Abstract
Antibody-based therapeutics and vaccines are essential to combat COVID-19 morbidity and mortality following severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) infection. Multiple mutations in SARS-CoV-2 that could impair antibody defenses propagated in human-to-human transmission and spillover/spillback events between humans and animals. To develop prevention and therapeutic strategies, we formed an international consortium to map the epitope landscape on the SARS-CoV-2 Spike, defining and structurally illustrating seven receptor-binding domain (RBD)-directed antibody communities with distinct footprints and competition profiles. Pseudovirion-based neutralization assays reveal Spike mutations, individually and clustered together in variants, that impact antibody function among the communities. Key classes of RBD-targeted antibodies maintain neutralization activity against these emerging SARS-CoV-2 variants. These results provide a framework for selecting antibody treatment cocktails and understanding how viral variants might affect antibody therapeutic efficacy.
One-Sentence Summary:
SARS-CoV-2 Spike antibodies in three distinct epitope communities retain activity against SARS-CoV-2 variants.
Cell entry of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is mediated by its surface glycoprotein, Spike. The S1 subunit of Spike contains the N-terminal domain (NTD) and the receptor-binding domain (RBD), which mediates recognition of the host cell receptor angiotensin-converting enzyme 2 (ACE2). The S2 subunit drives fusion between virus and host cell membranes. Spike, particularly the S1 subunit, is the primary target of neutralizing antibodies against SARS-CoV-2 (1).
Since SARS-CoV-2 first emerged, recurrent mutations in Spike arose during both human-to-human transmission (2–4) and spillover/spillback events between humans and animals (5–8). Distinct Variants of Concern (VOCs) or Variants of Interest (VOIs), including those first identified in the UK (alpha, B.1.1.7), South Africa (beta, B.1.351), Brazil (gamma, P.1), India (delta, B.1.617.2) and California (epsilon, B.1.429) carry several mutations associated with enhancement of human-to-human transmission (9). In particular, the receptor-binding motif (RBM) mutations K417, L452, E484 and N501 affect ACE2-Spike interactions (10). Variations at positions N439 and S477 are frequently detected in patient samples (3, 11, 12), whereas others such as V367F, Y453F and F486L are associated with cross-species transmission (6, 8). The NTD is also highly mutable and is especially prone to deletions: ΔHV69–70 and ΔY144 are both seen in B.1.1.7 and ΔHV69–70 is in the mink-associated Cluster V (6). ΔLAL242–244 appears in B.1.351, and ΔFR157–158 is found in B.1.617.2 (9). The NTD point mutations S13I and W152C alter disulfide bonding and conformation of the B.1.429 NTD (13). (Fig. S1)
SARS-CoV-2 will continue to evolve. By understanding antibody footprints and the distinct ways by which antibodies target Spike, we may deduce optimal combinations of mAbs to prevent and treat infection by emerging variants and to minimize the risk of viral escape. We can also gauge the susceptibility of mapped antibodies to new mutations and predict whether newly identified mAbs might also be susceptible to viral escape. Thus, we sought to define functionally important groups in an array of therapeutic candidates, and to dissect how key mutations, both individually and combined as in VOCs, affect antibody-mediated neutralization in a pseudovirus neutralization assay.
The Coronavirus Immunotherapeutic Consortium (CoVIC) was formed to analyze candidate antibody therapeutics side-by-side in standardized assays (14) and now includes over 350 monoclonal antibodies (mAbs) directed against the SARS-CoV-2 Spike protein from 56 different partners across four continents (15). The panel includes antibodies derived from COVID-19 survivors, phage display, naïve libraries, in silico methods and other strategies, each elicited, evaluated and selected using distinct criteria. The panel thus represents a broader and deeper array of antibodies from which both fundamental information and therapeutic cocktails can be derived. With the goals of FAIR (findable, accessible, interoperable, reusable) data analysis and management as well as inclusion of otherwise inaccessible clinical candidates, candidate antibody therapeutics were blinded, and tested in multiple in vitro and in vivo assays with comparative data uploaded into a publicly accessible database (covic.lji.org).
We first measured the affinity of 269 CoVIC mAbs for D614-Hexapro Spike ectodomain trimers and monomeric RBD and NTD, and the ability of each of these mAbs to block ACE2-RBD binding (Fig. S2–S5, Table S1 and covic.lji.org). The panel, formed by candidates for therapeutic use, includes NTD- or S2-directed antibodies, but is dominated by those targeting the RBD. In contrast to previous studies that classified mAbs using germline or structural information (10, 16), the 186 RBD-reactive mAbs of CoVIC analyzed here were instead distinguished by a competition profile created by high-throughput surface plasmon resonance (HT-SPR). RBD-directed antibodies can be sorted into seven core “communities” (Fig. 1, Fig. S6A, Table S2) that are broadly defined by the competition profiles of each mAb to one another. Communities can be further divided into finer clusters and bins based on their discrete competition with other clusters and/or their ability to compete with ACE2 (Fig. 1, Table S1 and Table S2).
Fig. 1. The antigenic landscape of the SARS-CoV-2 receptor binding domain can be divided into seven binding communities.
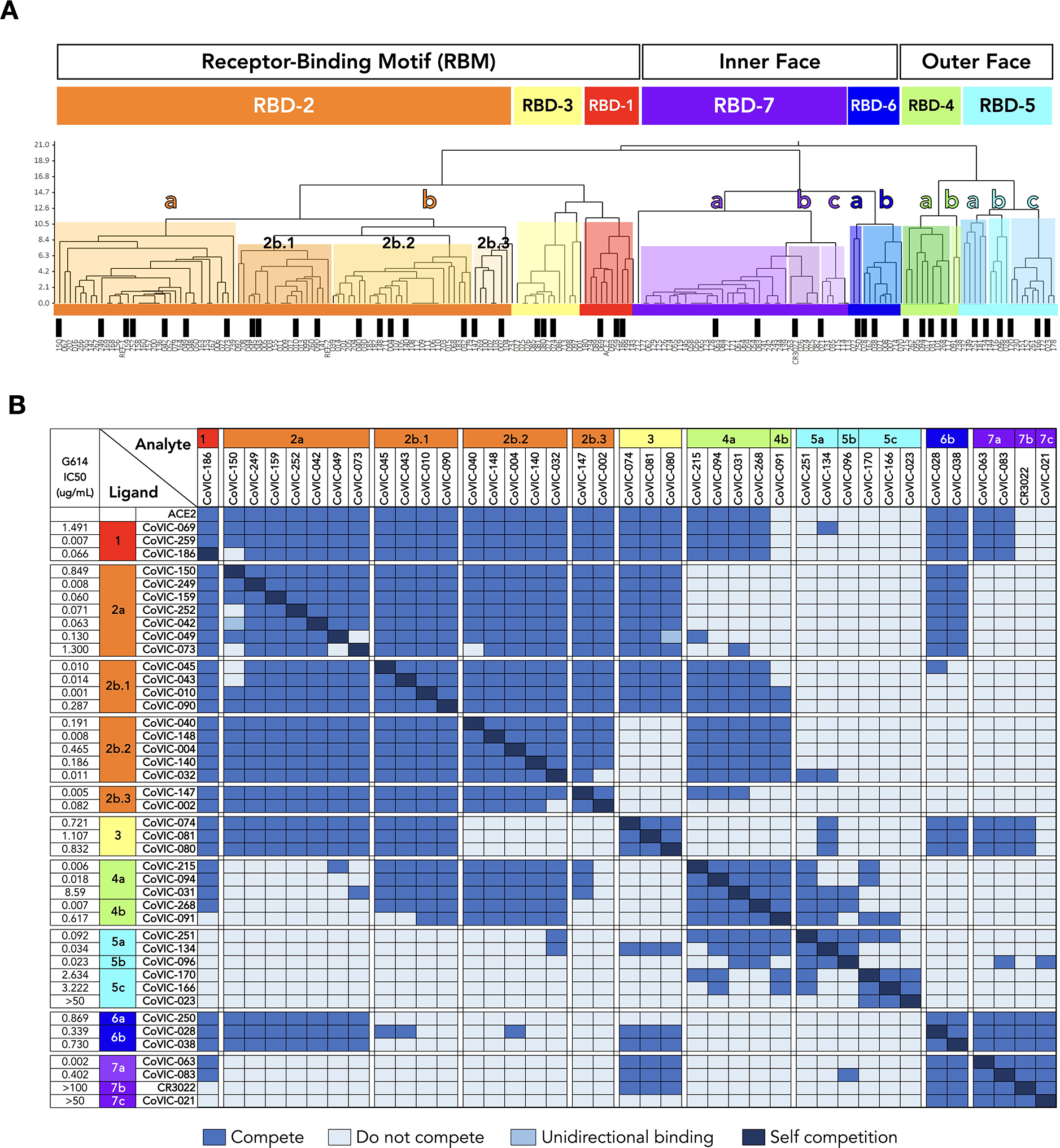
A. High-throughput SPR was used to determine the competitive relationship between 186 RBD-directed mAbs. The dataset was analyzed by Carterra Epitope software to sort competition profiles of clones into related clusters, which are represented as shared colored regions of the dendrogram. The RBD epitope landscape can be broadly divided into seven communities containing mAbs that bind the receptor-binding motif (RBD-1 through RBD-3), the outer face of the RBD (RBD-4 and RBD-5) or the inner face of the RBD (RBD-6 and RBD-7). Communities can be further divided into smaller clusters (e.g., RBD-2a and −2b) and bins (e.g., RBD-2b.1, −2b.2 and −2b.3) based on their discrete competition with other clusters and/or their ability to compete with ACE2 for Spike binding. Black bars indicate single clones that were used in further analyses. Table S1 lists additional metrics for the indicated mAbs (i.e., ACE2 blocking, kinetic analyses and germline information) and detailed information for the entire CoVIC panel is at covic.lji.org. B. Binary heat-map matrix demonstrating the competition profile for the finer clusters and bins for the subset of single clones indicated by black bars in panel A. The matrix here contains representative examples. The complete competition matrix for the study is in Table S2. RBD-2 can be divided into clusters “a” and “b”, which have varying ability to compete with mAbs in RBD-4 (e.g., RBD-2a mAbs do not compete while most RBD-2b mAbs do). Cluster RBD-2b can be divided into three smaller bins that vary in their competition with both RBD-3 and RBD-4 mAbs: those in 2b.1, but not 2b.2 or 2b.3, compete with RBD-3 mAbs whereas mAbs in 2b.1 and 2b.2, but not 2b.3, compete with RBD-4 mAbs. RBD-4 contains mAbs that do (RBD-4a) and do not (RBD-4b) compete with ACE2. RBD-5 and RBD-7 have clusters of mAbs with lower neutralizing potency (i.e., RBD-5c and RBD-7b and RBD-7c) relative to the other cluster in the same community (i.e., RBD-5a and RBD-5b and RBD-7a). Rows and columns indicate the immobilized mAb and injected analyte mAb, respectively. Table S2 shows the complete matrix for competition between all 186 mAbs.
To understand the position of each community relative to the others, we next mapped the footprints by negative-stain EM (NS-EM) for 25 example RBD-reactive mAbs chosen to span the range of communities and key clusters (Fig. 2,Table S3). To have a relatively agnostic view of antibody interactions with Spike, mAbs were not chosen based on germline origin, CDR feature or length, neutralization potency, particular antibody origin (e.g., human, mouse or in silico) or format (e.g., IgG, scFv-Fc, VHH-Fc, multivalent).
Fig. 2. Negative stain EM analysis of representatives from each RBD-directed community.
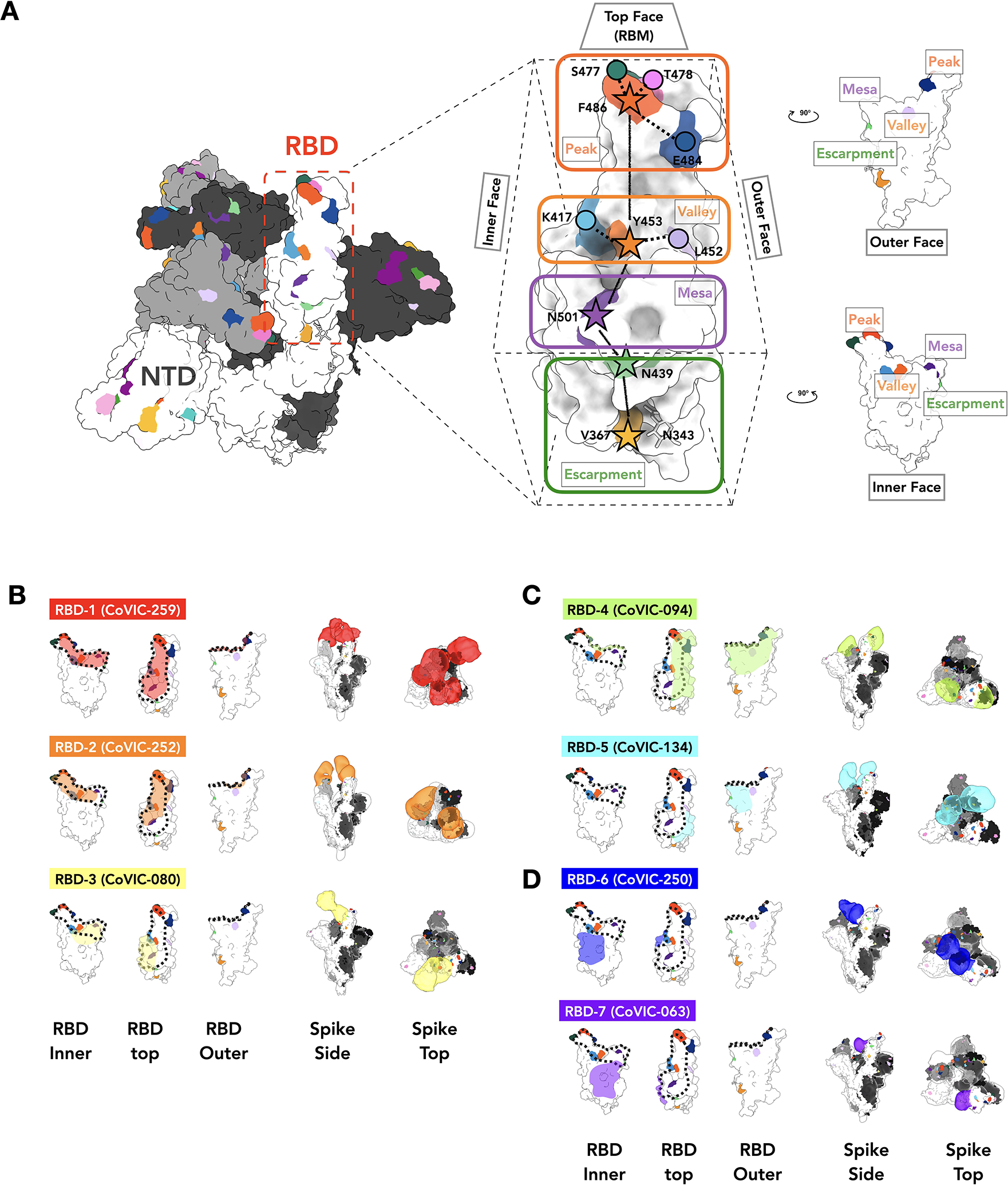
A. The location of important emerging mutations in RBD. The Spike trimer (adapted from PDB: 7A94 (39)) viewed from the top with one RBD “up” RBD, is shown, with individual Spike monomers colored white, gray and black. The RBM can be topologically divided into three subsections: the “Peak” that includes residues F486, S477, T478 and E484, the “Valley” including residues Y453, K417 and L452 and the “Mesa” with residue N501. Stars indicate residues on the central axis of RBD. The “Outer Face” (exposed in the RBD down/closed conformation), and “Inner Face” (buried inside the trimer in the RBD down/closed conformation) define the lateral faces of RBD and “Escarpment” (contains residues V367, N439 and glycan 343). B. NS-EM footprint of a representative antibody from each community mapped onto an RBD monomer. The colored shading corresponds to the community colors in Figure 1. The ACE2 binding site is outlined with a dotted line. Side and top views of Spike trimers show the Fab approach angle and binding stoichiometry for each representative. Table S3 shows NS-EM data for all 29 RBD-directed mAbs analyzed.
In parallel, we measured the neutralization activity of 41 RBD-directed mAbs (chosen to span the range of communities and key clusters) as well as a human ACE2-Fc format-based therapeutic candidate (CoVIC-069) fusion format. Neutralization was measured against pseudoviruses displaying the Spike protein bearing (a) the globally dominant G614 variation, (b) 15 single point mutations or deletions represented in circulating strains, (c) constellations of mutations found in four VOCs [B.1.1.1 (alpha), B.1.351 (beta), P.1 (gamma), and B.1.617.2 (delta)] and one VOI [B.1.429 (epsilon)] and (d) two pseudovariants carrying four mutations (termed 4×M, containing G261D, Y453F, F486L, and N501T) or five mutations (termed 5×M, carrying the 4×M mutations plus V367F) identified in human-mink spillover events (Fig. 3).
Fig. 3. RBD-5, −6 and −7 antibodies retain neutralization activity against pseudovirus bearing mutations singly or together in VOCs.
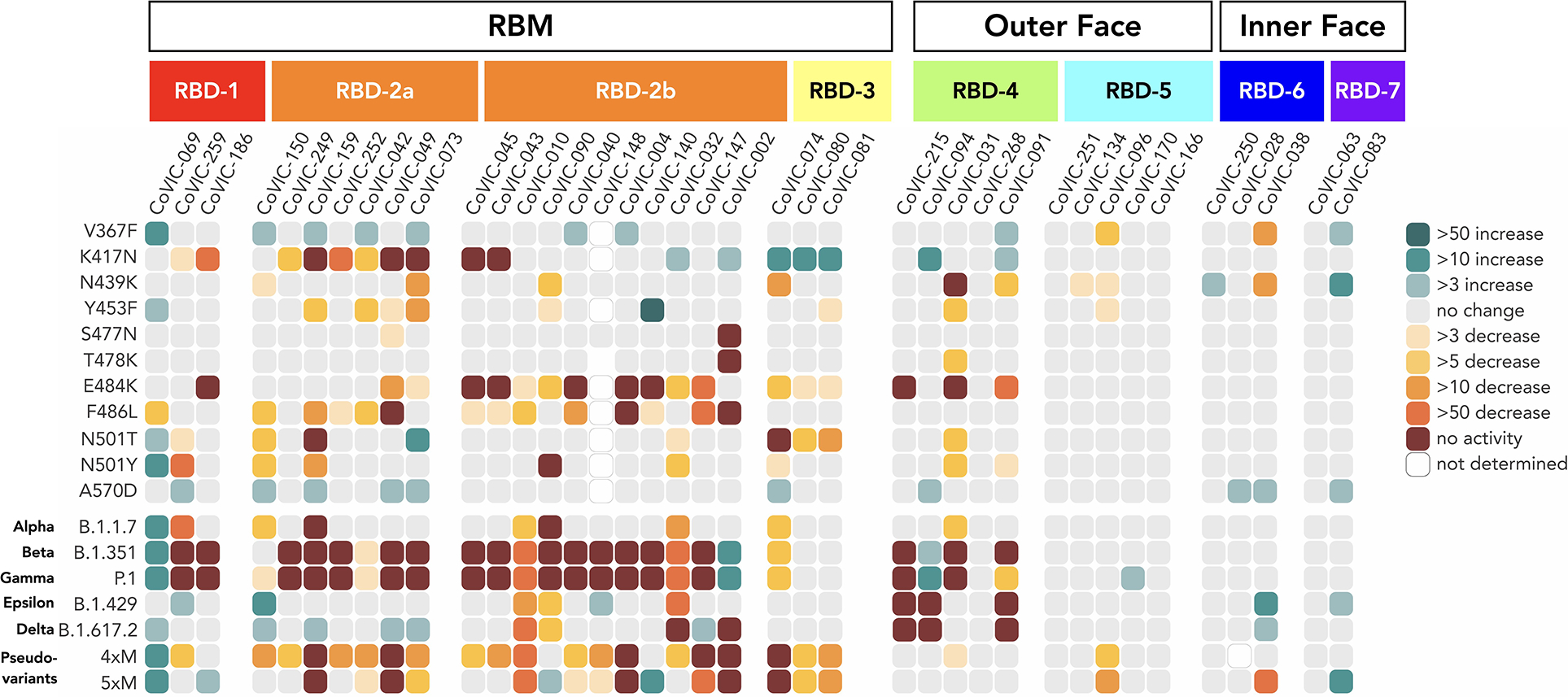
Fold-change differences in potency for 38 RBD-directed antibodies and an ACE2-Fc fusion (CoVIC-069) are shown in a heat map. In addition to VOCs, we also examined two pseudoviruses bearing clusters of mink-associated mutations: 4×M (G261D, Y453F, F486L and N501T) and 5×M (G261D, Y453F, F486L, N501T and V367F). Fig. S1 lists mutations represented in each variant. Fig. S10 shows neutralization curves for each virus-variant pair and Table S4 lists fold-change values corresponding to the heat map.
The mAbs in RBD-1 through −3 target the receptor-binding motif (RBM), compete with ACE2, and generally require the RBD to be in the “up” conformation for binding (footprints defined in Fig 2B, Table S3, covic.lji.org). Community RBD-1 contains hACE2-derived molecules and IgGs (e.g., CoVIC-259, EMD-24335) that largely overlap with the RBM (Fig. 2B, Fig. S6B, Table S3). The footprint for RBD-2 mAbs is shifted from the center of the ACE2 binding site towards the peak of the RBM (Fig. 2B, Fig. S6B, Table S3). RBD-2 is the largest community and can be divided further into clusters and then bins based on competition with other communities (Fig. 1). Cluster 2a antibodies (e.g., CoVIC-252, EMD-24339) bind towards the inner face of the RBD and its binding area overlaps highly with that of the therapeutic antibody REGN-10933 (17). Antibodies in 2b.1 (e.g., CoVIC-010, EMD-24343; similar to antibody COVA2–39 (18)) and 2b.2 (e.g., CoVIC-140, EMD-24383; similar to antibody C144 (16)) bind towards the outer face of the RBD and mAbs in bin 2b.3 (e.g., CoVIC-002, EMD-24345; similar to antibody S2E12 (19)) bind to the peak of the RBD (Fig. 2B, Fig. S7 and Table S3). Lastly, RBD-3 mAbs bind down from the center of the ACE2 binding site towards the RBD “mesa” (Fig. 2B, Table S2; e.g., CoVIC-080, EMD-24346; similar to antibody ADI-56046 (20)).
To simulate the authentic interactions between antibodies and Spike, intact IgGs were used for NS-EM structural analysis whenever possible. RBD-1 IgGs tend to fully occupy all three RBDs on one Spike and often crosslink two Spike trimers, whereas most RBD-2 IgGs tend to bind bivalently to a single Spike trimer (Fig. S8A, B, Fig. S9 and Table S3). RBD-3 IgGs can crosslink Spikes, and bivalent binding was also observed in some cases (Table S3 and Fig. S9).
General epitope position, and particularly RBM epitopes, is strongly associated with the propensity of particular Spike mutations to escape antibody-mediated neutralization (Fig. 3, Fig. S10 and Table S4). Neutralization by RBD-2a antibodies is heavily impacted by the K417N mutation, but rarely by the E484K mutation; those in RBD-2b are impacted by the E484K mutation but less so by K417N. Similarly, RBD-2a antibodies are resistant to the L452R mutation found in B.1.429 (epsilon) and B.1.617.2 (delta), while only some RBD-2b antibodies are sensitive to this mutation. Meanwhile, mAbs in RBD-3 are impacted by both N501T/Y and E484K mutations (Fig. 3, Fig. S10 and S11 and Table S4). In contrast to RBD-2 and −3, the susceptibility of neutralization activity of antibodies in RBD-1 to particular mutations is more variable (Fig. 2B, Fig. 3, Table S3, Table S4).
Regardless of the effect of particular single point mutations, nearly every RBD-1 or −2 mAb analyzed showed additive decreases in potency against pseudovirus carrying constellations of multiple mutations in the RBM (Fig. 3, Fig. S10 and Table S4). For B.1.351 and P.1, almost all RBD-1 and RBD-2 antibodies analyzed suffer a complete loss of neutralization activity. For example, CoVIC-249 and CoVIC-010 show moderate or no change in IC50 against the single point mutations K417N, E484K and N501Y, but CoVIC-249 loses all neutralization activity and CoVIC-010 potency falls by 1000-fold against B.1.351 (beta) and P.1 (gamma) which contain all three mutations. Many RBD-2 antibodies also lose activity against the 4×M mink pseudovariant that carries Y453F, F486L and N501T mutations (Fig. 3, Fig. S1B, Fig. S10 and Table S4).
In contrast, most RBD-1 and RBD-2 antibodies retain neutralization activity against B.1.1.7 (alpha), B.1.429 (epsilon) and B.1.617.2 (delta) variants, which each contain only one or two RBM-located mutations (N501Y, L452R or T478K/L452R respectively). Curiously, the V367F mutation identified in mink populations enhances neutralization by some RBD-2 mAbs and in some cases this mutation can offset decreases in potency resulting from other single point mutations. For example, CoVIC-040 has a 14- and 8-fold decrease in potency against the F486L mutation and the F486L-containing 4×M mink pseudovariant, respectively, but only a 4-fold decrease against the 5×M mink pseudovariant, which contains V367F in addition to the four mutations present in 4×M (Fig. 3, Fig. S10 and Table S4). V367 is adjacent to an N-linked glycan at position 343, which was recently implicated in providing a gating mechanism for the RBD (21). Substitution of valine with phenylalanine could alter the local environment of the N343 glycan moieties and enable the RBD to adopt a conformation more amenable to antibody interaction.
Antibodies in communities RBD-4 and RBD-5 bind to the outer face of the RBD and, like the Class 2 and Class 3 mAbs previously defined in (16), can do so in either the “up” or “down” RBD conformation without steric hindrance (Fig. 2C, Fig. S6, Fig. S12, and Table S3). The footprints of these groups largely overlap, but RBD-4 mAbs bind towards the outer edge of the RBM and can block ACE2 (e.g., CoVIC-094, EMD-24350; similar to antibody C002 (16)), whereas RBD-5 mAbs bind away from the RBM, towards the “S309” site and do not block ACE2 (e.g., CoVIC-134, EMD-24384; similar to antibody REGN-10987 (17)) (10) (Fig. 1B, Fig. 2C, Fig. S5, Fig. S6B, Tables S1–3). Some RBD-4 and RBD-5 IgGs can crosslink Spike trimers in solution (Fig. S8C and Table S3).
Interestingly, according to the five RBD-5 IgGs we imaged, only those IgGs that show Spike-cross linking tendency have potent neutralizing activity (Fig.1B, Fig. S13 and Table S3). A recent cryo-electron tomography study showed native Spike trimers on the SARS-CoV-2 virion surface tilt at variable degrees relative to the viral envelope (22). This finding provides a possibility for IgG-mediated Spike crosslinking on virions, and may contribute to the mechanism of neutralization of the RBD-5 mAbs in the absence of ACE2 blocking (Fig. S8D).
Most RBD-4 mAbs are impacted by E484K and/or L452R (represented in the B.1.429 variant) mutations (Fig. 3, Fig. S10, Table S4), and some are impacted by the N439K mutation, which is highly represented in sequences worldwide(3). RBD-5 mAbs, however, show broad resistance to nearly all mutations analyzed, with only two mAbs in this group showing moderate decreases in potency against V367F and N439K (Fig. 3, Fig. S10, Table S4).
RBD-6 (e.g., CoVIC-250, EMD-24352) and RBD-7 (e.g., CoVIC-063, EMD-24353) antibodies bind to the inner face of the RBD and access a previously described cryptic epitope (23, 24) (Fig. 2D, Fig. S6B and Table S3). Like Class 4 antibodies described in (16), binding of Spike by RBD-6 and RBD-7 antibodies requires two RBDs to be in the “up” configuration (Fig. S12). The representative IgGs in RBD-6 and RBD-7 each show stronger propensities to crosslink Spike trimers than RBM-directed antibodies (Fig. S9 and Table S3). RBD-6 and RBD-7 antibodies primarily vary in their competition with RBD-2a antibodies: the downward shift of the RBD-7 footprint on the inner face of the RBD relative to the RBD-6 footprint would allow simultaneous binding of RBD-2a antibodies with RBD-7, but not RBD-6, antibodies (Fig. 1B, Fig. 2D, Fig. S6B, Table S2). This cryptic RBD-6/7 site is also recognized by antibodies COVA1–16 (23) and CR3022 (24). Here, strategies of site recognition are further subdivided by competition subgroups, information useful for interpreting differences and antibody behavior and strategies for cocktail selection.
All RBD-6 and RBD-7a antibodies block ACE2, but antibodies in RBD-7b and 7c do not (Fig. 1B, Tables S1, S2 and S3). The representatives from the RBD-7b and 7c clusters (CR3022 and CoVIC-021, respectively) demonstrate poor neutralization of pseudoviruses in our assay. The distinct difference in neutralization behavior between 7a and 7b/7c suggests that at this cryptic epitope, competition with ACE2 is a determinant of neutralization (Fig. 1B, Table S4) (25). Importantly, due to their location away from the RBM, RBD-6 and RBD-7 antibodies are resistant to the mutations and variants analyzed (Fig. 3, Fig. S10, Table S4).
Previous reports identified a “supersite” as the primary target for neutralizing antibodies directed against the NTD (26). In addition to RBD-directed antibodies, we also analyzed four CoVIC NTD-directed antibodies by NS-EM and in neutralization assays. Together these four antibodies, grouped as NTD-1 through NTD-3, encompass the approximate boundaries of the supersite. The two NTD-1 antibodies bind from the top side of NTD to cover the NTD N-terminus and residue Y144 (Fig. 4, e.g., CoVIC-247, EMD-24355 and Table S3). The NTD-1 epitope overlaps with that of mAb 4A8 (27) and other “supersite” binders (28, 29). The NTD-2 antibody (CoVIC-245, EMD-24360) approaches from the front side of NTD and contacts Y144 as well as residues H69, V70, W152 and G261, all of which are deleted or substituted in emerging variants (Fig. 4 and Fig. S1). The NTD-2 footprint is similar to the footprint of antibodies in the “antigenic site V” group described in (26). The NTD-3 mAb (CoVIC-020, EMD-24356) binds to the left side of the NTD, proximal to the RBD of the adjacent monomer and in contact with residue W152 (Fig. 4B). The NTD-3 mAb represents a novel epitope and binding location of an anti-NTD antibody.
Fig. 4. NS-EM and neutralization analysis of mAb targeting NTD.
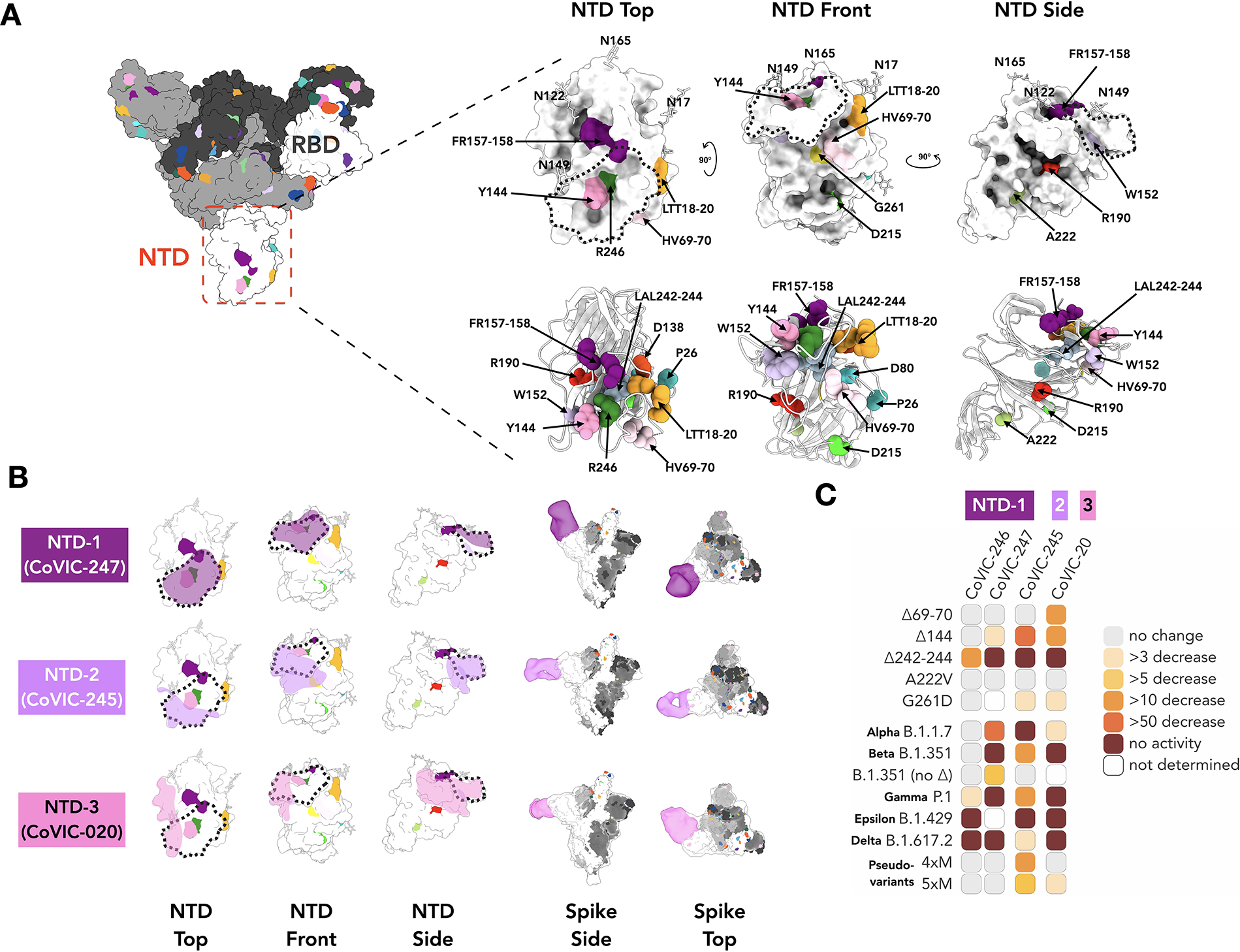
A. Surface and cartoon (adapted from PDB: 7A94 (39)) representation of the Spike NTD. The residue positions of mutations and deletions in circulating VOCs are indicated in three views of NTD. Fig. S1 lists mutations represented in each variant. B. Footprints for three NTD-targeted antibodies with the NTD “supersite”(26) indicated as a dotted line. The NTD-directed antibodies shown here define the approximate boundaries of the neutralizing epitope landscape. Additional NS-EM data are in Table S3. C. Fold-change in potency of pseudovirus neutralization experiments for each antibody-variant pair.
Unlike the RBD-directed antibodies, for which neutralization escape is strongly associated with antibody footprint, the NTD-directed antibodies are conformationally sensitive and affected by mutations outside of the discrete footprint. This finding is consistent with that for antibodies elicited by vaccines (30). Each of the four NTD mAbs analyzed exhibit a decreased or total loss of neutralization capacity for one or more of the NTD-located deletions (Δ69/70, ΔY144, Δ157–158 and Δ242–244) found in circulating VOCs, regardless of their binding location on NTD (Fig. 4C, Fig. S10 and Table S4). All NTD mAbs were impacted by P.1 (gamma), which lacks deletions and instead has several point mutations in the NTD. For B.1.429 (epsilon), altered disulfide bonding in the NTD arising from the S13I and W152C mutations (13) also abrogated mAb-mediated neutralization. Our results indicate that NTD mutations decrease not only neutralization potency but also the total fraction of virus neutralized (Fig. S10).
Several therapeutic antibody cocktails comprising pairs of different mAbs against Spike are currently under investigation for post-exposure treatment of COVID-19 (16, 17, 31, 32). However, the potency of some antibodies in these cocktails is compromised by emerging SARS-CoV-2 variants (33, 34). Meanwhile, exposure of virus to monoclonal or polyclonal antibodies can promote antibody-resistant mutations in Spike (34–37). Notably, SARS-CoV-2 variants that share critical mutations with B.1.1.7 (alpha) were isolated from an immunocompromised COVID-19 patient who received three rounds of convalescent plasma treatment, indicating that even a polyclonal therapeutic can drive evolution of resistant virus strains in unresolved infections (38).
Potency, variant-resistance and the ability to co-bind are important considerations when selecting antibodies for therapeutic cocktails. The analysis of the 186 RBD-directed mAbs presented here, each donated by different groups around the globe and each selected in different ways, describes discrete antibody communities, and functionally relevant sub-clusters and/or bins. This analysis provides a competition grid, and a framework for cocktail selection. Notably, combining this data with neutralization potency and mutational analysis can guide selection of broadly protective therapeutic cocktails.
Overall, antibodies from community RBD-1 through RBD-4 and those directed against the NTD are generally more potent than antibodies of other communities. The high potency and non-overlapping epitopes of RBD- and NTD-directed antibodies make them attractive as pairs for therapeutic cocktails. However, members of each of these groups are also highly susceptible to neutralization escape by mutations and deletions found in emerging VOCs. Indeed, a CoVIC bispecific antibody targeting the RBD-1 and NTD-1 sites could still neutralize single point mutations in the RBD (where the NTD arm could compensate), but was ineffective against B.1.351 (beta) and P.1 (gamma), which contain mutations that simultaneously escape both arms of the bispecific (Fig. S14).
In contrast, RBD-5, −6 and −7 antibodies often have lower potency but are more resistant to escape. Notably, the epitopes targeted by RBD-5, −6, and −7 antibodies have high sequence conservation among the Sarbecovirus subgenus of Betacoronavirus (Fig. S15). Enhanced potency for these communities might be achieved through engineering them as multivalent formats, making them key members of a variant-resistant cocktail that could also be suitable for treating other Sarbecovirus infections.
Taken together, the analysis presented here, made possible by broad participation of a few hundred therapeutic candidates in a global study, offers a detailed structural and competitive landscape of key antibody binding sites on Spike. The results of this effort can be used to predict and interpret effects of VOCs, and for strategic selection of durable therapeutics and cocktails against emerging variants.
Supplementary Material
Acknowledgments:
We are grateful for the multiple generous contributions of antibodies to the CoVIC study, with special thanks to Matt Beasley, Shahrad Daraeikia, Vincent Dussupt, Ben Kiefel, Sindy Liao, Chanjuan Liu, Letzibeth Mendez-Rivera, Pramila Rijal, Lisa Schimanski, Pete Smith, Tiong Tan, Alain Townsend, Jack Wang, Run Yan, and Luo Yang. We thank the electron microscope facility of La Jolla Institute for Immunology for the EM data collection.
Funding:
We gratefully acknowledge philanthropic support of the Overton family for this urgent study (Coronavirus Immunotherapeutic Consortium)
COVID-19 Therapeutics Accelerator INV-006133 (Coronavirus Immunotherapeutic Consortium)
Bill and Melinda Gates Foundation OPP1210938 (Coronavirus Immunotherapeutic Consortium)
GHR foundation (Coronavirus Immunotherapeutic Consortium)
NIH/NIAID grant U19 AI142790-S1 (Coronavirus Immunotherapeutic Consortium)
We also acknowledge philanthropic support of Carolee Lee, FastGrants from Emergent Ventures at the Mercatus Center, George Mason University for support of essential instrumentation, and T.S. for support of the Saphire laboratory efforts during the pandemic (EOS)
Early Postdoc Mobility Fellowship of the Swiss National Science Foundation P2EZP3_195680 (DZ)
Translating Duke Health Immunology Initiative (GT)
Footnotes
Competing interests: Authors declare that they have no competing interests.
Data and materials availability:
EM maps in this study have been uploaded to the EMDataResource. The EMDB access numbers are available in the main text and Table S3. Information concerning particular antibodies can be requested through the Coronavirus Immunotherapeutics Consortium at covic@lji.org
References and Notes
- 1.Piccoli L, Park Y-J, Tortorici MA, Czudnochowski N, Walls AC, Beltramello M, Silacci-Fregni C, Pinto D, Rosen LE, Bowen JE, Acton OJ, Jaconi S, Guarino B, Minola A, Zatta F, Sprugasci N, Bassi J, Peter A, De Marco A, Nix JC, Mele F, Jovic S, Rodriguez BF, Gupta SV, Jin F, Piumatti G, Lo Presti G, Pellanda AF, Biggiogero M, Tarkowski M, Pizzuto MS, Cameroni E, Havenar-Daughton C, Smithey M, Hong D, Lepori V, Albanese E, Ceschi A, Bernasconi E, Elzi L, Ferrari P, Garzoni C, Riva A, Snell G, Sallusto F, Fink K, Virgin HW, Lanzavecchia A, Corti D, Veesler D, Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology. Cell. 183, 1024–1042.e21 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Korber B, Fischer WM, Gnanakaran S, Yoon H, Theiler J, Abfalterer W, Hengartner N, Giorgi EE, Bhattacharya T, Foley B, Hastie KM, Parker MD, Partridge DG, Evans CM, Freeman TM, de Silva TI, Sheffield COVID-19 Genomics Group, McDanal C, Perez LG, Tang H, Moon-Walker A, Whelan SP, LaBranche CC, Saphire EO, Montefiori DC, Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell (2020), doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thomson EC, Rosen LE, Shepherd JG, Spreafico R, da Silva Filipe A, Wojcechowskyj JA, Davis C, Piccoli L, Pascall DJ, Dillen J, Lytras S, Czudnochowski N, Shah R, Meury M, Jesudason N, De Marco A, Li K, Bassi J, O’Toole A, Pinto D, Colquhoun RM, Culap K, Jackson B, Zatta F, Rambaut A, Jaconi S, Sreenu VB, Nix J, Zhang I, Jarrett RF, Glass WG, Beltramello M, Nomikou K, Pizzuto M, Tong L, Cameroni E, Croll TI, Johnson N, Di Iulio J, Wickenhagen A, Ceschi A, Harbison AM, Mair D, Ferrari P, Smollett K, Sallusto F, Carmichael S, Garzoni C, Nichols J, Galli M, Hughes J, Riva A, Ho A, Schiuma M, Semple MG, Openshaw PJM, Fadda E, Baillie JK, Chodera JD, Rihn SJ, Lycett SJ, Virgin HW, Telenti A, Corti D, Robertson DL, Snell G, Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated immunity. Cell (2021), doi: 10.1016/j.cell.2021.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.CDC, Emerging SARS-CoV-2 Variants (2021), (available at https://www.cdc.gov/coronavirus/2019-ncov/more/science-and-research/scientific-brief-emerging-variants.html).
- 5.Oreshkova N, Molenaar RJ, Vreman S, Harders F, Oude Munnink BB, Hakze-van der Honing RW, Gerhards N, Tolsma P, Bouwstra R, Sikkema RS, Tacken MG, de Rooij MM, Weesendorp E, Engelsma MY, Bruschke CJ, Smit LA, Koopmans M, van der Poel WH, Stegeman A, SARS-CoV-2 infection in farmed minks, the Netherlands, April and May 2020. Euro Surveill. 25 (2020), doi: 10.2807/1560-7917.ES.2020.25.23.2001005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.WHO | SARS-CoV-2 mink-associated variant strain – Denmark (2020) (available at https://www.who.int/csr/don/06-november-2020-mink-associated-sars-cov2-denmark/en/). [Google Scholar]
- 7.Oude Munnink BB, Sikkema RS, Nieuwenhuijse DF, Molenaar RJ, Munger E, Molenkamp R, van der Spek A, Tolsma P, Rietveld A, Brouwer M, Bouwmeester-Vincken N, Harders F, Hakze-van der Honing R, Wegdam-Blans MCA, Bouwstra RJ, GeurtsvanKessel C, van der Eijk AA, Velkers FC, Smit LAM, Stegeman A, van der Poel WHM, Koopmans MPG, Transmission of SARS-CoV-2 on mink farms between humans and mink and back to humans. Science (2020), doi: 10.1126/science.abe5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Dorp L, Tan CCS, Lam SD, Richard D, Owen C, Berchtold D, Orengo C, Balloux F, Recurrent mutations in SARS-CoV-2 genomes isolated from mink point to rapid host-adaptation. Cold Spring Harbor Laboratory (2020), p. 2020.11.16.384743. [Google Scholar]
- 9.CDC, SARS-CoV-2 Variants (2021), (available at https://www.cdc.gov/coronavirus/2019-ncov/cases-updates/variant-surveillance/variant-info.html).
- 10.Yuan M, Huang D, Lee C-CD, Wu NC, Jackson AM, Zhu X, Liu H, Peng L, van Gils MJ, Sanders RW, Burton DR, Reincke SM, Prüss H, Kreye J, Nemazee D, Ward AB, Wilson IA, Structural and functional ramifications of antigenic drift in recent SARS-CoV-2 variants. bioRxiv (2021), doi: 10.1101/2021.02.16.430500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen J, Wang R, Wang M, Wei G-W, Mutations Strengthened SARS-CoV-2 Infectivity. J. Mol. Biol. 432, 5212–5226 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hodcroft EB, Zuber M, Nadeau S, Crawford KHD, Bloom JD, Veesler D, Vaughan TG, Comas I, Candelas FG, Stadler T, Neher RA, Emergence and spread of a SARS-CoV-2 variant through Europe in the summer of 2020. medRxiv (2020), doi: 10.1101/2020.10.25.20219063. [DOI] [Google Scholar]
- 13.McCallum M, Bassi J, Marco AD, Chen A, Walls AC, Iulio JD, Tortorici MA, Navarro M-J, Silacci-Fregni C, Saliba C, Agostini M, Pinto D, Culap K, Bianchi S, Jaconi S, Cameroni E, Bowen JE, Tilles SW, Pizzuto MS, Guastalla SB, Bona G, Pellanda AF, Garzoni C, Van Voorhis WC, Rosen LE, Snell G, Telenti A, Virgin HW, Piccoli L, Corti D, Veesler D, SARS-CoV-2 immune evasion by variant B.1.427/B.1.429. bioRxiv (2021), doi: 10.1101/2021.03.31.437925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Collins FS, Woodcock J, Graham BS, Arvin A, Bieniasz P, Ho D, Alter G, Nussenzweig M, Burton D, Tavel J, Others, Therapeutic Neutralizing Monoclonal Antibodies: Report of a Summit sponsored by Operation Warp Speed and the National Institutes of Health (2020) (available at https://www.nih.gov/sites/default/files/research-training/initiatives/activ/20200909-mAb-summit-pub.pdf).
- 15.Coronavirus Immunotherapy Consortium (2020), (available at https://covic.lji.org/).
- 16.Barnes CO, Jette CA, Abernathy ME, Dam K-MA, Esswein SR, Gristick HB, Malyutin AG, Sharaf NG, Huey-Tubman KE, Lee YE, Robbiani DF, Nussenzweig MC, West AP Jr, Bjorkman PJ, SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature. 588, 682–687 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hansen J, Baum A, Pascal KE, Russo V, Giordano S, Wloga E, Fulton BO, Yan Y, Koon K, Patel K, Chung KM, Hermann A, Ullman E, Cruz J, Rafique A, Huang T, Fairhurst J, Libertiny C, Malbec M, Lee W-Y, Welsh R, Farr G, Pennington S, Deshpande D, Cheng J, Watty A, Bouffard P, Babb R, Levenkova N, Chen C, Zhang B, Romero Hernandez A, Saotome K, Zhou Y, Franklin M, Sivapalasingam S, Lye DC, Weston S, Logue J, Haupt R, Frieman M, Chen G, Olson W, Murphy AJ, Stahl N, Yancopoulos GD, Kyratsous CA, Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science. 369, 1010–1014 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu NC, Yuan M, Liu H, Lee C-CD, Zhu X, Bangaru S, Torres JL, Caniels TG, Brouwer PJM, van Gils MJ, Sanders RW, Ward AB, Wilson IA, An Alternative Binding Mode of IGHV3–53 Antibodies to the SARS-CoV-2 Receptor Binding Domain. Cell Rep. 33, 108274 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tortorici MA, Beltramello M, Lempp FA, Pinto D, Dang HV, Rosen LE, McCallum M, Bowen J, Minola A, Jaconi S, Zatta F, De Marco A, Guarino B, Bianchi S, Lauron EJ, Tucker H, Zhou J, Peter A, Havenar-Daughton C, Wojcechowskyj JA, Case JB, Chen RE, Kaiser H, Montiel-Ruiz M, Meury M, Czudnochowski N, Spreafico R, Dillen J, Ng C, Sprugasci N, Culap K, Benigni F, Abdelnabi R, Foo S-YC, Schmid MA, Cameroni E, Riva A, Gabrieli A, Galli M, Pizzuto MS, Neyts J, Diamond MS, Virgin HW, Snell G, Corti D, Fink K, Veesler D, Ultrapotent human antibodies protect against SARS-CoV-2 challenge via multiple mechanisms. Science. 370, 950–957 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wec AZ, Wrapp D, Herbert AS, Maurer DP, Haslwanter D, Sakharkar M, Jangra RK, Dieterle ME, Lilov A, Huang D, Tse LV, Johnson NV, Hsieh C-L, Wang N, Nett JH, Champney E, Burnina I, Brown M, Lin S, Sinclair M, Johnson C, Pudi S, Bortz R 3rd, Wirchnianski AS, Laudermilch E, Florez C, Fels JM, O’Brien CM, Graham BS, Nemazee D, Burton DR, Baric RS, Voss JE, Chandran K, Dye JM, McLellan JS, Walker LM, Broad neutralization of SARS-related viruses by human monoclonal antibodies. Science. 369, 731–736 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sztain T, Ahn S-H, Bogetti AT, Casalino L, Goldsmith JA, McCool RS, Kearns FL, Andrew McCammon J, McLellan JS, Chong LT, Amaro RE, A glycan gate controls opening of the SARS-CoV-2 spike protein. Cold Spring Harbor Laboratory (2021), p. 2021.02.15.431212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yao H, Song Y, Chen Y, Wu N, Xu J, Sun C, Zhang J, Weng T, Zhang Z, Wu Z, Cheng L, Shi D, Lu X, Lei J, Crispin M, Shi Y, Li L, Li S, Molecular Architecture of the SARS-CoV-2 Virus. Cell. 183, 730–738.e13 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu H, Wu NC, Yuan M, Bangaru S, Torres JL, Caniels TG, van Schooten J, Zhu X, Lee C-CD, Brouwer PJM, van Gils MJ, Sanders RW, Ward AB, Wilson IA, Cross-neutralization of a SARS-CoV-2 antibody to a functionally conserved site is mediated by avidity, doi: 10.1101/2020.08.02.233536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yuan M, Liu H, Wu NC, Lee C-CD, Zhu X, Zhao F, Huang D, Yu W, Hua Y, Tien H, Rogers TF, Landais E, Sok D, Jardine JG, Burton DR, Wilson IA, Structural basis of a shared antibody response to SARS-CoV-2. Science (2020), doi: 10.1126/science.abd2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huo J, Zhao Y, Ren J, Zhou D, Duyvesteyn HME, Ginn HM, Carrique L, Malinauskas T, Ruza RR, Shah PNM, Tan TK, Rijal P, Coombes N, Bewley KR, Tree JA, Radecke J, Paterson NG, Supasa P, Mongkolsapaya J, Screaton GR, Carroll M, Townsend A, Fry EE, Owens RJ, Stuart DI, Neutralization of SARS-CoV-2 by Destruction of the Prefusion Spike. Cell Host Microbe. 28, 445–454.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCallum M, De Marco A, Lempp FA, Tortorici MA, Pinto D, Walls AC, Beltramello M, Chen A, Liu Z, Zatta F, Zepeda S, di Iulio J, Bowen JE, Montiel-Ruiz M, Zhou J, Rosen LE, Bianchi S, Guarino B, Fregni CS, Abdelnabi R, Foo S-YC, Rothlauf PW, Bloyet L-M, Benigni F, Cameroni E, Neyts J, Riva A, Snell G, Telenti A, Whelan SPJ, Virgin HW, Corti D, Pizzuto MS, Veesler D, N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. Cell. 184, 2332–2347.e16 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chi X, Yan R, Zhang J, Zhang G, Zhang Y, Hao M, Zhang Z, Fan P, Dong Y, Yang Y, Chen Z, Guo Y, Zhang J, Li Y, Song X, Chen Y, Xia L, Fu L, Hou L, Xu J, Yu C, Li J, Zhou Q, Chen W, A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science. 369, 650–655 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cerutti G, Guo Y, Zhou T, Gorman J, Lee M, Rapp M, Reddem ER, Yu J, Bahna F, Bimela J, Huang Y, Katsamba PS, Liu L, Nair MS, Rawi R, Olia AS, Wang P, Chuang G-Y, Ho DD, Sheng Z, Kwong PD, Shapiro L, Potent SARS-CoV-2 Neutralizing Antibodies Directed Against Spike N-Terminal Domain Target a Single Supersite. Cold Spring Harbor Laboratory (2021), p. 2021.01.10.426120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCallum M, Marco AD, Lempp F, Tortorici MA, Pinto D, Walls AC, Beltramello M, Chen A, Liu Z, Zatta F, Zepeda S, di Iulio J, Bowen JE, Montiel-Ruiz M, Zhou J, Rosen LE, Bianchi S, Guarino B, Fregni CS, Abdelnabi R, Caroline Foo S-Y, Rothlauf PW, Bloyet L-M, Benigni F, Cameroni E, Neyts J, Riva A, Snell G, Telenti A, Whelan SPJ, Virgin HW, Corti D, Pizzuto MS, Veesler D, N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. bioRxiv (2021), doi: 10.1101/2021.01.14.426475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cao Y, Yisimayi A, Bai Y, Huang W, Li X, Zhang Z, Yuan T, An R, Wang J, Xiao T, Du S, Ma W, Song L, Li Y, Li X, Song W, Wu J, Liu S, Li X, Zhang Y, Su B, Guo X, Wei Y, Gao C, Zhang N, Zhang Y, Dou Y, Xu X, Shi R, Lu B, Jin R, Ma Y, Qin C, Wang Y, Feng Y, Xiao J, Xie XS, Humoral immune response to circulating SARS-CoV-2 variants elicited by inactivated and RBD-subunit vaccines. Cell Res. (2021), doi: 10.1038/s41422-021-00514-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baum A, Ajithdoss D, Copin R, Zhou A, Lanza K, Negron N, Ni M, Wei Y, Mohammadi K, Musser B, Atwal GS, Oyejide A, Goez-Gazi Y, Dutton J, Clemmons E, Staples HM, Bartley C, Klaffke B, Alfson K, Gazi M, Gonzalez O, Dick E Jr, Carrion R Jr, Pessaint L, Porto M, Cook A, Brown R, Ali V, Greenhouse J, Taylor T, Andersen H, Lewis MG, Stahl N, Murphy AJ, Yancopoulos GD, Kyratsous CA, REGN-COV2 antibodies prevent and treat SARS-CoV-2 infection in rhesus macaques and hamsters. Science. 370, 1110–1115 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clinical trials of monoclonal antibodies to prevent COVID-19 now enrolling (2020), (available at https://www.nih.gov/news-events/news-releases/clinical-trials-monoclonal-antibodies-prevent-covid-19-now-enrolling).
- 33.Wang Z, Schmidt F, Weisblum Y, Muecksch F, Barnes CO, Finkin S, Schaefer-Babajew D, Cipolla M, Gaebler C, Lieberman JA, Oliveira TY, Yang Z, Abernathy ME, Huey-Tubman KE, Hurley A, Turroja M, West KA, Gordon K, Millard KG, Ramos V, Silva JD, Xu J, Colbert RA, Patel R, Dizon J, Unson-O’Brien C, Shimeliovich I, Gazumyan A, Caskey M, Bjorkman PJ, Casellas R, Hatziioannou T, Bieniasz PD, Nussenzweig MC, mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature (2021), doi: 10.1038/s41586-021-03324-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang P, Nair MS, Liu L, Iketani S, Luo Y, Guo Y, Wang M, Yu J, Zhang B, Kwong PD, Graham BS, Mascola JR, Chang JY, Yin MT, Sobieszczyk M, Kyratsous CA, Shapiro L, Sheng Z, Huang Y, Ho DD, Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 593, 130–135 (2021). [DOI] [PubMed] [Google Scholar]
- 35.Weisblum Y, Schmidt F, Zhang F, DaSilva J, Poston D, Lorenzi JC, Muecksch F, Rutkowska M, Hoffmann H-H, Michailidis E, Gaebler C, Agudelo M, Cho A, Wang Z, Gazumyan A, Cipolla M, Luchsinger L, Hillyer CD, Caskey M, Robbiani DF, Rice CM, Nussenzweig MC, Hatziioannou T, Bieniasz PD, Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. Elife. 9 (2020), doi: 10.7554/eLife.61312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu Z, VanBlargan LA, Rothlauf PW, Bloyet L-M, Chen RE, Stumpf S, Zhao H, Errico JM, Theel ES, Ellebedy AH, Fremont DH, Diamond MS, Whelan SPJ, Landscape analysis of escape variants identifies SARS-CoV-2 spike mutations that attenuate monoclonal and serum antibody neutralization. bioRxiv (2020), doi: 10.1101/2020.11.06.372037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Greaney AJ, Starr TN, Gilchuk P, Zost SJ, Binshtein E, Loes AN, Hilton SK, Huddleston J, Eguia R, Crawford KHD, Dingens AS, Nargi RS, Sutton RE, Suryadevara N, Rothlauf PW, Liu Z, Whelan SPJ, Carnahan RH, Crowe JE Jr, Bloom JD, Complete Mapping of Mutations to the SARS-CoV-2 Spike Receptor-Binding Domain that Escape Antibody Recognition. Cell Host Microbe. 29, 44–57.e9 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kemp SA, Collier DA, Datir R, Ferreira I, Gayed S, Jahun A, Hosmillo M, Rees-Spear C, Mlcochova P, Lumb IU, Roberts DJ, Chandra A, Temperton N, Sharrocks K, Blane E, Briggs J, van Gils MJ, Smith K, Bradley JR, Smith C, Doffinger R, Ceron-Gutierrez L, Barcenas-Morales G, Pollock DD, Goldstein RA, Smielewska A, Skittrall JP, Gouliouris T, Goodfellow IG, Gkrania-Klotsas E, Illingworth C, McCoy LE, Gupta RK, Neutralising antibodies in Spike mediated SARS-CoV-2 adaptation. medRxiv (2020), doi: 10.1101/2020.12.05.20241927. [DOI] [Google Scholar]
- 39.Benton DJ, Wrobel AG, Xu P, Roustan C, Martin SR, Rosenthal PB, Skehel JJ, Gamblin SJ, Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion. Nature. 588, 327–330 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hsieh C-L, Goldsmith JA, Schaub JM, DiVenere AM, Kuo H-C, Javanmardi K, Le KC, Wrapp D, Lee AG, Liu Y, Chou C-W, Byrne PO, Hjorth CK, Johnson NV, Ludes-Meyers J, Nguyen AW, Park J, Wang N, Amengor D, Lavinder JJ, Ippolito GC, Maynard JA, Finkelstein IJ, McLellan JS, Structure-based design of prefusion-stabilized SARS-CoV-2 spikes. Science. 369, 1501–1505 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seydoux E, Homad LJ, MacCamy AJ, Parks KR, Hurlburt NK, Jennewein MF, Akins NR, Stuart AB, Wan Y-H, Feng J, Whaley RE, Singh S, Boeckh M, Cohen KW, McElrath MJ, Englund JA, Chu HY, Pancera M, McGuire AT, Stamatatos L, Analysis of a SARS-CoV-2-Infected Individual Reveals Development of Potent Neutralizing Antibodies with Limited Somatic Mutation. Immunity. 53, 98–105.e5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chao G, Lau WL, Hackel BJ, Sazinsky SL, Lippow SM, Wittrup KD, Isolating and engineering human antibodies using yeast surface display. Nat. Protoc. 1, 755–768 (2006). [DOI] [PubMed] [Google Scholar]
- 43.Miller A, Carr S, Rabbitts T, Ali H, Multimeric antibodies with increased valency surpassing functional affinity and potency thresholds using novel formats. MAbs. 12, 1752529 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beasley MD, Niven KP, Winnall WR, Kiefel BR, Bacterial cytoplasmic display platform Retained Display (ReD) identifies stable human germline antibody frameworks. Biotechnol. J. 10, 783–789 (2015). [DOI] [PubMed] [Google Scholar]
- 45.Yao H, Sun Y, Deng Y-Q, Wang N, Tan Y, Zhang N-N, Li X-F, Kong C, Xu Y-P, Chen Q, Cao T-S, Zhao H, Yan X, Cao L, Lv Z, Zhu D, Feng R, Wu N, Zhang W, Hu Y, Chen K, Zhang R-R, Lv Q, Sun S, Zhou Y, Yan R, Yang G, Sun X, Liu C, Lu X, Cheng L, Qiu H, Huang X-Y, Weng T, Shi D, Jiang W, Shao J, Wang L, Zhang J, Jiang T, Lang G, Qin C-F, Li L, Wang X, Rational development of a human antibody cocktail that deploys multiple functions to confer Pan-SARS-CoVs protection. Cell Res. 31, 25–36 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zost SJ, Gilchuk P, Case JB, Binshtein E, Chen RE, Nkolola JP, Schäfer A, Reidy JX, Trivette A, Nargi RS, Sutton RE, Suryadevara N, Martinez DR, Williamson LE, Chen EC, Jones T, Day S, Myers L, Hassan AO, Kafai NM, Winkler ES, Fox JM, Shrihari S, Mueller BK, Meiler J, Chandrashekar A, Mercado NB, Steinhardt JJ, Ren K, Loo Y-M, Kallewaard NL, McCune BT, Keeler SP, Holtzman MJ, Barouch DH, Gralinski LE, Baric RS, Thackray LB, Diamond MS, Carnahan RH, Crowe JE Jr, Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature. 584, 443–449 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zost SJ, Gilchuk P, Chen RE, Case JB, Reidy JX, Trivette A, Nargi RS, Sutton RE, Suryadevara N, Chen EC, Binshtein E, Shrihari S, Ostrowski M, Chu HY, Didier JE, MacRenaris KW, Jones T, Day S, Myers L, Eun-Hyung Lee F, Nguyen DC, Sanz I, Martinez DR, Rothlauf PW, Bloyet L-M, Whelan SPJ, Baric RS, Thackray LB, Diamond MS, Carnahan RH, Crowe JE Jr, Rapid isolation and profiling of a diverse panel of human monoclonal antibodies targeting the SARS-CoV-2 spike protein. Nat. Med. (2020), doi: 10.1038/s41591-020-0998-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tomic MT, Espinoza Y, Martinez Z, Pham K, Cobb RR, Snow DM, Earnhart CG, Pals T, Syar ES, Niemuth N, Kobs DJ, Farr-Jones S, Marks JD, Monoclonal Antibody Combinations Prevent Serotype A and Serotype B Inhalational Botulism in a Guinea Pig Model. Toxins. 11 (2019), doi: 10.3390/toxins11040208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu L, Wang P, Nair MS, Yu J, Rapp M, Wang Q, Luo Y, Chan JF-W, Sahi V, Figueroa A, Guo XV, Cerutti G, Bimela J, Gorman J, Zhou T, Chen Z, Yuen K-Y, Kwong PD, Sodroski JG, Yin MT, Sheng Z, Huang Y, Shapiro L, Ho DD, Potent neutralizing antibodies directed to multiple epitopes on SARS-CoV-2 spike. Nature (2020), doi: 10.1038/s41586-020-2571-7. [DOI] [PubMed] [Google Scholar]
- 50.Wan J, Xing S, Ding L, Wang Y, Gu C, Wu Y, Rong B, Li C, Wang S, Chen K, He C, Zhu D, Yuan S, Qiu C, Zhao C, Nie L, Gao Z, Jiao J, Zhang X, Wang X, Ying T, Wang H, Xie Y, Lu Y, Xu J, Lan F, Human-IgG-Neutralizing Monoclonal Antibodies Block the SARS-CoV-2 Infection. Cell Rep. 32, 107918 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schaefer W, Regula JT, Bähner M, Schanzer J, Croasdale R, Dürr H, Gassner C, Georges G, Kettenberger H, Imhof-Jung S, Schwaiger M, Stubenrauch KG, Sustmann C, Thomas M, Scheuer W, Klein C, Immunoglobulin domain crossover as a generic approach for the production of bispecific IgG antibodies. Proc. Natl. Acad. Sci. U. S. A. 108, 11187–11192 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li K, Horn GQ, Alam SM, Tomaras GD, Dennison SM, Titrationanalysis: A Tool for High-throughput Analysis of Binding Kinetics Data for Multiple Label-Free Platforms. Biophys. J. 120, 265a–266a (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Punjani A, Rubinstein JL, Fleet DJ, Brubaker MA, cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods. 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 54.Pettersen EF, Goddard TD, Huang CC, Meng EC, Couch GS, Croll TI, Morris JH, Ferrin TE, UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang C, Wang Y, Zhu Y, Liu C, Gu C, Xu S, Wang Y, Zhou Y, Wang Y, Han W, Hong X, Yang Y, Zhang X, Wang T, Xu C, Hong Q, Wang S, Zhao Q, Qiao W, Zang J, Kong L, Wang F, Wang H, Qu D, Lavillette D, Tang H, Deng Q, Xie Y, Cong Y, Huang Z, Development and structural basis of a two-MAb cocktail for treating SARS-CoV-2 infections. Nat. Commun. 12, 264 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG, Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
EM maps in this study have been uploaded to the EMDataResource. The EMDB access numbers are available in the main text and Table S3. Information concerning particular antibodies can be requested through the Coronavirus Immunotherapeutics Consortium at covic@lji.org


