Abstract
Background:
Asthma exacerbations are a serious public health concern due to high health care resource utilization, work/school productivity loss, impact on quality of life, and risk of mortality. The genetic basis of asthma exacerbations has been studied in several populations, but no prior study has performed a multi-ancestry meta-analysis of genome-wide association studies (meta-GWAS) for this trait. We aimed to identify common genetic loci associated with asthma exacerbations across diverse populations and to assess their functional role in regulating DNA methylation and gene expression.
Methods:
A meta-GWAS of asthma exacerbations in 4,989 Europeans, 2,181 Hispanics/Latinos, 1,250 Singaporean Chinese, and 972 African Americans analyzed 9.6 million genetic variants. Suggestively associated variants (p≤5×10−5) were assessed for replication in 36,477 European and 1,078 non-European asthma patients. Functional effects on DNA methylation were assessed in 595 Hispanic/Latino and African American asthma patients and in publicly available databases. The effect on gene expression was evaluated in silico.
Results:
126 independent variants were suggestively associated with asthma exacerbations in the discovery phase. Two variants independently replicated: rs12091010 located at vascular cell adhesion molecule-1/exostosin like glycosyltransferase-2 (VCAM1/EXTL2) (discovery: odds ratio (ORT allele) =0.82, p=9.05×10−6 and replication: ORT allele=0.89, p=5.35×10−3) and rs943126 from pantothenate kinase1 (PANK1) (discovery: ORC allele=0.85, p=3.10×10−5 and replication: ORC allele=0.89, p=1.30×10−2). Both variants regulate gene expression of genes where they locate and DNA methylation levels of nearby genes in whole blood.
Conclusions:
This multi-ancestry study revealed novel suggestive regulatory loci for asthma exacerbations located in genomic regions participating in inflammation and host defense.
Keywords: Asthma exacerbations, EXTL2, GWAS, single nucleotide polymorphism, PANK1
INTRODUCTION
Asthma is a common chronic inflammatory airway disorder affecting over 300 million people worldwide. The disparities in asthma prevalence across populations reflect a complex interplay between environmental exposures (i.e., air pollution and viral infections), behavioral and socioeconomic factors (i.e., treatment adherence and healthcare access), and genetic ancestry, which is a complex trait measured by background whole-genome variation that tracks with geographic and historical factors as well as the aforementioned factors influencing asthma prevalence (1,2).
Asthma exacerbations are defined as worsening of respiratory symptoms requiring hospitalization, unscheduled/emergency asthma care, and/or use of systemic corticosteroids (3). Prevention of asthma exacerbations is a major public health priority due to their associated consequences on health (i.e., decreased quality of life, accelerated decline in lung function, or mortality), school attendance, work productivity, and healthcare costs (1,4,5). To date, the best predictor of future exacerbations is the occurrence of one in the previous year (6). Thus, identifying potential biomarkers to guide the reduction and prevention of exacerbations is a priority for therapeutics development and for precision medicine of asthma.
With the advent of high-throughput sequencing and genotyping technologies, the study of the genetic contributions to asthma exacerbations has shifted from hypothesis-driven, limited candidate-gene strategies to genome-wide association studies (GWAS) (7).(7–14) Pharmacogenomics studies of asthma exacerbations as an outcome of treatment response have identified five suggestive associations for asthma exacerbations despite inhaled corticosteroids (CMTR1 (9), APOBEC3B-APOBEC3C (8), and CACNA2D3-WNT5A (11)), or long-acting beta2-agonists (TBX3 and EPHA7) (10). Beyond pharmacogenomics, other studies have focused on asthma exacerbations independently of treatment. In European-descent populations, CDHR3, CTNNA3, and HLA-DQB1 have been associated with severe asthma exacerbations (7,13). More recently, the representation of ethnically diverse populations has increased in GWAS of asthma exacerbations. A meta-analysis of GWAS in Hispanic/Latino children identified a single nucleotide polymorphism (SNP) at FLJ22447 that modulated KCNJ2-AS1 expression in nasal epithelium through DNA methylation (12). In Hispanic/Latinos and African Americans, a genome-wide significant locus for asthma with exacerbations regulated LINC01913 lung gene expression and DNA methylation levels of the PKDCC gene in whole blood (14). However, none of those studies has approached the search for genetic determinants of asthma exacerbations independently of treatment from a multi-ancestry framework.
To improve our understanding on genetic and biological mechanisms of asthma exacerbations across multiple populations, we conducted the first multi-ancestry meta-analysis of GWAS of asthma exacerbations independently of treatment and attempted to validate previous associations. Then, we conducted in silico and in vivo downstream analyses to assess the potential functional effects of the associated SNPs over DNA methylation and gene expression.
METHODS
Study design and study populations
We performed a two-stage study to identify genetic variants associated with asthma exacerbations, defined as a binary variable based on the presence of emergency care, hospitalizations, or administration of systemic corticosteroids because of asthma. We also considered a definition of moderate exacerbations (3), comprising unscheduled general practitioner or pulmonary specialist visits and school absence since no information on the former variables was available for some studies. A period of 6 to 24 months or ever was considered depending on the data available for each study (Table S1–S2). In the discovery phase, we performed a multi-ancestry meta-analysis of GWAS of asthma exacerbations in 9,392 patients with asthma from 12 studies, including 4,989 European-descents from nine studies, 2,181 Hispanics/Latinos, 1,250 Singaporean Chinese, and 972 African Americans. We attempted to replicate the findings from the discovery phase in a total of 37,555 participants with asthma, including 36,477 Europeans from seven studies, 877 Latinos from two studies, and 201 Filipinos from one study (Table S2). A detailed description of each study is available in the Supporting Information. All studies included were approved by their respective Institutional review boards, and written informed consent was provided by participants or their parents/caregivers. All methods followed the Declaration of Helsinki guidelines.
Assessment of genetic ancestry was performed using principal component analysis. The Haplotype Reference Consortium (r1.1 2016) (15) was used as the reference imputation panel for most studies, except for Avon Longitudinal Study of Parents and Children (ALSPAC) and Singapore Cross Sectional Genetic Epidemiology Study (SCSGES), which used the phase 3 of the 1000 Genomes Project (1KGP) (16). Genotyping and imputation procedures for the discovery and replication studies are detailed in the Supporting Information and Tables S1–S2.
Association analysis
Association between genetic variants and asthma exacerbations was tested using logistic regression models including age, sex, and principal components from the genotype matrix (if needed to correct for population stratification) (Table S1). Analyses were conducted separately for each study using PLINK 2.0 (17), EPACTS 3.2.6 (18) or rvtests 2.1.0 (19). Results were filtered with the EasyQC software (20) to retain variants with a minor allele frequency (MAF)≥1% and imputation quality R2≥0.3, absolute value of the beta coefficient <10, standard error of the beta included in the interval [0,10], and minor allele cut-off ≥6.
In the discovery phase, genetic variants that were available in at least two ethnic-specific studies were meta-analyzed with METASOFT (21), using fixed-effects or random-effects models based on the heterogeneity among studies (measured by the Cochran’s Q test p-value). Ethnic-specific results were then combined in a multi-ancestry meta-analysis. Independent variants (r2≤0.8) with suggestive association at p≤5×10−5 (22) within 1 Megabase were identified with GCTA-COJO v1.93.2 (23) using the 1KGP reference (16). These variants were evaluated in the replication stage, following the same procedures as in the discovery phase. Evidence of replication was considered if the variants showed consistent direction of effects with the discovery stage at p≤0.05.
Assessment of shared genetic basis of asthma exacerbations with other traits
To identify groups of genes previously associated with other traits, we used a Gene-Set Enrichment Analysis (GSEA), as implemented in FUMA GWAS (24) via the GENE2FUNC algorithm, and queried the GWAS catalog (25). SNPs with p≤1×10−4 in the discovery phase of the meta-analysis of GWAS were mapped to the closest gene using the UCSC Table Browser tool (26). A false discovery rate (FDR) of 5% was used to declare significance.
To estimate the pairwise genome-wide genetic correlations (Rg) between asthma exacerbations and other traits, we compared our findings with publicly-available GWAS summary statistics via LD score regression using LDHub (27). Since most of the GWAS have been conducted in European populations, the analysis was restricted to predominantly European-descent individuals to maximize the statistical power. A Bonferroni-corrected significance threshold of p<0.05/711 traits=6.48×10−5 was applied.
Sensitivity analysis
In order to assess the robustness of the genetic associations, we conducted sensitivity analyses for the time-dependent probability occurrence of exacerbations, the effect of Body Mass Index (BMI), obesity, asthma severity, and age group. Moreover, we evaluated the association of the variants with asthma susceptibility, as detailed in the Supporting Information. Studies from the discovery stage that had covariate data available were considered.
Methylation profiling and quality control
Whole blood DNA methylation from Hispanics/Latinos and African Americans was profiled using the Infinium HumanMethylation450 BeadChip or the Infinium Methylation EPIC BeadChip arrays. Briefly, low-quality probes and samples, outliers of DNA methylation, and samples with sex mismatch or mixed genotype distributions on the control SNP probes were excluded. Standard background correction, dye-bias correction, inter-array normalization, and probe-type bias adjustment were performed, and beta values were transformed to M-values for better statistical performance. Quality control is detailed in the Supporting Information.
Functional assessment of associated SNPs
DNA methylation quantitative trait loci (meQTL) analyses were conducted using fastQTL (28) for CpG sites within 1 Mb of SNPs with MAF≥0.01 in at least 10 samples, separately in 139 Mexican Americans and 241 Puerto Ricans from Genes-Environments & Admixture in Latino Americans (GALA II) and 215 African Americans from the Study of African Americans, Asthma, Genes & Environments (SAGE) studies. Linear regression models were corrected for asthma exacerbations status, age, sex, genetic ancestry, ReFACTor components as a proxy of cell heterogeneity, and methylation batch (when appropriate). The results from Mexican Americans and Puerto Ricans assayed with different methylation arrays were then meta-analyzed for each sub-ethnic group with METASOFT (21). SNP-CpG pairs were considered significant at Storey q-value <0.05. In silico evidence of functional effects of variants on gene expression and DNA methylation was assessed using QTLbase (29), Genotype-Tissue Expression (GTEx) v8 Portal (30), PhenoScanner v2 (31) and eFORGE-TF (32). Long-distance chromatin interactions were determined using the ChiCP tool (33).
Validation of previous associations
A literature search for all studies reporting genetic loci significantly associated with asthma exacerbations was conducted, as described in the Supporting Information. Association results in the discovery stage were extracted and significance threshold was defined as p = 0.05/number of tested SNPs to adjust for multiple testing.
RESULTS
Characteristics of the patients
In the discovery phase, we analyzed 2,781 exacerbators and 6,611 non-exacerbators; 53.1% were predominantly Europeans, 23.2% Hispanics/Latinos, 13.3% Singaporean Chinese, and 10.3% African Americans. The percentage of exacerbators ranged from 9.1% to 65.2% in Europeans, and reached 58.8% in Hispanics/Latinos, 46.1% in African Americans, and 3.4% in Singaporeans. The replication phase included 37,555 individuals with asthma (3,030 exacerbators and 34,525 non-exacerbators) where most participants were of European-descent (97.1%), followed by Latinos (2.3%) and Filipinos (0.5%). The percentage of exacerbators ranged from 4.8% to 65.2% in Europeans, reached approximately 43% in Latinos, and 1.3% in Filipinos (Table S1–S2). Regarding sex, 51.7% and 42.9% of participants were male in the discovery and replication phases, respectively.
Discovery phase
The Quantile-quantile plots did not show major genomic inflation due to population stratification in each individual study (Figure S1), the combined results from individuals of European descent (Figure S2), or the multi-ancestry meta-analysis (Figure S3). In the multi-ancestry meta-analysis of 9,634,748 variants, 447 SNPs exhibited suggestive association (Table S3). The most significant association was the intronic SNP rs6888198 within the cadherin-12 (CDH12) gene at chromosome 5p14.3 (odds ratio [OR] for C allele: 1.37, 95% confidence interval [CI]: 1.23–1.54, p=1.95×10−8) (Figure 1, Figure S4).
Figure 1. Manhattan plot of the results of the discovery stage of the multi-ancestry meta-analysis of GWAS of asthma exacerbations (represented as -log10 p-value on the y-axis) along the chromosome position of the variants analyzed (x-axis).
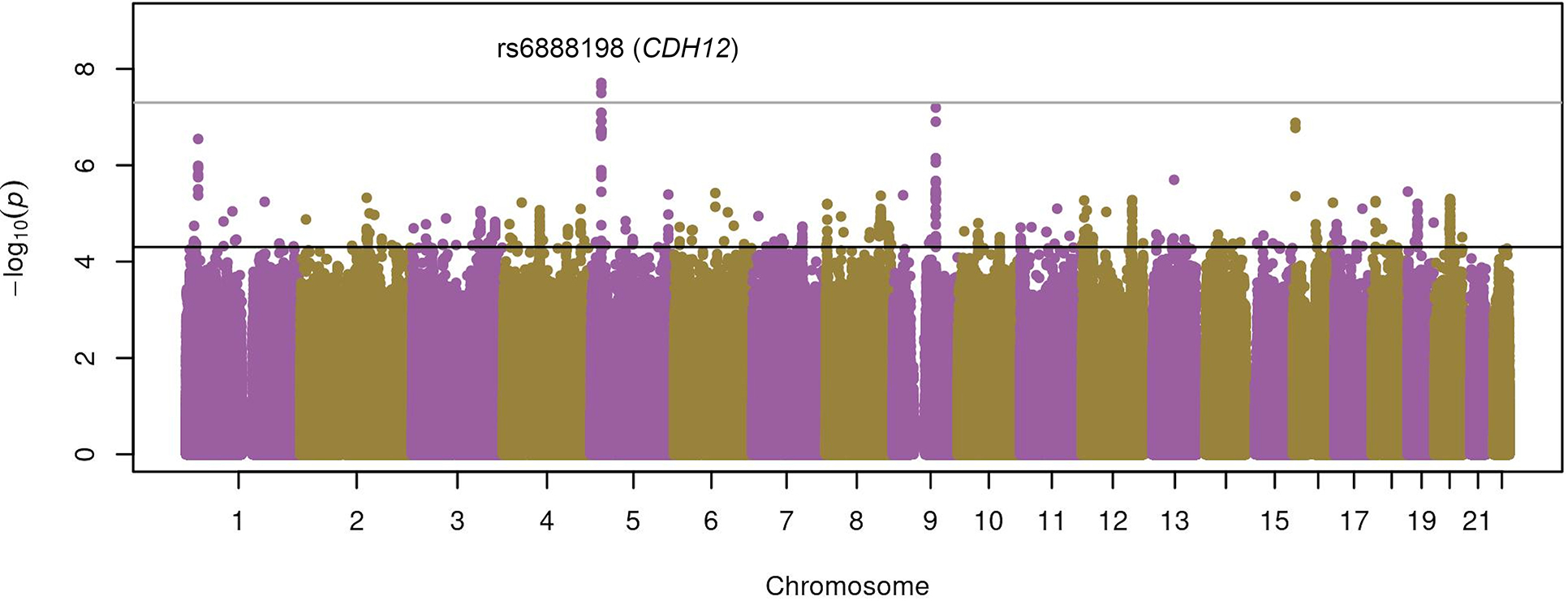
The suggestive (p=5×10−5) and genome-wide (p=5×10−8) significance thresholds are indicated by the black line and dark gray lines.
Replication phase
Fifteen of the 126 independent variants identified in the discovery phase were not available for replication since they were mostly present in African Americans and Hispanics/Latinos (Table S3). Two of the 106 variants present in more than one ethnic group were consistently associated with asthma exacerbations (Table 1): rs12091010 [VCAM1/EXTL2, OR for T allele: 0.89 (0.82–0.97), p=5.35×10−3] (Figure 2) and rs943126 [PANK1, OR for C allele: 0.92 (0.86–0.98), p=1.30×10−2] (Figure 3). In the meta-analysis across both phases, these variants reached an association p-value of 4.23×10−7 and 4.93×10−6, respectively. From five variants that were present only in non-Europeans in the replication stage, none exhibited p<0.05 in any other population group (Table S4). Even though rs6888198 reached genome-wide significance in the discovery and showed consistent effects among Europeans in the replication phase, this SNP had opposite effects in Latinos and Filipinos, which resulted in the lack of replication in the multi-ancestry replication phase (Table 1, Figure S5).
Table 1.
Association results for the top hit in the discovery stage and sentinel variants with significant and consistent effects in the discovery and replication phases, and the meta-analysis across both phases.
| Discovery | Replication | Meta-analysis (discovery and replication) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|||||||||||
| ID† | rsID | Closest gene | OR (95% CI) | P | Cochran’s Q P | OR (95% CI) | P | Cochran’s Q P | OR (95% CI) | P | Cochran’s Q P |
|
| |||||||||||
| 1:101210560:C:T | rs12091010 | EXTL2 | 0.82 (0.75–0.90) | 9.05E-06 | 4,83E-01 | 0.89 (0.82–0.97) | 5.35E-03 | 4.92E-01 | 0.86 (0.81–0.91) | 4.23E-07 | 4.47E-01 |
| 5:22659406:T:C | rs6888198 | CDH12 | 1.37 (1.23–1.54) | 1.95E-08 | 5,82E-01 | 1.02 (0.90–1.15) | 7.72E-01 | 7.18E-01 | 1.24 (1.05–1.45) | 2.41E-06 | 1.89E-02 |
| 10:91376299:T:C | rs943126 | PANK1 | 0.85 (0.78–0.92) | 3.10E-05 | 8,01E-02 | 0.92 (0.86–0.98) | 1.30E-02 | 3.87E-01 | 0.89 (0.85–0.94) | 4.93E-06 | 7.91E-02 |
The variant identifier corresponds to chromosomal position (hg19) followed by non-tested allele and tested allele.
Abbreviations: 95% CI: 95% confidence interval; OR: Odds ratio; P: P-value.
Figure 2. Forest plot of the association results for rs12091010 (VCAM1/EXTL2) in the meta-analysis of GWAS of asthma exacerbations.
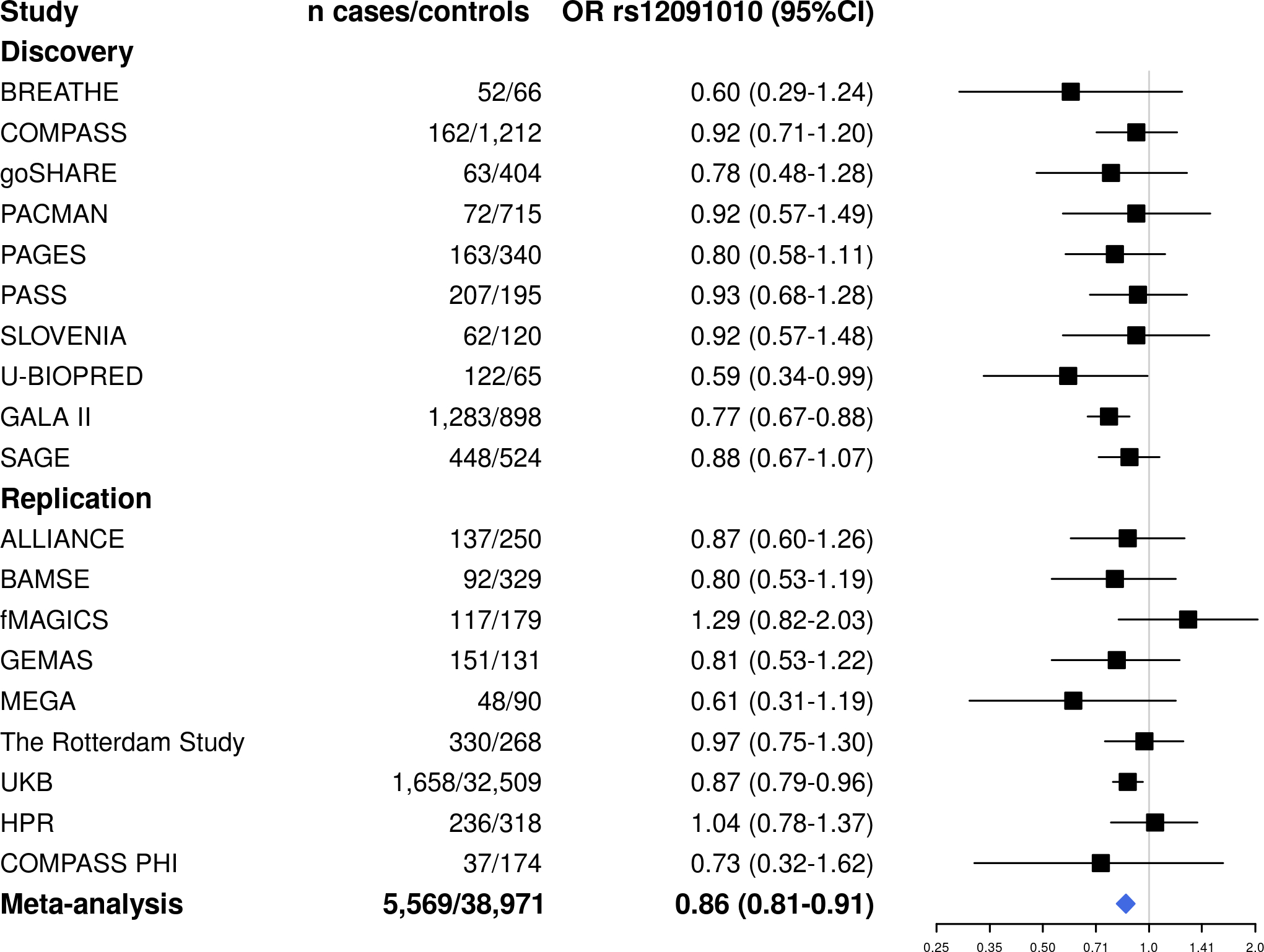
ALSPAC (discovery), SCSGES (discovery), and the subset of samples from BREATHE genotyped with the Illumina Infinium CoreExome-24 BeadChip (replication) had no genotyped or imputed data for rs12091010.
Figure 3. Forest plot of the association results for rs943126 (PANK1) in the meta-analysis of GWAS of asthma exacerbations.
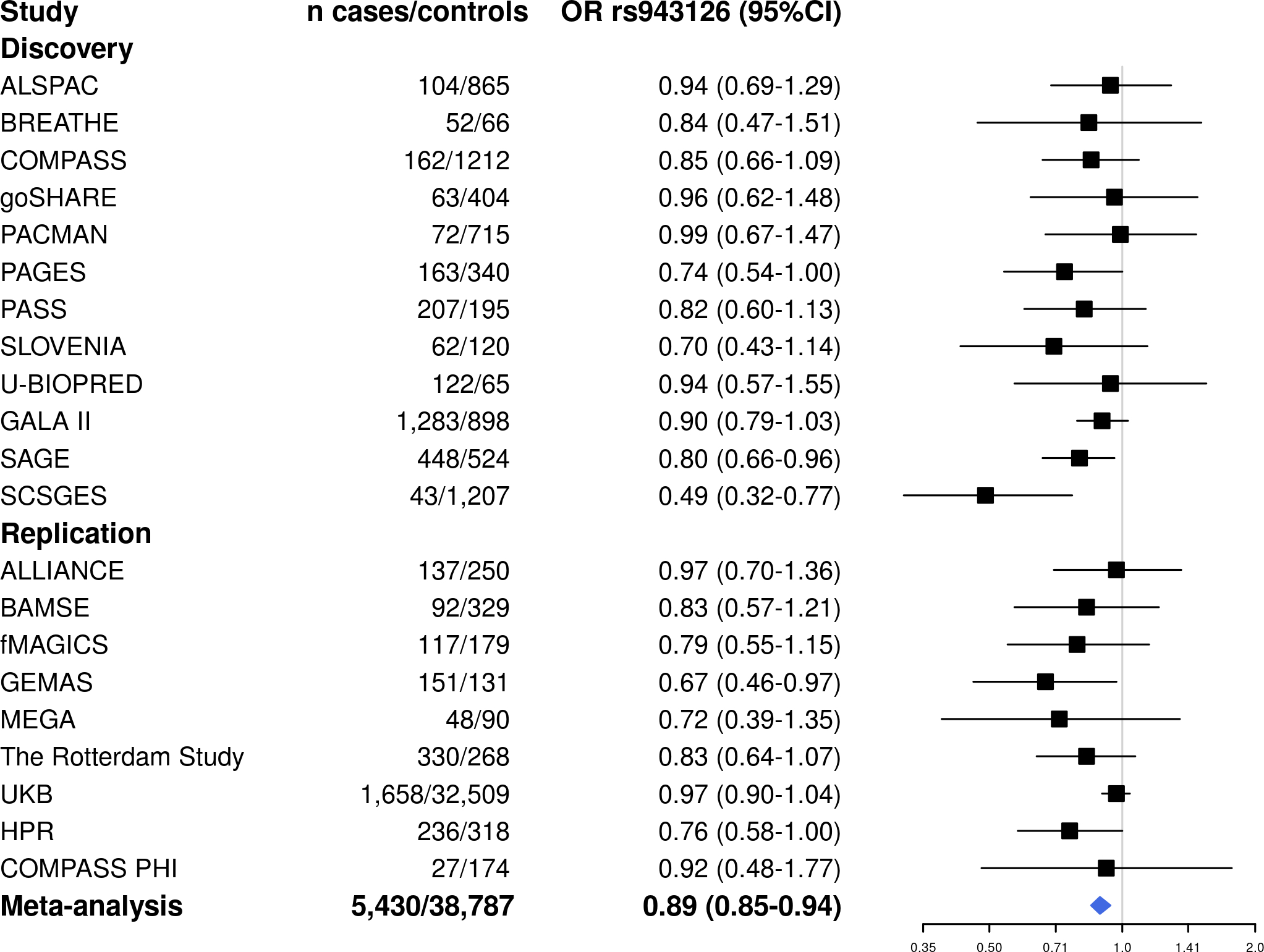
The subset of samples from BREATHE genotyped with the Illumina Infinium CoreExome-24 BeadChip (replication) had no available genotyped or imputed data for rs943126.
Gene-set enrichment and genome-wide genetic correlation analysis
Enrichment analysis of associations from the multi-ancestry discovery GWAS including 959 SNPs associated with asthma exacerbations at p≤1×10−4 revealed significant enrichment in several traits, including treatment response (min p=2.77×10−6), neurological conditions (min p=4.62×10−5), obesity (min p=6.52×10−5), or waist-to-hip ratio (min p=1.88×10−7) (Table S5).
A total of 16 traits exhibited genetic correlation with asthma exacerbations at p<0.05 (Table S6), including wheeze or whistling in the last year (Rg=0.47, p=1.01×10−2), emphysema/chronic bronchitis (Rg=0.55, p=3.89×10−2), asthma (Rg=0.32, p=3.99×10−2), and BMI (Rg=0.19, p=4.76×10−2). However, the associations did not remain significant after Bonferroni correction.
Sensitivity analysis
To assess the robustness of associations that replicated across stages to the time-dependent probability of occurrence of exacerbations, stratified analyses were performed in European-descents from the discovery stage that reported exacerbations for 6 vs. 12 months. Consistent effects per period were observed across periods (Table 3).
Table 3.
Sensitivity analysis for rs12091010 and rs943126 in individuals from the discovery stage.
| Exacerbations in the last 6 months | Exacerbations in the last 12 months | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| European-descent populations | European-descent populations | Multi-ancestry meta-analysis | |||||||
|
| |||||||||
| rsID | OR (95% CI) | P | Cochran’s Q P | OR (95% CI) | P | Cochran’s Q P | OR (95% CI) | P | Cochran’s Q P |
|
| |||||||||
| rs12091010 | 0.84 (0.67–1.04) | 1.08×10−1 | 5.14 ×10−1 | 0.86 (0.72–1.03) | 9.13 ×10−2 | 6.25 ×10−1 | 0.82 (0.74–0.90) | 3.45 ×10−5 | 4.84 ×10−1 |
| rs943126 | 0.78 (0.64–0.96) | 2.13 ×10−2 | 8.61 ×10−1 | 0.88 (0.74–1.04) | 1.26 ×10−1 | 8.28 ×10−1 | 0.85 (0.78–0.93) | 3.29 ×10−4 | 7.50 ×10−2 |
Abbreviations: 95% CI: 95% confidence interval; OR: Odds ratio; P: P-value.
Since the post-GWAS analyses revealed significant enrichment/correlation at p<0.05 with fat mass/distribution, the association of rs12091010 and rs943126 after additional adjustment by BMI/obesity was examined in individuals from the discovery phase with BMI data available. Moreover, the effect of asthma severity alone or combined with BMI/obesity on the genetic association exacerbations was evaluated. The effects sizes of the genetic association after additional adjustment by these variables remained consistent with the effects reported in the discovery stage (Table S7).
We next investigated if the observed effects could differ across age groups in those studies that analyzed exclusively children or adults, but the effect sizes remained consistent across age groups (Table S8). Moreover, to assess if the effects could be driven by the underlying asthma syndrome rather than asthma exacerbations and no significant association with asthma was found in results from the UK Biobank or the Michigan Genomics Initiative (Table S9).
Functional exploration of variants associated with asthma exacerbations
We next assessed for association DNA methylation in whole blood at 525, and 538 CpG sites with rs12091010 and rs943126, respectively. A total of 7 and 1 SNP-CpG pairs for rs943126 and rs12091010 exhibited Storey q<0.05, respectively (Table 2, Table S10). Two of these replicated consistently in Europeans for rs943126 (cg25770176 and cg00475140). In silico analyses, revealed 10 SNP-CpGs pairs, 3 of which showed consistent effects in Hispanics/Latinos and African Americans at Storey q<0.05 (Tables S11–S12) including the previous two pairs and rs943126-cg03948048. The 8 significant CpG sites in minority children showed significant enrichment (q<0.001) in transcription factor (TF) motifs in lung (Table S13). Besides, the T allele of rs12091010 was associated with decreased EXTL2 expression in whole blood from Europeans, according to PhenoScanner (31). The C allele of rs943126 was associated with increased expression of PANK1 in whole blood from Europeans (Table S14). Both variants showed evidence of long-range chromatin interaction with several genes in lymphoblastoid cells, including VCAM1 and EXTL2 for rs12091010 and PANK1 for rs943126 (Table S15).
Table 2.
Results from the meQTL analysis in whole blood in the GALA II and SAGE studies for genome-wide significant hit in the discovery and two SNPs that were replicated.
| Mexican Americans | Puerto Ricans | African Americans | Meta-analysis | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||||||
| SNP-CpG pair | Position (hg19) | Closest genes | Coef | SE | P | Coef | SE | P | Coef | SE | P | Coef | SE | P | Cochran’s Q P | Storey q |
|
| ||||||||||||||||
| rs943126-cg26800131 | 91574784 | KIF20B | −0.21 | 0.11 | 2.99E-04 | −0.07 | 0.04 | 6.99E-02 | −0.07 | 0.04 | 8.87E-02 | −0.18 | 0.10 | 1.85E-06 | 4.45E-03 | 9.95E-04 |
| rs943126-cg14920044 | 91296311 | SLC16A12 | 0.09 | 0.05 | 9.77E-02 | 0.24 | 0.06 | 1.68E-05 | 0.12 | 0.06 | 4.68E-02 | 0.15 | 0.03 | 4.61E-06 | 1.24E-01 | 1.24E-03 |
| rs943126-cg20654695 | 91444521 | KIF20B/PANK1 | −0.10 | 0.08 | 2.12E-01 | −0.06 | 0.04 | 9.04E-02 | −0.15 | 0.03 | 2.04E-05 | −0.10 | 0.02 | 9.96E-06 | 1.98E-01 | 1.79E-03 |
| rs943126-cg25770176 | 91405685 | PANK1 | −0.09 | 0.03 | 1.43E-03 | 0.00 | 0.09 | 2.78E-01 | −0.07 | 0.02 | 3.00E-03 | −0.07 | 0.02 | 1.86E-05 | 1.49E-01 | 2.50E-03 |
| rs12091010-cg05612904 | 101491636 | DPH5 | −0.07 | 0.04 | 6.99E-02 | −0.21 | 0.11 | 2.99E-04 | −0.09 | 0.04 | 2.72E-02 | −0.10 | 0.02 | 2.31E-05 | 9.52E-02 | 1.20E-02 |
| rs943126-cg00475140 | 91404454 | PANK1 | −0.21 | 0.07 | 1.39E-03 | −0.13 | 0.08 | 1.05E-01 | −0.19 | 0.09 | 3.19E-02 | −0.18 | 0.04 | 4.28E-05 | 5.25E-01 | 4.60E-03 |
| rs943126-cg15620114 | 91296457 | SLC16A12 | 0.09 | 0.08 | 2.64E-01 | 0.27 | 0.09 | 3.87E-03 | 0.20 | 0.08 | 7.76E-03 | 0.18 | 0.05 | 1.53E-04 | 5.70E-01 | 1.32E-02 |
| rs943126-cg04957662 | 91411382 | KIF20B/PANK1 | −0.34 | 0.79 | 6.69E-01 | −1.16 | 0.29 | 1.33E-04 | −0.77 | 0.26 | 2.89E-03 | −1.02 | 0.27 | 1.72E-04 | 3.14E-01 | 1.32E-02 |
Abbreviations: Coef: Coefficient of the regression; P: P-value; SE: Standard error; Storey q: Storey q-value.
Validation of previous associations
We next examined 47 previous genetic loci for asthma exacerbations (7,8,12,13,34–36) and moderate-to-severe asthma (37) for association with asthma exacerbations in the discovery phase. A total of 5 variants had p<0.05 in Europeans, 2 in Hispanics/Latinos, 5 in African Americans, and 1 in Singaporean Chinese (Table S16). These were in loci previously associated with asthma exacerbations (GSDMB, RAD50, HLA-DQB1, ADAM33, VDR, and CDHR3) or moderate-to-severe asthma (IKZF3, TSLP, MUC5AC, C11orf30, SMAD3, and WDR36). However, none of the SNPs exceeded the stringent Bonferroni-corrected threshold for significance (p=0.05/47=1.06×10−3).
DISCUSSION
To our knowledge, this is the first multi-ancestry meta-analysis of GWAS of asthma exacerbations independently of treatment including European, Hispanic/Latino, Asian, and African American patients with asthma. In our combined analysis of 46,947 individuals with asthma, two regulatory SNPs were significantly and consistently associated with asthma exacerbations in most of the studies included in the discovery and replication phases, independently of the type of exacerbation and the time period for which the exacerbation status was assessed. The SNP rs120910109 was located in the intergenic region of the VCAM1/EXTL2 genes whereas rs943126 was harbored within an intron 1 of PANK1.
VCAM1 encodes a surface protein predominantly expressed in endothelial cells that modulates leukocyte adhesion and trans-endothelial migration in response to pro-inflammatory cytokines, and lipopolysaccharide (LPS) among other factors (38,39). VCAM1 is involved in cancer progression and several immunological disorders, including asthma (38). In the ovalbumin mice model, anti-VCAM1 reduced airway hyperresponsiveness and eosinophilic inflammation (40). On the other hand, EXTL2 encodes an enzyme that controls glycosaminoglycan (GAG) biosynthesis via transference of N-acetylgalactosamine and N-acetylglucosamine to the glycosaminoglycan-protein linkage region (41). Decreased EXTL2 causes an over-accumulation of GAGs (42) that can promote inflammation in injured areas (43,44). Moreover, in bone marrow-derived macrophages from EXTL2−/− mice, there is overproduction of key molecules involved in inflammation and extracellular matrix remodeling, including tumor necrosis factor α (TNFα) and several matrix metalloproteinases (43). In a scenario of overaccumulation of GAGs under the loss of EXTL2 in macrophages, GAGs act as inflammatory mediators with strong Toll-like receptor 4 (TLR4) agonist capacity (44). Interestingly, genetic variation both VCAM1 and EXTL2 is associated with blood cell counts, and multiple sclerosis, according to the GWAS catalog (25).
PANK1 catalyzes coenzyme A biosynthesis, regulated by the transcription factor peroxisome proliferator-activating receptor α (PPAR-α) (45), a key anti-inflammatory factor in asthma (46). A decrease in PPAR-α expression is accompanied by a decrease in the expression of PANK1 and miR-107, which is encoded within the intron 5 of PANK1. TLR4 can also downregulate miR-107. In turn, this leads to a higher cyclin-dependent kinase 6 (CDK6) expression and subsequently increases the adhesion of macrophages in response to LPS (45). Bioproducts from bacterial infections, such as LPS, can trigger an inflammatory response and increase airway hyperresponsiveness and risk of asthma exacerbations (47,48). Moreover, p53 can regulate cell cycle progression via upregulation of PANK1 after DNA damage (49) and metabolism (50).
To prioritize gene targets, we assessed the functional capacity of relevant SNPs (51). Both rs12091010 and rs943126 exhibited an association with DNA methylation at several nearby CpG sites in whole blood from African Americans and Hispanics/Latinos with asthma. Additionally, the SNPs rs12091010 and rs943126 were associated with EXTL2 and PANK1 gene expression in whole blood from Europeans. Specifically, the T allele of rs12091010, located at 6 kb downstream of the 3’ UTR of VCAM1 and 150 kb upstream of the transcription start site of EXTL2, was associated with lower odds of having asthma exacerbations and decreased EXTL2 expression (31) The T allele is more common among Latinos/Admixed Americans, followed by Europeans, Africans, and East Asians (Figure S6). The T allele of rs943126 at PANK1, which is less common among Europeans than the rest of populations (Figure S7), was associated with a higher risk of asthma exacerbations in the combined analysis of the discovery and replication phases and with decreased gene expression of PANK1 in whole blood from Europeans. However, these eQTL effects were not validated in the GTEx data (30).
In the discovery phase, the most significant association was located at the intronic SNP rs6888198 (CDH12), but no evidence of replication was found in the second stage (p>0.05) despite the consistency of the direction of the effect across study phases. Interestingly, rs6888198 showed variable MAF among populations, with the largest MAF among Africans and Latinos (Figure S8). CDH12 has been associated with angiogenesis and progression of several types of cancers (52–54). Specifically, in colorectal cancer, it has been suggested that CDH12 increases cancer cell migration by promoting epithelial-mesenchymal transition via activation of the Snail transcription factor pathway. CDH12 expression is positively modulated by the chemotactic factor CCL2 (53,54), whose levels increases in blood and airway smooth muscle from asthma patients compared to healthy controls (55).
We also attempted to assess previously associated loci for asthma exacerbations or moderate-to-severe asthma for association with asthma exacerbations in multiple ethnic groups. Although several variants showed association at p<0.05, none surpassed the stringent Bonferroni correction, which could be due to differences in study design, phenotype definition, ethnicity, and clinical characteristics, among others. Of note, none of the previous findings was initially described in Asian or African populations, which highlights the need to increase ethnic diversity in genomic studies of asthma exacerbations.
Our study has several limitations. First, the VCAM1/EXTL2 and PANK1 loci did not surpass a stringent Bonferroni threshold of 4.7×10−4 (p=0.05/106 variants) in the replication stage nor the genome-wide significance in the combined analysis from all studies. Second, these loci exhibited modest effects sizes, which could impact the clinical relevance of these loci. Third, the history of asthma exacerbations was based on retrospective questionnaires in all cohorts but COMPASS, a randomized, prospective clinical trial. Fourth, to bring together large sample sizes necessary to map susceptibility variants, we considered studies where asthma exacerbations were reported for the previous 6 to 24 months or ever, which may have introduced some heterogeneity in the phenotype. Moreover, the replication stage comprised mostly European individuals, which hindered our capability to replicate associations driven in the discovery stage by non-Europeans. Despite these limitations, our findings exhibited consistent effects for the VCAM1/EXTL2 and PANK1 loci independent of the time period assessed. Future studies in adequately powered and phenotypically harmonised cohorts should untangle the role of these loci in the time-to-first exacerbation, the annual number of exacerbations, or the temporal distance among events, explore other epigenetic mechanisms known to be involved in asthma (e.g., histone modifications or miRNAs) (56), and the biological function of these genes. Moreover, although asthma exacerbation risk is influenced by sex in an age-dependent manner (57), and our analyses were corrected for sex, future genome-wide gene-by-sex interaction scans may reveal the influence of sex on the genetic susceptibility to exacerbations. On the other hand, we acknowledge several study strengths. Firstly, we leveraged clinical and genetic data from 46,947 asthma patients from different ethnicities from 18 independent studies. Our study had statistical power ≥80% to detect associations with MAF> 17% and relative risk (RR)>1.20 in the discovery stage and for variants with MAF≥1%, and was powered at 80% to detect associations with larger effect sizes (RR≥1.85). Second, we identified novel, biologically plausible genetic factors of asthma exacerbations demonstrated by transcriptomics and epigenomics studies and evidence for prior literature. Moreover, we accounted for blood cell-type heterogeneity to overcome the limitations of analyzing mixed cell types tissues (56,58). Third, we evaluated previous genetic signals from asthma exacerbations in populations from several ancestries.
We identified suggestive loci for asthma exacerbations with consistent genetic effects across individuals from varying ancestral backgrounds using a multi-ancestry approach. We also demonstrated that these loci are biologically functional and regulate RNA expression and adjacent CpG site DNA methylation as meQTL in whole blood cells. Our findings highlight VCAM1, EXTL2 and PANK1 as functional loci for asthma exacerbations applicable to people across different ancestral backgrounds warranting future investigation of these novel genomic mechanisms underlying asthma exacerbations.
Supplementary Material
KEY MESSAGES.
A large multi-ancestry meta-analysis of GWAS of asthma exacerbations revealed two novel susceptibility loci located close to PANK1 and at the intergenic region of VCAM1 and EXTL2. These loci decreased PANK1 and EXTL2 gene expression in whole blood, respectively. Both genetic variants were associated with DNA methylation levels at CpG sites nearby. Our results identified two gene targets for asthma exacerbations that should be further explored to assess their specific role in asthma.
ACKNOWLEDGMENTS
The authors thank the patients, families, recruiters, health care providers, and community clinics for their participation in the studies analyzed in this manuscript. The authors also acknowledge the contribution of the high-performance compute cluster Wynton HPC underlying UCSF’s Research Computing Capability to the results of this research and Sandra Salazar for her support as GALA II and SAGE study coordinator. The authors thank the Centro Nacional de Genotipado-Plataforma de Recursos Biomoleculares-Instituto de Salud Carlos III (CeGen-PRB3-ISCIII; www.cegen.org) for providing the genotyping services. The authors also acknowledge all the families who took part in ALSPAC, the midwives for their help in recruiting them, and the whole ALSPAC team, which includes interviewers, computer and laboratory technicians, clerical workers, research scientists, volunteers, managers, receptionists, and nurses.
FINANCIAL SUPPORT
This work was funded by the Spanish Ministry of Science and Innovation MCIN/AEI/10.13039/501100011033, and the European Regional Development Fund “ERDF A way of making Europe” by the European Union (SAF2017-83417R), by MCIN/AEI/10.13039/501100011033 (PID2020-116274RB-I00) and by the Allergopharma-EAACI award 2021. This study was also supported by the SysPharmPedia grant from the ERACoSysMed 1st Joint Transnational Call from the European Union under the Horizon 2020.
GALA II and SAGE studies were supported by the Sandler Family Foundation, the American Asthma Foundation, the RWJF Amos Medical Faculty Development Program, Harry Wm. and Diana V. Hind Distinguished Professor in Pharmaceutical Sciences II, the National Heart, Lung, and Blood Institute of the National Institutes of Health (R01HL117004, R01HL128439, R01HL135156, X01HL134589, R01HL141992, and R01HL141845), National Institute of Health and Environmental Health Sciences (R01ES015794 and R21ES24844); the National Institute on Minority Health and Health Disparities (NIMHD) (P60MD006902, R01MD010443, and R56MD013312); the National Institute of General Medical Sciences (NIGMS) (RL5GM118984); the Tobacco-Related Disease Research Program (24RT-0025 and 27IR-0030); and the National Human Genome Research Institute (NHGRI) (U01HG009080) to EGB. The PACMAN study was funded by a strategic alliance between GlaxoSmithKline and Utrecht Institute for Pharmaceutical Sciences. The SLOVENIA study was financially supported by the Slovenian Research Agency (research core funding No. P3-0067) and from SysPharmPediA grant, co-financed by the Ministry of Education, Science and Sport Slovenia (MIZS) (contract number C3330-16-500106). The SHARE Bioresource (GoSHARE) and SHARE have ongoing funding from NHS Research Scotland and were established by funding from The Wellcome Trust Biomedical Resource [Grant No. 099177/Z/12/Z]. Genotyping of samples from BREATHE, PAGES, and GoSHARE was funded by AC15/00015 and conducted at the Genotyping National Centre (CeGEN) CeGen-PRB3-ISCIII; supported by ISCIII and European Regional Development Fund (ERDF) (PT17/0019). ALSPAC was supported by the UK Medical Research Council and Wellcome (102215/2/13/2) and the University of Bristol. The Swedish Heart-Lung Foundation, the Swedish Research Council, and Region Stockholm (ALF project and database maintenance) funded the BAMSE study. The PASS study was funded by the NHS Chair of Pharmacogenetics via the UK Department of Health. U-BIOPRED was funded by the Innovative Medicines Initiative (IMI) Joint Undertaking, under grant agreement no. 115010, resources for which are composed of financial contribution from the European Union’s Seventh Framework Programme (FP7/2007–2013) and kind contributions from companies in the European Federation of Pharmaceutical Industries and Associations (EFPIA). Genotyping of samples from GEMAS and MEGA studies was funded by the Spanish Ministry of Science and Innovation (SAF2017-87417R) at the Spanish National Cancer Research Centre, in the Human Genotyping lab, a member of CeGen, PRB3, and was supported by grant PT17/0019, of the PE I+D+i 2013–2016, funded by ISCIII and ERDF. The genotyping of GEMAS was also partially funded by Fundación Canaria Instituto de Investigación Sanitaria de Canarias (PIFIISC19/17). The Rotterdam Study was funded by Erasmus Medical Center and Erasmus University Rotterdam; Netherlands Organization for the Health Research and Development (ZonMw); the Research Institute for Diseases in the Elderly (RIDE); the Ministry of Education, Culture and Science, the Ministry for Health, Welfare and Sports, the European Commission (DG XII), and the Municipality of Rotterdam. ALLIANCE Cohort was funded by grants from the German Federal Ministry of Education and Research (Bundesministerium für Bildung und Forschung, BMBF) as part of the German Centre for Lung Research (DZL) funding. The Hartford-Puerto Rico study was funded by the U.S. National Institutes of Health (grant HL07966 to JCC).
MP-Y was funded by the Ramón y Cajal Program (RYC-2015-17205) by MCIN/AEI/10.13039/501100011033 and by the European Social Fund “ESF Investing in your future”. MP-Y and JV were supported by CIBER de Enfermedades Respiratorias, Instituto de Salud Carlos III, Spain (CB/06/06/1088). EH-L was supported by a fellowship awarded by MCIN/AEI/10.13039/501100011033 and by “ESF Investing in your future” (PRE2018-083837).
JP-G was supported by a fellowship awarded by Spanish Ministry of Universities (FPU19/02175). AE-O reports funding from the Spanish Ministry of Science, Innovation, and Universities (MICIU) and Universidad de La Laguna (ULL). NH-P was supported by a Medium-Term Research Fellowship by the European Academy of Allergy and Clinical Immunology (EAACI) and a Long-Term Research Fellowship by the European Respiratory Society (ERS) (LTRF202101-00861).
UP and MG were supported by the Ministry of Education, Science and Sport of the Republic of Slovenia, grant PERMEABLE (contract number C3330-19-252012). SCSGES results were contributed by authors FTC and YYS. FTC has received research support from the Singapore Ministry of Education Academic Research Fund, Singapore Immunology Network (SIgN), National Medical Research Council (NMRC) (Singapore), Biomedical Research Council (BMRC) (Singapore), and the Agency for Science Technology and Research (A*STAR) (Singapore); Grant Numbers: N-154-000-038-001, R-154-000-191-112, R-154-000-404-112, R-154-000-553-112, R-154-000-565-112, R-154-000-630-112, R-154-000-A08-592, R-154-000-A27-597, R-154-000-A91-592, R-154-000-A95-592, R-154-000-B99-114, BMRC/01/1/21/18/077, BMRC/04/1/21/19/315, SIgN-06-006, SIgN-08-020, NMRC/1150/2008, and H17/01/a0/008. F.T.C. has received consulting fees from Sime Darby Technology Centre; First Resources Ltd; Genting Plantation, and Olam International, outside the submitted work. YYS has received research support from the NUS Resilience & Growth Postdoctoral Fellowships with grant number: R-141-000-036-281. QY conducted the analysis from Hartford-Puerto Rico and United Kingdom Biobank studies. QY was funded by the U.S. National Institutes of Health (HL138098).
All funding agencies had no role in the study design, data collection, and analysis, decision to publish, or preparation of the manuscript. The views expressed are those of the authors and not necessarily those of the any funder, National Health Service (NHS), the National Institute for Health Research (NIHR) or the United Kingdom Department of Health.
ABBREVIATIONS
- 1KGP
1000 Genomes Project
- CDK6
Cyclin-dependent kinase 6
- CI
Confidence interval
- GAG
Glycosaminoglycan
- GTEx
Genotype-Tissue Expression
- GWAS
Genome-wide association study
- LPS
Lipopolysaccharide
- MAF
Minor allele frequency
- meQTL
Methylation quantitative trait loci
- OR
Odds ratio
- PPAR-α
Peroxisome proliferator-activating receptor α
- RR
Relative risk
- SNP
Single nucleotide polymorphism
- TLR4
Toll-like receptor 4
- TNFα
Tumor necrosis factor α
Footnotes
CONFLICT OF INTEREST
AE-O received grants from the Spanish Ministry of Science, Innovation, and Universities (MICIU) and Universidad de La Laguna (ULL). EH-L, and MP-Y report funding from the Spanish Ministry of Science and Innovation (MCIN/AEI/10.13039/501100011033) and by “ESF Investing in your future” by the European Union. JP-G reports funding from the Spanish Ministry of Universities. MP-Y and FLD report grants from MCIN/AEI/10.13039/501100011033 and the European Regional Development Fund “ERDF A way of making Europe” by the European Union. MP-Y reports grant support from GlaxoSmithKline, Spain paid to Fundación Canaria Instituto de Investigación Sanitaria de Canarias (FIISC) for a project outside the submitted work. MP-Y and JV reports grants from Instituto de Salud Carlos III, Madrid, Spain. JV also reports funding by ISCIII and the European Regional Development Fund “ERDF A way of making Europe”. JMH-P has received fees from CSL Behring, GSK, Astra-Zeneca, laboratorios Menarini, Boehringer Ingelheim, FAES, laboratorios Esteve, Laboratorios Ferrer, Mundipharma, Laboratorios Rovi, Roche, Novartis, GRIFOLS, Pfizer, Acthelion-Jansen, Chiesi y Laboratorios Bial for the realization of courses, talks, consultancies, and other activities related to his professional activity. FTC has received research support from the Singapore Ministry of Education Academic Research Fund, Singapore Immunology Network (SIgN), National Medical Research Council (NMRC) (Singapore), Biomedical Research Council (BMRC) (Singapore), and the Agency for Science Technology and Research (A*STAR) (Singapore). FTC has received consulting fees from Sime Darby Technology Centre; First Resources Ltd; Genting Plantation, and Olam International, outside the submitted work. YYS has received research support from the NUS Resilience & Growth Postdoctoral Fellowships. UP and MG received grants from the Ministry of Education, Science and Sport from Slovenia, the Slovenian Research Agency. M-JC received grants from the Instituto de Salud Carlos III. DH received grant support from by NIHR for work on NIHR Alder Hey Clinical Research Facility, received payment for medicolegal report writing not related to asthma or pharmacogenomics for UK family court as an expert in pediatric clinical pharmacology. FJ-B received fees from ALK, Astra-Zeneca (AZ), Bial, Chiesi, Gebro Pharma, GlaxoSmithKline (GSK), Menarini, Rovi, Roxall, Sanofi, Stallergenes-Greer and Teva. G-B received fees from AZ, GSK, Boehringer-Ingelheim, Novartis, Chiesi and Sanofi. JC received research materials from Pharmavite and GSK and Merck in order to provide medications free of cost to participants in NIH-funded studies, unrelated to the current work. VO received grants from the National Heart, Lung, and Blood Institute, has participated in Data Safety Monitoring Boards for Regeneron and Sanofi, and participated as a Chair of the section on Genetics and Genomics of the American Thoracic Society. MVK has received grants from the German Federal Ministry of Education and Research, fees from Allergopharma GmbH, Sanofi Aventis GmbH, Infectopharm GmbH, Vertex GmbH and Leti GmbH, has participated in Data Safety Monitoring Boards for Sanofi Aventis GmbH and is the president of the German-Swiss-Austrian Society of Pediatric Pulmonology (GPP). NHP received support from the Instituto de Salud Carlos III, the European Social Funds from the European Union “ESF invests in your future”, the European Academy of Allergy and Clinical Immunology and the European Respiratory Society. MP has received grants from NHS Chair of Pharmacogenetic grant from UK Department of Health, has received partnership funding for the following: MRC Clinical Pharmacology Training Scheme (co-funded by MRC and Roche, UCB, Eli Lilly and Novartis); Joint PhD funding from EPSRC and AZ, and grant funding from Vistagen Therapeutics. He has also unrestricted educational grant support for the UK Pharmacogenetics and Stratified Medicine Network from Bristol-Myers Squibb and UCB. He has developed an HLA genotyping panel with MC Diagnostics, but does not benefit financially from this. MP is part of the IMI Consortium ARDAT (www.ardat.org). SQ has received fees from GSK, AZ, Sanofi, Teva, Novartis, and Chiesi. SJ-HV has received grants from SysPharmPediA EraNet. VdP has received fees from AZ and GSK. VP has received fees from Sanofi, AZ, Chiesi, MSD, and Boehringer Ingelheim, grant support from MSD, Chiesi Institutional, and Menarini. EvM has received grants from the German Federal Ministry of Education and Research and the Bavarian State Ministry of Health and Care, royalties/licenses from Elsevier GmbH, Georg Thieme Verlag, Springer-Verlag GmbH and Elsevier Ltd. EvM has recieved fees from the Chinese University of Hongkong, European Commission, HiPP GmbH & Co KG, AZ, Imperial College London, Massachusetts Medical Society, Springer-Verlag GmbH, Elsevier Ltd., Böhringer Ingelheim International GmbH, European Respiratory Society (ERS), Universiteit Utrecht, Faculteit Diergeneeskunde, Universität Salzburg, Springer Medizin Verlag GmbH, Japanese Society of Pediatric Allergy and Clinical Immunology (JSPACI), Klinikum Rechts der Isar, University of Colorado, Paul-Martini-Stiftung, Astra Zeneca, Imperial College London, Childreńs Hospital Research Institute of Manitoba, Kompetenzzentrum für Ernährung (Kern), OM Pharma S.A., Swedish Pediatric Society for Allergy and Lung Medicine, Chinese College of Allergy and Asthma (CCAA), Verein zur Förderung der Pneumologie am Krankenhaus Großhansdorf e.V., Pneumologie Developpement, Mondial Congress & Events GmbH & Co. KG, American Academy of Allergy, Asthma & Immunology, Imperial College London, Margaux Orange, Volkswagen Stiftung, Böhringer Ingelheim International GmbH, European Respiratory Society (ERS), Universiteit Utrecht, Faculteit Diergeneeskunde, Österreichische Gesellschaft f. Allergologie u. Immunologie, Massachusetts Medical Society, OM Pharma S. A., Hanson Wade Ltd., iKOMM GmbH, DSI Dansk Borneastma Center, American Thoracic Society, HiPP GmbH & Co KG, Universiteit Utrecht, Faculteit Bètawetenschappen. EvM has patents No. PCT/EP2019/085016, EP21189353.2. 2021. and PCT/US2021/016918. 2021. pending, royalties paid to ProtectImmun for patent EP2361632 and patents EP1411977, EP1637147, and EP 1964570 licensed to ProtectImmun. EvM is a member of the EXPANSE Scientific Advisory Board, Member of the BEAMS External Scientific Advisory Board (ESAB), Member of the Editorial Board of “The Journal of Allergy and Clinical Immunology: In Practice”, Member of the Scientific Advisory Board of the Children’s Respiratory and Environmental Workgroup (CREW), Member of the International Scientific & Societal Advisory Board (ISSAB) of Utrecht Life Sciences (ULS), University of Utrecht, Member of External Review Panel of the Faculty of Veterinary Science, University of Utrecht, Member of the Selection Committee for the Gottfried Wilhelm Leibniz Programme (DFG), Member of the International Advisory Board of Asthma UK Centre for Applied Research (AUKCAR), Member of the International Advisory Board of “The Lancet Respiratory Medicine”, Member of the Scientific Advisory Board of the CHILD (Canadian Healthy Infant Longitudinal Development) study, McMaster University, Hamilton, Canada, Asthma UK Centre for Applied Research and the Pediatric Scientific Advisory Board Iceland. The other authors declare no conflict of interest.
IMPACT STATEMENT
A large multi-ancestry meta-analysis of GWAS of asthma exacerbations revealed two novel susceptibility loci located close to PANK1 and at the intergenic region of VCAM1 and EXTL2. These loci decreased PANK1 and EXTL2 gene expression in whole blood, respectively. Both genetic variants were associated with DNA methylation levels at CpG sites nearby. Our results identified two gene targets for asthma exacerbations that should be further explored to assess their specific role in asthma.
DATA AVAILABILITY
All data necessary to evaluate the conclusions of this manuscript are reported in the main text and/or the Supporting Information. Genome-wide genotyping data for GALA II and SAGE are available at the database of Genotypes and Phenotypes (dbGaP) (Study Accession phs001274.v2.p1 and phs000092.v1.p1, respectively). The summary statistics of the multi-ancestry discovery phase are available at the Zenodo repository: 10.5281/zenodo.5513443.
REFERENCES
- 1.Global Initiative for Asthma. Global strategy for asthma management and prevention. 2021.http://ginasthma.org/ (accessed 21 Sep 2020).
- 2.Hernandez-Pacheco N, Flores C, Oh SS, Burchard EG, Pino-Yanes M. What Ancestry Can Tell Us About the Genetic Origins of Inter-Ethnic Differences in Asthma Expression. Curr Allergy Asthma Rep 2016;16:53. [DOI] [PubMed] [Google Scholar]
- 3.Reddel HK, Taylor DR, Bateman ED, Boulet LP, Boushey H a., Busse WW et al. Asthma control and exacerbations - Standardizing endpoints for clinical asthma trials and clinical practice. Am J Respir Crit Care Med 2009;180:59–99. [DOI] [PubMed] [Google Scholar]
- 4.Calhoun WJ, Haselkorn T, Miller DP, Omachi TA. Asthma exacerbations and lung function in patients with severe or difficult-to-treat asthma. J Allergy Clin Immunol 2015;136:1125–7.e4. [DOI] [PubMed] [Google Scholar]
- 5.Chipps BE, Haselkorn T, Rosén K, Mink DR, Trzaskoma BL, Luskin AT. Asthma Exacerbations and Triggers in Children in TENOR: Impact on Quality of Life. J Allergy Clin Immunol Pract 2018;6:169–176.e2. [DOI] [PubMed] [Google Scholar]
- 6.Puranik S, Forno E, Bush A, Celedón JC. Predicting severe asthma exacerbations in children. Am. J. Respir. Crit. Care Med. 2017;195:854–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Herrera-Luis E, Hernandez-Pacheco N, Vijverberg SJ, Flores C, Pino-Yanes M. Role of genomics in asthma exacerbations. Curr Opin Pulm Med 2019;25:101–112. [DOI] [PubMed] [Google Scholar]
- 8.Hernandez-Pacheco N, Farzan N, Francis B, Karimi L, Repnik K, Vijverberg SJ et al. Genome-wide association study of inhaled corticosteroid response in admixed children with asthma. Clin Exp Allergy 2019;49:789–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dahlin A, Denny J, Roden DM, Brilliant MH, Ingram C, Kitchner TE et al. CMTR1 is associated with increased asthma exacerbations in patients taking inhaled corticosteroids. Immunity, Inflamm Dis 2015;3:350–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Slob EMA, Richards LB, Vijverberg SJH, Longo C, Koppelman GH, Pijnenburg MWH et al. Genome-wide association studies of exacerbations in children using long-acting beta2-agonists. Pediatr Allergy Immunol 2021;32:1197–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hernandez-Pacheco N, Vijverberg SJ, Herrera-Luis E, Li J, Sio YY, Granell R et al. Genome-wide association study of asthma exacerbations despite inhaled corticosteroid use. Eur Respir J 2021;57. doi: 10.1183/13993003.03388-2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yan Q, Forno E, Herrera-Luis E, Pino-Yanes M, Qi C, Rios R et al. A genome-wide association study of severe asthma exacerbations in Latino children and adolescents. Eur Respir J 2021;57:2002693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yan Q, Forno E, Herrera-Luis E, Pino-Yanes M, Yang G, Oh S et al. A genome-wide association study of asthma hospitalizations in adults. J Allergy Clin Immunol 2021;147:933–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herrera-Luis E, Espuela-Ortiz A, Lorenzo-Diaz F, Keys KL, Mak ACY, Eng C et al. Genome-wide association study reveals a novel locus for asthma with severe exacerbations in diverse populations. Pediatr Allergy Immunol 2021;32:106–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCarthy S, Das S, Kretzschmar W, Delaneau O, Wood AR, Teumer A et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet 2016;48:1279–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.1000 Genomes Project Consortium, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM et al. A global reference for human genetic variation. Nature 2015;526:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang CC, Chow CC, Tellier LCAMC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 2015;4:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kang HM. EPACTS (Efficient and Parallelizable Association Container Toolbox). https://genome.sph.umich.edu/wiki/EPACTS. 2016.
- 19.Zhan X, Hu Y, Li B, Abecasis GR, Liu DJ. RVTESTS: an efficient and comprehensive tool for rare variant association analysis using sequence data: Table 1. Bioinformatics 2016;32:1423–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Winkler TW, Day FR, Croteau-Chonka DC, Wood AR, Locke AE, Mägi R et al. Quality control and conduct of genome-wide association meta-analyses. Nat Protoc 2014;9:1192–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han B, Eskin E. Random-effects model aimed at discovering associations in meta-analysis of genome-wide association studies. Am J Hum Genet 2011;88:586–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hammond RK, Pahl MC, Su C, Cousminer DL, Leonard ME, Lu S et al. Biological constraints on GWAS SNPs at suggestive significance thresholds reveal additional BMI loci. Elife 2021;10. doi: 10.7554/eLife.62206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: A tool for genome-wide complex trait analysis. Am J Hum Genet 2011;88:76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Watanabe K, Taskesen E, Van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun 2017;8:1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buniello A, MacArthur JAL, Cerezo M, Harris LW, Hayhurst J, Malangone C et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res 2019;47:D1005–D1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karolchik D, Hinrichs AS, Furey TS, Roskin KM, Sugnet CW, Haussler D et al. The UCSC Table Browser data retrieval tool. Nucleic Acids Res 2004;32:D493–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng J, Erzurumluoglu AM, Elsworth BL, Kemp JP, Howe L, Haycock PC et al. LD Hub: a centralized database and web interface to perform LD score regression that maximizes the potential of summary level GWAS data for SNP heritability and genetic correlation analysis. Bioinformatics 2017;33:272–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ongen H, Buil A, Brown AA, Dermitzakis ET, Delaneau O. Fast and efficient QTL mapper for thousands of molecular phenotypes. Bioinformatics 2016;32:1479–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zheng Z, Huang D, Wang J, Zhao K, Zhou Y, Guo Z et al. QTLbase: an integrative resource for quantitative trait loci across multiple human molecular phenotypes. Nucleic Acids Res 2020;48:D983–D991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Consortium GTEx. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science 2020;369:1318–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kamat MA, Blackshaw JA, Young R, Surendran P, Burgess S, Danesh J et al. PhenoScanner V2: an expanded tool for searching human genotype-phenotype associations. Bioinformatics 2019;35:4851–4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Breeze CE, Reynolds AP, van Dongen J, Dunham I, Lazar J, Neph S et al. eFORGE v2.0: updated analysis of cell type-specific signal in epigenomic data. Bioinformatics 2019;35:4767–4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schofield EC, Carver T, Achuthan P, Freire-Pritchett P, Spivakov M, Todd JA et al. CHiCP: a web-based tool for the integrative and interactive visualization of promoter capture Hi-C datasets. Bioinformatics 2016;32:2511–2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tse SM, Krajinovic M, Chauhan BF, Zemek R, Gravel J, Chalut D et al. Genetic determinants of acute asthma therapy response in children with moderate-to-severe asthma exacerbations. Pediatr Pulmonol 2019;54:378–385. [DOI] [PubMed] [Google Scholar]
- 35.Leiter K, Franks K, Borland ML, Coleman L, Harris L, Le Souëf PN et al. Vitamin D receptor polymorphisms are associated with severity of wheezing illnesses and asthma exacerbations in children. J Steroid Biochem Mol Biol 2020;201:105692. [DOI] [PubMed] [Google Scholar]
- 36.Tsai C-H, Wu AC, Chiang B-L, Yang Y-H, Hung S-P, Su M-W et al. CEACAM3 decreases asthma exacerbations and modulates respiratory syncytial virus latent infection in children. Thorax 2020;75:725–734. [DOI] [PubMed] [Google Scholar]
- 37.Shrine N, Portelli MA, John C, Soler Artigas M, Bennett N, Hall R et al. Moderate-to-severe asthma in individuals of European ancestry: a genome-wide association study. Lancet Respir Med 2019;7:20–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kong D-H, Kim Y, Kim M, Jang J, Lee S. Emerging Roles of Vascular Cell Adhesion Molecule-1 (VCAM-1) in Immunological Disorders and Cancer. Int J Mol Sci 2018;19:1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hortelano S, López-Fontal R, Través PG, Villa N, Grashoff C, Boscá L et al. ILK mediates LPS-induced vascular adhesion receptor expression and subsequent leucocyte trans-endothelial migration. Cardiovasc Res 2010;86:283–292. [DOI] [PubMed] [Google Scholar]
- 40.Lee J-H, Sohn J-H, Ryu SY, Hong C-S, Moon KD, Park J-W. A novel human anti-VCAM-1 monoclonal antibody ameliorates airway inflammation and remodelling. J Cell Mol Med 2013;17:1271–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kitagawa H, Shimakawa H, Sugahara K. The tumor suppressor EXT-like gene EXTL2 encodes an alpha1, 4-N-acetylhexosaminyltransferase that transfers N-acetylgalactosamine and N-acetylglucosamine to the common glycosaminoglycan-protein linkage region. The key enzyme for the chain initiation of he. J Biol Chem 1999;274:13933–13937. [DOI] [PubMed] [Google Scholar]
- 42.Nadanaka S, Zhou S, Kagiyama S, Shoji N, Sugahara K, Sugihara K et al. EXTL2, a member of the EXT family of tumor suppressors, controls glycosaminoglycan biosynthesis in a xylose kinase-dependent manner. J Biol Chem 2013;288:9321–9333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pu A, Mishra MK, Dong Y, Ghorbanigazar S, Stephenson EL, Rawji KS et al. The glycosyltransferase EXTL2 promotes proteoglycan deposition and injurious neuroinflammation following demyelination. J Neuroinflammation 2020;17:220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nadanaka S, Hashiguchi T, Kitagawa H. Aberrant glycosaminoglycan biosynthesis by tumor suppressor EXTL2 deficiency promotes liver inflammation and tumorigenesis through Toll-like 4 receptor signaling. FASEB J 2020;34:8385–8401. [DOI] [PubMed] [Google Scholar]
- 45.Hennessy EJ, Sheedy FJ, Santamaria D, Barbacid M, O’Neill LAJ. Toll-like receptor-4 (TLR4) down-regulates microRNA-107, increasing macrophage adhesion via cyclin-dependent kinase 6. J Biol Chem 2011;286:25531–25539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Banno A, Reddy AT, Lakshmi SP, Reddy RC. PPARs: Key Regulators of Airway Inflammation and Potential Therapeutic Targets in Asthma. Nucl Recept Res 2018;5. doi: 10.11131/2018/101306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kumari A, Dash D, Singh R. Lipopolysaccharide (LPS) exposure differently affects allergic asthma exacerbations and its amelioration by intranasal curcumin in mice. Cytokine 2015;76:334–342. [DOI] [PubMed] [Google Scholar]
- 48.Hadjigol S, Netto KG, Maltby S, Tay HL, Nguyen TH, Hansbro NG et al. Lipopolysaccharide induces steroid-resistant exacerbations in a mouse model of allergic airway disease collectively through IL-13 and pulmonary macrophage activation. Clin Exp Allergy 2020;50:82–94. [DOI] [PubMed] [Google Scholar]
- 49.Böhlig L, Friedrich M, Engeland K. p53 activates the PANK1/miRNA-107 gene leading to downregulation of CDK6 and p130 cell cycle proteins. Nucleic Acids Res 2011;39:440–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang L, Zhang B, Wang X, Liu Z, Li J, Zhang S et al. P53/PANK1/miR-107 signalling pathway spans the gap between metabolic reprogramming and insulin resistance induced by high-fat diet. J Cell Mol Med 2020;24:3611–3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.El-Husseini ZW, Gosens R, Dekker F, Koppelman GH. The genetics of asthma and the promise of genomics-guided drug target discovery. Lancet Respir Med 2020;8:1045–1056. [DOI] [PubMed] [Google Scholar]
- 52.Bankovic J, Stojsic J, Jovanovic D, Andjelkovic T, Milinkovic V, Ruzdijic S et al. Identification of genes associated with non-small-cell lung cancer promotion and progression. Lung Cancer 2010;67:151–159. [DOI] [PubMed] [Google Scholar]
- 53.Ma J, Zhao J, Lu J, Wang P, Feng H, Zong Y et al. Cadherin-12 enhances proliferation in colorectal cancer cells and increases progression by promoting EMT. Tumour Biol 2016;37:9077–9088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhao J, Li P, Feng H, Wang P, Zong Y, Ma J et al. Cadherin-12 contributes to tumorigenicity in colorectal cancer by promoting migration, invasion, adhersion and angiogenesis. J Transl Med 2013;11:288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Singh SR, Sutcliffe A, Kaur D, Gupta S, Desai D, Saunders R et al. CCL2 release by airway smooth muscle is increased in asthma and promotes fibrocyte migration. Allergy 2014;69:1189–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Potaczek DP, Harb H, Michel S, Alhamwe BA, Renz H, Tost J. Epigenetics and allergy: from basic mechanisms to clinical applications. Epigenomics 2017;9:539–571. [DOI] [PubMed] [Google Scholar]
- 57.Network BTS and SIG. British guideline on the management of asthma. SIGN 158. 2019.https://www.brit-thoracic.org.uk/quality-improvement/guidelines/asthma/ [Google Scholar]
- 58.Jaffe AE, Irizarry RA. Accounting for cellular heterogeneity is critical in epigenome-wide association studies. Genome Biol 2014;15:R31. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data necessary to evaluate the conclusions of this manuscript are reported in the main text and/or the Supporting Information. Genome-wide genotyping data for GALA II and SAGE are available at the database of Genotypes and Phenotypes (dbGaP) (Study Accession phs001274.v2.p1 and phs000092.v1.p1, respectively). The summary statistics of the multi-ancestry discovery phase are available at the Zenodo repository: 10.5281/zenodo.5513443.


