrane Domain Coordinates Triggering through Regulation of Bilayer Immersion and CD3 Subunit Associations. Immunity 2018, 49 (5), 829–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Pullen RH III; Abel SM Catch Bonds at T Cell Interfaces: Impact of Surface Reorganization and Membrane Fluctuations. Biophys. J 2017, 113 (1), 120–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Hui KL; Balagopalan L; Samelson LE; Upadhyaya A Cytoskeletal Forces during Signaling Activation in Jurkat T-Cells. Mol. Biol. Cell 2015, 26 (4), 685–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Hwang W; Mallis RJ; Lang MJ; Reinherz EL The αβ TCR Mechanosensor Exploits Dynamic Ectodomain Allostery to Optimize its Ligand Recognition Site. Proc. Natl. Acad. Sci. U. S. A 2020, 117 (35), 21336–21345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Davenport AJ; Cross RS; Watson KA; Liao Y; Shi W; Prince HM; Beavis PA; Trapani JA; Kershaw MH; Ritchie DS; et al. Chimeric Antigen Receptor T Cells form Nonclassical and Potent Immune Synapses Driving Rapid Cytotoxicity. Proc. Natl. Acad. Sci. U. S. A 2018, 115 (9), E2068–E2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Chang ZL; Lorenzini MH; Chen X; Tran U; Bangayan NJ; Chen YY Rewiring T-Cell Responses to Soluble Factors with Chimeric Antigen Receptors. Nat. Chem. Biol 2018, 14 (3), 317–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Chang ZL; Chen YY CARs: Synthetic Immunoreceptors for Cancer Therapy and Beyond. Trends Mol. Med 2017, 23 (5), 430–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Lindner SE; Johnson SM; Brown CE; Wang LD Chimeric Antigen Receptor Signaling: Functional Consequences and Design Implications. Sci. Adv 2020, 6 (21), eaaz3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Yang P; Wang Y; Yao Z; Gao X; Liu C; Wang X; Wu H; Ding X; Hu J; Lin B; et al. Enhanced Safety and Antitumor Efficacy of Switchable Dual Chimeric Antigen Receptor-Engineered T Cells against Solid Tumors through a Synthetic Bifunctional PD-L1-Blocking Peptide. J. Am. Chem. Soc 2020, 142 (44), 18874–18885. [DOI] [PubMed] [Google Scholar]
- (44).Kim MS; Ma JSY; Yun H; Cao Y; Kim JY; Chi V; Wang D; Woods A; Sherwood L; Caballero D; et al. Redirection of Genetically Engineered CAR-T Cells Using Bifunctional Small Molecules. J. Am. Chem. Soc 2015, 137 (8), 2832–2835. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


